Abstract
The NCI-60 human tumor cell line panel has proved to be a useful tool for the global cancer research community in the search for novel chemotherapeutics. The publicly available cell line characterization and compound screening data from the NCI-60 assay have significantly contributed to the understanding of cellular mechanisms targeted by new oncology agents. Signature sensitivity/resistance patterns generated for a given chemotherapeutic agent against the NCI-60 panel have long served as fingerprint presentations that encompass target information and the mechanism of action associated with the tested agent. We report the establishment of a new public NCI-60 resource based on the cell line screening of a large and growing set of 175 FDA-approved oncology drugs (AOD) plus >825 clinical and investigational oncology agents (IOA), representing a diverse set (>250) of therapeutic targets and mechanisms. This data resource is available to the public (https://ioa.cancer.gov) and includes the raw data from the screening of the IOA and AOD collection along with an extensive set of visualization and analysis tools to allow for comparative study of individual test compounds and multiple compound sets.
Introduction
For more than two decades, molecular target-based drug discovery through biochemical screening and rational drug design have been central strategies of drug discovery programs in both the academic and pharmaceutical communities. Recently, two independent studies analyzed the approaches used during successful drug discovery projects resulting in FDA-approved drugs, since 1999. Both studies found significant contributions from phenotypic screening and system-based approaches (1, 2). A related study focusing on oncology drugs approved by the FDA between 1999 and 2013 found that a phenotypic screen (mechanism-informed or de novo) played a major role in lead compound discovery and/or candidate selection in 19 of 48 (39.5%) programs, including five of 16 (31%) that produced first-in-class drugs (3). As oncology treatment has moved to targeted therapeutic agents based upon the known genetic and biological aberrations of the disease, the overlap between target-based drug discovery and phenotypic screening has become defined in terms of well-characterized cell line models of drug response (4, 5). Studies seeking to delineate the pharmacodynamics of drugs in cancer cells have increasingly focused on the identification and impact of the underlying genomic factors influencing drug response (6–10).
The NCI-60 human tumor cell line panel has proved to be a useful global cancer research community tool in the armamentarium of cancer drug discovery and has facilitated the understanding of molecular mechanisms for new oncology agents. The NCI-60 cell line panel includes nine cancer types [leukemia, non–small cell lung (NSCLC), colon, central nervous system, melanoma, ovarian, renal, prostate, and breast] and has been used to profile potential oncology chemotherapeutic agents for more than 20 years (11). Extensive genomic and proteomic profiling of the NCI-60 cell lines makes this panel among the best characterized collection of human cancer cell lines including studies of mutations, amplifications and deletions, proteomics, methylome, miRNA, exosomes, and more (12–16). Characterization data for the NCI-60 cell line and data from the NCI-60 screening assay are publicly available (https://dtp.cancer.gov/databases_tools/bulk_data.htm; https://discover.nci.nih.gov/rsconnect/cellminercdb/). Many groups have used the NCI-60 cell line panel for further studies or have done metadata analyses of the NCI-60 data from NCI. These include: https://web.expasy.org/cellosaurus/, https://www.cbioportal.org/, https://cancer.sanger.ac.uk/cosmic, https://discover.nci.nih.gov/rsconnect/cellminercdb/ and others.
This article describes a new NCI-60 resource based upon data obtained from the cell-based screening of a training set of 175 FDA-approved oncology drugs (AOD) plus >825 investigational oncology agents (IOA) and tool compounds. This accumulated set of drugs and investigational agents represents a diverse range of therapeutic targets and mechanisms (>250 in number) with multiple compound examples for many of the targets. The NCI-60 data from this set are available through a public website with tools to allow investigators to study how public NSC compounds relate to this training set of compounds of known and assigned mechanisms and to allow suppliers of compounds to the NCI-60 screen to study their data in the same way.
Materials and Methods
Chemistry
The IOAs and AODs are procured through internal/external synthesis and through their acquisition from external vendors. All sample lots of compounds in the IOA collection undergo rigorous 1H NMR and LC/MS examination to ensure structural integrity and >95% purity by high pressure liquid chromatography (HPLC). In some cases, additional analytic techniques are employed to maintain the integrity of the collection (e.g., chiral HPLC, optical rotation, X-ray crystallography, and 13C NMR). Proton NMR spectra and LC/MS data showing > 95% purity for the representative agents discussed in this account are found in the Supplementary Data.
Cell culture
NCI-60 cell lines are maintained by the NCI Developmental Therapeutics Program Tumor Repository. For each lot of cells, the Repository performed Applied Biosystems AmpFLSTR Identifiler testing with PCR amplification to confirm consistency with the published Identifiler short tandem repeat profile for the given cell line (17, 18). Each cell line was tested for Mycoplasma when it was accepted into the repository; routine Mycoplasma testing of lots was not performed. Cells were kept in continuous culture for no more than 20 passages. The optimal seeding density for each of the cell lines was determined prior to performing concentration–response studies (11, 19, 20). The NCI-60 screen was performed as described at: https://dtp.cancer.gov/discovery_development/nci-60/default.htm. Briefly, the NCI-60 human tumor lines were grown in RPMI1640 medium supplemented with 5% FBS and 2 mmol/L l-glutamine. For experiments, cells were inoculated into 96-well plates in 100 μL of complete medium at plating densities ranging from 5,000 to 40,000 cells/well depending on the doubling time of individual lines. The plates were incubated at 37°C in humidified 5% CO2/95% air for 24 hours. Compounds were formulated in DMSO. The plates were incubated for 48 hours. For staining, sulforhodamine B (SRB) solution (100 μL) at 0.4% (w/v) in 1% acetic acid was added to each well, and plates were incubated for 10 minutes at room temperature. The SRB was solubilized, and the absorbance at 515 nm was read. Using the absorbance measurements [time zero, (Tz), control growth, (C), and test growth (Ti)], the percent cell growth was calculated. Growth inhibition of 50% (GI50) is calculated from [(Ti − Tz)/(C − Tz)] × 100 = 50, which is the compound concentration resulting in a 50% reduction in the net protein increase (as measured by SRB staining) in control cells. The NCI-60 screen is performed in duplicate and mean values are reported.
NCI-60 compound screen
The IOA and AOD compounds were tested in the 2-day, 5-concentration standard NCI-60 screen, and the mean growth 50% inhibition (GI50) values are tabulated in Supplementary Table S1 (Supplementary Data). A public website (https://ioa.cancer.gov) has been established to make available up-to-date data related to the collection of NCI IOAs and FDA-AODs. The website includes both chemical data (structural and calculated property information for each drug/agent) and biological assay data (concentration–response data from the NCI-60 screen and calculated endpoint assay results [GI50, total growth inhibition and lethality (LC50)]. In addition, visualization and interactive tools are available which allow investigators to examine and compare NCI-60 data for individual compounds or sets of agents using mean graph or heat map formats and to run various COMPARE analyses, including concentration–response endpoints, mean graphs, COMPARE correlations, and correlation maps.
Several data presentation formats and analytic tools were developed during the establishment of the original NCI-60 screening assay to look at the relative cell line responses to specific drugs/compounds across the NCI-60 cell line panel while factoring out differences in overall potency of the compounds: the focus was the pattern of relative sensitivities to a compound among the cell lines (21). The mean graph is a bar graph where the center line (“zero”) represents the average sensitivity of all 60 cell lines, and the responses of the individual cell lines are presented relative to that center line. Because NCI-60 concentration data are tracked as the log(concentration), the values are log units, which further helps normalize the response patterns. The COMPARE algorithm looks for correlations in the graph patterns among compound data in the 60 cell lines and was built around the Pearson correlation coefficient, which produces a dimensionless result, further factoring out considerations of relative potency among compounds.
The relative patterns in the NCI-60 are often easier to see in a mean graph presentation than in a heat map. In addition, the correlation map tool can visualize correlations among all members of a compound set. The compounds are represented by nodes (circles) on the map, and the distances between nodes represent the correlations between the response patterns with more similar patterns being closer together. This presentation can make it easier to visualize the similarities than trying to interpret a set of mean graphs, a grid of correlation values or a colored heat map of correlation values.
The formal statistical considerations regarding a Pearson correlation are not applicable because the NCI-60 cell lines are not a random sampling of cell lines, and the compounds submitted to the NCI-60 screen are not a random sampling of chemical structures. Specifically, the level of correlation required for potential biological significance for the NCI-60 cell lines diverges from the critical values calculated for a Pearson correlation with 59 degrees of freedom. The correlation values that indicate potential biological significance are based on empirical evidence: COMPARE was used to predict compounds with possible similar mechanisms to compounds of known mechanism assayed in cell-free systems, and the likelihood of the prediction being borne out declined almost to zero as correlations decreased toward 0.6 (22). The cutoff used here for potential biological significance is 0.7 and correlations at 0.8 and above are considered strong. In contrast, the critical value for a Pearson correlation with 60 degrees of freedom is about 0.3, but at that level the relatedness among the IOA compounds is meaninglessly high as are the relatedness of most compounds assayed in the NCI-60.
Data availability
The data generated in this study are available within the article and its Supplementary Data and at ioa.cancer.gov.
Results
Several main themes emerged from the study of the NCI-60 patterns obtained from the evaluation of the IOA and AOD collection: (i) targeted inhibitors within a biological pathway tend to show a high correlation in GI50 patterns through COMPARE analysis forming clusters representing agents with a given mechanism as well as between agents sharing targets within respective biochemical pathways, (ii) NCI-60 response patterns can suggest unexpected “off-target” activities for a given investigation agent, and (iii) the activity of agents against the NCI-60 cell lines correlates with the gene expression patterns for the agent's target.
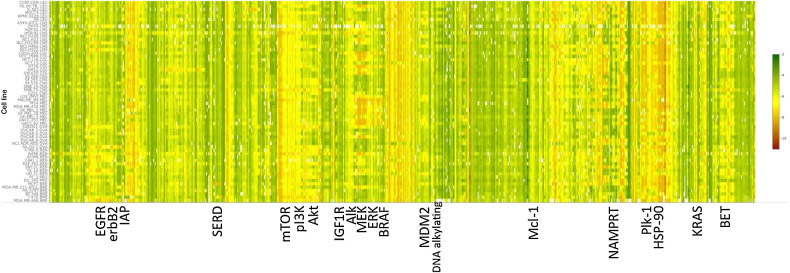
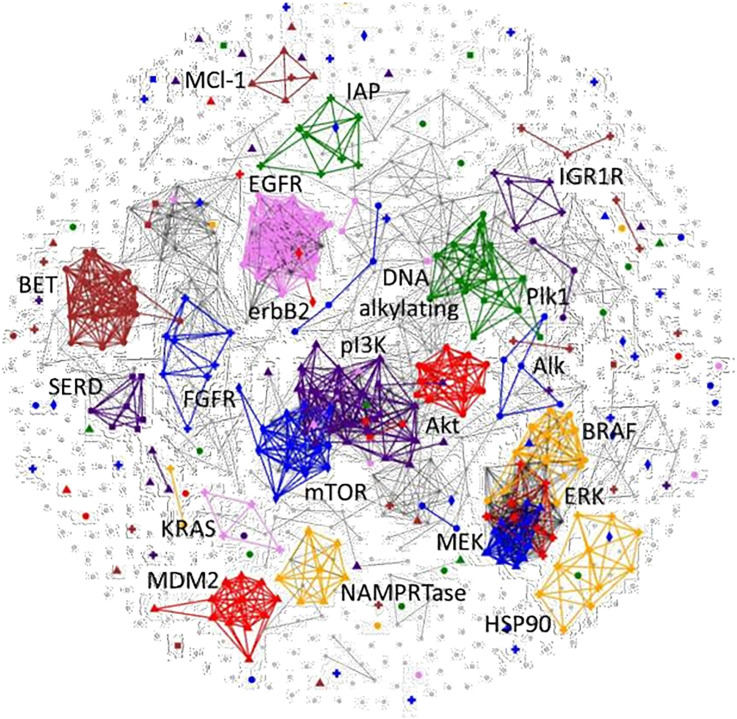
A heat map view of the hierarchical clustering of the NCI-60 growth response patterns (GI50s) from the 955 drugs/agents of the AOD and IOA set is shown in Fig. 1. The patterns observed for the drugs/agents in this view comprise the complete spectrum of activity from agents where all of the cell lines were found to be sensitive to those where complete resistance occurred. The clustering of these patterns by similarity (Pearson correlation) leads to multiple agents being grouped together based on similar response patterns that are consistent with their associated therapeutic targets. Not unexpectedly, the overall potency levels observed varies greatly among some of the compounds of the targeted groups of response patterns. Using the COMPARE algorithm of the GI50 patterns associated with the entire AOD/IOA compound set produces a 955×955 matrix grid of Pearson correlations. A useful visualization of this matrix grid is the correlation map view (Fig. 2) where a connection is drawn between any two agents showing a response pattern correlation above a selected threshold value (e.g., 0.7 for the figure shown). When viewed in this manner, similarities between the patterns produced by agents associated with a given molecular target form clustered groups, the more highly correlated the response patterns the closer together are the nodes. This presentation provides the same information as the clustered heat map in a manner that can be easier to interpret.
Figure 1.
Heat map view of hierarchical clustering of NCI-60 growth response Pearson correlation patterns. Color representations of GI50 values ranging from 10−12 M (red) to 10−2 M (green). Select agent-associated primary targets indicated for representative larger compound sets.
Figure 2.
Correlation map of GI50 response patterns for all compounds of the IOA set. The compounds are represented by nodes (circles) on the map and the distances between nodes represent the correlations (minimum 0.7) between the response patterns with more similar patterns being closer together. Select agent-associated primary targets are indicated for representative larger compound sets. These highlighted targets are assigned a unique color/symbol combination which is listed in the legend and are represented with larger symbols.
Correlations of agents targeting elements within signaling pathways
Agents that target different proteins within a biochemical pathway tend to show a high correlation in GI50 patterns through COMPARE analysis forming clusters representing agents similar NCI-60 meanGraph patterns indicating similar mechanisms of action as well as between agents sharing targets within a biochemical pathway. For example, COMPARE evaluation of agents with mechanisms of action targeting components within the PI3K/AKT/mTOR pathway form correlation map clusters linked together by both target and pathway (Fig. 2). Only three of 15 of the AKT inhibitors in the IOA set are not found in the connected cluster with a minimum COMPARE correlation of 0.7 (most at 0.85), including the multikinase inhibitors perifosine and AT-13148 and the inactive enantiomer of the Pfizer inhibitor, PF-4176340. In addition, the AKT cluster includes both allosteric (e.g., MK-2206) and competitive inhibitors, highlighting the fact that the NCI-60 is a functional cell assay with a cell growth inhibition/cell death readout. More than 75% of the PI3K inhibitors (32/42 agents) in the IOA set, including the selective PI3K alpha and PI3K beta inhibitors, form a highly connective cluster at a 0.7 COMPARE correlation with the target and pathway. An examination of the heat map view showed that at least half of the nonconnected PI3K outliers were universally inactive in the cell line assay. Finally, 12 of 20 of the mTOR inhibitors form a highly connective cluster at the 0.7 COMPARE correlation level, with half of the nonconnective mTOR singletons representative of the rapamycin class of inhibitors.
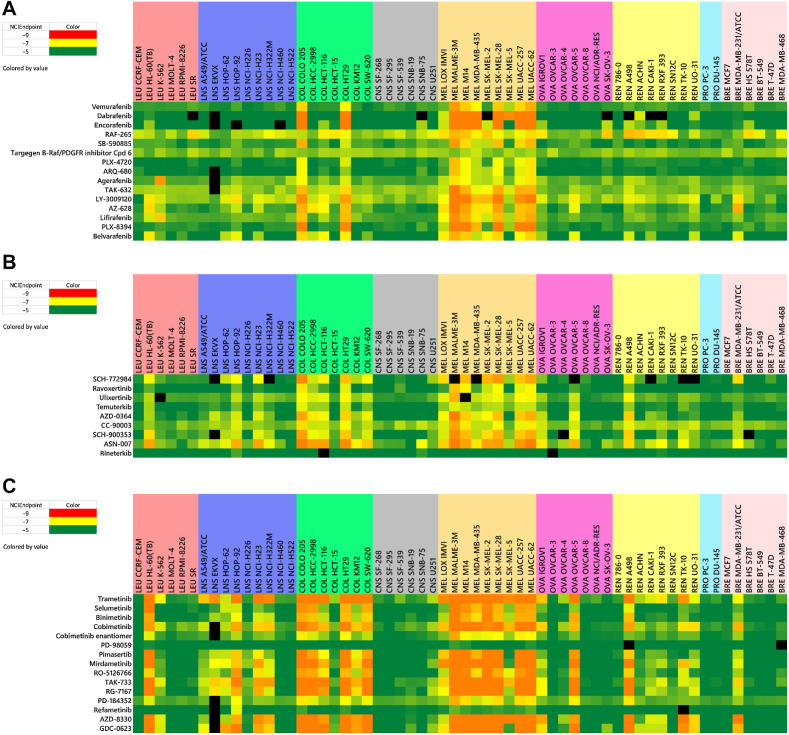
A similar situation results in the case of the 37 agents associated with targets within the BRAF/MEK/ERK pathway, which form a highly connective correlation map cluster (13/15 BRAF, 14/15 MEK, 8/9 ERK inhibitors) using a 0.75 COMPARE correlation constraint (most at 0.85). Examination of the heat graph view of the NCI-60 patterns for the BRAF inhibitors shows consistent sensitivity among almost all of the melanoma and at least two of the colon cancer lines (COLO 205, HT29; Fig. 3A). In general, the ERK inhibitors display sensitivity patterns similar to the BRAF agents, with additional activity observed against a leukemia (HL-60), ovarian (OVCAR-5), and renal (A498) cell lines and three other colon cell lines (HCC-298, HCT-116, SW-620; Fig. 3B). The MEK inhibitors as shown on the correlation map are active against all of the cell lines that are sensitive to the BRAF and ERK inhibitors as well as one additional colon line (KM12), a renal line (TK10), a breast line (MDA-MB-231) and three other NSCLC lines (HOP-92, NCI-H23, and NCI-H322; Fig. 3C). Other targeted investigational agents which cluster with correlations of ≥0.75 include NAMPRTase, HSP90, BET bromodomain, FGFR, IAP, PLK-1, IGF1R, EGFR, MDM2 inhibitors and DNA-alkylators (Fig. 2).
Figure 3.
Heat maps for BRAF inhibitors (A), ERK inhibitors (B), and MEK inhibitors (C). NCIEndpoint is color representation of GI50 values ranging from 10−9 M (red) to 10−5 M (green); cells are black when values were not obtained.
Off-target effects
In addition to enabling comparisons between drug sensitivity profiles that support common mechanisms of action, a second emerging theme from the study of NCI-60 patterns is the elucidation of unexpected “off-target” activities for investigational agents. Response patterns for agents that fall outside the “signature” pattern anticipated for a given mechanism of action can be used to suggest alternative targets that may be affected by the test compound. Examples of this can be seen in the NCI-60 data for agents targeting: (i) the BTK pathway, (ii) BET, (iii) BCR-ABL, (iv) ALK, TRK, IGF1R.
Bruton's tyrosine kinase (BTK) is a member of the TEC family of non-receptor tyrosine kinases and is a target that has been exploited in the treatment of B-cell malignant disease and inflammation. Ibrutinib was the first BTK inhibitor approved by the FDA for treatment of mantle cell lymphoma in 2013 and leukemia in 2014 (23). Ibrutinib is a pyrazolopyrimidine-based compound that contains an α, β-unsaturated acyl amide which forms a covalent bond with the Cys-481 near the active site of BTK. Several other structurally related irreversible BTK inhibitors have either been approved (acalabrutinib, zanubrutinib) or are in clinical testing (including tirabrutinib, branebrutinib, orelabrutinib, evobrutinib, and spebrutinib). The NCI-60 heat map profiles of these agents along with several competitive reversible inhibitors (fenebrutinib, vecabrutinib, ARQ-531, and RN-486) from the current IOA set are shown in Supplementary Fig. S1 (Supplementary Data). The NCI-60 response patterns for these agents, both the irreversible and reversible prototypes (acalabrutinib, tirabrutinib, fenebrutinib, evobrutinib, spebrutunub, orelabrutinib, rizabrutinib, vecabrutunub, branebrutinib, and RN-486), show minimal or no response among the tested cell lines. However, three BTK inhibitors (ibrutinib, zanubrutinib, and acalabrutinib) display a distinct pattern in the NCI-60 assay that is highly correlated (>0.72) to the sensitivity patterns for the EGFR inhibitors (e.g., nazartinib, osimertinib). In a report discussing the pharmacology of acalabrutinib, it was found that the agent was inactive in an EGFR recombinant kinase inhibition assay in contrast to ibrutinib, which was found to be a potent inhibitor (24). Subsequent findings found acalabrutinib to be a weak inhibitor of EGFR (IC50 7.5 μmol/L), consistent with the potency levels observed in the NCI-60 (25). Although zanubrutinib has been reported to be a more selective inhibitor with respect to EGFR (relative to ibrutinib), the high COMPARE correlation to the EGFR pattern suggests that EGFR target inhibitory activity remains a component of the response pattern (26). The NCI-60 patterns observed for both spebrutinib and RN-486 are more complex, leading to possibility that additional targets may be involved in the pharmacology associated with these agents. Finally, the distinct sensitivity pattern associated with ARQ-531 along with a high COMPARE correlation with members of the RAS/BRAF/MEK/ERK pathway further suggest its identification as a dual BTK/MEK inhibitor (27). From an examination of the response patterns associated with the BTK inhibitors, the NCI-60 assay exclusively provides information regarding the off-target activities associated with these agents.
A similar situation occurs with the bromodomain and extraterminal (BET) family of proteins that are involved in the binding of acetylated lysine residues to histones and are a target of interest in oncology. Small-molecule inhibitors of BET regulation of gene expression include the triazolo-benzodiazepines iBET-762, JQ-1, and the clinical candidate birabresib in addition to molibresib, AZD-5153, GSK-1210151A, PLX-51105, and PFI-1. These agents inhibit protein function by binding in a 2:1 stoichiometry to the two bromodomain proteins required for the process. Researchers at AstraZeneca working to improve the potency of the androgen receptor modulator AZD-3514, discovered that the underlying mechanism of this agent, was to act as a bivalent BET inhibitor, engaging two BRD-containing proteins to simultaneously inhibit BET regulation of gene expression (28). Other androgen receptor inhibitors including enzalutamide, JNJ-63576, apalutamide, and darolutamide are not associated with the BET bromodomain inhibitors. Systematic synthesis of analogs aiming to improve upon the BET potency of AZD-3514 while maintaining favorable pharmacokinetic properties resulted in the identification of the potent bivalent BET clinical candidate AZD-5153 (29). The COMPARE correlation map view of the NCI-60 patterns of the entire IOA set indeed shows the BET bromodomain clusters with the androgen modulator AZD-3514 with correlations uniformly ≥0.70 (Supplementary Fig. S2; Supplementary Data). The corresponding heat map view of this compound set shows AZD-3514 to be of relatively lower potency in the NCI-60 cell line assay, consistent with its level of bromodomain binding affinity (IC50 = 4.12 × 10−5 μmol/L, Reaction Biology; refs. 30, 31). Another agent in this cluster at a minimal correlation of 0.65 is the Hedgehog (SMO) inhibitor HPI-1. In fact, bromodomain inhibition has been observed to be an effective strategy for targeting Hedgehog-driven tumors (32). In support of this “off-target” finding for HPI-1, this agent was found to be active in a bromodomain AlphaScreen binding assay (IC50 = 1.29 × 10−5 μmol/L, Reaction Biology; refs. 30, 31).
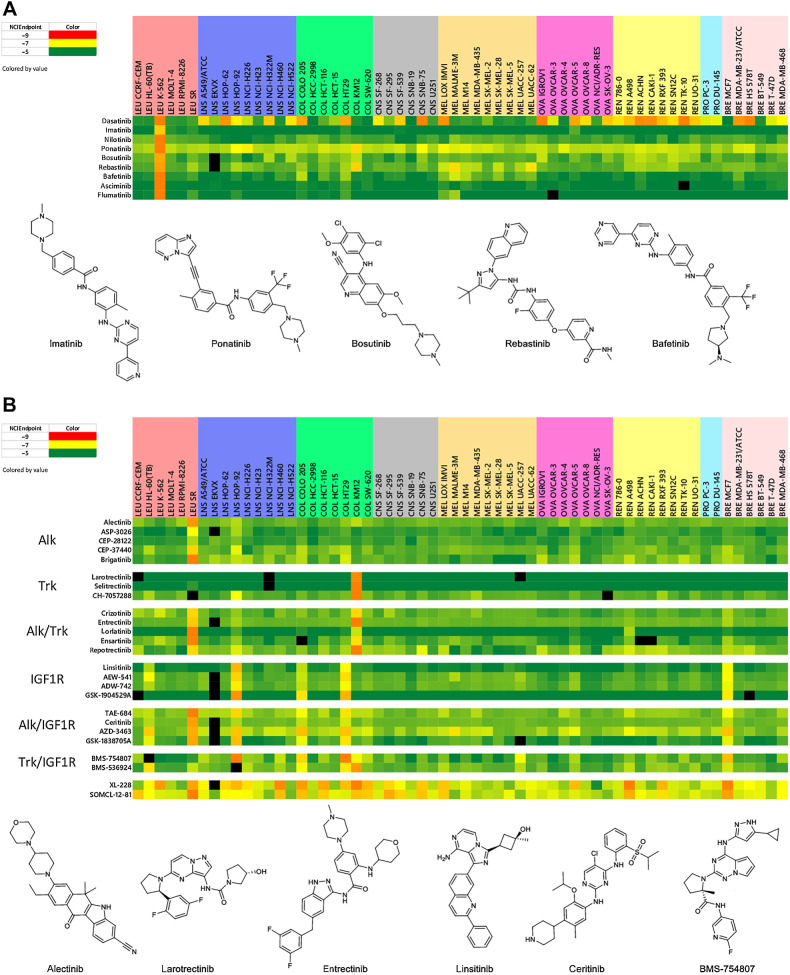
The absence of c-ABL in the fusion oncoprotein BCR-ABL leads to an abnormal tyrosine kinase that is responsible for chronic myeloid leukemia (CML; ref. 33). The discovery and development of the BCR-ABL inhibitor imatinib in 2001 revolutionized the treatment of CML. By binding at the ATP-binding site of the BCR-ABL kinase, blocking enzymatic activity and preventing antiapoptotic signaling in the cell (34). The ultimate development of cellular resistance to imatinib led to the FDA approval of several second-generation inhibitors (dasatinib, nilotinib, ponatinib, bosutinib) and the discovery of several third-generation blockers (rebastinib, bafetanib), including the selective allosteric inhibitor, ABL-001 (35). A heat map view of the sensitivity patterns produced upon testing these agents in the NCI-60 cell line assay is shown in Fig. 4A. Not surprisingly, the K-562 leukemia cell line which expresses the BCR-ABL fusion gene is sensitive to all of the BCR-ABL inhibitors (36). Because no other cell lines in the NCI-60 express BCR-ABL, it is likely that responses by other cell lines observed for this set of compounds are due to inhibition of other targets. In fact, the finding of TRK inhibitory activity for the BCR-ABL inhibitors ponatinib, rebastinib, and bosutinib was suggested by the sensitivity of the KM12 colon cell line (which highly expresses the NTRK fusion gene) to these agents. In addition, the observation of cell sensitivity in the colon HCT-116 and HT29 lines and in most of the melanoma lines in response to treatment with both bafetinib and rebastinib suggested the association of a BRAF inhibitory profile for these agents. The possibility of BRAF inhibitory activity for bafetinib and rebastinib was confirmed in two ways: (i) a finding of 1.8% and 15.8% enzyme activity relative to DMSO control for these agents (at 1 μmol/L), respectively, in a BRAF kinase profile assay (Reaction Biology; refs. 30, 31), and (ii) the finding of COMPARE correlations of 0.78 and 0.80, respectively, for bafetinib and rebastinib with the selective BRAF inhibitor vemurafenib when the algorithm is run without the K-562 and KM12 cell lines.
Figure 4.
Heat maps for second- and third-generation BCR-ABL inhibitors (A) and ALK, TRK, ALK/TRK, IGF1R, ALK/IGF1R, and TRK/IGF1R inhibitors (B). NCIEndpoint is color representation of GI50 values ranging from 10−9 M (red) to 10−5 M (green); cells are black when values were not obtained.
A fourth example of using NCI-60 response patterns to identify selective and dual inhibitors relates to the study of compounds designed to target the ALK, TRK, and IGF1R receptors (Fig. 4B). For the cell lines in the NCI-60 screen, only the SR leukemia line expresses the ALK fusion protein, leading to a response pattern of solely the SR line for ALK selective inhibitors, such as alectinib (CH-542802), ASP-3026, CEP-28122, CEP-37440, and brigatinib (AP-26113). Similarly, singular expression of NTRK fusion gene by the KM12 colon cell line leads to a related one-cell KM12 line pattern in the NCI-60 assay for the selective TRK inhibitors in the IOA set, including larotrectinib, selitrectnib, and CH-7057288. Dual ALK/TRK inhibitors are readily identified by their NCI-60 sensitivity patterns that target both the SR and KM12 lines and include crizotinib, entrectinib, lorlatinib (PF-06463922), ensartinib, and reprotrectinib (TPX-005). The insulin-like growth factor receptor, IGF-1R is another target where selective inhibitors have a distinctive response pattern upon screening in the NCI-60 panel. The selective IGF-1R inhibitors, linsitinib (ASP-7487), AEW-541, ADW-742, and GSK-1904529A, produce a distinct 5-cell line GI50 pattern in the assay, including the HL-60 leukemia, HOP-92 NSCLC, COLO-205 and HT-29 colon, and the MCF-7 breast cell lines. For ALK/IGF-1R dual inhibitors, such as TAE-684, ceritinib (LDK-378), AZD-3463, and GSK-1838705A, the 5-cell line IGF-1R GI50 pattern is supplemented by the addition of the SR line. Alternatively, the addition of the sensitive KM12 line to the IGF-1R pattern was observed for the dual TRK/IGF-1R inhibitors BMS-754807 and BMS-536924. Not surprisingly, the NCI-60 response patterns for the purported ALK inhibitor SOMCl-12-81 and IGF-1R inhibitor XL-228 suggest that these agents more appropriately belong to the class of nondescriptive multikinase inhibitors.
Confluence of screening activities with genetic status of the NCI-60 cell lines
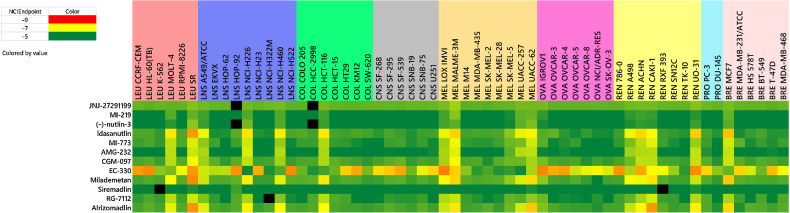
The extensive genomic characterization of the NCI-60 cell lines leads to a discussion of a third theme associated with response patterns generated from the testing of targeted inhibitors in the screen. Although systematic study of gene expression patterns and drug sensitivity data have yielded mixed results, there are several examples where significant correlations between these patterns are obtained (37). Expression of the mutant BRAF V600E gene in several of the melanoma lines and two colon lines in the NCI-60 cell line panel correlated with the response pattern obtained with several compounds specifically designed to inhibit the mutant BRAF V600E protein (Fig. 3A). Previously discussed was the association of the ALK fusion protein with the SR leukemia cell line as well as the NTRK fusion gene with the KM12 colon cell line (Fig. 4B). Another gene that has been characterized as wild-type (WT) or mutant for each of the NCI-60 cell lines is TP53 (38). Cell lines in the NCI-60 panel that express TP53 WT include the SR leukemia, A-549 and NCI-H460 NSCLC, HCT-116 colon, LOXIMVI, MALME-3M, SK-MEL-2, UACC-257, and UACC-62 melanoma, A498, ACHN, CAKI-1, UO-31 renal, and MCF-7 breast cancer cell lines. Not surprisingly, these are the lines that are sensitive to the MDM2-p53 inhibitors in the IOA collection that have been in clinical trials, including idasanutlin, AMG-232, CGM-097, MI-773, RG-7112, milademetan, and alrizomadin (Fig. 5). A grid COMPARE analysis of all these agents based on their NCI-60 GI50s show a range of Pearson correlation s from 0.86 to 0.97. Interestingly, another clinical MDM2 inhibitor (siremadlin) that was less potent in the NCI-60 cell line screen, had Pearson correlations averaging 0.71 to the other MDM2 inhibitors in the IOA set. Several alternative-targeted agents including the antipsychotic drug fluspirelene (39) and natural products including flavopiridol (40) have been reported to have off-target MDM2 inhibitory activity. In the NCI-60 cell line assay, fluspirelene and flavopiridol produced significant responses in the cell lines expressing TP53 WT, although COMPARE analysis of the GI50 sensitivity patterns with the MDM2 inhibitors in the IOA collection showed low Pearson correlations, likely due to the significant sensitivity observed among many other cell lines in the screen.
Figure 5.
Heat maps for TP53-expressing cell lines. NCIEndpoint is color representation of GI50 values ranging from 10−9 M (red) to 10−5 M (green); cells are black when values were not obtained.
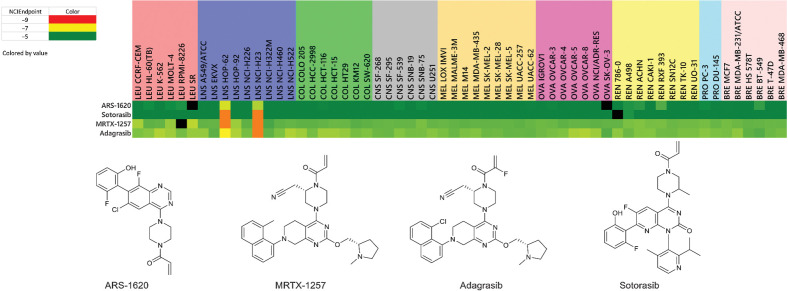
Finally, another gene of interest in the NCI-60 cell lines is KRAS. Specifically, the KRAS G12C mutation has been found to be expressed in two of the NCI-60 cell line panel NSCLC lines, HOP-62 and NCI-H23 (12). As expected, these two cell lines are highly responsive to the selective KRAS G12C inhibitors in the IOA collection, including ARS-1620, sotorasib, MRTX-1257, and adagrasib (Fig. 6).
Figure 6.
Heat maps for KRAS inhibitors. NCIEndpoint is color representation of GI50 values ranging from 10−9 M (red) to 10−5 M (green); cells are black when values were not obtained.
Discussion
Other cell line panel studies that incorporate larger numbers (∼1,000) of human cell lines, including the Cancer Cell Line Encyclopedia (CCLE; ref. 4) and the Cancer Genome Project (CGP; ref. 8), have been used to study the correlations between drug sensitivity profiles and the underlying genomic characteristics of the cancer cells. It has been argued that larger cell line panels are necessary to fully capture the genetic diversity associated with patient tumors that respond at lower rates to treatment with newer targeted therapies (as opposed to more traditional cytotoxic drugs; ref. 41). However, analyses comparing drug response data from the larger panel CCLE and CGP studies have found inconsistencies and poor correlations, likely due to differences in experimental protocols, drug concentrations tested, and analysis tools (42). Other reports have indicated that acceptable consistency in drug response profiling can be achieved between these two studies when differences in analytic methodology and experimental procedures are taken into account (43). Moreover, several cell line drug sensitivity studies have focused on metrics other than potency, including the evaluation of the slope of the concentration–response curve, AUC, and maximum effect to reveal systematic variations in responses (44, 45).
An alternative assessment of the data from these cancer tumor cell line screens focuses on the sensitivity/resistance patterns generated for a given chemotherapeutic agent over the entire range of cell lines examined (independent of potency). These patterns of activity were first defined through application of the Pearson correlation COMPARE algorithm on results of tested agents in the NCI-60 cell line screen more than 20 years ago (46). These sensitivity/resistance patterns serve as fingerprint presentations that encompass target information and the mechanism of action associated with the tested agents. Moreover, a statistical inspection of the respective patterns generated from the CCLE and CGP studies found them to be correlative, even with some discrepancy obtained with the individual compound-response measurements (47).
For many years, the NCI has maintained a growing collection of FDA-approved small-molecule oncology drugs (AOD) that are available individually or as a plated set for use by external cancer researchers (https://dtp.cancer.gov/organization/dscb/obtaining/default.htm). A hierarchical clustering of the NCI-60 growth inhibition sensitivity patterns of these drugs revealed high correlations between those agents with similar mechanism of action (48). Over the last decade, the Developmental Therapeutics Program at NCI has worked to build an additional compound library, the IOA set, through the acquisition of small-molecule investigational clinical and preclinical oncology agents (955), representing a diverse range of therapeutic targets (currently 235 in number) that fully encompass the hallmarks of cancer (49, 50). In general, the compounds chosen for this set are the result of highly optimized medicinal chemistry programs that tend to select for agents with high cell permeability properties.
The evaluation of this expanded set of investigational oncology agents in the NCI-60 cell line assay has allowed for establishment of a phenotypic systems–based resource (https://ioa.cancer.gov) for public use by the cancer research community. This data resource includes all of the raw data from the screening of the IOA and AOD collection along with visualization and analysis tools to allow for comparative study of individual test compounds and multiple compound sets. In addition, suppliers of compounds to the NCI-60 cell line screen can use their data when using the visualization and interactive tools to work with a partial or complete set of the AOD and IOA agents. For example, use of the correlation map view following the running of the COMPARE algorithm with a test compound will readily identify agents that display response patterns that meet a defined minimum correlation criteria for any agents in the AOD or IOA set. Evaluation of the 367 members of a published kinase inhibitor set in this manner produced confirmation of several highly correlative target inhibitor classes of agents (including EGFR and BRAF inhibitors; ref. 30).
This interactive resource can also be used to formulate hypotheses regarding unexpected “off-target” activity for a given test compound. Several examples have been offered herein whereby sensitivity patterns that deviate somewhat from the “signature” pattern anticipated for a given target have been used to suggest possible secondary mechanisms of action. For instance, as discussed previously, several agents of BCR-ABL, ALK, and IGF-1R inhibitor classes that have been confirmed as dual TRK inhibitors, produce responses in the colon KM12 cell line in addition to the NCI-60 patterns associated with their primary mechanism of action. It is a reasonable question to ask whether there are other targeted compounds to which the KM12 cell line responds and whether those agents also inhibit TRK kinase. A check of the IOA library found several examples of this type, including the EGFR inhibitor PF-6459988 and the FGFR inhibitor AZD4547, both of which have been confirmed to have TRK inhibition as a secondary activity. Interestingly, there are many members of the MEK family of inhibitors in the IOA library that also have a pattern that includes sensitivity of the KM12 cell line. Three MEK inhibitors, including the FDA-approved selumetinib, binimetinib, and PD-184352 are not cytotoxic to the KM12 colon cell line, suggesting the possibility of higher MEK selectivity.
Signature sensitivity patterns for some targeted agents can be delineated on the basis of the genomic underpinnings of many of the NCI-60 cell lines. For example, the BRAF inhibitor pattern among most of the melanoma and two of the colon cell lines aligns completely with the cell lines that harbor the BRAF V600E mutation. The NTRK fusion protein is present exclusively in the KM12 colon line and the same is true for ALK fusion protein in the SR lymphoma line. The expression of TP53 WT by a number of cell lines in the NCI-60 set leads to a distinct response pattern for the MDM2 inhibitors in the library. Finally, mutant KRAS leads to a distinct pattern for two NSCLC lines for inhibitors of KRAS G12C.
There are clearly limitations to the use of this resource as a tool for delineating mechanisms of action for a test agent. For example, several classes of targeted agents show flat response patterns, sometimes due to relative inactivity in all of the cell lines in the 2-day NCI-60 screen. As shown, the selective BTK inhibitors are such a class of agents. However, in this case, the relative inactivity of the agent class allows for the ready identification of secondary patterns as is the case for the dual BTK/EGFR inhibitors.
For more than two decades, the NCI-60 cell line screen has been an informative and useful component of the anticancer discovery arena. It is hoped that the targeted AOD and IOA resource described herein provides a further enhancement to this extensively utilized chemotherapeutic discovery tool.
Supplementary Material
Analytical Data
Table of GI50 data
Figures S1 and S2
Acknowledgments
This project was funded in part with federal funds from the NCI, NIH under contract no. 75N91019D00024/75N91020F00003.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This article is featured in Selected Articles from This Issue, p. 1243
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Authors' Disclosures
D.G. Wishka reports other support from NCI during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
J. Morris: Conceptualization, resources, formal analysis, supervision, visualization, writing–original draft, writing–review and editing. M.W. Kunkel: Software, formal analysis, validation, visualization, writing–review and editing. S.L. White: Resources, data curation. D.G. Wishka: Resources, data curation. O.D. Lopez: Resources, data curation. L. Bowles: Resources, data curation. P. Sellers Brady: Resources, data curation. P. Ramsey: Resources, data curation. J. Grams: Resources, data curation. T. Rohrer: Resources, data curation. K. Martin: Resources, data curation. T.S. Dexheimer: Data curation, supervision. N.P. Coussens: Resources, supervision. D. Evans: Resources, supervision. P. Risbood: Resources, data curation. D. Sonkin: Formal analysis, visualization. J.D. Williams: Resources, data curation, writing–review and editing. E.C. Polley: Formal analysis, visualization. J.M. Collins: Formal analysis, supervision. J.H. Doroshow: Conceptualization, resources, supervision. B.A. Teicher: Conceptualization, formal analysis, supervision, methodology, writing–review and editing.
References
- 1. Swinney DC, Anthony J. How were new medicines discovered? Nat Rev Drug Discov 2011;10:507–19. [DOI] [PubMed] [Google Scholar]
- 2. Eder J, Sedrani R, Wiesmann C. The discovery of first-in-class drugs: origins and evolution. Nat Rev Drug Discov 2014;13:577–87. [DOI] [PubMed] [Google Scholar]
- 3. Moffat JG, Rudolph J, Bailey D. Phenotypic screening in cancer drug discovery - past, present and future. Nat Rev Drug Discov 2014;13:588–602. [DOI] [PubMed] [Google Scholar]
- 4. Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483:603–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sharma SV, Haber DA, Settleman J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat Rev Cancer 2010;10:241–53. [DOI] [PubMed] [Google Scholar]
- 6. Basu A, Bodycombe NE, Cheah JH, Price EV, Liu K, Schaefer GI, et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell 2013;154:1151–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Garnett MJ, McDermott U. The evolving role of cancer cell line-based screens to define the impact of cancer genomes on drug response. Curr Opin Genet Dev 2014;24:114–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 2012;483:570–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Iorio F, Knijnenburg TA, Vis DJ, Bignell GR, Menden MP, Schubert M, et al. A landscape of pharmacogenomic interactions in cancer. Cell 2016;166:740–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rees MG, Seashore-Ludlow B, Cheah JH, Adams DJ, Price EV, Gill S, et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat Chem Biol 2016;12:109–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shoemaker RH. The NCI-60 human tumour cell line anticancer drug screen. Nat Rev Cancer 2006;6:813–23. [DOI] [PubMed] [Google Scholar]
- 12. Ikediobi ON, Davies H, Bignell G, Edkins S, Stevens C, O'Meara S, et al. Mutation analysis of 24 known cancer genes in the NCI-60 cell line set. Mol Cancer Ther 2006;5:2606–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blower PE, Verducci JS, Lin S, Zhou J, Chung JH, Dai Z, et al. MicroRNA expression profiles for the NCI-60 cancer cell panel. Mol Cancer Ther 2007;6:1483–91. [DOI] [PubMed] [Google Scholar]
- 14. Liu H, D'Andrade P, Fulmer-Smentek S, Lorenzi KKW, Weinstein JN, et al. mRNA and microRNA expression profiles of the NCI-60 integrated with drug activities. Mol Cancer Ther 2010;9:1080–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Park ES, Rabinovsky R, Carey M, Hennessy BT, Agarwal R, Liu W, et al. Integrative analysis of proteomic signatures, mutations, and drug responsiveness in the NCI-60 cancer cell line set. Mol Cancer Ther 2010;9:257–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Abaan OD, Polley EC, Davis SR, Zhu YJ, Bilke S, Walker RL, et al. The exomes of the NCI-60 panel: a genomic resource for cancer biology and systems pharmacology. Cancer Res 2013;73:4372–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Holbeck SL, Camalier R, Crowell JA, Govindharajulu JP, Hollingshead M, Anderson LW, et al. The National Cancer Institute ALMANAC: a comprehensive screening resource for the detection of anticancer drug pairs with enhanced therapeutic activity. Cancer Res 2017;77:3564–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Monks A, Scudiero D, Skehan P, Shoemaker R, Paull K, Vistica D, et al. Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines. J Natl Cancer Inst 1991;83:757–66. [DOI] [PubMed] [Google Scholar]
- 19. Lorenzi PL, Reinhold W, Varma S, Hutchinson AA, Pommier Y, Chanock SJ, et al. DNA fingerprinting of the NCI-60 cell line panel. Mol Cancer Ther 2009;8:713–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ritz C, Baty F, Streibig JC, Gerhard D. Dose-response analysis using R. PLoS One 2015;10:e0146021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Paull KD, Shoemaker RH, Hodes L, Monks A, Scudiero DA, Rubinstein L, et al. Display and analysis of patterns of differential activity of drugs against human tumor cell lines: development of mean graph and COMPARE algorithm. J Natl Cancer Inst 1989;81:1088–92. [DOI] [PubMed] [Google Scholar]
- 22. Paull KD, Lin CM, Malspeis L, Hamel E. Identification of novel antimitotic agents acting at the tubulin level by computer-assisted evaluation of differential cytotoxicity data. Cancer Res 1992;52:3892–900. [PubMed] [Google Scholar]
- 23. Timofeeva N, Gandhi V. Ibrutinib combinations in CLL therapy: scientific rationale and clinical results. Blood Cancer J 2021;11:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Barf T, Covey T, Izumi R, van de Kar B, Gulrajani M, van Lith B, et al. Acalabrutinib (ACP-196): a covalent bruton tyrosine kinase inhibitor with a differentiated selectivity and in vivo potency profile. J Pharmacol Exp Ther 2017;363:240–52. [DOI] [PubMed] [Google Scholar]
- 25. Liclican A, Serafini L, Xing W, Czerwieniec G, Steiner B, Wang T, et al. Biochemical characterization of tirabrutinib and other irreversible inhibitors of Bruton's tyrosine kinase reveals differences in on- and off - target inhibition. Biochim Biophys Acta Gen Subj 2020;1864:129531. [DOI] [PubMed] [Google Scholar]
- 26. Guo Y, Liu Y, Hu N, Yu D, Zhou C, Shi G, et al. Discovery of zanubrutinib (BGB-3111), a novel, potent, and selective covalent inhibitor of Bruton's tyrosine kinase. J Med Chem 2019;62:7923–40. [DOI] [PubMed] [Google Scholar]
- 27. Reiff SD, Mantel R, Smith LL, Greene JT, Muhowski EM, Fabian CA, et al. The BTK inhibitor ARQ 531 targets ibrutinib-resistant CLL and Richter transformation. Cancer Discov 2018;8:1300–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Loddick SA, Ross SJ, Thomason AG, Robinson DM, Walker GE, Dunkley TP, et al. AZD3514: a small molecule that modulates androgen receptor signaling and function in vitro and in vivo. Mol Cancer Ther 2013;12:1715–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bradbury RH, Callis R, Carr GR, Chen H, Clark E, Feron L, et al. Optimization of a series of bivalent triazolopyridazine based bromodomain and extraterminal inhibitors: the discovery of (3R)-4-[2-[4-[1-(3-Methoxy-[1,2,4]triazolo[4,3-b]pyridazin-6-yl)-4-piperidyl]phen oxy]ethyl]-1,3-dimethyl-piperazin-2-one (AZD5153). J Med Chem 2016;59:7801–17. [DOI] [PubMed] [Google Scholar]
- 30. Elkins JM, Fedele V, Szklarz M, Abdul Azeez KR, Salah E, Mikolajczyk J, et al. Comprehensive characterization of the published kinase inhibitor set. Nat Biotechnol 2016;34:95–103. [DOI] [PubMed] [Google Scholar]
- 31. Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat Biotechnol 2011;29:1039–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tang Y, Gholamin S, Schubert S, Willardson MI, Lee A, Bandopadhayay P, et al. Epigenetic targeting of Hedgehog pathway transcriptional output through BET bromodomain inhibition. Nat Med 2014;20:732–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hai A, Kizilbash NA, Zaidi SH, Alruwaili J, Shahzad K. Differences in structural elements of Bcr-Abl oncoprotein isoforms in chronic myelogenous leukemia. Bioinformation 2014;10:108–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Deininger MW, Druker BJ. Specific targeted therapy of chronic myelogenous leukemia with imatinib. Pharmacol Rev 2003;55:401–23. [DOI] [PubMed] [Google Scholar]
- 35. Carofiglio F, Trisciuzzi D, Gambacorta N, Leonetti F, Stefanachi A, Nicolotti O. Bcr-Abl allosteric inhibitors: where we are and where we are going to. Molecules 2020;25:4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McGahon AJ, Brown DG, Martin SJ, Amarante-Mendes GP, Cotter TG, Cohen GM, et al. Downregulation of Bcr-Abl in K562 cells restores susceptibility to apoptosis: characterization of the apoptotic death. Cell Death Differ 1997;4:95–104. [DOI] [PubMed] [Google Scholar]
- 37. Scherf U, Ross DT, Waltham M, Smith LH, Lee JK, Tanabe L, et al. A gene expression database for the molecular pharmacology of cancer. Nat Genet 2000;24:236–44. [DOI] [PubMed] [Google Scholar]
- 38. Leroy B, Girard L, Hollestelle A, Minna JD, Gazdar AF, Soussi T. Analysis of TP53 mutation status in human cancer cell lines: a reassessment. Hum Mutat 2014;35:756–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Patil SP, Pacitti MF, Gilroy KS, Ruggiero JC, Griffin JD, Butera JJ, et al. Identification of antipsychotic drug fluspirilene as a potential p53-MDM2 inhibitor: a combined computational and experimental study. J Comput Aided Mol Des 2015;29:155–63. [DOI] [PubMed] [Google Scholar]
- 40. Qin JJ, Li X, Hunt C, Wang W, Wang H, Zhang R. Natural products targeting the p53-MDM2 pathway and mutant p53: recent advances and implications in cancer medicine. Genes Dis 2018;5:204–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Seashore-Ludlow B, Rees MG, Cheah JH, Cokol M, Price EV, Coletti ME, et al. Harnessing connectivity in a large-scales-molecule sensitivity dataset. Cancer Discov 2015;5:1210–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Haibe-Kains B, El-Hachem N, Birkbak NJ, Jin AC, Beck AH, Aerts HJ, et al. Inconsistency in large pharmacogenomic studies. Nature 2013;504:389–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cancer Cell Line Encyclopedia Consortium; Genomics of Drug Sensitivity in Cancer Consortium. Pharmacogenomic agreement between two cancer cell line data sets. Nature 2015;528:84–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fallahi-Sichani M, Honarnejad S, Heiser LM, Gray JW, Sorger PK. Metrics other than potency reveal systematic variation in responses to cancer drugs. Nat Chem Biol 2013;9:708–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jenkins JL. Drug discovery: rethinking cellular drug response. Nat Chem Biol 2013;9:669–70. [DOI] [PubMed] [Google Scholar]
- 46. Weinstein JN, Myers TG, O'Connor PM, Friend SH, Fornace AJ Jr, Kohn KW, et al. An information-intensive approach to the molecular pharmacology of cancer. Science 1997;275:343–9. [DOI] [PubMed] [Google Scholar]
- 47. Weinstein JN, Lorenzi PL. Cancer: discrepancies in drug sensitivity. Nature 2013;504:381–3. [DOI] [PubMed] [Google Scholar]
- 48. Holbeck SL, Collins JM, Doroshow JH. Analysis of food and drug administration-approved anticancer agents in the NCI-60 panel of human tumor cell lines. Mol Cancer Ther 2010;9:1451–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 50. Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov 2022;12:31–46. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analytical Data
Table of GI50 data
Figures S1 and S2
Data Availability Statement
The data generated in this study are available within the article and its Supplementary Data and at ioa.cancer.gov.