Abstract
Purpose
Nonerythrocytic αII-spectrin (SPTAN1) variants have been previously associated with intellectual disability and epilepsy. We conducted this study to delineate the phenotypic spectrum of SPTAN1 variants.
Methods
We carried out SPTAN1 gene enrichment analysis in the rare disease component of the 100,000 Genomes Project and screened 100,000 Genomes Project, DECIPHER database, and GeneMatcher to identify individuals with SPTAN1 variants. Functional studies were performed on fibroblasts from 2 patients.
Results
Statistically significant enrichment of rare (minor allele frequency < 1 × 10–5) probably damaging SPTAN1 variants was identified in families with hereditary ataxia (HA) or hereditary spastic paraplegia (HSP) (12/1142 cases vs 52/23,847 controls, p = 2.8 × 10–5). We identified 31 individuals carrying SPTAN1 heterozygous variants or deletions. A total of 10 patients presented with pure or complex HSP/HA. The remaining 21 patients had developmental delay and seizures. Irregular αII-spectrin aggregation was noted in fibroblasts derived from 2 patients with p.(Arg19Trp) and p.(Glu2207del) variants.
Conclusion
We found that SPTAN1 is a genetic cause of neurodevelopmental disorder, which we classified into 3 distinct subgroups. The first comprises developmental epileptic encephalopathy. The second group exhibits milder phenotypes of developmental delay with or without seizures. The final group accounts for patients with pure or complex HSP/HA.
Keywords: Developmental delay, Developmental epileptic encephalopathy, Hereditary ataxia, Hereditary spastic paraplegia, SPTAN1
Introduction
The αII-spectrin gene, SPTAN1 (OMIM 182810), encodes a membrane scaffolding protein that plays an important role in the maintenance of integrity of myelinated axons, axonal development, and synaptogenesis.1 Heterozygous SPTAN1 pathogenic variants have been previously reported with variable phenotypes, most frequently causing mild to severe developmental epileptic encephalopathy (DEE) and developmental delay (DD)2 and rarely with hereditary motor neuropathy and autosomal recessive hereditary spastic paraplegia (HSP).3,4 A mouse model harboring αII-spectrin missense variant (p.Arg1098Gln) was reported to develop progressive ataxia with global neurodegeneration and seizures.5 On the basis of these findings, we carried out a SPTAN1 gene enrichment analysis in the 100,000 Genomes Project (100K GP)6 and identified a statistically significant enrichment for rare probably damaging variants in hereditary ataxia (HA) and HSP groups. In this study, we present an extended phenotypic spectrum of neurologic syndromes caused by pathogenic variations of SPTAN1 gene.
Materials and Methods
Patients
Our initial cohort comprised 100K GP neurology patients.6 All 100K GP genomes were previously screened for single nucleotide variants, small insertions/deletions, structural variants (SVs) or copy number variants (CNVs), and short tandem repeats in relevant genes from the PanelApp virtual gene panels (Genomics England).7 We then screened DECIPHER database cohort for patients carrying single nucleotide variants and/or SVs/CNVs in SPTAN1 gene.8 Additional families were subsequently recruited through GeneMatcher.9 All coding variants reported in this article are with reference to SPTAN1 RefSeq: NM_001130438.3 transcript. All procedures adhered to the principles set out in the Declaration of Helsinki and all patients/their guardians included in the study consented to participation according to ethical approval of the recruiting center.
Gene enrichment analysis
Case-control gene enrichment analysis was performed within the rare disease component of the 100K GP. Cases were defined as all 100K GP probands recruited under HA/HSP, whereas controls were all remaining probands recruited into the 100K GP except for those with neurologic and neurodevelopmental disorders or metabolic disorders. Enrichment of SPTAN1 rare, probably damaging variants in cases compared with controls was assessed via a two-sided Fisher exact test. The contributing variants were defined as rare (minor allele frequency < 1 × 10–5) and either protein-truncating variants or missense variants predicted to be pathogenic by 2 in silico tools (Combined Annotation Dependent Depletion [CADD]10 and Polymorphism Phenotyping [PolyPhen]11).
Functional studies
Fibroblasts derived from patient 1 (p. Arg19Trp) and patient 29 (p. Glu2207del) were used to test the functional effects of SPTAN1 variants on protein expression compared with that of healthy unrelated controls. Western blot analysis, immunocytochemistry, and confocal microscopy were performed as previously described.3
Structural modeling of SPTAN1 missense variants
Three-dimensional protein modeling was used to evaluate the effect of reported SPTAN1 missense variants. Although the crystal structure for full-length αII-spectrin is unknown, crystal structures of the N-terminal tetramerization site and 2 spectrin repeat unit of chicken brain αII-spectrin have been solved (Protein Data Bank: 3F31 and 3Fb2).12,13 We used the Protein Homology/analogY Recognition Engine V 2.0 (Phyre2) predicted models for C-terminal and spectrin repeats 13 to 20 of αII-spectrin protein.14 DynaMut software was used to predict variant effect.15 For simulating amino acid substitutions and visualization, UCSF Chimera built-in tools were used.16 In addition, in silico pathogenicity prediction analysis of all missense variants identified in the study and those previously reported in literature was conducted.
Results
SPTAN1 heterozygous damaging variants are enriched in families with HSP or HA
SPTAN1 was investigated as a candidate gene for HA or HSP using gene enrichment analysis in the rare disease component of the 100K GP, which has a total of 35,422 rare disease families, including 1142 HA/HSP probands as cases and 23,847 non-neurologic/non-metabolic unrelated individuals as controls. A case-control analysis revealed a statistically significant enrichment of rare probably damaging heterozygous variants of SPTAN1 in probands with HA or HSP (12/1142 cases vs 52/23,847 controls, p = .00002846, odds ratio = 4.8594, 95% CI = 2.5867-9.1290) (Supplemental Table 1). None of the SPTAN1 variants found in controls were protein-truncating variants and none overlapped with any of the missense variants described in the study.
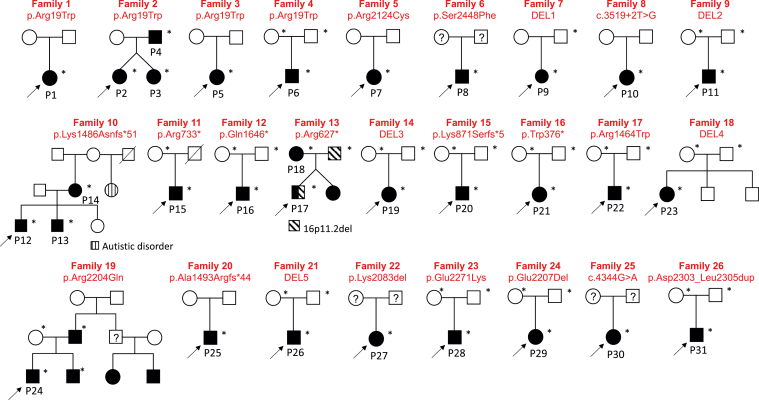
Subsequently, we screened 100K GP neurology cohort (16,014 individuals with neurodevelopmental disorders) for probably damaging SPTAN1 variants in families with spasticity and ataxia in addition to the previously described phenotypes of seizures and/or intellectual disability (ID). We identified 11 patients from 9 families (Table 1, Figures 1 and 2). Patients 1 to 4 had pure HSP phenotype and shared the same SPTAN1 variant, p.(Arg19Trp). Later-onset and a more complex phenotype was noted in patient 8 who harbored p.(Ser2448Phe) variant. Although patient 7 was recruited under early-onset dystonia phenotype, she presented with abnormal eye movements, ataxia, myoclonus, and dyspraxia and had SPTAN1 variant, p.(Arg2124Cys). Patient 10, who had pure ataxia, harbored a heterozygous splice alteration in SPTAN1 (NC_000009.12[SPTAN1_v001]:c.3519+2T>G). SPTAN1 gene was also screened for CNVs/SVs in the 100K GP and 1 deletion was identified. Patient 9 carried a large heterozygous in-frame deletion (DEL1), encompassing exons 25 to 27 (Supplemental Figure 1) and presented with pure HA. Both patients 9 and 10 carried sporadic SPTAN1 variants because the de novo nature could not be confirmed owing to the unavailability of family members. Additional 3 probands with SPTAN1 variants presenting with seizures, ID, and ataxia/spasticity were identified.
Table 1.
Clinical and genetic findings of 31 patients reported in this study
| General Information | Family 1 |
Family 2 |
Family 3 |
Family 4 |
Family 5 |
Family 6 |
Family 7 |
Family 8 |
||
|---|---|---|---|---|---|---|---|---|---|---|
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | Patient 7 | Patient 8 | Patient 9 | Patient 10 | |
| Sex/ethnicity | F/Asia | F/Europe | F/Europe | M/Europe | F/Europe | M/Europe | F/NA | M/Asia | F/NA | M/Europe |
| Age at last examination, y | 40 | 25 | 25 | 50 | 17 | 15 | 32 | 50 | 72 | 54 |
| Genetic findings | ||||||||||
| c.DNA | c.55C>T | c.55C>T | c.55C>T | c.55C>T | c.6370C>T | c.7343C>T | arr{hg38}9q34.11(128,597,905-128,602,892)x1 | NC_000009.12(SPTAN1_v001):c.3519+2T>G exon 25 | ||
| Protein | p.Arg19Trp | p.Arg19Trp | p.Arg19Trp | p.Arg19Trp | p.Arg2124Cys | p.Ser2448Phe | p.(Asp1139_Lys1193)del (DEL 1) |
p.? | ||
| Inheritance | Sporadic | AD | Sporadic | De novo | Sporadic | Unknown | Sporadic | Sporadic | ||
| Phenotypic category | Pure HSP/ataxia | Pure HSP/ataxia | Complex HSP | Pure HSP | Pure HSP/ataxia | Complex HSP | Pure HA | Pure HA | ||
| Initial symptoms | Spastic gait | Spastic gait | Spastic gait | Spastic gait | Learning disability/ spastic ataxia | Spastic gait | Spastic gait | Spastic gait | Ataxia | Ataxia |
| Age of onset, y | 8 | NA | NA | NA | 10 | 8 | NA | NA | 36 | 35 |
| SZ (age of onset) | – | – | – | – | Generalized (10 y) | – | – | – | – | – |
| Response to therapy | / | / | / | / | VAL; controlled | / | / | / | / | / |
| EEG | NA | NA | NA | NA | Normal | NA | NA | NA | NA | NA |
| Developmental history | ||||||||||
| ID | – | – | – | – | – | – | – | + | – | – |
| Learning disability | – | – | – | – | + | – | – | + | – | – |
| Motor delay | – | – | – | – | – | – | – | – | – | – |
| Language delay | – | – | – | – | – | – | – | – | – | – |
| Microcephaly | – | – | – | – | – | – | – | – | – | – |
| Neurologic findings | ||||||||||
| Ataxia | + | + | + | + | + | – | + | – | + | + |
| Spasticity | + | + | + | + | + | + | + | + | – | – |
| Extensor planter reflex | + | + | – | – | – | – | – | |||
| Abnormal eye movement | – | + | + | + | Nystagmus | – | + | Nystagmus | – | Nystagmus, esotropia |
| UL weakness | + | + | + | + | – | – | – | – | – | |
| LL weakness | + | + | + | + | + | + | – | – | – | – |
| LL hyperreflexia | + | + | + | + | + | + | – | – | – | – |
| Ankle clonus | + | + | + | – | – | – | ||||
| Bladder dysfunction | + | + | + | + | – | – | + | – | – | |
| Amyotrophy | – | + | + | + | – | – | – | – | – | |
| Myoclonus | – | – | – | – | – | – | + | + | – | – |
| Brain MRI | Normal | NA | NA | NA | Subcortical white mater hyper intensities | Normal | NA | Cerebellar atrophy | NA | Cerebellar atrophy marked in vermis |
| Other clinical features | – | Impaired vibratory sensation | – | – | – | Mild hearing loss, atrophic left kidney | Head tremors, dyspraxia, postural tremors | Fasciculations, impaired proprioception, pes cavus, macular dystrophy | Apraxia, adult-onset sensorineural hearing loss | Areflexia and decreased vibration sense in LL, vertical ophthalmoparesis |
| General Information | Family 9 |
Family 10 |
Family 11 |
Family 12 |
Family 13 |
Family 14 |
Family 15 |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| Patient 11 | Patient 12 | Patient 13 | Patient 14 | Patient 15 | Patient 16 | Patient 17 | Patient 18 | Patient 19 | Patient 20 | |
| Sex/ethnicity | M/Europe | M/Europe | M/Europe | F/Europe | M/Europe | M/NA | M/Europe | F/Europe | F/Europe | M/Europe |
| Age at last examination, y | 19 | 9 | 7 | 26 | 13 | 2 | 2 | 32 | 2 | 6 |
| Genetic findings | ||||||||||
| c.DNA | arr{hg38}9q34.11 (128,609,213-128,613,675)x1 | c.4458delA | c.2197C>T | c.4936C>T | c.1879C>T | arr{hg38}9q34.11 (128591376-128600369)x1 | c.2612del | |||
| Protein | p.(Ile1563_?)del (DEL2) | p.Lys1486Asnfs∗51 | p.Arg733∗ | p.Gln1646∗ | p.Arg627∗ | p.(Asp1003_Lys1193)del (DEL3) |
p.Lys871Serfs∗5 | |||
| Inheritance | De novo | AD | sporadic | De novo | AD | De novo | De novo | |||
| Phenotypic category | DD/ataxia | DD | DD | DD | DD | DD | DD | |||
| Initial symptoms | Ataxia | Speech delay | Speech delay | ID | ID | DD | Axial hypotonia | Axial hypotonia | DD | ID |
| Age of onset | 15 mo | 2 y | 2 y | 3 y | NA | Birth | 5 mo | 3 mo | 9 mo | 1 y |
| SZ (age of onset) | Febrile SZ | – | – | – | – | – | – | – | – | One febrile generalized tonic-clonic SZ (6 y) |
| Response to therapy | / | / | / | / | / | / | / | / | / | No therapy, controlled |
| EEG | NA | NA | NA | NA | NA | Increased excitability | NA | Normal | NA | Normal |
| Developmental history | ||||||||||
| ID | + | Mild | Mild | – | Mild | + | + | – | ? | – |
| Learning disability | + | + | + | + | + | + | + | + | ? | Mild |
| Motor delay | + | – | – | – | – | + | + | + | + | Mild |
| Speech delay | + | + | + | – | – | + | + | NA | + | Moderate |
| Microcephaly | – | – | – | – | + | – | – | – | – | |
| Neurologic findings | ||||||||||
| Ataxia | Severe | – | – | – | – | – | + | + | – | – |
| Spasticity | – | – | – | – | – | – | – | – | ||
| Extensor planter reflex | – | – | – | – | – | – | – | – | – | – |
| Abnormal eye movement | Convergent strabismus | – | – | – | – | – | Nystagmus | Nystagmus | Strabismus | – |
| UL weakness | – | – | – | – | – | – | – | – | – | – |
| LL weakness | – | – | – | – | – | – | + | + | – | – |
| LL hyperreflexia | – | – | – | – | – | – | – | – | – | – |
| Ankle clonus | – | – | – | – | – | – | – | – | – | – |
| Bladder dysfunction | + | – | – | – | – | – | – | – | – | – |
| Amyotrophy | NA | – | – | – | – | – | – | – | – | – |
| Myoclonus | – | – | – | – | – | – | – | – | – | – |
| Brain MRI | Severe cerebellar atrophy and slightly dilated fourth ventricle | NA | NA | NA | NA | Nonspecific small gliosis in the right anterior border zone | Cerebellar atrophy | Severe cerebellar atrophy | NA | NA |
| Other clinical features | Pneumococcal meningitis aged 15 months, hypermetropia | Attention deficit Hyperactivity disorder | – | Basedow disease in remission | obesity; dyslipidemia; testicular hypoplasia (MC4R disease) | epicanthus, low-set ears, high philtrum, finger pads, sickle feet, sandal furrow | 16p11.2 microdeletion involving PRRT2 | – | hypotonia, poor motor planning and body awareness, tongue tie | obesity, constipation, mild myopia |
| General Information | Family 16 |
Family 17 |
Family 18 |
Family 19 |
Family 20 |
Family 21 |
|---|---|---|---|---|---|---|
| Patient 21 | Patient 22 | Patient 23 | Patient 24 | Patient 25 | Patient 26 | |
| Sex/Ethnicity | F/Europe | M/NA | F/Europe | M/Europe | M/NA | M/Europe |
| Age at last examination, y | 8 | 9 | 5 | 10 | 9 | 8 |
| Genetic findings | ||||||
| c.DNA | c.1127G>A | c.4390C>T | arr{hg38}9q34.11 (128,582,754-128,587,726)x1 | c.6611G>A | c.4476del | arr{hg38}9q34.11 (128,587,422-128,600,316)x1 |
| Protein | p.Trp376∗ | p.Arg1464Trp | p.(Asn571_?)del (DEL4) | p.Arg2204Gln | p.Ala1493Argfs∗44 | p.(Ala927_Lys1193) del (DEL5) |
| Inheritance | De novo | De novo | De novo | AD | Sporadic | De novo |
| Phenotypic category | DD | DD/SZ | DD/SZ | DD/SZ | DD/SZ | DD/SZ |
| Initial symptoms | Speech delay | SZ | Hypotonia | DD | ID | SZ |
| Age of onset | 2 y | NA | 2 mo | 2 y | 2 y | 3 mo |
| SZ (age of onset) | – | Febrile and generalized myoclonic (3 y) | + (2 y) | Myoclonic absence (5 y) | Absence and generalized tonic-clonic (6 y) | Infantile spasms (3 mo) |
| Response to therapy | / | NA | CBZ, controlled | NA | Refractory | Partially controlled |
| EEG | NA | NA | NA | NA | Typical absence, continuous spikes and waves during sleep aspect | Vertex sharp waves, right and left independent parietal waves during sleep |
| Developmental history | ||||||
| ID | – | + | + | + | + | + |
| Learning disability | + | + | + | + | + | + |
| Motor delay | – | + | + | + | – | + |
| Speech delay | + | + | + | + | + | + |
| Microcephaly | + | + | – | – | – | + |
| Neurologic findings | ||||||
| Ataxia | – | + | + | – | – | – |
| Spasticity | – | NA | – | – | – | – |
| Extensor planter reflex | – | – | – | – | – | – |
| Abnormal eye movement | – | – | Strabismus | – | – | – |
| UL weakness | – | NA | – | – | – | – |
| LL weakness | – | NA | – | – | – | – |
| LL hyperreflexia | – | – | – | – | – | – |
| Ankle clonus | – | – | – | – | – | – |
| Bladder dysfunction | – | – | – | – | – | – |
| Amyotrophy | – | – | – | – | – | – |
| Myoclonus | – | – | – | – | – | – |
| Brain MRI | Normal | NA | Mild cerebral atrophy | Normal | Normal | Mild delay in myelination |
| Other clinical features | Dyspraxia, hyperlaxity | – | Hypotonia, sleep problems, constipation | – | ADHD | Poor proprioception, hypotonia |
| General Information | Family 22 |
Family 23 |
Family 24 |
Family 25 |
Family 26 |
|---|---|---|---|---|---|
| Patient 27 | Patient 28 | Patient 29 | Patient 30 | Patient 31 | |
| Sex/ethnicity | F/Europe | M/Europe | F/Africa | F/Europe | M/India |
| Age at last examination, y | 16 | 12 | 20 | 7 | 1 |
| Genetic findings | |||||
| c.DNA | c.6247_6249del | c.6811G>A | c.6619_6621del | c.4344G>A | c.6908_6916dup |
| Protein | p.Lys2083del | p.Glu2271Lys | p.Glu2207del | p.Gln1448= (splice) exon 38 |
p.Asp2303_Leu2305dup |
| Inheritance | Unknown | De novo | De novo | Unknown | De novo |
| Phenotypic category | DEE | DEE | DEE | DEE | DEE |
| Initial symptoms | SZ | SZ | Hypotonia | Axial hypotonia | SZ |
| Age of onset | 2 mo | 4 mo | 8 mo | Birth | Birth |
| SZ (age of onset) | Absence SZ (2 mo) | Myoclonic jerks, dystonic spasms (4 mo) | Tonic, oral automatisms, upward deviation of gaze | West syndrome (4 mo) | Generalized tonic-clonic (birth) |
| Response to therapy | NA | Refractory | Controlled | Refractory | Refractory |
| EEG | NA | Slow-wave activity, loss of normal rhythms, temporal spike, sharp wave discharges | Diffuse slow activity, irregular low to medium amplitude | Hypsarrhythmia | Multifocal epilepsy |
| Developmental history | |||||
| ID | + | + | + | severe | NA |
| Learning disability | + | + | + | severe | NA |
| Motor delay | + | + | + | severe | severe |
| Speech delay | + | + | + | severe | NA |
| Microcephaly | – | + | + | + | severe |
| Neurologic findings | |||||
| Ataxia | + | – | – | NA | NA |
| Spasticity | – | – | + | NA | + |
| Extensor planter reflex | – | – | + | – | + |
| Abnormal eye movement | Strabismus | – | Strabismus | Nystagmus | Nystagmus |
| UL weakness | – | – | + | + | + |
| LL weakness | – | – | + | + | + |
| LL hyperreflexia | – | + | – | – | + |
| Ankle clonus | – | – | – | – | + |
| Bladder dysfunction | – | – | – | – | – |
| Amyotrophy | – | – | + | – | – |
| Myoclonus | – | – | – | – | – |
| Brain MRI | NA | Cerebellar atrophy, delayed myelination, thin corpus callosum | NA | Delayed myelination, progressive brain and pontocerebellar atrophy | Delayed myelination, cerebral atrophy, thin corpus callosum |
| Other clinical features | – | Rod cone retinal dystrophy, GERD, scoliosis, dislocated hip | Visual impairment, dysmorphism, scoliosis | – | – |
All variants are reported with reference to RefSeq NM_001130438.3.
AD, autosomal dominant; ADHD, attention deficit hyperactivity disorder; CBZ, carbamazepine; DD, developmental delay; DEE, developmental epileptic encephalopathy; EEG, electroencephalography; F, female; GERD, gastroesophageal reflux; HA, hereditary ataxia; HSP, hereditary spastic paraplegia; ID, intellectual disability; LL, lower limbs; M, male; MRI, magnetic resonance imaging; NA, not available; SZ, seizures; UL, upper limbs; VAL, valproic acid; /, not applicable; –, absence of manifestation; +, presence of manifestation.
Figure 1.
Pedigrees of reported families with SPTAN1 variants showing disease segregation.
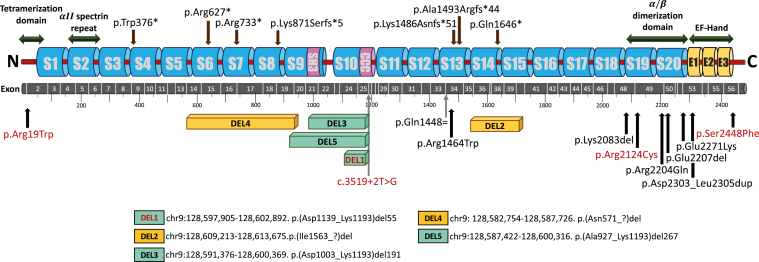
Figure 2.
Schematic structure of SPTAN1 gene and its coding protein highlighting variants identified in this study. Coding exon numbers (NM_001130438.3) are reported on the gray bar. Truncating variants are indicated on the top. Missenses, in-frame deletion/insertion, and splice variants are on the bottom. Deletion 1, 3, and 5 (green) remain in frame, whereas predictions for deletions 2 and 4 (orange) are not available. p.Gln1448= (c.4344G>A) is predicted to affect exon 33 donor splice site, based on maxENTScan (predicting splice sites using ‘Maximum Entropy Principle’) (maxENT score wild-type 6.99 → 3.84 mutant). The splice altering variant (NC_000009.12(SPTAN1_v001):c.3519+2T>G) predicted to alter exon 25 canonical donor splice site (maxENT score wild-type 10.28 → 2.63 mutant). Variants identified in patients presenting with HSP/HA are highlighted in red. All other variants are represented in black.
By screening DECIPHER database,8 4 additional variants were identified; 1 missense variant in patient 22 and 3 de novo microdeletions in the SPTAN1 gene. DEL2 in patient 11 is a 4.46-kilobase deletion that removes exons 36 to 40. This patient presented with ataxia and severe DD. Patients 23 and 26, who carried DEL4 (exons 14-20) and DEL5 (exons 20-27), respectively, presented with DD and seizures.
A total of 13 additional SPTAN1 families were identified through GeneMatcher.9 Patients 5 and 6 shared the same de novo missense variant, p.(Arg19Trp), and HSP phenotype as patients 1 to 4. Nevertheless, patient 5 presented with complex HSP, learning disability, and seizures. Three frameshift variants were identified in 5 patients (patients 12-14, 20, and 25) with DD/Seizures. Dominant inheritance was noted in 3 patients (patients 12-14). Five patients (patients 15-18 and 21) with ID/seizures carried nonsense SPTAN1 variants. The last variant was an in-frame deletion (DEL3), which was identified in patient 19, a 2-year-old presented with DD. This deletion is almost 9 kilobase, encompasses exons 22 to 27, and overlaps with DEL5. All variants reported in this study were classified according to the guidelines of the American College of Medical Genetics and Genomics and Association for Molecular Pathology.17 (Supplemental Table 2).
Clinical phenotypes
Detailed clinical information was collated for 31 individuals from 26 unrelated families carrying heterozygous variants in SPTAN1 (Table 1). Common phenotypes including ID/learning disability and motor delay were reported in 73.5% and 58.8% of our cohort, respectively. Remarkably, around half of the patients (15/31) manifested ataxia, and seizures were reported in almost one-third of the cases (12/31).
Recently reported phenotypes of HSP/HA were identified in a subgroup of our patients. A total of 8 individuals from 6 families presented with pure HSP/HA and further 2 families with complex HSP (Supplemental Video 1). All patients with HSP showed typical features. Most of them had lower limb hyperreflexia and/or ataxia (6/8) whereas none had sensory abnormalities. In contrast, 2 patients with HA showed a pure phenotype. Abnormal eye movement, a common condition in patients with ataxia, was noted in 70% of this HSP/HA group. Severe phenotype of DEE was reported in 5 patients. All had seizures in early months of life and had severe DD. The remaining 16 patients from 13 families presented with varying degrees of DD.
Functional consequences of SPTAN1 variants in patient-derived fibroblasts
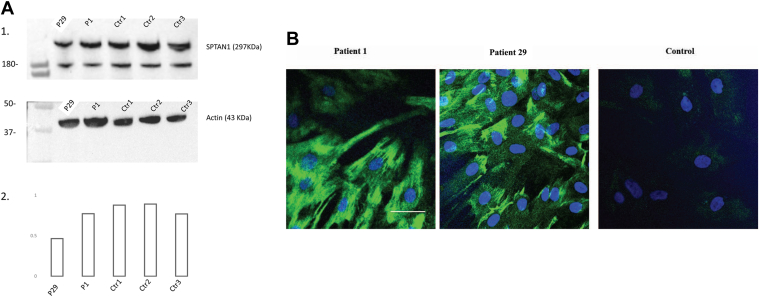
We analyzed fibroblasts derived from 2 patients with the missense variants p.(Arg19Trp) and p.(Glu2207del) (Figure 3). Although western blot did not show a quantitative difference in protein expression between patient 1 and controls, a quantitative reduction of protein expression in patient 29 was noted. Nevertheless, a high immunofluorescence brightness and intense immunoreactivity of αII-spectrin was observed in both the studied patients. In patient 1, αII-spectrin was ubiquitously expressed throughout the fibroblast cells, whereas there was a more localized aggregation of αII-spectrin in the plasma membrane with a relatively higher immunofluorescence brightness observed in the fibroblasts of patient 29.
Figure 3.
Representative images of αII-spectrin protein expression and staining pattern in fibroblast cells derived from 2 patients and unrelated controls. A. Western blot. 1. Western blotting of protein extracted from fibroblast cell lines of patients 1 and 29 and 3 wild-type age-matched controls. 2. Densitometric analysis of western blot using BioRad Image Lab software after relative normalization to actin as a housekeeping protein. The analysis showed no change in protein expression in patient 1 but showed a quantitative reduction of protein expression in patient 29. B. Immunocytochemical staining of αII-spectrin expression in primary fibroblasts of patients 1 and 29 and unrelated control individual with Alexa Fluor 488 conjugated secondary antibody (green) and Hoescht 33342 nuclear staining (blue). Scale bar represents 50 μm. Immunocytochemical staining showed high immunofluorescence brightness and intense immunoreactivity and aggregation of αII-spectrin in both studied patients compared with the healthy unrelated control. Ctr, control.
Structural modeling of SPTAN1 missense variants
The effect of missense variants on αII-spectrin protein structure (Q13813-1) was investigated using homology modeling of experimentally validated models12, 13, 14 (Supplemental Figure 2). Variant p.(Arg19Trp) had the most deleterious effect on protein structure. This variant is located within the N-terminal tetramerization domain and results in steric clashes with 2 leucine residues in the beta chain. Less severe effect was noted in modeling of p.(Arg1464Trp) and p.(Arg2204Gln) variants. Structural modeling showed no structural effect for p.(Glu2271Lys), p.(Arg2124Cys), and p.(Ser2448Phe); however these would likely affect αII-spectrin heterodimerization with its partner, β-spectrin. Overall, most (4/6) of the reported missense variants led to protein destabilization (Supplemental Table 3). In addition, according to in silico pathogenicity prediction, almost all missense variants identified in this study are potentially pathogenic (Supplemental Table 4).
Discussion
Identification of an enrichment of SPTAN1 heterozygous variants in patients presenting with HA and HSP confirms SPTAN1 involvement in a wide phenotypic spectrum. We suggest that SPTAN1 is a genetic cause of neurodevelopmental disorders, with 3 phenotypic subgroups (Table 1). The first group comprises patients with DEE presenting with severe phenotype (OMIM 613477). DEE was identified in 5 families, consistent with previous reports.2 A total of 16 patients manifested milder phenotype of DD with or without childhood-onset seizures forming the second phenotypic group. The final group consists of patients (n = 10) with pure or complex HSP/HA.
Involvement of SPTAN1 variants in peripheral nervous system abnormalities was previously reported for heterozygous variants causing hereditary motor sensory neuropathy3 and biallelic variants associated with autosomal recessive HSP.4 The phenotype of HA and HSP is further supported by a previously reported SPTAN1 mouse model presenting with unsteady gait and spasticity5 (Supplemental Video 2) and the recently published study reporting de novo and dominant variants of SPTAN1 in patients with ataxia and patients with spastic paraplegia.18
Phenotypic heterogeneity may be explained by the involvement of SPTAN1 pathogenic variants in different mechanisms of pathogenicity as it was described with other structural proteins.19 Syrbe et al2 concluded that variants in the last 2 αII-spectrin repeats are associated with severe phenotype due to protein aggregation with dominant negative effect. This mechanism is supported by our observation in αII-spectrin immunocytochemistry experiment performed on fibroblast of patient 29. However, further experiments on multiple cell lines would be imperative to support this hypothesis.
We noted that the excess of truncating variants in the milder category of our cohort (DD +/– seizures) is in agreement with the proposed mechanism of quantitative defect of αII-spectrin protein leading to a milder phenotype.20 We suggest that truncating variants are responsible for a mild DD with or without epilepsy.
In our HSP/HA group, we have detected accumulation of αII-spectrin in fibroblasts of patient 1 with the recurrent p.(Arg19Trp) variant, indicating abnormal protein function. Such interesting finding adds further evidence to Van de Vondel et al18 report of the recurrent p.(Arg19Trp) variant detected in 7 families with spastic paraplegia. All other identified variants in this group of our patients were missense variants except for 1 splice altering variant. All are predicted to have a moderate protein effect except for p.(Arg19Trp), which is localized at an essential position of the N-terminal tetramerization domain.1 An arginine to tryptophan change, p.(Arg35Trp), at the N-terminus has been reported previously in erythrocytic α-spectrin gene (SPTA1), where it prevented N-terminal domain of α-spectrin to form heterotetramers with its beta partner. We suggest a similar mechanism for SPTAN1 variant, p.(Arg19Trp).21 In a previous report, we showed that ataxia and/or HSP cases may be accounted by hypomorphic pathogenic variants in genes known to manifest with severe phenotypes when mutated.22 The p.(Arg2124Cys) and p.(Ser2448Phe) variants identified in families 5 and 6 are missense variants that are predicted to introduce mild structural alterations in the αII-spectrin protein potentially explaining the milder neurologic impairment. In family 8, with mild and late-onset ataxia, a splice altering variant (c.3519+2T>G) was detected. The resulting predicted in-frame deletion rather than a loss of function might explain the mild neurologic phenotype in this family.
It is interesting to notice that αII-spectrin forms heterotetramers with each of the 4 nonerythrocytic β-spectrins 1 to 4 at precise membrane domains.23 The first 3 β-spectrin genes (SPTBN1, SPTBN2, and SPTBN4) are responsible of multiple neurologic disorders, depending on the gene and inheritance pattern.19 Particularly, βIII spectrin (SPTBN2) is specific to Purkinje cells and is involved in a relatively pure late-onset ataxia phenotype.24 αII-Spectrin broad distribution in the neurons is therefore a plausible explanation of the pleiotropic consequences of SPTAN1 variants.
Molecular diagnosis of patients with neurodevelopmental disorders is often challenging owing to both phenotypic and genetic heterogenicity. We were able to expand the phenotypic and genetic spectrum of SPTAN1 variants, shedding light on the critical role that αII-spectrin has in maintaining brain health.
Data Availability
De-identified data are available upon request. Data requests can be made via email to the corresponding author and will be pending data use agreements.
Conflict of Interest
The authors declare no conflicts of interest.
Acknowledgments
H.M., J.V., and H.H. are supported by an Medical Research Council strategic award, MR/S005021/1, to establish International Centre for Genomic Medicine in Neuromuscular Diseases. H.M. is supported by Wellcome Trust grant 220906/Z/20/Z. H.H. is funded by the Medical Research Council (MR/S01165X/1, MR/S005021/1, G0601943), NIHR University College London Hospitals Biomedical Research Centre, Rosetree Trust UK, Ataxia UK, Multiple System Atrophy Trust, Brain Research UK, Sparks GOSH Charity, Muscular Dystrophy UK, and Multiple System Atrophy Trust. R.But. is supported by the Penelope Rare and Undiagnosed Disease Program at the University of Utah with funding from the Center for Genomic Medicine and with support from Matt Velinder (Department of Human Genetics, University of Utah) and Rong Mao and Pinar Bayrak-Toydemir (ARUP Laboratories). B.M. is supported by NKFIH K138669. This research was made possible through access to the data and findings generated by the 100,000 Genomes Project. The 100,000 Genomes Project is managed by Genomics England Limited (a wholly owned company of the Department of Health and Social Care). The 100,000 Genomes Project is funded by the National Institute for Health and Care Research and NHS England. The Wellcome Trust, Cancer Research UK, and the Medical Research Council have also funded the research infrastructure. The 100,000 Genomes Project uses data provided by patients and collected by the National Health Service as part of their care and support. This study makes use of data generated by the DatabasE of genomiC varIation and Phenotype in Humans using Ensembl Resources (DECIPHER) community. A full list of centers which contributed to the generation of the data is available at https://deciphergenomics.org/about/stats and via email from contact@deciphergenomics.org. Funding for the DECIPHER project was provided by Wellcome. We are thankful to the Deciphering Developmental Disorders Study for the invaluable collaboration. The Deciphering Developmental Disorders Study (Cambridge South Research Ethics Committee approval 10/H0305/83 and the Republic of Ireland Research Ethics Committee GEN/284/12) presents independent research commissioned by the Health Innovation Challenge Fund (grant number HICF-1009-003), a parallel funding partnership between the Wellcome Trust and Department of Health and the Wellcome Trust Sanger Institute (grant number WT098051). We also thank Lise Larrieu and Morgane Pointaux (Molecular Genetics’ Laboratory, Institut Universitaire de Recherche Clinique, CHU de Montpellier and PhyMedExp Univ Montpellier, Montpellier, France) for their excellent technical assistance. We appreciate the help of Grenenko Cecile (Service de Neurologie Pédiatrique, Centre Hospitalier Universitaire Amiens-Picardie, Amiens, France) and Jedraszak Guillaume new (Laboratoire de Génétique Constitutionnelle, Centre Hospitalier Universitaire Amiens-Picardie, Amiens, France) in the study. We are thankful to Béatrice Desnous (Reference Center for Inherited Metabolic Diseases, Marseille, France) for his contribution to clinical investigation. Finally, we are grateful to all the patients and their families.
Author Information
Conceptualization: H.M., H.H.; Data Curation: H.M., M.B., M.St.; Funding Acquisition: M.Ha., H.H.; Investigation: R.M., S.E., M.O., M.Su., S.Ban., J.C.-S., T.W., M.R., J.A.B., M.N., B.Co., R.A.J., T.B., M.He., J.G.-A., I.K., D.W., F.G., M.M., A.G., G.B.-L., H.G.-M., P.G., B.Ch., S.Bac., R.Bui., V.M., M.Z., R.But., D.H., G.B.-L., A.S., B.M., K.S., M.S., V.K.G., V.M.S.; Methodology: K.Z., E.C., V.C., C.R., E.G.; Software: D.M.; Resources: M.S.; Supervision: M.St., M.K., J.V., H.H.; Writing-original draft: H.M., M.B.; Writing-review and editing: R.M., S.E., M.O., M.Su., S.Ban., J.C.-S., T.W., R.M., J.A.B., M.N., B.Co., R.A.J., T.B., H.M., J.G.-A., I.K., D.W., F.G., M.M., A.G., G.B.-L., H.G.-M., P.G., B.Ch., S.Bac., R.Bui., M.He., V.M., M.Z., R.But., D.H., G.B.-L., A.S., B.M., K.S., M.S., V.K.G., V.M.S., A.G., K.Z., E.C., V.C., C.R., E.G., D.M., M.Ha., M.St., M.K., J.V., H.H.
Ethics Declaration
The study was carried out as per the ethical approval from the University College London Hospital Research Ethics Committee, reference: 07/Q0512/26. All institutions involved in this research received local ethical approval. All patients’ data were de-identified. Informed consent was obtained from all participants, their parents, or legal representatives. The authors have received and archived written patient consent for publishing the video of the individual included in this study. Experiments involving mice were approved by the local Ethical Committee in Wroclaw (Poland) under permission number 78/2018 and by the Institutional Animal Care and Use Committee at Yale University, which is accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care.
Footnotes
Additional Information
The online version of this article (https://doi.org/10.1016/j.gim.2022.09.013) contains supplementary material, which is available to authorized users.
Contributor Information
Heba Morsy, Email: heba.morsy@ucl.ac.uk.
Henry Houlden, Email: h.houlden@ucl.ac.uk.
Genomics England Research Consortium:
J.C. Ambrose, P. Arumugam, E.L. Baple, M. Bleda, F. Boardman-Pretty, J.M. Boissiere, C.R. Boustred, H. Brittain, M.J. Caulfield, G.C. Chan, C.E.H. Craig, L.C. Daugherty, A. de Burca, A. Devereau, G. Elgar, R.E. Foulger, T. Fowler, P. Furió-Tarí, J.M. Hackett, D. Halai, A. Hamblin, S. Henderson, J.E. Holman, T.J.P. Hubbard, K. Ibáñez, R. Jackson, L.J. Jones, D. Kasperaviciute, M. Kayikci, L. Lahnstein, K. Lawson, S.E.A. Leigh, I.U.S. Leong, F.J. Lopez, F. Maleady-Crowe, J. Mason, E.M. McDonagh, L. Moutsianas, M. Mueller, N. Murugaesu, A.C. Need, C.A. Odhams, C. Patch, D. Perez-Gil, D. Polychronopoulos, J. Pullinger, T. Rahim, A. Rendon, P. Riesgo-Ferreiro, T. Rogers, M. Ryten, K. Savage, K. Sawant, R.H. Scott, A. Siddiq, A. Sieghart, D. Smedley, K.R. Smith, A. Sosinsky, W. Spooner, H.E. Stevens, A. Stuckey, R. Sultana, E.R.A. Thomas, S.R. Thompson, C. Tregidgo, A. Tucci, E. Walsh, S.A. Watters, M.J. Welland, E. Williams, K. Witkowska, S.M. Wood, and M. Zarowiecki
Members of The Genomics England Research Consortium
J.C. Ambrose, P. Arumugam, E.L. Baple, M. Bleda, F. Boardman-Pretty, J.M. Boissiere, C.R. Boustred, H. Brittain, M.J. Caulfield, G.C. Chan, C.E.H. Craig, L.C. Daugherty, A. de Burca, A. Devereau, G. Elgar, R.E. Foulger, T. Fowler, P. Furió-Tarí, J.M. Hackett, D. Halai, A. Hamblin, S. Henderson, J.E. Holman, T.J.P. Hubbard, K. Ibáñez, R. Jackson, L.J. Jones, D. Kasperaviciute, M. Kayikci, L. Lahnstein, K. Lawson, S.E.A. Leigh, I.U.S. Leong, F.J. Lopez, F. Maleady-Crowe, J. Mason, E.M. McDonagh, L. Moutsianas, M. Mueller, N. Murugaesu, A.C. Need, C.A. Odhams, C. Patch, D. Perez-Gil, D. Polychronopoulos, J. Pullinger, T. Rahim, A. Rendon, P. Riesgo-Ferreiro, T. Rogers, M. Ryten, K. Savage, K. Sawant, R.H. Scott, A. Siddiq, A. Sieghart, D. Smedley, K.R. Smith, A. Sosinsky, W. Spooner, H.E. Stevens, A. Stuckey, R. Sultana, E.R.A. Thomas, S.R. Thompson, C. Tregidgo, A. Tucci, E. Walsh, S.A. Watters, M.J. Welland, E. Williams, K. Witkowska, S.M. Wood, M. Zarowiecki
Additional Information
References
- 1.Morrow J.S., Stankewich M.C. The spread of spectrin in ataxia and neurodegenerative disease. J Exp Neurol. 2021;2(3):131–139. [PMC free article] [PubMed] [Google Scholar]
- 2.Syrbe S., Harms F.L., Parrini E., et al. Delineating SPTAN1 associated phenotypes: from isolated epilepsy to encephalopathy with progressive brain atrophy. Brain. 2017;140(9):2322–2336. doi: 10.1093/brain/awx195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beijer D., Deconinck T., De Bleecker J.L., et al. Nonsense mutations in alpha-II spectrin in three families with juvenile onset hereditary motor neuropathy. Brain. 2019;142(9):2605–2616. doi: 10.1093/brain/awz216. [DOI] [PubMed] [Google Scholar]
- 4.Leveille E., Estiar M.A., Krohn L., et al. SPTAN1 variants as a potential cause for autosomal recessive hereditary spastic paraplegia. J Hum Genet. 2019;64(11):1145–1151. doi: 10.1038/s10038-019-0669-2. [DOI] [PubMed] [Google Scholar]
- 5.Miazek A., Zalas M., Skrzymowska J., et al. Age-dependent ataxia and neurodegeneration caused by an αII spectrin mutation with impaired regulation of its calpain sensitivity. Sci Rep. 2021;11(1):7312. doi: 10.1038/s41598-021-86470-1. Published correction appears in Sci Rep. 2021;11(1):18218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies M.D., Elbahy L., Fowler T., et al. Data from: The national genomics research and healthcare knowledgebase. figshare. 2017 doi: 10.6084/m9.figshare.4530893.v5. [DOI] [Google Scholar]
- 7.Martin A.R., Williams E., Foulger R.E., et al. PanelApp crowdsources expert knowledge to establish consensus diagnostic gene panels. Nat Genet. 2019;51(11):1560–1565. doi: 10.1038/s41588-019-0528-2. [DOI] [PubMed] [Google Scholar]
- 8.Firth H.V., Richards S.M., Bevan A.P., et al. DECIPHER: database of chromosomal imbalance and phenotype in humans using Ensembl resources. Am J Hum Genet. 2009;84(4):524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36(10):928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adzhubei I.A., Schmidt S., Peshkin L., et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mehboob S., Song Y., Witek M., et al. Crystal structure of the nonerythroid alpha-spectrin tetramerization site reveals differences between erythroid and nonerythroid spectrin tetramer formation. J Biol Chem. 2010;285(19):14572–14584. doi: 10.1074/jbc.M109.080028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans R.J., Davies D.R., Bullard J.M., et al. Structure of PolC reveals unique DNA binding and fidelity determinants. Proc Natl Acad Sci U S A. 2008;105(52):20695–20700. doi: 10.1073/pnas.0809989106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10(6):845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodrigues C.H., Pires D.E., Ascher D.B. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018;46(W1):W350–W355. doi: 10.1093/nar/gky300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pettersen E.F., Goddard T.D., Huang C.C., et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 17.Richards S., Aziz N., Bale S., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van de Vondel L., De Winter J., Beijer D., et al. De novo and dominantly inherited SPTAN1 mutations cause spastic paraplegia and cerebellar ataxia. Mov Disord. 2022;37(6):1175–1186. doi: 10.1002/mds.28959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veitia R.A., Caburet S., Birchler J.A. Mechanisms of Mendelian dominance. Clin Genet. 2018;93(3):419–428. doi: 10.1111/cge.13107. [DOI] [PubMed] [Google Scholar]
- 20.Gartner V., Markello T.C., Macnamara E., et al. Novel variants in SPTAN1 without epilepsy: an expansion of the phenotype. Am J Med Genet A. 2018;176(12):2768–2776. doi: 10.1002/ajmg.a.40628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morlé L., Morlé F., Roux A.F., et al. Spectrin Tunis (Sp alpha I/78), an elliptocytogenic variant, is due to the CGG----TGG codon change (Arg----Trp) at position 35 of the alpha I domain. Blood. 1989;74(2):828–832. [PubMed] [Google Scholar]
- 22.Benkirane M., Marelli C., Guissart C., et al. High rate of hypomorphic variants as the cause of inherited ataxia and related diseases: study of a cohort of 366 families. Genet Med. 2021;23(11):2160–2170. doi: 10.1038/s41436-021-01250-6. [DOI] [PubMed] [Google Scholar]
- 23.Leterrier C. A dual role for βII-spectrin in axons. Proc Natl Acad Sci U S A. 2019;116(31):15324–15326. doi: 10.1073/pnas.1909789116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ikeda Y., Dick K.A., Weatherspoon M.R., et al. Spectrin mutations cause spinocerebellar ataxia type 5. Nat Genet. 2006;38(2):184–190. doi: 10.1038/ng1728. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
De-identified data are available upon request. Data requests can be made via email to the corresponding author and will be pending data use agreements.