Abstract
Aspergillus is one of the largest and diverse genera of fungi with huge economical, biotechnological, and social significance. Taxonomically, Aspergillus is divided into six subgenera comprising 27 sections. In this study, 235 strains of Aspergillus subgenus Circumdati (section: Candidi, Circumdati, Flavi, Flavipedes, Nigri, and Terrei) preserved at the Korean Agricultural Culture Collection (KACC) were analyzed and re-identified using a combined dataset of partial β-tubulin (BenA), Calmodulin (CaM) gene sequences and morphological data. We confirmed nineteen species to be priorly reported in Korea (A. neotritici, A. terreus, A. floccosus, A. allahabadii, A. steynii, A. westerdijkiae, A. ochraceus, A. ostianus, A. sclerotiorum, A. luchuensis, A. tubingensis, A. niger, A. welwitschiae, A. japonicus, A. nomius, A. tamarii, A. parasiticus, A. flavi, and A. oryzae). Among the studied strains, three species (A. subalbidus, A. iizukae, and A. uvarum), previously unreported or not officially documented, were discovered in Korea, to the best of our knowledge. We have given a detailed description of the characteristic features of the three species, which remain uncharted in Korea.
Keywords: Aspergillus subgenus circumdati, unrecorded species, A. subalbidus, A. iizukae, A. uvarum
1. Introduction
The genus Aspergillus is one of the most ubiquitous and cosmopolitan filamentous fungi of the order Eurotiales. Species belonging to this genus are ecologically abundant and can be found in the air, soil, vegetation as well as indoor environments [1,2]. Several of the Aspergillus species are economically, biotechnologically, and medically important due to their ability to produce enzymes, organic acids, antibiotics, and other bioactive metabolites [3]. Nevertheless, some of the species are also frequently reported for their detrimental effects such as food spoilage, mycotoxin production, and as causal agents of mycoses [4]. The genus was first introduced in 1729 and has more than one thousand recorded taxa in the database “Index Fungorum.” According to a recent research on the Aspergillus taxonomy, the genus comprises – six subgenera (namely, Aspergillus, Circumdati, Cremei, Fumigati, Nidulantes, and Polypaecilum), 27 sections, 75 series with 446 species [5].
Species delimitation is a vital aspect of taxonomic research and precise identification of strains is essential for targeted applications as well as linking of research taking place across the world. In this regard, microbial resource centers play an important role in phenetic analysis and conservation of microbial strains of potential value in industry, medicine, environment, agriculture, and other scientific purposes [6]. Among such institutions around the world, Korean Agricultural Culture Collection (KACC) is a major research organization specializing in identification and long-term storage of fungal biodiversity which can serve as a potential source of useful fungal strains. It was established as a part of the Rural Development Administration (RDA) in the year 1995 and the collection currently preserves 14,079 strains of fungi from 3346 species covering all major fungal taxonomic groups, and among these, 9.2% (n = 1297) belong to Aspergillus.
In earlier days, Aspergillus strains were identified based on their morphology and deposited in KACC. In the last two decades, morphology-based identification was often found to be misleading, especially within the Aspergillus sections due to the occurrence of cryptic species [7]. Therefore, for accurate identification of Aspergillus, a polyphasic approach has been proposed which includes morphological analysis as well as molecular analysis, ecology, and extrolite profiling [8]. Basically, current identification and phylogeny of Aspergillus can be majorly relied on DNA barcodes which include internal transcribed spacer (ITS) region, Calmodulin (CaM), β-tubulin (BenA), and the RNA polymerase II second largest subunit (RPB2) [2].
At present, to improve the quality of the KACC resources, focus has shifted towards re-identification of the conserved strains using molecular techniques mainly based on DNA barcodes mentioned above, in addition to their morphological characteristics. In this context, a subset of Aspergillus strains stored in the KACC from 1995 to 2022 was studied using their sequence data as well as morphological characteristics. The identification of all strains was based on partial β-tubulin (BenA) and Calmodulin (CaM) gene sequences. To date, 81 different Aspergillus species have been described from Korea [9,10]. This study aimed to re-identify Korean strains of Aspergillus subgenus Circumdati preserved in KACC and provide a description of hitherto unrecorded species in Korea based on their morphological and molecular characteristics. This study complements existing knowledge on the diversity of Aspergillus species in Korea.
2. Materials and methods
2.1. Strains
A total of 235 strains belonging to genus Aspergillus subgenus Circumdati in KACC were studied. All the reagents and media used in the study were procured from Merck, Seoul, South Korea and Oxoid, Basingstoke, UK. The strains were isolated from diverse ecological niches in Korea. All the strains were revived in 4 mL of Malt extract broth, and subsequently transferred to malt extract agar (MEA). The strains examined in the study have been listed in Table 1.
Table 1.
Aspergillus subgenus Circumdati strains used in this study.
| Section | Re-identified scientific name | Scientific name by depositor | KACC number | Deposited year | Substrate | Region | Location | RDA GeneBank accession no. |
|
|---|---|---|---|---|---|---|---|---|---|
| CaM | BenA | ||||||||
| Candidi | A. subalbidus a | A. candidus | 46481 | 2011 | Meju | Gyeongsangbuk-do | Chilgok-gun | RDA0061631 | RDA0061627 |
| A. candidus | 46482 | 2011 | Meju | Gyeonggi-do | Icheon-si | RDA0061632 | RDA0061628 | ||
| A. neotritici | A. candidus | 44245 | 2005 | Rice | Seoul | Seongbuk-gu | RDA0061630 | RDA0061629 | |
| A. tritici | 46483 | 2011 | Meju | Gyeonggi-do | Yeoju-si | RDA0062637 | RDA0062638 | ||
| A. tritici | 48027 | 2015 | Nuruk | Gyeongsangnam-do | Jinju-si | RDA0062591 | RDA0062592 | ||
| Terrei | A. allahabadii | A. allahabadii | 49964 | 2021 | Soil | Jeollanam-do | Damyang-gun | RDA0062655 | RDA0062656 |
| A. floccosus | A. floccosus | 49740 | 2020 | Soil | Jeollabuk-do | Buan-gun | RDA0062653 | RDA0062654 | |
| A. terreus | A. terreus | 47277 | 2013 | Rice straw | Jeollanam-do | Haenam-gun | RDA0062649 | RDA0062650 | |
| A. terreus | 47390 | 2014 | Air | Korea | Anseong-si | RDA0062647 | RDA0062648 | ||
| A. terreus | 48446 | 2017 | Water | Busan | Saha-gu, | RDA0062651 | RDA0062652 | ||
| Flavipedes | A. iizukae | A. flavipes | 43789 | 2008 | Creosote contaminated soil | Seoul | Sangam-dong | RDA0064667 | RDA0064670 |
| A. iizukae | 48444 | 2017 | Crab | Busan | Saha-gu | RDA0064668 | RDA0064672 | ||
| A. iizukae | 48864 | 2019 | Soil | Gyeongsangnam-do | Miryang-si | RDA0064669 | RDA0064671 | ||
| Circumdati | A. ochraceus | A. ochraceus | 46484 | 2011 | Meju | Jeollanam-do | Haenam-gun | RDA0064675 | RDA0064694 |
| A. ochraceus | 46485 | 2011 | Meju | Gyeonggi-do | Icheon-si | RDA0064676 | RDA0064695 | ||
| A. ochraceus | 46486 | 2011 | Meju | Gyeonggi-do | Yeoju-si | RDA0064677 | RDA0064696 | ||
| A. ochraceus | 47132 | 2013 | Soybean | Korea | unknown | RDA0064682 | RDA0064701 | ||
| A. ochraceus | 47273 | 2013 | Rice straw | Jeollabuk-do | Sunchang-gun | RDA0064685 | RDA0064704 | ||
| A. ochraceus | 47384 | 2014 | Air | Gyeongsangbuk-do | Chilgok-gun | RDA0064687 | RDA0064706 | ||
| A. ostianus | A. ostianus | 47134 | 2013 | Soybean | Korea | unknown | RDA0064711 | RDA0064702 | |
| A. sclerotiorum | A. sclerotiorum | 47387 | 2014 | Air | Gyeongsangbuk-do | Chilgok-gun | RDA0064688 | RDA0064707 | |
| A. sclerotiorum | 48445 | 2017 | Rice | Jeollanam-do | Gwangju-si | RDA0064691 | RDA0064710 | ||
| A. steynii | A. ochraceus | 46821 | 2012 | Air | Gyeongsangbuk-do | Chilgok-gun | RDA0064679 | RDA0064698 | |
| A. steynii | 47388 | 2014 | Air | Gyeonggi-do | Icheon-si | RDA0064689 | RDA0064708 | ||
| A. westerdijkiae | A. ochraceus | 46822 | 2012 | Wheat straw | Jeollanam-do | Damyang-gun | RDA0064680 | RDA0064699 | |
| A. ochraceus | 46823 | 2012 | Soybean | Gyeonggi-do | Yangpyeong-gun | RDA0064681 | RDA0064700 | ||
| A. westerdijkiae | 46487 | 2011 | Meju | Jeollabuk-do | Sunchang-gun | RDA0064678 | RDA0064697 | ||
| A. westerdijkiae | 47139 | 2013 | Soybean | Korea | unknown | RDA0064684 | RDA0064703 | ||
| A. westerdijkiae | 47280 | 2013 | Rice straw | Jeollanam-do | Damyang-gun | RDA0064686 | RDA0064705 | ||
| A. westerdijkiae | 47394 | 2014 | Air | Gyeonggi-do | Yongin-si | RDA0064690 | RDA0064709 | ||
| Nigri | A. japonicus | A. japonicus | 42845 | 2007 | Soil | Gyeonggi-do | Suwon-si | RDA0064715 | RDA0064763 |
| A. japonicus | 43791 | 2008 | Creosote contaminated soil | Seoul | Sangam-dong | RDA0064716 | RDA0064764 | ||
| A. aculeatus | 47127 | 2013 | Soybean | Chungcheongnam-do | Gongju-si | RDA0065389 | RDA0065410 | ||
| A. japonicus | 48322 | 2017 | Unknown | Gyeongsangbuk-do | Sangju-si | RDA0064754 | RDA0064798 | ||
| A. luchuensis | A. luchuensis | 41731 | 1995 | Nuruk | Chungcheongbuk-do | Boeun-gun | RDA0064713 | RDA0064761 | |
| A. luchuensis | 46420 | 2011 | Traditional yeast | Gyeongsangbuk-do | Andong-si | RDA0064722 | RDA0064770 | ||
| A. luchuensis | 46490 | 2011 | Meju | Gyeonggi-do | Yeoju-si | RDA0064724 | RDA0064772 | ||
| A. luchuensis | 46491 | 2011 | Meju | Jeollabuk-do | Sunchang-gun | RDA0064725 | RDA0064773 | ||
| A. welwitschiae | 46492 | 2011 | Meju | Jeollanam-do | Haenam-gun | RDA0065375 | RDA0065396 | ||
| A. niger | 46493 | 2011 | Meju | Gyeonggi-do | Icheon-si | RDA0065376 | RDA0065397 | ||
| A. niger | 46494 | 2011 | Meju | Gangwon-do | Hoengseong-gun | RDA0065377 | RDA0065398 | ||
| A. luchuensis | 46516 | 2011 | Nuruk | Korea | unknown | RDA0064727 | RDA0064775 | ||
| A. luchuensis | 46879 | 2012 | Meju | Gyeonggi-do | Yongin-si | RDA0064729 | RDA0064777 | ||
| A. luchuensis | 46958 | 2012 | Nuruk | Gyeonggi-do | Hwaseong-si | RDA0064733 | RDA0064781 | ||
| A. luchuensis | 48309 | 2017 | Meju | Korea | Yangpyeong | RDA0064744 | RDA0064788 | ||
| A. luchuensis | 48310 | 2017 | Meju | Korea | Hoengseong | RDA0064745 | RDA0064789 | ||
| A. luchuensis | 48312 | 2017 | Meju | Korea | Yongin-si | RDA0064746 | RDA0064790 | ||
| A. luchuensis | 48313 | 2017 | Meju | Korea | Yongin-si | RDA0064747 | RDA0064791 | ||
| A. luchuensis | 48314 | 2017 | Meju | Korea | Yongin-si | RDA0064748 | RDA0064792 | ||
| A. luchuensis | 48315 | 2017 | Meju | Korea | Yongin-si | RDA0064749 | RDA0064793 | ||
| A. luchuensis | 48316 | 2017 | Meju | Korea | Yongin-si | RDA0064750 | RDA0064794 | ||
| A. luchuensis | 48318 | 2017 | Meju | Korea | Yongin-si | RDA0064751 | RDA0064795 | ||
| A. luchuensis | 48319 | 2017 | Meju | Korea | Anseong-gun | RDA0064752 | RDA0064796 | ||
| A. luchuensis | 48320 | 2017 | Meju | Korea | Yongin-si | RDA0064753 | RDA0064797 | ||
| A. luchuensis | 49930 | 2021 | Soy bean paste | Jeollabuk-do | Wanju-gun | RDA0064758 | RDA0064802 | ||
| A. luchuensis | 49931 | 2021 | Soy bean paste | Jeollabuk-do | Wanju-gun | RDA0064759 | RDA0064803 | ||
| A. niger | A. niger | 40279 | 1997 | Unknown | Korea | unknown | RDA0064712 | RDA0064760 | |
| A. niger | 42589 | 2007 | Man | Korea | Seoul | RDA0064714 | RDA0064762 | ||
| A. niger | 44333 | 2009 | Sputum | Korea | Seoul | RDA0064719 | RDA0064767 | ||
| A. tubingensis | 46498 | 2011 | Meju | Gangwon-do | Hoengseong-gun | RDA0065382 | RDA0065403 | ||
| A. luchuensis | 46880 | 2012 | Meju | Gyeonggi-do | Yangpyeong-gun | RDA0065380 | RDA0065401 | ||
| A. luchuensis | 48321 | 2017 | Meju | Gyeonggi-do | Yangpyeong-gun | RDA0065381 | RDA0065402 | ||
| A. tubingensis | A. niger | 40278 | 1997 | Unknown | Korea | unknown | RDA0065393 | RDA0065414 | |
| A. niger | 40280 | 1997 | Unknown | Korea | unknown | RDA0065394 | RDA0065415 | ||
| A. niger | 41018 | 2002 | Soil | Korea | unknown | RDA0065392 | RDA0065413 | ||
| A. niger | 43547 | 2008 | White pine | Korea | Daejeon | RDA0065395 | RDA0065416 | ||
| A. tubingensis | 43792 | 2008 | Ground soil | Seoul | Sangam-dong | RDA0064717 | RDA0064765 | ||
| A. tubingensis | 44304 | 2008 | Unknown | Chungcheongnam-do | Gongju-si | RDA0064718 | RDA0064766 | ||
| A. tubingensis | 44334 | 2009 | Sputum | Korea | Seoul | RDA0064720 | RDA0064768 | ||
| A. tubingensis | 44335 | 2009 | Sputum | Korea | Seoul | RDA0064721 | RDA0064769 | ||
| A. tubingensis | 46446 | 2011 | Campbell early | Chungcheongbuk-do | Yeongdong-gun | RDA0064723 | RDA0064771 | ||
| A. tubingensis | 46883 | 2012 | Unknown | Gyeonggi-do | Yeoju-si | RDA0064732 | RDA0064780 | ||
| A. tubingensis | 47137 | 2013 | Soybean | Korea | unknown | RDA0064734 | RDA0064782 | ||
| A. aculeatus | 47267 | 2013 | Rice straw | Jeollabuk-do | Sunchang-gun | RDA0065378 | RDA0065399 | ||
| A. tubingensis | 47278 | 2013 | Rice straw | Gyeonggi-do | Anseong-si | RDA0064735 | RDA0064783 | ||
| A. tubingensis | 47391 | 2014 | Air | Jeollabuk-do | Sunchang-gun | RDA0064737 | RDA0064785 | ||
| A. luchuensis | 48311 | 2017 | Meju | Korea | Yeoju-si | RDA0065391 | RDA0065412 | ||
| A. luchuensis | 48317 | 2017 | Meju | Jeollabuk-do | Sunchang-gun | RDA0065379 | RDA0065400 | ||
| A. tubingensis | 49868 | 2021 | Plastic debris | Jeollanam-do | Gwangju-si | RDA0064757 | RDA0064801 | ||
| A. uvarum | A. uvarum | 48630 | 2018 | Decayed tree | Seoul | Gwanak-gu | RDA0064755 | RDA0064799 | |
| A. welwitschiae | A. niger | 46495 | 2011 | Meju | Gyeonggi-do | Gimpo-si | RDA0065384 | RDA0065405 | |
| A. welwitschiae | 46496 | 2011 | Meju | Chungcheongbuk-do | Goesan-gun | RDA0064726 | RDA0064774 | ||
| A. niger | 46497 | 2011 | Meju | Gyeonggi-do | Yangpyeong-gun | RDA0065383 | RDA0065404 | ||
| A. tubingensis | 46499 | 2011 | Meju | Jeollabuk-do | Sunchang-gun | RDA0065385 | RDA0065406 | ||
| A. welwitschiae | 46881 | 2012 | Meju | Gyeonggi-do | Yeoju-si | RDA0064730 | RDA0064778 | ||
| A. welwitschiae | 46882 | 2012 | Meju | Jeollanam-do | Haenam-gun | RDA0064731 | RDA0064779 | ||
| A. niger | 47249 | 2013 | Onion | Jeollanam-do | Muan-gun | RDA0065388 | RDA0065409 | ||
| A. niger | 47272 | 2013 | Rice straw | Jeollanam-do | Haenam-gun | RDA0065387 | RDA0065408 | ||
| A. awamori | 47376 | 2014 | Air | Gyeonggi-do | Yongin-si | RDA0065386 | RDA0065407 | ||
| A. niger | 47383 | 2014 | Air | Jeollabuk-do | Sunchang-gun | RDA0065390 | RDA0065411 | ||
| A. welwitschiae | 48728 | 2019 | Groundnut | Jeollabuk-do | Wanju-gun | RDA0064756 | RDA0064800 | ||
| Flavi | A. flavus* | A. flavus | 45443 | 2010 | Meju | Chungcheongbuk-do | Cheongju-si | RDA0064859 | RDA0064879 |
| A. nomiae | A. nomius | 49741 | 2020 | Soil | Jeollanam-do | Jangseong-gun | RDA0064848 | RDA0064868 | |
| A. oryzae* | A. oryzae | 46469 | 2011 | Meju | Korea | unknown | RDA0064866 | RDA0064886 | |
| A. oryzae | 46471 | 2011 | Meju | Gyeonggi-do | Icheon-si | RDA0064865 | RDA0064885 | ||
| A. oryzae | 46640 | 2012 | Meju | Korea | unknown | RDA0064863 | RDA0064883 | ||
| A. oryzae | 46641 | 2012 | Meju | Korea | unknown | RDA0064864 | RDA0064884 | ||
| A. oryzae | 46810 | 2012 | Meju | Jeollabuk-do | Sunchang-gun | RDA0064862 | RDA0064882 | ||
| A. oryzae | 47133 | 2013 | Soybean | Korea | unknown | RDA0064861 | RDA0064881 | ||
| A. oryzae | 47488 | 2013 | Unknown | Korea | Seoul | RDA0064860 | RDA0064880 | ||
| A. oryzae | 48141 | 2016 | Air, Indoor | Gyeonggi-do | Bucheon-si | RDA0064867 | RDA0064887 | ||
| A. parasiticus | A. parasiticus | 46037 | 2011 | Meju environment | Gyeongsangbuk-do | Andong-si | RDA0064856 | RDA0064876 | |
| A. parasiticus | 46475 | 2011 | Meju environment | Jeollanam-do | Gangjin-gun | RDA0064857 | RDA0064877 | ||
| A. parasiticus | 46901 | 2012 | Peanut | Gyeonggi-do | Yeoju-si | RDA0064858 | RDA0064878 | ||
| A. tamarii | A. parasiticus | 46034 | 2011 | Silkworm | Gyeonggi-do | Suwon-si | RDA0064855 | RDA0064875 | |
| A. tamarii | 46476 | 2011 | Meju | Jeollabuk-do | Jeongeup-si | RDA0064849 | RDA0064869 | ||
| A. tamarii | 46477 | 2011 | Meju | Gyeongsangbuk-do | Goryeong-gun | RDA0064850 | RDA0064870 | ||
| A. tamarii | 46478 | 2011 | Meju | Gyeonggi-do | Icheon-si | RDA0064851 | RDA0064871 | ||
| A. tamarii | 46479 | 2011 | Meju | Gyeonggi-do | Suwon-si | RDA0064852 | RDA0064872 | ||
| A. tamarii | 46480 | 2011 | Meju | Gyeonggi-do | Icheon-si | RDA0064853 | RDA0064873 | ||
| A. tamarii | 47276 | 2013 | Rice straw | Gangwon-do | Yangyang-gun | RDA0064854 | RDA0064874 | ||
aThe unrecorded Aspergillus species in Korea are represented in bold.
*Only nine representative strains were taken from section Flavi.
2.2. DNA extraction, PCR amplification, and sequencing
Genomic DNA was extracted from the strains grown on MEA using the DNeasy® Plant Mini kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. Fragments of the BenA (primers Bt2a and Bt2b) and CaM (primers CMD5 and CMD6) genes were amplified as outlined by Glass and Donaldson [11], and Hong et al. [12]. The PCR products were sequenced bidirectionally at Macrogen Inc., South Korea, using the same primers used for PCR. Consensus sequences were computed from forward and reverse sequences using DNA STAR Lasergene SeqMan Pro version 10.0.1 (DNASTAR, Inc. Madison, WI).
2.3. Phylogenetic analyses
The newly generated sequences were supplemented with reference (preferably ex-type) sequences retrieved from previously published studies [5]. Alignment of the sequences was performed using the CLUSTAL W program [13] and were manually edited with MEGA version 7.0 (University Park, PA) [14]. The maximum likelihood (ML) method was used for the phylogenetic analysis. For ML analysis, the data were first analyzed using the nucleotide substitution model and the best substitution pattern was then used to construct the ML tree with MEGA version 7.0 [14]. To determine the support for each clade, a bootstrap analysis was performed with 1000 replications. The sequence of Aspergillus calidoustus CBS 121601T was used as an outgroup. The reference sequences used in this analysis have been listed in Table 2. Sequences generated in this study were deposited to KACC-GeneBank (http://genebank.rda.go.kr).
Table 2.
Reference sequences of Aspergillus species used in the phylogenetic analyses.
| Section | Species | Strain no. | Country | GenBank Accession no. |
|
|---|---|---|---|---|---|
| CaM | BenA | ||||
| Candidi | A. campestris | NRRL 13001 T | USA | EF669535 | EU014091 |
| A. candidus | NRRL 303 T | Unknown | EF669550 | EU014089 | |
| A. dobrogensis | CCF 4651 T | Romania | LT558722 | LT627027 | |
| A. magnus | UAMH 1324 T | Canada | ON164619 | ON164570 | |
| A. neotritici | CCF 3853 T | Czech Republic | HE661598 | FR775327 | |
| A. pragensis | CCF 3962 T | Czech Republic | FR751452 | HE661604 | |
| A. subalbidus | NRRL 312 T | Brazil | EF669551 | MN969366 | |
| A. taichungensis | DTO 031-C6 T | Taiwan | EU076310 | MN969367 | |
| A. tenebricus | DTO 337-H7 T | South Africa | ON164623 | ON164584 | |
| Terrei | A. allahabadii | NRRL 4539 T | India | EF669559 | EF669531 |
| A. floccosus | CBS 116.37 T | China | KP987066 | FJ491714 | |
| A. terreus | NRRL 255 T | USA | EF669544 | EF669519 | |
| Flavipedes | A. flavipes | NRRL 302 T | Unknown | EF669549 | EU014085 |
| A. iizukae | NRRL 3750 T | Japan | EF669555 | EU014086 | |
| Circumdati | A. elegans | NRRL 4850 T | USA | EF661390 | EF661349 |
| A. ochraceus | NRRL 398 T | Unknown | EF661381 | EF661322 | |
| A. ostianus | NRRL 420 T | Unknown | EF661385 | EF661324 | |
| A. sclerotiorum | NRRL 415 T | USA | EF661384 | EF661337 | |
| A. steynii | NRRL 35675 T | India | EF661378 | EF661347 | |
| A. westerdijkiae | NRRL 3174 T | South Africa | EF661360 | EF661329 | |
| Nigri |
A. aculeatus
A. japonicus |
NRRL 5094 T CBS 114.51 T |
Unknown Unknown |
EF661148 FN594551 |
HE577806 HE577804 JX500062 |
| A. luchuensis | KACC 46772 T | Japan | JX500071 | ||
| A. niger | NRRL 326 T | USA | EF661154 | EF661089 | |
| A. tubingensis | NRRL 4875 T | Unknown | EF661151 | EF661086 | |
| A. uvarum | ITEM 4834 T | Italy | AM745755 | AM745751 | |
| A. welwitschiae | CBS 139.54 T | Namibia | KC480196 | MN969369 | |
| Flavi | A. flavus | NRRL 1957 T | South Pacific Islands | EF661508 | EF661485 |
| A. nomiae | NRRL 13137 T | USA | AY017588 | AF255067 | |
| A. oryzae | NRRL 447 T | Unknown | EF661506 | EF661483 | |
| A. parasiticus | NRRL 502 T | USA | AY017584 | EF661481 | |
| A. tamarii | NRRL 20818 T | Unknown | EF661526 | EF661474 | |
| Outgroup | A. calidoustus | CBS 121601 T | Netherlands | HE616559 | FJ624456 |
CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; CCF: Culture Collection of Fungi at the Department of Botany of Charles University in Prague; ITEM: Microbial Culture Collection, Institute of Sciences of Food Production, Bari, Italy; KACC: Korean Agricultural Culture Collection, Wanju, Republic of Korea; NRRL: ARS culture collection, Peoria, IL, USA; T: ex-type strain.
2.4. Phenotypic analysis
The strains were three-point inoculated on Czapek Yeast extract agar (CYA), Dichloran 18% Glycerol agar (DG18), MEA, and yeast extract sucrose agar (YES) [15]. Media preparation, inoculation, and incubation were performed as described by Samson et al. [2] and all Petri dishes were incubated at 25 °C for 7 d. After 7 d of incubation, colony diameters were measured and colony characteristics were recorded (presence of soluble pigments, exudates, obverse and reverse colony colors, color of conidia). Microscopic examination was performed on colonies grown on MEA using Zeiss Axio imager A1 light microscope equipped with Axio cam ICc3 camera (ZEISS, Seoul, South Korea). Slides were prepared with lactic acid, which was used as the mounting fluid, and ethanol was, at times used to remove excess conidia. The size, shape, and pigmentation of conidia and conidiophores were recorded.
3. Results and discussion
3.1. Phylogenetic analyses
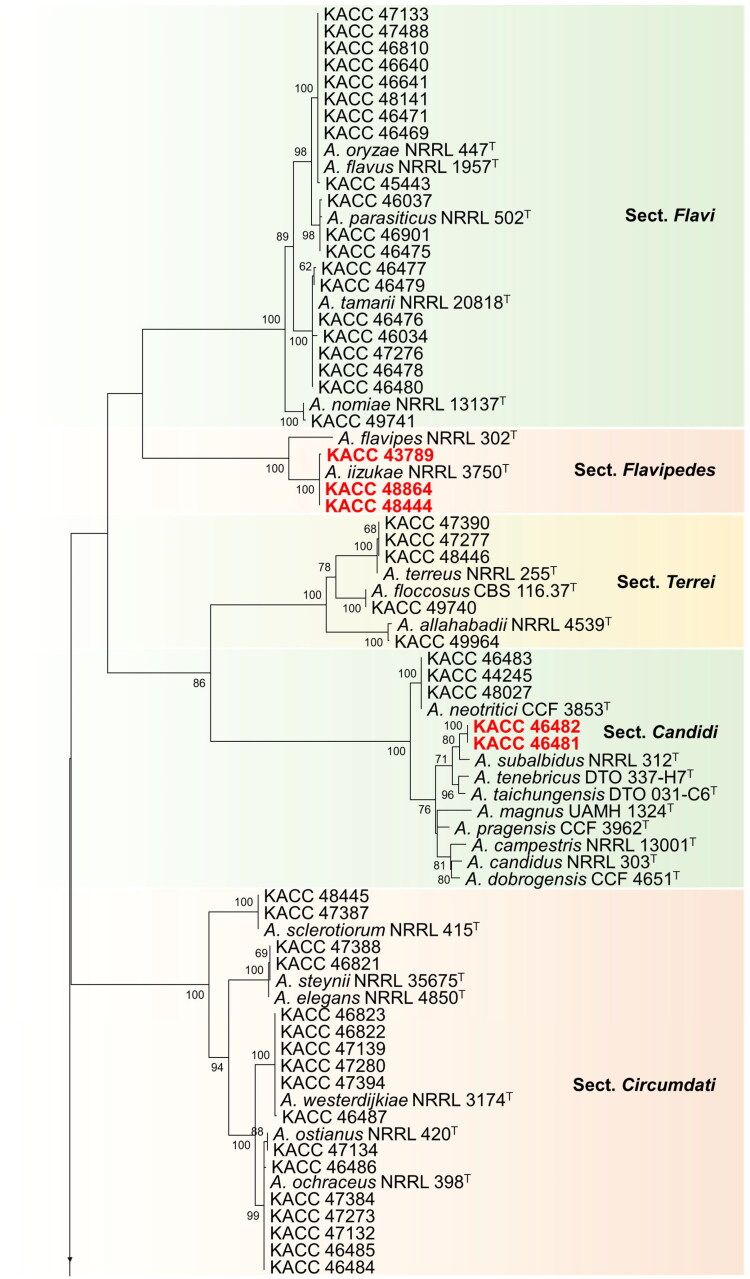
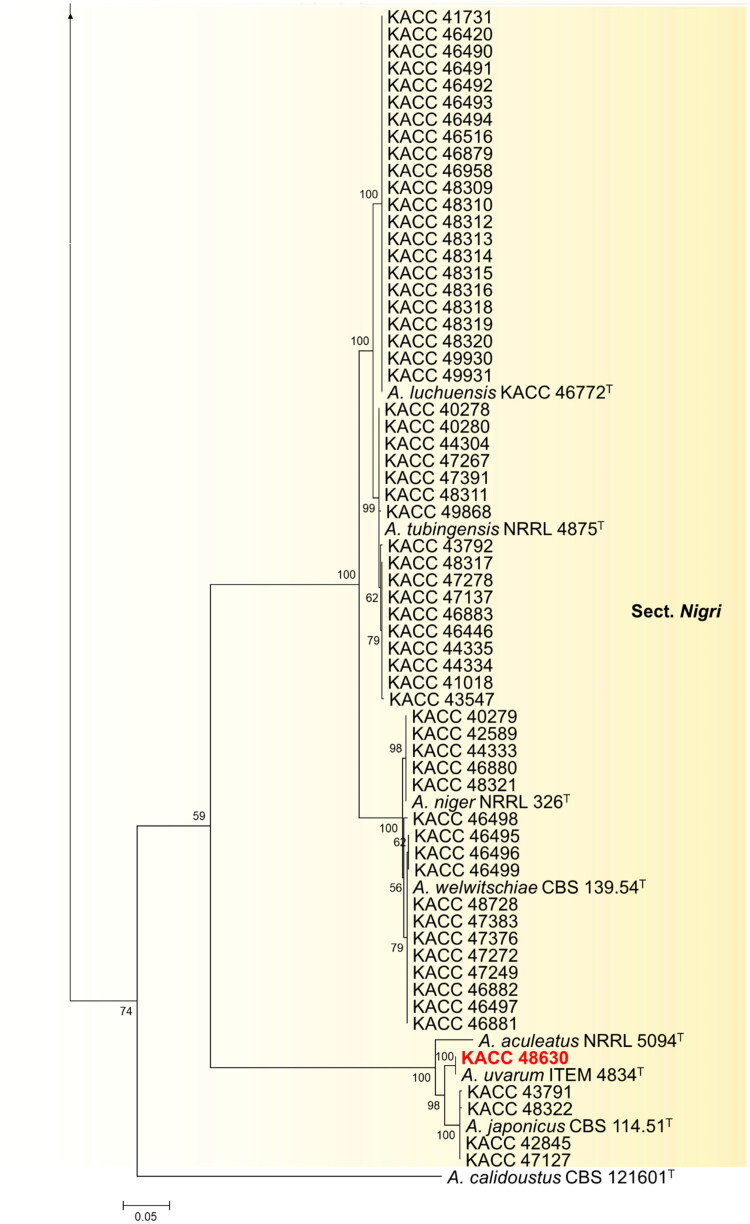
In this study, phylogenetic position of strains belonging to subgenus Circumdati was studied using concatenated data on BenA and CaM sequences (Figure 1). In the section Flavi, most of the strains belonging to A. flavus and A. oryzae exhibited highly similar BenA and CaM gene sequences. Therefore, we selected a few representative strains (9 out of 133) of A. flavus and A. oryzae for further phylogenetic analysis. Information about these strains is given in Table 1. In total, the concatenated alignment included 144 sequences: 111 derived from strains of the KACC and the others from publicly available (ex-) type species. The total length of the aligned data set was 1187 characters. The most optimal substitution model was K2 + G + I for the BenA and CaM data set.
Figure 1.
Phylogenetic position of Aspergillus subgenus Circumdati strains from the KACC based on a combined data set of partial BenA and CaM sequences. Bootstrap values >50 are presented at the nodes. The unrecorded species are represented in bold and red in color. Ex-type strains are denoted by the symbol “T.” A. calidoustus was used as the outgroup.
During our studies, the 235 strains taken were spread across 22 different Aspergillus species, of which 19 species have been previously reported from Korea. Three species constituted by six strains (highlighted in red bold text) were not previously described from Korea (Figure 1).
In the section Flavi, 40 species have been reported worldwide [5,16–18]. Among them, A. flavus, A. nomiae, A. parasiticus, A. tamarii, and A.oryzae have been recorded in Korea [9,19]. One hundred and forty-four strains from this study, belonging to – the section Flavi in Korea clustered with type strains of A. flavus, A. oryzae, A. nomiae, A. parasiticus, and A. tamarii and identified according to their closest type strain. In this section, A. flavus/oryzae complex contributed to a huge number of strains (n = 133). Since their BenA and CaM gene sequences were highly similar, only a few (n = 9) representative strains were selected and shown in Figure 1. Aflatoxin production was used to differentiate the strains between A. flavus and A. oryzae (data not shown), as the two species cannot be differentiated based on their BenA and CaM gene sequences.
In the section Flavipedes, all 3 strains used in this study were clustered with A. iizukae. Nineteen (19) species have been reported in the section Flavipedes [5,20,21]. In Korea, 4 species namely A. capensis, A. flavipes, A. polyporicola, and A. spelaeus have been recorded already [9]. We are adding a new record of A. iizukae in this section which was not previously reported in Korea.
Among 20 known species form the section Terrei [5,6,21], four of them, viz., A. alabamensis, A. allahabadii, A. floccosus, and A. terreus have been recorded in Korea [9]. Five strains of the section Terrei in Korea clustered along with type strains of A. allahabadii, A. floccosus, A. terreus and are therefore identified as A. allahabadii, A. floccosus, A. terreus respectively.
In the section Candidi of subgenus Circumdati, 5 strains were found to be clustered into two groups with A. neotritici and A. subalbidus. Nine species have been recorded in this section [5,22]. Recently, A. tritici was synonymized as A. neotritici [22]. In Candidi, A. candidus, A. pragensis and A. neotritici have been recorded in Korea [9]. A. subalbidus has not yet been recorded in Korea, which is included in the current report.
In the section Circumdati, 17 strains were found to cluster into five groups, comprising of A. ochraceus, A. ostianus, A. sclerotiorum, A. steynii, and A. westerdijkiae as nearest type strains. Thirty species have been recorded in the section [5,23,24], of which, A. ochraceus, A. ostianus, A. insulicola, A. sclerotiorum, A. steynii, and A. westerdijkiae are recorded in Korea [9].
In the section Nigri of subgenus Circumdati, 61 strains grouped into six clusters, with A. japonicus, A. luchuensis, A. niger, A. tubingensis, A. uvarum, and A. welwitschiae as their nearest neighbors. Until now, 32 species have been recorded in this section [5,24,25]. Among them, A. aculeatus, A. brunneoviolaceus, A. costaricaensis, A. floridensis, A. japonicus, A. niger, A. tubingensis, A. luchuensis, and A. welwitschiae are recorded in Korea [9,26]. A. uvarum have not been recorded in Korea, and we are reporting new record of A. uvarum in Korea in this article.
In the past decade, the classification of Aspergillus species has been developed to be based on a combination of molecular data, physiology, morphology, and/or extrolite data [12,27–29]. This approach was used by Houbraken et al. [5] to clarify the taxonomic position of Aspergillus, Pencillium, Talaromyces, and related genera. In this study, the strains used were documented and preserved at KACC well before the overview paper of Houbraken et al. [5] and the identification of these strains would have been most likely based on the morphological characters such as growth rate, color of the colony, thermotolerance, and size of conidial heads and conidia. However, currently, morphological features alone are understood to be inadequate to identify species because of morphological characteristics have been found to vary even with respect to change in their ecological habitats [30,31]. In our study, six strains were found to be clustered with unrecorded species of Korea. Till date, there have been few reports on undescribed Aspergillus species in Korea despite the genus having a cosmopolitan distribution. On the other hand, there has been a rise of several new Aspergillus species worldwide [10].
Among the three hitherto unrecorded species from Korea, the strains KACC 43789, KACC 48444, and KACC 48864 were phylogenetically close to the type strain A. iizukae NRRL255T belonging to the section Flavipedes (Figure 1). Morphological characters of the strains were also consistent with those of A. iizukae described by Hubka et al. [32]. The species was first described by Sugiyama in 1967 [33]. Aspergillus section Flavipedes endured a reexamination study of species limits using advanced species delimitation methods, and the revised section harbors 19 species with most of the species being reported from soil. Though the most common species from the section are ecologically diverse, occurring in the indoor environment, clinical samples, food and feed, droppings and other less common substrates/environments [20], in this study, the isolates were found to have mainly originated from soil and food. A. iizukae have been reported to produce diphenyl derivatives, namely iizukines A (1) and B (2); flavonolignans, namely Silybin A (1), silybin B (2), isosilybin A (3); oxidative enzymes, such as laccase, manganese peroxidase, and lignin peroxidase [34–36].
Based on their phylogeny, KACC 46481 and KACC 46482 were clustered with A. subalbidus NRRL 312T (ex-type strain), in section Candidi (Figure 1). Strains KACC 46481 and KACC 46482 were morphologically similar to A. subalbidus, as described by Visagie et al. [15]. Phylogenetically, A. subalbidus forms a separate clade, closely related to A. tenebricus, A. taichungensis, and A. neotritici (Figure 1). Recently, Aspergillus section Candidi underwent a monographic study, with the revised section Candidi hosting nine species. Members in this section have been reported from house dust, soil, herbivore dung, indoor air, and cave environments, and occasionally from clinical specimens [22]. To our best knowledge, this is first study to isolate A. subalbidus from meju, thereby revealing its significance as a fungus colonizing food environment.
As shown in Figure 1, strain KACC 48630 aligned with A. uvarum ITEM 4834 (ex-type strain) in section Nigri. Morphologically, the isolated strain represents similar characters with type strain ITEM 4834 of A. uvarum described by Perrone et al. [37]. These include sporulation with dark brown conidia; uniseriate conidiophores; globose to elliptical vesicle, 20–30 µm; and conidia globose to subglobose, spinose, 3–4 µm. Moreover, section Nigri, known as black aspergilli includes species with smooth conidiophores and hyaline or pigmentation below the vesicle; globose, subglobose, and pyriform vesicles; typically radiating conidial heads; or divergent columns in certain species [38]. These aspergilli have been isolated from soil samples, air environments, contaminated materials and plants [24,25,39]. In general, 30 species have been accepted in this section [5]. Two additional new species, A. oxumiae and A. hydei, were reported in soil cultivated with Agave sisalana, and from air under the tree Quercus variabilis [24,25]. The species A. uvarum is a rare member of the group of black aspergilli, which has a high significance in the industry due to its ability to produce secalonic acid, commonly produced by black aspergilli; and geodin, erdin, and dihydrogeodin, which are not produced by any other black aspergilli [37]. Based on macro- and micro-morphological characters and phylogenetic concordance between BenA & CaM gene phylogenies, we present here three undescribed species of Korea named A. iizukae, A. subalbidus, and A. uvarum.
3.2. Taxonomy
Aspergillus iizukae Sugiyama, J. Fac. Sci. Univ. Tokyo, Section 3: 390 (1967) [MB#326636] [33]
Colony characteristics: Colonies on CYA at 25 °C attain 21–22 mm diameter in 7 d, velutinous dull white with granular surface, no soluble pigment, reverse light brown. On MEA, the colonies were velutinous to floccose with granular surface, irregularly or radially wrinkled, light yellowish-brown sporulation, no soluble pigment, reverse strong yellowish brown and attains 20–21 mm diameter in 7 d. Colonies on YES attain 23–24 mm diameter after 7 d at 25 °C; light yellowish sporulation at center with white mycelium at margins, reverse pale yellow. On DG18, colonies were slow growth, clear white mycelium, reverse white, and reached 8–9 mm in diameter after 7 d at 25 °C.
Micromorphology: Conidial heads biseriate, stipes hyaline, smooth-walled, long <1000 um. Vesicles pyriform, 14–20 µm. Metulae covering one half to entire surface of the vesicle, 3–6 × 2–3 μm. Phialides 5–7 × 2–3.5 μm. Conidia globose, smooth, connectives sometimes remain on free conidia 2–3 μm (Figure 2).
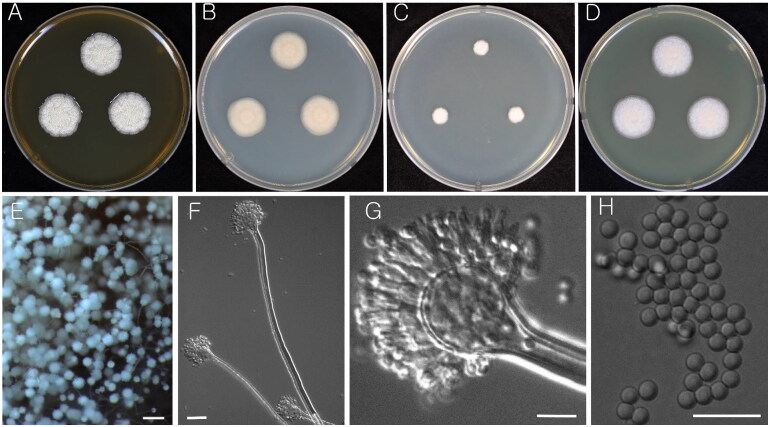
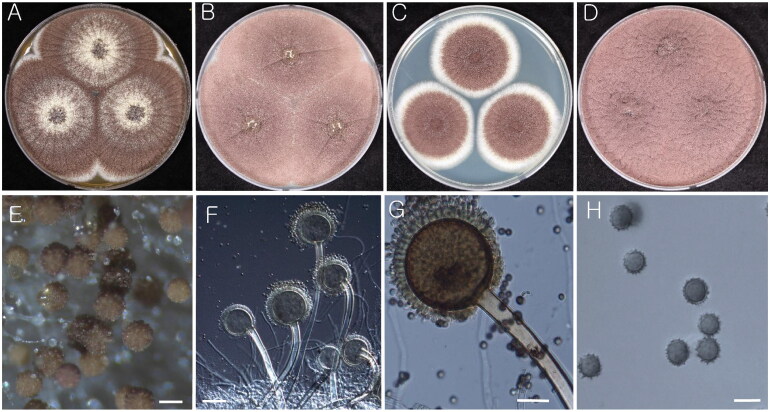
Figure 2.
Morphology of Aspergillus iizukae (KACC 43789). (A–D) Colonies grown on MEA, CYA, DG18, and YES media after 7 d at 25 °C from left to right. (E) Conidial heads on MEA. (F & G) Conidiophores with conidial head. (H) Conidia. Scale bars: E = 100 µm, F = 20 µm, G, H = 10 µm.
Strains examined: KACC 43789, KACC 48864, and KACC 48444
Remarks: A. iizukae is closely related to A. capensis. Recent species delimitation study proposed that A. capensis is synonymized with A. iizukae [20].
Aspergillus subalbidus Visagie, Hirooka & Samson, Studies in Mycology 78: 101 (2014) [MB#809190] [15].
Colony characteristics: On CYA, Colony mycelium and sporulation were found to be white, no soluble pigment present and reverse turned into light brown and eventually reached 17–19 mm in diameter after 7 d at 25 °C. On MEA, the colony surface was floccose, white mycelial areas and sporulation, soluble pigment absent and turned reverse yellow-orange, and further reached 16–22 mm in diameter after 7 d at 25 °C. On DG18, colony mycelium and sporulation were white in color and reverse white with 16–19 mm in diameter after 7 d at 25 °C. Colonies on YES were floccose after 7 d at 25 °C; conidia white, reverse centrally yellowish orange, fading into light yellow toward margin.
Micromorphology: Conidial heads biseriate, sometimes reduced Penicillium-like structures present, stipes hyaline, smooth-walled, 100–300 × 4–7 µm. Vesicles globose to subglobose, 6–13 µm. covering 100% of the head; Metulae 4–6 × 2–6 μm; Phialides ampulliform, 6–9 × 2.5–3.5 μm; Conidia globose to subglobose, smooth, 3–4 μm (Figure 3).
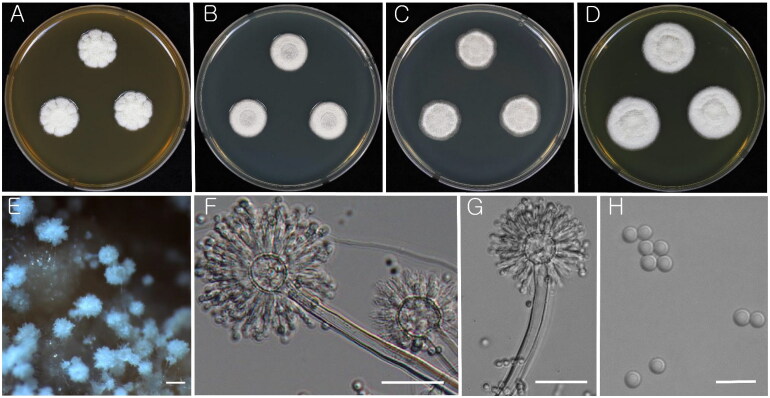
Figure 3.
Morphology of Aspergillus subalbidus (KACC 46482). (A–D) Colonies grown on MEA, CYA, DG18, and YES media after 7 d at 25 °C from left to right. (E) Conidial head on MEA. (F & G) Conidiophores with conidial head. (H) Conidia. Scale bars: E = 100 µm, F, G = 20 µm, H = 10 µm.
Strains examined: KACC 46481 and KACC 46482
Remarks: A. subalbidus is morphologically almost identical to A. candidus [15]. Phylogenetically, it forms a separate clade closely related to A. tenebricus, A. taichungensis, and A. neotritici (Figure 1).
Aspergillus uvarum G. Perrone, Varga & Kozak., International Journal of Systematic and Evolutionary Microbiology 58: 1036 (2008) [MB#510962] [37]
Colony characteristics: On CYA, colonies initially appeared white with flat mycelia and then turned brown–black, followed by reverse white, wrinkled, becoming dull yellow with black colony centers by age, and eventually reached 60–62 mm in diameter after 7 d at 25 °C. On MEA, colonies were dark brown-black, with sporulation, widespread, and turned reverse yellow-orange, and further reached 61–63 mm in diameter after 7 d at 25 °C. Colonies on YES were overgrown (90 mm plates) after 7 d at 25 °C; conidia brown–black, Abundant conidiogenesis, mycelium inconspicuous; reverse light yellow and wrinkled. On DG18, colonies were dark brown with clear white mycelium at margins, reverse white and reached 43–44 mm in diameter after 7 d at 25 °C.
Micromorphology: Conidial heads uniseriate, smooth-walled, 500–1000 × 5–10 µm. Vesicles globose to elliptical, 50–60 µm. Fertile over the entire surface. Phialides 5–7 × 4–5 µm. Conidia brown-black, globose to sub-globose, 4–5 µm, conspicuously spinose at maturity with spines projecting on the surface with 0.59 µm (Figure 4).
Figure 4.
Morphology of Aspergillus uvarum (KACC 48630). (A–D) Colonies grown on MEA, CYA, DG18, and YES media after 7 d at 25 °C from left to right. (E) Conidial head on MEA. (F & G) Conidiophores with conidial head. (H) Conidia. Scale bars: E = 100 µm, F= 50 µm, G = 20 µm, H = 5 µm.
Strain examined: KACC 48630
Remarks: A. uvarum is closely related to A. japonicus and A. aculeatus both morphologically and at a molecular level. Both species have echinulate conidial surface, uniseriate like A. uvarum [40]. However, A. japonicus has larger vesicle and similar conidial size to A. uvarum whereas A. aculeatus has a larger vesicle and ellipsoidal conidial shape [37]. KACC 48630 was well differentiated from type strains of A. japonicus and A. aculeatus. The strain KACC 48630 was deposited as A. uvarum in the year 2018. This species was later listed as an unrecorded species within Korea with strain CNUFC YB6 [41], albeit the publication was not effective due to lacking mycological specifics. Therefore, the authors here describe the species officially as unrecorded and publish the species effectively. This paper, therefore, is designed to serve as the official record of A. uvarum in Korea.
Microbial resource centers employ several methods to ensure purity of documented strains as any lapses in storage can negate research progress on particular strains [42]. KACC employs a minimum of two different preservation conditions and some of the strains used in this study have been in storage for more than 25 years. All strains in this study successfully revived and showed morphology that is typical of Aspergillus. Our study focused on re-identification and describing unrecorded species of Aspergillus from various environments, resolving taxonomic problems, and providing high quality organisms to conserve at KACC. The modern culture collection also has the goal to give access to high quality biological materials with associated information. The data presented in this study reinforces the importance of fungal collections and reassess their strain identification using current techniques.
Funding Statement
This study was financially supported by Ministry of Science and ICT [Grant number M3H9A1081254].
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Tsang CC, Tang JY, Lau SK, et al. Taxonomy and evolution of Aspergillus, Penicillium and Talaromyces in the omics era–past, present and future. Comput Struct Biotechnol J. 2018;16:197–210. doi: 10.1016/j.csbj.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Samson RA, Visagie CM, Houbraken J, et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud Mycol. 2014;78(1):141–173. doi: 10.1016/j.simyco.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JW, Kim SH, You YH, et al. Four unrecorded Aspergillus species from the rhizosphere soil in South Korea. Mycobiology. 2021;49(4):346–354. doi: 10.1080/12298093.2021.1944461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kocsubé S, Perrone G, Magistà D, et al. Aspergillus is monophyletic: evidence from multiple gene phylogenies and extrolites profiles. Stud Mycol. 2016;85(1):199–213. doi: 10.1016/j.simyco.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houbraken J, Kocsubé S, Visagie CM, et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): an overview of families, genera, subgenera, sections, series and species. Stud Mycol. 2020;95:5–169. doi: 10.1016/j.simyco.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barros Correia AC, Barbosa RN, Frisvad JC, et al. The polyphasic re-identification of a Brazilian Aspergillus section Terrei collection led to the discovery of two new species. Mycol Prog. 2020;19(9):885–903. doi: 10.1007/s11557-020-01605-4. [DOI] [Google Scholar]
- 7.Balajee SA, Nickle D, Varga J, et al. Molecular studies reveal frequent misidentification of Aspergillus fumigatus by morphotyping. Eukaryot Cell. 2006;5(10):1705–1712. doi: 10.1128/EC.00162-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stengel A, Stanke KM, Quattrone AC, et al. Improving taxonomic delimitation of fungal species in the age of genomics and phenomics. Front Microbiol. 2022;13:847067. doi: 10.3389/fmicb.2022.847067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korea . NLoSo: National List of Species of Korea. Incheon, South Korea: National Institute of Biological Resources; 2022. [Google Scholar]
- 10.Pangging M, Nguyen TT, Lee HB.. Seven undescribed Aspergillus species from different niches in Korea. Mycobiology. 2022;50(4):189–202. doi: 10.1080/12298093.2022.2116158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glass NL, Donaldson GC.. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;61(4):1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong SB, Go SJ, Shin H-D, et al. Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia. 2005;97(6):1316–1329. doi: 10.3852/mycologia.97.6.1316. [DOI] [PubMed] [Google Scholar]
- 13.Thompson JD, Gibson TJ, Plewniak F, et al. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar S, Stecher G, Tamura K.. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Visagie CM, Hirooka Y, Tanney JB, et al. Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Stud Mycol. 2014;78(1):63–139. doi: 10.1016/j.simyco.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silva JJ, Fungaro MHP, Wang X, et al. Deep genotypic species delimitation of Aspergillus section flavi isolated from Brazilian foodstuffs and the description of Aspergillus annui sp. nov. and Aspergillus saccharicola sp. nov. JoF. 2022;8(12):1279. doi: 10.3390/jof8121279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh P, Callicott KA, Orbach MJ, et al. Molecular analysis of S-morphology aflatoxin producers from the United States reveals previously unknown diversity and two new taxa. Front Microbiol. 2020;11:1236. doi: 10.3389/fmicb.2020.01236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilchrist CLM, Lacey HJ, Vuong D, et al. Comprehensive chemotaxonomic and genomic profiling of a biosynthetically talented Australian fungus, Aspergillus burnettii sp. nov. Fungal Genet Biol. 2020;143:103435. doi: 10.1016/j.fgb.2020.103435. [DOI] [PubMed] [Google Scholar]
- 19.Hong SB, Kim DH, Samson RA.. Aspergillus associated with Meju, a fermented soybean starting material for traditional soy sauce and soybean paste in Korea. Mycobiology. 2015;43(3):218–224. doi: 10.5941/MYCO.2015.43.3.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jurjević Ž, Houbraken J, Sklenář F, et al. Re-examination of species limits in Aspergillus section Flavipedes using advanced species delimitation methods and description of four new species. Stud Mycol. 2021;99(1):100120. doi: 10.1016/j.simyco.2021.100120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang XC, Zhuang WY.. New species of Aspergillus (Aspergillaceae) from tropical islands of China. J Fungi. 2022;8(3):225. doi: 10.3390/jof8030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glässnerová K, Sklenář F, Jurjević Ž, et al. A monograph of Aspergillus section Candidi. Stud Mycol. 2022;102(1):1–51. doi: 10.3114/sim.2022.102.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Bedak OA, Moubasher AH.. Aspergillus gaarensis, a new addition to section Circumdati from soil of lake El-Gaar in Wadi-El-Natron, Egypt. SIF. 2020;5(1):59–65. doi: 10.5943/sif/5/1/5. [DOI] [Google Scholar]
- 24.Crous PW, Wingfield MJ, Chooi YH, et al. Fungal planet description sheets: 1042-1111. Persoonia. 2020;44(1):301–459. doi: 10.3767/persoonia.2020.44.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doilom M, Guo J-W, Phookamsak R, et al. Screening of phosphate-solubilizing fungi from air and soil in Yunnan, China: four novel species in Aspergillus, Gongronella, Penicillium, and Talaromyces. Front Microbiol. 2020;11:585215. doi: 10.3389/fmicb.2020.585215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong S-B, Lee M, Kim D-H, et al. Aspergillus luchuensis, an industrially important black Aspergillus in east Asia. PLOS One. 2013;8(5):e63769. doi: 10.1371/journal.pone.0063769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frisvad JC, Larsen TO, De Vries R, et al. Secondary metabolite profiling, growth profiles and other tools for species recognition and important Aspergillus mycotoxins. Stud Mycol. 2007;59:31–37. doi: 10.3114/sim.2007.59.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frisvad JC, Hubka V, Ezekiel C, et al. Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Stud Mycol. 2019;93(1):1–63. doi: 10.1016/j.simyco.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen A, Frisvad JC, Sun B, et al. Aspergillus section Nidulantes (formerly Emericella): polyphasic taxonomy, chemistry and biology. Stud Mycol. 2016;84(1):1–118. doi: 10.1016/j.simyco.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geiser D, Klich M, Frisvad JC, et al. The current status of species recognition and identification in Aspergillus. Stud Mycol. 2007;59(1):1–10. doi: 10.3114/sim.2007.59.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balajee S, Houbraken J, Verweij P, et al. Aspergillus species identification in the clinical setting. Stud Mycol. 2007;59(1):39–46. doi: 10.3114/sim.2007.59.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hubka V, Nováková A, Kolařík M, et al. Revision of Aspergillus section Flavipedes: seven new species and proposal of section Jani sect. nov. Mycologia. 2015;107(1):169–208. doi: 10.3852/14-059. [DOI] [PubMed] [Google Scholar]
- 33.Sugiyama J. Mycoflora in core samples from stratigraphic drillings in middle Japan. The genus Aspergillus. J Fac Sci Univ Tokyo, Sect. 3, Botany. 1967;9:377–405. [Google Scholar]
- 34.Liu D, Yan L, Ma L, et al. Diphenyl derivatives from coastal saline soil fungus Aspergillus iizukae. Arch Pharm Res. 2015;38(6):1038–1043. doi: 10.1007/s12272-014-0371-z. [DOI] [PubMed] [Google Scholar]
- 35.El-Elimat T, Raja HA, Graf TN, et al. Flavonolignans from Aspergillus iizukae, a fungal endophyte of milk thistle (Silybum marianum). J Nat Prod. 2014;77(2):193–199. doi: 10.1021/np400955q. [DOI] [PubMed] [Google Scholar]
- 36.Noman E, Al-Gheethi A, Talip BA, et al. Oxidative enzymes from newly local strain Aspergillus iizukae EAN605 using pumpkin peels as a production substrate: optimized production, characterization, application and techno-economic analysis. J Hazard Mater. 2020;386:121954. doi: 10.1016/j.jhazmat.2019.121954. [DOI] [PubMed] [Google Scholar]
- 37.Perrone G, Varga J, Susca A, et al. Aspergillus uvarum sp. nov., an uniseriate black Aspergillus species isolated from grapes in Europe. Int J Syst Evol Microbiol. 2008;58(4):1032–1039. doi: 10.1099/ijs.0.65463-0. [DOI] [PubMed] [Google Scholar]
- 38.Gams W, Christensen M, Onions AH, et al. Infrageneric taxa of Aspergillus. Adv Penicillium Aspergillus Syst. 1986;102:55–62. [Google Scholar]
- 39.Fungaro MHP, Ferranti LS, Massi FP, et al. Aspergillus labruscus sp. nov., a new species of Aspergillus section Nigri discovered in Brazil. Sci Rep. 2017;7(1):6203. doi: 10.1038/s41598-017-06589-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silva DM, Batista LR, Rezende EF, et al. Identification of fungi of the genus Aspergillus section Nigri using polyphasic taxonomy. Braz J Microbiol. 2011;42(2):761–773. doi: 10.1590/S1517-83822011000200044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang KH, Lee HB.. Four undescribed fungal species belonging to Amphisphaeriales and Eurotiales in Korea. KSM Newslett. 2022;34(2):94. [Google Scholar]
- 42.Abd-Elsalam KA, Yassin MA, Moslem MA, et al. Culture collections, the new herbaria for fungal pathogens. Fungal Divers. 2010;45(1):21–32. doi: 10.1007/s13225-010-0063-z. [DOI] [Google Scholar]