Abstract
Background
As an inevitable event after kidney transplantation, ischemia‒reperfusion injury (IRI) can lead to a decrease in kidney transplant success. The search for signature genes of renal ischemia‒reperfusion injury (RIRI) is helpful in improving the diagnosis and guiding clinical treatment.
Methods
We first downloaded 3 datasets from the GEO database. Then, differentially expressed genes (DEGs) were identified and applied for functional enrichment analysis. After that, we performed three machine learning methods, including random forest (RF), Lasso regression analysis, and support vector machine recursive feature elimination (SVM-RFE), to further predict candidate genes. WGCNA was also executed to screen candidate genes from DEGs. Then, we took the intersection of candidate genes to obtain the signature genes of RIRI. Receiver operating characteristic (ROC) analysis was conducted to measure the predictive ability of the signature genes. Kaplan‒Meier analysis was used for association analysis between signature genes and graft survival. Verifying the expression of signature genes in the ischemia cell model.
Results
A total of 117 DEGs were screened out. Subsequently, RF, Lasso regression analysis, SVM-RFE and WGCNA identified 17, 25, 18 and 74 candidate genes, respectively. Finally, 3 signature genes (DUSP1, FOS, JUN) were screened out through the intersection of candidate genes. ROC analysis suggested that the 3 signature genes could well diagnose and predict RIRI. Kaplan‒Meier analysis indicated that patients with low FOS or JUN expression had a longer OS than those with high FOS or JUN expression. Finally, we validated using the ischemia cell model that compared to the control group, the expression level of JUN increased under hypoxic conditions.
Conclusions
Three signature genes (DUSP1, FOS, JUN) offer a good prediction for RIRI outcome and may serve as potential therapeutic targets for RIRI intervention, especially JUN. The prediction of graft survival by FOS and JUN may improve graft survival in patients with RIRI.
Keywords: Renal ischemia‒reperfusion injury, Random forest, Support vector machine recursive feature elimination
1. Introduction
As the preferred renal replacement therapy option for patients with end-stage renal disease (ESRD), renal transplantation improves the quality of life of patients compared to long-term dialysis [1]. However, the graft undergoes donor organ isolation, transport, storage, implantation into the recipient, and reperfusion during transplantation, which will inevitably lead to IRI [2]. In the perioperative stage following kidney transplantation, IRI represents a serious clinical challenge for clinicians. RIRI induces a series of clinical events, such as delayed graft function (DGF), graft rejection, chronic rejection, and chronic graft dysfunction, leading to an impact on long-term graft outcome [3]. Alleviating IRI can effectively lower the incidence of dialysis-requiring renal impairment following surgery [4]. IRI occurs in all dead donor kidney transplants [5]. In addition, IRI is the determining cause of critical acute kidney injury (AKI), a clinical syndrome characterized by rapid kidney failure and significant fatality rates [6,7]. Therefore, it is critical to gain a deeper knowledge of the molecular biological changes involved in the RIRI process and to develop a novel model to better anticipate the occurrence of IRI.
Several possible mechanisms may underlie RIRI, including calcium overload, damage caused by reactive oxygen species (ROS) and oxidative stress [8]. In response to these pathophysiological processes, current therapeutic options are oriented toward the suppression of the inflammatory response, cell apoptosis, oxidative stress, and renal fibrosis [[9], [10], [11]]. Nevertheless, currently, no effective therapeutic options are available to protect against RIRI. The main reason for this dilemma is the complexity of the mechanism of RIRI and a lack of good molecular targets for treatment, leading to uncertainty in renal transplantation outcomes. Currently, there are few studies about signature genes and RIRI, and the available studies have mainly focused on the relationship between DGF and RIRI. Wu J et al. utilized machine learning methods to identify DGF-related hub genes and built a robust prediction model for DGF and graft survival, which may be used to guide early prevention and tailored therapy of various postoperative problems following renal transplantation [12]. DGF is only one element of the complications associated with RIRI. The above study is not comprehensive and may overlook other key factors. Therefore, our study aimed to comprehensively analyse the signature genes of RIRI and their effects on transplanted kidneys (graft survival). A previous study mainly concentrated on constructing experimental RIRI mouse models to detect differentially expressed genes using just six mice [13]. A limited sample size will reduce statistical power for some observations. Accordingly, we comprehensively considered the limitations of the above studies to further search for molecular targets for alleviating IRI in renal transplantation.
The emergence of machine learning can unlock large biomedical and patient datasets, which is beneficial to new paradigms for the diagnosis and treatment of various diseases [14]. Typically, basic prediction tasks may be accomplished with conventional models (such as logistic regression), but sophisticated problems demand more elaborate models (e.g., neural networks) [15]. Thus, to identify efficient and effective signature genes for RIRI, RF, Lasso regression analysis and SVM-RFE were utilized to explore the datasets. Yang et al. identified 11 postmenopausal osteoporosis mRNA biomarkers using Lasso regression analysis and the Boruta algorithm and used classifiers, including SVM, decision tree and RF, to construct the model [16]. Wang et al. screened 12 autophagy-related genes (ATGs) for endometriosis by WCGNA, which would benefit research in endometriosis [17]. Currently, the wide applicability and powerful application of machine learning and WGCNA have been used by many researchers to identify disease signature genes. Therefore, we will work on the combination of machine learning and WGCNA with biological data studies of RIRI with the aim of uncovering the signature genes of RIRI.
To identify the signature genes of RIRI, we retrieved renal transplantation patient gene expression matrices from the Gene Expression Omnibus (GEO) database and screened and validated the signature genes of RIRI using a series of bioinformatics methods. In addition, we explored the correlation between signature genes and graft survival. Taken together, further identification of signature genes for RIRI may provide potential therapeutic targets for RIRI intervention, which is helpful for the improvement of RIRI clinical treatment.
2. Methods
2.1. Data acquisition and grouping
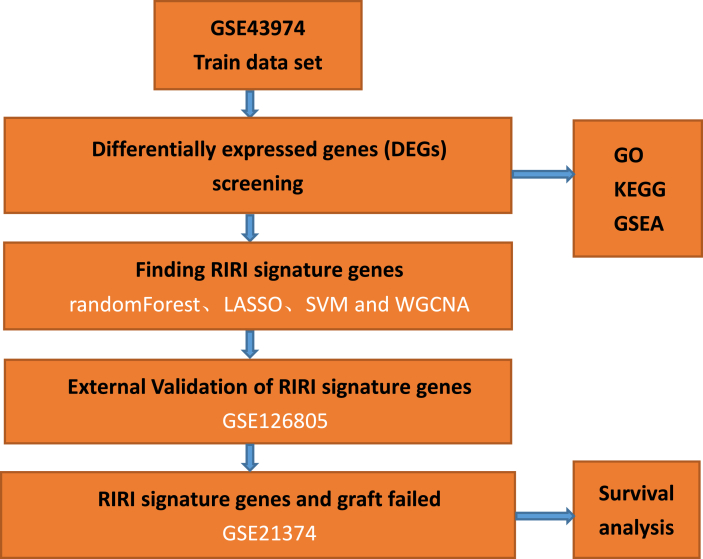
The detailed research process is shown in Fig. 1. As shown in Table 1, we systematically gathered 3 datasets that contained mRNA expression profiles from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). The GSE43974 dataset was used as the training set to find the signature genes of RIRI, while the GSE126805 dataset was used as the test set for external validation of the signature genes. The GSE21374 dataset was used to explore the correlation between signature genes and graft survival. Each dataset was divided into treatment and control groups, with the control and treatment groups indicating before and after ischemia‒reperfusion of the kidney graft, respectively. The R package “limma” was used to normalize the expression of all genes in different datasets.
Fig. 1.
The data analysis flowchart reveals our workflow.
Table 1.
mRNA expression profile datasets in the GEO database.
2.2. Identification and visualization of DEGs
To explore the difference between the treatment and control groups of the GSE43974 dataset at the genetic level, the R package “limma” was used to search DEGs, which were visualized by the R package “pheatmap”. |log2FC|≥0.585 and P < 0.05 were considered the thresholds for DEG filtering.
2.3. Function and pathway enrichment analysis
“ClusterProfiler” was utilized to perform GO, KEGG, and GSEA analyses to investigate the potential biological functions and pathways of DEGs. According to the categories of molecular function (MF), biological process (BP), and cellular component (CC), GO terms with P < 0.05 and Q < 0.05 were selected and presented accordingly in bubble plots. Similarly, the enriched KEGG pathways were shown in bubble plots and filtered out using P < 0.05 and Q < 0.05. The transcriptome data of patients in the treatment and control groups were submitted to GSEA, and the KEGG pathways were enriched with the DEGs in both groups. P < 0.05 and an FDR of 25 % were used to filter the DEGs in both groups.
2.4. Identification and validation of signature genes for RIRI
After obtaining DEGs, to further judge signature genes among them, we conducted RF, Lasso regression analysis, SVM-RFE, and WGCNA on the training dataset. RF was performed by the R package “randomForest”, and a mean decrease Gini >2 was set as the threshold to filter candidate genes. LASSO regression analysis was utilized by the R package “glmnet”. The penalization coefficient (log λ value) was calculated underlying the partial likelihood deviance of 10-fold cross-validation. The optimal log λ value was determined based on the minimum binomial deviance. SVM-RFE was applied by the R packages “e1071”, “kernlab”, and “caret”. The value of the variable corresponding to the minimum root mean square error (RMSE) represented the number of candidate genes screened. WGCNA was performed by the R package “WGCNA”. The candidate genes screened through the above four methods were intersected for the signature genes by the R package “VennDiagram”. To assess the projected values of signature genes as biomarkers for RIRI, ROC curves and the area under the ROC curves (AUC) were computed through the R package “pROC” in the GSE43974 dataset and the GSE126805 dataset.
2.5. Correlation between signature genes and graft survival
Patients were divided into high-expression and low-expression groups by the median gene expression. Analysis of the correlation between signature genes and graft survival was carried out by ROC curves and Kaplan–Meier survival analysis. According to the expression level of signature genes, patients were divided into high and low expression groups in the GSE21374 dataset. Kaplan–Meier survival analysis was utilized to assess differences in graft survival between the high and low expression groups by the “survival” R package in the GSE21374 dataset.
2.6. Simulated ischemia
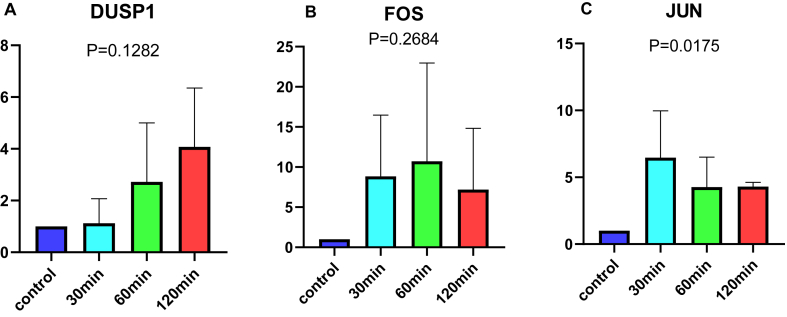
We isolated oxygen by covering the surface of the HK-2 cell culture medium with a layer of paraffin oil. This is a common biological model of cell ischemia. Samples (n = 4) were collected at 30 min, 60 min, and 120 min of hypoxia. The expression levels of DUSP1, FOS, and JUN were measured using PCR. The differences between groups were compared using the Kruskal‒Wallis test, and a P value of less than 0.05 indicated statistically significant differences.
3. Results
3.1. Determination of DEGs between the treatment and control groups
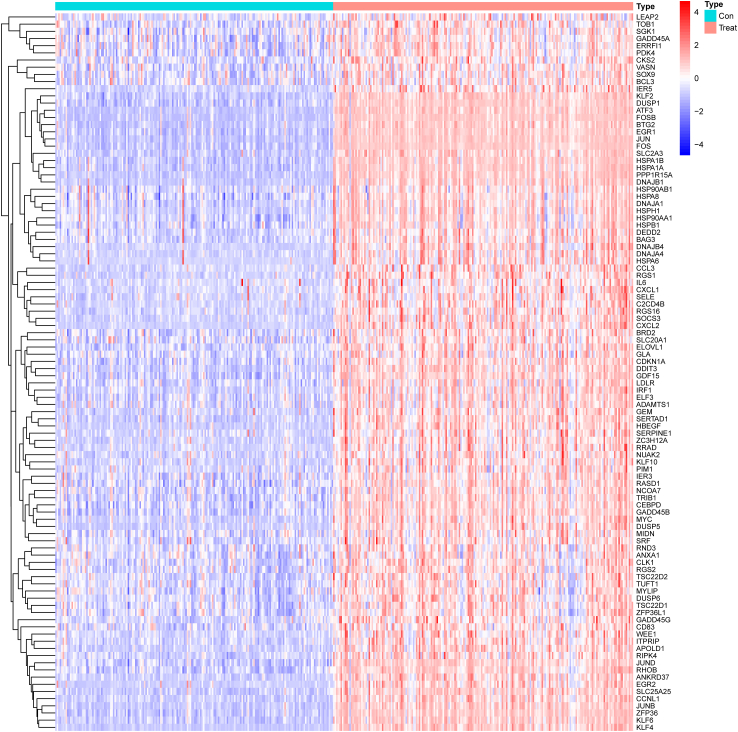
To investigate the key genes that may modulate RIRI progression, we performed a differential analysis based on the expression matrix of the treated and control groups in the GSE43974 dataset. Then, 117 DEGs were identified based on the threshold (|log2FC|≥0.585 and P < 0.05). Several DEGs between the treated and control groups are displayed in the heatmap (Fig. 2).
Fig. 2.
Heatmap of DEGs between the treated and control groups. The patients are represented in two different colors. In DEGs, red and blue indicate upregulated and downregulated expression levels, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Function and pathway enrichment analysis of DEGs
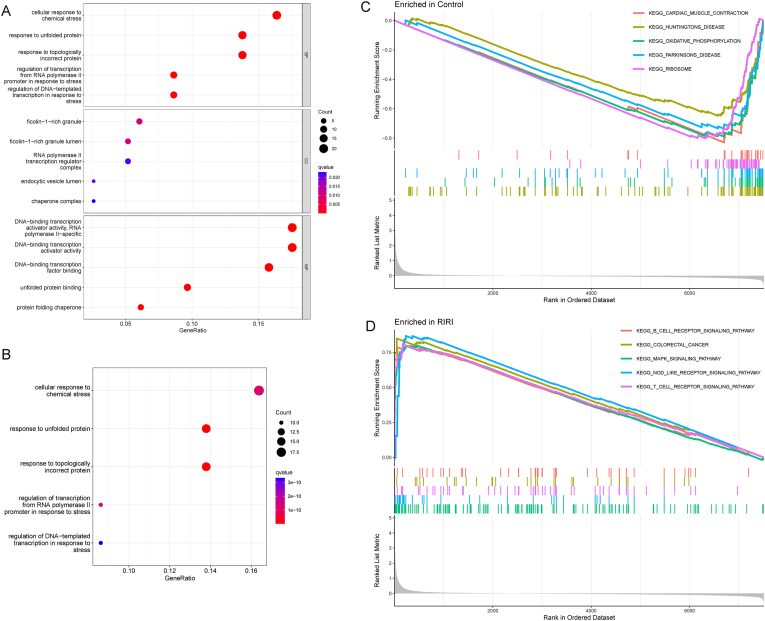
To clarify the biological role of DEGs in modulating RIRI development, we applied GO enrichment, KEGG pathway analysis, and GSEA. As shown in Fig. 3A, cellular response to chemical stress and response to unfolded protein were enriched through GO analysis. Additionally, KEGG analysis showed that three pathways were enriched significantly, namely, cellular response to chemical stress, response to unfolded protein, and response to topologically incorrect protein (Fig. 3B). Some disease-associated pathways belonged to the control group, including cardiac muscle contraction, Huntington's disease, oxidative phosphorylation, Parkinson's disease, and ribosome (Fig. 3C). GSEA revealed that the B-cell receptor signaling pathway, MAPK signaling pathway, NOD-like signaling pathway, and T-cell receptor signaling pathway were enriched in the RIRI group (Fig. 3D). Taken together, DEGs may have the potential to influence the outcomes of RIRI through stress and immune pathways.
Fig. 3.
Function and pathway enrichment analysis. (A) GO biological process enrichment analysis. BP: biological process, CC: cellular component, MF: molecular function. (B) KEGG pathway analysis. (C–D) GSEA of the enriched pathways in the control (C) and RIRI (D) groups.
3.3. Identification of signature genes for RIRI
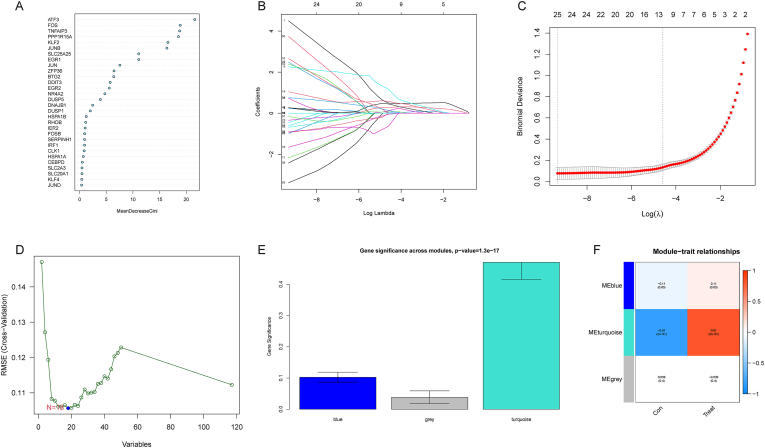
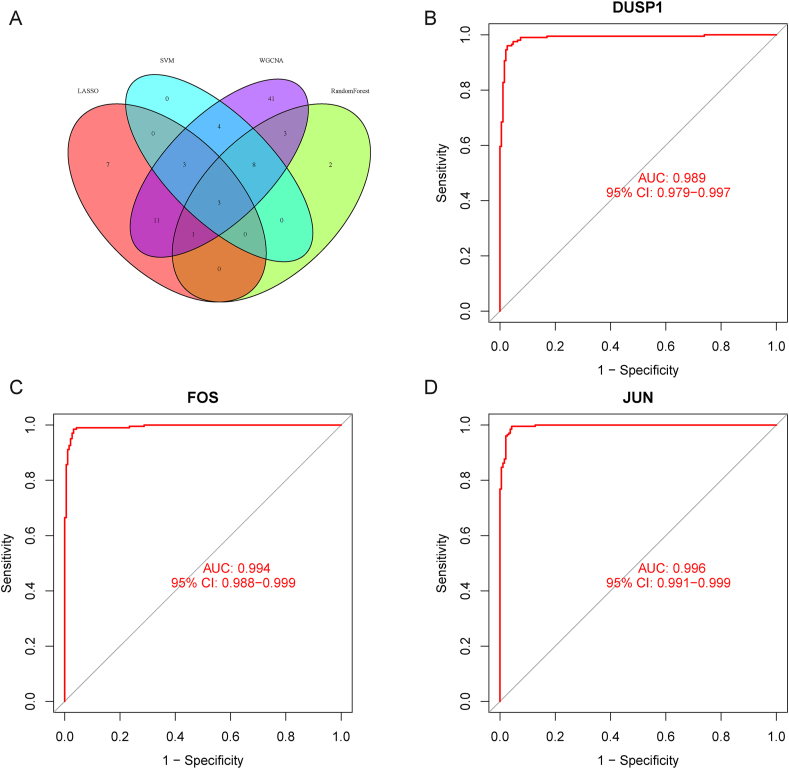
For the sake of further selecting diagnostic genes for RIRI, DEGs were subjected to 4 statistical methods to determine the signature genes. First, we obtained 17 candidate genes by RF based on the filtering conditions set by the Gini coefficient (Fig. 4A). Then, with the help of Lasso regression analysis, 25 candidate genes were identified according to the optimum log λ value (Fig. 4B–C). Additionally, 18 candidate genes were selected by SVM-RFE corresponding to the minimum value of RMSE (Fig. 4D). Moreover, as shown in Fig. 4E and F, genes in the turquoise module had the most related genes (P < 0.05). Genes in the turquoise module were positively correlated with the clinical traits of the treatment group and negatively correlated with those of the control group. Seventy-four candidate genes in the turquoise module were identified. Finally, 3 overlapping candidate genes, DUSP1, FOS, and JUN, were chosen based on the above 4 methods as signature genes for RIRI (Fig. 5A). Overall, 3 signature genes were selected that might be critical genes involved in the progression of RIRI. However, further validation is required in future studies.
Fig. 4.
Selection of signature genes for RIRI. (A) RF screening for candidate genes. The candidate genes were ranked according to the mean decrease Gini. (B–C) Lasso regression screening for candidate genes. (B) The upper horizontal coordinate indicates the number of variables with nonzero coefficients, the lower horizontal coordinate indicates the log λ value, and the vertical coordinate indicates the change in the coefficients of the variables. (C) The vertical coordinate indicates binomial deviance. The horizontal coordinate's significance is identical to that of the horizontal coordinate in diagram B. (D) SVM-RFE screening for candidate genes. The horizontal coordinate represents the variable values, and the vertical coordinate represents the cross-validation errors. (E–F) WGCNA screening for the module that contains the candidate genes. Darker colors indicate a stronger correlation between genes in modules and traits. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 5.
The predicted values of signature genes in the training dataset. (A) The Venn diagram shows the intersection of 3 signature genes from the 4 screening methods. (B–D) ROC curves of 3 signature genes (B: DUSP1; C: FOS; D: JUN). The horizontal coordinate represents the 1-specificity, and the vertical coordinate represents the sensitivity.
3.4. Validation of signature genes for RIRI
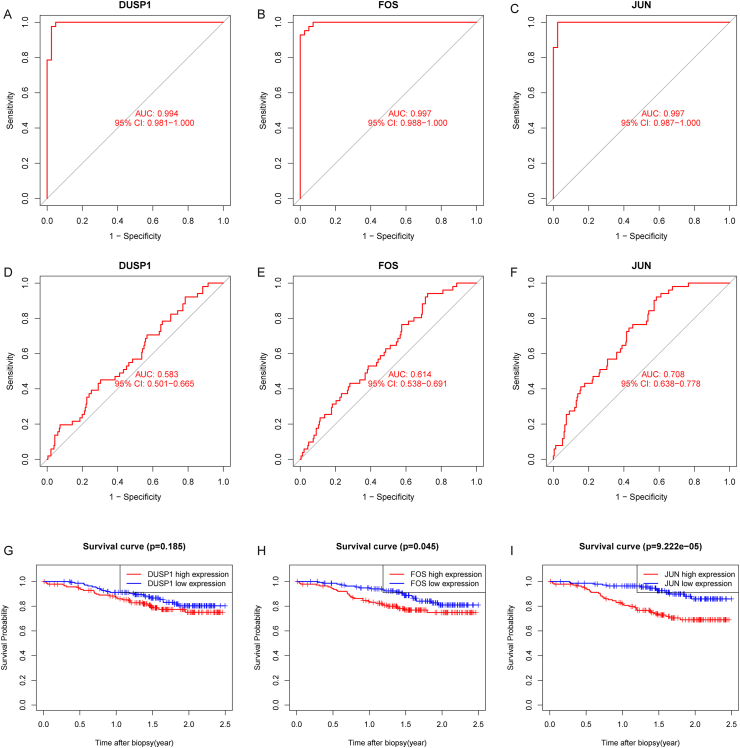
Underlying the above preliminary analysis, we further investigated the sensitivity and specificity of the 3 signature genes. The ROC curve showed that the AUC values of DUSP1, FOS, and JUN were 0.989 (95 % CI: 0.979−0.997), 0.994 (95 % CI: 0.988−0.999), and 0.996 (95 % CI: 0.991−0.999), respectively. The training dataset exhibited a strong diagnostic ability (Fig. 5B–D). Synchronously, Fig. 6A–C demonstrates the predictive efficiency of the 3 signature genes, with AUC values of DUSP1, FOS, and JUN of 0.994 (95 % CI: 0.981−1.000), 0.997 (95 % CI: 0.988−1.000), and 0.997 (95 % CI: 0.987−1.000), respectively, in the test dataset. In conclusion, the 3 signature genes may have potential diagnostic value for RIRI. We validated using the ischemia cell model that compared to the control group, the expression level of JUN increased under hypoxic conditions (Fig. 7A–C).
Fig. 6.
Diagnostic performance of the signature genes and correlation between the signature genes and graft survival. (A–F) ROC curves of 3 signature genes (A–C: in the GSE126805 dataset; D–F: in the GSE21374 dataset). (G–I) Kaplan–Meier survival revealed a notable difference in graft survival between the high- and low-expression groups (G: DUSP1; H: FOS; I: JUN).
Fig. 7.
The effect of ischemic duration on the expression levels of DUSP1, FOS, and JUN. N = 4. (A: DUSP1; B: FOS; C: JUN).
3.5. Correlation between signature genes and graft survival
To further explore the value of the 3 signature genes for RIRI research, we sought to determine the correlation between the 3 signature genes and graft survival. The diagnostic value in graft survival of the signature genes was assessed by ROC analysis, and Kaplan‒Meier analysis was used to evaluate graft survival differences between the high- and low-expression levels of signature genes. The AUC values of DUSP1, FOS, and JUN were 0.583 (95 % CI: 0.501−0.665), 0.614 (95 % CI: 0.538−0.691), and 0.708 (95 % CI: 0.638−0.778), respectively, in the GSE21374 dataset, which indicated that the 3 signature genes had good predictive efficacy for graft survival (Fig. 6D–F). Subsequently, patients with low FOS or JUN expression had a longer OS than those with high FOS or JUN expression (P < 0.05; Fig. 6HI). However, DUSP1 did not exhibit a significant effect on graft survival. (P > 0.05; Fig. 6G).
4. Discussion
As an unavoidable adverse event after renal transplantation, RIRI is a hurdle to be overcome in future research. Current strategies to prevent and treat RIRI are limited to restricting the preoperative diet, improving organ preservation conditions, reducing organ damage before and after ischemia, and applying cellular and pharmacological therapies [18]. In fact, after years of research and interventions, RIRI remains a problem in clinical practice, and new ideas are necessary to deepen the understanding of RIRI. Developments in genomics have led to tremendous advances in numerous areas of medical science. A study identified several candidate genes for RIRI with multiomics and bioinformatics and demonstrated their potential as biomarkers with treatment models, which suggested the potential of screening for signature genes to improve RIRI clinical management [19]. Therefore, our study incorporated 3 independent datasets to explore the signature genes of RIRIs.
First, we mined DEGs using the GSE43974 dataset as a training set and screened 3 signature genes (DUSP1, FOS, and JUN) with the RF, Lasso regression analysis, SVM-RFE, and WGCNA methods. Then, the GSE126805 dataset was used as an external independent validation set to verify these signature genes. In addition, we also explored the correlation between these signature genes and graft survival in the GSE21374 dataset. In this way, we hypothesized that these 3 signature genes may modulate RIRI progression, and targeted intervention of these genes may provide new ideas for the treatment of RIRI.
Chemical stress reactions, mainly oxidative stress, have always been the main pathway mediating the occurrence of IRI. Liu et al. found that inhibition of Brd4 to block the PI3K-AKT pathway reduced the FoxO4-mediated oxidative stress response and reduced the damage caused by RIRI [20]. Under pathological circumstances, cells may lose their ability to maintain proteostasis, which leads to a buildup of unfolded and improperly folded proteins in the endoplasmic reticulum (ER) and triggers the unfolded protein response (UPR) or ER stress [21]. Unfortunately, the UPR can induce apoptotic cell death in the event of chronic damage [22]. A study showed that the knockdown of CHOP, a transcriptional regulator of the UPR, ameliorated postischemic microcirculatory recovery [23], which might validate the regulation of the UPR as a therapeutic target for RIRI. A constructed analysis of differential lncRNA and mRNA expression in hepatic ischemia‒reperfusion injury suggests that mRNA may affect the molecular mechanisms and functions of hepatic ischemia‒reperfusion injury via pathways involving response to topologically incorrect protein [24]. Su et al. found effective alleviation of RIRI by inhibiting the expression of PANX1 and thereby blocking the MAPK signaling pathway [25]. Meanwhile, nod-like receptor protein 3 (NLRP3) inflammatory vesicles are activated in RIRI through the mROS-TXNIP-NLRP3 axis [26]. In addition, T cells are aggregated and activated during RIRI to activate adaptive immune responses [27]. In contrast, there are few reports on B cells in RIRI, and there is not yet sufficient evidence for their role in RIRI.
The protein encoded by DUSP1 (dual specificity phosphatase 1) is a phosphatase with dual specificity for tyrosine and threonine [28]. DUSP1 inactivates the MAP kinases ERK and JNK by dephosphorylating them, thus playing a role in physiological processes, including the stress response, inflammation, cycle arrest, and apoptosis, by regulating the MAPK signaling pathway. The typical MAPK signaling pathway comprises MAPK, MAPK kinase (MAPKK), and MAPKK kinase (MAPKK), and recent research has centered on ERK1/2, JNKs, and P38 isoforms [29]. Upregulation of DUSP1 inhibits the activation of JNK, thereby inhibiting apoptosis [30]. Currently, DUSP1 is mainly limited in the direction of myocardial ischemia‒reperfusion injury (MIRI) in IRI studies. Jin et al. found that MIRI led to the downregulation of DUSP1, which resulted in increased phosphorylation of JNK, triggering mitochondrial fission and mitochondrial autophagy and ultimately leading to extensive cell death [31]. Tan et al. showed that miR-378a-3p mediated the TRIM55/DUSP1/JNK axis to inhibit MIRI-induced cardiomyocyte apoptosis [32]. Although DUSP1 has rarely been reported in RIRI, an in vitro cellular model of ER-induced stress demonstrated that inhibition of the JNK pathway reduces ROS-dependent oxidative stress, thereby decreasing renal IR apoptosis [33]. In addition, p-JNK and p-ERK protein levels were reduced in the RIRI mouse model [34]. Taken together, we hypothesize that DUSP1 may influence the MAPK signaling pathway, which ultimately results in severe apoptosis and contributes to the development of RIRI.
FOS (c-fos) is a member of the FOS family of transcription factors, encoding leucine zipper proteins involved in dimerization and DNA binding [35]. Various studies have demonstrated that FOS is involved in IRI development, such as MIRI [36] and cerebral IRI (CIRI) [37]. Previous studies suggested the possible involvement of FOS in the development of RIRI. For example, in a study of renal transplant rejection, upregulation of ET-1 was associated with the promotion of IRI-activated FOS expression in renal tubular epithelial cells [38,39]. FOS is a crucial gene in the physiological and pathological processes of IRI-AKI, and its primary function is to modulate the body's immune microenvironment [40,41]. JUN (c-jun) is a member of the JUN family of transcription factors, which encodes a protein initially identified in transformed cells carrying the replication-deficient avian sarcoma virus 17 (ASV 17) genome and shares similarities with the yeast transcriptional regulatory protein Gcn4 [42]. Activator protein 1 (AP-1) is a dimer composed of the FOS gene family and the JUN gene family [43]. As subunits of AP-1, the relative abundance of FOS and JUN plays a decisive role in AP-1 affecting cellular functions [44]. The cellular processes of differentiation, proliferation, survival, and apoptosis are all impacted by AP-1. AP-1 is regulated by various stimuli, including oxidative stress [45], cytokines, growth factors [44], and oncogene activation [46], and through this regulation, it regulates various cellular activities, including differentiation, proliferation, survival, and apoptosis [44]. Various stimuli can affect AP-1 activity by activating the MAPK cascade to regulate AP-1 subunits (especially JUN), such as phosphorylating c-jun and enhancing the DNA binding of c-jun to ATF [47]. Thus, influencing the MAPK signaling pathway by altering stimuli to mediate AP-1 may be effective in inhibiting IRI, as verified by the studies of Shen et al. and Tan et al. [48,49]. In terms of RIRI, AP-1 induces RIRI. Reznik et al. performed a transcriptome analysis of renal grafts reperfused after warm ischemia and found that the transcript levels of the grafts were centered on AP-1 transcription factors and heat shock proteins instead of an overall change in transcript levels [50]. Gerhardt et al. performed a transcriptome analysis of different mononuclear transcriptomic analyses of renal proximal tubular cells from RIRI at different time periods and revealed significant activation of the AP-1 signaling pathway in late RIRI [51].
IRI induces various types of immune responses, renal functional load, graft hypoxia, and fibrotic response to reduce the long-term survival of grafts [52]. The complement system is continuously activated by IRI and imbalance, thus promoting the progression of acute kidney injury (AKI) to chronic kidney disease (CKD). The imbalanced complement system leads to irreversible fibrosis and aging of the kidney through the induction of the production of various cytokines and leukocyte chemotaxis, which results in decreased graft survival [53]. The reduction in functional renal units by IRI leads to an overload of renal function, which leads to the accumulation of toxic metabolites, exacerbates oxidative stress and inflammatory responses, and ultimately leads to graft inactivation [52]. To prevent RIRI from interfering with graft survival, current measures focus on reducing early harmful hazards and improving long-term graft survival, as evidenced by ischemic preconditioning, machine perfusion, gas-protective agents, metabolic, anti-inflammatory measures, antioxidant therapy, and suppression of innate immune responses [3]. Nevertheless, improving kidney graft survival remains a difficult task in transplantation sessions. To this end, our study mined 3 signature genes that yielded promising results in the association between RIRI and graft survival. The signature genes also had good specificity and sensitivity for predicting the survival of grafts, which reflected the reliability of our study. However, the survival curves for DUSP1 had no significance, and given this, we speculate that it is the lack of sensitivity of the data due to insufficient sample size.
However, there are several limitations of our study that need to be addressed. First, the signature genes were searched through retrospective data from public databases, and prospective data are needed to validate their clinical applicability. Second, the sample size of our study is limited, which will lead to bias in the results of the study. In addition, the sample quality of the public database is uncontrollable, which brings about confounding factors that will affect the results of our research to a certain extent. Therefore, collecting more cohort samples and strictly controlling the impact of confounding factors is the direction of our future research.
5. Conclusions
In this study, we identified 3 signature genes (DUSP1, FOS, JUN) based on machine learning and WGCNA. Good predictive performance indicates their accurate diagnostic ability in RIRI, suggesting that they will be potential therapeutic targets against RIRI. FOS and JUN were accurate in predicting the graft survival of RIRI patients. The probability of graft survival can be predicted by FOS and JUN, which is expected to enable patients to receive appropriate treatment immediately before RIRI progresses. Collectively, these results may contribute to new insights into the diagnosis and treatment of RIRI.
Author contribution statement
Zechao Lu: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper. Senkai Xu: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper. Haiqin Liao: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper. Yixin Zhang: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper. Zeguang Lu: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper. Zhibiao Li: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper. Yushu Chen: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper. Feng Guo: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper. Fucai Tang: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data. Zhaohui He: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Data availability statement
The gene expression data of patients were downloaded from GEO (https://www.ncbi.nlm.nih.gov/geo).
Ethical statement
The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The present study was supported by the Guangdong Basic and Applied Basic Research Foundation (contract no. 2020A1515010152) and the Public Health Research Project in Futian District, Shenzhen (grant no. FTWS2020064 and FTWS2021066).
Contributor Information
Fucai Tang, Email: tangfc@mail.sysu.edu.cn.
Zhaohui He, Email: hechh9@mail.sysu.edu.cn.
References
- 1.Hariharan S., Israni A.K., Danovitch G. Long-term survival after kidney transplantation. N. Engl. J. Med. 2021;385:729–743. doi: 10.1056/NEJMra2014530. [DOI] [PubMed] [Google Scholar]
- 2.Cavaille-Coll M., Bala S., Velidedeoglu E., Hernandez A., Archdeacon P., Gonzalez G., Neuland C., Meyer J., Albrecht R. Summary of fda workshop on ischemia reperfusion injury in kidney transplantation. Am. J. Transplant. 2013;13:1134–1148. doi: 10.1111/ajt.12210. [DOI] [PubMed] [Google Scholar]
- 3.Salvadori M., Rosso G., Bertoni E. Update on ischemia-reperfusion injury in kidney transplantation: pathogenesis and treatment. World J. Transplant. 2015;5:52–67. doi: 10.5500/wjt.v5.i2.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zarbock A., Kellum J.A. Remote ischemic preconditioning and protection of the kidney--a novel therapeutic option. Crit. Care Med. 2016;44:607–616. doi: 10.1097/CCM.0000000000001381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan J.S., Sheikh-Hamad D. Mitochondrial dysfunction in acute kidney injury and sex-specific implications. Med Res Arch. 2019;7 doi: 10.18103/mra.v7i2.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li D.D., Li N., Cai C., Wei C.M., Liu G.H., Wang T.H., Xu F.R. A molecular network-based pharmacological study on the protective effect of panax notoginseng rhizomes against renal ischemia-reperfusion injury. Front. Pharmacol. 2023;14 doi: 10.3389/fphar.2023.1134408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka S., Inoue T., Hossack J.A., Okusa M.D. Nonpharmacological, biomechanical approaches to control inflammation in acute kidney injury. Nephron Clin. Pract. 2017;137:277–281. doi: 10.1159/000477218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu M.Y., Yiang G.T., Liao W.T., Tsai A.P., Cheng Y.L., Cheng P.W., Li C.Y., Li C.J. Current mechanistic concepts in ischemia and reperfusion injury. Cell. Physiol. Biochem. 2018;46:1650–1667. doi: 10.1159/000489241. [DOI] [PubMed] [Google Scholar]
- 9.Liao X., Lv X., Zhang Y., Han Y., Li J., Zeng J., Tang D., Meng J., Yuan X., Peng Z., Tao L., Xie Y. Fluorofenidone inhibits uuo/iri-induced renal fibrosis by reducing mitochondrial damage. Oxid. Med. Cell. Longev. 2022;2022 doi: 10.1155/2022/2453617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shan Y., Chen D., Hu B., Xu G., Li W., Jin Y., Jin X., Jin X., Jin L. Allicin ameliorates renal ischemia/reperfusion injury via inhibition of oxidative stress and inflammation in rats. Biomed. Pharmacother. 2021;142 doi: 10.1016/j.biopha.2021.112077. [DOI] [PubMed] [Google Scholar]
- 11.Shi M., Wang J., Bi F., Bai Z. Diosmetin alleviates cerebral ischemia-reperfusion injury through keap1-mediated nrf2/are signaling pathway activation and nlrp3 inflammasome inhibition. Environ. Toxicol. 2022;37:1529–1542. doi: 10.1002/tox.23504. [DOI] [PubMed] [Google Scholar]
- 12.Wu J., Zhang F., Zheng X., Zhang J., Cao P., Sun Z., Wang W. Identification of renal ischemia reperfusion injury subtypes and predictive strategies for delayed graft function and graft survival based on neutrophil extracellular trap-related genes. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1047367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su M., Hu X., Lin J., Zhang L., Sun W., Zhang J., Tian Y., Qiu W. Identification of candidate genes involved in renal ischemia/reperfusion injury. DNA Cell Biol. 2019;38:256–262. doi: 10.1089/dna.2018.4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goecks J., Jalili V., Heiser L.M., Gray J.W. How machine learning will transform biomedicine. Cell. 2020;181:92–101. doi: 10.1016/j.cell.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duan Y., Xie E., Liu C., Sun J., Deng J. Establishment of a combined diagnostic model of abdominal aortic aneurysm with random forest and artificial neural network. BioMed Res. Int. 2022;2022 doi: 10.1155/2022/7173972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang C., Ren J., Li B., Jin C., Ma C., Cheng C., Sun Y., Shi X. Identification of gene biomarkers in patients with postmenopausal osteoporosis. Mol. Med. Rep. 2019;19:1065–1073. doi: 10.3892/mmr.2018.9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J., Cong S., Wu H., He Y., Liu X., Sun L., Zhao X., Zhang G. Identification and analysis of potential autophagy-related biomarkers in endometriosis by wgcna. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.743012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saat T.C., van den Akker E.K., Ijzermans J.N., Dor F.J., de Bruin R.W. Improving the outcome of kidney transplantation by ameliorating renal ischemia reperfusion injury: lost in translation? J. Transl. Med. 2016;14:20. doi: 10.1186/s12967-016-0767-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perco P., Pleban C., Kainz A., Lukas A., Mayer B., Oberbauer R. Gene expression and biomarkers in renal transplant ischemia reperfusion injury. Transpl. Int. 2007;20:2–11. doi: 10.1111/j.1432-2277.2006.00376.x. [DOI] [PubMed] [Google Scholar]
- 20.Liu H., Wang L., Weng X., Chen H., Du Y., Diao C., Chen Z., Liu X. Inhibition of brd4 alleviates renal ischemia/reperfusion injury-induced apoptosis and endoplasmic reticulum stress by blocking foxo4-mediated oxidative stress. Redox Biol. 2019;24 doi: 10.1016/j.redox.2019.101195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inagi R., Ishimoto Y., Nangaku M. Proteostasis in endoplasmic reticulum--new mechanisms in kidney disease. Nat. Rev. Nephrol. 2014;10:369–378. doi: 10.1038/nrneph.2014.67. [DOI] [PubMed] [Google Scholar]
- 22.Hetz C., Zhang K., Kaufman R.J. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 2020;21:421–438. doi: 10.1038/s41580-020-0250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong B., Zhou H., Han C., Yao J., Xu L., Zhang M., Fu Y., Xia Q. Ischemia/reperfusion-induced chop expression promotes apoptosis and impairs renal function recovery: the role of acidosis and gpr4. PLoS One. 2014;9 doi: 10.1371/journal.pone.0110944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hua Y., Xu Y., Li X., Yin B., Lu S., Wang C., Ke S., Qian B., Yu H., Bai M., Ma Y. Integrative analysis of the roles of lncrnas and mrnas in ischaemic preconditioning to alleviate liver ischaemia-reperfusion injury in mice. Biochem. Biophys. Res. Commun. 2022;627:30–38. doi: 10.1016/j.bbrc.2022.08.041. [DOI] [PubMed] [Google Scholar]
- 25.Su L., Jiang X., Yang C., Zhang J., Chen B., Li Y., Yao S., Xie Q., Gomez H., Murugan R., Peng Z. Pannexin 1 mediates ferroptosis that contributes to renal ischemia/reperfusion injury. J. Biol. Chem. 2019;294:19395–19404. doi: 10.1074/jbc.RA119.010949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wen Y., Liu Y.R., Tang T.T., Pan M.M., Xu S.C., Ma K.L., Lv L.L., Liu H., Liu B.C. Mros-txnip axis activates nlrp3 inflammasome to mediate renal injury during ischemic aki. Int. J. Biochem. Cell Biol. 2018;98:43–53. doi: 10.1016/j.biocel.2018.02.015. [DOI] [PubMed] [Google Scholar]
- 27.Eltzschig H.K., Eckle T. Ischemia and reperfusion--from mechanism to translation. Nat. Med. 2011;17:1391–1401. doi: 10.1038/nm.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orekhov A.N., Oishi Y., Nikiforov N.G., Zhelankin A.V., Dubrovsky L., Sobenin I.A., Kel A., Stelmashenko D., Makeev V.J., Foxx K., Jin X., Kruth H.S., Bukrinsky M. Modified ldl particles activate inflammatory pathways in monocyte-derived macrophages: transcriptome analysis. Curr. Pharmaceut. Des. 2018;24:3143–3151. doi: 10.2174/1381612824666180911120039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cargnello M., Roux P.P. Activation and function of the mapks and their substrates, the mapk-activated protein kinases. Microbiol. Mol. Biol. Rev. 2011;75:50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen J., Zhang Y., Yu H., Shen B., Liang Y., Jin R., Liu X., Shi L., Cai X. Role of dusp1/mkp1 in tumorigenesis, tumor progression and therapy. Cancer Med. 2016;5:2061–2068. doi: 10.1002/cam4.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin Q., Li R., Hu N., Xin T., Zhu P., Hu S., Ma S., Zhu H., Ren J., Zhou H. Dusp1 alleviates cardiac ischemia/reperfusion injury by suppressing the mff-required mitochondrial fission and bnip3-related mitophagy via the jnk pathways. Redox Biol. 2018;14:576–587. doi: 10.1016/j.redox.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan J., Shen J., Zhu H., Gong Y., Zhu H., Li J., Lin S., Wu G., Sun T. Mir-378a-3p inhibits ischemia/reperfusion-induced apoptosis in h9c2 cardiomyocytes by targeting trim55 via the dusp1-jnk1/2 signaling pathway. Aging (Albany NY) 2020;12:8939–8952. doi: 10.18632/aging.103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong D., Wang H., Liu M., Li X., Huang M., Zhou H., Lin S., Lin Z., Yang B. Ganoderma lucidum polysaccharide peptide prevents renal ischemia reperfusion injury via counteracting oxidative stress. Sci. Rep. 2015;5 doi: 10.1038/srep16910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu D., Shang H., Liu Y. Stanniocalcin-1 protects a mouse model from renal ischemia-reperfusion injury by affecting ros-mediated multiple signaling pathways. Int. J. Mol. Sci. 2016;17 doi: 10.3390/ijms17071051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milde-Langosch K. The fos family of transcription factors and their role in tumourigenesis. Eur. J. Cancer. 2005;41:2449–2461. doi: 10.1016/j.ejca.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 36.Bao Y., Qiao Y., Yu H., Zhang Z., Yang H., Xin X., Chen Y., Guo Y., Wu N., Jia D. Mirna-27a transcription activated by c-fos regulates myocardial ischemia-reperfusion injury by targeting atad3a. Oxid. Med. Cell. Longev. 2021;2021 doi: 10.1155/2021/2514947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meng Q., Ye C., Lu Y. Mir-181c regulates ischemia/reperfusion injury-induced neuronal cell death by regulating c-fos signaling. Pharmazie. 2020;75:90–93. doi: 10.1691/ph.2020.9856. [DOI] [PubMed] [Google Scholar]
- 38.Tejchman K., Nowacki A., Kotfis K., Skwirczynska E., Kotowski M., Zair L., Ostrowski M., Sienko J. The role of endothelins, il-18, and ngal in kidney hypothermic machine perfusion. Biomedicines. 2021;9 doi: 10.3390/biomedicines9040417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chareandee C., Herman W.H., Hricik D.E., Simonson M.S. Elevated endothelin-1 in tubular epithelium is associated with renal allograft rejection. Am. J. Kidney Dis. 2000;36:541–549. doi: 10.1053/ajkd.2000.9795. [DOI] [PubMed] [Google Scholar]
- 40.You R., Heyang Z., Ma Y., Xia P., Zheng H., Lin J., Ji P., Chen L. Identification of biomarkers, immune infiltration landscape, and treatment targets of ischemia-reperfusion acute kidney injury at an early stage by bioinformatics methods. Hereditas. 2022;159:24. doi: 10.1186/s41065-022-00236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He S., He L., Yan F., Li J., Liao X., Ling M., Jing R., Pan L. Identification of hub genes associated with acute kidney injury induced by renal ischemia-reperfusion injury in mice. Front. Physiol. 2022;13 doi: 10.3389/fphys.2022.951855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meng Q., Xia Y., jun C. At the crossroad of the signaling network. Protein Cell. 2011;2:889–898. doi: 10.1007/s13238-011-1113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin G., Yu B., Liang Z., Li L., Qu S., Chen K., Zhou L., Lu Q., Sun Y., Zhu X. Silencing of c-jun decreases cell migration, invasion, and emt in radioresistant human nasopharyngeal carcinoma cell line cne-2r. OncoTargets Ther. 2018;11:3805–3815. doi: 10.2147/OTT.S162700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papoudou-Bai A., Hatzimichael E., Barbouti A., Kanavaros P. Expression patterns of the activator protein-1 (ap-1) family members in lymphoid neoplasms. Clin. Exp. Med. 2017;17:291–304. doi: 10.1007/s10238-016-0436-z. [DOI] [PubMed] [Google Scholar]
- 45.Checa J., Aran J.M. Reactive oxygen species: drivers of physiological and pathological processes. J. Inflamm. Res. 2020;13:1057–1073. doi: 10.2147/JIR.S275595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sankpal N.V., Mayfield J.D., Willman M.W., Fleming T.P., Gillanders W.E. Activator protein 1 (ap-1) contributes to epcam-dependent breast cancer invasion. Breast Cancer Res. 2011;13:R124. doi: 10.1186/bcr3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shaulian E., Karin M. Ap-1 as a regulator of cell life and death. Nat. Cell Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 48.Shen W.C., Liang C.J., Huang T.M., Liu C.W., Wang S.H., Young G.H., Tsai J.S., Tseng Y.C., Peng Y.S., Wu V.C., Chen Y.L. Indoxyl sulfate enhances il-1beta-induced e-selectin expression in endothelial cells in acute kidney injury by the ros/mapks/nfkappab/ap-1 pathway. Arch. Toxicol. 2016;90:2779–2792. doi: 10.1007/s00204-015-1652-0. [DOI] [PubMed] [Google Scholar]
- 49.Tan J., Liu D., Lv X., Wang L., Zhao C., Che Y., Xie Q., Cui X. Mapk mediates inflammatory response and cell death in rat pulmonary microvascular endothelial cells in an ischemia-reperfusion model of lung transplantation. J. Heart Lung Transplant. 2013;32:823–831. doi: 10.1016/j.healun.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 50.Reznik A., Plotnikova O., Skvortsov A., Skoblov M., Reznik O., Baranova A. Reperfusion activates ap-1 and heat shock response in donor kidney parenchyma after warm ischemia. Biomed Res Int 2018. 2018 doi: 10.1155/2018/5717913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gerhardt L., Liu J., Koppitch K., Cippa P.E., Mcmahon A.P. Single-nuclear transcriptomics reveals diversity of proximal tubule cell states in a dynamic response to acute kidney injury. Proc. Natl. Acad. Sci. U.S.A. 2021;118 doi: 10.1073/pnas.2026684118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao H., Alam A., Soo A.P., George A., Ma D. Ischemia-reperfusion injury reduces long term renal graft survival: mechanism and beyond. EBioMedicine. 2018;28:31–42. doi: 10.1016/j.ebiom.2018.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Franzin R., Stasi A., Fiorentino M., Stallone G., Cantaluppi V., Gesualdo L., Castellano G. Inflammaging and complement system: a link between acute kidney injury and chronic graft damage. Front. Immunol. 2020;11:734. doi: 10.3389/fimmu.2020.00734. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The gene expression data of patients were downloaded from GEO (https://www.ncbi.nlm.nih.gov/geo).