Abstract
With the increasing development of artificial intelligence, large language models (LLMs) have been utilized to solve problems in natural language processing tasks. More recently, LLMs have shown unique potential in numerous applications within medicine but have been particularly investigated for their ability in clinical reasoning. Although the diagnostic accuracy of LLMs in forming differential diagnoses has been reviewed in general internal medicine applications, much is unknown in autoinflammatory disorders. From the nature of autoinflammatory diseases, forming a differential diagnosis is challenging due to the overlapping symptoms between disorders and even more difficult without genetic screening. In this work, the diagnostic accuracy of the Generative Pre-Trained Transformer Model-4 (GPT-4), GPT-3.5, and Large Language Model Meta AI (LLaMa) were evaluated in clinical vignettes of Deficiency of Interleukin-1 Receptor Antagonist (DIRA) and Familial Mediterranean Fever (FMF). We then compared these models to a control group including one internal medicine physician. It was found that GPT-4 did not significantly differ in correctly identifying DIRA and FMF patients compared to the internist. However, the physician maintained a significantly higher accuracy than GPT-3.5 and LLaMa 2 for either disease. Overall, we explore and discuss the unique potential of LLMs in diagnostics for autoimmune diseases.
Keywords: DIRA, Deficiency of Interleukin-1 receptor antagonist, Familial Mediterranean fever, FMF, Artificial intelligence
Highlights
-
•
Artificial intelligence models have potential as an alternative diagnostic tool for identifying autoinflammatory diseases.
-
•
GPT-4's ability did not significantly differ compared to an internist in identifying FMF.
-
•
LLaMa 2 and GPT-3.5 had a lower accuracy in identifying DIRA compared to that of an internist.
-
•
GPT-4 and GPT-3.5 had an accuracy above 50 % for FMF and 20 % for DIRA, respectively.
List of abbreviations:
- AI
Artificial Intelligence
- DIRA
Deficiency of Interleukin-1 Receptor Antagonist
- FMF
Familial Mediterranean Fever
- GPT-4
Generative Pre-Trained Transformer-4
- GPT-3
Generative Pre-Trained Transformer-3
- LLaMa
Large Language Model Meta AI
- LLM
Large Language Model
1. Introduction
The rapid development of artificial intelligence (AI) has led to the creation of large language models (LLMs). The foundations of LLMs are based on deep learning architectures trained on large datasets involving articles, websites, and other forms of text. This has led to the formation of commercial models such as OpenAI's Generative Pre-Trained Transformer-3 (GPT-3) and Meta's Large Language Model Meta AI (LLaMa) to solve natural language processing tasks [1,2]. Within medicine, these models have been thoroughly investigated in numerous applications. For instance, they have helped automating electronic health records and assisting in systematic reviews [3,4]. More recently, though, LLMs have been thoroughly investigated for complex diagnostic reasoning.
Given the recent success of LLMs in scoring near or above the passing threshold of the United States Medical Licensing Exam [5], current studies have evaluated the accuracy of these models in forming a differential diagnosis [[6], [7], [8]]. In a previously reported study by Kottlors et al. [6], GPT-4 scored acceptable to clinical professionals in imaging patterns (68.8 %–93.8 %, respectively). In another investigation by Kanjee et al. [7], authors found that GPT-4 was able to identify a correct diagnosis in 64 % of cases and 39 % as its primary diagnosis.
However, even with the thorough testing of LLMs in determining their diagnostic accuracy, much is still unknown of their abilities in challenging cases. In particular, autoinflammatory disorders are complex cases for their nature of overlapping symptoms such as systemic inflammation, increased acute phase reactants, acute arthritis, and recurrent fevers [9]. Additionally, without thorough genetic screening, forming a diagnosis of these diseases is even more difficult. In this work, we assess the accuracy of GPT-4, compared with GPT-3.5 and LLaMa 2, in forming a diagnosis for the Deficiency of Interleukin-1 Receptor Antagonist (DIRA) and Familial Mediterranean Fever (FMF).
DIRA is an ultra-rare autoinflammatory disorder that is caused by an autosomal recessive mutation in the IL1RN gene and therefore cannot encode IL-1Ra proteins [9]. This leads to abnormal activity of IL-1β and IL-1ɑ and causes an inflammatory state [9]. The primary symptoms of DIRA include systemic inflammation, multifocal osteomyelitis, and elevated acute phase reactants [10].
In comparison, FMF is an autoinflammatory disorder caused by an autosomal recessive mutation in the MEFV gene, resulting in lower production of pyrin [11]. This eventually causes an increase in IL-1β and forms an inflammatory response. The common symptoms of FMF are recurrent episodes of fever, seriotitis, abdominal pain, chest pain, and skin lesions [11].
Within this study, we prepared clinical vignettes of DIRA and FMF patients obtained from the PubMed search engine. Then, we evaluated and recorded the results from GPT-4, GPT-3.5, LLaMa 2, and compared them to an American Board Certified Internal Medicine Physician. Subsequently, we analyzed and discussed the scope of the results.
2. Materials and methods
2.1. Study design
Diagnostic accuracies were assessed of GPT-3.5 (OpenAI, San Francisco, CA, USA), GPT-4 (OpenAI, San Francisco, CA, USA), and LLaMa 2 (Meta AI, Menlo Park, CA, USA) models. This study was conducted at the California University of Science and Medicine, School of Medicine, Department of Medical Education, Colton, California, USA. A blind test was conducted on a certified internist to evaluate the accuracy compared to AI models. The clinical vignettes of DIRA and FMF were obtained from published manuscripts, and therefore did not require individual consent or approval from an Institutional Review Board.
GPT-3.5 (chat.openai.com) was accessed on August 11, 2023, GPT-4 (chat.openai.com) on August 12, 2023, and LLaMa 2 (labs.perplexity.ai, 70 b model) on August 13, 2023. For all models, there were no hyperparameters specifically tuned for clinical diagnosis.
2.2. Clinical vignettes
All case vignettes were obtained from the PubMed database. We used the following key-word index: “Familial Mediterranean Fever” or “FMF” and “Deficiency of Interleukin-1 Receptor Antagonist” or “DIRA.” Language restrictions were applied to only English. Gray literature was not included in the search, and all article types included were case reports. We obtained a total of twenty written vignettes for both DIRA [[12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23]] and FMF [[24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42]], respectively (n = 40).
For all written clinical vignettes, the format followed the history of present illness including all of the following information: Age, Sex, Clinical Presentation, Related Family and Patient History, Physical Examination, and any Laboratory Results. Each vignette had only one correct diagnosis. However, to determine the accuracy of models, we allowed up to 10 differential diagnoses and sorted them into 3 categories: primary choice (top choice), top five, and top ten. All 40 patient vignettes have been provided in Supplementary Material 1. We then analyzed each element, including age, sex, chief complaint, consanguinity, and laboratory results.
2.3. Data extraction
Due to the potential of LLMs to recall previous information within the same conversation, each vignette was shown in differing conversations. Additionally, the order of vignettes displayed were randomized through the covariate adaptive randomization technique through GraphPad Prism (version 10.0.2) [43].
Before extraction, J.P·P typed the following text into all models: “I am conducting an experiment on the accuracy of differential diagnoses in clinical cases to assess how you compare with other models. I am going to list a clinical case below, and want you to give me your top ten diagnoses. Do not list any explanations. Please only list ten diagnoses. Here is the case below: (clinical vignette).” The models then listed their ten diagnoses and the data was recorded. For the control group, the internist was given the same randomized vignettes tested on the AI models. The reference section was omitted for both groups.
2.4. Statistical analyses
Data was collected using Excel and statistical analysis was completed on GraphPad Prism (version 10.0.2). Due to the moderate sample size of each group, a Fisher's Exact test was conducted on categorical data. For continuous datasets, a Mann Whitney U Test was conducted to evaluate significance. Unless stated otherwise, a P-value below 0.05 was considered statistically significant.
3. Results
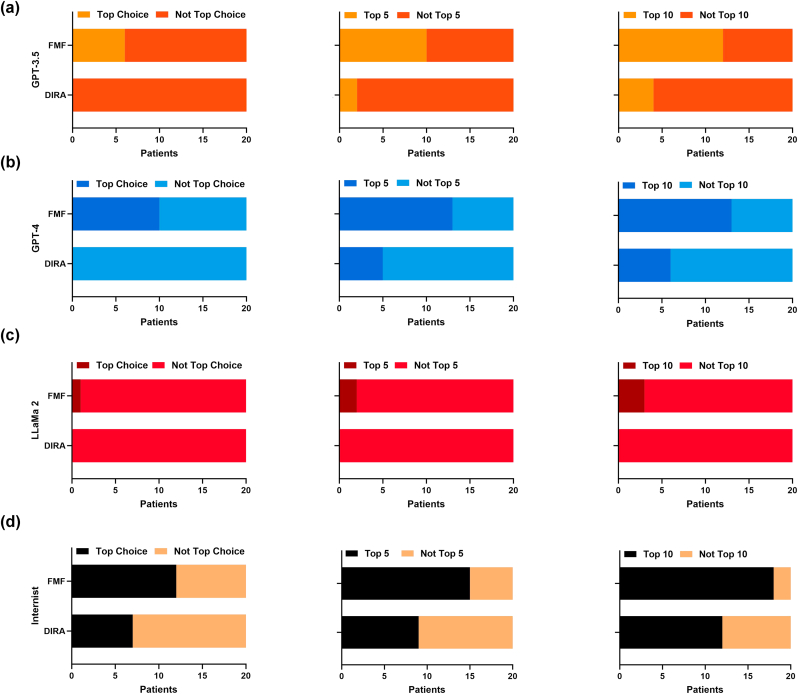
Within this pilot study, we report 40 clinical vignettes tested on GPT-4, GPT-3.5, and LLaMa 2 compared to an internal medicine physician. The results of each model's diagnostic accuracy has been displayed in Fig. 1.
Fig. 1.
(a) Diagnostic accuracy of GPT-3.5 at its primary choice (top choice), top five, and top ten diagnoses. (b) Diagnostic accuracy of GPT-4 at its primary choice (top choice), top five, and top ten diagnoses. (c) Diagnostic accuracy of LLaMa 2 at its primary choice (top choice), top five, and top ten diagnoses. (d) Diagnostic accuracy of the internist at its primary choice (top choice), top five, and top ten diagnoses.
Overall, all models had a higher accuracy identifying FMF compared to DIRA. The internist identified 90 %, GPT-4 identified 65 %, GPT-3.5 identified 60 %, and LLaMa 2 identified 17.6 % of FMF patients. Conversely, the internist identified 60 %, GPT-4 identified 30 %, GPT-3.5 identified 25 %, and LLaMa 2 identified 0 % of DIRA patients.
The internist performed significantly higher than GPT-4 in its primary prediction (35 % vs. 0 %, P = 0.0083), but did not differ significantly at its top five (45 % vs. 25 %, P = 0.3203, ns) and top ten prediction (60 % vs. 30 %, P = 0.1110, ns) for DIRA. In comparison, the two did not significantly differ at their primary (60 % vs. 50 %, P = 0.7512, ns), top five (75 % vs. 65 %, P = 0.7311, ns), and top ten prediction (90 % vs. 65 %, P = 0.1274, ns) for FMF.
Compared to GPT-3.5, the internist performed significantly higher at its primary (35 % vs. 0 %, P = 0.0083), top five (45 % vs. 10 %, P = 0.0310), and top ten prediction (60 % vs. 16.67 %, P = 0.0046) for DIRA. For FMF, both did not differ significantly at its primary (60 % vs. 30 %, P = 0.1110, ns), top five (75 % vs. 50 %, P = 0.1908, ns), and top ten prediction (90 % vs. 60 %, P = 0.0648, ns).
The internist also performed significantly higher than LLaMa 2 at its primary (35 % vs. 0 %, P = 0.0083), top five (45 % vs. 0 %, P = 0.0012), and top ten prediction (60 % vs. 0 %, P < 0.0001) for DIRA. Compared to FMF, the internist also performed higher than LLaMa 2 at its primary (60 % vs. 0 %, P = 0004), top five (75 % vs. 10 %, P < 0.0001), and top ten prediction (90 % vs. 15 %, P < 0.0001).
When comparing GPT-4 to the GPT-3.5 model, there were no significant differences in accuracy in their primary prediction (50 % vs. 30 %, P = 0.3332, ns), top five (65 % vs. 50 %, P = 0.5231, ns), and top ten prediction (65 % vs. 60 %, P = >0.9999, ns) for FMF patients. For DIRA patients, there were no significant differences between the models in their primary (0 % vs. 0 %, P = >0.9999, ns), top five (25 % vs. 10 %, P = 0.4075, ns), and top ten prediction (30 % vs. 16.67 %, P = 0.4716, ns).
Moreover, comparing GPT-4 to LLaMa 2, there was a significant difference at its primary (50 % vs. 5 %, P = 0.0033), top five (65 % vs. 10 %, P = 0.0008), and top ten prediction (65 % vs. 15 %, P = 0.0031) for FMF. For DIRA, it was not significant at its primary prediction (0 % vs. 0 %, P = >0.9999, ns). However, it was significant at its top five prediction (25 % vs. 0 %, P = 0.0471) and top ten prediction (30 % vs. 0 %, P = 0.0202).
Additionally, GPT-3.5 and LLaMa 2 did not differ significantly at its primary prediction (30 % vs. 5 %, P = 0.0915), but was significant at its top five (50 % vs. 10 %, P = 0.0138), and its top ten prediction (60 % vs. 15 %, P = 0.0079) for FMF. For DIRA, both models did not differ significantly at its primary (0 % vs. 0 %, P = >0.9999, ns), top five (10 % vs. 0 %, P = 0.4872, ns), and top ten prediction (16.67 % vs. 0 %, P = 0.1140, ns).
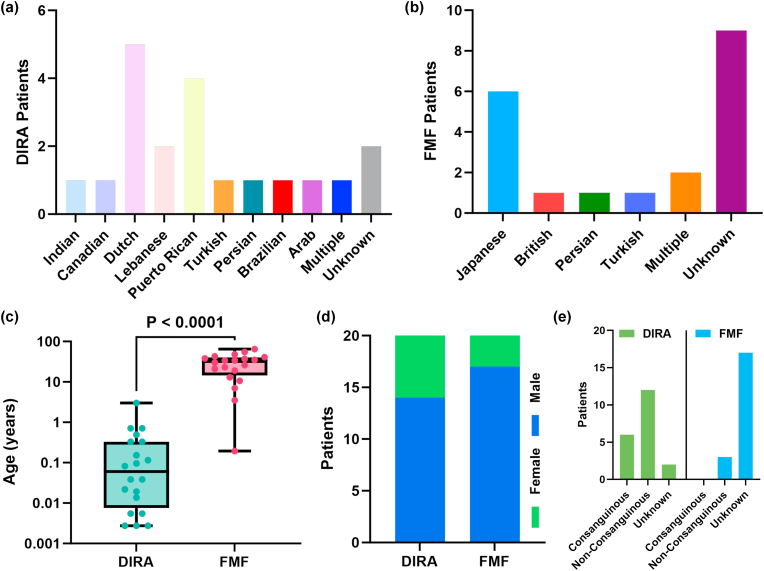
All elements involved in the clinical vignette were statistically compared between both DIRA and FMF patients shown in Fig. 2. All values have been presented as the mean ± standard error of the mean. DIRA patients were 0.3084 ± 0.1506 years old, which was significantly younger compared to FMF patients that were 29.16 ± 3.863 years (P < 0.0001) shown in Fig. 2c. For ethnicity, DIRA patients were much more diverse (10 total) compared to FMF (5 total), shown in Fig. 2a–b. For sex, there was no significant difference between both disease groups (P = 0.4506, ns). Although the majority of FMF vignettes did not have a reported consanguinity of patients, the majority of DIRA patients were non-consanguineous.
Fig. 2.
(a) Ethnicities of DIRA patients. (b) Ethnicities of FMF patients. (c) Age of DIRA patients vs. Age of FMF patients. (d) Sex of DIRA and FMF patients. (e) Consanguinity of DIRA and FMF patients.
From the ultra-rarity of DIRA, there are no guidelines or official criteria for diagnosis. However, there is an official criteria for FMF reported by Livneh et al. [44]. We have listed all of the major and minor criterias from the clinical presentation of all vignettes in Table 1 and Table 2, respectively.
Table 1.
Major criteria or typical attacks from FMF in all clinical vignettes. Simplified criteria model obtained from Bashardoust et al. [47].
| Major Criteria | n = 20 | Percentage (%) |
|---|---|---|
| Generalized Peronstitis | 0 | 0 |
| Pericarditis or Unilateral Pleuritis | 2 | 10 |
| Fever Alone | 19 | 95 |
| Incomplete Abdominal Attack | 16 | 80 |
| Monoarthritis (hip, knee, ankle) | 1 | 5 |
Table 2.
Minor criteria or incomplete attacks from FMF in all clinical vignettes. Simplified criteria model obtained from Bashardoust et al. [47].
| Minor Criteria | n = 20 | Percentage (%) |
|---|---|---|
| Chest | 6 | 30 |
| Joint | 4 | 20 |
| Favorable Response to Colchicine | 1 | 5 |
| Exertional Leg Pain | 0 | 0 |
4. Discussion
In this study, we evaluated the diagnostic accuracy of the GPT-4, GPT-3.5, and LLaMa 2 models in 40 clinical vignettes of DIRA and FMF compared to an internal medicine physician.
Given the overall results of all cases, there were no AI models that predicted DIRA as its primary prediction. All of the correct predictions were either in the top five or top ten diagnoses. The symptoms of DIRA suggest similarities to Chronic Recurrent Multifocal Osteomyelitis and Neonatal-Onset Multisystem Inflammatory Disease. Given that DIRA is a monogenic form of CRMO, the overlapping similarities with DIRA may be leading to a lower certainty of DIRA within AI models [45]. At the same time, NOMID has similar symptoms to DIRA such as systemic inflammation, skin rashes, and joint pain, which could also be leading to incorrect diagnoses by the model [46]. Without thorough genetic screening, it would be a difficult task for LLMs to differentiate rare autoinflammatory disorders such as DIRA from other similar disorders.
For FMF, the major elements of clinical presentation were abdominal pain and fever and the minor elements were chest and joint pain. These unique symptoms assisted in achieving a higher diagnostic accuracy of all FMF patients compared to DIRA patients for GPT-4 and GPT-3.5. Even though GPT-4 had a lower accuracy than the internist, the difference remained not significant. With additional training data and tuning of hyperparameters adjusted to the unique elements of FMF, AI models could potentially have higher accuracy in diagnosis.
However, this study has multiple limitations. Due to the rarity of DIRA, there are a low number of reported research articles and case reports for it. This reduced the sample size of the study to 20 vignettes per each disease. This study demonstrated the capability of AI models in challenging rheumatological diseases without any genetic screening. A higher sample of AI models and physicians will be investigated in the future.
To the best of our knowledge, this is the first study that evaluates the diagnostic accuracy of LLMs in autoinflammatory disorders, specifically DIRA and FMF. GPT-4 was able to correctly identify up to 65 % of FMF patients, not significantly different from an internist with 90 % identification. For DIRA, GPT-4 was able to correctly identify 30 % of patients, while the internist at 60 % identification. However, the internist performed significantly higher in both diseases compared to GPT-3.5 and LLaMa 2. Given the preliminary metrics of LLMs for their diagnostic accuracy, further testing is required to determine the accuracy of models in more difficult and similar rheumatological diseases.
Funding
Not applicable.
Author Contributions
J.P.P. - Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project Administration, Validation, Visualization, and Writing - original draft. K.P.P. - Project Administration, Supervision, Writing - original draft, and Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Not applicable.
Handling Editor: Dr Y Renaudineau
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jtauto.2023.100213.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- 1.Introducing ChatGPT OpenAI. 2022. https://openai.com/blog/chatgpt
- 2.Introducing LlaMa . Meta AI; 2023. A Foundational, 65-Billion-Parameter Large Language Model.https://ai.meta.com/blog/large-language-model-llama-meta-ai/ [Google Scholar]
- 3.Yang X., Chen A., PourNejatian N., Shin H.C., Smith K.E., Parisien C., Compas C.B., Martin C., Costa A., Flores M.G., Zhang Y., Magoc T., Harle C.A., Lipori G., Mitchell D.A., Hogan W.R., Shenkman L., Bian J., Wu Y. A large language model for electronic health records. Npj Digital Medicine. 2022;5(1) doi: 10.1038/s41746-022-00742-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qureshi R., Shaughnessy D.T., Gill K.a.R., Robinson K.A., Li T., Agai E. Are ChatGPT and large language models “the answer” to bringing us closer to systematic review automation? Syst. Rev. 2023;12(1) doi: 10.1186/s13643-023-02243-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kung T.H., Cheatham M., Medenilla A., Sillos C., De Leon L., Elepaño C., Madriaga M., Aggabao R., Diaz-Candido G., Maningo J., Tseng V. Performance of ChatGPT on USMLE: potential for AI-assisted medical education using large language models. PLOS Digital Health. 2023;2(2) doi: 10.1371/journal.pdig.0000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kottlors J., Bratke G., Rauen P., Kabbasch C., Persigehl T., Schlamann M., Lennartz S. Feasibility of differential diagnosis based on imaging patterns using a large language model. Radiology. 2023;308(1) doi: 10.1148/radiol.231167. [DOI] [PubMed] [Google Scholar]
- 7.Kanjee Z., Crowe B., Rodman A. Accuracy of a generative artificial intelligence model in a complex diagnostic challenge. JAMA. 2023;330(1):78. doi: 10.1001/jama.2023.8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirosawa T., Harada Y., Yokose M., Sakamoto T., Kawamura R., Shimizu T. Diagnostic accuracy of differential-diagnosis lists generated by generative pretrained transformer 3 chatbot for clinical vignettes with common chief complaints: a pilot study. Int. J. Environ. Res. Publ. Health. 2023;20(4):3378. doi: 10.3390/ijerph20043378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galozzi P., Punzi L., Sfriso P. Clinical overlapping in autoinflammatory diseases: the role of gene duplication. Front. Immunol. 2017;8 doi: 10.3389/fimmu.2017.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reddy S.T., Song J., Geoffrey R., Lorier R., Suchi M., Broeckel U., Hessner M.J., Verbsky J.W. An autoinflammatory disease due to homozygous deletion of the IL1RN locus. N. Engl. J. Med. 2009;360(23):2438–2444. doi: 10.1056/nejmoa0809568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Onen F. Familial mediterranean fever. Rheumatol. Int. 2005;26(6):489–496. doi: 10.1007/s00296-005-0074-3. [DOI] [PubMed] [Google Scholar]
- 12.Mendonça L.O., Malle L., Donovan F.X., Chandrasekharappa S.C., Sanchez G.a.M., Garg M., Tedgård U., Castells M., Saini S.S., Dutta S., Goldbach-Mansky R., Suri D., De Jesus A.A. Deficiency of interleukin-1 receptor Antagonist (DIRA): report of the first Indian patient and a novel deletion affecting IL1RN. J. Clin. Immunol. 2017;37(5):445–451. doi: 10.1007/s10875-017-0399-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aksentijevich I., Masters S.L., Ferguson P.J., Dancey P., Frenkel J., Van Royen-Kerkhoff A., Laxer R.M., Tedgård U., Cowen E.W., Pham T., Booty M.G., Estes J.D., Sandler N.G., Plass N., Stone D.L., Turner M.L., Hill S., Butman J.A., Schneider R.…Goldbach-Mansky R. An autoinflammatory disease with deficiency of the interleukin-1–receptor antagonist. N. Engl. J. Med. 2009;360(23):2426–2437. doi: 10.1056/nejmoa0807865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sözeri B., Gerçeker-Türk B., Yıldız-Atıkan B., Mir S., Berdeli A. A novel mutation of interleukin-1 receptor antagonist (il1rn) in a dira patient from Turkey: diagnosis and treatment. Turk. J. Pediatr. 2018;60(5):588. doi: 10.24953/turkjped.2018.05.020. [DOI] [PubMed] [Google Scholar]
- 15.Kuemmerle-Deschner J., Welzel T., Hoertnagel K., Tsiflikas I., Hospach A., Liu X., Schlipf S., Hansmann S., Samba S.D., Griesinger A., Benseler S.M., Weber A.N. New variant in the IL1RN-gene (DIRA) associated with late-onset, CRMO-like presentation. Rheumatology. 2020;59(11):3259–3263. doi: 10.1093/rheumatology/keaa119. [DOI] [PubMed] [Google Scholar]
- 16.Sakran W., Shalev S.A., El-Shanti H., Uziel Y. Chronic recurrent multifocal osteomyelitis and deficiency of interleukin-1–receptor antagonist. Pediatr. Infect. Dis. J. 2013;32(1):94. doi: 10.1097/inf.0b013e3182700cc1. [DOI] [PubMed] [Google Scholar]
- 17.Schnellbacher C., Ciocca G., Menendez R., Aksentijevich I., Goldbach-Mansky R., Duarte A.M., Rivas-Chacon R. Deficiency of interleukin-1 receptor antagonist responsive to anakinra. Pediatr. Dermatol. 2012;30(6):758–760. doi: 10.1111/j.1525-1470.2012.01725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ziaee V., Youssefian L., Faghankhani M., Jazayeri A., Saeidian A.H., Vahidnezhad H., Uitto J. Homozygous IL1RN mutation in siblings with deficiency of interleukin-1 receptor antagonist (DIRA) J. Clin. Immunol. 2020;40(4):637–642. doi: 10.1007/s10875-020-00767-w. [DOI] [PubMed] [Google Scholar]
- 19.Brau-Javier C.N., Gonzales-Chavez J., Toro J.R. Chronic cutaneous pustulosis due to a 175-kb deletion on chromosome 2q13. Arch. Dermatol. 2012;148(3):301. doi: 10.1001/archdermatol.2011.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thacker P.G., Binkovitz L.A., Thomas K.B. Deficiency of interleukin-1-receptor antagonist syndrome: a rare auto-inflammatory condition that mimics multiple classic radiographic findings. Pediatr. Radiol. 2011;42(4):495–498. doi: 10.1007/s00247-011-2208-y. [DOI] [PubMed] [Google Scholar]
- 21.Mendonça L.O., Grossi A., Caroli F., De Oliveira R.A., Kalil J., Castro F.F.M., Pontillo A., Ceccherini I., De Barros M.M.S.B., Gattorno M. A case report of a novel compound heterozygous mutation in a Brazilian patient with deficiency of Interleukin-1 receptor antagonist (DIRA) Pediatr. Rheumatol. 2020;18(1) doi: 10.1186/s12969-020-00454-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abdwani R., Abdalla E., Masilhi B.A., Shalaby A., Al-Maawali A. Novel mutation in interleukin 1 receptor antagonist associated with chronic diarrhoea in infancy. J. Paediatr. Child Health. 2022;58(1):186–188. doi: 10.1111/jpc.15440. [DOI] [PubMed] [Google Scholar]
- 23.Minkis K., Aksentijevich I., Goldbach-Mansky R., Magro C.M., Scott R.A., Davis J.G., Sardana N., Herzog R. Interleukin 1 receptor antagonist deficiency presenting as infantile pustulosis mimicking infantile pustular psoriasis. Arch. Dermatol. 2012;148(6) doi: 10.1001/archdermatol.2011.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hotta Y., Kawasaki T., Kotani T., Okada H., Ikeda K., Yamane S., Yamada N., Sekoguchi S., Isozaki Y., Nagao Y., Murotani M., Oyamada H. Familial mediterranean fever without fever. Intern. Med. 2020;59(10):1267–1270. doi: 10.2169/internalmedicine.3175-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cliff-Patel N., Yusuf B., Hamdani S., Ziauddin V. Familial Mediterranean fever: a differential diagnosis for the surgical abdomen. JRSM Open. 2022;13(9) doi: 10.1177/20542704221123433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ariga H., Kunisaki R., Ojima T., Suzuki S., Okada K., Kashimura J. Familial Mediterranean fever with colonic lesions: a case report. DEN Open. 2023;4(1) doi: 10.1002/deo2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kübra A., Sarici S.U., Kolukisa G., Altun D. Familial mediterranean fever with neonatal onset: case report. Case Reports in Pediatrics. 2020:1–3. doi: 10.1155/2020/6649525. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maggio M.C., Castiglia M.S., Corsello G. Familial Mediterranean Fever: an unusual cause of liver disease. Ital. J. Pediatr. 2019;45(1) doi: 10.1186/s13052-019-0712-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kasamaki K., Kusano C., Ikehara H., Suzuki S., Esaki M., Irie A., Hayashi K., Okuno H., Moriyama M., Gotoda T. Familial mediterranean fever with small bowel stenosis. Internal Medicine. 2019 doi: 10.2169/internalmedicine.2293-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans N., Ray J., Prather C.M. Atypical Autosomal-Dominant inheritance of familial Mediterranean fever. ACG Case Reports J. 2021;8(3) doi: 10.14309/crj.0000000000000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsumoto S., Urayoshi S., Yoshida Y. Familial Mediterranean fever in which Crohn's disease was suspected: a case report. BMC Res. Notes. 2014;7(1) doi: 10.1186/1756-0500-7-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi M., Ebe T., Kohara T., Inagaki M., Isonuma H., Hibiya I., Mori T., Watanabe K., Ikemoto H. Periodic fever compatible with familial mediterranean fever. Intern. Med. 1992;31(7):893–898. doi: 10.2169/internalmedicine.31.893. [DOI] [PubMed] [Google Scholar]
- 33.Malik A., Malik J., Javaid M., Khan H.S., Mohsin M., Shoaib M. A case of aortic dissection in familial Mediterranean fever. J. Cardiol. Cases. 2021;24(6):296–299. doi: 10.1016/j.jccase.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashida K., Terada E., Nagayama A., Sakamoto S., Hasuzawa N., Nomura M. Necessity of utilizing physiological glucocorticoids for managing familial Mediterranean fever. Am. J. Case Reports. 2020 doi: 10.12659/ajcr.920983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toshida M., Konishi Y., Ikenouchi A., Okamoto N., Yoshimura R. Colchicine-Resistant Familial Mediterranean Fever with Depressive State Successfully Treated with Escitalopram. Cureus. 2021 doi: 10.7759/cureus.15145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malek A., Zeraati T., Sadr-Nabavi A., Vakili N., Abbaszadegan M.R. Cardiac tamponade: a rare manifestation of familial Mediterranean fever. Case Reports in Rheumatology. 2022:1–5. doi: 10.1155/2022/8334375. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hosoi T., Ishii K., Tozaka N., Kishida D., Sekijima Y., Tamaoka A. Familial Mediterranean fever is important in the differential diagnosis of recurrent aseptic meningitis in Japan. Intern. Med. 2020;59(1):125–128. doi: 10.2169/internalmedicine.3432-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Darwish W.M., Darwish S., Darwish M., Darwish B.F. M680I/M694V heterozygous mutation in early onset familial Mediterranean fever. J. Med. Cases. 2021;12(9):351–354. doi: 10.14740/jmc3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Atikel Y.Ö., Derinkuyu B.E., Bakkaloglu S.A. Unusual presentation of familial Mediterranean fever with co‐existing polyarteritis nodosa and acute post‐streptococcal glomerulonephritis. Clinical Case Reports. 2022;10(7) doi: 10.1002/ccr3.6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asakura K., Yanai S., Nakamura S., Kawaski K., Eizuka M., Ishida K., Endo M., Sugai T., Migita K., Matsumoto T. Familial Mediterranean fever mimicking Crohn disease. Medicine. 2018;97(1):e9547. doi: 10.1097/md.0000000000009547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Erten Ş., Erzurum C., Altunoglu A. Three family members with familial mediterranean fever carrying the M694V mutation showed different clinical presentations. Intern. Med. 2012;51(13):1765–1768. doi: 10.2169/internalmedicine.51.7537. [DOI] [PubMed] [Google Scholar]
- 42.Iwata K., Toma T., Yachie A. Atypical familial mediterranean fever presenting with recurrent migratory polyarthritis. Intern. Med. 2019 doi: 10.2169/internalmedicine.3001-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin Y., Zhu M., Su Z. The pursuit of balance: an overview of covariate-adaptive randomization techniques in clinical trials. Contemp. Clin. Trials. 2015;45:21–25. doi: 10.1016/j.cct.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 44.Livneh A., Langevitz P., Zemer D., Zaks N., Kees S., Lidar T., Migdal A., Padeh S., Pras M. Criteria for the diagnosis of familial mediterranean fever. Arthritis Rheum. 1997;40(10):1879–1885. doi: 10.1002/art.1780401023. [DOI] [PubMed] [Google Scholar]
- 45.Rao A.P., Mallya P.P., Ranjani S., Raghuram J. Chronic recurrent multifocal osteomyelitis - a case series from India. Indian J. Orthop. 2018;52(6):672–677. doi: 10.4103/ortho.ijortho_464_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goldbach-Mansky R. Current status of understanding the pathogenesis and management of patients with NOMID/CINCA. Curr. Rheumatol. Rep. 2011;13(2):123–131. doi: 10.1007/s11926-011-0165-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bashardoust B. Familial Mediterranean fever; diagnosis, treatment, and complications. PubMed. 2015;4(1):5–8. https://pubmed.ncbi.nlm.nih.gov/28197466 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.