Abstract
Only one out of four mammalian arrestin subtypes, arrestin-3, facilitates the activation of JNK family kinases. Here we describe two different protocols used for elucidating the mechanisms involved. One is based on reconstitution of signaling modules from purified proteins: arrestin-3, MKK4, MKK7, JNK1, JNK2, and JNK3. The main advantage of this method is that it unambiguously establishes which effects are direct because only intended purified proteins are present in these assays. The key drawback is that the upstream-most kinases of these cascades, ASK1 or other MAPKKKs, are not available in purified form, limiting reconstitution to incomplete two-kinase modules. The other approach is used for analyzing the effects of arrestin-3 on JNK activation in intact cells. In this case, signaling modules include ASK1 and/or other MAPKKKs. However, as every cell expresses thousands of different proteins their possible effects on the readout cannot be excluded. Nonetheless, the combination of in vitro reconstitution from purified proteins and cell-based assays makes it possible to elucidate the mechanisms of arrestin-3-dependent activation of JNK family kinases.
Keywords: c-Jun N-terminal kinase (JNK), scaffold, activation, arrestin, biphasic dependence
INTRODUCTION
Mitogen-activated protein kinases (MAPKs) are expressed in virtually all eukaryotes, and the principles of the organization of MAP cascades are conserved from yeast to mammals (Widmann et al., 1999). MAPK signaling modules consist of at least three kinases, MAP kinase kinase kinase (MAP3K), MAP kinase kinase (MAP2K), and effector MAP kinase, that sequentially phosphorylate and activate each other (Widmann et al., 1999). Mammals express 21 MAP3Ks, 7 MAP2Ks, and 11 MAPKs (Johnson, 2011). All kinases demonstrate substrate specificity, which is further enhanced by scaffold proteins that bind particular kinases to organize specific signaling modules and to localize them to appropriate cellular compartments (Burack and Shaw, 2000; Dhanasekaran et al., 2007). Considering the fairly high protein phosphatase activity in the cytoplasm, activation of effector MAP kinases would be rarely achieved without scaffolds to bring into close proximity all kinases of a particular three-tiered activation cascade.
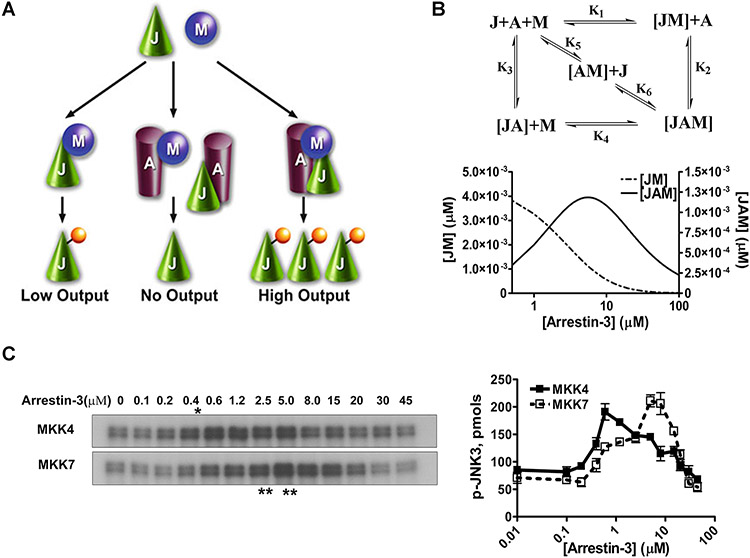
In the case of simple scaffolds that do not activate kinases or their substrates, but act by bringing them into proximity, optimal signaling is achieved at a certain level of expression, whereas both lower and higher scaffold levels result in lower signal output (Levchenko et al., 2000; Levchenko et al., 2004). Thus, the dependence of signaling on scaffold concentration is biphasic or bell-shaped: when the scaffold level is low, its amount is insufficient to organize all kinases into signaling modules, whereas when the concentration of scaffold exceeds optimal, it promotes the formation of incomplete and therefore unproductive complexes containing only a single kinase ( Fig. 2.12.1A ).
FIGURE 2.12.1.
Arrestin-3-mediated JNK3α2 activation by MKK4/7. (A) A three-state model showing the scaffolding mechanism of the two-kinase signaling module. A, J, and M designate arrestin-3, JNK3α2, and upstream kinases MKK4/7, respectively. Kinases can exist in three states: (a) interacting in solution, (b) bound to the scaffold to form incomplete complexes containing a single kinase, and (c) simultaneously assembled by arrestin-3 to form a complete signaling complex. (B) Six affinity constants (k1 through k6) describe the indicated binding equilibria. Calculated concentrations of JM (dotted line, left y axis) and JAM (solid line, right y axis) complexes at different arrestin-3 concentrations (KinTek Explorer 3.0; all six Kd values were set at 5 μM). (C) Representative autoradiograms showing JNK3α2 phosphorylated by MKK4 (upper panel) or MKK7 (lower panel) at the indicated concentration of arrestin-3 (10-sec incubation). The optimal arrestin-3 concentrations are indicated (*, for MKK4; **, for MKK7). (D) The effect of arrestin-3 concentration on JNK3α2 phosphorylation by both MKK4 and MKK7 is biphasic. The bands from the gels were excised and the radioactivity measured in a Tri-Carb liquid scintillation counter to quantify the incorporation of [32P]phosphate from [γ-32P]ATP into JNK3α2. Data from (Zhan et al., 2013b).
The activation of MAPKs of c-Jun N-terminal kinase (JNK) family is regulated by several specialized scaffolds, including JIP [JNK-interacting proteins (Willoughby et al., 2003)]. Arrestin-3 (a.k.a. β-arrestin2 or hTHY-ARRX), is one of two vertebrate nonvisual arrestins that act as a scaffold for the activation of JNK3 (McDonald et al., 2000). It was originally suggested that only receptor-bound arrestin-3 is active and that only JNK3, an isoform with limited expression in the nervous system and the heart, but not the ubiquitous JNK1 and JNK2 isoforms, is activated via an arrestin-3-dependent mechanism. The original work also suggested that arrestin-3 directly binds only two kinases in this cascade: the upstream-most MAP3K, ASK1, and the downstream MAPK, JNK3. It was proposed that out of two MAP2Ks necessary to activate JNK3, only MKK4 associates with this complex via its interactions with ASK1 and JNK3 (McDonald et al., 2000). However, full activation of any JNK family kinase requires dual phosphorylation in the activation loop: tyrosine by MKK4 and threonine by MKK7 (Fig. 2.12.2) (Gupta et al., 1996; Lawler et al., 1998).
Figure 2.12.2.
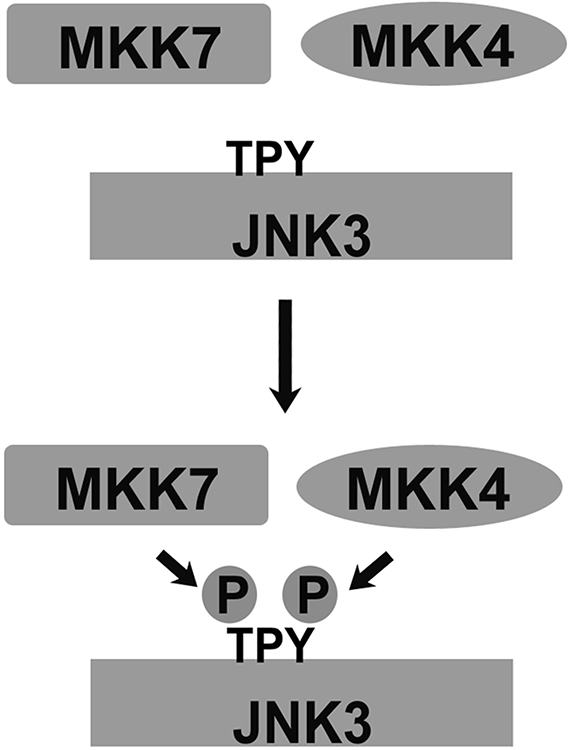
Full activation of JNKs requires dual phosphorylation. For full activity, JNK family kinases must be phosphorylated at Thr (by MKK7) and Tyr (by MKK4). Reprinted with permission from (Zhan et al., 2013a).
Subsequent studies (Miller et al., 2001; Song et al., 2009; Seo et al., 2011; Zhan et al., 2011b; Breitman et al., 2012; Perry-Hauser et al., 2022) revealed that free arrestin-3 facilitates JNK3 activation, that GPCR binding-deficient form of arrestin-3 facilitates JNK3 activation in cells (Song et al., 2009; Breitman et al., 2012), and that arrestin-3 directly binds to MKK4 (Zhan et al., 2011b) and MKK7 (Zhan et al., 2013a). All JNK isoforms are activated via phosphorylation of conserved Tyr and Thr residues by MKK4 and MKK7, respectively (Gupta et al., 1996; Lawler et al., 1998). Thus, the finding that arrestin-3 facilitates activation of JNK1 and JNK2 in vitro and in intact cells (Kook et al., 2014), acting in all cases as a simple scaffold (Kook et al., 2014), was no surprise. Interestingly, arrestin-3 demonstrates different affinities for MKK4 and MKK7 (Zhan et al., 2013a), and for different JNK isoforms (Kook et al., 2014). Arrestin-3 has higher affinity for MKK4 than for MKK7, with JNK3 binding further increasing arrestin-3 affinity for MKK4 while decreasing the affinity for the more abundant MKK7 (Zhan et al., 2013a). In cells this might serve to equalize the chances for these two MKKs to phosphorylate JNK3. Higher affinity for JNK2 than JNK1 (Kook et al., 2014) suggests that arrestin-3 plays a greater role in the regulation of the former of the two ubiquitous JNK subtypes. As the arrestin-3 affinity for the kinases of different pathways determines its optimal concentration for scaffolding particular signaling modules (Zhan et al., 2013a), the arrestin-3 expression level can direct signaling in this group of cascades to certain modules, leading to the activation of some JNK isoforms to a greater extent than others (Kook et al., 2014).
Described in this unit are methods for the purification of MKK4 and MKK7 in active and inactive forms, as well as purification of JNK1, JNK2, and JNK3. Detailed in Basic Protocol 1 are assays wherein individual MKK-JNK signaling modules are reconstituted from purified proteins in the absence and presence of pure arrestin-3. Also discussed are cell-based assays for characterizing arrestin-3-dependent activation of particular JNK isoforms by individual upstream kinases (Basic Protocol 2), and methods for testing whether arrestin-3 functions as a simple scaffold (Basic Protocol 3). Provided in Support Protocols 1, 2, 3, and 4 are methods of expression, purification, and activation of tagged and tag-free MKK4, MKK7, JNK1α1, and JNK2α2, respectively.
The in vitro and cell-based assays described in this unit can be used to identify and test small molecule inhibitors of particular kinase cascades resulting in activation of different JNK isoforms. While the throughput of these assays is too low for large-scale screening, they are useful for lead validation and for defining structure-activity relationships.
BASIC PROTOCOL 1
CONSTRUCTION OF ARRESTIN 3-SCAFFOLDED MKK4/7-JNK1/2/3 SIGNALING MODULES IN VITRO USING PURIFIED PROTEINS
As mammalian cells express thousands of different proteins, any effect of one protein on another in the intact cell might be mediated by intermediaries, the number and nature of which are unknown. Even co-immunoprecipitation of two proteins only shows that they can be present in the same complex, providing no information on whether they interact. Direct protein-protein interactions and their effects on the activity of the proteins involved can only be proven by demonstrating that particular purified proteins bind (this often affects functional properties of one or both partners). Described below are assays developed to examine the ability of purified MKK4 and MKK7 to phosphorylate individual purified JNK isoforms in the absence and presence of pure arrestin-3. The authors have employed both [γ-32P]-ATP (quantified by autoradiography and scintillation counting) and immunoblot (using phospho-JNK antibody) to measure JNK phosphorylation. Described below is the [γ-32P]-ATP measurement in JNK3 assays, while Alternate Protocol 1 provides a method for immunoblot detection of the activation of JNK1/2.
Materials
Purified proteins
Ice
Bradford Assay (Bio-Rad)
Kinase mix (see Table 2.12.1)
Antibodies: phospho-JNK antibody (rabbit, #9251, Cell Signaling Technology); Pan-JNK antibody (rabbit, #9252, Cell Signaling Technology)
Arrestin-3 solutions (see Table 2.12.2)
10 mM ATP (solution in distilled water; Sigma, cat. no. A2383)
ATP, [γ-32P] (PerkinElmer)
Sodium dodecyl sulfate (SDS) buffer (Sigma, cat. no. S3401)
10% SDS-PAGE gel (appendix 3b; Gallagher and Sasse, 2001)
Coomassie blue
Scintillation cocktail (Scintisafe Econo 2, Fisher)
Ultracentrifuge
1.5 ml tubes
Vortex mixer
30°C water bath
Gel dryer (e.g., Bio-Rad)
Autoradiography cassette
Scintillation vials
Scintillation counter
Shaking platform (room temperature)
Additional reagents and equipment for Protein Analysis by SDS-PAGE and Detection by Coomassie Blue or Silver Staining (see Gallagher and Sasse, 2001)
Table 2.12.1.
Preparing the Kinase Mixa
| Volume (each) | Volume (20×) | |
|---|---|---|
| 10× Kinase assay buffer (see recipe) | 2 μl | 40 μl |
| 100 mM DTT | 0.4 μl | 8 μl |
| JNK3α2 (12 μM stock) | 0.8 μl | 16 μl |
| p-MKK4 or p-MKK7 (10 μM stocks) | 0.1 μl | 2 μl |
| Total | 3.3 μl | 66 μl |
Antibodies: phospho-JNK antibody (rabbit, Cell Signaling Technology, cat. no. 9251); total JNK antibody (rabbit, Cell Signaling Technology, cat. no. 9252).
Table 2.12.2.
Preparing Arrestin-3 Solutions
| Tube Number | A1 | A2 | A3 | A4 | A5 | A6 | A7 | A8 | A9 | A10 | A11 | A12 | A13 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Arrestin-3 concentration (μM) | 0 | 0.1 | 0.2 | 0.4 | 0.6 | 1.2 | 2 | 4 | 8 | 12 | 20 | 30 | 45 |
| Arrestin-3 (μl) | 0 | 2a | 4 | 8 | 12 | 2.4 | 4 | 8 | 2.4 | 3.6 | 6 | 9.2 | 13.2 |
| Tris buffer (μl)b | 14.7 | 12.7 | 10.7 | 6.7 | 2.7 | 12.3 | 10.7 | 6.7 | 12.3 | 11.1 | 8.7 | 5.5 | 1.5 |
Arrestin stock concentrations: For A2-A5, 1 μM; For A6-A8, 10 μM; For A9-A13, 65 μM.
Tris buffer: 10 mM Tris·Cl, pH 7.5, and 100 mM NaCl.
Measurement of JNK3 phosphorylation using the incorporation of 32P
Remove purified proteins from −80°C storage and thaw on ice before use. After protein samples are completely thawed, ultracentrifuge them for 1 hr at 300,000 × g (TLA120.1 rotor in Optima TLX ultracentrifuge, Beckman), 4°C, to remove aggregated proteins. Carefully transfer the supernatants into 1.5-ml clean tubes. Determine the protein concentration using the Bradford assay.
Purified arrestin-3, GST-MKK4, GST-MKK7, His-JNK1α1, His-JNK2α2, and His-JNK3α2. The protocol for purifying arrestin-3 has been described previously (Zhan et al., 2011a; Vishnivetskiy et al., 2014). Protocols for the purification of the other proteins are described in the Support Protocols in the present unit, as are those for studying the activation of MKK4 (p-MKK4) and MKK7 (p-MKK7).
-
2.
Prepare the kinase mix (Table 2.12.1).
The kinase reactions are performed in a volume of 20 μL. To avoid pipetting small volumes, use a stock tube to premix the kinases with buffer. Table 2.12.1 shows the recipe for the kinase mix needed for 20 reactions.
-
3.
Prepare arrestin-3 solutions. Dilute arrestin-3 protein with Tris buffer as shown in Table 2.12.2. Note that arrestin-3 is unstable at lower than 100 mM ionic strength.
-
4.
Add 3.3 μL kinase mix into each tube (18 μL total volume at this point), mix well by gentle vortexing, and then incubate the samples for 15 min in a 30°C water bath.
-
5.
Prepare 1 mM ATP mixture containing [γ-32P]-ATP. For 20 reactions, add 4 μL of ATP (10 mM) and 4 μL of [γ-32P]-ATP to 32 μL distilled water at room temperature.
-
6.
Initiate the reaction by adding 2 μL of the 1 mM ATP mixture containing [γ-32P]-ATP (prepared in step 5), terminating the reaction by the addition of 20 μL SDS buffer.
At these concentrations of kinases at 30°C the reaction is linear for 10 to 15 sec.
-
7.
Load 10 μL samples from each sample tube onto a 10% SDS-PAGE gel (appendix 3b; Gallagher and Sasse, 2001). Electorphorese the gels. Stain the gels with Coomassie blue after electrophoresis.
-
8.
Dry the SDS-PAGE gel in a gel dryer (e.g., Bio-Rad), then place the dry gel onto an autoradiography cassette, and expose overnight. Adjust the exposure time to achieve clear differentiation between band intensities in various lanes (Fig. 2.12.1C).
-
9.
Quantify the samples using scintillation counting. Cut the JNK3 bands from the SDS-PAGE gel and place them into scintillation vials. After adding 5 mL scintillation cocktail, incubate the samples for 30 min at 25°C (or room temperature) on a shaker. Quantify radioactivity with the scintillation counter. Dilute the ATP mix containing [γ-32P]-ATP to a final concentration of 10 nM/μL as the standards (5 μL) to calculate the specific activity of the ATP mix (Fig. 2.12.1). Based on the specific activity of the ATP, calculate absolute amounts of phosphate incorporation into JNK bands (pmols in the lower panel of Fig. 2.12.1B)
ALTERNATE PROTOCOL 1
ARRESTIN-3 MEDIATED JNK1/2 ACTIVATION BY MKK4/7 (MEASUREMENTS OF JNK1/2 PHOSPHORYLATION USING IMMUNOBLOTTING WITH PHOSPHO-JNK ANTIBODIES)
This procedure describes immunoblotting as an alternative (to 32P incorporation; Basic Protocol 1) way of measuring in vitro JNK phosphorylation.
Additional Materials (also see Basic Protocol 1)
PVDF membrane (Millipore)
Enhanced chemiluminescence reagent (e.g., SuperSignal West Pico from Pierce)
X-ray film
Quantity One software (Bio-Rad) or equivalent
Follow step 1 of Basic Protocol 1.
Prepare the kinase mix (Table 2.12.3; ten reactions).
Prepare the arrestin solutions (see Table 2.12.4).
Add 3.3 μl kinase mix into each tube (18 μl total volume at this point) and mix well by gentle vortexing. Incubate the mixed samples for 15 min in a 30°C water bath.
Prepare 1 mM of the ATP mixture. For 10 reactions, add 2 μl of ATP (10 mM) to 18 μl of distilled water.
Initiate the assay by adding 2 μl of 1 mM ATP solution, terminating the reaction by the addition of 20 μl SDS buffer.
Table 2.12.3.
Preparing the Kinase Mix (JNK1 or JNK2)
| Volume (each) | Volume (10×) | |
|---|---|---|
| 10× Kinase assay buffer | 2 μl | 20 μl |
| 100 mM DTT | 0.4 μl | 4 μl |
| JNK1α1 or JNK2α2 (12 μM stock) | 0.8 μl | 8 μl |
| p-MKK4 or p-MKK7 (10 μM stock) | 0.1 μl | 1 μl |
| Total | 3.3 μl | 33 μl |
Table 2.12.4.
Preparing Arrestin-3 Solutions
| Tube number | A1 | A2 | A3 | A4 | A5 | A6 | A7 |
|---|---|---|---|---|---|---|---|
| Arrestin-3 concentration (μM) | 0 | 0.2 | 0.5 | 1 | 5 | 10 | 20 |
| Arrestin-3 (μl)a | 0 | 4 | 10 | 1 | 5 | 10 | 10 |
| Tris buffer (μl) | 14.7 | 10.7 | 4.7 | 13.7 | 9.7 | 4.7 | 4.7 |
Arrestin stock concentrations: For A2-A3, 1 μM; For A4-A6, 20 μM; For A7, 40 μM.
At 30°C the reactions are linear for 10 to 15 sec.
-
7.
Load 5 μl and 10 μl of reaction samples onto 10% SDS-PAGE gels (appendix 3b; Gallagher and Sasse, 2001) for immunoblot with pan-JNK and phospho-JNK antibodies, respectively.
-
8.
Transfer to a PVDF membrane.
-
9.
Block with 1% non-fat dry milk in TBST (see recipe).
-
10.
Wash away dry milk (3 washes with TBST (see recipe), at least 5 min each, with 2-3 brief rinses with TBST between washes).
-
11.
Incubate membranes with pan-JNK and phospho-JNK primary antibodies (Cell Signaling Technology) to check for equal input of JNK proteins and the phosphorylation of JNK, respectively. Incubation works best at 4oC overnight, but in many cases 1 h at room temperature is sufficient.
-
12.
Wash away primary antibodies, as in 10.
-
13.
Incubate with appropriate peroxidase-conjugated secondary antibodies (usually for 1 h at room temperature).
-
14.
Wash away secondary antibodies, as in 10.
-
15.
Develop immunoblots with enhanced chemiluminescence reagent (e.g., SuperSignal West Pico from Pierce) according to the manufacturer’s instructions.
-
16.
Blot away liquid and expose blots to X-ray film in the dark room. Adjust the exposure time to obtain a clear differentiation between band intensities in the various lanes while avoiding saturation. All of the bands should be different shades of gray.
-
17.
Quantify the bands on the X-ray film using Quantity One software (Bio-Rad) or equivalent. Plot phospho-JNK band intensity (arbitrary units) as a function of arrestin-3 concentration (Fig. 2.12.3)
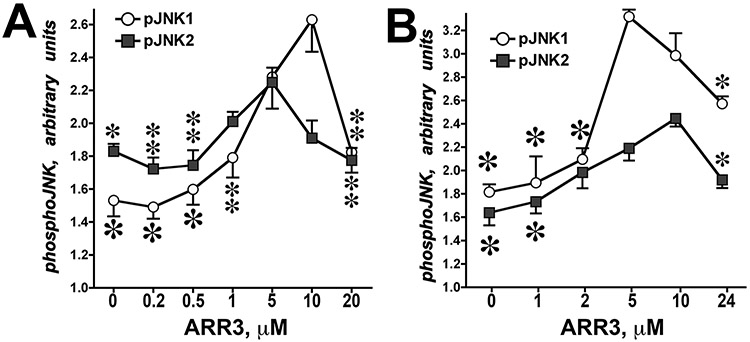
Figure 2.12.3.
Arrestin-3-mediated JNK1/2 activation by MKK4/7. In vitro phosphorylation of JNK1α1 and JNK2α2 in the presence of MKK4 (A) or MKK7 (B) yielded bell-shaped curves as functions of arrestin-3 concentration. Means ± SD of three independent experiments are shown. ANOVA analysis with arrestin-3 as main factor demonstrated significance of arrestin-3 concentration in the presence of MKK4 and MKK7 for both JNK1α1 and JNK2α2 (p < 0.001). * –p <0.001, ** –p <0.01, * –p <0.05 to maximal values (at 5 or 10 μM of arrestin-3, respectively) according to Bonferroni/Dunn post-hoc test with correction for multiple comparisons. Data from Kook et al. (2014).
BASIC PROTOCOL 2
THE ROLE OF ARRESTIN-3 IN JNK ACTIVATION IN INTACT CELLS
While direct interactions between proteins and their functional effects can only be proven using pure proteins (Basic Protocol 1), these interactions and their biological function must be studied in intact cells. Detailed below are protocols for cell-based assays that can be used to study arrestin-3-dependent activation of JNK family kinases.
Materials
COS-7 (African green monkey fibroblast) cells (ATCC). Other cell types can be used, but the levels of endogenous arrestins in them are higher than in COS-7. Arrestin-2/3 double knockout HEK293 cells (Alvarez-Curto et al., 2016) are the best, as in these cells the only arrestin present is the one you express.
Dulbecco’s modified Eagle’s medium (DMEM) with 4.5 g/liter glucose, l-glutamine and sodium pyruvate (CORNING cellgro, cat. no. 10-013-CV)
Fetal bovine serum, Qualified (Life Technologies, cat. no. 2017-07)
Penicillin/Streptomycin (Life Technologies, cat. no. 15140-122)
Opti-MEM I Reduced Serum Medium, no phenol red (Invitrogen, cat. no. 11058-021)
pcDNA3-HA-ASK1 and pcDNA3
pcDNA3-Arrestin-3
FuGENE HD Transfection Reagent (Promega, cat. no. E231A). Other transfection reagents can be used according to manufactirers’ instructions.
Growth medium
Serum-free medium
Ice
Dulbecco’s Phosphate-buffered saline (DPBS) without calcium and magnesium (CORNING cellgro, cat. no. 21-031-CV)
Lysis buffer (see recipe)
Bio-Rad Protein Assay Dye Reagent Concentrate (Bio-Rad, cat. no. 500-0006)
- Antibodies for immunoblot:
- F4C1: Mouse monoclonal antibody that detects the epitope DGVVLVD, a sequence that is present in all known mammalian arrestins (Donoso et al., 1990) or F431: Rabbit polyclonal antibody that detects the same epitope as F4C1 (Song et al., 2011)
- Phospho-SAPK/JNK (Thr183/Tyr185) antibody (Cell Signaling Technology, cat. no. 9251), SAPK/JNK antibody (Cell Signaling Technology, cat. no. 9252), HA-Tag (6E2) Mouse antibody (Cell Signaling Technology, cat. no. 2367), Monoclonal anti-Flag M2 antibody (Sigma, cat. no. F3165)
2× SDS Sample buffer (Sigma)
8% SDS-PAGE gel
0.05% Trypsin-EDTA (Life Technologies, cat. no. 25300-054)
37°C humidified incubator with 5% CO2
6-well plates
1.5-ml microcentrifuge tubes
Cell scraper
95°C heat block (Isotemp 125D, Fisher Scientific)
Electrophoresis equipment
PVDF membrane (Millipore)
Additional reagents and the equipment needed for performing SDS-PAGE electrophoresis (appendix 3b; Gallagher and Sasse, 2001), immunoblotting (described in Zhan et al., 2011b; Zhan et al., 2013a)., and measuring the protein concentration (appendix 3a; Olson and Markwell, 2007)
NOTE: Add all protease inhibitors immediately before use, especially PMSF, which is unstable in aqueous solutions.
ASK-1/MKK7/MKK4-induced JNK activation
To measure arrestin-3 effects on ASK1/MKK7/MKK4-induced activation of JNK family kinases in COS-7 cells, increasing amounts of HA-ASK1, Flag-MKK7β1, or HA-MKK4 are co-transfected into COS-7 cells with or without a fixed amount of arrestin-3.
Cell culture and transfection
-
1.
Maintain COS-7 African green monkey cells (or other cell lines) in a 37°C humidified incubator with 5% CO2 in DMEM supplemented with 10% heat-inactivated FBS, penicillin, and streptomycin.
-
2.
Plate 2.5 × 105 COS-7 cells in 2 mL per well of a 6-well plate one day before transfection to ensure the cells are 80% to 90% confluent at the time of transfection.
-
3.
To a sterile 1.5-ml microcentrifuge tube, add 125 μl Opti-MEM prewarmed to 37°C.
-
4a.
For ASK-1-induced JNK activation: Mix with Opti-MEM different amounts of pcDNA3-HA-ASK1 and empty pcDNA3 to a final total DNA concentration of 2 μg per transfection (Table 2.12.5).
-
4b.
For Arrestin-3 effect on ASK-1 induced JNK activation: Mix each concentration of pcDNA3-HA-ASK1 and pcDNA3 (total DNA 1.5 μg) with Opti-MEM, and then add 0.5 μg of pcDNA3-Arrestin-3 into each tube (Table 2.12.6).
-
5.
For a 3:1 FuGENE HD:DNA ratio, add 6 μl of FuGENE HD directly to the diluted DNA.
-
6.
Incubate the FuGENE HD and DNA mixture for 15 min at room temperature.
-
7.
Add each DNA-FuGENE HD complex to a separate well in a 6-well plate containing 2 mL of cells in growth medium.
-
8.
Gently mix the sample by rocking the plate back and forth.
-
9.
Return the cells into a CO2 incubator and incubate for 24 hr.
-
10.
Replace growth medium with 2 mL serum-free medium and incubate overnight at 37°C in a humidified incubator with 5% CO2 in tissue culture incubator.
Table 2.12.5.
Transfection to Determine ASK1-Dependent JNK Activation
| pcDNA3-HA-ASK1a | 0 μg | 0.05 μg | 0.1 μg | 0.25 μg | 0.5 μg | 1 μg | 1.5 μg |
|---|---|---|---|---|---|---|---|
| pcDNA3 | 2.0 μg | 1.95 μg | 1.9 μg | 1.75 μg | 1.5 μg | 1 μg | 0.5 μg |
| FuGENE HD | 6 μl | 6 μl | 6 μl | 6 μl | 6 μl | 6 μl | 6 μl |
| Opti-MEM | 125 μl | 125 μl | 125 μl | 125 μl | 125 μl | 125 μl | 125 μl |
For MKK7-induced JNK activation, add pcDNA3-Flag-MKK7β1 instead of pcDNA3-HA-ASK1.
Table 2.12.6.
Transfection to Determine ASK1- and Arrestin-3-Dependent JNK Activation
| pcDNA3-HA-ASK1a | 0 μg | 0.05 μg | 0.1 μg | 0.25 μg | 0.5 μg | 1 μg | 1.5 μg |
|---|---|---|---|---|---|---|---|
| pcDNA3 | 1.5 μg | 1.45 μg | 1.4 μg | 1.25 μg | 1.0 μg | 0.5 μg | 0 μg |
| pcDNA3-arrestin-3 | 0.5 μg | 0.5 μg | 0.5 μg | 0.5 μg | 0.5 μg | 0.5 μg | 0.5 μg |
| FuGENE HD | 6 μl | 6 μl | 6 μl | 6 μl | 6 μl | 6 μl | 6 μl |
| Opti-MEM | 125 μl | 125 μl | 125 μl | 125 μl | 125 μl | 125 μl | 125 μl |
For MKK7-induced JNK activation, add pcDNA3-Flag-MKK7β1 instead of pcDNA3-HA-ASK1.
Prepare the sample
-
11.
Take a 6-well culture plate from the incubator and place it on ice.
-
12.
Remove the medium by aspiration and wash the transfected COS-7 cells twice in each well with 1 ml cold phosphate-buffered saline (PBS), removing PBS by aspiration each time.
-
13.
Add 150 μL lysis buffer per well in a 6-well plate and collect lysates using a cell scraper.
-
14.
Heat the cell lysates for 10 min at 95°C and cool to room temperature.
-
15.
Measure the protein concentration (e.g., using Bio-Rad Protein Assay Dye Reagent Concentrate; also see appendix 3a; Olson and Markwell, 2007).
Immunoblot
-
16.
Use 10 μg of cell lysate protein for immunoblot with phospho-JNK and HA antibody and 5 μg for immunoblot with the arrestin-3 antibody. Mix the cell lysates with 2× SDS-sample buffer at a 1:1 ratio.
-
17.
Load the samples onto an 8% SDS-PAGE gel and electrophorese until the dye reaches the bottom of the gel.
-
18.
Transfer the sample to a PVDF membrane and develop with phospho-JNK and JNK antibody to monitor the activation of endogenous JNKs and HA-tag, F4C1 or F431 antibody or any other arrestin-specific antibody to check the expression of HA-ASK1 and arrestin-3, respectively (Figs. 2.12.4 and 2.12.5). Follow western blot procedures described in Alternate protocol 1 (steps 9-17).
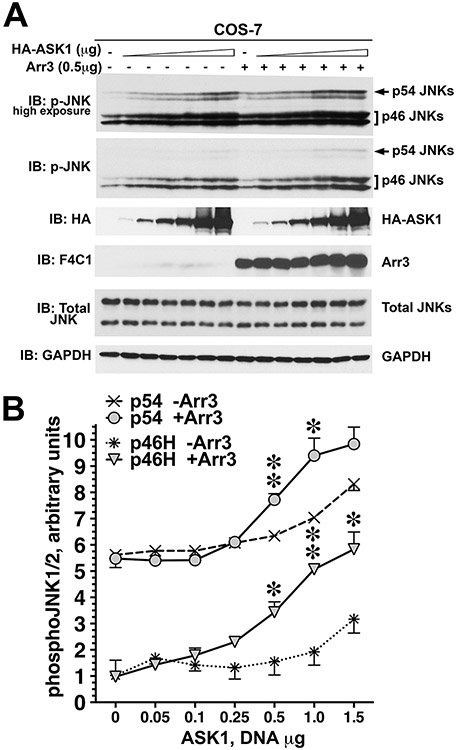
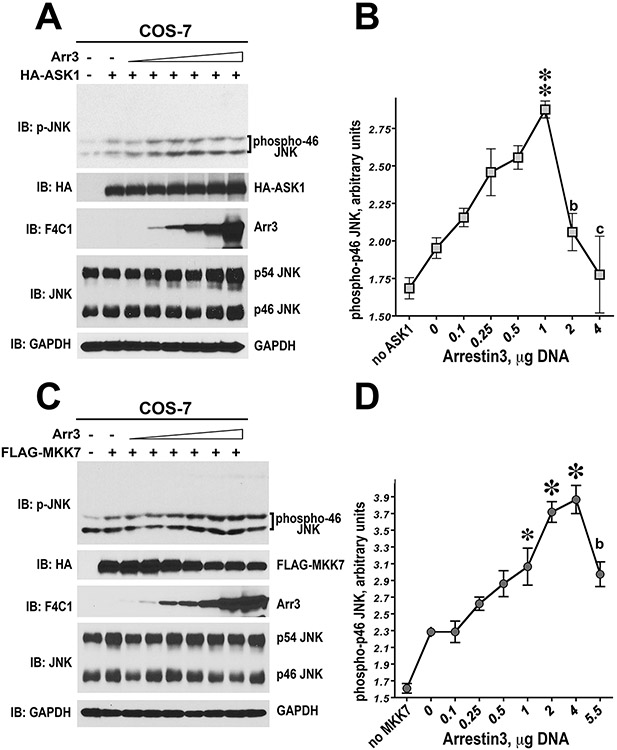
Figure 2.12.4.
Arrestin-3 promotes JNK1/2 activation induced by the expression of ASK1 in intact cells. (A) Representative immunoblot showing phosphorylation of endogenous JNK1/2 isoforms with or without arrestin-3 in COS-7 cells expressing varying amounts of ASK1. Upper p-JNK blot is the same as lower blot exposed for longer time to visualize p54 isoforms. (B) Quantification of phosphorylation of JNK p46H and p54 isoforms with or without arrestin-3. ANCOVA with arrestin-3 as factor and ASK1 concentration as co-variate showed significant effect of ASK1 concentration on the level of p46H and p54 phosphorylation (p < 0.0001). * –p < 0.05, ** –p < 0.01 to - Arr3, Student’s t-test for individual points. Data from Kook et al. (2014).
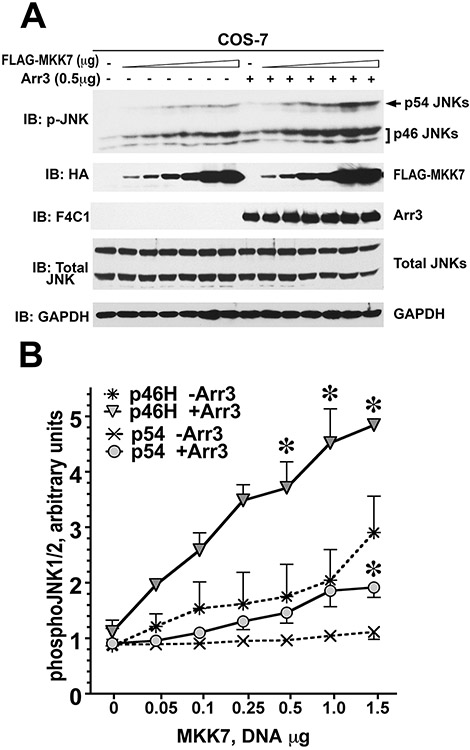
Figure 2.12.5.
Arrestin-3 enhances MKK7-dependent phosphorylation of endogenous JNK1/2 in intact cells. (A) Representative immunoblot showing phosphorylation of endogenous JNK1/2 isoforms with or without arrestin-3 in COS-7 cells expressing varying amounts of MKK7. (B) Quantification of phosphorylation of JNK p46H and p54 isoforms with or without arrestin-3. ANCOVA with arrestin-3 as factor and MKK7 concentration as co-variate showed significant effect of MKK7 concentration of the level of p46H and p54 phosphorylation (p <0.0001). The presence of arrestin-3 significantly affected the level of p46H, but not p54, phosphorylation across MKK7 concentrations (F(1,38) = 8.592 p = 0.0057). * –p < 0.05 to -Arr3, Student’s t-test for individual points. Data from Kook et al. (2014).
ALTERNATE PROTOCOL 2
MKK4-INDUCED JNK ACTIVATION
In COS-7 cells, pcDNA3-HA-MKK4 tends to express lower amounts of protein than pcDNA3-HA-ASK1. For this reason, more pcDNA3-MKK4-HA is used for transfection to ensure similar levels of expression. For materials, see Basic Protocol 2.
Transfection
All procedures are identical to those described in Basic Protocol 2 for ASK1-induced JNK activation except as follows:
Alternative step 4a: For MKK4-induced JNK activation, mix different amounts of pcDNA3-HA-MKK4 with empty pcDNA3 (total DNA 4.5 μg) with Opti-MEM (Table 2.12.7).
Table 1.12.7.
Transfection to Determine MKK4-Dependent JNK Activation
| pcDNA3-HA-MKK4 | 0 μg | 0.2 μg | 0.5 μg | 1.0 μg | 2.0 μg | 4.0 μg |
|---|---|---|---|---|---|---|
| pcDNA3 | 4.5 μg | 4.3 μg | 4.0 μg | 3.5 μg | 2.5 μg | 0.5 μg |
| FuGENE HD | 13.5 μl | 13.5 μl | 13.5 μl | 13.5 μl | 13.5 μl | 13.5 μl |
| Opti-MEM | 125 μl | 125 μl | 125 μl | 125 μl | 125 μl | 125 μl |
Alternative for step 4b: To examine the effect of arrestin-3 on MKK4-induced JNK activation, mix different amounts of pcDNA3-HA-MKK4 and empty pcDNA3 (total DNA 4.0 μg) with Opti-MEM, and then add to each tube 0.5 μg of pcDNA3-arrestin-3 (Table 2.12.8).
Table 2.12.8.
Transfection to Determine MKK4- and Arrestin-3-Dependent JNK Activation
| pcDNA3-HA-MKK4 | 0 μg | 0.2 μg | 0.5 μg | 1.0 μg | 2.0 μg | 4.0 μg |
|---|---|---|---|---|---|---|
| pcDNA3 | 4.0 μg | 3.8 μg | 3.5 μg | 3.0 μg | 2.0 μg | 0 μg |
| pcDNA3-arrestin-3 | 0.5 μg | 0.5 μg | 0.5 μg | 0.5 μg | 0.5 μg | 0.5 μg |
| FuGENE HD | 13.5 μl | 13.5 μl | 13.5 μl | 13.5 μl | 13.5 μl | 13.5 μl |
| Opti-MEM | 125 μl | 125 μl | 125 μl | 125 μl | 125 μl | 125 μl |
Alternative step 4c: To evaluate the regulation of peptide-mini-scaffold T1A on MKK4-induced JNK activation, mix different amounts of pcDNA3-HA-MKK4, and empty pcDNA3 (total DNA 4.0 μg) with Opti-MEM, and then add to tubes contains either empty pcDNA3, pcDNA3-YFP, pcDNA3-YPF-TIA, or pcDNA3-arrestin3 (Table 2.12.9) .
Table 2.12.9.
Transfection to Detect Biphasic Effect of Arrestin-3 on ASK1-Stimulated JNK Phosphorylationa (Fig. 2.12.6 A,B)
| pcDNA3-Arrestin-3 | 0 μg | 0 μg | 0.1 μg | 0.25 μg | 0.5 μg | 1.0 μg | 2.0 μg | 4.0 μg |
|---|---|---|---|---|---|---|---|---|
| pcDNA3-HA-ASK1 | 0 μg | 0.5 μg | 0.5 μg | 0.5 μg | 0.5 μg | 0.5 μg | 0.5 μg | 0.5 μg |
| pcDNA3 | 4.5 μg | 4.0 μg | 3.9 μg | 3.75 μg | 3.5 μg | 3.0 μg | 2.0 μg | 0 μg |
| FuGENE HD | 13.5 μl | 13.5 μl | 13.5 μl | 13.5 μl | 13.5 μl | 13.5 μl | 13.5 μl | 13.5 μl |
| Opti-MEM | 125 μl | 125 μl | 125 μl | 125 μl | 125 μl | 125 μl | 125 μl | 125 μl |
These DNA amounts were calculated for 6-well culture plate (equivalent to 35-mm dish). If plates of a different size are used the amount of total DNA should be changed in proportion to the plate area.
BASIC PROTOCOL 3
BIPHASIC EFFECT OF ARRESTIN-3 ON ASK1- AND MKK7-STIMULATED JNK PHOSPHORYLATION IN CELLS
To examine the concentration-dependence of arrestin-3 expression levels on ASK1- and MKK7-stimulated JNKs phosphorylation in COS-7 cells, the expression of ASK1 or MKK7 is kept constant while the expression of arrestin-3 is varied (Table 2.12.10).
Table 2.12.10.
Transfection to Detect Biphasic Effect of Arrestin-3 on MKK7-Stimulated JNK Phosphorylationa (Fig. 2.12.6 C,D)
|
pcDNA3- Arrestin-3 |
0 μg | 0 μg | 0.1 μg | 0.25 μg | 0.5 μg | 1.0 μg | 2.0 μg | 4.0 μg | 5.5 μg |
|---|---|---|---|---|---|---|---|---|---|
| pcDNA3-Flag-MKK7 | 0 μg | 0.08 μg | 0.08 μg | 0.08 μg | 0.08 μg | 0.08 μg | 0.08 μg | 0.08 μg | 0.08 μg |
| pcDNA3 | 5.58 μg | 5.5 μg | 5.4 μg | 5.25 μg | 5.0 μg | 4.5 μg | 3.5 μg | 1.5 μg | 0 μg |
| FuGENE HD | 18 μl | 18 μl | 18 μl | 18 μl | 18 μl | 18 μl | 18 μl | 18 μl | 18 μl |
| Opti-MEM | 125 μl | 125 μl | 125 μl | 125 μl | 125 μl | 125 μl | 125 μl | 125 μl | 125 μl |
These DNA amounts were calculated for 6-well culture plate (equivalent to 35-mm dish). If plates of a different size are used the amount of total DNA should be changed in proportion to the plate area.
SUPPORT PROTOCOL 1
EXPRESSION, PURIFICATION, AND ACTIVATION OF GST-MKK4
JNK3 is mainly activated by phosphorylation of the activation loop at a Thr-Pro-Tyr (TPY) motif by the MAP2Ks MKK4 and MKK7. The activation loop can be phosphorylated at Tyr-223 by activated MKK4.
Materials
Plasmid: pGEX4T1-MKK4 vector is a construct encoding full-length wild type Homo sapiens mitogen-activated protein kinase kinase 4 (MAP2K4) (GenBank accession number NM_003010) with an N-terminal-cleavable GST–tag (Yan et al., 2011; Zhan et al., 2011c); this plasmid is available upon request from Dr. K.N. Dalby (Division of chemical Biology & Medicinal Chemistry; BME 6.202B; 1 University Station C0850, Austin, Texas 78712-1074; dalby@austin.utexas.edu)
BL21 (DE3) (Novagen): prepare electrocompetent cells as described previously (see Gonzales et al., 2013)
Luria Broth (LB) medium (Sigma) containing 50 μg/mL ampicillin sodium (Sigma)
Terrific Broth (TB) medium (Sigma) containing 50 μg/mL ampicillin sodium (Sigma)
1 M Isopropyl β-d-1-thiogalactopyranoside (IPTG) solution in sterile water (Sigma)
Liquid nitrogen
Buffer A (see recipe)
1 M MgCl2 solution (Sigma)
Lysozyme lyophilized enzyme (MP Biomedicals)
100% (v/v) Glycerol
10% (v/v) Triton in water (Sigma)
Glutathione Sepharose High Performance (Amersham Biosciences); on the day of the experiment, pack the required beads in a glass Econo-Column, wash and equilibrate in buffer A
Buffer B (see recipe)
Bradford reagent (Bio-Rad)
GST-MEKK1c (C terminal 320 amino acids corresponding to the catalytic domain) is necessary for activating GST-MKK4, GST-MKK7-His6 [the protocol for GST-MEKK1c vector construction and protein purification has been described previously (see Khokhlatchev et al., 1997; Gallagher et al., 2002)]; this plasmid is available upon request from Dr. K.N. Dalby (Division of Chemical Biology & Medicinal Chemistry; BME 6.202B; 1 University Station C0850, Austin, Texas 78712-1074; dalby@austin.utexas.edu)
50 mM ATP (Roche) in 25 mM HEPES buffer (pH 7.5)
Buffer C (see recipe)
Buffer D (see recipe)
Shaking incubator
Sonicator
Econo-Column Chromatography glass columns, 2.5 × 20 cm (98 ml) (Bio-Rad)
Amicon Ultra-15 concentrator
Gel filtration column: 120 ml HiLoad 16/60 Superdex 200 prep grade column (Amersham)
Additional reagents and equipment for transforming the pGEX4T1-MKK4 vector into BL21 (DE3) electrocompetent cells (House et al., 2004) and measuring the protein concentration (Olson and Markwell, 2007)
Expression of GST-MKK4
Day 1: Transform the pGEX4T1-MKK4 vector into BL21 (DE3) electrocompetent cells following the published protocol for DNA electroporation (House et al., 2004).
Day 2: Inoculate a single colony of freshly transformed cells in a 30 ml culture of Luria Broth (LB) containing 50 μg/mL ampicillin. Incubate with shaking overnight at 37°C.
Day 3: Dilute the culture 100-fold into TB (Terrific Broth) containing 50 μg/mL ampicillin, and incubate with shaking at 37°C until the OD600 reaches 0.6 to 0.7. Induce GST-MKK4 expression by adding 25 to 50 μM IPTG, and continue shaking for 20 hr at 25°C.
Day 4: Pellet the cells by centrifuging for 15 min at 8000 × g, 4°C. Remove the supernatant and immediately freeze the cell pellet in liquid nitrogen and store up to one week at −80°C.
Purification of GST-MKK4
Day 5
-
5.
Resuspend the frozen, wet cells in 150 mL buffer A containing 0.2 mg/mL lysozyme, 1 mM MgCl2, and 20% (v/v) glycerol. Incubate the mixture for 30 min at 4°C, then add Triton X-100 to a final concentration of 1% (v/v), and incubate for 30 min at 4°C.
-
6.
Sonicate the cell lysate (power level 5, 50% duty cycle) for 5 to 10 min at 4°C (using 5-sec pulses at 5-sec intervals and with careful monitoring of the temperature using a temperature probe).
If the cell lysate is unusually viscous, the sonication time can be extended with careful monitoring of the temperature (do not exceed 4°C) until it becomes less viscous.
-
7.
Centrifuge the lysate for 30 min at 12,000 × g, 4°C, and mix the supernatant with 10 mL of Glutathione Sepharose High Performance resin equilibrated with buffer A. Shake gently for 1.5 hr at 4°C.
-
8.
Wash the beads with 150 mL buffer A in a glass Econo-column (98 mL).
-
9.
Elute the GST-tagged proteins with 20 mL buffer B containing 20% (v/v) glycerol. Adjust the pH of the GST elution buffer to 7.5 as glutathione decreases the pH to ~2.0. Adjust the pH of all the buffers after adding glycerol. Collect the eluted protein and dialyze overnight into 4 liters of buffer S containing 20% (v/v) glycerol at 4°C with slow stirring.
-
10.
Measure the protein concentration using the Bradford reagent following the manufacturer’s protocol. Immediately freeze the samples (100-μL aliquots at around 20 μM concentration) in liquid nitrogen and store up to 6 months at −80°C for later activation.
The estimated yield is ~20 to 30 mg of pure GST-MKK4 per 1 liter of cells. The estimated purity by 12% SDS/PAGE is generally around 85% (Zhan et al., 2011b; Zhan et al., 2013b).
Activation of GST-MKK4
Day 6
-
11.
Incubate 4 μM GST-MKK4 and 2 μM GST-MEKK1c for 60 min at 30°C in the presence of 4 mM ATP in 10 mL of activation buffer C.
-
12.
Concentrate the 10 mL reaction mixture to 2 to 3 mL using an Amicon Ultra-15 concentrator (10,000 molecular weight cutoff). Centrifuge for 10 to 15 min at 3000 × g (4400 rpm), 4°C.
-
13.
Purify the concentrated reaction mixture using a gel filtration column (120 mL HiLoad 16/60 Superdex 200 prep grade column) that has been equilibrated with buffer D containing 10% (v/v) glycerol.
This column separates the active GST-MKK4 away from the constitutively active GST-MEKK1c.
-
14.
The pure, activated GST-MKK4 fractions that were eluted from the gel filtration column do not require further dialysis so they can be frozen immediately after increasing the glycerol concentration to 20%. The GST-MKK4 activation, purification, and freezing must be performed in one day to minimize the possibility of GST tag cleavage. The tendency of GST-MKK4 to precipitate makes it essential to maintain the concentration of active and inactive GST-MKK4 at less than 20 μM. Store the activated GST-MKK4 in buffer D containing 20% (v/v) glycerol in 0.025-mL aliquots up to 6 months at −80°C until further use.
-
15.
Measure the protein concentration (e.g., using Bradford reagent following the manufacturer’s protocol; also see appendix 3a; Olson and Markwell, 2007.
At least 65% of GST-MKK4 should be recovered as pure activated GST-MKK4. The estimated purity by 12% SDS/PAGE is ~85% (Zhan et al., 2011b; Zhan et al., 2013b).
SUPPORT PROTOCOL 2
EXPRESSION, PURIFICATION, AND ACTIVATION OF GST-MKK7-HIS6
MKK7 is responsible for Thr-221 phosphorylation of JNK3 activation loop, and once JNK3 is threonine phosphorylated, then MKK4 can phosphorylate JNK3 on Tyr-223 (Lisnock et al., 2000).
Additional Materials (also see Support Protocol 1)
Plasmid: pGEX-MKK7-His6 vector is a construct encoding full length, wild-type Mus musculus mitogen-activated protein kinase kinase 7 (GenBank accession number NM_011944) with N-terminal cleavable GST–tag and C-terminal cleavable His-tag (Madsen et al., 2011; Zhan et al., 2013b); this plasmid is available upon request from Dr. K.N. Dalby (Division of Chemical Biology & Medicinal Chemistry; BME 6.202B; 1 University Station C0850; Austin, Texas 78712-1074; dalby@austin.utexas.edu)
Buffer E (see recipe)
NaCl (Biological grade MP Biomedical)
Ni-NTA beads (Qiagen); on the day of the experiment, pack the required beads in a glass Econo-Column, wash and, equilibrate in buffer E
Expression of GST-MKK7-His
Day 1: Transform the pGEX 4T1 vector containing DNA encoding full-length MKK7 into BL21 (DE3) electro-competent cells, using the published protocol for DNA electroporation (House et al., 2004).
Day 2: From a single colony of freshly transformed cells, inoculate a 30 ml culture of Luria Broth (LB) containing 50 μg/mL ampicillin. Incubate the culture with shaking at 250 rpm overnight at 37°C.
Day 3: Dilute the culture 100-fold into TB (Terrific Broth) medium containing 50 μg/mL ampicillin. Incubate at 37°C with shaking at 250 rpm. Once the OD600 of the culture reaches 0.6, induce expression by adding 25 μM IPTG. Continue shaking for 20 hr at 25°C.
Day 4: Pellet the cells by centrifuging for 12 min at 7000 × g, 4°C, remove the supernatant, and freeze the pellets immediately in liquid nitrogen. Store up to one week at −80°C.
Purification of GST-MKK7-His
Day 5
-
5.
Resuspend the frozen, wet cells in 150 mL buffer E containing 0.5 M NaCl, 0.2 mg/mL lysozyme, 1 mM MgCl2, and 20% (v/v) glycerol. Incubate the mixture for 30 min at 4°C, and then add Triton X-100 to a final concentration of 1% (v/v). Incubate at 4°C for another 30 min.
-
6.
Sonicate the cell lysate for 5 to 10 min at 4°C (using 5-sec pulses at 5-sec intervals and careful monitoring of the temperature using a temperature probe). If the cell lysate is unusually viscous, the sonication time can be extended with careful monitoring of the temperature (do not exceed 4°C) until it becomes less viscous. Remove the cell debris by centrifuging for 30 min at 17,600 × g, 4°C.
-
7.
Agitate the supernatant with Ni-NTA beads for 1 hr at 4°C. After washing the beads with 150 mL buffer E containing 10 mM imidazole and 20% glycerol, elute the GST-MKK7-His6-tagged proteins with 25 mL buffer E (pH 8.0) containing 200 mM imidazole and 20% glycerol.
-
8.
Collect the eluted protein and dialyze overnight at 4°C into buffer S containing 20% glycerol.
-
9.
Measure the protein concentration (e.g., using Bradford reagent following the manufacturer’s protocol; also see appendix 3a; Olson and Markwell, 2007) Immediately freeze the samples (100-μL aliquots of around 20 μM concentration) in liquid nitrogen and store up to 6 months at −80°C for later activation.
The estimated yield is ~20 mg of pure GST-MKK7-His6 per liter of culture with an estimated purity by 12% SDS/PAGE of more than 95% (Madsen et al., 2011; Zhan et al., 2013b). To enhance the solubility and stability of GST-MKK7-His6, dialyze at 4°C into storage buffer containing 590 mM sucrose instead of 20% glycerol. Note that the sucrose must be removed before activation by dialyzing it back into buffer S containing 20% glycerol.
Activation of GST-MKK7-His6
Day 6
-
10.
Incubate 4 μM GST-MKK7-His and 2 μM GST-MEKK1 for 60 min at 30°C in the presence of 4 mM ATP in 10 mL activation buffer C.
-
11.
Purify the reaction mixture using a Ni affinity column that separates the active GST-MKK7-His6 from GST-MEKK1. Dilute the reaction mixture in 100 mL buffer E containing 20% glycerol, and agitate the diluted reaction mixture with Ni-NTA beads for 1 hr at 4°C. After washing the beads with 100 ml buffer E containing 10 mM imidazole and 20% glycerol, elute the activated GST-MKK7-His6-tagged proteins with 10 mL buffer E (pH 8.0) containing 200 mM imidazole and 20% glycerol.
-
12.
Measure the protein concentration (e.g., using Bradford reagent following the manufacturer’s protocol; also see appendix 3a; Olson and Markwell, 2007).
-
13.
Store the activated GST-MKK7-His6 in buffer S containing 20% glycerol up to 6 months at −80°C until further use.
At least 60% of GST-MKK7-His6 should be recovered as pure active GST-MKK7-His6 with the estimated purity by 12% SDS/PAGE being more than 95% (Madsen et al., 2011; Zhan et al., 2013b). The tendency of GST-MKK7-His6 to precipitate requires that the concentration of active and inactive GST-MKK7-His6 be less than 20 μM.
SUPPORT PROTOCOL 3
EXPRESSION, PURIFICATION, AND ACTIVATION OF TAG-LESS JNK1A1
Additional Materials (also see Support Protocols 1 and 2)
Plasmid: pET28a(+)Tev-JNK1α1 vector is a construct encoding full length wild-type Homo sapien mitogen-activated protein kinase 8 (GenBank accession number NM_002750) with N-terminal Tev (Tobacco Etch Virus ) cleavable His-tag (Yan et al., 2011); this plasmid is available upon request from Dr. K.N. Dalby (Division of Medicinal Chemistry; BME 6.202B; 1 University Station C0850; Austin, Texas 78712-1074; dalby@austin.utexas.edu)
Luria Broth (LB) medium containing 30 μg/mL kanamycin (Sigma)
Imidazole
Buffer F (see recipe)
Buffer G (see recipe)
TEV (Tobacco Etch Virus ) protease[Express and purify TEV protease as previously described (Abramczyk et al., 2011)]; plasmid is available in Addgene, plasmid 8827: pRK793, TEV protease, S219V mutant
CaCl2
Mono Q HR 10/10 anion-exchange column (Amersham)
HiPrep 26/10 desalting column (Amersham)
Expression of His6-Tev-JNK1α1
Day 1: Electroporate pET28a(+)Tev-JNK1 into the E. coli strain BL21(DE3) using a published protocol for DNA electroporation (House et al., 2004).
Day 2: Use cells from a single colony to inoculate 30 mL Luria Broth (LB) medium containing 30 μg/mL kanamycin. Grow overnight at 37°C.
Day 3: Dilute the culture 100-fold into Luria Broth medium containing 30 μg/mL kanamycin and grow at 37°C to an OD600 of 0.6. Induce the cells by adding 500 μM IPTG and culture them for 5 hr at 30°C. Pellet the cells by centrifuging 12 min at 7000 × g, 4°C. Remove the supernatant and immediately freeze the resultant pellets in liquid nitrogen. Store up to one week at −80°C.
Purification of His6-Tev-JNK1α1
Day 4
-
4.
Lyse the cells in 150 mL buffer E containing 0.5 M NaCl and 0.2 mg/mL lysozyme. Incubate the mixture for 30 min at 4°C, and then add Triton X-100 to a final concentration of 1% (v/v). Incubate for 30 min at 4°C.
-
5.
Sonicate the cell lysate (power level 5, 50% duty cycle) for 5 to 10 min at 4°C (using 5-sec pulses at 5-sec intervals and careful monitoring of the temperature using a temperature probe). If the cell lysate is unusually viscous, the sonication time can be extended with careful monitoring of the temperature (do not exceed 4°C) until it becomes less viscous. Remove cell debris by centrifuging for 30 min at 17,600 × g, 4°C.
-
6.
Agitate the supernatant with Ni-NTA beads for 1 hr at 4°C. After washing the beads with 150 mL buffer E containing 10 mM imidazole, elute the His6-tagged proteins with 25 mL buffer E containing 200 mM imidazole.
-
7.
Apply the eluted proteins to a Mono Q HR 10/10 anion-exchange column equilibrated with buffer F. Develop the column over 15 to 17 column volumes with a linear gradient of 0 to 0.5 M NaCl.
Proteins were detected by absorbance at 280 nm by a UV detector and His6-JNK1α1 fractions were identified by 10% SDS/PAGE.
-
8.
Collect eluted fractions of His6-JNK1α1 and either process to cleave the His6-tag or dialyze into buffer S containing 10% glycerol.
-
9.
Measure the protein concentration (e.g., using Bradford reagent following the manufacturer’s protocol). Immediately freeze the sample in liquid nitrogen and store up to 6 months at −80°C for later use.
The estimated yield of is ~13 mg of pure His-tagged JNK1α1 per liter of culture. The estimated purity by 12% SDS/PAGE is greater than 95% (Yan et al., 2011).
TEV cleavage of His6-Tev-JNK1α1
-
10.
Following Mono Q HR 10/10 anion-exchange chromatography dialyze overnight the collected fractions of His6-Tev-JNK1α1 against cleavage buffer G.
Day 5
-
11.
Incubate the dialyzed protein with TEV enzyme (1.5 %) and 2.5 mM CaCl2 for 2 hr at 30°C. Estimate the cleavage by 10% SDS/PAGE and extend the incubation time until 100% cleavage is achieved.
-
12.
After cleavage, desalt the reaction mixture by using HiPrep 26/10 desalting column (Amersham) equlibrated in buffer F.
-
13.
Filter the protein through 25 mm syringe filter with 0.45 μm nylon membrane and load on a Mono Q HR 10/10 anion-exchange column equilibrated in buffer F. Develop the column over 15 to 17 column volumes with a linear gradient of 0 to 0.5 M NaCl.
-
14.
Collect the eluted fractions of tagless JNK1α1 and dialyze them into buffer S.
-
15.
Measure the protein concentration using Bradford reagent following the manufacturer’s protocol. Immediately freeze in liquid nitrogen and store up to 6 months at −80°C for later use.
Typically, 80% of the tagless JNKα1 is recovered after cleavage, with the estimated purity by 12% SDS/PAGE being greater than 95% (Yan et al., 2011).
Activation of tagless JNK1α1
Day 6
-
16.
Incubate 2 μM tagless JNK1α1, 100 nM active GST-MKK4, and 400 nM active GST- MKK7 for 60 min at 30°C in the presence of 4 mM ATP in 10 ml activation buffer C.
-
17.
Desalt the reaction mixture using HiPrep 26/10 desalting column (Amersham) equilibrated in buffer D.
-
18.
Pool the eluted protein fractions from the desalting column, then concentrate them to 2 to 3 ml using an Amicon Ultra-15 concentrator (10,000 molecular weight cutoff). Centrifuge for 10 to 15 min at 3000 × g, 4°C.
-
19.
Purify the concentrated reaction mixture using a gel filtration column (120 ml HiLoad 16/60 Superdex 200 prep grade column) that has been equilibrated with buffer D.
-
20.
Measure the protein concentration, e.g., using Bradford reagent following the manufacturer’s protocol.
-
21.
Store the activated, tagless JNK1α1 in buffer S containing 10% glycerol up to 6 months at −80°C until further use.
Typically, 60% of the activated JNK1α1 is recovered after activation with the estimated purity by 12% SDS/PAGE being more than 95% (Yan et al., 2011). The high tendency of JNK1α1 to precipitate requires that the concentration of active and inactive JNK1α1 be maintained at less than 20 μM.
SUPPORT PROTOCOL 4
Expression, Purification and Activation of Tag-Less JNK2α2
Additional Materials (also see Support Protocols 1, 2, and 3)
Plasmid pET28a(+)-JNK2α2 vector is a construct encoding full-length wild-type Homo sapien mitogen-activated protein kinase 9 (GenBank accession number NM_002752) with N-terminal Thr (Thrombin) cleavable His-tag (Madsen et al., 2011); this plasmid is available upon request from Dr. K.N. Dalby (Division of Chemical Biology & Medicinal Chemistry; BME 6.202B; 1 University Station C0850; Austin, Texas 78712-1074; dalby@austin.utexas.edu)
Thrombin protease (Novagen)
Expression of His6-Thr-JNK2α2
Day 1: Electroporate pET28a(+)-JNK2 into the E. coli strain BL21(DE3) following the published DNA electroporation protocol (House et al., 2004).
Day 2: Use cells from single colony to inoculate 30 ml Luria-Broth media containing 30 μg/ml kanamycin. Grow overnight at 37°C.
Day 3: Dilute the culture 100-fold into Luria-Broth medium containing 30 μg/mL kanamycin. Grow the colony at 37°C to an OD600 of 0.6. Induce the cells by adding 500 μM IPTG and culture them for 3 to 5 hr at 30°C. Pellet the cells by centrifuging for 12 min at 7000 × g, 4°C, and then freeze the pellets immediately in liquid nitrogen and store up to one week at −80°C.
Purification of His6-Thr-JNK2α2
Day 4
-
4.
Lyse the cells in 150 mL buffer E containing 0.5 M NaCl and 1% Triton X-100.
-
5.
Sonicate cell lysate (power level 5, 50% duty cycle) for 10 min at 4°C (using 5-sec pulses at 5-sec intervals with careful monitoring of the temperature using a temperature probe to ensure it remains below 4°C).
-
6.
Centrifuge the lysate for 30 min at 20,100 × g, 4°C, and agitate the supernatant with Ni-NTA beads for 1 hr at 4°C.
-
7.
After washing the beads in a glass Econo-column (98 ml) with 150 mL buffer E containing 10 mM imidazole, elute the His6-tagged proteins with 25 mL buffer E (pH 8.0) containing 200 mM imidazole.
-
8.
Apply the eluted proteins to a Mono Q HR 10/10 anion-exchange column equilibrated in buffer F. Develop the column over 15 to 17 column volumes with a linear gradient of 0 to 0.5 M NaCl.
-
9.
Collect the eluted fractions of His6-Thr-JNK2α2 that correspond to the middle area of the Mono Q protein chromatogram and either process them to cleave the His6-tag or dialyze into buffer S containing 10 % glycerol. Measure the protein concentration, e.g., using Bradford reagent following manufacturer’s protocol.
-
10.
Immediately freeze in liquid nitrogen and store up to 6 months at −80°C for later use (the expected purity ~95%).
-
11.
Assess the purity by 12% SDS/PAGE (the expected purity ~95%).
Thrombin cleavage of His6-Thr-JNK2α2
-
12.
Dialyze the collected fractions of His6-Thr-JNK2α2 after Mono Q HR 10/10 anion-exchange chromatography against cleavage buffer G overnight.
Day 5
-
13.
Incubate with mild agitation the dialyzed protein with thrombin (1 U thrombin/mg protein) and 2.5 mM CaCl2 for 3 to 5 hr at room temperature.
-
14.
Estimate the cleavage by 10% SDS/PAGE, extending the incubation time as needed to achieve 100% cleavage.
-
15.
After cleavage, desalt the reaction mixture using HiPrep 26/10 desalting column (Amersham) equilibrated in buffer F.
-
16.
Filter the protein through 25-mm syringe filter with 0.45-μm nylon membrane and load onto a Mono Q HR 10/10 anion-exchange column equilibrated in buffer F. Develop the column over 15 to 17 column volumes with a linear gradient of 0 to 0.5 M NaCl.
-
17.
Collect the eluted fractions of tagless JNK2α2 and dialyze them into buffer S containing 10% glycerol. Measure the protein concentration, e.g., using Bradford reagent following the manufacturer’s protocol.
-
18.
Immediately freeze in liquid nitrogen and store up to 6 months at −80°C for later use.
The estimated yield is ~35 mg of pure tagless JNK2α2 per liter of culture. The estimated purity by 12% SDS/PAGE is greater than 95% (Madsen et al., 2011).
Activation of tagless JNK2α2
Day 6
-
19.
Incubate 2 μM tagless JNK2α2, 100 nM active GST- MKK4, and 400 nM active GST- MKK7 for 60 min at 30°C in the presence of 4 mM ATP in 10 mL activation buffer C.
-
20.
Desalt the reaction mixture using HiPrep 26/10 desalting column (Amersham) equilibrated in buffer D. Pool the eluted protein fractions from the desalting column, then concentrate them to 2 to 3 ml using an Amicon Ultra-15 concentrator (10,000 molecular weight cutoff). Centrifuge for 10 to 15 min at 3000 × g, 4°C.
-
21.
Purify the concentrated reaction mixture using a gel filtration column (120 mL HiLoad 16/60 Superdex 200 prep grade column) that has been equilibrated with buffer D.
-
22.
Measure the protein concentration, e.g., using Bradford reagent following the manufacturer’s protocol.
-
23.
Store the activated, tagless JNK2α2 in buffer S containing 10% glycerol up to 6 months at −80°C until further use.
At least 50% of tagless JNK2α2 should be recovered as active tagless JNK2α2. The estimated purity by 12% SDS/PAGE is greater than 95% (Madsen et al., 2011).
After activation, the gel filtration chromatography may show two different species of activated JNK2α2. These two species should be collected separately, and their activities tested against any known JNK substrate.
REAGENTS AND SOLUTIONS
Use deionized, distilled water in all recipes and protocol steps. All protein purification and activation buffers need to be freshly prepared in the same day of usage. For common stock solutions, see appendix 2a; for suppliers, see suppliers appendix.
Buffer A, pH 7.3
10 mM Na2HPO4
1.8 mM KH2PO4
140 mM NaCl
2.7 mM KCl
0.1% (v/v) 2-mercaptoethanol
0.1 mM TPCK
0.1 mM PMSF
1 mM benzamidine
Store up to 12 hours at 4°C
Buffer B, pH 7.5
50 mM Tris·Cl, pH 7.5
20 mM reduced glutathione
0.1% (v/v) 2-mercaptoethanol
0.1 mM TPCK
0.1 mM PMSF
1 mM benzamidine
Store up to 12 hours at 4°C
Buffer C, pH 7.5
25 mM HEPES
20 mM MgCl2
0.1 mM EDTA
0.1 mM EGTA
2 mM TCEP
Store up to 12 hours at 4°C
Buffer D, pH 7.5
25 mM HEPES
100 mM KCl
0.1 mM EDTA
0.1 mM EGTA
2 mM TCEP
Store up to 12 hours at 4°C
Buffer E, pH 8
20 mM Tris·Cl, pH 8
0.03% Brij-30
0.1% (v/v) 2-mercaptoethanol
5 mM imidazole
1 mM benzamidine
0.1 mM PMSF
0.1 mM TPCK
Store up to 12 hours at 4°C
Buffer F, pH 8.0
20 mM Tris·Cl, pH 8.0
0.03% (v/v) Brij-30
0.1% (v/v) 2-mercaptoethanol
Store up to one week, in closed bottle, at 4°C
Buffer G (pH 8.4)
20 mM Tris·Cl, pH 8.4
150 mM NaCl
0.1 % (v/v) 2-mercaptoethanol
Store up to one week, in closed bottle, at 4°C
Buffer S, pH 7.5
25 mM HEPES
50 mM KCl
0.1 mM EDTA
0.1 mM EGTA
2 mM DTT
Store up to 24 hours at 4°C
Kinase assay buffer, 10×
100 mM HEPES, pH 7.4
50 mM MgCl2
1 M NaCl
Store up to 1 month at room temperature
Lysis buffer
1% of 10% (w/v) Sodium dodecyl sulfate (SDS) solution (Bio-Rad, cat. no. 161-0416)
10 mM Tris·Cl, pH 7.5
10 mM of 500 nM sodium fluoride (NaF) stock solution
100 μM of 100 mM sodium orthovanadate (Na3VO4) solution
2 mM EDTA
2 mM of 1 M benzamidine (BA) stock solution
1 mM of 100 mM phenylmethanesulfonylfluoride (PMSF; fresh solution in DMSO)
TBST
10 mM Tris-HCl, pH 8.0
150 mM NaCl
0.05% Tween 20
COMMENTARY
Background Information
Two types of experiments necessary to study arrestin-3-dependent activation of JNK family kinases are described in this unit. One is performed in intact cells (Basic Protocol 2) whereas the other involves reconstitution of signaling modules from purified proteins (Basic Protocol 1). Arrestin-3-dependent JNK activation was initially studied in intact cells overexpressing JNK3 and other components of the cascade (ASK1 and/or MKK4) (McDonald et al., 2000). It was extensively studied in the same cell-based paradigm with kinase and arrestin-3 overexpression (Song et al., 2009; Seo et al., 2011; Breitman et al., 2012; Zhan et al., 2016; Perry-Hauser et al., 2022). The main limitation of this approach is that it is inadequate for proving that protein-protein interactions detected by co-immunoprecipitation, or the functional effects of expressed arrestin-3 and/or kinases involved, are direct rather than mediated by unknown intermediaries among the thousands of proteins expressed in all cells.
While the reconstitution of MKK-JNK signaling module with pure proteins is perfectly suited for the identification of direct interactions and effects, this approach became feasible only when methods for the purification and activation MKK4, MKK7, and different JNK isoforms, were developed (Support Protocols 1-4). As ASK1, the most upstream kinase, is unavailable in purified form, only the two-kinase part of the cascade can be reconstructed. Despite limitations, this approach has already revealed novel findings. First, it demonstrated the direct binding of MKK4 (Zhan et al., 2011b), and then MKK7 (Zhan et al., 2013b), to arrestin-3. Second, this approach demonstrated that free arrestin-3 in the absence of any GPCRs acts as a scaffold for the MKK4-JNK3 (Zhan et al., 2011b) and MKK7-JNK3 (Zhan et al., 2013b) modules. Third, it has been reported that JNK3 binding affects the affinity of arrestin-3 for MKKs, increasing the binding of MKK4 and decreasing that of MKK7 (Zhan et al., 2013b). Fourth, in vitro reconstitution demonstrated that arrestin-3 binds not only JNK3 with limited expression in a few cell types, but also ubiquitous JNK1 and JNK2 (Kook et al., 2014). Finally, experiments with pure proteins made it possible to identify the main JNK3-binding site in the N-domain of arrestin-3, as well as supporting sites in its C-domain (Zhan et al., 2014). The in vitro data were used to design experiments in intact cells to prove that these phenomena occur in native cell environments (Zhan et al., 2013b, 2016; Kook et al., 2014; Perry-Hauser et al., 2022).
Critical Parameters
Purification of MKK4 and MKK7
The GST tag can be cleaved on the resin or in solution. It is imperative to complete the purification process in one day using efficient protease inhibitors and to perform all purification steps in the cold room at 4°C.
Reconstitution in vitro
For the in vitro reconstruction of MKK-JNK cascades the concentration of each kinase should be kept constant at 50 nM for MKKs and 0.5 μM for JNKs. Changes in kinase concentrations shift the optimum concentration of arrestin-3.
Although purified arrestin-3, MKK4/7, and JNK1/2/3 survive freeze-thaw cycles fairly well, after thawing the samples on ice the aggregated inactive proteins should be removed by ultra-centrifugation at 356,000 × g for 60 min at 4°C. Re-measure the protein concentrations after carefully transferring the supernatants to new tubes, taking care to avoid any traces of pellet.
The role of arrestin-3 in JNK activation in intact cells
The most critical parameter in executing these protocols is the quality of starting cell cultures and DNA for transfection. The purified DNA must be transfection-quality. This is the case for most DNA purification kits. After prolonged storage (more than 6 months) at −20°C and/or multiple freeze-thaw cycles the DNA yields expression levels that may occasionally be lower than anticipated based on previous experiments. When this occurs a fresh batch of DNA should be prepared. For the most reproducible results the cell cultures must have the same cell numbers to maintain a consistent cell density at the time of transfection for each experiment. All of the transfection tables shown above are based on 6-well cell culture plate. To transfect cells in different formats, the number of cells and the amounts of medium, DNA, and FugeneHD (or another transfection reagent) should be altered in proportion to the difference in surface area of culture plate. To keep the amount of DNA and FugeneHD for transfection constant in all experiments, an empty vector must be used to balance the total DNA amount.
Because the activation of JNKs is detected by a phopho-JNK antibody, the addition of phosphatase inhibitors (NaF and Na3VO4) to the cell lysis buffer is essential for preventing JNK dephosphorylation.
Troubleshooting
Purification of MKK4, MKK7, JNK1, and JNK2
The manufacturer’s protocols for the Glutathione Sepharose High Performance (Amersham Biosciences), Ni-NTA beads (Qiagen), 120 ml HiLoad 16/60 Superdex 200 prep grade column (Amersham) and Mono Q HR 10/10 anion exchange column (Amersham) contain comprehensive troubleshooting instructions for these purification methods.
Reconstitution in vitro
Because of the modest binding affinities (micromolar) (Zhan et al., 2011b; Perry et al., 2019) of arrestin-3 for the kinase components, the arrestin-dependent increase in JNK activation is modest (~2 to 3-fold). If the protein concentrations in the reconstructed system are miscalculated, the observed effects of arrestin could be even smaller. In fact, arrestin-3 acts as a simple scaffold by bringing the two kinases together without activating either (Zhan et al., 2011b; Zhan et al., 2013b; Kook et al., 2014). Therefore, while lower arrestin-3 concentrations facilitate JNK phosphorylation by MKKs, higher concentrations inhibit it, apparently by shifting the equilibrium to incomplete (and therefore unproductive) complexes arrestin-3-MKK and arrestin-3-JNK (Zhan et al., 2011b; Zhan et al., 2013b; Kook et al., 2014). To address this problem, calculate protein concentrations carefully and optimize the arrestin concentration, which is much lower for MKK4 than for MKK7 (Zhan et al., 2013b).
Another problem that is often encountered is the strong background in the [γ-32P]ATP assay. Experience suggests that the addition of 1-2 mM EDTA into the SDS-PAGE buffer and gel substantially decreases the noise caused by free [γ -32P]ATP.
The role of arrestin-3 in JNK activation in intact cells
Expression of HA-ASK1, HA-MKK4, Flag-MKK7 and arrestin3 in COS-7 cells may vary depending on the state of cells (confluence, passage number, etc.), the quality of the DNA, and the number of co-expressed proteins. The investigator should therefore test the expression of each protein with different amounts of DNA to determine the optimal range of each DNA for transfection, using the tables provided in this unit as a guide. Arrestin-3-dependent JNK3 activation is also observed without ASK1 overexpression, where only endogenous upstream kinases are involved (Perry-Hauser et al., 2022).
Anticipated Results
Reconstitution in vitro
The strategy employed for reconstituting MAPK cascades can be readily applied to other signaling systems involving three to four protein components that interact with one another (Fig. 2.12.1). The biphasic dependence of the output on scaffold concentration should be observed if the assays are carefully designed. The optimum concentration of the scaffold protein is determined chiefly by the affinities of the binding partners for each other. Therefore, this type of assay can be used to compare the binding affinities of different scaffold proteins for the kinases. It also can be used to evaluate the effects of dynamic movements (conformations) of scaffold proteins in signaling pathways.
The role of arrestin-3 in JNK activation in intact cells
Phospho-JNK (Thr183/Tyr185) antibody is used to detect endogenous levels of p46 and p54 JNK isoforms dually phosphorylated at threonine 183 and tyrosine 185. The level of the phosphorylation of endogenous JNK1/2 isoforms (p46 and p54) in cells transfected with empty pcDNA3 vector can be used as a control for the basal JNK activity. Depicted on Figure 2.12.4 is the gradual increase of HA-ASK1 expression in COS-7 cells with the increase of DNA concentration to be used for transfection procedure. Increasing amounts of HA-ASK1 (Fig. 2.12.4) or Flag-MKK7 (Fig. 2.12.5) result in a concentration-dependent increase of the phosphorylation levels of the p46 and p54 JNK isoforms. Anisomycin treatment inhibits protein synthesis and activates cellular stress pathways. Among other effects, anisomycin induces phosphorylation of endogenous JNKs via an arrestin-independent mechanism. Therefore, anisomycin-treated cells can be used as a source of doubly phosphorylated JNKs western blotting. Serial dilutions of anisomycin (1 μg/mL)-treated HEK-A cell lysates can be used to ensure that the intensity of ppJNK bands in all samples is in the linear range. To evaluate the arrestin-3 effect, the phosphorylation of p46 and p54 JNK isoforms induced by increasing amounts of HA-ASK1 (Fig. 2.12.4) or Flag-MKK7 (Fig. 2.12.5) can be quantified and compared with or without arrestin3 co-expression.
Time Considerations
Reconstitution in vitro
Once the proteins are purified the experiments can be completed fairly quickly. However, an extra hour is needed for high-speed centrifugation of thawed proteins to eliminate aggregates (see Critical Parameters) and subsequent measurements of protein concentrations in the supernatants. As a 10-sec reaction time is recommended, the reactions must be initiated and terminated individually in sequence. Appropriate reaction times can be adjusted based on the activity of upstream kinases. A 5- to 60-sec time scale should work in similar assay settings for JNKs and other MAP kinases. After premixing the kinases with arrestin it is important to incubate the mixtures for at least 15 min at 30°C to allow for equilibration of free proteins and complexes before the addition of ATP.
The role of arrestin-3 in JNK activation in intact cells
The experiment should be planned to allow plating of the cells one day before transfection. Four days are needed for the entire process of transfection, subsequent culture to express proteins in cells, and sample preparation. The procedure can be halted after cell lysis. Samples should be stored at −20°C or −80°C and used later for immunoblotting. The quality of frozen samples is maintained for many days, but could deteriorate after prolonged storage.
Figure 2.12.6.
Arrestin-3 enhances ASK1- and MKK7-dependent phosphorylation of endogenous JNK1/2 in intact cells. (A,C) Representative immunoblots showing phosphorylation of endogenous JNK1/2 isoforms in cells expressing ASK1 (A) or MKK7 (C) in the presence of increasing concentrations of arrestin-3. (B,D) Quantification of the levels of p46H JNK isoform phosphorylation. One-way ANOVA analysis with arrestin-3 concentration as factor yielded significant effect of arrestin-3 on JNK phosphorylation both in the presence of ASK1 and MKK7 (p <0.0001). ** –p < 0.01, * –p <0.05 to the value at 0 arrestin-3; b –p < 0.01, c –p < 0.001 to the maximal value (at 1 or 4 μg of arrestin-3 DNA), according to Bonferroni/Dunn post-hoc test. Data from Kook et al. (2014).
Table 2.12.8.1.
Transfection to evaluate the effect of T1A on JNK Activation by MKK4 (Figure 2.12.6 C) a
| pcDNA3-HA-MKK4 | 0 μg | 0.2 μg | 0.5 μg | 1.0 μg |
|---|---|---|---|---|
| pcDNA3 | 4.0 μg | 3.8 μg | 3.5 μg | 3.0 μg |
| pcDNA-YFP | 0 μg | 0.2 μg | 0 μg | 0 μg |
| pcDNA-YFP | 0 μg | 0 μg | 0.35 μg | 0 μg |
| pcDNA3-arrestin-3 | 0 μg | 0 μg | 0 μg | 0.5 μg |
| FuGENE HD | 13.5 μl | 13.5 μl | 13.5 μl | 13.5 μl |
| Opti-MEM | 125 μl | 125 μl | 125 μl | 125 μl |
For MKK7 induced JNK activation, add pcDNA3-Flag-MKK7 instead of pcDNA3-HA-MKK4
Acknowledgements
NIH grants EY011500, GM122491 (VVG); NS065868 and DA030103 (EVG); GM059802 and Welch Foundation (F-1390) (KND); TSK was supported by a postdoctoral trainee fellowship from Cancer Prevention Research Institute of Texas (CPRIT). VVG was also supported by Cornelius Vanderbilt Endowed Chair (Vanderbilt University).
Literature Cited
- Abramczyk O, Tavares CD, Devkota AK, Ryazanov AG, Turk BE, Riggs AF, Ozpolat B, and Dalby KN 2011. Purification and characterization of tagless recombinant human elongation factor 2 kinase (eEF-2K) expressed in Escherichia coli. Protein Expr. Purif 79:237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Curto E, Inoue A, Jenkins L, Raihan SZ, Prihandoko R, Tobin AB, Milligan G 2016. Targeted Elimination of G Proteins and Arrestins Defines Their Specific Contributions to Both Intensity and Duration of G Protein-coupled Receptor Signaling. J Biol Chem 291:27147–27159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitman M, Kook S, Gimenez LE, Lizama BN, Palazzo MC, Gurevich EV, and Gurevich VV 2012. Silent scaffolds: Inhibition of c-Jun N-terminal kinase 3 activity in the cell by a dominant-negative arrestin-3 mutant. J. Biol. Chem 287:19653–19664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burack WR and Shaw AS 2000. Signal transduction: Hanging on a scaffold. Curr. Opin. Cell Biol 12:211–216. [DOI] [PubMed] [Google Scholar]
- Dhanasekaran DN, Kashef K, Lee CM, Xu H, and Reddy EP 2007. Scaffold proteins of MAP-kinase modules. Oncogene 26:3185–3202. [DOI] [PubMed] [Google Scholar]
- Donoso LA, Gregerson DS, Smith L, Robertson S, Knospe V, Vrabec T, and Kalsow CM 1990. S-antigen: Preparation and characterization of site-specific monoclonal antibodies. Curr. Eye Res 9:343–355. [DOI] [PubMed] [Google Scholar]
- Gallagher S (electrophoresis, staining) and Sasse J (staining) 2001. Protein Analysis by SDS-PAGE and Detection by Coomassie Blue or Silver Staining. Curr. Protoc. Pharmacol 2:3B:A.3B.1–A.3B.10. [DOI] [PubMed] [Google Scholar]
- Gallagher ED, Xu S, Moomaw C, Slaughter CA, and Cobb MH 2002. Binding of JNK/SAPK to MEKK1 is regulated by phosphorylation. J. Biol. Chem 277:45785–45792. [DOI] [PubMed] [Google Scholar]
- Gonzales MF, Brooks T, Pukatzki SU, and Provenzano D 2013. Rapid protocol for preparation of electrocompetent Escherichia coli and Vibrio cholerae. J. Vis. Exp [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Barrett T, Whitmarsh AJ, Cavanagh J, Sluss HK, Dérijard B, and Davis RJ 1996. Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J 15:2760–2770. [PMC free article] [PubMed] [Google Scholar]
- House BL, Mortimer MW, and Kahn ML 2004. New recombination methods for Sinorhizobium meliloti genetics. Appl. Environ. Microbiol 70:2806–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GL 2011. Defining MAPK interactomes. ACS Chem. Biol 6:18–20. [DOI] [PubMed] [Google Scholar]
- Khokhlatchev A, Xu S, English J, Wu P, Schaefer E, and Cobb MH 1997. Reconstitution of mitogen-activated protein kinase phosphorylation cascades in bacteria. Efficient synthesis of active protein kinases. J. Biol. Chem 272:11057–11062. [DOI] [PubMed] [Google Scholar]
- Kook S, Zhan X, Kaoud TS, Dalby KN, Gurevich VV, and Gurevich EV 2014. Arrestin-3 binds JNK1α1 and JNK2α2 and facilitates the activation of these ubiquitous JNK isoforms in cells via scaffolding. J. Biol. Chem 289:in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler S, Fleming Y, Goedert M, and Cohen P 1998. Synergistic activation of SAPK1/JNK1 by two MAP kinase kinases in vitro. Curr. Biol. 8:1387–1390. [DOI] [PubMed] [Google Scholar]
- Levchenko A, Bruck J, and Sternberg PW 2000. Scaffold proteins may biphasically affect the levels of mitogen-activated protein kinase signaling and reduce its threshold properties. Proc. Natl. Acad. Sci. U.S.A 97:5818–5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko A, Bruck J, and Sternberg PW 2004. Regulatory modules that generate biphasic signal response in biological systems. Syst. Biol. (Stevenage) 1:139–148. [DOI] [PubMed] [Google Scholar]
- Lisnock J, Griffin P, Calaycay J, Frantz B, Parsons J, O’Keefe SJ, and LoGrasso P 2000. Activation of JNK3α1 Requires both MKK4 and MKK7: Kinetic characterization of in vitro phosphorylated JNK3α1. Biochemistry 39:3141–3148. [DOI] [PubMed] [Google Scholar]
- Madsen JA, Kaoud TS, Dalby KN, and Brodbelt JS 2011. 193-nm photodissociation of singly and multiply charged peptide anions for acidic proteome characterization. Proteomics 11:1329–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald PH, Chow CW, Miller WE, Laporte SA, Field ME, Lin FT, Davis RJ, and Lefkowitz RJ 2000. Beta-arrestin 2: A receptor-regulated MAPK scaffold for the activation of JNK3. Science 290:1574–1577. [DOI] [PubMed] [Google Scholar]
- Miller WE, McDonald PH, Cai SF, Field ME, Davis RJ, and Lefkowitz RJ 2001. Identification of a motif in the carboxyl terminus of beta -arrestin2 responsible for activation of JNK3. J. Biol. Chem 276:27770–27777. [DOI] [PubMed] [Google Scholar]
- Olson BJ and Markwell J 2007. Assays for Determination of Protein Concentration. Curr. Protoc. Pharmacol 38:3A:A.3A.1–A.3A.29. [DOI] [PubMed] [Google Scholar]
- Perry NA, Kaoud TS, Ortega OO, Kaya AI, Marcus DJ, Pleinis JM, Berndt S, Chen Q, Zhan X, Dalby KN, Lopez CF, Iverson TM, Gurevich VV 2019. Arrestin-3 scaffolding of the JNK3 cascade suggests a mechanism for signal amplification. Proc Natl Acad Sci U S A 116:810–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry-Hauser NA, Kaoud TS, Stoy H, Zhan X, Chen Q, Dalby KN, Iverson TM, Gurevich VV, Gurevich EV 2022. Short Arrestin-3-Derived Peptides Activate JNK3 in Cells. Int J Mol Sci 23:8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J, Tsakem EL, Breitman M, and Gurevich VV 2011. Identification of arrestin-3-specific residues necessary for JNK3 activation. J. Biol. Chem 286:27894–27901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Coffa S, Fu H, and Gurevich VV 2009. How does arrestin assemble MAPKs into a signaling complex? J. Biol. Chem 284:685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Vishnivetskiy SA, Seo J, Chen J, Gurevich EV, and Gurevich VV 2011. Arrestin-1 expression in rods: Balancing functional performance and photoreceptor health. Neuroscience 174:37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnivetskiy SA, Zhan X, Chen Q, Iverson TI, and Gurevich VV 2014. Arrestin expression in E. coli and purification. Curr. Protocols Pharmacol in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmann C, Gibson S, Jarpe MB, and Johnson GL 1999. Mitogen-activated protein kinase: Conservation of a three-kinase module from yeast to human. Physiol. Rev 79:143–180. [DOI] [PubMed] [Google Scholar]
- Willoughby EA, Perkins GR, Collins MK, and Whitmarsh AJ 2003. The JNK-interacting protein-1 scaffold protein targets MAPK phosphatase-7 to dephosphorylate JNK. J. Biol. Chem 278:10731–10736. [DOI] [PubMed] [Google Scholar]
- Yan C, Kaoud T, Lee S, Dalby KN, and Ren P 2011. Understanding the specificity of a docking interaction between JNK1 and the scaffolding protein JIP1. J. Phys. Chem. B 115:1491–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Gimenez LE, Gurevich VV, and Spiller BW 2011a. Crystal structure of arrestin-3 reveals the basis of the difference in receptor binding between two non-visual arrestins. J. Mol. Biol/ 406:467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Kaoud TS, Dalby KN, and Gurevich VV 2011b. Non-visual arrestins function as simple scaffolds assembling MKK4- JNK3α2 signaling complex. Biochemistry 50:10520–10529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Kaoud TS, Dalby KN, and Gurevich VV 2011c. Nonvisual arrestins function as simple scaffolds assembling the MKK4-JNK3alpha2 signaling complex. Biochemistry 50:10520–10529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Kaoud TS, Kook S, Dalby KN, and Gurevich VV 2013a. JNK3 binding to arrestin-3 differentially affects the recruitment of upstream MAP kinase kinases. J. Biol. Chem/ 288:28535–28547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Kaoud TS, Kook S, Dalby KN, and Gurevich VV 2013b. JNK3 enzyme binding to arrestin-3 differentially affects the recruitment of upstream mitogen-activated protein (MAP) kinase kinases. J. Biol. Chem 288:28535–28547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Perez A, Gimenez LE, Vishnivetskiy SA, and Gurevich VV 2014. Arrestin-3 binds the MAP kinase JNK3α2 via multiple sites on both domains. Cell Signal. 26:766–776. [DOI] [PMC free article] [PubMed] [Google Scholar]