Abstract
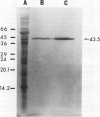
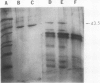
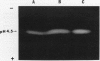
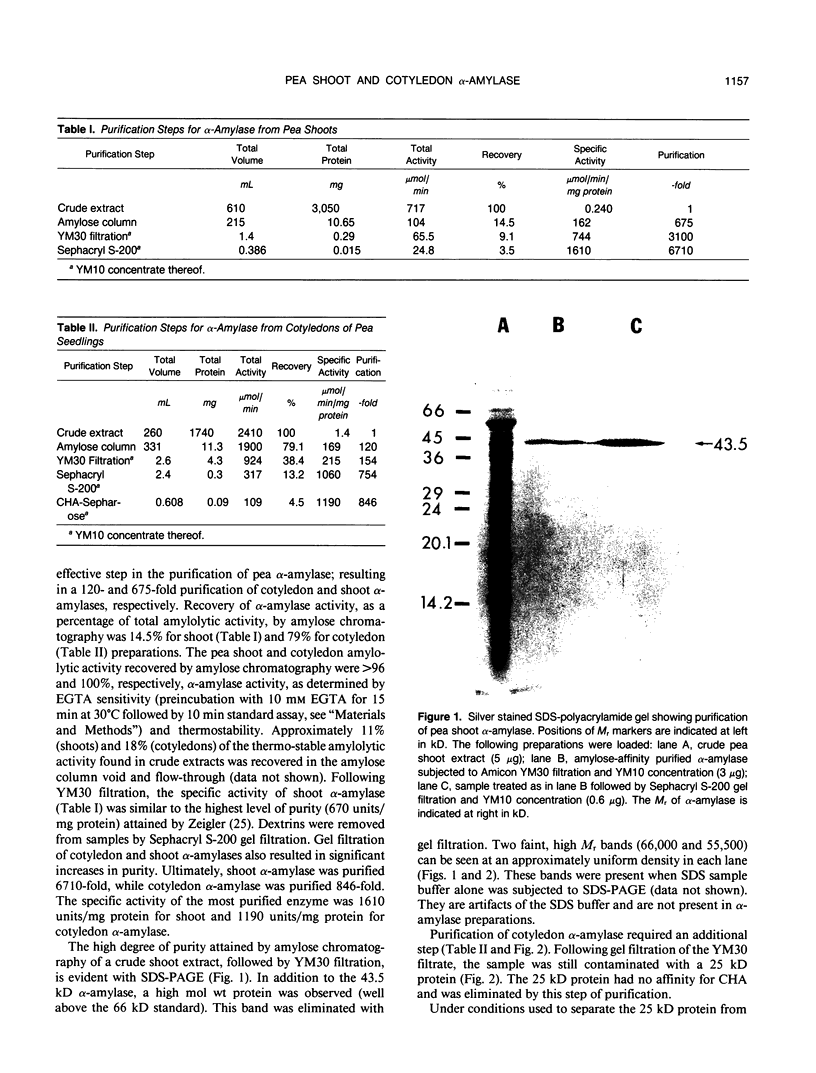
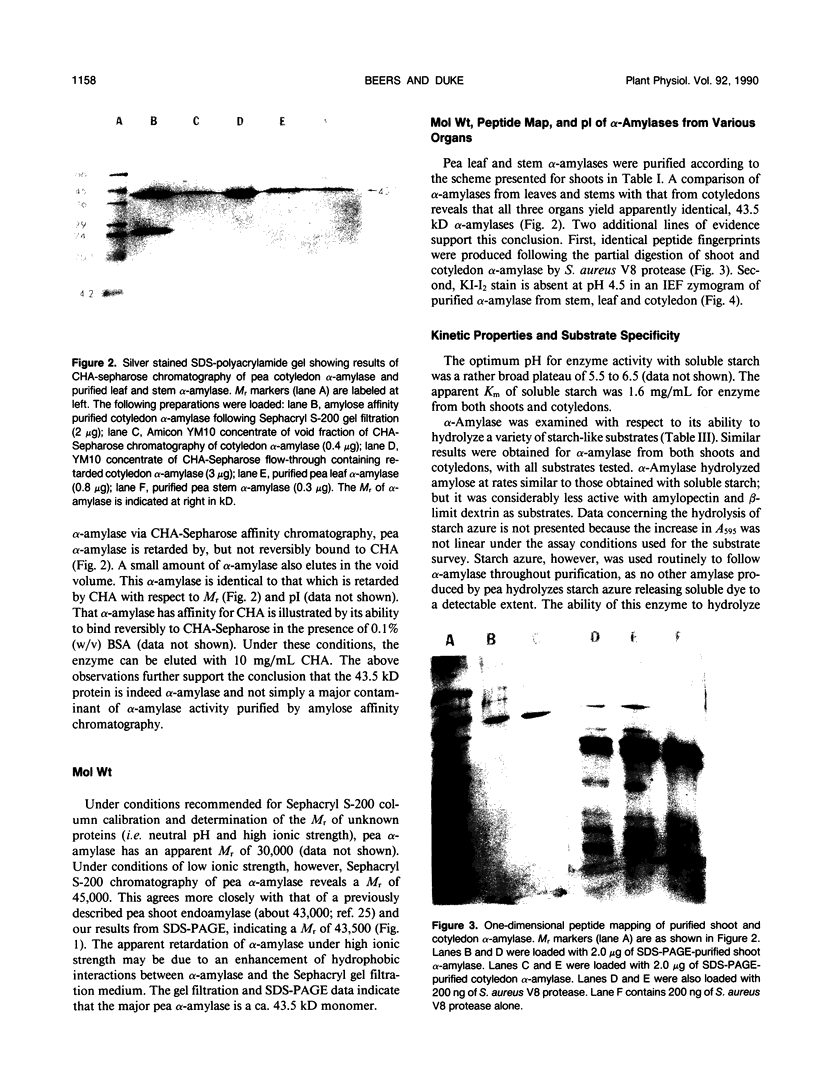
The most abundant α-amylase (EC 3.2.1.1) in shoots and cotyledons from pea (Pisum sativum L.) seedlings was purified 6700-and 850-fold, respectively, utilizing affinity (amylose and cycloheptaamylose) and gel filtration chromatography and ultrafiltration. This α-amylase contributed at least 79 and 15% of the total amylolytic activity in seedling cotyledons and shoots, respectively. The enzyme was identified as an α-amylase by polarimetry, substrate specificity, and end product analyses. The purified α-amylases from shoots and cotyledons appear identical. Both are 43.5 kilodalton monomers with pls of 4.5, broad pH activity optima from 5.5 to 6.5, and nearly identical substrate specificities. They produce identical one-dimensional peptide fingerprints following partial proteolysis in the presence of SDS. Calcium is required for activity and thermal stability of this amylase. The enzyme cannot attack maltodextrins with degrees of polymerization below that of maltotetraose, and hydrolysis of intact starch granules was detected only after prolonged incubation. It best utilizes soluble starch as substrate. Glucose and maltose are the major end products of the enzyme with amylose as substrate. This α-amylase appears to be secreted, in that it is at least partially localized in the apoplast of shoots. The native enzyme exhibits a high degree of resistance to degradation by proteinase K, trypsin/chymostrypsin, thermolysin, and Staphylococcus aureus V8 protease. It does not appear to be a high-mannose-type glycoprotein. Common cell wall constituents (e.g. β-glucan) are not substrates of the enzyme. A very low amount of this α-amylase appears to be associated with chloroplasts; however, it is unclear whether this activity is contamination or α-amylase which is integrally associated with the chloroplast.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beers E. P., Duke S. H. Localization of alpha-Amylase in the Apoplast of Pea (Pisum sativum L.) Stems. Plant Physiol. 1988 Aug;87(4):799–802. doi: 10.1104/pp.87.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Chao S. E., Scandalios J. G. Developmentally dependent expression of tissue specific amylases in maize. Mol Gen Genet. 1972;115(1):1–9. doi: 10.1007/BF00272211. [DOI] [PubMed] [Google Scholar]
- Cleveland D. W. Peptide mapping in one dimension by limited proteolysis of sodium dodecyl sulfate-solubilized proteins. Methods Enzymol. 1983;96:222–229. doi: 10.1016/s0076-6879(83)96020-2. [DOI] [PubMed] [Google Scholar]
- Doehlert D. C., Duke S. H. Specific Determination of alpha-Amylase Activity in Crude Plant Extracts Containing beta-Amylase. Plant Physiol. 1983 Feb;71(2):229–234. doi: 10.1104/pp.71.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duke S. H., Schrader L. E., Henson C. A., Servaites J. C., Vogelzang R. D., Pendleton J. W. Low root temperature effects on soybean nitrogen metabolism and photosynthesis. Plant Physiol. 1979 May;63(5):956–962. doi: 10.1104/pp.63.5.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fett W. F., Dunn M. F. Exopolysaccharides Produced by Phytopathogenic Pseudomonas syringae Pathovars in Infected Leaves of Susceptible Hosts. Plant Physiol. 1989 Jan;89(1):5–9. doi: 10.1104/pp.89.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen J. V., Hanson A. D., Chandler P. C. Water stress enhances expression of an alpha-amylase gene in barley leaves. Plant Physiol. 1986 Feb;80(2):350–359. doi: 10.1104/pp.80.2.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakefuda G., Duke S. H. Characterization of Pea Chloroplast D-Enzyme (4-alpha-d-Glucanotransferase). Plant Physiol. 1989 Sep;91(1):136–143. doi: 10.1104/pp.91.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakefuda G., Duke S. H. Electrophoretic transfer as a technique for the detection and identification of plant amylolytic enzymes in polyacrylamide gels. Plant Physiol. 1984 May;75(1):278–280. doi: 10.1104/pp.75.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Morrissey J. H. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal Biochem. 1981 Nov 1;117(2):307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- Swain R. R., Dekker E. E. Seed germination studies. I. Purification and properties of an alpha-amylase from the cotyledons of germinating peas. Biochim Biophys Acta. 1966 Jul 6;122(1):75–86. doi: 10.1016/0926-6593(66)90092-0. [DOI] [PubMed] [Google Scholar]
- Trimble R. B., Maley F. Optimizing hydrolysis of N-linked high-mannose oligosaccharides by endo-beta-N-acetylglucosaminidase H. Anal Biochem. 1984 Sep;141(2):515–522. doi: 10.1016/0003-2697(84)90080-0. [DOI] [PubMed] [Google Scholar]
- Yomo H., Varner J. E. Control of the formation of amylases and proteases in the cotyledons of germinating peas. Plant Physiol. 1973 Apr;51(4):708–713. doi: 10.1104/pp.51.4.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler P., Beck E. Exoamylase activity in vacuoles isolated from pea and wheat leaf protoplasts. Plant Physiol. 1986 Dec;82(4):1119–1121. doi: 10.1104/pp.82.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler P. Partial purification and characterization of the major endoamylase of mature pea leaves. Plant Physiol. 1988 Mar;86(3):659–666. doi: 10.1104/pp.86.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]