Abstract
Fungal pathogens have been designated by the World Health Organization as microbial threats of the highest priority for global health. It remains a major challenge to improve antifungal efficacy at the site of infection while avoiding off-target effects, fungal spreading, and drug tolerance. Here, we develop a nanozyme-based microrobotic platform that directs localized catalysis to the infection site with microscale precision to achieve targeted fungal killing. Using electromagnetic field frequency modulation and fine-scale spatiotemporal control, structured iron oxide nanozyme assemblies are formed that display tunable dynamic shape transformation and catalysis activation. The catalytic activity varies depending on the motion, velocity, and shape providing controllable reactive oxygen species (ROS) generation. Unexpectedly, nanozyme assemblies bind avidly to fungal (Candida albicans) cell surfaces to enable concentrated accumulation and targeted ROS-mediated killing in situ. By exploiting these tunable properties and selective binding to fungi, localized antifungal activity is achieved using in vivo-like cell spheroid and animal tissue infection models. Structured nanozyme assemblies are directed to C. albicans-infected sites using programmable algorithms to perform precisely guided spatial targeting and on-site catalysis resulting in rapid fungal eradication. This nanozyme-based microrobotics approach provides a uniquely effective and targeted therapeutic modality for pathogen elimination at the infection site.
Keywords: Candida albicans, biofilms, iron oxide, microrobots, assemblies
Graphical Abstract
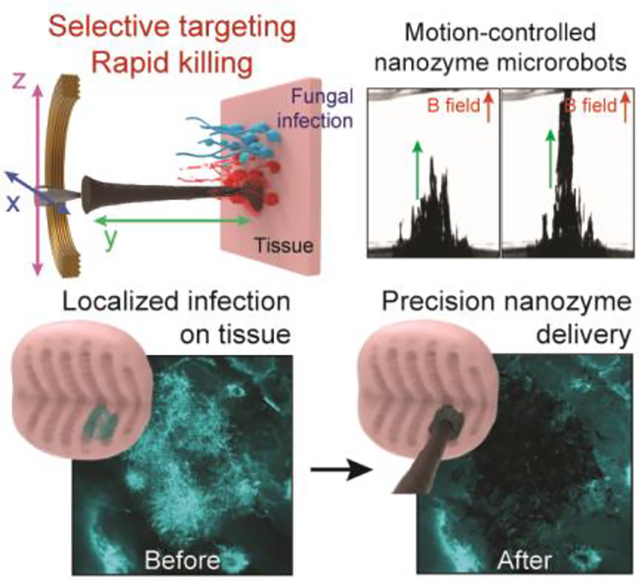
We develop a microrobotic platform using iron oxide nanoparticle-based nanozymes to achieve precise spatial targeting and localized catalysis for rapid fungal killing. These nanozyme-microrobots demonstrate programmable motions, controllable catalysis, and selective binding to fungi. Using tissue infection models, we show localized delivery of nanozymes and targeted fungal eradication, providing a unique antifungal modality for pathogen killing at the infection site.
1. Introduction
Fungi are notoriously resistant to treatments and are a major public-health concern worldwide.[1,2] Recently, the World Health Organization (WHO) has designated fungi such as Candida albicans as microbial threats of the highest priority for global health despite current antifungal therapies, which indicates alternative approaches are urgently needed.[3,4] Nanomaterials have gained increased attention for potential applications as antifungal agents.[5,6] However, the killing potency of nano-drugs is often limited requiring prolonged (hours-long) treatment time while lacking fungal targeting specificity. Hence, drug delivery with enhanced selectivity to the infection site and rapid fungal eradication remain major challenges to avoid off-target effects, as well as to prevent drug resistance development and pathogen spreading.[4]
Recent progress in catalytic nanoparticles (termed nanozymes) with magnetic properties has led to new treatments against bacterial biofilms with precision and efficacy that could be leveraged for treatment of fungal infections.[7–13] Specifically, iron oxide nanoparticles (IONP) have been employed against bacterial pathogens associated with several biofilm infections due to high peroxidase-like activity that activates hydrogen peroxide to generate reactive oxygen species (ROS) with potent biofilm killing effects.[14,15] Furthermore, the high magnetic susceptibility of IONP permits precise controllability to enhance localized delivery of nanoparticles and antibiofilm efficacy.[7,13] These properties combined with proven biocompatibility, facile synthesis/scalability, and flexible surface functionalization/coating render IONP as an attractive nanomaterial for biomedical applications.[16–18] However, nanozyme-based treatment against fungal infections remains underexplored.
Candida albicans is one of the most common and widespread fungal organisms causing human infections.[2] It is an opportunistic pathogen that forms highly localized biofilm infections on both implanted surfaces and soft tissues that could be targeted using the nanozyme approach.[19,20] Recently, we combined the catalytic and magnetic properties of iron oxide nanoparticles to create small scale antibiofilm robots that demonstrated effective killing and removal of bacterial pathogens.[7,13] Magnetic robotic actuation is attractive because it allows tether-free controlled motion and enables a wide variety of robotic locomotion strategies to precisely deliver catalytic action,[21–23] which could be leveraged for spatial targeting of localized fungal infections.
Here, we create microrobotic assemblies composed of nanozymes using fine-scale spatiotemporal control for precise targeting of infection sites and localized catalysis to achieve effective fungal killing on-site. Using electromagnetic field frequency and positioning modulation, we assemble morphology-adjustable nanozyme microrobots that can freely extend, retract, roll, vibrate and glide with high spatiotemporal precision. The catalytic activity of these nanozyme assemblies varies depending on the motion, velocity and shape providing on-site reactive oxygen species (ROS) generation. Furthermore, we unexpectedly find that nanozyme assemblies bind avidly to fungal cells (C. albicans), more specifically than to human (gingival) cells. This selective binding interaction to fungi promotes localized accumulation of high amounts of nanozymes and targeted ROS generation in situ, potentiating the antifungal effect. We demonstrate directed delivery and catalytic killing of fungal cells within 10 minutes through cell-nanozyme binding and spatial targeting using cell spheroid and tissue infection models. The nanozyme-based robotics approach results in rapid and precise C. albicans eradication, providing a uniquely effective and targeted antifungal modality for pathogen killing at the infection site.
2. Results and discussion
2.1. Preparation and assembly of nanozyme microrobotic assemblies
We leveraged the efficient catalysis of iron oxide nanoparticles (termed nanozymes)[24] and their magnetic properties to create shape-tunable assemblies using electromagnetic field frequency modulation and fine-scale spatiotemporal positioning, while preserving their catalytic activity (Figure 1). Conceptually, we combine nanozyme properties with robotic principles of controllability and automation to develop nanozyme-based microrobots capable of targeting and treating localized fungal infections at multiple length scales (Figure 1A).
Figure 1. Assembly, control, and functional properties of robotic nanozyme assemblies and their mode of action.
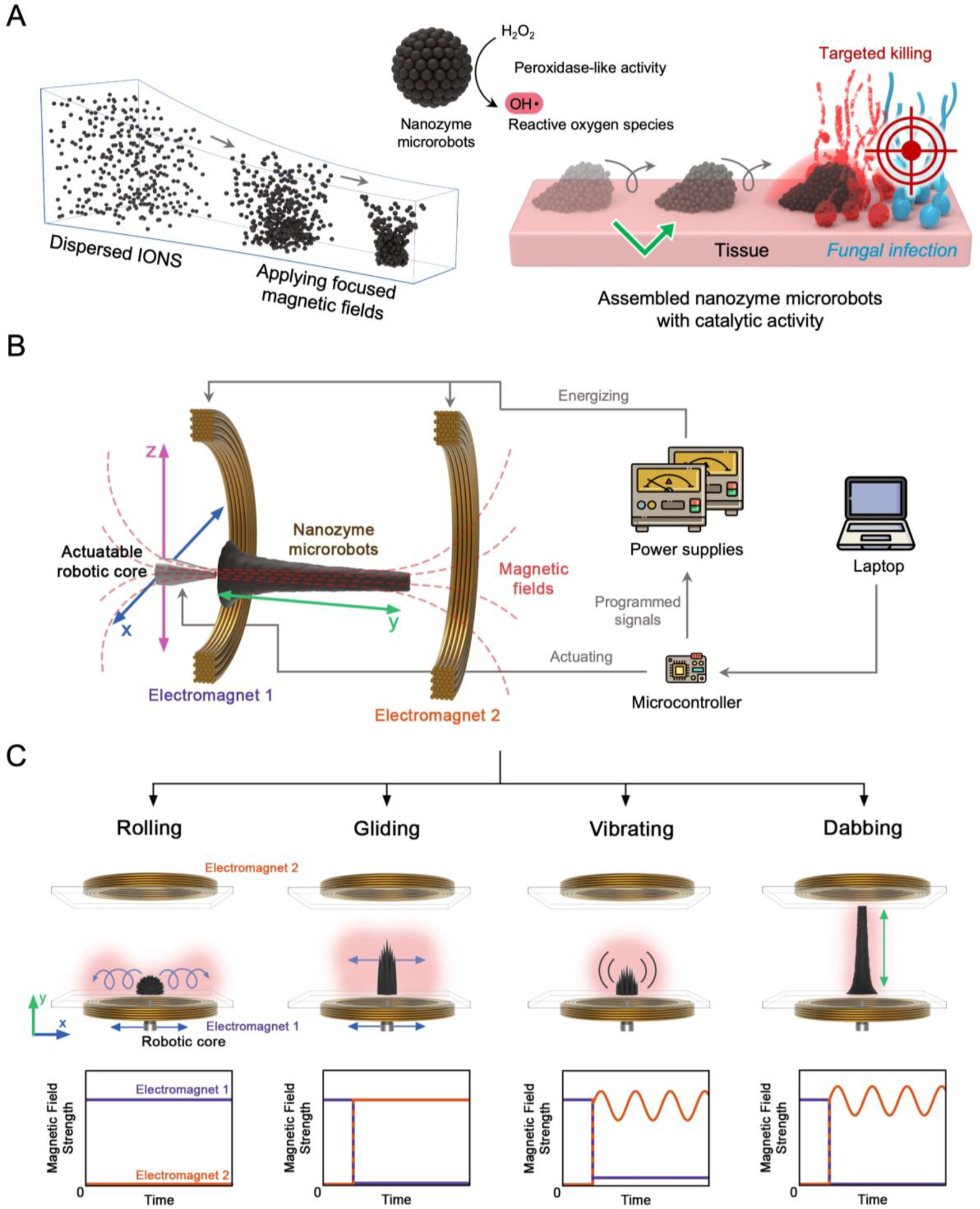
(A) On-site assembly of individual nanozymes into catalytically active superstructures. The motion dynamics, morphology, and the location of catalysis of the structured assemblies can be controlled creating nanozyme microrobots for targeting fungal infection. (B) Electromagnet cores guide the nanozyme microrobot assemblies with controllable morphology, position, and motion using programmed algorithms. (C) Programmable dynamic motions via magnetic field modulation enable controlled catalytic activities and targeted treatment.
Our approach is based on on-site assembly of individual nanozymes into a set of specific reconfigurable superstructures using a custom-built electromagnet array with multi-axis motion. The experimental setup is shown schematically in Figure 1B. Two coaxially arranged programmable electromagnets are located on either side of a customized container, and an iron core is positioned at the center of electromagnet-1. IONPs made in-house are building blocks for nanozyme-microrobots (Figure S1, Supporting Information). The aqueous IONP dispersions are transferred to the container in the working space between the electromagnets. Each of the nanozyme-microrobot configurations is initiated by applying a magnetic field to the dispersion, which is produced by passing direct current through electromagnet-1 on the side with the actuatable core. The magnetic field generated from electromagnet-1 induces the magnetization of the iron core and thus the localized magnetic field density near the core within the container, which induces a sea-urchin-shaped nanozyme assembly next to the iron core. This collection step initializes each of the formations and motions used in these studies.
The shape and motion of the nanozyme microrobots are controlled according to the applied magnetic field from electromagnets, which is harmonized via a programmable microcontroller. According to the ratio of the magnetic field strength generated by the electromagnet-1 and electromagnet-2, the nanozyme assemblies can be variably extended along the y-axis from the core side, while oscillating components of the magnetic field enable both structural vibration, as well as enhanced controllability of extension. In addition, the x-z position of the nanozyme assemblies is coupled with the position of the iron core, which can also be adjusted by the microcontroller. This controllability allows abundant automated multimodal three-dimensional (3D) motions of nanozyme microrobots.
Although catalytic activities of dispersed IONP have been well-studied, little is known about the catalysis of structured, mesoscale nanozyme assemblies. We therefore first study various magnetic field configurations and corresponding motions as well as the catalytic properties of nanozyme microrobots using the custom-built electromagnetic device. We develop 4 different dynamic motions including rolling, vibrating, gliding, and dabbing (Figure 1C), which can generate ROS dynamically at different spatial regions and deliver nanozymes through shape transformations. The rolling motion is induced by translating the initial sea-urchin-like structure across the container surface. For this motion, the position of the electromagnet core is varied while electromagnet-1 is continuously energized with a constant current. The characteristic rolling motion is caused by a balance between container wall friction, which anchors domains of the structured assemblies near the wall, and recirculation of nanozyme domains on the outer surface of the assembly. The gliding motion is induced by energizing electromagnet-2 after the initial collection step, which causes extension of the structured assemblies along the central axis of the electromagnets. In this form, the entire high aspect ratio structure glides along the surface as the electromagnet core is actuated. The rolling and gliding motions can be used for convective fluidic mixing and distribution of generated ROS within the spatial range of actuation.
To achieve the vibration motion, electromagnet-2 is energized with a bias current as well as an oscillating component. This causes the structural ‘spikes’ to rapidly oscillate alternately inward and outward relative to the central axis. Lastly, the dabbing motion is characterized by controllable linear extension along the y-axis. This motion is enabled by applying a simultaneous bias field and oscillating field to electromagnet-2 after an initial x-z plane positioning step. The vibration and dabbing motions are useful for precisely delivering nanozyme catalysis to specific target locations on distal surfaces.
Next, we investigate peroxidase-like catalytic activity of nanozyme microrobots with three representative dynamic motions (rolling, vibrating, and gliding) using the colorimetric TMB (3,3′,5,5′-tetramethylbenzidine) assay. Nanozyme can oxidize colorless TMB to blue-colored oxidized TMB in the presence of H2O2 via ROS generation from H2O2 catalysis. The oxidized TMB can be quantitatively determined by measuring the absorbance values at 652 nm.[24]
To determine the catalytic activity according to the three representative dynamic motions of nanozyme assemblies with the same amount of iron oxide nanoparticles (1.0 mg mL−1), linear velocities from 0 to 12 mm s−1 with 10 mm path lengths along the x-axis are tested for the rolling and gliding motions, and frequencies from 0 to 16 Hz are considered for the vibrating motion (Figure 2 A,B). We find that faster linear velocities and higher frequencies lead to more vigorous catalytic reactions for all dynamic motions, whereas minimal catalysis is observed in static conditions (Figure 2 C–E). Hence, dynamic motions activate ROS generation by the nanozyme assemblies and increase proportionally to linear velocities, leading to significantly enhanced catalytic activity. In the case of rolling and gliding, the highest catalytic activity is about 7 and 6 times greater than that of the vibrating nanozyme microrobot, respectively (Figure 2 F–H).
Figure 2. Catalytic properties of the nanozyme microrobots.
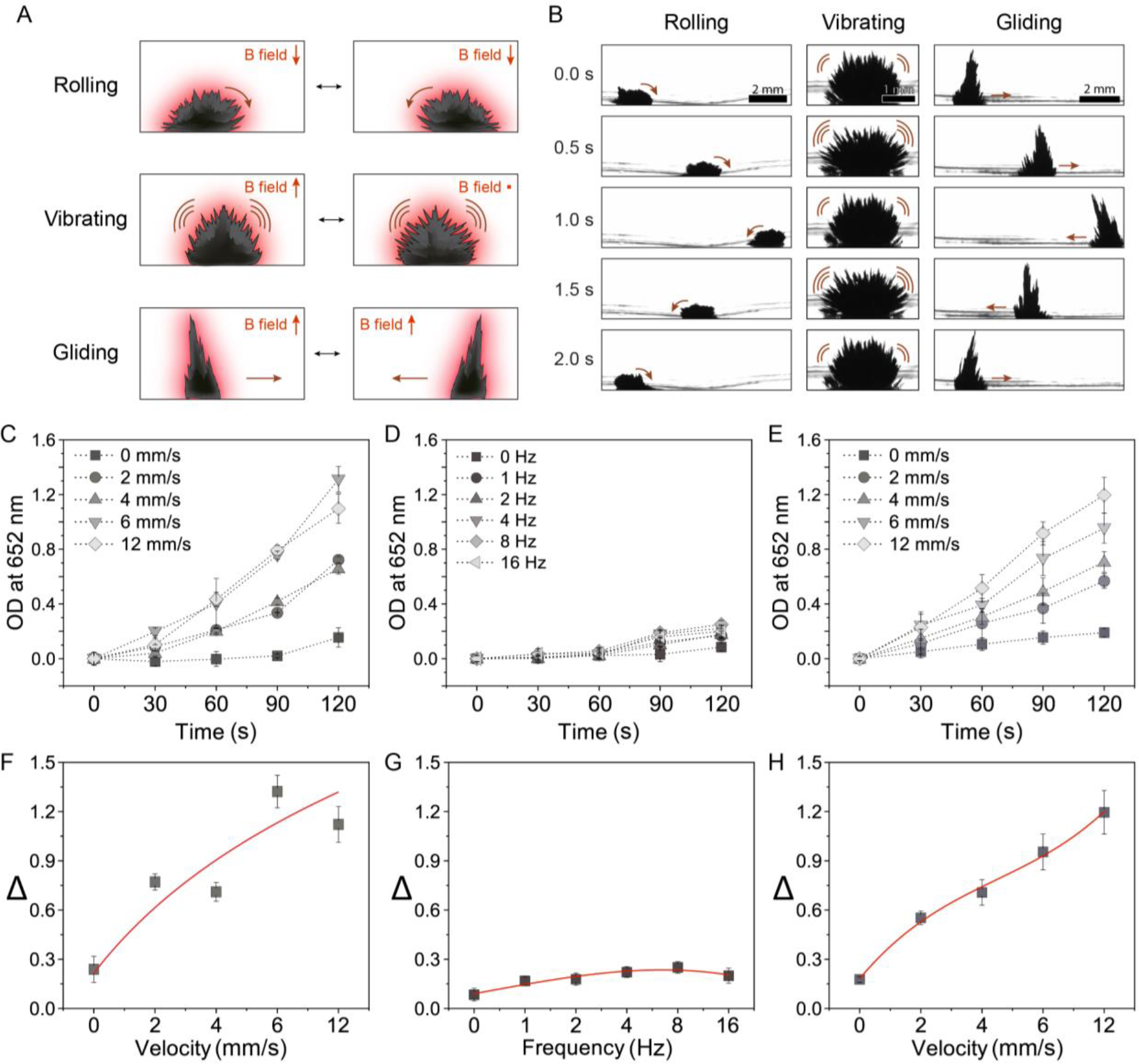
(A) Catalytic activity in situ generated by motion dynamics. The TMB assay demonstrates the generation of ROS on-site from H2O2 by the catalytically active (peroxidase-like) nanozyme microrobots. (B) The location of the catalysis is controlled by the shape and motion of the nanozyme assemblies. (1 mg mL−1 IONPs). Catalytic activity dynamics of (C) rolling, (D) vibrating, and (E) gliding motions. (F) Velocity dependent catalysis of rolling. (G) Frequency dependent catalysis of vibrating. (H) Velocity dependent catalysis of gliding. The rolling motion shows efficient catalytic activity. Data are mean ± standard deviation (n=3).
It is noteworthy that increasing vibration frequency results in only incremental enhancement of catalytic activity despite creating more pronounced structural movements, indicating that sub-millimeter-scaled amplitude of the vibrating motions may not be enough to generate sufficient mass transport (Figure S2, Supporting Information). However, centimeter-scale movements of rolling and gliding motions generate fluid convection for mixing and significantly enhance ROS production (Figure S3 and Movie S2, Supporting Information). In addition, the shape of the nanozyme microrobot assemblies remains intact during lateral gliding motion. But in rolling motion, as the assembly circulates, nanoparticles located within the structure are continually exposed to the interface, possibly causing slightly greater catalytic activity. Furthermore, the dynamic motion of the assembly of IONPs augments the diffusion and mass transfer within the chamber based on mechanical agitation. This shifts the equilibrium of peroxidase-like catalytic reactions occurring on the IONP surface, resulting in enhanced ROS generation. Interestingly, the catalytic activity enhanced by rolling motion saturates above 6 mm s−1 (Figure 2F and Figure S4, Supporting Information). For gliding motion, the catalytic activity rises linearly up to 12 mm s−1 before the superstructure loses structural stability at higher linear velocities (Figure 2H).
Altogether, we develop nanozyme microrobots with shape-tunable and motion-enhanced catalysis. Given the limited surface area compared to freely dispersed IONPs, the nanozyme microrobots display reduced catalytic activity when in static state, suppressing unnecessarily occurring catalytic reactions. On the other hand, nanozyme microrobots can perform localized catalytic reactions when set in motion providing an effective approach to locally generate and directly deliver ROS to targeted spatial locations through the dynamic motions, thereby avoiding off-target effects.
2.2. Antifungal activity of nanozyme-based microrobots
Next, we assess whether the nanozyme assemblies with motion-induced ROS can kill fungal cells. We use C. albicans as a model pathogen because it is the most common fungus in the human mycobiome and is associated with a variety of fungal infections.[2,25,26] To investigate killing efficacy, we test the nanozyme-based microrobots against planktonic cells and biofilms. We select the ‘rolling nanozyme microrobot’ as the most efficient motion to generate antimicrobial catalysis (diagram in Figure 3A). We find that ROS generation for 5 min effectively killed planktonic C. albicans (>10 times more effective than 0.5% H2O2), while causing complete fungal eradication within 10 min (undetectable viable cells) (Figure 3B, left). ROS exhibit antimicrobial activity against bacterial and fungal cells by causing oxidative damage of intracellular biomolecules, including deoxyribonucleic acid (DNA), enzymes and other proteins.[27] For example, the iron-sulphur cluster decomposes upon ROS exposure, which can disrupt metabolic pathways and lead to cell death. ROS can also cause cell membrane damage while producing reactive aldehydes that are toxic to the microbial cells.
Figure 3. Fungal killing and binding by nanozymes.
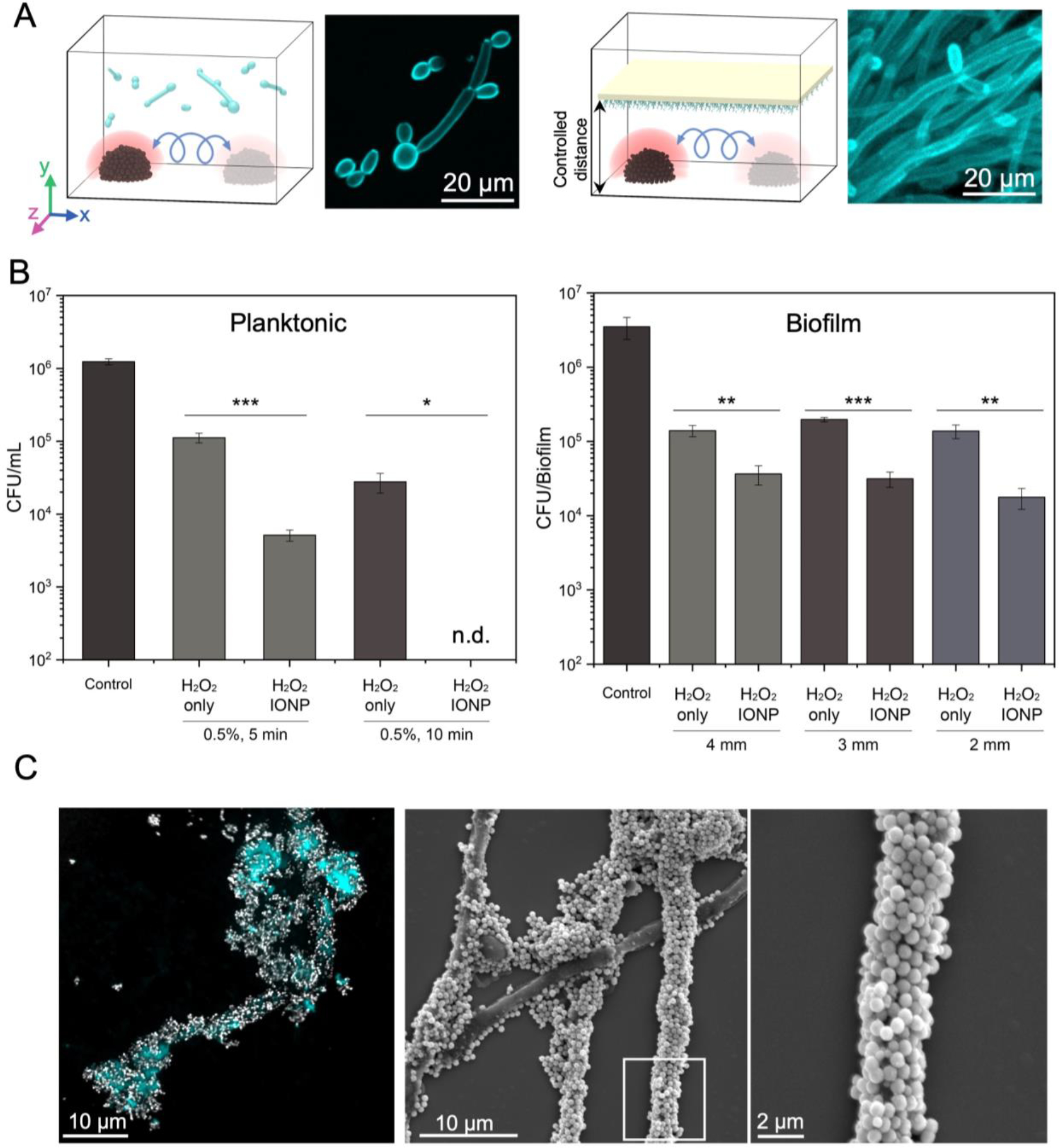
(A) Experimental design for killing planktonic fungal cells (left) and biofilms (right) using convective mixing. The rolling nanozyme microrobots create convective ROS mixing and dispersion against planktonic cells or biofilm surfaces at fixed distances. (B) Cell viability counts show killing of planktonic cells and biofilms of C. albicans at specific distances (2, 3 and 4 mm from the outer surface of the nanozyme microrobot) via catalytic activation of 0.5% H2O2. (C) Confocal images and SEM micrographs demonstrate the binding of nanozymes on C. albicans cells. Data are mean ± standard deviation; * P < 0.05, ** P< 0.01, *** P < 0.001 by one-way analysis of variance with Tukey’s multiple-comparison test (n=12), and n.d. stands for not detectable.
Given the enrichment of ROS generation by the dynamic motion, we compared the antifungal killing activity of ‘rolling nanozyme microrobot’ to the same amount of nanozymes dispersed in solution or in static assembled mode (Figure S5, Supporting Information). We find that nanozyme treatment in static mode or dispersed phase results in only a modest killing effect (<1-log reduction) indicating that the dynamic motion of the nanozyme microrobot is important for increased antifungal potency.
We then test the antifungal efficacy of nanozyme-microrobots against pre-formed C. albicans biofilms without physical contact (distal killing). We find that the biofilm killing efficacy of nanozyme treatment is increased compared to H2O2 only. However, as the distance between nanozyme assemblies and biofilm is decreased from 4 to 2 mm, the increase in killing efficacy is not statistically significant despite there being more dead cells (Figure 3B, right). Nanozymes without H2O2 (IONP only) have minimal antifungal effect confirming the catalytic, ROS-mediated killing mechanism (Figure S6A and S6B, Supporting Information). The reduced antibiofilm efficacy using the distal (non-contact) nanozyme-microrobotic approach (i.e., assemblies 2–4 mm away from the biofilm surface) could be due to several reasons, including enhanced antimicrobial tolerance of biofilms (compared to planktonic cells) as well as the limited ROS diffusion distance.[28–30] To address these limitations, closer proximity or direct surface contact may be required for effective antifungal killing in a biofilm state.
Unexpectedly, we observe large amounts of nanozymes bound on C. albicans cells when attempting to achieve closer or direct contact to the fungal surface. The nanozymes adhere with remarkably high affinity to both yeast and hyphal forms of C. albicans coating nearly the entire fungal surface, as shown using confocal fluorescence and scanning electron microscopy (SEM) (Figure 3C, left and middle). Magnified SEM images show nanozymes forming a coating wrapping around the fungal hyphae (Figure 3C, right). To further investigate the mechanism of nanozyme binding on the surface of C. albicans, we use C. albicans mutants with defects in (i) hyphal morphogenesis (ΔΔefg1, a master regulator of C. albicans hyphal formation); (ii) cell-wall adhesins (ΔΔbcr1, a master regulator of hyphal wall adhesins, and ΔΔals1/ΔΔals3 double mutant); (iii) cell-wall iron binding proteins (ΔΔrim101); and (iv) cell-wall mannan polysaccharides (ΔΔpmr1, ΔΔoch1, ΔΔmnt1/ΔΔmnt2). Among Candida mutants, we find significantly reduced binding affinity of nanozymes in a mutant strain defective in cell wall O-mannan biosynthesis (ΔΔmnt1/ΔΔmnt2)[31] (Figure S7, Supporting Information), suggesting that O-mannan polysaccharides on the fungal surface are critical for nanozyme-fungal interactions. All other mutant strains do not impact nanoparticle binding despite their unique morphological differences compared to the wild type. Notably, the nanozymes bind poorly to tissue suggesting higher affinity to C. albicans but less binding to human cells (Figure S8, Supporting Information). Such selective binding to the fungal cells indicates the possibility to directly target them for localized delivery and catalytic killing in situ.
The data indicate antifungal killing against planktonic C. albicans cells through nanozyme-microrobots via active mixing and convection of catalytic products, but close proximity may be required for antibiofilm activity. Given that the nanozymes show exceptional binding capacity to the fungal surface, delivering nanozymes directly to the infection sites may potentiate killing via in situ delivery-catalysis and lead to effective and precise antifungal targeting.
2.3. Nanozyme microrobot-directed delivery for in situ catalysis killing
To effectively use the nanozyme microrobots for direct delivery to the C. albicans biofilm surface, we adapt the ‘rolling nanozyme microrobot’ to traverse the fungal biofilm in direct contact, thereby “painting” a specific location for targeted antifungal action. By adjusting the number of strokes or passes, we precisely control the amount of fungal biofilm coating from light to dense layers of nanozyme (Figure 4A). Visually, this creates striking swathes of localized layering as shown in Figure 4A. Close-up images show the boundary between the coated and non-coated areas demonstrating precision delivery of nanozymes via this controlled robotic approach.
Figure 4. Precision-guided delivery of nanozyme for site-specific catalytic killing of fungal biofilms.
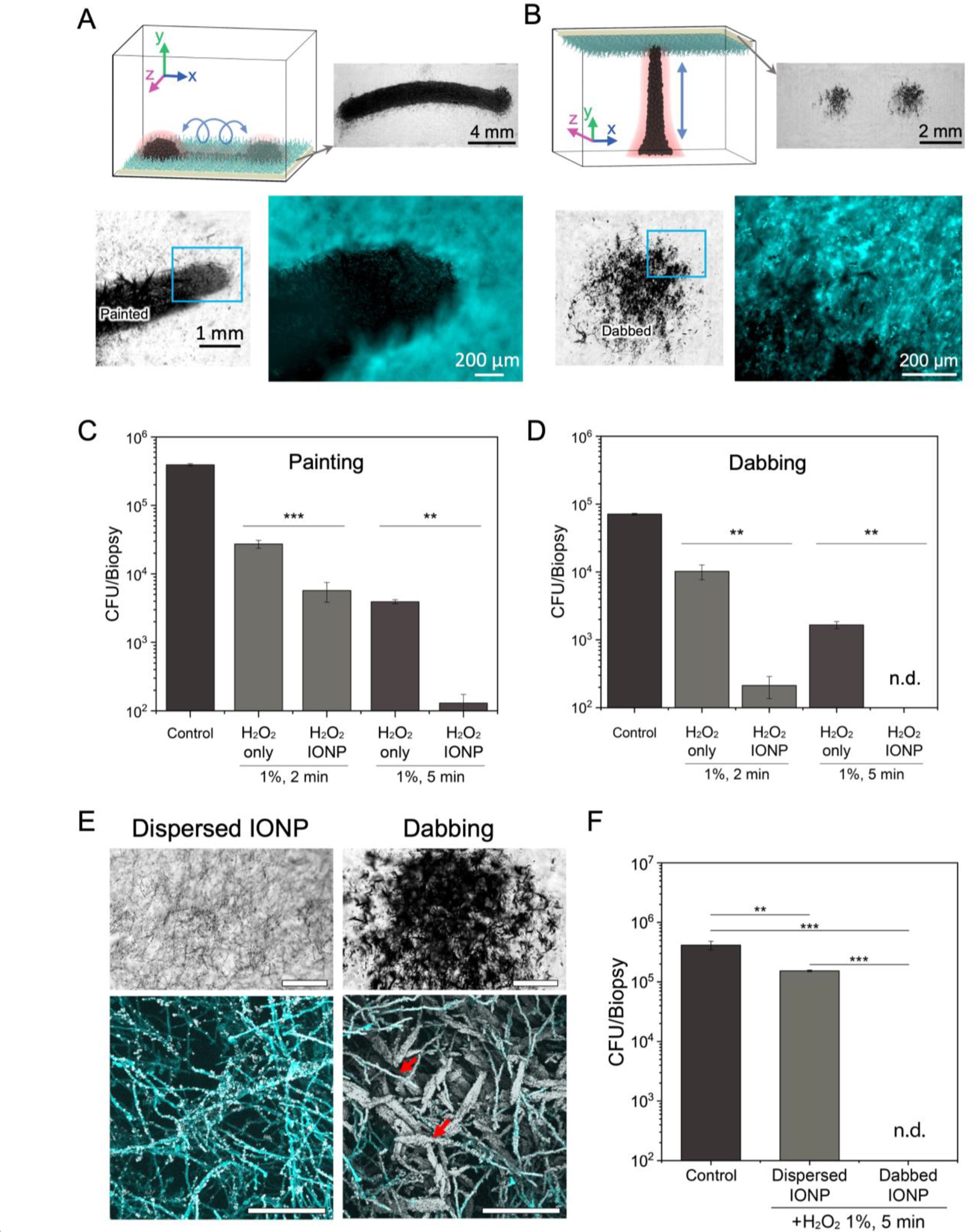
(A) Experimental design for precise delivery of nanozymes to fungal biofilms using a ‘painting nanozyme microrobot’. (B) Experimental design for targeting specific areas with submillimeter precision using a ‘dabbing nanozyme microrobot’. (C) Cell viability counts show eradication of C. albicans cells within painted areas by the painting microrobot. (D) Cell viability counts show complete killing of C. albicans cells within dabbed areas by the dabbing microrobot. (E) Coating efficiency of dispersed IONP treatment and IONP dabbing. Bright field microscopy (upper, scale bar: 500 μm) and confocal microscopy images (lower, scale bar:100 μm) show clear differences in IONP coating density on C. albicans biofilms. IONP dabbing delivers a greater amount of IONPs to the target area than dispersed IONP treatment. Hyphae of C. albicans are heavily coated with IONPs (red arrows). (F) Cell viability counts show limited killing of C. albicans cells by dispersed IONP treatment. Data are mean ± standard deviation; ** P< 0.01, *** P < 0.001 by one-way analysis of variance with Tukey’s multiple-comparison test (n=12 for panel C, n=8 for panel D and n=8 for panel F), and n.d. stands for not detectable.
To increase targeting precision, we devise a control algorithm to cause a linear extension-retraction motion to create “dabbing” dynamics for more subtle yet highly localized delivery, whereby the size and diameter of the dabbed region can be controlled (Figure 4B and Figure S9, Supporting Information). By varying the timing, electromagnet-1 and -2 currents, and the oscillating field frequency and magnitude, we are able to control the extension of the nanozyme assemblies in a fashion similar to a linearly extending robotic arm.
Next, we test the biofilm killing efficacy of dynamic nanozyme assemblies providing directed, controlled delivery of catalysis for in situ killing via ‘painting nanozyme-microrobot’. The data show that the nanozymes are delivered onto the targeted area on the fungal biofilm, and the cell-bound nanozymes remain highly catalytic (Figure S10, Supporting Information). We find that the ROS generation in situ significantly reduces the number of viable cells within the targeted area of fungal biofilms in 2 min (~10 times more effective than H2O2 alone (Figure 4C), while causing nearly complete fungal killing with negligible viable cells within 5 min, indicating high precision and efficacy of the painting approach.
We further investigate the possibilities to spatially target localized fungal pathogens within the biofilm through precision-guided ‘dabbing nanozyme microrobot’. Similarly, the data reveal that the targeted areas do not harbor viable fungal cells after 5 min treatment, whereas fungal areas in proximity but outside the targeted area remain viable (Figure 4D). As expected, nanozymes without H2O2 (IONP only) show minimal effect on biofilms (Figure S6C and S6D, Supporting Information). Furthermore, we find that the dabbing motion efficiently delivers large amounts of IONP locally, forming a dense coating and large deposits of nanozymes onto C. albicans surfaces (Figure 4E). In contrast, dispersed IONPs sparsely bind to the fungal surface. Notably, the dabbed nanozyme accumulation results in more effective killing (no detectable viable cells) than dispersed nanozyme solution treatment (<1-log reduction than control) (Figure 4F). We also evaluate the efficacy of the nanozyme-targeting approach against a biofilm-forming clinical isolate of C. albicans (UR#5).[32] The data show that the ‘dabbing nanozyme microrobot’ can also eradicate the clinical isolate in the targeted biofilm area within 10 min (Figure S11A, Supporting Information).
Collectively, the data indicate remarkable precision and efficacy in a highly controllable manner achieving bulk and localized fungal killing. These painting and dabbing nanozyme-microrobots represent a uniquely effective way to provide antifungal activity on targeted surfaces. Additionally, the rapid (within 10 min) and high killing efficacy (complete eradication) obtained in this work represent a significant improvement over the antifungal performance (> 1.5 h treatment time, < 98.85% of antifungal efficacy) recently reported for other nanozyme-based antifungal approaches.[33–35]
2.4. Precision and efficacy using experimental models
To further assess the antifungal precision and efficacy, we develop a host cell-fungal (human gingival spheroid and C. albicans) model that allows testing fungal cell capturing-killing to validate nanozyme-binding properties and catalytic antifungal activity of the microrobots (Figure 5). In this model, 3D spheroids of human gingival cells are premixed with C. albicans to emulate co-existence of host cells and fungal pathogens (diagram in Figure 5A). Then, a nanozyme microrobot is magnetically guided to capture and remove the fungal cells without disturbing the human tissue-like spheroid. The captured fungi are transported to a chamber to demonstrate catalytic killing in the presence of hydrogen peroxide.
Figure 5. Precision capturing and killing of fungal aggregates using nanozyme-microrobotics strategy.
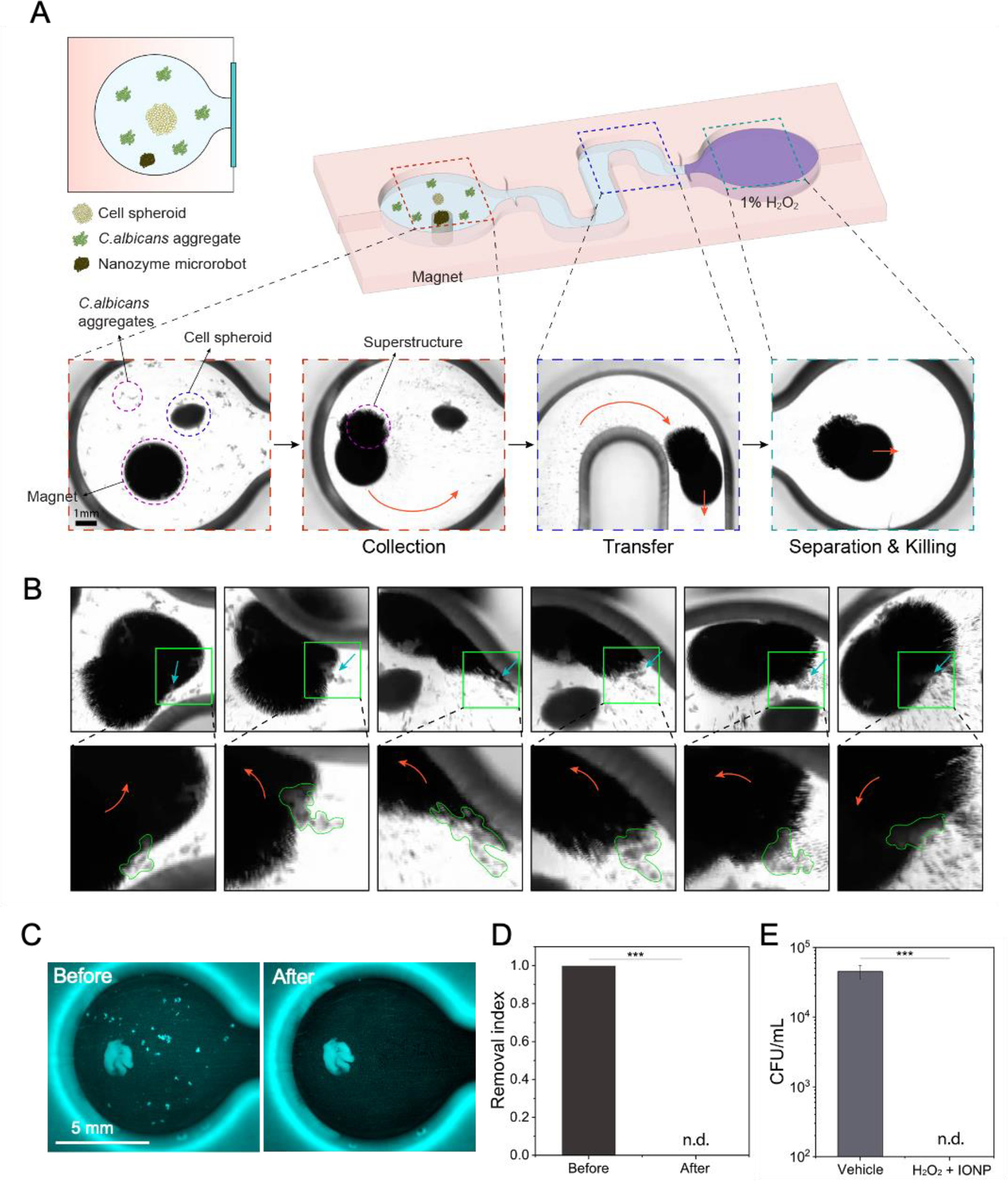
(A) A schematic of the experimental platform for testing C. albicans capture and killing in the presence of cell spheroid using nanozyme microrobots. (B) Fungal binding, dragging, and engulfing by the nanozyme microrobot. Close-up images show the fungal aggregates marked by green lines. (C) Before and after fluorescence imaging shows fungal aggregates effectively removed without binding or disturbing the cell spheroid by a magnetically controlled nanozyme microrobot. (D) Quantitative image analyses show complete removal of fungal aggregates. (E) Cell viability counts show eradication of the targeted and captured fungal aggregates via catalytic activation of H2O2. Data are mean ± standard deviation; *** P < 0.001 by Student’s t-test (n=4), and n.d. stands for not detectable.
In the demonstration, we find that the nanozyme assembly could relocate through magnetically controlled motions to capture the fungal aggregates surrounding the spheroid tissue. Time-lapse images show the nanozyme assembly can bind, capture, and engulf fungal aggregates while moving across the surface (Figure 5B). The nanozyme-microrobot minimally binds to the spheroid tissue allowing effective and targeted fungal binding and removal. We also quantitatively assess the effectiveness of fungal capturing by comparing the amount of fungal cells before and after the treatment using computational imaging analysis (Figure 5C and Figure S12, Supporting Information). The data show that the nanozyme robotic approach effectively separates and removes the fungal cells from the mixture (Figure 5D). The captured C. albicans cells are transferred to a separate chamber containing H2O2 to demonstrate catalytic killing. After 10 min exposure, the fungi are recovered and assessed for viability; the data show complete eradication with no detectable viable cells (Figure 5E).
Next, we explore directed delivery in a palatal mucosal tissue model with localized C. albicans growth to mimic clinical characteristics of fungal infection on soft tissues. To effectively use the nanozyme-microrobots for precise delivery, we develop an automated and programmable delivery system. Our system enables control of nanozyme assemblies with three translational degrees-of-freedom. Two of the directions are controlled by servo motors which reposition the electromagnet cores controlling the center of the nanozyme microrobot, while the third degree-of-freedom is controlled by modulating the currents to precisely and continuously extend the assemblies along the direction of the central axis of the coils.
To create the precise dabbing motion, we first apply a current to the coil opposite the target to collect nanozymes. While holding the nanozymes against the substrate, we adjust the center of the electromagnet core in the flat, facial plane of the electromagnetic coil. Once centered in a position aligned with the target in the z-direction, we apply current to the coil behind the target, which causes a linear extension of the structured assembly. However, the assembly typically adjusts shape in abrupt, discrete steps as the nanoparticles reorganize during extension, even when current is adjusted continuously. To overcome this discrete, step-like extension, we apply a continuously modulating sine wave at 15 Hz superimposed on a bias signal to create a smooth, controllable, and repeatable extension. The high frequency modulated wave aids in smooth morphological rearrangement of the nanoparticle assemblies during the extension and retraction phases, and results in higher aspect ratio superstructures. This results in the dabbing motion used to precisely target tissue surfaces (Figure 6A, Figure S9, and Movie S3, Supporting Information). The time-lapse and the dabbing show remarkable precision and localized nanozyme delivery, whereby the amount and size can be controlled by the number of dabs and concentration of nanozymes (Figure 6B–C). Higher nanozyme concentration and more dabs result in an increase in coated area.
Figure 6. Precision programming and automation for the dabbing nanozyme microrobot.
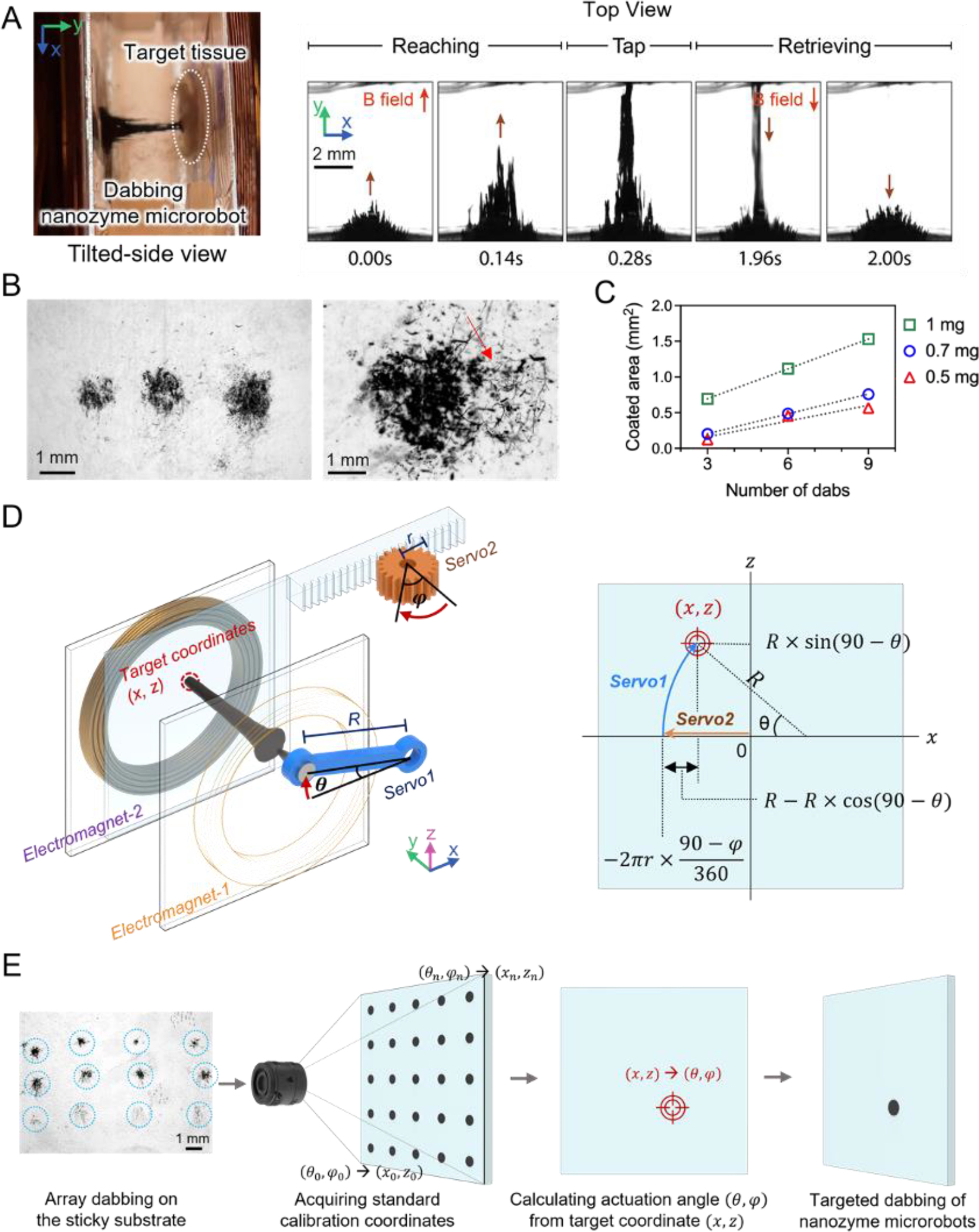
(A) The shape of the dabbing superstructure extending and tapping the targeted surface as visualized via a stereoscope. (B) Controllable dabbing and precision targeting provide localized delivery of tunable amounts of nanozyme. C. albicans coated with nanozymes on the targeted area (right image). (C) The area of biofilm coated with nanozymes depends on the IONP concentration. (D) Targeting location is determined by converting servo motor rotation positions to x-z coordinates. (E) A nanozyme dabbing test array aids fine-tuning of the positioning accuracy.
To automate nanozyme delivery, we program an algorithm to convert the servo motor positioning angles into the coordinates of the target. The x-axis is controlled by a rack and pinion gear system, while the y-position is determined by a radially sweeping arm (Figure 6D and Figure S13, Supporting Information). To achieve sub-millimeter dabbing motions and further improve precision delivery of nanozymes via the dabbing method, we calibrate the nanozyme assemblies to correct for minor positioning errors introduced by sources such as gear positioning and backlash (Figure 6E). Nanozyme assemblies are initially tested on an adhesive substrate during this calibration step. The resulting ‘hitmap’ is used for error correction before nanozyme delivery to tissues.
In a clinical setting, Candida infection is characterized by localized white plaques (thrush) on the epithelial tissue comprised mostly of hyphal forms.[36] To test the precision and efficacy of the nanozyme-based robotics approach, we use an experimental model to recapitulate the anatomical features of oral mucosa and localized fungal infection. We first create a primary whole-organ culture using explant murine oral mucosa with localized infection sites using C. albicans inoculated on predetermined areas on the palatal tissue (Figure 7A). The data show that C. albicans colonized the mucosal tissue forming a focal infection harboring mostly hyphal cells adhered to the mucosal epithelium (Figure 7B), similar to those found clinically. We then apply the automated delivery via nanozyme-microrobots using dabbing algorithms established in Section 2.4 above towards localized C. albicans infection sites (diagram in Figure 7C). Spatial coordinates of the targeted sites are extracted using image analysis and used to deliver nanozymes via the guided ‘dabbing approach’. Imaging analyses show controllable dosage of nanozymes delivered on-site to the specimens depending upon the number of dabs applied (Figure 7D). Fluorescence images show that the focal C. albicans infection site was coated by nanozymes whereas the surrounding epithelial tissue was devoid of nanozymes, suggesting high spatial delivery precision (Figure 7E). Catalytic activation kills the fungal cells on-site, resulting in significant reduction of viable fungi on the mucosal tissue (Figure 7F). The IONP alone group has no antifungal effect (Figure S6E, Supporting Information).
Figure 7. Precision targeting and antifungal killing on murine mucosa using dabbing nanozyme-microrobots.
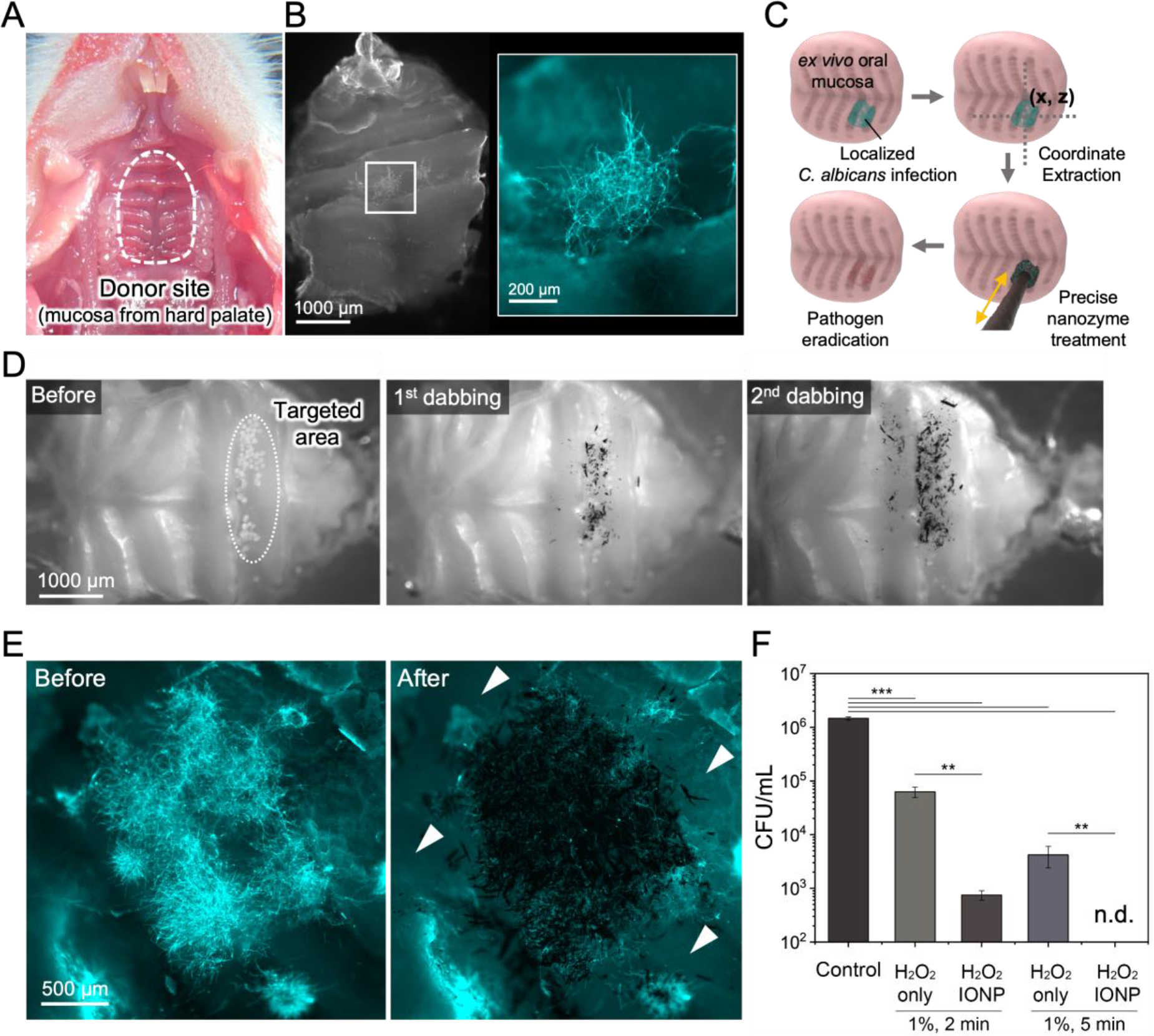
(A) Explant of murine oral mucosal tissue harvested from the palate. (B) Focal C. albicans infection developed on the oral mucosa characterized by localized hyphal accumulation. (C) A schematic diagram of the coordinate extraction and precision-guided treatment using the dabbing nanozyme microrobot. (D) Bright field images showing sequential nanozyme dabbing. (E) Fluorescence imaging shows high spatial precision for targeting the site of fungal infection. (F) Cell viability counts show effective killing of targeted fungal cells on mucosa via catalytic activation of H2O2 on site. Data are mean ± standard deviation; ** P< 0.01, *** P < 0.001 by one-way analysis of variance with Tukey’s multiple-comparison test (n=8), and n.d. stands for not detectable.
The biocompatibility and biosafety of IONP nanozymes are crucial factors that determine their potential for future clinical application. IONP nanozymes have been thoroughly investigated in our previous studies, which strongly supports biocompatibility both in vitro and in vivo.[9, 37] To further ensure biocompatibility and biosafety, we test whether the dabbing of nanozyme superstructures can affect the integrity of the soft tissue surface. Using the ex vivo palatal mucosal tissue model, which is directly exposed to dabbing nanozyme treatment and then collected for histological analysis, we find that IONP dabbing does not cause physical damage on mucosa tissue when compared to the control group (Figure S14, Supporting Information).
3. Conclusion
In summary, we employ microrobotic techniques to control the morphology, position, and on-site catalysis of nanozyme assemblies and demonstrate rapid killing with microscale precision against C. albicans, a widespread opportunistic fungal pathogen associated with many human infections. We study the catalytic activity of assembled nanozymes under various motions including rolling, vibrating, and gliding and apply this knowledge to achieve precision-guided antifungal treatment using biofilm and tissue infection models. Unexpectedly, we find that the nanozyme assemblies bind avidly to the surface of C. albicans and exploit this selective adhesion property to promote localized delivery and enhanced killing. The adhesion specificity and robotic motion routines are combined to create uniquely effective treatments for C. albicans infections. We demonstrate three key features of the nanozyme robotics approach: (1) controllable nanozyme catalysis through tunable dynamic motions and velocity promotes antifungal effects while avoiding unmitigated free radical production, (2) precise spatial targeting allows localized accumulation of high amounts of nanozymes to the site of infection and targeted ROS generation in situ, potentiating the biofilm killing activity and minimizing off-target effects, and (3) rapid fungal eradication due to high binding affinity and in situ killing of C. albicans. These findings may lead to high precision nanozyme-based therapies to treat fungal infections, which have been increasingly recognized as a major global health threat.
While our studies demonstrate remarkable potency and precision against fungal cells and tissue infections, there are some limitations, but also opportunities to motivate further studies. Although colorimetric indicators such as TMB give a measure of time-integrated ROS generation, they lack a direct correspondence to the real-time position of ROS. Thus, using TMB as an indicator does not directly relate to enhanced killing efficacy of cells. Future investigations analyzing time-lapsed cell killing concomitantly with direct ROS measurement may lead to better understanding of antifungal activity. Our work also reveals the interplay between localized nanozyme-enhanced generation of ROS, consumption of the source reactant, and the limited diffusion distance from the site of catalysis, emphasizing the importance of the on-site catalytic activity for killing efficacy. Additional characterization of the organization dynamics of superstructures, the influence of nanoparticle size/surface chemistry and spatial interactions with C. albicans cell wall may reveal novel mechanistic insights on the relation of in situ catalytic activity and microbial killing. The exact reasons for nanozyme binding to fungal surfaces are unknown although O-linked mannans appear to be at least one of the modulatory factors. Detailed studies on cell wall genetics and composition are needed to assess the binding affinity and adhesion mechanisms of nanozymes to fungal surfaces, and whether other microbes can be similarly targeted.
Other nanomaterials, including metal-, carbon-, and lipid-based nanoparticles, have been investigated for their potential applications as antifungal agents.[5,6] These materials can also improve the efficacy of conventional antifungal agents, reduce the development of drug resistance, and enhance drug delivery. However, to the best of our knowledge, current antifungal nano-drugs require longer treatment time and are unable to provide such a combination of precise spatial targeting and effective in situ killing at the site of fungal infection (a common clinical trait), which may also prevent off-target effects. In addition, iron oxide nanoparticles (including Food and Drug Administration-approved formulations) have been shown to be biocompatible, non-toxic, and safe for humans at therapeutic dosages, which can facilitate clinical translation.
Altogether, this study demonstrates that microrobotic techniques for assembly and control of nanozyme constructs provide unique spatial targeting approaches for enhancing the precision and efficacy of antifungal treatment. We expect our findings will be broadly applicable to advancing nanozyme-based robotic assemblies for controllable catalytic reactions and precise delivery of reaction products from the scale of a single nanozyme to that of an assembled superstructure, which can be exploited for treatment of other microbial infections. Further in-depth analysis using in vivo and pre-clinical models could evaluate the clinical feasibility of this approach.
4. Experimental Section/Methods
Synthesis and Characterization of Iron Oxide Nanoparticles:
The iron oxide nanoparticles (IONPs) were synthesized by a modified solvothermal method via reducing FeCl3 at high temperature.[17,24] The reaction mixture was prepared by dissolving 975 mg of FeCl3 in 30 mL of ethylene glycol. 1.5 g of sodium acetate was added as a reductant and solvent. Before the heat treatment, the mixture was stirred for 30 min for homogenization. Subsequently, the obtained homogeneous mixture was transferred to a custom-built 50 mL Teflon-lined stainless-steel autoclave and heated at 200 °C for 14 h. The final products were washed 3 times with Milli-Q water (MilliporeSigma, Burlington, MA, USA) and ethanol. Transmission electron microscopy (TEM) photographs were taken using a FEI Tecnai T12 microscope (FEI, Hillsboro, OR) at an accelerating voltage of 100 kV. The final concentration of IONPs was determined by inductively coupled plasma-optical emission spectrometry (ICP-OES) using a Spectro Genesis ICP (SPECTRO Analytical Instruments GmbH, Boschstr, Germany). ICP-OES was calibrated with calibration standards containing known amounts of elements. The diameters of ions are measured using the “imfindcircles” function in MATLAB, which detects circles of a consistent size. The average diameter is found to be 307.8 nm, with a standard deviation of 79.0 nm. We used this nanoparticle size because our preliminary antifungal activity analysis showed that 300 nm IONPs were capable of C. albicans eradication compared to 200 or 500 nm nanoparticles (Figure S15, Supplemental data).
Magnetic Field Control Device:
The magnetic fields for the nanozyme microrobots were generated by two electromagnets coaxially assembled at a distance of 8 mm. Each electromagnet was independently energized using a programmable DC power supply (Sorensen XTR60-14, AMETEK Programmable Power, San Diego, USA). An iron core (2 mm in diameter) was mounted on a servo motor to generate the high-gradient magnetic fields at a focused area. The position of the iron core is controlled using a micro servo motor. Between the two electromagnets, a slidable chamber (15 × 7.3 × 19 mm3) was designed to accommodate the IONP dispersion and the position of the chamber was controlled by another servo motor. The electromagnets and servo motors were controlled and harmonized by a microcontroller (Arduino Nano, Arduino, Somerville, MA, USA). The Arduino integrated development environment software was used for system coding.
To control the motion of nanozyme microrobots, the micro servo motor connected to the iron core was programmed to move between 65° and 115°, which created an up-and-down arc-shaped iron core movement to control the z-coordinate of the nanozyme microrobot. The second micro servo motor (FS90) was used and programmed to control x-axis coordinates with a pinion gear (5 mm radius) attached to the rack of the slidable container for left-to-right motion. A combination of the micro servo motors with the arm and pinion gear allowed high spatial precision (0.087 mm of x-axis, ~0.244 mm of y-axis) and repeatability in position and timing with linear velocities ranging from 0 to 12 mm s−1. Actuation in all experiments was performed via the application of a magnetic field and servo programmed to achieve the desired motion pattern. The geometries of the slidable container and the body of the device were developed and designed with Onshape, a computer-aided design software (Onshape Inc., Cambridge, USA). The slidable container and the body of the device were fabricated using a low-force stereolithography (SLA) 3D printer (Form 3B, Formlabs Inc., MA, USA) with a biocompatible resin (Surgical Guide). 3D printed parts were rinsed for 20 min in 99.9% isopropanol and then photopolymerized for 30 min under ultraviolet light (405 nm light at 70 °C) (FormCure, Formlabs Inc., MA, USA).
Dynamic motions of Nanozyme Microrobots:
We created four distinct nanozyme microrobot motions: rolling, vibrating, gliding and dabbing, named based on the observed dynamics of the nanozyme superstructures. For each motion, nanozymes were first collected from the solution to the wall of the chamber by applying current to the electromagnet-1 for 1.2 s (50 mT, at the center of the chamber). To generate the rolling motion, a side-to-side motion was activated by moving the iron core (on the same side of the electromagnet-1) with the servo motor while maintaining the current at a constant value.
The vibrating motion was activated by applying an oscillating field to the electromagnet-2 after the nanozyme collection step. The static field (50 mT, at the center of the container) from the electromagnet-1 was switched off while an oscillating sine wave (varying from 0 to 16 Hz) was generated by electromagnet-2 to produce the vibrating motion. The strength of the magnetic fields was measured to be 12 to 20 mT, at the center of the container. For the gliding motion, the electromagnet-2 was energized by a constant current to reshape the magnetic field such that the nanozyme superstructure was forced to extend along the central axis of the electromagnets. The strength of the magnetic field was measured to be 50 mT at the center of the container. This extended structure was translated from side-to-side with a servo motor. The velocity of the side-to-side motion varied by controlling the angular velocity of the servo motor. To create the dabbing motion, two servo motors were used to reposition the iron core to the targeted position while maintaining a static current. This positioning step was followed by applying a constant bias field with a superimposed sinusoidal field (15 Hz) to the electromagnet-2. This enables smooth extension of the superstructure. The strength of the magnetic fields was measured to be 10 to 18 mT, at the center of the container. Once the nanozymes were delivered to the targeted tissue or substrate, the electromagnet-2 was de-energized such that the superstructure fully retracted.
Catalytic Properties of Nanozyme Microrobots:
To measure the catalytic properties of nanozyme microrobots, 952.2 μL of 0.1 M sodium acetate (NaAc) buffer (pH 4.5) was added into the actuation chamber followed by 4 μL of TMB (25 mg mL−1 in dimethylformamide) to reach a final concentration of 0.1 mg mL−1. Then, 40 μL of dispersed IONPs was added into the chamber at a final concentration of 1 mg mL−1. Immediately before starting the motions (i.e., rolling, vibrating, or gliding), 3.4 μL of H2O2 (30%, v/v) was added into the mixture (final concentration: 0.1%). At 30, 60, 90 and 120 s, the mixture was transferred to a cuvette and the absorbance at 652 nm was measured using a spectrophotometer (Beckman Coulter, Inc., Fullerton, CA, USA).
C. albicans Biofilm on Model Surfaces and Non-Surface Attached Fungal Aggregates:
Individual 3D-printed acrylic sheets were created as model surfaces with dimensions of 18.0 × 18.0 × 0.4 mm3. 3D printing was done using a low-force stereolithography (SLA) 3D printer (Form 3B, Formlabs Inc., MA, USA) with a biocompatible Surgical Guide resin. 3D printed parts were rinsed for 20 min in 99.9% isopropanol and then photopolymerized for 30 min under ultraviolet light (405 nm light at 70 °C) (FormCure, Formlabs Inc., MA, USA). The 3D-printed surfaces were sterilized in an autoclave for 15 min at 121 °C, and then coated with filter-sterilized, clarified whole human saliva, which was designed to mimic the denture (abiotic) surface in the oral cavity. C. albicans SN250, a biofilm-forming strain, was grown in ultrafiltered (10-kDa cutoff; Millipore, MA, USA) tryptone-yeast extract (UFTYE, pH 5.5) broth containing 1% (w/v) glucose at 37 °C and 5% CO2 to mid-exponential phase (6 h) following the protocol described previously.[38] The saliva-coated 3D-printed surfaces were inoculated with ~105 CFU of actively growing C. albicans cells (yeast form, mid-exponential phase) per milliliter in UFTYE (pH 7.0) containing 1% (w/v) sucrose at 37 °C with 5% CO2 for 12 h. Non-surface attached aggregates of C. albicans were prepared following established protocols.[39] Briefly, planktonic C. albicans cells (105 CFU mL−1, yeast form, mid-exponential phase) were resuspended in filter-sterilized, clarified whole human saliva supplemented with 1% (w/v) sucrose and were incubated at 37 °C for 60 min to form fungal aggregates. C. albicans was stained with concanavalin A (ConA) lectin conjugated with tetramethylrhodamine (555/580 nm; Molecular Probes Inc., Eugene, OR, USA).
Planktonic Fungal Killing by Nanozyme Microrobots:
C. albicans SN250 was grown to mid-exponential phase (yeast form, 6 h) in UFYTE (pH 7.0) broth with 1% (wt/vol) glucose as described above. C. albicans cells (yeast and hyphal forms) were harvested by centrifugation (6,000 g, 10 min, 4 °C) as described previously[40] and resuspended in 0.1 M NaAC (pH 4.5). IONPs (1 mg mL−1) were premixed with 0.5% or 1% H2O2 for 10 s for catalytic activation and transferred into the actuation chamber. Then, 100 μL of C. albicans cell suspension was added into the chamber and the IONPs were actuated by the magnetic field to generate rolling motion. The treatments were performed with 0.5% H2O2 (for 5 and 10 min), and in the presence of 1% H2O2 (for 2 and 5 min). After the treatment, the C. albicans cells were serial diluted and the number of viable cells (CFU, colony forming unit) was determined. The detection limit is the minimal number of viable counts that can be reliably quantified via the CFU assay.[41]
Non-Contact Convection-Based Fungal Biofilm Killing:
IONP nanozymes were pre-mixed with H2O2 for catalytic activation as described above. The IONP- H2O2 mixture (final concentration of H2O2: 0.5%) was transferred into the actuation chamber. Then, C. albicans biofilm preformed on a 3D-printed substrate was inserted into the chamber and positioned at 2, 3 or 4 mm away from the side wall of the chamber where the nanozyme assembly was formed. The nanozyme microrobot was actuated by the magnetic field to generate rolling motions. After the 5-min treatment, C. albicans biofilms were harvested from the 3D-printed substrate by sonication. Samples were then homogenized, serially diluted and plated on Sabouraud dextrose agar to determine the number of viable cells (CFU, colony forming unit).[41]
Fungal Biofilm killing by Nanozyme Painting:
IONP nanozymes were pre-mixed with H2O2 and transferred to the actuation chamber. Next, C. albicans biofilm preformed on a 3D-printed substrate was inserted into the chamber. The IONP microrobot was rolled over the biofilm once in less than 10 s by the magnetic field to avoid physical removal of biofilm by the nanozyme microrobot. After painting, C. albicans biofilm was incubated to reach a total treatment time of 2 or 5 min in the presence of 1% H2O2. Only the painted areas of C. albicans biofilms were collected from 3D-printed substrates using sterilized dental mini-spatulas under a stereo zoom microscope (Carl Zeiss Microscopy GmbH, Jena, Germany), and then serially diluted for CFU counting.[41]
Fungal Biofilm killing by Nanozyme Dabbing:
IONP nanozymes were pre-mixed with H2O2 and were transferred to the actuation chamber. Preformed C. albicans biofilm on 3D-printed surfaces was inserted into the chamber (8 mm away from the side of the chamber where the nanozyme microrobot was formed). Biofilms were treated with dabbing motions in less than 20 s to form a localized treatment area. Dabbed spots were incubated in the presence of 1% H2O2 for 2 or 5 min. Only the biofilm within the dabbed areas was collected using sterilized dental mini-spatulas under a stereo zoom microscope system and serially diluted for CFU counting.[41]
Culture of Human Gingival Cells Spheroids and Explant Mucosal Tissue:
Human gingival tissue-derived mesenchymal stem cells (GMSCs) were isolated from deidentified gingival tissues obtained during third molar (wisdom tooth) extraction at the University of Pennsylvania under the approved Institutional Review Board (IRB) protocol (IRB#816238).[42,43] GMSCs were expanded up to passage five in Minimum Essential Medium Alpha (MEM α) media (containing 15% FBS, 2 mM L-glutamine, 100 μM ascorbic acid, 100 U mL−1 penicillin, and 100 μg mL−1 streptomycin) before being used in the experiments as described previously.[44] Cells were seeded at a density of 106 mL−1 in ultra-low attachment tubes and grown for 3 days in a humidified tissue culture incubator (37 °C, 5% CO2) to form 3D spheroid. To model a localized C. albicans infection on the oral mucosal tissue, a primary whole-organ explant culture of murine oral mucosa was used as described previously.[45] Briefly, mucosal tissue (4 mm × 2 mm) was harvested from the palate of C57BL/6 mice and cultivated in MEM α media (15% FBS, 2 mM L-glutamine, 100 μM ascorbic acid, 100 U mL−1 penicillin, and 100 μg mL−1 streptomycin) at 37 °C with 5% CO2 for 24 h. The explant cultures were then used to create focal C. albicans infection as detailed in the section “Focal Candida Infection on Explant Mucosal Tissue Culture”.
Precise Fungal Capturing and Killing by Nanozyme Microrobots:
To test the precise fungal capturing and killing by nanozyme microrobots, a custom-built dual chamber device and an in vivo host-fungal (human gingival cells and C. albicans) model was developed (diagram in Figure 6A). The 3D-printed device has two chambers (10 mm in diameter, 5-mm deep) connected by a canal (3.5-mm wide, 60-mm long) with a gate on each side. A 3D human gingival spheroid (~106 cells) and C. albicans aggregates (~104 CFU) were inoculated and premixed in the first chamber to mimic the oral environment where the host cells and fungal pathogens co-exist. Nanozyme microrobots were magnetically actuated by a neodymium magnet to precisely capture the fungal cells. Then, the captured fungal cells were separated from the mixture by the nanozyme microrobots and transported through the canal to the second chamber (containing 1% H2O2), which served as a catalytic “killing pool” via H2O2 activation. After killing for 10 min, the fungal aggregates were collected and serial diluted to determine the number of viable cells (CFU).[41]
Focal Candida Infection on Explant Mucosal Tissue Culture:
A primary explant culture of murine oral mucosa was used to create a focal Candida infection that mimics localized infection of the oral cavity by C. albicans. The tissue preparation procedure was conducted under the University of Pennsylvania’s Institutional Animal Care and Use Committee (IACUC) protocol (IACUC#806682). To establish a localized C. albicans infection on tissue specimens, we used hydroxyapatite beads (80-μm particle size, Bio-Rad Laboratories, USA) as micro-carriers for fungal cells, which allowed inoculating fungal cell onto the mucosal tissue with microscale precision. Briefly, saliva-coated hydroxyapatite beads (sHA) were pre-incubated with ~106 CFU mL−1 C. albicans (yeast form) in adsorption buffer (50 mM KCl, 1.0 mM KPO4, 1.0 mM CaCl2, 0.1 mM MgCl2, pH 6.5) for 60 min to allow binding of fungal cells. The beads were washed three times with adsorption buffer to remove unbound fungal cells. Then, the beads were carefully delivered to a predefined site on the mucosal tissue using a microinjection pipette tip under a microscope (Zeiss Axio Zoom V16). Approximately 20 beads were deposited on each tissue specimen. After the inoculation, tissues were incubated in fresh MSC medium at 37 °C with 5% CO2 for 12 h to establish a focal Candida infection. For histological analysis, the tissue samples were collected after treatment and fixed in 4% paraformaldehyde (PFA), followed by embedding in OCT compound (Sakura Finetek, Torrance, CA, USA). Frozen sections were prepared and slides were stained with H&E staining.
Qualitative and quantitative imaging analysis:
Brightfield and fluorescence images were taken with a Zeiss Axio Zoom.V16 upright stereo zoom microscope system (Carl Zeiss Microscopy GmbH, Jena, Germany) with 1× objective (numerical aperture, 0.25). For fluorescence imaging, Samples were stained with Concanavalin A-tetramethylrhodamine conjugate (Molecular Probes) to visualize C. albicans cells as described previously.[38] Additionally, high-resolution confocal microscopy was performed using a Zeiss LSM 800 upright confocal microscope (Zeiss, Germany) equipped with a 40 × water immersion objective (numerical aperture = 1.2) and a 10 × objective (numerical aperture = 0.7). Computational image processing and quantitative analysis were conducted as described previously.[39] Briefly, fluorescence images were imported using ImageJ Fiji software (https://imagej.net/software/fiji/). After denoising, images were segmented based on the fluorescent intensity using the Otsu algorithm and a particle size-based filtering was applied to only include C. albicans aggregates in the images. Regions of interest were created for each C. albicans aggregate and the total surface area (in μm2) was calculated. The Removal Index (RI) was calculated to evaluate the effectiveness of C. albicans removal by nanozyme microrobots, which is defined as . Data were presented as not detectable when <2% of fungal cells were detected after the treatment.
Statistical Analysis:
Statistical analyses were performed with GraphPad Prism 8.0 (GraphPad Software, CA). Data are represented as mean ± standard deviation. Two-sided Student’s t-test was used to compare mean between two experimental groups. Comparisons of mean between multiple groups were performed using a two-sided one-way analysis of variance (ANOVA) with post hoc Tukey’s test. A P value less than 0.05 was considered significant. At least three independent experiments were performed unless otherwise stated.
Supplementary Material
Acknowledgements
M.J.O. and S.Y. contributed equally to this work. This work was supported in part by the National Institute for Dental and Craniofacial Research (NIDCR) grant R01 DE025848 (H. Koo) and R56 DE029985 (E. Steager). M.J.O. is a recipient of the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education (No. NRF-2021R1A6A3A03044553). Z.R. is supported by the National Institute of Dental and Craniofacial Research Postdoctoral Training Program under Award R90DE031532.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the funders.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Min Jun Oh, Biofilm Research Laboratories, Levy Center for Oral Health, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Chemical and Biomolecular Engineering, School of Engineering & Applied Sciences, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Orthodontics, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Seokyoung Yoon, Biofilm Research Laboratories, Levy Center for Oral Health, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Radiology, Perelman School of Medicine, University of Pennsylvania, Pennsylvania, Philadelphia, PA 19104, USA.
Alaa Babeer, Biofilm Research Laboratories, Levy Center for Oral Health, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Endodontics, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Oral Biology, King Abdulaziz University, Jeddah 21589, KSA.
Yuan Liu, Biofilm Research Laboratories, Levy Center for Oral Health, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Preventive & Restorative Sciences, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Zhi Ren, Biofilm Research Laboratories, Levy Center for Oral Health, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Center for Innovation & Precision Dentistry, School of Dental Medicine and School of Engineering & Applied Sciences, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Orthodontics, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Zhenting Xiang, Biofilm Research Laboratories, Levy Center for Oral Health, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Orthodontics, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Yilan Miao, Biofilm Research Laboratories, Levy Center for Oral Health, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Orthodontics, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
David P. Cormode, Department of Radiology, Perelman School of Medicine, University of Pennsylvania, Pennsylvania, Philadelphia, PA 19104, USA Department of Bioengineering, School of Engineering & Applied Sciences, University of Pennsylvania, Philadelphia, PA 19104, USA.
Chider Chen, Department of Oral and Maxillofacial Surgery and Pharmacology, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA, 19104, USA.
Edward Steager, Biofilm Research Laboratories, Levy Center for Oral Health, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Center for Innovation & Precision Dentistry, School of Dental Medicine and School of Engineering & Applied Sciences, University of Pennsylvania, Philadelphia, PA 19104, USA; GRASP Laboratory, School of Engineering & Applied Sciences, University of Pennsylvania, Philadelphia, PA 19104, USA.
Hyun Koo, Biofilm Research Laboratories, Levy Center for Oral Health, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Orthodontics, School of Dental Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Center for Innovation & Precision Dentistry, School of Dental Medicine and School of Engineering & Applied Sciences, University of Pennsylvania, Philadelphia, PA 19104, USA.
References
- [1].Vallabhaneni S, Mody RK, Walker T, Chiller T, Infect Dis Clin North Am. 2016, 30, 1–11. [DOI] [PubMed] [Google Scholar]
- [2].Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, White TC, Sci. Transl. Med. 2012, 4, 165rv13. [DOI] [PubMed] [Google Scholar]
- [3].WHO, WHO fungal priority pathogens list to guide research, development and public health action, 2022, ISBN; 978-92-4-006024-1. [Google Scholar]
- [4].Perfect JR, Nat. Rev. Drug Discovery 2017, 16, 603–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sousa F, Ferreira D, Reis S, Costa P, Pharmaceuticals 2020, 13, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].León-Buitimea A, Garza-Cervantes JA, Gallegos-Alvarado DY, Osorio-Concepción M, Morones-Ramírez JR, Pathogens 2021, 10, 1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hwang G, Paula AJ, Hunter EE, Liu Y, Babeer A, Karabucak B, Stebe K, Kumar V, Steager E, Koo H, Sci. Rob. 2019, 4, eaaw2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Li Y, Zhu W, Li J, Chu H, Colloids Surf., B 2021, 198, 111465. [DOI] [PubMed] [Google Scholar]
- [9].Huang Y, Liu Y, Shah S, Kim D, Simon-Soro A, Ito T, Hajfathalian M, Li Y, Hsu J, Nieves LM, Alawi F, Naha PC, Cormode DP, Koo H, Biomaterials 2020, 268, 120581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Naha PC, Liu Y, Hwang G, Huang Y, Gubara S, Jonnakuti V, Simon-Soro A, Kim D, Gao L, Koo H, Cormode DP, ACS Nano 2019, 13, 4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Su Y, Ding M, Dong H, Hu Y, Yang D, Shao J, Huang B, Mater. Chem. Front. 2022, 6, 2596. [Google Scholar]
- [12].Liu Y, Huang Y, Kim D, Ren Z, Oh MJ, Cormode DP, Hara AT, Zero DT, Koo H, Nano Lett. 2021, 21, 9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Oh MJ, Babeer A, Liu Y, Ren Z, Wu J, Issadore DA, Stebe KJ, Lee D, Steager E, Koo H, ACS Nano 2022, 16, 11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wu JJX, Wang XY, Wang Q, Lou ZP, Li SR, Zhu YY, Qin L, Wei H, Chem. Soc. Rev. 2019, 48, 1004. [DOI] [PubMed] [Google Scholar]
- [15].Huang Y, Ren J, Qu X, Chem. Rev. 2019, 119, 4357–4412. [DOI] [PubMed] [Google Scholar]
- [16].Cormode DP, Gao L, Koo H, Trends in biotechnology 2018, 36, 15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Deng H, Li X, Peng Q, Wang X, Chen J, Li Y, Angew. Chem. Int. Ed. 2005, 44, 2782–2785. [DOI] [PubMed] [Google Scholar]
- [18].Sheng J, Wu Y, Ding H, Feng K, Shen Y, Zhang Y, Gu N, Adv. Mater. 2023, 2211210. 10.1002/adma.202211210 [DOI] [PubMed] [Google Scholar]
- [19].Poulain D, Crit. Rev. Microbiol. 2015, 41, 208–217. [DOI] [PubMed] [Google Scholar]
- [20].Cavalheiro M, Teixeira MC, Front. Med. 2018, 5, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ebrahimi N, Bi C, Cappelleri DJ, Ciuti G, Conn AT, Faivre D, Habibi N, Hošovský A, Iacovacci V, Khalil ISM, Magdanz V, Misra S, Pawashe C, Rashidifar R, Soto-Rodriguez PED, Fekete Z, Jafari A, Adv. Funct. Mater. 2020, 31, 2005137. [Google Scholar]
- [22].Dong Y, Wang L, Zhang Z, Ji F, Chan TKF, Yang H, Chan CPL, Yang Z, Chen Z, Chang WT, Chan JYK, Sung JJY, Zhang L, Sci. Adv, 2022, 8, abq8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ma X, Wang L, Wang P, Liu Z, Hao J, Wu J, Chu G, Huang M, Mair LO, Huang C, Chem. Eng. J. 2022, 431, 133971. [Google Scholar]
- [24].Gao L, Zhuang J, Nie L, Zhang J, Zhang Y, Gu N, Wang T, Feng J, Yang D, Perrett S, Yan X, Nat. Nanotechnol. 2007, 2, 577. [DOI] [PubMed] [Google Scholar]
- [25].Mishra K, Bukavina L, Ghannoum M, Front. Microbiol. 2021, 12, 636131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].d’Enfert C, Kaune AK, Alaban LR, Chakraborty S, Cole N, Delavy M, Kosmala D, Marsaux B, Frois-Martins R, Morelli M, Rosati D, Valentine M, Xie Z, Emritloll Y, Warn PA, Bequet F, Bougnoux ME, Bornes S, Gresnigt MS, Hube B, Jacobsen ID, Legrand M, Leibundgut-Landmann S, Manichanh C, Munro CA, Netea MG, Queiroz K, Roget K, Thomas V, Thoral C, Van den Abbeele P, Walker AW, Brown AJP. FEMS Microbiol. Rev. 2021, 45, 1–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Imlay JA, Mol. Microbiol. 2006, 59, 1073–1082. [DOI] [PubMed] [Google Scholar]
- [28].Koppenol WH, Free Radical Biol J. Med. 1986, 1, 281. [Google Scholar]
- [29].Attri P, Kim YH, Park DH, Park JH, Hong YJ, Uhm HS, Kim K-N, Fridman A, Choi EH, Sci. Rep. 2015, 5, 9332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Guo Q, Yue Q, Zhao J, Wang L, Wang H, Wei X, Liu J, Jia J, Chem Commun. 2011, 47, 11906–11908. [DOI] [PubMed] [Google Scholar]
- [31].Munro CA, Bates S, Buurman ET, Hughes HB, Maccallum DM, Bertram G, Atrih A, Ferguson MAJ, Bain JM, Brand A, Hamilton S, Westwater C, Thomson LM, Brown AJP, Odds FC, Gow NAR, J. Biol. Chem. 2005, 280, 1051–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Xiang Z, Wakade RS, Ribeiro AA, Hu W, Bittinger K, Simon-Soro A, Kim D, Li J, Krysan DJ, Liu Y, Koo H, mBio, 2023, 14, e02769–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wang B, Zhang W, Pan Q, Tao J, Li S, Jiang T, Zhao x., Nano Lett. 2023, 23, 1327–1336. [DOI] [PubMed] [Google Scholar]
- [34].Li X, Xu Y, Ouyang D, Ye K, Chen Y, Li Q, Xia Q, Wu X, Yang Y, Biochem. Eng. J. 2023, 119, 108791. [Google Scholar]
- [35].Ji Y, Han Z, Ding H, Xu X, Wang D, Zhu Y, An F, Tang S, Zhang H, Deng J, Zhou Q, ACS Appl. Mater. Interfaces 2021, 13, 17289–17299. [DOI] [PubMed] [Google Scholar]
- [36].Desai JV, J. Fungi 2018, 4, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gao L, Liu Y, Kim D, Li Y, Hwang G, Naha PC, Cormode DP, Koo H, Biomaterials 2016, 101, 272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Falsetta ML, Klein MI, Colonne PM, Scott-Anne K, Gregoire S, Pai CH, Gonzalez-Begne M, Watson G, Krysan DJ, Bowen WH, Koo H Infection and Immunity 2014, 82, 1968–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Negrini TC, Ren Z, Miao Y, Kim D, Simon-soro Á, Liu Y, Koo H, Arthur RA, Front. Cell. Infect. Microbiol. 2022, 12, 993640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kim H, Dhall A, Liu Y, Bawazir M, Koo H, Hwang G, MBio 2021, 12, e00651–00621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].National Committee for Clinical Laboratory Standards, Reference method for broth dilution antifungal susceptibility testing of conidium-forming filamentous fungi: proposed standard M38-A (NCCLS, Wayne, 2002) [Google Scholar]
- [42].Arora P, L W, Huang X, Yu W, Huang R, Jiang Q, Chen C, Int. J. Mol. Sci. 2022, 23, 4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Li W, Huang X, Yu W, Xu Y, Huang R, Park J, Moshaverinia A, Arora P, Chen C, J. Dent. Res. 2022, 101, 802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ansari S, Chen C, Xu X, Annabi N, Zadeh HH, Wu BM, Khademhosseini A, Shi S, Moshaverinia A, Ann. Biomed. Eng. 2016, 44, 1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Shamir ER, Ewald AJ, Nat. Rev. Mol. Cell Biol. 2014, 15, 647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


