Abstract
Purpose:
To demonstrate that random forest models trained on a large national sample can accurately predict relevant outcomes and may ultimately contribute to future clinical decision support tools in IR.
Materials and Methods:
Patient data from years 2012–2014 of the National Inpatient Sample were used to develop random forest machine learning models to predict iatrogenic pneumothorax after computed tomography–guided transthoracic biopsy (TTB), in-hospital mortality after transjugular intrahepatic portosystemic shunt (TIPS), and length of stay > 3 days after uterine artery embolization (UAE). Model performance was evaluated with area under the receiver operating characteristic curve (AUROC) and maximum F1 score. The threshold for AUROC significance was set at 0.75.
Results:
AUROC was 0.913 for the TTB model, 0.788 for the TIPS model, and 0.879 for the UAE model. Maximum F1 score was 0.532 for the TTB model, 0.357 for the TIPS model, and 0.700 for the UAE model. The TTB model had the highest AUROC, while the UAE model had the highest F1 score. All models met the criteria for AUROC significance.
Conclusions:
This study demonstrates that machine learning models may suitably predict a variety of different clinically relevant outcomes, including procedure-specific complications, mortality, and length of stay. Performance of these models will improve as more high-quality IR data become available.
As more data become available with the widespread adoption of electronic health records (EHRs) and multi-institution registries, the development of robust machine learning algorithms for clinical outcome prediction becomes increasingly feasible. Furthermore, the continuing standardization of EHR interfaces is making it possible to efficiently integrate third-party artificial intelligence (AI) applications for point-of-care decision support (1). Traditionally, clinical risk calculators have been developed outside of the EHR using scoring systems or linear models validated on a limited sample of patients. Modern risk calculators using machine learning offer the potential to uncover nonlinear associations missed by these older models. They can be tailored to individual patient populations and can adapt automatically with access to new data.
In clinical medicine, the focus of many new AI applications has been on computer vision using deep learning, a tool that has broad implications in fields that rely on the identification of images, such as radiology, pathology, dermatology, and ophthalmology (2,3). However, the literature suggests that AI can also be harnessed to forecast outcomes, such as mortality, length of stay (LOS), and readmission, to help clinicians provide patients with accurate prognoses and plan for anticipated complications (4–6). Random forests are a type of supervised machine learning model used for binary classification problems. Recently published work has demonstrated that it is possible to develop random forests with data from a single institution to predict outcomes after procedures in interventional radiology (IR). One study presented a model developed to predict survival following yttrium-90 radio-embolization using only baseline factors obtained before treatment (7). Another group used random forests to classify treatment response after transarterial chemoembolization (8). AI solutions, if properly integrated into the clinical workflow, may help interventional radiologists with planning before the procedure by providing predictions for a wide variety of procedures. The purpose of this study was to demonstrate that random forest models trained on a large national sample can accurately predict these outcomes and may ultimately contribute to future clinical decision support tools in IR.
MATERIALS AND METHODS
This was a retrospective study using de-identified data from a national database. The data did not contain any of the 18 personal health identifiers designated by the Health Insurance Portability and Accountability Act, and therefore this study did not qualify as human subjects research as defined by the institutional review board.
Materials
The National Inpatient Sample (NIS) is the “largest publicly available all-payer inpatient health care database in the United States,” compiled annually by the Agency for Healthcare Research and Quality as part of the Healthcare Cost and Utilization Project (HCUP) (9). The sample of hospitals participating in the Healthcare Cost and Utilization Project encompasses > 95% of the US population. This study examined patients from the NIS presenting in 2012–2014.
Inclusion and Exclusion Criteria
This study used machine learning to predict outcomes after 3 different procedures that have varying incidence in the available data. The following outcomes were investigated:
Iatrogenic pneumothorax from a computed tomography–guided transthoracic biopsy (TTB)
In-hospital mortality after transjugular intrahepatic portosystemic shunt (TIPS)
LOS > 3 days after uterine artery embolization (UAE)
All patients undergoing each of the 3 selected procedures were queried using the associated International Classification of Diseases, 9th Revision, Clinical Modification procedure codes: 3326 (TTB), 39.1 (TIPS), 68.24 (UAE with coils), and 68.25 (UAE without coils). Features used as inputs for all 3 of the models are listed in Tables E1–E3 (available online on the article’s Supplemental Material page at www.jvir.org). Features corresponding to historical clinical diagnoses were selected separately for each model; features with missing data in > 1% of rows were excluded. For the TTB model, 84 binary features indicating history of a previously diagnosed medical condition met the inclusion criteria. For the TIPS model, 68 such features were included, and 46 were included for the UAE model. The outcome of interest for each model was extracted from the original dataset and stored as a reference. Subsequently, patients were excluded from analysis if a data element for any of the selected features or outcomes was absent.
Patient Characteristics
Following application of the inclusion and exclusion criteria to the original data, 12,046 patients who underwent TTB, 2,263 patients who underwent TIPS, and 826 patients who underwent UAE were identified. Of the patients who underwent TTB, 937 (7.78%) had a reported iatrogenic pneumothorax. Of the TIPS patients who underwent TIPS, 184 (8.13%) died during their admission. Of the UAE patients who underwent UAE, 231 (28.0%) had an LOS > 3 days. Patient demographic information is presented in Table 1.
Table 1.
Patient Demographic and Clinical Information
| Data Element | TTB | TIPS | UAE |
|---|---|---|---|
| Age, y, median | 69 (1–90) | 57 (0–89) | 44 (17–90) |
| (range) | |||
| Medical comorbidities, median (range) | 6(0–22) | 6 (0–19) | 2 (0–17) |
| Sex | |||
| Female | 5,761 (47.8%) | 807 (35.7%) | 826 (100%) |
| Male | 6,285 (52.2%) | 1,455 (64.3%) | 0 (0%) |
| Race | |||
| White | 8,954 (74.3%) | 1,674 (74.0%) | 270 (32.7%) |
| Black | 1,643 (13.6%) | 121 (5.3%) | 368 (44.5%) |
| Hispanic | 791 (6.6%) | 309 (13.7%) | 105 (12.7%) |
| Asian/Pacific Islander | 275 (2.3%) | 32 (1.4%) | 36 (4.4%) |
| Native American | 69 (0.6%) | 31 (1.4%) | 4 (0.5%) |
| Other | 314 (2.6%) | 95 (4.2%) | 43 (5.2%) |
| National ZIP income quartile* | |||
| 1 | 3,816 (31.7%) | 710 (31.4%) | 226 (27.4%) |
| 2 | 3,289 (27.3%) | 623 (27.5%) | 154 (18.6%) |
| 3 | 2,764 (22.9%) | 536 (23.7%) | 205 (24.8%) |
| 4 | 2,177 (18.1%) | 393 (17.4%) | 241 (29.2%) |
| No. patients with outcome of interest† | 937 (7.78%) | 184 (8.13%) | 231 (28.0%) |
| No. patients | 12,046 | 2263 | 826 |
Note–Demographic and comorbidity information for the patients in each of the 3 clinical problems.
TIPS = transjugular intrahepatic portosystemic shunt; TTB = transthoracic biopsy; UAE = uterine artery embolization.
Quartile 1 is the highest (richest) national income quartile by ZIP code, and quartile 4 is the lowest (poorest) quartile.
Iatrogenic pneumothorax, death, and long length of stay, respectively.
Data Partitioning and Resampling
Before analysis, the data were randomly partitioned into training (50%), tuning (25%), and testing (25%) sets. Final counts are included in Table E4 (available online on the article’s Supplemental Material page at www.jvir.org). It is known that classification using machine learning may suffer when the outcome frequency in the training set is imbalanced (10). To address this issue, the training set was resampled using the Synthetic Minority Oversampling Technique (SMOTE) (11), which has been validated for clinical outcome prediction (12).
Random Forest Model
Random forests rely on an ensemble of decision trees to classify inputs. This is generally done to improve the performance of the random forest model compared with individual decision trees, which are prone to overfitting on the training data. In this study, 3 different random forest classifiers were developed from the training sets following the use of SMOTE. The models were then evaluated on the tuning set so that hyperparameters could be adjusted for optimal performance.
Each decision tree within a random forest is trained on a random sample of the original data using a random subset of features. At every branching point, the feature that minimizes the entropy (maximizes information gain) of the resulting nodes is selected (13). In essence, the decision tree attempts to separate the data into groups that have the least heterogeneity of outcomes (Fig 1 shows an example). The branching continues until all of the features have been used or all of the tree’s terminal nodes have reached maximum internal concordance. During model evaluation, outputs across all of the decision trees are averaged to calculate a final probability of the outcome; the threshold probability can then be set to generate a prediction.
Figure 1.
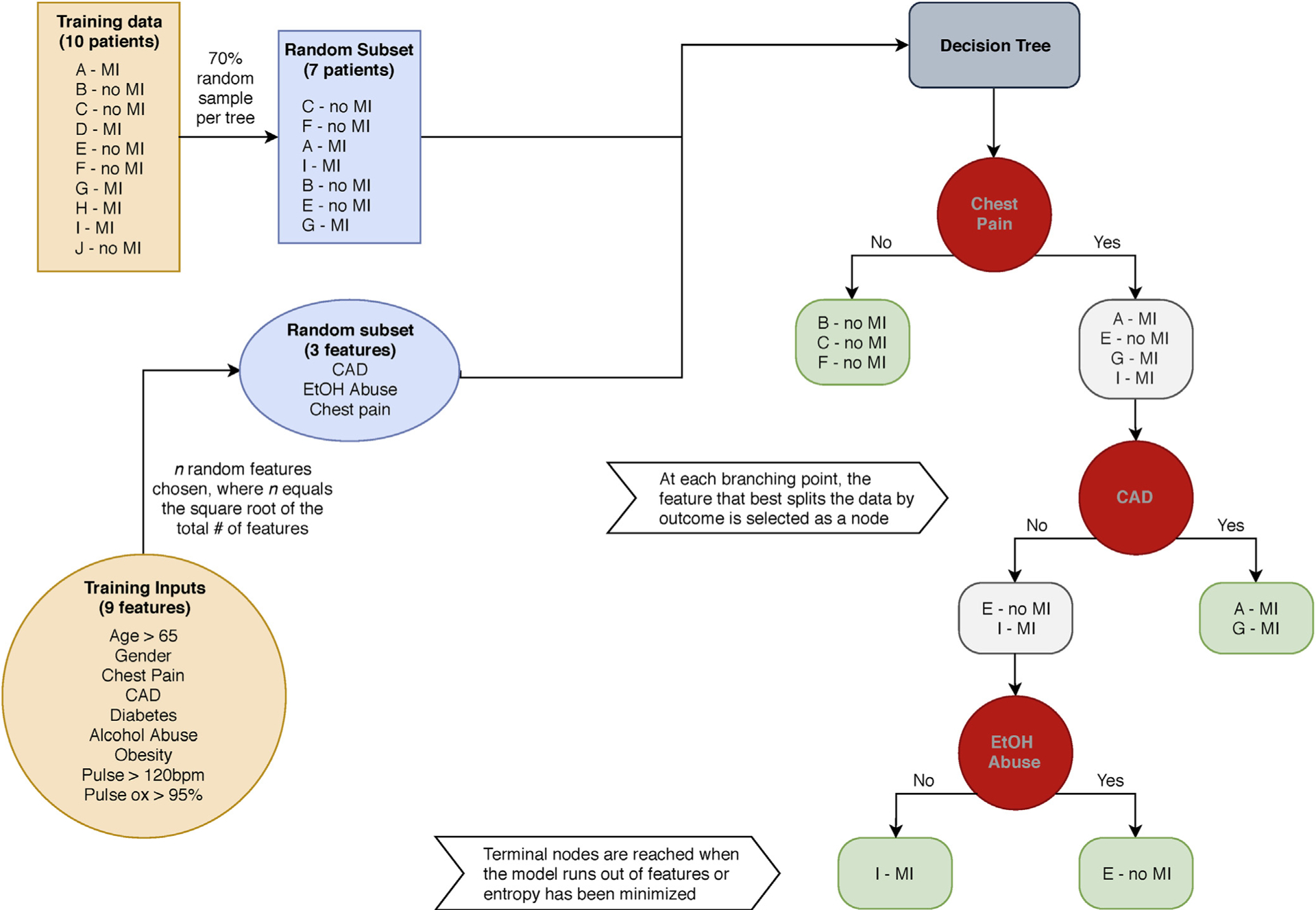
Decision tree example. Pictured is a hypothetical example of training a single decision tree within a random forest model for predicting myocardial infarction (MI). CAD = coronary artery disease; EtOH = alcohol.
Model Evaluation
The trained TTB, TIPS, and UAE models were used to generate predictions on the testing data for iatrogenic pneumothorax, mortality, and extended LOS, respectively. Predictions from the models were then compared against the reference outcomes. Receiver operating characteristic (ROC) curves and probability histograms were plotted for each of the models. Performance was evaluated using area under the receiver operating characteristic curve (AUROC) and maximum F1 score. The following standard guidelines for AUROC interpretation were used: < 0.70, poor; 0.70–0.80, acceptable; 0.80–0.90, excellent; > 0.90, outstanding (14). The associated threshold and specificity for a 90% fixed sensitivity were determined. Additionally, precision and recall corresponding to the maximum F1 score were calculated. Training and evaluation speeds were recorded for each model.
Software
Data preprocessing and analysis were conducted using Version 0.6.2 of the Julia programming language (15). Resampling, model development, and plotting were implemented using (PredictMD), an open-source machine learning toolkit, designed by the authors to provide a uniform interface for developing and testing clinical prediction tools ("BenchmarkTools.jl"). Runtimes were assessed using the BenchmarkTools.jl package (16). All computations were performed on a consumer-grade laptop computer (2 GHz Intel Core i5, 8 GB DDR3 RAM, 256 GB SSD; Intel Corp, Santa Clara, California).
RESULTS
The final hyperparameters for each random forest model are shown in Table 2. The number of trees was 1,000 for all models. SMOTE oversampling percentage was hand-tuned to generate sufficient data to train the models. For each forest, the number of features per tree was set to integer value closest to the square root of the total number of features.
Table 2.
Model Hyperparameters
| TTB | TIPS | UAE | |
|---|---|---|---|
| SMOTE oversampling percentage | 300% | 1,000% | 800% |
| Total number of features in random forest | 95 | 81 | 58 |
| Total number of trees in random forest | 1,000 | 1,000 | 1,000 |
| Number of features per individual tree | 10 | 9 | 8 |
Note–Hyperparameters for each of the 3 random forest models. Hyperparameters describe the process used for constructing the random forest models. The SMOTE oversampling percentage describes how many synthetic data points were created from the original dataset. For example, a SMOTE oversampling percentage of 300% (respectively, 1,000% and 800%) means that for each patient in the original dataset who had the outcome of interest, 3 (respectively, 10 and 8) synthetic data points were created. Each random forest consisted of 1,000 trees. Each individual tree was trained on a subset of features (selected randomly) and a subset of patients (70% of the total patients, selected randomly). There was no maximum node depth (ie, there was no limit to the size of any individual tree).
SMOTE = Synthetic Minority Oversampling Technique; TIPS = transjugular intrahepatic portosystemic shunt; TTB transthoracic biopsy; UAE = uterine artery embolization.
The performance of the models on the testing set is shown in Table 3. AUROC was 0.913 for the TTB model, 0.788 for the TIPS model, and 0.879 for the UAE model. Maximum F1 score was 0.532 for the TTB model, 0.357 for the TIPS model, and 0.700 for the UAE model. The TTB model had the highest AUROC, while the UAE model had the highest F1 score. Based on the established guidelines for AUROC interpretation, the TTB model was “outstanding,” the TIPS model was “acceptable,” and the UAE model was “excellent” (14). Model training and evaluation speeds are presented in Table 4. Additional features per tree was related to longer training times and slower evaluation speeds.
Table 3.
Model Performance Metrics
| TTB | TIPS | UAE | |
|---|---|---|---|
| AUROC | 0.913 | 0.788 | 0.879 |
| Maximum F1 score | 0.532 | 0.376 | 0.700 |
| Precision (at maximum F1 score) | 0.426 | 0.279 | 0.563 |
| Recall (at maximum F1 score) | 0.709 | 0.576 | 0.915 |
| Sensitivity | 90.0% | 90.0% | 90.0% |
| Specificity (at sensitivity of 90%) | 82.4% | 45.3% | 68.0% |
| Threshold (at sensitivity of 90%) | 0.209 | 0.103 | 0.195 |
Note–Performance metrics for each of the random forest models when evaluated on the testing set. AUROC is a good overall summary of each model’s performance. The maximum F1 score is useful for evaluating the performance of each model on imbalanced data (ie, when there are far more patients without the outcome of interest than with the outcome of interest). The F1 score is defined as the harmonic mean of precision (positive predictive value) and recall (sensitivity). Precision and recall values corresponding to the maximum F1 score have also been provided. Threshold refers to the classifier value that fixed sensitivity at 90%. The corresponding specificity was computed and is reported.
AUROC = area under the receiver operating characteristic curve; TIPS = transjugular intrahepatic portosystemic shunt; TTB = transthoracic biopsy; UAE = uterine artery embolization.
Table 4.
Model Training and Evaluation Speeds
| TTB | TIPS | UAE | |
|---|---|---|---|
| Time to train random forest, s | 8.667 | 5.435 | 3.340 |
| Time to run random forest | 0.0126 | 0.0124 | 0.0077 |
Note–Time required to train the random forests and to evaluate them on a new patient. Smaller values correspond to faster speeds.
TIPS = transjugular intrahepatic portosystemic shunt; TTB = transthoracic biopsy; UAE = uterine artery embolization
The distribution of probability outputs by each model were plotted as classifier histograms (Fig E1a–c [available online on the article’s Supplemental Material page at www.jvir.org]). These figures demonstrate that, among the 3 models, the TTB model does the best job of separating the positive classes from the negative classes. This is consistent with the fact that it has the highest AUROC value. For each model, an ROC curve was plotted (Fig 2a–c). Comparison of the 3 ROC curves shows that the ROC curve of the TTB model is closest to the theoretical ideal classifier, which again is consistent with the result that it has the highest AUROC.
Figure 2.
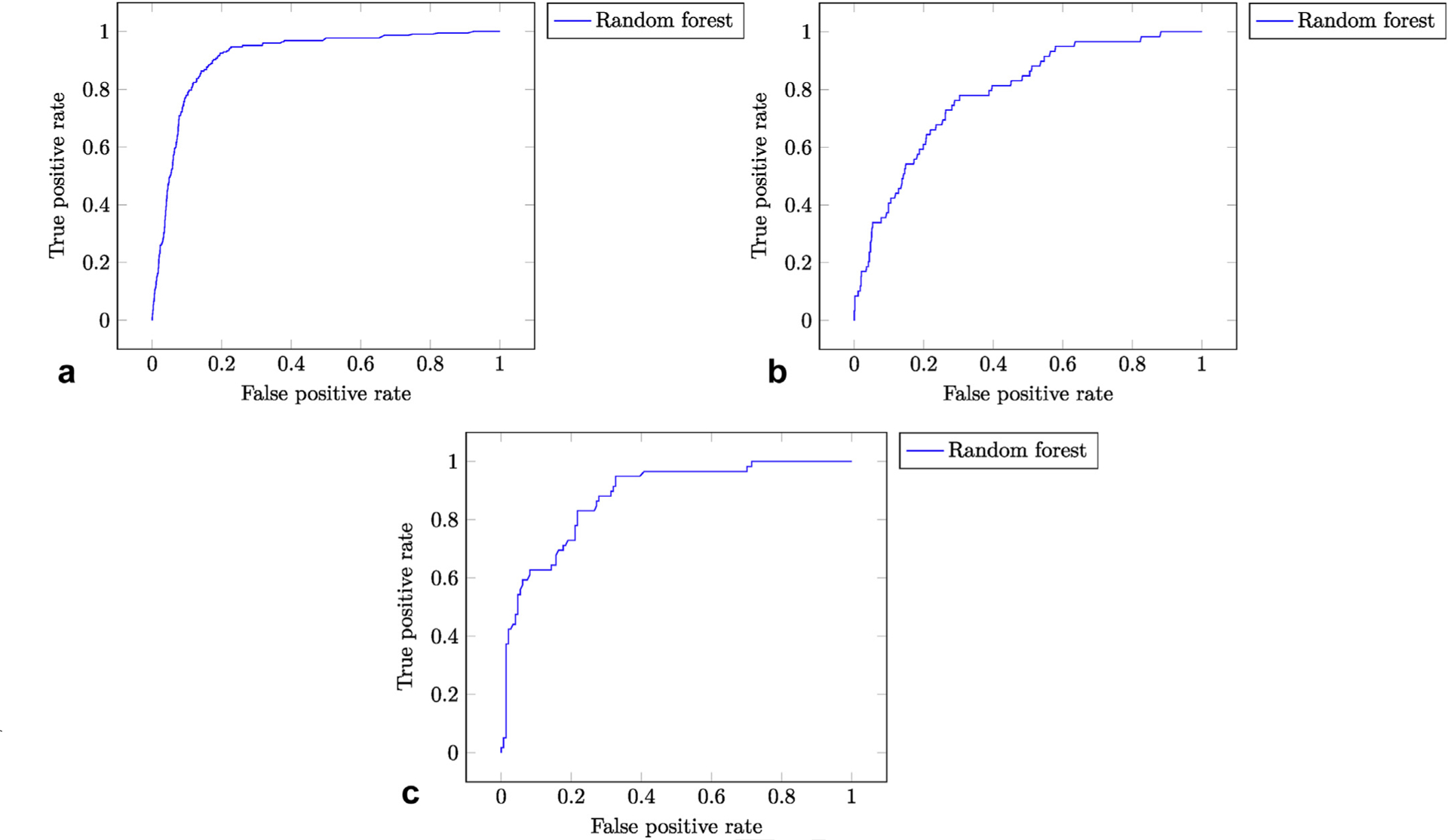
(a–c) ROC curves. The true positive rate (sensitivity) is plotted against the false positive rate (1 – specificity) over various thresholds. The theoretical ideal classifier corresponds to a point in the top left corner (sensitivity 100%, specificity 100%). (a) TTB ROC curve. (b) TIPS ROC curve. (c) UAE ROC curve.
DISCUSSION
This study presents a framework for developing and evaluating random forest models for the prediction of outcomes after IR procedures. The results demonstrate that these models may be effective at predicting outcomes after procedures when trained on a large national dataset. This is particularly exciting considering that all of the model inputs are features that were available before admission. Based on AUROC, the TTB model is the most accurate (0.913), which may be due in part to the fact that it was trained on substantially more data than the other 2 models. F1 scores provide an indication of how well a model predicts instances of a minority class; the high F1 score (0.700) (and corresponding precision and recall) of the UAE model is likely a result of less class imbalance in the original dataset.
For the TTB, TIPS, and UAE models, it is possible to achieve a high sensitivity for the predicted outcome without completely sacrificing specificity, albeit with vastly different results (specificities of 82.4%, 45.3%, and 68.0%, respectively). In these examples, sensitivity was fixed at 90% because importance was given to identifying patients at high risk for developing the studied outcomes. Users seeking to identify low-risk patients (eg, to screen for appropriate outpatient candidates) would adjust the predictive threshold to maximize specificity. All the possible pairs of sensitivity and specificity for each model are represented by points on the ROC curves (Fig 2a–c).
The use of the SMOTE algorithm was essential in addressing the problem that arises from training a classification model on imbalanced data. When predicting negative outcomes with low incidence, models tend to be biased toward the majority outcome, thus underpredicting instances of the minority outcome. Although using SMOTE can decrease the effect of this bias, the classifier’s ability to predict minority outcomes may still be limited when applied to imbalanced testing data, as demonstrated by the variation in maximum F1 score between models.
Prior work in identifying patients at high risk for pneumothorax following TTB focused on determining individual risk factors, such as age, emphysema, lesion depth and size, and needle passes, as independent predictors for developing a pneumothorax (17–19). The only published application of machine learning to this problem has been directed toward automatically detecting pneumothoraces on chest radiographs after biopsy using artificial neural networks (3). The results demonstrate that a machine learning model does not require technical, procedure-specific information to accurately predict this outcome, as suggested in previous studies; however, integration of these features into the TTB model may improve its already “outstanding” performance.
Several popular tools currently exist for predicting outcomes in patients with cirrhosis, such as the Model for End-Stage Liver Disease (MELD) score and the Child-Pugh score. These have also been repurposed to predict 30-day mortality following TIPS (AUROC of 0.878 and 0.822, respectively) (20) as well as in-hospital mortality in the setting of acute upper gastrointestinal bleeding (AUROC of 0.810 and 0.796, respectively) (21). However, both MELD and Child-Pugh scores suffer from an absence of demographic factors and medical comorbidities and have not been optimized specifically for patients with TIPS. The random forests in this study do not perform as well as MELD or Child-Pugh score in predicting mortality, but they are still able to make predictions using only demographic factors and medical comorbidities. Trivedi et al (22) published an analysis of national trends and outcomes following TIPS using the years 2003–2012 of the NIS and concluded that demographic, socioeconomic, and clinical factors “may aid clinicians in better assessing preprocedural risk.” This study implements the insightful recommendation of the authors by providing a computerized clinical decision support tool based on those factors.
UAE is a safe and effective alternative to hysterectomy for the treatment of symptomatic fibroid disease as well as emergent uterine bleeding, especially in the setting of postpartum hemorrhage (23,24). It has been shown to decrease hospital LOS by > 4 days and results in fewer major complications compared with hysterectomy (23). Despite these advantages, in some cases UAE is not curative, and surgical intervention becomes necessary. One study showed that classification of patients undergoing UAE for postpartum hemorrhage based on uterine artery staining on angiography can help identify patients at increased risk for UAE failure (24), but otherwise there has been a dearth of information published regarding patient selection for UAE in both the gynecology and the radiology literature. This study presents a methodology for developing models for patient selection and risk prediction in UAE. Identification of patients at high risk for extended LOS before the procedure can be used to improve patient selection, inform shared decision making, and anticipate utilization of hospital resources.
Random forests are well-studied machine learning models with multiple areas of strength. As demonstrated in Table 4, random forests offer competitive speeds during both the training and the evaluation phases. Additionally, the ensemble structure of random forests allows the training phase to be parallelized over multiple central processing unit cores or even distributed over multiple computers, making them suitable for use in a real-time setting (eg, an EHR) where performance is prioritized (13). Random forests also excel at processing high-dimensional data with a large number of input features (25). To increase the portability of these models, clinical applications of machine learning can be programmed as web-based calculators that allow providers to access the tools from any computer, tablet, or mobile device. As computerized clinical decision support becomes more common, these tools will be developed into applications that use open interoperability standards to integrate directly into commercial EHR systems and automatically display outcome predictions in the patient chart (1).
Random forests discover and exploit nonlinear relationships among the input variables. These nonlinearities help make random forests more accurate than linear models. However, there is no intuitive method for visualizing these relationships and thereby understand the relative importance of contributing features, which has led some authors to describe random forests as black boxes (26). This is a limitation of random forests that is shared by other nonlinear models, such as the widely used pooled cohort equations for predicting atherosclerotic cardiovascular disease (27).
The incidence of pneumothorax is lower in the NIS sample (7.78%) compared with published figures (9%–54%) (17). This may be due in part to providers not documenting clinically negligible pneumothoraces or miscoding them as noniatrogenic. This highlights the importance of consistent documentation when mining large national databases and suggests that the TTB model is effective only at predicting a documented iatrogenic pneumothorax. Fortunately, this outcome likely represents a clinically relevant event that can impact patient management (eg, longer observation, additional imaging, chest tube placement).
One salient limitation of using the NIS database for TIPS outcome prediction is the lack of laboratory data, which are included in the aforementioned scoring tools (MELD and Child-Pugh). Ultimately, any model that attempts to predict mortality after TIPS should integrate demographic, clinical, and laboratory data. If trained on a sample that includes laboratory values, it is expected that the performance of this machine learning model would increase substantially and surpass the predictive capabilities of linear scoring tools alone for mortality after TIPS.
The analysis here is limited owing to its inclusion of patients undergoing UAE for all causes. Machine learning algorithms require a sufficiently large sample to accurately train models; the low volume of reported UAEs in the NIS necessitated the inclusion of both elective and nonelective procedures. Performance may be biased by differences in demographics and comorbidities between patients from elective and nonelective groups. Decision support tools must be developed using specific data related to their population of interest before they can be integrated into clinical workflow.
The features and sample sizes in these models were limited owing to the use of a general national database of inpatient admissions. This underscores the underlying need to develop a multicenter registry of IR procedures, which can be leveraged to enhance clinical decision support. The methodology in this study may then be applied to the multicenter registry to develop more powerful random forest models.
In conclusion, this study showed that random forest models may be used to predict a variety of different clinically relevant, postprocedural outcomes. Accuracy of these models is influenced by multiple factors, including dataset size, imbalance in outcome incidence, available features, and proportion of missing data. Ultimately, results from this investigation encourage the application of machine learning methods to IR decision support tools through the use of high-quality data.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded in part by National Institutes of Health Grant U54GM115677. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
ABBREVIATIONS
- AI
artificial intelligence
- AUROC
area under the receiver operating characteristic curve
- EHR
electronic health record
- LOS
length of stay
- MELD
Model for End-Stage Liver Disease
- NIS
National Inpatient Sample
- ROC
receiver operating characteristic
- SMOTE
Synthetic Minority Oversampling Technique
- TIPS
transjugular intrahepatic portosystemic shunt
- TTB
transthoracic biopsy
- UAE
uterine artery embolization
Footnotes
None of the authors have identified a conflict of interest.
From the SIR 2019 Annual Scientific Meeting.
Figure E1 and Tables E1n, can be found by accessing the online version of this article on www.jvir.org and clicking on the Supplemental Material tab.
REFERENCES
- 1.Bloomfield RA, Polo-Wood F, Mandel JC, Mandl KD. Opening the Duke electronic health record to apps: implementing SMART on FHIR. Int J Med Inform 2017; 99:1–10. [DOI] [PubMed] [Google Scholar]
- 2.Saria S, Butte A, Sheikh A. Better medicine through machine learning: what’s real, and what’s artificial? PLoS Med 2018; 15:e1002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor AG, Mielke C, Mongan J. Automated detection of moderate and large pneumothorax on frontal chest x-rays using deep convolutional neural networks: a retrospective study. PLoS Med 2018; 15:e1002697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes S, Hamrock E, Toerper M, Siddiqui S, Levin S. Real-time prediction of inpatient length of stay for discharge prioritization. J Am Med Inform Assoc 2016; 23:e2–e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldstein BA, Pencina MJ, Montez-Rath ME, Winkelmayer WC. Predicting mortality over different time horizons: which data elements are needed? J Am Med Inform Assoc 2017; 24:176–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalagara S, Eltorai AEM, Durand WM, DePasse JM, Daniels AH. Machine learning modeling for predicting hospital readmission following lumbar laminectomy. J Neurosurg Spine 2019; 30:344–352. [DOI] [PubMed] [Google Scholar]
- 7.Ingrisch M, Schöppe F, Paprottka KJ, et al. Prediction of 90 Y-radioembolization outcome from pre-therapeutic factors with random survival forests. J Nucl Med 2018; 59:769–773. [DOI] [PubMed] [Google Scholar]
- 8.Abajian A, Murali N, Savic LJ, et al. Predicting treatment response to intra-arterial therapies for hepatocellular carcinoma with the use of supervised machine learning—an artificial intelligence concept. J Vasc Interv Radiol 2018; 29:850–857.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cost Healthcare and Project Utilization. Overview of the National Inpatient Sample. Available at: https://www.hcup-us.ahrq.gov/nisoverview.jsp. Accessed June 10, 2019. [Google Scholar]
- 10.Japkowicz N, Stephen S. The class imbalance problem: a systematic study. Intell Data Anal 2002; 6:429–449. [Google Scholar]
- 11.Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: Synthetic Minority Over-sampling Technique. J Artif Intell Res 2002; 16:321–357. [Google Scholar]
- 12.Ho KC, Speier W, El-Saden S, et al. Predicting discharge mortality after acute ischemic stroke using balanced data. AMIA Annu Symp Proc 2014; 2014:1787–1796. [PMC free article] [PubMed] [Google Scholar]
- 13.Van Essen B, Macaraeg C, Gokhale M, Prenger R. Accelerating a random forest classifier: multi-core, GP-GPU, or FPGA?. In: Proceedings of the 2012 IEEE 20th International Symposium on Field-Programmable Custom Computing Machines, FCCM 2012. IEEE; 2012. p. 232–239. [Google Scholar]
- 14.Hosmer DW Jr, Lemeshow S, Sturdivant RX. Area under the receiver operating characteristic curve. Applied Logistic Regression. 3rd ed. Hoboken, NJ: John Wiley & Sons; 2013. p. 173–182. [Google Scholar]
- 15.Bezanson J, Edelman A, Karpinski S, Shah VB. Julia: a fresh approach to numerical computing. SIAM Rev 2017; 59:65–98. [Google Scholar]
- 16.Chen J, Revels J. Robust benchmarking in noisy environments. In: Proceedings of the 20th Annual IEEE High Performance Extreme Computing Conference; 2016. [Google Scholar]
- 17.Boskovic T, Stanic J, Pena-Karan S, et al. Pneumothorax after transthoracic needle biopsy of lung lesions under CT guidance. J Thorac Dis 2014; 6(Suppl 1):S99–S107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moreland A, Novogrodsky E, Brody L, et al. Pneumothorax with prolonged chest tube requirement after CT-guided percutaneous lung biopsy: incidence and risk factors. Eur Radiol 2016; 26:3483–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim WH, Park CM, Yoon SH, et al. Time-dependent analysis of incidence, risk factors and clinical significance of pneumothorax after percutaneous lung biopsy. Eur Radiol 2018; 28:1328–1337. [DOI] [PubMed] [Google Scholar]
- 20.Gaba RC, Couture PM, Bui JT, et al. Prognostic capability of different liver disease scoring systems for prediction of early mortality after transjugular intrahepatic portosystemic shunt creation. J Vasc Interv Radiol 2013; 24: 411–420.e4. [DOI] [PubMed] [Google Scholar]
- 21.Peng Y, Qi X, Dai J, Li H, Guo X. Child-Pugh versus MELD score for predicting the in-hospital mortality of acute upper gastrointestinal bleeding in liver cirrhosis. Int J Clin Exp Med 2015; 8:751–757. [PMC free article] [PubMed] [Google Scholar]
- 22.Trivedi PS, Rochon PJ, Durham JD, Ryu RK. National trends and outcomes of transjugular intrahepatic portosystemic shunt creation using the Nationwide Inpatient Sample. J Vasc Interv Radiol 2016; 27:838–845. [DOI] [PubMed] [Google Scholar]
- 23.Pinto I, Chimeno P, Romo A, et al. Uterine fibroids: uterine artery embolization versus abdominal hysterectomy for treatment—a prospective, randomized, and controlled clinical trial. Radiology 2003; 226:425–431. [DOI] [PubMed] [Google Scholar]
- 24.Ueshima E, Sugimoto K, Okada T, et al. Classification of uterine artery angiographic images: a predictive factor of failure in uterine artery embolization for postpartum hemorrhage. Jpn J Radiol 2018; 36:394–400. [DOI] [PubMed] [Google Scholar]
- 25.Darst BF, Malecki KC, Engelman CD. Using recursive feature elimination in random forest to account for correlated variables in high dimensional data. BMC Genet 2018; 19:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan L, Liu G, Mao X, et al. Development of prediction models using machine learning algorithms for girls with suspected central precocious puberty: retrospective study. JMIR Med Inform 2019; 7:e11728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goff DC, Lloyd-Jones DM, Bennett G, et al. Reply: 2013 ACC/AHA guideline on the assessment of cardiovascular risk. J Am Coll Cardiol 2014; 63:2886. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


