Abstract
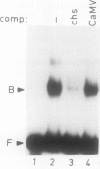
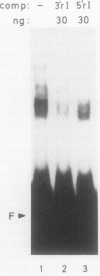
A positive regulatory element directing maximal expression of the Antirrhinum majus chalcone synthase promoter was characterized by protein-DNA-interaction studies and cis deletion analysis. The positive regulatory element consists of a 47 base pair direct repeat between positions −564 and −670 and provides three binding sites for nuclear protein factors from Nicotiana tabacum and Antirrhinum majus. Oligonucleotide competition assays revealed that the same factor(s) interact(s) with all three binding sites. Transient expression of chimeric chalcone synthase-neomycin phosphotransferase II genes in parsley protoplasts demonstrated that both halves of the 47 base pair repeat element are required for its in vivo function. A possible role of redundant binding sites for the positive regulatory function of the 47 base pair repeat element is discussed.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baumruker T., Sturm R., Herr W. OBP100 binds remarkably degenerate octamer motifs through specific interactions with flanking sequences. Genes Dev. 1988 Nov;2(11):1400–1413. doi: 10.1101/gad.2.11.1400. [DOI] [PubMed] [Google Scholar]
- Dangl J. L., Hauffe K. D., Lipphardt S., Hahlbrock K., Scheel D. Parsley protoplasts retain differential responsiveness to u.v. light and fungal elicitor. EMBO J. 1987 Sep;6(9):2551–2556. doi: 10.1002/j.1460-2075.1987.tb02543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferl R. J., Nick H. S. In vivo detection of regulatory factor binding sites in the 5' flanking region of maize Adh1. J Biol Chem. 1987 Jun 15;262(17):7947–7950. [PubMed] [Google Scholar]
- Giuliano G., Pichersky E., Malik V. S., Timko M. P., Scolnik P. A., Cashmore A. R. An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7089–7093. doi: 10.1073/pnas.85.19.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P. J., Kay S. A., Chua N. H. Sequence-specific interactions of a pea nuclear factor with light-responsive elements upstream of the rbcS-3A gene. EMBO J. 1987 Sep;6(9):2543–2549. doi: 10.1002/j.1460-2075.1987.tb02542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P. J., Yong M. H., Cuozzo M., Kano-Murakami Y., Silverstein P., Chua N. H. Binding site requirements for pea nuclear protein factor GT-1 correlate with sequences required for light-dependent transcriptional activation of the rbcS-3A gene. EMBO J. 1988 Dec 20;7(13):4035–4044. doi: 10.1002/j.1460-2075.1988.tb03297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller W., Hahlbrock K. Highly purified "flavanone synthase" from parsley catalyzes the formation of naringenin chalcone. Arch Biochem Biophys. 1980 Apr 1;200(2):617–619. doi: 10.1016/0003-9861(80)90395-1. [DOI] [PubMed] [Google Scholar]
- Johnson P. F., Landschulz W. H., Graves B. J., McKnight S. L. Identification of a rat liver nuclear protein that binds to the enhancer core element of three animal viruses. Genes Dev. 1987 Apr;1(2):133–146. doi: 10.1101/gad.1.2.133. [DOI] [PubMed] [Google Scholar]
- Kaulen Hildegard, Schell Jeff, Kreuzaler Fritz. Light-induced expression of the chimeric chalcone synthase-NPTII gene in tobacco cells. EMBO J. 1986 Jan;5(1):1–8. doi: 10.1002/j.1460-2075.1986.tb04169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlemeier C., Cuozzo M., Green P. J., Goyvaerts E., Ward K., Chua N. H. Localization and conditional redundancy of regulatory elements in rbcS-3A, a pea gene encoding the small subunit of ribulose-bisphosphate carboxylase. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4662–4666. doi: 10.1073/pnas.85.13.4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipphardt S., Brettschneider R., Kreuzaler F., Schell J., Dangl J. L. UV-inducible transient expression in parsley protoplasts identifies regulatory cis-elements of a chimeric Antirrhinum majus chalcone synthase gene. EMBO J. 1988 Dec 20;7(13):4027–4033. doi: 10.1002/j.1460-2075.1988.tb03296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odell J. T., Nagy F., Chua N. H. Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. 1985 Feb 28-Mar 6Nature. 313(6005):810–812. doi: 10.1038/313810a0. [DOI] [PubMed] [Google Scholar]
- Pfeifer K., Prezant T., Guarente L. Yeast HAP1 activator binds to two upstream activation sites of different sequence. Cell. 1987 Apr 10;49(1):19–27. doi: 10.1016/0092-8674(87)90751-3. [DOI] [PubMed] [Google Scholar]
- Reimold U., Kröger M., Kreuzaler F., Hahlbrock K. Coding and 3' non-coding nucleotide sequence of chalcone synthase mRNA and assignment of amino acid sequence of the enzyme. EMBO J. 1983;2(10):1801–1805. doi: 10.1002/j.1460-2075.1983.tb01661.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss B., Sprengel R., Will H., Schaller H. A new sensitive method for qualitative and quantitative assay of neomycin phosphotransferase in crude cell extracts. Gene. 1984 Oct;30(1-3):211–217. doi: 10.1016/0378-1119(84)90122-7. [DOI] [PubMed] [Google Scholar]
- Ryder T. B., Hedrick S. A., Bell J. N., Liang X. W., Clouse S. D., Lamb C. J. Organization and differential activation of a gene family encoding the plant defense enzyme chalcone synthase in Phaseolus vulgaris. Mol Gen Genet. 1987 Dec;210(2):219–233. doi: 10.1007/BF00325687. [DOI] [PubMed] [Google Scholar]
- Schreier P. H., Seftor E. A., Schell J., Bohnert H. J. The use of nuclear-encoded sequences to direct the light-regulated synthesis and transport of a foreign protein into plant chloroplasts. EMBO J. 1985 Jan;4(1):25–32. doi: 10.1002/j.1460-2075.1985.tb02312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Lefert P., Becker-André M., Schulz W., Hahlbrock K., Dangl J. L. Functional architecture of the light-responsive chalcone synthase promoter from parsley. Plant Cell. 1989 Jul;1(7):707–714. doi: 10.1105/tpc.1.7.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Lefert P., Dangl J. L., Becker-André M., Hahlbrock K., Schulz W. Inducible in vivo DNA footprints define sequences necessary for UV light activation of the parsley chalcone synthase gene. EMBO J. 1989 Mar;8(3):651–656. doi: 10.1002/j.1460-2075.1989.tb03422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer H., Bonas U., Saedler H. Transposon-induced alterations in the promoter region affect transcription of the chalcone synthase gene of Antirrhinum majus. Mol Gen Genet. 1988 Jan;211(1):49–55. doi: 10.1007/BF00338392. [DOI] [PubMed] [Google Scholar]
- Staiger D., Kaulen H., Schell J. A CACGTG motif of the Antirrhinum majus chalcone synthase promoter is recognized by an evolutionarily conserved nuclear protein. Proc Natl Acad Sci U S A. 1989 Sep;86(18):6930–6934. doi: 10.1073/pnas.86.18.6930. [DOI] [PMC free article] [PubMed] [Google Scholar]