Abstract
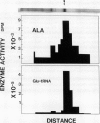
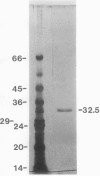
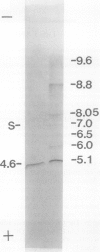
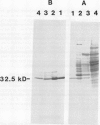
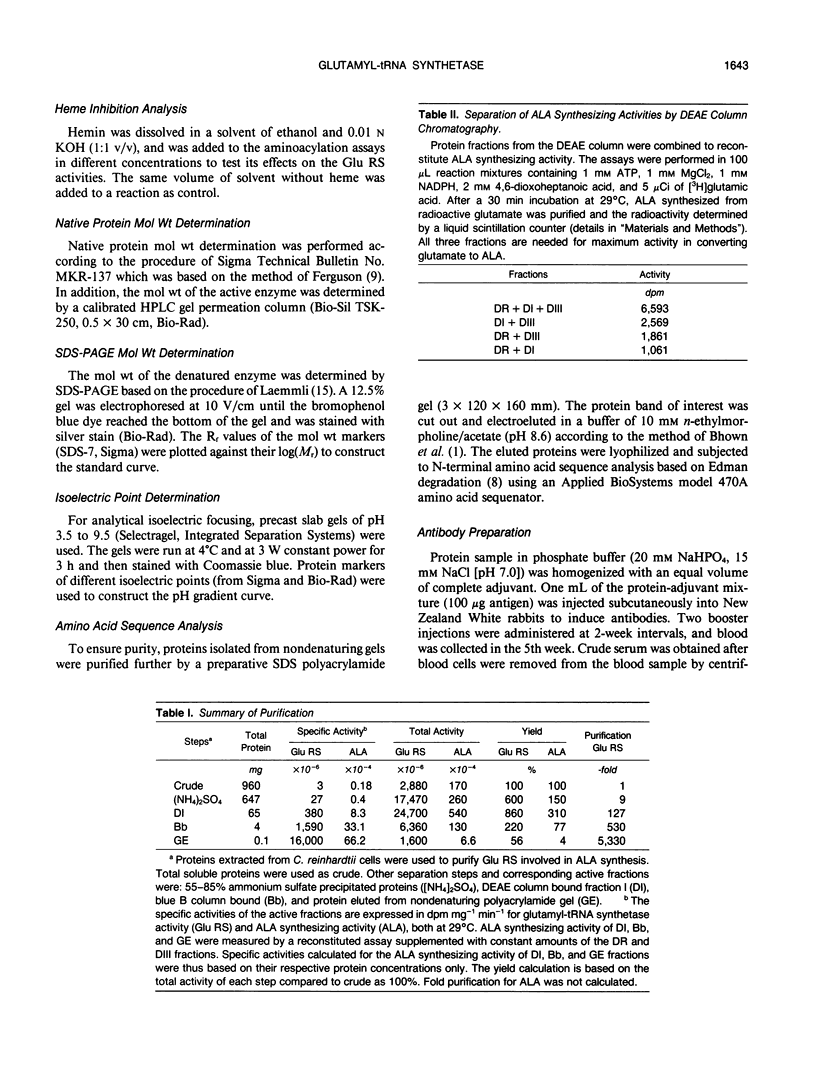
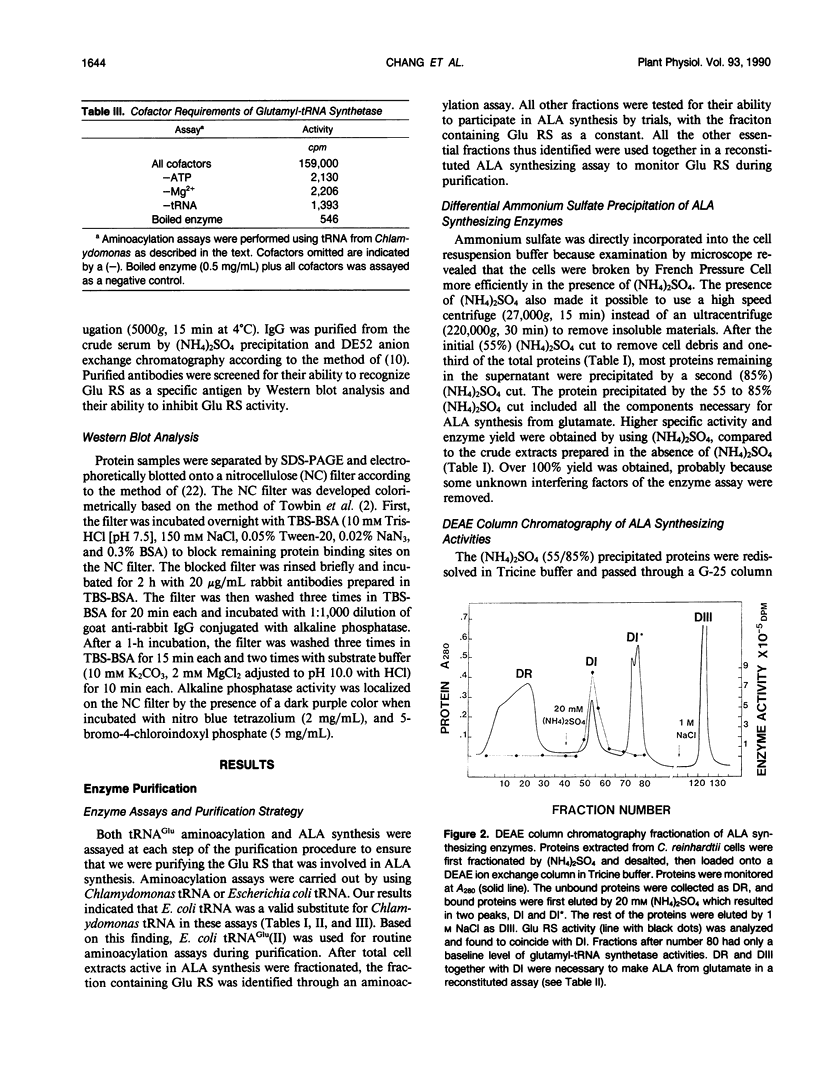
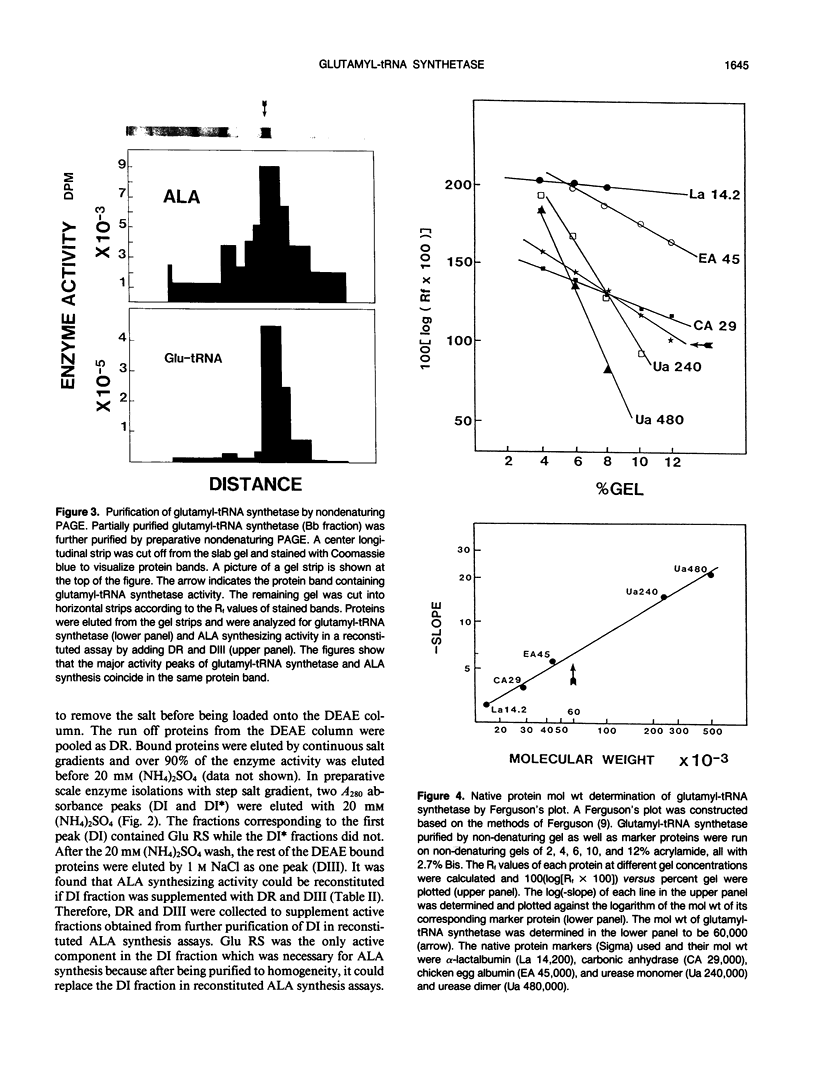
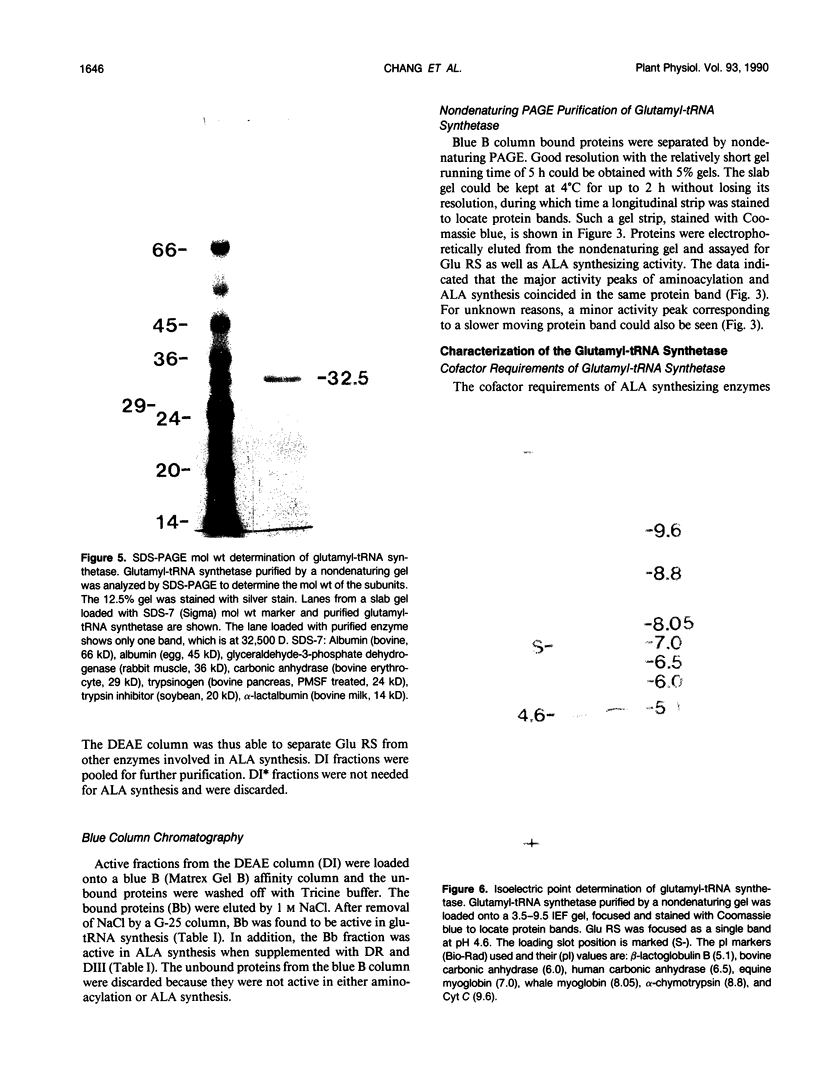
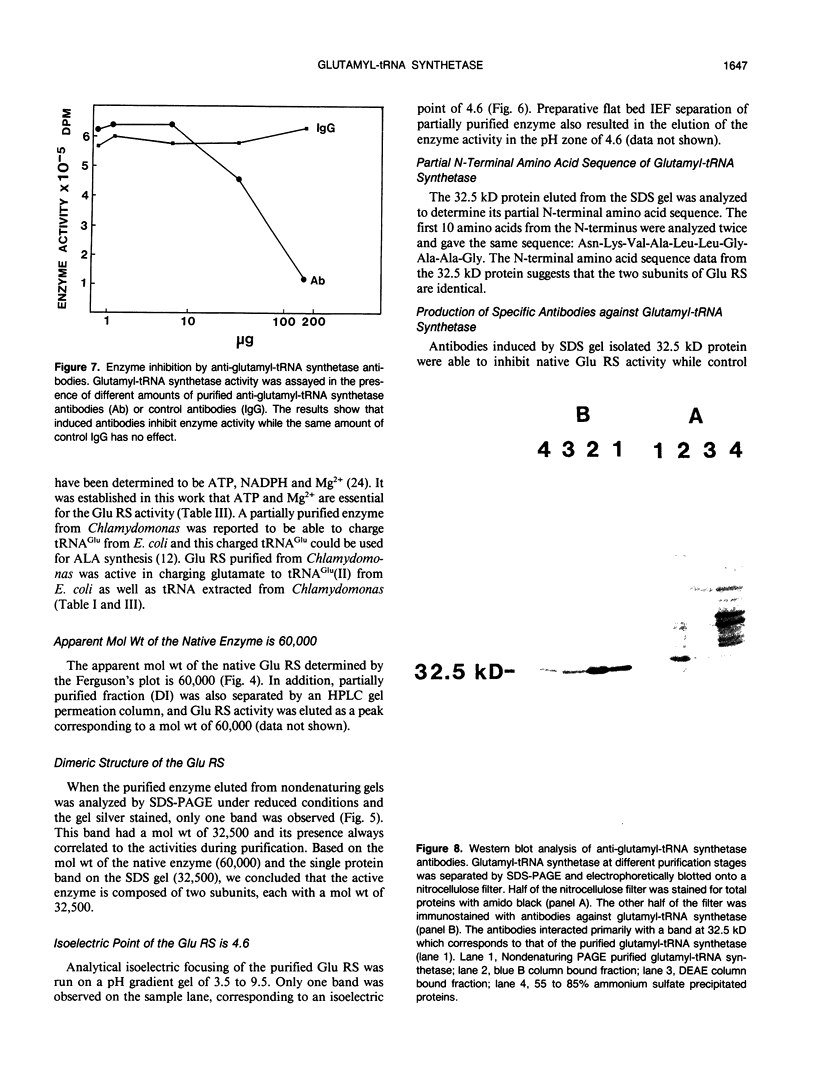
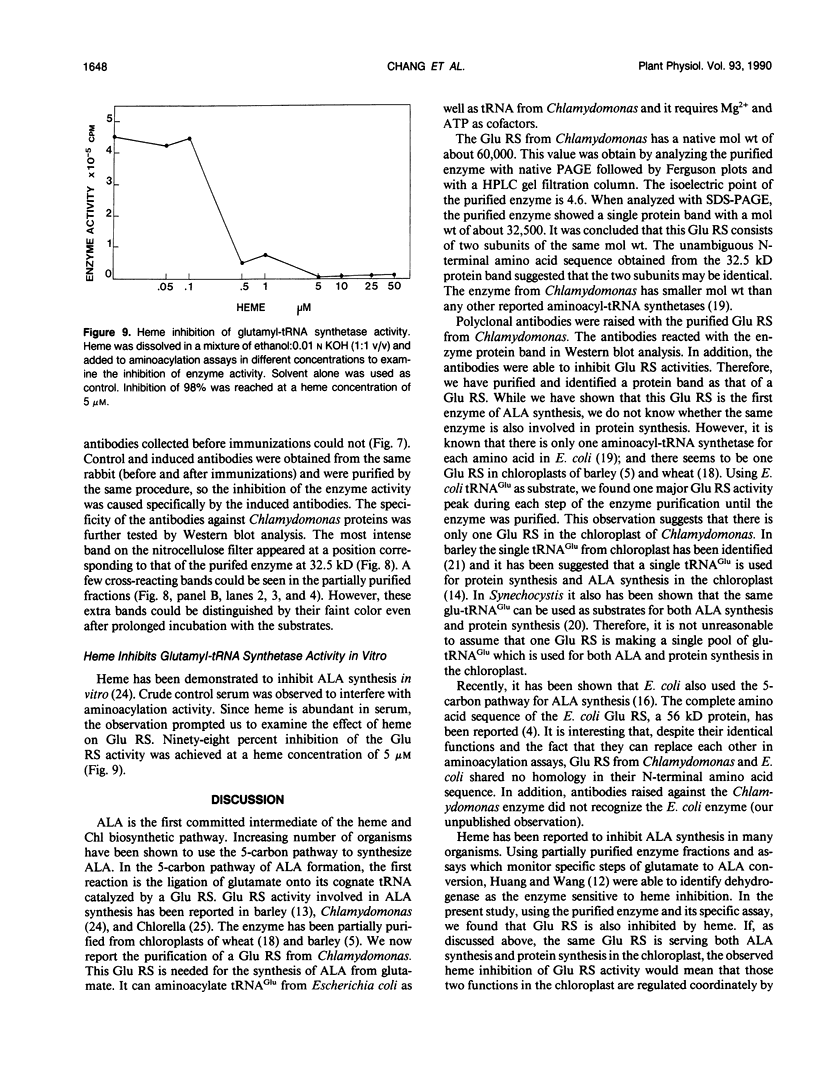
Chlorophyll biosynthesis starts with the synthesis of glutamyl-tRNA (glu-tRNA) by a glutamyl-tRNA synthetase (Glu RS). The glu-tRNA is subsequently transformed to δ-aminolevulinic acid (ALA), which is a committed and regulated precursor in the chlorophyll biosynthetic pathway. The Glu RS from a green alga, Chlamydomonas reinhardtii, was purified and shown to be able to synthesize glu-tRNA and to participate in ALA synthesis in a coupled enzyme assay. Physical and chemical characterization of the purified Glu RS indicated that the enzyme had been purified to homogeneity. The purified enzyme has a native molecular weight of 60,000, an isoelectric point of 4.6, and it formed a single band of 32,500 daltons when analyzed by a silver stained denaturing gel. The N-terminal amino acid sequence of the 32,500 dalton protein was determined to be Asn-Lys-Val-Ala-Leu-Leu-Gly-Ala-Ala-Gly. The molecular weight analyses together with the unambiguous N-terminal amino acid sequence obtained from the purified enzyme suggested that the native enzyme was composed of two identical subunits. Polyclonal antibodies raised against the purified and denatured enzyme were able to inhibit the activity of the native enzyme and to interact specifically with the 32,500 dalton band on Western blots. Thus, the antibodies provided an additional linkage for the structural and functional identities of the enzyme. In vitro experiments showed that over 90% of the glu RS activity was inhibited by 5 micromolar heme, which suggested that Glu RS may be a regulated enzyme in the chlorophyll biosynthetic pathway.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bhown A. S., Mole J. E., Hunter F., Bennett J. C. High-sensitivity sequence determination of proteins quantitatively recovered from sodium dodecyl sulfate gels using an improved electrodialysis procedure. Anal Biochem. 1980 Mar 15;103(1):184–190. doi: 10.1016/0003-2697(80)90254-7. [DOI] [PubMed] [Google Scholar]
- Blake M. S., Johnston K. H., Russell-Jones G. J., Gotschlich E. C. A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on Western blots. Anal Biochem. 1984 Jan;136(1):175–179. doi: 10.1016/0003-2697(84)90320-8. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Breton R., Sanfaçon H., Papayannopoulos I., Biemann K., Lapointe J. Glutamyl-tRNA synthetase of Escherichia coli. Isolation and primary structure of the gltX gene and homology with other aminoacyl-tRNA synthetases. J Biol Chem. 1986 Aug 15;261(23):10610–10617. [PubMed] [Google Scholar]
- Chen M. W., Jahn D., Schön A., O'Neill G. P., Söll D. Purification and characterization of Chlamydomonas reinhardtii chloroplast glutamyl-tRNA synthetase, a natural misacylating enzyme. J Biol Chem. 1990 Mar 5;265(7):4054–4057. [PubMed] [Google Scholar]
- Edman P., Begg G. A protein sequenator. Eur J Biochem. 1967 Mar;1(1):80–91. doi: 10.1007/978-3-662-25813-2_14. [DOI] [PubMed] [Google Scholar]
- FERGUSON K. A. STARCH-GEL ELECTROPHORESIS--APPLICATION TO THE CLASSIFICATION OF PITUITARY PROTEINS AND POLYPEPTIDES. Metabolism. 1964 Oct;13:SUPPL–SUPPL1002. doi: 10.1016/s0026-0495(64)80018-4. [DOI] [PubMed] [Google Scholar]
- Gorevic P. D., Prelli F. C., Frangione B. Immunoglobulin G (IgG). Methods Enzymol. 1985;116:3–25. doi: 10.1016/s0076-6879(85)16003-9. [DOI] [PubMed] [Google Scholar]
- Huang D. D., Wang W. Y. Chlorophyll biosynthesis in Chlamydomonas starts with the formation of glutamyl-tRNA. J Biol Chem. 1986 Oct 15;261(29):13451–13455. [PubMed] [Google Scholar]
- Huang D. D., Wang W. Y., Gough S. P., Kannangara C. G. delta-Aminolevulinic acid-synthesizing enzymes need an RNA moiety for activity. Science. 1984 Sep 28;225(4669):1482–1484. doi: 10.1126/science.6206568. [DOI] [PubMed] [Google Scholar]
- Kannangara C. G., Gough S. P., Bruyant P., Hoober J. K., Kahn A., von Wettstein D. tRNA(Glu) as a cofactor in delta-aminolevulinate biosynthesis: steps that regulate chlorophyll synthesis. Trends Biochem Sci. 1988 Apr;13(4):139–143. doi: 10.1016/0968-0004(88)90071-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Li J. M., Brathwaite O., Cosloy S. D., Russell C. S. 5-Aminolevulinic acid synthesis in Escherichia coli. J Bacteriol. 1989 May;171(5):2547–2552. doi: 10.1128/jb.171.5.2547-2552.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mau Y. H., Wang W. Y., Tamura R. N., Chang T. E. Identification of an intermediate of delta-aminolevulinate biosynthesis in Chlamydomonas by high-performance liquid chromatography. Arch Biochem Biophys. 1987 May 15;255(1):75–79. doi: 10.1016/0003-9861(87)90295-5. [DOI] [PubMed] [Google Scholar]
- Ratinaud M. H., Thomes J. C., Julien R. Glutamyl-tRNA synthetases from wheat. Isolation and characterization of three dimeric enzymes. Eur J Biochem. 1983 Oct 3;135(3):471–477. doi: 10.1111/j.1432-1033.1983.tb07675.x. [DOI] [PubMed] [Google Scholar]
- Schimmel P. Aminoacyl tRNA synthetases: general scheme of structure-function relationships in the polypeptides and recognition of transfer RNAs. Annu Rev Biochem. 1987;56:125–158. doi: 10.1146/annurev.bi.56.070187.001013. [DOI] [PubMed] [Google Scholar]
- Schneegurt M. A., Rieble S., Beale S. I. The tRNA Required for in Vitro delta-Aminolevulinic Acid Formation from Glutamate in Synechocystis Extracts : Determination of Activity in a Synechocystis in Vitro Protein Synthesizing System. Plant Physiol. 1988 Dec;88(4):1358–1366. doi: 10.1104/pp.88.4.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schön A., Kannangara C. G., Gough S., Söll D. Protein biosynthesis in organelles requires misaminoacylation of tRNA. Nature. 1988 Jan 14;331(6152):187–190. doi: 10.1038/331187a0. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. Y., Huang D. D., Stachon D., Gough S. P., Kannangara C. G. Purification, Characterization, and Fractionation of the delta-Aminolevulinic Acid Synthesizing Enzymes from Light-Grown Chlamydomonas reinhardtii Cells. Plant Physiol. 1984 Mar;74(3):569–575. doi: 10.1104/pp.74.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. Y., Wang W. L., Boynton J. E., Gillham N. W. Genetic control of chlorophyll biosynthesis in Chlamydomonas. Analysis of mutants at two loci mediating the conversion of protoporphyrin-IX to magnesium protoporphyrin. J Cell Biol. 1974 Dec;63(3):806–823. doi: 10.1083/jcb.63.3.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K., Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969 Aug 25;244(16):4406–4412. [PubMed] [Google Scholar]
- Weinstein J. D., Mayer S. M., Beale S. I. Formation of delta-Aminolevulinic Acid from Glutamic Acid in Algal Extracts : Separation into an RNA and Three Required Enzyme Components by Serial Affinity Chromatography. Plant Physiol. 1987 Jun;84(2):244–250. doi: 10.1104/pp.84.2.244. [DOI] [PMC free article] [PubMed] [Google Scholar]