Abstract
MR1-restricted mucosal-associated invariant T (MAIT) cells play a unique role in the immune system. These cells develop intra-thymically through a three-stage process but the events that regulate this are largely unknown. Here, using bulk and single-cell RNAseq-based transcriptomic analysis in mice and humans, we studied the changing transcriptional landscape that accompanies transition through each stage. Many transcripts were sharply modulated during MAIT cell development, including SLAM family members, chemokine receptors, and transcription factors. We also demonstrate that stage-3 ‘mature’ MAIT cells comprise distinct sub-populations including newly arrived-transitional stage-3 cells, IFN-γ-producing MAIT1 cells and IL-17-producing MAIT17 cells. Moreover, the validity and importance of several transcripts detected in this study is directly demonstrated using specific mutant mice. For example, MAIT cell intrathymic maturation was found to be halted in SLAM-associated protein (SAP)-deficient and Cxcr6-deficient mouse models, providing clear evidence for their role in modulating MAIT cell development. These data underpin a model that maps the changing transcriptional landscape and identifies key factors that regulate the process of MAIT cell differentiation, with many parallels between mice and humans.
One sentence summary
Characterizing MAIT cell development led to the identification of key regulators that specify MAIT cell fate in the thymus.
Introduction
Mucosal-associated invariant T (MAIT) cells are a population of unconventional T cells that detect vitamin-B2 (riboflavin) metabolites presented by the monomorphic antigen-presenting molecule MR1(1, 2). Most MAIT cells express a semi-invariant T cell receptor (TCR) comprising a TRAV1–2/TRAJ33 TCR-α chain, paired with a limited array of TCR-β chains (enriched for TRBV19 and TRBV13 in mice, and TRBV6 and TRBV20 in humans). MAIT cells are highly abundant in humans, representing up to 10% of circulating T cells and up to 45% of T cells in some tissues such as liver(1). However, for reasons that are unclear, they are also highly variable between individuals. While the consequences of low or high MAIT cells are uncertain in humans; in mice, many studies have demonstrated that MAIT cell deficiencies can be important in immunity to bacterial and viral infections(1, 3, 4) and in autoimmune and inflammatory diseases(1, 5). It is therefore critical that we understand how the development and homeostasis of MAIT cells is regulated.
MAIT cells are known to develop in the thymus through a three-stage process in mice and humans(6). In mice, the first stage is defined as CD24+CD44−, the second is CD24−CD44− and the third stage is CD24−CD44+. In humans, the corresponding stages are defined as: CD27−CD161−; CD27+CD161−; and CD27+CD161+ cells(6). The progression through each stage is thought to depend on the interaction of developing MAIT cells with MR1 expressed on cortical thymocytes(7). MR1 is not only important for their initial selection towards the MAIT cell lineage, but also for their subsequent maturation in the thymus(6). The progression from stage 1 to stage 2 also depends on the microRNA (miRNA) processing enzyme Drosha(6), microRNA miR-181a/b-1(8), and the presence of undefined microbial factors as this process is impaired in germ-free mice(6). The progression from stage 2 to stage 3 is dependent on the transcription factor promyelocytic leukemia zinc finger (PLZF) and the cytokine IL-18(6). Stage 3 MAIT cells are closest in phenotype to mature MAIT cells in the periphery, although further extrathymic maturation of these cells is required for them to reach a fully mature phenotype(6, 9). Thus, the transition from stage 2 and stage 3 represent key events in MAIT cell maturation where MAIT cells not only enter their final maturation stage, but also diversify into functionally distinct subsets. A major subset of stage 3 cells in the thymus (MAIT17) produce IL-17 and expresses the transcription factor RORγt, while another subset (MAIT1) produces interferon-γ and expresses the transcription factor T-bet(6, 10). Thus, it seems that either within, or just prior to, stage 3 of MAIT cell development there are some important differentiation events that contribute to the divergent MAIT cell populations and it is likely that these cells do not all group into one transcriptionally homogeneous population.
In this study, we have carried out bulk and single cell RNAseq transcriptomic analysis of MAIT cells from both mice and humans at each stage of their development. These results have revealed many genes that are sharply up or downregulated as MAIT cells develop and there are substantial commonalities between the two species. Based on this information, we have used a series of gene-deleted mice to identify factors that control development of this unique arm of the immune system. Furthermore, we have also dissected stage 3 MAIT cells into transcriptionally distinct subpopulations which has led us to construct a more detailed developmental model that reflects the functional bifurcation events that see MAIT cells branch into two distinct lineages. Collectively, our findings provide a major advance in our understanding of how MAIT cells develop and differentiate and the factors that control this process.
Results
Differentially expressed genes associated with intrathymic mouse MAIT cell development.
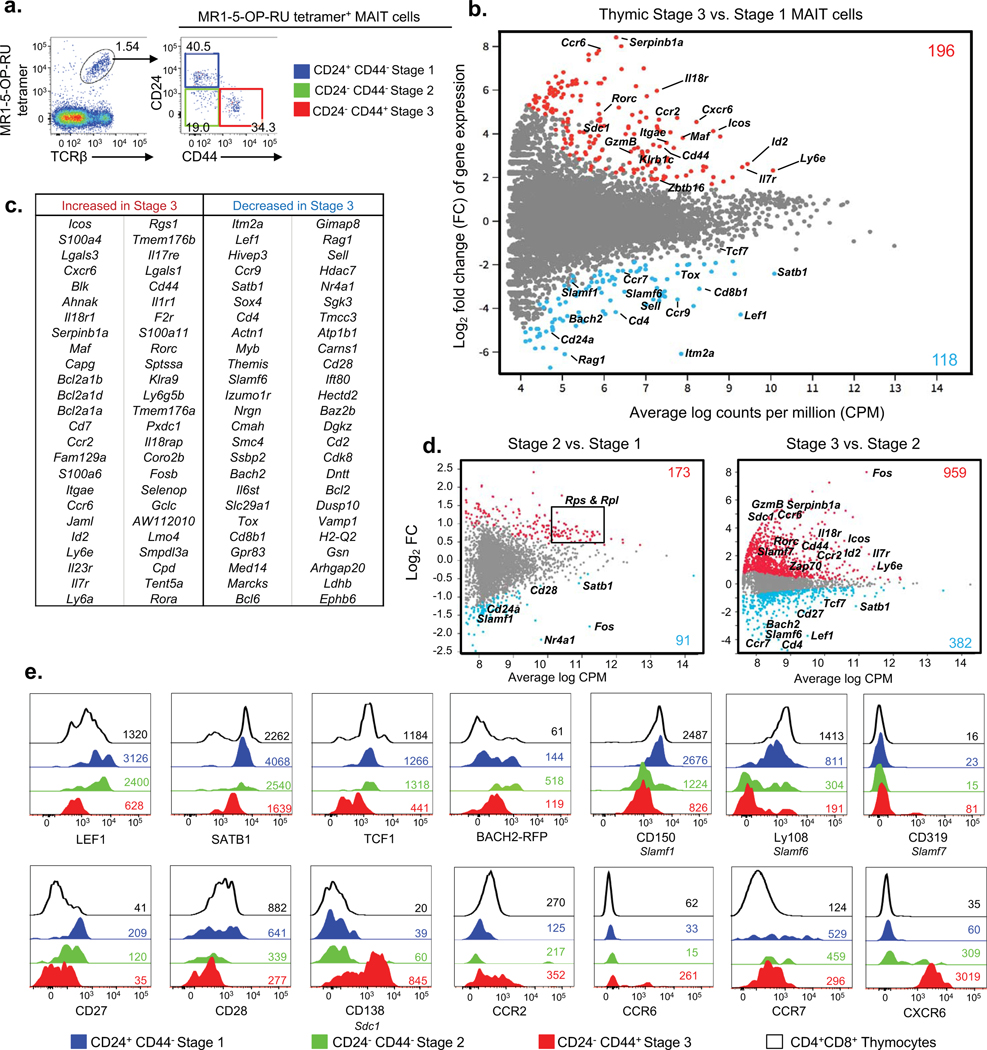
We previously described a three-stage intra-thymic development pathway for MAIT cells in mice and humans. In mice, the pathway begins with CD24+CD44− stage 1 MAIT cells, which develop into CD24−CD44− stage 2 MAIT cells and then into CD24−CD44+ stage 3 MAIT cells(6) (Fig. 1a). To investigate the molecular mechanisms that underpin this process, we compared the transcriptomes of thymic immature stage 1 and mature stage 3 MAIT cells from mouse thymus using bulk (100 cell) RNAseq. We identified 314 differentially expressed genes (DEGs) between stages 1 and 3, of which 196 were significantly upregulated and 118 significantly downregulated, between stages 1 and 3 (Fig. 1b). A list of the top 50 DEGs is shown in (Fig. 1c) and a full list of these genes can be found in Table S1. As a strong validation of the RNAseq-based approach, DEGs included several markers that we had previously identified for each population(6); thus, stage 1 cells expressed higher: Cd24a, Cd4 and Cd8b, Cd69, Sell (CD62L); while stage 3 cells expressed higher Cd44, Icos, Il18r (CD218), Itgae (CD103), Il7r (CD127), Zbtb16 (PLZF), Klrb1c (NK1.1) and Rorc (RORγt). Importantly, these data revealed many additional genes that were upregulated (such as Cxcr6, Ccr6, Maf, Sdc1 (Syndecan 1, CD138), GzmB (Granzyme B) and Ly6e) or downregulated (such as Slamf1, Slamf6, Tcf7, Satb1, Tox, Ccr7, Ccr9 and Lef1) as MAIT cells mature in the thymus (Figs. 1b and c). To determine whether these transcriptional changes were associated with transitions between stage 1 and 2 or stage 2 and 3, we used single cell RNAseq to separately probe these populations separately because stage 2 cells are quite rare and difficult to isolate sufficient numbers for bulk RNAseq. These data demonstrated that the transition from stage 1 to 2 is accompanied by 173 transcripts that increased and 91 genes that decreased (Fig. 1d). Interestingly, the majority of transcripts (70 of 184) that increased encode ribosomal proteins (Rps and Rpl), suggesting that this transition is associated with increased cellular activity and protein production, consistent with a recent study showing that stage 2 is linked to increased proliferation(8). Gene set testing performed on the differentially expressed genes using Gene Ontology (GO) and KEGG databases also highlighted the heavy enrichment of ribosomal activity pathways between stage 2 and stage 1 (Fig. S2). Additionally, the transition from stage 1 to stage 2 was associated with changes in some important immunological genes, including increased: Zbtb16 (PLZF), Il18r (CD218), Il23r and Ccr2; and decreased: SlamF1, Satb1, Cd28, and Cd24a (already validated as the surface marker that separates stage 1 from stage 2 cells). A comparison of stage 2 to stage 3 shows that most of the changes identified in association with the bulk RNAseq comparison of stage 1 and stage 3 (Fig. 1b) were associated with this transition.
Figure 1. Transcriptomic analysis of mouse MAIT cell development.
a. Flow cytometric analysis of 3 stages of mouse thymic MAIT cells post MR1–5-OP-RU tetramer enrichment. MAIT cell stages are defined with CD24 and CD44; stage 1 (S1, CD24+CD44−) in blue, stage 2 (S2, CD24−CD44−) in green and stage 3 (S3, CD24−CD44+) in red. b. MD plot showing gene expression comparison of bulk-cell purified stage 3 versus stage 1 MAIT cells. Colored dots indicate genes significantly up-regulated in stage 3 (red) and stage 1 (blue). Colored numbers represent the total number of differentially expressed genes (DEG). (c) Table lists the top 50 most DEGs within each subset. b, c. Data are from 3 pooled biological replicates each generated from a pool of thymi from five mice. d. Gene expression comparison of single-cell purified stage 2 versus stage 1 (left) and stage 3 versus stage 2 (right) MAIT cells. e. Phenotypic analysis of thymic MAIT cell stages for expression of transcription factors LEF1 (encoded by gene Lef1), SATB1 (Satb1), TCF1 (Tcf7), Bach2 (Bach2); SLAM molecules CD150 (Slamf1), Ly108 (Slamf6), and CD319 (Slamf7); costimulation receptors CD27 (Cd27) and CD28 (Cd28); glycoprotein CD138 (Sdc1) and chemokine receptors CCR2 (Ccr2), CCR6 (Ccr6), CCR7 (Ccr7) and CXCR6 (Cxcr6). Histograms depict stage 1 MAIT cells in blue, stage 2 MAIT cells in green, stage 3 MAIT cells in red, CD4+CD8+ double positive (DP) thymocytes in black. Coloured numbers indicate mean fluorescence intensity values (MFI) of each marker for the respective cell population. a, e. Data are representative of at least 2 independent experiments with a total of 2 separate samples (pools of 3 thymi).
To further validate these transcriptional data, we performed flow cytometry analysis with a selection of antibodies and confirmed that transcription factors LEF1, SATB1 and TCF1 (Tcf7) were downregulated as MAIT cells develop, whereas BACH2 transiently increased between stage 1 and 2 and decreased at stage 3 (Fig. 1e). Surface receptors CD138, CCR2, CCR6 and CXCR6 were upregulated, whereas CCR7, CD27, CD28, CD150 (Slamf1) and Ly108 (Slamf6) were downregulated at the protein level, between stages 1, 2 and 3 (Fig. 1e). Given that Slamf1 and Slamf6 were clearly downregulated as MAIT cells mature, we also examined Slamf7 (CD319), another slam family member that showed the opposite trend in our transcriptome analysis. This protein was clearly detected on a small subset of stage 3 thymic MAIT cells, reflecting heterogeneity in stage 3 (Fig. 1e). Accordingly, MAIT cells undergo major changes in gene and protein expression as they mature from stage 1 to stage 2, and even more-so between stages 2 and 3. Furthermore our data identify a number of candidate genes that might be important for the development of MAIT cells within the thymus.
Separation of mature stage 3 MAIT cells into functionally distinct subsets.
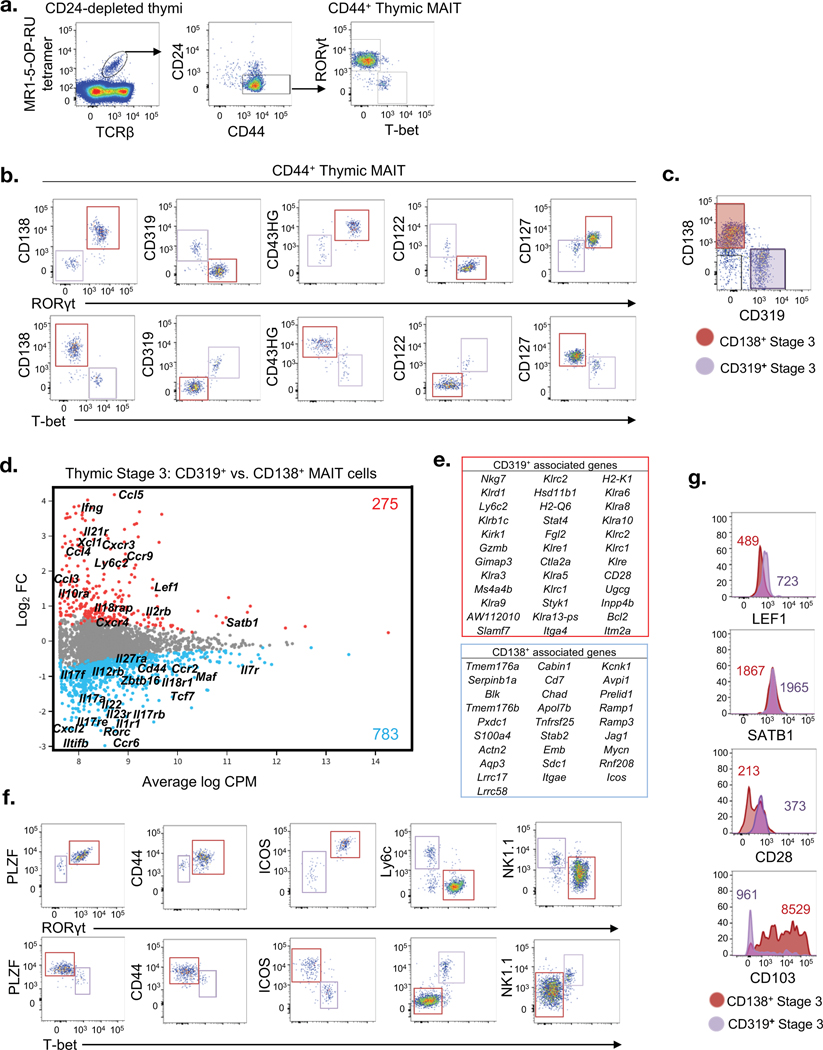
As we have previously reported(6, 10), thymic stage 3 mouse MAIT cells include 2 functionally distinct subsets, defined as IFN-γ-producing T-bet+ MAIT1 and IL-17 producing RORγt+ MAIT17 cells (Fig. 2a). To compare these cell types at the transcriptomic level, we avoided cell fixation and permeabilization and/or activation by identifying cell surface markers to separate them. We investigated several possible candidate markers that are differentially expressed by stage 3 MAIT cells (Fig. 1c; Table S1), including some that are expressed by related NKT1 and NKT17 cells(11, 12). These included: CD138 (Syndecan 1), CD43 (high glycosylation (HG) variant), CD122 (IL-2Rβ), CD127 (IL-7Rα) and CD319 (Slamf7) (Fig. 1e). We tested these markers against RORγt and T-bet intracellular staining (Fig. 2b). RORγt+ cells stained higher for CD138, CD43HG, and CD127, whereas T-bet+ cells stained higher for CD319 and CD122. The combination of CD319 versus CD138 provided a clear and mutually exclusive separation, allowing us to identify MAIT1 and MAIT17 cells respectively, within the stage 3 MAIT cell population without fixation and permeabilization (Fig. 2c). We also observed that some stage 3 MAIT cells did not express CD319 or CD138, which we surmised may encompass uncommitted stage 3 cells.
Figure 2. Bifurcation and transcriptomic comparison of mature mouse MAIT cells.
a. Flow cytometric analysis of mature mouse thymic MAIT cells post HSA-complement mediated depletion showing RORγt and T-bet expression in CD44+ stage 3 MAIT cells. b. Panel of markers showing expression of CD138 (Sdc1), CD319 (Slamf7), CD43HG (Spn), CD122 (Il2rb) and CD127 (Il7r) relative to RORγt and T-bet in stage 3 MAIT cells. (c) Flow cytometry profile of stage 3 MAIT cells co-stained with CD138 and CD319. d. Gene expression comparison of purified CD319+ versus CD138+ stage 3 MAIT cells. Colored dots indicate genes significantly up-regulated in CD319+ cells (red) and CD138+ cells (blue). Colored numbers represent the total number of DEGs. e. Tables list genes enriched corresponding to CD319+ (red, top) and CD138+ (blue, bottom) MAIT cell subsets. f. Flow cytometry analysis for expression of PLZF, CD44, ICOS, Ly6c and NK1.1 relative to RORγt+ and T-bet+ mature MAIT cells. g. LEF1, SATB1, CD28 and CD103 expression on CD138+ and CD319+ stained mature MAIT cells. Coloured numbers indicate mean fluorescence intensity values (MFI) of each marker for the respective CD138+ or CD319+ MAIT cells. a-c, f-g. Data are representative of at least 2 independent experiments.
We isolated CD319+ (MAIT1) and CD138+ (MAIT17) single cells and performed single cell RNAseq comparison of these two mature thymic MAIT cell populations (Fig. 2d). Over 1000 genes were differentially expressed between the mature CD138+ and CD319+ stage 3 MAIT cell subsets, 783 that were more highly expressed by the MAIT17 cells and 275 genes that were more highly expressed by the MAIT1 cells (Fig. 2d and e). As expected from our cytometry-based analysis in Figure 2b and c, CD319+ MAIT1 cells had higher gene expression of genes encoding CD319 (Slamf7), CD122 (Il2rb) and IFN-γ (Ifng), while CD138+ MAIT17 cells expressed higher levels of the genes encoding CD138 (Sdc1), CD127 (Il7ra), RORγt (Rorc) and IL-17A (Il17a) (Fig. 2d and e). In addition to these genes, MAIT1 cells were distinguished by higher expression of genes encoding natural killer (NK) family members including Klr family members and many other genes including Lef1, Nkg7, Klrb1c, Ly6c2, GzmB, Bcl2, Stat4, Ms4a4b and Cd28 (Fig. 2d and e). In contrast, MAIT17 cells had higher expression of genes encoding IL-17-related molecules such as Il17a, Il17f, Il22, Il23r, Il17rb, Il17re and also higher expression of Blk, Maf, Icos, Zbtb16, Tnfrs25 and Cd44 (Fig. 2d). Flow cytometry-based analysis of MAIT1 and MAIT17 subsets was used to validate some of these results (Fig. 2f). For example, RORγt+ MAIT17 cells expressed moderately higher PLZF and CD44 and much higher levels of ICOS compared to T-bet+ MAIT1 cells. Conversely, Ly6c and NK1.1 (Kkrb1c) were positive on many (but not all) MAIT1 cells, while most MAIT17 cells did not express these markers. Using CD138 and CD319 as surrogate markers, LEF1 expression and CD28 were confirmed as being lower on MAIT17 cells, while SATB1, which was expressed slightly higher in MAIT1 cells at the transcript level (Fig. 2d), showed no clear difference at the protein level (Fig. 2g). Additionally, MAIT17 cells expressed much higher levels of CD103 (Itgae) compared to MAIT1 (Fig. 2g). Taken together, these data demonstrate that MAIT1 and MAIT17 cells are transcriptionally and functionally distinct populations that emerge as MAIT cells develop in the thymus.
Lineage diversification within stage 3 of MAIT cell development
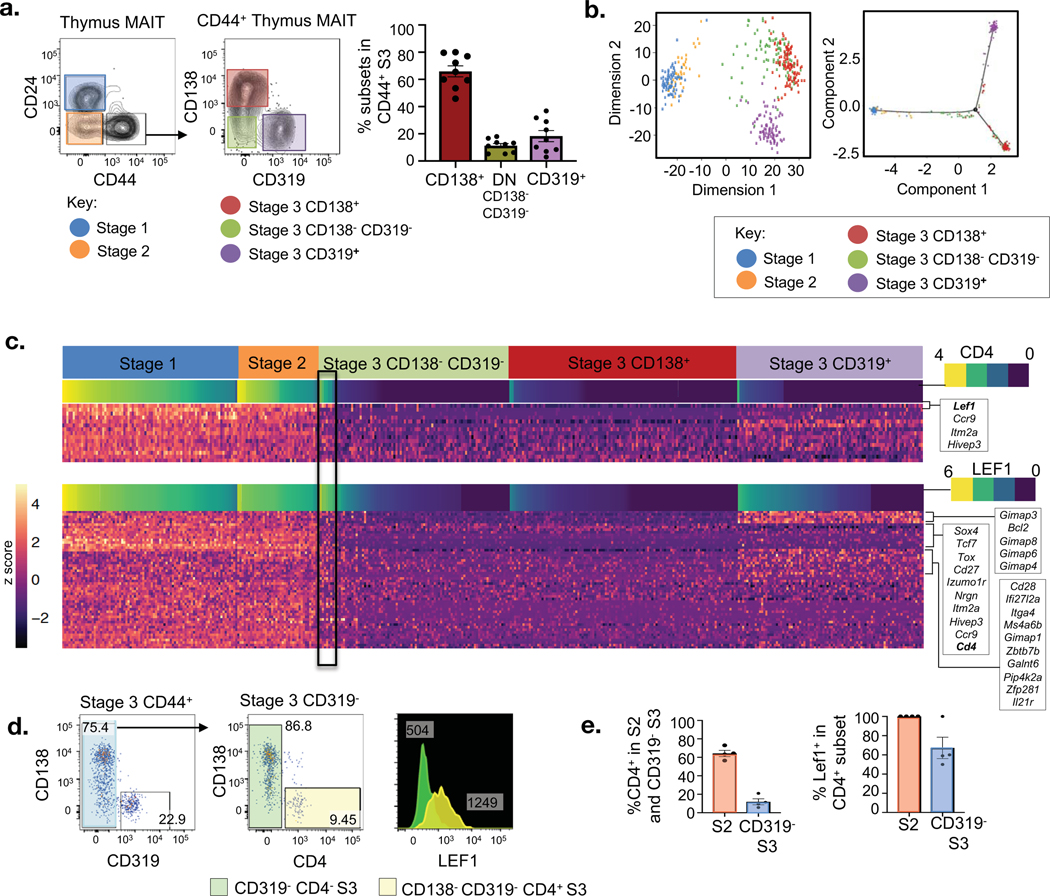
Given that MAIT1 and MAIT17 cells exhibit a highly divergent transcriptional program, we sought to determine whether this happens before, at, or after they reach stage 3 of MAIT cell development in the thymus. Using the combination of cell surface CD138 and CD319 expression, it was clear that stage 1 and 2 MAIT cells (Fig. 1d), and some stage 3 cells (Fig. 3a), lacked expression of these markers. This raised the possibility that MAIT1 and MAIT17 cells emerge after MAIT cells arrive at stage 3 and moreover, that some newly arrived stage 3 cells may lack MAIT1 and MAIT17 transcriptomic signatures and will more closely resemble less mature stage 2 cells. To investigate these subsets of stage 3 cells in greater detail, we compared stage 3 (CD24−CD44+) cells, sorted into three subpopulations: CD138−CD319+, CD138+CD319−, CD138−CD319−; alongside immature stage 1 (CD24+CD44−) and stage 2 (CD24−CD44−) cells, using single cell RNAseq analysis. Principal component analysis and Monocle analysis of these populations provided a clear demonstration of the marked differences between thymus-derived CD319+ (MAIT1) and CD138+ (MAIT17) stage 3 cells (Fig. 3b). Moreover, it demonstrated that the CD138− CD319− stage 3 cells were heterogeneous, with some sitting between the CD138+ and CD319+ populations based on principle component analysis dimension 2, while others appeared to merge with the CD138+ subset (Fig. 3b). There was less overlap between CD138−CD319− stage 3 cells and the CD319+ subset, although some CD319 cells merged with the CD138−CD319− subpopulation in the middle (Fig. 3b). This principal component analysis also showed that immature stage 1 and stage 2 cells were clearly distinct from most stage 3 cells including CD138+, CD319+ and CD138−CD319− cells, although some CD138−CD319− cells fell between immature stage 1 and stage 2 cells and CD138+ and CD319+ cells on dimension 1 (Fig. 3b). Additionally, Pseudotime analysis carried out using Monocle on DDRtree clustering of the cell subsets also showed two trajectories branching from an intermediate CD138−CD319− subpopulation towards terminally differentiated (CD138+ and CD319+) stage 3 subsets (Fig. 3b and Fig. S3a). Thus, CD138−CD319− cells may include candidates for newly-arrived stage 3 MAIT cells that share similarities with stage 2 precursors.
Figure 3. Lineage diversification within stage 3 of MAIT cell development.
a. Flow cytometry profile of mouse MAIT cell subsets present within the mouse thymus single-cell sorted for RNA-sequencing analysis. Graph depicts percentages of each stage 3 subpopulation across 8 samples. b. (Left) Principal component analysis (PCA) on the respective sorted subsets in (a), and (Right) Monocole DDRtree showing the clustering of respective MAIT cell subsets. c. Heatmap showing the differentially expressed genes across MAIT cell subsets ordered by decreasing Cd4 (top) and Lef1 (bottom) expression, grouped across developmental subsets. (n = 363 in total, n = 75 stage 1, n = 34 stage 2, n = 81 CD138− CD319− DN stage 3, n = 95 CD138+ stage 3, n = 78 CD319 stage 3). d. Flow cytometry analysis mature CD44+ MAIT cells for CD138 and CD319 expression (left). Analysis of CD4+ subset (yellow) from mature CD44+ CD319− MAIT cells (middle). Histogram overlay analysis of LEF1 expression between CD319− and CD138−CD319− CD4+ stage 3 MAIT cells (right). e. Scatter plot graph shows percentage of CD4+ subset within stage 2 and CD319− stage 3 MAIT cells. Graph shows percentage LEF1+ cells within CD4+ S2 and CD4+ S3 MAIT cell populations.
Heat map analysis of the top genes that separate each of the populations is shown in Figure 3c and the full heat map is provided in Figure S1. The columns are grouped into each of the different MAIT cell subsets as described above, and further organised for each subset based on CD4 expression level, since CD4 is highly expressed by stage 1 and 2 cells, but is absent from most mature stage 3 cells(6). This heatmap analysis again highlights the many similarities between stage 1 and 2 MAIT cells (Fig. 1d) and how a major transcriptomic change is apparent when comparing these to the majority of stage 3 cells. Similar to Figure 2d and e, this data also reflects the extensive transcriptomic differences between CD319+ MAIT1 and CD138+ MAIT17 cells (Fig. 3c). It was also apparent that many of the stage 3 CD138−CD319− cells more closely resembled CD138+ MAIT17 stage 3 cells, but consistent with the principal component analysis (Fig. 3b), a subset of the CD138−CD319− cells, outlined by the black rectangle (Fig. 3c), appeared to be more similar to stage 2 cells in their overall transcript signature. For example, these cells express Cd4 at mid to high levels, in contrast to most other stage 3 cells that do not express Cd4. As Lef1 was found to be the top gene most positively associated to Cd4, a second heatmap ordered via Lef1 expression illustrated a similar subset of cells within this black rectangle (Fig. 3c). These Cd4 and Lef1 expressing stage 3 cells also expressed higher levels of many other genes that are more in line with the stage 2 signature than other stage 3 cells, including Tcf7, Itm2a and Satb1 and they showed little expression of MAIT1 or MAIT17 related genes (Fig. S1). Importantly, CD4 and LEF1 expressing CD138−CD319− stage 3 cells were almost entirely encompassed within the cells in the centre of the principal component and Monocle analysis (Fig. 3b), and Cd4 and Lef1 also showed a similar expression trajectory, when Pseudotime is overlaid onto Monocle analysis (Fig. S3b). To exclude the possibility that the CD4+LEF1+ cells were contaminating stage 2 cells during the single cell RNAseq sorting, we used flow cytometry to examine stage 3 MAIT cells, and confirmed that a clear subset (~10%) of CD4+ cells was present within the CD138−CD319− population (Fig. 3d). Moreover, intracellular transcription factor staining revealed that these also expressed LEF1, in contrast to most other stage 3 cells (Fig. 3d and 3e). Thus, these data suggest that CD4+LEF1+CD319−CD138− stage 3 cells are the next stage beyond stage 2 and represent newly arrived stage 3 cells that have not yet committed to either the MAIT1 or MAIT17 lineages. In order to directly investigate this developmental sequence, we reanalysed experiments from our earlier study(6) examining stage 3 cells from very young (2 week-old mice). We report here that a much higher percentage of stage 3 cells were CD4+ compared to in older mice (Fig. S4a).
Aside from the CD4+LEF1+CD138−CD319− stage 3 cells, which also seem to be a less mature subset of stage 3 cells based on their DEG profile (Fig. S4b), the remaining population of CD4−LEF1−CD138−CD319− stage 3 cells contained a mix of cells whose characteristics were more in common with CD138+ MAIT17 cells than with CD319+ MAIT1 cells. This is clearly seen using MD plot analysis where relatively few genes were differentially expressed between LEF− CD138−CD319− stage 3 cells and CD138+ MAIT17 cells (Fig. S4c), while many genes were differentially expressed between LEF1+ CD138−CD319− stage 3 cells and CD138+ MAIT17 cells (Fig. S4d). Of the many genes that distinguished CD4−LEF1−CD138−CD319− cells from CD319+ cells, some of the most noteworthy included: Slamf7, GzmA, Ccl5, Bcl2 (all increased); and Ccr6, Ccr7, Icos, (decreased) and some of the IL-17-associated genes such as Rorc, Il17ra, Il17re. The enrichment of these lineage specific signatures were recapitulated in KEGG pathway analysis performed between these subets (Fig. S2b). Of the few genes that distinguished CD4−LEF1−CD138−CD319− and CD138+ cells, notable genes included increased Klf4 and proto-oncogene transcription factors Fos, FosB, and Jun (Fig. S4c and d).
Taken together, these data highlight a high degree of diversity within stage 3 of MAIT cell development. These data suggest there are distinct subpopulations of stage 3 MAIT cells, the earliest arrivals being the CD4+LEF1+ stage 3 cells and the most functionally mature being the CD319+ MAIT1 or CD138+ MAIT17 cells, as well as a population of CD4−LEF1−CD138−CD319− cells that comprises a mixture of cells, including some resembling CD138+ MAIT17 cells which we propose represent putative precursors of mature CD138+ MAIT17 cells. Conversely, we were unable to detect a clear population of CD4−CD319−CD138− cells that might reflect MAIT1 precursors, suggesting that this transition happens quite rapidly, and/or that CD319 is an early marker associated with the differentiation of this functionally distinct sub-lineage. The detection of some CD319+ cells in the middle range of the principal component analysis plot (Fig. 3b) supports this possibility.
Factors that regulate MAIT cell differentiation.
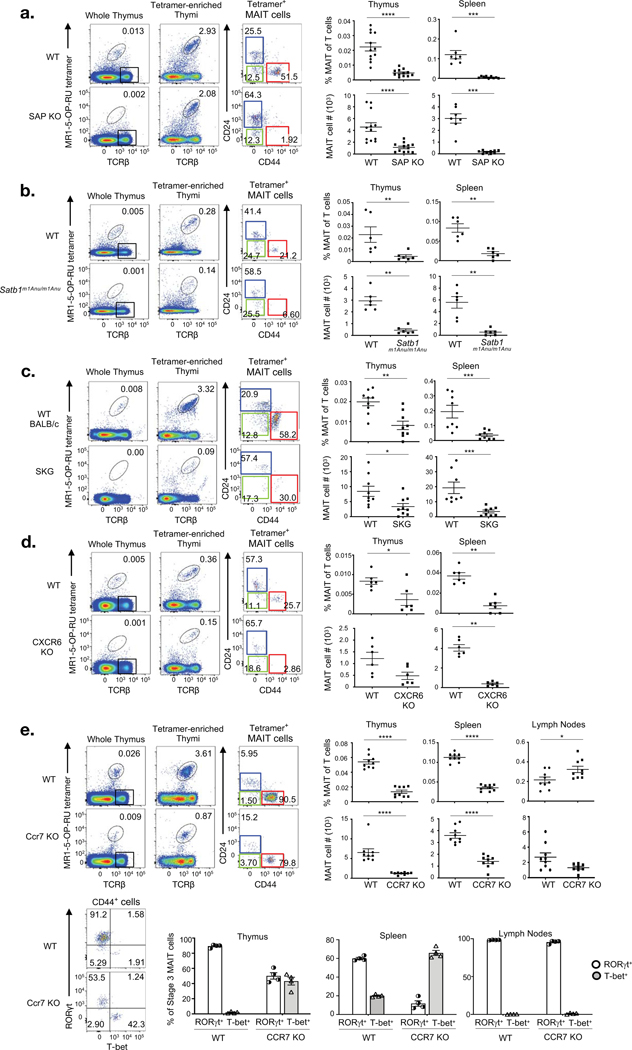
As many of the genes that were sharply modulated as MAIT cells mature are known to be important in the development or function of at least some types of T cells, we selected a panel of these genes to examine their importance in MAIT cell maturation. Because SLAM family members were differentially expressed between immature stage 1 MAIT cells and mature stage 3 MAIT cells (Fig. 1), we investigated MAIT cell development in mice lacking the SLAM adaptor protein (SAP) that transmits SLAM-based intracellular signals(13). This analysis revealed a key role for slam family members in regulating MAIT cell development beyond stage 1, such that stage 2 MAIT cells were numerically diminished and stage 3 MAIT cells were essentially absent, with the few residual stage 3 cells enriched for CD4+ (Fig. S4a), supporting the hypothesis that these could be early stage 3 cells. Furthermore, we were unable to detect MAIT cells in spleens of SAP-deficient (SAP KO) mice (Fig. 4a). Conventional TCRβ+ MR1-tetramer− cells were present in moderately higher numbers in these SAP-deficient samples (Fig. S5a), while as expected, these mice also had a reduction in CD1d-restricted NKT cells, and only a slight decrease in the percentage, but not number, of γδ T cells (Fig. S5b).
Figure 4. Identification of SAP, SATB1, Zap70, CXCR6, and CCR7 as key factors that regulate MAIT cell development with gene-deleted mice.
a-e. 3 panels depicting analysis of MAIT cells WT control mice and SAP KO (a), Satb1m1Anu/m1Anu (b), SKG (c), CXCR6 KO (d) and CCR7 KO (e) mice. Left panel of flow cytometry analysis show detection of MAIT cells from whole and MR1–5-OP-RU tetramer enriched thymus of respective mice, analysis of MR1-tetramer enriched MAIT cells for CD24 and CD44 expression. Scatter graphs depict absolute numbers and percentage of MAIT cells of T cells in thymus and spleen (a-e) and lymph nodes (e) of the respective mouse strains. Bar graphs in (e) show the proportion of RORγt and T-bet subset of stage 3 MAIT cells in WT and CCR7 KO thymus, spleen and lymph nodes. Data are representative of 2 independent experiments with a total of 6–9 mice per group (a-e; mean ± SEM) *P<0.1 **P<0.01 ***P<0.001, ****P<0.0001 (Mann-Whitney rank sum U test (a-e)).
SATB1 is a chromatin organising protein with crucial functions in T cells, including in Foxp3+ T-regulatory (T-reg) cell development and function(14) and NKT cell development(15). This gene was differentially expressed as MAIT cells develop, with highest expression in stage 1 MAIT cells (Fig. 1b–d). We examined a novel Satb1m1Anu/m1Anu mouse model that is unlike Satb1-deficient mice, which are runted and typically die by 3 weeks after birth(16). Mice homozygous for the Satb1m1Anu allele are healthy and have a normal lifespan but exhibit abnormalities in T cell development (Fig. S6 and manuscript under preparation). Satb1m1Anu/m1Anu mice show a major defect in MAIT cell development beyond stage 2 with a marked reduction of stage 3 MAIT cells (Fig. 4b). MAIT cells were also markedly diminished in the spleens of Satb1m1Anu/m1Anu mice (Fig. 4b). As anticipated, Satb1m1Anu/m1Anu mice also had a major reduction in CD1d-restricted NKT cells(15), but no significant change in γδ T cells, aside from a moderately reduced number of these in spleen (Fig. S5b). Conventional TCRβ+ MR1-tetramer− cell numbers were not significantly different in Satb1m1Anu/m1Anu samples, apart from a verified increase in the CD4+ Foxp3+ T-reg compartment (Fig. S5a and S6).
Next we examined Zap70-deficient (SKG) mice(17) because we have previously shown that MAIT cell development requires ongoing MR1-dependent signalling, suggesting a role for TCR signalling, and Zap70-deficient mice were previously reported to lack NKT cells(18). We found that indeed, MAIT cells are diminished in thymus and spleen of the Zap70-deficient mice, although all subsets were still detectable (Fig. 4c), supporting a role for TCR signalling not only in the initial emergence of Stage 1 MAIT cells, but also in MAIT cell maturation to stage 3. Interestingly, we did not detect a change in thymus NKT cells, and furthermore, these cells were more frequent in spleens of Zap70-deficient mice. TCRβ+ MR1 tetramer− T cells in thymus were present in normal numbers in Zap70-deficient mice (Fig. S5).
As several chemokine receptors were strongly modulated as MAIT cells mature, we examined whether signalling through some representative chemokine receptors is important for MAIT cell maturation. CXCR6-deficient mice exhibited a near complete block in MAIT cell development between stages 2 and 3, and no MAIT cells could be detected in spleen or lymph nodes of CXCR6-deficient mice (Fig. 4d). CXCR6-deficient mice also had a moderate reduction in the number of CD1d-restricted NKT cells in thymus and a major reduction of these cells in spleen (Fig. S5b). In contrast, CCR7-deficient mice had a partial defect in MAIT cell development with a large reduction in the percentage and number of MAIT cells in thymus and spleen, but not lymph nodes. Interestingly, CCR7 KO mice retained a clear population of MAIT cells at stage 3, that on further analysis showed to be heavily biased towards Tbet+ MAIT1 cells compared to MAIT cells from wild-type mice (Fig. 4e), demonstrating that CCR7 is selectively required for the differentiation of Rorγt+ MAIT17 subset. This effect was even more pronounced for MAIT cells in spleens of CCR7-deficient mice, but no difference was observed for these cells in lymph nodes (Fig. 4e). As an aside, these results also suggest that CD138+ MAIT cells might preferentially home to, or expand in, lymph nodes and that CCR7 is not required for their presence in this site. CCR7 KO mice also had a major reduction in CD1d-restricted NKT cells and, to a lesser extent, γδ T cells (Fig. S5b). Conventional TCRβ+ MR1-tetramer− cells were present in normal numbers in CXCR6 and CCR7 deficient samples (Fig. S5a). Interestingly, mice deficient in Bach2, a repressor of effector differentiation regulated by PLZF(19), exhibited no change in MAIT cell percentage or number despite its elevated expression in early stage MAIT cells (Fig. S7).
Differentially expressed genes associated with human MAIT cell development.
In humans, there are important parallels with mouse MAIT cell development, including the existence of a 3-stage development pathway where stage 1 MAIT cells are CD27−CD161−; stage 2 MAIT cells are CD27+CD161−, while stage 3 MAIT cells are CD27+/−CD161+ (Fig. 5a)(6).
Figure 5. Transcriptomic analysis of human MAIT cell development.
a. Flow cytometric analysis of 3 stages of human Vα7.2+ thymic MAIT cells of CD3+ cells pre- and post TRAV1–2 enrichment. MAIT cell stages are defined with CD27 and CD161; stage 1 (S1, CD27−CD161−) in blue, stage 2 (S2, CD27+CD161−) in green and stage 3 (S3, CD27+/−CD161+) in red. b. MD plot showing gene expression comparison of bulk-cell purified stage 3 versus stage 1 human MAIT cells. Colored dots indicate genes significantly up-regulated in stage 3 (red) and stage 1 (blue). Colored numbers represent the total number of differentially expressed genes (DEG). c. Table lists the top 50 most DEGs within each subset. d. Gene expression comparison of purified stage 2 versus stage 1 human MAIT cells, and stage 3 versus stage 2 human MAIT cells. b-d. Data are representative from 2 separate sorts, with a total of 8 donor samples. (e) Phenotypic analysis of human thymic MAIT cell stages for expression of transcription factors SATB1, LEF1, TCF1, CD1 molecules CD1a, b and d, HLA-A,B,C, and chemokine receptors CCR5 and CCR6. Histograms depict stage 1 MAIT cells in blue, stage 2 MAIT cells in green, stage 3 MAIT cells in red, CD4+CD8+ double positive (DP) thymocytes in black. f. Flow cytometry analysis of CD4+ (yellow) and CD4− (pink) CD161+ thymic MAIT cells for LEF1 expression. Scatter plots show percentage CD4+ cells within CD161+ thymic MAIT cells and percentage LEF1+ cells within this CD4+ population. (e,f) Data indicate mean ± SEM and are representative of at least 3 experiments using 3–7 separate donor samples.
To explore the changing transcriptomic landscape during human MAIT cell development, we carried out RNAseq analysis of different stages of thymic MAIT cells. A comparison of stage 1 to stage 3 revealed 625 differentially expressed genes, 390 of which were increased and 235 were decreased as MAIT cells mature (Fig. 5b). The 50 most significant of each category are listed in Figure 5c and the full list of differentially expressed genes is provided in Table S2 and a heatmap is provided in Figure S8. Our previous study identified higher protein expression of CD218 (IL18R), CD161 (KLRB1), CD27, and PLZF (ZBTB16) in stage 3 MAIT cells compared to stage 1 MAIT cells(6) and this was also reflected by an increase in gene expression of these molecules (Figs. 5b and c). Additionally, we detected a significant increase in SLAMF7, GZMA (granzyme A), GZMK, CXCR6, CCR5, CCR6, IL2RB (CD122) and IL7RA (CD127), ABCB1 encoding the ATP-binding cassette-multi-drug efflux protein 1 (MDR1) that is prominently expressed on peripheral blood MAIT cells(20, 21), and CEBPD encoding for the bZIP transcription factor C/EBPδ recently shown to be important for MAIT cell trafficking(22). Conversely, there were many genes that were downregulated in stage 3 human MAIT cells, including: RAG1, SATB1, Lef1, Tcf7, CCR9, BACH2, CD8β and CD4 (Figs. 5b and c). It is also noteworthy that many of these were similarly modulated between stages 1 and 3 in mouse MAIT cells, including ZBTB16, CXCR6, CCR6, CCR9, SLAMF7, IL2RB, IL7R, IL18R, RAG1, SATB1, LEF1, TCF7, BACH2, CD8β and CD4 (Fig. 1 b–d and Fig. 5b–c).
We next investigated where most of these transcriptional changes occurred by comparing stage 1 to stage 2, and stage 2 to stage 3 (Fig. 5d). This revealed that only 84 genes were significantly modulated (51 up, 33 down) between stage 1 to stage 2, while a much greater change took place between stage 2 and stage 3 (205 up and 148 down). As expected, one of the genes that increased between stage 1 and 2 was CD27, which is consistent with its use as a marker to separate these stages. Also increased between stages 1 and 2 were HLA-A, B and C genes and CCL5, and decreased were RAG1, RAG2, and CD1e, which is consistent with the relative maturity of stage 2 cells. As expected, most of the changes between stages 2 and 3 were the same as those detected between stages 1 and 3. Accordingly, these data reveal that most changes in gene expression occur between stages 2 and stage 3 in humans, which is similar to that in mice (Fig. 1), further validating the three-stage thymic development pathway we originally proposed(6).
Using flow cytometry, we were able to validate results from these RNAseq studies, showing a decrease in expression of SATB1, LEF1 and TCF1 transcription factors and CD1 proteins (CD1a, b and d) as human MAIT cells mature from stages 1 to 3 (Fig. 5e). Conversely, human MAIT cells upregulate expression of HLA class I, CCR5 and CCR6 molecules as they mature intrathymically (Fig. 5e–f). These findings, in conjunction with previously validated factors (CD27, CD161, CD218, PLZF)(6) provide assurance that the modulated transcripts detected in our RNAseq based assay are reflective of actual changes in protein expression as MAIT cells mature. Considering that we detected a CD4+ LEF1+ population that appear to be early stage 3 cells in mice, and that these proteins were similarly expressed in human MAIT cells, we looked for the presence of a comparable population in humans. Indeed, we found that approximately 15% of human stage 3 cells expressed CD4, and these were all LEF1+ (Fig. 5g). This suggests that a similar population within stage 3 cells expresses analogous factors in both mice and human thymus.
Further transcriptional modulation associated with extrathymic MAIT cell differentiation.
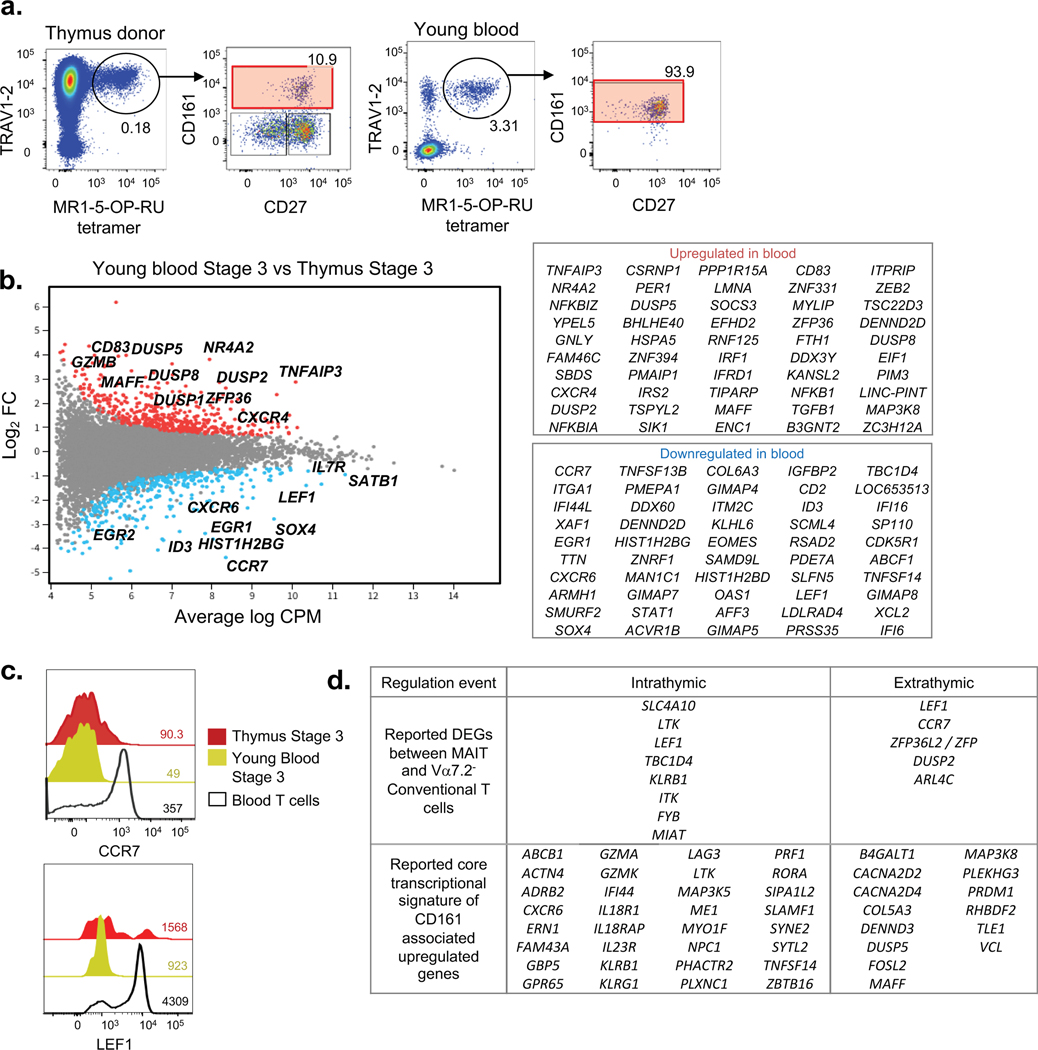
We have previously documented that human MAIT cells appear to undergo further, extrathymic maturation after leaving the thymus, increasing their capacity to produce cytokines and switching from CD8αβ to CD8αα(6). Hence, we carried out RNAseq analysis on sorted stage 3 MAIT cells from human thymus and blood samples from the same donors. These data revealed considerable diversity between mature thymic versus extrathymic MAIT cells, even despite the blood coming from very young thymus-blood matched donors (Fig. 6a–b). Of those that increased, some of the immunologically relevant genes were: TGFB1, NFκB1, IRF1, CD83, CXCR4 and SOCS3 while those that decreased included: LEF1, CCR7, CXCR6, EOMES, CD2, ID3, IL7R, SOX4, TOX2 and SATB1. Of note, we have recently profiled SATB1 expression in human T cells where thymic T cell progenitors have the highest levels of SATB1 that is subsequently downregulated in mature peripheral CD4+ and CD8+ T cells(23). We validated via flow cytometry that CCR7 and LEF1 expression decreased between thymus and young blood stage 3 MAIT cells (Fig. 6c). It has previously been reported that CD8αα MAIT cells arise extrathymically as most stage 3 cells in the thymus are CD8αβ(6, 9). We found that CD8β was reduced on average in peripheral MAIT cells but the difference did not quite reach significance in our analysis. Taken together, these data reveal that human MAIT cells continue to mature and differentiate after leaving the thymus, which may be a result of extrathymic exposure to antigen, cytokines, or other factors. Previous studies have examined the transcriptional profile of human MAIT cells, defined as CD161-associated genes in one study(24) or Vα7.2+ CD161+ T cells in another(25). We found considerable overlap of these reported genes with the DEGs revealed in our RNA-seq based study on purified MAIT cells and could further segregate these gene regulation events in a temporal manner. Thus we grouped the genes that stood out in these signatures into two stages: those that arise intra-thymically between stages 1 and 3 and those that arise after MAIT cells leave the thymus (Fig. 6d).
Figure 6. Transcriptomic analysis of extrathymic MAIT cell development.
a. Flow cytometry profile with CD27 and CD161 expression of Vα7.2+ MAIT cells of CD3+ cells from young (thymus donor) and adult peripheral blood sample. b. MD plot showing gene expression comparison of bulk-cell purified stage 3 MAIT cells from young peripheral blood vs thymus sample and associated list of most differentially regulated genes. Data derived from 5 young blood and 3 thymus donor samples. c. Flow cytometry analysis of CCR7 and LEF1 expression on stage 3 thymus MAIT cells, stage 3 young blood MAIT cells, and young blood conventional T cells. Data are representative of at least 3 experiments using 3 separate donor samples. d. Classification of known DEGs between MAIT and conventional T cells, and known core transcriptional signature of CD161-associated upregulated genes into intrathymic (between thymic stage 3 vs stage 1) and extrathymic (between young blood and thymus) regulation events.
We also carried out RNAseq based comparison of MAIT cells from mouse spleen (essentially all stage 3) to stage 3 MAIT cells from mouse thymus. This analysis revealed surprisingly few genes (19) that were differentially expressed between these populations, 5 that were increased and 14 that were decreased. Immunologically relevant genes amongst those that were increased include DTX1 (Deltex1) and CCL5, whereas amongst those that were decreased include LY6A, KLRA3 and KLRA9. The main message to arise through these data seems to be that there is remarkable consistency between the transcriptome of stage 3 MAIT cells in mouse thymus and MAIT cells in spleen (Figure S9).
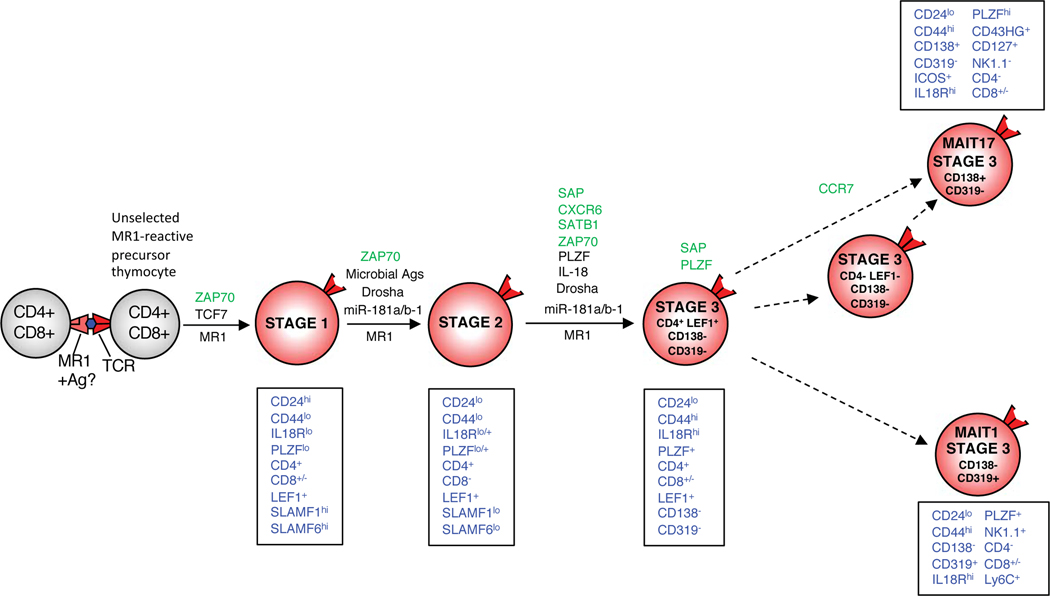
In summary, these data have provided a major advance to our current understanding of MAIT cell development, identifying a new population of CD4+ LEF1+ stage 3 cells that appears to be present in both mice and humans, and many genes that are modulated through the three-stage progression that are similar between mice and humans. This suggests that these conserved transcriptional changes play a key role in MAIT cell development in both species. We have integrated the information derived from our findings in this paper into a new model of MAIT cell development, as illustrated in Figure 7.
Figure 7. A new model of MAIT cell development.
Dashed lines depict likely pathways based on transcriptome analysis. Phenotypic signature of each stage is shown in blue text in boxes. Factors that regulate each differentiation step are depicted above arrow for each relevant steps: Green text represents newly defined factors that support the relevant step, red text represents newly defined factors that inhibit the relevant step, black text represents previously defined factors, from this study. This model is based on mouse MAIT cell development.
Discussion
We have previously presented a model of MAIT cell development that progressed through three distinct stages in mice and humans. Here, using these stages as a starting point, we have mapped the transcriptional changes that occur as MAIT cells progress through each stage, in both species. Our results have highlighted a greater level of complexity in the stage 3 phase of MAIT cell development, allowing many key advancements to the existing model developed in our previous publication(6). This has resulted in a new model (Fig. 7) that integrates some of the key findings from this paper.
Our findings have confirmed that stages 1 and 2 are clearly separate populations with numerous transcriptional changes, and there are many more transcriptional changes that separate stages 2 and 3. This is true for both mice and humans and indeed, many of the changes align between both species which is quite reassuring that mouse MAIT cell development represents a viable model for understanding this process in humans. We also identified an unexpected degree of heterogeneity in mouse stage 3 cells and with the power of single-cell RNAseq, we were able to separate these into at least 4 distinct subpopulations. Of these, we have identified a new stage 3 subpopulation, defined as CD4+ LEF1+ stage 3 cells, that seem to bridge the gap between stage 2 and the bulk of stage 3 cells This is illustrated by principle component analysis and is experimentally supported by its enrichment in stage 3 cells from very young mice and PLZF null mice. At the other end of the stage 3 spectrum we have populations of functionally mature MAIT1 and MAIT17 cells that are capable of IFN-γ or IL-17A production, respectively. While we had previously identified these two populations(6, 10), here, we demonstrate that they are remarkably distinct at the transcriptomic level and furthermore, that they can be readily separated using CD138 and CD319 cell-surface markers. A comparison of the genes expressed by MAIT1 and MAIT17 cells to those recently published for NKT1 cells and NKT17 cells, highlights considerable overlap between these functionally similar subsets(12). Moreover, a recent study focused on tissue resident MAIT cells showed that RORγt+ MAIT17 cells and RORγt− MAIT cells, (enriched for MAIT1 cells), identified from the lung, liver and spleen shared similarity to NKT1 and NKT17 cells isolated from the same tissue, although clear differences were observed between tissues for each cell type(26). In our study, we focussed on MAIT cell subsets in the thymus to understand where this transcriptional bifurcation is likely to occur during intra-thymic MAIT cell development. We propose that MAIT1 and MAIT17 cells arise in the thymus, within stage 3, from a subset that did not label with anti-CD138 or CD319. These cells are themselves diverse, including the CD4+LEF1+ population that more closely resembles stage 2 cells, and CD4−LEF1−CD138−CD319− cells, which most closely resemble MAIT17 cells. Thus, we have proposed a model pathway that integrates these stage 3 subpopulations into the earlier 3 stage pathway of MAIT cell development(6) (Fig. 7). This pathway postulates that stage 2 cells (which express CD4 and LEF1) first arrive as CD4+LEF1+ stage 3 cells. These CD4+ stage 3 cells then transition to either CD319+ MAIT1 or CD138+ MAIT17 cells. The CD138−CD319−CD4−LEF1− stage 3 population may contain intermediates for both MAIT1 and MAIT17 cells, although as a population they more closely resembled the MAIT17 lineage and may reflect a continuum with MAIT17 cells.
We were able to validate many of the transcriptional changes observed by antibody mediated detection of the relevant proteins, for example, SLAM family members (Slamf1, Slamf6, Slamf7) varied widely at the level of transcripts and this was verified by antibody staining of the relevant MAIT cell subsets. Importantly, we were also able to directly test the biological significance of some of the key factors detected in this study, demonstrating a critical role that several of these gene products play in the process of MAIT cell development. An initial report suggested that the SLAM-SAP-Fyn pathway was not critical for the development of MAIT cells because SAP-deficient patients contained normal frequencies of MAIT cells(27). However, the expression of SLAMF1 and SLAMF6 in early MAIT cell precursors, and of SLAMF7 (CD319) by MAIT1 cells, prompted us to re-examine the role of SLAM family members using SAP deficient mice, revealing that MAIT cell numbers are drastically reduced in these mice. The dissimilarities in MAIT cell frequency observed between species is likely due to the large expansion of MAIT cells that occurs in humans but not mice(6, 28–31). Furthermore, while this paper was under review, another study has been recently published that also demonstrated downregulation of expression of SLAMF6 as MAIT cells developed and showed impaired MAIT cell development in SAP-deficient mice(32). CCR7 was recently shown to be expressed by RAG2+ early MAIT cell precursors(33) and we validated this finding here using transcriptome and flow cytometry analysis. Furthermore, we demonstrate that a deficiency in CCR7 results in a reduction of thymic and splenic MAIT cells and a bias towards MAIT1 cells in these organs. CCR7 might be required for thymocyte migration as previous studies showed that NKT cell development is impaired in CCR7 KO mice, likely due to diminished thymocyte migration into the thymic medulla(34). We also showed that mice that lack Zap70 have a drastic reduction in thymic MAIT cells suggesting a role for TCR signalling in MAIT cell development, although these mice have reduced thymocyte survival and this might be caused by diminished Bcl-XL, RORγ and TCF1/LEF1 in these mice(35, 36). Notwithstanding the lack of formal validation for the role of LEF1 with LEF1-deficient mouse models, we recently demonstrated that Tcf7, encoding TCF1 is essential for MAIT cell development with virtually no detectable MAIT cells in Tcf7 KO mice(37), indicating a Tcf7-dependent step at the earliest stage of MAIT cell development (Fig. 7). Given that TCF1 and LEF1 are similarly regulated as MAIT cells mature, and that they have a similar role in conventional T cell development(38) and both play coordinated roles in NKT cell development(39, 40), we suggest that similar to TCF1, LEF1 will also be a key factor in regulating MAIT cell development.
Our data identifies many other genes that are differentially expressed as MAIT cell develop in the thymus and indeed the data from this manuscript may serve as a basis for many new experiments that explore the development, function and differentiation of these cells. For example, c-maf is upregulated in stage 3 thymic MAIT cells, specifically MAIT17 cells, and it was recently reported to regulate the expression of known IL-17 effector program genes (Blk, Rorc and Il17a) in IL-17 producing mouse γδ T cells(41). We demonstrate here that these same genes are preferentially expressed by MAIT17 cells compared to MAIT1 cells and less mature MAIT cells. Thus, we have markedly expanded the list of known factors that control MAIT cell development before stage 1 (SKG); between stages 2 and 3 (SLAM family members via SAP; CXCR6; SATB1); and the functional branchpoint within stage 3 (CCR7).
It was encouraging that there were many homologous/orthologous genes that are similarly regulated in both human and mouse MAIT cell maturation through stages 1–3, in terms of transcription factor and surface protein expression. As some key examples: PLZF, IL-7 receptor (CD127), IL-18 receptor (CD218), CD161 (NK1.1/KLRB1), SATB1, LEF1 and CD4 undergo similar regulation between stage 1, 2 and 3 cells, in both mice and humans. Considering the large evolutionary gap between humans and mice, and that many genes in humans are simply not expressed or differentially regulated in mice and vice versa, we feel that the two models from each species are perhaps surprisingly well aligned and information gained from mouse models is valuable for understanding intrathymic MAIT cell development in both species. We note that these observations build on previous findings that human thymic stage 3 MAIT cells do not show the same clear bifurcation in the thymus as mouse stage 3 MAIT cells, but the bifurfication event in human MAIT cell development appears to occur post-thymically, and seem to be influenced by different tissue microenvironments(42–44).
In summary, this combined transcriptomic based study has mapped in fine detail the molecular changes that occur in MAIT cells as they develop in the thymus of mice and humans. Many of these have been validated by flow cytometry and we have established the functional significance of several of these as regulators of MAIT cell development using gene knockout mice. The results provide a new and far more detailed model of how MAIT cells develop in mice and humans and provides a framework upon which future studies of these cells, and the factors that control their generation, will be founded.
Methods
Mice
C57BL/6 wild-type (WT), BALB/c WT, CXCR6-GFP(45) (C57BL/6 background), Satb1m1Anu/m1Anu (C57BL/6 background), Bach2CD4-cre (46) (C57BL/6 background) and SKG(17) (BALB/c background) mice were bred in-house at the Department of Microbiology and Immunology Animal House, University of Melbourne. Satb1m1Anu/m1Anu mice were generated from the Satb1m1Anu mutation. The Satb1m1Anu allele (NM_001163630.1:c.1500T>A), which encodes leucine instead of phenylalanine at amino acid 393 of SATB1, was identified in a screen for immunological regulators based on the chemical mutagen, N-ethyl-N-nitrosourea(47). The Satb1m1Anu mutation was identified by exome sequencing(48) of 3 littermate mice that exhibited decreased CD44 expression on CD4+ and CD8+ T cells (manuscript in preparation). Similar phenotypic abnormalities in mice with T cell-specific ablation of SATB1 expression were observed in Satb1m1Anu/m1Anu mice ((14), Fig. S6). SAP KO (Sh2d1a−/−) mice(49) and littermate controls are housed in the University of Queensland Animal Facility. CCR7 KO mice were sourced from JAX (006621) and were bred and maintained at the University of Adelaide Animal Facility behind a specific pathogen free (SPF) barrier in IVC caging. CCR7 WT controls were age and sex matched with WT C57BL/6J that were bred and maintained in the same room and on the same rack. All mice used were aged between 6 to 10 weeks old at time of sacrifice. All procedures using mice were approved by the University of Melbourne, University of Queensland or University of Adelaide Animal Ethics Committees, The Australian National University Animal Experimentation Ethics Committee, or by the Walter and Eliza Hall Institute Animal Ethics Committee. Single cell suspensions from mouse thymus, spleen and lymph nodes were prepared as previously described(10).
Human blood and tissue
Adult peripheral human blood samples were obtained from the Australian Red Cross Blood Service under agreement number 13–04VIC-07. Young human peripheral blood samples and matching thymus were obtained from the Royal Children’s Hospital (RCH), Victoria, Australia. These thymic and matched peripheral blood data were derived from 8 donors between the age of 2 months to 4 years old.
Experiments were conducted in accordance with University of Melbourne Human Research and Ethics committee guidelines (reference numbers 1035100 and 1443540) and RCH Human Research Ethics Committee Approval (reference number 24131 G). Blood mononuclear cells were isolated by Ficoll-Paque Plus™ density gradient centrifugation (GE Healthcare). Donor thymi were cut into small pieces and passed through a 70-micron cell strainer into ice-cold RPMI-1640 medium containing 2mM EDTA before being washed into PBS + 2% Fetal Calf Serum (FACS buffer).
Magnetic bead enrichment of thymic MAIT cells.
Using 5-OP-RU formed in situ from synthetic 5-A-RU and methylglyoxal, mouse and human MR1 tetramers were generated and biotinylated as previously described (50–52). Biotinylated MR1–5-OP-RU monomers were tetramerized with streptavidin conjugated to either PE (SA-PE) (BD Pharmingen) or Brilliant Violet 421 (SA-BV) (Biolegend). Single cell suspensions of mouse thymus were prepared and stained with PE-mouse MR1–5-OP-RU tetramers prior to magnetic bead enrichment using anti-PE microbeads as per manufacturer’s instructions (Miltenyi Biotec). One independent enriched sample represents 2–3 pooled thymi unless otherwise specified. Single cell suspensions of human thymus were enriched for TRAV1.2+ cells by staining for TRAV1.2-PE antibody, followed by magnetic bead enrichment using anti-PE microbeads (Miltenyi Biotec).
Flow cytometry, sorting and intracellular transcription factor staining.
Mouse and human cells were stained with viability dye 7-aminoactinomycin D (7-AAD; Sigma) and cell surface antibodies concurrently with MR1-tetramers, at room temperature (RT) for 30 minutes. List of antibodies used are detailed in Table S4. Transcription factors were assessed by staining with antibodies after the cells were surface-stained and permeabilized with the eBioscience Foxp3 Fixation/Permeabilization kit, according to the manufacturer’s instructions. Apart from intracellular analysis, mouse cells were analysed unfixed after staining, whereas human cells were fixed with 1% paraformaldehyde (PFA). Analysis was carried out using a BD LSR Fortessa equipped with a 561nm yellow-green laser and data processed using FlowJo software (Treestar). Mouse cells are gated on B220− lymphocytes and human cells on CD14−CD19− lymphocytes after dead cell and doublet exclusion. Flow cytometric sorting for live, unfixed bulk (100 cells)- or single cell were performed using a BD FACS AriaIII cell sorter. Flow cytometry gating strategy is shown in Figure S10.
Cell sort for RNA sequencing
Bulk-cell sorting involved sorting 3 separate samples for 100 cells as biological and technical repeats. Bulk and single MAIT cells were flow sorted into a chilled 384-well PCR plate (Greiner #785290) containing 1.2μl of primer/lysis mix [20 nM indexed polydT primer (custom made, IDT), 1:6.000.000 dilution of ERCC RNA spike-in mix (Ambion #4456740), 1 mM dNTPs (NEB #N0446S), 1.2 units SUPERaseIN Rnase Inhibitor (Thermo Fisher #AM2696), 0.2% Triton X-100 solution (Sigma Aldrich #93443–100ml), DEPC water (Thermo Fisher#AM9920)] using a BD FACSAria III flow cytometer (BD Biosciences, San Jose, CA, USA). Sorted plates were sealed, centrifuged for 1 min at 3000 rpm and immediately frozen upside down at −80°C until further processing using an adapted CelSeq2 protocol(53).
cDNA generation
Sorted plates were thawed on ice and briefly centrifuged. To lyse the cells and anneal the mRNA capture primer the plate was incubated at 65C for 5 min and immediately chilled on ice for at least 2 min before adding 0.8μl reverse transcription reaction mix [in 2μl RT reaction: 1 x First-Strand Buffer (Invitrogen #18064–014), 20 mM DTT (Invitrogen#18064–014), 4 units RNaseOUT (Invitrogen #10777–019), 10 units SuperScript II (Invitrogen #18064–014)]. The plate was incubated at 42°C for 1 hr, 70°C for 10 min and chilled to 4°C to generate first strand cDNA.
Pooling
Samples were pooled by placing the plate upside down on a trough [Nalgene #VP 792D] using a home-made 3D printed adapter to hold plate in place. The combined plate/trough stack was place in a plate centrifuge and spun at 1500 rpm for 1 min. Using a P1000 pipette the sample was transferred into a fresh 1.5 ml Eppendorf LoBind tube [Eppendorf #0030108051].
Exonuclease 1 treatment and clean-up
Exo I [NEB #M0293L; 20 U/μl] was added to the pooled sample at a final concentration 1 U/ul concentration and gently mixed by pipetting. In a heat block the sample was incubated at 37C for 30 min, followed by 80°C for 10 min and placed on ice. Once samples reached room temperature a 1.2X NucleoMag NGS Clean-up and Size select magnetic beads (Macherey-Nagel #7449970.5) clean-up was performed according to manufactures instruction. To reduce the amount of beads for each 100μl pooled sample 20μl beads and 100μl bead binding buffer (20% PEG8000, 2.5M NaCl, pH5.5) was added. The cDNA was eluted in 17 μl DEPC water and transferred to a fresh 0.2 ml PCR tube.
Second strand DNA synthesis
3μl of second strand reaction mix was added to the sample [1x NEBNext Second Strand Synthesis buffer (NEB #E6111S), NEBNext Second Strand Synthesis Enzyme Mix: 2.4 units DNA Polymerase I (E. coli), 2 units RNase H, 10 units E. coli DNA Ligase (NEB #E6111S), DEPC water (Thermo Fisher #AM9920)]. The sample was gently mixed by pipetting and incubated at 16°C for 2 hrs in a thermal cycle with unheated lid to generate double stranded cDNA followed by a 1.2X NucleoMag NGS Clean-up and Size select magnetic beads (Macherey-Nagel #7449970.5) clean-up according to manufactures instruction. The cDNA was eluted in 6.4 μl DEPC water and kept with beads for the following IVT reaction.
In Vitro Transcription (IVT)
9.6μl of IVT reaction mix [1.6μl of each of the following: ATP,GTP,CTP,UTP solution, 10X T7 buffer, T7 enzyme (MEGAscript T7 Transcription Kit – Ambion #AM1334)] was added and incubated at 37C for 13 hrs and then chilled and kept at 4C. To remove leftover primers 6μl ExoSAP-IT For PCR Product Clean-Up (Affymetrix #78200) was added and the sample was incubated at 37°C for 15 min and then chilled and kept at 4C.
RNA fragmentation and clean-up
Chemical heat fragmentation was performed by adding 5.5μl of 10X Fragmentation buffer (RNA fragmentation reagents #AM8740) to the sample and incubation in pre-heated thermal cycler at 94C for 2.5 min followed by immediately chill on ice and addition of 2.75μl of Fragmentation Stop buffer (RNA fragmentation reagents #AM8740). The fragmented amplified RNA was purified using 1.8X RNAClean XP beads (Beckman coulter #A63987) according to manufacturers instruction and eluted in 6ul DEPC water of which 5μl (no beads) were transferred to a fresh tube for library preparation.
5’-tagged-random-hexamer reverse transcription (ranhexRT)
The fragmented RNA was transcribed into cDNA using 5’-tagged random hexamer primers 9 (GCCTTGGCACCCGAGAATTCCANNNNNN) introducing a partial Illumina adapter as also described in CEL-Seq2(53). To remove RNA secondary structure and anneal the mRNA capture primer 1μl of tagged random hexamer (100μM) and 0.5μl of 10mM dNTPs (dNTP solution set NEB #N0446S) were added to the sample and incubated at 65C for 5 min and immediately chilled on ice for at least 2 min before adding 4μl reverse transcription reaction mix [in 10μl RT reaction: 1x Fist Strand buffer (Invitrogen #18064–014), 20 mM DTT (Invitrogen #18064–014), 4 units RNaseOUT (Invitrogen #10777–019), 10 units SuperScript II (Invitrogen #18064–014)].
Library amplification and cleanup
The PCR primers introduce the full-length adaptor sequence required for Illumina sequencing (for details see Illumina small RNA PCR primers). PCR was performed in 12.5μl using half of the ranhexRT sample as a template [1X KAPA HiFi HotStart ReadyMix (KapaBiosystems #KK2602), 400 nM each primer]. The final PCR amplified library was submitted to two consecutive 1x NucleoMag NGS Clean-up and Size select magnetic beads (Macherey-Nagel #7449970.5) according to manufacturer’s instructions. The final library was eluted in 20μl of 10 mM Trizma hydrochloride solution (Sigma-Aldrich #T2319–1L).
RNA sequencing data analysis
CelSeq2 RNA sequencing reads were mapped to either the GRCm38 mouse genome or GRCh38 human genome using the Subread aligner (54) and assigned to genes using scPipe(55) with ENSEMBL v86 annotation. Gene counts were exported as a matrix by scPipe with UMI-aware counting and imported into R(56). For 100 cell bulk samples, cells were removed from further analysis if they failed to achieve 5000 total counts. In single cell samples, cells were removed using the detect_outlier function from the scPipe. In 100 cell bulk samples, genes were filtered out if they failed to achieve 5 counts per million in 3 samples and in single cells genes were required to be expressed in at least 5 samples and an average of 2 or more counts in those samples. Heatmaps were generated on normalised expression values using heatmap2 from the gplots package with row normalisation. Dimensionality reduction was performed on normalised log2-count-per-million expression values with size factors from computeSumFactor in scran(57). Mean-Difference (MD) plots were generated using limma(58) and Glimma(59). Differential expression analyses used negative binomial GLMs and likelihood ratio tests from edgeR(60, 61). Genes with a false discovery rate (FDR)(62) < 0.0 5 were deemed differentially expressed. We used monocle2 (v2.12.0)(63) to estimate single-cell trajectories and create subsequent visualisations. Starting from the scran-normalized data, the 1000-most highly variable genes were selected and used as input to construct a Discriminative Dimensionality Reduction with Trees (DDRTree). The DDRTree was then used to estimate a pseudotime trajectory by the orderCells() function in monocle2, rooting the tree using the Stage 1 cells. Gene set testing was performed on the differentially expressed genes using goana(64) and kegga from the limma package for Gene Ontology(65) and KEGG(66) pathway database respectively.”
Supplementary Material
Supplementary Figure 1. Full heatmap data for Figure 3c.
Supplementary Figure 2. Gene ontology (GO) and KEGG pathway enrichment analysis in MAIT cell development.
Supplementary Figure 3. Trajectory analysis on sorted single cells.
Supplementary Figure 4. Mean-difference (MD) profiles of differentially expressed genes (DEG) of Lef1+ stage 3 MAIT cells compared to other subsets.
Supplementary Figure 5. Conventional TCRβ+, NKT and γδT cell compartments in SAP, Satb1m1Anu/m1Anu, SKG, Cxcr6 KO and Ccr7 KO mice.
Supplementary Figure 6. Abnormal Foxp3+ T-reg phenotype of mice homozygous for the Satb1m1Anu allele.
Supplementary Figure 7. MAIT cell development appear to be normal in Bach2 deficient mice.
Supplementary Figure 8: Full heatmap data for Figure 5.
Supplementary Figure 9. DEGs between bulk splenic and thymic mature MAIT cells.
Supplementary Figure 10. Flow cytometry gating strategies and representative sorting purities.
Supplementary table 1. Full list of differentially expressed genes from bulk (100-cell) purified thymic stage 3 vs stage 1 mouse MAIT cells.
Supplementary table 2. Full list of top differentially expressed genes between stage 3 and stage 1 human MAIT cells.
Supplementary table 3. List of antibodies used.
Acknowledgements
The authors thank the staff in the Doherty Institute, University of Queensland, University of Adelaide, the Australian National University and Walter and Eliza Hall Institute Animal Facilities and Biological Resource Facilities (BRF) for animal husbandry assistance. We thank Tina Luke and staff at the Doherty Institute Flow Cytometry Facility, Simon Monard and the staff at the WEHI Flow Cytometry Facility, and Vanta Jameson and staff at the Melbourne Brain Centre Flow Cytometry Facility, for flow cytometry support. We also thank Stephen Turner and Adam Uldrich for helpful discussions.
Funding
This work was supported by the National Health and Medical Research Council of Australia (NHMRC): 1083942, 1113293, 1140126 to D.I.G; NHMRC 1122277, 1054925 to G.T.B; NHMRC 1071916 to K.K.; NHMRC 1016953, 1113904, 585490 and 1081858, and National Institutes of Health (U19-AI100627) to CCG. H-F.K is supported by an NHMRC ECF Fellowship (1160333); D.I.G. H-F.K and D.G.P are also supported by the Australian Research Council CE140100011. D.G.P is supported by a CSL Centenary Fellowship; K.K is supported by an NHMRC Senior Research Fellowship (1102792); A.K. is supported by an NHMRC Fellowship (1139607) and D.F, D.I.G and G.T.B are supported by NHMRC Senior Principal Research Fellowships (1117017, 1117766 and 1135898 respectively). This study was made possible through Victorian State Government Operational Infrastructure Support and Australian Government NHMRC Independent Research Institute Infrastructure Support scheme.
Footnotes
Competing interests:
J.Mc. and D.P.F. are named inventors on a patent application (PCT/AU2013/000742, WO2014005194) and J.Y.W.M., Z.C., J.Mc. and D.P.F. are named inventors on another patent application (PCT/AU2015/050148, WO2015149130) involving MR1 ligands for MR1-restricted MAIT cells owned by University of Queensland, Monash University and University of Melbourne. The remaining authors declare no competing financial interests.
Data and material availability
The RNA sequencing profiling data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) repository with the accession number GSE137350.
References
- 1.Godfrey DI, Koay HF, McCluskey J, Gherardin NA, The biology and functional significance of MR1-restricted T cells. Nature Immunology in press, (2019). [DOI] [PubMed] [Google Scholar]
- 2.Kjer-Nielsen L, Corbett AJ, Chen Z, Liu L, Mak JY, Godfrey DI, Rossjohn J, Fairlie DP, McCluskey J, Eckle SB, An overview on the identification of MAIT cell antigens. Immunol Cell Biol 96, 573–587 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Meermeier EW, Harriff MJ, Karamooz E, Lewinsohn DM, MAIT cells and microbial immunity. Immunol Cell Biol 96, 607–617 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ussher JE, Willberg CB, Klenerman P, MAIT cells and viruses. Immunology and cell biology, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rouxel O, Lehuen A, Mucosal-associated invariant T cells in autoimmune and immune-mediated diseases. Immunology and cell biology 96, 618–629 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Koay HF, Gherardin NA, Enders A, Loh L, Mackay LK, Almeida CF, Russ BE, Nold-Petry CA, Nold MF, Bedoui S, Chen Z, Corbett AJ, Eckle SB, Meehan B, d’Udekem Y, Konstantinov IE, Lappas M, Liu L, Goodnow CC, Fairlie DP, Rossjohn J, Chong MM, Kedzierska K, Berzins SP, Belz GT, McCluskey J, Uldrich AP, Godfrey DI, Pellicci DG, A three-stage intrathymic development pathway for the mucosal-associated invariant T cell lineage. Nat Immunol 17, 1300–1311 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Seach N, Guerri L, Le Bourhis L, Mburu Y, Cui Y, Bessoles S, Soudais C, Lantz O, Double-positive thymocytes select mucosal-associated invariant T cells. Journal of immunology 191, 6002–6009 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Winter SJ, Kunze-Schumacher H, Imelmann E, Grewers Z, Osthues T, Krueger A, MicroRNA miR-181a/b-1 controls MAIT cell development. Immunology and cell biology, (2018). [DOI] [PubMed] [Google Scholar]
- 9.Leeansyah E, Loh L, Nixon DF, Sandberg JK, Acquisition of innate-like microbial reactivity in mucosal tissues during human fetal MAIT-cell development. Nature communications 5, 3143 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahimpour A, Koay HF, Enders A, Clanchy R, Eckle SB, Meehan B, Chen Z, Whittle B, Liu L, Fairlie DP, Goodnow CC, McCluskey J, Rossjohn J, Uldrich AP, Pellicci DG, Godfrey DI, Identification of phenotypically and functionally heterogeneous mouse mucosal-associated invariant T cells using MR1 tetramers. The Journal of experimental medicine 212, 1095–1108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cameron G, Godfrey DI, Differential surface phenotype and context-dependent reactivity of functionally diverse NKT cells. Immunol Cell Biol in press, (2018). [DOI] [PubMed] [Google Scholar]
- 12.Engel I, Seumois G, Chavez L, Samaniego-Castruita D, White B, Chawla A, Mock D, Vijayanand P, Kronenberg M, Innate-like functions of natural killer T cell subsets result from highly divergent gene programs. Nat Immunol 17, 728–739 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nichols KE, Hom J, Gong SY, Ganguly A, Ma CS, Cannons JL, Tangye SG, Schwartzberg PL, Koretzky GA, Stein PL, Regulation of NKT cell development by SAP, the protein defective in XLP. Nature medicine 11, 340–345 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitagawa Y, Ohkura N, Kidani Y, Vandenbon A, Hirota K, Kawakami R, Yasuda K, Motooka D, Nakamura S, Kondo M, Taniuchi I, Kohwi-Shigematsu T, Sakaguchi S, Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nature immunology 18, 173–183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kakugawa K, Kojo S, Tanaka H, Seo W, Endo TA, Kitagawa Y, Muroi S, Tenno M, Yasmin N, Kohwi Y, Sakaguchi S, Kowhi-Shigematsu T, Taniuchi I, Essential Roles of SATB1 in Specifying T Lymphocyte Subsets. Cell reports 19, 1176–1188 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Alvarez JD, Yasui DH, Niida H, Joh T, Loh DY, Kohwi-Shigematsu T, The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes & development 14, 521–535 (2000). [PMC free article] [PubMed] [Google Scholar]
- 17.Sakaguchi N, Takahashi T, Hata H, Nomura T, Tagami T, Yamazaki S, Sakihama T, Matsutani T, Negishi I, Nakatsuru S, Sakaguchi S, Altered thymic T-cell selection due to a mutation of the ZAP-70 gene causes autoimmune arthritis in mice. Nature 426, 454–460 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Iwabuchi K, Iwabuchi C, Tone S, Itoh D, Tosa N, Negishi I, Ogasawara K, Uede T, Onoe K, Defective development of NK1.1+ T-cell antigen receptor alphabeta+ cells in zeta-associated protein 70 null mice with an accumulation of NK1.1+ CD3- NK-like cells in the thymus. Blood 97, 1765–1775. (2001). [DOI] [PubMed] [Google Scholar]
- 19.Mao AP, Constantinides MG, Mathew, Zuo, Chen, Weirauch MT, Bendelac A, Multiple layers of transcriptional regulation by PLZF in NKT-cell development. Proc Natl Acad Sci U S A 113, 7602–7607 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dusseaux M, Martin E, Serriari N, Peguillet I, Premel V, Louis D, Milder M, Le Bourhis L, Soudais C, Treiner E, Lantz O, Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood 117, 1250–1259 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Fergusson JR, Ussher JE, Kurioka A, Klenerman P, Walker LJ, High MDR-1 expression by MAIT cells confers resistance to cytotoxic but not immunosuppressive MDR-1 substrates. Clinical and experimental immunology 194, 180–191 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee CH, Zhang HH, Singh SP, Koo L, Kabat J, Tsang H, Singh TP, Farber JM, C/EBPdelta drives interactions between human MAIT cells and endothelial cells that are important for extravasation. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nussing S, Koay HF, Sant S, Loudovaris T, Mannering SI, Lappas M, Konstantinov DUY,IE, Berzins SP, Rimmelzwaan GF, Turner SJ, Clemens EB, Godfrey DI, Nguyen TH, Kedzierska K, Divergent SATB1 expression across human life span and tissue compartments. Immunology and cell biology, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fergusson JR, Smith KE, Fleming VM, Rajoriya N, Newell EW, Simmons R, Marchi E, Bjorkander S, Kang YH, Swadling L, Kurioka A, Sahgal N, Lockstone H, Baban D, Freeman GJ, Sverremark-Ekstrom E, Davis MM, Davenport MP, Venturi V, Ussher JE, Willberg CB, Klenerman P, CD161 defines a transcriptional and functional phenotype across distinct human T cell lineages. Cell Rep 9, 1075–1088 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park D, Kim HG, Kim M, Park T, Ha HH, Lee DH, Park KS, Park SJ, Lim HJ, Lee CH, Differences in the molecular signatures of mucosal-associated invariant T cells and conventional T cells. Sci Rep 9, 7094 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salou M, Legoux F, Gilet J, Darbois A, du Halgouet A, Alonso R, Richer W, Goubet AG, Daviaud C, Menger L, Procopio E, Premel V, Lantz O, A common transcriptomic program acquired in the thymus defines tissue residency of MAIT and NKT subsets. The Journal of experimental medicine 216, 133–151 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin E, Treiner E, Duban L, Guerri L, Laude H, Toly C, Premel V, Devys A, Moura IC, Tilloy F, Cherif S, Vera G, Latour S, Soudais C, Lantz O, Stepwise development of MAIT cells in mouse and human. PLoS biology 7, e54 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ben Youssef G, Tourret M, Salou M, Ghazarian L, Houdouin V, Mondot S, Mburu Y, Lambert M, Azarnoush S, Diana JS, Virlouvet AL, Peuchmaur M, Schmitz T, Dalle JH, Lantz O, Biran V, Caillat-Zucman S, Ontogeny of human mucosal-associated invariant T cells and related T cell subsets. The Journal of experimental medicine 215, 459–479 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gherardin NA, Souter MNT, Koay HF, Mangas KM, Seemann T, Stinear TP, Eckle SBG, Berzins SP, d’Udekem Y, Konstantinov IE, Fairlie DP, Ritchie DS, Neeson PJ, Pellicci DG, Uldrich AP, McCluskey J, Godfrey DI, Human blood MAIT cell subsets defined using MR1 tetramers. Immunology and Cell Biology 96, 507–525 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lepore M, Kalinicenko A, Colone A, Paleja B, Singhal A, Tschumi A, Lee B, Poidinger M, Zolezzi F, Quagliata L, Sander P, Newell E, Bertoletti A, Terracciano L, De Libero G, Mori L, Parallel T-cell cloning and deep sequencing of human MAIT cells reveal stable oligoclonal TCRbeta repertoire. Nature communications 5, 3866 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Tilloy F, Treiner E, Park SH, Garcia C, Lemonnier F, de la Salle H, Bendelac A, Bonneville M, Lantz O, An invariant T cell receptor alpha chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted alpha/beta T cell subpopulation in mammals. The Journal of experimental medicine 189, 1907–1921 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Legoux F, Gilet J, Procopio E, Echasserieau K, Bernardeau K, Lantz O, Molecular mechanisms of lineage decisions in metabolite-specific T cells. Nature immunology 20, 1244–1255 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Hogquist KA, CCR7 defines a precursor for murine iNKT cells in thymus and periphery. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cowan JE, McCarthy NI, Parnell SM, White AJ, Bacon A, Serge A, Irla M, Lane PJ, Jenkinson EJ, Jenkinson WE, Anderson G, Differential Requirement for CCR4 and CCR7 during the Development of Innate and Adaptive alphabetaT Cells in the Adult Thymus. J Immunol, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palacios EH, Weiss A, Distinct roles for Syk and ZAP-70 during early thymocyte development. J Exp Med 204, 1703–1715 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo J, Hawwari A, Li H, Sun Z, Mahanta SK, Littman DR, Krangel MS, He YW, Regulation of the TCRalpha repertoire by the survival window of CD4(+)CD8(+) thymocytes. Nat Immunol 3, 469–476 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Mielke LA, Liao Y, Clemens EB, Firth MA, Duckworth B, Huang Q, Almeida FF, Chopin M, Koay HF, Bell CA, Hediyeh-Zadeh S, Park SL, Raghu D, Choi J, Putoczki TL, Hodgkin PD, Franks AE, Mackay LK, Godfrey DI, Davis MJ, Xue HH, Bryant VL, Kedzierska K, Shi W, Belz GT, TCF-1 limits the formation of Tc17 cells via repression of the MAF-RORgammat axis. J Exp Med, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu S, Zhou X, Steinke FC, Liu C, Chen SC, Zagorodna O, Jing X, Yokota Y, Meyerholz DK, Mullighan CG, Knudson CM, Zhao DM, Xue HH, The TCF-1 and LEF-1 transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity 37, 813–826 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berga-Bolanos R, Zhu WS, Steinke FC, Xue HH, Sen JM, Cell-autonomous requirement for TCF1 and LEF1 in the development of Natural Killer T cells. Mol Immunol 68, 484–489 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carr T, Krishnamoorthy V, Yu S, Xue HH, Kee BL, Verykokakis M, The transcription factor lymphoid enhancer factor 1 controls invariant natural killer T cell expansion and Th2-type effector differentiation. The Journal of experimental medicine 212, 793–807 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zuberbuehler MK, Parker ME, Wheaton JD, Espinosa JR, Salzler HR, Park E, Ciofani M, The transcription factor c-Maf is essential for the commitment of IL-17-producing gammadelta T cells. Nat Immunol 20, 73–85 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang XZ, Jo J, Tan AT, Sandalova E, Chia A, Tan KC, Lee KH, Gehring AJ, De Libero G, Bertoletti A, IL-7 licenses activation of human liver intrasinusoidal mucosal-associated invariant T cells. Journal of immunology 190, 3142–3152 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Gibbs A, Leeansyah E, Introini A, Paquin-Proulx D, Hasselrot K, Andersson E, Broliden K, Sandberg JK, Tjernlund A, MAIT cells reside in the female genital mucosa and are biased towards IL-17 and IL-22 production in response to bacterial stimulation. Mucosal immunology 10, 35–45 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sobkowiak MJ, Davanian H, Heymann R, Gibbs A, Emgard J, Dias J, Aleman S, Kruger-Weiner C, Moll M, Tjernlund A, Leeansyah E, Sallberg Chen M, Sandberg JK, Tissue-resident MAIT cell populations in human oral mucosa exhibit an activated profile and produce IL-17. European journal of immunology, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Unutmaz D, Xiang W, Sunshine MJ, Campbell J, Butcher E, Littman DR, The primate lentiviral receptor Bonzo/STRL33 is coordinately regulated with CCR5 and its expression pattern is conserved between human and mouse. Journal of immunology 165, 3284–3292 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Kometani K, Nakagawa R, Shinnakasu R, Kaji T, Rybouchkin A, Moriyama S, Furukawa K, Koseki H, Takemori T, Kurosaki T, Repression of the transcription factor Bach2 contributes to predisposition of IgG1 memory B cells toward plasma cell differentiation. Immunity 39, 136–147 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Nelms KA, Goodnow CC, Genome-wide ENU mutagenesis to reveal immune regulators. Immunity 15, 409–418 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Andrews TD, Whittle B, Field MA, Balakishnan B, Zhang Y, Shao Y, Cho V, Kirk M, Singh M, Xia Y, Hager J, Winslade S, Sjollema G, Beutler B, Enders A, Goodnow CC, Massively parallel sequencing of the mouse exome to accurately identify rare, induced mutations: an immediate source for thousands of new mouse models. Open Biol 2, 120061 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Czar MJ, Kersh EN, Mijares LA, Lanier G, Lewis J, Yap G, Chen A, Sher A, Duckett CS, Ahmed R, Schwartzberg PL, Altered lymphocyte responses and cytokine production in mice deficient in the X-linked lymphoproliferative disease gene SH2D1A/DSHP/SAP. Proc Natl Acad Sci U S A 98, 7449–7454 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mak JY, Xu W, Reid RC, Corbett AJ, Meehan BS, Wang H, Chen Z, Rossjohn J, McCluskey J, Liu L, Fairlie DP, Stabilizing short-lived Schiff base derivatives of 5-aminouracils that activate mucosal-associated invariant T cells. Nature communications 8, 14599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corbett AJ, Eckle SB, Birkinshaw RW, Liu L, Patel O, Mahony J, Chen Z, Reantragoon R, Meehan B, Cao H, Williamson NA, Strugnell RA, Van Sinderen D, Mak JY, Fairlie DP, Kjer-Nielsen L, Rossjohn J, McCluskey J, T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature 509, 361–365 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Eckle SB, Birkinshaw RW, Kostenko L, Corbett AJ, McWilliam HE, Reantragoon R, Chen Z, Gherardin NA, Beddoe T, Liu L, Patel O, Meehan B, Fairlie DP, Villadangos JA, Godfrey DI, Kjer-Nielsen L, McCluskey J, Rossjohn J, A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells. The Journal of experimental medicine 211, 1585–1600 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hashimshony T, Senderovich N, Avital G, Klochendler A, de Leeuw Y, Anavy L, Gennert D, Li S, Livak KJ, Rozenblatt-Rosen O, Dor Y, Regev A, Yanai I, CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome biology 17, 77 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao Y, Smyth GK, Shi W, The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 41, e108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tian L, Su S, Dong X, Amann-Zalcenstein D, Biben C, Seidi A, Hilton DJ, Naik SH, Ritchie ME, scPipe: A flexible R/Bioconductor preprocessing pipeline for single-cell RNA-sequencing data. PLoS Comput Biol 14, e1006361 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.R. C. Team. (R Foundation for Statistical Computing, Vienna, Austria, 2016). [Google Scholar]
- 57.Lun AT, McCarthy DJ, Marioni JC, A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res 5, 2122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK, limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Su S, Law CW, Ah-Cann C, Asselin-Labat ML, Blewitt ME, Ritchie ME, Glimma: interactive graphics for gene expression analysis. Bioinformatics 33, 2050–2052 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCarthy DJ, Chen Y, Smyth GK, Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40, 4288–4297 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinson MD, McCarthy DJ, Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Benjamini Y, Hochberg Y, Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 57, 289–300 (1995). [Google Scholar]
- 63.Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, Trapnell C, Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 14, 979–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Young MD, Wakefield MJ, Smyth GK, Oshlack A, Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11, R14 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, Richter J, Rubin GM, Blake JA, Bult C, Dolan M, Drabkin H, Eppig JT, Hill DP, Ni L, Ringwald M, Balakrishnan R, Cherry JM, Christie KR, Costanzo MC, Dwight SS, Engel S, Fisk DG, Hirschman JE, Hong EL, Nash RS, Sethuraman A, Theesfeld CL, Botstein D, Dolinski K, Feierbach B, Berardini T, Mundodi S, Rhee SY, Apweiler R, Barrell D, Camon E, Dimmer E, Lee V, Chisholm R, Gaudet P, Kibbe W, Kishore R, Schwarz EM, Sternberg P, Gwinn M, Hannick L, Wortman J, Berriman M, Wood V, de la Cruz N, Tonellato P, Jaiswal P, Seigfried T, White R, Gene Ontology C, The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res 32, D258–261 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kanehisa M, Goto S, KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Full heatmap data for Figure 3c.
Supplementary Figure 2. Gene ontology (GO) and KEGG pathway enrichment analysis in MAIT cell development.
Supplementary Figure 3. Trajectory analysis on sorted single cells.
Supplementary Figure 4. Mean-difference (MD) profiles of differentially expressed genes (DEG) of Lef1+ stage 3 MAIT cells compared to other subsets.
Supplementary Figure 5. Conventional TCRβ+, NKT and γδT cell compartments in SAP, Satb1m1Anu/m1Anu, SKG, Cxcr6 KO and Ccr7 KO mice.
Supplementary Figure 6. Abnormal Foxp3+ T-reg phenotype of mice homozygous for the Satb1m1Anu allele.
Supplementary Figure 7. MAIT cell development appear to be normal in Bach2 deficient mice.
Supplementary Figure 8: Full heatmap data for Figure 5.
Supplementary Figure 9. DEGs between bulk splenic and thymic mature MAIT cells.
Supplementary Figure 10. Flow cytometry gating strategies and representative sorting purities.
Supplementary table 1. Full list of differentially expressed genes from bulk (100-cell) purified thymic stage 3 vs stage 1 mouse MAIT cells.
Supplementary table 2. Full list of top differentially expressed genes between stage 3 and stage 1 human MAIT cells.
Supplementary table 3. List of antibodies used.
Data Availability Statement
The RNA sequencing profiling data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) repository with the accession number GSE137350.