Abstract
Extracellular vesicles (EVs) play a crucial role in intercellular communication, participating in the paracrine trophic support or in the propagation of toxic molecules, including proteins. RTP801 is a stress‐regulated protein, whose levels are elevated during neurodegeneration and induce neuron death. However, whether RTP801 toxicity is transferred trans‐neuronally via EVs remains unknown. Hence, we overexpressed or silenced RTP801 protein in cultured cortical neurons, isolated their derived EVs (RTP801‐EVs or shRTP801‐EVs, respectively), and characterized EVs protein content by mass spectrometry (MS). RTP801‐EVs toxicity was assessed by treating cultured neurons with these EVs and quantifying apoptotic neuron death and branching. We also tested shRTP801‐EVs functionality in the pathologic in vitro model of 6‐Hydroxydopamine (6‐OHDA). Expression of RTP801 increased the number of EVs released by neurons. Moreover, RTP801 led to a distinct proteomic signature of neuron‐derived EVs, containing more pro‐apoptotic markers. Hence, we observed that RTP801‐induced toxicity was transferred to neurons via EVs, activating apoptosis and impairing neuron morphology complexity. In contrast, shRTP801‐EVs were able to increase the arborization in recipient neurons. The 6‐OHDA neurotoxin elevated levels of RTP801 in EVs, and 6‐OHDA‐derived EVs lost the mTOR/Akt signalling activation via Akt and RPS6 downstream effectors. Interestingly, EVs derived from neurons where RTP801 was silenced prior to exposing them to 6‐OHDA maintained Akt and RPS6 transactivation in recipient neurons. Taken together, these results suggest that RTP801‐induced toxicity is transferred via EVs, and therefore, it could contribute to the progression of neurodegenerative diseases, in which RTP801 is involved.
Keywords: 6‐OHDA, extracellular vesicles, mTOR signalling, neuron, RTP801/REDD1, toxicity
1. METHODS
1.1. Cell cultures
HEK293 cells were cultured in DMEM medium supplemented with 10% foetal bovine serum (FBS) and penicillin/streptomycin (all from Thermo Fisher Scientific) in a 5% CO2 atmosphere at 37°C. EVs‐depleted FBS was obtained from 18 h of ultracentrifugation at 100,000 × g in a 70.1 Ti rotor and was used to prepare complete media (10% FBS).
Rat primary cortical cultures were prepared as previously described (Lesuisse & Martin, 2002; Solana‐Balaguer et al., 2023). Briefly, cortices from embryonic (E18) Sprague‐Dawley rats were dissected out, dissociated in 0.05% trypsin, and plated at a density of 750 cells/mm2 on poly‐L‐lysine‐coated plates or at a density of 300 cells/mm2 on poly‐L‐lysine‐coated coverslips. Neurons were maintained in Neurobasal medium supplemented with B27 (1:50), 2 mM GlutaMAX, and penicillin/streptomycin (all from Thermo Fisher Scientific). One‐third of the medium was replaced every 7 days. Cell cultures were maintained in a 5% CO2 atmosphere at 37°C.
1.2. Lentiviral preparation
Lentiviruses to knockdown the expression of RTP801 were produced in HEK293 cells transfected with a pLL3.7 vector expressing RTP801 (shRTP801, 5′‐AAGACTCCTCATACCTGGATG‐3′) or scramble (shCtr, 5′‐ GTGCGTTGCTAGTACCAAC‐3′) shRNA (sequence validated in (Malagelada et al., 2006)). Lentiviruses to overexpress RTP801 were produced in HEK293 cells transfected with a FUGW vector expressing only eGFP or eGFP‐RTP801 (RTP801 tagged with eGFP at the N‐terminal, sequence in (Malagelada et al., 2006)). Transfections were carried out using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer's instructions. Seventy‐two hours post‐transfection, cell media was collected and centrifuged to remove debris. Virus‐containing media was incubated with PEG 6000 8.5% (Panreac) and NaCl 0.35 M for at least 90 min at 4°C. Lentiviruses were finally concentrated by centrifuging the media at 7500 × g for 15 min and resuspended in sterile 1X PBS/Ca2+.
1.3. Transfection, lentiviral infection, and treatments
HEK293 cells were transfected with pEGFP‐C3 or pEGFP‐RTP801‐C3 and rat primary cortical cultures with a pCMS‐eGFP plasmid (Addgene). Transfections were done using Lipofectamine 2000 (Thermo Fisher Scientific) following the manufacturer's protocol.
Rat primary cortical neurons were transduced at DIV11 with lentiviral particles containing eGFP or eGFP‐RTP801 constructs. 48 h later (DIV13), cultures were harvested, and EVs were isolated from the culture media. For loss of function experiments, neurons were transduced at DIV6‐7 with lentiviral particles containing an shRNA construct against RTP801 (shRTP801) or a scrambled control sequence (shCT) as previously described (Canal et al., 2016). Six days later (DIV13‐14) cultures were harvested and EVs were isolated.
Half of the medium was kept aside right before the transduction and was used to replace all the media 24 h prior EVs isolation, to avoid lentivirus contamination during EVs isolation.
6‐Hydroxydopamine (6‐OHDA, Tocris Bioscience) treatments were performed in cortical primary neurons after 13–14 days of differentiation at a concentration of 50 μM in H2O for 16 h, as previously described (Romaní‐Aumedes et al., 2014). One‐third of the media was replaced right before exposure to the toxin.
1.4. Extracellular vesicles isolation, characterization, labeling, and treatment
EVs were purified from depleted‐DMEM media or Neurobasal medium as previously described with minor modifications (Danzer et al., 2012). To compare the number, size, and content of EVs produced between different conditions, the exact same volume of culture media was subjected to the EVs isolation method (10 mL of neurobasal medium per P100 seeded plate). Briefly, cell culture was collected and centrifuged at 300 × g for 5 min at room temperature to remove cell debris and centrifuged again for 20 min at 2500 × g in a centrifuge Beckman GPR and 4°C. After, cell media was centrifuged for 30 min at 10,000 × g in a SS‐34 rotor in a centrifuge Sorvall (4°C) to remove other microvesicles of bigger diameter. Finally, the supernatant was ultracentrifuged at 4°C for 2 h at 100,000 x g in a 70.1 Ti rotor, in an Ultracentrifuge Optima L‐100XP with Polypropylene Centrifuge Tubes (Beckman 355630), final supernatant was discarded, and the resulting pellet was washed with 1X PBS‐Ca2+ and ultracentrifuged again for 1 h at 100,000 × g (4°C). The resulting pellet contained the EV fraction. For western blot (WB) analysis, the pellet was resuspended in loading buffer (Thermo Fisher Scientific) and loaded into the WB gel. For EVs quantification, the pellet was resuspended in 1X PBS, and vesicle size and quantity were determined using the Nanosight nanoparticle tracking LM10 system (NTA) (Malvern). For transmission electron microscopy (TEM) observation, the pellet was resuspended in 1X PBS‐2% paraformaldehyde (PFA) and then subjected to negative staining. Briefly, the suspension was adsorbed on copper grids for 25 min and contrasted with 2% uranyl acetate for 2 min and finally washed in water. Samples were observed with JEOL J1010 80 kV microscope.
For protein concentration analysis, EVs were resuspended in 1X PBS and protein concentration was measured by MicroBCA™ (Thermo Fisher Scientific).
For the generation of red fluorescent‐labelled EVs, the EVs in 1X PBS were incubated with 200 nM of the fluorogenic probe MemGlow‐560 (Cytoskeleton Inc.) for 1 h at RT, as previously described (Solana‐Balaguer et al., 2023). The excess of dye was removed with two centrifuges at 10,000 × g for 15 min at 4°C. The resulting supernatant contained the labelled EVs. For EVs internalization assay, neuronal cultures were treated with labelled EVs for 4 h. As a negative control, the same protocol was followed using the MemGlow‐560 suspension without EVs (only 1X PBS).
1.5. Western blotting
Whole‐cell extracts were collected and processed as previously described (Malagelada et al., 2006). The following primary antibodies were used (1:1000 it not stated otherwise): rabbit monoclonal anti‐GFP (Cell Signalling, #2956), rabbit polyclonal anti‐RTP801 (1:500, Proteintech, #10638‐1‐AP), mouse monoclonal anti‐TSG101 (Abcam, #ab83), mouse monoclonal anti‐Flotillin‐1 (BD Bioscience, #610821), mouse monoclonal anti‐Alix (Cell Signalling Technology, #2171), rabbit polyclonal anti‐VGLUT1 (Synaptic Systems, #135303), rabbit polyclonal anti‐Calnexin (Abcam, #ab22595), rabbit polyclonal anti‐Nedd4 (Santa Cruz Biotechnologies, #sc‐25508), rabbit polyclonal anti‐Phospho‐Akt‐Ser473 (Cell Signalling Technology, #4060S), rabbit polyclonal anti‐Phospho‐RPS6‐Ser235/236 (Cell Signalling Technology, #4858S), rabbit polyclonal anti‐Akt (Cell Signalling Technology, #4691S), mouse monoclonal anti‐RPS6 (Cell Signalling Technology, #2317), rabbit polyclonal anti‐Phospho‐p70S6K (Cell Signalling Technology, #9208), rabbit polyclonal anti‐p70S6K (Cell Signalling, #9202), rabbit monoclonal anti‐Phospho‐4EBP1 (Cell Signalling Technology, #2855), rabbit monoclonal anti‐4EBP1 (Cell Signalling Technology, #9644).
Although RTP801 antibody (Proteintech, #10638‐1‐AP) show several non‐specific bands, the specific 34 KDa band has been widely validated using shRNAs against RTP801 (Canal et al., 2016; Malagelada et al., 2006, 2008, 2011; Martín‐Flores et al., 2016, 2020; Pérez‐Sisqués, Martín‐Flores et al., 2021; Pérez‐Sisqués, Sancho‐Balsells et al., 2021; Pérez‐Sisqués, Solana‐Balaguer et al., 2021) or with a RTP801 KO model (Pérez‐Sisqués, Martín‐Flores et al., 2021). In the case of GFP antibody (Cell Signalling, #2956), in samples with GFP overexpression, apart from the 27 KDa band, other upper bands can be observed due to GFP protein aggregation (Namba et al., 2022).
The loading control was obtained by incubation with an anti‐α‐actin‐Peroxidase antibody (1:100,000; Merk, #A3854). Horseradish peroxidase‐conjugated goat anti‐mouse and anti‐rabbit secondary antibodies (1:10,000) were obtained from Thermo Fisher Scientific (1:10,000, #31430 and #31460, respectively). Chemiluminescent images were acquired using a Chemidoc imager (BioRad) or a LAS‐4000 Imager (Fujifilm, Valhalla, NY, USA) and quantified by computer‐assisted densitometric analysis (ImageJ). All the blots used for the figures are shown in Figure S11.
1.6. Immunofluorescence and image analyses
Rat primary cortical neurons were fixed with 4% PFA in 1X PBS and stained as previously described (Martín‐Flores et al., 2015). The following primary antibodies were used: rabbit polyclonal anti‐RTP801 antibody (1:200; Proteintech, #10638‐1‐AP), mouse monoclonal anti‐MAP2 (1:500, Abcam, #ab11268), chicken polyclonal anti‐GFP (1:500, Synaptic Systems, #132006) and rabbit polyclonal anti‐cleaved caspase‐3 (1:200; Cell Signalling Technologies, #9661). Goat anti‐mouse and anti‐rabbit secondary antibodies conjugated to Alexa 488 or Alexa 555 or Alexa 633 were purchased from Thermo Fisher Scientific (1:500). For nuclear staining, Hoechst 33342 (1:5000; Thermo Fisher Scientific, #H3570) was used.
For EVs uptake assay, images were acquired with ZEISS LSM880 confocal microscope as z‐stacks (five slices in a range of 5 µm) with a 63× magnification. Orthogonal views (YZ and XZ) were obtained with ImageJ to show the colocalizing EVs inside the neuron.
For the toxicity and branching analysis, images were acquired with a ZEISS Axio Observer Z1 fluorescence microscopy with a 20× magnification. Using Cell Profiler cell image analysis software (www.cellprofiler.org, Broad Institute) (Carpenter et al., 2006; Stirling et al., 2021), nuclei and neuronal somas were identified as independent objects. A mask with neuronal soma was used to obtain only neuronal nuclei. Cleaved caspase‐3 positive cells were identified as independent objects and were related with neuronal nuclei, with the RelateObjects module, to distinguish nuclei positive or negative for cleaved caspase‐3. Measures of mean intensity of cleaved caspase‐3 per neuron were obtained. Nuclei classification into viable or apoptotic was based on intensity, intensity distribution, size and shape, texture, and granularity parameters, using machine learning in Cell Profiler Analyst software (Jones et al., 2008; Stirling et al., 2021), as described in (Solana‐Balaguer et al., 2023).
For the branching analysis, analyses were performed as previously described (Solana‐Balaguer et al., 2023). Briefly, neurites were enhanced and turned into a binary image. From this image, and using the neuronal soma as input, neurons were identified as independent objects. Neurons touching the border of the image were discarded. Neuron objects were used to mask the binary image of enhanced neurites. From this image, the morphological skeleton was generated with Morph module. Measurements of endpoints (termini branches), trunks (primary dendrites), non‐trunk (intermediate branches), and total tree length were obtained per neuron, using the MeasureObjectSkeleton module. Neurons from five fields per coverslip were analysed and the mean of all neurons per field was acquired. At least three independent cultures were analysed.
1.7. Proteomics
Samples were processed and analysed at the Proteomics Platform of Navarrabiomed‐IdiSNA Center for Biomedical Research. For sample preparation, protein extracts were diluted in Laemmli sample buffer (4%) and were then loaded into a 0.75‐mm‐thick polyacrylamide gel containing a 4% stacking gel cast over 12.5% resolving gel. To concentrate the entire proteome at the stacking/resolving gel interface, the run was stopped as soon as the front entered 3 mm of the resolving gel. Gel was then stained using Coomassie Brilliant Blue and bands were excised and digested using 1:20 trypsin solution at 37°C for 16 h as previously described (Shevchenko et al., 2007). Peptide fragments were purified and concentred using C18 Zip Tip Solid Reverse Phase columns (Millipore). Samples were then separated by reverse phase LC–MS/MS using an UltiMate 3000 UHPLC System (ThermoFisher) fitted with a column in an acetonitrile gradient coupled to the Orbitrap Exploris 480 MS (ThermoFisher). Mass range was set to 375–1500 ppm. All the other acquisition parameters were set as previously described (Andrés‐Benito et al., 2019). The MaxQuant computing platform v.1.6.17.0 (Cox & Mann, 2008). and the environment‐integrated Andromeda search engine (Cox et al., 2011) were used to process the raw files. For peptide identification, a target‐decoy search strategy (Elias & Gygi, 2010) was performed against a target/decoy version of the rat UniProt database without isoforms with a maximum peptide mass of 7500 Da. The false discovery rate limit was set to 1% on both the peptide and protein identification levels. The Perseus software v.1.6.14.0 (Tyanova et al., 2016) was used for statistical and differential expression analyses. Only proteins with at least two identified peptides were considered for further analyses. The option ‘two samples t‐test’ was used to compare experimental conditions. Here, comparisons were considered to be statistically different if the following conditions were met: (i) Benjamini‐Hochberg adjusted p‐values under 0.05 and a (ii) log2 fold‐change over 0.3 and under −0.3. R (v.4.2.1) packages ComplexHeatmap (Gu et al., 2016), EnhancedVolcano and mixOmics (Rohart et al., 2017), were used for multivariate data analysis and visualization.
1.8. Statistics
All experiments were performed at least in three independent neuronal cultures, and data of each replicate is reported as mean ± SEM. Normal distribution was considered when all the data passed one of the following normality tests: D'Agostino‐Pearson, Shapiro–Wilk, and Kolmogorov–Smirnov. When normality was assumed, parametric statistical analyses were performed. When two conditions were compared, analyses were performed using the unpaired two‐sided t‐test (95% confidence). Before pairs of comparisons, we performed the F test to compare variances. T‐test with Welch's correction was performed when variances were unequal. When multiple groups were compared, one‐way ANOVA or two‐way ANOVA with Bonferroni's post hoc test was performed. To detect significant outlier values, Grubb's and ROUT tests were used. Values of P < 0.05 were considered statistically significant. The statistics used in each experiment are specified in figure legends.
2. BACKGROUND
Cell‐to‐cell transmission is a common mechanism for the progression of several neurodegenerative disorders including Parkinson's disease (PD) or Alzheimer's disease (AD) (Lee et al., 2010). Mounting evidence indicates the existence of prion‐like transmission of protein aggregates in interconnected brain areas, such as alpha(α)‐synuclein, or amyloid‐beta(β) (reviewed in Guo & Lee, 2014; Zhang et al., 2018). For example, the pathological staging process in PD proposed by Braak and co‐workers (Braak et al., 2004) is based on the caudo‐rostrally distribution of α‐synuclein aggregates in post‐mortem brains and supports the hypothesis that α‐synuclein may spread to neighbour cells. Therefore, transneural propagation of toxic proteins could contribute to the early stages of neurodegenerative disorders. However, the precise mechanisms of this neuron‐to‐neuron communication and the specific effects that exert on the recipient cells are not yet completely elucidated.
EVs act as vehicles for transcellular communication in general and have a crucial role in the central nervous system (CNS) (Quek & Hill, 2017; Solana‐Balaguer et al., 2023). There are different types of EVs, distinguished by size, biogenesis, and composition. Among them, exosomes are 50–100/150 nm vesicles (depending on the method used for the isolation and characterization, reviewed in Erdbrügger and Lannigan (2016)), that are delivered into the extracellular space upon fusion to the plasma membrane of endosomal intermediates called multivesicular bodies (MVBs). In contrast, microvesicles are 0.1–1 µm released by outward budding from the plasma membrane (Chuo et al., 2018; van Niel et al., 2018). EVs transport active proteins, lipids, RNAs, and signalling complexes that modulate gene and protein expression in the receiving cells (reviewed in Budnik et al., 2016; Frühbeis et al., 2013). EVs are biologically active, and, in neurons, they can be both protective and toxic (Bellingham et al., 2012; Montecalvo et al., 2012). Moreover, EVs have a crucial role in neuronal development and connectivity (Chivet et al., 2014; Sharma et al., 2019). In the mammalian nervous system, cortical neurons secrete EVs in a process that depends on synaptic glutamatergic activity and post‐synaptic calcium rise (Lachenal et al., 2011). Recently, EVs have been proposed as vehicles to transfer toxic proteins trans‐synaptically, such as α‐synuclein or amyloid‐β, which are associated with neurodegenerative disorders (Emmanouilidou et al., 2010; Rajendran et al., 2006).
RTP801, also known as REDD1, is a toxic protein for neurons. RTP801 is sufficient to induce cell death in NGF‐differentiated PC12 cells (Malagelada et al., 2006; Shoshani et al., 2002) and cultured sympathetic (Malagelada et al., 2006) and cortical neurons (Canal et al., 2016; Romaní‐Aumedes et al., 2014). RTP801 is coded by the stress‐responsive gene DDIT4, whose expression is induced in response to hypoxia, nutrient deprivation (Sofer et al., 2005), dexamethasone, thapsigargin, tunicamycin, heat shock (Wang et al., 2003) or cigarette smoke in lung cells (Yoshida et al., 2010). In the CNS, RTP801 expression is increased in response to ischemia (Shoshani et al., 2002), β‐amyloid peptide (Kim et al., 2003; Morel et al., 2009), mutant huntingtin (mhtt) (Martín‐Flores et al., 2016), and both PD‐mimetic toxins 1‐methy‐4‐phenyl‐1,2,3,6‐tetrahydropyridine (MPTP) and 6‐OHDA (Malagelada et al., 2006). Importantly, RTP801 is not only accumulated in cellular and animal pathologic models, but also in samples from patients suffering from neurodegenerative diseases. In fact, RTP801 protein is elevated in dopaminergic neurons from the substantia nigra pars compacta in sporadic and mutant parkin PD patients (Malagelada et al., 2006; Romaní‐Aumedes et al., 2014), in postmortem striata and neuron‐derived iPS cells from Huntington´s disease (HD) patients (Martín‐Flores et al., 2020, 2016) and lymphocytes and postmortem hippocampi from AD patients (Damjanac et al., 2009; Pérez‐Sisqués, Sancho‐Balsells et al., 2021). Remarkably, DDIT4 is one of the top three common upregulated transcripts in PD and HD (Labadorf et al., 2018). Some studies show that RTP801 modulates cytotoxicity in vitro in the context of neurodegeneration (Kim et al., 2003; Malagelada et al., 2008; Morel et al., 2009). In line with this, downregulation of RTP801 in AD and HD mouse models prevents cognitive deficits and neuroinflammation (Pérez‐Sisqués, Sancho‐Balsells et al., 2021; Pérez‐Sisqués, Solana‐Balaguer et al., 2021).
The action mechanism of RTP801 involves a sequential regulation of the mechanistic target of rapamycin (mTOR) and Akt survival kinases. mTOR exists in two complexes, each one with different specificity over substrates. The mTORC1 complex regulates protein translation by phosphorylating substrates such as ribosomal protein S6 kinase/S6 (RPS6) and 4EBP1 (Hara et al., 2002; Thedieck et al., 2007) and the mTORC2 complex regulates survival signals by phosphorylating Akt at Serine residue 473, among other substrates (Jacinto et al., 2006; Sarbassov et al., 2005). In PD, RTP801 acts upstream of tuberous sclerosis complex 1 and 2 (TSC1/TSC2), which in turn inhibits mTOR (Brugarolas et al., 2004). As consequence, RPS6/4EBP1 cannot be phosphorylated by mTORC1, and protein translation is tuned down. But most importantly, the neuronal survival kinase Akt cannot be phosphorylated by mTORC2 and so it is unable to enhance pro‐survival signals, thereby triggering neuronal death (Malagelada et al., 2008).
Therefore, given that RTP801 protein is upregulated in many neurodegenerative diseases and have a pro‐apoptotic role that might cause neuron death, in this study we aimed to investigate whether RTP801‐induced toxicity in culture could be transferred to other neurons via EVs, as a potential mechanism of neurodegeneration spreading.
Our results reveal that RTP801 is present in neuron‐derived EVs and promotes their loading with pro‐apoptotic proteins, which could contribute to transfer toxicity to neurons. Transneuronal RTP801 impairs neuron arborization and activates apoptosis in recipient neurons. Moreover, neuron‐derived EVs exerted a trophic effect on neurons, activating the mTOR/Akt signalling pathway, an effect that was lost via RTP801 when EVs were derived from 6‐OHDA‐stressed neurons.
3. RESULTS
3.1. RTP801 is present and released in EVs
To investigate whether endogenous RTP801 is present in neuronal EVs, neuronal culture media (DIV14) was subjected to sequential ultracentrifugation. The presence of EVs within the exosomal range was confirmed using NTA, with a size average of 135 nm (Figure 1a). We also confirmed the expected size and shape of small EVs using TEM (Figure 1b). We observed by WB that the small EVs fraction (sEVs or P100K) was enriched in EVs markers such as Alix, Flotillin‐1 and TSG101, whereas they showed little signal for the negative control calnexin, in comparison with neuronal lysates or larger vesicles (lEVS or P10K). Furthermore, we detected that endogenous RTP801 was present in low levels in EVs derived from primary neurons (Figure 1c). To test whether these EVs could be internalized by neurons, we labelled them with a fluorogenic membrane probe (Memglow‐560, in red) and treated cortical cultured neurons with these labelled EVs. We confirmed the presence of internalized EVs under confocal microscope. Negative control using vehicle, confirmed that Memglow‐560 specifically binds to EVs’‐membrane, as no fluorescent signal was detected in the absence of the vesicles (1X PBS) (Figure 1d).
FIGURE 1.
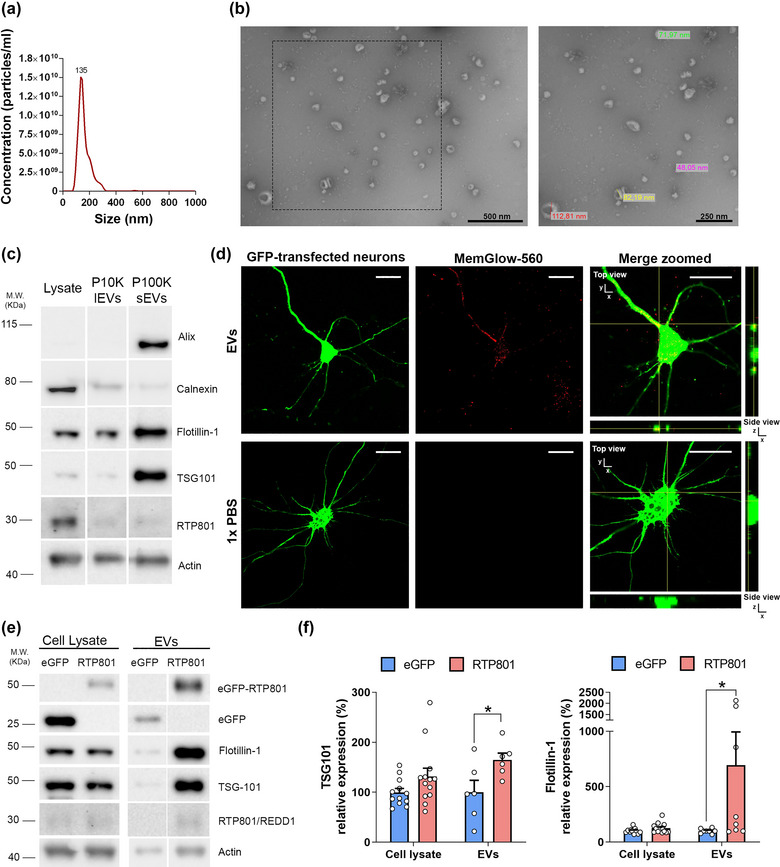
RTP801 is present in EVs and its overexpression increases EVs release. For EVs characterization, EVs were isolated from the culture media of cortical neurons at DIV13 following a sequential ultracentrifugation protocol. (a) Size distribution of nanoparticles by NanoSight particle tracking analysis (NTA). (b) Transmission electron micrographs of the vesicles show particles with the characteristic morphology and size of EVs. (c) WB analysis of total cell lysates, large EVs (P10K or lEVs) and small EVs (P100K or sEVs). Membranes were probed against common EVs markers (Alix, TSG‐101 and Flotillin‐1), EVs negative marker (Calnexin), and RTP801. Actin is used as a loading control for the lysates. (d) Cultured neurons were transfected at DIV12 with an eGFP plasmid (in green). EVs were isolated from sister neuronal cultures and labeled with the fluorogenic cell membrane probe MemGlow‐560 (in red). EVs particles were administered to eGFP‐cultured neurons with a ratio of 400 EVs:cell. MemGlow‐560 specifically binds to EVs, as no fluorescent signal was detected in 1X PBS samples. Orthogonal views show EVs internalization into neurons. Scale bar of 5 µm. (e) Cortical primary neurons (DIV11) were transduced with lentiviral particles containing eGFP empty vector (as control) or eGFP‐RTP801 constructs. 48 h after, EVs were isolated and total protein content was analysed by WB. Membranes were probed against RTP801 and eGFP, TSG‐101, and flotillin‐1 as EVs markers and actin as a loading control for the lysates. Representative immunoblots are shown. (f) Graphs show values obtained by densitometric analysis of WB data relative to the total protein content of the cell lysate. Values represent culture replicates of at least three independent neuronal cultures (mean ± SEM). Data were analysed by unpaired T‐test with Welch's correction in the cell lysate and in the EVs fraction (*P < 0.05 vs. control EVs).
Next, to confirm that ectopic RTP801 could be incorporated in EVs, we transduced cortical cultured neurons with lentiviral particles containing the ectopic form of RTP801 tagged with eGFP at the N‐terminal (eGFP‐RTP801). We also transduced sister cultures with lentiviral particles only containing eGFP as control. EVs released in the culture media were collected 48 h after transduction and the EVs‐producing neurons were lysed. EVs per each condition were isolated starting from the exact volume of culture media collected from eGFP or eGFP‐RTP801 overexpressing neurons and then resolved in a WB. We observed that ectopic RTP801 (with a molecular weight of around 55 KDa due to the eGFP tag) was found and enriched in EVs (Figure 1e).
We also observed ectopic control protein eGFP in the EVs fraction. Furthermore, ectopic RTP801 elevated TSG101 and Flotillin‐1 in the EVs fraction, suggesting an increase in the EVs release compared to the control eGFP condition (Figure 1e and f). NTA measurements confirmed that ectopic RTP801 enhanced EVs release (4.38 × 1010 ± 1.329 × 109particles/mL in eGFP‐overexpressing cultures (Figure S1A) and 1.56 × 1011 ± 4.24 × 1010particles/mL in RTP801‐overexpressing cultures (Figure S1B); P‐value = 0.0076 Student's T‐test). Although the increased amount of EVs in eGFP‐RTP801 condition elevated the levels of actin in the EVs fraction, it must be mentioned that actin is a proper loading control for the cell lysates but not for the EVs fraction since actin, and other cytoskeletal proteins, can be loaded also in EVs (Yoshioka et al., 2013). In fact, the increased amount of EVs is also reflected as an increase in actin levels.
We confirmed this finding in HEK293T cells, a proliferating non‐neuronal cell model, where overexpression of RTP801 does not induce cell death (Canal et al., 2014). Like our results in neurons, RTP801 was present in the EVs fraction and induced an elevation of TSG101 and Flotillin‐1 (Figure S2A and B). NTA measurements corroborated that ectopic RTP801 increased the release of EVs in the cultured media (1.11 × 1011 ± 2.46 × 1010 particles/mL in eGFP‐overexpressing cultures (Figure S1C) and 2.36 × 1011 ± 1.69 × 1010 particles/mL in eGFP‐RTP801‐overexpressing cultures (Figure S1D); P‐value = 0.0273 Student's T‐test). Moreover, we observed that ectopic RTP801 was promoting the presence of the endogenous RTP801 in the EVs fractions in both neurons (Figure 1e) and HEK293 cells (Figure S2A). Altogether, these results support that ectopic RTP801 increases the release of EVs compared to the control conditions.
3.2. RTP801 modulates the protein cargo of neuron‐derived EVs
Next, to investigate whether RTP801 could influence EVs protein cargo, we overexpressed or silenced RTP801 in cultured cortical neurons. Neurons were transduced with lentiviral particles containing eGFP or eGFP‐RTP801 constructs or with scrambled shRNA (shCT) or shRNA against RTP801 (shRTP801). Then, we isolated EVs from the respective culture media (eGFP‐, RTP801‐, shCT‐ or shRTP801‐EVs) and assessed EVs protein content by MS/proteomics (Figure 2).
FIGURE 2.
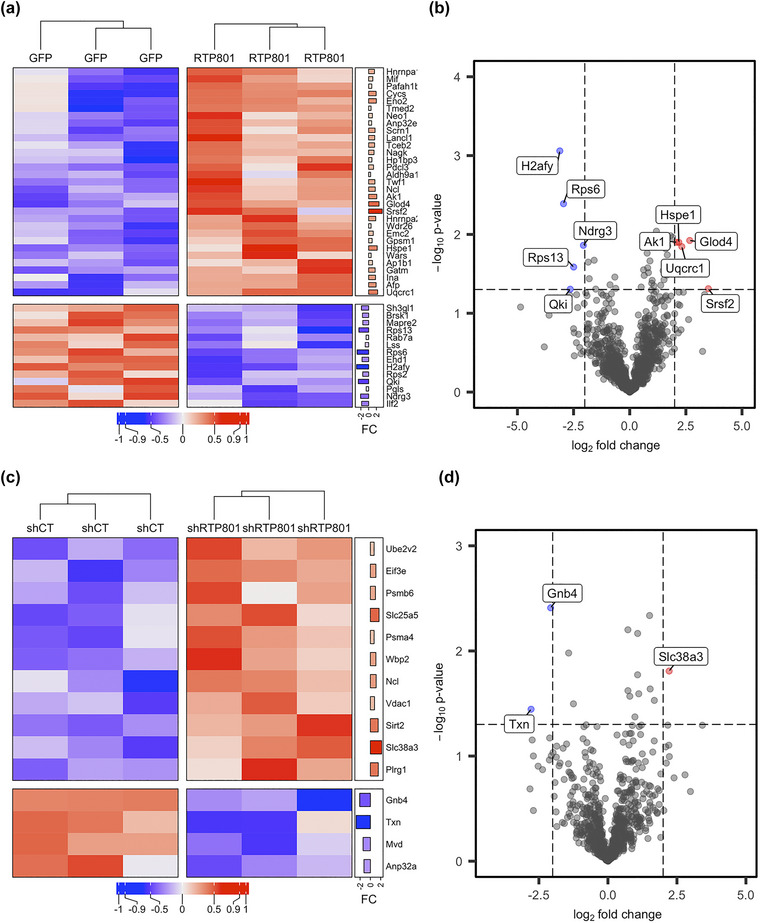
Modulation of RTP801 expression alters protein cargo of neuronal‐derived EVs in vitro. (a) Heatmap showing the differentially expressed proteins in GFP and RTP801‐derived EVs. Significantly overexpressed proteins in the RTP801 group in comparison to the GFP group are depicted in red, whereas proteins that are underrepresented in the RTP801 group are shown in blue. In the right annotation the log2 fold change (FC) is displayed as a bar plot for each of the proteins. (b) The volcano plot shows the whole set of proteins detected in the comparison between GFP and RTP801 experimental groups. The most overexpressed proteins being significant (‐log10 p‐value > = 1.2 and log2 fold change < −2.5) are plotted in red and the most downregulated proteins (‐log10 p‐value > = 1.2 and log2 fold change > 2.5) in blue. (c) Heatmap showing the differentially expressed proteins in shCT and shRTP801‐derived EVs. Significantly overexpressed proteins in the shRTP801 group in comparison to the shCT group are depicted in red, whereas proteins that are underrepresented in the shRTP801 group are shown in blue. In the right annotation, the log2 fold change (FC) is displayed as a barplot for each of the proteins. (d) The volcano plot shows the whole set of proteins detected in the comparison between shCT and shRTP801 experimental groups. The most overexpressed proteins being significant (‐log10 p‐value > = 1.2 and log2 fold change < −2.5) are plotted in red and the most downregulated proteins (‐log10 p‐value > = 1.2 and log2 fold change > 2.5) in blue. In both cases, these proteins are also tagged with the gene symbol. In both cases these proteins are also tagged with the gene symbol. Note that the heatmaps (a) and (c) only show the differentially expressed proteins between groups, while the whole set of proteins is used for the representation in volcano plots (b) and (d).
PLS‐DA plots revealed that both RTP801 overexpression and silencing changed the protein composition of EVs, as judged by sample group clustering in both comparisons (Figure S3). Pairwise comparisons of the proteomic data were used to identify enriched proteins and showed differentially expressed proteins in RTP801‐derived EVs (Figure 2a and b) and in shRTP801‐ derived EVs (Figure 2c and d).
Volcano plots showed that the most overexpressed proteins in RTP801‐EVs were SrSf2 (Serine/arginine‐rich splicing factor 2), Glod4 (Glyoxalase domain‐containing protein 4), Uqcrc1 (Cytochrome b‐c1 complex subunit 1), Hspe1 (10 kDa heat shock protein) and Ak1 (Adenylate kinase isoenzyme 1). SrSf2, Uqcrc1 and Hspe1 have been linked to apoptotic processes, as shown in Table 1. Ak1 has been described to have a neuropathogenic role in Aβ‐mediated tau phosphorylation (Park et al., 2012). The most downregulated proteins in RTP801‐EVs were H2afy (Core histone macro‐H2A.1), RPS6 (40S ribosomal protein S6), Qki (protein quaking), Rps13 (40S ribosomal protein S13), and Ndgr3 (protein NDGR3). Strikingly, H2afy and Ndgr3 have been linked to anti‐apoptotic processes (Table 1), and RPS6 is a substrate of mTOR activation which regulates protein translation, that in turn, is associated with cell survival (Hara et al., 2002; Kim et al., 2002; Thedieck et al., 2007) (Figure 2b).
TABLE 1.
Significantly altered proteins involved in apoptosis found in EVs derived from RTP801‐overexpressing neurons compared to the control. Pro‐apoptotic proteins are shown in red, anti‐apoptotic proteins in green, and proteins with a dual role as pro‐ and anti‐apoptotic in orange. Up‐arrows indicate that the protein is upregulated and down‐arrows that the protein is downregulated (Cas, abbreviated form os caspase).
| Protein ID | Protein name | Gene name | FC | Described function in apoptosis | References | |
|---|---|---|---|---|---|---|
| ▲ | Q6PDU1 | Serine/arginine‐rich splicing factor 2 | SrSf2 | 3.50 | SrSf2 down‐regulation inhibits apoptotic pathways in renal cancer | Kędzierska et al. (2016) |
| SrSf2 is necessary for the survival of hematopoietic cells and its loss results in apoptosis | Komeno et al. (2015) | |||||
| SrSf2 promotes the pre‐mRNA splicing of apoptosis‐related genes, such as Cas8, Cas9, c‐Flip and Bcl‐x, in response to DNA‐damaging agents | Li and Wang (2021) | |||||
| ▲ | Q68FY0 | Cytochrome b‐c1 complex subunit 1, mitochondrial | Uqcrc1 | 2.32 | Uqcrc1 regulates cytocrome c (cyt‐c)‐induced apoptosis | Hung et al. (2021) |
| ▲ | P26772 | 10 kDa heat shock protein, mitochondrial | Hspe1 | 2.19 | The function of Hspe1 and Hspd1 in apoptosis is complex, as they can regulate both positively and negatively the intrinsic pathway | Samali et al. (1999) |
| Lau et al. (1997) | ||||||
| Lin et al. (2001) | ||||||
| ▲ | P62898 | Cytochrome c, somatic | Cycs | 1.92 | Cycs triggers the activation of apoptosis‐protease activating factor 1, which is necessary for the maturation of Cas9 and Cas3 | Garrido et al. (2006) |
| In neurons, the block of cyt‐c activity prevents apoptosis | Neame et al. (1998) | |||||
| ▲ | P02773 | Alpha‐fetoprotein | Afp | 1.91 | Afp induces apoptosis in tumour cells by the activation of Cas3 | Dudich et al. (1999) |
| Knock‐down of Afp inhibits hepatocellular carcinoma cell proliferation and tumour growth by inducing apoptosis | Chen et al. (2020) | |||||
| ▲ | A7VJC2 | Heterogeneous nuclear ribonucleoproteins A2/B1 | Hnrnpa2b1 | 1.82 | Loss of hnrnpa triggers apoptosis in ovarian cancer | Yang et al. (2020) |
| Knock‐down of hnrnpa2B1 promotes apoptosis of breast cancer cells | Yin et al. (2020) | |||||
| ▲ | P23565 | Alpha‐internexin | Ina | 1.77 | Over‐expression of Ina in PC12 cells results in apoptosis‐like cell death | Chien et al. (2005) |
| High levels of Ina in Purkinje cells lead to degeneration and death of these neurons | Ching et al. (1999) | |||||
| ▲ | P13383 | Nucleolin | Ncl | 1.70 | Ncl inhibits apoptosis in B‐cell lymphomas | Wise et al. (2013) |
| Ncl is a negative regulator of H2O2 induced apoptosis | Zhang et al. (2010) | |||||
| Ncl antisense oligonucleotides induce apoptosis | Wu et al. (2012) | |||||
| Chemical inhibition of human Ncl induces early apoptosis in càncer cells | Bellone et al. (2022) | |||||
| ▲ | Q9QX69 | LanC‐like protein 1 | Lancl1 | 1.56 | Over‐expression of LanCL1 preserves cell viability after oxygen and glucose deprivation in neuronal HT22 cells | Xie et al. (2018) |
| Over‐expression of LanCL1 protects motor neurons from apoptosis in mice models of amyotrophic lateral sclerosis | Tan et al. (2020) | |||||
| ▲ | P04256 | Heterogeneous nuclear ribonucleoprotein A1 | Hnrnpa1 | 1.48 | HnRNP A1 is a nucleo‐cytoplasmic shuttling protein which is known to either up‐ or down‐regulate the IRES‐dependent translation of varios mRNAs that encode proteins related to apoptosis and proliferation, such as XIAP, c‐Myc, FGF2, Apaf‐a, cyclin‐D1 and Bcl‐xL. | Lewis et al. (2007) |
| Lewis et al. (2007) | ||||||
| Jo et al. (2008) | ||||||
| Cammas et al. (2007) | ||||||
| Bevilacqua et al. (2010) | ||||||
| Knock‐down of hnRNP A1 in mature neurons induce neuronal dysfunction and cell death | Anees et al. (2021) | |||||
| ▲ | Q4KLJ8 | Phosducin‐like protein 3 | Pdcl3 | 1.25 | Pdcl3 regulates the activation of caspases during apoptosis | Wilkinson et al. (2004) |
| ▲ | P30904 | Macrophage migration inhibitory factor | Mif | 1.08 | Over‐expression of Mif leads to a deficient deletion of apoptotic cells in photocarcinogenesis | Honda et al. (2009) |
| Knock‐down of Mif increases apoptotic cells in perivascular‐resident macrophage‐like melanocytes | Zhang et al. (2017) | |||||
| In cortical neurons, after a traumatic brain injury, Mif promotes neuronal cell death in vivo | Ruan et al. (2021) | |||||
| Mif controls cytokine release in neuroinflammation and inhibition of MIF significantly reduced cytokine production and cell death. | Nasiri et al. (2020) | |||||
| ▲ | P97603 | Neogenin | Neo1 | 0.98 | Inhibition of neogenin promotes neuronal survival after spinal cord injury | Chen and Shifman (2019) |
| Neogenin overexpression induces apoptosis in cerebellar neurons | Fujita et al. (2008) | |||||
| ▲ | P63004 | Platelet‐activating factor acetylhydrolase IB subunit alpha | Pafah1b1 | 0.80 | Deletion of Pafah1b1 in mice provokes germ cell apoptosis | Feng et al. (2022) |
| ▲ | F1LTR1 | WD repeat‐containing protein 26 | Wdr26 | 0.62 | Over‐expression of WDR26 reduces cell death and cas‐3 mediated apoptosis in H2O2 treated cells | Liu et al. (2022) |
| WDR6 inhibits apoptosis of cardiomyocytes after oxidative stress | Feng et al. (2012) | |||||
| ▲ | Q63524 | Transmembrane emp24 domain‐containing protein 2 | Tmed2 | 0.62 | Tmed2 is related to apoptosis in MM.1S and RPMI 8226 cell lines | Ge et al. (2020) |
| ▼ | P85971 | 6‐phosphogluconolactonase | Pgls | −0.62 | Pgls knockdown increases ROS and apoptosis rate in hepatocellular carcinoma cell lines | Li et al. (2021) |
| ▼ | B2DD29 | Serine/threonine‐protein kinase BRSK1 | Brsk1 | −1.58 | Loss of Brsk1 results in neuronal apoptosis and reduces the number of neural progenitors | Dhumale et al. (2018) |
| ▼ | Q641Z6 | EH domain‐containing protein 1 | Ehd1 | −1.69 | Ehd1 knockout mice show increased number of apoptotic cells in the lens epithelium | Arya et al. (2015) |
| ▼ | Q7TP98 | Interleukin enhancer‐binding factor 2 | Ilf2 | −1.80 | Ilf2 overexpression increases cell viability and decreases cell apoptosis, whereas knockdown shows opposite trends in human vascular smooth muscle cells. | Wei et al. (2021) |
| Ilf2 downregulation increases the number of apoptotic cells in liver cancer cells | Cheng et al. (2016) | |||||
| ▼ | O35964 | Endophilin‐A2 | Sh3gl1 | −1.85 | Endophilin A2 protects muscle cells from H2O2 induced apoptosis | Liu et al. (2016) |
| ▼ | Q6AYR2 | Protein NDRG3 | Ndrg3 | −2.06 | NDRG3 promotes cell proliferation and anti‐apoptosis during hypoxia | Lee et al. (2015) |
| Knockdown NDRG3 promotes cell apoptosis via ERK1/2 signalling pathway in gastric cancer cells | Liu et al. (2022) | |||||
| ▼ | Q02874 | Core histone macro‐H2A.1 | H2afy | −3.11 | H2AFY knockdown suppresses cell proliferation and promotes apoptosis of HCC cells in vitro. | Huang et al. (2021) |
In contrast, in the case of shRTP801‐EVs, the most overexpressed protein was Slc38a3 (sodium‐coupled neutral amino acid transporter 3) and the most downregulated were Gnb4 (guanine‐nucleotide‐binding protein subunit beta‐4) and Txn (thioredoxin) (Figure 2d). All three proteins have been associated with apoptotic processes, and remarkably, Slc38a3, the most enriched one, has been linked to anti‐apoptotic roles, as shown in Table 2.
TABLE 2.
Significantly altered proteins involved in apoptosis in EVs derived from neurons where RTP801 was silenced with an shRNA compared to the control. Pro‐apoptotic proteins are shown in red, anti‐apoptotic proteins in green, and proteins with a dual role as pro‐ and anti‐apoptotic in orange. Up‐arrows indicate that the protein is upregulated and down‐arrows that the protein is downregulated (Cas, abbreviated form of caspase).
| Protein ID | Protein name | Gene name | FC | Described function in apoptosis | References | |
|---|---|---|---|---|---|---|
| ▲ | Q9JHZ9 | Sodium‐coupled neutral amino acid transporter 3 | Slc38a3 | 2.22 | Slc38a3 promotes metastasis by activation of PDK1/Akt | (Wang et al., 2017) |
| Slc38Aa3 levels increase during stress | (Krokowski et al., 2013) | |||||
| Slc38a3 knock‐down disrupts mTOR/S6K pathway | (Chan et al., 2016) | |||||
| ▲ | Q09073 | ADP/ATP translocase 2 | Slc25a5 | 1.68 | Inhibition of Slc25a5 in neuroblastoma reduces cell viability | (Seneviratne et al., 2023) |
| Slc25a5 inhibits apoptosis | (Chevrollier et al., 2011) | |||||
| ▲ | Q9WUC8 | Pleiotropic regulator 1 | Plrg1 | 1.53 | Plrg1 deficiency increases the rate of apoptosis | (Kleinridders et al., 2009) |
| ▲ | Q5RJQ4 | NAD‐dependent protein deacetylase sirtuin‐2 | Sirt2 | 1.51 | Sirt2 knock‐down inhibits cell proliferation and promotes cell apoptosis in multiple myeloma | (Ding & Hao, 2021) |
| Sirt2 inhibition sensitizes lung cancer cells to etoposide‐mediated apoptosis | (Hoffmann et al., 2014) | |||||
| Sirt‐2 induced apoptosis in MPP+‐treated neuronal cells | (Liu et al., 2014) | |||||
| ▲ | P13383 | Nucleolin | Ncl | 1.12 | Ncl inhibits apoptosis in B‐cell lymphomas | (Wise et al., 2013) |
| Ncl is a negative regulator of H2O2 induced apoptosis | (Zhang et al., 2010) | |||||
| Ncl antisense oligodeoxynucleotides induce apoptosis | (Wu et al., 2012) | |||||
| Chemical inhibition of human Ncl induced early apoptosis in cancer cells | (Bellone et al., 2022) | |||||
| ▲ | Q8R478 | WW domain‐binding protein 2 | Wbp2 | 1.07 | Downregulation of Wbp2 results in apoptosis of glioma cells | (Gao et al., 2020) |
| (Chen et al., 2018) | ||||||
| ▲ | P28073 | Proteasome subunit beta type‐6 | Psmb6 | 0.98 | Psmb6 along with Psmb5 inhibition induce cytotoxicity | (Britton et al., 2009) |
| ▲ | Q9Z2L0 | Voltage‐dependent anion‐selective channel protein 1 | Vdac1 | 0.79 | Vdac1 prevents cell death during hypoxic conditions. | (Brahimi‐Horn et al., 2012) |
| Overexpression of Vdac1 induce apoptosis in many cell types | (Weisthal et al., 2014) | |||||
| (Abu‐Hamad et al., 2008) | ||||||
| (Ghosh et al., 2007) | ||||||
| (Godbole et al., 2003) | ||||||
| (Zaid et al., 2005) | ||||||
| ▼ | P49911 | Acidic leucine‐rich nuclear phosphoprotein 32 family member A | Anp32a | −1.25 | Anp32a knockdown induces apoptosis in myeloid leukaemia cell lines | (Yang et al., 2018) |
| Anp32a activates the Apaf apoptosome and therefore induces cas‐9 dependent apoptosis | (Hill et al., 2004) | |||||
| (Jiang et al., 2003) | ||||||
| ▼ | O35353 | Guanine nucleotide‐binding protein subunit beta‐4 | Gnb4 | −2.07 | Ectopic Gnb4 induces or inhibits apoptosis dependent on the cell line | (Wang et al., 2018) |
| ▼ | P11232 | Thioredoxin | Txn | −2.78 | Txn‐1 attenuates neuronal apoptosis in ischemia conditions | (Ma et al., 2012) |
| (Zhou et al., 2009) | ||||||
| Txn depletion induces oxidative stress | (Yang et al., 2011) | |||||
Taking into account all the differentially expressed proteins (Figure 2a and c) and based on a bibliographic research of its apoptotic role (Tables 1 and 2), we confirmed our first observation that RTP801‐EVs cargo was enriched in proteins involved in apoptosis and seemed to have higher levels of pro‐apoptotic proteins (in red, such as cytochrome b‐c1, cytochrome c and alpha‐internexin). The same cargo showed lower levels of anti‐apoptotic proteins (in green, such as core histone macro‐H2A, NDRG3 protein, and endophilin), in comparison with its control eGFP‐EVs. In contrast, EVs derived from RTP801‐silenced neurons (shRTP801‐EVs) showed high levels of anti‐apoptotic proteins (in green, such as sodium‐coupled neutral aminoacid transporter 3, ADP/ATP traslocase 2 or pleiotropic regulator 1) and lower levels of other apoptotic‐related proteins (Figure 2a and c, Tables 1 and 2).
Therefore, although we did not get statistically significant alterations in pathway analyses, our bibliographic research of the differentially expressed proteins points towards an apoptotic role of RTP801‐EVs. The fact that elevated levels of RTP801 have been reported to be toxic to neurons (Canal et al., 2016; Romaní‐Aumedes et al., 2014), and that EVs derived from neurons overexpressing RTP801 contain high levels of this protein along with apoptotic‐related proteins, may suggest that RTP801 toxicity could be transferred via EVs.
3.3. Overexpression of RTP801 promotes apoptosis and impairs neuron arborization complexity via EVs in cultured cortical neurons
To evaluate the transneuronal pro‐apoptotic role of RTP801 or other RTP801‐induced apoptotic proteins, we used Evs from RTP801 overexpressing neurons (RTP801‐Evs) or RTP801 silenced neurons (shRTP801‐Evs), along with its controls (eGFP‐Evs and shCT‐Evs, respectively). We treated sister cortical neuronal cultures with three different doses per conditions: 400, 800 or 1600 EVs per cell (EVs:cell ratio). After 24 h, untreated (UT) or EVs‐treated neuronal cultures were fixed and immunostained with antibodies against MAP2 and cleaved caspase‐3 (ClvCas3), and nuclei were stained with Hoechst 33342 (Figure 3a and b). Neurons (MAP2+ cells, in green) were segregated as ClvCas3 positive (ClvCas3+, in red) or negative (ClvCas3‐), and neuronal nuclei were into viable, condensed, or fragmented, considering the last two as apoptotic. To apply this classification, we used a machine learning pipeline (Figure 3c), which showed an accuracy of 95,65% (Figure 3d). We observed that ClvCas3 intensity levels were increased in neurons treated at the highest dose of RTP801‐EVs (Figure 3e and f), which highly suggests their putative pro‐apoptotic role. Not only ClvCas3 levels were increased but also the number of neurons MAP2+ expressing ClvCas3 (Figure S4A). No changes were observed with the treatment with the other EVs (shCT‐EVs, shRTP801‐EVs, or eGFP‐EVs), or at lower doses of treatment (Figures 3e and f and S4A).
FIGURE 3.
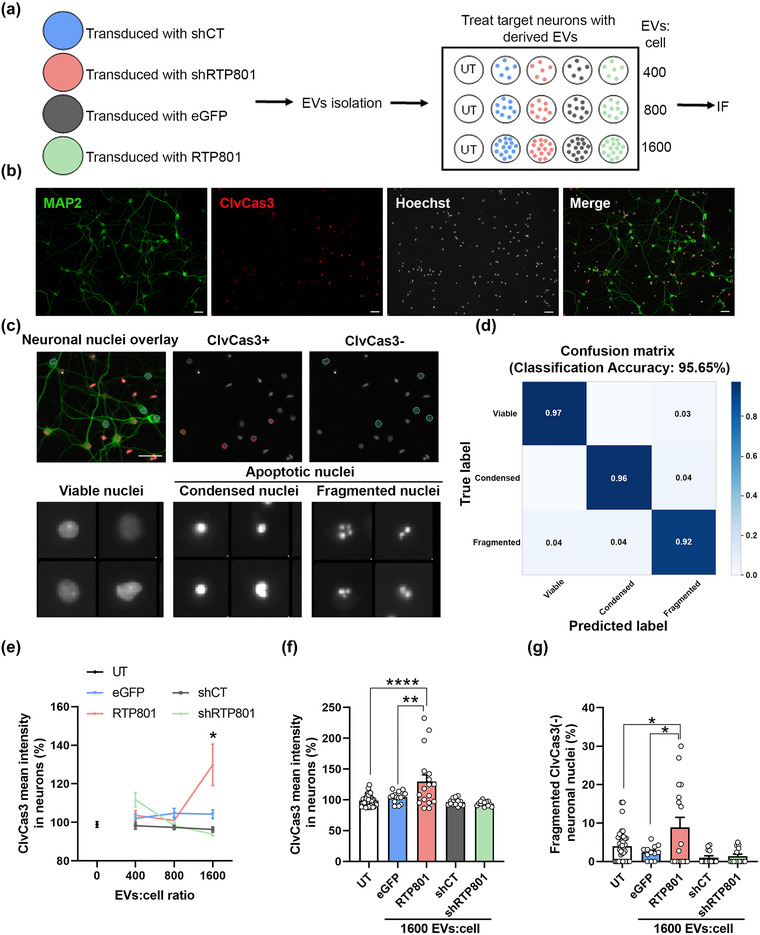
EVs derived from RTP801 overexpressing neurons induce neuron cell death via apoptosis. (a) Schematic representation of the experimental procedure. Cultured cortical neurons were transduced with lentiviral particles containing eGFP, eGFP‐RTP801, shCT and shRTP801 constructs. EVs were isolated from the culture medium and used to treat sister cortical neurons at three different doses: 400, 800 and 1,600 EVs:cell. After 24 h, neurons were fixed, and immunofluorescence against active caspase‐3 was performed. (b) Representative images of neuronal cultures at DIV14. MAP2 (in green) is used to identify neurons and ClvCas3 (in red) to identify cells expressing cleaved caspase‐3. Scale bar of 50 µm. (c) Neuronal nuclei were classified by phenotype: ClvCas3(+) (red outline) represents neurons expressing ClvCas3 and ClvCas3(‐) (blue outline) represents neurons with no active caspase‐3. Scale bar of 50 μm. Neuronal nuclei were then classified by machine learning into viable or apoptotic. Apoptotic nuclei were subclassified as condensed and fragmented. (d) Confusion matrix obtained with the classification training with CellProfiler Analyst. (e) ClvCas3 mean intensity in neurons. Data of the 3 doses of EVs‐treatment. (f) Data of ClvCas3 mean intensity only at the highest EVs dose (1,600 EVs:cell). (g) Quantification of neurons undergoing the last step of the apoptotic process (Fragmented ClvCas(‐) neuronal nuclei). At least neurons of 5 different fields were analysed per replicate. Values represent culture replicates of at least three independent neuronal cultures (mean ± SEM). Data was analysed by One‐way ANOVA (*P < 0.05, **P < 0.01, ****P < 0.0001).
Interestingly, we observed a reduction in the number of MAP2+ cells with the RTP801‐EVs treatment (Figure S4B). MAP2 protein is sensitive to proteases such as caspases and calpains, involved in neuron death (Johnson et al., 1991; Sun et al., 2009, 2008); hence, it is logical to lose MAP2 signalling when a neuron is in the last stages of the apoptotic process. In line with this, after the RTP801‐EVs treatment we observed an increase in the total number of apoptotic nuclei, contrary to shRTP801‐EVs treatment, which caused a decrease in the apoptotic nuclei (Figure S4C), suggesting a potential pro‐survival effect of RTP801 silencing. Moreover, considering MAP2+ cells, we also observed an increase in the number of neurons with fragmented chromatin and negative for ClvCas3 (Figures 3g and S4), combinatory readouts of last stage of the apoptotic process (Takemoto et al., 2003; Tawa et al., 2004), confirming the toxicity of RTP801 at the highest dose of RTP801‐EVs treatment (Figures 3g and S5).
Together, our findings strongly suggest that ectopic RTP801 is transferred via EVs and, along with a pro‐apoptotic cargo induces apoptosis in recipient neurons.
To further gain insight into the toxic effect of RTP801‐EVs, we next evaluated neuron arborization complexity after 24 h of treatment. To do so, neuron skeleton was extracted to quantify primary dendrites, intermediate branches, termini branches (endpoints), and total tree length (the length of all the branches in a neuron) (Figure 4a). RTP801‐EVs treatment (at a ratio of 1,600 EVs:cell) reduced primary dendrites, intermediate branches, and endpoints in mature neurons. Strikingly, shRTP801‐EVs at the same dose produced the opposite effect in cultured sister neurons. Indeed, shRTP801‐EVs increased the total tree length with no effect in the primary dendrites (Figures 4b and c and S6). These results further confirm the toxicity of RTP801‐EVs and hint the potential neurotrophic function of shRTP801‐EVs.
FIGURE 4.
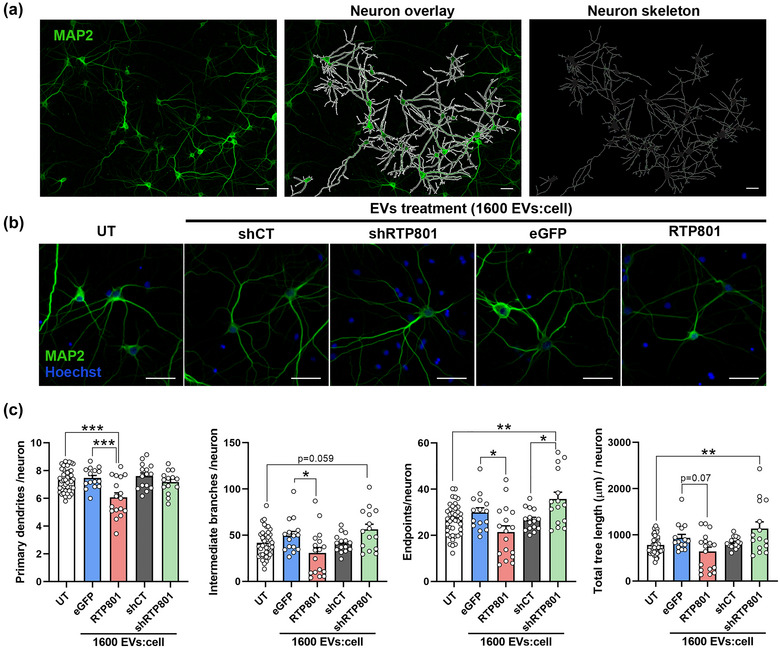
Treatment with EVs derived from RTP801 overexpressing neurons reduces neuron arborization while EVs derived from shRTP801 increase neuron morphology's complexity. (a) Neurons were untreated (UT) or treated with EVs derived from eGFP, eGFP‐RTP801, shCT or shRTP801 transduced neurons, at a dose of 1,600 EVs:cell. Immunocytochemistry of MAP2 was performed to visualize cultured neurons. Using CellProfiler software, neurons not touching the borders of the image were identified as independent objects, and neuron skeleton was obtained. (b) Representative images of neuron arborization in UT neurons and neurons treated with EVs derived from eGFP, RTP801, shCT or shRTP801 transduced neurons. (c) Morphological assessment of neurons. Primary dendrites, intermediate branches, termini branches (endpoints), and the total tree length were analysed. Scale bar of 50 μm. At least neurons of 5 different fields were analysed per replicate. Values represent culture replicates of at least three independent neuronal cultures (mean ± SEM). Data was analysed by One‐way ANOVA (*P < 0.05, **P < 0.01, ***P < 0.001).
3.4. 6‐OHDA elevates RTP801 protein levels in the EVs
Next, to test the transneuronal toxicity of endogenous RTP801 via EVs, we turned to the 6‐OHDA in vitro model, a PD neurotoxin that mediates neuronal cell death via RTP801 (Malagelada et al., 2006).
To test our hypothesis, cortical neurons at DIV14 were exposed to 50 µM 6‐OHDA for 16 h. We showed that 6‐OHDA exposure induced a reduction in cell viability in our cultures (Figure S7). Then, released EVs and cell lysates were collected for analyses. NTA analysis revealed that 6‐OHDA increased the number of extracellular particles in the treated cultures versus the non‐treated ones (∅ = 3.60 × 1010 ± 9,00 × 109 particles/mL (Figure S1E) versus 6‐OHDA = 1.52 × 1011 ± 3.88 × 1009 particles/mL (Figure S1F); P‐value = 0.0036 Student's T‐test). No variation in the vesicles size was found (145.05 ± 10.11 nm in untreated cultures vs. 134.54 ± 7.31 nm in 6‐OHDA treated cultures). By TEM, we confirmed that 6‐OHDA induced cortical neurons to release more EVs, and that these particles had the expected size and shape of exosomes and small microvesicles (Figure 5a). The EVs protein content was examined by WB analysis and showed that 6‐OHDA exposure increased the levels of the EVs markers NEDD4 (Putz et al., 2008), TSG101, and Flotilin‐1, in the EVs fraction. In this line, TSG101 and Flotillin‐1 were decreased in the neuronal lysates (Figure 5b and c). Importantly, 6‐OHDA elevated RTP801 protein levels in both cell lysates and EVs fractions (Figure 5b and c). These results indicate that 6‐OHDA increases the release of EVs and elevates the endogenous levels of RTP801 in EVs, suggesting a toxic propagation of RTP801.
FIGURE 5.
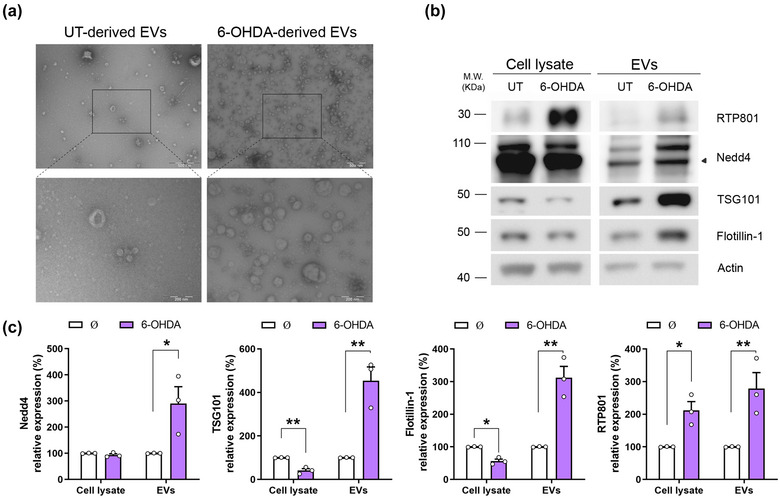
6‐OHDA exposure increases RTP801 protein levels in EVs and elevates their secretion in cortical primary cultures. (a) Rat cortical neurons at DIV14 were treated with the toxin 6‐OHDA 50 µM for 16 h or untreated (UT) as control. Afterward, cell media was collected, and EVs were isolated by sequential centrifugations. EVs suspension was negatively stained and observed by TEM. Electron micrographs show the presence of vesicles between 30 and 110 nm. Scale bars are 500 nm and 200 nm in magnification micrographs (b) Sister cultures were treated with 6‐OHDA 50 μM for 16 h or not (∅) and total protein content was analysed by WB. Membranes were probed against RTP801 and NEDD4, TSG101, and Flotillin‐1 as EVs markers and actin as a loading control. Graphs show values obtained by densitometric analysis of WB data relative to the total protein content of the cell lysate. Values represent culture replicates of at least three independent neuronal cultures (mean ± SEM). Representative immunoblots are shown. Data were analysed by Student's T‐test in the cell lysate and the EVs fraction (*P < 0.05, **P < 0.01).
3.5. 6‐OHDA‐induced EVs do not mediate pro‐survival signals in recipient cortical neurons
As the previous results demonstrated that RTP801 mediates neuron death via EVs, here we investigated whether the increased levels of endogenous RTP801 in 6‐OHDA‐derived EVs could mediate transcellular toxicity and affect intracellular signalling in recipient naïve neurons.
First, EVs were isolated from cortical neuronal cultures exposed or not to 6‐OHDA, and sister neuronal cultures were treated with these isolated EVs for 4 h, 16 h, or 24 h, at a ratio of 400 EVs:cell (Figure 6a). We observed that after 24 h, control (CT) EVs stimulated the mTOR/Akt survival pathway. We observed that CT‐EVs increased the phosphorylation of the Ser235/236 residues in RPS6, as a readout of mTORC1 activity, and the phosphorylation of Ser473 residue of Akt, as a readout of mTORC2 activity (Figure 6b and c). No changes were observed at earlier time‐points. In contrast, 6‐OHDA‐derived EVs were not able to stimulate either phosphorylation (Figure 6b and c). Importantly, 6‐OHDA‐derived EVs treatment did not compromised cell viability, supporting the notion that in the unlikely case that 6‐OHDA is co‐purified in EVs, this is not active anymore (Figure S8).
FIGURE 6.
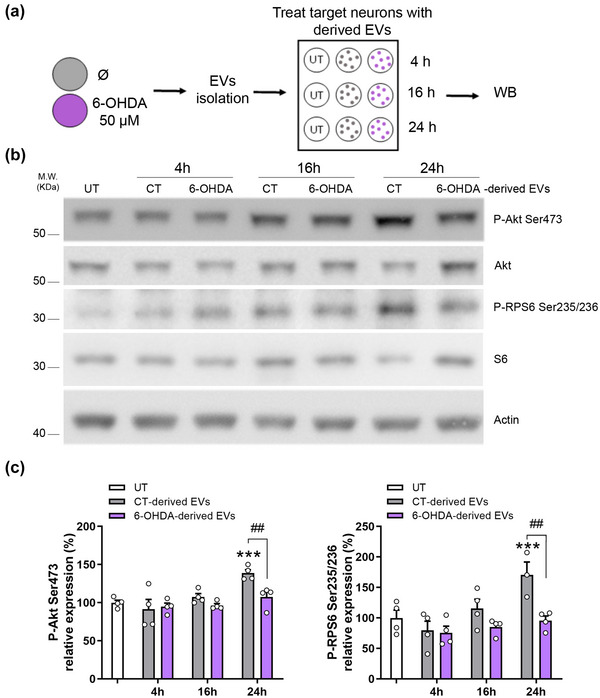
Neuron‐derived EVs activate the mTOR/Akt survival signalling via the phosphorylation of Ser473 residue of Akt and Ser235/236 residues of RPS6. (a) Schematic representation of the experimental procedure. Cultured cortical neurons were exposed to 6‐OHDA 50 µM or not (Ø) and 16 h later, EVs were isolated from the cultured medium and were administered to target cortical neurons at a dose of 400 EVs:cell. After 4, 16, or 24 h of treatment, cell extracts were collected and subjected to WB. (b) Representative immunoblots show phosho‐Ser473‐Akt, phospho‐Ser235/236‐RPS6, total Akt, total RPS6, and actin as a loading control. (c) Densitometric analysis of phospho‐Ser473‐Akt levels and phospho‐Ser235/246‐RPS6 levels. Values represent culture replicates of at least three independent neuronal cultures (mean ± SEM). Data were analysed by One‐way ANOVA with Bonferroni's multiple comparison test (***P < 0.001 vs UT and ##P < 0.01).
To prove that 6‐OHDA‐induced RTP801 mediated the loss of mTOR/Akt survival pathway activation via EVs, we first exposed or not to 6‐OHDA cultures transduced with shCT or shRTP801. Consistent with our previous work, we observed an increase in RTP801 levels in control neurons exposed to 6‐OHDA but not in neurons with a partial knockdown of RTP801 (Figure S9).
We next isolated EVs from these conditions, and from non‐transduced neuronal cultures (CT), and these vesicles from the six different conditions were used to treat naïve sister neuronal cultures. 24 h later, recipient neuron lysates were analysed by WB (Figure 7a). Like our previous results (Figure 6), EVs derived from control, shCT or shRTP801‐expressing neurons elevated levels of phospho‐Ser473‐Akt and phospho‐Ser235/236‐RPS6. However, EVs derived from neurons exposed to 6‐OHDA (CT and shCT) did not exert this activation. Strikingly, when RTP801 was silenced (shRTP801‐neurons), EVs could activate of phospho‐Ser473‐Akt and phospho‐Ser235/236‐RPS6 (Figure 7b and c). We also tested the effect over other mTOR/Akt readouts, such as phospho‐Ser371‐p70S6K and phospho‐T37/46‐4EBP‐1, but no difference was observed between conditions (Figure S10). Our findings are consistent with the trophic effect previously observed at the level of neuron arborization of EVs derived from shRTP801‐expressing neurons.
FIGURE 7.
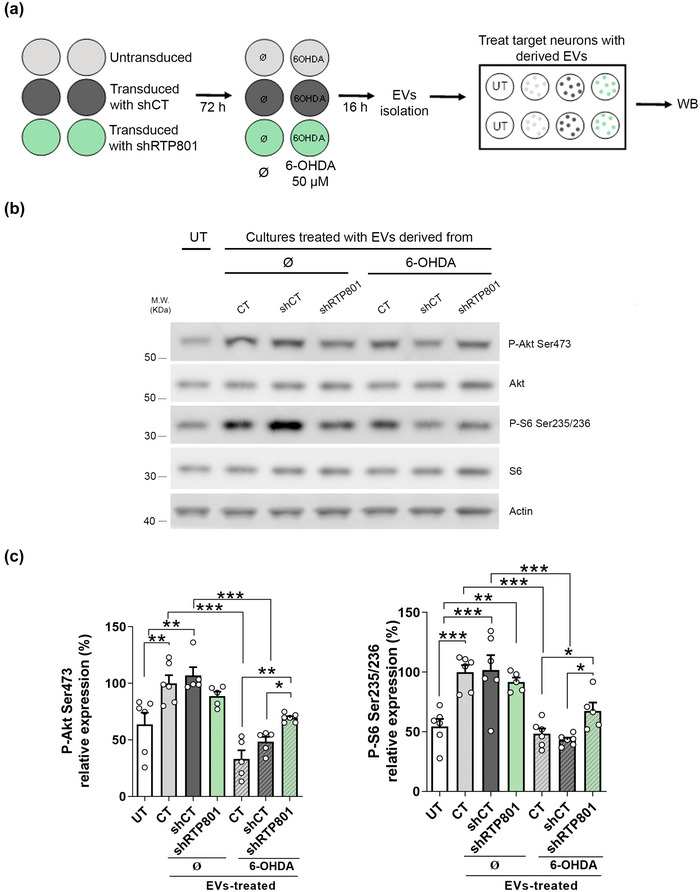
6‐OHDA‐induced RTP801 counteract EVs trophic effect in recipient neurons. (a) Rat cortical neurons at DIV10 were untransduced (CT) or transduced with shCT or shRTP801. Seventy‐hours later, cells were exposed to 6‐OHDA 50 µM or not (∅) for 16 h. After that, EVs were isolated from the culture media by sequential centrifugations and EVs were administrated to cortical cultures (400 EVs:cell), and a control condition without EVs treatment was performed as a negative control (UT). Twenty‐four hours later, cells were harvested, and protein extracts were subjected to western blotting. (b) Membranes were probed against phosho‐Ser473‐Akt, Phospho‐Ser235/236‐RPS6, total Akt, total RPS6 and actin as a loading control. Representative immunoblots are shown. (c) Densitometric analysis of phospho‐Ser473‐Akt levels and phospho‐Ser235/246‐RPS6 levels. Values represent culture replicates of at least three independent neuronal cultures (mean ± SEM). Data were analysed by One‐way ANOVA followed by Bonferroni's posthoc test. (***P < 0.005, **P < 0.01, *P < 0.05).
These results confirm that RTP801 toxicity can be transferred via neuron‐derived EVs and in pathological conditions, RTP801 elevation can specifically block EVs‐induced activation of Akt and RPS6 in recipient neurons.
4. DISCUSSION
This study describes for the first time that endogenous RTP801 toxicity can be transferred via EVs to recipient neurons. We found that endogenous RTP801 is present in EVs, and ectopic expression led to an enrichment of RTP801 in EVs in primary neurons and cell lines. Both RTP801 overexpression and silencing altered the proteomic profile of EVs. Overexpressing of RTP801 promoted the loading of apoptotic‐related proteins whereas RTP801 knockdown increased in the levels of anti‐apoptotic proteins. RTP801‐EVs were able to transfer toxicity to recipient neurons, impairing neuron arborization and inducing apoptotic death. Moreover, in cultured neurons, endogenous RTP801 was elevated in response to 6‐OHDA, and the EVs isolated from these stressed neurons lost the ability to activate the mTOR/Akt pathway via Akt and RPS6. In line with this, the knockdown of RTP801 in EV‐producing cells exposed to 6‐OHDA rescued this activation of mTOR/Akt pathway in recipient neurons.
Intercellular transfer of bioactive molecules by EVs has been widely investigated in the nervous system (Budnik et al., 2016; Frühbeis et al., 2013; Holm et al., 2018). One of the mechanisms for the progression of neurodegenerative diseases is the spreading of disease‐causing proteins from one brain area across other interconnected regions (Moreira et al., 2021). EVs have been suggested to play a role in the propagation of aggregation‐prone proteins, as several pathogenic proteins, such as β‐amyloid (Rajendran et al., 2006), Tau (Wang et al., 2017), SOD1 (Basso et al., 2013), and α‐synuclein (Emmanouilidou et al., 2010) can be released through this pathway. Our data revealed that both endogenous and ectopic RTP801 are present in EVs isolated from cultured cortical neurons.
We also observed that expression of RTP801 increased EVs release in both HEK293T cells and cultured neurons. However, the mechanism by which RTP801 could potentiate EVs release is not known yet. There is evidence that RTP801 induces an elevation of cytoplasmic calcium levels from mitochondrial reservoirs, as RTP801 is associated with ROS mitochondrial production (Shoshani et al., 2002) and endoplasmic reticulum stress (Zhang et al., 2018). Another potential mechanism is that RTP801 could affect autophagy and therefore EVs release since these two cellular events are tightly intertwined (Baixauli et al., 2014). In this line, RTP801 elevation in mice overexpressing α‐synuclein A53T and exposed to chronic restraint stress is associated with autophagy inhibition (Zhang et al., 2018).
Our proteomic results indicated that overexpression or knock‐down of RTP801 changed the protein content of neuron‐derived EVs. Increasing evidence shows that EVs’ molecular composition is highly influenced by environmental challenges. Indeed, intracellular stress can modulate the specific sorting of proteins and other components, and this, modify the biological response induced in recipient cells (Eldh et al., 2010; Frühbeis et al., 2013; Malenka & Bear, 2004). In line with these observations, and the fact that RTP801 induces neuron‐death (Canal et al., 2016; Romaní‐Aumedes et al., 2014), we found that EVs derived from neurons overexpressing RTP801 (RTP801‐EVs) contained higher levels of pro‐apoptotic proteins (such as cytochrome b‐c1, cytochrome c and alpha‐internexin) and lower levels of anti‐apoptotic proteins (such as core histone macro‐H2A, NDRG3 protein, and endophilin). When the apoptotic cascade is initiated, cytochrome c is released to the cytosol (Garrido et al., 2006; Hung et al., 2021). Other apoptotic‐related proteins, such as alpha‐internexin (Chien et al., 2005; Ching et al., 1999), phosducin‐like protein 3 (Wilkinson et al., 2004), neogenin (Chen & Shifman, 2019; Fujita et al., 2008), and transmembrane protein 2 (Ge et al., 2020), might be activated also in response to RTP801 to accelerate the cell death.
Not only apoptotic‐related proteins were found enriched, but also proteins involved in neurodegeneration such as adenylate kinase 1, which is involved in the neuropathology of AD (Park et al., 2012). Moreover, RPS6 was found in low levels in RTP801‐EVs, in line with the inhibitory role of RTP801 of the mTOR pathway (Hara et al., 2002; Kim et al., 2002; Thedieck et al., 2007).
Strikingly, in shRTP801‐EVs we found an enrichment of proteins associated with anti‐apoptotic processes (such as sodium‐coupled neutral aminoacid transporter 3, ADP/ATP traslocase 2 or pleiotropic regulator 1), highly indicating a potential trophic role of these EVs derived from RTP801 silenced neurons.
In line with our proteomic results, RTP801‐EVs treatment increased the levels of active caspase‐3 in neurons, highly suggesting that RTP801‐EVs were activating apoptosis in recipient cells. We also observed a visible effect on late apoptosis, since a higher number of neurons presented fragmented nuclei with no cleaved caspase‐3. In this line, Takemoto et al. (2003) showed that caspase‐3 inactivation preceded nuclear fragmentation (Takemoto et al., 2003). Moreover, the toxic effect of RTP801‐EVs treatment was also observed by the loss of MAP2+ cells, and the increase in apoptotic total nuclei. Hence our observations support that RTP801‐induced protein cargo in EVs trigger apoptotic neuron death in a dose‐dependent manner, but we cannot discard that, other molecules, such as lipids or RNAs sensitive to RTP801 levels, could contribute to this effect as well.
Neuron‐derived EVs have been shown to provide trophic support to neurons under nutrient deprivation conditions, preventing neuron complexity loss (Solana‐Balaguer et al., 2023). Here we showed that RTP801‐EVs significantly impaired dendritic arborization in recipient neurons, further confirming RTP801 toxicity role via EVs and supporting the apoptotic role of RTP801‐EVs. Dendritic branching defects in mature neurons have devastating consequences on neuronal function and survival and are common features in patients suffering from neurodegenerative diseases (Kweon et al., 2017). The fact that increased levels of RTP801 are found in PD, AD and HD human postmortem brains, and contribute to neuron impairment in disease mouse models (Pérez‐Sisqués, Sancho‐Balsells et al., 2021; Pérez‐Sisqués, Solana‐Balaguer et al., 2021), suggests that RTP801 could be an active player in spreading neurodegeneration via EVs and therefore, a potential future target in the treatment of neurodegeneration.
Importantly, EVs derived from neurons with silenced RTP801 (shRTP801‐EVs) led to an increased dendritic arborization complexity in recipient neurons. This suggests that RTP801 is negatively modulating neuron branching in mature neurons. RTP801 is a negative regulator of mTORC1 (Tan & Hagen, 2013), and Nakai et al. (2020) described that the activation of mTORC1 induced neurite outgrowth and branching in cultured cortical and hippocampal neurons, while the inhibition abolished these effects (Nakai et al., 2020). In line with our results, Ota et al. (2014) reported an atrophy of primary apical dendrites in neurons overexpressing RTP801 present at cortical layer V, measured by a decrease in spine density (Ota et al., 2014). Furthermore, other upregulated proteins in shRTP801‐EVs could be contributing to the trophic support. For instance, SLC28A levels are upregulated by BDNF and contribute to dendritic growth and cortical neuron branching (Burkhalter et al., 2007). In addition, sirtuin‐2 delivery to neurons via exosomes has been shown to enhance mitochondria ATP production (Chamberlain et al., 2021), which could explain the increase in morphology complexity. Furthermore, nucleolin is involved in subcellular mRNA localization, and therefore its function is crucial to promote neuron growth (Perry et al., 2016).
Our previous work showed that RTP801 protein is involved in PD pathogenesis (Canal et al., 2016; Romaní‐Aumedes et al., 2014). Here, we also found that RTP801 was enriched in EVs derived from cortical neurons treated with the neurotoxin 6‐OHDA. Hence, our results suggest that oxidative stress induced by 6‐OHDA can contribute to EVs release potentiation, exacerbating neurodegeneration in PD. Importantly, although 6‐OHDA treatment caused a reduction in cell viability, 6‐OHDA‐derived EVs did not induce cell death, which indicates that we are not co‐isolating 6‐OHDA along with the EVs.
As abovementioned, EVs not only act as vehicles to transfer toxic proteins but also can exert a neuroprotective function (Harrison et al., 2016; Jarmalavičiūtė et al., 2015). Our findings indicate that neural EVs play a trophic role by promoting specifically the phosphorylation of Ser235/236 residues of RPS6 and the Ser473 residue of Akt, readouts of the mTORC 1 and 2 kinase activities, respectively. Neuron‐derived EVs have been shown to modulate synaptic plasticity activating these effectors via TrkB‐mediated signalling (Solana‐Balaguer et al., 2023). Moreover, oligodendroglia derived EVs induced neuroprotection by the phosphorylation of Akt and ERK under oxidative stress or starvation (Fröhlich et al., 2014). Furthermore, Tassew et al. (2017) reported that fibroblasts derived EVs were able to activate mTOR in cortical neurons by Wnt recruiting (Tassew et al., 2017). Interestingly, we observed that 6‐OHDA‐induced EVs were not able to promote mTOR/Akt pro‐survival signals in recipient neurons. On the contrary, when RTP801 expression was suppressed, the derived EVs (shRTP801‐EVs) could partially restore the phosphorylation of both Akt and RPS6. Hence, these data suggest that 6‐OHDA‐induced RTP801 impairs this EVs‐induced signalling cascade transneuronally. Therefore, our results indicate that RTP801 interferes with the ability of EVs to promote the signalling and the trophic support to recipient neurons.
4.1. Limitations of the study
Our cultured cortical neurons were seeded without antimitotics, to avoid neuron toxicity (Shi & Mitchison, 2017). In this line, we demonstrated in our previous study that the presence of astrocytes in our cultures is less than 10% (Solana‐Balaguer et al., 2023). However, we cannot discard the presence of some astrocyte‐derived EVs, and their minor effect into the total bulk of neuron‐derived EVs. Moreover, although our study indicates that RTP801 toxicity is transferred via EVs, it is difficult to isolate whether the detrimental effects are solely caused by RTP801 or by the combination of the pro‐apoptotic proteins loaded in the EVs. Our proteomics results suggest that is the combination of both situations. Furthermore, we faced the limitation of RTP801 detection due to its rapid turnover (2‐5 protein half‐life (Kimball et al., 2008; Malagelada et al., 2010)) and due to EVs also get integrated into the endolysosomal system after 24 h. Thus, further studies will be required to understand RTP801 toxicity transmission via EVs, and the likely role of glia in this intricate regulation.
5. CONCLUSIONS
Our results indicate that the modulation of RTP801 expression alters the protein composition of neuron‐derived EVs. Furthermore, high levels of RTP801 in EVs exert a toxic effect on recipient neurons triggering apoptosis and branching alterations. In contrast, silencing RTP801 in EVs may have a trophic effect on neurite complexity. Moreover, we showed that under pathological conditions, RTP801‐enriched EVs lost the mTOR/Akt signalling activation. Altogether, our results highly suggest that RTP801, along with other RTP801‐induced apoptotic proteins in EVs contribute to neural cell death and neurodegeneration in vitro.
AUTHOR CONTRIBUTIONS
Júlia Solana‐Balaguer: Conceptualization; data curation; formal analysis; investigation; methodology; resources; Software; writing—original draft; writing—review and editing.Núria Martín‐Flores: Conceptualization; data curation; formal analysis; investigation; methodology; resources; Software; writing—review and editing. Pol Garcia‐Segura: Conceptualization; data curation; formal analysis; investigation; resources; software; writing—review and editing. Genís Campoy‐Campos: Formal analysis; investigation; methodology; resources; writing—review and editing. Leticia Pérez‐Sisqués: Data curation; formal analysis; investigation; writing—review and editing. Almudena Chicote‐González: Conceptualization; formal analysis; investigation; methodology; writing—review and editing. Joaquín Fernández‐Irigoyen: Formal analysis; investigation; methodology; writing—review and editing. Enrique Santamaría: Formal analysis; investigation; methodology; resources; writing—review and editing. Esther Pérez‐Navarro: Formal analysis; investigation; resources; writing—review and editing. Jordi Alberch: Formal analysis; investigation; resources; writing—review and editing. Cristina Malagelada: Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; supervision; writing—review and editing
CONFLICT OF INTEREST STATEMENT
All authors declare to have no competing interests.
Supporting information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
ACKNOWLEDGEMENTS
We are grateful to M. Calvo and J.M. Rebled (Centres Científics i Tecnològics, Universitat de Barcelona) for help in confocal and electronic microscopy, respectively. We thank Dr. Hernando del Portillo, Joan Seguí (Institut d'Investigació Germans Trias i Pujol, IGTP) and Amable Bernabé (Instituto de Ciencia de Materiales de Barcelona, ICMAB) for technical assistance from the NanoSight measurements. We thank Dr. J. Romaní for technical assistance. We thank Dr. Miguel Segura (Vall d'Hebron Research Institute, VHIR) for technical advice and helpful discussion. Supported in part by grants from Ministerio de Ciencia e Innovación /AEI/10.13039/501100011033/ and ‘FEDER’: SAF2017‐88812‐R and PID2020‐119236RB‐I00, from CM; PID2020‐119386RB‐I00 from JA and PID2019‐106447RB‐I00 from EP‐N. JS‐B was supported by a FPU grant from the Spanish Ministry of Science and Innovation (grant #FPU18/00194). NM‐F was supported by a FPI grant from the Spanish Ministry of Science and Innovation (grant IBES‐2015‐072727). PG‐S was supported by a FPU grant from the Spanish Ministry of Science and Innovation (grant #FPU21/02928). GC‐C was funded by a FI‐2021 grant from the Agència de Gestió d'Ajuts Universitaris i de Recerca (AGAUR) (grant #FI‐B‐00378). AC‐G was supported by a Michael J. Fox Foundation grant (MJFF‐000858). Supported also by the crowdfunding campaign ‘SOS recerca en Parkinson’ via Goteo.org and Portal d'Avall, S.L.
Solana‐Balaguer, J. , Martín‐Flores, N. , Garcia‐Segura, P. , Campoy‐Campos, G. , Pérez‐Sisqués, L. , Chicote‐González, A. , Fernández‐Irigoyen, J. , Santamaría, E. , Pérez‐Navarro, E. , Alberch, J. , & Malagelada, C. (2023). RTP801 mediates transneuronal toxicity in culture via extracellular vesicles. Journal of Extracellular Vesicles, 12, e12378. 10.1002/jev2.12378
Present addresses
Núria Martín‐Flores, Cell and Developmental Biology, University College London, London, UK.
Leticia Pérez‐Sisqués, Centre for Craniofacial and Regenerative Biology, King's College, London, UK.
Júlia Solana‐Balaguer and Núria Martín‐Flores contributed equally to this work.
Contributor Information
Júlia Solana‐Balaguer, Email: juliasolana@ub.edu.
Núria Martín‐Flores, Email: n.flores@ucl.ac.uk.
Cristina Malagelada, Email: cristina.malagelada@ub.edu.
DATA AVAILABILITY STATEMENT
Mass‐spectrometry data and search results files were deposited in the Proteome Xchange Consortium via the JPOST partner repository (https://repository.jpostdb.org) (Okuda S, Watanabe Y, Moriya Y, Kawano S, Yamamoto T, Matsumoto M, Takami T, Kobayashi D, Araki N, Yoshizawa AC et al.: jPOSTrepo: an international standard data repository for proteomes. Nucleic Acids Res 2017, 45(D1):D1107‐D1111) with the identifier PXD040082 for ProteomeXchange and JPST002037 for jPOST (for reviewers: https://repository.jpostdb.org/preview/170310131463ea007f81c18; Access key: 3507).
REFERENCES
- Abu‐Hamad, S. , Zaid, H. , Israelson, A. , Nahon, E. , & Shoshan‐Barmatz, V. (2008). Hexokinase‐I protection against apoptotic cell death is mediated via interaction with the voltage‐dependent anion channel‐1: Mapping the site of binding. The Journal of Biological Chemistry, 283(19), 13482–13490. 10.1074/jbc.M708216200 [DOI] [PubMed] [Google Scholar]
- Andrés‐Benito, P. , Gelpi, E. , Povedano, M. , Ausín, K. , Fernández‐Irigoyen, J. , Santamaría, E. , & Ferrer, I. (2019). Combined transcriptomics and proteomics in frontal cortex area 8 in frontotemporal lobar degeneration linked to C9ORF72 expansion. Journal of Alzheimer's Disease: JAD, 68(3), 1287–1307. 10.3233/JAD-181123 [DOI] [PubMed] [Google Scholar]
- Anees, A. , Salapa, H. E. , Thibault, P. A. , Hutchinson, C. , Hammond, S. A. , & Levin, M. C. (2021). Knock‐down of heterogeneous nuclear ribonucleoprotein A1 results in neurite damage, altered stress granule biology, and cellular toxicity in differentiated neuronal cells. eNeuro, 8(6), ENEURO.0350–ENEU21.2021. 10.1523/ENEURO.0350-21.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya, P. , Rainey, M. A. , Bhattacharyya, S. , Mohapatra, B. C. , George, M. , Kuracha, M. R. , Storck, M. D. , Band, V. , Govindarajan, V. , & Band, H. (2015). The endocytic recycling regulatory protein EHD1 Is required for ocular lens development. Developmental Biology, 408(1), 41–55. 10.1016/j.ydbio.2015.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baixauli, F. , López‐Otín, C. , & Mittelbrunn, M. (2014). Exosomes and autophagy: Coordinated mechanisms for the maintenance of cellular fitness. Frontiers in Immunology, 5, 403. 10.3389/fimmu.2014.00403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso, M. , Pozzi, S. , Tortarolo, M. , Fiordaliso, F. , Bisighini, C. , Pasetto, L. , Spaltro, G. , Lidonnici, D. , Gensano, F. , Battaglia, E. , Bendotti, C. , & Bonetto, V. (2013). Mutant copper‐zinc superoxide dismutase (SOD1) induces protein secretion pathway alterations and exosome release in astrocytes: Implications for disease spreading and motor neuron pathology in amyotrophic lateral sclerosis. The Journal of Biological Chemistry, 288(22), 15699–15711. 10.1074/jbc.M112.425066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellingham, S. A. , Guo, B. B. , Coleman, B. M. , & Hill, A. F. (2012). Exosomes: Vehicles for the transfer of toxic proteins associated with neurodegenerative diseases? Frontiers in Physiology, 3, 124. 10.3389/fphys.2012.00124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellone, M. L. , Fiengo, L. , Cerchia, C. , Cotugno, R. , Bader, A. , Lavecchia, A. , De Tommasi, N. , & Piaz, F. D. (2022). Impairment of nucleolin activity and phosphorylation by a trachylobane diterpene from Psiadia punctulata in cancer cells. International Journal of Molecular Sciences, 23(19), 11390. 10.3390/ijms231911390/S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua, E. , Wang, X. , Majumder, M. , Gaccioli, F. , Yuan, C. L. , Wang, C. , Zhu, X. , Jordan, L. E. , Scheuner, D. , Kaufman, R. J. , Koromilas, A. E. , Snider, M. D. , Holcik, M. , & Hatzoglou, M. (2010). eIF2alpha phosphorylation tips the balance to apoptosis during osmotic stress. The Journal of Biological Chemistry, 285(22), 17098–17111. 10.1074/jbc.M110.109439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak, H. , Ghebremedhin, E. , Rüb, U. , Bratzke, H. , & Del Tredici, K. (2004). Stages in the development of Parkinson's disease‐related pathology. Cell and Tissue Research, 318(1), 121–134. 10.1007/s00441-004-0956-9 [DOI] [PubMed] [Google Scholar]
- Brahimi‐Horn, M. C. , Ben‐Hail, D. , Ilie, M. , Gounon, P. , Rouleau, M. , Hofman, V. , Doyen, J. , Mari, B. , Shoshan‐Barmatz, V. , Hofman, P. , Pouysségur, J. , & Mazure, N. M. (2012). Expression of a truncated active form of VDAC1 in lung cancer associates with hypoxic cell survival and correlates with progression to chemotherapy resistance. Cancer Research, 72(8), 2140–2150. 10.1158/0008-5472.CAN-11-3940/650241/AM/EXPRESSION-OF-A-TRUNCATED-ACTIVE-FORM-OF-VDAC1-IN [DOI] [PubMed] [Google Scholar]
- Britton, M. , Lucas, M. M. , Downey, S. L. , Screen, M. , Pletnev, A. A. , Verdoes, M. , Tokhunts, R. A. , Amir, O. , Goddard, A. L. , Pelphrey, P. M. , Wright, D. L. , Overkleeft, H. S. , & Kisselev, A. F. (2009). Selective inhibitor of proteasome's caspase‐like sites sensitizes cells to specific inhibition of chymotrypsin‐like sites. Chemistry & Biology, 16(12), 1278–1289. 10.1016/j.chembiol.2009.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugarolas, J. , Lei, K. , Hurley, R. L. , Manning, B. D. , Reiling, J. H. , Hafen, E. , Witters, L. A. , Ellisen, L. W. , & Kaelin, W. G., Jr (2004). Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes & Development, 18(23), 2893–2904. 10.1101/gad.1256804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budnik, V. , Ruiz‐Cañada, C. , & Wendler, F. (2016). Extracellular vesicles round off communication in the nervous system. Nature Reviews. Neuroscience, 17(3), 160–172. 10.1038/nrn.2015.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhalter, J. , Fiumelli, H. , Erickson, J. D. , & Martin, J. L. (2007). A critical role for system A amino acid transport in the regulation of dendritic development by brain‐derived neurotrophic factor (BDNF). The Journal of Biological Chemistry, 282(8), 5152–5159. 10.1074/jbc.M608548200 [DOI] [PubMed] [Google Scholar]
- Cammas, A. , Pileur, F. , Bonnal, S. , Lewis, S. M. , Lévêque, N. , Holcik, M. , & Vagner, S. (2007). Cytoplasmic relocalization of heterogeneous nuclear ribonucleoprotein A1 controls translation initiation of specific mRNAs. Molecular Biology of the Cell, 18(12), 5048–5059. 10.1091/mbc.e07-06-0603/SUPPL_FILE/SUPPLEMENTALFIGSTABLLES.PDF [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canal, M. , Martín‐Flores, N. , Pérez‐Sisqués, L. , Romaní‐Aumedes, J. , Altas, B. , Man, H. Y. , Kawabe, H. , Alberch, J. , & Malagelada, C. (2016). Loss of NEDD4 contributes to RTP801 elevation and neuron toxicity: Implications for Parkinson's disease. Oncotarget, 7(37), 58813–58831. 10.18632/oncotarget.11020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canal, M. , Romaní‐Aumedes, J. , Martín‐Flores, N. , Pérez‐Fernández, V. , & Malagelada, C. (2014). RTP801/REDD1: A stress coping regulator that turns into a troublemaker in neurodegenerative disorders. Frontiers in Cellular Neuroscience, 8, 313. 10.3389/fncel.2014.00313/BIBTEX [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter, A. E. , Jones, T. R. , Lamprecht, M. R. , Clarke, C. , Kang, I. H. , Friman, O. , Guertin, D. A. , Chang, J. H. , Lindquist, R. A. , Moffat, J. , Golland, P. , & Sabatini, D. M. (2006). CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biology, 7(10), 1–11. 10.1186/gb-2006-7-10-r100/FIGURES/4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain, K. A. , Huang, N. , Xie, Y. , LiCausi, F. , Li, S. , Li, Y. , & Sheng, Z. H. (2021). Oligodendrocytes enhance axonal energy metabolism by deacetylation of mitochondrial proteins through transcellular delivery of SIRT2. Neuron, 109(21), 3456–3472.e8. 10.1016/j.neuron.2021.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, K. , Busque, S. M. , Sailer, M. , Stoeger, C. , Bröer, S. , Daniel, H. , Rubio‐Aliaga, I. , & Wagner, C. A. (2016). Loss of function mutation of the Slc38a3 glutamine transporter reveals its critical role for amino acid metabolism in the liver, brain, and kidney. Pflugers Archiv: European Journal of Physiology, 468(2), 213–227. 10.1007/s00424-015-1742-0/METRICS [DOI] [PubMed] [Google Scholar]
- Chen, J. , & Shifman, M. I. (2019). Inhibition of neogenin promotes neuronal survival and improved behavior recovery after spinal cord injury. Neuroscience, 408, 430–447. 10.1016/j.neuroscience.2019.03.055 [DOI] [PubMed] [Google Scholar]
- Chen, S. , Zhang, Y. , Wang, H. , Zeng, Y. Y. , Li, Z. , Li, M. L. , Li, F. F. , You, J. , Zhang, Z. M. , & Tzeng, C. M. (2018). WW domain‐binding protein 2 acts as an oncogene by modulating the activity of the glycolytic enzyme ENO1 in glioma. Cell Death & Disease, 9(3), 347. 10.1038/s41419-018-0376-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T. , Dai, X. , Dai, J. , Ding, C. , Zhang, Z. , Lin, Z. , Hu, J. , Lu, M. , Wang, Z. , Qi, Y. , Zhang, L. , Pan, R. , Zhao, Z. , Lu, L. , Liao, W. , & Lu, X. (2020). AFP promotes HCC progression by suppressing the HuR‐mediated Fas/FADD apoptotic pathway. Cell Death & Disease, 11(10), 1–15. 10.1038/s41419-020-03030-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, S. , Jiang, X. , Ding, C. , Du, C. , Owusu‐Ansah, K. G. , Weng, X. , Hu, W. , Peng, C. , Lv, Z. , Tong, R. , Xiao, H. , Xie, H. , Zhou, L. , Wu, J. , & Zheng, S. (2016). Expression and critical role of interleukin enhancer binding factor 2 in hepatocellular carcinoma. International Journal of Molecular Sciences, 17(8), 1373. 10.3390/ijms17081373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevrollier, A. , Loiseau, D. , Reynier, P. , & Stepien, G. (2011). Adenine nucleotide translocase 2 is a key mitochondrial protein in cancer metabolism. Biochimica et Biophysica Acta, 1807(6), 562–567. 10.1016/j.bbabio.2010.10.008 [DOI] [PubMed] [Google Scholar]
- Chien, C. L. , Liu, T. C. , Ho, C. L. , & Lu, K. S. (2005). Overexpression of neuronal intermediate filament protein alpha‐internexin in PC12 cells. Journal of Neuroscience Research, 80(5), 693–706. 10.1002/jnr.20506 [DOI] [PubMed] [Google Scholar]
- Ching, G. Y. , Chien, C. L. , Flores, R. , & Liem, R. K. (1999). Overexpression of alpha‐internexin causes abnormal neurofilamentous accumulations and motor coordination deficits in transgenic mice. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 19(8), 2974–2986. 10.1523/JNEUROSCI.19-08-02974.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivet, M. , Javalet, C. , Laulagnier, K. , Blot, B. , Hemming, F. J. , & Sadoul, R. (2014). Exosomes secreted by cortical neurons upon glutamatergic synapse activation specifically interact with neurons. Journal of Extracellular Vesicles, 3, 24722. 10.3402/jev.v3.24722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuo, S. T. , Chien, J. C. , & Lai, C. P. (2018). Imaging extracellular vesicles: Current and emerging methods. Journal of Biomedical Science, 25(1), 1–10. 10.1186/s12929-018-0494-5/FIGURES/3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, J. , & Mann, M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.‐range mass accuracies and proteome‐wide protein quantification. Nature Biotechnology, 26(12), 1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Cox, J. , Neuhauser, N. , Michalski, A. , Scheltema, R. A. , Olsen, J. V. , & Mann, M. (2011). Andromeda: A peptide search engine integrated into the MaxQuant environment. Journal of Proteome Research, 10(4), 1794–1805. 10.1021/pr101065j_SI_002.ZIP [DOI] [PubMed] [Google Scholar]
- Damjanac, M. , Page, G. , Ragot, S. , Laborie, G. , Gil, R. , Hugon, J. , & Paccalin, M. (2009). PKR, a cognitive decline biomarker, can regulate translation via two consecutive molecular targets p53 and Redd1 in lymphocytes of AD patients. Journal of Cellular and Molecular Medicine, 13(8B), 1823–1832. 10.1111/j.1582-4934.2009.00688.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danzer, K. M. , Kranich, L. R. , Ruf, W. P. , Cagsal‐Getkin, O. , Winslow, A. R. , Zhu, L. , Vanderburg, C. R. , & McLean, P. J. (2012). Exosomal cell‐to‐cell transmission of alpha synuclein oligomers. Molecular Neurodegeneration, 7, 42. 10.1186/1750-1326-7-42/FIGURES/7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhumale, P. , Menon, S. , Chiang, J. , & Püschel, A. W. (2018). The loss of the kinases SadA and SadB results in early neuronal apoptosis and a reduced number of progenitors. PLoS One, 13(4), e0196698. 10.1371/journal.pone.0196698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, T. , & Hao, J. (2021). Sirtuin 2 knockdown inhibits cell proliferation and RAS/ERK signaling, and promotes cell apoptosis and cell cycle arrest in multiple myeloma. Molecular Medicine Reports, 24(5), 760. 10.3892/mmr.2021.12400/HTML [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudich, E. , Semenkova, L. , Dudich, I. , Gorbatova, E. , Tochtamisheva, N. , Tatulov, E. , Nikolaeva, M. , & Sukhikh, G. (1999). Alpha‐fetoprotein causes apoptosis in tumor cells via a pathway independent of CD95, TNFR1 and TNFR2 through activation of caspase‐3‐like proteases. European Journal of Biochemistry, 266(3), 750–761. 10.1046/j.1432-1327.1999.00868.x [DOI] [PubMed] [Google Scholar]
- Eldh, M. , Ekström, K. , Valadi, H. , Sjöstrand, M. , Olsson, B. , Jernås, M. , & Lötvall, J. (2010). Exosomes communicate protective messages during oxidative stress; possible role of exosomal shuttle RNA. PLoS ONE, 5(12), 1–8. 10.1371/journal.pone.0015353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias, J. E. , & Gygi, S. P. (2010). Target‐decoy search strategy for mass spectrometry‐based proteomics. Methods in Molecular Biology (Clifton, N.J.), 604, 55–71. 10.1007/978-1-60761-444-9_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmanouilidou, E. , Melachroinou, K. , Roumeliotis, T. , Garbis, S. D. , Ntzouni, M. , Margaritis, L. H. , Stefanis, L. , & Vekrellis, K. (2010). Cell‐produced alpha‐synuclein is secreted in a calcium‐dependent manner by exosomes and impacts neuronal survival. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 30(20), 6838–6851. 10.1523/JNEUROSCI.5699-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdbrügger, U. , & Lannigan, J. (2016). Analytical challenges of extracellular vesicle detection: A comparison of different techniques. Cytometry. Part A: The Journal of the International Society for Analytical Cytology, 89(2), 123–134. 10.1002/cyto.a.22795 [DOI] [PubMed] [Google Scholar]
- Feng, Y. , Chen, D. , Wang, T. , Zhou, J. , Xu, W. , Xiong, H. , Bai, R. , Wu, S. , Li, J. , & Li, F. (2022). Sertoli cell survival and barrier function are regulated by miR‐181c/d‐Pafah1b1 axis during mammalian spermatogenesis. Cellular and Molecular Life Sciences: CMLS, 79(9), 1–20. 10.1007/s00018-022-04521-w/FIGURES/6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Y. , Zhang, C. , Luo, Q. , Wei, X. , Jiang, B. , Zhu, H. , Zhang, L. , Jiang, L. , Liu, M. , & Xiao, X. (2012). A novel WD‐repeat protein, WDR26, inhibits apoptosis of cardiomyocytes induced by oxidative stress. Free Radical Research, 46(6), 777–784. 10.3109/10715762.2012.678840/SUPPL_FILE/IFRA_A_678840_SM0001.PDF [DOI] [PubMed] [Google Scholar]
- Fröhlich, D. , Kuo, W. P. , Frühbeis, C. , Sun, J. J. , Zehendner, C. M. , Luhmann, H. J. , Pinto, S. , Toedling, J. , Trotter, J. , & Krämer‐Albers, E. M. (2014). Multifaceted effects of oligodendroglial exosomes on neurons: Impact on neuronal firing rate, signal transduction and gene regulation. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 369(1652), 20130510. 10.1098/rstb.2013.0510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frühbeis, C. , Fröhlich, D. , Kuo, W. P. , Amphornrat, J. , Thilemann, S. , Saab, A. S. , Kirchhoff, F. , Möbius, W. , Goebbels, S. , Nave, K. A. , Schneider, A. , Simons, M. , Klugmann, M. , Trotter, J. , & Krämer‐Albers, E. M. (2013). Neurotransmitter‐triggered transfer of exosomes mediates oligodendrocyte‐neuron communication. PLoS Biology, 11(7), e1001604. 10.1371/journal.pbio.1001604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frühbeis, C. , Fröhlich, D. , Kuo, W. P. , & Krämer‐Albers, E. M. (2013). Extracellular vesicles as mediators of neuron‐glia communication. Frontiers in Cellular Neuroscience, 7, 182. 10.3389/fncel.2013.00182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, Y. , Taniguchi, J. , Uchikawa, M. , Endo, M. , Hata, K. , Kubo, T. , Mueller, B. K. , & Yamashita, T. (2008). Neogenin regulates neuronal survival through DAP kinase. Cell Death and Differentiation, 15(10), 1593–1608. 10.1038/cdd.2008.92 [DOI] [PubMed] [Google Scholar]
- Gao, J. , Dai, C. , Yu, X. , Yin, X. B. , & Zhou, F. (2020). microRNA‐485‐5p inhibits the progression of hepatocellular carcinoma through blocking the WBP2/Wnt signaling pathway. Cellular Signalling, 66, 109466. 10.1016/j.cellsig.2019.109466 [DOI] [PubMed] [Google Scholar]
- Garrido, C. , Galluzzi, L. , Brunet, M. , Puig, P. E. , Didelot, C. , & Kroemer, G. (2006). Mechanisms of cytochrome c release from mitochondria. Cell Death and Differentiation, 13(9), 1423–1433. 10.1038/sj.cdd.4401950 [DOI] [PubMed] [Google Scholar]
- Ge, X. , Jiang, W. , Jiang, Y. , Lv, X. , Liu, X. , & Wang, X. (2020). Expression and importance of TMED2 in multiple myeloma cells. Cancer Management and Research, 12, 12895–12903. 10.2147/CMAR.S278570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, X. , Jiang, W. , Jiang, Y. , Lv, X. , Liu, X. , & Wang, X. (2020). Expression and importance of TMED2 in multiple myeloma cells. Cancer Management and Research, 12, 12895–12903. 10.2147/CMAR.S278570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, T. , Pandey, N. , Maitra, A. , Brahmachari, S. K. , & Pillai, B. (2007). A role for voltage‐dependent anion channel Vdac1 in polyglutamine‐mediated neuronal cell death. PLoS ONE, 2(11), e1170. 10.1371/journal.pone.0001170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godbole, A. , Varghese, J. , Sarin, A. , & Mathew, M. K. (2003). VDAC is a conserved element of death pathways in plant and animal systems. Biochimica et Biophysica Acta, 1642(1–2), 87–96. 10.1016/s0167-4889(03)00102-2 [DOI] [PubMed] [Google Scholar]
- Gu, Z. , Eils, R. , & Schlesner, M. (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics (Oxford, England), 32(18), 2847–2849. 10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- Guo, J. L. , & Lee, V. M. (2014). Cell‐to‐cell transmission of pathogenic proteins in neurodegenerative diseases. Nature Medicine, 20(2), 130–138. 10.1038/nm.3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara, K. , Maruki, Y. , Long, X. , Yoshino, K. , Oshiro, N. , Hidayat, S. , Tokunaga, C. , Avruch, J. , & Yonezawa, K. (2002). Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell, 110(2), 177–189. 10.1016/s0092-8674(02)00833-4 [DOI] [PubMed] [Google Scholar]
- Harrison, E. B. , Hochfelder, C. G. , Lamberty, B. G. , Meays, B. M. , Morsey, B. M. , Kelso, M. L. , Fox, H. S. , & Yelamanchili, S. V. (2016). Traumatic brain injury increases levels of miR‐21 in extracellular vesicles: Implications for neuroinflammation. FEBS Open Bio, 6(8), 835–846. 10.1002/2211-5463.12092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, M. M. , Adrain, C. , Duriez, P. J. , Creagh, E. M. , & Martin, S. J. (2004). Analysis of the composition, assembly kinetics and activity of native Apaf‐1 apoptosomes. The EMBO Journal, 23(10), 2134–2145. 10.1038/sj.emboj.7600210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, G. , Breitenbücher, F. , Schuler, M. , & Ehrenhofer‐Murray, A. E. (2014). A novel sirtuin 2 (SIRT2) inhibitor with p53‐dependent pro‐apoptotic activity in non‐small cell lung cancer. The Journal of Biological Chemistry, 289(8), 5208–5216. 10.1074/jbc.M113.487736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, M. M. , Kaiser, J. , & Schwab, M. E. (2018). Extracellular vesicles: Multimodal envoys in neural maintenance and repair. Trends in Neurosciences, 41(6), 360–372. 10.1016/j.tins.2018.03.006 [DOI] [PubMed] [Google Scholar]
- Honda, A. , Abe, R. , Yoshihisa, Y. , Makino, T. , Matsunaga, K. , Nishihira, J. , Shimizu, H. , & Shimizu, T. (2009). Deficient deletion of apoptotic cells by macrophage migration inhibitory factor (MIF) overexpression accelerates photocarcinogenesis. Carcinogenesis, 30(9), 1597–1605. 10.1093/carcin/bgp160 [DOI] [PubMed] [Google Scholar]
- Huang, Y. , Huang, S. , Ma, L. , Wang, Y. , Wang, X. , Xiao, L. , Qin, W. , Li, L. , & Yuan, X. (2021). Exploring the prognostic value, immune implication and biological function of H2AFY Gene in hepatocellular carcinoma. Frontiers in Immunology, 12, 723293. 10.3389/fimmu.2021.723293/FULL [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung, Y. C. , Huang, K. L. , Chen, P. L. , Li, J. L. , Lu, S. H. , Chang, J. C. , Lin, H. Y. , Lo, W. C. , Huang, S. Y. , Lee, T. T. , Lin, T. Y. , Imai, Y. , Hattori, N. , Liu, C. S. , Tsai, S. Y. , Chen, C. H. , Lin, C. H. , & Chan, C. C. (2021). UQCRC1 engages cytochrome c for neuronal apoptotic cell death. Cell Reports, 36(12), 109729. 10.1016/j.celrep.2021.109729 [DOI] [PubMed] [Google Scholar]
- Hung, Y. C. , Huang, K. L. , Chen, P. L. , Li, J. L. , Lu, S. H. , Chang, J. C. , Lin, H. Y. , Lo, W. C. , Huang, S. Y. , Lee, T. T. , Lin, T. Y. , Imai, Y. , Hattori, N. , Liu, C. S. , Tsai, S. Y. , Chen, C. H. , Lin, C. H. , & Chan, C. C. (2021). UQCRC1 engages cytochrome c for neuronal apoptotic cell death. Cell Reports, 36(12), 109729. 10.1016/j.celrep.2021.109729 [DOI] [PubMed] [Google Scholar]
- Jacinto, E. , Facchinetti, V. , Liu, D. , Soto, N. , Wei, S. , Jung, S. Y. , Huang, Q. , Qin, J. , & Su, B. (2006). SIN1/MIP1 maintains rictor‐mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell, 127(1), 125–137. 10.1016/j.cell.2006.08.033 [DOI] [PubMed] [Google Scholar]
- Jarmalavičiūtė, A. , Tunaitis, V. , Pivoraitė, U. , Venalis, A. , & Pivoriūnas, A. (2015). Exosomes from dental pulp stem cells rescue human dopaminergic neurons from 6‐hydroxy‐dopamine‐induced apoptosis. Cytotherapy, 17(7), 932–939. 10.1016/j.jcyt.2014.07.013 [DOI] [PubMed] [Google Scholar]
- Jiang, X. , Kim, H. E. , Shu, H. , Zhao, Y. , Zhang, H. , Kofron, J. , Donnelly, J. , Burns, D. , Ng, S. C. , Rosenberg, S. , & Wang, X. (2003). Distinctive roles of PHAP proteins and prothymosin‐alpha in a death regulatory pathway. Science (New York, N.Y.), 299(5604), 223–226. 10.1126/science.1076807/SUPPL_FILE/JIANG.SOM.PDF [DOI] [PubMed] [Google Scholar]
- Jo, O. D. , Martin, J. , Bernath, A. , Masri, J. , Lichtenstein, A. , & Gera, J. (2008). Heterogeneous nuclear ribonucleoprotein A1 regulates cyclin D1 and c‐myc internal ribosome entry site function through Akt signaling. The Journal of Biological Chemistry, 283(34), 23274–23287. 10.1074/jbc.M801185200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, G. V. , Litersky, J. M. , & Jope, R. S. (1991). Degradation of microtubule‐associated protein 2 and brain spectrin by calpain: A comparative study. Journal of Neurochemistry, 56(5), 1630–1638. 10.1111/j.1471-4159.1991.tb02061.x [DOI] [PubMed] [Google Scholar]
- Jones, T. R. , Kang, I. H. , Wheeler, D. B. , Lindquist, R. A. , Papallo, A. , Sabatini, D. M. , Golland, P. , & Carpenter, A. E. (2008). CellProfiler analyst: Data exploration and analysis software for complex image‐based screens. BMC Bioinformatics [Electronic Resource], 9(1), 1–16. 10.1186/1471-2105-9-482/FIGURES/8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kędzierska, H. , Popławski, P. , Hoser, G. , Rybicka, B. , Rodzik, K. , Sokół, E. , Bogusławska, J. , Tański, Z. , Fogtman, A. , Koblowska, M. , & Piekiełko‐Witkowska, A. (2016). Decreased expression of SRSF2 splicing factor inhibits apoptotic pathways in renal cancer. International Journal of Molecular Sciences, 17(10), 1598. 10.3390/ijms17101598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. H. , Sarbassov, D. D. , Ali, S. M. , King, J. E. , Latek, R. R. , Erdjument‐Bromage, H. , Tempst, P. , & Sabatini, D. M. (2002). mTOR interacts with raptor to form a nutrient‐sensitive complex that signals to the cell growth machinery. Cell, 110(2), 163–175. 10.1016/s0092-8674(02)00808-5 [DOI] [PubMed] [Google Scholar]
- Kim, J. R. , Lee, S. R. , Chung, H. J. , Kim, S. , Baek, S. H. , Kim, J. H. , & Kim, Y. S. (2003). Identification of amyloid beta‐peptide responsive genes by cDNA microarray technology: Involvement of RTP801 in amyloid beta‐peptide toxicity. Experimental & Molecular Medicine, 35(5), 403–411. 10.1038/emm.2003.53 [DOI] [PubMed] [Google Scholar]
- Kim, J. R. , Lee, S. R. , Chung, H. J. , Kim, S. , Baek, S. H. , Kim, J. H. , & Kim, Y. S. (2003). Identification of amyloid beta‐peptide responsive genes by cDNA microarray technology: Involvement of RTP801 in amyloid beta‐peptide toxicity. Experimental & Molecular Medicine, 35(5), 403–411. 10.1038/emm.2003.53 [DOI] [PubMed] [Google Scholar]
- Kimball, S. R. , Do, A. N. D. , Kutzler, L. , Cavener, D. R. , & Jefferson, L. S. (2008). Rapid turnover of the mTOR complex 1 (mTORC1) repressor REDD1 and activation of mTORC1 signaling following inhibition of protein synthesis. The Journal of Biological Chemistry, 283(6), 3465–3475. 10.1074/jbc.M706643200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinridders, A. , Pogoda, H. M. , Irlenbusch, S. , Smyth, N. , Koncz, C. , Hammerschmidt, M. , & Brüning, J. C. (2009). PLRG1 is an essential regulator of cell proliferation and apoptosis during vertebrate development and tissue homeostasis. Molecular and Cellular Biology, 29(11), 3173–3185. 10.1128/MCB.01807-08/SUPPL_FILE/MCBSUPPLEMENTAL_MATERIAL_1.DOC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeno, Y. , Huang, Y. J. , Qiu, J. , Lin, L. , Xu, Y. , Zhou, Y. , Chen, L. , Monterroza, D. D. , Li, H. , DeKelver, R. C. , Yan, M. , Fu, X. D. , & Zhang, D. E. (2015). SRSF2 is essential for hematopoiesis, and its myelodysplastic syndrome‐related mutations dysregulate alternative pre‐mRNA splicing. Molecular and Cellular Biology, 35(17), 3071–3082. 10.1128/MCB.00202-15/SUPPL_FILE/ZMB999100943SO1.XLSX [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokowski, D. , Han, J. , Saikia, M. , Majumder, M. , Yuan, C. L. , Guan, B. J. , Bevilacqua, E. , Bussolati, O. , Bröer, S. , Arvan, P. , Tchórzewski, M. , Snider, M. D. , Puchowicz, M. , Croniger, C. M. , Kimball, S. R. , Pan, T. , Koromilas, A. E. , Kaufman, R. J. , & Hatzoglou, M. (2013). A self‐defeating anabolic program leads to β‐cell apoptosis in endoplasmic reticulum stress‐induced diabetes via regulation of amino acid flux. The Journal of Biological Chemistry, 288(24), 17202–17213. 10.1074/jbc.M113.466920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kweon, J. H. , Kim, S. , & Lee, S. B. (2017). The cellular basis of dendrite pathology in neurodegenerative diseases. BMB Reports, 50(1), 5–11. 10.5483/bmbrep.2017.50.1.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labadorf, A. , Choi, S. H. , & Myers, R. H. (2018). Evidence for a pan‐neurodegenerative disease response in Huntington's and Parkinson's disease expression profiles. Frontiers in Molecular Neuroscience, 10, 430. 10.3389/fnmol.2017.00430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachenal, G. , Pernet‐Gallay, K. , Chivet, M. , Hemming, F. J. , Belly, A. , Bodon, G. , Blot, B. , Haase, G. , Goldberg, Y. , & Sadoul, R. (2011). Release of exosomes from differentiated neurons and its regulation by synaptic glutamatergic activity. Molecular and Cellular Neurosciences, 46(2), 409–418. 10.1016/j.mcn.2010.11.004 [DOI] [PubMed] [Google Scholar]
- Lau, S. , Patnaik, N. , Sayen, M. R. , & Mestril, R. (1997). Simultaneous overexpression of two stress proteins in rat cardiomyocytes and myogenic cells confers protection against ischemia‐induced injury. Circulation, 96(7), 2287–2294. 10.1161/01.cir.96.7.2287 [DOI] [PubMed] [Google Scholar]
- Lee, D. C. , Sohn, H. A. , Park, Z. Y. , Oh, S. , Kang, Y. K. , Lee, K. M. , Kang, M. , Jang, Y. J. , Yang, S. J. , Hong, Y. K. , Noh, H. , Kim, J. A. , Kim, D. J. , Bae, K. H. , Kim, D. M. , Chung, S. J. , Yoo, H. S. , Yu, D. Y. , Park, K. C. , & Yeom, Y. I. (2015). A lactate‐induced response to hypoxia. Cell, 161(3), 595–609. 10.1016/j.cell.2015.03.011 [DOI] [PubMed] [Google Scholar]
- Lee, S. J. , Desplats, P. , Sigurdson, C. , Tsigelny, I. , & Masliah, E. (2010). Cell‐to‐cell transmission of non‐prion protein aggregates. Nature Reviews. Neurology, 6(12), 702–706. 10.1038/nrneurol.2010.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesuisse, C. , & Martin, L. J. (2002). Long‐term culture of mouse cortical neurons as a model for neuronal development, aging, and death. Journal of Neurobiology, 51(1), 9–23. 10.1002/neu.10037 [DOI] [PubMed] [Google Scholar]
- Lewis, S. M. , Veyrier, A. , Hosszu Ungureanu, N. , Bonnal, S. , Vagner, S. , & Holcik, M. (2007). Subcellular relocalization of a trans‐acting factor regulates XIAP IRES‐dependent translation. Molecular Biology of the Cell, 18(4), 1302–1311. 10.1091/mbc.e06-06-0515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. , Chen, J. , Li, Y. , Wu, B. , Ye, Z. , Tian, X. , Wei, Y. , Hao, Z. , Pan, Y. , Zhou, H. , Yang, K. , Fu, Z. , Xu, J. , & Lu, Y. (2021). 6‐Phosphogluconolactonase promotes hepatocellular carcinogenesis by activating pentose phosphate pathway. Frontiers in Cell and Developmental Biology, 9, 753196. 10.3389/fcell.2021.753196/BIBTEX [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, K. , & Wang, Z. (2021). Splicing factor SRSF2‐centric gene regulation. International Journal of Biological Sciences, 17(7), 1708–1715. 10.7150/ijbs.58888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, K. M. , Lin, B. , Lian, I. Y. , Mestril, R. , Scheffler, I. E. , & Dillmann, W. H. (2001). Combined and individual mitochondrial HSP60 and HSP10 expression in cardiac myocytes protects mitochondrial function and prevents apoptotic cell deaths induced by simulated ischemia‐reoxygenation. Circulation, 103(13), 1787–1792. 10.1161/01.cir.103.13.1787 [DOI] [PubMed] [Google Scholar]
- Liu, L. , Arun, A. , Ellis, L. , Peritore, C. , & Donmez, G. (2014). SIRT2 enhances 1‐methyl‐4‐phenyl‐1,2,3,6‐tetrahydropyridine (MPTP)‐induced nigrostriatal damage via apoptotic pathway. Frontiers in Aging Neuroscience, 6, 184. 10.3389/fnagi.2014.00184/ABSTRACT [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Gao, M. , Ma, M. M. , Tang, Y. B. , Zhou, J. G. , Wang, G. L. , Du, Y. H. , & Guan, Y. Y. (2016). Endophilin A2 protects H2O2‐induced apoptosis by blockade of Bax translocation in rat basilar artery smooth muscle cells. Journal of Molecular and Cellular Cardiology, 92, 122–133. 10.1016/j.yjmcc.2016.02.004 [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Xia, J. , Zheng, R. , & Shao, S. (2022). High expression of NDRG3 suppresses cell apoptosis and promotes the cell proliferation and migration in gastric cancer. Asian Journal of Surgery, 45(10), 2019–2020. 10.1016/j.asjsur.2022.04.064 [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Zhao, Y. , Li, K. , Miao, S. , Xu, Y. , & Zhao, J. (2022). WD‐40 repeat protein 26 protects against oxidative stress‐induced injury in astrocytes via Nrf2/HO‐1 pathways. Molecular Biology Reports, 49(2), 1045–1056. 10.1007/s11033-021-06925-6/METRICS [DOI] [PubMed] [Google Scholar]
- Ma, Y. H. , Su, N. , Chao, X. D. , Zhang, Y. Q. , Zhang, L. , Han, F. , Luo, P. , Fei, Z. , & Qu, Y. (2012). Thioredoxin‐1 attenuates post‐ischemic neuronal apoptosis via reducing oxidative/nitrative stress. Neurochemistry International, 60(5), 475–483. 10.1016/j.neuint.2012.01.029 [DOI] [PubMed] [Google Scholar]
- Malagelada, C. , Jin, Z. H. , & Greene, L. A. (2008). RTP801 is induced in Parkinson's disease and mediates neuron death by inhibiting Akt phosphorylation/activation. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 28(53), 14363. 10.1523/JNEUROSCI.3928-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagelada, C. , Jin, Z. H. , Jackson‐Lewis, V. , Przedborski, S. , & Greene, L. A. (2010). Rapamycin protects against neuron death in in vitro and in vivo models of Parkinson's disease. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 30(3), 1166–1175. 10.1523/JNEUROSCI.3944-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagelada, C. , López‐Toledano, M. A. , Willett, R. T. , Jin, Z. H. , Shelanski, M. L. , & Greene, L. A. (2011). RTP801/REDD1 regulates the timing of cortical neurogenesis and neuron migration. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 31(9), 3186–3196. 10.1523/JNEUROSCI.4011-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagelada, C. , Ryu, E. J. , Biswas, S. C. , Jackson‐Lewis, V. , & Greene, L. A. (2006). RTP801 is elevated in Parkinson brain substantia nigral neurons and mediates death in cellular models of Parkinson's disease by a mechanism involving mammalian target of rapamycin inactivation. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 26(39), 9996–10005. 10.1523/JNEUROSCI.3292-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagelada, C. , Ryu, E. J. , Biswas, S. C. , Jackson‐Lewis, V. , & Greene, L. A. (2006). RTP801 is elevated in Parkinson brain substantia nigral neurons and mediates death in cellular models of Parkinson's disease by a mechanism involving mammalian target of rapamycin inactivation. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 26(39), 9996–10005. 10.1523/JNEUROSCI.3292-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malenka, R. C. , & Bear, M. F. (2004). LTP and LTD: An embarrassment of riches. Neuron, 44(1), 5–21. 10.1016/j.neuron.2004.09.012 [DOI] [PubMed] [Google Scholar]
- Martín‐Flores, N. , Pérez‐Sisqués, L. , Creus‐Muncunill, J. , Masana, M. , Ginés, S. , Alberch, J. , Pérez‐Navarro, E. , & Malagelada, C. (2020). Synaptic RTP801 contributes to motor‐learning dysfunction in Huntington's disease. Cell Death & Disease, 11(7), 1–15. 10.1038/s41419-020-02775-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín‐Flores, N. , Romaní‐Aumedes, J. , Rué, L. , Canal, M. , Sanders, P. , Straccia, M. , Allen, N. D. , Alberch, J. , Canals, J. M. , Pérez‐Navarro, E. , & Malagelada, C. (2015). RTP801 is involved in mutant huntingtin‐induced cell death. Molecular Neurobiology, 53(5), 2857–2868. 10.1007/s12035-015-9166-6 [DOI] [PubMed] [Google Scholar]
- Martín‐Flores, N. , Romaní‐Aumedes, J. , Rué, L. , Canal, M. , Sanders, P. , Straccia, M. , Allen, N. D. , Alberch, J. , Canals, J. M. , Pérez‐Navarro, E. , & Malagelada, C. (2016). RTP801 is involved in mutant huntingtin‐induced cell death. Molecular Neurobiology, 53(5), 2857–2868. 10.1007/s12035-015-9166-6 [DOI] [PubMed] [Google Scholar]
- Montecalvo, A. , Larregina, A. T. , Shufesky, W. J. , Stolz, D. B. , Sullivan, M. L. , Karlsson, J. M. , Baty, C. J. , Gibson, G. A. , Erdos, G. , Wang, Z. , Milosevic, J. , Tkacheva, O. A. , Divito, S. J. , Jordan, R. , Lyons‐Weiler, J. , Watkins, S. C. , & Morelli, A. E. (2012). Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood, 119(3), 756–766. 10.1182/blood-2011-02-338004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira, R. , Mendonça, L. S. , & Pereira de Almeida, L. (2021). Extracellular vesicles physiological role and the particular case of disease‐spreading mechanisms in polyglutamine diseases. International Journal of Molecular Sciences, 22(22), 12288. 10.3390/ijms222212288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel, M. , Couturier, J. , Pontcharraud, R. , Gil, R. , Fauconneau, B. , Paccalin, M. , & Page, G. (2009). Evidence of molecular links between PKR and mTOR signalling pathways in Abeta neurotoxicity: Role of p53, Redd1 and TSC2. Neurobiology of Disease, 36(1), 151–161. 10.1016/j.nbd.2009.07.004 [DOI] [PubMed] [Google Scholar]
- Morel, M. , Couturier, J. , Pontcharraud, R. , Gil, R. , Fauconneau, B. , Paccalin, M. , & Page, G. (2009). Evidence of molecular links between PKR and mTOR signalling pathways in Abeta neurotoxicity: Role of p53, Redd1 and TSC2. Neurobiology of Disease, 36(1), 151–161. 10.1016/j.nbd.2009.07.004 [DOI] [PubMed] [Google Scholar]
- Nakai, H. , Tsumagari, R. , Maruo, K. , Nakashima, A. , Kikkawa, U. , Ueda, S. , Yamanoue, M. , Saito, N. , Takei, N. , & Shirai, Y. (2020). mTORC1 is involved in DGKβ‐induced neurite outgrowth and spinogenesis. Neurochemistry International, 134, 104645. 10.1016/j.neuint.2019.104645 [DOI] [PubMed] [Google Scholar]
- Namba, S. , Kato, H. , Shigenobu, S. , Makino, T. , & Moriya, H. (2022). Massive expression of cysteine‐containing proteins causes abnormal elongation of yeast cells by perturbing the proteasome. G3 (Bethesda, Md.), 12(6), jkac106. 10.1093/g3journal/jkac106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasiri, E. , Sankowski, R. , Dietrich, H. , Oikonomidi, A. , Huerta, P. T. , Popp, J. , Al‐Abed, Y. , & Bacher, M. (2020). Key role of MIF‐related neuroinflammation in neurodegeneration and cognitive impairment in Alzheimer's disease. Molecular Medicine (Cambridge, Mass), 26(1), 1–12. 10.1186/s10020-020-00163-5/FIGURES/5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neame, S. J. , Rubin, L. L. , & Philpott, K. L. (1998). Blocking cytochrome c activity within intact neurons inhibits apoptosis. The Journal of Cell Biology, 142(6), 1583–1593. 10.1083/jcb.142.6.1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota, K. T. , Liu, R. J. , Voleti, B. , Maldonado‐Aviles, J. G. , Duric, V. , Iwata, M. , Dutheil, S. , Duman, C. , Boikess, S. , Lewis, D. A. , Stockmeier, C. A. , DiLeone, R. J. , Rex, C. , Aghajanian, G. K. , & Duman, R. S. (2014). REDD1 is essential for stress‐induced synaptic loss and depressive behavior. Nature Medicine, 20(5), 531–535. 10.1038/nm.3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, H. , Kam, T. I. , Kim, Y. , Choi, H. , Gwon, Y. , Kim, C. , Koh, J. Y. , & Jung, Y. K. (2012). Neuropathogenic role of adenylate kinase‐1 in Aβ‐mediated tau phosphorylation via AMPK and GSK3β. Human Molecular Genetics, 21(12), 2725–2737. 10.1093/hmg/dds100 [DOI] [PubMed] [Google Scholar]
- Pérez‐Sisqués, L. , Martín‐Flores, N. , Masana, M. , Solana‐Balaguer, J. , Llobet, A. , Romaní‐Aumedes, J. , Canal, M. , Campoy‐Campos, G. , García‐García, E. , Sánchez‐Fernández, N. , Fernández‐García, S. , Gilbert, J. P. , Rodríguez, M. J. , Man, H. Y. , Feinstein, E. , Williamson, D. L. , Soto, D. , Gasull, X. , Alberch, J. , & Malagelada, C. (2021). RTP801 regulates motor cortex synaptic transmission and learning. Experimental Neurology, 342, 113755. 10.1016/j.expneurol.2021.113755 [DOI] [PubMed] [Google Scholar]
- Pérez‐Sisqués, L. , Sancho‐Balsells, A. , Solana‐Balaguer, J. , Campoy‐Campos, G. , Vives‐Isern, M. , Soler‐Palazón, F. , Anglada‐Huguet, M. , López‐Toledano, M. Á. , Mandelkow, E. M. , Alberch, J. , Giralt, A. , & Malagelada, C. (2021). RTP801/REDD1 contributes to neuroinflammation severity and memory impairments in Alzheimer's disease. Cell Death & Disease, 12(6), 1–13. 10.1038/s41419-021-03899-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐Sisqués, L. , Solana‐Balaguer, J. , Campoy‐Campos, G. , Martín‐Flores, N. , Sancho‐Balsells, A. , Vives‐Isern, M. , Soler‐Palazón, F. , Garcia‐Forn, M. , Masana, M. , Alberch, J. , Pérez‐Navarro, E. , Giralt, A. , & Malagelada, C. (2021). RTP801/REDD1 is involved in neuroinflammation and modulates cognitive dysfunction in Huntington's disease. Biomolecules, 12(1), 34. 10.3390/biom12010034/S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, R. B. , Rishal, I. , Doron‐Mandel, E. , Kalinski, A. L. , Medzihradszky, K. F. , Terenzio, M. , Alber, S. , Koley, S. , Lin, A. , Rozenbaum, M. , Yudin, D. , Sahoo, P. K. , Gomes, C. , Shinder, V. , Geraisy, W. , Huebner, E. A. , Woolf, C. J. , Yaron, A. , Burlingame, A. L. , … & Fainzilber, M. (2016). Nucleolin‐mediated RNA localization regulates neuron growth and cycling cell size. Cell Reports, 16(6), 1664–1676. 10.1016/j.celrep.2016.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putz, U. , Howitt, J. , Lackovic, J. , Foot, N. , Kumar, S. , Silke, J. , & Tan, S. S. (2008). Nedd4 family‐interacting protein 1 (Ndfip1) is required for the exosomal secretion of Nedd4 family proteins. The Journal of Biological Chemistry, 283(47), 32621–32627. 10.1074/jbc.M804120200 [DOI] [PubMed] [Google Scholar]
- Quek, C. , & Hill, A. F. (2017). The role of extracellular vesicles in neurodegenerative diseases. Biochemical and Biophysical Research Communications, 483(4), 1178–1186. 10.1016/j.bbrc.2016.09.090 [DOI] [PubMed] [Google Scholar]
- Rajendran, L. , Honsho, M. , Zahn, T. R. , Keller, P. , Geiger, K. D. , Verkade, P. , & Simons, K. (2006). Alzheimer's disease beta‐amyloid peptides are released in association with exosomes. Proceedings of the National Academy of Sciences of the United States of America, 103(30), 11172–11177. 10.1073/pnas.0603838103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohart, F. , Gautier, B. , Singh, A. , & Lê Cao, K. A. (2017). mixOmics: An R package for 'omics feature selection and multiple data integration. PLoS Computational Biology, 13(11), e1005752. 10.1371/journal.pcbi.1005752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaní‐Aumedes, J. , Canal, M. , Martín‐Flores, N. , Sun, X. , Pérez‐Fernández, V. , Wewering, S. , Fernández‐Santiago, R. , Ezquerra, M. , Pont‐Sunyer, C. , Lafuente, A. , Alberch, J. , Luebbert, H. , Tolosa, E. , Levy, O. A. , Greene, L. A. , & Malagelada, C. (2014). Parkin loss of function contributes to RTP801 elevation and neurodegeneration in Parkinson's disease. Cell Death & Disease, 5(8), e1364. 10.1038/cddis.2014.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan, Z. , Lu, Q. , Wang, J. E. , Zhou, M. , Liu, S. , Zhang, H. , Durvasula, A. , Wang, Y. , Wang, Y. , Luo, W. , & Wang, Y. (2021). MIF promotes neurodegeneration and cell death via its nuclease activity following traumatic brain injury. Cellular and Molecular Life Sciences: CMLS, 79(1), 39. 10.1007/s00018-021-04037-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samali, A. , Cai, J. , Zhivotovsky, B. , Jones, D. P. , & Orrenius, S. (1999). Presence of a pre‐apoptotic complex of pro‐caspase‐3, Hsp60 and Hsp10 in the mitochondrial fraction of jurkat cells. The EMBO Journal, 18(8), 2040–2048. 10.1093/emboj/18.8.2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov, D. D. , Guertin, D. A. , Ali, S. M. , & Sabatini, D. M. (2005). Phosphorylation and regulation of Akt/PKB by the rictor‐mTOR complex. Science (New York, N.Y.), 307(5712), 1098–1101. 10.1126/science.1106148/SUPPL_FILE/SARBASSOV.SOM.PDF [DOI] [PubMed] [Google Scholar]
- Seneviratne, J. A. , Carter, D. R. , Mittra, R. , Gifford, A. , Kim, P. Y. , Luo, J. S. , Mayoh, C. , Salib, A. , Rahmanto, A. S. , Murray, J. , Cheng, N. C. , Nagy, Z. , Wang, Q. , Kleynhans, A. , Tan, O. , Sutton, S. K. , Xue, C. , Chung, S. A. , Zhang, Y. , … & Marshall, G. M. (2023). Inhibition of mitochondrial translocase SLC25A5 and histone deacetylation is an effective combination therapy in neuroblastoma. International Journal of Cancer, 152(7), 1399–1413. 10.1002/ijc.34349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, P. , Mesci, P. , Carromeu, C. , McClatchy, D. R. , Schiapparelli, L. , Yates, J. R., 3rd , Muotri, A. R. , & Cline, H. T. (2019). Exosomes regulate neurogenesis and circuit assembly. Proceedings of the National Academy of Sciences of the United States of America, 116(32), 16086–16094. 10.1073/pnas.1902513116.SD11.XLSX [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko, A. , Tomas, H. , Havlis, J. , Olsen, J. V. , & Mann, M. (2007). In‐gel digestion for mass spectrometric characterization of proteins and proteomes. Nature Protocols, 1(6), 2856–2860. 10.1038/nprot.2006.468 [DOI] [PubMed] [Google Scholar]
- Shi, J. , & Mitchison, T. J. (2017). Cell death response to anti‐mitotic drug treatment in cell culture, mouse tumor model and the clinic. Endocrine‐related Cancer, 24(9), T83–T96. 10.1530/ERC-17-0003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoshani, T. , Faerman, A. , Mett, I. , Zelin, E. , Tenne, T. , Gorodin, S. , Moshel, Y. , Elbaz, S. , Budanov, A. , Chajut, A. , Kalinski, H. , Kamer, I. , Rozen, A. , Mor, O. , Keshet, E. , Leshkowitz, D. , Einat, P. , Skaliter, R. , & Feinstein, E. (2002). Identification of a novel hypoxia‐inducible factor 1‐responsive gene, RTP801, involved in apoptosis. Molecular and Cellular Biology, 22(7), 2283–2293. 10.1128/MCB.22.7.2283-2293.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofer, A. , Lei, K. , Johannessen, C. M. , & Ellisen, L. W. (2005). Regulation of mTOR and cell growth in response to energy stress by REDD1. Molecular and Cellular Biology, 25(14), 5834–5845. 10.1128/MCB.25.14.5834-5845.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solana‐Balaguer, J. , Campoy‐Campos, G. , Martín‐Flores, N. , Pérez‐Sisqués, L. , Sitjà‐Roqueta, L. , Kucukerden, M. , Gámez‐Valero, A. , Coll‐Manzano, A. , Martí, E. , Pérez‐Navarro, E. , Alberch, J. , Soriano, J. , Masana, M. , & Malagelada, C. (2023). Neuron‐derived extracellular vesicles contain synaptic proteins, promote spine formation, activate TrkB‐mediated signalling and preserve neuronal complexity. Journal of Extracellular Vesicles, 12(9), e12355. 10.1002/jev2.12355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling, D. R. , Carpenter, A. E. , & Cimini, B. A. (2021). CellProfiler Analyst 3.0: Accessible data exploration and machine learning for image analysis. Bioinformatics (Oxford, England), 37(21), 3992–3994. 10.1093/bioinformatics/btab634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling, D. R. , Swain‐Bowden, M. J. , Lucas, A. M. , Carpenter, A. E. , Cimini, B. A. , & Goodman, A. (2021). CellProfiler 4: Improvements in speed, utility and usability. BMC Bioinformatics [Electronic Resource], 22(1), 1–11. 10.1186/s12859-021-04344-9/FIGURES/6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, M. , Zhao, Y. , Gu, Y. , & Xu, C. (2009). Inhibition of nNOS reduces ischemic cell death through down‐regulating calpain and caspase‐3 after experimental stroke. Neurochemistry International, 54(5‐6), 339–346. 10.1016/j.neuint.2008.12.017 [DOI] [PubMed] [Google Scholar]
- Sun, M. , Zhao, Y. , & Xu, C. (2008). Cross‐talk between calpain and caspase‐3 in penumbra and core during focal cerebral ischemia‐reperfusion. Cellular and Molecular Neurobiology, 28(1), 71–85. 10.1007/s10571-007-9250-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto, K. , Nagai, T. , Miyawaki, A. , & Miura, M. (2003). Spatio‐temporal activation of caspase revealed by indicator that is insensitive to environmental effects. The Journal of Cell Biology, 160(2), 235–243. 10.1083/jcb.200207111/VIDEO-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, C. Y. , & Hagen, T. (2013). mTORC1 dependent regulation of REDD1 protein stability. PLoS ONE, 8(5), e63970. 10.1371/journal.pone.0063970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, H. , Chen, M. , Pang, D. , Xia, X. , Du, C. , Yang, W. , Cui, Y. , Huang, C. , Jiang, W. , Bi, D. , Li, C. , Shang, H. , Worley, P. F. , & Xiao, B. (2020). LanCL1 promotes motor neuron survival and extends the lifespan of amyotrophic lateral sclerosis mice. Cell Death and Differentiation, 27(4), 1369–1382. 10.1038/s41418-019-0422-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassew, N. G. , Charish, J. , Shabanzadeh, A. P. , Luga, V. , Harada, H. , Farhani, N. , D'Onofrio, P. , Choi, B. , Ellabban, A. , Nickerson, P. E. B. , Wallace, V. A. , Koeberle, P. D. , Wrana, J. L. , & Monnier, P. P. (2017). Exosomes mediate mobilization of autocrine Wnt10b to promote axonal regeneration in the injured CNS. Cell Reports, 20(1), 99–111. 10.1016/j.celrep.2017.06.009 [DOI] [PubMed] [Google Scholar]
- Tawa, P. , Hell, K. , Giroux, A. , Grimm, E. , Han, Y. , Nicholson, D. W. , & Xanthoudakis, S. (2004). Catalytic activity of caspase‐3 is required for its degradation: Stabilization of the active complex by synthetic inhibitors. Cell Death and Differentiation, 11(4), 439–447. 10.1038/sj.cdd.4401360 [DOI] [PubMed] [Google Scholar]
- Thedieck, K. , Polak, P. , Kim, M. L. , Molle, K. D. , Cohen, A. , Jenö, P. , Arrieumerlou, C. , & Hall, M. N. (2007). PRAS40 and PRR5‐like protein are new mTOR interactors that regulate apoptosis. PLoS ONE, 2(11), e1217. 10.1371/journal.pone.0001217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova, S. , Temu, T. , Sinitcyn, P. , Carlson, A. , Hein, M. Y. , Geiger, T. , Mann, M. , & Cox, J. (2016). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nature Methods, 13(9), 731–740. 10.1038/nmeth.3901 [DOI] [PubMed] [Google Scholar]
- van Niel, G. , D'Angelo, G. , & Raposo, G. (2018). Shedding light on the cell biology of extracellular vesicles. Nature Reviews. Molecular Cell Biology, 19(4), 213–228. 10.1038/nrm.2017.125 [DOI] [PubMed] [Google Scholar]
- Wang, B. , Li, D. , Rodriguez‐Juarez, R. , Farfus, A. , Storozynsky, Q. , Malach, M. , Carpenter, E. , Filkowski, J. , Lykkesfeldt, A. E. , & Kovalchuk, O. (2018). A suppressive role of guanine nucleotide‐binding protein subunit beta‐4 inhibited by DNA methylation in the growth of anti‐estrogen resistant breast cancer cells. BMC Cancer, 18(1), 817. 10.1186/s12885-018-4711-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Balaji, V. , Kaniyappan, S. , Krüger, L. , Irsen, S. , Tepper, K. , Chandupatla, R. , Maetzler, W. , Schneider, A. , Mandelkow, E. , & Mandelkow, E. M. (2017). The release and trans‐synaptic transmission of Tau via exosomes. Molecular Neurodegeneration, 12(1), 5. 10.1186/s13024-016-0143-y/FIGURES/9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Fu, L. , Cui, M. , Wang, Y. , Xu, Y. , Li, M. , & Mi, J. (2017). Amino acid transporter SLC38A3 promotes metastasis of non‐small cell lung cancer cells by activating PDK1. Cancer Letters, 393, 8–15. 10.1016/j.canlet.2017.01.036 [DOI] [PubMed] [Google Scholar]
- Wang, Z. , Malone, M. H. , Thomenius, M. J. , Zhong, F. , Xu, F. , & Distelhorst, C. W. (2003). Dexamethasone‐induced gene 2 (dig2) is a novel pro‐survival stress gene induced rapidly by diverse apoptotic signals. The Journal of Biological Chemistry, 278(29), 27053–27058. 10.1074/jbc.M303723200 [DOI] [PubMed] [Google Scholar]
- Wei, L. , Yang, C. , Wang, G. , Li, K. , Zhang, Y. , Guan, H. , Sun, Z. , & Zhong, C. (2021). Interleukin enhancer binding factor 2 regulates cell viability and apoptosis of human brain vascular smooth muscle cells. Journal of Molecular Neuroscience: MN, 71(2), 225–233. 10.1007/s12031-020-01638-0/METRICS [DOI] [PubMed] [Google Scholar]
- Weisthal, S. , Keinan, N. , Ben‐Hail, D. , Arif, T. , & Shoshan‐Barmatz, V. (2014). Ca(2+)‐mediated regulation of VDAC1 expression levels is associated with cell death induction. Biochimica et Biophysica Acta, 1843(10), 2270–2281. 10.1016/j.bbamcr.2014.03.021 [DOI] [PubMed] [Google Scholar]
- Wilkinson, J. C. , Richter, B. W. , Wilkinson, A. S. , Burstein, E. , Rumble, J. M. , Balliu, B. , & Duckett, C. S. (2004). VIAF, a conserved inhibitor of apoptosis (IAP)‐interacting factor that modulates caspase activation. The Journal of Biological Chemistry, 279(49), 51091–51099. 10.1074/jbc.M409623200 [DOI] [PubMed] [Google Scholar]
- Wise, J. F. , Berkova, Z. , Mathur, R. , Zhu, H. , Braun, F. K. , Tao, R. H. , Sabichi, A. L. , Ao, X. , Maeng, H. , & Samaniego, F. (2013). Nucleolin inhibits Fas ligand binding and suppresses Fas‐mediated apoptosis in vivo via a surface nucleolin‐Fas complex. Blood, 121(23), 4729–4739. 10.1182/blood-2012-12-471094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C. D. , Chou, H. W. , Kuo, Y. S. , Lu, R. M. , Hwang, Y. C. , Wu, H. C. , & Lin, C. T. (2012). Nucleolin antisense oligodeoxynucleotides induce apoptosis and may be used as a potential drug for nasopharyngeal carcinoma therapy. Oncology Reports, 27(1), 94–100. 10.3892/or.2011.1476/HTML [DOI] [PubMed] [Google Scholar]
- Xie, Z. , Cao, B. Q. , Wang, T. , Lei, Q. , Kang, T. , Ge, C. Y. , Gao, W. J. , & Hui, H. (2018). LanCL1 attenuates ischemia‐induced oxidative stress by Sirt3‐mediated preservation of mitochondrial function. Brain Research Bulletin, 142, 216–223. 10.1016/j.brainresbull.2018.07.017 [DOI] [PubMed] [Google Scholar]
- Yang, L. , Wu, D. , Wang, X. , & Cederbaum, A. I. (2011). Depletion of cytosolic or mitochondrial thioredoxin increases CYP2E1‐induced oxidative stress via an ASK‐1‐JNK1 pathway in HepG2 cells. Free Radical Biology & Medicine, 51(1), 185–196. 10.1016/j.freeradbiomed.2011.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. , Lu, B. , Sun, X. , Han, C. , Fu, C. , Xu, K. , Wang, M. , Li, D. , Chen, Z. , Opal, P. , Wen, Q. , Crispino, J. D. , Wang, Q. F. , & Huang, Z. (2018). ANP32A regulates histone H3 acetylation and promotes leukemogenesis. Leukemia, 32(7), 1587–1597. 10.1038/s41375-018-0010-7 [DOI] [PubMed] [Google Scholar]
- Yang, Y. , Wei, Q. , Tang, Y. , Wang, Y. , Luo, Q. , Zhao, H. , He, M. , Wang, H. , Zeng, Q. , Lu, W. , Xu, J. , Liu, T. , & Yi, P. (2020). Loss of hnRNPA2B1 inhibits malignant capability and promotes apoptosis via down‐regulating Lin28B expression in ovarian cancer. Cancer Letters, 475, 43–52. 10.1016/j.canlet.2020.01.029 [DOI] [PubMed] [Google Scholar]
- Yin, D. , Kong, C. , & Chen, M. (2020). Effect of hnRNPA2/B1 on the proliferation and apoptosis of glioma U251 cells via the regulation of AKT and STAT3 pathways. Bioscience Reports, 40(7), BSR20190318. 10.1042/BSR20190318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, T. , Mett, I. , Bhunia, A. K. , Bowman, J. , Perez, M. , Zhang, L. , Gandjeva, A. , Zhen, L. , Chukwueke, U. , Mao, T. , Richter, A. , Brown, E. , Ashush, H. , Notkin, N. , Gelfand, A. , Thimmulappa, R. K. , Rangasamy, T. , Sussan, T. , Cosgrove, G. , … & Tuder, R. M. (2010). Rtp801, a suppressor of mTOR signaling, is an essential mediator of cigarette smoke‐induced pulmonary injury and emphysema. Nature Medicine, 16(7), 767–773. 10.1038/nm.2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka, Y. , Konishi, Y. , Kosaka, N. , Katsuda, T. , Kato, T. , & Ochiya, T. (2013). Comparative marker analysis of extracellular vesicles in different human cancer types. Journal of Extracellular Vesicles, 2(1). 10.3402/jev.v2i0.20424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaid, H. , Abu‐Hamad, S. , Israelson, A. , Nathan, I. , & Shoshan‐Barmatz, V. (2005). The voltage‐dependent anion channel‐1 modulates apoptotic cell death. Cell Death and Differentiation, 12(7), 751–760. 10.1038/sj.cdd.4401599 [DOI] [PubMed] [Google Scholar]
- Zhang, B. , Wang, H. , Jiang, B. , Liang, P. , Liu, M. , Deng, G. , & Xiao, X. (2010). Nucleolin/C23 is a negative regulator of hydrogen peroxide‐induced apoptosis in HUVECs. Cell Stress & Chaperones, 15(3), 249–257. 10.1007/s12192-009-0138-5/METRICS [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W. , Zheng, J. , Meng, J. , Neng, L. , Chen, X. , & Qin, Z. (2017). Macrophage migration inhibitory factor knockdown inhibit viability and induce apoptosis of PVM/Ms. Molecular Medicine Reports, 16(6), 8643–8648. 10.3892/mmr.2017.7684/HTML [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Chu, S. F. , Wang, S. S. , Jiang, Y. N. , Gao, Y. , Yang, P. F. , Ai, Q. D. , & Chen, N. H. (2018). RTP801 is a critical factor in the neurodegeneration process of A53T α‐synuclein in a mouse model of Parkinson's disease under chronic restraint stress. British Journal of Pharmacology, 175(4), 590–605. 10.1111/bph.14091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Nie, S. , & Chen, L. (2018). Targeting prion‐like protein spreading in neurodegenerative diseases. Neural Regeneration Research, 13(11), 1875–1878. 10.4103/1673-5374.239433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, F. , Gomi, M. , Fujimoto, M. , Hayase, M. , Marumo, T. , Masutani, H. , Yodoi, J. , Hashimoto, N. , Nozaki, K. , & Takagi, Y. (2009). Attenuation of neuronal degeneration in thioredoxin‐1 overexpressing mice after mild focal ischemia. Brain Research, 1272, 62–70. 10.1016/j.brainres.2009.03.023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Data Availability Statement
Mass‐spectrometry data and search results files were deposited in the Proteome Xchange Consortium via the JPOST partner repository (https://repository.jpostdb.org) (Okuda S, Watanabe Y, Moriya Y, Kawano S, Yamamoto T, Matsumoto M, Takami T, Kobayashi D, Araki N, Yoshizawa AC et al.: jPOSTrepo: an international standard data repository for proteomes. Nucleic Acids Res 2017, 45(D1):D1107‐D1111) with the identifier PXD040082 for ProteomeXchange and JPST002037 for jPOST (for reviewers: https://repository.jpostdb.org/preview/170310131463ea007f81c18; Access key: 3507).


