Diffuse midline gliomas (DMGs) harboring loss of H3K27me3 due to H3K27M/I mutations or EZHIP overexpression comprise a World Health Organization (WHO) defined subtype of pediatric diffuse high-grade gliomas (H3K27-altered DMG), which carries a poor prognosis with median overall survival (OS) of 12 months [2, 3]. H3K27M-mutant DMGs (H3K27M-DMGs) additionally develop somatic alterations in driver genes (eg, TP53, PPM1D, ATRX, PIK3CA, ACVR1) during tumor evolution. DNA methylation profiling of H3K27M-DMGs reveals distinct clustering in comparison to other pediatric-type diffuse high-grade gliomas. Despite the generally poor prognosis, long-term survivors (LTS) have been reported with distinct molecular profiles such as somatic FGFR1 mutations [6] and rare cases with non-diffuse (circumscribed) histology patterns [4]. Here, we aim to identify clinical, genomic, and epigenomic characteristics of LTS patients with H3K27M-DMG.
Through a comprehensive multi-site and literature case review, we identified 85 patients with confirmed H3K27M-DMG (by DNA sequencing or IHC) and LTS, defined as OS of at least 36 months from initial diagnosis (Online Resource [OR] Table S1). We extracted available demographic, diagnostic, genomic, therapeutic, and clinical outcome data and performed DNA methylation analysis when tissue was available (30 samples from 26 patients). A control cohort of 453 patients with confirmed H3K27M-DMG and OS < 18 months (short-term survival [STS]) with detailed histology, demographic, and CNS tumor location from Pratt et al. [4] was utilized. We further assembled two molecular cohorts with 258 H3K27M-DMG patients (208 STS patients) with clinical and genomic profiles (see OR) and 20 STS H3K27M-DMG patients (MNP2.0 cohort) with clinical, genomic, and tumor DNA methylation profiles.
The median age of our LTS cohort was 13.2 years (interquartile range [IQR] 7–30 years), 64.7% of patients were females, and the median OS was 51.6 months (IQR 40.7–63.8 months). Clinically, patients received a variety of therapy types of various durations and timing throughout disease courses (Fig. 1a). At diagnosis, LTS patients had a greater frequency of non-infiltrative/circumscribed tumors (18.7% vs 1.7%, P < 0.0001) and were more frequently diagnosed with tumors in the thalamus (42.3% vs 14.7%, P < 0.0001) compared to STS patients. No statistical association was found between H3.1 vs H3.3K27M and survival (12.3% STS vs 14.2% LTS with H3.1, P = 0.85; OR Fig. S1).
Fig. 1.
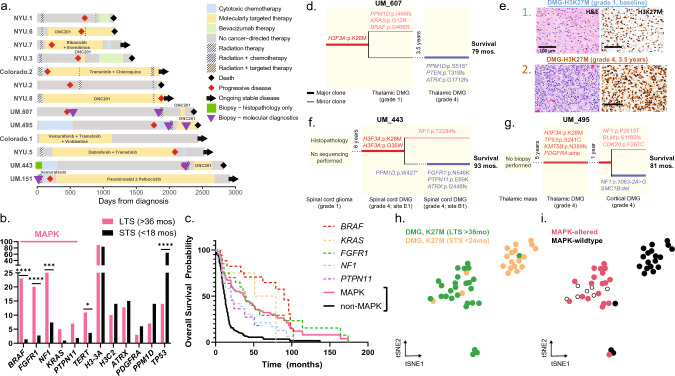
Characteristics of LTS patients. a Swimmer plot for 13 LTS patients demonstrates wide variety in timing and types of therapies. b Bar plot comparing oncogene mutation frequencies in LTS (n = 55) and molecular control (OS < 18 months, n = 208) cohorts highlights the higher frequency of MAPK pathway alterations in the LTS cohort. c Kaplan–Meier curve for combined LTS and molecular control cohort (n = 310) reveals that MAPK pathway genetic alterations are associated with improved OS at 36 months and shows differential survival among MAPK oncogenes. d Clonal map demonstrates molecular evolution of UM-607 from time of initial and subsequent biopsies, with thin lines for sub-clonal (< 10%) and thick lines for clonal branching events. e Histology stains demonstrate histologic evolution from low- to high-grade. f–g Clonal maps demonstrate the molecular evolution of UM-443 and UM-495 at various timepoints and tumor samples. h–i Joint analysis using t-SNE dimensionality reduction of LTS (23 patients, 27 tumors) and STS (20 patients; MNP2.0 cohort) H3K27M-DMGs reveals two clusters separating tumors from LTS/STS patients (h) and tumors with/without MAPK pathway alterations (i)
Evaluation of the genomic landscape of LTS patients (OR Fig. S2) revealed a high frequency of MAPK pathway alterations (69.0%, 38/55) compared to STS patients (12.0%, 25/208, P < 0.001). Clonality of mutations is included for five patients with available variant allele frequency (VAF) data (sub-clonal defined as VAF < 10%) in OR Table S2. The most frequently mutated MAPK pathway genes in LTS patients were BRAF (23.6%, n = 13/55), NF1 (23.6%, n = 13/55), and FGFR1 (21.8%, n = 12/55; Fig. 1b, OR Table S1). In addition, seven LTS patients presented with MAPK-associated mutations in PTPN11 (9.1%, n = 5/55) and KRAS (5.5%, n = 3/55). In contrast, individual MAPK genes were rarely mutated in STS patients (7.7% NF1, 2.4% FGFR1, 1.4% BRAF, 1.4% PTPN11, 1.0% KRAS). Moreover, we observed a striking absence of somatic TP53 mutations in LTS patients (16.4%, 9/55 vs 65.4%, 136/208, P < 0.0001). A combined survival analysis of our LTS and molecular control cohort (n = 310) revealed that MAPK pathway alterations are associated with LTS (MAPK-altered vs wildtype, OS at 36 months 51.3% vs 7.6%; Fig. 1c). A univariate and multivariate (sex, age, H3.1/H3.3, TP53, MAPK) survival analysis that was restricted to our molecular control cohort (n = 258 patients) further demonstrated that MAPK pathway alterations are associated with LTS (OS at 36 months 7.4% vs 0%; OR Figs. S3, S4). This combined retrospective molecular cohort may be a closer reflection of the prevalence of patients with LTS and MAPK alterations in the general H3K27M-DMG population (5–10%).
We further investigated the tumor evolution of three cases with multiple biopsies for histology review and molecular profiling (Fig. 1d–g). UM-607 was diagnosed with a tectal-thalamic H3K27M-DMG with diffuse, low-grade histology (see Fig. 1d inlay “1”, Fig. 1e) but developed progression 3.5 years later, and re-biopsy showed high-grade features (Fig. 1d inlay “2”, Fig. 1e) and a significantly altered tumor genomic profile, ultimately surviving 79 months from diagnosis. UM-443 had a spinal cord glioma initially diagnosed as low-grade glioma (LGG) on histology review with no tumor sequencing performed, and later was interpreted as high-grade on histology and with H3K27M mutation with intra-tumoral genomic heterogeneity as evidenced by phylogenetic analysis (Fig. 1f and OR Fig. S5). UM-495 presented with a thalamic mass, presumed to be LGG on imaging so biopsy was deferred; however, biopsy five years later confirmed H3K27M-DMG, and subsequent biopsy one year later of a cortical metastasis revealed additional molecular alterations (Fig. 1g). As these phylogenetic trees demonstrate, H3K27M mutations were the founding events in all patients and MAPK alterations were either clonal or sub-clonal. Also, both patients (UM-607, UM-443) with initial low-grade histology lacked TP53 mutations.
Finally, DNA methylation-based classification of LTS DMGs (26 patients; n = 30 tumors) against 40 reference gliomas, glioneuronal tumors, and neuronal tumors described by Capper et al. [1] showed closest association with H3K27M-DMGs (OR Fig. S6). Three LTS samples mapped closest to healthy brain tissues and were presumably of low tumor content. We next performed a joint analysis of LTS (n = 23 patients) and STS H3K27M-DMG samples (n = 20 patients, MNP2.0 cohort) and discovered two clusters that separate LTS and STS patients (Fig. 1h). Strikingly, MAPK pathway mutations were exclusively seen in the LTS methylation cluster (Fig. 1i). The LTS cluster included three STS cases, all with MAPK alterations. This data suggests that the LTS DNA methylation cluster is potentially defined by MAPK alterations. DNA methylation of multi-timepoint LTS samples from UM cases revealed that all map into the MAPK H3K27M-DMG cluster (OR Fig. S7).
In summary, we identify enrichment of alterations in MAPK pathway genes in patients with LTS compared to those with typical survival. Furthermore, H3K27M-DMGs with MAPK alterations demonstrate a unique DNA methylation signature, even in some cases without a MAPK in initial sample, thus raising the possibility of a unique evolutionary trajectory that selects for subsequent MAPK alteration (eg, UM-443 and UM-495). The current WHO classification includes the entity “diffuse low-grade glioma, MAPK pathway-altered”, which is defined by genetic alterations in BRAF and FGFR1. Moreover, circumscribed astrocytic gliomas include “high-grade astrocytomas with piloid features” with recurrent MAPK (NF1, FGFR1) alterations [3, 5]. Our data demonstrates distinct clinical outcomes, DNA methylation patterns, MAPK mutations, and absence of TP53 mutations that supports a distinct LTS-associated subtype of H3K27M-DMG, which we propose as “diffuse midline glioma, H3K27M-mutant and MAPK pathway-altered”. As a few MAPK-wildtype cases clustered with the “MAPK-altered” group, methylation analysis should be considered with other clinical and molecular diagnostic elements. Our study also confirms that a subset of H3K27M-DMGs show circumscribed histology, and this contradiction with the diagnostic category “diffuse” midline glioma may need to be re-addressed in the future.
Limitations of our study include a relatively small sample of patients with LTS and clinical data limited by availability in the literature in many cases. Further investigation into the biology of H3K27M/MAPK-altered tumors is warranted to confirm our findings. Overall, our results suggest that numerous factors contribute to LTS in patients with H3K27M-DMG, particularly low-grade and non-infiltrative histology, thalamic/non-brainstem location, absence of somatic TP53 mutations, and enrichment of MAPK pathway alterations. These findings further support the use of biopsy in these patients for clarification of H3K27M-DMG subtype, prognosis, and treatment.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to acknowledge the families and patients who participated in donation of their samples to help future patients with DMG. We would like to acknowledge the International DIPG/DMG Registry for support of this study.
Author contributions
Conceptualization: CK, SMW, DP; methodology: CK, SMW, HJR, SJ; formal analysis and investigation: CK, SMW, DP, HJR, SJ; writing—original draft preparation: CK, SMW, HJR, SJ; writing—review and editing: all authors; funding acquisition: CK, SMW, SV, AMC; supervision: CK, SMW.
Funding
Open access funding provided by EPFL Lausanne. National Institutes of Health Grant R01-NS119231 (CK). National Institutes of Health Grant R01-NS124607 (CK). National Institutes of Health Grant R01-NS110572 (SV). National Institutes of Health Grant R01-CA261926 (SV). DOD Grant CA201129P1 (CK). University of Michigan Chad Carr Pediatric Brain Tumor Center (CK, SV). The Evans Family (CK). ChadTough Defeat DIPG Foundation (CK, SV). Catching Up with Jack (CK). Pediatric Brain Tumor Foundation (CK). Prayers From Maria Foundation (CK). Michael Miller Memorial Foundation (CK). Morgan Behen Golf Classic (CK). Yuvaan Tiwari Foundation (CK). Sontag Foundation (SV). Alex Lemonade Stand Foundation (SV). Hyundai Hope Foundation (SV). National Institutes of Health Clinical Sequencing Exploratory Research Award Grant 1UM1HG006508 for PEDS-MIONCOSEQ Study (AC). École Polytechnique Fédérale de Lausanne (SMW).
Data availability
All tumor somatic sequencing data generated from MiOncoSeq has been uploaded to the Database of Genotypes and Phenotypes (dbGaP) [accession number phs000673.v1.p1]. For all other patients, genetic results were used as reported by the literature or treating physician. Publicly available data that support the findings of this study were obtained from Pratt et al. [4] and the MNP2.0 study. All other data generated in this study are available upon reasonable request.
Declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethics approval
This research study was conducted retrospectively from data obtained for clinical or prior research purposes. The IRB at the University of Michigan Medical School approved this study with a HIPAA authorization waiver.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Holly J. Roberts, Sunjong Ji, Sebastian M. Waszak, and Carl Koschmann contributed equally to this work.
Contributor Information
Sebastian M. Waszak, Email: sebastian.waszak@epfl.ch
Carl Koschmann, Email: ckoschma@med.umich.edu.
References
- 1.Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555:469–474. doi: 10.1038/nature26000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khuong-Quang D-A, Buczkowicz P, Rakopoulos P, Liu X-Y, Fontebasso AM, Bouffet E, et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol. 2012;124:439–447. doi: 10.1007/s00401-012-0998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol. 2021;23:1231–1251. doi: 10.1093/neuonc/noab106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pratt D, Natarajan SK, Banda A, Giannini C, Vats P, Koschmann C, et al. Circumscribed/non-diffuse histology confers a better prognosis in H3K27M-mutant gliomas. Acta Neuropathol. 2018;135:299–301. doi: 10.1007/s00401-018-1805-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryall S, Zapotocky M, Fukuoka K, Nobre L, Guerreiro Stucklin A, Bennett J, et al. Integrated molecular and clinical analysis of 1,000 pediatric low-grade gliomas. Cancer Cell. 2020;37:569–583.e5. doi: 10.1016/j.ccell.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schüller U, Iglauer P, Dorostkar MM, Mawrin C, Herms J, Giese A, et al. Mutations within FGFR1 are associated with superior outcome in a series of 83 diffuse midline gliomas with H3F3A K27M mutations. Acta Neuropathol. 2021;141:323–325. doi: 10.1007/s00401-020-02259-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All tumor somatic sequencing data generated from MiOncoSeq has been uploaded to the Database of Genotypes and Phenotypes (dbGaP) [accession number phs000673.v1.p1]. For all other patients, genetic results were used as reported by the literature or treating physician. Publicly available data that support the findings of this study were obtained from Pratt et al. [4] and the MNP2.0 study. All other data generated in this study are available upon reasonable request.