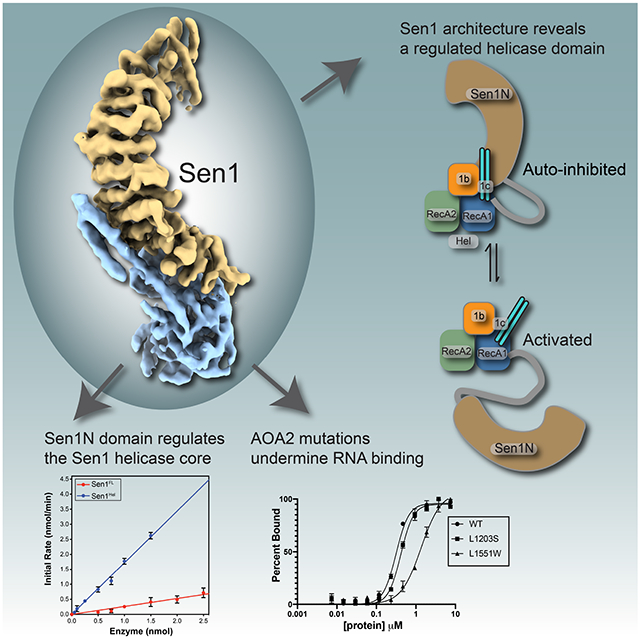
SUMMARY
The Senataxin (SETX, Sen1 in yeasts) RNA-DNA hybrid resolving helicase regulates multiple nuclear transactions including DNA replication, transcription, and DNA repair, but the molecular basis for Sen1 activities is ill-defined. Here, Sen1 cryo-EM reconstructions reveal an elongated inchworm-like architecture. Sen1 is comprised of an amino terminal helical repeat Sen1 N-terminal (Sen1N) regulatory domain that is flexibly linked to its C-terminal SF1B helicase motor core (Sen1Hel) via an intrinsically disordered tether. In an autoinhibited state, the Sen1Sen1N domain regulates substrate engagement by promoting occlusion of the RNA substrate binding cleft. The X-ray structure of an activated Sen1Hel engaging single-stranded RNA and ADP-SO4 shows the enzyme encircles RNA and implicates a single nucleotide power stroke in the Sen1 RNA translocation mechanism. Together, our data unveil dynamic protein-protein and protein-RNA interfaces underpinning helicase regulation and inactivation of human SETX activity by RNA-binding deficient mutants in Ataxia with Oculomotor Apraxia 2 neurodegenerative disease.
Keywords: Sen1, SETX, Senataxin, cryo-EM, X-ray crystallography, RNA-DNA hybrid, R-loop, Helicase, SF1B, DNA repair, transcription
Graphical Abstract
eTOC Blurb
Sen1/SETX helicases resolve RNA-DNA hybrids to regulate transcription, DNA replication and DNA repair. Appel et al solve cryo-EM and X-ray crystal structures of Sen1-RNA complexes showing how the enzyme is auto-regulated, and report how Sen1/SETX can be inactivated by AOA2 RNA binding cleft mutants.
INTRODUCTION
R-loops form when a nascent transcribed RNA pairs with its DNA template to displace the non-template DNA strand that becomes extruded as single stranded DNA (ssDNA). R-loops can occur during normal transcription, when topoisomerase reactions are perturbed, during replication-transcription collisions, and also at DNA double strand breaks1-3. Multiple lines of evidence implicate the RNA-DNA hybrid resolving Senataxin (SETX, Sen1 in yeast) helicase in the regulation of R-loops1-3. Yeast Sen1 participates in RNA processing, DNA replication, transcription termination, and DNA repair3-5. DNA repair roles are consistent with human SETX deficient cells showing marked sensitivity to DNA damaging agents including mitomycin C, camptothecin, H2O26 and the anticancer Top2 poison etoposide7. Sen1/SETX orthologs also cooperate with the Mre11-Rad50-Nbs1 complex in double strand break (DSB) repair in yeast8 and mammalian cells9, and human SETX limits illegitimate DNA end-joining of DSB ends produced in actively transcribed genes 7. A role for Sen1 activity in resolving RNA-DNA hybrids during S-phase is consistent with its functions in preventing genome instability arising from replication/transcription collisions10. Autosomal recessive mutations in human Senataxin (SETX) cause Ataxia-Oculomotor-Apraxia 2 (AOA2), whereas dominant SETX mutations are linked to Amyotrophic Lateral Sclerosis Type 4 (ALS4)11,12 (Supplementary Table 1). How SETX mutations cause disease remains poorly defined.
The Sen1 polypeptide broadly segments by homology into N- and C-terminal halves. The N-terminal Sen1Sen1N region is a protein-protein interaction domain4. Deletion of the Sen1N region of budding yeast Sen1 results in both heat and cold temperature sensitive growth phenotypes, whereas the catalytic core is essential. Both the N- and C-terminal Sen1 domains are requisite for robust transcription termination function13. Sen1 binds the RNA polymerase II large subunit (Rpb1) through its N-terminal region4, regulates transcriptional termination, modulates genome-wide of RNA polymerase II distribution on protein-coding and non-coding genes14. Sen1 also directly binds Nrd1-Nab315 and this Nrd1–Nab3–Sen1 (NNS) complex functions in a poly(A)-independent mechanism of transcription termination of non-coding snoRNA, snRNAs and cryptic unstable transcripts16-18. In this context it is hypothesized Sen1 helicase activity promotes displacement of Pol II from the DNA template14,18.
Consistent with its roles in promoting genome stability during replication, Sen1Sen1N binds Ctf419, a DNA replisome protein-protein interaction hub20,21. Sen1Sen1N mutants impair association with the replisome and confer a synthetic lethal phenotype in the context of deletion of RNase H1 (RNH1) and RNase H2 (RNH201) RNA-DNA hybrid metabolism genes and show increased genome instability and recombination. The C-terminal superfamily I (SF1) helicase domain (Sen1Hel)22 has ATP-stimulated DNA and RNA dependent 5′-3′ DNA translocation and unwinding activities23-25. Sen1Sen1N also regulates helicase and RNA-DNA stimulated ATPase activities23,24. The X-ray crystal structure of the ADP-bound catalytic core of S. cerevisiae Sen1 revealed the enzyme is structurally related to the eukaryotic nonsense mediated decay Upf1 protein23. However, despite the broadly implicated roles for Sen1 in DNA and RNA metabolism, the molecular basis for Sen1 cis- and trans- regulation and the underlying mechanisms controlling its nucleic acid binding and catalytic activities remain diffusely characterized due to a lack of detailed molecular characterization of Sen1 RNA/DNA and protein-protein interaction interfaces.
Precisely how Sen1 translocates along RNA or DNA, recognizes, unwinds RNA-DNA and DNA-DNA hetero duplexes, and how Sen1 is regulated remains undefined. Here, we report biochemical and structural characterization of Chaetomium thermophilum Sen1 (CtSen1). Our results reveal the enzyme motor core is flexibly linked to a dynamically bound helical repeat regulatory domain. We define key elements of the Sen1 catalytic mechanism, and an unexpected mode of in cis autoinhibition characterized by potential allosteric control of the enzyme–nucleic acid binding surface.
RESULTS
Cryo-EM Structure of Sen1FL
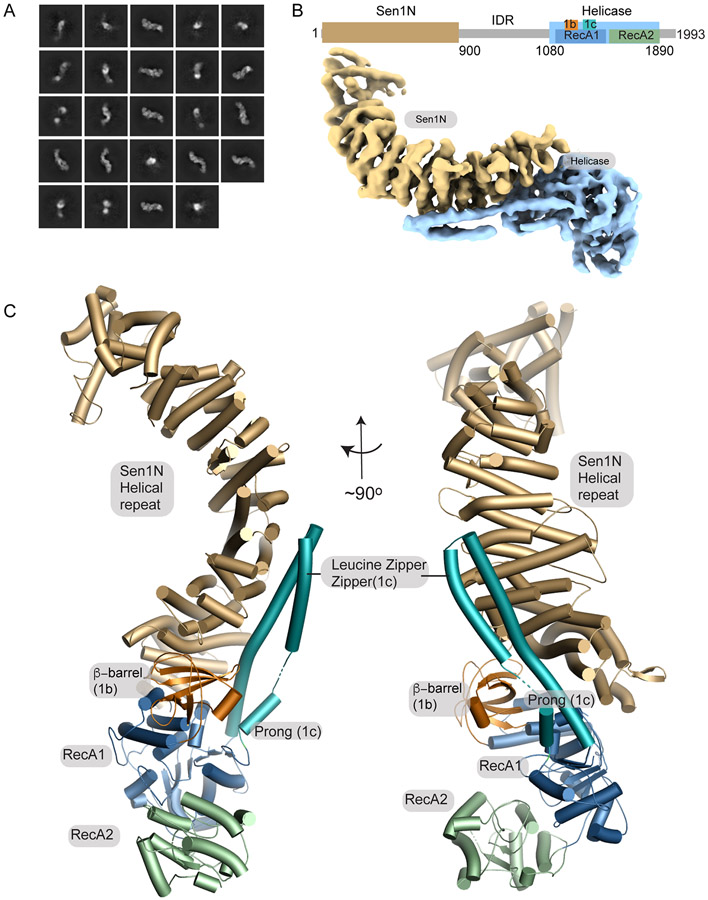
We employed a selectable high yield YFP-fusion expression approach26 to express and purify the full-length CtSen1 (Sen1FL, aa 1-1993) in mammalian HEK293F cells (Supplementary Figure 1A). CtSen1 contains the conserved Sen1N domain and SF1B helicase motor core, but like some other fungal species15, lacks Nrd1-interaction motifs (NIMs). This expression system yields milligram quantities of pure protein suitable for biophysical analysis (Supplementary Figure 1B). Limited tryptic digestion of Sen1FL yields two structurally ordered halves corresponding to the N-terminal domain (Sen1Sen1N), and carboxyl-terminal helicase core (Sen1Hel) (Supplementary Figure 1C). By contrast, a central intrinsically disordered region (IDR) of low-complexity sequence was proteolytically labile, consistent with protein order-disorder predictions (Supplementary Figure 1D).
To understand the basis for Sen1 catalytic activities and regulation we examined its molecular architecture using cryo-electron microscopy (cryo-EM) and determined its structure to 7.4 Å (4.6 Å masked FSC). 2D class averages for Sen1FL revealed a highly asymmetric and elongated architecture resembling the sideview of an inchworm (Figures 1A, Supplementary Figure 2, and Table 1). Overall, the Sen1FL structure is characterized by a prominent protruding Sen1Sen1N helical repeat region that is comprised of 46 α-helices (Figure 1B and 1C, Supplementary Figure 4). The ordered Sen1Sen1N domain in the cryo-EM reconstruction corresponds to the N-terminal 900 residues of the protein. A ~180 amino acid linker (aa 901-1080) between the structured domains is not visible and is likely disordered as expected from our proteolysis results. The three-lobed Sen1Hel nucleic acid motor is bound to the convex face of the helical repeat and is defined by its two closely juxtaposed RecA homology domains (RecA1 and RecA2) (Figure 1C). Two RecA1 insertions include the β-barrel (1b) and helical "Prong" (1c) that are also found in the Upf127 and Ighmbp228 helicases. An extended interface between Sen1Sen1N and Sen1Hel involves eight Sen1Sen1N helical repeats and both the RecA1 core, and its 1b and 1c insertions. An additional Sen1-specific coiled-coiled (leucine-zipper) insertion extends from the 1c-Prong and engages the Sen1Sen1N helical repeats (Figure 1C). We refer to this element as the leucine-zipper-zipper (LZZ), as it appears to “zipper" up the helical repeat helices, thereby reinforcing the Sen1Sen1N-Sen1Hel juncture.
Figure 1. Cryo-EM structure of Sen1FL.
(A) 2D cryo-EM class averages of full-length Sen1 show an asymmetric elongated assembly.
(B) Cryo-EM volume of Sen1FL is displayed showing the Sen1N domain in brown, and the helicase core in blue.
(C) The Sen1N helical repeat (tan). Extensive interactions with helical repeat are mediated by RecA1 1b (orange) and 1c (turquoise) insertion elements. The LZZ (RecA1 1c insertion) reinforces the interface.
Table 1.
Cryo-EM data collection and refinement statistics
| Sen1FL | Sen1Sen1N | Sen1N-PP-C | |
|---|---|---|---|
| EMDB | EMD-29439 | EMD-29440 | EMD-29426 |
| PDB ID | 8FTK | n/a | 8FTH |
| Data collection and processing | |||
| Microscope | Titan Krios | Titan Krios | Titan Krios |
| Detector | K2 summit | K2 summit | Gatan3 Bioquantum |
| Magnification | 130,000 | 130,000 | 130,000 |
| Voltage(kV) | 300 | 300 | 300 |
| Electron exposure (e–/Å2) | 60 | 60 | 40 |
| Defocus range (μm) | −1.4 to −2.2 | −1.4 to −2.2 | −1.0 to −2.7 |
| Pixel size (Å) | 0.53 | 0.53 | 0.67 |
| Symmetry imposed | C1 | C1 | C1 |
| Number of micrographs | 5089 | 5089 | 6379 (set1) + 3655 (set2) |
| Initial particle images (no.) | 1351862 | 1351862 | 3400325 (set1) + 3273289 (set2) |
| Final particle images (no.) | 29391 | 74760 | 471395 |
| Map resolution - unmaksed (Å) (FSC = 0.143) | 7.40 | 6.86 | 3.69 |
| Map resolution -masked (Å) (FSC = 0.143) | 4.56 | 4.49 | 3.17 |
| Refinement | |||
| Initial models used (PDB code) | AlphaFold (1-900), CtSen1Hel X-ray | No model refined | AlphaFold (1-900), CtSen1Hel X-ray |
| Map sharpening B factor (Å2) | 122.6 | 142.5 | |
| Model composition | Polyalanine | ||
| Nonhydrogen atoms | 7840 | 9778 | |
| Protein residues | 1584 | 1263 | |
| ADP nucleotides | 0 | 1 | |
| B factors (Å2) (mean) | |||
| Protein | 219.90 | 56.70 | |
| Nucleotide | n/a | 40.90 | |
| R.M.S. deviations | |||
| Bond lengths (Å) | 0.002 | 0.008 | |
| Bond angles (°) | 0.573 | 1.266 | |
| Map-model CC | |||
| CC (mask) | 0.81 | 0.75 | |
| CC (volume) | 0.80 | 0.72 | |
| CC (peaks) | 0.74 | 0.61 | |
| Validation | |||
| Molprobity score | 1.60 | 1.60 | |
| Clashscore | 4.21 | 4.89 | |
| Poor rotamers (%) | 0.00 | 0.49 | |
| Ramachandran plot | |||
| Outliers (%) | 0.00 | 0.00 | |
| Allowed (%) | 5.86 | 4.87 | |
| Favored (%) | 94.14 | 95.13 | |
Sen1Hel is regulated by Sen1Sen1N
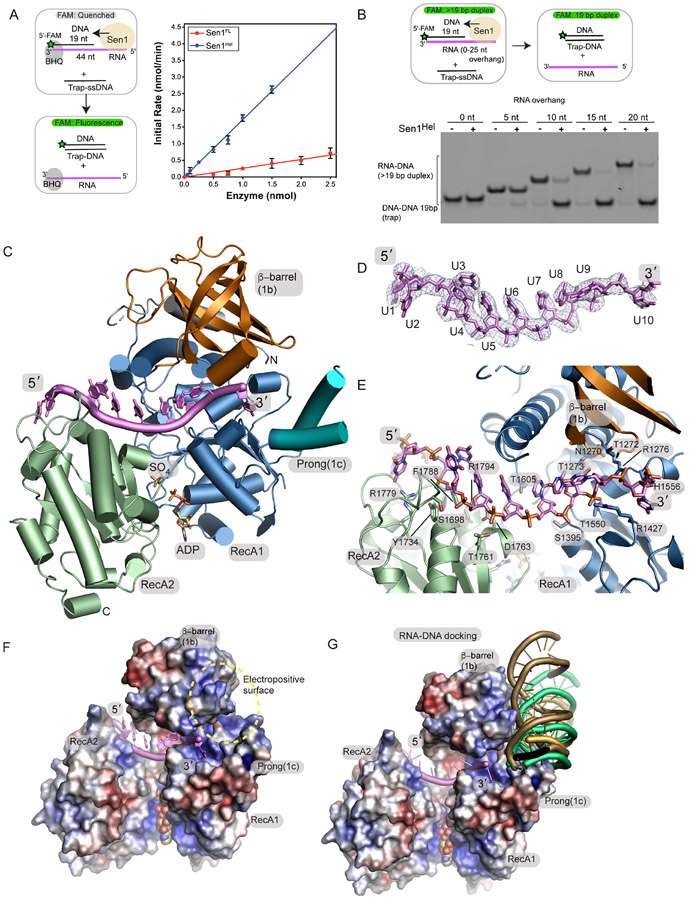
We next evaluated CtSen1 catalytic activity using an RNA-DNA helicase assay that monitors Sen1 displacement of black hole quencher (BHQ) modified 44 nucleotide (nt) RNA strand paired to a 5′-FAM labeled 19 nt DNA in the presence of an unlabeled 19 nt trap (Figure 2A). Sen1FL had a rate of RNA/DNA unwinding (kobs) of 0.25 ± 0.01 min−1. By comparison, purified Sen1Hel domain had a 6.8-fold higher activity (kobs = 1.70 ± 0.03 min−1). Together with the proteolysis data, these results suggest that Sen1Sen1N is physically tethered to Sen1Hel via a flexible linker peptide, and that Sen1Sen1N negatively regulates Sen1Hel helicase activity in cis. Consistent with this notion, from our inspection of the micrographs, 2D classifications, and ab initio reconstructions, in addition to a full-length enzyme assembly (Figure 1C), two additional species were also prominent in our ab initio EM reconstructions (Supplementary Figure 2). One of these structures may correspond the helicase core of the protein, but the lack of discernable secondary structure in these envelopes could not allow us to conclusively model this species as the catalytic core. However, we also determined and refined a Sen1Sen1N envelope to 6.9 Å (4.5 Å masked FSC) (Supplementary Figures 2D and 2F). In this structure, the overall fold for Sen1Sen1N is maintained as observed in the full-length reconstruction, but the helicase domain is absent. Together, limited protease digestion, disorder predictions, helicase activity measurements, and EM observations suggest that Sen1Sen1N reversibly engages Sen1Hel, and that deletion of the Sen1Sen1N domain activates the enzyme.
Figure 2. Crystal structure of the Sen1Hel–ssRNA–ADP–SO4 complex.
(A) Left: RNA-DNA unwinding substrates. Right: Sen1FL and Sen1Hel at the amounts indicated were incubated with 50 nmol of Sub7 (Table S2). Reactions were initiated by addition of a mixture containing ATP (1 mM) and MgCl2 (2 mM). FAM signal was detected at 520 nm every 10 sec over 30 min at 25 °C. Initial rates were determined by linear fit of the first 5 min of the unwinding reaction. Error bars are SD from 5 replicates.
(B) Top: gel-based RNA-DNA unwinding assay. Bottom: Effect of RNA overhang length on Sen1Hel RNA-DNA unwinding activity. Sub1-Sub5 (Supplementary Table 3) at 20 nM, Sen1Hel (50 nM), 19-Trap DNA (Table 1) at 200 nM, ATP (1 mM), MgCl2 (2 mM) were incubated for 15 min at 37 °C.
(C) X-ray structure of the Sen1Hel–RNA–ADP–SO4 complex. The extended LZZ coiled coil is not shown.
(D) Composite Omit 2Fo-Fc electron density at 1.0 σ, carved 2.0 Å around the RNA chain.
(E) Sen1Hel RNA-protein interactions show an extensive RNA binding interface.
(F) APBS surface electrostatic surface representation (blue=electropositive, red= electronegative) of the Sen1 RNA binding surface. Sen1 encircles the RNA via its RecA1 and RecA2 domains.
(G) Representative HADDOCK RNA-DNA poses displayed show secondary putative RNA-DNA hybrid binding electropositive surface flanks the ssRNA binding site and is assembled by the 1b and 1c RecA1 insertions.
Sen1Hel RNA binding mechanism
To better define the basis for Sen1 substrate engagement we tested Sen1FL and Sen1Hel association with RNA-DNA hybrid substrates bearing increasing 5′-RNA overhangs using electrophoretic mobility shift assays (EMSAs) (Supplementary Figures 3A-3B). Both proteins bound a 44 nt ssRNA, whereas binding of RNA-DNA hybrids requires a minimal flanking 5′-RNA overhang of 10-15 nt, consistent with the known 5′-3′ polarity of Sen1 homologs25 (Supplementary Figure 3B). Also, like ScSen1, CtSen1 also harbors both DNA-DNA and RNA-DNA unwinding activity (Supplementary Figure 3C). In a gel-based helicase assay monitoring transfer of a labeled DNA strand from an RNA-DNA to a smaller DNA-DNA trap, we tested the RNA length requirement for Sen1 RNA-dependent DNA unwinding activity (Figure 2B). Sen1Hel also displays a strong preference for unwinding RNA-DNA substrates with greater than or equal to a 10 nt 5′ RNA overhang flanking the RNA-DNA heteroduplex. These helicase activity and RNA binding data map an activated catalytic core of the protein Sen1Hel and indicate that stable RNA binding and robust RNA-DNA duplex unwinding requires a nucleic acid binding footprint of greater than or equal to 10 nt of 5′-overhanging ssRNA.
We next trapped a Sen1Hel RNA binding and translocation intermediate by varying ssRNA length and various nucleotides (ATP, ADP, AMP-PNP) in crystallization trials. Monoclinic crystals of the activated Sen1Hel core complexed with ADP and a 15 nt ssRNA substrate diffract to 3.0 Å resolution (Figure 2C-D, Table 2). The ssRNA binding groove localizes to the juncture of the RecA1-RecA2-β-barrel interface and accommodates 10 ordered ribonucleotides of the co-crystallized 15 nt poly-uridine (poly-U) substrate. In one molecule of the crystallographic asymmetric unit, ADP and a sulphate (SO4) ion bind at the RecA1-RecA2 interface. In the second Sen1 polypeptide, the ATP binding pocket is occupied by two sulphate molecules, in a conformation that is overall like the ADP-SO4 bound state. In this later molecule we surmise that as both the P-loop and the γ-phosphate binding site are bound with sulphate anions, that closure of the ATP-binding groove was stabilized during crystallization. For structural discussions we focused our analysis on ADP-SO4 bound molecule as in this configuration, ADP-SO4 mimics a hydrolyzed ATP, prior to release of inorganic phosphate29.
Table 2.
X-ray data collection and refinement statistics
| PDB code: 8FTM | SEN1Hel-RNA-ADP-SO4 |
| Wavelength (Å) | 1.000 |
| Resolution range (Å) | 50-3.0 |
| Space group | P21 |
| Unit cell | a=56.705 b=107.323 c=162.029 β=99.64 |
| Total reflections | 258925 |
| Multiplicity | 6.9 (6.9) |
| Completeness (%) | 99.1 (99.5) |
| Mean I/sigma(I) | 11.4 (1.3) |
| CC1/2 | 0.999 (0.872) |
| CC* | 1.000 (0.965) |
| R-meas | 0.121 (0.725) |
| R-work | 0.212 (0.310) |
| R-free | 0.251 (0.348) |
| Number of non-hydrogen atoms | 11842 |
| Protein residues | 1445 |
| RNA nucleotides | 22 |
| Ligands (ADP, SO4) | 10 |
| Water molecules | 6 |
| Protein residues | 1428 |
| RMS(bonds) | 0.002 |
| RMS(angles) | 0.52 |
| Ramachandran favored (%) | 96.01 |
| Ramachandran allowed/outliers (%) | 3.99/0.00 |
| Average B-factor (overall) | 91.08 |
| Protein | 90.87 |
| RNA | 96.11 |
| Ligands (ADP, SO4) | 102.20 |
| Water | 55.71 |
Statistics for the highest-resolution shell are shown in parentheses.
The Sen1Hel RNA binding interface is defined by a basic amino acid and Ser-Thr rich groove that traverses the paired Sen1 RecA homology motor domains (Figure 2E, Supplementary Figure 5). Sen1 utilizes a dual-grip configuration for ssRNA binding, similar to other helicases22,30. Within the 5′ grip, eight conserved residues (R1779, F1788, R1794, Y1734, S1698, T1761, D1763, and T1605) clasp six nucleotides (U1-U6). Contacts to the 5′-terminal end of the substrate kink and wrap the RNA around the distal end of RecA2. In the second grip, the four 3′-terminal nucleotides (U7-U10) are engaged exclusively by the RecA1 core (S1395, T1550, R1427, T1273, T1272, R1276 and H1556) and the RecA1-β-barrel insertion (N1270). A surface electrostatic view shows how Sen1Hel envelops ssRNA utilizing an electropositive RNA binding channel (Figure 2F). Close positioning of the 3′ terminus of ordered RNA next to the 1c-Prong suggests this motif is important for RNA-DNA strand separation during helicase unwinding. In addition, an electropositive surface proximal to the emergence point of the last ordered RNA 3′-nucleobase is appropriately positioned to accommodate the RNA-DNA hybrid that will be unwound as the enzyme translocates in the 5′ to 3′ direction. Consistent with this notion, docking using an RNA-DNA hybrid substrate (from PDB: 2QK9) in HADDOCK31 identifies possible duplex binding poses at this site (Figure 2G).
The Sen1 RNA translocation power stroke
The SF1B superfamily of 5′-3′ helicases and translocases have been broadly classified into two branches, the Pif1-like subfamily (e.g. Pif1, RecD) and the Upf1-like subfamily to which Sen1/SETX belongs22. A great deal of our understanding of SF1B helicases 5′-3′ translocation comes from seminal studies on the Pif1-like enzyme RecD232. Similar to RecD232, and the SF1A superfamily enzyme UvrD33 we hypothesize that ATP dependent motor domain ratcheting drives Sen1 RNA translocation. In this model, alternating nucleic acid grips operates in Sen1 to guide ssRNA binding and RNA translocation via tight and weak ssRNA binding within the RecA1 and RecA2 domains.
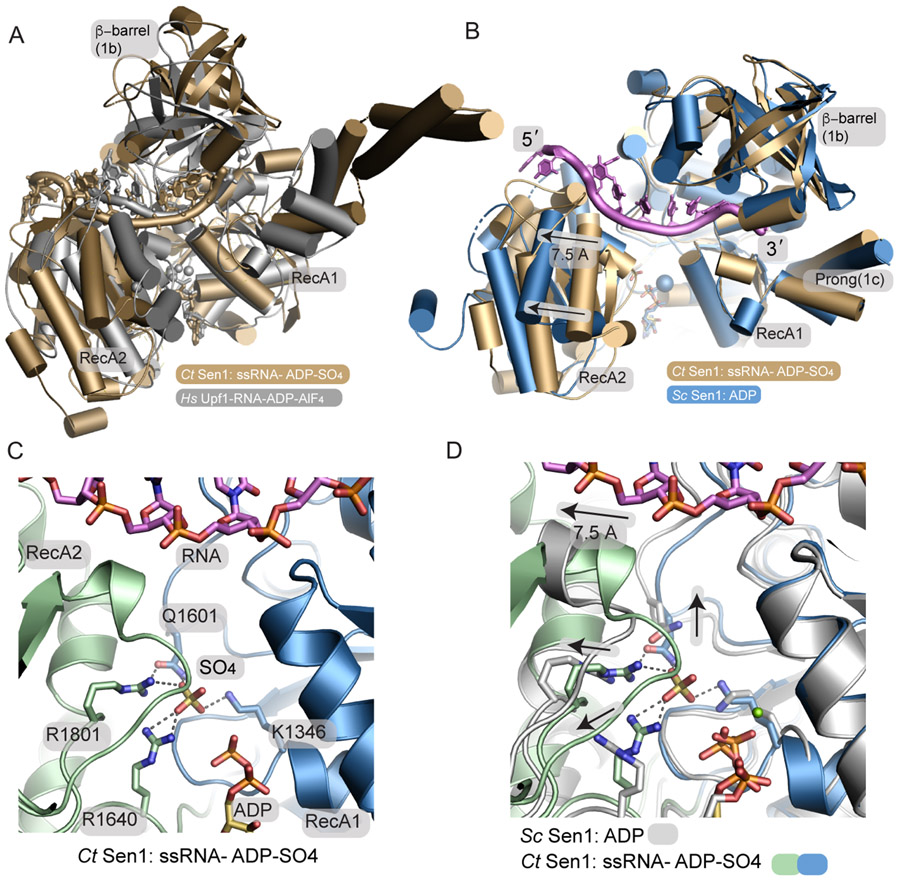
Compared to other structurally characterized helicases, Sen1 is most closely related to Upf127 and Ighmbp228, though it should be noted neither Upf1 nor Ighmbp2 have Sen1N-like N-terminal domains. Structures of Upf1 have been solved bound to non-hydrolyzable AMP-PNP, phosphate, and ADP bound states in the absence of RNA34. In addition, a Upf1-RNA-ADP-AlF4− transition state mimic has been trapped27. A structural overlay of the Sen1-ADP-SO4 complex with the Upf1-RNA-ADP- AlF4− transition state mimic shows how Sen1 has generally similar overall RNA binding characteristics (Figure 3A). The exception to this is the orientation of the β-barrel that in Upf1 in a distinct conformation compared to the Sen1. Separation of the RecA1-RecA2 domains of Sen1 is also greater compared to Upf1, possibly reflecting differences in a transition state mimic versus the post-hydrolyzed ADP-SO4 state of the Sen1-RNA complex.
Figure 3. A phosphate sensor is a lynchpin in the Sen1 power stroke.
(A) Structural superposition of the Sen1Hel–RNA–ADP–SO4 complex (tan) with the HsUpf1-RNA-ADP-AlF4 complex (grey).
(B) Structural superposition of the Sen1-RNA-ADP-S04 complex with ScSen1-ADP-Mg2+ complex, RSCB:5MZN. Close superposition of the RecA1 domains is observed. A 7.5 Å translocation is coincident with phosphate release. The blue sphere marks the position of Mg2+ in the ScSen1 -ADP-Mg2+ complex.
(C) Sen1 active site. Four residues from the RecA1 and RecA2 domains bind SO4, a mimic of the hydrolyzed phosphate prior to phosphate release.
(D) Active site structural superposition of ScSen1-ADP and CtSen1-RNA-ADP-SO4 complexes colored as indicated. Arrows show structural rearrangements of conserved gamma-phosphate sensing residues upon phosphate (SO4 mimic) release.
To better understand the Sen1 RNA translocation mechanism, we analyzed nucleotide-dependent conformational changes in the enzyme. Structural superposition of our CtSen1-RNA-ADP-SO4 quaternary complex with a previously determined ScSen1-ADP complex that crystallized in absence of nucleic acid23 (RCSB PDB 5MZN) reveals elaborate global Sen1Hel conformational changes that stem from local rearrangements in the nucleotide binding site (Figure 3B). In this superposition the RecA1 domains are well aligned overall, but we observe a concerted ~6-8 Å rigid body translation and a −30° rotation of the RecA2 domain relative to the RecA1 core. In the RNA-ADP-SO4 bound state determined here, the sulphate ion γ-phosphate mimic acts as a lynchpin that maintains interdomain contacts and is coordinated at the nexus of the RecA1 K1346 (Motif I, P-loop), Q1601 (Motif III), and RecA2 R1640 (Motif IIIa) and R1801 (Motif VI) (Figure 3C and Supplementary Figure 5). By comparison, in the ADP-bound RNA-free state of ScSen1, absence of the γ-phosphate is coincident with the collapse of this interdomain salt-bridging network. Local side chain rearrangements and a global shift in the RecA2 domain are positioned to rachet bound RNA (Figure 3C and 3D). Overall, the conformational differences observed are appropriate to drive a stepwise 1 nt helicase ratcheting during the helicase power stroke by pulling the RecA2 bound 5′-end away from RecA1 and facilitate 5′-3′ RNA translocation coincident with phosphate release. In addition, RecA1 Arg 1427 (Motif 1b) is positioned to serve as a ratchet “pawl” akin to nucleobase stacking interactions observed for mycobacterial AdnAB helicase35,36 (Figure 2E).
Determinants of Sen1 autoinhibition
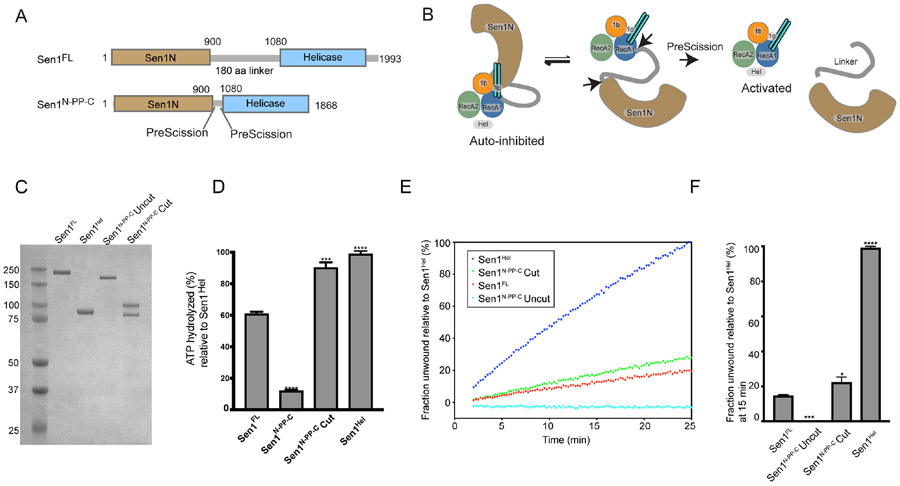
As the Sen1Sen1N domain negatively regulates catalytic activity of the full-length protein (Figure 2A), we hypothesized the full-length structure characterized here reflects an auto-inhibited state of the enzyme. We further reasoned that tethering of Sen1Sen1N to the Sen1Hel domain via a flexible linker facilitates reversible binding of the Sen1N domain to the helicase core (Figures 4A and 4B). To evaluate regulatory roles of the Sen1Sen1N and the IDR linker, we engineered a Sen1 internal truncation (Sen1N-PP-C) that replaces the native ~180 amino acid linker with a tandem PreScission-protease cleavable tether (Figure 4A-C). This design is facilitated by the close juxtaposition of the C-terminal end of Sen1Sen1N and the N-terminal flank of Sen1Hel. Absent nucleic acid, Sen1Hel harbors no detectable ATPase activity under the conditions examined (Supplementary Figure 3D). However, the addition of 30 nt ssRNA (30U) 44 nt ssRNA (44U) (but not 15 nt ssRNA, 15U) all stimulated Sen1 ATPase hydrolysis (Supplementary Figure 3D, Supplementary Table 2). Likewise, a helicase substrate RNA-DNA hybrid stimulates ATPase activity as expected (Figure 4D). Strikingly, truncation of the linker results in marked attenuation of Sen1 nucleic acid stimulated ATPase (Figure 4D) and RNA-DNA unwinding activities (Figures 4E and 4F) compared to Sen1FL or Sen1Hel. However, Sen1N-PP-C autoinhibition is reversible, as protease cleavage liberates the catalytic domain from Sen1Sen1N as engineered (Figure 4C), and partially restores ATPase and helicase activities (Figures 4D-F). Thus, covalent tethering of Sen1Sen1N facilitates robust Sen1 autoinhibition. Moreover, interdomain linker length is tuned for reversible suppression of Sen1 helicase and ATPase activities as shortening of the linker length results in robust autoinhibition.
Figure 4. The Sen1N domain is autoinhibitory.
(A) Design of a PreScission Protease cleavable Sen1.
(B) Schematic of activation of PreScission Protease cleavable Sen1N-PP-C.
(C) Purified Sen1 proteins or PreScission Protease cleaved protein (Sen1N-PP-C – Cut) used in panels D–F.
(D) Substrate stimulated ATP hydrolysis of mutant Sen1 proteins was monitored in a phosphate release assay. Sen1FL, Sen1N-PP-C (Uncut), PreScission Protease-cleaved Sen1N-PP-C (Cut) and Sen1Hel (5 nM) were incubated with Sub6 (Supplementary Table 3) at 5 μM and ATP (1 mM) and MgCl2 (2 mM) for 15 min at 37 °C. Error bars are SD from 3 replicates, ***p<0.001, **** p<0.0001.
(E) Helicase assay of indicated Sen1 protein (1 nM) was performed as in Figure 2A and normalized to unwound fraction of Sen1Hel for comparison.
(F) Comparison of unwound fractions at 25 min from experiments in “E”. Error bars are SD from 3 replicates. Statistical analysis was done using paired t-test relative to Sen1FL. *p<0.05, ***p<0.001, ****p<0.0001.
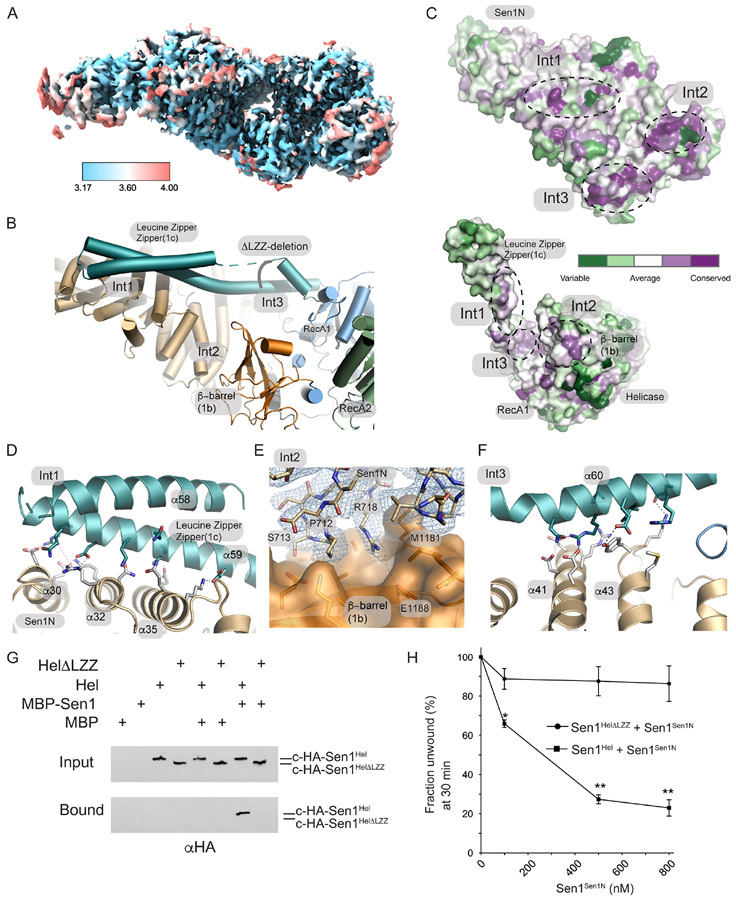
A detailed analysis of enzyme conformational changes associated with auto-inhibition was initially precluded as the resolution of our Sen1FL reconstruction was limited (Supplementary Figure 2). We reasoned that the Sen1N-PP-C internal deletion construct might also be of utility in structural determination of the autoinhibited state given this construct is strongly autoinhibited. Indeed Sen1N-PP-C particles yielded a 3.69 Å (3.17 Å masked FSC) cryo-EM map suitable for tracing the Sen1 polypeptide, in the presence of ADP (Figure 5A, Supplementary Figure 6). The Sen1N-PP-C structure shows how a 3-point contact of the Sen1Sen1N domain stabilizes an inhibited conformation of Sen1Hel. The three Sen1Sen1N- Sen1Hel interaction interfaces (Int1-Int3, Figure 5B) contribute to an extensive (2686 Å2 buried solvent accessible surface) interaction interface between the domains. An analysis of the conservation of the Sen1Sen1N domain surface from 39 Sen1 homologs using ConSurf37 shows how the Int1-Int3 surfaces on the helical repeat that are utilized for Sen1Hel engagement all correspond to conserved regions of the protein (Figure 5C). A major feature of the Sen1Hel-Sen1Sen1N interaction interface is the extended LZZ RecA1 insertion (aa 1457-1525) that binds to the Sen1Sen1N helical array at Int1 (Figure 5B, Supplementary Figure 4 and Supplementary Figure 5). Several complimentary salt bridging interactions are formed from LZZ helix α59 to the Sen1Sen1N domain helices α30, α32 and α35 (Figure 5D). Int2 involves a direct interaction with the Sen1 β-barrel (1b) insertion and is composed of a mixture of salt bridging interactions (e.g. R718 to E1188) and hydrophobic interactions (Figure 5E). A core conserved element of Int2 is a short helical element initiated by P712 that binds in a shallow pocket on the β-barrel domain. Like Int1, Int3 involves the 1c-prong-LZZ binding (α60) from the helicase domain via a series of complementary salt bridges with the Sen1Sen1N helical repeat (α41 and α43) (Figure 5F). Overall, the inter-domain interactions are mediated by an array reversible salt-bridging contacts that secure the catalytic domain in a conformation that appears incompatible with RNA-DNA substrate engagement.
Figure 5. Cryo-EM structure of Sen1N-PP-C.
(A) Local resolution of Sen1N-PP-C, calculated in using the Local Resolution Estimation in CryoSparc, and displayed using a blue (high-resolution, 3.17 Å) to red (low-resolution, 4.0 Å) color-coded surface.
(B) Three interaction interfaces (Int1-Int3) mediate Sen1Sen1N interaction with Sen1Hel. The LZZ from Sen1Hel binds both Int1 and Int3 on Sen1Sen1N. A ΔLZZ internal deletion construct is marked.
(C) Consurf analysis of the Sen1Sen1N surface reveals conserved surface interaction interfaces (Int1-Int3, dotted lines) are involved in interactions with Sen1Hel. Top: Sen1N domain interaction interfaces that bind the helicase domain. Bottom: Helicase domain interaction surfaces that bind the Sen1N domain.
(D) Molecular details of the Int1 interdomain Sen1Sen1N - Sen1Hel interaction interface.
(E) Cryo-EM density for Sen1Sen1N Int2 region displayed contoured at 11.0 σ overlaid upon a molecular surface diagram of the β-barrel domain (orange).
(F) Molecular details of the Int3 interdomain Sen1Sen1N - Sen1Hel interaction interface.
(G) MBP-pulldowns show the LZZ is critical for the Sen1Sen1N–Sen1Hel interaction.
(H) Trans inhibition of Sen1Hel by Sen1Sen1N. Sen1Hel (1 nM) was pre-incubated with Sen1Sen1N at the indicated concentrations for 5 min on ice. Reactions were initiated by addition of 50 nM Sub7 (Supplementary Table 3) and a mixture containing ATP (1 mM) and MgCl2 (2 mM). The percentage of unwound ssDNA was calculated from the FAM signal at 30 min. Error bars are SD from 3 replicates, **p<0.01, *p<0.05.
To directly test functional roles of the Sen1Sen1N-Sen1Hel interface, we probed interactions between Sen1Sen1N and the Sen1Hel helicase core. C-terminally hemagglutinin-tagged helicase domain (c-HA-Sen1Hel) bound to column immobilized MBP-Sen1Sen1N, but not to MBP controls (Figure 5G). However, a Sen1Hel mutant with an internal deletion mutation within the LZZ ( Sen1Hel-ΔLZZ, Δ1457-1525, Figure 5H) that deletes all Int1 interdomain contacts and a portion of helicase domain residues participating in Int3 fails to bind Sen1Sen1N, consistent with a critical role for the LZZ in mediating the interdomain interaction. Given the direct interaction between regulatory and catalytic domains, we next tested if Sen1Sen1N can modulate Sen1Hel helicase activity in trans. Titration of purified Sen1Sen1N suppresses Sen1Hel unwinding activity to ~25% of WT at 800 nM Sen1Sen1N (Figure 5H). Furthermore, deletion of the LZZ does not impair helicase activity, but renders the catalytic core immune to Sen1Sen1N mediated auto-inhibition in trans. Thus, we conclude that direct binding of the Sen1Sen1N to the catalytic core is stabilized by the LZZ, and that the LZZ facilitates autoinhibition.
Sen1Sen1N regulates Sen1Hel substrate engagement
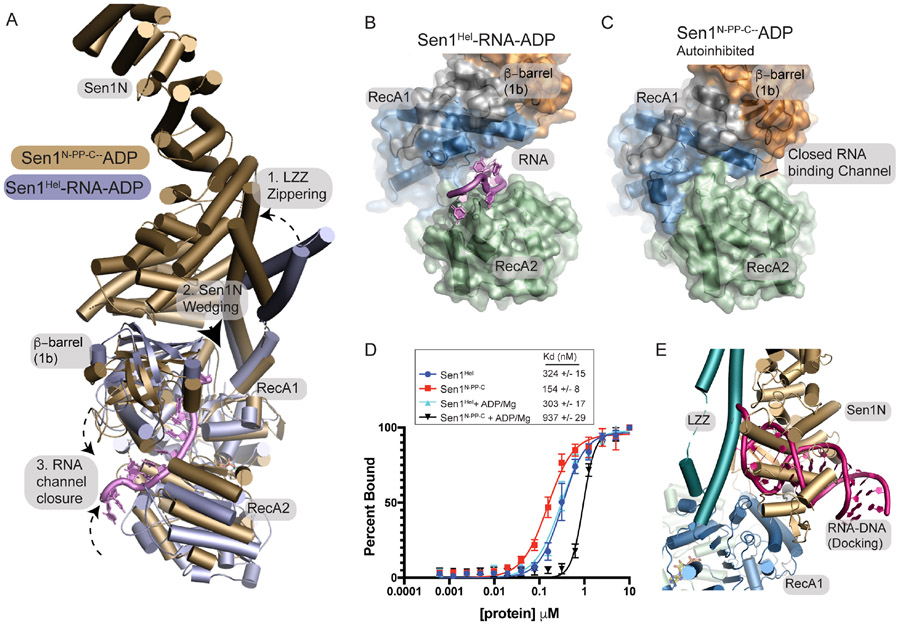
To better understand the mechanism of helicase regulation, we examined structural superpositions of the autoinhibited Sen1N-PP-C–ADP structure with the Sen1Hel–RNA–ADP complex. A morph between these two states reveals a complex cascade of rearrangements in Sen1Hel that is coincident with Sen1Sen1N binding (Supplementary Movie 1). The salient features of this transition include: 1) LZZ “zippering” of the Sen1Sen1N domain and conformational change of the LZZ upon binding Sen1Sen1N, 2) Wedging of the Sen1Sen1N domain between the RecA1 β-barrel and RecA1 core domain junction, and 3) Closure of the RNA binding channel (Figure 6A). In the RNA bound crystal structure, the LZZ insertion is ordered for the two molecules in the asymmetric unit but stabilized by crystal contacts. Compared to the RNA bound state, the LZZ undergoes significant reorganization when engaged with Sen1Sen1N, consistent with the key role for the LZZ in stabilizing Sen1Hel–Sen1Sen1N interactions (Figure 6A). Sen1Sen1N wedging pushes the β-barrel towards the RNA binding channel. Coincident with this motion, an array of conformational changes are associated with complete rearrangement of the RNA binding cleft. Visualization of the Sen1 molecular surface shows how the ssRNA binding channel is completely blocked in the Sen1N-PP-C-ADP complex structure (Figures 6B and 6C). Structural comparisons of the RNA-bound and inhibited state indicates that blockage of the RNA binding channel is achieved by a series of conformational changes in the RNA binding loops that reorganize to engage the β-barrel, and sterically occlude the ssRNA substrate binding cleft.
Figure 6. Mechanism of Sen1 auto-inhibition.
(A) Structural superposition of Sen1N-PP-C and the Sen1Hel RNA complex reveals a cascade of conformational rearrangements associated with autoinhibition.
(B) Surface diagram of the Sen1-RNA complex and Sen1N-PP-C shows alterations in the RNA binding channel in the closed state “C”.
(D) Fluorescence polarization ssRNA binding. Binding to ssRNA was conducted using enzyme titration and monitoring fluorescence polarization from Sub8 (Supplementary Table 3. Error bars reflect SD from 3 replicates.
(E) Superposition of an example HADDOCK docking binding pose and the autoinhibited Sen1N-PP-C state.
To test if autoinhibition regulates substrate binding, we compared equilibrium substrate binding of Sen1Hel and Sen1N-PP-C to a 40 nt ssRNA using fluorescence polarization (FP) (Figure 6D). Consistent with the structural observations, RNA binding was significantly impaired in the autoinhibited Sen1N-PP-C protein construct (Figure 6D), but only in the presence of ADP nucleotide. In the absence of nucleotide, Sen1N-PP-C binds ~2-fold more strongly than the isolated helicase domain to a 40 nt ssRNA. We speculate that the Sen1N bound catalytic domain can bind to a short ssRNA, by engaging an opened RNA binding cleft. Overall, our structural and biochemical results show that direct interactions of the Sen1Sen1N with Sen1Hel suppresses RNA/DNA dependent ATPase and RNA-DNA unwinding activity of Sen1Hel and reveal that autoregulation of the enzyme involves dynamic intramolecular binding of Sen1Hel to Sen1Sen1N that stabilizes nucleotide dependent states which are incompatible with RNA substrate engagement. Interestingly, we also note that a proposed RNA-DNA hybrid binding site determined by nucleic acid docking (Figure 2G) overlaps completely Sen1Sen1N domain in the autoinhibited state, suggesting that Sen1Sen1N might also compete directly with RNA-DNA substrate binding by mimicking the heteroduplex region of an RNA-DNA hybrid (Figure 6E).
AOA2 mutants impair Sen1 RNA binding and helicase activity
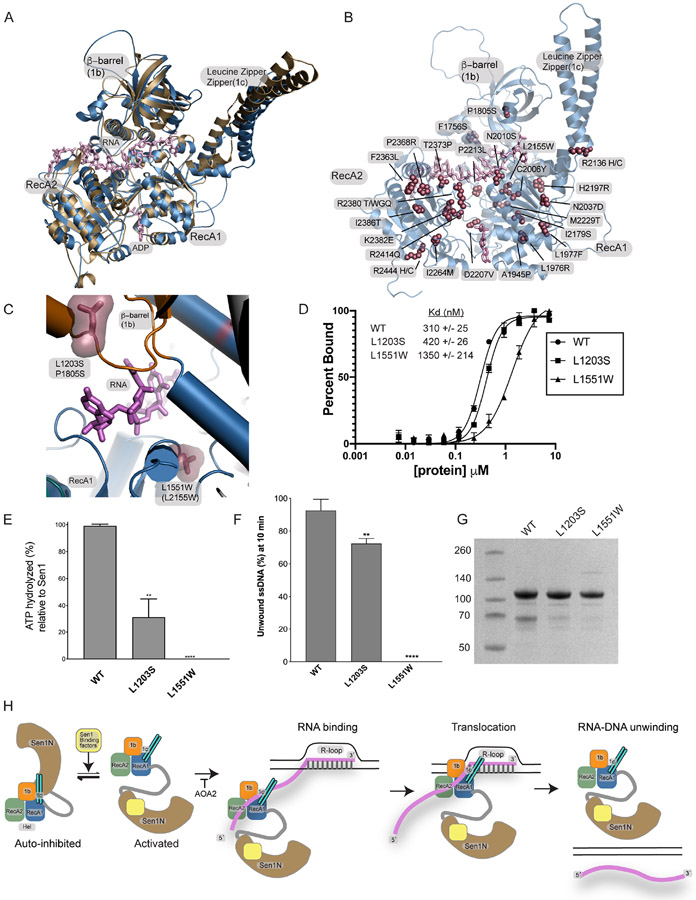
We analyzed human AOA2 catalytic core mutants in the context of the CtSen1Hel-RNA complex structure (Figure 7, Supplementary Figure 4, and Supplementary Table 1). An AlphaFold model38 of the hSETX catalytic core shows close similarity to our CtSen1-RNA complex structure (Figure 7A). We used both structures to make predictions of the potential impacts of SETX missense variants (Figure 7B, Supplementary Table 1). Many of the mutations (F1756S, A1945P, L1976R, L1977F, N2037D, I2179S, H2197R, M2229T, I2264M, I2386T, R2444H, R2444C) would be predicted to impact protein folding. Several mutants map directly to the ssRNA binding region or proximal to structural elements contacting ssRNA (P1805S, C2006Y, L2155W, N2010S, P2213L, T2373P) or the predicted RNA/DNA hybrid binding site near the base of the LZZ (R2136H, R2136C). A third class of mutations are found close to the ATP binding site, and therefore may impact ATP binding and/or hydrolysis (D2207V, R2414Q). Given the proximity to the RecA1-RecA2 interface and their participation in structural salt bridging interactions close to the ATP binding site, 5 substitutions (R2380T, R2380W, R2380G, R2380Q, K2382E), might impact protein folding and/or ATP regulated conformational changes. Lastly, two mutations cluster to a surface on the RecA2 domain and may mark a protein binding surface of unknown function (F2363L, P2368R).
Figure 7. AOA2 mutants Sen1 RNA binding mechanism.
(A) Structural overlay of an Alphafold model of human SETX helicase domain (blue) with the CtSen1 RNA complex (tan).
(B) Alphafold model of the hSETX helicase domain showing the location of mapped AOA2 and ALS4 mutations
(C) Two AOA2 mutations map to the RNA binding cleft. Human equivalent positions are shown below the CtSen1 numbering in parentheses and the CtSen1 structure is displayed.
(D) Fluorescence polarization ssRNA binding. Binding to ssRNA was conducted using enzyme titration and monitoring fluorescence polarization from Sub8 (Supplementary Table 3). Error bars reflect SD from 3 replicates.
(E) ATPase activity. Proteins (5 nM) were incubated with Sub6 (Supplementary Table 3) and ATP (1 mM) at 5 μM for 15 min at 37 °C. The L1551W mutant had no measurable activity. Error bars are SD from 3 replicates, **p<0.01, ****p<0.0001.
(F) RNA—DNA unwinding activity. Proteins (1 nM) were incubated with 50 nM Sub7 (Supplementary Table 3), ATP (1 mM) and MgCl2 (2 mM). FAM signal at 10 min was used to compare the unwinding activities of Sen1Hel mutants. The L1551W mutant had no measurable activity. Error bars are SD from 3 replicates, **p<0.01, ****p<0.0001.
(G) Purified Sen1Hel proteins used in panels B–D.
(H) Model for Sen1 autoinhibition and RNA translocation.
Our attention focused on two mutations that map to the RNA binding cleft characterized here. P1805S (L1203S in CtSen1) and L2155W (L1551W in CtSen1) mutations directly flank the structurally characterized RNA binding channel (Figure 7C). In FP ssRNA binding assays using a FAM-labeled substrate, compared to WT Sen1Hel (Kd= 310 nM), Sen1Hel-L1203S (Kd= 420 nM) had a modest impact on substrate equilibrium binding (Figure 7D). By comparison, the L1551W substitution that we would predict to distort the base of the ssRNA binding cleft bound ssRNA weakly (Kd=1350 nM) (Figure 7D). We find that both mutations impair substrate stimulated ATPase activity (Figure 7E), and that the L1551W mutant has a severe impact on RNA-DNA unwinding activity (Figure 7F). Thus, we conclude that the integrity of the ssRNA binding cleft characterized here is compromised in a subset of AOA2 mutations linked to neurodegenerative disease.
DISCUSSION
Herein, we define the global architecture of the Sen1 RNA-DNA helicase. Our results establish that interactions of Sen1Sen1N with Sen1Hel regulates helicase activity in cis (Figure 7H). The N-terminal helical autoregulatory domain directly engages the Sen1 catalytic core through extensive molecular interactions. Sen1Sen1N deletion activates the protein in vitro, and the Sen1Sen1N helical repeats suppress Sen1 catalytic activity through nucleotide dependent allosteric regulation of the helicase RNA-DNA hybrid binding channel. This regulatory mechanism involves both modulation of ATPase activity and RNA binding and translocation, with the major effect being on the later. It is also probable that Sen1Sen1N binding to Sen1Hel restricts the interdomain motions regulated by ATP-induced conformational changes, thereby rendering the catalytic core arthritic, preventing molecular motions that underpin the Sen1 RNA-DNA unwinding chemistry. A critical feature of this interaction is flexible tethering of Sen1Sen1N to Sen1Hel, and the length of the intrinsically disordered tether is an important regulator of helicase inhibition. The IDR region is comprised of “low complexity” sequence and predicted to be disordered (Supplementary Figure 1D). Notably, there are stretches of conserved sequences present in the IDR. In addition to serving as a flexible regulatory tether, the interdomain linker might also be directly involved in regulating enzyme activation by binding regulatory partners, or to nucleic acid directly.
Our Sen1 structures also provide insights into Sen1 mutations mapping to the N-terminal domain that impact protein-protein interaction and have pleiotropic effects on Sen1 transcriptional regulation and gene expression. The Sen1-K128E substitution blocks interaction with Rnt1p ribonuclease and Sen1-R302W impairs binding to Rbp1p, the large subunit of yeast RNA polymerase II39. Sen1-K128E and Sen1-R302W variants differentially affect Sen1 functions in RNA processing and transcription respectively39. An additional sen1-3 allele Sen1Sen1N triple mutation (W773A, E774A, W777A) most likely disrupts Sen1Sen1N protein folding and specifically impacts Sen1 recruitment to the DNA replisome and protein-protein interactions with Ctf4 and Mrc119. Thus, the collective data point to a complex regulatory role for the Sen1N region in regulating protein-protein interactions and modulating Sen1 catalytic activity. We hypothesize that Sen1Sen1N interactions with protein partners such as RNA polymerase or Ctf4 facilitate both Sen1 recruitment to RNA-DNA targets, and regulation of Sen1 catalytic activities. That the majority of autoinhibitory contacts from the Sen1N domain are assembled by salt-bridging networks is consistent with a role for these interactions in mediating a reversible regulatory switch. Consistent with this hypothesis the equivalent region to the CtSen1 LZZ is important for mediating yeast Sen1 transcriptional termination activity and interaction with RNA polymerase23. Thus, the Sen1Sen1N and Sen1Hel domain surfaces participating in autoinhibition are likely involved in regulating transition to the active helicase state depending on protein binding partner context. Sen1 is most like the nonsense mediated decay Upf1 RNA helicase. With respect to the autoregulatory mechanism, in Upf1, an N-terminal CH domain dynamically regulates activity of the core helicase domain in concert with the trans binding factor Upf227,40. We posit that Sen1Sen1N binding factors recruit Sen1 to RNA-DNA hybrids and may regulate Sen1Sen1N- helicase dynamics.
The X-ray structures of the Sen1 catalytic core further reveal conserved determinants for its RNA binding and translocation mechanism. Like other nucleic acid helicases22,33, the Sen1 helicase mechanism involves conformational changes governed by ATP binding and hydrolysis, as well as dynamic remodeling of the RNA-DNA binding surface. While the ssRNA binding site is well defined in our crystal structures, it remains an open question as to how the enzyme engages the duplex portion of the substrate. Docking analysis suggests that a suitable site for RNA-DNA hybrid engagement is found on the β-barrel SF1B insertion. The positioning of this site is conspicuous as it directly overlaps with the Sen1Sen1N binding site on the catalytic domain observed in the autoinhibited conformation. Thus, we hypothesize that helicase regulation also involves direct mimicry of the nucleic acid binding surface by Sen1Sen1N. Like budding yeast Sen1, CtSen1 also unwinds both RNA-DNA and DNA-DNA duplexes. Consistent with these observations, inspection of the Sen1-RNA complex structure reveals that no specific protein-nucleic acid contacts appear suited to differentiate RNA from DNA. Sen1 DNA helicase activity may be relevant to DNA replication and repair roles for Sen1 homologs, consistent with recruitment of the protein into DNA replication and repair machineries8,9,19. More work is needed to assess the structural and multifunctional roles for Sen1 RNA-DNA and DNA-DNA unwinding activities.
Limitations of this study
There are limitations to the current study. It is possible that regulation of vertebrate SETX via the N-terminal domain is conserved with Sen1. However, the inability to produce suitable quantities of full-length human SETX has precluded detailed analysis of the conservation of this mechanism to date. In our analysis of ATP-dependent conformational control of Sen1 function we have inferred mechanism using states from two different species captured in different crystalline states. Additional work is required to further characterize the nature of Sen1 interactions with regulatory factors, to decipher the basis for additional Sen1 and vertebrate SETX RNA-DNA and DNA-DNA unwinding reaction and ATP-dependent translocation intermediates, and to define the impacts of AOA2 and ALS4 mutations on these RNA-DNA transactions.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, R. Scott Williams (williamsrs@niehs.nih.gov).
Materials Availability
All materials generated by this study are listed in the Key Resources table and are available from the Lead Contact without restriction.
Key Resource Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit Anti-MBP | GeneTex | GTX124267 |
| Mouse Anti-HA | GeneTex | GTX628489 |
| IRDye680 Goat Anti Rabbit | LI-COR | 926-68071 |
| IRDye800 Goat Anti Mouse | LI-COR | 926-32210 |
| Bacterial and virus strains | ||
| BL21-AI E. coli | Invitrogen | Cat# C607003 |
| HEK 293 Freestyle | Thermo Fisher | Cat# R79007 |
| Chemicals, peptides, and recombinant proteins | ||
| L-Arabinose | GoldBio | A-300-25 |
| Lipofectamine ™ 2000 Transfection Reagent | Thermo Fisher | 11668019 |
| Recombinant CtSen1FL (full length) | This paper | N/A |
| Recombinant CtSen1N-PP-C (1-900-PreScission-PreScission-1080-1868) | This paper | N/A |
| Recombinant CtSen1Sen1N (1-900) | This paper | N/A |
| Recombinant CtSen1Hel (1087-1890) | This paper | N/A |
| Recombinant CtSen1HelΔLZZ (1087(Δ1457-1525)-1890) | This paper | N/A |
| Recombinant CtSen1Hel-1878 (1087-1878) | This paper | N/A |
| Recombinant CtSen1Hel mutants (L1203S and L1551W) | This paper | N/A |
| Critical commercial assays | ||
| QuickChange Site-Directed Mutagenesis Kit | Agilent | 200519 |
| Q5 Mutagenesis Kit | NEB | E0552S |
| Malachite Green Phosphate Assay Kit | Cayman | 10009325 |
| Deposited data | ||
| CtSen1FL, cryo-EM map and refined coordinates | This paper | EMD: 29439, PDB: 8FTK |
| CtSen1Sen1N, cryo-EM map | This paper | EMD: 29440 |
| CtSen1N-PP-C, cryo-EM map and refined coordinates | This paper | EMD: 29426, PDB: 8FTH |
| CtSen1Hel-RNA-ADP-SO4, X-ray data and refined coordinates | This paper | PDB: 8FTM |
| Uncropped gels, Mendeley data | This paper | doi:10.17632/djm6wdc5tg.1 |
| Oligonucleotides | ||
| CtSen1Hel L1203S-fwd (AGTAAGGATCGCGGTAGTACCGAAGGCGACATC) | This paper | N/A |
| CtSen1Hel L1203S-rev (GATGTCGCCTTCGGTACTACCGCGATCCTTACT) | This paper | N/A |
| CtSen1Hel L1551W-fwd (GTTCTGTGTGCCACGTGGAGTGGCAGCGGCCAC) | This paper | N/A |
| CtSen1Hel L1551W-rev (GTGGCCGCTGCCACTCCACGTGGCACACAGAAC) | This paper | N/A |
| CtSen1Hel-1878-fwd (GTGGAAATGCACGACTAATGACGTACGAGTGAA) | This paper | N/A |
| CtSen1Hel-1878-rev (TTCACTCGTACGTCATTAGTCGTGCATTTCCAC) | This paper | N/A |
| CtSen1Hel-cHA-fwd (GCCGGATTATGCGTAATGAAAGGGTGGGCGCGC) | This paper | N/A |
| CtSen1Hel-cHA-rev (ACATCATACGGATATGGCATCGCCGGCGGGAT) | This paper | N/A |
| Substrate oligos for RNA-DNA unwinding assays | Supplementary Table 3 | N/A |
| Substrate oligos for ATPase assays | Supplementary Table 2 | N/A |
| Substrate oligos for EMSAs | Supplementary Table 3 | N/A |
| Substrate RNAs for crystallization | Supplementary Table 2 | N/A |
| Substrate RNA for fluorescence polarization RNA binding | Supplementary Table 2 | N/A |
| Recombinant DNA | ||
| CtSen1FL (full length) | This paper | N/A |
| CtSen1N-PP-C (1-900-PreScission-PreScission-1080-1868) | This paper | N/A |
| CtSen1Sen1N (1-900) | This paper | N/A |
| CtSen1Hel (1087-1890) | This paper | N/A |
| CtSen1HelΔLZZ (1087(Δ1457-1525)-1890) | This paper | N/A |
| CtSen1Hel-1878 (1087-1878) | This paper | N/A |
| CtSen1Hel mutants (L1203S and L1551W) | This paper | N/A |
| Software and algorithms | ||
| PHENIX | Adams et al.47 | N/A |
| Coot | Emsley and Cowtan45 | N/A |
| HKL-2000 | HKL Research Inc. | N/A |
| PyMOL | Schrödinger | N/A |
| Prism 8.0 | GraphPad | N/A |
| TOPAZ | Bepler et al.44 | N/A |
| Emperia Imaging Software | LI-COR | N/A |
| AlphaFold | Senior et al.46 | N/A |
| CryoSparc v2.7 | Punjani et al.42 and Punjani et al.43 | N/A |
| MotionCorr2 | Zheng et al.41 | N/A |
| Smartscope | Bouvette et al.48 | N/A |
Data and Code Availability
The data presented in this paper will be shared by the lead contact upon request. Uncropped gels are available at Mendeley at doi:10.17632/djm6wdc5tg.1. Cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB). X-ray data and refined molecular models have been deposited in the RSCB Protein Data Bank (PDB). EMDB and PDB Accession codes are available in the key resources table. All datasets will be publicly accessible at the date of publication.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental Model and Study Participant Details Bacterial strain
BL21-AI cells (Life Technologies) were transformed with the constructs for protein expression overnight at 15°C in Terrific Broth. Induction of expression was carried out by the primary addition of 0.1% (w/v) final concentration L-Arabinose (GoldBio).
Mammalian cell lines
HEK 293 Freestyle (HEK293F) cells were used to express YFP fusion full length Sen1 proteins. Stable cell lines were created by Lipofectamine 2000 (Thermo Fisher) followed by rounds of blasticidin selection.
METHOD DETAILS
Chaetomium thermophilum Sen1 construct designs
The Chaetomium thermophilum Sen1 homolog CtSen1FL (UniProt G0S163_CHATD), CtSen1Sen1N (aa 1-900), CtSen1Hel (aa 1080-1890), Sen1Hel-ΔLZZ (aa 1080-(Δ1457-1525)-1890), and CtSen1N-PP-C (CtSen1 aa 1-900-LEVLFQGPLEVLFQGP-1080-1868) were all ordered from Genewiz/Azentra as synthetic genes in pUC57-Kan vector. The synthetic plasmids were designed to contain a Tobacco Etch Virus (TEV) recognition site directly upstream of the gene and the entirety was flanked by AttL recombination sites for LR subcloning into Gateway destination vectors for expression as N-terminally-tagged fusion proteins. CtSen1FL and CtSen1N-PP-C were codon optimized for mammalian expression and subcloned into the ampicillin resistant (AmpR) plasmid pcDNA6.2 N-EmGFP-DEST (Invitrogen). CtSen1Sen1N, CtSen1Hel, Sen1Hel-ΔLZZ were codon optimized for bacterial expression and subcloned into pDEST-His-MBP (AmpR, Addgene). All plasmids were confirmed by DNA sequencing (Genewiz/Azentra). QuikChange site-directed mutagenesis (Stratagene) was used to create the crystallization construct CtSen1Hel-1878, the CtSen1Hel mutants L1203S and L1551W, and the CtSen1Hel and CtSen1Hel-ΔLZZ C-terminal-HA fusions.
Protein expression and purification
HEK293F cells stably overexpressing N-terminal YFP fusion proteins, CtSen1FL and CtSen1N-PP-C, were created by Lipofectamine 2000 (Thermo Fisher) transfection followed by rounds of blasticidin selection. Protein was isolated from suspension culture via anti-GFP/YFP single domain camelid nanobody (sdAB) as previously described26 and released from the nanobody by on-column TEV protease digestion in Sen1 storage buffer (20 mM Tris pH 7.9, 200 mM NaCl, 2 mM MgCl2, 1 mM TCEP). Protein was further purified by size-exclusion chromatography in Sen1 storage buffer followed by high resolution anion exchange chromatography on a Cytiva MonoQ 10/100GL column in ion exchange buffers (Buffer A: 20 mM Tris pH 7.9, 1 mM TCEP and Buffer B: 20 mM Tris pH 7.9, 1 M NaCl, 1 mM TCEP). Sen1FL elutes at ~140 mM NaCl. Protein was then buffer exchanged into Sen1 storage buffer using Millipore spin 10,000 MW cutoff concentrators. For experiments where CtSen1N-PP-C was digested with PreScission protease (GE Healthcare/Cytiva) to yield separate N-terminal and C-terminal domains, 1 μL of 0.8 mg mL−1 protease was added to 15 μL of 2 mg mL−1 protein and incubated for 1 hour at room temperature.
CtSen1Sen1N, CtSen1Hel, CtSen1HelL1203S, CtSen1HelL1551W, CtSen1Hel-1878, and Sen1Hel-ΔLZZ proteins were expressed as N-terminal His-MBP fusions. BL21-AI cells were transformed with constructs for protein expression overnight at 15°C in Terrific Broth. Induction of expression was carried out by the primary addition of 0.1% (w/v) final concentration L-Arabinose. Following amylose affinity chromatography performed in Sen1 storage buffer for binding and (plus 30 mM maltose) for protein elution, proteins were subjected to overnight TEV protease digestion for MBP tag removal prior to ion exchange chromatography in ion exchange buffers (see above). For pull-down assays using MBP-CtSen1Sen1N, TEV digestion was omitted. CtSen1Hel constructs and mutants were purified by cation exchange chromatography (Cytiva HiTrap SP column) in Buffer A and Buffer B, described above and polished by gel filtration (Cytiva Superdex 200 Analytical 10/300 GL) in Sen1 storage buffer. CtSen1Sen1N constructs, both MBP-tagged and TEV digested, were purified by anion exchange chromatography (Cytiva HiTrap Q column) and polished by gel filtration as described for the CtSen1Hel proteins. Final purity was assessed by SDS-PAGE and fractions pooled and concentrated for subsequent experiments.
Limited proteolysis
CtSen1FL was subjected to limited proteolysis, on ice, in 10 mL reactions containing 1 mg mL−1 CtSen1FL combined with 1 mg mL−1, 0.1 mg mL−1 or 0.01 mg mL−1 trypsin. Reactions were quenched at 5, 15, 30, or 60 minutes by the addition of 2 mM PMSF and Laemmli SDS-PAGE dye followed by incubation at 95°C for 10 minutes. Trypsinized protein was resolved by SDS-PAGE and individual bands excised for peptide identification by LC-MS/MS spectrophotometry. Gel bands were digested using a Progest robotic digester (Genomic Solutions, Ann Arbor, WI). Minced gel bands were incubated twice for 15 min in 100 μL of 25 mM ammonium bicarbonate/50% (v/v) acetonitrile. The gel was then dehydrated by a 20-min incubation in 100 μL of acetonitrile followed by drying under a nitrogen stream. Then, 270 ng of trypsin (Promega) was added followed by an 8 min incubation at 37 °C. The supernatants from the digests were collected and the gel was re-extracted three times: once with 50 μL of water for 20 min and twice with 20-min incubations in 50 μL of 5% (v/v) formic acid/50% (v/v) acetonitrile. All of these extractions were pooled, lyophilized, and resuspended in 10 μL of 1:1 water with 0.1% formic acid:acetonitrile with 0.1% formic acid. Samples were spotted (0.3 μL) onto a stainless-steel target. A 33% saturated solution of alpha-cyano-hydroxycinnamic acid in 50:50 (v/v) 0.1% formic acid: 0.1% formic acid in acetonitrile was added (0.7 μL) to the sample and mixed on target. MALDI-TOF and MALDI-TOF/TOF (top 6 ions selected for MS/MS) experiments were performed on an Applied Biosystems 4800 Plus MALDI TOF/TOF Analyzer in positive ion reflector modes. The MS was first calibrated externally using the Bruker Daltonics Peptide Calibration Mixture and the MS/MS calibrated externally using the fragment ions of the angiotensin I M+Sen ion (m/z 1298.68). If detected in the sample, autolytic tryptic peaks were used for further internal calibration in the MS mode. A focus mass of m/z 1600 was used for the MS acquisitions. For MS/MS 1000 kV was used for the collision energy. Data were processed and searched inside the Protein Pilot software using a MASCOT search engine. Searches were against the Swiss-Prot/UniProt database and search parameters included tryptic enzyme specificity with up to 2 missed cleavages, 0.06 Da mass tolerance for the MS and a 0.1 Da mass tolerance for the MS/MS, and variable methionine oxidation.
MBP pull-down assays
In Micro Bio-Spin columns (Bio-Rad) 200 μL reactions were assembled with 1 μM of indicated MBP-CtSen1Sen1N fusion or MBP protein in binding buffer (10 mM HEPES pH 7.5, 20 mM NaCl, 0.5 mM TCEP, 0.1 mg ml−1 BSA), 1 μM of CtSen1Hel-c-HA or 1 μM of Sen1Hel-ΔLZZ-c-HA was added followed by 20 μL of amylose resin (NEB). Reactions were mixed on a nutator at 4°C overnight, centrifuged at 1000 x g for 1 min, washed with 100 μL binding buffer, eluted with 20 μL of 50 mM maltose. Samples were resolved on a NuPage (Thermo Fisher) 4–12% Bis–Tris SDS-PAGE followed by immunoblotting with probes for MBP and HA (GeneTex).
Fluorescence polarization RNA Binding Assay
CtSen1 RNA binding was monitored using the change in fluorescence polarization (FP) of a 5'-FAM-labeled single stranded RNA oligonucleotide (Supplementary Table 2). In a 50 μL reaction, 10 nM of labeled RNA substrate was mixed with CtSen1Hel protein at concentrations ranging from 0.01 μM to 10 μM in binding buffer (20 mM HEPES pH 7.5, 50 mM NaCl, 1 mM TCEP, 0.2 mg mL−1 BSA, 10% glycerol). Binding reactions were incubated in black, flat bottom, half-area 96 well plates (Corning Costar) for 20 minutes at room temperature, and FP measurements were collected at room temperature with the POLARstar Omega microplate reader (BMG Labtech) using excitation and emission wavelengths of 485 and 520 nm, respectively. Equilibrium binding dissociation constants (Kd) for the RNA interactions were calculated by fitting data expressed as a fraction bound ratio from FP measurements, using the “specific binding with Hill slope” model in GraphPad Prism.
Preparation of CtSen1FL and CtSen1N-PP-C cryo-EM specimens
Purified CtSen1FL and CtSen1N-PP-C were resolved over a Source Q high resolution anion exchange column (Cytiva) followed by buffer exchange into Sen1 storage buffer. For CtSen1FL sample preparation, protein was concentrated to 1.1 mg mL−1 and deposited onto CF-1.2/1.3-3CU-50 C-Flat™ grids (Protochips Inc.) after glow discharging (30 sec, 15 mA) both sides of the grids using a Pelco EasiGlow. 3 μL of Sen1 storage buffer was deposited on the back side of the grid followed by 3 μL of sample on the front side. Grids were plunge-frozen in liquid ethane using the Vitrobot (FEI) plunge-freeze device at 95% relative humidity, chamber temperature 12°C. For CtSen1N-PP-C sample preparation, 5 mM ADP was included in the 0.2 mg mL−1 protein solution. UltrAuFoil R1.2/1.3 300 mesh gold grids (Quantifoil) were rendered hydrophilic using the Tergeo plasma cleaner (Pie Scientific) and 3 μL protein-ADP sample were deposited onto the grids and blotted for 3 seconds using an Automatic Plunge Freezer (Leica).
Cryo-EM data collection, processing and model building of CtSen1FL
Grids of CtSen1FL were imaged using an FEI Titan Krios transmission electron microscope operating at 300 kV equipped with a K2 summit direct electron detector 130,000 x magnification to a final pixel size of 0.53 Å/pixel using SerialEM 3.7 data collection software. Movies were recorded in counting mode at 130,000 x magnification (Table 1). Motion and drift corrections were performed using MotionCor241. Cryo-EM data processing including CTF corrections were performed in CryoSparc v2.742,43. Following initial automated particle picking and 2D classification, a TOPAZ44 neural network model facilitated acquisition of high-quality particles for downstream analysis (Supplementary Figure 2A). 2D classifications and ab initio modeling provided three low-resolution volumes that were used as input maps for CryoSparc heterogenous refinement. These volumes corresponded to Sen1FL, Sen1Sen1N and possibly Sen1Hel. Additional rounds of 2D classification ab initio reconstruction and heterogenous refinement in CryoSparc, followed by a final non-uniform refinement yielded a 4.6 Å (masked) resolution cryo-EM reconstruction of Sen1FL (Figure 1A-1C, Supplementary Figure 2C and 2E, and Methods). A second volume class corresponding to Sen1Sen1N was also refined to 4.5 Å (masked) (Supplementary Figures 2D and 2F), whereas the Sen1Hel was poorly defined, but broadly resembled the Sen1Hel core structure. The Sen1FL cryo-EM map was traced in Coot45 using AlphaFold model templates46 for the Sen1Sen1N region (aa 1-900). The refined X-ray structure for the Sen1Hel (below) determined herein was used as an initial model to fit the catalytic core. For Sen1Hel, rigid body refinement indicated that the RecA2 domain is found in an opened conformation compared to the RNA bound X-ray structure. An all polyalanine model was refined using real-space refinement in PHENIX47 (Table 1).
Cryo-EM data collection, processing and model building of Sen1N-PP-C
Screening of cryo-EM grids for data collection was conducted using Smartscope48. Grids of Sen1N-PP-C were imaged on an FEI Titan Krios transmission electron microscope operating at 300 kV using a Gatan 3 Bioquantum detector at 130,000 x magnification to a final pixel size of 0.67Å/pixel using SerialEM 3.7 data collection software. Movies were recorded in counting mode at 130,000 x magnification (Table 1). Motion and drift were corrections were performed using MotionCor241. Cryo-EM data processing including CTF corrections were performed in CryoSparc v2.742,43. Two datasets (6379 micrographs and 3655 micrographs) were collected from the same grid and combined (Supplementary Figure 6A). Following initial automated particle picking and 2D classification, a TOPAZ44 neural network model facilitated acquisition of high-quality particles for downstream analysis. 2D classifications, ab initio reconstructions, were followed by rounds of heterogenous refinement, and non-uniform refinement in CryoSparc yielded a 3.17 Å map suitable for model building and refinement (Supplementary Figure 6).
The Sen1N-PP-C cryo-EM map was traced using AlphaFold model templates46 for the Sen1Sen1N region. Sen1 residues 1-255 are poorly defined in the Sen1N-PP-C cryo-EM map and were not modeled. A polyalanine model was built in Coot for residues 244-333 due to static structural disorder. The map was of sufficiently good quality to trace most of the protein chain of Sen1Sen1N (aa 334-888), and Sen1Hel (aa 1102-1844). Regions of the map that had poorly defined surface loops defined were fit as polyalanine or excluded. No cryo-EM density was observed for the engineered PreScission protease cleavable interdomain linker. Real space refinement performed in PHENIX47 with iterative model building in Coot45 produced a final model with good refinement statistics (Table 1).
Crystallization and structure determination of CtSen1Hel-1878
A truncated version of the helicase core (CtSen1Hel-1878, aa 1080-1878) was used for crystallization. Crystals of the CtSen1Hel-1878-RNA-ADP-SO4 complex were grown using the sitting-drop vapor diffusion method by mixing 200 nL of precipitant with 200 nL of protein mixture (10 mg ml−1 CtSen1Hel-1878, 8 mM ADP, 40 μM NaF, 10 nt polyribouridine (10 rU) at 1.5 X molar ratio to protein, in Sen1 storage buffer (see above). Crystals were obtained in 200 mM sodium sulfate and 20% (w/v) PEG3350. Crystals grew overnight at 4°C and were transferred to a cryoprotectant containing 2 mM ADP, 16% PEG3350, 26% (v/v) ethylene glycol, then flash frozen in liquid nitrogen. High-resolution datasets were collected at the Advanced Photon Source on beamline 22-ID. X-ray diffraction data were processed and scaled using HKL200049 (Table 2). Initial structures were solved using molecular replacement with the S. cerevisiae Sen1 protein (5MZN). Iterative building in45 and refined in PHENIX47 (Table 2).
Electrophoretic mobility shift assays
All oligonucleotides were supplied by Integrated DNA Technologies (Supplementary Table 2). Reactions (10 μL) were performed in a buffer containing 10 mM Tris-HCl pH 7.5 and 50 mM NaCl. 10 nM RNA-DNA substrate (Sub4, Supplementary Tables 4 and 5) was incubated with Sen1FL, Sen1N-PP-C and Sen1Hel (50, 150, 500 and 1000 nM) on ice for 15 min. Samples were resolved on 6% TBE gels and FAM-labeled reaction products were scanned at 520 nm on a Typhoon FLA 9000 scanner (GE Healthcare). The bands were quantified using ImageQuant.
ATPase assay
Reactions (50 μL) were performed in a buffer containing 10 mM Tris pH 7.5, 50 mM NaCl, 7.5 μM ZnCl2, 1 mM DTT and 0.1 mg mL−1 BSA. Sen1 proteins (5 nM) were incubated with Sub6 (5000 nM) (Supplementary Tables 4 and 5), ATP (1 mM) and MgCl2 (2 mM) for 15 min at 37 °C. Samples were quenched with EDTA (10 mM). The hydrolysis products were detected using colorimetric Malachite Green Phosphate Assay Kit (Cayman). Absorbance was measured at 620 nm. Free phosphate concentrations were calculated from the calibration curve and used to determine the percentage of hydrolyzed ATP.
RNA-DNA unwinding assays
Reactions were performed in a buffer containing 10 mM Tris pH 7.5, 50 mM NaCl, 7.5 μM ZnCl2, 1 mM DTT and 0.1 mg mL−1 BSA. Gel-based unwinding assays were performed in 10 μl reactions containing Sen1Hel (50 nM), Sub1-Sub5 (20 nM) (Supplementary Tables 4 and 5), and 19-Trap DNA (200 nM) used to trap unwound FAM 19-DNA (Supplementary Table 2). Reactions were initiated by addition of a mixture containing ATP (1 mM) and MgCl2 (2 mM) and allowed to proceed for 15 min at 37°C. Reactions were stopped by addition of EDTA (10 mM). Proteins were removed by treatment with SDS (1%) and Proteinase K (0.1 mg mL−1). Samples were resolved on 15% TBE gels and FAM-labeled reaction products were visualized on a Typhoon FLA 9000 scanner (GE Healthcare). For fluorescence-based unwinding assays, the Sub7 was used as RNA-DNA substrate. Reactions were performed in 50 μL reactions in 5 replicates. Enzymes at the indicated concentrations were incubated with 50 nM Sub7 (Supplementary Tables 4 and 5). Reactions were initiated by addition of a mixture containing ATP (1 mM) and MgCl2 (2 mM). The signal was detected at 520 nm every 10 sec over 30 min at room temperature. The initial rates were calculated from the linear fit of the percentage of unwound RNA-DNA during the first 5 min of the reaction. Initial rates were plotted against the amount of enzyme (x-axis). Rates of reactions were calculated from the slope of the curves. For mutational analysis, Sen1Hel mutants (1 nM) were incubated with Sub7 (50 nM) for 10 min at RT.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analysis was performed using GraphPad Prism 8. The test performed, the sample size (n) and the number of independent replicates for each experiment are depicted in the figure legends.
Supplementary Material
Supplementary Video 1 Structural morph between Sen1-RNA-ADP-SO4 complex and the autoinhibited Sen1NPPC- ADP state, Related to Figure 6. As the Sen1N domain (tan) interacts with the by RecA1 1b (orange) and 1c (turquoise) insertion elements, conformational change is relayed to the RNA binding cleft. In this ADP bound auto-inhibited state, closure of the RNA binding channel sterically occludes RNA access.
HIGHLIGHTS.
Sen1 (Senataxin, SETX) RNA-DNA helicase structure resembles an elongated inchworm.
Sen1 is auto-regulated by the Sen1N domain that directly binds its helicase core.
Sen1 helicase core RNA complex structures show the enzyme encircles RNA.
AOA2 RNA binding cleft mutations undermine RNA-DNA unwinding activity.
INCLUSION AND DIVERSITY.
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. We support inclusive, diverse, equitable conduct of research.
ACKNOWLEDGMENTS
The research was supported by the US National Institute of Health Intramural Program, US National Institute of Environmental Health Sciences (NIEHS) 1Z01ES102765 (to R.S.W.). We thank Dr. Lars Pedersen of the NIEHS Collaborative X-ray Crystallography Group for data collection support and the Advanced Photon Source (APS) Southeast Regional Collaborative Access Team (SER-CAT) for beamline access. Use of the APS was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. We thank Dr. Lars Pedersen and Dr. Robin Stanley for expert comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Garcia-Muse T, and Aguilera A (2019). R Loops: From Physiological to Pathological Roles. Cell 179, 604–618. 10.1016/j.cell.2019.08.055. [DOI] [PubMed] [Google Scholar]
- 2.Crossley MP, Bocek M, and Cimprich KA (2019). R-Loops as Cellular Regulators and Genomic Threats. Mol Cell 73, 398–411. 10.1016/j.molcel.2019.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Groh M, Albulescu LO, Cristini A, and Gromak N (2017). Senataxin: Genome Guardian at the Interface of Transcription and Neurodegeneration. J Mol Biol 429, 3181–3195. 10.1016/j.jmb.2016.10.021. [DOI] [PubMed] [Google Scholar]
- 4.Ursic D, Chinchilla K, Finkel JS, and Culbertson MR (2004). Multiple protein/protein and protein/RNA interactions suggest roles for yeast DNA/RNA helicase Sen1p in transcription, transcription-coupled DNA repair and RNA processing. Nucleic Acids Res 32, 2441–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, and Brow DA (2006). Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell 24, 735–746. [DOI] [PubMed] [Google Scholar]
- 6.Suraweera A, Becherel OJ, Chen P, Rundle N, Woods R, Nakamura J, Gatei M, Criscuolo C, Filla A, Chessa L, et al. (2007). Senataxin, defective in ataxia oculomotor apraxia type 2, is involved in the defense against oxidative DNA damage. J Cell Biol 177, 969–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen S, Puget N, Lin YL, Clouaire T, Aguirrebengoa M, Rocher V, Pasero P, Canitrot Y, and Legube G (2018). Senataxin resolves RNA:DNA hybrids forming at DNA double-strand breaks to prevent translocations. Nat Commun 9, 533. 10.1038/s41467-018-02894-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rawal CC, Zardoni L, Di Terlizzi M, Galati E, Brambati A, Lazzaro F, Liberi G, and Pellicioli A (2020). Senataxin Ortholog Sen1 Limits DNA:RNA Hybrid Accumulation at DNA Double-Strand Breaks to Control End Resection and Repair Fidelity. Cell Rep 31, 107603. 10.1016/j.celrep.2020.107603. [DOI] [PubMed] [Google Scholar]
- 9.Yuce O, and West SC (2013). Senataxin, defective in the neurodegenerative disorder ataxia with oculomotor apraxia 2, lies at the interface of transcription and the DNA damage response. Mol Cell Biol 33, 406–417. 10.1128/MCB.01195-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zardoni L, Nardini E, Brambati A, Lucca C, Choudhary R, Loperfido F, Sabbioneda S, and Liberi G (2021). Elongating RNA polymerase II and RNA:DNA hybrids hinder fork progression and gene expression at sites of head-on replication-transcription collisions. Nucleic Acids Res 49, 12769–12784. 10.1093/nar/gkab1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moreira MC, Klur S, Watanabe M, Nemeth AH, Le Ber I, Moniz JC, Tranchant C, Aubourg P, Tazir M, Schols L, et al. (2004). Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat Genet 36, 225–227. [DOI] [PubMed] [Google Scholar]
- 12.Lavin MF, Gueven N, and Grattan-Smith P (2008). Defective responses to DNA single- and double-strand breaks in spinocerebellar ataxia. DNA Repair (Amst) 7, 1061–1076. [DOI] [PubMed] [Google Scholar]
- 13.Chen X, Muller U, Sundling KE, and Brow DA (2014). Saccharomyces cerevisiae Sen1 as a model for the study of mutations in human Senataxin that elicit cerebellar ataxia. Genetics 198, 577–590. 10.1534/genetics.114.167585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, and Brow DA (2006). Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell 24, 735–746. 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Chun Y, Buratowski S, and Tong L (2019). Identification of Three Sequence Motifs in the Transcription Termination Factor Sen1 that Mediate Direct Interactions with Nrd1. Structure 27, 1156–1161 e1154. 10.1016/j.str.2019.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thiebaut M, Kisseleva-Romanova E, Rougemaille M, Boulay J, and Libri D (2006). Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1-nab3 pathway in genome surveillance. Mol Cell 23, 853–864. 10.1016/j.molcel.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 17.Steinmetz EJ, Conrad NK, Brow DA, and Corden JL (2001). RNA-binding protein Nrd1 directs poly(A)-independent 3'-end formation of RNA polymerase II transcripts. Nature 413, 327–331. 10.1038/35095090. [DOI] [PubMed] [Google Scholar]
- 18.Porrua O, Hobor F, Boulay J, Kubicek K, D'Aubenton-Carafa Y, Gudipati RK, Stefl R, and Libri D (2012). In vivo SELEX reveals novel sequence and structural determinants of Nrd1-Nab3-Sen1-dependent transcription termination. EMBO J 31, 3935–3948. 10.1038/emboj.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Appanah R, Lones EC, Aiello U, Libri D, and De Piccoli G (2020). Sen1 Is Recruited to Replication Forks via Ctf4 and Mrc1 and Promotes Genome Stability. Cell Rep 30, 2094–2105 e2099. 10.1016/j.celrep.2020.01.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simon AC, Zhou JC, Perera RL, van Deursen F, Evrin C, Ivanova ME, Kilkenny ML, Renault L, Kjaer S, Matak-Vinkovic D, et al. (2014). A Ctf4 trimer couples the CMG helicase to DNA polymerase alpha in the eukaryotic replisome. Nature 510, 293–297. 10.1038/nature13234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villa F, Simon AC, Ortiz Bazan MA, Kilkenny ML, Wirthensohn D, Wightman M, Matak-Vinkovic D, Pellegrini L, and Labib K (2016). Ctf4 Is a Hub in the Eukaryotic Replisome that Links Multiple CIP-Box Proteins to the CMG Helicase. Mol Cell 63, 385–396. 10.1016/j.molcel.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singleton MR, Dillingham MS, and Wigley DB (2007). Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem 76, 23–50. [DOI] [PubMed] [Google Scholar]
- 23.Leonaite B, Han Z, Basquin J, Bonneau F, Libri D, Porrua O, and Conti E (2017). Sen1 has unique structural features grafted on the architecture of the Upf1-like helicase family. EMBO J 36, 1590–1604. 10.15252/embj.201696174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han Z, Libri D, and Porrua O (2017). Biochemical characterization of the helicase Sen1 provides new insights into the mechanisms of non-coding transcription termination. Nucleic Acids Res 45, 1355–1370. 10.1093/nar/gkw1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin-Tumasz S, and Brow DA (2015). Saccharomyces cerevisiae Sen1 Helicase Domain Exhibits 5'- to 3'-Helicase Activity with a Preference for Translocation on DNA Rather than RNA. J Biol Chem 290, 22880–22889. 10.1074/jbc.M115.674002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schellenberg MJ, Petrovich RM, Malone CC, and Williams RS (2018). Selectable high-yield recombinant protein production in human cells using a GFP/YFP nanobody affinity support. Protein Sci 27, 1083–1092. 10.1002/pro.3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakrabarti S, Jayachandran U, Bonneau F, Fiorini F, Basquin C, Domcke S, Le Hir H, and Conti E (2011). Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol Cell 41, 693–703. 10.1016/j.molcel.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 28.Lim SC, Bowler MW, Lai TF, and Song H (2012). The Ighmbp2 helicase structure reveals the molecular basis for disease-causing mutations in DMSA1. Nucleic Acids Res 40, 11009–11022. 10.1093/nar/gks792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lacabanne D, Wiegand T, Wili N, Kozlova MI, Cadalbert R, Klose D, Mulkidjanian AY, Meier BH, and Bockmann A (2020). ATP Analogues for Structural Investigations: Case Studies of a DnaB Helicase and an ABC Transporter. Molecules 25. 10.3390/molecules25225268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao Y, and Yang W (2020). Different mechanisms for translocation by monomeric and hexameric helicases. Curr Opin Struct Biol 61, 25–32. 10.1016/j.sbi.2019.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Zundert GCP, Rodrigues J, Trellet M, Schmitz C, Kastritis PL, Karaca E, Melquiond ASJ, van Dijk M, de Vries SJ, and Bonvin A (2016). The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J Mol Biol 428, 720–725. 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 32.Saikrishnan K, Powell B, Cook NJ, Webb MR, and Wigley DB (2009). Mechanistic basis of 5'-3' translocation in SF1B helicases. Cell 137, 849–859. 10.1016/j.cell.2009.03.036. [DOI] [PubMed] [Google Scholar]
- 33.Lee JY, and Yang W (2006). UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell 127, 1349–1360. 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng Z, Muhlrad D, Lim MK, Parker R, and Song H (2007). Structural and functional insights into the human Upf1 helicase core. EMBO J 26, 253–264. 10.1038/sj.emboj.7601464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Unciuleac MC, Meir A, Xue C, Warren GM, Greene EC, and Shuman S (2021). Clutch mechanism of chemomechanical coupling in a DNA resecting motor nuclease. Proc Natl Acad Sci U S A 118. 10.1073/pnas.2023955118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warren GM, Meir A, Wang J, Patel DJ, Greene EC, and Shuman S (2022). Structure-activity relationships at a nucleobase-stacking tryptophan required for chemomechanical coupling in the DNA resecting motor-nuclease AdnAB. Nucleic Acids Res 50, 952–961. 10.1093/nar/gkab1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, and Ben-Tal N (2016). ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 44, W344–350. 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finkel JS, Chinchilla K, Ursic D, and Culbertson MR (2010). Sen1p performs two genetically separable functions in transcription and processing of U5 small nuclear RNA in Saccharomyces cerevisiae. Genetics 184, 107–118. 10.1534/genetics.109.110031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ozgur S, Buchwald G, Falk S, Chakrabarti S, Prabu JR, and Conti E (2015). The conformational plasticity of eukaryotic RNA-dependent ATPases. Febs J 282, 850–863. 10.1111/febs.13198. [DOI] [PubMed] [Google Scholar]
- 41.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332. 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296. 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 43.Punjani A, Zhang H, and Fleet DJ (2020). Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat Methods 17, 1214–1221. 10.1038/s41592-020-00990-8. [DOI] [PubMed] [Google Scholar]
- 44.Bepler T, Morin A, Rapp M, Brasch J, Shapiro L, Noble AJ, and Berger B (2019). Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat Methods 16, 1153–1160. 10.1038/s41592-019-0575-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta crystallographica. Section D, Biological crystallography 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Senior AW, Evans R, Jumper J, Kirkpatrick J, Sifre L, Green T, Qin C, Zidek A, Nelson AWR, Bridgland A, et al. (2020). Improved protein structure prediction using potentials from deep learning. Nature 577, 706–710. 10.1038/s41586-019-1923-7. [DOI] [PubMed] [Google Scholar]
- 47.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Echols N, Headd JJ, Hung LW, Jain S, Kapral GJ, Grosse Kunstleve RW, et al. (2011). The Phenix software for automated determination of macromolecular structures. Methods 55, 94–106. 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bouvette J, Huang Q, Riccio AA, Copeland WC, Bartesaghi A, and Borgnia MJ (2022). Automated systematic evaluation of cryo-EM specimens with SmartScope. Elife 11. 10.7554/eLife.80047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Otwinowski Z, and Minor W (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276, 307–326. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Video 1 Structural morph between Sen1-RNA-ADP-SO4 complex and the autoinhibited Sen1NPPC- ADP state, Related to Figure 6. As the Sen1N domain (tan) interacts with the by RecA1 1b (orange) and 1c (turquoise) insertion elements, conformational change is relayed to the RNA binding cleft. In this ADP bound auto-inhibited state, closure of the RNA binding channel sterically occludes RNA access.
Data Availability Statement
The data presented in this paper will be shared by the lead contact upon request. Uncropped gels are available at Mendeley at doi:10.17632/djm6wdc5tg.1. Cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB). X-ray data and refined molecular models have been deposited in the RSCB Protein Data Bank (PDB). EMDB and PDB Accession codes are available in the key resources table. All datasets will be publicly accessible at the date of publication.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.