Summary
Tooth enamel secreted by ameloblasts (AM) is the hardest material in the human body, acting as a shield to protect the teeth. However, the enamel is gradually damaged or partially lost in over 90% of adults and cannot be regenerated due to a lack of ameloblasts in erupted teeth. Here we use single-cell combinatorial indexing RNA sequencing (sci-RNA-seq) to establish a spatiotemporal single-cell census for the developing human tooth and identify regulatory mechanisms controlling the differentiation process of human ameloblasts. We identify key signaling pathways involved between the support cells and ameloblasts during fetal development and recapitulate those findings in human ameloblast in vitro differentiation from iPSCs. We furthermore develop a disease model of Amelogenesis imperfecta in a 3D organoid system and show AM maturation to mineralized structure in vivo. These studies pave the way for future regenerative dentistry.
Graphical Abstract
eTOC blurb
Alghadeer et al. identify, using advanced sequencing techniques, key insights into the development and regeneration of tooth enamel. By identifying regulatory mechanisms and successfully differentiating human ameloblasts in vitro, this study may define tools for future therapies targeting enamel-related genetic diseases and advancements in regenerative dentistry.
Introduction
Omnivores and herbivores alike have an outer, calcified enamel layer that protects the living tooth structures. Some animals can regenerate this layer due to active dental epithelial stem cells that produce new functional ameloblast. However, adult human does not have active enamel-secreting ameloblasts nor the stem cell niche to regenerate them, and therefore is incapable of regenerating lost enamel layer resulting in failed tooth function and loss of this critical organ in adult human body1,2. Today we do not know how to generate the regenerative human cell type, Ameloblast.
In addition to injury and damage, congenital genetic diseases such as Amelogenesis Imperfecta can contribute to enamel loss3. Ameloblasts are dental epithelial cells that interact with ectomesenchyme derived odontoblasts to produce the protective shell. Ameloblasts secrete enamel protein matrix and deposit minerals to achieve hard and mature tooth enamel during human development4. During tooth eruption in humans, the ameloblasts undergo apoptosis1,5. Odontoblasts produce the inner protective tooth layer, dentin6. Although tooth development has been studied over several years7, most of these excellent developmental and molecular studies have been conducted using murine models8–12 which presents several challenges when applied to human development13–15. For example, mice do not have secondary teeth, instead mouse incisors undergo continuous regeneration due to a population of epithelial stem cells in the labial cervical loop that allows for continued enamel formation throughout life16. This regenerative process does not occur in adult human teeth. Mouse molar development, however, shares some similarities with human teeth, but significant differences between the two exist as well. For instance, the shape and number of cusp and the timing and sequence of molar development also vary between the mouse and human teeth17. While studying tooth development in mice can provide valuable insights into the fundamental mechanisms of tooth differentiation10, it is important to fully understand the unique aspects of human dentation and to develop effective treatments for human dental disorders10.
In addition to ameloblast lineage, enamel organ also contains multiple populations of support cells, including the stellate reticulum and the inner and outer enamel epithelium18. These support cells are thought to be essential for ameloblast function19–21; however, it is not understood how they are mechanistically involved in ameloblast differentiation and functional maturation. Animal studies have suggested several pathways in driving and regulating this communication, such as the hedgehog HH22, NOTCH19, and FGF23 pathways. However, the temporal regulation and the extent to which these pathways originate from support cells are not clearly understood since these cells are poorly studied in humans. Dissecting human tooth development at the single-cell level can capture the patterns of gene expression that characterize small populations of support cells that are involved in the differentiation.
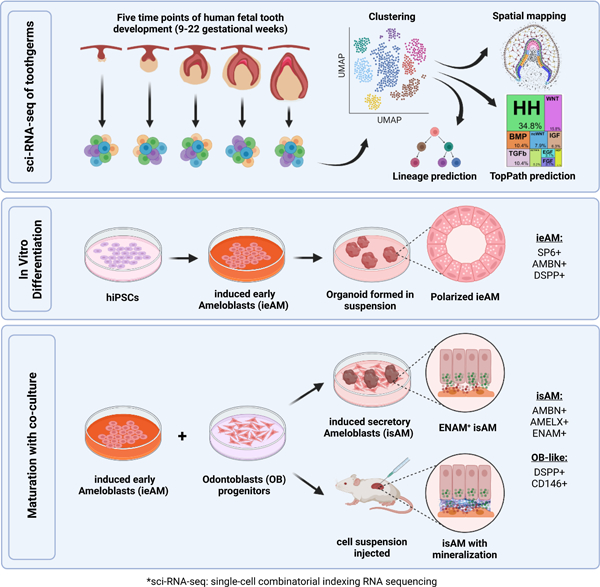
In order to facilitate the regeneration of human tooth structures in the future, we have utilized single-cell combinatorial indexing RNA sequencing (sci-RNA-seq)24 technology to study human fetal tooth development at 9–22 gestational weeks (gw). Through computational and RNAscope MultiPlex analysis, we established a spatiotemporal single-cell census for developing human teeth that includes both the epithelial and mesenchymal cell types. Our computational studies established human-specific transcriptional profiles for the celltypes of the developing tooth and identified branches in the predicted developmental trajectories, as well as previously undescribed populations of epithelial support tissues. Further, we elucidated critical signaling pathways governing the fate of early ameloblasts (ieAM) to study the etiology of Amelogenesis imperfecta. We developed a maturation co-culture protocol to generate induced secretory ameloblasts (isAM); Through 3D organoid cultures in vitro and transplantation experiments in vivo, these isAM organoids exhibited mineralization (calcium deposition) and expressed essential enamel proteins such as Ameloblastin, Amelogenin, and Enamelin. Hence, we have coined the term Ameloblast Organoid to describe this new class of organoids. These findings contribute to our understanding of dental tissue differentiation and provide insights for disease modeling and regenerative approaches.
Results
A single-cell census of the developing human fetal odontogenic tissues
To better understand early oral differentiation and to dissect how the epithelial and mesenchymal cell lineages acquire the odontogenic competence, we analyzed the developmental gene expression profiles of human fetal stages by single-cell sequencing. In humans, oral tissue development begins around 6gw and starts as a thickening in the oral epithelium18,25,26, giving rise to all primary teeth and salivary gland tissue. Individual teeth develop independently as an extension of the main dental lamina and progress through a series of morphological stages (bud, cap, & bell) within bony crypts of the jaws27. Additionally, each developing tooth is surrounded by thick fibrous tissue called the dental follicle28. The dental follicle and the tissue it contains comprise the toothgerm29 (Figure 1A). The oral epithelium will also give rise to the salivary glands (Figure 1A). Like teeth, salivary glands derive from the invagination of a thickened sheet of oral epithelium into the underlying mesenchyme, known as the initial bud stage30. We collected toothgerm and salivary gland samples from five human fetal age groups (Figure 1A–1B and S1A–S1C). These age groups represented the following developmental stages for tooth differentiation: the cap stage (9–11gw and 12–13gw), the early bell stage (14–16gw), and the late bell stage (17–19gw and 20–22gw) (Figure 1A–1E)18,31. We also collected submandibular salivary glands (SMSG) from three matched timepoints (12–13gw, 14–16gw, 17–19gw) that cover the pseudo-glandular and canalicular stages for salivary gland development32 (Figure 1A).
Figure 1. Single-cell census of developing human fetal jaws, teeth, and salivary glands using sci-RNA-seq.
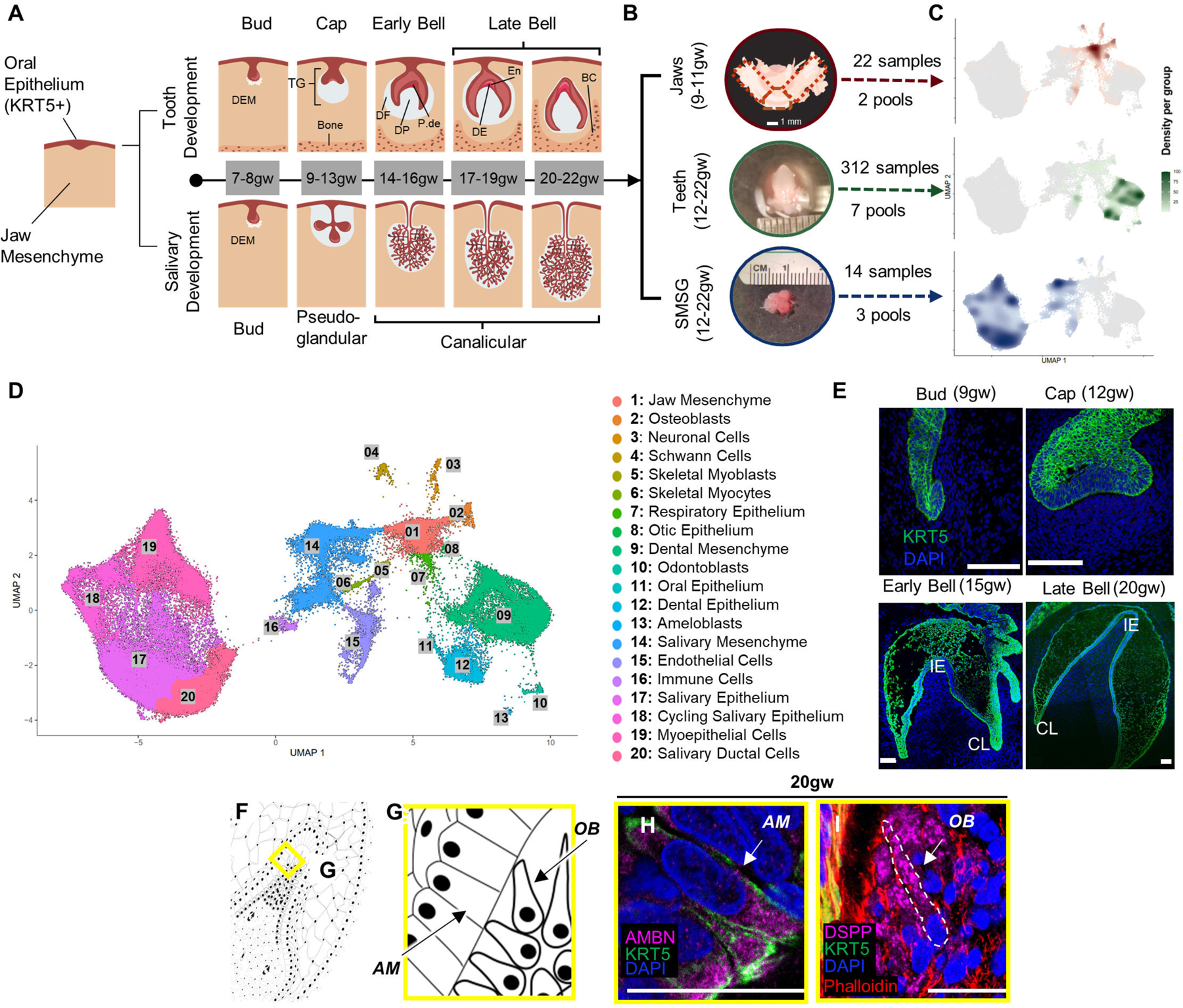
(A) Stepwise development of oral epithelium (red) and dental ectomesenchyme (grey) leading to tooth and salivary gland formation. TG: toothgerm, DF: dental follicle, DP: dental papilla, P-de: pre-dentin, De: dentin, En: enamel matrix. (B) Dissection of toothgerms and salivary glands from fetal jaw tissue. (C) Density plots showing tissue types in UMAP coordinate. (D) UMAP graph with 20 annotated clusters. (E) Immunofluorescence staining of toothgerms with Krt5 (green) and nuclear stain DAPI (blue). Abbreviations: incisal edge (IE), cervical loop (CL). (F, G) Simplified illustration. Immunofluorescence of toothgerms with DSPP (odontoblast marker) and AMBN (ameloblast marker) at 20gw (H, I). Scale bars: 50μm.
Single-cell sequencing data of the tissue samples were analyzed using Monocle324,33 and visualized in uniform manifold approximation and projection (UMAP) space (Figure 1D). The distribution of the cells from each tissue origin was identified by using density plots based on tissue type (Figure 1C) or by individual samples (Figure S1D). Utilizing a graph-based clustering algorithm, we annotated 20 major clusters based on key marker genes (Figure 1D; Figure S1E; File S1) from PanglaoDB34 and other sources in literature. The major cell types in salivary gland samples include salivary mesenchyme, salivary epithelium, cycling salivary epithelium, myoepithelium, and ductal cells (Figures 1C–1D and S1E)35. In the jaw samples (9–11gw) (Figures 1C–1D and S1E), we identified mesenchymal progenitors, osteoblasts, neuronal, Schwann cells, muscle, respiratory epithelium, otic epithelium, and oral epithelium (Figures 1C–1D and S1E). The major cell types in tooth samples include dental mesenchyme, epithelium, odontoblasts, and ameloblasts. The cell types observed in all samples include endothelial and immune cells. The present manuscript focuses on the gene expression and signaling pathways governing tooth development.
To confirm the timing of the tooth morphological stages, we performed immunohistochemistry on tissue sections. As expected, all the enamel organ derived tissues were visualized by KRT5 (green) (Figure 1E). The two critical cell types, odontoblasts and ameloblasts secrete the mineralized protective layers that cover the soft dental pulp. Odontoblasts were identified by the expression of dentin sialophosphoprotein (DSPP), while ameloblasts expressed ameloblastin (AMBN) and enamelin (ENAM) proteins (Figures 1F–1I and S4G-S4K’).
Spatial localization of sci-RNA-seq defined clusters identifies subodontoblasts in humans and suggests they can give rise to preodontoblasts
To dissect the odontoblast lineage, we subset the developing jaw mesenchyme, dental ectomesenchyme, and odontoblast cells and embedded the data into a UMAP space (Figure 1D and Figure 2A). This analysis yielded six transcriptionally defined clusters: dental papilla (DP), preodontoblast (POB), odontoblast (OB), subodontoblast (SOB), dental ectomesenchyme (DEM), and dental follicle (DF) (Figure 2A)10 (Figure 2B; Figure S2A–S2B; File S1 and File S2: meta-analytic estimators).
Figure 2. Dental Mesenchyme Developmental Trajectory.
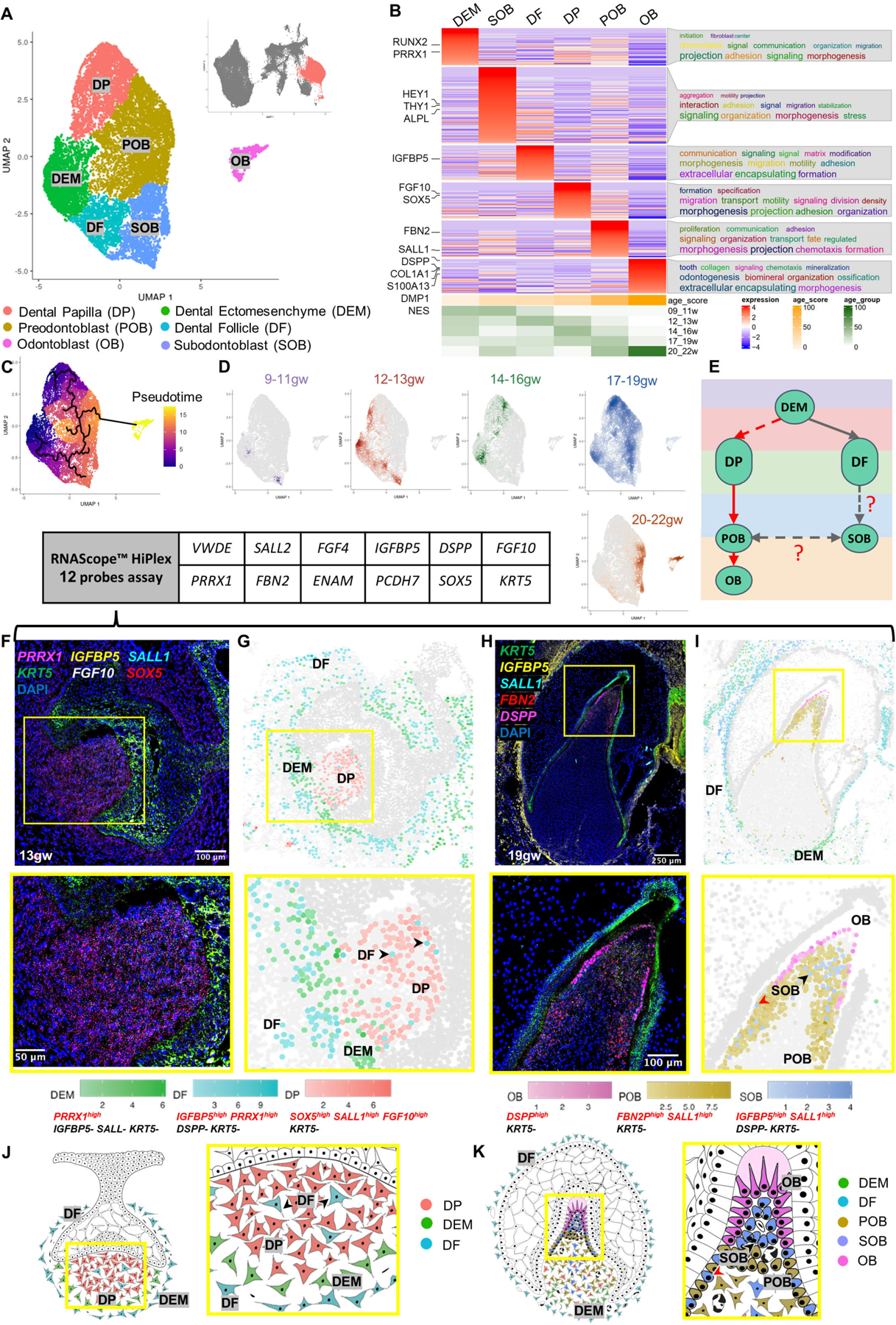
(A) UMAP graph showing subclustered dental mesenchyme derived cells from molar and incisor toothgerms. Six transcriptionally defined clusters are identified: DP, POB, OB, SOB, OB, DEM, and DF. (B) Custom heatmap reveals marker genes, associated GO-terms, and age scores per cluster. (C) Pseudotime trajectory analysis suggests two branches (DP and DF) in dental mesenchyme. (D) Real-time density plots indicate cell migration from early progenitors to differentiated cell types. (E) Simplified differentiation trajectory tree illustrating common DEM progenitor giving rise to DP and DF. (F) RNAScope in situ hybridization image and inset showing marker probes at 13gw. (G) RNAScope map for marker combinations corresponding to dental mesenchyme clusters at 13gw. (H) RNAScope in situ hybridization image and inset showing marker probes at 19gw. (I) RNAScope map for marker combinations corresponding to dental mesenchyme clusters at 19gw. (J) Diagram illustrating the developing dental mesenchyme cell types in the human toothgerm.
Furthermore, to evaluate the potential function of each cluster, we performed gene ontology analysis using ViSEAGO36, which uses data mining to establish semantic links between highly expressed genes in a given cluster. This analysis shows that DP and DEM are characterized by signaling, morphogenesis, initiation and specification, supporting their role as precursor populations. In contrast, POB is characterized by their proliferation and fate determination. The enriched genes in DF were related to matrix modification and encapsulation. Genes expressed in SOB are connected to aggregation, motility and projections, characteristics of a cell type sensing and influencing its environment, while OB shows GO-terms toward odontogenesis, tooth organization, and mineralization (Figure 2B).
Progenitor sources and cells’ progression towards differentiation were assessed by pseudotime trajectory analysis that revealed the presence of two progenitor sources within the developing dental mesenchyme: the DP that gives rise to POB and, subsequently, OB; and the DF predicted to give rise to SOB, which is predicted by the algorithm to have a capacity to transition to POB (Figure 2C). Pseudotime analysis is supported by real-time density plots that show reduced progenitor type cell population density as the toothgerm develops, indicating fate commitment to OB lineage begins after 13gw in human fetal development and is largely complete by 20gw (Figure 2D). Broad expression of dental ectomesenchyme marker PRRX1 is observed in the DEM, DF and DP (Figure S2C), supporting previous findings37 that a shared cranial neural crest progenitor gives rise to both DP and DF. Thus, we propose a simplified trajectory of both the odontoblast and dental follicle lineages (red and gray arrowheads in Figure 2E), with a shared PRRX1+ DEM progenitor giving rise to both DF and DP.
To localize and validate the computationally identified clusters in human fetal tissue, we performed multiplex RNAScope in situ hybridization on toothgerms at early (13gw) and late (19gw) tooth development (Figure 2F and 2H). Each cell was identified by characterization of 2–4 expression markers (AND/OFF logic). After performing multiplex signal quantification per cell, we converted the RNAScope images into spatial datasets of single cells (Figure 2G and 2I; S2E–S2F). In agreement with the sci-RNA-seq data (Figure 2B; File S1; File S3–RNAScope_logic_tables.xlsx), dental mesenchyme-derived cell types display spatiotemporally specific expression patterns. At 13gw (Figures 2F–2G), the dental pulp consists of cells positive for markers in DP cluster (Fig.2A, F-G), with DEM localized to the apical pulp. The surprising presence of sparse DF type cells within the early dental pulp (13gw, black arrowheads in Figures 2G and 2J; identified by multiplex spatial in situs) supports the pseudotime trajectory suggesting a subgroup of DF as progenitors for SOB, and that this fate commitment occurs prior to 13gw. This connection has been previously suggested, but it has not been conclusively confirmed using lineage analysis in mouse incisors and molars38. Apical papilla is shown in mouse in vivo lineage tracing to produce OB and pulp cells39. Further lineage analysis experiments in human are required to test these hypotheses. At 13gw the developing toothgerm is surrounded by DF cells (Figures 2G and 2J), a pattern that persists to late tooth development 19gw (Figures 2H–2I, 2K). By 19gw, we observe that the dental pulp contains a mixed population of SOB and POB with smaller contributions from DP and DEM. OB are localized at the incisal edge (Figures 2H–2I, 2K). We observe SOB directly beneath the OB (black arrowhead, Figure 2I and 2K) and, intriguingly, intermingled with POB at the pulpal periphery (red arrowhead, Figure 2I and 2K). This finding supports the pseudotime trajectory (Figure 2C), indicating it is possible that SOB can give rise to OB following injury, as seen previously in mouse models6,40. SOB represents a small portion of the pulpal cell population (Figure 2H–2I), suggesting that OBs are mainly derived from POB while SOB serves as a reserve (Figure 2C; 2E; 2H; 2I; 2K; S2D). Lineage tracing studies are necessary to validate this finding in vivo and further dissect SOB’s role in odontoblast development and repair. In summary, we have spatially validated the key computationally identified clusters in human odontoblast development in fetal tissue using high resolution, multiplex RNAscope analysis.
To further assess the validity of the proposed clusters in the OB lineage, we compared our data to previously published datasets for the dental mesenchyme in adult human molars10, and for postnatal odontoblasts of mouse molars39 (Figure S2G–S2H). We identified the enriched marker genes in each cluster for each dataset to compare the annotated cell types and, as expected, found significant overlaps between human fetal and adult OB, as well as mouse fetal OB (though more overlap was observed between human samples, suggesting species differences; Figure S2G–S2H)(File S2: list of overlapping genes). Go term analysis for the overlapped genes in OB in both datasets were related to odontogenesis and mineralization (Figure S2I–S2J). We also noted a significant overlap between SOB and POB with the peri-odontoblastic layer of adult human teeth, confirming that they share similar anatomical position in relation to OB (Figure S2H). Importantly, SOB significantly overlapped with the apical papilla cluster in mouse molars, suggesting similarity between these two cell types (Figure S2G). As mentioned above, apical papilla is shown in mouse in vivo lineage tracing to produce OB and pulp cells39, supporting the hypothesis that human SOB is also a bipotent cell type (Fig.2E). Future studies are required to test these hypotheses in human.
Spatial localization of sci-RNA-seq defined clusters in dental epithelium identifies stage-specific support cell types and cervical loop stem cells
To further analyze the subtypes of the dental epithelium, we subset oral epithelium, dental epithelium, and ameloblast clusters and validated these spatially in the human fetal tissues by multiplex RNAscope, as done above for odontoblast lineage (Figures 2 and 3). The subset yielded 13 unique clusters that we identified by collating highly expressed cluster-specific genes (Figure 3B and S3A; File S1 and File S2: meta-analytic estimators); oral epithelium (OE), dental epithelium (DE), enamel knot (EK), enamel epithelium (outer enamel epithelium/inner enamel epithelium, OEE/IEE), cervical loop (CL), inner and outer stratum intermedium (SII, SIO), inner and outer stellate reticulum (SRI and SRO), pre-ameloblasts (PA) and two AMBN expressing ameloblast clusters (early ‘eAM’ and secretory ‘sAM’; Figure 3A–3D; File S1). The identity of these clusters aligned with their likely real-time appearance as represented by a real-time distribution of cells (Figure S3B). Moreover, GO analysis (Figure 3B) indicated cell type-specific roles in tooth development in agreement with the annotations. For example, the OE cluster identified proper stratified epithelium, including keratinization, keratinocyte differentiation, and cornification41, while the DE shows epithelial organization and differentiation, indicative of its function in reorganizing to form the tooth bud42.
Figure 3. Ameloblast Developmental Trajectory.
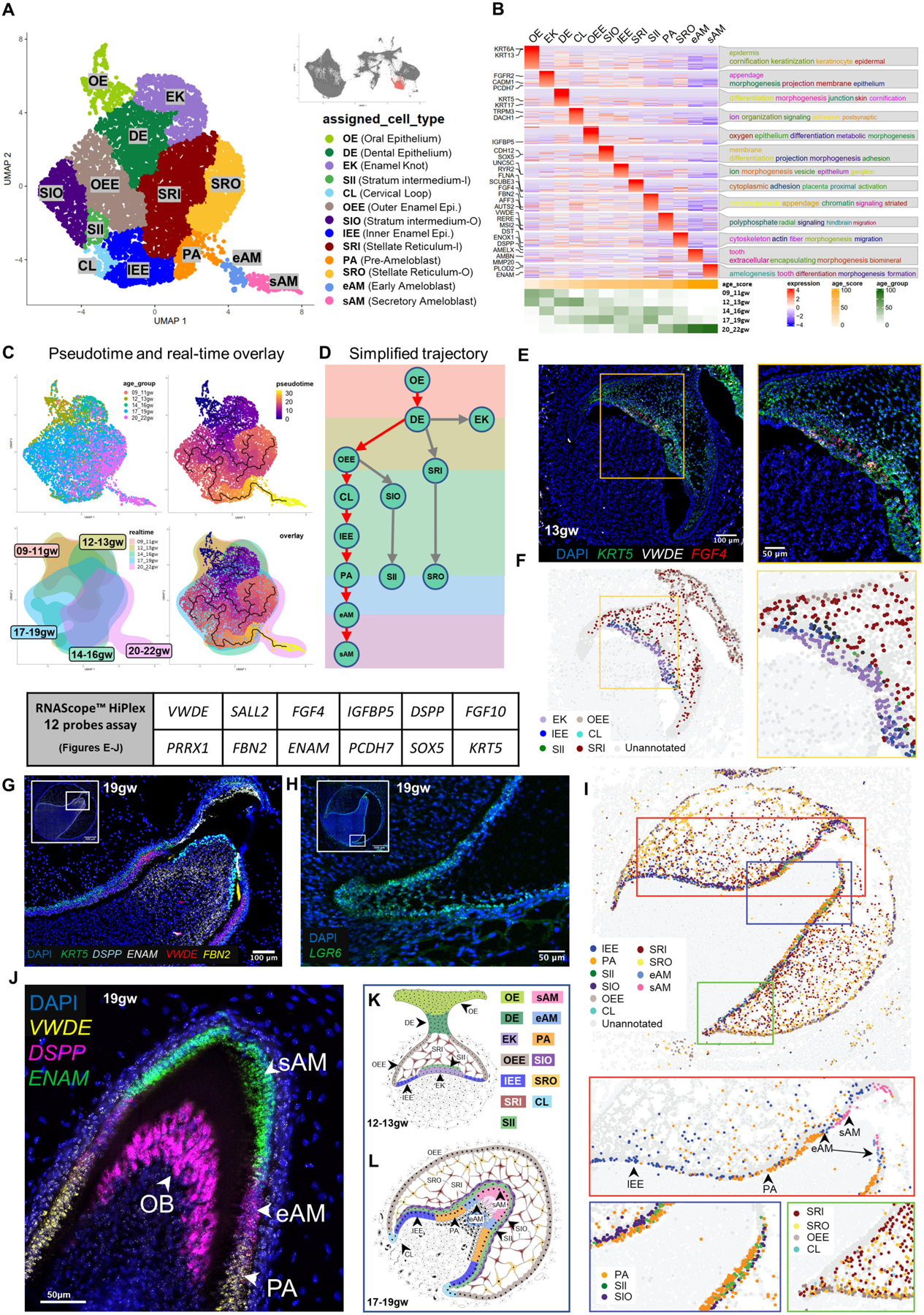
(A) UMAP graph showing subclustered dental epithelium derived cells from molar and incisor toothgerms. Thirteen transcriptionally defined clusters are identified, including OE, DE, EK, OEE, IEE, CL, SII, SIO, SR, SRI, PA, eAM, and sAM. (B) Custom heatmap reveals marker genes, associated GO-terms, and age scores per cluster. (C) Pseudotime trajectory analysis and real-time overlay suggest the DE gives rise to three branch lineages. (D) Simplified differentiation trajectory tree illustrating separate lineages originating from the DE. (E) RNAScope in situ hybridization image and inset for VWDE and FGF4 probes at 13gw of incisor. (F) RNAScope map of individual dental epithelium-derived clusters at 13gw of incisor. (G) RNAScope in situ hybridization image and inset for DSPP, ENAM, VWDE, and FBN2 probes at 19gw of central incisor. (H) RNAScope in situ hybridization image for LGR6 marking the CL at 19gw. (I) RNAScope map of individual dental epithelium derived clusters at 19gw. (J) RNAScope in situ hybridization of 19gw lateral incisor showing the transition of PA to eAM to sAM. (K) Diagram of developing dental epithelium derived cell types at 12–13gw. (L) Diagram of developing dental epithelium derived cell types at 17–19gw.
To identify the predicted developmental trajectory of the dental epithelial lineages, we performed pseudotime analysis (Figure 3C) summarized by the simplified tree graphs (Figure 3D, ameloblast trajectory with red arrows). The trajectory analysis suggests that the OE directly gives rise to DE. The DE then gives rise to the EK and SR lineages and the OEE lineage, which gives rise to SI, IEE/PA, and eAM/sAM.
To validate the bioinformatic findings, and to spatially localize the predicted clusters, we performed multiplex RNAScope in situ hybridization at multiple timepoints in human fetal tissue. We used combinations of cluster-specific markers identified by transcriptional analysis to map cells from each cluster in the fetal tissues (Figure 3E, 3G, 3H and 3J; Figure S3C; File S3–RNAScope_logic_tables.xlsx). Computational pseudo-spatial mapping of these cells validated the spatial identity of the predicted clusters (Figure 3A) in human fetal tissue, and identified insights on EK, support cells, and CL potential role (Figure 3F and 3I).
The EK is a signaling center that has previously been identified in mouse tooth development and is thought to organize epithelial budding or folding during cap and bell stage transitions43–45. Primary EK emerges during the cap stage, followed by secondary EK formation in the bell stage. We identified a cluster consistent with EK in human fetal development. Real-time distribution showed that this cluster appeared at 9–11gw (early cap stage) and again at 14–16gw (early bell stage) (Figures 3B; S3B and S3D; 3E–3F), in line with the expected appearance of primary and secondary EK, respectively. EK are essential signaling centers in tooth morphogenesis, plausibly playing a role in determining crown shape7. Accordingly, GO terms identified in association with EK cluster included morphogenesis and appendage development (Figure 3B). These findings identify the human EK population at the transcriptional level and thereby will lead to further understanding of the initiation of human tooth morphogenesis and toothgerm type determination46.
We identified two types of support cells in enamel organ using sci-RNA seq/spatial multiplex in situ analysis, SR and SI. SR are support cells with a star-shaped appearance in histological sections47, which are thought to provide nutrients to the developing ameloblasts18, while SI are thought to support ameloblast differentiation47 (Figures 3A–3B, 3G, 3I, 3K–3L). Our transcriptomic and in situ analysis identified spatially localized two subgroups of SR, inner SR (SRI) closer to the inner surface of the toothgerm and outer SR (SRO) (Figures 3A–3D; 3K-L and S3A), as well as two human SI sub-clusters that appear at 12gw and persist to later development (Figures 3A–3B; 3G, 3I, 3K–3L and S3A). Inner SI (SII) represents the cell layer closer to ameloblasts lineage, and outer SI (SIO) represents the parallel cells adjacent to SII (Figures 3G, 3I, 3K–3L and S3E). Go term analysis suggests increased adhesion in SII, but cell migration and Integrin mediated signaling in SIO. Pathway analyses suggest HH and WNT involvement in SII and TGFβ in SIO (Figure S3D–S3E). These data indicate SII and SIO support cells may have precise signaling capacity to the specific, nearby epithelial cells in ameloblast lineage. Further studies are required to test this hypothesis.
Through sci-RNA-Seq and RNAScope analysis, we identified the inner and outer enamel epithelium (IEE and OEE)10,47. During the early bell stage, the OEE serves as the basal cells on the periphery of the tooth organ, giving rise to the SI, CL, and PA lineages (17–19gw) (Figures 3E–3G, 3I, 3K–3L). Additionally, we localized a small population of LGR6+ cells to the cervical loop (CL) where the OEE and IEE meet, a region previously reported to contain epithelial stem cells in the regenerating adult mouse incisor48. Importantly, our trajectory analysis predicts that stem cells in the CL can generate the ameloblast lineage, indicating a stage-specific role for the CL in human tooth development. While traditionally associated with later root development, our findings suggest that during early fetal development, the CL plays a role in generating the ameloblast lineage as the tooth crown expands. Further investigations are necessary to elucidate the precise role of CL cells in determination of root number and morphology.
Moreover, we found from our data that the transition stages of ameloblast lineage PA, eAM and sAM can be marked precisely by three specific genes: VWDE precisely marks PA (Figure 3J)10, low DSPP expression (lower than OB) marks eAM9, while ENAM marks sAM (Figure 3I, 3J and 3L; Figure S4K–S4M).
Sci-RNA-seq predicts spatio-temporal expression patterns of critical signaling pathways in ameloblasts and facilitates the development of human iPSC-derived early ameloblasts (ieAM) in vitro
To understand the signaling pathways involved in ameloblast differentiation, we compiled a comprehensive analysis pipeline based on ligand-receptor interactions and downstream transcriptional activity (Figure S5A). Briefly, a talklr49 R package was used to identify specific ligand-receptor communications between the cell types at each developmental time point. DEsingle50 and scMLnet51 programs were used to evaluate the downstream signaling activity by establishing multilayer networks between ligands and receptors and between transcription factors and their differentially expressed targets. Finally, activity scores were assigned to each pathway, which represent a percentage (0–100%) of the overall activity for all pathways included in the analysis (TopPath).
Using TopPath, we identified the most active pathways and their associated ligands in ameloblast lineage (Figures 4A-B; S5A–S5C). Our findings revealed that during OE to DE transition, dental mesenchyme secretes BMP, ACTIVIN, and noncanonical WNT ligands, while canonical WNT ligands are secreted within the OE. Similarly, during DE to OEE transition, WNT ligands are secreted from DE and EK, while BMP and FGF ligands originate mainly from the dental mesenchyme. In the OEE to IEE transition, the condensed dental papilla (dental mesenchyme) primarily influences the ameloblast lineage through BMP secretion. Notably, support cells SRI play a significant role by secreting ligands for the TGFβ pathway. Other support cell types, such as SII and SIO, exhibit stage-specific signaling behavior by secreting ncWNT/HH/EGF and FGF ligands, respectively. Mesoderm-derived POB and OB interact significantly with epithelial clusters and secrete FGF and BMP ligands during the PA to eAM transition or the transition to sAM. In the maturation phase from eAM to sAM, WNT ligands are primarily secreted from eAM cells.
Figure 4. Signaling Pathway Analysis of the Ameloblast Trajectory.
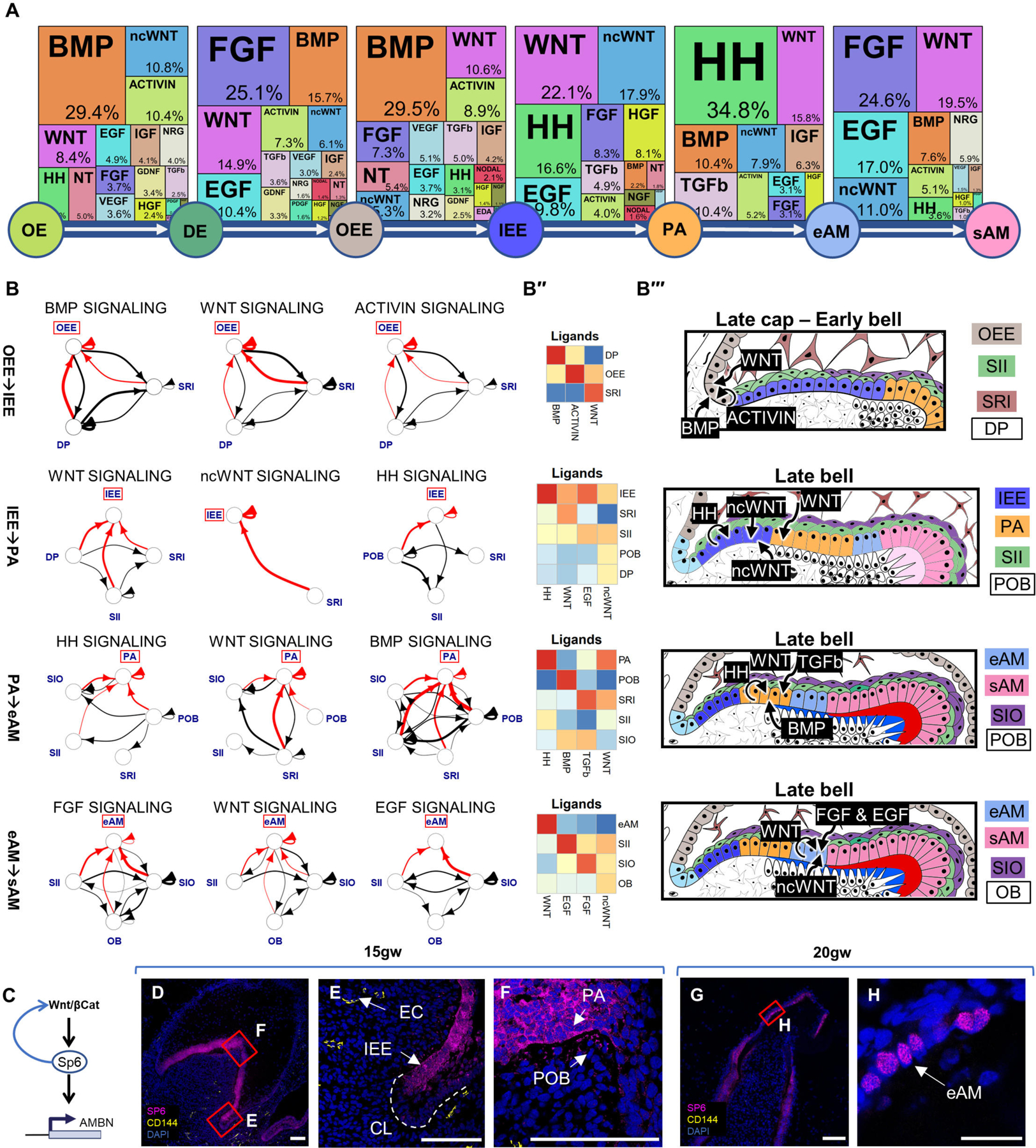
(A) Identification of the most active signaling pathways involved in ameloblast differentiation: BMP, WNT, HH, and FGF. (B) Sources of critical signaling ligands for the top pathways at each developmental stage, originating from both dental epithelium and mesenchyme-derived tissues. (B’’) Heatmaps showing pathway ligand gene expression averaged per cluster. (B’’’) Diagrams illustrating the suggested ligand sources for each pathway at different stages of tooth development. (C) Proposed involvement of the WNT pathway in activating SP6 expression, leading to AMBN expression. Immunofluorescence staining of SP6 in 15gw toothgerm mainly in cytosol of IEE (D-F) and 20gw toothgerm (G-H), mainly localized to the nuclei of AM. Scale bars: 50μm.
Our analysis indicates that support cells SI and SR play a critical role in IEE to PA differentiation by secreting ncWNT/EGF and WNT ligands (Figures 4A–4B’; 4B’’’ and S5B–S5C). During the late stages of ameloblast differentiation (PA to eAM), HH ligands are secreted by PA and SII, while TGFβ ligands are secreted by SRI and SIO. In the final maturation stage (eAM to sAM), EGF is secreted by SII and FGF is secreted by SIO. Interestingly, WNT activity in the OEE to IEE transition is associated with the emergence of SP6 expression in IEE (Figure 4C). WNT pathway has been shown to induce expression of the transcription factor SP652–54, which in turn was found to act on AMBN/AMELX promoters55 (Figures 4C–4F). We found SP6 initially localized in the cytoplasm of IEE/PA, SP6 later translocates to the nuclei coinciding with AMBN expression in eAM/sAM (Figure 4H). These findings support the hypothesis that SP6 expression, induced by the WNT pathway in the IEE transition stage, becomes functional in the eAM stage by translocating to the nucleus and inducing AMBN expression. Future loss-of-function analysis is required to validate this hypothesis. The data suggest that WNT, TGFβ, HH, FGF, and BMP pathways are the most active pathways in ameloblast development compared to the 25 pathways analyzed.
We utilized sci-RNA-seq data (Figure 4A–4B) to develop an iPSC-derived ieAM differentiation protocol (Figure 5A). We first optimized iPSC differentiation into OE56–58. At day 10, OE markers were upregulated, pluripotency markers downregulated, and neuroepithelial/early mesodermal markers unchanged (Figure 5B: Figure S6A–B). OE cells were differentiated into early-stage ameloblasts by inducing BMP4, TGFβ1, WNT/CHIR99021, EGF, and HH/SAG pathways (Figures 4A; 5A, S5B). BMP pathway was transiently inhibited using LDN. This induced remarkable epithelial morphological changes and high expression of early ameloblast marker AMBN at day 16 (Figure 5C; S6C), representing ieAM differentiation stage (Figure S4M).
Figure 5. Human Induced Pluripotent Stem Cells (HiPSC) Derived Pre-Ameloblast Differentiation Protocol Guided by sci-RNA-seq.
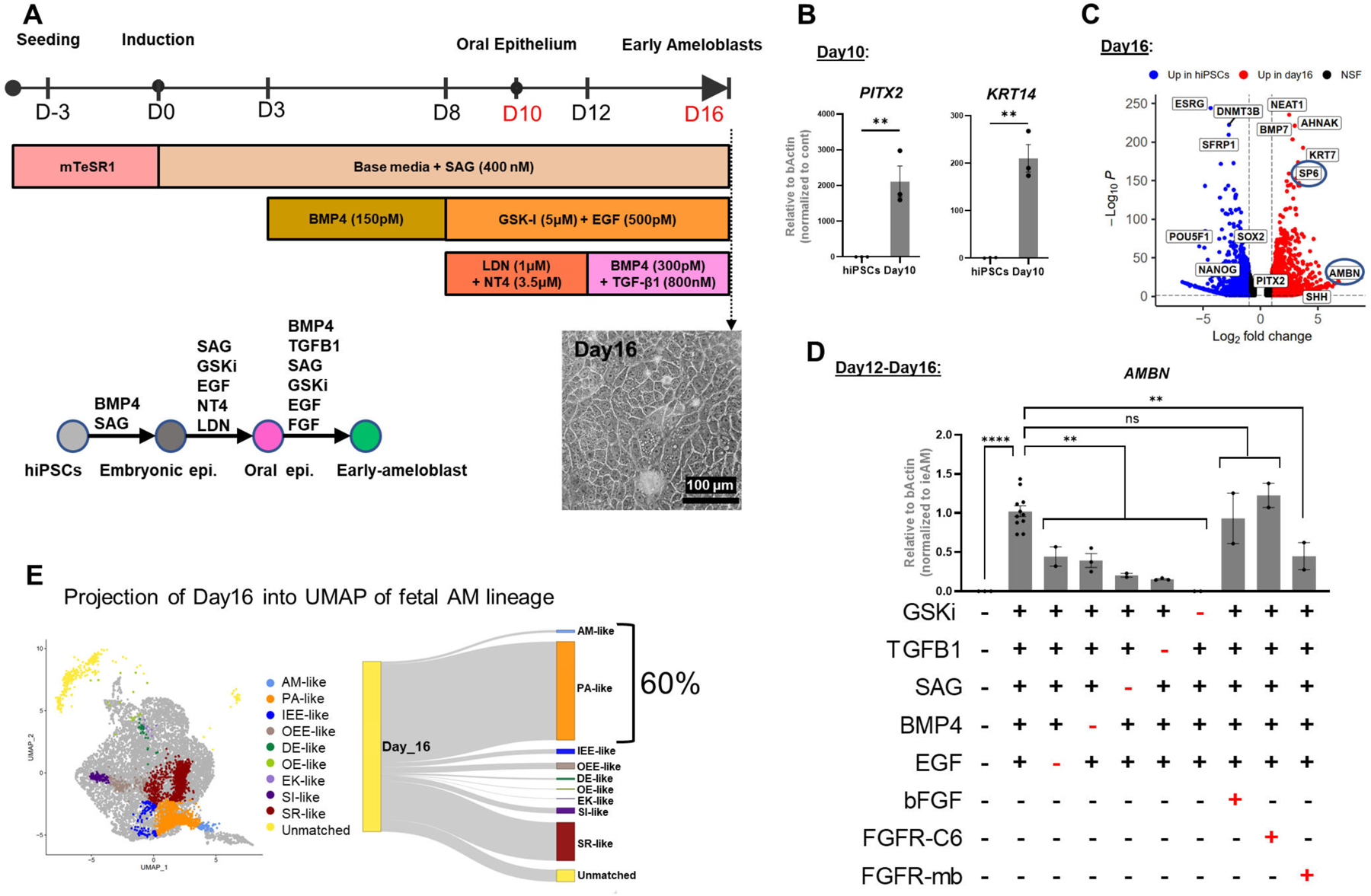
(A) Schematic of the 16-day differentiation protocol targeting signaling pathways using growth factors and small molecules. (B) QRT-PCR analysis showing upregulated expression of oral epithelium markers PITX2 and KRT14 at Day 10 of differentiation. (C) Bulk RNA-seq analysis demonstrating upregulation of ameloblast markers SP6 and AMBN at Day 16 of differentiation compared to undifferentiated hiPSC control. (D) Evaluation of pathway efficiency during differentiation by removing each agonist and or adding FGFR-mb to inhibit the FGFR1/2c pathway and assessing AMBN expression in QRT-PCR. Each performed in duplicates or more. (E) Projection of Day 16 differentiated cells onto in vivo dental epithelium-derived cell types, showing that 60% of the cells resemble the gene expression pattern of PA and eAM. Error bars represent SEM. Statistical significance was determined using one-way ANOVA; ***p < 0.001; ****p < 0.0001.
To dissect which pathways were essential for the differentiation from day10 to day16, we eliminated EGF, SAG, BMP4, or TGFβ1 independently and found that the expression of AMBN was significantly reduced in each case (Figure 5D). However, eliminating GSKi completely abolished AMBN expression, suggesting that WNT signaling is a master regulator upstream to the other pathways (Figures 4C-H; 5D). Our pathway prediction pipeline also highlighted the involvement of the FGF pathway but adding bFGF or FGFR1/2c-specific agonist (FGFR-C6)59 did not significantly affect AMBN expression. We postulated that endogenous FGF ligands secreted by cells in culture saturate the receptors, rendering exogenous ligands ineffective. To validate this hypothesis, we utilized a computationally designed protein, FGFR1/2 mini-binder (FGFR-mb)60,59, specifically inhibiting FGFR1/2c splice variant activity. Addition of FGFR1/2c-minibinder significantly reduced AMBN expression, confirming the requirement of the FGFR pathway, particularly the FGFR1/2c splice variant, in ameloblast differentiation (Figure 5D). We further confirmed that the FGFR1c splice variant is more abundant than the FGFR1b splice variant during day10 to day16 of ameloblast differentiation (Figure S6K). These findings underscore the importance of AI-designed mini-proteins in dissecting signaling pathway requirements and suggest their potential use alongside genetic perturbations in iPSC-derived differentiation studies.
To analyze the efficiency of the differentiation, we performed sci-RNA-seq on Day10 and Day16 of iPSC derived ameloblast differentiation (day10-OE and day16-early ameloblasts) and compared the gene expression data to the fetal tissue gene expression data. Our initial clustering and trajectory analysis indicated three major clusters at day 10 and six clusters at day16 (Figure S6D–S6E). Sequencing identified a significant overlap between human fetal and iPSC-derived ameloblasts in 2D culture. A survey of relevant markers to the dental epithelium (Figures S6F–S6G) showed the kinetics of their differential expression across the proposed trajectory (Figures S6D–S6E). Utilizing the markers for the oral/dental epithelial progenitors45,61, enamel epithelium62, and ameloblasts63 we were able to identify all the differentiated cell types (Figure S6E, S6J). Interestingly, FGF2 expression in the sequenced day10/day16 is highly abundant supporting our early hypothesis about the possible self-saturation of FGFR pathway (Figure S6L).
To compare the in vivo and in vitro datasets, we employed the projection method in Seurat 4.0 and the integration method in LIGER software packages64,65. A small subset of cells in the day 16 sample exhibited characteristics of OE-like, DE-like, SR-like, and SI-like cells. However, the majority (60%) of cells belonged to the PA and AM-like clusters, indicating a predominant differentiation toward the ameloblast lineage (Figure 5E). River plot analysis using LIGER revealed the correspondence between annotated clusters from the fetal dental epithelial lineage and the in vitro day 16 differentiation clusters (Figure S6H). The fetal OE cluster aligned with cluster 1 (d16_1) in the in vitro differentiation, while DE, SR, and OEE clusters from the in vivo samples matched mainly with cluster 2 (d16_2), and the SI cluster matched cluster 4 (d16_4) (Figure S6E). Pre-ameloblast and ameloblast clusters corresponded to clusters 5 and 6 (d16_5, d16_6), respectively, which encompassed 47% of total cells (Figure S6I). Furthermore, analysis of AMBN production, revealed that 25% of cells in the day 16 differentiated samples were capable of producing AMBN (Figure S6C) [7]. These findings indicate that our day 16 iPSC-derived early ameloblast-like cells (iAM) share similarities with fetal pre-ameloblasts and early ameloblasts, demonstrating the ability of our 2D differentiation procedure to generate pre-ameloblasts and early differentiated ameloblast-like states, namely ieAM.
Amelogenesis imperfecta mutant ieAM show defects in polarity
Developing iAM differentiation protocols allows us to study the etiology of tooth diseases (amelogenesis imperfecta) resulting from ameloblast defects. To analyze mutant ameloblast phenotypes, we first developed an ameloblast organoid system to study this polarized cell type in a 3D model. To generate AMBN expressing iAM cells with apical-basal polarity (as seen with AM in vivo) we grew the 16d iAM cells in suspension to form spheroids (7days, Figure 6A-D). We then performed immunofluorescence staining for SP6, AMBN, and ZO1 and DSPP (Figure 6B-D) and observed that the fetal eAM marker, transcription factor SP6 is expressed in all the differentiated cells and is exclusively localized to the nucleus, suggesting that eAM is enriched cell type in the organoids. As seen in vivo AM, the nucleus is located towards the basal side of the cell (Figure 6C-D). Interestingly the ameloblasts in the produced organoids are in an early developmental stage based on their expression patterns, nuclear SP6, apical ZO-1, high AMBN and low DSPP66 (Figure 3G and 3J; S4G–S4K’, S4M; Fig.6A-D). Hence, we call them iPSC-derived early ameloblasts, ieAM.
Figure 6. Characterization of ieAM and formation of ieAM organoids.
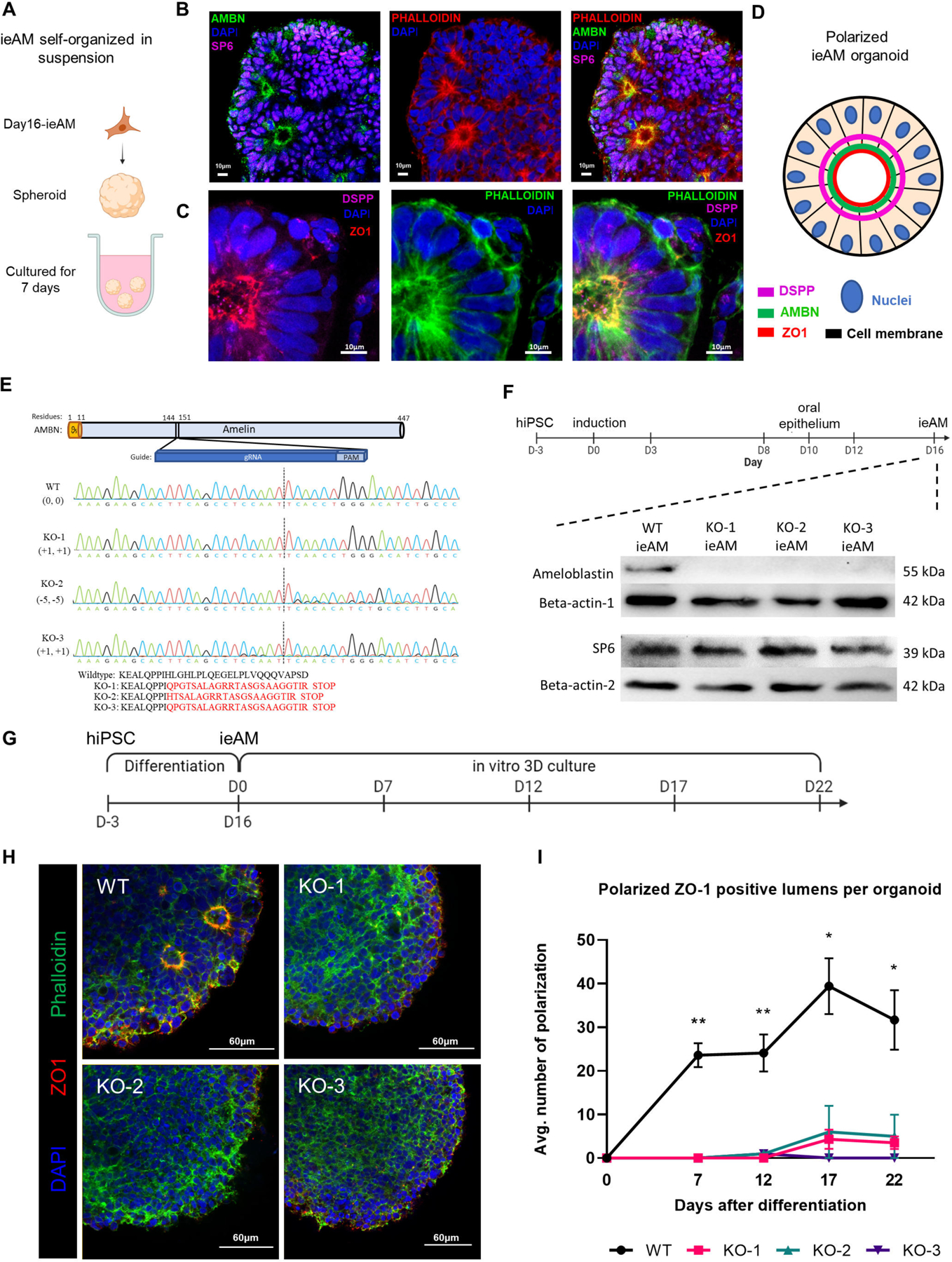
(A) Schematic of iAM organoids formation while cultured in suspension in ultra-low attachment plate. The formed iAM organoids express SP6 in the nuclei and AMBN (B), DSPP and ZO-1 (C) toward the apical side of the polarized ameloblasts. (D) A diagram simplifying the ameloblast organoid polarized structure toward a central lumen marked by ZO1, DSPP and AMBN. The markers observed indicate that the ameloblasts are in early stage of development (ieAM; high expression of AMBN and low expression of DSPP). (E) Protein structure of ameloblastin with guide RNA location indicated, as well as DNA sequencing chromatograms to compare the wild-type AMBN with the three AMBN mutant DNA sequences. The protein sequences below show that the three mutant cell lines have an early stop codon in their AMBN gene. KO-2 and KO-3 have a small population (<5%) of +1 and −11 respectively. (F) Western blot analysis showing AMBN protein knocked out. SP6 protein is a transcription factor of the AMBN gene, which acts as a marker for early-ameloblasts. The timeline of ieAM differentiation is shown above, indicating that the Western blot analysis was done on day 16. (G) Timeline of the ieAM differentiation plus in vitro 3D organoids culturing. (H) Representative confocal microscopy images showing cross-sections of in vitro 3D-cultured organoids derived from wild-type and mutant early-ameloblast lines stained with DAPI (blue), Phalloidin (F-actin, green) and ZO-1 (tight junction protein 1, red). Scale: 60μm. (I): Quantification of polarized ZO-1 positive lumens among in vitro 3D-cultured wild-type and mutant organoids at various time points after the 16-day ameloblast differentiation. *p ≤ 0.05 and ** p ≤ 0.01
We next used the ieAM organoid model to study human ameloblast diseases67 in vitro. CRISPR/Cas9 was employed to generate three AMBN knockout iPSC lines (Figure 6E–6F). The number of polarized ameloblasts in the differentiating organoids was quantified over 22 days, revealing significantly lower counts in the AMBN KO-mutant cell lines compared to wild-type ieAM (Figure 6G–6I). These findings support prior research indicating the role of AMBN in maintaining cell polarity and validate the ieAM organoid model for studying amelogenesis imperfecta and gene function67,68. Specifically, our results highlight the requirement of AMBN for early ameloblast cellular polarity.
Ameloblast maturation in vivo
To assess the maturation potential of ieAM cells in vivo, we injected differentiated cells (day16, 2D-cultured cells) intramuscularly into adult SCID mice, and after 8 weeks, the presence of injected cells was confirmed by human nuclear antigen staining (Figure S7B). Subsequent analysis of serial sections revealed that the injected iAM cells exhibited a more mature phenotype, as indicated by the expression of definitive ameloblast markers such as AMELX, AMBN, DSPP, and KRT14, as well as their ability to produce calcified material demonstrated by Alizarin red and Von Kossa staining (Figure S7A–S7H). However, these cells were negative for ENAM, indicating that they did not fully mature.
Ameloblast and odontoblast co-culture allows further maturation in vitro
Since close contact between AM and OB is critical for tooth development, we developed an organoid model of the two cell types. We co-cultured the induced early ameloblast organoids with primary human dental pulp stem cells (DPSCs) to assess the interaction level between the two cell types and the effects on ameloblast maturation. We found that simple co-culture in suspension can eliminate transient DSPP and induce AMELX expression in ieAM organoids and induce strong DSPP expression in the odontoblast organoids, (as observed in the developing human tooth; Figure S4C–S4C’’, S4K–S4K’), as well as induction of calcified matrix (Figure 7A–D).
Figure 7: Ameloblast and Odontoblast Co-culture Allows Further Maturation of ieAM into isAM.
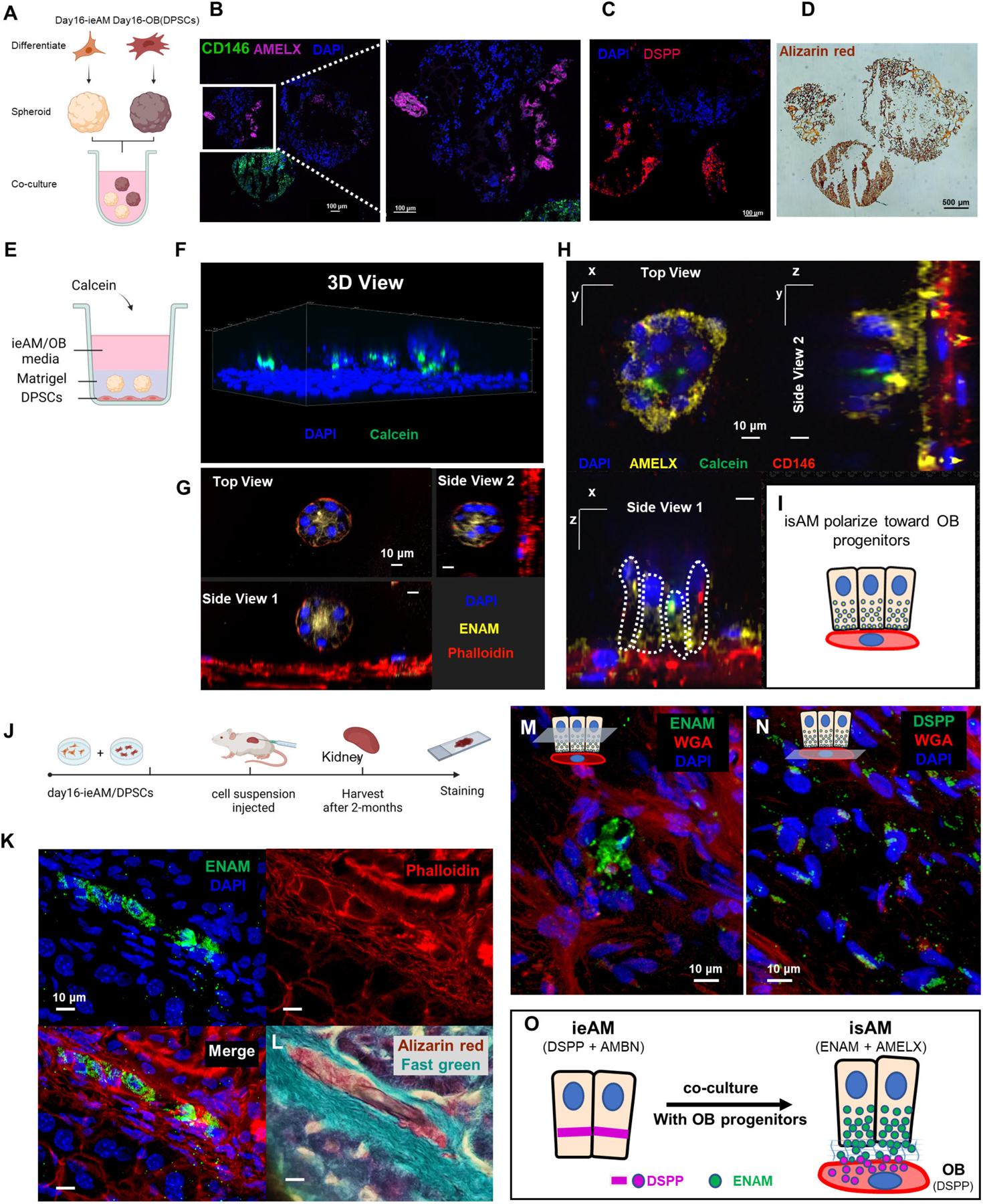
(A) Schematic representation of the co-culture experiment between iAM organoids and DPSC organoids in suspension culture. The organoids were formed separately and then combined for 14 days in iAM base media. Immunofluorescence analysis revealed the expression of AMELX in iAM organoids (B) and the expression of CD146 (mesenchymal marker) and DSPP in DPSC/OB organoids (C). Alizarin red staining (D) indicated positive calcification in both organoid types, with DPSC/OB organoids showing more calcifications. (E) Schematic representation of the co-culture experiment between DPSCs as a monolayer and iAM embedded in Matrigel above it. Calcein, a fluorescent dye that binds to calcium, was added to the media containing a mixture of iAM base media and odontogenic media. (F) Three-dimensional reconstructed image from confocal images of the co-cultured organoids, showing association with Calcein and expression of ENAM at the center after 7 days (G). After 14 days, the organoids close to CD146-expressing DPSC/OB started to revert polarity towards DPSCs/OB while expressing AMELX (H), as depicted in the simplified diagram (I). (J) Schematic representation of the in vivo mouse experiment, involving the injection of day16-ieAM combined with DPSCs beneath the capsule of the right kidney in adult SCID mice. Kidneys were dissected and cryosectioned for further analysis. (K) Immunofluorescence staining showing ENAM-positive isAM in engrafted areas beneath the capsule. (L) Alizarin red staining indicating calcifications associated with isAM. (M) Immunofluorescence staining showing ENAM-positive isAM and high DSPP-positive DPSCs/OB (N). (O) Summary model proposing the interaction between ieAM and DPSCs/OB leading to the maturation of isAM.
After confirming the maturation of induced ameloblasts (ieAM) in the presence of DPSC derived OB-lineage, we cocultured the cells in a layered approach. DPSCs were plated at the bottom of a flat bottom plate, and ieAM organoids were embedded in a Matrigel layer above the DPSCs (Figure 7E). Calcein was added to a mixed media of ieAM and odontogenic media. Using 3D reconstructed confocal images we identified the association of ieAM with calcein, indicating their ability to undergo mineralization/calcification (Figure 7F). Furthermore, the co-cultured ieAM expressed ENAM and AMELX (Figure 7G-H) and reverted their polarity towards the differentiating OB-lineage (Figure 7H–7I), demonstrating their maturation to secretory state (isAM). This 3D organoid system replicates the normal cell-to-cell interface observed in tooth development, where enamel proteins are secreted towards OB (Figure S4K–S4K’), thus paving the way for the development of human tooth organoids in vitro.
Kidney capsule injections of co-cultured ieAM and DPSCs show mineralization and Ameloblastin, Amelogenin, and Enamelin secretion
Upon confirming that ieAM and DPSCs interact and mature together in co-culture, we investigated their behavior in vivo settings. We transplanted the cells (day16-ieAM and DPSCs) into adult SCID mouse kidney capsules (Figure 7J). After 8 weeks the injected cells were identified by human nuclear antigen staining (Figure S7L–M), showing the survival of AM and OB-lineages. Importantly, we identified ENAM+ cells (isAM) in the graft (Figure 7K, 7M, S7N) with calcified material (Figure 7L). The region was surrounded by high DSPP+ cells (DSPP/OB) (Figure 7N, S7N), suggesting a formation of AM/OB interface in vivo. Based on the serial section analysis (Figure 7G and Figure 7L, S7N) we propose that the mode of interaction is polarized isAM facing OB-lineage, as depicted in the model shown in (Figure 7O). The transplanted cocultured cells show mineralization, and AMBN, AMELX and ENAM secretion.
Discussion
Ameloblasts and odontoblasts are two critical cell types that secrete enamel and dentin, the protective tooth coverings required for functional tooth. While odontoblasts have limited regenerative potential in adults, the absence of ameloblasts poses a challenge for enamel regeneration. Through single-cell sequencing, we characterized the cell types and molecular interactions during human tooth development and guided by these insights, we developed a differentiation protocol to generate early ameloblast organoids (ieAM) from human iPSCs, enabling investigations into the etiology of human dental diseases. Ameloblastin mutant (resulting in amelogenesis imperfecta) culminates in a dramatic cellular polarity defect in ieAM organoids, resulting in mechanistic insights of the disease. Finally, we used this information to develop an ameloblast organoid that expresses mature ameloblast markers and secretes mineralized calcium in vitro and in vivo.
Through sci-RNA-seq analysis, we discovered transcriptionally defined subgroups within the epithelial and mesenchymal lineages, including previously unreported support cell types (SRI, SRO, SIO, and SII) involved in human tooth development. Notably, we found SRI support cells to produce a TGFβ ligand during early tooth development, potentially facilitating the differentiation of inner enamel epithelium (IEE) to preameloblasts (PA). Additionally, SII support cells secreted EGF, while SIO support cells secreted FGF ligands during the later maturation of ameloblasts. Our findings contribute to a deeper understanding of the role of support cells in the patterning and development of ameloblasts. In the mesenchymal lineage, we identified two main branches, the dental papilla (DP) and dental follicle (DF) precursors, both of which are primarily present during early fetal stages and diminish after 20gw. A subset of DF cells differentiates into SOBs, which exhibit transcriptional characteristics suggesting their potential involvement in regeneration after injury40. Furthermore, our study linked DLX3, a well-known disease gene in tooth development69,70, as a key marker for SOBs, emphasizing the need for further investigations into the function of DLX3 using disease-in-a-dish approaches.
Our studies confirmed the crucial roles of WNT, BMP, and FGF pathways, known to be disrupted in hypodontia and tooth agenesis, in different stages of tooth development. Specifically, our analysis revealed that BMP4 signaling is critical during the early transitions from oral epithelium (OE) to dental epithelium (DE) and from DE to outer enamel epithelium (OEE), explaining the correlation between BMP4 mutations and tooth agenesis71. Additionally, we identified the importance of FGF signaling in ameloblast maturation, shedding light on how disruptions in FGF signaling can lead to enamel irregularities72. Our pathway analysis provides insights into the crosstalk that drives tooth development73–75. This comprehensive understanding will facilitate further investigations into the interplay between pathways and enable the development of more effective strategies for mitigating or reversing tooth loss. Furthermore, our use of AI-designed receptor-specific mini-binder proteins offers a simplified method to identify specific signaling pathway requirements in differentiation processes, as demonstrated in our study on FGFR1/2c pathway requirement in ameloblast maturation. This method has broad applicability for analyzing various signaling pathways in normal and disease organoids.
Our studies have revealed new insights and potential approaches to address the challenge of enamel regeneration in adult humans. By conducting the single-cell analysis of the cervical loop (CL) in human fetal teeth, we have uncovered its role in early tooth development. While extensively studied in mouse incisors, where it contributes to continuous growth, a parallel function of the CL in human tissue remained unclear. Classically, the CL is known to give rise to Hertwig’s Epithelial Root Sheath, which initiates root formation. Intriguingly, our analysis identified a role for the CL in giving rise to human ameloblasts in early tooth development, as the crown expands before the root formation. Our findings provide a basis for future studies to develop CL-like cells with ameloblast lineage potential.
Finally, the present work characterized the molecular basis for human ameloblast differentiation. We have used this knowledge to develop an assay for differentiating human iPSC-derived early ameloblasts in a dish (ieAM). Comparing fetal data to the ieAM differentiation suggests that ieAM shares high similarity with fetal pre-ameloblasts and early ameloblasts. In addition, the ieAM cells showed a significant increase in maturation, including calcifications, when tested in vivo. Co-culturing ieAM and OB lineage in vitro or in vivo accelerates maturation to isAM stage with polarity, apical secretion of enamel proteins and calcification toward the OB lineage cells, as seen in fetal development.
Hence, we have developed a chemically defined serum-free differentiation protocol to generate human dental epithelium, and their subsequent differentiation into enamel organ-like 3D organoids. This developed organoid has potential for future dental therapies. Furthermore, it can be used to study dental diseases such as Amelogenesis Imperfecta that will guide the field toward therapeutic approaches.
Limitations of the Study
In this study, we analyzed single cell RNA sequencing data and multiplex RNAscope in situ data from human fetal dental tissues at multiple timepoints. Due to limitations in the number of human fetal dental cells, we had to pool samples from multiple donors. As a result, retrospective lineage tracing methods were not feasible. Genetic recorder systems for tracing lineage or assessing clonality are potentially possible in the future studies.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Hannele Ruohola-Baker. (hannele@uw.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The data generated in this study can be downloaded in raw and processed forms from the NCBI Gene Expression Omnibus under accession number (GSE184749). Upon publication, raw RNAScope data will be made publicly available on dryad.org (Dryad research data repository).
The custom R codes used to generate some of the results in this paper are available in https://github.com/Ruohola-Baker-lab/Tooth_sciRNAseq (DOI: 10.5281/zenodo.8076442).
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Fetal Human Tissue collection and dissection
This study is approved by the Institutional Review Boards (IRB) at University of Washington for the use of human fetal tissues: BDRL (CR000000131) and Ruohola-Baker Laboratory (STUDY00005235). Fetal craniofacial tissues were collected from Birth Defect Research Laboratory (BDRL), University of Washington, which was obtained under informed consent from donors, and transferred to Ruohola-Baker laboratory submerged in Hank’s Balanced Salt Solution (HBSS) media (Gibco, #14025092) on ice. Toothgerms and salivary glands were dissected in cold RNase free Phosphate-Buffered Saline (PBS) (Invitrogen, #AM9624) within six hours from the initial dissection at BDRL. To extract the toothgerms, a vertical cut was made at the midline of the upper/lower jaw for orientation, then a horizontal cut was made from the right side of the midline along the top of the alveolar ridge to expose one toothgerm at a time. The first two toothgerms from the midline were the incisors, the next toothgerm was the canine, and the last two toothgerms were the molars. The same procedure was followed to extract toothgerms on the left side of the jaw. The submandibular salivary glands were harvested from the distal end of the lower jaw. The toothgerms from 9–11gw were too small for dissection and not useable for sequencing; therefore, these jaws were cut into two posterior sections and one anterior section to separate molars from the incisors and canines at these timepoints. The extracted tissues were transferred into an Eppendorf tube and snapped-frozen using liquid nitrogen. The frozen samples were stored at −80°C until nuclei extraction.
Cell lines and in vitro differentiation
Briefly, hiPSCs (WTC-11 human induced pluripotent stem cells) (Coriell, #GM25256) were seeded on 12-well plates coated with growth factor-reduced Matrigel (Corning, #356231) and cultured in mTeSR1 stem cell medium (StemCell Technologies, #85850) until cells reach confluency with medium changes daily. On the first day of differentiation (deemed Day 0), stem cell media is replaced with ameloblast base media consisted of either EpiCult-C media (StemCell Technologies, #05630) or RPMI 1640 Medium (Thermo, #11875093) mixed with EpiLife (Thermo, #MEPI500CA) at 1:1 ratio, supplemented with 0.1x supplement S7 (Thermo, #S0175), 0.1uM β-mercaptoethanol (BME) (Sigma, #M7522) and 400um smoothened agonist (SAG) (Selleckchem, # S7779). On day 3 of differentiation 150pM of bone morphogenic protein-4 (BMP4) (rndsystems, #314-BP-010) is continuously added daily till day 7. At day 8, the base media is supplemented with 1uM of BMP-I inhibitor (LDN-193189) (Tocris, # 6053), 5uM of GSK3-Inhibitor (CHIR99021) (Selleckchem, # 4423), 500pM epidermal growth factor (EGF) (rndsystems, #236-EG) and 3.5μM of Neurotrophin-4 (NT4) (rndsystems, #268-N4). The cultures were then harvested at day 10 at an oral epithelium stage, or extended to day 16 by adding 300pM BMP4, and 800nM transforming growth factor beta 1(TGFβ1) (rndsystems, #7754-BH) for the early ameloblast stage at day16. For testing FGFR signaling requirement for the maturation process we added 50nM purified FGFR-mb (see below) to the media at day 12 and harvested the samples at day 16 of the differentiation.
METHOD DETAILS
Nuclei extraction
Frozen tissues were carefully transferred to a stack of chilled aluminum foil kept on dry ice to prevent thawing. The folded foil encapsulating the tissues were placed on a block of dry ice and the foil was pounded with a pestle to pulverize the tissues into powder. 1mL of lysis buffer that contains nuclei buffer (10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2, pH 7.4), 0.1% IGEPAL CA-630, 1% SUPERase In RNase inhibitor (20 U/μL, Thermo), and 1% BSA (20 mg/mL, NEB) were added onto the tissue powder and transferred to a 1.5mL tubes. Samples were incubated in the lysis buffer for 1 hour on ice. The samples were pipetted up and down with pre-cut 1000uL pipette tip to disassociate the tissue further. The dissociated tissues were passed through 70 um cell strainers (Corning) into a 50mL conical tube. The strainers were rinsed with lysis buffer to minimize nuclei loss. The samples were centrifuged to pellet the nuclei at 500g for 5 minutes at 4°C and the supernatant was discarded. The samples resuspended again in 1ml lysis buffer, transferred into new 15mL tubes, pelleted again and the supernatant was discarded. The pellets were resuspended in 50ul of nuclei buffer, and 5 mL of 4% Paraformaldehyde (PFA) (EMS) diluted in RNase free PBS, was added to fix the nuclei for 15 minutes on ice. The tubes were flicked gently every 5 minutes to reduce clumping of nuclei. The fixed nuclei were pelleted at 500g for 3 minutes at 4°C and the PFA waste was discarded. The pelleted nuclei washed in nuclei wash buffer (cell lysis buffer without IGEPAL) and then centrifuged again at 500g for 5 minutes 4°C, and the supernatant was discarded. Finally, the pellets were resuspended again in nuclei wash buffer and then flash-frozen in liquid nitrogen before storing in −80°C.
For nuclei extraction from the differentiation culture, the cells were treated with StemPro Accutase (Thermo, #A1110501) for 7min to detach the cells and transfer them into 15mL tube, then incubated in trypsin (Thermo, #25300054) for another 7min to prevent re-clumping. The cells were span down to remove trypsin after inactivation with more media. The pellet was treated with nuclei lysis buffer and the same steps for nuclei extraction protocol were followed.
Sci-RNA-seq
Single-cell combinatorial-indexing RNA-sequencing (sci-RNA-seq) protocol is described previously24. sci-RNA-seq relies on the following steps, (i) thawed nuclei were permeabilized with 0.2% TritonX-100 (Sigma, #T9284) (in nuclei wash buffer) for 3 min on ice, and briefly sonicated to reduce nuclei clumping; (ii) nuclei distributed across 96-well plates; (iii) A first molecular index is introduced to the mRNA of cells within each well, with in situ reverse transcription (RT) incorporating the unique molecular identifiers (UMIs); (iv) All cells were pooled and redistributed to multiple 96-well plates in limiting numbers (e.g., 10 to 100 per well) and a second molecular index is introduced by hairpin ligation;(v) Second strand synthesis, tagmentation, purification and indexed PCR; (vi) Library purification and sequencing is performed.
All libraries were sequenced on one NovaSeq platform (Illumina). Base calls, downstream sequence processing and single-cell digital-expression matrix generation steps were similar as described in sci-RNA-seq3 paper24. STAR76 v.2.5.2b54 aligner used with default settings and gene annotations (GRCh38-primary-assembly, gencode.v27). Uniquely mapping reads were extracted, and duplicates were removed using the UMI sequence, reverse transcription index, hairpin ligation adaptor index and read 2 end-coordinate (that is, reads with identical UMI, reverse transcription index, ligation adaptor index and tagmentation site were considered duplicates).
Data Analysis
All low-quality reads were removed from the data (including jaws, toothgerms and salivary glands samples from all time points) by setting UMI cutoff to greater than 200 and removing all mitochondrial reads (QC table: Figure S1F). Following Monocle3 workflow24,33,77, data underwent normalization by size factor, preprocessing, dimension reduction (UMAP algorithm78), and unsupervised graph-based clustering analysis (Leiden Algorithm 79,80). Certain clusters from the initial analysis were selected for further sub-clustering, and the previous analysis repeated. Pseudotime analysis also was done with Monocle3 following the default workflow, which include learning the graph, ordering the cells, and plotting the trajectory over UMAP. PanglaoDB34, a curated single-cell gene expression database was utilized to explore the consensus of cell type markers used across publicly available single-cell datasets.
Top marker genes
Each dataset or subset was analyzed with monocle’s top_maker function to find potential marker genes. All non-protein coding genes, ribosomal and mitochondrial genes were excluded from the input genes, and only the top 100 genes sorted by marker score were included in the results.
Heatmap and GO-terms enrichment
ComplexHeatmap package81 was used to generate custom heatmaps that integrate GO-terms per clusters. ViSEAGO package36 used to generate the GO-terms, and simplifyEnrichment package82 used to extract keywords from the top 100 GO-terms (by p value) per cluster. The top 50 marker genes in each cluster were utilized as the input for ViSEAGO. The keywords generated by simplify Enrichment, were filtered to eliminate redundant and irrelevant words, and only the very top words are displayed on the heatmap.
Pseudotime analysis
Pseudotime analysis was done using monocle3 and the density outline of each time point were overlayed on the UMAP graph, to give a better indication of the temporal presence of each cluster.
Comparison of enriched overlapping genes between datasets
The following datasets were downloaded from GEO: GSE14612310, and GSE18938139, and processed and normalized using Seurat 4.0 package64. The function “FindAllMarkers” was used with default settings to generate all enriched genes for each cluster with each dataset. The lists were filtered by p value < 0.05. To visualize the number of overlapping genes, the UpSetR package83 was used to generate upset plots. The hypergeometric statical test was used to calculate the significance of the overlaps, and Bonferroni correction was applied for the adjusted p value. Go term analysis and graph was produced by ShinyGO84.
Top pathway analysis
We analyzed the stages of ameloblast development as identified in (Figure 3D). OEE and CL were combined as one OEE cluster to increase the statistical power (Figure 5A). To analyze those stages in a thorough and reproducible manner, we compiled a comprehensive analysis pipeline that evaluates pathway activity based on ligand receptor interaction and downstream activity. The workflow for our analysis is shown in (Figure S5A). The first step in the analysis is selecting the appropriate input for each stage of the differentiation to be analyzed. At each stage, we consider the progenitor cells and the target cell type to be differentiated into, as well as all the support cell types that are present in the same stage and that are likely to send the signals. The second step is to analyze all the potential ligand-receptor interactions between the selected cell types, but only focus on in-coming interactions toward the progenitor cells of interest. For this part of the analysis, we used a software, talklr package49, which uses an information-theoretic approach to identify and rank ligand-receptor interactions with high cell type-specificity. We further filtered talklr output by selecting those ligand-receptor pairs that fall within the major signaling pathway of interest (TGFβ, BMP, GDF, GDNF, NODAL, ACTIVIN, WNT, ncWNT, EGF, NRG, FGF, PDGF, VEGF, IGF, INSULIN, HH, EDA, NGF, NT, FLT3, HGF, NRXN, OCLN). The third step of the workflow is to obtain the differentially expressed genes (DEGs) between the progenitor cells of interest and their differentiated cell type. This set of genes can be used to evaluate the downstream activity and can be linked to specific ligand-receptor pairs. We used DEsingle package50 with FDR threshold set to 0.1 to obtain DEGs. The top marker genes for the progenitor cells were also excluded from DEGs in this analysis, to ensure more weight is given to the differentiated cell type. The fourth step is to generate a multilayer network that models the upstream interactions (obtained from step #2) and the downstream interactions that includes transcription factors (TF) and their target genes (DEGs obtained from step #3). We used the R package scMLnet51 to generate multilayered network interactions that consists of a top layer for ligands, a layer for receptors, a layer for TFs and a layer for TF-targets. The fifth step is to implement a scoring system to evaluate the connectivity of each part of the multilayered network obtained from the previous step, to determine which path is probably more active. We started by assigning fold-change values (obtained in step #3) to target genes at the lowest level. At the next level, the TF layer, we assigned the mean values of all the connected TF-targets to each TF. Normalization of the scores to the interaction database depth is done after each step, to ensure the scores remain comparable with each category of interactions. At the receptor layer, we calculated the sum of the values of all the connected TFs to each receptor. At the ligand layer, we calculated the sum of the values of all the connected receptors to each ligand. And finally, all ligands that fall within the same pathway family are aggregated together. The sixth step of our pipeline is to rank pathways based on the percentage of activity compared to the overall combined activity scores of all pathways evaluated in this analysis. The results indicate the most active pathways or the most active ligands that are key drivers of the differentiation at a specific stage of development (Figure 5A).
Differential expression
DEsingle package50 was used to calculate differential expression (DE) between clusters. DEsingle was designed for single-cell RNA sequencing, and it employs Zero-Inflated Negative Binomial model to estimate the proportion of real and dropout zeros. Our cutoff for DE genes were set to include genes with False Discovery Rate (FDR) < 0.1 and more than twofold change.
Multilayer network analysis
To generate a multilayer network that models the upstream interactions and the downstream interactions that includes transcription factors (TF) and their target genes, we used the R package scMLnet51. A custom wrapper code was developed to integrate talklr and DEsingle results with scMLnet.
Signaling interaction
In our study, we used talklr package49 to identify ligand-receptor interaction changes between two adjacent tooth developmental stages. talklr uses an information-theoretic approach to identify ligand-receptor interactions with high cell type-specificity. Ligand-receptor interaction score is defined as Li*Rj, the product of expression levels for the ligand in cell type i and the receptor in cell type j. We normalize interaction scores by dividing Li*Rj with the sum of interaction scores across all n2 cell-cell interactions. talklr uses the Kullback-Leibler divergence to quantify how much the observed interaction score distribution differs from the reference distribution. The reference distribution is the equi-probable distribution where every possible interaction has probability, when the aim is to identify cell type-specific ligand-receptor pairs in a single condition. Compared to existing methods such as cellPhoneDB85 or singleCellSignalR86 the unique strength of talklr is that it can automatically uncover changes in ligand-receptor re-wiring between two conditions (e.g., different time points, disease vs. normal), where the reference distribution is the observed interaction scores in the baseline condition. The parameters we used were 0.001 for expression threshold, which was determined by calculating the level of expression of the 20th quantile of the aggregated clusters, and 1e-06 for the pseudo-count value which was determined by the minimum averaged expression value in the set. We considered the interactions among the top 100 ligand-receptor pairs returned by talklr, and we further prioritized them by selecting those that are known to be from physically proximal cell types.
Datasets projection analysis
Seurat 4.0 package64 was used to project the in vitro differentiation sample into the UMAP space of the fetal ameloblasts sample. The dataset in monocle object format that contains the precomputed PCA and UMAP was converted into Seurat object. The projection was done with the default parameters. Graph-based clustering was performed on the projected data by calculating the nearest neighbor cluster center of the fetal sample. Package ‘networkD3’87 was used to create the river plot showing the proportions of the classified cells.
Datasets integration analysis
LIGER package65 was used to integrate the fetal dental epithelium lineage dataset with the differentiation datasets to facilitate the cell type label transfer between the sets. The following integration parameters were used: k = 25, lambda = 10, and these settings were determined by utilizing the built-in function that suggests the best values that suit our datasets. For the river plot generation, the minimum fraction of the branching streams was set to 0.25, and the minimum number of cells set to 50. Clusters that have no out- or ingoing connection were eliminated from the graph for clarity.
RNA Fluorescence in situ Hybridization (FISH) and analysis
A 12-probe RNAScope HiPlex assay (Advanced Cell Diagnostics, Inc.) including probes against 13 transcripts differentially expressed between cell type clusters in mesenchyme- and epithelial-derived lineages were selected to distinguish cell populations: VWDE, SALL1, FGF4, IGFBP5, FGF10, PRRX1, FBN2, ENAM, PCDH7, SOX5, KRT5, and either DSPP or LGR6. Fresh frozen tissue sections from 13gw and 19gw were assayed according to the manufacturer’s protocol. Briefly, the fresh-frozen tissue sections were fixed using 4% paraformaldehyde in 1X PBS, dehydrated, and treated with the Protease IV kit component. The first four probes were imaged after completing the manufacturer’s specified hybridization steps, counterstaining, and coverslipping. Images of tissue sections were obtained using a Nikon Ti2 with an Aura light engine (Lumencor, Beaverton, OR), and BrightLineSedat filter set optimized for DAPI, FITC, TRITC, Cy5 & Cy7 (Semrock, Rochester, NY: LED-DA/FI/TR/Cy5/Cy7–5X5M-A-000) or a Yokogawa CSU-X1 spinning disk confocal microscope (Yokogawa Corporation, Sugar Land, TX) with a Celesta light engine (Lumencor), ORCA-Fusion scientific CMOS camera (Hamamatsu Corp, Bridgewater, NJ), and a HS-625 high speed emission filter wheel (Finger Lakes Instrumentation, Lima, NY). Coverslips were removed, the first four fluorophores were cleaved, and the process was repeated for probes 5–8 and then probes 9–12 (File S4). Images were analyzed using Fiji (ImageJ2 v2.3.0) and QuPath (v0.3.0) quantitative pathology and bioimage analysis freeware88. Briefly, The DAPI channel images for imaging rounds two and three were aligned to the DAPI image for imaging round one using the BigDataViewer > BigWarp plugin in Fiji. Matching reference points were identified across the DAPI images and the resultant landmark tables were used in a custom .groovy script (File S5) to align the FITC, Cy3, Cy5, and Cy7 images from the three rounds of imaging. Images were uniformly background corrected and scaled as indicated in File S4. Cellular segmentation was performed in QuPath and positive signal foci and clusters were identified as subcellular detections. Parameters were set to allow for detection of foci while avoiding false positive detection events using positive and negative control images. From QuPath, the coordinates and the number of spots estimated (sum of individual puncta and estimated number of transcripts for clustered signal) for each segmented cell were processed using custom R scripts to map cell locations and expression levels. Out of the transcripts assayed by RNAScope, probe set criteria (File S3) used to identify a given cell population in RNAScope data was selected based on differential expression across the cell types identified in the sci-RNA-seq data at corresponding time points (Figure S3C). Cells matching expression criteria for a cluster’s probe set were designated by cluster color and mapped spatially.
De novo FGFR-Miniprotein expression
The gene encoding the designed FGFR-mb protein sequence was synthesized and cloned into modified pET-29b(+) E. coli plasmid expression vectors (GenScript, N-terminal 8-His tag followed by a TEV cleavage site). The sequence of the N-terminal tag is MSHHHHHHHHSENLYFQSGGG, which is followed immediately by the sequence of the designed protein. Plasmids were transformed into chemically competent E. coli Lemo21 cells (NEB). The protein expression was performed using Studier autoinduction medium supplemented with antibiotic, and cultures were grown overnight. Then, IPTG was added to a final concentration of 500 mM and the cells were grown overnight at 22 °C for expression. The cells were collected by spinning at 4,000g for 10 min and then resuspended in lysis buffer (300 mM NaCl, 30 mM Tris-HCL (pH 8.0), with 0.25% CHAPS for cell assay samples) with DNase and protease inhibitor tablets. The cells were lysed with a sonicator (Qsonica Sonicators) for 4 min in total (2 min each time, 10s on, 10s off) with an amplitude of 80%. The soluble fraction was clarified by centrifugation at 20,000g for 30 min. The soluble fraction was purified by immobilized metal affinity chromatography (Qiagen) followed by FPLC SEC (Superdex 75 10/300 GL, GE Healthcare). The protein samples were characterized by SDS–PAGE, and purity was greater than 95%. Protein concentrations were determined by absorbance at 280 nm measured with a NanoDrop spectrophotometer (Thermo Scientific) using predicted extinction coefficients.
RNA extraction and QRT-PCR analysis
RNA was extracted using Trizol (Life Technologies) according to manufacturer’s instructions. RNA samples were treated with Turbo DNase (Thermo Fisher Scientific) and quantified using Nanodrop ND-1000. Reverse transcription was performed using iScript cDNA Synthesis Kit (Bio-Rad). 10 ng of cDNA was used to perform QRT-PCR using SYBR Green (Applied Biosystems) on a 7300 real time PCR system (Applied Biosystems). The PCR conditions were set up as the following: stage 1 as 50 °C for 2 mins, stage 2 as 95 °C for 10mis, 95 °C for 15 sec, 60 °C for 1 min (40 Cycles). ß-actin was used as an endogenous control. The primer sequences used in this work are available in File S6.
Development of Ameloblast Organoid
The day16 differentiated ieAM cells were trypsinized using TrypLE (Thermo Scientific) and re-plated in in 24-well ultra-low attachment plate (Corning, #4441) containing an ameloblast base medium with 10 μM ROCKi (Y-27632, Selleckchem, #S1049). The organoid cultures were maintained at 37°C in 5% CO2, and the medium was changed every 3-days until further analysis.
Co-culture protocol for ameloblast and odontoblast organoid
The day16 differentiated ieAM cells were cultured in ultra-low attachment 12-well plate for a week in ameloblast base medium. The odontogenic organoids were made in a similar manner in a separate plate by culturing DPSCs (isolated from primary molar sample of young patient89) in odontogenic differentiation medium containing DMEM (Gibco, #11995073) ascorbic acid (Sigma, #A8960), β-Glycerophosphate (Sigma, #35675), and dexamethasone (Sigma, #D2915), 10% FBS (Gibco, #10437028) and 1% Penicillin/Streptomycin (Gibco, #15140122). The two types of organoids were co-cultured in the same wells for two weeks, supplemented with a 1:1 mixture of both odontogenic and ameloblasts base media at 37°C in 5% CO2. The co-culture was sampled later for further analysis.
Co-culture protocol for monolayer
The DPSCs were plated as monolayer mixed in 25% (v/v) of Matrigel (Corning, #356231) diluted in odontogenic media in a glass-bottomed 24-well plate (Corning, #3603). The following day, ieAM cells suspended in the ameloblast base medium and 10 μM ROCKi (Y-27632, Selleckchem, #S1049) were added on top of the DPSCs monolayer and then incubated for 24 hours at 37°C in 5% CO2. The formed organoids were supplemented with fresh media (1:1 mixture ameloblast and odontogenic media) containing Calcein solution (Sigma, #C0875) (1uM, 1:1000) on every three consecutive days. The co-culture was sampled on the 14th day for further analysis.
Cryosectioning and Immunostaining for the organoids
The organoids were imbedded in OCT compound (Tissue-Tek, # 4583) and slowly frozen on a metal block chilled on dry ice. Frozen organoids were cut using Cryostat (Leica CM1850) to create 10μm slices and fixed on glass slides (Fiserbrand, #12–55015) for staining. The organoid sections were fixed in 4% paraformaldehyde (PFA) for 10–15min at RT and later washed thrice with 1X PBS for 5 min each. Slides were then immersed in 0.5% TritonX 100 at RT for 5 minutes to facilitate permeabilization. Later blocking was done for 1hour at RT in a humidified chamber with a blocking buffer consisting of 5% goat serum (VWR, #101098–382), 3% bovine serum albumin (BSA) (VWR, #9048–46-8) and 0.1% Triton X-100 (Sigma, #T9284). The organoids were incubated in primary antibodies (File S6) overnight at 4℃ in a humidified chamber. After 3×5 minute washing in PBS in a coplin jar, the slides were transferred to a humidified chamber with secondary antibodies. Secondary antibodies and Phalloidin (File S6) were applied for 1hour at RT in the same blocking agent, followed by rinsing the slides with PBS 3×5min in coplin jar. The slides were incubated in autofluorescence quenching solution (Vector Labs, #SP-8400) for 5 min at RT under dark conditions and rinsed 1x with PBS. DAPI (Thermo Fisher, #D1306) was applied for 10 minutes at room temperature in PBS. Slides were then rinsed with PBS for 10 minutes in a coplin jar. Slides were then mounted with Vectashield (Vector Labs, #H-1700) and stored at 4℃ for imaging.
Whole-mount immunostaining analysis
The organoids were collected in a 2ml tube after two weeks and washed thoroughly with 1x PBS before fixation. The organoids were fixed in 4% paraformaldehyde (PFA) for 10–15min at RT on a rocker. Later the fixed organoids were washed thrice with 1X PBS for 5 min each. The organoids were then immersed in 0.5% TritonX 100 at RT on a rocker for 5 minutes. Later blocking was done for 1hour at RT on a rocker with a blocking buffer consisting of 5% goat serum (VWR, #101098–382), 3% BSA (VWR, #9048–46-8) and 0.1% Triton X-100 (Sigma, #T9284). The organoids were incubated overnight in the primary antibodies (File S6) at 4°C on a rocker. After 3×5min washing in PBS, the organoids were incubated with secondary antibodies (File S6) for an hour at RT on a rocker. The primary and secondary antibodies were prepared in the blocking agent consisting of 0.1% Triton X-100 and 3% goat serum. Followed by washing the organoids with PBS 3×5min on a rocker. The organoids were incubated in autofluorescence quenching solution (Vector Labs, #SP-8400) for 5 min at RT under dark conditions on a rocker and rinsed 1x with PBS. Followed by incubation in 200 ul of 1xPBS containing DAPI (Thermo Fisher, #D1306) for 10 min. The organoids were then rinsed with PBS, mounted with Vectashield (Vector Labs, #H-1700), and stored at 4℃ for imaging.
AMBN mutant generation using CRISPR
One million WTC11 iPSC were electroporated with Cas9 (0.3uM, Sigma) and gRNA targeting AMBN (1.5uM, Synthego) as RNP complex using Amaxa nucleofector (Human Stem Cell kit 2) in presence of ROCK inhibitor. Individual colonies were hand-picked and plated into 96 well plates. DNA was extracted using Quick Extract DNA extraction solution (Epicentre #QE09050) and nested PCR was performed using Phusion Flash polymerase (ThermoFisher, #F631S). The PCR product was isolated from the gel and purified using the Monarch DNA Gel Extraction Kit (NEB, #T1020S) and sent for Sanger sequencing analysis (through Genewiz). gRNA sequence: CTTCAGCCTCCAATTCACCT. PCR primer sequences: F: TTACGAGCAATGGTGGTCCC, R: AGTGCCCTGCAAATTCGTTT.
Growing and plating AMBN WT and mutant lines
Three mutant hiPSC colonies (KO-1, KO-2 & KO-3) with +1 reading frame at the AMBN gene were selected based on the Sanger sequencing result. Prior to plating the cells for the differentiation experiment, the cell concentration for all 4 hiPSC lines (WT, KO-1, KO-2 and KO-3) were counted using Cytosmart cell counter and adjusted accordingly to ensure equal plating. 10 μM ROCKi (Y-27632, Selleckchem, #S1049) was added to each hiPSC line then plated on Matrigel-coated 6-well flat bottom tissue culture plate with mTeSR1 media (StemCell Technologies, #85850). The cells were then allowed to undergo differentiation to an early-ameloblast stage till day 16.
Conforming KO at the protein level
The cells were subjected to lysis directly on the plate by using lysis buffer containing 20 mM Tris-HCl pH 7.5, 150 mM NaCl, 15% Glycerol, 1% Triton x-100, 1 M ß-Glycerolphosphate, 0.5 M NaF, 0.1 M Sodium Pyrophosphate, Orthovanadate, PMSF, and 2% SDS. Subsequently, 25 U of Benzonase Nuclease (EMD Chemicals, Gibbstown, NJ) and 100x phosphatase inhibitor cocktail were added to the lysate. To prepare the sample for analysis, 4x Laemli sample buffer (Bio-Rad, #1610747), comprising 900 μl of sample buffer and 100 μl β-Mercaptoethanol (Sigma, #M7522), was added to the lysate, which was then heated for 5 mins at 95°C. After that, 30μl of protein sample was loaded onto SDS-PAGE using a Protean TGX pre-casted gradient gel, with a range of 4%−20% (Bio-Rad, #17000546) and transferred to the Nitro-Cellulose membrane (Bio-Rad, #1620115) using a semi-dry transfer system (Bio-Rad). The membranes were then blocked for 1 hour with 5% BSA, corresponding to the primary antibodies (File S6): AMBN (Santa Cruz #sc-271012, 1:500), SP6 (Atlas #HPA024516, 1:1000), β-Actin (Cell Signaling #13E5, 1:10,000) in 5% BSA, and incubated overnight at 4°C with the primary antibodies on a rocker. Next day, the membranes were washed with 1X TBST (3 times, 10 min interval). After incubation with primary antibodies, the membranes were exposed to anti-rabbit IgG HRP conjugate secondary antibody (Bio-Rad, ##1721019)(1:10,000) repared in 5% milk for an hour and incubated at room temperature. All the membranes were washed with 1× TBST (3 times, 10 min of interval) after secondary antibody incubation and developed using immobilon-luminol reagent assay (EMP Millipore) on Bio-Rad ChemiDoc Imager.
Injection of iPSC-derived ameloblast-like cells into mouse muscles
hiPSCs (WTC11) were allowed to undergo differentiation for the early ameloblast stage at day16 using the basal supplements mentioned above cultured in Matrigel. 1 × 106 ieAM cells were resuspended in Matrigel supplemented with a cocktail of prosurvival factors90 and injected into the femoral muscle of SCID-Beige mice (Charles River, Wilmington, MA). Mice were kept under BioSafety containment Level 2. Mice were sacrificed and femoral muscles were harvested after 2 months and were dissected at the site of injection (left leg muscle) to perform further analysis. Experiment was performed in compliance with ethical regulations, IACUC protocol #4152–01. After dissection, left leg muscles were embedded in embedding cryo-mold (Polysciences, #18986–1) with minimum amount of Tissue-Tek O.C.T. compound (Sakura, catalog number: 4583) to cover the muscle region. The embedded tissue was then snap-frozen by placing on a cold-resistant beaker of 2-methylbutane solution (EMD, #MX0760–1) into a slurry of liquid nitrogen for 5-mins, which allows fast cooling to −80 °C. The snap-frozen samples are then placed in a −80 °C freezer for storage. The cryostat and blade are both pre-chilled to −20°C before cryo-sectioning. 10μm-thick sections were made on pre-chilled Superfrost Plus microscope slides (Fisherbrand, #12–550-15) and then stored in a −80 °C.
Injection of iPSC-derived ameloblast-like and OB-like cells into mouse kidney capsules
hiPSCs (WTC11) were allowed to undergo differentiation for the early ameloblast at day16 using the basal supplements mentioned above. DPSCs were cultured and expanded in DMEM media for 6 days. On the day of transplantation, the DPSCs were trypsinized with 0.05% trypsin, pelleted down in a 1.5 mL microcentrifuge tube and the supernatant was aspirated. ieAM-day16 cells were treated with trypLE for 5-min and then aspirated off. The cells were then scraped off as clumps from the plate, added to the DPSC pellet, and allowed to gravity settle for 15-minutes. The combined cell clump was transferred to a beveled, kinked PE50 tubing (BD Intramedic, 427517) using a p200 pipette and centrifuged at 300g for 3-mins to concentrate the cells toward the end of the tubing. NOD-SCID mice were anesthetized via 1.5–2% isoflurane inhalation. Mice were shaved and the site was sterilized with betadine and alcohol wipes prior to making a 1–2 cm dorsal flank incision. Kidneys were externalized using a cotton swab and the capsule was nicked near the caudal end using a needle tip (22 gauge). The beveled PE50 tubing was inserted beneath the capsule and cellular material was implanted under the control of a Hamilton syringe. A cotton swab was used to clot and seal the opening in the capsule to hold the implant in place. Next, the kidney was returned to the abdominal cavity, the peritoneum was sutured shut with absorbable sutures, and the skin was closed with surgical staples. Experiment was performed in compliance with ethical regulations, IACUC protocol #4152–01. Mice were sacrificed at 8 weeks post-transplantation through CO2 inhalation and kidneys were excised, fixed in 4% paraformaldehyde for 1 hour, and transferred to 30% sucrose solution overnight. Kidneys were then embedded in embedding cryo-mold (Polysciences, #18986–1) with Tissue-Tek O.C.T. compound (Sakura, catalog number: 4583). The embedded tissue was then snap-frozen by placing on a cold-resistant beaker of 2-methylbutane solution (EMD. #MX0760–1) into a slurry of liquid nitrogen for 5-mins, which allows fast cooling to −80 °C. The frozen samples are then placed in a −80 °C freezer for storage. The cryostat and blade are both pre-chilled to −20°C before cryo-sectioning. 10 μm-thick sections were made on pre-chilled Superfrost Plus microscope slides (Fisherbrand, #12–550-15) and then stored in a −80 °C.
Calcification assays: Von Kossa and Alizarin Red Staining
Identification of mineralization was performed on tissue sections stained with Von Kossa and Alizarin Red S. Frozen leg muscle sections (10μm) were fixed with 4% paraformaldehyde (EMS, #15710) in H2O at room temperature for 12min. Rinse the section with deionized distilled water thrice for 5min each. Sections were incubated in with 5% silver nitrate solution (SIGMA-ALDRICH #209139) placed under ultraviolet light for 1 hour. The section was rinsed with several changes of deionized distilled water for 5min each and later incubated in 5% Sodium Thiosulfate solution (SIGMA-ALDRICH #217263) for 5 minutes to remove un-reacted silver. Similarly, sections were stained with 2% Alizarin red S solution (pH4.2) (Sigma, #A5533) for 1 hour in the dark. The slides were thoroughly with deionized distilled water for 5min each followed by counterstaining the sections with nuclear fast red stain (EMS, # 26078–05) or Fast Green FCF Solution 0.2% Aqueous (EMS, #26053–02) for 5 minutes. Rinsed in deionized distilled water briefly for 5mins each the slides were successfully transferred into coplin jars to perform dehydration step through graded alcohol and clear the slides in CitriSolv solution (Decon, #1601). Slides were then mounted with Vectashield (Vector Labs, #H-1400–10) and stored at room temperature for imaging.
Cryosectioning of fetal samples
Jaw tissues were fixed with 4% PFA overnight at 4°C followed by 30% sucrose (Sigma, #RDD023) treatment until the tissue sank to the bottom of the tube. The tissue is then imbedded in OCT compound (Tissue-Tek, # 4583) and slowly frozen on a metal block chilled on dry ice. Frozen samples were cut using Cryostat (Leica CM1850) to create 10μm slices of tissue and fixed on glass slides (Fiserbrand, #12–55015) for staining.
Immunofluorescence staining and Confocal Imaging of toothgerms
Fetal jaw segments containing toothgerms embedded in O.C.T. were cryosectioned to 10-micron thick sections. The slides were stored at −80℃ after cryosectioning and warmed at room temperature prior to staining. Tissues were fixed in 4% paraformaldehyde (PFA) then immersed in 1X PBS for 3×5 minute washes. Antigen retrieval was performed using 10X Citrate Buffer (Sigma-Aldrich, # C9999) in a capped coplin jar microwaved for ~45 seconds followed by 15-minutes incubation in microwave. Slides were then allowed to be washed in PBS at room temperature for 7 minutes. Slides were blocked for 90 minutes at room temperature in a humidified chamber with a blocking buffer consisting of 0.1% Triton X-100 and 5% BSA (VWR, #9048–46-8). All the antibodies used in this study and their concentrations are listed in File S6. The primary antibodies were incubated overnight at 4℃ in a humidified chamber. After 3×5 minute washing in PBS in a coplin jar, the slides were transferred to a humidified chamber with secondary antibodies. Secondary antibodies were applied for 75 minutes at room temperature in the same blocking agent. Slides were then rinsed with PBS 4×10 minute washes in a coplin jar. DAPI (Thermo Fisher, #D1306) was applied for 10 minutes at room temperature in PBS. Slides were then rinsed with PBS for 10 minutes in a coplin jar. Slides were then mounted with Vectashield (Vector Labs, #H-1700) and stored at 4℃ for imaging. Confocal Imaging was done on a Leica TCS-SPE Confocal microscope using a 40x objective and Leica Software. Images were processed with Fiji software distribution of ImageJ v2.3.091,92. NIS-Elements (RRID:SCR_014329) was used for 3D reconstruction.
Graphics and illustrations
The illustrations in the graphical abstract and in Figure 6, 7 and S7 was created with BioRender.com.
QUANTIFICATION AND STATISTICAL ANALYSIS
Organoids timepoint analysis & quantification
ieAM organoids were stained every 5 days, starting from 7 days after initial plating with DAPI, Phalloidin, and ZO1, and were observed using a confocal laser scanning microscope (Leica TCS SP8). For WT and mutant lines, individual ieAM organoids were scanned from top to bottom along the z-directions, and the numbers of apical-basolateral polarized lumens observed were counted. The apical-basolateral polarized lumen was enclosed with the apical surfaces of individual polarized ieAM cells, which were denoted by high expression of ZO1 and Phalloidin. At each time point, the counts of apical-basolateral polarized lumens of individual ieAM organoids were summed up. The average number of apical-basolateral polarized lumens per ieAM organoids was calculated by dividing the total counts of apical-basolateral polarized lumens by the total numbers of ieAM organoids, which were then reported as a line graph. For lumen to be counted as such, at least six cells needed to be in contact.
Statistical analysis
All quantifications show the mean, and error bars are +− SEM. Ordinary one-way ANOVA was used for multiple comparisons. A two-tailed, unpaired t-test was used for comparing groups of two using GraphPad Prism. P-values < 0.05, 0.01, 0.001, 0.0001 are indicated with *, **, *** and ***, respectively.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| AMBN | Santa Cruz | sc-271012; RRID: AB_10613795 |
| AMELX | Santa Cruz | sc-365284; RRID: AB_10843799 |
| DSPP | Santa Cruz | sc-73632; RRID: AB_2230660 |
| ENAM | Thermo Fisher | PA5–25734; RRID: AB_2543234 |
| KRT14 | Thermo Fisher | LL002; RRID: AB_306091 |
| KRT5 | Sigma-Aldrich | HPA059479; RRID:AB_2684034 |
| ZO-1 | Invitrogen | 33–9100; RRID: AB_87181 |
| anti-GFP | Invitrogen | A-1112; RRID:AB_221569 |
| SP6 | Atlas Antibodies | HPA024516; RRID: AB_10960551 |
| VIMENTIN | Cell Signalling | 5741S; RRID: AB_10695459 |
| Human Nuclei | Millipore | MAB1281; RRID:AB_94090 |
| CD144 | BD Biosciences | 555661, RRID: AB_396015 |
| CD146 (MCAM) | Abcam | ab75769; RRID: AB_2143375 |
| DAPI | Thermo Fisher | D1306, RRID: AB_2629482 |
| Mouse IgG (Alexa Flour 488) | Thermo Fisher | A11001; RRID: AB_2534069 |
| Rabbit IgG (Alexa Flour 488) | Thermo Fisher | A-32731; RRID:AB_2633280 |
| Mouse IgG (Alexa Flour 568) | Thermo Fisher | A-11004; RRID:AB_2534072 |
| Rabbit IgG (Alexa Flour 568) | Thermo Fisher | A-11036, RRID:AB_10563566 |
| Mouse IgG (Alexa Flour 647) | Thermo Fisher | A32728, RRID:AB_2633277 |
| Rabbit IgG (Alexa Flour 647) | Thermo Fisher | A32733, RRID:AB_2633282 |
| See File S for concentrations | N/A | N/A |
| Bacterial and virus strains | ||
| chemically competent E. coli Lemo21 | NEB | C2528J |
| Biological samples | ||
| Human fetal jaw tissues | UW Birth Defect Research Laboratory (BDRL) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Matrigel | Corning | #356231; |
| β-mercaptoethanol (BME) | Sigma | #M7522 |
| Smoothened agonist (SAG) | Selleckchem | # S7779 |
| Bone morphogenic protein-4 (BMP4) | rndsystems | #314-BP-010 |
| BMP-I inhibitor (LDN-193189) | Tocris | # 6053 |
| GSK3-Inhibitor (CHIR99021) | Selleckchem | # 4423 |
| Epidermal growth factor (EGF) | rndsystems | #236-EG |
| Supplement S7 | Thermo Fisher | #S0175 |
| Neurotrophin-4 (NT4) | rndsystems | #268-N4 |
| Transforming growth factor beta 1(TGFβ1) | rndsystems | #7754-BH |
| ROCKi (Y-27632) | Selleckchem | #S1049 |
| Ascorbic acid | Sigma | #A8960 |
| β-Glycerophosphate | Sigma | #35675 |
| Dexamethasone | Sigma | #D2915 |
| FGFR-mb | Cao et al., 202260 | NA |
| Calcein | Sigma | #C0875 |
| Alizarin red S stain | Sigma | #A5533 |
| Sodium Thiosulfate | Sigma | #217263 |
| Silver Nitrate | Sigma | #209139 |
| Paraformaldehyde | EMS | #15710 |
| Tissue-Tek O.C.T. | Sakura | #4583 |
| 2-methylbutane | EMD | #MX0760–1 |
| RPMI 1640 Medium | Thermo Fisher | #11875093 |
| EpiCult-C media | StemCell Technologies | #05630 |
| EpiLife | Thermo Fisher Scientific | #MEPI500CA |
| mTeSR1 stem cell medium | StemCell Technologies | #85850 |
| Critical commercial assays | ||
| RNAscope Intro Pack for HiPlex12 Reagents Kit (488, 550,650,750) - Human | Advanced Cell Diagnostics, Inc | 324442 |
| Deposited data | ||
| Raw and analyzed data | This paper | GEO: GSE184749 |
| RNAscope raw and processed data | This paper | https://datadryad.org/stash/share/TiKnbN_96SrgYmHHJ7kHa4kH8AYjAFcMXG0BsMlVOeE |
| Human reference genome NCBI build 38, GRCh38 | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.38/ |
| Experimental models: Cell lines | ||
| WTC-11 human induced pluripotent stem cells) | Coriell | #GM25256 |
| Dental Pulp Stem Cells | Macrin et al., 201989 | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: CB17.Cg-PrkdcscidLystbg-J/Crl | Charles River Laboratories | RRID: IMSR_CRL:250 |
| Oligonucleotides | ||
| See File S for all primers | N/A | N/A |
| Recombinant DNA | ||
| pET-29b(+) | GeneScript | N/A |
| Software and algorithms | ||
| Seurat 4.0 package | Hao et al., 202164 | https://satijalab.org/seurat/ |
| networkD3 | Allaire et al., 201787 | https://cran.r-project.org/package=networkD3 |
| Monocle3 | Cao et al., 201924; Qiu77 et al., 2017; Trapnell et al., 201433 | https://cole-trapnell-lab.github.io/monocle3/ |
| STAR | (Dobin et al., 2013)76 | https://github.com/alexdobin/STAR |
| Fiji (ImageJ2 v2.3.0) | Schindelin et al., 201291; Schindelin et al., 201592 | https://imagej.net/software/fiji/ |
| UMAP algorithm | McInnes et al., 201878 | https://github.com/lmcinnes/umap |
| Leiden Algorithm | Levine et al., 201579; Traag et al., 201980. | https://github.com/vtraag/leidenalg |
| UpSetR | Conway et al., 201783 | https://github.com/hms-dbmi/UpSetR |
| ComplexHeatmap R package | Gu et al., 201681 | https://github.com/jokergoo/ComplexHeatmap |
| ViSEAGO R package | Brionne et al., 201936 | https://bioconductor.org/packages/release/bioc/html/ViSEAGO.html |
| simplifyEnrichment R package | Gu and Hübschmann, 202182 | https://github.com/jokergoo/simplifyEnrichment |
| talklr R package | Wang, 202049 | https://github.com/yuliangwang/talklr |
| DEsingle R package | Miao et al., 201850 | https://github.com/miaozhun/DEsingle |
| scMLnet R package | Cheng et al., 202151 | https://github.com/SunXQlab/scMLnet |
| LIGER R package | Welch et al., 201965 | https://github.com/welch-lab/liger |
| QuPath (v0.3.0) | Bankhead et al., 201788 | https://qupath.github.io/ |
| BioRender | BioRender | https://www.biorender.com/ |
| NIS-Elements | Nikon | RRID:SCR_014329 |
| Custom R codes | This paper | https://github.com/Ruohola-Baker-lab/Tooth_sciRNAseq.(DOI: 10.5281/zenodo.8076442) |
Highlights.
Single-Cell census of human tooth development (9–22gw) define the major cell types.
TopPath analysis identify key signaling pathways in human ameloblast development.
Human ameloblast differentiation achieved from iPSCs for future enamel studies.
3D organoid system models Amelogenesis imperfecta, advancing enamel therapies.
Acknowledgment
We thank the Ruohola-Baker lab members for their helpful discussions. We thank Chris Cavanaugh, Fatima Al-Shimmary, Christopher Kelley, Aaron Liu, Infencia Xavier, Dibyo Maiti, Mickey Kim, Gwen L. Tilmes, and Anoushka Amath for their technical assistance. We are grateful to Profs. Carol Ware and Irma Thesleff for their inspiring discussions and guidance. We acknowledge Kimberly A. Aldinger, Ian G. Phelps, Jennifer C. Dempsey, Kevin Lee, and Lucy Cort from BDRL for their support in sample processing and troubleshooting nuclei isolation. We appreciate the technical support and guidance provided by the BBI in sequencing project design. This work is supported by grants from the National Institutes of Health 1P01GM081619, R01GM097372, R01GM97372-03S1 and R01GM083867 and the NHLBI Progenitor Cell Biology Consortium (U01HL099997; UO1HL099993) for HRB. This work was supported by the BBI grants for JM, YW and HRB. This work was also supported by Dr. Douglass L. Mourell Research Fund for HRB, HZ, AA, and YTZ. The Birth Defects Research Laboratory was supported by NIH award number 5R24HD000836, to IAG and DD, from the Eunice Kennedy Shriver National Institute of Child Health and Human Development. AA was supported by Imam Abdulrahman bin Faisal University, and Saudi Arabian Cultural Mission (SACM) to the USA. SHD, YTZ and DDE were supported by National Institutes of Health T90DE021984, and SHD was also supported by ARCS. MCR and work conducted in the ISCRM Genomics Core were supported by a generous gift from the John H. Tietze Foundation.
Inclusion and Diversity
We support inclusive, diverse, and equitable conduct of research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests, except as follows. AA, SHD, YTZ, DDE, YW, HZ, JM, DB and HR-B are co-inventors on a patent application entitled ‘A Method to Direct the Differentiation of Human Induced Pluripotent Stem Cells into Early Ameloblasts’ (PCT/US2022/053517 filed 12/20/2022). We support inclusive, diverse, and equitable conduct of research.
Supplemental information. Figures S1-S8
File S1. Marker genes used to identify cell types related to Figure.1D; Figure 2A and Figure 3A in separate excel sheets.
File S2: Meta-analytic estimators for mesenchymal and epithelial lineage related to Figure 2B and Figure 3B in separate excel sheets.
File S3: RNAScope_logic_tables.xlsx – Criteria used to locate cell types using RNAScope in situ hybridization signals related to Figure 3–4
File S4: QuPath_Settings.xlsx – Settings used to quantify RNAScope HiPlex in situ hybridization signal using QuPath software, related to STAR methods.
File S5: align-to-R1-batched.groovy – Script used to uniformly align the FITC, Cy3, Cy5, and Cy7 images from the three rounds of RNAScope HiPlex in situ hybridization imaging in Fiji, related to STAR methods.
File S6: Antibody list and primer list, related to STAR methods.
References
- 1.Park SJ, Bae HS, Cho YS, Lim SR, Kang SA, and Park JC (2013). Apoptosis of the reduced enamel epithelium and its implications for bone resorption during tooth eruption. Journal of molecular histology 44, 65–73. 10.1007/s10735-012-9465-4. [DOI] [PubMed] [Google Scholar]
- 2.Fugolin APP, and Pfeifer CS (2017). New Resins for Dental Composites. Journal of dental research 96, 1085–1091. 10.1177/0022034517720658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith CEL, Poulter JA, Antanaviciute A, Kirkham J, Brookes SJ, Inglehearn CF, and Mighell AJ (2017). Amelogenesis Imperfecta; Genes, Proteins, and Pathways. Frontiers in physiology 8, 435. 10.3389/fphys.2017.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jernvall J, and Thesleff I (2012). Tooth shape formation and tooth renewal: evolving with the same signals. Development (Cambridge, England) 139, 3487–3497. 10.1242/dev.085084. [DOI] [PubMed] [Google Scholar]
- 5.Yajima-Himuro S, Oshima M, Yamamoto G, Ogawa M, Furuya M, Tanaka J, Nishii K, Mishima K, Tachikawa T, Tsuji T, and Yamamoto M (2014). The junctional epithelium originates from the odontogenic epithelium of an erupted tooth. Scientific reports 4, 4867. 10.1038/srep04867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruch JV, Lesot H, and Bègue-Kirn C (1995). Odontoblast differentiation. The International journal of developmental biology 39, 51–68. [PubMed] [Google Scholar]
- 7.Yu T, and Klein OD (2020). Molecular and cellular mechanisms of tooth development, homeostasis and repair. Development (Cambridge, England) 147. 10.1242/dev.184754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balic A, and Thesleff I (2015). Tissue Interactions Regulating Tooth Development and Renewal. Current topics in developmental biology 115, 157–186. 10.1016/bs.ctdb.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Chiba Y, Saito K, Martin D, Boger ET, Rhodes C, Yoshizaki K, Nakamura T, Yamada A, Morell RJ, Yamada Y, and Fukumoto S (2020). Single-Cell RNA-Sequencing From Mouse Incisor Reveals Dental Epithelial Cell-Type Specific Genes. Frontiers in cell and developmental biology 8, 841. 10.3389/fcell.2020.00841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krivanek J, Soldatov RA, Kastriti ME, Chontorotzea T, Herdina AN, Petersen J, Szarowska B, Landova M, Matejova VK, Holla LI, et al. (2020). Dental cell type atlas reveals stem and differentiated cell types in mouse and human teeth. Nat Commun 11, 4816. 10.1038/s41467-020-18512-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharir A, Marangoni P, Zilionis R, Wan M, Wald T, Hu JK, Kawaguchi K, Castillo-Azofeifa D, Epstein L, Harrington K, et al. (2019). A large pool of actively cycling progenitors orchestrates self-renewal and injury repair of an ectodermal appendage. Nature cell biology 21, 1102–1112. 10.1038/s41556-019-0378-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thesleff I (2014). Current understanding of the process of tooth formation: transfer from the laboratory to the clinic. Australian dental journal 59 Suppl 1, 48–54. 10.1111/adj.12102. [DOI] [PubMed] [Google Scholar]
- 13.Fresia R, Marangoni P, Burstyn-Cohen T, and Sharir A (2021). From Bite to Byte: Dental Structures Resolved at a Single-Cell Resolution. Journal of dental research 100, 897–905. 10.1177/00220345211001848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hovorakova M, Lesot H, Peterka M, and Peterkova R (2018). Early development of the human dentition revisited. Journal of anatomy 233, 135–145. 10.1111/joa.12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balic A (2019). Concise Review: Cellular and Molecular Mechanisms Regulation of Tooth Initiation. Stem cells (Dayton, Ohio) 37, 26–32. 10.1002/stem.2917. [DOI] [PubMed] [Google Scholar]
- 16.Harada H, Mitsuyasu T, Toyono T, and Toyoshima K (2002). Epithelial stem cells in teeth. Odontology 90, 1–6. 10.1007/s102660200000. [DOI] [PubMed] [Google Scholar]
- 17.Kavanagh KD, Evans AR, and Jernvall J (2007). Predicting evolutionary patterns of mammalian teeth from development. Nature 449, 427–432. 10.1038/nature06153. [DOI] [PubMed] [Google Scholar]
- 18.Nanci A, and TenCate AR (2018). Ten Cate’s oral histology : development, structure, and function [Google Scholar]
- 19.Harada H, Ichimori Y, Yokohama-Tamaki T, Ohshima H, Kawano S, Katsube K, and Wakisaka S (2006). Stratum intermedium lineage diverges from ameloblast lineage via Notch signaling. Biochemical and biophysical research communications 340, 611–616. 10.1016/j.bbrc.2005.12.053. [DOI] [PubMed] [Google Scholar]
- 20.Maas R, and Bei M (1997). The genetic control of early tooth development. Critical reviews in oral biology and medicine : an official publication of the American Association of Oral Biologists 8, 4–39. 10.1177/10454411970080010101. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura M, Bringas P Jr., and Slavkin HC (1991). Inner enamel epithelia synthesize and secrete enamel proteins during mouse molar occlusal “enamel-free area” development. Journal of craniofacial genetics and developmental biology 11, 96–104. [PubMed] [Google Scholar]
- 22.Koyama E, Wu C, Shimo T, Iwamoto M, Ohmori T, Kurisu K, Ookura T, Bashir MM, Abrams WR, Tucker T, and Pacifici M (2001). Development of stratum intermedium and its role as a Sonic hedgehog-signaling structure during odontogenesis. Developmental dynamics : an official publication of the American Association of Anatomists 222, 178–191. 10.1002/dvdy.1186. [DOI] [PubMed] [Google Scholar]
- 23.Takamori K, Hosokawa R, Xu X, Deng X, Bringas P Jr., and Chai Y (2008). Epithelial fibroblast growth factor receptor 1 regulates enamel formation. Journal of dental research 87, 238–243. 10.1177/154405910808700307. [DOI] [PubMed] [Google Scholar]
- 24.Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, et al. (2019). The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502. 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Paula F, Teshima THN, Hsieh R, Souza MM, Nico MMS, and Lourenco SV (2017). Overview of Human Salivary Glands: Highlights of Morphology and Developing Processes. Anatomical record (Hoboken, N.J. : 2007) 300, 1180–1188. 10.1002/ar.23569. [DOI] [PubMed] [Google Scholar]
- 26.Jussila M, and Thesleff I (2012). Signaling networks regulating tooth organogenesis and regeneration, and the specification of dental mesenchymal and epithelial cell lineages. Cold Spring Harbor perspectives in biology 4, a008425. 10.1101/cshperspect.a008425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radlanski RJ, Renz H, Tsengelsaikhan N, Schuster F, and Zimmermann CA (2016). The remodeling pattern of human mandibular alveolar bone during prenatal formation from 19 to 270mm CRL. Annals of anatomy = Anatomischer Anzeiger : official organ of the Anatomische Gesellschaft 205, 65–74. 10.1016/j.aanat.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Wise GE, Marks SC Jr., and Zhao L (1998). Effect of CSF-1 on in vivo expression of c-fos in the dental follicle during tooth eruption. European journal of oral sciences 106 Suppl 1, 397–400. 10.1111/j.1600-0722.1998.tb02205.x. [DOI] [PubMed] [Google Scholar]
- 29.Kardos TB, and Hubbard MJ (1981). Rapid dissection of rodent molar-tooth germs. Laboratory animals 15, 371–373. 10.1258/002367781780952870. [DOI] [PubMed] [Google Scholar]
- 30.Cha S (2017). Salivary gland development and regeneration : advances in research and clinical approaches to functional restoration [Google Scholar]
- 31.Nelson SJ (2020). Wheeler’s dental anatomy, physiology, and occlusion [Google Scholar]
- 32.Quirós-Terrón L, Arráez-Aybar LA, Murillo-González J, De-la-Cuadra-Blanco C, Martínez-Álvarez MC, Sanz-Casado JV, and Mérida-Velasco JR (2019). Initial stages of development of the submandibular gland (human embryos at 5.5–8 weeks of development). Journal of anatomy 234, 700–708. 10.1111/joa.12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, and Rinn JL (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nature biotechnology 32, 381–386. 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franzén O, Gan LM, and Björkegren JLM (2019). PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database : the journal of biological databases and curation 2019. 10.1093/database/baz046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ehnes DD, Alghadeer A, Hanson-Drury S, Zhao YT, Tilmes G, Mathieu J, and Ruohola-Baker H (2022). Sci-Seq of human fetal salivary tissue introduces human transcriptional paradigms and a novel cell population. Frontiers in Dental Medicine, 49. 10.3389/fdmed.2022.887057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brionne A, Juanchich A, and Hennequet-Antier C (2019). ViSEAGO: a Bioconductor package for clustering biological functions using Gene Ontology and semantic similarity. BioData mining 12, 16. 10.1186/s13040-019-0204-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chai Y, Jiang X, Ito Y, Bringas P Jr., Han J, Rowitch DH, Soriano P, McMahon AP, and Sucov HM (2000). Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development (Cambridge, England) 127, 1671–1679. [DOI] [PubMed] [Google Scholar]
- 38.Seidel K, Marangoni P, Tang C, Houshmand B, Du W, Maas RL, Murray S, Oldham MC, and Klein OD (2017). Resolving stem and progenitor cells in the adult mouse incisor through gene co-expression analysis. eLife 6. 10.7554/eLife.24712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jing J, Feng J, Yuan Y, Guo T, Lei J, Pei F, Ho TV, and Chai Y (2022). Spatiotemporal single-cell regulatory atlas reveals neural crest lineage diversification and cellular function during tooth morphogenesis. Nat Commun 13, 4803. 10.1038/s41467-022-32490-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harada M, Kenmotsu S, Nakasone N, Nakakura-Ohshima K, and Ohshima H (2008). Cell dynamics in the pulpal healing process following cavity preparation in rat molars. Histochemistry and cell biology 130, 773–783. 10.1007/s00418-008-0438-3. [DOI] [PubMed] [Google Scholar]
- 41.Adams D (1976). Keratinization of the oral epithelium. Annals of the Royal College of Surgeons of England 58, 351–358. [PMC free article] [PubMed] [Google Scholar]
- 42.Ahtiainen L, Uski I, Thesleff I, and Mikkola ML (2016). Early epithelial signaling center governs tooth budding morphogenesis. The Journal of cell biology 214, 753–767. 10.1083/jcb.201512074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thesleff I, Keränen S, and Jernvall J (2001). Enamel knots as signaling centers linking tooth morphogenesis and odontoblast differentiation. Advances in dental research 15, 14–18. 10.1177/08959374010150010401. [DOI] [PubMed] [Google Scholar]
- 44.Vaahtokari A, Aberg T, Jernvall J, Keränen S, and Thesleff I (1996). The enamel knot as a signaling center in the developing mouse tooth. Mechanisms of development 54, 39–43. 10.1016/0925-4773(95)00459-9. [DOI] [PubMed] [Google Scholar]
- 45.Yu W, Sun Z, Sweat Y, Sweat M, Venugopalan SR, Eliason S, Cao H, Paine ML, and Amendt BA (2020). Pitx2-Sox2-Lef1 interactions specify progenitor oral/dental epithelial cell signaling centers. Development (Cambridge, England) 147. 10.1242/dev.186023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hanson-Drury S, Patni AP, Lee DL, Alghadeer A, Zhao YT, Ehnes DD, Vo VN, Kim SY, Jithendra D, Phal A, et al. (2023). Single Cell RNA Sequencing Reveals Human Tooth Type Identity and Guides In Vitro hiPSC Derived Odontoblast (iOB). Frontiers in Dental Medicine Volume 4. 10.3389/fdmed.2023.1209503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu H, Yan X, Pandya M, Luan X, and Diekwisch TG (2016). Daughters of the Enamel Organ: Development, Fate, and Function of the Stratum Intermedium, Stellate Reticulum, and Outer Enamel Epithelium. Stem cells and development 25, 1580–1590. 10.1089/scd.2016.0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang JY, Wang C, Jin C, Yang C, Huang Y, Liu J, McKeehan WL, D’Souza RN, and Wang F (2013). Self-renewal and multilineage differentiation of mouse dental epithelial stem cells. Stem cell research 11, 990–1002. 10.1016/j.scr.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y (2020). talklr uncovers ligand-receptor mediated intercellular crosstalk. bioRxiv, 2020.2002.2001.930602. 10.1101/2020.02.01.930602. [DOI] [Google Scholar]
- 50.Miao Z, Deng K, Wang X, and Zhang X (2018). DEsingle for detecting three types of differential expression in single-cell RNA-seq data. Bioinformatics (Oxford, England) 34, 3223–3224. 10.1093/bioinformatics/bty332. [DOI] [PubMed] [Google Scholar]
- 51.Cheng J, Zhang J, Wu Z, and Sun X (2021). Inferring microenvironmental regulation of gene expression from single-cell RNA sequencing data using scMLnet with an application to COVID-19. Briefings in bioinformatics 22, 988–1005. 10.1093/bib/bbaa327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aurrekoetxea M, Irastorza I, García-Gallastegui P, Jiménez-Rojo L, Nakamura T, Yamada Y, Ibarretxe G, and Unda FJ (2016). Wnt/β-Catenin Regulates the Activity of Epiprofin/Sp6, SHH, FGF, and BMP to Coordinate the Stages of Odontogenesis. Frontiers in cell and developmental biology 4, 25. 10.3389/fcell.2016.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haro E, Delgado I, Junco M, Yamada Y, Mansouri A, Oberg KC, and Ros MA (2014). Sp6 and Sp8 transcription factors control AER formation and dorsal-ventral patterning in limb development. PLoS genetics 10, e1004468. 10.1371/journal.pgen.1004468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ibarretxe G, Aurrekoetxea M, Crende O, Badiola I, Jimenez-Rojo L, Nakamura T, Yamada Y, and Unda F (2012). Epiprofin/Sp6 regulates Wnt-BMP signaling and the establishment of cellular junctions during the bell stage of tooth development. Cell and tissue research 350, 95–107. 10.1007/s00441-012-1459-8. [DOI] [PubMed] [Google Scholar]
- 55.Rhodes CS, Yoshitomi Y, Burbelo PD, Freese NH, Nakamura T, Chiba Y, and Yamada Y (2021). Sp6/Epiprofin is a master regulator in the developing tooth. Biochemical and biophysical research communications 581, 89–95. 10.1016/j.bbrc.2021.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ochiai H, Suga H, Yamada T, Sakakibara M, Kasai T, Ozone C, Ogawa K, Goto M, Banno R, Tsunekawa S, et al. (2015). BMP4 and FGF strongly induce differentiation of mouse ES cells into oral ectoderm. Stem cell research 15, 290–298. 10.1016/j.scr.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 57.Suga H, Kadoshima T, Minaguchi M, Ohgushi M, Soen M, Nakano T, Takata N, Wataya T, Muguruma K, Miyoshi H, et al. (2011). Self-formation of functional adenohypophysis in three-dimensional culture. Nature 480, 57–62. 10.1038/nature10637. [DOI] [PubMed] [Google Scholar]
- 58.Tanaka J, Ogawa M, Hojo H, Kawashima Y, Mabuchi Y, Hata K, Nakamura S, Yasuhara R, Takamatsu K, Irié T, et al. (2018). Generation of orthotopically functional salivary gland from embryonic stem cells. Nat Commun 9, 4216. 10.1038/s41467-018-06469-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edman NI, Redler RL, Phal A, Schlichthaerle T, Srivatsan SR, Etemadi A, An SJ, Favor A, Ehnes D, Li Z, et al. (2023). Modulation of FGF pathway signaling and vascular differentiation using designed oligomeric assemblies. bioRxiv, 2023.2003.2014.532666. 10.1101/2023.03.14.532666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cao L, Coventry B, Goreshnik I, Huang B, Sheffler W, Park JS, Jude KM, Marković I, Kadam RU, Verschueren KHG, et al. (2022). Design of protein-binding proteins from the target structure alone. Nature 605, 551–560. 10.1038/s41586-022-04654-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun Z, Yu W, Sanz Navarro M, Sweat M, Eliason S, Sharp T, Liu H, Seidel K, Zhang L, Moreno M, et al. (2016). Sox2 and Lef-1 interact with Pitx2 to regulate incisor development and stem cell renewal. Development (Cambridge, England) 143, 4115–4126. 10.1242/dev.138883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakamura T, Jimenez-Rojo L, Koyama E, Pacifici M, de Vega S, Iwamoto M, Fukumoto S, Unda F, and Yamada Y (2017). Epiprofin Regulates Enamel Formation and Tooth Morphogenesis by Controlling Epithelial-Mesenchymal Interactions During Tooth Development. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research 32, 601–610. 10.1002/jbmr.3024. [DOI] [PubMed] [Google Scholar]
- 63.Seidel K, Ahn CP, Lyons D, Nee A, Ting K, Brownell I, Cao T, Carano RA, Curran T, Schober M, et al. (2010). Hedgehog signaling regulates the generation of ameloblast progenitors in the continuously growing mouse incisor. Development (Cambridge, England) 137, 3753–3761. 10.1242/dev.056358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e3529. 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Welch JD, Kozareva V, Ferreira A, Vanderburg C, Martin C, and Macosko EZ (2019). Single-Cell Multi-omic Integration Compares and Contrasts Features of Brain Cell Identity. Cell 177, 1873–1887.e1817. 10.1016/j.cell.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.D’Souza RN, Cavender A, Sunavala G, Alvarez J, Ohshima T, Kulkarni AB, and MacDougall M (1997). Gene expression patterns of murine dentin matrix protein 1 (Dmp1) and dentin sialophosphoprotein (DSPP) suggest distinct developmental functions in vivo. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research 12, 2040–2049. 10.1359/jbmr.1997.12.12.2040. [DOI] [PubMed] [Google Scholar]
- 67.Visakan G, Su J, and Moradian-Oldak J (2022). Ameloblastin promotes polarization of ameloblast cell lines in a 3-D cell culture system. Matrix biology : journal of the International Society for Matrix Biology 105, 72–86. 10.1016/j.matbio.2021.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liang T, Hu Y, Smith CE, Richardson AS, Zhang H, Yang J, Lin B, Wang SK, Kim JW, Chun YH, et al. (2019). AMBN mutations causing hypoplastic amelogenesis imperfecta and Ambn knockout-NLS-lacZ knockin mice exhibiting failed amelogenesis and Ambn tissue-specificity. Molecular genetics & genomic medicine 7, e929. 10.1002/mgg3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duverger O, and Morasso MI (2018). Pleiotropic function of DLX3 in amelogenesis: from regulating pH and keratin expression to controlling enamel rod decussation. Connective tissue research 59, 30–34. 10.1080/03008207.2017.1408602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Duverger O, Zah A, Isaac J, Sun HW, Bartels AK, Lian JB, Berdal A, Hwang J, and Morasso MI (2012). Neural crest deletion of Dlx3 leads to major dentin defects through down-regulation of Dspp. The Journal of biological chemistry 287, 12230–12240. 10.1074/jbc.M111.326900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu M, Wang H, Fan Z, Xie C, Liu H, Liu Y, Han D, Wong SW, and Feng H (2019). BMP4 mutations in tooth agenesis and low bone mass. Archives of oral biology 103, 40–46. 10.1016/j.archoralbio.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marangoni P, Charles C, Ahn Y, Seidel K, Jheon A, Ganss B, Krumlauf R, Viriot L, and Klein OD (2019). Downregulation of FGF Signaling by Spry4 Overexpression Leads to Shape Impairment, Enamel Irregularities, and Delayed Signaling Center Formation in the Mouse Molar. JBMR plus 3, e10205. 10.1002/jbm4.10205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu J, Saiyin W, Xie X, Mao L, and Li L (2020). Ablation of Fam20c causes amelogenesis imperfecta via inhibiting Smad dependent BMP signaling pathway. Biology direct 15, 16. 10.1186/s13062-020-00270-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Malik Z, Alexiou M, Hallgrimsson B, Economides AN, Luder HU, and Graf D (2018). Bone Morphogenetic Protein 2 Coordinates Early Tooth Mineralization. Journal of dental research 97, 835–843. 10.1177/0022034518758044. [DOI] [PubMed] [Google Scholar]
- 75.Yu M, Wong SW, Han D, and Cai T (2019). Genetic analysis: Wnt and other pathways in nonsyndromic tooth agenesis. Oral diseases 25, 646–651. 10.1111/odi.12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, and Trapnell C (2017). Reversed graph embedding resolves complex single-cell trajectories. Nature methods 14, 979–982. 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McInnes L, Healy J, and Melville J (2018). Umap: Uniform manifold approximation and projection for dimension reduction. arXiv preprint arXiv:1802.03426 [Google Scholar]
- 79.Levine JH, Simonds EF, Bendall SC, Davis KL, Amir el AD, Tadmor MD, Litvin O, Fienberg HG, Jager A, Zunder ER, et al. (2015). Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell 162, 184–197. 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Traag VA, Waltman L, and van Eck NJ (2019). From Louvain to Leiden: guaranteeing well-connected communities. Scientific reports 9, 5233. 10.1038/s41598-019-41695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gu Z, Eils R, and Schlesner M (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics (Oxford, England) 32, 2847–2849. 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 82.Gu Z, and Hübschmann D (2021). simplifyEnrichment: an R/Bioconductor package for Clustering and Visualizing Functional Enrichment Results. bioRxiv, 2020.2010.2027.312116. 10.1101/2020.10.27.312116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Conway JR, Lex A, and Gehlenborg N (2017). UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics (Oxford, England) 33, 2938–2940. 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ge SX, Jung D, and Yao R (2019). ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics (Oxford, England) 36, 2628–2629. 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Efremova M, Vento-Tormo M, Teichmann SA, and Vento-Tormo R (2020). CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nature protocols 15, 1484–1506. 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 86.Cabello-Aguilar S, Alame M, Kon-Sun-Tack F, Fau C, Lacroix M, and Colinge J (2020). SingleCellSignalR: inference of intercellular networks from single-cell transcriptomics. Nucleic Acids Research 48, e55. 10.1093/nar/gkaa183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Allaire J, Ellis P, Gandrud C, Kuo K, Lewis B, Owen J, Russell K, Rogers J, Sese C, and Yetman C (2017). Package ‘networkD3’. D3 JavaScript Network Graphs from R [Google Scholar]
- 88.Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, McQuaid S, Gray RT, Murray LJ, Coleman HG, et al. (2017). QuPath: Open source software for digital pathology image analysis. Scientific reports 7, 16878. 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Macrin D, Alghadeer A, Zhao YT, Miklas JW, Hussein AM, Detraux D, Robitaille AM, Madan A, Moon RT, Wang Y, et al. (2019). Metabolism as an early predictor of DPSCs aging. Scientific reports 9, 2195. 10.1038/s41598-018-37489-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, et al. (2007). Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nature biotechnology 25, 1015–1024. 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- 91.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nature methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schindelin J, Rueden CT, Hiner MC, and Eliceiri KW (2015). The ImageJ ecosystem: An open platform for biomedical image analysis. Molecular reproduction and development 82, 518–529. 10.1002/mrd.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study can be downloaded in raw and processed forms from the NCBI Gene Expression Omnibus under accession number (GSE184749). Upon publication, raw RNAScope data will be made publicly available on dryad.org (Dryad research data repository).
The custom R codes used to generate some of the results in this paper are available in https://github.com/Ruohola-Baker-lab/Tooth_sciRNAseq (DOI: 10.5281/zenodo.8076442).


