Summary
Background
Paediatric Huntington disease with highly expanded mutations (HE-PHD; >80 CAG repeats) presents atypically, compared to adult-onset Huntington disease (AOHD), with neurodevelopmental delay, epilepsy, abnormal brain glucose metabolism, early striatal damage, and reduced lifespan. Since genetic GLUT-1 deficiency syndrome shows a symptom spectrum similar to HE-PHD, we investigated the potential role of the two main glucose transporters, GLUT-1 and GLUT-3, in HE-PHD.
Methods
We compared GLUT-1 and GLUT-3 protein expression in HE-PHD, juvenile-onset (JOHD), and AOHD brains (n = 2; n = 3; n = 6) and periphery (n = 3; n = 2; n = 2) versus healthy adult controls (n = 6; n = 6). We also investigated mitochondrial complexes and hexokinase-II protein expression.
Findings
GLUT-1 and GLUT-3 expression were significantly lower in HE-PHD frontal cortex (p = 0.009, 95% [CI 13.4, 14.7]; p = 0.017, 95% [CI 14.2, 14.5]) versus controls. In fibroblasts, GLUT-1 and GLUT-3 expression were lower compared to controls (p < 0.0001, 95% [CI 0.91, 1.09]; p = 0.046, 95% [CI 0.93, 1.07]). In the frontal cortex, this occurred without evidence of extensive neuronal degeneration. Patients with HE-PHD had deregulated mitochondrial complex expression, particularly complexes II-III, levels of which were lower in frontal cortex versus controls (p = 0.027, 95% [CI 17.1, 17.6]; p = 0.002, 95% CI [16.6, 16.9]) and patients with AOHD (p = 0.052, 95% [CI 17.0, 17.6]; p = 0.002, 95% [CI 16.6, 16.7]). Hexokinase-II expression was also lower in HE-PHD frontal cortex and striatum versus controls (p = 0.010, 95% [CI 17.8, 18.2]; p = 0.045, 95% [CI 18.6, 18.7]) and in frontal cortex versus patients with AOHD (p = 0.013, 95% [CI 17.7, 18.1]). Expression JOHD levels were consistently different to those of HE-PHD but similar to those of AOHD.
Interpretation
Our data suggest a dysfunctional hypometabolic state occurring specifically in paediatric Huntington disease brains.
Funding
‘5 × 1000’ Personal Income Tax donation to LIRH Foundation; Italian Ministry of HealthRC2301MH04 and RF-2016-02364123 to CSS.
Keywords: Glucose transporters, Glucose metabolism, Paediatric-onset Huntington disease, Brain dysfunction, Frontal cortex, Fibroblasts
Research in context.
Evidence before this study
Huntington disease (HD) is a rare, dominantly transmitted, neurodegenerative disease caused by an expanded CAG mutation in the HTT gene. Glucose brain metabolism is affected in HD and glucose uptake into brain cells reduces progressively before neurological symptoms occur. However, the mechanisms underlying these alterations are not known. We hypothesised that impaired glucose transport may be involved. We reviewed the available literature by searching PubMed for studies published prior to December 2022 that contained the following terms: ‘glucose transporters’ OR ‘GLUT-1’ AND ‘Huntington disease’, OR ‘juvenile Huntington disease’ OR ‘paediatric Huntington disease’. We found a few references combining ‘glucose transporters’ or ‘GLUT-1’ and HD, but no references combining ‘glucose transporters’ or ‘GLUT-1’ and juvenile or paediatric HD. Deregulation of glucose metabolism is a hallmark of several neurodegenerative disorders, as well as aberrant brain neurodevelopment, micromalformations, and epilepsy. These are all conditions that have been associated with HE-PHD, the most aggressive form of HD, characterised by a faster disease progression and a shorter life span compared to adult-onset HD. Interestingly, certain paediatric HD clinical features are also observed in patients with GLUT-1 deficiency syndrome, where genetic defects in this glucose transporter result in a chronic shortage of glucose in the brain, suggesting glucose transport or metabolism may be altered in paediatric HD. To our knowledge, there are no studies examining glucose transporters or metabolism in paediatric HD.
Added value of this study
This study assessed the involvement of glucose transporters in paediatric HD pathogenesis. We found significantly lower protein expression of the main glucose transporter, GLUT-1, in the brain and periphery of patients with HE-PHD, as well as a deregulation of the mitochondrial machinery involved in glucose metabolism. A direct relationship between GLUT-1 function, glucose metabolism, neurodegeneration and neurodevelopment remains poorly understood. Together, these data in HE-PHD may enable the development of new biomarkers and therapeutic strategies in paediatric HD.
Implications of all the available evidence
Our findings suggest that abnormal energy metabolism in HE-PHD is mediated by a significant reduction in expression of glucose transporters, especially GLUT-1, in both the brain and periphery, which occurs prior to neurodegeneration. Such a biological change may interfere with key pathological mechanisms affecting neurodevelopment and neurodegeneration, thus representing a critical therapeutic target for paediatric HD. This is crucial given the paucity of disease-modifying treatments against this devastating disorder.
Introduction
Huntington disease (HD) is one of nine autosomal dominant, rare, neurodegenerative, polyglutamine (polyQ) disorders characterised by pathological expansions of a trinucleotide CAG repeat region encoding polyQ. In HD, the CAG region is located in the HTT gene, which encodes the polyQ region in the huntingtin protein.1 HD typically manifests between 30 and 40 years of age with a movement disorder (typically chorea), cognitive dysfunction (e.g., abnormal executive functions) and behavioural changes, which are associated with an early progressive neurodegeneration of the cortex and striatum.2
Normal CAG repeat lengths are usually stably inherited, whereas mutant expansions (>36 repeats) can show instability when transmitted to subsequent generations. A mutant CAG repeat further increases the risk of somatic CAG expansion and can lead to earlier disease onset, at a juvenile age (i.e., <20 years). Highly expanded (HE) mutations (i.e., >80 CAG repeats) cause paediatric onset of disease (i.e., during the first decade of life).3
Patients with HE paediatric HD (HE-PHD) often exhibit neurodevelopmental delay or regression and a particularly severe phenotype, resulting in a shorter lifespan when compared to juvenile-onset HD (JOHD), which has relatively shorter CAG repeat lengths (i.e., <73),3 or adult-onset Huntington disease (AOHD).3 HE-PHD is also associated with a high rate of infantile epilepsy,3,4 liver steatosis,5 striatal volume loss with preserved brain cortex,3,6 and severe striatal glucose hypometabolism.7
Abnormal glucose metabolism is a hallmark of several neurodegenerative diseases and is due, at least in part, to altered expression of glucose transporters (GLUTs),8 which control glucose uptake into the brain. GLUT-1 and 3 are the major transporters of glucose across the blood–brain barrier and into neurons.9
GLUT-1 deficiency syndrome is a rare genetic neurometabolic disorder caused by mutations in the SCL2A1 gene, which results in impaired glucose transport into the brain.10 Clinically, GLUT-1 deficiency syndrome manifests with neurodevelopmental delay, microcephaly, a high rate of infantile epilepsy, cognitive impairment and varying degrees of spasticity, ataxia, and dystonia11 – a spectrum of symptoms that shares several similarities with HE-PHD.3,12,13
We hypothesised that expression of GLUT-1 and GLUT-3 may underlie HE-PHD pathology and differ from AOHD. We therefore compared GLUT-1 and GLUT-3 expression in the brain and peripheral tissues of patients with HE-PHD, JOHD and AOHD, and control subjects. Furthermore, we postulated that defects in glucose uptake are associated with impaired brain energy metabolism in patients with HD. Thus, we evaluated expression of mitochondrial complexes and hexokinase-II (HK-II) in the brains of the same four populations.
Methods
Study outcomes
Our primary outcome was to compare protein expression of the main glucose transporters (e.g., GLUT-1, GLUT-3) and associated cargo protein (Rab11-A) in the brain and peripheral tissues of patients with HE-PHD, JOHD and AOHD, and control subjects. Our secondary outcome was to compare protein expression of mitochondrial complexes and HK-II, in the same four cohorts. Additional outcomes included brain and peripheral gene expression of SLC2A1 and SLC2A3, which encode GLUT-1 and GLUT-3 (all four cohorts); GLUT-1 localization in the brain (HE-PHD, AOHD and control cohorts only); and cell counts/neurodegeneration in the brain (HE-PHD cohort only).
Study population and tissue acquisition
HE-PHD was defined as HD manifesting with paediatric onset and a mutant HTT expansion length >80 CAG repeats,3 a threshold associated with childhood-onset of disease.3 JOHD was defined as HD manifesting with early age of onset, retrospectively indicated in the approximate range of 18–25 years and HTT expansion length >55 CAG repeats. AOHD was defined as HD manifesting in adulthood at age > 30 years and a mutant HTT expansion length of ≤55 CAG repeats.14 (Table 1). The CAG-Age Product (CAP) score, considered a predictor of HD progression and a commonly used measure of cumulative exposure to the effects of mutant (CAG expanded) Huntingtin, was calculated for all patients.15
Table 1.
Characteristics of HE-PHD, JHD, AOHD patients and controls involved in fibroblast and brain tissue analyses.
| Cohort | Code | Centre | Sex | Tissue | Expanded CAG repeatExpn | Age of onset (years) | Age/Age of death (years) | CAP at death | Vonsattel grade | HD phase | First main symptoms |
|---|---|---|---|---|---|---|---|---|---|---|---|
| HD86b | LUMC | F | FrCx, Striatum | 86 | 4 | 19 | 994 | 3 | Final | Bradykinesia, Dystonia | |
| HD252 | LUMC | F | FrCx, Striatum | 83 | 6 | 17 | 839 | 3 | Final | Rigidity | |
| HE-PHD | HD707-02a | LIRH | M | Fibroblasts | 114 | 3.5 | 5 | NA | NA | Initial | Ataxia, Dystonia |
| HD130-05a, b | LIRH | F | Fibroblasts | 95 | 2 | 6 | NA | NA | Moderate | Dystonia | |
| HD379-05a, b | LIRH | M | Fibroblasts | 87 | 5 | 7 | NA | NA | Moderate | Dystonia | |
| HD123 | LUMC | M | FrCx, Striatum | 59 | 19–22 | 38 | 963 | 2/3 | Final | Rigidity, chorea | |
| HD208 | LUMC | M | FrCx, Striatum | 68 | 21–23 | 37 | 1270 | 3 | Final | Dystonia, chorea | |
| JOHD | HD247 | LUMC | F | FrCx | 56 | 22–25 | 44 | 983 | 2/3 | Final | Ataxia, chorea |
| HD83-08 | LIRH | F | Fibroblasts | 56 | 20–24 | 35 | NA | NA | Advanced | Dystonia, chorea | |
| HD701-01 | LIRH | F | Fibroblasts | 62 | 18 | 30 | NA | NA | Advanced | Dystonia, chorea | |
| HD255 | LUMC | M | FrCx, Striatum | 55 | 30–33 | 45 | 960 | 3 | Final | Chorea | |
| HD246 | LUMC | M | FrCx, Striatum | 44 | 35 | 47 | 486 | 3 | Advanced | Cognitive changes | |
| HD234 | LUMC | M | FrCx | NK | 31 | 46 | NK | 3 | Final | Behavioural changes, chorea | |
| AOHD | T4161 | LIRH | M | FrCx | 46 | >35 | 56 | 691 | 3 | Final | Chorea |
| T1991 | LIRH | M | FrCx | 48 | >35 | 53 | 688 | 3 | Final | Chorea | |
| HD249c | LUMC | F | FrCx, Striatum | 46 | 35 | 60 | 740 | 3 | Final | Chorea | |
| HD364-06 | LIRH | F | Fibroblasts | 41 | 48 | 50 | NA | NA | Initial | Chorea | |
| HD598-01 | LIRH | M | Fibroblasts | 40 | 54 | 62 | NA | NA | Initial | Chorea | |
| E15-02c | LUMC | M | Striatum | NK | NA | 54 | NA | NA | NA | NA | |
| E16-07c | LUMC | F | FrCx | 18/19 | NA | 61 | NA | NA | NA | NA | |
| E14-138 | LUMC | M | FrCx, Striatum | 17/17 | NA | 48 | NA | NA | NA | NA | |
| T4452 | LIRH | M | FrCx | 15/20 | NA | 67 | NA | NA | NA | NA | |
| T4233 | LIRH | M | FrCx | 16/17 | NA | 74 | NA | NA | NA | NA | |
| Controls | T4434 | LIRH | F | FrCx | 18/27 | NA | 79 | NA | NA | NA | NA |
| CTR1 | OPBG | F | Fibroblasts | NK | NA | NA | NA | NA | NA | NA | |
| CTR2 | OPBG | F | Fibroblasts | NK | NA | NA | NA | NA | NA | NA | |
| CTR3 | LIRH | F | Fibroblasts | NK | NA | NA | NA | NA | NA | NA | |
| CTR4 | LIRH | M | Fibroblasts | NK | NA | NA | NA | NA | NA | NA | |
| CTR5 | LIRH | F | Fibroblasts | NK | NA | NA | NA | NA | NA | NA | |
| CTR6 | LIRH | M | Fibroblasts | NK | NA | 65 | NA | NA | NA | NA |
AOHD, adult-onset Huntington disease; CAP, CAG age product; F, female; FrCx, frontal cortex; HD, Huntington disease; HE-PHD, highly expanded paediatric Huntington disease; LIRH, Lega Italiana Ricerca Huntington; LUMC, Leiden University Medical Center; JHD, juvenile Huntington disease; M, male; NA, not applicable; NK, not known; OPBG, Ospedale Pediatrico Bambino Gesù.
Detailed clinical and genetic description in Graziola et al.1
Individual with documented epilepsy.
Sample used for IHC experiments.
Post-mortem brain tissue samples of the frontal cortex and striatum were taken from donor brains of deceased patients with either HE-PHD (n = 2), JOHD (n = 3) or AOHD (n = 6), and healthy adult controls (n = 6) (age-matched to AOHD only, owing to the lack of available donor brains from healthy children) and collected at the Department of Pathology, Leiden University Medical Center (LUMC), Leiden, the Netherlands (Table 1).
Human fibroblast cell lines were generated at the IRCCS Bambino Gesú Children’s Hospital (OPBG), Rome, Italy following skin punch biopsies taken from three patients with HE-PHD, two patients with JOHD, two patients with AOHD (all recruited from the Lega Italiana Ricerca Huntington [LIRH] Foundation outpatient clinic) and six age-matched controls (four recruited from the LIRH Foundation and two recruited from the OPBG; Table 1). These patients had also been entered into the ENROLL-HD study, the world’s largest observational study for patients with HD and their families, at the LIRH Foundation site.3
Sample size determination
Owing to the rarity of HD, especially HE-PHD, no formal sample size calculations were performed. Indeed, HE-PHD is so rare that its prevalence has yet to be determined. Furthermore, obtaining HE-PHD brain specimens is particularly challenging since it relies on brain donations from deceased children, which is generally considered an exceptional event. Our available sample size of 30 subjects, including patients and healthy controls (brain donors, n = 17; fibroblast donors, n = 13), was therefore based on the number of samples obtainable within a reasonable timeframe for analysis.
Analysis of protein and transcripts
Deep frozen human brain samples and fibroblasts were prepared in radioimmunoprecipitation (RIPA) buffer (50 mM Tris-HCl, 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 1 mM EDTA, 0.1% SDS, pH 7.4) and phosphatase and protease inhibitors (Sigma–Aldrich). Detailed laboratory procedures for protein expression analysis have been previously described.16 Briefly, 20 μg of protein preparations from patients and controls were separated via SDS-PAGE using Criterion™ TGX Stain-Free™ precast gels (Bio-Rad Laboratories or ThermoFisher) and transferred to a nitrocellulose or polyvinylidene difluoride membranes by Trans-Blot Turbo Transfer System (Bio-Rad Laboratories). Membranes were probed with the following primary antibodies: GLUT-1 (Ab15309, Abcam, 1:1000, RRID: AB_301844), GLUT-3 (Ab15311, Abcam, 1:1000, RRID: AB_301846), Rab11-A (sc-166912, Santa Cruz Biotec., 1:1000, RRID: AB_10611645), HK-II (sc-130358, Santa Cruz, 1:1000, RRID:AB_2295219), OXPHOS (Ab110411, Abcam, 1:5000, RRID:AB_2756818), NDUFUB8 (NBP2-75586, NOVUS, 1:5000), and VDAC (PA1-954A, Invitrogen, 1:1000, RRID:AB_2304154). GAPDH (MA5-15738, Invitrogen, 1:1000, RRID: AB_10977387) and anti-Vinculin (V9264, Sigma–Aldrich, 1:1000, RRID: AB_10603627) were the housekeeping proteins used for normalization in brain tissue and fibroblasts, respectively. Immunodetection was performed with horseradish peroxidase (HRP)-conjugated secondary antibodies anti-rabbit (1:10000; L005661, Bio-Rad Laboratories) or anti-mouse (1:10000; L005662, Bio-Rad Laboratories or 1:30000, Jackson Immuno-Research). Blots were then imaged by the ChemiDoc MP imaging system using Chemiluminescence settings. Western blot results were quantified and visualised as percentage of variation relative to controls using Image Lab 6.1 software (Bio-Rad Laboratories). All experiments were performed in triplicate. Total RNA was isolated from patients’ and controls’ fibroblasts using the Total RNA Purification Plus Kit (Norgen Biotek Corp, Canada) and, for each sample, 2 μg of total RNA was reverse transcribed according to the manufacturer’s protocol for M-MLV reverse transcriptase (Promega Italia, Italy). cDNAs were amplified (TaqMan assays) in triplicate with primers for SLC2A1 (Hs00892681_m1), SLC2A3 (Hs00359840_m1), Rab11-A (Hs00366449_g1), Rab11-B (Hs00188448_m1), and GAPDH (Hs99999905_m1) conjugated with fluorochrome FAM (Applied Biosystems Italia). The level of expression was measured by real-time quantitative reverse transcriptase PCR (qRT-PCR) using cycle threshold (Ct). The Ct was obtained by subtracting the Ct value of the gene of interest from the Ct value of the housekeeping gene (GAPDH). Data were analysed using the 2−ΔΔCt method and reported as fold difference relative to controls. The analysis was performed using the QuantStudio™ 12K FlexSoftware v2.2 (Applied Biosystems). All PCR reactions were performed using a QuantStudio 12K Flex Real-Time PCR System (Applied Biosystems).
Immunohistochemistry and cell counts
Formalin-Fixed-Paraffin-Embedded 5 μm brain sections were cut and stained using haematoxylin and eosin (H&E). Immunostaining was performed for GLUT-1 (Ab15309, Abcam, 1:500, RRID: AB_301844) and NeuN (AB104225, Abcam, 1:500, RRID: AB_10711153). Sections were deparaffinised and rehydrated, and subsequent antigen retrieval was performed in citrate buffer (pH 6.0) using a pressure cooker (15 min). Afterwards, endogenous enzyme block was performed by a 10-min incubation in 3% H2O2 in demineralised water, and non-specific antibody block was performed by a 1-h incubation in 1% BSA in PBS-T. Primary antibody incubation was performed overnight (GLUT-1 at room temperature, NeuN at 4 °C), followed by incubation with an anti-rabbit HRP secondary antibody (sc-2030, Santa-Cruz, 1:200) for 1 h at room temperature. Visualization of immunostaining was performed with chromogen 3,30’-diaminobenzidine (DAB). Finally, sections were counterstained with haematoxylin, dehydrated and cover slipped. The slides were digitised using an automatic bright field microscope (Philips Ultra Fast Scanner, Philips, Netherlands) for microscopic evaluation and taking pictures. For easy visualization purposes of GLUT-1 immunopositivity, a binary mask of the DAB signal was established by use of free Image-J software (colour deconvolution, grey scale 8-bit, threshold grey value 0–188, binary mask).
The total number of neurons in HE-PHD donors was estimated by stereological analyses of NeuN-stained slides by use of free QuPath software (positive cell detection, optical density sum, cell intensity threshold: DAB OD max) (Supplementary Fig. S1). Per slide, the mean number of neurons per mm2 was calculated in three separate areas by analyzing cortical layers I to VI. Cell counts were performed by a single individual (HSB).
Fibroblast cell lines
Human cultured fibroblasts were obtained from skin biopsy of patients and aged matched controls. Human fibroblasts were cultured in Dulbecco’s Modified Eagle Medium high glucose (4.5 g/l) supplemented with 10% foetal bovine serum, 50 μg/ml uridine and 110 mg/l sodium pyruvate. To analyse cell cycle status, 5 × 105 fibroblasts from patients and controls were plated and harvested at about 80% confluence. 2 × 106 cells were washed with PBS and centrifuged for 5 min at 1500 rpm at 4 °C. The obtained pellet was fixed with a cold solution of methanol/acetone 2:1 (v/v) by gently vortexing, and incubated over night at 4 °C. The next day, cells were centrifuged at 1100 rpm for 5 min at 4 °C and the pellet was re-suspended in a solution containing propidium iodide 500 μg/ml and RNAse 1 mg/ml and incubated for 30 min at room temperature. After incubation, cells were analysed by flow cytometry (BD LSRFortessa X-20, BD Biosciences).
Statistics
Patient data were expressed as mean ± standard error of the mean (SEM). For brain tissue, densitometry data were first log-transformed and then analysed using Analysis of Variance (ANOVA) to assess the overall differences among groups, i.e., controls, HE-PHD, JOHD, and AOHD. Subsequently, post-hoc comparisons were conducted using the Tukey multiple comparisons of means test or the Wilcoxon rank sum exact test, in case of deviation from normality. Normality was assessed using the Shapiro–Wilk normality test. Multiplicity was not considered in this study. All statistical tests were two-tailed, and the level of significance was set at p < 0.05. Statistical analyses were performed using the R software (version 4.2.2, R Foundation for Statistical Computing, Vienna, Austria).
Ethics
Samples were provided to the LIRH Foundation for research purposes. This study was performed in accordance with the ethical principles outlined in the World Medical Association Declaration of Helsinki. Pseudo-anonymization of all donors was preserved by using a coded system for the tissue samples. Clinical study protocols and informed consent forms for patients and healthy controls were approved by the Institutional Review Board of the LIRH Foundation on 28th October 2022 (n. 10.281022).
Role of the funding source
The funding source had no role in study design, data collection and interpretation, analysis, writing of this report, or in the decision to submit the paper for publication.
Results
Study population and tissue acquisition
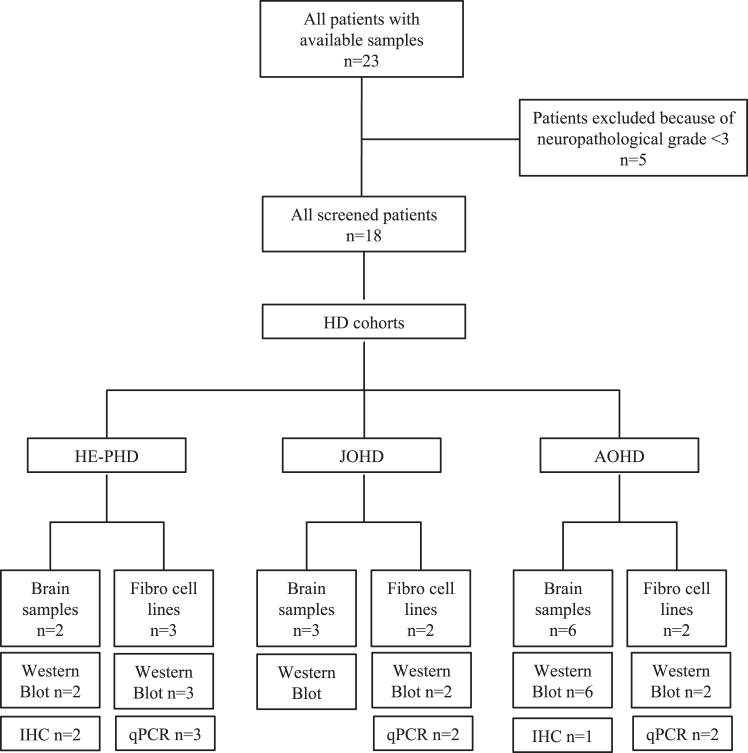
In total, 17 deceased donors were identified for the brain tissue analyses (HE-PHD, n = 2; JOHD, n = 3; AOHD, n = 6; healthy adult controls, n = 6) and 13 individuals participated in the fibroblast analyses (HE-PHD, n = 3; JOHD, n = 2; AOHD, n = 2; healthy adult controls, n = 6) (Fig. 1; Table 1).
Fig. 1.
Study design, participants, and procedures. IHC = immunohistochemistry.
All patients in the HE-PHD cohort (n = 5) showed childhood onset of disease at <10 years of age (Table 1). The HTT mutation was inherited maternally in one patient and transmitted paternally in all other patients.
Post-mortem delay (<12 h) was comparable between controls, patients with HE-PHD, JOHD and AOHD. All striatal samples from patients with HD were categorised as Vonsattel neuropathological grade 3 (Table 1). Further clinical and neuropathological characteristics of the HE-PHD brain donors are also included in Table 1. Case description can be found in Supplementary Materials.
Patients of any sex were eligible for inclusion in this study. Sex was self-reported by participants or, for deceased donors, documented per the patient’s medical records.
Low GLUT-1 expression in HE-PHD tissues
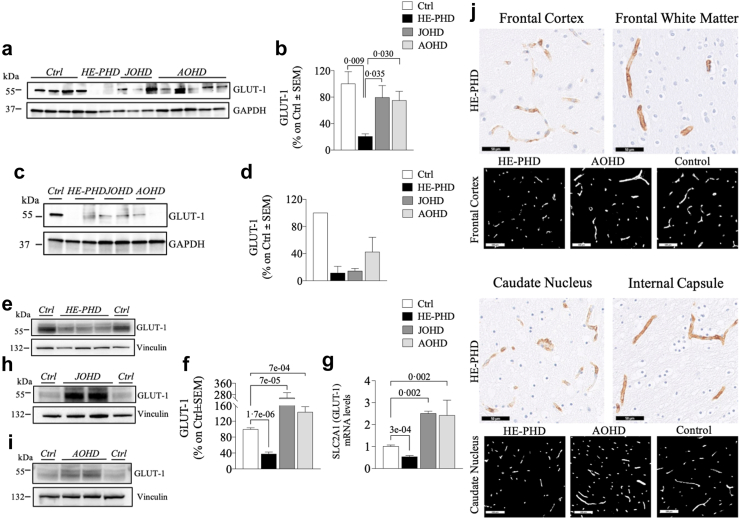
The ANOVA test for GLUT-1 protein expression revealed a statistically significant difference among groups in the frontal cortex (F (3, 10) = 5.93, p = 0.014), but not in the striatum (F (3, 4) = 3.054, p = 0.15). In the frontal cortex of donors with HE-PHD, the GLUT-1 protein expression level was ∼4 or 5 times lower than in the JOHD, AOHD, or control samples (Fig. 2a and b; Table 2). The protein expression level of GLUT-1 was also lower in the striatum of donors with HE-PHD than in all other groups, although this difference was not statistically significant (Fig. 2c and d; Table 2). In contrast, neither JOHD nor AOHD samples showed any significant difference in GLUT-1 expression in the frontal cortex or in the striatum when compared to the control samples (Tukey test, frontal cortex: JOHD p = 0.884, 95% CI [−1.25, 0.77], AOHD p = 0.704, 95% CI [1.21, −0.57]; striatum: JOHD p = 0.407, 95% CI [−5.81, 2.35], AOHD p = 0.698, 95% CI [−2.99, 5.17]) (Fig. 2a–d). The expression levels of GLUT-1 protein and SLC2A1 mRNA were 2 to ∼7 times statistically significantly lower in peripheral fibroblasts of donors with HE-PHD than in all other groups (Fig. 2e–i; Table 3). On the other hand, GLUT-1 protein and SLC2A1 mRNA levels were statistically significantly greater in peripheral fibroblasts of donors with JOHD (Wilcoxon rank sum exact test, protein W = 132, p = 0.0002, 95% CI [0.65, 2.35]; mRNA W = 126, p = 0.0003, 95% CI [1.23, 1.77]) and AOHD (Wilcoxon rank sum exact test, protein W = 123, p = 0.0016, 95% CI [0.17, 0.62]; mRNA W = 100, p = 0.0332, 95% CI [0.05, 2.49]) than in controls (Fig. 2e–i).
Fig. 2.
GLUT-1 in brain tissues and fibroblasts of patients with HE-PHD, JOHD and AOHD and controls. Representative Western blot images and densitometric evaluation of GLUT-1 protein levels normalised to GAPDH or Vinculin protein levels in the FrCx (a, b) striatum (c, d) and fibroblasts (e, f, h, i) of patients with HE-PHD, JOHD and AOHD and healthy adult controls. All densitometric values are reported as a percentage of controls (set at 100%) and are the mean ± SEM of three independent experiments (p < 0.05, one-way ANOVA with Tukey multiple comparisons of means or Wilcoxon rank sum exact post-hoc tests). qRT-PCR analysis of SLC2A1 (GLUT-1) total mRNA in fibroblasts of patients with HE-PHD, JOHD and AOHD and controls (g). Relative mRNA levels were normalised using GAPDH and were calculated as 2−ΔCt. Results are the mean ± SEM of three independent experiments (p < 0.05, one-way ANOVA with Tukey multiple comparisons of means post-hoc test). IHC staining of GLUT-1 in the FrCx (white matter) of HE-PHD donor HD252 (j). Smaller length and lower branching complexity of immune-positive capillaries in the FrCx of HE-PHD donor HD252, as compared to AOHD donor HD249 and healthy control donor E16-07. Scale bars: 100 μm. GLUT-1 IHC staining in the striatum (caudate nucleus, internal capsule) of HE-PHD donor (HD86) (j). Binary representation of the DAB signal, illustrating smaller length and lower branching complexity of immune-positive capillaries in the caudate nucleus of HD86, as compared to AOHD donor HD249 and healthy control donor E15-02. Scale bars 50 μm. The use of the same loading control in different figures serves as a representative image. Each individual protein has been normalized against its respective loading control.
Table 2.
Summary of average log-transformed protein expression levels ± SEM, Confidence Intervals (lower, upper), and [p-values] in the frontal cortex and striatum of highly expanded paediatric (HE-PHD) compared to juvenile onset HD (JOHD), adult onset HD (AOHD) and control (Ctrl) individuals.
| Protein | Cohort | Frontal cortex | Striatum |
|---|---|---|---|
| GLUT1 | HE-PHD | 12.5 ± 0.19 (10.1, 14.9) | 9.83 ± 1.30 (−6.67, 26.3) |
| Ctrl | 14.0 ± 0.19 (13.4, 14.7) [p = 0.009] | 12.8 ± 0.12 (11.2, 14.4) [p = 0.129] | |
| JOHD | 13.8 ± 0.27 (12.7, 15.0) [p = 0.035] | 11.0 ± 0.07 (10.1, 12.0) [p = 0.65] | |
| AOHD | 13.7 ± 0.21 (13.1, 14.3) [p = 0.030] | 11.7 ± 0.56 (4.57, 18.8) [p = 0.376] | |
| GLUT3 | HE-PHD | 13.1 ± 0.44 (7.50, 18.7) | 13.3 ± 0.20 (10.7, 15.9) |
| Ctrl | 14.3 ± 0.05 (14.2, 14.5) [p = 0.017] | 14.5 ± 0.02 (14.3, 14.7) [p = 0.022] | |
| JOHD | 13.0 ± 0.70 (10.0, 16.1) [p = 0.987] | 13.3 ± 0.21 (10.7, 16.0) [p = 0.999] | |
| AOHD | 13.7 ± 0.21 (13.2, 14.3) [p = 0.440] | 13.8 ± 0.15 (11.9, 15.7) [p = 0.309] | |
| Rab11-A | HE-PHD | 16.3 ± 0.02 (16.1, 16.5) | 16.5 ± 0.49 (10.3, 22.7) |
| Ctrl | 17.4 ± 0.08 (17.1, 17.6) [p = 0.011] | 17.6 ± 0.01 (17.6, 17.6) [p = 0.170] | |
| JOHD | 16.9 ± 0.34 (15.5, 18.4) [p = 0.175] | 17.3 ± 0.31 (13.3, 21.3) [p = 0.377] | |
| AOHD | 17.4 ± 0.09 (17.1, 17.6) [p = 0.010] | 17.3 ± 0.12 (15.8, 18.7) [p = 0.404] | |
| HK-II | HE-PHD | 16.7 ± 0.21 (14.0, 19.4) | 16.7 ± 0.59 (9,2, 24.2) |
| Ctrl | 18.0 ± 0.12 (17.8, 18.2) [p = 0.010] | 18.6 ± 0.01 (18.6, 18.7) [p = 0.045] | |
| JOHD | 17.5 ± 0.44 (15.6, 19.4) [p = 0.131] | 17.7 ± 0.22 (14.8, 20.5) [p = 0.283] | |
| AOHD | 17.9 ± 0.06 (17.7, 18.1) [p = 0.013] | 17.9 ± 0.14 (16.0, 19.7) [p = 0.193] | |
| Complex I | HE-PHD | 16.9 ± 0.62 (9.0, 24.8) | 18.1 ± 0.11 (17.4, 20.1) |
| Ctrl | 17.6 ± 0.19 (17.0, 18.3) [p = 0.155] | 18.7 ± 0.01 (18.7, 18.8) [p = 0.230] | |
| JOHD | 17.9 ± 0.21 (17.4, 18.5) [p = 0.049] | 18.4 ± 0.03 (18.0, 18.9) [p = 0.633] | |
| AOHD | 17.8 ± 0.06 (17.6, 18.0) [p = 0.064] | 18.8 ± 0.11 (17.4, 20.1) [p = 0.173] | |
| Complex II | HE-PHD | 16.7 ± 0.15 (14.8, 18.6) | 15.8 ± 0.19 (13.3, 18.2) |
| Ctrl | 17.4 ± 0.09 (17.1, 17.6) [p = 0.027] | 17.3 ± 0.01 (17.3, 17.4) [p = 0.024] | |
| JOHD | 17.2 ± 0.13 (16.6, 17.7) [p = 0.150] | 17.0 ± 0.33 (12.8, 21.2) [p = 0.048] | |
| AOHD | 17.3 ± 0.11 (17.0, 17.6) [p = 0.052] | 16.7 ± 0.19 (14.3, 19.2) [p = 0.099] | |
| Complex III | HE-PHD | 15.6 ± 0.41 (10.3, 20.8) | 17.6 ± 0.43 (12.1, 23.1) |
| Ctrl | 16.7 ± 0.05 (16.6, 16.9) [p = 0.002] | 19.1 ± 0.01 (19.1, 19.2) [p = 0.029] | |
| JOHD | 16.1 ± 0.21 (15.2, 17.0) [p = 0.061] | 18.4 ± 0.03 (17.9, 18.8) [p = 0.215] | |
| AOHD | 16.6 ± 0.01 (16.6, 16.7) [p = 0.002] | 18.3 ± 0.03 (18.0, 18.7) [p = 0.246] | |
| Complex IV | HE-PHD | 16.0 ± 0.75 (6.48, 25.6) | 16.2 ± 0.51 (9.76, 22.6) |
| Ctrl | 17.6 ± 0.11 (17.2, 17.9) [p = 0.217] | 18.9 ± 0.01 (18.9, 19.1) [p = 0.010] | |
| JOHD | 17.1 ± 0.94 (13.1, 21.2) [p = 0.501] | 17.5 ± 0.15 (15.6, 19.3) [p = 0.120] | |
| AOHD | 17.8 ± 0.13 (17.4, 18.1) [p = 0.123] | 17.8 ± 0.28 (14.3, 21.4) [p = 0.058] | |
| Complex V | HE-PHD | 15.7 ± 0.14 (13.9, 17.5) | 16.8 ± 0.31 (12.8, 20.8) |
| Ctrl | 16.2 ± 0.13 (15.8, 16.6) [p = 0.161] | 18.2 ± 0.02 (18.2, 28.5) [p = 0.012] | |
| JOHD | 16.2 ± 0.21 (15.3, 17.0) [p = 0.246] | 17.4 ± 0.02 (17.2, 17.6) [p = 0.172] | |
| AOHD | 16.2 ± 0.14 (13.9, 17.5) [p = 0.156] | 17.3 ± 0.07 (16.3, 18.2) [p = 0.277] |
Assessment of highly expanded (HE) Paediatric Huntington disease (PHD) compared with control (Ctrl), Juvenile-Onset (JOHD), and Adult-Onset HD (AOHD) individuals.
Table 3.
Summary of average protein and gene expression levels ± SEM, Confidence Intervals (lower, upper), and [p-values] in fibroblast cell lines of highly expanded paediatric (HE-PHD) compared to juvenile onset HD (JOHD), adult onset HD (AOHD) and control (Ctrl) individuals.
| Protein | Cohort | Levels |
|---|---|---|
| GLUT1 | HE-PHD | 0.38 ± 0.17 (0.27, 0.48) |
| Ctrl | 1.00 ± 0.04 (0.91, 1.09) [p = 1.7e-06] | |
| JOHD | 2.59 ± 0.35 (1.68, 3.49) [p = 7e-05] | |
| AOHD | 1.43 ± 0.14 (1.08, 1.79) [p = 7e-04] | |
| Gene exp. | ||
| SLC2A1 | HE-PHD | 0.54 ± 0.05 (0.41, 0.67) |
| Ctrl | 1.02 ± 0.08 (0.83, 1.21) [p = 3e-04] | |
| JOHD | 2.51 ± 0.09 (2.25, 2.76) [p = 0.002] | |
| AOHD | 2.43 ± 0.69 (0.65, 4.20) [p = 0.002] | |
| Protein | ||
| GLUT3 | HE-PHD | 0.87 ± 0.04 (0.79, 0.97) |
| Ctrl | 1.05 ± 0.03 (0.93, 1.07) [p = 0.046] | |
| JOHD | 1.09 ± 0.09 (0.99, 1.20) [p = 0.002] | |
| AOHD | 1.12 ± 0.09 (0.89, 1.34) [p = 0.045] | |
| Gene exp. | ||
| SLC2A3 | HE-PHD | 0.89 ± 0.12 (0.61, 1.19) |
| Ctrl | 1.11 ± 0.18 (0.67, 1.55) [p = 0.359] | |
| JOHD | 1.18 ± 0.17 (0.75, 1.62) [p = 0.204] | |
| AOHD | 1.70 ± 0.58 (0.19, 3.20) [p = 0.573] | |
| Protein | ||
| Rab11-A | HE-PHD | 1.06 ± 0.06 (0.93, 1.19) |
| Ctrl | 1.00 ± 0.05 (0.89, 1.11) [p = 0.434] | |
| JOHD | 1.14 ± 0.09 (1.04, 1.23) [p = 0.840] | |
| AOHD | 1.06 ± 0.08 (0.85, 1.26) [p = 1] | |
| Gene exp. | ||
| Rab11-A | HE-PHD | 0.93 ± 0.08 (0.73, 1.13) |
| Ctrl | 1.00 ± 0.32 (0.92, 1.08) [p = 0.443] | |
| JOHD | 0.87 ± 0.06 (0.71, 1.02) [p = 0.569] | |
| AOHD | 1.66 ± 0.52 (0.33, 2.99) [p = 0.175] |
Assessment of highly expanded (HE) Paediatric Huntington disease (PHD) compared with control (Ctrl), Juvenile-Onset (JOHD), and Adult-Onset HD (AOHD) individuals.
GLUT-1 localization was predominantly found in endothelial cells lining parenchymal capillaries of HE-PHD, AOHD and control brain tissues. In the frontal cortex of HE-PHD donors, a patchy lack of GLUT-1 immunopositivity and reduced expression was observed, as compared to the adjacent subcortical white matter within the same donor (Fig. 2j). Also, in the frontal cortex, a smaller length and less branching complexity of capillaries was visualized in donors with HE-PHD, as compared to donors with AOHD and control (Fig. 2j). GLUT-1 visualization in the caudal neostriatum of HE-PHD donors revealed areas with low or no immunopositivity, as compared to the adjacent capsular white matter showing a regular signal (Fig. 2j). In addition, the length and branching complexity of GLUT-1 positive capillaries in the striatum appeared lower as compared to donors with AOHD and control (Fig. 2j).
Limited neurodegeneration in the frontal cortex of patients with HE-PHD
Microscopic analysis of the frontal cortex revealed limited signs of degeneration (i.e., eccentric nucleus and eosinophilic cytoplasm) and neuronal loss in one donor with HE-PHD (HD86; mean neuronal cell count, 349 per mm2), and normal structure and architecture of the neocortex in the other (HD252; mean neuronal cell count 493 per mm2) (Supplementary Fig. S2a and b). Conversely, in both patients there was remarkable neuronal loss in the striatum, in line with Vonsattel grade 3.
Low GLUT-3 and Rab11-A expression in HE-PHD tissues
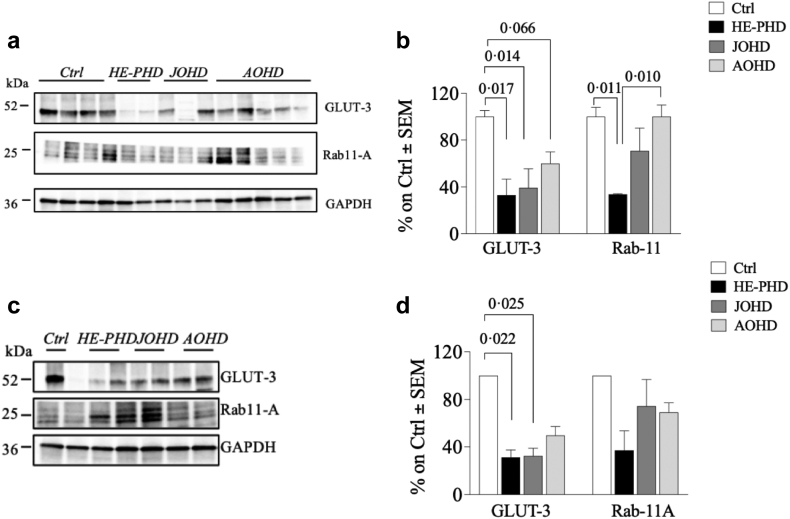
The protein expression levels of GLUT-3 were globally significantly different among groups, both in the frontal cortex (ANOVA test, F (3, 10) = 7.05, p = 0.0079) and in the striatum (ANOVA test, F (3, 4) = 11.6, p = 0.019). Compared to controls, protein expression levels of GLUT-3 were significantly lower in both the frontal cortex and striatum of donors with HE-PHD (Fig. 3a–d; Table 2). GLUT-3 was also lower in donors with AOHD compared to controls in the frontal cortex (Tukey test, p = 0.066, 95% CI [−1.28, 0.14]) (Fig. 3a–d), and lower in donors with JOHD compared to controls in both the frontal cortex (Tukey test, p = 0.014, 95% CI [−6.26, −5.32) and striatum (Tukey test, p = 0.025, 95% CI [−2.09, −0.21]). In peripheral fibroblasts, SLC2A3 gene expression was not statistically significantly different between any group (Table 3), whereas its protein expression was statistically significantly lower in donors with HE-PHD when compared to donors with JOHD, AOHD and controls (Supplementary Fig. S1a–e; Table 3).
Fig. 3.
Evaluation of GLUT-3 and Rab11-A levels in brain tissues of patients with HE-PHD, JOHD, AOHD and controls. Representative Western blot images and densitometric evaluation of GLUT-3 and Rab11-A protein levels normalised to GAPDH protein levels in the FrCx (a, b) and striatum (c, d) of patients with HE-PHD, JOHD and AOHD and healthy adult controls. All densitometric values are reported as a percentage of controls (set at 100%) and are the mean ± SEM of three experiments (p < 0.05, one-way ANOVA with Tukey multiple comparisons of means post-hoc test). The use of the same loading control in different figures serves as a representative image. Each individual protein has been normalized against its respective loading control.
Expression levels of the cargo protein Rab11-A were significantly lower in the frontal cortex of donors with HE-PHD compared to both controls (Tukey test, p = 0.011, 95% CI [−1.91, −0.25]) and AOHD (Tukey test, p = 0.010, 95% CI [−1.87, −0.27]) (Fig. 3a and b). Data collected in the striatum did not show any differences in Rab11-A protein levels between the HE-PHD cohort and any other cohort (Fig. 3c and d). Rab11-A protein levels in fibroblasts also did not differ between the HE-PHD cohort and any other cohort (Supplementary Fig. S1a, d–g; Table 3).
Impairment in glucose uptake is associated with defects of mitochondrial machinery in patients with HE-PHD
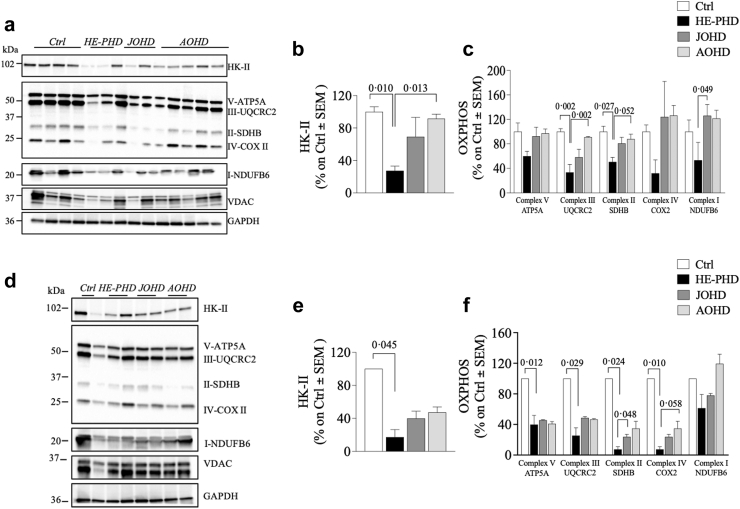
Mitochondrial complex expression was dysregulated in the brains of patients with HD. In particular, levels of complex II and complex III subunits were significantly lower in the frontal cortex of donors with HE-PHD, compared to controls (Tukey test, p = 0.027, 95% CI [−1.22, −0.074] and p = 0.002, 95% CI [−1.82, −0.48], respectively) and compared to patients with AOHD (Tukey test, p = 0.052, 95% CI [−1.09, 0.005], and p = 0.002, 95% CI [−1.73, −0.43], respectively) (Fig. 4a, c; Table 2). Furthermore, the expression level of the complex I subunit in HE-PHD was statistically significantly lower compared to donors with JOHD (Tukey test, p = 0.049, 95% CI [−2.10, −0.006]), and numerically lower compared to donors with AOHD, following a trend towards statistical significance (Tukey test, p = 0.064, 95% CI [−1.87, 0.048]) (Fig. 4a, c; Table 2). We also evaluated levels of complex IV and V (ATP synthase) and voltage-dependent anion-selective channel, a protein that plays a key role in maintaining high rates of oxidative phosphorylation.17 No statistically significant differences between groups were observed for either protein in the frontal cortex.
Fig. 4.
Evaluation of HK-II and mitochondrial machinery complexes (OXPHOS) levels in brain tissues of patients with HE-PHD, JOHD, AOHD and controls. Representative Western blot images and densitometric evaluation of HK-II and OXPHOS protein levels normalised to GAPDH protein levels in the FrCx (a–c) and striatum (d–f) of patients with HE-PHD, JOHD and AOHD and healthy adult controls. All densitometric values are reported as a percentage of controls (set at 100%) and are presented as the mean ± SEM of three independent experiments (p < 0.05, one-way ANOVA with Tukey multiple comparisons of means post-hoc test). The use of the same loading control in different figures serves as a representative image. Each individual protein has been normalized against its respective loading control.
Deregulation of mitochondrial complexes expression was also observed in the striatum, with statistically significantly lower levels of all complexes, except complex I, seen in patients with HE-PHD compared to controls (Fig. 4d, f; Table 2).
The HE-PHD cohort showed statistically significantly lower levels of HK-II protein expression in the frontal cortex and striatum, compared to controls (Tukey test, p = 0.010, 95% CI [−2.30, −0.33], and p = 0.045, 95% CI [−3.78, −0.06], respectively) and in the frontal cortex compared to AOHD (Tukey test, p = 0.013, 95% CI [−2.18, −0.27]) (Fig. 4a, b, d, e; Table 2). In contrast, no difference in HK-II protein expression in the frontal cortex was observed between the AOHD and control cohorts (Tukey test, p = 0.982, 95% CI [0.86, −0.67]; striatum, p = 0.443, 95% CI [2.62, −1.10]). In peripheral fibroblasts, HK-II protein expression differed only between the JOHD and control cohorts (Tukey test, p = 0.014, 95% CI [0.07, 0.77]) (Supplementary Fig. S1h–k).
Discussion
Neurodegenerative conditions, such as Alzheimer, Huntington, and Parkinson diseases, are associated with brain glucose hypometabolism, which is contributed to by impaired GLUT-1 and GLUT-3 glucose transporters.8 Interestingly, dysregulation in GLUT transporters has also been observed in aberrant brain neurodevelopment,18 micromalformations19 and epilepsy, all conditions that have been associated with HD.3,4,20,21
Patients with HE-PHD manifest signs and symptoms that are uncommon to AOHD, including high frequency of epileptic seizures, early onset hypokinetic-rigid syndrome/dystonia and developmental delay.3,4 Interestingly, certain HE-PHD clinical features, such as epilepsy, are also observed in patients with GLUT-1 deficiency syndrome, where genetic defects in this transporter result in a chronic shortage of glucose in the brain.22 This suggests that alterations in glucose transport or metabolism may also occur in HE-PHD and could explain some of its unique symptoms, compared to AOHD.
In the context of HD, this study demonstrates reduced protein expression of GLUT-1 and GLUT-3 in the frontal cortex and striatum of brains from patients with HE-PHD, compared to brains from patients affected since adulthood, including JOHD, and from controls. Our results are partly in line with others, who have reported significantly lower GLUT-1 and GLUT-3 protein expression in the striatum of late-stage AOHD (classified as neuropathological Vonsattel grade 3), compared to earlier-stage AOHD (neuropathological Vonsattel grades 1 and 2).20 However, they found no evidence of reduced GLUT-1 and GLUT-3 expression in AOHD cortex,23 whereas in our frontal cortex samples we found statistically significantly lower GLUT-1 expression in patients with HE-PHD versus controls, and statistically significantly lower GLUT-3 expression in all cohorts (HE-PHD, JOHD and AOHD) versus controls.
Furthermore, we identified reduced protein levels of Rab11-A in HE-PHD frontal cortex. Rab11-A is a small GTPase that has a critical role in regulating the trafficking of GLUT-3 to the neuronal cell surface,24 experiments in HD cell models have shown reduced neuronal GLUT-325 and Rab11-A expression levels and highlights the relevance of altered glucose transportation in models with HE mutations.
In HE-PHD brain samples, low levels of GLUT-1 and GLUT-3 were observed alongside low levels of mitochondrial complexes involved in energy metabolism. In the frontal cortex, significant differences versus controls and versus AOHD were seen for complexes II and III and versus JOHD for complex I. Results from in vitro studies have revealed calcium abnormalities in mitochondria from patients and transgenic mice with HD, and that these defects are a direct effect of HE polyQ.26 Our findings in brain tissues affected by HE HTT mutations, therefore, corroborate these in vitro observations.
Furthermore, we also observed significantly lower levels of HK-II in HE-PHD but not AOHD brains compared to control. Hexokinases catalyse the first committed step of glucose metabolism, by phosphorylating glucose to glucose-6-phosphate.27 HK-II also protects cells from death during hypoxia and functions as a sensor of glucose availability, inducing apoptosis in response to glucose depletion.28
Our finding, that expression levels of GLUT-1, GLUT-3, mitochondrial complexes and HK-II were significantly and selectively lower in HE-PHD frontal cortex and striatum, shows that a disease-relevant biological dysfunction (i.e., hypometabolic state) occurs without any substantial neuronal loss in the frontal cortex of these patients. Relatively preserved brain cortex has been observed previously in HE-PHD by magnetic resonance imaging (MRI), magnetic resonance spectroscopy and neuropathological studies,3 and is in line with findings from ongoing studies using volumetric MRI in paediatric-onset patients (Sabatini, personal communication). Nevertheless, the disease pathophysiology of patients with HE HTT mutations, concurs with a more severe disease and worse prognosis.
To validate the findings from our study on brain tissues of patients with HE-PHD, we conducted an analysis of metabolic profiles in fibroblasts obtained from patients with HE-PHD. Our results revealed significantly reduced levels of GLUT-1 in both gene and protein expression, and a somewhat less pronounced decrease in GLUT-3 protein expression. However, no significant differences were observed in gene expression when compared to the healthy control. This discrepancy between findings in the brain tissues versus fibroblasts is likely explained by the fact that GLUT-3 is not the main transporter of glucose in peripheral cells such as fibroblasts.29 While our data emphasize that the primary pathology in HE-PHD is found in the brain, they also highlight the probable existence of a peripheral component to the disease, as also suggested by our recent study which identified liver steatosis in patients with HE-PHD.5 The evidence of peripheral biological abnormalities may represent an important observation considering currently ongoing experimental therapies aimed at lowering mutated HTT in both the brain and periphery.30
Since a direct relationship between GLUT function, neurodegeneration and neurodevelopment is still poorly understood, additional studies in HD and other neurodegenerative diseases are therefore needed due to its potential role in HD pathogenesis20 and therapy perspectives.31 In fact, growing evidence suggests an important modulatory role for glucose transport in the HD mitochondrial deregulation, emphasizing that abnormal energy metabolism could interfere with brain neurodevelopment and may represent a critical therapeutic target for HD, as also suggested for adulthood disease.32 For instance, the GLUT-1 role in HE-PHD pathology suggests that a ketogenic diet could be beneficial for patients with HE-PHD when initiated in the early stages of the disease.11 The high-fat ketogenic diet may result in permanent ketosis and provide the brain with an alternative energy source. The beneficial effects of the ketogenic, normocaloric, with low carbohydrate intake diet, currently used in epilepsy and movement disorders, are well documented in children with GLUT-1 deficiency and may offer new therapeutic approaches to HE-PHD. This is especially important given that there is no validated experimental protocol so far to at least address any disease-modifying treatment against HE-PHD.
We are aware that our study has a number of limitations, the main one being that our HE-PHD brain donor cohort is limited to only two patients. This is due to the extremely rare occurrence of this variant. HD itself is a rare disease and only a small proportion of about 6% patients have juvenile-onset disease,14 and an even smaller proportion have HE-PHD. Thus, the opportunity to acquire a donor brain from a patient with HE-PHD is exceptionally rare. Likewise, owing to a general lack of donor brains from healthy controls, especially child donors, it was not possible to age-match our healthy control donor brain cohort to our HE-PHD cohort. It is therefore possible that some of the differences we observed between patients with HE-PHD and controls may have been due to differences in age rather than pathophysiology. Another limitation is due to the analysis of frontal cortex only, while an extensive study of several parts of brain and cortical regions might have offered additional insights. Unfortunately, we cannot provide a fine stratification of brain areas in our current context. This argues again that more cases and more extensive examination of the cerebral cortex are required to draw any firm conclusions.
An additional limitation is due to the clinical stratification of our adult cohort, specifically concerning the retrospective reconstruction of the approximate age of onset of JOHD. To limit such a bias, we included in the JOHD cohort only those patients with mutation size >55 CAG repeats, a mutation size which is widely believed to be associated with JOHD.14 Notwithstanding these limitations, our findings shed light on the pathology of this devastating disease variant and highlight differences in potential biological mechanisms underlying HE-PHD and AOHD. Such differentiation may be important not only from a therapeutic perspective but also to ensure appropriate inclusion of paediatric patients into Huntington disease clinical trials.14
Finally, our study highlighted a difference in GLUT-1 expression between HE-PHD and JOHD. Such a biological difference, in addition to the CAG mutation length, suggests there may be a need to reconsider the old classification of HD variants and abandon the terminology “juvenile-onset”, which currently includes some adult-onset patients and instead replace this with the term “paediatric-onset” HD, to represent the category of patients that appears clinically and biologically different from adulthood HD.
Contributors
AT contributed to conceptualization, methodology, formal analysis, validation and editing; HB contributed to methodology, formal analysis, investigation and editing; FP contributed to methodology, project administration, data curation and editing. AT, HB and FP contributed equally to this work. MDN contributed to methodology, data curation, editing. TM contributed to methodology, formal analysis, software and editing. MPA contributed to methodology, data curation, editing. MZ and SP contributed to methodology, investigation, editing. RC, SdB, MP, and WvR-M contributed to conceptualization, data curation, supervision, editing and had direct access to the data and verified the data. FS contributed to conceptualization, funding acquisition, resources, supervision, writing, and editing and had direct access to the data and verified the data. All authors reviewed and approved the final version of the manuscript.
Data sharing statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request. We will consider data requests from qualified scientific or medical researchers conducting legitimate scientific research. Data will be made available to researchers whose proposal has been approved by an independent review committee. We will share anonymized individual patient level data collected during the study, as well as associated study documents, including the clinical study protocol, statistical analysis plan and informed consent forms. Requests for such data can be made immediately upon publication (no end date). Data will be shared by email to ferdinando.squitieri@lirh.it.
Declaration of interests
FS received remuneration as member of the scientific advisory board of PTC Therapeutics and Prilenia. Leiden University Medical Center (LUMC) receives grants from the European Huntington’s Disease Network (EHDN) and Cure HD Initiative (CHDI), participates in an EU Horizon 2020 project: Innovative Medicines Initiative (IMI) 2 (IDEA_FAST), and participates in clinical trials sponsored by PRILENIA, PTC Therapeutics, WAVE and VICO Therapeutics. The aforementioned sponsors had no role in the design, execution, interpretation, or writing of this manuscript. S.T. de Bot is a board member of the scientific advisory board of the Vereniging van Huntington (HD patient association); unpaid. W.v.R.M. declares to have patents on exon skipping approaches for neurological disorders. In the past, some of these patents have been licensed to ProQR therapeutics. W.v.R.M. further discloses being ad hoc consultant for Accure Therapeutics and Herbert Smith Freehills. Remuneration for these activities is paid to the LUMC. She is on the scientific advisory board of the European Huntington Disease Network, the Dutch Brain Foundation, the Dutch Ataxia Foundation. She is co-director of the Dutch Centre of RNA Therapeutics, board member of the 1 Mutation 1 Medicine network and core member of the Ataxia Advisory Committee for Therapeutics.
Acknowledgements
We are grateful to all the patients and their families for participating in our study, and for offering all information, data and updates on the disease in these patients. FS is CSO of the Italian League for Huntington (LIRH) Foundation. We also thank Dr Abigail Woollard, a professional medical writer funded by the LIRH Foundation, for providing editorial revisions to the draft manuscript. HB and SdB are members of the European Reference Network for Rare Neurological Diseases - Project ID No 739510.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104849.
Appendix ASupplementary data
References
- 1.MacDonald M.E., Ambrose C.M., Duyao M.P., et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72(6):971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Tabrizi S.J., Langbehn D.R., Leavitt B.R., et al. Biological and clinical manifestations of Huntington’s disease in the longitudinal TRACK-HD study: cross-sectional analysis of baseline data. Lancet Neurol. 2009;8(9):791–801. doi: 10.1016/S1474-4422(09)70170-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fusilli C., Migliore S., Mazza T., et al. Biological and clinical manifestations of juvenile Huntington’s disease: a retrospective analysis. Lancet Neurol. 2018;17(11):986–993. doi: 10.1016/S1474-4422(18)30294-1. [DOI] [PubMed] [Google Scholar]
- 4.Cloud L.J., Rosenblatt A., Margolis R.L., et al. Seizures in juvenile Huntington’s disease: frequency and characterization in a multicenter cohort. Mov Disord. 2012;27(14):1797–1800. doi: 10.1002/mds.25237. [DOI] [PubMed] [Google Scholar]
- 5.Squitieri F., Monti L., Graziola F., Colafati G.S., Sabatini U. Early liver steatosis in children with pediatric Huntington disease and highly expanded CAG mutations. Park Relat Disord. 2022;103:102–104. doi: 10.1016/j.parkreldis.2022.08.027. [DOI] [PubMed] [Google Scholar]
- 6.Tereshchenko A., Magnotta V., Epping E., et al. Brain structure in juvenile-onset Huntington disease. Neurology. 2019;92(17):e1939–e1947. doi: 10.1212/WNL.0000000000007355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou X.Y., Lu J.Y., Zuo C.T., Wang J., Sun Y.M. Juvenile-onset hypokinetic-rigid Huntington’s disease: a case with dual-tracer positron emission tomography. Quant Imaging Med Surg. 2021;11(1):479–482. doi: 10.21037/qims-19-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Głuchowska K., Pliszka M., Szablewski L. Expression of glucose transporters in human neurodegenerative diseases. Biochem Biophys Res Commun. 2021;540:8–15. doi: 10.1016/j.bbrc.2020.12.067. [DOI] [PubMed] [Google Scholar]
- 9.Benarroch E.E. Brain glucose transporters: implications for neurologic disease. Neurology. 2014;82(15):1374–1379. doi: 10.1212/WNL.0000000000000328. [DOI] [PubMed] [Google Scholar]
- 10.Seidner G., Alvarez M.G., Yeh J.I., et al. GLUT-1 deficiency syndrome caused by haploinsufficiency of the blood-brain barrier hexose carrier. Nat Genet. 1998;18(2):188–191. doi: 10.1038/ng0298-188. [DOI] [PubMed] [Google Scholar]
- 11.Leen W.G., Klepper J., Verbeek M.M., et al. Glucose transporter-1 deficiency syndrome: the expanding clinical and genetic spectrum of a treatable disorder. Brain. 2010;133(Pt 3):655–670. doi: 10.1093/brain/awp336. [DOI] [PubMed] [Google Scholar]
- 12.Ribaï P., Nguyen K., Hahn-Barma V., et al. Psychiatric and cognitive difficulties as indicators of juvenile huntington disease onset in 29 patients. Arch Neurol. 2007;64(6):813–819. doi: 10.1001/archneur.64.6.813. [DOI] [PubMed] [Google Scholar]
- 13.Nopoulos P.C., Aylward E.H., Ross C.A., et al. Smaller intracranial volume in prodromal Huntington’s disease: evidence for abnormal neurodevelopment. Brain. 2011;134(1):137–142. doi: 10.1093/brain/awq280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quarrell O.W.J., Nance M.A., Nopoulos P., et al. Defining pediatric huntington disease: time to abandon the term Juvenile Huntington disease? Mov Disord. 2019;34(4):584–585. doi: 10.1002/mds.27640. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y., Long J.D., Mills J.A., et al. Indexing disease progression at study entry with individuals at-risk for Huntington disease. Am J Med Genet B Neuropsychiatr Genet. 2011;156(7):751–763. doi: 10.1002/ajmg.b.31232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Domenico F., Tramutola A., Barone E., et al. Restoration of aberrant mTOR signaling by intranasal rapamycin reduces oxidative damage: focus on HNE-modified proteins in a mouse model of down syndrome. Redox Biol. 2019;23 doi: 10.1016/j.redox.2019.101162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klepinin A., Ounpuu L., Mado K., et al. The complexity of mitochondrial outer membrane permeability and VDAC regulation by associated proteins. J Bioenerg Biomembr. 2018;50(5):339–354. doi: 10.1007/s10863-018-9765-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tramutola A., Lanzillotta C., Di Domenico F., et al. Brain insulin resistance triggers early onset Alzheimer disease in down syndrome. Neurobiol Dis. 2020;137 doi: 10.1016/j.nbd.2020.104772. [DOI] [PubMed] [Google Scholar]
- 19.Ghosh C., Myers R., O’Connor C., et al. Cortical dysplasia in rats provokes neurovascular alterations, GLUT1 dysfunction, and metabolic disturbances that are sustained post-seizure induction. Mol Neurobiol. 2022;59(4):2389–2406. doi: 10.1007/s12035-021-02624-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barnat M., Capizzi M., Aparicio E., et al. Huntington’s disease alters human neurodevelopment. Science. 2020;369(6505):787–793. doi: 10.1126/science.aax3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hickman R.A., Faust P.L., Rosenblum M.K., Marder K., Mehler M.F., Vonsattel J.P. Developmental malformations in Huntington disease: neuropathologic evidence of focal neuronal migration defects in a subset of adult brains. Acta Neuropathol. 2021;141(3):399–413. doi: 10.1007/s00401-021-02269-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Vivo D.C., Trifiletti R.R., Jacobson R.I., Ronen G.M., Behmand R.A., Harik S.I. Defective glucose transport across the blood-brain barrier as a cause of persistent hypoglycorrhachia, seizures, and developmental delay. N Engl J Med. 1991;325(10):703–709. doi: 10.1056/NEJM199109053251006. [DOI] [PubMed] [Google Scholar]
- 23.Gamberino W.C., Brennan W.A. Glucose transporter isoform expression in Huntington’s disease brain. J Neurochem. 1994;63(4):1392–1397. doi: 10.1046/j.1471-4159.1994.63041392.x. [DOI] [PubMed] [Google Scholar]
- 24.McClory H., Williams D., Sapp E., et al. Glucose transporter 3 is a rab11-dependent trafficking cargo and its transport to the cell surface is reduced in neurons of CAG140 Huntington’s disease mice. Acta Neuropathol Commun. 2014;2:179. doi: 10.1186/s40478-014-0178-7. BioMed Central Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solìs-Maldonado M., Mirò M.P., Acuña A.I., et al. Altered lactate metabolism in Huntington's disease is dependent on GLUT3 expression. CNS Neurosci Ther. 2018;24(4):343–352. doi: 10.1111/cns.12837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panov A.V., Gutekunst C.A., Leavitt B.R., et al. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat Neurosci. 2002;5(8):731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- 27.Han R., Liang J., Zhou B. Glucose metabolic dysfunction in neurodegenerative diseases—new mechanistic insights and the potential of hypoxia as a prospective therapy targeting metabolic reprogramming. Int J Mol Sci. 2021;22(11):5887. doi: 10.3390/ijms22115887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mergenthaler P., Kahl A., Kamitz A., et al. Mitochondrial hexokinase II (HKII) and phosphoprotein enriched in astrocytes (PEA15) form a molecular switch governing cellular fate depending on the metabolic state. Proc Natl Acad Sci U S A. 2012;109(5):1518–1523. doi: 10.1073/pnas.1108225109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simpson I.A., Dwyer D., Malide D., Moley K.H., Travis A., Vannucci S.J. The facilitative glucose transporter GLUT3: 20 years of distinction. Am J Physiol Metab. 2008;295(2):E242–E253. doi: 10.1152/ajpendo.90388.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhattacharyya A., Trotta C.R., Narasimhan J., et al. Small molecule splicing modifiers with systemic HTT-lowering activity. Nat Commun. 2021;12(1):1–12. doi: 10.1038/s41467-021-27157-z. 2021 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braz B.Y., Wennagel D., Ratié L., et al. Treating early postnatal circuit defect delays Huntington's disease onset and pathology in mice. Science. 2022;377(6613) doi: 10.1126/science.abq5011. [DOI] [PubMed] [Google Scholar]
- 32.Saft C., Zange J., Andrich J., et al. Mitochondrial impairment in patients and asymptomatic mutation carriers of Huntington's disease. Mov Disord. 2005;20(6):674–679. doi: 10.1002/mds.20373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.