Abstract
Background
Circulating osteoprogenitors (COP) are a population of cells in the peripheral circulation that possess functional and phenotypical characteristics of multipotent stromal cells (MSCs). This population has a solid potential to become an abundant, accessible, and replenishable source of MSCs with multiple potential clinical applications. However, a comprehensive functional characterization of COP cells is still required to test and fully develop their use in clinical settings.
Methods
This study characterized COP cells by comparing them to bone marrow-derived MSCs (BM-MSCs) and adipose-derived MSCs (ASCs) through detailed transcriptomic and proteomic analyses.
Results
We demonstrate that COP cells have a distinct gene and protein expression pattern with a significantly stronger immune footprint, likely owing to their hematopoietic lineage. In addition, regarding progenitor cell differentiation and proliferation pathways, COP cells have a similar expression pattern to BM-MSCs and ASCs.
Conclusion
COP cells are a unique but functionally similar population to BM-MSCs and ASCs, sharing their proliferation and differentiation capacity, thus presenting an accessible source of MSCs with strong potential for translational regenerative medicine strategies.
Keywords: circulating osteoprogenitor cell, circulating osteogenic precursor, multilineage stromal cells, adipose-derived stromal cells
Graphical Abstract
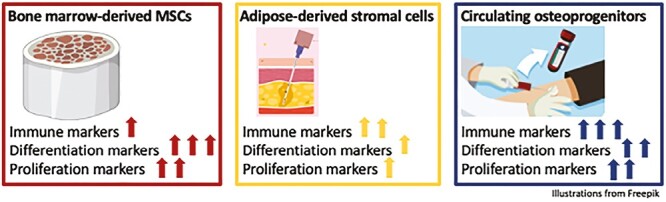
Significance Statement.
There is growing evidence that circulating osteoprogenitors (COP) cells can differentiate into several mesenchymal tissues, including bone, muscle, and cartilage. However, whether these cells belong to the multipotent stromal cells (MSCs) group – a finding that would open multiple therapeutic uses – remains unknown. In this study, we thoroughly characterized these cells by comparing their transcriptome and genomic characteristics to two of the most used MSCs in clinical practice: bone marrow-derived and adipocyte-derived MSCs. This paper represents a significant step in understanding the biology of COP cells, which could have substantial clinical implications in the future.
Introduction
The bone marrow stroma contains cells with a stem-cell-like character that allows them to differentiate into bone, cartilage, adipocytes, and hematopoietic supporting tissues.1 These bone marrow stromal cells have demonstrated their efficacy in bone tissue engineering and regenerative medicine-based applications, including age-related musculoskeletal diseases.2,3 However, their use in clinical settings has been limited by collection risk, expansion challenges, and high cost.4 Since the beginning of the 20th century, another population of cells with the capacity for connective tissue formation was identified.5 However, their specific differentiation capacity remained unknown until the end of the century when cells bearing markers of osteogenesis were isolated from the blood of patients with cancer undergoing stem cell mobilization treatment,6 and evidence of in vivo bone formation came shortly after.7 Since then, the concept of circulating mesenchymal progenitor cells has developed significantly.
Initially, the cells were assumed to be bone marrow multipotent stromal cells (MSCs) which had been stimulated to circulate in response to an osteogenic demand, a theory supported by their absence or extreme rarity in skeletally mature adults,8,9 as well as an increase in their number in response to bone fracture,10 and pubertal long bone growth.11 In all these cases, the cells adhered to the characteristics which were broadly used to identify MSCs in the bone marrow,that is, presence of the markers CD105, CD73, and CD90, as well as absence of the markers of hematopoietic lineage, CD45, CD14, CD34, CD11b, CD79α, or CD19,12 further supporting the theory of bone marrow origin.
More recently, another population of circulating cells with bone formation markers and a capacity for osteogenesis that appeared to stem from the hematopoietic lineage was soon identified. Cells bearing the markers CD34, CD45, and CD14 have since been shown to form mesodermal tissues in vitro and in vivo,13-15 casting doubt on the lineage of COP cells in humans. While traditionally, the mesenchymal and hematopoietic progenitor lineages have been considered separately, evidence has proposed a common ancestor for the two.16,17 Since then, a wide range of COP cell characterizations has been described in the literature, both with and without hematopoietic markers.15
Since their discovery, the concept of COP cells has developed significantly. They appear to represent a heterogeneous group of cells, with at least 2 major populations present, one which aligns with widely used definitions for the bone marrow MSC, and another which bears markers of hematopoietic lineage.18,19 The MSC-like population of cells appears only to be present in times of pathological or physiological bone formation, such as pubertal growth11 and fracture healing,10,20 as potential bone marrow MSCs that are induced to circulate in injury.21,22 The hematopoietic COP cells appear to be at a steady state in circulation in healthy individuals,23 but their level increases or decreases in conditions of bone formation24,25 or bone loss26 and is associated with bone mineral density and lean mass in adults.27
While significant advances have been made in the last 2 decades, there are still many outstanding questions regarding the identity of COP cells as mesenchymal progenitors, particularly in the case of the hematopoietic population. It is not yet known how similar they are to the more well-known bone marrow (BM)-derived MSCs (BM-MSCs) or other mesenchymal precursor populations such as adipose-derived stromal cells (ASCs).28,29 Their capacity for proliferation and differentiation15has a significant bearing on their potential and suitability for use in cellular and regenerative therapies, and there have been few studies showing clear comparisons between the other cells being investigated for these purposes. Therefore, this study aimed to compare differential genetic and protein expression in primary human hematopoietic COP cells (CD45+/CD34+) with equivalent BM-MSCs and ASCs. We hypothesized that COP cells have a similar genetic and proteomic profile to other well-known MSC populations but with more accessible collection methods in clinical settings and a strong potential for translational regenerative medicine strategies in the future.
Methods
Acquisition and Isolation of Primary Progenitor Cells
Circulating Osteoprogenitor Cells
COP cells were isolated from 16 healthy donors and characterized as previously described.15 Buffy coat samples were acquired from the Australian Red Cross blood service (ARCBS), as a waste component of therapeutic red blood cell products, with sex and date of birth provided. Donors were aged between 18 and 75, in good health, and were required to pass a medical history screening that excludes people with communicable, cardiovascular, and several other diseases.
The whole blood taken from donors by the ARCBS is centrifuged without density-gradient separation solutions on site, and the buffy coats provided for research were contaminated with red blood and platelets. To combat this, the buffy coats underwent Ficoll density-gradient centrifugation, to collect the purified PBMCs. The sample was diluted with a double volume of phosphate-buffered saline (PBS) supplemented with 5% fetal bovine serum (Bovogen Biologicals, Cat. No: FBSAU-2108E). Then the blood PBS mix was carefully layered on 12 mL of Ficoll Paque solution (GE Healthcare Companies, GE17-1440-02) in 50 mL centrifuge tubes, ensuring clean separation between the sample and the Ficoll. This was centrifuged at 400g for 40 minutes with the centrifuge brakes off, and the buffy coat was removed carefully from the interface of the Ficoll solution. The buffy coat was diluted 10:1 with PBS and centrifuged at 100g for 10 minutes, 3 times to remove contaminant platelets and aspirated Ficoll solution.
The acquired peripheral blood mononuclear cells (PBMCs) underwent fluorescence-activated cell sorting (FACS) to isolate COP cells. Briefly, cells were incubated with fragment crystallizable domain receptor (FCR) blocking reagent (Miltenyi Biotec, Cat. No: 130-059-901) before being incubated with fluorochrome-conjugated antibodies CD45-Fluorescein isothiocyanate (BD Biosciences, Cat. No: 555482), CD34-Allophycocyanin (BD Biosciences, Cat. No: 340441) and ALP-brilliant violet 421 Allophycocyanin (BD Biosciences, Cat. No: 752998) at a concentration of 1:100 v/v at 4°C in the dark. The cells were washed 3 times in PBS, resuspended in polypropylene FACS tubes, and passed through a 70 µm cell strainer to ensure cell aggregates did not interrupt the sorting process. Immediately before sorting, 5 µl of 7-AAD viability dye (BD-Biosciences, Cat. No: 559925) was added to each tube.
FACS was performed on a 4 laser (405 nm, 488 nm, 561 nm, and 633 nm) BD FACSAria III cytometer, and cells were collected into sterile 1.5 mL tubes with 1 mL of recovery media (low glucose Dulbecco’s modified eagle medium, with 15% FBS, 1% penicillin/streptomycin and 2.5 mM l-glutamine). To expedite the sorting process to minimize the stress placed on the collected cells, a 70-micron nozzle was used, and the highest flow speed possible was used, decided on by the cytometer conflict rate. First, forward and side scatter were used to select the leukocyte fraction of the PBMCs, then a process of doublet discrimination was used to remove aggregate cells. The fluorescence gating was used to select COP cells, first 7-AAD negative (live) cells were selected, with CD45+, CD34+, and then ALP+ cells then isolated (Supplementary Fig. 1). The ultimate proportion of COP cells in the PBMCs was 0.4%-1%, in line with previous studies. Fluorescence minus one, single color, and unstained controls were used to set up gates at the beginning of each isolation.
Once isolated, the COP cells were counted and plated at a density of 1.25 × 105 cells per cm2 in sterile tissue culture flasks precoated with 1 µg of fibronectin per cm2 in the media described earlier. We have previously shown that initial plating on fibronectin is required to successfully expand COP cells.15 Following the first passage, fibronectin is not needed. Once the COP cells reached 70%-80% confluence, they were detached by incubation with sterile triple cell detachment solution (Gibco, Cat. No: 12604013) for 8 minutes at room temperature, washed via centrifugation at 200g for 5 minutes, counted and replated on standard cell culture flasks at a density of 0.5 × 105.
Bone Marrow-Derived Multipotent Stromal Cells
In brief, bone marrow (BM) was aspirated from vertebral bodies (5-8 mL) of 16 spinal orthopedic surgery patients as per the Institutional Review Board (IRB) of the Medical University of South Carolina. Bone marrow was collected in EDTA-coated vacutainer tubes and then filtered through a 100 μm filter to remove bone fragments and cell clumps. The BM underwent Ficoll density gradient separation as described above within 1 hour of surgical collection, and the mononuclear cell layer was collected from the gradient. The cells were incubated with immunomagnetic beads attached to a CD271 antibody (Miltenyi Biotec Inc., 133-099-023) for 15 minutes at room temperature before being passed through a magnetic column, positively selecting the MSCs as previously described and characterized.30 These were frozen at −80°C and transported on dry ice under monitored temperature control to the Melbourne lab for analyses. These were cultured in low-glucose DMEM (as described for COP cell culture above), beginning at the same seeding density, through 2 passages, for 7 days until adequate numbers of cells were collected for both proteomics and transcriptomics.
Adipose-Derived Stromal Cells
Sixteen primary ASC samples were commercially acquired for research use (LACell, New Orleans, LA) from donors undergoing liposuction as described and characterized previously.31 These were transported on dry ice under monitored temperature control to the Melbourne lab for expansion and analysis. When received, the cells underwent 2 passages over 7 days in the conditions described for COP cell culture, beginning at the same plating density above until adequate cells were collected for analysis through trypsinization.
Proteomics
Passage 2 COP cells, BM-MSCs, and ASCs were harvested via incubation with trypsin and washed 3 times in serum-free PBS to remove contaminant protein from cell culture media additives. Then cells were lysed in 9M urea, 50 mM Tris-HCl with 100 units/mL of nuclease at pH 8 at room temperature with regular pipette mixing for 30 minutes, then centrifuged at 20 000g for 15 minutes to remove debris with the supernatants transferred to low-protein retention tubes for mass spectrometry. Protein was reduced in 1 mM dithiothreitol and alkylated in 5.5 mM iodoacetamide. The urea concentration was decreased to 1.6 M with 50 mM ammonium bicarbonate. The samples were digested with Lys-C at a 1:50 protease: protein ratio for 3 hours at room temperature followed by trypsin digestion overnight at 37°C at a 1:50 protease:protein ratio. The digestion was acidified to 1% formic acid. The resulting peptides were desalted using C18 Stage Tips. Tips were conditioned with 80% acetonitrile (ACN), 5% formic acid (FA), equilibrated and loaded in 95% water, 5% FA. The peptides were desalted by washing with 95% water, 5% formic acid 5 times, and eluted with 80% ACN, 5% FA. The peptides were dried in a SpeedVac and stored at −80°C.
Liquid Chromatography and Mass Spectrometry Data Acquisition Parameters
Peptides were separated and analyzed on an EASY nLC 1200 System (ThermoScientific) in-line with the Orbitrap Fusion Lumos Tribrid Mass Spectrometer (Thermo Fisher Scientific) with instrument control software v.4.2.28.14. Two µg of tryptic peptides were pressure loaded onto C18 reversed-phase column (Acclaim PepMap RSLC, 75 µm × 50 cm (C18, 2 µm, 100 Å) Thermo Fisher Scientific cat. #164536) using a gradient of 5% to 40% B in 180 minutes (Solvent A: 5% acetonitrile/0.1% formic acid; Solvent B: 80% acetonitrile/0.1% formic acid) at a flow rate of 300 nL/minute.
Mass spectra were acquired in data-dependent mode with a high resolution (60 000) FTMS survey scan, mass range of m/z 375-1500, followed by tandem mass spectra (MS/MS) of the most intense precursors with a cycle time of 3 seconds. The automatic gain control target value was 4.0e5 for the survey MS scan. HCD fragmentation was performed with a precursor isolation window of 1.6 m/z, a maximum injection time of 50 ms, and HCD collision energy of 35%. Monoisotopic-precursor selection was set to “peptide” with no Apex detection. Precursors within 10 ppm mass tolerance were dynamically excluded from resequencing for 15 seconds. Advanced peak determination was not enabled. Precursor ions with undetermined charge states, 1, or >5 were excluded.
For protein identification and quantification, the LC-MS/MS data were searched using the MaxQuant (MQ) v.1.6.3.3 platform, and the protein intensities were normalized using the label-free quantification (LFQ) algorithm.32-34 Data were searched against a human SwissProt reviewed database with 42 312 proteins (March 2021) and a database of common contaminants. The false discovery rate (FDR), determined using a reversed database strategy, was set at 1% at the protein and peptide level. Fully tryptic peptides with a minimum of 7 residues were required, including cleavage between lysine and proline. Two missed cleavages were permitted. The “Label-Free Quantitation” (LFQ) feature was used with “Match between runs” enabled for those features that had spectra in at least one of the runs. The “stabilize large ratios” feature was enabled, and “fast LFQ” was disabled. A 4.5 ppm mass tolerance was used. A minimum ratio count of 2 was required for quantification with at least one unique peptide. Parameters included static modification of cysteine with carbamidomethyl and variable protein N-terminal acetylation and oxidation of methionine.
The protein groups text files from the MQ search results were processed in Perseus.35 Common contaminants, reverse matches, and proteins identified only by a modified peptide were removed and the LFQ protein intensities were log2 transformed. The histograms illustrated the protein intensities were normalized and had similar distributions. Quantitative values were filtered to keep proteins that were quantified in 70% of measurements. A 2-sample Welch’s t-test was performed for each group (COP, MSC, and ADASC), as well as comparing for age (O/Y) and gender (M/F). The threshold for change was P < .05. Label-free proteomics data underwent analysis at the Monash University bioinformatics platform, with the data uploaded onto the LFQ-analyst software for processing.36 Identification of significantly regulated proteins was performed via student’s t-test with a permutation-based FDR cutoff of 0.05 and fold change threshold of S0 = 0.1.42 The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identified PXD035803.
RNA Sequencing
Passage 2 COP cells, BM-MSCs, and ASCs were trypsinized and washed twice in serum-free PBS before undergoing RNA and miRNA extraction with the miRNeasy Minikit (Qiagen, USA), with additional on-column DNase treatment, according to the manufacturer’s instructions. RNA was collected from the same donor cells as used for proteomics. Briefly, the cell pellets collected by centrifugation are incubated in 700 µl of Qiazol lysis reagent (Qiagen) for 5 minutes at room temperature before being passed through a QIAShredder lysis column. Lysates were then mixed with 140 µl of chloroform (Sigma-Aldrich, USA), mixed by vigorous shaking for 15 seconds, and incubated at room temperature for 3 minutes before being centrifuged at 12 000g for 15 minutes at 4OC. The upper aqueous phase generated by centrifugation is collected and mixed with 1.5 volumes of 100% ethanol and added to an RNA spin column (Qiagen). This was centrifuged for 15 seconds at 10 000g and the liquid that flowed through the column discarded. The column membrane is then washed with the supplied RWT buffer with 15 seconds centrifugation at 10 000g, before being incubated with a DNase solution for 15 minutes at room temperature and rewashed with RWT buffer. The membrane was then further washed with the supplied RPE buffer twice, with two 10 000g centrifugations, once for 15 seconds, then once for 2 minutes. The membrane was then dried with maximum speed centrifugation for 1 minute, and the captured RNA was eluted in 30 µL of nuclease-free water by centrifugation for 1 minute at 10 000g. RNA quality was assessed via Qubit fluorometer (Invitrogen, USA), and Agilent TapeStation electrophoresis. At least 2 µg of total RNA underwent library preparation and sequencing. The RNA underwent secondary quality control by an AATI fragment analyzer before sequencing to ensure no degradation of the samples.
Libraries were prepared using MGIEasy and mRNA library preparation chemistry systems. Sequencing was performed on an MGITech MGISEQ2000RS system, with V2 MGIEasy chemistry, on 3 sequencing lanes with at least 400 million raw reads per lane. Read sequences were output as FASTQ, which were mapped to a human genome index file downloaded from the University of California, Santa Cruz (UCSC) genome browser in March 2021 with the package ‘Rsubread, on R version 4.0.5 using the RStudio software (version 1.4.1106). Mapped read sequences underwent quality control, with Phred quality scores greater than 30 (1 in 1000 chance of incorrect base call) deemed adequate.
Quantification and Statistical Analysis
Transcriptomics Preprocessing
Before quality control, transcriptome was filtered for lowly expressed genes using the edgeR package. Library size and distribution of data were investigated and normalized by library sizes. MDS plots were generated to determine the greatest variation source in the data. Normalization for composition bias was performed, and differential expression analyses were obtained by contrast using limma-voom.37
Proteomics Preprocessing
Before normalization, proteomic data were filtered for high-confidence protein observations. In addition, contaminants, proteins only identified by a single peptide, and proteins not identified/quantified consistently across the experiment have been removed. Remaining missing values were imputed using MNAR method, assuming the missingness was due to low expression for such proteins, which are then normalized using the VSN method. Both imputations and VSN were conducted by the DEP package.
Statistical Analyses for the Transcriptome and Proteome
We first profiled transcriptome and proteome patterns according to cell type. Preprocesses transcriptome and proteome data (described earlier) were used in the analyses. We used contrast analysis and moderated Bayesian statistics as implemented in limma. Expression (logFC) of transcripts and proteins were contrasted between cell types accounted for covariates (age and sex). Results with an FDR < 0.05 were deemed significant.
Integration of Transcriptome and Proteome Profile
We have applied 2 distinct methods for integration analyses, namely, horizontal multiple integrations (DIABLO-mixOmics) described in detail here,38,39 and multicontrast pathway enrichment analyses (mitch) described in detail here.40 The transcriptomic results have been uploaded to the Sequence Read Archive at the National Center for Biotechnology Information (NCBI) and can be found with the identifier PRJNA987312 (https://www.ncbi.nlm.nih.gov/sra/PRJNA987312).
Secondary Statistical Analyses
Group means were compared through one-way ANOVA for significance, and individual group differences were identified through Tukey post hoc testing. An alpha value of significance was set at 0.95, with P-values less than .05 considered statistically significant. In both gene and protein analyses, fold regulation cutoffs of 2-fold were used to assess differential expression. All statistical tests were performed using the R software (version 4.0.5) using the RStudio user interface (version 1.4.1106) and visualized using the ggplot2 package.
Results
Transcriptome of COP Cells Is Distinct From That of BM-MSCs and ASCs With a Stronger Immune Profile
A total of 17 459 unique mRNAs and 6104 unique proteins were expressed across the dataset, with overwhelming concordance between transcription and translation, with only 63 proteins expressed without an associated mRNA (Fig. 1A). The 3 populations clustered strongly on multidimensional scaling analysis, with multidimensional scaling (MDS) vector 1 accounting for 71% of the separation between the populations, and dimension 2 accounting for 8%. BM-MSCs and ASCs clustered more closely, with COP cells being further removed from the other cells (Fig. 1B). The 3 cell types had a high level of similarity in expressed genes (Fig. 1C), with COP cells coexpressing 74.8% of the same genes as ASCs and BM-MSCs, and ASCs and BM-MSCs having 90.4% similarity.
Figure 1.
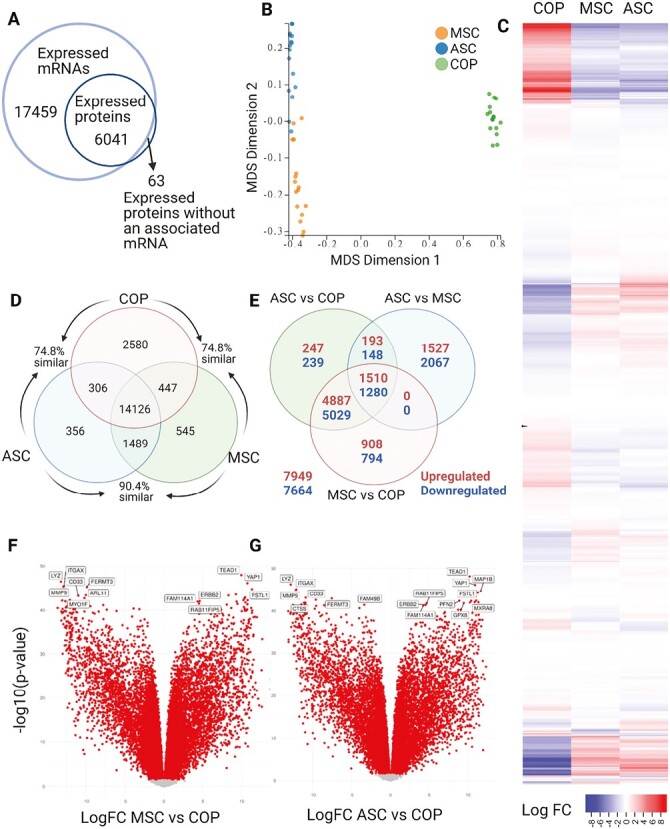
Comparative transcriptomics. (A) Concordance between mRNA and protein. (B) Multidimensional scaling analyses comparing the 3 cell types. (C) Heatmap of differential expression of mRNA across full transcriptome. (D) Venn diagram describing unique mRNA expression. (E) Venn diagram showing results of contrast analysis. (F) Volcano plot of mRNA expression of ASCs vs. COP cells. (G) Volcano plot of mRNA expression of ASCs vs. COP cells.
The 3 cell types coexpressed 14 126 genes, with COP cells expressing the most unique genes with 2580. The BM-MSCs expressed 545 unique genes and ASCs 356. COP cells coexpressed 306 genes with ASCs, and 447 with BM-MSCs, while the ASCs and BM-MSCs shared expression of 1489 that were not expressed by COP cells (Fig. 1D). The contrast analysis identified a large number of differentially expressed genes (DEGs, Fig. 1E) (adj. P-value < .05), with 7669 DEGs between adipose and marrow, 14 477 DEGs between adipose and COP, and 14 408 DEGs between marrow and COP (Fig. 1E). Among the topmost variable genes (ie, those with the largest effect sizes), several were associated with immune system activity, including LYZ (lysozyme), CYBB (Cytochrome B-245 Beta Chain), MMP9 (Matrix Metallopeptidase 9), HLA-DRA (Major Histocompatibility Complex, Class II, DR Alpha), ITGAX (Integrin Subunit Alpha X), and CD33 (CD33 Molecule; Fig. 1F and 1G).
Proteome of COP Cells Shows a Stronger Immune Profile Although Closely Aligned to BM-MSCs and ASCs
The 3 cell types remained clearly clustered on principle component analysis (PCA); however, there was more separation between the populations, with all 3 spreading evenly along component 1 (Fig. 2A), which accounted for 79.1% of the variation, but with COP cells further separated on component 2, accounting for 12% of the variation (Fig. 2B).
Figure 2.
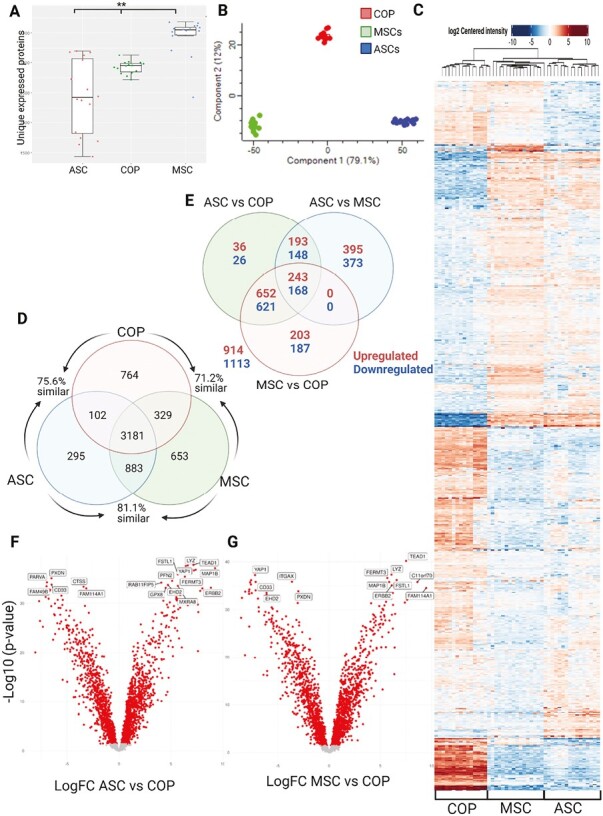
Comparative proteomics. (A) Unique expressed proteins between the cell types. **P = .01. (B) PCA plot clustering by cell type. (C) Heatmap of differential expression of proteins across full proteome. (D) Venn diagram describing unique protein expression. (E) Venn diagram showing results of contrast analysis. (F) Volcano plot of protein expression of ASCs vs. COP cells. (G) Volcano plot of protein expression of ASCs vs. COP cells.
As in the transcriptome, there was significant similarity in protein expression (Fig. 2C), with COP cells showing 75.6% similarity to ASCs and 71.2% similarity to MSCs, while ASCs and MSCs were more similar at 81.1% (Fig. 2D). Again, there was a large amount of differential regulation across the expressed proteins, with 2027 individual proteins differentially expressed in the sample (Fig. 2E). Contrast analyses revealed 1520 proteins DE between ASCs and BM-MSCs, 2087 proteins differentially expressed between adipose and COP cells, and 2074 proteins DE between BM-MSCs and COP cells (Fig. 2F and G). Among the proteins with the greatest differential expression were TAGLN (Transgelin), COL1A1 (Collagen Type I Alpha 1 Chain), FUCA1 (Alpha-L-Fucosidase 1), CYBB (Cytochrome B-245 Beta Chain), IFI30 (IFI30 Lysosomal Thiol Reductase), HLA-DRA (Major Histocompatibility Complex, Class II, DR Alpha).
Transcriptome and Proteome of the 3 Cell Types Are Strongly Correlated
An intersection analysis of the differentially expressed transcripts and proteins revealed that an overwhelming number of proteins overlap with transcripts, with only 310 proteins out of 2387 being divergent (Fig. 3A). The top proteins and transcripts are strongly correlated, and the correlation direction depends on cell type (Fig. 3B and Supplementary Fig. 2). A multi-omics signature selected from component 1 is shown in Fig. 4A. The importance (ie, largest contribution to the coefficients) of each protein/transcript is represented by the length of the bar (ie, its loading coefficient value). The combination of the coefficient sign (positive/negative) and the colors indicate that component 1 discriminates cell types highlighting which cell has the largest expression in each selected marker. Finally, a clustered image map of the top-selected variables for both proteome and transcriptome is shown in Fig. 4B, highlighting the inverse association between COP and Adipose/Marrow, according to cell type.
Figure 3.
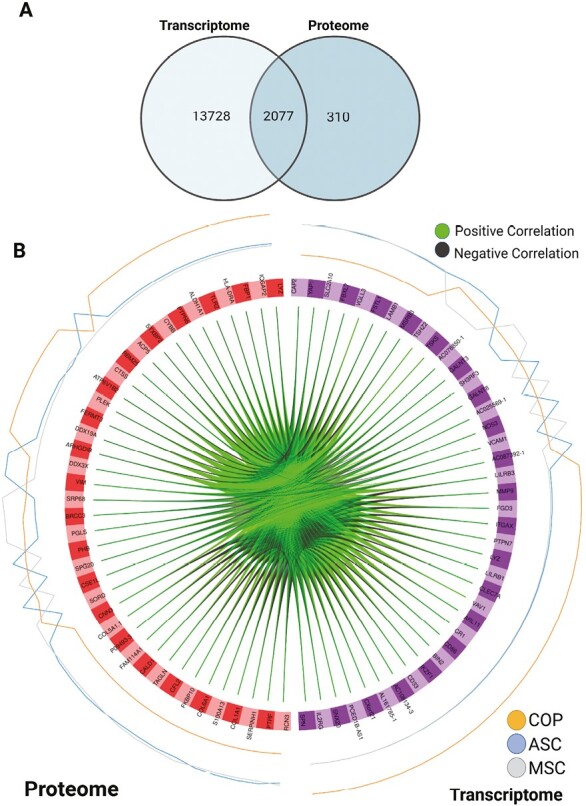
Interaction results. (A) Venn diagram of results from the intersection analysis. (B) Circus plot from multiblock sPLS-DA. The plot represents the top 30 genes with a correlation greater than 0.7 between variables (cells) of different types, represented on the side quadrants. The internal connecting lines show the positive/negative correlations. The outer lines show the expression levels of each cell type in each group. This plot enables to visualize the cross-correlations between data types (proteome vs. transcriptome), and the nature of these correlations (positive/negative). Each node represents a selected variable with colors indicating their type (green = proteins, purple = transcripts). The color of the edges represents positive or negative correlations.
Figure 4.
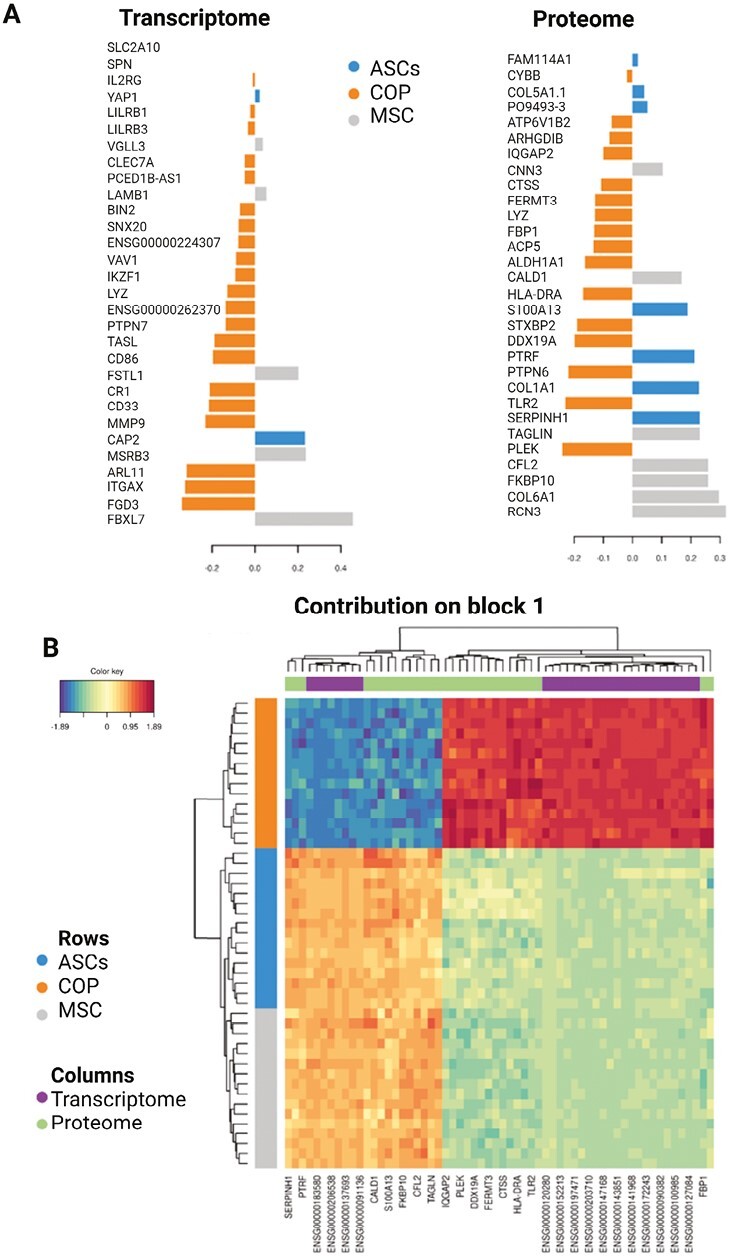
Continuation of interaction results. (A) Loading plot for the variables selected by multiblock sPLS-DA performed on the cells on component 1. This plot shows the multi-omics signature on component 1, separated by data type (Proteome/Transcriptome). The most important variables (according to the absolute value of their coefficients) are ordered from bottom to top. Colors indicate the class for which the median expression value is the highest for cell type. (B) Clustered Image Map for the variables selected by multiblock sPLS-DA performed on the cells on component 1. The CIM represents samples in rows (indicated by their cell subtype on the left side of the plot) and selected features in columns (indicated by their data type at the top of the plot). According to the CIM, component 1 accurately classifies COP cells from the ASCs and MSCs with a group of overexpressed transcripts and proteins.
Pathway Analysis Shows That COP Cells Have an Enrichment of Immune-Associated Functions
Pathway analysis of the transcriptome showed that COP cells had significantly greater expression of genes associated with immune cell physiology, with both adaptive and innate immunity represented. The “immunoregulatory interactions between a lymphoid and non-lymphoid cell,” and the “ROS and RNS production in phagocytes” pathways had the largest increase in expression over the other cells; however, several other immune pathways were also represented (Fig. 5A). The most underrepresented pathways in the COP cell transcriptome compared with the other cells were the “collagen biosynthesis and modifying enzymes” and “intraflagellar transport” pathways (Fig. 5A). The proteome pathway analysis revealed a similar overrepresentation of immune pathways in COP cells, with the “immune system” “innate immune system,” and “neutrophil degranulation” pathways overexpressed in COP cells (Fig. 5B); however, the magnitude of these differences was smaller than in the transcriptome. The BM-MSCs and ASCs had greater expression of proteins associated with control of RNA transcription and translation; however again, the magnitude of these changes was smaller between the 3 cell types (Fig. 5B).
Figure 5.
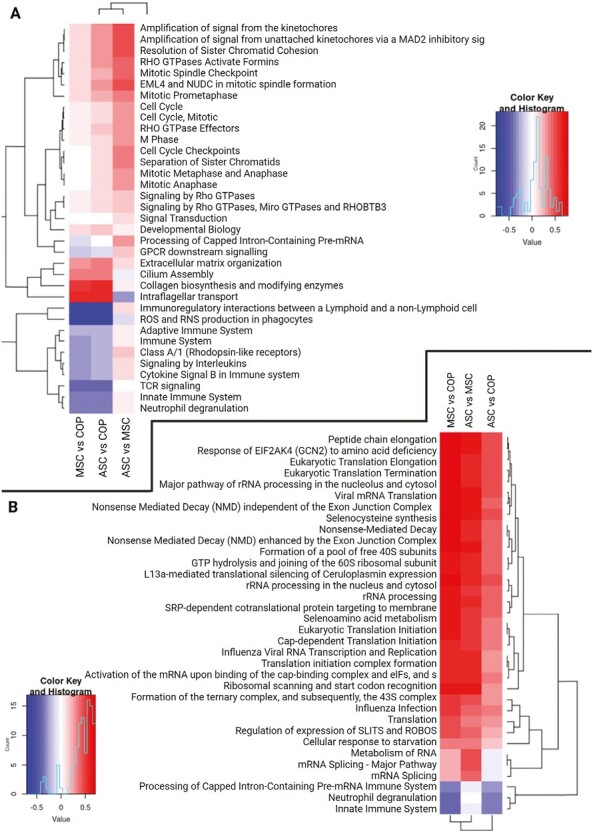
Pathway analyses for transcriptome and proteome results. Red represents enrichment of pathway, and blue represents a decrease in representation of the pathway. (A) Transcriptome pathway enrichment. (B) Proteome pathway enrichment.
Circulating Osteoprogenitor Cells Have a Similar Profile of Differentiation Genes to BM-MSCs and ASCs
To enable comparisons between the differentiation capacity of the cells, genes from the stem cell differentiation (GO:0048863) gene ontology were used to filter the dataset. Of the filtered genes, the 3 cell types expressed 34, with 20 of those differentially expressed (>1 logFC) between them (Fig. 6A-6C). Of these, only 9 were ultimately expressed by the cells at the protein level (Fig. 6D), with BM-MSCs broadly having the strongest expression, followed by COP, with ASCs having the lowest expression, except beta-catenin 1, which was not expressed by COP cells (Fig. 6E).
Figure 6.
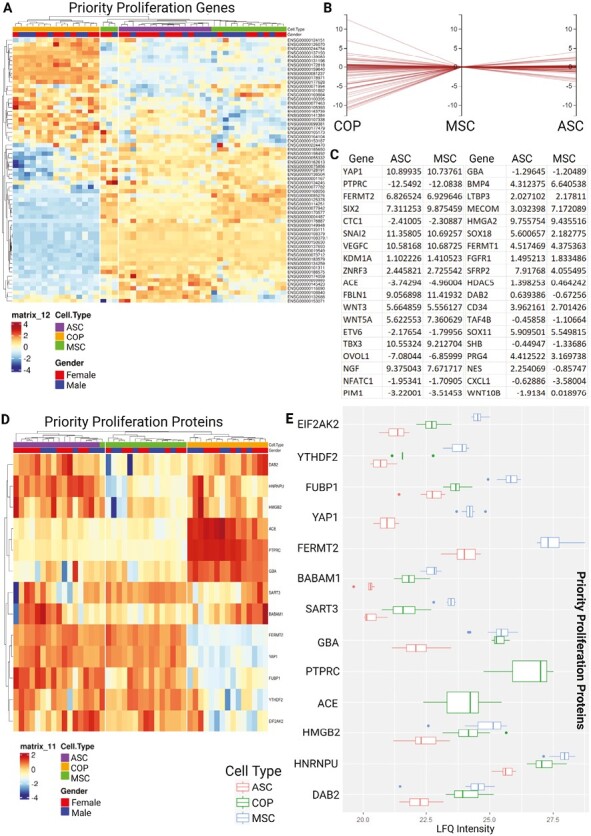
Expression of priority differentiation genes and proteins, from the stem cell differentiation gene ontology. (A) Heat map showing differential regulation of the 34 expressed genes from the stem cell differentiation gene ontology. (B) Horizontal coordinates chart showing flog fold change between cell types with MSC expression as the reference. (C) Table showing the regulation of all proteins with greater than log1-fold differentiation relative to COP cells for ASCs and MSCs (20/34). (D) Heat map of differential protein expression across priority proteins by cell type. (E) Relative expression graph comparing protein LFQ expression between cell types.
Circulating Osteoprogenitor Cells Have a Similar Profile of Stem Cell Proliferation Genes to BM-MSCs and ASCs
A similar process was undertaken to evaluate the proliferation capacity of the cells, with genes from the stem cell proliferation (GO:0072089) gene ontology used to filter the dataset. Of these, the 3 cells expressed 62 in total, with 38 of those differentially expressed (>1 logFC) between the cell types (Fig. 7A-7C). At the protein level, only 13 of these RNAs were ultimately expressed by the cells (Fig. 7D), with a similar pattern of expression to that seen in the transcriptome, with BM-MSCs broadly having the strongest expression, followed by COP, and ASCs having the lowest expression. The exception to this pattern of expression was in the angiotensin-converting enzyme (ACE) and protein tyrosine phosphatase receptor type C (PTPRC) proteins, which were only expressed by COP cells, and fermitin family homolog 2 (FERMT2), which was not expressed by COP cells (Fig. 7E).
Figure 7.
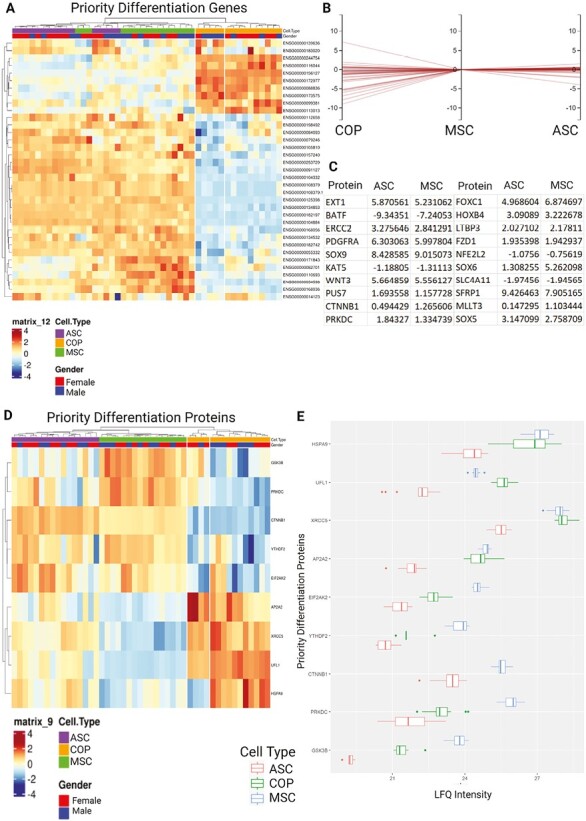
Expression of priority proliferation genes and proteins, from the stem cell proliferation gene ontology. The cells expressed 62 of the 96 genes in the ontology. (A) Heat map showing differential regulation of the 62 expressed genes from the stem cell proliferation gene ontology. (B) Horizontal coordinates chart showing log fold change between cell types with MSC expression as the reference. (C) Table showing the regulation of all proteins with greater than log1-fold differentiation relative to COP cells for ASCs and MSCs (38/62). (D) Heat map of differential protein expression across priority proteins by cell type. (E) Relative expression graphs comparing protein LFQ expression between cell types.
Discussion
In this study, we performed a comprehensive characterization of the COP cells showing that they have a distinct profile of gene and protein expression but have key similarities in physiologically relevant domains with other better-known MSCs populations. Overall, COP cells appear to be both immune and progenitor, with mixed physiology likely contributing to both.
The key differentiating factor between the three cell types is the strong immune physiology of COP cells. COP cells express a significant number of genes and proteins associated with innate immune system function, phagocytosis, and adaptive immune system priming, such as HLA-DRA and CYBB, which were overexpressed in both the proteome and transcriptome of COP cells. Given the origins of COP cells in a subpopulation of the PBMCs, and their identification through markers associated with the lymphocyte lineage, this is an unsurprising finding. The overexpression of HLA-DRA and CYBB strongly suggests macrophage physiology, implying similar functionality in vivo. HLA-DRA is a strong indicator of functionality as an antigen-presenting cell similar to macrophages and dendritic cells, and CYBB is associated with the respiratory burst physiology of phagocytes. Given their circulating nature, much of this physiology centers on cellular homing to sites of repair or injury, which necessitates sensitivity to cytokinetic stimuli, inflammation, and local extracellular matrix adherence, which are unlikely to be required by BM-MSCs and ASCs, which are largely tissue embedded in adults, only differentiating and migrating locally.
This is supported by the strong overexpression of integrin subunit alpha X (ITGAX), as well as strong expression of integrin subunit beta 2, which together form the leukocyte-associated marker αXβ2, which mediates phagocytosis, as well as endothelial adhesion and migration in monocytes and other leukocytes. These physiologies are reflected in the pathway and integration analyses, where COP cells overexpress genes and proteins in various immune-associated pathways, particularly around phagocyte activity and immune signaling. However, it is essential to note that while COP cells had the strongest immune footprint, both BM-MSCs and ASCs also have strong immune physiology acting as immunomodulatory and immunosuppressive cells.41 Indeed, most of their physiological effects in a therapeutic setting are centered on their immune function rather than their stem/progenitor roles, as they stimulate growth and differentiation of cells locally, rather than proliferating and generating new tissue themselves, as well as having a strong immunomodulatory effect.41 This has led to their licensing for use in steroid-refractory graft-versus-host disease, in which they limit immune recognition of foreign tissues, evidence of their strong immunomodulatory physiology.42 Interestingly, the ASC is commonly considered an even more powerful modulator of the immune system due to a greater level of cytokine secretion.43 Given that COP cells possess a significantly stronger immune phenotype than either BM-MSCs or ASCs, it is possible that they may exceed both in their immunosuppressive effects; however, this remains to be determined empirically.
The immune phenotype of COP cells also raises questions about their lineage. They have been shown to originate from the bone marrow,20,22 which supported the MSCs as their parent cell. However, increasing evidence suggests that they more likely descend from the hematopoietic stem cell (HSC). This is reflected in their expression of hematopoietic markers, their strong immune physiology, and their ability to circulate freely. Interestingly, while often considered as separate, cells with an immune phenotype have frequently been implicated in mesenchymal progenitor physiology. A subset of CD34+ cells in the bone marrow have been shown to strongly differentiate into osteoblasts and form bone,44 and macrophage-MSC crosstalk is a critical component of tissue repair, with each regulating the other to generate effective healing.45 Macrophage-mediated recruitment of MSCs is a crucial step in the intramembranous bone formation required for fracture healing, and signaling from the MSCs polarizes the macrophages to an anti-inflammatory, anabolic M2 phenotype, allowing for ongoing restoration of local tissues.46 Given that COP cells are a subset of the monocyte population, it could be that their role in fracture healing is to trigger MSC migration locally, after which they contribute to tissue deposition as osteoblast-like cells following differentiation.
While the bulk of the differences between the three cell types was explained by variation in markers of immune physiology, there were substantial similarities between the cells. Analyzing the genes and proteins in the MSCs differentiation and proliferation physiologies reveals a similar expression profile in functionally related genes. Broadly, there was a pattern of BM-MSCs having the greatest expression of these markers, followed by COP cells then finally ASCs, potentially suggesting a matching strength of function. This could make COP cells a good candidate for ongoing investigation into therapeutics, as they are more readily harvested than BM-MSCs, but with greater potential differentiation and proliferation capacity than ASCs. In addition to the markers of mesenchymal differentiation and proliferation shared by all 3 cells, COP cells also strongly express the PTPRC and ACE proteins, which are associated with the proliferation of hematopoietic cells, potentially further reflecting an HSC origin. COP cells also lacked expression of the FERMT2 protein, unlike MSCs and ASCs. FERMT2 (or kindlin-2) has been associated with MSC fate determination and proliferation.47 In addition, the similarity in expression in these priority genes and proteins implies a similar progenitor cell physiology between the three cell types.
While this study provides substantial evidence regarding the physiology of COP cells, several outstanding questions remain. Our results suggest that COP cells could represent a high-potential target for cellular therapies and regenerative medicine; however, future studies should seek to evaluate their immune activity and secretome. Given their phenotypic similarity to macrophages, their immunoreactive function could range from lytic, inflammatory M1 polarized cells, to immunosuppressive, and regenerative M2 macrophages. While both BM-MSCs and ASCs have an immunosuppressive effect, this must be identified in COP cells for effective translation.
While the experiments underpinning this work are methodologically robust, there are some caveats to their interpretation. First, while effort was made to analyze cells in a state as close to in situ as possible, they still required a small number of passages in culture to obtain sufficient numbers for such large analyses. It is likely at this time that there were changes to their expression of genes and proteins in culture, making the data gained more challenging to apply to a physiologically natural state. With rare or small cell populations such as COP cells and BM-MSCs in particular, or those requiring plastic adherence as a step in their isolation, obtaining naïve cells in numbers great enough for robust analysis is challenging. However, these models of cultured cells have been used extensively in basic discovery with success, and the data gained should provide relevant insights for future work. In addition, the cells were obtained at 3 different collection sites in the US and Australia, providing potential batch effects. We minimized this as much as possible by ensuring all samples were run simultaneously by the same technicians at the same facilities. We further aimed to combat this with a large number of samples, and consistent processing within the progenitor cell subgroups, with the cells in each of the 3 groups, as collecting the three cell types from single donors was not practicable or possible.
The large scale of the analyses contained here also increases the risk of false-positive findings due to statistical noise. While we used a large number of different samples and robust correction for multiple comparisons in the bioinformatic processing of the data, there is always a chance of some results being due to chance. However, the strong correlation between the proteome and transcriptome seen in the study also provides increased support for the findings herein.
Conclusion
Our comprehensive characterization of COP cells provides solid evidence of the potential of these cells to become an abundant, accessible, and replenishable source of MSCs with multiple potential clinical applications for the repair and regeneration of acute and chronically damaged tissues.
Supplementary Material
Acknowledgements
Under Director Lauren E. Ball, Ph.D. the Proteomic Analysis was performed at the Mass Spectrometry Facility, a University Shared Research Resource at the Medical University of South Carolina, using instrumentation acquired through the NIH shared instrumentation grant program (S10 OD010731-Orbitrap Elite Mass Spectrometer or Orbitrap Fusion Lumos ETD/UVD MS (S10 OD025126).The authors acknowledge the support of Monash Bioinformatics Platform for this work.
Contributor Information
Jack Feehan, Department of Medicine - Western Health, The University of Melbourne, Melbourne, Victoria (VIC), Australia; Australian Institute for Musculoskeletal Science (AIMSS), Western Health, Victoria University and University of Melbourne, Melbourne, Victoria (VIC), Australia; Institute for Health and Sport, Victoria University, Melbourne, Victoria (VIC), Australia.
Macsue Jacques, Institute for Health and Sport, Victoria University, Melbourne, Victoria (VIC), Australia.
Dmitry Kondrikov, Department of Pathology and Laboratory Medicine, The Medical University of South Carolina, Charleston, SC, USA.
Nir Eynon, Institute for Health and Sport, Victoria University, Melbourne, Victoria (VIC), Australia; Australian Regenerative Medicine Institute, Monash University, Clayton, Victoria, Australia.
Tissa Wijeratne, Australian Institute for Musculoskeletal Science (AIMSS), Western Health, Victoria University and University of Melbourne, Melbourne, Victoria (VIC), Australia; Institute for Health and Sport, Victoria University, Melbourne, Victoria (VIC), Australia.
Vasso Apostolopoulos, Australian Institute for Musculoskeletal Science (AIMSS), Western Health, Victoria University and University of Melbourne, Melbourne, Victoria (VIC), Australia; Institute for Health and Sport, Victoria University, Melbourne, Victoria (VIC), Australia.
Jeffrey M Gimble, Center for Stem Cell Research and Regenerative Medicine, Tulane University School of Medicine, New Orleans, LA, USA.
William D Hill, Department of Pathology and Laboratory Medicine, The Medical University of South Carolina, Charleston, SC, USA; Department of Veterans Affairs, Ralph H Johnson VA Medical Center, Charleston, SC, USA.
Gustavo Duque, Department of Medicine - Western Health, The University of Melbourne, Melbourne, Victoria (VIC), Australia; Australian Institute for Musculoskeletal Science (AIMSS), Western Health, Victoria University and University of Melbourne, Melbourne, Victoria (VIC), Australia; Institute for Health and Sport, Victoria University, Melbourne, Victoria (VIC), Australia; Bone, Muscle and Geroscience Research Group, Research Institute of the McGill University Health Centre, Montreal, Quebec, Canada; Dr. Joseph Kaufmann Chair in Geriatric Medicine, Department of Medicine, McGill University, Montreal, Quebec, Canada.
Funding
This research was supported through an Australian Government Research Training Program Scholarship and Stipend to J.F., and seed grants from the Australian Institute for Musculoskeletal Science (AIMSS) to J.F. and G.D. This work was supported by the National Institutes of Health’s (NIH) National Institute on Aging (NIA) R01 AG067510-01A1(WDH), P01 AG036675 (WDH), the Medical University of South Carolina (W.D.H.) and supported in part by the Department of Veterans Affairs (Merit Award 1I01CX000930-01, W.D.H.). The contents of this publication do not represent the views of the Department of Veterans Affairs or the United States Government.
Conflict of Interest
Tissa Wijeratne is the Director and Chair of the Department of Neurology, Western Health. Jeffrey M. Gimble declared cofounder, co-owner, and employee of Obatala Sciences and Talaria Antibodies; inventor on patents filed and stock holder in Obatala Sciences; consultant/scientific adviser for Mitosense; honoraria in 2023 from the American Orthopedic Society of Sports Medicine. William D. Hill declared employment with Medical University of South Carolina; research funding and travel reimbursement from NIH. The other authors declared no potential conflict of interests.
Author Contributions
Conceptualization: J.F., W.H., G.D. Methodology: J.F., D.K., G.D. Investigation: J.F., M.J., D.K. Resources: T.W., J.G., W.H.; Writing - Original Draft: J.F. Writing - Review and Editing: M.J., D.K., N.E., T.W., V.A., J.G., W.H., G.D. Visualization: J.F., M.J. Supervision: V.A., W.H., G.D. Funding Acquisition: T.W., W.H., G.D.
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethical Approval and Consent to Participate
The research related to human use has been complied with all the relevant national regulations and institutional policies and in accordance with the tenets of the Helsinki Declaration. The use of bone marrow cells was approved by the Institutional Review Board (IRB) of Augusta University. Protocols for the collection of adipose-derived stem cells were reviewed and approved by the Western Institutional Review Board, Louisiana State University. The use of peripheral blood samples was approved by the Western Health Human Research Ethics Committee, which issued a waiver of ethics review as donors’ consent to research use at the time of donation.
References
- 1. Charbord P, Livne E, Gross G, et al. Human bone marrow mesenchymal stem cells: a systematic reappraisal via the genostem experience. Stem Cell Rev Rep. 2011;7(1):32-42. 10.1007/s12015-010-9125-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Polymeri A, Giannobile WV, Kaigler D.. Bone marrow stromal stem cells in tissue engineering and regenerative medicine. Horm Metab Res. 2016;48(11):700-713. 10.1055/s-0042-118458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mahindran E, Law JX, Ng MH, Nordin F.. Mesenchymal stem cell transplantation for the treatment of age-related musculoskeletal frailty. Int J Mol Sci. 2021;22(19):10542. 10.3390/ijms221910542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lavrentieva A, Hoffmann A, Lee-Thedieck C.. Limited potential or unfavorable manipulations? strategies toward efficient mesenchymal stem/stromal cell applications. Front Cell Dev Biol. 2020;8:316. 10.3389/fcell.2020.00316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maximow AA. Cultures of blood leucocytes. From lymphocyte and monocyte to connective tissue. Gustav Fischer; 1928. [Google Scholar]
- 6. Fernández M, Simon V, Herrera G, et al. Detection of stromal cells in peripheral blood progenitor cell collections from breast cancer patients. Bone Marrow Transplant. 1997;20(4):265-271. 10.1038/sj.bmt.1700890 [DOI] [PubMed] [Google Scholar]
- 7. Kuznetsov SA, Mankani MH, Gronthos S, et al. Circulating skeletal stem cells. J Cell Biol. 2001;153(5):1133-1140. 10.1083/jcb.153.5.1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kuznetsov SA, Mankani MH, Leet AI, et al. Circulating connective tissue precursors: extreme rarity in humans and chondrogenic potential in guinea pigs. Stem Cells. 2007;25(7):1830-1839. 10.1634/stemcells.2007-0140 [DOI] [PubMed] [Google Scholar]
- 9. Zvaifler NJ, Marinova-Mutafchieva L, Adams G, et al. Mesenchymal precursor cells in the blood of normal individuals. Arthritis Res. 2000;2(6):477-488. 10.1186/ar130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alm JJ, Koivu HM, Heino TJ, et al. Circulating plastic adherent mesenchymal stem cells in aged hip fracture patients. J Orthop Res. 2010;28(12):1634-1642. 10.1002/jor.21167 [DOI] [PubMed] [Google Scholar]
- 11. Eghbali-Fatourechi GZ, Lamsam J, Fraser D, et al. Circulating osteoblast-lineage cells in humans. N Engl J Med. 2005;352(19):1959-1966. 10.1056/NEJMoa044264 [DOI] [PubMed] [Google Scholar]
- 12. Dominici M, Le Blanc K, Mueller I, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8(4):315-317. 10.1080/14653240600855905 [DOI] [PubMed] [Google Scholar]
- 13. Kuwana M, Okazaki Y, Kodama H, et al. Human circulating CD14+ monocytes as a source of progenitors that exhibit mesenchymal cell differentiation. J Leukoc Biol. 2003;74(5):833-845. 10.1189/jlb.0403170 [DOI] [PubMed] [Google Scholar]
- 14. Dalle Carbonare L, Valenti MT, Zanatta M, Donatelli L, Lo Cascio V.. Circulating mesenchymal stem cells with abnormal osteogenic differentiation in patients with osteoporosis. Arthritis Rheum. 2009;60(11):3356-3365. 10.1002/art.24884 [DOI] [PubMed] [Google Scholar]
- 15. Feehan J, Nurgali K, Apostolopoulos V, Duque G.. Development and validation of a new method to isolate, expand, and differentiate circulating osteogenic precursor (COP) cells. Bone Rep. 2021;15:101109. 10.1016/j.bonr.2021.101109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mehrotra M, Williams CR, Ogawa M, LaRue AC.. Hematopoietic stem cells give rise to osteo-chondrogenic cells. Blood Cells Mol Dis. 2013;50(1):41-49. 10.1016/j.bcmd.2012.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ogawa M, LaRue AC, Mehrotra M.. Hematopoietic stem cells are pluripotent and not just “hematopoietic.”. Blood Cells Mol Dis. 2013;51(1):3-8. 10.1016/j.bcmd.2013.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Feehan J, Nurgali K, Apostolopoulos V, Al Saedi A, Duque G.. Circulating osteogenic precursor cells: building bone from blood. EBioMedicine. 2019;39:603-611. 10.1016/j.ebiom.2018.11.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Feehan J, Kassem M, Pignolo RJ, Duque G.. Bone from blood: characteristics and clinical implications of circulating osteogenic progenitor (COP) cells. J Bone Miner Res. 2021;36(1):12-23. 10.1002/jbmr.4204 [DOI] [PubMed] [Google Scholar]
- 20. Kumagai K, Vasanji A, Drazba JA, Butler RS, Muschler GF.. Circulating cells with osteogenic potential are physiologically mobilized into the fracture healing site in the parabiotic mice model. J Orthop Res. 2008;26(2):165-175. 10.1002/jor.20477 [DOI] [PubMed] [Google Scholar]
- 21. Otsuru S, Tamai K, Yamazaki T, Yoshikawa H, Kaneda Y.. Circulating bone marrow-derived osteoblast progenitor cells are recruited to the bone-forming site by the CXCR4/stromal cell-derived factor-1 pathway. Stem Cells. 2008;26(1):223-234. 10.1634/stemcells.2007-0515 [DOI] [PubMed] [Google Scholar]
- 22. Otsuru S, Tamai K, Yamazaki T, Yoshikawa H, Kaneda Y.. Bone marrow-derived osteoblast progenitor cells in circulating blood contribute to ectopic bone formation in mice. Biochem Biophys Res Commun. 2007;354(2):453-458. 10.1016/j.bbrc.2006.12.226 [DOI] [PubMed] [Google Scholar]
- 23. Gunawardene P, Al Saedi A, Singh L, et al. Age, gender, and percentage of circulating osteoprogenitor (COP) cells: The COP Study. Exp Gerontol. 2017;96:68-72. 10.1016/j.exger.2017.06.004 [DOI] [PubMed] [Google Scholar]
- 24. Egan KP, Duque G, Keenan MA, Pignolo RJ.. Circulating osteogenic precursor cells in non-hereditary heterotopic ossification. Bone. 2018;109:61-64. 10.1016/j.bone.2017.12.028 [DOI] [PubMed] [Google Scholar]
- 25. Suda RK, Billings PC, Egan KP, et al. Circulating osteogenic precursor cells in heterotopic bone formation. Stem Cells. 2009;27(9):2209-2219. 10.1002/stem.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pirro M, Leli C, Fabbriciani G, et al. Association between circulating osteoprogenitor cell numbers and bone mineral density in postmenopausal osteoporosis. Osteoporos Int. 2010;21(2):297-306. 10.1007/s00198-009-0968-0 [DOI] [PubMed] [Google Scholar]
- 27. Feehan J, Smith C, Tripodi N, et al. Higher levels of circulating osteoprogenitor cells are associated with higher bone mineral density and lean mass in older adults: a cross-sectional study. JBMR Plus. 2021;5(11):e10561. 10.1002/jbm4.10561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gimble JM, Katz AJ, Bunnell BA.. Adipose-derived stem cells for regenerative medicine. Circ Res. 2007;100(9):1249-1260. 10.1161/01.RES.0000265074.83288.09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bourin P, Bunnell BA, Casteilla L, et al. Stromal cells from the adipose tissue-derived stromal vascular fraction and culture expanded adipose tissue-derived stromal/stem cells: a joint statement of the International Federation for Adipose Therapeutics and Science (IFATS) and the International Society for Cellular Therapy (ISCT). Cytotherapy. 2013;15(6):641-648. 10.1016/j.jcyt.2013.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eisa NH, Sudharsan PT, Herrero SM, et al. Age-associated changes in microRNAs affect the differentiation potential of human mesenchymal stem cells: novel role of miR-29b-1-5p expression. Bone. 2021;153:116154. 10.1016/j.bone.2021.116154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yu G, Floyd ZE, Wu X, Halvorsen YD, Gimble JM.. Isolation of human adipose-derived stem cells from lipoaspirates. Methods Mol Biol. 2011;702:17-27. 10.1007/978-1-61737-960-4_2 [DOI] [PubMed] [Google Scholar]
- 32. Tyanova S, Temu T, Cox J.. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016;11(12):2301-2319. 10.1038/nprot.2016.136 [DOI] [PubMed] [Google Scholar]
- 33. Cox J, Hein MY, Luber CA, et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13(9):2513-2526. 10.1074/mcp.M113.031591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cox J, Mann M.. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367-1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- 35. Tyanova S, Temu T, Sinitcyn P, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13(9):731-740. 10.1038/nmeth.3901 [DOI] [PubMed] [Google Scholar]
- 36. Shah AD, Goode RJA, Huang C, Powell DR, Schittenhelm RB.. LFQ-Analyst: an easy-to-use interactive web platform to analyze and visualize label-free proteomics data preprocessed with MaxQuant. J Proteome Res. 2020;19(1):204-211. 10.1021/acs.jproteome.9b00496 [DOI] [PubMed] [Google Scholar]
- 37. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Singh A, Shannon CP, Gautier B, et al. DIABLO: an integrative approach for identifying key molecular drivers from multi-omics assays. Bioinformatics. 2019;35(17):3055-3062. 10.1093/bioinformatics/bty1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tenenhaus A, Philippe C, Guillemot V, et al. Variable selection for generalized canonical correlation analysis. Biostatistics. 2014;15(3):569-583. 10.1093/biostatistics/kxu001 [DOI] [PubMed] [Google Scholar]
- 40. Kaspi A, Ziemann M.. Multi-contrast pathway enrichment for multi-omics and single-cell profiling data. BMC Genomics. 2020;21(1):447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ankrum JA, Ong JF, Karp JM.. Mesenchymal stem cells: immune evasive, not immune privileged. Nat Biotechnol. 2014;32(3):252-260. 10.1038/nbt.2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu X, Jiang J, Gu Z, et al. Mesenchymal stromal cell therapies: immunomodulatory properties and clinical progress. Stem Cell Res Ther. 2020;11(1):345. 10.1186/s13287-020-01855-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ribeiro A, Laranjeira P, Mendes S, et al. Mesenchymal stem cells from umbilical cord matrix, adipose tissue and bone marrow exhibit different capability to suppress peripheral blood B, natural killer and T cells. Stem Cell Res Ther. 2013;4(5):125. 10.1186/scrt336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen JL, Hunt P, McElvain M, et al. Osteoblast precursor cells are found in CD34+ cells from human bone marrow. Stem Cells. 1997;15(5):368-377. 10.1002/stem.150368 [DOI] [PubMed] [Google Scholar]
- 45. Lu D, Xu Y, Liu Q, Zhang Q.. Mesenchymal stem cell-macrophage crosstalk and maintenance of inflammatory microenvironment homeostasis. Front Cell Dev Biol. 2021;9:681171. 10.3389/fcell.2021.681171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pajarinen J, Lin T, Gibon E, et al. Mesenchymal stem cell-macrophage crosstalk and bone healing. Biomaterials. 2019;196:80-89. 10.1016/j.biomaterials.2017.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen Z, Shen K, Zheng Z, et al. Kindlin-2 promotes chondrogenesis and ameliorates IL-1beta-induced inflammation in chondrocytes cocultured with BMSCs in the direct contact coculture system. Oxid Med Cell Longev. 2022;2022:3156245. 10.1155/2022/3156245 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


