Abstract
In healthcare, the development and deployment of insufficiently fair systems of artificial intelligence can undermine the delivery of equitable care. Assessments of AI models stratified across sub-populations have revealed inequalities in how patients are diagnosed, given treatments, and billed for healthcare costs. In this Perspective, we outline fairness in machine learning through the lens of healthcare, and discuss how algorithmic biases (in data acquisition, genetic variation and intra-observer labelling variability, in particular) arise in clinical workflows and the healthcare disparities that they can cause. We also review emerging technology for mitigating biases via disentanglement, federated learning and model explainability, and their role in the development of AI-based software as a medical device.
Introduction
In healthcare, the development and deployment of insufficiently fair systems of artificial intelligence (AI) can undermine the delivery of equitable care. Assessments of AI models stratified across subpopulations have revealed inequalities in how patients are diagnosed, treated and billed. In this Perspective, we outline fairness in machine learning through the lens of healthcare, and discuss how algorithmic biases (in data acquisition, genetic variation and intra-observer labelling variability, in particular) arise in clinical workflows and the resulting healthcare disparities. We also review emerging technology for mitigating biases via disentanglement, federated learning and model explainability, and their role in the development of AI-based software as a medical device.
With the proliferation of AI algorithms in healthcare, there are growing ethical concerns regarding the disparate impact that the models may have on under-represented communities1,2,3,4,5,6,7,8. Audit studies have shown that AI algorithms may discover spurious causal structures in the data that correlate with protected-identity status. These correlations imply that some AI algorithms may use protected-identity statuses as a shortcut to predict health outcomes3,9,10. For instance, in pathology images, the intensity of haematoxylin and eosin (H&E) stains can predict ethnicity on the Cancer Genome Atlas (TCGA), owing to hospital-specific image-acquisition protocols9. On radiology images, convolutional neural networks (CNNs) may underdiagnose and misdiagnose underserved groups (in particular, Hispanic patients and patients on Medicaid in the United States) at a disproportionate rate compared with White patients, and capture implicit information about patient race10,11,12,13. Despite the large disparities in performance, there is a lack of regulation on how to train and evaluate AI models on diverse and protected subgroups. With an increasing number of algorithms receiving approval from the United States Food and Drug Administration (FDA) as AI-based software as a medical device (AI-SaMD), AI is poised to penetrate routine clinical care over the next decade by replacing or assisting human interpretation for disease diagnosis and prognosis, and for the prediction of treatment responses. However, if left unchecked, algorithms may amplify existing healthcare inequities that have already impacted underserved subpopulations14,15,16.
In this Perspective, we discuss current challenges in the development of fair AI for medicine and healthcare, through diverse viewpoints spanning medicine, machine learning and their intersection in guiding public policy on the development and deployment of AI-SaMD. Discussions of AI-exacerbated healthcare disparities have primarily debated the usage of race-specific covariates in risk calculators and have overlooked broader and systemic inequities that are often implicitly encoded in the processes generating medical data. These inequalities are not easily mitigated by ignoring race5,17,18,19,20. And vice versa, conventional bias-mitigation strategies in AI may fail to translate to real-world clinical settings, because protected health information may include sensitive attributes, such as race and gender, and because data-generating processes across healthcare systems are heterogeneous and often capture different patient demographics, causing data mismatches in model development and deployment21.
We begin by providing a succinct overview of healthcare disparities, fair machine learning and fairness criteria. We then outline current inequities in healthcare systems and their varied data-generating processes (such as the absence of genetic diversity in biomedical datasets and differing image-acquisition standards across hospitals), and their connections to fundamental machine-learning problems (in particular, dataset shift, representation learning and robustness; Fig. 1). By understanding how inequities can drive disparities in the performance of AI algorithms, we highlight federated learning, representation learning and explainability as emerging research areas for the mitigation of bias and for improving the evaluation of fairness in the deployment lifecycle for AI-SaMD. We provide a glossary of terms in Box 1.
Fig. 1 |.
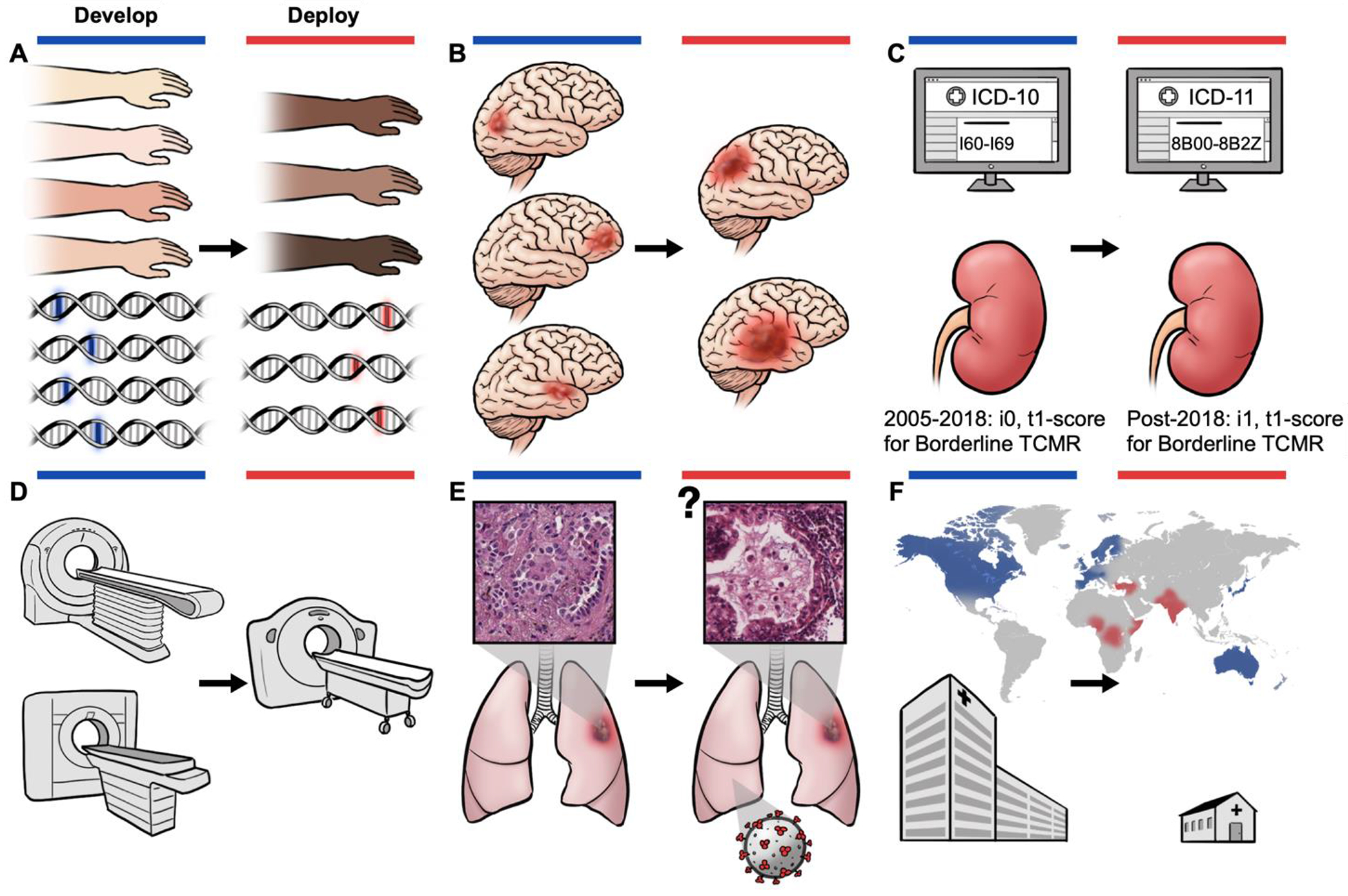
Connecting healthcare disparities and dataset shifts to algorithm fairness. A, Population shift as a result of genetic variation and of other population-specific phenotypes across subpopulations. Current AI algorithms for the diagnosis of skin cancer using dermoscopic and macroscopic photographs may be developed on datasets that underrepresent darker skin types, which may exacerbate health disparities of some geographic regions138,139,140,388. In developing algorithms using datasets that overrepresent individuals with European ancestry, the prevalence of certain mutations may also differ in the training and test distributions. This is the case for disparities in EGFR-mutation frequencies across European and Asian populations146. B, Population shifts and prevalence shifts resulting from disparities in social determinants of health. Differences in healthcare access may result in delayed referrals, and cause later-stage disease diagnoses and worsened mortality rates18,34. C, Concept shift as a result of the ongoing refinement of medical-classification systems, such as the recategorization of strokes, which was previously defined under diseases of the circulatory system in ICD-10 and is now defined under neurological disorders in ICD-11127,128. In other taxonomies, such as the Banff-classification system for renal allograft assessment, which updates the diagnostic criteria approximately every two years, the use of the post-2018 Banff criteria for borderline cases of T-cell-mediated rejection (TCMR), all i0,t1-score biopsies would be classified as ‘normal’189. D, Acquisition shift as a result of differing data-curation protocols (associated with the use of different MRI/CT scanners, radiation dosages, sample-preparation protocols or image-acquisition parameters) may induce batch effects in the data9,279. E, Novel or insufficiently understood occurrences, such as interactions between the SARS-CoV-2 virus and lung cancer, may arise in new types of dataset shift such as open set label shift389. F, Global-health challenges in the deployment of AI-SaMDs in low-and-middle-income countries can lead to resource constraints for AI-SaMDs, such as limitations in GPU resources, a lack of digitization of medical records and other health data, as well as dataset-shift barriers such as differing demographics, disease prevalence, classification systems, and data-curation protocols. Group fairness criteria may also be difficult to satisfy when AI-SaMD deployment faces constraints in the access of protected health information.
Box 1 |. Glossary of terms.
Health disparities: Group-level inequalities as a result of socioeconomic factors and social determinants of health, such as insurance status, education level, average income in ZIP code, language, age, gender, sexual identity or orientation, and BMI.
Race: An evolving human construct categorizing human populations. It differs from ancestry, ethnicity, nationality and other taxonomies. Race is usually self-reported.
Protected or sensitive attributes: Patient-level metadata which predictive algorithms should not discriminate against.
Protected subgroup: Patients belonging to the same category in a protected attribute.
Disparate treatment: Intentional discrimination against protected subgroups. Disparate treatment can result from machine-learning algorithms that include sensitive-attribute information as direct input, or that have confounding features that explain the protected attribute.
Disparate impact: Unintentional discrimination as a result of disproportionate impact on protected subgroups.
Algorithm fairness: A concept for defining, quantifying and mitigating unfairness from machine-learning predictions that may cause disproportionate harm to individuals or groups of individuals. Fairness is a formalization of the minimization of disparate treatment and impact. There are multiple criteria for the quantification of fairness, yet they typically involve the evaluation of differences in performance metrics (such as accuracy, true positive rate, false positive rate, or risk measures; larger differences would indicate larger disparate impacts) across protected subgroups, as defined in Box 2.
AI-SaMD (artificial-intelligence-based software as a medical device): a categorization of medical devices undergoing regulation by the United States Food and Drug Administration (FDA).
Model auditing: Post-hoc quantitative evaluation for the assessment of violations of fairness criteria. This is often coupled with explainability techniques for attributing specific features to algorithm fairness.
Dataset shift: A mismatch in the distribution of data in the source and target datasets (or of the training and testing datasets).
Domain adaptation: Correction of dataset shifts in the source and target datasets. Typically, domain-adaptation methods match the datasets’ input spaces (via importance weighting or related techniques) or feature spaces (via adversarial learning).
Federated learning: A form of privacy-preserving distributed learning that trains neural networks on local clients and sends updated weight parameters to a centralized server without sharing the data.
Fair representation learning: Learning intermediate feature representations that are invariant to protected attributes.
Disentanglement: A property of intermediate feature representations in deep neural networks, in which individual features control independent sources of variation in the data.
Fairness and machine learning
Understanding health disparities and inequities
Healthcare disparities can lead to differences in healthcare quality, access to healthcare and health outcomes across patient subgroups. These disparities are deeply shaped by both historical and current socioeconomic inequities22,23,24,25. Although they are often viewed at observable group-level characteristics—such as race, gender, age and ethnicity—the sources of these disparities encompass a wider range of observed and latent risk factors, including body mass index, education, insurance type, geography and genetics. As formalized by the United States Department of Health and Human Services, most of these factors are defined within the five domains of social determinants of health: economic stability, education access and quality, healthcare access and quality, neighbourhood and built environment, and social and community context. These factors are commonly attributed to disparate health outcomes and to mistrust in the healthcare system26,27,28,29,30,31,32. For instance, in the early 2000s, reports by the United States Surgeon General documented the disparities in tobacco use and in access to mental healthcare as experienced by different racial and ethnic groups33. In the epidemiology of maternal mortality in the United States, fatality rates for Black women are substantially higher than for White women, owing to economic instability, the lack of providers accepting public insurance and poor healthcare access (as exemplified by counties that do not offer obstetric care, also known as maternal care deserts)34,35,36.
Definition of fairness
The axiomatization of fairness is a collective societal problem that has existed beyond the evaluation of healthcare disparities. In legal history, fairness was a central problem in the development of non-discrimination laws (for example, titles VI and VII of the Civil Rights Act of 1964, which prohibit discrimination based on legally protected classes, such as race, colour, sex and national origin) in federal programs and employment46. In the Griggs versus Duke Power Company case in 1971, the Supreme Court of the United States prohibited the use of race (and other implicit variables) in hiring decisions, even if discrimination was not intended47. Naturally, fairness spans many human endeavours, such as diversity hiring in recruitment, the distribution of justice in governance, the development of moral machines in autonomous vehicles48,49,50, and more recently the revisiting of historical biases of existing algorithms in healthcare and the potential deployment of AI algorithms in AI-SaMD11,12,18,19,20,51,52,53,54. Frameworks for understanding and implementing fairness in AI have been largely aimed at learning neutral models that are invariant to protected class identities when predicting outcomes (disparate treatment), and that have non-discriminatory impact on protected subgroups with equalized outcomes (disparate impact)55,56,57.
Formally, for a sample with features X and with a target label Y, we define A as a protected attribute that denotes a sensitive characteristic about the population of the sample that a model P(Y|X) should be non-discriminatory against when predicting Y. To mitigate disparate treatment in algorithms, an intuitive (but naive) fairness strategy is ‘fairness through unawareness’; that is, knowledge of A is denied to the model.
Although removing race would seemingly debias the eGFR equation, for many applications denying protected-attribute information can be insufficient to satisfy guarantees of non-discrimination and of fairness. This is because there can be other input features that may be unknown confounders that correlate with membership to a protected group58,59,60,61,62. A canonical counterexample to ‘fairness through unawareness’ is the 1998 COMPAS algorithm, a risk tool that excluded race as a covariate in predicting criminal recidivism. The algorithm was contended to be fair in the mitigation of disparate treatment63. However, despite not using race as a covariate, a retrospective study found that COMPAS assigned medium-to-high risk scores to Black defendants twice as often than to White defendants (45% versus 23%, respectively)4. This example illustrates how differing notions of fairness can be in conflict with one another, and has since motivated the ongoing development of formal definitions of evaluation criteria for group fairness for use in supervised-learning algorithms56,64,65,66 (Box 2). For example, for the risk of re-offense, fairness via predictive parity was satisfied for White and Black defendants, whereas fairness via equalized odds was violated, owing to unequal FPRs. And, for the eGFR-based prediction of CKD, removing race can correct the overestimation of kidney function and lower the false-negative rate (FNR), yet may also lead to the underestimation of kidney function and to an increase in the FPR. Hence, depending on the context, fairness criteria can have different disparate impact. Different practical applications may thus be better served by different fairness criteria.
Box 2 |. Brief background on fairness criteria.
There are three commonly used fairness criteria for binary classification tasks, described below by adapting the notation used in refs. 56,57,64,65,66,391.
We note denote our data distribution for samples , labels , and protected subgroups . We note denote the model parameterized by that produces scores 1, and the threshold used to derive a classifier where .
For the problem of placing patients on the kidney transplant waitlist, we use represent patient covariates (e.g. - age, body size, Serum Creatinine, Serum Cystatin C), denotes self-reported race categories , be a model that produces probability risk score for needing a transplant, and be our classifier as a threshold policy that qualifies patients for the waitlist () if the risk score is greater than a therapeutic threshold . As a regression task, is equivalent to current equations for estimating glomeruli filtration rate (eGFR), with corresponding to clinical guidelines for recommending kidney transplantation if eGFR value is less than 40,41,42. Though the true prevalence for patients in subgroup that require kidney transplantation is unknown, in this simplified but illustrative example, we make assumptions that: 1) 43, and 2) all non-waitlisted patients that develop kidney failure are patients that would have needed a transplant (measurable outcome as false negative rate, FNR).
Demographic parity.
Demographic parity asserts that the fraction of positive predictions made by the model should be equal across protected subgroups, e.g. – the proportion of Black and White that qualify for the waitlist is equal65,391. Hence, for subgroups and , the predictions should satisfy the independence criterion via the constraint . The independence criterion reflects the notion that decisions should be made independently of the subgroup identity. However, demographic parity only equalizes the positive predictions and does not consider if the prevalence of transplant need across subgroups is the different. After removing disparate treatment, Black patients may still be more at-risk for kidney failure than White patients and thus should have greater proportion of patients qualifying for the waitlist. Enforcing demographic parity in this scenario would mean equalizing the positive predictions made by the model, resulting in equal treatment rates but disparate outcomes between Black and White patients.
Predictive parity.
Predictive parity asserts that the predictive positive values (PPVs) and predictive negative values (PNVs) should be equalized across subgroups65,391. Hence, the sufficiency criteria should be satisfied via the constraint for , which implies scores should have consistent meaning and correspond to observable risk across groups (also known as calibration by group). For example, under calibration, amongst patients with risk score , 20% of patients have need for transplantation to preclude kidney failure. When the risk distributions across subgroups differ, threshold policies may cause miscalibration. For example, suppose at risk score , and . This threshold policy would under-qualify certain Black patients (higher FNR, lower PNV) and over-qualify certain White patients according to their implied thresholds. Models are often naturally calibrated when trained with protected attributes392, however, this leads back to the ethical issues of introducing race in risk calculators.
Equalized odds.
Equalized odds asserts that the true positive rates (TPRs) and false positive rates (FPRs) should be equalized across protected subgroups56,65,391. Hence, the separability criteria should be satisfied via the respective TPR and FPR constraints and 2. Therefore, differently from demographic parity, equalized odds enforces similar error rates across all subgroups. However, as emphasized by Barocas, Hardt, & Narayanan391, “who bears the cost of misclassification”? In satisfying equalized odds with post-processing techniques, often group-specific thresholds need to be set for each population, which is not feasible if the ROC curves do not intersect. Moreover, an important limitation of fairness criteria is the impossibility to satisfy all criterion unless under certain scenarios. For example, when the prevalence differs from subgroups, equalized odds and predictive parity cannot be satisfied at the same time. This can be seen in the following expression:
which was previously used to highlight the impossibility of satisfying equalized odds and predictive parity in recividism prediction65. In equalizing FNR to satisfy separation for two subgroups, sufficiency is violated as from our preposition and thus PPV cannot also be equivalent. In other words, at the cost of equalizing underdiagnosis, we would over-qualify Black or White patients.
Techniques for the mitigation of fairness
To reduce violations of group fairness, bias-mitigation techniques can be used to adapt existing algorithms with pre-processing steps that blind, augment or reweight the input space to satisfy group fairness55,67,68,69,70,71,72; with in-processing steps that construct additional optimization constraints or with regularization-loss terms that penalize non-discrimination73,74,75,76,77,78; and with post-processing steps that apply corrections to calibrate model predictions across subgroups56,57,64,79,80.
Pre-processing
Algorithmic biases in healthcare often stem from historical inequities that create spurious associations linking protected class identity to disease outcome in the dataset, in particular when the underlying causal factors stem from factors that span social determinants of health. By training algorithms on health data that have internalized such biases, the distribution of outcomes across ethnicities may be skewed; for example, underserved Hispanic and Black patients have more delayed referrals, which may result in more high-grade and invasive phenotypes at the time of cancer diagnosis. Such sources of labelling prejudice are known as ‘negative legacy’ or as sample-selection bias73. To mitigate this form of bias, data pre-processing techniques such as importance weighting (Fig. 2) can be applied. Importance weighting reweights infrequent samples belonging to protected subgroups67,68,70,71,72. Similarly, resampling aims to correct for sample-selection bias by obtaining fairer subsamples of the original training dataset and can be intuitively applied to correct for the under-representation of subgroups81,82,83,84,85. For tabular-structured data, blinding, data transformation and other techniques can also be used to directly eliminate proxy variables that encode protected attributes. However, these techniques may be subject to high variance and sensitivity under dataset shift, may be sensitive to outliers and data paucity in subgroups, and may overlook joint relationships between multiple proxy variables62,86,87,88.
Fig. 2 |. Strategies for mitigating disparate impact.
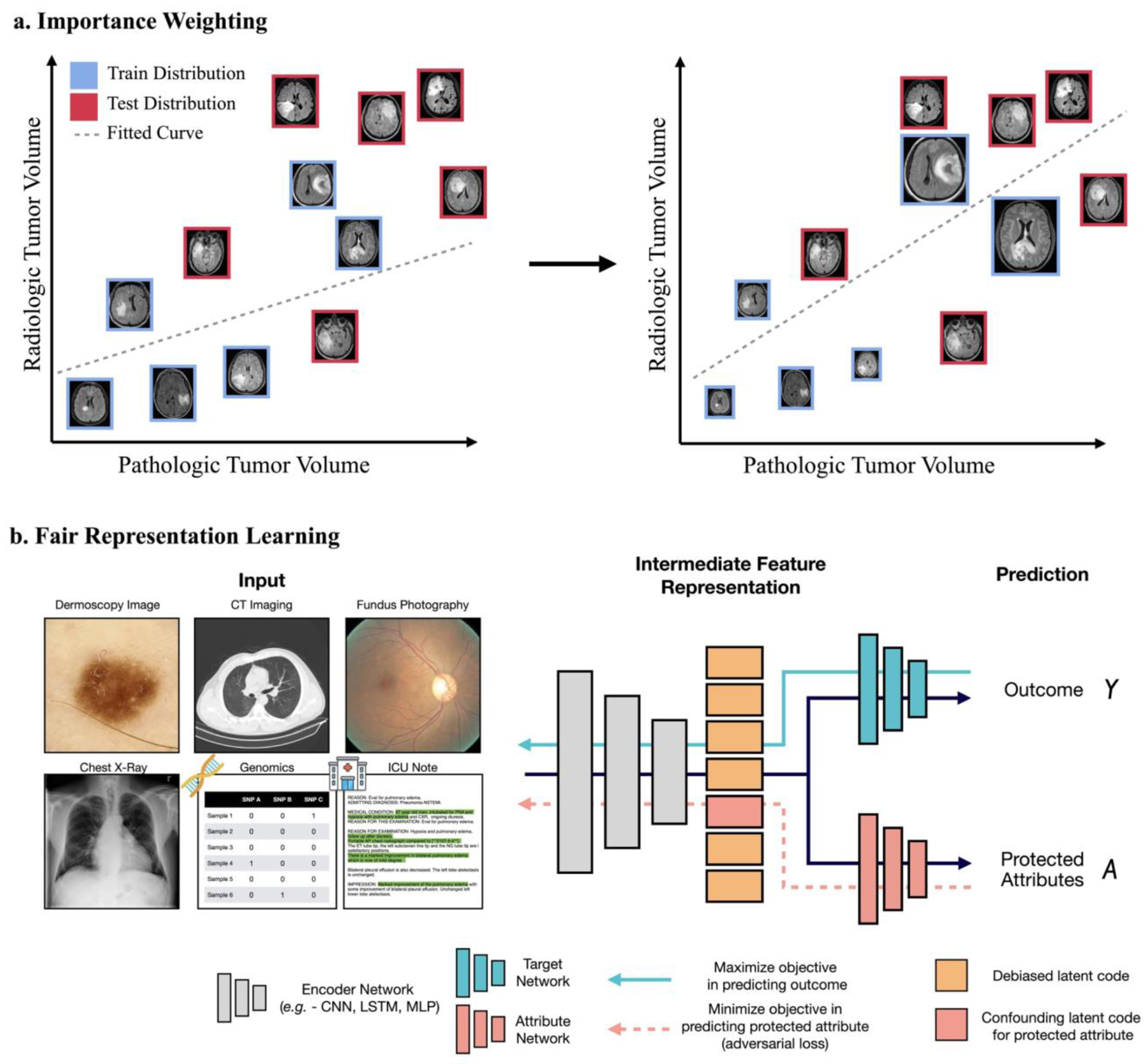
a, For under-represented samples in the training and test datasets, importance weighting can be applied to reweight the infrequent samples so that their distribution matches in the two datasets. The schematic shows that, before importance reweighting, a model that overfits to samples with a low tumour volume in the training distribution (blue) underfits a test distribution that has more cases with large tumour volumes. For the model to better fit the test distribution, importance reweighting can be used to increase the importance of the cases with large tumour volumes (denoted by larger image sizes). b, To remove protected attributes in the representation space of structured data (CT imaging data or text data such as intensive care unit (ICU) notes), deep-learning algorithms can be further supervised with the protected attribute used as a target label, so that the loss function for the prediction of the attribute is maximized. Such strategies are also referred to as ‘debiasing’. Clinical images can include subtle biases that may leak protected-attribute information, such as age, gender and self-reported race, as has been shown for fundus photography and chest radiography. Y and A denote, respectively, the model’s outcome and a protected attribute. LSTM, long short-term memory; MLP, multilayer perceptron.
In-processing
Biased data-curation protocols may induce correlations between protected attributes and other features, which may be implicitly captured during model development. For example, medical images (such as radiographs, pathology slides and fundus photographs) can leak protected attributes, which can become ‘shortcuts’ to model predictions9,12,89,90. To mitigate the effect of confounding variables, in-processing techniques adopt a non-discrimination term within the model to penalize learning discriminative features of a protected attribute73,74,75,91,92. For instance, a logistic-regression model can be modified to include non-discrimination terms by computing the covariance of the protected attribute with the signed distance of the sample’s feature vectors to the decision boundary, or by modifying the decision-boundary parameters to maximize fairness (by minimizing disparate impact or mistreatment), subject to accuracy constraints74. For deep-learning models, such as CNNs, adversarial-loss terms (inspired by the minimax objective of a generative adversarial network; GAN) can be used to make the internal feature representations invariant to protected subgroups93 (Fig. 2). Modifications to stochastic gradient descent can also be made to weigh fairness constraints in online learning frameworks94. A limitation of in-processing approaches is that the learning objective is made non-convex when including these additional non-discriminatory terms. Moreover, metrics such as the FPR and the FNR can be sensitive to the shape of risk distributions and to label prevalence across subgroups, which may result in reduced overall performance57,95,96.
Post-processing
Post-processing refers to techniques that modify the output of a trained model (such as probability scores or decision thresholds) to satisfy group-fairness metrics. To achieve equalized odds, one can simply pick thresholds for each group such that the model achieves the same operating point across all groups. However, when the receiver operating curves do not intersect, or when the desired operating point does not correspond to an intersection point, this approach requires systematically worsening the performance for select subgroups using a randomized decision rule56. This implies that the performance on select groups may need to be artificially reduced to satisfy equalized odds. Hence, the difficulty of the task could vary between different groups11, which may raise ethical concerns. For survival models and other rule-based systems that assess risk using discrete scores (for example, by defining high cardiovascular risk as a systolic blood pressure higher than 130 mmHg; ref. 97), probability thresholds for each risk category can be selected to satisfy predictive parity. In this case, the proportion of positive-labelled samples in each category is equalized across subgroups (known as calibration; Box 2). To calibrate continuous risk scores, such as predicted probabilities, a logit transform can be applied to the predicted probabilities and then a calibration curve can be fitted for each group. This ensures that the risk scores have the same meaning regardless of group membership96. However, as was found for the COMPAS algorithm, satisfying both predictive parity and equalized odds may be impossible.
Targeted data collection
In practice, increasing the size of the dataset mitigates biases98. Although audits of publicly available and commercial AI algorithms have revealed large performance disparities, collecting data for under-represented subgroups can be an effective stopgap81,99. However, targeted data collection may require surveillance, and hence pose ethical and privacy concerns. Also, there are practical limitations in the collection of protected health information, as well as stringent data-interoperability standards100.
Healthcare disparities arising from dataset shift
Many domain-specific challenges in healthcare preclude the adoption of bias-mitigation techniques for reducing harm from AI algorithms. In particular, benchmarking these techniques in real-world healthcare applications has shown that optimizing fairness parity can result in worse model performance or in suboptimal calibration across subgroups. This is often described as an accuracy–fairness trade-off95,96,101,102,103. Benchmarks such as MEDFAIR, which evaluated 11 fairness techniques across 10 diverse medical-imaging datasets, found that current state-of-the-art methods do not outperform ‘fairness through unawareness’ with statistical significance. Also, fairness techniques often make strong assumptions about the learning scenario, such as the training and test data being independently and identically distributed, an assumption which is often violated when using data from hospitals in different geographies or when employing different data-curation protocols86,104,105,106,107. Another assumption is the availability of clean and protected attributes at test time, which is a re-occurring challenge for the development of fairness methods when working in healthcare applications that limit access to protected health information108,109,110,111,112,113,114,115. Moreover, because genetic ancestry is causally associated with many genetic traits and diseases, there are many clinical problems for which including protected attributes such as ancestry (rather than self-reported race, which is a social construct shaped by historical inequities) may promote fairness.
Many healthcare disparities in medical AI can be understood as arising from fundamental machine-learning challenges, such as dataset shift. Dataset shift can arise from differences in population demographics, genetic ancestry, image-acquisition techniques, disease prevalence and social determinants of health among other factors, and can cause disparate performance at test time31,53,106,116,117,118,119,120,121. Specifically, dataset shift occurs when there is a mismatch between the distributions of the training and test datasets during algorithm development, (that is, and ), and may lead to disparate performance at the subgroup level86,107,119,122,123. Thus, in addition to the above types of bias mitigation strategy, methods for quantifying and mitigating dataset shift such as group distributionally robust optimization (GroupDRO) are commonly used across many fairness studies103,108,123,124,125.
Group unfairness via dataset shift (also known as subpopulation shift) is particularly central in ‘black box’ AI algorithms developed for structured data such as images and text. When developing or using such algorithms, practitioners are often unaware of domain-specific cues that would ‘leak’ subgroup identity present in the input data121. For instance, an AI algorithm trained on cancer-pathology data from the United States and deployed on data from Turkey can misdiagnose Turkish cancer patients, owing to domain shifts from variations in H&E staining protocols and to population shifts from an imbalanced ethnic-minority representation126. Likewise, hospitals operating with different International Classification of Disease (ICD) taxonomies can lead to concept shifts in how algorithms are evaluated127,128. Overall, algorithms sensitive to dataset shifts can exacerbate healthcare disparities and underperform on fairness metrics.
Challenges in the deployment of fair AI-SaMD
In this section, we examine several broad and systematic challenges in the deployment of fair AI in medicine and healthcare. We discuss common dataset shifts in the settings of genomics, medical imaging, electronic medical records (EMRs) and other clinical data (Figs. 1, 3 and 4). Specifically, we examine several types of dataset shift and the impact of this failure mode on healthcare disparities129. Supplementary Table 1 provides examples. For a formal introduction to the topic, we refer to refs. 116,129.
Fig. 3 |. Genetic drift as population shift.
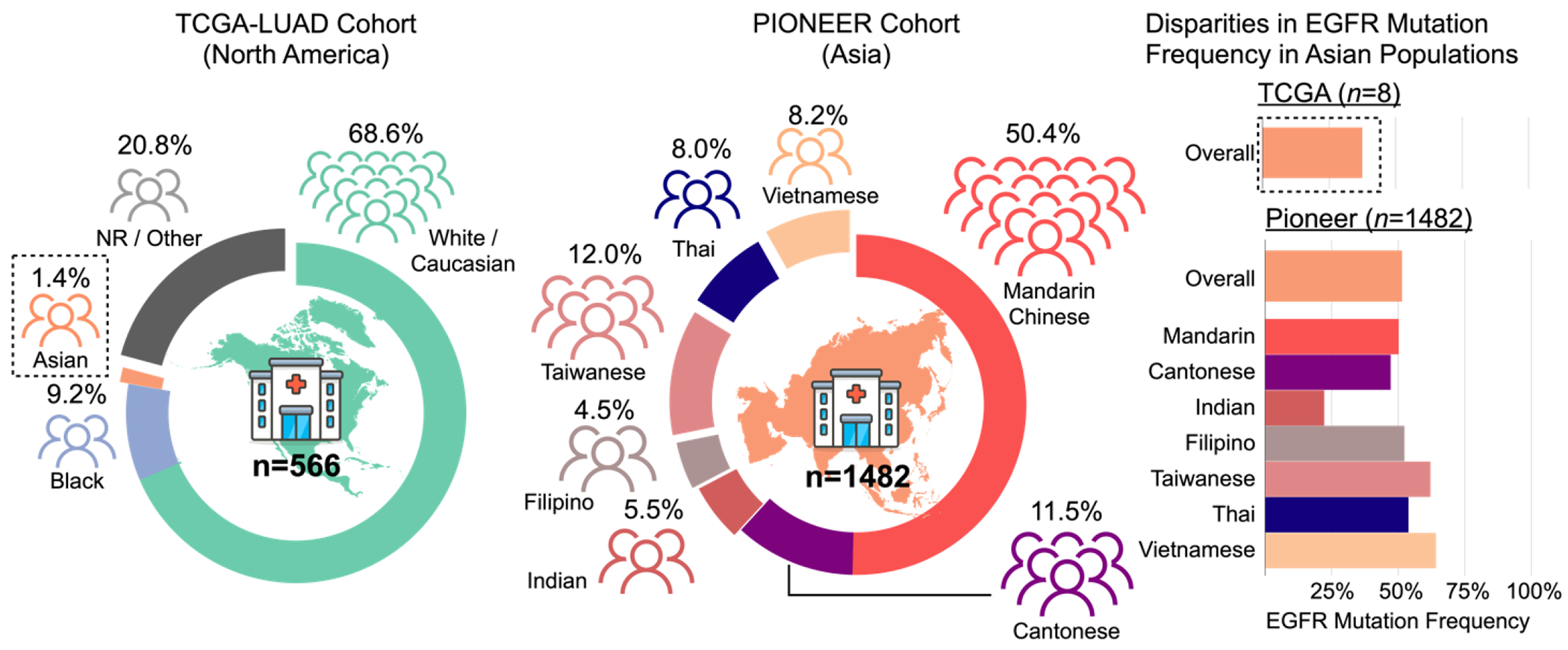
Demographic characteristics and gene-mutation frequencies for EGFR in patients with lung adenocarcinoma in the TCGA-LUAD and PIONEER cohorts. Of the 566 patients with lung adenocarcinoma in the TCGA, only 1.4% (n = 8) self-reported as ‘Asian’; in the PIONEER cohort, 1,482 patients did. The PIONEER study included a more fine-grained characterization of self-reported ethnicity and nationality: Mandarin Chinese, Cantonese, Taiwanese and Vietnamese, Thai, Filipino and Indian. Because of the underrepresentation of Asian patients in the TCGA, the mutation frequency for EGFR, which is commonly used in guiding the use of tyrosine kinase inhibitors as treatment, was only 37.5% (n = 3). For the PIONEER cohort, the overall EGFR-mutation frequency for all Asian patients was 51.4% (n = 653), and different ethnic subpopulations had different EGFR-mutation frequencies.
Fig. 4 |. Dataset shifts in the deployment of AI-SaMDs for a clinical-grade AI algorithms.
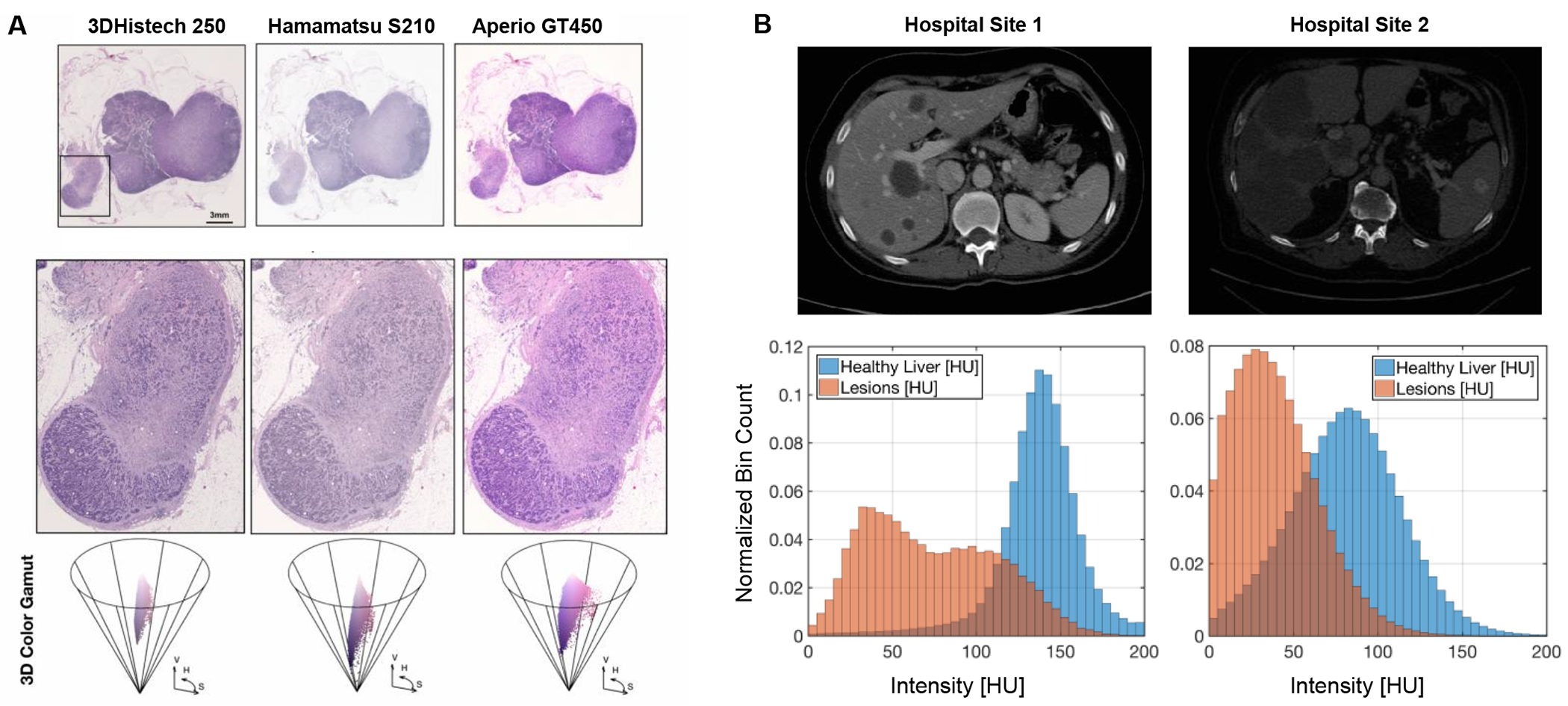
a, Examples of site-specific H\&E stain variability under different whole slide scanners, resulting in variable histologic tissue appearance. b, Example of variations in CT scans acquired at two different centers. The histograms shows the radiointensity in normal liver tissue and in the liver lesions. Due to differences in the acquisition protocols, there might be significant overlap between CT values of normal liver from one center and tumor values from another center.
Lack of representation in biomedical datasets
In the development and integration of AI-based computer-aided diagnostic systems in healthcare, the vast majority of models are trained on datasets that over-represent individuals of European ancestry, often without the consideration of algorithm fairness. For instance, in TCGA, across 8,594 tumour samples from 33 cancer types, 82.0% of all cases are from White patients, 10.1% are from Black or African American people, 7.5% are from Asians and 0.4% are from highly under-reported minorities (Hispanics, Native Americans, Native Hawaiians and other Pacific Islanders; denoted as ‘other’ in TCGA; Fig. 3)130. The CAMELYON16/17 challenge, which was used in validating the first ‘clinical-grade’ AI models for the detection of lymph-node metastases from diagnostic pathology images, was sourced entirely from the Netherlands131,132. In dermatology, amongst 14 publicly available skin-image datasets, a meta-analysis found that 11 of the datasets (78.6%) originated from North America, Europe and Oceania, and involved limited reporting of ethnicity (1.3%) and Fitzpatrick skin type (2.1%) as well as severe under-representation of darker-skinned patients133. Similarly, a study of 94 ophthalmological datasets found that 10 of them (10.6%) originated from Africa or the Middle East and that 2 of them (2.1%) were from South America, and that the datasets generally omitted age, sex and ethnicity (74%)134. Owing to such disparities, differences in performance across algorithms may in part result from a lack of publicly available independent cohorts that are large and diverse. Table 1 lists biomedical datasets that report sex and race demographics.
Table 1:
Reported demography data were obtained for all patient populations in the original dataset. Model auditing may use only certain subsets, owing to missing labels or to insufficient samples for evaluation in the case of extremely under-represented minorities, or owing to targeting different protected attributes (such as age, income and geography). Dashes denote demographic data that were not made publicly available or acquired.
| Dataset | Modalities | Number of patients | Female patients (%) | White (%) | Black (%) | Asian (%) | Hispanic or Latino (%) | Pacific Islander or Native Hawaiian (%) | American Indian or Alaskan Native (%) | Unknown or other (%) | Audit refs. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| TCGA 356 | Pathology, MRI/CT, genomics | 10,953 | 48.5 | 67.5 | 7.9 | 5.9 | 0.3 | 0.01 | 0.2 | – | 9 |
| UK Biobank 141 | Genomics | 503,317 | 54.4 | 94.6 | 1.6 | 2.3 | – | – | – | 1.5 | 142 |
| PIONEER 146 | Genomics | 1,482 | 43.4 | – | – | 100 | – | – | – | – | N/A |
| eMerge Network172,173 | Genomics | 20,247 | – | 77.7 | 16.1 | 0.1 | – | 0.2 | 0.2 | 4.5 | 229 |
| NHANES 357 | Lab measurements | 15,560 | 50.4 | 33.9 | 26.3 | 10.5 | 22.7 | – | – | 6.5 | 339,54 |
| Undisclosed EMR data 2 | EMRs, billing transactions | 49,618 | 62.9 | 87.7 | 12.3 | – | – | – | – | – | |
| OAI358,359 | Limb X-rays | 4,172 | 57.4 | 70.9 | 29.1 | – | – | – | – | – | 3,359 |
| SIIM-ISIC360,361,362 | Dermoscopy | 2,056 | 48 | – | – | – | – | – | – | – | 361,362 |
| NIH AREDS363,364 | Fundus photography | 4,203 | 56.7 | 97.7 | 1.4 | 8 | 2 | 1.2 | 1 | – | 163 |
| RadFusion 365 | EMRs, CT | 1,794 | 52.1 | 62.6 | – | – | – | – | – | 37.4 | 365 |
| CPTAC 366 | Pathology, proteomics | 2,347 | 39.5 | 36.5 | 3.2 | 10 | 2.3 | 0.1 | 0.4 | 49.1 | N/A |
| MIMIC367,368 | Chest X-rays, EMRs, waveforms | 43,005 | 44.1 | 68.2 | 9.2 | 2.9 | 4 | 0.2 | 0.2 | – | 10,95,125,336,337,124 |
| CheXpert 310 | Chest X-rays | 64,740 | 41 | 67 | 6 | 13 | – | – | – | 11.3 | 336,369 |
| NIH NLST369,370 | Chest X-rays, spiral CT | 53,456 | 41 | 90.8 | 4.4 | 2 | 1.7 | 0.4 | 0.4 | 2 | N/A |
| RSPECT 371 | CT | 270 | 53 | 90 | 10 | – | – | – | – | – | N/A |
| DHA 372 | Limb X-rays | 691 | 49.2 | 52 | 48.2 | – | – | – | – | – | N/A |
| EMBED 373 | Mammography | 115,910 | 100 | 38.9 | 41.6 | 6.5 | 5.6 | 1 | – | 11.3 | N/A |
| Optum 372 | EMRs, billing transactions | 5,802,865 | 56.1 | 67 | 7.5 | 2.8 | 7.5 | – | – | 15.2 | 95 |
| eICU-CRD374,375 | EMRs | 200,859 | 46 | 77.3 | 10.6 | 1.6 | 3.7 | – | 0.9 | 5.9 | 375 |
| Heritage Health376,377,378,379 | EMRs | 172,731 | 54.4 | – | – | – | – | – | – | – | 170,270,379,363 |
| Pima Indians Diabetes380,381 | Population health study | 768 | 100 | – | – | – | – | – | 100 | – | 381,271 |
| Warfarin382,383 | Drug relationship | 5,052 | – | 55.3 | 8.9 | 30.3 | – | – | – | 5.4 | 383 |
| Infant Health (IDHP)384,385 | Clinical measures | 985 | 50.9 | 36.9a | 52.5 | – | 10.7 | – | – | – | 170,386 |
| DrugNet 386 | Clinical measures | 293 | 29.4 | 8.5a | 33.8 | – | 52.9 | – | – | – | 387 |
= Grouping of White and unknown/other.
AREDS, Age-Related Eye Disease Study; CPTAC, Clinical Proteomic Tumor Analysis Consortium; DHA, Digital Hand Atlas; eICU-CRD, Electronic ICU Collaborative Research Database; EMBED, Emory Breast Imaging Dataset; EMR, electronic medical record; ICU, intensive care unit; NLST, National Lung Screening Trial; N/A, not applicable; OAI, Osteoarthiritis Initative; RSPECT, RNA Pulmonary Embolism CT Dataset; SIIM-ISIC, International Skin Imaging Collaboration.
Studies that have audited AI applications in healthcare that do consider group-fairness criteria have shown that algorithms developed using problematic ethnicity-skewed datasets provide worse outcomes for under-represented populations. CNNs trained on publicly available chest X-ray datasets (such as, Medical Information Mart for Intensive Care (MIMIC)-CXR, CheXpert and National Institutes of Health (NIH) ChestX-ray15) underdiagnose underserved populations; that is, the likelihood is greater of the algorithms incorrectly predicting ‘no symptomatic conditions (findings)’ for female patients, Black patients, Hispanic patients and patients with Medicaid insurance11. These patient populations are systematically underserved and are therefore under-represented in the datasets. Thus, these algorithms may be biased by population shifts (healthcare disparities, owing to worse social determinants of health) and prevalence shifts, because a greater proportion of underserved patients are diagnosed with ‘no finding’135. Follow-up discussions to ref. 11 have proposed bias-mitigation strategies via pre-processing and post-processing techniques (such as, importance reweighting and calibration). However, the absence of diversity in the datasets makes it difficult to select thresholds for each subgroup that would balance underdiagnosis rates, which may reduce overall model performance53,121,136. Biases such as population and prevalence shifts are also heavily influenced by how the dataset is stratified into training-validation-test splits, and should also be taken into account when studying disparities13.
Inclusion of ancestry and genetic variation
Ancestry is a crucial determining factor of the mutational landscape and the pathogenesis of diseases137,138,139,140. Our understanding of many diseases has been developed using biobank repositories that predominantly represent individuals with European ancestry. Additionally, the prevalence of certain mutations is only detectable via high-throughput sequencing of large and representative cohorts141,142,143,144,145. For instance, individuals with Asian ancestry are known to have a high prevalence of mutations in the epidermal growth-factor receptor (EGFR; discovered by other high-sequencing efforts), as detected in the PIONEER cohort, which enrolled 1,482 Asian patients146 (Fig. 3). However, owing to the absence of genetic diversity in datasets such as TCGA, many such common genomic alterations may be undetectable, despite being extensively used to discover molecular subtypes and despite having helped to redefine World Health Organization (WHO) taxonomies for cancer classification147,148.
Therefore, studies that control for social determinants of health may nevertheless be affected by population shift from genetic variation, and hence may manifest population-specific phenotypes. For instance, many cancer types have well-known disparities explained by biological determinants in which, even after controlling for socioeconomic status and access to healthcare, there exist population-specific genetic variants and gene-expression profiles that contribute substantially towards disparities in clinical outcomes and treatment responses149,150,151,152,153,154,155,156,157,158,159. Glioblastomas, for instance, demonstrate sex differences in clinical features (in the left temporal lobe for males and in the right temporal lobe for females), in genetic features (the association of neurofibromatosis type 1 (NF1) inactivation with tumour growth and in whether variations in the isocitrate dehydrogenase 1 (IDH1) sequence are a prognostic marker) and in outcomes (worse survival and treatment responses in men)160,161. In aggressive cancer types such as triple negative breast cancer (TNBC), there is mounting evidence that ancestry-specific innate immune variants contribute to the higher incidence of TNBC and mortality in people of African ancestry155,156,159. In prostate cancer, diagnostic gene panels (such as OncotypeDX, developed with patients of predominantly European ancestry) predict poorer prognosis (higher risk) in people of European descent than African Americans; this introduces the notion of population-specific gene signatures that could shed light on a complex aetiology162. In ophthalmology, there are known phenotypic variations across ethnicities, such as melanin concentration within uveal melanocytes (a higher concentration leads to darker fundus pigmentation), retinal-vessel appearance as a function of retinal arteriolar calibre size, and optic-disc size163. And, for transgender women, there may be novel histological findings following complications with gender-affirming surgery164.
It may thus be beneficial to include protected attributes such as sex, ethnicity and ancestry into AI algorithms, especially when the target label is strongly associated with the protected attribute. An example is integrating histology and patient-sex information in AI algorithms, as it can improve predictions of the origin of a tumour in metastatic cancers of unknown primary. This could be used as an assistive tool in recommending diagnostic immunohistochemistry for difficult-to-diagnose cases. Although the sex of a patient can be viewed as a sensitive attribute, not including this information may result in unusual diagnoses, such as predicting the prostate as the origin of a tumour in cases of cancer of unknown primary in women165. As another example, the prediction of mutations from whole-slide images via deep learning could become a low-cost screening approach for inferring genetic aberrations without the need for high-throughput sequencing. It could be used to predict biomarkers (such as microsatellite instability) that guide the use of immune-checkpoint inhibition therapy166, or of EGFR status to guide the selection of a tyrosine kinase inhibitor in the treatment of lung cancer167. However, if the approach were to be trained on TCGA and evaluated on the PIONEER cohort, it may predict a low EGFR-mutation frequency in Asian patients and lead to incorrect cancer-treatment strategies for this population. In this particular instance, using protected class information such as ancestry as a conditional label may improve performance on mutation-prediction tasks. Yet disentangling genetic variation and measuring the contribution of ancestry towards phenotypic variation in the tissue microenvironment is currently precluded by the lack of suitable, large and publicly available datasets. Moreover, it is generally unclear where and when protected attributes can be used to improve fairness outcomes. And bias-mitigation strategies that consider the inclusion of protected attributes are few94,168,169,170,171.
The importance of developing diverse data biobanks is well-known in the context of genome-wide association studies, where variations in linkage disequilibrium structures and minor allele frequencies across ancestral populations can contribute to worsening the performance of polygenic-risk models for under-represented populations172,173,174,175,176,177. Indeed, a fixed-effect meta-analysis found that polygenic-risk models for schizophrenia trained only on European populations performed worse for East Asian populations, owing to differing allele frequencies175. Additionally, cross-ancestry association studies that include populations from divergent ancestries have uncovered new diseased loci174.
Image acquisition and measurement variation
Variations in image acquisition and measurement technique can also leak protected class information. In this type of covariate shift (also known as domain shift or acquisition shift), institution-specific protocols and other non-biological factors that affect data acquisition can induce variability in the acquired data129,178. For example, X-ray, mammography or computed tomography (CT) images are affected by radiation dosage. Similarly, in pathology, heterogeneities in tissue preparation and in staining protocols as well as scanner-specific parameters for slide digitization can affect model performance in slide-level cancer-diagnostic tasks (Fig. 4).
Domain shift as a result of site-specific or region-specific factors that correlate with demographic characteristics may introduce spurious associations with ethnicity. For example, an audit study assessing site-specific stain variability of pathology slides in TCGA found shifts in stain intensity in the only site (the University of Chicago) that had a greater prevalence of patients of African ancestry9. Hence, clinical-grade AI algorithms in pathology may be learning inadvertent cues for ethnicity owing to institution-specific staining patterns. In this instance of domain shift, variable staining intensity can be corrected using domain adaptation and optimal transport techniques that adapt the test distribution to the training dataset. This can be performed on either the input space or the representation space. For instance, deep-learning techniques using GANs can learn stain features as a form of ‘style transfer’, in which a GAN can be used to pre-process data at deployment time to match the training distribution179,180. Other in-processing techniques such as adversarial regularization can be leveraged to learn domain-invariant features using semi-supervised learning and samples from both the training and test distributions. However, a practical limitation in both mitigation strategies is that the respective style-transfer or gradient-reversal layers would need to be fine-tuned with data from the test distribution for each deployment site, which can be challenging owing to data interoperability between institutions and to regulatory requirements for AI-SaMDs14. In some applications, understanding sources of shift presents a challenge for the development of bias-mitigation strategies that remove unwanted confounding factors. For instance, CNNs can reliably predict race in chest X-ray and other radiographs after controlling for image-acquisition factors, removing bone-density information and severely degrading image quality12.
Evolving dataset shifts over time
Dataset shifts can also occur as a result of changes in diagnostic criteria and in labelling paradigms across populations. This is known as concept shift or concept drift181,182,183,184,185,186, and it involves a change in the conditional distributions P(X|Y) or P(Y|X) while the marginal distributions P(X) and P(Y) remain unchanged. Concept shift is similar to other temporal dataset shifts187,188 (such as label shift), in that an increased prevalence in disease Y (for example, pneumonia) causing X (for instance, cough) does not change the causal relationship P(X|Y). However, in concept shift, the relationship between X and Y changes. This can occur if the criteria for diagnosing disease Y are revised over time or are incongruent between populations. Frequently studied examples of concept shift include the migration from ICD-8 to ICD-9, which refactored the coding for surgical procedures; the subsequent migration from ICD-9 to ICD-10, which resulted in a large spike in opioid-related inpatient stays127,128; and the more recent recategorization of ICD-10 to ICD-11, which recategorized strokes as neurological disorders rather than cardiovascular diseases.
Taxonomies and classification systems for many diseases undergo constant evolution, owing to new scientific discoveries and to research findings from randomized control trials. These changes may cause substantial variation in the way AI-SaMDs are evaluated across countries. An example of this is the assessment of kidney transplantation using the Banff classification (which since 1991 has established diagnostic criteria for renal-allograft assessment). Since its original development, the Banff classification has been subject to several major revisions: the establishment of a diagnosis based on antibody-mediated rejection (ABMR) in 1997, the specification of chronic ABMR on the basis of a transplant glomerulopathy biomarker in 2005, and the requirement of evidence of antibody interactions with the microvascular endothelium as a prerequisite for diagnosing chronic ABMR in 2013 (which resulted in a doubling of the diagnosis rate)189. Other notable examples are the shift from the Fuhrman nuclear grading system to the WHO/International Society of Urological Pathology grading system in renal cell carcinomas, the refined WHO taxonomy of diffuse gliomas to include molecular subtyping, the ongoing refinement of American College of Cardiology/American Heart Association guidelines for defining hypertension severity and the 17 varying diagnostic criteria for Behcet’s disease that have been proposed around the world97,190,191,192. As AI algorithms in medicine are often trained on large repositories of historical data (to overcome data paucity), numerous pitfalls may be affecting AI-SaMDs: they may have poor stability in adapting to changing disease prevalence, may be trained and deployed across healthcare systems with different labelling systems and may be trained with datasets affected by historical biases or that do not include data for under-represented populations. The fairness of an AI-SaMD under concept shift has been rarely analysed, yet it is well documented that many international disease-classification systems have poor intra-observer agreement, which suggests that an algorithm trained in one country may not be evaluated under the same labelling paradigm in another country189,190. To mitigate label shifts and concept shifts, some guidelines have emphasized the importance of guaranteeing model stability to how the data were generated118, and the use of reactive or proactive approaches for intervening on temporal dataset shifts in early-warning systems such as those for the prediction of sepsis117,184. However, at the moment there are relatively few strategies for the mitigation of concept shift in AI-SaMDs122,193,194,195.
Variations in self-reported race
As with concept shift across train and test distributions, different geographic regions and countries may collect protected attribute data with varying levels of stringency and granularity, which complicates the incorporation of race as a covariate in evaluations of fairness of medical AI. In addition to historical inequities that have confounded race in elucidating biological differences, another challenge is the active evolution of the understanding of race itself196. As discussions regarding race and ethnicity have moved more into the mainstream, the medical community has begun to realize that the taxonomies of the past do not adequately represent the groups of people that they purport to. Indeed, it is now accepted that race is a social construct and that there is greater genetic variability within a particular race than there are between races197,198,199. As such, the categorization of patients by race can obscure culture, history, socioeconomic status and other confounders of fairness; indeed, they may all separately or synergistically influence a particular patient’s health200,201. These factors can also vary by location: the same person may be considered of different races in different geographic locations, as exemplified by self-reported Asian ethnicity in the TCGA and PIONEER cohorts, and by self-reported race in the COMPAS algorithm146,201.
Ideally, discussions should centre explicitly around each component of race and include ancestry, a concept that has a clear definition—the geographic origins of one’s ancestors—and that is directly connected to the patient’s underlying genetic admixture and hence to many traits and diseases. However, introducing this granularity in fairness evaluations has clear drawbacks in terms of the power of subgroup analyses, as ancestry DNA testing is not routinely performed on patients at most institutions. In practice, institutions would often fall back on the traditional ‘dropdown menu’ for selecting only a single race or ethnicity. Performing fairness evaluations without explicitly considering these potential confounders of race may mean that the AI system is sensitive to unaccounted-for factors200.
Paths forward
Using federated learning to increase data diversity
Federated learning is a distributed-learning paradigm in which a network of participants uses their own computing resources and local data to collectively train a global model stored on a server202,203,204,205. Unlike machine learning performed over a centralized pool of training data, in federated learning users in principle retain oversight of their own data and must only share the update of weight parameters or gradient signals (with privacy-preserving guarantees) from their locally trained model with the central server. In this way, algorithms can be trained on large and diverse datasets without sharing sensitive information. Federated learning has been applied to a variety of clinical settings to overcome data-interoperability standards that would usually prohibit sensitive health data from being shared and to tackle low-data regimes of clinical machine-learning tasks, for example for the prediction of rare diseases98,206,207,208,209,210,211,212,213,214,215,216,217,218,219,220. Federated learning applied to EMRs has satisfied privacy-preserving guarantees for transferring sensitive health data and has enabled the development of early-warning systems for hospitalization, sepsis and other preventive tasks215,221. In radiology, federated learning under various network architectures, privacy-preserving protocols and adversarial attacks has leveraged multi-institutional collaboration to aid the validation of AI algorithms for prostate segmentation, brain-cancer detection, the monitoring of the progression of Alzheimer’s disease using magnetic resonance imaging (MRI) and the classification of paediatric chest X-ray images211,214,222,223,224. In pathology, federated learning has been used to assess the robustness of the performance of weakly supervised algorithms for the analysis of whole-slide images under various privacy-preserving noise levels in diagnostic and prognostic tasks225. It has also been used to overcome low sample sizes in the development of AI models for COVID-19 pathology, and in independent test-cohort evaluation226,227,228.
Regarding fairness, federated learning for the development of decentralized AI-SaMDs can be naturally extended to address many of the cases of dataset shift and to mitigate disparate impact via model development on larger and more diverse patient populations229. In developing polygenic risk scores, decentralized information infrastructures have been shown to harmonize biobank protocols and to enable tangible material transfer agreements across multiple hospitals230, which can then enable model development on large and diverse biobank datasets229,231. Federated learning applied to multi-site domain adaptation across distributed clients would naturally mitigate many instances of dataset shift232,233,234,235,236. In particular, methods such as federated multi-target domain adaptation address practical scenarios, such as when centralized label data are made available only at the client (source) and the unlabelled data are distributed across multiple clients as targets237. The application of federated learning in fairness may allow for new fairness criteria, such as client-based fairness that equalizes model performance at only the client-level238,239, as well as novel formulations of existing bias-mitigation strategies that may not require centralizing information about protected attributes for evaluating fairness criteria240,241,242,243,244,245,246.
Operationalizing fairness principles across healthcare ecosystems
Although federated learning may overcome data-interoperability standards and enable the training of AI-SAMDs with diverse cohorts, the evaluation of AI biases in federated-learning settings would need to be extensively studied. Despite numerous technical advances in improving the communication efficiency, robustness and security of parameter updates, one central statistical challenge is learning from data that are not independent and/or not identically distributed (known as non-i.i.d data). This arises because of the sometimes-vast differences in local-data distribution at contributing sites, which can lead to the divergence of local model weights during training following synchronized initiation204,247,248,249. Accordingly, the performance of federated-learning algorithms (including that of the well-known FedAvg algorithm250,251) that use averaging to aggregate local model-parameter updates deteriorates substantially when applied to non-i.i.d. data252. Such statistical challenges may produce further disparate impact depending on the heterogeneity of the data distributions across clients. For instance, in using multi-site data in the TCGA invasive breast carcinoma (BRCA) cohort as individual clients for federated learning, a majority of parameter updates would come from clients that over-represent individuals with European ancestry, with only one parameter update coming from a single client that has majority representation for African ancestry. This problem can be decomposed into two phenomena, known as local drift (when clients are biased towards minimizing the loss objective of their own distribution) and global drift (when the server is updating diverging gradients from clients with of mismatched data distributions). As a result, many federated-learning approaches developed for learning in non-i.i.d. scenarios inherently adapt dataset shift and bias-mitigation techniques to resolve local and global drift in tandem with evaluating fairness236,237,239,245,246,253,254. Without bias-mitigation strategies, federated models would still be subject to persisting biases found in centralized models, such as the problems of site-specific image-stain variability, intra-observer variability, and under-representation of ethnic minorities in the multi-site TCGA-BRCA cohort9. Federated learning may enable the fine-tuning of AI-SaMDs at each deployment site; however, the evaluation of race and ethnicity for healthcare applications via federated models has yet to be benchmarked. Additionally, because race and ethnicity and other sensitive information are typically isolated in separate databases, there may be logistic barriers to accessing such protected attribute data at each site.
The difficulty of operationalizing fairness principles for the much simpler AI development and deployment life cycles of centralized models will also affect the practical adoption of fair federated-learning paradigms. In current organizational structures, the roles and responsibilities created for implementing fairness principles are typically isolated into ‘practitioner’ or data-regulator roles (which design AI fairness checklists for guiding the ethical development of algorithms in the organization) and ‘engineer’ or data-user roles (which follow the checklist during algorithm implementation255). Such binary partitioning of the roles may lead to poor practices, as fairness checklists are often too broad or abstractive, and are not co-designed with engineers to address problem-specific and technical challenges for the achievement of fairness256. For federated-learning paradigms for the development and global deployment of AI-SaMDs, the design of fairness checklists would require interdisciplinary collaboration from all relevant healthcare roles (in particular, clinicians, ethics practitioners, engineers and researchers), as well as further involvement from stakeholders at participating institutions so as to identify potential site-specific biases that may be propagated during parameter sharing or as a result of accuracy–fairness trade-offs carried out at inference time255.
Overall, although federated learning presents an opportunity to evaluate AI algorithms on diverse biomedical data at a global scale, it faces unknown challenges in the design of global fairness checklists that consider the burdens and patient preferences of each region. For instance, a federated scheduling algorithm for patient follow-ups calibrated to set a high threshold to maximize fairness criteria may not account for substantial differences in burden (which could be much higher at a low-resource setting)257. As with the problems of label shift or concept shift that may occur at various sites, there may be additional complexity arising from culture-specific or regional factors affecting access to protected information, and from definitions and criteria for fairness from differing moral and ethical philosophies48. Navigating such ethical conflicts may involve considering the preferences of diverse stakeholders, particularly of under-represented populations.
Fair representation learning
By focusing on learning intermediate representations that retain discriminative features from the input space X without any features correlating with A, typically via an adversarial-loss term (Fig. 1), fair representation learning is orthogonal to causality258, model robustness and many other subjects in machine learning, and shares techniques and goals with adversarial learning for debiasing representations. Inspired by the minimax objective in GANs, fair representation learning has been used to learn domain-invariant features of the distributions of the training and test datasets (; ref. 179), treatment-invariant representations for producing counterfactual explanations259,260,261, and race-invariant or ethnicity-invariant features to remove disparate impact in deep-learning models67,101,262,263,264,265,266,267,268.
Although fair-representation methods are typically supervised, training fair AI algorithms in an unsupervised manner would allow representations to be freely transferred to other domains without constraints on the downstream classifiers (such as being fair or enabling greater applications of fair algorithms without access to protected attributes93,269). One prominent example is the method known as learned adversarially fair and transferable representation (LAFTR)270, which first modified the GAN minimax objective with an adversarial-loss term to make the latent feature representation invariant to the protected class. LAFTR also showed that such representations are transferable (as examined in the Charlson Comorbidity Index prediction task in the Heritage Health dataset, in which LAFTR was able to transfer to other tasks without leaking sensitive attributes93,269). Across other tasks in medicine, LAFTR can be extended as a privacy-preserving machine-learning approach that allows for the transfer of useful intermediate features. This could advance multi-institutional collaboration in the fine-tuning of algorithms without leaking sensitive information. Similar to LAFTR is unsupervised fair clustering, which aims at learning attribute-invariant cluster assignments (which can also be done via adversarial learning for debiasing representations)271,272. Still, a main limitation in many of these unsupervised fairness approaches is that they depend on having the protected attribute at hand during training, which may not be possible in many clinical settings in which protected class identity is secured. Moreover, in assessing the accuracy–fairness trade-off, adding additional regularization components may decrease representation quality and thus lower performance in downstream fairness tasks103.
Despite access to protected attributes possibly constraining model training, geographical data in the client identities may be used as proxy variables for subgroup identity. This may inform the development of fairness techniques without access to sensitive information. Decentralized frameworks have shown that federated learning in combination with fair representation learning can be used to learn federated, adversarial and debiasing representations with similar privacy-preserving and transferable properties as LAFTR125,273,274,275,276,277,278,279,280,281. Using client identities as proxy variables for protected attributes in adversarial regularization may hold in certain scenarios273, as geographical location is more closely linked to genetic diversity than ethnicity282.
Debiased representations via disentanglement
Disentanglement in generative models can also be used to further promote fairness in learned representations without requiring access to protected attributes. It aims at disentangling independent and easy-to-interpret factors of data in latent space, and has allowed for the isolation of sources of variation in objects, such as colour, pose, position and shape283,284,285,286,287. BetaVAE is a method to quantify disentanglement in deep generative models. It uses a variational autoencoder (VAE) bottleneck for unsupervised learning, followed by the generation of a disentanglement score by training a linear classifier to predict the fixed factor of variation from the representation287. Disentangled VAEs with adversarial-loss components have been used to disentangle size, skin colour and eccentricity in dermoscopy images, as well as causal health conditions and anatomical factors in physiological waveforms288,289,290,291.
With regards to fairness and dataset shifts, disentanglement can be viewed as a form of data pre-processing for debiasing representations in downstream fairness tasks and for providing flexibility in terms of allowing data users to isolate and truncate specific latent codes that correspond to protected attributes in representation space255,292,293. The evaluation of unsupervised VAE-based disentangled models has shown that disentanglement scores correlate with fairness metrics, benchmarked on numerous fairness-classification tasks without the need for protected attribute information294. Disentanglement would be particularly advantageous in settings where the latent code information for including and excluding protected attributes needs to be flexibly adapted; for example, in pathology, where ethnicity may be excluded when predicting cancer stage yet included when predicting mutation status293. Disentanglement-like methods have been used to cluster faces without latent code information according to dominant features such as skin colour and hair colour272,295,296. In chest X-ray images, they have also been shown to mitigate hidden biases in the detection of SARS-CoV-2297. In combination with federated learning, FedDis and related frameworks have been used to isolate sensitive attributes in non-i.i.d. MRI lesion data; images were disentangled according to shape and appearance features, with only the shape parameter shared between clients254,298,299,300,301,302 (Fig. 5).
Fig. 5 |. A decentralized framework that integrates federated learning with adversarial learning and disentanglement.
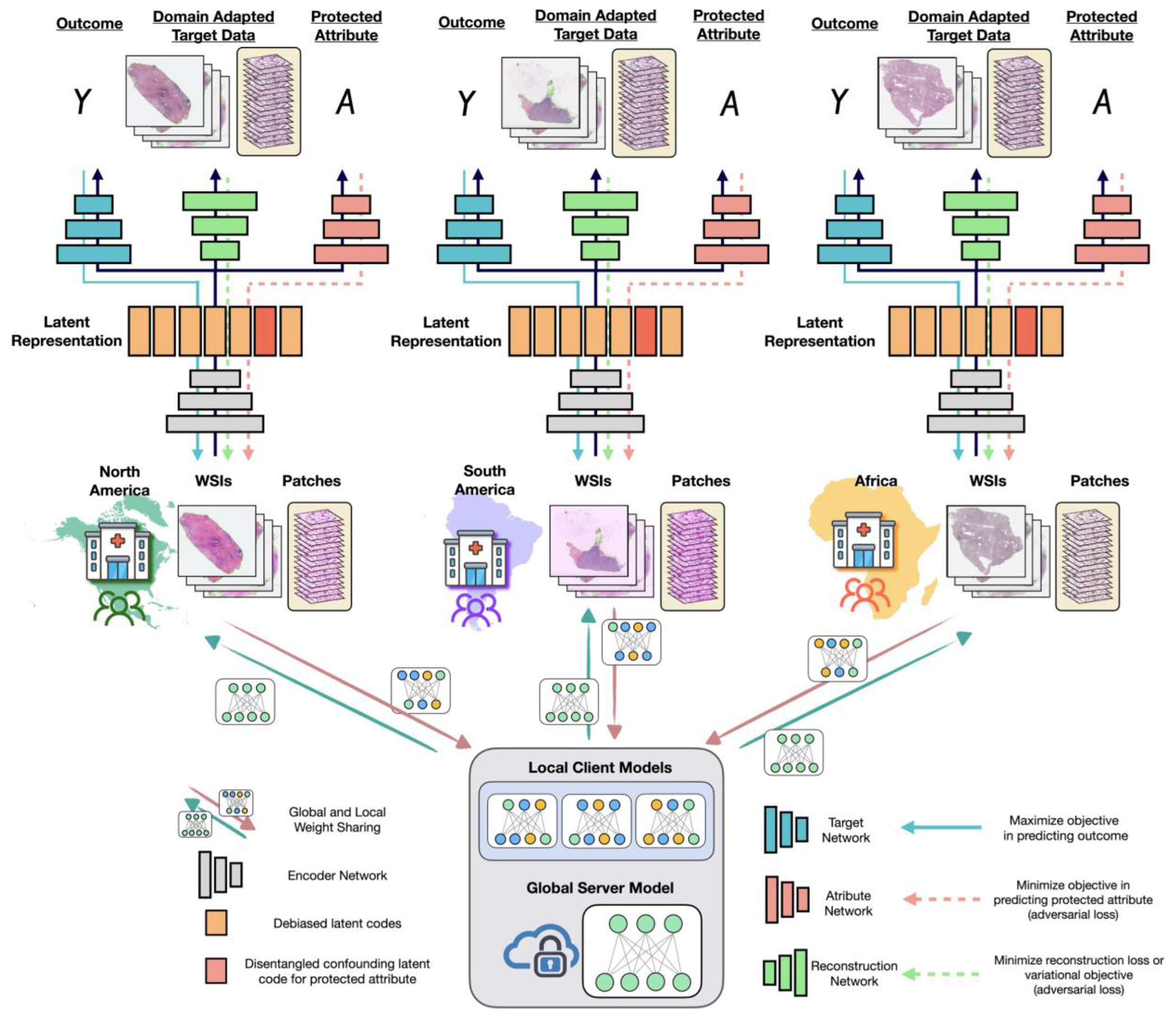
In addition to aiding the development of algorithms using larger and more diverse patient populations, federated learning can be integrated with many techniques in representation learning and in unsupervised domain adaptation that can learn in the presence of unobserved protected attributes. In federated learning, global and local weights are shared between the global server and the local clients (such as different hospitals in different countries), each with different datasets of whole-slide images (WSIs) and image patches. Different domain-adaptation methods can be used with federated learning. In federated adversarial and debiasing (FADE), the client IDs were used as protected attributes, and adversarial learning was used to debias the representation so that it did not vary with geographic region273 (red). In FedDis, shape and appearance features in brain MRI scans were disentangled, with only the shape parameter shared between clients298 (orange). In federated adversarial domain adaptation (FADA), disentanglement and adversarial learning were used to further mitigate domain shifts across clients236 (red and orange). Federated learning can also be used in combination with style transfer, synthetic data generation, and image normalization. In these cases, domain-adapted target data or features would need to be shared, or other techniques employed (green)236,239,299,300,301,302,331,390. Y and A denote, respectively, the model’s outcome and a protected attribute.
Disentangling the roles of data regulators and data users in life cycles of AI-SaMDs
Within current development and deployment life cycles of AI-SaMDs and other AI algorithms, the adaptability of unsupervised fair-representation and disentanglement methods would allow the refinement of the distribution of responsibilities in organizational structures by adding the role of ‘data producers’; that is, those who produce ‘cleaned up’ versions of the input for more informative downstream tasks, as proposed previously255. In this setting, the roles of ‘data users’ and ‘data regulators’ would be separate and compatible with conventional model-development pipelines without the need to consider additional fairness constraints255. Moreover, ‘data producers’ would also quantify the potential accuracy–fairness trade-offs when using regularization components to achieve debiased and disentangled representations. It has been hypothesized that such an approach could pave a path forward for a three-party governance model that simplifies communication overhead when discussing concerns of accuracy–fairness trade-offs, and that adapts to test populations without the complexities of federated learning, which also needs access to protected attributes255. Still, fair representation learning has yet to be benchmarked against competitive self-supervised learning methods (particularly, contrastive learning), and more evaluations of fair representation learning in clinical settings are also needed303. Future work on understanding disentanglement and on adapting it to robust self-supervised learning paradigms would contribute to improving fairness in transfer-learning tasks and serve as a privacy-preserving measure for clinical machine-learning tasks.
Algorithm interpretability for building fair and trustworthy AI
In current regulatory frameworks for the development of AI-SaMDs, algorithm interpretability is pivotal in medical decision-making and in model auditing so as to understand the sources of unfairness and to detect dataset shifts304. Trust in AI algorithms is also an important consideration in current regulation of AI-SaMDs. As a conceptualization for the machine-learning community, and now broadly advocated by regulatory bodies, trust is the fulfilment of a contract in human–AI collaboration. Such contracts are AI functionalities that are anticipated to have known vulnerabilities305. For instance, model correctness is a contract that anticipates patterns that distinguish the model’s correct and incorrect cases available to the user. There are many different types of contract (concerning technical robustness, safety, non-discrimination and transparency, in particular) outlined by the European Commission’s ethical guidelines for trustworthy AI306,307. Similarly, the FDA, in its Good Machine Learning Practices guidelines outlined bias assessment and interpretability as contracts in its action plan for developing trust in AI-SaMDs16. In the remainder of this section, we discuss current applications of algorithm interpretability, and their relevance to fairness in medicine and healthcare.
Interpretability methods and model auditing
For post-hoc interpretability in deep-learning architectures, class-activation maps (CAMs, or saliency mapping) are a commonly used technique for finding sensitive input features that would explain the decision made by an algorithm. CAMs compute the partial derivatives of the predictions with respect to the pixel intensities computed during the back-propagation step of the neural network. The derivatives are then used to produce a visualization of the informative pixel regions308. To produce more fine-grained visualizations, extensions of these methods (such as Grad-CAM) instead attribute how neurons of an intermediate feature layer in a CNN would affect its output, such that the attributions for these intermediate features can be up-sampled to the original image size and viewed as a mask to identify discriminative image regions309. CAM-based methods have gained widespread adoption in the clinical interpretability of the output of CNNs, because salient regions (rather than low-level pixel intensities) would refer to high-level image features. Because these techniques can be applied without modifying the neural networks, they have been used in preclinical applications such as the detection of skin lesions, the localization of disease in chest X-ray images and the segmentation of organs in CT images3,89,310,311,312,313,314,315,316.
However, saliency-mapping techniques may not be accurate, understandable to humans or actionable, and thus insufficiently trustworthy by practitioners in medical support, decision-making, biomarker discovery and model auditing317,318,319. Indeed, interpretability via saliency mapping is typically qualitative but not sufficiently quantifiable to evaluate group differences. An audit of Grad-CAM interpretability in natural images and in echocardiograms found that saliency maps can be misleading and often equivalent to results from simple edge detectors304,320. In the diagnosis of chest radiographs, saliency-mapping techniques are insufficiently accurate if used to localize pathological features314,321. In cancer prognosis, although useful in highlighting important regions-of-interest in histology tissue, it is unclear how highly attributed pixel regions can be used by clinicians for patient stratification without further post-hoc assessment322. And in the problem of unknown dataset shifts of radiology images leaking self-reported race, model auditing via saliency mapping was ineffective in determining any explainable anatomic landmarks or image-acquisition factors associated with the misdiagnoses12. Still, saliency mapping can be used to detect spurious bugs or artefacts in the input feature space (although they do not always detect them in ‘contaminated’ models323,324). Overall, although the visual appeal of saliency mapping may be informative for some uses in medical interpretation, its many limitations preclude its use for enhancing trust into AI algorithms and for the assessment of fairness in their clinical deployment305,325.
Techniques such as Shapley additive explanations (SHAP) and Integrated Gradients have also found utility in explaining machine-learning predictions and dataset shift across a variety of applications326,327,328. In SHAP, feature importance values are computed by decomposing the model output into a set of attribution units, with each attribution relating to the influence of its respective feature on the model output. In healthcare applications, these methods have been used to predict drug-combination synergies in transcriptomic data, to corroborate the prognostic value of hallmark genes and morphological biomarkers in cancer survival, and to identify novel risk factors in EMR data for mortality prediction322,329,330,331,332. As with saliency mapping, SHAP-based and Integrated-Gradients-based methods have limitations in post-hoc explainability in that they can be sensitive to the choice of ‘baseline’ input for conveying the absence of a signal, to estimation and convergence strategies and other parameters, and these assumptions should be carefully considered when used in clinical applications. Differently, these techniques attribute features at both a global level across the entire dataset (for assessing overall feature importance) and a local level for individual samples (for explaining individual predictions), and thus can be intuitively extended to measure global and individual fairness criteria and other model-auditing applications333. For instance, as group-fairness metrics are computed as performance differences of model outputs across protected subgroups, SHAP can be directly used to decompose the difference in model outputs into attributions that quantify the impact of influential features on disparity measures334. Other analyses have found that greater disparity measures correlate with larger SHAP values for the biased features following mitigation strategies335. In model-auditing applications, on the MIMIC dataset, an analysis examining mortality prediction models using Integrated Gradients found disparities in feature importance across ethnicity, gender and age subgroups, with ethnicity often ranked as one of the most important features across a diverse set of models and explainability techniques336,337,338. On National Health and Nutrition Examination Survey (NHANES), a graphical model variation of SHAP could discover relationships between race and access to food programs in predicting 15-year survival rates339. In other studies, however, SHAP and Integrated Gradients have led to disparities in measuring the faithfulness of explanations on the OpenXAI fairness benchmark compared to other attribution methods, which alludes to the sensitivity of baseline choice and other parameters in these class of methods340. Still, SHAP can be used in many other ways in fairness beyond feature interpretability, such as attributing influential data points that would influence model performance in data-resource allocation, attributing model-performance change to different types of distribution shift, and attributing the contribution of participants in fair federated learning83,100,244,335,341,342,343.
Fitting algorithm design to comply with regulatory requirements for explainability
Although algorithm-interpretability techniques are model-agnostic, the efficacy and usage of these techniques vary across different types of data modalities and model architectures, and thus have important implications in the regulatory assessment of human–AI trust for AI-SaMDs and in the choice of algorithm design. For instance, structured modalities such as imaging data can be difficult to interpret and trust by clinical and machine-learning practitioners, as feature attributions computed for influential pixels in a CNN are less meaningful in explaining quantifiable disparity measures. On the other hand, SHAP-based and Integrated-Gradients-based methods have been extensively validated to perform well on regulatory genomics data329,344. As such, regulatory agencies may enforce specific contracts for building trustworthy AI (such as quantifying feature importance for fairness-disparity measures). These contracts may also inform the design of algorithms that would see clinical deployment, potentially inspiring new types of deep-learning approaches with different interpretability mechanisms345. For instance, instead of CNNs, advancements in computer vision have proposed the usage of Transformers that process images as a sequence of ‘visual tokens’ and that naturally produce prototypical explanations346. In this setting, instead of attributing individual pixels, attributions (in the form of attention weights) are given to semantic high-level visual concepts, such as the ‘blue crown’ and ‘blue back’ attributes that co-occur frequently in blue jay birds, or tumour-lymphocyte colocalization in histology slides as a biomarker of favourable cancer prognosis346,347. In contrast with the poor localization performance of saliency mapping in chest X-ray images, high-attention regions identified within histology slides can have strong agreement with clinical annotations of the anatomical regions of interest346,348. With increasing progress being made in large pretrained models (also known as ‘foundation models’) with capabilities such as zero-shot classification, unsupervised object segmentation and chain-of-thought reasoning, new definitions and evaluation strategies need to be devised to understand the extent of trust and explainability, especially when the models are used by patients349,350,351.
The development of handcrafted and human-interpretable features may overcome challenges in model explainability. Indeed, statistical contour-based and image-based cell features have been shown to predict molecular signatures, such as immune-checkpoint protein expression and homologous-recombination deficiency352. As an approach for mitigating and explaining unfairness, handcrafted features for predicting prostate-cancer recurrence have corroborated stromal morphology with aggressive cancer phenotypes, and helped elucidate population-specific phenotypes for African American patients353,354,355. Approaches for medical imaging not based on deep learning may have niche applications, for instance in the context of small yet high-dimensional clinical-trial datasets. However, simpler approaches in these settings may fulfill trust better by providing more coherent and actionable clinical interpretability to the user, or by assisting in further post-hoc analyses of deep-learning-based features. We believe that features obtained via deep learning and handcrafted features will be needed in the ongoing assessment of harm by AI-SaMDs via interpretability techniques.
Outlook
Medicine ought to account for genetic ancestry, socioeconomic status, access to care and other health correlates in a fair and productive manner. However, AI algorithms can exacerbate healthcare disparities, owing to insufficient genetic diversity in biomedical datasets, to variabilities in medical-data-generating processes across healthcare systems and to other forms of dataset shifts that may cause disparate performance during the deployment of AI-SaMDs. When discussing and addressing these concerns by developing bias-mitigating strategies in AI-SaMDs, the opinions and expertise of clinical practitioners and other suitable stakeholders in the healthcare ecosystem, as well as those of machine-learning researchers and engineers, must be included. In this Perspective, we have argued that federated learning, fair representation learning and model interpretability can be used to incorporate fairness into the design and training of AI models. We hope that this contribution also serves as a gateway to understand the domain-specific and shared challenges of the stakeholders contributing to advancing healthcare equity.
Supplementary Material
Footnotes
A suitable bijective map (e.g. the sigmoid function) can be used to transform the score from to [0,1], such that it can be interpreted as a probability score, e.g., . Whole for regression tasks, it is often suffices to leave as the identity map. Here, we drop as it is implicitly specified by the task of interest.
Related criterion such as equality of opportunity relaxes this notion to only consider equalized TPRs. Equality of odds can be similarly defined for TNRs and FNRs.
References
- 1.Buolamwini J & Gebru T Gender shades: intersectional accuracy disparities in commercial gender classification. In Conf. on Fairness, Accountability and Transparency 77–91 (PMLR, 2018). [Google Scholar]
- 2.Obermeyer Z, Powers B, Vogeli C & Mullainathan S Dissecting racial bias in an algorithm used to manage the health of populations. Science 366, 447–453 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Pierson E, Cutler DM, Leskovec J, Mullainathan S & Obermeyer Z An algorithmic approach to reducing unexplained pain disparities in underserved populations. Nat. Med 27, 136–140 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Hooker S Moving beyond ‘algorithmic bias is a data problem’. Patterns 2, 100241 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCradden MD, Joshi S, Mazwi M & Anderson JA Ethical limitations of algorithmic fairness solutions in health care machine learning. Lancet Digit. Health 2, e221–e223 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Mhasawade V, Zhao Y & Chunara R Machine learning and algorithmic fairness in public and population health. Nat. Mach. Intell 3, 659–666 (2021). [Google Scholar]
- 7.Currie G & Hawk KE Ethical and legal challenges of artificial intelligence in nuclear medicine. In Seminars in Nuclear Medicine (Elsevier, 2020). [DOI] [PubMed] [Google Scholar]
- 8.Chen IY et al. Ethical machine learning in healthcare. Annu. Rev. Biomed. Data Sci 4, 123–144 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howard FM et al. The impact of site-specific digital histology signatures on deep learning model accuracy and bias. Nat. Commun 12, 4423 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seyyed-Kalantari L, Liu G, McDermott M, Chen IY & Ghassemi M CheXclusion: fairness gaps in deep chest X-ray classifiers. In BIOCOMPUTING 2021: Proc. Pacific Symposium 232–243 (World Scientific, 2020). [PubMed] [Google Scholar]
- 11.Seyyed-Kalantari L, Zhang H, McDermott M, Chen IY & Ghassemi M Underdiagnosis bias of artificial intelligence algorithms applied to chest radiographs in under-served patient populations. Nat. Med 27, 2176–2182 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gichoya JW et al. AI recognition of patient race in medical imaging: a modelling study. Lancet Digit. Health 4, E406–E414 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glocker B, Jones C, Bernhardt M & Winzeck S Algorithmic encoding of protected characteristics in chest X-ray disease detection models. EBioMedicine 89, 104467 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Proposed Regulatory Framework for Modifications to Artificial Intelligence. Machine Learning (AI/ML)-Based Software as a Medical Device (SaMD) (US FDA, 2019). [Google Scholar]
- 15.Gaube S et al. Do as AI say: susceptibility in deployment of clinical decision-aids. NPJ Digit. Med 4, 31 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Artificial Intelligence/Machine Learning (AI/ML)-Based Software as a Medical Device (SAMD) Action Plan (US FDA, 2021). [Google Scholar]
- 17.Vyas DA et al. Challenging the use of race in the vaginal birth after cesarean section calculator. Women’s Health Issues 29, 201–204 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Vyas DA, Eisenstein LG & Jones DS Hidden in plain sight—reconsidering the use of race correction in clinical algorithms. N. Engl. J. Med 383, 874–882 (2020). [DOI] [PubMed] [Google Scholar]
- 19.van der Burgh AC, Hoorn EJ & Chaker L Removing race from kidney function estimates. JAMA 325, 2018 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Diao JA et al. Clinical implications of removing race from estimates of kidney function. JAMA 325, 184–186 (2021). 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caton S & Haas C Fairness in machine learning: a survey Preprint at 10.48550/arXiv.2010.04053 (2020). [DOI] [Google Scholar]
- 22.Adler NE, Glymour MM & Fielding J Addressing social determinants of health and health inequalities. JAMA 316, 1641–1642 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Phelan JC & Link BG Is racism a fundamental cause of inequalities in health? Annu. Rev. Socio 41, 311–330 (2015). [Google Scholar]
- 24.Yehia BR et al. Association of race with mortality among patients hospitalized with coronavirus disease 2019 (COVID-19) at 92 US hospitals. JAMA Netw. Open 3, e2018039 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez L, Hart LH & Katz MH Racial and ethnic health disparities related to COVID-19. JAMA 325, 719–720 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Bonvicini KA LGBT healthcare disparities: what progress have we made? Patient Educ. Couns 100, 2357–2361 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Yamada T et al. Access disparity and health inequality of the elderly: unmet needs and delayed healthcare. Int. J. Environ. Res. Public Health 12, 1745–1772 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moy E, Dayton E & Clancy CM Compiling the evidence: the national healthcare disparities reports. Health Aff 24, 376–387 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Balsa AI, Seiler N, McGuire TG & Bloche MG Clinical uncertainty and healthcare disparities. Am. J. Law Med 29, 203–219 (2003). [PubMed] [Google Scholar]
- 30.Marmot M Social determinants of health inequalities. Lancet 365, 1099–1104 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Maness SB et al. Social determinants of health and health disparities: COVID-19 exposures and mortality among African American people in the United States. Public Health Rep 136, 18–22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seligman HK, Laraia BA & Kushel MB Food insecurity is associated with chronic disease among low-income NHANES participants. J. Nutr 140, 304–310 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thun MJ, Apicella LF & Henley SJ Smoking vs other risk factors as the cause of smoking attributable deaths: confounding in the courtroom. JAMA 284, 706–712 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Tucker MJ, Berg CJ, Callaghan WM & Hsia J The Black–White disparity in pregnancy-related mortality from 5 conditions: differences in prevalence and case-fatality rates. Am. J. Public Health 97, 247–251 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gadson A, Akpovi E & Mehta PK Exploring the social determinants of racial/ethnic disparities in prenatal care utilization and maternal outcome. In Seminars in Perinatology 41, 308–317 (Elsevier, 2017). [DOI] [PubMed] [Google Scholar]
- 36.Wallace M et al. Maternity care deserts and pregnancy-associated mortality in louisiana. Women’s Health Issues 31, 122–129 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burchard EG et al. The importance of race and ethnic background in biomedical research and clinical practice. N. Engl. J. Med 348, 1170–1175 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Phimister EG Medicine and the racial divide. N. Engl. J. Med 348, 1081–1082 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Bonham VL, Green ED & Pérez-Stable EJ Examining how race, ethnicity, and ancestry data are used in biomedical research. JAMA 320, 1533–1534 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eneanya ND, Yang W & Reese PP Reconsidering the consequences of using race to estimate kidney function. JAMA 322, 113–114 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Zelnick LR, Leca N, Young B & Bansal N Association of the estimated glomerular filtration rate with vs without a coefficient for race with time to eligibility for kidney transplant. JAMA Netw. Open 4, e2034004 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chadban SJ et al. KDIGO clinical practice guideline on the evaluation and management of candidates for kidney transplantation. Transplantation 104, S11–S103 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Wesselman H et al. Social determinants of health and race disparities in kidney transplant. Clin. J. Am. Soc. Nephrol 16, 262–274 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanis JA HNME & Johansson H A brief history of frax. Arch. Osteoporos 13, 118 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lewiecki EM, Wright NC & Singer AJ Racial disparities, frax, and the care of patients with osteoporosis. Osteoporos. Int 31, 2069–2071 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Civil Rights Act of 1964. Title VII, Equal Employment Opportunities https://en.wikipedia.org/wiki/Civil_Rights_Act_of_1964 (1964)
- 47.Griggs v. Duke Power Co https://en.wikipedia.org/wiki/Griggs_v._Duke_Power_Co (1971).
- 48.Awad E et al. The moral machine experiment. Nature 563, 59–64 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Feller A, Pierson E, Corbett-Davies S & Goel S A computer program used for bail and sentencing decisions was labeled biased against blacks. it’s actually not that clear. The Washington Post (17 October 2016). [Google Scholar]
- 50.Dressel J & Farid H The accuracy, fairness, and limits of predicting recidivism. Sci. Adv 4, eaao5580 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Char DS, Shah NH & Magnus D Implementing machine learning in health care—addressing ethical challenges. N. Engl. J. Med 378, 981–983 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bernhardt M, Jones C & Glocker B Potential sources of dataset bias complicate investigation of underdiagnosis by machine learning algorithms. Nat. Med 28, 1157–1158 (2022). [DOI] [PubMed] [Google Scholar]
- 53.Mukherjee P et al. Confounding factors need to be accounted for in assessing bias by machine learning algorithms. Nat. Med 28, 1159–1160 (2022). [DOI] [PubMed] [Google Scholar]
- 54.Diao JA, Powe NR & Manrai AK Race-free equations for eGFR: comparing effects on CKD classification. J. Am. Soc. Nephrol 32, 1868–1870 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feldman M, Friedler SA, Moeller J, Scheidegger C & Venkatasubramanian S Certifying and removing disparate impact In Proc. 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 259–268 (2015). [Google Scholar]
- 56.Hardt M, Price E & Srebro N Equality of opportunity in supervised learning. In Adv. Neural Information Processing Systems (2016). [Google Scholar]
- 57.Corbett-Davies S & Goel S The measure and mismeasure of fairness: a critical review of fair machine learning Preprint at 10.48550/arXiv.1808.00023 (2018). [DOI] [Google Scholar]
- 58.Calders T, Kamiran F & Pechenizkiy M Building classifiers with independency constraints In Int. Conf. Data Mining Workshops 13–18 (IEEE, 2009). [Google Scholar]
- 59.Chen J, Kallus N, Mao X, Svacha G & Udell M Fairness under unawareness: assessing disparity when protected class is unobserved In Proc. Conf. Fairness, Accountability, and Transparency 339–348 (2019). [Google Scholar]
- 60.Zliobaite I, Kamiran F & Calders T Handling conditional discrimination In 11th Int. Conf. Data Mining 992–1001 (IEEE, 2011). [Google Scholar]
- 61.Dwork C, Hardt M, Pitassi T, Reingold O & Zemel R Fairness through awareness In Proc. 3rd Innovations in Theoretical Computer Science Conf. 214–226 (2012). [Google Scholar]
- 62.Pedreshi D, Ruggieri S & Turini F Discrimination-aware data mining In Proc. 14th ACM SIGKDD Int. Conf. Knowledge Discovery and Data Mining 560–568 (2008). [Google Scholar]
- 63.Angwin J, Larson J, Mattu S & Kirchner L In Ethics of Data and Analytics 254–264 (Auerbach, 2016). [Google Scholar]
- 64.Kleinberg J, Mullainathan S & Raghavan M Inherent trade-offs in the fair determination of risk scores In 8th Innovations in Theoretical Computer Science Conf. (ITCS; 2017) [Google Scholar]
- 65.Chouldechova A Fair prediction with disparate impact: a study of bias in recidivism prediction instruments. Big Data 5, 153–163 (2017). [DOI] [PubMed] [Google Scholar]
- 66.Joseph M, Kearns M, Morgenstern JH & Roth A Fairness in learning: classic and contextual bandits. In Adv. Neural Information Processing Systems (2016). [Google Scholar]
- 67.Celis LE & Keswani V Improved adversarial learning for fair classification Preprint at 10.48550/arXiv.1901.10443 (2019). [DOI] [Google Scholar]
- 68.Kamiran F & Calders T Data preprocessing techniques for classification without discrimination. Knowl. Inf. Syst 33, 1–33 (2012). [Google Scholar]
- 69.Calmon FP, Wei D, Vinzamuri B, Ramamurthy KN & Varshney KR Optimized pre-processing for discrimination prevention In Proc. 31st Int. Conf. Neural Information Processing Systems 3995–4004 (2017). [Google Scholar]
- 70.Krasanakis E, Spyromitros-Xioufis E, Papadopoulos S & Kompatsiaris Y Adaptive sensitive reweighting to mitigate bias in fairness-aware classification In Proc. 2018 World Wide Web Conf. 853–862 (2018). [Google Scholar]
- 71.Jiang H & Nachum O Identifying and correcting label bias in machine learning In Int. Conf. Artificial Intelligence and Statistics 702–712 (PMLR, 2020). [Google Scholar]
- 72.Chai X et al. Unsupervised domain adaptation techniques based on auto-encoder for non-stationary eeg-based emotion recognition. Comput. Biol. Med 79, 205–214 (2016). [DOI] [PubMed] [Google Scholar]
- 73.Kamishima T, Akaho S, Asoh H & Sakuma J Fairness-aware classifier with prejudice remover regularizer In Joint Eur. Conf. Machine Learning and Knowledge Discovery in Databases 35–50 (Springer, 2012). [Google Scholar]
- 74.Zafar MB, Valera I, Rogriguez MG & Gummadi KP Fairness constraints: mechanisms for fair classification. In Artificial Intelligence and Statistics 962–970 (PMLR, 2017). [Google Scholar]
- 75.Goel N, Yaghini M & Faltings B Non-discriminatory machine learning through convex fairness criteria In 32nd AAAI Conference on Artificial Intelligence (2018). [Google Scholar]
- 76.Goh G, Cotter A, Gupta M & Friedlander MP Satisfying real-world goals with dataset constraints. In Adv. Neural Information Processing Systems (2016). [Google Scholar]
- 77.Agarwal A et al. A reductions approach to fair classification In Int. Conf. Machine Learning 60–69 (PMLR, 2018). [Google Scholar]
- 78.Corbett-Davies S, Pierson E, Feller A, Goel S & Huq A Algorithmic decision making and the cost of fairness In Proc. 23rd ACM SIGKDD Int. Conf. Knowledge Discovery and Data Mining 797–806 (2017). [Google Scholar]
- 79.Pleiss G, Raghavan M, Wu F, Kleinberg J & Weinberger KQ On fairness and calibration. In Adv. Neural Information Processing Systems (2017). [Google Scholar]
- 80.Chouldechova A, Benavides-Prado D, Fialko O & Vaithianathan R A case study of algorithm assisted decision making in child maltreatment hotline screening decisions In Conf. Fairness, Accountability and Transparency 134–148 (PMLR, 2018). [Google Scholar]
- 81.Abernethy J, Awasthi P, Kleindessner M, Morgenstern J & Zhang J Active sampling for min-max fairness In Int. Conf. Machine Learning 53–65, (PMLR, 2022). [Google Scholar]
- 82.Iosifidis V & Ntoutsi E Dealing with bias via data augmentation in supervised learning scenarios. In Proc. Int. Workshop on Bias in Information, Algorithms, and Systems (eds. Bates J et al. ) (2018). [Google Scholar]
- 83.Vodrahalli K, Li K & Malik J Are all training examples created equal? An empirical study Preprint at 10.48550/arXiv.1811.12569 (2018). [DOI] [Google Scholar]
- 84.Barocas S & Selbst AD Big data’s disparate impact. Calif. Law Rev 104, 671 (2016). [Google Scholar]
- 85.O’Neil C Weapons of Math Destruction: How Big Data Increases Inequality and Threatens Democracy (Crown, 2016). [Google Scholar]
- 86.Rezaei A, Liu A, Memarrast O & Ziebart BD Robust fairness under covariate shift In Proc. AAAI Conf. Artificial Intelligence 35, 9419–9427 (2021). [Google Scholar]
- 87.Alabi D, Immorlica N & Kalai A Unleashing linear optimizers for group-fair learning and optimization In Conf. Learning Theory 2043–2066 (PMLR, 2018). [Google Scholar]
- 88.Kearns M, Neel S, Roth A & Wu ZS Preventing fairness gerrymandering: auditing and learning for subgroup fairness In Int. Conf. Machine Learning 2564–2572 (PMLR, 2018). [Google Scholar]
- 89.Poplin R et al. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat. Biomed. Eng 2, 158–164 (2018). [DOI] [PubMed] [Google Scholar]
- 90.Babenko B et al. Detection of signs of disease in external photographs of the eyes via deep learning. Nat. Biomed. Eng 6, 1370–1383 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kamishima T, Akaho S & Sakuma J Fairness-aware learning through regularization approach In 2011 IEEE 11th Int. Conf. Data Mining Workshops 643–650 (IEEE, 2011). [Google Scholar]
- 92.Zafar MB, Valera I, Gomez Rodriguez M & Gummadi KP Fairness beyond disparate treatment & disparate impact: Learning classification without disparate mistreatment In Proc. 26th Int. Conf. World Wide Web 1171–1180 (2017). [Google Scholar]
- 93.Zemel R, Wu Y, Swersky K, Pitassi T & Dwork C Learning fair representations In Int. Conf. Machine Learning 325–333 (PMLR, 2013). [Google Scholar]
- 94.Kim M, Reingold O & Rothblum G Fairness through computationally-bounded awareness. In Adv. Neural Information Processing Systems (2018). [Google Scholar]
- 95.Pfohl SR, Foryciarz A & Shah NH An empirical characterization of fair machine learning for clinical risk prediction. J. Biomed. Inform 113, 103621 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Foryciarz A, Pfohl SR, Patel B & Shah N Evaluating algorithmic fairness in the presence of clinical guidelines: the case of atherosclerotic cardiovascular disease risk estimation. BMJ Health Care Inf 29, e100460 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Muntner P et al. Potential US population impact of the 2017 ACC/AHA high blood pressure guideline. Circulation 137, 109–118 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen I, Johansson FD & Sontag D Why is my classifier discriminatory? In Adv. Neural Information Processing Systems (2018). [Google Scholar]
- 99.Raji ID & Buolamwini J Actionable auditing: investigating the impact of publicly naming biased performance results of commercial AI products In Proc. 2019 AAAI/ACM Conf. AI, Ethics, and Society 429–435 (2019). [Google Scholar]
- 100.Rolf E, Worledge T, Recht B & Jordan MI Representation matters: assessing the importance of subgroup allocations in training data In Int. Conf. Machine Learning 9040–9051 (2021). [Google Scholar]
- 101.Zhao H & Gordon G Inherent tradeoffs in learning fair representations. In Adv. Neural Information Processing Systems 32, 15675–15685 (2019). [Google Scholar]
- 102.Pfohl S et al. Creating fair models of atherosclerotic cardiovascular disease risk In Proc. 2019 AAAI/ACM Conf. AI, Ethics, and Society 271–278 (2019). [Google Scholar]
- 103.Pfohl SR Recommendations for Algorithmic Fairness Assessments of Predictive Models in Healthcare: Evidence from Large-scale Empirical Analyses PhD thesis, Stanford Univ. (2021). [Google Scholar]
- 104.Singh H, Singh R, Mhasawade V & Chunara R Fairness violations and mitigation under covariate shift In Proc. 2021 ACM Conf. Fairness, Accountability, and Transparency 3–13 (2021). [Google Scholar]
- 105.Biswas A & Mukherjee S Ensuring fairness under prior probability shifts In Proc. 2021 AAAI/ACM Conf. AI, Ethics, and Society 414–424 (2021). [Google Scholar]
- 106.Giguere S et al. Fairness guarantees under demographic shift In Int. Conf. Learning Representations (2021). [Google Scholar]
- 107.Mishler A & Dalmasso N Fair when trained, unfair when deployed: observable fairness measures are unstable in performative prediction settings Preprint at 10.48550/arXiv.2202.05049 (2022). [DOI] [Google Scholar]
- 108.Duchi J & Namkoong H Learning models with uniform performance via distributionally robust optimization. Ann. Stat 49, 1378–1406 (2021). [Google Scholar]
- 109.Hashimoto T, Srivastava M, Namkoong H & Liang P Fairness without demographics in repeated loss minimization In Int. Conf. Machine Learning 1929–1938 (PMLR, 2018). [Google Scholar]
- 110.Wang S et al. Robust optimization for fairness with noisy protected groups. In Adv. Neural Information Processing Systems 33, 5190–5203 (2020). [Google Scholar]
- 111.Coston A et al. Fair transfer learning with missing protected attributes In Proc. 2019 AAAI/ACM Conf. AI, Ethics, and Society 91–98 (2019). [Google Scholar]
- 112.Schumann C et al. Transfer of machine learning fairness across domains In NeurIPS AI for Social Good Workshop (2019). [Google Scholar]
- 113.Lahoti P et al. Fairness without demographics through adversarially reweighted learning. In Adv. Neural Information Processing Systems 33, 728–740 (2020). [Google Scholar]
- 114.Yan S, Kao H. t. & Ferrara E Fair class balancing: enhancing model fairness without observing sensitive attributes In Proc. 29th ACM Int. Conf. Information and Knowledge Management 1715–1724 (2020). [Google Scholar]
- 115.Zhao T, Dai E, Shu K & Wang S Towards fair classifiers without sensitive attributes: exploring biases in related features In Proc. 15th ACM Int. Conf. Web Search and Data Mining 1433–1442 (2022). [Google Scholar]
- 116.Quinonero-Candela J, Sugiyama M, Lawrence ND & Schwaighofer A Dataset Shift in Machine Learning (MIT Press, 2009). [Google Scholar]
- 117.Subbaswamy A, Schulam P & Saria S Preventing failures due to dataset shift: learning predictive models that transport In 22nd Int. Conf. Artificial Intelligence and Statistics 3118–3127 (PMLR, 2019). [Google Scholar]
- 118.Subbaswamy A & Saria S From development to deployment: dataset shift, causality, and shift-stable models in health AI. Biostatistics 21, 345–352 (2020). [DOI] [PubMed] [Google Scholar]
- 119.Guo LL et al. Evaluation of domain generalization and adaptation on improving model robustness to temporal dataset shift in clinical medicine. Sci. Rep 12, 2726 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Singh H, Singh R, Mhasawade V & Chunara R Fair predictors under distribution shift In NeurIPS Workshop on Fair ML for Health (2019). [Google Scholar]
- 121.Bernhardt M, Jones C & Glocker B Investigating underdiagnosis of ai algorithms in the presence of multiple sources of dataset bias. Nat. Med 28, 1157–1158 (2022). [DOI] [PubMed] [Google Scholar]
- 122.Ghosh A & Shanbhag A FairCanary: rapid continuous explainable fairness In Proc. AAAI/ACM Conf. AI, Ethics, and Society (2022). [Google Scholar]
- 123.Sagawa S, Koh PW, Hashimoto TB & Liang P Distributionally robust neural networks In Int. Conf. Learning Representations (2020). [Google Scholar]
- 124.Yang Y, Zhang H, Katabi D & Ghassemi M Change is hard: a closer look at subpopulation shift In Int. Conf. Machine Learning (2023). [Google Scholar]
- 125.Zong Y, Yang Y & Hospedales T MEDFAIR: benchmarking fairness for medical imaging In Int. Conf. Learning Representations (2023). [Google Scholar]
- 126.Lipkova J et al. Deep learning-enabled assessment of cardiac allograft rejection from endomyocardial biopsies. Nat. Med 28, 575–582 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tedeschi P & Griffith JR Classification of hospital patients as ‘surgical’ Implications of the shift to ICD-9-CM. Med. Care 22, 189–192 (1984). [DOI] [PubMed] [Google Scholar]
- 128.Heslin KC et al. Trends in opioid-related inpatient stays shifted after the US transitioned to ICD-10-CM diagnosis coding in 2015. Med. Care 55, 918–923 (2017). [DOI] [PubMed] [Google Scholar]
- 129.Castro DC, Walker I & Glocker B Causality matters in medical imaging. Nat. Commun 11, 3673 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gao J et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bejnordi BE et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 318, 2199–2210 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Campanella G et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med 25, 1301–1309 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wen D et al. Characteristics of publicly available skin cancer image datasets: a systematic review. Lancet Digit. Health 4, e64–e74 (2021). [DOI] [PubMed] [Google Scholar]
- 134.Khan SM et al. A global review of publicly available datasets for ophthalmological imaging: barriers to access, usability, and generalisability. Lancet Digit. Health 3, e51–e66 (2021). [DOI] [PubMed] [Google Scholar]
- 135.Mamary AJ et al. Race and gender disparities are evident in COPD underdiagnoses across all severities of measured airflow obstruction. Chronic Obstr. Pulm. Dis 5, 177 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Seyyed-Kalantari L, Zhang H, McDermott M, Chen IY & Ghassemi M Reply to: ‘potential sources of dataset bias complicate investigation of underdiagnosis by machine learning algorithms’ and ‘confounding factors need to be accounted for in assessing bias by machine learning algorithms’. Nat. Med 28, 1161–1162 (2022). [DOI] [PubMed] [Google Scholar]
- 137.Landry LG, Ali N, Williams DR, Rehm HL & Bonham VL Lack of diversity in genomic databases is a barrier to translating precision medicine research into practice. Health Aff 37, 780–785 (2018). [DOI] [PubMed] [Google Scholar]
- 138.Gusev A et al. Atlas of prostate cancer heritability in European and African-American men pinpoints tissue-specific regulation. Nat. Commun 7, 10979 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hinch AG et al. The landscape of recombination in African Americans. Nature 476, 170–175 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shriver MD et al. Skin pigmentation, biogeographical ancestry and admixture mapping. Hum. Genet 112, 387–399 (2003). [DOI] [PubMed] [Google Scholar]
- 141.Sudlow C et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 12, e1001779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Puyol-Anton E et al. Fairness in cardiac MR image analysis: an investigation of bias due to data imbalance in deep learning based segmentation. Med. Image Comput. Computer Assist. Intervention 24, 413–423 (2021). [Google Scholar]
- 143.Kraft SA et al. Beyond consent: building trusting relationships with diverse populations in precision medicine research. Am. J. Bioeth 18, 3–20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.West KM, Blacksher E & Burke W Genomics, health disparities, and missed opportunities for the nation’s research agenda. JAMA 317, 1831–1832 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mahal BA et al. Racial differences in genomic profiling of prostate cancer. N. Engl. J. Med 383, 1083–1085 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shi Y et al. A prospective, molecular epidemiology study of EGFR mutations in asian patients with advanced non–small-cell lung cancer of adenocarcinoma histology (PIONEER). J. Thorac. Oncol 9, 154–162 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Spratt DE et al. Racial/ethnic disparities in genomic sequencing. JAMA Oncol 2, 1070–1074 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang G et al. Characterization of frequently mutated cancer genes in chinese breast tumors: a comparison of chinese and TCGA cohorts. Ann. Transl. Med 7, 179 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zavala VA et al. Cancer health disparities in racial/ethnic minorities in the United States. Br. J. Cancer 124, 315–332 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhang W, Edwards A, Flemington EK & Zhang K Racial disparities in patient survival and tumor mutation burden, and the association between tumor mutation burden and cancer incidence rate. Sci. Rep 7, 13639 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ooi SL, Martinez ME & Li CI Disparities in breast cancer characteristics and outcomes by race/ethnicity. Breast Cancer Res. Treat 127, 729–738 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Henderson BE, Lee NH, Seewaldt V & Shen H The influence of race and ethnicity on the biology of cancer. Nat. Rev. Cancer 12, 648–653 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gamble P et al. Determining breast cancer biomarker status and associated morphological features using deep learning. Commun. Med 1, 1–12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Borrell LN et al. Race and genetic ancestry in medicine—a time for reckoning with racism. N. Engl. J. Med 384, 474–480 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Martini R, Newman L & Davis M Breast cancer disparities in outcomes; unmasking biological determinants associated with racial and genetic diversity. Clin. Exp. Metastasis 39, 7–14 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Martini R et al. African ancestry–associated gene expression profiles in triple-negative breast cancer underlie altered tumor biology and clinical outcome in women of African descent. Cancer Discov. 12, 2530–2551 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Herbst RS et al. Atezolizumab for first-line treatment of PD-L1–selected patients with NSCLC. N. Engl. J. Med 383, 1328–1339 (2020). [DOI] [PubMed] [Google Scholar]
- 158.Clarke MA, Devesa SS, Hammer A & Wentzensen N Racial and ethnic differences in hysterectomy-corrected uterine corpus cancer mortality by stage and histologic subtype. JAMA Oncol 8, 895–903 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yeyeodu ST, Kidd LR & Kimbro KS Protective innate immune variants in racial/ethnic disparities of breast and prostate cancer. Cancer Immunol. Res 7, 1384–1389 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yang W et al. Sex differences in gbm revealed by analysis of patient imaging, transcriptome, and survival data. Sci. Transl. Med 11, eaao5253 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Carrano A, Juarez JJ, Incontri D, Ibarra A & Cazares HG Sex-specific differences in glioblastoma. Cells 10, 1783 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Creed JH et al. Commercial gene expression tests for prostate cancer prognosis provide paradoxical estimates of race-specific risk. Cancer Epidemiol. Biomark. Prev 29, 246–253 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Burlina P, Joshi N, Paul W, Pacheco KD & Bressler NM Addressing artificial intelligence bias in retinal diagnostics. Transl. Vis. Sci. Technol 10, 13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kakadekar A, Greene DN, Schmidt RL, Khalifa MA & Andrews AR Nonhormone-related histologic findings in postsurgical pathology specimens from transgender persons: a systematic review. Am. J. Clin. Pathol 157, 337–344 (2022). [DOI] [PubMed] [Google Scholar]
- 165.Lu MY et al. AI-based pathology predicts origins for cancers of unknown primary. Nature 594, 106–110 (2021). [DOI] [PubMed] [Google Scholar]
- 166.Kather JN et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med 25, 1054–1056 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Echle A et al. Deep learning in cancer pathology: a new generation of clinical biomarkers. Br. J. Cancer 124, 686–696 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Dwork C, Immorlica N, Kalai AT & Leiserson M Decoupled classifiers for fair and efficient machine learning In Conf. Fairness, Accountability and Transparency (PMLR, 2018). [Google Scholar]
- 169.Lipton Z, McAuley J & Chouldechova A Does mitigating ml’s impact disparity require treatment disparity? In Adv. Neural Information Processing Systems (2018). [Google Scholar]
- 170.Madras D, Creager E, Pitassi T & Zemel R Fairness through causal awareness: learning causal latent-variable models for biased data In Proc. Conf. Fairness, Accountability, and Transparency 349–358 (2019). [Google Scholar]
- 171.Lohaus M, Kleindessner M, Kenthapadi K, Locatello F & Russell C Are two heads the same as one? Identifying disparate treatment in fair neural networks. In Adv. Neural Information Processing Systems (2022). [Google Scholar]
- 172.McCarty CA et al. The emerge network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med. Genet 4, 1–11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Gottesman O et al. The electronic medical records and genomics (emerge) network: past, present, and future. Genet. Med 15, 761–771 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Duncan L et al. Analysis of polygenic risk score usage and performance in diverse human populations. Nat. Commun 10, 3328 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lam M et al. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat. Genet 51, 1670–1678 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Martin AR et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet 51, 584–591 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Manrai AK et al. Genetic misdiagnoses and the potential for health disparities. N. Engl. J. Med 375, 655–665 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Dehkharghanian T et al. Biased data, biased AI: deep networks predict the acquisition site of TCGA images. Diagn. Pathol 18, 1–12 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ganin Y & Lempitsky V Unsupervised domain adaptation by backpropagation In Int. Conf. Machine Learning 1180–1189 (PMLR, 2015). [Google Scholar]
- 180.Shaban MT, Baur C, Navab N & Albarqouni S StainGAN: stain style transfer for digital histological images In 2019 IEEE 16th Int. Symp. Biomedical Imaging (ISBI 2019) 953–956 (IEEE, 2019). [Google Scholar]
- 181.Widmer G & Kubat M Learning in the presence of concept drift and hidden contexts. Mach. Learn 23, 69–101 (1996). [Google Scholar]
- 182.Schlimmer JC & Granger RH Incremental learning from noisy data. Mach. Learn 1, 317–354 (1986). [Google Scholar]
- 183.Lu J et al. Learning under concept drift: a review. IEEE Trans. Knowl. Data Eng 31, 2346–2363 (2018). [Google Scholar]
- 184.Guo LL et al. Systematic review of approaches to preserve machine learning performance in the presence of temporal dataset shift in clinical medicine. Appl. Clin. Inform 12, 808–815 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Barocas S et al. Designing disaggregated evaluations of AI systems: choices, considerations, and tradeoffs In Proc. 2021 AAAI/ACM Conf. AI, Ethics, and Society 368–378 (2021). [Google Scholar]
- 186.Zhou H, Chen Y & Lipton ZC Evaluating model performance in medical datasets over time In Proc. Conf. Health, Inference, and Learning (2023). [Google Scholar]
- 187.Scholkopf B et al. On causal and anticausal learning In Int. Conf. Machine Learning (2012). [Google Scholar]
- 188.Lipton Z, Wang Y-X & Smola A Detecting and correcting for label shift with black box predictors In Int. Conf. Machine Learning 3122–3130 (PMLR, 2018). [Google Scholar]
- 189.Loupy A, Mengel M & Haas M Thirty years of the international banff classification for allograft pathology: the past, present, and future of kidney transplant diagnostics. Kidney Int 101, 678–691 (2022). [DOI] [PubMed] [Google Scholar]
- 190.Delahunt B et al. Gleason and Fuhrman no longer make the grade. Histopathology 68, 475–481 (2016). [DOI] [PubMed] [Google Scholar]
- 191.Davatchi F et al. The saga of diagnostic/classification criteria in Behcet’s disease. Int. J. Rheum. Dis 18, 594–605 (2015). [DOI] [PubMed] [Google Scholar]
- 192.Louis DN et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 131, 803–820 (2016). [DOI] [PubMed] [Google Scholar]
- 193.Bifet A & Gavalda R Learning from time-changing data with adaptive windowing In Proc. 2007 SIAM International Conference on Data Mining 443–448 (SIAM, 2007). [Google Scholar]
- 194.Nigenda D et al. Amazon SageMaker Model Monitor: a system for real-time insights into deployed machine learning models In Proc. 28th ACM SIGKDD Conf. Knowledge Discovery and Data Mining (2022). [Google Scholar]
- 195.Miroshnikov A, Kotsiopoulos K, Franks R & Kannan AR Wasserstein-based fairness interpretability framework for machine learning models. Mach. Learn 111, 3307–3357 (2022). [Google Scholar]
- 196.Board AE AAA statement on race. Am. Anthropol 100, 712–713 (1998). [Google Scholar]
- 197.Oni-Orisan A, Mavura Y, Banda Y, Thornton TA & Sebro R Embracing genetic diversity to improve black health. N. Engl. J. Med 384, 1163–1167 (2021). [DOI] [PubMed] [Google Scholar]
- 198.Calhoun A The pathophysiology of racial disparities. N. Engl. J. Med 384, e78 (2021). [DOI] [PubMed] [Google Scholar]
- 199.Sun R et al. Don’t ignore genetic data from minority populations. Nature 585, 184–186 (2020). [DOI] [PubMed] [Google Scholar]
- 200.Lannin DR et al. Influence of socioeconomic and cultural factors on racial differences in late-stage presentation of breast cancer. JAMA 279, 1801–1807 (1998). [DOI] [PubMed] [Google Scholar]
- 201.Bao M et al. It’s COMPASlicated: the messy relationship between RAI datasets and algorithmic fairness benchmarks In 35th Conf. Neural Information Processing Systems Datasets and Benchmarks (2021). [Google Scholar]
- 202.Hao M et al. Efficient and privacy-enhanced federated learning for industrial artificial intelligence. IEEE Trans. Ind. Inf 16, 6532–6542 (2019). [Google Scholar]
- 203.Yang Q, Liu Y, Chen T & Tong Y Federated machine learning: concept and applications. ACM Trans. Intell. Syst. Technol 10, 1–19 (2019). [Google Scholar]
- 204.Bonawitz K et al. Practical secure aggregation for privacy-preserving machine learning In Proc. 2017 ACM SIGSAC Conf. Computer and Communications Security 1175–1191 (2017). [Google Scholar]
- 205.Bonawitz K et al. Towards federated learning at scale: system design. In Proc. Mach. Learn. Syst 1, 374–388 (2019). [Google Scholar]
- 206.Brisimi TS et al. Federated learning of predictive models from federated electronic health records. Int. J. Med. Inform 112, 59–67 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Huang L et al. Patient clustering improves efficiency of federated machine learning to predict mortality and hospital stay time using distributed electronic medical records. J. Biomed. Inform 99, 103291 (2019). [DOI] [PubMed] [Google Scholar]
- 208.Xu J et al. Federated learning for healthcare informatics. J. Healthc. Inform. Res 5, 1–19 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Chakroborty S, Patel KR & Freytag A Beyond federated learning: fusion strategies for diabetic retinopathy screening algorithms trained from different device types. Invest. Ophthalmol. Vis. Sci 62, 85–85 (2021). [Google Scholar]
- 210.Ju C et al. Federated transfer learning for EEG signal classification In 42nd Annu. Int. Conf. IEEE Engineering in Medicine and Biology Society 3040–3045 (IEEE, 2020). [DOI] [PubMed] [Google Scholar]
- 211.Li W et al. Privacy-preserving federated brain tumour segmentation. In Int. Workshop on Machine Learning in Medical Imaging 133–141 (Springer, 2019). [Google Scholar]
- 212.Kaissis G et al. End-to-end privacy preserving deep learning on multi-institutional medical imaging. Nat. Mach. Intell 3, 473–484 (2021). [Google Scholar]
- 213.Rieke N et al. The future of digital health with federated learning. NPJ Digit. Med 3, 119 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Sheller MJ et al. Federated learning in medicine: facilitating multi-institutional collaborations without sharing patient data. Sci. Rep 10, 12598 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Choudhury O et al. Differential privacy-enabled federated learning for sensitive health data In Machine Learning for Health (ML4H) Workshop at NeurIPS (2019). [Google Scholar]
- 216.Kushida CA et al. Strategies for de-identification and anonymization of electronic health record data for use in multicenter research studies. Med. Care 50, S82–S101 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.van der Haak M et al. Data security and protection in cross-institutional electronic patient records. Int. J. Med. Inform 70, 117–130 (2003). [DOI] [PubMed] [Google Scholar]
- 218.Veale M & Binns R Fairer machine learning in the real world: mitigating discrimination without collecting sensitive data. Big Data Soc. 4, 2053951717743530 (2017). [Google Scholar]
- 219.Fiume M et al. Federated discovery and sharing of genomic data using beacons. Nat. Biotechnol 37, 220–224 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Sadilek A et al. Privacy-first health research with federated learning. NPJ Digit. Med 4, 132 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Duan R, Boland MR, Moore JH & Chen Y ODAL: a one-shot distributed algorithm to perform logistic regressions on electronic health records data from multiple clinical sites. In BIOCOMPUTING 2019: Proc. Pacific Symposium 30–41 (World Scientific, 2018). [PMC free article] [PubMed] [Google Scholar]
- 222.Sarma KV et al. Federated learning improves site performance in multicenter deep learning without data sharing. J. Am. Med. Inform. Assoc 28, 1259–1264 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Silva S et al. Federated learning in distributed medical databases: meta-analysis of large-scale subcortical brain data In 2019 IEEE 16th International Symposium on Biomedical Imaging 270–274 (IEEE, 2019). [Google Scholar]
- 224.Roy AG, Siddiqui S, Polsterl S, Navab N & Wachinger C BrainTorrent: a peer-to-peer environment for decentralized federated learning Preprint at 10.48550/arXiv.1905.06731 (2019). [DOI]
- 225.Lu MY et al. Federated learning for computational pathology on gigapixel whole slide images. Med. Image Anal 76, 102298 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Dou Q et al. Federated deep learning for detecting COVID-19 lung abnormalities in CT: a privacy-preserving multinational validation study. NPJ Digit. Med 4, 60 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Yang D et al. Federated semi-supervised learning for COVID region segmentation in chest CT using multinational data from China, Italy, Japan. Med. Image Anal 70, 101992 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Vaid A et al. Federated learning of electronic health records to improve mortality prediction in hospitalized patients with COVID-19: machine learning approach. JMIR Med. Inform 9, e24207 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Li S, Cai T & Duan R Targeting underrepresented populations in precision medicine: a federated transfer learning approach Preprint at 10.48550/arXiv.2108.12112 (2023). [DOI] [PMC free article] [PubMed]
- 230.Mandl KD et al. The genomics research and innovation network: creating an interoperable, federated, genomics learning system. Genet. Med 22, 371–380 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Cai M et al. A unified framework for cross-population trait prediction by leveraging the genetic correlation of polygenic traits. Am. J. Hum. Genet 108, 632–655 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Liang J, Hu D & Feng J Do we really need to access the source data? Source hypothesis transfer for unsupervised domain adaptation In Int. Conf. Machine Learning 6028–6039 (PMLR, 2020). [Google Scholar]
- 233.Song L, Ma C, Zhang G & Zhang Y Privacy-preserving unsupervised domain adaptation in federated setting. IEEE Access 8, 143233–143240 (2020). [Google Scholar]
- 234.Li X et al. Multi-site fMRI analysis using privacy-preserving federated learning and domain adaptation: ABIDE results. Med. Image Anal 65, 101765 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Peterson D, Kanani P & Marathe VJ Private federated learning with domain adaptation In Federated Learning for Data Privacy and Confidentiality Workshop in NeurIPS (2019). [Google Scholar]
- 236.Peng X, Huang Z, Zhu Y & Saenko K Federated adversarial domain adaptation In Int. Conf. Learning Representations (2020). [Google Scholar]
- 237.Yao C-H et al. Federated multi-target domain adaptation In Proc. IEEE/CVF Winter Conference on Applications of Computer Vision 1424–1433 (2022). [Google Scholar]
- 238.Li T, Sanjabi M, Beirami A & Smith V Fair resource allocation in federated learning In Int. Conf. Learning Representations (2020). [Google Scholar]
- 239.Mohri M, Sivek G & Suresh AT Agnostic federated learning In Int. Conf. Machine Learning 4615–4625 (PMLR, 2019). [Google Scholar]
- 240.Ezzeldin YH, Yan S, He C, Ferrara E & Avestimehr S FairFed: enabling group fairness in federated learning In Proc. AAAI Conf. Artificial Intelligence (2023). [Google Scholar]
- 241.Papadaki A, Martinez N, Bertran M, Sapiro G & Rodrigues M Minimax demographic group fairness in federated learning In ACM Conf. Fairness, Accountability, and Transparency 142–159 (2022). [Google Scholar]
- 242.Chen D, Gao D, Kuang W, Li Y & Ding B pFL-Bench: a comprehensive benchmark for personalized federated learning In 36th Conf. Neural Information Processing Systems Datasets and Benchmarks Track (2022). [Google Scholar]
- 243.Chai J & Wang X Self-supervised fair representation learning without demographics. In Adv. Neural Information Processing Systems (2022). [Google Scholar]
- 244.coj/>Jiang M et al. Fair federated medical image segmentation via client contribution estimation In Proc. IEEE/CVF Conf. Computer Vision and Pattern Recognition 16302–16311 (2023). [Google Scholar]
- 245.Jiang M, Wang Z & Dou Q Harmofl: harmonizing local and global drifts in federated learning on heterogeneous medical images In Proc. AAAI Conf. Artificial Intelligence 1087–1095 (2022). [Google Scholar]
- 246.Xu YY, Lin CS and Wang YCF Bias-eliminating augmentation learning for debiased federated learning In Proc. IEEE/CVF Conf. Computer Vision and Pattern Recognition 20442–20452 (2023). [Google Scholar]
- 247.Zhao Y et al. Federated learning with non-IID data Preprint at 10.48550/arXiv.1806.00582 (2018). [DOI]
- 248.Konečný J et al. Federated learning: strategies for improving communication efficiency Preprint at 10.48550/arXiv.1610.05492 (2016). [DOI]
- 249.Lin Y, Han S, Mao H, Wang Y & Dally WJ Deep gradient compression: reducing the communication bandwidth for distributed training In Int. Conf. Learning Representations (2018). [Google Scholar]
- 250.McMahan B, et al. Communication-efficient learning of deep networks from decentralized data. In Artificial Intelligence and Statistics 1273–1282 (PMLR, 2017). [Google Scholar]
- 251.Li T et al. Federated optimization in heterogeneous networks. In Proc. Mach. Learn. Syst 2, 429–450 (2020). [Google Scholar]
- 252.Sattler F, Wiedemann S, Muller K-R & Samek W Robust and communication-efficient federated learning from non-iid data. In IEEE Trans. Neural Netw Learn. Syst. 31, 3400–3413 (2019). [DOI] [PubMed] [Google Scholar]
- 253.Abay A et al. Mitigating bias in federated learning Preprint at 10.48550/arXiv.2012.02447 (2020). [DOI]
- 254.Luo Z, Wang Y, Wang Z, Sun Z & Tan T Disentangled federated learning for tackling attributes skew via invariant aggregation and diversity transferring In Int. Conf. Machine Learning 14527–14541 (PMLR, 2022). [Google Scholar]
- 255.McNamara D, Ong CS & Williamson RC Costs and benefits of fair representation learning In Proc. 2019 AAAI/ACM Conference on AI, Ethics, and Society 263–270 (2019). [Google Scholar]
- 256.Madaio MA, Stark L, Wortman Vaughan J & Wallach H Co-designing checklists to understand organizational challenges and opportunities around fairness in AI In Proc. 2020 CHI Conf. Human Factors in Computing Systems (2020). [Google Scholar]
- 257.Jung K et al. A framework for making predictive models useful in practice. J. Am. Med. Inform. Assoc 28, 1149–1158 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Pogodin R et al. Efficient conditionally invariant representation learning In Int. Conf. Learning Representations (2023). [Google Scholar]
- 259.Louizos C et al. Causal effect inference with deep latent-variable models. In Adv. Neural Information Processing Systems (2017). [Google Scholar]
- 260.Shi C, Blei D & Veitch V Adapting neural networks for the estimation of treatment effects. In Adv. Neural Information Processing Systems (2019). [Google Scholar]
- 261.Yoon J, Jordon J & Van Der Schaar M GANITE: estimation of individualized treatment effects using generative adversarial nets In Int. Conf. Learning Representations (2018). [Google Scholar]
- 262.Rezaei A, Fathony R, Memarrast O & Ziebart B Fairness for robust log loss classification In Proc. AAAI Conf. Artificial Intelligence 34, 5511–5518 (2020). [Google Scholar]
- 263.Petrović A, Nikolić M, Radovanović S, Delibašić B & Jovanović M FAIR: Fair adversarial instance re-weighting. Neurocomputing 476, 14–37 (2020). [Google Scholar]
- 264.Sattigeri P, Hoffman SC, Chenthamarakshan V & Varshney KR Fairness GAN: generating datasets with fairness properties using a generative adversarial network. IBM J. Res. Dev 63, 3:1–3:9 (2019). [Google Scholar]
- 265.Xu D, Yuan S, Zhang L & Wu X FairGAN: fairness-aware generative adversarial networks In 2018 IEEE International Conference on Big Data 570–575 (IEEE, 2018). [Google Scholar]
- 266.Xu H, Liu X, Li Y, Jain A & Tang J To be robust or to be fair: towards fairness in adversarial training In Int. Conf. Machine Learning 11492–11501 (PMLR, 2021). [Google Scholar]
- 267.Wadsworth C, Vera F & Piech C Achieving fairness through adversarial learning: an application to recidivism prediction In FAT/ML Workshop (2018). [Google Scholar]
- 268.Adel T, Valera I, Ghahramani Z & Weller A One-network adversarial fairness In Proc. AAAI Conf. Artificial Intelligence 33, 2412–2420 (2019). [Google Scholar]
- 269.Madras D, Creager E, Pitassi T & Zemel R Learning adversarially fair and transferable representations In Int. Conf. Machine Learning 3384–3393 (PMLR, 2018). [Google Scholar]
- 270.Madras D, Creager E, Pitassi T & Zemel R Learning adversarially fair and transferable representations In Proc. 35th Int. Conf. Machine Learning (eds. Dy J & Krause A) 3384–3393 (PMLR, 2018). [Google Scholar]
- 271.Chen X, Fain B, Lyu L & Munagala K Proportionally fair clustering In Proc. 36th Int. Conf. Machine Learning (eds. Chaudhuri K & Salakhutdinov R) 1032–1041 (PMLR, 2019). [Google Scholar]
- 272.Li P, Zhao H & Liu H Deep fair clustering for visual learning In Proc. IEEE/CVF Conf. Computer Vision and Pattern Recognition 9070–9079 (2020). [Google Scholar]
- 273.Hong J et al. Federated adversarial debiasing for fair and transferable representations In Proc. 27th ACM SIGKDD Conf. Knowledge Discovery and Data Mining 617–627 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Qi T et al. FairVFL: a fair vertical federated learning framework with contrastive adversarial learning. In Adv. Neural Information Processing Systems (2022). [Google Scholar]
- 275.Chen Y, Raab R, Wang J & Liu Y Fairness transferability subject to bounded distribution shift. In Adv. Neural Information Processing Systems (2022). [Google Scholar]
- 276.An B, Che Z, Ding M & Huang F Transferring fairness under distribution shifts via fair consistency regularization. In Adv. Neural Information Processing Systems (2022). [Google Scholar]
- 277.Giguere S et al. Fairness guarantees under demographic shift In Int. Conf. Learning Representations (2022). [Google Scholar]
- 278.Schrouff J et al. Diagnosing failures of fairness transfer across distribution shift in real-world medical settings. In Adv. Neural Information Processing Systems (2022). [Google Scholar]
- 279.Lipkova J et al. Personalized radiotherapy design for glioblastoma: integrating mathematical tumor models, multimodal scans, and Bayesian inference. In IEEE Trans. Med. Imaging 38, 1875–1884 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Cen LP et al. Automatic detection of 39 fundus diseases and conditions in retinal photographs using deep neural networks. Nat. Commun 12, 4828 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Lézoray O, Revenu M & Desvignes M Graph-based skin lesion segmentation of multispectral dermoscopic images In IEEE Int. Conf. Image Processing 897–901 (2014). [Google Scholar]
- 282.Manica A, Prugnolle F & Balloux F Geography is a better determinant of human genetic differentiation than ethnicity. Hum. Genet 118, 366–371 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Hadad N, Wolf L & Shahar M A two-step disentanglement method In Proc. IEEE Conf. Computer Vision and Pattern Recognition 772–780 (2018). [Google Scholar]
- 284.Achille A & Soatto S Emergence of invariance and disentanglement in deep representations. J. Mach. Learn. Res 19, 1947–1980 (2018). [Google Scholar]
- 285.Chen RT, Li X, Grosse R & Duvenaud D Isolating sources of disentanglement in variational autoencoders. In Adv. Neural Information Processing Systems (2018). [Google Scholar]
- 286.Kim H & Mnih A Disentangling by factorising In Int. Conf. Machine Learning 2649–2658 (PMLR, 2018). [Google Scholar]
- 287.Higgins I et al. beta-VAE: learning basic visual concepts with a constrained variational framework In Int Conf. Learning Representations (2017). [Google Scholar]
- 288.Sarhan MH, Eslami A, Navab N & Albarqouni S Learning interpretable disentangled representations using adversarial VAEs. In Domain Adaptation and Representation Transfer and Medical Image Learning with Less Labels and Imperfect Data 37–44 (Springer, 2019). [Google Scholar]
- 289.Gyawali PK et al. Learning to disentangle inter-subject anatomical variations in electrocardiographic data. In IEEE Trans. Biomedical Engineering (IEEE, 2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Bing S, Fortuin V & Ratsch G On disentanglement in Gaussian process variational autoencoders In 4th Symp. Adv. Approximate Bayesian Inference (2021). [Google Scholar]
- 291.Xu Y, He H, Shen T & Jaakkola TS Controlling directions orthogonal to a classifier In Int. Conf. Learning Representations (2022). [Google Scholar]
- 292.Cisse M & Koyejo S Fairness and representation learning. In NeurIPS Invited Talk 2019; https://cs.stanford.edu/~sanmi/documents/Representation_Learning_Fairness_NeurIPS19_Tutorial.pdf (2019).
- 293.Creager E et al. Flexibly fair representation learning by disentanglement In Int. Conf. Machine Learning 1436–1445 (PMLR, 2019). [Google Scholar]
- 294.Locatello F et al. On the fairness of disentangled representations. In Adv. Neural Information Processing Systems (2019). [Google Scholar]
- 295.Lee J, Kim E, Lee J, Lee J & Choo J Learning debiased representation via disentangled feature augmentation. In Adv. Neural Information Processing Systems 34, 25123–25133 (2021). [Google Scholar]
- 296.Zhang YK, Wang QW, Zhan DC & Ye HJ Learning debiased representations via conditional attribute interpolation In Proc. IEEE/CVF Conf. Computer Vision and Pattern Recognition 7599–7608 (2023). [Google Scholar]
- 297.Tartaglione E, Barbano CA & Grangetto M End: entangling and disentangling deep representations for bias correction In Proc. IEEE/CVF Conf. Computer Vision and Pattern Recognition 13508–13517 (2021). [Google Scholar]
- 298.Bercea CI, Wiestler B, Rueckert D & Albarqouni S FedDis: disentangled federated learning for unsupervised brain pathology segmentation Preprint at 10.48550/arXiv.2103.03705 (2021). [DOI]
- 299.Ke J, Shen Y & Lu Y Style normalization in histology with federated learning In 2021 IEEE 18th Int. Symp. Biomedical Imaging 953–956 (IEEE, 2021). [Google Scholar]
- 300.Pfohl SR, Dai AM & Heller K Federated and differentially private learning for electronic health records In Machine Learning for Health (ML4H) Workshop at NeurIPS (2019). [Google Scholar]
- 301.Xin B et al. Private FL-GAN: differential privacy synthetic data generation based on federated learning In 2020 IEEE Int. Conf. Acoustics, Speech and Signal Processing 2927–2931 (IEEE, 2020). [Google Scholar]
- 302.Rajotte J-F et al. Reducing bias and increasing utility by federated generative modeling of medical images using a centralized adversary In Proc. Conf. Information Technology for Social Good 79–84 (2021). [Google Scholar]
- 303.Chen T, Kornblith S, Norouzi M & Hinton G A simple framework for contrastive learning of visual representations In Int. Conf. Machine Learning 1597–1607 (PMLR, 2020). [Google Scholar]
- 304.Shad R, Cunningham JP, Ashley EA, Langlotz CP & Hiesinger W Designing clinically translatable artificial intelligence systems for high-dimensional medical imaging. Nat. Mach. Intell 3, 929–935 (2021). [Google Scholar]
- 305.Jacovi A, Marasovic A, Miller T & Goldberg Y Formalizing trust in artificial intelligence: prerequisites, causes and goals of human trust in AI In Proc. 2021 ACM Conf. Fairness, Accountability, and Transparency 624–635 (2021). [Google Scholar]
- 306.Floridi L Establishing the rules for building trustworthy AI. Nat. Mach. Intell 1, 261–262 (2019). [Google Scholar]
- 307.High-Level Expert Group on Artificial Intelligence. Ethics Guidelines for Trustworthy AI (European Commission, 2019). [Google Scholar]
- 308.Simonyan K, Vedaldi A & Zisserman A Deep inside convolutional networks: visualising image classification models and saliency maps In Workshop at Int. Conf. Learning Representations (2014). [Google Scholar]
- 309.Selvaraju RR et al. Grad-CAM: visual explanations from deep networks via gradient-based localization In Proc. IEEE Int. Conf. Computer Vision 618–626 (2017). [Google Scholar]
- 310.Irvin J et al. CheXpert: a large chest radiograph dataset with uncertainty labels and expert comparison In Proc. AAAI Conf. Artificial Intelligence 33, 590–597 (2019). [Google Scholar]
- 311.Sayres R et al. Using a deep learning algorithm and integrated gradients explanation to assist grading for diabetic retinopathy. Ophthalmology 126, 552–564 (2019). [DOI] [PubMed] [Google Scholar]
- 312.Patro BN, Lunayach M, Patel S & Namboodiri VP U-CAM: visual explanation using uncertainty based class activation maps In Proc. IEEE/CVF Int. Conf. Computer Vision 7444–7453 (2019). [Google Scholar]
- 313.Grewal M, Srivastava MM, Kumar P & Varadarajan S RADNET: radiologist level accuracy using deep learning for hemorrhage detection in CT scans In 2018 IEEE 15th Int. Symp. Biomedical Imaging 281–284 (IEEE, 2018). [Google Scholar]
- 314.Arun NT et al. Assessing the validity of saliency maps for abnormality localization in medical imaging. In Medical Imaging with Deep Learning (2020). [Google Scholar]
- 315.Schlemper J et al. Attention-gated networks for improving ultrasound scan plane detection. In Medical Imaging with Deep Learning (2018). [Google Scholar]
- 316.Schlemper J et al. Attention gated networks: learning to leverage salient regions in medical images. Med. Image Anal 53, 197–207 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Mittelstadt B, Russell C & Wachter S Explaining explanations in AI In Proc. Conf. Fairness, Accountability, and Transparency 279–288 (2019). [Google Scholar]
- 318.Kindermans P-J et al. in Explainable AI: Interpreting, Explaining and Visualizing Deep Learning 267–280 (Springer, 2019). [Google Scholar]
- 319.Kaur H et al. Interpreting interpretability: understanding data scientists’ use of interpretability tools for machine learning In Proc. 2020 CHI Conf. Human Factors in Computing Systems (2020). [Google Scholar]
- 320.Adebayo J et al. Sanity checks for saliency maps. In Adv. Neural Information Processing Systems (2018). [Google Scholar]
- 321.Saporta A et al. Benchmarking saliency methods for chest X-ray interpretation. Nat. Mach. Intell 4, 867–878 (2022). [Google Scholar]
- 322.Chen RJ et al. Pan-cancer integrative histology-genomic analysis via multimodal deep learning. Cancer Cell 40, 865–878 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.DeGrave AJ, Janizek JD & Lee S-I AI for radiographic COVID-19 detection selects shortcuts over signal. Nat. Mach. Intell 3, 610–619 (2021). [Google Scholar]
- 324.Adebayo J, Muelly M, Liccardi I & Kim B Debugging tests for model explanations. In Adv. Neural Information Processing Syst 33, 700–712 (2020). [Google Scholar]
- 325.Lee MK & Rich K Who is included in human perceptions of AI?: Trust and perceived fairness around healthcare AI and cultural mistrust In Proc. 2021 CHI Conf. Human Factors in Computing Systems (2021). [Google Scholar]
- 326.Sundararajan M, Taly A & Yan Q Axiomatic attribution for deep networks In Int. Conf. Machine Learning 3319–3328 (PMLR, 2017). [Google Scholar]
- 327.Lundberg SM & Lee S-I A unified approach to interpreting model predictions In Proc. 31st Int. Conf. Neural Information Processing Systems 4768–4777 (2017). [Google Scholar]
- 328.Ghassemi M, Oakden-Rayner L & Beam AL The false hope of current approaches to explainable artificial intelligence in health care. Lancet Digit. Health 3, e745–e750 (2021). [DOI] [PubMed] [Google Scholar]
- 329.Kim GB, Gao Y, Palsson BO & Lee SY DeepTFactor: a deep learning-based tool for the prediction of transcription factors. Proc. Natl Acad. Sci. USA 118, e2021171118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Lundberg SM et al. Explainable machine-learning predictions for the prevention of hypoxaemia during surgery. Nat. Biomed. Eng 2, 749–760 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Qiu W et al. Interpretable machine learning prediction of all-cause mortality. Commun. Med 2, 125 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Janizek JD et al. Uncovering expression signatures of synergistic drug responses via ensembles of explainable machine-learning models. Nat. Biomed. Eng 10.1038/s41551-023-01034-0 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Wexler J, Pushkarna M, Robinson S, Bolukbasi T & Zaldivar A Probing ML models for fairness with the What-If tool and SHAP: hands-on tutorial In Proc. 2020 Conference on Fairness, Accountability, and Transparency 705 (2020). [Google Scholar]
- 334.Lundberg SM Explaining quantitative measures of fairness In Fair & Responsible AI Workshop @ CHI2020; https://scottlundberg.com/files/fairness_explanations.pdf (2020). [Google Scholar]
- 335.Cesaro J & Cozman FG Measuring unfairness through game-theoretic interpretability In Machine Learning and Knowledge Discovery in Databases: International Workshops of ECML PKDD (2019). [Google Scholar]
- 336.Meng C, Trinh L, Xu N & Liu Y Interpretability and fairness evaluation of deep learning models on MIMIC-IV dataset. Sci. Rep 12, 7166 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Panigutti C, Perotti A, Panisson A, Bajardi P & Pedreschi D FairLens: auditing black-box clinical decision support systems. Inf. Process. Manag 58, 102657 (2021). [Google Scholar]
- 338.Röösli E, Bozkurt S & Hernandez-Boussard T Peeking into a black box, the fairness and generalizability of a MIMIC-III benchmarking model. Sci. Data 9, 24 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Pan W, Cui S, Bian J, Zhang C & Wang F Explaining algorithmic fairness through fairnessaware causal path decomposition In Proc. 27th ACM SIGKDD Conf. Knowledge Discovery and Data Mining 1287–1297 (2021). [Google Scholar]
- 340.Agarwal C et al. Openxai: towards a transparent evaluation of model explanations. In Adv. Neural Information Processing Systems 35, 15784–15799 (2022). [Google Scholar]
- 341.Zhang H, Singh H, Ghassemi M & Joshi S “Why did the model fail?”: attributing model performance changes to distribution shifts In Int. Conf. Machine Learning (2023). [Google Scholar]
- 342.Ghorbani A & Zou J Data Shapley: equitable valuation of data for machine learning In Int. Conf. Machine Learn. 97, 2242–2251 (2019). [Google Scholar]
- 343.Pandl KD, Feiland F, Thiebes S & Sunyaev A Trustworthy machine learning for health care: scalable data valuation with the Shapley value In Proc. Conf. Health, Inference, and Learning 47–57 (2021). [Google Scholar]
- 344.Prakash EI, Shrikumar A & Kundaje A Towards more realistic simulated datasets for benchmarking deep learning models in regulatory genomics. In Machine Learning in Computational Biology 58–77 (2022). [Google Scholar]
- 345.Oktay O et al. Attention U-Net: learning where to look for the pancreas. In Medical Imaging with Deep Learning (2018). [Google Scholar]
- 346.Dosovitskiy A et al. An image is worth 16x16 words: transformers for image recognition at scale In Int. Conf. Learning Representations (2020). [Google Scholar]
- 347.Lu MY Data-efficient and weakly supervised computational pathology on whole-slide images. Nat. Biomed. Eng 5, 555–570 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Yufei C et al. Bayes-MIL: a new probabilistic perspective on attention-based multiple instance learning for whole slide images In Int. Conf. Learning Representations (2023). [Google Scholar]
- 349.Van Gansbeke W, Vandenhende S, Georgoulis S & Van Gool L Unsupervised semantic segmentation by contrasting object mask proposals In Proc. IEEE/CVF Int. Conf. Computer Vision 10052–10062 (2021). [Google Scholar]
- 350.Radford A et al. Learning transferable visual models from natural language supervision In Int. Conf. Machine Learning 8748–8763 (2021). [Google Scholar]
- 351.Wei J et al. Chain of thought prompting elicits reasoning in large language models. In Adv. Neural Information Processing Systems (2022). [Google Scholar]
- 352.Javed SA, Juyal D, Padigela H, Taylor-Weiner A & Yu L Additive MIL: intrinsically interpretable multiple instance learning for pathology. In Adv. Neural Information Processing Systems (2022). [Google Scholar]
- 353.Diao JA et al. Human-interpretable image features derived from densely mapped cancer pathology slides predict diverse molecular phenotypes. Nat. Commun 12, 1613 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Bhargava HK et al. Computationally derived image signature of stromal morphology is prognostic of prostate cancer recurrence following prostatectomy in african american patients. Clin. Cancer Res 26, 1915–1923 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Curtis JR et al. Population-based fracture risk assessment and osteoporosis treatment disparities by race and gender. J. Gen. Intern. Med 24, 956–962 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Liu J et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 173, 400–416 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Foley RN, Wang C & Collins AJ Cardiovascular risk factor profiles and kidney function stage in the US general population: the NHANES III study. In Mayo Clinic Proc 80, 1270–1277 (Elsevier, 2005). [DOI] [PubMed] [Google Scholar]
- 358.Nevitt M, Felson D & Lester G The osteoarthritis initiative. Protocol for the cohort study 1; https://nda.nih.gov/static/docs/StudyDesignProtocolAndAppendices.pdf (2006).
- 359.Vaughn IA, Terry EL, Bartley EJ, Schaefer N & Fillingim RB Racial-ethnic differences in osteoarthritis pain and disability: a meta-analysis. J. Pain 20, 629–644 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Rotemberg V et al. A patient-centric dataset of images and metadata for identifying melanomas using clinical context. Sci. Data 8, 34 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Kinyanjui NM et al. Estimating skin tone and effects on classification performance in dermatology datasets Preprint at 10.48550/arXiv.1910.13268 (2019). [DOI]
- 362.Kinyanjui NM et al. Fairness of classifiers across skin tones in dermatology In Int. Conf. Medical Image Computing and Computer-Assisted Intervention 320–329 (2020). [Google Scholar]
- 363.Chew EY et al. The Age-Related Eye Disease Study 2 (AREDS2): study design and baseline characteristics (AREDS2 report number 1). Ophthalmology 119, 2282–2289 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Joshi N & Burlina P AI fairness via domain adaptation Preprint at 10.48550/arXiv.2104.01109 (2021). [DOI]
- 365.Zhou Y et al. RadFusion: benchmarking performance and fairness for multi-modal pulmonary embolism detection from CT and EMR Preprint at 10.48550/arXiv.2111.11665 (2021). [DOI]
- 366.Edwards NJ et al. The CPTAC data portal: a resource for cancer proteomics research. J. Proteome Res 14, 2707–2713 (2015). [DOI] [PubMed] [Google Scholar]
- 367.Johnson AE et al. MIMIC-III, a freely accessible critical care database. Sci. Data 3, 160035 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Boag W, Suresh H, Celi LA, Szolovits P & Ghassemi M Racial disparities and mistrust in end-of-life care In Machine Learning for Healthcare Conf. 587–602 (PMLR, 2018). [Google Scholar]
- 369.Prosper AE et al. Association of inclusion of more black individuals in lung cancer screening with reduced mortality. JAMA Netw. Open 4, e2119629 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.National Lung Screening Trial Research Team. et al. The National Lung Screening Trial: overview and study design. Radiology 258, 243–253 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Colak E et al. The RSNA pulmonary embolism CT dataset. Radiol. Artif. Intell 3, e200254 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Gertych A, Zhang A, Sayre J, Pospiech-Kurkowska S & Huang H Bone age assessment of children using a digital hand atlas. Comput. Med. Imaging Graph 31, 322–331 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Jeong JJ et al. The EMory BrEast imaging Dataset (EMBED): a racially diverse, granular dataset of 3.4 million screening and diagnostic mammographic images. Radiol. Artif. Intell 5.1, e220047 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Pollard TJ et al. The eICU collaborative research database, a freely available multi-center database for critical care research. Sci. Data 5, 1–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Sheikhalishahi S, Balaraman V & Osmani V Benchmarking machine learning models on multicentre eICU critical care dataset. PLoS ONE 15, e0235424 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.El Emam K et al. De-identification methods for open health data: the case of the heritage health prize claims dataset. J. Med. Internet Res 14, e33 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Madras D, Pitassi T & Zemel R Predict responsibly: improving fairness and accuracy by learning to defer. In Adv. Neural Information Processing Systems (2018). [Google Scholar]
- 378.Louizos C, Swersky K, Li Y, Welling M & Zemel R The variational fair autoencoder In Int. Conf. Learning Representations (2016). [Google Scholar]
- 379.Raff E & Sylvester J Gradient reversal against discrimination Preprint at 10.48550/arXiv.1807.00392 (2018). [DOI]
- 380.Smith JW, Everhart J, Dickson W, Knowler W & Johannes R Using the ADAP learning algorithm to forecast the onset of diabetes mellitus. In Proc. Symp. Computer Applications in Medical Care 261–265 (1988). [Google Scholar]
- 381.Sharma S et al. Data augmentation for discrimination prevention and bias disambiguation In Proc. AAAI/ACM Conf. AI, Ethics, and Society 358–364 (Association for Computing Machinery, 2020). [Google Scholar]
- 382.International Warfarin Pharmacogenetics Consortium. et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N. Engl. J. Med 360, 753–764 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Kallus N, Mao X & Zhou A Assessing algorithmic fairness with unobserved protected class using data combination In Proc. 2020 Conf. Fairness, Accountability, and Transparency 110 (Association for Computing Machinery, 2020). [Google Scholar]
- 384.Gross RT Infant Health and Development Program (IHDP): Enhancing the Outcomes of Low Birth Weight, Premature Infants in the United States, 1985–1988 (Inter-university Consortium for Political and Social Research, 1993); https://www.icpsr.umich.edu/web/HMCA/studies/9795 [Google Scholar]
- 385.Madras D, Creager E, Pitassi T & Zemel R Fairness through causal awareness: learning causal latent-variable models for biased data In Proc. Conf. Fairness, Accountability, and Transparency 30, 349–358 (Association for Computing Machinery, 2019). [Google Scholar]
- 386.Weeks MR, Clair S, Borgatti SP, Radda K & Schensul JJ Social networks of drug users in high-risk sites: finding the connections. AIDS Behav 6, 193–206 (2002). [Google Scholar]
- 387.Kleindessner M, Samadi S, Awasthi P & Morgenstern J Guarantees for spectral clustering with fairness constraints In Int. Conf. Machine Learning 3458–3467 (2019). [Google Scholar]
- 388.Daneshjou R et al. Disparities in dermatology AI performance on a diverse, curated clinical image set. Sci. Adv 8, eabq6147 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Garg S, Balakrishnan S & Lipton ZC Domain adaptation under open set label shift. In Adv. Neural Information Processing Systems (2022). [Google Scholar]
- 390.Pham TH, Zhang X & Zhang P Fairness and accuracy under domain generalization In Int. Conf. Learning Representations (2023). [Google Scholar]
- 391.Barocas S, Hardt M & Narayanan A Fairness in machine learning. NIPS Tutor 1, 2017 (2017). [Google Scholar]
- 392.Liu LT, Simchowitz M & Hardt M The implicit fairness criterion of unconstrained learning In Int. Conf. Machine Learning 4051–4060 (PMLR, 2017). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


