Abstract
Many analytical methods used in gut microbiome research focus on either single bacterial taxa or the whole microbiome, ignoring multibacteria relationships (microbial cliques). We present a novel analytical approach to identify microbial cliques within the gut microbiome of children at 9–11 years associated with prenatal lead (Pb) exposure. Data came from a subset of participants (n = 123) in the Programming Research in Obesity, Growth, Environment and Social Stressors cohort. Pb concentrations were measured in maternal whole blood from the second and third trimesters of pregnancy. Stool samples collected at 9–11 years old underwent metagenomic sequencing to assess the gut microbiome. Using a novel analytical approach, Microbial Co-occurrence Analysis (MiCA), we paired a machine learning algorithm with randomization-based inference to first identify microbial cliques that were predictive of prenatal Pb exposure and then estimate the association between prenatal Pb exposure and microbial clique abundance. With second-trimester Pb exposure, we identified a two-taxa microbial clique that included Bifidobacterium adolescentis and Ruminococcus callidus and a three-taxa clique that also included Prevotella clara. Increasing second-trimester Pb exposure was associated with significantly increased odds of having the two-taxa microbial clique below the median relative abundance (odds ratio (OR) = 1.03, 95% confidence interval (CI) [1.01–1.05]). Using a novel combination of machine learning and causal inference, MiCA identified a significant association between second-trimester Pb exposure and the reduced abundance of a probiotic microbial clique within the gut microbiome in late childhood.
Keywords: Metal exposure, Gut microbiome, Machine learning, Causal inference, Microbial co-occurrence, Exposome, Probiotic
Short abstract
Emerging evidence suggests that the prenatal period may be an important window of metal exposure for the development of gut microbiome into childhood. Using a combination of machine learning and the causal inference framework, we evaluated the association between prenatal metal exposure and gut bacterial clique in late childhood.
Introduction
Human metal exposure has been long recognized as a public health threat, and recent studies suggest that one potential mechanism for its adverse health effects is through the gut microbiome.1,2 A growing body of animal and human studies have shown that exposure to heavy metals (e.g., As, Cd, and Pb) can alter the gut microbiome composition and metabolic function, reduce diversity, and select for antibiotic resistance.2−8 Human Pb exposure has been linked to shifts in the microbiome throughout the life course.9,10 Sitarik et al. found that prenatal Pb exposure measured in baby teeth was associated with decreased abundance of several Bacteroides species and an increased abundance of the pro-inflammatory genus Collinsella in the gut microbiome at 1 and 6 months old.11 Likewise, Shen et al. found that prenatal Pb exposure measured in maternal blood was associated with an increased abundance of Fusobacteriota in the gut microbiome of children 6–7 years old.12 A study out of our group (Eggers et al.) also found that Pb exposure in the second and third trimesters of pregnancy was associated with decreased abundance of several short-chain fatty-acid-producing bacteria in children 9–11 years of age.13 Taken together, these studies indicate that the prenatal period may be a particularly critical window of Pb exposure on the development of the human gut microbiome in childhood. Moreover, the link between prenatal Pb exposure and increased abundance of pro-inflammatory and potentially pathogenic bacteria and decreased abundance of probiotic bacteria suggests that these prenatal Pb exposures may lead to poor health outcomes later in childhood via these alterations to the gut microbiome. This association may be due to changes in the maternal gut microbiome or programming of the immune system during development. However, further investigation is needed to clarify the underlying biological mechanisms.
Each of these highlighted studies and the vast majority of epidemiological microbiome studies have used a whole microbiome and/or single taxa approach for investigation. They identified single taxa or the whole microbiome, as measured using diversity as an indicator associated with a specific exposure or outcome of interest. However, current literature from microbial ecology shows that bacteria (and other microbiome members) biochemically interact with one another and their host organisms at levels between 1-on-1 and the whole microbiome, i.e., groups. For instance, many gut microbes are unable to be cultured in the lab without other bacteria in coculture.14 In most cases, these bacteria do not need to be cocultured with everything from the gut microbiome, just one or two others. This group of bacteria within the microbiome, or as we call it, a "microbial clique", is overlooked by conventional microbiome epidemiology methods. There are studies of the human microbiome that use network analysis to investigate co-occurring microbes within the human microbiome;15,16 however, they are based on correlations and can be difficult to use inside of an epidemiologic framework to understand associations and account for confounding variables. Furthermore, the results of network analyses are often difficult to interpret. Unsupervised clustering analyses are used to identify clusters based on relative abundances.17 However, as the size of the clusters increases, interpretations of the clusters and understanding of the interrelation between the taxa become difficult; therefore, deriving any biological plausibility becomes challenging.
The ability to assess associations between exposures or outcomes of epidemiologic interest and microbial cliques within the human microbiome is an important gap in the field. However, finding microbial cliques associated with an outcome of interest is challenging because of (1) considerable computational complexity as the number of taxa increases and (2) limitations of the sample size in most human microbiome studies. Multiple methods exist where multiordered microbial cliques can be prespecified or hard-coded in the models; however, such strategies might ignore many plausible and informative combinations and could be underpowered due to restrictions on sample size.18−20 On the other hand, microbial cliques can be potentially discovered using projection-based dimensionality reduction techniques. However, the interpretations of the final products can be challenging to interpret and are often qualitative.21,22 Microbial cliques can be constructed through threshold-based relative abundance, which might aid in interpretation. Such threshold-based construction carries considerable similarity with toxicological threshold-based interactions.23−25 Tree-based machine learning models can provide a natural and computationally efficient solution to such construction.20 Even with a substantial number of taxa, these models can create multiple threshold-based combinations of taxa predictive of the outcome of interest. Still, the challenge remains in interpretability since most machine learning models are generally black-box, creating a tension between prediction quality and meaningful interpretability. Moreover, a highly predictive machine learning model may not be ideal for assessing associations.26 Thus, there is a need for methods that can investigate novel microbial cliques while reducing data dimensionality and assessing associations in an interpretable way.
In this study, we aim to identify microbial cliques within the gut microbiome of children 9–11 years old, in association with prenatal Pb exposure. To accomplish this, we used a novel statistical approach called Microbial Co-occurrence Analysis (MiCA), which combined interpretable machine learning and causal inference frameworks to first identify microbial cliques and then test their associations with prenatal Pb exposure.
Methods
Study Design
Data came from the PROGRESS cohort, based in Mexico City, Mexico. The study enrolled 948 pregnant women who gave birth through the Mexican Social Security System. Pregnant women completed study visits in the second trimester (2T) and the third trimester (3T) and at birth. The offspring were followed and completed study visits every 6 months during infancy and every two years after that. Surveys, physical exams, and psychological and behavioral assessments were conducted during each study visit. Biological specimens, including blood, were also collected. In addition, stool samples were collected for microbiome analysis from a subset of participants (n = 123) at ages 9–11 years old. The study protocol for PROGRESS was reviewed and approved by the Institutional Review Board (IRB) at the Icahn School of Medicine at Mount Sinai (ISMMS), and all three committees (Research, Ethics in Research, and Biosafety) were included in the IRB at the National Institute of Public Health in Cuernavaca, Mexico.
Pb Exposure Measurement
Maternal whole blood was drawn during 2T and 3T, and Pb exposure analysis was performed as previously described.27 Pb concentration was measured using inductively coupled plasma mass spectrometry (ICP-MS) at the trace metals laboratory at ISMMS.
Gut Microbiome Sample Collection and Processing
Microbiome sample collection was conducted as previously described.13 Briefly, whole stool samples were collected at home by participants and stored in the refrigerator until they were picked up by the PROGRESS field team and delivered to ABC Hospital in Mexico City for aliquoting, following the FAST protocol.28 Aliquots were stored at −70 °C and shipped to the Microbiome Translational Center at ISMMS, where they underwent DNA extraction and library prep in two separate batches (n = 50 and n = 73). Shotgun metagenomic sequencing was performed for each batch separately using an Illumina HiSeq. Sequencing reads were trimmed for quality using Trimmomatic,29 and bowtie230 was used to remove human reads. MetaPhlAn231 and StrainPhlAn32 were then used to determine microbial taxonomy, and HUMAnN233 was used with MetaCyc34 to profile microbial gene pathways.
Covariates
Several relevant covariates were considered in this analysis, including child sex, child age at the time of stool sample collection, maternal socioeconomic status (SES) during pregnancy, maternal age at birth, maternal body mass index (BMI) during pregnancy, and microbiome analysis batch. Maternal height and weight were measured at 2T and were used to calculate BMI. Maternal SES during pregnancy was assessed based on the 1994 Mexican Association of Intelligence Agencies Market and Opinion (AMAI) rule 13*6, where families were categorized into six levels of SES based on 13 questions about household characteristics. Most families in PROGRESS were low to middle SES; therefore, the six levels were condensed into three: lower, middle, and higher.35
Statistical Analysis
All analyses were conducted in R version 4.0.3; the false discovery rate was implemented to correct the raw p-values from multiple comparison errors. Any two-tailed p-value less than 0.05 was considered statistically significant.
Data Processing
Pb concentrations were log2 transformed to better meet distributional assumptions. Microbiome count data were converted to relative abundances for all analyses. The analysis included only those taxa with at least 5% relative abundance in both batches to account for analytical batch effects. The relative abundance data were not rescaled to reflect the contribution of the original distribution of the whole taxa. Further, all models were controlled for a batch indicator variable. Further modeling approaches to correct batch effects are described in the following subsection.
Microbiome Co-occurrence Analysis (MiCA)
MiCA was conducted in two stages to identify microbial cliques associated with prenatal Pb exposures. The first part of this algorithm used a machine learning-based prediction framework to discover microbial cliques predictive of Pb exposure. The next stage restored the directionality and estimated the association between Pb exposure and the joint-relative abundance of the discovered cliques using a causal inference (or simply a classical association-based) framework.
The microbial cliques were searched using repeated hold-out signed iterated random forest (rh-SiRF), where the outcome was prenatal Pb exposure and the predictors were relative abundances of the selected taxa. The SiRF (signed iterative random forest) algorithm combined a state-of-the-art predictive tool called “Iterative Random Forests” with Random Intersection Trees (RIT) to search for combinations of taxa predictive of Pb exposure.36−38 Instead of searching for all possible combinations, SiRF can tease out the most prevalent taxa combinations on the decision path. The algorithm begins with a simple random forest (RF) and then sequentially reweighs the predictive taxa to fit the iterative RFs. From the reweighed RF, decision rules are extracted and fed into a generalization of the RIT to discover microbial cliques from the decision paths. This algorithm introduces a bagging step to assess the “stability” of the discovered cliques estimated through many bootstrapped iterations. Therefore, the higher the stability of a discovered clique, the better. On top of the SIRF algorithm, we introduced a repeated hold-out step that randomly partitions the data in training and testing sets for better generalizability.39 The whole rh-SiRF algorithm is repeated 1000 times, although iterating more than 100 times should suffice.
The rh-SiRF discovers microbial cliques through decision paths, representing a collective form associated with the outcome rather than a particular functional form. Thus, the discovered cliques include information about directionality and relative abundance thresholds. Microbial cliques can include taxa with abundance thresholds in opposite directions and can include multiple (more than two) taxa. We fitted the SiRF algorithm in three ways: (1) trained the model on one batch and then tested it on another batch, (2) trained the model on randomly chosen 60% of the data and then tested it on the remaining 40%, and finally (3) repeated the rh-SiRF algorithm over 300 times with training and test data partitioning 60–40% irrespective of the batches. Microbial cliques were chosen based on having a stability of more than 75%, having a prevalence of more than 10%, being common to all data partitioning strategies, and having a higher than random chance of occurrence among the 300 repeated holdouts (i.e., the relative frequency of occurrence was more than 1%). While fitting the SiRF algorithm on the first two data partitioning strategies (i.e., training on one batch and testing on another and 60–40% data splitting), we calculated the exposure co-occurrence list to find the important cliques based on mutual co-occurrence. The idea of exposure co-occurrence list follows from the heuristics of distributed word representations (popularly known as word embedding) and is widely applied in tasks related to Natural Language Processing.40,41
For the next stage of association analysis, the discovered microbial cliques were extracted as indicator functions with respect to their median relative abundances. For example, a microbial clique of A and B, denoted by A-B-, implies that a lower relative abundance of A and B is predictive of prenatal Pb exposure, whereas A+B+ means that a higher relative abundance of A and B is predictive of prenatal Pb exposure. For ease of interpretation and generalizability, we converted A-B- to an indicator function with respect to their median relative abundances; i.e., for an individual, the indicator would be nonzero if both the taxa A and B had below median relative abundance. Otherwise, it would be zero. Note that microbial cliques discovered through this algorithm might have different directionalities (A+B- or A-B+). Further, the number of taxa in a clique can be more than two.
We implemented a randomization-based inference built upon the Rubin causal model to estimate the association between prenatal Pb exposure and the odds of microbial clique abundance below the median. First, a matched-sampling strategy was utilized to obtain similar covariate distributions between the binarized clique, below and above median relative abundance. Given the covariates, we assumed that this approach of covariate-balancing42 could create potential “exchangeable” groups so that the clique was hypothetically and randomly assigned to each individual, and those covariates did not confound the clique assignment. Next, due to the small sample size and to prevent discarding a large number of samples, a subclass matching with the propensity score43 was used to construct similar groups of microbial cliques with above and below median relative abundance. Finally, love plots of the differences in standardized means in covariates were used to examine the extent of covariate balancing (setting the threshold for the standardized mean difference to 0.1).44,45 We used logistic regression with matched microbial clique as the outcome and prenatal Pb concentration as the exposure after adjusting for covariates. Moreover, without relying on asymptotic arguments, we conducted randomization-based inference to construct the null randomization distribution of the test statistic by considering 105 possible exposure assignments and estimated the randomization-based p-value. We also estimated 95% Fisher Confidence Intervals (CIs) based on the randomized p-value under this framework.46 A schematic of the MiCA algorithm and codes with illustration on a simulated data set is provided on GitHub through a vignette(https://github.com/vishalmidya/MiCA-Microbial-Co-occurrence-Analysis/blob/main/MiCA-vignette.md).
In addition to the previously described steps to eliminate batch effects (5% relative abundance in both batches and multiple SiRF training and testing approaches), we also included the batch indicator in covariate balancing, as well as a covariate in all models. Any missing data in the covariates or exposures were imputed using the predictive mean matching implementation of the MICE package in R.47
Policy Relevant Pb Concentration Analysis
We conducted an exploratory analysis using policy-relevant Pb concentration thresholds to estimate the odds of having a below median relative abundance of the microbial clique at Pb concentrations that was easily interpretable. We dichotomized the sample at the United States guideline level for child Pb poisoning (3.5 μg/dL),48 the Mexican guideline for child Pb poisoning (5 μg/dL),49 and the study median of prenatal Pb exposure. Odds ratios were estimated for having a below median relative abundance of each member of the microbial clique with respect to dichotomized Pb exposure (above vs below the policy-relevant thresholds). Like previous analyses, odds ratios were estimated by using logistic regressions after covariate-balancing through subclass-based matching. Since this is an exploratory analysis, we estimated the 95% CIs based on distributional asymptotic arguments. All models were further adjusted by covariates.
Gene Pathway Analysis
To interpret the functionality of the cliques, we studied the gene pathways associated (1) with each individual clique member and (2) shared by all clique members using Venn diagrams. We extracted the gene pathways to understand their joint functionality.
Sensitivity Analyses
We conducted multiple sensitivity analyses. First, we repeated the association analyses (1) without the randomization-based causal inference framework (i.e., without any covariate balancing or matching) and (2) without imputing any missing covariate data. Second, the randomization-based causal inference framework was repeated using separate thresholds (25th and 40th percentile) for microbial clique abundance rather than the 50th percentile for each clique member. Third, the models were further adjusted for postnatal child Pb exposure at 12 and 24 months. Fourth, we estimated the Pearson correlations for the taxa within cliques to understand whether relative abundances of the taxa were associated with clique formation. Fifth, similar to the policy-relevant Pb Concentration analysis, we conducted an exploratory study using Pb concentration thresholds to identify the quantile with the highest odds with respect to the indicator of microbial clique abundance. Finally, we dichotomized the sample with gradually increasing percentile thresholds of prenatal Pb exposure, considering all exposures at or above the threshold vs below the threshold, and sequentially estimated the associations with microbial clique relative abundance all below the median.
Results
Study Population
The study population comprised 49 females and 74 males with an average age of 9.7 years. The 5th and 95th percentiles of observed Pb concentration for 2T and 3T were 10.86 μg/L to 89.18 μg/L and 11.78 μg/L to 77.06 μg/L, respectively. The mean Pb concentration at 2T was 33.6 μg/L and 34.9 μg/L at 3T. Mothers with lower SES during pregnancy were more likely to have higher blood Pb concentrations in both trimesters. Further, the number of children above the United States guideline for child Pb poisoning (≥3.5 μg/dL)48,50 based on second-trimester Pb exposure was 42 (34.1%). In contrast, it was 18 (14.6%) based on the Mexican guideline for child Pb poisoning (≥5 μg/dL).49 The descriptive statistics of Pb exposure and covariates from the study population are provided in Table 1.
Table 1. Descriptive Statistics of Pb Exposure and Covariates from the Study Population.
| total | |
|---|---|
| Exposure | N = 123 |
| Second trimester Pb (μg/L) median (IQR) | 25.9 (21.6) |
| Third trimester Pb (μg/L) median (IQR) | 27.6 (25.7) |
| Covariates | |
| Child sex | N = 123 |
| Male n (%) | 74 (60.2) |
| Female n (%) | 49 (39.8) |
| Maternal SES in pregnancy | |
| Lower n (%) | 66 (53.6) |
| Medium n (%) | 45 (36.6) |
| Higher n (%) | 12 (9.8) |
| Maternal age at birth (years) mean (SE) | 28.5 (0.5) |
| Maternal BMI in pregnancy (kg/m2) mean (SE) | 27.2 (0.4) |
| Child age at gut microbial sample collection (years) mean (SE) | 9.7 (0.7) |
MiCA
In the first stage of MiCA, the rh-SiRF identified three separate 2-taxa cliques predictive of Pb concentration in 2T, which included (1) Bifidobacterium adolescentis and Ruminococcus callidus, (2) B. adolescentis and Paraprevotella clara, and (3) R. callidus and P. clara. All three cliques had a stability of more than 75% and a prevalence of more than 10% and were common to all three data partitioning techniques. Next, using the exposure co-occurrence list, we scored how many times each of the taxa occurred within the sets of detected cliques (1) when the model was trained on one batch and then tested on another batch and (2) when the model was trained on randomly chosen 60% of the data and then tested on the remaining 40%. In the first case, B. adolescentis occurred six times, whereas R. callidus occurred four times, forming a clique with a co-occurrence score of 6/4. At the same time, the score was 6/5 for the second case. Finally, among these three cliques, the clique of B. adolescentis and R. callidus had the highest score (6/4 and 6/5) in the exposure co-occurrence list, and therefore, we denoted it as the primary clique (Supplementary Table 1).
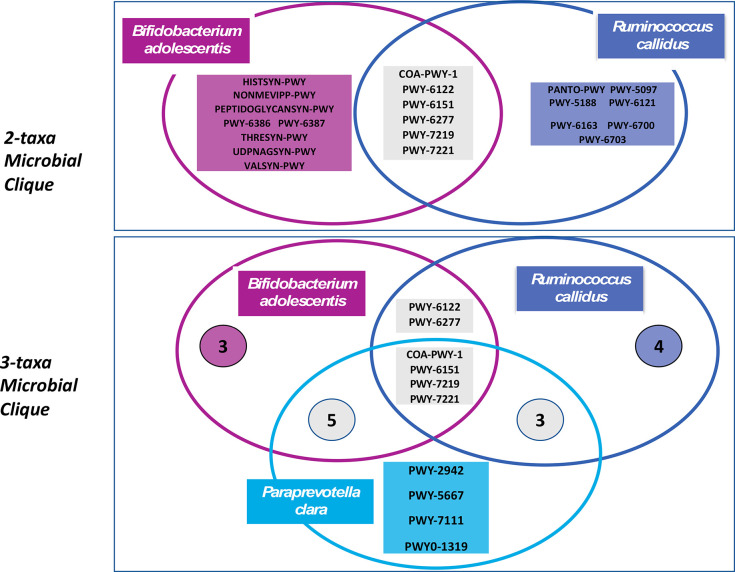
Further, based on the commutativity of three cliques, we hypothesized a 3-taxa clique comprising B. adolescentis, P. clara, and R. callidus (Figure 1). However, for 3T, no clique was common to all three data partitionings (Supplementary Table 2). In the sections below, we studied the primary 2-taxa clique of B. adolescentis and R. callidus and the 3-taxa clique of B. adolescentis, P. clara, and R. callidus. Lower relative abundance of all taxa in both the cliques was predictive of 2T Pb concentrations, implying that having both or all three of the bacteria co-occur at low or no abundance is predictive of higher 2T Pb concentration. No microbial cliques were identified as predictive of 3T Pb concentration. The details from the data partitioning for 2T Pb and 3T Pb are presented in Supplementary Tables 1 and 2.
Figure 1.
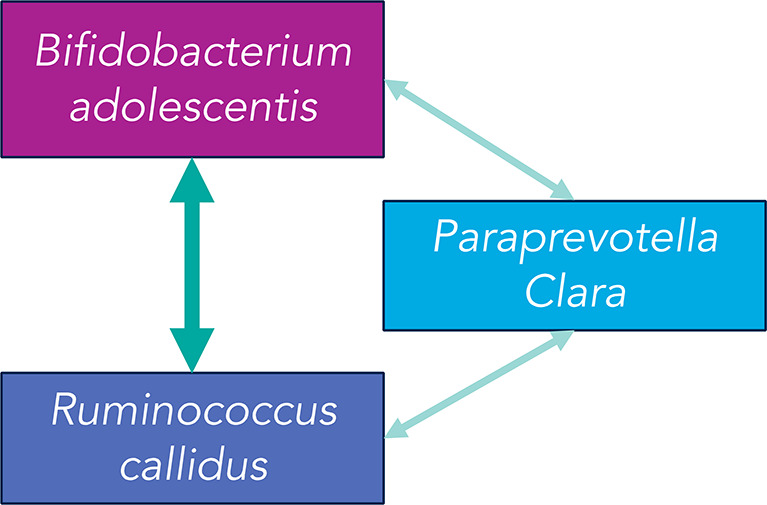
Visual representation of the bacterial taxa in the microbial cliques identified using Microbiome Co-occurrence Analysis (MiCA). The width of the arrows represents the relative frequency of occurrence of the corresponding 2-taxa clique.
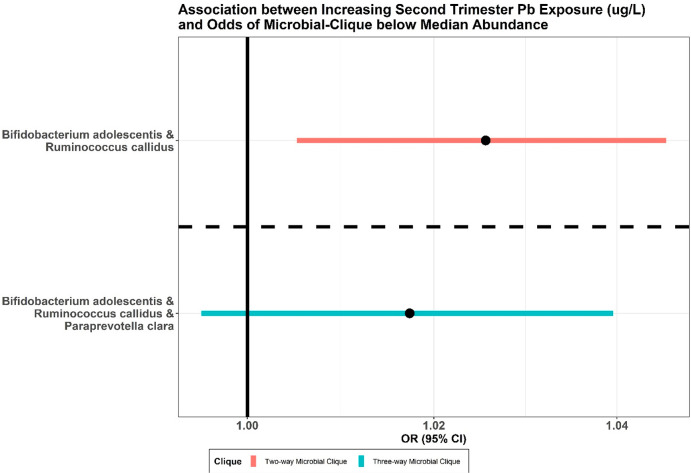
In the second stage of MiCA, using a causal inference framework, we found significantly increased odds (odds ratio (OR) = 1.03, 95% Fisher Confidence Interval (FCIs): [1.01, 1.05], and randomization-based p-value = 0.02) of having both R. callidus and B. adolescentis below median relative abundance with increasing Pb concentration in 2T (Figure 2). We also found increased odds (OR = 1.02, 95% FCIs: [0.99, 1.04], and randomization-based p-value = 0.16) of all bacteria in the 3-taxa clique (P. clara, R. callidus, and B. adolescentis) having below median relative abundance with increasing 2T Pb concentration, although not statistically significant. Note that covariates were balanced and adjusted in both models. The distribution of propensity scores and the love plot of covariate balancing after subclass matching are presented in Supplementary Figures 1 and 2. Finally, we used the false discovery rate to correct the randomization-based p-values of multiple comparison errors. The adjusted p-values for the 2-taxa and 3-taxa cliques were 0.04 and 0.16, respectively.
Figure 2.
Adjusted odds ratios for below median abundance (reduced probiotic protection) of all members in 2-taxa (red) and 3-taxa (teal) microbial cliques with a one-unit increase in second-trimester prenatal Pb concentration.
Pb Concentration Thresholds
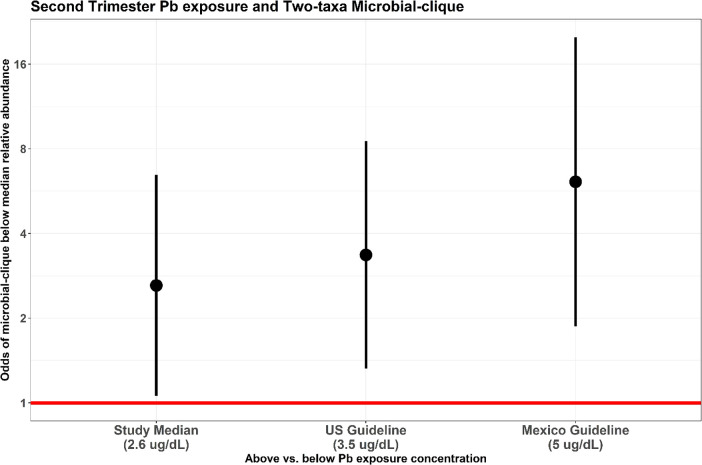
In an exploratory analysis (Figure 3), we found that those with higher 2T Pb concentration (≥2.6 μg/dL, i.e., the study median) had higher odds (OR = 2.61, 95% CIs: [1.06, 6.45]) of having a below median relative abundance of the 2-taxa clique (B. adolescentis and R. callidus). Similarly, those with 2T Pb concentration at or above the United States guideline for child Pb poisoning (≥3.5 μg/dL) and the Mexican guideline (≥5 μg/dL) had higher odds (OR = 3.36, 95% CIs: [1.32, 8.51] and OR = 6.11, 95% CIs: [1.87, 19.93], respectively) of having a below median relative abundance of the same 2-taxa clique. There is an increasing trend in the odds of below median relative abundance of the two-taxa clique, as the 2T Pb concentration cutoff threshold increases. In other words, children whose mothers had increased 2T Pb concentration, including levels below the US and Mexico guidelines for child Pb poisoning, have increased odds of having a low abundance of the 2-taxa clique late in childhood.
Figure 3.
Odds (95% CI) of having below median relative abundance (reduced probiotic protection) of both members of the 2-taxa clique with respect to 2T Pb concentration above vs below the cutoff. Pb concentration cutoffs shown are at the study median (2.6 μg/dL), the current United States guideline for child Pb poisoning (3.5 μg/dL), and the current Mexico guideline for child Pb poisoning (5 μg/dL).
Gene Pathways
In an analysis of the gene pathways belonging to the members of the 2-taxa and 3-taxa microbial cliques identified by MiCA, we examined the gene pathways from each bacterium (Figure 4). The pathways that are highly abundant in both B. adolescentis and R. callidus were related to nucleic acid biosynthesis and coenzyme A biosynthesis, essential functions of cellular life. On the other hand, the pathways that were not common between B. adolescentis and R. callidus in the 2-taxa clique were related more to amino acid biosynthesis and energy metabolism. Similar trends were present when considering P. clara as part of the 3-taxa clique. The gene pathways from each taxon is in Supplementary Table 3.
Figure 4.
Venn diagrams depicting gene pathways from each taxon within the 2- and 3-taxa microbial cliques. The gene pathways were noted in rectangular boxes. For the 3-taxa microbial clique, numbers in circles denote the number of pathways in that particular subset.
Exploratory and Sensitivity Analyses
We repeated the association analysis without the causal inference framework and any imputations. The effect sizes did not alter more than 5%, and the model-based asymptotic p-values remained reasonably similar to the randomization-based p-values (Supplementary Table 4). Further, estimates remained practically unchanged (<5%) after repeating the analysis without imputing any missing covariate (Supplementary Table 5). Under the causal inference framework, the directionality of the associations (ORs) for having both 2-taxa and 3-taxa cliques below the 25th or 40th percentile relative abundance remained positively associated with increasing Pb concentration in 2T, although it was not statistically significant (Supplementary Table 6). After adjustment for child Pb exposure at 12 and 24 months, the estimated associations did not change by more than 5% (Supplementary Table 7). The estimated Spearman correlation coefficients between the taxa were very small (−0.06 to 0.1), implying relative abundance of the taxa may not be a factor in forming the cliques (similar results were obtained for Pearson correlation as well). Lastly, in an exploratory analysis of 2T Pb concentration, we found that the odds of the 2-taxa clique with below median relative abundance were highest for those with Pb concentrations in the 50–75th percentile or above (overall aggregated OR = 2.76, 95% CI: [1.13, 6.75]). Likewise, the same Pb concentrations were associated with the highest odds of below median relative abundance for the 3-taxa clique, although the aggregated odds were not statistically significant (mean OR = 1.95, 95% CI: [0.73, 5.21]). For 2- and 3-taxa cliques, there was an increasing trend in the odds of below median relative abundance as the cutoff threshold of 2T Pb concentration gradually increased (Supplementary Figure 3).
Discussion
This study presents a novel approach in microbiome analysis that detects microbial clique(s) predictive of an outcome of interest and estimates the association between that outcome and the clique(s). We used MiCA to identify 2-taxa and 3-taxa microbial cliques in the gut microbiome of children 9–11 years of age, which were negatively causally associated with prenatal Pb exposure. We further explored policy-relevant thresholds for 2T Pb exposure and found significantly increased odds of having below median relative abundance (reduced probiotic protection) of the 2-taxa microbial clique at and below the current child Pb poisoning guidelines for the United States and Mexico. We also investigated the gene function pathways within the cliques to shed light on their potentially interactive functions.
The microbial clique members, B. adolescentis, R. callidus, and P. clara, play various beneficial roles in the human gut microbiome. P. clara is the most recently identified, with comparatively little known about its health benefits;51 however, a recent study in dialysis patients found that increased abundance of P. clara was associated with reduced constipation.52R. callidus is a short-chain fatty-acid-producing bacteria with anti-inflammatory function.53−55 Low abundance of R. callidus has been associated with Parkinson’s disease,56 colitis and Crohn’s disease,54,57,58 liver disease,55 and obesity.15B. adolescentis is a crucial human gut microbe that acts as a starch degrader and Gamma-aminobutyric acid (GABA) producer and helps enhance the intestinal barrier.59−63B. adolescentis is commonly used as a probiotic supplement and has been linked to the prevention and alleviation of many detrimental health conditions, including liver disease,61,64 colitis,65,66 viral infection,67,68 arthritis,69 type 2 diabetes,62 anxiety, depression, and other mental health disorders.66,70 Additionally, B. adolescentis is known to modify the overall composition of the gut microbiome, perhaps its most relevant feature for this analysis, increasing the abundance of other probiotic or beneficial bacteria within the microbiome and amplifying its beneficial effects.61−63 Thus, our finding of B. adolescentis as a member of both the 2-taxa and 3-taxa microbial clique, in combination with other potentially beneficial bacterial taxa, is highly consistent with previous literature.61−63 When considering the findings of our analysis in the context of this previous evidence, it is clear that prenatal Pb exposure, particularly in the second trimester of pregnancy, has the potential to lead to several detrimental health outcomes via alterations in the gut microbiome, specifically by reducing the abundance of these 2-taxa and 3-taxa microbial cliques.
The reduced abundance of these probiotic microbial cliques happens not only above the guideline for blood Pb concentrations for child Pb poisoning in both Mexico and the United States but also below that at the study median of 2.6 μg/dL. Occupational Pb exposure guidelines in the United States only require medical monitoring of employees with blood Pb concentrations above 40 μg/dL,71 more than 10× the concentrations observed in this study. Children of mothers with blood Pb levels below the child Pb poisoning guidelines during the second trimester of pregnancy are less likely to have these beneficial gut bacteria in late childhood, and mothers with occupational Pb exposure are likely at an even greater risk. Because a reduced abundance of B. adolescentis and R. callidus has been associated with IBD, liver disease, and reductions in mental health55,57,61,65,70 the current United States and Mexico guidelines for Pb exposure, while better than past guidelines and procedures, are insufficient to protect against these detrimental health outcomes.
To better understand the potential roles of each taxon within the microbial cliques, we examined the top 20 most abundant gene pathways from each taxon within the cliques. Approximately half of the gene pathways for each taxon were shared with the other clique members, and the other half were unique to that specific taxon. In general, the redundant genes within the clique were key pathways needed for all cellular life, and the unique gene pathways included functions that were more specific to each taxa’s metabolism. This indicates that each member of the microbial group provides unique and potentially complementary functions. Moreover, these taxa are included in the clique not because they are redundant in function and potentially fill the same niche with regard to their association with prenatal Pb exposure but because they are different.
MiCA provides several statistical advantages over other, more traditional microbiome analysis methods (results previously shown using the same data13). The amalgamation of interpretable machine learning algorithms with causal inference tools serves as both predictive and associative models. Searching for cliques is difficult when the number of taxa is high (which is the usual scenario); therefore, the usage of machine learning algorithms significantly reduces the computational complexity and the associative regression models provide interpretability. MiCA does not rely heavily on highly abundant or prevalent taxa within the study samples. As demonstrated in this analysis, MiCA can identify associations with bacteria in low abundance together. MiCA also does not rely on correlations between clique members; thus, cliques can be discovered in association with exposure or an outcome of interest even when the taxa within the cliques are not highly correlated within the study sample as a whole. MiCA can also detect cliques in multiple directions with respect to the threshold in a single rh-SiRF analysis. The rh-SiRF step also serves as a major tool for higher prediction accuracy and therefore selects only a few key cliques. Hence, the association tests are highly focused on only a few relationships, reducing the need to correct for multiple comparisons. However, the most significant advantage of MiCA is that it analyzes the gut microbiome using a different biological framework than any other epidemiologic analytical tool we know of, i.e., cliques instead of single taxa or the whole microbiome.
While this study presents a novel analytic approach and adds new information about the relationship between Pb exposure and the human gut microbiome, there are some limitations to consider. Limitations of this analysis include using a relatively small sample size with samples processed in multiple batches. However, we took various precautions to reduce batch effects in our estimates and still found statistical significance with a small sample size and conservative estimates. Another limitation of this analysis is that we used maternal blood concentration to estimate prenatal Pb exposure in children, which is not a direct exposure estimate. Thus, it is possible that the mechanism of this association may work through maternal exposure rather than prenatal child exposure. For instance, it may be that the maternal Pb exposures alter the maternal gut microbiome during pregnancy, which is then vertically transferred to the offspring at birth rather than prenatal child Pb exposure priming the child microbiome composition later in childhood. It may also be the case that maternal and child Pb exposures are correlated throughout childhood, and maternal transmission continues to influence the microbiome of the child during childhood. This study is limited in understanding the underlying biochemical mechanism between Pb exposure and the co-occurring bacteria. Further investigation is needed in vitro and animal models to better elucidate these mechanisms.
Future analyses could include other biological matrices to estimate Pb exposure, for instance, baby teeth, which can measure direct prenatal exposure starting in the second trimester of pregnancy. As a potential future direction, we hope to develop MiCA further to include multiple metal exposures and other potential predictors that may influence the microbiome, including diet and other biological and microbial ecological factors. Future reverse translational studies should be conducted using animal and in vitro models to clarify the biochemical mechanisms driving the causal associations identified in this study.
Acknowledgments
The authors would like to acknowledge the entire PROGRESS study team as well as the participants. We would also like to thank Dr. Jeremiah Faith and the Microbiome Translational Center at the Icahn School of Medicine at Mount Sinai.
Data Availability Statement
The data that was used in this study can be made accessible to researchers upon appropriate request with the following restrictions to ensure the privacy of human subjects. Note that access to the data is limited due to a data-sharing agreement approved by the IRB at Mount Sinai. Researchers who are interested in accessing PROGRESS data must send their resume/CV as well as CITI training certificates to the IRB chair, Ilene Wilets (Ilene.wilets@mssm.edu). They must also submit a data analysis plan to the Principal Investigators for PROGRESS: Robert O. Wright (robert.wright@mssm.edu), Martha Tellez-Rojo (mmtellez@insp.mx), and Andrea Baccarelli (andrea.baccarelli@columbia.edu). Once this process is completed, the PROGRESS data analyst, Nia McRae (nia.x.mcrae@mssm.edu), will send a deidentified data set via Box, a secure data-sharing platform. The Sequence Read Archive database accession number is PRJNA975184.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.est.3c04346.
Supplementary Table 1: MiCA for Pb exposure at 2T; Supplementary Table 2: MiCA for Pb exposure at 3T; Supplementary Table 3: gene pathways; Supplementary Table 4: odds ratio and 95% CIs from association analysis without any covariate balancing or matching; Supplementary Table 5: association estimates after repeating the analysis without imputing any missing covariate data; Supplementary Table 6: association estimates after cutoff thresholds being set at the 25th or 40th percentile relative abundance; Supplementary Table 7: association estimates after adjusting for child Pb exposure at 12 and 24 months; Supplementary Figure 1: distribution of propensity scores after covariate balancing; Supplementary Figure 2: love plot of covariate balancing after subclass matching; Supplementary Figure 3: exploratory analysis of 2T Pb concentration (PDF)
The authors declare the following competing financial interest(s): M.A. is an employee and equity holder of Linus Biotechnology Inc., a start-up company of Mount Sinai Health System. The company develops tools for the detection of autism spectrum disorder and related conditions. The following authors report no competing interests: V.M., J.M.L., C.G., L.A.T.-O., J.K.G., R.O.W., M.M.T.-R., and S.E.
Special Issue
Published as part of the Environmental Science & Technologyvirtual special issue “The Exposome and Human Health”.
Supplementary Material
References
- Claus S. P.; Guillou H.; Ellero-Simatos S. The Gut Microbiota: A Major Player in the Toxicity of Environmental Pollutants?. NPJ. Biofilms Microbiomes 2016, 2, 16003. 10.1038/npjbiofilms.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; Brejnrod A. D.; Ernst M.; Rykær M.; Herschend J.; Olsen N. M. C.; Dorrestein P. C.; Rensing C.; Sorensen S. J. Heavy Metal Exposure Causes Changes in the Metabolic Health-Associated Gut Microbiome and Metabolites. Environ. Int. 2019, 126, 454–467. 10.1016/j.envint.2019.02.048. [DOI] [PubMed] [Google Scholar]
- Brabec J. L.; Wright J.; Ly T.; Wong H. T.; McClimans C. J.; Tokarev V.; Lamendella R.; Sherchand S.; Shrestha D.; Uprety S.; Dangol B.; Tandukar S.; Sherchand J. B.; Sherchan S. P. Arsenic Disturbs the Gut Microbiome of Individuals in a Disadvantaged Community in Nepal. Heliyon 2020, 6 (1), e03313 10.1016/j.heliyon.2020.e03313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton J.; Massart S.; Vandamme P.; De Brandt E.; Pot B.; Foligné B. Ecotoxicology inside the Gut: Impact of Heavy Metals on the Mouse Microbiome. BMC Pharmacol. Toxicol. 2013, 14, 62. 10.1186/2050-6511-14-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi L.; Bian X.; Gao B.; Tu P.; Ru H.; Lu K. The Effects of an Environmentally Relevant Level of Arsenic on the Gut Microbiome and Its Functional Metagenome. Toxicol. Sci. 2017, 160 (2), 193–204. 10.1093/toxsci/kfx174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers S.; Safdar N.; Kates A.; Sethi A. K.; Peppard P. E.; Kanarek M. S.; Malecki K. M. C. Urinary Lead Level and Colonization by Antibiotic Resistant Bacteria: Evidence from a Population-Based Study. Environ. Epidemiol. 2021, 5 (6), e175 10.1097/EE9.0000000000000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B.; Chi L.; Mahbub R.; Bian X.; Tu P.; Ru H.; Lu K. Multi-Omics Reveals That Lead Exposure Disturbs Gut Microbiome Development, Key Metabolites, and Metabolic Pathways. Chem. Res. Toxicol. 2017, 30 (4), 996–1005. 10.1021/acs.chemrestox.6b00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisanian M.; Holladay S. D.; Karpuzoglu E.; Kerr R. P.; Williams S. M.; Stabler L.; McArthur J. V.; Tuckfield R. C.; Gogal R. M. Exposure of Juvenile Leghorn Chickens to Lead Acetate Enhances Antibiotic Resistance in Enteric Bacterial Flora. Poult. Sci. 2014, 93 (4), 891–897. 10.3382/ps.2013-03600. [DOI] [PubMed] [Google Scholar]
- Eggers S.; Safdar N.; Sethi A. K.; Suen G.; Peppard P. E.; Kates A. E.; Skarlupka J. H.; Kanarek M.; Malecki K. M. C. Urinary Lead Concentration and Composition of the Adult Gut Microbiota in a Cross-Sectional Population-Based Sample. Environ. Int. 2019, 133, 105122. 10.1016/j.envint.2019.105122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laue H. E.; Moroishi Y.; Jackson B. P.; Palys T. J.; Madan J. C.; Karagas M. R. Nutrient-Toxic Element Mixtures and the Early Postnatal Gut Microbiome in a United States Longitudinal Birth Cohort. Environ. Int. 2020, 138, 105613. 10.1016/j.envint.2020.105613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitarik A. R.; Arora M.; Austin C.; Bielak L. F.; Eggers S.; Johnson C. C.; Lynch S. V.; Kyun Park S.; Hank Wu K.-H.; Yong G. J. M.; Cassidy-Bushrow A. E. Fetal and Early Postnatal Lead Exposure Measured in Teeth Associates with Infant Gut Microbiota. Environ. Int. 2020, 144, 106062. 10.1016/j.envint.2020.106062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y.; Laue H. E.; Shrubsole M. J.; Wu H.; Bloomquist T. R.; Larouche A.; Zhao K.; Gao F.; Boivin A.; Prada D.; Hunting D. J.; Gillet V.; Takser L.; Baccarelli A. A. Associations of Childhood and Perinatal Blood Metals with Children’s Gut Microbiomes in a Canadian Gestation Cohort. Environ. Health Perspect. 2022, 130 (1), 17007. 10.1289/EHP9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers S.; Midya V.; Bixby M.; Gennings C.; Torres-Olascoaga L. A.; Walker R. W.; Wright R. O.; Arora M.; Téllez-Rojo M. M. Prenatal Lead Exposure Is Negatively Associated with the Gut Microbiome in Childhood. Front. Microbiol. 2023, 14, 1193919. 10.3389/fmicb.2023.1193919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart E. J. Growing Unculturable Bacteria. J. Bacteriol. 2012, 194 (16), 4151–4160. 10.1128/JB.00345-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas L. R.; Bernabé B. P.; Priyadarshini M.; Fei N.; Park S. J.; Brown L.; Plange-Rhule J.; Nelson D.; Toh E. C.; Gao X.; Dong Q.; Sun J.; Kliethermes S.; Gottel N.; Luke A.; Gilbert J. A.; Layden B. T. Decreased Microbial Co-Occurrence Network Stability and SCFA Receptor Level Correlates with Obesity in African-Origin Women. Sci. Rep. 2018, 8 (1), 17135. 10.1038/s41598-018-35230-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust K.; Sathirapongsasuti J. F.; Izard J.; Segata N.; Gevers D.; Raes J.; Huttenhower C. Microbial Co-Occurrence Relationships in the Human Microbiome. PLOS Comput. Biol. 2012, 8 (7), e1002606 10.1371/journal.pcbi.1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y.; Zhang L.; Peterson C. B.; Do K.-A.; Jenq R. R. Performance Determinants of Unsupervised Clustering Methods for Microbiome Data. Microbiome 2022, 10 (1), 25. 10.1186/s40168-021-01199-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson E. A.Statistical and Machine Learning Methods for Pattern Identification in Environmental Mixtures. Ph.D. Thesis, Columbia University, 2021. [Google Scholar]
- Joubert B. R.; Kioumourtzoglou M.-A.; Chamberlain T.; Chen H. Y.; Gennings C.; Turyk M. E.; Miranda M. L.; Webster T. F.; Ensor K. B.; Dunson D. B. Powering Research through Innovative Methods for Mixtures in Epidemiology (PRIME) Program: Novel and Expanded Statistical Methods. Int. J. Environ. Res. Public. Health 2022, 19 (3), 1378. 10.3390/ijerph19031378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midya V.; Alcala C. S.; Rechtman E.; Gregory J. K.; Kannan K.; Hertz-Picciotto I.; Teitelbaum S. L.; Gennings C.; Rosa M. J.; Valvi D.. Machine Learning Assisted Discovery of Interactions between Pesticides, Phthalates, Phenols, and Trace Elements in Child Neurodevelopment. Environ. Sci. Technol. 2023; 10.1021/acs.est.3c00848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellavia A.; Dickerson A. S.; Rotem R. S.; Hansen J.; Gredal O.; Weisskopf M. G. Joint and Interactive Effects between Health Comorbidities and Environmental Exposures in Predicting Amyotrophic Lateral Sclerosis. Int. J. Hyg. Environ. Health 2021, 231, 113655. 10.1016/j.ijheh.2020.113655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampa E.; Lind L.; Lind P. M.; Bornefalk-Hermansson A. The Identification of Complex Interactions in Epidemiology and Toxicology: A Simulation Study of Boosted Regression Trees. Environ. Health 2014, 13 (1), 57. 10.1186/1476-069X-13-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennings C.; Schwartz P.; Carter W. H.; Simmons J. E. Detection of Departures from Additivity in Mixtures of Many Chemicals with a Threshold Model. J. Agric. Biol. Environ. Stat. 1997, 2 (2), 198–211. 10.2307/1400403. [DOI] [Google Scholar]
- Hamm A. K.; Hans Carter W. Jr; Gennings C. Analysis of an Interaction Threshold in a Mixture of Drugs and/or Chemicals. Stat. Med. 2005, 24 (16), 2493–2507. 10.1002/sim.2110. [DOI] [PubMed] [Google Scholar]
- Yeatts S. D.; Gennings C.; Wagner E. D.; Simmons J. E.; Plewa M. J. Detecting Departure From Additivity Along a Fixed-Ratio Mixture Ray With a Piecewise Model for Dose and Interaction Thresholds. J. Agric. Biol. Environ. Stat. 2010, 15 (4), 510–522. 10.1007/s13253-010-0030-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmueli G. To Explain or to Predict?. Stat. Sci. 2010, 25 (3), 289–310. 10.1214/10-STS330. [DOI] [Google Scholar]
- Heiss J. A.; Téllez-Rojo M. M.; Estrada-Gutiérrez G.; Schnaas L.; Amarasiriwardena C.; Baccarelli A. A.; Wright R. O.; Just A. C. Prenatal Lead Exposure and Cord Blood DNA Methylation in PROGRESS: An Epigenome-Wide Association Study. Environ. Epigenetics 2020, 6 (1), dvaa014 10.1093/eep/dvaa014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano K. A.; Dill-McFarland K. A.; Kasahara K.; Kerby R. L.; Vivas E. I.; Amador-Noguez D.; Herd P.; Rey F. E. Fecal Aliquot Straw Technique (FAST) Allows for Easy and Reproducible Subsampling: Assessing Interpersonal Variation in Trimethylamine-N-Oxide (TMAO) Accumulation. Microbiome 2018, 6 (1), 91. 10.1186/s40168-018-0458-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M.; Lohse M.; Usadel B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinforma. Oxf. Engl. 2014, 30 (15), 2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B.; Salzberg S. L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9 (4), 357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong D. T.; Franzosa E. A.; Tickle T. L.; Scholz M.; Weingart G.; Pasolli E.; Tett A.; Huttenhower C.; Segata N. MetaPhlAn2 for Enhanced Metagenomic Taxonomic Profiling. Nat. Methods 2015, 12 (10), 902–903. 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- Truong D. T.; Tett A.; Pasolli E.; Huttenhower C.; Segata N. Microbial Strain-Level Population Structure and Genetic Diversity from Metagenomes. Genome Res. 2017, 27 (4), 626–638. 10.1101/gr.216242.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa E. A.; McIver L. J.; Rahnavard G.; Thompson L. R.; Schirmer M.; Weingart G.; Lipson K. S.; Knight R.; Caporaso J. G.; Segata N.; Huttenhower C. Species-Level Functional Profiling of Metagenomes and Metatranscriptomes. Nat. Methods 2018, 15 (11), 962–968. 10.1038/s41592-018-0176-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R.; Billington R.; Keseler I. M.; Kothari A.; Krummenacker M.; Midford P. E.; Ong W. K.; Paley S.; Subhraveti P.; Karp P. D. The MetaCyc Database of Metabolic Pathways and Enzymes - a 2019 Update. Nucleic Acids Res. 2020, 48 (D1), D445–D453. 10.1093/nar/gkz862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders A. P.; Gennings C.; Tamayo-Ortiz M.; Mistry S.; Pantic I.; Martinez M.; Estrada-Gutierrez G.; Espejel-Nuñez A.; Olascoaga L. T.; Wright R. O.; Téllez-Rojo M. M.; Arora M.; Austin C. Prenatal and Early Childhood Critical Windows for the Association of Nephrotoxic Metal and Metalloid Mixtures with Kidney Function. Environ. Int. 2022, 166, 107361. 10.1016/j.envint.2022.107361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S.; Kumbier K.; Brown J. B.; Yu B. Iterative Random Forests to Discover Predictive and Stable High-Order Interactions. Proc. Natl. Acad. Sci. U. S. A. 2018, 115 (8), 1943–1948. 10.1073/pnas.1711236115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumbier K.; Basu S.; Brown J. B.; Celniker S.; Yu B.. Refining Interaction Search through Signed Iterative Random Forests. arXiv Prepr. 2018, arXiv:1810.07287; 10.48550/arXiv.1810.07287 (accessed 2023–01–22). [DOI] [Google Scholar]
- Shah R. D.; Meinshausen N. Random Intersection Trees. J. Mach. Learn. Res. 2014, 15 (1), 629–654. [Google Scholar]
- Tanner E. M.; Bornehag C.-G.; Gennings C. Repeated Holdout Validation for Weighted Quantile Sum Regression. MethodsX 2019, 6, 2855–2860. 10.1016/j.mex.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Xu L.; Tian F.; Jiang L.; Zhong X.; Chen E.. Word Embedding Revisited: A New Representation Learning and Explicit Matrix Factorization Perspective. Twenty-Fourth International Joint Conference on Artificial Intelligence, 2015.
- Lin J.Scalable Language Processing Algorithms for the Masses: A Case Study in Computing Word Co-Occurrence Matrices with MapReduce. Proceedings of the 2008 Conference on Empirical Methods in Natural Language Processing, 2008.
- Greifer N.Covariate Balance Tables and Plots: A Guide to the Cobalt Package. https://cloud.r-project.org/web/packages/cobalt/vignettes/cobalt.html (accessed 2023–05–08).
- Ho D.; Imai K.; King G.; Stuart E. A. MatchIt: Nonparametric Preprocessing for Parametric Causal Inference. J. Stat. Softw. 2011, 42, 1–28. 10.18637/jss.v042.i08. [DOI] [Google Scholar]
- Linden A. Graphical displays for assessing covariate balance in matching studies. J. Eval. Clin. Pract. 2015, 21 (2), 242–247. 10.1111/jep.12297. [DOI] [PubMed] [Google Scholar]
- Zhang Z.; Kim H. J.; Lonjon G.; Zhu Y. Balance Diagnostics after Propensity Score Matching. Ann. Transl. Med. 2019, 7 (1), 16. 10.21037/atm.2018.12.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbens G. W.; Rubin D. B. Fisher’s Exact p-Values for Completely Randomized Experiments. Causal Inference Stat. Soc. Biomed. Sci. 2015, 57–82. 10.1017/CBO9781139025751.006. [DOI] [Google Scholar]
- van Buuren S.; Groothuis-Oudshoorn K. Mice: Multivariate Imputation by Chained Equations in R. J. Stat. Softw. 2011, 45, 1–67. 10.18637/jss.v045.i03. [DOI] [Google Scholar]
- CDC Blood Lead Reference Value; https://www.cdc.gov/nceh/lead/data/blood-lead-reference-value.htm (accessed 2023–05–08).
- DOF - Official Gazette of the Federation. https://dof.gob.mx/nota_detalle.php?codigo=5495551&fecha=30/08/2017#gsc.tab=0 (accessed 2023–05–10).
- Betts K. S. CDC Updates Guidelines for Children’s Lead Exposure. Environ. Health Perspect. 2012, 120 (7), a268–a268. 10.1289/ehp.120-a268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morotomi M.; Nagai F.; Sakon H.; Tanaka R. Paraprevotella Clara Gen. Nov., Sp. Nov. and Paraprevotella Xylaniphila Sp. Nov., Members of the Family “Prevotellaceae” Isolated from Human Faeces. Int. J. Syst. Evol. Microbiol. 2009, 59 (8), 1895–1900. 10.1099/ijs.0.008169-0. [DOI] [PubMed] [Google Scholar]
- Peng Y.; Zeng Y.; Zheng T.; Xie X.; Wu J.; Fu L.; Lu F.; Zhang L.; Chen Y.; Liu X.; Wang L. Effects of Tiaopi Xiezhuo Decoction on Constipation and Gut Dysbiosis in Patients with Peritoneal Dialysis. Pharm. Biol. 2023, 61 (1), 531–540. 10.1080/13880209.2023.2193595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Tapia M.; Hernández-Velázquez I.; Pichardo-Ontiveros E.; Granados-Portillo O.; Gálvez A.; R Tovar A.; Torres N. Consumption of Cooked Black Beans Stimulates a Cluster of Some Clostridia Class Bacteria Decreasing Inflammatory Response and Improving Insulin Sensitivity. Nutrients 2020, 12 (4), 1182. 10.3390/nu12041182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satokari R.; Fuentes S.; Mattila E.; Jalanka J.; de Vos W. M.; Arkkila P. Fecal Transplantation Treatment of Antibiotic-Induced, Noninfectious Colitis and Long-Term Microbiota Follow-Up. Case Rep. Med. 2014, 2014, 913867. 10.1155/2014/913867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng S.; Chen J.; Zhang Y.; Qin Q.; Li W.; Yan S.; Wang Y.; Li T.; Gao X.; Tang L.; Li A.; Ding S. Structural and Functional Alterations of Gut Microbiota in Males With Hyperuricemia and High Levels of Liver Enzymes. Front. Med. 2021, 8, 779994. 10.3389/fmed.2021.779994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov V. A.; Saltykova I. V.; Zhukova I. A.; Alifirova V. M.; Zhukova N. G.; Dorofeeva Y. B.; Tyakht A. V.; Kovarsky B. A.; Alekseev D. G.; Kostryukova E. S.; Mironova Y. S.; Izhboldina O. P.; Nikitina M. A.; Perevozchikova T. V.; Fait E. A.; Babenko V. V.; Vakhitova M. T.; Govorun V. M.; Sazonov A. E. Analysis of Gut Microbiota in Patients with Parkinson’s Disease. Bull. Exp. Biol. Med. 2017, 162 (6), 734–737. 10.1007/s10517-017-3700-7. [DOI] [PubMed] [Google Scholar]
- Al-Amrah H.; Saadah O. I.; Mosli M.; Annese V.; Al-Hindi R.; Edris S.; Alshehri D.; Alatawi H.; Alatawy M.; Bahieldin A. Composition of the Gut Microbiota in Patients with Inflammatory Bowel Disease in Saudi Arabia: A Pilot Study. Saudi J. Gastroenterol. Off. J. Saudi Gastroenterol. Assoc. 2023, 29, 102. 10.4103/sjg.sjg_368_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S.; Denman S. E.; Morrison M.; Yu Z.; Dore J.; Leclerc M.; McSweeney C. S. Dysbiosis of Fecal Microbiota in Crohn’s Disease Patients as Revealed by a Custom Phylogenetic Microarray. Inflamm. Bowel Dis. 2010, 16 (12), 2034–2042. 10.1002/ibd.21319. [DOI] [PubMed] [Google Scholar]
- Altaib H.; Kozakai T.; Badr Y.; Nakao H.; El-Nouby M. A. M.; Yanase E.; Nomura I.; Suzuki T. Cell Factory for γ-Aminobutyric Acid (GABA) Production Using Bifidobacterium Adolescentis. Microb. Cell Factories 2022, 21 (1), 33. 10.1186/s12934-021-01729-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duranti S.; Ruiz L.; Lugli G. A.; Tames H.; Milani C.; Mancabelli L.; Mancino W.; Longhi G.; Carnevali L.; Sgoifo A.; Margolles A.; Ventura M.; Ruas-Madiedo P.; Turroni F. Bifidobacterium Adolescentis as a Key Member of the Human Gut Microbiota in the Production of GABA. Sci. Rep. 2020, 10 (1), 14112. 10.1038/s41598-020-70986-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Lv L.; Ye J.; Fang D.; Shi D.; Wu W.; Wang Q.; Wu J.; Yang L.; Bian X.; Jiang X.; Jiang H.; Yan R.; Peng C.; Li L. Bifidobacterium Adolescentis CGMCC 15058 Alleviates Liver Injury, Enhances the Intestinal Barrier and Modifies the Gut Microbiota in D-Galactosamine-Treated Rats. Appl. Microbiol. Biotechnol. 2019, 103 (1), 375–393. 10.1007/s00253-018-9454-y. [DOI] [PubMed] [Google Scholar]
- Qian X.; Si Q.; Lin G.; Zhu M.; Lu J.; Zhang H.; Wang G.; Chen W. Bifidobacterium Adolescentis Is Effective in Relieving Type 2 Diabetes and May Be Related to Its Dominant Core Genome and Gut Microbiota Modulation Capacity. Nutrients 2022, 14 (12), 2479. 10.3390/nu14122479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.; Kong Q.; Cui S.; Li X.; Gu Z.; Zhao J.; Zhang H.; Chen W.; Wang G. Bifidobacterium Adolescentis Isolated from Different Hosts Modifies the Intestinal Microbiota and Displays Differential Metabolic and Immunomodulatory Properties in Mice Fed a High-Fat Diet. Nutrients 2021, 13 (3), 1017. 10.3390/nu13031017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X.; Liu D.; Gao Q.; Ni J.; Qian L.; Ni Y.; Fang Q.; Jia W.; Li H. Bifidobacterium Adolescentis Alleviates Liver Steatosis and Steatohepatitis by Increasing Fibroblast Growth Factor 21 Sensitivity. Front. Endocrinol. 2021, 12, 773340. 10.3389/fendo.2021.773340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L.; Qi Y.; Qu S.; Chen X.; Li A.; Hendi M.; Xu C.; Wang L.; Hou T.; Si J.; Chen S. B. Adolescentis Ameliorates Chronic Colitis by Regulating Treg/Th2 Response and Gut Microbiota Remodeling. Gut Microbes 2021, 13 (1), 1–17. 10.1080/19490976.2020.1826746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H.-M.; Lee K.-E.; Kim D.-H. The Preventive and Curative Effects of Lactobacillus Reuteri NK33 and Bifidobacterium Adolescentis NK98 on Immobilization Stress-Induced Anxiety/Depression and Colitis in Mice. Nutrients 2019, 11 (4), 819. 10.3390/nu11040819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. J.; Lee D. K.; Park J. E.; Park I. H.; Seo J. G.; Ha N. J. Antiviral Activity of Bifidobacterium Adolescentis SPM1605 against Coxsackievirus B3. Biotechnol. Biotechnol. Equip. 2014, 28 (4), 681–688. 10.1080/13102818.2014.945237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D.; Breiman A.; le Pendu J.; Uyttendaele M. Anti-Viral Effect of Bifidobacterium Adolescentis against Noroviruses. Front. Microbiol. 2016, 7, 864. 10.3389/fmicb.2016.00864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z.; Yang B.; Ross R. P.; Stanton C.; Shi G.; Zhao J.; Zhang H.; Chen W. Protective Effects of Bifidobacterium Adolescentis on Collagen-Induced Arthritis in Rats Depend on Timing of Administration. Food Funct. 2020, 11 (5), 4499–4511. 10.1039/D0FO00077A. [DOI] [PubMed] [Google Scholar]
- Lee H. J.; Hong J. K.; Kim J.-K.; Kim D.-H.; Jang S. W.; Han S.-W.; Yoon I.-Y. Effects of Probiotic NVP-1704 on Mental Health and Sleep in Healthy Adults: An 8-Week Randomized, Double-Blind. Placebo-Controlled Trial. Nutrients 2021, 13 (8), 2660. 10.3390/nu13082660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Committee on Potential Health Risks from Recurrent Lead Exposure of DOD Firing-Range Personnel; Committee on Toxicology; Board on Environmental Studies and Toxicology; Division on Earth and Life Studies; National Research Council . Occupational Standards and Guidelines for Lead. In Potential Health Risks to DOD Firing-Range Personnel from Recurrent Lead Exposure; National Academies Press: Washington, DC, US, 2012. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that was used in this study can be made accessible to researchers upon appropriate request with the following restrictions to ensure the privacy of human subjects. Note that access to the data is limited due to a data-sharing agreement approved by the IRB at Mount Sinai. Researchers who are interested in accessing PROGRESS data must send their resume/CV as well as CITI training certificates to the IRB chair, Ilene Wilets (Ilene.wilets@mssm.edu). They must also submit a data analysis plan to the Principal Investigators for PROGRESS: Robert O. Wright (robert.wright@mssm.edu), Martha Tellez-Rojo (mmtellez@insp.mx), and Andrea Baccarelli (andrea.baccarelli@columbia.edu). Once this process is completed, the PROGRESS data analyst, Nia McRae (nia.x.mcrae@mssm.edu), will send a deidentified data set via Box, a secure data-sharing platform. The Sequence Read Archive database accession number is PRJNA975184.