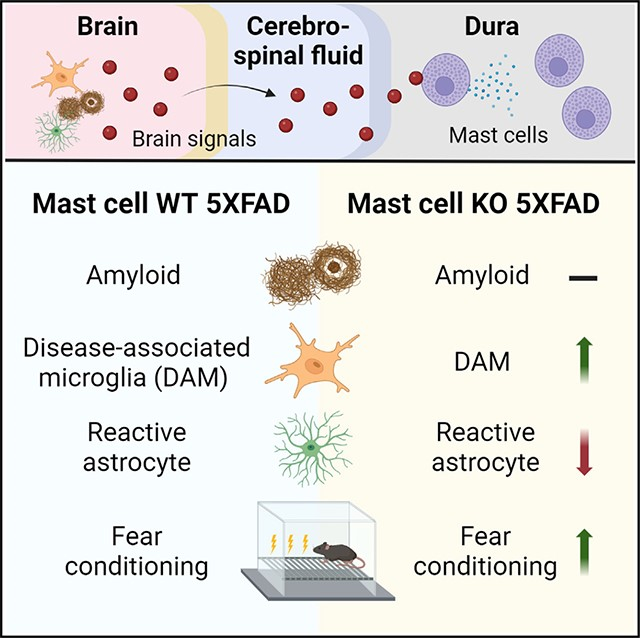
SUMMARY
Emerging evidence suggests that peripheral immune cells contribute to Alzheimer’s disease (AD) neuropathogenesis. Among these, mast cells are known for their functions in allergic reactions and neuroinflammation; however, little is known about their role in AD. Here, we crossed 5XFAD mice with mast cell-deficient strains and observed the effects on AD-related neuropathology and cognitive impairment. We found that mast cell depletion improved contextual fear conditioning in 5XFAD mice without affecting cued fear conditioning, anxiety-like behavior, or amyloid burden. Furthermore, mast cell depletion led to an upregulation of transcriptomic signatures for putatively protective disease-associated microglia and resulted in reduced markers indicative of reactive astrocytes. We hypothesize a system of bidirectional communication between dural mast cells and the brain, where mast cells respond to signals from the brain environment by expressing immune-regulatory mediators, impacting cognition and glial cell function. These findings highlight mast cells as potential therapeutic targets for AD.
In brief
Lin et al. show that depletion of mast cells restores cognitive function (contextual fear conditioning), increases disease-associated microglia gene expression, reduces GFAP levels, and alters astrocytic morphology. These data implicate mast cells as potential targets for treating AD.
Graphical abstract
INTRODUCTION
Alzheimer’s disease (AD) is the most common cause of dementia, yet the etiology and neuropathogenic events leading to cognitive impairment are not fully understood. Although the key neuropathological hallmarks of AD include abundant β-amyloid plaques, tau tangles, and neuroinflammation,1 emerging evidence suggests that a variety of brain border and peripheral immune cells also impact AD neuropathogenesis.2,3 Indeed, it has been hypothesized that mast cells, one class of immune cell, and their secreted inflammatory signals contribute to neurodegenerative diseases.4–7 Mast cells are tissue-resident innate immune cells with established roles in allergic reactions, which secrete a variety of immune-regulatory mediators, including histamine, prostaglandins, proteases, and cytokines.8,9 The role of mast cells within the neuroimmune axis is poorly understood. However, CNS-associated mast cells have been reported to be detrimental in rodent models of hypoxic-ischemic encephalopathy, stroke, and encephalitis via their release of proteases and cytokines.10–14 Moreover, mast cell-derived mediators, potentially histamine and tryptase, trigger microglial activation in rodent models of CNS inflammation15–20 and modulate astrocytic responses in vitro.21,22 Although the potential role of mast cells in AD neuropathogenesis has been largely unexplored, indirect evidence suggests a deleterious effect of mast cells in AD.23
Here, we investigated the contribution of mast cells to AD neuropathology and cognitive deficits with mechanistic focus on glial-mediated neuroinflammation. To this end, we crossed the 5XFAD mouse model24 with the mast cell-deficient Cpa3Cre/+ strain25,26 and evaluated AD neuropathology and assessed cognitive behavior. We show that mast cell deficiency improved contextual fear conditioning (FC) in 5XFAD mice without affecting cued FC, anxiety-like behaviors, or levels of β-amyloid. Importantly, we observed an enhanced transcriptional profile of disease-associated microglia and reduced markers indicative of reactive astrocytes in brains of mast cell-deficient 5XFAD mice. These studies show that mast cells are associated with cognitive deficits and regulate AD-related glial responses using the 5XFAD mouse model.
RESULTS
Meningeal mast cells scan cerebrospinal fluid contents and are required for meningeal expression of genes encoding immune-relevant effector proteins
Recent studies provide compelling evidence of communication between brain border immune cells and cells of the CNS.27,28 We first attempted to determine whether meningeal mast cells scan the contents of cerebrospinal fluid (CSF) to indirectly receive information about the status of the brain parenchyma. To test this, we introduced into CSF a fluorophore-conjugated antibody probe (FITC-labeled anti-Kit antibody) that binds to mast cells and then isolated dura mater to inspect the samples for the fluorescent signal. Our results showed positive labeling of mast cells, particularly those located near lymphatic sinuses (Figure 1A), indicating that mast cells are able to scan CSF contents and thereby indirectly monitor the brain environment.
Figure 1. Dural mast cells can scan cerebrospinal fluid contents and are required for meningeal expression of genes encoding immune-relevant effector proteins.
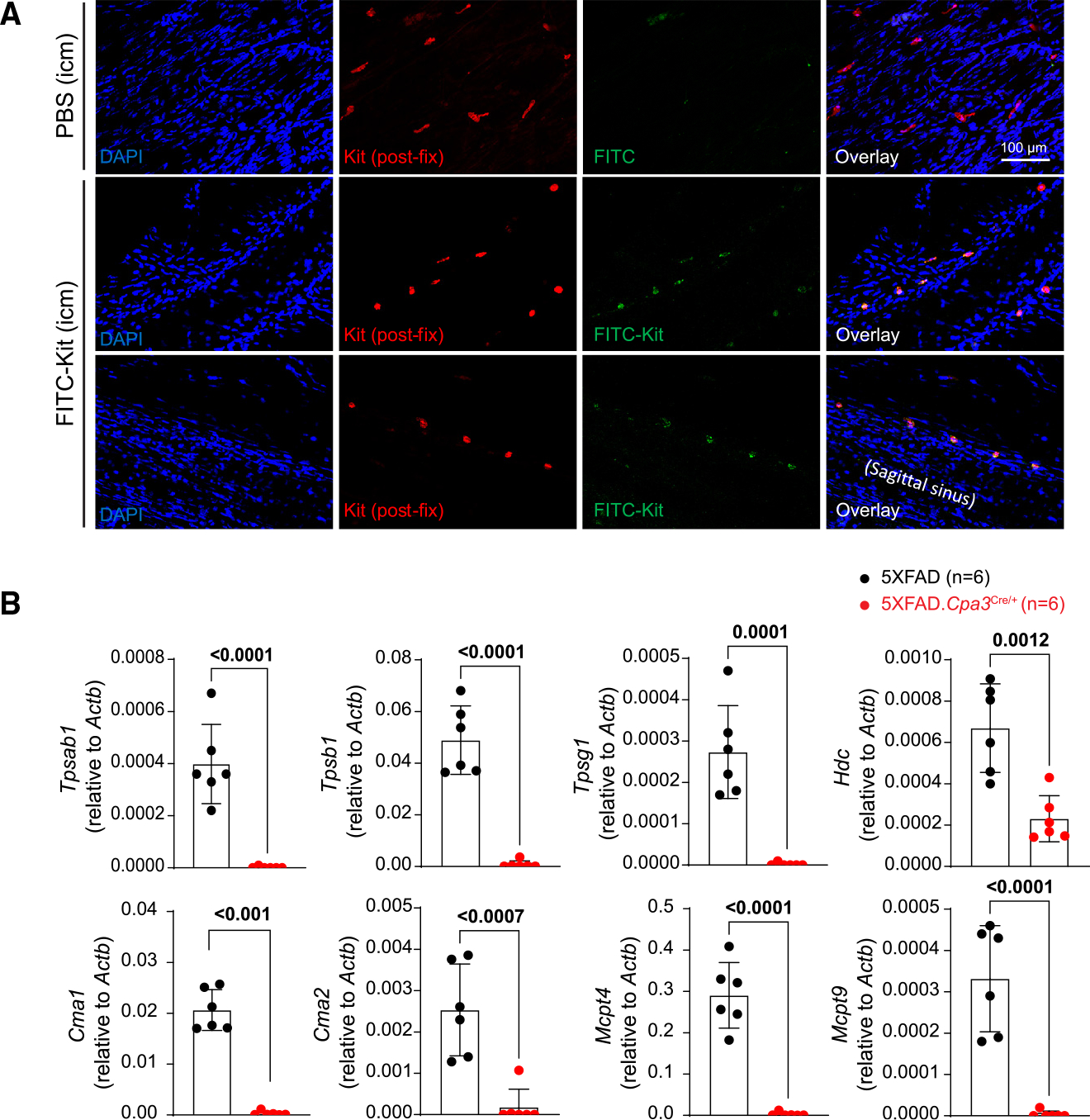
(A) Immunofluorescence images of dura harvested from mice intracisternally injected with PBS or FITC-labeled anti-KIT antibody. The tissues were then fixed and stained with anti-Kit antibody detected by secondary antibody conjugated with Alexa Fluor 568. Data are from one representative experiment of two (n = 1–2 mice/group).
(B) Quantitative PCR analysis of mast cell mediator genes in dura samples harvested from 12- to 13-month-old female 5XFAD and 5XFAD.Cpa3Cre/+ mice (n = 6/group). Bar graphs depict mean ± SD with individual mice as points. Unpaired t tests were performed to determine significance. See also Figure S1.
We next investigated whether mast cells are abnormally activated in 5XFAD amyloidosis model mice. To this end, we conducted immunofluorescent staining, flow cytometry, and quantitative PCR (qPCR) analyses to compare characteristics of dural mast cells between wild-type (WT) and 5XFAD mice aged 8–10 months (Figures S1A–S1E). We found no differences between groups in mast cell density, morphology, degranulation as detected by CD107a positivity, or gene expression (Figures S1A–S1E).
To directly assess the contribution of mast cells to AD neuropathogenesis, we crossed 5XFAD mice with the mast cell-deficient Cpa3Cre/+ strain. The Cpa3Cre/+ mouse contains an insertion of Cre recombinase into the Cpa3 gene, which encodes the mast cell-specific enzyme carboxypeptidase A3. Cpa3Cre/+ mice are specifically devoid of mast cells.25 We verified that 5XFAD.Cpa3Cre/+ mice did not have mast cells in the ear pinna or dura (Figure S1F). Our qPCR results show that mast cell deficiency greatly reduced gene expression of histidine decarboxylase, tryptase, chymase 1, chymase 2, and mast cell proteases 4 and 9 in dura of 5XFAD mice (Figure 1B). These results indicate that mast cells are required for the meningeal expression of histamine and several immune-relevant proteases.
Mast cell deficiency improves contextual fear conditioning in 5XFAD mice without affecting cued fear conditioning or anxiety-like behavior
A recent study reported that treating AD mice with masitinib, a tyrosine kinase inhibitor that modulates mast cell activity, ameliorated spatial memory deficits in an AD mouse model.29 To examine whether mast cell deficiency improves spatial cognition, we subjected 1-year-old female WT, Cpa3Cre/+, 5XFAD, and 5XFAD.Cpa3Cre/+ mice to contextual FC and cued FC protocols (Figure 2A). We did not observe any baseline differences in freezing behavior during the habituation period (Figure 2B). In the contextual FC test, 5XFAD mice displayed significantly reduced freezing behavior compared to WT mice and mast cell-deficient 5XFAD.Cpa3Cre/+ mice (Figure 2C; Cpa3 genotype x 5XFAD genotype interaction: p = 0.043). In contrast, 5XFAD. Cpa3Cre/+ mice froze at levels that were similar to those of WT mice, suggesting that mast cells likely contribute to fear-based memory deficits in 5XFAD mice (Figure 2C). 3–4 h after the contextual test, animals were placed in an altered FC chamber and allowed to explore for 3 min, followed by a 3-min tone (cued test). In contrast to the contextual FC tests, we observed no differences among these four groups (Figure 2D).
Figure 2. Depletion of mast cells restores contextual fear conditioning without affecting anxiety-like behavior in 5XFAD mice.
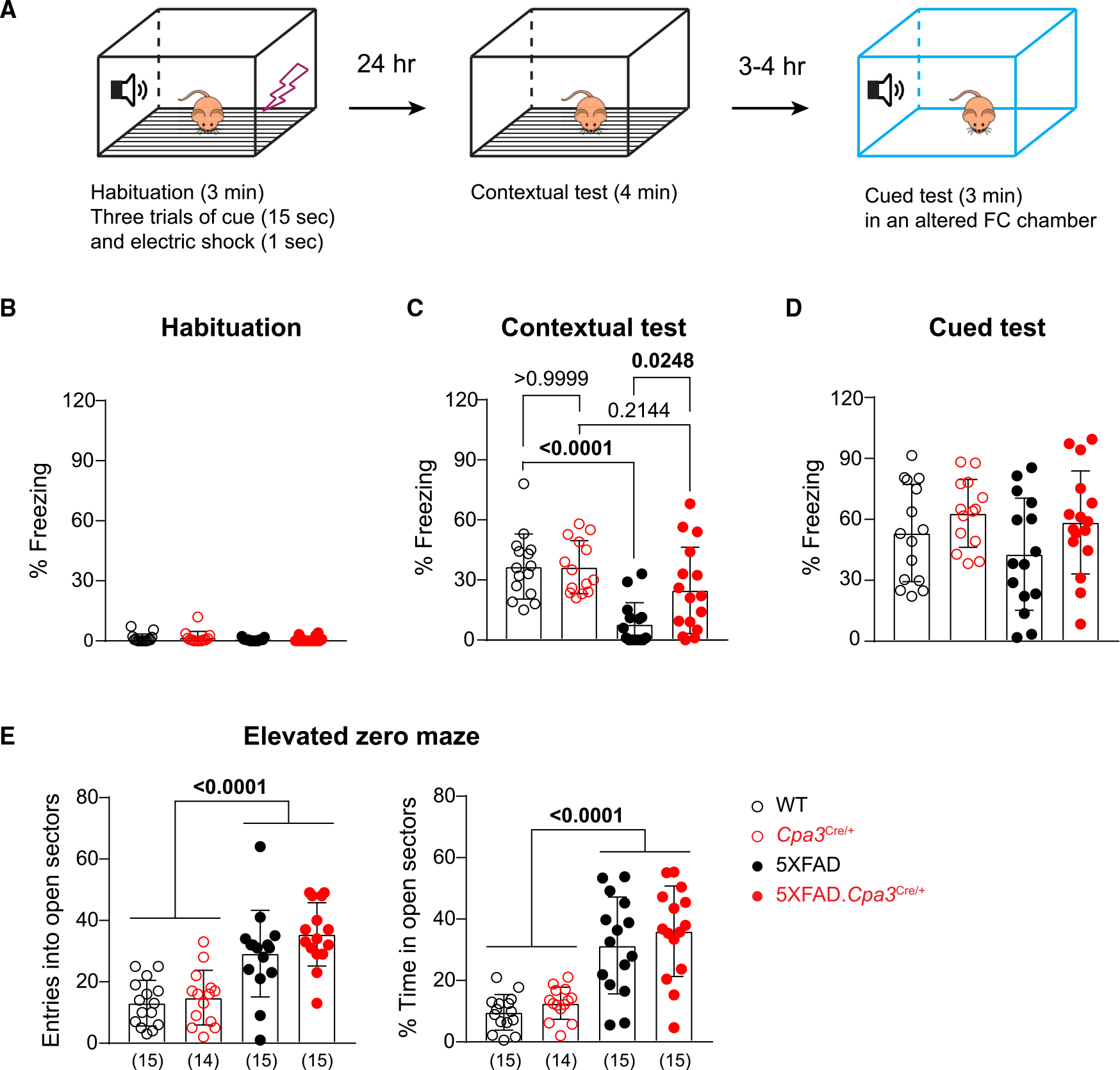
1-year-old female WT, Cpa3Cre/+, 5XFAD, and 5XFAD.Cpa3Cre/+ mice were evaluated in the fear conditioning (FC) test and elevated zero maze.
(A) Illustration of the 2-day FC test procedure.
(B–D) The freezing frequency during habituation (B), contextual FC test (C), and cued FC test (D).
(E) Number of entries and percent time spent in the open sectors in elevated zero maze. The number of mice is specified in the panels. Bar graphs depict mean ± SD with individual mice as points. Two-way ANOVA with Tukey’s multiple comparison tests were performed to determine significance. See also Figure S2.
Mice lacking mast cells due to Kit mutations have been reported to exhibit enhanced anxiety-like behaviors.30,31 Thus, we next examined whether the restored contextual freezing responses of 5XFAD.Cpa3Cre/+ mice (versus 5XFAD mice) was related to altered levels of anxiety. For this purpose, we evaluated the mice for approach-avoidance conflicts in an elevated zero maze.32 Consistent with the literature, 5XFAD mice displayed a decreased anxiety phenotype; i.e., they were more likely to explore the open sectors33 (Figure 2E; Cpa3 genotype x 5XFAD genotype interaction: p = 0.4172, 5XFAD genotype main effect: p < 0.0001). Notably, 5XFAD.Cpa3Cre/+ and 5XFAD mice showed similar numbers of entry and time spent in the open sectors (Figure 2E). Thus, improved contextual FC in 5XFAD.Cpa3Cre/+ mice was unlikely to be due to enhanced anxiety.
Mast cell deficiency does not alter amyloid pathology in the brains of 5XFAD mice
We next investigated whether the observed improvement in cognitive function in 5XFAD.Cpa3Cre/+ mice was linked to changes in β-amyloid pathology. We harvested brains from 7- to 9-month-old 5XFAD and control mice of both sexes, with and without mast cells. Cortical tissues were dissected and processed for tris-buffered saline (TBS)-soluble fraction and formic acid (FA)-soluble fraction of cell lysates. Amyloid monomers and oligomers are soluble in TBS, while amyloid fibrils derived from plaques are TBS insoluble but soluble in FA. ELISA assays revealed that the Aβ levels were comparable between the 5XFAD and 5XFAD.Cpa3Cre/+ mice, regardless of sex, strain, or lysate fraction (Figure S2A). We next assessed amyloid plaque burden in the cortex and hippocampus of female 5XFAD and 5XFAD. Cpa3Cre/+ mice. For this purpose, we used the 3D6 antibody, which recognizes the first five amino acids of Aβ peptide for immunofluorescence staining.34 Consistent with our ELISA assays, we found that amyloid plaque density and Aβ burden were similar between 5XFAD and 5XFAD.Cpa3Cre/+ mice (Figures S2B–S2C). Thus, the observed improvement in cognitive function could not be attributed to β-amyloid levels in the mast cell-deficient 5XFAD mice.
Membrane-bound amyloid precursor protein (APP) can be processed by α- or β-secretases followed by γ-secretase.35 To assess the effects of mast cell deficiency on APP processing, we examined APP metabolites in cortical brain lysates prepared from 8- to 9-month-old female and male mice. Western blot analyses showed that the levels of full-length APP, APP C-terminal fragment β (CTFβ), and secreted APP N-terminal fragments (sAPPα and sAPPβ) were unchanged in the 5XFAD.Cpa3Cre/+ mice compared to 5XFAD mice (Figure S2D). Collectively, these results indicate that mast cell deficiency does not alter either amyloid deposition or APP processing.
Depletion of mast cells increases disease-associated microglial transcriptional profile and multiple biological pathways
Microglia undergo dramatic morphological and transcriptional changes in response to β-amyloid and play multi-faceted and complex roles in neurodegeneration.36 We next investigated whether mast cell deficiency regulates microglial activity in 8-month-old female 5XFAD and 5XFAD.Cpa3Cre/+ mice (n = 4/genotype). To systematically profile the transcription of microglia, we conducted single-cell RNA sequencing (scRNA-seq) using the 10x Genomics Chromium system. To avoid the stress and other potential confounding effects caused by cell sorting, we utilized total brain cells harvested from the cortical tissue with viability rates 95%–99%.
The scRNA-seq identified eight primary brain-resident and -infiltrating cells, with microglia (n = 31,351) being the largest cell type (Figure S3A). Neurons (n = 926) and astrocytes (n = 2,076) were not well represented, probably because the brain preparation methods we employed were not optimal to dissociate cells with long processes. The microglia were partitioned into nine transcriptional subclusters and could be grouped into three microglial states: homeostatic (subclusters 1, 3, 4, and 5; feature genes: Tmem119, P2ry12), interferon (IFN)-responsive (subcluster 7; feature genes: Isg15, Stat1), and the disease-associated ones (subclusters 0, 2, 6, and 8; feature genes: Apoe, Csf1) (Figure 3A). We found an increase in the frequency of disease-associated microglia (DAM) and a reduction in frequency of homeostatic microglia in 5XFAD.Cpa3Cre/+ mice relative to 5XFAD mice (Figure 3B). Remarkably, multiple DAM signature genes were significantly upregulated in a mast cell-deficient environment (Figure 3C). These genes included stage 1 (B2m, Apoe, Ctsb, Ctsd, Lyz2, Gnas) and stage 2 (Cst7, Lpl, Csf1, Ccl6, Axl, Itgax, Spp1) DAM genes37,38 (Table S1). Silvin et al. previously integrated the scRNA-seq datasets from Keren-Shaul et al. and Van Hove et al. to arrive at eight DAM-conserved genes. Notably, five (Fabp5, Igf1, Itgax, Spp1, Gm1673) of these eight genes were significantly increased in microglia harvested from 5XFAD.Cpa3Cre/+ mice (Table S1).39,40 In addition, several major histocompatibility complex (MHC; B2m, Cd74, H2-Ab1, H2-Aa) and lysosomal (Cd68, Ctsb, Ctsd, Lyz2, Cst7) genes were upregulated, while the microglial homeostatic genes Crybb1, Txnip, and Glul were downregulated in microglia harvested from 5XFAD.Cpa3Cre/+ mice (Figure 3C).
Figure 3. Depletion of mast cells increases transcriptomic profiles of disease-associated microglia and impacts multiple biological pathways.
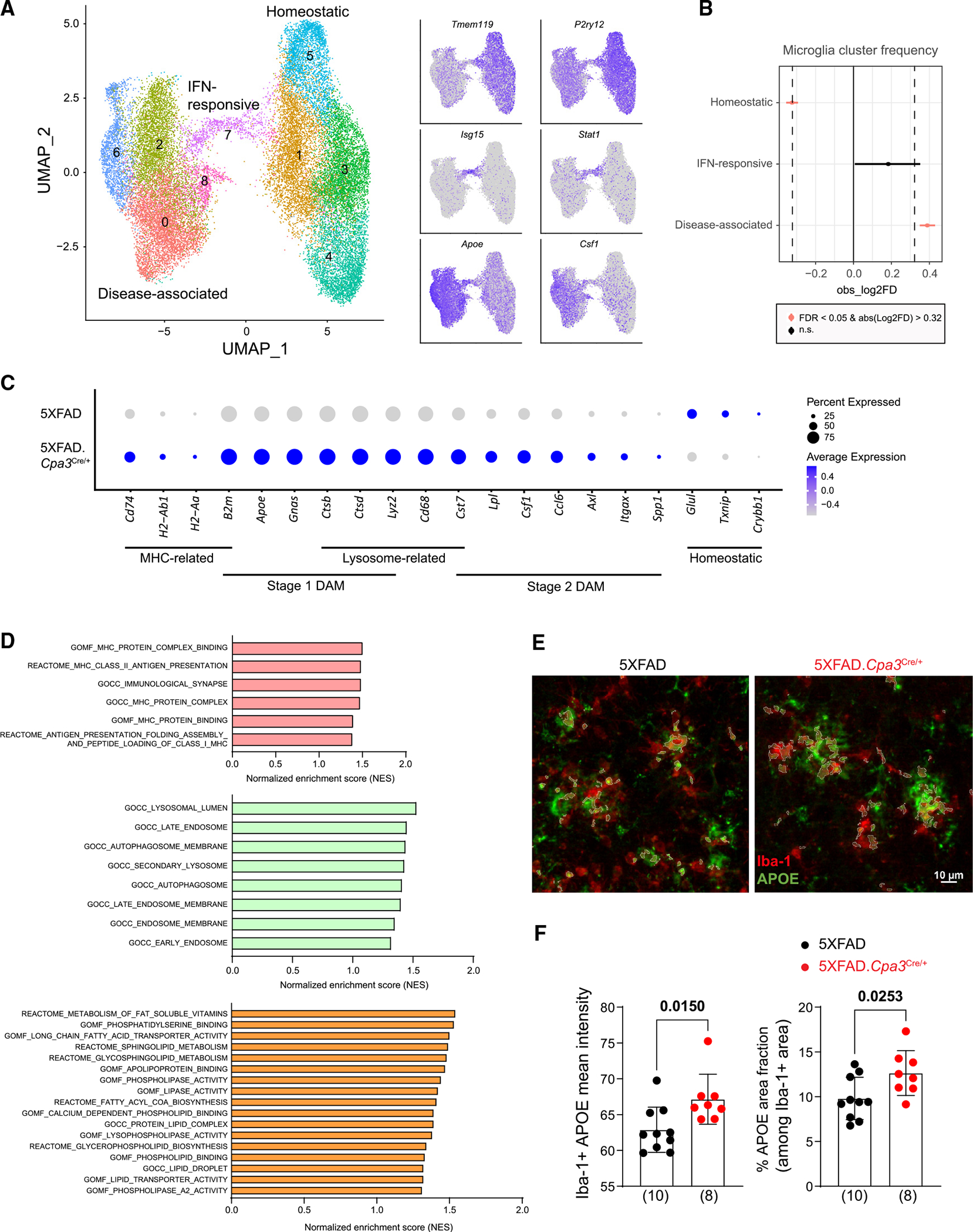
(A) Brain cells were isolated from the cortical regions of eight 8-month-old female 5XFAD and 5XFAD.Cpa3Cre/+ mice (n = 4/genotype under cold conditions and single-cell sequenced). UMAP plot of 31,351 single-cell RNA microglia profiles were partitioned into nine transcriptional subclusters and grouped into three microglial states based on their feature genes: homeostatic (Tmem119, P2ry12), IFN-responsive (Isg15, Stat1), and disease-associated ones (Apoe, Csf1).
(B) Changes in frequency of microglial states in 5XFAD and 5XFAD.Cpa3Cre/+ brains.
(C) Dot plot of DAM, MHC, lysosomal, and homeostatic genes of disease-associated state of microglia in (A). Expression level (color scale) of marker genes in two genotypes and the percentage of cells expressing them (dot size).
(D) GSEA analysis identified several MHC, endo-lysosome, and lipid pathways that were upregulated in mast cell-deficient disease-associated microglia subclusters compared with mast cell-sufficient microglia (false discovery rate q values < 0.25).
(E) Brain cortical sections were labeled with Iba-1 (red) and APOE (green) antibodies. Representative images of the visual area of isocortex from 8- to 9-month-old female 5XFAD (n = 10) and 5XFAD.Cpa3Cre/+ (n = 8) mice.
(F) Quantification of mean fluorescence intensity and coverage of APOE among Iba-1-positive microglia. Bar graphs depict mean ± SD with individual mice as points. Unpaired t test was performed to determine significance. IFN, interferon; MHC, major histocompatibility complex; GSEA; gene set enrichment analysis. See also Figure S3 and Table S1.
To explore the pathways enriched in microglia from mast cell-deficient mice, we analyzed the enrichment of functional categories among genes whose expression was affected in disease-associated subclusters in 5XFAD.Cpa3Cre/+ mice. Gene set enrichment analysis (GSEA) revealed several modules related to MHC binding and assembly, indicating that mast cells might affect antigen presentation by microglia (Figure 3D). GSEA analysis also uncovered enrichments in endo-lysosomal, endoplasmic reticulum, and Golgi apparatus pathways in 5XFAD. Cpa3Cre/+ mice, reflecting the potential effects of mast cells on microglial intracellular transport (Figures 3D and S3B).
Notably, GSEA analysis revealed differences in lipid-related gene expression between microglia obtained from mast cell-sufficient and -deficient 5XFAD brains (Figure 3D). Apoe, a DAM signature gene and an AD risk gene, plays a critical role in lipid transport. The expression of Apoe was significantly upregulated in microglia from 5XFAD.Cpa3Cre/+ mice (Figure 3C). To validate APOE protein expression, we performed immunohistochemistry to co-stain APOE and the microglial marker Iba-1 in the visual area of isocortex (Figures 3E and 3F). We found that the mean fluorescence intensity of microglial APOE significantly increased in 5XFAD.Cpa3Cre/+ mice. In addition, the mast cell-deficient 5XFAD mice demonstrated a significantly greater proportion of brain area that exhibited double positivity for APOE and Iba-1. Collectively, these data indicated that mast cell deficiency upregulated the transcriptomic signature of DAM and regulate multiple biological pathways.
Depletion of mast cells unexpectedly increases proinflammatory cytokine expression in myeloid populations of 5XFAD mice
We next investigated the impact of mast cell deficiency on proinflammatory cytokines associated with neuroinflammation. Surprisingly, electrochemiluminescence assays revealed a trend toward higher levels of two proinflammatory cytokines, TNF and IL-1β, in the cortical tissues of 5XFAD.Cpa3Cre/+ mice compared to their 5XFAD counterparts (Figure S3C). These findings were confirmed by scRNA-seq, which revealed elevated expression of Tnf and Il1b in 5XFAD.Cpa3Cre/+ disease-associated subclusters (Figure S3D). Tnf and Il1b are not signature genes of DAM. In fact, the cells that highly expressed these cytokines were enriched in subclusters 0 and 8, whereas the DAM signature genes (e.g., Ccl6) were primarily located in subclusters 2 and 6 (Figures 3A and S3E), suggesting that these proinflammatory cytokine-producing cells might not be actual DAMs.
Recently, Silvin et al. identified a distinct population of disease inflammatory macrophages (DIMs) that are ontogenetically and functionally different from previously identified DAMs.40 Unlike DAMs, DIMs are monocyte derived and express high levels of Tnf and Il1b. Silvin et al. integrated three scRNA-seq datasets and established a list of 24 genes that are highly expressed in DIMs compared to DAMs. We then compared this DIM-conserved gene list with the feature genes of our microglial subclusters. Interestingly, subclusters 2 and 6, which highly expressed DAM signature genes, only showed a few DIM genes (Figure S3F). In contrast, subclusters 8 and 0 contained more differentially expressed genes that were DIM conserved, including Tnf and Il1b. These transcriptional profiles suggest that some DIMs might be present in our disease-associated subclusters and could be potential sources of TNF and IL-1β detected in 5XFAD.Cpa3Cre/+ mice.
Mast cell deficiency reduces GFAP expression and alters astrocyte morphology in 5XFAD mice
In the brains of 5XFAD mice, astrocytes become reactive in response to β-amyloid and microglial activation.33 To examine whether mast cell deficiency affects astrocytes, we assessed the expression of the reactive cytoskeleton protein GFAP and morphology. We collected cortical brain tissue from 8- to 9-month-old female AD and non-AD mice and prepared samples for western blot and immunohistochemistry. We observed a significant reduction in GFAP levels in 5XFAD.Cpa3Cre/+ mice compared to 5XFAD animals (Figure 4A; Cpa3 genotype x 5XFAD genotype interaction: p = 0.0002). Confocal microscopy showed that GFAP immunostaining intensity, coverage, and circularity were significantly decreased in the cortex of 5XFAD.Cpa3Cre/+ mice (Figures 4B and 4C). We next performed z stack imaging and used NIS Element software to examine individual astrocytes. Consistently, GFAP-positive astrocytes displayed a significant reduction of circularity and shape factor, indicating an alteration in astrocyte morphology in the absence of mast cells (Figures 4D and 4E).
Figure 4. Mast cell deficiency reduces GFAP expression and alters astrocyte morphology in 5XFAD mice.
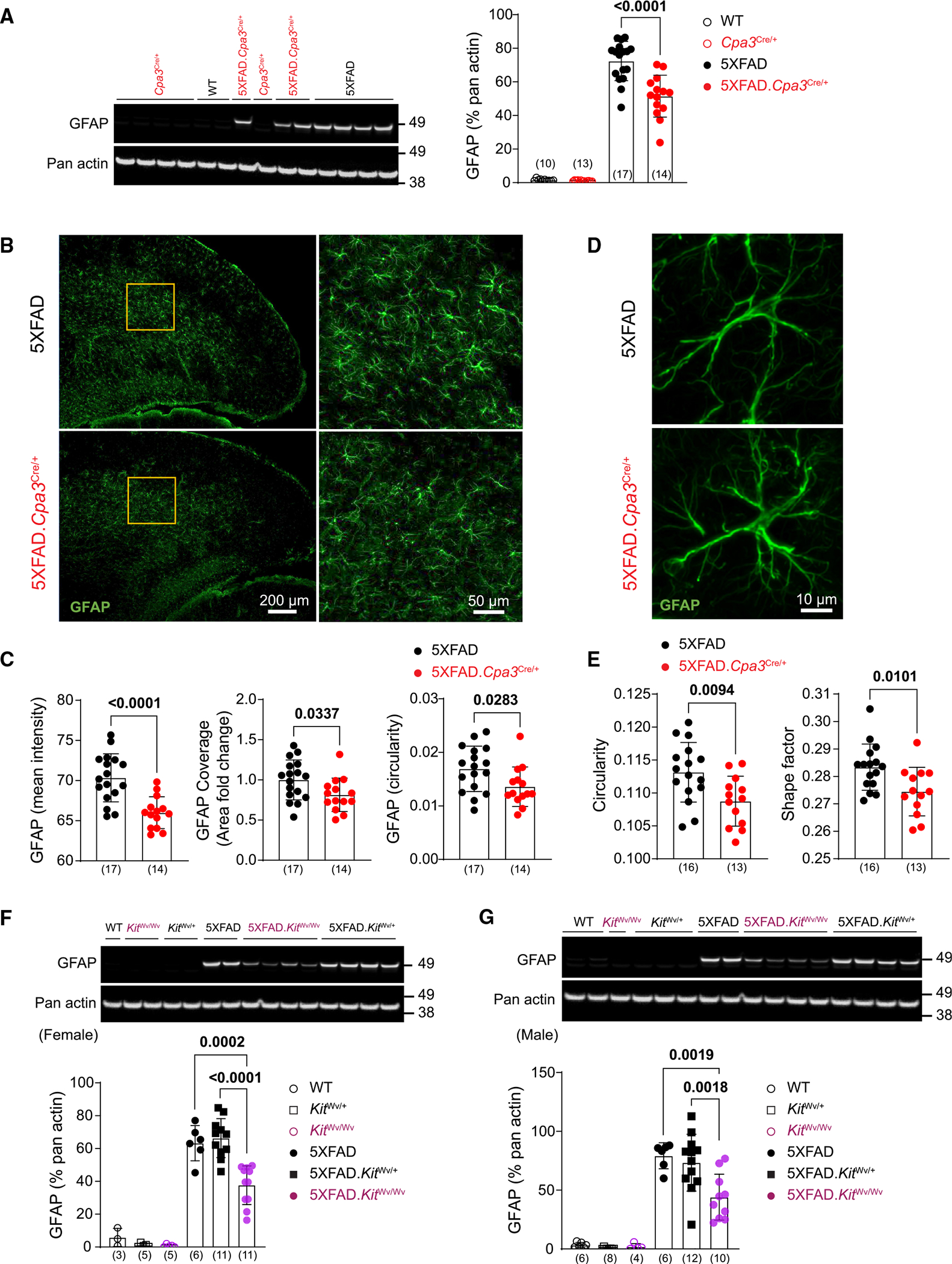
(A) Representative western blots and protein levels for reactive astrocyte marker GFAP measured from cortical homogenates of 8- to 9-month-old female Cpa3Cre cohort mice.
(B) Brain cortical sections were labeled with GFAP (green) antibody. Representative images of cortex from 8- to 9-month-old female 5XFAD and 5XFAD.Cpa3Cre/+ mice. The details of GFAP-positive astrocytes in the inserts are shown.
(C) Quantification of mean fluorescence intensity, area fold change, and circularity of GFAP-positive cells in (B).
(D) Representative images of a single astrocyte in the visual cortex of 8- to 9-month-old female 5XFAD and 5XFAD.Cpa3Cre/+ mice.
(E) Quantification of GFAP-positive astrocyte circularity and shape factor in (D).
(F and G) Representative western blots and protein levels for astrogliosis marker GFAP measured from cortical homogenates of 7- to 9-month-old female (F) or 8- to 10-month-old male (G) KitWv cohort mice. GFAP, glial fibrillary acidic protein. Bar graphs depict mean ± SD with individual mice as points. The number of mice is specified in the panels. Two-way ANOVA with Tukey’s multiple comparison tests (A, F, and G) and unpaired t test (C and E) were performed to determine significance. See also Figure S4.
To confirm that mast cell deficiency reduced markers of reactive astrocytes, we further crossed 5XFAD mice with the widely used mast cell-deficient mouse line, KitWv/Wv strain, and repeated the experiments (Figure S4A). Consistent with the results obtained from 5XFAD.Cpa3Cre/+ mice, GFAP levels were significantly reduced in both female and male 5XFAD.KitWv/Wv mice (Figures 4F and 4G; Kit genotype x 5XFAD genotype interaction: p = 0.0023 and p = 0.0243, respectively). Furthermore, z stack immunofluorescence analysis revealed that astrocytes harvested from male 5XFAD.KitWv/Wv exhibited decreased circularity and shape factor compared to their counterparts from Kit heterozygous 5XFAD mice (Figures S4B and S4C). Thus, using two distinct genetic methods of mast cell depletion (Cpa3Cre/+ and KitWv), we demonstrated that the absence of mast cells reduced expression of GFAP, a marker for reactive astrocytes, and altered astrocyte morphology in the brains of 5XFAD mice.
DISCUSSION
Mast cells are primarily known for their role in triggering IgE-dependent allergic reactions. Nonetheless, limited evidence has suggested a potential link between mast cells and Alzheimer’s disease.23 Here, we directly assessed the role of mast cells in AD pathogenesis by knocking out mast cells in 5XFAD mice. We show that mast cell depletion improved contextual FC without affecting cued FC, anxiety-like behavior, or amyloid pathology, and it elevated neuroprotective DAM transcriptomic signatures and reduced markers indicative of reactive astrocytes. We also provide evidence that mast cells in dura mater are capable of scanning the CSF for signals from the brain environment and respond by expressing immune-regulatory mediators that impact brain glial cells and cognition. This aligns well with recent reports of communication between meningeal immune cells and brain cells.28,41 Reports have shown that mast cells respond to amyloid by releasing inflammatory mediators.42,43 Based on our findings, we hypothesize a system of bidirectional communication between dural mast cells and the CNS, whereby mast cells scan the CSF for signals from the brain environment and respond by expressing immune-regulatory mediators, impacting cognition and brain glial cell function. AD pathology can compromise the blood-brain barrier allowing peripheral mediators, including those released by mast cells, to penetrate the brain parenchyma and act on brain-resident microglia and astrocytes.44. It has been shown that mast cell-derived mediators, such as histamine and tryptase, trigger microglial activation in rodent models of CNS inflammation.17–19 DAMs express a neuroprotective gene signature implicated in remyelination and tissue repair.40,45 In mast cell-deficient 5XFAD mice, we found an increased DAM cluster frequency, elevated DAM transcriptional profile, and enriched biological pathways. These data, coupled with our meningeal gene expression results, suggest release of histamine, tryptase, and other mediators from mast cells may inhibit the formation of neuroprotective DAMs.
Not only does mast cell deficiency enhance the neuroprotective microglia phenotype, but it may also confer beneficial effects by modulating astrocyte reactivity. Using two distinct mast cell-deficient 5XFAD strains, we showed that depletion of mast cells reduced markers of reactive astrocytes. Reactive astrocytes play a detrimental role in driving cognitive impairment in AD models.46,47 One group reported that blocking astrocyte reactivity protects human neurons in vitro and improved spatial memory in two AD mouse models.48 In addition, several other research labs demonstrated a correlation between decreased astrocyte reactivity, as evidenced by the downregulation of GFAP, and improved cognition in mice with amyloidosis.49–53 Collectively, our observations on glial responses in mast cell-depleted 5XFAD mice suggest that mast cells contribute to AD pathogenesis by suppressing neuroprotective DAMs and accelerating reactive astrogliosis.
The elevated DAM gene profile and reduced reactive astrocyte markers may contribute to the restored hippocampus-dependent contextual FC in mast cell-deficient 5XFAD mice. Notably, cued FC was unaffected, indicating that the improvement in context fear was not due to an overall enhanced freezing response. The fact that only context fear was restored, which is known to rely on cortico-hippocampal communications, suggests that mast cell deficiency may be beneficial for context processing, as fear responses depend on amygdala-brainstem pathways. Moreover, we did not observe any changes in anxiety-like behavior between mast cell-sufficient and -deficient 5XFAD mice. In conclusion, these results indicated that mast cell deficiency could improve hippocampus-dependent cognitive function, and this improvement was not due to enhanced freezing responses or anxiety-like behavior.
Recent AD clinical trials and mouse studies have included drugs that regulate mast cells, such as cromolyn, imatinib (Gleevec), and masitinib.54–56 Cromolyn stabilizes mast cells and is an approved asthma medication,57,58 although the mechanism of action is still unclear. Imatinib and masitinib are tyrosine kinase inhibitors that target Kit signaling, which is required for mast cell survival. Both drugs demonstrate therapeutic potential for treating human systemic mastocytosis, a rare disease resulting from abnormal accumulation and activation of mast cells.54,59,60 Interestingly, Li et al. reported that masitinib treatment improved cognitive performance of AD mice without impacting amyloid loads, potentially due to its effects on mast cells.29 Notably, a phase 3 clinical trial investigating masitinib for the treatment of mild-to-moderate AD patients demonstrated cognitive and functional improvements.61
In conclusion, we show that depletion of mast cells improves contextual FC in 5XFAD mice without affecting cued FC, anxiety-like behavior, or amyloid burden. Mast cell depletion also led to upregulation of transcriptomic signatures for putatively protective DAM and resulted in reduced markers indicative of reactive astrocytes. Based on our findings, we hypothesize a system of bidirectional communication between dural mast cells and the CNS, whereby mast cells scan the CSF for signals from the brain environment and respond by expressing immune-regulatory mediators that impact cognition and brain glial cell function. In summary, we provide the direct evidence that mast cells impact cognitive decline in 5XFAD mice by regulating AD-related glial function, suggesting that mast cells may serve as promising therapeutic targets for AD.
Limitations of the study
In this study, we employed two genetic models to demonstrate that mast cell deficiency enhances cognition and induces changes in glial responses. However, it is important to acknowledge that the use of global knockout animals (5XFAD.Cpa3Cre/+ mice) leaves open the possibility of physiological compensation due to permanent mast cell depletion, which may contribute to the observed phenotypes. Additionally, previous findings have indicated that Cpa3Cre/+ mice exhibit a reduced number of splenic basophils, suggesting that other cell types expressing Cpa3 could potentially play a role in the observed effects. Although this in vivo study demonstrates the impact of mast cell deficiency on glial cells and cognitive function, the specific mechanism through how mast cells exert their influence on the brain remains yet to be elucidated. We hypothesized that dura mast cell-derived mediators may play a crucial role in penetrating the brain parenchyma and orchestrating these effects. Nonetheless, further investigations are required to provide a comprehensive understanding of the mechanism of action. Lastly, it is worth noting that our analysis was limited to the cortex and hippocampus, and therefore, the impact of mast cell deficiency on other brain regions remains unknown.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Rudolph E. Tanzi (tanzi@helix.mgh.harvard.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the Key resources table. All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Alexa Fluor 647 anti-mouse CD206 (MMR) Antibody | BioLegend | Cat# 141712; RRID: AB_10900420 |
| Alexa Fluor 700 anti-mouse Ly-6C Antibody | BioLegend | Cat# 128024: RRID: AB_10643270 |
| Alexa Fluor® 488 anti-mouse Ly-6G Antibody | Biolegend | Cat# 127626; RRID: AB_2561340 |
| APC/Cy7 anti-mouse/human CD11b Antibody | BioLegend | Cat# 101226; RRID: AB_830642 |
| BD Pharmingen™ APC Rat Anti-Mouse CD19 | BD Biosciences | Cat# 550992; RRID: AB_10893995 |
| Brilliant Violet 421™ anti-mouse F4/80 Antibody | Biolegend | Cat# 123137; RRID: AB_2563102 |
| Brilliant Violet 510™ anti-mouse I-A/I-E Antibody | Biolegend | Cat# 107635; RRID: AB_2561397 |
| Brilliant Violet 711™ anti-mouse CD45 Antibody | BioLegend | Cat# 103147; RRID: AB_2564383 |
| Cy5 AffiniPure Donkey Anti-Goat IgG (H + L) | Jackson ImmunoResearch | Cat# 705-175-147; RRID: AB_2340415 |
| Donkey anti-Goat IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 568, Thermo Fisher Scientific | Thermo Fisher Scientific | Cat# A-11057; RRID: AB_2534104 |
| Donkey anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488, Thermo Fisher Scientific | Thermo Fisher Scientific | Cat# A-21206; RRID: AB_2535792 |
| Donkey anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 568, Thermo Fisher Scientific | Thermo Fisher Scientific | Cat# A10042; RRID: AB_2534017 |
| Donkey Anti-Rat IgG (H + L) Antibody, Alexa Fluor 488 Conjugated, Molecular Probes | Thermo Fisher Scientific | Cat# A-21208; RRID: AB_2535794 |
| FITC anti-mouse CD117 (c-Kit) Antibody | BioLegend | Cat#105805; RRID: AB_10060183 |
| Goat anti-mouse Iba-1 | FUJIFILM (Wako) | Cat# 011-27991; RRID: AB_2935833 |
| Goat Anti-mouse IgG (H + L), F(ab’)2 Fragment (Alexa Fluor 647 Conjugate) antibody | Cell Signaling Technology | Cat# 4410; RRID: AB_1904023 |
| Goat Anti-Mouse IgG1 (HRP) preadsorbed | Abcam | Cat# ab98693; RRID: AB_10674928 |
| Goat anti-Rabbit IgG (H + L) Cross-Adsorbed Secondary Antibody, HRP | Thermo Fisher Scientific | Cat# G-21234; RRID: AB_2536530 |
| Goat anti-Rat IgG (H + L) Cross-Adsorbed Secondary Antibody, HRP | Thermo Fisher Scientific | Cat# A18871; RRID: AB_2535648 |
| Human/Mouse CD117/c-kit Antibody | R&D | Cat# AF1356; RRID: AB_354750 |
| Lab Vision™ Actin, pan Ab-5, Mouse Monoclonal Antibody | Fisher Scientific | Cat# MS-1295-P1; RRID: AB_63316 |
| Mouse monoclonal anti-Aβ1-5, clone 3D6 | Lilly | N/A, gift |
| PE anti-mouse FcεRIα Antibody | BioLegend | Cat# 134307; RRID: AB_1626104 |
| PE/Cyanine7 anti-mouse CD117 (c-Kit) Antibody | Biolegend | Cat# 105813; RRID: AB_313222 |
| PE/Cyanine7 anti-mouse CD19 Antibody | Biolegend | Cat# 115520; RRID: AB_313655 |
| PerCP/Cyanine5.5 anti-mouse CD107a (LAMP-1) Antibody | BioLegend | Cat# 121626; RRID: AB_2572055 |
| Purified anti-mouse CD16/32 Antibody (clone 93) | Biolegend | Cat# 101301; RRID: AB_312800 |
| Purified anti-β-Amyloid, 1–16 Antibody (6E10) | BioLegend | Cat# 803001; RRID: AB_2564653 |
| Rabbit anti-APP C terminus (C66) | In-house | https://pubmed.ncbi.nlm.nih.gov/34010653/ |
| Rat anti-human/mouse GFAP Monoclonal Antibody (2.2B10) | Thermo Fisher Scientific | Cat# 13–0300; RRID: AB_2532994 |
| Recombinant rabbit Anti-Apolipoprotein E (APOE) antibody [EPR19378] | Abcam | Cat# ab183596; RRID: AB_2832971 |
| sAPPβ-sw (6A1) Anti-Human Mouse IgG MoAb | IBL-America | Cat# JP10321; RRID: AB_1630822 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| 2-mercaptoethanol | Sigma-Aldrich | Cat# M3148-25ML |
| 10X PBS Solution pH 7.4 | FISHER | Cat# P0496 |
| Absolute Ethanol (200 proof) | Fisher Scientific | Cat# BP2818-4 |
| Bovine serum albumin (BSA) | Fisher | Cat# BP-1600-100 |
| Collagenase D (100 mg) | Sigma-Aldrich | Cat# 11088858001 |
| DAPI solution | Thermo Fisher Scientific | Cat# 62248 |
| Deoxyribonuclease I | Worthington | Cat# LS002004 |
| Dispase II | Millipore Sigma | Cat# D4693 |
| EDTA (0.5 M), pH 8.0, RNase-free | Thermo Fisher Scientific | Cat# AM9261 |
| Ethylene glycol | Fisher Scientific | Cat# BP230-1 |
| FLUKA Formic acid, for mass spectrometry, ~98% | Thomas Scientific | Cat# 94318-250ML |
| Glycerol | Fisher Scientific | Cat# G31-500 |
| Halt phosphatase inhibitor cocktail | Thermo Fisher Scientific | Cat# 1862495 |
| HBSS with calcium and magnesium | Thermo | Cat# 24020117 |
| HBSS without calcium and magnesium | Gibco | Cat# 14170 |
| HistoPrep Xylene | Fisher Scientific | Cat# HC-700-1GAL |
| Hoechst 33342 Solution (20 mM) | Thermo Fisher Scientific | Cat# 62249 |
| Hyaluronidase from bovine testes | Millipore Sigma | Cat# H3506-100MG |
| M-PER™ Mammalian Protein Extraction Reagent | Thermo Fisher Scientific | Cat# 78501 |
| Normal donkey serum | Sigma-Aldrich | Cat# D9663, RRID:AB_2810235 |
| OneComp eBeads™ Compensation Beads | Thermo Fisher Scientific | Cat# 01-1111-42 |
| Papain from papaya latex | Sigma-Aldrich | Cat# P4762-100MG |
| Paraformaldehyde (32%) | Electron Microscopy Sciences | Cat# 50-980-495 |
| Percoll | GE Healthcare | Cat# 17-0891-01 |
| Pierce ECL Western Blotting Substrate | Thermo Fisher Scientific | Cat# 32106 |
| PowerTrack SYBR Green Master Mix | Thermo Fisher Scientific | Cat# A46110 |
| ProLong™ Gold Antifade Mountant | Thermo Fisher Scientific | Cat# P36934 |
| QIAzol Lysis Reagent | Qiagen | Cat# 79306 |
| RBC Lysis Buffer (10X) | BioLegend | Cat# 420301 |
| Restore Western Blot Stripping Buffer | Thermo Fisher Scientific | Cat# 21059 |
| Richard-Allan Scientific Cytoseal XYL | Thermo Fisher Scientific | Cat# 8312-4 |
| RNAprotect Tissue Reagent | Qiagen | Cat# 76104 |
| RPMI 1640 Medium, no phenol red | Thermo Fisher Scientific | Cat# 11835030 |
| Sodium azide | Sigma-Aldrich | Cat# 71289-5G |
| Sodium Chloride (Crystalline/Certified ACS) | Fisher Scientific | Cat# S271-3 |
| Sucrose | Millipore Sigma | Cat# 84097 |
| SuperBlock™ T20 (TBS) Blocking Buffer | Thermo Fisher Scientific | Cat# 37536 |
| SuperSignal West Femto Maximum | Thermo Fisher Scientific | Cat# 34096 |
| Sensitivity Substrate | ||
| TBS, Tris-Buffered Saline, 10X Solution, pH 7.4 | Fisher Scientific | Cat# BP24711 |
| Toluidine blue dye | Sigma-Aldrich | Cat#198161-5G |
| Triton X-100 | Sigma-Aldrich | Cat# TX1568-1 |
| Xpert Prestained Protein Marker | GenDEPOT | Cat# P8502-050 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Human/rat beta amyloid (40) ELISA Kit | Wako | Cat# 294-62501 |
| Human/rat beta amyloid (42) ELISA Kit | Wako | Cat# 290-62601 |
| LIVE/DEAD™ Fixable Yellow Dead Cell Stain Kit, for 405 nm excitation | Thermo Fisher Scientific | Cat# L34968 |
| Pierce BCA Protein Assay Kit | Thermo Fisher Scientific | Cat# 23225 |
| RNeasy Lipid Tissue Mini Kit | Qiagen | Cat# 74804 |
| RNeasy Plus Mini Kit | Qiagen | Cat# 74134 |
| SuperScript™ IV VILO™ Master Mix with ezDNase™ Enzyme | Thermo Fisher Scientific | Cat# 11766050 |
| V-PLEX Proinflammatory Panel1 (mouse) Kit (5 Plate) | Meso Scale Diagnostics | K15048D-2 |
|
| ||
| Deposited data | ||
|
| ||
| Raw sequencing data files for single-cell RNA sequencing | This paper | GEO accession: GSE240687 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| 5XFAD and WT Mice | The Jackson Laboratory | Cat# 034848-JAX, RRID:MMRRC_034848-JAX |
| C57BL/6J-KitW-v/J Mice | The Jackson Laboratory | Cat# JAX:000049, RRID:IMSR_JAX:000049 |
| CreMaster Mice | Rodewald Lab | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| Adobe Illustrator (version 25.3.1) | Adobe | RRID:SCR_010279 |
| EthoVision XT (version 9.0) | Noldus Information Technology | RRID:SCR_000441 |
| FACSDiva Software (version 6.1.3) | BD Biosciences | RRID:SCR_001456 |
| FIJI-ImageJ | NIH | RRID:SCR_003070 |
| FlowJo (version 10.7.1) | Tree Star | RRID:SCR_008520 |
| GraphPad Prism 7 and 8 | GraphPad Software | RRID:SCR_002798 |
| NIS-Elements Advanced Research software (version 5.42.01) | Nikon | https://www.microscope.healthcare.nikon.com |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Female and male Cpa3Cre/+ (the Rodewald Lab), KitWv/+(The Jackson Laboratory), 5XFAD transgenic mice (The Jackson Laboratory), and WT littermates were used for experiments. Cpa3Cre/+ or KitWv/+ were crossed to 5XFAD transgenic mice and used for experiments at 3–12 months of age. All 5XFAD mice were heterozygotes with respect to the transgene. Mice were group-housed in our SPF facility and kept on a 12 h light/dark cycle with ad libitum access to food and water. All procedures in this study were conducted in accordance with approvals granted by the Institutional Animal Care and Use Committee (IACUC) at Massachusetts General Hospital.
METHOD DETAILS
Intracisternal injection
The administration of intracisternal magna (icm) injection was performed following the protocol previously described.62 Prior to the procedure, the mouse received a subcutaneous dose of pre-emptive buprenorphine (Buprenex) at a dosage of 0.1 mg/kg. The mouse was placed on a stereotactic frame (Stoelting, Wood dale IL USA) and the surgical site was prepared by shaving, disinfecting, and dissecting the cervical muscles. The cisterna magna membrane was then identified using microscopy and punctured using a Hamilton syringe (Gastight #1701) fitted with a 34-gauge needle. Either 10 μL of FITC-conjugated anti-Kit antibody (BioLegend) or sterile PBS was slowly injected into the cisterna magna over a period of 1 min while the mouse was under 1.5–2.0% isoflurane anesthesia. Following the injection, the surgical site was sutured, and the mouse allowed to recover.
Tissue harvest and sample preparation
Mice were deeply anesthetized with 3–4% isoflurane, and perfused transcardially with 25–30 mL PBS. Isolated brains were bisected longitudinally. Hemispheres were separated for biochemical analyses, flow cytometry, and histology. In some experiments, ear pinnae were harvested for histology, and skull caps/dura were harvested for histology.
For biochemical analyses, cortical tissues were dissected from the right hemisphere, weighed, and frozen on dry ice according to our published methods with some modifications.56 Cortical tissues were homogenized with 10X volume tris-buffered saline (TBS) buffer solution (mL; based on weight in mg) containing 1X Halt protease inhibitor cocktail (Thermo Fisher Scientific) and 5 mM EDTA. We first processed the crude brain lysates by ultracentrifugation at 100,000 × g for 30 min at 4°C. The supernatant was collected as TBS-soluble brain samples (TBS fractions). The pellet was further extracted in 200 μL of 70% formic acid (FA) followed by sonication and subsequent ultracentrifugation at 100,000 × g for 30 min at 4°C. The supernatant was collected as FA-soluble samples (FA fractions). We also processed the crude brain lysates by adding 3X volume Mammalian Protein Extraction Reagent (MPER) (Thermo Fisher Scientific) with 1X Halt protease inhibitor cocktail, followed by centrifugation at 12,000 rpm for 20 min at 4°C. The supernatant was transferred to clean eppendorf tubes and collected as total proteins (MPER fractions). Bicinchoninic acid (BCA) assay (Thermo Fisher Scientific) was performed to determine protein concentrations.
For quantitative RT-PCR, mouse dura, cortical, or hippocampal samples were stored in RNAprotect Tissue Reagent (Qiagen) and stored at −20°C for immediate stabilization of RNA.
For flow cytometry, the dura was dissected and incubated in the dura digestion mix (0.66 mg/mL collagenase D, 800 μg/ml dispase II, 85 μg/ml hyaluronidase, and 125 U/ml DNase I) in Hanks′ Balanced Salt Solution (HBSS) with gentle shaking (120 rpm) at 37°C for 30 min. After incubation, the suspension was mixed with 10 mL neutralization buffer (2% FCS and 1 mM EDTA in PBS) and placed on ice. The tissues were further broken down by pipetting. Cells were then filtered and centrifuged at 400 × g for 8 min at 4°C. The Supernatant was carefully aspirated and the cells were resuspended in FACS buffer (0.5% BSA, 2 mM EDTA, and 0.02% sodium azide in PBS) for surface staining.
For histology, the left hemisphere of mouse brain was fixed in 15 mL 4% paraformaldehyde (Electron Microscopy Sciences) in cold PBS for 48 h at 4°C and then transferred into a 30% sucrose solution at 4°C until sedimented. Sagittal or coronal sections (40 μm) were cut from an ice-cooled block using a sliding microtome (Leica). Brain sections were stored in a cryoprotective reagent containing 28.5% ethylene glycol (Fisher Scientific) and 23.8% glycerol (Fisher Scientific) at −20°C until staining. In some experiments, the brain and left ear pinna were collected and fixed in 4% paraformaldehyde in PBS for 24–48 h at 4°C. The tissues were then embedded in paraffin and sectioned coronally (5 μm) at the Massachusetts General Hospital DF/HCC Pathology Core facility. Skull caps were dissected and fixed in 4% paraformaldehyde in PBS for 24 h at 4°C before transferring to the storage buffer (10X PBS plus 0.02% sodium azide).
To perform single-cell sequencing, the cortex from both hemispheres was dissected and placed in 0.66 mg/mL pre-warmed papain/RPMI medium in C-tubes (Miltenyi Biotec). The tissues were dissociated using a Gentle MACS dissociator (Miltenyi Biotec) for 37 s (program 1), followed by incubation at 37°C for 15 min under rotation (120 rpm). Next, 0.66 mg/mL pre-warmed collagenase D/RPMI medium was added, and the tissues were dissociated again for 31 s (program 2) using the Gentle MACS dissociator. Next, DNase I (Worthington) was added to the final concentration of 125 U/ml, and the mixture was incubated for another 10 min at 37°C under rotation. The tissues were then dissociated for 61 s (program 3) using the Gentle MACS dissociator. The cell suspensions were incubated for 15 min at 37°C under rotation. After the final incubation, we mixed the suspension with neutralization buffer (2% FCS and 1 mM EDTA in PBS) and broke down the tissues by pipetting. The cells were filtered and centrifuged at 400 × g for 8 min at 4°C. The supernatant was aspirated, and the cells were resuspended in 27% percoll (GE Healthcare). The cells were centrifuged again at 850 × g for 40 min at 4°C with a deceleration of 4, and then washed and lysed for red blood cells (BioLegend). The cell suspension was resuspended in cold FACS buffer without azide and the viable cells were counted before library preparation.
Single-cell RNA-sequencing
Single-cell RNA-seq library construction was performed on the Chromium 10x instrument using Chromium single cell 3′ reagent v3.0 kit (10x Genomics), followed by sequencing on Illumina NextSeq 2000 instrument, which resulted in approximately 100 million reads per sample.
Single-cell RNA-seq data analysis
Initial processing of scRNA-Seq sequencing data was performed using CellRanger (v4.0.0) (https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome).63 In brief, reads were aligned to mm10 mouse reference genome (mapping rate of ~90%) followed by the generation of read counts per gene in each cell. Further analysis was performed using Seurat 3.2.3 package (https://satijalab.org/seurat/)64. We filtered out cells with <200 expressed genes and genes expressed in <3 cells, followed by the exclusion of cells with high content of mitochondrial transcripts (>10% of total reads). A total of 44,835 cells were analyzed. Counts across all cells for each sample were normalized using NormalizeData function in Seurat. Using the FindVariableFeatures function, 2000 features were selected to be used in Principal Component Analysis (PCA). All individual samples were integrated using Seurat canonical correlation analysis (CCA) method. Integration anchors were determined using FindIntegrationAnchors function and then used in IntegrateData function. UMAP plots and cell clusters were generated using RunUMAP and FindClusters functions, followed by manual cell type annotation based on type-specific gene markers. In addition, cell types corresponding to individual cell clusters were inferred using the ELeFHAnt method (https://doi.org/10.1101/2021.09.07.459342) by the annotation transfer from the Tabula Muris65 brain dataset as a reference. Feature plots for individual genes were generated using FeaturePlot functions. Differentially expressed genes between conditions within cell populations of interest were identified using FindMarkers function. Comparison of cell fractions within cell populations was performed using scProportionTest package (https://github.com/rpolicastro/scProportionTest).66 Pathway enrichment among genes affected by mast cell deficiency in cell subpopulations was analyzed with GSEA package67 using the mouse GSEA MSigDB database.
Quantitative RT-PCR
Mouse dura samples were transferred from RNAprotect Tissue Reagent (Qiagen) to 350 μL Buffer RLT Plus (Qiagen) with 2-mercaptoethanol. The tissues were disrupted and homogenized by TissueRuptor II (Qiagen) with increasing speed for 3–5 min. Genomic DNA was removed and total RNA was isolated using RNeasy Plus Mini Kit (Qiagen) according to the manufacturer’s instructions. The RNA concentrations were 25–70 ng/μL for dura samples. Around 200 ng total RNA was subjected to cDNA synthesis using SuperScript IV VILO Master Mix with ezDNase enzyme kit (Invitrogen). cDNA was 10-fold diluted and mixed with PowerTrack SYBR Green Master Mix (Thermo Fisher Scientific) at 1:5 ratio. Forward and reverse primers (Integrated DNA Technologies) were used at 0.5 μM and added to the above cDNA-SYBR Green mix in a 96-well Hard-Shell PCR plate (BioRad). Quantitative real-time PCR was performed on a BioRad CFX96 Real-Time system (BioRad). Expression of genes was determined relative to Actb by the ΔCT method.
The following primers were used for quantitative RT-PCR.
| Target | Forward primer | Reverse primer |
|---|---|---|
|
| ||
| Actb | CATTGCTGACAGGATGCAGAAGG | TGCTGGAAGGTGGACAGTGAGG |
| Cma1 | TCTCCTGGGTTCCAGCACCAAA | GCCGACAGGTAGTTCTCAGAAG |
| Cma2 | GTCTTCCAAGTTCAATGACATCGT | ACATCGTCCCAGGCTTGGCAAA |
| Hdc | GAGTACGCTGACTCCTTCACCT | CAGAGTTGGCATGTCGGAGGTA |
| Mcpt4 | CGACTATAACCTCCAGGTCTGC | GAGGAGATTCGGGTGAAGACTG |
| Mcpt9 | GAGAGGTAGAACTGAAAATCGTGG | CAGTAGAGGTCCTCCAGAGTCT |
| Tpsab1 | ATGACCACCTGATGACTGTGAGCCAG | AGGAACGGAGGTCATCCTGGATGTG |
| Tpsb1 | CGACATTGATAATGACGAGCCTC | ACAGGCTGTTTTCCACAATGG |
| Tpsg1 | TAGCCTCCGTCTGCACAAGGTG | GGAGACAGTGTGACCGTAAGCT |
Immunoblotting
Ten to 30 μg protein of cortical lysates were loaded and separated by 4–12% gradient Bis/Tris gels (Life Technologies) and transferred to PVDF membranes using iBlot 2 Transfer Stack and iBlot 2 Gel Transfer Device (Invitrogen). Blots were incubated in the SuperBlock Blocking Buffer (Thermo Fisher Scientific) for 1 h at room temperature with shaking. Blots were then incubated with primary antibody overnight at 4°C with gentle shaking. Primary antibodies were used at the following dilutions: mouse anti-Aβ1–16 (1:1,000, BioLegend), rabbit anti-APP C terminus antibody (C66; 1:1,000, in-house), mouse anti-sAPPβ-sw (1:1,000, IBL-America), rat anti-GFAP (1:1,000, Life Technologies), mouse anti-pan actin (1:2,000, Thermo Scientific). Blots were washed at least four times before incubation with appropriate HRP-conjugated secondary antibodies at room temperature for 45–60 min with shaking. After 4–5 washes, Pierce ECL Western Blotting Substrate or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific) was applied. The images were captured by Odyssey Platform (LI-COR) and analyzed by FIJI-ImageJ software (National Institutes of Health). In some experiments, the blots were stripped in Restore Western Blot Stripping Buffer (Thermo Scientific) at room temperature for 15 min with shaking before re-probing with another primary antibody.
ELISA and electrochemiluminescence assay
Levels of Aβ peptides in the TBS and FA brain lysates were determined by Wako Human/Rat β Amyloid ELISA Kits (Fujifilm) according to the manufacturer’s instructions. In brief, 100 μL diluted TBS and FA samples were incubated in the pre-coated 96-well plate at 4°C with gentle shaking. On the next day, the wells were washed, and the manufacturer-supplied HRP-conjugated detection antibody was added. The mixed solutions were incubated for 1 h at 4°C. After washing, TMB solution was added, and the reaction was terminated by adding the Stop solution. The values of absorbance at 405 nm were obtained using a BioTek multi-mode reader.
Levels of TNF and IL-1β in the MPER brain lysates were determined by V-PLEX Proinflammatory Panel 1 (mouse) Kit (Meso Scale Discovery) according to the manufacturer’s instructions. In brief, 50 μL of diluted MPER sample or calibrator were added to the MSD MULTI-SPOT 96-well 10-spot plates coated with an array of cytokine capture antibodies. The plate was incubated for 2 h with shaking (1,000 rpm). A volume of 25 μL of the detection antibody solution was added and incubated for 2 h at room temperature with shaking (1,000 rpm). The plates were washed before adding MSD Read Buffer, and immediately read on a Meso QuickPlex SQ 120 system (Meso Scale Discovery). Cytokine levels generated by MSD assays were analyzed by the software MSD Discovery Workbench (Meso Scale Discovery) and normalized to total tissue weights.
Immunohistochemistry and image analysis
Toluidine blue staining of mouse ear pinnae and brain sections
Toluidine blue working solution was made by dissolving 50 mg toluidine blue O powder (Sigma) in 70% ethanol and mixing with 1% sodium chloride, pH 2.3. Mouse ear sections were deparaffinized in 3 changes of xylene and decreasing concentrations of ethanol before staining with acidic toluidine blue working solution for 15–30 s followed by distilled water rinses. Sections were dehydrated with ethanol, cleared with xylene, and mounted with Cytoseal Xyl (Thermo Fisher Scientific). Mouse ear sections were imaged under 10X objective and brain sections were imaged under 20X objective on a Nikon Eclipse Ci microscope that was equipped with a DS-Ri2 camera and NIS-Elements Advanced Research imaging software (Nikon).
Immunofluorescence of dura mater
Dura was carefully dissected from the skull but still kept connected at the center of the sagittal sinus. Skull-connected dura was washed in PBS and blocked in Block-Perm solution (2% donkey serum/1% BSA/0.1%Triton X-100/0.05% Tween 20 in 10X PBS) for 1 h at room temperature with shaking. Dura was then incubated with primary antibodies overnight at 4°C: goat anti-Kit polyclonal antibody (1:200; R&D) with or without rabbit anti-TPSAB1 (tryptase) polyclonal antibody (1:200; Cell Signaling). Dura was washed before incubation with the combination of some of the following antibodies: Alexa Fluor 568 donkey anti-goat IgG (1:500; Thermo Fisher Scientific), Cy5 AffiniPure Donkey Anti-Goat IgG (H + L) (1:200; Jackson ImmunoResearch), or Alexa Fluor 488 donkey anti-rabbit IgG (1:500; Thermo Fisher Scientific) for 1 h at room temperature with shaking. After washing in PBS, nuclei were stained by 2 nM Hoechst 33342 Solution (Thermo Fisher Scientific) for 10 min at room temperature with shaking. Dura was completely dissected from the skull and mounted with Invitrogen ProLong Gold Antifade Mounting reagent (Thermo Fisher Scientific). The dura sections were imaged under a 20X objective (EC Plan-NEOFLUAR 20X/0,5) using the Zeiss Axio Imager 2 fluorescent microscope, with the Apotome 3 inserted and the slide scanner function enabled. The microscope was equipped with the ORCA-Flash4.0 V3 Digital CMOS camera (Hamamatsu) and the imaging software StereoInvestigator (MBF Biosciences). The experimenters were blind to the genotype of the individual animal while taking the images.
For the assessment of Kit immunofluorescence signals in the dura, contours of the left and right parietal regions of the dura mater were drawn and a z stack slide scan image was taken at 5 μm intervals and a depth of 30 μm. The max projection of the 3-D image was exported in a TIFF file for analysis via NIS-Elements version AR 5.42.01. Through this analysis software, measurements such as cell count, area fraction, circularity (defined as (4*π*Area)/(Perimeter2)), and shape factor (defined as (4*π*Area)/(Convex hull perimeter)2) were obtained from each image. Values from each section were averaged to generate a mean result for each animal. See also Table S2 for GA3 and analysis parameters.
Immunofluorescence of floating brain sections
Sagittal or coronal brain sections were transferred from cryoprotectant reagents to the Netwell Insert (Corning) placed in 12-well plates or plastic trays filled with PBS. Brian sections were blocked in 5% donkey serum/0.3%Triton X-100/PBS solution in 24-well plates for 1–2 h at room temperature with shaking. Sections were then incubated with primary antibodies overnight at 4°C with shaking. The following primary antibodies were used: goat anti-Iba-1 polyclonal antibody (1:500; FUJIFILM), rabbit anti-APOE monoclonal antibody (EPR19378; 1:1,000; abcam); rat anti-GFAP monoclonal antibody (clone 2.2B10; 1:600; Thermo Fisher Scientific), or mouse anti-Aβ 3D6 antibody (1:2,000, a gift from Lilly). On the following day, brain sections were washed in PBS and incubated with appropriate fluorochrome-conjugated secondary antibodies for 2 h at room temperature with shaking. The following secondary antibodies were used: donkey anti-rat Alexa Fluor 488 (1:600; Thermo Fisher Scientific), donkey anti-rabbit Alexa Fluor 488 (1:600; Thermo Fisher Scientific), donkey anti-rabbit Alexa Fluor 568 (1:600; Thermo Fisher Scientific), and goat anti-mouse IgG (H + L), F(ab’)2 Fragment Alexa Fluor 647 (1:600; Cell Signaling Technology). Nuclei were stained by 2 nM Hoechst 33342 Solution (Thermo Fisher Scientific) for 10 min at room temperature with shaking. Brain sections were mounted with Invitrogen ProLong Gold Antifade Mounting reagent (Thermo Fisher Scientific). Stained sections were imaged under 10X objective on a Nikon A1 confocal microscope equipped with NIS-Elements Advanced Research software (Nikon) or under a 20X objective (EC Plan-NEOFLUAR 20x/0,5) with the Zeiss Axio Imager 2 fluorescent microscope. The Zeiss microscope was equipped with the ORCA-Flash4.0 V3 Digital CMOS camera (Hamamatsu) and the imaging software StereoInvestigator (MBF Biosciences). With the Apotome 3 inserted and the slide scanner function enabled, visual area of the isocortex were selected and a z stack slide scan image was taken at 1 μm intervals were taken at a depth of 35–40 μm. The max projection of the 3-D image was exported in a TIFF file for analysis via NIS-Elements version AR 5.42.01.
For the assessment of GFAP immunofluorescence signals taken by Nikon A1 confocal microscope, four to six sagittal brain sections were imaged for each animal. Similar regions encompassing portions of the rear cortex were manually drawn with the circle tool of FIJI-ImageJ software (National Institutes of Health). The mean fluorescence intensity, percent area (area occupied by all GFAP signals divided by the total area), and circularity were calculated in the cortex for each section using the Analyze plugin (measurements: area, shape description, integrated density, area fraction, and mean gray value). Values from each section were averaged to generate a mean result for each animal.
For the assessment of Iba-1, APOE, and GFAP immunofluorescence signals taken by the Zeiss microscope, three to six sections of the visual cortical area were imaged for each animal. Measurements including mean intensity, area fraction, circularity (defined as (4×π×Area)/(Perimeter2)), and shape factor (defined as (4×π×Area)/(Convex hull perimeter)2) were obtained from each 20X image. Values from each section were averaged to generate a mean plaque density or burden for each animal. The experimenters were blind to the genotype of the individual animal while taking the images and analyzing the data.
For the assessment of Aβ plaque density and burden, four to six sagittal sections spanning the cortex and hippocampus were imaged for each animal. Similar regions encompassing portions of the cortex or hippocampus were drawn with the circle tool of FIJI-ImageJ software (National Institutes of Health). The amyloid plaque density (number of plaques of a certain size range in a region of interest) and plaque burden (area occupied by all plaques divided by the total area) were calculated in the cortex and hippocampus for each section using the Analyze Particles plugin (Size: 30–50 μm2, 50–100 μm2, 100–200 μm2, and larger than 200 μm2; circularity: 0–1). Values from each section were averaged to generate a mean plaque density or burden for each animal.
Flow cytometry
Fc receptor blocking was performed (clone 93; BioLegend) in FACS buffer (0.5% BSA, 2 mM EDTA, and 0.02% sodium azide in PBS) for 5 min at 4°C before surface staining (4°C for 30 min). The following anti-mouse antibodies from BioLegend were used for surface staining: APC/Cyanine7 anti-CD11b, PE/Cyanine7 anti-mouse CD19 Antibody, Brilliant Violet 711 anti-CD45, Alexa Fluor 647 anti-mouse CD206 (MMR) Antibody, Brilliant Violet 421 anti-mouse F4/80 Antibody, PE anti-mouse FcεRIα, Alexa Fluor 700 anti-mouse Ly-6C Antibody, Alexa Fluor 488 anti-mouse Ly-6G Antibody, Brilliant Violet 510 anti-mouse I-A/I-E Antibody, and PE/Cyanine7 anti-mouse Kit. The APC Rat Anti-Mouse CD19 was from BD Pharmingen. Live cells were distinguished by LIVE/DEAD Fixable Yellow Dead Cell Stain Kit (Thermo Fisher Scientific). Flow cytometry was performed on BD LSRFortessa X-20 flow cytometer. Flow cytometry data were analyzed with FlowJo software (Tree Star).
Behavior tests
One-year-old female WT, Cpa3Cre/+, 5XFAD, and 5XFAD.Cpa3Cre/+ mice were used for fear conditioning test and elevated zero maze with genotypes blinded to the researcher. Both tests were conducted during daylight hours (9:00 a.m. - 4:00 p.m.). Before each test, all animals were handled with mouse tunnels (Bio-Serv) for three consecutive days (10–15 min each day) by the experimenter. The tunnels were used to transfer mice during behavioral testing. Mice in their home cages were transported to the behavior room at least 30 min before each test.
Contextual and cued fear conditioning (FC)
The training and testing sessions of contextual and cued fear conditioning were conducted and recorded in our animal behavior room. The FC chamber was inside a sound-attenuating box with a dual (visible/I.R.) light, a speaker, and a camera. The FC chamber and metal grids were thoroughly cleaned at the start of testing and between every mouse with 70% ethanol and paper towels to eliminate olfactory cues. On the morning of the first day, mice were placed individually in the FC chamber and allowed habituation and exploration of the context for 3 min. The baseline freezing time and episodes were determined during this period. Mice then experienced three pairs of cue aversive stimuli, consisting of tone (15 s; 5 kHz; 90 dB)–shock (1 s; 0.7 mA) pairings, separated by an interval of 1 min. Mice were moved to a clean cage 45 s after the last shock. After all mice from the same cage had completed their trials, mice were moved back to their home cages. The next morning, mice were tested for conditioned fear of the training context for 4 min (contextual test), in the same chamber without any cue stimuli. Contextual FC freezing time were recorded during this 4-min period. Three to 4 h after the context test, animals were placed in an altered FC chamber and allowed to explore for 3 min, followed by a 3-min tone stimulus (cued test). Cued FC freezing time were determined during this 3-min period. Mice were removed from the chamber 1 min after the cue.
Elevated zero maze
To assess anxiety-like behavior, mice were subjected to elevated zero maze. The maze was elevated 50 cm from the ground, consisting of two open sectors without walls and two sectors enclosed by a 15.5-cm-high wall. Mice were placed individually at the entrance of a closed sector and allowed to explore undisturbed for 5 min. At the start and between trials, the maze was cleaned with 70% ethanol and paper towels. Time spent in the closed/open sectors and the number of entries into open sectors were calculated by the Noldus Ethovision software (Wageningen, Netherlands).
QUANTIFICATION AND STATISTICAL ANALYSIS
The data in each figure represents biologically independent replicates. No statistical methods were used to predetermine sample sizes. Statistical analysis was performed using the GraphPad Prism (GraphPad Software). Data were analyzed by two-way ANOVA followed by Tukey’s multiple comparisons tests or unpaired two-tailed Student’s t tests. Number of animals is specified in the panels. All p values and statistical tests are indicated in the figure.
Supplementary Material
Highlights.
Mast cell depletion restores cognition in 5XFAD mice without altering β-amyloid
Mast cell deficiency elevates levels of disease-associated microglia in 5XFAD mice
Absence of mast cells reduces markers indicative of reactive astrocytes
ACKNOWLEDGMENTS
We thank Drs. Thorsten B. Feyerabend and Hans-Reimer Rodewald from the Deutsches Krebsforschungszentrum (DKFZ), Germany, for providing Cpa3Cre/+ mice and helpful suggestions. We thank Donna Romano, Se Hoon Choi, Luisa Quinti, Joseph Park, Ana Griciuc, Nanda Navalpur Shanmugam, Deepak Vijaya Kumar, Anna Maaser-Hecker, Can “Martin” Zhang, Nolan Shen, Sherri Zhen, Junseok Bae, Ethan Liu, and other current and former members in the Genetics and Aging Research Unit at Massachusetts General Hospital. We thank Meng Chen, Frank Zamudio, Xin Jiang, and Evan Bordt for helping to set up animal behavior tests. The research was supported by grants from the Cure Alzheimer’s Fund (R.E.T.), JPB Foundation (R.E.T.), and NIH (T32 AG000222 to C.-C. L., P30DK040561 to R.I.S.). We thank the MGH Pathology Flow, Image and Mass Cytometry Core, MGH NextGen Sequencing Core, and the DF/HCC Pathology Core.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
DECLARATION OF INTERESTS
R.E.T. is a consultant to and shareholder in AZTherapeutics. R.I.S. is a shareholder in Feromics.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.113141.
REFERENCES
- 1.Knopman DS, Amieva H, Petersen RC, Chételat G, Holtzman DM, Hyman BT, Nixon RA, and Jones DT (2021). Alzheimer disease. Nat. Rev. Dis. Primers 7, 33. 10.1038/s41572-021-00269-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen X, and Holtzman DM (2022). Emerging roles of innate and adaptive immunity in Alzheimer’s disease. Immunity 55, 2236–2254. 10.1016/J.IMMUNI.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jorfi M, Maaser-Hecker A, and Tanzi RE (2023). The neuroimmune axis of Alzheimer’s disease. Genome Med. 15, 6. 10.1186/S13073-023-01155-W. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones MK, Nair A, and Gupta M (2019). Mast cells in neurodegenerative disease. Front. Cell. Neurosci. 13, 171–179. 10.3389/fncel.2019.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forsythe P (2019). Mast Cells in Neuroimmune Interactions. Trends Neurosci. 42, 43–55. 10.1016/j.tins.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Carthy E, and Ellender T (2021). Histamine, Neuroinflammation and Neurodevelopment: A Review. Front. Neurosci. 15, 680214. 10.3389/FNINS.2021.680214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kempuraj D, Mentor S, Thangavel R, Ahmed ME, Selvakumar GP, Raikwar SP, Dubova I, Zaheer S, Iyer SS, and Zaheer A (2019). Mast cells in stress, pain, blood-brain barrier, neuroinflammation and alzheimer’s disease. Front. Cell. Neurosci. 13, 54–11. 10.3389/fncel.2019.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graham AC, Temple RM, and Obar JJ (2015). Mast cells and influenza A virus: Association with allergic responses and beyond. Front. Immunol. 6, 238. 10.3389/fimmu.2015.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pejler G (2020). Novel Insight into the in vivo Function of Mast Cell Chymase: Lessons from Knockouts and Inhibitors. J. Innate Immun. 12, 357–372. 10.1159/000506985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelissen S, Lemmens E, Geurts N, Kramer P, Maurer M, Hendriks J, and Hendrix S (2013). The role of mast cells in neuroinflammation. Acta Neuropathol. 125, 637–650. 10.1007/s00401-013-1092-y. [DOI] [PubMed] [Google Scholar]
- 11.Jin Y, Silverman AJ, and Vannucci SJ (2007). Mast cell stabilization limits hypoxic-ischemic brain damage in the immature rat. Dev. Neurosci. 29, 373–384. 10.1159/000105478. [DOI] [PubMed] [Google Scholar]
- 12.Arac A, Grimbaldeston MA, Nepomuceno ARB, Olayiwola O, Pereira MP, Nishiyama Y, Tsykin A, Goodall GJ, Schlecht U, Vogel H, et al. (2014). Evidence that meningeal mast cells can worsen stroke pathology in mice. Am. J. Pathol. 184, 2493–2504. 10.1016/j.ajpath.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sayed BA, Christy AL, Walker ME, and Brown MA (2010). Meningeal Mast Cells Affect Early T Cell Central Nervous System Infiltration and Blood-Brain Barrier Integrity through TNF: A Role for Neutrophil Recruitment? J. Immunol. 184, 6891–6900. 10.4049/jimmunol.1000126. [DOI] [PubMed] [Google Scholar]
- 14.Hsieh JT, Rathore APS, Soundarajan G, and St. John, A.L. (2019). Japanese encephalitis virus neuropenetrance is driven by mast cell chymase. Nat. Commun. 10, 706. 10.1038/s41467-019-08641-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, Dong H, Li N, Zhang S, Sun J, Zhang S, and Qian Y (2016). Activated brain mast cells contribute to postoperative cognitive dysfunction by evoking microglia activation and neuronal apoptosis. J. Neuroinflammation 13, 127. 10.1186/s12974-016-0592-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong H, Zhang X, Wang Y, Zhou X, Qian Y, and Zhang S (2017). Suppression of Brain Mast Cells Degranulation Inhibits Microglial Activation and Central Nervous System Inflammation. Mol. Neurobiol. 54, 997–1007. 10.1007/S12035-016-9720-X. [DOI] [PubMed] [Google Scholar]
- 17.Dong H, Wang Y, Zhang X, Zhang X, Qian Y, Ding H, and Zhang S (2019). Stabilization of Brain Mast Cells Alleviates LPS-Induced Neuroinflammation by Inhibiting Microglia Activation. Front. Cell. Neurosci. 13, 191. 10.3389/FNCEL.2019.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Sha H, Zhou L, Chen Y, Zhou Q, Dong H, and Qian Y (2020). The Mast Cell Is an Early Activator of Lipopolysaccharide-Induced Neuroinflammation and Blood-Brain Barrier Dysfunction in the Hippocampus. Mediators Inflamm. 2020, 8098439. 10.1155/2020/8098439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sandhu JK, and Kulka M (2021). Decoding mast cell-microglia communication in neurodegenerative diseases. Int. J. Mol. Sci. 22, 1093. 10.3390/ijms22031093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang S, Zeng X, Yang H, Hu G, and He S (2012). Mast cell tryptase induces microglia activation via protease-activated receptor 2 signaling. Cell. Physiol. Biochem. 29, 931–940. 10.1159/000171029. [DOI] [PubMed] [Google Scholar]
- 21.Kempuraj D, Thangavel R, Selvakumar GP, Ahmed ME, Zaheer S, Raikwar SP, Zahoor H, Saeed D, Dubova I, Giler G, et al. (2019). Mast Cell Proteases Activate Astrocytes and Glia-Neurons and Release Interleukin-33 by Activating p38 and ERK1/2 MAPKs and NF-κB. Mol. Neurobiol. 56, 1681–1693. 10.1007/s12035-018-1177-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng X, Zhang S, Xu L, Yang H, and He S (2013). Activation of protease-activated receptor 2-mediated signaling by mast cell tryptase modulates cytokine production in primary cultured astrocytes. Mediators Inflamm. 2013, 140812. 10.1155/2013/140812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaik-Dasthagirisaheb YB, and Conti P (2016). The Role of Mast Cells in Alzheimer’s Disease. Adv. Clin. Exp. Med. 25, 781–787. 10.17219/acem/61914. [DOI] [PubMed] [Google Scholar]
- 24.Oakley H, Cole SL, Logan S, Maus E, Shao P, Craft J, Guillozet-Bongaarts A, Ohno M, Disterhoft J, Van Eldik L, et al. (2006). Intraneuronal beta-Amyloid Aggregates, Neurodegeneration, and Neuron Loss in Transgenic Mice with Five Familial Alzheimer’s Disease Mutations: Potential Factors in Amyloid Plaque Formation. J. Neurosci. 26, 10129–10140. 10.1523/JNEUROSCI.1202-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feyerabend TB, Weiser A, Tietz A, Stassen M, Harris N, Kopf M, Radermacher P, Möller P, Benoist C, Mathis D, et al. (2011). Cre-mediated cell ablation contests mast cell contribution in models of antibody- and T cell-mediated autoimmunity. Immunity 35, 832–844. 10.1016/j.immuni.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 26.Reber LL, Marichal T, and Galli SJ (2012). New models for analyzing mast cell functions in vivo. Trends Immunol. 33, 613–625. 10.1016/j.it.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rustenhoven J, Drieu A, Mamuladze T, de Lima KA, Dykstra T, Wall M, Papadopoulos Z, Kanamori M, Salvador AF, Baker W, et al. (2021). Functional characterization of the dural sinuses as a neuroimmune interface. Cell 184, 1000–1016.e27. 10.1016/J.CELL.2020.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rustenhoven J, and Kipnis J (2022). Brain borders at the central stage of neuroimmunology. Nature 612, 417–429. 10.1038/S41586-022-05474-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li T, Martin E, Abada YS, Boucher C, Cès A, Youssef I, Fenaux G, Forand Y, Legrand A, Nachiket N, et al. (2020). Effects of Chronic Masitinib Treatment in APPswe/PSEN1dE9 Transgenic Mice Modeling Alzheimer’s Disease. J. Alzheimer’s Dis. 76, 1339–1345. 10.3233/JAD-200466. [DOI] [PubMed] [Google Scholar]
- 30.Chikahisa S, Kodama T, Soya A, Sagawa Y, Ishimaru Y, Séi H, and Nishino S (2013). Histamine from Brain Resident MAST Cells Promotes Wakefulness and Modulates Behavioral States. PLoS One 8, e78434. 10.1371/journal.pone.0078434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nautiyal KM, Ribeiro AC, Pfaff DW, and Silver R (2008). Brain mast cells link the immune system to anxiety-like behavior. Proc. Natl. Acad. Sci. USA. 105, 18053–18057. 10.1073/pnas.0809479105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calhoon GG, and Tye KM (2015). Resolving the neural circuits of anxiety. Nat. Neurosci. 18, 1394–1404. 10.1038/nn.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jawhar S, Trawicka A, Jenneckens C, Bayer TA, and Wirths O (2012). Motor deficits, neuron loss, and reduced anxiety coinciding with axonal degeneration and intraneuronal Aβ aggregation in the 5XFAD mouse model of Alzheimer’s disease. Neurobiol. Aging 33, 196.e29–196.e40. 10.1016/j.neurobiolaging.20. [DOI] [PubMed] [Google Scholar]
- 34.Johnson-Wood K, Lee M, Motter R, Hu K, Gordon G, Barbour R, Khan K, Gordon M, Tan H, Games D, et al. (1997). Amyloid precursor protein processing and Aβ42 deposition in a transgenic mouse model of Alzheimer disease. Proc. Natl. Acad. Sci. USA. 94, 1550–1555. 10.1073/pnas.94.4.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanzi RE, and Bertram L (2005). Twenty years of the Alzheimer’s disease amyloid hypothesis: a genetic perspective. Cell 120, 545–555. 10.1016/J.CELL.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 36.Paolicelli RC, Sierra A, Stevens B, Tremblay ME, Aguzzi A, Ajami B, Amit I, Audinat E, Bechmann I, Bennett M, et al. (2022). Microglia states and nomenclature: A field at its crossroads. Neuron 110, 3458–3483. 10.1016/J.NEURON.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, David E, Baruch K, Lara-Astaiso D, Toth B, Itzkovitz S, Colonna M, Schwartz M, and Amit I (2017). A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 169, 1276–1290.e17. 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 38.Krasemann S, Madore C, Cialic R, Baufeld C, Calcagno N, El Fatimy R, Beckers L, O’Loughlin E, Xu Y, Fanek Z, et al. (2017). The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity 47, 566–581.e9. 10.1016/J.IMMUNI.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Hove H, Martens L, Scheyltjens I, De Vlaminck K, Pombo Antunes AR, De Prijck S, Vandamme N, De Schepper S, Van Isterdael G, Scott CL, et al. (2019). A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat. Neurosci. 22, 1021–1035. 10.1038/S41593-019-0393-4. [DOI] [PubMed] [Google Scholar]
- 40.Silvin A, Uderhardt S, Piot C, Da Mesquita S, Yang K, Geirsdottir L, Mulder K, Eyal D, Liu Z, Bridlance C, et al. (2022). Dual ontogeny of disease-associated microglia and disease inflammatory macrophages in aging and neurodegeneration. Immunity 55, 1448–1465.e6. 10.1016/J.IMMUNI.2022.07.004. [DOI] [PubMed] [Google Scholar]
- 41.Alves De Lima K, Rustenhoven J, and Kipnis J (2020). Meningeal Immunity and Its Function in Maintenance of the Central Nervous System in Health and Disease. Annu. Rev. Immunol. 38, 597–620. 10.1146/annurev-immunol-102319-1034. [DOI] [PubMed] [Google Scholar]
- 42.Niederhoffer N, Levy R, Sick E, Andre P, Coupin G, Lombard Y, and Gies JP (2009). Amyloid β peptides trigger CD47-dependent mast cell secretory and phagocytic responses. Int. J. Immunopathol. Pharmacol. 22, 473–483. 10.1177/039463200902200224. [DOI] [PubMed] [Google Scholar]
- 43.Liu J, Liu S, Zeng L, and Tsilioni I (2023). Amyloid Beta Peptides Lead to Mast Cell Activation in a Novel 3D Hydrogel Model. Int. J. Mol. Sci. 24, 12002. 10.3390/ijms241512002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sweeney MD, Sagare AP, and Zlokovic BV (2018. Mar). Blood-brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat Rev Neurol 14 (3), 133–150. 10.1038/nrneurol.2017.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deczkowska A, Keren-Shaul H, Weiner A, Colonna C, Schwartz., Amit I. Disease-Associated Microglia: A Universal Immune Sensor of Neurodegeneration. Cell. 2018. May 17;173(5):1073–1081. doi: 10.1016/j.cell.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 46.Price BR, Johnson LA, and Norris CM (2021). Reactive astrocytes: The nexus of pathological and clinical hallmarks of Alzheimer’s disease. Ageing Res. Rev. 68, 101335. 10.1016/j.arr.2021.101335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smit T, Deshayes NAC, Borchelt DR, Kamphuis W, Middeldorp J, and Hol EM (2021). Reactive astrocytes as treatment targets in Alzheimer’s disease—Systematic review of studies using the APPswePS1dE9 mouse model. Glia 69, 1852–1881. 10.1002/glia.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park JS, Kam TI, Lee S, Park H, Oh Y, Kwon SH, Song JJ, Kim D, Kim H, Jhaldiyal A, et al. (2021). Blocking microglial activation of reactive astrocytes is neuroprotective in models of Alzheimer’s disease. Acta Neuropathol. Commun. 9, 78. 10.1186/s40478-021-01180-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ceyzériat K, Ben Haim L, Denizot A, Pommier D, Matos M, Guillemaud O, Palomares MA, Abjean L, Petit F, Gipchtein P, et al. (2018). Modulation of astrocyte reactivity improves functional deficits in mouse models of Alzheimer’s disease. Acta Neuropathol. Commun. 6, 104. 10.1186/S40478-018-0606-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Furman JL, Sama DM, Gant JC, Beckett TL, Murphy MP, Bachstetter AD, Van Eldik LJ, and Norris CM (2012). Targeting astrocytes ameliorates neurologic changes in a mouse model of Alzheimer’s disease. J. Neurosci. 32, 16129–16140. 10.1523/JNEUROSCI.2323-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ben Haim L, Ceyzériat K, Carrillo-de Sauvage MA, Aubry F, Auregan G, Guillermier M, Ruiz M, Petit F, Houitte D, Faivre E, et al. (2015). The JAK/STAT3 pathway is a common inducer of astrocyte reactivity in Alzheimer’s and Huntington’s diseases. J. Neurosci. 35, 2817–2829. 10.1523/JNEUROSCI.3516-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reichenbach N, Delekate A, Plescher M, Schmitt F, Krauss S, Blank N, Halle A, and Petzold GC (2019). Inhibition of Stat3-mediated astrogliosis ameliorates pathology in an Alzheimer’s disease model. EMBO Mol. Med. 11, e9665. 10.15252/EMMM.201809665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sompol P, Furman JL, Pleiss MM, Kraner SD, Artiushin IA, Batten SR, Quintero JE, Simmerman LA, Beckett TL, Lovell MA, et al. (2017). Calcineurin/NFAT Signaling in Activated Astrocytes Drives Network Hyperexcitability in Aβ-Bearing Mice. J. Neurosci. 37, 6132–6148. 10.1523/JNEUROSCI.0877-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dubreuil P, Letard S, Ciufolini M, Gros L, Humbert M, Castéran N, Borge L, Hajem B, Lermet A, Sippl W, et al. (2009). Masitinib (AB1010), a potent and selective tyrosine kinase inhibitor targeting KIT. PLoS One 4, e7258. 10.1371/journal.pone.0007258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ettcheto M, Cano A, Sanchez-López E, Verdaguer E, Folch J, Auladell C, and Camins A (2021). Masitinib for the treatment of Alzheimer’s disease. Neurodegener. Dis. Manag. 11, 263–276. 10.2217/NMT-2021-0019. [DOI] [PubMed] [Google Scholar]
- 56.Zhang C, Griciuc A, Hudry E, Wan Y, Quinti L, Ward J, Forte AM, Shen X, Ran C, Elmaleh DR, and Tanzi RE (2018). Cromolyn Reduces Levels of the Alzheimer’s Disease-Associated Amyloid β-Protein by Promoting Microglial Phagocytosis. Sci. Rep. 8, 1144. 10.1038/s41598-018-19641-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burgher LW, Elliott RM, and Kass I (1971). A perspective on the role of cromolyn sodium as an antiasthmatic agent. Chest 60, 210–213. 10.1378/chest.60.3.2. [DOI] [PubMed] [Google Scholar]
- 58.Yazid S, Sinniah A, Solito E, Calder V, and Flower RJ (2013). Anti-Allergic Cromones Inhibit Histamine and Eicosanoid Release from Activated Human and Murine Mast Cells by Releasing Annexin A1. PLoS One 8, e58963. 10.1371/journal.pone.0058963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lortholary O, Chandesris MO, Bulai Livideanu C, Paul C, Guillet G, Jassem E, Niedoszytko M, Barete S, Verstovsek S, Grattan C, et al. (2017). Masitinib for treatment of severely symptomatic indolent systemic mastocytosis: a randomised, placebo-controlled, phase 3 study. Lancet 389, 612–620. 10.1016/S0140-6736(16)31403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Piris-Villaespesa M, and Alvarez-Twose I (2020). Systemic Mastocytosis: Following the Tyrosine Kinase Inhibition Roadmap. Front. Pharmacol. 11, 443. 10.3389/fphar.2020.00443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dubois B, López-Arrieta J, Lipschitz S, Doskas T, Spiru L, Moroz S, Venger O, Vermersch P, Moussy A, Mansfield CD, et al. (2023). Masitinib for mild-to-moderate Alzheimer’s disease: results from a randomized, placebo-controlled, phase 3, clinical trial. Alzheimer’s Res. Ther. 15, 39. 10.1186/S13195-023-01169-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Herisson F, Frodermann V, Courties G, Rohde D, Sun Y, Vandoorne K, Wojtkiewicz GR, Masson GS, Vinegoni C, Kim J, et al. (2018). Direct vascular channels connect skull bone marrow and the brain surface enabling myeloid cell migration. Nat. Neurosci. 21, 1209–1217. 10.1038/S41593-018-0213-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. (2017). Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049. 10.1038/NCOMMS14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LM, Yeung B, Rogers AJ, McElrath JM, Blish CA, Gottardo R, Smibert P, Satija R. Integrated analysis of multimodal single-cell data. Cell. 2021. Jun 24;184(13):3573–3587.e29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tabula Muris Consortium; Overall coordination; Logistical coordination; Organ collection and processing; Library preparation and sequencing; Computational data analysis; Cell type annotation; Writing group; Supplemental text writing group; Principal investigators (2018). Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372. 10.1038/S41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miller SA, Policastro RA, Sriramkumar S, Lai T, Huntington TD, Ladaika CA, Kim D, Hao C, Zentner GE, and O’Hagan HM (2021). LSD1 and Aberrant DNA Methylation Mediate Persistence of Enteroendocrine Progenitors That Support BRAF-Mutant Colorectal Cancer. Cancer Res. 81, 3791–3805. 10.1158/0008-5472.CAN-20-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. (2003). PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273. 10.1038/NG1180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the Key resources table. All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Alexa Fluor 647 anti-mouse CD206 (MMR) Antibody | BioLegend | Cat# 141712; RRID: AB_10900420 |
| Alexa Fluor 700 anti-mouse Ly-6C Antibody | BioLegend | Cat# 128024: RRID: AB_10643270 |
| Alexa Fluor® 488 anti-mouse Ly-6G Antibody | Biolegend | Cat# 127626; RRID: AB_2561340 |
| APC/Cy7 anti-mouse/human CD11b Antibody | BioLegend | Cat# 101226; RRID: AB_830642 |
| BD Pharmingen™ APC Rat Anti-Mouse CD19 | BD Biosciences | Cat# 550992; RRID: AB_10893995 |
| Brilliant Violet 421™ anti-mouse F4/80 Antibody | Biolegend | Cat# 123137; RRID: AB_2563102 |
| Brilliant Violet 510™ anti-mouse I-A/I-E Antibody | Biolegend | Cat# 107635; RRID: AB_2561397 |
| Brilliant Violet 711™ anti-mouse CD45 Antibody | BioLegend | Cat# 103147; RRID: AB_2564383 |
| Cy5 AffiniPure Donkey Anti-Goat IgG (H + L) | Jackson ImmunoResearch | Cat# 705-175-147; RRID: AB_2340415 |
| Donkey anti-Goat IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 568, Thermo Fisher Scientific | Thermo Fisher Scientific | Cat# A-11057; RRID: AB_2534104 |
| Donkey anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488, Thermo Fisher Scientific | Thermo Fisher Scientific | Cat# A-21206; RRID: AB_2535792 |
| Donkey anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 568, Thermo Fisher Scientific | Thermo Fisher Scientific | Cat# A10042; RRID: AB_2534017 |
| Donkey Anti-Rat IgG (H + L) Antibody, Alexa Fluor 488 Conjugated, Molecular Probes | Thermo Fisher Scientific | Cat# A-21208; RRID: AB_2535794 |
| FITC anti-mouse CD117 (c-Kit) Antibody | BioLegend | Cat#105805; RRID: AB_10060183 |
| Goat anti-mouse Iba-1 | FUJIFILM (Wako) | Cat# 011-27991; RRID: AB_2935833 |
| Goat Anti-mouse IgG (H + L), F(ab’)2 Fragment (Alexa Fluor 647 Conjugate) antibody | Cell Signaling Technology | Cat# 4410; RRID: AB_1904023 |
| Goat Anti-Mouse IgG1 (HRP) preadsorbed | Abcam | Cat# ab98693; RRID: AB_10674928 |
| Goat anti-Rabbit IgG (H + L) Cross-Adsorbed Secondary Antibody, HRP | Thermo Fisher Scientific | Cat# G-21234; RRID: AB_2536530 |
| Goat anti-Rat IgG (H + L) Cross-Adsorbed Secondary Antibody, HRP | Thermo Fisher Scientific | Cat# A18871; RRID: AB_2535648 |
| Human/Mouse CD117/c-kit Antibody | R&D | Cat# AF1356; RRID: AB_354750 |
| Lab Vision™ Actin, pan Ab-5, Mouse Monoclonal Antibody | Fisher Scientific | Cat# MS-1295-P1; RRID: AB_63316 |
| Mouse monoclonal anti-Aβ1-5, clone 3D6 | Lilly | N/A, gift |
| PE anti-mouse FcεRIα Antibody | BioLegend | Cat# 134307; RRID: AB_1626104 |
| PE/Cyanine7 anti-mouse CD117 (c-Kit) Antibody | Biolegend | Cat# 105813; RRID: AB_313222 |
| PE/Cyanine7 anti-mouse CD19 Antibody | Biolegend | Cat# 115520; RRID: AB_313655 |
| PerCP/Cyanine5.5 anti-mouse CD107a (LAMP-1) Antibody | BioLegend | Cat# 121626; RRID: AB_2572055 |
| Purified anti-mouse CD16/32 Antibody (clone 93) | Biolegend | Cat# 101301; RRID: AB_312800 |
| Purified anti-β-Amyloid, 1–16 Antibody (6E10) | BioLegend | Cat# 803001; RRID: AB_2564653 |
| Rabbit anti-APP C terminus (C66) | In-house | https://pubmed.ncbi.nlm.nih.gov/34010653/ |
| Rat anti-human/mouse GFAP Monoclonal Antibody (2.2B10) | Thermo Fisher Scientific | Cat# 13–0300; RRID: AB_2532994 |
| Recombinant rabbit Anti-Apolipoprotein E (APOE) antibody [EPR19378] | Abcam | Cat# ab183596; RRID: AB_2832971 |
| sAPPβ-sw (6A1) Anti-Human Mouse IgG MoAb | IBL-America | Cat# JP10321; RRID: AB_1630822 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| 2-mercaptoethanol | Sigma-Aldrich | Cat# M3148-25ML |
| 10X PBS Solution pH 7.4 | FISHER | Cat# P0496 |
| Absolute Ethanol (200 proof) | Fisher Scientific | Cat# BP2818-4 |
| Bovine serum albumin (BSA) | Fisher | Cat# BP-1600-100 |
| Collagenase D (100 mg) | Sigma-Aldrich | Cat# 11088858001 |
| DAPI solution | Thermo Fisher Scientific | Cat# 62248 |
| Deoxyribonuclease I | Worthington | Cat# LS002004 |
| Dispase II | Millipore Sigma | Cat# D4693 |
| EDTA (0.5 M), pH 8.0, RNase-free | Thermo Fisher Scientific | Cat# AM9261 |
| Ethylene glycol | Fisher Scientific | Cat# BP230-1 |
| FLUKA Formic acid, for mass spectrometry, ~98% | Thomas Scientific | Cat# 94318-250ML |
| Glycerol | Fisher Scientific | Cat# G31-500 |
| Halt phosphatase inhibitor cocktail | Thermo Fisher Scientific | Cat# 1862495 |
| HBSS with calcium and magnesium | Thermo | Cat# 24020117 |
| HBSS without calcium and magnesium | Gibco | Cat# 14170 |
| HistoPrep Xylene | Fisher Scientific | Cat# HC-700-1GAL |
| Hoechst 33342 Solution (20 mM) | Thermo Fisher Scientific | Cat# 62249 |
| Hyaluronidase from bovine testes | Millipore Sigma | Cat# H3506-100MG |
| M-PER™ Mammalian Protein Extraction Reagent | Thermo Fisher Scientific | Cat# 78501 |
| Normal donkey serum | Sigma-Aldrich | Cat# D9663, RRID:AB_2810235 |
| OneComp eBeads™ Compensation Beads | Thermo Fisher Scientific | Cat# 01-1111-42 |
| Papain from papaya latex | Sigma-Aldrich | Cat# P4762-100MG |
| Paraformaldehyde (32%) | Electron Microscopy Sciences | Cat# 50-980-495 |
| Percoll | GE Healthcare | Cat# 17-0891-01 |
| Pierce ECL Western Blotting Substrate | Thermo Fisher Scientific | Cat# 32106 |
| PowerTrack SYBR Green Master Mix | Thermo Fisher Scientific | Cat# A46110 |
| ProLong™ Gold Antifade Mountant | Thermo Fisher Scientific | Cat# P36934 |
| QIAzol Lysis Reagent | Qiagen | Cat# 79306 |
| RBC Lysis Buffer (10X) | BioLegend | Cat# 420301 |
| Restore Western Blot Stripping Buffer | Thermo Fisher Scientific | Cat# 21059 |
| Richard-Allan Scientific Cytoseal XYL | Thermo Fisher Scientific | Cat# 8312-4 |
| RNAprotect Tissue Reagent | Qiagen | Cat# 76104 |
| RPMI 1640 Medium, no phenol red | Thermo Fisher Scientific | Cat# 11835030 |
| Sodium azide | Sigma-Aldrich | Cat# 71289-5G |
| Sodium Chloride (Crystalline/Certified ACS) | Fisher Scientific | Cat# S271-3 |
| Sucrose | Millipore Sigma | Cat# 84097 |
| SuperBlock™ T20 (TBS) Blocking Buffer | Thermo Fisher Scientific | Cat# 37536 |
| SuperSignal West Femto Maximum | Thermo Fisher Scientific | Cat# 34096 |
| Sensitivity Substrate | ||
| TBS, Tris-Buffered Saline, 10X Solution, pH 7.4 | Fisher Scientific | Cat# BP24711 |
| Toluidine blue dye | Sigma-Aldrich | Cat#198161-5G |
| Triton X-100 | Sigma-Aldrich | Cat# TX1568-1 |
| Xpert Prestained Protein Marker | GenDEPOT | Cat# P8502-050 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Human/rat beta amyloid (40) ELISA Kit | Wako | Cat# 294-62501 |
| Human/rat beta amyloid (42) ELISA Kit | Wako | Cat# 290-62601 |
| LIVE/DEAD™ Fixable Yellow Dead Cell Stain Kit, for 405 nm excitation | Thermo Fisher Scientific | Cat# L34968 |
| Pierce BCA Protein Assay Kit | Thermo Fisher Scientific | Cat# 23225 |
| RNeasy Lipid Tissue Mini Kit | Qiagen | Cat# 74804 |
| RNeasy Plus Mini Kit | Qiagen | Cat# 74134 |
| SuperScript™ IV VILO™ Master Mix with ezDNase™ Enzyme | Thermo Fisher Scientific | Cat# 11766050 |
| V-PLEX Proinflammatory Panel1 (mouse) Kit (5 Plate) | Meso Scale Diagnostics | K15048D-2 |
|
| ||
| Deposited data | ||
|
| ||
| Raw sequencing data files for single-cell RNA sequencing | This paper | GEO accession: GSE240687 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| 5XFAD and WT Mice | The Jackson Laboratory | Cat# 034848-JAX, RRID:MMRRC_034848-JAX |
| C57BL/6J-KitW-v/J Mice | The Jackson Laboratory | Cat# JAX:000049, RRID:IMSR_JAX:000049 |
| CreMaster Mice | Rodewald Lab | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| Adobe Illustrator (version 25.3.1) | Adobe | RRID:SCR_010279 |
| EthoVision XT (version 9.0) | Noldus Information Technology | RRID:SCR_000441 |
| FACSDiva Software (version 6.1.3) | BD Biosciences | RRID:SCR_001456 |
| FIJI-ImageJ | NIH | RRID:SCR_003070 |
| FlowJo (version 10.7.1) | Tree Star | RRID:SCR_008520 |
| GraphPad Prism 7 and 8 | GraphPad Software | RRID:SCR_002798 |
| NIS-Elements Advanced Research software (version 5.42.01) | Nikon | https://www.microscope.healthcare.nikon.com |


