Summary
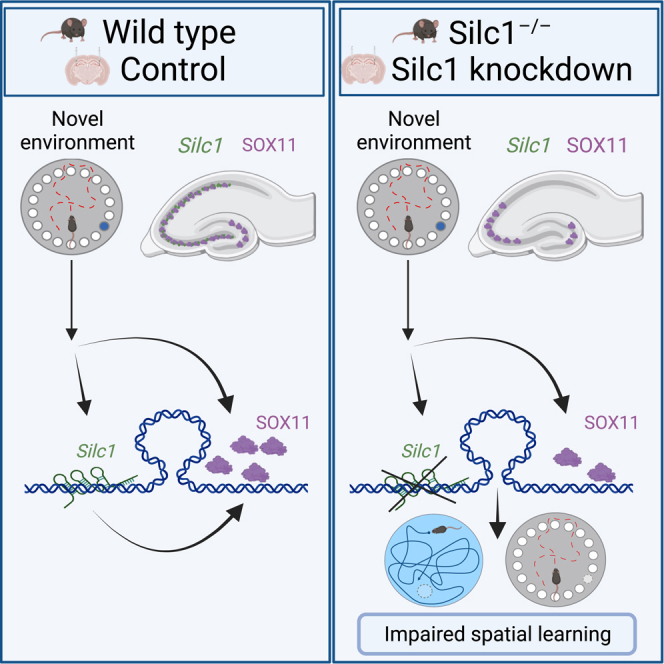
Long noncoding RNAs (lncRNAs) are expressed in many brain circuits and types of neurons; nevertheless, their functional significance for normal brain functions remains elusive. Here, we study the functions in the central nervous system of Silc1, an lncRNA we have shown previously to be important for neuronal regeneration in the peripheral nervous system. We found that Silc1 is rapidly and strongly induced in the hippocampus upon exposure to novelty and is required for efficient spatial learning. Silc1 production is important for induction of Sox11 (its cis-regulated target gene) throughout the CA1–CA3 regions and proper expression of key Sox11 target genes. Consistent with its role in neuronal plasticity, Silc1 levels decline during aging and in models of Alzheimer’s disease. Overall, we describe a plasticity pathway in which Silc1 acts as an immediate-early gene to activate Sox11 and induce a neuronal growth-associated transcriptional program important for learning.
Keywords: long non-coding RNAs, lncRNAs, hippocampus, Sox11, Silc1, learning
Graphical abstract
Highlights
-
•
Silc1 is rapidly and strongly induced upon stimulation in the hippocampus
-
•
Silc1 production is important for expression of Sox11 and Sox11 target genes
-
•
Silc1 is required for efficient spatial learning
-
•
Silc1 levels decline during aging and in models of Alzheimer’s disease
Perry et al. report the role of Silc1 as an immediate-early gene, induced following exposure to a novel environment in the hippocampus, specifically in the CA3 region. Silc1 activates Sox11 to induce a neuronal growth-associated transcriptional program that is crucial for efficient memory formation in mice.
Introduction
Long noncoding RNAs (lncRNAs) are products of pervasive transcription of eukaryotic genomes. While tens of thousands of lncRNA genes have now been annotated in the mammalian genome,1 the functions of the vast majority of them, if any, remain unclear. Cells of the nervous system express a particularly rich repertoire of lncRNA genes, and some are indicated to play particularly important roles in neurogenesis and/or functioning of the nervous system (reviewed in Hezroni et al.2). Some of them are also affected by various neurological diseases, like the GOMAFU lncRNA (also known as MIAT and RNCR2), which is important for neuronal development and is a risk factor and biomarker for schizophrenia.3 The molecular mechanisms underlying learning and memory formation remain only partially understood. Several lncRNAs have been found to accumulate in the synaptic compartment in response to neural activity, and some have been implicated in the process via analysis of genetic models. Loss of the Carip lncRNA, which binds specifically to Ca2+/calmodulin-dependent protein kinase IIβ (CaMKIIβ), affects phosphorylation of AMPA and NMDA receptors and causes dysfunction of synaptic transmission, attenuated long-term potentiation, and impaired spatial memory formation.4 Other lncRNAs, such as Neat1, have been studied using transient perturbations and reported to affect memory formation.5 In vitro studies using hippocampal neurons indicated that the ADEPTR lncRNA is transported synaptically, and loss of its function suppresses activity-dependent changes in synaptic transmission and the structural plasticity of dendritic spines.6 Furthermore, an experience-induced lncRNA called ADRAM has been found to be expressed in the infralimbic prefrontal cortex of adult male mice in response to fear-related learning. ADRAM acts as a scaffold and combinatorial guide, recruiting the brain-enriched chaperone protein 14-3-3 to the promoter of the memory-associated immediate-early gene Nr4a2. It is also required for fear extinction memory.7 The lncRNA Gas5 has been reported to regulate activity-dependent trafficking and clustering in dendrites through its interaction with the RNA binding proteins G3bp2 and Caprin. Cell-type-specific, state-dependent, and synapse-specific knockdown (KD) of the Gas5 variant leads to impaired fear extinction memory.8
We have recently characterized lncRNA expression during regeneration in the peripheral nervous system and characterized two lncRNAs induced by sciatic nerve crush and regulation of neurite outgrowth.9 We have further demonstrated that one of these, Silc1, an lncRNA conserved in sequence throughout mammals, is required for timely regeneration in vivo, and its loss in Silc1−/− mice is associated with reduced expression of Sox11, a transcription factor with well-established roles in neurogenesis and neuroregeneration in the adult brain and in the regenerating dorsal root ganglia (DRGs).9,10 Silc1 is transcribed from within a large gene desert flanking the Sox11 gene, which lies ∼200 kb upstream of it, and the two loci appear in spatial proximity to each other in various chromatin capture datasets.9,11,12 The human ortholog, SILC1, was recently studied in neuroblastoma cells, where SOX11 is a lineage dependence factor.13,14
Sox11, a member of the SoxC family of transcription factors, has so far been primarily studied in the context of embryonic neurogenesis, where it has been proposed to have overlapping targets with other SoxC transcription factors (TFs), Sox4 and Sox12, as well as with other members of the Sox family.15 Sox11−/− mice die shortly after birth,16 consistent with its requirement for proliferation and growth of neuronal cells of various types.17,18,19 Conditional loss of Sox11 in the neuronal lineage (obtained using tamoxifen-inducible Nestin-driven Cre) demonstrated that loss of Sox11 reduces neurogenesis in the embryo and adult.20 Specific ablation of Sox11 in the subgranular zone (SGZ) of the dentate gyrus (DG) in the hippocampus reduced the number of DCX+ or NeuroD1+ cells.20 In addition to proliferation and differentiation, loss of Sox11 also affects axonal growth in embryonic sensory neurons in vivo and in vitro17 and axonal growth in adult DRG neurons upon injury.21
Little is known about the roles of Sox11 in post-mitotic neurons in the brain, where, although its expression is modest, it is one of the most abundantly expressed Sox genes. Sox11 levels in the adult brain are particularly high in the SGZ of the adult hippocampus,22,23 which is one of the two sites of ongoing adult neurogenesis.24 At early postnatal stages, Sox11 is particularly high in late neuroblasts and immature granule and pyramidal neurons, and then its levels decline in mature neurons.25 Sox11 mRNA is broadly induced in the DG upon electroconvulsive stimulation,26,27 suggesting a role in neuronal activity. A recent study has further examined this increase and noted that it also occurs when mice are placed in a novel environment, specifically in mature neurons in the granule layer of the DG,28 where its expression is sparse under standard (familiar) housing conditions. This “novelty”-induced increase was associated with a subset of Fos+ cells28 which experienced stronger neuronal activation. Notably, SOX11+ cells were not observed in the CA subfields in that study. Several target genes of SOX11 are known and include Dcx, which is expressed in a domain tightly overlapping with Sox11.23
Because it is presently impossible to deduce the function of lncRNAs from their sequences or structures, co-expression with annotated protein-coding genes is often used as a first and readily available method to predict the function of lnRNAs.29,30,31 Nevertheless, it remains unclear whether lncRNAs are indeed often co-expressed with their target genes in the genetic circuits in which they are involved because correlated expression domains can simply result from co-regulation by other factors. Another approach for deducing functional connections is genomic proximity to other genes of interest,32 although lncRNAs produced from loci near other genes often do not appear to regulate their expression.33,34
Therefore, the current study sought to explore the function of Silc1 in the central nervous system, characterize its relationship with Sox11, and identify its role in neuronal activity.
Results
Silc1 is broadly expressed in the central nervous system
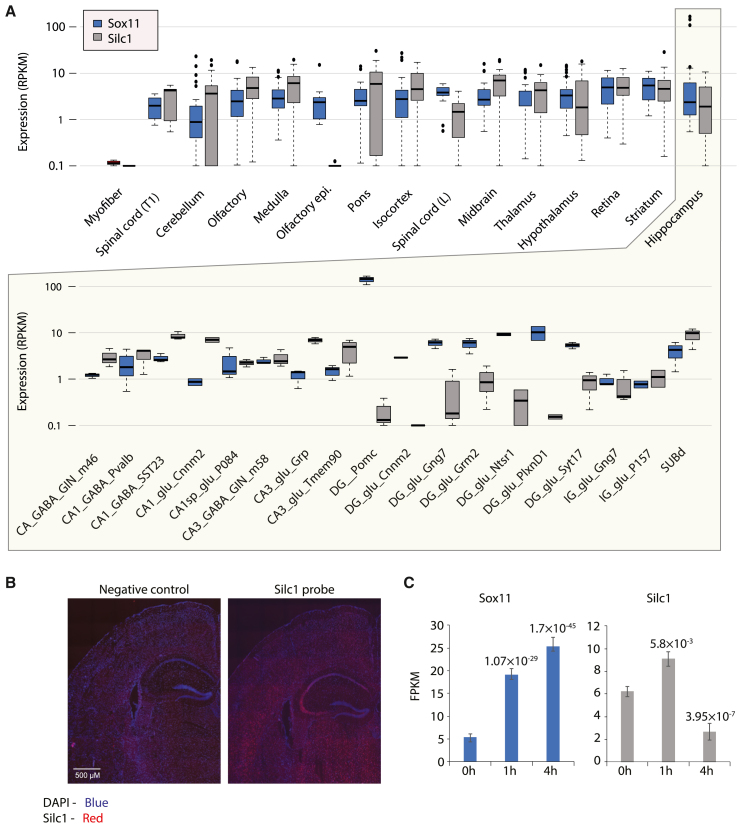
In our initial description of Silc1, we noted that, in contrast to Sox11, which is expressed at higher levels at embryonic stages, Silc1 was detected almost exclusively in postnatal samples of nervous systems and that, in those postnatal samples, Silc1 expression was substantial and largely comparable with that of Sox11.9 We also noted that, while Silc1 was induced by ∼10-fold following sciatic nerve injury, the RNA sequencing (RNA-seq)-derived expression levels in the injured DRG were on par with its steady-state levels in the central nervous system, where it was not induced in various injury models.9 We therefore wanted to first examine where Silc1 and Sox11 are expressed in the postnatal brain. We re-analyzed the RNA-seq data from the NeuroSeq atlas, a set of genetically defined cellular populations isolated by a combination of fluorescent proteins and microdissection from the mature brain35 (Figure 1A). Across the 517 samples in this dataset, Sox11 was detected in 467 (90%) and Silc1 in 504 (97%) with at least 5 reads per million in 40% of the samples for Sox11 and 80% of the samples for Silc1, suggesting that both genes are rather broadly present in the mature brain. Across the samples, Sox11 and Silc1 exhibited a significant but overall modest correlation (Spearman R = 0.25, p = 2.46 × 10−8). Sox11 expression was an order of magnitude higher in a single population in the hippocampus, POMC+ neuronal progenitors, where Silc1 was barely detectable (RPKM < 0.3; Figure 1A), but both genes were expressed at similar and consistent levels of 3–5 RPKM across most other populations, with a notable lack of Silc1 expression in the olfactory epithelium (Figure 1A). Across all mouse genes in the NeuroSeq dataset, Silc1 was most closely correlated (R = 0.57, p < 10−16) with Thy1, a marker for neuronal maturation and cessation of neurite outgrowth,36 suggesting that Silc1 is predominantly expressed in mature neurons. Consistent with the RNA-seq data, fluorescence in situ hybridization (FISH) analysis using RNAscope showed broad expression of Silc1 throughout the adult brain (Figure 1B).
Figure 1.
Expression domains of Silc1 and Sox11
(A) Expression levels of Sox11 (blue) and Silc1 (gray) in sorted populations from different regions of the CNS. Data are from the Allen brain atlas. The bottom panel shows the different populations of cells in the hippocampus.
(B) Fluorescent in situ hybridization (FISH) assay using RNAscope for brain sections from a WT mouse. A negative control was performed in parallel as an indicator of background staining. Tissues were hybridized with Silc1 probe (red), counterstained with DAPI (blue), and imaged using a 100× oil immersion objective. Scale bar, 500 μm. Silc1 is expressed in the cortical area and in the hippocampus.
(C) Quantification of the indicated genes in RNA-seq data from the DG at the indicated time points following electroconvulsive stimulation using RNA-seq data from Su et al.27 The p values were computed for comparison with the “0 h” time point using DeSeq237 and adjusted for multiple testing (n = 3).
See also Figure S1.
We conclude that a small population of neuronal progenitors in the hippocampus expresses exceptionally high levels of Sox11 and no Silc1, closely resembling the embryonic progenitors. In other parts of the mature brain, Silc1 and Sox11 are expressed, but with modestly correlated patterns that can even be anti-correlated when considering individual regions (see below).
Silc1 and Sox11 are differentially induced by neuronal activity in the hippocampus
To focus on a particular region in the adult brain where Silc1 may play a relevant role, we examined public RNA-seq data and literature for evidence of changes in the expression of Silc1 and/or Sox11 in various physiological settings.
We noted that, upon electroconvulsive stimulation, Sox11 mRNA has been shown previously shown to be specifically induced in the DG.26,28 Indeed, examining RNA-seq-based expression in the DG upon stimulation indicated that Silc1 and Sox11 are induced at early time points, followed by a decline of Silc1 to significantly below basal levels (Figure 1C). We further examined ATAC-seq data from the same study and from the hippocampus of mice stimulated with kainic acid38 and found several specific activity-induced enhancers in the gene deserts flanking Sox11, which also overlapped binding sites for the AP-1 complex (Figure S1A), suggesting a plausible regulatory route for neural activity-driven induction of the two genes.
Interestingly, when zooming into the hippocampus cell populations of the NeuroSeq dataset, Silc1 and Sox11 expression patterns were strongly anticorrelated (Spearman R = −0.45, p = 8.2 × 10−4; Figure 1A), with Sox11 mRNA predominantly expressed in the DG and Silc1 in the CA3 region. Similar results were observed in a HippoSeq dataset (Figure S1B).39 Therefore, while, on the tissue level, Sox11 and Silc1 appear to be regulated by neuronal activation, similar to their co-induction in the DRG,9 the baseline expression patterns of the two genes in the hippocampus appear to be strikingly different.
Several studies examined neuronal activity during exposure to a novel environment and found that neuronal activity was significantly increased in the DG. A mechanism for novelty-driven memory acquisition in the DG has been proposed by direct projection of ventral mossy cells to dorsal dentate granule cells. This dorsoventral interaction has been proposed as crucial for novelty-dependent memory formation.40
Silc1 and Sox11 are immediate-early genes that are upregulated in mice following exposure to a novel environment
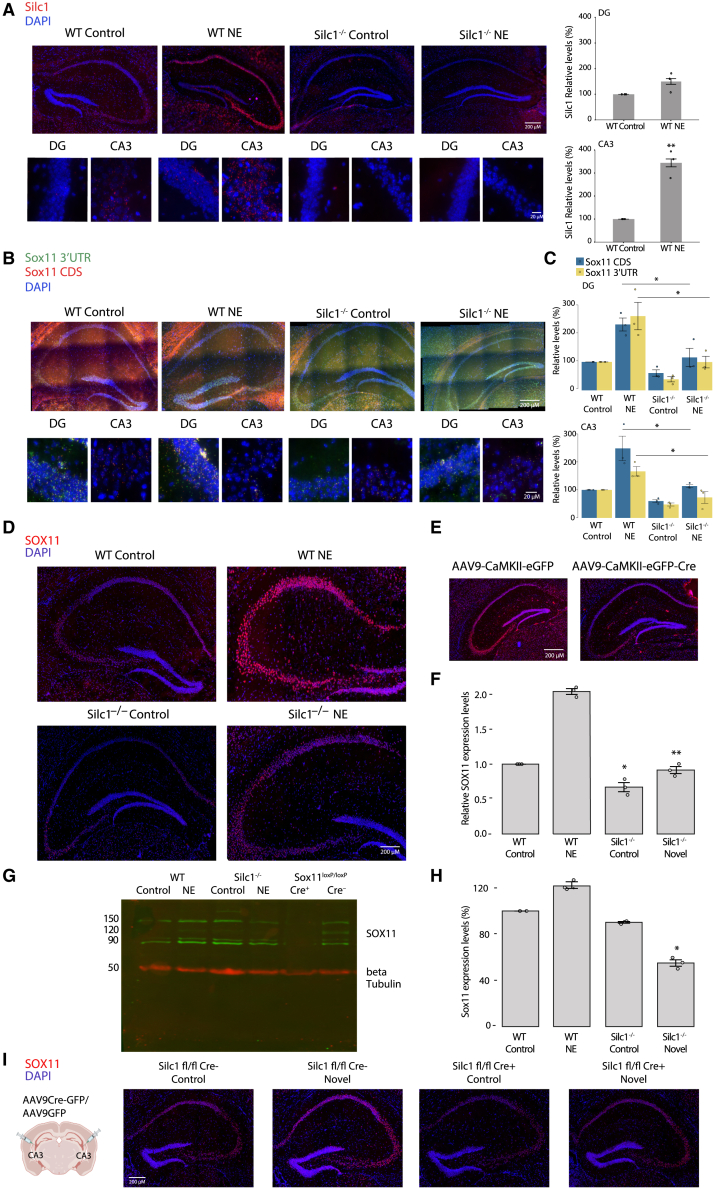
Because an intact Silc1 locus was associated with a response of mice to stimuli, we used exposure to the Barnes maze setting41 as a novel environment (NE) paradigm and assessed expression at several time points (0.5, 1, 2, and 6 h). We first used RNAscope to map expression of Silc1 and Sox11 in the home cage (HC) and after NE exposure. We observed the highest induction of Silc1 1 h post NE exposure (Figure S2A), similar to the response of other immediate-early genes, like Fos and Arc;42 therefore, we continued to use the NE setting in subsequent experiments. Because the Sox11 coding sequence (CDS) and 3′ UTR are not always observed strictly in the same cells or regions,43 we used separate sets of probes targeting the CDS and the 3′ UTR to uncover potential subtleties that might be critical after NE exposure. Consistent with the publicly available data (Figure 1), under control conditions, Silc1 was higher in the CA3 than in the DG, whereas Sox11 mRNA was more abundant in the DG. Following NE, the expression of both RNAs increased in both regions (Figures 2A–2C and S2A). When examining SOX11 protein, surprisingly, the most notable increase was in the CA3 region (Figure 2D). We validated the specificity of this signal by Cre injection into the hippocampus of Sox11 fl/fl mice from Bhattaram et al.44 (Figures 2E and 2G). Strikingly, in Silc1−/− mice, there was a significant reduction in Sox11 mRNA and protein levels in under the HC and NE conditions that was noted in the DG and CA3 regions (Figures 2B and 2D–2F). Consistent with previous reports, Sox11 CDS and 3′ UTR expression patterns did not always overlap, yet loss of Silc1 resulted in a concordant decrease in both signals in the DG and CA3 regions (Figure 2B). Furthermore, we observed a substantial reduction in the protein expression of SOX11 in the CA3. These results were verified by a western blot analysis with SOX11 antibody using proteins extracted from the hippocampus of WT and Silc1−/− mice under HC and NE conditions. SOX11 protein levels were significantly reduced in Silc1−/− mice upon stimulus in the NE setting (Figures 2G and 2H). Assessing the colocalization of Fos+ cells and Sox11 in the DG and CA3 regions indicated low rates of cells with prominent colocalization (Figure S3A), suggesting that Sox11 expression is not limited to the Fos+ cells that presumably experienced the strongest activation.
Figure 2.
Expression of Silc1 and Sox11 under control and novel environment (NE) conditions in WT and Silc1−/− mice
(A) RNAscope FISH assay on hippocampal sections from mice with the indicated genotype. Tissues were counterstained with a Silc1 probe (red) and DAPI (blue) and imaged using 20× (scale bar, 200 μm) and 100× oil immersion objectives (scale bar, 20 μm). NE exposure was performed using the Barnes maze setting, and the hippocampus was extracted for coronal sections after 1 h of stimulus. 12 images of non-overlapping fields per biological repeat were quantified; 3 biological repeats. Mean ± SEM is shown. The p value was calculated using unpaired two-sample t test; ∗∗p < 0.005.
(B) As in (A) with tissues hybridized with Sox11 CDS (red) and 3′ UTR (green) probes and counterstained with DAPI (blue).
(C) As in (A) for the number of green and red dots. The p value was calculated using unpaired two-sample t test; ∗p < 0.05.
(D) Immunostaining with anti-SOX11 (red) and DAPI (blue) in hippocampi of WT and Silc1−/− mice under HC and NE conditions. Imaging was performed using a 20× objective (scale bar, 200 μm).
(E) As in (A) for Sox11fl/fl mice injected stereotaxically in the CA3 region with Cre-GFP- or GFP-expressing AAV9 viruses. Imaging was done using a 20× objective (scale bar, 200 μm). SOX11 levels were significantly reduced after Cre injection, which indicates the specificity of the SOX11 antibody used for staining.
(F) Quantification of 3 biological repeats of hippocampus staining. Mean ± SEM, ∗p < 0.05, ∗∗p < 0.005, unpaired two-sample t test.
(G) Western blot with SOX11 and β-tubulin antibodies from mice with the indicated genotype, using whole-hippocampus protein extract. Hippocampus protein extract from Sox11fl/fl mice that were stereotaxically injected in the CA3 region using AAV9 Cre-GFP or AAV9 GFP were used as a specificity control for the antibodies.
(H) Western blot quantification. SOX11 expression levels were normalized to β-tubulin levels. n = 3. Mean ± SEM is shown; ∗p < 0.05, unpaired two-sample t test.
(I) As in (A) for hippocampus sections from Silc1fl/fl mice that were stereotaxically injected in the CA3 region using AAV9 Cre-GFP or AAV9 GFP. Two weeks after injection, the mice were exposed to an NE, and the hippocampus was extracted for coronal sections after 1 h of NE exposure. Imaging was performed using a 20× objective (scale bar, 200 μm).
See also Figure S2.
Conditional depletion of Silc1 in the adult brain leads to reduced SOX11 expression
Silc1−/− mice lack the Silc1 promoter and never express Silc1, so the observations of changes in SOX11 in Silc1−/− mice may reflect changes in embryonic brain development or postnatal brain maturation. To address this, we generated Silc1 conditional mice by inserting loxP sites at regions flanking the Silc1 promoter (using the same CRISPR guide RNAs used for generation of Silc1−/− mice).
For specific Silc1 reduction in the adult hippocampus, AAV9 Cre-GFP or AAV9 GFP was injected into the CA3 region of the hippocampus of Silc1fl/fl mice. A local reduction of Silc1 levels was detected only around the Cre injection site (Figure S2B). In addition, SOX11 protein levels were assessed using immunostaining and indicated a specific reduction in SOX11 following Cre injection only in cells that expressed GFP (Figures 2I and S3B). Taken together, the findings demonstrated that Silc1 lncRNA, Sox11 mRNA, and SOX11 protein are induced in the CA3 region when mice are exposed to an NE and that Silc1 depletion leads to a reduction of Sox11 mRNA and protein in the hippocampus.
Knockdown of Silc1 in the adult hippocampus
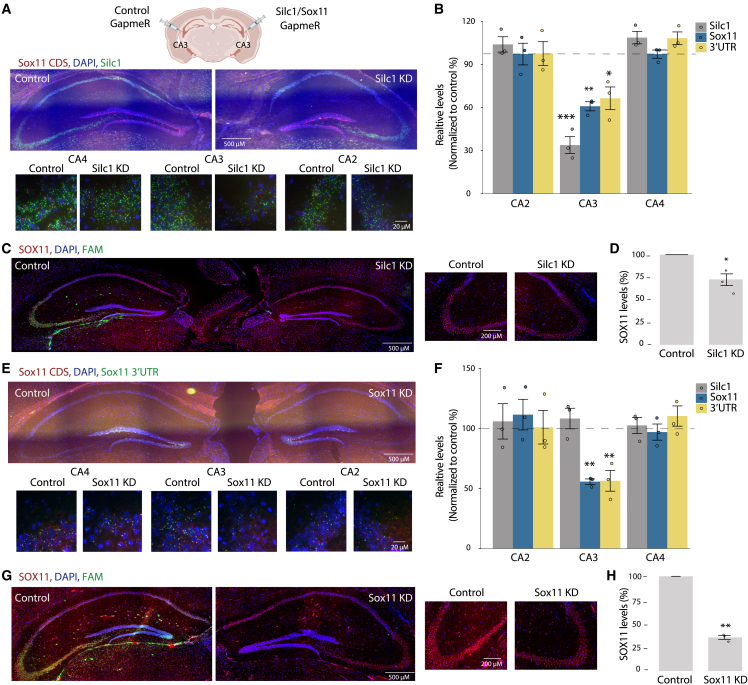
When using Silc1−/− or Silc1fl/fl mice, it is impossible to separately test the relative contribution of the Silc1 promoter, which they are lacking, and the role of transcription of Silc1 and/or its RNA product. To tease apart these effects, we first attempted to use CRISPR-Cas9 to introduce a polyadenylation signal into the first exon of Silc1, but this only partially reduced Silc1 expression in animals carrying homozygous insertions (Figure S3C). Therefore, we opted for using GapmeRs, antisense nucleotides, to degrade the Silc1 RNA product, potentially also affecting its transcription without altering the DNA of the locus.45,46 We first used a primary DRG culture to select GapmeRs that effectively reduce the expression levels of Silc1 or Sox11 (with separate GapmeRs targeting Sox11 CDS and its 3′ UTR). Efficient GapmeRs were selected by transfection into cultured DRG neurons (Figure S3D). We then introduced these GapmeRs specifically into the CA3 region of the hippocampus by stereotaxic injection and assessed the expression of Silc1 and Sox11 by RNAscope. Five days after injection of the Silc1-targeting GapmeR, we observed a local reduction in the expression of Silc1, which coincided with reduced levels of the CDS and 3′ UTR of Sox11 as well as of SOX11 protein (Figures 3A–3D). GapmeRs targeting Sox11, on the other hand, reduced Sox11 levels (further confirming the specificity of our detection reagents) but did not notably affect the expression of Silc1 (Figures 3E–3H and S4A–S4D). These results indicate that transcription through the Silc1 locus or the Silc1 RNA product in mature adult neurons is essential for regulation of Sox11 levels.
Figure 3.
Silc1 and Sox11 KD by injections of GapmeRs to the CA3 region
(A) Control Fluorescein amidites (FAM)-labeled (left) or Silc1-targeting (right) GapmeRs were injected into the CA3 region. 5 days later, the hippocampus was extracted for coronal sections. RNAscope analysis of Silc1 and Sox11 expression was performed using Silc1 (green) and Sox11 CDS (red) probes and DAPI. Imaging was done using 20× (scale bar, 200 μm) and 100× oil immersion objectives (scale bar, 20 μm).
(B) RNAscope quantification of the number of green and red dots, normalized to control GapmeR, performed using IMARIS software. 12 images of non-overlapping fields were quantified per biological repeat; 3 biological repeats. Mean ± SEM is shown. The p value was calculated using an unpaired two-sample t test; ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.001.
(C) Immunostaining with anti-SOX11 (red) and DAPI (blue) in hippocampi of Silc1 KD mice. The FAM signal marks the injection site of the control GapmeR. Imaging was performed using a 20× objective (scale bar, 200 μm).
(D) Quantification of 3 biological repeats of hippocampus staining, normalized to injection of control GapmeR. Mean ± SEM; ∗p < 0.05, unpaired two-sample t test.
(E) As in (A) using control (left) or Sox11 CDS (right) GapmeRs.
(F) As in (B) using control or Sox11 CDS GapmeRs.
(G) As in (C) for Sox11 KD mice.
(H) As in (D) for Sox11 KD mice.
See also Figure S3.
Overexpression of the Silc1 RNA sequence does not affect Sox11 or downstream genes
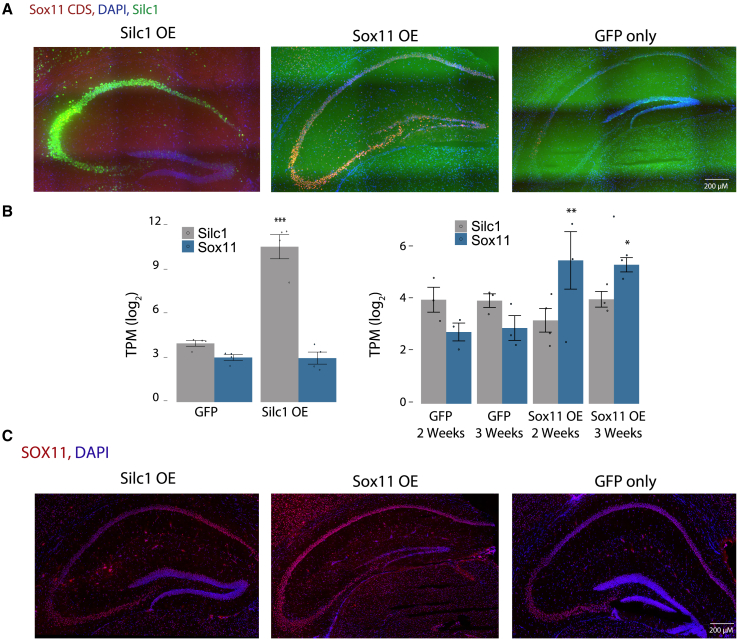
We have demonstrated previously that, in the DRG, Silc1 regulates Sox11 expression strictly in cis, and overexpression of Silc1 cDNA did not alter Sox11 levels.9 Therefore, we assessed whether that is also the case in the hippocampus. In addition, we sought a signature for changes in gene expression following upregulation of Sox11 in the CA3, where it has not been studied to date. To that end, we used AAV9 injected into the CA3 region to overexpress SOX11 in the hippocampus. The AAV vector expresses a GFP mRNA either as a control or fused to the Sox11 CDS or the Silc1 sequence, driven by the neuron-specific CaMKIIα promoter.47 As depicted in Figures 4A and 4B, 2 or 3 weeks after introduction, the GFP signal diffused to other regions of the hippocampus, and we observed a significant increase in the expression of Silc1 or Sox11 but no substantial or significant cross-regulation between the two genes (Figures 4A and 4B). Silc1 overexpression did not affect SOX11 protein levels (Figure 4C). We then injected AAV9 viruses overexpressing Silc1 into the CA3 region of Silc1−/− mice. Silc1 levels were rescued but without any notable effect on Sox11 transcript or protein levels (Figures S4E and S4F). These data suggest that, as in the DRG, the overall presence of the Silc1 RNA sequence in the cell does not affect Sox11 levels; moreover, the function of the Silc1 RNA product, if any, is only relevant when it is produced from its endogenous locus.
Figure 4.
OE of Sox11 and Silc1 in the hippocampus
(A) AAV9-Silc1, AAV9-Sox11, or AAV9-GFP was injected into the CA3 region for Silc1 or Sox11 OE. After 2–3 weeks, the hippocampus was extracted for coronal sections. RNAscope analysis of Silc1 and Sox11 expression using Silc1 (green) and Sox11 CDS (red) probes and DAPI. Imaging was done using 20× (scale bar, 200 μm) and 100× oil immersion objectives (scale bar, 20 μm).
(B) RNA-seq quantification of Silc1 and Sox11; 3 biological repeats. Mean ± SEM is shown. The p values were calculated using unpaired two-sample t test; ∗p < 0.05,∗∗p < 0.005, ∗∗∗p < 0.001.
(C) Immunostaining with anti-SOX11 (red) and DAPI (blue) in hippocampi of Silc1 and Sox11 OE mice. Imaging was performed using a 20× objective (scale bar, 200 μm).
See also Figure S4.
Silc1 is required for efficient spatial learning
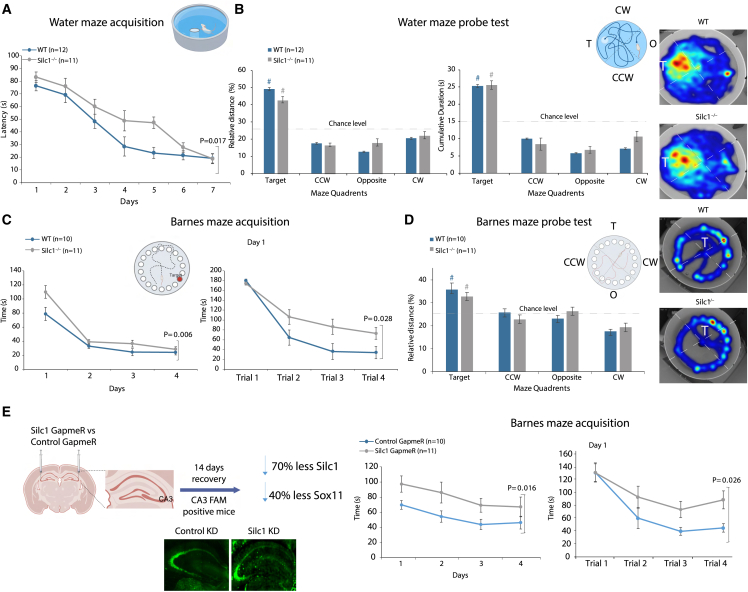
Because the hippocampus is associated with spatial memory acquisition, we then turned to examining the consequences of a lack of Silc1 on learning and memory formation using two well-established spatial learning paradigms, the Morris water maze (MWM) and the less stress-provoking Barnes maze.41,48
MWM training consisted of 7 daily sessions, each comprised of four trials (from different starting points). In each training trial, mice were allowed to swim either until they located the platform or until 90 s elapsed. The latency (in seconds) to reach the platform was recorded. Silc1−/− mice exhibited impaired spatial learning (Figure 5A); however, in the memory recall test (probe trial) performed 24 h following the last training session, no significant differences were observed between the genotypes in either index (quadrant total distance and cumulative duration) (Figure 5B). A similar pattern of results was observed in the Barnes maze. Wild-type (WT) and Silc1−/− mice (different cohorts) were trained for a total of 4 days, with four trials each day. The latency (in seconds) to enter the escape tunnel was recorded. As in the MWM, Silc1−/− mice exhibited impaired spatial learning of the Barnes maze, which was particularly pronounced on the first day but did not differ from the WT in the probe trial (Figures 5C and 5D). In addition, mice (different cohort) underwent fear conditioning and were tested for hippocampus-dependent contextual memory as well as amygdala-dependent cue memory (Figure S5A–S5C). No differences were noted between the genotypes during the conditioning phase or either of the memory tests.
Figure 5.
Silc1−/− reduction delays spatial learning
(A) Escape latencies (in seconds) over MWM training sessions; compared with the WT (n = 12), Silc1−/− (n = 11) mice exhibited significantly slower learning (gene [main effect]: F(1,22) = 6.712, p = 0.017).
(B) Probe test session (24 h after the last acquisition session). Left: relative distance swum (%). center: cumulative duration (seconds) per quadrant. Right: grouped heatmaps (cumulative duration). Probe test data indicated that Silc1−/− and WT mice explored the target quadrant significantly more than chance level (#; one-sample t test: relative distance [%]: WT t(11) = 8.737; p = 0.000. Silc1−/− t(10) = 10.021; p = 0.000. Cumulative duration (seconds): WT t(11) = 7.359; p = 0.000. Silc1−/− t(10) = 8.400; p = 0.000) in a similar manner (WT vs. Silc1−/−: relative distance [%]: t(21) = 1.849; p = 0.079. Cumulative duration (seconds): t(21) = 0.137; p = 0.893).
(C) Escape latencies (seconds) in the Barnes maze over daily training sessions (left) and within trials of day 1 (right) compared with WT (n = 10) and Silc1−/− mice (n = 11) exhibited significantly slower learning (gene [main effect]: over days F(1,19) = 9.484; p = 0.006. Day1 F(1,19) = 5.682; p = 0.028).
(D) Probe test session (24 h after the last acquisition session). Left: relative distance swum (%). Right: grouped heatmaps (cumulative duration). Probe test data indicated that Silc1−/− and WT mice explored the target quadrant significantly more than chance level (#: one-sample t test: relative distance [%]: WT t(9) = 3.697; p = 0.005. Silc1−/− t(10) = 4.246; p = 0.002) in a similar manner (WT vs. Silc1−/−: relative distance [%]: t(19) = 0.882; p = 0.389).
(E) Barnes maze escape latencies (in seconds) over daily training sessions (left) and within trials of day 1 (right) of WT C57BL/6 mice injected with Silc1 GapmeR (n = 12) or control GapmeR (n = 10) indicated that knockdown of Silc1 impairs learning (gene [main effect]: over days F(1,20) = 6.943 = 0.016. Day 1 F(1,20) = 5.801; p = 0.026).
Data represent mean ± SEM (error bars). All datasets were analyzed by two-way ANOVA for gene (between subjects), daily training sessions/trials (within subjects with repeated measures), and their interaction (gene × training). Images were generated using BioRender. See also Figure S5.
Finally, we further evaluated the role of Silc1 in learning, utilizing mice with site-specific CA3 knockdown. Silc1 GapmeR or control GapmeR was injected into C57BL/6 mice. This treatment led to a reduction in the expression of Silc1 (approximately –70%) and of Sox11 (approximately –30%). Two weeks after injection, we assessed their learning in the Barnes maze and observed a similar pattern of effects as in Silc1−/− mice (Figure 5E).
Collectively, the data from these behavioral assessments indicate that lack of Silc1 delays spatial learning without affecting long-term memory.
SOX11 drives a specific gene expression program in the adult hippocampus, which is affected by loss of Silc1
While the Sox11-driven transcriptional program has been studied extensively in other settings, it is not known which transcription is sensitive to Sox11 levels in mature adult neurons. To characterize these changes, we used RNA-seq to measure gene expression in the hippocampus of Sox11flox/flox mice injected with an AAV vector driving expression of Cre (or GFP control) compared with the WT mouse hippocampus overexpressing Sox11 fused with GFP (or GFP control). Two or three weeks post injection, the hippocampus was extracted and subjected to RNA-seq. Three weeks after the injection, we observed a reduction in GFP and Sox11 transcript levels, possibly because of cell toxicity caused by SOX11 overexpression (as observed in other systems49) or silencing of the transgene (Figure 4B). In the RNA-seq data after 3 weeks of SOX11 overexpression (OE), the 144 significantly reduced genes (fold change < 0.5 and adjusted p < 0.05) were significantly enriched with genes downregulated in neurodegenerative diseases: Huntington’s disease (Enrichr analysis, adjusted p = 1 × 10−13) and Parkinson’s disease (p = 2.85 × 10−8).
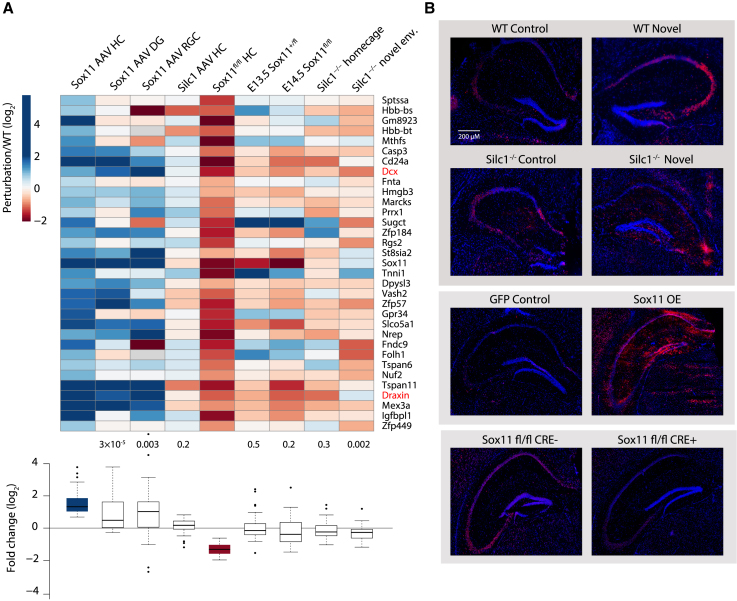
Therefore, we focused on the RNA-seq data from the hippocampus extracts that were collected 2 weeks after AAV introduction for OE or one week after Cre injection. Among the 14,181 expressed genes (average FPKM ≧ 0.5; Table S1), we focused on genes that changed significantly (p < 0.05) in opposite directions by at least 50%. Under these definitions, 33 genes were positively regulated by Sox11 in the adult hippocampus (Figure 6A), including the well-characterized targets Dcx22 and Mex3a,50 and only 7 were negatively regulated. These data are consistent with SOX11 acting predominantly as an activator, and therefore, we focused on the positively regulated genes in further analyses. These genes were also significantly upregulated in published datasets of AAV-mediated SOX11 induction in the DG28 and retinal ganglion cells (RGCs).51 While some of the genes were also downregulated in the dentate neuroepithelium at embryonic day 13.5 (E13.5) in embryos lacking SOX11, specifically in the telencephalon,52 the difference was not significant when considering these genes as a group. Further supporting the 33 genes being tightly associated with Sox11 activity, Enrichr5 found that they were enriched with genes co-expressed with human SOX11 in the ARCHS4 database (adjusted p = 2 × 10−5; odds ratio, 16.84). Hereafter, we refer to these genes as “Sox11 targets.”
Figure 6.
Characterization of genes regulated by Sox11 and Silc1 by RNA-seq
(A) Top: heatmap of changes in gene expression relative to the respective controls of the 33 genes that were upregulated following AAV-mediated Sox11 OE in the hippocampus and downregulated following AAV-mediated introduction of Cre into the hippocampus of Sox11fl/fl mice (p < 0.05, fold change of at least 50%). Bottom: boxplots of the same changes, with p values computed using a two-sided one-sample t test.
(B) Immunostaining with anti-Draxin (red) and DAPI (blue) in the hippocampi of WT and Silc1−/− mice under HC and NE conditions (Sox11 OE and Cre- or control-injected Sox11fl/fl mice). Imaging was performed using a 20× objective (scale bar, 200 μm).
See also Figures S5 and S6 and Table S1.
Consistent with its inability to lead to changes in SOX11 levels, AAV-mediated induction of Silc1 did not significantly affect the expression of Sox11 targets (Figure 6A). In contrast, these genes were significantly reduced in the hippocampus of Silc1−/− mice following NE exposure relative to WT mice. The seven negatively regulated targets were supported in the other datasets to a lesser extent and did not change significantly in the Silc1−/− hippocampus (Figure S5D).
Another 1,047 genes were downregulated with the same criteria (25% reduction and p < 0.05; Table S1) in the E13.5 Sox11-null dentate neuroepithelium but did not meet our bidirectional change criteria in the adult hippocampus (“embryonic Sox11 targets”; Figure S5E). Notably, a major fraction of the embryonic Sox11 targets was substantially induced upon Sox11 OE in the DG and RGC but to a much lesser extent in the other section of the hippocampus. While some of the 1,047 genes were downregulated upon Cre injection, the fold changes were overall modest. In addition, in the Silc1−/− hippocampus, although the overall reduction was significant, the median fold change was not substantial; i.e., close to zero. Therefore, we conclude that, whereas Sox11 is required for proper expression of many genes in the embryo and regulates their expression in immature neurons, only a small subset of genes remains sensitive to SOX11 levels in mature neurons in the hippocampus. We suggest that, in these cells, the expression levels of SOX11 are much lower than in the embryo or in the DG SGZ; therefore, even Sox11 OE via an AAV, which upregulates Sox11 by ∼30-fold, still results in lower Sox11 levels than those present in the E13.5 embryo. Alternatively, post-translational modifications of SOX11 in the embryo and SGZ differ from those in mature neurons and limit SOX11 activity. Nevertheless, a small but notable set of specific Sox11 targets, including Dcx and Draxin, remains sensitive to increases and decreases in Sox11 levels throughout the mature hippocampus. Crucially, these genes are significantly reduced in the Silc1−/− hippocampus following exposure to an NE (NE condition). We validated changes in DRAXIN protein levels using immunostaining and obtained a pattern of results similar to those of the RNA-seq analysis (Figure 6B).
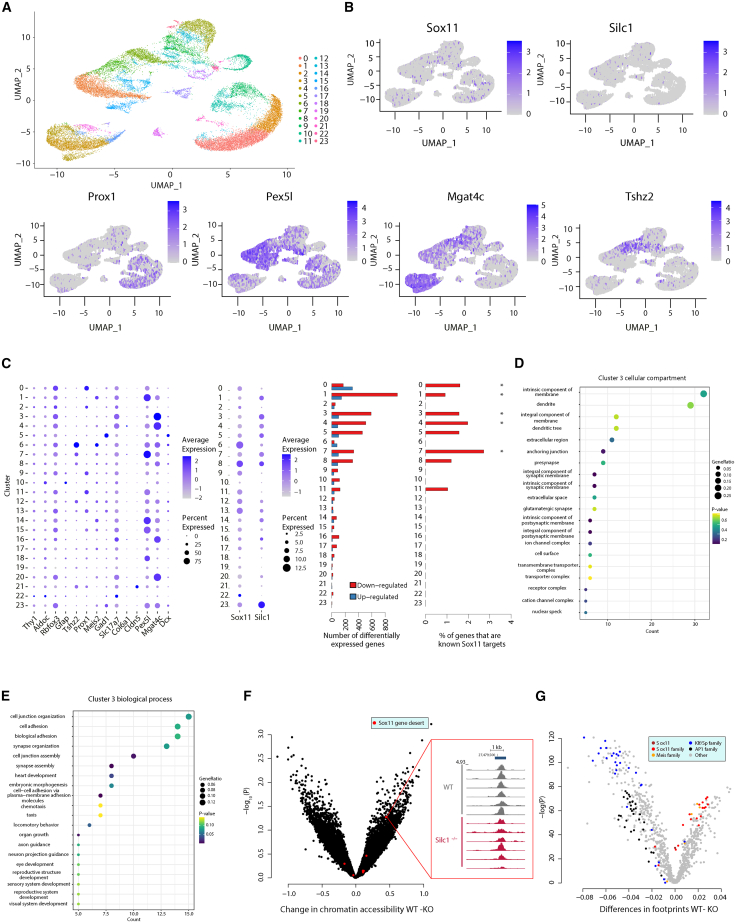
To obtain a higher resolution of the changes in gene expression that occur specifically in the CA3 region in Silc1−/− mice placed in an NE, we performed single-nucleus RNA-seq (snRNA-seq) on hippocampi of two WT and two Silc1−/− mice. Processing and clustering of the data (STAR Methods) identified 24 clusters of cells (Figure 7A) with a clear separation between DG (Prox1+), CA3 (Mgat4c+), and CA1 (Pex5l+) cells (Figures 7B, S6A, and S6B). Consistent with their relatively low expression, Silc1 and Sox11 were detected in a minority of the cells, but Silc1 was evidently expressed predominantly outside of the DG, whereas Sox11 was detected in cells from different regions (Figure 7B). The low fraction of cells in which Sox11 mRNA was detectable (<10% in most clusters) precluded analysis of its differential expression in the snRNA-seq data, and so we focused on more abundant mRNAs that were differentially expressed in individual clusters. Gene expression was mostly downregulated, and, as expected, the largest changes in gene expression occurred in clusters where Silc1 expression was highest: 1, 3–5, and 7–8, all of which had low Prox1 expression, suggesting that they corresponded to cells outside of the DG, and of which clusters 3 and 4 had the highest Mgat4 expression, suggesting that these are CA3 cells (Figures 7B and 7C). Notably, four of these clusters were enriched with genes positively regulated by Sox11 in other studies28,51,52 (“known” targets in Figure 7C), suggesting that at least some of the changes in gene expression occur through changes in Sox11 activity.
Figure 7.
Differences in cell-type-specific gene expression and chromatin accessibility in the Silc1−/− hippocampus
(A) UMAP visualization of the snRNA-seq expression data, with color coding of the 22 clusters of cells.
(B) Expression levels of Sox11, Silc1, and selected marker genes projected onto the UMAP visualization.
(C) Left: dot plot of the expression levels and the fraction of cells in which each indicated gene was expressed in each indicated cluster. Right: numbers of significantly differentially expressed genes in each cluster and the fraction of these that overlap with the “known” Sox11 targets from other studies.28,51,52
(D and E) GO cellular compartment (D) and biological process (E) terms enriched in the genes significantly downregulated in Silc1−/− cells compared with WT cells in cluster 3.
(F) Volcano plot of the difference in read coverage (x axis) and statistical significance (y axis) at the 31,764 peaks called by MACS2 using the entire dataset. Peaks in the ∼2-Mb gene desert flanking Sox11 are shown in red. The inset shows ATAC-seq read coverage in the most differential Sox11-proximal peak, which is one of the peaks highlighted in Figure S1.
(G) Changes in TF footprints for each TF family present in the JASPAR database. Motifs assigned to the indicated families are marked in one of five colors.
To characterize the processes affected by the changes in gene expression, we performed Gene Ontology (GO) analysis on the genes differentially expressed in individual clusters. Several clusters were enriched with genes related to synaptic transmission and axon guidance and/or encoding proteins with dendritic localization (Figures 7D, 7E, and S6C), some of which were also Sox11 targets, such as Syn3 (significantly reduced in cluster 1), Sestd1 (significantly reduced in cluster 7), and Lzts1 (significantly reduced in cluster 8).
Collectively, the data suggest that, in the adult hippocampus, Sox11 drives a specific gene expression program containing a subset of the Sox11 targets in the developing brain; expression of this program is affected by loss of Silc1, specifically in the regions outside of the DG where Silc1 expression is high. The affected genes encode proteins related to synaptic transmission and axon guidance, providing a potential mechanism for how loss of Silc1 affects learning. It thus appears that a subset of a regulatory program used during development is co-opted during memory formation in the hippocampus.
Loss of Silc1 results in reduction of chromatin binding at Sox family binding sites
We presumed that the changes in gene expression are driven by changes in chromatin and used ATAC-seq53 to profile accessible chromatin in the hippocampus of WT and Silc1−/− mice placed in an NE (six biological replicates). We first quantified accessibility at 31,764 peaks by jointly analyzing the full dataset by MACS254 (Figure 7F; Table S2). No peak was differentially accessible between WT and Silc1−/− mice when accounting at a false discovery rate (FDR) of less than 0.05. The peaks in the two gene deserts flanking Sox11 did not appear to change their accessibility (Figure 7F), with the notable exception of an AP-1-bound peak (the central peak in the magnified panel in the figure). To evaluate whether there were changes in TF binding within the accessible regions, we analyzed TF footprinting with TOBIAS.55 This analysis (Table S3) implicated numerous TFs as differentially binding accessible genome regions, notably indicating reduced binding to TF binding sites of the Sox family (Figure 7G), consistent with reduced protein expression of Sox11, which is the main Sox factor expressed in the hippocampus (Figure S5F). Notably, no other Sox gene was significantly differentially expressed in individual clusters in the snRNA-seq data.
Discussion
Sox11 has been studied in many systems, including the retina, but in the forebrain, it has been studied almost exclusively in the context of the developing brain or immature neurons in the SGZ. The view emerging from multiple studies is that, in these systems, Sox11 regulates targets shared in part with those of Sox4 and supports further maturation of neurons that recently exited the cell cycle. Surprisingly, we find that, upon exposure to an NE, Sox11 is prominently induced in mature parts of the hippocampus. Among the genes differentially expressed upon loss of Silc1, which presumably affects predominantly mature neurons because it does not appear to be expressed in immature ones, we find a small but prominent set of Sox11 targets that are also regulated by Sox11 in the context of immature neurons and normally also predominantly expressed in neuronal progenitors and immature neurons in the SGZ, such as Dcx, Draxin,56 and Prrx1,57 and directly or indirectly regulate genes involved in synaptic transmission. The design principle underlying this apparent re-use of a master regulator of neuronal maturation in the context of plasticity of mature neurons will be an interesting subject for future studies. Interestingly, the loss of some of the Sox11 targets prominently regulated is related to abnormal mossy fiber formation (St8sia258). Notably, among the Sox11 targets, loss of Dcx as well as P311, encoded by the Nrep gene, leads to defects in spatial learning.59,60
Combined with our previous observations in the peripheral nervous system (PNS), this study solidifies the notion that Sox11 has at least two main regulatory regimens. The first is active in immature neurons, in the embryo and in the SGZ. These are cells that exited the cell cycle but undergo neuronal growth, which is apparently dependent on high SOX11 levels. Presumably, a variety of enhancer elements are supporting this high Sox11 expression because neither Silc1 nor other lncRNAs in the large gene deserts flanking Sox11 appear to be expressed in these cells. The second regimen is activated in cells that cease their growth and possibly coincides with expression of Thy1, which is the gene most tightly correlated with Silc1 in the mouse NeuroSeq expression atlas. Under basal conditions, this program supports minimal expression of Sox11, which is likely not required for the steady-state activity of these neurons. Upon activation, likely via activity of the AP-1 TFs, Sox11 is induced quite potently in this regimen. This activation can result from injury signaling in the PNS or from exposure to an NE in the CA subfields in the hippocampus. Timely and potent induction of Sox11 under these conditions depends on Silc1 transcription or the RNA product because it is sensitive to perturbation by antisense oligonucleotides. Our findings support the notion that ncRNAs in general and lncRNAs in particular have a critical role in different stages of neural circuit development, activity-dependent circuit remodeling (learning and memory), and circuit maladapations underlying neurological and neuropsychiatric disorders.61,62
As a regulator of transcription in the nervous system, Silc1 joins the ranks of other lncRNAs shown to mediate transcriptional control in the context of learning, including Gm12371, ADRAM, and Neat1.7,63,64 Notably, while Silc1 has some effect on the basal level of Sox11 expression in the nervous system, which is low and lower than that of Silc1, it appears to be particularly important for timely induction of Sox11 expression upon different physiological cues. Therefore, it is reminiscent of other cis-acting RNAs,32 whose modes of action are largely unknown. A common denominator of these RNAs is relatively high expression under unstimulated conditions and efficient splicing, which has been linked to more efficient enhancer activity in broad regions flanking the spliced lncRNAs.65,66,67 It is therefore possible that transcription of Silc1 and its splicing enables proper positioning of the broad Sox11 locus in a nuclear and chromatin environment that facilitates a stronger response to stimulus, such as that occurring when mice are exposed to an NE. Indeed, we found a region in the Silc1−/− hippocampus that is differentially accessible and corresponds to AP-1 regulated enhancer. However, these changes were variable between mice and did not reach statistical significance (Figure 7F). Development and availability of methods that will allow measurement of chromatin accessibility in situ can potentially shed further light on whether this enhancer becomes specifically less accessible in in the Silc1−/− hippocampus in cells that normally activate Sox11 expression following exposure to an NE.
The roles of Silc1 in neuropathology can be an interesting topic for further study. Timely learning is impaired in aging and neurodegenerative diseases. In gene expression data collected from the hippocampus and cortex from C57BL/6 background mice used in our study,68 Silc1 expression was reduced in both brain regions during the course of aging, with further reduction in an Alzheimer’s disease (AD) model, which expresses five familial AD mutations under the control of a Thy1 mini-gene. In control mice, Silc1 expression was strongly negatively correlated with age in both brain regions (hippocampus: R = −0.58, p = 6.42 × 10−6; cortex: R = −0.47, p = 5.08 × 10−4) (Figure S7). At most ages, a further reduction was found in AD mice relative to controls (Figure S7). These results show that Silc1 continues to be broadly and abundantly expressed in the forebrain throughout life and that its loss might be associated with aging- and AD-related cognitive decline.
Limitations of the study
We focused here on the interplay between Sox11 and Silc1 in the hippocampus and on their roles in spatial memory acquisition, but notably, the two genes, and in particular Silc1, are broadly expressed in the adult brain, suggesting that, under certain conditions, Silc1 can also be important beyond the hippocampus, and this can be explored in future studies. We focused on the hippocampus because electroconvulsive stimulation of the whole brain most prominently activates Sox11 in the DG,26 and we are not aware of other physiological conditions where Sox11 is transcriptionally activated in the adult brain. It is possible that other conditions and regions require activation of the “immature neuron” transcriptional program, including Dcx, Draxin, and the other Sox11 targets we describe here.
We do not presently know the mechanism by which Silc1 facilitates activation of Sox11 in the CNS and PNS. In both systems, accessibility of the Sox11 promoter does not appear to change during Sox11 activation; to the extent measurable by genome-wide ATAC-seq, the promoter appears to be highly accessible in all neuronal tissues. The paucity of cell lines in which Silc1 is endogenously expressed hinders some experimental approaches to its perturbation. Silc1 is not expressed in mouse cell lines, in the mouse embryo, or in commonly used primary hippocampus cultures obtained from embryonic or early postnatal stages. We managed to successfully obtain and characterize, by qPCR and RNA-seq, primary cultures of adult hippocampal neurons but found that those cells lost all expression of Silc1 or Sox11 (Figure S3C). While Neuro2a cells express some Sox11 and can be induced by CRISPR activation to express Silc1,9 these levels of induction are insufficient to drive levels of Sox11 expression comparable with those in the brain. Therefore, we are presently limited to experimental manipulations that can be performed in the adult brain, such as introduction of GapmeRs and AAVs as performed here.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-SOX11 | Merck Millipore | Cat#ABN105 |
| Mouse anti-NeuN | Merck Millipore | Cat#MAB377; RRID: AB_2298772 |
| Rabbit anti-Draxin | Abcam | Cat#ab117452; RRID: AB_10898861 |
| Mouse anti-Beta-tubulin | Sigma | Cat#T4026; RRID: AB_477577 |
| AzureSpectra 700 Goat-anti-mouse secondary antibody | Azure biosystem | Cat#AC2129 |
| AzureSpectra 800 Goat-anti-rabbit secondary antibody | Azure biosystem | Cat#AC2134 |
| Donkey anti-Mouse Alexa 594 | Molecular Probes | Cat#A21203; RRID: AB_141633 |
| Goat anti Rabbit Alexa 647 | Abcam | Cat#ab150079; RRID: AB_2722623 |
| Bacterial and virus strains | ||
| AAV9 pENN.AAV.CamkII.HI.GFP-Cre.WPRE.SV40 | Addgene | 105551-AAV9 |
| AAV-9 pENN.AAV.CamKII0.4.eGFP.WPRE.rBG | Addgene | 105541-AAV9 |
| AAV-9 Silc1 | This study | N/A |
| AAV-9 Sox11 | This study | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Papain | Sigma | P4762 |
| Dispase II | Roche | 165859 |
| Percoll | Sigma | P1644 |
| Poly L lysin | Sigma | P4832 |
| Laminin | Life | 23017–015 |
| Collagenase type II | Worthington | CSL-2 |
| Critical commercial assays | ||
| SENSE mRNA-Seq Library Prep Kit V2 | Lexogen | 001.96 |
| ACD RNAscope Fluorescent Multiplex Assay | ACDBio | 320850 |
| 10x Genomics Chromium Single Cell Kit Version 3.1 | 10x Genomics | |
| Nextera XT DNA Library Preparation Kit | Illumina | FC-131-1096 |
| NovaSeq 6000 SP Flow Cell | Illumina | 20028401 |
| Deposited data | ||
| Raw and analyzed RNA-seq data | This study | GEO: GSE216643 |
| Raw and analyzed ATAC-seq data | This study | GEO: GSE216643 |
| Raw and analyzed scRNA-seq data | This study | GEO: GSE216643 |
| Analyzed data of RNA-seq from the NeuroSeq atlas | Sugino et al.35 | GEO: GSE79238 |
| Analyzed data of RNA-seq–based expression in the DG upon stimulation and ATAC-seq | Sun et al.26 von Wittgenstein et al.28 | GEO: GSE140180 |
| Analyzed data of ATAC-seq upon kainic acid stimulation | Fernandez-Albert.et al.38 | GEO: GSE125068 |
| Analyzed data from Hipposeq | Cembrowski et al.39 | GEO: GSE74985 |
| Experimental models: Organisms/strains | ||
| Mouse: C57black6 Ola HSD | Harlan Laboratories | N/A |
| Mouse: Silc1−/− mice | Perry et al.9 | N/A |
| Mouse: Silc1 loxP mice | This paper | N/A |
| Mouse: Silc1 polyA mice | This paper | N/A |
| Mouse: Sox11 loxP mice | Prof. Veronique Lefebvre lab | N/A |
| Mouse: C57black6 Ola HSD | Harlan Laboratories | N/A |
| Oligonucleotides | ||
| For a list of oligonucleotides used in this study see Table S4 | This study | N/A |
| For a list of GapmerRs used in this study see Table S5 | This study | N/A |
| Recombinant DNA | ||
| Silc1 pcDNA3.1(+) vector | Perry et al.9 | N/A |
| pLenti-CMV-GFP-Sox11 vector | Addgene | 120387 |
| pENN.AAV.CamKII0.4.eGFP.WPRE.rBG | Addgene | 105541 |
| Software and algorithms | ||
| STAR | Dobin et al.69 | https://github.com/alexdobin/STAR |
| RSEM | Li and Dewey70 | https://deweylab.github.io/RSEM/ |
| Cell Ranger 7.1.0 | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/ |
| Bowtie2 | Langmead and Salzberg71 | https://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Fiji (ImageJ) analysis software | NIH | https://fiji.sc/ |
| IMARIS (v7.7.2) software | Oxford instruments | https://imaris.oxinst.com/ |
| Other | ||
| Silc1 probes- RNAscope 2.5vs. probe ״Mm GM9866״ | ACD | 536709 |
| Sox11 CDS probes- RNAscope 2.5vs. probe | ACD | 440811 |
| Sox11 3′UTR probes- RNAscope 2.5vs. probe | ACD | 805071 |
| Fos probes- RNAscope 2.5vs. probe | ACD | 316921 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Igor Ulitsky (igor.ulitsky@weizmann.ac.il).
Materials availability
This study did not generate new unique reagents.
Experimental model and study participant details
Animals
The study was conducted following the guidelines of the Weizmann Institutional Animal Care and Use Committee (IACUC). C57black6 Ola HSD male mice were purchased from Harlan Laboratories (Rehovot, Israel). All other mouse strains were bred and maintained at the Veterinary Resources Department of the Weizmann Institute. For the behavioral tests and GapmeR injections, we used 8-week-old male C57BL/6 OlaHsd mice. For all other experiments, we used both male and female mice.
DRG cultures
Adult mouse DRGs were dissociated for neuron cultures with 100 U of papain followed by 1 mg/mL collagenase-II and 1.2 mg/mL dispase. The ganglia were then triturated in HBSS, 10 mM glucose, and 5 mM HEPES (pH 7.35). Neurons were recovered through percoll, plated on laminin, and grown in F12 medium for 48 h.72 Adult male mice DRG cultures were transfected with GapmeRs using DharmaFect 4 (Dharmacon). 72 h after transfection total RNA was extracted to ensure knockdown.
Method details
Generation of Silc1 polyA mice
Mice carrying a Silc1polyA allele were generated using the CRISPR/Cas9 system for insertion of transcription terminator by standard procedures at the Weizmann transgenic core facility using a single guide RNA (sgRNA) targeted to exon 1 (chr12:27160402, mm10 assembly). gRNA sequence was designed using CHOPCHOP73 and ordered from IDT (gRNA sequence: GTGCTTGGCACTGCTTGGCA). For homologous recombination, an ssODN (200 nt) containing two homology arms (50 nt each), a short poly(A) site (49 nt), and two MAZ sites74 was synthetized by IDT (Table S4). The poly(A)/MAZ insertion was detected by PCR amplifications. Sequences of primers used for genotyping appear in Table S4. Lines were bred and maintained on C57BL/6 background at the Veterinary Resources facility of the Weizmann Institute. All the experiments were done on 6–8 weeks old mice from F3 generation.
Generation of Silc1 conditional mice
Mice carrying the Silc1fl conditional alleles were generated using the CLICK system75 using a long single-stranded DNA (lssDNA), by standard procedures at the Weizmann transgenic core facility. Two single guide RNAs (sgRNAs) targeted to sites before the Silc1 promoter (chr12:27160159, mm10 assembly) and after exon 1 (chr12:27161872, mm10 assembly) were designed using CHOPCHOP73 and ordered from IDT9). 12 μg of the lssDNA were also ordered from IDT. Silc1fl/fl mice were identified by genotyping and sequencing using primers flanking the loxP insertion sites. Sequences of primers used for genotyping appear in Table S4. Lines were bred and maintained on C57BL/6 background at the Veterinary Resources facility of the Weizmann Institute. All the experiments were done on 6–8 weeks old mice.
Morris water maze
The water maze76 consisted of a circular tank (120 cm diameter) filled with 25(±2)°C water clouded with milk powder with an escape platform located in one of the four maze quadrants’ center, submerged 0.5 cm below the water surface. The testing room provided only distal visual-spatial cues for orientation. Acquisition phase - The mice underwent 4 trials per day with an inter-trial interval of 2 min, over 7 consecutive days. In each trial, the mice were required to find the platform within 90 s. The escape latency in each trial was recorded. Each mouse was allowed to remain on the platform for 15 s and was then removed from the maze. If the mouse did not find the platform in the allocated time, it was manually placed on the platform for 15 s. Probe test - Memory was assessed 24 h after the last trial. The escape platform was removed, and mice were allowed to search for it for 1 min. The time spent (sec)and distance (cm) swimming in each of the different quadrants of the pool, were monitored using an automated tracking system (Ethovision XT, Noldus, the Netherlands).
Barnes circular maze
The test was performed as previously described.41 The apparatus used was an elevated circular platform (0.90 m in diameter) with 20 holes (5 cm diameter) around the perimeter of the platform, one of which was connected to a dark escape recessed chamber (target box). The maze was positioned in a room with large, simple visual cues attached to the surrounding walls. The acquisition consisted of four daily trials for 4 days, separated by a 15 min intertrial interval. Each mouse was positioned in the center of the maze in an opaque cylinder for 1 min, which was gently lifted and removed to start the session. The mice were allowed to find the target box for 3 min. At the end of the 3 min, if the mouse failed to find the recessed escape box, it was gently guided to the chamber and allowed to stay in it for 1 min. The location of the escape box was kept constant with respect to the distal visual cues. An animal was considered to enter the escape chamber when the animal’s entire body was inside the chamber and no longer visible on the maze surface. The escape latency in each trial was recorded. Recall was tested 24 h after the last training session (day 5); in this probe trial, the target hole was closed, and the time spent (sec) and distance (cm) walking in each of the different quadrants of the maze were collected using an automated tracking system (Ethovision XT, Noldus, the Netherlands).
Fear conditioning
The test was performed as previously described.77 A computer-controlled fear-conditioning system (Ethovision XT, Noldus, the Netherlands) monitors the procedure while measuring inactivity (freezing) behavior. 1. Conditioning: conditioning takes place on day 2 in one 5-min training session. Mice were placed in the chamber to explore the context for 2 min. Then we applied a conditioned stimulus (CS) for 30 s, 3,000 Hz, pulsed 10 Hz, 80 dB, and a co-terminating foot shock (delivered through the metal grid floor) for as an unconditional stimulus (US): 0.7 mA, 2 s, constant current. The CS–US pairing was repeated twice with a fixed inter-trial interval (ITI) of 60 s. The US is delivered through the metal grid floor. Mice were removed from this chamber 1 min after the last CS-US pairing and put back in their home cage. A constant auditory background noise (white noise, 62 dB) was presented throughout the experiment. 2. Testing: Context-dependent memory was tested 24 h after the conditioning by re-exposure to the conditioning box for 5 min without any CS or US. The Cue dependent memory was tested 1 h after the Context test by exposure to the CS in the same temporal patern as in the conditioning, yet in different environmental conditions ([black Plexiglas box (instead of clear), black Plexiglas floor (instead of metal grid), no illumination, no background noise, cleaning solution: acetic acid 10% (instead of alcohol 10%)].
Western blot and immunofluorescence
Brain sections were fixed with 4% paraformaldehyde for 3 h followed by overnight in 30% sucrose with overhead rotation. Tissue was frozen in Tissue-Tek O.C.T compound (Sakura 4583) blocks and sectioned using a Leica cryostat (CM3050) at 10 μm thickness. Blocking and permeabilization were done with 5% donkey serum, 2% BSA, and 0.1% Triton X-100 in PBS. Primary antibodies were diluted in a permeabilization buffer. Antibodies used: Sox11 antibody, anti-rabbit (ABN105) from Millipore, NeuN antibody, anti-mouse (MAB377) from Millipore and Draxin antibody, anti-rabbit (ab117452) from Abcam. Secondary antibodies: Donkey anti-Mouse Alexa 594 (Molecular Probes A21203) and Goat anti Rabbit Alexa 647 (Abcam ab150079). Nuclei were stained using DAPI (Thermo Fisher Scientific). Imaging was done using a Leica DM4000 B microscope with Leica DFC365 FX CCD camera and Leica application suite (LAS) X software. Western blots were carried out as previously described.72 For Westerns, the samples were resolved on 10% SDS PAGE, transferred to nitrocellulose, and incubated with primary antibodies overnight. Antibodies used: Sox11 antibody, anti-rabbit (ABN105) from Millipore and beta-tubulin antibody, anti-mouse (T4026) from Sigma. AzureSpectra fluorescent 700 anti-mouse and 800 anti-rabbit (Azure biosystem) were used as the secondary antibodies for fluorescent quantification of Western blots. Blots were imaged on an Azure Imager system.
RNAscope FISH
Brains were immediately frozen on dry ice in a tissue-freezing medium. Brains were sliced on a cryostat (Leica CM 1950) into 8-μm sections, adhered to SuperFrost Plus slides (VWR), and immediately stored at −80°C until use. Samples were processed according to the ACD RNAscope Fluorescent Multiplex Assay manual using Silc1 probes- RNAscope 2.5vs. probe “Mm GM9866” Cat No. 536709, Sox11 CDS probe Cat No. 440811, Sox11 3′UTR probe Cat No. 805071 and Fos probe Cat No. 316921. Imaging was performed on a Nikon-Ti-E inverted fluorescence microscope with a 100× oil-immersion objective and a Photometrics Pixis 1024 CCD camera using MetaMorph software as previously described.78
RNA extraction and sequencing
Total RNA was extracted from the hippocampus using the TRIREAGENT (MRC) according to the manufacturer’s protocol. Strand-specific mRNA-seq libraries were prepared from 1 μg total RNA using the SENSE-mRNA-Seq-V2 (Lexogen), according to the manufacturers’ protocol and sequenced on a NextSeq 500 machine or Novaseq 6000 machine to obtain 75 nt and 150 nt single- or paired-end reads. All RNA-seq dataset is deposited in GEO database with the accession GSE216643.
Quantitative reverse-transcription PCR (qRT-PCR)
Reverse transcription was done using qScript Flex cDNA synthesis kit (Quanta Biosciences), using random primers. Quantitative PCR was performed in a ViiA 7 Real-Time PCR System (Thermo) in a 10 μL reaction mixture containing 0.1 μM forward and reverse primers, fast SYBR master mix (Applied Biosystems), and template cDNA. A reaction containing DDW instead of cDNA was used as a no-template control and was amplified for each primer pair. Only samples free of DNA contamination were further analyzed. The gene-specific primer pairs used for mRNA expression level analysis are listed in Table S4.
ATAC-seq
ATAC-seq was performed as described53 with minor adjustments for brain tissue. Briefly, hippocampus tissue was extracted in 500 μL Nuclear extraction buffer (10mM Tris, 10mM NaCl, 3mM MgCl2, 0.1% Igepal, 0.1% Tween, protease inhibitor cocktail) for 5 min on ice, then a 21g needle on a 1 mL syringe was used to shear the tissue through the needle 5 times. NeuN-positive nuclei were separated by fluorescence-activated cell sorting. Libraries were sequenced with 50 bp paired-end mode on NovaSeq6000.
Microinfusion of antisense LNA GapmeR
8 weeks old C57BL/6J male mice (Envigo, Israel) were anesthetized with isoflurane and placed in a stereotactic frame (n = 3 per group). The skull was exposed to antiseptic conditions and a small craniotomy was made with a thin drill over the hippocampus. Antisense LNA GapmeRs (custom designed, 3′-FAM-labeled, Qiagen, Table S5) were bilaterally microinfused using a 2 μL calibrated micropipette (Hamilton syringes ga 25/70mm/pst3), which was pulled to create a long narrow shank. 1 μL was infused slowly by pressure infusion into the CA3 region (from bregma +3.1 mm anteroposterior, ±2.8 mm mediolateral and +3.2 mm dorsoventral axis), the micropipette was kept in place for 30 s to ensure adequate diffusion. The wound was sutured with sterile nylon material.
AAV9 vectors cloning and virus generation
AAV9 expression of GFP-Cre from CamKII promoter: Addgene number- 105551-AAV9 (plasmid: pENN.AAV.CamkII.HI.GFP-Cre.WPRE.SV40) and AAV9 control virus - 105541-AAV9 (plasmid: pENN.AAV.CamKII0.4.eGFP.WPRE.rBG).
AAV9 plasmids were used for Silc1 and Sox11 overexpression. We cloned Silc1 from the Silc1 pcDNA3.1(+) vector9 and Sox11 from pLenti-CMV-GFP-Sox11 vector (Addgene: #120387) into pENN.AAV.CamKII0.4.eGFP.WPRE.rBG plasmid downstream of the GFP sequence. Recombinant AAV9 plasmids were produced by transfecting HEK293T cells using the AAVpro helper-free systems. AAV9 viral preparations were purified using the AAVpro Purification Kit (Takara Bio. Inc., Cat#6666).
Microinfusion of AAV9 viruses
8 weeks-old C57BL/6J male mice (Envigo, Israel, n = 3 per group) received bilateral stereotaxic injections of AAV9 Silc1, AAV9 Sox11 or AAV9 GFP control into the hippocampus CA3 regions (titer of 1012 vg/mL, 0.2 μL Min). The virus was delivered using a 2 μL Hamilton syringe connected to a motorized nano-injector. To allow diffusion of the solution into the brain tissue, the needle was left in place for 4 min after the injection (from bregma +3.1 mm anteroposterior, ±2.8 mm mediolateral and +3.2 mm dorsoventral axis). The wound was sutured with sterile nylon material. The mice recovered from the surgery for a period of 2–3 weeks before the hippocampus was extracted for RNA extraction or for sections. The slides were screened for GFP signal at the injection site and mice that did not show fluorescent labeling at the aimed injection location were excluded from the data. AAV9 Cre-GFP and AAV9 GFP were injected into Soxfl/fl mice and Silc1fl/fl Mice using the same parameters.
Single-nucleus snRNA sequencing
Mice were sacrificed and the hippocampus of each mouse was dissected and placed in a lysis buffer (10mM Tris pH 7.4, 10mM NaCl, 3mM MgCl2, 0.1% NP-40 and 1:1000 RNasIN), according to 10x genomics Nuclei isolation for single cell sequencing protocol. The pellet containing the nuclei was resuspended in a wash buffer (PBS, 1%BSA and 1:1000 RNasIN). 1000/μl nuclei from each sample were taken for library preparation using 10x Genomics Chromium Single Cell Kit Version 3.1 according to manufacturer protocol. All libraries were pooled and sequenced on NovaSeq 6000 to an average depth of approximately 200,000,000 reads per sample.
Quantification and statistical analysis
Quantification of immunofluorescence
The immunofluorescence staining was quantified using Fiji (ImageJ) analysis software.
RNAscope quantification
RNAscope analysis was done using IMARIS (v7.7.2) software.
RNA-seq data analysis
RNA-seq reads were mapped to the mouse genome (mm10 assembly) using STAR69 to generate read coverage tracks visualized using the UCSC genome browser. RSEM70 with RefSeq annotations was used to call differential expression between samples with data collected in this study and data from public datasets on OEof Sox11 in the hippocampus: SOX11 induction in the DG28 – SRP229390; SOX11 induction and in RGCs51 – SRP290800; dentate neuroepithelium at E13.5 in the embryos lacking SOX1152 – SRP285830; aging hippocampus and cortex – SRP309056. Differential expression between conditions was called using DESeq2 with default parameters.37
ATAC-seq data analysis
ATAC-seq reads were mapped to the mouse genome using Bowtie271 and peaks were called using all the samples together using MACS2.54 The peaks were then adjusted to a fixed width of 140 nt around the peak summit and differential read coverage between the six WT and the six Silc1−/− samples was called using HOMER79 with the default DESeq2 parameters. The data were also processed using TOBIAS55 with default parameters to compute differential TF footprints.
snRNA-seq data analysis
snRNA-seq data were processed with Cell Ranger 7.1.0 with default parameters, ‘--expect-cells 60000’ option, and mm10-2020-A transcritome build. Data were then loaded into and further analyzed with Seurat version 4.0.4. After filtering cells with less than 300 or more than 5000 genes or with more than 10% mitochondrial RNA, 4,521-12,733 cells were used further per sample. These were normalized with Seurat using default parameters, the 2,000 most variable features were identified, the data were scale, PCA was applied with 30 top components and UMAP with 5 dimensions. Doublets were then identified using DoubletFinder80 with a prior of 10%, and 4,099–11,460 cells were retained.
The four samples were merged with defaulted parameters, and processed again as described above, neighbors were identified using the first 10 dimensions and clustered with shared nearest neighbor (SNN) modularity optimization as implemented in Seurat with resolution of 0.5. Differential expression was computed by Seurat FindMarkers function with default parameters and logfc.threshold set to 0.1 and min.pct = 0.1 comparing the cells from the WT and the Silc1−/− backgrounds. GO analysis was run using clusterProfiler 4.0 separately on the with logfc<(–0.2) in each cluster.81
Acknowledgments
We thank members of the Ulitsky laboratory for helpful discussions and comments on the manuscript; Alan Monziani for assistance with Gene Ontology analysis of the single-cell data, Liat Fellus Alyagor and Dana Hirsch for help with RNAscope experiments and analysis; and Dieter Chichung Lie, Yaniv Ziv, Ofer Yizhar, and Ivo Spiegel for helpful discussions. Sox11 loxP mice were a kind gift from Prof. Veronique Lefebvre (Leonard and Madlyn Abramson Pediatric Research Center, Philadelphia, PA, USA). This research was funded by the European Research Council Consolidator grant lncImpact (863589), the Kekst Family Institute for Medical Genetics, the Nella and Leon Benoziyo Center for Neurological Diseases, the Weizmann-Brazil Center for Research on Neurodegeneration, and the Gladys Monroy and Larry Marks Center for Brain Disorders. R.B.-T.P. is the incumbent of the Arlyn Imberman Research Fellow Chair.
Author contributions
R.B.-T.P. and I.U. conceived the study. R.B.-T.P. carried out all experiments with assistance from M. Tsoory with behavioral experiments and M. Tamasov, who performed the stereotaxic injections. I.U. analyzed the high-throughput sequencing data, and R.B.-T.P. analyzed all other data. R.B.-T.P. and I.U. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: September 23, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.113168.
Contributor Information
Rotem Ben-Tov Perry, Email: rotem.ben-tov-perry@weizmann.ac.il.
Igor Ulitsky, Email: igor.ulitsky@weizmann.ac.il.
Supplemental information
Data and code availability
-
•
All RNA-seq, scRNA-seq, ATAC-seq data generated in this study have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper also analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Frankish A., Diekhans M., Jungreis I., Lagarde J., Loveland J.E., Mudge J.M., Sisu C., Wright J.C., Armstrong J., Barnes I., et al. GENCODE 2021. Nucleic Acids Res. 2021;49:D916–D923. doi: 10.1093/nar/gkaa1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hezroni H., Perry R.B.T., Ulitsky I. Long Noncoding RNAs in Development and Regeneration of the Neural Lineage. Cold Spring Harb. Symp. Quant. Biol. 2019;84:165–177. doi: 10.1101/sqb.2019.84.039347. [DOI] [PubMed] [Google Scholar]
- 3.Barry G., Briggs J.A., Vanichkina D.P., Poth E.M., Beveridge N.J., Ratnu V.S., Nayler S.P., Nones K., Hu J., Bredy T.W., et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry. 2014;19:486–494. doi: 10.1038/mp.2013.45. [DOI] [PubMed] [Google Scholar]
- 4.Cui X., Zhang R., Yang Y., Wu E., Tang Y., Zhao Z., Li C., Yang L., Teng X., Ye Y., et al. Identification and characterization of long non-coding RNA Carip in modulating spatial learning and memory. Cell Rep. 2022;38 doi: 10.1016/j.celrep.2022.110398. [DOI] [PubMed] [Google Scholar]
- 5.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grinman E., Nakahata Y., Avchalumov Y., Espadas I., Swarnkar S., Yasuda R., Puthanveettil S.V. Activity-regulated synaptic targeting of lncRNA ADEPTR mediates structural plasticity by localizing Sptn1 and AnkB in dendrites. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abf0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei W., Zhao Q., Wang Z., Liau W.-S., Basic D., Ren H., Marshall P.R., Zajaczkowski E.L., Leighton L.J., Madugalle S.U., et al. ADRAM is an experience-dependent long noncoding RNA that drives fear extinction through a direct interaction with the chaperone protein 14-3-3. Cell Rep. 2022;38 doi: 10.1016/j.celrep.2022.110546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liau W.-S., Zhao Q., Bademosi A., Gormal R., Gong H., Marshall P.R., Periyakaruppiah A., Madugalle S.U., Zajackowski E.L., Leighton L.J., et al. Fear extinction is regulated by long noncoding RNA activity at the synapse. bioRxiv. 2022 doi: 10.1101/2022.03.30.486308. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perry R.B.-T., Hezroni H., Goldrich M.J., Ulitsky I. Regulation of Neuroregeneration by Long Noncoding RNAs. Mol. Cell. 2018;72:553–567.e5. doi: 10.1016/j.molcel.2018.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsang S.M., Oliemuller E., Howard B.A. Regulatory roles for SOX11 in development, stem cells and cancer. Semin. Cancer Biol. 2020;67:3–11. doi: 10.1016/j.semcancer.2020.06.015. [DOI] [PubMed] [Google Scholar]
- 11.Malysheva V., Mendoza-Parra M.A., Blum M., Gronemeyer H., Malysheva V., Gronemeyer H. Highly Dynamic Chromatin Interactions Drive Neurogenesis through Gene Regulatory Networks. bioRxiv. 2018 doi: 10.1101/303842. Preprint at. [DOI] [Google Scholar]
- 12.Bonev B., Mendelson Cohen N., Szabo Q., Fritsch L., Papadopoulos G.L., Lubling Y., Xu X., Lv X., Hugnot J.-P., Tanay A., Cavalli G. Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell. 2017;171:557–572.e24. doi: 10.1016/j.cell.2017.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Decaesteker B., Louwagie A., Loontiens S., De Vloed F., Roels J., Vanhauwaert S., De Brouwer S., Sanders E., Denecker G., D’haene E., et al. SOX11 is a lineage-dependency factor and master epigenetic regulator in neuroblastoma. bioRxiv. 2020 doi: 10.1101/2020.08.21.261131. Preprint at. [DOI] [Google Scholar]
- 14.Ye M., Ma J., Liu B., Liu X., Ma D., Dong K. Linc01105 acts as an oncogene in the development of neuroblastoma. Oncol. Rep. 2021;46:205–1538. doi: 10.3892/or.2021.8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bergsland M., Ramsköld D., Zaouter C., Klum S., Sandberg R., Muhr J. Sequentially acting Sox transcription factors in neural lineage development. Genes Dev. 2011;25:2453–2464. doi: 10.1101/gad.176008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sock E., Rettig S.D., Enderich J., Bösl M.R., Tamm E.R., Wegner M. Gene targeting reveals a widespread role for the high-mobility-group transcription factor Sox11 in tissue remodeling. Mol. Cell Biol. 2004;24:6635–6644. doi: 10.1128/MCB.24.15.6635-6644.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin L., Lee V.M., Wang Y., Lin J.S., Sock E., Wegner M., Lei L. Sox11 regulates survival and axonal growth of embryonic sensory neurons. Dev. Dyn. 2011;240:52–64. doi: 10.1002/dvdy.22489. [DOI] [PubMed] [Google Scholar]
- 18.Potzner M.R., Tsarovina K., Binder E., Penzo-Méndez A., Lefebvre V., Rohrer H., Wegner M., Sock E. Sequential requirement of Sox4 and Sox11 during development of the sympathetic nervous system. Development. 2010;137:775–784. doi: 10.1242/dev.042101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thein D.C., Thalhammer J.M., Hartwig A.C., Crenshaw E.B., 3rd, Lefebvre V., Wegner M., Sock E. The closely related transcription factors Sox4 and Sox11 function as survival factors during spinal cord development. J. Neurochem. 2010;115:131–141. doi: 10.1111/j.1471-4159.2010.06910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y., Lin L., Lai H., Parada L.F., Lei L. Transcription factor Sox11 is essential for both embryonic and adult neurogenesis. Dev. Dyn. 2013;242:638–653. doi: 10.1002/dvdy.23962. [DOI] [PubMed] [Google Scholar]
- 21.Jankowski M.P., Miller L., Koerber H.R. Increased expression of transcription factor SRY-box containing gene 11 (Sox11) enhances neurite growth by regulating neurotrophic factor responsiveness. Neuroscience. 2018;382:93–104. doi: 10.1016/j.neuroscience.2018.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haslinger A., Schwarz T.J., Covic M., Lie D.C. Expression of Sox11 in adult neurogenic niches suggests a stage-specific role in adult neurogenesis. Eur. J. Neurosci. 2009;29:2103–2114. doi: 10.1111/j.1460-9568.2009.06768.x. [DOI] [PubMed] [Google Scholar]
- 23.Mu L., Berti L., Masserdotti G., Covic M., Michaelidis T.M., Doberauer K., Merz K., Rehfeld F., Haslinger A., Wegner M., et al. SoxC transcription factors are required for neuronal differentiation in adult hippocampal neurogenesis. J. Neurosci. 2012;32:3067–3080. doi: 10.1523/JNEUROSCI.4679-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ming G.-L., Song H. Adult neurogenesis in the mammalian central nervous system. Annu. Rev. Neurosci. 2005;28:223–250. doi: 10.1146/annurev.neuro.28.051804.101459. [DOI] [PubMed] [Google Scholar]
- 25.Hochgerner H., Zeisel A., Lönnerberg P., Linnarsson S. Conserved properties of dentate gyrus neurogenesis across postnatal development revealed by single-cell RNA sequencing. Nat. Neurosci. 2018;21:290–299. doi: 10.1038/s41593-017-0056-2. [DOI] [PubMed] [Google Scholar]
- 26.Sun W., Park K.W., Choe J., Rhyu I.J., Kim I.H., Park S.K., Choi B., Choi S.-H., Park S.H., Kim H. Identification of novel electroconvulsive shock-induced and activity-dependent genes in the rat brain. Biochem. Biophys. Res. Commun. 2005;327:848–856. doi: 10.1016/j.bbrc.2004.12.050. [DOI] [PubMed] [Google Scholar]
- 27.Su Y., Shin J., Zhong C., Wang S., Roychowdhury P., Lim J., Kim D., Ming G.-L., Song H. Neuronal activity modifies the chromatin accessibility landscape in the adult brain. Nat. Neurosci. 2017;20:476–483. doi: 10.1038/nn.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Wittgenstein J., Zheng F., Wittmann M.-T., Balta E.-A., Ferrazzi F., Schäffner I., Häberle B.M., Valero-Aracama M.J., Koehl M., Miranda C.J., et al. Sox11 is an Activity-Regulated Gene with Dentate-Gyrus-Specific Expression Upon General Neural Activation. Cereb. Cortex. 2020;30:3731–3743. doi: 10.1093/cercor/bhz338. [DOI] [PubMed] [Google Scholar]
- 29.Guttman M., Donaghey J., Carey B.W., Garber M., Grenier J.K., Munson G., Young G., Lucas A.B., Ach R., Bruhn L., et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guttman M., Amit I., Garber M., French C., Lin M.F., Feldser D., Huarte M., Zuk O., Carey B.W., Cassady J.P., et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan X., Hu Z., Feng Y., Hu X., Yuan J., Zhao S.D., Zhang Y., Yang L., Shan W., He Q., et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell. 2015;28:529–540. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gil N., Ulitsky I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat. Rev. Genet. 2020;21:102–117. doi: 10.1038/s41576-019-0184-5. [DOI] [PubMed] [Google Scholar]
- 33.Hezroni H., Ben-Tov Perry R., Gil N., Degani N., Ulitsky I. Regulation of neuronal commitment in mouse embryonic stem cells by the Reno1/Bahcc1 locus. EMBO Rep. 2020;21 doi: 10.15252/embr.202051264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramos A.D., Andersen R.E., Liu S.J., Nowakowski T.J., Hong S.J., Gertz C., Salinas R.D., Zarabi H., Kriegstein A.R., Lim D.A. The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell. 2015;16:439–447. doi: 10.1016/j.stem.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sugino K., Clark E., Schulmann A., Shima Y., Wang L., Hunt D.L., Hooks B.M., Tränkner D., Chandrashekar J., Picard S., et al. Mapping the transcriptional diversity of genetically and anatomically defined cell populations in the mouse brain. Elife. 2019;8 doi: 10.7554/eLife.38619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bradley J.E., Ramirez G., Hagood J.S. Roles and regulation of Thy-1, a context-dependent modulator of cell phenotype. Biofactors. 2009;35:258–265. doi: 10.1002/biof.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Love M.I., Huber W., Anders S. Differential analysis of count data--the DESeq2 package. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernandez-Albert J., Lipinski M., Lopez-Cascales M.T., Rowley M.J., Martin-Gonzalez A.M., Del Blanco B., Corces V.G., Barco A. Immediate and deferred epigenomic signatures of in vivo neuronal activation in mouse hippocampus. Nat. Neurosci. 2019;22:1718–1730. doi: 10.1038/s41593-019-0476-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cembrowski M.S., Wang L., Sugino K., Shields B.C., Spruston N. Hipposeq: a comprehensive RNA-seq database of gene expression in hippocampal principal neurons. Elife. 2016;5 doi: 10.7554/eLife.14997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fredes F., Shigemoto R. The role of hippocampal mossy cells in novelty detection. Neurobiol. Learn. Mem. 2021;183 doi: 10.1016/j.nlm.2021.107486. [DOI] [PubMed] [Google Scholar]
- 41.Pitts M.W. Barnes Maze Procedure for Spatial Learning and Memory in Mice. Bio. Protoc. 2018;8 doi: 10.21769/BioProtoc.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gallo F.T., Katche C., Morici J.F., Medina J.H., Weisstaub N.V. Immediate Early Genes, Memory and Psychiatric Disorders: Focus on c-Fos, Egr1 and Arc. Front. Behav. Neurosci. 2018;12:79. doi: 10.3389/fnbeh.2018.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Struebing F.L., Wang J., Li Y., King R., Mistretta O.C., English A.W., Geisert E.E. Differential Expression of and mRNA Isoforms in the Injured and Regenerating Nervous Systems. Front. Mol. Neurosci. 2017;10:354. doi: 10.3389/fnmol.2017.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhattaram P., Penzo-Méndez A., Sock E., Colmenares C., Kaneko K.J., Vassilev A., Depamphilis M.L., Wegner M., Lefebvre V. Organogenesis relies on SoxC transcription factors for the survival of neural and mesenchymal progenitors. Nat. Commun. 2010;1:9. doi: 10.1038/ncomms1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee J.-S., Mendell J.T. Antisense-Mediated Transcript Knockdown Triggers Premature Transcription Termination. Mol. Cell. 2020;77:1044–1054.e3. doi: 10.1016/j.molcel.2019.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai F., Damle S.S., Ling K.K., Rigo F. Directed RNase H Cleavage of Nascent Transcripts Causes Transcription Termination. Mol. Cell. 2020;77:1032–1043.e4. doi: 10.1016/j.molcel.2019.12.029. [DOI] [PubMed] [Google Scholar]
- 47.Wang X., Zhang C., Szábo G., Sun Q.-Q. Distribution of CaMKIIα expression in the brain in vivo, studied by CaMKIIα-GFP mice. Brain Res. 2013;1518:9–25. doi: 10.1016/j.brainres.2013.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Animal Behavior (2022). (Academic Press).
- 49.Norsworthy M.W., Bei F., Kawaguchi R., Wang Q., Tran N.M., Li Y., Brommer B., Zhang Y., Wang C., Sanes J.R., et al. Sox11 Expression Promotes Regeneration of Some Retinal Ganglion Cell Types but Kills Others. Neuron. 2017;94:1112–1120.e4. doi: 10.1016/j.neuron.2017.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oliemuller E., Newman R., Tsang S.M., Foo S., Muirhead G., Noor F., Haider S., Aurrekoetxea-Rodríguez I., Vivanco M.d., Howard B.A. SOX11 promotes epithelial/mesenchymal hybrid state and alters tropism of invasive breast cancer cells. Elife. 2020;9 doi: 10.7554/eLife.58374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang K.-C., Bian M., Xia X., Madaan A., Sun C., Wang Q., Li L., Nahmou M., Noro T., Yokota S., et al. Posttranslational Modification of Sox11 Regulates RGC Survival and Axon Regeneration. eNeuro. 2021;8 doi: 10.1523/ENEURO.0358-20.2020. ENEURO.0358.20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abulaiti X., Wang A., Zhang H., Su H., Gao R., Chen J., Gao S., Li L. Disrupted mossy fiber connections from defective embryonic neurogenesis contribute to SOX11-associated schizophrenia. Cell. Mol. Life Sci. 2022;79:180. doi: 10.1007/s00018-022-04206-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bentsen M., Goymann P., Schultheis H., Klee K., Petrova A., Wiegandt R., Fust A., Preussner J., Kuenne C., Braun T., et al. ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation. Nat. Commun. 2020;11:4267. doi: 10.1038/s41467-020-18035-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang S., Su Y., Shinmyo Y., Islam S.M., Naser I.B., Ahmed G., Tamamaki N., Tanaka H. Draxin, a repulsive axon guidance protein, is involved in hippocampal development. Neurosci. Res. 2010;66:53–61. doi: 10.1016/j.neures.2009.09.1710. [DOI] [PubMed] [Google Scholar]
- 57.Shimozaki K., Clemenson G.D., Jr., Gage F.H. Paired related homeobox protein 1 is a regulator of stemness in adult neural stem/progenitor cells. J. Neurosci. 2013;33:4066–4075. doi: 10.1523/JNEUROSCI.4586-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Angata K., Long J.M., Bukalo O., Lee W., Dityatev A., Wynshaw-Boris A., Schachner M., Fukuda M., Marth J.D. Sialyltransferase ST8Sia-II assembles a subset of polysialic acid that directs hippocampal axonal targeting and promotes fear behavior. J. Biol. Chem. 2004;279:32603–32613. doi: 10.1074/jbc.M403429200. [DOI] [PubMed] [Google Scholar]
- 59.Taylor G.A., Rodriguiz R.M., Greene R.I., Daniell X., Henry S.C., Crooks K.R., Kotloski R., Tessarollo L., Phillips L.E., Wetsel W.C. Behavioral characterization of P311 knockout mice. Genes Brain Behav. 2008;7:786–795. doi: 10.1111/j.1601-183X.2008.00420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Corbo J.C., Deuel T.A., Long J.M., LaPorte P., Tsai E., Wynshaw-Boris A., Walsh C.A. Doublecortin Is Required in Mice for Lamination of the Hippocampus But Not the Neocortex. J. Neurosci. 2002;22:7548–7557. doi: 10.1523/JNEUROSCI.22-17-07548.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Soutschek M., Schratt G. Non-coding RNA in the wiring and remodeling of neural circuits. Neuron. 2023;111:2140–2154. doi: 10.1016/j.neuron.2023.04.031. [DOI] [PubMed] [Google Scholar]
- 62.Mercer T.R., Dinger M.E., Sunkin S.M., Mehler M.F., Mattick J.S. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. USA. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Butler A.A., Johnston D.R., Kaur S., Lubin F.D. Long noncoding RNA NEAT1 mediates neuronal histone methylation and age-related memory impairment. Sci. Signal. 2019;12 doi: 10.1126/scisignal.aaw9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Raveendra B.L., Swarnkar S., Avchalumov Y., Liu X.-A., Grinman E., Badal K., Reich A., Pascal B.D., Puthanveettil S.V. Long noncoding RNA GM12371 acts as a transcriptional regulator of synapse function. Proc. Natl. Acad. Sci. USA. 2018;115:E10197–E10205. doi: 10.1073/pnas.1722587115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gil N., Ulitsky I. Production of Spliced Long Noncoding RNAs Specifies Regions with Increased Enhancer Activity. Cell Syst. 2018;7:537–547.e3. doi: 10.1016/j.cels.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tan J.Y., Biasini A., Young R.S., Marques A.C. Splicing of enhancer-associated lincRNAs contributes to enhancer activity. Life Sci. Alliance. 2020;3 doi: 10.26508/lsa.202000663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tan J.Y., Marques A.C. The activity of human enhancers is modulated by the splicing of their associated lncRNAs. PLoS Comput. Biol. 2022;18 doi: 10.1371/journal.pcbi.1009722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Forner S., Kawauchi S., Balderrama-Gutierrez G., Kramár E.A., Matheos D.P., Phan J., Javonillo D.I., Tran K.M., Hingco E., da Cunha C., et al. Systematic phenotyping and characterization of the 5xFAD mouse model of Alzheimer’s disease. Sci. Data. 2021;8:270. doi: 10.1038/s41597-021-01054-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rishal I., Michaelevski I., Rozenbaum M., Shinder V., Medzihradszky K.F., Burlingame A.L., Fainzilber M. Axoplasm isolation from peripheral nerve. Dev. Neurobiol. 2010;70:126–133. doi: 10.1002/dneu.20755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Labun K., Montague T.G., Gagnon J.A., Thyme S.B., Valen E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016;44:W272–W276. doi: 10.1093/nar/gkw398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ballarino M., Cipriano A., Tita R., Santini T., Desideri F., Morlando M., Colantoni A., Carrieri C., Nicoletti C., Musarò A., et al. Deficiency in the nuclear long noncoding RNA causes myogenic defects and heart remodeling in mice. EMBO J. 2018;37 doi: 10.15252/embj.201899697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miyasaka Y., Uno Y., Yoshimi K., Kunihiro Y., Yoshimura T., Tanaka T., Ishikubo H., Hiraoka Y., Takemoto N., Tanaka T., et al. CLICK: one-step generation of conditional knockout mice. BMC Genom. 2018;19:318. doi: 10.1186/s12864-018-4713-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nunez J. Morris Water Maze Experiment. J. Vis. Exp. 2008 doi: 10.3791/897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sharma V., Cohen N., Sood R., Ounallah-Saad H., Gal-Ben-Ari S., Rosenblum K. Trace Fear Conditioning: Procedure for Assessing Complex Hippocampal Function in Mice. Bio. Protoc. 2018;8:e2475. doi: 10.21769/BioProtoc.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bahar Halpern K., Itzkovitz S. Single molecule approaches for quantifying transcription and degradation rates in intact mammalian tissues. Methods. 2016;98:134–142. doi: 10.1016/j.ymeth.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 79.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McGinnis C.S., Murrow L.M., Gartner Z.J. DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst. 2019;8:329–337.e4. doi: 10.1016/j.cels.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All RNA-seq, scRNA-seq, ATAC-seq data generated in this study have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper also analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.