Abstract
The treatment of advanced prostate cancer remains a formidable challenge due to the limited availability of effective treatment options. Therefore, it is imperative to identify promising druggable targets that provide substantial clinical benefits and to develop effective treatment strategies to overcome therapeutic resistance. Cyclosporin A (CsA) showed an anticancer effect on prostate cancer in cultured cell and xenograft models. E2F8 was identified as a master transcription factor that regulated a clinically significant CsA specific gene signature. The expression of E2F8 increased during prostate cancer progression and high levels of E2F8 expression are associated with a poor prognosis in patients with prostate cancer. MELK was identified as a crucial upstream regulator of E2F8 expression through the transcriptional regulatory network and Bayesian network analyses. Knockdown of E2F8 or MELK inhibited cell growth and colony formation in prostate cancer cells. High expression levels of E2F8 and androgen receptor (AR) are associated with a worse prognosis in patients with prostate cancer compared with low levels of both genes. The inhibition of E2F8 improved the response to AR blockade therapy. These results suggested that CsA has potential as an effective anticancer treatment for prostate cancer, while also revealing the oncogenic role of E2F8 and its association with clinical outcomes in prostate cancer. These results provided valuable insight into the development of therapeutic and diagnostic approaches for prostate cancer.
Keywords: prostate cancer, oncogene, E2F8, cyclosporin A, prognosis
Introduction
De novo or recurrent metastatic prostate cancers are initially amenable to androgen deprivation therapy (ADT). However, the majority of these lesions inevitably relapse and evolve into incurable and lethal castration-resistant prostate cancer (CRPC) (1–4). CRPC is extremely resistant to all types of currently available therapeutic regimens, posing a formidable clinical challenge (5,6). Therefore, it is imperative to identify promising druggable targets that yield significant clinical benefits and to develop effective treatment strategies that overcome therapeutic resistance.
Cyclosporin A (CsA) is a potent immunosuppressive agent that has been widely used in organ transplantation (7,8). Previous studies have demonstrated that CsA exerts antitumor or chemosensitizing activity against different types of cancer, including prostate cancer (9,10). In addition, CsA has been investigated in clinical trials for its potential to treat several cancers (11–13). However, the mechanism of antitumor action of CsA is poorly understood, particularly in the context of prostate cancer. Therefore, further research is needed to elucidate the mechanism of action of CsA in prostate cancer, which may lead to identifying potential targets for therapeutic intervention.
E2F8 is a member of the atypical E2F family that plays a crucial role in embryonic development (14). Emerging evidence demonstrates that E2F8 functions as an oncogene by mediating the hallmarks of cancer, including sustaining proliferative signaling and resisting cell death (15–17). However, it remains to be elucidated whether E2F8 is a promising therapeutic target for the treatment of prostate cancer. In addition, the signaling mechanism regulating E2F8 expression remains elusive.
The present study performed transcriptomic analyses to assess the mechanism of the antitumor action of CsA in prostate cancer. E2F8 was identified as a master transcription factor that induced oncogenic phenotypes and determined clinical outcome in prostate cancer. The results will provide insight into the development of E2F8-targeted therapy for the treatment of prostate cancer.
Materials and methods
Cell culture and reagents
PC-3 (cat. no. CRL-1435), LNCaP (cat. no. CRL-1740), DU145 (cat. no. HTB-81) and 22Rv1 (cat. no. CRL-2505) prostate cancer cell lines were purchased from the American Type Culture Collection. Cells were cultured in RPMI 1640 (PC-3, LNCaP and 22Rv1) or DMEM (DU145) containing 10% fetal bovine serum, penicillin (100 U/ml) and streptomycin (100 µg/ml). All cell culture reagents were obtained from HyClone (Cytiva). All other reagents not specified were supplied by MilliporeSigma. All cell lines tested negative for mycoplasma contamination using the Mycoplasma PCR Detection kit (Intron Biotechnology, Inc.). These cell lines have been authenticated in the three years using short tandem repeat analysis.
Tumor xenograft experiments
Male Balb/C nude mice (4–5 weeks old; 18–20 g; n=10) were purchased from Charles River Laboratories Japan, Inc. Mice were housed in laminar flow cabinets under specific pathogen-free condition (37°C; 12-h light/dark cycle; 60% relative humidity and free access to food and water). Mice were anesthetized by the inhalation of isoflurane (Terrell; Piramal Critical Care Inc.) in oxygen (2 l/min): Induction with 4% isoflurane for 2 min in an anesthetic induction chamber, followed by maintenance with 2% isoflurane for 1–2 min after transferring to a nose cone. The 22Rv1 cells (5×106 cells; 100 µl of cell suspension) were subcutaneously injected into the right flank of each mouse. The humane endpoints were when the largest tumor size was >20 mm in diameter. None of the mice reached the endpoints of the present study. When the tumor reached ~180 mm3, mice were randomly divided into two groups (five in each group) and intraperitoneally injected with 20 mg/kg CsA or DMSO, every other day for 14 days. After 14 days, mice were humanely sacrificed under overdosed isoflurane. Mice were placed into a chamber filled with vapor of the anesthetic isoflurane until respiration ceased (within 2 min) and the tumors were excised. The tumor volume was calculated using the following formula: V=(L × W2) × 0.5 (V, volume of tumor; L, length of tumor; W, width of tumor). All animal experiments were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of the Asan Institute for Life Sciences at the ASAN Medical Center, University of Ulsan College of Medicine, Seoul (approval no. 2021-13-234).
MTT assay
After cells were transfected with the short interfering (si)RNAs for 48 h, MTT assay was performed to assess cell growth according to the manufacturer's instructions (MilliporeSigma). The assay was quantitated by measuring the absorbance at 570 nm on a BioTek SynergyMx microplate reader (BioTek Instruments, Inc.).
Colony formation assay
PC-3 (2–5×103 cells/well), LNCaP (8×103 cells/well), DU145 (1–3×103 cells/well) and 22Rv1 (2–5×103 cells/well) cells were seeded into 6-well plates. Cells were treated with CsA (10 µM) or siRNAs for 9 days (once every 3 days). The cells were fixed with 3.7% paraformaldehyde and stained with 0.5% crystal violet (MilliporeSigma) for 15 min at room temperature. The number of colonies, defined as >50 cells/colony, was counted using ImageJ (1.8.0 172; National Institutes of Health).
Microarray experiment
PC-3 cells were treated with 10 µM CsA in 0.1% ethanol (vehicle) for 24 h. Microarray experiments were performed as we previously described (18,19). The microarray data are available through the Gene Expression Omnibus database (GSE109505; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE109505). Rigorous data preprocessing and single channel array normalization (SCAN) were performed and microarray probes were mapped to gene symbols as previously described (20–22). Of the 20,661 mapped genes, 17,629 protein-coding genes were selected for further analyses. The internal clusters were validated through hierarchical clustering and principal component analysis (PCA) (20,21).
Collection of public microarray data and analysis of the prostate cancer-specific transcriptional interactome
The GSE67157 and GSE109505 datasets were used to construct the PC-3 cell-specific interactome. Rigorous preprocessing, such as quality control testing, normalization and batch effect adjustment, was conducted as previously described (21,23,24). Of the common 11,506 genes from two datasets, 5,000 genes of high variance were selected for implementing the Algorithm for the Reconstruction of Gene Regulatory Networks (ARACNe) (20,21). The list of human transcription factors (TFs) was obtained from the Animal Transcription Factor Database 2.0 (AnimalTFDB 2.0) (23) and used for Master Regulator Analysis (MRA). ARACNe preprocessing and MRA-Fisher's exact test (FET) analysis were run in geWorkbench software version 2.6.0 (http://wiki.c2b2.columbia.edu/workbench/i-ndex.php/Home) as described in our previous reports (20,21,25,26). GSE3325 and GSE35988 data were used to assess the changes in gene expression profiles during prostate cancer progression. A more detailed description is provided in Fig. S1 and its legend.
Significance analysis of microarrays (SAM) and gene set enrichment analysis (GSEA)
SAM was used to identify differentially expressed genes (DEGs) from the GSE67157, GSE109505, GSE3325 and GSE35988 data (27). A tuning parameter, delta of 0.4, was optimized to give the cutoff for significance with the estimation of the false discovery rate (FDR) q-value threshold of 0.01. GSEA (Hallmark Gene Set from the Molecular Signature Database) was performed to obtain a biological interpretation of clinically significant CsA-specific DEGs (28). Universal concept signature scores were calculated for E2F8 target genes and concept signature enrichment analysis performed for deep functional assessment of the pathways. A more detailed description is given in Fig. S1 and its legend.
Reverse transcription (RT)PCR and western blotting
PC-3, LNCaP, DU145 and 22Rv1 cells were treated with CsA at the indicated concentrations for various times. Total RNA was extracted using the RNeasy Mini Kit (Qiagen GmbH) and reverse transcribed with SuperScriptIII First-Strand (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturers' instructions. RT-PCR and real-time PCR were performed using specific primers for E2F8 (16,21), MELK (29), β-actin (30) or 18S (31). The primer sequences are provided in Table SI. The thermocycling conditions for PCR were: 95°C for 5 min, followed by 31 cycles of 95°C for 45 sec, 51°C (MELK and β-actin) or 54°C (E2F8) for 45 sec, and 72°C for 45 sec. PCR products were separated by 1% agarose gel electrophoresis and visualized with SYBR Safe DNA Gel Stain (Invitrogen; Thermo Fisher Scientific, Inc.). Agarose gel electrophoresis images were acquired with Gel Documentation XR System (Bio-Rad Laboratories, Inc.). Real-time PCR was conducted using StepOne Real-Time PCR system (Thermo Fisher Scientific Inc.).
For western blotting, the crude extracts were prepared by incubation with RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 0.5 M EDTA) containing protease and phosphatase inhibitor cocktails (MilliporeSigma). The protein concentrations were determined by BCA assay kit (Thermo Fisher Scientific, Inc.). The samples (10–30 µg for each) were resolved in 7% (E2F8), 8% (MELK), or 10% (E2F1 and β-tubulin) SDS-PAGE gels and transferred onto NC membranes (Bio-Rad Laboratories, Inc.). The membranes were blocked using 5% w/v skimmed milk (BD Difco; BD Biosciences) in Tris-buffered saline containing 0.1% Tween-20 (TBST) for 1 h at room temperature. The membranes were probed with the anti-E2F8 (1:2,000; cat. no. A303-039A; Bethyl Laboratories, Inc.), anti-MLEK (1:1,000; cat. no. 2274S; Cell Signaling Technology, Inc.), anti-E2F1 (1:500; cat. no. sc-193; Santa Cruz, Inc.), and anti-β-tubulin (1:5,000; cat. no. T4026; MilliporeSigma, Inc.) antibodies for 1 h at room temperature. After washing three times with TBST, the membranes were incubated with a goat anti-Rabbit IgG-HRP antibody (1:5,000; cat. no. A120-101P, Bethyl Laboratories, Inc.) or a goat anti-Mouse IgG-HRP antibody (1:5,000; cat. no. A90-116P, Bethyl Laboratories, Inc.) for 1 h at room temperature. The signals were determined by the enhanced chemiluminescence reaction (ECL; Amersham; Cytiva). X-ray films were scanned and analyzed using EPSON Scan Software (EPSON Expression 11000XL, Seiko Epson Corporation). The data shown are representative of at least four independent experiments. Full scan images are shown in Appendix S1.
siRNA transfection
Cells were transfected with 50 nM siControl (21) or siRNAs against E2F8 [siE2F8-1 and siE2F8-2 (21)] or MELK [siMELK-1 (29) and siMELK-2 (32)] for 48 h at 37°C using Lipofectamine® RNAiMAX reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The siRNAs were obtained from Genolution Pharmaceuticals Inc. (Seoul, Korea). The siRNA sequences are provided in Table SII. The time interval between transfection and subsequent experiments was 48 h or 72 h for MTT assay, 9 days for colony formation assay (see also Figure legends).
Bayesian network analysis
The Cancer Genome Atlas (TCGA)-Prostate Adenocarcinoma (PRAD) gene expression data were obtained from Xena Functional Genomics Explorer (https://tcga.xenahubs.net/download/TCGA.PRAD.sampleMap/HiSeqV2.gz). The E2F8 gene signature, which was inferred in MRA-FET analysis, was extracted and the continuous value of expression levels in each gene was discretized to equal-width bins (8 bins) using the unsupervised discretization method available in the Information-Theoretic Measures (Infotheo) R version 4.1.1 (http://www.R-project.org/) (11). The fast greedy equivalence search (FGES)-discrete algorithm was used to identify E2F8-interacting genes (33). A more detailed description is given in Fig. S1 and its legend.
Coexpression analysis
The gene expression data of TCGA-primary tumor samples were downloaded using the TCGA Biolinks package (ver. 2.14.1, http://bioconductor.org/packages/release/bioc/html/TCGAbiolinks.html). Before normalization, duplicated FFPE samples were removed. Gene reads were normalized in counts per million (CPM) using the edgeR package (ver. 3.28.1, http://bioconductor.org/packages/release/bioc/html/edgeR.html) and log2 transformation (CPM+1). The Ensembl gene IDs were mapped into HGNC symbols using the biomaRt package (ver. 2.42.1, http://bioconductor.org/packages/release/bioc/html/biomaRt.html) and the expression data of androgen receptor (AR), MELK and E2F8 extracted for coexpression analysis.
The coordinated TF activities associated with therapeutic response
TCGA-PRAD RNA-seq raw bam files (n=554) were downloaded from NCI Genomic Data Commons (GDC) data portal (https://portal.gdc.cancer.gov/). Patient response data for chemotherapy or radiotherapy were provided by Xena Functional Genomics Explorer (https://xenabrowser.net/hub/) GDC Hub (34). When the therapeutic response data were mapped to the available PRAD RNA-seq samples, 313 patients were identified as having a complete response (CR), 34 patients as partial response (PR), 29 patients as progressive disease (PD) and 27 patients as stable disease (SD). Integrated System for Motif Activity Response Analysis (ISMARA, http://ismara.unibas.ch/mara/) was used to infer regulatory networks and sample-specific TF motif activities from gene expression data (35). ISMARA allowed the mapping of transcriptomic profiles to a lower-dimensional inferred TF activity space, largely preserving the relationships between samples. The TF activities driving expression changes were calculated and the TF activity differences between therapeutic response and non-response patients were examined. The TF activities significantly associated with therapy response were defined by P-value <0.05 and absolute mean activity difference >0.008.
Statistical analysis
The Kaplan-Meier survival curve and log-rank test were used to determine overall survival curves as previously described (36,37). Using median gene expression values from cBioPortal transcriptomic data as a bifurcating point, the samples were divided into high- and low-expression groups and the survival rates were compared between the two groups. The Cox proportional hazards model was applied to estimate hazard ratios (HRs) and 95% confidence intervals (CIs). Pearson correlation was used to determine the correlations of the expression levels among E2F8, MELK, or AR from the TCGA-PRAD and cBioPortal data. A comparison of means among experimental groups was performed using one-way ANOVA followed by Bonferroni's multiple comparison test. P<0.05 was considered to indicate a statistically significant difference.
Results
CsA inhibits prostate cancer growth in vitro and in vivo
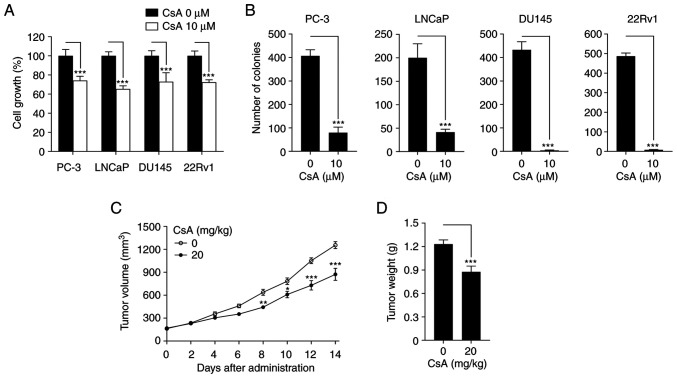
To determine the antitumor activity of CsA in prostate cancer, MTT and colony formation assays were first performed. CsA suppressed cell growth and colony formation in PC-3, LNCaP, DU145 and 22Rv1 prostate cancer cells (Fig. 1A and B). In addition, CsA inhibited tumor volume and weight in 22Rv1 cell xenograft mouse model (Fig. 1C and D). These results demonstrate that CsA has an anticancer activity against prostate cancer.
Figure 1.
CsA inhibits prostate cancer growth. (A) PC-3, LNCaP, DU145 and 22Rv1 cells were treated with 10 µM CsA for 48 h prior to MTT analysis. Cell growth was expressed as a relative value compared with that of vehicle-treated group which was set to 100%. The data were expressed as the mean ± SEM (n=4) ***P<0.005. (B) Each cell was treated with 10 µM CsA for 9 days (once every 3 days), after which colony formation was assessed. The data were expressed as the mean ± SEM (n=3) ***P<0.005. (C) Tumor volumes were recorded twice (every other day) for 14 days. The figures show the mean ± SEM (n=5) *P<0.05; **P<0.01; ***P<0.005. (D) At 14 days after xenograft implantation, the mice were sacrificed to determine tumor weight. The figures show the mean ± SEM (n=5) ***P<0.005. CsA, cyclosporin A.
To understand the antitumor mechanism of action of CsA, microarray experiments we performed using CsA-treated PC-3 cells (GSE109505). Hierarchical clustering analysis and PCA showed that CsA-treated and untreated cells were clustered into two discrete groups (Figs. S1, S2A and S2B), indicating that CsA induces a distinct change in gene expression profiles. Analysis using SAM found that CsA significantly affected the expression levels of 3,319 genes (Fig. S2C). Among these 3,319 differentially expressed genes (DEGs), 2,500 (593+1,907) genes were downregulated in CsA-treated cells, whereas 819 (278+541) genes were upregulated in CsA-treated PC-3 cells (Fig. S2D).
To assess the clinical significance of the 3,319 DEGs, they were compared with the DEGs derived from the transcriptomic data of patients with prostate cancer (GSE3325). First 3,654 DEGs between metastatic and nonmetastatic (benign or primary) cancer were obtained: 1,398 genes were upregulated and 2,256 genes were downregulated in patients with metastatic cancer (Fig. S2D; leftmost circles). It was identified that 871 (593+278) DEGs showed anti-similarity (or inverse correlation) between the 3,319 DEGs from GSE109505 and the 3,654 DEGs from GSE3325 (Fig. S2C and D). These 871 anti-similar DEGs were termed the ‘clinically significant CsA-induced gene expression (CCI) signature’. GSEA revealed that this CCI signature is associated with 19 hallmark pathways (Fig. S2E). In particular, CsA activates cell death-related pathways and inhibits cell cycle-related pathways, providing the mechanistic explanation for the anticancer activity of CsA (Fig. 1).
E2F8 is identified as a master regulator that is associated with a poor prognosis in patients with prostate cancer
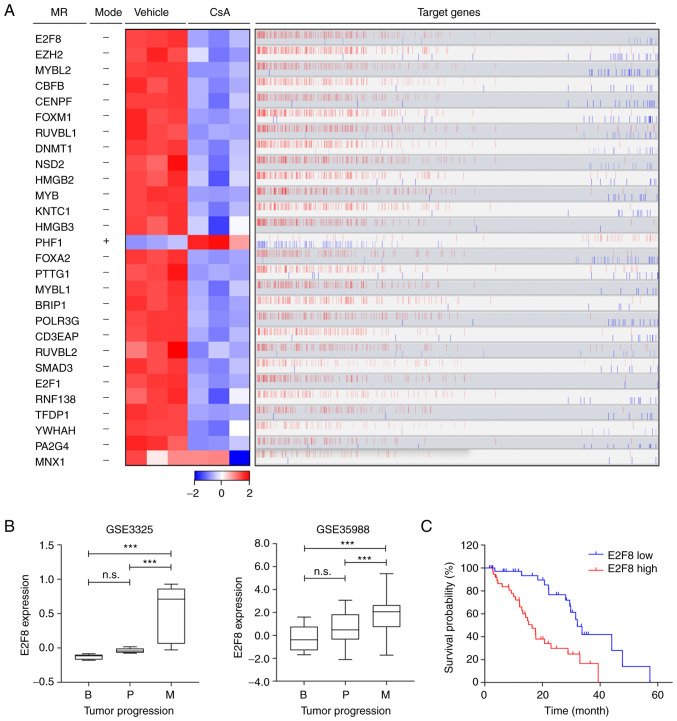
To elucidate the molecular mechanism by which CsA regulates the CCI signature, the prostate cancer cell-specific transcriptional interactome was analyzed using ARACNe and MRA algorithms. These identified 28 transcription factors as master regulators (MRs) that control the CCI signature (Fig. 2A and Table I): 27 MRs were markedly downregulated in CsA-treated PC-3 cells, whereas only one MR (PHF1) was upregulated (Fig. 2A; heatmap). Overall, the expression patterns of these MRs correlated with those of their target genes (Fig. 2A; red bars). Based on ‘markers in the intersection set’ (Table I), E2F8 was identified as a top MR that affected the majority of genes belonging to the CCI signature (216 target genes out of 871; Figs. 2A and S3). These 216 target genes of E2F8 were mainly associated with cell cycle or proliferation pathways (Fig. S3). CsA markedly reduced E2F8 expression (Fig. 2A), which correlated positively with the expression levels of the most target genes (bar plot in Fig. 2A). These results suggested that E2F8 mainly acts as a transcriptional activator.
Figure 2.
E2F8 is identified as a master regulator in prostate cancer cells. (A) A total of 28 TFs are identified as MRs that regulate the CCI signature. The heatmap shows the changes in gene expression levels of MRs between vehicle- and CsA-treated PC-3 cells. The mode explains whether CsA positively (+) or negatively (−) affects the expression of MRs. The bar graph shows positively (red) or negatively (blue) correlated target genes of the MRs (Spearman's correlation). The data were visualized using R 3.6.3 software (https://cran.r-project.org/src/base/R-3) (left) and geWorkbench software 2.6.0 (http://wiki.c2b2.columbia.edu/workbench/i-ndex.php/Home) (right). (B) The expression levels of E2F8 in patients with prostate cancer (GSE3325 and GSE35988) were represented in the box plots. The x-axis indicates three different stages of prostate cancer and y-axis indicates the normalized expression levels. ***P<0.005; n.s., not significant. (C) Survival curve for patients with prostate cancer based on the expression levels of E2F8 from cBioPortal transcriptomic data. CsA, cyclosporin A; TFs, transcription factors; CCI, clinically significant CsA-induced gene expression; MRs, master regulators; B, benign prostate hyperplasia; P, primary tumor; M, metastatic tumor.
Table I.
List of 28 MRs that control the CCI signature.
| GeneID | Symbol | Description | Markers in intersection seta | FET P-valueb | Markers in regulonc | Moded | Fold Change | q-value |
|---|---|---|---|---|---|---|---|---|
| 79733 | E2F8 | E2F transcription factor 8 | 216 | 4.11×10−56 | 439 | - | 0.71 | 1.00×10−16 |
| 2146 | EZH2 | enhancer of zeste 2 polycomb repressive complex 2 subunit | 198 | 1.09×10−29 | 509 | - | 0.78 | 1.00×10−16 |
| 4605 | MYBL2 | MYB proto-oncogene like 2 | 196 | 9.37×10−44 | 434 | - | 0.73 | 1.00×10−16 |
| 865 | CBFB | core-binding factor beta subunit | 190 | 4.32×10−30 | 498 | - | 0.82 | 1.00×10−16 |
| 1063 | CENPF | centromere protein F | 183 | 9.56×10−36 | 429 | - | 0.66 | 1.00×10−16 |
| 2305 | FOXM1 | forkhead box M1 | 183 | 1.63×10−31 | 445 | - | 0.73 | 1.00×10−16 |
| 8607 | RUVBL1 | RuvB like AAA ATPase 1 | 173 | 7.29×10−7 | 656 | - | 0.83 | 1.00×10−16 |
| 1786 | DNMT1 | DNA methyltransferase 1 | 170 | 2.87×10−26 | 440 | - | 0.74 | 1.00×10−16 |
| 7468 | NSD2 | nuclear receptor binding SET domain protein 2 | 162 | 1.39×10−16 | 512 | - | 0.83 | 1.00×10−16 |
| 3148 | HMGB2 | high mobility group box 2 | 161 | 5.27×10−23 | 422 | - | 0.66 | 1.00×10−16 |
| 4602 | MYB | MYB proto-oncogene, transcription factor | 158 | 4.24×10−16 | 487 | - | 0.72 | 1.00×10−16 |
| 9735 | KNTC1 | kinetochore associated 1 | 151 | 2.48×10−36 | 319 | - | 0.75 | 1.00×10−16 |
| 3149 | HMGB3 | high mobility group box 3 | 135 | 3.35×10−23 | 335 | - | 0.83 | 1.00×10−16 |
| 5252 | PHF1 | PHD finger protein 1 | 135 | 2.33×10−11 | 394 | + | 1.16 | 1.07×10−5 |
| 3170 | FOXA2 | forkhead box A2 | 132 | 9.60×10−6 | 513 | - | 0.76 | 1.00×10−16 |
| 9232 | PTTG1 | pituitary tumor-transforming 1 | 131 | 3.00×10−9 | 395 | - | 0.73 | 1.00×10−16 |
| 4603 | MYBL1 | MYB proto-oncogene like 1 | 127 | 8.34×10−31 | 272 | - | 0.75 | 1.00×10−16 |
| 83990 | BRIP1 | BRCA1 interacting protein C-terminal helicase 1 | 126 | 1.10×10−6 | 479 | - | 0.73 | 1.00×10−16 |
| 10622 | POLR3G | RNA polymerase III subunit G | 123 | 4.26×10−7 | 457 | - | 0.82 | 1.00×10−16 |
| 10849 | CD3EAP | CD3e molecule associated protein | 121 | 4.19×10−15 | 333 | - | 0.84 | 1.00×10−16 |
| 10856 | RUVBL2 | RuvB like AAA ATPase 2 | 120 | 5.93×10−7 | 403 | - | 0.89 | 1.06×10−6 |
| 4088 | SMAD3 | SMAD family member 3 | 116 | 2.87×10−7 | 407 | - | 0.82 | 1.00×10−16 |
| 1869 | E2F1 | E2F transcription factor 1 | 109 | 3.46×10−15 | 304 | - | 0.82 | 1.00×10−16 |
| 51444 | RNF138 | ring finger protein 138 | 98 | 6.15×10−7 | 343 | - | 0.80 | 1.00×10−16 |
| 7027 | TFDP1 | transcription factor Dp-1 | 98 | 3.28×10−12 | 291 | - | 0.77 | 1.00×10−16 |
| 7533 | YWHAH | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein eta | 89 | 7.85×10−9 | 277 | - | 0.83 | 1.00×10−16 |
| 5036 | PA2G4 | proliferation-associated 2G4 | 80 | 3.40×10−7 | 254 | - | 0.84 | 1.00×10−16 |
| 3110 | MNX1 | motor neuron and pancreas homeobox 1 | 49 | 1.01×10−7 | 128 | - | 0.77 | 1.00×10−16 |
Makers in intersection set, the number of markers found in the intersection of the signature and the regulon of the candidate MR.
FET P-value, the P-value from Fisher's exact test. It shows how much significantly the marker (gene) belongs to the signature set and the regulon of the MR.
Markers in regulon, the number of markers (genes) found to be first neighbor of the master regulator in the loaded network.
Mode, plus or minus mode means that the MR expression positively or negatively correlates with CsA. MR, master regulator; FET, Fisher's exact test; CsA, cyclosporin A.
To assess the clinical relevance of the MRs, the data of patients with prostate cancer (GSE3325 and GSE35988) were analyzed. Among these 28 MRs, the expression levels of 10 MRs were markedly upregulated during prostate cancer progression (E2F8 in Fig. 2B and other nine MRs in S4). Kaplan-Meier analysis showed that high expression levels of these 10 MRs are associated with worse prognosis in patients with prostate cancer (E2F8 in Fig. 2C and other nine MRs in S5). In particular, E2F8 showed the highest hazard ratio (HR=3.028; P=0.0002; Figs. 2C and S5B). These results indicated that E2F8 acts as a clinically significant MR crucial for governing a large portion of the CCI signature. In addition, the findings suggested that E2F8 serves as a useful prognosis marker for prostate cancer.
E2F8 serves as a therapeutic target for prostate cancer
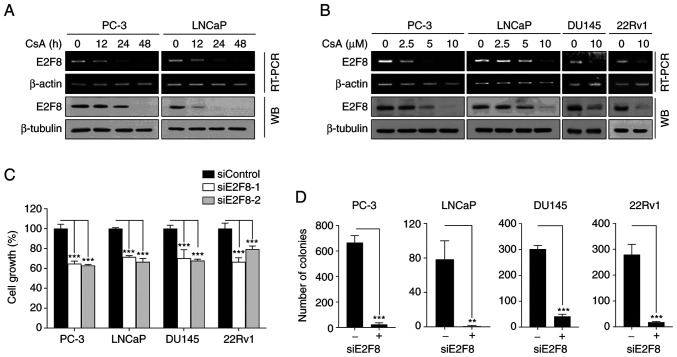
To confirm whether CsA suppressed E2F8 expression, RT-PCR, real-time PCR and western blot analysis we performed in various types of prostate cancer cells. CsA inhibited E2F8 mRNA (Figs. 3A and B and S6A) and protein expression (Fig. 3A and B) in a time- and concentration-dependent manner. As PC-3 and DU145 cells do not express functional AR, the results suggested that CsA suppressed E2F8 expression through an AR-independent mechanism.
Figure 3.
CsA downregulates E2F8 expression in prostate cancer cells. (A) PC-3 and LNCaP cells were treated with 10 µM CsA for the indicated times prior to RT-PCR and western blot analysis. (B) PC-3, LNCaP, DU145 and 22Rv1 cells were treated with the indicated concentrations of CsA for 48 h prior to RT-PCR and western blot analysis. (C) Each cell was transfected with 50 nM siE2F8-1 or siE2F8-2 for 48 h prior to MTT assay. Cell viability was expressed as a relative value compared with that of siControl which was set to 100%. The data were expressed as the mean ± SEM (n=6) ***P<0.005. (D) Each cell was transfected with 50 nM siE2F8-1 for 9 days (once every 3 days), after which colony formation assessed. The data were expressed as the mean ± SEM (n=3) **P<0.01, ***P<0.005. CsA, cyclosporin A; RT-PCR, reverse transcription PCR; si, short interfering.
To assess the role of E2F8 in prostate cancer, the effect of siRNAs against E2F8 (siE2F8-1 and −2) were examined using MTT and colony formation assays. The siE2F8s effectively inhibited E2F8 expression in all tested cell lines (Fig. S6B). It was found that E2F8 knockdown suppressed cell growth and colony formation in all tested cells (Fig. 3C and D). These results suggested that E2F8 represents an attractive therapeutic target for prostate cancer.
MELK is crucial for regulating E2F8 expression in prostate cancer
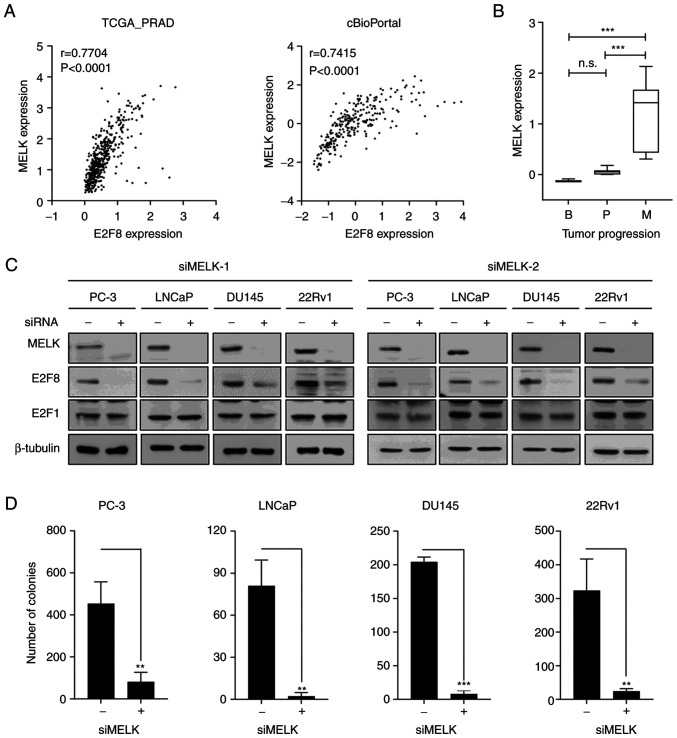
To elucidate the molecular mechanism by which CsA suppresses E2F8 expression, Bayesian network analysis was employed, which is an effective approach to model causal relationships between observed biological data and gene expression data (38,39). The FGES-discrete algorithm identified MELK as an E2F8-interacting gene. Analysis of TCGA-PRAD and cBioPortal transcriptomic data showed the strong positive correlation between E2F8 and MELK expression in patients with prostate cancer (r=0.7704, P<0.0001 and r=0.7415, P<0.0001, respectively; Fig. 4A). In addition, analysis of the GSE3325 data showed that MELK is markedly upregulated during prostate cancer progression (Fig. 4B). Whether MELK causally regulates E2F8 expression was then examined. The siRNA-based knockdown of MELK (siMELK-1 and −2) markedly reduced E2F8 expression in all tested cells (Fig. 4C). In contrast to E2F8, siMELKs did not affect the expression of E2F1, one of the identified MRs (Fig. 2 and Table I), which plays a crucial role in prostate cancer growth (20,40). Altogether, these results indicate that MELK is a crucial upstream signaling molecule for controlling E2F8 expression in prostate cancer cells.
Figure 4.
MELK is crucial for regulating E2F8 expression in prostate cancer cells. (A) Pearson correlation analysis showed coexpression correlation between MELK and E2F8 in TCGA-PRAD and cBioPortal transcriptomic data. (B) MELK expression levels in patients with prostate cancer (GSE3325) were represented in the box plots. ***P<0.005, n.s., not significant. (C) Each cell was transfected with 50 nM siMELK-1 or siMELK-2 for 48 h prior to western blot analysis. (D) Each cell was transfected with 50 nM siMELK-1 for 9 days (once every 3 days), after which colony formation assessed. The data were expressed as the mean ± SEM (n=3). **P<0.01; ***P<0.005. TCGA-PRAD, The Cancer Genome Atlas-Prostate Adenocarcinoma; r, Pearson correlation coefficient; si, short interfering.
To determine whether CsA suppresses MELK expression, RT-PCR, real-time PCR and western blot analysis were performed in various prostate cancer cells. CsA inhibited MELK expression at the protein level (Fig. S7A), but not at the mRNA level (Fig. S7A and B), suggesting that CsA regulated MELK at the posttranscriptional or translational level. To assess the functional importance of MELK in prostate cancer cells, whether forced suppression of MELK expression affects prostate cancer growth was investigated using MTT and colony formation assays. The siMELKs inhibited cell growth and colony formation in various prostate cancer cells (Figs. 4D, S7C and S7D). Altogether, these results indicate that the MELK-E2F8 signaling axis plays a crucial role in prostate cancer biology.
E2F8-targeted therapy demonstrates clinical significance in prostate cancer
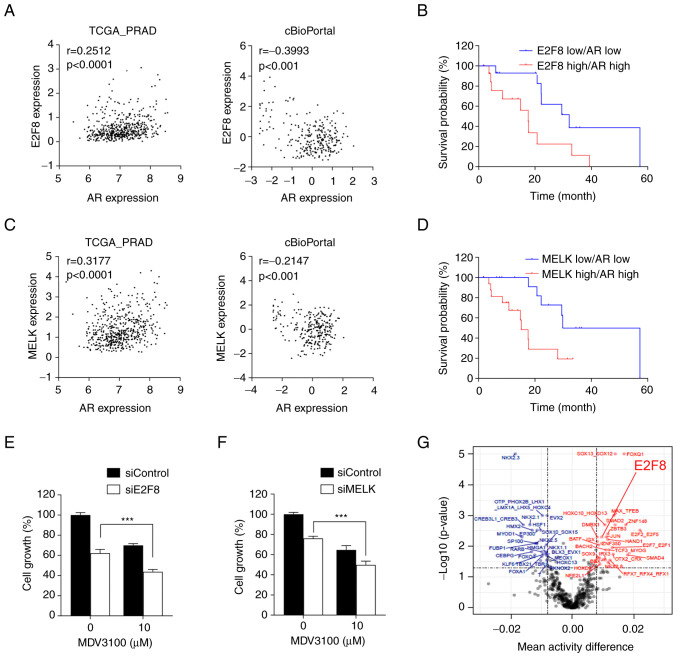
AR is a crucial driver of CRPC progression and its expression is frequently upregulated during prostate cancer progression (41–44). The present study showed that E2F8 expression increases during prostate cancer progression and exerts oncogenic activity (Figs. 2 and 3). Based on these observations, the interplay between AR and E2F8 signaling was examined. The analysis of TCGA-PRAD and cBioPortal transcriptomic data found that there was no significant coexpression correlation between E2F8 and AR or between MELK and AR expression levels (Fig. 5A and C).
Figure 5.
Clinical significance of E2F8 in prostate cancer. (A and C) Coexpression correlation between E2F8 and AR or between MELK and AR in TCGA-PRAD and cBioPortal transcriptomic data. (B and D) Survival curve for patients with prostate cancer based on the expression levels of E2F8 and AR or those of MELK and AR from cBioPortal data. (E) LNCaP cells were treated with 50 nM siE2F8-1, 10 µM MDV300, or both for 72 h prior to MTT assay. The data were expressed as the mean ± SEM (n=4-6). ***P<0.005. (F) LNCaP cells were treated with 50 nM siMELK-1, 10 µM MDV300, or both for 72 h prior to MTT assay. The data were expressed as the mean ± SEM (n=4-6) ***P<0.005. (G) Inferred TF motif activity differences between patients with CR (highlighted in blue) and SD (highlighted in red). The x-axis denotes the mean TF activity differences and the y-axis indicates-log10 (P-values). The vertical dotted line represents an absolute mean TF activity difference of 0.008 and the horizontal dotted line means the P-value 0.05 for significant TFs. AR, androgen receptor; TCGA-PRAD, The Cancer Genome Atlas-Prostate Adenocarcinoma; r, Pearson correlation coefficient; TF, transcription factor; si, short interfering; TCGA-PRAD, The Cancer Genome Atlas-Prostate Adenocarcinoma.
Nonetheless, Kaplan-Meier analysis showed that overall survival was greatly reduced in patients with prostate cancer with high levels of both E2F8 and AR expression, compared with those with low levels of both genes (HR=4.33, P=0.0019; Fig. 5B). Similar results were obtained from patients with prostate cancer with high levels of both MELK and AR expression, compared with those with low levels of both genes (HR=3.3, P=0.0041; Fig. 5D). Altogether, these results suggest that the MELK-E2F8 axis and AR signaling act independently, but their additive effects worsen the clinical outcomes of prostate cancer. Hence. simultaneous inhibition of E2F8 and AR could be a promising therapeutic strategy for patients with prostate cancer with concurrent overexpression of these molecules.
To investigate the translatability of the findings, LNCaP cells (AR-positive, AR-V7-negative cells) were treated with siE2F8 and the AR antagonist MDV3100. The treatment with siE2F8 increased the sensitivity of prostate cancer cells to MDV3100 (Fig. 5E), suggesting a potential therapeutic strategy for improving the efficacy anti-androgen therapy. In addition, siE2F8 had a similar effect as siMELK in enhancing the efficacy of MDV3100 (Fig. 5F). Finally, the ISMARA algorithm showed that E2F8 activity is markedly higher in patients with SD than in those with CR (Fig. 5G), suggesting that E2F8 is a predictive marker for therapeutic response in patients with prostate cancer.
Discussion
The present study described five main findings: i) CsA exhibits antitumor properties against prostate cancer in cultured cell and xenograft models; ii) E2F8 is a master regulator in controlling the CCI signature; iii) E2F8 is upregulated during tumor progression and the high expression levels of E2F8 are associated with a poor prognosis in patients with prostate cancer; iv) MELK is a crucial upstream regulator of E2F8 expression; and v) the inhibition of E2F8 or MELK sensitizes prostate cancer cells to AR blockade therapy. Considering that CsA has been investigated in clinical trials (11–13), the results provided a promising basis for future research aimed at the development of prostate cancer therapy and prognosis in clinic.
The present study demonstrated that CsA reduces E2F8 expression levels, although the pattern of reduction differs among prostate cancer cell lines. CsA reduced E2F8 protein expression in LNCaP cells faster than in PC-3 cells (Fig. 3A). These findings suggest that the cellular rewiring determining E2F8 expression levels is cell type- or context-dependent. Furthermore, the mechanism underlying E2F8 overexpression, which is commonly observed in patients with prostate cancer, may vary among the individuals. Further research into the various regulatory mechanisms of E2F8 expression can contribute to an improved understanding of prostate cancer and the development of diagnostic and therapeutic strategies toward precision oncology.
Accumulating evidence indicates that MELK is upregulated in various types of human cancer, including advanced prostate cancer (45) and acts as an oncogenic driver, making it a potential therapeutic target (46). In line with these findings, the present study showed that knockdown of MELK suppressed cell growth and colony formation in prostate cancer cells. In addition, it found that high expression levels of both MELK and AR are associated with a poor prognosis in patients with prostate cancer, similar to those of both E2F8 and AR. In addition, siE2F8 had a similar effect to siMELK in enhancing the efficacy of MDV3100. Collectively, these findings suggested that the MELK-E2F8 signaling axis has oncogenic potential and cooperates with AR signaling to promote prostate carcinogenesis.
Survival analysis revealed that the combined high expression of both E2F8 and AR predicted a poor prognosis in patients with prostate cancer. However, there was no significant coexpression correlation between E2F8 and AR. In addition, when the overlap between the 216 target genes of E2F8 and the 149 target genes of AR (47) were compared, it was found that only six genes (CDK1, CHAF1A, IGF2BP3, MCM2, RPA3 and STIL) were commonly shared, suggesting that both TFs act independently in prostate cancer. These results suggested that in significant portion of patient with prostate cancer, E2F8 and AR signaling acted independently. However, in certain cases where both genes are concurrently overexpressed, their combined effects exacerbate the clinical outcomes of prostate cancer. Exploring further investigations into the interaction between E2F8 and AR signaling may provide to new avenues for prostate cancer research. In particular, considering the findings that CsA regulates E2F8 expression in an AR-independent manner, it is plausible to investigate the possibility that E2F8 mediates the non-AR-driven evolution of CRPC.
In conclusion, the present study demonstrated that CsA suppressed MELK-mediated E2F8 expression, leading to its antitumor activity against prostate cancer. High expression level of E2F8 was associated with a poor prognosis in patients with prostate cancer and inhibiting E2F8 enhanced AR blockade therapy. Therefore, CsA may serve as an effective anticancer agent for prostate cancer, while E2F8 represents a promising target for diagnosis and treatment of this disease.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by a grant from the National Research Foundation of Korea grant funded by the Korea government (MSIT; grant nos. 2018R1A4A1023822 and 2020R1A2C1102574), a grant from the Education and Research Encouragement Fund of Seoul National University Hospital (grant no. 0320210340).
Availability of data and materials
All data supporting the findings of this study are included in this published article. All additional information are available from the corresponding author on reasonable request.
Authors' contributions
SL, JNC, and JHJ designed the present study. DYL, SL, YSK, SP, SMB, EAC performed the experiments. DYL, SL, JNC, and JHJ confirm the authenticity of all the raw data. JNC, EJP, HHP, SYK and IS contributed to data analysis and interpretation. SL, DYL, SP, SYK, JNC, and JHJ wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All animal experiments were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of the Asan Institute for Life Sciences at the ASAN Medical Center, University of Ulsan College of Medicine, Seoul (approval no. 2021-13-234).
Patient consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
References
- 1.Carceles-Cordon M, Kelly WK, Gomella L, Knudsen KE, Rodriguez-Bravo V, Domingo-Domenech J. Cellular rewiring in lethal prostate cancer: The architect of drug resistance. Nat Rev Urol. 2020;17:292–307. doi: 10.1038/s41585-020-0298-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Attard G, Parker C, Eeles RA, Schröder F, Tomlins SA, Tannock I, Drake CG, de Bono JS. Prostate cancer. Lancet. 2016;387:70–82. doi: 10.1016/S0140-6736(14)61947-4. [DOI] [PubMed] [Google Scholar]
- 3.Litwin MS, Tan HJ. The diagnosis and treatment of prostate cancer: A review. JAMA. 2017;317:2532–2542. doi: 10.1001/jama.2017.7248. [DOI] [PubMed] [Google Scholar]
- 4.Sandhu S, Moore CM, Chiong E, Beltran H, Bristow RG, Williams SG. Prostate cancer. Lancet. 2021;398:1075–1090. doi: 10.1016/S0140-6736(21)00950-8. [DOI] [PubMed] [Google Scholar]
- 5.Yap TA, Smith AD, Ferraldeschi R, Al-Lazikani B, Workman P, de Bono JS. Drug discovery in advanced prostate cancer: Translating biology into therapy. Nat Rev Drug Discov. 2016;15:699–718. doi: 10.1038/nrd.2016.120. [DOI] [PubMed] [Google Scholar]
- 6.Ku SY, Gleave ME, Beltran H. Towards precision oncology in advanced prostate cancer. Nat Rev Urol. 2019;16:645–654. doi: 10.1038/s41585-019-0237-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsuda S, Koyasu S. Mechanisms of action of cyclosporine. Immunopharmacology. 2000;47:119–125. doi: 10.1016/S0162-3109(00)00192-2. [DOI] [PubMed] [Google Scholar]
- 8.Tedesco D, Haragsim L. Cyclosporine: A review. J Transplant. 2012;2012:230386. doi: 10.1155/2012/230386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Periyasamy S, Hinds T, Jr, Shemshedini L, Shou W, Sanchez ER. FKBP51 and Cyp40 are positive regulators of androgen-dependent prostate cancer cell growth and the targets of FK506 and cyclosporin A. Oncogene. 2010;29:1691–1701. doi: 10.1038/onc.2009.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee CR, Chun JN, Kim SY, Park S, Kim SH, Park EJ, Kim IS, Cho NH, Kim IG, So I, et al. Cyclosporin A suppresses prostate cancer cell growth through CaMKKβ/AMPK-mediated inhibition of mTORC1 signaling. Biochem Pharmacol. 2012;84:425–431. doi: 10.1016/j.bcp.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Krishnamurthy A, Dasari A, Noonan AM, Mehnert JM, Lockhart AC, Leong S, Capasso A, Stein MN, Sanoff HK, Lee JJ, et al. Phase Ib results of the rational combination of selumetinib and cyclosporin A in advanced solid tumors with an expansion cohort in metastatic colorectal cancer. Cancer Res. 2018;78:5398–5407. doi: 10.1158/0008-5472.CAN-18-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flores C, Fouquet G, Moura IC, Maciel TT, Hermine O. Lessons to learn from low-dose cyclosporin-A: A new approach for unexpected clinical applications. Front Immunol. 2019;10:588. doi: 10.3389/fimmu.2019.00588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isshiki Y, Tanaka H, Suzuki Y, Yoshida Y. Cyclosporine is a potential curative treatment option for advanced thymoma. Exp Hematol Oncol. 2017;6:13. doi: 10.1186/s40164-017-0073-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Ran C, Li E, Gordon F, Comstock G, Siddiqui H, Cleghorn W, Chen HZ, Kornacker K, Liu CG, et al. Synergistic function of E2F7 and E2F8 is essential for cell survival and embryonic development. Dev Cell. 2008;14:62–75. doi: 10.1016/j.devcel.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park SA, Platt J, Lee JW, López-Giráldez F, Herbst RS, Koo JS. E2F8 as a novel therapeutic target for lung cancer. J Natl Cancer Inst. 2015;107:djv151. doi: 10.1093/jnci/djv151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng Q, Wang Q, Zong WY, Zheng DL, Wen YX, Wang KS, Teng XM, Zhang X, Huang J, Han ZG. E2F8 contributes to human hepatocellular carcinoma via regulating cell proliferation. Cancer Res. 2010;70:782–791. doi: 10.1158/0008-5472.CAN-09-3082. [DOI] [PubMed] [Google Scholar]
- 17.Lee DY, Chun JN, Cho M, So I, Jeon JH. Emerging role of E2F8 in human cancer. Biochim Biophys Acta Mol Basis Dis. 2023;1869:166745. doi: 10.1016/j.bbadis.2023.166745. [DOI] [PubMed] [Google Scholar]
- 18.Kim SH, Lee S, Piccolo SR, Allen-Brady K, Park EJ, Chun JN, Kim TW, Cho NH, Kim IG, So I, Jeon JH. Menthol induces cell-cycle arrest in PC-3 cells by down-regulating G2/M genes, including polo-like kinase 1. Biochem Biophys Res Commun. 2012;422:436–441. doi: 10.1016/j.bbrc.2012.03.131. [DOI] [PubMed] [Google Scholar]
- 19.Chun JN, Kim SY, Park EJ, Kwon EJ, Bae DJ, Kim IS, Kim HK, Park JK, Lee SW, Park HH, et al. Schisandrin B suppresses TGFβ1-induced stress fiber formation by inhibiting myosin light chain phosphorylation. J Ethnopharmacol. 2014;152:364–371. doi: 10.1016/j.jep.2014.01.024. [DOI] [PubMed] [Google Scholar]
- 20.Lee S, Chun JN, Kim SH, So I, Jeon JH. Icilin inhibits E2F1-mediated cell cycle regulatory programs in prostate cancer. Biochem Biophys Res Commun. 2013;441:1005–1010. doi: 10.1016/j.bbrc.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 21.Lee S, Park YR, Kim SH, Park EJ, Kang MJ, So I, Chun JN, Jeon JH. Geraniol suppresses prostate cancer growth through down-regulation of E2F8. Cancer Med. 2016;5:2899–2908. doi: 10.1002/cam4.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chun JN, Park S, Lee S, Kim JK, Park EJ, Kang M, Kim HK, Park JK, So I, Jeon JH. Schisandrol B and schisandrin B inhibit TGFβ1-mediated NF-κB activation via a Smad-independent mechanism. Oncotarget. 2017;9:3121–3130. doi: 10.18632/oncotarget.23213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 24.Lee S, Piccolo SR, Allen-Brady K. Robust meta-analysis shows that glioma transcriptional subtyping complements traditional approaches. Cell Oncol (Dordr) 2014;37:317–329. doi: 10.1007/s13402-014-0190-8. [DOI] [PubMed] [Google Scholar]
- 25.Floratos A, Smith K, Ji Z, Watkinson J, Califano A. geWorkbench: An open source platform for integrative genomics. Bioinformatics. 2010;26:1779–1780. doi: 10.1093/bioinformatics/btq282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Dalla Favera R, Califano A. ARACNE: An algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7((Suppl 1)):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kig C, Beullens M, Beke L, Van Eynde A, Linders JT, Brehmer D, Bollen M. Maternal embryonic leucine zipper kinase (MELK) reduces replication stress in glioblastoma cells. J Biol Chem. 2017;292:12786. doi: 10.1074/jbc.A113.471433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim SH, Kim SY, Park EJ, Kim J, Park HH, So I, Kim SJ, Jeon JH. Icilin induces G1 arrest through activating JNK and p38 kinase in a TRPM8-independent manner. Biochem Biophys Res Commun. 2011;406:30–35. doi: 10.1016/j.bbrc.2011.01.094. [DOI] [PubMed] [Google Scholar]
- 31.Kuang J, Yan X, Genders AJ, Granata C, Bishop DJ. An overview of technical considerations when using quantitative real-time PCR analysis of gene expression in human exercise research. PLoS One. 2018;13:e0196438. doi: 10.1371/journal.pone.0196438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, Wang Y, Shen F, Xu Y, Zhang Y, Zou X, Zhou J, Chen Y. Maternal embryonic leucine zipper kinase: A novel biomarker and a potential therapeutic target of cervical cancer. Cancer Med. 2018;7:5665–5678. doi: 10.1002/cam4.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramsey J, Glymour M, Sanchez-Romero R, Glymour C. A million variables and more: the fast greedy equivalence search algorithm for learning high-dimensional graphical causal models, with an application to functional magnetic resonance images. Int J Data Sci Anal. 2017;3:121–129. doi: 10.1007/s41060-016-0032-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38:675–678. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balwierz PJ, Pachkov M, Arnold P, Gruber AJ, Zavolan M, van Nimwegen E. ISMARA: Automated modeling of genomic signals as a democracy of regulatory motifs. Genome Res. 2014;24:869–884. doi: 10.1101/gr.169508.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park YR, Chun JN, So I, Kim HJ, Baek S, Jeon JH, Shin SY. Data-driven analysis of TRP channels in cancer: Linking variation in gene expression to clinical significance. Cancer Genomics Proteomics. 2016;13:83–90. [PubMed] [Google Scholar]
- 37.Park S, Lim JM, Chun JN, Lee S, Kim TM, Kim DW, Kim SY, Bae DJ, Bae SM, So I, et al. Altered expression of fucosylation pathway genes is associated with poor prognosis and tumor metastasis in non-small cell lung cancer. Int J Oncol. 2020;56:559–567. doi: 10.3892/ijo.2019.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu J, Smith VA, Wang PP, Hartemink AJ, Jarvis ED. Advances to Bayesian network inference for generating causal networks from observational biological data. Bioinformatics. 2004;20:3594–3603. doi: 10.1093/bioinformatics/bth448. [DOI] [PubMed] [Google Scholar]
- 39.Wang YX, Huang H. Review on statistical methods for gene network reconstruction using expression data. J Theor Biol. 2014;362:53–61. doi: 10.1016/j.jtbi.2014.03.040. [DOI] [PubMed] [Google Scholar]
- 40.Chun JN, Cho M, Park S, So I, Jeon JH. The conflicting role of E2F1 in prostate cancer: A matter of cell context or interpretational flexibility? Biochim Biophys Acta Rev Cancer. 2020;1873:188336. doi: 10.1016/j.bbcan.2019.188336. [DOI] [PubMed] [Google Scholar]
- 41.Taplin ME, Bubley GJ, Shuster TD, Frantz ME, Spooner AE, Ogata GK, Keer HN, Balk SP. Mutation of the androgen-receptor gene in metastatic androgen-independent prostate cancer. N Engl J Med. 1995;332:1393–1398. doi: 10.1056/NEJM199505253322101. [DOI] [PubMed] [Google Scholar]
- 42.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shafi AA, Yen AE, Weigel NL. Androgen receptors in hormone-dependent and castration-resistant prostate cancer. Pharmacol Ther. 2013;140:223–238. doi: 10.1016/j.pharmthera.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 44.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, Montgomery B, Taplin ME, Pritchard CC, Attard G, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuner R, Fälth M, Pressinotti NC, Brase JC, Puig SB, Metzger J, Gade S, Schäfer G, Bartsch G, Steiner E, et al. The maternal embryonic leucine zipper kinase (MELK) is upregulated in high-grade prostate cancer. J Mol Med (Berl) 2013;91:237–248. doi: 10.1007/s00109-012-0949-1. [DOI] [PubMed] [Google Scholar]
- 46.Jurmeister S, Ramos-Montoya A, Sandi C, Pértega-Gomes N, Wadhwa K, Lamb AD, Dunning MJ, Attig J, Carroll JS, Fryer LG, et al. Identification of potential therapeutic targets in prostate cancer through a cross-species approach. EMBO Mol Med. 2018;10:e8274. doi: 10.15252/emmm.201708274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharma NL, Massie CE, Ramos-Montoya A, Zecchini V, Scott HE, Lamb AD, MacArthur S, Stark R, Warren AY, Mills IG, Neal DE. The androgen receptor induces a distinct transcriptional program in castration-resistant prostate cancer in man. Cancer Cell. 2013;23:35–47. doi: 10.1016/j.ccr.2012.11.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are included in this published article. All additional information are available from the corresponding author on reasonable request.