Abstract
The emergence of the Omicron variant in November 2021, has caused panic worldwide due to the rapid evolution and the ability of the virus to escape the immune system. Since, several Omicron sublineages (BA.1 to BA.5) and their descendent recombinant lineages have been circulating worldwide. Furthermore, in December 2022, a new Omicron subvariant XBB.1.5 characterized by an unusual mutation in the spike protein evolved in the United States and rapidly spread to the other continents. Our study reports on the first cases of XBB.1.5 sublineage among indigenous Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-COV-2) positive cases detected through the influenza sentinel surveillance system in Niger. All influenza suspected cases were tested for both influenza and SARS-COV-2 using the Centre for Disease Control and prevention (CDC) Influenza SARS-COV-2 Multiplex quantitative Reverse-Transcription Polymerase Chain Reaction (qRT-PCR) Assay. SARS-COV-2 positive samples with cycle threshold ≤28 were selected for whole genome sequencing subsequently using the Oxford Nanopore Midnight protocol with rapid barcoding on a MinIon MK1B device. A total of 51 SARS-COV-2 positive samples were confirmed between December 2022 and March 2023. We successfully obtained 19 sequences with a predominance of the XBB.1/XBB.1.5 sublineages (73.7 %). In addition, a recombinant XBD sequence was also first-ever identified in early March 2023. Our findings support the need to strengthen the influenza sentinel surveillance for routine Coronavirus Disease 2019 (COVID-19) surveillance and SARS-COV-2 variants monitoring in Niger.
Keywords: Severe acute respiratory syndrome Coronavirus-2, XBB.1.5 sublineage, Influenza sentinel surveillance system, Niger
Graphical abstract
The World Health Organization (WHO) has called XBB.1.5 the most transmissible Omicron strain so far. In the U.S., it has spread like wildfire in the New England area, where infections rose over a short period of time to almost 94 % of cases as reported by the Centers for Disease Control and Prevention (CDC) at the beginning of February 2023.
Highlights
-
•
First identification of the SARS-COV-2/XBB.1.5 lineage in Niger.
-
•
High prevalence of XBB/XBB.1.5 among sequences identified from January 2023.
-
•
First detection of the SARS-COV-2 recombinant variant XBD in Niger.
-
•
Importance to strengthen and extend the existing influenza sentinel surveillance.
-
•
Crucial need for continuous SARS-COV-2 genomic surveillance in Niger.
Abbreviations
- SARS-COV-2
Severe Acute Respiratory Syndrome Coronavirus 2
- CDC
Centre for Disease Control and prevention
- qRT-PCR
quantitative Reverse-Transcription Polymerase Chain Reaction
- COVID-19
Coronavirus Disease 2019
- VOC
variant of concern
- WHO
World Health Organization
- TAG-VE
Technical Advisory Group on Virus Evolution
- ILI
Influenza Like Illness
- Ct
Threshold cycle
- SARI
Severe Acute Respiratory Infections
- IRR
International Reagents Resources
- RNA
Ribonucleic Acid
- N
nucleocapsid
- ONT
Oxford Nanopore Technologies
- cDNA
complementary Deoxyribonucleic Acid
- bp
base pairs
- kb
kilobases
- ng
nanograms
- ML
Maximum-Likelihood
- UKHSA
UK Health Security Agency
1. Introduction
COVID-19 is an acute respiratory infection caused by the SARS-COV-2 which belongs to the Coronaviridae family [1]. Since the advent of the COVID-19 pandemic, many variants of SARS-COV-2 have been reported globally. A spike gene (S-gene) deletion (Δ69–70) identified in the N-terminal region of the SARS-COV-2 Alpha variant which emerged in late 2020, resulted in primer erosion and Spike (S)-gene amplification failure in various reverse transcription–polymerase chain reaction (RT-PCR) assays [2,3]. Genomic sequencing of SARS-COV-2 isolates, which alongside S-gene's mutations target amplification assays, has facilitated early monitoring of emerging variants and their epidemiologic characteristics to inform public health decisions regarding vaccine policy, the use of therapeutics, and health care capacity [4]. The virus has evolved continuously to give rise to new variants [5]. As of 27 October 2022, the Omicron (B.1.1.529 has become the most widespread and dominant variant globally and was considered as the only circulating variant of concern (VOC) by the World Health Organization (WHO) [6,7]. First identified in 26 November 2021 in South Africa [8], the Omicron variant was associated with a high transmissibility and a significant number of mutations [9]. The distinguishing characteristics of the Omicron variant compared to the prior VOCs, was its ability to escape the immune system's defense and therefore rendering available COVID-19 vaccines less effective [10,11]. Due to its rapid evolution, several Omicron sublineages (BA.1 to BA.5) and their descendent recombinant lineages (XA to XW) rapidly emerged worldwide [4,11].
Since December 2022, a new sublineage XBB.1.5 also known as “Kraken”, has emerged in the United States and rapidly spread to the other continents [12]. The XBB.1.5 was highly transmissible and identified in 38 countries between 22 October 2022 and 11 January 2023 [13]. However, most of the generated sequences data were from the USA [14]. Therefore, on 11 January 2023, the WHO and its Technical Advisory Group on Virus Evolution (TAG-VE) recommended that countries prioritize studies to address uncertainties relating to the spread of the XBB.1.5, antibody escape, and disease severity due to infection with the XBB.1.5 subvariant [13]. The XBB.1.5 spread much faster than previous XBB/BQ, and was dominant globally as of 21 January 2023 [[15], [16], [17]]. It caused hospitalisations and exhibited elevated immune escape properties compared to the prior VOCs, due to a rarely seen amino-acid change in the spike protein, called F486P [[18], [19], [20]].
In Niger Republic, although the genetic evolution of SARS-COV-2 has not been well studied, recent establishment of sequencing capacity enabled to monitor the circulation dynamic of the Omicron variant which has been first detected on 22 December 2021. Since then, SARS-COV-2 positive cases detected either through the COVID-19 national surveillance or influenza sentinel surveillance are routinely sequenced for better understanding of the recent molecular epidemiology of the virus.
This study reports the first cases of XBB.1.5 sublineage among SARS-COV-2 positive cases detected mainly through the influenza sentinel surveillance system in Niger.
2. Materials and methods
2.1. Study design
Since the detection of the index SARS-COV-2 case in Niger, the existing influenza sentinel surveillance program allowed also the routine monitoring of SARS-COV-2. This surveillance program system includes eight sentinel sites distributed in three provinces, of which four are in Niamey, the capital city. For instance, nasopharyngeal swab samples were collected from both influenza like illness (ILI) and severe acute respiratory infections (SARI) cases based on the WHO case definition. For continuous monitoring of the circulation dynamics of the previously characterized SARS-COV-2 variants and rapid identification of the emerging variants, all positive samples with threshold cycle (Ct) value < 30 were subjected to genomic sequencing.
2.2. Molecular testing
Ribonucleic Acid (RNA) from all influenza-suspected specimens was manually extracted using the QIAamp Viral RNA Mini kit (Qiagen, Germany). The extracted RNA were tested for both influenza viruses and SARS-COV-2 by using the CDC Influenza-SARS-COV-2 Multiplex qRT-PCR Assay (Catalog No. FluSC2PPB-RUO) provided through the International Reagents Resources (IRR). The oligonucleotide primers and probe for detection of SARS-COV-2 targeted the carboxy-terminal portion of the nucleocapsid (N) gene.
2.3. Sequencing
SARS-COV-2-positive samples were subjected to whole genome sequencing using the Oxford Nanopore Midnight protocol with rapid barcoding on a MinIon MK1B device (Oxford Nanopore Technologies (ONT)). Briefly, complementary Deoxyribonucleic Acid (cDNA) synthesis was performed on the extracted RNA using the LunaScript RT mastermix (New England Biolabs) and overlapping amplicons of 1200-base pairs (bp) covering the cover the 30-kilobases (kb) SARS-COV-2 genome, were generated by multiplex PCR using the Midnight primer pools. Amplicons from each sample were pooled in a single tube and barcoded using the Oxford Nanopore Rapid Barcoding Kit according to the manufacturer's protocol. Barcoded samples were pooled in equal volume and purified using beads. The purified libraries were normalized to 800 ng (ng), adapter-ligated and loaded on a primed R9.4.1 flow cell [21].
2.4. Bioinformatics analysis
The base-called sequencing data were analyzed using the EDGE COVID-19 web-based platform with default parameters [22]. Features such as base coverage and lineage determination for the characterized samples were noted and the consensus SARS-COV-2 sequences were downloaded for further analyses. For lineage confirmation, the consensus sequences were submitted to the Nextclade Web-based platform (v 2.12.0) [23]. A Maximum-Likelihood (ML) tree was inferred using a dataset of 50 sequences including the newly characterized sequences from Niger and previously available XBB.1.5 sequences from Africa with the IQ-TREE software [24] for 1000 Bootstrap replications. The ML tree was rooted on midpoints.
3. Results
3.1. Molecular testing
Overall, 51 SARS-COV-2-positive samples were confirmed through the influenza sentinel surveillance system between December 2022 and March 2023. A total of 23 (45.1 %) samples were eligible for sequencing with Ct value ≤ 28 and 19 sequences have been successfully obtained and submitted to GISAID. The 19 newly characterized sequences included two Omicron sublineages, notably BA.5 and BA.3; 14 XBB.1/XBB.1.5 sublineages and a XBD recombinant variant. The XBB.1/XBB.1.5 sublineages were predominated from early January to mid-March 2023 (73.7 %) and all isolates originated from the capital city Niamey and the Dosso region. In addition, the XBD sequence was also identified in early March 2023 (Table 1).
Table 1.
Characteristics of SARS-COV-2/XBB1.5 sequences generated in this study.
| GISAID |
Date of collection | Lineage | % Coverage | Origin |
|---|---|---|---|---|
| Accession Number | ||||
| >hCoV-19/Niger/GRP12905/22 | 06/12/2022 | XBB.1 | 84.97 | Niamey |
| >hCoV-19/Niger/GRP13123/22 | 16/12/2022 | XBB.1 | 86.31 | Niamey |
| >hCoV-19/Niger/GRP13348/23 | 03/01/2023 | XBB.1 | 89.05 | Niamey |
| >hCoV-19/Niger/GRP13532/23 | 12/01/2023 | XBB.1.5 | 88.38 | Dosso |
| >hCoV-19/Niger/GRP12271/22 | 24/11/2022 | XBB.1 | 80.11 | Niamey |
| >hCoV-19/Niger/GRP12794/22 | 29/11/2022 | XBB.1 | 81.09 | Niamey |
| >hCoV-19/Niger/GRP13586/23 | 19/01/2023 | XBB.1 | 83.41 | Dosso |
| >hCoV-19/Niger/GRP13845/23 | 07/02/2023 | XBB.1.5 | 91.81 | Niamey |
| >hCoV-19/Niger/GRP13894/23 | 09/02/2023 | XBB.1.5 | 99.39 | Dosso |
| >hCoV-19/Niger/GRP13927/23 | 14/02/2023 | XBB.1.5 | 97.7 | Niamey |
| >hCoV-19/Niger/GRP14247/23 | 02/03/2023 | XBB.1 | 87.67 | Niamey |
| >hCoV-19/Niger/GRP14371/23 | 09/03/2023 | XBD | 71.57 | Niamey |
| >hCoV-19/Niger/GRP14103/23 | 21/02/2023 | XBB.1 | 93.87 | Niamey |
| >hCoV-19/Niger/GRP14435/23 | 16/03/2023 | XBB.1 | 80.90 | Niamey |
| >hCoV-19/Niger/CE59/23 | 02/03/2023 | XBB.1 | 93.00 | Niamey |
3.2. Phylogenetic analysis
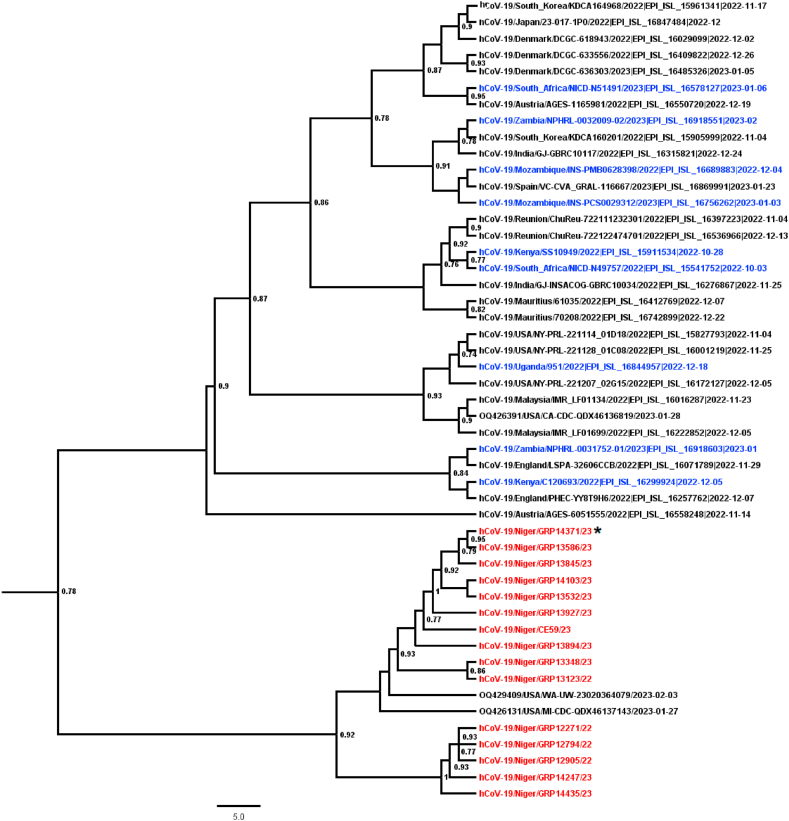
The Phylogenetic analysis showed that the different XBB.1/XBB.1.5 strains from Niger grouped together and clustered with two previous sequences from the USA collected between January and February 2023 (Gisaid accession numbers ‘‘OQ42940/USA/WA-UW-23020364079″ and ‘‘OQ42613/USA/MI-CDC-QDX46137143) (Fig. 1). In addition, the branch's order pointed to a recent introduction of XBB.1.5 in Niger, probably from the USA, while it was different from the virus previously identified in the other African countries from December 2022. In addition, the XBD isolate grouped with the XBB1/XBB.1.5 sequences from Niger, exhibiting a local virus evolution (Fig. 1).
Fig. 1.
Phylogenetic Maximum-Likelihood (ML) tree based on near-complete genome sequences of XBB.1.5 circulating in Niger. The ML tree was inferred using the IQ-TREE software [24] for 1000 Bootstrap replications and rooted on midpoints. Sequences from Niger are color-coded in red while the previous XBB.1.5 sequences from Africa were highlighted in blue. The recombinant XBD sequence was highlighted with the Asterix sign.
4. Discussion
The XBB.1.5 Omicron subvariant is a sublineage of the XBB variant, a recombinant of two BA.2 sublineages characterized by a large expansion globally and was associated with an unusually accelerated rate of increase in cases of COVID-19 and an ability to partially evade the antibody-mediated protection acquired by vaccination or prior infection [25,26]. To date, the XBB.1.5 is of considerable concern as it rapidly spread in USA since October 2022 [27], became the predominant Omicron variant in North America and Europe from early 2023 [26]. Thus, it was classified by the WHO as a VOC [13,[15], [16], [17], [18], [19], [20]].
In Niger, a decreasing prevalence of COVID-19 cases was noted from the second semester of 2022 and could be associated with a low notification of suspected cases or asymptomatic circulation of more adapted variants. Therefore, a continuous genomic surveillance of SARS-COV-2-positive cases confirmed through the influenza sentinel surveillance system led to the first identification of sequences of the XBB.1/XBB.1.5 Omicron subvariants in the country from December 2022.
The high prevalence recorded in our study in Niger for the XBB.1/XBB.1.5 isolates among sequences characterized from early January to mid-March 2023, coincided with its rapid spread and increased prevalence in North America and Europe [26,28,29]. As of 11 January 2023, the UK Health Security Agency (UKHSA) reported a weekly increase of 38.9 % for the XBB.1.5 Omicron subvariant compared to the recently dominant BQ1.1 sublineage of Omicron in the UK [30]. These data suggested a rapid global spread of the XBB.1.5. Unfortunately, preliminary data reported during that period on transmission advantage and immune escape from previous infection and vaccination for XBB.1 subvariants showed that they escaped neutralizing antibodies more effectively than to the BA.5 variants after both monovalent and bivalent mRNA vaccine boosting [31]. However, continuous practice of COVID-19 appropriate behaviour, and booster vaccines along with strong immunity could help minimize severity and fight the infection. In addition, contingency plans that focus on vaccination campaigns needs to be prioritize [26]. With their ability to evade neutralizing antibodies in recipients of vaccines [32,33] or produced monoclonal antibodies [34], the newer XBB.1/XBB.1.5 Omicron subvariants can re-infect people who were previously infected with earlier Omicron subvariants. However, the risk of severe hospitalization and death associated with the XBB.1.5 is lower in vaccinated people is lower than in unvaccinated people [35,36]. Therefore, getting vaccinated should still be encouraged before the licensing of specific vaccines against XBB.1.5 or broad‐spectrum vaccines. Nevertheless, more clinical studies assessing the severity of XBB.1.5 in unvaccinated people could be supported, particularly in older age groups, people with immunosuppression and underlying heart, kidney, and lung disease [37], in the African context where the vaccination coverage is still low [38]. The rapid transmissibility and effective immune escape capacity of the XBB.1.5 remind to us the necessary to promote global vaccination with existing vaccines and emphasize the importance of boosters, particularly among the elderly and children.
The phylogenetic data showed that the newly characterized XBB.1.5 recombinant sequences were the first detected from Niger and grouped with an isolate recently identified from the USA in February 2023; suggesting a recent introduction of the XBB.1.5 in the country. Although the virus was previously identified in the continent [39], the newly characterized sequences from Niger grouped with no previously available sequences from Africa, suggesting the XBB.1.5 introduction in Niger from the USA where it was first identified since October 2022 [13] and a local virus evolution from the XBB variant. In addition, no XBB.1.5 sequences were available from the neighbour West African country prior to those generated in our study. The newly characterized sequences could be also useful in future sub-regional, regional or global phylodynamic studies assessing the epidemiological patterns that have driven the XBB.1.5 rapid spread, more sub-regional. Numerous other recombinant sub-variants have evolved during the post-Omicron period, such as the XBD identified also in our study. However, the fact that it grouped with the XBB1/XBB.1.5 sequences from Niger showed also that the SARS-COV-2 Omicron XBB variant is slowly experiencing a local evolution. Thus, more experimental studies could be promoted to better understand their pathophysiology [40]. In addition, the current genomic surveillance of SARS-COV-2 variants in Niger could be strengthened by extending the existing influenza sentinel surveillance system to new sites throughout the country for rapid identification of probable hotspots in the community. To support the development of strategies to enhance preparedness of the health systems and guard against detrimental effects on population health, periodical genomic surveillance in confirmed patients and arriving or outbound travellers, is utmost important for assessing the emergence of new hybrid variants such as the XBB.1.16 [41] and the transmission dynamics, virulence and severity factors of the newly identified Omicron subvariants.
5. Conclusions
Although the XBB.1.5 is a super variant in the context of morphology and pathogenesis as it spreads much faster than previous XBB/BQ, causes hospitalisations [42], and has achieved immune evasiveness by acquiring a rare type of mutation called F486P in its receptor binding domain [43], appropriate and effective usage of available resources is crucial to minimize severity and mortality. Although the clinical impact and health outcomes of XBB.1.5 infection have been found to be lower in vaccinated people in western countries, more clinical studies are warranted in Africa where the vaccination coverage is still low, to evaluate the severity of the XBB.1.5 infection in unvaccinated people. Our findings point also to the need to strengthen the influenza sentinel surveillance program for continuous monitoring of the circulation dynamics of previously identified SARS-COV-2 lineages and rapid identification of emerging variants in Niger. Furthermore, the available sequencing capacities should be reinforced at the country level for genomic surveillance of pathogens of public health importance in order to implement rapid and appropriate control measures, if necessary.
Ethical approval
The National Ethics Committee of Niger approved the protocol for influenza sentinel surveillance system as a less than minimal risk for research by reference number 06/2009/CCNE. Written informed consent was obtained from all patients or legal guardians prior to enrolment. All methods including the use of human samples, were performed in accordance with the Declaration of Helsinki.
Funding
This work was supported by Agence Française de Développement through the AFROSCREEN project (grant agreement CZZ3209), coordinated by ANRS | Maladies infectieuses émergentes in partnership with Institut Pasteur and IRD.
We would additionally like to thank members from the AFROSCREEN Consortium (https://www.afroscreen.org/en/network/) for their work and support on genomic surveillance in Africa.
Data availability statement
All data generated or analyzed during this study are included in this published article. The characterized XBB/XBB 1.5 sequences from Niger have been deposited to GISAID under the following accession numbers: >hCoV-19/Niger/GRP12905/22; >hCoV-19/Niger/GRP13123/22, >hCoV-19/Niger/GRP13348/23, >hCoV-19/Niger/GRP13532/23, >hCoV-19/Niger/GRP12271/22; >hCoV-19/Niger/GRP12794/22; >hCoV-19/Niger/GRP13586/23; >hCoV-19/Niger/GRP13845/23; >hCoV-19/Niger/GRP13894/23; >hCoV-19/Niger/GRP13927/23; >hCoV-19/Niger/GRP14247/23; >hCoV-19/Niger/GRP14371/23; >hCoV-19/Niger/GRP14103/23; >hCoV-19/Niger/GRP14435/23; >hCoV-19/Niger/CE59/23.
CRediT authorship contribution statement
Adamou Lagare: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Project administration, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. Martin Faye: Conceptualization, Data curation, Formal analysis, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. Moussa Issa: Data curation, Formal analysis, Investigation, Methodology. Oumou Hamidou: Data curation, Investigation, Methodology. Baruani Bienvenu: Funding acquisition, Resources, Supervision. Abdoulkarim Mohamed: Formal analysis, Investigation, Supervision. Balki Aoula: Formal analysis, Investigation, Supervision. Katoumi Moumouni: Formal analysis, Investigation, Supervision. Fatima Hassane: Formal analysis, Investigation, Supervision. Younoussa Adamou Otto: Formal analysis, Methodology, Supervision. Didier D.K. Tambwe: Funding acquisition, Resources, Supervision, Writing – original draft. Elh Ibrahim Tassiou: Funding acquisition, Resources, Supervision, Writing – original draft. Haoua Seini: Funding acquisition, Resources, Writing – original draft. Ousmane Faye: Funding acquisition, Validation, Visualization, Writing – original draft. Ronan Jambou: Conceptualization, Funding acquisition, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Adamou Lagare reports equipment, drugs, or supplies was provided by Institut Pasteur.
Acknowledgements
We are grateful to all stakeholders from influenza sentinel sites for their commitment and acknowledge technical support from World Health Organization, John Hopkins University, CDC Africa. Further, we thank all the laboratories that have shared SARSCoV-2 sequence data in GISAID that we included as comparison data in our analysis.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e20916.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Pal M., Berhanu G., Desalegn C., Kandi V. Severe acute respiratory Syndrome coronavirus-2 (SARS-CoV-2): an update. Cureus. 2020;12(3) doi: 10.7759/cureus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meng B., Kemp S.A., Papa G., et al. COVID-19 Genomics UK (COG-UK) Consortium. Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.109292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark C., Schrecker J., Hardison M., Taitel M.S. Validation of reduced S-gene target performance and failure for rapid surveillance of SARS-CoV-2 variants. PLoS One. 2022;17 doi: 10.1371/journal.pone.0275150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambrou A.S., Shirk P., Steele M.K., et al. Strain surveillance and emerging variants bioinformatic working group; strain surveillance and emerging variants NS3 working group. Genomic surveillance for SARS-CoV-2 variants: predominance of the delta (B.1.617.2) and omicron (B.1.1.529) variants—United States, june 2021–january 2022. MMWR Morb. Mortal. Wkly. Rep. 2022;71:206–211. doi: 10.15585/mmwr.mm7106a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Low neutralization of SARS-CoV-2 Omicron BA.2.75.2, BQ.1.1 and XBB.1 by parental mRNA vaccine or a BA.5 bivalent booster. Kurhade C, Zou J, Xia H, et al. Nat. Med. 2022;29:344–347. doi: 10.1038/s41591-022-02162-x. [DOI] [PubMed] [Google Scholar]
- 6.Gong W., Parkkila S., Wu X., Aspatwar A. SARS-CoV-2 variants and COVID-19 vaccines: current challenges and future strategies. Int. Rev. Immunol. 2022:1–22. doi: 10.1080/08830185.2022.2079642. [DOI] [PubMed] [Google Scholar]
- 7.Velavan T.P., Ntoumi F., Kremsner P.G., Lee S.S., Meyer C.G. Emergence and geographic dominance of Omicron subvariants XBB/XBB.1.5 and BF.7 - the public health challenges. Int. J. Infect. Dis. 2023;128:307–309. doi: 10.1016/j.ijid.2023.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kandeel M., Mohamed M.E.M., Abd El-Lateef H.M., Venugopala K.N., El-Beltagi H.S. Omicron variant genome evolution and phylogenetics. J. Med. Virol. 2022;94(4):1627–1632. doi: 10.1002/jmv.27515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ren S.Y., Wang W.B., Gao R.D., Zhou A.M. Omicron variant (B.1.1.529) of SARS-CoV-2: mutation, infectivity, transmission, and vaccine resistance. World J Clin Cases. 2022;10(1):1–11. doi: 10.12998/wjcc.v10.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mistry P., Barmania F., Mellet J., Peta K., Strydom A., Viljoen I.M., James W., Gordon S., Pepper M.S. SARS-CoV-2 variants, vaccines, and host immunity. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.809244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tallei T.E., Alhumaid S., AlMusa Z., Fatimawali Kusumawaty D., Alynbiawi A., Alshukairi A.N., Rabaan A.A. Update on the omicron sub-variants BA.4 and BA.5. Rev. Med. Virol. 2023;33(1) doi: 10.1002/rmv.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parums D.V. Editorial: the XBB.1.5 ('Kraken') subvariant of omicron SARS-CoV-2 and its rapid global spread. Med. Sci. Mon. Int. Med. J. Exp. Clin. Res. 2023;29 doi: 10.12659/MSM.939580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.World Health Organization (WHO) 2023. Technical Advisory Group on Virus Evolution (TAG-VE)https://www.who.int/docs/default-source/coronaviruse/11jan2023_xbb15_rapid_risk_assessment.pdf Available at: (Assessed on 23 March 2023) [Google Scholar]
- 14.Ma K.C., Shirk P., Lambrou A.S., Hassell N., Zheng X.Y., Payne A.B., Ali A.R., Batra D., Caravas J., Chau R., Cook P.W., Howard D., Kovacs N.A., Lacek K.A., Lee J.S., MacCannell D.R., Malapati L., Mathew S., Mittal N., Nagilla R.R., Parikh R., Paul P., Rambo-Martin B.L., Shepard S.S., Sheth M., Wentworth D.E., Winn A., Hall A.J., Silk B.J., Thornburg N., Kondor R., Scobie H.M., Paden C.R. Genomic surveillance for SARS-CoV-2 variants: circulation of omicron lineages - United States, january 2022-may 2023. MMWR Morb. Mortal. Wkly. Rep. 2023;72(24):651–656. doi: 10.15585/mmwr.mm7224a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yue C., Song W., Wang L., et al. Enhanced Transmissibility of XBB.1.5 is contributed by both strong ACE2 binding and antibody evasion. Immunology. 2023 doi: 10.1101/2023.01.03.522427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Browne E. Why COVID's XBB.1.5 ‘Kraken’ variant is so contagious. Sci. Am. 2023 https://www.scientificamerican.com/article/why-covids-xbb-1-5-kraken-variant-is-so-contagious/ Available at: Assessed on 23 March 2023. [Google Scholar]
- 17.Centers for Disease Control and Prevention (CDC) 2023. COVID Data Tracker.https://covid.cdc.gov/covid-data-tracker/#variant-proportions Available at: (Assessed on 23 March 2023) [Google Scholar]
- 18.Callaway E. Coronavirus variant XBB.1.5 rises in the United States – is it a global threat? Nature. 2023;613(7943):222–223. doi: 10.1038/d41586-023-00014-3. [DOI] [PubMed] [Google Scholar]
- 19.Graham F. Daily briefing: is subvariant XBB.1.5 a global threat? Nature. 2023 Jan doi: 10.1038/d41586-023-00052-x. [DOI] [PubMed] [Google Scholar]
- 20.Hotez P. XBB.1.5 emerges in the Americas: what it means to the region. Lancet Reg Health Am. 2023;18 doi: 10.1016/j.lana.2023.100433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tegally H., Moir M., Everatt J., Giovanetti M., Scheepers C., Wilkinson E., Subramoney K., Makatini Z., Moyo S., Amoako D.G., et al. Emergence of SARS-CoV-2 omicron lineages BA.4 and BA.5 in South Africa. Nat. Med. 2022;28(9):1785–1790. doi: 10.1038/s41591-022-01911-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lo C.C., Shakya M., Connor R., Davenport K., Flynn M., Gutiérrez A.M.Y., Hu B., Li P.E., Jackson E.P., Xu Y., Chain P.S.G. Edge COVID-19: a web platform to generate submission-ready genomes from SARS-CoV-2 sequencing efforts. Bioinformatics. 2022;38(10):2700–2704. doi: 10.1093/bioinformatics/btac176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aksamentov I., Roemer C., Hodcroft E., Nextclade Neher R. Clade assignment, mutation calling and quality control for viral genomes. J. Open Source Softw. 2021;6:3773. doi: 10.21105/joss.03773. [DOI] [Google Scholar]
- 24.Trifinopoulos J., Nguyen L.T., von Haeseler A., Minh B.Q. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44(W1):W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia S., Wang L., Zhu Y., Lu L., Jiang S. Origin, virological features, immune evasion and intervention of SARS-CoV-2 Omicron sublineages. Signal Transduct. Targeted Ther. 2022;7(1):1–7. doi: 10.1038/s41392-022-01105-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Velavan T.P., Ntoumi F., Kremsner P.G., Lee S.S., Meyer C.G. Emergence and geographic dominance of Omicron subvariants XBB/XBB.1.5 and BF.7 - the public health challenges. Int. J. Infect. Dis. 2023 Mar;128:307–309. doi: 10.1016/j.ijid.2023.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The Lancet Regional Health-Americas Another variant, XBB.1.5: will it be a tsunami for the Americas? Lancet Reg Health Am. 2023 Feb;18 doi: 10.1016/j.lana.2023.100452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.World Health Organization (WHO) 2023. XBB.1.5 Rapid Risk Assessment.https://www.who.int/docs/default-source/coronaviruse/11jan2023_xbb15 _rapid_ risk_assessment.pdf Available at: [Google Scholar]
- 29.European Centre for Disease Prevention and Control (ECDC) 2023. Threat Assessment Brief: Implications for the EU/EEA of the Spread of the SARS-CoV-2 Omicron XBB.1.5 Sub-lineage.https://www.ecdc.europa.eu/en/publications-data/covid-19-threat-assessment-brief-implications-spread-omicron-xbb Available at: [Google Scholar]
- 30.UK Health Security Agency (HCSA) 2023. Technical Briefing 49. SARS-CoV-2 Variants of Concern and Variants under Investigation in England.https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1128554/variant-technical-briefing-49-11-january-2023.pdf Available at: [Google Scholar]
- 31.Miller J., Hachmann N.P., Collier A.Y., et al. Substantial neutralization escape by SARS-CoV-2 Omicron variants BQ.1.1 and XBB.1. N. Engl. J. Med. 2023 doi: 10.1056/NEJMc2214314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X., Chen L.L., Ip J.D., Chan W.M., Hung I.F., Yuen K.Y., et al. Omicron sublineage recombinant XBB evades neutralising antibodies in recipients of BNT162b2 or CoronaVac vaccines. Lancet Microbe. 2022 doi: 10.1016/S2666-5247(22)00335-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y., Long Y., Wang F., et al. Characterization of SARS-CoV-2 recombinants and emerging Omicron sublineages. Int. J. Med. Sci. 2023;20(1):151–162. doi: 10.7150/ijms.79116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yue C., Song W., Wang L., Jian F., Chen X., Gao F., et al. Enhanced transmissibility of XBB.1.5 is contributed by both strong ACE2 binding and antibody evasion. bioRxiv. 2023 doi: 10.1101/2023.01.03.522427. 2023.01.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizrahi B., Sudry T., Flaks‐Manov N., et al. Long covid outcomes at one year after mild SARS‐CoV‐2 infection: nationwide cohort study. BMJ. 2023;380 doi: 10.1136/bmj-2022-072529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feikin D.R., Higdon M.M., Abu‐Raddad L.J., et al. Duration of effectiveness of vaccines against SARS‐CoV‐2 infection and COVID‐19 disease: results of a systematic review and meta‐regression. Lancet. 2022;399(10328):924–944. doi: 10.1016/S0140-6736(22)00152-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klein E.Y., Fall A., Norton J.M., Eldesouki R.E., Abdullah O., Han L., Yunker M., Mostafa H.H. Severity outcomes associated with SARS-CoV-2 XBB variants, an observational analysis. J. Clin. Virol. 2023 Aug;165 doi: 10.1016/j.jcv.2023.105500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turyasingura N., James W.G., Vermund S.H. COVID-19 vaccine equity in Africa. Trans. R. Soc. Trop. Med. Hyg. 2023 doi: 10.1093/trstmh/trac130. trac130. doi: 10.1093/trstmh/trac130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelleni M.T. Evolution of SARS CoV-2 omicron subvariants BF.7 and XBB.1.5: time to follow Africa and abort all COVID restrictions. J. Infect. 2023 doi: 10.1016/j.jinf.2023.01.027. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chatterjee S., Bhattacharya M., Nag S., Dhama K., Chakraborty C. A detailed overview of SARS-CoV-2 omicron: its sub-variants, mutations and pathophysiology, clinical characteristics, immunological landscape, immune escape, and therapies. Viruses. 2023;15(1):167. doi: 10.3390/v15010167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khairnar K., Tomar S.S. COVID-19 genome surveillance: a geographical landscape and mutational mapping of SARS-CoV-2 variants in Central India over two years. medRxiv. 2023 doi: 10.1101/2023.03.22.23287566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mohanty A., Rohilla R., Mehta R., Padhi B.K., Sah R. XBB.1.5 an emerging threat: correspondence. Int. J. Surg. 2023;109(4):1050–1051. doi: 10.1097/JS9.0000000000000261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Omicron's XBB.1.5 sub-variant explained. Should India be worried of this strain? https://www.livemint.com/science/health/omicrons-xbb-1-5-sub-variant-explained-should-india-be-worried-of-this-strain-11672789266832.html (Accessed 6 August 2023).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article. The characterized XBB/XBB 1.5 sequences from Niger have been deposited to GISAID under the following accession numbers: >hCoV-19/Niger/GRP12905/22; >hCoV-19/Niger/GRP13123/22, >hCoV-19/Niger/GRP13348/23, >hCoV-19/Niger/GRP13532/23, >hCoV-19/Niger/GRP12271/22; >hCoV-19/Niger/GRP12794/22; >hCoV-19/Niger/GRP13586/23; >hCoV-19/Niger/GRP13845/23; >hCoV-19/Niger/GRP13894/23; >hCoV-19/Niger/GRP13927/23; >hCoV-19/Niger/GRP14247/23; >hCoV-19/Niger/GRP14371/23; >hCoV-19/Niger/GRP14103/23; >hCoV-19/Niger/GRP14435/23; >hCoV-19/Niger/CE59/23.