Abstract
Multi-task learning (MTL) methods have been extensively employed for joint localization and classification of breast lesions on ultrasound images to assist in cancer diagnosis and personalized treatment. One typical paradigm in MTL is a shared trunk network architecture. However, such a model design may suffer information-sharing conflicts and only achieve suboptimal performance for individual tasks. Additionally, the model relies on fully-supervised learning methodologies, imposing heavy burdens on data annotation. In this study, we propose a novel joint localization and classification model based on attention mechanisms and a sequential semi-supervised learning strategy to address these challenges. Our proposed framework offers three primary advantages. First, a lesion-aware network with multiple attention modules is designed to improve model performance on lesion localization. An attention-based classifier explicitly establishes correlations between the two tasks, alleviating information-sharing conflicts while leveraging location information to assist in classification. Second, a two-stage sequential semi-supervised learning strategy is designed for model training to achieve optimal performance on both tasks and substantially reduces the need for data annotation. Third, the asymmetric and modular model architecture allows for the flexible interchangeability of individual components, rendering the model adaptable to various applications. Experimental results from two different breast ultrasound image datasets under varied conditions have demonstrated the effectiveness of the proposed method. Furthermore, we conduct comprehensive investigations into the impacts of various factors on model performance, gaining in-depth insights into the mechanism of our proposed framework. The code is available at https://github.com/comp-imaging-sci/lanet-bus.git.
Keywords: Breast ultrasound, Breast tumor classification, Breast tumor localization, Multi-task learning, Semi-supervised learning, Attention mechanism
1. Introduction
Breast neoplasm is a commonly seen disease among women between 35–55 years of age (Pyakurel et al., 2014; Nothacker et al., 2009). It has various subtypes ranging from benign cysts, high-risk lesions, and pre-cancer lesions to malignant cancers. Early cancer diagnosis can significantly improve treatment outcomes and reduce breast cancer mortality rates. Medical imaging techniques, including mammography, magnetic resonance imaging (MRI), and ultrasound imaging (Nothacker et al., 2009; Lee et al., 2010), have been employed in the noninvasive identification of early-stage breast cancer cells (Wang, 2017). Mammography, while the gold standard for breast cancer screening, exhibits relatively low sensitivity in patients with dense breasts (Nothacker et al., 2009; Lee et al., 2010). MRI is usually used along with mammography to screen patients with elevated cancer risk, mitigating the high false positive rate and avoiding unnecessary invasive biopsies (den Dekker et al., 2021). To complement these imaging techniques, previous studies have shown that breast ultrasound can inspect certain changes in breast tissue that can be harder to see in mammograms (Gordon and Goldenberg, 1995; Kolb et al., 1998; Buchberger et al., 1999; Berg et al., 2008). Due to its low cost, widespread availability, and radiation-free safety, breast ultrasound is a ubiquitous breast imaging modality used extensively in diagnostic examinations. It is generally the first scan performed for young (< 30 years old), pregnant, or lactating patients with breast concerns (Lee et al., 2010).
Manually analyzing large volumes of breast ultrasound screening images is time-consuming and substantially increases the overall workload for radiologists. Intrinsic characteristics of breast ultrasound images also bring additional challenges to image analysis, such as similarities in texture and contrast between lesions and surrounding tissue, lesion tissue heterogeneity, and user-dependent scan quality (Hooley et al., 2013). Computer-aided diagnosis (CAD) systems have been developed to address these challenges and assist in clinical applications such as localization and classification (Shan et al., 2016; Wu et al., 2019; Yap et al., 2018; Shin et al., 2018; Han et al., 2017). Lesion classification aims to stratify lesions into different subtypes to support appropriate treatment planning, while localizing and segmenting lesions can facilitate the classification task to achieve better diagnosis accuracy (Vakanski et al., 2020; Tang et al., 2021; Abbas et al., 2020; Cao et al., 2017; Shin et al., 2018; Zhou et al., 2021). Instead of handling these tasks separately, multi-task learning (MTL) techniques have been designed for joint lesion segmentation and classification (Wu et al., 2019; Yap et al., 2018; Shin et al., 2018; Han et al., 2017; Zhou et al., 2021; Crawshaw, 2020; Rasaee and Rivaz, 2021; Chowdary et al., 2022; Singh et al., 2019). One typical paradigm of MTL models employs a network architecture that encompasses a shared feature extractor along with two appended task-specific branches for lesion segmentation and classification, respectively. Such shared trunk-based models are trained using a hybrid task-related loss function to achieve optimal performance for both tasks. The network design and training process allows inter-task information sharing, therefore improving data efficiency and reducing the risk of overfitting.
However, training MTL models is more intricate and challenging than training single-task models. Due to the possible information-sharing conflicts between tasks with distinct objectives, increasing the model performance on one task might compromise the other. Balancing information sharing across tasks is critical in ensuring overall model performance (Li et al., 2022). Several methods, such as task-specific loss weighting (Crawshaw, 2020; Liu et al., 2019) and gradient demodulation (Crawshaw, 2020; Sinha et al., 2018) have been proposed to address this problem by balancing the contribution of each task to the hybrid loss.
Recently, many studies have been conducted to employ attention mechanisms to mitigate information-sharing conflicts in MTL for segmentation and classification using BUS images (Zhang et al., 2021; Vakanski et al., 2020; Han et al., 2020). An attention module employs the information bottleneck method to highlight crucial input features while downplaying the less relevant aspects (Tishby and Zaslavsky, 2015; Liu et al., 2019; Ronneberger et al., 2015), thus yielding task-specific representation and minimizing interference during model optimization. Furthermore, attention mechanisms have been explored to aggregate multi-scale features to capture expressive combinations of local and global information to enhance lesion segmentation performance (Xu et al., 2023; Lyu et al., 2023; Xu et al., 2022; Zhou et al., 2021; Chen et al., 2019).
Traditionally, MTL models, including attention-based ones, are trained under fully-supervised learning paradigms. Such training paradigms require large datasets with both class labels and pixel-level tumor segmentation annotations. Particularly, obtaining the latter is usually time-consuming and expensive. Semi-supervised learning (SSL) techniques have recently gained growing attention in training deeplearning (DL) based models, because they leverage unlabeled data and reduce the requirement for a large amount of labeled data (Yang et al., 2021; Han et al., 2020; Mittal et al., 2021; Zhai et al., 2022; Kim et al., 2021; Farooq et al., 2023; Chen et al., 2019). SSL methods enable the training of models using samples without complete and explicit labels, which combine the advantages of both supervised and unsupervised learning and enhance the model’s learning capability. In recent years, a variety of SSL techniques have been proposed, including pseudo-labeling (Liu et al., 2021b; Dai et al., 2019; Liu et al., 2021b), contrastive learning (Mittal et al., 2021; Farooq et al., 2023), and generative SSL (Imran and Terzopoulos, 2019; Han et al., 2020). Among these, pseudo-labeling is widely used because of its simplicity and efficiency. However, a majority of these SSL methods have primarily targeted single-task applications. Designing specific SSL strategies tailored to MTL models remains an area lacking in-depth understanding.
In this study, we propose a novel joint localization and classification framework integrating the attention mechanism and the semi-supervised learning strategy to address these challenges. The framework comprises a feature extractor (FEX), an auxiliary lesion-aware network (LA-Net) for lesion localization, and an attention-based classifier (AC) for lesion classification. With the proposed two-stage sequential semi-supervised training strategy, the designed framework effectively mitigates the concerns of information-sharing conflicts and data annotation burdens, thereby achieving optimal model performance on both tasks. Experimental results from two BUS image datasets demonstrate the effectiveness of the proposed method. The effects of various network factors on model performance are thoroughly investigated to gain a deep understanding of the designed framework. The main contributions of this study can be summarized as follows:
The proposed auxiliary LA-Net, including multiple attention modules that capture discriminative features within the extracted representations, facilitates precise tumor localization. A channel attention module (CAM) and a spatial attention module (SAM) are designed in the LA-Net to exploit multi-scale and location-related features to improve lesion localization performance. A mask-attention module (MAM) is designed in the attention-based classifier to bridge the two task-specific branches and refine the classifier’s performance using the localization information captured by the LA-Net. This design enhances the features specific to the tumor region and downplays the possible negative effects of ultrasound image noise and contrast limitations. In this way, our method alleviates potential information-sharing conflicts and achieves optimal model performance on both localization and classification tasks.
A sequential semi-supervised learning strategy based on a pseudo-labeling paradigm (Lee et al., 2013) is proposed for model training. This training strategy has two stages: (1) training the LA-Net using location labels and (2) utilizing the pre-trained LA-Net within our framework to enhance learning stability. The MAM-assisted learning strategy facilitates direct optimization of the AC using the localization information captured by the LA-Net. It enables LA-Net to learn discriminative features by leveraging classification outcomes even in the absence of location labels. This strategy permits utilizing large amounts of BUS images without location labels to improve model performance while concurrently reducing the need for lesion location annotations.
Our modular framework is designed as an asymmetric U-Net-like architecture (Ronneberger et al., 2015). It facilitates flexible configuration of feature extraction trunks and task-specific branches. In this study, FEX is designed with ResNet (He et al., 2016) to demonstrate model efficacy. Other powerful deep convolutional neural networks (CNNs) like EfficientNet (Tan and Le, 2019) can also be integrated into the framework as the encoder. This extensibility allows the integration of various encoders to extract hierarchical multi-scale features enriched with local and global lesion-related information from input BUS images. Such representative features alleviate the challenges due to substantial intra-class variation and inter-class similarity in BUS images.
The remainder of the paper is organized as follows. Section 2 introduces the related background knowledge of multi-task learning, vision attention mechanisms, and semi-supervised learning. Section 3 describes the network architecture and training strategy of the proposed method. Section 4 describes the datasets and implementation details of the proposed method. The experimental results are shown in Section 5, and the discussion and conclusion are described in Sections 6 and 7, respectively.
2. Related work
2.1. Multi-task learning
Most MTL methods applied to BUS images are based on a design of shared trunk architecture. This architecture consists of a shared trunk network responsible for extracting features from input images and two appended branches for lesion classification and localization tasks. The model is usually optimized by a hybrid loss combining both classification and localization losses. This design aims to learn the features by leveraging both classification- and localization-related information, thus reducing the risk of overfitting and improving task performance (Crawshaw, 2020). For example, Zhou et al. employed a VNet-based encoder–decoder network for segmentation task, while the intermediate feature maps generated by the encoder were reused by a lightweight network for the classification task (Zhou et al., 2021). Chowdary et al. employed a residual U-Net architecture for segmentation and shared intermediate feature maps with a two-layer fully-connected (FC) network for classification (Chowdary et al., 2022). Rasaee et al. designed an asymmetric encoder–decoder architecture (Rasaee and Rivaz, 2021). The feature extraction trunk and classification branch were designed as a ResNet50 network (He et al., 2016). A segmentation branch with three upsampling blocks was appended to the feature extraction trunk to extract the feature map from the last convolutional layer for lesion segmentation. Singh et al. revisited the U-Net architecture to incorporate atrous convolutional layers and a channel weighting block for segmentation (Singh et al., 2019). The subsequent segmentation predictions were further revised through adversarial training (Goodfellow et al., 2014). After adversarial training, the statistical boundary features of the segmented tumor contours were used to distinguish tumor classes. Mishra et al. proposed a U-Net-like architecture for segmentation and fed the intermediate features of all encoder and decoder blocks into a residual CNN for classification (Mishra et al., 2022).
To avoid laborious annotation of pixel-level segmentation, some methods simplify pixel-wise lesion segmentation to object localization in the form of bounding boxes (Cao et al., 2017; Shin et al., 2018). For instance, Cao et al. investigated the performance of several popular object localization methods such as YOLO (Redmon et al., 2016) and SSD (Liu et al., 2016) applied to BUS images (Cao et al., 2017). Shin et al. employed Faster-RCNN (Ren et al., 2015) for joint localization and classification of breast tumors in the BUS image dataset (Shin et al., 2018). Beyond shared-trunk-based MTL methods, other alternative MTL methods have been proposed, including cross-talk, prediction distillation, and task routing. More details on those methods can be found in Crawshaw’s survey (Crawshaw, 2020).
Within shared trunk-based MTL methods, effective mitigation of possible information-sharing conflicts is critical for achieving optimal model performance in both tasks (Li et al., 2022). Task-specific loss weighting is one solution to balance the contribution of each task during model optimization by assigning different weights to individual task losses (Crawshaw, 2020; Liu et al., 2019). For example, Liu et al. introduced an adaptive weighting method to dynamically balance task weights guided by the rate of change in the loss of each task (Liu et al., 2019). Gradient demodulation is another solution to equalize gradient magnitudes across tasks during model optimization to balance learning efficiency and ensure that each task is optimized with equal importance (Crawshaw, 2020; Sinha et al., 2018). For instance, Sinha et al. employed adversarial training to align the gradients from different tasks to boost model performance (Sinha et al., 2018).
2.2. Vision attention mechanism
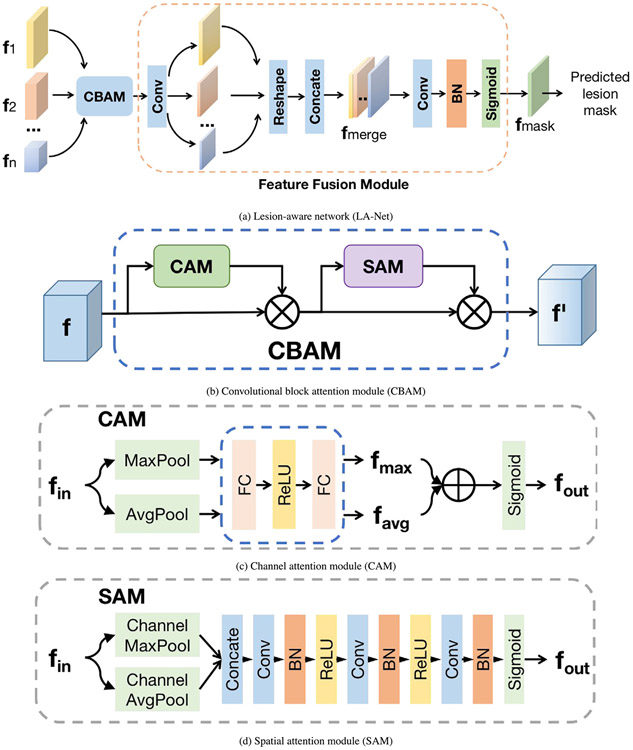
Vision attention mechanisms, inspired by the human visual system’s neuronal structure (Itti et al., 1998), have been applied in computer vision tasks to enhance feature extraction by emphasizing discriminative task-specific information, thereby improving model performance (Liu et al., 2019). Within the domain of BUS image processing, attention mechanisms have gained growing attention (Xu et al., 2022; Zhang et al., 2021; Ma et al., 2020; Singh et al., 2020). For instance, Xu et al. proposed a self-attention module on top of a U-Net architecture to utilize contextual information to improve both breast tumor segmentation and classification performance (Xu et al., 2022). Zhang et al. proposed an MTL framework with soft and hard attention modules to guide the model to focus on tumor regions and thus enhancing model performance (Zhang et al., 2021). Woo et al. developed a convolutional block attention module with versatile applicability to various CNNs, leveraging channel and spatial information from input features to achieve improved model performance (Woo et al., 2018).
Previous studies have demonstrated the potency of multi-scale features extracted from diverse hierarchical layers of CNNs, which offer rich local and global information and significantly benefit overall model performance (Tang et al., 2021; Lin et al., 2016). The incorporation of these multi-scale features with specific attention mechanisms is emerging as a prominent strategy in designing MTL methods. For example, Lyu et al. proposed a pyramid attention network combining spatial and channel attention mechanisms with multi-scale features for segmentation task (Lyu et al., 2023). The multi-scale feature pyramid was achieved by a depth-wise separable small-size convolution strategy. Xu et al. designed a regional attention module on top of a U-Net-like segmentation network (Xu et al., 2023). It aims to decode the multi-scale information captured by the segmentation task to guide the classifier in learning class-specific features from tumor, peritumoral, and background regions.
These methods have shown improved performance for both classification and segmentation tasks. However, they predominantly stem from a U-Net framework (Ronneberger et al., 2015) with symmetric encoder and decoder architecture. Such a design may constrain the flexible and smooth adaptation of encoders and decoders to diverse applications. For example, the off-the-shelf CNNs pretrained on large-scale datasets cannot be directly integrated into these methods and employed as encoders.
2.3. Semi-supervised learning
Semi-supervised learning (SSL) methods are widely used to alleviate the necessity for large-scale labeled datasets (Yang et al., 2021; Han et al., 2020; Mittal et al., 2021; Zhai et al., 2022; Kim et al., 2021). These methods utilize large amounts of unlabeled data to learn useful characteristics of input data distributions, thereby enhancing model performance without the need for additional annotations. One type of widely-used SSL method is pseudo-labeling (Liu et al., 2021b; Dai et al., 2019; Liu et al., 2021b). This approach assumes that predictions of unlabeled data with high prediction confidence are highly likely to be correct. These high-confidence predictions can be used as surrogate ground-truth labels during training. Dai et al. proposed a sequential training strategy that exploits unlabeled annotations to train a 3-dimensional U-Net network for brain tumor segmentation using MRI data (Dai et al., 2019). The network is initially trained on an unlabeled dataset with massive automatically generated pseudo labels, followed by fine-tuning through transfer learning on a small dataset with manually annotated labels. Liu et al. revised a 3-dimensional VNet (Milletari et al., 2016) with a deep attentive module to focus on important information from extracted multi-scale features to achieve accurate breast cancer segmentation on 3D ultrasound data (Liu et al., 2021b).
Other popular SSL methods can be classified as contrastive learning (Mittal et al., 2021; Farooq et al., 2023) and generative SSL (Imran and Terzopoulos, 2019; Han et al., 2020). The idea of contrastive learning is to learn useful representations by comparing similar and dissimilar examples. For example, Mittal et al. introduced a dual-branch model with a consistency regularization technique to combine a GAN-based network for segmentation and a multi-label teacher network to filter out false positive segmentation predictions (Mittal et al., 2021). Generative SSL methods leverage generative models (e.g., GAN (Goodfellow et al., 2014)) to capture the underlying distribution of data samples and synthesize additional labeled data for training. Han et al. adopted a generative adversarial network (GAN)-based model for lesion segmentation with a semi-supervised learning strategy, exploiting unlabeled BUS images to improve the quality of generated segmentation (Han et al., 2020). A more detailed review of SSL methods can be found in the literature (Van Engelen and Hoos, 2020; Yang et al., 2022).
3. Methods
3.1. Proposed auxiliary attention-based joint classification and localization framework
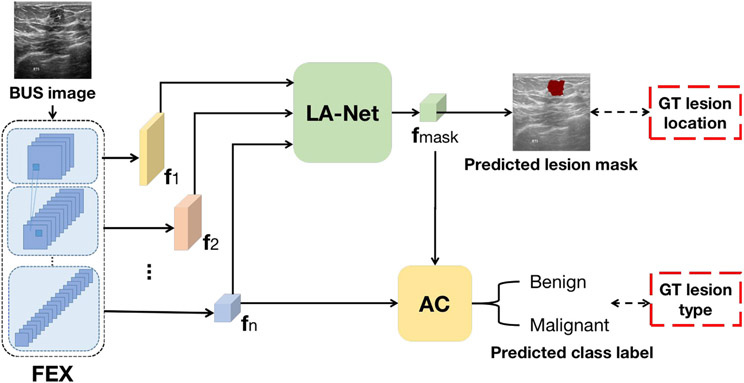
The proposed joint classification and localization framework is shown in Fig. 1. The framework consists of a feature extractor (FEX), an attached attention-based classifier (AC) for classification, and an auxiliary lesion-aware network (LA-Net) for lesion localization. Given BUS images, FEX extracts multi-scale image feature maps that capture local and global information, which are fed into LA-Net for lesion localization and AC for lesion classification. The lesion-aware feature map () obtained by the LA-Net is further leveraged by the AC to improve the performance of lesion-type classification. Notably, the AC explicitly correlates the classification and localization branches through an attention module, which takes both of the FEX and of the LA-Net as inputs. Such a design alleviates the potential conflicts and ensures balanced optimization for both localization and classification branches. The model architecture is discussed below in terms of each network.
Fig. 1.
The proposed framework for joint localization and classification in BUS images. FEX: Feature extractor; AC: Attention-based classifier; LA-Net: Lesion-aware network; {}: Extracted multi-scale image feature maps by FEX; : Lesion-aware feature maps produced by LA-Net. GT denotes ground truth.
3.1.1. Feature extractor (FEX)
The FEX is structured hierarchically with a series of convolutional blocks. Given a 2-dimensional BUS image , FEX extracts multi-scale feature maps {} using each convolutional block. The extracted feature maps serve as inputs for LA-Net, while only the topmost extracted feature map is used as input for AC. FEX can be described as:
| (1) |
where represents the mapping function of FEX parameterized by .
3.1.2. Lesion-aware network (LA-net)
As shown in Fig. 2(a), the LA-Net architecture comprises a convolutional block attention module (CBAM) and a feature fusion module. The motivation for this design is driven by the versatility of integrating CBAM into diverse CNNs, seamlessly enhancing both classification and localization performance (Woo et al., 2018). In addition, CBAM can be trained end-to-end along with the base CNNs (Woo et al., 2018). The extracted multi-scale feature maps {} from the FEX are first exploited by CBAM and then fused by the feature fusion module to predict lesion location. As shown in Fig. 2(b), CBAM includes a channel attention module (CAM, shown in Fig. 2(c)) and a spatial attention module (SAM, shown in Fig. 2(d)) to sequentially process the input feature maps. Both CAM and SAM rely on an information bottleneck design (Tishby and Zaslavsky, 2015) to effectively compress the input feature maps and selectively highlight the discriminative channel-wise and spatial relationships in the input features.
Fig. 2.
(a) Network architecture of lesion-aware network (LA-Net), including a CBAM and a feature fusion module. {} are multi-scale feature maps extracted from FEX. (b) Architecture of the CBAM module. CAM denotes the channel attention module, SAM denotes the spatial attention module, and denotes element-wise multiplication. (c) Architecture of the CAM module. MaxPool involves applying maximum pooling to a feature map along the width and height axes to output a vector; AvgPool represents average pooling done in the same way. denotes element-wise addition. (d) Architecture of the SAM module. Channel MaxPool applies maximum pooling to a multi-channel feature map along the channel axis to output a single-channel feature map. Channel AvgPool applies average pooling similarly.
The architecture of the feature fusion module is illustrated in Fig. 2(a). The first convolutional layer compresses the multiple channels of the input feature map into a single channel, distilling the learned knowledge to emphasize regions of interest (ROIs) relevant to the lesion. Each compressed intermediate feature map is resized to match the dimension of feature and concatenated into a merged feature map with dimensions . Here denotes the size of . The merged feature map is processed by a sequence of a convolutional layer, a batch normalization (BN) layer, and a sigmoid activation layer. The output is the lesion-aware feature map , with each pixel reflecting its probability of belonging to either the lesion or background. The predicted lesion location mask is determined via a binary function with a confidence threshold of 0.5.
| (2) |
| (3) |
where represents the probability of each pixel on belonging to the lesion, represents the mapping function of the LA-Net, which is parameterized by trainable parameters and binarize(.) is a binary function.
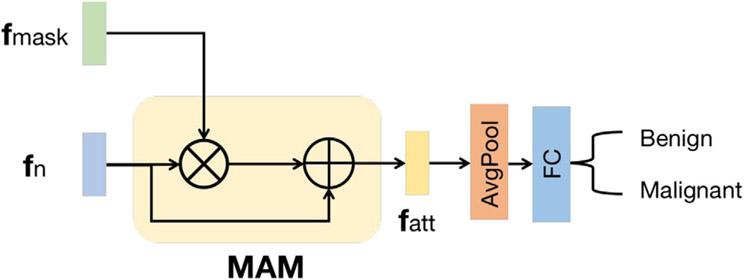
3.1.3. Attention-based classifier (AC)
The AC includes a mask-attention module (MAM) and a classification head, as shown in Fig. 3. The feature map extracted from the last layer of the FEX is enriched by the lesion-aware feature map through MAM. The MAM module consists of a feature element-wise multiplication layer and a skip-connected element-wise addition layer. It directly enhances the features for classification by leveraging the location information captured by LA-Net. This design enables the learned features to emphasize the lesion region and downplay negative impacts due to the noise or artifacts in the non-lesion regions of BUS images. The classification head has an average pooling layer and a fully-connected layer. It employs the enhanced feature map to predict the probability of an input BUS image belonging to each of the K classes. The classification process can be described as:
| (4) |
| (5) |
where is the mapping function of the classifier parameterized by , denotes element-wise multiplication, is the enhanced feature map, and represents the probability of an input BUS image belonging to each of the K classes.
Fig. 3.
Network architecture of attention-based classifier (AC). is the top extracted feature map of the FEX, is the lesion-aware feature map learned by LA-Net, is the enhanced feature map, MAM is the mask attention module in which represents element-wise multiplication, and represents element-wise addition.
3.2. Sequential semi-supervised learning strategy
A two-stage sequential semi-supervised training strategy is introduced to train the framework to achieve optimal classification and localization performance. In the first stage, the FEX and LA-Net are jointly trained only using images with lesion location labels. The loss function measures the geometric disparity between the predicted and ground-truth (GT) lesion locations:
| (6) |
where represents the probabilities of pixels on belonging to the lesion, and represents the GT lesion location mask.
In the second stage, the FEX, LA-Net, and AC are jointly trained in a semi-supervised learning strategy using all training samples, including those without lesion location labels. Notably, the well-trained LA-Net and FEX in the first stage are utilized in the second stage to improve training stability. A hybrid loss employed for model training in this stage can be described as:
| (7) |
where measures the discrepancy between the GT class labels and predicted class labels from classifier ; is defined as a hybrid localization loss that integrates the losses from training samples with and without location labels, denoted and ; is a weighting factor to regulate the contribution of and ; and , , and represent the trainable parameters of FEX, AC, and LA-Net, respectively.
Particularly, in represents the probabilities of input data belonging to each of the K classes and is a one-hot GT label vector with K elements. In , has the same meaning as in Eq. (3), in mirrors but is predicted from unlabeled data. is the pseudo lesion location label determined by applying a binarize function on when the image’s predicted class probability is higher than a predefined classification confidence threshold . The parameter is a coefficient controlling the contributions from losses of and . It reduces the contribution of samples without location labels in the localization loss during model training and mitigates the negative impact of potentially inaccurate pseudo labels. The hybrid loss design in the two-stage training strategy optimizes the AC using the localization information captured by the LA-Net. Furthermore, it allows the LA-Net to learn discriminative features guided by classification outcomes when the image’s location label is absent. In this study, cross-entropy was used for , , and . The detailed training process is described in Algorithm 1.
4. Dataset & method implementation
4.1. Dataset
The proposed method was evaluated on two breast ultrasound image datasets. Notably, in this study, we referred to image samples with both a lesion location and lesion-type labels as fully-labeled samples, while samples annotated only with lesion-type labels were termed partly-labeled samples. Accordingly, a dataset containing only fully-labeled samples was classified as a fully-labeled dataset, while a dataset comprising both fully-labeled and partly-labeled samples was categorized as a partly-labeled dataset.
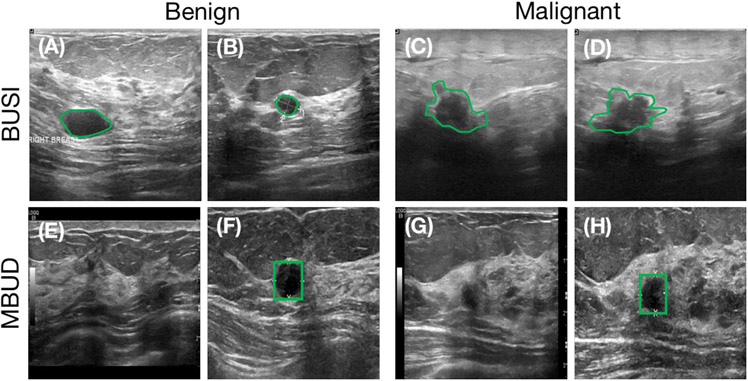
The first utilized dataset was the Breast Ultrasound Image (BUSI) Dataset, a publicly available resource provided by Al-Dhabyani et al. (2020). This database consists of 780 images collected from 600 patients, each of which includes labeled lesion segmentation contour and lesion type. Consequently, this dataset is considered fully-labeled. Further details regarding this dataset are shown in Table 1, with four example images shown in the top row of Fig. 4 (panels A to D).
Table 1.
Details of the BUSI dataset.
| Class label | Training images | Testing images |
|---|---|---|
| Benign | 397 | 40 |
| Malignant | 170 | 40 |
Fig. 4.
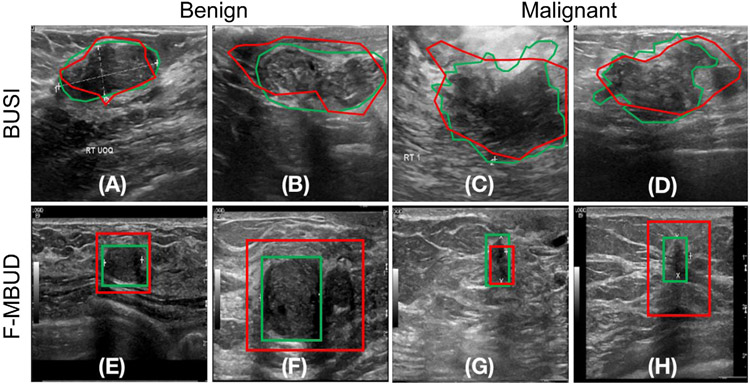
Example images from the BUSI dataset (top row) and MBUD dataset (bottom row). The green contours indicate annotated lesion masks. (A) and (B): BUSI dataset examples with benign lesions. (C) and (D): BUSI dataset examples with malignant lesions. (E) and (F): MBUD dataset examples with benign lesions. Lesion locations are not available for this image (E). (G) and (H): MBUD dataset examples with malignant lesions. Lesion locations are not available for this image (H).
The second dataset, termed the Mayo Clinic Breast Ultrasound Data (MBUD), was collected from the Department of Radiology at Mayo Clinic. This dataset comprises 160 scanning videos collected from 136 patients using a LOGIQ E9 ultrasound system. A total of 22202 two-dimensional images were extracted from these videos and annotated with class labels. Bounding boxes were employed to annotate lesion locations in 384 of these images. Therefore, the MBUD dataset is considered partly-labeled. Four example images from this dataset are presented in the bottom row of Fig. 4 (panels E to H). Specifically, the subset of MBUD images with both location and class labels was regarded as a fully-labeled dataset, denoted as F-MBUD. A comprehensive overview of the MBUD and F-MBUD datasets is shown in Table 2.
Table 2.
Details of the MBUD dataset. The numbers within brackets indicate the counts of images in the fully-labeled subset F-MBUD dataset.
| Class label | Training images | Testing images |
|---|---|---|
| Benign | 13440 (220) | 2435 (36) |
| Malignant | 4954 (108) | 1373 (20) |
4.2. Network architecture
The standard residual neural networks (ResNets) (He et al., 2016) are used as the FEX in the proposed framework. ResNet consists of multiple residual blocks with shortcut connections between nonadjacent convolutional layers. This architecture design addresses the issues of gradient vanishing and improves training stability. ResNet has gained popularity in BUS image classification tasks due to its effectiveness and has demonstrated promising performance in previous studies (Ding et al., 2022; Mishra et al., 2021). Four multi-scale feature maps {} are extracted from the intermediate residual blocks of FEX with dimensions of {}, respectively. Here, denotes the number of channels in the ith extracted feature map, is the size of the ith extracted feature map, and . Each residual block down-samples the feature map size by half: . Within LA-Net, CBAM is applied individually to each feature map extracted from FEX, following the original CBAM network configurations described in Woo et al. (2018). In AC, MAM is parameter-free and the FC layer has two neurons corresponding to binary classification.
4.3. Settings for framework training and testing
The BUSI dataset and MBUD dataset were employed to train and evaluate our framework separately. The total number of images used for model training and testing is shown in Table 1 (BUSI) and Table 2 (MBUD). The training images were randomly divided into a training subset (85%) and a validation subset (15%). The parameters of pretrained ResNets on the ImageNet dataset (Deng et al., 2009) were used to initialize the FEX (Wightman, 2019). For initializing the LA-Net’s parameters, the Xavier initialization scheme was applied (Glorot and Bengio, 2010).
During each training epoch, a mini-batch of 8 images was randomly sampled from the training subset. The input images were sequentially preprocessed with data augmentation techniques, including random rotation of [90°, 180°, 270°], random horizontal and vertical flips, and random color jittering. The preprocessed images were resized to 256 × 256 pixels using bilinear interpolation. Subsequently, the resized images were normalized to the range of [0, 1] and then served as the model input. The loss functions defined in Section 3.2 were calculated to optimize the model parameters using the Adam stochastic gradient algorithm (Kingma and Ba, 2015) with an initial learning rate of . Noted that in cases where the input images lacked location labels, pseudo location labels were generated by setting the classification confidence threshold to 0.8 as described in Section 3.2. The weighing coefficients and in Eq. (7) were set to 0.1 and 0.5, respectively.
| Algorithm 1: Minibatch training of the proposed framework | |
|---|---|
|
|
The framework was trained with the training subset for 100 epochs. After each epoch, the trained model was evaluated with the validation subset. The model weights with the highest validation accuracy were further assessed using the testing images, which have both class and location labels, as shown in Table 1 (BUSI) and Table 2 (MBUD). To assess the training stability, the procedure described above was repeated three times. The proposed framework was implemented by use of PyTorch 1.7.0 (Paszke et al., 2019). The training and validation processes were executed using Nvidia GeForce GTX 1080ti GPUs.
4.4. Other methods for comparison
In this study, we have implemented and compared the proposed method with two comparative MTL methods, four single-task classification methods, and two single-task segmentation methods. The four single-task classification methods include ResNet18 (R18) (He et al., 2016), ResNet50 (R50) (He et al., 2016), EfficientNet (EB0) (Tan and Le, 2019), and vision transformer (ViT) (Dosovitskiy et al., 2020)). The two classic single-task segmentation methods are U-Net (Ronneberger et al., 2015) and DeepLabV3 (Chen et al., 2017)). Additionally, we evaluated two recently proposed MTL methods: RMTL (Rasaee and Rivaz, 2021) and attention gated network (AGN) (Schlemper et al., 2019)). RMTL is a typical shared-trunk MTL method tailored for BUS images. Similar to our method, it can employ various CNN-based networks as its feature extraction backbone. In the experimental setup, both ResNet50 and ResNet18 served as the feature extraction backbone for our method and RMTL, resulting in methods named R50+LA-Net, R18+LA-Net, R50+RMTL and R18+RMTL, respectively.
4.5. Performance evaluation metrics
The evaluation of the proposed method encompasses both classification and localization performance. Accuracy, precision, recall, -score, and area under the ROC curve (AUC) were employed as the metrics to evaluate the classification performance. Two commonly-used segmentation metrics, intersection over union (IoU) and dice score, were employed to evaluate the localization performance. When using the MBUD dataset with bounding boxes as location labels, the predicted lesion mask obtained from the LA-Net was transformed into a bounding box before evaluating the localization performance. The metrics are defined as follows:
| (8) |
where means the number of true positive samples, means the number of false negative samples, means the number of false positive samples, means the number of true negative samples, means the predicted lesion area, means the GT lesion area, means the intersection area of and , and means the union area of and .
5. Experimental results
5.1. Performance of our model trained with partly-labeled MBUD dataset
The MBUD dataset described in Section 4.1 was employed to evaluate model performance for classification and localization. The study compared the proposed method to eight comparative methods, as described in Section 4.4. Specifically, the single-task classification methods, including ViT, EB0, R18, and R50, were directly trained using all samples with complete class labels within the MBUD dataset. Meanwhile, two single-task segmentation methods (U-Net and DeepLabV3) and two MTL methods (AGN and RMTL) were trained on the fully-labeled F-MBUD subset. All methods were evaluated using the MBUD testing set, as shown in Table 2.
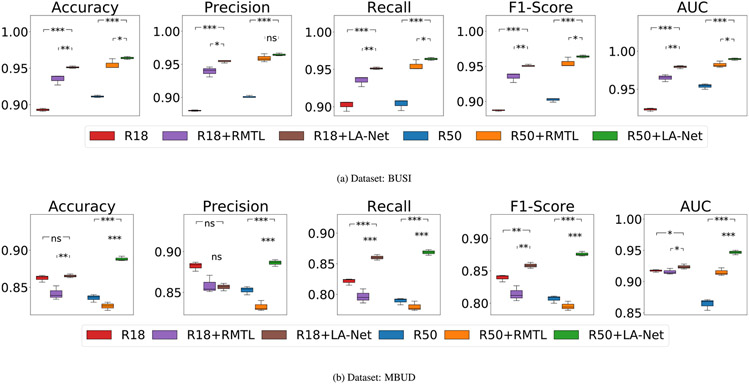
The classification results are shown in Table 3. The proposed method (#11), using ResNet50 as the FEX, achieved superior classification outcomes compared to all other comparative MTL and single-task classification methods. Particularly, the comparative MTL methods (#5 to #7 in Table 3) can only be trained using the fully-labeled F-MBUD subset, but not the large amounts of images without location labels (a total of 18066 images) in the MBUD dataset. They showed lower classification performance than our method and even the other four single-task classification methods. It indicates the susceptibility of these methods to overfitting due to the limited training data. Comparatively, our methods trained on the F-MBUD subset (#8 and #9 of Table 3) still outperformed AGN and RMTL methods. It demonstrates the capacity of our method in mitigating overfitting risk even when trained on the small F-MBUD dataset. These results show the effectiveness of our network design in improving classification performance and reducing overfitting by exploiting the limited lesion location information in the dataset and being trained with the semi-supervised learning strategy.
Table 3.
Classification performance of the proposed method and comparison methods on the partly-labeled MBUD dataset.
| ID | Method | Dataset | Accuracy | Precision | Recall | -Score | AUC | p-value |
|---|---|---|---|---|---|---|---|---|
| #1 | ViT | MBUD | 0.847 ± 0.048 | 0.863 ± 0.044 | 0.804 ± 0.065 | 0.821 ± 0.063 | 0.913 ± 0.034 | *** |
| #2 | EB0 | MBUD | 0.868 ± 0.007 | 0.879 ± 0.044 | 0.845 ± 0.051 | 0.849 ± 0.004 | 0.912 ± 0.018 | *** |
| #3 | R18 | MBUD | 0.862 ± 0.012 | 0.882 ± 0.014 | 0.821 ± 0.014 | 0.839 ± 0.014 | 0.917 ± 0.005 | *** |
| #4 | R50 | MBUD | 0.836 ± 0.013 | 0.853 ± 0.013 | 0.789 ± 0.014 | 0.807 ± 0.015 | 0.864 ± 0.023 | *** |
| #5 | AGN | F-MBUD | 0.783 ± 0.044 | 0.790 ± 0.089 | 0.731 ± 0.041 | 0.744 ± 0.046 | 0.834 ± 0.034 | *** |
| #6 | R18+RMTL | F-MBUD | 0.577 ± 0.080 | 0.645 ± 0.015 | 0.636 ± 0.024 | 0.575 ± 0.082 | 0.719 ± 0.102 | *** |
| #7 | R50+RMTL | F-MBUD | 0.710 ± 0.084 | 0.739 ± 0.179 | 0.674 ± 0.093 | 0.662 ± 0.029 | 0.789 ± 0.092 | *** |
| #8 | R18+LA-Net (Ours) | F-MBUD | 0.759 ± 0.071 | 0.750 ± 0.043 | 0.766 ± 0.038 | 0.750 ± 0.062 | 0.829 ± 0.016 | *** |
| #9 | R50+LA-Net (Ours) | F-MBUD | 0.827 ± 0.090 | 0.816 ± 0.097 | 0.816 ± 0.054 | 0.814 ± 0.081 | 0.901 ± 0.030 | *** |
| #10 | R18+LA-Net (Ours) | MBUD | 0.865 ± 0.006 | 0.857 ± 0.011 | 0.860 ± 0.011 | 0.858 ± 0.011 | 0.924 ± 0.010 | – |
| #11 | R50+LA-Net (Ours) | MBUD | 0.888 ± 0.010 | 0.886 ± 0.010 | 0.869 ± 0.011 | 0.876 ± 0.009 | 0.947 ± 0.013 | – |
Notes: In this experiment, both ResNet18 and ResNet50 were used as FEXs in our framework. The input image size was 256 × 256 pixels. The results are represented as a mean value with a ±95% confidence interval. The p-value shows the t-test result of comparing the AUC of a comparative method with our method’s. The null hypothesis is that the AUC of the comparative method is larger than or equal to our method’s. The symbols *, **, and * * * indicate that the p-value is less than 0.05, 0.01, and 0.001, respectively.
The localization results are shown in Table 4. Our method, using ResNet18 as the FEX, demonstrated better localization performance than all other comparative methods. This result indicates that the depth of the FEX network has varying impacts on model performance in terms of localization and classification; however, a deeper network does not necessarily guarantee better performance. Interestingly, the localization performance of our method trained on the small yet fully-labeled F-MBUD subset (#8 in Table 4) was better than that achieved with the large yet partly-labeled MBUD dataset (#6 in Table 4). This phenomenon may arise due to the inclusion of large amounts of images without location labels in model training, and a more in-depth analysis of this aspect is discussed in Section 5.3.2.
Table 4.
Localization performance of the proposed method and comparison methods on the partly-labeled MBUD dataset.
| ID | Network | Dataset | IoU | Dice | p-value |
|---|---|---|---|---|---|
| #1 | U-Net | F-MBUD | 0.445 ± 0.019 | 0.549 ± 0.009 | * |
| #2 | DeepLabV3 | F-MBUD | 0.441 ± 0.013 | 0.559 ± 0.020 | * |
| #3 | AGN | F-MBUD | 0.442 ± 0.022 | 0.526 ± 0.016 | * |
| #4 | R18+RMTL | F-MBUD | 0.377 ± 0.032 | 0.521 ± 0.030 | *** |
| #5 | R50+RMTL | F-MBUD | 0.383 ± 0.055 | 0.517 ± 0.057 | ** |
| #6 | R18+LA-Net (Ours) | MBUD | 0.410 ± 0.014 | 0.532 ± 0.019 | ** |
| #7 | R50+LA-Net (Ours) | MBUD | 0.382 ± 0.012 | 0.506 ± 0.022 | *** |
| #8 | R18+LA-Net (Ours) | F-MBUD | 0.449 ± 0.022 | 0.580 ± 0.035 | – |
| #9 | R50+LA-Net (Ours) | F-MBUD | 0.406 ± 0.035 | 0.534 ± 0.040 | – |
Notes: In this experiment, Both ResNet18 and ResNet50 were used as the FEXs. The input image size was 256 × 256 pixels. The p-value shows the t-test result of comparing the IoU of a comparison method with our method’s. The null hypothesis is that the IoU of the comparison method is larger than or equal to our method’s. The meaning of the p-values is the same as described in Table 3.
5.2. Performance of our method trained with fully-labeled datasets
In order to individually investigate the effectiveness of the designed framework architecture, we trained our method and other comparative methods described in Section 4.4 using the same fully-labeled datasets and compared their performance. Two fully-labeled training datasets, BUSI and F-MBUD, were employed for model training. The classification results are shown in Table 5. When using ResNet50 as the FEX, our method achieved significantly higher performance than all other comparison methods in terms of all classification evaluation metrics on both datasets. This result indicates that the integration of LA-Net and AC designs enables the utilization of localization information to improve classification performance. In addition, our model employing ResNet50-based FEX outperformed that using ResNet18 as the FEX in terms of classification performance in both datasets. It indicates that the depth of the FEX network influences the classification performance. A more detailed discussion can be found in Section 5.3.3 below. Interestingly, the classification performance of the comparative RMTL method trained with the F-MBUD dataset was even lower than that of the vanilla ResNets. One possible reason is that RMTL training demands pixel-level segmented lesion contours, while the F-MBUD dataset contains only lesion bounding box annotations.
Table 5.
Classification performance of the proposed method and comparison methods on the fully-labeled BUSI and F-MBUD datasets.
| Dataset | Method | Accuracy | Precision | Recall | -Score | AUC | p-value |
|---|---|---|---|---|---|---|---|
| BUSI | ViT | 0.878 ± 0.030 | 0.884 ± 0.014 | 0.878 ± 0.030 | 0.877 ± 0.032 | 0.953 ± 0.019 | *** |
| EB0 | 0.919 ± 0.017 | 0.920 ± 0.017 | 0.919 ± 0.017 | 0.919 ± 0.017 | 0.961 ± 0.007 | *** | |
| R18 | 0.886 ± 0.017 | 0.899 ± 0.030 | 0.886 ± 0.017 | 0.885 ± 0.017 | 0.964 ± 0.012 | *** | |
| R50 | 0.939 ± 0.001 | 0.940 ± 0.003 | 0.939 ± 0.001 | 0.939 ± 0.001 | 0.967 ± 0.004 | *** | |
| AGN | 0.898 ± 0.047 | 0.901 ± 0.042 | 0.898 ± 0.047 | 0.898 ± 0.047 | 0.933 ± 0.020 | – | |
| R18+RMTL | 0.935 ± 0.017 | 0.939 ± 0.019 | 0.935 ± 0.017 | 0.935 ± 0.017 | 0.965 ± 0.011 | *** | |
| R50+RMTL | 0.955 ± 0.017 | 0.959 ± 0.014 | 0.955 ± 0.017 | 0.955 ± 0.017 | 0.982 ± 0.010 | * | |
| R18+LA-Net (Ours) | 0.951 ± 0.001 | 0.955 ± 0.006 | 0.951 ± 0.001 | 0.951 ± 0.001 | 0.980 ± 0.005 | *** | |
| R50+LA-Net (Ours) | 0.964 ± 0.001 | 0.964 ± 0.003 | 0.964 ± 0.001 | 0.964 ± 0.001 | 0.989 ± 0.004 | – | |
| F-MBUD | ViT | 0.905 ± 0.026 | 0.913 ± 0.075 | 0.889 ± 0.039 | 0.894 ± 0.014 | 0.970 ± 0.007 | * |
| EB0 | 0.881 ± 0.026 | 0.879 ± 0.031 | 0.859 ± 0.031 | 0.867 ± 0.029 | 0.915 ± 0.018 | *** | |
| R18 | 0.893 ± 0.001 | 0.880 ± 0.001 | 0.902 ± 0.017 | 0.887 ± 0.003 | 0.923 ± 0.005 | *** | |
| R50 | 0.911 ± 0.001 | 0.901 ± 0.004 | 0.904 ± 0.018 | 0.902 ± 0.007 | 0.954 ± 0.009 | ** | |
| AGN | 0.890 ± 0.030 | 0.891 ± 0.030 | 0.890 ± 0.030 | 0.890 ± 0.030 | 0.938 ± 0.016 | *** | |
| R18+RMTL | 0.833 ± 0.026 | 0.818 ± 0.027 | 0.830 ± 0.030 | 0.822 ± 0.026 | 0.910 ± 0.011 | *** | |
| R50+RMTL | 0.887 ± 0.026 | 0.876 ± 0.027 | 0.905 ± 0.032 | 0.882 ± 0.026 | 0.907 ± 0.014 | *** | |
| R18+LA-Net (Ours) | 0.939 ± 0.026 | 0.929 ± 0.015 | 0.915 ± 0.016 | 0.921 ± 0.003 | 0.979 ± 0.012 | – | |
| R50+LA-Net (Ours) | 0.946 ± 0.001 | 0.947 ± 0.002 | 0.935 ± 0.001 | 0.941 ± 0.001 | 0.969 ± 0.001 | – |
Notes: Two different networks, ResNet18 and ResNet50, were used as the FEXs in our method. The input image size was 256 × 256 pixels. The results are represented as a mean value with a ±95% confidence interval. The p-value shows the t-test result of comparing the AUC of a comparison method with our method’s. The null hypothesis of the AUC of the comparison method is larger than or equal to our method’s. The meaning of the p-values is the same as described in Table 3.
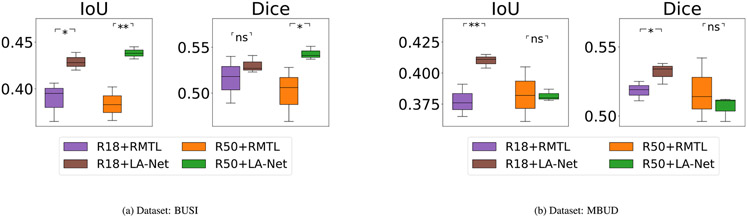
The localization performance of our method and comparative methods are shown in Table 6. When using the F-MBUD dataset, the proposed method using ResNet18-based FEX achieved superior localization performance compared to all other comparison methods, including our method using ResNet50-based FEX. Interestingly, on the BUSI dataset, our method significantly outperformed the other RMTL methods, yet its performance was lower than the other two single-task segmentation methods (U-Net and DeepLabV3). This phenomenon might be attributed to the properties of lesion localization labels. In the BUSI dataset, all lesions are accurately delineated rather than being roughly marked with bounding boxes. Therefore, the extracted lesion-relevant information more precisely captures the intrinsic characteristics of the lesion, rendering it suitable for training single-task segmentation networks. However, there is a possibility of information conflict in the proposed multi-task learning method that might potentially compromise its localization performance. The features learned for classification may not necessarily align with the requirements for accurate localization and even have a detrimental effect on it. On the contrary, in the F-MBUD dataset, the lesion localization labels are in the form of bounding boxes, which poses challenges for traditional single-task segmentation methods in capturing intrinsic lesion shape information. Our method, which leverages classification information to facilitate lesion localization, can alleviate the negative effect of this form of annotation and achieve better localization performance. These results demonstrate the significant effects of lesion localization/segmentation labels on the model performance. Additionally, the varied performance achieved by our method using ResNet50 and ResNet18 as FEXs also implies the possible impact of FEX network depth on localization performance. Eight examples of BUS images for the two datasets are shown in Fig. 5, containing the ground truth and predicted lesion masks.
Table 6.
Localization performance of the proposed method and the comparison methods on the fully-labeled BUSI and F-MBUD datasets.
| Dataset | Network | IoU | Dice | p-value |
|---|---|---|---|---|
| BUSI | U-Net | 0.46S ± 0.032 | 0.580 ± 0.049 | ns |
| DeepLabV3 | 0.495 ± 0.016 | 0.593 ± 0.016 | ns | |
| AGN | 0.424 ± 0.022 | 0.526 ± 0.016 | * | |
| R18+RMTL | 0.388 ± 0.052 | 0.515 ± 0.063 | *** | |
| R50+RMTL | 0.384 ± 0.045 | 0.501 ± 0.074 | *** | |
| R18+LA-Net (Ours) | 0.429 ± 0.024 | 0.530 ± 0.023 | – | |
| R50+LA-Net (Ours) | 0.438 ± 0.016 | 0.543 ± 0.018 | – | |
| F-MBUD | U-Net | 0.445 ± 0.019 | 0.549 ± 0.009 | * |
| DeepLabV3 | 0.441 ± 0.013 | 0.559 ± 0.020 | * | |
| AGN | 0.442 ± 0.024 | 0.543 ± 0.022 | * | |
| R18+RMTL | 0.377 ± 0.032 | 0.521 ± 0.030 | *** | |
| R50+RMTL | 0.383 ± 0.055 | 0.517 ± 0.057 | ** | |
| R18+LA-Net (Ours) | 0.449 ± 0.022 | 0.580 ± 0.035 | – | |
| R50+LA-Net (Ours) | 0.406 ± 0.035 | 0.534 ± 0.040 | – |
Notes: Two difference networks, ResNet18 and ResNet50, were used as FEXs in our method. The input image size was 256 × 256 pixels. The results are represented as a mean value with a ±95% confidence interval. The p-value shows the t-test result of comparing the IoU of the comparison method with our method’s. The null hypothesis is that the IoU of the comparison method is larger than or equal to our method’s. Here ns represents p-value > 0.05 (no significant improvement). The symbols *, ** and * * * indicate the p-value is less than 0.05, 0.01 and 0.01, respectively.
Fig. 5.
Examples of predicted lesion masks using our method in the BUSI dataset (top row) and F-MBUD dataset (bottom row). The green contours indicate the GT lesion locations, while the red contours show the predicted lesion locations. (A) and (B) are two benign examples from the BUSI dataset. (C) and (D) are two malignant examples from the BUSI dataset. (E) and (F) are two benign examples from the F-MBUD dataset. (G) and (H) are two malignant examples from the F-MBUD dataset.
5.3. Ablation studies
5.3.1. Impact of attention modules on model performance
An ablation experiment was conducted to investigate the impact of each attention module (CAM, SAM, and MAM) on the overall model performance. After individually removing each attention module, the model was re-trained while maintaining the parameter settings described in Section 4.3. Both the fully-labeled BUSI dataset and partly-labeled MBUD dataset were used for model training. Table 7 and Table 8 demonstrated the model performance with respect to classification and localization, respectively. The original model containing all three attention modules achieved superior classification and localization performance compared to the other ablated models. Particularly, the classification performance of the model without MAM was significantly reduced, while the model without CAM or SAM showed greatly degraded localization performance. The results demonstrate that the three different attention modules contribute differently to the model performance. Specifically, CAM and SAM in the LA-Net exploit the essential channel-wise and lesion-related spatial relationships in the input features, therefore playing an important role in localization performance. Differently, MAM is crucial for leveraging lesion localization information to assist in the classification.
Table 7.
Impacts of three attention modules on classification performance.
| Dataset | CAM | SAM | MAM | Accuracy | Precision | Recall | -Score | AUC | p-value |
|---|---|---|---|---|---|---|---|---|---|
| BUSI | ✗ | ✗ | ✗ | 0.939 ± 0.001 | 0.940 ± 0.003 | 0.939 ± 0.001 | 0.939 ± 0.001 | 0.967 ± 0.004 | *** |
| ✗ | ✓ | ✓ | 0.939 ± 0.001 | 0.939 ± 0.001 | 0.939 ± 0.001 | 0.939 ± 0.001 | 0.973 ± 0.017 | ** | |
| ✓ | ✗ | ✓ | 0.939 ± 0.001 | 0.944 ± 0.007 | 0.939 ± 0.001 | 0.939 ± 0.001 | 0.975 ± 0.011 | ** | |
| ✓ | ✓ | ✗ | 0.911 ± 0.018 | 0.920 ± 0.004 | 0.911 ± 0.018 | 0.910 ± 0.017 | 0.968 ± 0.008 | *** | |
| ✓ | ✓ | ✓ | 0.964 ± 0.001 | 0.965 ± 0.002 | 0.964 ± 0.001 | 0.964 ± 0.001 | 0.989 ± 0.004 | – | |
| MBUD | ✗ | ✗ | ✗ | 0.836 ± 0.013 | 0.853 ± 0.013 | 0.789 ± 0.014 | 0.807 ± 0.015 | 0.864 ± 0.023 | *** |
| ✗ | ✓ | ✓ | 0.824 ± 0.005 | 0.813 ± 0.008 | 0.799 ± 0.003 | 0.805 ± 0.004 | 0.912 ± 0.034 | ** | |
| ✓ | ✗ | ✓ | 0.856 ± 0.024 | 0.864 ± 0.017 | 0.822 ± 0.026 | 0.835 ± 0.027 | 0.906 ± 0.040 | ** | |
| ✓ | ✗ | ✓ | 0.845 ± 0.018 | 0.861 ± 0.019 | 0.801 ± 0.020 | 0.818 ± 0.020 | 0.883 ± 0.050 | ** | |
| ✓ | ✓ | ✓ | 0.888 ± 0.008 | 0.886 ± 0.010 | 0.869 ± 0.011 | 0.876 ± 0.009 | 0.947 ± 0.009 | – |
Notes: The attention module marked with ✗means its removal from our method in the ablation experiment. In this experiment, ResNet50 was used as FEX, and the input image size was 256 × 256 pixels. The outcomes are represented as the mean value ±95% confidence interval. The null hypothesis and meaning of p-values remain consistent with the description in Table 3.
Table 8.
Impacts of three attention modules on localization performance.
| Dataset | CAM | SAM | MAM | IoU | Dice | p-value |
|---|---|---|---|---|---|---|
| BUSI | ✗ | ✓ | ✓ | 0.411 ± 0.024 | 0.521 ± 0.018 | ** |
| ✓ | ✗ | ✓ | 0.413 ± 0.014 | 0.515 ± 0.005 | ** | |
| ✓ | ✓ | ✗ | 0.419 ± 0.010 | 0.528 ± 0.007 | ** | |
| ✓ | ✓ | ✓ | 0.438 ± 0.016 | 0.543 ± 0.018 | – | |
| MBUD | ✗ | ✓ | ✓ | 0.341 ± 0.039 | 0.466 ± 0.056 | ** |
| ✓ | ✗ | ✓ | 0.358 ± 0.024 | 0.468 ± 0.029 | ** | |
| ✓ | ✗ | ✓ | 0.384 ± 0.051 | 0.510 ± 0.059 | ns | |
| ✓ | ✓ | ✓ | 0.406 ± 0.035 | 0.534 ± 0.040 | – |
Notes: The symbol ✗means this attention module was removed in our framework. In this experiment, ResNet50 was used as the FEX, and the input image size was 256 × 256 pixels. The results are represented as a mean value with a ±95% confidence interval. The null hypothesis and meaning of the p-values remain consistent with the description in Table 6.
5.3.2. Impact of sequential semi-supervised learning strategy
To explore the gain achieved by using the proposed sequential semi-supervised learning (s-SSL) strategy, an ablation study was conducted using the MBUD dataset. Our method was trained separately using two approaches: employing the partly-labeled MBUD dataset with the s-SSL strategy and using only the fully-labeled F-MBUD subset without s-SSL. For comparison, the RMTL method was also trained with and without the adapted s-SSL strategy. As shown in Table 9, using s-SSL significantly improved the classification performance of both our method and the RMTL method. This success highlights the effectiveness of the s-SSL strategy in MTL by efficiently exploiting the potential of images without location labels. Conversely, this s-SSL strategy decreased the localization performance of both our method and the RMTL method, as seen in Table 10. This phenomenon implies that there is a trade-off that both our and the RMTL models tend to sacrifice the localization performance to pursue optimal classification performance when large amounts of class labels rather than lesion location labels are used for model training. One potential explanation is that the distribution of lesion locations in the unlabeled images varies from that of the labeled images. These MTL methods might exploit inconsistent lesion location information from unlabeled images, subsequently impacting their localization performance when tested on the labeled images. Note that our method demonstrated a more moderate decrease in localization performance when compared to the RMTL method. This result indicates the efficacy of our method in managing and mitigating the inherent optimization conflicts between the two tasks.
Table 9.
Impact of sequential semi-supervised learning strategy on classification performance.
| Method | s-SSL | Accuracy | Precision | Recall | -Score | AUC | p-value |
|---|---|---|---|---|---|---|---|
| R18+RMTL | ✗ | 0.577 ± 0.080 | 0.645 ± 0.015 | 0.636 ± 0.024 | 0.575 ± 0.082 | 0.719 ± 0.102 | *** |
| ✓ | 0.842 ± 0.023 | 0.859 ± 0.027 | 0.797 ± 0.029 | 0.814 ± 0.029 | 0.916 ± 0.012 | – | |
| R50+RMTL | ✗ | 0.710 ± 0.085 | 0.739 ± 0.179 | 0.674 ± 0.093 | 0.662 ± 0.029 | 0.789 ± 0.092 | ** |
| ✓ | 0.825 ± 0.014 | 0.833 ± 0.016 | 0.780 ± 0.020 | 0.795 ± 0.018 | 0.915 ± 0.016 | – | |
| R18+LA-Net | ✗ | 0.759 ± 0.071 | 0.750 ± 0.043 | 0.766 ± 0.038 | 0.750 ± 0.062 | 0.829 ± 0.016 | *** |
| ✓ | 0.865 ± 0.006 | 0.857 ± 0.011 | 0.860 ± 0.011 | 0.858 ± 0.011 | 0.924 ± 0.010 | – | |
| R50+LA-Net | ✗ | 0.827 ± 0.090 | 0.816 ± 0.097 | 0.816 ± 0.055 | 0.814 ± 0.081 | 0.901 ± 0.030 | * |
| ✓ | 0.888 ± 0.008 | 0.886 ± 0.010 | 0.869 ± 0.011 | 0.876 ± 0.009 | 0.947 ± 0.009 | – |
Notes: This experiment was conducted on the partly-labeled MBUD dataset. The symbols ✓ and ✗show whether the method is trained with or without our sequential semi-supervised learning strategy. ResNet50 was used as FEX, and the input image size was 256 × 256 pixels. The outcomes are represented as mean value ±95% confidence interval. The null hypothesis and meaning of p-values remain consistent with the description in Table 3.
Table 10.
Impact of sequential semi-supervised learning strategy on localization performance.
| Dataset | s-SSL | IoU | Dice | p-value |
|---|---|---|---|---|
| R18+RMTL | ✗ | 0.377 ± 0.032 | 0.518 ± 0.017 | ns |
| ✓ | 0.255 ± 0.042 | 0.367 ± 0.038 | – | |
| R50+RMTL | ✗ | 0.383 ± 0.055 | 0.517 ± 0.058 | ns |
| ✓ | 0.310 ± 0.026 | 0.415 ± 0.023 | – | |
| R18+LA-Net | ✗ | 0.449 ± 0.022 | 0.580 ± 0.037 | ns |
| ✓ | 0.410 ± 0.014 | 0.532 ± 0.019 | – | |
| R50+LA-Net | ✗ | 0.406 ± 0.035 | 0.534 ± 0.040 | ns |
| ✓ | 0.382 ± 0.012 | 0.506 ± 0.022 | – |
Notes: This experiment was conducted on the partly-labeled MBUD dataset. The symbols ✓ and ✗show whether the method is trained with or without the sequential semi-supervised learning strategy. ResNet50 was used as FEX, and the input image size was 256 × 256 pixels. The outcomes are represented as mean value ±95% confidence interval. The null hypothesis and meaning of p-values remain consistent with the description in Table 6.
5.3.3. Impact of FEX network depth
To assess the influence of the FEX architecture, the performance of models with ResNet18-based and ResNet50-based FEXs was evaluated on both BUSI and MUBD datasets and compared to the vanilla ResNet18 and ResNet50. As demonstrated in Fig. 6, our method was effective in improving classification performance using both ResNet architectures on both datasets. Using ResNet50 as the FEX generally achieved superior performance than that of using ResNet18. For comparison, the same experiment was also conducted for the RMTL method, which similarly employs ResNets as its feature extraction backbone. While RMTL showed higher performance over the Vanilla ResNet on the BUSI dataset, it suffered degraded performance when using the MBUD dataset, as shown in Fig. 6(b). One possible reason for this discrepancy is that the RMTL method suffers high overfitting issues when it can only be trained with a small number of fully-labeled samples. Our method, however, significantly alleviates the overfitting problem by fully utilizing a large number of images without location labels.
Fig. 6.
Ablation study to investigate the impact of FEX network depth on classification performance using (a) BUSI and (b) MBUD datasets. ResNet18 (R18) and ResNet50 (R50) were used as FEXs in our method and the backbone of the RMTL method. The input image size was 256 × 256 pixels. The statistical significance symbol indicates the t-test result of the selected pair of methods. The null hypothesis is that the average metric of the left method is less than or equal to the right method. The meaning of p-values is the same as that described in Table 3.
Similarly, Fig. 7(a) shows that our method consistently led to higher localization performance than the RMTL method on the BUSI dataset. However, Fig. 7(b) exhibits divergent outcomes for the MBUD dataset. Our method using ResNet50 as the FEX did not notably outperform the RMTL method, as well as our method with ResNet18-based FEX, in terms of localization performance. These results imply that adopting a deeper network as the FEX in our method might be inadvertently influenced by large-scale unlabeled images, therefore compromising the model’s capacity to predict accurate true location distribution and leading to the similar trade-off discussed in Section 5.3.2.
Fig. 7.
Ablation study to investigate the impact of FEX network depth on localization performance using (a) BUSI and (b) MBUD datasets. The network and parameter settings are the same as Fig. 6.
5.3.4. Impact of the number of partial-labeling training samples
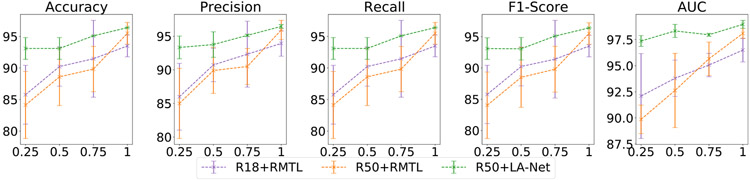
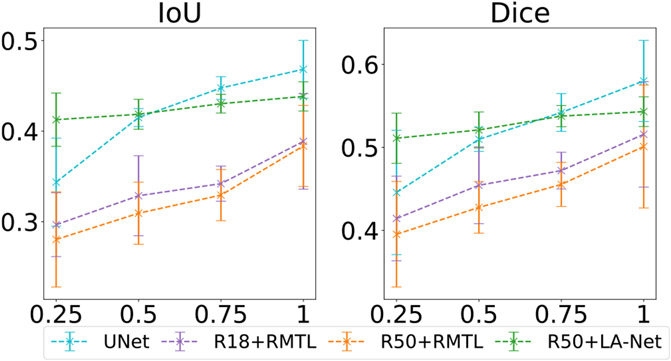
We conducted experiments to understand the impact of the number of partly-labeled training samples on model performance. As defined in Section 4.1, partly-labeled samples represent those with lesion-type labels but no corresponding location labels. Using the fully-labeled BUSI dataset, we randomly retained 25%, 50%, and 75% of the location labels in the dataset and discarded the rest to create four partly-labeled datasets. Our method was individually trained with these four partly-labeled datasets. For comparison, a single-task segmentation method (U-Net) and two RMTL methods were trained using the fully-labeled images from each of the four partly-labeled datasets. As shown in Fig. 8, our method achieved greater classification performance than the three compared methods across all four partly-labeled datasets. The classification performance showed improvement with an increased number of fully-labeled training samples. Notably, our method showed less performance degradation along with the decrease of partly-labeled training samples. A similar trend was observed in the localization performance of our method, as shown in Fig. 9. These results indicate that our method can enhance model performance and reduce the risk of overfitting for datasets with varying amounts of location labels compared to the RMTL methods.
Fig. 8.
Classification performance of the proposed method on BUSI-derived datasets with varied numbers of location labels. The numbers 25%, 50%, 75%, and 100% on the x-axis represent the ratio of training samples with location labels compared to the original BUSI dataset in which all samples have both classification and localization labels. Each point represents the mean of each metric at the given labeling ratio, and the error bar shows its 95% confidence interval. In this experiment, ResNet50 was used as the FEX, and the training image size was 256 × 256 pixels.
Fig. 9.
Localization performance of the proposed method on BUSI-derived datasets with varied numbers of location labels. The detailed description is the same as Fig. 8.
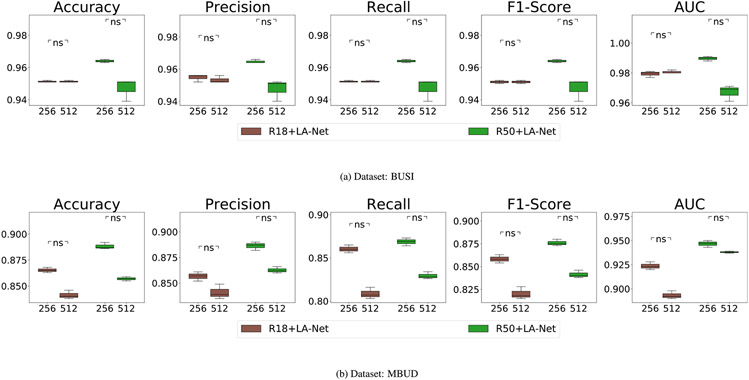
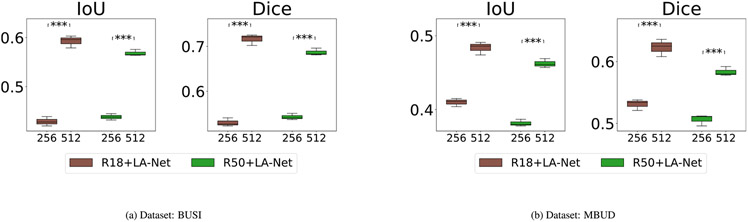
5.3.5. Impact of training image size
The influence of training image size on model performance was explored using the BUSI and MBUD datasets. In this experiment, two different image sizes were tested, including 256 × 256 and 512 × 512 pixels. Both ResNet18 and ResNet50 were used as the FEX in our framework. As shown in Fig. 10, the classification performance achieved by using images with an input size of 256 × 256 pixels was generally higher than that of using images with larger images of 512 × 512 pixels. However, Fig. 11 demonstrates that using larger images of 512 × 512 pixels usually led to significantly better localization performance. These results indicate that the trade-off persists in balancing classification and localization performance and can also be influenced by the sizes of images used.
Fig. 10.
Ablation study to investigate the classification performance of using different training image sizes on (a) BUSI and (b) MBUD dataset. The numbers shown on the x-axis represent the input image sizes, either 356 × 256 or 512 × 512 pixels. The null hypothesis and meaning of p-values remain consistent with the description in Fig. 6.
Fig. 11.
Ablation study to investigate the localization performance of using different input sizes on (a) BUSI and (b) MBUD dataset. The detailed description is the same as Fig. 10.
5.4. Classification result interpretation through visualization of the discriminative region
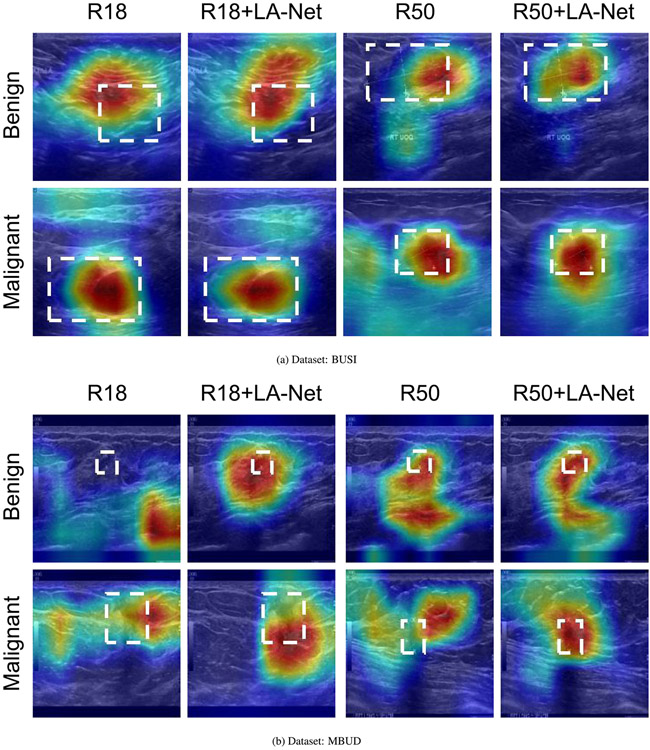
Class activation mapping (Selvaraju et al., 2017; Zhou et al., 2016) has been broadly employed to interpret classification networks across various applications, including BUS image analysis (Byra et al., 2022; Ding et al., 2022). This technique enhances the interpretability of model performance by visualizing the areas that contribute most to the classification decision. Gradient-weighted class activation mapping (Grad-CAM) (Selvaraju et al., 2017) is an effective and applicable class activation mapping technique for highlighting potential ROIs based on the gradient score of each class. In this study, Grad-CAM was utilized to identify the discriminative lesion regions within BUS images. The proposed method and the vanilla ResNet18 and ResNet50 were subjected to Grad-CAM analysis. The generated Grad-CAM examples are shown in Fig. 12. It is observed that the attention regions recognized by our method had a higher degree of overlap with the GT lesion locations. These examples indicate that the LA-Net effectively assists the feature extraction process, enabling the FEX to extract more discriminative information from lesion areas. Consequently, our framework reduces the interference from the noisy background and improves the overall classification performance.
Fig. 12.
Class activation maps generated by the ResNet18- and ResNet50-based FEXs of our methods and vanilla ResNets on the BUSI dataset (panel a) and MBUD dataset (panel b). The white dashed rectangles indicate the GT lesion locations. The regions with warmer colors represent higher confidence corresponding to the target class label.
6. Discussion
The MTL framework proposed in this study integrates the advantages of attention mechanisms and a semi-supervised learning strategy to address the challenges for joint localization and classification of breast tumors on BUS images. Multiple attention modules are designed in the framework to explicitly highlight the discriminative features within the lesion region and suppress the negative impact of high image noise and low contrast inherent to BUS images. A mask attention module is a key component of our design, which connects the two task-specific branches to balance the importance of individual tasks and alleviate the potential information-sharing conflicts during model training. The proposed sequential semi-supervised learning strategy enables the model to leverage partly-labeled datasets effectively, greatly reducing the burdens of data annotation. Additionally, the framework is modularized so that each component can be flexibly adapted with suitable architecture to satisfy various demands.
We conducted complete experiments to validate the model performance and compare it against eight state-of-the-art methods using two different BUS image datasets. The comparative analysis results showed the effectiveness of our method in improving model performance across both localization and classification tasks. Furthermore, we also conducted an in-depth exploration of various network-related factors that might have an influence on model performance, including attention modules, the number of location labels, the depth of the feature extractor network, and the input image size. Our ablation studies demonstrated that three attention modules (CAM, SAM, and MAM) have different contributions to the overall model performance. The designed two-stage s-SSL strategy enables our method to leverage large amounts of partly-labeled images to significantly enhance the model performance. Additionally, the insights gained about network depth and training image size can serve as valuable guidance for adapting the method to diverse applications.
Given the inherent flexibility of our modular framework, one future research topic is to investigate the potential improvement of model performance and generalities by upgrading the current network architecture design. The modern network architectures, such as Vision Transformer (Dosovitskiy et al., 2020) and Swin Transformer (Liu et al., 2021a), will be employed as the FEX in our framework to understand their impacts on model performance and robustness when adapted to small BUS datasets. Also, the performance of cutting-edge attention modules, such as non-local attention module (Wang et al., 2018) and multi-head attention module (Vaswani et al., 2017), will be investigated to understand their abilities to address the issues of information-sharing conflict. Recently, the analysis of 3D BUS scanning videos has attracted growing interest in the community (Zhou et al., 2021; Liu et al., 2021b; Dai et al., 2019). Integrating appropriate temporal attention modules (Yan et al., 2019) into the proposed framework to capture the temporal information holds the promise for addressing evolving challenges in 3D BUS data analysis.
Regarding the improvement of model training strategies, more advanced semi-supervised learning techniques will be explored to further reduce the reliance on data annotation, such as generative-based methods (Zhou, 2018; Yang et al., 2021; Zhai et al., 2022; Fan et al., 2021), consistency regularization methods (Mittal et al., 2021), and self-supervised learning (He et al., 2022; Caron et al., 2020, 2021; Chen et al., 2020b, 2021). Particularly, recent cutting-edge self-supervised learning methods, such as masked-autoencoder (MAE) (He et al., 2022) and masked image modeling (SimMIM) (Xie et al., 2022), leverage pretext tasks to learn useful representations from large-scale unlabeled images. The learned representations can be fine-tuned on small task-specific datasets for downstream classification or localization tasks. These representation learning paradigms are extensively studied in various imaging applications (Zhou et al., 2022; Chen et al., 2022). However, there are two major challenges to adapt these designs to efficiently capture the inherent structure of medical images, especially breast ultrasound images with high noise and textural similarities between the lesions and normal tissues. First, the presence of spurious correlations between image slices in the ultrasound scanning data hinders the model from capturing robust representations during self-supervised learning. Second, self-supervised learning usually necessitates extensive data to acquire complete data distribution and ensure the generalizability of learned features to unseen data. Learning precise data distribution and lesion-relevant features becomes particularly challenging when working with relatively small BUS datasets. Our modular asymmetric framework is adaptable to various encoders, facilitating the extraction of multi-scale features embedded with both local and global lesion information from input BUS images. An ongoing project is to explore more powerful encoders as feature extractors, and train them with self-supervised methods to tackle the aforementioned challenges. Concurrently, we will continuously collect imaging data with low correlations to mitigate the negative effects due to data relevancy.
Another interesting research direction involves extending the applications of the proposed framework to multi-modal ultrasound data, such as color Doppler and shear wave elastography data (Chang et al., 2013). Multimodal data provides complementary but limited information because it captures tumor phenotypes from diverse perspectives. Combining multimodal data in BUS analysis holds the potential to enhance model performance on both tasks. However, various challenges, such as redundancy, uncertainty, and heterogeneity among multimodal data, as well as the imbalanced patient cohorts, necessitate careful consideration when designing an effective and robust method. Furthermore, we will explore other advanced methods to further refine model interpretability, such as counterfactual explanation (Goyal et al., 2019) and concept whitening (Chen et al., 2020a), These approaches are aimed at providing transparent and safe predictions to support clinical decision-making.
7. Conclusions
The proposed method is capable of processing partly-labeled BUS images for joint localization and classification tasks. It is designed to address information-sharing conflicts inherent in shared-trunk-based MTL methods, to alleviate difficulties of time-consuming data annotation in clinical practice, and to suppress the negative effects of the intrinsic characteristics of BUS images. Extensive experiments conducted on two BUS datasets demonstrated the efficiency, robustness, and generality of the proposed method. The proposed method can potentially be applied to real-time clinical applications. It also holds the promise to be applied to a number of different problems, such as joint localization and recognition of prostate cancers and renal lesions, for which ultrasound images are commonly employed.
Acknowledgment
This work was supported by the Department of Defense (DoD) through the Breast Cancer Research Program (BCRP) under Award No. E01 W81XWH-21-1-0062. Opinions, interpretations, conclusions, and recommendations are those of the author and are not necessarily endorsed by the Department of Defense. This work was partly supported by NIH awards R56DE033344, R01EB034249, R01CA233873, Cancer Center at Illinois seed grant, and Jump ARCHES award. The authors acknowledge Michael Wu and Ethan Xiao for helping with data analysis and manuscript proofreading.
Footnotes
CRediT authorship contribution statement
Zong Fan: Conceptualization, Formal analysis, Methodology, Software, Writing – original draft, Validation, Visualization. Ping Gong: Resources, Data curation, Validation. Shanshan Tang: Resources, Data curation. Christine U. Lee: Resources, Data curation. Xiaohui Zhang: Writing review & editing, Visualization. Pengfei Song: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing. Shigao Chen: Conceptualization, Resources, Supervision, Writing – review & editing. Hua Li: Conceptualization, Funding acquisition, Supervision, Writing – review & editing, Project administration.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
The authors do not have permission to share data.
References
- Abbas A, Abdelsamea MM, Gaber MM, 2020. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl. Intell 1–11. 10.1007/s10489-020-01829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Dhabyani W, Gomaa MM, Khaled H, Fahmy AA, 2020. Dataset of breast ultrasound images. Data Brief 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg WA, Blume JD, Cormack JB, Mendelson EB, Lehrer D, Bóhm-Vélez M, Pisano ED, Jong RA, Evans WP, Morton MJ, et al. , 2008. Combined screening with ultrasound and mammography vs mammography alone in women at elevated risk of breast cancer. JAMA 299 (18), 2151–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger W, DeKoekkoek-Doll P, Springer P, Obrist P, Dünser M, 1999. Incidental findings on sonography of the breast: Clinical significance and diagnostic workup. AJR. Am. J. Roentgenol 173 (4), 921–927. [DOI] [PubMed] [Google Scholar]
- Byra M, Dobruch-Sobczak K, Piotrzkowska-Wróblewska H, Klimonda Z, Litniewski J, 2022. Explaining a deep learning based breast ultrasound image classifier with saliency maps. J. Ultrasonography 22, 70–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Duan L, Yang G, Yue T, Chen Q, Fu H, Xu Y, 2017. Breast tumor detection in ultrasound images using deep learning. In: International Workshop on Patch-Based Techniques in Medical Imaging. Springer, pp. 121–128. [Google Scholar]
- Caron M, Misra I, Mairal J, Goyal P, Bojanowski P, Joulin A, 2020. Unsupervised learning of visual features by contrasting cluster assignments. Adv. Neural Inf. Process. Syst 33, 9912–9924. [Google Scholar]
- Caron M, Touvron H, Misra I, Jégou H, Mairal J, Bojanowski P, Joulin A, 2021. Emerging properties in self-supervised vision transformers. In: Proceedings of the IEEE/CVF International Conference on Computer Vision. pp. 9650–9660. [Google Scholar]
- Chang JM, Won J-K, Lee K-B, Park IA, Yi A, Moon WK, 2013. Comparison of shear-wave and strain ultrasound elastography in the differentiation of benign and malignant breast lesions. Am. J. Roentgenol 201 (2), W347–W356. [DOI] [PubMed] [Google Scholar]
- Chen Z, Bei Y, Rudin C, 2020a. Concept whitening for interpretable image recognition. Nat. Mach. Intell 2, 772–782. [Google Scholar]
- Chen S, Bortsova G, García-Uceda Juárez A, Tulder G.v., Bruijne M.d., 2019. Multi-task attention-based semi-supervised learning for medical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, pp. 457–465. [Google Scholar]
- Chen Z, Du Y, Hu J, Liu Y, Li G, Wan X, Chang T-H, 2022. Multi-modal masked autoencoders for medical vision-and-language pre-training. In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2022: 25th International Conference, Singapore, September 18–22, 2022, Proceedings, Part V. Springer, pp. 679–689. [Google Scholar]
- Chen T, Kornblith S, Swersky K, Norouzi M, Hinton GE, 2020b. Big self-supervised models are strong semi-supervised learners. Adv. Neural Inf. Process. Syst 33, 22243–22255. [Google Scholar]
- Chen L-C, Papandreou G, Schroff F, Adam H, 2017. Rethinking atrous convolution for semantic image segmentation. arXiv preprint arXiv:1706.05587. [Google Scholar]
- Chen X, Xie S, He K, 2021. An empirical study of training self-supervised vision transformers. In: Proceedings of the IEEE/CVF International Conference on Computer Vision. pp. 9640–9649. [Google Scholar]
- Chowdary J, Yogarajah P, Chaurasia P, Guruviah V, 2022. A multi-task learning framework for automated segmentation and classification of breast tumors from ultrasound images. Ultrason. Imaging 44, 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawshaw M., 2020. Multi-task learning with deep neural networks: A survey. arXiv abs/2009.09796 [Google Scholar]
- Dai C, Mo Y, Angelini E, Guo Y, Bai W, 2019. Transfer learning from partial annotations for whole brain segmentation. In: Domain Adaptation and Representation Transfer and Medical Image Learning with Less Labels and Imperfect Data: First MICCAI Workshop, DART 2019, and First International Workshop, MIL3ID 2019, Shenzhen, Held in Conjunction with MICCAI 2019, Shenzhen, China, October 13 and 17, 2019, Proceedings 1. Springer, pp. 199–206. [Google Scholar]
- den Dekker BM, Bakker MF, de Lange SV, Veldhuis WB, van Diest PJ, Duvivier KM, Lobbes MB, Loo CE, Mann RM, Monninkhof EM, et al. , 2021. Reducing false-positive screening MRI rate in women with extremely dense breasts using prediction models based on data from the DENSE trial. Radiology 301 (2), 283–292. [DOI] [PubMed] [Google Scholar]
- Deng J, Dong W, Socher R, Li L, Li K, Fei-Fei L, 2009. ImageNet: A large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition. pp. 248–255. [Google Scholar]
- Ding W, Wang J, Zhou W, Zhou S, Chang C, Shi J, 2022. Joint localization and classification of breast cancer in B-mode ultrasound imaging via collaborative learning with elastography. IEEE J. Biomed. Health Inf. [DOI] [PubMed] [Google Scholar]
- Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, Dehghani M, Minderer M, Heigold G, Gelly S, et al. , 2020. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv: 2010.11929 [Google Scholar]
- Fan Z, He S, Ruan S, Wang X, Li H, 2021. Deep learning-based multi-class COVID-19 classification with X-ray images. In: Linte CA, Siewerdsen JH (Eds.), Medical Imaging 2021: Image-Guided Procedures, Robotic Interventions, and Modeling, Vol. 11598. SPIE, International Society for Optics and Photonics, 1159813. 10.1117/12.2582261. [DOI] [Google Scholar]
- Farooq MU, Ullah Z, Gwak J, 2023. Residual attention based uncertainty-guided mean teacher model for semi-supervised breast masses segmentation in 2D ultrasonography. Comput. Med. Imaging Graph 102173. [DOI] [PubMed] [Google Scholar]
- Glorot X, Bengio Y, 2010. Understanding the difficulty of training deep feedforward neural networks. In: Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics, Vol. 9. JMLR Workshop and Conference Proceedings, pp. 249–256, URL: http://proceedings.mlr.press/v9/glorot10a.html. [Google Scholar]
- Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville AC, Bengio Y, 2014. Generative adversarial networks. arXiv abs/1406.2661 [Google Scholar]
- Gordon PB, Goldenberg SL, 1995. Malignant breast masses detected only by ultrasound. A retrospective review. Cancer 76 (4), 626–630. [DOI] [PubMed] [Google Scholar]
- Goyal Y, Wu Z, Ernst J, Batra D, Parikh D, Lee S, 2019. Counterfactual visual explanations. arXiv abs/1904.07451. [Google Scholar]
- Han L, Huang Y, Dou H, Wang S, Ahamad S, Luo H, Liu Q, Fan J, Zhang J, 2020. Semi-supervised segmentation of lesion from breast ultrasound images with attentional generative adversarial network. Comput. Methods Programs Biomed 189, 105275. [DOI] [PubMed] [Google Scholar]
- Han S, Kang H, Jeong J-Y, Park MH, Kim W, Bang W-C, Seong YK, 2017. A deep learning framework for supporting the classification of breast lesions in ultrasound images. Phys. Med. Biol 62 19, 7714–7728. [DOI] [PubMed] [Google Scholar]
- Han Z, Wei B, Hong Y, Li T, Cong J, Zhu X, Wei H, Zhang W, 2020. Accurate screening of COVID-19 using attention-based deep 3D multiple instance learning. IEEE Trans. Med. Imaging 39 (8), 2584–2594. 10.1109/TMI.2020.2996256. [DOI] [PubMed] [Google Scholar]
- He K, Chen X, Xie S, Li Y, Dollár P, Girshick R, 2022. Masked autoencoders are scalable vision learners. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. pp. 16000–16009. [Google Scholar]
- He K, Zhang X, Ren S, Sun J, 2016. Deep residual learning for image recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition. CVPR. pp. 770–778. [Google Scholar]
- Hooley RJ, Scoutt LM, Philpotts LE, 2013. Breast ultrasonography: State of the art. Radiology 268 3, 642–659. [DOI] [PubMed] [Google Scholar]
- Imran A-A-Z, Terzopoulos D, 2019. Semi-supervised multi-task learning with chest X-ray images. In: Machine Learning in Medical Imaging: 10th International Workshop, MLMI 2019, Held in Conjunction with MICCAI 2019, Shenzhen, China, October 13, 2019, Proceedings 10. Springer. pp. 151–159. [Google Scholar]
- Itti L, Koch C, Niebur E, 1998. A model of saliency-based visual attention for rapid scene analysis. IEEE Trans. Pattern Anal. Mach. Intell 20 (11), 1254–1259. [Google Scholar]
- Kim J, Kim HJ, Kim C, Lee J-H, Kim K, Park YM, Kim HW, Ki SY, Kim YM, Kim WH, 2021. Weakly-supervised deep learning for ultrasound diagnosis of breast cancer. Sci. Rep 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingma DP, Ba J, 2015. Adam: A method for stochastic optimization. CoRR abs/1412.6980 [Google Scholar]
- Kolb TM, Lichy J, Newhouse JH, 1998. Occult cancer in women with dense breasts: Detection with screening US–diagnostic yield and tumor characteristics. Radiology 207 (1), 191–199. [DOI] [PubMed] [Google Scholar]
- Lee CH, Dershaw DD, Kopans D, Evans P, Monsees B, Monticciolo D, Brenner RJ, Bassett L, Berg W, Feig S, et al. , 2010. Breast cancer screening with imaging: Recommendations from the society of breast imaging and the ACR on the use of mammography, breast MRI, breast ultrasound, and other technologies for the detection of clinically occult breast cancer. J. Am. College Radiol 7 (1), 18–27. [DOI] [PubMed] [Google Scholar]
- Lee D-H, et al. , 2013. Pseudo-label: The simple and efficient semi-supervised learning method for deep neural networks. In: Workshop on Challenges in Representation Learning, Vol. 3, no. 2. ICML, p. 896. [Google Scholar]
- Li K, Li H, Anastasio MA, 2022. A task-informed model training method for deep neural network-based image denoising. In: Mello-Thoms CR, Taylor-Phillips S (Eds.), Medical Imaging 2022: Image Perception, Observer Performance, and Technology Assessment, Vol. 12035. SPIE, International Society for Optics and Photonics, 1203510. 10.1117/12.2613181. [DOI] [Google Scholar]
- Lin T-Y, Dollár P, Girshick RB, He K, Hariharan B, Belongie SJ, 2016. Feature pyramid networks for object detection. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition. CVPR. pp. 936–944. [Google Scholar]
- Liu W, Anguelov D, Erhan D, Szegedy C, Reed S, Fu C-Y, Berg AC, 2016. Ssd: Single shot multibox detector. In: European Conference on Computer Vision. Springer. pp. 21–37. [Google Scholar]
- Liu S, Johns E, Davison AJ, 2019. End-to-end multi-task learning with attention. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition. CVPR. pp. 1871–1880. [Google Scholar]
- Liu Z, Lin Y, Cao Y, Hu H, Wei Y, Zhang Z, Lin S, Guo B, 2021a. Swin transformer: Hierarchical vision transformer using shifted windows. In: Proceedings of the IEEE/CVF International Conference on Computer Vision. pp. 10012–10022. [Google Scholar]
- Liu Y, Yang Y, Jiang W, Wang T, Lei B, 2021b. Semi-supervised attention-guided VNet for breast cancer detection via multi-task learning. In: Image and Graphics: 11th International Conference, ICIG 2021, Haikou, China, August 6–8, 2021 Proceedings, Part II 11. Springer, pp. 559–570. [Google Scholar]
- Lyu Y, Xu Y, Jiang X, Liu J, Zhao X, Zhu X, 2023. AMS-PAN: Breast ultrasound image segmentation model combining attention mechanism and multi-scale features. Biomed. Signal Process. Control 81, 104425. [Google Scholar]
- Ma B, Zhao Y, Yang Y, Zhang X, Dong X, Zeng D, Ma S, Li S, 2020. MRI image synthesis with dual discriminator adversarial learning and difficulty-aware attention mechanism for hippocampal subfields segmentation. Comput. Med. Imaging Graph 86, 101800. 10.1016/j.compmedimag.2020.101800, URL: https://www.sciencedirect.com/science/article/pii/S0895611120300951. [DOI] [PubMed] [Google Scholar]
- Milletari F, Navab N, Ahmadi S-A, 2016. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In: 2016 Fourth International Conference on 3D Vision. 3DV, Ieee, pp. 565–571. [Google Scholar]
- Mishra AK, Roy P, Bandyopadhyay S, Das SK, 2021. Breast ultrasound tumour classification: A machine learning—Radiomics based approach. Expert Syst. 38 (7), e12713. [Google Scholar]
- Mishra AK, Roy P, Bandyopadhyay S, Das SK, 2022. A multi-task learning based approach for efficient breast cancer detection and classification. Expert Syst. 39 (9), e13047. [Google Scholar]
- Mittal S, Tatarchenko M, Brox T, 2021. Semi-supervised semantic segmentation with high- and low-level consistency. IEEE Trans. Pattern Anal. Mach. Intell 43, 1369–1379. [DOI] [PubMed] [Google Scholar]
- Nothacker M, Duda V, Hahn M, Warm M, Degenhardt F, Madjar H, Weinbrenner S, Albert U-S, 2009. Early detection of breast cancer: Benefits and risks of supplemental breast ultrasound in asymptomatic women with mammographically dense breast tissue. A systematic review. BMC Cancer 9 (1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, Killeen T, Lin Z, Gimelshein N, Antiga L, et al. , 2019. Pytorch: An imperative style, high-performance deep learning library. Adv. Neural Inf. Process. Syst 32. [Google Scholar]
- Pyakurel D, Karki S, Agrawal C, 2014. A study on microvascular density in breast carcinoma. J. Pathol. Nepal 4 (7), 570–575. [Google Scholar]
- Rasaee H, Rivaz H, 2021. Explainable AI and susceptibility to adversarial attacks: A case study in classification of breast ultrasound images. In: 2021 IEEE International Ultrasonics Symposium. IUS, IEEE, pp. 1–4. [Google Scholar]
- Redmon J, Divvala S, Girshick R, Farhadi A, 2016. You only look once: Unified, real-time object detection. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. pp. 779–788. [Google Scholar]
- Ren S, He K, Girshick R, Sun J, 2015. Faster R-CNN: Towards real-time object detection with region proposal networks. Adv. Neural Inf. Process. Syst 28. [DOI] [PubMed] [Google Scholar]
- Ronneberger O, Fischer P, Brox T, 2015. U-Net: Convolutional networks for biomedical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, pp. 234–241. [Google Scholar]
- Schlemper J, Oktay O, Schaap M, Heinrich MP, Kainz B, Glocker B, Rueckert D, 2019. Attention gated networks: Learning to Leverage Salient Regions in medical images. Med. Image Anal 53, 197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D, 2017. Gradcam: Visual explanations from deep networks via gradient-based localization. In: Proceedings of the IEEE International Conference on Computer Vision. pp. 618–626. [Google Scholar]
- Shan J, Alam SK, Garra B, Zhang Y, Ahmed T, 2016. Computer-aided diagnosis for breast ultrasound using computerized BI-RADS features and machine learning methods. Ultrasound Med. Biol 42 (4), 980–988. [DOI] [PubMed] [Google Scholar]
- Shin SY, Lee S, Yun ID, Kim SM, Lee KM, 2018. Joint weakly and semi-supervised deep learning for localization and classification of masses in breast ultrasound images. IEEE Trans. Med. Imaging 38 (3), 762–774. [DOI] [PubMed] [Google Scholar]
- Singh VK, Abdel-Nasser M, Akram F, Rashwan HA, Sarker MMK, Pandey N, Romani S, Puig D, 2020. Breast tumor segmentation in ultrasound images using contextual-information-aware deep adversarial learning framework. Expert Syst. Appl 162, 113870. [Google Scholar]
- Singh VK, Rashwan HA, Abdel-Nasser M, Sarker M, Kamal M, Akram F, Pandey N, Romani S, Puig D, 2019. An efficient solution for breast tumor segmentation and classification in ultrasound images using deep adversarial learning. arXiv preprint arXiv:1907.00887 [Google Scholar]
- Sinha A, Chen Z, Badrinarayanan V, Rabinovich A, 2018. Gradient adversarial training of neural networks. arXiv preprint arXiv: 1806.08028 [Google Scholar]
- Tan M, Le QV, 2019. EfficientNet: Rethinking model scaling for convolutional neural networks. arXiv abs/1905.11946 [Google Scholar]
- Tang P, Yang X, Nan Y, Xiang S, Liang Q, 2021. Feature pyramid nonlocal network with transform modal ensemble learning for breast tumor segmentation in ultrasound images. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 68 (12), 3549–3559. [DOI] [PubMed] [Google Scholar]
- Tishby N, Zaslavsky N, 2015. Deep learning and the information bottleneck principle. In: 2015 Ieee Information Theory Workshop. Itw, IEEE, pp. 1–5. [Google Scholar]
- Vakanski A, Xian M, Freer PE, 2020. Attention-enriched deep learning model for breast tumor segmentation in ultrasound images. Ultrasound Med. Biol 46 (10), 2819–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Engelen JE, Hoos HH, 2020. A survey on semi-supervised learning. Mach. Learn 109 (2), 373–440. [Google Scholar]
- Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I, 2017. Attention is all you need. Adv. Neural Inf. Process. Syst 30. [Google Scholar]
- Wang L., 2017. Early diagnosis of breast cancer. Sensors 17 (7), 1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Girshick R, Gupta A, He K, 2018. Non-local neural networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. pp. 7794–7803. [Google Scholar]
- Wightman R., 2019. PyTorch image models. 10.5281/zenodo.4414861, GitHub repository, GitHub, https://github.com/rwightman/pytorch-image-models. [DOI] [Google Scholar]
- Woo S, Park J, Lee J-Y, Kweon IS, 2018. Cbam: Convolutional block attention module. In: Proceedings of the European Conference on Computer Vision. ECCV, pp. 3–19. [Google Scholar]
- Wu G-G, Zhou L-Q, Xu J-W, Wang J-Y, Wei Q, Deng Y-B, Cui X-W, Dietrich CF, 2019. Artificial intelligence in breast ultrasound. World J. Radiol 11 (2), 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Zhang Z, Cao Y, Lin Y, Bao J, Yao Z, Dai Q, Hu H, 2022. Simmim: A simple framework for masked image modeling. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. pp. 9653–9663. [Google Scholar]
- Xu M, Huang K, Qi X, 2022. Multi-task learning with context-oriented self-attention for breast ultrasound image classification and segmentation. In: 2022 IEEE 19th International Symposium on Biomedical Imaging. ISBI, pp. 1–5. [Google Scholar]
- Xu M, Huang K, Qi X, 2023. A regional-attentive multi-task learning framework for breast ultrasound image segmentation and classification. IEEE Access. [Google Scholar]
- Yan C, Tu Y, Wang X, Zhang Y, Hao X, Zhang Y, Dai Q, 2019. STAT: Spatial-temporal attention mechanism for video captioning. IEEE Trans. Multimed 22 (1), 229–241. [Google Scholar]
- Yang X, Song Z, King I, Xu Z, 2021. A survey on deep semi-supervised learning. arXiv abs/2103.00550 [Google Scholar]
- Yang X, Song Z, King I, Xu Z, 2022. A survey on deep semi-supervised learning. IEEE Trans. Knowl. Data Eng. [Google Scholar]
- Yap MH, Goyal M, Osman FM, Martí R, Denton E, Juette A, Zwiggelaar R, 2018. Breast ultrasound lesions recognition: End-to-end deep learning approaches. J. Med. Imaging 6 (1), 011007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai D, Hu B, Gong X, Zou H, Luo J, 2022. ASS-GAN: Asymmetric semi-supervised GAN for breast ultrasound image segmentation. Neurocomputing 493, 204–216. [Google Scholar]
- Zhang G, Zhao K, Hong Y, Qiu X, Zhang K, Wei B, 2021. SHA-MTL: Soft and hard attention multi-task learning for automated breast cancer ultrasound image segmentation and classification. Int. J. Comput. Assist. Radiol. Surg 16, 1719–1725. [DOI] [PubMed] [Google Scholar]
- Zhou Z-H, 2018. A brief introduction to weakly supervised learning. Natl. Sci. Rev 5 (1), 44–53. [Google Scholar]
- Zhou Y, Chen H, Li Y, Liu Q, Xu X, Wang S, Shen D, Yap P-T, 2021. Multi-task learning for segmentation and classification of tumors in 3D automated breast ultrasound images. Med. Image Anal 70, 101918. [DOI] [PubMed] [Google Scholar]
- Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A, 2016. Learning deep features for discriminative localization. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. pp. 2921–2929. [Google Scholar]
- Zhou L, Liu H, Bae J, He J, Samaras D, Prasanna P, 2022. Self pre-training with masked autoencoders for medical image analysis. arXiv preprint arXiv:2203.05573 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors do not have permission to share data.