Abstract
The Escherichia coli outer membrane TonB-dependent transporters for iron complexes and cobalamins recognize their multiple and diverse substrates with high specificity and affinity. The X-ray crystallographic structures of several transporters show that the substrate-binding surfaces are comprised of residues from the internal globular domain and multiple extracellular loops. The extracellular loops on the N-terminal half of the transmembrane beta-barrel of the cobalamin transporter BtuB participate in binding of the cofactor calcium atoms and undergo substantial conformation changes upon substrate binding. The functional relevance of the five C-terminal loops was examined by examining the effects of short in-frame deletions. Each loop contributed in different ways to the binding of BtuB substrates. Deletions in loops 7, 8, 9, and 11 strongly decreased cobalamin binding and transport, whereas deletions in loops 8, 9, and 10 affected binding and entry of phage BF23. None of the loops were essential for the action of colicin E1 or E3, which is consistent with the crystallographic observation that the colicin E3 receptor-binding domain can contact almost all of the loops. A deletion in loop 9 or 11 eliminated the ability of cobalamin to inhibit the action of colicin E1. These phenotypes show that there are multiple independent binding elements and point out similarities and differences in binding properties among the TonB-dependent transporters.
TonB-dependent transporters (TBDT) are present in many gram-negative bacteria and carry out the high-affinity and energy-dependent uptake of several organometallic complexes across the outer membrane (OM). Their substrates include cobalamins, such as vitamin B12 (cyanocobalamin [CNCbl]), and ferric iron bound to siderophores, heme, hemophores, or host iron- or heme-binding proteins (reviewed in references 17 and 32). These physiologically valuable substrates are too large to enter efficiently through the general porin channels and are often present at low levels in natural environments. Energy for the uptake process is coupled to these OM proteins by the TonB complex, consisting of the transenvelope protein TonB and the integral cytoplasmic membrane proteins ExbB and ExbD (reviewed in reference 33).
To date, the structures of four TBDT from Escherichia coli have been determined by X-ray crystallography. These are the transporters for ferrichrome FhuA (19, 29), for enterobactin FepA (5), for ferric dicitrate FecA (18), and for cobalamin BtuB (13). These proteins are very similar in basic structure, although they differ considerably in size and sequence. The membrane-spanning portion of all four proteins consists of a barrel with 22 antiparallel, moderately amphipathic β-strands. The β-strands in the barrel are connected by very short turns on the periplasmic face and by relatively long loops on the extracellular face which extend some distance above the membrane. The pore of the barrel is occluded by insertion of a conserved N-terminal globular domain, called the hatch, plug, or cork. Substrate-binding residues of all four TBDT occur in the hatch domain and in multiple loops.
The 594-residue BtuB protein is the TBDT for uptake of CNCbl, as well as several lethal agents, the E colicins, colicin A, and phage BF23. Although uptake of CNCbl requires TonB function, entry of the colicins instead requires the Tol system, and phage BF23 is independent of both the TonB and Tol systems (1, 24). Binding of CNCbl to BtuB strongly depends on the presence of calcium (3, 13). Binding of Ca and CNCbl results in major changes in the conformation of extracellular loops 2 through 5. The binding pocket for CNCbl is formed from residues in two apices of the hatch and in barrel loops 2 to 4 and 9 to 11. The structure of the complex of BtuB with the receptor-binding domain of colicin E3 (E3R135) was described previously (26). The broad interface was formed by contact of 27 colicin E3R residues with 29 BtuB residues, which were located in the apices of the hatch and in loops 3 to 5 and 7 to 11. BtuB residues 229, 276, 289, 497, and 579 in loops 3, 4, 9, and 11, as well as residues in the hatch domain, were found to interact with both CNCbl and colicin E3.
Despite their use of different translocation mechanisms and the marked differences in their structures, all of the BtuB substrates display mutual competition for binding to BtuB. This competition could result from the binding of all substrates to the same or overlapping sites on BtuB, as seen for CNCbl and colicin E3R. It is also possible that the binding of one substrate could elicit conformational changes that lead to occlusion of separate binding surfaces for other substrates. Previous genetic attempts to investigate the mechanism of substrate competition and to identify BtuB residues important for substrate binding have not been informative (Kadner, unpublished observations). The recent structural analyses suggest that changes in any one residue in the extensive binding interfaces might not prevent substrate binding.
In this study we aimed to examine the contribution of extracellular loops to BtuB function. Our experiments were initiated before the structure of BtuB was known. Attempts to align BtuB to the structures of FhuA and FepA yielded consistent predictions for the positions of only the five C-terminal loops of the barrel, and hence these five loops were targeted for deletion analysis. The subsequently determined structures showed that the N-terminal loops play several critical roles in BtuB function. The phenotypes of mutants indicated the presence of overlapping and independent binding determinants which could not be deduced from the structures. Comparison of the effect of loop deletions in several TBDT with the structural information revealed common and specific features of the substrate-binding process.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmids.
E. coli strain JM109 [F′ traD136 lacIq Δ(lacZ)M15 proAB e14− (McrA−) thi gyrA96 endA1 hsdR17 (rk− mk+) relA1 supE44 recA1] was used as a host for plasmid maintenance and as a recipient for transformation of ligation products. Strain RK5016 [Δlac(U169) araD139 rpsL gyrA thi non metE argH recA ΔbtuB] (22) was used for assays of the BtuB phenotype. The btuB deletion variants were cloned into plasmid pAG1, a derivative of pUC8 carrying the btuB gene expressed from its native promoter (20). They were subcloned by restriction fragment exchange into plasmid pKO3 (27) with a temperature-sensitive replicon to examine mutant phenotypes when btuB was expressed at a low copy number. Expression from pKO3 derivatives was examined following cell growth at 25 or 30°C.
Bacteria were grown in Luria-Bertani (LB) medium or minimal medium A (30) supplemented with ampicillin or chloramphenicol for plasmid maintenance. Liquid cultures were grown in a gyratory incubator at 250 rpm at 35°C or in a shaking water bath at 25°C. Solid media contained 1.8% agar. L-soft agar (0.8% agar in LB medium) was used for assays of colicin and bacteriophage BF23 killing zones.
For DNA manipulation we used standard methods (36). The nucleotide sequence of each btuB deletion was verified by automated DNA sequencing at the University of Virginia Biomolecular Resource Facility.
Mutagenesis of btuB was performed by a two-step PCR-based method (23). PCRs were carried out by using Vent DNA polymerase (New England Biolabs) as recommended by the manufacturer. Oligonucleotides were obtained from QIAGEN, and their sequences are available upon request. Briefly, two complementary mutagenic oligonucleotides were used in separate PCRs performed with upstream and downstream primers annealing to the 5′ and 3′ ends of the btuB gene and with plasmid pAG1 as the template. The mutagenic oligonucleotides were designed to replace each targeted 15- to 30-bp sequence of btuB with a 6-bp SphI restriction site encoding the amino acids Ala-Cys. The products of the first reactions were used with the btuB 5′ and 3′ primers in a second PCR to obtain a full-length btuB variant. The product was digested with appropriate enzymes and ligated into pAG1 digested with the same enzymes. Plasmids in ampicillin-resistant transformants were screened for the presence of the introduced SphI site.
Phenotype assays.
The CNCbl growth phenotype was determined for strain RK5016 carrying plasmid pAG1 or pKO3btuB with btuB deletion alleles. Strains were streaked on minimal medium A plates supplemented with methionine (100 μg/ml) or CNCbl at a concentration of 0.1 to 5,000 nM (1). The growth response is reported below as the lowest concentration of CNCbl that allowed formation of colonies that were comparable in size to those of the same strain grown with methionine.
(i) [57Co]cyanocobalamin transport and binding assays.
[57Co]CNCbl was prepared as described previously (6). Transport of [57Co]CNCbl by strain RK5016 carrying pKO3 plasmids expressing btuB deletion mutations was tested as previously described (21). Briefly, [57Co]CNCbl (600 cpm/pmol) at a final concentration of 10 nM was added to 10 ml of washed cells at an optical density at 650 nm of 0.2 in 0.1 M phosphate buffer (pH 6.6) with 5% glucose and 0.1 mM CaCl2. Uptake suspensions were incubated in 50-ml flasks at 30°C. At intervals 1-ml portions (1.3 × 108 cells) were filtered (Millipore HA; pore size, 0.45 μm) and washed with 20 ml of 0.1 M LiCl. At 30 min, 5 μM unlabeled CNCbl was added to the remaining cells, and sampling was continued in order to measure release of the label. The radioactivity retained on dried filters was determined in a scintillation counter with ReadySafe scintillation fluid. Uptake is expressed below in picomoles per 109 cells, and the means and standard deviations were calculated from two or three independent experiments.
Binding of [57Co]CNCbl to outer membranes was assessed as described previously (21). Outer membranes were prepared by Sarkosyl extraction of total membranes from cells of the ΔbtuB strain RK5016 expressing the btuB variants from high-copy-number plasmid pAG1 grown in minimal medium with methionine and were frozen at −20°C. Protein concentrations were determined by using a modification of the Lowry assay (39). For the binding assay, 3.5 μg of outer membrane protein in 750 μl (final volume) of 0.1 M phosphate buffer (pH 6.6) containing 5% glucose and 0.1 mM CaCl2 was mixed with [57Co]CNCbl (600 or 2,000 cpm/pmol) at concentrations ranging from 1 to 50 nM. Following incubation at 37°C for 30 min in 10-ml polypropylene tubes, triplicate samples were filtered as described above and washed twice with 10 ml of 0.1 M LiCl. The filters were dried, and the retained radioactivity was measured as described above. Binding is expressed below in picomoles per milligram of protein. The means and standard deviations were calculated from three independent experiments. The binding of [57Co]CNCbl to outer membranes from strain RK5016 carrying vector plasmid pBR322 was subtracted. Binding data were analyzed by using the GraphPad Prism program.
(ii) Colicin and BF23 assays.
Colicins E1 and E3 were extracted from mitomycin C-induced E. coli strains carrying plasmids pColE1 and pColE3 (RK4431 and RK4433, respectively) by a protocol modified from the protocol of Schwartz and Helinski (38) and were used without further purification. The colicin titer (expressed in arbitrary killing units) was determined from the maximal dilution that gave complete killing of the indicator strain on spot plates. For spot assays of colicin or phage BF23 sensitivity, cells were inoculated into L-soft agar lawns on LB agar plates with the appropriate antibiotic; 5 mM CaCl2 was added for assays of the BF23 response. Serial 10-fold dilutions of colicins E1 and E3 or of phage BF23 were spotted in 5-μl drops onto the lawns, and the plates were incubated overnight. The response is expressed below as the negative logarithm of the last dilution that resulted in complete clearing.
The E1 lysis and E3 survival assays developed by Cavard (9-11) were modified for use with 96-well microtiter plates (Immulon II; Dynatech). For the colicin E1 assay, 130-μl portions of cells were added to wells containing 10 μl of serial fourfold dilutions of colicin and incubated for 10 min in a rotating incubator at 30°C. Sodium dodecyl sulfate (SDS) was added to a final concentration of 0.05% to initiate lysis of E1-intoxicated cells. The decrease in turbidity after 15 min was measured with a microplate reader and was used to calculate the percentage of lysed cells. For the colicin E3 assay, which did not render cells susceptible to SDS-induced lysis, 10-μl portions of cells at an A595 of 1.0 were treated with serial dilutions of colicin E3, and after 20 min of incubation the samples were diluted sevenfold with LB medium. The cells were allowed to grow for 2 h, and the extent of killing was calculated from the growth inhibition. To measure substrate competition, dilutions of CNCbl were incubated with cells for 15 min before addition of colicins.
(iii) BF23 adsorption.
Assays of BF23 adsorption have been described previously (4). Briefly, a 200-μl sample containing 2 × 106 PFU of phage BF23 was incubated with 200 μl of log-phase cells (A595, 1.0) for 10 min at 37°C in LB medium containing 5 mM CaCl2. Twenty-five microliters of the mixture was diluted 100-fold into LB medium containing 50 μl of chloroform and vortexed for 10 s. Cells along with any adsorbed or injected phage were removed by centrifugation at 8,000 rpm in a clinical centrifuge at 4°C. The number of unadsorbed phage remaining in the supernatant was determined by plating with the indicator strain RK5173, with correction for the small number of phage that were removed by the btuB null strain. Assays were performed three times in duplicate.
BtuB levels in outer membrane fraction.
RK5016 cells carrying the mutant btuB genes on derivatives of plasmid pKO3btuB or pAG1 were grown in minimal medium at the appropriate temperature. The OM fraction was prepared by Sarkosyl solubilization of total membranes as previously described (21, 25). Proteins in the total cell extract or the OM fraction were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and were visualized by Coomassie blue staining or by Western immunoblotting with monoclonal antibody BtuB-1, as previously described (7). The proportion of BtuB relative to the amount of the major OM proteins was estimated by densitometry of scanned gels.
Surface exposure of the BtuB proteins was detected by reaction of the introduced cysteine residues with a sulfhydryl reagent unable to penetrate the cytoplasmic membrane. Cells carrying loop deletion mutations in pAG1 derivatives were incubated with biotin maleimide [N-biotinoyl-N′-(6-maleimideohexanoyl)-hydrazide]. Whole-cell suspensions were boiled for 5 min in SDS-PAGE sample buffer without a reducing agent. Proteins were resolved by SDS-PAGE on a 10% polyacrylamide gel and transferred to a nitrocellulose membrane. The membrane was blocked overnight in 3% bovine serum albumin in phosphate-buffered saline-Tween. The membrane was incubated with horseradish peroxidase-labeled Neutravidin, washed extensively, developed with a chemiluminescence substrate, and exposed to X-ray film. In addition, intact biotinylated cells were probed by using an enzyme-linked immunosorbent assay format with the enzyme-labeled Neutravidin to show that the Cys residue was exposed to the cell exterior.
RESULTS
Mutagenesis strategy for deletions in loops 7 to 11.
Because this project was started before the BtuB structure was known, we used several molecular modeling approaches to align the BtuB sequence on the structures of FepA and FhuA for prediction of the location of its extracellular loops. These methods provided consistent identification of the hatch and barrel domains and of the five loops at the C-terminal end of the barrel, but predictions of the other loops were inconsistent. The crystal structure of BtuB (13) confirmed the predicted locations of the five C-terminal loops and showed that the assignments of the other loops were unreliable. Of the N-terminal loops, loops 1 and 6 are very short, and loops 2 through 5 play key roles in substrate binding. C-terminal loops 7 to 11 have less obvious functions, and genetic analysis of these loops should complement the structural features.
The five C-terminal extracellular loops of BtuB, designated L7 to L11, were targeted for deletion in seven mutations which removed four to nine residues. Two nonoverlapping deletions in L7 and L11 were used in order to provide fuller coverage without the use of large deletions. Deleted residues were replaced with the coding region for Ala-Cys to facilitate screening by insertion of a unique SphI site, to add flexible residues, and to detect cell surface exposure of the unique Cys residue. The sequences of the mutations were confirmed, and the btuB variants were cloned into the high-copy-number plasmid pAG1 and the low-copy-number plasmid pKO3. Attempts to transfer some of the mutations into the chromosomal btuB locus were unsuccessful. Sequences just beyond the 3′ end of the btuB gene cannot be stably carried on plasmids (22), which limited our ability to carry out allelic transfer by homologous recombination and required the use of plasmid vectors. Most of the BtuB deletions, described in Table 1, are named for the affected loops; the one exception is the Δnlfd variant, which removed the highly conserved NLFD motif at the start of L11.
TABLE 1.
BtuB loop deletion variants
| Variant | Loopa
|
Deleted residues
|
Amt of BtuB protein (% of OM protein)b | ||
|---|---|---|---|---|---|
| Size (residues) | Position | Position | Sequence | ||
| Vector | NA | NAc | NA | 0 | |
| BtuB+ | NA | NA | NA | 46.1 ± 3.6 | |
| ΔL7-1 | 17 | 396-412 | 400-405 | QLYGFY | 47.3 ± 3.4 |
| ΔL7-2 | 17 | 396-412 | 406-410 | GNPNL | 40.4 ± 3.6 |
| ΔL8 | 15 | 439-452 | 444-448 | IDYDD | 47.6 ± 8.8 |
| ΔL9 | 8 | 488-497 | 494-499 | PLLRRA | 51.6 ± 8.4 |
| ΔL10 | 9 | 530-538 | 533-538 | SYPYQT | 39.1 ± 5.9 |
| ΔL11 | 15 | 567-581 | 572-580 | DYETVYGYQ | 44.2 ± 3.5 |
| Δnlfd | 15 | 567-581 | 567-570 | NLFD | 22.2 ± 1.2 |
The sizes and positions of the extracellular loops were determined from the crystal structure.
The amounts of BtuB in the outer membrane were estimated by densitometry of Coomassie blue-stained SDS-PAGE electropherograms of outer membrane proteins extracted from cells of strain RK5016 (ΔbtuB) carrying plasmid pAG1 derivatives expressing the different loop deletion variants. The amount of BtuB is expressed as a percentage of the intensity of staining of it and the major OM proteins.
NA, not applicable.
BtuB protein expression levels.
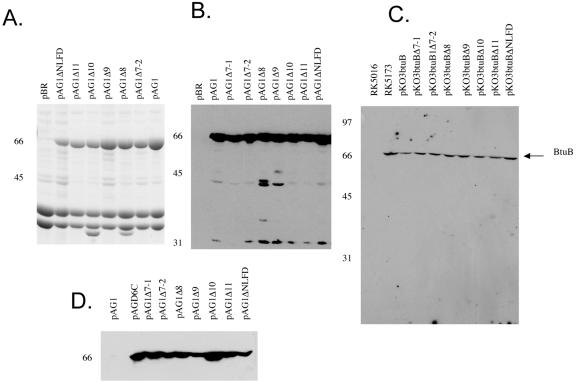
The expression levels of the variant BtuB proteins were compared on Coomassie blue-stained SDS-PAGE electropherograms of the OM fraction of cells carrying the high-copy-number pAG1 derivatives (Fig. 1A) and by Western immunoblotting of extracts from cells carrying the pAG1 (Fig. 1B) or pKO3 (Fig. 1C) derivatives. The Western analyses of whole-cell and OM proteins revealed a single immunoreactive band for each variant at the same mobility as wild-type BtuB, indicating that all variants were effectively inserted into the OM. The amount of BtuB was compared to the content of the major OM proteins OmpA and porins OmpC/F by densitometry of the Coomassie blue-stained OM proteins (Fig. 1A and Table 1). When expressed at high copy numbers, most of BtuB variants were present at comparable high levels like wild-type BtuB, in the range from 39 to 52% of the level of the total major OM proteins; the only exception was the level of the Δnlfd variant, which was about one-half that of the other variants. When the proteins were expressed from the low-copy-number pKO3 plasmid at 25°C, immunoblots indicated that most of the BtuB variants were present at levels comparable to the level of BtuB in the haploid RK5173 cells, corresponding to about 200 molecules per cell (15, 35). Again, the level of Δnlfd was usually about 40% that of the other variants. Although most loop deletions had little or no effect on incorporation into the OM, the decreased level of the Δnlfd variant suggests that these residues, which fold into an unusual conformation (12), may contribute to insertion or the stability of BtuB in the OM.
FIG. 1.
Expression of btuB loop deletion variants. Cells of strain RK5016 carrying derivatives of plasmid pAG1 or pKO3 with different loop deletions were processed for identification of BtuB proteins as described in Materials and Methods. (A) Coomassie blue-stained electropherogram of outer membranes from cells carrying high-copy-number pAG1 derivatives. BtuB is the band migrating at 66 kDa. (B) Western immunoblot of an extract of the same cells as in panel A. (C) Western immunoblot of an extract of cells carrying derivatives of low-copy-number plasmid pKO3. (D) Biotin maleimide labeling of cells carrying derivatives of pAG1. As described in Materials and Methods, whole cells were labeled with biotin maleimide, washed, resolved by SDS-PAGE, transferred to a nitrocellulose membrane, exposed to horseradish peroxidase-labeled Neutravidin, and developed by chemiluminescence. The positions of molecular weight markers are indicated on the left of each panel.
To confirm the surface presentation of the variant proteins, cells were treated with a biotin maleimide derivative which does not cross the cytoplasmic membrane but covalently labels OM and periplasmic proteins containing accessible cysteine residues (8). Wild-type BtuB lacks Cys residues and was not labeled (Fig. 1D). BtuB D6C, with a Cys residue in the Ton box on the periplasmic face, was labeled. Each of the loop deletion proteins having a Cys residue at the site of the deletion was labeled to approximately the same extent as BtuB D6C. The biotin moiety on each deletion protein was accessible to Neutravidin in intact cells, indicating that there was proper exposure of the Cys to the cell exterior (data not shown). The decreased labeling of the Δnlfd variant could have reflected a decrease in the amount of the protein or the accessibility to the Cys residue.
CNCbl utilization, transport, and binding.
Growth assays measure the ability of CNCbl to support the growth of a metE btuB strain in the absence of methionine. A metE strain lacking the cobalamin-independent homocysteine methyltransferase MetE requires methionine unless cobalamin is available to allow functioning of the cobalamin-dependent MetH enzyme (34). This growth assay responds to very low levels of CNCbl uptake because as few as 25 molecules can support cell doubling (14). A metE btuB+ strain shows a full growth response at around 0.1 nM CNCbl, whereas a btuB-null strain requires roughly 5,000 nM CNCbl. The growth responses of strain RK5016 carrying pKO3 derivatives with btuB loop deletions showed that the ΔL10 and Δnlfd variants had a behavior similar to that of the wild type (Table 2). Deletions in loops 7, 8, and 9 responded at around 5 nM CNCbl, and the ΔL11 variant required around 500 nM CNCbl. Thus, changes in four of the five loops impaired but did not eliminate CNCbl uptake.
TABLE 2.
Functional properties of the BtuB loop deletion variants
| BtuB varianta | Growth response (nM)b | CNCbl uptake (pmol at 30 min)c | BF23 titerd | BF23 adsorption (% of input)e | Colicin E1 responseg
|
||
|---|---|---|---|---|---|---|---|
| Titerf | Maximum killing | E1(0:5) | |||||
| Vector | 5,000 | 0.4 | R | 0 | R | 0 | NA |
| BtuB+ | 0.1 | 21.9 | 6 | 97 ± 6 | 5 | 68 | 30 |
| ΔL7-1 | 5 | 2.2 | 6 | 98.1 ± 1.7 | 4 | 32 | 26 |
| ΔL7-2 | 5 | 3.0 | 6 | 94.9 ± 6 | 4 | 56 | 240 |
| ΔL8 | 5 | 1.7 | R | 1 ± 3 | 4 | 41 | 118 |
| ΔL9 | 5 | 2.7 | R | 7 ± 8 | 4 | 65 | 46 |
| ΔL10 | 0.1 | 13.3 | R | 1.4 ± 3.6 | 3 | 57 | 840 |
| ΔL11 | 500 | 0.7 | 5 | 70.8 ± 5.9 | 4 | 41 | 112 |
| Δnlfd | 0.1 | 12.5 | 5 | 77.5 ± 10 | 5 | 71 | 30 |
Strain RK5016 variants carrying btuB alleles in low-copy-number plasmid pKO3 were grown at 25°C, as described in Materials and Methods. The vector plasmid was pKO3 without an insert.
The growth response is expressed as the concentration of CNCbl that allowed growth of cells comparable to that with methionine supplementation. The CNCbl concentrations differed in 10-fold steps. The growth response to all variants was TonB dependent (data not shown).
The CNCbl uptake results are taken from the data in Fig. 2 and represent the amount of labeled CNCbl taken up at 30 min.
Serial dilutions of BF23 were spotted on lawns of the different strains on LB medium plates with 2.5 mM CaCl2. The titer is reported as the negative logarithm of the lowest concentration that gave a zone of clear lysis. R, resistant to phage killing at the highest concentration tested.
Adsorption of phage BF23 was measured as described in Materials and Methods and is expressed as the percentage of input phage that were adsorbed in 10 min; the data are means ± standard deviations.
The titer is the response to colicin E1, as defined above.
The values for the maximum extent of cell killing and the concentration of colicin E1 (in killing units) that conferred half-maximal killing [E1 (0.5)] were taken from the data in Fig. 4.
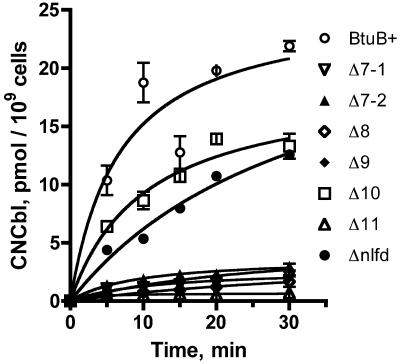
The rate of uptake of [57Co]CNCbl was measured in cells expressing the deletion variants from the low-copy-number plasmid pKO3 (Fig. 2). Consistent with the growth phenotypes, two patterns of transport activity were seen. The ΔL10 and Δnlfd variants displayed levels of uptake activity that were about 60% of that conferred by the wild-type allele. The mutants with deletions in loops 7, 8, 9, and 11 displayed greatly decreased uptake activity, down to 10% or less of the wild-type level. They retained a level of transport activity that was higher than that of the ΔbtuB strain carrying the vector. The binding and uptake phases were affected similarly in the mutants.
FIG. 2.
Uptake of [57Co]CNCbl by cells expressing BtuB loop deletion variants. The strains used carry derivatives of the low-copy-number plasmid pKO3. The uptake values are averages from replicate experiments.
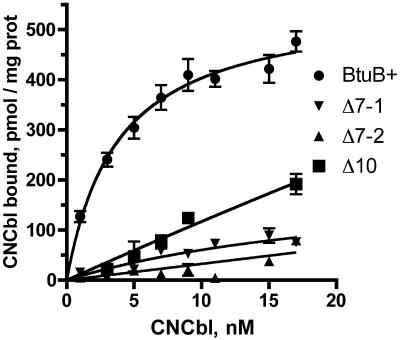
The levels of binding of labeled CNCbl to OM from cells expressing some transport-proficient BtuB variants from the high-copy-number plasmid pAG1 were determined at various CNCbl concentrations and plotted as the amount bound versus the free concentration remaining (Fig. 3). Wild-type BtuB showed a hyperbolic binding process; nonlinear least-squares curve fitting indicated a binding constant of 2.8 nM, which was consistent with previous measurements (21). Binding to the ΔL10 variant showed about a 10-fold-higher binding constant (30 nM) and about 75% of the capacity of the wild-type protein. These binding values were consistent with the transport rates of this mutant. The level of CNCbl binding to the two ΔL7 deletions was too low to determine reliably, and other mutants were not examined. Thus, all five loops affect the binding and transport of CNCbl, although L10 plays a lesser role.
FIG. 3.
Binding of [57Co]CNCbl by outer membranes containing BtuB loop deletion variants. Binding values (in picomoles per milligram of OM protein) were determined for outer membranes by using cells carrying pAG1 derivatives expressing wild-type BtuB, ΔL10, ΔL7-1, and ΔL7-2. All binding data are averages, and the values are fit to a single hyperbolic binding curve. prot, protein.
Phage BF23 adsorption.
The response to phage BF23 was determined in cells expressing the loop deletion variants from pKO3. When dilutions of phage BF23 were spotted onto lawns of the various strains, strains with both deletions in loop 7 were as susceptible as the wild-type strain (Table 2). Both deletions in loop 11, including Δnlfd, showed decreased susceptibility by about 1-log dilution. Deletions in loops 8, 9, and 10 conferred resistance to the highest concentration of BF23 tested. When the BtuB variants were expressed from the high-copy-number plasmid pAG1, only the loop 8 deletion showed resistance, indicating that the other loop deletions did not completely abolish phage binding.
Phage adsorption was measured from the number of BF23 phage remaining in the medium after exposure to cells for 10 min, followed by removal of adsorbed phage. The deletions in loop 7 showed the same degree of adsorption as cells expressing wild-type BtuB showed (Table 2). The two deletions in loop 11 showed moderately decreased adsorption, consistent with the moderate decrease in phage susceptibility. Deletions in loops 8, 9, and 10 eliminated or greatly decreased adsorption, indicating that the resistance seen in the plate assay resulted from loss of phage binding to BtuB. Thus, residues in loops 8 to 10 are crucial for BF23 adsorption.
Response to colicins E1 and E3.
The effects of the loop deletions on susceptibility to colicins E1 and E3 were compared. These BtuB-dependent colicins differ in their translocation pathways and the sequences of their receptor-binding domains. Spot tests with serial colicin E1 dilutions revealed only small decreases in the susceptibility of the variants, suggesting that multiple loops contribute independently to colicin binding (Table 2). The cells with deletions in loops 7, 8, 9, and 11 required about 10 times more colicin E1 for formation of a zone of clearing than the parent strain required, and deletion of loop 10 conferred about a 100-fold decrease in the titer. There was no impairment of the response to colicin E3, which was assayed with cells carrying the pAG1 derivatives (data not shown).
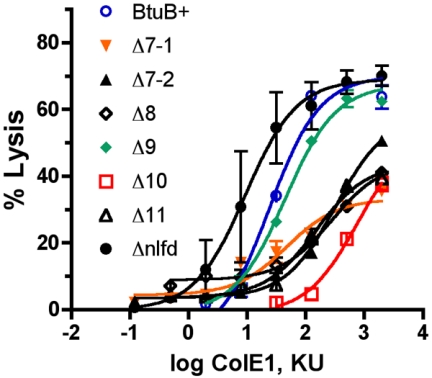
The rapid cell lysis assay developed by Cavard (9) is based on the finding that cells intoxicated with pore-forming colicins, such as colicin E1, undergo lysis upon addition of SDS. This assay was adapted to microtiter plates. Cultures in microtiter plates were exposed to colicin E1 dilutions for 10 min before SDS was added. The extent of cell lysis after 10 min was determined from the decrease in turbidity as measured with a plate reader. Data from two experiments with triplicate values were averaged and plotted against the input colicin amount, expressed in arbitrary killing units. Data were fit to a hyperbolic curve by using the GraphPad Prism program (Fig. 4), which yielded values for the maximal percentage of lysis and the colicin E1 concentration that gave half-maximal lysis (Table 2).
FIG. 4.
Responses of cells expressing BtuB loop deletion variants to colicin E1. Cells carrying derivatives of plasmid pKO3 with btuB variants were exposed to different dilutions of colicin E1 (ColE1) for 10 min before SDS was added. The extent of cell lysis after 10 min was determined with a microplate reader. The no-lysis value was obtained from the turbidity in the absence of colicin but with SDS, and the 100% lysis value was the value obtained with medium alone. Data were fit by using the GraphPad Prism program. KU, killing units.
All of the BtuB loop deletion variants were susceptible to colicin E1, which is consistent with the spot assay. The ΔL9 and Δnlfd variants gave response curves similar to the wild-type BtuB curves (Fig. 4). The other loop deletions were less susceptible to colicin E1; ΔL10 was the least sensitive, requiring around 300-fold more colicin for equivalent lysis. These results indicated that no single loop was required for colicin E1 binding but each loop except L9 contributed.
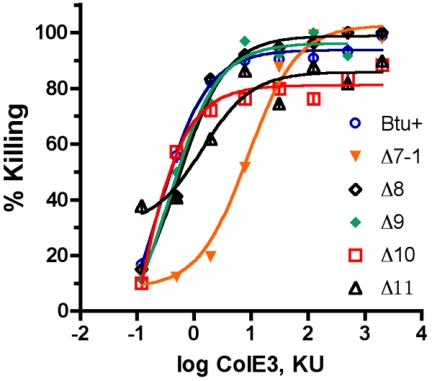
The response to colicin E3 was also examined by a microplate assay. Colicin E3 inhibits protein synthesis and does not sensitize cells to lysis in the presence of SDS. Cells were exposed for 20 min to serial dilutions of the colicin and then diluted with LB medium. The increase in turbidity following 2 h of incubation reflected the number of viable cells remaining after exposure to the colicin and thus provided a measure of cell killing. The results (Fig. 5) revealed that most of the loop deletion variants showed a dose-response curve similar to that of the wild type; the only exception was the ΔL7-1 variant, which required about a 30-fold-increased level of colicin for equivalent killing.
FIG. 5.
Responses of cells expressing BtuB loop deletion variants to colicin E3. Cells carrying derivatives of plasmid pKO3 with btuB variants were exposed to different dilutions of colicin E3 (ColE3) for 10 min before sevenfold dilution with LB growth medium. The extent of cell killing after 20 min was calculated from the inhibition of growth as measured with a microplate reader. KU, killing units.
Competition by CNCbl against colicin E1 killing.
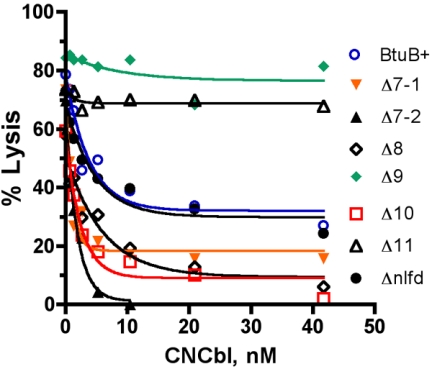
The binding of the E colicins to BtuB is competitively inhibited by CNCbl (15). The abilities of increasing concentrations of CNCbl to inhibit killing by colicin E1 were compared by performing the lysis assay for the BtuB loop deletion variants. Competition was examined over a range of colicin E1 concentrations, and the data for an intermediate level are shown (Fig. 6). Three different patterns of inhibition by CNCbl were seen. With wild-type BtuB and Δnlfd, CNCbl conferred partial inhibition against colicin E1, with half-maximal protection at around 3 nM CNCbl. The ΔL9 and ΔL11 variants showed essentially no protection by CNCbl against colicin E1, suggesting that these deletions removed residues that are important for the ability of CNCbl to interfere with colicin E1 binding. In contrast, for the deletions in loops 7, 8, and 10, CNCbl was a more effective inhibitor than it was in the wild type. Inhibition was more extensive and occurred with higher affinity. The simplest interpretation of these findings is that multiple surfaces bind colicin E1 independently. Binding of colicin to L11 is inhibited by CNCbl, but binding to the other loops is not. Absence of L11 results in a lack of inhibition by CNCbl, and the absence of other loops makes colicin binding even more dependent on L11 and thus more susceptible to inhibition by CNCbl.
FIG. 6.
Ability of CNCbl to protect against colicin E1-induced cell lysis. The colicin E1 lysis assay was performed by using a range of concentrations of colicin and CNCbl. The data show the ability of increasing concentrations of CNCbl to block lysis at an intermediate concentration of colicin (125 killing units). The cells carried a plasmid pKO3 derivative with the btuB loop deletions indicated.
DISCUSSION
The Btu system offers an excellent opportunity for analysis of transport across the outer and cytoplasmic membranes. Structures have been determined for each Btu transporter component, the outer membrane transporter BtuB (13), the periplasmic cobalamin-binding protein BtuF (2), and the ABC permease comprised of BtuC and BtuD in the cytoplasmic membrane (28). The BtuB structures in the absence and presence of calcium, calcium and CNCbl, or the receptor-binding domain of colicin E3 reveal the location of ligand-binding residues (12, 13, 26). The present functional analysis of the contribution of some extracellular loops of BtuB in substrate binding complements this structural information. Future studies could extend this approach to loops 2 to 5, whose identity was not available until the crystallographic analysis was completed. The BtuB crystal structures identified residues that contribute to substrate binding. Calcium-binding residues are present in loops 2 to 4. Hydrogen bonds and polar or van der Waals contacts to CNCbl are formed by residues distributed in the hatch apices and in loops 2 to 4 and 9 to 11 (12, 13). The colicin E3 receptor-binding domain is much larger than CNCbl and makes a close approach to 29 residues of BtuB in the hatch apices and in loops 3 to 5 and 7 to 11 (26). Colicin E3 contacts almost all of the BtuB loops, since loops 1 and 6 are very short. Five residues in loops 3, 4, 9, and 11 are contacted by both colicin E3 and CNCbl. The overlapping binding sites of colicin E3 and CNCbl provide a basis for the competitive binding of these molecules.
The short in-frame deletions in loops 7 to 11 of BtuB described here removed several residues involved in CNCbl contact, such as R497 in L9, Y531 in L10, and Y579 in L11. The phenotypes of these mutants showed that loops 7, 8, 9, and 11 contribute strongly to CNCbl binding and transport. A deletion in L10 had a definite but small effect on CNCbl binding. Although all five C-terminal loops contribute to CNCbl binding and transport, none of the deletions completely abrogated CNCbl uptake. It is not known whether the inhibitory effect of deletions in loops 7 and 8, which were not observed to contact CNCbl, reflect their participation in weak interactions with CNCbl or result from some misalignment of other loops in these deletions.
The colicin E3R domain makes a close approach to residues in the hatch apices and most extracellular loops. In light of the large number of contacts, it is not surprising that single amino acid substitutions did not prevent colicin binding. No loop deletion blocked colicin E1 action and caused at most a 2-log decrease in the titer for colicin E1. No loop deletion except ΔL7-1 had a major effect on susceptibility to colicin E3. Either each loop makes a small contribution to colicin binding or multiple binding surfaces act independently. In contrast, deletions in loops 8 to 10 resulted in resistance to BF23, and ΔL8 conferred resistance even when BtuB was overexpressed. Thus, phage BF23 binding differs from binding of the E colicins. The ability of FhuA to mediate entry of phage T5 was partially impaired by deletion of loop 8 (16), indicating that the interactions of these two closely related phages with their respective receptors occur at the same location on the surface but require different extents of the surface.
Investigation of the competition between colicin E1 and CNCbl was facilitated by the use of a rapid and more quantitative assay system. Deletion of loop 9 or 11 abrogated this competition. Both of these loops contain residues, R497 in loop 9 and Y579 in loop 11, that contact CNCbl and colicin E3R. When either loop was disrupted, binding of CNCbl was greatly decreased but colicin E1 action was little affected, thereby accounting for the lack of protection by CNCbl. A deletion in loop 10 caused a relatively small defect in CNCbl binding but caused the greatest decrease in colicin E1 action. The ΔL10 deletion removed residues 534 and 536, which are contact sites for colicin E3. Assuming that the binding extent of colicin E1 is similar to that of colicin E3, the removal of a surface that contributes to appreciable binding of colicin but not CNCbl could account for the greater degree of protection by CNCbl and colicin E1 that was seen. The deletions in loops 7 and 8 also conferred a higher level of protection by CNCbl against colicin E1. These deletions strongly decreased binding of CNCbl, and thus we expected that CNCbl could no longer compete effectively with colicin E1.
Comparison to similar deletions in the loops of other TBDT showed some differences in their contributions to substrate binding. For FecA, deletions in loops 7, 8, and 11 strongly impaired ferric citrate binding, whereas deletions in loops 9 and 10 had lesser effects (37). Binding of ferrichrome by FhuA was only modestly affected by deletions in loops 7 through 10, although loop 11 was essential (16). For FepA, deletions in loops 7 and 8 eliminated binding and transport of ferric enterobactin (31). Thus, loop 11 plays a crucial role in the function of all TBDT examined. This loop starts with the highly conserved NLFD motif. In our experiments with BtuB, deletion of this motif seemed to decrease the amount of BtuB in the OM by about one-half and had a corresponding effect on all BtuB phenotypes. We suggest that this motif contributes to OM insertion but not to substrate binding or transport by BtuB.
In conclusion, multiple loops in BtuB form overlapping and apparently independent binding surfaces for specific substrates. Some of the same BtuB residues are involved in functional contact with CNCbl and colicin E3R, which shows the remarkable ability of at least these two very different substrates to be recognized by the same constellation of binding residues of BtuB.
Acknowledgments
We appreciate the advice and comments of Clive Bradbeer, Nazir Barekzi, and David Chimento.
This work was supported by research grant GM19078 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Bassford, P. J., Jr., and R. J. Kadner. 1977. Genetic analysis of components involved in vitamin B12 uptake in Escherichia coli. J. Bacteriol. 132:796-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borths, E. L., K. P. Locher, A. T. Lee, and D. C. Rees. 2002. The structure of Escherichia coli BtuF and binding to its cognate ATP binding cassette transporter. Proc. Natl. Acad. Sci. USA 99:16642-16647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradbeer, C., and A. Gudmundsdottir. 1990. Interdependence of calcium and cobalamin binding by wild-type and mutant BtuB protein in the outer membrane of Escherichia coli. J. Bacteriol. 172:4919-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradbeer, C., and M. L. Woodrow. 1976. Transport of vitamin B12 in Escherichia coli: energy dependence. J. Bacteriol. 128:99-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchanan, S. K., B. S. Smith, L. Venkatramani, D. Xia, L. Esser, M. Palnitkar, R. Chakraborty, D. van der Helm, and J. Deisenhofer. 1999. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 6:56-63. [DOI] [PubMed] [Google Scholar]
- 6.Cadieux, N., C. Bradbeer, and R. J. Kadner. 2000. Sequence changes in the Ton box region of BtuB affect its transport activities and interaction with TonB protein. J. Bacteriol. 182:5954-5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cadieux, N., and R. J. Kadner. 1999. Site-directed disulfide bonding reveals an interaction site between energy-coupling protein TonB and BtuB, the outer membrane cobalamin transporter. Proc. Natl. Acad. Sci. USA 96:10673-10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cadieux, N., P. G. Phan, D. S. Cafiso, and R. J. Kadner. 2003. Differential substrate-induced signaling through the TonB-dependent transporter BtuB. Proc. Natl. Acad. Sci. USA 100:10688-10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavard, D. 1976. Sensitization of colicin K-treated bacteria by sodium dodecyl sulfate: presence of free colicin in colicin K-treated cultures of Escherichia coli. Antimicrob. Agents Chemother. 9:639-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cavard, D., and C. Lazdunski. 1981. Involvement of BtuB and OmpF proteins in binding and uptake of colicin A. FEMS Lett. 12:311-316. [Google Scholar]
- 11.Cavard, D., and C. Lazdunski. 1994. Rescue by vitamin B12 of Escherichia coli cells treated with colicins A and E allow measurement of the kinetics of colicin binding on BtuB. FEMS Microbiol. Lett. 116:37-42. [DOI] [PubMed] [Google Scholar]
- 12.Chimento, D. P., R. J. Kadner, and M. C. Wiener. 2003. The Escherichia coli outer membrane cobalamin transporter BtuB: structural analysis of calcium and substrate binding, and identification of orthologous transporters by sequence/structure conservation. J. Mol. Biol. 332:999-1014. [DOI] [PubMed] [Google Scholar]
- 13.Chimento, D. P., A. K. Mohanty, R. J. Kadner, and M. C. Wiener. 2003. Substrate-induced transmembrane signaling in the cobalamin transporter BtuB. Nat. Struct. Biol. 10:394-401. [DOI] [PubMed] [Google Scholar]
- 14.DiGirolamo, P. M., R. J. Kadner, and C. Bradbeer. 1971. Isolation of vitamin B12 transport mutants of Escherichia coli. J. Bacteriol. 106:751-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DiMasi, D. R., J. S. White, C. A. Schnaitman, and C. Bradbeer. 1973. Transport of vitamin B12 in Escherichia coli: common receptor sites for vitamin B12 and the E colicins on the outer membrane of the cell envelope. J. Bacteriol. 115:506-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Endriss, F., and V. Braun. 2004. Loop deletions indicate regions important for FhuA transport and receptor functions in Escherichia coli. J. Bacteriol. 186:4818-4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faraldo-Gomez, J. D., and M. S. P. Sansom. 2003. Acquisition of siderophores in gram-negative bacteria. Nat. Rev. Mol. Cell Biol. 4:105-116. [DOI] [PubMed] [Google Scholar]
- 18.Ferguson, A. D., R. Chakraborty, B. S. Smith, L. Esser, D. van der Helm, and J. Deisenhofer. 2002. Structural basis of gating by the outer membrane transporter FecA. Science 295:1715-1719. [DOI] [PubMed] [Google Scholar]
- 19.Ferguson, A. D., E. Hofmann, J. W. Coulton, K. Diederichs, and W. Welte. 1998. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science 282:2215-2220. [DOI] [PubMed] [Google Scholar]
- 20.Gudmundsdottir, A., P. E. Bell, M. D. Lundrigan, C. Bradbeer, and R. J. Kadner. 1989. Point mutations in a conserved region (TonB box) of Escherichia coli outer membrane protein BtuB affect vitamin B12 transport. J. Bacteriol. 171:6526-6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gudmundsdottir, A., C. Bradbeer, and R. J. Kadner. 1988. Altered binding and transport of vitamin B12 resulting from insertion mutations in the Escherichia coli btuB gene. J. Biol. Chem. 263:14224-14230. [PubMed] [Google Scholar]
- 22.Heller, K., B. J. Mann, and R. J. Kadner. 1985. Cloning and expression of the gene for the vitamin B12 receptor protein in the outer membrane of Escherichia coli. J. Bacteriol. 161:896-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.James, R., C. Kleanthous, and G. R. Moore. 1996. The biology of E colicins: paradigms and paradoxes. Microbiology 142:1569-1580. [DOI] [PubMed] [Google Scholar]
- 25.Koester, W., A. Gudmundsdottir, M. D. Lundrigan, A. Seiffert, and R. J. Kadner. 1991. Deletions or duplications in the BtuB protein affect its level in the outer membrane of Escherichia coli. J. Bacteriol. 173:5639-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurisu, G., S. D. Zakharov, M. V. Zhalina, S. Bano, V. Y. Eroukova, T. I. Rokitskaya, Y. N. Antonenko, M. C. Wiener, and W. A. Cramer. 2003. The structure of BtuB with bound colicin E3 R-domain implies a translocon. Nat. Struct. Biol. 10:948-954. [DOI] [PubMed] [Google Scholar]
- 27.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Locher, K. P., A. T. Lee, and D. C. Rees. 2002. The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science 296:1091-1098. [DOI] [PubMed] [Google Scholar]
- 29.Locher, K. P., B. Rees, R. Koebnik, A. Mitschler, L. Moulinier, J. P. Rosenbusch, and D. Moras. 1998. Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell 95:771-778. [DOI] [PubMed] [Google Scholar]
- 30.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Newton, S. M. C., J. D. Igo, D. C. Scott, and P. E. Klebba. 1999. Effect of loop deletions on the binding and transport of ferric enterobactin by FepA. Mol. Microbiol. 32:1153-1165. [DOI] [PubMed] [Google Scholar]
- 32.Nikaido, H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Postle, K., and R. J. Kadner. 2003. Touch and go: tying TonB to transport. Mol. Microbiol. 49:869-882. [DOI] [PubMed] [Google Scholar]
- 34.Roth, J. R., J. G. Lawrence, and T. A. Bobik. 1996. Cobalamin (coenzyme B12): synthesis and biological significance. Annu. Rev. Microbiol. 50:137-181. [DOI] [PubMed] [Google Scholar]
- 35.Sabet, S. F., and C. A. Schnaitman. 1973. Purification and properties of the colicin E3 receptor of Escherichia coli. J. Biol. Chem. 248:1797-1806. [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 37.Sauter, A., and V. Braun. 2004. Defined inactive FecA derivatives mutated in functional domains of the outer membrane transport and signaling protein of Escherichia coli K-12. J. Bacteriol. 186:5303-5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwartz, S. A., and D. R. Helinski. 1971. Purification and characterization of colicin E1. J. Biol. Chem. 246:6318-6327. [PubMed] [Google Scholar]
- 39.Stoscheck, C. M. 1990. Quantitation of protein. Methods Enzymol. 182:50-68. [DOI] [PubMed] [Google Scholar]