Abstract
The dendritic-cell-specific intercellular adhesion molecule nonintegrin (DC-SIGN) CD209 is a receptor for Escherichia coli K-12 that promotes bacterial adherence and phagocytosis. However, the ligand of E. coli for DC-SIGN has not yet been identified. In this study, we found that DC-SIGN did not mediate the phagocytosis of several pathogenic strains of E. coli, including enteropathogenic E. coli, enterohemorrhagic E. coli, enterotoxigenic E. coli, and uropathogenic E. coli, in dendritic cells or HeLa cells expressing human DC-SIGN antigen. However, we showed that an outer core lipopolysaccharide (LPS) (rough) mutant, unlike an inner core LPS (deep rough) mutant or O-antigen-expressing recombinant of E. coli K-12 was phagocytosed. These results demonstrate that the host cells expressing DC-SIGN can phagocytose E. coli in part by interacting with the complete core region of the LPS molecule. These results provide a mechanism for how O antigen acts as an antiphagocytic factor.
Dendritic cells (DCs) are professional phagocytic cells that play an essential role in host defense against invading pathogens and help control and maintain innate and adaptive immunity. Microbial pathogens could subvert functions of DCs, such as antigen presentation, production of different chemokines and cytokines, and phagocytosis of microorganisms (1, 31).
Lipopolysaccharide (LPS) is a uniquely bacterial glycolipid. It consists of three structurally distinct domains; lipid A, the membrane anchor and endotoxic portion of LPS; the core saccharide consisting of a branched chain of nonrepeating hexose and heptose sugars; and the O-antigen side chain, a repeating unit of sugars that extends into the extracellular milieu. The core can be further divided into the outer core (semivariable composition of sugars and linkages among bacterial species and strains) and the inner core (considerably conserved among bacterial species and strains). Most enteric bacterial pathogens contain O antigens as a component of their LPS, and O antigens can promote resistance to serum killing and phagocytosis, enhancing the pathogenicity of members of the family Enterobacteriaceae, such as Escherichia coli, Shigella spp., and Salmonella (2, 6, 20, 21, 28).
DCs uniquely express a C-type lectin called DC-specific intercellular adhesion molecule-grabbing nonintegrin, CD209 (dendritic-cell-specific intercellular adhesion molecule nonintegrin [DC-SIGN]). DCs can interact with microorganisms by using DC-SIGN. Bacteria, such as Helicobacter pylori and certain strains of Klebsiella pneumoniae, interact with DC-SIGN through LPS structures that contain Lex [Galβ1-4(Fucα1-3)GlcNAcβ] or mannose sugar residues (18, 34, 35, 38). Mycobacterium tuberculosis uses its mannose-capped cell wall component to interact with DC-SIGN to promote its own internalization, which in turn inhibits DC functions (9, 34). DC-SIGN also serves as a receptor for the gp120 antigen of human immunodeficiency virus type 1 (HIV-1) and acts as a carrier for HIV-1, delivering virus to target cells, such as CD4 lymphocytes (7, 8, 19). Although signal transduction mechanisms for how DC-SIGN mediates the uptake of microorganisms are not yet clear, the immunoreceptor tyrosine-based activation motif (ITAM)-like motif on the cytoplasmic domain might play a role in the uptake of bacteria or viruses (35).
Recently, we demonstrated that DC-SIGN promotes adherence and phagocytosis of a nonpathogenic E. coli K-12 strain in both DCs and HeLa cells expressing human DC-SIGN antigen (HeLa-DC-SIGN) (39). In this study, we investigated whether DC-SIGN also served as a receptor for pathogenic E. coli and defined a region of the core LPS as a ligand on E. coli for DC-SIGN.
MATERIALS AND METHODS
Bacterial strains, antibodies, reagents, and culture cell lines.
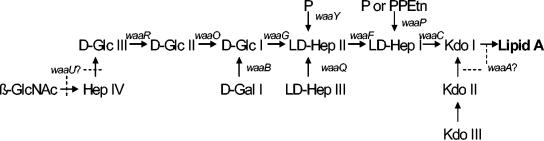
E. coli HB101, a laboratory-derived isolate (hybrid of E. coli K-12) that lacks O antigen, has recently been demonstrated to bind DC-SIGN (39). E. coli K-12 strain CS180 contains core LPS but lacks O antigen (29). Two derivatives of CS180, CS1861 (CS180 harboring pSS37, a plasmid containing all of the genes necessary for the expression of the Shigella dysenteriae 1 O antigen) and a deep rough mutant CS2429 (waaC) (lacking both O antigen and most of the core) were used to assess the role of LPS in bacterium-DC interactions (11, 13, 29). Strains CS2198, CS2210, CS2274, and CS2774, isogenic mutants of CS180, contain mutations in the waaR, waaP, waaR waaB (double mutant), and waaQ genes, respectively (12, 24-27) (Fig. 1). These strains produce intermediate-length core LPS and were used to define the minimum E. coli K-12 LPS structure necessary to act as a DC-SIGN ligand.
FIG. 1.
Structures of inner and outer core regions of E. coli K-12 LPS and the genes involved in their synthesis. Genes involved in the biosynthesis of core LPS are shown at their approximate site of action (solid line). The sites, which are variably substituted, are indicated by broken lines. Abbreviations: GlcNAc, N-acetylglucosamine; Glc, glucose; Hep, heptose; Gal, galactose; P, phosphate; PPEtn, phosphoethanolamine; Kdo, 2-keto-3-deoxyoctonate.
Enteropathogenic E. coli (EPEC) (ATCC 49106), enterohemorrhagic E. coli (EHEC) (ATCC 438950), and enterotoxigenic E. coli (ETEC) (BAA-454) were purchased from American Type Culture Collection. The uropathogenic E. coli (UPEC) (116) strain was isolated from the urinary tract of a patient during an outbreak in a nursing home in New Zealand (unpublished data). E. coli GR-12, an UPEC, was originally isolated from a patient with pyelonephritis (33). GR-12-SMB-316, a derivative from E. coli GR-12, is a rough strain, in which the genes for O-antigen syntheses have been deleted (2). Bacteria were cultured on Luria-Bertani medium (LB) supplemented with 1.5% agar at 37°C overnight.
Anti-CD209 monoclonal antibody (MAb), specific for DC-SIGN, was purchased from Pharmingen, San Diego, Calif. YTH71.3., an anti-CD66 antibody, which recognizes CEACAM1, CEACAM6, and CEACAM3 (CD66acd) (23), was purchased from Harlan Bioproducts (Indianapolis, Ind.). Mannan, the ligand antagonist of human mannose receptor, was purchased from Sigma (Steinheim, Germany).
HeLa-DC-SIGN cells were constructed by transfecting HeLa cells with human DC-SIGN cDNA and selected for surface antigen expression as described previously (22, 30).
Preparation of DCs.
Peripheral blood mononuclear cells (PBMC) were isolated from buffy coats obtained from the Indiana Blood Center (Indianapolis, Ind.) by density gradient centrifugation over FicollPaqueplus (1.077 g/ml; Pharmacia, Piscataway, N.J.) (17). Acquisition of human blood was approved by the Institutional Review Board (IRB) and Study Committees at Indiana University School of Medicine (IUPUI and Clarian). Buffy coats were diluted 1:4 with phosphate-buffered saline (PBS) and loaded in a 1:1 (vol/vol) ratio on Ficoll and centrifuged without stopping for 30 min. PBMC were washed four times with PBS, and monocytes were purified from PBMC using CD14 microbeads (Miltenyi Biotec, Auburn, Calif.) as previously described (17). To increase purity, cells were passed over a second CD14 microbead column. The final purity of the isolated monocytes was >98% as assessed by labeling with a fluorescein isothiocyanate (FITC)-conjugated CD14 antibody (Caltag Laboratories, Burlingame, Calif.) and flow cytometric analysis. Purified CD14+ monocytes (5 × 105 cells/ml) were cultured for 6 days to promote differentiation of immature monocyte-derived DCs in culture medium consisting of RPMI 1640 (BioWhittaker, Walkersville, Md.), 10% heat-inactivated fetal bovine serum (HyClone, Logan, Utah), 100 U of penicillin per ml, and 100 μg of streptomycin per ml in the presence of 20 ng of recombinant human granulocyte-macrophage colony-stimulating factor (Immunex, Seattle, Wash.) per ml and 10 ng of recombinant human interleukin 4 (Peprotech, Rock Hill, N.J.) per ml. The DCs derived from these cultured monocytes display typical dendrites, and in mixed lymphocyte cultures, they promote activation of alloreactive T cells. The phenotypes of these cells are HLA-DR+ CD1a+ CD86+ CD40+ CD14− (17). Upon LPS stimulation, these DCs express CD83 (40, 41).
Adherence and phagocytosis assays.
The assays for adherence and phagocytosis have been described previously (3-5). Briefly, DCs and HeLa cells were plated into the wells of 24-well plates, and each well was covered with a coverslip. DCs were suspended in RPMI 1640 with 2% fetal calf serum (FCS) at a concentration of 4 × 105/ml. A 0.5-ml sample of each of these cell suspensions was added to each well in 24-well plates, and after the addition of 50 μl of bacterial suspensions at a concentration of 4 × 107 CFU/ml, the cells were incubated at 37°C for 2.5 h in the presence of 5% CO2. The DC monolayers were washed twice with PBS with a cytospin and fixed with 2% paraformaldehyde in PBS containing Giemsa stain. The number of associated bacteria (adherent and internalized) per DC was determined by microscopy by counting the bacteria associated with 100 cells on the coverslips. For the HeLa cells, the associated bacteria were quantified by washing the cells three times with RPMI 1640 with 2% FCS and plating the culture after the cells were lysed by 0.5% saponin (Calbiochem Corp., San Diego, Calif.).
To determine the internalization of bacteria, gentamicin, which kills extracellular bacteria but cannot penetrate into host cells, was added into each well to a final concentration of 100 μg/ml, and the cultures were incubated for 90 min. The cells were suspended in PBS containing 0.5% saponin, diluted, and plated on LB plates. The level of internalization of bacteria in DCs and HeLa cells was calculated by determining the CFU recovered from lysed cells. All experiments were performed in triplicate, and data were expressed as means ± standard errors. Statistical significance was calculated using the Student's t test. It should be noted that the infection time selected in our experiment is based on our preliminary experiments and a balance between the numbers of bacteria killed by host cells and the time needed for bacterial entry.
RESULTS
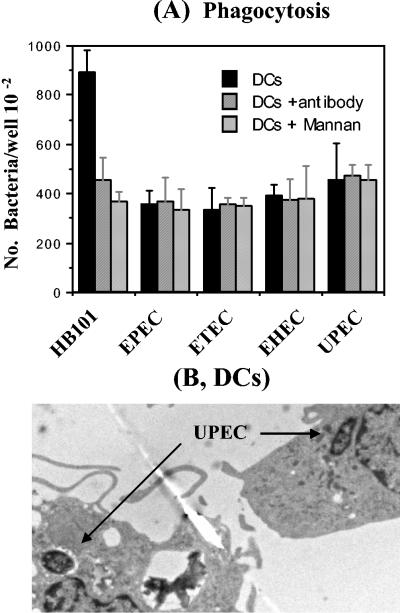
Neither anti-DC-SIGN antibody nor mannan inhibit the phagocytosis of pathogenic E. coli by DCs. We recently showed that DCs phagocytose E. coli HB101, an avirulent E. coli strain. We also showed that this interaction was reduced by an anti-DC-SIGN MAb and by mannan. To investigate whether a similar interaction occurs between pathogenic E. coli and DCs, we performed the same phagocytosis assay with four pathogenic E. coli strains, EPEC, ETEC, EHEC, and UPEC strains. All of these pathogenic E. coli strains were phagocytosed by DCs to some extent, although there was less uptake than with HB101 (Fig. 2A). Phagocytosis of UPEC (Fig. 2B) and HB101 (39) was confirmed by electron microscopy. In contrast, phagocytosis of these pathogenic E. coli by DCs was not reduced by the addition of anti-DC-SIGN antibody or mannan, suggesting that DCs use different phagocytic mechanisms to internalize HB101 and the pathogenic E. coli.
FIG. 2.
DCs phagocytose pathogenic E. coli in a DC-SIGN-independent manner. Four pathogenic E. coli strains were examined for phagocytosis by DCs alone or in the presence of anti-DC-SIGN or mannan. DCs were incubated for 2.5 h with these E. coli strains, and the internalization of E. coli by DCs was measured by killing the extracellular bacteria with gentamicin (100 μg/ml [final concentration]) (A) and confirmed by electron microscopy (B), showing the phagocytosed UPEC. The number of phagocytosed bacteria was determined by counting CFU recovered after gentamicin treatment.
DC-SIGN does not mediate phagocytosis of pathogenic E. coli.
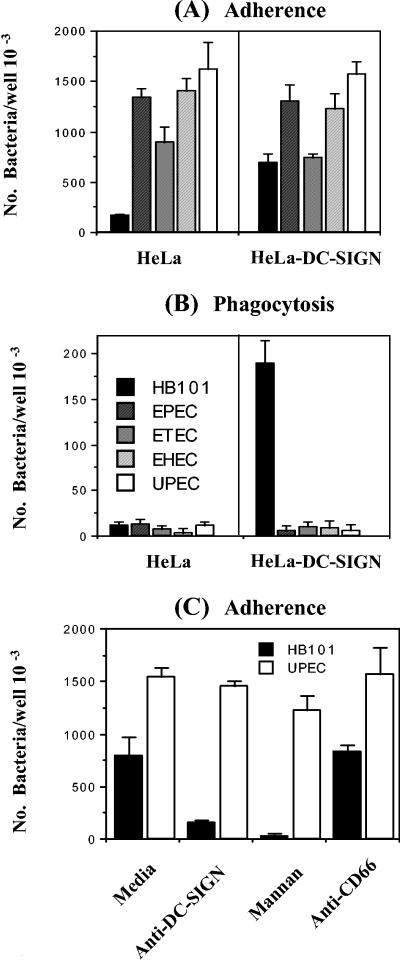
To further investigate the interaction of DCs with pathogenic E. coli, a stably transfected DC-SIGN HeLa cell line (HeLa-DC-SIGN) was used to test its ability to bind and phagocytose the panel of pathogenic E. coli strains. E. coli HB101 bound to and was phagocytosed by HeLa-DC-SIGN (Fig. 3A and B), but not to HeLa cells, as shown previously (39). In contrast, all pathogenic E. coli strains bound to both HeLa and HeLa-DC-SIGN. Surprisingly, none of the pathogenic E. coli strains could be taken up by HeLa-DC-SIGN. In addition, the UPEC-mediated adherence was not inhibited by anti-DC-SIGN antibody or mannan (Fig. 3C), which blocked the interaction of HB101 with HeLa-DC-SIGN (39). These results demonstrate that DC-SIGN is not a receptor for these pathogenic strains of E. coli.
FIG. 3.
HeLa-DC-SIGN cells phagocytose E. coli HB101, but not the pathogenic E. coli strains. The adherence (A) and phagocytosis (B) of these pathogenic E. coli strains with HeLa and HeLa-DC-SIGN were performed as described previously (39). E. coli HB101 serves as a positive control. Anti-DC-SIGN antibody (5 μg/ml) and mannan (0.5 mg/ml) were also used to inhibit the adherence of HB101 and UPEC with HeLa-DC-SIGN (C). Anti-DC66 antibody (5 μg/ml) was used as a negative control. The phagocytic rates values for HB101 and UPEC were statistically significantly different (P < 0.001).
The core region of LPS is required to mediate the E. coli-DC interaction.
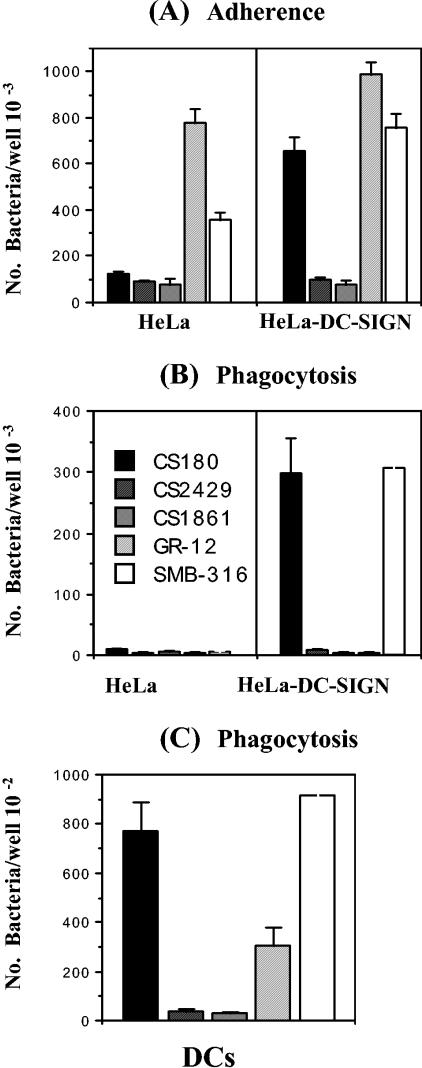
DC-SIGN belongs to the mannose receptor family, and interactions with bacteria usually occur through the mannose-rich components on the bacterial surface. Several bacteria target mannose receptors by using the mannose components on O antigens (35, 38). As described above, E. coli HB101 used in this study lacks O antigen. To examine which domains of the E. coli LPS may interact with DC-SIGN, two sets of strains were used to examine their interaction with DCs and HeLa-DC-SIGN. The first set includes three E. coli K-12 strains: CS180, a naturally rough strain; its isogenic deep rough waaC mutant, CS2429, containing only lipid A and two 2-keto-3-deoxyoctonate (KDO) molecules (Fig. 1); and CS1861, the CS180 strain expressing the full-length O antigen of S. dysenteriae 1 from pSS37 (11, 13, 29). The second set of strains consists of a smooth UPEC (GR-12) and its rough isogenic derivative, SMB-316. Strains CS180 and SMB-316 promoted typical DC-SIGN-mediated adherence and phagocytosis (Fig. 4A and B), but strains CS2429 and CS1861 were resistant to both DCs (Fig. 4C) and HeLa-DC-SIGN (Fig. 4B). Strain GR-12 was also completely resistant to phagocytosis via HeLa-DC-SIGN (Fig. 4B). It was phagocytosed to a certain level by DCs (Fig. 4C). This phagocytosis was probably not mediated by DC-SIGN for the reasons described above. This result demonstrated that the ligand of E. coli K-12 for DC-SIGN is in the core region of LPS and suggests a role for O antigen in the bacterial resistance of phagocytosis by DCs.
FIG. 4.
HeLa-DC-SIGN and DCs cannot phagocytose the deep rough mutant of E. coli and the E. coli expressing O antigen. The adherence (A) and internalization (B) of E. coli K-12 CS180, its derivatives, CS2429 and CS1861, E. coli UPEC GR-12 and its derivative by HeLa, HeLa-DC-SIGN, and DCs (C) were performed as described in the legend to Fig. 2. The number of phagocytosed bacteria was determined by the recovery of bacteria from the gentamicin protection assay.
Examination of the sugars responsible for core LPS-DC-SIGN interaction.
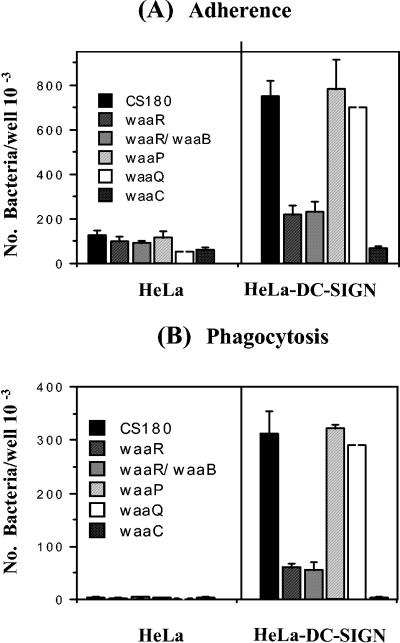
To examine which sugar residues in the core LPS are responsible for the interaction with DC-SIGN in E. coli, additional E. coli K-12 core LPS mutants, waaR, waaR waaB, waaP, and waaQ mutants, were used. The waaC mutant and strain CS180 were used here as the negative and positive controls, respectively. All of these strains were tested for their ability to adhere to and be phagocytosed by HeLa and HeLa-DC-SIGN. As shown in Fig. 5, the waaR and waaR waaB mutants demonstrated the same level of intermediate resistance to phagocytosis by HeLa-DC-SIGN as the waaC mutant did. However, the waaP and waaQ mutants behaved like their isogenic parent strain, CS180, in that they were fully sensitive to phagocytosis by HeLa-DC-SIGN. These results in combination with data from rough strain SMB-316 strongly suggest that it is the core LPS itself, rather than other components, that bind to DC-SIGN. It is possible that the ligand could be other outer membrane proteins, since the O antigen blocks access to many surface structures which are exposed in rough mutants, and the deep rough defects in the LPS core might result in defective (or decreased) expression of other outer membrane proteins, which could be the ligand. However, because the waaR mutant is also resistant to the phagocytosis and is not a deep rough mutant, we conclude that the ligand for DC-SIGN is from the core LPS region.
FIG. 5.
Determination of the epitopes on core LPS binding to DC-SIGN. The adherence (A) and internalization (B) of the core LPS mutants (the waaR, waaP, waaB, waaQ, and waaC mutants and the waaR waaB double mutant) and their parent strain, CS180 (Fig. 1), were examined and measured as described in the legend to Fig. 2.
DISCUSSION
Our previous study demonstrated that DC-SIGN could be a receptor for the phagocytosis of E. coli HB101 (39). Transfected HeLa cells expressing DC-SIGN, but not untransfected HeLa cells, were able to bind and avidly internalize E. coli. This interaction was blocked by mannan, an antagonist of human mannose receptors, including DC-SIGN, and by antibody specific for DC-SIGN (39).
In this study, we examined the roles of DCs and DC-SIGN in interactions with several pathogenic E. coli strains, including EPEC, EHEC, ETEC, and UPEC and found that all pathogenic E. coli strains were phagocytosed to some degree by DCs. However, unlike nonpathogenic E. coli HB101, phagocytosis of these pathogenic E. coli strains was not mediated directly by DC-SIGN. Since E. coli HB101 is a rough strain that does not contain the O-antigen polysaccharide on its LPS, which most pathogenic E. coli strains possess, we speculated that the ligand(s) of DC-SIGN receptors in E. coli could be sterically hindered in these pathogenic E. coli strains by O antigens.
E. coli K-12 (HB101 and CS180 strains used in this study) are rough strains, lacking the polysaccharide O-side chain. Therefore, if E. coli LPS is involved in binding to DC-SIGN, it should be the core or lipid A region. To test the hypothesis, we used a deep rough E. coli K-12 strain, CS2429, which lacks the O antigen and the majority of the core region, and strain CS1861, which covers the core region with the O antigen of S. dysenteriae 1. It should be noted that E. coli and S. dysenteriae are very closely related genetically and have homologous sequences for many genes involved in the synthesis of LPS (15, 16, 36). We also used a pathogenic E. coli, UPEC (GR-12) and its O-antigen deletion mutant. Our findings demonstrate that the core region of LPS is required to mediate the E. coli-DC-SIGN interaction.
We attempted to determine which specific sugar residues in the core region mediate the binding between DC-SIGN and E. coli K-12 using several core LPS mutants as shown in Fig. 1. Although we were unable to acquire all of the core LPS mutants to systematically analyze their involvement, based on the results in Fig. 5, we propose the following scenario. The fact that the waaP and waaQ mutants did not influence the adherence and phagocytosis by HeLa-DC-SIGN suggests that the inner core branch constituents added by the WaaP enzyme (a branch phosphoethanolamine or phosphate) and the WaaQ enzyme (a branch heptose III) are not involved in the interaction. The most significant results were the results for the waaR mutant and waaR waaB double mutant, which are resistant to phagocytosis by HeLa-DC-SIGN, indicating that it is these specific sugar residues that determine the core-DC-SIGN interaction, since both mutants are rough, rather than deep rough, mutants. This result also indicated that d-Gal I might not participate in the interaction with DC-SIGN. Because waaU is proposed to encode Hep IV transferase, it is not well documented whether the core LPS of E. coli K-12 contains β-GlcNAc and Hep IV, although there are some biochemical data to support the existence of the Hep IV position at the terminal Glc in the chain (10, 37). Therefore, the specific molecule in core LPS that is responsible for interacting with DC-SIGN could be either β-GlcNAc or d-Glc III. Moreover, a recent report on core LPS of Neisseria meningitidis (32) and our preliminary data from Neisseria gonorrhoeae (data not shown) suggested that the epitope may be β-GlcNAc that binds to DC-SIGN. We are in the process of analyzing each step in the biosynthesis of the LPS core to determine the precise biochemical ligand for DC-SIGN.
One major role of O antigens in the pathogenicity of members of the family Enterobacteriaceae, such as E. coli, Shigella, Klebsiella, and Salmonella, is promoting resistance to serum killing and phagocytosis (2, 6, 20, 21, 28), but the mechanism of how these gram-negative enteric bacilli resist phagocytosis by host cells is not clear (2, 6). Our results provide a potential mechanism, suggesting that O antigen functions as an antiphagocytic factor simply by shielding the ligand necessary for host cell contact. Very interestingly, a similar observation was obtained for plasminogen activation in Yersinia pestis and Salmonella enterica (14). Since the LPS cores of members of the family Enterobacteriaceae are similar, our results suggest that DC-SIGN may also serve as a receptor for LPS cores of Salmonella, Yersinia, and Shigella. As indicated above, some core LPS mutants from either N. meningitidis (32) or N. gonorrhoeae (data not shown) bind to DC-SIGN.
Acknowledgments
We thank Ines Chen and Margaret Bauer for useful suggestions and editorial comments on the manuscript.
This work was supported in part by PHS grant R01AI 47736 to T. Chen.
REFERENCES
- 1.Agrawal, A., J. Lingappa, S. H. Leppla, S. Agrawal, A. Jabbar, C. Quinn, and B. Pulendran. 2003. Impairment of dendritic cells and adaptive immunity by anthrax lethal toxin. Nature 424:329-334. [DOI] [PubMed] [Google Scholar]
- 2.Burns, S. M., and S. I. Hull. 1998. Comparison of loss of serum resistance by defined lipopolysaccharide mutants and an acapsular mutant of uropathogenic Escherichia coli O75:K5. Infect. Immun. 66:4244-4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, T., S. Bolland, I. Chen, J. Parker, M. Pantelic, F. Grunert, and W. Zimmermann. 2001. The CGM1a (CEACAM3/CD66d) mediated phagocytic pathway of Neisseria gonorrhoeae expressing opacity (Opa) proteins is also the pathway to cell death. J. Biol. Chem. 276:17413-17419. [DOI] [PubMed] [Google Scholar]
- 4.Chen, T., and E. Gotschlich. 1996. CGM1a antigen of neutrophils, a receptor of gonococcal opacity proteins. Proc. Natl. Acad. Sci. USA 93:14851-14856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, T., F. Grunert, A. Medina-Marino, and E. Gotschlich. 1997. Several carcinoembryonic antigens (CD66) serve as receptors for gonococcal opacity proteins. J. Exp. Med. 185:1557-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cortes, G., N. Borrell, B. de Astorza, C. Gomez, J. Sauleda, and S. Alberti. 2002. Molecular analysis of the contribution of the capsular polysaccharide and the lipopolysaccharide O side chain to the virulence of Klebsiella pneumoniae in a murine model of pneumonia. Infect. Immun. 70:2583-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engering, A., S. J. Van Vliet, T. B. Geijtenbeek, and Y. Van Kooyk. 2002. Subset of DC-SIGN+ dendritic cells in human blood transmits HIV-1 to T lymphocytes. Blood 100:1780-1786. [DOI] [PubMed] [Google Scholar]
- 8.Geijtenbeek, T. B., D. S. Kwon, R. Torensma, S. J. van Vliet, G. C. van Duijnhoven, J. Middel, I. L. Cornelissen, H. S. Nottet, V. N. KewalRamani, D. R. Littman, C. G. Figdor, and Y. van Kooyk. 2000. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 100:587-597. [DOI] [PubMed] [Google Scholar]
- 9.Geijtenbeek, T. B., S. J. Van Vliet, E. A. Koppel, M. Sanchez-Hernandez, C. M. Vandenbroucke-Grauls, B. Appelmelk, and Y. Van Kooyk. 2003. Mycobacteria target DC-SIGN to suppress dendritic cell function. J. Exp. Med. 197:7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heinrichs, D. E., J. A. Yethon, and C. Whitfield. 1998. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol. Microbiol. 30:221-232. [DOI] [PubMed] [Google Scholar]
- 11.Klena, J., R. S. Ashford II, and C. A. Schnaitman. 1992. Role of Escherichia coli K-12 rfa genes and the rfp gene of Shigella dysenteriae 1 in generation of lipopolysaccharide core heterogeneity and attachment of O antigen. J. Bacteriol. 174:7297-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klena, J. D., E. Pradel, and C. A. Schnaitman. 1992. Comparison of lipopolysaccharide biosynthesis genes rfaK, rfaL, rfaY, and rfaZ of Escherichia coli K-12 and Salmonella typhimurium. J. Bacteriol. 174:4746-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klena, J. D., and C. A. Schnaitman. 1993. Function of the rfb gene cluster and the rfe gene in the synthesis of O antigen by Shigella dysenteriae 1. Mol. Microbiol. 9:393-402. [DOI] [PubMed] [Google Scholar]
- 14.Kukkonen, M., M. Suomalainen, P. Kyllonen, K. Lahteenmaki, H. Lang, R. Virkola, I. M. Helander, O. Holst, and T. K. Korhonen. 2004. Lack of O-antigen is essential for plasminogen activation by Yersinia pestis and Salmonella enterica. Mol. Microbiol. 51:215-225. [DOI] [PubMed] [Google Scholar]
- 15.Lan, R., M. C. Alles, K. Donohoe, M. B. Martinez, and P. R. Reeves. 2004. Molecular evolutionary relationships of enteroinvasive Escherichia coli and Shigella spp. Infect. Immun. 72:5080-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lan, R., and P. R. Reeves. 2002. Escherichia coli in disguise: molecular origins of Shigella. Microbes Infect. 4:1125-1132. [DOI] [PubMed] [Google Scholar]
- 17.Li, G., Y. J. Kim, C. Mantel, and H. E. Broxmeyer. 2003. P-selectin enhances generation of CD14+CD16+ dendritic-like cells and inhibits macrophage maturation from human peripheral blood monocytes. J. Immunol. 171:669-677. [DOI] [PubMed] [Google Scholar]
- 18.Maeda, N., J. Nigou, J. L. Herrmann, M. Jackson, A. Amara, P. H. Lagrange, G. Puzo, B. Gicquel, and O. Neyrolles. 2003. The cell surface receptor DC-SIGN discriminates between Mycobacterium species through selective recognition of the mannose caps on lipoarabinomannan. J. Biol. Chem. 278:5513-5516. [DOI] [PubMed] [Google Scholar]
- 19.McDonald, D., L. Wu, S. M. Bohks, V. N. KewalRamani, D. Unutmaz, and T. J. Hope. 2003. Recruitment of HIV and its receptors to dendritic cell-T cell junctions. Science 300:1295-1297. [DOI] [PubMed] [Google Scholar]
- 20.Morona, R., C. Daniels, and L. Van Den Bosch. 2003. Genetic modulation of Shigella flexneri 2a lipopolysaccharide O antigen modal chain length reveals that it has been optimized for virulence. Microbiology 149:925-939. [DOI] [PubMed] [Google Scholar]
- 21.Murray, G. L., S. R. Attridge, and R. Morona. 2003. Regulation of Salmonella typhimurium lipopolysaccharide O antigen chain length is required for virulence: identification of FepE as a second Wzz. Mol. Microbiol. 47:1395-1406. [DOI] [PubMed] [Google Scholar]
- 22.Nobile, C., A. Moris, F. Porrot, N. Sol-Foulon, and O. Schwartz. 2003. Inhibition of human immunodeficiency virus type 1 Env-mediated fusion by DC-SIGN. J. Virol. 77:5313-5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pantelic, M., I. Chen, J. Parker, P. Zhang, F. Grunert, and T. Chen. 2004. Retinoic acid treated HL60 cells express CEACAM1 (CD66a) and phagocytose Neisseria gonorrhoeae. FEMS Immunol. Med. Microbiol. 42:261-266. [DOI] [PubMed] [Google Scholar]
- 24.Parker, C. T., A. W. Kloser, C. A. Schnaitman, M. A. Stein, S. Gottesman, and B. W. Gibson. 1992. Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. J. Bacteriol. 174:2525-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parker, C. T., E. Pradel, and C. A. Schnaitman. 1992. Identification and sequences of the lipopolysaccharide core biosynthetic genes rfaQ, rfaP, and rfaG of Escherichia coli K-12. J. Bacteriol. 174:930-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pradel, E., C. T. Parker, and C. A. Schnaitman. 1992. Structures of the rfaB, rfaI, rfaJ, and rfaS genes of Escherichia coli K-12 and their roles in assembly of the lipopolysaccharide core. J. Bacteriol. 174:4736-4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pradel, E., and C. A. Schnaitman. 1991. Effect of rfaH (sfrB) and temperature on expression of rfa genes of Escherichia coli K-12. J. Bacteriol. 173:6428-6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russo, T. A., B. A. Davidson, U. B. Carlino-MacDonald, J. D. Helinski, R. L. Priore, and P. R. Knight III. 2003. The effects of Escherichia coli capsule, O-antigen, host neutrophils, and complement in a rat model of Gram-negative pneumonia. FEMS Microbiol. Lett. 226:355-361. [DOI] [PubMed] [Google Scholar]
- 29.Schnaitman, C. A., and J. D. Klena. 1993. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol. Rev. 57:655-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sol-Foulon, N., A. Moris, C. Nobile, C. Boccaccio, A. Engering, J. P. Abastado, J. M. Heard, Y. van Kooyk, and O. Schwartz. 2002. HIV-1 Nef-induced upregulation of DC-SIGN in dendritic cells promotes lymphocyte clustering and viral spread. Immunity 16:145-155. [DOI] [PubMed] [Google Scholar]
- 31.Starnbach, M. N., and R. J. Collier. 2003. Anthrax delivers a lethal blow to host immunity. Nat. Med. 9:996-997. [DOI] [PubMed] [Google Scholar]
- 32.Steeghs, L., U. Uronen-Hansson, S. Van Vliet, A. Van Mourik, N. Klein, Y. Van Kooyk, R. Callard, J. Van De Winkel, and P. Van Der Ley. 2004. Lipopolysaccharide-mediated targeting of Neisseria meningitidis to dendritic cells: binding of lgtB LPS to DC-SIGN, p. 120-121. Abstr. 14th Int. Pathog. Neisseria Meet., Milwaukee, Wis., 5 to 10 September 2004.
- 33.Svanborg Eden, C., R. Hull, S. Falkow, and H. Leffler. 1983. Target cell specificity of wild-type E. coli and mutants and clones with genetically defined adhesins. Prog. Food Nutr. Sci. 7:75-89. [PubMed] [Google Scholar]
- 34.Tailleux, L., O. Schwartz, J. L. Herrmann, E. Pivert, M. Jackson, A. Amara, L. Legres, D. Dreher, L. P. Nicod, J. C. Gluckman, P. H. Lagrange, B. Gicquel, and O. Neyrolles. 2003. DC-SIGN is the major Mycobacterium tuberculosis receptor on human dendritic cells. J. Exp. Med. 197:121-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Kooyk, Y., and T. B. Geijtenbeek. 2003. DC-SIGN: escape mechanism for pathogens. Nat. Rev. Immunol. 3:697-709. [DOI] [PubMed] [Google Scholar]
- 36.Wang, L., W. Qu, and P. R. Reeves. 2001. Sequence analysis of four Shigella boydii O-antigen loci: implication for Escherichia coli and Shigella relationships. Infect. Immun. 69:6923-6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitfield, C., N. Kaniuk, and E. Frirdich. 2003. Molecular insights into the assembly and diversity of the outer core oligosaccharide in lipopolysaccharides from Escherichia coli and Salmonella. J. Endotoxin Res. 9:244-249. [DOI] [PubMed] [Google Scholar]
- 38.Zamze, S., L. Martinez-Pomares, H. Jones, P. R. Taylor, R. J. Stillion, S. Gordon, and S. Y. Wong. 2002. Recognition of bacterial capsular polysaccharides and lipopolysaccharides by the macrophage mannose receptor. J. Biol. Chem. 277:41613-41623. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, P., O. Schwartz, M. Pantelic, G. Li, H. Chang, C. Nobile, S. Hong, J. Klena, and T. Chen. Submitted for publication.
- 40.Zhou, L. J., and T. F. Tedder. 1996. CD14+ blood monocytes can differentiate into functionally mature CD83+ dendritic cells. Proc. Natl. Acad. Sci. USA 93:2588-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou, L. J., and T. F. Tedder. 1995. Human blood dendritic cells selectively express CD83, a member of the immunoglobulin superfamily. J. Immunol. 154:3821-3835. [PubMed] [Google Scholar]