Abstract
The human aryl hydrocarbon receptor (AHR) undergoes continuous shuttling between nucleus and cytoplasm. Binding to exogenous or endogenous ligands promotes its rapid nuclear import. The proposed mechanism for the ligand-dependent import is based on exposing the bipartite nuclear localisation signal (NLS) to members of the importin (IMP) superfamily. Among this, the molecular interactions involved in the basal import still need to be clarified. Utilizing fluorescently fused AHR variants, we recapitulated and characterized AHR localization and nucleo-cytoplasmic shuttling in living cells. Analysis of AHR variants carrying NLS point mutations demonstrated a mandatory role of first (13RKRRK17) and second (37KR-R40) NLS segments on the basal import of AHR. Further experiments indicated that ligand-induced import is mainly regulated through the first NLS, while the second NLS is supportive but not essential. Additionally, applying IMPα/β specific inhibitors, ivermectin (IVM) and importazole (IPZ), slowed down the ligand-induced import and, correspondingly, decreased the basal nuclear accumulation of the receptor. In conclusion, our data show that ligand-induced and basal nuclear entry of AHR rely on the same mechanism but are controlled uniquely by the two NLS components.
Subject terms: Biochemistry, Biotechnology, Cell biology, Molecular biology
Introduction
The aryl hydrocarbon receptor (AHR) is a ligand-dependent transcription factor, which was initially discovered due to its contribution in mediating the toxicity of exogenous environmental toxicants, like 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)1 and benzo[a]pyrene (B[a]P)2. Exposure to AHR ligands, as for example the synthetic flavonoid β-naphthoflavone (BNF)3, induces the expression of xenobiotic metabolizing enzymes (XMEs). These include phase I XMEs such as cytochrome P450-dependent monooxygenase 1A1 (CYP1A1) and phase II XMEs such as glutathione S-transferase α (GST-α)4, indicating AHR’s role in xenobiotic metabolism5.
More current studies discovered essential physiological roles of AHR by demonstrating that many compounds from endogenous origins, including indole, heme, and tryptophan metabolites6,7, are able to bind and activate the receptor. As for example, the endogenous indirubin (IND) is generated from dietary tryptophan by the intestinal and urinary microbiome8. It is one of the most potent AHR ligands and exhibits anti-inflammatory effects in vivo9. Furthermore, current research reveals that AHR has a more complex role in cell biology. In fact, AHR interacts with other signaling pathways involved in proliferation, apoptosis, and cell cycle regulation10,11.
AHR activation is firmly linked with nuclear transition that depends on a sequence of positively charged amino acids, the nuclear localization signal (NLS)12. The NLS comprises two segments (bipartite NLS) and is located within the conserved bHLH (basic helix–loop–helix) domain in the N-terminal part of the protein13. According to the classical AHR pathway, unliganded AHR is mainly located in the cytoplasm bound by a chaperon complex consisting of two molecules of HSP9014 and single molecules of co-chaperone p2315 and hepatitis x-associated protein-2 (XAP2)16. Most AHR ligands are able to cross the cell membrane through simple diffusion due to their hydrophobicity17. Ligand binding to cytosolic AHR triggers a conformational change that exposes the NLS to a nuclear transporter in order to permit nuclear import18. On the molecular level, members of the importin (IMP) superfamily IMPβ1 or its adaptor protein IMPα can recognize the revealed NLS18,19. Thereafter, AHR along with the IMPα/β1 heterodimer reaches the nucleus through the nuclear pore complexes (NPCs). On the nuclear side of NPCs, IMPβ1 binds to RanGTP (Ras-related nuclear protein) subsequently leading to a release of the NLS-cargo20.
In the nucleus, AHR releases its chaperons21 and builds a stable dimer with the AHR nuclear translocator (ARNT)22,23. The AHR/ARNT heterodimer recruits several co-factors24, binds its target sequences, called XRE (xenobiotic response element)25, and regulates the expression of AHR target genes. Ultimately, AHR is exported from the nucleus to the cytoplasm to undergo proteasomal degradation marking the end of activation26. This nuclear export is established through CRM1 (chromosome region maintenance 1 also known as exportin) that binds the nuclear export signal (NES) of the receptor27. In a previous report, we showed that the molecular interactions performed by the receptor in the nucleus, ARNT dimerization and DNA binding, do not alter the nuclear retention after ligand binding28.
Analyzing AHR translocation in vitro, however, discloses many aspects that are still not completely understood. For example, AHR undergoes constitutive shuttling between the nucleus and cytoplasm29,30, which neither leads to any activation31, nor demands HSP90 dissociation32. Although most of the effort given on AHR trafficking intends to clarify the ligand-induced transition, the basal shuttling requests further investigation. On this stage, the research done in the last decade revealed many nuclear import inhibitors targeting IMPs33. The best characterized ones are importazole (IPZ)34 and ivermectin (IVM)35,36. The former revealed interferences with the IMPα/β1 mediated-nuclear entry of NF-κB33 and the latter showed import suppressing properties on the hypoxia induced factor (HIF)37. Both import inhibitors differ markedly in their mechanism. While IVM inhibits the binding between IMPα and the NLS38,39, IPZ interferes with IMPβ1 and RanGTP binding40.
In the current study, we aimed to elucidate the mechanism of AHR translocation in the basal and ligand-induced state in living cells. We confirmed faithful recapitulation of endogenous AHR localization for our EYFP-AHR overexpression system, independent of cell cycle, expression level, or fusion with EYFP. Through point mutations over the entire NLS, importance of both parts of the bipartite NLS for AHR localization has been shown. But surprisingly, these effects are not transferable on the ligand-induced import, which can still occur with 2nd NLS mutants, confirming that the 1st NLS of AHR is mandatory for the nuclear import. Application of the nuclear transport inhibitors IVM and IPZ resulted in a clear reduction of nuclear accumulation of ligand-free and ligand-bound AHR. In summary, our data demonstrated that ligand-induced and basal nuclear entry are two different processes that rely on the same molecular mechanism.
Results
EYFP-AHR in MCF-7ΔAHR faithfully recapitulates AHR localization
In our study, we aimed to investigate the nucleo-cytoplasmic translocation of unliganded and ligand bound AHR. To avoid any confounding effects through endogenous AHR, AHR-knockout MCF-7 cells (MCF-7ΔAHR) have been used41. Transient transfection of MCF-7ΔAHR cells with plasmids coding for EYFP-AHRWT or EYFP-AHRmut allowed real-time tracking of AHR’s subcellular localization in living cells as described previously28,31.
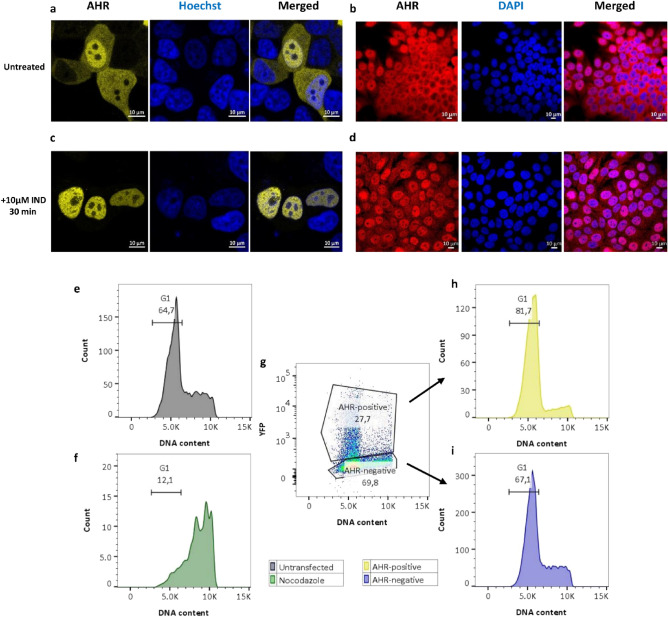
Notably, localization of AHRWT in MCF-7ΔAHR cells can be defined in three different states: accumulated in the nucleus, equally distributed or predominantly cytoplasmic (Figs. 1a, 2h and Supplementary Fig. S1). Antibody staining of endogenous AHR in fixed MCF-7WT revealed a similar distribution (Fig. 1b). Treating cells for 30 min with 10 µM IND, however, captured only predominantly nuclear accumulated AHR in both cases (Fig. 1c,d). To assure functional output of EYFP-AHRWT translocation, we verified CYP1A1 and CYP1B1 mRNA induction after IND treatment with qPCR (Supplementary Fig. S2).
Figure 1.
AHR intracellular distribution in MCF-7 cells. Representative images of EYFP-AHRWT transiently transfected in MCF-7ΔAHR cells before (a) and after (c) 30 min treatment with 10 µM indirubin (IND). Immunofluorescence images of AHR in MCF-7WT cells before (b) and after (d) 30 min treatment with 10 µM IND. Nuclei are marked either with Hoechst (a,c) or with DAPI (b,d). Cell cycle analysis of control cells without (e) and with nocodazole (f). EYFP-AHR transfected cells were sorted for expression level (g) and subpopulations individually analyzed for cell cycle for EYFP-AHR-positive (h) and for EYFP-AHR-negative cells (i). Total number of analyzed cells: 7624 (e), 1149 (f), 18,192 (g), 4441 (h) and 11,181 (i).
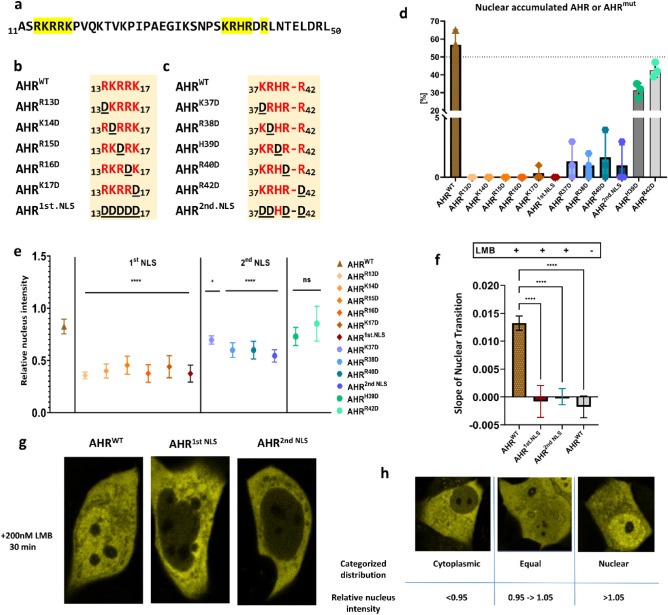
Figure 2.
Role of the nuclear localization signal (NLS) on the basal shuttling of AHR. The N-terminal bHLH domain of AHR is indicated as amino acid sequence (a). Graphical representation of mutated amino acids in 1st NLS (b) and 2nd NLS (c). The percentage of cells with nuclear accumulated AHRWT or AHRmut for 100 randomly chosen cells in the basal state (d) according to our classification (h). The ratio of the mean fluorescence intensity within the nucleus and the mean fluorescence intensity of the cells in the cytoplasmic state is given as relative nuclear intensity (e). Slopes of the nuclear transition of AHRWT or AHRmut after treatment with 200 nM leptomycin B (LMB) for 30 min (f). Values represent the mean ± S.D of n = 100 (d), n = 10 (e), n = 14 for AHRWT and n = 5 for AHRmut or unstimulated AHRWT (f). Statistical analysis was performed with a one-way ANOVA, Dunnett's post-test, *p < 0.05, ****p < 0.0001.
To exclude an influence of the cell cycle on AHR localization, we determined the cell cycle phase of EYFP-AHRWT transfected MCF-7ΔAHR cells by measuring DNA content. After 48 h of cultivation, roughly 65% of MCF 7ΔAHR cells were in G1 phase (Fig. 1e). With nocodazole as positive control for cell cycle analysis, we shifted the population to mainly G2 phase (Fig. 1f). Interestingly, cells that have been transfected, but were negative for EYFP-AHR expression, exhibited the same distribution as untransfected cells (Fig. 1i). On the other side, the great majority of EYFP-AHR positive cells were in G1 phase (Fig. 1g,h). To further elucidate whether changed cell cycle progression is a result of EYFP-AHR overexpression, we similarly analyzed the cell cycle of cells transiently transfected with EYFP alone. Interestingly, these cells exhibited changed cell cycle distribution as well, leading to the hypothesis, that transfection itself alters cell cycle progression (Supplementary Fig. S3). Overall, transfection of EYFP-AHRWT in MCF-7ΔAHR faithfully recapitulates AHR localization, independent of expression level and cell cycle.
Impact of individual amino acids of the NLS on the basal import of AHR
Since we sought to investigate the nuclear transition of AHR, we decided to address the impact of NLS components on the basal import of AHR first. Therefore, we mutated every positively charged amino acid of the bipartite NLS individually and generated mutants for the first and the second part of the NLS as well (Fig. 2a–c).
We evaluated the effect of individual amino acids on the intracellular distribution of AHR in MCF-7ΔAHR cells by categorizing at least 100 cells into one of the three described states (Fig. 2h and Supplementary Fig. S4). Consistently, switching the positively charged NLS amino acid arginine to aspartate (AHRR13D, AHRK14D, AHRR15D, AHRR16D, AHRK17D, AHR1st.NLS, AHRK37D, AHRR38D, AHRR40D, and AHR2nd.NLS) shifted the AHR compartmentalization to be solely cytoplasmic. On the other hand, similar substitution of the other two positions (AHRH39D and AHRR42D) did not alter the typical cellular distribution (Fig. 2d and Supplementary Figs. S5 and S6).
Next, we analyzed the relative nuclear intensity (Fig. 2h) of basal EYFP-AHRWT and EYFP-AHRmut, respectively, thus informing about the nuclear retention of AHR in the absence of ligands (Fig. 2e). Our results exhibited a significant decrease in the relative nuclear intensity of NLS mutants compared with primary cytosolic EYFP-AHRWT cells (Fig. 2e and Supplementary Fig. S5). Interestingly, mutations in the first part of the NLS seemed to have a higher impact on the nuclear retention than mutations on the second part of the NLS. Again, EYFP-AHRH39D and EYFP-AHRR42D revealed a comparable nuclear intensity to the wild type (Fig. 2e).
Moreover, we tested whether the basal import can still occur in first and second NLS full mutants after inhibiting nuclear export using leptomycin B (LMB) by measuring the change of relative nuclear intensity over time (Supplementary Fig. S7). Remarkably, the basal import for both mutants was completely repressed after treating the cells with 200 nM LMB for 30 min (Fig. 2f,g and supplementary Fig. S8).
The ligand-induced nuclear entry of AHR depends primarily on the first part of NLS
Next, we studied the ligand-induced nuclear import of NLS mutants. After treatment with either IND or BNF, we tracked the subcellular location of fluorescent fusion proteins over 15 min.
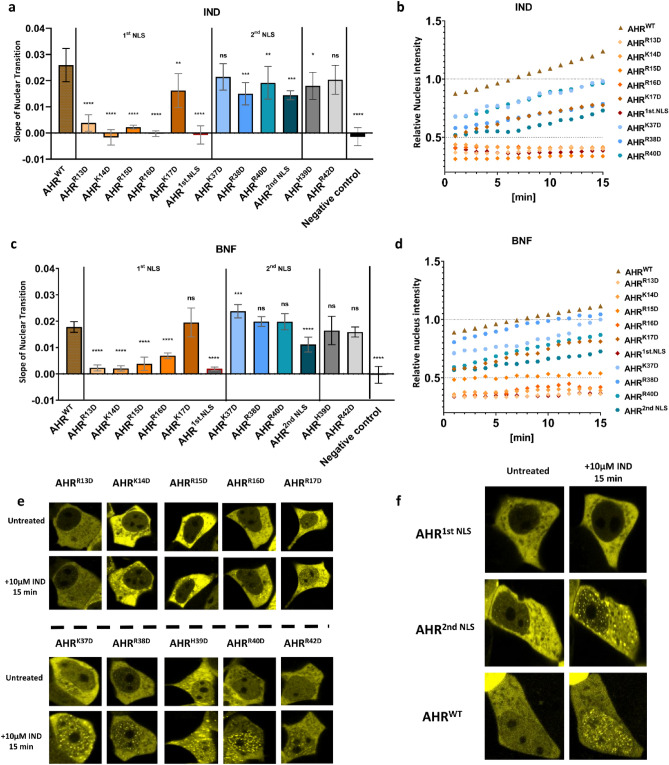
In response to ligands, all mutants with point mutations in the 1st NLS, except EYFP-AHRK17D, revealed a clear reduction of nuclear transition slopes compared to the wild type protein (Fig. 3a–d). In contrast, mutants with point mutations in the 2nd NLS resulted only in a slight reduction of nuclear import after stimulation with IND and showed no effect on the import rate for BNF treatment (Fig. 3a–d). When mutating the entire first or second NLS, respectively, the effect on the nuclear transition slope was similar: mutating the 1st NLS blocked the nuclear transition entirely, whereas a mutation of the 2nd NLS only reduced the rate of nuclear import (Fig. 3a–f). EYFP-AHRH39D and EYFP-AHRR42D had no to very limited effect on the nuclear transition for IND and BNF, respectively, supporting the results from the basal shuttling that these amino acids do not contribute to NLS function (Fig. 3a–e). Interestingly, independent of import rate, nuclear accumulation can take place of ligand activated EYFP-AHRWT or EYFP-AHRmut, leading to the formation of protein clusters within the nucleus (Fig. 3e, f and supplementary Fig. S3).
Figure 3.
The nuclear entry of AHR depends primarily on the first part of nuclear localization signal (NLS). Slopes of the nuclear transition of EYFP-AHRWT or EYFP-AHRmut after treatment with 10 µM indirubin (IND) (a) or 10 µM β-naphthoflavone (BNF) (c) for 15 min. Negative control refers to untreated sample transfected with AHRWT. Mean of time-lapse measurements after stimulation with 10 μM IND (b) or 10 μM BNF (d) for 15 min. Representative images of 1st and 2nd NLS point mutants before and after treatment with IND (e). Snapshots of AHRWT, AHR1st.NLS and AHR2nd.NLS before and after treatment with IND (f). Data show the mean ± S.D of n = 12 for AHRWT, n = 5 for AHRmut and NC (a–c). Statistical analysis was performed with a one-way ANOVA, Dunnett's post-test, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
IPZ and IVM reduce nuclear accumulation of AHR
Taken together, the results of basal and ligand-dependent nuclear import for the individual amino acids of the NLS indicate different roles for the two parts of the NLS and we hypothesized a difference in import mechanism for unliganded and ligand-bound AHR. We next intended to study the influence of importin specific inhibitors IPZ and IVM on the nucleo-cytoplasmic translocation of AHR.
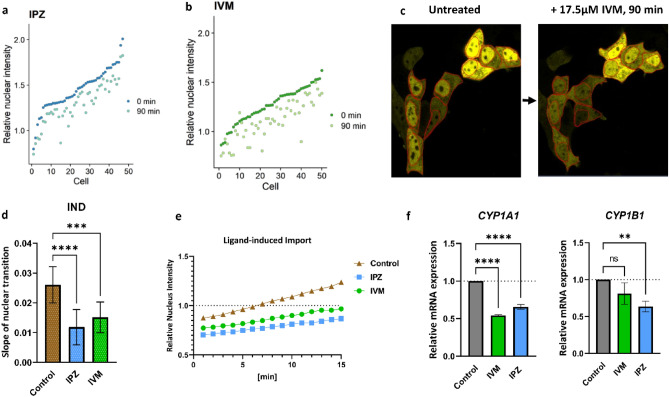
MCF-7ΔAHR cells transiently transfected with EYFP-AHRWT were incubated either with IPZ or IVM for 90 min. Image analysis elicited a significant reduction of the nuclear EYFP-AHRWT staining. This was also accompanied with an increase of cytoplasmic EYFP-AHRWT in studied cells. Therefore, the relative nuclear intensity diminished to 80% during this treatment (Fig. 4a–c and Supplementary Fig. S9).
Figure 4.
The inhibitors of importin (IMP) α/β mediated import importazole (IPZ) and ivermectin (IVM) reduce the nuclear accumulation of AHR. MCF-7ΔAHR cells were transfected with EYFP-AHRWT and treated with IPZ or IVM for 90 min at concentrations of 10 µM and 17.5 µM, respectively. Relative nuclear intensity of around 50 cells (sorted according to 0 min value) at the time of treatment (0 min) and after 90 min of treatment with IPZ (a) and IVM (b), respectively. Representative images of cells before and after incubation with 17.5 µM IVM for 90 min (c). Slopes of nuclear transition (d) and mean of time-lapse measurements (e) after incubation with 10 µM IPZ or 7.5 µM IVM for 22 h followed by co-treatment with 10 µM indirubin (IND) for 15 min. Data is expressed as mean + /− S.D. for 12 cells. Control implies to cells treated with ligand only. One-way ANOVA, Dunnett's post-test, ***p < 0.001, ****p < 0.0001. MCF-7WT cells were treated with IPZ or IVM for 24 h at concentrations of 10 µM and 7.5 µM, respectively. Thereafter, cells were co-treated with 2.5 µM IND for 2 h. Relative CYP1A1 and CYP1B1 mRNA levels determined by qPCR (f). Values were standardized against HPRT and normalized to a sample treated with DMSO and ligand (mean + /− S.D.; one-way ANOVA, Dunnett’s post-test, **p < 0.01, ****p < 0.0001).
Further analysis examined the influence of IPZ and IVM on the ligand-induced nuclear import of AHR. In response to IND stimulation, EYFP-AHRWT live tracking exhibited remarkable deceleration of around 50% of nuclear import for both IPZ and IVM compared with the control (Fig. 4d,e). Taken together, these results indicate that basal and ligand-induced nuclear import are both affected by application of both import inhibitors.
To evaluate whether suppressing the AHR nuclear entry will influence AHR activity, we determined the functional output of the AHR after ligand stimulation either with or without preincubation with import inhibitors.
Compared to ligand stimulated control without import inhibitors, IVM treatment resulted in a notable decrease of CYP1A1 mRNA levels and a slight decrease of CYP1B1 mRNA expression (Fig. 4f). On the other hand, IPZ resulted in a significant decrease in the expression of both CYP1A1 and CYP1B1 (Fig. 4f).
Discussion
For import and export through the nuclear envelope, proteins larger than 40 kDa require specific signals in the form of amino acid sequences42. These signals permit recognition by nuclear transporters like the IMPα/β1 heterodimer for nuclear import or CRM1 for nuclear export. AHR possesses NLS and NES clusters, which unsurprisingly results in the ability of dynamic shuttling. Certainly, in the literature AHR is repeatedly defined as a cytoplasmic protein, while the nuclear entry requests ligand binding that induces a conformational change unmasking the NLS43,44,45. This theory ignores the nuclear retention of unliganded receptor detected in various in vitro systems confirming the existence of basal shuttling. Additionally, a nuclear trapped AHR shares a similar ability to bind ligands and induce CYP1A1 expression in comparison to wild type AHR29. From our point of view, a mainly cytoplasmic localization could be attributed to high export efficiency or impeded import in individual cells. One popular theory links the basal nuclear accumulation of AHR with the presence of endogenous ligands in cell culture medium46. Nevertheless, a study of our lab showed that a part of ligand binding domain of AHR, namely the amino acid H291, is essential to bind endogenous ligands but is not mandatory for the nucleo-cytoplasmic shuttling47
Our first results confirmed that AHR subcellular localization of endogenous or overexpressed AHR varies between neighboring cells. From a technical aspect, cells in 2D cell culture are normally not synchronized and follow their own cell cycle progression. This offers the possibility that the noticed receptor staining might be related to various cell cycle stages. Especially since the cell cycle influences many physiological processes. In fact, in MCF-7 cells it has been shown that AHR physically interacts with proteins implicated in the cell cycle, like cyclin-dependent kinase 4 (CDK4)48 or the inhibitor of CDK2, p2749. More interesting, knocking out p27 resulted in increasing level of nuclear AHR50. In comparison to our results, it is interesting to note that the increase of detected cells in G1 phase in AHR positive cells might be related to AHR function. However, transfecting the MCF-7 cells with EYFP leads to a similar increase in the G1 phase, referring to a transfection-related effect on the cell cycle in general. Concisely, in our experiments typical distribution of AHR has been observed independent of cell cycle, expression level, or fusion with EYFP. Based on these data, we could make sure that we our system is suitable to study AHR localization.
In a previous study of our lab, we examined the DNA binding motif which has one overlapping amino acid with the 2nd NLS (EYFP-AHRR40D). This substitution impairs DNA binding and basal import, thereby changing the intracellular distribution28. Interestingly, this point mutation does not interfere with the ligand-induced import, which led us to hypothesize that two different import mechanisms for the AHR nuclear entry exist28.
Initially, Ikuta and co-workers identified the minimal NLS of AHR as 13RKRR16 and 37KRH39 by analyzing the intracellular distribution12. Our results expand the NLS sequence to include Arg17 along with Arg40 considering the localization of unliganded AHR. At the same time, His39 and Arg42 do not seem to influence the nuclear import in general. Technically, the methods used about 2 decades ago covered only short AHR variants involving the NLS and neighboring amino acids12,30. But this is not sufficient for full characterization of the AHR, since the C-terminus has an effect on its translocalization31. In particular, our experiments highlight a central role of the 1st NLS for the basal nuclear accumulation and the ligand-induced import of the receptor. Point mutations in the 2nd NLS lead to predominant cytoplasmic AHR staining, pointing to a diminished basal import, with only a slight reduction of the ligand-induced import.
Nuclear import can occur through IMPβ1 alone or with help of the adaptor protein IMPα51,52. According to Petrulis et al., murine AHR can be bound by IMPβ1 directly18. In combination with the apparently differing mechanisms for basal and ligand-induced import, we wanted to elucidate whether IMPα is mandatory for AHR translocation. IVM and IPZ are both specific inhibitors for IMP mediated import. IVM impedes binding between IMPα and NLS, thereby disrupting IMPα/β1 mediated transport38,39. IPZ disrupts IMPβ1 RanGTP binding, through which all IMPβ1 associated imports are inhibited40. Both inhibitors clearly reduce the basal nuclear accumulation of EYFP-AHR and diminish the ligand-induced import rate indicating similar mechanisms for both import processes.
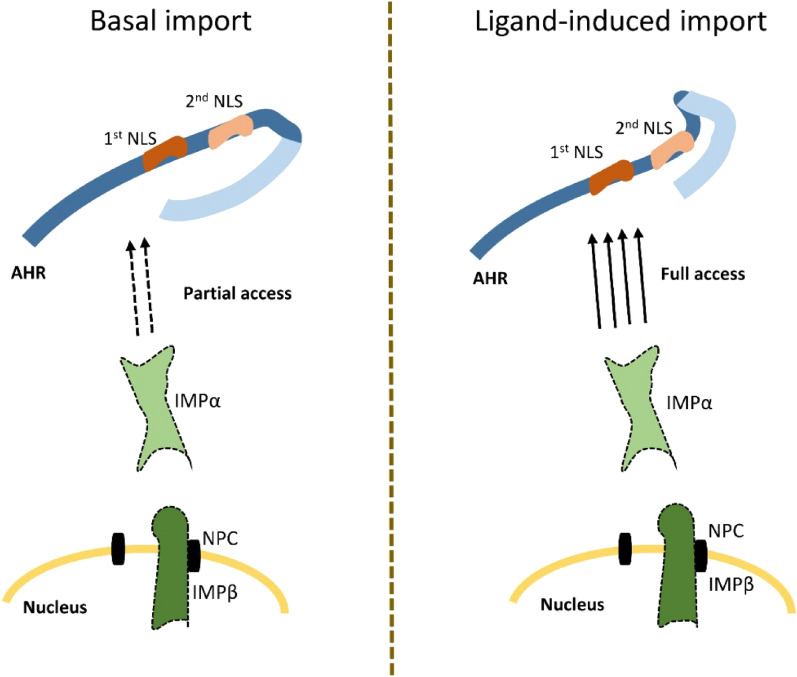
According to our results, we suggest the following hypothesis: In the unliganded state, blocking of both NLS parts is only partial, thereby allowing IMPα binding only at a diminished rate. The part of the AHR chaperone complex that is responsible for the shielding still needs to be determined. After ligand binding, the conformational change facilitates rapid IMPα recruitment without impediments. Interestingly, ligand-induced import is mainly implemented through the 1st NLS. The 2nd NLS is complementary but not essential. In conclusion, we show that ligand-independent nuclear import is mediated by the same molecular mechanism as the ligand-dependent import (Fig. 5).
Figure 5.
Nuclear import of AHR is mediated by importin (IMP) α/β1. The basal import occurs at a diminished rate because of a partial blocking of the 1st and 2nd nuclear localisation signal (NLS) through the c-terminal part of AHR or parts of the chaperon complex. Upon ligand binding, a conformational change grants full access for IMPα to the 1st NLS, thereby increasing import rate drastically. NPC, nuclear pore complex.
Methods
Plasmids
The plasmid pEYFP-AHR-C1 has been used previously28 and encodes the human AHR starting with the amino acid alanine from position 11. GenScript (GenScript Biotech, Leiden, Netherlands) generated the modified variants of pEYFP-AHR that contain mutations at specific amino acids AHRR13D, AHRK14D, AHRR15D, AHRR16D, AHRK17D, AHR1st.NLS, AHRK37D, AHRR38D, AHRH39D, AHRR40D, AHRR42D and AHR2nd.NLS.). AHRWT and AHRmut have been validated by sequencing (Eurofins, Germany).
Cell culture
MCF-7 cell line was purchased from ATCC (Manassas, VA, USA). AHR knockout MCF-7 cells (MCF-7∆AHR) were generated in our lab using a CRISPR-Cas9 based approach41. Both cell lines were cultured in DMEM supplemented with 10% (v/v) FCS, 100 U/ml penicillin, 100 mg/ml streptomycin, and 2 mM L-glutamine. All cell culture media and supplements were bought from Pan-Biotech (Aidenbach, Germany). Cells were maintained in 5% CO2 at 37 °C and humidified atmosphere. Cells were routinely inspected for absence of mycoplasma contamination.
Reagents
Β-naphthoflavone (BNF), dimethyl sulfoxide (DMSO) and ivermectin (IVM) were acquired from Sigma-Aldrich (Sigma-Aldrich Chemie GmbH, Munich, Germany). Importazole (IPZ) was obtained from BIOZOL (Biozol Diagnostica Vertrieb GmbH, Eching, Germany). Indirubin (IND) was purchased from Enzo Life Sciences (Enzo Life Sciences GmbH, Lörrach, Germany). Leptomycin B (LMB) was deliviered from Santa Cruz Biotechnology (Santa Cruz Biotechnology Inc., TX, USA). DMSO was used as vehicle for BNF, IND, IPZ and IVM, while ethanol (Carl Roth, Germany) was used as a vehicle for LMB. The end-concentration does not exceed 0.1% when using DMSO or ethanol. All chemicals were ordered at the highest available purity.
Transient transfection
MCF-7 or MCF-7∆AHR cells were seeded either on 6-well plates (both from Techno Plastic Products AG, Trasadingen, Switzerland) or on glass bottom dishes (In VitroScientific, Sunyvale, CA, USA). After 24 h, cells on multiwell plates were transfected with lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) and an appropriate DNA amount as indicated by the manufacturer’s instructions. Meanwhile, cells on glass bottom dishes were transfected with Xfect (Takara Bio Europe SAS, Saint-Germain-en-Laye, France) and an appropriate DNA amount as stated by the manufacturer’s instructions. In both cases, transfection medium was refreshed after incubation for 4 h.
Cell treatment with import inhibitors
To investigate the influence of import inhibitors on the basal localization of AHR, EYFP-AHRWT transiently transfected MCF-7ΔAHR cells were incubated with IPZ or IVM at concentration of 10 µM and 17.5 µM, respectively, for 90 min. To study the impact on ligand induced import, cells were incubated with 10 µM IPZ or 7.5 µM IVM after medium change after transfection for nearly 22 h and then co-treated with 10 µM IND for 15 min. To measure the effect of import inhibitors on the transcriptional activity of AHR, total RNA levels of MCF-7WT cells were gathered after incubating the cells for 22 h with either IPZ or IVM at concentration of 10 µM and 7.5 µM, respectively, and afterwards stimulated with 2.5 µM IND for 2 h.
Immunofluorescence staining
Cover glasses were coated with Poly-L-Lysin 0.01% solution (Sigma-Aldrich, Germany) according to the manufacturer’s instructions. Cells were grown on cover glasses with a cell density of 2.5 × 105/ml for 48 h and then fixed in 3.7% formaldehyde (15 min), permeabilized with 0.2% Triton X-100 (Sigma-Aldrich, Germany) in PBS (10 min). Finally, cells were blocked with 5% fetal bovine serum (FBS) in PBS for 1 h at room temperature. After blocking, the coverslips were incubated with primary antibody for 90 min, washed and then incubated with the secondary antibody for 1 h. The antibodies were diluted in 1.5% FBS in PBS solution. Slides were mounted in Vectashield HardSet Mounting Medium with DAPI (VECTOR LABORATORIES, INC., Burlingame, CA, USA). The used primary antibody is anti-AHR (sc-5579, Santa Cruz Biotechnology, 1:25) and secondary antibody is goat anti rabbit, Alexa Fluor™ 594 (A-11012, 1:400), purchased from Invitrogen, Thermo Fisher Scientific, Germany.
Cell cycle analysis by flow cytometry
Cells grown on glass bottom dishes were transfected using Xfect as described above. After 24 h, cells were detached from culture flasks by using trypsin (0.5%-EDTA 0.2%; Pan-Biotech (Aidenbach, Germany)) solution for 20 min. After cell fixation with ice cold 80% ethanol for 1 h, cells were washed with PBS and stained with propidium iodide (PI)/RNAse solution (Thermo Fisher Scientific, Germany) according to the manufacturer’s protocol. Stained cells were analyzed by fluorescence-activated cell-sorting (FACS) (BD FACSAria III). For data analysis, FlowJo (V.10.7.1, BD Biosciences) was used. For analysis, samples were analyzed to discriminate doublets (FSC-A versus FSC-W). Cell cycle analysis was performed using the PI-A intensity on a linear scale of either AHR-YFP-positive or -negative cells. To determine the intensity of G2-phase and verify cell cycle analysis, nocodazole (0.6 µg/ml; Sigma-Aldrich, Germany) synchronized cells were analyzed as positive control.
Gene expression analysis
Total RNA was isolated form cells using RNeasy Midi kit and QIAshredder (both from QIAGEN GmbH, Hilden, Germany). Thereafter, purity and concentration of isolated RNA were determined using a plate reader device (Infinite M200 PRO, Tecan Trading AG, Männedorf, Switzerland). 1 µg or 500 ng of the extracted RNA was reversely transcribed into cDNA by using the high-capacity cDNA Reverse transcription kit (Applied Biosystems, Foster City, CA USA).
Transcript levels were determined with fast SYBR Green mix (Applied Biosystems, Foster City, CA, USA) and quantitative PCR (qPCR) device 7500 Fast Real-Time PCR instrument (Applied Biosystems, Foster City, CA, USA). Hypoxanthine–guanine phosphoribosyl transferase (HPRT) was used as reference gene.
The primer sequences are listed in Table 1.
Table 1.
Primer sequences used.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| CYP1A1 | 5′-CCAAGAGTCCACCCTTCCCAGCT-3′ | 5′-GAGGCCAGAAGAAACTCCGTGGC-3′ |
| CYP1B1 | 5′-TGGATTTGGAGAACGTACCG-3′ | 5′-CCACGACCTGATCCAATTCT-3’ |
| HPRT | 5′-GTTCTGTGGCCATCTGCTTAG-3′ | 5′-GCCCAAAGGGAACTGATAGTC-3′ |
On-line confocal microscopy
As described in28, for cell monitoring the confocal microscope LSM 700 (Carl Zeiss Jena GmbH, Jena, Germany) was used. During the measurement of living cells, suitable cell culture conditions like 5% CO2 and 37 °C were maintained. The relative nuclear intensity refers to the mean fluorescence intensity of the nucleus divided by the mean fluorescence intensity of the whole cell. This was determined by outlining the nucleus and whole cells as region of interest (ROI) by using ZEN 2012 blue edition (Carl Zeiss Jena GmbH). The slope of nuclear transition was measured after plotting the relative nucleus intensity against time of the treatment. Representative examples of the analysis of AHR localization, time-lapse measurements and relative nuclear intensity are in (supplementary Figs. S1, S4 and S7).
Statistics
Data analysis and graphing were performed with GraphPad Prism (Graph Pad, La Jolla, CA, USA) and R-Studio (R-Tools Technology Inc, Canada). Statistical analysis was done using paired Dunnett’s multiple comparisons test, one-way ANOVA, *p < 0.01, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Supplementary Information
Author contributions
R.H., J.K. designed the experimental program. R.H. performed the experimental work, R.S. contributed to the confocal microscopy and J.K., M.M. planned and contributed to the FACS analysis. Data analysis and figures creation were done by R.H. and J.K. Manuscript writing was done by R.H. with contributions from A.L. and J.K. The final version was approved by all authors.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-47066-z.
References
- 1.Beischlag TV, Luis Morales J, Hollingshead BD, Perdew GH. The aryl hydrocarbon receptor complex and the control of gene expression. Crit. Rev. Eukaryot. Gene Expr. 2008;18(3):207–250. doi: 10.1615/CritRevEukarGeneExpr.v18.i3.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luch A. Nature and nurture: Lessons from chemical carcinogenesis. Nat. Rev. Cancer. 2005;5(2):113–125. doi: 10.1038/nrc1546. [DOI] [PubMed] [Google Scholar]
- 3.Zhou X, et al. β-naphthoflavone activation of the Ah receptor alleviates irradiation-induced intestinal injury in mice. ANTIGE. 2020;9(12):1264. doi: 10.3390/antiox9121264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nebert DW, et al. Role of the aromatic hydrocarbon receptor and [Ah] gene battery in the oxidative stress response, cell cycle control, and apoptosis. Biochem. Pharmacol. 2000;59(1):65–85. doi: 10.1016/S0006-2952(99)00310-X. [DOI] [PubMed] [Google Scholar]
- 5.Esser C, Rannug A. The aryl hydrocarbon receptor in barrier organ physiology, immunology, and toxicology. Pharmacol. Rev. 2015;67(2):259–279. doi: 10.1124/pr.114.009001. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen LP, Bradfield CA. The search for endogenous activators of the aryl hydrocarbon receptor. Chem. Res. Toxicol. 2008;21(1):102–116. doi: 10.1021/tx7001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei GZ, et al. Tryptophan-metabolizing gut microbes regulate adult neurogenesis via the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. U.S.A. 2021;118(27):e2021091118. doi: 10.1073/pnas.2021091118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meijer L, Guyard N, Skaltousnis LA, Eisenbrand G. Indirubin, the Red Shade of Indigo. Life in Progress Edition; 2006. [Google Scholar]
- 9.Qi T, Li H, Li S. Indirubin improves antioxidant and anti-inflammatory functions in lipopolysaccharide-challenged mice. Oncotarget. 2017;8(22):36658–36663. doi: 10.18632/oncotarget.17560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tappenden DM, Hwang HJ, Yang L, Thomas RS, LaPres JJ. The Aryl-hydrocarbon receptor protein interaction network (AHR-PIN) as identified by tandem affinity purification (TAP) and mass spectrometry. J. Toxicol. 2013;2013:279829. doi: 10.1155/2013/279829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denison MS, Soshilov AA, He G, DeGroot DE, Zhao B. Exactly the same but different: Promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicol. Sci. 2011;124(1):1–22. doi: 10.1093/toxsci/kfr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikuta T, Eguchi H, Tachibana T, Yoneda Y, Kawajiri K. Nuclear localization and export signals of the human aryl hydrocarbon receptor. J. Biol. Chem. 1998;273(5):2895–2904. doi: 10.1074/jbc.273.5.2895. [DOI] [PubMed] [Google Scholar]
- 13.Greb-Markiewicz B, Kolonko M. Subcellular localization signals of bHLH-PAS proteins: Their significance, current state of knowledge and future perspectives. Int. J. Mol. Sci. 2019;20(19):4746. doi: 10.3390/ijms20194746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perdew GH. Association of the Ah receptor with the 90-kDa heat shock protein. J. Biol. Chem. 1988;263(27):13802–13805. doi: 10.1016/S0021-9258(18)68314-0. [DOI] [PubMed] [Google Scholar]
- 15.Kazlauskas A, Poellinger L, Pongratz I. Evidence that the co-chaperone p23 regulates ligand responsiveness of the dioxin (Aryl hydrocarbon) receptor. J. Biol. Chem. 1999;274(19):13519–13524. doi: 10.1074/jbc.274.19.13519. [DOI] [PubMed] [Google Scholar]
- 16.Meyer BK, Pray-Grant MG, Vanden Heuvel JP, Perdew GH. Hepatitis B virus X-associated protein 2 is a subunit of the unliganded aryl hydrocarbon receptor core complex and exhibits transcriptional enhancer activity. Mol. Cell. Biol. 1998;18(2):978–988. doi: 10.1128/MCB.18.2.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stevens EA, Mezrich JD, Bradfield CA. The aryl hydrocarbon receptor: a perspective on potential roles in the immune system. Immunology. 2009;127(3):299–311. doi: 10.1111/j.1365-2567.2009.03054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petrulis JR, Kusnadi A, Ramadoss P, Hollingshead B, Perdew GH. The hsp90 co-chaperone XAP2 alters Importin β recognition of the bipartite nuclear localization signal of the Ah receptor and represses transcriptional activity. J. Biol. Chem. 2003;278(4):2677–2685. doi: 10.1074/jbc.M209331200. [DOI] [PubMed] [Google Scholar]
- 19.Ikuta T, Kobayashi Y, Kawajiri K. Phosphorylation of nuclear localization signal inhibits the ligand-dependent nuclear import of aryl hydrocarbon receptor. Biochem. Biophys. Res. Commun. 2004;317(2):545–550. doi: 10.1016/j.bbrc.2004.03.076. [DOI] [PubMed] [Google Scholar]
- 20.Kosyna FK, Depping R. Controlling the gatekeeper: Therapeutic targeting of nuclear transport. Cells. 2018;7(11):221. doi: 10.3390/cells7110221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsuji N, et al. The activation mechanism of the aryl hydrocarbon receptor (AhR) by molecular chaperone HSP90. FEBS Open Bio. 2014;4:796–803. doi: 10.1016/j.fob.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barouki R, Aggerbeck M, Aggerbeck L, Coumoul X. The aryl hydrocarbon receptor system. Drug. Metabol. Drug. Interact. 2012;27(1):3–8. doi: 10.1515/dmdi-2011-0035. [DOI] [PubMed] [Google Scholar]
- 23.Schulte KW, Green E, Wilz A, Platten M, Daumke O. Structural basis for aryl hydrocarbon receptor-mediated gene activation. Structure. 2017;25(7):1025–1033.e3. doi: 10.1016/j.str.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 24.Hestermann EV, Brown M. Agonist and chemopreventative ligands induce differential transcriptional cofactor recruitment by aryl hydrocarbon receptor. Mol. Cell. Biol. 2003;23(21):7920–7925. doi: 10.1128/MCB.23.21.7920-7925.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swanson HI, Chan WK, Bradfield CA. DNA binding specificities and pairing rules of the Ah receptor, ARNT, and SIM proteins. J. Biol. Chem. 1995;270(44):26292–26302. doi: 10.1074/jbc.270.44.26292. [DOI] [PubMed] [Google Scholar]
- 26.Davarinos NA, Pollenz RS. Aryl hydrocarbon receptor imported into the nucleus following ligand binding is rapidly degraded via the cytosplasmic proteasome following nuclear export. J. Biol. Chem. 1999;274(40):28708–28715. doi: 10.1074/jbc.274.40.28708. [DOI] [PubMed] [Google Scholar]
- 27.Marlowe, J., & Puga, A., Novel AHR Interactions, in Comprehensive Toxicology (Second Edition). 93–115 (Elsevier. 2010)
- 28.Haidar R, et al. The role of DNA-binding and ARNT dimerization on the nucleo-cytoplasmic translocation of the aryl hydrocarbon receptor. Sci. Rep. 2021;11(1):18194. doi: 10.1038/s41598-021-97507-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richter CA, Tillitt DE, Hannink M. Regulation of subcellular localization of the aryl hydrocarbon receptor (AhR) Arch. Biochem. Biophys. 2001;389(2):207–217. doi: 10.1006/abbi.2001.2339. [DOI] [PubMed] [Google Scholar]
- 30.Ikuta T, et al. Nucleocytoplasmic shuttling of the aryl hydrocarbon receptor. J. Biochem. 2000;127(3):503–509. doi: 10.1093/oxfordjournals.jbchem.a022633. [DOI] [PubMed] [Google Scholar]
- 31.Tkachenko A, et al. The Q-rich/PST domain of the AHR regulates both ligand-induced nuclear transport and nucleocytoplasmic shuttling. Sci. Rep. 2016;6:32009. doi: 10.1038/srep32009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heid SE, Pollenz RS, Swanson HI. Role of heat shock protein 90 dissociation in mediating agonist-induced activation of the aryl hydrocarbon receptor. Mol. Pharmacol. 2000;57(1):82–92. [PubMed] [Google Scholar]
- 33.Jans DA, Martin AJ, Wagstaff KM. Inhibitors of nuclear transport. Curr. Opin. Cell. Biol. 2019;58:50–60. doi: 10.1016/j.ceb.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 34.Kuusisto HV, Jans DA. Hyper-dependence of breast cancer cell types on the nuclear transporter Importin β1. Biochim. Biophys. Acta Mol. Cell Res. 2015;1853(8):1870–1878. doi: 10.1016/j.bbamcr.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 35.Tay MYF, et al. Nuclear localization of dengue virus (DENV) 1–4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin. Antiviral Res. 2013;99(3):301–306. doi: 10.1016/j.antiviral.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 36.Götz V, et al. Influenza A viruses escape from MxA restriction at the expense of efficient nuclear vRNP import. Sci. Rep. 2016;6(1):23138. doi: 10.1038/srep23138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kosyna FK, Nagel M, Kluxen L, Kraushaar K, Depping R. The importin α/β-specific inhibitor ivermectin affects HIF-dependent hypoxia response pathways. Biol. Chem. 2015;396(12):1357–1367. doi: 10.1515/hsz-2015-0171. [DOI] [PubMed] [Google Scholar]
- 38.Wagstaff KM, Rawlinson SM, Hearps AC, Jans DA. An AlphaScreen®-based assay for high-throughput screening for specific inhibitors of nuclear import. J. Biomol. Screen. 2011;16(2):192–200. doi: 10.1177/1087057110390360. [DOI] [PubMed] [Google Scholar]
- 39.Wagstaff KM, Sivakumaran H, Heaton SM, Harrich D, Jans DA. Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 2012;443(3):851–856. doi: 10.1042/BJ20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soderholm JF, et al. Importazole, a small molecule inhibitor of the transport receptor importin-β. ACS Chem. Biol. 2011;6(7):700–708. doi: 10.1021/cb2000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tarnow P, et al. Characterization of Quinoline yellow dyes as transient aryl hydrocarbon receptor agonists. Chem. Res. Toxicol. 2020;33(3):742–750. doi: 10.1021/acs.chemrestox.9b00351. [DOI] [PubMed] [Google Scholar]
- 42.Cautain B, Hill R, de Pedro N, Link W. Components and regulation of nuclear transport processes. Febs j. 2015;282(3):445–462. doi: 10.1111/febs.13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choudhary M, Malek G. The aryl hydrocarbon receptor: a mediator and potential therapeutic target for ocular and non-ocular neurodegenerative diseases. Int. J. Mol. Sci. 2020;21(18):6777. doi: 10.3390/ijms21186777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guerrina N, Traboulsi H, Eidelman DH, Baglole CJ. The aryl hydrocarbon receptor and the maintenance of lung health. Int. J. Mol. Sci. 2018;19(12):3882. doi: 10.3390/ijms19123882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shivanna B, Chu C, Moorthy B. The aryl hydrocarbon receptor (AHR): A novel therapeutic target for pulmonary diseases? Int. J. Mol. Sci. 2022;23(3):1516. doi: 10.3390/ijms23031516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Veldhoen M, Hirota K, Christensen J, O'Garra A, Stockinger B. Natural agonists for aryl hydrocarbon receptor in culture medium are essential for optimal differentiation of Th17 T cells. J. Exp. Med. 2009;206(1):43–49. doi: 10.1084/jem.20081438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tkachenko A, et al. Nuclear transport of the human aryl hydrocarbon receptor and subsequent gene induction relies on its residue histidine 291. Arch. Toxicol. 2018;92(3):1151–1160. doi: 10.1007/s00204-017-2129-0. [DOI] [PubMed] [Google Scholar]
- 48.Barhoover MA, Hall JM, Greenlee WF, Thomas RS. Aryl hydrocarbon receptor regulates cell cycle progression in human breast cancer cells via a functional interaction with cyclin-dependent kinase 4. Mol. Pharmacol. 2010;77(2):195–201. doi: 10.1124/mol.109.059675. [DOI] [PubMed] [Google Scholar]
- 49.Kolluri SK, Weiss C, Koff A, Göttlicher M. p27(Kip1) induction and inhibition of proliferation by the intracellular Ah receptor in developing thymus and hepatoma cells. Genes Dev. 1999;13(13):1742–1753. doi: 10.1101/gad.13.13.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elson DJ, et al. The cyclin-dependent kinase inhibitor p27(Kip1) interacts with the aryl hydrocarbon receptor and negatively regulates its transcriptional activity. FEBS Lett. 2022;596(16):2056–2071. doi: 10.1002/1873-3468.14434. [DOI] [PubMed] [Google Scholar]
- 51.Lu J, et al. Types of nuclear localization signals and mechanisms of protein import into the nucleus. Cell Commun. Signal. 2021;19(1):60. doi: 10.1186/s12964-021-00741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Palmeri D, Malim MH. Importin beta can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin alpha. Mol Cell Biol. 1999;19(2):1218–1225. doi: 10.1128/MCB.19.2.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.