Abstract
Toll-like and interleukin-1/18 receptor/resistance (TIR) domain–containing proteins function as important signaling and immune regulatory molecules. TIR domain–containing proteins identified in eukaryotic and prokaryotic species also exhibit NAD+ hydrolase activity in select bacteria, plants, and mammalian cells. We report the crystal structure of the Acinetobacter baumannii TIR domain protein (AbTir-TIR) with confirmed NAD+ hydrolysis and map the conformational effects of its interaction with NAD+ using hydrogen-deuterium exchange-mass spectrometry. NAD+ results in mild decreases in deuterium uptake at the dimeric interface. In addition, AbTir-TIR exhibits EX1 kinetics indicative of large cooperative conformational changes, which are slowed down upon substrate binding. Additionally, we have developed label-free imaging using the minimally invasive spectroscopic method 2-photon excitation with fluorescence lifetime imaging, which shows differences in bacteria expressing native and mutant NAD+ hydrolase-inactivated AbTir-TIRE208A protein. Our observations are consistent with substrate-induced conformational changes reported in other TIR model systems with NAD+ hydrolase activity. These studies provide further insight into bacterial TIR protein mechanisms and their varying roles in biology.
Keywords: toll/interleukin-1 receptor (TIR), nicotinamide adenine dinucleotide (NAD), hydrolase, bacterial pathogenesis, innate immunity
Bacterial toll-like and interleukin-1/18 receptor (TIR) resistance proteins have been identified based on sequence motifs conserved with human TIR proteins (1, 2). Uropathogenic Escherichia coli CFT073 and Brucella Spp. express soluble TIR domain–containing proteins that act as virulence factors by physically interacting with and disrupting host TIR domain–containing signaling complexes to promote pathogenicity and modulate immunity (3, 4, 5, 6). TIR domain–containing proteins also have been identified both in several additional disease-causing bacteria as well as in nonpathogenic and commensal bacteria, raising questions regarding potential roles they might have apart from their originally described roles in pathogenicity and modulation of mammalian TLR signaling (2, 3, 7, 8, 9, 10).
In a recent discovery, TIR domain–containing proteins, which are conserved across biology in bacteria, plants, and animals, were found to exhibit enzymatic function in NAD+ hydrolysis (11, 12). NAD+ is an essential cofactor and critical regulator of cellular and metabolic function (13). In humans, TIR-mediated NAD+ hydrolase activity is involved in axonal degradation via the TIR domain–containing adaptor protein Sterile alpha and TIR motif–containing protein SARM (14, 15). In plants and bacteria, TIR domain–containing proteins with NAD+ hydrolase activity are involved in immune protection against pathogens (12, 16, 17). Several pathogenic bacterial TIR proteins with confirmed roles in virulence exhibit NAD+ hydrolase activity. Characterized bacterial TIR domain–containing proteins include Staphylococcus aureus (TirS) (18, 19), Pseudomonas aeruginosa (PumA) (20), Uropathogenic E. coli CFT073 (UPEC-TcpC) (3, 4, 21), Brucella melitensis (TcpB) (4, 22), and Acinetobacter baumannii (AbTir-TIR) (11). Bacterial TIR domain–containing proteins TcpC and TcpB were originally shown to disrupt mammalian TIR adaptor–mediated innate immune signaling pathways (4, 21), whereas, TirS expression and activity in HEK293T reduces mammalian cell viability and NAD levels compared with catalytically inactive mutants (11). Several bacterial TIRs also have been identified as part of antiphage response mechanisms, involving abortive infection and bacteria self-killing as a mechanism to disrupt and protect against phage infection and replication (2).
The cleavage of NAD+ can result in a variety of products, depending on the particular bacterial TIR employed. Initial characterization of NADase activity by mammalian SARM1 showed the cleavage of NAD+ into nicotinamide (Nam), ADP ribose (ADPR), and cyclic ADPR (cADPR) (11, 12). A subset of bacterial TIRs, including AbTIR from A. baumannii, BtpA-TIR from Brucella sp., and TcpO from the archaea Methanobrevibacter olleyae, however, were found to produce a novel noncanonical variant of cyclic ADPR (v-cADPR or N7-cADPR), which was later classified as 2′cADPR (11, 23). The bacterial TIR from the antiphage defense system Thoersis, ThsB, produces the cADPR variant 3′cADPR from NAD+ to activate the Thoersis ThsA to deplete cellular NAD+ levels through its sirtuin 2 domain, leading to cell death and termination of phage replication (16, 23, 24). It remains to be seen whether the various NAD cleavage products, besides 3′cADPR play roles in signaling pathways within bacteria.
NAD+ hydrolase activity also has been identified in nonpathogenic and commensal bacteria (9, 11, 12, 17). This newly discovered TIR-mediated enzymatic function lends additional insight and opens new questions regarding the function TIR domain–containing protein may play in bacterial-host interactions. To better understand the molecular mechanisms underlying enzymatically active bacterial TIR proteins, we have performed structure-function studies characterizing the A. baumannii TIR protein (AbTir-TIR).
A. baumannii is a Gram-negative bacterium and a leading cause of hospital-acquired infections. Due to the emergence of multidrug-resistant A. baumannii strains that are resistant to nearly all antibiotics classes, it is a bacterial pathogen of global concern. Immune responses against the highly prevalent multidrug-resistant strain A. baumannii CN40 include inflammasome and caspase activation via type I IFN signaling responses mediated by the TIR domain–containing adapter-inducing interferon-β (25). AbTir-TIR exhibits NAD+ hydrolase activity (11). Upon cleavage of NAD, AbTir-TIR catalyzes production of the novel cADPR variant, 2′cADPR, whose potential functions in metabolism and signaling in host-bacterial interactions have yet to be fully characterized (11). To better understand the mechanism of bacterial TIR NAD+ hydrolase activity and function, we pursued molecular studies of AbTir-TIR. Accordingly, we have used X-ray crystallography to determine the atomic structure of AbTir-TIR and have confirmed its enzymatic activity using liquid chromatography, mass spectrometry, and colorimetric assays. In addition, we have used 2-photon excitation with fluorescence lifetime imaging (2p-FLIM) to image live bacteria label-free that express AbTir-TIR and hydrogen-deuterium exchange mass spectrometry (HDX-MS) to map the kinetics and conformational changes induced by the interaction of NAD+ with AbTir-TIR.
Understanding bacterial TIR protein–mediated regulation of host innate immune responses in the context of NAD+ hydrolase activity could lead to novel antimicrobial strategies for limiting infection, controlling inflammation and modulating immunity and cellular processes.
Results
A. baumannii TIR protein is conserved among bacteria and humans
A GenBank blastP search of the Protein Database (PDB) using A. baumannii Tir-TIR domain identified several other bacterial TIR domain–containing proteins with homology to AbTir-TIR, including structurally characterized TcpB from Brucella spp. (PDB ID 4LQC) and PdTIR Paracoccus denitrificans PdTIR (PDB 3H16) (Fig. S1). Surprisingly, this search did not identify structures of TIR proteins with NAD+ hydrolase activity from plants or animals (12, 16). A sequence alignment of AbTir-TIR with bacterial TIR proteins with identified NAD+ hydrolase activity shows the conservation of the C-Helix WxxxE motif identified in Brucella TcpB to be important for microtubule binding. This motif includes a highly conserved Tryptophan (W) and a Glutamic acid (Glu, E) found at the carboxy terminus that is involved in substrate interactions and necessary for enzymatic activity in most hydrolases (11, 23, 26). This region is highly conserved across plant, bacteria, and mammalian extracellular TIR proteins (Fig. S1) (11, 26, 27). The C-helix WxxxE motif and surrounding region include the NAD+-binding site, TIR interactions, and autoregulation site in huSARM1 and plant NLR RUN1 (27, 28).
AbTir-TIR domain is observed to dimerize in solution and in the crystal lattice
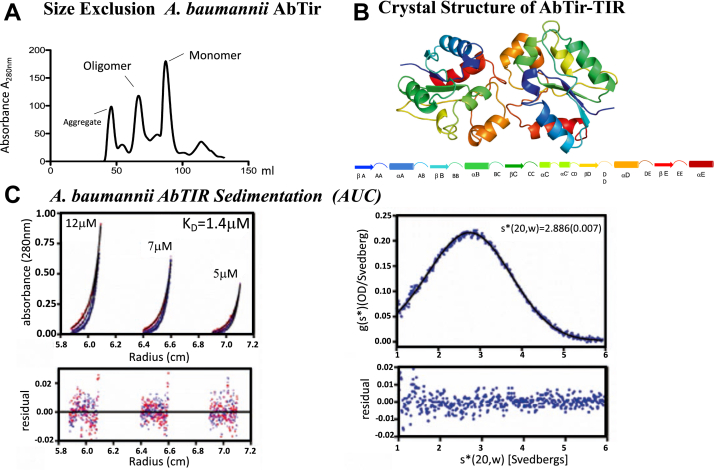
The expressed and purified AbTir-TIR domain is observed in solution as a weakly self-associated dimer by size exclusion, analytical ultracentrifugation (AUC), and within the asymmetric crystal lattice (Fig. 1, A–C). TIR domain self-association is consistent with observations for other prokaryotic and eukaryotic TIR domain–containing protein in the absence of NAD (22, 29, 30, 31). The size-exclusion chromatogram (SEC) of AbTir-TIR indicated the formation of higher-ordered oligomeric assemblies. The sample separated by SEC exhibits both a single monomer peak and higher-ordered species consistent with dimer and soluble aggregate species based on comparison with the size-exclusion standard (Fig. 1A). Soluble aggregate and oligomer fractions were confirmed to migrate at a molecular weight similar to monomer peak fractions during SDS-PAGE analysis (data not shown). SEC-purified monomer peak fractions were concentrated and screened for crystallization using commercial screens (Fig. S2). Small broom–like crystals were identified, reproduced, and optimized using streak seeding (Fig. S2). Single crystals were isolated and rasterized to identify single crystal diffraction, and data was collected using the highly automated microfocus beamline NSLS-II-17-ID AMX (32). To further confirm the dimer species observed by SEC and in the crystal of AbTir-TIR, we performed in solution sedimentation AUC analysis on the isolated AbTir-TIR monomer SEC peak fraction species (33) (https://core.uconn.edu/resources/biophysics#au-software). Sedimentation equilibrium acquired on purified AbTir-TIR samples at varying concentrations and speeds indicate weak dimerization with a KD of 1.4 μM. Sedimentation velocity performed on concentrated monomer fraction (AbTIR) at 10 μM 45K rpm and analyzed using the program DCDT+, indicates a sedimentation coefficient, s20,w, of 2.8S, which is consistent with a dimer observed in the crystal (Fig. 1C) (34, 35).
Figure 1.
AbTir-TIR is observed as a dimer in solution by SEC, AUC, and in the crystal.A, size-exclusion chromatograms showing isolation of stabilized oligomers of full-length AbTir-TIRE208A. B, the crystal structure of AbTir-TIRwt domain is observed as a dimer is shown as a colored-coded cartoon ribbon representation. The AbTir-TIR domain shown consists of five alternating alpha helix and beta-strand secondary structures that are gradient colored starting at the amino terminus in blue to the carboxyl terminal in red (A–E, bottom left to right). C, analytical ultracentrifugation (AUC) sedimentation profiles of A. baumannii AbTir-TIR. AUC indicates that AbTir-TIR forms a weakly associated dimer in solution. Analysis of sedimentation equilibrium profiles performed at the indicated AbTir-TIR concentrations 12, 7, and 5 μM acquired at 23, 26, and 29K rpm indicate dimerization with a KD of1.4 μM. Sedimentation velocity performed on concentrated monomer fraction (AbTir-TIR) at 10 μM 45K rpm and analyzed using DCDT+, indicates a sedimentation coefficient, s20,w, of 2.8S, consistent with a dimer observed in crystal (34, 35). AbTir-TIR, Acinetobacter baumannii TIR domain protein; AUC, analytical ultracentrifugation; SEC, size-exclusion chromatography.
Recombinant AbTir-TIR is enzymatically active
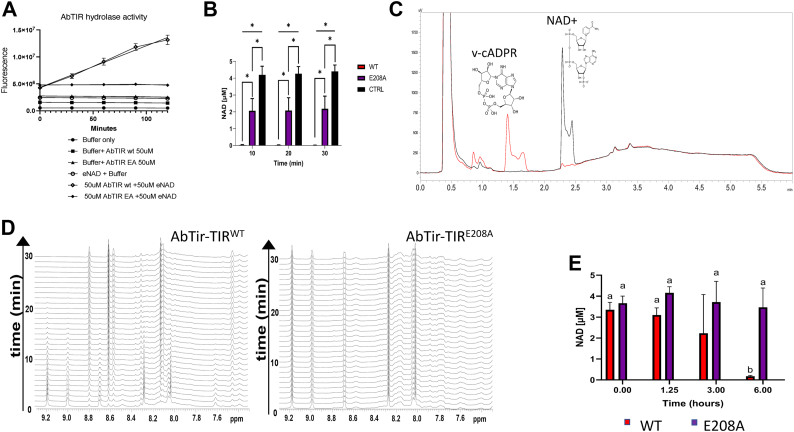
Several assays confirm the enzymatic activity of expressed and purified AbTir-TIR protein. The NAD+ hydrolase activity of recombinant purified AbTir-TIR protein was assessed using ethano-NAD and the EnzyChrom NAD+ assay (Fig. 2, A, B and E). Ethano-NAD (εNAD), which yields a fluorescent product upon cleavage, was used to assess the enzymatic activity of purified recombinant AbTir-TIRwt in comparison with enzymatic inactivated mutant AbTir-TIRE208A (Fig. 2A) (11, 36). Only AbTir-TIRwt shows fluorescence of εNAD, indicating NAD+ cleavage in comparison to inactivated mutant AbTir-TIRE208A and buffer controls. In a similar fashion the EnzyChrom NAD/NADH assay also shows a reduction of NAD by purified AbTIRwt in comparison with inactivated mutant AbTir-TIRE208A. Purified AbTir-TIRwt and AbTir-TIRE208A were incubated with 5 μM of NAD for 10 to 30 min, and the levels of NAD+ remaining in the samples were measured. Incubation with AbTir-TIRwt resulted in a significant decrease in the levels of NAD+ compared with AbTir-TIRE208A (two-way ANOVA, p < 0.0001).LC-MS trace analysis revealed that recombinant AbTir-TIRwt is capable of hydrolyzing NAD+ into ADPR as well as cADPR products (11) (Fig. 2C). One-dimensional NMR spectra also showed NAD+ hydrolase activity of AbTir-AbTIRWT in comparison with inactive mutant AbTir-AbTIRE208A (Fig. 2E) (37).
Figure 2.
NAD+ Hydrolase activity of AbTir-TIR expressing bacteria and purified recombinant protein.A, recombinant purified AbTir-TIRwt cleaves εNAD. Fluorescence of εNAD in the presence or absence of buffer, AbTir-TIRwt, AbTir-TIRE208A measured over time. Buffer only (●), buffer + AbTir-TIRwt (■), buffer + AbTir-TIRwt (▲), εNAD + AbTir-TIRwt (▼), εNAD + AbTir-TIRE208A (◆), εNAD + buffer (○). B, NAD+ concentration overtime of purified recombinant active (AbTir-TIRwt), inactivated (AbTir-TIRE208A) with buffer control. Asterisk (∗) indicates a statistically significant p-value of p ≤ 0.05. C, HPLC trace showing generation of novel cyclic ADP ribose by AbTir-TIRwt protein used in crystallization. HPLC traces of NAD+ protein with AbTir-TIR protein used for crystallization at 0 min (black) and after 60 min (red) incubation. D, direct visualization of NAD+ catalysis by NMR. One-dimensional NMR spectra of 1 mM NAD+ collected every minute for AbTir-TIRWT (left) and AbTir-TIRE208A. Catalysis by AbTir-TIRWT can be observed by the disappearance of the nicotinamide resonances (i.e., 9.2 and 9.0 ppm) that are not observed in the presence of the inactive mutant. E, time course of NAD+ concentration of bacteria expressing active (AbTir-TIRwt) and inactivated (AbTir-TIRE208A). εNAD, ethano-NAD; AbTir-TIR, Acinetobacter baumannii TIR domain protein.
Besides promoting NAD cleavage in vitro, AbTir-TIRwt reduces cellular levels of NAD+ upon recombinant expression in E. coli. AbTir-TIRwt and E208A. NAD+ levels in bacterial lysates from cells expressing AbTir-TIRwt and AbTir-TIRE208A were measured 0 to 6 h, following IPTG induction of AbTIR Tir-TIRwt and E208A expression. Expression of catalytically active AbTir-TIRwt promoted significantly greater levels of NAD+ loss in bacterial lysates than did expression of AbTir-TIRE208A (two-way ANOVA, p < 0.0001).
To evaluate AbTir-TIR activity in vivo we developed a label-free assay using the intrinsic fluorescence of NAD(P)H and lifetime (FLT) imaging with 2p-FLIM using live bacteria–expressing AbTIR. The average FLTs of bacteria induced or not to express enzymatically active (AbTIRwt/+IPTG) or inactivated mutant protein (AbTIRE208A/+IPTG) were imaged and quantitated as histograms (Fig. S3, A–E). Differences are observed for bacteria induced to express active AbTIR wt compared with inactivated mutant AbTIRE208A. There were also differences seen compared with uninduced bacteria, which may be due to induction with IPTG, affecting bacteria growth and metabolism. FLTs are independent of the bacteria concentration and protein expression level. Changes in the average FLTs are known to reflect differences in bound and unbound forms of NAD(P)H (38, 39). Overall, these results indicate AbTir-TIR protein used for structural studies was enzymatically active or inactivated due to mutations, as similarly described in other reports (11, 23).
The AbTir-TIR crystal structure comparison with other bacterial TIR–containing proteins
The structure of A. baumannii TIR domain (AbTir-TIR) was determined to using X-ray crystallography (Table 1). The secondary structure architecture is typical of most TIR domains comprised of five alternating beta (β)-strands and alpha (α) helices (lettered A-E) as initially described (40, 41) (Fig. 1B). Conserved loops, helices, individual residues, and motifs have been identified and functionally characterized to be important for protein interactions and signaling within several TIR domain–containing proteins and NAD+ hydrolases (42). The AbTir-TIR is observed as dimeric in solution by SEC and AUC and in the crystal lattice. TIR domain self-association and dimerization, previously shown for SARM TIR hydrolase activity, is consistent with oligomerization of other reported bacterial TIR structures (21, 22, 30, 43, 44).
Table 1.
X-ray diffraction and data collection
| AbTIRwt (aa134–269) PDB code 8G83 | |
|---|---|
| No. of crystals | 1 |
| Beamline | 17-ID-1 AMX, NSLS-II |
| Wavelength (Å) | 0.920072 |
| Detector | EIGER |
| Crystal to detector distance(mm) | 248.844 |
| Rotation range per image | 0.2, 0–180 |
| No. of images | 1–900 (truncated 200–800) |
| Space group | P21 21 21 |
| Unit-cell parameters(Å,°) | 44.20 76.19 97.33, 90, 90, 90 |
| Molecules per asymmetric unit | 2 |
| Resolution limits (Å) | 28.86 3.031 (3.139–3.031) |
| Total reflections | 13,278 (1261) |
| No. of unique observations | 6668(639) |
| Completeness (%) | 98.01 (95.09) |
| Multiplicity | 2.0 (2.0) |
| Rmerge | 0.05218 (0.308) |
| Rmeas | 0.0738 (0.435) |
| Rp.i.m. | 0.05218 (0.308) |
| Mean I/sigma I | 10.06 (2.32) |
| Rwork | 0.2276 (0.3156) |
| Rfree | 0.2743 (0.2529) |
| CC (work) | 0.952 (0.760) |
| CC (free) | 0.934 (0.908) |
| No. of nonhydrogen atoms | 2129 |
| Macromolecules | 2122 |
| Protein residues | 277 |
| RMS (bonds) | 0.006 |
| RMS (angles) | 0.79 |
| Ramachandran favored (%) | 94.14 |
| Ramachandran allowed (%) | 5.86 |
| Ramachandran outliers (%) | 0.00 |
| Rotamer outliers (%) | 0.00 |
| Clashscore | 4.58 |
| Average B-factor | 56.44 |
| macromolecules | 56.46 |
| solvent | 50.00 |
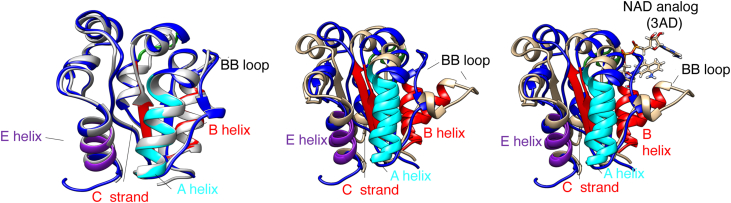
Previously reported bacterial TIR domain–containing structures include Brucella TcpB/BtpA (4LQC, 4LZP, 4C7M) and P. denitrificans PdTIR (3H16). Most TIR domains are observed as oligomeric species, either dimer and/or tetramer. Individual and TIR domain interactions and loop positions are similar among other reported bacterial TIR domain structures with a RMSD of 1.663 Å (TcpB-4LQC), 1.782 Å (BtpA/TcpB 4LZP), and 2.669 Å (PtDTIR -3H16) for 134, 134, 132, 130, and 137 atom pairs, respectively (Fig. S4). The X-ray structures of AbTir-TIRwt (8G83) and recently reported AbTir-TIR structure (7UWG) are similar (23) (Fig. 3-left panel). Both apo crystal structures have similar positions for beta strands, alpha helices, and intervening loops with an overall RMSD of 0.839 Å. In particular, similar positions for individual residues contained within the WxxxE motif are observed. Both W204 and E208 which are important for selectivity and enzymatic function are observed, solvent-exposed, and facing outward in the absence of substrate. By contrast, large conformational changes are observed throughout the entirety of the TIR domain structure when comparing apo unliganded X-ray crystal structures (8G83 and 7UWG) with NAD analog–bound (3AD) cryo-EM structure AbTir-TIR-3AD 7UXU (Fig. 3 middle and right panel). Comparing apo X-ray and substrate analog bound cryo-EM structures and RMSD of 4.834 is observed (8G83 and 7UXU). Several secondary structures and loops undergo repositioning upon binding substrate analog. These include conserved residues known to be important for hydrolase activity. For example, both W204 and E208 contained within the WxxxE loop are reoriented inward facing and involved in binding to substrate analog.
Figure 3.
Least squares comparison of unliganded and NAD analog bound X-ray and cryo-EM AbTir-TIR domains. Least squares comparison of crystal structures of AbTIr-TIR 8G83 (blue) and 7UWG (gray), left panel. Least squares comparison of X-ray AbTir-TIR 8G83 (blue) and cryoEM structure 7UXU (tan) without (middle panel) and with NAD analog 3AD shown (right panel). HDX-MS peptides with changes in EX1 kinetics and conformations upon binding εNAD are colored as shown in Figure 4. A helix (cyan) FVRPLAETLQQL, B helix, BC loop and C beta-strand (red) RQKIDSGLRNSKYGTVVL, and E helix IAHQLAD (purple). εNAD, ethano-NAD; AbTir-TIR, Acinetobacter baumannii TIR domain protein; HDX-MS, hydrogen-deuterium exchange mass spectrometry.
AbTir-TIR exhibits unique kinetics and large conformational changes, which are slowed down in the presence of substrate
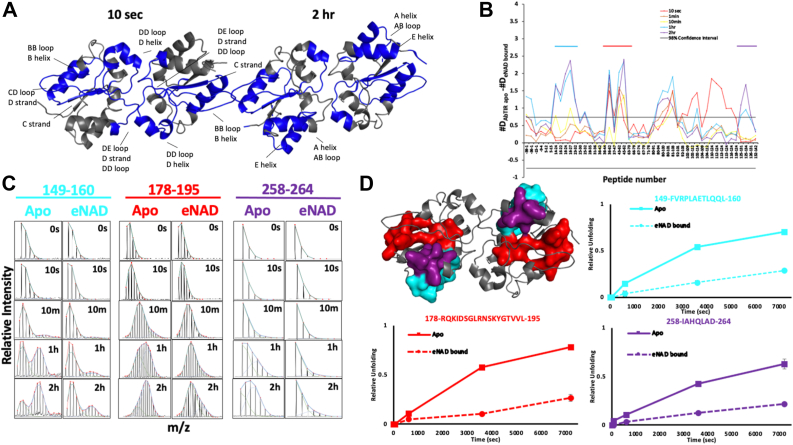
We sought to further characterize AbTir-TIR’s structural dynamics and the conformational consequence of its interaction with NAD+ substrate using HDX-MS. HDX-MS allows in-solution mapping of interfaces, interactions, and allosteric effects by providing comparative peptide-level kinetics of deuterium uptake at global time scales. Sequencing reactions of the AbTir-TIRwt recombinant protein used for crystallization resulted in near complete peptide coverage for AbTir-TIRwt (Fig. S5). The peptide deuteration levels were evaluated in the presence and absence of NAD+ and the observed differences in peptide deuteration were mapped onto the crystal structure of AbTir-TIR (Fig. 4A). Overall, the addition of NAD+ resulted in decreases in deuterium uptake by AbTir-TIR across multiple deuterium incubation time points (Fig. 4B). Notably, the deuterium uptake at the DD and EE loops located at the dimeric interface observed in X-ray structures 8G83 and 7UWG were significantly decreased at the earliest time point (10 s). In addition, several AbTir-TIR peptides exhibited EX1-like exchange kinetics in the absence of NAD+. These regions are highlighted by cyan, red, and purple bars in Figure 4B and color-coded accordingly in Figure 4, C and D. The stacked isotopic envelops for representative peptides in these regions (Fig. 4C) display progressive bimodal envelops characteristic of EX1 kinetic behavior in the unliganded form. When mapped onto the structures of AbTir-TIR, these areas, correspond to the amino-terminal A helix (FVRPLAETLQQL), the central B helix, the BC loop, the C β-strand (RQKIDSGLRNSKYGTVVL), and the carboxyl-terminal E helix (IAHQLAD) (Figs. 3, 4, C and D and Fig. S1) (23). These peptides were found to colocalize, and the rate of formation of the high m/z species in the bimodal envelop was observed to be similar for all three regions. These observations reflect large, slow, and cooperative conformational changes of AbTir-TIR in the absence of NAD+. Upon substrate binding, the progression of bimodal behavior is significantly reduced, suggesting that NAD+ restricts or decreases the rate of these conformational changes. The NAD+ analog 3AD–bound cryo-EM structure (7UXU) exhibits a large conformational change in the BB loop and B helix region as well as unique molecular assemblies in comparison to unliganded X-ray structures. Peptides exhibiting reduced deuterium uptake, large conformational changes, and unique kinetics when mapped on the monomers of unliganded (8G83 and 7UWG) or ligand-bound form of AbTir (7UXU), with the exception of the B helix are not in direct contact with the substrate and instead map to molecular assembly TIR interfaces, loops, core β strands, and helices (Figs. 3 and 4A (7, 23, 45).
Figure 4.
HDX-MS of AbTir-TIRwtexhibit EX1 kinetics and conformations upon binding NAD+. Substrate binding results in large decreases in deuterium uptake in AbTir-TIRwt. Significant EX1 exchange kinetics consistent with large conformational changes are observed for AbTir-TIRwt, in the absence of NAD and weakened upon ligand binding. A, regions with significant decreases in deuterium uptake after 10s and 2 h incubation are mapped on the structure of AbTir-TIR. B, the difference plot (Apo – εNAD-bound) in absolute deuterium uptake at each incubation time point (color coded) and for each peptide (x-axis). C, stacked spectra of representative spectra showing the signature bimodal isotopic envelops of EX1 kinetic regime of exchange. Apo- and NAD-bound states are shown. Peptides are color coded as in panel D. D, surface and ribbon representation of AbTir-TIRwt. Surface representation shows peptides displaying bimodal EX1 behavior. Plots show the progressive accumulation of the high m/z species deconvolved from the bimodal isotopic envelop as a function of time of deuterium incubation for the apo- (solid) and εNAD-bound (dashed) AbTir-TIRwt. εNAD, ethano-NAD; AbTir-TIR, Acinetobacter baumannii TIR domain protein; HDX-MS, hydrogen-deuterium exchange mass spectrometry.
Discussion
Our structural studies of the AbTir-TIR are consistent with results reported by Manik et al. and reveal similarities and differences among bacterial, plant, and mammalian TIR proteins (7, 8, 21, 22, 23). A least-squares comparison with other bacterial TIR domain counterparts exhibits overall similarities of loops and interactions. Consistent with bacterial TIRs reported to date, the crystal structure of AbTir-TIR retains a similar overall core TIR domain fold with dimerization interactions as well as confirmed NAD+ hydrolase activity (22, 26). Several NAD hydrolase TIR domains have been observed as dimeric and tetrameric species in the absence of NAD (22, 44, 46). A noncleavable NAD analog (3AD)-bound cryo-EM structure does, however, exhibit a unique TIR domain interface scaffold compared with apo AbTIR crystal structures (23).
NAD+ hydrolase activity of recombinant AbTir-TIR used for structural studies was confirmed by several different methods. LC/MS analysis indicates the ability of the recombinant AbTir-TIRwt to hydrolyze NAD+ and produce novel cADPR products (11, 16, 17, 28). Additionally, inactivated mutant AbTir-TIRE208A has significantly reduced enzymatic activity. It has been postulated that the NAD hydrolase activity of bacterial TIR domain proteins characterized to be virulence factors could be a contributing factor in their virulence (11). This NADase-mediated virulence may be particularly potent for the expression of TIR-only constructs if other domains in these proteins act to regulate TIR NADase function, as the N-terminal Armadillo/HEAT motif (ARM) does for SARM1 (47, 48, 49). Indeed, Manik et al. (23) showed stronger NADase activity for AbTirTIR than for a full-length AbTir construct. These differences could be mediated by molecular crowding agents or additional motifs contained within the TIR domain–containing proteins, some of which have been found in plants, animals, and bacteria that help stabilize, scaffold, oligomerize, and regulate active and inactive conformations (8, 31).
The bacterial TIR NAD hydrolase activity and its effect on virulence using bacteria model systems that naturally express them at the germline level has not been thoroughly evaluated. Questions regarding the enzymatic role of bacterial TIR hydrolases in bacteria themselves, in the disruption of mammalian TIR signaling pathways and their effect on host responses remain to be fully characterized.
To assess the effect of AbTir-TIR expression directly in bacteria that produce them we used label-free 2p-FLIM live imaging. 2p-FLIM is a minimally invasive spectroscopic method used increasingly in biological, biomedical, and cancer applications (38, 39, 50, 51, 52, 53, 54, 55). 2p-FLIM uses the intrinsic fluorescence of NAD(P)H to monitor cellular function and biomedical applications, including tissue morphology and high-density protein arrays (56, 57, 58, 59, 60). Variances in FLTs are observed in bacteria expressing active (AbTir-TIRwt) and inactivated (AbTir-TIRE208A). FLT imaging is reflective of the molecular environment of NADH and independent of the protein concentration and intensity (61, 62, 63). Differences among nonexpressing (-IPTG) and expressing (+IPTG) active AbTIRwt and inactive AbTIRE208A FLTs could be influenced by a number of factors including induction efficiency, toxicity of bacterial TIR hydrolase expression, lac I operon control, leaky expression, autoinduction, TIR overexpression related toxicity, NAD+ processing and salvage pathways, or some other unaccounted phenomenon.
Changes in FLTs may reflect differences between bound and unbound free forms of NAD(P)H among active or inactivated AbTir-TIR. It is currently unclear the effect this may have in the bacteria (38, 39). These current studies do not address observations resulting from bacterial TIR–induced toxicity, IPTG-driven metabolic differences, or leaky plasmid expression. Although, we have not fully characterized the molecular pathways resulting in AbTir-TIR–induced NAD(P)H differences further studies using bacterial isolates and strains that naturally express or are bacterial TIR domain hydrolases may help bear this out. 2p-FLIM may prove useful as a minimally invasive live, label-free method for evaluating microbial infection in bacteria, plants, and animals. Future studies characterizing bacterial isolates and strains that naturally express are absent or have naturally induced TIR domain–containing hydrolase protein expression may better reflect biology.
Sequence comparison reveals conservation of the C-Helix WxxxE motif that is important for TcpB microtubule binding and includes the highly conserved Glu (E) residue, which is critical for the enzymatic function of nearly all NAD+ hydrolases (11). Without NAD+ bound, this region including the highly conserved Glu (208) and Trp (W204) identified to be important for cADPR production are observed facing away from the potential ligand-binding region. Positioning of the Glu (E208) and Trp (W204) are consistent with observations reported by Manik et al. HDX-MS substrate mediated conformational changes are also consistent with differences observed in X-ray structures and 3AD (NAD+ analog)-bound cryo-EM structures. HDX-MS studies reveal that a large part of AbTir-TIR’s exchange behavior is governed by large conformational changes displaying EX1 kinetics, which are altered upon ligand binding. These HDX-MS observations suggest that AbTir-TIR undergoes large conformational rearrangement and sampling, indicative of a heterogenous and complex conformational landscape in its native state. The WxxxE motif is not seen to be differentially protected from deuterium uptake by the addition of substrate and is not part of the region undergoing EX1-type conformational changes. Indeed, the conformational changes described by the HDX-MS are distinct in comparison with observations of X-ray and NAD analog–bound cryo-EM AbTIR assemblies (8G83, 7UWG, and 7UXU). A NAD analog–bound (3AD) cryo-EM structure of AbTIR exhibits a different interface and unique oligomeric scaffold compared with apo forms AbTIR X-ray crystal structures. Substrate-induced conformational changes and kinetics observed in solution by HDX-MS at the TIR interfaces, loop, and secondary structure may be reflective of the transition from an unbound inactive state to an active substrate-bound assembly state. It will be interesting if an apo AbTIR cryo-EM structure or ligand-bound crystal structure exhibits similar or different TIR interactions.
Bioinformatics analysis of the AbTir-TIR domain identified regions that are highly conserved among bacterial and select human TIR domain–containing proteins (11, 26, 27, 42). Direct targeting of regions and residues involved in mediating conformational change and substrate binding may provide insight into novel therapeutics to mitigate TIR-mediated pathogenesis. Indeed, the several NAD analogs as well as the anti-sepsis TIR–specific small molecule antagonist TAKEDA-242 which protects against lethal influenza binds specifically to the Cys747 located at position 2 within the WxC747xxE motif of TLR-TIR4 C helix. Small-molecule targeting of the WxxxE motif and surrounding region, as observed by TAK-242 protection against lethal influenza infection, may provide protection against additional microbial infections or diseases (64). This cysteine is observed highly conserved in several TIR-TIR interfaces involving the C Helix WxxxE motif. Changes in neighbor secondary structure elements, interactions, and the highly conserved “box2” residues may affect BB Loop position among different TIR domains.
Experimental procedures
Protein expression, purification, AUC, and crystallization
The protein expression construct containing StrepTag-AbTir-TIR-HisTag within the pet30a+ vector was obtained from Dr Kow Essuman (11). The construct for mutant AbTir-TIRE208A was created using PCR-based site-directed mutagenesis using the AbTir-TIRwt construct as a template. Recombinant AbTir-TIR wt or E208A protein was expressed, purified, crystallized, and data were collected similar to previously described (21, 22). A starter culture (10–100 ml) of bacteria containing pet30a+ plasmid in LB with 50 μg/ml kanamycin was grown overnight at either 18 °C or 30 °C and then inoculated into a Fernbach or Erlenmeyer flask containing 0.5-1L LB and incubated with shaking (200–250 rpm). At an absorbance at 600 nm of (A600 nm) between 0.6 and 1 IPTG (0.5 mM) was added and cells were incubated with shaking for an additional 3 h at 30 °C or 6 h at 18 °C, depending on the experiment. Bacteria were harvested by centrifugation at 10 to 15,000 relative centrifugal force for 20 to 30 min at 4 °C. Bacteria were resuspended using a buffer containing 25 mM Hepes or Tris at pH 7.2 with 150 mM NaCl using 25 to 50 ml of resuspension buffer for 1 L of culture and were frozen at −80 °C for later use. Frozen pellets were thawed on ice or overnight at 4 °C and lysed by sonication 2 to 3 times for 1 min at 50% output power and 50:50 duty cycle on ice. Lysates were clarified by centrifugation at 10 to 12,000 relative centrifugal force for 20 to 30 min at 4 °C with supernatant further clarified using a 0.4 or 0.8 μm filter (PALL corporation) and immediately poured over a 3 to 5 ml nickel-nitrilotriacetic acid column at 4 °C. The column was washed with 5 to 10 column volumes of resuspension buffer, and protein was eluted stepwise using resuspension buffer with increasing concentrations of imidazole (0.02 M, 0.2 M, and 0.5 M) at 4 °C. Elution fractions containing AbTIR were determined by SDS-PAGE and then further purified by size exclusion using a S200 10/30 column.
After nickel purification, AbTir-TIR was concentrated using an amicon 15 kDa molecular weight cut-off and further purified using SEC using a S200 column in buffer containing 25 mM Hepes or Tris–HCl pH7.2, 150 mM NaCl. After affinity and size-exclusion purification yields were typically between 0.2 and 0.4 mg/ml for each 1 L of bacteria growth culture. Yields were affected by expression time and temperature as well as the temperature of lysate during thawing, sonication, and purification.
Initial crystals of AbTir-TIRwt were identified from commercial screens using vapor diffusion method and further optimized by microseeding and streak-seeding to yield long, thin crystal clusters. Despite the success of seeding to reproduce crystals, attempts to further optimize crystals using various strategies including varying seed stock, batch or sparse matrix rescreening, liquid–liquid diffusion, or macro seeding failed to markedly improve crystal size or singularity. Minor improvements in crystal length were observed intermittently and were usually associated with reduced nucleation events. Crystallization of AbTir-TIRwt with NAM or NAD+ substrate either by soaking or cocrystallization methods produced similar starburst-like crystals but these did not yield single-crystal diffraction beyond ∼4 Å and no complete datasets were collected.
AUC sedimentation equilibrium measurements on AbTir-TIR samples prepared at 12, 7, and 5 μM were performed using a Beckman Coulter Optima XL-I centrifuge at rotor speeds of 23, 26, and 29K rpm in the same buffer used for size exclusion. Data were acquired at 280 nm after 6, 12, and 18 h of centrifugation, and the resulting nine scans were globally analyzed using Heteroanalysis (https://core.uconn.edu/resource/biophysics/#au-software) with single species and monomer-dimer model. Based on the distribution of the residuals and the square root of the variance of the fit, the latter model was more appropriate. Sedimentation velocity was performed on AbTir-TIRwt prepared at 10 μM at 45K rpm. Analysis of the scans acquired at 280 nm using DCDT+ yielded a single species with a sedimentation coefficient, s20,w, of 2.8S (34, 35).
Structure determination
The structure of AbTir-TIRwt was determined by molecular replacement using the TcpB TIR domain structure (4LQC) as a template model (22, 65). An initial Cα model was built using autobuild (Phenix), followed by manual building in Coot (65, 66). Iterative manual building of discontinuous loops and termini followed Phenix refinement, resulting in the AbTir-TIR model.
NAD+ hydrolase activity assays
The ability of AbTir-TIR protein to cleave NAD+ was confirmed by HPLC metabolite measurements, Etheno-NAD+ Assay, EnzyChrom NAD/NADH Assay (Bioassay systems), and one-dimensional NMR. HPLC metabolite measurement was performed in the presence of NAD+ incubated for 30 and 60 min using recombinant purified AbTir-TIRwt protein used for crystallization as described previously (11). Etheno-NAD+ Assay was performed using Nicotinamide 1, εNAD Sigma N263 was incubated with recombinant AbTir-TIR protein at 50 uM in 10 mM Hepes pH 7.5, 150 mM NaCl buffer with a reaction volume of 100 ul. Fluorescence was read using a Molecular Devices Spectramax plus plate reader (at Ex/Em = 310/410) (36). NAD levels and hydrolase activity of recombinant AbTir-TIRwt and E208A were measured using the EnzyChrom NAD/NADH Assay (Bioassay systems). AbTIR-TIRwt and E208A were incubated with 5 μM NAD for 10 to 30 min. Means (N = 6) + SEM were analyzed by two-way ANOVA to compare differences in protein type over time, followed by Tukey’s post hoc analysis; ∗ indicates p < 0.0001. AbTir-TIRwt reduces cellular levels of NAD+ upon recombinant expression in E. coli AbTIR-TIRwt and E208A. To improve protein expression yields and mitigate toxicity, bacteria was grown at 18 °C expression induced with IPTG and bacteria harvested at either 3 or 6 h after induction. Means (N = 4) + SEM were analyzed by two-way ANOVA to compare differences in protein type over time, followed by Sidak’s post hoc analysis. Letters indicate statistically significant difference (p < 0.001). Measurement of hydrolase activity of AbTir-TIR expressed in T7 cells was performed by measuring NAD+ levels in cells before and after expression of AbTir-TIRwt and E208A using the EnzyChrom NAD/NADH Assay (Bioassay systems). Briefly, T7 cells transformed with pet30A+ vectors encoding for StrepTag-AbTir-TIR-HisTagwt and E208A were grown to mid-log phase (A600 = 0.6–0.8), induced with 0.5 mM IPTG and incubated with shaking for 0 to 6 h. Cells were normalized by A600 measurements, homogenized in NAD extraction buffer (supplied), and measured for NAD+ levels according to the manufacturer’s instructions. NADase activity of recombinant, purified AbTir-TIR was further verified by one-dimensional NMR spectra that followed NADase activities, a total volume of 330 ml of 1 mM NAD+ was used with 20 ml D2O. Reactions were followed for 30 min using 16 scans for a total of 30 spectra by adding 300 mM of either AbTir-TIRWT or AbTir-TIRE208A. Data were collected using a Varian 900 spectrometer at the Rocky Mountain 900 facility.
2-photon excitation with fluorescence lifetime imaging
NAD hydrolase activity of bacteria containing AbTir-TIRwt or AbTir-TIRE208A plasmid was further analyzed by 2p-FLIM. Samples were grown and induced to express AbTir-TIRwt or AbTir-TIRE208A as described for protein expression and hydrolase activity experiments. Bacteria were measured by absorbance at 600 nm (A600) and adjusted by dilution to the same concentration prior to imaging. A customized confocal microscope (based on ISS Q2 laser scanning nanoscope) with single-molecule detection sensitivity was used for performing 2p-FLIM. The excitation source is a pulsed femtosecond laser (Calmar, 780 nm, 90 fs pulse width, and 50 MHz repetition rate) equipped with an ISS excitation power control unit. An incident wavelength of 780 nm was used for exciting NAD(P)H in cells. The excitation light was reflected by a dichroic mirror to a high-numerical aperture water objective (60×; numerical aperture, 1.2) and focused onto the sample. The fluorescence was collected by single photon counting avalanche photodiodes through a dichroic beam splitter, Chroma short pass (750SP), and 460/55 band-pass filters, thus eliminating the scattered excitation light and collecting fluorescence from the NAD(P)H in the region of interest. The imaging in the ISS Q2 setup was performed with Galvo-controlled mirrors with related electronics and optics controlled through the 3X-DAC control card. The software module in ISS VistaVision for data acquisition and processing and the time-correlated single photon counting module from Becker & Hickl (SPC-150) facilitate FLIM measurements and analyses. Statistical analysis of data was performed using GraphPad prism 8.0 (https://www.graphpad.com/) and Origin 2021 software (https://www.originlab.com/).
Hydrogen-deuterium exchange mass spectrometry
The uptake of deuterium following exposure to deuterated water is monitored by LC-MS in a bottom-up approach. Undeuterated (control) samples were first acquired to obtain a sequence coverage map using an experimental workflow as follows: 2 μl of 50 μM AbTIR in 25 mM Tris, 150 mM NaCl pH 7.4 was diluted with 98 μl ice-cold quench containing 50 mM glycine pH 2.5, 1 M guanidine-HCl, resulting in a 100 μl reaction. The experimental workflow for the deuterated samples for H/D exchange reactions was as follows: 2 μl of apo 50 μM AbTIR or in complex with 10 mM εNAD was incubated in 48 μl D2O buffer containing 25 mM Tris, 150 mM NaCl, pH 7.0 at 25 °C. The resulting 50 μl deuteration reactions were quenched at various times (10 s, 1 min, 10 min, 1 h, and 2 h) with 100 μl ice-cold quench (100 mM glycine pH 2.5, 1.5 M Guanidine) prior to injection. Using a LEAP autosampler for high-throughput acquisition, deuterated and undeuterated (control) samples were injected into a Waters HDX system equipped with an Acquity M-class Ultra-Performance Liquid Chromatography with in-line pepsin digestion. The resulting peptides were trapped on an Acquity Ultra-Performance Liquid Chromatography BEH C18 peptide trap and separated using an Acquity Ultra-Performance Liquid Chromatography BEH C18 column, followed by injection into a Waters Synapt G2Si mass spectrometer with ion mobility capabilities. Resulting peptides were identified using the ProteinLynx Global Server 3.0.3 from Waters. Samples at all deuterium incubation time points (including undeuterated controls) were acquired in triplicate. The centroid mass shift of peptides identified by ProteinLynx Global Server 3.0.3 was tracked as a function of deuterium incubation times using DynamX 3.0 software (Waters)(https://www.waters.com/waters/library.htm?locale=en_US&lid=134832928).
Data availability
The crystal structure of A. baumannii Tir-TIR (AbTIR-TIR) was deposited in the PDB under the accession code 8G83. All other data is contained within the manuscript or will be shared upon request (Greg Snyder gsnyder@ihv.umaryland.edu).
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This research used AMX beamline resources of the National Synchrotron Light Source II, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under Contract No. DE-SC0012704. The Center for BioMolecular Structure (CBMS) is primarily supported by the National Institutes of Health, National Institute of General Medical Sciences (NIGMS) through a Center Core P30 Grant (P30GM133893), and by the DOE Office of Biological and Environmental Research (KP1607011). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
E. K., J. O. O., Y. W., G. G., J. C., C. T., S. B., S. N., A. S. S., J. S. R., E. E., A. H. S, T. O., K. E., J. M., A. D., K. R., M. L. D. S, D. D., and G. A. S. investigation; J. O. O., G. G., J. C., C. T., S. B., S. N., A. H. S, T. O., K. R., M. L. D. S, D. D., and G. A. S. visualization; J. O. O., G. G., J. C., C. T., S. B., S. N., D. B., J. S. R., E. E., A. H. S, T. O., K. E., J. M., A. D., K. R., M. L. D. S, D. D., and G. A. S. formal analysis; J. O. O., D. B., J. S. R., E. E., K. R., M. L. D. S, D. D., and G. A. S. writing-original draft; D. B., J. S. R., E. E., K. E., J. M., A. D., K. R., M. L. D. S, D. D., and G. A. S. methodology; D. B., K. R., M. L. D. S, D. D., and G. A. S. supervision; D. B., K. E., J. M., and A. D. writing-review and editing; K. R., M. L. D. S, D. D., and G. A. S. conceptualization.
Funding and additional information
Funding was provided to C. P. and M. L. D. S., R25GM119970 Bridges to the Doctorate: a partnership Between Towson University and University of Maryland School of Medicine and to T. O. and M. L. D. S., R25GM058264 facilitating the seamless transition from community college to Towson University. The Institute of Human Virology, Summer High School Opportunity Research Experience (SHORE) provided funding to G. A. S., G. G., C. T., and S. B. The late Hon. U.S. Rep. Elijah Cummings and the University of Maryland Baltimore, Summer Bioscience Internship Program and the Baltimore City Youthworks Program provided funding to J. C. Funding was provided to K. R. under NIH/NIAID R01-AI172487.
Reviewed by members of the JBC Editorial Board. Edited by Alex Toker
Supporting information
References
- 1.Turner J.D. A bioinformatic approach to the identification of bacterial proteins interacting with Toll-interleukin 1 receptor-resistance (TIR) homology domains. FEMS Immunol. Med. Microbiol. 2003;37:13–21. doi: 10.1016/S0928-8244(03)00095-6. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Q., Zmasek C.M., Cai X., Godzik A. TIR domain-containing adaptor SARM is a late addition to the ongoing microbe-host dialog. Dev. Comp. Immunol. 2011;35:461–468. doi: 10.1016/j.dci.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waldhuber A., Snyder G.A., Römmler F., Cirl C., Müller T., Xiao T.S., et al. A comparative analysis of the mechanism of Toll-like receptor-disruption by TIR-containing protein C from Uropathogenic Escherichia coli. Pathogens. 2016;5:25. doi: 10.3390/pathogens5010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cirl C., Wieser A., Yadav M., Duerr S., Schubert S., Fischer H., et al. Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins. Nat. Med. 2008;14:399–406. doi: 10.1038/nm1734. [DOI] [PubMed] [Google Scholar]
- 5.Radhakrishnan G.K., Splitter G.A. Biochemical and functional analysis of TIR domain containing protein from Brucella melitensis. Biochem. Biophys. Res. Commun. 2010;397:59–63. doi: 10.1016/j.bbrc.2010.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radhakrishnan G.K., Yu Q., Harms J.S., Splitter G.A. Brucella TIR domain-containing protein mimics properties of the toll-like receptor adaptor protein TIRAP. J. Biol. Chem. 2009;284:9892–9898. doi: 10.1074/jbc.M805458200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi Y., Kerry P.S., Nanson J.D., Bosanac T., Sasaki Y., Krauss R., et al. Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules. Mol. Cell. 2022;82:1643–1659.e10. doi: 10.1016/j.molcel.2022.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maruta N., Burdett H., Lim B.Y.J., Hu X., Desa S., Manik M.K., et al. Structural basis of NLR activation and innate immune signalling in plants. Immunogenetics. 2022;74:5–26. doi: 10.1007/s00251-021-01242-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiAntonio A., Milbrandt J., Figley M.D. The SARM1 TIR NADase: mechanistic similarities to bacterial phage defense and Toxin-Antitoxin systems. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.752898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weagley J.S., Zaydman M., Venkatesh S., Sasaki Y., Damaraju N., Yenkin A., et al. Products of gut microbial Toll/interleukin-1 receptor domain NADase activities in gnotobiotic mice and Bangladeshi children with malnutrition. Cell Rep. 2022;39 doi: 10.1016/j.celrep.2022.110738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Essuman K., Summers D.W., Sasaki Y., Mao X., Yim A.K.Y., DiAntonio A., et al. TIR domain proteins are an Ancient family of NAD(+)-Consuming Enzymes. Curr. Biol. 2018;28:421–430.e4. doi: 10.1016/j.cub.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Essuman K., Summers D.W., Sasaki Y., Mao X., DiAntonio A., Milbrandt J. The SARM1 Toll/interleukin-1 receptor domain possesses intrinsic NAD(+) cleavage activity that promotes Pathological axonal degeneration. Neuron. 2017;93:1334–1343.e5. doi: 10.1016/j.neuron.2017.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williamson D.H., Lund P., Krebs H.A. The redox state of free nicotinamide-adenine dinucleotide in the cytoplasm and mitochondria of rat liver. Biochem. J. 1967;103:514–527. doi: 10.1042/bj1030514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerdts J., Brace E.J., Sasaki Y., DiAntonio A., Milbrandt J. SARM1 activation triggers axon degeneration locally via NAD⁺ destruction. Science. 2015;348:453–457. doi: 10.1126/science.1258366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerdts J., Summers D.W., Sasaki Y., DiAntonio A., Milbrandt J. Sarm1-mediated axon degeneration requires both SAM and TIR interactions. J. Neurosci. 2013;33:13569–13580. doi: 10.1523/JNEUROSCI.1197-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bayless A.M., Nishimura M.T. Enzymatic functions for Toll/interleukin-1 receptor domain proteins in the plant immune system. Front. Genet. 2020;11:539. doi: 10.3389/fgene.2020.00539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wan L., Essuman K., Anderson R.G., Sasaki Y., Monteiro F., Chung E.H., et al. TIR domains of plant immune receptors are NAD. Science. 2019;365:799–803. doi: 10.1126/science.aax1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patot S., Imbert P., Baude J., Martins Simões P., Campergue J.B., Louche A., et al. The TIR homologue lies near resistance genes in staphylococcus aureus, coupling modulation of virulence and antimicrobial susceptibility. PLoS Pathog. 2017;13:e1006092. doi: 10.1371/journal.ppat.1006092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Askarian F., van Sorge N.M., Sangvik M., Beasley F.C., Henriksen J.R., Sollid J.U.E., et al. A Staphylococcus aureus TIR domain protein virulence factor blocks TLR2-mediated NF-kappaB signaling. J. Innate Immun. 2014;6:485–498. doi: 10.1159/000357618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imbert P.R., Louche A., Luizet J.B., Grandjean T., Bigot S., Wood T.E., et al. A Pseudomonas aeruginosa TIR effector mediates immune evasion by targeting UBAP1 and TLR adaptors. EMBO J. 2017;36:1869–1887. doi: 10.15252/embj.201695343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snyder G.A., Cirl C., Jiang J., Chen K., Waldhuber A., Smith P., et al. Molecular mechanisms for the subversion of MyD88 signaling by TcpC from virulent uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 2013;110:6985–6990. doi: 10.1073/pnas.1215770110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snyder G.A., Deredge D., Waldhuber A., Fresquez T., Wilkins D.Z., Smith P.T., et al. Crystal structures of the Toll/Interleukin-1 receptor (TIR) domains from the Brucella protein TcpB and host adaptor TIRAP reveal mechanisms of molecular mimicry. J. Biol. Chem. 2014;289:669–679. doi: 10.1074/jbc.M113.523407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manik M.K., Shi Y., Li S., Zaydman M.A., Damaraju N., Eastman S., et al. Cyclic ADP ribose isomers: production, chemical structures, and immune signaling. Science. 2022;377:eadc8969. doi: 10.1126/science.adc8969. [DOI] [PubMed] [Google Scholar]
- 24.Ofir G., Herbst E., Baroz M., Cohen D., Millman A., Doron S., et al. Antiviral activity of bacterial TIR domains via immune signalling molecules. Nature. 2021;600:116–120. doi: 10.1038/s41586-021-04098-7. [DOI] [PubMed] [Google Scholar]
- 25.Li Y., Guo X., Hu C., Du Y., Guo C., Di W., et al. Type I IFN operates pyroptosis and necroptosis during multidrug-resistant A. baumannii infection. Cell Death Differ. 2018;25:1304–1318. doi: 10.1038/s41418-017-0041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sikowitz M.D., Cooper L.E., Begley T.P., Kaminski P.A., Ealick S.E. Reversal of the substrate specificity of CMP N-glycosidase to dCMP. Biochemistry. 2013;52:4037–4047. doi: 10.1021/bi400316p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Felix C., Kaplan Türköz B., Ranaldi S., Koelblen T., Terradot L., O'Callaghan D., et al. The Brucella TIR domain containing proteins BtpA and BtpB have a structural WxxxE motif important for protection against microtubule depolymerisation. Cell Commun. Signal. 2014;12:53. doi: 10.1186/s12964-014-0053-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horsefield S., Burdett H., Zhang X., Manik M.K., Shi Y., Chen J., et al. NAD+ cleavage activity by animal and plant TIR domains in cell death pathways. Science. 2019;365:793–799. doi: 10.1126/science.aax1911. [DOI] [PubMed] [Google Scholar]
- 29.Bratkowski M., Burdett T.C., Danao J., Wang X., Mathur P., Gu W., et al. Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease. Neuron. 2022;110:3711–3726.e16. doi: 10.1016/j.neuron.2022.08.017. [DOI] [PubMed] [Google Scholar]
- 30.Snyder G.A., Sundberg E.J. Molecular interactions in interleukin and toll-like receptor signaling pathways. Curr. Pharm. Des. 2013;20:1244–1258. doi: 10.2174/13816128113199990069. [DOI] [PubMed] [Google Scholar]
- 31.Li S., Manik M.K., Shi Y., Kobe B., Ve T. Toll/interleukin-1 receptor domains in bacterial and plant immunity. Curr. Opin. Microbiol. 2023;74 doi: 10.1016/j.mib.2023.102316. [DOI] [PubMed] [Google Scholar]
- 32.Miller M.S., Maheshwari S., Shi W., Gao Y., Chu N., Soares A.S., et al. Getting the most out of Your crystals: data collection at the new high-Flux, microfocus MX beamlines at NSLS-II. Molecules. 2019;24:496. doi: 10.3390/molecules24030496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson M.L., Correia J.J., Yphantis D.A., Halvorson H.R. Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophys. J. 1981;36:575–588. doi: 10.1016/S0006-3495(81)84753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stafford W.F. Boundary analysis in sedimentation transport experiments: a procedure for obtaining sedimentation coefficient distributions using the time derivative of the concentration profile. Anal. Biochem. 1992;203:295–301. doi: 10.1016/0003-2697(92)90316-y. [DOI] [PubMed] [Google Scholar]
- 35.Philo J.S. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal. Biochem. 2006;354:238–246. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 36.Schultz M.B., Lu Y., Braidy N., Sinclair D.A. Assays for NAD+-Dependent Reactions and NAD+ Metabolites. Methods Mol. Biol. 2018;1813:77–90. doi: 10.1007/978-1-4939-8588-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee E., Redzic J.S., Nemkov T., Saviola A.J., Dzieciatkowska M., Hansen K.C., et al. Human and bacterial Toll-interleukin receptor domains exhibit distinct dynamic features and functions. Molecules. 2022;27:4494. doi: 10.3390/molecules27144494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berezin M.Y., Achilefu S. Fluorescence lifetime measurements and biological imaging. Chem. Rev. 2010;110:2641–2684. doi: 10.1021/cr900343z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Becker W. Fluorescence lifetime imaging--techniques and applications. J. Microsc. 2012;247:119–136. doi: 10.1111/j.1365-2818.2012.03618.x. [DOI] [PubMed] [Google Scholar]
- 40.Xu Y., Tao X., Shen B., Horng T., Medzhitov R., Manley J.L., et al. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature. 2000;408:111–115. doi: 10.1038/35040600. [DOI] [PubMed] [Google Scholar]
- 41.Rock F.L., Hardiman G., Timans J.C., Kastelein R.A., Bazan J.F. A family of human receptors structurally related to Drosophila Toll. Proc. Natl. Acad. Sci. U. S. A. 1998;95:588–593. doi: 10.1073/pnas.95.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toshchakov V.Y., Neuwald A.F. A survey of TIR domain sequence and structure divergence. Immunogenetics. 2020;72:181–203. doi: 10.1007/s00251-020-01157-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan S.L., Low L.Y., Hsu S., Li S., Liu T., Santelli E., et al. Molecular mimicry in innate immunity: crystal structure of a bacterial TIR domain. J. Biol. Chem. 2009;284:21386–21392. doi: 10.1074/jbc.C109.007591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alaidarous M., Ve T., Casey L.W., Valkov E., Ericsson D.J., Ullah M.O., et al. Mechanism of bacterial interference with TLR4 signaling by Brucella Toll/interleukin-1 receptor domain-containing protein TcpB. J. Biol. Chem. 2014;289:654–668. doi: 10.1074/jbc.M113.523274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sporny M., Guez-Haddad J., Khazma T., Yaron A., Dessau M., Shkolnisky Y., et al. Structural basis for SARM1 inhibition and activation under energetic stress. Elife. 2020;9 doi: 10.7554/eLife.62021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaplan-Turkoz B., Koelblen T., Felix C., Candusso M.P., O'Callaghan D., Vergunst A.C., et al. Structure of the Toll/interleukin 1 receptor (TIR) domain of the immunosuppressive Brucella effector BtpA/Btp1/TcpB. FEBS Lett. 2013;587:3412–3416. doi: 10.1016/j.febslet.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 47.Figley M.D., Gu W., Nanson J.D., Shi Y., Sasaki Y., Cunnea K., et al. SARM1 is a metabolic sensor activated by an increased NMN/NAD. Neuron. 2021;109:1118–1136.e11. doi: 10.1016/j.neuron.2021.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang Y., et al. The NAD+-mediated self-inhibition mechanism of pro-neurodegenerative SARM1. Nature. 2020;588:658–663. doi: 10.1038/s41586-020-2862-z. [DOI] [PubMed] [Google Scholar]
- 49.Bratkowski M., Xie T., Thayer D.A., Lad S., Mathur P., Yang Y.S., et al. Structural and mechanistic regulation of the Pro-degenerative NAD hydrolase SARM1. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.107999. [DOI] [PubMed] [Google Scholar]
- 50.Lakowicz J.R. Third ed. Springer; New York, NY: 2006. Principles of Fluorescence Spectroscopy, Chapter 22. [Google Scholar]
- 51.Ray K., Lakowicz J.R. Metal-enhanced fluorescence lifetime imaging and spectroscopy on a modified SERS substrate. J. Phys. Chem. C Nanomater. Interfaces. 2013;117 doi: 10.1021/jp404590j. 15790–15797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fixler D., Nayhoz T., Ray K. Diffusion reflection and fluorescence lifetime imaging microscopy Study of Fluorophore-Conjugated Gold Nanoparticles or Nanorods in solid phantoms. ACS Photonics. 2014;1:900–905. doi: 10.1021/ph500214m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Penjweini R., Roarke B., Alspaugh G., Gevorgyan A., Andreoni A., Pasut A., et al. Single cell-based fluorescence lifetime imaging of intracellular oxygenation and metabolism. Redox Biol. 2020;34 doi: 10.1016/j.redox.2020.101549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Niehorster T., Löschberger A., Gregor I., Krämer B., Rahn H.J., Patting M., et al. Multi-target spectrally resolved fluorescence lifetime imaging microscopy. Nat. Methods. 2016;13:257–262. doi: 10.1038/nmeth.3740. [DOI] [PubMed] [Google Scholar]
- 55.Barnoy E.A., Fixler D., Popovtzer R., Nayhoz T., Ray K. An ultra-sensitive dual-mode imaging system using metal-enhanced fluorescence in solid phantoms. Nano Res. 2015;8:3912–3921. doi: 10.1007/s12274-015-0891-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun Y., Hatami N., Yee M., Phipps J., Elson D.S., Gorin F., et al. Fluorescence lifetime imaging microscopy for brain tumor image-guided surgery. J. Biomed. Opt. 2010;15 doi: 10.1117/1.3486612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Coda S., Thompson A.J., Kennedy G.T., Roche K.L., Ayaru L., Bansi D.S., et al. Fluorescence lifetime spectroscopy of tissue autofluorescence in normal and diseased colon measured ex vivo using a fiber-optic probe. Biomed. Opt. Express. 2014;5:515–538. doi: 10.1364/BOE.5.000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Skala M.C., Riching K.M., Gendron-Fitzpatrick A., Eickhoff J., Eliceiri K.W., White J.G., et al. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc. Natl. Acad. Sci. U. S. A. 2007;104:19494–19499. doi: 10.1073/pnas.0708425104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sarder P., Maji D., Achilefu S. Molecular probes for fluorescence lifetime imaging. Bioconjug. Chem. 2015;26:963–974. doi: 10.1021/acs.bioconjchem.5b00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun Y., Hays N.M., Periasamy A., Davidson M.W., Day R.N. Monitoring protein interactions in living cells with fluorescence lifetime imaging microscopy. Methods Enzymol. 2012;504:371–391. doi: 10.1016/B978-0-12-391857-4.00019-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cao R., Wallrabe H., Periasamy A. Multiphoton FLIM imaging of NAD(P)H and FAD with one excitation wavelength. J. Biomed. Opt. 2020;25:1–16. doi: 10.1117/1.JBO.25.1.014510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang S., Heikal A.A., Webb W.W. Two-photon fluorescence spectroscopy and microscopy of NAD(P)H and flavoprotein. Biophys. J. 2002;82:2811–2825. doi: 10.1016/S0006-3495(02)75621-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blacker T.S., Mann Z.F., Gale J.E., Ziegler M., Bain A.J., Szabadkai G., et al. Separating NADH and NADPH fluorescence in live cells and tissues using FLIM. Nat. Commun. 2014;5:3936. doi: 10.1038/ncomms4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shirey K.A., Lai W., Brown L.J., Blanco J.C.G., Beadenkopf R., Wang Y., et al. Select targeting of intracellular Toll-interleukin-1 receptor resistance domains for protection against influenza-induced disease. Innate Immun. 2019;26:26–34. doi: 10.1177/1753425919846281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The crystal structure of A. baumannii Tir-TIR (AbTIR-TIR) was deposited in the PDB under the accession code 8G83. All other data is contained within the manuscript or will be shared upon request (Greg Snyder gsnyder@ihv.umaryland.edu).