The EGFR–ERK pathway is one of the most important signaling cascades in cell survival and proliferation. The study reports that CDK2 regulates the ERK pathway via USP37. Importantly, combined CDK1/2 and EGFR inhibitors have a synergetic anticancer effect through the downregulation of ERK1/2 stability and activity.
Abstract
The EGFR-RAS-ERK pathway is one of the most important signaling cascades in cell survival, growth, and proliferation. Aberrant activation of this pathway is a common mechanism in various cancers. Here, we report that CDK2 is a novel regulator of the ERK pathway via USP37 deubiquitinase (DUB). Mechanistically, CDK2 phosphorylates USP37, which is required for USP37 DUB activity. Further, USP37 deubiquitinates and stabilizes ERK1/2, thereby enhancing cancer cell proliferation. Thus, CDK2 is able to promote cell proliferation by activating USP37 and, in turn, stabilizing ERK1/2. Importantly, combined CDK1/2 and EGFR inhibitors have a synergetic anticancer effect through the downregulation of ERK1/2 stability and activity. Indeed, our patient-derived xenograft (PDX) results suggest that targeting both ERK1/2 stability and activity kills cancer cells more efficiently even at lower doses of these two inhibitors, which may reduce their associated side effects and indicate a potential new combination strategy for cancer therapy.
Introduction
The RAS–ERK signaling cascade is evolutionarily conserved and activated by growth factors binding to receptor tyrosine kinase (RTK), such as EGFR, which results in the activation of the small G-protein RAS (Chou et al., 2010; Du et al., 2019). Subsequently, RAF, MEK, and ERK are successively activated in a phosphorylation-dependent cascade (Aoki et al., 2011; May and Hill, 2008). The RAS–ERK pathway regulates various physiological processes, including cell proliferation, survival, differentiation, and transformation (Aoki et al., 2011; Roux and Blenis, 2004). Overactivation of the RAS–ERK pathway occurs in >30% of human malignancies, including lung, colon, and head and neck squamous cell carcinoma (HNSCC; Mainardi et al., 2018; Yoshida et al., 2006; Zhao and Adjei, 2014). The extracellular signal-regulated kinases ERK1 and ERK2 (hereafter ERK1/2), central regulators of the RAS–ERK pathway, are protein–serine/threonine kinases that contribute to the RAS–ERK signaling module (Lavoie et al., 2020). ERK1/2 are direct downstream effectors of the dual-specificity MAPK/ERK kinases MEK1 and MEK2, which are activated following growth factors and mitogen stimulus. After activation by the RAS–RAF–MEK kinases cascade, ERK1/2 phosphorylates downstream effectors to participate in the regulation of numerous physiological processes, including cell adhesion, transcription, survival, proliferation, growth, and differentiation (Caunt et al., 2015; Roskoski Jr, 2012). Hyperactivation of ERK1/2 is highly implicated in tumorigenesis as a result of abundant mutations and overexpression of upstream regulators, including RAS, RAF, and MEK (Frémin and Meloche, 2010; Kaplan et al., 2011; Yu et al., 2015). In addition, reactivation of ERK1/2 is a primary reason for chemoresistance after treatment with EGFR, KRAS, BRAF, and MEK inhibitors (Fedele et al., 2018; Morris et al., 2013; Sanchez et al., 2019; Tadesse et al., 2020; Yu et al., 2015; Zhang et al., 2021a), suggesting that targeting ERK1/2 protein levels may be a potential strategy for the treatment of cancers with high ERK status.
Cyclin-dependent kinases (CDKs), a large family of serine/threonine protein kinases, play pivotal roles in regulating cell progression, cell cycle, survival, and proliferation (Asghar et al., 2015; Hochegger et al., 2008). CDK4/6 and CDK1/2 are commonly dysregulated in various types of cancers (Musgrove et al., 2011; Otto and Sicinski, 2017). CDK4/6 inhibitors have been FDA-approved to kill cancers, while CDK1/2 inhibitors for cancer treatment are still undergoing clinical trials (Lim et al., 2016; O’Leary et al., 2016; Sherr et al., 2016; Zhang et al., 2021c).
How ERK1/2 is activated is well-established. MEK1/2 directly phosphorylates ERK1/2 at Tyr204/187 and Thr202/185, which is required for ERK1/2 activation (Canagarajah et al., 1997; Roskoski, 2019; Zhang et al., 1994). Additionally, it was recently reported that K63-linked polyubiquitination of ERK1/2 promotes the interaction between MEK1/2 and ERK1/2, which is vital for MEK1/2-mediated ERK1/2 activation (Zhu et al., 2021). Intriguingly, MEKK1 functions as an E3 ligase and promotes ubiquitination and degradation of ERK1/2 (Lu et al., 2002). However, the regulation of ERK1/2 protein stability remains to be further explored. In this study, we found that the CDK2-USP37 axis is a novel regulator for ERK1/2 through ERK1/2 stabilization. Mechanistically, CDK2 phosphorylates USP37 at serine 628 (Ser628) and then activates USP37 deubiquitinase activity. Further, USP37 removes the polyubiquitinated chains from ERK1/2 and prevents its degradation. Thus, CDK2 upregulates ERK1/2 stability via USP37.
Results
Combined CDK1/2 and EGFR inhibitors create a synergistic anticancer effect
Epidermal growth factor receptor (EGFR), a key upstream molecule of RAS/RAF/MEK/ERK pathways, plays a pivotal role in regulating normal cell proliferation, survival, and differentiation (Kong et al., 2021). Aberrant regulation of EGFR resulting in the activation of the RAS/RAF/MEK/ERK cascade is a common mechanism in various cancers and is always related to poor survival (Hirata and Kiyokawa, 2019; Noordhuis et al., 2009). Several EGFR inhibitors have shown survival benefits for patients carrying EGFR mutations (Bianco et al., 2007; Troiani et al., 2012). However, the reactivation of the RAS–ERK pathway is a major cause of chemoresistance following treatment with EGFR inhibitors (Corcoran et al., 2012; Ercan et al., 2012; Hazar-Rethinam et al., 2018), the mechanism for which remains unclear. Hence, it is urgent to further understand the regulatory mechanism of the EGFR–RAS–ERK cascade.
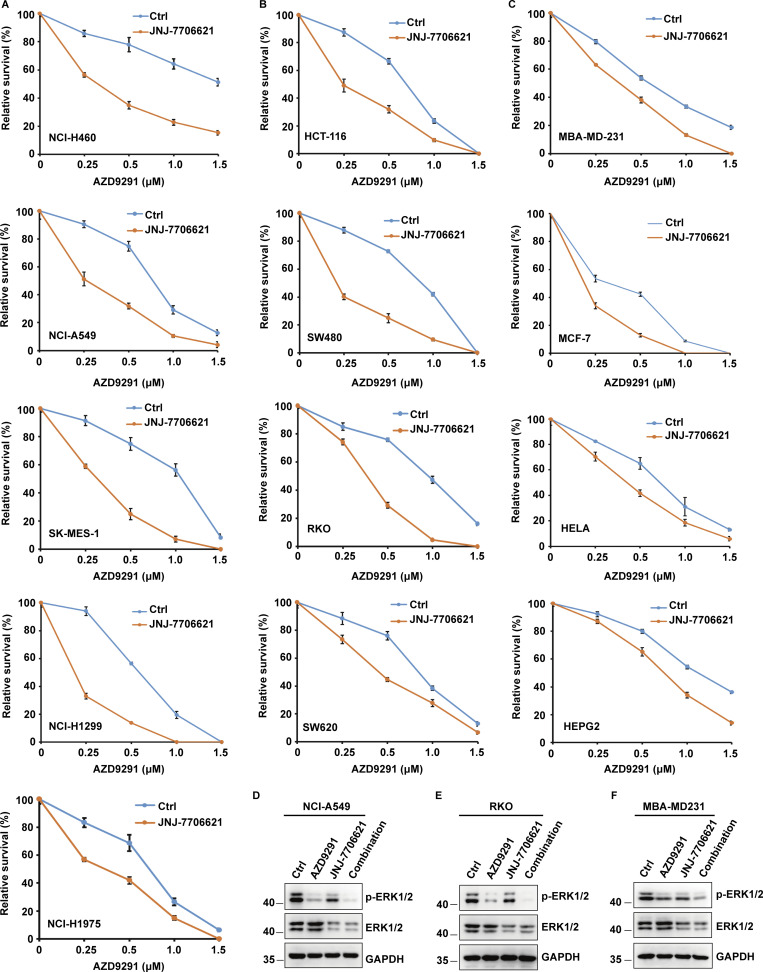
Intriguingly, we unexpectedly found that the lower dosage of CDK1/2 inhibitors significantly enhanced the anticancer effect of EGFR inhibitors in various cancer cells (Fig. 1, A–C). To identify the mechanism, ERK1/2 signaling was detected by Western blot following the use of CDK1/2 inhibitors with or without EGFR inhibitors. We found that treatment with EGFR inhibitors markedly decreased the level of ERK1/2 phosphorylation (Fig. 1, D–F). Surprisingly, treating cells with CDK1/2 inhibitors resulted in a sharp decrease in the ERK1/2 protein level (Fig. 1, D–F). In addition, the combination of EGFR and CDK1/2 inhibitors enhanced the decrease in both phosphorylation and protein levels of ERK1/2 (Fig. 1, D–F). Taken together, our results suggest that CDK1/2 may regulate ERK1/2 protein levels.
Figure 1.
Combined CDK1/2 inhibitors and EGFR inhibitors present a synergistic anticancer effect. (A) Several lung cancer cell lines were utilized to perform colony formation as indicated. Data were represented as the means ± SD of three independent experiments. (B) Several colon cell lines were utilized to perform colony formation as indicated. Data were represented as the means ± SD of three independent experiments. (C) Various cancer cell lines were utilized to perform colony formation as indicated. Data were represented as the means ± SD of three independent experiments. (D–F) NCI-A549 (D), RKO (E), or MBA-MD231 (F) cells were respectively treated with JNJ-7706621 (CDK1/2i, 10 μM), AZD9291 (EGFRi, 10 μM), or JNJ-7706621 (CDK1/2i, 10 μM) + AZD9291 (EGFRi, 10 μM) for 10 h before harvest. The samples were immunoblotted with indicated antibodies. Source data are available for this figure: SourceData F1.
CDK2 regulates ERK1/2 protein level
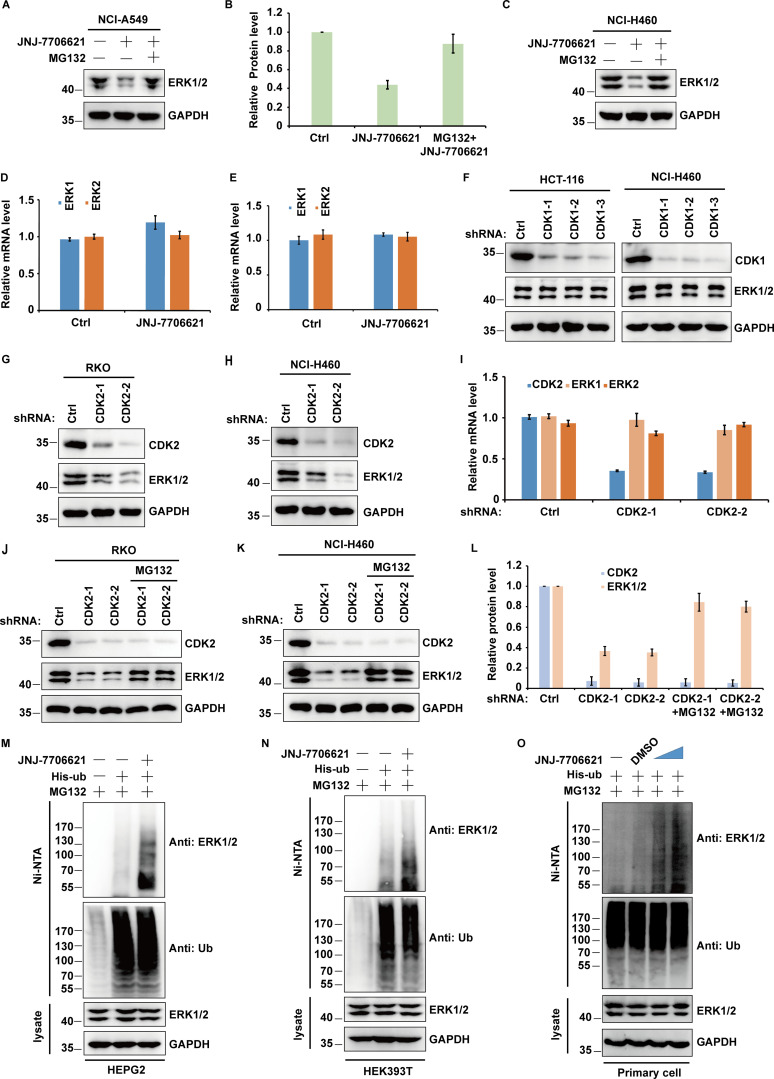
Given that treating cells with CDK1/2 inhibitors (CDK1/2i) reduced the ERK1/2 protein level, we further studied the role of CDK1/2 in regulating ERK1/2 stability. As shown in Fig. 2, A–C, treatment with the proteasome inhibitor MG132 was able to antagonize CDK1/2i-induced ERK1/2 downregulation. In addition, CDK1/2i treatment had no effect on the mRNA level of ERK1/2 (Fig. 2, D and E). Further, the depletion of CDK2 but not CDK1 significantly decreased ERK1/2 protein levels but not mRNA levels (Fig. 2, F–I). Meanwhile, decreased ERK1/2 levels could be reversed by the addition of proteasome inhibitor MG132 (Fig. 2, J–L). In addition, CDK1/2i treatment significantly increased ERK1/2 ubiquitination in different cell lines (Fig. 2, M–O). Taken together, these results suggested that CDK2 upregulates ERK1/2 protein levels by impairing ERK1/2 polyubiquitination in a proteasome-dependent manner.
Figure 2.
CDK2 regulates ERK1/2 protein level. (A) NCI-A549 cells were treated with JNJ-7706621 (10 μM) alone or combined with MG132 (30 μM) for 10 h before harvest. Half of the cells were lysed, and then Western blot was performed with the indicated antibodies. (B) Relative ERK1/2 protein levels were quantified with GAPDH from A as internal standards. Immunoreactive bands of at least three independent experiments were quantified using ImageJ software. Data were represented as means ± SD. (C) NCI-H460 cells were treated with JNJ-7706621 alone or combined with MG132 (30 μM) for 10 h before harvest. Half of the cells were lysed and then Western blot was performed with the indicated antibodies. (D) The mRNAs were extracted from the rest of the cells from A and then subjected to qRT–PCR. β-actin served as an endogenous control. Data were represented as the means ± SD of three independent experiments. (E) The mRNAs were extracted from the rest of the cells from C and subjected to qRT–PCR. β-actin served as an endogenous control. Data were represented as the means ± SD of three independent experiments. (F) HCT-116 and NCI-H460 cells stably expressing control or CDK1 shRNAs as indicated were harvested, and then cell lysates were blotted with the indicated antibodies. (G and H) RKO (G) and NCI-H460 (H) cells stably expressing control or CDK2 shRNAs as indicated were harvested, and then cell lysates were blotted with the indicated antibodies. (I) The mRNAs were extracted from the rest of the cells from H and then subjected to qRT-PCR. β-actin served as an endogenous control. Data were represented as the means ± SD of three independent experiments. (J and K) RKO (J) and NCI-H460 (K) cells stably expressing control or two different CDK2 shRNAs were treated with vehicle or MG132 for 10 h before harvest. Western blot was performed with the indicated antibodies. (L) Relative ERK1/2 or CDK2 protein levels were quantified with GAPDH from K as internal standards. Immunoreactive bands of at least three independent experiments were quantified using ImageJ software. Data were represented as means ± SD. (M and N) The various cells transfected with indicated constructs were treated with JNJ-7706621 (10 μM) alone or combined with MG132 (30 μM) for 10 h before harvest. Covalently modified proteins were purified on NiNTA-agarose under denatured conditions and then blotted with indicated antibodies. (O) The primary cells of the basal cells transfected with indicated constructs were treated with JNJ-7706621 alone or combined with MG132 for 10 h before harvest. Covalently modified proteins were purified on NiNTA-agarose under denatured conditions and then blotted with indicated antibodies. Source data are available for this figure: SourceData F2.
USP37 interacts with ERK1/2
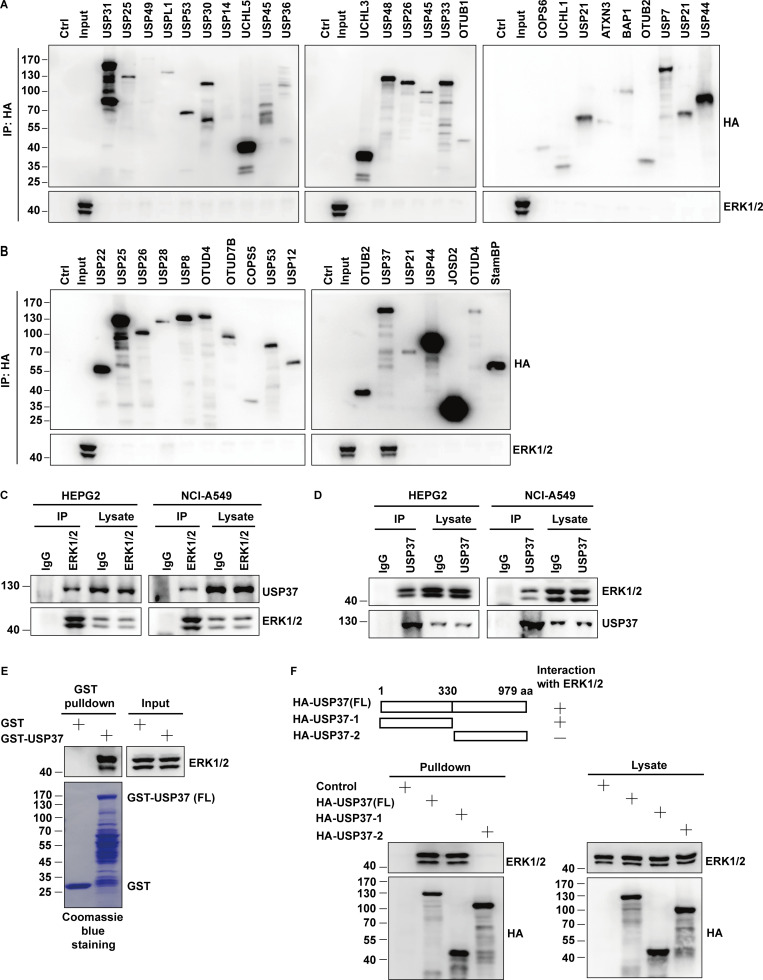
Since deubiquitinases (DUBs) play a vital role in removing polyubiquitinated chains from proteins and preventing protein degradation (Hochstrasser, 1996; Wilkinson, 2000), we screened deubiquitinase families to identify DUBs that deubiquitinate and stabilize ERK1/2 and may play a role in connecting CDKs and ERK1/2. A panel of deubiquitinases were overexpressed in HEK293T cells individually, and ERK1/2 interacting proteins were examined. We found that USP37 is a novel binding partner of ERK1/2 (Fig. 3, A and B). A coimmunoprecipitation assay was then performed to confirm the interaction between endogenous USP37 and ERK1/2. As shown in Fig. 3 C, immunoprecipitation of endogenous ERK1/2 pulled down USP37 proteins in different cancer cell lines. The interaction between USP37 and ERK1/2 was further confirmed using a reciprocal coimmunoprecipitation assay (Fig. 3 D). In addition, the binding between USP37 and ERK1/2 was further confirmed using bacterially expressed GST-USP37 proteins (Fig. 3 E). Moreover, the segment of USP37 responsible for binding with ERK1/2 was mapped. As shown in Fig. 3 F, the N-terminal region of USP37 (1–330 aa) was critical for the interaction between USP37 and ERK1/2.
Figure 3.
USP37 interacts with ERK1/2. (A and B) HEK293T cells were transfected indicated deubiquitinases (DUBs) plasmid. After transfection for 48 h, cells were lysed and purified using anti-HA-agarose beads. Then coimmunoprecipitating endogenous ERK1/2 was detected by anti-ERK1/2 antibody. (C and D) The cancer cell lysates were subject to immunoprecipitation with control IgG, anti-ERK1/2 (C), or anti-USP37 (D) antibodies. The immunoprecipitates were then blotted with the indicated antibodies. (E) HEK293T cell lysates were incubated with GST or GST-USP37 proteins. After washing, proteins bound on sepharose were blotted with indicated antibodies. (F) HEK293T cells were transfected with the indicated constructs. Cells were lysed and cell lysates were purified on anti-HA-agarose, and then coimmunoprecipitating endogenous ERK1/2 was detected by anti-ERK1/2 antibody. Source data are available for this figure: SourceData F3.
USP37 deubiquitinates and stabilizes ERK1/2
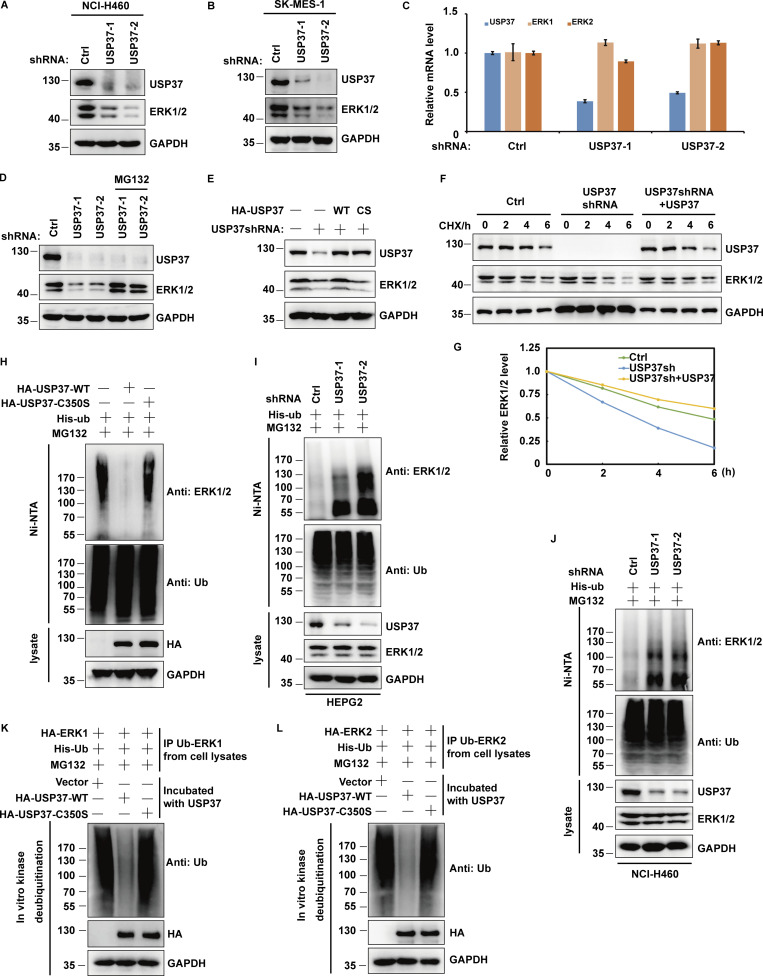
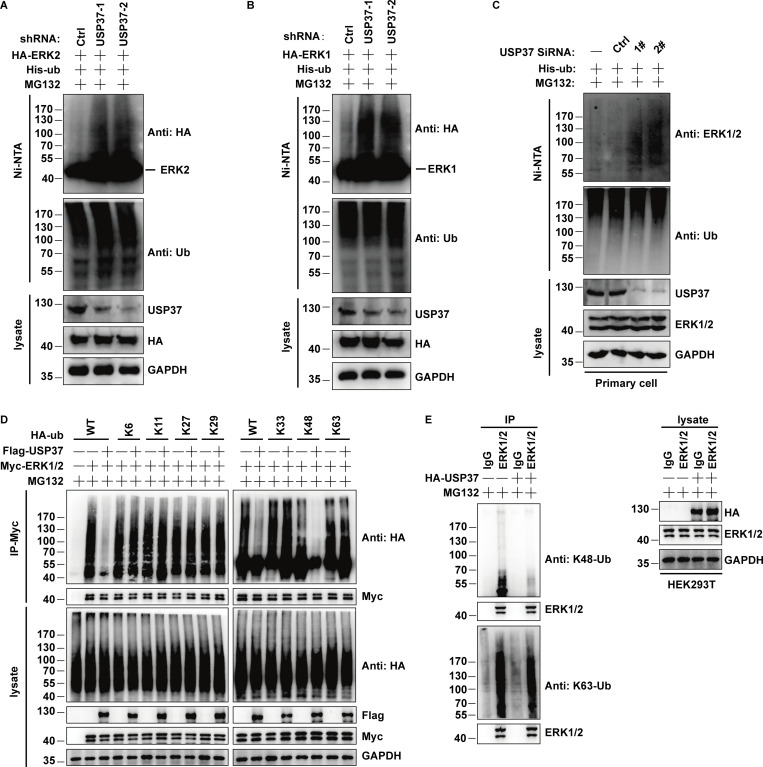
Given that USP37 is a deubiquitinase and interacts with ERK1/2, it is possible that USP37 could deubiquitinate and stabilize ERK1/2. First, depletion of USP37 in NCI-H460 or SK-MES-1 cells was employed to detect the ERK1/2 protein level. As shown in Fig. 4, A–C, the knockdown of USP37 decreased ERK1/2 protein levels but not ERK1/2 mRNA levels. In addition, the decrease in ERK1/2 protein levels could be reversed using the proteasome inhibitor MG132 (Fig. 4 D). Furthermore, ERK1/2 proteins could be rescued by reconstituted shRNA-resistant wild-type USP37 (USP37 WT) but not by the catalytically inactive mutant (USP37 CS) in USP37-depleted cells (Fig. 4 E). To further confirm whether USP37 plays a pivotal role in regulating ERK1/2 stability, we treated cells with cycloheximide (CHX) and then examined the half-life of ERK1/2. ERK1/2 stability was dramatically decreased in USP37-depleted cells, while reconstitution of USP37 recovered ERK1/2 protein stability (Fig. 4, F and G).
Figure 4.
USP37 deubiquitinates and stabilizes ERK1/2. (A) Cells stably expressing control or USP37 shRNAs were harvested. The cells were lysed and Western blot was performed with the anti-ERK1/2, anti-USP37, and anti-GAPDH antibodies. (B) Cells stably expressing control or USP37 shRNAs were collected and then half of the cells were lysed and Western blot was performed with the indicated antibodies. (C) The mRNAs were extracted from the rest of the cells from B and subjected to qRT-PCR. β-actin served as an endogenous control. Data were represented as the means ± SD of three independent experiments. (D) Cells stably expressing control or USP37 shRNAs were treated with vehicle or MG132 for 10 h before harvest. Cells were lysed and then Western blot was performed with the indicated antibodies. (E) Cells stably expressing control or USP37 shRNAs were transfected with the HA-USP37-WT (wild type) or HA-USP37-C350S (catalytically inactive mutant) for 48 h. Cells were lysed and then Western blot was performed with the indicated antibodies. (F) Cells expressing control, USP37 shRNA, USP37 shRNA together with shRNA-resistant USP37 were treated with cycloheximide (CHX) and then harvested at the indicated four-point times (0, 2, 4, and 6 h). The samples were immunoblotted with indicated antibodies. (G) Quantification of the ERK1/2 protein levels relative to GAPDH shown in F using ImageJ software. (H) Cells transfected with indicated constructs were treated with MG132 for 12 h before being harvested. Covalently modified proteins were purified on NiNTA-agarose under denatured conditions and then blotted with indicated antibodies. (I and J) The cell of HEPG2 (C) or NCI-H460 (D) stably expressing control or USP37shRNAs were transfected with His-ub and then were treated with MG132 for 12 h before being harvested. Covalently modified proteins purified on NiNTA-agarose under denatured conditions. Ubiquitinated ERK1/2 was detected by anti-ERK1/2 antibody. (K and L) Deubiquitination of ERK1 or ERK2 in vitro by USP37. Ubiquitinated ERK1/2 protein was incubated with purified USP37 WT or USP37 CS in vitro and then blotted with the indicated antibodies. Source data are available for this figure: SourceData F4.
Next, we examined whether USP37 deubiquitinates ERK1/2. As shown in Fig. 4 H, overexpression of wild-type USP37, but not the USP37 CS mutant, decreased ERK1/2 ubiquitination. On the contrary, the depletion of USP37 resulted in a significant increase in ERK1/2 ubiquitination (Fig. 4, I and J; and Fig. S1, A–C). In addition, USP37 primarily removed the K48-linked poly-Ub chains from ERK1/2 (Fig. S1 D). The result was further confirmed using anti-K48-linkage-specific polyubiquitin antibodies (Fig. S1 E).
Figure S1.
Related to Fig. 4. USP37 removes K48-linked ubiquitin chains of ERK1/2. (A and B) Cells stably expressing control or USP37shRNAs were transfected with HA-ERK2 or HA-ERK1 combined with His-ub and then treated with MG132 for 12 h before being harvested. Covalently modified proteins purified on NiNTA-agarose under denatured conditions. Ubiquitinated ERK1/2 was detected by anti-HA antibody. (C) Primary cells of the basal cells transfected with vector or USP37siRNAs constructs were treated with MG132 (30 μM) for 10 h before harvest. Covalently modified proteins were purified on NiNTA-agarose under denatured conditions and then blotted with indicated antibodies. (D) Ubiquitin chain type on ERK1/2. ERK1/2 expression vectors and indicated HA-tag ubiquitin were transfected into USP37 expression cells. Cell lysates were boiled and immunoprecipitated with anti-HA beads and immunoblotted as indicated. (E) HEK293T cells transfected with the USP37 constructs were treated with MG132 for 10 h before harvest. ERK1/2 was immunoprecipitated with anti-ERK1/2 monoclonal antibodies and immunoblotted with anti-ubiquitin (linkage-specific-k48) antibodies or anti-ubiquitin (linkage-specific-k63) antibodies. Source data are available for this figure: SourceData FS1.
To confirm whether USP37 directly deubiquitinates ERK1/2, an in vitro deubiquitination assay was performed. Fig. 4, K and L shows that wild-type USP37, but not the USP37 CS mutant, deubiquitinated ERK1/2 in vitro. Taken together, these results indicated that USP37 is a deubiquitinase for ERK1/2 and stabilizes ERK1/2.
USP37 regulates cell proliferation and tumor growth through ERK1/2
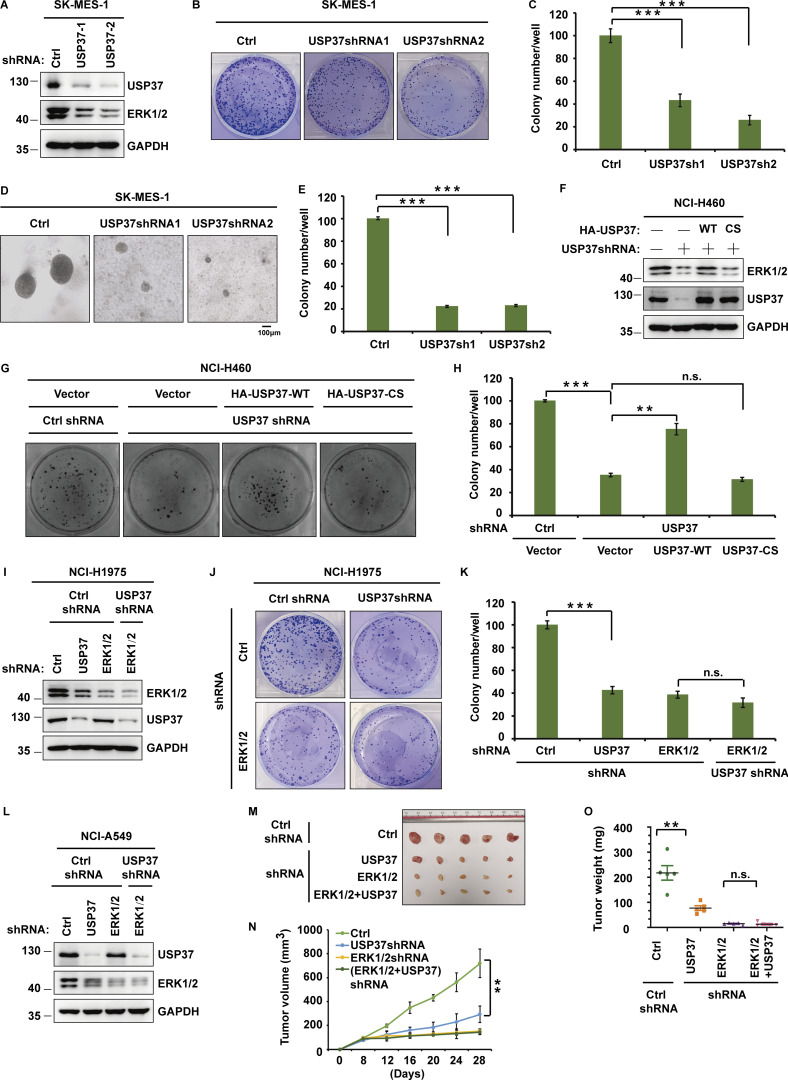
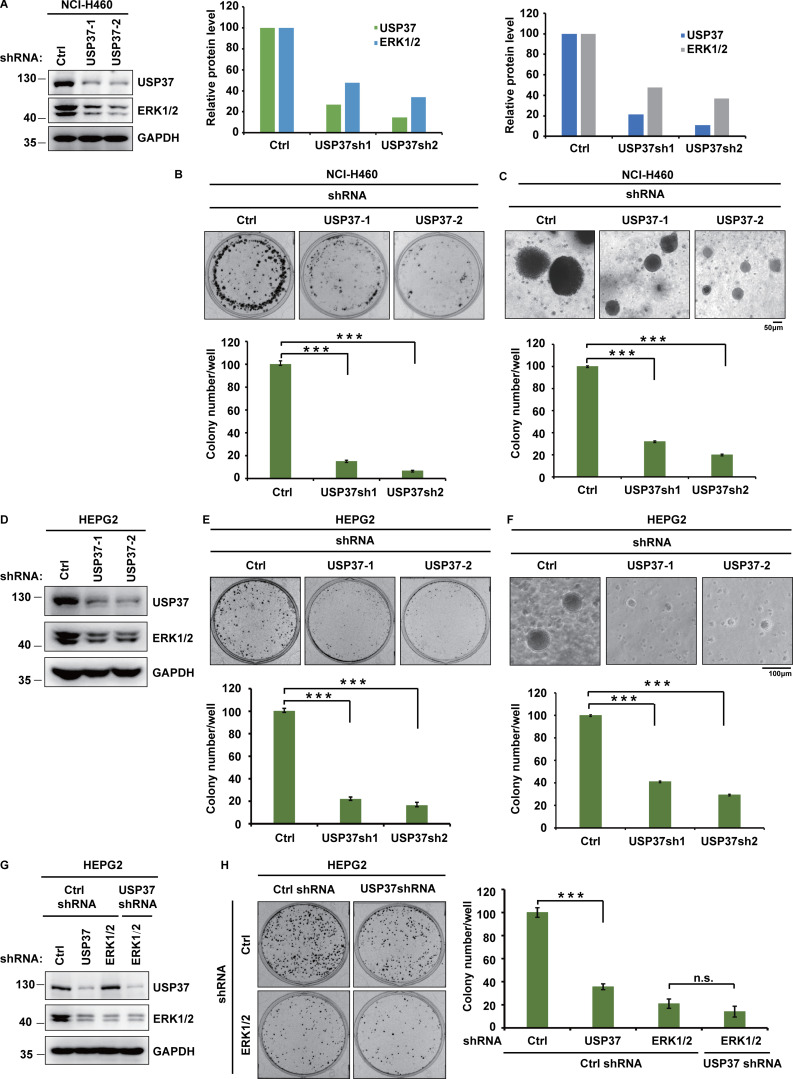
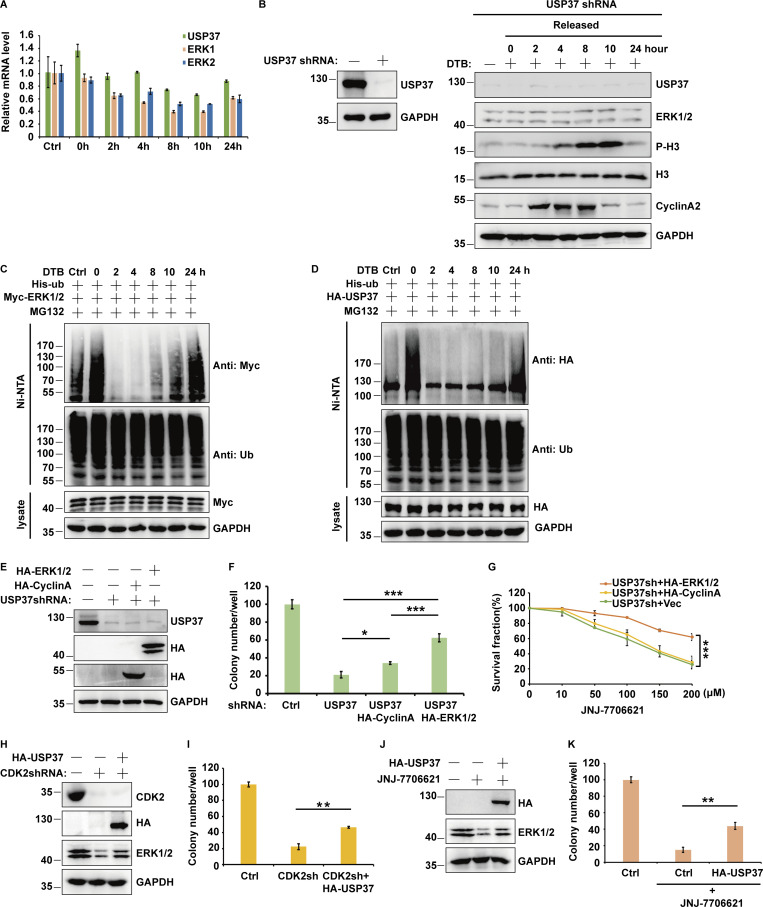
Since the ERK signaling pathway is critical for cancer cell proliferation (Lavoie et al., 2020; Sun et al., 2015), we next studied whether USP37 functions as a cancer-promoting protein by regulating the ERK1/2 pathway. As shown in Fig. 5, A–E and Fig. S2, A–F, depletion of USP37 with two different shRNAs decreased the proliferation of various cancer cells. Furthermore, reconstituting USP37-depleted cells with USP37 WT rescued these phenotypes, but the USP37 CS mutant did not (Fig. 5, F–H), suggesting that the deubiquitinase activity of USP37 is essential for its proliferation ability. In addition, the knockdown of USP37 did not further decrease cell proliferation in ERK1/2-depleted NCI-H1975 cells, indicating that USP37 regulates cancer cell growth and proliferation in an ERK1/2-dependent manner (Fig. 5, I–K; and Fig. S2, G and H). To investigate the biological function of the USP37-ERK1/2 axis in lung cancer cells in vivo, shRNAs were stably transfected into NCI-A549 cells, which were subcutaneously implanted into nude mice and monitored for tumor growth. Mice bearing USP37 shRNA-expressing NCI-A549 cells showed decreased tumor growth (Fig. 5, L and M). 28 d after tumor cell implantation, a threefold decrease in tumor volume (Fig. 5, M and N) and a threefold decrease in tumor weights was observed in tumors formed by USP37-depleted NCI-A549 cells (Fig. 5 O). Moreover, the depletion of USP37 did not further inhibit tumor growth in the ERK1/2 knockdown group (Fig. 5, L–O). These results indicate that USP37 regulates cell proliferation and tumorigenesis in an ERK1/2-dependent manner.
Figure 5.
USP37 regulates cell proliferation and tumor growth through ERK1/2. (A) Cells stably expressing control or USP37 shRNAs were lysed and cell lysates were then blotted with the indicated antibodies. (B) Cells from A were utilized to perform a colony formation assay. (C) Quantitation of the colonies is shown in B. Data were represented as the means ± SD of three independent experiments. Statistical analyses were performed with a two-tailed paired t test. ***P < 0.001 (Control versus USP37sh1), ***P < 0.001 (Control versus USP37sh2). (D) Cells from A were utilized to perform soft agar colony formation assay. Scale bars represent 100 μm. (E) Quantification of the colonies shown in D. Data was represented as the means ± SD of three independent experiments. Statistical analyses were performed with a two-tailed paired t test. ***P < 0.001 (Control versus USP37sh1), ***P < 0.001 (Control versus USP37sh2). (F) Cells stably expressing control or USP37 shRNAs were transfected with the HA-USP37-WT (wild type) or HA-USP37-C350S (catalytically inactive mutant). Cells were lysed and then Western blot was performed with the indicated antibodies. (G) Cells from F were utilized to perform a colony formation assay. (H) Quantitation of the colonies is shown in G. Data were represented as the means ± SD of three independent experiments. Statistical analyses were performed with two-tailed paired t test. **P < 0.01, ***P < 0.001; N.S. stands for no significant change. (I) Cells stably expressing the control shRNA, USP37shRNA, ERK1/2shRNA, or USP37shRNA together with ERK1/2shRNA were lysed, and cell lysates were blotted with the indicated antibodies. (J) Cells from I were utilized to perform a colony formation assay. (K) Quantitation of the colonies is shown in J. Data were represented as the means ± SD of three independent experiments. Statistical analyses were performed with two-tailed paired t test. ***P < 0.001; N.S. stands for no significant change. (L) Cells stably expressing the control shRNA, USP37shRNA, ERK1/2shRNA, or USP37shRNA together with ERK1/2shRNA were lysed, and cell lysates were blotted with the indicated antibodies. (M–O) Cells from L were subcutaneously injected into nude mice (2 × 106 cells). Mice were sacrificed after 4 wk. Tumor images were acquired as shown in M. Tumor volumes (N) and tumor weights (O) were measured. n = 5; data points in N represent mean tumor volume ± SD. Data points in O represent mean tumor weight ± SD. Statistical analyses were performed with a two-tailed unpaired t test. **P <0.01; N.S. stands for no significant change. Source data are available for this figure: SourceData F5.
Figure S2.
Related to Fig. 5. USP37 regulates cell proliferation through ERK1/2. (A) Cells stably expressing as indicated USP37 shRNAs were lysed and cell lysates were then blotted with the indicated antibodies. Light panel: Immunoreactive bands were quantified using ImageJ software. Relative USP37 and ERK1/2 protein levels were quantified with GAPDH from Fig. S2 A as internal standards. Right panel: Immunoreactive bands were quantified using ImageJ software. Relative USP37 and ERK1/2 protein levels were quantified with GAPDH from Fig. 5 A as internal standards. (B) Cells from A were performed colony formation assay as indicated. Upper panel: The representative images of colony formation. Lower panel: Quantitation of the colonies. Data were represented as the means ± SD of three independent experiments. Statistical analyses were performed with two-tailed paired t test. ***P < 0.001 (Control versus USP37sh1), ***P < 0.001 (Control versus USP37sh2). (C) Cells from A were utilized to perform soft agar colony formation assay. Scale bars represent 50 μm. Upper panel: the representative images of colony formation. Lower panel: Quantitation of the colonies. Data was represented as the means ± SD of three independent experiments. Statistical analyses were performed with two-tailed paired t test. ***P < 0.001 (Control versus USP37sh1), ***P < 0.001 (Control versus USP37sh2). (D) Cells stably expressing the control or USP37 shRNAs were lysed and cell lysates were then blotted with the indicated antibodies. (E) Cells from D were utilized to perform colony formation assay as indicated. Upper panel: The representative images of colony formation. Lower panel: Quantitation of the colonies. Data were represented as the means ± SD of three independent experiments. Statistical analyses were performed with two-tailed paired t test. ***P < 0.001 (Control versus USP37sh1), ***P < 0.001 (Control versus USP37sh2). (F) Cells from D were utilized to perform soft agar colony formation assay. Scale bars represent 100 μm. Upper panel: The representative images of colony formation. Lower panel: Quantitation of the colonies. Data were represented as the means ± SD of three independent experiments. Statistical analyses were performed with two-tailed paired t test. ***P < 0.001 (Control versus USP37sh1), ***P < 0.001 (Control versus USP37sh2). (G) Cells stably expressing the control shRNA, USP37 shRNA, ERK1/2 shRNA, or USP37shRNA together with ERK1/2 shRNA were lysed, and cell lysates were blotted with the indicated antibodies. (H) Cells from G were utilized to performed colony formation assay. Left panel: The representative images of colony formation. Right panel: Quantitation of the colonies. Data was represented as the means ± SD of three independent experiments. Statistical analyses were performed with two-tailed paired t test. ***P < 0.001 (Control versus USP37sh), n.s (ERK1/2sh versus ERK1/2sh together with USP37sh). Source data are available for this figure: SourceData FS2.
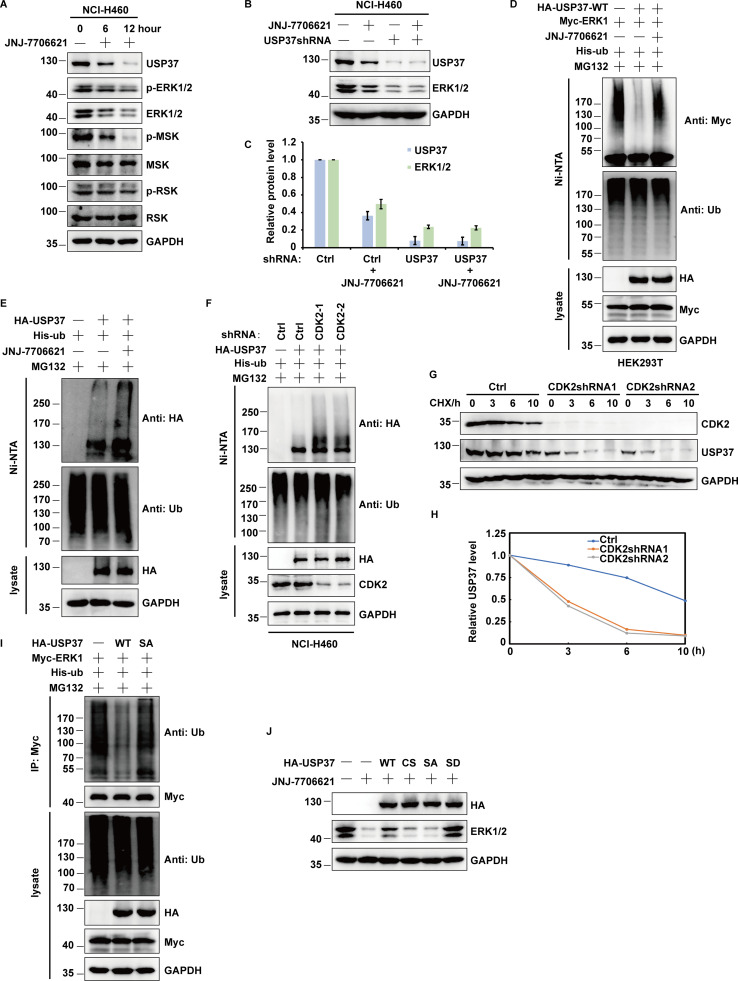
CDK2 regulates ERK1/2 stability by phosphorylating and activating USP37
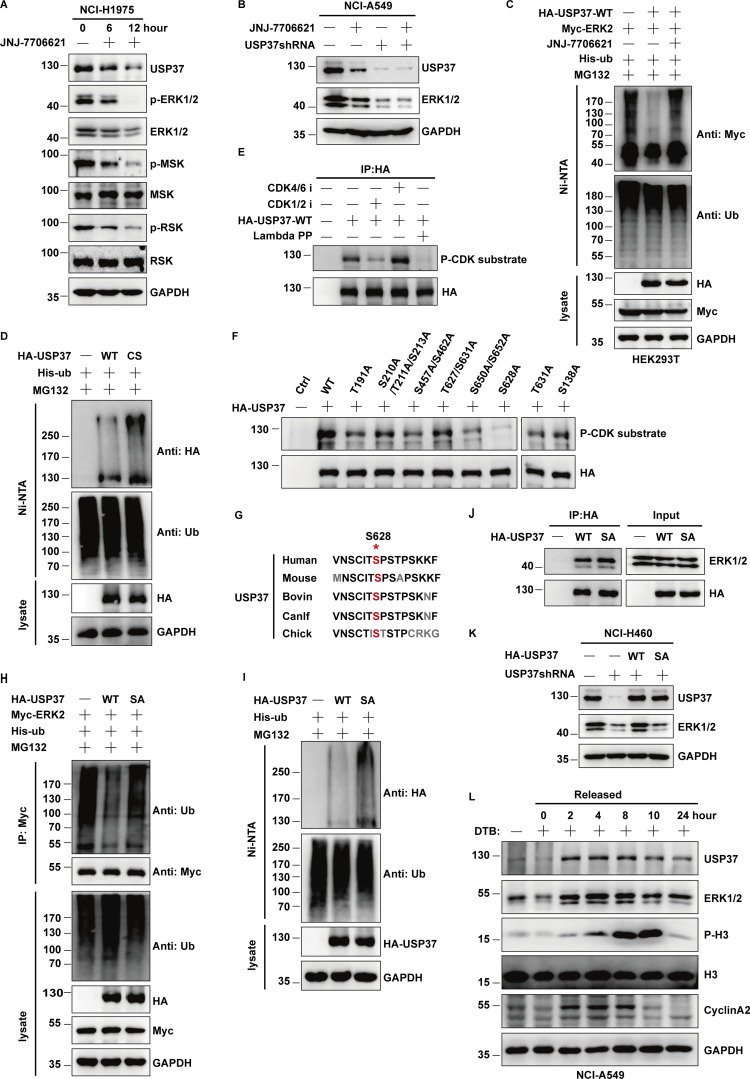
Previous reports reported that USP37 is a substrate of CDK2 (Huang et al., 2011). This suggests that CDK2 regulates ERK1/2 protein stability via USP37. To test this hypothesis, different cancer cells were treated with CDK1/2 inhibitors and the ERK signaling pathway was examined. As shown in Fig. 6 A and Fig. S3 A, CDK1/2 inhibitors significantly decreased ERK1/2 protein levels and attenuated the ERK signaling cascade. Additionally, CDK1/2 inhibitors decreased ERK1/2 protein levels in control cells but not USP37-depleted cells, suggesting that CDK2 may regulate ERK1/2 protein levels through USP37 (Fig. 6 B; and Fig. S3, B and C). Since USP37 could deubiquitinate and stabilize ERK1/2, we next examined whether CDK2 could affect the USP37-mediated deubiquitination of ERK1/2. As shown in Fig. 6 C and Fig. S3 D, overexpression of USP37 resulted in an evident decrease in polyubiquitination of ERK1/2, while CDK1/2 inhibitors reversed this function.
Figure 6.
CDK2 regulates ERK1/2 stability by phosphorylating and activating USP37. (A) Cells were untreated or treated with JNJ-7706621 for three different point times (0, 6, and 12 h) before harvest, and then cell lysates were blotted with the indicated antibodies. (B) Cells stably expressing control shRNA or USP37shRNA were with/without the JNJ-7706621 before harvest, and then cell lysates were blotted with the indicated antibodies. (C) Cells transfected with as indicated constructs were treated with as indicated. Covalently modified proteins purified on NiNTA-agarose under denatured conditions. Ubiquitinated ERK1/2 was detected by anti-Myc antibody. (D) Cells transfected with as indicated plasmids were treated with MG132 for 10 h before being harvested. Covalently modified proteins purified on NiNTA-agarose under denatured conditions. Ubiquitinated USP37 was detected by anti-HA antibody. (E) HA-USP37 WT was transiently transfected into HEK293T cells. These cells were subsequently treated with vehicle, CDK1/2i, or CDK4/6i for 12 h. Cells were lysed and purified using anti–HA-agarose beads. One of the samples was additionally treated with Lambda PPase as indicated. The immunoprecipitates were then blotted with the indicated antibodies. (F) USP37 WT or several USP37 SA mutant plasmids were transfected into HEK293T cells, respectively. Cell lysates were immunoprecipitated with HA beads and immunoblotted as indicated. (G) Sequence alignment of USP37 in various species. *: phosphorylation site. (H) HEK293T cells were transfected with Myc-ERK2 plasmid together with His-ub, and then treated with as indicated. Cell lysates were purified with anti-Myc beads and immunoblotted with indicated antibodies. (I) Cells transfected with the indicated constructs were treated with MG132 for 10 h before being harvested. Covalently modified proteins purified on NiNTA-agarose under denatured conditions. Ubiquitinated USP37 was detected by anti-HA antibody. (J) Cells transfected with the indicated constructs. After transfection 48 h, cells were lysed and purified using anti–HA-agarose beads. The immunoprecipitates were then blotted with the indicated antibodies. (K) Cells stably expressing control shRNA or USP37shRNA were transfected with the indicated constructs were lysed, and cell lysates were blotted with the indicated antibodies. (L) Cells were treated with double thymidine block (DTB) and then released as indicated time points, and then the cell lysates were blotted with the indicated antibodies. Source data are available for this figure: SourceData F6.
Figure S3.
Related to Fig. 6. CDK2 regulates USP37 stability by phosphorylating and activating USP37. (A) Cells were treated with/without JNJ-7706621 for the three different point times (0, 6, and 12 h) before harvest, and then cell lysates were blotted with the indicated antibodies. (B) Cells stably expressing control shRNA or USP37 shRNA were treated with/without JNJ-7706621 before harvest, and then cell lysates were blotted with the indicated antibodies. (C) Immunoreactive bands of at least three independent experiments were quantified using ImageJ software. Relative ERK1/2 and USP37 protein levels were quantified with GAPDH from B as internal standards. Data were represented as means ± SD. (D) Cells transfected with the indicated constructs were treated with/without JNJ-7706621, and then treated with MG132 for 10 h before harvested. Covalently modified proteins purified on NiNTA-agarose under denatured conditions. Ubiquitinated ERK1/2 was detected by anti-Myc antibody. (E) Cells transfected with the indicated constructs were with/without NJ-7706621 and meanwhile treated with MG132 for 10 h before being harvested. Covalently modified proteins were purified on NiNTA-agarose under denatured conditions. Ubiquitinated USP37 was detected by anti-HA antibody. (F) Cells stably expressing control or CDK2shRNAs were transfected with His-ub, and then were treated with MG132 for 12 h before harvested. Covalently modified proteins purified on NiNTA-agarose under denatured conditions. Ubiquitinated USP37 was detected by anti-HA antibody. (G) Cells expressing Control, CDK2shRNA1, and CDK2shRNA2 were treated with cycloheximide (CHX) and then harvested at the indicated four-point times (0, 3, 6, and 10 h). The samples were immunoblotted with indicated antibodies. (H) Quantification of the USP37 protein levels relative to GAPDH shown in G using ImageJ software. (I) HEK293T cells were transfected with Myc-ERK1 plasmid together with His-ub, and then treated with as indicated. Cell lysates were purified with anti-Myc beads and immunoblotted with indicated antibodies. (J) Cells were transfected with the indicated constructs and treated with JNJ-7706621 before harvest, and cell lysates were blotted with the indicated antibodies. Source data are available for this figure: SourceData FS3.
Intriguingly, we found that CDK1/2i treatment also markedly decreased USP37 protein levels but led to a significant increase in USP37 polyubiquitination (Fig. 6, A and B; and Fig. S3, A–C and E). Furthermore, the knockdown of CDK2 significantly increased USP37 polyubiquitination (Fig. S3 F). In addition, a CHX chasing assay showed that the depletion of CDK2 dramatically decreased USP37 stability (Fig. S3, G and H), suggesting that CDK1/2 might regulate USP37 protein levels and prevent its degradation. Moreover, a previous study showed that USP37 could deubiquitinate and stabilize itself (Huang et al., 2011). Hence, we hypothesized that CDK1/2 might regulate ERK1/2 and USP37 protein levels by governing USP37 activity. Next, we examined whether USP37 could deubiquitinate itself. As shown in Fig. 6 D, compared with cells expressing USP37 WT, more ubiquitinated USP37 was detected in cells expressing the USP37 CS mutant. In addition, our results coincided with a previous study that found USP37 to be a substrate of CDK kinases (Huang et al., 2011; Fig. 6 E). CDK1/2 inhibitors, but not CDK4/6 inhibitors, inhibited USP37 phosphorylation (Fig. 6 E).
Next, several candidate sites from the PhoshpoSite Plus database (https://www.phosphosite.org/homeAction.action) were examined along with a previous study (Huang et al., 2011), which conformed the CDK substrate motif (SP/TP sites). As shown in Fig. 6 F, the S628A mutant of USP37 markedly decreased phospho-CDK substrate signaling, suggesting that S628 is a major site for CDK1/2 mediation of USP37 phosphorylation. Furthermore, the S628 site of USP37 is highly conserved across different species (Fig. 6 G). Compared with USP37 wild type, the USP37 SA (S628A) mutant failed to deubiquitinate ERK1/2 and USP37 itself (Fig. 6, H and I; and Fig. S3 I), which may explain why CDK1/2i also decreased USP37 protein levels (Fig. 6, A and B; and Fig. S3, A–C). However, USP37 phosphorylation did not affect its binding to ERK1/2 (Fig. 6 J). Additionally, complemented USP37 WT, but not the USP37 SA mutant, restored the ERK1/2 protein level in USP37-depleted cells (Fig. 6 K). Moreover, overexpression of USP37 WT or USP37 SD, but not the USP37 CS or USP37 SA mutants, restored the ERK1/2 protein level in cells treated with CDK1/2i (Fig. S3 J).
The kinase activity of CDK2 is cell cycle-dependent and is critical for G1/S transition and cell cycle progression (Hamada et al., 1996; Hoffmann et al., 1994). Therefore, cells released from double thymidine block (DTB) treatment were employed to examine USP37 and ERK1/2 expression at different cell cycle phases. As shown in Fig. 6 L and Fig. S4 A, there was a high correlation between the USP37 and ERK1/2 protein levels but not at the mRNA level during cell cycle progression. However, depletion of USP37 abolished the differential expression of ERK1/2 in different phases of the cell cycle (Fig. S4 B), suggesting that the cell cycle regulates ERK1/2 expression in a USP37-dependent manner. In addition, high polyubiquitination of ERK1/2 or USP37 was correlated with low ERK1/2 or USP37 protein levels during cell cycle progression (Fig. 6 L; and Fig. S4, C and D). These findings suggest that the CDK2-mediated phosphorylation of USP37 exerted a vital function in regulating USP37 DUB activity and the USP37-mediated deubiquitination and stabilization of ERK1/2.
Figure S4.
Related to Fig. 6. CDK2 through USP37 regulates cell growth. (A) The mRNAs were extracted from the rest of the cells from Fig. 6 L, and then subjected to qRT–PCR. β-actin served as an endogenous control. Data was represented as the means ± SD of three independent experiments. (B) Cells stably expressing USP37shRNAs were treated with double thymidine block (DTB) and then released as indicated time points, and then cell lysates were blotted with the indicated antibodies. (C and D) Cell were transfected with the indicated constructs. Cells were treated with double thymidine block (DTB) and then released as indicated time points, and then were treated with MG132 for 12 h before harvested. Covalently modified proteins purified on NiNTA-agarose under denatured conditions. Ubiquitinated ERK1/2 (C) or USP37 (D) was detected by anti-Myc antibody or anti-HA antibody. (E) Cells stably expressing control shRNA or USP37shRNA were transfected with the indicated constructs were lysed, and cell lysates were blotted with the indicated antibodies. (F and G) Cells from E were utilized to perform colony formation (F) or CCK8 assays (G) as indicated. Data were represented as the means ± SD of three independent experiments. Statistical analyses were performed with two-tailed paired t test. *P < 0.05, **P < 0.01, ***P < 0.001. (H) Cells stably expressing control shRNA or CDK2shRNA were transfected with the indicated constructs were lysed, and cell lysates were blotted with the indicated antibodies. (I) Cells from H were utilized to perform colony formation as indicated. Data were represented as the means ± SD of three independent experiments. Statistical analyses were performed with two-tailed paired t test. **P < 0.01. (J) Cells stably expressing control or USP37 and treated with JNJ7706621 were lysed and cell lysates were blotted with the indicated antibodies. (K) Cells from J were utilized to perform colony formation as indicated. Data were represented as the means ± SD of three independent experiments. Statistical analyses were performed with two-tailed paired t test. **P < 0.01. Source data are available for this figure: SourceData FS4.
It has been reported that USP37 is also a deubiquitinase for cyclin A, which regulates G1/S transition and cell cycle progression (Huang et al., 2011). Hence, USP37 may regulate cell proliferation via both ERK1/2 and cyclin A. As shown in Fig. S4, E and F, overexpression of ERK1/2 restored cell growth in USP37-depleted cells, while overexpression of cyclin A had mild rescue efficiency. Moreover, we found that overexpression of ERK1/2 but not Cyclin A in USP37-shRNA-depleted cancer cells led to cells’ resistance to the CDK1/2 inhibitor (Fig. S4 G). In addition, reconstitution of USP37 partially rescued ERK1/2 protein levels or tumor cell growth in either CDK2-knockdown or CDK1/2i-treated cells (Fig. S4, H–K).
USP37 positively correlates with ERK1/2 in clinical cancer samples
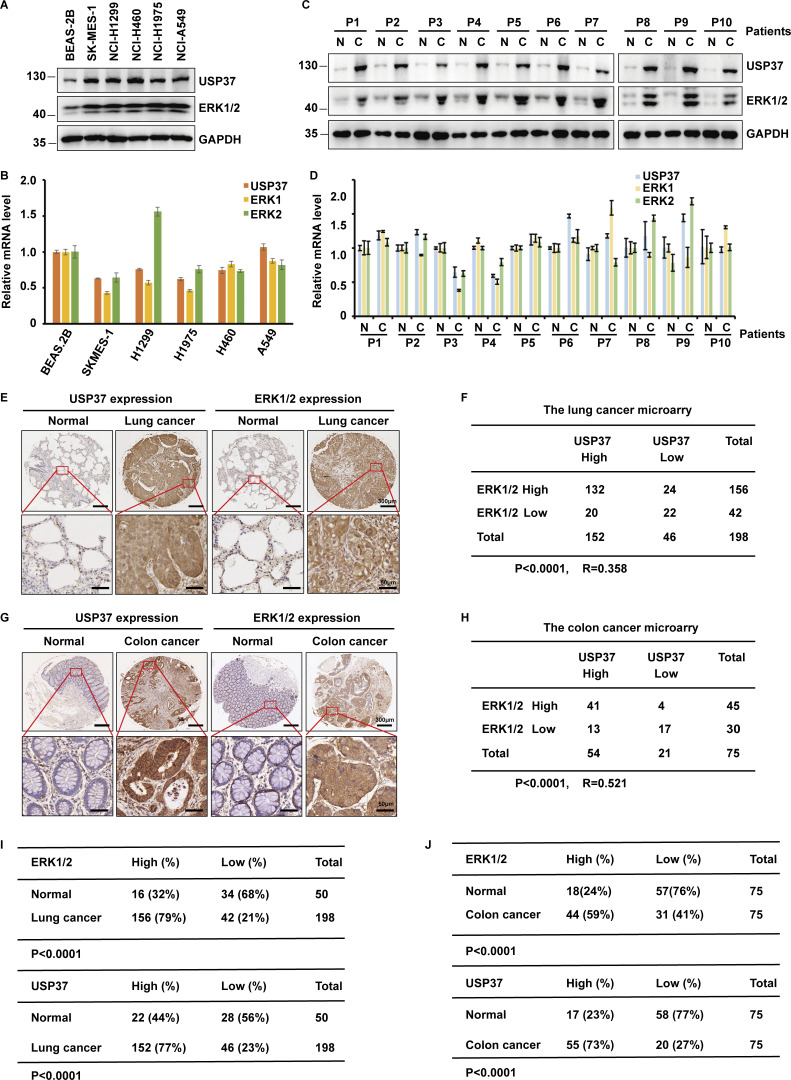
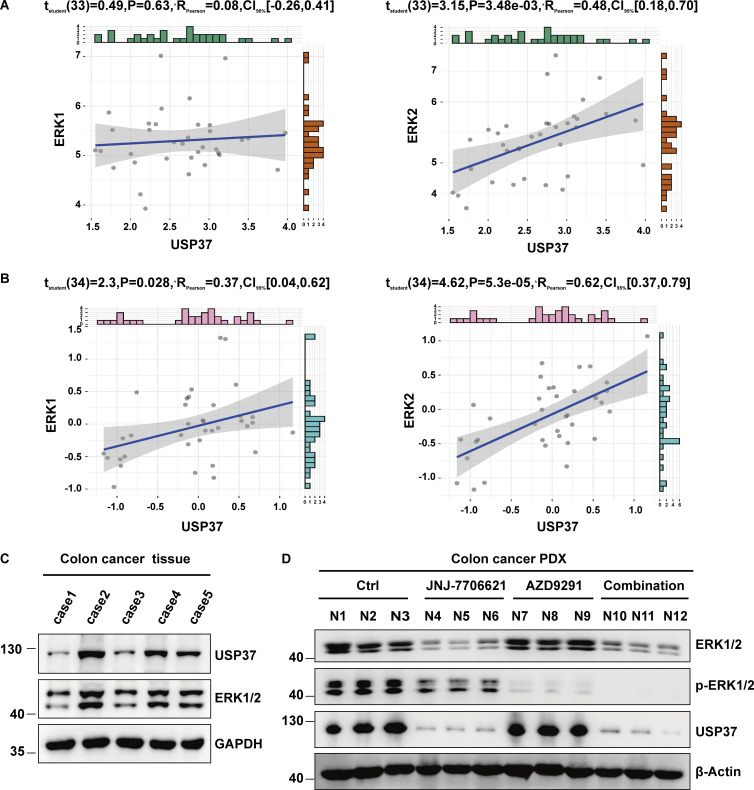
ERK1/2 has been reported as tumor-promoting proteins that are overactivated or excessively expressed in various cancers (Samatar and Poulikakos, 2014). However, the mechanism of ERK1/2 upregulation in cancers remains unknown. We first examined the expression of USP37 and ERK1/2 in multiple lung cancer cell lines and lung cancer tissues. As shown in Fig. 7, A–D, protein levels but not mRNA levels of ERK1/2 and USP37 were highly correlated and much higher in lung cancer cell lines and lung cancer tissues. Furthermore, we performed immunohistochemical staining of ERK1/2 and USP37 using the lung cancer and colon cancer microarrays. As shown in Fig. 7, E–J, upregulations of USP37 and ERK1/2 were observed in cancer samples. In addition, the expressions of USP37 and ERK1/2 positively correlated in lung and colon cancer samples (Fig. 7, I and J). Given that the transcriptomics and proteomics data of cancer cell lines has been published (Nusinow et al., 2020), data from Cancer Cell Line Encyclopedia (CCLE; https://sites.broadinstitute.org/ccle/) were utilized to analyze the correlation between USP37 and ERK1/2 at the mRNA or protein level. Fig. S5, A and B shows a higher correlation between USP37 and ERK1/2 in protein than mRNA expression in cancer cell lines. Taken together, our results suggest that USP37 and ERK1/2 expressions are highly correlated in lung and colon cancers.
Figure 7.
USP37 is positively correlated to ERK1/2 in clinical cancer samples. (A) Cell lysates from several lung cancer cell lines were blotted with USP37 and ERK1/2 antibodies. Lysates from normal lung epithelial cell line BEAS-2B were used as a control. (B) The mRNAs were extracted from the rest of the cancer cells from A and subjected to qRT-PCR. β-actin served as an endogenous control. Data was represented as the means ± SD of three independent experiments. (C) Lung cancer tissue and corresponding normal lung tissues were blotted with USP37 and ERK1/2 antibodies. Lysates from corresponding normal lung tissues were used as a control. (D) The mRNAs were extracted from the rest of the tissue from C and subjected to qRT-PCR. β-actin served as an endogenous control. Data was represented as the means ± SD of three independent experiments. (E) Representative images of immunohistochemical staining of USP37 and ERK1/2 in normal and lung cancer tissue microarray. Scale bars, 300 or 50 μm. (F) Correlation study of ERK1/2 and USP37 in lung cancer. Statistical analyses were performed with χ2 test. R, Pearson correlation coefficient. (G) Representative images of immunohistochemical staining of USP37 and ERK1/2 in normal and colon cancer. Scale bars, 300 or 50 μm. (H) Correlation study of ERK1/2 and USP37 in colon tissue microarray. Statistical analysis was performed with χ2 test. R, Pearson correlation coefficient. (I) Quantification of ERK1/2 and USP37 protein levels in normal and lung cancer tissue microarray. Statistical analyses were performed with χ2 test. (J) Quantification of ERK1/2 and USP37 protein levels in normal and colon tissue microarray. Statistical analyses were performed with χ2 test. Source data are available for this figure: SourceData F7.
Figure S5.
Related to Figs. 7 and 8. Combined CDK1/2 and EGFR inhibitors have a synergetic anticancer effect in vivo. (A and B) The date of gene expression from the CCLE database or proteomics from https://depmap.org. The cell lines were classified according to the site of the disease. The correlation between ERK1 or ERK2 with USP37 was analyzed in isotype cell line respectively. P < 0.05 was considered statistically significant. Pearson correlation coefficient was used to represent the size of the correlation. (C) Tissue derived from five colon cancer patients was examined for ERK1/2 and USP37 protein levels by Western blot. The highest ERK1/2 colon cancer tissue among these five colon cancer tissues was employed to construct xenograft (PDX) models derived from patients. (D) ERK1/2, p-ERK1/2, and USP37 protein levels from the colon cancer PDX xenografts (Fig. 8 F) were examined by Western blot. Source data are available for this figure: SourceData FS5.
Combined CDK1/2 and EGFR inhibitors have a synergetic anticancer effect in vivo
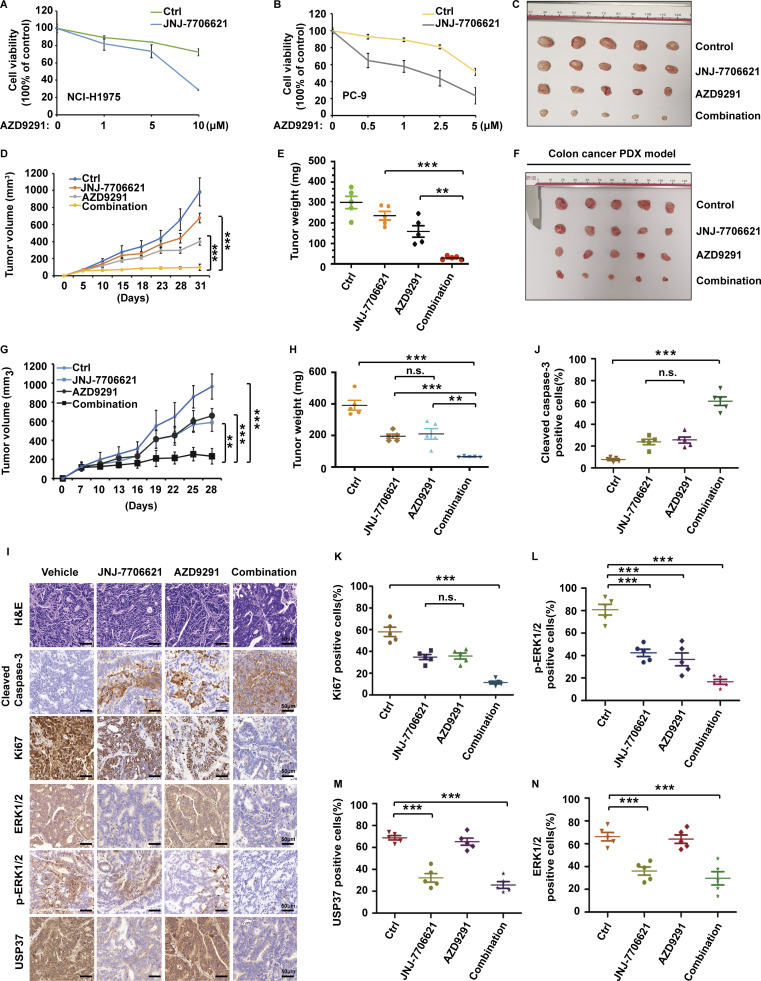
CDK2 upregulates ERK1/2 stability through USP37. Meanwhile, EGFR inhibitors could decrease ERK1/2 activity. We hypothesized that a combination of CDK2 and EGFR inhibition might obtain a synergetic anticancer effect through the downregulation of ERK1/2 stability and activity. CDK1/2 and EGFR inhibitors were examined to determine if their combination produced synergistic killing effects on cancer cells with EGFR mutations. As shown in Fig. 8, A and B, CDK1/2 inhibitor led cancer cells with EGFR mutants to more sensitize to EGFR inhibition in vitro. To further confirm this synergistic cell-killing effects in vivo, we performed a xenograft assay using the NCI-H1975 cell line, which carries the EGFR L858R mutant. As shown in Fig. 8, C–E, treatment with a CDK1/2 inhibitor or EGFR inhibitor alone showed slight growth inhibition; however, combination treatment dramatically suppressed the tumor growth (Fig. 8, C–E).
Figure 8.
Combined CDK1/2 and EGFR inhibitors have a synergetic anti-cancer effect in vivo. (A and B) NCI-H1975 (A) or PC-9 (B) cells were utilized to performed CCK-8 assays as indicated. Data were represented as the means ± SD of three independent experiments. (C–E) JNJ-7706621 at 10 mg/kg (twice/week), AZD9291 at 2 mg/kg (twice/week), or the combination was administered twice a week via intraperitoneal injection to Nu/Nu mice bearing the NCI-H1975 cell line xenografts (n = 5/group). Combination treatment led to a statistically significant decrease in tumor growth compared with either single-agent treatment. Mice were sacrificed after 4 wk. Tumor images were acquired as shown in C. Tumor volumes (D) and tumor weights (E) were measured. Data points in D represent mean tumor volume ± SD, n = 5. Data points in E represent mean tumor weight ± SD, n = 5. Statistical analyses were performed with two-tailed paired t test. **P < 0.01; N.S. stands for no significant change. (F–H) Tumor growth assay of colon PDX xenografts in Nu/Nu mice treated with vehicle, JNJ-7706621 (10 mg/kg, twice/week), AZD9291 (2 mg/kg, twice/week), or the combination for the models in vivo. Mice were sacrificed after 4 wk. Tumor images were acquired as shown in F. Tumor volumes (G) and tumor weights (H) were measured. Data points in G represent mean tumor volume ± SD, n = 5. Data points in H represent mean tumor weight ± SD, n = 5. Statistical analyses were performed with two-tailed paired t test. **P < 0.01, ***P < 0.01; N.S. stands for no significant change. (I–N) Representative images of H&E, Cleaved caspase3, Ki67, USP37, ERK1/2, and p-ERK1/2 staining using colon PDX tissues from I. Scale bars, 50 μm. (J–N) Quantification of cleaved caspase3 (J), Ki67 (K), p-ERK1/2 (L), USP37 (M), and ERK1/2 (N). Statistical analyses were performed with two-tailed paired t test. **P < 0.01, ***P < 0.01; N.S. stands for no significant change.
Next, we examined USP37 and ERK1/2 expression in several colon cancer tissues. As shown in Fig. S5 C shows a good correlation between USP37 and ERK1/2 expression in these colon cancer tissues, and the case2 sample showed higher USP37 and ERK1/2 expression, which was utilized to establish a patient-derived xenograft (PDX) model. As shown in Fig. 8, F–N and Fig. S5 D, CDK1/2 or EGFR inhibitors alone resulted in a slight inhibition in tumor growth and phosphor-ERK1/2. However, combination treatment significantly decreased tumor growth and phosphor-ERK1/2 and promoted cancer cell apoptosis (Fig. 8, F–N and Fig. S5 D).
These results suggest that CDK1/2 and EGFR inhibitors possess synergetic anticancer effects through the downregulation of ERK1/2 stability and activity. Dual targeting of both ERK1/2 stability and activity killed cancer cells more efficiently, even in lower doses of these two inhibitors, which could reduce the chemotherapeutic side effects, and indicates a potential new combination strategy for clinical cancer therapy.
Discussion
The EGFR–RAS–ERK pathway is one of the most important signaling cascades promoting cell survival, growth, and proliferation (Chou et al., 2010). Overactivation of this pathway is widely observed in various cancers (Aoki et al., 2011; Lavoie et al., 2020). Additionally, cell cycle–dependent kinases (CDKs) and their partner cyclins are other vital modulators involved in cell survival and proliferation (Ding et al., 2020). The aberrances of CDKs, especially CDK1/2/4/6, are highly implicated in tumorigenesis, and targeting CDK proteins is a promising strategy for cancer therapy (Otto and Sicinski, 2017; Susanti and Tjahjono, 2021; Zhang et al., 2021b, 2021c). Even though CDK kinases and the EGFR–RAS–ERK signaling cascade are two important regulators controlling cell growth, the relationship between CDKs and the EGFR–RAS–ERK signaling pathway is unclear. Overactive EGFR due to frequent mutations or overexpression is observed in various cancers, including lung cancer, colon cancer, and pancreatic cancer (Normanno et al., 2006). Several EGFR inhibitors have a great effect on inhibiting tumor growth and benefit patients with EGFR-overactivated cancers by downregulating the RAS–ERK signaling pathway (He et al., 2021; Huang and Fu, 2015). However, like other chemotherapeutic drugs, chemoresistance is the major cause of EGFR inhibitor therapy failure (Chong and Jänne, 2013; Misale et al., 2014). Reactivation of ERK1/2 has been reported to be one of the main factors resulting in chemoresistance in EGFR inhibitor therapies (Aoki et al., 2011; Ercan et al., 2012; Huang and Fu, 2015). Hence, it is necessary to clarify the ERK1/2 regulation further.
This study found that CDK2 is a novel modulator of ERK1/2 and that USP37, a deubiquitinase, is the link between CDK2 and the ERK signaling cascade. Our study resulted in a model of the phosphorylation-deubiquitination cascade referred to as the CDK2–USP37 axis that regulates the deubiquitination and stabilization of ERK1/2. Mechanistically, depletion of CDK2 or treatment with CDK1/2 inhibitors decreased ERK1/2 protein levels through USP37, which deubiquitinates and stabilizes ERK1/2. Meanwhile, USP37 is phosphorylated and activated by CDK2, promoting the removal of polyubiquitin chains from ERK1/2 and resulting in the stabilization of ERK1/2.
USP37 promotes cancer cell growth via ERK1/2 in vitro and in vivo. Importantly, the combination of CDK1/2 and EGFR inhibitors results in a synergetic anticancer effect through the downregulation of ERK1/2 stability and activity. Our findings suggest that a chemotherapeutic “double strike” theory is necessary to kill cancer cells. Mechanistically, EGFR inhibitor treatment downregulates the RAS–ERK signaling cascade by inhibiting the phosphorylation of key pathway components. However, CDK1/2 inhibitors hindered the RAS–ERK signaling cascade by destabilizing ERK1/2 protein levels. Hence, ERK signaling is extremely downregulated by the synergistic effects of these dual strikes. In our PDX models shown in Fig. 8, F–N, we used a lower dose of EGFR inhibitors (AZD9291) and CDK1/2 inhibitors (JNJ-7706621) compared with previous reports. However, CDK1/2 or EGFR inhibitors alone showed slight inhibition in colon cancer PDX growth, while combined low doses of CDK1/2 and EGFR inhibitors significantly decreased colon cancer PDX proliferation. These results suggested that dual strikes for targeting both ERK1/2 stability and activity can kill cancer cells more efficiently even in lower doses, which could reduce the chemotherapeutic side effects associated with larger doses. Thus, this method is a potential strategy for clinical cancer therapy. More importantly, MAPK signaling pathway-targeting therapies, including EGFR, KRAS, BRAF, and MEK inhibitors, directly or indirectly target ERK1/2 phosphorylation and activation, while CDK1/2 inhibitors can directly target ERK1/2 on the protein level. However, reactivation of ERK1/2 by an alternative compensation pathway is a major cause of chemoresistance after EGFR, KRAS, BRAF, and MEK inhibitor treatments (Fedele et al., 2018; Morris et al., 2013; Sanchez et al., 2019; Tadesse et al., 2020; Yu et al., 2015; Zhang et al., 2021a). Meanwhile, CDK1/2 inhibitors may be a potential choice for recurring cancers by downregulating ERK1/2 protein levels.
Materials and methods
Cell culture
Human lung cancer cells (NCI-A549, NCI-H460, SK-MES-1, NCI-H1299, and NCI-H1975), normal lung cells (BEAS-2B), and breast cancer cells (MBA-MD-231 and MCF-7) were purchased from the American Type Culture Collection (ATCC). Human colon cancer cells (HCT-116, SW620, SW480, and RKO), cervical cancer cells (Hela), liver cancer cells (HEPG2), and HEK293T cells were also purchased from ATCC. SW480 was cultured in L-15 supplemented with 10% fetal bovine serum (FBS). Human lung cancer cells were cultured in RPMI-1640 supplement with 10% FBS. Unless otherwise stated, all other cells were maintained in DMEM supplemented with FBS.
Plasmids, reagents, and antibodies
DUB plasmids were purchased from Addgene. HA-FLAG-USP37 was subcloned into PLVX3 or pGEX-4T-2 vectors (Clontech). USP37 T191A, S210A/T211A/S213A, S457A/S462A, T627/S613A, S650A/S652A, S628A, T631A, S138A, and C350S mutants were established by a two-step mutation method. All the vectors were confirmed using DNA sequencing.
MG132, cycloheximide (CHX), puromycin, polybrene, streptavidin-linked agarose, anti-FLAG M2 agarose, and anti-HA M2 agarose were from Sigma-Aldrich. CDK1/2 inhibitors (JNJ-7706621), EGFR inhibitors (AZD9291), and CDK4/6 inhibitors (PD 0332991) were purchased from Selleck.
The following antibodies were used in this research: anti-GAPDH (60004-1-lg), anti-β-actin (66009-1-Ig; Proteintech), anti-USP37 (18465-I-AP; Proteintech), anti-p44/42 MAPK (ERK1/2; 9102S; CST), anti-phospho-p44/42 MAPK (ERK1/2 and Thr202/Tyr204; 9101S; CST), anti-CDK1 (19532-I-AP; Proteintech), anti-CDK2 (10112-I-AP; Proteintech), anti-Histone-H3 (17168-I-AP; Proteintech), anti-phosphor-Histone-H3 (ab5176; Abcam), anti-cyclinA2 (67955S; CST), anti-P-CDK substrate (9477S; CST), anti-RSK (9333S; CST), anti-MSK (3489S; CST), anti-Phospho-p90RSK (Ser380; 9341S; CST), anti-Phospho-MSK1 (Thr581; 9595S; CST), anti-HA-tag (3724S; CST), anti-Myc (2276S; CST), anti-ub (P4D1; Santa Cruz Biotechnology), anti-K63-linkage Specific Polyubiquitinatin (12930S; CST), anti-K48-linkage Specific Polyubiquitinatin (ab140601; Abcam), anti-Cleaved-caspase3 (9664S; CST), and anti-Ki67 (34330SF; CST). All antibodies were obtained from formal commercial channels.
Lentivirus packaging and infection
USP37 short hairpin RNAs (shRNAs), ERK1shRNAs, ERK2shRNAs, CDK1shRNAs, and CDK2shRNAs were purchased from miaoling bio website (http://www.miaolingbio.com/):
USP37 shRNA #1: 5′-GGAGTACACAGAAGCTGAAGC-3′
USP37 shRNA #2: 5′-GGAGATAAGTAAGAGAGATGC-3′
ERK1 shRNA #1: 5′-TCATCGGCATCCGAGACATTC-3′
ERK2 shRNA #1: 5′-GCACCAACCATCGAGCAAATG-3′
CDK1 shRNA #1: 5′-GGATGTGCTTATGCAGGATTC-3′
CDK1 shRNA #2: 5′-GGGTCAGCTCGTTACTCAACT-3′
CDK1 shRNA #3: 5′-GGTCAGTACATGGATTCTTCA-3′
CDK2 shRNA #1: 5′-GGCCAGGAGTTACTTCTATGC-3′
CDK2 shRNA #2: 5′-CTCCTGGGCTGCAAATATTAT-3′
USP37siRNA#1: 5′-AAGCGTGGTTTACTTACAA-3′
USP37siRNA#2: 5′-GGAGTGCACATATGGCAAT-3′.
Plko.1 lentiviral or plvx3 constructs and packaging plasmids (pMD2G and pSPAX2) were used to package the virus into HEK293T cells by using Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific). Viral supernatant was collected twice (24 and 48 h) after the cotransfection of lentiviral vectors and packaging plasmids (pMD2G and pSPAX2). Various cancer or HEK293T cells were infected with viral supernatant with the addition of polybrene (8 μg/ml; Sigma-Aldrich), which enhances infection efficiency. Stable cells were selected with media containing puromycin (2 μg/ml; Sigma-Aldrich), which was screened in 5% culture at 37°C for about 7–14 d. The stable cell lines were determined by immunoblotting.
RNA extraction, reverse transcription, and real-time PCR
RNA samples were extracted with RNAiso Plus reagent (Takara), and reverse transcription was performed according to the manufacturer’s instructions. Real-time polymerase chain reaction (PCR) was performed using Power SYBR Green PCR Master Mix (Takara). Each data point was repeated three times with independent samples and normalized with β-actin gene as the control.
The 2−ΔΔCt method was used for quantification of gene expression.
USP37 sequence (5′->3′) Forward primer: 5′-GGAACACTCTTCTGGTGGCA-3′; Reverse primer: 5′-GTGAGAGGAAAGGGGCACTC-3′.
ERK1 sequence (5′->3′) Forward primer: 5′-GGCCCGAAACTACCTACAGT-3′; Reverse primer: 5′-TAGGTAGGTCATCCAGCTCCA-3′.
ERK2 sequence (5′->3′) Forward primer: 5′-CCAGAGAACCCTGAGGGAGA-3′; Reverse primer: 5′-TGTTGAGCAGCAGGTTGGAA-3′.
β-actin sequence (5′->3′) Forward primer: 5′-CATCCTGCGTCTGGACCT-3′; Reverse primer: 5′-GTACTTGCGCTCAGGAGGAG-3′;
CDK2 sequence (5′->3′) Forward primer: 5′-CCAGGAGTTACTTCTATGCCTGA-3′; Reverse primer: 5′-TTCATCCAGGGGAGGTACAAC-3′.
Coimmunoprecipitation and Western blotting
For transient transfection and coimmunoprecipitation assays, constructs encoding HA-tagged ERK1 or HA-tagged ERK2 were transiently cotransfected into HEK293T cells for 48 h before harvest. The transfected cells were lysed with NETN buffer (20 mM Tris–HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, and 0.5% Nonidet P-40) containing 1× protease inhibitors cocktail (Roche) on ice for 30 min. After the removal of cell debris by centrifugation at 12,000 rpm for 15 min, the soluble fractions were collected and incubated with HA beads for 4 h at 4°C. Beads were washed five times with NETN buffer and boiled in 1× SDS loading buffer for 10 min. Then, the proteins isolated were subjected to SDS-PAGE gels for electrophoresis separation. The separated proteins were transferred to 0.45 μm PVDF membrane (Thermo Fisher Scientific). The membrane was blocked by 5% non-fat milk in TBST buffer (TBST with 0.1% Tween 20) for 30 min at room temperature and washed three times, for 10 min each, with TBST. Then the membrane was incubated with the primary antibodies at 4°C overnight. The membrane was washed three times with TBST, incubated with the anti-rabbit IgG secondary antibody (Proteintech) or anti-mouse IgG secondary (SA00001-2; Proteintech) at room temperature for 1 h, and after washing, visualized under a chemiluminescence imaging system (5200CE; Tanon) by an enhanced chemiluminescence (ECL) detection kit (89880; Thermo Fisher Scientific). Finally, bands of Western blot were quantified by using ImageJ 1.53a (1.8.0_172; Java) software.
Protein stability assay
To examine ERK1/2 or USP37 protein stability, cycloheximide (CHX, 0.1 mg/ml) was added to the cell culture medium and cells were harvested at the indicated times. The harvested cells were lysed in NETN buffer (20 mM Tris–HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, and 0.5% Nonidet P-40) containing 1× protease inhibitors cocktail (Roche) on ice for 30 min. After the removal of cell debris by centrifugation at 12,000 rpm for 15 min, the soluble fractions were boiled in reducing SDS sample buffer. Then, the lyses were analyzed by Western blot with antibodies. Finally, quantification of the ERK1/2 or USP37 protein levels relative to GAPDH was done using ImageJ 1.53a (1.8.0_172; Java) software.
Deubiquitination of ERK1/2 in vivo and in vitro
For the in vivo ubiquitination assay, HEK293T cells were transfected with HA-USP37 wild-type (WT) or mutant Cys 350 to Ser (CS) and His-ub. After 2 d, cells were treated with MG132 (30 μM) for 12 h before harvesting. Cells were lysed in a buffer containing 8 M Urea, 0.1 M NaH2PO4, 300 mM NaCl, and 0.01 M Tris (pH 8.0). Lysates were incubated with Ni-NTA agarose beads (Sigma-Aldrich) for 1–2 h at room temperature. Beads were washed five times with lysis buffer. Input samples and beads were boiled in loading buffers and analyzed using SDS-PAGE and immunoblotting.
For the in vitro deubiquitination assay, HA-ERK2 was coexpressed with His-ub in HEK293T cells. After treatment as described above, ubiquitinated ERK2 was purified from the cell extracts with anti-HA-antibody-coated agarose beads in HA-lysis buffer (50 mM Tris–HCl pH 7.8, 137 mM NaCl, 10 mM NaF, 1 mM EDTA, 1% Triton X -100, 0.2% Sarkosyl, 1 mM DTT, 10% glycerol and fresh proteinase inhibitor). After extensive washing with the HA-lysis buffer, the proteins were eluted with HA-peptides. The HEK293T cells were transfected with HA-USP37 wild-type (WT) or mutant Cys 350 to Ser (CS mutant) and purified using HA antibody-coated agarose beads. The purified HA-ERK2 proteins were incubated with purified USP37 proteins in deubiquitination buffer (50 mM Tris–HCl pH 8.0, 50 mM NaCl, 1 mM EDTA, 10 mM DTT, 5% glycerol) for 2 h at 37°C. After the reaction, deubiquitination was analyzed by Western blot with indicated antibodies.
Soft agar colony-formation assay
Cancer cells were infected with lentivirus encoding the indicated shRNAs. After selection with puromycin, the indicated stable knockdown cells (1,000 cells per well in a 6-well plate) were plated in 0.8% (wt/vol) low-melting temperature agarose in 1.2% (wt/vol) agarose base layer, both of which contained complete medium. After 2 wk, colonies were visualized and counted at room temperature under a light microscope (DMi3000 B; Leica) using a 4× objective lens (numerical aperture, 0.70).
Colony formation assay
Drug sensitivity was determined using the colony formation assay. Various cancer cells (1,000 cells per well) were inoculated into a six-well plate and cultured for 1 d, then treated with CDK1/2 inhibitors, EGFR inhibitors, or both at indicated doses, and incubated for 2 wk to allow colony formation. Colonies were stained with Giemsa and quantified. Colonies were visualized and counted at room temperature under a camera (Z30, EXPEED6; Nikon).
Tissue microarray
Cancer and adjacent normal tissue samples were purchased from Alenabio (Lun-2085a) and Shanghai Outdo Biotech Company (HLugA060PG02, HRec-Ade150CS-02). Immunohistochemical staining was carried out with the IHC Select HRP/DAB kit (cat. DAB50; Millipore) using USP37 (dilution 1:200) or ERK1/2 (dilution 1:200) antibodies. The immunostaining was scored by pathologists in a blinded manner. A four-tier grading system (0 = negative, 1 = weak, 2 = moderate, and 3 = strong staining intensity) was used, as previously described (Yuan et al., 2010). Meanwhile, the “negative” and “weak” staining samples were grouped into “low” group and “moderate,” respectively, and “strong staining intensity” stanning samples were grouped into “high” group.
PDX studies
The PDX model was obtained from the Anorectal Surgery Department of Shanghai East Hospital, Shanghai, China. PDX tumor fragments (3 × 3 × 3 mm3) were implanted in both armpits of female nude mice purchased from the Beijing Vital River Laboratory Animal Technology Co., Ltd. Approximately 6 d after PDX inoculation, the nude mice were randomly assigned to treatment groups. The tumors were allowed to grow to an average volume of 150–250 mm3 as detected by caliper measurements. Animals were treated via intraperitoneal injections two times per week. The control mice received saline, while the treatment groups received JNJ-7706261 (CDK1/2 inhibitors) at 10 mg/kg, AZD991 (EGFR inhibitors) at 2 mg/kg, or a combination thereof. Mice were 8 wk old and treatment group sizes included at least five to eight mice per group. In the PDX studies, tumor volume was measured every 2 d with calipers and the tumor volume (TV) was calculated as TV = (D × d2/2), where “D” is the largest and “d” is the smallest superficial visible diameter of the tumor mass. All results were documented as mm3. Tumor weight was measured when the tumors were subcutaneously removed from the nude mouse.
Statistical analysis
For cell survival assay, drug sensitivity analysis, and soft agar, data are represented as the mean ± SD of three independent experiments. For positive cells assay, data are represented as the mean ± SD of three independent experiments. For the animal study, data are represented as the mean ± SD of five mice. Statistical analysis was performed with a two-tailed paired Student’s t test or χ2 test. Statistical significance is represented in figures by *P < 0.05; **P < 0.01; ***P < 0.001; N.S. stands for no significant change.
Online supplemental material
Fig. S1 contains additional information in support of Fig. 4 and shows that USP37 removes K48-linked ubiquitin chains of ERK1/2. Fig. S2 contains additional information in support of Fig. 5 and manifests that USP37 regulates cancer cell proliferation through ERK1/2. Fig. S3 (in support of Fig. 6) indicates that CDK2 regulates USP37 stability by phosphorylating and activating USP37. Fig. S4 (in support of Fig. 6) shows that CDK2 through USP37 regulates cell growth. Fig. S5 has additional data that support Figs. 7 and 8, showing that combined CDK1/2 and EGFR inhibitors have a synergetic anticancer effect in vivo.
Supplementary Material
is the source file for Fig. 1.
is the source file for Fig. 2.
is the source file for Fig. 3.
is the source file for Fig. 4.
is the source file for Fig. 5.
is the source file for Fig. 6.
is the source file for Fig. 7.
is the source file for Fig. S1.
is the source file for Fig. S2.
is the source file for Fig. S3.
is the source file for Fig. S4.
is the source file for Fig. S5.
Acknowledgments
We gratefully acknowledge funding from the “National Key R&D Program of China” (2022YFA1302803to J. Yuan), the National Natural Science Foundation of China (82002985 to Y. Chen, 82172994 to Y. Chen, 82225035 to J. Yuan, 32090032 to J. Yuan, and 32070713 to J. Yuan), Shanghai Pujiang program (2020PJD070 to Y. Chen), China Postdoctoral Science Foundation (2020M681384 to Y. Chen), the Natural Science Foundation of Shanghai Municipality (22XD1422300 to J. Yuan), Shanghai Municipal Health Commission (2022XD053 to J. Yuan), and Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (2019-I2M-5-028 to F. Gong).
Author contributions: J. Yuan, Y. Chen, F. Gong, and J. Wu conceived the study and designed the experiments. J. Wu, Y. Chen, and R. Li performed the experiments. J. Wu wrote the paper. J. Yuan, Z. Lou, Y. Chen, and F. Gong revised the manuscript. Y. Guan and M. Chen performed bioinformatics analysis. R. Li and X. Yang performed mouse experiments. H. Yin, M. Jin, B. Huang, X. Ding, J. Yang, Z. Wang, Y. He, Q. Wang, J. Luo, P. Wang, Z. Mao, and M.S.Y. Huen provided the constructional suggestions and help for this paper.
Data availability
The datasets and unique materials generated during the current study are available from the corresponding author upon reasonable request.
References
- Aoki, K., Yamada M., Kunida K., Yasuda S., and Matsuda M.. 2011. Processive phosphorylation of ERK MAP kinase in mammalian cells. Proc. Natl. Acad. Sci. USA. 108:12675–12680. 10.1073/pnas.1104030108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asghar, U., Witkiewicz A.K., Turner N.C., and Knudsen E.S.. 2015. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat. Rev. Drug Discov. 14:130–146. 10.1038/nrd4504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco, R., Gelardi T., Damiano V., Ciardiello F., and Tortora G.. 2007. Rational bases for the development of EGFR inhibitors for cancer treatment. Int. J. Biochem. Cell Biol. 39:1416–1431. 10.1016/j.biocel.2007.05.008 [DOI] [PubMed] [Google Scholar]
- Canagarajah, B.J., Khokhlatchev A., Cobb M.H., and Goldsmith E.J.. 1997. Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell. 90:859–869. 10.1016/S0092-8674(00)80351-7 [DOI] [PubMed] [Google Scholar]
- Caunt, C.J., Sale M.J., Smith P.D., and Cook S.J.. 2015. MEK1 and MEK2 inhibitors and cancer therapy: The long and winding road. Nat. Rev. Cancer. 15:577–592. 10.1038/nrc4000 [DOI] [PubMed] [Google Scholar]
- Chong, C.R., and Jänne P.A.. 2013. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat. Med. 19:1389–1400. 10.1038/nm.3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, Y.-T., Lin H.-H., Lien Y.-C., Wang Y.-H., Hong C.-F., Kao Y.-R., Lin S.-C., Chang Y.-C., Lin S.-Y., Chen S.-J.. et al. 2010. EGFR promotes lung tumorigenesis by activating miR-7 through a Ras/ERK/Myc pathway that targets the Ets2 transcriptional repressor ERF. Cancer Res. 70:8822–8831. 10.1158/0008-5472.CAN-10-0638 [DOI] [PubMed] [Google Scholar]
- Corcoran, R.B., Ebi H., Turke A.B., Coffee E.M., Nishino M., Cogdill A.P., Brown R.D., Della Pelle P., Dias-Santagata D., Hung K.E., et al. 2012. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2:227–235. 10.1158/2159-8290.CD-11-0341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, L., Cao J., Lin W., Chen H., Xiong X., Ao H., Yu M., Lin J., and Cui Q.. 2020. The roles of cyclin-dependent kinases in cell-cycle progression and therapeutic strategies in human breast cancer. Int. J. Mol. Sci. 21:1960. 10.3390/ijms21061960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, R., Shen W., Liu Y., Gao W., Zhou W., Li J., Zhao S., Chen C., Chen Y., Liu Y., et al. 2019. TGIF2 promotes the progression of lung adenocarcinoma by bridging EGFR/RAS/ERK signaling to cancer cell stemness. Signal. Transduct. Target. Ther. 4:60. 10.1038/s41392-019-0098-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercan, D., Xu C., Yanagita M., Monast C.S., Pratilas C.A., Montero J., Butaney M., Shimamura T., Sholl L., Ivanova E.V., et al. 2012. Reactivation of ERK signaling causes resistance to EGFR kinase inhibitors. Cancer Discov. 2:934–947. 10.1158/2159-8290.CD-12-0103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedele, C., Ran H., Diskin B., Wei W., Jen J., Geer M.J., Araki K., Ozerdem U., Simeone D.M., Miller G., et al. 2018. SHP2 inhibition prevents adaptive resistance to MEK inhibitors in multiple cancer models. Cancer Discov. 8:1237–1249. 10.1158/2159-8290.CD-18-0444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frémin, C., and Meloche S.. 2010. From basic research to clinical development of MEK1/2 inhibitors for cancer therapy. J. Hematol. Oncol. 3:8. 10.1186/1756-8722-3-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada, K., Takuwa N., Zhou W., Kumada M., and Takuwa Y.. 1996. Protein kinase C inhibits the CAK-CDK2 cyclin-dependent kinase cascade and G1/S cell cycle progression in human diploid fibroblasts. Biochim. Biophys. Acta. 1310:149–156. 10.1016/0167-4889(95)00148-4 [DOI] [PubMed] [Google Scholar]
- Hazar-Rethinam, M., Kleyman M., Han G.C., Liu D., Ahronian L.G., Shahzade H.A., Chen L., Parikh A.R., Allen J.N., Clark J.W., et al. 2018. Convergent therapeutic strategies to overcome the heterogeneity of acquired resistance in BRAFV600E colorectal cancer. Cancer Discov. 8:417–427. 10.1158/2159-8290.CD-17-1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, J., Huang Z., Han L., Gong Y., and Xie C.. 2021. Mechanisms and management of 3rd-generation EGFR-TKI resistance in advanced non-small cell lung cancer (review). Int. J. Oncol. 59:90. 10.3892/ijo.2021.5270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata, E., and Kiyokawa E.. 2019. ERK activity imaging during migration of living cells in vitro and in vivo. Int. J. Mol. Sci. 20:679. 10.3390/ijms20030679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochegger, H., Takeda S., and Hunt T.. 2008. Cyclin-dependent kinases and cell-cycle transitions: Does one fit all? Nat. Rev. Mol. Cell Biol. 9:910–916. 10.1038/nrm2510 [DOI] [PubMed] [Google Scholar]
- Hochstrasser, M. 1996. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30:405–439. 10.1146/annurev.genet.30.1.405 [DOI] [PubMed] [Google Scholar]
- Hoffmann, I., Draetta G., and Karsenti E.. 1994. Activation of the phosphatase activity of human cdc25A by a cdk2‐cyclin E dependent phosphorylation at the G1/S transition. EMBO J. 13:4302–4310. 10.1002/j.1460-2075.1994.tb06750.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, L., and Fu L.. 2015. Mechanisms of resistance to EGFR tyrosine kinase inhibitors. Acta Pharm. Sin. B. 5:390–401. 10.1016/j.apsb.2015.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X., Summers M.K., Pham V., Lill J.R., Liu J., Lee G., Kirkpatrick D.S., Jackson P.K., Fang G., and Dixit V.M.. 2011. Deubiquitinase USP37 is activated by CDK2 to antagonize APC(CDH1) and promote S phase entry. Mol. Cell. 42:511–523. 10.1016/j.molcel.2011.03.027 [DOI] [PubMed] [Google Scholar]
- Kaplan, F.M., Shao Y., Mayberry M.M., and Aplin A.E.. 2011. Hyperactivation of MEK-ERK1/2 signaling and resistance to apoptosis induced by the oncogenic B-RAF inhibitor, PLX4720, in mutant N-RAS melanoma cells. Oncogene. 30:366–371. 10.1038/onc.2010.408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, L., Zhang Q., Mao J., Cheng L., Shi X., Yu L., Hu J., Yang M., Li L., Liu B., and Qian X.. 2021. A dual-targeted molecular therapy of PP242 and cetuximab plays an anti-tumor effect through EGFR downstream signaling pathways in colorectal cancer. J. Gastrointest. Oncol. 12:1625–1642. 10.21037/jgo-21-467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie, H., Gagnon J., and Therrien M.. 2020. ERK signalling: A master regulator of cell behaviour, life and fate. Nat. Rev. Mol. Cell Biol. 21:607–632. 10.1038/s41580-020-0255-7 [DOI] [PubMed] [Google Scholar]
- Lim, J.S., Turner N.C., and Yap T.A.. 2016. CDK4/6 inhibitors: Promising opportunities beyond breast cancer. Cancer Discov. 6:697–699. 10.1158/2159-8290.CD-16-0563 [DOI] [PubMed] [Google Scholar]
- Lu, Z., Xu, S., Joazeiro C., Cobb M.H., and Hunter T.. 2002. The PHD domain of MEKK1 acts as an E3 ubiquitin ligase and mediates ubiquitination and degradation of ERK1/2. Mol Cell. 9:945–956. 10.1016/s1097-2765(02)00519-1 [DOI] [PubMed] [Google Scholar]
- Mainardi, S., Mulero-Sánchez A., Prahallad A., Germano G., Bosma A., Krimpenfort P., Lieftink C., Steinberg J.D., de Wit N., Gonçalves-Ribeiro S., et al. 2018. SHP2 is required for growth of KRAS-mutant non-small-cell lung cancer in vivo. Nat. Med. 24:961–967. 10.1038/s41591-018-0023-9 [DOI] [PubMed] [Google Scholar]
- May, L.T., and Hill S.J.. 2008. ERK phosphorylation: Spatial and temporal regulation by G protein-coupled receptors. Int. J. Biochem. Cell Biol. 40:2013–2017. 10.1016/j.biocel.2008.04.001 [DOI] [PubMed] [Google Scholar]
- Misale, S., Di Nicolantonio F., Sartore-Bianchi A., Siena S., and Bardelli A.. 2014. Resistance to anti-EGFR therapy in colorectal cancer: From heterogeneity to convergent evolution. Cancer Discov. 4:1269–1280. 10.1158/2159-8290.CD-14-0462 [DOI] [PubMed] [Google Scholar]
- Morris, E.J., Jha S., Restaino C.R., Dayananth P., Zhu H., Cooper A., Carr D., Deng Y., Jin W., Black S., et al. 2013. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov. 3:742–750. 10.1158/2159-8290.CD-13-0070 [DOI] [PubMed] [Google Scholar]
- Musgrove, E.A., Caldon C.E., Barraclough J., Stone A., and Sutherland R.L.. 2011. Cyclin D as a therapeutic target in cancer. Nat. Rev. Cancer. 11:558–572. 10.1038/nrc3090 [DOI] [PubMed] [Google Scholar]
- Noordhuis, M.G., Eijsink J.J., Ten Hoor K.A., Roossink F., Hollema H., Arts H.J., Pras E., Maduro J.H., Reyners A.K., de Bock G.H., et al. 2009. Expression of epidermal growth factor receptor (EGFR) and activated EGFR predict poor response to (chemo)radiation and survival in cervical cancer. Clin. Cancer Res. 15:7389–7397. 10.1158/1078-0432.CCR-09-1149 [DOI] [PubMed] [Google Scholar]
- Normanno, N., De Luca A., Bianco C., Strizzi L., Mancino M., Maiello M.R., Carotenuto A., De Feo G., Caponigro F., and Salomon D.S.. 2006. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 366:2–16. 10.1016/j.gene.2005.10.018 [DOI] [PubMed] [Google Scholar]
- Nusinow, D.P., Szpyt J., Ghandi M., Rose C.M., McDonald E.R. III, Kalocsay M., Jané-Valbuena J., Gelfand E., Schweppe D.K., Jedrychowski M., et al. 2020. Quantitative proteomics of the cancer cell line Encyclopedia. Cell. 180:387–402.e16. 10.1016/j.cell.2019.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary, B., Finn R.S., and Turner N.C.. 2016. Treating cancer with selective CDK4/6 inhibitors. Nat. Rev. Clin. Oncol. 13:417–430. 10.1038/nrclinonc.2016.26 [DOI] [PubMed] [Google Scholar]
- Otto, T., and Sicinski P.. 2017. Cell cycle proteins as promising targets in cancer therapy. Nat. Rev. Cancer. 17:93–115. 10.1038/nrc.2016.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskoski, R. Jr.. 2012. ERK1/2 MAP kinases: Structure, function, and regulation. Pharmacol Res. 66:105–143. 10.1016/j.phrs.2012.04.005 [DOI] [PubMed] [Google Scholar]
- Roskoski, R. Jr.. 2019. Targeting ERK1/2 protein-serine/threonine kinases in human cancers. Pharmacol. Res. 142:151–168. 10.1016/j.phrs.2019.01.039 [DOI] [PubMed] [Google Scholar]
- Roux, P.P., and Blenis J.. 2004. ERK and p38 MAPK-activated protein kinases: A family of protein kinases with diverse biological functions. Microbiol. Mol. Biol. Rev. 68:320–344. 10.1128/MMBR.68.2.320-344.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samatar, A.A., and Poulikakos P.I.. 2014. Targeting RAS-ERK signalling in cancer: Promises and challenges. Nat. Rev. Drug Discov. 13:928–942. 10.1038/nrd4281 [DOI] [PubMed] [Google Scholar]
- Sanchez, I.M., Purwin T.J., Chervoneva I., Erkes D.A., Nguyen M.Q., Davies M.A., Nathanson K.L., Kemper K., Peeper D.S., and Aplin A.E.. 2019. In vivo ERK1/2 reporter predictively models response and resistance to combined BRAF and MEK inhibitors in melanoma. Mol. Cancer Ther. 18:1637–1648. 10.1158/1535-7163.MCT-18-1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr, C.J., Beach D., and Shapiro G.I.. 2016. Targeting CDK4 and CDK6: From discovery to therapy. Cancer Discov. 6:353–367. 10.1158/2159-8290.CD-15-0894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y., Liu W.-Z., Liu T., Feng X., Yang N., and Zhou H.-F.. 2015. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J. Recept. Signal. Transduct. Res. 35:600–604. 10.3109/10799893.2015.1030412 [DOI] [PubMed] [Google Scholar]
- Susanti, N.M.P., and Tjahjono D.H.. 2021. Cyclin-dependent kinase 4 and 6 inhibitors in cell cycle dysregulation for breast cancer treatment. Molecules. 26:4462. 10.3390/molecules26154462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadesse, S., Anshabo A.T., Portman N., Lim E., Tilley W., Caldon C.E., and Wang S.. 2020. Targeting CDK2 in cancer: Challenges and opportunities for therapy. Drug Discov. Today. 25:406–413. 10.1016/j.drudis.2019.12.001 [DOI] [PubMed] [Google Scholar]
- Troiani, T., Martinelli E., Capasso A., Morgillo F., Orditura M., De Vita F., and Ciardiello F.. 2012. Targeting EGFR in pancreatic cancer treatment. Curr. Drug Targets. 13:802–810. 10.2174/138945012800564158 [DOI] [PubMed] [Google Scholar]
- Wilkinson, K.D. 2000. Ubiquitination and deubiquitination: Targeting of proteins for degradation by the proteasome. Semin. Cell Dev. Biol. 11:141–148. 10.1006/scdb.2000.0164 [DOI] [PubMed] [Google Scholar]
- Yoshida, T., Hisamoto T., Akiba J., Koga H., Nakamura K., Tokunaga Y., Hanada S., Kumemura H., Maeyama M., Harada M., et al. 2006. Spreds, inhibitors of the Ras/ERK signal transduction, are dysregulated in human hepatocellular carcinoma and linked to the malignant phenotype of tumors. Oncogene. 25:6056–6066. 10.1038/sj.onc.1209635 [DOI] [PubMed] [Google Scholar]
- Yu, Z., Ye S., Hu G., Lv M., Tu Z., Zhou K., and Li Q.. 2015. The RAF-MEK-ERK pathway: Targeting ERK to overcome obstacles to effective cancer therapy. Future Med. Chem. 7:269–289. 10.4155/fmc.14.143 [DOI] [PubMed] [Google Scholar]
- Yuan, J., Luo K., Zhang L., Cheville J.C., and Lou Z.. 2010. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell. 140:384–396. 10.1016/j.cell.2009.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F., Strand A., Robbins D., Cobb M.H., and Goldsmith E.J.. 1994. Atomic structure of the MAP kinase ERK2 at 2.3 A resolution. Nature. 367:704–711. 10.1038/367704a0 [DOI] [PubMed] [Google Scholar]
- Zhang, F., Tang X., Fan S., Liu X., Sun J., Ju C., Liang Y., Liu R., Zhou R., Yu B., et al. 2021a. Targeting the p300/NONO axis sensitizes melanoma cells to BRAF inhibitors. Oncogene. 40:4137–4150. 10.1038/s41388-021-01834-1 [DOI] [PubMed] [Google Scholar]
- Zhang, J., Xu D., Zhou Y., Zhu Z., and Yang X.. 2021b. Mechanisms and implications of CDK4/6 inhibitors for the treatment of NSCLC. Front. Oncol. 11:676041. 10.3389/fonc.2021.676041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M., Zhang L., Hei R., Li X., Cai H., Wu X., Zheng Q., and Cai C.. 2021c. CDK inhibitors in cancer therapy, an overview of recent development. Am. J. Cancer Res. 11:1913–1935 [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y., and Adjei A.A.. 2014. The clinical development of MEK inhibitors. Nat. Rev. Clin. Oncol. 11:385–400. 10.1038/nrclinonc.2014.83 [DOI] [PubMed] [Google Scholar]
- Zhu, G., Herlyn M., and Yang X.. 2021. TRIM15 and CYLD regulate ERK activation via lysine-63-linked polyubiquitination. Nat. Cell Biol. 23:978–991. 10.1038/s41556-021-00732-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
is the source file for Fig. 1.
is the source file for Fig. 2.
is the source file for Fig. 3.
is the source file for Fig. 4.
is the source file for Fig. 5.
is the source file for Fig. 6.
is the source file for Fig. 7.
is the source file for Fig. S1.
is the source file for Fig. S2.
is the source file for Fig. S3.
is the source file for Fig. S4.
is the source file for Fig. S5.
Data Availability Statement
The datasets and unique materials generated during the current study are available from the corresponding author upon reasonable request.