Abstract
Control of microorganisms such as Bacillus cereus spores is critical to ensure the safety and a long shelf life of foods. A bifunctional single chain antibody has been developed for detection and binding of B. cereus T spores. The genes that encode B. cereus T spore single-chain antibody and streptavidin were connected for use in immunoassays and immobilization of the recombinant antibodies. A truncated streptavidin, which is smaller than but has biotin binding ability similar to that of streptavidin, was used as the affinity domain because of its high and specific affinity with biotin. The fusion protein gene was expressed in Escherichia coli BL21 (DE3) with the T7 RNA polymerase-T7 promoter expression system. Immunoblotting revealed an antigen specificity similar to that of its parent native monoclonal antibody. The single-chain antibody-streptavidin fusion protein can be used in an immunoassay of B. cereus spores by applying a biotinylated enzyme detection system. The recombinant antibodies were immobilized on biotinylated magnetic beads by taking advantage of the strong biotin-streptavidin affinity. Various liquids were artificially contaminated with 5 × 104 B. cereus spores per ml. Greater than 90% of the B. cereus spores in phosphate buffer or 37% of the spores in whole milk were tightly bound and removed from the liquid phase by the immunomagnetic beads.
Antibodies bind antigens, including microorganisms, with high specificity and have been used in immunoassays for the rapid detection of pathogens. The use of antibodies may shorten the time required for microbial enrichment and isolation from a few days to a few hours. Several immunoadsorption approaches have been used for detection of microorganisms in food systems. Pathogens can be bound by dye-conjugated free antibodies and can subsequently be counted by fluorescence microscopy (14) or flow cytometric technology (25). Target microorganisms can also be trapped by an immobilized antibody and detected by enzyme-linked immunosorbent assaying (ELISA) (26). Recently, immunomagnetic separation technology (11) has broadened the use of antibodies in detection or isolation of food-borne pathogens (22, 36). These immunomagnetic beads are able to bind the target microorganisms, thus allowing collection of the bead-bound microbes simply by applying a magnetic field. These magnetically recovered microorganisms have been detected by luminescence assaying (39) or PCR (8) or have been simply identified or counted from selective medium (36).
Traditionally, antibodies can be obtained only from immunized animals; however, recent progress in molecular biology has made it possible to produce monoclonal antibody fragments from bacteria (35). To date, most of the antibody fragments produced from recombinant technology have been single-chain antibodies, consisting only of the variable-region domains of the heavy and light chains of the parent antibody and a short peptide linker used to connect these two domains. An effector protein can be genetically fused with the single-chain antibody to allow expression as a bifunctional antibody. For example, single-chain antibodies have been fused with alkaline phosphatase and used for diagnosis and immunoassays (5). Some affinity tails such as the FLAG tag (23), strep tag (33), His tag (34), calmodulin (28), or streptavidin (7) can be attached to the single-chain antibodies for direct detection by commercially available detection systems and for recovery of recombinant antibodies from the cell lysate by affinity chromatography.
Spore-forming bacteria such as Bacillus cereus may cause food-borne illness or spoilage and are problematic because they can survive mild heat treatment. Detection and control in food processing are exacerbated for bacterial spores because they typically are present in low numbers and are metabolically inactive. A procedure to concentrate and detect low numbers of these metabolically inactive yet significant organisms would be useful. In the present study, a truncated streptavidin gene (3) was amplified by PCR to introduce unique restriction enzyme sites. It was connected with the gene of single-chain anti-B. cereus spore antibody (19) to form a fusion protein gene. This bifunctional single-chain antibody gene was expressed by Escherichia coli. Both native and recombinant monoclonal antibodies revealed similar antigen specificities. This streptavidin-conjugated antibody can be immobilized on the surface of biotinylated magnetic beads, and its spore binding ability was demonstrated.
MATERIALS AND METHODS
Bacterial spores and cells.
E. coli JM109 {endA1 recA1 gyrA96 thi hsdR17 (rk− mk+) relA1 supE44 Δ(lac-proAB) [F′ traD36 proAB lacIqZΔM15]} was provided by Promega (Madison, Wis.). E. coli BL21 (DE3), which carries the T7 RNA polymerase gene under lacUV5 promoter control, was purchased from Novagen (Madison, Wis.). The competent E. coli cells used for gene transformation were prepared by a simple polyethylene glycol-dimethyl sulfoxide protocol (6). Spores of B. cereus T were prepared on fortified nutrient agar sporulation medium (15). After collection and washing, the spore suspension was stored at −20°C. The numbers of spores were enumerated on Trypticase soy agar (Difco, Detroit, Mich.) plates and by direct microscopic counting.
DNA manipulation and sequencing.
Most of the gene cloning procedures were based on the protocols described by Maloy (24). The DNA fragments generated from PCR or restriction enzyme digestion were purified by a diatomaceous earth-based protocol. The DNA sequences of PCR products and the fusion protein gene were obtained by the cycle sequencing method (20) and were detected by a nonradioactive silver-staining protocol (2). The DNA-sequencing-grade Taq DNA polymerase and nucleotides were purchased from Promega. For accuracy, both strands of the DNA were sequenced.
Construction of expression vectors. (i) Plasmid DNA and oligonucleotides.
The plasmid pGEM-3Z, which was used for general cloning and sequencing purposes, was obtained from Promega. The pET22b(+)-derived plasmid pET22IgTag (19) was used as the single-chain antibody gene source. This plasmid contains a T7/lac promoter (37), a pelB signal sequence (16) for protein relocation, the complete anti-B. cereus single-chain antibody gene, and the T7 transcription terminator. The oligonucleotides used for PCR primers or DNA sequencing were synthesized at the Molecular Biology Center, North Carolina State University, Genosys Biotechnologies, Inc. (Woodlands, Tex.), or GIBCO BRL (Gaithersburg, Md.).
(ii) Modification of streptavidin gene.
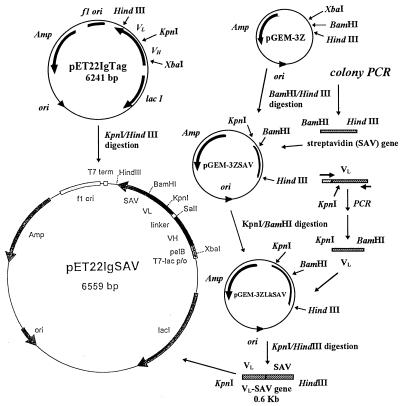
The plasmid pUC8-SZ (a gift from C. E. Argarana), which contained the complete streptavidin gene (1), was modified by colony PCR (9) with primers STREP5 (5′-CATCGGATCCGGCATCACCGGCACCTGGTACAAC) and STREP3 (5′-GAGGAAGCTTACGGCTTCACCTTGGTGAAGGT). Colonies of pUC8-SZ transformants were picked from a Luria-Bertani (LB)–ampicillin plate and were used as the gene source. Each colony was suspended with 50 μl of distilled water in a microcentrifuge tube and then heated in boiling water for 5 min to lyse the cells. The cell lysate was centrifuged at 12,000 × g for 5 min, and 10 μl of the supernatant containing the target DNA was mixed with a deoxynucleoside triphosphate mixture (Boehringer Mannheim, Indianapolis, Ind.), 10× PCR buffer (Sigma, St. Louis, Mo.), and 30 pmol each of primers STREP5 and STREP3 for PCR. The amplification solution was heated in a Perkin-Elmer Thermal Cycler 480 (Norwalk, Conn.) at 95°C for 6 min, and then the Taq DNA polymerase was added. The target gene was amplified by 40 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min. The STREP5 and STREP3 primers introduced BamHI and HindIII restriction enzyme sites at the boundaries of the biotin binding-domain gene. The STREP3 primer also encoded a stop codon (TAA) for the C-terminal end of the streptavidin gene for further fusion protein construction. After digestion of the PCR-modified streptavidin genes with BamHI and HindIII, this DNA fragment was cloned into the pGEM-3Z cloning vector to form pGEM-3ZSAV plasmid (see Fig. 1).
FIG. 1.
Construction of the streptavidin-conjugated single-chain antibody fusion gene and expression vector.
(iii) Preparation of the streptavidin-conjugated single-chain antibody construct.
For connection of the streptavidin gene with the single-chain antibody gene, the light-chain, variable-region gene of single-chain antibody in pET22IgTag was modified by PCR with primers Lklinker (5′-ATCGGTCGACGG TGG TGG TGGTTCCGG TGG TGG TGG T TCCGG TGG TGG TGG T TCC AAATTGTTCTCACCCA) and LkBamHI (5′-CCCAAGCTTACTGGATGGTGGGAAGATGGATCC). This PCR-derived DNA, containing a unique BamHI site at the 3′-end of the light-chain variable gene, was digested with KpnI and BamHI and then ligated with pGEM-3ZSAV to form pGEM-3ZLkSAV. The streptavidin-conjugated single-chain antibody gene was assembled by insertion of the 0.6-kb KpnI/HindIII-digested pGEM-3ZLkSAV DNA fragment into the 5.9-kb fragment from a KpnI/HindIII digest of pET22IgTag to form pET22IgSAV (Fig. 1).
Expression and recovery of fusion protein.
Plasmid DNA, which contained the fusion protein gene (VH and VL plus core streptavidin), was transformed into E. coli BL21 (DE3). Transformants were inoculated in LB medium (24) containing ampicillin (100 μg/ml). The overnight culture was transferred into plasmid medium (24) containing ampicillin (500 μg/ml) and incubated at 37°C for 5 h with shaking, and then protein expression was induced by the addition of 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG; GIBCO BRL) and by incubation for 2 h at 37°C. The cellular location of the fusion protein was analyzed using the procedures as previously described (19). The periplasmic and cytoplasmic proteins were extracted by osmotic shock and cell lysis (lysozyme treatment and sonication) procedures, respectively. The insoluble cellular fraction, containing membrane proteins and inclusion bodies, was solubilized overnight in 6 M guanidine-HCl in 0.1 M Tris buffer (pH 8.0) at 4°C. The guanidine-HCl-solubilized fraction was centrifuged at 10,000 × g for 10 min at 4°C to remove any precipitate before dialyzing against phosphate buffer (0.02 M Na-phosphate, 0.8% NaCl [pH 7.8]). The dialyzed guanidine-HCl-solubilized fraction was centrifuged to remove any possible precipitate and stored at −20°C as the final solubilized cellular fraction. Each fraction recovered from the cell culture was mixed with 2× sodium dodecyl sulfate (SDS) sample buffer (0.25 M Tris [pH 6.8], 30% [wt/vol] sucrose, 2% [wt/vol] SDS, 20% β-mercaptoethanol, 2 mM EDTA, and 0.1% [wt/vol] bromophenol blue), heated at 100°C for 5 min, and then analyzed by SDS-6 or 12% polyacrylamide gel electrophoresis (PAGE). For observation of formation of fusion protein oligomer, the protein sample was heated at a low temperature (55 to 60°C) in the presence of 2× SDS sample buffer prior to electrophoresis (3).
Western blotting and N-terminal sequence analysis of the streptavidin-conjugated single-chain antibody.
The procedure of protein blotting was modified from the original procedures described by Towbin et al. (40). Proteins on the polyacrylamide gel were transferred to Immobilon P membranes (Millipore, Bedford, Mass.), and 1% bovine serum albumin (BSA) in 0.02 M Na phosphate, 0.8% NaCl (pH 7.0) (PBS) was used as a blocking agent. Rabbit antistreptavidin antiserum (Sigma catalog no. S6390) was the primary antibody. Bound primary antibody was probed with goat anti-rabbit immunoglobulin G (IgG) horseradish peroxidase conjugate (Sigma catalog no. A8275). The enzyme activity was detected with a 3-amino-9-ethylcarbazole (AEC) chromogen kit (Biomeda, Foster City, Calif.). Proteins in the final solubilized cellular fraction were electrophoretically blotted to a protein-sequencing-grade polyvinylidene difluoride membrane (Bio-Rad, Hercules, Calif.) and stained with Coomassie brilliant blue R-250. The protein band that represented the streptavidin-conjugated, single-chain antibody was sequenced. The first seven amino acids of this fusion protein were determined by a microsequencing method at the Medical Center, University of North Carolina at Chapel Hill.
Purification of native monoclonal antibody.
The parent, native monoclonal antibody against B. cereus spores (31) was purified from tissue culture supernatant by thiophilic gel affinity chromatography as described by Hutchens and Porath (13). The purified monoclonal antibody was dialyzed against phosphate buffer and concentrated with an Amicon Centricon-10 ultrafiltration concentrator (Amicon, Beverly, Mass.).
Determination of protein concentrations.
The protein concentrations of purified native monoclonal antibody and the final solubilized cellular fraction from the E. coli culture were determined by the Bradford dye-binding method (4) with bovine gamma globulin (Bio-Rad) as the standard. The percentage of fusion protein present in the final solubilized cellular fraction was quantitated from the SDS-PAGE gel by using the Personal Densitometer SI and FragmeNT software (Molecular Dynamics, Sunnyvale, Calif.).
Dot blot immunoassay.
The immunoassay protocol was modified from the method described by Phillips (29). Fifty microliters of a B. cereus T spore suspension (approximately 108 spores/ml) was applied into each well of a dot blot apparatus (Bio-Rad) and filtered through a 0.45-μm-pore-size Immobilon P membrane (Millipore). The nonspecific binding sites of spores and the blotting membrane were blocked by 1% BSA in PBS. One hundred microliters of antibody solution was added (approximately 2 to 4 μg of antibody), and the mixture was incubated for 40 min at room temperature. Unbound antibody was removed by washing with TPBS (0.05% Tween 20 [vol/vol] in PBS). The bound native monoclonal antibody was determined with the biotinylated anti-mouse IgG–avidin-horseradish peroxidase conjugate detection system (Biomeda). Bound streptavidin-conjugated single-chain antibody was probed directly by applying biotinylated horseradish peroxidase (Sigma catalog no. P2907). To reduce the background signal from the membrane, 4-chloro-1-naphthol (Bio-Rad) was chosen as the peroxidase substrate.
Functional assays.
The antigen specificities of native monoclonal and streptavidin-conjugated single-chain antibodies were determined by the dot blot immunoassay method described above. The final concentrations of both types of antibodies were adjusted by PBS.
(i) Spore binding experiment.
The spore-binding specificities of both native monoclonal antibody and streptavidin-conjugated single-chain antibody were tested by using different species of spores. Spores of Bacillus subtilis A, B. subtilis subsp. globigii, Bacillus megaterium, Bacillus stearothermophilus, and Clostridium perfringens spores were obtained and prepared as described by Quinlan and Foegeding (31).
(ii) Competition experiment.
The ability of streptavidin-conjugated single-chain antibody to bind B. cereus spores was tested by a dot blot immunoassay with streptavidin-conjugated single-chain antibody (2 μg of total protein/ml) in the presence of native anti-B. cereus spore monoclonal antibody as a competitor protein. The binding of streptavidin-conjugated single-chain antibody was detected by biotinylated horseradish peroxidase. The concentrations of competitor ranged from 0.02 μg/ml (0.125 nM) to 100 μg/ml (0.625 mM).
Preparation of biotinylated matrices.
Magnetic beads that were coated covalently with sheep anti-mouse IgG antibodies (M-280; Dynal, Oslo, Norway) were used as the solid support for immobilization of single-chain antibodies. N-hydroxysuccinimidobiotin (Sigma) was used as the biotinylation reagent. Approximately 400 μl of beads was washed three times with 1 ml of PBS, and then the beads were resuspended in 250 μl of 0.12 M borate buffer (pH 8.8). Fifty microliters of N-hydroxysuccinimidobiotin–dimethyl sulfoxide solution (10 mg/ml) was added, and the suspension was mixed on a horizontal sample mixer (Dynal) at room temperature for 4 h.
The residual N-hydroxysuccinimidobiotin in the bead suspension was removed by washing the beads three times with 3 volumes of PBS. The cleaned biotinylated beads were stored in a storage solution (0.1% BSA in PBS, filtered by a 0.22-μm-pore-size membrane, with 0.02% [wt/vol] sodium azide added) at 4°C. The beads were checked by a simple enzyme-linked assay to validate the presence of functional biotin groups. Approximately 20 μl of biotinylated beads was transferred into a microcentrifuge tube and washed with 200 μl of PBS, and then the beads were resuspended in 200 μl of 1% BSA in PBS for 40 min to block the nonspecific binding sites. After the BSA-blocked beads were washed with 200 μl of TPBS three times, 200 μl of streptavidin-conjugated horseradish peroxidase (Sigma) in TPBS was added and mixed with the beads at room temperature for 20 min. Unbound enzyme was removed by washing the beads with 200 μl of TPBS three times. One milliliter of o-phenylenediamine–H2O2 (21) was added as the peroxidase substrate. After the reaction suspension was mixed for 15 min, the substrate solution in the biotinylated beads turned yellow while the nonbiotinylated beads that were used as a control were colorless.
Immobilization of recombinant antibody.
The biotinylated beads in storage solution were mixed with streptavidin-conjugated single-chain antibody (0.1 mg/ml) in a microcentrifuge tube overnight at 4°C or for 1 h at room temperature by using a horizontal sample mixer. The beads were cleaned by being washed three times with storage solution. The single-chain antibody-coated magnetic beads were resuspended in storage solution at a level of 6 × 105 beads per μl of suspension (10 μg/μl) and stored at 4°C. Nonbiotinylated beads treated by the same procedure were used as a control. For qualitative assay of the immobilized streptavidin-conjugated single-chain antibody on the beads, an ELISA method was used. The details of procedure were similar to the detection of the biotinyl group given previously. A rabbit antistreptavidin antiserum was used to document that the beads were coated with streptavidin fusion proteins. Bound primary antibody was detected by goat antirabbit antibody conjugated to horseradish peroxidase with o-phenylenediamine–H2O2 as the substrate.
Evaluation of spore-binding ability by using immobilized single-chain antibody.
The B. cereus spore stock suspension was diluted with filter-sterilized (0.22-μm-pore-size membrane) 0.1% BSA in PBS or pasteurized whole milk (purchased from a local supermarket and then stored at 4°C for various periods of time) to a final spore concentration of approximately 5 × 104 CFU/ml. One milliliter of diluted spores was transferred into a siliconized sterile microcentrifuge tube for the spore binding test. Approximately 50 μl of antibody-coated beads was mixed with the spore suspension at room temperature (23°C) or at 4°C for 1 h with a horizontal sample mixer at a speed of 25 rpm. The magnetic particle concentrator for the microcentrifuge tube (MPC-E-1; Dynal) was used to collect the magnetic beads. The supernatant was removed carefully, and the spores present were defined as the unbound fraction. The pelleted beads were washed three times by resuspending the bead pellet with 500 μl of sterilized PBS and vortexing for each wash. Spores in the washing solutions were designated the bound but removable fraction. The buffer-washed beads were resuspended in 500 μl of PBS, and the spore fraction remaining was designated the bound fraction.
The spore number was determined by the spread plating method with Trypticase soy agar plates. One hundred microliters of serial dilutions from each fraction was plated directly. Mannitol yolk polymyxin agar plates (10) were used as the selective medium for identification of B. cereus from milk testing solutions. Biotinylated beads without fusion protein were used as a negative control. The number of B. cereus spores used in the spore-binding tests was calibrated by a Trypticase soy agar plate count for each test.
RESULTS AND DISCUSSION
Construction of the streptavidin-conjugated single-chain antibody gene and expression vector.
The interaction of streptavidin and biotin is one of the strongest noncovalent affinities known in biology. It can be used not only as a reporter for use in immunoassays but also as a domain for bioselective immobilization (41). For this reason, it was chosen as the affinity domain in this study. Native streptavidin is sensitive to the action of proteolytic enzymes at both the N and the C termini (3). However, enzymatically truncated streptavidins, which are also called core streptavidins and which include residues 12 through 140 of native streptavidin, still have strong biotin-binding ability. Like streptavidin, core streptavidin can associate into tetramers (3, 7, 32), but these are less prone to aggregation than is streptavidin (32). In this case, the PCR-modified core streptavidin gene encoded 120 amino acids, including the 16th to the 135th amino acid of native streptavidin, and a new stop codon. Because Taq DNA polymerase, a nonproofreading DNA polymerase, was used in these PCR modifications, screening of the correct product is critical for construction of a bifunctional single-chain antibody. A base substitution error was detected by DNA sequencing of the PCR-modified core streptavidin gene segment. Fortunately, only a silent mutation (GGC to GGT; the 68th amino acid in the wild-type streptavidin gene [1]) was detected, and the primary structure of core streptavidin protein was identical to that originally reported (1). In this case, the streptavidin-conjugated single-chain antibody structure included VH-linker-VL-core streptavidin. Thus, the PCR-derived VL-core streptavidin gene fragments were assembled into the pET22IgTag vector (which contained VH, VL, and strep tag peptide genes) to form a new pET22IgSAV streptavidin-conjugated single-chain antibody expression vector (Fig. 1).
Expression and recovery of bifunctional single-chain antibody from E. coli.
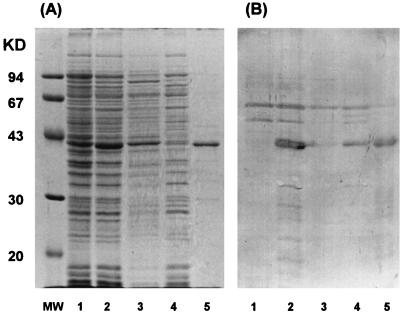
The T7 RNA polymerase-T7 promoter expression system is a tightly controlled bacterial expression system. It has been used for production of core streptavidin, which is potentially lethal to host cells (32). In the present study, the streptavidin-fusion antibody was expressed successfully within a few hours by this system. Although the fusion protein was produced mainly as an insoluble form, the active bifunctional fusion antibody was recovered by simple denaturation, renaturation, and dialysis operations. The SDS-PAGE and Western blotting analyses (Fig. 2) showed that most of the single-chain antibody existed in the final solubilized cellular fraction, while a small amount of the fusion protein was detected in the soluble cytoplasmic fraction. Densitometry data of the final solubilized cellular fraction revealed that approximately 70% of the soluble protein was streptavidin-conjugated single-chain antibody. The concentration of soluble streptavidin-conjugated single-chain antibody present was approximately 3 to 7 mg of fusion protein/liter of culture. N-terminal sequence analysis indicated that this protein was mature fusion protein. The high expression rate of the T7 RNA polymerase-T7 promoter system may be the cause for expression of the fusion protein mainly in an insoluble form (18).
FIG. 2.
Protein fractionation analysis of streptavidin-conjugated single-chain anti-B. cereus spore T antibody. (A) SDS-12% PAGE; (B) Western blot. Fusion protein was probed with rabbit anti-streptavidin antiserum (Sigma S6390) and anti-rabbit IgG peroxidase conjugate (Sigma A8275). Lanes: MW, molecular weight standards; 1, total cell sample (without IPTG induction); 2, total cell sample (with 0.2 mM IPTG induction); 3, osmotic shock fraction; 4, soluble fraction from cell lysis; 5, final solubilized cellular fraction.
Characteristics of streptavidin.
The expressed streptavidin-conjugated recombinant antibody retained many of the characteristics of native streptavidin. Not only did this fusion protein bind biotin and the B. cereus spore antigen, but the core streptavidin also associated into a tetramer or higher order oligomers. The results from SDS-6% PAGE suggested that the core streptavidin-conjugated fusion protein remained as an oligomer when the final solubilized cellular fraction in 2× SDS sample buffer mixture (1:1) was heated at 55°C (Fig. 3, lane 2). However, SDS-PAGE results showed that by heating at 100°C, the apparent molecular mass of the main protein in the final solubilized cellular fraction was shifted from 200,000 to 45,000 Da (39,800 Da; calculated from primary structure) (Fig. 3, lane 1). These data suggest that the protein associates into an oligomer probably composed of 4 or 5 U.
FIG. 3.
Formation of streptavidin-conjugated single-chain antibody oligomers; final solubilized cellular fraction and 2× SDS sampling buffer heated at 100°C for 5 min (lane 1) or 55°C for 5 min (lane 2). MW, molecular weight standards.
Binding properties of the monoclonal antibody and the single-chain antibody.
Because of the bifunctional nature of the fusion protein, the spore binding ability can be detected simply by applying biotinylated horseradish peroxidase. Six different species of Bacillus and Clostridium spores were used to compare the antigen specificity of this streptavidin-conjugated single-chain antibody with that of its parent monoclonal antibody. The two types of antibodies exhibited similar spore-binding behaviors, with the exception that the single-chain antibody showed slight cross-reaction with C. perfringens spores (Fig. 4). The unexpected cross-reaction with B. subtilis A spores observed with the strep tag (a 10-amino-acid peptide)-conjugated single-chain antibody that has the same primary structure for the antigen-binding domains (19) was not detected in this single-chain antibody. A simple competitive dot blotting immunoassay demonstrated that the recombinant and parent monoclonal antibodies bind to the same or very similar epitopes. The data showed that binding of streptavidin-conjugated single-chain antibody (2 μg/ml) to the antigen was inhibited by native monoclonal antibody when the concentration of parent monoclonal antibody was ≥2 μg/ml. The BSA control at any concentration did not inhibit binding. Hence, the two types of antibodies (recombinant single chain and native) competed for the same antigens on B. cereus spores. A control assay with BSA confirmed that specific competition, rather than nonspecific blocking, occurred between the native and recombinant antibodies. Thus, they must have similar tertiary structures in the variable region domains, and the fusion with streptavidin must not have significantly altered the conformation of the antigen-binding domain. Furthermore, the formation of multimeric complexes might increase the apparent affinity of single-chain antibody (17) or stabilize the conformation of the single-chain antibody favoring antigen binding and mimic the antigen-binding behavior of native bivalent or multivalent antibodies.
FIG. 4.
Results (shown in triplicate) of dot blot immunoassays indicating antigen specificity testing. (A) Native monoclonal anti-B. cereus spore T antibody; (B) streptavidin-conjugated single-chain anti-B. cereus T spore antibody.
Spore-binding tests.
Spore binding was tested in phosphate buffer and in whole-milk systems. The performance of the immunomagnetic beads in the phosphate buffer system is indicated in Table 1. Controls, including the original magnetic beads with and without added fusion protein and biotinylated beads that were not coated with fusion protein, were tested to determine the degree of nonspecific binding. The results of controls without added fusion protein indicated a low level (<1%) of nonspecific binding of the spores. Nonbiotinylated original beads mixed with the streptavidin-conjugated single-chain antibody control were used as a control to evaluate interaction of the fusion protein directly with the beads. The results indicated that fusion protein did interact directly with the beads, resulting in low levels of specific (3.2%) and nonspecific (32.4%) interaction with the spores. These data coincided with the result of an ELISA for qualitative assaying of immobilized fusion protein on the beads. In the ELISA, original (nonbiotinylated) magnetic beads and the biotinylated beads turned the substrate faint yellow (A490, ∼0.3). Fusion protein immobilization on the biotinylated beads was documented by the substrate turning orange (A490, ∼2.0). Nonbiotinylated beads with added fusion protein turned the substrate solution light yellow (A490, ∼0.8), indicating some nonspecific binding.
TABLE 1.
Fractions of B. cereus sporesa recovered from the phosphate buffer system by the single-chain antibody-coated immunomagnetic beads
| Type of beads | Fusion protein immobilization treatment | Incubation temp (°C) | % of sporesb
|
Total (%) | ||
|---|---|---|---|---|---|---|
| Unbound | Bound but removed by washing | Bound | ||||
| Biotinylatedc | + | 23 | 1.7 (0.6) | 0.3 (0.2) | 98.0 (0.8) | 100 |
| Biotinylated | + | 4 | 9.3 (3.4) | 0.7 (0.2) | 90.0 (3.6) | 100 |
| Biotinylated | − | 4 | 99.3 (0.5) | 0.6 (0.4) | 0.1 (0.08) | 100 |
| Biotinylated | − | 23 | 98.6 (0.6) | 1.3 (0.6) | 0.1 (0.6) | 100 |
| Originald | − | 23 | 94.3 (3.4) | 5.0 (3.0) | 0.7 (0.5) | 100 |
| Original | + | 23 | 64.4 (7.8) | 32.4 (7.1) | 3.2 (1.6) | 100 |
Spore concentration, 5 × 104 spores/ml; incubation time, 60 min.
Averages from three trials (numbers in parentheses are standard deviations).
Commercial sheep anti-mouse IgG-coated magnetic beads which were biotinylated as described in Materials and Methods.
Commercial sheep anti-mouse IgG-coated magnetic beads.
The immunomagnetic beads can remove B. cereus spores nearly quantitatively from a phosphate buffer system at room or refrigeration temperature within 1 h (Table 1). Only 2 to 9% of the original B. cereus spores were in the sample solution after mixing the immunomagnetic beads with the spore suspension for 1 h. Colony counts indicated that ≥90% of spores were removed from the 0.1% BSA–PBS buffer system. Almost all of the captured spores were tightly bound to the immunomagnetic beads. Less than 1% of the bead-bound spores were released from the matrix after three rounds of PBS washing. To determine their spore-binding ability in a food, pasteurized whole milk was used as a test system. The results given in Table 2 show that some of the spore-binding ability of immunomagnetic beads could be inhibited by whole milk. However, the immunomagnetic beads did specifically bind approximately 37% of the B. cereus spores in the presence of complex food components. These results indicated that immobilized streptavidin-conjugated single-chain antibody showed specific spore binding and removal ability in both buffer and whole-milk systems; however, the spore removal ability decreased to approximately 37 to 40% in a whole-milk system. Most of the milk lipids, approximately 4% in whole milk, are present in 2- to 3-μm globules that are surrounded by a membrane (38). The presence of these fat particles may interfere with the effective contact of spores with the antigen binding site. However, the performance of immobilized Pseudomonas aeruginosa recombinant antibody decreased from 95 to 75% when the sample system was changed from PBS to fat-free milk (27). Thus, it is also possible that the soluble proteins in milk blocked some antigens on the spore or some antigen-binding sites on the surfaces of antibody-coated beads and thereby reduced the efficiency of specific binding. These data suggest that it is possible to use immobilized recombinant antibody fragments as a bioprocessing aid to concentrate organisms from food for microbiological evaluation.
TABLE 2.
Fractions of B. cereus sporesa recovered from pasteurized whole milkb by the single-chain-antibody-coated immunomagnetic beads
| Type of beads | Incubation (°C) | % of sporesc
|
Total (%) | ||
|---|---|---|---|---|---|
| Unbound | Bound but removed by washing | Bound | |||
| Immunomagneticd | 4 | 62.3 (11.5) | 0.7 (0.3) | 37.0 (11.2) | 100 |
| Controle | 4 | 99.0 (0.1) | 0.9 (0.1) | 0.1 (0.05) | 100 |
Spore concentration, 5 × 104 spores/ml; incubation time, 60 min.
The aerobic plate count of milk sample was <2 × 102 CFU/ml before spiking with B. cereus spores.
Values are averages of three trials (numbers in parentheses are standard deviations).
Streptavidin-conjugated single-chain antibodies were immobilized on biotinylated magnetic beads.
The beads were biotinylated by N-hydroxysuccinimidobiotin, but the streptavidin-conjugated antibody was not added.
This study has demonstrated that active anti-B. cereus spore single-chain antibody conjugated to streptavidin can be expressed and recovered efficiently within 24 h. The recombinant fusion antibody functioned essentially in the same manner as the native antibody in immunoassays for pathogen detection and also exhibited the biotin binding characteristics of native streptavidin. Because the techniques to increase the productivity of soluble recombinant antibody (18) and scale-up and optimization of fermentation for mass production of single chain antibody (12, 23, 30) are available already, it may be possible to use this recombinant antibody in food processing and food testing or related industries. It could be applied to rapid immunoassay detection for bioselective concentration and potentially for removal of B. cereus spores or of other pathogens from liquid food systems to enhance food safety or to reduce the severity of required preservation processes.
ACKNOWLEDGMENTS
This study was supported by USDA National Research Initiative in Food Safety through competitive grant no. 93-03930 and by the Dairy Management, Inc., through the Southeast Dairy Foods Research Center.
Footnotes
Paper no. FSR97-45 of the Journal Series of the Department of Food Science, North Carolina State University, Raleigh.
REFERENCES
- 1.Argarana C E, Kuntz I D, Birken S, Axel R, Cantor C R. Molecular cloning and nucleotide sequence of the streptavidin gene. Nucleic Acids Res. 1986;14:1871–1882. doi: 10.1093/nar/14.4.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassam B J, Caetano-Anolles G, Gresshoff P M. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem. 1991;196:80–83. doi: 10.1016/0003-2697(91)90120-i. [DOI] [PubMed] [Google Scholar]
- 3.Bayer E A, Ben-Hur H, Hiller Y, Wilchek M. Postsecretory modifications of streptavidin. Biochem J. 1989;259:369–376. doi: 10.1042/bj2590369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Carrier A, Ducancel F, Settiawan N B, Cattolico L, Maillere B, Leonetti M, Drevet P, Menez A, Boulain J. Recombinant antibody-alkaline phosphatase conjugates for diagnosis of human IgGs: application to anti-HBsAg detection. J Immunol Methods. 1995;181:177–186. doi: 10.1016/0022-1759(94)00344-v. [DOI] [PubMed] [Google Scholar]
- 6.Chung C T, Niemela S L, Miller R H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubel S, Breitling F, Kontermann R, Schmidt T, Skerra A, Little M. Bifunctional and multimeric complexes of streptavidin fused to single chain antibodies (scFv) J Immunol Methods. 1995;178:201–209. doi: 10.1016/0022-1759(94)00257-w. [DOI] [PubMed] [Google Scholar]
- 8.Fluit A C, Torensma R, Visser M J C, Aarsman C J M, Poppelier M J J G, Keller B H I, Klapwijk P, Verhoef J. Detection of Listeria monocytogenes in cheese with the magnetic immuno-polymerase chain reaction assay. Appl Environ Microbiol. 1993;59:1289–1293. doi: 10.1128/aem.59.5.1289-1293.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gussow D, Clackson T. Direct clone characterization from plaques and colonies by the polymerase chain reaction. Nucleic Acids Res. 1989;17:4000. doi: 10.1093/nar/17.10.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harmon S M, Goepfert J M, Bennet R W. Bacillus cereus. In: Vanderzant C, Splittstoesser D F, editors. Compendium of methods for the microbiological examination of foods. 3rd ed. Washington, D.C: American Public Health Association; 1992. pp. 593–604. [Google Scholar]
- 11.Haukanes B, Kvam C. Application of magnetic beads in bioassays. Bio/Technology. 1993;11:60–63. doi: 10.1038/nbt0193-60. [DOI] [PubMed] [Google Scholar]
- 12.Huston J S, Mudgett-Hunter M, Tai M-S, McCartney J, Warren F, Haber E, Oppermann H. Protein engineering of single-chain Fv analogs and fusion proteins. Methods Enzymol. 1991;203:46–88. doi: 10.1016/0076-6879(91)03005-2. [DOI] [PubMed] [Google Scholar]
- 13.Hutchens T W, Porath J. Thiophilic adsorption of immunoglobulins-analysis of conditions optimal for selective immobilization and purification. Anal Biochem. 1986;159:217–226. doi: 10.1016/0003-2697(86)90331-3. [DOI] [PubMed] [Google Scholar]
- 14.Insalata N F, Mahnke C W, Dunlap W G. Rapid, direct fluorescent-antibody method for the detection of salmonellae in food and feeds. Appl Microbiol. 1972;24:645–649. doi: 10.1128/am.24.4.645-649.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson K M, Nelson C L, Busta F F. Germination and heat resistance of Bacillus cereus spores from strains associated with diarrheal and emetic food-borne illnesses. J Food Sci. 1982;47:1268–1271. [Google Scholar]
- 16.Keen N T, Tamaki S. Structure of two pectate lyase genes from Erwinia chrysanthemi EC16 and their high level expression in Escherichia coli. J Bacteriol. 1986;168:595–606. doi: 10.1128/jb.168.2.595-606.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kipriyanov S M, Little M, Kropshofer H, Breitling F, Gotter S, Dubel S. Affinity enhancement of a recombinant antibody: formation of complexes with multiple valency by a single-chain Fv fragment-core streptavidin fusion. Prot Eng. 1996;9:203–211. doi: 10.1093/protein/9.2.203. [DOI] [PubMed] [Google Scholar]
- 18.Kipriyanov S M, Moldenhauer G, Little M. High level production of soluble single chain antibodies in small-scale Escherichia coli cultures. J Immunol Methods. 1997;200:69–77. doi: 10.1016/s0022-1759(96)00188-3. [DOI] [PubMed] [Google Scholar]
- 19.Koo K, Foegeding P M, Swaisgood H E. Construction and expression of a bifunctional single-chain antibody against Bacillus cereus spores. Appl Environ Microbiol. 1998;64:2490–2496. doi: 10.1128/aem.64.7.2490-2496.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kretz K, Callen W, Hedden V. Cycle sequencing. In: Dieffenbach C W, Dveksler G S, editors. PCR primer: a laboratory manual. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 527–536. [Google Scholar]
- 21.Liddell J E, Cryer A. A practical guide to monoclonal antibodies. West Sussex, United Kingdom: John Wiley and Sons; 1991. p. 156. [Google Scholar]
- 22.Lund A, Hellemann A L, Vartdal F. Rapid isolation of K88+Escherichia coli by using immunomagnetic particles. J Clin Microbiol. 1988;26:2572–2575. doi: 10.1128/jcm.26.12.2572-2575.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malby R L, Caldwell J B, Gruen L C, Harley V R, Ivancic N, Kortt A A, Lilley G G, Power B E, Webster R G, Colman P M, Hudson P J. Recombinant antineuraminidase single chain antibody: expression, characterization, and crystallization in complex with antigen. Proteins. 1993;16:57–63. doi: 10.1002/prot.340160107. [DOI] [PubMed] [Google Scholar]
- 24.Maloy S R. Experimental techniques in bacterial genetics. Boston, Mass: Jones and Bartlett; 1990. [Google Scholar]
- 25.McClelland R G, Pinder A C. Detection of Salmonella typhimurium in dairy products with flow cytometry and monoclonal antibodies. Appl Environ Microbiol. 1994;60:4255–4262. doi: 10.1128/aem.60.12.4255-4262.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merino S, Camprubi S, Tomas J M. Detection of Aeromonas hydrophila in food with an enzyme-linked immunosorbent assay. J Appl Bacteriol. 1993;74:149–154. doi: 10.1111/j.1365-2672.1993.tb03008.x. [DOI] [PubMed] [Google Scholar]
- 27.Molloy P, Brydon L, Porter A J, Harris W J. Separation and concentration of bacteria with immobilized antibody fragments. J Appl Bacteriol. 1995;78:359–365. doi: 10.1111/j.1365-2672.1995.tb03418.x. [DOI] [PubMed] [Google Scholar]
- 28.Neri D, de Lalla C, Petrul H, Neri P, Winter G. Calmodulin as a versatile tag for antibody fragments. Bio/Technology. 1995;13:373–377. doi: 10.1038/nbt0495-373. [DOI] [PubMed] [Google Scholar]
- 29.Phillips T M. Analytical techniques in immunochemistry. New York, N.Y: Marcel Dekker, Inc.; 1992. pp. 263–300. [Google Scholar]
- 30.Plückthun A. Antibodies from Escherichia coli. Handb Exp Pharmacol. 1994;113:269–315. [Google Scholar]
- 31.Quinlan J J, Foegeding P M. Monoclonal antibodies for use in detection of Bacillus and Clostridium spores. Appl Environ Microbiol. 1997;63:482–487. doi: 10.1128/aem.63.2.482-487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sano T, Cantor C R. Expression of a cloned streptavidin gene in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:142–146. doi: 10.1073/pnas.87.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt T G M, Skerra A. One-step affinity purification of bacterially produced proteins by means of the “strep tag” and immobilized recombinant core streptavidin. J Chromatogr Ser A. 1994;676:337–345. doi: 10.1016/0021-9673(94)80434-6. [DOI] [PubMed] [Google Scholar]
- 34.Skerra A, Pfitzinger I, Pluckthun A. The functional expression of antibody Fv fragments in Escherichia coli: improved vectors and a generally applicable purification technique. Bio/Technology. 1991;9:273–278. doi: 10.1038/nbt0391-273. [DOI] [PubMed] [Google Scholar]
- 35.Skerra A, Pluckthun A. Assembly of a functional immunoglobulin Fv fragment in Escherichia coli. Science. 1988;240:1038–1041. doi: 10.1126/science.3285470. [DOI] [PubMed] [Google Scholar]
- 36.Skjerve E, Rorvik L M, Olsvik O. Detection of Listeria monocytogenes in foods by immunomagnetic separation. Appl Environ Microbiol. 1990;56:3478–3481. doi: 10.1128/aem.56.11.3478-3481.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 38.Swaisgood H E. Characteristics of edible fluids of animal origin: milk. In: Fennema O R, editor. Food chemistry. 2nd ed. New York, N.Y: Marcel Dekker, Inc.; 1985. pp. 791–828. [Google Scholar]
- 39.Torensma R, Visser M J C, Aarsman C J M, Groebbe-Heij A, Poppelier M J J G, van Beurden R, Fluit A C, Verhoef J. Monoclonal antibodies that identify gram-negative bacteria using the magnetic immunoluminescence assay. J Microbiol Methods. 1992;15:135–142. [Google Scholar]
- 40.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walsh M K, Swaisgood H E. An Escherichia coli plasmid vector system for production of streptavidin fusion proteins: expression and bioselective adsorption of streptavidin-β-galactosidase. Biotechnol Bioeng. 1994;44:1348–1354. doi: 10.1002/bit.260441111. [DOI] [PubMed] [Google Scholar]