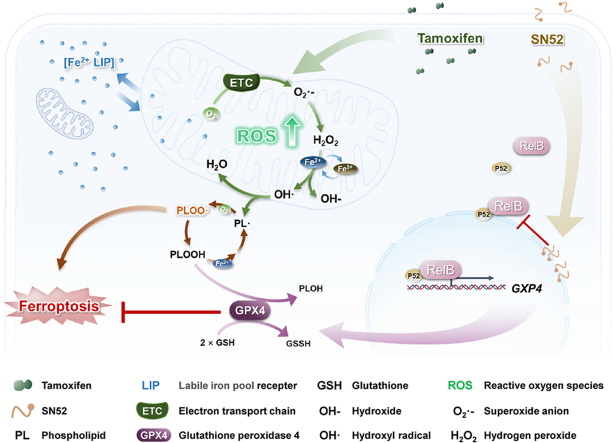
Abstract
Tamoxifen (TAM) resistance remains a major obstacle in the treatment of advanced breast cancer (BCa). In addition to the competitive inhibition of the estrogen receptor (ER) signaling pathway, damping of mitochondrial function by increasing reactive oxygen species (ROS) is critical for enhancing TAM pharmacodynamics. Here, we showed that RelB contributes to TAM resistance by inhibiting TAM-provoked ferroptosis. TAM-induced ROS level promoted ferroptosis in TAM-sensitive cells, but the effect was alleviated in TAM-resistant cells with high constitutive levels of RelB. Mechanistically, RelB inhibited ferroptosis by transcriptional upregulating glutathione peroxidase 4 (GPX4). Consequently, elevating RelB and GPX4 in sensitive cells increased TAM resistance, and conversely, depriving RelB and GPX4 in resistant cells decreased TAM resistance. Furthermore, suppression of RelB transcriptional activation resensitized TAM-resistant cells by enhancing ferroptosis in vitro and in vivo. The inactivation of GPX4 in TAM-resistant cells consistently resensitized TAM by increasing ferroptosis-mediated cell death. Together, this study uncovered that inhibition of ferroptosis contributes to TAM resistance of BCa via RelB-upregulated GPX4.
Keywords: Tamoxifen resistance, Breast cancer, ROS, Ferroptosis, GPX4, RelB
Graphical abstract
1. Introduction
Tamoxifen (TAM) is the first line of a therapeutic drug to treat premenopausal women diagnosed with estrogen receptor-postive (ER+) breast cancer (BCa) [1]. TAM was initially designed to block the interaction between estrogen and ER for treating ER+ BCa [2]. In addition, mounting evidence has demonstrated that TAM also perturbs mitochondrial function due to increased reactive oxygen species (ROS) [3,4]. Thus, TAM not merely induces cell death in ER+ BCa cells but also inhibits many other types of tumor cell independence of ER [5,6]. As a result, BCa patients have received therapeutic benefits from TAM. Unfortunately, most patients eventually develop more aggressive phenotypes that were resistant to TAM [7]. The dysfunction of ER signaling is thought to be the main consequence of TAM resistance. Nevertheless, several studies highlighted that TAM resistance in some patients was not due to the inactivation of or loss of ER [8,9]. In this regard, the deficiency of the mitochondrial function by excessive ROS is involved in BCa TAM resistance [3,10,11].
Increasing ROS to the threshold level of cell death is the principle for the most convenient radio- and chemo-therapeutic strategies [12,13]. To date, programmed cell death is widely recognized to be involved in anticancer treatment, such as apoptosis and necroptosis [14,15]. In addition, ferroptosis is a newly discovered iron-dependent regulated cell death involved in cancer progression and therapeutic response [[16], [17], [18]]. Ferroptosis resulted from extensive phospholipids peroxidation and led to membrane structure instability. It is caused by hydroxyl radicals generated through the Fenton reaction [19]. Therefore, increasing ROS generation and/or reprogramming iron metabolism are essential for promoting ferroptosis [20]. Consequently, the cellular antioxidant defense systems prevent ROS-induced ferroptosis by inhibiting iron-dependent lipid peroxidation [21,22]. Among antioxidant enzymes, glutathione peroxidase 4 (GPX4) is critical to block ferroptosis via glutathione-dependent inhibition of lipid peroxidation [23]. Furthermore, recent evidence revealed that FSP1, a glutathione-independent ferroptosis inhibitor, can prevent lipid peroxidation via activation of CoQ oxidoreductase [21,24]. Mechanistically, multiple signaling pathways are relevant to the inhibition of ferroptosis in cancer progression and therapeutic response, including NF-κB, PI3K/AKT/mTOR, and Stat3 [[25], [26], [27]].
NF-κB, a redox-sensitive transcription factor, participates in the adaptive activation of antioxidant response for activating the pro-survival pathways [28]. In particular, the activation of the NF-κB pathway plays a crucial role in cancer progression and therapeutic resistance [29]. Numerous studies have demonstrated that the NF-κB activation contributes to BCa endocrine therapy resistance, such as TAM [30,31]. The NF-κB pathway contributes to inhibition of apoptosis, pyroptosis, and necrosis. Moreover, recent studies have shown that the NF-κB pathway is also implicated in regulating ferroptosis [32]. In addition to the well-documented canonical NF-κB pathway, the RelB-activated noncanonical NF-κB pathway also contributes to cancer progression [33]. Previously, we reported that RelB enhances prostate cancer radioresistance and immune evasion by upregulating MnSOD and PD-L1 [34,35]. Recently, we showed that the constitutive level of RelB increased in advanced BCa, and repression of RelB transcriptional activation suppressed BCa tumorigenesis [36].
This present study aims to examine whether ferroptosis inhibition is involved in BCa TAM resistance. We found that TAM-induced ROS enhanced ferroptosis in BCa cells, and vice versa; activation of GPX4 resensitized the cells to TAM by inhibiting ferroptosis. Mechanistically, RelB activation inhibited ferroptosis by upregulating GPX4, and consistently suppression of RelB-activated GPX4 resensitized the resistant BCa cells to TAM.
2. Results
2.1. ROS-induced ferroptosis relevant to TAM-induced cell death
To investigate the TAM-resistant (TAMR) mechanisms, we used ER+ BCa cell lines (MCF7 and T47D) to establish their TAMR cell lines (MCF7/TAMR and T47D/TAMR). Additionally, triple-negative breast cancer (TNBC) MDA-MB-231 cell line characterized TAM insensitive [37], was included as an innate TAMR control. After TAM treatment, MTT and colony survival assays confirmed the TAMR phenotype. Although the growth rates of TAMR lines were equivalent to their parental cell lines in the untreated condition (Fig. S1A), the half-inhibitory concentration (IC50) of TAM significantly increased in TAMR cell lines compared to TAM-sensitive (TAMS) cell lines (Figs. S1B and C). Colony survival assay further confirmed the TAMR phenotype (Figs. S1D and E). In parallel, ERα decreased but drug-resistant marker BCRP increased in TAMR cells (Fig. S1F).
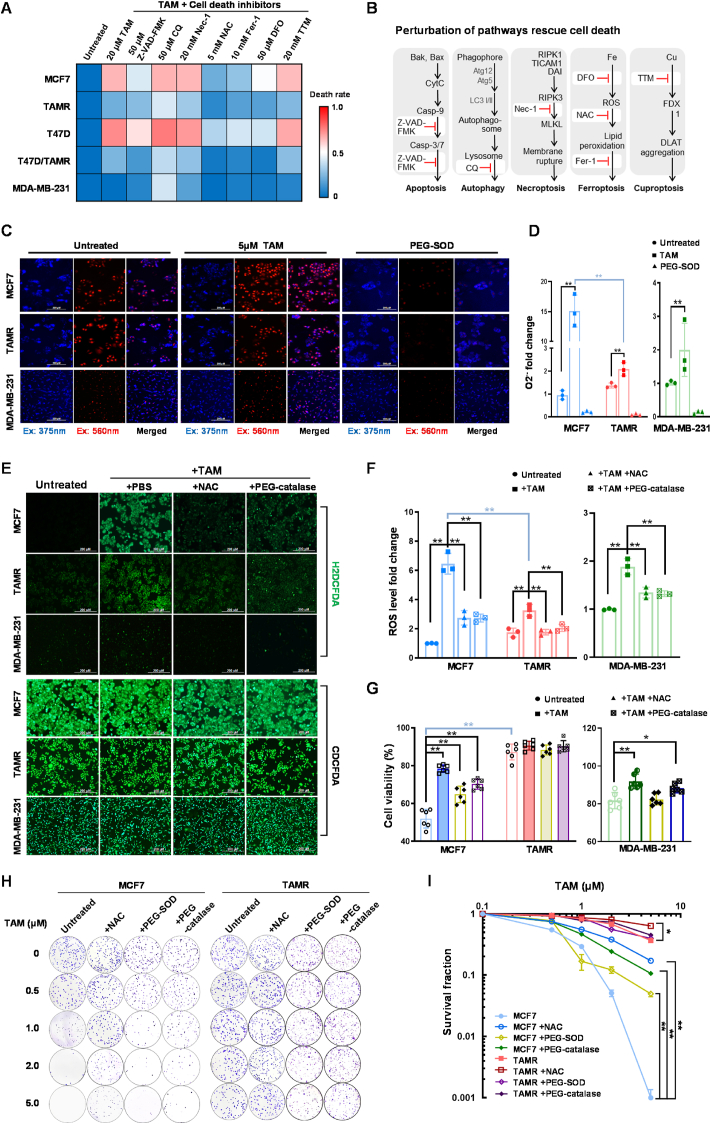
The programmed cell death mechanisms involve apoptosis, necroptosis, autophagy, ferroptosis, and cuproptosis [38]. Numerous studies have reported that TAM efficiently induced apoptotic cell death in BCa [39,40]. To examine whether other mechanisms are implicated in TAM-induced cell death, we pretreated both TAMS and TAMR BCa cell lines with multiple cell death inhibitors to block TAM-induced cytotoxicity (Fig. 1A and B). Intriguingly, the pan-caspase inhibitor (Z-VAD-FMK) was essential but not sufficient to rescue the cells against TAM, suggesting that the apoptotic pathway is not only responsible for TAM-induced cell death. Notably, inhibitors of ferroptosis including deferoxamine (DFO), ferrostatin-1 (Fer-1), and N-acetylcysteine (NAC) also contributed to protect the cell survival, indicating that ferroptosis is involved in TAM resistance. However, there were no significant protective effects observed with other types of cell death inhibitors, such as necrostatin-1 (Nec-1), chloroquine (CQ), and tetrathiomolybdate (TTM), which target necroptosis, autophagy, and cuproptosis, respectively.
Fig. 1.
TAM-induced cytotoxicity by increasing ROS level. (A) Multiple cell death inhibitors were used to examine their protective effects on survival of BCa cells against Tamoxifen (TAM), including 20 mM necrostatin-1 (Nec-1), 10 mM ferrostatin-1 (Fer-1), 5 mM N-acetylcysteine (NAC), 50 μM deferoxamine (DFO), 50 μM Z-VAD-FMK, 50 μM chloroquine (CQ) or 20 μM tetrathiomolybdate (TTM). The cells were pretreated with the inhibitors for 12 h and then treated with 20 μM TAM for 48 h. MTT assay was performed to quantify cell survival as indicated in a heatmap (n = 3). (B) Schematic diagrams of apoptosis, autophagy, necroptosis, cuproptosis, and ferroptosis. Inhibitors targeting the relative cell death pathways are marked in red colors. (C, D) Superoxide anions in the treated cells were quantified using a DHE fluorescent probe (DHE fluorescence indicated as blue color and its oxidative products indicated as red color). PEG-SOD was used as a negative control to remove superoxide anions (n = 3). (E, F) The levels of ROS were quantified using an H2DCFDA probe with CDCFDA normalization. NAC and PEG-catalase were used as negative controls to remove specific types of ROS (n = 3). (G) The cytotoxicity of TAM was estimated using the MTT assay. (H, I) TAM-mediated cytotoxicity was estimated by colony survival assay (n = 3). Data are shown as mean ± SD, * (p < 0.05) and **(p < 0.01); t-test (D-right), two-way ANOVA (D-left, F–H). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Subsequently, we quantified the levels of ROS in TAM-treated BCa cells. Firstly, cellular superoxide anion (O2•−) was determined using dihydroethidium (DHE) probe. TAM significantly increased the levels O2•− in TAMS cells, but its effect was attenuated in TAMR cells (Fig. 1C and D; Figs. S2A and B). In addition, we used a mitochondrial superoxide indicator (MitoSOX) with Hoechst dye normalization to quantify O2•− in mitochondria. TAM induced mitochondrial O2•− anion was removed by PEG-SOD (Figs. S2C and D). In addition, we used a H2DCFDA probe with CDCFDA dye normalization to quantify total ROS in cells. TAM continuously increased the radicals in TAMS cells compared to TAMR cells, and the effects were further abolished by adding antioxidant agents, such as NAC and PEG-catalase (Fig. 1E and F; Figs. S2E and F). Furthermore, we included a DAF-FM DA probe to quantify the levels of nitric oxide (NO) in TAM-treated cells. Consistently, TAM induced NO production in TAMS cells, but its effect was alleviated in TAMR cells, suggesting that NO is involved in TAM-induced cytotoxicity (Figs. S2G and H).
Moreover, we assessed the effects of antioxidant agents on the protection of BCa cells by counteracting TAM-induced ROS. As expected, TAM increased the higher levels of cytotoxicity in TAMS cells compared to TAMR cells. PEG-catalase efficiently protected the cells against TAM by removing hydrogen peroxide and total ROS, whereas a less protective effect was observed in cells pretreated with PEG-SOD to eliminate O2•− anion (Fig. 1G–I; Figs. S3A–J). Together, these results suggested that TAM-induced cell death is partly mediated via ROS-provoked ferroptosis, regardless of its impact on ER suppression. Thus, the elimination of ROS contributes to enhancing cell tolerance to TAM treatment.
2.2. TAM-stimulated ferroptosis in BCa cells
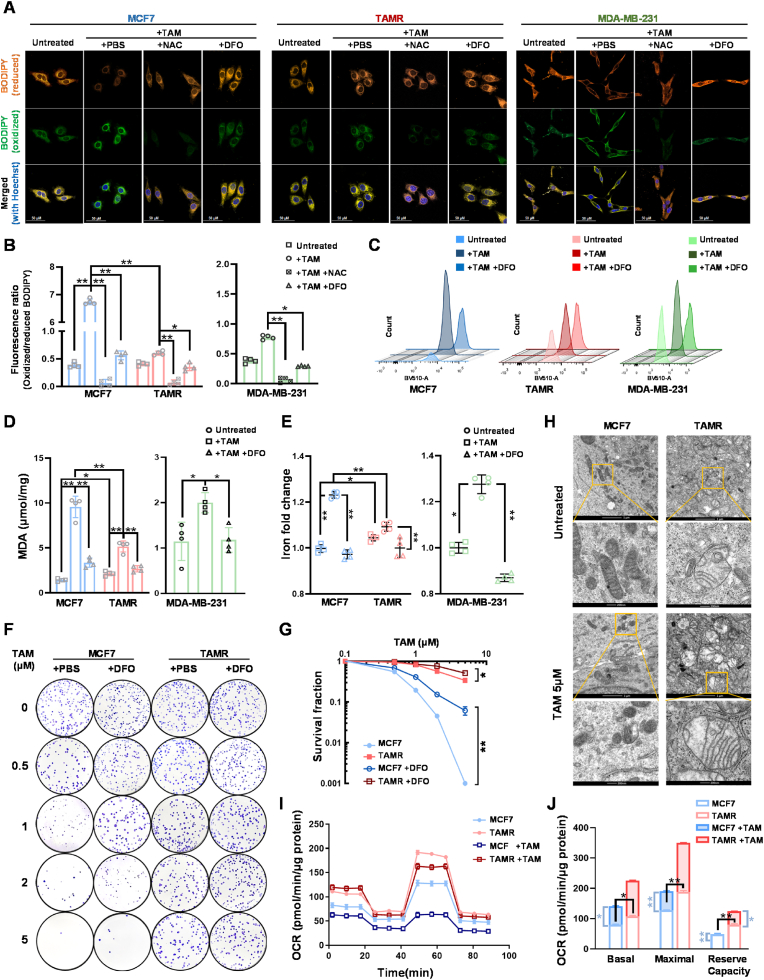
To examine whether TAM-induced ferroptosis in BCa cells, we used BODIPY C11, a fluorescent probe specifically sensitive to lipid peroxidation, to estimate the level of TAM-stimulated cell membrane oxidation. The relative lipid peroxidation was examined using spectrophotometry, flow cytometer, and confocal microscopy. After TAM treatment, increases in oxidized lipid forms in TAMS cells vs. TAMR cells were detected by confocal microscopy, following the emission of the BODIPY dye shifting from 590 nm (orange) to 510 nm (green) upon oxidation. The results indicated that 12 h after TAM treatment efficiently induced lipid peroxidation in TAMS cells, although the initial lipid peroxidation in TAMR cells was higher than in TAMS cells (Figs. S4A and B). Notably, iron chelator DFO efficiently abolished the TAM-induced lipid peroxidation, indicating that ferroptosis is involved in TAM-mediated cell death (Fig. 2A; Fig. S4C). Oxidized and reduced fluorescent intensities were further measured by spectrophotometry. The results showed that the ratio of oxidized/reduced BODIPY significantly increased in TAMS cells compared to TAMR cells (Fig. 2B; Fig. S4D). Furthermore, oxidized lipids in the living cells were quantified by flow cytometry at 510 nm and confirmed that the number of cells with high levels of lipid oxidation increased in TAMS cells vs. TAMR cells, which was blocked by DFO (Fig. 2C; Fig. S4E). Malondialdehyde (MDA), a marker of a metabolite of intracellular lipid oxidation, was also used to verify TAM-induced ferroptosis. TAM increased the level of MDA in TAMS cells, and its effect was eliminated in TAMR cells or DFO-pretreated cells (Fig. 2D; Fig. S4F).
Fig. 2.
TAM-enhanced ferroptosis in BCa cells. (A) After treatment, lipids and relative oxidized forms in living cells were imaged using a BODIPY 581/591 probe conjugated with dual fluorescent dyes under confocal microscopy. DFO served as a control to reduce the amount of oxidized lipids. (B) In addition, the levels of lipids and oxidative lipides were quantified by spectrophotometer using a microplate reader. The fluorescence ratio of oxidized/reduced BODIPY is indicated in a bar graph (n = 4). (C) Furthermore, TAM-induced lipid peroxidation was quantified by flow cytometry with the BODIPY 581/591 probe. (D) TAM-induced lipid peroxidation was validated using an MDA assay (n = 4). (E) The contents of iron ion in the TAM-treated cells were measured using an iron colorimetric assay (n = 4). (F, G) After DFO-TAM treatment, the cell survival rates in different types of cell lines were quantified by colony survival assay (F). The DFO protective effect on cell survival against TAM was calculated in (G) (n = 3). (H) Mitochondrial damage in TAM-treated cells was examined by TEM. (I, J) Mitochondrial OCR in the treated cells was measured using a Seahorse XF-96 analyzer. Supplemental reagents including oligomycin, FCCP and Antimycin A/Rotenone were automatically injected into the analyzing plates to determine OCR of basal, ATP-linked, maximal and background image, respectively (I). OCR in the different periods were calculated in (J). Data are shown as mean ± SD, * (p < 0.05) and **(p < 0.01); one-way ANOVA (B-, D-, E-right), two-way ANOVA (B-, D-, E-left, G, J).
Furthermore, we used 2,2′-bipyridine to measure intracellular ferrous (Fe2+) concentration, a bidentate nitrogen-based ligand selectively binding to Fe2+ ions but not ferric (Fe3+) ions. TAM increased Fe2+ levels in TAMS cells but not in TAMR cells. As expected, pretreating with DFO efficiently alleviated Fe2+ levels in TAMS cells (Fig. 2E; Fig. S4G). Moreover, the results from cell survival experiments showed that although DFO was not toxic to cells, it could rescue cell survival from TAM-induced cytotoxicity in TAMS cells (Fig. 2F and G; Figs. S5A–H). Nevertheless, DFO provided only slight protection in TAMR cells, and no protection found in ER− cells, indicating other mechanisms involved in TAM-induced cell death, like ER inactivation.
Since ferroptosis is relevant to mitochondria dysfunction via membrane damage by lipid peroxidation [17,41], we further analyzed the mitochondria morphology in TAM-treated cells using a transmission electron microscope (TEM). Expectedly, after TAM treatment, mitochondria in TAMS cells seemed to disappear from normal structure; typically flattened irregular shape without cristae compared to those in the untreated cells. In contrast, more intact mitochondria presence in TAMR cells showed a normal enlarged oval shape with intact inner membranes and expanded cristae. TAM appeared less toxic effect on damaging mitochondrial structure in TAMR cells, indicating the capability of the cells to maintain redox homeostasis (Fig. 2H). In addition, mitochondrial membrane potential (ΔΨm) was analyzed using a JC-1 assay to quantify the ratio of aggregated form to monomer. TAM reduced assembly of JC-1 in TAMS cells compared to slight effects on TAMSR cells, indicating that TAM induced mitochondrial damage in TAMS cells (Figs. S6A and B).
To further assess mitochondrial function, we measured the mitochondrial oxygen consumption rate (OCR) using a Seahorse XF Analyzer. Under untreated condition, TAMR cells exhibited higher basal mitochondrial respiration than TAMS cells. TAM decreased the basal, maximal respiration and spare respiratory capacity in TAMS cells, while its effect on the respiratory capacity of TAMR cells was limited (Fig. 2I and J; Figs. S6C–F). The results indicated that TAM impaired mitochondrial respiration by compromising their maximal respiration and spare respiratory capacity, which was potentially linked to membrane damage caused by lipid peroxidation. In contrast, TAMR cells could maintain mitochondrial structure, maximal respiration, and spare respiratory capacity. Together, these results suggested that TAM impaired mitochondrial function by inducing ferroptosis in BCa cells.
2.3. RelB activation inhibits ferroptosis in TAMR cells
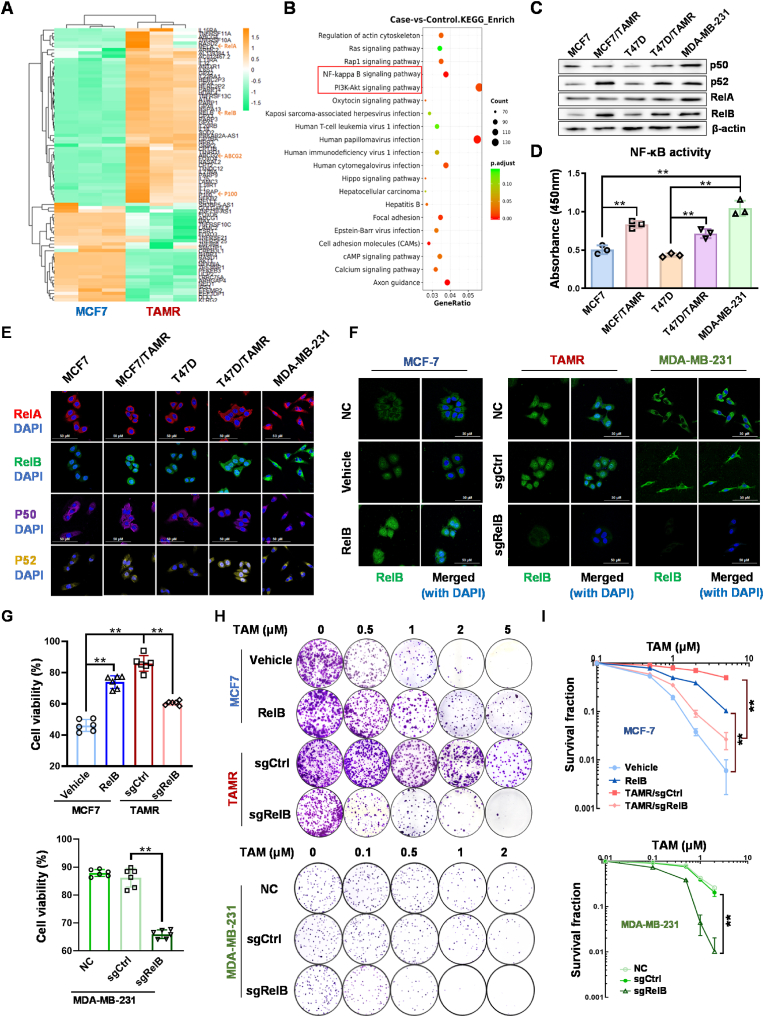
To investigate the mechanism underlying TAM-induced ferroptosis, we performed RNA-seq to analyze transcriptomes in TAMR cells vs. TAMS cells (Fig. 3A). The data assessment by the enhanced KEGG signaling pathway revealed multiple pathways relevant to TAMR phenotype, including PI3K-Akt and NF-κB (Fig. 3B). Since PI3K-Akt is thought to be an upstream signaling in regulating the NF-κB pathway [42,43], we predicted that the NF-κB pathway plays a predominant role in developing the BCa TAMR phenotype. In this regard, we examined the NF-κB DNA-binding activity and the expression levels of NF-κB family members in TAMS cells vs. TAMR cells. Intriguingly, compared to the p50:RelA dimer-based canonical NF-κB pathway, the activation of the p52:RelB dimer-based noncanonical NF-κB pathway mainly contributes to increasing the DNA-binding activity (Fig. 3C and D). Additionally, the nuclear levels of the p52:RelB dimer were higher in TAMR cells than TAMS cells, but the p50:RelA dimer was unlike (Fig. 3E).
Fig. 3.
Activation of RelB in TAMR cells. (A) The profiles of transcripts in TAMS cells vs. TAMR were analyzed by RNA-seq. The members of the NF-κB family are indicated by arrows (n = 3). (B) The potential signal pathways relevant to TAM resistance were assessed by the KEGG pathway enrichment analysis. The PI3K-AKT-NF-κB axis is indicated in a red box. (C) The protein levels of the NF-κB family members in TAMS cells vs. TAMR cells were quantified by immunoblotting. (D) The relative NF-κB DNA binding activities were measured using a ELISA kit with a standard NF-κB probe (n = 3). (E) Nuclear translocation of the NF-κB family members was imaged by confocal microscopy with a DAPI co-localization. (F) The nuclear RelB levels in RelB-overexpressed and TAMR cells vs. TAMS cells were quantified by confocal microscopy. (G) TAM-induced cytotoxicity in RelB-overexpressed TAMS cells and RelB-deprived TAMR cells was estimated by MTT assay (n = 6). (H, I) Furthermore, cell survival rates in the RelB-manipulated cells were confirmed by colony survival assay (n = 3). Data are shown as mean ± SD, * (p < 0.05) and **(p < 0.01); one-way ANOVA (D, G), two-way ANOVA (I). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To verify whether the noncanonical NF-κB pathway fosters BCa TAM resistance, we asymmetrically manipulated RelB expression via either ectopically expressing RelB in TAMS cells or knocking out endogenous RelB in TAMR cells using CRISPR/Cas9 gene edition (Fig. S7A). Correspondingly, the relative nuclear RelB was confirmed by confocal microscopy (Fig. 3F; Fig. S7B). Notably, the cell viability and survival rate were correlated to the levels of nuclear RelB. Elevating RelB in TAMS cells led to increased TAM resistance. Conversely, the depriving RelB in TAMR cells resulted in resensitized the cells to TAM, suggesting that RelB contributes to BCa TAM resistance (Fig. 3G–I; Fig. S7C-L).
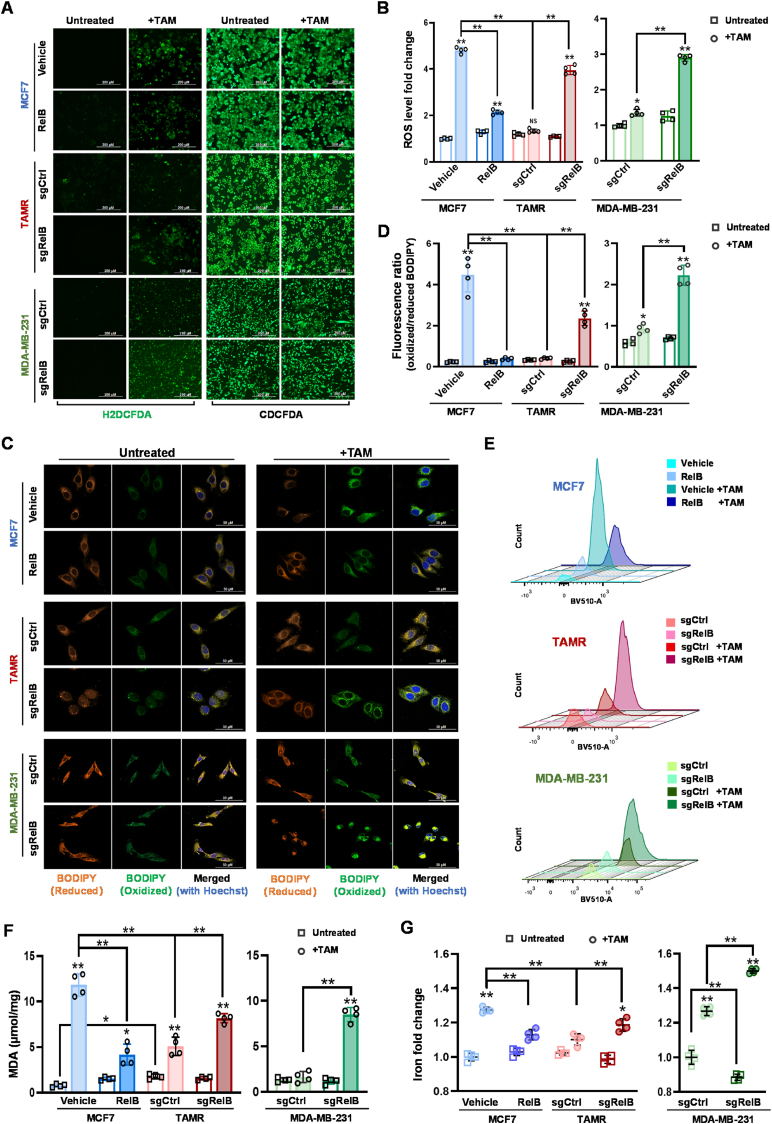
To examine whether the RelB enhances BCa TAM resistance by suppressing ROS-induced ferroptosis, we verified ROS-induced ferroptosis in RelB-manipulated BCa cells. Under untreated condition, increasing RelB decreased ROS in TAMS cells. In contrast, knockout of RelB increased ROS in TAMR cells (Fig. 4A and B; Figs. S8A and B). Consequently, the rise of RelB inhibited TAM-induced lipid peroxidation in TAMS cells. Conversely, the deprivation of RelB enhanced TAM-induced lipid peroxidation in TAMR cells (Fig. 4C–E; Figs. S8C–E). Additionally, the MDA measurement further confirmed that RelB was sufficient to suppress lipid peroxidation induced by TAM (Fig. 4F; Fig. S8F). In parallel, intracellular Fe2+ was susceptible to TAM treatment, while the level of Fe2+ in the cells could be manipulable via the administration of RelB (Fig. 4G; Fig. S8G).
Fig. 4.
The effect of RelB on the induction of ferroptosis. (A, B) The levels of ROS in RelB-manipulated MCF7 cells were measured using a H2DCFDA probe with CDCFDA normalization (A). TAM-induced ROS in each cell line was plotted (B) (n = 4). (C) TAM-induced lipid peroxidation in RelB-manipulated cells was imaged by confocal microscopy with a BODIPY 581/591 probe. (D) After TAM-treatment, amounts of lipids and oxidative lipids in RelB-manipulated cells were measured by spectrophotometer. The ratio of oxidized/reduced BODIPY is indicated in a bar graph (n = 4). (E) TAM-induced lipid peroxidation in RelB-manipulated cells was confirmed by flow cytometry. (F) Furthermore, lipid peroxidation generated in RelB-manipulated cells was validated by MDA assay (n = 4). (G) The contents of iron ion in RelB-manipulated cells were measured using an iron colorimetric assay (n = 4). Data are shown as mean ± SD, * (p < 0.05) and **(p < 0.01); two-way ANOVA.
2.4. The reverse of TAMR phenotype via blockage of RelB nuclear translocation
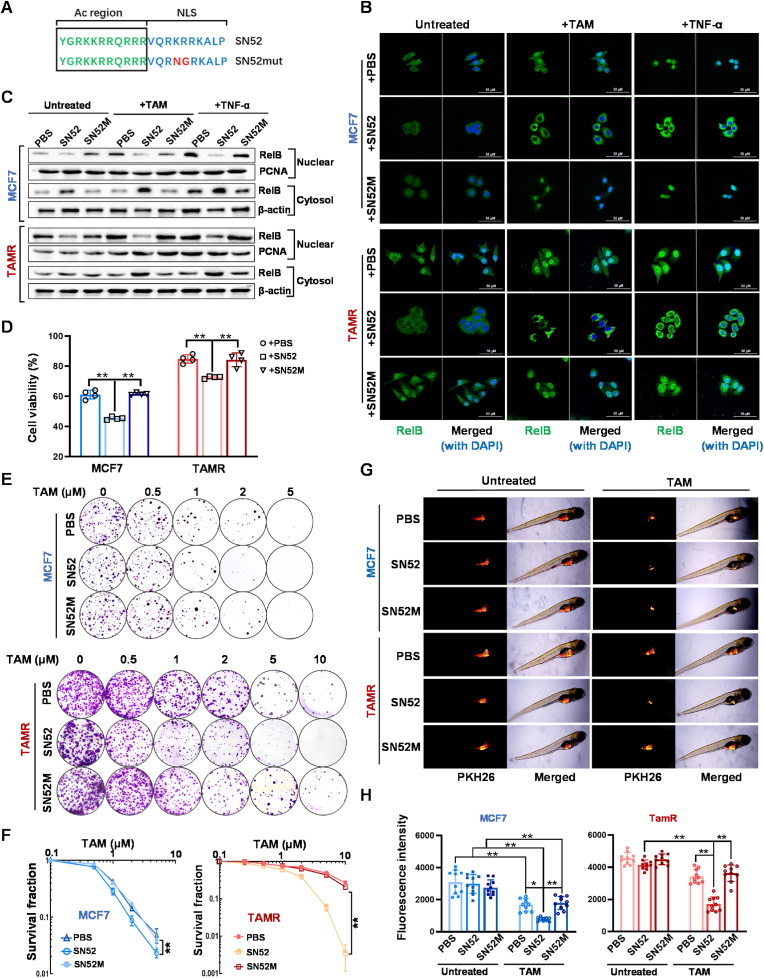
We have previously designed an SN52 peptide that specifically prevents nuclear translocation of p52:RelB dimer [44]. In this study, we further modified the SN52 inhibitor by improving its cell permeability via acetylation. In parallel, SN52 M, a mutant-type peptide, was included as a control (Fig. 5A). BCa cells were pretreated with SN52 and followed by TAM or TNF-α treatment. The effect of SN52 on RelB nuclear translocation was examined by confocal microscopy. As expected, SN52 sufficiently blocked TAM- or TNF-α-induced RelB nuclear translocation, but SN52 M was unlike (Fig. 5B), which was confirmed by immunoblotting (Fig. 5C). Intriguingly, the results of cytotoxic analyses showed that SN52 remarkably resensitized the resistant cells to TAM irrespective of a slightly increasing the cytotoxicity in TAMS cells (Fig. 5D–F).
Fig. 5.
Resensitization of TAM via SN52-inhibited RelB nuclear translocation. (A) a SN52 peptide and its mutant control SN52 M with an acetylation (Ac) region for improving cell permeability were redesigned to inhibit RelB nuclear translocation. The Ac-region provided cell permeability, and the nuclear localization signal (NLS) was designed to compete with p52. The mutant NLS indicated in SN52 M served as a control. (B) Nuclear RelB levels in SN52-pretreated cells were imaged by confocal microscopy. (C) The levels of cytoplasmic and nuclear RelB in SN52-pretreated cells were measured by immunoblotting. (D) The effect of SN52 on enhanced TAM cytotoxicity was estimated by MTT assay (n = 4). (E, F) SN52 effect on cell survival in TAMS cells vs. TAMR cells was confirmed by colony survival assay (n = 3). (G, H) Furthermore, the SN52 effect on tumor growth was validated using a zebrafish xenograft tumor model (n = 10). Data are shown as mean ± SD, * (p < 0.05) and **(p < 0.01); two-way ANOVA.
In addition, the SN52-pretreated cells were labeled with a PKH26 fluorescent dye and then microinjected into zebrafish embryos to verify the effect of SN52 in vivo. TAM was added to the nutrient solutions 36 h after injection. Tumors formed as the fish grownup and were tracked daily by fluorescent imaging. Although TAM appeared no toxic effect on zebrafish growth and survival, TAM significantly reduced the tumor growth in the group injected with TAMS cells. In contrast, the drug efficiency was alleviated in the group injected with TAMR cells. Notably, SN52 resensitized the resistant group to TAM, suggesting that the suppression of RelB by SN52 is sufficient to reverse the BCa TAMR phenotype (Fig. 5G and H).
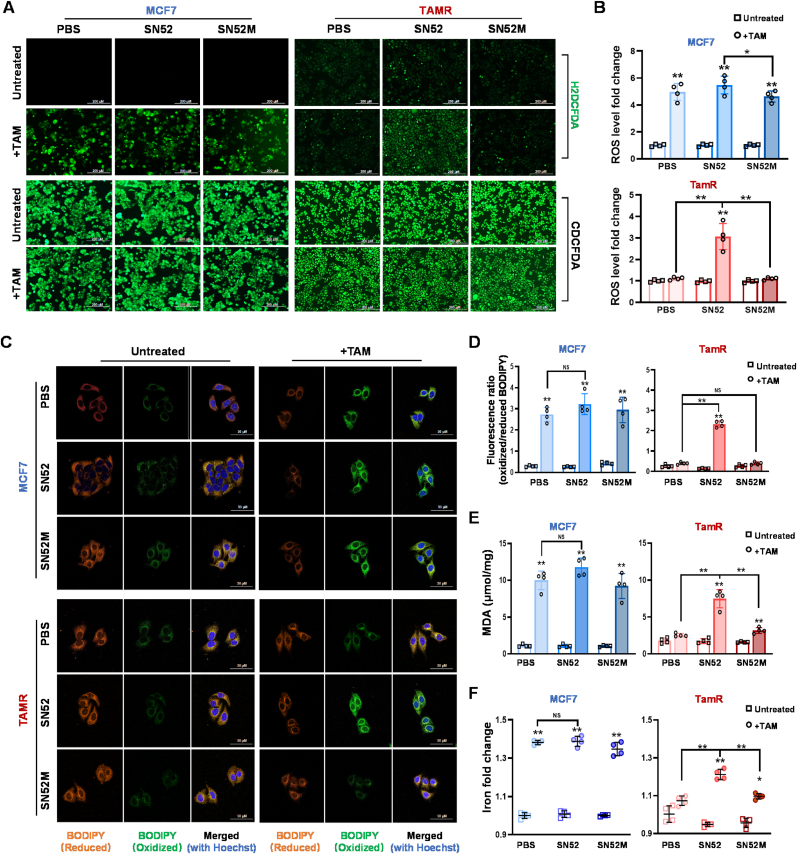
Moreover, we examined whether the SN52-mediated TAM re-sensitization was due to increased ferroptosis. As expected, SN52 increased ROS levels in TAMR cells after TAM treatment irrespective of no effect on ROS generation in TAMS cells (Fig. 6A and B). Additionally, SN52 elevated lipid peroxidation levels in TAMR cells, even if no further increase in ferroptosis was observed in TAMS cells (Fig. 6C and D). Consistently, SN52 highly increased MDA and Fe2+ concentrations in TAMR cells compared to no effect observed in TAMS cells (Fig. 6E and F).
Fig. 6.
Inhibition of ferroptosis via SN52 suppressing RelB. (A, B) The ROS levels in TAM-treated cells were quantified using an H2DCFDA probe (A). ROS fold changes in SN52-pretreated cells as indicated in (B) (n = 4). (C) Oxidized lipids in SN52/TAM-treated cells were imaged using a BODIPY fluorescent probe under confocal microscopy. (D) The ratio of oxidized/reduced BODIPY in the cells were analyzed by spectrophotometer (n = 4). (E) The levels of lipid peroxidation in the cells were quantified by MDA assay (n = 4). (F) The contents of iron ions in the cells were measured using an iron quantification kit (n = 4). Data are shown as mean ± SD, * (p < 0.05) and **(p < 0.01), two-way ANOVA.
2.5. RelB-activated GPX4 in TAMR cells
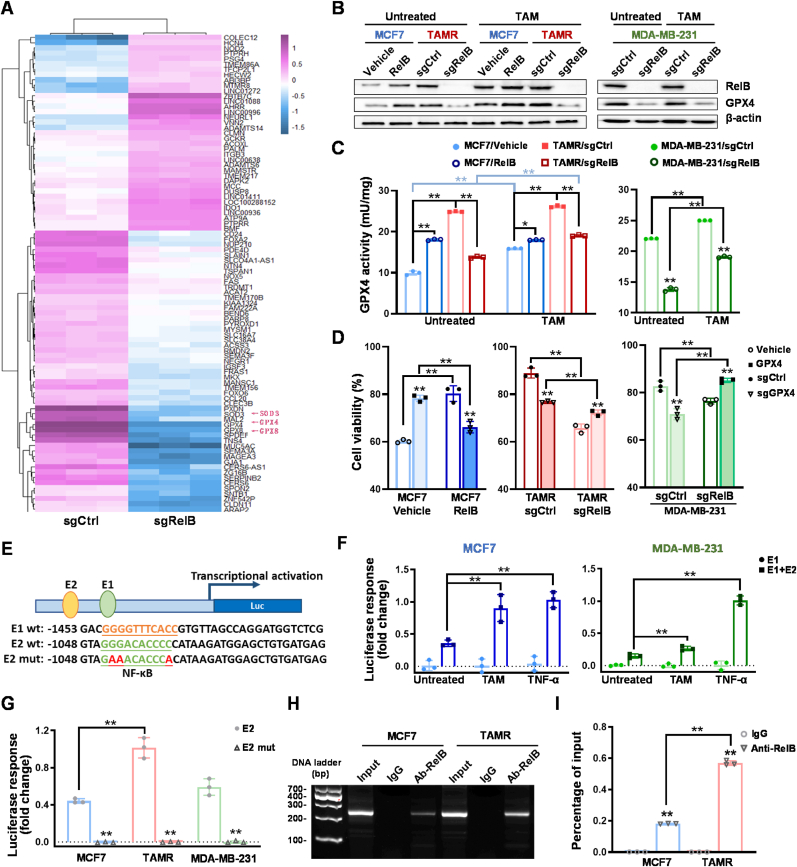
To discover RelB-regulated transcripts involved in BCa TAM resistance, we knocked out RelB in MDA-MB-231 cells and analyzed the profiles of transcripts by RNA-Seq. Compared to the transcripts in control cells, the mRNA levels of antioxidants decreased in the RelB-deprived cells, such as GPX4 (Fig. 7A), which was confirmed by immunoblotting (Fig. 7B). Consistently, RelB deprivation decreased the GPX4 enzymic activities in both TAMS and TAMR cells (Fig. 7C). To verify whether RelB directly regulate the expression of GPX4, we ectopically expressed GPX4 in TAMS cells and RelB-deprived TAMR cells (Fig. S9A); conversely, knocked out GPX4 in RelB-overexpressed cells and TAMR cells (Fig. S9B). Cytotoxicity showed that the elevation of GPX4 in RelB-deprived cells enhanced the cell resistance to TAM (Fig. 7D; Figs. S9C–F). In contrast, the deprivation of GPX4 in TAMR cells with high levels of RelB resensitized the cells to TAM (Fig. 7D; Fig. S9G-N).
Fig. 7.
RelB-upregulated GPX4 expression. (A) The transcriptomes in RelB KO vs. control MDA-MB-231 cells were analyzed by RNA-seq. The antioxidants including GPX4 are indicated by red arrows. (B) After TAM treatment, the protein levels of GPX4 in the RelB-manipulated cells were quantified by immunoblotting. (C) The relative GPX4 activity in TAM-treated cells was measured (n = 3). (D) After TAM treatment, TAM-treated cytotoxicity was estimated by MTT assay (n = 3). (E) A 5′-flanking region of the human GPX4 gene containing two putative NF-κB elements (E1 and E2) with a core promoter was cloned to drive the luciferase reporter expression. The E1 and E2 sites were further mutated as indicated by red letters. (F) TNF-α- and TAM-induced enhancer responses in TAMS and TAMR cells were estimated by β-gal-normalized luciferase reporter activities (n = 3). (G) The E2 site for enhancer response was determined using its mutant construct (n = 3). (H, I) Furthermore, the E2 site was precipitated using a RelB antibody and quantified by standard PCR (H) and qPCR (I) (n = 3). Chromatins without immunoprecipitation served as an input control, and IgG-precipitated chromatins served as a negative control. Data are shown as mean ± SD, * (p < 0.05) and **(p < 0.01), two-way ANOVA. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To elucidate how RelB regulates GPX4 expression, we identified two putative NF-κB elements (E1, E2) located in the 5′-flanking region of the human GPX4 gene. We constructed a luciferase reporter driven by the NF-κB enhancer fragments, including E1 + E2, E1 alone, or mutant E2 (Fig. 7E). TAM and TNF-α induced the reporter activity driven by the E1 + E2 fragment. However, no induction was observed in a truncated fragment only carrying E1, indicating that the E2 site is essential for the transcriptional activation (Fig. 7F). Consistently, the induction of reporter response was alleviated by mutating the E2 site (Fig. 7G). Accordingly, a RelB antibody was able to pull-down the E2 fragment, which was higher in chromatin derived from TAMR cells than TAMS cells (Fig. 7H and I). These results suggested that RelB-upregulated GPX4 contributed to TAM resistance by inhibiting ferroptosis.
2.6. RelB-activated GPX4 enhancing TAM resistance in mouse xenograft tumors
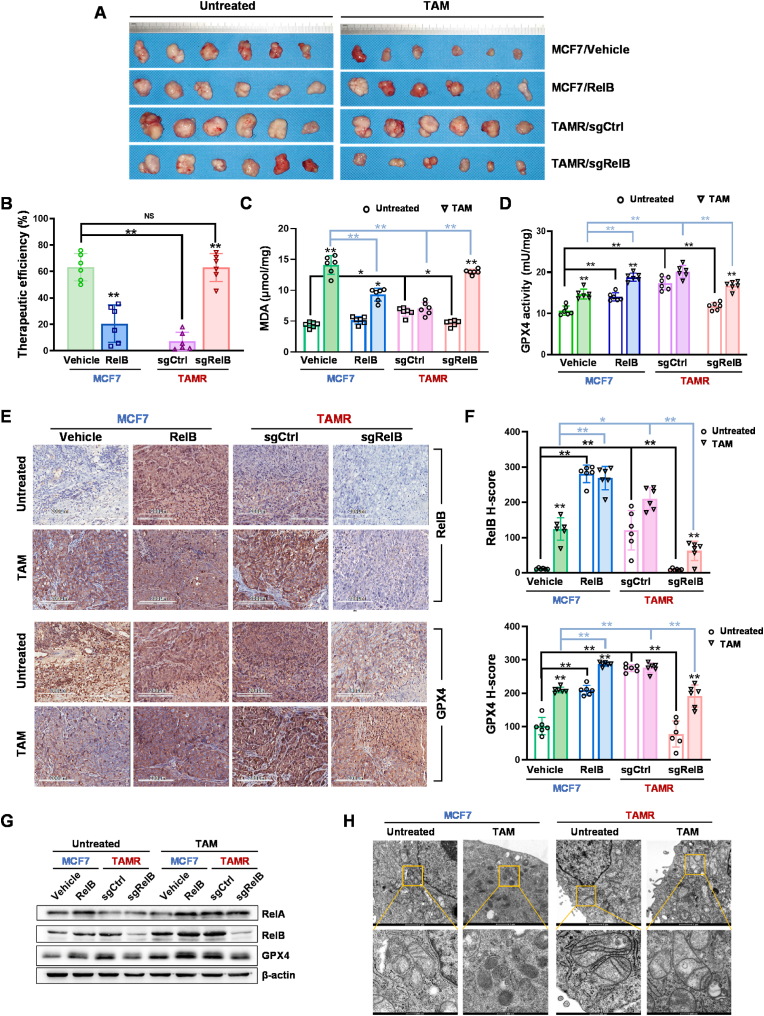
To validate the RelB-mediated GPX4 activation involved in the TAM resistance, we established a mouse xenograft tumor model by subcutaneously injecting MCF7 cells with different levels of RelB into mammary fat pads of female nude mice and measured tumor volume every other day. The tumor growth rate was significantly higher in the groups injected with RelB-overexpressed and TAMR cells than in the control group injected with wild-type MCF7 cells. However, the tumor growth slowed down in the RelB-deprived group. Mice were treated with TAM when tumor volume reached 500 mm3. Compared to the control group, the TAM therapeutic efficacy decreased in the RelB-overexpressed and TAMR groups, but the therapeutic efficacy was restored in the RelB-deprived TAMR group (Fig. 8A and B; Figs. S10A–D). TAM significantly induced MDA levels in the tumor tissues, but the induction was declined in RelB-overexpressed groups. Conversely, the level of MDA was decreased in TAMR groups, but it was recovered by depriving RelB (Fig. 8C). Consistently, the levels of GPX4 in tumor tissues were highly correlated to the levels of RelB manipulated in the injection cells. The GPX4 activity was adaptively induced by treating with TAM, and further increased in RelB-overexpressed and TAMR groups. Conversely, the GPX4 activity was decreased in the RelB-deprived TAMR groups (Fig. 8D).
Fig. 8.
Validation of RelB-enhanced TAM resistance in vivo. MCF7 cells with different levels of RelB were subcutaneously injected into the right mammary fat pads of female nude mice to form xenograft tumors (n = 6 in each group). Mice were treated with TAM (4 mg/kg) when tumor volume reached to 500 mm3. (A) The excised tumor tissues were photographed. (B) TAM therapeutic efficacies were assessed in 4 groups based on changed tumor volumes (n = 6). (C) The levels of lipid peroxidation in tumor tissues were quantified by MDA assay (n = 6). (D) The GPX4 enzymic activities in the tumor tissues were measured in each group (n = 6). (E, F) The levels of GPX4 and RelB in the tumor tissues quantified by IHC (E). The H-scores in each group are indicated in bar graphs (F) (n = 6). (G) further the levels of GPX4 and RelB expression in the tumor tissues were confirmed by immunoblotting (n = 6). (H) Mitochondrial structure in tumor tissues was examined by TEM. Data are shown as mean ± SD, * (p < 0.05) and **(p < 0.01), two-way ANOVA.
The relative protein levels in the tumor tissues were examined by IHC and further confirmed by immunoblotting. The deprivation of RelB led to increased RelA levels in tumor tissues, although no significant influence on RelA expression was observed in the RelB-overexpressed or TAMR groups. RelB and GPX4 were adaptively responsive to TAM and further increased in the TAMR groups, but their levels were alleviated by depleting RelB in TAMR cells (Fig. 8E–G). Correspondingly, mitochondria were impaired in TAM-treated tumor tissues, but the mitochondrial structure was intact in the TAMR groups (Fig. 8H). Taken together, these results validated that the RelB-GPX4 axis contributes to BCa TAM resistance by inhibiting ferroptosis, as illustrated in Graphical Abstract.
3. Discussion
BCa is the most common malignant tumor and the leading cause of cancer death in women globally [45]. Because ER is the key therapeutic vulnerability for ER+ BCa, TAM is the longstanding endocrinal drug for treating patients who express ER [1,46]. Unfortunately, TAM resistance and tumor recurrence remain insuperable obstacles in conquering advanced BCa [7]. The mechanisms underlying TAM resistance are still incompletely defined. In this regard, it is widely recognized that the reactivation of ER is associated with TAM resistance [47]. In addition, increasing evidence demonstrated other mechanisms involved in the TAM resistance irrespective of ER, such as activation of ERK or PI3K signaling pathways [9,48,49].
Indeed, in addition to targeting ER, TAM has been shown to preferentially accumulate in the cell membrane, particularly it is capable of impairing mitochondrial function [3]. TAM induces ROS generation by enhancing the proton permeability or inactivating mitochondria antioxidant [50,51]. Thus, the dysfunction of antioxidants is implicated in BCa TAM resistance. Here, we used a lipid-soluble BODIPY 581/591 probe to assess lipid peroxidation in live cells. The results showed that TAM-induced excessive ROS attacked the cell membrane leading to increased lipid peroxidation and changing the morphology of mitochondria. Nevertheless, the probe is capable to penetrate cell membranes and freely diffuse across the cytosolic compartments, and distribute throughout the cytoplasm, including nuclei. In addition, ferroptosis inhibitor DFO efficiently counteracted TAM-induced lipid peroxidation. Thus, the present study uncovered that ferroptosis is critical for TAM-mediated cell death beyond apoptosis.
Hydroxyl radical (OH•), a potent oxidative radical, is a leading factor causing lipid peroxidation. Deprivation of the antioxidant defense system led to the increasing generation of toxic OH• [52], thereby enhancing the cell membrane destruction by catalyzing lipid peroxidation relying on iron via the Fenton reaction [17,22]. Ferroptosis is a non-apoptotic form of programmed cell death mediated by iron-dependent lipid peroxidation. GPX4, a GSH-dependent peroxidase, functioned as a key ferroptosis inhibitor to defense lipid peroxidation [23,53]. We found that TAM enhanced ferroptosis via ROS-induced lipid peroxidation, which was reduced in TAMR cells with high levels of GPX4. Consequently, silencing of GPX4 led to TAM re-sensitization in the resistant BCa cells. The finding provides proof-of-concept evidence that GPX4-inhibited ferroptosis is involved in BCa TAM resistance. In addition to ROS, TAM also induced NO production in sensitive BCa cells, another critical reactive species that functions as a cytotoxic inducer and/or adaptive signaling stimulator. Recent studies have demonstrated that NO fosters ferroptosis by depleting GSH [54,55]. Thus, we predict that the high NO level may induce both apoptosis and ferroptosis by promoting mitochondrial damage and depleting GSH.
Provoking redox-sensitive signaling pathways is thought to contribute to cellular defense against oxidative stress via adaptive activation of prosurvival signaling pathways, such as Ras/MAPK/ERK, PI3K/Akt, and NF-κB [29,56]. Numerous factors participate in the regulation of ferroptosis by influencing GPX4 function through either transcriptional regulation or post-translational regulation [57]. Transcription factors Nrf2, Stat3, and ELK1 have been shown to regulate GPX4 expression via cis/trans-regulatory manners [58,59]. Additionally, ubiquitination-mediated GPX4 degradation enhanced ferroptosis in triple-negative BCa cancers [60,61]. Here, we revealed that RelB upregulated GPX4 in BCa TAMR cells. The NF-κB enhancer element identified in the GPX4 gene was responsive to ferroptosis inhibition, suggesting that the RelB-activated noncanonical NF-κB pathway plays a predominant role in TAMR via inhibition of ferroptosis by upregulating GPX4. Consequently, an NF-κB inhibitor SN52 efficiently resensitized TAM by blocking RelB nuclear translocation. These results are consistent with our previous finding that prompted the importance of the noncanonical NF-κB pathway in BCa progression [36].
In summary, the results from this study suggest that ferroptosis is involved in BCa TAM resistance. Thus, inhibition of ferroptosis can resensitize TAM to treat aggressive BCa cells irrespective of the ER signaling pathway. Activating the NF-κB pathway is critical for acquiring TAM resistance. In particular, the RelB-based noncanonical NF-κB pathway plays a vital role in BCa TAM resistance. RelB activation inhibits ferroptosis by transcriptionally upregulating GPX4. Overall, this study, for the first time, demonstrates that RelB contributes to BCa TAM resistance via the upregulation of GPX4.
4. Methods
4.1. Cell culture and gene manipulation
Human breast adenocarcinoma cell lines MCF7, T47D, and MDA-MB-231 were purchased from the Chinese Academy of Sciences Cell Bank of Type Culture Collection (CBTCCCAS, China). All cell lines were identified using STR profiling (provided in Supplemental Materials) and cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco, USA) supplemented with 10 % fetal bovine serum (FBS, Gibco), 100 U/mL penicillin, and 100 g/mL streptomycin (Beyotime, China) in a humidified atmosphere of 5 % CO2 at 37 °C. The TAMR variant of MCF7 and T47D cell lines was established by exposing MCF7 and T47D to gradually increasing concentrations of TAM (Sigma, USA). RelB and GPX4 were ectopically expressed in TAMS cells using a Lipofectamine 2000 transfection reagent (Invitrogen, USA). Conversely, RelB and GPX4 were knocked out in TAMR and MDA-MB-231 cells using a CRISPR/Cas9-mediated gene-edition system [36]. Briefly, BCa cells were transfected with a Cas9-single guide RNA (sgRNA) expression plasmid targeting RelB (sgRNA, 5′-GACGAATACATTAAGGAGAA-3′) and GPX4 (sgRNA, 5′-AGCCCCGCCGCGATGAGCCT-3′). Single-cell colonies with distinct levels of RelB and GPX4 were selected using puromycin or G418 (Sigma) and further validated by western blots.
4.2. Cell survival analysis
TAM-mediated cytotoxicity was analyzed by MTT assay and colony survival assay. For the MTT assay, cells were plated into 96-well plates at 2000 cells/well in the completed media for 12 h allowing cell adhesion. The cells were treated with a series of dilutions of TAM. After 48 h, an MTT reagent (Sigma) was applied to the cells and cell viability was determined using a microplate reader (Thermo, USA) at 562 nm. Multiple cell death inhibitors were used to pretreat cells for 30 min before TAM treatment, including 100 μM DFO (Abcam, USA), 20 mM necrostatin-1 (Abcam), 10 mM ferrostatin-1 (Abcam), 5 mM NAC adjusted to pH of 7.4 (Abcam), 50 μM Z-VAD-FMK (Abcam), 50 μM chloroquine (Abcam), and 20 μM TTM (MCE, USA). For colony survival assay, cells were seeded into 6-well plates at the density of 200 cells/well. After 24 h of culture, the cells were treated with dose-escalated TAM. Fourteen days after the treatment, formed colonies were stained with 1 % crystal violet dye (Aladdin, China) for 30 min. The cell surviving fractions were calculated based on the ratio of the number of colonies formed to the number of cells efficiently plated.
4.3. ROS quantification
The levels of ROS in TAM-treated BCa cells were measured using multiple fluorescent probes, as previously described by other groups [62,63]. BCa cells were cultured into a 6-well plate within 2 ml medium to 60 % confluence. The cells were pretreated with ferroptosis inhibitors for 30 min and then treated with TAM for 6 h. To analyze the level of superoxide anion, the cells were incubated with 5 μM dihydroethidium (DHE) probe (Sigma) for 30 min at 37 °C. The probe specifically reacts with superoxide to produce 2-hydroxyethidium, which can be quantified from 370/420 nm to 535/610 nm. In addition, the cells were incubated with 5 μM MitoSOX Red probe (Sigma) to measure mitochondrial superoxide at 396/610 nm. The cells were stained with a Hoechst dye (Sigma) to control the cell numbers. To verify TAM-induced superoxide, the cells were pretreated with 100 U/m polyethylene glycol-conjugated superoxide dismutase (PEG-SOD, Sigma, S9549) for 30 min to specifically remove superoxide anions in live cells.
Furthermore, the cells were incubated with 10 μM 2′,7′-dichlorofluorescein diacetate (H2DCFDA) probe (Invitrogen) to quantify total ROS normalized with nuclei image stained by Hoechst dye. The probe mainly reacts with the two radicals to form DCF that can be detected at 488/525 nm. Additionally, 2 μM 5-(and-6)-Carboxy-2′,7′-dichlorodihydrofluorescein diacetate (CDCFDA, Invitrogen), an oxidation-insensitive dye, was included to exclude DCF images enhanced by uptake of ester cleavage. To verify TAM-induced the radical generation, the cells were pretreated with 5 mM NAC and 1000 U/mL PEG-catalase (Sigma, C4963) for 30 min to eliminate the OH• and ONOO- radicals. The fluorescence image was analyzed using a fluorescence microscope (Nikon, Japan), and the fluorescent intensity was quantified using an M200 multifunction microplate reader (Tecan, Germany). The detail information of all probes used in this study is listed in Table S1.
4.4. NO quantification
The levels of NO in BCa cells were measured using a DAF-FM DA probe (MCE). The cells were pre-incubated with 5 μM DAF-FM DA for 30 min to load the probe, followed by treatment with 5 μM TAM or 100 μM sodium nitroprusside (SNP, Sigma) as the positive control for 15 min. Subsequently, the cells were incubated with fresh Hank's buffer for 20 min to complete de-esterification. Nuclear staining was achieved using Hoechst dye. The fluorescence image was analyzed by Nikon fluorescence microscope at 488/515 nm, and fluorescent intensity was further quantified by M200 microplate reader at 488/515.
4.5. Quantification of lipid peroxidation
The level of lipid peroxidation was quantified using a BODIPY 581/591 C11 probe (Invitrogen) as described previously [64]. Briefly, BCa cells were treated with TAM for different times (0, 3, 6, 12, 24 h), and followed by incubation with 1 μM BODIPY for 30 min. To remove the effect of TAM, the cells were pretreated with 5 μM NAC for 30 min prior to TAM treatment. The fluorescent density was measured by simultaneously acquiring green fluorescence (484/510 nm) and red fluorescence (581/610 nm). The level of lipid peroxidation was estimated based on a ratio of the oxidized form (green) to reduced form (red). For imaging lipid peroxidation, after incubation with C11-BODIPY and Hoechst, the cells were placed in glass-bottom dishes and observed using an LSM 710 confocal microscopy (Zeiss, Germany). Lipid peroxidation was further quantified using a FACSAira II SORP flow cytometer (BD, USA).
Furthermore, lipid peroxidation was measured by MDA assay. After TAM treatment, BCa cells or tissues were lysed using a RIPA reagent and centrifuged at 10,000 g for 10 min to remove the cell debris. The level of lipid peroxidation was measured using an MDA assay kit (Beyotime). Briefly, 100 μL of cell supernatant was mixed with 200 μL of thiobarbituric acid and then heated at 95 °C for 40 min. The absorbance of the reaction mixture was measured (532 nm) at room temperature using a multifunction microplate reader (Tecan).
4.6. Iron ion detection
The level of intracellular levels of ferrous (Fe2+) ions was determined using an iron assay kit (Abcam) according to the manufacturer's protocols. Briefly, cell lysate was preincubated with an assay buffer for 30 min at 37 °C and then incubated with an iron probe for 60 min. The optical density was immediately measured using a multifunction microplate reader (Tecan) at 593 nm.
4.7. Mitochondrial morphology
TAM-mediated alteration of mitochondrial morphology was examined by TEM. For analyzing mitochondrial structure in tumors, the tissue samples were fixed in 2.5 % glutaraldehyde (Sigma) at 4 °C overnight. After washing with PBS, the tissues were immersed in PBS containing 2 % osmium tetroxide for 2 h at room temperature. For the preparation of TEM slices, the cell and tissue samples were embedded and then sliced using an RMC ultramicrotome. The sections (100 nm) were double-stained with uranyl acetate and lead citrate and then examined using a Tecnai Spirit TEM (FEI, USA) at an acceleration voltage of 120 kV.
4.8. JC-1 assay
Mitochondrial membrane potential (ΔΨm) in TAM-treated cells was analyzed using a JC-1 assay kit (Invitrogen). After treatment, cells were harvested and washed with 1 × PBS. The collected cells were incubated within a 1 μM JC-1 solution in darkness at 37 °C for 15 min. After washing with 1 × PBS, red fluorescence (aggregated form) and green fluorescence (monomer) were acquired using a flow cytometer. ΔΨm was assessed using the ratio of red-to-green fluorescence.
4.9. Quantification of oxygen consumption
Mitochondrial respiration was assessed by measurement of OCR using a Seahorse XF Analyzer (Agilent, USA). The cells were transferred into a 96-well XF96 plate at 5 × 103 cells/well and subjected to TAM treatment for 12 h. Cartridge plates for metabolic stress injections were hydrated for 24 h at 37 °C without CO2 in calibrant solution. One hour before the assay, the culture medium in the XF96 plate was replaced by a Seahorse Assay medium. OCR was measured under four following conditions: basal, 1 μM oligomycin, 0.5 μM FCCP, and 0.5 μM rotenone +0.5 μM antimycin. OCR were normalized by the total protein.
4.10. RNA sequencing (RNA-seq)
Total RNA was isolated from BCa cells using an RNA isolation kit (Invitrogen), and mRNA was converted into cDNA libraries with adapters for sequencing. Next-generation sequencing (NGS) was performed on the Illumina Hiseq 4000 platform with a 150 bp paired-end module by Fcmacs Biotech Co., Ltd (Nanjing, China). Raw reads were filtered by removing reads containing adapter, poly-N, and low-quality reads for subsequent analysis. The expression level of each gene was quantified using featureCounts, and the resulting counts were normalized to fragments per kilobase of transcript per million mapped reads (FPKM). Differential expression analysis was performed using the DESeq2 package in R, which uses a negative binomial distribution to model the count data. Genes with a corrected p-value less than 0.05 and an absolute value of log2 (fold change) greater than or equal to 1 were considered significantly differentially expressed. To visualize the expression patterns of the differentially expressed genes, custom scripts in R software were used for clustering and heatmap analysis (https://www.r-project.org). In addition, the altered mRNA expression profile in shRelB cells vs. shCtrl cells was analyzed using KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment through the DAVID bioinformatics tool (https://david.ncifcrf.gov).
4.11. Blockade of RelB nuclear translocation
Previously, we designed a SN52 peptide to block nuclear translocation of the p52∶RelB dimer via conjugation of a consensus sequence of nuclear localization signal (NLS) derived from p52 to a cell permeable region. To improve its permeability for in vivo study, we redesigned SN52 by conjugating the NLS region with an acetylation (Ac) region. The peptides were synthesized and purified by Nanjing Peptide Biotech LTD, China.
4.12. Immunoblotting
Total proteins were extracted from BCa cells and tumor tissues using a protein extraction kit (Keygen, China). Cytoplasmic and nuclear proteins were prepared using a nuclear-cytosol extraction kit (Beyotime) according to the manufacturer's instructions. The protein extracts (50–100 μg) were separated on SDS-PAGE gels and then transferred to PVDF membranes (Sigma). The membranes were blocked in 5 % skim milk for 2 h and then incubated with primary antibodies overnight at 4 °C. Membrane-bound primary antibodies were detected using the appropriate secondary antibodies at room temperature for 2 h. Enhanced chemiluminescence (Bio-Rad, USA) was applied for the visualization of images. All antibodies used in this study are listed in Table S1.
4.13. Immunofluorescence
Confocal microscopy was performed to examine the cellular localization of proteins using the relative antibodies (Table S1). After washing with PBS, the cells were fixed in 4 % paraformaldehyde for 15 min and permeabilized with ice-cold 0.5 % Triton X-100 for 20 min at room temperature. The samples were subjected to probing with the appropriate primary and secondary antibodies. The nuclei were counterstained with DAPI (Cell Signaling Technology). The fluorescence was visualized and captured with confocal microscopy (Zeiss).
4.14. Measurement of GPX4 activity
To measure GPX4 activity [65], cell or tissue samples were homogenized in lysis buffer containing 100 mM Tris/Base (pH 8.0), 2.0 mM EDTA, 1.5 mM NaN3, 0.1 % Triton X-100, and 1 mM PMSF. After centrifugation at 10,000 g for 10 min at 4 °C, the supernatants were collected and the protein concentration was quantified. The GPX4 activity in the extracts was measured in reaction buffer (100 mM Tris/Base [pH 8.0], 2 mM EDTA, 1.5 mM NaN3, 0.1 % Triton X-100) containing glutathione reductase (1.5 U/ml), glutathione (3 mM), and NADPH (0.2 mM). The kinetic measurements were initiated after a 5-min incubation period, and the absorbance at 340 nm was measured every 20 s for a total of 5 min using an M200 multifunction microplate reader (Tecan). During the first 100 s, the nonspecific rate of oxidation of NADPH was monitored and recorded. GPX4-specific enzymatic reaction was initiated by adding 30 μM PCOOH substrate (Sigma), and the GPX4-dependent oxidation of NADPH was monitored for the next 200 s. The GPX4 activity was determined by calculating the rate of NADPH loss over time and normalized to the protein concentration. The molar extinction coefficient of NADH at 340 nm (ε340) is 6270 M−1 cm−1. The enzymatic activity of GPX4 was calculated using the following equation: [GPX4 activity] (mU/mg) = (SlopePCOOH - SlopeBackground) × [total reaction volume] (L) × 109/[ε340 × d × protein (mg)] = ΔSlope × 57.996/mg protein.
4.15. Luciferase reporter assay
The NF-κB enhancer of the human GPX4 gene was subcloned into the pGL4 vector to drive luciferase reporter response. The reporter construct was co-transfected into BCa cells with a pSV-β-galactosidase (β-gal) expression plasmid. The transfected cells were washed twice with ice-cold PBS and lysed in culture dishes using a lysis buffer. The luciferase activity was measured using a luciferase reporter assay kit (Promega, USA) with a luminometer (Berthold Tech., Germany). Additionally, the β-gal activity was quantified using a β-galactosidase enzyme assay kit (Promega, USA). The luciferase reporter response was estimated by normalizing the luciferase activity to the β-gal activity.
4.16. Chromatin immunoprecipitation (ChIP)
The RelB-mediated cis/trans transcriptional regulation of the GPX4 gene was assessed using a ChIP kit (Cell Signaling Tech.). Briefly, BCa cells derived chromatins were sonicated to yield DNA fragments (200–500 bp). A RelB antibody (Cell Signaling Tech) was used to pull down the truncated chromatin. No antibody pulldown served as the input control and pulldown with rabbit IgG (Cell Signaling Tech) served as a negative antibody control. The precipitated enhancer fragments were amplified by quantitative PCR (qPCR). E1 forward primer, 5′-CAGGATGGGGAGTCACTGAC-3′, E1 reverse primer: 5′-GGCTGATCTCGAACTCCTGA-3’; E2 forward primer, 5′-CTACAGGAGCCCTCGACCA-3′, E2 reverse primer, 5′-GCTCTAGAAAGATATGTGGCAAC-3’. The amount of pulldown DNA fragments was quantified by normalizing with the input control.
4.17. Zebrafish tumor xenograft model
A zebrafish tumor experimental model was used to verify SN52-mediated tumor suppression by inhibiting RelB activation in vivo (the protocol No. IACUC-2006003 was approved by Nanjing Medical University). BCa cells were labeled with PKH26 fluorescent dye (Invitrogen) in FBS-free media. After washing with PBS, 200 cells were microinjected into the perivitelline cavity of zebrafish embryos (hpf) and allowed to incubate at 28 °C for 1 h and then turned to 34 °C to continue the cultivation. To inhibit RelB activation, 24 h after cell injection, the nutrient solution was replaced with 100 μg/mL SN52 or SN52 M control. After 12 h, 10 μM TAM was added to the zebrafish nutrient solution to inhibit tumor growth. Three days after treatment, the tumors formed in adult fishes were imaged on a fluorescent microscope.
4.18. Mouse tumor xenograft model
A mouse tumor experimental model was used to verify the role of RelB-GPX4 axis in BCa TAMR (the protocol No. IACUC-2102009 was approved by Nanjing Medical University). For tumor formation, 5 × 106 BCa cells with different levels of RelB were subcutaneously injected into the left mammary fat pads of five-week-old female nude (BALB/c) mice (Beijing Vital River Lab Animal Tech Co. Ltd, China). Tumor volumes were measured using digital calipers every other day and calculated using a standard formula (V = 0.52 × AB2, A and B represent the diagonal tumor lengths). After tumor volume reached 500 mm3, Mice were randomly divided into two groups for treating with 4 mg/kg of TAM and saline control. The mice were sacrificed when tumor volume reached 2500 mm3 and tumor tissues were excised for IHC and immunoblotting analyses.
4.19. Immunohistochemistry (IHC)
Tumor tissues were fixed paraffin-embedded slides and dewaxed by xylene and then soaked in 5 % BSA buffer for 1 h. The slides were incubated with diluted primary antibodies followed by biotinylated secondary antibody. After washing the slides, a DAB Substrate Kit (Cell Signaling Tech.) was used to observe immunostaining images under a microscope. The intensity of IHC staining was scored as negative (score 0), weak (score 1), medium (score 2), and strong (score 3). The “H-score” was calculated using “∑pi (i + 1)” for all slides, in which “pi” represented the percentage of positive cells and “i” represented the staining intensity.
4.20. Statistical analysis
Data were presented as the mean ± standard deviation (SD) of at least three replicates. Significant differences between two groups were analyzed by unpaired Student's t-test. One-way or two-way ANOVA with Šídák's multiple comparisons test was applied for multiple-group experiments using Prism 9.0 (GraphPad). Statistical significance was accepted at p < 0.05 (*) and p < 0.01 (**).
Funding
This study was supported by the National Natural Science Foundation of China (No. 81572742 to Y Xu; and No. 81872365 to J Tang.
Author contributions
Z.X. and Y.X. contributed to the conception and study design. Z.X., X.W., W.S., F.X., H.K., W.H., and Y.Z. developed methodology, performed experiments and acquired data. Z.X., W.S. and Y.X, analyzed and interpreted data. Z.X., and Y.X. wrote and revised the manuscript. Q.J. and J.T. provided administrative and material supports. All authors discussed the results approved the manuscript submission.
Ethics declarations
The Research Committee and the Institutional Animal Care of and Use of Nanjing Medical University approved animal studies conducted in this study (No. IACUC-2006003, No. IACUC-2102009).
Declaration of competing interest
The authors declare no potential competing interests.
Acknowledgements
We would like to thank Drs. Daret St. Clair and William St. Clair, University of Kentucky Markey Cancer Center, for their many kind suggestions. We also gratefully acknowledge Dr. Hongbing Shen, Nanjing Medical University Cancer Research Center, for providing necessary research facilities.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102952.
Contributor Information
Qin Jiang, Email: jqin710@vip.sina.com.
Jinhai Tang, Email: jhtang@njmu.edu.cn.
Yong Xu, Email: yxu4696@njmu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Cardoso F., Paluch-Shimon S., Senkus E., Curigliano G., Aapro M.S., Andre F., et al. 5th ESO-ESMO international consensus guidelines for advanced breast cancer (ABC 5) Ann. Oncol. 2020;31(12):1623–1649. doi: 10.1016/j.annonc.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dackus G., Jozwiak K., Sonke G.S., van der Wall E., van Diest P.J., Siesling S., et al. Adjuvant aromatase inhibitors or tamoxifen following chemotherapy for perimenopausal breast cancer patients. J. Natl. Cancer Inst. 2021;113(11):1506–1514. doi: 10.1093/jnci/djab091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tomkova V., Sandoval-Acuna C., Torrealba N., Truksa J. Mitochondrial fragmentation, elevated mitochondrial superoxide and respiratory supercomplexes disassembly is connected with the tamoxifen-resistant phenotype of breast cancer cells. Free Radic. Biol. Med. 2019;143:510–521. doi: 10.1016/j.freeradbiomed.2019.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Vacurova E., Trnovska J., Svoboda P., Skop V., Novosadova V., Reguera D.P., et al. Mitochondrially targeted tamoxifen alleviates markers of obesity and type 2 diabetes mellitus in mice. Nat. Commun. 2022;13(1):1866. doi: 10.1038/s41467-022-29486-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hard G.C., Williams G.M., Iatropoulos M.J. Tamoxifen and liver cancer. Lancet. 1993;342(8868):444–445. doi: 10.1016/0140-6736(93)92865-q. [DOI] [PubMed] [Google Scholar]
- 6.Abdelmalek C.M., Hu Z., Kronenberger T., Kublbeck J., Kinnen F.J.M., Hesse S.S., et al. Gefitinib-tamoxifen hybrid ligands as potent agents against triple-negative breast cancer. J. Med. Chem. 2022;65(6):4616–4632. doi: 10.1021/acs.jmedchem.1c01646. [DOI] [PubMed] [Google Scholar]
- 7.Bi M., Zhang Z., Jiang Y.Z., Xue P., Wang H., Lai Z., et al. Enhancer reprogramming driven by high-order assemblies of transcription factors promotes phenotypic plasticity and breast cancer endocrine resistance. Nat. Cell Biol. 2020;22(6):701–715. doi: 10.1038/s41556-020-0514-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanker A.B., Sudhan D.R., Arteaga C.L. Overcoming endocrine resistance in breast cancer. Cancer Cell. 2020;37(4):496–513. doi: 10.1016/j.ccell.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu Y., Liu Y., Zhang C., Chu J., Wu Y., Li Y., et al. Tamoxifen-resistant breast cancer cells are resistant to DNA-damaging chemotherapy because of upregulated BARD1 and BRCA1. Nat. Commun. 2018;9(1):1595. doi: 10.1038/s41467-018-03951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Javadov S., Jang S., Chapa-Dubocq X.R., Khuchua Z., Camara A.K. Mitochondrial respiratory supercomplexes in mammalian cells: structural versus functional role. J. Mol. Med. (Berl.) 2021;99(1):57–73. doi: 10.1007/s00109-020-02004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandez-Caggiano M., Kamynina A., Francois A.A., Prysyazhna O., Eykyn T.R., Krasemann S., et al. Mitochondrial pyruvate carrier abundance mediates pathological cardiac hypertrophy. Nat. Metab. 2020;2(11):1223–1231. doi: 10.1038/s42255-020-00276-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trachootham D., Alexandre J., Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat. Rev. Drug Discov. 2009;8(7):579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 13.Fu L.H., Wan Y., Qi C., He J., Li C., Yang C., et al. Nanocatalytic theranostics with glutathione depletion and enhanced reactive oxygen species generation for efficient cancer therapy. Adv. Mater. 2021;33(7) doi: 10.1002/adma.202006892. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y., Gao W., Shi X., Ding J., Liu W., He H., et al. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature. 2017;547(7661):99–103. doi: 10.1038/nature22393. [DOI] [PubMed] [Google Scholar]
- 15.Koren E., Fuchs Y. Modes of regulated cell death in cancer. Cancer Discov. 2021;11(2):245–265. doi: 10.1158/2159-8290.CD-20-0789. [DOI] [PubMed] [Google Scholar]
- 16.Stockwell B.R. Ferroptosis turns 10: emerging mechanisms, physiological functions, and therapeutic applications. Cell. 2022;185(14):2401–2421. doi: 10.1016/j.cell.2022.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E., et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galluzzi L., Vitale I., Aaronson S.A., Abrams J.M., Adam D., Agostinis P., et al. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ. 2018;25(3):486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen Z., Liu T., Li Y., Lau J., Yang Z., Fan W., et al. Fenton-reaction-acceleratable magnetic nanoparticles for ferroptosis therapy of orthotopic brain tumors. ACS Nano. 2018;12(11):11355–11365. doi: 10.1021/acsnano.8b06201. [DOI] [PubMed] [Google Scholar]
- 20.Tang D., Chen X., Kang R., Kroemer G. Ferroptosis: molecular mechanisms and health implications. Cell Res. 2021;31(2):107–125. doi: 10.1038/s41422-020-00441-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bersuker K., Hendricks J.M., Li Z., Magtanong L., Ford B., Tang P.H., et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575(7784):688–692. doi: 10.1038/s41586-019-1705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stockwell B.R., Friedmann Angeli J.P., Bayir H., Bush A.I., Conrad M., Dixon S.J., et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171(2):273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang W.S., SriRamaratnam R., Welsch M.E., Shimada K., Skouta R., Viswanathan V.S., et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156(1–2):317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doll S., Freitas F.P., Shah R., Aldrovandi M., da Silva M.C., Ingold I., et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. 2019;575(7784):693–698. doi: 10.1038/s41586-019-1707-0. [DOI] [PubMed] [Google Scholar]
- 25.Schmitt A., Xu W., Bucher P., Grimm M., Konantz M., Horn H., et al. Dimethyl fumarate induces ferroptosis and impairs NF-kappaB/STAT3 signaling in DLBCL. Blood. 2021;138(10):871–884. doi: 10.1182/blood.2020009404. [DOI] [PubMed] [Google Scholar]
- 26.Yi J., Zhu J., Wu J., Thompson C.B., Jiang X. Oncogenic activation of PI3K-AKT-mTOR signaling suppresses ferroptosis via SREBP-mediated lipogenesis. Proc. Natl. Acad. Sci. U. S. A. 2020;117(49):31189–31197. doi: 10.1073/pnas.2017152117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ouyang S., Li H., Lou L., Huang Q., Zhang Z., Mo J., et al. Inhibition of STAT3-ferroptosis negative regulatory axis suppresses tumor growth and alleviates chemoresistance in gastric cancer. Redox Biol. 2022;52 doi: 10.1016/j.redox.2022.102317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim B., Woodward W.A., Wang X., Reuben J.M., Ueno N.T. Inflammatory breast cancer biology: the tumour microenvironment is key. Nat. Rev. Cancer. 2018;18(8):485–499. doi: 10.1038/s41568-018-0010-y. [DOI] [PubMed] [Google Scholar]
- 29.Zhao H., Wu L., Yan G., Chen Y., Zhou M., Wu Y., et al. Inflammation and tumor progression: signaling pathways and targeted intervention. Signal Transduct. Targeted Ther. 2021;6(1):263. doi: 10.1038/s41392-021-00658-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Azuma K., Ikeda K., Suzuki T., Aogi K., Horie-Inoue K., Inoue S. TRIM47 activates NF-kappaB signaling via PKC-epsilon/PKD3 stabilization and contributes to endocrine therapy resistance in breast cancer. Proc. Natl. Acad. Sci. U. S. A. 2021;118(35) doi: 10.1073/pnas.2100784118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Douglass J.D., Dorfman M.D., Fasnacht R., Shaffer L.D., Thaler J.P. Astrocyte IKKbeta/NF-kappaB signaling is required for diet-induced obesity and hypothalamic inflammation. Mol. Metabol. 2017;6(4):366–373. doi: 10.1016/j.molmet.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jomen W., Ohtake T., Akita T., Suto D., Yagi H., Osawa Y., et al. Iron chelator deferasirox inhibits NF-kappaB activity in hepatoma cells and changes sorafenib-induced programmed cell deaths. Biomed. Pharmacother. 2022;153 doi: 10.1016/j.biopha.2022.113363. [DOI] [PubMed] [Google Scholar]
- 33.Liu Z., Mar K.B., Hanners N.W., Perelman S.S., Kanchwala M., Xing C., et al. A NIK-SIX signalling axis controls inflammation by targeted silencing of non-canonical NF-kappaB. Nature. 2019;568(7751):249–253. doi: 10.1038/s41586-019-1041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y., Xu Z., Ding J., Tan C., Hu W., Li Y., et al. HZ08 suppresses RelB-activated MnSOD expression and enhances Radiosensitivity of prostate Cancer cells. J. Exp. Clin. Cancer Res. 2018;37(1):174. doi: 10.1186/s13046-018-0849-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., Zhu S., Du Y., Xu F., Sun W., Xu Z., et al. RelB upregulates PD-L1 and exacerbates prostate cancer immune evasion. J. Exp. Clin. Cancer Res. 2022;41(1):66. doi: 10.1186/s13046-022-02243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang M., Zhang Y., Xu Z., Qian P., Sun W., Wang X., et al. RelB sustains endocrine resistant malignancy: an insight of noncanonical NF-kappaB pathway into breast Cancer progression. Cell Commun. Signal. 2020;18(1):128. doi: 10.1186/s12964-020-00613-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maqbool S.N., Lim S.C., Park K.C., Hanif R., Richardson D.R., Jansson P.J., et al. Overcoming tamoxifen resistance in oestrogen receptor-positive breast cancer using the novel thiosemicarbazone anti-cancer agent, DpC. Br. J. Pharmacol. 2020;177(10):2365–2380. doi: 10.1111/bph.14985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao M., Qin R., Huang W., Zhu H.P., Peng F., Han B., et al. Targeting regulated cell death (RCD) with small-molecule compounds in triple-negative breast cancer: a revisited perspective from molecular mechanisms to targeted therapies. J. Hematol. Oncol. 2022;15(1):44. doi: 10.1186/s13045-022-01260-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drago J.Z., Chandarlapaty S., Jhaveri K. Targeting apoptosis: a new paradigm for the treatment of estrogen receptor-positive breast cancer. Cancer Discov. 2019;9(3):323–325. doi: 10.1158/2159-8290.CD-19-0050. [DOI] [PubMed] [Google Scholar]
- 40.Kahlson M.A., Dixon S.J. Copper-induced cell death. Science. 2022;375(6586):1231–1232. doi: 10.1126/science.abo3959. [DOI] [PubMed] [Google Scholar]
- 41.D'Herde K., Krysko D.V. Ferroptosis: oxidized PEs trigger death. Nat. Chem. Biol. 2017;13(1):4–5. doi: 10.1038/nchembio.2261. [DOI] [PubMed] [Google Scholar]
- 42.Liu W., Wang S., Sun Q., Yang Z., Liu M., Tang H. DCLK1 promotes epithelial-mesenchymal transition via the PI3K/Akt/NF-kappaB pathway in colorectal cancer. Int. J. Cancer. 2018;142(10):2068–2079. doi: 10.1002/ijc.31232. [DOI] [PubMed] [Google Scholar]
- 43.Long X., Wong C.C., Tong L., Chu E.S.H., Ho Szeto C., Go M.Y.Y., et al. Peptostreptococcus anaerobius promotes colorectal carcinogenesis and modulates tumour immunity. Nat Microbiol. 2019;4(12):2319–2330. doi: 10.1038/s41564-019-0541-3. [DOI] [PubMed] [Google Scholar]
- 44.Xu Y., Fang F., St Clair D.K., Sompol P., Josson S., St Clair W.H. SN52, a novel nuclear factor-kappaB inhibitor, blocks nuclear import of RelB:p52 dimer and sensitizes prostate cancer cells to ionizing radiation. Mol. Cancer Therapeut. 2008;7(8):2367–2376. doi: 10.1158/1535-7163.MCT-08-0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 46.Ponde N.F., Zardavas D., Piccart M. Progress in adjuvant systemic therapy for breast cancer. Nat. Rev. Clin. Oncol. 2019;16(1):27–44. doi: 10.1038/s41571-018-0089-9. [DOI] [PubMed] [Google Scholar]
- 47.Najim O., Seghers S., Sergoynne L., Van Gaver H., Papadimitriou K., Wouters K., et al. The association between type of endocrine therapy and development of estrogen receptor-1 mutation(s) in patients with hormone-sensitive advanced breast cancer: a systematic review and meta-analysis of randomized and non-randomized trials. Biochim. Biophys. Acta Rev. Canc. 2019;1872(2) doi: 10.1016/j.bbcan.2019.188315. [DOI] [PubMed] [Google Scholar]
- 48.Li D., Ji H., Niu X., Yin L., Wang Y., Gu Y., et al. Tumor-associated macrophages secrete CC-chemokine ligand 2 and induce tamoxifen resistance by activating PI3K/Akt/mTOR in breast cancer. Cancer Sci. 2020;111(1):47–58. doi: 10.1111/cas.14230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peng W.X., Huang J.G., Yang L., Gong A.H., Mo Y.Y. Linc-RoR promotes MAPK/ERK signaling and confers estrogen-independent growth of breast cancer. Mol. Cancer. 2017;16(1):161. doi: 10.1186/s12943-017-0727-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu L., Zou P., Zheng L., Linarelli L.E., Amarell S., Passaro A., et al. Tamoxifen reduces fat mass by boosting reactive oxygen species. Cell Death Dis. 2015;6:e1586. doi: 10.1038/cddis.2014.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu Y., Zou X., Dean A.E., Brien J.O., Gao Y., Tran E.L., et al. Lysine 68 acetylation directs MnSOD as a tetrameric detoxification complex versus a monomeric tumor promoter. Nat. Commun. 2019;10(1):2399. doi: 10.1038/s41467-019-10352-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou Y., Fan S., Feng L., Huang X., Chen X. Manipulating intratumoral Fenton chemistry for enhanced chemodynamic and chemodynamic-synergized multimodal therapy. Adv. Mater. 2021;33(48) doi: 10.1002/adma.202104223. [DOI] [PubMed] [Google Scholar]
- 53.Friedmann Angeli J.P., Schneider M., Proneth B., Tyurina Y.Y., Tyurin V.A., Hammond V.J., et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 2014;16(12):1180–1191. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mao G., Xin D., Wang Q., Lai D. Sodium molybdate inhibits the growth of ovarian cancer cells via inducing both ferroptosis and apoptosis. Free Radic. Biol. Med. 2022;182:79–92. doi: 10.1016/j.freeradbiomed.2022.02.023. [DOI] [PubMed] [Google Scholar]
- 55.Yu W., Jia F., Fu J., Chen Y., Huang Y., Jin Q., et al. Enhanced transcutaneous chemodynamic therapy for melanoma treatment through cascaded fenton-like reactions and nitric oxide delivery. ACS Nano. 2023;17(16):15713–15723. doi: 10.1021/acsnano.3c02964. [DOI] [PubMed] [Google Scholar]
- 56.Moloney J.N., Cotter T.G. ROS signalling in the biology of cancer. Semin. Cell Dev. Biol. 2018;80:50–64. doi: 10.1016/j.semcdb.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 57.Dai C., Chen X., Li J., Comish P., Kang R., Tang D. Transcription factors in ferroptotic cell death. Cancer Gene Ther. 2020;27(9):645–656. doi: 10.1038/s41417-020-0170-2. [DOI] [PubMed] [Google Scholar]
- 58.Qiang Z., Dong H., Xia Y., Chai D., Hu R., Jiang H. Nrf2 and STAT3 alleviates ferroptosis-mediated IIR-ALI by regulating SLC7A11. Oxid. Med. Cell. Longev. 2020;2020 doi: 10.1155/2020/5146982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei S., Yu Z., Shi R., An L., Zhang Q., Zhang Q., et al. GPX4 suppresses ferroptosis to promote malignant progression of endometrial carcinoma via transcriptional activation by ELK1. BMC Cancer. 2022;22(1):881. doi: 10.1186/s12885-022-09986-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ding Y., Chen X., Liu C., Ge W., Wang Q., Hao X., et al. Identification of a small molecule as inducer of ferroptosis and apoptosis through ubiquitination of GPX4 in triple negative breast cancer cells. J. Hematol. Oncol. 2021;14(1):19. doi: 10.1186/s13045-020-01016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li K., Lin C., Li M., Xu K., He Y., Mao Y., et al. Multienzyme-like reactivity cooperatively impairs glutathione peroxidase 4 and ferroptosis suppressor protein 1 pathways in triple-negative breast cancer for sensitized ferroptosis therapy. ACS Nano. 2022;16(2):2381–2398. doi: 10.1021/acsnano.1c08664. [DOI] [PubMed] [Google Scholar]
- 62.Murphy M.P., Bayir H., Belousov V., Chang C.J., Davies K.J.A., Davies M.J., et al. Guidelines for measuring reactive oxygen species and oxidative damage in cells and in vivo. Nat. Metab. 2022;4(6):651–662. doi: 10.1038/s42255-022-00591-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kalyanaraman B., Darley-Usmar V., Davies K.J., Dennery P.A., Forman H.J., Grisham M.B., et al. Measuring reactive oxygen and nitrogen species with fluorescent probes: challenges and limitations. Free Radic. Biol. Med. 2012;52(1):1–6. doi: 10.1016/j.freeradbiomed.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoshida M., Minagawa S., Araya J., Sakamoto T., Hara H., Tsubouchi K., et al. Involvement of cigarette smoke-induced epithelial cell ferroptosis in COPD pathogenesis. Nat. Commun. 2019;10(1):3145. doi: 10.1038/s41467-019-10991-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stolwijk J.M., Falls-Hubert K.C., Searby C.C., Wagner B.A., Buettner G.R. Simultaneous detection of the enzyme activities of GPx1 and GPx4 guide optimization of selenium in cell biological experiments. Redox Biol. 2020;32 doi: 10.1016/j.redox.2020.101518. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.