Abstract
The biofilm matrix is a fortress; sheltering bacteria in a protective and nourishing barrier that allows for growth and adaptation to various surroundings. A variety of different components are found within the matrix including water, lipids, proteins, extracellular DNA, RNA, membrane vesicles, phages, and exopolysaccharides. As part of its biofilm matrix, Pseudomonas aeruginosa is genetically capable of producing three chemically distinct exopolysaccharides – alginate, Pel, and Psl – each of which has a distinct role in biofilm formation and immune evasion during infection. The polymers are produced by highly conserved mechanisms of secretion, involving many proteins that span both the inner and outer bacterial membranes. Experimentally determined structures, predictive modelling of proteins whose structures are yet to be solved, and structural homology comparisons give us insight into the molecular mechanisms of these secretion systems, from polymer synthesis to modification and export. Here, we review recent advances that enhance our understanding of P. aeruginosa multiprotein exopolysaccharide biosynthetic complexes, and how the glycoside hydrolases/lyases within these systems have been commandeered for antimicrobial applications.
Keywords: Pseudomonas aeruginosa, biofilm, exopolysaccharide, alginate, Pel, Psl, exopolysaccharide secretion system, glycoside hydrolases, biologics
This comprehensive review highlights recent advances in the field of Pseudomonas aeruginosa biofilm exopolysaccharide biosynthesis and discusses the potential use of glycoside hydrolases for antibiofilm therapies.
Introduction
Pseudomonas aeruginosa is a ubiquitous and versatile bacterium able to survive in a variety of different environmental niches. In addition to infecting plants, insects, and animals, P. aeruginosa is an opportunistic human pathogen notorious for establishing chronic lung infections in individuals with cystic fibrosis (CF), as well as burn wound, chronic urinary catheter, diabetic foot ulcer, corneal, and mechanical ventilation-related infections (Morrison and Wenzel 1984, Gellatly and Hancock 2013). Isolation of persistent strains from chronic infections reveals mechanisms that allow P. aeruginosa to adopt a biofilm-associated lifestyle, which confers tolerance to antibiotic treatment and aids in evasion of the host immune response (Marvig et al. 2015, Rossi et al. 2021).
A biofilm is a highly structured community of bacterial cells encased in a self-produced extracellular matrix. Biofilms can refer to surface-attached biofilms, surface-associated aggregates, free-floating aggregates, mats, and flocs, each of which can be comprised of either a single species or multiple microorganisms (Sauer et al. 2022). The matrix of the biofilm can be composed of extracellular DNA (eDNA), proteins, RNA, exopolysaccharides, water, lipids, and/or extracellular membrane vesicles (Flemming and Wingender 2010, Chiba et al. 2022). Although each of these components is intriguing to explore, herein we focus on the exopolysaccharide component of the biofilm matrix, how they function to aid P. aeruginosa in establishing persistent and antibiotic tolerant infections, how they are produced, and how enzyme therapeutics can degrade exopolysaccharides for antibiofilm applications.
Pseudomonas aeruginosa is genetically capable of producing three biofilm exopolysaccharides – alginate, Pel, and Psl – each of which have a role in chronic infections and confer advantages to the bacteria against eradication. Alginate is secreted as a high molecular weight, anionic, random polymer composed of 1–4 linked β-d-mannuronate, O-acetylated β-d-mannuronate, and α-l-guluronate residues (Evans and Linker 1973). In chronic CF lung infections, P. aeruginosa converts to a mucoid phenotype and overproduces alginate (Govan and Deretic 1996). This conversion is associated with a decline in lung function, poor patient prognoses, and a significant increase in patient mortality (Lyczak et al. 2000, Pritt et al. 2007, Li et al. 2017). Pel is a cationic polymer that has a cell-associated form and secreted, cell-free form (Jennings et al. 2015). Mass spectrometry and NMR studies recently determined that the cell-free form is predominantly composed of a dimeric repeat of α-1,4-linked galactosamine (GalN) and N-acetylgalactosamine (GalNAc) (Le Mauff et al. 2022). While it is currently not known if the composition of the two forms of Pel are identical, Pel-binding antibodies and lectins have been reported to detect both the cell-associated and secreted forms (Razvi et al. 2023a), demonstrating that there has to be some similarity between the two forms. Psl is a neutral polymer composed of repeating pentasaccharides of d-mannose, d-glucose, and l-rhamnose (Byrd et al. 2009). There are also two forms of Psl, a low molecular weight cell-free form and a higher molecular weight cell-associated form. Similar to Pel, it is not currently known whether the two forms are chemically identical.
The mechanisms used to control the ratios of cell-free to cell-associated Pel and Psl are only now being uncovered. Studies on the bifunctional glycoside hydrolase/deacetylase enzyme, PelA, in the P. aeruginosa pel biosynthetic operon have demonstrated that the hydrolase activity of PelA is required for generating cell-free Pel (Razvi et al. 2023a). Generation of the cell-free form of Pel has implications for biofilm biomechanics and virulence (Razvi et al. 2023a). This is discussed in more detail below (see sections ‘Pel biosynthesis’ – ‘Polymer modification’). Studies on the Sia signal transduction system involved in c-di-GMP production have revealed that the Sia system impacts cell-associated Psl. The biological consequences of altering the ratios of cell-associated to cell-free forms of Psl are yet to be determined (Dreifus et al. 2022).
Role of exopolysaccharides in biofilms
Alginate
Although P. aeruginosa is the most infamous pathogen in CF lung infections, individuals with CF encounter a plethora of microbes throughout their lifetime, including: Aspergillus species, respiratory syncytial virus, influenza virus, Burkholderia species, and Staphylococcus aureus (Gilligan 2014). While the conversion to a mucoid phenotype and the overproduction alginate is associated with poor CF patient outcomes, alginate is exceptionally beneficial not only to P. aeruginosa but also to the other pulmonary pathogens that P. aeruginosa may encounter. For example, mucoid P. aeruginosa is significantly more tolerant to antibiotic treatment and more persistent against the host immune response (Simpson et al. 1988, Hentzer et al. 2001, Song et al. 2003, Høiby et al. 2011, Goltermann and Tolker-Nielsen 2017, Rowe III et al. 2023). Alginate-overproducing Pseudomonas also forms a highly structured biofilm architecture (Hentzer et al. 2001) and these biofilms maintain their hydration and resist desiccation (Chang et al. 2007). Pseudomonas aeruginosa is typically aggressive towards other bacteria such as S. aureus (Hotterbeekx et al. 2017, Camus et al. 2021), however, conversion to a mucoid phenotype in response to the CF lung environment renders P. aeruginosa more cooperative and docile. Pseudomonas aeruginosa isolates from CF patients that are coinfected with S. aureus are less competitive towards S. aureus compared to isolates from monoinfected CF patients (Limoli et al. 2017). Specifically, P. aeruginosa conversion to an alginate overproducing mucoid phenotype reduces 2-heptyl-4-hydroxyquinolone-N-oxide (HQNO) and rhamnolipid production, thus preventing S. aureus killing (Limoli et al. 2017, Price et al. 2020). Exogeneous addition of alginate to nonmucoid strains of P. aeruginosa was also found to protect S. aureus in coculture via reduced production of pyocyanin, rhamnolipids, and HQNO, amongst other siderophores and quorum sensing molecules (Price et al. 2020).
Psl has also been identified as a critical matrix component of mucoid biofilms, as deletion of Psl genes in an alginate-overproducing P. aeruginosa strain resulted in decreased biofilm formation (Ma et al. 2012). However, in the presence of elevated calcium, P. aeruginosa produces calcium-cross-linked mucoid biofilms that can form in the absence of Psl (Jacobs et al. 2022). Furthermore, compared to mucoid biofilms grown without calcium present, calcium-gelled mucoid biofilms are thicker (Sarkisova et al. 2005) with individual cells being suspended in the alginate gel instead of densely packed aggregates (Jacobs et al. 2022). Although mucoid biofilms were able to limit penetration of tobramycin, calcium-gelled mucoid biofilms were significantly more protected from tobramycin treatment as the cross-linked alginate further impeded penetration of the antibiotic (Jacobs et al. 2022). Fluorescence microscopy of explanted CF lung samples shows presence of individual P. aeruginosa cells suspended in a matrix material, providing evidence for these calcium-cross-linked mucoid biofilms in the CF airway (Jacobs et al. 2022).
Pel
Pel is required for formation of pellicle biofilms at the air–liquid interface and colony wrinkling (Razvi et al. 2023a). Under reduced availability of oxygen, Pel production provides P. aeruginosa a growth advantage in pellicle biofilms when in competition with a non-Pel producing strain (Madsen et al. 2015). However, this competitive advantage is lost in colony biofilms. Pel is critical for initiating and maintaining cell-to-cell interactions which influence aggregate formation (Colvin et al. 2011). In vitro, the positively charged polymer binds eDNA through ionic interactions, forming the structural core in the stalk of the P. aeruginosa biofilm (Jennings et al. 2015). While, in the context of chronic P. aeruginosa infections in the CF lung, alginate is frequently discussed as the exopolysaccharide of primary interest, both Pel and Psl have been detected in the sputum from CF patients that tested positive for P. aeruginosa aggregates (Jennings et al. 2021). The pH of the CF airway is mildly acidic, making the partially deacetylated Pel positively charged, and thus able to interact with host-derived anionic polymers in the CF airway, such as DNA and mucin (Coakley et al. 2003, Jennings et al. 2021). Although Pel has been shown to increase tolerance to antibiotic treatment (Goltermann and Tolker-Nielsen 2017), Pel bound to eDNA was found to enhance tolerance to antibiotic and mucolytic treatments compared to Pel not bound to eDNA (Jennings et al. 2021), thus highlighting the importance of targeting Pel when treating P. aeruginosa biofilm infections.
The two forms of Pel appear to play distinct roles within the biofilm with the cell-free form modulating P. aeruginosa biofilm biomechanics – stiffness, wettability, cohesiveness, and buoyancy – and decreasing virulence in Drosophila melanogaster and Caenorhabditis elegans infection models (Razvi et al. 2023a). The mechanism(s) by which the Pel polymer modulates virulence have not yet been determined. Pel has also recently been shown to interact with CdrA, a biofilm matrix adhesion that promotes biofilm aggregation (Borlee et al. 2010, Reichhardt et al. 2020). Deletion of CdrA in a Pel-dominant strain of P. aeruginosa reduces biofilm aggregation in comparison to a Psl-dominant or Pel/Psl redundant strain of P. aeruginosa, thus highlighting that exopolysaccharides do not function in isolation to generate biofilms (Reichhardt et al. 2020).
In mixed biofilms, the naturally Pel producing P. aeruginosa strain PA14 perseveres in nutrient-starved conditions and cells remain on their attached surface, while the Psl-producing P. aeruginosa strain PAO1 was more likely to disperse in search of a new surface (Kasetty et al. 2021). Thus, Pel production appears to influence P. aeruginosa survival strategies during starvation, although the mechanism by which this occurs is unclear. PA14 is also significantly more successful in invading resident PAO1 biofilms, compared to PAO1 invasion of PA14 resident biofilms, demonstrating that Pel production confers an advantage to exploiting already established biofilms (Kasetty et al. 2021).
Psl
Although alginate production contributes to coexistence between P. aeruginosa and S. aureus (Limoli et al. 2017), Psl production antagonizes the growth of S. aureus in vitro (Lui et al. 2022). Treatment of S. aureus with cell-free Psl from P. aeruginosa spent media was found to kill S. aureus by disrupting the cell envelope, leading to cell lysis. Purified cell-free Psl was sufficient to kill S. aureus in liquid culture, while the individual monosaccharides that Psl is composed of (d-mannose, d-glucose, and l-rhamnose) had no effect on S. aureus growth, demonstrating that structural integrity of the Psl polymer is required for S. aureus killing. Moreover, Psl production in clinical isolates of P. aeruginosa is positively correlated with S. aureus killing, suggesting that Psl influences how P. aeruginosa coexists with other pulmonary pathogens (Lui et al. 2022).
The cell-associated form of Psl is associated with the P. aeruginosa cell surface in a helical pattern and contributes to cell-to-cell interactions and early biofilm attachment (Ma et al. 2009). As P. aeruginosa moves across a surface, it deposits a trail of Psl that influences motility and surface attachment in bacteria that encounter the trail (Zhao et al. 2013). This is most likely because Psl can function as an external signal to stimulate c-di-GMP production and therefore biofilm formation (Irie et al. 2012). Psl promotes resistance to the biofilm inhibitor polysorbate 80 while mutation in the Psl promoter, and therefore an inability to produce Psl, results in sensitivity (Zegans et al. 2012). Psl also promotes biofilm tolerance to colistin, polymyxin B, tobramycin, and ciprofloxacin antibiotics (Billings et al. 2013, Goltermann and Tolker-Nielsen 2017). Ionic interactions are proposed to partly contribute to sequestration of antibiotics by Psl, as addition of NaCl increased the efficacy of the antibiotics (Billings et al. 2013). However, as Psl is neutral in charge (Byrd et al. 2009), Psl may interact with other biofilm matrix components that could provide a negative charge (Billings et al. 2013). In coculture biofilms, the Psl-mediated protection against antibiotics is extended to Psl-deficient P. aeruginosa, S. aureus, and Escherichia coli (Billings et al. 2013). Production of Psl also allows PAO1 to outcompete PA14 in a dual biofilm and in a dispersal regime (Kasetty et al. 2021).
During infection, Psl contributes to more efficient colonization of lung epithelia (Jones and Wozniak 2017, Ma et al. 2006) and decreases killing by phagocytes (Mishra et al. 2012). As described above, 50% of mucoid clinical isolates from CF sputum were found to produce Psl (Jones and Wozniak 2017), demonstrating the complexity of P. aeruginosa biofilms in infection. In total, 99% of P. aeruginosa keratitis isolates were found to produce Psl (Zegans et al. 2016). These Psl-producing isolates were susceptible to opsonophagocytic killing mediated by an anti-Psl monoclonal antibody, demonstrating a need for Psl-targeting therapeutics.
Psl has also been shown to bind to a number of extracellular matrix proteins, including ecotin and CdrA (Reichhardt et al. 2018, Tseng et al. 2018). Ecotin has been shown to bind to Psl in both a cell-free system and in biofilms (Tseng et al. 2018). As a serine protease inhibitor (Chung et al. 1983), ecotin significantly inhibits neutrophil elastase activity when matrix associated (Tseng et al. 2018). Ecotin protects both planktonic and biofilm P. aeruginosa cells from elastase-mediated killing, highlighting the Psl-ecotin interaction as a mechanism by which P. aeruginosa can increase its tolerance to the host immune response (Tseng et al. 2018). In contrast, CdrA was found to increase biofilm formation and autoaggregation in a Psl-dependent manner (Borlee et al. 2010). In a developing P. aeruginosa biofilm, CdrA is required for Psl localization around the bacterial aggregate as a tightly associated shell, and thus is important for biofilm integrity through its interaction with Psl (Borlee et al. 2010). In the absence of any exopolysaccharides, CdrA–CdrA interactions promote bacterial aggregation, however CdrA-only biofilms are susceptible to degradation by the P. aeruginosa-produced protease LasB (Reichhardt et al. 2018). Interaction with Psl protects CdrA from proteolytic degradation, enhancing biofilm integrity (Reichhardt et al. 2018). Similarly, Psl can interact with eDNA from various sources including salmon sperm, human neutrophils, and S. aureus, potentially increasing P. aeruginosa survival within a human host or a multispecies biofilm (Wang et al. 2015). This interaction forms a skeleton-like web in P. aeruginosa air–liquid interface pellicle and flow-cell biofilms and confers eDNA protection from DNAse I degradation (Wang et al. 2015). Psl has also been shown to interact with antimicrobial peptides, suggesting that the innate host immune response may detect and respond to biofilm infections using these peptides (Chin et al. 2017).
Mechanisms of exopolysaccharide biosynthesis in P. aeruginosa
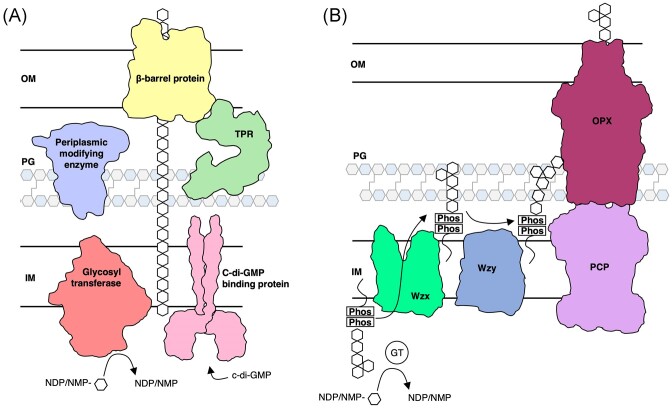
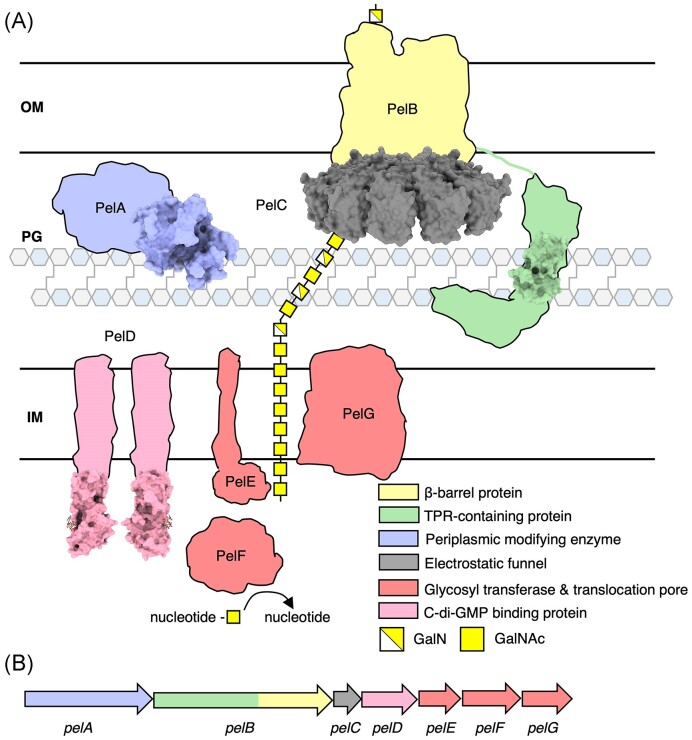
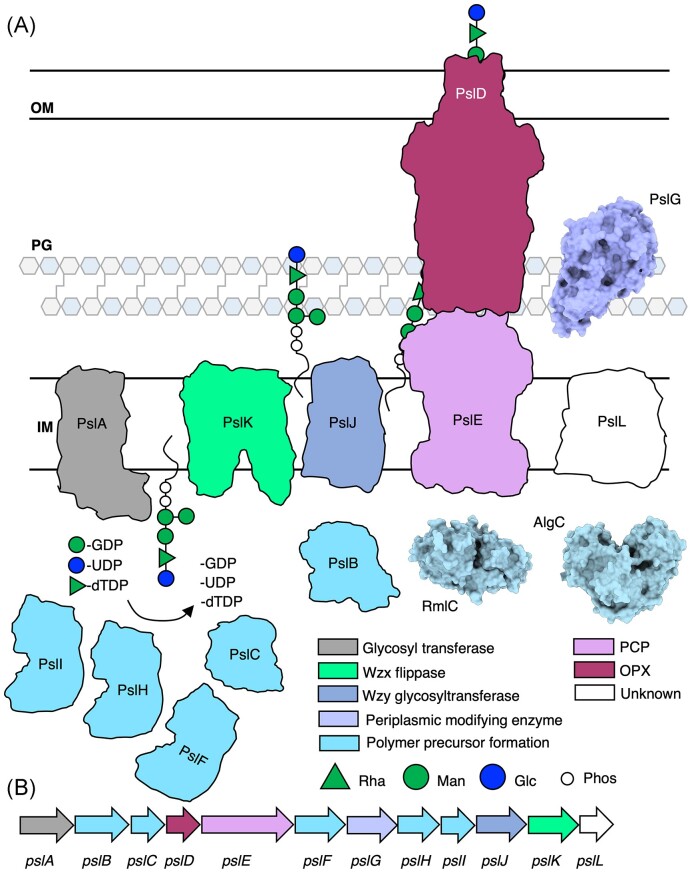
Although alginate and Pel are chemically distinct and each have their own roles in biofilm formation and infection (Table 1) (Franklin et al. 2011), the mechanisms by which they are synthesized are similar, allowing for comparisons to be made across the two systems. Alginate and Pel are synthesized and secreted via a synthase-dependent mechanism, while Psl is synthesized and secreted via a Wzx/Wzy-dependent mechanism (Fig. 1). Synthase-dependent secretion systems minimally consist of a cyclic-di-GMP binding protein that regulates polymer synthesis, a glycosyl transferase (GT) and translocation pore that synthesizes the polymer and translocates it to the periplasm, one or more periplasmic polymer modifying enzymes, an outer membrane-linked tetratricopeptide repeat (TPR)-containing protein, and a β-barrel outer membrane (OM) porin that exports the polymer (Fig. 1A) (Whitney and Howell 2013, Low and Howell 2018, Whitfield et al. 2020c). In contrast, the Wzx/Wzy-dependent secretion system consists of cytoplasmic GTs that generate a polysaccharide repeat unit, a Wzx flippase that flips the initiated lipid-anchored polysaccharide repeat unit from the cytoplasm to the inner membrane (IM), a periplasmic Wzy GT that extends the repeat unit to form a polymer, a polysaccharide copolymerase protein (PCP) and an outer membrane polysaccharide export protein (OPX) that form a translocation pathway to export the polymer through the periplasm and across the OM (Fig. 1B) (Whitney and Howell 2013; Tables 2–4).
Table 1.
Role of exopolysaccharides in Pseudomonas biofilms.
| Exopolysaccharide | Role or function | Reference |
|---|---|---|
| Alginate | ||
| Tolerance to antibiotics | Hentzer et al. (2001), Høiby et al. (2011), Jacobs et al. (2022), Goltermann and Tolker-Nielsen (2017) | |
| Persistence against host immune response | Simpson et al. (1988), Song et al. (2003), Rowe III et al. (2023) | |
| Biofilm architecture | Hentzer et al. (2001), Hay et al. (2009), Jacobs et al. (2022) | |
| Biofilm attachment | Hay et al. (2009) | |
| Increased biofilm thickness due to cross-linking with calcium | Jacobs et al. (2022), Sarkisova et al. (2005) | |
| Maintenance of hydration | Chang et al. (2007) | |
| Resistance to desiccation | Chang et al. (2007) | |
| Contributes to coinfection with S. aureus | Limoli et al. (2017), Price et al. (2020) | |
| Pel | ||
| Pellicle formation/colony wrinkling | Madsen et al. (2015), Razvi et al. (2023a) | |
| Competitive advantage in pellicle biofilms under limited oxygen conditions | Madsen et al. (2015) | |
| Cell-to-cell interactions/aggregation | Colvin et al. (2011) | |
| Biofilm structure | Jennings et al. (2015) | |
| Tolerance to antibiotic treatment | Jennings et al. (2015), Goltermann and Tolker-Nielsen (2017) | |
| Invasion of existing biofilms | Kasetty et al. (2021) | |
| Perseverance in nutrient-starved conditions | Kasetty et al. (2021) | |
| Biofilm biomechanics | Razvi et al. (2023a) | |
| Psl | ||
| Antagonizes S. aureus growth by lysis | Lui et al. (2022) | |
| Early biofilm attachment | Ma et al. (2009), Zhao et al. (2013) | |
| Cell-to-cell interactions | Ma et al. (2009) | |
| Influences motility | Zhao et al. (2013) | |
| Stimulates c-di-GMP | Irie et al. (2012) | |
| Resistance to polysorbate 80 and antibiotics | Zegans et al. (2012), Billings et al. (2013), Goltermann and Tolker-Nielsen (2017) | |
| Competitive advantage against Pel-overproducing P. aeruginosa in early colonization, biofilm growth, and dispersal | Kasetty et al. (2021) | |
| Colonization of lung epithelia | Jones and Wozniak (2017), Ma et al. (2006) | |
| Persistence against host immune response | Mishra et al. (2012), Pestrak et al. (2019) | |
| Interacts with matrix proteins CdrA and ecotin for biofilm integrity | Borlee et al. (2010), Reichhardt et al. (2018), Tseng et al. (2018) | |
| Interacts with eDNA for biofilm/pellicle integrity | Wang et al. (2015) | |
| Interacts with antimicrobial peptides | Chin et al. (2017) | |
| Dispersal in nutrient-starved conditions | Kasetty et al. (2021) |
Figure 1.
Synthase- and Wzx/Wzy-dependent secretion systems in Gram-negative bacteria. (A) In the synthase-dependent pathway, sugars are polymerized and translocated across the IM by a GT (salmon) in response to c-di-GMP binding a c-di-GMP receptor protein (pink). Once in the periplasm, the sugar polymer can be modified by periplasmic modifying enzymes (periwinkle). Transport across the OM is facilitated by a ß-barrel protein (yellow) and TPR-containing protein (green). (B) The Wzx/Wzy-dependent pathway, a sugar polymer repeat unit is assembled on a undecaprenyl-phosphate lipid carrier by GTs. A Wzx flippase (bright green) translocates the repeat unit across the IM where a Wzy glycosyltransferase (dark blue) assembles repeat units to form the polymer. The polymer is transported across the periplasm and OM by a PCP protein and an OPX protein. IM, inner membrane; PG, peptidoglycan; OM, outer membrane; NDP, nucleotide disphosphate; NMP, nucleotide monophosphate; and Phos, phosphate molecule.
Table 2.
Structures and structural predictions of alginate biosynthetic proteins.
| Protein | UniProtKB | PDB code for structure | PDB code for structurally similar protein from DALI search | Function or predicted function | Fold or domains (residues)a | Reference |
|---|---|---|---|---|---|---|
| AlgD | P11759 | 1MFZ, 1MUU, 1MV8 | GDP-mannose 6-dehydrogenase | Rossmann fold (1–202), connecting helix (203–234), (235–436) | Snook et al. (2003) | |
| Alg8 | Q52463 | 7SP8 | Mannuronan synthase, GT | GT-A fold | Maloney et al. (2022) | |
| Alg44 | Q9HY69 | 4RT1, 4RT0 | 5C22 | C-di-GMP-binding protein | PilZ domain (1–122), TM helix (152–185), membrane fusion protein (186–389) | Whitney et al. (2015), Kim et al. (2016) |
| AlgK | P96956 | 3E4B, 7ULA | Alginate export | TPR-containing protein | Keiski et al. (2010), Gheorghita et al. (2022a) | |
| AlgE | P18895 | 3RBH, 4AZL, 4B61, 4AFK | Alginate export | β-barrel porin | Whitney et al. (2011), Tan et al. (2014) | |
| AlgG | Q51371 | 4NK8, 4OZZ, 4OZY | C-5 mannuronan epimerase | Right-handed parallel β-helix fold | Wolfram et al. (2014) | |
| AlgX | Q51372 | 4KNC, 7ULA | O-acetyltransferase | SGNH hydrolase-like domain (27–347), CBM domain (348–474) | Riley et al. (2013), Gheorghita et al. (2022a) | |
| AlgL | Q06749 | 4OZV, 4OZW, 7SA8 | Alginate lyase | (α/α)5 toroid fold | Gheorghita et al. (2022b) | |
| AlgI | Q51392 | 6BUG | O-acetyltransferase | MBOAT | Ma et al. (2018) | |
| AlgJ | Q51393 | 4O8V | O-acetyltransferase | TM helices (1–78), SGNH hydrolase-like | Baker et al. (2014) | |
| AlgF | Q06062 | 6D10, 6CZT | Alginate acetylation, protein–protein interactions | β-sandwich fold (38–119, 124–212) | Low et al. (2023) | |
| AlgA | P07874 | 2 × 65, 2PFW | PMI activity, GMP activity | Rossmann-like fold (1–272), left β-helix (279–318), cupin domain (358–463) | Pelissier et al. (2010) | |
| AlgC | P26276 | 1K2Y, 1K35, 2FKF, 2FKM, 2H4L, 2H5A | Phosphomannomutase | Mixed α/β fold (1–153, 154–256, 257–368), TATA-box binding protein-like superfamily (369–463) | Regni et al. (2002, 2006a, b) |
Numbering for P. aeruginosa protein.
Table 3.
Structures and structural predictions of Pel biosynthetic proteins.
| Protein | UniProtKB | PDB code for structure | PDB code for structurally similar protein from DALI search | Function or predicted function | Fold or domains (residues)a | Reference |
|---|---|---|---|---|---|---|
| PelA | Q9HZE4 | 5TCB | 4LY4, 6EZN, 7DD9 | Glycoside hydrolase, deacetylase | Hydrolase domain (47–303), reductase domain (304–506), deacetylase domain (520–800), β-jelly roll (840–946) | Shaik et al. (2011), Wild et al. (2018), Wang et al. (2021) |
| PelB | Q9HZE5 | 5WFT | 6TZK | Pel export | TM helix (19–36), TPR-containing protein (37–877), b-barrel porin (878–1193) | Marmont et al. (2017b), Acheson et al. (2019) |
| PelC | Q9HZE6 | 5T10, 5T11, 5T0Z | Electrostatic funnel | Mixed α/β fold | Marmont et al. (2017a) | |
| PelD | Q9HZE7 | 4DN0, 4DMZ, 4EUV, 4EU0, 4ETZ, 4ETX | C-di-GMP binding protein | GAF domain (181–304), GGDEF domain (321–444) | Li et al. (2012), Whitney et al. (2012) | |
| PelE | Q9HZE8 | 4JHR | Protein interaction | TPR-like protein | Pan et al. (2013) | |
| PelF | Q9HZE9 | 4XSO | GT | GT-B fold | Wang et al. (2016a) | |
| PelG | Q9HZF0 | 6IDS | 12 transmembrane helices | EPS-E subgroup of the MOP transporter superfamily | Kusakizako et al. (2019) | |
| PA4068 | Q9HWW0 | 6WJB, 6WJ9, 6WJA | UDP-N-acetylglucosamine C4-epimerase | N-terminal Rossmann fold (1–173), C-terminal substrate-binding α/β domain (174–309) | Marmont et al. (2020) |
Numbering for P. aeruginosa protein.
Table 4.
Structures and structural predictions of Psl biosynthetic proteins.
| Protein | UniProtKB | PDB code for structure | PDB code for structurally similar protein from DALI search | Function or predicted function | Fold or domains (residues)a | Reference |
|---|---|---|---|---|---|---|
| PslA | Q9I1N8 | 3NKL, 5W7L | Polymer polymerization, initiating GT that adds Psl repeat unit to lipid carrier | TM domain (1–147), Rossmann fold (147–273), bacterial sugar transferase (273–478) | Ray et al. (2018) | |
| PslB | Q9I1N7 | 2 × 65, 2PFW | PMI activity, GMP activity | Rossmann-like fold (9–216), left β-helix (217–368), cupin domain (369–488) | Pelissier et al. (2010) | |
| PslC | Q9I1N6 | 7SP8 | GT | GT-A fold | Maloney et al. (2022) | |
| PslD | Q9I1N5 | 2J58 | OPX | Antiparallel β-sandwich with α helix (46–134), mixed α/β fold (135–220) | Dong et al. (2006) | |
| PslE | Q9I1N4 | 7NII | PCP | TM domain (5–39, 457–484), protein kinase domain (458–662) | Yang et al. (2021) | |
| PslF | Q9I1N3 | 6KIH | GT | GT-B fold | Li et al. (2020) | |
| PslG | Q9I1N2 | 5BXA, 4ZN2 | Glycoside hydrolase | TM helix (1–31), (β/α)8 TIM barrel fold (31–359), β sandwich fold (360–442) | Baker et al. (2015), Yu et al. (2015) | |
| PslH | Q9I1N1 | 6KIH | GT-B fold | Li et al. (2020) | ||
| PslI | Q9I1N0 | 6KIH | GT-B fold | Li et al. (2020) | ||
| PslJ | Q9I1M9 | 7TPG | Wzy GT | GT-C fold | Ashraf et al. (2022) | |
| PslK | Q9I1M8 | 6NC6 | Wzx flippase | MATE-like, MurJ-like | Kuk et al. (2019) | |
| PslL | Q9I1M7 | O-acyltransferase, unknown |
Numbering for P. aeruginosa protein.
In all three exopolysaccharide biosynthetic systems in P. aeruginosa, there is an enzyme that can cleave the polymer; AlgL, PelA, and PslG are periplasmic polymer modifying enzymes that can degrade alginate, Pel, and Psl, respectively. Each enzyme is implicated in biofilm formation or cell viability during polymer production. As we will discuss in this review, each system’s polymer degrading enzyme has a distinct role, even though they share a similar core function. This core function is being explored as a potential antimicrobial therapy that is designed to degrade the exopolysaccharides during P. aeruginosa biofilm infections (see section ‘Glycoside hydrolases and lyases as potential therapeutics’).
Despite their differences in mechanism of polymer production, the processes of generating alginate, Pel, and Psl can be broken down into the following steps: (1) precursor formation, (2) polymer polymerization, (3) polymer modification, and (4) polymer secretion. We will not focus on how sigma factors, levels of cyclic-dimeric guanosine monophosphate (c-di-GMP), small DNAs or other signals regulate P. aeruginosa exopolysaccharide biosynthesis, except where it pertains to the c-di-GMP receptors present in the biosynthetic systems, as the regulation of exopolysaccharide biosynthesis has recently been reviewed elsewhere (Ma et al. 2022). In the following sections, we discuss recent advances that enhance our understanding of P. aeruginosa exopolysaccharide biosynthesis and engineering of glycoside hydrolases/lyases for antibiofilm therapeutics. At the time of the last review discussing P. aeruginosa exopolysaccharide biosynthesis (Franklin et al. 2011), only four experimentally determined structures were reported in the alginate system and none were reported in the Pel and Psl systems. To date, experimentally determined structures for 15 exopolysaccharide biosynthesis proteins have now been reported: 10 from the alginate system, four from the Pel system, and one from the Psl system. Moreover, our use of the AlphaFold structural prediction artificial intelligence program (Jumper et al. 2021) alongside the DALI structural homology search software (Holm and Rosenström 2010, Holm 2022) has given us insight into the structure and function of individuals proteins and protein complexes in these systems. Thus, our focus will be predominantly on structural advances that have been made since the last review (Franklin et al. 2011), although we will touch on many aspects of P. aeruginosa exopolysaccharide biosynthesis.
Alginate biosynthesis
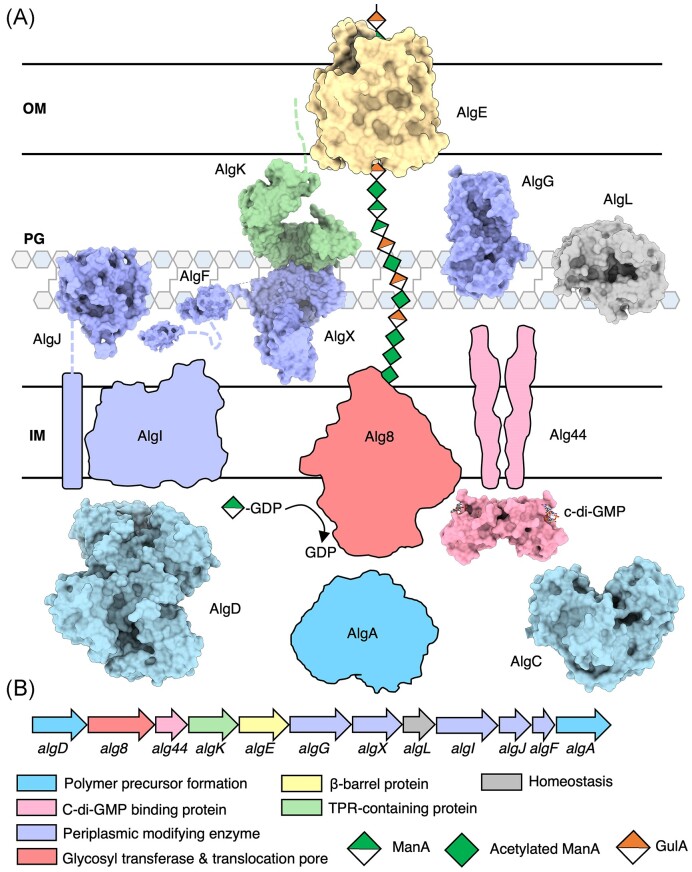
The 12 genes involved in alginate biosynthesis, except for algC, are organized in a single operon: algD-8-44-K-E-G-X-L-I-J-F-A (Fig. 2) (Jain and Ohman 2004). The promoter located upstream of algD is under the control of the sigma factor AlgU/T (Martin et al. 1993, Boucher et al. 1997), which also regulates hundreds of genes in P. aeruginosa unrelated to alginate production (Hershberger et al. 1995, Yu et al. 1995, 1996, Schurr et al. 1996, Wood and Ohman 2009, 2012). Typically, when P. aeruginosa undergoes conversion to a mucoid phenotype in the CF lung, the antisigma factor that is responsible for regulating AlgU/T, MucA, is mutated (mucA22) (Martin et al. 1993, Boucher et al. 1997). Thus, AlgU/T is constitutively active, leading to constitutive expression of the alginate operon. Evidence for two internal promoters within the operon, upstream of algG and algI, has been demonstrated suggesting a mechanism for P. aeruginosa to tailor alginate modifications under varying conditions (Paletta and Ohman 2012). Although it is well-understood how alginate acetylation and epimerization confer advantages to P. aeruginosa, it is still not known what environmental triggers influence the bacteria to alter the degree of polymer modifications or which sigma factors are involved in regulating the internal promoters.
Figure 2.
Model of the alginate biosynthetic complex in P. aeruginosa. (A) Model of the alginate biosynthetic complex colour coded by function. Structures of AlgD (PDB: 1MFZ, 1MUU, 1MV8) (Snook et al. 2003), AlgC (PDB: 1K2Y, 1K35) (Regni et al. 2002), the cytoplasmic domain of Alg44 (PDB: 4RT0, 4RT1,) (Whitney et al. 2015), cytoplasmic domain of AlgJ (PDB: 4O8V) (Baker et al. 2014), AlgF (PDB: 6D10, 6CZT), AlgX (PDB: 4KNC) (Riley et al. 2013), AlgG (PDB: 4NK8, 4OZZ, 4OZY) (Wolfram et al. 2014), AlgL (PDB: 4OZV, 4OZW, 7SA8) (Gheorghita et al. 2022b), AlgK (PDB: 3E4B) (Keiski et al. 2010), and AlgE (PDB: 3RBH, 4AZL, 4B61, 4AFK) (Whitney et al. 2011, Tan et al. 2014) have been experimentally determined and are drawn to scale and shown in a surface representation. (B) Alginate operon in P. aeruginosa colour coded by proposed function. IM, inner membrane; PG, peptidoglycan; and OM, outer membrane.
Alginate biosynthesis is initiated with precursor formation in the cytoplasm by AlgA, AlgC, and AlgD. In response to c-di-GMP binding the PilZ domain of Alg44, the polymerase Alg8 synthesizes a homopolymer of d-mannuronate, which is translocated to the periplasm (Jain and Ohman 2004, Remminghorst and Rehm 2006a, b, Merighi et al. 2007, Whitney et al. 2015). Once in the periplasm, AlgI, AlgJ, AlgF, and AlgX are involved in acetylating the polymer at O2 and/or O3 hydroxyl. Acetylation maintains the P. aeruginosa biofilm architecture and reduces killing by macrophages and susceptibility to enzymatic degradation (Whitfield et al. 2015, Farrell and Tipton 2012). AlgG can epimerize nonacetylated mannuronate residues to l-guluronate, improving biofilm cohesion (Whitfield et al. 2015, Grant et al. 1973, Simpson et al. 1988, Nivens et al. 2001). Finally, the polymer is exported out of the cell by the OM porin AlgE with the help of the TPR-containing protein AlgK. The alginate lyase, AlgL, maintains the homeostasis of the periplasm during polymer biosynthesis by degrading aberrant or improperly exported polymer (Fig. 2) (Gheorghita et al. 2022b). Deletion of either algX, algG, or algK results in secretion of small uronic acid AlgL degradation products (Jain et al. 2003, Robles-Price et al. 2004), suggesting that formation of a transenvelope complex is required for successful export of alginate polymer.
Precursor formation
For alginate production, the cytoplasmic proteins AlgA, AlgC, and AlgD perform four chemical reactions that are required to generate the GDP-mannuronic acid precursor:
In the first reaction, AlgA’s phosphomannose isomerase (PMI) activity converts fructose-6-phosphate to mannose-6-phosphate (reaction 1), which is then converted to mannose-1-phosphate by AlgC’s phosphomannomutase activity (reaction 2). AlgA uses its GDP-mannose pyrophosphorylase (GMP) activity to add GDP to mannose (reaction 3), which is then finally converted to GDP-mannuronate by AlgD’s GDP-mannose dehydrogenase activity (reaction 4) (Darzins et al. 1986, Deretic et al. 1987a, b, Zielinski et al. 1991, Franklin et al. 2011).
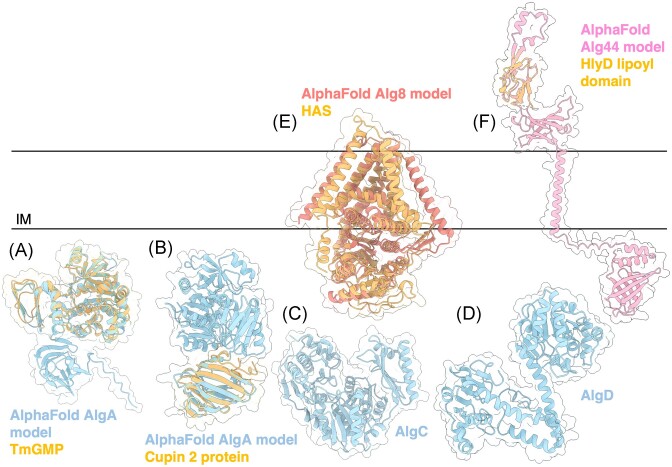
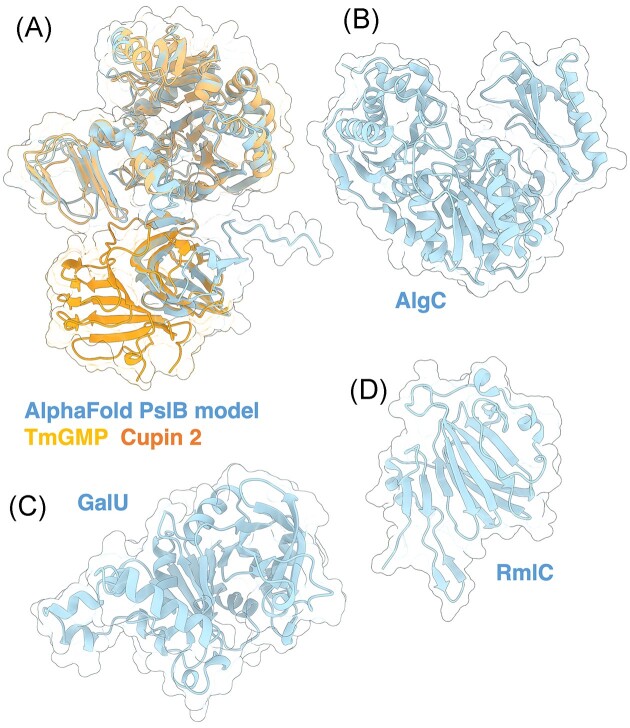
AlgA is a bifunctional enzyme with two predicted domains: a PMI domain and GMP domain (Darzins et al. 1986). Although the structure of AlgA has not yet been solved, comparison of its AlphaFold predicted model to other 3D structures using the DALI server demonstrates that AlgA has three domains, two of which resemble the GMP enzyme from Thermotoga maritima (TmGMP) (Fig. 3A) (PDB: 2 × 65) (Holm and Rosenström 2010, Pelissier et al. 2010, Jumper et al. 2021). The N-terminal domain of TmGMP has a Rossmann-like fold characteristic of proteins that bind or generate sugar-nucleotide molecules, while its C-terminal domain has an α-helix followed by a left-handed parallel β helix. TmGMP functions as a homodimer, however, it is not clear if AlgA behaves similarly. The third AlgA domain is a C-terminal Cupin 2 barrel that is predicted to have PMI activity and resembles the RmlC-like Cupin (SFRI_3101) from Shewanella frigidmarina NCIMB 400 (Fig. 3B) (PDB: 2PFW). Cupins are a functionally diverse family of proteins and have been found in other PMIs, including a PMI from Candida albicans essential in cell wall biosynthesis (Cleasby et al. 1996, Dunwell et al. 2004) (PDB: 1PMI).
Figure 3.
Structures and predicted AlphaFold models of P. aeruginosa alginate biosynthesis proteins. (A) Superimposition of P. aeruginosa AlgA AlphaFold model (light blue) and the crystal structure of T. maritima GMP TmGMP monomer (orange) (PDB: 2 × 65) with a Cα RMSD of 1.179 Å. (B) Superimposition of P. aeruginosa AlgA AlphaFold model (light blue) and the crystal structure of Shewanella frigidimarina Cupin 2 conserved barrel domain protein (orange) (PDB: 2PFW) with a Cα RMSD of 0.843 Å. (C) Crystal structure of P. aeruginosa AlgC (PDB: 1K2Y). (D) Crystal structure of a monomer of P. aeruginosa AlgD (PDB: 1MV8). (E) Superimposition of P. aeruginosa Alg8 AlphaFold model (salmon) with the crystal structure of the hyaluronan synthase (HAS) from Paramecium bursaria Chlorella virus CZ-2 (orange) (PDB: 7SP8) with a Cα RMSD of 2.932 Å. (F) Superimposition of a monomer of P. aeruginosa Alg44 AlphaFold model (pink) with the crystal structure of the lipoyl domain from the membrane fusion protein HlyD from E. coli (orange) (PDB: 5C22) with a Cα RMSD of 0.774 Å. IM; inner membrane.
AlgC has four domains, three of which consist of a mixed α/β structure with a four-stranded β-sheet sandwiched between two α-helices (Fig. 3C) (Regni et al. 2002) (PDB: 1K2Y). The fourth domain is a member of the TATA-box binding protein-like fold superfamily, consisting of a four-stranded antiparallel β-sheet flanked by two α-helices and two β-strands (Regni et al. 2002). Together, AlgC forms a compact heart-shaped structure. Analysis of the active site and sequence comparisons to other phosphomannomutase/phosphoglucomutase enzymes revealed residues important for activity: the active site Ser108 involved in phosphoryl group transfer; a metal binding loop (residues 242–246) that chelates a Mg2+ ion that is required for activity; a sugar binding loop (residues 324–328); and the distal phosphate binding site residue, Arg421, that is thought to interact with the bis-phosphorylated reaction intermediate (Regni et al. 2002).
The structure of AlgD has also been determined, revealing two domains connected by a long α-helix (Fig. 3D) (Snook et al. 2003) (PDB: 1MV8). Both domains have a Rossmann fold with a central, twisted β-sheet flanked by α-helices. The structure was complexed with NADH and GDP-mannuronic acid, revealing that AlgD forms a domain swapped dimer, with each polypeptide contributing to the active site (Snook et al. 2003, Franklin et al. 2011). Given that AlgA, AlgC, and AlgD are responsible for generating GDP-mannuronate, it is possible that these enzymes form a cytoplasmic subcomplex that can channels the polymer precursor molecule between the various active sites and ultimately to the polymerization machinery, however, this has not yet been experimentally determined.
Polymerization
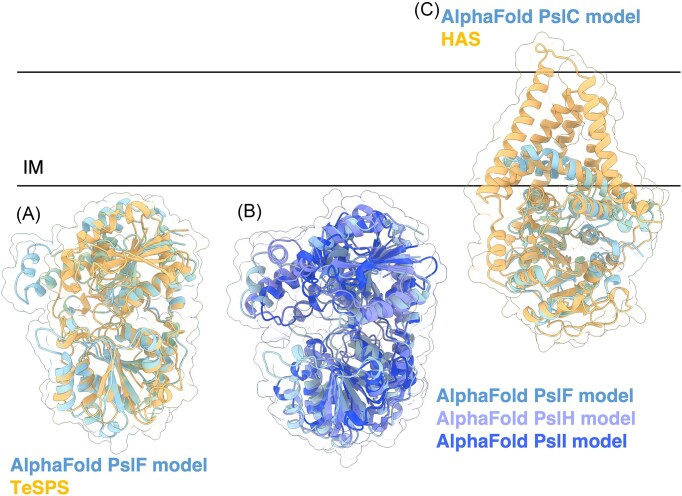
Once GDP-mannuronate is generated, the IM GT Alg8 can polymerize polymannuronate (polyM) in response to c-di-GMP binding to the PilZ domain containing protein Alg44 (Merighi et al. 2007, Oglesby et al. 2008). Deletion of either alg8 or alg44 completely abrogates alginate production and results in no alginate being secreted (Oglesby et al. 2008). Coimmunoprecipitation (co-IP) studies show that Alg8 and Alg44 interact in vivo, forming a functional unit that is required for alginate production (Moradali et al. 2015). The structure of Alg8 is yet to be determined, but it is predicted to have a GT-A fold with alternating α-helices and β-sheets (Breton et al. 2012) and is a member of the GT-2 superfamily, as defined by the carbohydrate-active enzyme database (CAZy; http://www.cazy.org/) (Drula et al. 2022). Alg8 has four transmembrane (TM) domains, a cytoplasmic GT domain, and two canonical DXD motifs, although only one is required for in vivo polymerase activity (Remminghorst and Rehm 2006a, Oglesby et al. 2008). Results from the DALI server show that the AlphaFold predicted model of Alg8 resembles hyaluronan synthase (HAS) from Paramecium bursaria Chlorella virus CZ-2 (Fig. 3E) (Holm and Rosenström 2010, Jumper et al. 2021, Maloney et al. 2022) (PDB: 7SP8). HAS displays a cytoplasmic GT-A fold that packs against five TM helices and three amphipathic interface helices. One of the interface helices (residues 331–357) contains the QXXRW active site motif that is characteristic of membrane-embedded processive family-2 GTs and is involved in holding the growing polymer chain in the active site (Saxena and Brown 1997). Above the QXXRW motif, HAS forms a TM channel through which the polymer is translocated upon substrate binding. UDP-N-acetylglucosamine (UDP-GlcNAc)-bound HAS shows that the side chain of Gln338 in the QXXRW motif hydrogen bonds with the UDP-GlcNAc diphosphate hydrogen. Although this motif is conserved in other synthase-dependent GTs, including the PNAG synthase PgaC, the chitin synthase ChsB, and the cellulose synthase BscA, this motif is LXXRW in Alg8. It is unclear if the Leu residue in the Alg8 LXXRW motif performs a similar role with GDP-mannuronate. Furthermore, UDP-GlcNAc-bound HAS shows a partially hydrolysed substrate, giving us insight into the mechanism of hyaluronic acid polymerization and membrane translocation. In a resting state, HAS has a priming loop that occludes the active site. When HAS binds UDP-GlcNAc, the priming loop is displaced from the active site, inducing a conformational change whereby the GT domain undergoes a 10° rigid body rotation towards the IM, and UDP-GlcNAc is hydrolyzed. Residue W342 binds to GlcNAc, the priming loop returns to the active site and the GT domain relaxes away from the IM, generating the primed state of the enzyme. This process continues with repeating UPD-GlcNAc and UDP-glucuronic acid, with polymer translocation coupled to substrate binding (Maloney et al. 2022). As Alg8 and HAS are both members of the GT-2 family, structural and mechanistic insights from HAS are informative, however, the HAS priming loop is not present in the AlphaFold model of Alg8; this region in Alg8 is predicted to be structured and form a β-hairpin. The AlphaFold model of Alg8 is of high confidence and there is low sequence conservation between Alg8 and HAS in this region; 5/18 residues are conserved, 3/18 residues have similar chemical properties, and the remaining 10/18 residues are not conserved, suggesting that Alg8 may use a different mechanism of polymer elongation and translocation. Although HAS is not known to have interaction partners that are required for its activity, post-translational modifications have been reported to influence human HAS activity (Melero-Fernandez de Mera et al. 2019, Kasai et al. 2020). To date, it is not known whether Alg8 is post-translationally modified or if post-translational modifications influence Alg8 activity.
The structure of the cytoplasmic PilZ domain of Alg44 has been solved suggesting that the protein functions as a homodimer, with each PilZ domain binding a dimer of c-di-GMP (Whitney et al. 2015) (PDB: 4RT0). Alg44 is sometimes referred to as a copolymerase (Rehman et al. 2013, Moradali et al. 2015, Moradali et al. 2017), but it is still unclear how Alg44 and Alg8 interact, how Alg44 binding ci-d-GMP influences Alg8 to allow for polymer synthesis, if Alg44 assists Alg8 in polymer translocation into the periplasm, and how this might occur. In addition to its role in polymerization, Alg44 has been proposed to coordinate alginate biosynthesis proteins across the inner and OMs by interacting with AlgK and AlgX via its periplasmic domain (Moradali et al. 2015, Rehman et al. 2013). Alg44’s periplasmic domain is predicted to be a membrane fusion protein domain like the multidrug efflux pump protein, MexA (Remminghorst and Rehm 2006a), which is involved in linking the inner and OMs. The DALI server demonstrates that Alg44’s AlphaFold predicted model resembles the membrane fusion protein HlyD from E. coli (Fig. 3F) (Kim et al. 2016) (PDB: 5C22). HlyD is part of the Hly translocator protein complex, a type I secretion system, which uropathogenic strains of E. coli use to secrete cytotoxin α-hemolysin (Kim et al. 2016). HlyD mediates toxin secretion by acting as a periplasmic adaptor protein that spans across the inner and OMs (Kim et al. 2016). HlyD has two domains, an α-helical domain and a lipoyl domain. HlyD is proposed to function as a hexamer, forming a cogwheel-like structure to allow for symmetrical interactions between the TolC OM channel protein and the IM ABC transporter HlyB. Currently, it is unclear how a homodimer of Alg44’s periplasmic domain might interact with other alginate proteins.
Polymer modification
After translocation to the periplasm, d-mannuronate residues can be epimerized to l-guluronate by AlgG (Franklin et al. 1994). The crystal structure of Pseudomonas syringae AlgG has been solved, demonstrating that the C5 epimerase adopts a right-handed parallel β-helix fold with a carbohydrate-binding/sugar hydrolysis domain (Fig. 4A) (Wolfram et al. 2014) (PDB: 4NK6). Site-directed mutagenesis and functional studies in P. syringae AlgG suggest a mechanism of epimerization where His319 is the catalytic base, Arg345 neutralizes the anionic intermediate, and water is the catalytic acid (Wolfram et al. 2014). Examining the surface electrostatic potential and residue conservation revealed a ∼49 Å long region below the active site that is highly electropositive and highly conserved, suggesting that this is an extended substrate binding site. Indeed, in vitro ligand binding studies with mannuronate oligomers of up to 12 residues demonstrated that AlgG binding affinity increases with ligand length, up to nine residues, leading to the suggestion that the AlgG substrate binding site can accommodate alginate polymers of at least nine residues in length (Wolfram et al. 2014). A S272N mutation in P. aeruginosa does not completely abrogate alginate biosynthesis but results in a polymer lacking l-guluronate residues (Chitnis and Ohman 1990). The effects of a catalytically inactive AlgG on the ability of P. aeruginosa to tolerate antibiotic treatment or evade the host immune response has not yet been assessed. To date, no AlgG interaction partners have been directly identified, however mutual stability analyses suggest that AlgG may interact with Alg8 (Rehman et al. 2013).
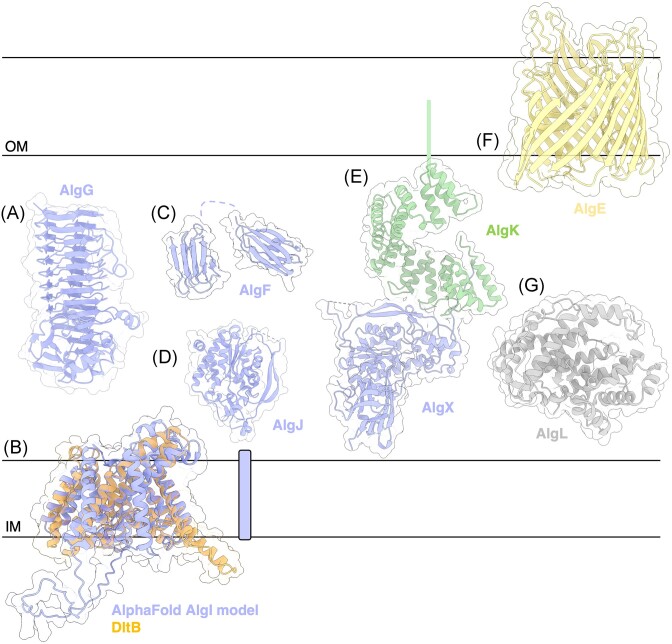
Figure 4.
Structures and predicted AlphaFold models of P. aeruginosa alginate modification and export proteins. (A) Crystal structure of P. syringae pv. Tomato AlgG (PDB: 4NK6). (B) Superimposition of P. aeruginosa AlgI AlphaFold model (periwinkle) and the crystal structure of Streptococcus thermophilus O-acyltransferase DltB (orange) (PDB: 6BUG) with a Cα RMSD of 1.788 Å. (C) CS-Rosetta determined structure of P. aeruginosa AlgF (N-terminus PDB: 6CZT; C-terminus: 6D10). (D) Crystal structure of P. aeruginosa AlgJ (PDB: 4O8V). (E) Crystal structure of P. aeruginosa AlgL (4OZV). (F) Crystal structure of the P. putida AlgKX complex (PDB: 7ULA). (G) Crystal structure of P. aeruginosa AlgE (PDB: 3RBH). (H) Crystal structure of P. aeruginosa AlgL (PDB: 4OZV). IM, inner membrane; OM, outer membrane.
In addition to epimerization, d-mannuronate residues can also be O-acetylated via the acetylation complex AlgIJFX (Franklin and Ohman 1993, 1996, 2002, Jain and Ohman 2004, Chanasit et al. 2020). A combination of co-IP, mutual stability studies, and in vitro isothermal titration calorimetry with the acetylation machinery in P. aeruginosa has demonstrated that AlgI interacts with AlgJ and AlgF, and that AlgF interacts with AlgJ and AlgX, suggesting that AlgIJFX form a complex in vivo (Chanasit et al. 2020, Low et al. 2023). Deletion of alg8 was found to destabilize AlgI in P. aeruginosa, implying that Alg8 and AlgI interact (Chanasit et al. 2020). Furthermore, co-IP studies also revealed that AlgX and Alg44 interact (Moradali et al. 2015). Thus, the O-acetylation machinery and polymerization machinery may form an IM polymerization/modification complex, with alginate O-acetylation linked to polymerization.
The source of the acetyl group used to modify alginate is still unknown, although acetyl-CoA has been suggested (Franklin et al. 2011). In the IM, the membrane bound O-acyl transferase (MBOAT) family enzyme AlgI is thought to transfer an acetyl group from a cytoplasmic donor into the periplasm (Franklin and Ohman 1996, 2002). How this occurs is still unknown, thus future studies into the structure of AlgI would help to elucidate the mechanism AlgI uses to transfer the acetyl group. To gain some insight, we used the DALI server and found that the AlphaFold predicted structure of AlgI resembles the MBOAT DltB from Streptococcus thermophilus involved in d-alanylation of cell wall teichoic acid polymer (Fig. 4B) (Ma et al. 2018) (PDB: 6BUG). The crystal structure of DltB reveals a ring of 11 TM helices that create a highly conserved funnel which extends into the lipid bilayer and mediates transfer of a d-alanyl group to the polymer. In comparison, AlgI is predicted to have two additional TM helices at the C-terminus, which are connected by a long, unstructured cytoplasmic loop. At the bottom of the extracellular funnel in DltB is the catalytic His336, which is highly conserved across MBOATs and required for d-alanylation in vivo (Ma et al. 2018). The DltB tunnel is formed by three long TM helices and a small horizontal helix H12 in the central core of the protein. A conserved Trp285 on helix H12 occludes the tunnel, keeping it in a closed conformation. It is proposed that helix H12 is a mobile structure that may undergo a conformational change leading to tunnel opening and subsequent transfer for the d-alanyl group from DltC to DltB. Comparison to DltB suggests that AlgI also has a funnel that allows an acetyl group to travel across a lipid bilayer. Furthermore, the helix H12 structure is conserved in the AlphaFold AlgI model, suggesting that AlgI may use a similar mechanism for acetyl group transfer. However, DltB requires interaction with its cytoplasmic d-alanyl donor protein, DltC, as mutation of the DltB–DltC binding site abolishes d-alanylation in vivo (Ma et al. 2018). To date, no cytoplasmic AlgI binding partners have been identified but we hypothesize that the large, unstructured cytoplasmic loop observed in the AlgI AlphaFold model may be involved in coordinating interaction with an acetyl group donor protein (Fig. 4B).
To initiate polymer acetylation, it is hypothesized that AlgI relays an acetyl group to the SGNH hydrolase-like acetyltransferase enzyme AlgJ (Fig. 4D) (Baker et al. 2014) (PDB: 4O8V). Using site-directed mutagenesis and in vitro enzymatic assays, the catalytic triad residues Asp193, His195, and Ser297 were found to be required for P. aeruginosa AlgJ acetylesterase activity. Since AlgJ does not bind polyM oligosaccharides in vitro (Baker et al. 2014), it is thought to be an intermediary enzyme that transfers the acetyl group to AlgX (Riley et al. 2013) (PDB: 4KNC, 7ULA) via interaction with the bilobed β-sandwich protein AlgF (Fig. 4C) (PDB: 6D10, 6CZT) (Low et al. 2023). AlgF is proposed to act as a protein–protein interaction mediator to facilitate acetyl group transfer between the AlgJ and AlgX (Low et al. 2023). AlgX has been shown to directly bind to and acetylate polyM oligosaccharides in vitro (Baker et al. 2014), thus positioning it as the terminal acetyltransferase enzyme in the acetyl group relay. AlgX has a catalytic triad composed of Asp174, His176, and Ser269, which are required for acetylesterase activity in vitro and alginate acetylation in vivo (Riley et al. 2013). In addition to an SGNH hydrolase-like acetyltransferase domain similar to AlgJ, AlgX also has a carbohydrate-binding (CBM) domain, which is assumed to bind alginate (Riley et al. 2013). However, AlgX’s CBM has not yet been assessed in isolation for its ability to bind polyM oligosaccharides.
Deletion of either algI, algF, or algJ results in secretion of unacetylated alginate, revealing that acetylation is not a requirement for alginate polymerization and secretion (Franklin and Ohman 1996). However, acetylation-deficient P. aeruginosa display abrogated cell adhesion and biofilm architecture (Tielen et al. 2005). Unlike the rest of the acetylation machinery, deletion of algX results in secretion of small uronic acid degradation products generated by AlgL, suggesting that AlgX is required for proper export of alginate. An interaction between AlgX and the TPR-containing export protein AlgK was detected previously (Hay et al. 2012), but it was only recently that the molecular details of this interaction have been uncovered.
The structure of AlgX in complex with AlgK has been determined by X-ray crystallographic methods, providing insight into how modification and export are coordinated for polymer production (Fig. 4E) (Gheorghita et al. 2022a). The AlgKX complex (PDB: 7ULA) reveals that the N-terminus of AlgX interacts with TPRs nine and ten of AlgK. Complex interaction is mediated by a highly conserved, hydrophobic patch on the N-terminus of AlgX that is buried in a deep, conserved hydrophobic groove on AlgK (Gheorghita et al. 2022a). The AlgKX interaction generates an electropositive passageway through which alginate can traverse towards the OM porin AlgE after acetylation by AlgX (Fig. 4F) (Whitney et al. 2011) (PDB: 3RBH). Thus, the structure demonstrates that acetylation immediately precedes export and, therefore, implies an order in which alginate modifications occur. Furthermore, in vitro ligand binding analyses demonstrated that the AlgKX complex is capable of binding both polyM and polymannuronate-guluronate (polyMG) alginate oligosaccharide ligands. When AlgKX complex formation was disrupted in vivo, P. aeruginosa alginate secretion and biofilm attachment was abrogated, revealing that AlgX has a role in alginate export (Gheorghita et al. 2022a).
Polymer secretion
Comparison to other OM porins within Gram-negative synthase-dependent secretion systems show that the ß-barrel porin is fused to a periplasmic TPR domain, suggesting that AlgK and AlgE interact to facilitate polymer export (Keiski et al. 2010). Moreover, deletion of algE destabilizes AlgK (Rehman et al. 2013) and deletion of algK impairs the proper localization of AlgE (Keiski et al. 2010), further emphasizing that AlgE and AlgK form an OM secretion complex. Within the E. coli cellulose system, the structure of the OM porin and TPR-containing protein BscC (Acheson et al. 2019) (PDB: 6TZK) provides precedence for how an AlgEK complex may be oriented. Molecular dynamics simulations (Tan et al. 2014) and predictive structural modelling by AlphaFold (Gheorghita et al. 2022a) both reveal a model of the AlgEK complex that further extends the electropositive pathway from the active site of AlgX, across the interior of AlgK, and through AlgE.
AlgE forms an 18-stranded antiparallel β-barrel with an electropositive pore constriction (PDB: 3RBH) (Fig. 4F). The crystal structure of AlgE suggests that the porin adopts a closed conformation where the electropositive core constriction is occluded on both sides by an extracellular loop L2 and a periplasmic loop T8. L2 is thought to prevent the surrounding LPS from interfering with alginate export, while deletion of the T8 loop resulted in a threefold increase in in vitro iodide efflux, implying that that T8 regulates passage of molecules through the porin (Whitney et al. 2011). AlgE is thought to require interaction with AlgK to induce the T8 loop into an open conformation to allow alginate export (Tan et al. 2014). However, it is still unclear if interaction with AlgK induces an open position in AlgE’s T8 loop as AlphaFold is limited in its ability to predict conformational changes (Gheorghita et al. 2022a). Deletion of algE abrogates alginate polymer secretion and only free uronic acids, presumably AlgL degradation products, are secreted (Rehman and Rehm 2013).
For alginate to be secreted to the extracellular matrix, energy must be used to drive polymer export through AlgE. Molecular dynamics studies on AlgE and polyMG to determine whether a push or pull force influences alginate polymer migration through AlgE, suggest that AlgE itself does not influence the directionality of alginate transport, although breathing motions of the protein may facilitate polymer translocation. Thus, it was concluded that that the energy required for this process must be provided by the IM polymerization machinery (Tan et al. 2014).
Homeostasis
Various roles for AlgL have been proposed, including regulating the length of secreted alginate (Boyd et al. 1993, May and Chakrabarty 1994), aiding in biofilm detachment (Boyd and Chakrabarty 1994), and being completely dispensable for alginate production (Wang et al. 2016b). Previously, AlgL has been described to be a polymer modifying enzyme, similar to AlgG and AlgX, however, recent work has demonstrated another role for the enzyme. The structure of AlgL revealed an (α/α)5 toroid fold (Fig. 4G) (PDB: 4OZV), with a highly conserved and highly electropositive binding groove (Gheorghita et al. 2022b). Structural comparison with other polysaccharide lyase family 5 (PL5) members and in vitro enzymatic assays identified key residues: K66, W205, Y259, and H202, Y246, R249 involved in substrate binding and catalysis, respectively. In vivo analyses in P. aeruginosa determined that both an algL deletion and mutation of its catalytic residues, Y256 and R249, results in growth defects, abnormal cellular morphologies, and cell lysis in a genetic background where the alginate operon is isolated from its native system of regulation by AlgU/T (Gheorghita et al. 2022b). Moreover, an algL deletion was found to directly result in accumulation of alginate within the periplasm. Thus, suggesting that AlgL functions to maintain the homeostasis of the periplasm during alginate production. Growth defects and periplasmic retention of alginate were mitigated with constitutive expression of the AlgU/T regulon, revealing that a gene or genes within the regulon compensate for the loss of algL in a muca22 background. The alternative alginate lyases PA1167 and/or PA1784 have been proposed to perform this function, acting as a fail-safe to ensure cell viability during alginate production (Gheorghita et al. 2022b). Co-IP studies determined that AlgL does not interact with other alginate proteins, revealing that it does not need to interact with the alginate biosynthetic complex to performs its function as a periplasmic homeostasis enzyme (Wang et al. 2016b, Gheorghita et al. 2022b).
Pel biosynthesis
The seven genes required for Pel biosynthesis are in a single operon: pelA-B-C-D-E-F-G (Fig. 5) (Friedman and Kolter 2004a). The promoter upstream of pelA is regulated by the transcription factor FleQ in response to c-di-GMP; FleQ acts as both a transcriptional activator and repressor by binding to two distinct sites at the pelA promoter: boxes 1 and 2 (Baraquet et al. 2012). In the absence of c-di-GMP, FleQ represses transcription via distortion of the pelA promoter at box 2, while FleQ bound to c-di-GMP at boxes 1 and 2 relieves the DNA distortion and leads to pel expression (Baraquet et al. 2012).
Figure 5.
Model of the Pel biosynthetic complex in P. aeruginosa. (A) Model of the Pel biosynthetic complex colour coded by function. Structures of the cytoplasmic domain of PelD (PDB: 4DN0) (Whitney et al. 2012), the hydrolase domain PelA (PDB: 5TCB), PelC (PDB: 5T11, 5T10, 5T0Z) (Marmont et al. 2017a), and the complete TPR motifs R9 and R10 of PelB (PDB: 5WFT) (Marmont et al. 2017b) have been experimentally determined and are drawn to scale and shown in a surface representation. (B) Pel operon in P. aeruginosa colour coded by function. IM, inner membrane; PG, peptidoglycan; and OM, outer membrane.
Biosynthesis of Pel is produced by a synthase-dependent secretion system and is regulated by c-di-GMP binding to the GGDEF domain of PelD (Whitney et al. 2012). The cytoplasmic glycosyltransferase PelF is believed to bind to the sugar nucleotide precursor UDP-GalNAc and polymerize a GalNAc homopolymer, which is then translocated to the periplasm via PelG. Once in the periplasm, Pel is modified by the bifunctional deacetylase/hydrolase enzyme PelA (Colvin et al. 2013). Modification by partial deacetylation renders Pel positively charged, allowing it to be guided by the negatively charged funnel protein PelC for export through the porin PelB (Fig. 5) (Marmont et al. 2017a). Interaction between the TPR-containing domain of PelB and PelA enhances PelA’s deacetylase activity, attenuates PelA’s hydrolase activity, and is required for Pel-dependent biofilm formation (Marmont et al. 2017b). PelA’s hydrolase activity has recently been shown to be important for the formation of secreted cell-free Pel, which is involved in biofilm biomechanics and virulence modulation (Razvi et al. 2023a).
Although we focus on Pel production in P. aeruginosa, Pel biosynthetic loci have recently been identified in Gram-positive bacteria, including Bacillus, Streptococcus, Bifidobacterium, and Clostridium species (Bundalovic-Torma et al. 2020, Whitfield et al. 2020a). Bacillus cereus ATCC 10987 was found to produce a Pel-like polysaccharide and deletion of genes within the pel operon resulted in no biofilm formation. Bioinformatics analyses have identified > 900 syntenically conserved pel operons in a wide variety of Gram-positive bacterial genera, demonstrating that it is a phylogenetically widespread biofilm matrix determinant (Whitfield and Howell 2021), and thus providing evidence that Pel may be a common biofilm exopolysaccharide component for many bacteria.
Precursor formation
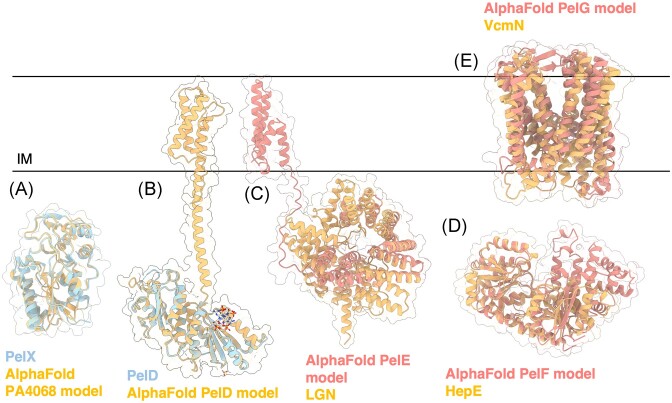
Unlike the alginate system, the pel operon in P. aeruginosa does not encode genes related to polymer precursor formation suggesting that Pel sugar-nucleotide precursors are generated from more general metabolic pathways (Franklin et al. 2011). This is not entirely unprecedented; algC is not associated with the rest of the alginate operon (Fig. 2B) as it is required for precursor production in other pathways, including the Psl exopolysaccharide and LPS production (Goldberg et al. 1993, Coyne et al. 1994, Byrd et al. 2009). However, in other bacteria with pel operons such as Curvibacter delicatus, Acidovorax citrulli, Legionella maceachernii, and Pseudomonas protogens, there is a pelX gene adjacent to the pel operon that has been shown to encode a UDP-N-acetylglucosamine C4-epimerase required for Pel-dependent biofilm formation in P. protogens (Marmont et al. 2020). The structure of PelX in complex with UDP-N-acetylglucosamine (PDB: 6WJB, 6WJ9) and UDP-N-acetylgalactosamine (PDB: 6WJA) determined by X-ray crystallographic methods revealed an N-terminal Rossmann fold and C-terminal α/β substrate-binding domain. PelX belongs to the short-chain dehydrogenase/reductase (SDR) superfamily and has the characteristic GXXGXXG NAD+ binding motif and SX24YX3K catalytic triad. Mutation of the catalytic Ser121 and Tyr146 rendered PelX catalytically inactive. Although pelX is not present in P. aeruginosa, Marmont et al. (2020) suggest that the poorly characterized PA4068 gene may be functionally analogous to pelX. PA4068 is annotated as a probable UDP-N-acetylglucosamine epimerase belonging to the SDR superfamily and its AlphaFold model strongly resembles PelX with a Cα RMSD of 0.345 Å (Fig. 6A). The striking similarity between PA4068 and PelX suggests that PA2068 may function as the UDP-N-acetylglucosamine C4-epimerase required for Pel precursor formation in P. aeruginosa (Marmont et al. 2020).
Figure 6.
Structures and predicted AlphaFold models of P. aeruginosa Pel biosynthesis proteins. (A) Superimposition of P. aeruginosa PA4068 AlphaFold model (blue) with the crystal structure of PelX from P. protogens (orange) (PDB: 6WJB) with a Ca RMSD of 0.345 Å. (B) Crystal structure of a monomer of P. aeruginosa PelD (blue) bound to c-di-GMP (PDB: 4DN0) superimposed with the AlphaFold model of PelD (orange). (C) Superimposition of P. aeruginosa PelE AlphaFold model (salmon) with the crystal structure of LGN from Mus musculus (orange) (PDB: 4JHR) with a Ca RMSD of 4.023 Å. (D) Superimposition of P. aeruginosa PelF AlphaFold model (salmon) with the crystal structure of HepE from Anabaena sp. (orange) (PDB: 4XSO) with a Ca RMSD of 1.251 Å. (E) Superimposition of P. aeruginosa PelG AlphaFold model (salmon) with the crystal structure of VcmN from Vibrio cholerae (orange) (PDB: 6IDS) with a Ca RMSD of 5.077 Å. IM, inner membrane.
Polymerization
Through a series of co-IPs in P. aeruginosa, PelD, PelE, and PelF were found to form a stable IM complex that mediates Pel polymerization (Whitfield et al. 2020b). Although co-IP with PelG was unsuccessful, PelD, PelE, PelF, and PelG were found to directly interact via bacterial two-hybrid (BACTH) assay in E. coli and PelE was found to require PelD and PelG for stability (Whitfield et al. 2020b). As proteins that interact and form complexes often display codependence for stability, it was concluded that PelDEFG form an IM complex that functions to polymerize Pel and translocate it into the periplasm (Whitfield et al. 2020b).
PelD is an IM c-di-GMP binding protein, with an N-terminal coiled-coil motif that is proposed to mediate dimerization and a C-terminal cytoplasmic GAF domain and degenerate GGDEF domain (Fig. 6B) (Whitney et al. 2012) (PDB: 4DN0). Although GAF domains are capable of binding cyclic nucleotides (Martinez et al. 2002), the GGDEF domain was found to bind dimeric c-di-GMP at an RXXD motif. This motif generally functions as an allosteric inhibition site for diguanylate cyclases; however, point mutation of the motif (R367A or D370) demonstrated that it is required for c-di-GMP binding in vitro and biofilm attachment and pellicle formation in vivo (Whitney et al. 2012). When c-di-GMP binding to PelD is abrogated by a R367A mutation, assembly of the PelDEFG complex is not affected. Thus, binding of c-di-GMP is thought to result in a conformational change or structural rearrangements between the proteins to activate Pel biosynthesis. Deletion of any one of these proteins in P. aeruginosa results in loss of Pel production as indicated by smooth colony morphologies, loss of Congo red stain binding, and loss of pellicle formation (Whitfield et al. 2020b).
PelE is predicted to be an IM protein with two TM helices and a C-terminal cytoplasmic domain (Franklin et al. 2011, Whitfield et al. 2020b). Its AlphaFold predicted model reveals a C-terminal α-helical TPR-like structure that resembles LGN from Mus musculus involved in asymmetrical cell division (Fig. 6C) (Pan et al. 2013) (PDB: 4JHR). TPR and TPR-like motifs are well-known for their ability to act as protein scaffolds and mediators of protein–protein interactions across organisms (Perez-Riba and Itzhaki 2019). Through a series of BACTH assays, Whitfield et al. (2020b) proposed that PelE functions as an intermediary that facilitates a stronger interaction between PelD and PelG. Mutual stability studies also showed that PelD and PelG enhance the stability of PelE, as PelE levels were significantly reduced in strains lacking pelD or pelG.
PelF is a cytoplasmic protein predicted to be the GT of the system (Franklin et al. 2011) and has been shown to specifically bind UDP with micromolar affinity using isothermal titration calorimetry, suggesting that PelF utilizes UDP-sugar nucleotides for Pel polymerization (Jennings et al. 2015). Moreover, these studies further provide evidence that PA2068, analogous to pelX, may function as the UDP-sugar epimerase required for Pel precursor formation (Marmont et al. 2020). Although it is a cytoplasmic protein, PelF localizes to the IM through interaction with PelD, PelE, and/or PelG (Whitfield et al. 2020b). Structural homology analysis using the AlphaFold predicted model of PelF and the DALI server show that PelF is like the glucosyltransferase HepE from Anabaena sp. involved in heterocyst envelope polysaccharide formation (Fig 6D) (Wang et al. 2016a) (PDB: 4XSO). Each subunit of HepE displays a GT-B fold with two β/α/β Rossmann-fold domains and an interdomain binding pocket. The crystal structure of HepE in complex with UDP-glucose reveals that residues Gly235, Arg267, Phe266, and Ile2670 interact with the uracil moiety of UDP; Glu296 and Arg292 interact with the ribose; Gly14, Arg292, and Lys213 interact with the pyrophosphate moiety; and Phe290, Glu288, Gly291, His121, Asn151, and Asn174 interact with the glucosyl moiety. In vitro, HepE was found to hydrolyze its sugar donor UDP-glucose to UDP. This activity was increased 4-fold in the presence of its acceptor, mannose. Similarly, PelF was shown to use UDP-glucose as a substrate by various in vitro assays, including UDP-glucose consumption and measurement of intracellular UDP-glucose levels in PelF-deficient and PelF-producing P. aeruginosa strains (Ghafoor et al. 2013). Recent data have revealed that the cell-free form of Pel does not contain glucose (Le Mauff et al. 2022), whether glucose is present in the cell-associated form of the polymer is yet to be determined. Mutation of the HepE catalytic residues His212 and Glu288 and residues involved in acceptor recognition – Glu16, Asp122, Arg208, and Phe239 – significantly abrogated in vitro enzymatic activity (Wang et al. 2016a). These catalytic residues are conserved in PelF as His214 and Glu405. Mutation of Glu405 was found to reduce PelF in vivo Congo red binding activity and pellicle formation compared to WT PelF. Additionally, mutation of residues predicted to be involved in acceptor recognition, Arg325 and Lys330 (Arg208 and Lys213 in HepE), completely abrogated in vivo PelF Congo red binding activity and demonstrated reduced pellicle formation (Ghafoor et al. 2013). HepE was found to have low activity in the presence of its nucleotide sugar substrate, however, it was proposed that this might be due to absence of its activating partner, HepD, a neighbouring putative GT (Wang et al. 2016a). Thus, PelF may similarly require interaction with the remaining inner complex proteins for activation of Pel polymerization.
PelG belongs to the multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily and was previously predicted to be part of the multidrug and toxic compound extrusion (MATE) (Jagessar et al. 2019) transporter subgroup (Franklin et al. 2011). However, more recent annotations in the Transporter Classification Database (Saier et al. 2021) suggests that PelG is part of the putative exopolysaccharide exporter (EPS-E) subgroup. At present, little is known about this MOP subgroup. The DALI server suggests that the PelG AlphaFold model is structurally similar to the MATE transporter VcmN from Vibrio cholerae (Fig. 6E) (Kusakizako et al. 2019) (PDB: 6IDS). VcmN couples H+ with its substrate transport mechanism via rearrangements in its hydrogen bonding network that induce a conformational change in one of its TM helices, TM1. In a substrate-bound state, Asp53 in TM1 is deprotonated, thus TM1 adopts a straight conformation and VcmN is in an outward-open form. Proton binding to Asp35 causes a rearrangement of the hydrogen bonding network and induces a bend in TM1 at Pro20, which reduces the volume of the substrate-binding site and extrudes the substrate. VcmN employs an antiporter mechanism, therefore, binding of the proton and the substrate are mutually exclusive events. With the proton still bound, VcmN adopts an occluded form that is an intermediary between outward-open and inward-open forms. VcmN then converts to an inward-open form, the proton is released, and a substrate enters the substrate-binding site. VcmN converts to a substrate-bound occluded form and is ready to adopt an outward-open form to restart the transport cycle (Kusakizako et al. 2019). Although the mechanisms for transport of small molecules and polymers must differ, structural similarities to MATE transporters suggest that PelG functions as the translocon required for Pel translocation into the periplasm. It is unlikely that PelG couples Pel translocation with H+, as the Asp35 in VcmN responsible for proton-binding is not conserved in PelG and there are no residues within the vicinity that could perform this function. Examination of the AlphaFold model of PelG reveals a series of conserved aromatic residues that stretch from the cytoplasmic to periplasmic face of the protein. These residues form a potential pore for export of the polymer that is reminiscent of the pore found in the cellulase synthase BcsA (Morgan et al. 2012).
Polymer modification
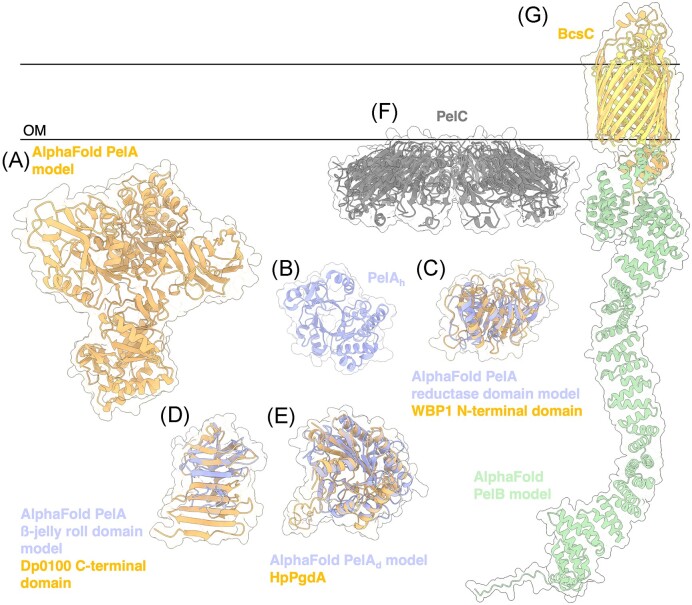
PelA is a large multidomain bifunctional enzyme that modifies Pel by hydrolysis and deacetylation (Colvin et al. 2013, Razvi et al. 2023a). Previous bioinformatics analysis suggest that PelA has four domains: an N-terminal hydrolase domain, reductase domain, deacetylase domain, and a β-jelly roll domain at the C-terminal end (Colvin et al. 2013). The AlphaFold model of PelA confirms this domain composition (Fig. 7A).
Figure 7.
Structures and predicted AlphaFold models of P. aeruginosa Pel modification and export proteins. (A) AlphaFold model of P. aeruginosa PelA. (B) Crystal structure of the P. aeruginosa PelA hydrolase domain (PelAh) (PDB: 5TCB). (C) Superimposition of P. aeruginosa PelA reductase domain AlphaFold model (periwinkle) with the CryoEM structure of WBP1 from Saccharomyces cerevisiae (orange) (PDB: 6EZN) with a Cα RMSD of 4.650 Å. (D) Superimposition of P. aeruginosa PelA β-jelly roll AlphaFold model (periwinkle) with the crystal structure of the C-terminal domain of Dp0100 alginate lyase from Defluviitalea phaphyphila (orange) (PDB: 6JP4) with a Cα RMSD of 4.379 Å. (E) Superimposition of P. aeruginosa PelA deacetylase (PelAd) domain AlphaFold model (periwinkle) with the crystal structure of HpPgdA (PDB: 4LY4) peptidoglycan deacetylase from Helicobacter pylori (orange) with a Cα RMSD of 3.100 Å. F, Crystal structure of Paraburkholderia phytofirmans PelC (PDB: 5T10). G, Superimposition of P. aeruginosa PelB AlphaFold model (yellow and green) with the crystal structure of BcsC (orange) (PDB: 6TZK) from E. coli with a Cα RMSD of 3.119 Å. OM, outer membrane.
To date, only the structure of the hydrolase domain (PelAh) has been solved, revealing an α/β TIM barrel (Fig. 7B) (Le Mauff et al. 2019) (PDB: 5TCB), a versatile fold used by many enzyme families that catalyze different reactions (Wierenga 2001). In vitro studies determined that PelAh is an endo-acting α-1,4-N-acetylgalactosaminidase that utilizes a retaining mechanism of catalysis (Le Mauff et al. 2019). Hydrolysis with net retention of configuration is most commonly achieved through a two-step, double displacement mechanism in which the glycosyl enzyme intermediate is formed and hydrolyzed by acid/base catalysis medicated by the carboxylic side chains of aspartic or glutamic acid residues that are ∼5.5 Å apart (Koshland 1953). In PelAh the distance between the catalytic Glu218 and Asp160 residues is ∼ 5.5 Å. Mutation of the catalytic residue Glu218 renders PelAh inactive and unable to hydrolyze Pel-dependent biofilms (Baker et al. 2016). The ability of PelA’s hydrolase activity to disrupt and inhibit biofilms is discussed in greater detail below. In vivo in P. aeruginosa, PelAh was recently found to be involved in biofilm dispersion and generating the secreted form of Pel (Cherny and Sauer 2020, Razvi et al. 2023a). The inactive E218A PelAh mutant of P. aeruginosa produced increased adherent biofilm biomass compared to the wild-type (WT) PelAh complemented strain of P. aeruginosa (Razvi et al. 2023a). Furthermore, E218A PelAhP. aeruginosa demonstrated altered biofilm biomechanical properties including buoyancy, cohesion, stiffness, wrinkling, and wettability, and resulted in increased virulence in two animal models of infection (Razvi et al. 2023a). Specifically, the E218A mutation in PelAh decreased pellicle buoyancy and cohesion, increased flocculation of bacterial cells, decreased colony wrinkling, increased surface wettability, and decreased biofilm stiffness through decreased ionic interactions within the biofilm (Razvi et al. 2023a).
When the reductase domain AlphaFold model of PelA was analyzed by DALI, the cryo-electron microscopy (cryoEM) structure of the yeast oligosaccharyltransferase complex was listed as a top hit (Wild et al. 2018) (PDB: 6EZN). The reductase domain shows structural similarity to the N-terminal domain of WW Domain Binding Protein 1 (WBP1) within subcomplex III (Fig. 7C). WBP1 is noncatalytic and is proposed to be involved in substrate binding (Wild et al. 2018), thus suggesting that the reductase domain of PelA may interact with Pel and help orient the substrate on the enzyme for catalysis.
Analysis by DALI determined that the AlphaFold model of PelA’s ß-jelly roll domain is similar to the C-terminal ß-sandwich domain of the polysaccharide lyase 39 family (PL39) alginate lyase from Defluviitalea phaphyphila, Dp0100 (Fig. 7D) (PDB:6JP4) (Ji et al. 2019). The crystal structure of Dp0100 with a pentamer of mannuronate identified the N-terminal (α/α)6 toroid fold domain as the substrate-binding and catalytic domain (PDB:6JPN). The role of the C-terminal ß-sandwich domain was not investigated, and it is unclear if this domain is capable of binding alginate. However, the ß-jelly roll fold is common in many CBM families within CAZy (Drula et al. 2022), suggesting that PelA’s ß-jelly roll domain, like its reductase domain, may be involved in binding Pel and orienting the polymer for modification.
The AlphaFold predicted model of the deacetylase domain (PelADA) strongly resembles the HpPgdA peptidoglycan deacetylase from Helicobacter pylori (Shaik et al. 2011) (Fig. 7E) (PDB: 4LY4). A single domain of the tetrameric HpPgdA resembles a noncanonical TIM-barrel fold (Shaik et al. 2011). The HpPgdA active site coordinates a zinc ion and is thought to be a member of the carbohydrate esterase 4 (CE4) superfamily of enzymes. Similarly, PelADA has been proposed to function as a metal dependent carbohydrate esterase as addition of the metal chelator EDTA greatly reduced activity of the enzyme, although it is still not known, which metal is required for optimal PelADA activity (Colvin et al. 2013). Comparison to other carbohydrate esterase enzymes revealed the conserved catalytic residue Asp528 and the D-H-H metal coordination triad composed of Asp530, His600, and His605. Mutation of any of these residues to alanine results in a 10-fold reduction of in vitro activity. Furthermore, PelA deacetylase activity-deficient mutant strains resulted in abrogated biofilm and pellicle formation, loss of the characteristic wrinkly colony morphology, and loss of Pel secretion in vivo in P. aeruginosa, demonstrating that Pel deacetylation is required for Pel secretion (Colvin et al. 2013, Razvi et al. 2023b). As recent structural characterization of the Pel polymer has shown that it is comprised predominantly of GalN-GalNAc repeating units (Le Mauff et al. 2022), PelADA is proposed to deacetylate every other GalNAc residue. NMR analyses and molecular dynamics simulations performed on synthetic α-1,4-GalNAc7 polymer revealed that the polymer adopts an elongated, linear structure that is stabilized by a nonconventional inter-residue hydrogen bond (Zhang et al. 2020). Thus, the carbon C2 acetyl groups are on either side of the polymer in an alternating fashion, suggesting that PelADA may deacetylate one face of the polymer in a processive manner (Zhang et al. 2020, Le Mauff et al. 2022).
Polymer secretion
Once partially deacetylated, positively charged Pel is guided by PelC for export (Fig. 5F) (Marmont et al. 2017a) (PDB: 5T11, 5T10, 5T0Z). PelC is a lipoprotein that localizes to the OM, with deletion of pelC resulting in abrogated pellicle formation and biofilm attachment (Vasseur et al. 2007, Marmont et al. 2017a). Using X-ray crystallographic methods, PelC was determined to have a mixed α/β fold and form a dodecamer ring-like complex that is ∼120 Å in diameter with a 32 Å pore. Consistent with the PelC crystal structures, preliminary negative-stain electron microscopy of PelC purified from P. aeruginosa also revealed a ring-like structure that is ∼120 Å in diameter, providing evidence to suggest that PelC functions as an oligomeric complex in vivo. The negatively charged concave pore of PelC is critical for biofilm formation as charge-switching mutations were found to significantly reduce pellicle formation. Thus, it is proposed that the PelC ring attracts modified Pel towards its pore to funnel the polysaccharide to the β-barrel protein PelB, forming a novel secretion complex not previously identified in other synthase-dependent systems (Marmont et al. 2017a).
The AlphaFold predicted model of PelB strongly resembles the previously described synthase-dependent cellulose export protein, BcsC from E. coli (Fig. 5G) (Acheson et al. 2019) (PDB: 6TZK). Unlike the AlgE porin, no periplasm-facing loop is observed in the β-barrel of either BcsC or the predicted PelB model, suggesting that the pore of PelB remains constitutively in an open conformation. Although it is predicted to have 19 TPR motifs, only a small segment, TPR motifs (R9 and R10) and two partial TRP motifs (R8 and R11), of PelB’s TPR domain has been structurally determined (Marmont et al. 2017b) (PDB: 5WFT). PelB’s 19 TPR motifs are ~200 Å, and could, therefore, span the width of the periplasm and interact with the IM Pel proteins to coordinate polymer production, as suggested for AlgK in the alginate system. However, to date, no IM Pel proteins have been identified to interact with PelB’s TPR domain. Additionally, bioinformatics analysis using Phobius (Madeira et al. 2019), and previous work from the Villoux lab (Vasseur et al. 2007) suggests that the N-terminal residues 19–36 form a TM helix, possibly inserting the N-terminus of PelB into the IM. Unlike AlgE, AlgK, and BcsC, PelB is not predicted to have a N-terminal signal sequence for processing to the periplasm/OM, further suggesting that this region may be required for insertion into the IM. If this is the case, how PelB is processed and oriented appropriately in both membranes, and the potential periplasmic chaperones associated with this process, is currently unknown.
Given that PelC is thought to associate with PelB at the OM to guide Pel for export, it is also not clear how PelB’s TPR domain is oriented with respect to PelC. An interaction between PelB and PelC has yet to be demonstrated, but this may be due to the size of the interaction interface. It has been proposed that Trp149 on the membrane proximal surface of PelC is involved in orienting PelC against the OM and participating in stacking interactions with periplasm-facing aromatic residues on PelB. Indeed, mutation of PelC Trp149 resulted in abrogated pellicle formation and biofilm attachment. To accommodate the PelB–PelC interaction, it has been suggested that there is an unstructured region ~120 residues in length between the porin and TPR domains of PelB that could thread through the 32 Å pore of PelC (Marmont et al. 2017a). Although the AlphaFold model of PelB suggests that this region is structured and forms part of the TPR domain (Fig. 5G), analysis of PelC and the PelB AlphaFold model does allow for the TPR region of PelB to thread through the pore of PelC. Using structural visualization software, the membrane proximal convex surface of the PelC dodecamer can be positioned against the periplasm-facing pore of PelB’s ß-barrel domain to generate a model of the Pel OM secretion complex. This complex demonstrates that PelB’s TPR domain enters the PelC pore at the membrane proximal end, raising the question if PelB’s TPRs are also involved in binding Pel and guiding it for export, as was observed with the TPR-containing protein AlgK in the alginate system (Gheorghita et al. 2022a).
Similar to how polymer modification and export are coupled in the alginate system via formation of the AlgKX complex, PelB interacts with PelA to modulate its enzymatic activities and influence biofilm formation (Marmont et al. 2017b). Although we do not yet have structural insights into the PelAB complex, mutagenesis analysis revealed that PelB interacts with PelA via its TPR motifs R9-14, facilitating localization of PelA to the OM (Marmont et al. 2017b). PelB’s TPR domain was found to interact with full length PelA, but not its isolated hydrolase domain, suggesting an interaction with the deacetylase domain. This interaction is required for pellicle formation and biofilm attachment (Marmont et al. 2017b). Furthermore, interaction with PelB attenuated PelA’s hydrolase and enhanced deacetylase activity in vitro (Marmont et al. 2017b). Marmont et al. (2017b) suggest that interaction with PelB induces a conformational change in PelA that allows for more efficient deacetylation of Pel, given that deacetylation is a requirement for Pel export. PelA hydrolase activity is responsible for generating the secreted form of Pel thus, an interaction between PelA and PelB may result in an altered cell-associated to cell-free Pel ratio, consequently influencing biofilm biomechanics and virulence (Razvi et al. 2023a). How an optimal balance between PelA deacetylase and hydrolase activity is achieved via interaction with PelB is not clear. No other PelA interaction partners which may also influence PelA’s enzymatic activities have been identified to date.
Psl biosynthesis
Except for algC, galU, and rmlC, the genes involved in Psl biosynthesis are organized on a single operon in P. aeruginosa: pslA-B-C-D-E-F-G-H-I-J-K-L (Fig. 8) (Friedman and Kolter 2004b, Matasukawa and Greenberg 2004, Jackson et al. 2004). Transcriptional regulation of the Psl operon is coordinated by the alternative sigma factor RpoS (Irie et al. 2010), a general stress response regulator that has been previously linked to biofilm formation (Adams and McLean 1999, Battesti et al. 2011). The quorum-sensing transcription factor LasR was also found to bind to a promoter region upstream of the Psl operon (Gilbert et al. 2009). LasR is part of a two-component system that produces the acyl-homoserine lactone (HSL) 3-oxo-C12-HSL, mediating virulence, pathogenesis, suppression of the host immunity, and antagonism against other pathogens such as Staphylococcus epidermidis (Singh et al. 2017, Santajit et al. 2020).
Figure 8.
Model of the Psl biosynthetic complex in P. aeruginosa. (A) Model of the Psl biosynthetic complex colour coded by function. The structures of PslG (PDB: 5BXA, 5BX9) (Baker et al. 2015), AlgC (PDB: 1K2Y, 1K35) (Regni et al. 2002), and RmlC (PDB: 2IXJ, 2IXK, 2IXH, 2IXI) (Dong et al. 2007) have been experimentally determined and are drawn to scale and shown in surface representations. (B) Psl operon in P. aeruginosa colour coded by proposed function. IM, inner membrane; PG, peptidoglycan; and OM, outer membrane.
Psl biosynthesis is initiated in the cytoplasm, with generation of the GDP-mannose, UDP-glucose, and dTDP-rhamnose sugar precursors by AlgC, PslB, GalU, and RmlC (Franklin et al. 2011). PslC, PslF, PslH, and PslI are proposed to be involved in Psl polymerization taking the activated sugar precursors and forming the pentasaccharide repeating until of Psl (Franklin et al. 2011). PslA is thought to assembly Psl oligosaccharide repeating units onto an isoprenoid lipid that is cytoplasmically facing and this repeating unit is then flipped into the periplasm by the predicted Wzx flippase PslK (Franklin et al. 2011). Once in the periplasm, the predicted Wzy GT PslJ will add the next Psl repeat unit onto a pre-existing unit to polymerize Psl (Whitfield et al. 2020). PslE and PslD are the predicted PCP and OPX proteins, respectively, which are proposed to translocate Psl across the periplasm and out of the bacteria (Franklin et al. 2011, Whitfield et al. 2020).
Precursor formation
For Psl production, the cytoplasmic proteins PslB, AlgC, GalU, and RmlC are required to generate the precursor molecules GDP-mannose, UDP-glucose, and dTDP-rhamnose. To generate GDP-mannose, PslB’s predicted PMI activity converts fructose-6-phosphate to mannose-6-phosphate (reaction a1), which is then converted to mannose-1-phosphate by AlgC’s phosphomannomutase activity (reaction a2). PslB then uses its GMP activity to add GDP to mannose-1-phosphate (reaction 3a) (Byrd et al. 2009, Franklin et al. 2011).
PslB, like AlgA, is predicted to be a bifunctional enzyme with a PMI domain and GMP domain and its AlphaFold predicted model also has structural similarities to TmGMP and Cupin 2 (Fig. 9A). To generate UDP-glucose, AlgC’s phosphoglucomutase activity is thought to produce glucose-1-phosphate (reaction b1) (Fig. 9B), to which GalU can then add a UDP (reaction b2) (Fig. 9C) (Regni et al. 2002, Byrd et al. 2009).
Figure 9.
Structures and predicted AlphaFold models of P. aeruginosa enzymes involved in Psl precursor formation. (A) Superimposition of the P. aeruginosa PslB AlphaFold model (blue) with the crystal structure of T. maritima GMP TmGMP monomer (orange) (PDB: 2 × 65) with a Cα RMSD of 1.403 Å and the crystal structure of S. frigidimarina Cupin 2 conserved barrel domain protein (dark orange) (PDB: 2PFW) with a Cα RMSD of 0.801 Å. (B) Crystal structure of P. aeruginosa AlgC (PDB: 2FKF). (C) P. aeruginosa GalU AlphaFold model. (D) Crystal structure of a monomer of P. aeruginosa RmlC (PDB: 2IXH).
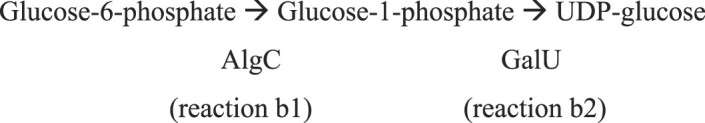
dTDP-rhamnose is thought to come from the Rml pathway, as ∆rmlC strains of P. aeruginosa do not produce Psl and are attachment-defective (Fig. 9D) (Byrd et al. 2009, Dong et al. 2007).
Using the precursor sugars, the GTs PslF, PslH, PslI, and PslC are thought to generate the repeating unit of the Psl polysaccharide (Fig. 10) (Franklin et al. 2011). PslF, PslH, and PslI share structural similarity with the sucrose-phosphate synthase from Thermosynechococcus elongatus (TeSPS), an enzyme involved in sucrose synthesis (Fig. 10A and B) (Li et al. 2020) (PDB: 6KIH). TeSPS has a GT-B fold with two Rossmann-type domains: a N-terminal A domain and C-terminal B domain. Mass spectrometry analysis of the enzyme demonstrated that it can synthesize sucrose-6-phosphate from fructose-6-phosphate and UDP-glucose, providing evidence that PslF/PslH/PslI bind sugar nucleotides.
Figure 10.
Structures and predicted AlphaFold models of P. aeruginosa Psl biosynthesis proteins. (A) Superimposition of the P. aeruginosa PslF AlphaFold model (blue) with crystal structure of TeSPS from T. elongatus (orange) (PDB: 6KIH) with a Cα RMSD of 2.54 Å. (B) Superimposition of the P. aeruginosa PslF (blue), PslH (periwinkle), and PslI (dark blue) AlphaFold models. Superimposition of PslH and PslI with PslF generates a Cα RMSD of 3.618 Å and 3.290 Å, respectively. (C) Superimposition of the P. aeruginosa PslC AlphaFold model (blue) with the cryoEM structure of HAS from P. bursaria Chlorella virus CZ-2 (orange) (PDB: 7SP8) with Cα RMSD of 2.836 Å. IM, inner membrane.
Like Alg8, PslC’s Alphafold predicted model strongly resembles the GT-A HAS from P. bursaria Chlorella virus CZ-2 (Fig. 10C) (Maloney et al. 2022) (PDB: 7SP8). Unlike Alg8 and HAS, PslC is not predicted to have any TM helices, however, it is predicted to retain the amphipathic interface helices, suggesting that it associates with the IM. PslC likely interacts with other Psl biosynthetic machinery in the IM, such as PslA, to coordinate transfer of the repeating unit to the isoprenoid lipid. As PslC is not a membrane-embedded GT, it lacks the QXXRW motif characteristic of this class of GT enzymes. Thus, PslF, PslH, PslI, and PslC are likely involved in generating the Psl repeat unit, however, it is unclear which enzyme is responsible for binding each of the three nucleotide sugars.
Polymerization
PslA, PslJ, PslK, and PslL are all predicted to have IM spanning domains and have been proposed to form an IM Psl polymerization complex (Fig. 11) (Franklin et al. 2011). Based on the AlphaFold model, PslA is predicted to have four TM helices, followed by two additional domains, an N- and C-terminal domain (Fig. 11A). The N-terminal domain has a Rossmann fold characteristic of proteins that bind sugar-nucleotide moieties and resembles the N-terminal domain of UDP-D-quinovosamine 4-dehydrogenase from Vibrio fischeri (PDB: 3NKL) (Fig. 11B). UDP-D-quinovosamine is converted to N-acetyl-d-quinovosamine found in the lipopolysaccharide (LPS) produced by some Gram-negative bacteria, including P. aeruginosa (Bowser et al. 1974, Sonesson et al. 1989, Ramos 2004). This N-terminal domain of PslA is most likely involved in binding the Psl repeat unit. The C-terminal domain of PslA strongly resembles the phosphoglycosyl transferase PglC from Campylobacter concisus (PDB: 5W7L) (Fig. 11C) (Ray et al. 2018). PglC has a distinct architecture with a re-entrant membrane helix formed by a helix-break-helix motif, allowing it to sit in the IM ~14 Å deep (Ray et al. 2018). Although PslA is predicted to be inserted into the IM via its N-terminal TM helices, the AlphaFold model demonstrates that the re-entrant helix is also present, suggesting that proximity to the IM is important for the function of PslA’s C-terminal domain. PglC has an α-helix-associated β-hairpin motif with the catalytic dyad, Asp93 and Glu94, that catalyzes the first membrane-committed step in bacterial N-linked protein glycosylation (Lukose et al. 2017, Ray et al. 2018). This catalytic dyad is also present in PslA as Asp372 and Glu373, although mutagenesis studies have not yet confirmed that these residues are required for PslA activity. In general, phosphoglycosyl transferase’s function to transfer a phosphosugar from a nucleoside disphosphate (NDP) sugar moiety to a polyprenol-phosphate acceptor in the membrane (Lukose et al. 2017). Ray et al. (2018) argue that the tactical placement of PglC’s active site at the membrane interface allows for coordination of the hydrophobic properties of the polyprenol lipid anchor and the soluble phosphosugar. Thus, the structural similarity of PslA to PglC suggests that PslA functions as the initiating GT to assemble the Psl repeat unit onto a lipid carrier on the cytoplasmic face of the IM.
Figure 11.
Structures and predicted AlphaFold models of P. aeruginosa Psl biosynthesis proteins, continued. (A) P. aeruginosa PslA AlphaFold model. (B) Superimposition of the P. aeruginosa PslA AlphaFold model (grey) with the crystal structure of the N-terminal domain of UDP-d-quinovosamine 4-dehydrogenase from V. fischeri (orange) (PDB: 3NKL) with a CαRMSD of 1.337 Å. (C) Superimposition of the C-terminus of P. aeruginosa PslA AlphaFold model (grey) with the crystal structure of the phosphoglycosyl transferase PglC from C. concisus (orange) (PDB: 5W7L) with a Cα RMSD of 1.603 Å. IM, inner membrane. (D) Superimposition of the P. aeruginosa PslJ AlphaFold model (navy) with the cryoEM structure of O-antigen ligase WaaL from Cupriavidus metallidurans (orange) (PDB: 7TPG) with a Cα RMSD of 4.42 Å. (D) Superimposition of the P. aeruginosa PslK AlphaFold model (green) with the crystal structure of the lipid II flippase MurJ from Thermosipho africanus (orange) (PDB: 6NC6) with a Cα RMSD of 2.04 Å. (F) AlphaFold model of P. aeruginosa PslL. (G) Crystal structure of P. aeruginosa PslG (PDB: 5BXA). IM, inner membrane.
The AlphaFold predicted model of PslJ resembles the cryoEM structure of O-antigen ligase WaaL from Cupriavidus metallidurans (PDB: 7TPG) (Ashraf et al. 2022) (Fig. 11D). WaaL is a ligase and catalyzes the final step in LPS biosynthesis with the addition of O-antigen to the lipid A core oligosaccharide (Ashraf et al. 2022). WaaL is an IM protein with 12 TM helices, a short cytoplasmic helix, and four short α-helical periplasmic segments, two of which are amphipathic and are parallel to the membrane. Overall, the enzyme displays a GT-C fold with two cavities on the periplasmic side; one cavity that binds the undecaprenyl pyrophosphate lipid carrier with O-antigen, and the other is proposed to bind lipid A. PslJ is similarly predicted to have two cavities; one that may bind the isoprenoid lipid carrier and the other binding the Psl repeat unit. The structures of apo- and undecaprenyl pyrophosphate-found C. metallidurans WaaL allowed Ashraf et al. (2022) to propose a mechanism. The lipid-linked O-antigen enters the cytoplasm-facing cavity of WaaL and is stabilized by Arg288 and Arg215. The lipid A core oligosaccharide enters the periplasm-facing cavity between the first three TM helices, on the opposite side of the structure from the O-antigen binding cavity. The catalytic residue, H338, required for O-antigen extension, first abstracts a proton from the leading hydroxyl group on the outer core of the lipid A core oligosaccharide. The oxygen is then able to perform a nucleophilic attack on the C1 carbon of the lipid-linked O-antigen sugar ring to cleave the sugar-phosphate bond (Ashraf et al. 2022). Mutation of the catalytic His338 was shown to abolish enzyme activity in E. coli, however, this residue is not conserved in PslJ. Although experimental data from WaaL give us insight into how PslJ may likely function as the GT that adds Psl repeat units to the lipid carrier once the lipid carrier has been flipped over to the periplasmic side, PslJ most likely uses a different set of residues to accomplish this role.
The predicted flippase PslK shares structural similarity with the lipid II flippase MurJ from Thermosipho africanus (PDB: 6NC6) (Kuk et al. 2019) (Fig. 11E). MurJ is an IM protein with 14 TM helices; helices 1–6 form the N-lobe and helices 7–12 form the C-lobe. Between the N- and C-lobes is a large central cavity that is highly electropositive (Kuk et al. 2019). TM helices 13 and 14 form a hydrophobic groove that connects to the central cavity. Similarly, PslK is predicted to have 14 TM helices with a highly electropositive central cavity between its predicted N- and C-lobes and a hydrophobic groove between TM helices 13 and 14. The crystal structure of MurJ shows coordination of Na+ by the C-lobe residues Asp235, Asn374, Asp378, and Thr394. Mutation of these residues resulted in perturbations in protein folding, thus how Na+ coordination affects MurJ function was not able to be assessed. However, Kuk et al. (2022) propose that MurJ uses an alternating-access mechanism that exchanges lipid II with Na+ and Cl− ions.
While MurJ is in an inward-open conformation, lipid II enters the central cavity and its disphosphate moiety is bound by Arg24, Asp25 in TM8 and Arg255 in TM1 – the putative substrate-binding triad. Its lipid tail interacts with the hydrophobic groove formed by TH helices 13 and 14. A thin gate formed by Glu57 and Arg352 occludes the lipid II headgroup from the cytoplasm. These interactions cause a bend in the C-lobe and MurJ adopts an inward occluded conformation. Na+ binding to the C-lobe primes MurJ to adopt an outward-facing conformation and the lipid II headgroup is released into the periplasmic space, while the tail remains associated with MurJ. Cl− enters the substrate binding site and engages the Arg24, Asp25, and Arg255 triad, resetting MurJ to an inward closed apo conformation. Release of the ions into the cytoplasm completes the transport cycle and MurJ adopts an inward open conformation (Kuk et al. 2022).
In PslK, the catalytic triad is present as Arg22, Glu23, and Lys259. A neighbouring residue Arg258 may also function to interact with the lipid II diphosphate moiety. The Arg in the thin gate is conserved as Arg352 in PslK, however, the opposing acidic residue (Glu57 in MurJ) is not conserved. It is possible that a neighbouring acidic residue, Glu47, may be able to compensate. Of the Na+ coordinating residues, only Glu239 (Asp235 in MurJ) and Asn374 are conserved in PslK. Thus, PslK may employ a similar mechanism to flip the lipid carrier with a Psl repeat unit to the periplasmic face of the IM. Although parallels can be drawn between the two proteins, it is unclear if PslK similarly uses Na+ and Cl− ions to flip the Psl-lipid moiety from the cytoplasm to the periplasm.
The function of PslL is currently unknown. Sequence similarity analysis predicted it to be an O-acyltransferase within the NolL superfamily (Franklin et al. 2011), though Psl is not known to be acetylated or modified by addition of small functional groups. Searching for existing structures with structural homology to PslL’s AlphaFold model (Fig. 11F) did not provide any clear insight into PslL’s potential role in Psl biosynthesis. The AlphaFold model predicts 10 TM helices arranged in a manner that generates a pore through the middle. Between helices 5 and 6 is a periplasmic β-hairpin. Additionally, it currently unknown whether PslL has any interaction partners within the Psl system, which may provide clues into its function. Thus, the role of PslL in Psl biosynthesis remains to be determined.
Polymer modification
Like alginate and Pel, the Psl system also has an enzyme that can modify the polymer by cleavage – PslG. The crystal structure of PslG has been determined (Baker et al. 2015) (PDB: 5BXA, 5BX9), revealing two domains: an N-terminal (β/α)8 TIM barrel fold and C-terminal β-sandwich fold (Fig. 11G). Bioinformatics analysis and the AlphaFold model suggests that PslG is anchored to the IM by a single N-terminal TM domain. Subcellular fractionation of PslG from P. aeruginosa demonstrates that PslG is enriched in the IM (Baker et al. 2015). The N-terminal (β/α)8 TIM barrel fold domain of PslG forms a highly conserved and highly electronegative ∼32 Å long groove that is involved in substrate binding and catalysis. Indeed, this groove was found to bind mannose in vitro via an intrinsic tryptophan fluorescence assay. Furthermore, PslG was shown to hydrolyze Psl in vitro and mutation of its predicted catalytic residues Asp165 and Asp276 abrogated hydrolase activity. How PslG’s hydrolase activity could be used for antibiofilm applications is discussed below.
In a Psl-overproducing strain of P. aeruginosa, neither PslG nor its hydrolase activity were required for Psl production or biofilm formation in microtitre plates, although its overexpression resulted in reduced Psl and biofilm formation (Baker et al. 2015). However, Zhang et al. (2022) recently demonstrated that loss of PslG in a non-Psl-overproducing strain results in lack of ring formation in pellicle biofilms and increased microcolony formation compared to the parental strain. Furthermore, loss of PslG results in significantly higher maximum pellicle thickness compared to the parental strain and impairs the cell division process by causing daughter cells to become physically linked to each other via Psl on the bacterial periphery (Zhang et al. 2022). Surprisingly, although PslG-deficient mutants were found to produce less Psl than the parental strain, their Psl had a stronger effect on stimulating intracellular c-di-GMP production and the PslG-deficient mutants demonstrated increased c-di-GMP levels in daughter cells after division (Zhang et al. 2022). Thus, this recent work demonstrates that PslG and its ability to hydrolyze Psl has wide-spread implications for P. aeruginosa cell fate and biofilm formation.
Polymer secretion
The predicted PCP protein, PslE, demonstrates structural similarity to the tyrosine-protein kinase Wzc from E. coli that function as the PCP in capsular polysaccharide biosynthesis (PDB: 7NII) (Yang et al. 2021) (Fig. 12A). Wzc is regarded as a master regulator of polymerization and translocation in polymer production. The cryo-EM structure reveals that it functions as an octamer, forming a large central cavity using its TM helices, and has a C-terminal cytosolic protein tyrosine kinase (PTK) domain (Yang et al. 2021). The periplasmic helices of Wzc are thought to interact with the periplasmic region of the OPX protein Wza to form the translocation machinery. In comparison to Wzc, the AlphaFold predicted model of PslE has a substantially longer periplasmic region, which may also function as an oligomer to form a translocation pore that spans across the entire periplasm (Fig. 12A). Similar to Wzc, PslE has a PTK domain with conserved active site residues, however, PslE appears to lack the C-terminal Tyr rich tail of Wzc that is required for autophosphorylation (Yang et al. 2021). Autophosphorylation of this Tyr-rich tail causes disassembly of the octamer, while dephosphorylation via the phosphatase Wzb allows for reassembly (Yang et al. 2021). A phosphatase is yet to be discovered to interact with the Psl machinery. Thus, it is unclear if phosphorylation is the mechanism by which formation of the PslE translocation machinery is regulated, or if the PTK domain of PslE functions to phosphorylate a different target. PslE was found to interact with PslG and PslA via BACTH assay, suggesting that PslE acts as a periplasmic scaffold (PslG) and recruits proteins (PslA) to the biosynthetic complex (Wu et al. 2019). It is possible that PslE regulates the formation of this complex by phosphorylation.
Figure 12.
Structures and predicted AlphaFold models of P. aeruginosa Psl export proteins. (A) Superimposition of P. aeruginosa PslE AlphaFold model (purple) with the tyrosine-protein kinase Wzc from E. coli (orange) (PDB: 7NII) with a Cα RMSD of 5.40 Å. (B) Superimposition of P. aeruginosa PslD AlphaFold model (burgundy) with the crystal structure of Wza from E. coli (orange) (PDB: 2J58) with a Cα RMSD of 2.81 Å. IM, inner membrane; OM, outer membrane.
PslD is predicted to be the OPX protein of the system and shares structural similarity with the OPX protein Wza from E. coli (PDB: 2J58) (Dong et al. 2006) (Fig. 12B). Wza is split up into four domains: domain 1 has an antiparallel β-sandwich with an α-helix, domain 2 has a central five-stranded mixed β-sheet with three α-helices, domain 3 is similar to domain 2 with an additional β-strand, and domain 4 is an amphipathic helix (Dong et al. 2006). PslD has domains 1 and 2 but is lacking domain 3 and is, therefore, shorter than Wza (Fig. 12B). The first half of domain 4 (residues 222–236) forms a helix, however, the remaining half of domain 4 (residues 237–256) is unstructured in the AlphaFold2 model. Domain 4 is amphipathic; residues Val230, Tyr233, Leu234, Leu238, and Phe240 generate a hydrophobic face in the first half of domain 4, while residues Val246, Val248, Tyr250, and Val252 generate a hydrophobic face in the second half of domain 4. As amphipathic helices are known to interact with membranes (Giménez-Andrés et al. 2018), the amphipathic properties of this domain may allow it to interface with the membrane in some manner. However, analysis by DeepTMHMM (Hallgren et al. 2022) does not predict PslD to contain any TM/OM domains. Predictive modelling by Wu et al. (2019) confirms that PslD can be modelled onto Wza, however, they also determined that PslD lacks the OM barrel, making it unclear how Psl translocates the OM. Short periplasmic OPX proteins have been found in other Gram-negative bacteria, including Myxococcus xanthus, however, their genes are coupled with a β-barrel protein, forming a composite OPX/β-barrel protein translocon (Schwabe et al. 2022). Although PslD is a short OPX protein, the Psl operon does not encode a β-barrel protein.
Wza is thought to interact with Wzc to form a translocation complex that protects the capsular polysaccharide polymer as it transverses the periplasm and OM. Thus, PslD and PslE may similarly form a secretion complex to export Psl, although this complex will look structurally different compared to Wza/Wzc due to the differences in protein length. The AlphaFold model of PslE is long enough to span both the inner and OMs (Fig. 12A). Thus, it is possible that PslE helps to form a channel through the OM. However, results from the DALI search do not support this and analysis by DeepTMHMM (Hallgren et al. 2022) does not predict the tip of PslE to be inserted in the OM. Thus, it is unclear how PslE and PslD form the OPX/PCP complex required for polymer translocation and how Psl traverses the OM. However, Wu et al. (2019) have demonstrated that PslD and PslE interact via BACTH and that PslE helps localize PslD to the OM, suggesting that they form a complex for Psl secretion.
Glycoside hydrolases and lyases as potential therapeutics
The exopolysaccharide biosynthetic systems in P. aeruginosa contain proteins involved in polymer synthesis, modification, export, and degradation; the enzymes AlgL, PelAh, and PslG can cleave alginate, Pel, and Psl polymers, respectively. Using recombinant protein technology, these enzymes can be purified and applied to biofilms, resulting in the degradation of exopolysaccharide biofilm components. In this section, we discuss the clever use of bacterial glycoside hydrolases/lyases for antibiofilm applications.
Alginate
In 1993, Murata et al. (1993) discussed the future use of alginate lyases as a therapeutic to help clear CF patient airways and promote delivery of inhaled aerosol antimicrobials. The following year, treatment of CF sputum with alginate lyase was first reported, demonstrating that addition of exogenous enzyme isolated from mucoid P. aeruginosa to a sputum sample in vitro disrupted the alginate within the sputum and reduced its viscoelasticity (Mrsny et al. 1994). The viscosity of alginate hinders antibiotic penetration, with only a 2% suspension of P. aeruginosa alginate completely blocking diffusion of tobramycin and gentamicin (Hatch and Schiller 1998). Addition of exogenous AlgL was found to restore antibiotic diffusion through the alginate, demonstrating its potential as an adjunct to antibiotics (Hatch and Schiller 1998). Exogenous AlgL was also able to inhibit adherence of mucoid P. aeruginosa biofilms (Mai et al. 1993). Moreover, a combination of AlgL and gentamicin was more effective at clearing mucoid P. aeruginosa biofilms compared to antibiotic alone (Alkawash et al. 2006). Work has also been done to improve AlgL’s enzymatic activity and therefore enhance alginate degradation (Cho et al. 2016). A K97D/K321A AlgL mutant in combination with tazocin was significantly more effective at disrupting ciprofloxacin-resistant P. aeruginosa biofilms compared to the WT AlgL/tazocin combination (Cho et al. 2016).
As the concept of using alginate lyases for eradication of biofilms gained traction, other sources of enzymes were also explored. Alginate lyases from other Gram-negative bacteria have been investigated for their antibiofilm activity, including AlgL from Azotobacter vinelandii (Jang et al. 2016) and Flavobacterium (Bugli et al. 2013, Maiorana et al. 2014), A1-II and A1-III from Sphingomonas (Lampp and Griswold 2013, Blanco-Cabra et al. 2020), Aly08 from Vibrio (Li et al. 2019), and AlyA1 from Zobellia galanctanivorans (Blanco-Cabra et al. 2020). Although H. pylori and Aspergillus fumigatus are not known to produce alginate as part of their biofilm matrix, AlgL from Flavobacterium was investigated for its ability to enhance antibiotic killing of these bacteria (Maiorana et al. 2014, Bugli et al. 2016). Surprisingly, AlgL demonstrated a synergistic effect with clarithromycin again H. pylori biofilms, potentially due to biofilm matrix degradation and/or retention of H. pylori in the bacillary form, compared to the more antibiotic resistant coccoid form (Bugli et al. 2016, Ierardi et al. 2020). Similarly, Flavobacterium AlgL was found to significantly potentiate amphotericin and liposomal amphotericin antifungal effects against A. fumigatus (Maiorana et al. 2014). Coadministration of DNase with Flavobacterium AlgL was found to potentiate the antibiotic activity of tobramycin and amikacin on mucoid P. aeruginosa biofilms compared to antibiotic treatment alone (Alipour et al. 2009). AlgL activity has also been associated with biofilm inhibition as Pseudoalteromonas (AlyP1400) inhibited biofilm formation from a clinical isolate of P. aeruginosa in a dose-dependent manner while the heat-inactivated enzyme did not. Furthermore, AlyP1400 in combination with tobramycin or ciprofloxacin significantly reduced biofilm biomass and surface area, compared to antibiotic treatment alone (Daboor et al. 2021).
Recently, a commercially available mannuronate-specific alginate lyase from an unspecified source was used to develop AlgL-functionalized chitosan nanoparticles of ciprofloxacin (Patel et al. 2019). The nanoparticles were shown to enhance degradation of a mucoid P. aeruginosa biofilm in vitro, resulting in greatly reduced biofilm biomass, thickness, and density. The nanoparticles did not exhibit toxicity in human lung epithelial cells or rat lung tissue. Overall, Patel et al. (2019) demonstrate that this delivery strategy has potential as a therapeutic treatment.
In contrast, Lampp and Griswold (2013) reported that alginate lyase activity from Sphingomonas A1-III is not required to enhance the antibiotic activity of tobramycin against mucoid P. aeruginosa. They determined that antibiotic potentiation could be achieved with bovine serum albumin and is a nonspecific phenomenon that could be observed with proteins in general, although no other proteins were tested (Lampp and Griswold 2013). Similarly, treatment of P. aeruginosa biofilms and P. aeruginosa, P. fluorescens, and Klebsiella pneumoniae mixed species biofilms with AlgL from A. vinelandii did not degrade the biofilm matrix to a detectable extent (Christensen et al. 2001). It is unclear why this is the case as A. vinelandii AlgL has a preference for unacetylated polyM and it can also cleave polyMG and acetylated alginate (Ertesvåg et al. 1998). The authors concluded that other exopolysaccharides may be involved in biofilm formation or AlgL activity may have been inhibited or prevented. Specifically, the authors speculated that AlgL may have poor penetration into the biofilm, or its activity is attenuated by other biofilm components (Christensen et al. 2001).
Pel
The ability of PelAh to hydrolyze Pel makes the polymer a potential target for development of antimicrobial therapeutics. This isolated glycoside hydrolase domain can inhibit and disrupt P. aeruginosa biofilms via Pel hydrolysis, potentiate colistin killing of P. aeruginosa, and increase human neutrophil killing of P. aeruginosa (Baker et al. 2016). Hydrolysis of Pel-dependent biofilms and human neutrophil killing of P. aeruginosa is abrogated with a catalytically inactive E218A mutant of PelAh, demonstrating that its enzymatic activity is critical for this antibiofilm function. As the A. fumigatus galactosaminogalactan (GAG) exopolysaccharide is composed of galactosamine, PelAh has also been shown to disrupt and degrade A. fumigatus biofilms (Snarr et al. 2017). Furthermore, PelAh increases A. fumigatus sensitivity to posaconazole, amphotericin B, and caspofungin antifungals. When epithelial cell injury was tested using a chromium-51 release assay, PelAh was found to be protective against A. fumigatus-induced damage and demonstrated significantly reduced chromium-51 release compared to the untreated control (Snarr et al. 2017). In a mouse model of pulmonary invasive aspergillosis, mice that were treated intratracheally with a single dose of PelAh had a significant increase in survival compared to untreated mice (Ostapska et al. 2021). When PelAh was immobilized on a bacterial cellulose (BC)-based wound dressing, significantly more cells were detached and released from the BC membrane compared to the control (Szymańska et al. 2020). Analysis of the BC membrane by scanning electron microscopy showed that the control had P. aeruginosa cells and biofilm matrix components present, however, PelAh immobilization removed the biofilm, revealing more distinctly visible P. aeruginosa cells. Finally, BC-PelAh was found to be nontoxic to mouse fibroblast cells, suggesting that BC-PelAh wound dressings could be safely used for protection against P. aeruginosa biofilm formation (Szymańska et al. 2020).
Psl
Outside of P. aeruginosa, PslG’s role as a glycoside hydrolase has been cleverly exploited to cleave Psl produced during infection and act as an adjunct therapy alongside antibiotics. In a Psl-dependent biofilm, PslG was found to potentiate killing by colistin (Baker et al. 2016). Tobramycin is a commonly used anti-Pseudomonas antibiotic, however, it gets sequestered at the P. aeruginosa biofilm periphery, limiting its penetration through the biofilm and therefore its efficacy (Tseng et al. 2013). The positively charged antibiotic is thought to be sequestered by negatively charged biofilm components via ionic interactions, as tobramycin penetration increased with addition of Mn2+ cations in a dose-dependent manner (Tseng et al. 2013). Treatment with PslG was found to improve tobramycin penetration as Cy-5-labelled antibiotic localized throughout the biofilm structure and less was observed at the periphery compared to the untreated control (Pestrak et al. 2019). In a mouse wound model infected with P. aeruginosa, PslG was found to enhance the effectiveness of tobramycin treatment by significantly reducing the bacterial burden compared to tobramycin treatment alone (Pestrak et al. 2019). Furthermore, treatment with the glycoside hydrolase was not toxic to host cells in a mouse punch biopsy wound infection model and did not impede wound healing (Pestrak et al. 2019).
Although PslG is stable for 48 h in P. aeruginosa culture and human lung fibroblast cell culture, (Baker et al. 2016), concerns have been raised regarding the longevity and activity of biofilm-degrading enzymes in vivo due to their potential susceptibility to proteolysis (Thorn et al. 2021a). Thus, mechanisms to enhance PslG longevity in combination with antibiotics and allow for effective delivery of the therapeutic at the site of infection have been recently explored in the form of lipid-based liquid crystal nanoparticles (LCNPs) (Thorn et al. 2021b). LCNPs are antimicrobial carriers that release their cargo upon degradation by secreted bacterial lipases (Thorn et al. 2020). Compared to PslG alone, LCNPs protect PslG from proteolytic degradation and help to retain PslG’s ability to degrade P. aerugiosa biofilms postprotease treatment (Thorn et al. 2021b). When preformed P. aeruginosa biofilms were treated with LCNPs formulated with PslG and tobramycin, bacterial load was significantly reduced by up to 3000 times compared to the unformulated enzyme and drug combination, demonstrating the enhanced antimicrobial effect that LCNPs confer (Thorn et al. 2021b). In a C. elegans model of infection with P. aeruginosa, LCNP-formulated tobramycin had similar reductions in bacterial load compared to the LCNP-formulated tobramycin and PslG combination. Both treatments (LCNP-formulated tobramycin and LCNP-formulated tobramycin/PslG combination) resulted in similar C. elegans survival, suggesting that therapeutic delivery of PslG with LCNPs is critical in an in vivo model of infection (Thorn et al. 2021b). This study used a lower dose of PslG compared to other in vivo investigations (Pestrak et al. 2019), thus utilizing higher doses of PslG may improve the effect of the LCNP-formulated tobramycin/PslG combination (Thorn et al. 2021b).
When PslG was immobilized on medical grade microcatheters, it inhibited P. aeruginosa biofilm formation on the microcatheter compared to the control (Asker et al. 2021). In an in vivo central venous catheter infection rat model, the PslG-coated catheter showed significant inhibition of P. aeruginosa biofilm formation after 24 h compared to the uncoated control, demonstrating the potential of PslG to reduce medical device-associated infections (Asker et al. 2021).
As both Pel and Psl have been identified in CF sputum samples, the use of both PelAh and PslG glycoside hydrolases may prove to be an effective strategy in eradicating P. aeruginosa biofilm infections. Indeed, PelAh and PslG in combination has been shown to disrupt biofilms from both clinical and environmental isolates of P. aeruginosa, demonstrating that the enzymes are compatible with each other and can be utilized simultaneously (Baker et al. 2016). The PelAh–PslG treatment significantly increased P. aeruginosa susceptibility to tobramycin, polymyxin, colistin, and neomycin antibiotics. Furthermore, the hydrolase combination was found to enhance neutrophil function against P. aeruginosa by increasing neutrophil phagocytosis and reactive oxygen species production, and enhance the innate immune response by increasing complement C3 protein deposition in vitro (Pestrak et al. 2019). In mice, intratracheal PelAh–PslG treatment was found to be well-tolerated and did not induce pulmonary damage. When the PelAh–PslG combination was tested as an intratracheal prophylactic treatment in combination with antibiotics in a mouse model of acute P. aeruginosa infection, use of the glycoside hydrolase combination with ciprofloxacin significantly reduced the bacterial burden compared to hydrolase treatment or ciprofloxacin alone (Ostapska et al. 2022b). Treatment with the glycoside hydrolases alone without antibiotic did not decrease bacterial burden and mice experienced more bacteremia compared to a PelAh–PslG and antibiotic combination, suggesting that coadministration of antibiotics will be required to protect against bacterial dissemination that is induced by the glycoside hydrolases (Ostapska et al. 2022b). Overall, these studies highlight the therapeutic potential of glycoside hydrolase combination therapy as an adjunct to antibiotics.
Other microbial exopolysaccharides that can be targeted by glycoside lyases/hydrolases
The experimentally determined structures of the glycoside lyases/hydrolases AlgL, and the hydrolase domains of PelA and PslG have provided insight into not only how these enzymes function in polymer biosynthesis but also how they might be exploited for antibiofilm therapies. Outside of P. aeruginosa, other exopolysaccharide hydrolases have been identified with the potential for therapeutic applications. Dispersin B (DspB), from the periodontal pathogen Aggregatibacter actinomycetemcomitans (PDB: 1YHT), is a glycoside hydrolase that degrades the exopolysaccharide poly-β-1,6-N-acetyl-d-glucosamine (PNAG) and is found outside of the PNAG biosynthetic operon (Itoh et al. 2005, Ramasubbu et al. 2005). DspB was found to disrupt biofilms produced by E. coli and S. epidermidis. When DspB was absorbed into polyurethane material imitating a medical device, S. epidermidis biofilm formation was inhibited on DspB-treated polyurethane compared to untreated material (Donelli et al. 2007). Furthermore, a synergistic effect was observed with DspB and cefamandole nafate against oxacillin-resistant biofilm-forming S. epidermidis compared to the antibiotic alone (Donelli et al. 2007). Similarly, catheters that were coated with DspB and triclosan demonstrated synergistic antibiofilm effects against S. epidermidis, E. coli, and S. aureus (Darouiche et al. 2009). Commercialization of DspB (DispersinB®) by Kane Biotech is currently being pursued.
Outside of bacteria, the glycoside hydrolase Sph3 from the A. fumigatus GAG gtb3 gene cluster (PDB: 5D6T) is required for GAG exopolysaccharide biosynthesis and can hydrolyze purified and cell-associated GAG (Bamford et al. 2015, Le Mauff et al. 2019). Aspergillus fumigatus biofilms that are treated with exogenous Sph3 are disrupted and the GAG decoration on the surface of the hyphae is degraded (Snarr et al. 2017). This antibiofilm effect was also observed with four clinical A. fumigatus isolates, demonstrating that Sph3 activity is not limited to the laboratory strain. Sph3 was also found to potentiate posaconazole, amphotericin B, and caspofungin by increasing their intracellular penetration (Snarr et al. 2017). Furthermore, exogenous Sph3 protected epithelial cells from A. fumigatus-induced damage, as determined by an established chromium release damage assay (Snarr et al. 2017). In the context of infection, Sph3-treated mice that were intratracheally infected with A. fumigatus had no detectable lesions in the lung, compared to infected mice that were untreated (Snarr et al. 2017). In a mouse model of pulmonary invasive aspergillosis, mice that were treated intratracheally with a single dose of the hydrolase domain of Sph3 (Sph3h) had a significant increase in survival compared to untreated mice (Ostapska et al. 2021). Aspergillus fumigatus-infected mice that were treated with Sph3h also demonstrated a significantly lower fungal burden than the buffer-treated control mice. Interestingly, when a catalytically inactive variant of Sph3h D166AAC was evaluated in mice, a similar level of protection against A. fumigatus was observed, demonstrating that Sph3h enzymatic activity is not required for its antifungal activity (Ostapska et al. 2021). Thus, it was proposed that Sph3h may engage in lectin-like interactions with GAG, thus interfering with GAG’s function in infection and immune evasion (Ostapska et al. 2021).
Also in the A. fumigatus GAG gtb3 gene cluster is another glycoside hydrolase, Ega3 (PDB: 6OJ1, 6OJB). Ega3 was shown to disrupt both Pel- and GAG-dependent biofilms from P. aeruginosa and A. fumigatus, respectively, in a concentration-dependent manner (Bamford et al. 2019). Similar to PelAh and Sph3h, intratracheal treatment with the hydrolase domain of Ega3 (Ega3h) resulted in a significant increase in survival of A. fumigatus-infected mice (Ostapska et al. 2021). Ega3h-treated mice also had a significantly lower fungal burden than the buffer-treated control mice (Ostapska et al. 2021). Similar to the PelAh–PslG combination treatment, an Ega3h–PslG combination enhanced neutrophil function and the innate immune response by the same measures (Ostapska et al. 2022b). Ega3h–PslG was similarly well-tolerated by mice, however, this combination did not reduce the bacterial burden in a mouse model of acute P. aeruginosa infection when treated in conjunction with ciprofloxacin or ceftazidime, compared to antibiotic treatment alone (Ostapska et al. 2022b).
Other various glycoside hydrolases have also been explored, including amylase and cellulose that degrade chitosan and cellulose, respectively. For example, α-amylase from Bacillus subtilis and cellulase from Aspergillus niger were found to significantly reduce the biomass of preformed S. aureus and P. aeruginosa coculture biofilms and increase bacterial dispersal (Fleming et al. 2017). In a murine burn wound model, the combination of both enzymes significantly improved the efficacy of gentamicin sulphate for both S. aureus and P. aeruginosa infections (Fleming et al. 2017). On its own, α-amylase has also been shown to inhibit and disrupt biofilms from V. cholerae and methicillin-resistant S. aureus (Kalpana et al. 2012, Watters et al. 2016), thus demonstrating α-amylase’s potential as an antibiofilm therapeutic. However, there are concerns about α-amylase’s contradictory biofilm-formation effect (Lahiri et al. 2021). Various Streptococcus species have salivary α-amylase binding proteins that can contribute to oral biofilm formation (Lahiri et al. 2021).
Discussion and future directions
The biofilm matrix is composed of many different components. We now have evidence which reveals that these components can interact with each other and that these interactions have functional consequences on biofilm integrity. Both Pel and Psl have been shown to interact with eDNA and the biofilm matrix protein CdrA (Wang et al. 2015, Reichhardt et al. 2018), while alginate has been shown to interact with the extracellular lipase LipA in mucoid biofilms (Tielen et al. 2013). The alginate–LipA interaction heat stabilizes LipA and protects it from proteolytic degradation, potentially enhancing uptake of fatty acids and P. aeruginosa survival in harsh environments (Tielen et al. 2013). As our understanding of mixed species biofilms and their biofilm matrixomes has improved (Karygianni et al. 2020), the ability of P. aeruginosa exopolysaccharides to interact with biofilm matrix components from other coinfecting pathogens, such as S. aureus protein A (SpA) has also been investigated. Staphylococcus aureus protein A was found to bind to Psl and type IV pili on the P. aeruginosa surface, protecting P. aeruginosa from phagocytosis by neutrophils in vitro (Armbruster et al. 2016).
How P. aeruginosa exopolysaccharides interact with biofilm matrix components from other coinfecting pathogens, such as C. albicans, has not yet been reported. Thus, this gap in the current literature leaves us with a few questions, which we hope can be addressed as research on biofilm matrixomes expands and the methods used to investigate them evolve. These questions include: Which P. aeruginosa exopolysaccharides and coinfecting pathogen biofilm matrix components interact with each other? What are the consequences of these interactions on the respective pathogens? Are these interactions a mechanism for cooperation or competition in a mixed biofilm? How do these mechanisms evolve over time? In a model of infection, how does this impact the host and/or affect antimicrobial treatments? As coculture models have become more widely studied, P. aeruginosa has been recently shown to adhere to A. fumigatus hyphae in coculture when the fungi are producing the GAG exopolysaccharide (Ostapska et al. 2022a). Ostapska et al. showed that de-N-acetylated GAG produced from A. fumigatus can incorporate itself into the P. aeruginosa biofilm and increase P. aeruginosa tolerance to the antibiotic colistin. However, it is not clear if P. aeruginosa exopolysaccharides are interacting with GAG, or if other biofilm matrix components are responsible. In contrast, Pel produced from P. aeruginosa was only able to influence A. fumigatus biofilms when P. aeruginosa secreted products, including antifungal compounds, were removed by dialysis (Ostapska et al. 2022a). This suggested that the co-operative biofilm interaction between the two pathogens is unidirectional; P. aeruginosa secreted products display antifungal effects that overcome the Pel-mediated modulation of A. fumigatus biofilm formation.
Progress has been made in determining the structures of P. aeruginosa exopolysaccharide biosynthetic proteins and protein–protein interaction networks within each system. However, for many proteins within the three systems, their structures have not yet been experimentally determined and, therefore, we lack mechanistic insights into how these proteins function. For example, within the alginate system, structures of the IM proteins Alg44, Alg8, and AlgI have not yet been elucidated. It is still unclear how Alg44 and Alg8 interact, and if/how the IM acetylation machinery associates with the polymerization machinery to form an IM polymerization and modification complex. Furthermore, we do not understand how AlgI transfers the acetyl group from the cytoplasm to the periplasm, or how Alg8 translocates nascent polymer across the IM. Similarly, in the Pel system, formation of the PelDEFG IM complex has not been studied from a structural perspective, and within the Psl system the majority of the proteins have not yet been structurally determined or functionally characterized.
Although this review has showcased experimentally determined structures and/or AlphaFold models for all the proteins within each system, it is still unclear how the biosynthetic proteins within each system are organized to form a complex that coordinates polymer synthesis, modification, and secretion across the inner and OMs. Future work on the alginate, Pel, and Psl systems should take a more holistic approach and focus on the entirety of the complexes, or subcomplexes, to address how the proteins interact with each other. Determining interaction networks via co-IPs, BACTH, and mutual stability analyses allow us to associate proteins with each other, but these assays tend to lack residue-specific information and do not inform us of conformational changes that may occur upon interaction. Site-directed mutagenesis in conjugation with protein interaction assays can be used to validate predicted AlphaFold complex models if structural determination of complexes/subcomplexes is unsuccessful.
In conclusion, in this review we summarized the recent advancement made in studying P. aeruginosa biofilm exopolysaccharide secretion systems from a structural perspective. We highlight newly elucidated structures of biosynthetic proteins and insights into their function and role in polymer production. Advancements made in structural prediction software, in conjugation with structural homology searches, allowed us to generate hypotheses about how proteins whose structures have not yet been determined may function. We discussed how the exopolysaccharide degrading activity of lyases and glycoside hydrolases have been exploited for antibiofilm applications and their potential as antimicrobials. We also described protein–protein interaction networks that have been established, to shed light onto how these biosynthetic complexes are organized and coordinated. As we continue to study P. aeruginosa and its biofilm components, we hope to discover ways in which we can use our understanding of these systems to generate therapeutics targeted towards eradicating P. aeruginosa biofilm infections.
Contributor Information
Andreea A Gheorghita, Program in Molecular Medicine, Peter Gilgan Centre for Research and Learning, The Hospital for Sick Children, 686 Bay St, Toronto, ON M5G 0A4, Canada; Department of Biochemistry, University of Toronto, Medical Science Building, 1 King's College Cir, Toronto, ON M5S 1A8, Canada.
Daniel J Wozniak, Department of Microbial Infection and Immunity, The Ohio State University College of Medicine, 776 Biomedical Research Tower, 460 W 12th Ave, Columbus, OH 43210, United States; Department of Microbiology, The Ohio State University College, Biological Sciences Bldg, 105, 484 W 12th Ave, Columbus, OH 43210, United States.
Matthew R Parsek, Department of Microbiology, University of Washington, Health Sciences Bldg, 1705 NE Pacific St, Seattle, WA 98195-7735, United States.
P Lynne Howell, Program in Molecular Medicine, Peter Gilgan Centre for Research and Learning, The Hospital for Sick Children, 686 Bay St, Toronto, ON M5G 0A4, Canada; Department of Biochemistry, University of Toronto, Medical Science Building, 1 King's College Cir, Toronto, ON M5S 1A8, Canada.
Conflict of interest
The authors declare no competing interests.
Funding
This work was supported by grants from the Canadian Institute of Health Research (CIHR) to P.L.H. (FDN154327). P.L.H. was the recipient of a Tier 1 Canada Research Chair (2006–2020). D.J.W. and M.R.P. are supported by the NIH grants (R01AI077628, R01AI134895, and R01AI143916). A.A.G. is supported by graduate scholarships from Cystic Fibrosis Canada and The Hospital for Sick Children Foundation Student Scholarship Program.
References
- Acheson JF, Derewenda ZS, Zimmer J. Architecture of the cellulose synthase outer membrane channel and its association with the periplasmic TPR domain. Structure. 2019;27:1855–61.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams JL, McLean RJ. Impact of rpoS deletion on Escherichia coli biofilms. Appl Environ Microbiol. 1999;65:4285–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alipour M, Suntres ZE, Omri A. Importance of DNase and alginate lyase for enhancing free and liposome encapsulated aminoglycoside activity against Pseudomonas aeruginosa. J Antimicrob Chemother. 2009;64:317–25. [DOI] [PubMed] [Google Scholar]
- Alkawash MA, Soothill JS, Schiller NL. Alginate lyase enhances antibiotic killing of mucoid Pseudomonas aeruginosa in biofilms. Apmis. 2006;114:131–8. [DOI] [PubMed] [Google Scholar]
- Armbruster CR, Wolter DJ, Mishra Met al. Staphylococcus aureus protein A mediates interspecies interactions at the cell surface of Pseudomonas aeruginosa. mBio. 2016;7:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf KU, Nygaard R, Vickery ONet al. Structural basis of lipopolysaccharide maturation by the O-antigen ligase. Nature. 2022;604:371–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asker D, Awad TS, Raju Det al. Preventing Pseudomonas aeruginosa biofilms on indwelling catheters by surface-bound enzymes. ACS Appl Bio Mater. 2021;4:8248–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker P, Hill PJ, Snarr BDet al. Exopolysaccharide biosynthetic glycoside hydrolases can be utilized to disrupt and prevent Pseudomonas aeruginosa biofilms. Sci Adv. 2016;2:e1501632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker P, Ricer T, Moynihan PJet al. P. aeruginosa SGNH Hydrolase-like proteins AlgJ and AlgX have similar topology but separate and distinct roles in alginate acetylation. PloS Pathog. 2014;10:e1004334. 10.1371/journal.ppat.1004334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker P, Whitfield GB, Hill PJet al. Characterization of the Pseudomonas aeruginosa glycoside hydrolase PslG reveals that its levels are critical for Psl polysaccharide biosynthesis and biofilm formation. J Biol Chem. 2015;290:28374–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford NC, Le Mauff F, Subramanian ASet al. Ega3 from the fungal pathogen Aspergillus fumigatus is an endo-α-1,4-galactosaminidase that disrupts microbial biofilms. J Biol Chem. 2019;294:13833–13849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford NC, Snarr BD, Gravelat FNet al. Sph3 is a glycoside hydrolase required for the biosynthesis of galactosaminogalactan in Aspergillus fumigatus. J Biol Chem. 2015;290:27438–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraquet C, Murakami K, Parsek MRet al. The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the Pel operon promoter in response to c-di-GMP. Nucleic Acids Research. 2012;40:7207–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battesti A, Majdalani N, Gottesman S. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol. 2011;65:189–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billings N, Millan MR, Caldara Met al. Extracellular matrix component Psl provides fast-acting antibiotic defense in Pseudomonas aeruginosa biofilms. PloS Pathog. 2013;9:e1003526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Cabra N, Paetzold B, Ferrar Tet al. Characterization of different alginate lyases for dissolving Pseudomonas aeruginosa biofilms. Sci Rep. 2020;10:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borlee BR, Goldman AD, Murakami Ket al. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol. 2010;75:827–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher JC, Yu H, Mudd MHet al. Mucoid Pseudomonas aeruginosa in cystic fibrosis: characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect Immun. 1997;65:3838–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowser DV, Wheat RW, Foster JWet al. Occurrence of quinovosamine in lipopolysaccharides of Brucella species. Infect Immun. 1974;9:772–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd A, Chakrabarty AM. Role of alginate lyase in cell detachment of Pseudomonas aeruginosa. Appl Environ Microbiol. 1994;60:2355–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd A, Ghosh M, May TBet al. Sequence of the algL gene of Pseudomonas aeruginosa and purification of its alginate lyase product. Gene. 1993;131:1–8. [DOI] [PubMed] [Google Scholar]
- Breton C, Fournel-Gigleux S, Palcic MM. Recent structures, evolution and mechanisms of glycosyltransferases. Curr Opin Struct Biol. 2012;22:540–549. [DOI] [PubMed] [Google Scholar]
- Bugli F, Palmieri V, Torelli Ret al. In vitro effect of clarithromycin and alginate lyase against Helicobacter pylori biofilm. Biotechnol Progr. 2016;32:1584–91. [DOI] [PubMed] [Google Scholar]
- Bugli F, Posteraro B, Papi Met al. In vitro interaction between alginate lyase and amphotericin B against Aspergillus fumigatus biofilm determined by different methods. Antimicrob Agents Chemother. 2013;57:1275–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundalovic-Torma C, Whitfield GB, Marmont LSet al. A systematic pipeline for classifying bacterial operons reveals the evolutionary landscape of biofilm machineries. PloS Comput Biol. 2020;16:e1007721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd MS, Sadovskaya I, Vinogradov Eet al. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol Microbiol. 2009;73:622–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus L, Briaud P, Vandenesch Fet al. How bacterial adaptation to cystic fibrosis environment shapes interactions between Pseudomonas aeruginosa and Staphylococcus aureus. Front Microbiol. 2021;12:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanasit W, Gonzaga ZJC, Rehm BHA. Analysis of the alginate O-acetylation machinery in Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 2020;104:2179–2191. [DOI] [PubMed] [Google Scholar]
- Chang WS, van de Mortel M, Nielsen Let al. Alginate production by Pseudomonas putida creates a hydrated microenvironment and contributes to biofilm architecture and stress tolerance under water-limiting conditions. J Bacteriol. 2007;189:8290–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherny KE, Sauer K. Untethering and degradation of the polysaccharide matrix are essential steps in the dispersion response of Pseudomonas aeruginosa biofilms. J Bacteriol. 2020;202. 10.1128/JB.00575-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba A, Seki M, Suzuki Yet al. Staphylococcus aureus utilizes environmental RNA as a building material in specific polysaccharide-dependent biofilms. NPJ Biofilms Microbiomes. 2022;8:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin JSF, Sinha S, Nalaparaju Aet al. Pseudomonas aeruginosa Psl exopolysaccharide interacts with the antimicrobial peptide LG21. Water. 2017;9:681. 10.3390/w9090681. [DOI] [Google Scholar]
- Chitnis CE, Ohman DE. Cloning of Pseudomonas aeruginosa algG, which controls alginate structure. J Bacteriol. 1990;172:2894–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Huang X, Lan Piao Yet al. Molecular modeling and redesign of alginate lyase from Pseudomonas aeruginosa for accelerating CRPA biofilm degradation. Proteins. 2016;84:1875–87. [DOI] [PubMed] [Google Scholar]
- Christensen BE, Ertesvåg H, Beyenal Het al. Resistance of biofilms containing alginate-producing bacteria to disintegration by an alginate degrading enzyme (AlgL). Biofouling. 2001;17:203–10. [Google Scholar]
- Chung CH, Ives HE, Almeda Set al. Purification from Escherichia coli of a periplasmic protein that is a potent inhibitor of pancreatic proteases. J Biol Chem. 1983;258:11032–11038. [PubMed] [Google Scholar]
- Cleasby A, Wonacott A, Skarzynskp Tet al. The X-ray crystal structure of phosphomannose isomerase from Candida albicans at 1.7 Å resolution. Nat Struct Mol Biol. 1996;3:470–479. [DOI] [PubMed] [Google Scholar]
- Coakley RD, Grubb BR, Paradiso AMet al. Abnormal surface liquid pH regulation by cultured cystic fibrosis bronchial epithelium. Proc Natl Acad Sci USA. 2003;100:16083–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin KM, Alnabelseya N, Baker Pet al. PelA deacetylase activity is required for Pel polysaccharide synthesis in Pseudomonas aeruginosa. J Bacteriol. 2013;195:2329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin KM, Gordon VD, Murakami Ket al. The pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PloS Pathog. 2011;7:e1001264. 10.1371/journal.ppat.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne MJ, Russell KS, Coyle CLet al. The Pseudomonas aeruginosa algC gene encodes phosphoglucomutase, required for the synthesis of a complete lipopolysaccharide core. J Bacteriol. 1994;176:3500–07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daboor SM, Rohde JR, Cheng Z. Disruption of the extracellular polymeric network of Pseudomonas aeruginosa biofilms by alginate lyase enhances pathogen eradication by antibiotics. J Cystic Fibrosis. 2021;20:264–70. [DOI] [PubMed] [Google Scholar]
- Darouiche RO, Mansouri MD, Gawande P vet al. Antimicrobial and antibiofilm efficacy of triclosan and DispersinB® combination. J Antimicrob Chemother. 2009;64:88–93. [DOI] [PubMed] [Google Scholar]
- Darzins A, Frantz B, Vanags RIet al. Nucleotide sequence analysis of the phosphomannose isomerase gene (PMI) of Pseudomonas aeruginosa and comparison with the corresponding Escherichia coli gene manA. Gene. 1986;42:293–302. [DOI] [PubMed] [Google Scholar]
- Deretic V, Gill JF, Chakrabarty AM. Gene algD coding for GDP-mannose dehydrogenase is transcriptionally activated in mucoid Pseudomonas aeruginosa. J Bacteriol. 1987a;169:351–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Gill JF, Chakrabarty AM. Pseudomonas aeruginosa infection in cystic fibrosis: nucleotide sequence and transcriptional regulation of the algD gene. Nucl Acids Res. 1987b;15:4567–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donelli G, Francolini I, Romoli Det al. Synergistic activity of dispersin B and cefamandole nafate in inhibition of Staphylococcal biofilm growth on polyurethanes. Antimicrob Agents Chemother. 2007;51:2733–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Beis K, Nesper Jet al. The structure of Wza, the translocon group 1 capsular polysaccharides in Escherichia coli, defines a new class of membrane protein. Nature. 2006;444:226–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Major LL, Srikannathasan Vet al. RmlC, a C3’ and C5’ carbohydrate epimerase, appears to operate via an intermediate with an unusual twist boat conformation. J Mol Biol. 2007;365:146–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreifus JE, O'Neal L, Jacobs HMet al. The Sia system and c-di-GMP play a crucial role in controlling cell-association of Psl in planktonic P. aeruginosa. J Bacteriol. 2022;204. 10.1128/jb.00335-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drula E, Garron M-L, Dogan Set al. The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res. 2022;50:D571–D577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunwell JM, Purvis A, Khuri S. Cupins: the most functionally diverse protein superfamily?. Phytochemistry. 2004;65:7–17. [DOI] [PubMed] [Google Scholar]
- Ertesvåg H, Erlien F, Skjåk-Bræk Get al. Biochemical properties and substrate specificities of a recombinantly produced Azotobacter vinelandii alginate lyase. J Bacteriol. 1998;180:3779–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans LR, Linker A. Production and characterization of the slime polysaccharide of Pseudomonas aeruginosa. J Bacteriol. 1973;116:915–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrel EK, Tipton PA. Functional characterization of AlgL, an alginate lyase from Pseudomonas aeruginosa. Biochem. 2012;51:102569–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming D, Chahin L, Rumbaugh K. Glycoside hydrolases degrade polymicrobial bacterial biofilms in wounds. Antimicrob Agents Chemother. 2017;61. 10.1128/AAC.01998-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–33. [DOI] [PubMed] [Google Scholar]
- Franklin MJ, Chitnis CE, Gacesa Pet al. Pseudomonas aeruginosa AlgG is a polymer level alginate C5-mannuronan epimerase. J Bacteriol. 1994;176:1821–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin MJ, Nivens DE, Weadge JTet al. Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front Microbio. 2011;2:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin MJ, Ohman DE. Identification of algF in the alginate biosynthetic gene cluster of Pseudomonas aeruginosa which is required for alginate acetylation. J Bacteriol. 1993;175:5057–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin MJ, Ohman DE. Identification of algI and algJ in the Pseudomonas aeruginosa alginate biosynthetic gene cluster which are required for alginate O acetylation. J Bacteriol. 1996;178:2186–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin MJ, Ohman DE. Mutant analysis and cellular localization of the AlgI, AlgJ, and AlgF proteins required for O acetylation of alginate in Pseudomonas aeruginosa. J Bacteriol. 2002;184:3000–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman L, Kolter R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol. 2004;51:675–90. [DOI] [PubMed] [Google Scholar]
- Friedman L, Kolter R. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J Bacteriol. 2004;186:4457–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellatly SL, Hancock REW. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis. 2013;67:159–73. [DOI] [PubMed] [Google Scholar]
- Ghafoor A, Jordens Z, Rehm BHA. Role of PelF in pel polysaccharide biosynthesis in Pseudomonas aeruginosa. Appl Environ Microbiol. 2013;79:2968–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheorghita AA, Li YE, Kitova ENet al. Structure of the AlgKX modification and secretion complex required for alginate production and biofilm attachment in Pseudomonas aeruginosa. Nat Commun. 2022a;13:7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheorghita AA, Wolfram F, Whitfield GBet al. The Pseudomonas aeruginosa homeostasis enzyme AlgL clears the periplasmic space of accumulated alginate during polymer biosynthesis. J Biol Chem. 2022b;298:101560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert KB, Kim TH, Gupta Ret al. Global position analysis of the Pseudomonas aeruginosa quorum-sensing transcription factor LasR. Mol Microbiol. 2009;73:1072–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilligan PH. Infections in patients with cystic fibrosis: diagnostic microbiology update. Clin Lab Med. 2014;34:197–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giménez-Andrés M, Čopič A, Antonny B. The many faces of amphipathic helices. Biomolecules. 2018;8:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg JB, Hatano K, Pier GB. Synthesis of lipopolysaccharide O side chains by Pseudomonas aeruginosa PAO1 requires the enzyme phosphomannomutase. J Bacteriol. 1993;175:1605–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goltermann L, Tolker-Nielsen T. Importance of the exopolysaccharide matrix in antimicrobial tolerance of Pseudomonas aeruginosa aggregates. Antimicrob Agents Chemother. 2017;61:e02696–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govan JR, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant GT, Morris ER, Rees DAet al. Biological interactions between polysaccharides and divalent cations: the egg-box model. FEBS Lett. 1973;32:195–8. [Google Scholar]
- Hallgren J, Tsirigos KD, Damgaard Pedersen Met al. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. bioRxiv. 2022. 10.1101/2022.04.08.487609. [DOI]
- Hatch RA, Schiller NL. Alginate lyase promotes diffusion of aminoglycosides through the extracellular polysaccharide of mucoid Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1998;42:974–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay ID, Gatland K, Campisano Aet al. Impact of alginate overproduction on attachment and biofilm architecture of a supermucoid Pseudomonas aeruginosa strain. Appl Environ Microbiol. 2009;75:6022–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay ID, Schmidt O, Filitcheva Jet al. Identification of a periplasmic AlgK-AlgX-MucD multiprotein complex in Pseudomonas aeruginosa involved in biosynthesis and regulation of alginate. Appl Microbiol Biotechnol. 2012;93:215–27. [DOI] [PubMed] [Google Scholar]
- Hentzer M, Teitzel GM, Balzer GJet al. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J Bacteriol. 2001;183:5395–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberger CD, Ye RW, Parsek MRet al. The algT (algU) gene of Pseudomonas aeruginosa, a key regulator involved in alginate biosynthesis, encodes an alternative δ factor (σE). Proc Natl Acad Sci USA. 1995;92:7941–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høiby N, Ciofu O, Johansen HKet al. The clinical impact of bacterial biofilms. Int J Oral Sci. 2011;3:55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. Dali server: structural unification of protein families. Nucleic Acids Res. 2022;50:W210–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotterbeekx A, Kumar-Singh S, Goossens Het al. In vivo and In vitro interactions between Pseudomonas aeruginosa and Staphylococcus spp. Front Cell Infect Microbiol. 2017;7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ierardi E, Losurdo G, Mileti Aet al. The puzzle of coccoid forms of Helicobacter pylori: beyond basic science. Antibiotics. 2020;9:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie Y, Borlee BR, O’Connor JRet al. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2012;109:20632–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie Y, Starkey M, Edwards ANet al. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol Microbiol. 2010;78:158–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Wang X, Hinnebusch BJet al. Depolymerization of β-1,6-N-acetyl-D-glucosamin disrupts the integrity of diverse bacterial biofilms. J Bacteriol. 2005;187:382–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson KD, Starkey M, Kremer Set al. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J Bacteriol. 2004;186:4466–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs HM, O'Neal L, Lopatto Eet al. Mucoid Pseudomonas aeruginosa can produce calcium-gelled biofilms independent of the matrix components Psl and CdrA. J Bacteriol. 2022;204:e0056821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagessar KL, Mchaourab HS, Claxton DP. The N-terminal domain of an archaeal multidrug and toxin extrusion (MATE) transporter mediates proton coupling required for prokaryotic drug resistance. J Biol Chem. 2019;294:12807–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Franklin MJ, Ertesvåg Het al. The dual roles of AlgG in C-5-epimerization and secretion of alginate polymers in Pseudomonas aeruginosa. Mol Microbiol. 2003;47:1123–33. [DOI] [PubMed] [Google Scholar]
- Jain S, Ohman DE. Alginate Biosynthesis in Psuedomonas. Boston: Springer, 2004. [Google Scholar]
- Jang CH, Piao YL, Huang Xet al. Modeling and re-engineering of Azotobacter vinelandii alginate lyase to enhance its catalytic efficiency for accelerating biofilm degradation. PloS ONE. 2016;11:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings LK, Dreifus JE, Reichhardt Cet al. Pseudomonas aeruginosa aggregates in cystic fibrosis sputum produce exopolysaccharides that likely impede current therapies. Cell Rep. 2021;34:108782. 10.1016/j.celrep.2021.108782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings LK, Storek KM, Ledvina HEet al. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc Natl Acad Sci USA. 2015;112:11353–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji S, Dix SR, Aziz AAet al. The molecular basis of endolytic activity of a multidomain alginate lyase from Defluviitalea phaphyphila, a representative of a new lyase family, PL39. J Biol Chem. 2019;294:18077–091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CJ, Wozniak DJ. Psl produced by mucoid Pseudomonas aeruginosa contributes to the establishment of biofilms and immune evasion. mBio. 2017;8. 10.1128/mBio.00864-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel Aet al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalpana BJ, Aarthy S, Pandian SK. Antibiofilm activity of α-amylase from Bacillus subtilis S8-18 against biofilm forming human bacterial pathogens. Appl Biochem Biotechnol. 2012;167:1778–94. [DOI] [PubMed] [Google Scholar]
- Karygianni L, Ren Z, Koo Het al. Biofilm matrixome: extracellular components in structured microbial communities. Trends Microbiol. 2020;28:668–81. [DOI] [PubMed] [Google Scholar]
- Kasai K, Kuroda Y, Takabuchi Yet al. Phosphorylation of Thr328 in hyaluronan synthase 2 is essential for hyaluronan synthesis. Biochem Biophys Res Commun. 2020;533:732–38. [DOI] [PubMed] [Google Scholar]
- Kasetty S, Katharios-Lanwermeyer S, O’Toole GAet al. Differential surface competition and biofilm invasion of Pseudomonas aeruginosa PA14 and PAO1. J Bacteriol. 2021;203:e00265–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiski CL, Harwich M, Jain Set al. AlgK is a TPR-containing protein and the periplasmic component of a novel exopolysaccharide secretin. Structure. 2010;18:265–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-S, Song S, Lee Met al. Crystal structure of a soluble fragment of the membrane fusion protein HlyD in a Type I secretion system of Gram-negative bacteria. Structure. 2016;24:477–85. [DOI] [PubMed] [Google Scholar]
- Koshland D. Stereochemistry and the mechanism of enzymatic reactions. Biol Rev. 1953;28:416–36. [Google Scholar]
- Kuk ACY, Hao A, Guan Zet al. Visualizing conformation transitions of the Lipid II flippase MurJ. Nat Commun. 2019;10. 10.1038/s41467-019-09658-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuk ACY, Hao A, Lee SY. Structure and mechanism of the lipid flippase MurJ. Annu Rev Biochem. 2022;91:705–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusakizako T, Claxton DP, Tanaka Yet al. Structural basis of H+-dependent conformational change in a bacterial MATE transporter. Structure. 2019;27:293–301.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri D, Nag M, Banerjee Ret al. Amylases: biofilm inducer or biofilm inhibitor?. Front Cell Infect Microbiol. 2021;11:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampp JW, Griswold KE. Alginate lyase exhibits catalysis-independent biofilm dispersion and antibiotic synergy. Antimicrob Agents Chemother. 2013;57:137–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Mauff F, Bamford NC, Alnabelseya Net al. Molecular mechanism of Aspergillus fumigatus biofilm disruption by fungal and bacterial glycoside hydrolases. J Biol Chem. 2019;294:10760–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Mauff F, Razvi E, Reichhardt Cet al. The Pel polysaccharide is predominantly composed of a dimeric repeat of α-1,4 linked galactosamine and N-acetylgalactosamine. Commun Biol. 2022;5. 10.1038/s42003-022-03453-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wang Y, Li Xet al. Enhancing the thermo-stability and anti-biofilm activity of alginate lyase by immobilization on low molecular weight chitosan nanoparticles. IJMS. 4565, 2019;20. 10.3390/ijms20184565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Yao Y, Yang Get al. Co-crystal structure of Thermosynechococcus elongatus sucrose phosphate synthase with UDP and sucrose-6-phosphate provides insight into its mechanism of action involving an oxocarbenium ion and the glycosidic bond. Front Microbiol. 2020;11:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Chen J-H, Hao Yet al. Structures of the PelD cyclic diguanylate effector involved in pellicle formation in Pseudomonas aeruginosa PAO1. J Biol Chem. 2012;287:30191–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Kosorok MR, Farrell PMet al. Longitudinal development of mucoid Pseudomonas aeruginosa infection and lung disease progression in children with cystic fibrosis. JAMA. 2005;293:581–8. [DOI] [PubMed] [Google Scholar]
- Limoli DH, Whitfield GB, Kitao Tet al. Pseudomonas aeruginosa alginate overproduction promotes coexistence with Staphylococcus aureus in a model of cystic fibrosis respiratory Infection. mBio. 2017;8:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low KE, Gheorghita AA, Tammam SDet al. Pseudomonas aeruginosa AlgF is a protein-protein interaction mediator required for acetylation of the alginate exopolysaccharide. J Biol Chem. 2023;299:105314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low KE, Howell PL. Gram-negative synthase-dependent exopolysaccharide biosynthetic machines. Curr Opin Struct Biol. 2018;53:32–44. [DOI] [PubMed] [Google Scholar]
- Lui Y, Gloag ES, Hill PJet al. Interbacterial antagonism mediated by a released polysaccharide. J Bacteriol. 2022;204:e0007622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukose V, Walvoort MTC, Imperiali B. Bacterial phosphoglycosyl transferases: Initiators of glycan biosynthesis at the membrane interface. Glycobiology. 2017;27:820–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyczak JB, Cannon CL, Pier GB. Establishment of Pseudomonas aeruginosa infection: Lessons from a versatile opportunist. Microbes Infect. 2000;2:1051–60. [DOI] [PubMed] [Google Scholar]
- Ma D, Wang Z, Merrikh CNet al. Crystal structure of a membrane-bound O-acyltransferase. Nature. 2018;562:286–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Conover M, Lu Het al. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PloS Pathog. 2009;5:e1000354. 10.1371/journal.ppat.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Jackson KD, Landry RM, Parsek MR, Wozniak DJ. Analysis of Pseudomonas aeruginosa conditional variants reveals roles for the psl polysaccharide in adhesion and maintaining biofilm structure post attachment. J Bacteriol. 2006;188:8213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Wang S, Wang Det al. The roles of biofilm matrix polysaccharide Psl in mucoid Pseudomonas aeruginosa biofilms. FEMS Immunol Med Microbiol. 2012;65:377–80. [DOI] [PubMed] [Google Scholar]
- Ma LZ, Wang D, Liu Yet al. Regulation of biofilm exopolysaccharide biosynthesis and degradation in Pseudomonas aeruginosa. Annu Rev Microbiol. 2022;76:413–33. [DOI] [PubMed] [Google Scholar]
- Madeira F, Park YM, Lee Jet al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen JS, Lin YC, Squyres GRet al. Facultative control of matrix production optimizes competitive fitness in Pseudomonas aeruginosa PA14 biofilm models. Appl Environ Microbiol. 2015;81:8414–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai GT, McCormack JG, Seow WKet al. Inhibition of adherence of mucoid Pseudomonas aeruginosa by alginase, specific monoclonal antibodies, and antibiotics. Infect Immun. 1993;61:4338–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiorana A, Bugli F, Papi Met al. Adhesive properties of Aspergillus fumigatus biofilms probed by atomic force microscopy and effects of alginate lyase enzyme. Microscopie. 2014;22:37–42. [Google Scholar]
- Maloney FP, Kuklewicz J, Corey RAet al. Structure, substrate recognition and initiation of hyaluronan synthase. Nature. 2022;604:195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmont LS, Rich JD, Whitney JCet al. Oligomeric lipoprotein PelC guides Pel polysaccharide export across the outer membrane of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2017;114:2892–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmont LS, Whitfield GB, Pfoh Ret al. PelX is a UDP-N-acetylglucosamine C4-epimerase involved in Pel polysaccharide-dependent biofilm formation. J Biol Chem. 2020;295:11949–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmont LS, Whitfield GB, Rich JDet al. PelA and PelB proteins form a modification and secretion complex essential for Pel polysaccharide-dependent biofilm formation in Pseudomonas aeruginosa. J Biol Chem. 2017;292:19411–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DW, Schurr MJ, Mudd MHet al. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci USA. 1993;90:8377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez SE, Beavo JA, Hol WGJ. GAF domains: two-billion-year-old molecular switches that bind cyclic nucleotides. Mol Intervent. 2002;2:317–23. [DOI] [PubMed] [Google Scholar]
- Marvig RL, Sommer LM, Molin Set al. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat Genet. 2015;47:57–64. [DOI] [PubMed] [Google Scholar]
- Matasukawa M, Greenberg EP. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J Bacteriol. 2004;186:4449–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May TB, Chakrabarty AM. Pseudomonas aeruginosa: genes and enzymes of alginate synthesis. Trends Microbiol. 1994;2:151–7. [DOI] [PubMed] [Google Scholar]
- Melero-Fernandez de Mera RM, Arasu UT, Kärnä Ret al. Effects of mutations in the post-translational modification sites on the trafficking of hyaluronan synthase 2 (HAS2). Matrix Biol. 2019;80:85–103. [DOI] [PubMed] [Google Scholar]
- Merighi M, Lee VT, Hyodo Met al. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol Microbiol. 2007;65:876–95. [DOI] [PubMed] [Google Scholar]
- Mishra M, Byrd MS, Sergeant Set al. Pseudomonas aeruginosa Psl polysaccharide reduces neutrophil phagocytosis and the oxidative response by limiting complement-mediated opsonization. Cell Microbiol. 2012;14:95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradali MF, Donati I, Sims IMet al. Alginate polymerization and modification are linked in Pseudomonas aeruginosa. mBio. 2015;6:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradali MF, Ghods S, Rehm BHA. Activation mechanism and cellular localization of membrane-anchored alginate polymerase in Pseudomonas aeruginosa. Appl Environ Microbiol. 2017;83:e03499–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JLW, Strumillo J, Zimmer J. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature. 2013;493:181–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison AJ, Wenzel RP. Epidemiology of infections due to Pseudomonas aeruginosa. Rev Infect Dis. 1984;6:S627–42. [DOI] [PubMed] [Google Scholar]
- Mrsny RJ, Lazazzera BA, Daugherty ALet al. Addition of a bacterial alginate lyase to purulent CF sputum in vitro can result in the disruption of alginate and modification of sputum viscoelasticity. Pulm Pharmacol. 1994;7:357–66. [DOI] [PubMed] [Google Scholar]
- Murata K, Inose T, Hisano Tet al. Bacterial alginate lyase: enzymology, genetics and application. J Ferment Bioeng. 1993;76:427–37. [Google Scholar]
- Nivens DE, Ohman DE, Williams Jet al. Role of alginate and its O acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J Bacteriol. 2001;183:1047–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oglesby LL, Jain S, Ohman DE. Membrane topology and roles of Pseudomonas aeruginosa Alg8 and Alg44 in alginate polymerization. Microbiology. 2008;154:1605–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostapska H, Le Mauff F, Gravelat FNet al. Co-operative biofilm interactions between Aspergillus fumigatus and Pseudomonas aeruginosa through secreted galactosaminogalactan exopolysaccharide. J Fungi. 2022a;8:336. 10.3390/jof8040336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostapska H, Raju D, Corsini Ret al. Preclinical evaluation of recombinant microbial glycoside hydrolases as antibiofilm agents in acute pulmonary Pseudomonas aeruginosa Infection. Antimicrob Agents Chemother. 2022;66:e0005222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostapska H, Raju D, Lehoux Met al. Preclinical evaluation of recombinant microbial glycoside hydrolases in the prevention of experimental invasive aspergillosis. mBio. 2021;12:e02446–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paletta JL, Ohman DE. Evidence for two promoters internal to the alginate biosynthesis operon in Pseudomonas aeruginosa. Curr Microbiol. 2012;65:770–5. [DOI] [PubMed] [Google Scholar]
- Pan Z, Zhu J, Shang Yet al. An autoinhibited conformation of LGN reveals a distinct interaction mode between GoLoco motifs and TPR motifs. Structure. 2013;21:1007–17. [DOI] [PubMed] [Google Scholar]
- Patel KK, Tripathi M, Pandey Net al. Alginate lyase immobilized chitosan nanoparticles of ciprofloxacin for the improved antimicrobial activity against the biofilm associated mucoid P. aeruginosa infection in cystic fibrosis. Int J Pharm. 2019;563:30–42. [DOI] [PubMed] [Google Scholar]
- Pelissier MC, Lesley SA, Kuhn Pet al. Structural insights into the catalytic mechanism of bacterial guanosine-diphospho-D-mannose pyrophosphorylase and its regulation by divalent ions. J Biol Chem. 2010;285:27468–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riba A, Itzhaki LS. The tetratricopeptide-repeat motif is a versatile platform that enables diverse modes of molecular recognition. Curr Opin Struct Biol. 2019;54:43–9. [DOI] [PubMed] [Google Scholar]
- Pestrak MJ, Baker P, Dellos-Nolan Set al. Treatment with the Pseudomonas aeruginosa glycoside hydrolase PslG combats wound infection by improving antibiotic efficacy and host innate immune activity. Antimicrob Agents Chemother. 2019;63:e00234–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CE, Brown DG, Limoli DHet al. Exogenous alginate protects Staphylococcus aureus from killing by Pseudomonas aeruginosa. J Bacteriol. 2020;202:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritt B, O'Brien L, Winn W. Mucoid Pseudomonas in cystic fibrosis. Am J Clin Pathol. 2007;128:32–34. [DOI] [PubMed] [Google Scholar]
- Ramasubbu N, Thomas LM, Ragunath Cet al. Structural analysis of dispersin B, a biofilm-releasing glycoside hydrolase from the periodontopathogen Actinobacillus actinomycetemcomitans. J Mol Biol. 2005;349:475–86. [DOI] [PubMed] [Google Scholar]
- Ramos J-L. Pseudomonas: Volume 3 Biosynthesis of Macromolecules and Molecular Metabolism. Berlin: Springer Science & Business Media, 2004. [Google Scholar]
- Ray LC, Das D, Entova Set al. Membrane association of monotopic phosphoglycosyl transferase underpins function. Nat Chem Biol. 2018;14:538–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razvi E, DiFrancesco BR, Wasney GAet al. Small molecular inhibition of exopolysaccharide modification enzymes is a viable strategy to block bacterial biofilm formation. Microbiol Spectr. 2023;11:e0029623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razvi E, Whitfield GB, Reichhardt Cet al. Glycoside hydrolase processing of the Pel polysaccharide alters biofilm biomechanics and Pseudomonas aeruginosa virulence. NPJ Biofilms Microbiomes. 2023a;9. 10.1038/s41522-023-00375-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regni C, Schramm AM, Beamer LJ. The reaction of phosphohexomutase from Pseudomonas aeruginosa: structural insights into a simple processive enzyme. J Biol Chem. 2006a;281:15564–71. [DOI] [PubMed] [Google Scholar]
- Regni C, Shackelford GS, Beamer LJ. Complexes of the enzyme phosphomannomutase. /phosphoglucomutase with a slow substrate and an inhibitor. Acta Crystallogr F Struct Biol Cryst Commun. 2006b;62:722–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regni C, Tipton PA, Beamer LJ. Crystal structure of PMM/PGM: an enzyme in the biosynthetic pathway of P. aeruginosa virulence factors. Structure. 2002;10:269–79. [DOI] [PubMed] [Google Scholar]
- Rehman ZU, Rehm BHA. Dual roles of Pseudomonas aeruginosa AlgE in secretion of the virulence factor alginate and formation of the secretion complex. Appl Environ Microbiol. 2013;79:2002–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman ZU, Wang Y, Moradali MFet al. Insights into the assembly of the alginate biosynthesis machinery in Pseudomonas aeruginosa. Appl Environ Microbiol. 2013;79:3264–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichhardt C, Jacobs HM, Matwichuk Met al. The versatile Pseudomonas aeruginosa biofilm matrix protein CdrA promotes aggregation through different extracellular exopolysaccharide interactions. J Bacteriol. 2020;202:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichhardt C, Wong C, da Silva DPet al. CDRA interactions within the Pseudomonas aeruginosa biofilm matrix safeguard it from proteolysis and promote cellular packing. mBio. 2018;9. 10.1128/mBio.01376-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remminghorst U, Rehm BHA. Alg44, a unique protein required for alginate biosynthesis in Pseudomonas aeruginosa. FEBS Lett. 2006a;580:3883–8. [DOI] [PubMed] [Google Scholar]
- Remminghorst U, Rehm BHA. Bacterial alginates: from biosynthesis to applications. Biotechnol Lett. 2006b;28:1701–12. [DOI] [PubMed] [Google Scholar]
- Riley LM, Weadge JT, Baker Pet al. Structural and functional characterization of Pseudomonas aeruginosa AlgX: role of AlgX in alginate acetylation. J Biol Chem. 2013;288:22299–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles-Price A, Wong TY, Sletta Het al. AlgX is a periplasmic protein required for alginate biosynthesis in Pseudomonas aeruginosa. J Bacteriol. 2004;186:7369–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi E, la Rosa R, Bartell JAet al. Pseudomonas aeruginosa adaptation and evolution in patients with cystic fibrosis. Nat Rev Microbiol. 2021;19:331–42. [DOI] [PubMed] [Google Scholar]
- Rowe WJ III, Lebman DA, Ohman DE. Mechanism of resistance to phagocytosis and pulmonary persistence in mucoid Pseudomonas aeruginosa. Front Cell Infect Microbiol. 2023;13:1125901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH, Reddy VS, Moreno-Hagelsieb Get al. The Transporter Classification Database (TCDB): 2021 update. Nucelic Acids Res. 2021;49:D461–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santajit S, Seesuay W, Mahasongkram Ket al. Human single-chain variable fragments neutralize Pseudomonas aeruginosa quorum sensing molecule, 3O-C12-HSL, and Prevent cells from the HSL-mediated apoptosis. Front Microbiol. 2020;11:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkisova S, Patrauchan MA, Berglund Det al. Calcium-induced virulence factors associated with extracellular matrix of mucoid Pseudomonas aeruginosa biofilms. J Bacteriol. 2005;187:4327–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K, Stoodley P, Goeres DMet al. The biofilm life cycle: expanding the conceptual model of biofilm formation. Nat Rev Microbiol. 2022;20:608–20. 10.1038/s41579-022-00767-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena IM, Brown RM. Identification of cellulose synthase(s) in higher plants: Sequence analysis of processive -glycosyltransferases with the common motif “D, D, D35Q(R,Q)XRW.”. Cellulose. 1997;4:33–49. [Google Scholar]
- Schurr MJ, Yu H, Martinez-Salazar JMet al. Control of AlgU, a member of the σ(E)-like family of stress sigma factors, by the negative regulators mucA and mucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J Bacteriol. 1996;178:4997–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe J, Pérez-Burgos M, Herfurth Met al. Evidence for a widespread third system for bacterial polysaccharide export across the outer membrane comprising a composite OPX/β-barrel translocon. mBio. 2022;13. 10.1128/mbio.02032-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaik MM, Cendron L, Percudani Ret al. The structure of Helicobacter pylori HP0310 reveals an atypical peptidoglycan deacetylase. PLoS ONE. 2011;6:e19207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JA, Smith SE, Dean RT. Alginate inhibition of the uptake of Pseudomonas aeruginosa by macrophages. J Gen Microbiol. 1988;134:29–36. [DOI] [PubMed] [Google Scholar]
- Singh PK, Yadav VK, Kalia Met al. Pseudomonas aeruginosa auto inducer 3-oxo-C12-HSL exerts bacteriostatic effect and inhibits Staphylococcus epidermidis biofilm. Microbial Pathog. 2017;110:612–9. [DOI] [PubMed] [Google Scholar]
- Snarr BD, Baker P, Bamford NCet al. Microbial glycoside hydrolases as antibiofilm agents with cross-kingdom activity. Proc Natl Acad Sci USA. 2017;114:7124–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snook CF, Tipton PA, Beamer LJ. Crystal structure of GDP-mannose dehydrogenase: a key enzyme of alginate biosynthesis in P. aeruginosa. Biochemistry. 2003;42:4658–68. [DOI] [PubMed] [Google Scholar]
- Sonesson A, Jantzen E, Bryn Ket al. Chemical composition of a lipopolysaccharide from Legionella pneumophila. Arch. Microbiol. 1989;153:72–8. [DOI] [PubMed] [Google Scholar]
- Song Z, Wu H, Ciofu Oet al. Pseudomonas aeruginosa alginate is refractory to Th1 immune response and impedes host immune clearance in a mouse model of acute lung infection. J Med Microbiol. 2003;52:731–40. [DOI] [PubMed] [Google Scholar]
- Szymańska M, Karakulska J, Sobolewski Pet al. Glycoside hydrolase (PelAh) immobilization prevents Pseudomonas aeruginosa biofilm formation on cellulose-based wound dressing. Carbohydr Polym. 116625, 2020;246. 10.1016/j.carbpol.2020.116625. [DOI] [PubMed] [Google Scholar]
- Tan J, Rouse SL, Li Det al. A conformational landscape for alginate secretion across the outer membrane of Pseudomonas aeruginosa. Acta Crystallogr D Biol Crystallogr. 2014;70:2054–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorn CR, Clulow AJ, Boyd BJet al. Bacterial lipase triggers the release of antibiotics from digestible liquid crystal nanoparticles. J Controll Rel. 2020;319:168–82. [DOI] [PubMed] [Google Scholar]
- Thorn CR, Howell PL, Wozniak DJet al. Enhancing the therapeutic use of biofilm-dispersing enzymes with smart drug delivery systems. Adv Drug Deliv Rev. 2021a;179:113916. [DOI] [PubMed] [Google Scholar]
- Thorn CR, Raju D, Lacdao Iet al. Protective liquid crystal nanoparticles for targeted delivery of PslG: a biofilm dispersing enzyme. ACS Infect Dis. 2021b;7:2102–15. [DOI] [PubMed] [Google Scholar]
- Tielen P, Kuhn H, Rosenau Fet al. Interaction between extracellular lipase LipA and the polysaccharide alginate of Pseudomonas aeruginosa. BMC Microbiol. 159, 2013;13. 10.1186/1471-2180-13-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tielen P, Strathmann M, Jaeger KEet al. Alginate acetylation influences initial surface colonization by mucoid Pseudomonas aeruginosa. Microbiol Res. 2005;160:165–76. [DOI] [PubMed] [Google Scholar]
- Tseng BS, Reichhardt C, Merrihew GEet al. A biofilm matrix-associated protease inhibitor protects Pseudomonas aeruginosa from proteolytic attack. mBio. 2018;9:e00543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng BS, Zhang W, Harrison JJet al. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ Microbiol. 2013;15:2865–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasseur P, Soscia C, Voulhoux Ret al. PelC is a Pseudomonas aeruginosa outer membrane lipoprotein of the OMA family of proteins involved in exopolysaccharide transport. Biochimie. 2007;89:903–15. [DOI] [PubMed] [Google Scholar]
- Wang S, Liu X, Liu Het al. The exopolysaccharide Psl–eDNA interaction enables the formation of a biofilm skeleton in Pseudomonas aeruginosa. Environ Microbiol Rep. 2015;7:330–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X-P, Jiang Y-L, Dai Y-Net al. Structural and enzymatic analyses of a glucosyltransferase Alr3699/HepE involved in Anabaena heterocyst envelop polysaccharide biosynthesis. Glycobiology. 2016a;26:520–31. [DOI] [PubMed] [Google Scholar]
- Wang Y, Moradali MF, Goudarztalejerdi Aet al. Biological function of a polysaccharide degrading enzyme in the periplasm. Sci Rep. 2016b;6:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang J, Liu Xet al. Molecular and structural mechanisms of ZZ domain-mediated cargo selection by Nbr1. EMBO J. 2021;40:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters CM, Burton T, Kirui DKet al. Enzymatic degradation of in vitro Staphylococcus aureus biofilms supplemented with human plasma. Infect Drug Resist. 2016;9:71–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield C, Wear SS, Sande C. Assembly of bacterial capsular polysaccharides and exopolysaccharides. Annu Rev Microbiol. 2020c;74:521–43. [DOI] [PubMed] [Google Scholar]
- Whitfield GB, Howell PL. The matrix revisited: opening night for the Pel polysaccharide across eubacterial kingdoms. Microbiol Insights. 2021;14:117863612098858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield GB, Marmont LS, Bundalovic-Torma Cet al. Discovery and characterization of a Gram-positive Pel polysaccharide biosynthetic gene cluster. PLoS Pathog. 2020a;16:e1008281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield GB, Marmont LS, Howell PL. Enzymatic modifications of exopolysaccharides enhance bacterial persistence. Front Microbiol. 2015;6:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield GB, Marmont LS, Ostaszewski Aet al. Pel polysaccharide biosynthesis requires an inner membrane complex comprised of PelD, PelE, PelF, and PelG. J Bacteriol. 2020b;202:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney JC, Colvin KM, Marmont LSet al. Structure of the cytoplasmic region of PelD, a degenerate diguanylate cyclase receptor that regulates exopolysaccharide production in Pseudomonas aeruginosa. J Biol Chem. 2012;287:23582–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney JC, Hay ID, Li Cet al. Structural basis for alginate secretion across the bacterial outer membrane. Proc Natl Acad Sci USA. 2011;108:13083–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney JC, Howell PL. Synthase-dependent exopolysaccharide secretion in Gram-negative bacteria. Trends Microbiol. 2013;21:63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney JC, Whitfield GB, Marmont LSet al. Dimeric c-di-GMP is required for post-translational regulation of alginate production in Pseudomonas aeruginosa. J Biol Chem. 2015;290:12451–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierenga RK. The TIM-barrel fold: a versatile framework for efficient enzymes. FEBS Lett. 2001;492:193–98. [DOI] [PubMed] [Google Scholar]
- Wild R, Kowal J, Eyring Jet al. Structure of the yeast oligosaccharyltransferase complex gives insight into eukaryotic N-glycosylation. Science. 2018;359:545–50. [DOI] [PubMed] [Google Scholar]
- Wolfram F, Kitova EN, Robinson Het al. Catalytic mechanism and mode of action of the periplasmic alginate epimerase AlgG. J Biol Chem. 2014;289:6006–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LF, Ohman DE. Identification of genes in the σ22 regulon of Pseudomonas aeruginosa required for cell envelope homeostasis in either the planktonic or the sessile mode of growth. mBio. 2012;3:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LF, Ohman DE. Use of cell wall stress to characterize σ22 (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol Microbiol. 2009;72:183–201. [DOI] [PubMed] [Google Scholar]
- Wu H, Wang D, Tang Met al. The advance of assembly of exopolysaccharide Psl biosynthesis machinery in Pseudomonas aeruginosa. Microbiologyopen. 2019;8:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Liu J, Clarke BRet al. The molecular basis of regulation of bacterial capsule assembly by Wzc. Nat Commun. 2021;12:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Boucher JC, Hibler NSet al. Virulence properties of Pseudomonas aeruginosa lacking the extreme- stress sigma factor AlgU (σ(E)). Infect Immun. 1996;64:2774–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Schurr MJ, Deretic V. Functional equivalence of Escherichia coli σ(E) and Pseudomonas aeruginosa AlgU: E. coli rpoE restores mucoidy and reduces sensitivity to reactive oxygen intermediates in algU mutants of P. aeruginosa. J Bacteriol. 1995;177:3259–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Su T, Wu Het al. PslG, a self-produced glycosyl hydrolase, triggers biofilm disassembly by disrupting exopolysaccharide matrix. Cell Res. 2015;25:1352–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegans ME, DiGiandomenico A, Ray Ket al. Association of biofilm formation, Psl exopolysaccharide expression, and clinical outcomes in Pseudomonas aeruginosa keratitis: analysis of isolates in the steroids for corneal ulcers trial. JAMA Ophthalmol. 2016;134:383–9. [DOI] [PubMed] [Google Scholar]
- Zegans ME, Wozniak D, Griffin Eet al. Pseudomonas aeruginosa exopolysaccharide psl promotes resistance to the biofilm inhibitor polysorbate 80. Antimicrob Agents Chemother. 2012;56:4112–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wu H, Wang Det al. Intracellular glycosyl hydrolase PslG shapes bacterial cell fate, signaling, and the biofilm development of Pseudomonas aeruginosa. elife. 2022;11:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gómez-Redondo M, Jiménez-Osés Get al. Synthesis and structural analysis of Aspergillus fumigatus galactosaminogalactans featuring α-galactose, α-galactosamine and α- N-acetyl galactosamine linkages. Angewandte Chemie. 2020;132:12846–50. [DOI] [PubMed] [Google Scholar]
- Zhao K, Tseng BS, Beckerman Bet al. Psl trails guide exploration and microcolony formation in Pseudomonas aeruginosa biofilms. Nature. 2013;497:388–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski NA, Chakrabarty AM, Berry A. Characterization and regulation of the Pseudomonas aeruginosa algC gene encoding phosphomannomutase. J Biol Chem. 1991;266:9754–63. [PubMed] [Google Scholar]