Abstract
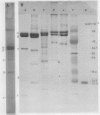
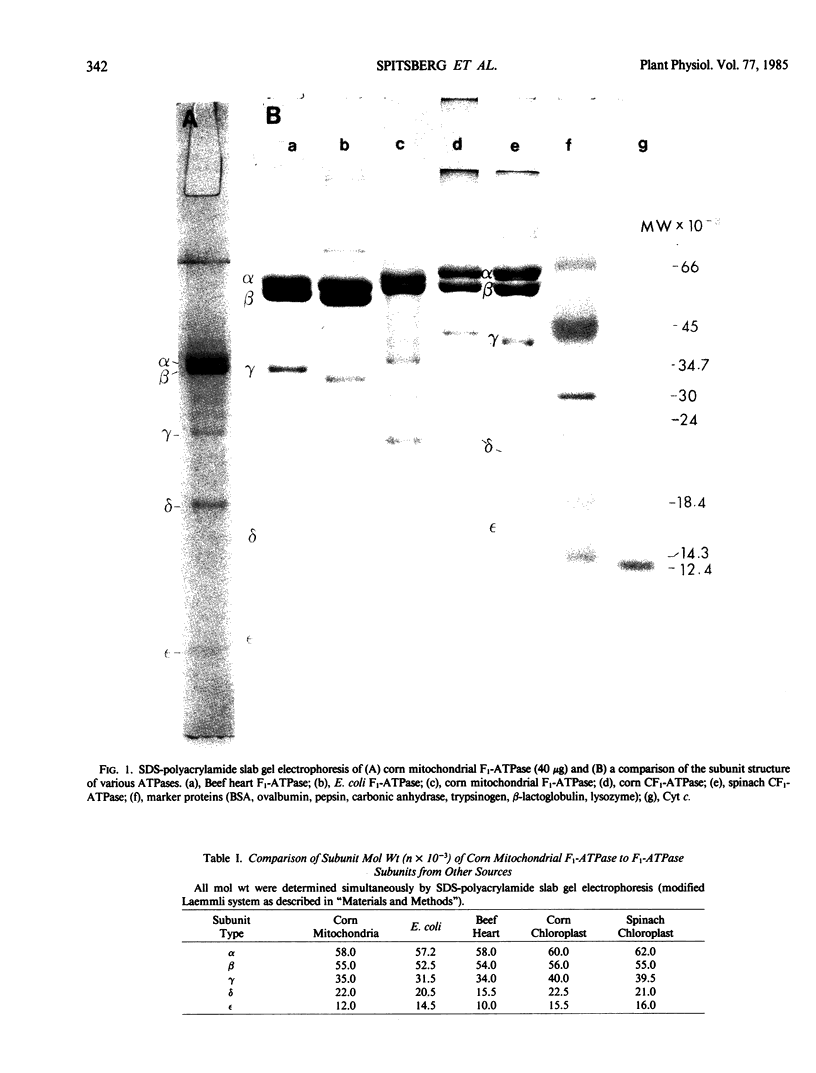
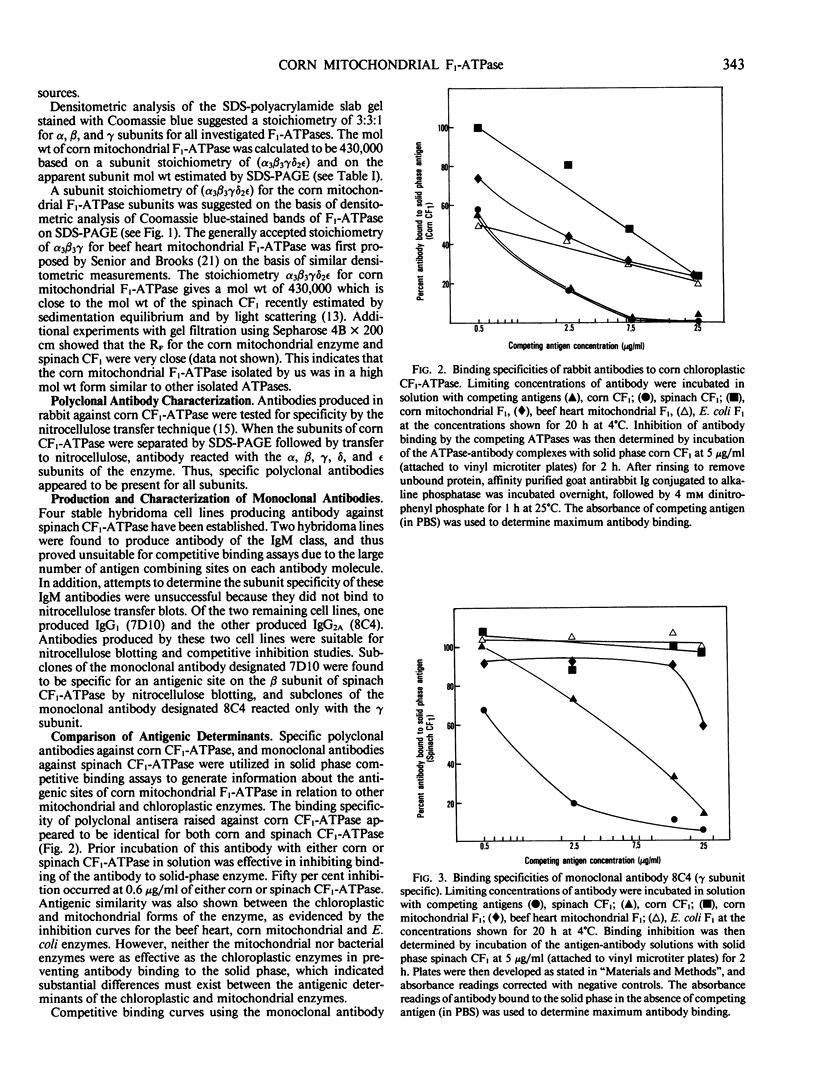
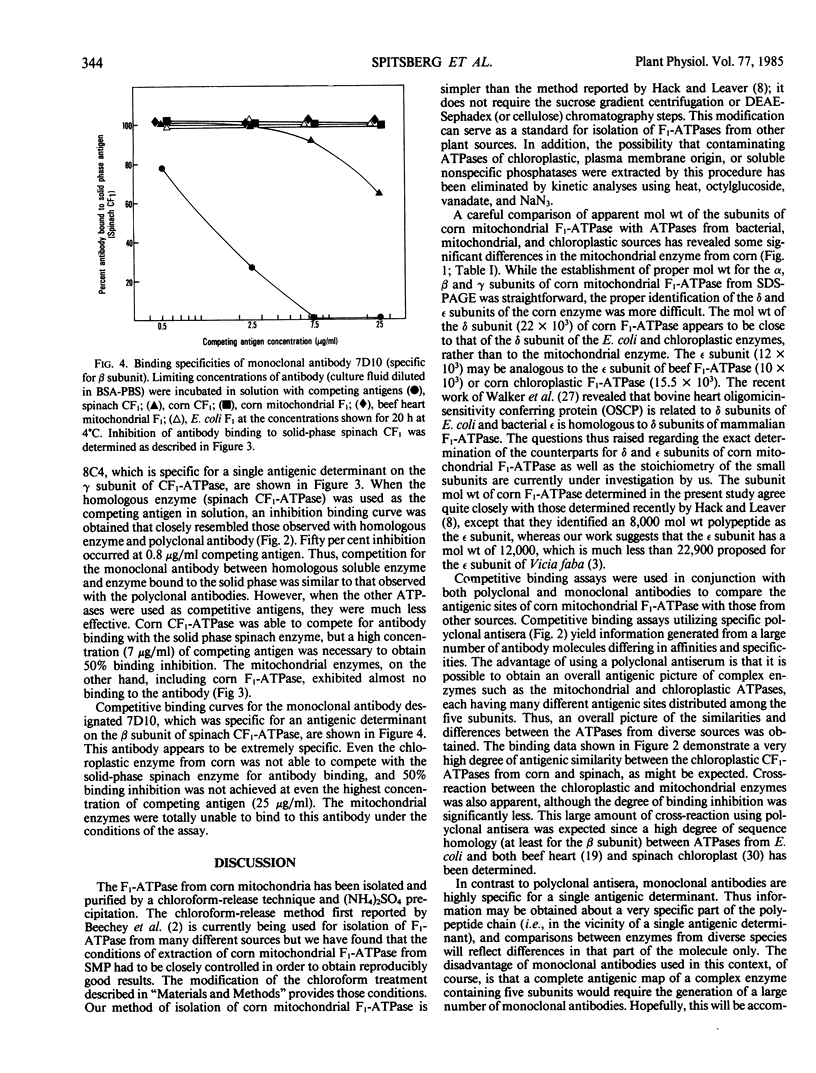
Corn mitochondrial F1-ATPase was purified from submitochondrial particles by chloroform extraction. Enzyme stored in ammonium sulfate at 4°C was substantially activated by ATP, while enzyme stored at −70°C in 25% glycerol was not. Enzyme in glycerol remained fully active (8-9 micromoles Pi released per minute per milligram), while the ammonium sulfate preparations steadily lost activity over a 2-month storage period. The enzyme was cold labile, and inactived by 4 minutes at 60°C. Treatment with octylglucoside resulted in complete loss of activity, while vanadate had no effect on activity. The apparent subunit molecular weights of corn mitochondrial F1-ATPase were determined by SDS-polyacrylamide gel electrophoresis to be 58,000 (α), 55,000 (β), 35,000 (γ), 22,000 (δ), and 12,000 (ε). Monoclonal and polyclonal antibodies used in competitive binding assays demonstrated that corn mitochondrial F1-ATPase was antigenically distinct from the chloroplastic CF1-ATPases of corn and spinach. Monoclonal antibodies against antigenic sites on spinach CF1-ATPase β and γ subunits were used to demonstrate that those sites were either changed substantially or totally absent from the mitochondrial F1-ATPase.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beechey R. B., Hubbard S. A., Linnett P. E., Mitchell A. D., Munn E. A. A simple and rapid method for the preparation of adenosine triphosphatase from submitochondrial particles. Biochem J. 1975 Jun;148(3):533–537. doi: 10.1042/bj1480533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutry M., Briquet M., Goffeau A. The alpha subunit of a plant mitochondrial F1-ATPase is translated in mitochondria. J Biol Chem. 1983 Jul 25;258(14):8524–8526. [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Gallagher S. R., Leonard R. T. Effect of vanadate, molybdate, and azide on membrane-associated ATPase and soluble phosphatase activities of corn roots. Plant Physiol. 1982 Nov;70(5):1335–1340. doi: 10.1104/pp.70.5.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giss B., Antoniou J., Smith G., Brumbaugh J. A method for culturing chick melanocytes: the effect of BRL-3A cell conditioning and related additives. In Vitro. 1982 Oct;18(10):817–826. doi: 10.1007/BF02796322. [DOI] [PubMed] [Google Scholar]
- Hack E., Leaver C. J. The alpha-subunit of the maize F(1)-ATPase is synthesised in the mitochondrion. EMBO J. 1983;2(10):1783–1789. doi: 10.1002/j.1460-2075.1983.tb01658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki Y., Asahi T. Purification and characterization of the soluble form of mitochondrial adenosine triphosphatase from sweet potato. Arch Biochem Biophys. 1983 Nov;227(1):164–173. doi: 10.1016/0003-9861(83)90359-4. [DOI] [PubMed] [Google Scholar]
- Kanazawa H., Futai M. Structure and function of H+-ATPase: what we have learned from Escherichia coli H+-ATPase. Ann N Y Acad Sci. 1982;402:45–64. doi: 10.1111/j.1749-6632.1982.tb25731.x. [DOI] [PubMed] [Google Scholar]
- Kearney J. F., Radbruch A., Liesegang B., Rajewsky K. A new mouse myeloma cell line that has lost immunoglobulin expression but permits the construction of antibody-secreting hybrid cell lines. J Immunol. 1979 Oct;123(4):1548–1550. [PubMed] [Google Scholar]
- O'Connor C. G., Ashman L. K. Application of the nitrocellulose transfer technique and alkaline phosphatase conjugated anti-immunoglobulin for determination of the specificity of monoclonal antibodies to protein mixtures. J Immunol Methods. 1982 Oct 29;54(2):267–271. doi: 10.1016/0022-1759(82)90068-0. [DOI] [PubMed] [Google Scholar]
- Penefsky H. S. Reversible binding of Pi by beef heart mitochondrial adenosine triphosphatase. J Biol Chem. 1977 May 10;252(9):2891–2899. [PubMed] [Google Scholar]
- Pick U., Bassilian S. The effects of octylglucoside on the interactions of chloroplast coupling factor 1 (CF1) with adenine nucleotides. Eur J Biochem. 1983 Jun 15;133(2):289–297. doi: 10.1111/j.1432-1033.1983.tb07461.x. [DOI] [PubMed] [Google Scholar]
- Runswick M. J., Walker J. E. The amino acid sequence of the beta-subunit of ATP synthase from bovine heart mitochondria. J Biol Chem. 1983 Mar 10;258(5):3081–3089. [PubMed] [Google Scholar]
- Ryrie I. J., Gallagher A. The yeast mitochondrial ATPase complex. Subunit composition and evidence for a latent protease contaminant. Biochim Biophys Acta. 1979 Jan 11;545(1):1–14. doi: 10.1016/0005-2728(79)90108-7. [DOI] [PubMed] [Google Scholar]
- Senior A. E., Brooks J. C. Studies on the mitochondrial oligomycin-insensitivt ATPase. I. An improved method of purification and the behavior of the enzyme in solutions of various depolymerizing agents. Arch Biochem Biophys. 1970 Sep;140(1):257–266. doi: 10.1016/0003-9861(70)90030-5. [DOI] [PubMed] [Google Scholar]
- Senior A. E., Brooks J. C. The subunit composition of the mitochondrial oligomycin-insensitive ATPase. FEBS Lett. 1971 Oct 1;17(2):327–329. doi: 10.1016/0014-5793(71)80178-3. [DOI] [PubMed] [Google Scholar]
- Senior A. E., Wise J. G. The proton-ATPase of bacteria and mitochondria. J Membr Biol. 1983;73(2):105–124. doi: 10.1007/BF01870434. [DOI] [PubMed] [Google Scholar]
- Spitsberg V. L., Blair J. E. Evidence supporting the identity of beef heart mitochondrial chloroform-released adenosine triphosphatase (ATPase) with coupling factor I. Biochim Biophys Acta. 1977 Apr 11;460(1):136–141. doi: 10.1016/0005-2728(77)90159-1. [DOI] [PubMed] [Google Scholar]
- Spitsberg V. L., Morris H. P., Chan S. H. Isolation and comparative studies of mitochondrial F1-ATPase from Morris hepatoma and rat liver. Arch Biochem Biophys. 1979 Jun;195(1):136–144. doi: 10.1016/0003-9861(79)90335-7. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Runswick M. J., Saraste M. Subunit equivalence in Escherichia coli and bovine heart mitochondrial F1F0 ATPases. FEBS Lett. 1982 Sep 20;146(2):393–396. doi: 10.1016/0014-5793(82)80960-5. [DOI] [PubMed] [Google Scholar]
- Younis H. M., Winget G. D., Racker E. Requirement of the delta subunit of chloroplast coupling factor 1 for photophosphorylation. J Biol Chem. 1977 Mar 10;252(5):1814–1818. [PubMed] [Google Scholar]
- Zurawski G., Bottomley W., Whitfeld P. R. Structures of the genes for the beta and epsilon subunits of spinach chloroplast ATPase indicate a dicistronic mRNA and an overlapping translation stop/start signal. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6260–6264. doi: 10.1073/pnas.79.20.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]