Abstract
In mammalian cells, besides nuclei, mitochondria are the only semi-autonomous organelles possessing own DNA organized in the form of nucleoids. While eukaryotic nuclear DNA compaction, chromatin compartmentalization and transcription are regulated by phase separation, our recent work proposed a model of mitochondrial nucleoid self-assembly and transcriptional regulation by multi-phase separation. Herein, we summarized the phase separation both in the nucleus and mitochondrial nucleoids, and did a comparison of the organization and activity regulating, which would provide new insight into the understanding of both architecture and genetics of nucleus and mitochondrial nucleoids.
INTRODUCTION
The compartmentation in cells is critical for carrying multiple functions (Boeynaems et al. 2018). It is well known that the inner membrane system segregates space in cells into organelles, such as the endoplasmic reticulum, Golgi apparatus, lysosomes, endosomes and so on. These organelles concentrate special factors to generate small reaction centers, which provide spatiotemporal control over cellular materials, metabolic and signals for cells in carrying multiple functions and pathways. Despite the inner membrane system, phase separation could also concentrate biomolecule independence of the membrane systems, which greatly expands the compartmentation in cells and reduces the threshold for generating small reaction centers (Banani et al. 2016, 2017; Riback et al. 2020).
Phase separation serves as a biomolecular condenser, which concentrates biomolecules to form droplet-like or gel-like structures (Banani et al. 2017; Shin and Brangwynne 2017). It is a kind of self-assembly driven by the intrinsic property of the biomolecules, such as weak interaction beneath intrinsically disordered regions (IDR) or multivalent effect between molecule interaction (Alberti et al. 2019). Not only proteins, DNAs, RNAs and even small molecules, such as dNTPs and drugs could be concentrated by co-phase separation (Klein et al. 2020; Wang et al. 2018), which provides the substrates, enzymes or cofactors for the efficient reaction in the small reaction centers generated by phase separation.
Phase separation provides the driving force to concentrate biomolecules into membraneless organelles, such as stress granules, cell junctions, signal transduction, transcription complex and so on (Banani et al. 2016). It is also reported to take part in the condensation of chromatin, enhancers, nuclear speckles, heterochromatin and even the architecture of nuclei, such as the generation of the nucleolus (Sabari et al. 2020). Nuclei are the largest organelle in cells and play critical important and multiple roles in DNA storage, replication and transcription, ribosome assembling, etc. The compartmentation by phase separation is critical for the nuclei in carrying multiple roles.
Mitochondria are the only semi-autonomous organelle in mammalian cells, owing a circle DNA, which plays critical roles in energy production, metabolism, apoptosis, and so on. Mitochondria DNA is organized into nucleoids by TFAM, one of the most abundant proteins in mitochondria. The crista plays a critical role in the compartmentation of mitochondria for metabolism and energy production (Cogliati et al. 2013; Frey and Mannella 2000). However, there are rare reports about the compartmentation in mitochondrial nucleoids, which share many commons with the nuclei, as well as some unique features. Recently, we reported that phase separation drives the self-assembly of the nucleoid, and regulates mitochondrial transcription, which provides a new model for mitochondria compartmentation (Long et al. 2021).
Herein, we summarized the compartmentation of nuclei and mitochondria in architecture assembling and discussed the phase separation from nuclei to mitochondrial nucleoids.
MAIN
Phase separation drives the chromatin condensation in nuclear architecture assembly
Nuclei are membrane organelles, which are responsible for chromatin-DNA storage, replication and transcription, etc. The chromatin-DNA is compacted by histones and condensates gradually from nucleosomes, 10 nm chromatin fiber, 30 nm chromatin fiber, euchromatin and heterochromatin to chromosomes and stored in the nuclei (Misteli 2020; Song et al. 2014). The chromatin-containing enhancers, especially super-enhancers could be gathered together with activators in a phase separation manner, which promotes transcription activity (Sabari et al. 2018). What’s more, the chromatin fiber could be locked by CTCF loops and clustered into droplets similarly driven by phase separation (Hansen et al. 2019). Many nuclear speckles acting as small reaction centers are generated by chromatin fiber, RNAs and so on, which are also reported to be driven by phase separation (Bi et al. 2019; Guo et al. 2019). The m6A-modification on RNA is reported to be critical for generating transcription activation speckles by phase separation (Cheng et al. 2021; Lee et al. 2021). The chromatin fiber could be further condensed by linker histone H1 and HP1a which also drives the special heterochromatin foci formation in a phase separation manner (Larson et al. 2017; Strom et al. 2017; Wang et al. 2020). The phase separation induces the aggregation of nucleosomes or chromatin fibers. What’s more, the condensation by phase separation in heterochromatin prevents the invasion of transcription factors, maintaining the inactivation status (Larson and Narlikar 2018; Sabari et al. 2020). In contrast, the euchromatin is enriched with transcription activation factors, RNA polymerase, RNAs and so on, but not H1 and HP1a, promoting gene activation and mRNA transcription.
The phase separation also provides a mechanical force for the architecture assembly and maintenance in cell nuclei. The chromatin is reported to be a gel-like structure, rather than liquid as the previous report (Strickfaden et al. 2020). The gel-like structure is also a feature of phase separation, which shows properties between solids and liquids. The gel-like structure of chromatin should also provide important mechanical support for the nucleus. The dynamic of the phase separation in chromatin may also regulate the mechanics of the nuclei, resulting in cell migration and invasion (Nava et al. 2020).
The nucleolus is the largest sub-organelle in the nuclei, which could be easily visualized under a normal microscope even in a bright field. It is a special reaction center without membrane segregation, which is responsible for rRNA transcription and ribosome assembly. It is reported to be a layered structure, starting from an inner core from rRNA transcription and proceeding toward the periphery (Brangwynne et al. 2011; Lafontaine et al. 2021). To keep the reaction smoothly and steadily, a multi-layered droplet-like liquid-liquid phase separation was found to drive the nucleolus formation (Feric et al. 2016; Lafontaine et al. 2021).
Phase separation drives the assembly of the mitochondrial nucleoid
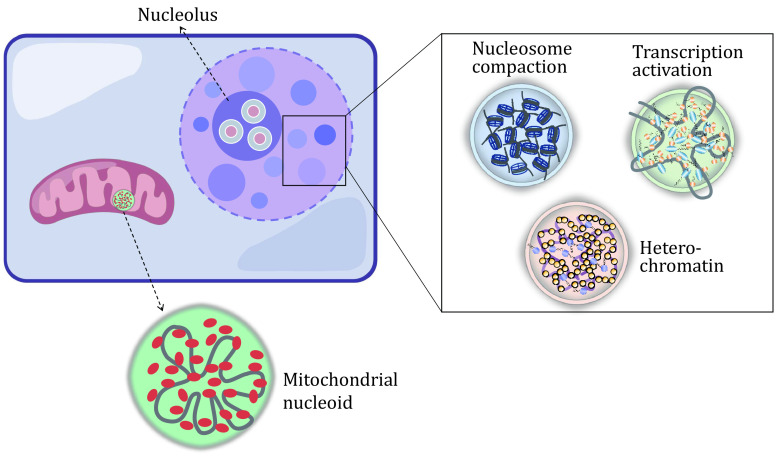
Unlike nuclei, the nucleoid is a membraneless organelle in mitochondria, lacking membranes to segregate it from the mitochondrial matrix. Mitochondria are thought to be derived from ancient bacteria (Roger et al. 2017), which possess a simple circle DNA, compacted by TFAM proteins into a nucleoid structure, similar to the nucleoid in bacteria (Kukat et al. 2015). Although the nucleoid is much more relaxed than chromatins, it shares many commons with nucleosomes. The high abundance of TFAM coated mtDNA in a ratio of 1:1000 to 1:2000 as reported (Ngo et al. 2014; Shen and Bogenhagen 2001; Takamatsu et al. 2002). The core of nucleoid constructed by TFAM-mtDNA, which could also be assembled easily in vitro in a droplet-like manner (Farge et al. 2014; Feric et al. 2021; Long et al. 2021). Our data suggest that the nucleoid is self-assembled by phase separation (Long et al. 2021), and the flexible linker domain between two HMG domains of TFAM contributes to the phase separation of TFAM in vitro. The interaction between TFAM and mtDNA provides another force as the multi-covalent effect in driving the droplet-like structure of nucleoid. The phase separation promotes the segregation of nucleoids from the mitochondrial matrix, but keeps the interaction between the two of them, which should be a critical feature for mitochondria (Fig. 1).
Figure 1.
Phase separation in chromatin assembly of the nuclei and the assembly of mitochondrial nucleoid. The schematic diagram represents the phase separation in chromatin condensation and nucleus architecture. The phase separation of TFAM (red) and DNA (gray) drives the nucleoid assembly in mitochondria
Phase separation regulates transcription in nuclei
Nuclear transcription should be carried out by multi-component transcription machinery, including hundreds of proteins (Guo et al. 2019; Steinbach et al. 2019). RNA Polymerase II (Pol II) plays a central role through its polymerase activity in mRNA generation, while the phase separation by the CTD domain also contributes to the recruitment of many other components for smooth reaction (Boehning et al. 2018; Guo et al. 2019; Lu et al. 2018). This CTD domain contains a conserved repeat of Y1S2P3T4S5P6S7, such as 52 repeats in humans, which served as an IDR for phase separation (Boehning et al. 2018). Other subunits necessary for transcription could be further recruited by the co-phase separation manner. The phosphorylation of CTD by CDK7 could switch the transcription from initiation to elongation by regulating the phase separation of Pol II (Boehning et al. 2018). The mediators were also reported to act in a similar manner of phase separation and recruit transcription activators to promote transcription or recruit repressors to inhibit transcription activity (Cho et al. 2018). The cis-elements, such as enhancers are also reported to be recruited by phase separation, recruiting activators and forming enhancer granules (Sabari et al. 2018). Moreover, transcribed RNAs could also regulate the feedback of transcription by phase separation, whereby RNAs initially stimulate condensates of transcription but then ultimately arrest the process by promoting the dissolution of these condensates (Henninger et al. 2020). The cleavage of pre-mRNA could also be observed as a droplet-like structure, which may be regulated by phase separation, too (Guo et al. 2019) (Fig. 2A).
Figure 2.
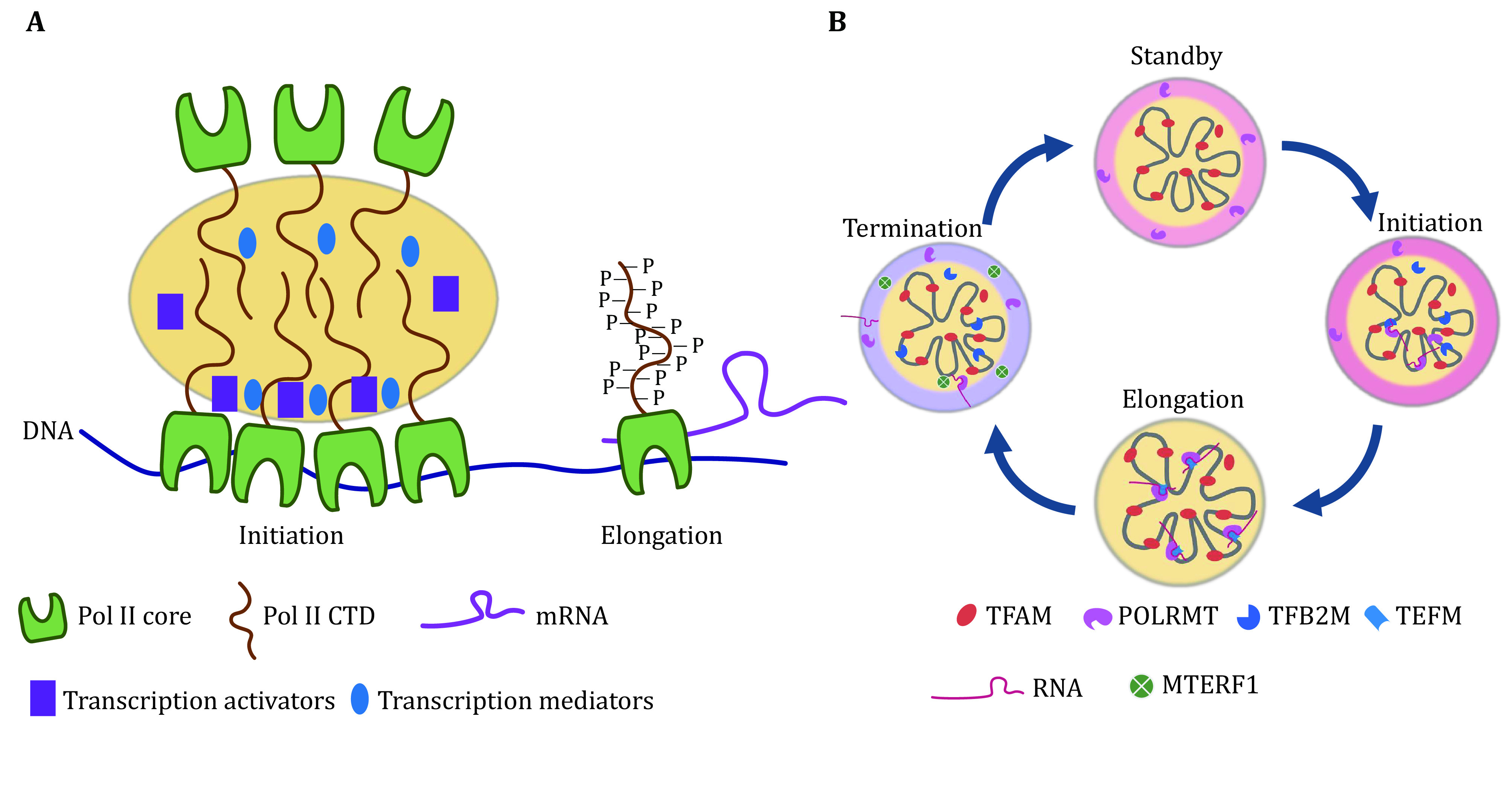
Phase separation drives transcription in nuclei and mitochondria. A The CTD tail (yellow) of Pol Ⅱ drives the phase separation, which recruits transcription activators (purple) and mediators (blue) in transcription initiation. The phosphorylation of CTD breaks the phase separation of Pol Ⅱ and promotes transcription elongation. B Phase separation in regulating transcription in mitochondria. The schematic diagram represents the phase separation in regulating mitochondrial transcription
Phase separation in the modulation of mitochondria transcription
Mitochondria have a series of special machinery in DNA replication and transcription, different from nuclear DNA replication and transcription (Hillen et al. 2018). Although mitochondria originated from an ancestral prokaryotic organism, they have developed a unique transcription network to satisfy complicated demands. Mitochondrial transcription is carried out by a mitochondrial RNA polymerase (mtRNAP or POLRMT) that is more similar to single-subunit RNA polymerases (RNAPs) in bacteriophages (Masters et al. 1987), but is not related to multi-subunit RNAPs, such as bacterial RNAP or eukaryotic RNA polymerase II (Pol II), both exhibiting phase behavior (Boehning et al. 2018; Ladouceur et al. 2020; Lu et al. 2018). The bacteriophage T7 RNA polymerase, the close relative of the mitochondrial RNA polymerase POLRMT, could work independently of other factors in transcription (Durniak et al. 2008). However, mitochondrial transcription is a multi-component transcription and requires the assistance of additional protein factors for each step of the transcription cycle (Gustafsson et al. 2016; Hillen et al. 2018).
For the transcription initiation, mitochondrial RNA polymerase POLRMT is not able to bind to the promoter by itself (Hillen et al. 2017). It relies on the help of TFAM and TFB2M in promoter melting. In the nucleus, the recruitment of multi-components relies on the phase separation of a CTD domain of Pol II with 26–52 repeats (Boehning et al. 2018; Lu et al. 2018). However, POLRMT lacks such a tail, which is also not found in T7 RNA polymerase in bacteria (Long et al. 2021). A similar pattern of phase separation could be found on the core complex of nucleoid as TFAM-DNA (Long et al. 2021). Mitochondria have developed a unique way of regulating nucleoid orchestration and transcription (Fig. 2B).
The phase separation of TFAM-DNA could recruit both TFB2M and POLRMT in a co-phase separation manner, and promote efficient transcription initiation. It is a special multi-phase separation, in which TFAM-DNA presents in the core structure and POLRMT coats the core nucleoid in a shield structure. Meanwhile, TFB2M shows more flexibility and could resident both in the core and shield. This kind of special multi-phase separation could concentrate POLRMT, but also prevent the touch of POLRMT to the other part of mtDNA, which may keep the transcription in standby or low activity status. What’s more, the co-phase separation of TFB2M could promote promoter melting by opening the double-strand DNA and promoting the touch of POLRMT to mtDNA promoters, resulting in breaking the multi-phase of POLRMT and promoting transcription. The phase separation also concentrates the substrate for transcription. NTPs could be recruited by the phase separation of TFAM-DNA, which keeps the transcription efficient.
The elongation factor TEFM plays critical a role in the transformation of transcription initiation to elongation, which could also be recruited by co-phase separation with TFAM-DNA. It further remodels the multi-phase separation of POLRMT and promotes the transcription machinery to go further alongside the mtDNA.
The transcription termination in mitochondrial is still not fully determined (Terzioglu et al. 2013; Yakubovskaya et al. 2010). The terminal factors as MTERF1 are reported to bind the leu-tRNA site and terminate the transcription from the HSP promoter (Yakubovskaya et al. 2010). The MTERF1 could also be recruited by TFAM-DNA in a multi-phase separation manner (Long et al. 2021). MTERF1 circling the core complex of the nucleoid is restricted in the outer layer without the touch of most of the mtDNA-TFAM core. This kind of phase separation may also restrict the activity of MTERF1 as non-engagement with the transcription complex. Moreover, MTERF1 could co-phase separation with POLRMT in the outer layer, which may further prevent the activity of free POLRMT in the outer layer.
Similar to the transcription center in nuclei, there are also some special RNA granules in the mitochondria. Mitochondrial RNA granules are clusters of RNA transcripts and ribosome assembling (Pearce et al. 2017). The granules known as MRGs, are enrichment with nascent RNAs and RNA-binding proteins (Jourdain et al. 2013). It is reported to be an RNA processing center, in which newly synthesized RNAs are cleaved and modified. Many other proteins are also identified in the RNA granules, including GRSF1, FASTKD protein family and so on (Jourdain et al. 2013), which may also be driven by phase separation. Moreover, the mitochondrial ribosome assembling process is also a mystery, which may also be happened in a center driven by multi-phase separation, similar to the nucleoids.
SUMMARY AND PERSPECTIVE
The compartmentation by phase separation provides new insight into the architecture and transcription modulation both in nuclei and the mitochondrial nucleoids. Phase separation drives chromatin condensation, nucleolus assembly and nucleus architecture construction. It also drives the mitochondria nucleoid assembly and transcription modulation.
However, there are still many questions that need to be solved. The question of how phase separation regulates chromatin dynamics in cell division and what has happened about the phase separation in control of heterochromatin building and rebuilding process in development is still uncovered. For phase separation in mitochondria, how does the transcription transform to translation in the phase separation model? Does mtDNA replication share a similar phase separation with transcription? There is still a lot of work to do as a new model and theory, and a lot of questions should be answered.
Conflict of interest
Qi Long, Yanshuang Zhou, Jingyi Guo, Hao Wu and Xingguo Liu declare that they have no conflict of interest.
Acknowledgements
This study is funded by the National Natural Science Foundation projects of China (92157202, 32025010, 32241002, 92254301, 32261160376, 31970709, 32070729, 32100619, 32170747), the National Key Research and Development Program of China (2022YFA1103800, 2019YFA0904500), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB0480000), the Key Research Program, CAS (ZDBS-ZRKJZ-TLC003), International Cooperation Program, CAS (154144KYSB20200006), CAS Project for Young Scientists in Basic Research (YSBR-075), Guangdong Province Science and Technology Program (2023A1515030231, 2022A1515110493, 2020B1212060052, 2021A1515012513, 2021B1515020096, 2022A1515012616, 2022A1515110951), Guangzhou Science and Technology Program (202102020827, 202102080066), Open research funds of the Sixth Affiliated Hospital of Guangzhou Medical University, Qingyuan People's Hospital (202301-203), Open Research Program of Key Laboratory of Regenerative Biology, CAS (KLRB202107, KLRB202203), and CAS Youth Innovation Promotion Association (to Y. W and K. C).
Compliance with Ethical Standards
Human and animal rights and informed consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- Alberti S, Gladfelter A, Mittag T Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell. 2019;176(3):419–434. doi: 10.1016/j.cell.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, Rosen MK Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18(5):285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, Rosen MK Compositional control of phase-separated cellular bodies. Cell. 2016;166(3):651–663. doi: 10.1016/j.cell.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Xu Y, Li T, Li X, Li W, Shao W, Wang K, Zhan G, Wu Z, Liu W, Lu JY, Wang L, Zhao J, Wu J, Na J, Li G, Li P, Shen X RNA targets ribogenesis factor WDR43 to chromatin for transcription and pluripotency control. Mol Cell. 2019;75(1):102–116. doi: 10.1016/j.molcel.2019.05.007. [DOI] [PubMed] [Google Scholar]
- Boehning M, Dugast-Darzacq C, Rankovic M, Hansen AS, Yu T, Marie-Nelly H, McSwiggen DT, Kokic G, Dailey GM, Cramer P, Darzacq X, Zweckstetter M RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat Struct Mol Biol. 2018;25(9):833–840. doi: 10.1038/s41594-018-0112-y. [DOI] [PubMed] [Google Scholar]
- Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, Schymkowitz J, Shorter J, Wolozin B, Van Den Bosch L, Tompa P, Fuxreiter M Protein phase separation: a new phase in cell biology. Trends Cell Biol. 2018;28(6):420–435. doi: 10.1016/j.tcb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Mitchison TJ, Hyman AA Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci USA. 2011;108(11):4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Xie W, Pickering BF, Chu KL, Savino AM, Yang X, Luo H, Nguyen DT, Mo S, Barin E, Velleca A, Rohwetter TM, Patel DJ, Jaffrey SR, Kharas MG N(6)-Methyladenosine on mRNA facilitates a phase-separated nuclear body that suppresses myeloid leukemic differentiation. Cancer Cell. 2021;39(7):958–972. doi: 10.1016/j.ccell.2021.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho WK, Spille JH, Hecht M, Lee C, Li C, Grube V, Cisse, II Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science. 2018;361(6400):412–415. doi: 10.1126/science.aar4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogliati S, Frezza C, Soriano ME, Varanita T, Quintana-Cabrera R, Corrado M, Cipolat S, Costa V, Casarin A, Gomes LC, Perales-Clemente E, Salviati L, Fernandez-Silva P, Enriquez JA, Scorrano L Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell. 2013;155(1):160–171. doi: 10.1016/j.cell.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durniak KJ, Bailey S, Steitz TA The structure of a transcribing T7 RNA polymerase in transition from initiation to elongation. Science. 2008;322(5901):553–557. doi: 10.1126/science.1163433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farge G, Mehmedovic M, Baclayon M, van den Wildenberg SM, Roos WH, Gustafsson CM, Wuite GJ, Falkenberg M In vitro-reconstituted nucleoids can block mitochondrial DNA replication and transcription. Cell Rep. 2014;8(1):66–74. doi: 10.1016/j.celrep.2014.05.046. [DOI] [PubMed] [Google Scholar]
- Feric M, Demarest TG, Tian J, Croteau DL, Bohr VA, Misteli T Self-assembly of multi-component mitochondrial nucleoids via phase separation. EMBO J. 2021;40(6):e107165. doi: 10.15252/embj.2020107165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016;165(7):1686–1697. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey TG, Mannella CA The internal structure of mitochondria. Trends Biochem Sci. 2000;25(7):319–324. doi: 10.1016/S0968-0004(00)01609-1. [DOI] [PubMed] [Google Scholar]
- Guo YE, Manteiga JC, Henninger JE, Sabari BR, Dall'Agnese A, Hannett NM, Spille JH, Afeyan LK, Zamudio AV, Shrinivas K, Abraham BJ, Boija A, Decker TM, Rimel JK, Fant CB, Lee TI, Cisse, II, Sharp PA, Taatjes DJ, Young RA Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature. 2019;572(7770):543–548. doi: 10.1038/s41586-019-1464-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson CM, Falkenberg M, Larsson NG Maintenance and expression of mammalian mitochondrial DNA. Annu Rev Biochem. 2016;85:133–160. doi: 10.1146/annurev-biochem-060815-014402. [DOI] [PubMed] [Google Scholar]
- Hansen AS, Hsieh TS, Cattoglio C, Pustova I, Saldana-Meyer R, Reinberg D, Darzacq X, Tjian R Distinct classes of chromatin loops revealed by deletion of an RNA-binding region in CTCF. Mol Cell. 2019;76(3):395–411. doi: 10.1016/j.molcel.2019.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henninger JE, Oksuz O, Shrinivas K, Sagi I, LeRoy G, Zheng MM, Andrews JO, Zamudio AV, Lazaris C, Hannett NM, Lee TI, Sharp PA, Cisse, II, Chakraborty AK, Young RA RNA-mediated feedback control of transcriptional condensates. Cell. 2020;184(1):207–225. doi: 10.1016/j.cell.2020.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen HS, Morozov YI, Sarfallah A, Temiakov D, Cramer P Structural basis of mitochondrial transcription initiation. Cell. 2017;171(5):1072–1081. doi: 10.1016/j.cell.2017.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen HS, Temiakov D, Cramer P Structural basis of mitochondrial transcription. Nat Struct Mol Biol. 2018;25(9):754–765. doi: 10.1038/s41594-018-0122-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdain AA, Koppen M, Wydro M, Rodley CD, Lightowlers RN, Chrzanowska-Lightowlers ZM, Martinou JC GRSF1 regulates RNA processing in mitochondrial RNA granules. Cell Metab. 2013;17(3):399–410. doi: 10.1016/j.cmet.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein IA, Boija A, Afeyan LK, Hawken SW, Fan M, Dall'Agnese A, Oksuz O, Henninger JE, Shrinivas K, Sabari BR, Sagi I, Clark VE, Platt JM, Kar M, McCall PM, Zamudio AV, Manteiga JC, Coffey EL, Li CH, Hannett NM, Guo YE, Decker TM, Lee TI, Zhang T, Weng JK, Taatjes DJ, Chakraborty A, Sharp PA, Chang YT, Hyman AA, Gray NS, Young RA Partitioning of cancer therapeutics in nuclear condensates. Science. 2020;368(6497):1386–1392. doi: 10.1126/science.aaz4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukat C, Davies KM, Wurm CA, Spåhr H, Bonekamp NA, Kühl I, Joos F, Polosa PL, Park CB, Posse V, Falkenberg M, Jakobs S, Kühlbrandt W, Larsson NG Cross-strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid. Proc Natl Acad Sci USA. 2015;112(36):11288–11293. doi: 10.1073/pnas.1512131112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladouceur AM, Parmar BS, Biedzinski S, Wall J, Tope SG, Cohn D, Kim A, Soubry N, Reyes-Lamothe R, Weber SC Clusters of bacterial RNA polymerase are biomolecular condensates that assemble through liquid-liquid phase separation. Proc Natl Acad Sci USA. 2020;117(31):18540–18549. doi: 10.1073/pnas.2005019117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DLJ, Riback JA, Bascetin R, Brangwynne CP The nucleolus as a multiphase liquid condensate. Nat Rev Mol Cell Biol. 2021;22(3):165–182. doi: 10.1038/s41580-020-0272-6. [DOI] [PubMed] [Google Scholar]
- Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, Narlikar GJ Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature. 2017;547(7662):236–240. doi: 10.1038/nature22822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson AG, Narlikar GJ The role of phase separation in heterochromatin formation, function, and regulation. Biochemistry. 2018;57(17):2540–2548. doi: 10.1021/acs.biochem.8b00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Wang R, Xiong F, Krakowiak J, Liao Z, Nguyen PT, Moroz-Omori EV, Shao J, Zhu X, Bolt MJ, Wu H, Singh PK, Bi M, Shi CJ, Jamal N, Li G, Mistry R, Jung SY, Tsai KL, Ferreon JC, Stossi F, Caflisch A, Liu Z, Mancini MA, Li W Enhancer RNA m6A methylation facilitates transcriptional condensate formation and gene activation. Mol Cell. 2021;81(16):3368–3385. doi: 10.1016/j.molcel.2021.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q, Zhou Y, Wu H, Du S, Hu M, Qi J, Li W, Guo J, Wu Y, Yang L, Xiang G, Wang L, Ye S, Wen J, Mao H, Wang J, Zhao H, Chan WY, Liu J, Chen Y, Li P, Liu X Phase separation drives the self-assembly of mitochondrial nucleoids for transcriptional modulation. Nat Struct Mol Biol. 2021;28(11):900–908. doi: 10.1038/s41594-021-00671-w. [DOI] [PubMed] [Google Scholar]
- Lu H, Yu D, Hansen AS, Ganguly S, Liu R, Heckert A, Darzacq X, Zhou Q Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature. 2018;558(7709):318–323. doi: 10.1038/s41586-018-0174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters BS, Stohl LL, Clayton DA Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell. 1987;51(1):89–99. doi: 10.1016/0092-8674(87)90013-4. [DOI] [PubMed] [Google Scholar]
- Misteli T The self-organizing genome: principles of genome architecture and function. Cell. 2020;183(1):28–45. doi: 10.1016/j.cell.2020.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo HB, Lovely GA, Phillips R, Chan DC Distinct structural features of TFAM drive mitochondrial DNA packaging versus transcriptional activation. Nat Commun. 2014;5:3077. doi: 10.1038/ncomms4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce SF, Rebelo-Guiomar P, D'Souza AR, Powell CA, Van Haute L, Minczuk M Regulation of mammalian mitochondrial gene expression: recent advances. Trends Biochem Sci. 2017;42(8):625–639. doi: 10.1016/j.tibs.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riback JA, Zhu L, Ferrolino MC, Tolbert M, Mitrea DM, Sanders DW, Wei M-T, Kriwacki RW, Brangwynne CP Composition-dependent thermodynamics of intracellular phase separation. Nature. 2020;581(7807):209–214. doi: 10.1038/s41586-020-2256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger AJ, Munoz-Gomez SA, Kamikawa R The origin and diversification of mitochondria. Curr Biol. 2017;27(21):R1177–R1192. doi: 10.1016/j.cub.2017.09.015. [DOI] [PubMed] [Google Scholar]
- Sabari BR, Dall'Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, Li CH, Guo YE, Day DS, Schuijers J, Vasile E, Malik S, Hnisz D, Lee TI, Cisse, II, Roeder RG, Sharp PA, Chakraborty AK, Young RA Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018;361(6400):eaar3958. doi: 10.1126/science.aar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari BR, Dall'Agnese A, Young RA Biomolecular condensates in the nucleus. Trends Biochem Sci. 2020;45(11):961–977. doi: 10.1016/j.tibs.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen EL, Bogenhagen DF Developmentally-regulated packaging of mitochondrial DNA by the HMG-box protein mtTFA during Xenopus oogenesis. Nucleic Acids Res. 2001;29(13):2822–2828. doi: 10.1093/nar/29.13.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, Brangwynne CP Liquid phase condensation in cell physiology and disease. Science. 2017;357(6357):eaaf4382. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- Song F, Chen P, Sun D, Wang M, Dong L, Liang D, Xu RM, Zhu P, Li G Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science. 2014;344(6182):376–380. doi: 10.1126/science.1251413. [DOI] [PubMed] [Google Scholar]
- Steinbach N, Hasson D, Mathur D, Stratikopoulos EE, Sachidanandam R, Bernstein E, Parsons RE PTEN interacts with the transcription machinery on chromatin and regulates RNA polymerase II-mediated transcription. Nucleic Acids Res. 2019;47(11):5573–5586. doi: 10.1093/nar/gkz272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickfaden H, Tolsma TO, Sharma A, Underhill DA, Hansen JC, Hendzel MJ Condensed chromatin behaves like a solid on the mesoscale in vitro and in living cells. Cell. 2020;183(7):1772–1784. doi: 10.1016/j.cell.2020.11.027. [DOI] [PubMed] [Google Scholar]
- Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH Phase separation drives heterochromatin domain formation. Nature. 2017;547(7662):241–245. doi: 10.1038/nature22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamatsu C, Umeda S, Ohsato T, Ohno T, Abe Y, Fukuoh A, Shinagawa H, Hamasaki N, Kang D Regulation of mitochondrial D-loops by transcription factor A and single-stranded DNA-binding protein. EMBO Rep. 2002;3(5):451–456. doi: 10.1093/embo-reports/kvf099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzioglu M, Ruzzenente B, Harmel J, Mourier A, Jemt E, Lopez MD, Kukat C, Stewart JB, Wibom R, Meharg C, Habermann B, Falkenberg M, Gustafsson CM, Park CB, Larsson NG MTERF1 binds mtDNA to prevent transcriptional interference at the light-strand promoter but is dispensable for rRNA gene transcription regulation. Cell Metab. 2013;17(4):618–626. doi: 10.1016/j.cmet.2013.03.006. [DOI] [PubMed] [Google Scholar]
- Wang J, Choi JM, Holehouse AS, Lee HO, Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D, Poser I, Pappu RV, Alberti S, Hyman AA A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell. 2018;174(3):688–699. doi: 10.1016/j.cell.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Hu M, Zuo MQ, Zhao J, Wu D, Huang L, Wen Y, Li Y, Chen P, Bao X, Dong MQ, Li G, Li P Rett syndrome-causing mutations compromise MeCP2-mediated liquid-liquid phase separation of chromatin. Cell Res. 2020;30(5):393–407. doi: 10.1038/s41422-020-0288-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakubovskaya E, Mejia E, Byrnes J, Hambardjieva E, Garcia-Diaz M Helix unwinding and base flipping enable human MTERF1 to terminate mitochondrial transcription. Cell. 2010;141(6):982–993. doi: 10.1016/j.cell.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]