ABSTRACT
Biofilm-producing Pseudomonas aeruginosa infections pose a severe threat to public health and are responsible for high morbidity and mortality. Phage-antibiotic combinations (PACs) are a promising strategy for combatting multidrug-resistant (MDR), extensively drug-resistant (XDR), and difficult-to-treat P. aeruginosa infections. Ten MDR/XDR P. aeruginosa strains and five P. aeruginosa-specific phages were genetically characterized and evaluated based upon their antibiotic susceptibilities and phage sensitivities. Two selected strains, AR351 (XDR) and I0003-1 (MDR), were treated singly and in combination with either a broad-spectrum or narrow-spectrum phage, phage EM-T3762627-2_AH (EM), or 14207, respectively, and bactericidal antibiotics of five classes in biofilm time-kill analyses. Synergy and/or bactericidal activity was demonstrated with all PACs against one or both drug-resistant P. aeruginosa strains (average reduction: −Δ3.32 log10 CFU/cm2). Slightly improved ciprofloxacin susceptibility was observed in both strains after exposure to phages (EM and 14207) in combination with ciprofloxacin and colistin. Based on phage cocktail optimization with four phages (EM, 14207, E20050-C (EC), and 109), we identified several effective phage-antibiotic cocktails for further analysis in a 4-day pharmacokinetic/pharmacodynamic in vitro biofilm model. Three-phage cocktail, EM + EC + 109, in combination with ciprofloxacin demonstrated the greatest biofilm reduction against AR351 (−Δ4.70 log10 CFU/cm2 from baseline). Of remarkable interest, the addition of phage 109 prevented phage resistance development to EM and EC in the biofilm model. PACs can demonstrate synergy and offer enhanced eradication of biofilm against drug-resistant P. aeruginosa while preventing the emergence of resistance.
KEYWORDS: bacteriophage, Pseudomonas aeruginosa, antimicrobial resistance, synergy, MDR, phage, cocktail, biofilms, antibiotics, antimicrobial combinations
INTRODUCTION
Pseudomonas aeruginosa represents one of the most challenging of nosocomial pathogens to face in clinical practice due to its overwhelming capacity to acquire resistance mechanisms and cause severe infection (1). Another important virulence factor is the biofilm structure, which plays a major role in antibiotic resistance by slowing antibiotic penetration and microorganism protection from the host immune system (2). Novel therapeutic alternatives such as bacteriophages are a potential solution to recalcitrant infections associated with increased antibiotic resistance (3). A recent case series described the use of phage therapy in combination with antibiotics to treat 10 patients with persistent, drug-resistant infections (4). Of those, five patients underwent phage therapy for P. aeruginosa infections, where three of the five patients were considered successes by the treatment team, while two of the five patients experienced treatment failure attributed to ventricular assist device infections, which were chronic (>1 year), biofilm-based infections.
While the use of bacteriophages may be helpful in these refractory multidrug-resistant (MDR) infections, it should be noted that bacterial susceptibility to phages is often limited to planktonic analyses, and previous studies have shown that planktonic isolates susceptible to a specific phage may not be susceptible in biofilm form (5). Therefore, it may be valuable to assess the impact of phage and phage-antibiotic combinations (PACs) in biofilm conditions to identify potential, effective treatment regimens for these difficult-to-treat infections. In addition, many biofilm studies evaluating phage cocktails and PACs are limited to static experiments conducted in microtiter plates, which may not provide a realistic environment for biofilm development and make it difficult to translate their results to clinical contexts (6 – 8). Our study utilizes biofilm time-kill analyses (TKAs) and an in vitro, dynamic pharmacokinetic/pharmacodynamic biofilm model [Centers for Disease Control and Prevention (CDC) biofilm reactor (CBR)] with humanized antibiotic exposures to assess the impact of PAC on MDR and extensively drug-resistant (XDR) P. aeruginosa isolates and to develop an effective phage-antibiotic cocktail.
RESULTS
Antimicrobial susceptibility and phage sensitivity testing
Of the 10 clinical strains, 9 had decreased or complete loss of OprD expression likely contributing to the carbapenem resistance observed; only strain I0003-1 displayed unchanged OprD expression (Table 1). Every strain with ciprofloxacin (CIP) resistance had mutations in the quinolone resistance-determining regions of gyrA, gyrB, parC, or parE, which likely contributed to the resistance seen in 8 of 10 strains. Furthermore, dysregulation of MexAB-OprM and MexCD-OprM expressions via repressor mutations, or presence of Class A, B, or D β-lactamases was present in the majority of the strains (9). Antimicrobial susceptibility testing was performed for all strains with CIP, amikacin (AMK), meropenem (MEM), colistin (COL), and aztreonam (AZT) as shown in Table 1. Two strains with varying antibiotic resistance profiles AR351 (XDR) and I0003-1 (MDR) were selected for further evaluation in biofilm TKAs with bactericidal antibiotics of five classes. Therefore, the antimicrobial susceptibilities of AR351 and I0003-1 were also determined in the presence of biofilm (Table 2). The biofilm MICs (MBICs) were generally higher than the planktonic MICs.
TABLE 1.
Strain characterization a
| Antibiotic resistance genetic traits | Phage multiplication d | Antibiotic MIC (µg/mL) e | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. aeruginosa strain | ST | OprD expected phenotype | Genes for aminoglycoside-modifying enzymes c | β-Lactamase gene(s) | MexAB-OprM and MexCD-OprM repressor mutations | QRDR mutations | Fosfomycin resistance b | EM | LL | 14207 | EC | 109 | MEM | COL | AMK | AZT | CIP |
| AR351 | 4020 | OprD decrease b | aph(3′)-IIb-like |
blaPDC-39
blaOXA-85 blaOXA-488 blaOXA-485 |
nalC | gyrA | fosA | High | High | None | High | Med | 32 | 1 | 64 | 64 | 1 |
| HFHS22 | 966 | OprD loss | aph(3′)-IIb-like, aac(3)-IIIa |
blaOXA-395
blaPDC-5 |
mexR nalC | gyrA parC | fosA | Med | Med | None | None | Low | 16 | 2 | 4 | 8 | 4 |
| K0001-1 | 2191 | OprD loss |
aph(3′)-Iib, ant(2″)-Ia, aac(6′)-Ib4 |
blaIMP-48
blaOXA-10 blaOXA-1135 blaPDC-5 |
mexR nalD | gyrA parC parE | fosA-like | Low | Low | None | Low | None | >64 | 2 | 4 | 64 | >8 |
| K0003-1 | 3390 | OprD loss |
aph(3′)-Iib, ant(2″)-Ia, aac(6′)-Ib |
blaOXA-10
blaOXA-1135 blaIMP-48 blaPDC-5 |
nalC nalD | gyrA parC parE | fosA-like | Low | None | None | None | None | >64 | 4 | >64 | 64 | >8 |
| AR352 | 111 | OprD loss | aph(3′)-IIb-like |
blaPDC-3
blaVIM-2 blaOXA-395 |
mexR | gyrA parC | fosA | Low | None | None | None | None | >64 | 1 | 4 | >256 | 1 |
| I0003-1 | 919 | No change | aph(3′)-IIb-like |
blaOXA-50
blaPDC-374 |
mexR nalC | None | fosA-like | High | High | Med | Med | High | 16 | 1 | 16 | 64 | 0.5 |
| G0002-2 | 389 | OprD loss | aph(3′)-IIb-like |
blaPDC-374
blaOXA-50 |
mexR nalC | gyrA | fosA-like | Med | High | None | Med | High | 32 | 2 | 1 | 64 | 2 |
| AR353 | 308 | OprD loss |
aph(3′)-IIb-like, aadA6, aadA1, ant(2″)-la, aac(6′)-ll, aac(6′)-lb4 |
bla
GES-1
bla OXA-2 bla OXA-488 blaPDC-19a |
mexR nalC | gyrA gyrB parC parE | fosA | Low | Low | Med | High | Low | 16 | 1 | >64 | 16 | 4 |
| AR357 | 235 | OprD loss |
aph(3′)-IIb-like,
aadA6, aadA2, aadA1, ant(2″)-la, aacA16 |
bla
VEB-1
bla OXA-10 bla OXA-488 blaPDC-35 |
mexR nalC | gyrA parC parE | fosA-like | None | None | None | Med | Low | 8 | 1 | >64 | 128 | 4 |
| AR356 | b | OprD loss | aac(6′)-Ib4, aadA16-like |
bla
KPC-2
blaPDC-42 |
mexR nalC nalD | gyrA gyrB parC parE | None detected | None | None | None | Low | None | 32 | 1 | 2 | >256 | .0625 |
Multilocus STs, resistance characteristics, and susceptibilities of MDR P. aeruginosa strains, including their sensitivity to each Pseudomonas phage. ST, sequence type; Med, medium; QRDR, quinolone resistance-determining regions; EM, phage EM-T3762627-2_AH; LL, phage LL-5504721-AH; 14207, 14207 phage; EC, phage E2005-C; 109, phage 109; MEM, meropenem; COL, colistin; AMK, amikacin; AZT, aztreonam; CIP, ciprofloxacin.Highlighted strains, AR351 and RI0003-1, were classified as XDR and MDR, respectively, according to CLSI breakpoints and were chosen for synergy analysis (10, 11).
Unassigned sequence type.
Previously identified resistance mechanisms (12).
High, medium, and low phage sensitivities were defined as phage counts of >107, between 103 and 107, and <103 PFU/mL, respectively.
Initial antimicrobial planktonic MICs prior to 24-h time-kill analyses.
TABLE 2.
MBIC and MIC values for AR351 and I0003-1 a
| Antimicrobial agent | Strain AR351 | Strain I0003-1 | ||
|---|---|---|---|---|
| MIC (μg/mL) | MBIC (μg/mL) | MIC (μg/mL) | MBIC (μg/mL) | |
| MEM | 32 | >64 | 16 | 32 |
| COL | 1 | 16 | 1 | 32 |
| AMK | 64 | >64 | 16 | 32 |
| CIP | 1 | 2 | 0.5 | 1 |
| AZT | 64 | >64 | 64 | >64 |
Comparison of planktonic and biofilm MICs for selected strains: AR351 and I0003-1.
Phage sensitivity testing was performed for all 10 strains with five phages, which facilitated identification of phages with narrow- and broad-spectrum activity (Table 1). Phage 14207 demonstrated the narrowest spectrum of activity (active against 2 of 10 strains), and phage EM-T3762627-2_AH (EM) demonstrated the broadest spectrum of activity (active against 8 of 10 strains). Both phages (14207 and EM) were selected for further evaluation in biofilm TKAs with bactericidal antibiotics based on their varying spectra of activity (Fig. 1 and 2). Subsequent DNA sequencing showed that lytic phages EM and phage LL-5504721-AH (LL) are related to the Pakpunavirus genus and are nearly identical to each other, as previously reported (GenBank ON169972) (13). Lytic phages EC and 109 were determined to be related to the Pbunavirus genus. Phage 14207 has not been sequenced. Phages 14207, EC, EM, and 109 were selected for further analysis in biofilm TKAs to identify the most effective phage-antibiotic cocktails. Phage LL was not included in the analysis due to nearly identical genetic similarity to phage EM.
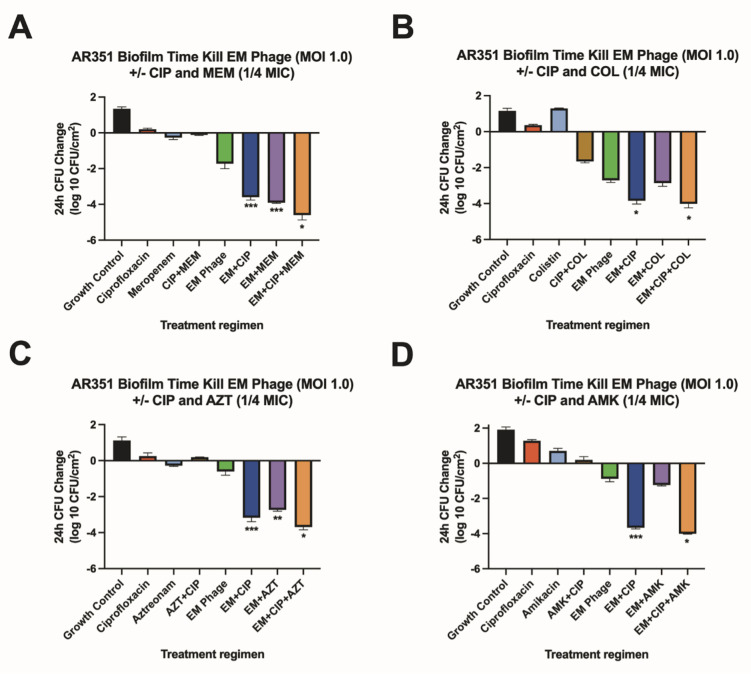
FIG 1.
Biofilm time-kill analyses for AR351. Biofilm time-kill analyses of CIP-COL, CIP-AZT, CIP-AMK, and CIP-MEM alone and in combination with EM-T3762626-2_AH (EM) phage against AR351 at 0.25× MIC (COL 0.25 µg/mL, AZT 16 µg/mL, AMK 16 µg/mL, CIP 0.25 µg/mL, and MEM 8 µg/mL) and a theoretical MOI of 1.0. Subinhibitory antibiotic concentrations are used to mimic the expected drug ineffectiveness that would otherwise not occur in this simplified in vitro system. The bars represent the difference in 24-h CFU/cm2 from baseline. (A) Strain AR351 versus ciprofloxacin (1/4 MIC) and/or meropenem (1/4 MIC) with and without phage EM (MOI 1.0). (B) Strain AR351 versus ciprofloxacin (1/4 MIC) and/or colistin (1/4 MIC) with and without phage EM (MOI 1.0). (C) Strain AR351 versus ciprofloxacin (1/4 MIC) and/ or aztreonam (1/4 MIC) with and without phage EM (MOI 1.0). (D) Strain RAR351 versus ciprofloxacin (1/4 MIC) and/or amikacin (1/4 MIC) with and without phage EM (MOI 1.0). Values are means ± standard deviations from two biological replicates. *Regimen(s) that demonstrated bactericidal activity. **Regimen(s) that demonstrated synergy. ***Regimen(s) that demonstrated both synergy and bactericidal activity. AMK, amikacin; AZT, aztreonam; CIP, ciprofloxacin; COL, colistin; EM, phage EM-T3762627-2_AH; MEM, meropenem; MIC, minimum inhibitory concentration; MOI, multiplicity of infection.
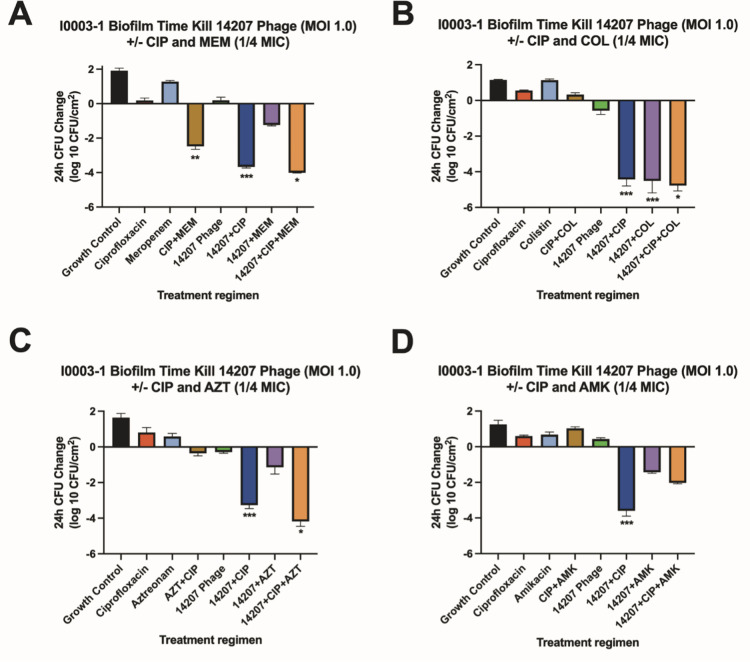
FIG 2.
Biofilm time-kill analyses for I0003-1. Biofilm time-kill analyses of CIP-COL, CIP-AZT, CIP-AMK, and CIP-MEM alone and in combination with 14207 phage against RI0003-1 at 0.25× MIC (COL 0.25 µg/mL, AMK 4 µg/mL, AZT 16 µg/mL, CIP 0.125 µg/mL, and MEM 4 µg/mL) and a theoretical MOI of 1.0. Subinhibitory antibiotic concentrations are used to mimic the expected drug ineffectiveness that would otherwise not occur in this simplified in vitro system. The bars represent the difference in 24-h CFU/cm2 from baseline. (A) Strain RI0003-1 versus ciprofloxacin (1/4 MIC) and/or meropenem (1/4 MIC) with and without phage 14207 (MOI 1.0). (B) Strain RI0003-1 versus ciprofloxacin (1/4 MIC) and/or colistin (1/4 MIC) with and without phage 14207 (MOI 1.0). (C) Strain RI0003-1 versus ciprofloxacin (1/4 MIC) and/ or aztreonam (1/4 MIC) with and without phage 14207 (MOI 1.0). (D) Strain RI0003-1 versus ciprofloxacin (1/4 MIC) and/or amikacin (1/4 MIC) with and without phage 14207 (MOI 1.0). Values are means ± standard deviations from two biological replicates. *Regimen(s) that demonstrated bactericidal activity. **Regimen(s) that demonstrated synergy. ***Regimen(s) that demonstrated both synergy and bactericidal activity. 14207, 14207 phage; AMK, amikacin; AZT, aztreonam; CIP, ciprofloxacin; COL, colistin; MEM, meropenem; MIC, minimum inhibitory concentration; MOI, multiplicity of infection.
Biofilm time-kill analyses
To investigate the impact of phage in addition to five different antimicrobial classes on biofilm eradication, we investigated two drug-resistant P. aeruginosa strains (AR351 and I0003-1) with varying antimicrobial susceptibility profiles and two phages (EM and 14207) with broad and narrow spectra of activity.
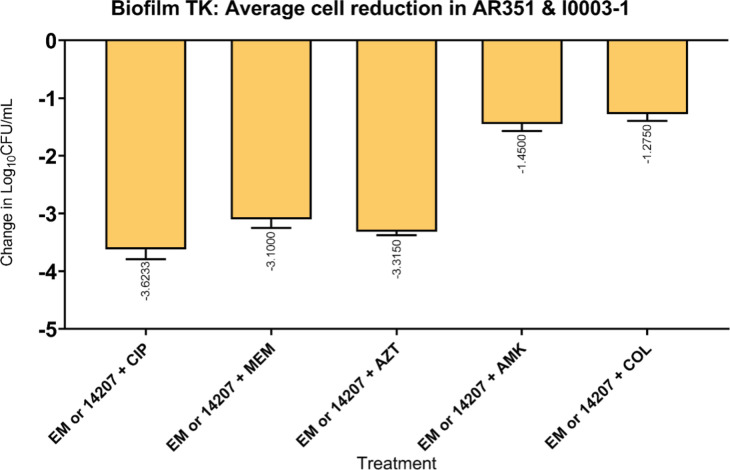
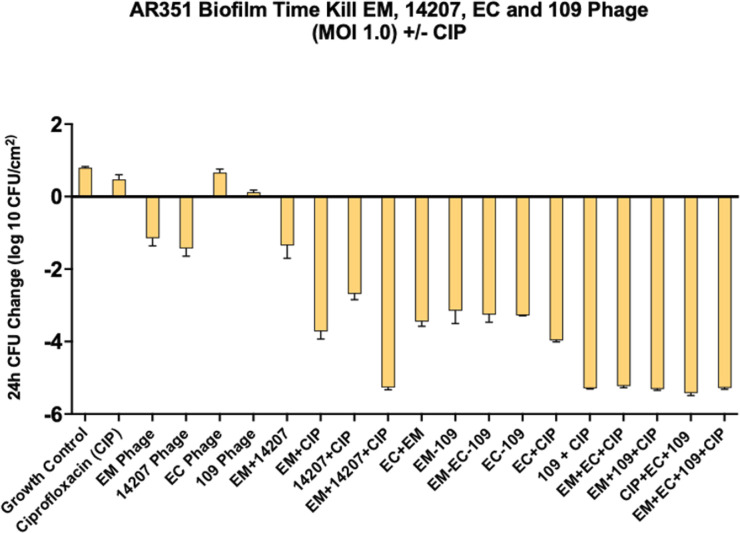
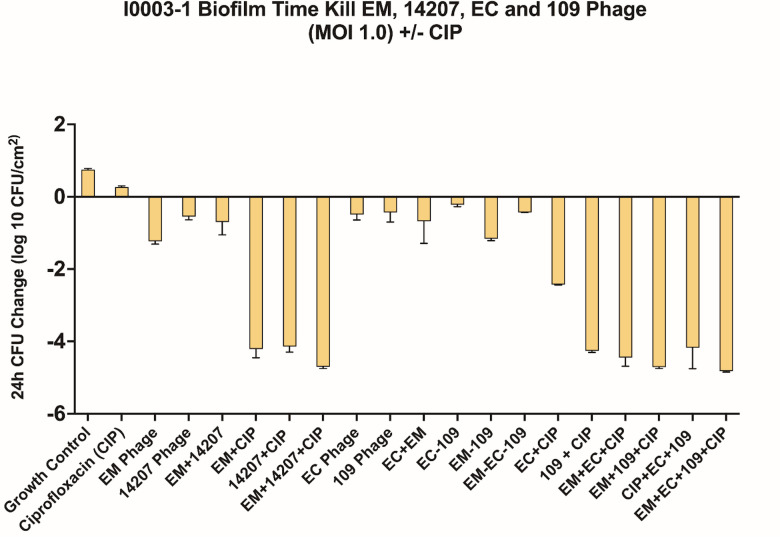
We used subinhibitory concentrations of antibiotics in order to reflect the treatment failure scenarios that we are attempting to model. As expected, antimicrobial-only regimens at subinhibitory concentrations had little to no effect on biofilm cell reduction. Similarly, phage-only regimens were bacteriostatic (average reduction: −Δ0.65 log10 CFU/cm2 from starting inoculum baseline), whereas PACs demonstrated synergy and/or bactericidal activity against both strains (average reduction: −Δ3.32 log10 CFU/cm2 from starting inoculum baseline). Against AR351, the most effective regimen was phage EM in combination with CIP and MEM (−Δ4.60 log10 CFU/cm2 from starting inoculum baseline), shown in Fig. 1. Against I0003-1, the most effective regimen was phage 14207 in combination with CIP and COL (−Δ4.77 log10 CFU/cm2 from starting inoculum baseline) (Fig. 2). Overall, PAC regimens demonstrated greater average biofilm cell reduction against strain AR351 (−Δ3.44 log10 CFU/cm2) compared to strain I0003-1 (−Δ3.19 log10 CFU/cm2). The most effective regimens against both strains with the largest biofilm cell reduction (−Δ4.60 to 4.77 log10 CFU/cm2) included CIP. In addition, among all phage plus single-antibiotic combination regimens, phage plus CIP was most effective at reducing biofilm cell count against both strains (average reduction: −Δ 4.14 log10 CFU/cm2) (Fig. 3); therefore, CIP was selected for further analysis in phage-antibiotic cocktail evaluations (Fig. 4 and 5) and biofilm models (Fig. 6). Strains AR351 and I0003-1 were evaluated in additional biofilm time-kills against CIP in combination with all phages (EM, 14207, EC, and 109) to identify the optimal phage-antibiotic cocktails for evaluation in biofilm models (Fig. 4 and 5). Phage LL was not considered for analysis in biofilm time-kills due to having nearly identical genetic similarity to phage EM, as reported previously (13). The phage-antibiotic cocktails that displayed the greatest average biofilm reduction against both strains were EM + EC + 109 + CIP (−Δ5.05 log10 CFU/cm2), EM + 109 + CIP (−Δ5.01 log10 CFU/cm2), and EM + 14207 + CIP (−Δ4.99 log10 CFU/cm2). Two phage-antibiotic cocktails, EM + EC + 109 + CIP and EM + 14207 + CIP, were selected for evaluation in biofilm models based on their performance in biofilm TKAs (Fig. 4 and 5). Phage-antibiotic cocktail EM + 109 + CIP was not evaluated due to its greater suspected potential for phage resistance compared to EM + EC + 109 + CIP. Based on its strong resistance profile, XDR strain AR351 was selected for biofilm model assessment as it represents a pathogen that might necessitate salvage therapy using PACs.
FIG 3.
Comparison of five antimicrobial classes plus phage (EM or 14207) against I0003-1 and AR351. Average biofilm cell reduction seen with single phage plus antibiotic combinations against AR351 and I0003-1 in biofilm time-kill analyses of EM or 14207, respectively, with CIP, MEM, AZT, AMK, and COL at 0.25× MIC and a theoretical MOI of 1.0. Subinhibitory antibiotic concentrations are used to mimic the expected drug ineffectiveness that would otherwise not occur in this simplified in vitro system. The bars represent the difference in 24-h CFU/cm2 from baseline. Values are means ± standard deviations from two biological replicates. 14207, 14207 phage; AMK, amikacin; AZT, aztreonam; CIP, ciprofloxacin; COL, colistin; EM, EM phage; MEM, meropenem; MIC, minimum inhibitory concentration; MOI, multiplicity of infection; TK, time-kill.
FIG 4.
Phage cocktail optimization against AR351. 109, 109 phage; 14207, 14207 phage; CIP, ciprofloxacin; EC, EC phage; EM, phage EM-T3762627-2_AH; MOI, multiplicity of infection.
FIG 5.
Phage cocktail optimization against I0003-1. 109, 109 phage; 14207, 14207 phage; CIP, ciprofloxacin; EC, EC phage; EM, phage EM-T3762627-2_AH; MOI, multiplicity of infection.
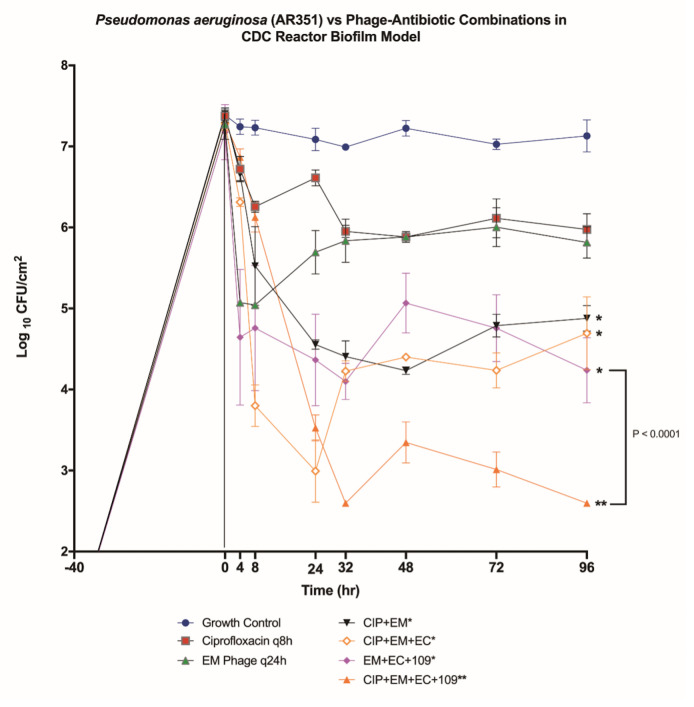
FIG 6.
Biofilm model for AR351 versus phage cocktail ± ciprofloxacin. In vitro PK/PD biofilm model results for all phage and CIP combinations against P. aeruginosa strain AR351. The error bars indicate standard deviation between repeats. *Regimens that met their definition for improvement. **Regimens that met their definition for enhancement.
Biofilm models
To determine the impact of humanized CIP pharmacokinetic exposures and phage on biofilm-embedded XDR P. aeruginosa strain AR351, we conducted a series of experiments with doses of CIP alone or in combination with phages in the pharmacokinetic/pharmacodynamic (PK/PD) biofilm model over 4 days. The quantitative changes in log10 CFU/cm2 for the evaluated regimens against P. aeruginosa strain AR351 are shown in Fig. 6. The most effective regimen was three-phage cocktail EM + EC + 109 once daily plus CIP 400 mg every 8 h (q8h), which together demonstrated bactericidal activity and enhancement at 96 h against AR351 [−Δ4.70 log10 CFU/cm2 from starting inoculum baseline, respectively (P < 0.0001)]. The phage cocktail, EM + EC + 109, without the addition of CIP demonstrated improvement compared to phage EM alone but did not meet the bactericidal definition [−Δ2.94 log10 CFU/cm2 from baseline (P < 0.0001)]. Phage-antibiotic combinations, EM + CIP and EM + EC + CIP, both demonstrated improved killing against AR351 compared to EM alone but did not reach bactericidal reduction after 96 h [−Δ2.52 and −Δ2.59 log10 CFU/cm2 from baseline, respectively (both P < 0.0001)]. Although demonstrating promise during phage-antibiotic cocktail evaluations (Fig. 4 and 5), phage-antibiotic cocktail regimen, EM + 14207 + CIP, did not reach bactericidal reduction after 96 h in the biofilm model (−Δ2.39 log10 CFU/cm2 from baseline, data not shown). Monotherapy regimens, CIP alone and EM alone, had lesser impact on biofilm cell reduction after 96 h (−Δ1.40 and −Δ1.45 log10 CFU/cm2 from baseline, respectively) (Fig. 6).
Pharmacokinetics
To ensure that the targeted CIP concentrations were obtained in the biofilm PK/PD model, antibiotic samples were quantified using bioassay analysis. The free AUC from 0- to 24-h (fAUC0–24) value for CIP 400 mg q8h in biofilm state was 46.29 µg·h/mL ± 0.69. The fC max value was 2.3 ± 0.21 µg/mL (target, 2.9 µg/mL), and the t 1/2 was 4.35 ± 0.69 (target, 5 h). Overall, the measured PK concentrations aligned with the target values.
Resistance testing
In biofilm, time-kill analyses, we observed CIP MIC reductions at 24 h in AR351 (1.0 to 0.5 μg/mL) and I0003-1 (0.5 to 0.125 μg/mL) after exposure to most phage-antibiotic combinations (Table 3). No evidence of bacterial resistance to EM or 14207 was observed at the end of 24 h in time-kill samples with PACs except when combined with AZT and CIP against AR351, whereas all phage-alone regimens developed resistance. Additionally, CIP resistance/elevated MICs were observed with CIP monotherapy regimens against AR351 (CIP MIC 1 to 2 µg/mL) and I0003-1 (CIP MIC 0.5 to 1.0 µg/mL), but none were observed in any of the PAC regimens at the end of 24 h.
TABLE 3.
Resistance evaluations for strains AR351 and I0003-1 a
| Treatment regimen | I0003-1: resistance at 24 h in biofilm TKAs | Treatment regimen | AR351: resistance at 24-h biofilm TKAs | ||
|---|---|---|---|---|---|
| CIP MIC | 14207 | CIP MIC | EM | ||
| Monotherapies | Monotherapies | ||||
| CIP | 2 | Not tested | CIP | 1 | Not tested |
| 14207 | Not tested | R | EM | Not tested | R |
| Multiple antibiotics | Multiple antibiotics | ||||
| MEM + CIP | 1 | Not tested | MEM + CIP | 1 | Not tested |
| AZT + CIP | 1 | Not tested | AZT + CIP | 1 | Not tested |
| AMK + CIP | 1 | Not tested | AMK + CIP | 1 | Not tested |
| COL + CIP | 1 | Not tested | COL + CIP | 1 | Not tested |
| PACs | PACs | ||||
| 14207 + CIP | 1 | S | EM + CIP | 0.5 | S |
| 14207 + COL + CIP | 0.125 | S | EM + COL + CIP | 0.5 | S |
| 14207 + AMK + CIP | 0.125 | S | EM + AMK + CIP | 0.5 | S |
| 14207 + MEM + CIP | 0.25 | S | EM + MEM + CIP | 0.5 | S |
| 14207 + AZT + CIP | 0.125 | S | EM + AZT + CIP | 1 | R |
Antibiotic and phage resistance testing results after 24 or 96 h for biofilm time-kill analyses and biofilm models, respectively.COL, colistin; MEM, meropenem; AMK, amikacin; CIP, ciprofloxacin; AZT, aztreonam; MIC, minimum inhibitory concentration; 14207, 14207 phage; EM, phage EM-T3762627-2_AH; EC, phage E2005-C; 109, 109 phage; PACs, phage-antibiotic combinations; S, sensitive defined as <10 CFU in spot; R, resistant defined as no visible effect of phage versus phosphate-buffered saline control.
In the PK/PD biofilm models for strain AR351, the potential for emergence of antibiotic resistance was evaluated by sampling at 96 h for each treatment regimen (Table 3). Resistance plating revealed CIP-non-susceptible mutants at 96 h (with CIP MIC of 8 µg/mL) for the phage-antibiotic combination model with CIP plus phages EM and EC. However, with the addition of phage 109 to EM + EC + CIP, the CIP MIC was preserved (baseline CIP MIC of 1 µg/mL observed at 96 h). EM phage resistance was detected in the model coupon samples for all regimens except the CIP combination regimen with phages EM, EC, and 109. Similarly, bacterial resistance to phage EC at 96 h was prevented only in the combination regimen with CIP and phages EM, EC, and 109. All regimens with phage 109, regardless of ciprofloxacin addition, developed resistance to phage 109.
DISCUSSION
Our study describes the use of PACs to control biofilms produced by XDR P. aeruginosa. There are several notable findings in the present study: (i) phage-antibiotic synergy (PAS) with CIP was demonstrated against both drug-resistant strains in biofilm TKAs, despite the difference in phage spectrum of activity (i.e., host range) between the two tested phages; (ii) enhancement was demonstrated with a phage cocktail in combination with CIP in an in vitro dynamic biofilm reactor model against an XDR P. aeruginosa strain; (iii) antibiotic resistance was prevented with the addition of phage 109 in the biofilm model; and (iv) phage resistance was prevented with the addition of CIP and phage 109 to the treatment regimen in the dynamic biofilm model.
These results expand on our previous work to explore the effect of PACs on MDR P. aeruginosa in planktonic TKAs (13). Some findings are consistent across these studies. For example, EM-MEM and EM-CIP were synergistic against AR351 (called 10266 in the previous study) in both planktonic and biofilm 24-h TKAs. However, while EM-CIP-COL was able to further reduce planktonic cell counts below the detection limit, the addition of COL did not show that benefit in biofilm TKAs. Among the strains and treatment combinations tested in TKAs, we have so far only taken P. aeruginosa AR351 and phage + CIP combinations into the 4-day PK/PD model, where EM + CIP continued to show enhanced activity relative to either agent alone, but did not succeed in reducing biofilm cell counts below detection limit until other phages were added to the treatment regimen. We also needed the 4-day model to detect the value of adding phage 109 to EM + EC + CIP (Fig. 4 versus 6).
Similar to our planktonic and biofilm TKAs, previous studies using static biofilm experiments have also reported synergy with phage plus meropenem, ciprofloxacin, or gentamicin against drug-resistant P. aeruginosa strains (7, 14). Specifically, one study observed impressive biofilm reduction (below detection limit) with phage EPA1, belonging to the Pakpunavirus genus, when combined with CIP (14). Another study observed complete eradication of bacterial cells with a T1-like siphovirus, phage Motto, when combined with CIP (7). Notably, our study examined phages from two genera (Pbunavirus and Pakpunavirus), in combination with CIP, and demonstrated significant biofilm reduction. However, the referenced studies relied on biofilms formed on the walls of microtiter plates, which are limited by their static nature. A recent study emphasized the importance of shear stress conditions on biofilm formation, which ultimately affects interaction with phages (8). Our biofilm TKA method incorporates some shear forces, and our 4-day model further incorporates continuous liquid flow and humanized antibiotic PK. At least two prior studies involving P. aeruginosa biofilms formed under shear showed that phage-only treatments did not produce persistent, substantial reductions in P. aeruginosa cell counts, although multiple treatments and cocktails yielded better results than single phages (15, 16). We saw similar results in our study, with the phage cocktail having a greater effect in the 4-day PK/PD model than phage EM alone but still not being sufficient to reduce biofilm counts below detection limit until we added CIP. Some animal studies have also shown that phage + antibiotic treatment led to greater reductions in in vivo biofilm CFU counts than either alone (17). We hypothesize that studies such as ours, which incorporate shear and longer treatment exposures, may be more realistic models for understanding how phage + antibiotic combinations will interact with biofilms while minimizing animal use. Although we have so far only tested a limited number of strains, the biofilm TKAs appear to be a very useful screening tool to identify PACs that might be most effective in higher-complexity, low-throughput models. However, the translatability of these various models will not be truly understood until robust clinical trial data become available.
Interestingly, both our previous planktonic and current biofilm data suggest that simple in vitro screens for antibiotic MIC and phage spectrum of activity do not necessarily predict the potential for PAS. In our prior planktonic study (13), we observed PAS in spite of pre-existing resistance to the antibiotic that was being used. In the current biofilm study, the activity of phage alone in biofilm TKAs was not predictive of PAS. Specifically, phage EM had a small but clear effect against P. aeruginosa AR351, whereas phage 14207 had no effect against P. aeruginosa I0003-1, CIP alone had little or no effect; however, both phages had similar and substantial effects against their respective target strains when combined with CIP. Previous studies have demonstrated excellent permeation of fluoroquinolones through P. aeruginosa biofilms in comparison to aminoglycosides and β-lactams, which might explain the added benefit of CIP and why the CIP MIC only increased by twofold in the MBIC analysis (Table 2) (18, 19). In future, it would be interesting to more systematically investigate the effects of PACs in which the individual agents have little antibiofilm activity on their own.
Our use of phage cocktails had a significant benefit in the 4-day model. Specifically, the addition of phage 109 thwarted resistance development to phages EM and EC as well as antimicrobial resistance to ciprofloxacin. A common approach to phage cocktail design is to emphasize both “breadth” and “depth” by combining phages with overlapping host ranges so as to maximize both the number of bacterial strains that are targeted (breadth) and the number that are targeted by more than one phage (depth), thereby limiting the strains’ ability to mutate and develop cross resistance to the phages in the cocktail (20). By that logic, adding phage 109 to EM + EC + CIP might have closed off a previously available genetic pathway by which AR351 could evade the activity of ciprofloxacin, EM, and EC. The specific genetic mechanism underlying this effect is not clear but is not likely related to phage receptor binding since all three phages belong to genera (Pakpunavirus and Pbunavirus) that use LPS O-antigen as cell surface receptors (21, 22).
One limitation of this study is the lack of antibiotic and phage dose-escalation and dose-staggering evaluations. Recent studies have described enhanced reduction in bacterial densities of P. aeruginosa biofilms in vitro when treatment with phages preceded antibiotics, so this may be an area of interest for future studies investigating the impact of PACs on P. aeruginosa biofilm eradication (23, 24). However, given current recommended treatment strategies, patients with drug-resistant infections will always be treated with antibiotics prior to phage therapy, so there is little clinical value to this suggested approach at present. Another limitation is that our study did not evaluate older biofilms against PACs. Previous studies have shown that the age of biofilm can impact the anti-biofilm activity of PACs (7). In future studies, older biofilms, grown for >24 h, should be tested, and the duration of the biofilm model should be extended beyond 4 days to assess long-term success of PACs on biofilm eradication in multiple strains and the development/prevention of bacterial and phage resistance. Further studies should also emphasize reaching the target area under the curve (AUC)/MIC ratio of 125 for CIP since reaching this target has been found to be predictive of the efficacy of fluoroquinolones in the treatment of Gram-negative infections (25). Strengths of this study include the utilization of a dynamic CBR model with an XDR clinical P. aeruginosa isolate against phage cocktails varying in their breadth and depth of activity. In addition, the current study measures the effect of PAC on antibiotic and phage resistance in P. aeruginosa biofilm, which emphasizes the positive impact of PAC on the prevention of ciprofloxacin resistance and effect of phage cocktail depth of activity on prevention of phage resistance.
In conclusion, our study contributes toward the growing body of evidence supporting the potential value of PACs for biofilm-related infections. Further investigation utilizing in vitro and in vivo methods to optimize phage cocktails in combination with antibiotics is warranted, specifically with regard to the contribution of phage cocktails to resistance management.
MATERIALS AND METHODS
Bacterial strains
The initial isolate screening included 10 well-characterized MDR P. aeruginosa strains isolated from patients hospitalized at Detroit Medical Center in Detroit, MichiganMI (12). The 10 strains were further fully characterized to describe mutations in genes commonly associated with resistance in P. aeruginosa (Table 1). For each strain, DNA was extracted using the MasterPure Gram Positive DNA Purification Kit (Lucigen) according to manufacturer’s instructions, and whole-genome sequencing was performed in a ONT MinION using the Rapid Barcoding Kit. The genomes were de novo assembled with Flye version 2.9, polished with Medaka version 1.6, and analyzed using ResFinder version 4.1 (Center for Genomic Epidemiology, https://www.genomicepidemiology.org/), PubMLST (https://pubmlst.org/organisms/pseudomonas-aeruginosa), Bacterial and Viral Bioinformatics Resource Center, and Basic Local Alignment Search Tool, NCBI). Mutation-associated resistance mechanisms were analyzed using PAO1 genome as comparator, as previously described (23). Sequences are available under Bioproject #PRJNA939146. Trypticase soy agar (Difco, Detroit, MI, USA) was used for growth and quantification of organisms. Phage host organisms, EM-T3762627-2 and LL-5504721, were kindly provided by J. Alexander (AdventHealth Orlando, Winter Springs, FL, USA), and host organisms Pa109 and EAMS2005-C307 were provided by R. Donlan at the US Centers for Disease Control and Prevention (Atlanta, GA, USA). Phage host organism ATCC 14207 was purchased commercially from ATCC (Manassas, VA, USA).
Antimicrobial agents and media
Mueller-Hinton broth II (MHB) (Difco) with 25-mg/L calcium and 12.5- mg/L magnesium was used for antibiotic susceptibility testing. Several antipseudomonal classes of antibiotics were tested, including carbapenems (MEM), monobactams (AZT), polymyxins (COL), aminoglycosides (AMK), and fluoroquinolones (CIP), which were purchased commercially from Sigma Chemical Company (St. Louis, MO, USA). Phage propagation and sensitivity testing used 100% brain heart infusion (BHI) broth (BD, Bacto, San Jose, CA, USA) with 0.5% agar (Oxoid, Lenexa, KS, USA) for overlays and 1.5% agar for underlays (26). Glucose supplemented tryptic soy broth (GSTSB) and MHB were utilized in biofilm TKAs.
Bacteriophages, quantification, and propagation
P. aeruginosa phages EM and LL were provided by J. Alexander (Orlando, FL, USA). P. aeruginosa phages phi109 (abbreviated here as 109), originally from the UK PHS Colindale phage typing set, and E2005-C (abbreviated here as EC), were kindly provided by the R. Donlan at the US Centers for Disease Control and Prevention. P. aeruginosa phage 14207 (ATCC 14207-B1) was purchased commercially from ATCC (Manassas, VA, USA). Based on a 70% intergenomic similarity threshold, EM (GenBank: ON169972.1) and LL belong to the Pakpunavirus genus, whereas EC (GenBank: OQ831729) and 109 (GenBank: OQ831730) belong to the Pbunavirus genus. Phage 14207 has not been sequenced. Phage-mediated lysis and release of phage progeny particles were confirmed by the formation of individual plaques on selected P. aeruginosa strains (26). Phages were quantified using the modified small-drop agar overlay method (26). The phages were further propagated until an adequate titer (~109 PFU/mL) was achieved to begin biofilm time-kill analysis. To begin, an underlay layer of BHI agar was poured into square petri plates. A 6-mL overlay of 100% BHI broth with 0.5% BHI agar was briefly combined with 80 µL of a host P. aeruginosa culture containing approximately 109 CFU/mL (McFarland 4.0) and poured atop the underlay layer. The overlay was briefly allowed to set, and following this, 500 µL of purified liquid bacteriophage was spread over top and incubated in a 37°C incubator overnight. The overlay agar was scraped into 3 mL of phosphate-buffered saline + 10-mM magnesium sulfate, then centrifuged at 1,000 rpm for 20 minutes at 4°C, and filtered twice (pore size: 0.22 µM) to remove bacteria and to obtain a cell-free lysate. The lysate was then stored at 2–8°C for experimental use.
Phage genome sequencing
Phages EC and 109 were newly sequenced for this study. Phage genomic DNA was isolated by treating the filtered lysates with 10 µg/mL each of DNase and RNase to remove bacterial nucleic acids, overnight precipitation in 10% PEG-8000 with 1 M NaCl, resuspension in 5-mM MgSO4, proteinase K treatment (0.1 mg/mL, 5-mM EDTA, 50°C for 30 minutes), organic extraction (phenol/chloroform/isoamyl alcohol, then chloroform/isoamyl alcohol) using PhaseLock gel tubes (Quantabio 2302830, Beverly, MA, USA), and sodium acetate-ethanol precipitation. A PCR-free genomic library was prepared and sequenced (Illumina MiSeq, PE150 reads). Trimmed reads (adapters removed, minimum base quality = 30) were assembled using Unicycler (version 0.5.0 + galaxy1, normal bridging mode, contigs ≥1,000 bp). Subsequent read-mapping to identify terminal repeats used untrimmed reads to ensure accurate identification of packaged genome termini. The main features of each phage genome are provided in Table 4. LL is nearly identical to phage EM, which we have used in previous studies (13). Genomes of 109 and E2005-C are 87.5% identical at the nucleotide level (Fig. S1) and were annotated using Cenote-Taker2 version 2.1.3 in annotation-only mode (27).
TABLE 4.
Genomic characteristics of the phages used in this study c
| Phage | Single-copy genome (kb) | Direct terminal repeats (bp) | GC% | Annotated genes (protein, tRNA) | Assigned genus a |
|---|---|---|---|---|---|
| LL | 93,607 | 1,192 | 49.6 | 195, 17 | Pakpunavirus |
| EM | 93,583 | 1,192 | 49.6 | 193, 17 | Pakpunavirus |
| 109 | 65,597 | No b | 55.0 | 103, 0 | Pbunavirus |
| EC | 66,591 | No b | 55.7 | 100 0 | Pbunavirus |
All listed genera are within viruses Duplodnaviria, Heunggongvirae, Uroviricota, and Caudoviricetes. Genus assignments for 109 and E2005-C are based on ≥70% similarity to representative members of the Pbunavirus genus (Fig. S1; VIRIDIC web version 1, default settings) (28).
Read mapping data indicated a sequence-specific packaging initiation site similar to Pa193 (NC_050148.1) and PA8P1 (NC_048806.1), but not fixed-length terminal repeats.
EM, phage EM-T3762627-2_AH; EC, phage E2005-C; 109, phage 109; LL, phage LL-5504721-AH; GC, guanine-cytosine percentage.
Phage sensitivity assay
Bacterial sensitivities to phages were tested using spot testing. First, all phage test suspensions were adjusted to the same value (~107) using their respective propagation strains. Then, 10-fold serial dilutions of phage were spotted onto 100% BHI broth with 0.5% BHI overlay plates containing McFarland 4.0 of the target bacteria (26). Plates contained 5 mL of overlay agar which was briefly mixed with 80 µL of McFarland 4.0 culture and left to dry for 10 minutes before spotting 5 µL of purified phage onto the bacterial lawn. The plates were incubated overnight following drop testing, and plaque counts of >107 , between 103 and 107, and <103 PFU/mL were classified as high, medium, and low phage sensitivity, respectively (29). No sensitivity to phage was defined as no visual detection of individual phage plaques and/or no bacterial lawn clearance.
In vitro antibiotic susceptibility testing
All 10 strains underwent antimicrobial susceptibility testing via the broth microdilution method per Clinical and Laboratory Standards Institute (CLSI) standards (Table 1) (10). Escherichia coli ATCC 25922 was used as the internal quality control strain. AR351 (XDR) and RI0003-1 (MDR) exhibiting mutations in MexAB-OprM repressor genes were selected for MBIC testing and biofilm time-kill analysis due to their varying resistance profiles (resistant to three to four selected antibiotic categories) and based on preliminary work (Table 1) (12, 13). MBIC testing was carried out using the pin-lid method as previously described (30). All MICs were performed per CLSI guidelines (10). Plates were incubated at 37°C for 18–24 h prior to reading the results with MIC reductions measured by serial twofold dilutions.
Biofilm time-kill analyses
Initial biofilm time-kill analyses were performed using CIP, AZT, AMK, MEM, and COL at 0.25× MIC values to simulate subinhibitory concentrations typically encountered in biofilm infections often leading to treatment failure. Based on spectrum of activity, broad-spectrum phage EM (i.e., wide host range) and narrow-spectrum phage 14207 (i.e., narrow host range) were selected for evaluation in biofilm time-kill analyses against strains AR351 and I0003-1, respectively (Fig. 1 and 2). Phage LL was not considered for analysis in biofilm time-kills because it is nearly identical to phage EM, as reported previously (13). Our previous evaluations identified an optimal multiplicity of infection (MOIinput) of 1, which represents the ratio of input phage particles to the target organism (13). Given the lack of guidance regarding optimal phage dosing in humans, an MOIinput of 1 was used throughout this study to optimize bacterial killing and demonstration of synergy. The experiment was performed in 24-well tissue culture-treated plates with 2 mL of 1% GSTSB broth and four sterile polystyrene beads in each well. Biofilms were established on the polystyrene beads before treatment. Plates were incubated in a shaker incubator at 37°C for 24 h to allow for biofilm growth on the beads in 1% GSTSB. After 24 h of incubation, GSTSB was aspirated and replaced with MHB. Phage and antibiotic were then added after the first (0 h) sampling. One bead was aseptically removed for sampling and processed, as previously described, at 0, 4, 8, and 24 h to create a growth curve (31). Bactericidal activity was defined as a ≥3-log10 CFU/cm2 reduction from baseline. Synergy between two agents was defined as a ≥2-log10 CFU/cm2 reduction compared with the most potent single agent. Synergy for triple combinations was defined as ≥2-log10 CFU/cm2 reduction compared with the most potent double combination regimen. Based on greatest average bacterial cell reduction in the initial biofilm time-kill experiments (Fig. 3), one antibiotic (CIP) was chosen for additional biofilm time-kills with phages EM, 14207, EC, and 109 (Fig. 4 and 5) to identify optimal phage-antibiotic cocktails to be evaluated in the biofilm model. Phage LL was not considered for analysis in biofilm time-kills because it is nearly identical to phage EM, as reported previously (13).
In vitro biofilm model
Strain AR351 was inoculated into TSA plates incubated at 37°C for 24 h and then suspended in normal saline solution to reach a concentration equivalent to 1.7 McFarland. The in vitro model consisted of a previously described CDC biofilm reactor (CBR) model (BioSurface Technologies, Bozeman, MT, USA) set up with polyurethane coupons inserted into eight rods, simulating human PK, to evaluate the in vitro activity of antimicrobials and phage (32). Briefly, A 40-h biofilm conditioning phase was carried out prior to evaluation of the phage-antimicrobial combinations and consisted of 24 h of incubation at 37°C of inoculated 1% gSTSB, followed by 16 h of a continuous flow with a 1/10 concentration of gSTSB performed with peristaltic pumps (Masterflex; Cole-Parmer Instrument Co., Chicago, IL, USA). After completion of conditioning and continuous flow phases, Mueller-Hinton broth supplemented with 25-mg/L calcium was used as medium for the experiment. Boluses of antimicrobials and phage were injected into the reactor after the biofilm conditioning phase was completed. Each CBR model allowed for eight rods with two polyurethane coupons per rod. The CBR was placed in a 37°C incubator throughout the procedure. Fresh medium (MHB) was continuously supplied and removed from the compartment along with the drugs and phage via a peristaltic pump set to simulate the half-life (t 1/2) of the drugs. The regimen evaluated was CIP exposed at the following dose simulation (assuming 30% protein binding and targeting an average half-life of 5 h): 400 mg q8h [fCmax (maximum concentration of the free, unbound fraction of drug in serum) = 2.9 mg/L] (33). The other regimens were phage EM every 24 h (q24h) (MOIinput = 1.0), phage EM q24h + CIP 400 mg q8h (MOIinput = 1.0), phage EM + EC q24h + CIP 400 mg q8h (MOIinput = 1.0), EM + EC + 109 q24h (MOIinput = 1.0), phage cocktail EM + EC + 109 q24h (MOIinput = 1.0) + CIP 400 mg q8h, phage cocktail EM + 14207 q24h (MOIinput = 1.0) + CIP 400 mg q8h, and growth control with clearance of media set to mimic a 5-h t 1/2, similar to that described for the drug experiments. All model experiments were completed in duplicate to ensure reproducibility.
Pharmacodynamic analysis
One rod from each model was aseptically removed at 0, 4, 8, 24, 48, 72, and 96 h. Two coupons were removed from each rod, and each coupon was rinsed twice in sterile normal saline solution to remove excess planktonic cells. Then, each coupon was placed into a sterile tube containing 10 mL of normal saline and processed, and colony counts were determined as previously described (34). The biofilm-embedded cell concentration (means and standard deviations in CFU counts per square centimeter) was computed for each coupon. The limit of detection of these methods of colony count determination was 2 log10 CFU/cm2. The total reduction in log10 CFU/cm2 over 96 h was determined by plotting time-kill curves based on the number of viable organisms over the time period. Bactericidal (99.9% kill) and bacteriostatic effects were defined as a ≥3-log10 CFU/cm2 reduction and a <3-log10 CFU/cm2 reduction in the colony count compared to the starting inoculum baseline, respectively. Enhancement and improvement of activity by the addition of a drug were defined as a ≥2-log10 CFU/cm2 increase and a 1- to 2-log10 CFU/cm2 increase in bacterial eradication from starting inoculum baseline compared to the level seen with the most active single agent of the combination, respectively (34). Antagonistic activity was defined as an increase in bacterial growth of ≥2 log10 CFU/cm2 in comparison to the most active single agent from the combination.
Pharmacokinetic analysis
Direct media samples were obtained through the injection port of the biofilm model at 0, 2, 4, 8, and 24 h to verify the attainment of target antibiotic concentrations. All samples were stored at −80°C until they were ready for PK assay. CIP concentrations were measured via bioassay using a concentration-zone standard curve created with disc diffusion against Escherichia coli ATCC 25922. Of note, the E. coli ATCC strain 25922 was not sensitive to any of the P. aeruginosa phages (alone or as a cocktail). Agar plates (Difco) were pre-swabbed with a 1.7-McFarland unit suspension of E. coli. Sterile discs were placed and filled with standard (1, 2, 4, and 5 µg/mL) or sample concentrations at 15 µL. The plates were incubated for 24 h at 37°C, and then zones of inhibition around each disc were measured using an automatic colony counter (Scan 1200; Interscience, Woburn, MA, USA). A standard curve was created using zones of inhibition of known concentrations (1, 2, 4, and 5 µg/mL), and sample timepoint zones of inhibition were plotted against this curve to obtain sample concentrations. The determination coefficient (r 2) was 0.98 for the standard curve. Intra-assay coefficients of variance were less than 5% for low, medium, and high standards (1, 2, and 4 mg/L), respectively. These concentrations were used to calculate half-lives and determine the area under the curve as well as peak concentrations using the trapezoidal methodology with PK Analyst software (version 1.10, provided by MicroMath Scientific Software located in Salt Lake City, UT, USA).
Resistance testing
The emergence of antibiotic and phage resistance was determined as previously described by using the 24-h biofilm TKA liquid sample and 96-h biofilm model liquid sample (26, 35 – 37). The double-drop method was used to perform phage resistance screening (26, 36). Phage resistance was scored from the spots as resistant (no visible effect of phage versus phosphate-buffered saline control), intermediate (bacteria present but phage activity visible), and sensitive (<10 CFU in spot) (36).
Statistical analysis
Statistical analysis was performed using one-way analysis of variance with Tukey’s multiple comparisons test (with P < 0.05 considered significant) using Prism 8 for macOS software (GraphPad version 8.4.3).
ACKNOWLEDGMENTS
The authors thank the Food and Drug Administration/CBER Facility for Biotechnology Resources (particularly Wells W. Wu and Chao-Kai Chou) for phage sequencing services and the team at Case Western Reserve University for characterization of the Pseudomonas aeruginosa strains. We thank Keith Kaye, MD, MPH, for providing K0001-1, K0003-1, I0003-1, and G0003-2 Pseudomonas aeruginosa strains from The Overcome Study (38).
D.H. drafted the main part of the manuscript; A.E. contributed to the parts of the manuscript; M.R., S.L., and A.J. provided support and conceptual advice at all stages of the manuscript preparation; all authors reviewed, edited, and suggested revisions to the manuscript, and read and agreed to the published version of the manuscript.
D.J.H., A.E., S.A., N.B., K.L., K.S., R.K., A.J.K., T.M., R.S., J.A., S.M.L, L.J.R., S.H.M., and R.A.B. have nothing to disclose; M.J.R. received research support, consulted or participated in a speaker bureau for AbbVie, Basilea, Entasis, La Jolla, Merck, Paratek, Shionogi, and T2 Biosystems, and is partially funded by NIAID R21AI163726.
Contributor Information
Michael J. Rybak, Email: m.rybak@wayne.edu.
Jared A. Silverman, Bill & Melinda Gates Medical Research Institute, Cambridge, Massachusetts, USA
DATA AVAILABILITY
Phage DNA sequence information is available through GenBank (accession numbers: ON169972, OQ831730, OQ831729). P. aeruginosa strain sequence information is available through GenBank (Bioproject number: PRJNA939146).
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aac.00578-23.
Intergenomic similarity matrix for phages E2005-C, 109, and one phage from each ICTV-recognized species within the Pbunavirus genus.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Del Giacomo P, Raffaelli F, Losito AR, Fiori B, Tumbarello M. 2022. XDR-Pseudomonas aeruginosa outside the ICU: is there still place for colistin Antibiotics (Basel) 11:193. doi: 10.3390/antibiotics11020193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Heidari R, Farajzadeh Sheikh A, Hashemzadeh M, Farshadzadeh Z, Salmanzadeh S, Saki M. 2022. Antibiotic resistance, biofilm production ability and genetic diversity of carbapenem-resistant Pseudomonas aeruginosa strains isolated from nosocomial infections in southwestern Iran. Mol Biol Rep 49:3811–3822. doi: 10.1007/s11033-022-07225-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schooley RT, Biswas B, Gill JJ, Hernandez-Morales A, Lancaster J, Lessor L, Barr JJ, Reed SL, Rohwer F, Benler S, Segall AM, Taplitz R, Smith DM, Kerr K, Kumaraswamy M, Nizet V, Lin L, McCauley MD, Strathdee SA, Benson CA, Pope RK, Leroux BM, Picel AC, Mateczun AJ, Cilwa KE, Regeimbal JM, Estrella LA, Wolfe DM, Henry MS, Quinones J, Salka S, Bishop-Lilly KA, Young R, Hamilton T. 2017. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob Agents Chemother 61:e00954-17. doi: 10.1128/AAC.00954-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aslam S, Lampley E, Wooten D, Karris M, Benson C, Strathdee S, Schooley RT. 2020. Lessons learned from the first 10 consecutive cases of intravenous bacteriophage therapy to treat multidrug-resistant bacterial infections at a single center in the United States. Open Forum Infect Dis 7:ofaa389. doi: 10.1093/ofid/ofaa389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Totten KMC, Patel R. 2022. Phage activity against planktonic and biofilm Staphylococcus aureus periprosthetic joint infection isolates. Antimicrob Agents Chemother 66:e0187921. doi: 10.1128/AAC.01879-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fong SA, Drilling A, Morales S, Cornet ME, Woodworth BA, Fokkens WJ, Psaltis AJ, Vreugde S, Wormald P-J. 2017. Activity of bacteriophages in removing biofilms of Pseudomonas aeruginosa isolates from chronic rhinosinusitis patients. Front Cell Infect Microbiol 7:418. doi: 10.3389/fcimb.2017.00418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Manohar P, Loh B, Nachimuthu R, Leptihn S. Phage-antibiotic combinations to control Pseudomonas aeruginosa-Candida two-species biofilms. Microbiology. doi: 10.1101/2022.08.18.504394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pires DP, Melo LDR, Azeredo J. 2021. Understanding the complex phage-host interactions in biofilm communities. Annu Rev Virol 8:73–94. doi: 10.1146/annurev-virology-091919-074222 [DOI] [PubMed] [Google Scholar]
- 9. Queenan AM, Bush K. 2007. Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev 20:440–458, doi: 10.1128/CMR.00001-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clinical and Laboratory Standards Institute . 2020. Performance standards for antimicrobial susceptibility testing; 30th informational supplement. Clinical and Laboratory Standards Institute, CLSI document MS100. Wayne, PA. [Google Scholar]
- 11. Magiorakos A-P, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 12. Mikhail S, Singh NB, Kebriaei R, Rice SA, Stamper KC, Castanheira M, Rybak MJ. 2019. Evaluation of the synergy of ceftazidime-avibactam in combination with meropenem, amikacin, aztreonam, colistin, or fosfomycin against well-characterized multidrug-resistant Klebsiella pneumoniae and Pseudomonas aeruginosa. Antimicrob Agents Chemother 63:e00779-19. doi: 10.1128/AAC.00779-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Holger DJ, Lev KL, Kebriaei R, Morrisette T, Shah R, Alexander J, Lehman SM, Rybak MJ. 2022. Bacteriophage-antibiotic combination therapy for multidrug-resistant Pseudomonas aeruginosa: in vitro synergy testing. J Appl Microbiol 133:1636–1649. doi: 10.1111/jam.15647 [DOI] [PubMed] [Google Scholar]
- 14. Akturk E, Oliveira H, Santos SB, Costa S, Kuyumcu S, Melo LDR, Azeredo J. 2019. Synergistic action of phage and antibiotics: parameters to enhance the killing efficacy against mono and dual-species biofilms. Antibiotics (Basel) 8:103. doi: 10.3390/antibiotics8030103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fu W, Forster T, Mayer O, Curtin JJ, Lehman SM, Donlan RM. 2010. Bacteriophage cocktail for the prevention of biofilm formation by Pseudomonas aeruginosa on catheters in an in vitro model system. Antimicrob Agents Chemother 54:397–404. doi: 10.1128/AAC.00669-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lehman SM, Donlan RM. 2015. Bacteriophage-mediated control of a two-species biofilm formed by microorganisms causing catheter-associated urinary tract infections in an in vitro urinary catheter model. Antimicrob Agents Chemother 59:1127–1137. doi: 10.1128/AAC.03786-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yilmaz C, Colak M, Yilmaz BC, Ersoz G, Kutateladze M, Gozlugol M. 2013. Bacteriophage therapy in implant-related infections: an experimental study. J Bone Joint Surg Am 95:117–125. doi: 10.2106/JBJS.K.01135 [DOI] [PubMed] [Google Scholar]
- 18. Walters MC, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. 2003. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother 47:317–323. doi: 10.1128/AAC.47.1.317-323.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shigeta M, Tanaka G, Komatsuzawa H, Sugai M, Suginaka H, Usui T. 1997. Permeation of antimicrobial agents through Pseudomonas aeruginosa biofilms: a simple method. Chemotherapy 43:340–345. doi: 10.1159/000239587 [DOI] [PubMed] [Google Scholar]
- 20. Abedon ST, Danis-Wlodarczyk KM, Wozniak DJ. 2021. Phage cocktail development for bacteriophage therapy: toward improving spectrum of activity breadth and depth. Pharmaceuticals 14:1019. doi: 10.3390/ph14101019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li G, Shen M, Yang Y, Le S, Li M, Wang J, Zhao Y, Tan Y, Hu F, Lu S. 2018. Adaptation of Pseudomonas aeruginosa to phage PaP1 predation via O-antigen polymerase mutation. Front Microbiol 9:1170. doi: 10.3389/fmicb.2018.01170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Danis-Wlodarczyk K, Olszak T, Arabski M, Wasik S, Majkowska-Skrobek G, Augustyniak D, Gula G, Briers Y, Jang HB, Vandenheuvel D, Duda KA, Lavigne R, Drulis-Kawa Z. 2015. Characterization of the newly isolated lytic bacteriophages KTN6 and KT28 and their efficacy against Pseudomonas aeruginosa biofilm. PLoS One 10:e0137015. doi: 10.1371/journal.pone.0137015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Suh GA, Lodise TP, Tamma PD, Knisely JM, Alexander J, Aslam S, Barton KD, Bizzell E, Totten KMC, Campbell JL, Chan BK, Cunningham SA, Goodman KE, Greenwood-Quaintance KE, Harris AD, Hesse S, Maresso A, Nussenblatt V, Pride D, Rybak MJ, Sund Z, van Duin D, Van Tyne D, Patel R, Antibacterial Resistance Leadership Group . 2022. Considerations for the use of phage therapy in clinical practice. Antimicrob Agents Chemother 66:e0207121. doi: 10.1128/AAC.02071-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chaudhry WN, Concepción-Acevedo J, Park T, Andleeb S, Bull JJ, Levin BR. 2017. Synergy and order effects of antibiotics and phages in killing Pseudomonas aeruginosa biofilms. PLoS One 12:e0168615. doi: 10.1371/journal.pone.0168615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dalhoff A. 2014. Pharmacokinetics and pharmacodynamics of aerosolized antibacterial agents in chronically infected cystic fibrosis patients. Clin Microbiol Rev 27:753–782. doi: 10.1128/CMR.00022-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mazzocco A, Waddell TE, Lingohr E, Johnson RP, Enumeration of Bacteriophages Using the Small Drop Plaque Assay System . 2009. Bacteriophages: methods and protocols, volume 1: isolation, characterization, and interactions, p 81–85. In Clokie MRJ, Kropinski AM (ed), Methods in molecular biology. Humana Press. doi: 10.1007/978-1-60327-164-6 [DOI] [PubMed] [Google Scholar]
- 27. Tisza MJ, Belford AK, Domínguez-Huerta G, Bolduc B, Buck CB. 2021. Cenote-taker 2 democratizes virus discovery and sequence annotation. Virus Evol 7:veaa100. doi: 10.1093/ve/veaa100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moraru C, Varsani A, Kropinski AM. 2020. VIRIDIC—a novel tool to calculate the intergenomic similarities of prokaryote-infecting viruses. Viruses 12:1268. doi: 10.3390/v12111268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Morrisette T, Lev KL, Kebriaei R, Abdul-Mutakabbir JC, Stamper KC, Morales S, Lehman SM, Canfield GS, Duerkop BA, Arias CA, Rybak MJ. 2020. Bacteriophage-antibiotic combinations for enterococcus faecium with varying bacteriophage and daptomycin susceptibilities. Antimicrob Agents Chemother 64:e00993-20. doi: 10.1128/AAC.00993-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. 1999. The calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol 37:1771–1776. doi: 10.1128/JCM.37.6.1771-1776.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kebriaei R, Lev KL, Shah RM, Stamper KC, Holger DJ, Morrisette T, Kunz Coyne AJ, Lehman SM, Rybak MJ. 2022. Eradication of biofilm-mediated methicillin-resistant staphylococcus aureus infections in vitro: bacteriophage-antibiotic combination. Microbiol Spectr 10:e0041122. doi: 10.1128/spectrum.00411-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barber KE, Smith JR, Ireland CE, Boles BR, Rose WE, Rybak MJ. 2015. Evaluation of ceftaroline alone and in combination against biofilm-producing methicillin-resistant staphylococcus aureus with reduced susceptibility to daptomycin and vancomycin in an in vitro pharmacokinetic/pharmacodynamic model . Antimicrob Agents Chemother 59:4497–4503. doi: 10.1128/AAC.00386-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. CIPRO(R), intravenous (ciprofloxacin) for intravenous infusion. 2013. Medication Guide. Bayer HealthCare Pharmaceuticals, Wayne, New Jersey. [Google Scholar]
- 34. Jahanbakhsh S, Singh NB, Yim J, Kebriaei R, Smith JR, Lev K, Tran TT, Rose WE, Arias CA, Rybak MJ. 2020. Impact of daptomycin dose exposure alone or in combination with β-lactams or rifampin against vancomycin-resistant enterococci in an in vitro biofilm model. Antimicrob Agents Chemother 64:e02074-19. doi: 10.1128/AAC.02074-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. O’Flynn G, Ross RP, Fitzgerald GF, Coffey A. 2004. Evaluation of a cocktail of three bacteriophages for biocontrol of Escherichia coli O157:H7. Appl Environ Microbiol 70:3417–3424. doi: 10.1128/AEM.70.6.3417-3424.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lehman SM, Mearns G, Rankin D, Cole RA, Smrekar F, Branston SD, Morales S. 2019. Design and preclinical development of a phage product for the treatment of antibiotic-resistant Staphylococcus aureus infections. Viruses 11:88. doi: 10.3390/v11010088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kebriaei R, Rice SA, Singh KV, Stamper KC, Dinh AQ, Rios R, Diaz L, Murray BE, Munita JM, Tran TT, Arias CA, Rybak MJ. 2018. Influence of Inoculum effect on the efficacy of daptomycin monotherapy and in combination with β-lactams against daptomycin-susceptible enterococcus faecium harboring LiaSR substitutions. Antimicrob Agents Chemother 62:e00315-18. doi: 10.1128/AAC.00315-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kaye KS, Marchaim D, Thamlikitkul V, Carmeli Y, Chiu C-H, Daikos G, Dhar S, Durante-Mangoni E, Gikas A, Kotanidou A, Paul M, Roilides E, Rybak M, Samarkos M, Sims M, Tancheva D, Tsiodras S, Kett D, Patel G, Calfee D, Leibovici L, Power L, Munoz-Price S, Stevenson K, Susick L, Latack K, Daniel J, Chiou C, Divine GW, Ghazyaran V, Pogue JM. 2023. Colistin monotherapy versus combination therapy for carbapenem-resistant organisms. NEJM Evid 2. doi: 10.1056/evidoa2200131 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Intergenomic similarity matrix for phages E2005-C, 109, and one phage from each ICTV-recognized species within the Pbunavirus genus.
Data Availability Statement
Phage DNA sequence information is available through GenBank (accession numbers: ON169972, OQ831730, OQ831729). P. aeruginosa strain sequence information is available through GenBank (Bioproject number: PRJNA939146).