Abstract
Growing evidence suggests that social relationship quality can influence age-related health outcomes, although how the quality of one’s relationships directly relates to the underlying aging process is less clear. We hypothesized that the absence of close relationships and lower support and higher strain within existing relationships would be associated with an accelerated epigenetic aging profile among older adults in the Health and Retirement Study. Adults (N = 3,647) aged 50–100 years completed ratings of support and strain in relationships with their spouse, children, other family members, and friends. They also provided a blood sample that was used for DNA methylation profiling to calculate a priori-specified epigenetic aging measures: Horvath, Hannum, PhenoAge, GrimAge, and Dunedin Pace of Aging methylation (DunedinPoAm38). Generalized linear models that adjusted for chronological age, sex, and race/ethnicity and applied a false discovery rate correction revealed that the absence of marital and friend relationships related to an older GrimAge and faster DunedinPoAm38. Among those with existing relationships, lower support from a spouse, child, other family, and friends and higher strain with friends related to an older PhenoAge and GrimAge and faster DunedinPoAm38. In secondary analyses that further adjusted for socioeconomic and lifestyle factors, lower support from other family members and friends was associated with greater epigenetic aging. Findings suggest that the absence of close relationships and lower support within existing relationships—particularly with family members and friends—relate to accelerated epigenetic aging in older adulthood, offering one mechanism through which social relationships might influence risk for age-related declines and disease.
Keywords: social relationships, social support, social strain, biological aging, epigenetic clock, DNA methylation
1. Introduction
A sizeable literature has established that the quality of one’s social relationships can have a significant impact on health and well-being across the lifespan.1,2 Current theoretical frameworks posit that social relationships promote health by fulfilling basic needs for social connection and by providing a buffering resource during times of stress; however, they can also be a source of conflict and strain.3–5 Researchers have identified that perceived social support and social strain are distinct dimensions of relationship quality that can influence health.3 Whereas social support is defined as the availability of resources, advice, understanding, or acceptance within relationships,3 social strain has been described as the presence of criticism, insensitivity, demands, or feelings of being let down by close others.6 Although relatively fewer studies have focused on social strain, both lower social support and greater strain have been concurrently and prospectively associated with multiple age-related conditions in middle- and older adulthood, including functional limitations,7,8 poorer physical and cognitive functioning,9–11 and incidence and progression of cardiovascular disease.12–14 Social support and strain have also been identified as reliable predictors of all-cause mortality1,15,16 and mortality from cancer,17,18 stroke,19 and cardiovascular disease.20 These findings suggest that social relationships influence aging, although how these qualities of relationships directly relate to the underlying aging process is less clear.
A growing body of evidence has linked social relationship quality to key biological aging processes such as inflammation and telomere shortening, pointing to potential pathways through which relationships may influence these aging-related outcomes. For instance, several studies found that lower social support was associated with elevated peripheral markers of inflammation including interleukin (IL)-6, IL-8, tumor necrosis factor-α (TNF-α), and C-reactive protein (CRP) in middle-aged and older adults,21,22 and increased activation of transcription factor nuclear factor-кB (NF-кB), which regulates the expression of inflammatory genes.23 Social strain has also been associated with greater circulating IL-6 and TNF-α24,25 and NF-кB signaling.23 However, a handful of studies did not find associations between social support and circulating CRP, an indicator of systemic inflammation.22 Prior research has also suggested that older adults who reported lower social support had shorter telomeres,26–28 the protective caps at the end of chromosomes that naturally shorten over time but are vulnerable to accelerated shortening and are considered a hallmark of aging.29
Epigenetic markers of aging offer another approach to track the underlying biological aging process. Epigenetic aging refers to age-related alterations to the epigenome (i.e., chemical compounds that modify DNA but do not change its coding sequence), which include histone modifications, chromatin remodeling, and changes in DNA methylation patterns.29 One of the most widely used approaches to measuring epigenetic aging, termed the ‘epigenetic clock,’ was developed by identifying distinct regions of the DNA that become hypo- or hyper-methylated with age, and correlates with chronological age across a range of cell types and tissues.30,31 More recent versions of the epigenetic clock, commonly referred to as “second generation” clocks, were developed based on DNA methylation patterns that are associated with multiple biomarkers and are predictive of phenotypic aging outcomes (e.g., morbidity and mortality; PhenoAge)32 and time to death (GrimAge).33 In contrast to the epigenetic clock measures, the Dunedin Pace of Aging methylation (DunedinPoAm38) measure was developed to estimate an individual’s rate of biological aging at a single point in time, based on data from the Dunedin cohort and changes in 18 biomarkers of organ-system integrity assessed over a 12-year period.34 Measures of epigenetic age are useful because they capture biological aging in a metric that is intuitive (i.e., years) to assess individuals’ rate of aging and identify those who are biologically older or younger than their chronological age.35,36 They also integrate multiple physiological systems into a single numerical measure of biological age that in turn predicts multiple age-related conditions, including frailty, cognitive decline, and cancer, as well as all-cause and specific-cause (e.g., cancer) mortality, and importantly, may be modifiable by interventions and show feasibility for clinical use.35,37
The present study extends the literature on social relationships and biological aging by investigating associations between social relationships and epigenetic markers of aging in a large, nationally representative sample of older adults in the Health and Retirement Study (HRS). Whereas most prior research on social relationships and biological aging has focused exclusively on general social support that is non-specific to relationship source or support that is aggregated across sources (e.g., spouse, family, friends), we aimed to examine both positive and negative dimensions of relationship quality in older adults’ relationships with their spouse, children, other family members, and friends. One exception to this is a study that assessed support and strain in marital, family, and friend relationships, finding that strain with family members was most robustly associated with a higher inflammatory burden in middle-aged and older adults.38 In addition, a recent study with a subset of HRS participants found that greater support from friends (but not change in support over time) was associated with a slower Dunedin pace of aging up to 10 years later.39 Given that not all participants in the HRS sample were married or had other types of close relationships, we first aimed to test whether the presence versus absence of these relationships was associated with epigenetic aging, and among those with existing relationships, whether social support and strain within these relationships related to epigenetic aging. We focused on both support and strain within different relationship types due to the changes in social ties that may occur with aging, whereby some relationships may become more salient and stable and fulfill different purposes or needs (e.g., intimacy, companionship, caregiving) in older adulthood. Finally, we focused on five established, a priori-identified epigenetic aging measures derived from DNA methylation profiling to provide insights into how relationship quality may influence the underlying process of aging at the molecular level. These measures are the most commonly assessed in previous research and include the two original “first generation” Horvath and Hannum clocks, as well as the newer “second generation” PhenoAge, GrimAge, DunedinPoAm38 measures.
Based on previous research linking social relationship quality to age-related conditions and other hallmarks of biological aging (i.e., inflammation and telomere length), we hypothesized that, overall, an absence of close relationships would be associated with an accelerated epigenetic aging profile relative to the presence of close relationships, which provide at least an opportunity for social contact and support in older adulthood. However, we would like to note that previous research has yielded mixed findings regarding parental status and health,40,41 with some studies suggesting that having children may confer a health benefit in older age.42–45 We also hypothesized that among those in existing relationships, lower support and higher strain would be associated with an accelerated epigenetic aging profile. Given that the extant literature is not sufficiently developed, we did not generate hypotheses about whether differences in the strength of associations between specific relationship types and epigenetic aging would be observed, although there is preliminary evidence to suggest that relationships with family and friends may be particularly relevant.
2. Methods
2.1. Ethics statement
This investigation has been conducted in accordance with the ethical standards and according to the Declaration of Helsinki and according to national and international guidelines and has been approved by the Institutional Review Board at the University of Michigan.
2.2. Participants
The present study used data from the University of Michigan Health and Retirement Study (HRS), a longitudinal, nationally representative study of nearly 20,000 U.S. adults over the age of 50. For this study, participants were 4,018 adults aged 50–100 years who provided a blood sample as part of the 2016 Venous Blood Study (VBS) that was used to assess epigenetic aging.46 HRS participants were excluded from the VBS if they were not community dwelling (i.e., they were incarcerated or residing in an assisted living setting of any type). The epigenetic aging subsample of the VBS was designed to be representative of the U.S. population when weighted. For the present analysis, 86 participants were missing data for at least one covariate. To retain participants, analyses included all participants who provided reports of their relationship status, support, or strain for each type of relationship. Thus, the statistical models for each relationship type had different sample sizes (see tables for details). The weighted sample had a mean age of 68.7 years and was 55.1% female. Participants self-identified as Hispanic (7.9%), non-Hispanic Black (9.3%), non-Hispanic of another race (3.3%), and non-Hispanic white (79.5%). The educational distribution of the participants included less than a high school education (13.1%), high school diploma or GED (30.2%), some college (26.0%), and college diploma or higher (30.7%).
2.3. Procedures
Participants in the HRS study completed core interviews every other year, and self-administered psychosocial questionnaires were also given to alternating random halves of the full sample every two years. As part of the self-administered psychosocial questionnaire, participants completed ratings of support and strain in their relationships with their spouse, children, other family members, and friends. If participants were missing social support and strain data from the 2016 or 2014 questionnaire, we used data from the 2012 or 2010 questionnaire, respectively. If participants were missing data from the 2012 or 2010 questionnaire, we used data from 2008 (when the social support and strain measures were first included in the questionnaire). Most social relationship data was obtained from the 2016 or 2014 questionnaire (81.9 to 87.2%), with smaller proportions obtained from 2012 or 2010 (11.5 to 14.8%) and 2008 (1.3 to 3.3%).‡Participants also provided a blood sample as part of the 2016 Venous Blood Study (VBS) and DNA methylation profiling was performed using the Illumina Infinium Methylation EPIC BeadChip (Illumina, San Diego, CA) to derive the epigenetic aging measures, as described in detail previously.46,47
2.4. Measures
2.4.1. Social relationship measures
Participants completed ratings of perceived support and strain within four types of relationships: their spouse (husband, wife, or partner with whom they live), child or children, other immediate family members (e.g., brothers, sisters, parents, cousins, grandchildren), and friends. For each relationship type, three items assessed support (“How much do they really understand the way you feel about things?”, “How much can you rely on them if you have a serious problem?”, and “How much can you open up to them if you need to talk about your worries?”) and four items assessed strain (“How often do they make too many demands on you?”, “How much do they criticize you?”, “How much do they let you down when you are counting on them?”, and “How much do they get on your nerves?”). Responses for each item ranged from 1 (not at all) to 4 (a lot) and items were averaged to create a composite score, with higher scores indicating greater support or strain. We created a relationship status variable for each type of relationship that was coded as ‘present’ (vs. absent) if participants reported having that type of relationship.
2.4.2. Epigenetic aging measures
The epigenetic aging measures for this study included the Horvath, Hannum, PhenoAge, and GrimAge clocks and DunedinPoAm38. The Horvath estimate of epigenetic age is based on DNA methylation levels at 353 cytosine-phosphate-guanine base pair (CpG) sites and was developed as a predictor of chronological age across multiple tissues and cell types.30 The Hannum estimate of epigenetic age is based on DNA methylation levels at 71 CpG sites and was developed as a predictor of chronological age in whole blood samples.31 Phenotypic epigenetic age—also referred to as PhenoAge—is estimated from DNA methylation levels at 513 CpG sites and was developed as a predictor of mortality risk based on 9 markers of tissue and immune function (albumin, creatinine, serum glucose, C-reactive protein [CRP], lymphocyte percent, mean (red) cell volume, red cell distribution width, alkaline phosphatase, and white blood cell count) and chronological age in whole blood samples.32 GrimAge is estimated from DNA methylation levels at 1,030 total CpG sites and was developed as a predictor of time to death based on 7 DNA methylation surrogates of plasma proteins associated with physiological risk and stress factors (adrenomedullin, beta-2 microglobulin, cystatin C, growth differentiation factor 15 [GDF-15], leptin, plasminogen activation inhibitor 1 [PAI-1], tissue inhibitor metalloproteinase 1 [TIMP-1]) and a DNA methylation-based estimator of smoking pack years.33 DunedinPoAm38 is estimated from DNA methylation levels at 46 CpG sites and was developed to estimate an individual’s rate of biological aging, expressed in years of epigenetic aging per chronological year. DunedinPoAm38 is based on a composite estimate of change in 18 biomarkers of organ-system integrity assessed over a 12-year period in the Dunedin cohort study.34
2.5. Covariates
Several variables that might affect epigenetic aging estimates were evaluated as covariates in the main analyses based on previous research,32,33,36,48,49 including age, biological sex (male as the reference group), and self-identified race/ethnicity (non-Hispanic Black, Hispanic, and non-Hispanic other race, with non-Hispanic white as the reference group). Secondary analyses also considered educational attainment (less than high school, high school diploma or GED, and some college, with a college diploma or higher as the reference group), BMI category (25 to < 30 as overweight, 30 to < 35 as obese I, and ≥ 35 as obese II, with < 25 as normal or underweight as the reference group), smoking status (current and past, with never as the reference group), and alcohol use (1–4 drinks per day, and 5+ drinks per day, with none as the reference group). Post-hoc sensitivity analyses evaluated physical activity as an additional covariate: Participants reported how often they engaged in mild, moderate, and vigorous physical activity, with responses ranging from 1 (hardly ever or never) to 4 (more than once a week or every day), and items were averaged to create a composite score representing the frequency of any type of physical activity. Given that variations in blood cell composition may influence the estimation of and account for some age-related differences in epigenetic aging,36,47 an additional set of post-hoc sensitivity analyses evaluated the percentage of monocyte, natural killer (NK) cell, B cell, and T cell (CD4 total, CD8 naïve, CD8 total) subsets assessed using flow cytometry and neutrophils assessed using hematology complete blood count.
2.6. Data analysis plan
To examine whether having a specific type of social relationship (spouse, children, other family, friends) was associated with epigenetic aging, we first conducted generalized linear models (GLMs) for each epigenetic aging measure that included participants’ relationship status for each relationship type (present vs. absent) as separate predictors, adjusting for chronological age, biological sex, and self-identified race/ethnicity. Next, to examine whether social relationship quality was associated with epigenetic aging, we conducted a second set of models that included each social support or strain measure as separate predictors, adjusting for chronological age, biological sex, and self-identified race/ethnicity. We then applied a 5% false discovery rate (FDR) correction for multiple testing50 across the five epigenetic aging measures for each social domain.
We performed an additional set of models as secondary analyses that further adjusted for educational attainment and lifestyle factors, including BMI category, smoking status, and alcohol use and post-hoc analyses that adjusted for physical activity and cell subsets. Observations were weighted to be nationally representative of community dwelling older U.S. adults using sampling weights provided by HRS. For participants who did not have specific weights for the 2016 Venous Blood Sample, weights from the 2016 HRS core interview were used. All analyses were conducted in R 4.1.3 “One Push-Up” using the tidyverse, jtools, and survey packages.51–53
3. Results
3.1. Preliminary analyses
Sample characteristics appear in Table 1. Approximately two thirds of participants in the sample were married (65.1%), and a majority had children (85.9%), other family members (93.4%), and friends (91.2%). On average, participants reported slightly higher support than strain for all relationship types. The average epigenetic age for the clock measures (Horvath, Hannum, PhenoAge, GrimAge) ranged from 54.0 to 67.2 and the average DunedinPoAm8 was 1.1 years of epigenetic age for each year of chronological age. Social support and strain variables were moderately correlated (r = .10 to .51; Supplemental Table 1). Epigenetic aging variables were low to highly correlated (r = .12 to .77; Supplemental Table 2), which is consistent with other studies.54,55
Table 1.
Sample characteristics for older adults in the Health and Retirement Study with epigenetic aging data (N = 3,647).
| Mean / % | SD | Range | |
|---|---|---|---|
| Chronological age, years | 68.7 | 9.3 | 50−100 |
| Biological sex | |||
| Female | 55.1 | ||
| Male | 44.9 | ||
| Self−identified race/ethnicity | |||
| Black, non−Hispanic | 9.3 | ||
| Hispanic | 7.9 | ||
| Other race, non−Hispanic | 3.3 | ||
| White, non−Hispanic | 79.5 | ||
| Educational attainment | |||
| Less than high school | 13.1 | ||
| High school diploma or GED | 30.2 | ||
| Some college | 26.0 | ||
| College diploma or higher | 30.7 | ||
| BMI category | |||
| Normal or underweight (<25) | 27.0 | ||
| Overweight (25 to < 30) | 37.3 | ||
| Obese I (30 to < 35) | 22.3 | ||
| Obese II (>35) | 13.4 | ||
| Smoking status | |||
| Never | 44.7 | ||
| Current | 10.4 | ||
| Former | 44.8 | ||
| Alcohol use | |||
| None | 56.2 | ||
| 1−4 drinks per day | 41.2 | ||
| 5+ drinks per day | 0.03 | ||
| Physical activity | 2.9 | 0.9 | 1−4 |
| Relationship status | |||
| Has a spouse | 65.1 | ||
| Has children | 85.9 | ||
| Has other family | 93.4 | ||
| Has friends | 91.2 | ||
| Social support | |||
| Spousal support | 3.4 | 0.7 | 1−4 |
| Child support | 3.2 | 0.8 | 1−4 |
| Other family support | 2.8 | 0.9 | 1−4 |
| Friend support | 3.0 | 0.8 | 1−4 |
| Social strain | |||
| Spousal strain | 2.0 | 0.7 | 1−4 |
| Child strain | 1.7 | 0.7 | 1−4 |
| Other family strain | 1.6 | 0.6 | 1−4 |
| Friend strain | 1.4 | 0.5 | 1−4 |
| Epigenetic aging variables | |||
| Horvath | 65.2 | 9.4 | 29.9−114.5 |
| Hannum | 54.0 | 8.9 | 32.0−107.8 |
| PhenoAge | 56.8 | 9.9 | 27.4−101.7 |
| GrimAge | 67.2 | 8.5 | 45.6−99.6 |
| DunedinPoAm38 | 1.1 | 0.1 | 0.8−1.4 |
Note. DunedinPoAm38 = Dunedin Pace of Aging methylation; BMI = body mass index.
3.2. Spousal relationship status and quality
Consistent with hypotheses, being married was associated with a younger GrimAge (unstandardized b = −0.822, SE = 0.219, p <.001) and a slower DunedinPoAm38 (b = −0.012, SE = 0.004, p = .004), and these associations remained statistically significant following FDR correction (Table 2). Being married was not associated with the Horvath, Hannum, or PhenoAge measures.
Table 2.
Generalized linear models with spousal relationship status, support, and strain predicting epigenetic aging.
| Horvath | Hannum | PhenoAge | GrimAge | DunedinPoAm38 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model | β | b(SE) | p | β | b(SE) | p | β | b(SE) | p | β | b(SE) | p | β | b(SE) | p |
| Spousal status (has a spouse) | |||||||||||||||
| Model 1 | −0.001 | −0.018 (0.2S0) | 0.94 | −0.001 | 0.021 (0.203) | 0.92 | −0.018 | 0.386 (0.282) | 0.18 | −0.045 | 0.822 (0.219) | <0.001* | 0.065 | −0.012 (0.004) | 0.004* |
| Model 2 | 0.002 | 0.031 (0.272) | 0.91 | 0.004 | 0.078 (0.205) | 0.70 | −0.011 | −0.233 (0.280) | 0.41 | 0.009 | 0.165 (0.217) | 0.45 | −0.015 | −0.003 (0.004) | 0.40 |
| Spousal support | |||||||||||||||
| Model 1 | 0.002 | 0.035 (0.228) | 0.88 | −0.028 | −0.380 (0.184) | 0.04 | −0.041 | −0.604 (0.248) | 0.02* | −0.026 | −0.333 (0.125) | 0.01* | −0.064 | −0.008 (0.003) | 0.02* |
| Model 2 | 0.002 | 0.030 (0.221) | 0.89 | −0.025 | 0.339 (0.172) | 0.06 | −0.036 | −0.528 (0.248) | 0.04 | 0.008 | 0.096 (0.123) | 0.44 | −0.035 | −0.005 (0.003) | 0.15 |
| Spousal strain | |||||||||||||||
| Model 1 | 0.001 | 0.014 (0.221) | 0.95 | 0.026 | 0.348 (0.142) | 0.02 | 0.012 | 0.167 (0.215) | 0.44 | −0.012 | 0.150 (0.136) | 0.28 | 0.026 | 0.003 (0.003) | 0.32 |
| Model 2 | −0.002 | −0.026 (0.221) | 0.91 | 0.022 | 0.294 (0.138) | 0.04 | 0.007 | 0.105 (0.218) | 0.63 | −0.017 | 0.208 (0.115) | 0.08 | 0.018 | 0.002 (0.003) | 0.46 |
Note, b = unstandardized coefficient. Bold font denotes statistically significant associations. Asterisks indicate statistically significant associations following false discovery rate correction. Model 1 adjusts for chronological age, biological sex, and self-identified race/ethnicity, Model 2 adjusts for Model 1 variables as well as educational attainment, body mass index category, smoking status, and alcohol use. The sample size for the spousal status models is n = 3,646 (Model 1) and n = 3,371 (Model 2), for the spousal support models is n = 2,714, and for the spousal strain models is n = 2,699.
Among those who were married, greater spousal support was associated with a younger Hannum age (b = −0.380, SE = 0.184, p = .04), PhenoAge (b = −0.604, SE = 0.248, p = .02), and GrimAge (b = −0.333, SE = 0.125, p = .01) and a slower DunedinPoAm38 (b = −0.008, SE = 0.003, p = .02), and associations with GrimAge and DunedinPoAm38 remained statistically significant following FDR correction (Figure 1; Table 2). In secondary analyses that further adjusted for educational attainment and lifestyle factors (BMI category, smoking status, and alcohol use), greater spousal support was associated with a younger PhenoAge (b = −0.528, SE = 0.248, p = .04; however, this association was reduced to non-significance following FDR correction. Spousal support was not associated with the Horvath measure.
Figure 1. Associations between social support and epigenetic aging in the Health and Retirement Study.
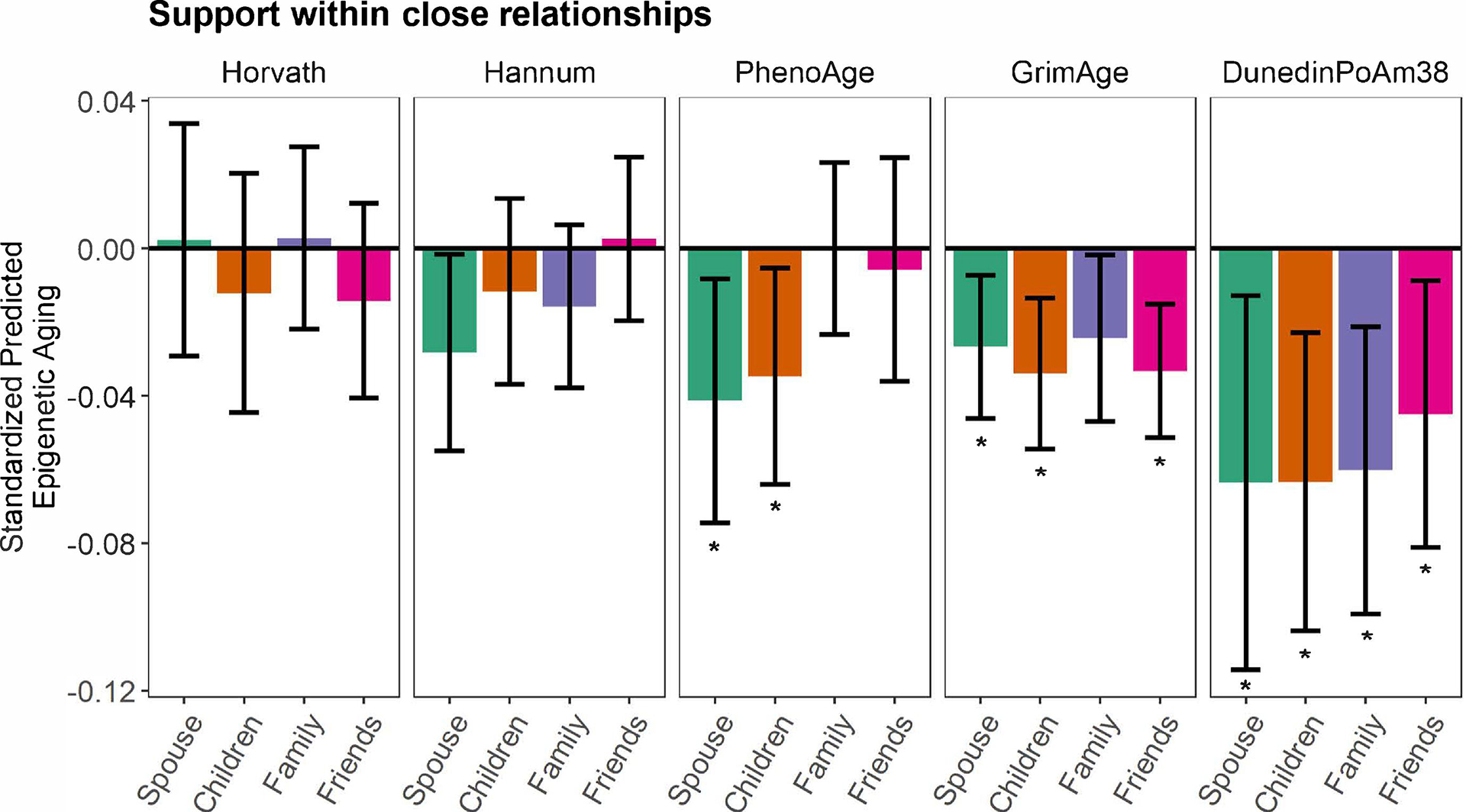
Generalized linear models (GLMs) with standardized coefficients (relationship and epigenetic aging variables were z-scored) showing associations between perceived support from one’s spouse, child, other family members, and friends and epigenetic aging. Models adjusted for chronological age, biological sex, and self-identified race/ethnicity. Error bars represent 95% confidence intervals for each point estimate. Asterisks denote associations that remained statistically significant following false discovery rate correction for multiple testing. Associations between support from friends and family members and GrimAge and DunedinPoAm38, respectively, remained statistically significant in secondary models that further adjusted for educational attainment and lifestyle factors.
Among those who were married, greater spousal strain was associated with an older Hannum age (b = 0.348, SE = 0.142, p = .02); however, this association was reduced to non-significance following FDR correction (Figure 2; Table 2). In secondary analyses that further adjusted for educational attainment and lifestyle factors, greater spousal strain was also associated with an older Hannum age (b = 0.294, SE = 0.138, p = .04), but the association was not significant following FDR correction. Spousal strain was not associated with the Horvath, PhenoAge, GrimAge, or DunedinPoAm38 measures.
Figure 2. Associations between social strain and epigenetic aging in the Health and Retirement Study.
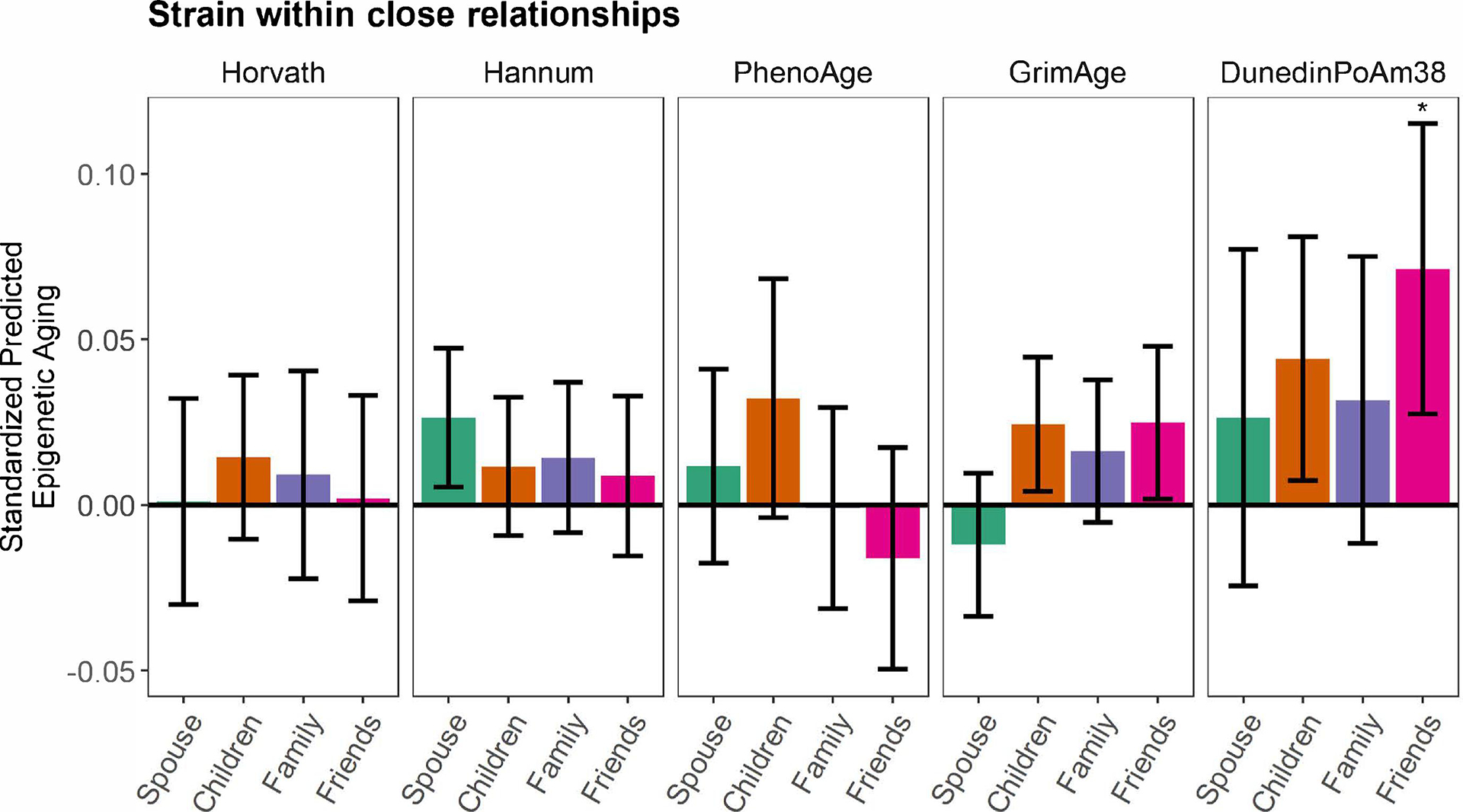
Generalized linear models (GLMs) with standardized coefficients (relationship and epigenetic aging variables were z-scored) showing associations between perceived strain with one’s spouse, child, other family members, and friends and epigenetic aging. Models adjusted for chronological age, biological sex, and self-identified race/ethnicity. Error bars represent 95% confidence intervals for each point estimate. Asterisks denote associations that remained statistically significant following false discovery rate correction for multiple testing. Social strain was not associated with epigenetic aging in secondary models that further adjusted for educational attainment and lifestyle factors.
3.3. Child relationship status and quality
Consistent with hypotheses, having a child was associated with a younger Hannum age (b = −0.582, SE = 0.286, p = .047) and GrimAge (b = −0.588, SE = 0.292, p = .0496); however, these associations were reduced to non-significance following FDR correction (Table 3). In secondary analyses that further adjusted for educational attainment and lifestyle factors, having a child was also associated with a younger GrimAge (b = −0.583, SE = 0.240, p = .02), but the association was not significant following FDR correction. Having a child was not associated with the Horvath, PhenoAge, or DunedinPoAm38 measures.
Table 3.
Generalized linear models with child relationship status, support, and strain predicting epigenetic aging.
| Horvath | Hannum | PhenoAge | GrimAge | DunedinPoAm38 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model | β | b(SE) | p | β | b(SE) | p | β | b(SE) | p | β | b(SE) | p | β | b(SE) | p |
| Child status (has a child) | |||||||||||||||
| Model 1 | −0.022 | −0.653 (0.413) | 0.12 | −0.021 | −0.582 (0.286) | 0.047 | −0.020 | 0.627 (0.406) | 0.13 | −0.022 | −0.588 (0.292) | 0.0496 | −0.026 | −0.007 (0.006) | 0.24 |
| Model 2 | 0.023 | −0.684 (0.427) | 0.12 | −0.020 | −0.570 (0.310) | 0.07 | 0.024 | −0.725 (0.415) | 0.09 | −0.022 | −0.583 (0.240) | 0.02 | −0.025 | −0.007 (0.005) | 0.20 |
| Child support | |||||||||||||||
| Model 1 | −0.012 | −0.156 (0.214) | 0.47 | −0.012 | −0.144 (0.159) | 0.37 | −0.035 | −0.464 (0.201) | 0.03* | −0.034 | −0.389 (0.120) | 0.002* | −0.063 | −0.008 (0.003) | 0.003* |
| Model 2 | −0.010 | −0.124 (0.216) | 0.57 | −0.008 | −0.102 (0.158) | 0.52 | −0.029 | −0.390 (0.203) | 0.06 | −0.022 | −0.247 (0.107) | 0.03 | −0.046 | −0.006 (0.002) | 0.01 |
| Child strain | |||||||||||||||
| Model 1 | 0.014 | 0.213 (0.187) | 0.26 | 0.012 | 0.163 (0.151) | 0.28 | 0.032 | 0.493 (0.283) | 0.09 | 0.024 | 0.319 (0.136) | 0.02 | 0.044 | 0.006 (0.003) | 0.02 |
| Model 2 | 0.011 | 0.163 (0.190) | 0.40 | 0.006 | 0.085 (0.144) | 0.56 | 0.024 | 0.373 (0.291) | 0.21 | 0.005 | 0.063 (0.110) | 0.57 | 0.015 | 0.002 (0.002) | 0.39 |
Note, b = unstandardized coefficient. Bold font denotes statistically significant associations. Asterisks indicate statistically significant associations following false discovery rate correction. Model 1 adjusts for chronological age, biological sex, and self-identified race/ethnicity. Model 2 adjusts for Model 1 variables as well as educational attainment, body mass index category, smoking status, and alcohol use. The sample size for the child status models is n = 3,676 (Model 1) and n = 3,602 (Model 2), for the child support models is n = 3,306, and for the child strain models is n = 3,314.
Among those who had a child, greater support from one’s child was associated with a younger PhenoAge (b = −0.464, SE = 0.201, p = .03) and GrimAge (b = −0.389, SE = 0.120, p = .002) and a slower DunedinPoAm38 (b = −0.008, SE = 0.003, p = .003), and the associations remained statistically significant following FDR correction (Figure 1; Table 3). In secondary analyses that further adjusted for educational attainment and lifestyle factors (BMI category, smoking status, and alcohol use), greater support from one’s child was associated with a younger GrimAge (b = −0.247, SE = 0.107, p = .03) and slower DunedinPoAm38 (b = −0.006, SE = 0.002, p = .01); however, the associations were reduced to non-significance following FDR correction. Support from one’s child was not associated with the Horvath or Hannum measures.
Among those who had a child, greater strain with one’s child was associated with an older GrimAge (b = 0.319, SE = 0.136, p = .02) and DunedinPoAm38 (b = 0.006, SE = 0.003, p = .02); however, the associations were reduced to non-significance following FDR correction (Figure 2; Table 3). Strain with one’s child was not significantly associated with the Horvath, Hannum, or PhenoAge measures in these models. In secondary models that further adjusted for educational attainment and lifestyle factors (BMI category, smoking status, and alcohol use), strain with one’s child was not associated with any of the epigenetic aging measures.
3.4. Other family member relationship status and quality
Consistent with hypotheses, having other family members was associated with a younger Hannum age (b = −0.691, SE = 0.341, p = .048); however, this association was reduced to non-significance following FDR correction (Table 4). In secondary analyses that further adjusted for educational attainment and lifestyle factors, having other family members was also associated with a younger Hannum age (b = −0.701, SE = 0.339, p = .045), but the association was not significant following FDR correction. Having other family members was not associated with the Horvath, PhenoAge, GrimAge, or DunedinPoAm38 measures.
Table 4.
Generalized linear models with other family relationship status, support, and strain predicting epigenetic aging.
| Horvath | Hannum | PheraoAge | GrimAge | DunedinPoAm38 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model | β | b(SE) | p | β | b(SE) | p | β | b(SE) | β | p | b(SE) | p | β | b(SE) | p |
| Other family status (has other family) | |||||||||||||||
| Model 1 | 0.018 | 0.683 (0.506) | 0.18 | −0.019 | −0.691 (0.341) | 0.048 | −0.021 | −0.825 (0.448) | 0.07 | −0.015 | −0.494 (0.419) | 0.24 | −0.033 | −0.012 (0.008) | 0.18 |
| Model 2 | 0.020 | 0.737 (0.513) | 0.16 | −0.020 | −0.701 (0.339) | 0.045 | −0.021 | −0.818 (0.453) | 0.08 | −0.008 | −0.266 (0.347) | 0.45 | −0.021 | −0.008 (0.008) | 0.35 |
| Other family support | |||||||||||||||
| Model 1 | 0.003 | 0.030 (0.137) | 0.83 | −0.016 | −0.164 (0.118) | 0.17 | 0.00003 | 0.0003 (0.136) | 0.998 | −0.024 | −0.237 (0.113) | 0.04 | − 0.060 | −0.006 (0.002) | −0.004* |
| Model 2 | 0.006 | 0.069 (0.136) | 0.61 | −0.012 | −0.125 (0.114) | 0.28 | 0.005 | 0.061 (0.136) | 0.66 | −0.019 | −0.187 (0.082) | 0.03 | −0.051 | −0.005 (0.002) | 0.002* |
| Other family strain | |||||||||||||||
| Model 1 | 0.009 | 0.138 (0.241) | 0.57 | 0.014 | 0.207 (0.167) | 0.22 | −0.001 | −0.015 (0.244) | 0.95 | 0.016 | 0.219 (0.148) | 0.15 | 0.032 | 0.005 (0.003) | 0.16 |
| Model 2 | 0.007 | 0.098 (0.238) | 0.68 | 0.010 | 0.146 (0.169) | 0.39 | −0.007 | −0.107 (0.258) | 0.68 | 0.002 | 0.024 (0.123) | 0.85 | 0.007 | 0.001 (0.003) | 0.72 |
Note, b = standardized coefficient. Bold font denotes statistically significant associations. Asterisks indicate statistically significant associations following false discovery rate correction. Model 1 adjusts for chronological age. biological sex, and self-identified race/ethnicity. Model 2 adjusts for Model 1 variables as well as educational attainment, body mats index category, smoking status, and alcohol use. The sample size for the other family status models is n = 3,710 (Model 1) and n = 3,635, (Model 2), for the other family support models is n = 3,370, and for the other family strain models is n = 3,569.
Among those who had other family members, greater family support was associated with a younger GrimAge (b = −0.237, SE = 0.113, p = .04) and a slower DunedinPoAm38 (b = −0.006, SE = 0.002, p = .004), and the association with DunedinPoAm38 remained statistically significant following FDR correction (Figure 1; Table 4). In secondary analyses that further adjusted for educational attainment and lifestyle factors, greater support from other family members was associated with a younger GrimAge (b = −0.187, SE = 0.082, p = .03) and a slower DunedinPoAm38 (b = −0.005, SE = 0.002, p = .002) and the association with DunedinPoAm38 remained statistically significant following FDR correction. Family support was not associated with the Horvath, Hannum, or PhenoAge measures.
Among those who had other family members, strain within family relationships was not associated with any of the epigenetic aging measures (Figure 2; Table 4).
3.5. Friend relationship status and quality
Consistent with hypotheses, having friends was associated with a younger GrimAge (b = −1.624, SE = 0.344, p <.001) and a slower DunedinPoAm38 (b = −0.019, SE = 0.006, p = .005), and these associations remained statistically significant following FDR correction (Table 5). In secondary analyses that further adjusted for educational attainment and lifestyle factors, having friends was associated with a younger GrimAge (b = −0.814, SE = 0.312, p = .01), but the association was reduced to non-significance following FDR correction. Having friends was not associated with the Horvath, Hannum, or PhenoAge measures.
Table 5.
Generalized linear models with friend relationship status, support, and strain predicting epigenetic aging.
| Horvath | Hannum | PheraoAge | GrimAge | DunedinPoAm38 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model | β | b(SE) | p | β | b(SE) | p | β | b(SE) | p | β | b(SE) | p | β | b(SE) | p |
| Friend status (has friends) | |||||||||||||||
| Model 1 | −0.005 | −0.167 (0.409) | 0.68 | −0.009 | −0.270 (0.312) | 0.39 | −0.013 | 0.453 (0.562) | 0.42 | −0.056 | −1.624 (0.344) | <0.001* | −0.060 | −0.019 (0.006) | 0.005* |
| Model 2 | −0.002 | −0.066 (0.408) | 0.87 | −0.004 | −0.110 (0.294) | 0.71 | −0.008 | 0.273 (0.594) | 0.65 | −0.028 | −0.814 (0.312) | 0.01 | −0.016 | −0.005 (0.006) | 0.44 |
| Friend support | |||||||||||||||
| Model 1 | −0.014 | −0.180 (0.170) | 0.29 | 0.002 | 0.030 (0.137) | 0.83 | −0.006 | −0.077 (0.204) | 0.71 | −0.033 | −0.379 (0.105) | <0.001* | −0.045 | −0.005 (0.002) | 0.02* |
| Model 2 | −0.013 | −0.163 (0.173) | 0.35 | 0.007 | 0.090 (0.145) | 034 | 0.002 | 0.024 (0.215) | 0.91 | −0.023 | −0.264 (0.090) | 0.006* | −0.030 | −0.004 (0.002) | 0.09 |
| Friend strain | |||||||||||||||
| Model 1 | 0.002 | 0.039 (0.306) | 0.90 | 0.009 | 0.163 (0.229) | 0.48 | −0.016 | 0.328 (0.347) | 0.35 | 0.025 | 0.432 (0.204) | 0.04 | 0.071 | 0.013 (0.004) | 0.003* |
| Model 2 | −0.002 | −0.029 (0.301) | 0.92 | 0.002 | 0.036 (0.214) | 0.87 | −0.025 | −0.511 (0.330) | 0.13 | −0.005 | −0.088 (0.165) | 0.60 | 0.024 | 0.004 (0.00−1) | 0.21 |
Note, b = unstandardized coefficient. Bold font denotes statistically significant associations. Asterisks indicate statistically significant associations following false discovery rate correction. Model 1 adjusts for chronological age, biological sex, and self-identified race/ethnicity. Model 2 adjusts for Model 1 variables as well as educational attainment, both’ mass index category, smoking status, and alcohol use. The sample size for the friend status models was n = 3,702 (Model l) and n = 3,627 (Model 2), for the friend support models is n = 3,475, and for the friend strain models is n = 3,473.
Among those who had friends, greater friend support was associated with a younger GrimAge (b = −0.379, SE = 0.105, p < .001) and slower DunedinPoAm38 (b = −0.005, SE = 0.002, p = .02), and these associations remained statistically significant following FDR correction (Figure 1; Table 5). In secondary analyses that further adjusted for educational attainment and lifestyle factors, the association with GrimAge remained statistically significant (b = −0.264, SE = 0.090, p = .006), including following FDR correction. Friend support was not associated with the Horvath, Hannum, or PhenoAge measures.
Among those who had friends, greater strain with friends was associated with an older GrimAge (b = 0.432, SE = 0.204, p = .04) and a faster DunedinPoAm38 (b = 0.013, SE = 0.004, p = .003), and the association with DunedinPoAm38 remained statistically significant following FDR correction (Figure 2; Table 5). Strain with friends was not associated with the Horvath, Hannum, or PhenoAge measures in these models. In secondary analyses that further adjusted for educational attainment and lifestyle factors, strain with friends was not associated with epigenetic aging.
3.6. Post-hoc sensitivity analyses with adjustment for physical activity and cell subsets
In post-hoc sensitivity analyses that further adjusted for physical activity, the pattern of findings remained the same with two exceptions (Supplemental Table 3). Specifically, the association between support from one’s children and a younger GrimAge (b = −0.169, SE = 0.103, p = .11) was reduced to non-significance and the association between support from friends and a younger GrimAge (b = −0.217, SE = 0.093, p = .03) was no longer statistically significant with the FDR correction.
In a second set of post-hoc sensitivity analyses that adjusted for cell subsets, the overall pattern of findings was similar, and the magnitude of associations with epigenetic aging increased for some of the relationship variables (Supplemental Table 4). Specifically, several associations that were statistically or marginally significant in previous models increased in magnitude such that they remained statistically significant following FDR correction. For instance, having other family members was associated with a younger Hannum age (b = −0.847, SE = 0.332, p = .02) and PhenoAge (b = 1.261, SE = 0.476, p = .01), and having friends was associated with a younger GrimAge (b = −0.795, SE = 0.259, p = .004). In addition, greater support from one’s child was associated with a younger PhenoAge (b = −0.544, SE = 0.202, p = .01) and slower DunedinPoAm38 (b = −0.006, SE = 0.002, p = .01), whereas greater strain with one’s spouse was associated with an older Hannum age (b = 0.415, SE = 0.136, p = .004). Also of note, the association between support from other family members and a slower DunedinPoAm38 (b = −0.005, SE = 0.002, p = .01) was reduced to marginal significance following FDR correction in models that adjusted for cell subsets.
4. Discussion
The present study investigated associations between social relationships and epigenetic aging in a nationally representative sample of older adults in the Health and Retirement Study. Specifically, we examined whether the absence of close relationships as well as lower perceived support and higher strain in existing relationships with one’s spouse, child, other family members, and friends were associated with an accelerated epigenetic aging profile. As hypothesized, older adults who did not have a spouse or friend relationships were biologically older based on GrimAge and DunedinPoAm38 estimates than their peers who were married or had friendships. Individuals who reported lower support—feeling less understood, that they could not rely upon, and/or that they could not open up—in relationships with their spouse, child, family members, or friends had an accelerated epigenetic aging profile based on PhenoAge, GrimAge, and DunedinPoAm38 estimates relative to same-aged peers who experienced greater support. Specifically, the difference in epigenetic age between older adults who reported the lowest and highest levels of support ranged from 1.02 to 1.83 years, depending on the relationship type and the epigenetic measure. In addition, individuals who reported that their friends made too many demands on them, criticized them, let them down, and/or got on their nerves had an accelerated aging profile based on DunedinPoAm38 estimates relative to same-aged peers who experienced less strain. It was somewhat surprising that social support and strain were less consistently or not at all associated with the “first generation” Horvath and Hannum clocks, although these measures were developed to predict chronological age alone and are likely capturing different aspects of the aging process than the “second generation” PhenoAge, GrimAge, and DunedinPoAm38 measures, which were developed based on biomarkers that are predictive of morbidity and mortality.48 Given that previous research has linked relationship quality to age-related conditions such as functional limitations,7,8 poorer physical and cognitive functioning,9–11 cardiovascular disease,12–14 and mortality,1,15,16 these findings suggest that the DNA methylation patterns captured by these “second generation” clocks may serve as a plausible mechanism through which relationship processes influence aging and health.
In secondary analyses that also adjusted for educational attainment and lifestyle factors (BMI, smoking status, alcohol use) that have been associated with epigenetic aging in prior research,32,33,36,49 relationship quality was more robustly associated with epigenetic aging than relationship status, with stronger effects for social support than for social strain. Specifically, lower support from friends and family members (other than a spouse or child) was associated with an older GrimAge and faster DunedinPoAm38, respectively, over and above these well-established health risk factors, although the effect sizes for associations with social support (β = −0.023 and −0.051, respectively) were typically smaller than the effect sizes for educational attainment (0.122 to 0.199) and lifestyle factors (BMI: 0.004 to 0.358; smoking status: 0.229 to 1.349; alcohol use: −0.029 to 0.157) in models that adjusted for all the factors. In post-hoc sensitivity analyses that further adjusted for physical activity, the magnitude of the association between support from friends and GrimAge was reduced slightly and did not remain statistically significant with FDR correction. Overall, the pattern of findings from these secondary analyses suggest that lifestyle factors may mediate associations between social relationship status and quality and epigenetic aging, although this remains to be empirically tested. Interestingly, adjustment for cell subsets strengthened some of the associations, such that having other family members was significantly associated with a younger Hannum age and PhenoAge and having friends was associated with a younger GrimAge. In addition, lower support from one’s child was associated with an older PhenoAge and faster DunedinPoAm38, whereas greater strain with one’s spouse was associated with an older Hannum age. These results suggest that individual variations in blood cell composition observed in whole blood (particularly neutrophils, B cells, and CD8 naïve cells)—which fluctuate in response to biological and psychosocial conditions and the aging process itself56–59—can influence estimation of epigenetic aging, and that not accounting for these variations may in some cases obscure associations with social factors.
These findings point to the importance of friend and family relationships for older adults and are consistent with prior research with population-based samples of middle-aged and older adults, which found that strain with family members was more strongly associated with a higher inflammatory burden than strain within other relationships38 and support from friends (but not other relationship types) predicted a slower DunedinPoAm38 approximately 10 years later in a smaller sample of HRS participants (effect size: β = −0.07).39 Our results that GrimAge and DunedinPoAm38 were most robustly associated with relationship quality are particularly noteworthy in a sample of older adults who may be experiencing or beginning to experience declines in their health, given that these measures are predictive of multiple age-related conditions such as declines in cognitive and physical function, cancer, and cardiovascular disease, as well as time to death.32,33 In addition, relationships with close others become more salient, and in some cases, more stable or involuntary (e.g., more difficult to choose to exit) in older adulthood as individuals who experience health declines may rely more on others for support.60,61 Therefore, the experience of being in close relationships—particularly with family and friends—that are characterized by lower support may have a particular influence on the health and well-being of older adults—and epigenetic aging may be one mechanism through which this occurs.
Results from this study contribute to a growing literature on the influence of social relationships on key biological aging processes. Previous research has linked social support and strain to peripheral markers of inflammation, activation of transcription factor NF-кB, which regulates the expression of inflammatory genes, and telomere length. These findings also extend an emerging literature on psychosocial stress and epigenetic aging, which has linked exposure to early life adversity62 and traumatic experiences63 to accelerated epigenetic aging in adulthood. Given that the absence of close relationships, as well as low social support and high strain have been considered forms of social stress, the presence and quality of social relationships may affect epigenetic aging through similar stress-related pathways; however, the specific cellular and molecular mechanisms through which stress may impact epigenetic aging are not well understood. In response to stress, chronic or repeated activation of the sympathetic nervous system (SNS) and the hypothalamus-adrenal-pituitary (HPA) axis releases neuroendocrine mediators (e.g., catecholamines, glucocorticoids) that interact with receptors on the surface of cells. Mounting evidence suggests that this stress signaling cascade can initiate multiple biological aging pathways within cells, including those that contribute to DNA damage, telomere attrition, cellular senescence, and inflammation.64 It will be important for future research to begin to delineate the specific biological pathways through which experiences of stress and social adversity (and associated neuroendocrine mediators) may modify DNA methylation and other epigenetic processes to alter rates of aging.
Our results should be considered in light of study limitations, which suggest directions for future research. Most notably, at this time, the Health and Retirement Study has measured epigenetic aging at a single timepoint, which limited the present analyses to concurrent associations and precluded the investigation of the influence of social relationship quality on changes in epigenetic aging over time. On account of this, we were also unable to test an alternative hypothesis that epigenetic aging, as a marker of an underlying aging process, might influence changes in social relationship status and quality. It will be important for future research to examine the directionally of the observed effects and to link these associations with age-related health outcomes at future timepoints. In addition, the social support measures for this study focused primarily on emotional support and did not address other forms of support, such as tangible or informational support. Although this study accounted for the potential contributions of lifestyle factors such as smoking, alcohol use, and physical activity to epigenetic aging, whether these and other behavioral and psychological factors (e.g., depression, stress appraisals)2 mediate associations between social relationship quality and epigenetic aging in older adulthood remains a question for future investigation. Finally, although the HRS 2016 Venous Blood Study includes several validated measures of epigenetic aging, the whole genome DNA methylation data are not currently available, which precluded an analysis of specific or novel DNA methylation sites that may be associated with social relationship quality beyond the epigenetic aging measures.
Despite these limitations, the present study extends the literature on social relationships and biological aging by demonstrating that the absence of close relationships as well as lower support and higher strain in existing relationships are associated with an accelerated epigenetic aging profile in older adults. Furthermore, lower support from family and friends was robustly associated with an accelerated aging profile, over and above well-established lifestyle factors such as smoking status and alcohol use. These findings suggest that epigenetic aging may be a plausible biological mechanism through which social relationship quality might influence aging and age-related health outcomes such as cancer, cardiovascular disease, dementia, and early mortality. In addition, this investigation involved a large, socioeconomically diverse, and nationally representative sample of community dwelling older adults in the United States, which increases generalizability of the findings. In light of emerging evidence that these epigenetic aging mechanisms may be sensitive to and modifiable by behavioral interventions,65,66 these results suggest that close relationship quality—particularly with family members and friends— may represent a behavioral target for intervention in older adulthood that has the potential to prevent, slow, or reverse accelerated aging and extend the healthspan (number of years free from age-related disease and disability) and lifespan.
Supplementary Material
Supplemental Information
Supplemental Table 1. Correlations between the relationship support and strain variables.
Supplemental Table 2. Correlations between the epigenetic aging variables.
Supplemental Table 3. Post-hoc sensitivity analysis with relationship status, support, and strain predicting epigenetic aging, adjusting for physical activity.
Supplemental Table 4. Post-hoc sensitivity analysis with relationship status, support, and strain predicting epigenetic aging, adjusting for cell subsets.
Highlights.
An absence of marital and friend relationships related to greater epigenetic aging.
Lower support from family members and friends related to greater epigenetic aging.
Lower social support more robustly related to epigenetic aging than social strain.
Epigenetic aging may be a mechanism through which relationships influence health.
Acknowledgements
The Health and Retirement Study is sponsored by the National Institute on Aging [U01AG009740] and is conducted by the University of Michigan.
Funding
This work was supported by the USC/UCLA Center on Biodemography and Population Health through a grant from the National Institute on Aging [P30AG017265]. This research was also supported by the National Institute on Aging [R25AG053227, K01AG065485, T32AG00037, R01AGG060110] and the UCLA Cousins Center for Psychoneuroimmunology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
The authors have no conflicts of interest to declare.
A post-hoc sensitivity analysis that excluded participants who provided social relationship data in 2008 suggested a similar overall pattern of findings.
References
- 1.Holt-Lunstad J, Smith TB, Layton JB. Social relationships and mortality risk: A meta-analytic review. PLoS Med. 2010;7(7):e1000316. doi: 10.1371/journal.pmed.1000316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uchino BN, Bowen K, de Grey RK, Mikel J, Fisher EB. Social support and physical health: Models, mechanisms, and opportunities. Principles and Concepts of Behavioral Medicine: A Global Handbook. Published online October 8, 2018:341–372. doi: 10.1007/978-0-387-93826-4_12 [DOI] [Google Scholar]
- 3.Cohen S Social relationships and health. American Psychologist. 2004;59(8):676–684. doi: 10.1037/0003-066X.59.8.676 [DOI] [PubMed] [Google Scholar]
- 4.Pietromonaco PR, Collins NL. Interpersonal mechanisms linking close relationships to health. American Psychologist. 2017;72(6):531–542. doi: 10.1037/amp0000129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birmingham WC, Holt-Lunstad J. Social aggravation: Understanding the complex role of social relationships on stress and health-relevant physiology. International Journal of Psychophysiology. 2018;131:13–23. doi: 10.1016/j.ijpsycho.2018.03.023 [DOI] [PubMed] [Google Scholar]
- 6.Brooks KP, Dunkel Schetter C. Social Negativity and Health: Conceptual and Measurement Issues. Soc Personal Psychol Compass. 2011;5(11):904–918. doi: 10.1111/j.1751-9004.2011.00395.x [DOI] [Google Scholar]
- 7.Newsom JT, Mahan TL, Rook KS, Krause N. Stable Negative Social Exchanges and Health. Health Psychology. 2008;27(1):78–86. doi: 10.1037/0278-6133.27.1.78 [DOI] [PubMed] [Google Scholar]
- 8.Mavandadi S, Rook KS, Newsom JT. Positive and negative social exchanges and disability in later life: An investigation of trajectories of change. Journals of Gerontology - Series B Psychological Sciences and Social Sciences. 2007;62(6):S361–S370. doi: 10.1093/geronb/62.6.S361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seeman T, Chen X. Risk and protective factors for physical functioning in older adults with and without chronic conditions: MacArthur studies of successful aging. Journals of Gerontology - Series B Psychological Sciences and Social Sciences. 2002;57(3):S135–S144. doi: 10.1093/geronb/57.3.S135 [DOI] [PubMed] [Google Scholar]
- 10.Tun PA, Miller-Martinez D, Lachman ME, Seeman T. Social strain and executive function across the lifespan: The dark (and light) sides of social engagement. Aging, Neuropsychology, and Cognition. 2013;20(3):320–338. doi: 10.1080/13825585.2012.707173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seeman TE, Miller-Martinez DM, Stein Merkin S, Lachman ME, Tun PA, Karlamangla AS. Histories of social engagement and adult cognition: midlife in the U.S. study. J Gerontol B Psychol Sci Soc Sci. 2011;66 Suppl 1. doi: 10.1093/geronb/gbq091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Vogli R, Chandola T, Marmot MG. Negative aspects of close relationships and heart disease. Arch Intern Med. 2007;167(18):1951–1957. doi: 10.1001/archinte.167.18.1951 [DOI] [PubMed] [Google Scholar]
- 13.Wang HX, Mittleman MA, Orth-Gomer K. Influence of social support on progression of coronary artery disease in women. Soc Sci Med. 2005;60(3):599–607. doi: 10.1016/J.SOCSCIMED.2004.05.021 [DOI] [PubMed] [Google Scholar]
- 14.Orth-Gomer K, Rosengren A, Wilhelmsen L. Lack of social support and incidence of coronary heart disease in middle- aged Swedish men. Psychosom Med. 1993;55(1):37–43. doi: 10.1097/00006842-199301000-00007 [DOI] [PubMed] [Google Scholar]
- 15.Kroenke CH, Quesenberry C, Kwan ML, Sweeney C, Castillo A, Caan BJ. Social networks, social support, and burden in relationships, and mortality after breast cancer diagnosis in the Life after Breast Cancer Epidemiology (LACE) Study. Breast Cancer Res Treat. 2013;137(1):261–271. doi: 10.1007/s10549-012-2253-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Birditt K, Antonucci TC. Life sustaining irritations? Relationship quality and mortality in the context of chronic illness. Soc Sci Med. 2008;67(8):1291–1299. doi: 10.1016/j.socscimed.2008.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinquart M, Duberstein PR. Associations of social networks with cancer mortality: A meta-analysis. Crit Rev Oncol Hematol. 2010;75(2):122–137. doi: 10.1016/j.critrevonc.2009.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boen CE, Barrow DA, Bensen JT, et al. Social relationships, inflammation, and cancer survival. Cancer Epidemiology Biomarkers and Prevention. 2018;27(5):541–549. doi: 10.1158/1055-9965.EPI-17-0836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanne D, Goldbourt U, Medalie JH. Perceived Family Difficulties and Prediction of 23-Year Stroke Mortality among Middle-Aged Men. Cerebrovasc Dis. 2004;18:277–282. doi: 10.1159/000080352 [DOI] [PubMed] [Google Scholar]
- 20.Barth J, Schneider S, von Känel R. Lack of social support in the etiology and the prognosis of coronary heart disease: A systematic review and meta-analysis. Psychosom Med. 2010;72(3):229–238. doi: 10.1097/PSY.0B013E3181D01611 [DOI] [PubMed] [Google Scholar]
- 21.Elliot AJ, Heffner KL, Mooney CJ, Moynihan JA, Chapman BP. Social Relationships and Inflammatory Markers in the MIDUS Cohort: The Role of Age and Gender Differences. J Aging Health. 2018;30(6):904–923. doi: 10.1177/0898264317698551 [DOI] [PubMed] [Google Scholar]
- 22.Uchino BN, Trettevik R, Kent de Grey RG, Cronan S, Hogan J, Baucom BRW. Social support, social integration, and inflammatory cytokines: A meta-analysis. Health Psychology. 2018;37(5):462–471. doi: 10.1037/hea0000594 [DOI] [PubMed] [Google Scholar]
- 23.Robles TF, Repetti RL, Reynolds BM, Chung PJ, Arevalo JMG, Cole SW. Family environments and leukocyte transcriptome indicators of a proinflammatory phenotype in children and parents. Dev Psychopathol. 2018;30(1):235–253. doi: 10.1017/S0954579417000591 [DOI] [PubMed] [Google Scholar]
- 24.Whisman MA, Sbarra DA. Marital adjustment and interleukin-6 (IL-6). Journal of Family Psychology. 2012;26(2):290–295. doi: 10.1037/a0026902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiecolt-Glaser JK, Loving TJ, Stowell JR, et al. Hostile marital interactions, proinflammatory cytokine production, and wound healing. Arch Gen Psychiatry. 2005;62(12):1377–1384. doi: 10.1001/archpsyc.62.12.1377 [DOI] [PubMed] [Google Scholar]
- 26.Rentscher KE, Carroll JE, Mitchell C. Psychosocial stressors and telomere length: A current review of the science. Annu Rev Public Health. 2020;41. doi: 10.1146/annurev-publhealth [DOI] [PubMed] [Google Scholar]
- 27.Carroll JE, Diez Roux AV, Fitzpatrick AL, Seeman T. Low Social Support Is Associated With Shorter Leukocyte Telomere Length in Late Life. Psychosom Med. 2013;75(2):171–177. doi: 10.1097/psy.0b013e31828233bf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zalli A, Carvalho LA, Lin J, et al. Shorter telomeres with high telomerase activity are associated with raised allostatic load and impoverished psychosocial resources. Proceedings of the National Academy of Sciences. 2014;111(12):4519–4524. doi: 10.1073/pnas.1322145111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194. doi: 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horvath S DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. doi: 10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hannum G, Guinney J, Zhao L, et al. Genome-wide Methylation Profiles Reveal Quantitative Views of Human Aging Rates. Mol Cell. 2013;49(2):359–367. doi: 10.1016/j.molcel.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levine ME, Lu AT, Quach A, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging. 2018;10(4):573–591. doi: 10.18632/aging.101414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu AT, Quach A, Wilson JG, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging. 2019;11(2):303–327. doi: 10.18632/aging.101684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belsky DW, Caspi A, Arseneault L, et al. Quantification of the pace of biological aging in humans through a blood test, the DunedinPoAm DNA methylation algorithm. Elife. 2020;9:1–56. doi: 10.7554/eLife.54870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferrucci L, Gonzalez-Freire M, Fabbri E, et al. Measuring biological aging in humans: A quest. Aging Cell. 2020;19(2). doi: 10.1111/acel.13080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crimmins EM, Thyagarajan B, Levine ME, Weir DR, Faul J. Associations of Age, Sex, Race/Ethnicity, and Education With 13 Epigenetic Clocks in a Nationally Representative U.S. Sample: The Health and Retirement Study. The Journals of Gerontology: Series A. 2021;76(6):1117–1123. doi: 10.1093/GERONA/GLAB016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fransquet PD, Wrigglesworth J, Woods RL, Ernst ME, Ryan J. The epigenetic clock as a predictor of disease and mortality risk: A systematic review and meta-analysis. Clin Epigenetics. 2019;11(1):1–17. doi: 10.1186/s13148-019-0656-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang YC, Schorpp K, Harris KM. Social support, social strain and inflammation: Evidence from a national longitudinal study of U.S. adults. Soc Sci Med. 2014;107:124–135. doi: 10.1016/J.SOCSCIMED.2014.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hillmann AR, Dhingra R, Reed RG. Positive social factors prospectively predict younger epigenetic age: Findings from the Health and Retirement Study. Psychoneuroendocrinology. 2023;148. doi: 10.1016/J.PSYNEUEN.2022.105988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nomaguchi K, Milkie MA. Parenthood and Well-Being: A Decade in Review. Journal of Marriage and Family. 2020;82(1):198–223. doi: 10.1111/JOMF.12646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Umberson D, Pudrovska T, Reczek C. Parenthood, Childlessness, and Well-Being: A Life Course Perspective. Journal of Marriage and Family. 2010;72(3):612–629. doi: 10.1111/J.1741-3737.2010.00721.X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Fletcher J. Parental status in later life and parents’ risk of cognitive impairment. SSM Popul Health. 2021;16:100968. doi: 10.1016/J.SSMPH.2021.100968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Modig K, Talbäck M, Torssander J, Ahlbom A. Payback time? Influence of having children on mortality in old age. J Epidemiol Community Health. 2017;71(5):424–430. doi: 10.1136/JECH-2016-207857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ning K, Zhao L, Franklin M, et al. Parity is associated with cognitive function and brain age in both females and males. Scientific Reports 2020 10:1. 2020;10(1):1–9. doi: 10.1038/s41598-020-63014-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Read SL, Grundy EMD. Fertility History and Cognition in Later Life. The Journals of Gerontology: Series B. 2017;72(6):1021–1031. doi: 10.1093/GERONB/GBW013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crimmins EM, Kim JK, Fisher J, Faul JD. HRS Epigenetic Clocks.; 2020.
- 47.Crimmins EM, Faul JD, Thyagarajan B, Weir DR. Venous Blood Collection and Assay Protocol in the 2016 Health and Retirement Study 2016 Venous Blood Study (VBS).; 2017. https://hrsdata.isr.umich.edu/sites/default/files/documentation/data-descriptions/HRS2016VBSDD.pdf
- 48.Oblak L, van der Zaag J, Higgins-Chen AT, Levine ME, Boks MP. A systematic review of biological, social and environmental factors associated with epigenetic clock acceleration. Ageing Res Rev. 2021;69:101348. doi: 10.1016/J.ARR.2021.101348 [DOI] [PubMed] [Google Scholar]
- 49.Beach SRH, Dogan M v., Lei MK, et al. Methylomic Aging as a Window onto the Influence of Lifestyle: Tobacco and Alcohol Use Alter the Rate of Biological Aging. J Am Geriatr Soc. 2015;63(12):2519–2525. doi: 10.1111/jgs.13830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society: Series B (Methodological). 1995;57(1):289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 51.Wickham H, Averick M, Bryan J, et al. Welcome to the Tidyverse. J Open Source Softw. 2019;4(43):1686. doi: 10.21105/joss.01686 [DOI] [Google Scholar]
- 52.Long J jtools: Analysis and Presentation of Social Scientific Data. Published online 2020.
- 53.Lumley T Analysis of complex survey samples. J Stat Softw. 2004;9:1–19. doi: 10.18637/jss.v009.i08 [DOI] [Google Scholar]
- 54.Belsky DW, Moffitt TE, Cohen AA, et al. Eleven Telomere, Epigenetic Clock, and Biomarker-Composite Quantifications of Biological Aging: Do They Measure the Same Thing? Am J Epidemiol. 2018;187(6):1220–1230. doi: 10.1093/AJE/KWX346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li X, Ploner A, Wang Y, et al. Longitudinal trajectories, correlations and mortality associations of nine biological ages across 20-years follow-up. Elife. 2020;9. doi: 10.7554/ELIFE.51507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hawkley LC, Cacioppo JT. Stress and the aging immune system. Brain Behav Immun. 2004;18(2):114–119. doi: 10.1016/J.BBI.2003.09.005 [DOI] [PubMed] [Google Scholar]
- 57.Dhabhar FS. Effects of stress on immune function: the good, the bad, and the beautiful. Immunologic Research 2014 58:2. 2014;58(2):193–210. doi: 10.1007/S12026-014-8517-0 [DOI] [PubMed] [Google Scholar]
- 58.Schedlowski M, Jacobs R, Stratmann G, et al. Changes of natural killer cells during acute psychological stress. J Clin Immunol. 1993;13(2):119–126. doi: 10.1007/BF00919268/METRICS [DOI] [PubMed] [Google Scholar]
- 59.Herbert TB, Cohen S. Stress and immunity in humans: A meta-analytic review. Psychosom Med. 1993;55(4):364–379. doi: 10.1097/00006842-199307000-00004 [DOI] [PubMed] [Google Scholar]
- 60.Uchino BN. Understanding the Links Between Social Support and Physical Health: A Life-Span Perspective With Emphasis on the Separability of Perceived and Received Support. Perspectives on Psychological Science. 2009;4(3):236–255. doi: 10.1111/j.1745-6924.2009.01122.x [DOI] [PubMed] [Google Scholar]
- 61.Wrzus C, Hänel M, Wagner J, Neyer FJ. Social network changes and life events across the life span: A meta-analysis. Psychol Bull. 2013;139(1):53–80. doi: 10.1037/a0028601 [DOI] [PubMed] [Google Scholar]
- 62.Palma-Gudiel H, Fañanás L, Horvath S, Zannas AS. Psychosocial stress and epigenetic aging. Int Rev Neurobiol. 2020;150:107–128. doi: 10.1016/BS.IRN.2019.10.020 [DOI] [PubMed] [Google Scholar]
- 63.Wolf EJ, Maniates H, Nugent N, et al. Traumatic stress and accelerated DNA methylation age: A meta-analysis. Psychoneuroendocrinology. 2018;92:123–134. doi: 10.1016/j.psyneuen.2017.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Polsky LR, Rentscher KE, Carroll JE. Stress-induced biological aging: A review and guide for research priorities. Brain Behav Immun. 2022;104:97–109. doi: 10.1016/J.BBI.2022.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brody GH, Yu T, Chen E, Beach SRH, Miller GE. Family-centered prevention ameliorates the longitudinal association between risky family processes and epigenetic aging. J Child Psychol Psychiatry. 2016;57(5):566–574. doi: 10.1111/jcpp.12495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Waziry R, Ryan CP, Corcoran DL, et al. Effect of long-term caloric restriction on DNA methylation measures of biological aging in healthy adults from the CALERIE trial. Nature Aging 2023 3:3. 2023;3(3):248–257. doi: 10.1038/s43587-022-00357-y [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Information
Supplemental Table 1. Correlations between the relationship support and strain variables.
Supplemental Table 2. Correlations between the epigenetic aging variables.
Supplemental Table 3. Post-hoc sensitivity analysis with relationship status, support, and strain predicting epigenetic aging, adjusting for physical activity.
Supplemental Table 4. Post-hoc sensitivity analysis with relationship status, support, and strain predicting epigenetic aging, adjusting for cell subsets.


