Abstract
Dyslipidemia is an independent and modifiable risk factor for aging and age-related disorders. Routine lipid panel cannot capture all individual lipid species in blood (i.e., blood lipidome). To date, a comprehensive assessment of the blood lipidome associated with mortality is lacking in large-scale community-dwelling individuals, especially in a longitudinal setting. Using liquid chromatograph-mass spectrometry, we repeatedly measured individual lipid species in 3,821 plasma samples collected at two visits (~ 5.5 years apart) from 1,930 unique American Indians in the Strong Heart Family Study. We first identified baseline lipids associated with risks for all-cause mortality and CVD mortality (mean follow-up period: 17.8 years) in American Indians, followed by replication of top hits in European Caucasians in the Malmö Diet and Cancer-Cardiovascular Cohort (n = 3,943, mean follow-up period: 23.7 years). The model adjusted age, sex, BMI, smoking, hypertension, diabetes, and LDL-c at baseline. We then examined the associations between changes in lipid species and risk of mortality. Multiple testing was controlled by false discovery rate (FDR). We found that baseline levels and longitudinal changes of multiple lipid species, e.g., cholesterol esters, glycerophospholipids, sphingomyelins, and triacylglycerols, were significantly associated with risks of all-cause or CVD mortality. Many lipids identified in American Indians could be replicated in European Caucasians. Network analysis identified differential lipid networks associated with risk of mortality. Our findings provide novel insight into the role of dyslipidemia in disease mortality and offer potential biomarkers for early prediction and risk reduction in American Indians and other ethnic groups.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11357-023-00793-7.
Keywords: All-cause mortality, CVD mortality, Longitudinal lipidomics, Metabolomics, American Indians, Strong Heart Study
Introduction
Dyslipidemia is defined as having a high level of plasma triglycerides, total cholesterol, or low-density lipoprotein cholesterol (LDL-c), and/or a low level of high-density lipoprotein cholesterol (HDL-c). Dyslipidemia is an independent and one of the most important modifiable risk factors for atherosclerosis cardiovascular diseases (ACVD) [1, 2], which are the major contributors to mortality worldwide [3]. According to the 2015–2018 National Health and Nutrition Examination Survey (NHANES), adults aged 40–59 years have the highest prevalence of high total cholesterol (15.7%), followed by those aged ≥ 60 years (11.4%), and lowest among those aged 20–39 years (7.5%) [4]. In addition, the prevalence of dyslipidemia varies greatly among different racial/ethnic groups [5, 6]. Non-Hispanic whites had the highest prevalence of dyslipidemia (30.4%), followed by African Americans (30%), Hispanic (29.3%) and Chinese Americans (23.9%) [7]. Moreover, there is a trend over the recent decades towards an increase in the prevalence of dyslipidemia among all ethnic groups [6, 8], including American Indians [9], paralleled to changes in lifestyle. Compared to other ethnic groups, few studies have reported the prevalence and incidence of dyslipidemia among American Indians [9], a minority group suffering from high rates of obesity, diabetes, cardiovascular diseases (CVD) and other metabolic disorders (e.g., chronic kidney disease) [10, 11].
Epidemiological studies have shown that individuals with metabolic disorders suffer from a higher mortality rate than those without [12, 13]. Deciphering the metabolic pathways underlying the association between dysregulated lipid metabolism and mortality may lead to novel biomarkers for risk prediction, stratification, and early intervention. However, routine lipid panel cannot capture all molecular lipids in blood (i.e., blood lipidome), and thus has limited ability to detect perturbed lipid metabolism associated with diseases and mortality. Lipidomics is an emerging high-throughput biochemical technique that can identify and quantify hundreds to thousands of molecular lipid species in a biospecimen. Using this technology, several studies have reported associations of altered blood lipid species, e.g., phosphatidylcholines, ceramides, and phosphatidylethanolamines, with all-cause or disease-specific mortality in human populations [14–19]. However, existing studies were almost exclusively cross-sectional and were largely limited by smaller sample size and/or low coverage of the blood lipidome. To date, no large-scale epidemiological studies have examined the relationship between plasma lipidome and mortality in American Indians. The temporal relationship between change in blood lipid species and risk of all-cause or CVD mortality is also lacking in any racial/ethnic group.
Leveraging the lipidomic data generated in 3,821 fasting blood samples from 1,930 unique American Indians attending two clinical examinations (mean follow-up: 5.5 years) in the Strong Heart Family Study (SHFS), here we report findings from the first large-scale longitudinal lipidomic profiling of all-cause and CVD mortality in American Indians, followed by replication in European Caucasians in the Malmö Diet and Cancer-Cardiovascular Cohort (MDC-CC). Our primary goals are to (1) identify individual lipid species associated with risk of all-cause and CVD mortality beyond traditional risk factors, and (2) examine the temporal relationship between longitudinal changes in plasma lipidome and risk of all-cause and CVD mortality in American Indians.
Materials and methods
Study populations
The Strong Heart Family Study (SHFS, 2001-ongoing), a sub-cohort of the Strong Heart Study (SHS, 1989-ongoing), is a family-based prospective study designed to identify genetic and lifestyle factors for CVD and its risk factors in American Indians [20–22]. Briefly, 2,786 tribal members (aged 14) in three geographic regions (Arizona, North/South Dakota, Oklahoma) were initially examined in 2001–2003 and re-examined in 2006–2009 (mean 5.5 years apart) using the same protocols. Information for demography, family history, medical records, and lifestyle was collected at each visit. A total of 1,930 individuals (mean age at baseline: 40.4) with complete information for clinical and lipidomic data was included in the current analysis. More information for covariate assessments was described elsewhere [23]. All participants provided informed consents. The SHFS protocols were approved by the Institutional Review Boards of the participating institutions and the American Indian tribes.
The Malmö Diet and Cancer-Cardiovascular Cohort (MDC-CC), a sub-cohort of the Malmö Diet and Cancer Study [24], is a prospective population-based cohort designed to study the epidemiology of carotid artery disease (CAD) and its risk factors in European Caucasians in Sweden [25]. A total of 3,943 participants (mean age at baseline: 57.7) with complete information for clinical phenotypes and lipidomic data was included in the current analysis. All MDC-CC participants provided written informed consent, and the study protocols were approved by the Ethics Committee at Lund University.
Study follow-up and ascertainment of mortality
In the SHFS, baseline information was collected in 2001–2003 and all living participants were followed through December 31, 2020. Detailed methods for ascertainment of deaths and causes of deaths in the SHFS have been described previously [20, 21, 26]. Briefly, death of a participant was identified from the Indian Health Service hospital records and direct contact with the family members. The cause of death was determined by physicians on the SHS Mortality Review Committee using medical records, autopsy reports, and informant interview. Information on mortality was retrieved from the mortality decision form used for cardiovascular disease surveillance in the SHS. The cause of death was determined independently by two members of the SHS Mortality Committee after reviewing information including death certificates and medical records including pathology reports and informant interviews, when needed. Death certificate codes were recorded according to the International Classification of Diseases Injuries, and Causes of Death, 9th Revision (ICD-9) [21]. CVD mortality was defined as death caused by myocardial infarction, stroke, sudden death from coronary heart disease (CHD), or congestive heart failure (ICD-9 codes 390–448).
In the MDC-CC, all subjects were followed from the baseline examination until death, emigration from Sweden, or December 31 2020, whichever came first [27]. Causes of death and the vital status of participants were retrieved through record linkage of the personal identification number and the Swedish Cause of Death Register and the National Tax Board [27, 28]. CVD mortality was defined as death caused by CVD on the basis of ICD-9 codes 390–459 or ICD-10 codes I00-I99.
Assessments of clinical covariates
In the SHFS, demographic information (age and sex), lifestyle habits (smoking/drinking status, physical activity), medical history, family history of illnesses, and use of prescription medications were collected using structured questionnaires [21, 22]. Smoking status was categorized as current versus non-current smokers (former and never smokers combined). Drinking status was categorized as current versus non-current drinkers (former and never drinkers combined). Anthropometric measures including height, weight and waist circumferences were obtained through physical examinations at each visit. Body mass index (BMI) was calculated as body weight in kilograms divided by the square of height in meters. Blood samples were collected into 10 ml EDTA tubes after an overnight fast. Fasting glucose and blood lipids, including total cholesterol, triglycerides, LDL-c and HDL-c were measured by standard laboratory methods [21]. Hypertension was defined as blood pressure levels ≥ 140/90 mmHg or use of antihypertension medications. Type 2 diabetes was defined as fasting plasma glucose ≥ 126 mg/dL or use of hypoglycemic medications. Estimated glomerular filtration rate (eGFR) was calculated using the chronic kidney disease (CKD) Epidemiology Collaboration (CKD-EPI) [29]. Physical activity was assessed by the mean number of steps per day calculated by averaging the total number of steps recorded each day during the 7-day period. Information on use of lipid-lowering drugs was also collected at each visit. Diet quality was assessed using the Alternative Healthy Eating Index (AHEI)-2010 [30].
In the MDC-CC, information on lifestyle and clinical factors, such as BMI, blood pressure, use of antihypertensive treatment, current smoking, clinical lipids, and blood glucose, were obtained using previously described methods [31].
Lipidomic data acquisition, pre-processing and quality control
In the SHFS, relative abundance of fasting plasma lipid species at two time points (mean 5.5 years apart) was quantified by untargeted liquid chromatography-mass spectrometry (LC–MS) as described previously [23]. After pre-processing and quality control, we obtained 1,542 lipids (518 known, 1,024 unknown, see Table S1) in 3,977 EDTA plasma samples (1,983 at baseline, 1,994 at follow-up). Outlier samples were detected by principal component analysis (PCA) and those beyond mean ± 5 standard deviation (SD) for any of the first three principal components were further removed. After further excluding outlier samples or those with missing covariates, 1,930 participants (1,930 at baseline, 1,891 at follow-up) with complete clinical and lipidomic data were included in the current analysis (Figure S1).
Lipidomic profiling in the MDC-CC was performed using citrate fasting plasma samples collected at enrollment (i.e., baseline) by mass spectrometry as previously described [32, 33]. Spectra were analyzed with in-house developed lipid identification software based on LipidXplorer [34]. Data processing, normalization and batch correction was performed as described previously [25, 33]. Lipids that were present in at least 70% of the participants were included in the subsequent statistical analysis, resulting in a total of 184 lipids [25, 33]. Missing values of the remaining lipids were imputed using the NIPALS algorithm [35]. Of the 518 known lipids measured in the SHFS, 178 lipids were also measured in the MDC-CC, and these lipids were used in the replication analysis (Table S1). No outliers were detected in the MDC-CC samples. A total of 3,943 participants (58.8% females) with available lipidomic data were included in the current study to replicate findings from the SHFS. The mean age of MDC-CC participants was 57.7 years at baseline and 81.3 years at follow-up (mean 23.7 years of follow-up).
Statistical analysis
Figure S1 illustrates the procedures for participants’ selection and statistical analyses. All continuous variables including lipids were standardized to zero mean and unit variance. Multiple testing was controlled by false discovery rate (FDR) using the Storey’s q-value method [36, 37].
Prospective association analysis
To identify baseline plasma lipid species predictive of risk for all-cause or CVD mortality, we constructed frailty Cox proportional hazards models in the SHFS. In this model, baseline level of each individual lipid was the independent variable and time to death was the outcome, adjusting for age, sex, BMI, smoking, hypertension, diabetes, LDL-c, and eGFR at baseline. The frailty Cox model is an extension of the Cox proportional hazard model [38]. The frailty term in the model accounts for unobserved heterogeneity and relatedness among family members in the SHFS. The putative lipids (raw P < 0.05) in the SHFS were validated in the MDC-CC using Cox proportional hazard model, adjusting for age, sex, BMI, smoking, hypertension, diabetes and LDL-c at baseline. The associations between the lipid species and all-cause mortality analysis were analyzed using the same method with the CVD status at baseline additionally adjusted in both SHFS and MDC-CC. The assumptions for proportional hazards model were checked using the Schoenfeld residuals. Replication was defined as lipids with P < 0.05 and consistent directions of association in both cohorts. Results from two cohorts were then combined by random-effects meta-analysis.
Class-level association analysis
To identify lipid classes associated with mortality in American Indians, we first conducted principal component analysis (PCA) to identify major lipid classes (e.g., the first principal component of lipids) in each lipid class, and then tested their associations with mortality by frailty Cox proportional hazards model, adjusting for age, sex, BMI, smoking, hypertension, diabetes, LDL-c and eGFR at baseline. The all-cause mortality analysis additionally adjusted for CVD status at baseline.
Repeated measurement analysis
To examine whether longitudinal change in plasma lipidome was associated with risk of all-cause or CVD mortality, we constructed frailty Cox proportional hazards model in the SHFS. In the model, time to all-cause or CVD mortality was the outcome and change in the relative abundance of lipids (between baseline and end of follow-up) was the predictor, adjusting for age, sex and smoking at baseline and changes in continuous variables (i.e., BMI, fasting glucose, systolic blood pressure, LDL-c, eGFR) plus baseline lipid. The all-cause mortality analysis additionally adjusted for CVD status at baseline. This analysis was performed in the SHFS only as MDC-CC only measured blood lipid species at baseline.
Differential lipid network analysis
To identify lipid networks (i.e., sets of lipids that are highly correlated) associated with all-cause or CVD mortality, we constructed lipid modules (subnetworks) using the Weighted Correlation Network Analysis (WGCNA) [39]. Briefly, signed weighted lipid co-regulation networks were constructed using all 1,542 baseline lipids among American Indians who died of all-causes, CVD and those who remained to be alive by end of study follow-up, separately. Lipid species were hierarchically clustered, and those with a high topological overlap similarity were grouped into a same module. Differential modular analysis was performed to dissect intra-module difference (i.e., difference of connectivity among lipids within a module) between individuals died of all causes or CVD and those who remained to be alive. To quantify the intra-module difference, we calculated modular differential connectivity (MDC) [40], i.e., the difference in the total connectivity of all lipid pairs for a specific lipid module between individuals died due to all causes or CVD and those who were alive. Gain of connectivity (GOC) was defined if MDC > 0 and loss of connectivity (LOC) if MDC < 0. Statistical significance of MDC was assessed by 1,000 permutation tests [41].
Sensitivity analysis
To examine whether physical activity (steps/day), use of lipid-lowering drugs (yes/no) or diet quality (assessed by the Alternate Healthy Eating Index-2010 [30]) affect our results, we additionally adjusted for these variables in the above described models. To examine the impact of diabetes on our results, we re-ran the analyses by excluding diabetic individuals at baseline (n = 356 in the all-cause mortality analysis, n = 283 in the CVD mortality analysis). The sensitivity analyses focused on putative lipids (P < 0.05) detected in both SHFS and MDC-CC.
Results
In the SHFS, 295 out of 1,930 participants died during follow-up (mean 17.8 years). Among these, 66 participants died of CVD. In the MDC-CC, 1,845 out of 3,943 participants died during follow-up (mean 23.7 years). A total of 566 (out of 1,845) died of CVD. The age-standardized all-cause and CVD mortality rates in the SHFS were 18.2% and 4.4%, respectively. The age-standardized all-cause and CVD mortality rates in the MDC-CC were 22.9% and 12.5%, respectively. Table 1 presents the baseline characteristics of participants in the two populations. In both cohorts, deceased individuals were more likely to be males, and had higher baseline levels of blood pressure, total cholesterol, triglycerides and fasting glucose compared to those who remained to be alive by end of the follow-up. Of note, current smoking, higher baseline levels of BMI and LDL-c, and lower baseline level of HDL-c were significantly associated with an increased risk of mortality in the MDC-CC, but not SHFS. The different findings between these two populations could be attributed to many factors, e.g., different ages of study participants, different lifestyles, different genetic makeups, different sample sizes, and differences in many unknown or unmeasured factors between SHFS and MDC-CC.
Table 1.
Baseline characteristics of participants in the SHFS and MDC-CC according to their vital status by end of study follow-up (mean ~ 17.8 years of follow-up in the SHFS, mean ~ 23.7 years of follow-up in the MDC-CC)
| Characteristics | SHFS | MDC-CC | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Living participants (n = 1,635) | All-cause death (n = 295) | P (vs. living participants) | CVD Death (n = 66) | P (vs. living participants) | Living participants (n = 2,098) | All-cause death (n = 1,845) | P (vs. living participants) | CVD Death (n = 566) | P (vs. living participants) | |
| Age (years) | 38.6 ± 13.2 | 50.2 ± 14.3 | < 2.0 × 10–16 | 53.0 ± 13.2 | 2.44 × 10–15 | 55.2 ± 5.4 | 60.5 ± 5.3 | < 2.0 × 10–16 | 61.3 ± 4.9 | < 2.0 × 10–16 |
| Female, n (%) | 1037 (63.4%) | 166 (56.3%) | 0.027 | 29 (43.9%) | 0.002 | 1346 (64.2%) | 973 (52.7%) | < 2.0 × 10–16 | 263 (46.5%) | < 2.0 × 10–16 |
| BMI (kg/m2) | 31.7 ± 7.4 | 32.5 ± 8.1 | 0.09 | 31.2 ± 7.2 | 0.584 | 25.3 ± 3.7 | 26.0 ± 4.0 | < 2.0 × 10–16 | 26.5 ± 4.3 | < 2.0 × 10–16 |
| Current smoking, n (%) | 653 (39.9%) | 112 (38.0%) | 0.51 | 26 (39.4%) | 0.931 | 374 (17.8%) | 519 (28.1%) | < 2.0 × 10–16 | 147 (26.0%) | < 2.0 × 10–16 |
| Hypertension, n (%) | 431 (26.4%) | 138 (46.8%) | 6.81 × 10–12 | 37 (56.1%) | 7.54 × 10–7 | 902 (43.0%) | 1171 (63.5%) | < 2.0 × 10–16 | 418 (73.9%) | < 2.0 × 10–16 |
| Type 2 diabetes, n (%) | 249 (15.2%) | 107 (36.3%) | 1.33 × 10–15 | 34 (51.5%) | 4.34 × 10–11 | 117 (5.6%) | 248 (13.4%) | < 2.0 × 10–16 | 90 (15.9%) | < 2.0 × 10–16 |
| Cardiovascular disease, n (%) | 14 (0.9%) | 6 (2.0%) | 0.081 | 1 (1.5%) | 0.582 | 33 (1.6%) | 113 (6.1%) | < 2.0 × 10–16 | 61 (10.8%) | < 2.0 × 10–16 |
| Systolic blood pressure (mmHg) | 121.3 ± 15.0 | 128.5 ± 16.5 | 1.98 × 10–14 | 131.9 ± 17.0 | 8.57 × 10–9 | 137.7 ± 17.8 | 146.5 ± 19.3 | < 2.0 × 10–16 | 150.9 ± 20.1 | < 2.0 × 10–16 |
| Total cholesterol (mg/dL) | 184.4 ± 33.8 | 189.3 ± 37.2 | 0.026 | 191.7 ± 38.9 | 0.103 | 235.6 ± 41.1 | 241.1 ± 42.0 | < 2.0 × 10–16 | 240.5 ± 40.6 | 0.012 |
| HDL-c (mg/dL) | 51.7 ± 14.3 | 53.0 ± 15.3 | 0.163 | 50.3 ± 12.9 | 0.387 | 55.4 ± 14.3 | 52.2 ± 14.2 | < 2.0 × 10–16 | 50.6 ± 13.7 | < 2.0 × 10–16 |
| LDL-c (mg/dL) | 101.5 ± 29.4 | 102.6 ± 32.6 | 0.597 | 102.3 ± 35.8 | 0.867 | 158.8 ± 37.3 | 164.1 ± 38.7 | < 2.0 × 10–16 | 164.7 ± 37.6 | 0.001 |
| Triglycerides (mg/dL) | 160.7 ± 102.3 | 178.2 ± 167.0 | 0.008 | 195.2 ± 88.9 | 3.0 × 10–4 | 108.4 ± 52.3 | 124.5 ± 59.0 | < 2.0 × 10–16 | 127.7 ± 60.3 | < 2.0 × 10–16 |
| Fasting glucose (mg/dL) | 105.5 ± 39.5 | 129.4 ± 66.1 | 2.03 × 10–12 | 149.2 ± 77.0 | 1.23 × 10–11 | 89.4 ± 13.8 | 97.6 ± 31.4 | < 2.0 × 10–16 | 99.5 ± 33.0 | < 2.0 × 10–16 |
Continuous variables were expressed as mean ± standard deviation and qualitative variables were expressed as n (%). P-values were obtained by generalized estimating equation (GEE) model in the SHFS to account for family relatedness and logistic regression in the MDC-CC. BMI Body mass index; HDL-c High-density lipoprotein cholesterol; LDL-c Low-density lipoprotein cholesterol
Baseline plasma lipid species significantly predicted risk of all-cause mortality
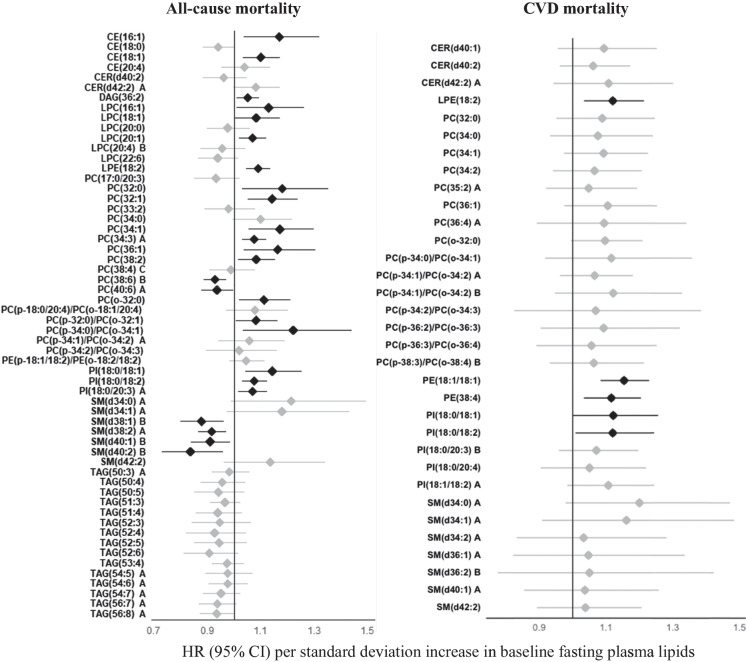
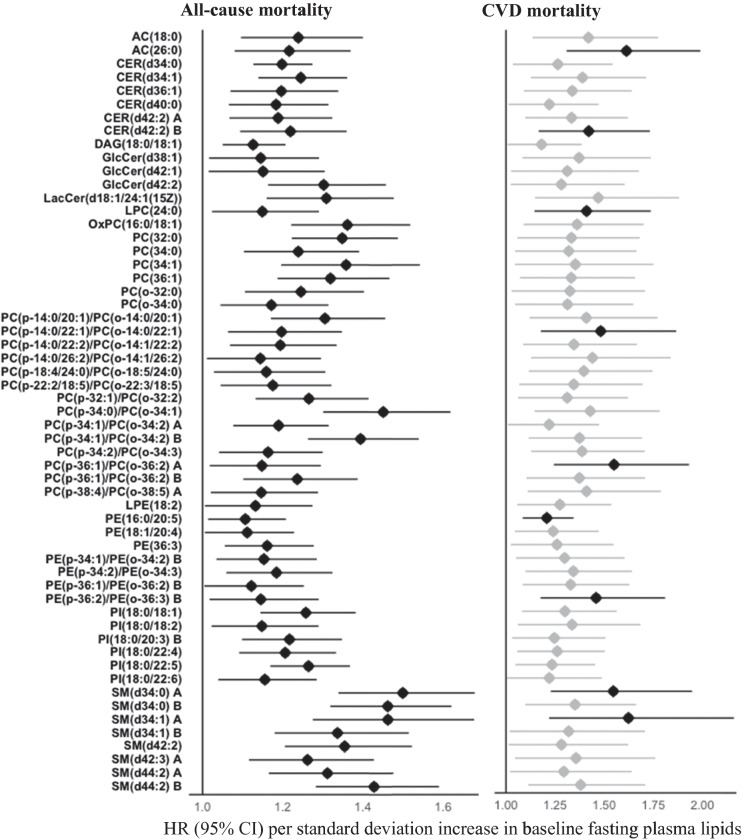
In the SHFS (discovery cohort), 552 baseline lipids (192 known) were associated with all-cause mortality at P < 0.05 (Fig. 1, Table S2). Of these, higher baseline levels of 120 lipids, including 9 acylcarnitines (ACs), 3 cholesterol esters (CEs), 6 fatty acids (FAs), 45 phosphatidylcholines (PCs), 17 phosphatidylethanolamines (PEs), 1 phosphatidylglycerol (PG), 9 phosphatidylinositols (PIs), 1 phosphatidylserine (PS), 3 diacylglycerols (DAGs), 16 sphingomyelins (SMs), 6 ceramides (CERs), 3 glucosylceramides (GlcCers), and 1 lactosylceramide (LacCer), were associated with an increased risk of all-cause mortality. In contrast, higher baseline levels of 72 lipids, including CE(18:0), LPE(20:6), 19 phosphatidylcholines, 1 diacylglycerol, 30 triacylglycerols, 2 ceramides, and 18 sphingomyelins, were associated with a reduced risk of all-cause mortality. After correction for multiple testing (q < 0.05), 183 known lipids (out of 192) remained to be statistically significant.
Fig. 1.
Associations between baseline plasma lipid species and risk of all-cause or CVD mortality in American Indians in the SHFS (P < 0.05). Hazard ratios (HRs) and 95% confidence intervals (CIs) were obtained by frailty Cox proportional hazard model, adjusting for age, sex, BMI, smoking, hypertension, diabetes, LDL-c, and eGFR at baseline. The all-cause mortality analysis additionally adjusted for CVD status at baseline. Family relatedness was accounted for by including a random effect term (i.e., a frailty term) in the model. Only the top known lipids in each class are shown. Lipids highlighted in black indicate replication in the MDC-CC. The letter A or B in the name of lipids indicates isomers
Of the 192 known lipid species identified in the SHFS (P < 0.05), 58 lipids were also measured in the MDC-CC. Of these, 15 baseline lipids, including CE(16:1), CE(18:1), LPE(18:2), 9 phosphatidylcholines (e.g., PC(32:0), PC(34:1), PC(38:2)) and 3 phosphatidylinositols, were positively, whereas 5 lipids, including PC(38:6) B and 4 sphingomyelins (i.e., SM(d38:1) B, SM(d38:2) A, SM(d40:1) B, SM(d40:2) B), were inversely associated with all-cause mortality (q < 0.05) in the MDC-CC with same directions of associations (Figure S2, Table S2).
Transethnic meta-analysis identified 25 lipids significantly associated with risk of all-cause mortality (P < 0.05) (Fig. 2). After correction for multiple testing (q < 0.05), a total of 11 lipids, including 2 cholesterol esters (i.e., CE(16:1), CE(18:1)), 5 phosphatidylcholines (i.e., LPC(20:1), PC(32:1), PC(34:1), PC(34:3) A, PC(36:1)), LPE(18:2) and 3 phosphatidylinositols (i.e., PI(18:0/18:1), PI(18:0/18:2), PI(18:0/20:3) A) were positively, whereas 4 lipids including PC(38:6) B and 3 sphingomyelins (i.e., SM(d38:2) A, SM(d38:1) B, SM(d40:2) B) were inversely associated with risk of all-cause mortality.
Fig. 2.
Associations between baseline plasma lipid species and risk of all-cause or CVD mortality in the meta-analysis. Hazard ratios (HRs) and 95% confidence intervals (CIs) were obtained by random-effects meta-analysis. Only known lipids with P < 0.05 in the SHFS and those also measured in the MDC-CC are shown. Significant lipids (P < 0.05) are highlighted in black. The letter A, B, or C in the name of lipids indicates isomers
Baseline plasma lipid species significantly predicted risk of CVD mortality
In the SHFS, 326 baseline lipids (105 known) were associated with CVD mortality at P < 0.05 (Fig. 1, Table S3). Of these, higher baseline levels of 103 lipids, including 3 acylcarnitines (i.e., AC(18:0), AC(24:0), AC(26:0)), 1 cholesterol ester (i.e., CE(22:5) B), 1 diacylglycerol (i.e., DAG(18:0/18:1)), 67 glycerophospholipids (e.g., 42 phosphatidylcholines (PCs), 16 phosphatidylethanolamines (PEs), 9 phosphatidylinositols (PIs)), and 31 sphingolipids (e.g., 15 sphingomyelins (SMs), 10 ceramides (CERs), 5 glucosylceramides (GlcCers), and 1 lactosylceramide (LacCer)), were associated with an increased risk of all-cause mortality. In contrast, two lipids including LPC(22:5) and LPG(17:0) were inversely associated with risk of CVD mortality. After correction for multiple testing (q < 0.05), 10 lipids (i.e., AC(26:0), CER(d42:2) B, GlcCer(d40:1), LPC(24:0), PC(p-14:0/22:1)/PC(o-14:0/22:1), PC(p-36:1)/PC(o-36:2) A, PE(16:0/20:5), PE(p-36:2)/PE(o-36:3) B, SM(d34:0) A, SM(d34:1) A) remained to be significantly associated with CVD mortality.
Of the 105 known lipid species identified in the SHFS, 33 lipids were also measured in the MDC-CC. Of these, 3 baseline lipids (i.e., LPE(18:2), PE(18:1/18:1), PE(38:4)) were positively associated with CVD mortality (P < 0.05) in the MDC-CC (Figure S3, Table S3). One lipid (i.e., PE(18:1/18:1)) remained to be significant at q < 0.05.
Transethnic meta-analysis found that higher baseline levels of 5 known lipids (i.e., LPE(18:2), PE(18:1/18:1), PE(38:4), PI(18:0/18:2), PI(18:0/18:1)) were associated with an increased risk of CVD mortality (P < 0.05) (Fig. 2). Of these, one lipid (i.e., PE(18:1/18:1)) remained to be statistically significant at q < 0.05.
Figure 3 shows the associations of 57 baseline lipids with the risk of both all-cause and CVD mortality (P < 0.05) in American Indians. Specifically, higher baseline levels of 2 acylcarnitines, 1 diacylglycerol (i.e., DAG(18:0/18:1)), 18 sphingolipids (e.g., sphingomyelins, ceramides, glucosylceramides, lactosylceramide) and 36 glycerophospholipids (e.g., phosphatidylcholines, phosphatidylethanolamines, phosphatidylinositols) were significantly associated with an increased risk of both all-cause and CVD mortality.
Fig. 3.
Associations between baseline plasma lipid species and risk of both all-cause and CVD mortality in American Indians in the SHFS (P < 0.05). Hazard ratios (HRs) and 95% confidence intervals (CIs) were obtained by frailty Cox proportional hazard model, adjusting for age, sex, BMI, smoking, hypertension, diabetes, LDL-c, and eGFR at baseline. The all-cause mortality analysis additionally adjusted for CVD status at baseline. Family relatedness was accounted for by including a random effect term (i.e., a frailty term) in the model. Only known lipids with P < 0.05 are shown. Lipids with q < 0.05 are highlighted in black. The letter A or B in the name of lipids indicates isomers
Lipid classes associated with all-cause and CVD mortality
Class-level association analysis showed that, after correction for multiple testing (q < 0.05), higher baseline levels of saturated acylcarnitines, ceramides, phosphatidylcholines, ether-phosphatidylcholines, sphingomyelins, and unsaturated phosphatidylinositols were associated with an increased risk of all-cause mortality. Higher baseline levels of saturated acylcarnitines and unsaturated ceramides, glucosylceramides and ether-phosphatidylcholines were associated with an increased risk of CVD mortality at q < 0.05. The associations of the saturated and unsaturated lipid classes with all-cause and CVD mortality are shown in Table S4.
Longitudinal changes in plasma lipid species significantly predicted risk of all-cause mortality
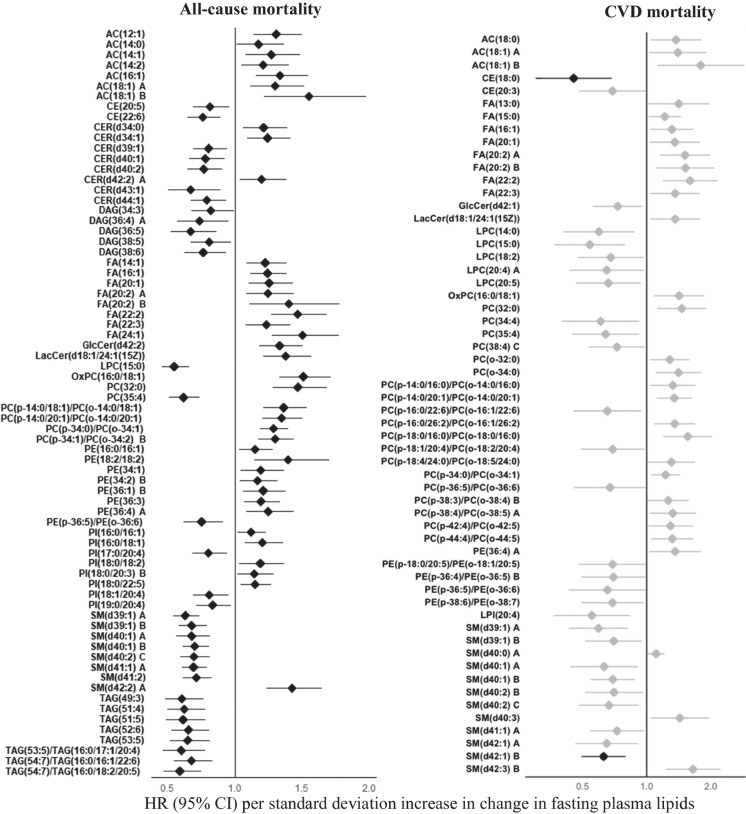
After adjusting for clinical factors and baseline lipids, longitudinal changes in 624 lipids (223 known) were significantly associated with risk of all-cause mortality at P < 0.05 (Fig. 4, Table S5). Of these, changes in 7 acylcarnitines, 10 fatty acids, 50 glycerophospholipids (e.g., 29 phosphatidylcholines, 14 phosphatidylethanolamines, 7 phosphatidylinositols) and 16 sphingolipids (e.g., 11 sphingomyelins, 3 ceramides, 1 glucosylceramide, 1 lactosylceramide) were positively associated with risk of all-cause mortality. Changes in 2 cholesterol esters, 46 glycerophospholipids (e.g., 42 phosphatidylcholines, 1 phosphatidylethanolamine, 3 phosphatidylinositols), 36 sphingolipids (e.g., 31 sphingomyelins, 5 ceramides), 56 glycerolipids (5 diacylglycerols, 51 triacylglycerols) were inversely associated with risk of all-cause mortality. All the 223 known lipids remained to be significant at q < 0.05.
Fig. 4.
Associations between longitudinal changes in plasma lipid species and risk of all-cause or CVD mortality in American Indians in the SHFS (P < 0.05). Hazard ratios (HRs) and 95% confidence intervals (CIs) were obtained by frailty Cox proportional hazard model, adjusting for age, sex and smoking at baseline as well as changes in continuous covariates (i.e., BMI, fasting glucose, systolic blood pressure, LDL-c, and eGFR). The all-cause mortality analysis additionally adjusted for CVD status at baseline. Family relatedness was accounted for by including a random effect term (i.e., a frailty term) in the model. Only top known lipids in each class are shown. Lipids with q < 0.05 are highlighted in black. The letter A, B or C in the name of lipids indicates isomers
Longitudinal change in plasma lipid species predicted risk of CVD mortality
Longitudinal changes in 199 lipids (58 known) were associated with risk of CVD mortality at P < 0.05 (Fig. 4, Table S5). Specifically, changes in 3 acylcarnitines, 8 fatty acids, 1 lactosylceramide, 3 sphingomyelins, 14 phosphatidylcholines, 1 phosphatidylethanolamine were positively, whereas changes in 2 cholesterol esters, 1 glucosylceramide, 11 phosphatidylcholines, 4 phosphatidylethanolamines, 1 phosphatidylinositol and 9 sphingomyelins were inversely associated with risk of CVD mortality. After correction for multiple testing (q < 0.05), changes in two known lipids (i.e., CE(18:0) and SM(d42:1) B) remained to be significantly associated with an increased risk of CVD mortality.
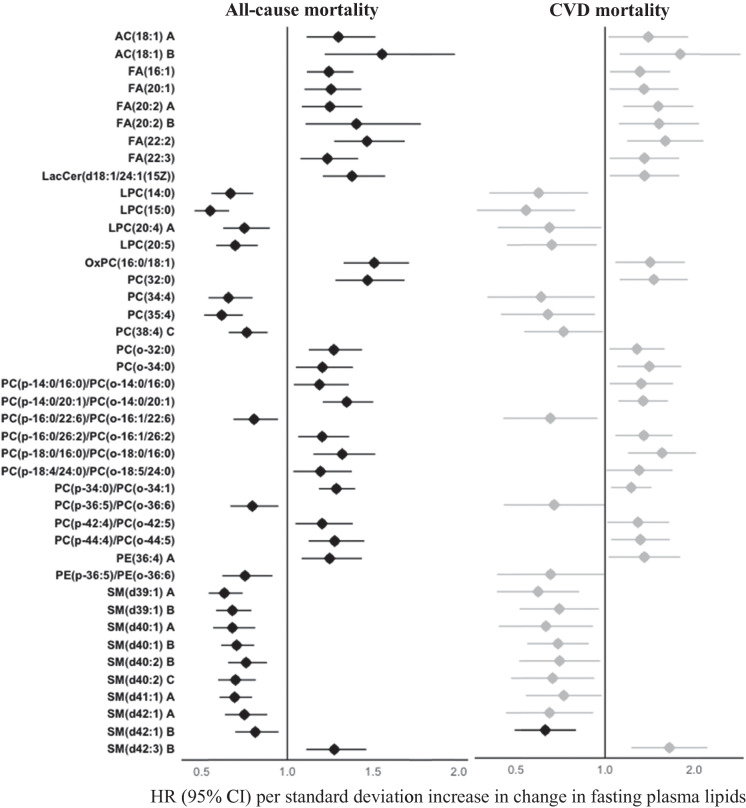
Of the 58 known lipids at P < 0.05, longitudinal changes in 42 known lipids were significantly associated with both all-cause and CVD mortality at P < 0.05 (Fig. 5). Of these, changes in 2 acylcarnitines, 6 fatty acids, 12 phosphatidylcholines, 1 phosphatidylethanolamine, 1 sphingomyelin, and 1 lactosylceramide were positively, whereas changes in 9 phosphatidylcholines, 9 sphingomyelins and 1 phosphatidylethanolamine were inversely associated with risk for both all-cause and CVD mortality.
Fig. 5.
Associations between longitudinal changes in plasma lipid species and risk of both all-cause and CVD mortality in American Indians in the SHFS (P < 0.05). Hazard ratios (HRs) and 95% confidence intervals (CIs) were obtained by frailty Cox proportional hazard model, adjusting for age, sex and smoking at baseline, and changes in continuous covariates (i.e., BMI, fasting glucose, systolic blood pressure, LDL-c, and eGFR). The all-cause mortality analysis additionally adjusted for CVD status at baseline. Family relatedness was accounted for by including a random effect term (i.e., a frailty term) in the model. Only known lipids with P < 0.05 are shown. Lipids with q < 0.05 are highlighted in black. The letter A, B, or C in the name of lipids indicates isomers
Differential lipid networks associated with risk of all-cause and CVD mortality
Network analysis in the SHFS identified 9, 11 and 12 lipid modules among participants who died of all causes, CVD and those who remained to be alive, respectively (Figure S4). One of the modules (i.e., module turquoise) exhibited significant difference in lipid connectivity when comparing American Indians who died of all-causes to those who remained to be alive by end of follow-up (Figure S5). Lipids in this module included ceramides, cholesterol ester, phosphatidylcholines, phosphatidylinositols, and sphingomyelins. Compared to those who remain to be alive, deceased individuals exhibited gain of connectivity (GOC) for lipids in the module turquoise (modular differential connectivity (MDC) = 202.2, P = 0.046). Hub lipids in this module included SM(d17:0/18:2) A, SM(d39:1) A, SM(d41:1) A and SM(d38:1) A. Regarding differential lipid networks for CVD mortality (Figure S6), we observed significant difference in lipid connectivity in another module (i.e., module green). Lipids in this module included cholesterol esters, fatty acids, glucosylceramides, phosphatidylcholines, and sphingomyelins. Compared to individuals who remained to be alive, those died of CVD exhibited gain of connectivity (GOC) for lipids in the module green (modular differential connectivity (MDC) = 92.9, P = 0.026). Hub lipids in this module included PC(p-14:0/26:2)/PC(o-14:1/26:2), GlcCer(d40:1), PC(p-16:1/26:2)/PC(o-16:2/26:2) and PC(p-16:0/26:2)/PC(o-16:1/26:2).
Sensitivity analysis
Additional adjustments for physical activity, use of lipid-lowering drugs, and diet quality (assessed by AHEI-2010) slightly attenuated the associations between the identified lipids and risk of all-cause or CVD mortality, but most of the observed associations remained to be statistically significant (Table S6). In addition, majority of the identified lipids remained to be significant after excluding diabetic participants (Table S6).
Discussion
In this large-scale longitudinal lipidomic analysis that included over 5,800 community-dwelling individuals in two prospective cohorts, we had several key findings. First, altered baseline levels of multiple lipid species were significantly associated with risk of all-cause and/or CVD mortality beyond known risk factors. Specifically, our prospective association analysis demonstrated that elevated baseline levels of 120 individual lipid species, including 9 acylcarnitines, 3 cholesterol esters, 3 diacylglycerols, 6 fatty acids, 73 glycerophospholipids, and 26 sphingolipids, and reduced baseline levels of 72 lipid species, including CE(18:0), 20 glycerophospholipids, 31 glycerolipids, and 20 sphingolipids, were significantly associated with an increased risk of all-cause mortality in American Indians, independent of known clinical factors. Of these, the associations of 20 lipids, including 2 unsaturated cholesterol esters (i.e., CE(16:1), CE(18:1)), 2 unsaturated lyso-glycerophospholipids (i.e., LPC(20:1), LPE(18:2)), 7 very long-chain phosphatidylcholines (e.g., PC(32:0), PC(34:1), PC(38:2)), 2 very long-chain ether-phosphatidylcholines (i.e., PC(o-32:0), PC(p-34:0)/PC(o-34:1)), 3 very long-chain unsaturated phosphatidylinositols (i.e., PI(18:0/18:1), PI(18:0/18:2), PI(18:0/20:3) A), and 4 very long-chain unsaturated sphingomyelins (i.e., SM(d38:1) B, SM(d38:2) A, SM(d40:1) B, SM(d40:2) B), were confirmed in European Caucasians in the MDC-CC (with same direction of association). Moreover, higher baseline levels of 103 lipid species, including 3 acylcarnitines, 1 cholesterol ester, 1 diacylglycerol, 67 glycerophospholipids (e.g., PCs, PEs, PIs), 31 sphingolipids (e.g., SMs, CERs, GlcCers) and lower baseline levels of 2 lyso-glycerophospholipids (i.e., LPC(22:5), LPG(17:0)), were significantly associated with an increased risk of CVD mortality in American Indians. Of these, 3 lipids (i.e., LPE(18:2), PE(18:1/18:1), PE(38:4)) were replicated in European Caucasians in the MDC-CC. Second, our repeated measurement analysis identified, for the first time, that longitudinal changes in fasting plasma lipidome significantly predicted the risk of all-cause and CVD mortality, independent of clinical factors and baseline lipids. In particular, changes in acylcarnitines, fatty acids, phosphatidylcholines, phosphatidylethanolamines, and sphingomyelins were significantly associated with both all-cause and CVD mortality in American Indians. Third, our network analysis identified differential lipid clusters (i.e., modules) associated with risk of all-cause and CVD mortality in American Indians. Together, these findings revealed the distinct lipidomic signatures associated with all-cause mortality or CVD mortality. The newly identified molecular lipid species may serve as potential novel biomarkers for risk stratification and early identification of at-risk individuals.
We observed significant associations between baseline glycerophospholipids (e.g., PCs, PEs, PIs, PC(P)/PC(O), PE(P)/PE(O)) and risks of all-cause and CVD mortality in both American Indians and European Caucasians. Longitudinal changes in glycerophospholipids (e.g., PCs, PEs) including ether-glycerophospholipids (e.g., PC(P)/PC(O), PE(P)/PE(O)) were also associated with risks of all-cause and CVD mortality in American Indians. These results corroborated previous studies demonstrating that some species of glycerophospholipids, e.g., PC(32:0), PC(34:1), PC(38:6), LPC(16:1), LPC(20:4), PC(O-34:1), PC(O-34:3), were associated with all-cause and CVD mortality in Caucasians and Asians [14, 16, 42–44]. Moreover, altered glycerophospholipids (e.g., PCs, PEs, PIs, PC(P)/PC(O), PE(P)/PE(O)) have been previously associated with diabetes [45], chronic kidney disease [46], CVD [47], biological aging and depression in various populations including American Indians [23, 48–50].
Glycerophospholipids (e.g., PCs, PEs, PIs, PC(P)/PC(O), PE(P)/PE(O)) are key components of cell membrane and involved in many cellular processes including cell signaling and metabolism [51]. PCs can be hydrolyzed to lysophosphatidylcholines via lipoprotein-associated phospholipase A2 (Lp-PLA2) [52], which mediates vascular inflammation and other biological processes [53]. An increased blood level of Lp-PLA2 has been associated with atherosclerotic plaque rupture and subsequent cardiovascular events [53]. PEs are essential for membrane integrity, cell division, and mitochondrial respiratory function [54]. Altered PEs may affect the stability and functions of membrane proteins [54]. PIs are involved in the hormone signal transduction, neurotransmitters and growth factors [55]. They can be phosphorylated to polyphosphoinositides (PPIs), which regulate vital cell signaling reactions and membrane homeostasis [56]. Ether-glycerophospholipids facilitate membrane signaling and lipids oxidation [57]. Abnormal alterations in any of these glycerophospholipids may affect membrane integrity and signaling, thereby contributing to diseases and mortality.
In addition to glycerophospholipids, altered baseline levels of multiple long-chain sphingolipids (e.g., SMs and CERs) and their longitudinal changes over time were also significantly associated with risks of all-cause and CVD mortality in American Indians. Of these, the inverse associations between baseline levels of four sphingomyelins (i.e., SM(d38:1) B, SM(d38:2) A, SM(d40:1) B, SM(d40:2) B) and risk of all-cause mortality were confirmed in European Caucasians. The observed associations of plasma ceramides in our study (e.g., CER(d34:1), CER(d36:1), CER(d42:2)) are in agreement with previous studies among Caucasians and Asians [14, 16, 44]. Moreover, some of the identified sphingolipids (e.g., SM(d40:2), SM(d36:3), SM(d42:2), SM(d42:3), CER(d34:0), CER(d40:0)) in the present study were also associated with risks of diabetes and depression in our previous studies of the same group of American Indians [23, 50]. Together, these results support previous studies demonstrating that sphingomyelins and ceramides were associated with coronary artery disease (CAD), myocardial infarction (MI) and CVD [44, 58, 59]. Sphingomyelins play crucial roles in regulating cell dysfunction, apoptosis and autophagy, all of which have been implicated in age-related diseases [60, 61]. Ceramides are involved in diverse biological processes including apoptosis, oxidative stress, inflammation and endoplasmic reticulum stress [62]. Altered metabolism in these sphingolipids may contribute to mortality through disturbing one or more of these biological pathways.
In line with previous studies showing that elevated levels of some long-chain unsaturated triacylglycerols (e.g., TAG(56:9), TAG(58:10), TAG(50:4), TAG(56:6)) were associated with reduced risks of diabetes, CVD and mortality [45, 63, 64], we found that higher baseline levels of some long-chain unsaturated triacylglycerols (e.g., TAG(56:9), TAG(52:6), TAG(50:5)) and their longitudinal changes were associated with a lower risk of all-cause mortality in American Indians. In addition, altered baseline levels of long-chain acylcarnitines and their longitudinal changes were associated with risks of both all-cause and CVD mortality in American Indians. Acylcarnitines are formed by the combination of free carnitines and acyl-coenzyme A produced by fatty acids [65]. Long-chain acylcarnitines have been shown to function as transporters in oxidative catabolism and modulators of multiple biological processes, such as energy metabolism, cellular stress, cardiac function, mitochondrial β-oxidation, insulin signaling, and inflammation [65]. Perturbed metabolism of acylcarnitines has been associated with a variety of metabolic disorders, such as CVD [66], diabetes [67], depression [68], neurological disorders[69] and cancers [70]. The observed associations between long-chain fatty acids and risks of all-cause and CVD mortality could be attributed to their roles in regulating membrane structure and function, intracellular signaling, immune response, and inflammation [71].
Our class-level association analysis showed that higher baseline levels of saturated lipids, such as acylcarnitines, ceramides, sphingomyelins, phosphatidylcholines, ether-phosphatidylcholines, and unsaturated lipids such as phosphatidylinositols, ether-phosphatidylcholines, ceramides and glucosylceramides were significantly associated with increased risks of all-cause or CVD mortality. These findings appear to corroborate previous research demonstrating that saturated or unsaturated fatty acids may exert lipotoxicity through mitochondrial β-oxidation or peroxidation [72–74].
Several limitations of our study should be noted. First, despite the large number of lipids detected in our study, many lipids are unknown and we were unable to distinguish isomeric lipids either. Additional experiments are needed to characterize these unknowns if considered of interest. Moreover, due to the use of different mass spectrometry platforms (and thus different lipid classes/species, and different resolution/coverage), only a small number of lipids detected in the SHFS were also available in the MDC-CC. As such, many putative lipids identified in American Indians were unable to be replicated in European Caucasians. In this regard, lipids detected in both populations should assure us of the robustness of the findings. Those identified in one but not the other could be attributed to the differences between the two populations (e.g., demographics, genetics, lifestyle, environmental exposures, etc.). Second, our findings were derived from American Indians with high prevalence of obesity and type 2 diabetes. However, all our analyses adjusted for BMI and diabetes, and excluding diabetic participants did not affect our results. Moreover, given the rising tides of obesity and diabetes in almost all racial/ethnic groups, our findings should be able to generalize to other populations. Third, although our statistical models controlled many known risk factors, we cannot exclude the possibility of residual confounding by unknown or unmeasured factors. Fourth, despite of the large sample size included in our study, we still do not have adequate power to examine the association between plasma lipidome and cause-specific mortality (e.g., cancer mortality). Finally, as all other observational studies, we cannot establish the causal relationship between the observed associations between perturbed lipid metabolism and risk of mortality.
However, our study has several strengths. First, the current analysis included nearly 6,000 participants in two community-based prospective cohorts comprising individuals with diverse backgrounds in demographic (e.g., age, gender, socioeconomic status), genetic, lifestyle (e.g., diet, physical activity) and environmental exposures. Despite these differences, many identified lipids could be replicated in both cohorts, signifying the robustness of our findings. Second, the longitudinal profiling of plasma lipidome in a large community-based prospective cohort represents the major strength of this study. To our knowledge, this represents the first of its kind in the fields of lipidomic studies and mortality in any racial/ethnic group. Third, our statistical models adjusted for a comprehensive list of covariates, including metabolic disorders (e.g., obesity, hypertension, diabetes and CVD). Moreover, we performed sensitivity analyses to examine the potential effects of physical activity, use of lipid-lowering medications, and diet quality on our results. Thus, lipids identified in our study should be independent of these risk factors. Finally, the high-resolution lipidomic platform allowed us to identify a large number of lipid species that may have been missed out in previous research and offers unprecedented opportunities for future investigations.
In summary, altered baseline levels of multiple molecular lipid species and their longitudinal changes over time are significantly associated with risks of all-cause and CVD mortality in American Indians. The newly identified lipid species provide novel insight into the role of dyslipidemia in disease mortality and offer potential biomarkers for risk stratification, early prevention, and risk reduction in American Indians and other ethnic groups.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the Strong Heart Study (SHS) participants, the Indian Health Service facilities, and the participating tribal communities for their extraordinary cooperation and involvement, which has contributed to the success of the SHS. The content expressed in this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Indian Health Service (IHS).
Author contributions
JZ and OF conceptualized and designed the study, obtained the funding and generated the data. GM conducted the statistical analyses. JZ and GM drafted the manuscript. All coauthors provided critical review of the manuscript and contributed to data interpretation.
Funding
This study was supported by the National Institute of Health (NIH) grant R01DK107532-01A1 (JZ). The Strong Heart Study (SHS) has been funded in whole or in part with federal funds from the National Heart, Lung, and Blood Institute, National Institute of Health, Department of Health and Human Services, under contract numbers 75N92019D00027, 75N92019D00028, 75N92019D00029, and 75N92019D00030. The study was previously supported by research grants: R01HL109315, R01HL109301, R01HL109284, R01HL109282, and R01HL109319 and by cooperative agreements: U01HL41642, U01HL41652, U01HL41654, U01HL65520, and U01HL65521. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data availability
The phenotype data used in the Strong Heart Family Study can be requested through the Strong Heart Study (https://strongheartstudy.org/). The lipidomic data in the SHS can be obtained from the corresponding author upon a reasonable request. Clinical and lipidomic data used in the Malmö Diet and Cancer-Cardiovascular Cohort can be requested through the MDC Steering Committee (https://www.malmo-kohorter.lu.se/malmo-cohorts).
Declarations
Ethics approval
The Strong Heart Family Study (SHFS) protocols were approved by the Institutional Review Boards of participating institutions and the American Indian tribes. The Malmö Diet and Cancer-Cardiovascular Cohort (MDC-CC) was approved by the Ethics Committee at Lund University.
Disclosures
The authors declare no conflicts of interest.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Newman WP, Freedman DS, Voors AW, et al. Relation of serum lipoprotein levels and systolic blood pressure to early atherosclerosis. The Bogalusa Heart Study. N Engl J Med. 1986;314(3):138–144. doi: 10.1056/NEJM198601163140302. [DOI] [PubMed] [Google Scholar]
- 2.Hurtubise J, McLellan K, Durr K, Onasanya O, Nwabuko D, Ndisang JF. The different facets of dyslipidemia and hypertension in atherosclerosis. Curr Atheroscler Rep. 2016;18(12):82. doi: 10.1007/s11883-016-0632-z. [DOI] [PubMed] [Google Scholar]
- 3.Murray CJ, Lopez AD. Mortality by cause for eight regions of the world: Global Burden of Disease Study. Lancet. 1997;349(9061):1269–1276. doi: 10.1016/S0140-6736(96)07493-4. [DOI] [PubMed] [Google Scholar]
- 4.(2020) QuickStats: prevalence of high total cholesterol* among adults aged ≥20 years. MMWR Morb Mortal Wkly Rep 69(22): 690. 10.15585/mmwr.mm6922a5. [DOI] [PMC free article] [PubMed]
- 5.Pu J, Romanelli R, Zhao B, et al. Dyslipidemia in special ethnic populations. Endocrinol Metab Clin North Am. 2016;45(1):205–216. doi: 10.1016/j.ecl.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frank AT, Zhao B, Jose PO, Azar KM, Fortmann SP, Palaniappan LP. Racial/ethnic differences in dyslipidemia patterns. Circulation. 2014;129(5):570–579. doi: 10.1161/CIRCULATIONAHA.113.005757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goff DC, Bertoni AG, Kramer H, et al. Dyslipidemia prevalence, treatment, and control in the Multi-Ethnic Study of Atherosclerosis (MESA): gender, ethnicity, and coronary artery calcium. Circulation. 2006;113(5):647–656. doi: 10.1161/CIRCULATIONAHA.105.552737. [DOI] [PubMed] [Google Scholar]
- 8.Wang S, Xu L, Jonas JB, You QS, Wang YX, Yang H. Prevalence and associated factors of dyslipidemia in the adult Chinese population. PLoS One. 2011;6(3):e17326. doi: 10.1371/journal.pone.0017326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deen JF, Adams AK, Fretts A, et al. Cardiovascular disease in American Indian and Alaska native youth: unique risk factors and areas of scholarly need. J Am Heart Assoc. 2017; 6(10). 10.1161/JAHA.117.007576. [DOI] [PMC free article] [PubMed]
- 10.Breathett K, Sims M, Gross M, et al. Cardiovascular health in American Indians and Alaska natives: a scientific statement from the American Heart Association. Circulation. 2020;141(25):e948–e959. doi: 10.1161/CIR.0000000000000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Narva AS. Reducing the burden of chronic kidney disease among American Indians. Adv Chronic Kidney Dis. 2008;15(2):168–173. doi: 10.1053/j.ackd.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 12.Ford ES. Risks for all-cause mortality, cardiovascular disease, and diabetes associated with the metabolic syndrome: a summary of the evidence. Diabetes Care. 2005;28(7):1769–1778. doi: 10.2337/diacare.28.7.1769. [DOI] [PubMed] [Google Scholar]
- 13.Guize L, Thomas F, Pannier B, Bean K, Jego B, Benetos A. All-cause mortality associated with specific combinations of the metabolic syndrome according to recent definitions. Diabetes Care. 2007;30(9):2381–2387. doi: 10.2337/dc07-0186. [DOI] [PubMed] [Google Scholar]
- 14.Sigruener A, Kleber ME, Heimerl S, Liebisch G, Schmitz G, Maerz W. Glycerophospholipid and sphingolipid species and mortality: the Ludwigshafen Risk and Cardiovascular Health (LURIC) study. PLoS One. 2014;9(1):e85724. doi: 10.1371/journal.pone.0085724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deelen J, Kettunen J, Fischer K, et al. A metabolic profile of all-cause mortality risk identified in an observational study of 44,168 individuals. Nat Commun. 2019;10(1):3346. doi: 10.1038/s41467-019-11311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin M, Zhu Q, Lai W, et al. Insights into the prognosis of lipidomic dysregulation for death risk in patients with coronary artery disease. Clin Transl Med. 2020;10(5):e189. doi: 10.1002/ctm2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bellis C, Kulkarni H, Mamtani M, et al. Human plasma lipidome is pleiotropically associated with cardiovascular risk factors and death. Circ Cardiovasc Genet. 2014;7(6):854–863. doi: 10.1161/CIRCGENETICS.114.000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker ME, Xanthakis V, Peterson LR, et al. Dietary patterns, ceramide ratios, and risk of all-cause and cause-specific mortality: the framingham offspring study. J Nutr. 2020;150(11):2994–3004. doi: 10.1093/jn/nxaa269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peterson LR, Xanthakis V, Duncan MS, et al. Ceramide remodeling and risk of cardiovascular events and mortality. J Am Heart Assoc. 2018; 7(10). 10.1161/JAHA.117.007931. [DOI] [PMC free article] [PubMed]
- 20.Howard BV, Lee ET, Cowan LD, et al. Rising tide of cardiovascular disease in American Indians. The Strong Heart Study. Circulation. 1999;99(18):2389–2395. doi: 10.1161/01.cir.99.18.2389. [DOI] [PubMed] [Google Scholar]
- 21.Lee ET, Welty TK, Fabsitz R, et al. The Strong Heart Study. A study of cardiovascular disease in American Indians: design and methods. Am J Epidemiol. 1990;132(6):1141–1155. doi: 10.1093/oxfordjournals.aje.a115757. [DOI] [PubMed] [Google Scholar]
- 22.North KE, Howard BV, Welty TK, et al. Genetic and environmental contributions to cardiovascular disease risk in American Indians: the strong heart family study. Am J Epidemiol. 2003;157(4):303–314. doi: 10.1093/aje/kwf208. [DOI] [PubMed] [Google Scholar]
- 23.Miao G, Zhang Y, Huo Z, et al. Longitudinal plasma lipidome and risk of type 2 diabetes in a large sample of american indians with normal fasting glucose: The Strong Heart Family Study. Diabetes Care. 2021;44(12):2664–2672. doi: 10.2337/dc21-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berglund G, Elmstähl S, Janzon L, Larsson SA. The malmo diet and cancer study. Design and feasibility. J Intern Med. 1993;233(1):45–51. doi: 10.1111/j.1365-2796.1993.tb00647.x. [DOI] [PubMed] [Google Scholar]
- 25.Ottosson F, Emami Khoonsari P, Gerl MJ, Simons K, Melander O, Fernandez C. A plasma lipid signature predicts incident coronary artery disease. Int J Cardiol. 2021;331:249–254. doi: 10.1016/j.ijcard.2021.01.059. [DOI] [PubMed] [Google Scholar]
- 26.Lee ET, Cowan LD, Welty TK, et al. All-cause mortality and cardiovascular disease mortality in three American Indian populations, aged 45–74 years, 1984–1988. The Strong Heart Study. Am J Epidemiol. 1998;147(11):995–1008. doi: 10.1093/oxfordjournals.aje.a009406. [DOI] [PubMed] [Google Scholar]
- 27.Drake I, Gullberg B, Sonestedt E, et al. Scoring models of a diet quality index and the predictive capability of mortality in a population-based cohort of Swedish men and women. Public Health Nutr. 2013;16(3):468–478. doi: 10.1017/S1368980012002789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hallengren E, Almgren P, Engström G, et al. Fasting levels of high-sensitivity growth hormone predict cardiovascular morbidity and mortality: the Malmö Diet and Cancer study. J Am Coll Cardiol. 2014;64(14):1452–1460. doi: 10.1016/j.jacc.2014.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JS, Chang PY, Zhang Y, Kizer JR, Best LG, Howard BV. Triglyceride and HDL-C dyslipidemia and risks of coronary heart disease and ischemic stroke by glycemic dysregulation status: The Strong Heart Study. Diabetes Care. 2017;40(4):529–537. doi: 10.2337/dc16-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiuve SE, Fung TT, Rimm EB, et al. Alternative dietary indices both strongly predict risk of chronic disease. J Nutr. 2012;142(6):1009–1018. doi: 10.3945/jn.111.157222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosvall M, Ostergren PO, Hedblad B, Isacsson SO, Janzon L, Berglund G. Occupational status, educational level, and the prevalence of carotid atherosclerosis in a general population sample of middle-aged Swedish men and women: results from the Malmö Diet and Cancer Study. Am J Epidemiol. 2000;152(4):334–346. doi: 10.1093/aje/152.4.334. [DOI] [PubMed] [Google Scholar]
- 32.Surma MA, Herzog R, Vasilj A, et al. An automated shotgun lipidomics platform for high throughput, comprehensive, and quantitative analysis of blood plasma intact lipids. Eur J Lipid Sci Technol. 2015;117(10):1540–1549. doi: 10.1002/ejlt.201500145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lauber C, Gerl MJ, Klose C, Ottosson F, Melander O, Simons K. Lipidomic risk scores are independent of polygenic risk scores and can predict incidence of diabetes and cardiovascular disease in a large population cohort. PLoS Biol. 2022;20(3):e3001561. doi: 10.1371/journal.pbio.3001561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herzog R, Schuhmann K, Schwudke D, et al. LipidXplorer: a software for consensual cross-platform lipidomics. PLoS One. 2012;7(1):e29851. doi: 10.1371/journal.pone.0029851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wold S, Sjöström M, Eriksson L. PLS-regression: a basic tool of chemometrics. Chemom Intell Lab Syst. 2001;58(2):109–130. doi: 10.1016/S0169-7439(01)00155-1. [DOI] [Google Scholar]
- 36.Storey JD. A direct approach to false discovery rates. J R Stat Soc Ser B (Stat Methodol) 2002;64(3):479–498. doi: 10.1111/1467-9868.00346. [DOI] [Google Scholar]
- 37.Storey John D, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henderson R, Oman P. Effect of Frailty on marginal regression estimates in survival analysis. J R Stat Soc Ser B (Stat Methodol) 1999;61(2):367–379. doi: 10.1111/1467-9868.00182. [DOI] [Google Scholar]
- 39.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langfelder P, Luo R, Oldham MC, Horvath S. Is my network module preserved and reproducible? PLoS Comput Biol. 2011;7(1):e1001057. doi: 10.1371/journal.pcbi.1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang B, Gaiteri C, Bodea LG, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell. 2013;153(3):707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tofte N, Suvitaival T, Ahonen L, et al. Lipidomic analysis reveals sphingomyelin and phosphatidylcholine species associated with renal impairment and all-cause mortality in type 1 diabetes. Sci Rep. 2019;9(1):16398. 10.1038/s41598-019-52916-w [DOI] [PMC free article] [PubMed]
- 43.Alshehry ZH, Mundra PA, Barlow CK, et al. Plasma lipidomic profiles improve on traditional risk factors for the prediction of cardiovascular events in type 2 diabetes mellitus. Circulation. 2016;134(21):1637–1650. doi: 10.1161/CIRCULATIONAHA.116.023233. [DOI] [PubMed] [Google Scholar]
- 44.Mundra PA, Barlow CK, Nestel PJ, et al. Large-scale plasma lipidomic profiling identifies lipids that predict cardiovascular events in secondary prevention. JCI Insight. 2018; 3(17). 10.1172/jci.insight.121326 [DOI] [PMC free article] [PubMed]
- 45.Razquin C, Toledo E, Clish CB, et al. Plasma lipidomic profiling and risk of type 2 diabetes in the PREDIMED trial. Diabetes Care. 2018;41(12):2617–2624. doi: 10.2337/dc18-0840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen H, Chen L, Liu D, et al. Combined clinical phenotype and lipidomic analysis reveals the impact of chronic kidney disease on lipid metabolism. J Proteome Res. 2017;16(4):1566–1578. doi: 10.1021/acs.jproteome.6b00956. [DOI] [PubMed] [Google Scholar]
- 47.Stegemann C, Pechlaner R, Willeit P, et al. Lipidomics profiling and risk of cardiovascular disease in the prospective population-based Bruneck study. Circulation. 2014;129(18):1821–1831. doi: 10.1161/CIRCULATIONAHA.113.002500. [DOI] [PubMed] [Google Scholar]
- 48.Zeng W, Beyene HB, Kuokkanen M, et al. Lipidomic profiling in the Strong Heart Study identified American Indians at risk of chronic kidney disease. Kidney Int. 2022 doi: 10.1016/j.kint.2022.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subedi P, Palma-Gudiel H, Fiehn O, et al. Lipidomics profiling of biological aging in American Indians: the Strong Heart Family Study. Geroscience. 2022 doi: 10.1007/s11357-022-00638-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miao G, Deen J, Struzeski JB, et al. Plasma lipidomic profile of depressive symptoms: a longitudinal study in a large sample of community-dwelling American Indians in the strong heart study. Mol Psychiatry. 2023 doi: 10.1038/s41380-023-01948-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Volinsky R, Kinnunen PK. Oxidized phosphatidylcholines in membrane-level cellular signaling: from biophysics to physiology and molecular pathology. FEBS J. 2013;280(12):2806–2816. doi: 10.1111/febs.12247. [DOI] [PubMed] [Google Scholar]
- 52.Norris PC, Gosselin D, Reichart D, Glass CK, Dennis EA. Phospholipase A2 regulates eicosanoid class switching during inflammasome activation. Proc Natl Acad Sci U S A. 2014;111(35):12746–12751. doi: 10.1073/pnas.1404372111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang F, Wang K, Shen J. Lipoprotein-associated phospholipase A2: The story continues. Med Res Rev. 2020;40(1):79–134. doi: 10.1002/med.21597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Calzada E, Onguka O, Claypool SM. Phosphatidylethanolamine Metabolism in Health and Disease. Int Rev Cell Mol Biol. 2016;321:29–88. doi: 10.1016/bs.ircmb.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wymann MP, Schneiter R. Lipid signalling in disease. Nat Rev Mol Cell Biol. 2008;9(2):162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- 56.Dickson EJ, Hille B. Understanding phosphoinositides: rare, dynamic, and essential membrane phospholipids. Biochem J. 2019;476(1):1–23. doi: 10.1042/BCJ20180022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Braverman NE. Moser AB (2012) Functions of plasmalogen lipids in health and disease. Biochim Biophys Acta. 1822;9:1442–1452. doi: 10.1016/j.bbadis.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 58.Jiang XC, Paultre F, Pearson TA, et al. Plasma sphingomyelin level as a risk factor for coronary artery disease. Arterioscler Thromb Vasc Biol. 2000;20(12):2614–2618. doi: 10.1161/01.atv.20.12.2614. [DOI] [PubMed] [Google Scholar]
- 59.Floegel A, Kühn T, Sookthai D, et al. Serum metabolites and risk of myocardial infarction and ischemic stroke: a targeted metabolomic approach in two German prospective cohorts. Eur J Epidemiol. 2018;33(1):55–66. doi: 10.1007/s10654-017-0333-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cutler RG, Mattson MP. Sphingomyelin and ceramide as regulators of development and lifespan. Mech Ageing Dev. 2001;122(9):895–908. doi: 10.1016/s0047-6374(01)00246-9. [DOI] [PubMed] [Google Scholar]
- 61.Taniguchi M. Okazaki T (2014) The role of sphingomyelin and sphingomyelin synthases in cell death, proliferation and migration-from cell and animal models to human disorders. Biochim Biophys Acta. 1841;5:692–703. doi: 10.1016/j.bbalip.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 62.Longato L, Ripp K, Setshedi M, et al. Insulin resistance, ceramide accumulation, and endoplasmic reticulum stress in human chronic alcohol-related liver disease. Oxid Med Cell Longev. 2012;2012:479348. doi: 10.1155/2012/479348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rhee EP, Cheng S, Larson MG, et al. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J Clin Invest. 2011;121(4):1402–1411. doi: 10.1172/JCI44442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fernandez C, Sandin M, Sampaio JL, et al. Plasma lipid composition and risk of developing cardiovascular disease. PLoS One. 2013;8(8):e71846. doi: 10.1371/journal.pone.0071846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCoin CS, Knotts TA, Adams SH. Acylcarnitines–old actors auditioning for new roles in metabolic physiology. Nat Rev Endocrinol. 2015;11(10):617–625. doi: 10.1038/nrendo.2015.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ruiz M, Labarthe F, Fortier A, et al. Circulating acylcarnitine profile in human heart failure: a surrogate of fatty acid metabolic dysregulation in mitochondria and beyond. Am J Physiol Heart Circ Physiol. 2017;313(4):H768–H781. doi: 10.1152/ajpheart.00820.2016. [DOI] [PubMed] [Google Scholar]
- 67.Sun L, Liang L, Gao X, et al. Early prediction of developing type 2 diabetes by plasma acylcarnitines: a population-based study. Diabetes Care. 2016;39(9):1563–1570. doi: 10.2337/dc16-0232. [DOI] [PubMed] [Google Scholar]
- 68.MahmoudianDehkordi S, Ahmed AT, Bhattacharyya S, et al. Alterations in acylcarnitines, amines, and lipids inform about the mechanism of action of citalopram/escitalopram in major depression. Transl Psychiatry. 2021;11(1):153. doi: 10.1038/s41398-020-01097-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ciavardelli D, Piras F, Consalvo A, et al. Medium-chain plasma acylcarnitines, ketone levels, cognition, and gray matter volumes in healthy elderly, mildly cognitively impaired, or Alzheimer's disease subjects. Neurobiol Aging. 2016;43:1–12. doi: 10.1016/j.neurobiolaging.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 70.Melone MAB, Valentino A, Margarucci S, Galderisi U, Giordano A, Peluso G. The carnitine system and cancer metabolic plasticity. Cell Death Dis. 2018;9(2):228. doi: 10.1038/s41419-018-0313-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zárate R, El Jaber-Vazdekis N, Tejera N, Pérez JA, Rodríguez C. Significance of long chain polyunsaturated fatty acids in human health. Clin Transl Med. 2017;6(1):25. doi: 10.1186/s40169-017-0153-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Garbarino J, Sturley SL. Saturated with fat: new perspectives on lipotoxicity. Curr Opin Clin Nutr Metab Care. 2009;12(2):110–116. doi: 10.1097/MCO.0b013e32832182ee. [DOI] [PubMed] [Google Scholar]
- 73.Comporti M. Lipid peroxidation. Biopathological significance. Mol Aspects Med. 1993;14(3):199–207. doi: 10.1016/0098-2997(93)90006-y. [DOI] [PubMed] [Google Scholar]
- 74.Adeva-Andany MM, Carneiro-Freire N, Seco-Filgueira M, Fernández-Fernández C, Mouriño-Bayolo D. Mitochondrial β-oxidation of saturated fatty acids in humans. Mitochondrion. 2019;46:73–90. doi: 10.1016/j.mito.2018.02.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The phenotype data used in the Strong Heart Family Study can be requested through the Strong Heart Study (https://strongheartstudy.org/). The lipidomic data in the SHS can be obtained from the corresponding author upon a reasonable request. Clinical and lipidomic data used in the Malmö Diet and Cancer-Cardiovascular Cohort can be requested through the MDC Steering Committee (https://www.malmo-kohorter.lu.se/malmo-cohorts).