Visual Abstract
Abstract
αβ T cell receptor (TCRαβ) T cells modified to express chimeric antigen receptors (CAR), are now available as authorized therapies for certain B-cell malignancies. However the process of autologous harvest and generation of patient-specific products is costly, with complex logistics and infrastructure requirements. Premanufactured banks of allogeneic donor–derived CAR T cells could help widen applicability if the challenges of HLA-mismatched T-cell therapy can be addressed. Genome editing is being applied to overcome allogeneic barriers, most notably, by disrupting TCRαβ to prevent graft-versus-host disease, and multiple competing editing technologies, including CRISPR/Cas9 and base editing, have reached clinical phase testing. Improvements in accuracy and efficiency have unlocked applications for a wider range of blood malignancies, with multiplexed editing incorporated to target HLA molecules, shared antigens and checkpoint pathways. Clinical trials will help establish safety profiles and determine the durability of responses as well as the role of consolidation with allogeneic transplantation.
Complex patient-specific manufacturing and variable potency due to poor T-cell fitness limit advances in autologous chimeric antigen receptor T therapies and support the potential attractiveness of having potent immune effector cell therapy that is readily available “off the shelf.” Edited by Associate Editor Helen Heslop and authored by leading experts, this review series focuses on several types of banked allogeneic immune cell therapies under development, highlighting their attributes and speculating on their anticipated future place in the therapeutic armamentarium.
Introduction
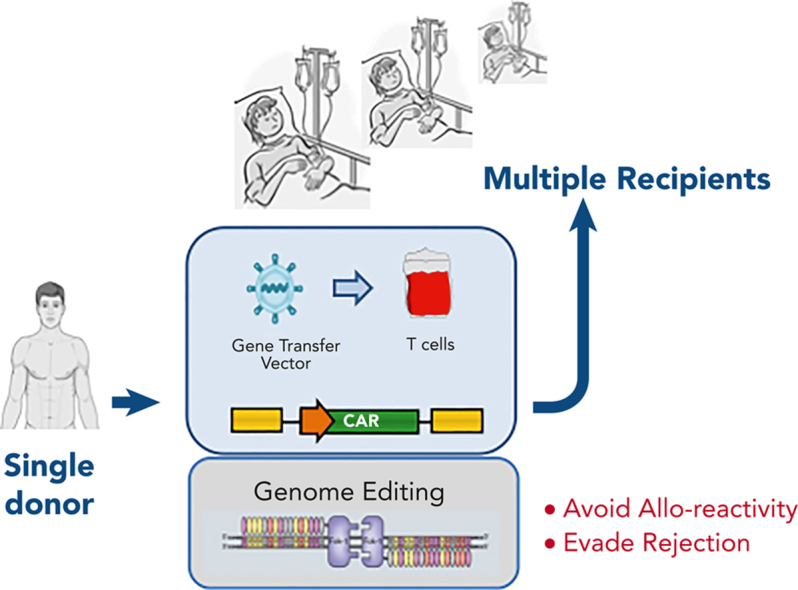
Autologous gene–modified T cells, including chimeric antigen receptor (CAR) T cells have been widely investigated through clinical trials, leading to approvals from the US Food and Drug Administration for new cell-based therapies against hematological malignancies. Anti-CD19 CAR T-cell therapy, tisagenlecleucel, was approved for refractory or relapsed (R/R) B-cell acute lymphoblastic leukemia (B-ALL) and non-Hodgkin lymphoma (NHL)1, 2, 3, 4 and axicabtagene ciloleucel for B-cell follicular lymphoma and NHL.5, 6, 7 Brexucabtagene autoleucel has been authorized for B-ALL and mantle cell lymphoma,8,9 and lisocabtagene maraleucel for B-cell NHL.10 For multiple myeloma, idecabtagene vicleucel and ciltacabtagene autoleucelare are available against R/R multiple myeloma.11, 12, 13, 14 A variety of additional autologous therapies are under commercial development or are being provided by academic centers.15,16 However, the infrastructure, logistics, and expertise required to generate these bespoke cell therapies are costly and difficult to replicate, especially in resource-scarce settings. Wider applications and routes to broader access at a reduced cost are under investigation through genome engineering of healthy donor-derived T cells, using genetic modification to overcome barriers associated with HLA-mismatched cell therapies. Ultimate ambitions aim to establish readymade universal cell banks for “off-the-shelf” affordable and accessible cell therapy without the risk of manufacturing delays, product failures,17 or accidental transduction of blasts.18 Premanufactured CAR T cell banks should also enable optimally timed therapeutic interventions earlier in the course of disease evolution as an alternative to intense chemotherapy. Critical issues to address in the allogeneic non-matched setting include the risk of graft-versus-host disease (GVHD) from the donor-derived T cells and host-mediated rejection from innate and adaptive humoral or cellular immune compartments. Third party, donor-derived, virus-specific T cells infused after allogeneic stem cell transplantation (SCT) can deliver immunity without significant GVHD,19 and such populations have been used as a starting material for CAR therapy.20,21 Alternatively, endogenous T cell receptor (TCR) expression can be disrupted by using antibody-derived protein expression blockers22 RNA interference,23 or genome editing. The latter, in combination with immunosuppressive conditioning of the host, has perhaps the greatest potential to provide strategic solutions for further improvements in allogeneic CAR T-cell therapies.
Genome editing technologies
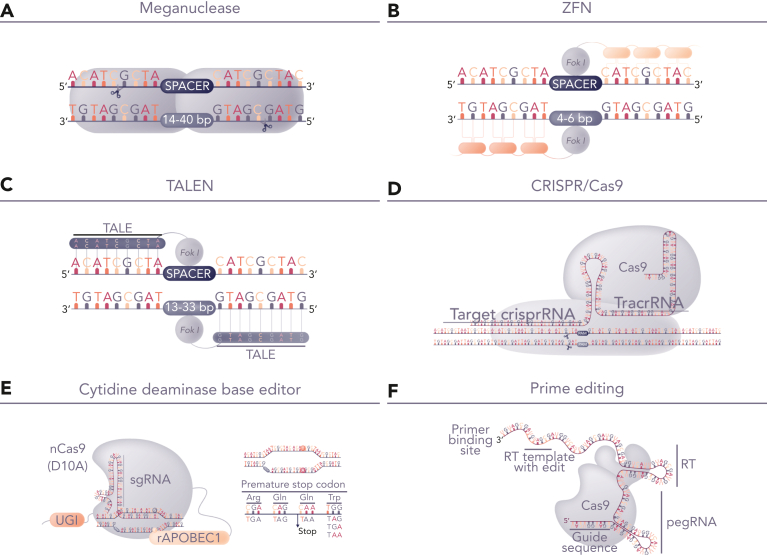
Protein-based DNA-recognition domains fused to nuclease enzymes, including zinc finger nucleases, homing endonucleases, and transcription activator-like effector nucleases (TALENs) have been investigated for the engineering of allogeneic CAR T cells for more than a decade (Figure 1). These platforms were constrained by limited targeting opportunities, although were suitable for disruption of TCR-related genes such as the TCR α-chain constant (TRAC) domain gene for initial proof of concept and modeling studies of CAR19-engineered T cells24,25 and T cells modified to express recombinant TCRs.26 Preclinical studies using TALENs for the multiplexed editing of CAR19 T cells demonstrated how messenger RNA (mRNA)–based delivery of the editors could be combined with the lentiviral expression of a CAR to generate allogeneic CAR19 T cells devoid of endogenous TCR.27 Subsequent variations included homing endonuclease–edited iterations with a site-specific insertion of a CAR transgene at the TRAC locus.28 Early hurdles of efficiency and reagent-mediated toxicity were addressed and translational clinical applications of TALENs were first investigated in patients with B-ALL (Table 1).29 Subsequently, the development of RNA-guided systems based on clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 for cell engineering has provided an ever larger toolbox30, 31, 32, 33 for laboratory and preclinical T-cell engineering studies.34, 35, 36, 37 Alternatives to precise DNA cleavage and gene insertion by CRISPR/Cas9 include disruption through targeted base conversion using CRISPR-guided cytidine deamination38 to introduce premature stop codons39 or carefully targeted adenosine deamination40 to modify critical splice sites. Most recently, prime editing has been added to the toolbox and incorporates an impaired Cas9 fused to murine reverse transcriptase and a prime-editing guide RNA for localized insertion, deletion, or base conversion without dsDNA breaks.41 Although iterations of this platform are still evolving,42,43 translational applications of other tools have been advanced to clinical phase applications and human studies, and T-cell therapies have offered a well-circumscribed arena for investigation, with the safeguards of ex vivo engineering and characterization as well as clear therapeutic objectives and defined safety readouts.
Figure 1.
Genome-editing platforms for T-cell modification. (A-D) Editing tools for highly specific dsDNA cleavage comprise either protein-based DNA recognition molecules including (A) homing endonucleases (meganuclease), (B) zinc finger nucleases, (C) TALENs, (D) RNA-guided nucleases exemplified by CRISPR/Cas9. After DNA cleavage, repair by nonhomologous end joining offers the prospect of gene-knockout with indel signatures and alternative homologous repair based repair, which can be exploited for site-specific transgene insertion. The latter requires delivery of template DNA flanked by homology arms and allows placement of CAR genes under the transcriptional control of endogenous transcriptional machinery. Alternatively, (E) base editors use nickase-restricted Cas9 variants, fused to cytidine deaminase or adenosine deaminase for highly targeted C>T or A>G base conversion. Cytidine base editor (CBE) has been used for multiplexed gene knockout, avoiding translocations usually encountered following nuclease activity. (F) Emerging prime editing offers the prospect of gene editing through localized template repair by fusing reverse transcriptase to deactivated Cas9. dsDNA, double-stranded DNA.
Table 1.
Trials of genome-edited T cells with published or interim clinical data available
| Study | Investigational product | Modifications | Indication | Lymphodepletion | Number | Toxicity | Outcomes |
|---|---|---|---|---|---|---|---|
| Great Ormond Street Hospital Special’s License |
UCART19 | LV-CAR19 TALEN ko of TRAC & CD52 | B-ALL | F,C, AntiCD52 | 2 | Grade2 GVHD | CR 100%29 |
| Servier/Allogene NCT02808442 NCT02746952 |
UCART19 | LV-CAR19 TALEN ko of TRAC & CD52 | B-ALL | F,C, ± AntiCD52 | 21 | Grade 3+ CRS 15%; Grade 3+ infections 39% | CR 67%68 |
| Allogene NCT04416984 NCT03939026 |
ALLO-501A ALLO-501 |
LV-CAR19 TALEN ko of TRAC & CD52 | LBCL | F,C, AntiCD52 | 47 | Grade 3+ CRS: 2%; Grade 3+ infections 24% | CR 50%113 |
| Allogene NCT04093596 |
Allo-715 | LV-anti BCMA TALEN ko of TRAC & CD52 | MM | F,C, AntiCD52 | 26 (DL3,4) | Grade 3 infections 13% | ORR 61%114 |
| Cellectis NCT04150497 |
UCART22 | LV-CAR22 TALEN ko of TRAC & CD52 | B-ALL | F,C, ± AntiCD52 | 9 | No Grade 3 CRS or infection | No CR115 |
| Precision Bio NCT03666000 |
PBCAR0191 | Arcus ko TRAC AAV site specific inserted CAR19 | B-ALL NHL | F, C | 27 | Grade 3+ CRS 6%; Grade 3+ infections 31%-80% | CR/CRi 62%-80%72,73 |
| CRISPR Tx NCT04035434 |
CTX110 | CRISPR/Cas9 TRAC & B2m AAV site specific inserted CAR19 | LBCL | F,C | 24 DL2+ |
Grade 3+ infections 9%; Grade 3+ ICANS 4% | CR 38%74 |
| Great Ormond Street Hospital NCT04557436 |
TT52CAR19 | CRISPR/Cas9 TRAC, CD52 LV CAR19 | B-ALL | F,C, AntiCD52 | 6 | NCT04557436, TT52CAR19' Grade 3+ ICANS 17% | CR/CRi66%70 |
| Beijing Chinese PLA General Hospital NCT03166878 |
U-Car | CRISPR/Cas9 TRAC, B2m LV CAR19 | DLBCL | F,C | 2 | No CR71 | |
| Zhejiang University Nanjing Bioheng Biotech |
CTA 101 | CRISPR/Cas9 TRAC,CD52 LV CAR19/22 | B-ALL | F,C, AntiCD52 | 6 | Grade 3 CRS 16% Grade 3 infections 50% |
CR/CRi83%116 |
| Gracell ChiCTR1900025311 |
GC027 | CRISPR TRAC CD7 LV-CAR7 | T-ALL | F,C ±Mel | 6 | Grade 3 CRS 100% Grade 3 infections 50% |
CR/CRi83%84 |
| CRISPR Tx NCT04502446 |
CTX130 | CRISPR/Cas9 TRAC, B2m, CD70 AAV-site specific | TCL | F,C | 15 | Grade 3+ infections 7%; | CR29%75 |
C, cyclophosphamide; DLBCL, diffuse large B-cell lymphoma; F, fludarabine; ICANS, immune cell associated neurotoxicity syndrome; LV, lentivirus; Mel, melphalan; TCL, T-cell lymphoma; ko, knockout.
Genome editing to prevent alloreactivity
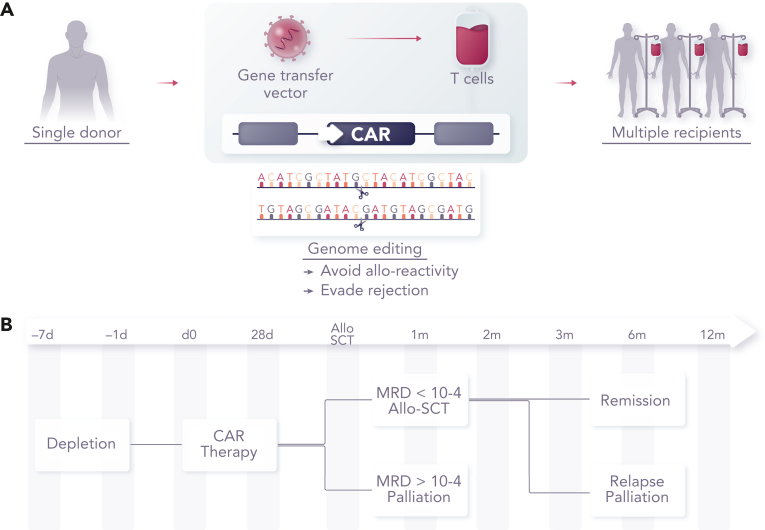
GVHD is mediated by antigen-specific αβ T-cell receptors (TCRαβs), and cell surface expression can be disrupted at the genomic level by targeting genes associated with the expression of the multimeric TCRαβ/CD3 complex (Figure 2). Notably, the TRAC locus has been targeted, and with the gene exhibiting allelic exclusion, disruption of the active allele is sufficient to efficiently prevent TCRαβ expression. Alternatively, editing the T cell receptor β chain constant (TRBC1/2) locus can result in the disruption of the β chain, and in either case, residual TCRαβ cells can be stringently depleted using commercially available magnetic bead depletion systems, resulting in T-cell products with redirected specificity through CAR (or recombinant TCR expression) and a greatly reduced alloreactivity. Similar TCRαβ disruption has also been reported using RNA interference,23,44 and by the restriction of components of the multimeric receptor complex at the protein level,22 as alternatives to genome editing. Efficient and stable disruption, combined with stringent removal of residual TCRαβ cells, has proven to be a successful mitigation against GVHD, and in human studies, (Table 1) the adoption of dose limits extrapolated from haploidentical allo-SCT for TCRαβ carriage of <5 × 104/kg has proven to be robust.45,46 The additional issue of how long allogeneic cells persist before host-mediated rejection or clearance with chemotherapy ahead of allo-SCT is also a relevant consideration, with complications akin to transfusion-associated GVHD being possible if residual TCRαβ T cells expand and mediate uninhibited multiorgan alloreactivity.47
Figure 2.
Concept of banked universal CAR T cells. (A) CAR T cells can be generated from healthy allogeneic donors for use in multiple recipients after genome editing to remove endogenous TCRαβ to prevent GVHD, disruption of HLA to reduce rejection or removal of CD52 to allow cells to persist in the presence of alemtuzumab, a serotherapy used as a part of augmented lymphodepletion. (B) Therapeutic effects sufficient to induce molecular remission are achievable within a period of 2 to 4 weeks, offering a bridge of consolidation with allogeneic SCT.
Editing to evade host-mediated rejection
Donor-derived allogeneic T cells are susceptible to host-mediated rejection after immune recognition, particularly in the context of a mismatched HLA. Preexisting anti-HLA antibodies could meditate rapid clearance of mismatched donor cells, and thus, screening and exclusion of previously transplanted or heavily transfused patients with anti-HLA antibodies is warranted. Alternative banks, with different HLA typing, may allow the circumvention of specific anti-HLA antibodies directed against a particular cell batch. Addressing cell-mediated responses by the host T and natural killer cells is generally managed by the administration of preparative conditioning chemotherapy ahead of cell infusion. In the autologous setting, host lymphodepletion using combinations of fludarabine and cyclophosphamide is thought to promote the expansion of infused CAR T cells through reduced homeostatic competition.48,49 More intense dosing regimens in the allogeneic setting aim to subdue host cellular responses against mismatched allogeneic T cells as well as provide a homeostatic advantage. However, greater lymphodepletion has increased the risk of complications associated with T-cell immunodeficiency and may extend to wider and more protracted cytopenia. Another strategy has been to include serotherapy based on an anti-CD52 conditioning antibody such as alemtuzumab, which is licensed for immunomodulation in multiple sclerosis50 and used widely in allo-SCT. In this context, the genetic disruption of CD52 as well as TRAC, allows allogeneic T cells to evade serotherapy effects while the host compartment has depleted.27 Conventionally, effects of alemtuzumab are generally borne for a period of 3 to 4 weeks, the period during which reactivation of viruses is common and can be problematic in the context of prolonged T- and B-cell immunodeficiency.51 Alternative serotherapy-based strategies could employ anti-CD3 monoclonal antibodies for energizing or depleting host CD3+ T cells while exploiting resistance of infused cells devoid of TCRαβ/CD3 on their surface.52 In addition, resistance to the lymphodepleting chemotherapy agent, fludarabine, through the disruption of deoxycytidine kinase in CAR T cells has also been investigated in preclinical studies,53 although it has not been assessed in clinical testing phases. There may also be applications of cellular therapies to counter host-mediated responses, for example, by using T cells expressing alloimmune defense receptors54 or signaling chimeric HLA molecules55 to target host alloreactive cells or more broadly by using CAR T cells through augmented lymphodepletion; a study investigating tandem anti-CD7 and CAR19 is currently underway.56
Another approach involves the removal of HLA molecules from the infused cells to promote immunological stealth, and achieving the disruption of HLA class I in T cells in models has been relatively straightforward through the targeting of the β2 microglobulin (B2m) gene locus.57 Whether the disruption of interactions between host CD8 T cells and donor HLA class I is sufficient to prevent rejection is under investigation in several studies (Table 1). It may be necessary to simultaneously prevent interactions between host CD4 T cells and HLA class II on the surface of activated incoming CAR T cells, and editing of HLA class II-associated transcription factors has also been modeled.58 There is also the possibility of responses mediated by natural killer cells against donor T cells lacking HLA expression as part of the missing-self immunity, and there are strategies proposed to mitigate against this through the expression of nonpolymorphic HLA molecules59 or by selectively retaining more conserved HLA molecules.60 Finally, risks associated with the evasion of immune surveillance by HLA-depleted CAR T cells, for instance, after the infection with T cell trophic viruses, have to be considered and factored into study designs, as done for other hypoimmunogenic cell therapies in development.61
Advanced editing for site-specific transgene insertion
Targeted CAR transgene integration might ameliorate vector-mediated variegation effects (discussed below) and may promote more physiologically regulated cell responses.62 In the context of engineering T cells with recombinant TCRαβ, orthotopically sited TCRα and TCRβ chains supported TCRαβ downregulation after antigenic stimulus, in a fashion similar to normal physiological responses mediated by T cells.63 Such control may be critical in preventing activation-induced cell death or exhaustion-related dysfunction. With respect to CAR transgenes, effective physiological control may have been ceded, given the inclusion of potent costimulatory domains, but animal studies have suggested that the CAR gene insertion at the TRAC locus under the control of endogenous TCR transcriptional machinery can provide improved cytotoxic activity and, in carefully calibrated models, may support reduced exhaustion.62 Non-integrating Adeno-associated viruses (AAV) were used to deliver homology-flanked templates for CAR insertion in such models, but advances in non-viral engineering processes have allowed CRISPR/Cas9 ribonucleoprotein (RNP)–mediated delivery of recombinant template DNA for similarly efficient homology-directed site-specific recombinant TCR63,64 or CAR integration.65,66 (Figure 3) Direct comparisons in human trials will be required to determine how targeted CAR integration and expression under the transcriptional control of endogenous promoters compares with conventional vector-mediated (nontargeted) transduction, and whether the inclusion of strong heterologous promoter elements influence CAR T-cell expansion, replicative capacity, and anti-tumor activity.
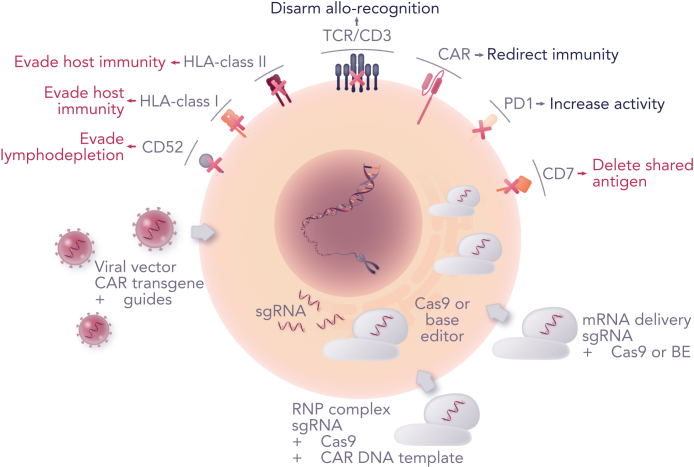
Figure 3.
Advanced engineering strategies. Combining CAR transfer with genome editing offers the prospect of enhanced CAR T cells. Viral vector CAR transduction has been combined with co-expressed guide RNAs and electroporation of mRNA coding for Cas9 or base editor (BE). Alternatively, Cas9/guide RNP complexes offer efficient editing, and a route to site-specific transgene insertion by homologous recombination if a suitable CAR template is provided. Other advances include the ability to remove shared antigens such as CD7 that would otherwise result in T-cell fratricide during manufacture, and simultaneous manipulation of checkpoint pathways to address exhaustion and promote activity.
Clinical trials of genome-edited CAR T cells
Formal trials of universal CAR19 (UCAR19) T cells in children (PALL study) and adults (CALM study) followed successful therapy in 2 infants with R/R B-ALL who had relapsed after a first allo-SCT.29 The prognosis for such children is known to be poor, with around 15% event–free survival in European cohorts of children who relapse after allo-SCT in B-ALL,67 and in both cases, the manufacture of autologous CAR19 T cells had not been possible. Both infants were conditioned with a combination of fludarabine, cyclophosphamide, and alemtuzumab and received a single dose of UCAR19 T cells. Bone marrow examination after 28 days revealed molecular remission on performing flow cytometry and molecular analysis. Interestingly, there was no notable CRS or ICANS, although the first infant developed skin GVHD over the coming weeks and required systemic steroid therapy. Subsequent second transplants after reduced-intensity conditioning using their original stem cell donors consolidated full donor chimerism, and both patients enjoyed complete immune recovery, including restoration of B-cell compartments and humoral immunity. Thus, time-limited–anti-leukemia effects of allogeneic CAR19 T cells had been sufficient to ensure remission, with deletion and non-persistence of infused cells after the conditioning for transplant was verified. Subsequent phase 1 trials were sponsored by Servier & Allogene, with trial sites in Europe and North America, whereby the pediatric study was structured as a bridge to allo-SCT, whereas the CALM study included optional allo-SCT. Interim analysis in 2020 reported that 7 children and 14 adults had been treated, with supportive data on safety and efficacy effects. CRS was common (91%), although more severe grade 3 to 4 CRS was reported in only 3 patients. GVHD only occurred in 2 patients, but 6 patients had grade 4 cytopenia. Two treatment-related deaths occurred; 1 caused by neutropenic sepsis in a patient with concurrent CRS and 1 by pulmonary hemorrhage in a patient with persistent cytopenia. Complete remission (CR) or CR with incomplete count recovery (CRi) by day 28 was 67% and the effects were poor in 4 subjects who omitted alemtuzumab.58,68
Subsequently, CRISPR/Cas9 technology was incorporated into a lentiviral configuration for the coupled expression of multiplexed single guide RNAs, and CAR19. Placement of specific guide RNAs under the control of polymerase III promoters into the vector 3′ long terminal repeat (LTR) sidestepped the need to manufacture guide RNA, and one round of lentiviral exposure ahead of Cas9 mRNA delivery by electroporation and magnetic column processing yielded CAR+TCR-CD52-CAR T cells.37 Cells were screened for karyotype aberrations and possible translocations between edited sites as well as guide-dependent off-target effects preidentified by digenome sequencing.69 Children in the United Kingdom without autologous CAR options were treated and followed a similar treatment protocol with fludarabine, cyclophosphamide, and alemtuzumab lymphodepletion after a single dose of cells. Four of the first 6 treated patients achieved remissions by day 28 and proceeded to allo-SCT. Two children later relapsed and 2 were in ongoing remission.70 An expected consequence of using intensified lymphodepletion, including alemtuzumab, was the frequency of viral reactivations and the depth of cytopenia in the first 3 to 4 weeks after therapy, but the strategy may also have dampened the severity of CRS through depletion of monocytes and macrophages, which may otherwise have generated interleukin-6 and other cytokines. Direct outcome comparisons with licensed autologous therapies are difficult in the absence of controlled studies, but in the context of refractory pediatric B-ALL, long-term disease-free survival in children who received augmented lymphodepletion appears broadly comparable for the small number treated to date.
Guo et al,71 in Beijing, China, applied allogeneic CAR19 T cells generated by lentiviral transduction and electroporation of CRISPR/Cas9 RNP complexes targeting TRAC and B2m. The products were enriched by CD3-mediated depletion and infused into 2 adult patients with R/R diffuse large B-cell lymphoma after the lymphodepletion with fludarabine and cyclophosphamide.71 There was evidence of expansion and biological activity without GVHD, although the disease progressed in both the subjects.
Precision Biosciences have used a proprietary homing endonuclease technology, Arcus, in the early-phase trials of allogeneic CAR19 T cells. Initially, T cells had a single TRAC edit and a sited CAR insertion (with a promoter cassette) and were depleted of residual TCRαβ for the use in patients with R/R B-ALL. Results from PBCAR0191 in 15 subjects noted no instances of GVHD and evidence of remissions by day 28 in 9 subjects, with subsequent allogeneic SCT in 4 subjects.72 In other patients, enhanced lymphodepletion with higher doses of fludarabine (120 mg/m2 in total) and cyclophosphamide (3000 mg/m2 in total) was investigated to improve expansion and persistence in subjects with CD19+ NHL or ALL.73 Although 70% of the responses were noted by day 28, by 6 months, the majority had relapsed. Further iterations (PBCAR19B) are now clinical investigation following the incorporation of an anti-B2m short hairpin RNA to disrupt HLA class I and a non-polymorphic HLA-E transgene to address the issue of host-mediated rejection (NCT04649112).
CRISPR Therapeutics is testing universal donor T cells with a CAR19 homology–flanked cassette integrated into the TRAC locus using AAV, with a second edit at the B2m site using CRISPR/Cas9. Preliminary results from patients with NHL reported a 38% remission rate with no GVHD and 1 case of severe neurotoxicity attributed to viral reactivation, but longer-term outcomes are awaited.74 A similar approach with additional disruption of CD70 is being tested in patients with T-cell lymphoma using anti-CD70 CAR inserted at the TRAC locus (discussed below).75
Genome editing to address fratricide between CAR T cells
When using CAR-T strategies against T-cell malignancies, we have to consider factors that were more easily resolved with anti-B-cell CAR therapies. First, one of the consequences of successful anti-T-cell effects after the infusion is the likelihood of viral reactivations and other complications during a period of deep T-cell lymphopenia. Unlike in the B-cell setting, where B-cell aplasia can be addressed by regular immunoglobulin replacement therapy, there are no ready solutions to T-cell deficiency. The kinetics and quality of T-cell reconstitution may be problematic and could necessitate allogeneic SCT to ensure timely restoration of the T-cell compartment.
Second, the fratricide effects between T cells designed to express receptors against T-cell antigens may preclude the generation of effector cells and critically impair yields by the end of manufacture. Antigens expressed on T cells, including the TCRαβ/CD3 complex, CD5, and CD7 have been targeted by CARs following the steps to disrupt expression by inhibitory or restriction proteins76 or by genome editing.77 Genome editing using TALENs to target TRAC was used to disrupt the assembly of the cell surface TCRαβ/CD3 complex ahead of the expression of an anti-CD3ε CAR in T cells.78 CRISPR/Cas9 editing of TRAC and CD7 for the generation of universal donor CAR7 T cells has also been described and could be used as an allogeneic non-matched donor therapy that avoids the need to harvest from heavily-treated patients and mitigates against the risk of transduction of leukemic blasts.79 In addition, base-edited CAR7 T cells generated by multiplexed cytidine deamination disruption of T cell receptor β chain and CD7 have been described as a universal platform with advantages over Cas9 editing in terms of translocations after multiple edits.80,81 As part of the extended multiplexed editing, PD1 disruption has been incorporated into CAR7-edited cells,81 although the role and method of checkpoint pathway manipulation as well as the most appropriate targets and optimal timing require further elucidation for cellular therapies in general.82
Human applications of anti-CD7 CAR T-cell therapies have been reported with encouraging results against refractory T-cell malignancies, with generally manageable toxicity profiles. In China, anti-CD7 CAR T cells, manufactured from previous or new allo-SCT donors were generated using a vector incorporating an endoplasmic reticulum retention element fused to a CD7-binder (rather than genome editing) to prevent fratricide. Eighteen of 20 patients with R/R T-ALL achieved complete remission and 7 patients proceeded to stem cell transplantation (NCT04689659).83 Moreover, also in China, Gracell84 has reported the application of donor-derived CRISPR/Cas9 CAR7 T cells edited at the TRAC and CD7 loci in T-ALL with 6 out of 6 treated patients achieving CR/CRi in interim reports. The company has also initiated studies of universal TRAC/CD7-disrupted T cells expressing dual moiety CARs against both CD7 and CD19, aiming to exploit the lymphodepleting effects of CAR7 to promote CAR19 effects against B-cell malignancy.56 An initial trial of base-edited CAR7 T cells for T-ALL is underway in London for children with refractory leukemia, aiming to secure remission ahead of allo-SCT (ISRCTN15323014). These universal donor cells have 3 genes disrupted (TRBC, CD7, and CD52) ahead of lentiviral transduction for CAR7 expression.85
Finally, in the context of T-cell lymphoma, allogeneic CAR T-cell therapy, CTX130TM, targets CD70 and includes CRISPR/Cas9 disruption of TRAC,B2m, and CD70. Interim results from 15 evaluable patients have recently been reported with 29% complete remissions, no significant GVHD, and acceptable toxicity profiles in the initial trial phase.75
Cell sources and manufacturing yields
Adult donor-derived T cells have been most widely used for starting cells using steady-state leukapheresis, an efficient and cost-effective method of sourcing large numbers of healthy donor mononuclear cells.86 Excess material can be cryopreserved and used in multiple production campaigns. Cells are generally activated without preseparation of T cells with anti-CD3 and anti-CD28 reagents, and T-cell expansion and dominance develop in the culture over several days. There may be advantages in isolating and transducing specific T-cell subsets, and this has been investigated in studies in an autologous setting.87 The identification and targeting of populations with optimal expansion and effector function would provide a route to more efficient use of materials, including vector stocks. For banking purposes, yields after engineering and any enrichment steps, such as depletion of residual TCRαβ T cells are dependent on the efficiency of the gene-transfer and editing steps as well as expansion kinetics. Longer periods of ex vivo manipulation and proliferation may increase yields but may lead to exhaustion and loss off effector populations, although in general, each manufacturing campaign can readily produce and bank dozens of single-infusion doses.
Alternative starting material includes umbilical cord blood T cells which have distinct ontogeny because naïve phenotype may have anti-tumor advantages, although cell numbers are often limited compared with peripheral blood collections.88, 89, 90 In the future, unlimited cells may be available through pluripotent stem cell engineering, as demonstrated in preclinical studies of CAR-engineered–T cell–derived induced pluripotent stem cells (iPS), including iterations with MHC disruption.91, 92, 93 However, transcriptional studies determined that the lymphocytes subsequently generated are not true TCRαβ-T cells and may resemble innate TCRγδ T cells more. Replicating the complexity of differentiation and maturation of the adaptive T-cell compartment and delivering TCRαβ T cell-like function will require further development, but in the interim, the first clinical applications of products that are currently available are underway. A clonal iPS–derived T-cell line (FT819) with a CAR19 incorporating a modified signaling domain and inserted at the TRAC locus is being investigated in combination with interleukin-2 in subjects with R/R B-cell malignancies (NCT04629729) with the first patient receiving the dose in 2021.94
Safety and risks of allogeneic genome-edited T cells
Similar to the autologous setting, complications and toxicities of allogeneic edited cells can arise because of the antigen-binding properties of the CAR and may include on- and off-target effects, and issues such as CRS and neurotoxicity may arise in the days and weeks after infusion. In the longer term, perhaps unforeseen consequences related to gene modification and the use of integrating vectors could also arise in a manner similar to the autologous context. Existing long-term experience, over a period of 2 decades with gamma-retroviral and lentiviral transduction, provides confidence that the transformation risk in T-cell studies is low.95,96 In contrast, gamma-retroviral gene addition to hematopoietic stem cells for the correction of various single-gene inherited immune disorders has been linked to malignant transformation.97, 98, 99 Studies subsequently mapped risks to the promoter/enhancer elements of viral LTRs, specific insertion sites, and transactivation of protooncogenes.98, 99, 100, 101 Although much of these risks were obviated by adopting self-inactivating configurations,102,103 recent reports of stem cell transformation in clinical lentiviral stem cell studies have again heightened concerns, although underlying predispositions and the role of chemotherapy may also be important.104 In the T-cell arena, extended tracking in CAR trials has reported clonal dominance (but not transformation) arising as a consequence of lentiviral integration sites in trials of CAR19-105 and CAR22-106 engineered T cells, and there are data mapping integration sites from a large number of subjects, including the earliest treated subject from more than 10 years ago.107 However, T-cell lymphoma has recently been reported in 2 patients receiving piggyBac transposon–modified CAR19 T cells. The underlying mechanisms have yet to be elucidated, but the experience highlights the risks of unexpected adverse events and the importance of careful long-term monitoring. Regarding genome editing, there have been extensive efforts to map on- and off-target nuclease effects and descriptions of chromosomal loss and gain, translocations, and other aberrations after cell manipulation. The quantification of predictable translocations after multiplexed nuclease-mediated–dsDNA cleavage has been reported for some products.27 Karyotype and fluorescence in situ hybridization analysis using a probe close to the TRAC locus on chromosome 14 had previously found that around 5% of the metaphase spreads exhibited abnormal karyotypes after TALEN modification for universal CAR19 T cells.29 Recently, such chromosome 14 abnormalities triggered a temporary regulatory hold after many concerns relating to a patient who had received Allogene Therapeutics’ ALLO-501A CAR-19 T cells trial.108 A similar frequency of karyotype aberrations was also reported after recombinant TCR lentiviral transfer in autologous T cells edited at TRAC and PD1 using CRISPR/Cas9, albeit without any overt consequences.109 Subsequently, additional unbiased investigations using the same guide RNA sequences reported frequent aneuploidy and truncations of chromosomes harboring the TRAC and PD1 target sites.110 Although CRISPR/Cas9 editing at the PD1 locus has suggested that PD-1 operates as a haploinsufficient suppressor of T-cell lymphomagenesis in some animal studies,111 there have been no reports of T-cell lymphomas in human trials involving PD1 editing.
For the lentiviral CRISPR/Cas9 product, TT52CAR19, in which duplex guide RNA cassettes are incorporated into lentiviral LTRs, karyotypes were reported normal at the end of production, but around 1% of cells carried predictable translocations quantified using droplet digital polymerase chain reaction.70 The application of base editors should largely address translocation risk from dsDNA breakage, and several studies have reported direct comparisons between Cas9 editing and base editing for gene disruption.80,112 Mapping sites of off-target base editor activity, whether guide-dependent or otherwise is challenging, and predicting the impact of any detected changes is difficult. The full impact of undesirable edits may perhaps only become apparent in the event of unexpected side events linked to populations of engineered cells, which would then be interrogated in detail. The role of vigilant monitoring, patient tracking, and sample retention over the long term is essential for early-stage therapeutic applications.
Summary
The ability to harvest, manipulate ex vivo, and then characterize and cryopreserve T cells has provided an attractive arena for early adoption of gene therapy technologies, including emerging genome editing platforms. Applications to generate ‘off-the-shelf’ CAR T-cell therapies that can be used without HLA matching are in clinical phase studies, and as the challenges of host immunity and rejection are addressed, wider deployment earlier in therapeutic hierarchies can be anticipated. In time, premanufactured, ready-to-use allogeneic CAR T cells have the potential to increase accessibility and provide reduced-cost alternatives to bespoke CAR therapies.
Conflict-of-interest disclosure: W.Q. has received research funding from Miltenyi, Cellectis & Servier related to T-cell editing and has filed patents related to the application of genome-edited T cells; holds stock in Autolus Therapeutics; and is an advisor for Tessa Therapeutics, Wugen, Novartis, Kite, and Virocell.
Acknowledgments
W.Q. is supported by MRC, Wellcome Trust and the National Institute of Health Research.
Authorship
Contribution: W.Q. wrote the manuscript.
References
- 1.Neelapu SS, Locke FL, Bartlett NL, et al. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N Engl J Med. 2017;377(26):2531–2544. doi: 10.1056/NEJMoa1707447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maude SL, Laetsch TW, Buechner J, et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N Engl J Med. 2018;378(5):439–448. doi: 10.1056/NEJMoa1709866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuster SJ, Bishop MR, Tam CS, et al. Tisagenlecleucel in adult relapsed or refractory diffuse large B-cell lymphoma. N Engl J Med. 2019;380(1):45–56. doi: 10.1056/NEJMoa1804980. [DOI] [PubMed] [Google Scholar]
- 4.Bach PB, Giralt SA, Saltz LB. FDA approval of tisagenlecleucel: promise and complexities of a $475000 cancer drug. JAMA. 2017;318(19):1861–1862. doi: 10.1001/jama.2017.15218. [DOI] [PubMed] [Google Scholar]
- 5.Locke FL, Ghobadi A, Jacobson CA, et al. Long-term safety and activity of axicabtagene ciloleucel in refractory large B-cell lymphoma (ZUMA-1): a single-arm, multicentre, phase 1-2 trial. Lancet Oncol. 2019;20(1):31–42. doi: 10.1016/S1470-2045(18)30864-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neelapu SS, Dickinson M, Munoz J, et al. Axicabtagene ciloleucel as first-line therapy in high-risk large B-cell lymphoma: the phase 2 ZUMA-12 trial. Nat Med. 2022;28(4):735–742. doi: 10.1038/s41591-022-01731-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouchkouj N, Zimmerman M, Kasamon YL, et al. FDA approval summary: axicabtagene ciloleucel for relapsed or refractory follicular lymphoma. Oncologist. 2022;27(7):587–594. doi: 10.1093/oncolo/oyac054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frey NV. Approval of brexucabtagene autoleucel for adults with relapsed and refractory acute lymphocytic leukemia. Blood. 2022;140(1):11–15. doi: 10.1182/blood.2021014892. [DOI] [PubMed] [Google Scholar]
- 9.Wang M, Munoz J, Goy A, et al. KTE-X19 CAR T-cell therapy in relapsed or refractory mantle-cell lymphoma. N Engl J Med. 2020;382(14):1331–1342. doi: 10.1056/NEJMoa1914347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abramson JS, Palomba ML, Gordon LI, et al. Lisocabtagene maraleucel for patients with relapsed or refractory large B-cell lymphomas (TRANSCEND NHL 001): a multicentre seamless design study. Lancet. 2020;396(10254):839–852. doi: 10.1016/S0140-6736(20)31366-0. [DOI] [PubMed] [Google Scholar]
- 11.Raje N, Berdeja J, Lin Y, et al. Anti-BCMA CAR T-cell therapy bb2121 in relapsed or refractory multiple myeloma. N Engl J Med. 2019;380(18):1726–1737. doi: 10.1056/NEJMoa1817226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munshi NC, Anderson LD, Jr., Shah N, et al. Idecabtagene vicleucel in relapsed and refractory multiple myeloma. N Engl J Med. 2021;384(8):705–716. doi: 10.1056/NEJMoa2024850. [DOI] [PubMed] [Google Scholar]
- 13.Berdeja JG, Madduri D, Usmani SZ, et al. Ciltacabtagene autoleucel, a B-cell maturation antigen-directed chimeric antigen receptor T-cell therapy in patients with relapsed or refractory multiple myeloma (CARTITUDE-1): a phase 1b/2 open-label study. Lancet. 2021;398(10297):314–324. doi: 10.1016/S0140-6736(21)00933-8. [DOI] [PubMed] [Google Scholar]
- 14.Sharma P, Kanapuru B, George B, et al. FDA approval summary: idecabtagene vicleucel for relapsed or refractory multiple myeloma. Clin Cancer Res. 2022;28(9):1759–1764. doi: 10.1158/1078-0432.CCR-21-3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacoby E, Bielorai B, Avigdor A, et al. Locally produced CD19 CAR T cells leading to clinical remissions in medullary and extramedullary relapsed acute lymphoblastic leukemia. Am J Hematol. 2018;93(12):1485–1492. doi: 10.1002/ajh.25274. [DOI] [PubMed] [Google Scholar]
- 16.Ortiz-Maldonado V, Rives S, Castella M, et al. CART19-BE-01: a multicenter trial of ARI-0001 cell therapy in patients with CD19(+) relapsed/refractory malignancies. Mol Ther. 2021;29(2):636–644. doi: 10.1016/j.ymthe.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bersenev A. CAR-T cell manufacturing: time to put it in gear. Transfusion. 2017;57(5):1104–1106. doi: 10.1111/trf.14110. [DOI] [PubMed] [Google Scholar]
- 18.Ruella M, Xu J, Barrett DM, et al. Induction of resistance to chimeric antigen receptor T cell therapy by transduction of a single leukemic B cell. Nat Med. 2018;24(10):1499–1503. doi: 10.1038/s41591-018-0201-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tzannou I, Papadopoulou A, Naik S, et al. Off-the-shelf virus-specific T cells to treat BK virus, human herpesvirus 6, cytomegalovirus, Epstein-Barr virus, and adenovirus infections after allogeneic hematopoietic stem-cell transplantation. J Clin Oncol. 2017;35(31):3547–3557. doi: 10.1200/JCO.2017.73.0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curran KJ, Sauter CS, Kernan NA, et al. Durable remission following “off-the-shelf” chimeric antigen receptor (CAR) T-cells in patients with relapse/refractory (R/R) B-cell malignancies. Biol Blood Marrow Transplant. 2020;26(suppl 3):S89. [Google Scholar]
- 21.Cruz CR, Micklethwaite KP, Savoldo B, et al. Infusion of donor-derived CD19-redirected virus-specific T cells for B-cell malignancies relapsed after allogeneic stem cell transplant: a phase 1 study. Blood. 2013;122(17):2965–2973. doi: 10.1182/blood-2013-06-506741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamiya T, Wong D, Png YT, Campana D. A novel method to generate T-cell receptor-deficient chimeric antigen receptor T cells. Blood Adv. 2018;2(5):517–528. doi: 10.1182/bloodadvances.2017012823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bunse M, Bendle GM, Linnemann C, et al. RNAi-mediated TCR knockdown prevents autoimmunity in mice caused by mixed TCR dimers following TCR gene transfer. Mol Ther. 2014;22(11):1983–1991. doi: 10.1038/mt.2014.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Torikai H, Reik A, Liu PQ, et al. A foundation for universal T-cell based immunotherapy: T cells engineered to express a CD19-specific chimeric-antigen-receptor and eliminate expression of endogenous TCR. Blood. 2012;119(24):5697–5705. doi: 10.1182/blood-2012-01-405365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooper LJ. Off-the-shelf T-cell therapy. Blood. 2010;116(23):4741–4743. doi: 10.1182/blood-2010-10-308379. [DOI] [PubMed] [Google Scholar]
- 26.Provasi E, Genovese P, Lombardo A, et al. Editing T cell specificity towards leukemia by zinc finger nucleases and lentiviral gene transfer. Nat Med. 2012;18(5):807–815. doi: 10.1038/nm.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poirot L, Philip B, Schiffer-Mannioui C, et al. Multiplex genome-edited T-cell manufacturing platform for “off-the-shelf” adoptive T-cell immunotherapies. Cancer Res. 2015;75(18):3853–3864. doi: 10.1158/0008-5472.CAN-14-3321. [DOI] [PubMed] [Google Scholar]
- 28.MacLeod DT, Antony J, Martin AJ, et al. Integration of a CD19 CAR into the TCR alpha chain locus streamlines production of allogeneic gene-edited CAR T cells. Mol Ther. 2017;25(4):949–961. doi: 10.1016/j.ymthe.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qasim W, Zhan H, Samarasinghe S, et al. Molecular remission of infant B-ALL after infusion of universal TALEN gene-edited CAR T cells. Sci Transl Med. 2017;9(374):eaaj2013. doi: 10.1126/scitranslmed.aaj2013. [DOI] [PubMed] [Google Scholar]
- 30.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31(3):230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 33.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schumann K, Lin S, Boyer E, et al. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc Natl Acad Sci U S A. 2015;112(33):10437–10442. doi: 10.1073/pnas.1512503112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren J, Liu X, Fang C, Jiang S, June CH, Zhao Y. Multiplex genome editing to generate universal CAR T cells resistant to PD1 inhibition. Clin Cancer Res. 2016;23(9):2255–2266. doi: 10.1158/1078-0432.CCR-16-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ren J, Zhang X, Liu X, et al. A versatile system for rapid multiplex genome-edited CAR T cell generation. Oncotarget. 2017;8(10):17002–17011. doi: 10.18632/oncotarget.15218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Georgiadis C, Preece R, Nickolay L, et al. Long terminal repeat CRISPR-CAR-coupled “Universal” T cells mediate potent anti-leukemic effects. Mol Ther. 2018;26(5):1215–1227. doi: 10.1016/j.ymthe.2018.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533(7603):420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Billon P. CRISPR-mediated base editing enables efficient disruption of eukaryotic genes through induction of stop codons. Mol Cell. 2017;67(6):1068–1079.e4. doi: 10.1016/j.molcel.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaudelli NM. Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage. Nature. 2017;551(7681):464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anzalone AV, Koblan LW, Liu DR. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol. 2020;38(7):824–844. doi: 10.1038/s41587-020-0561-9. [DOI] [PubMed] [Google Scholar]
- 42.Anzalone AV, Gao XD, Podracky CJ, et al. Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nat Biotechnol. 2022;40(5):731–740. doi: 10.1038/s41587-021-01133-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petri K, Zhang W, Ma J, et al. CRISPR prime editing with ribonucleoprotein complexes in zebrafish and primary human cells. Nat Biotechnol. 2022;40(2):189–193. doi: 10.1038/s41587-021-00901-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Al-Homsi A.-S., Anguille S, Deeren D, et al. Immunicy-1: targeting BCMA with cyad-211 to establish proof of concept of an shRNA-based allogeneic CAR T cell therapy platform. Blood. 2021;138:2817. [Google Scholar]
- 45.Bertaina A, Merli P, Rutella S, et al. HLA-haploidentical stem cell transplantation after removal of alphabeta+ T and B cells in children with nonmalignant disorders. Blood. 2014;124(5):822–826. doi: 10.1182/blood-2014-03-563817. [DOI] [PubMed] [Google Scholar]
- 46.Shah RM, Elfeky R, Nademi Z, et al. T-cell receptor alphabeta(+) and CD19(+) cell-depleted haploidentical and mismatched hematopoietic stem cell transplantation in primary immune deficiency. J Allergy Clin Immunol. 2018;141(4):1417–1426.e1411. doi: 10.1016/j.jaci.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 47.Kopolovic I, Ostro J, Tsubota H, et al. A systematic review of transfusion-associated graft-versus-host disease. Blood. 2015;126(3):406–414. doi: 10.1182/blood-2015-01-620872. [DOI] [PubMed] [Google Scholar]
- 48.Hirayama AV, Gauthier J, Hay KA, et al. The response to lymphodepletion impacts PFS in patients with aggressive non-Hodgkin lymphoma treated with CD19 CAR T cells. Blood. 2019;133(17):1876–1887. doi: 10.1182/blood-2018-11-887067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nissani A, Lev-Ari S, Meirson T, et al. Comparison of non-myeloablative lymphodepleting preconditioning regimens in patients undergoing adoptive T cell therapy. J Immunother Cancer. 2021;9(5):e001743. doi: 10.1136/jitc-2020-001743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coles AJ, Compston A, signatories Product licences for alemtuzumab and multiple sclerosis. Lancet. 2014;383(9920):867–868. doi: 10.1016/S0140-6736(14)60440-2. [DOI] [PubMed] [Google Scholar]
- 51.Chakrabarti S, Mackinnon S, Chopra R, et al. High incidence of cytomegalovirus infection after nonmyeloablative stem cell transplantation: potential role of Campath-1H in delaying immune reconstitution. Blood. 2002;99(12):4357–4363. doi: 10.1182/blood.v99.12.4357. [DOI] [PubMed] [Google Scholar]
- 52.Kuhn C, Weiner HL. Therapeutic anti-CD3 monoclonal antibodies: from bench to bedside. Immunotherapy. 2016;8(8):889–906. doi: 10.2217/imt-2016-0049. [DOI] [PubMed] [Google Scholar]
- 53.Valton J, Guyot V, Marechal A, et al. A multidrug resistant engineered CAR T cell for allogeneic combination immunotherapy. Mol Ther. 2015;23(9):1507–1518. doi: 10.1038/mt.2015.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mo F, Watanabe N, McKenna MK, et al. Engineered off-the-shelf therapeutic T cells resist host immune rejection. Nat Biotechnol. 2021;39(1):56–63. doi: 10.1038/s41587-020-0601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quach DH, Becerra-Dominguez L, Rouce RH, Rooney CM. A strategy to protect off-the-shelf cell therapy products using virus-specific T-cells engineered to eliminate alloreactive T-cells. J Transl Med. 2019;17(1):240. doi: 10.1186/s12967-019-1988-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ge J, Yang C, Sun J, et al. Preclinical results of an allogeneic, universal CD19/CD7-targeting CAR-T cell therapy (GC502) for B cell malignancies [abstract] Blood. 2021;138(suppl 1) Abstract 1722. [Google Scholar]
- 57.Torikai H, Reik A, Soldner F, et al. Toward eliminating HLA class I expression to generate universal cells from allogeneic donors. Blood. 2013;122(8):1341–1349. doi: 10.1182/blood-2013-03-478255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kagoya Y, Guo T, Yeung B, et al. Genetic ablation of HLA class I, class II, and the T-cell receptor enables allogeneic T cells to be used for adoptive T-cell therapy. Cancer Immunol Res. 2020;8(7):926–936. doi: 10.1158/2326-6066.CIR-18-0508. [DOI] [PubMed] [Google Scholar]
- 59.Gornalusse GG, Hirata RK, Funk SE, et al. HLA-E-expressing pluripotent stem cells escape allogeneic responses and lysis by NK cells. Nat Biotechnol. 2017;35(8):765–772. doi: 10.1038/nbt.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu H, Wang B, Ono M, et al. Targeted Disruption of HLA genes via CRISPR-Cas9 generates iPSCs with enhanced immune compatibility. Cell Stem Cell. 2019;24(4):566–578.e7. doi: 10.1016/j.stem.2019.02.005. [DOI] [PubMed] [Google Scholar]
- 61.Matheus F, Raveh T, Oro AE, Wernig M, Drukker M. Is hypoimmunogenic stem cell therapy safe in times of pandemics? Stem Cell Rep. 2022;17(4):711–714. doi: 10.1016/j.stemcr.2022.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eyquem J, Mansilla-Soto J, Giavridis T, et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543(7643):113–117. doi: 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schober K, Muller TR, Gokmen F, et al. Orthotopic replacement of T-cell receptor alpha- and beta-chains with preservation of near-physiological T-cell function. Nat Biomed Eng. 2019;3(12):974–984. doi: 10.1038/s41551-019-0409-0. [DOI] [PubMed] [Google Scholar]
- 64.Roth TL, Puig-Saus C, Yu R, et al. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature. 2018;559(7714):405–409. doi: 10.1038/s41586-018-0326-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oh SA, Senger K, Madireddi S, et al. High-efficiency nonviral CRISPR/Cas9-mediated gene editing of human T cells using plasmid donor DNA. J Exp Med. 2022;219(5):e20211530. doi: 10.1084/jem.20211530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kath J, Du W, Pruene A, et al. Pharmacological interventions enhance virus-free generation of TRAC-replaced CAR T cells. Mol Ther Methods Clin Dev. 2022;25:311–330. doi: 10.1016/j.omtm.2022.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuhlen M, Willasch AM, Dalle JH, et al. Outcome of relapse after allogeneic HSCT in children with ALL enrolled in the ALL-SCT 2003/2007 trial. Br J Haematol. 2018;180(1):82–89. doi: 10.1111/bjh.14965. [DOI] [PubMed] [Google Scholar]
- 68.Benjamin R, Graham C, Yallop D, et al. Genome-edited, donor-derived allogeneic anti-CD19 chimeric antigen receptor T cells in paediatric and adult B-cell acute lymphoblastic leukaemia: results of two phase 1 studies. Lancet. 2020;396(10266):1885–1894. doi: 10.1016/S0140-6736(20)32334-5. [DOI] [PubMed] [Google Scholar]
- 69.Kim D, Bae S, Park J, et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12(3):237–243. doi: 10.1038/nmeth.3284. 231 p following 243. [DOI] [PubMed] [Google Scholar]
- 70.Ottaviano G, Georgiadis C, Gkazi SA, et al. Phase 1 clinical trial of CRISPR-engineered CAR19 universal T cells for treatment of children with refractory B cell leukemia. Sci Transl Med. 2022;14(668) doi: 10.1126/scitranslmed.abq3010. [DOI] [PubMed] [Google Scholar]
- 71.Guo Y, Tong C, Su L, et al. CRISPR/Cas9 genome-edited universal CAR T cells in patients with relapsed/refractory lymphoma. Blood Adv. 2022;6(8):2695–2699. doi: 10.1182/bloodadvances.2021006232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jain N, Kantarjian H, Solomon SR, et al. Preliminary safety and efficacy of PBCAR0191, an allogeneic ‘off-the-shelf’ CD19-directed CAR-T for patients with relapsed/refractory (R/R) CD19+ B-ALL [abstract] Blood. 2021;138(suppl 1) Abstract 650. [Google Scholar]
- 73.Shah BD, Jacobson C, Solomon SR, et al. Allogeneic CAR-T PBCAR0191 with intensified lymphodepletion is highly active in patients with relapsed/refractory B-cell malignancies [abstract] Blood. 2021;138(suppl 1) Abstract 302. [Google Scholar]
- 74.McGuirk J, Bachier CR, Bishop MR, et al. A phase 1 dose escalation and cohort expansion study of the safety and efficacy of allogeneic CRISPR-Cas9–engineered T cells (CTX110) in patients (Pts) with relapsed or refractory (R/R) B-cell malignancies (CARBON) J Clin Oncol. 2021;39(suppl 15) TPS7570-TPS7570. [Google Scholar]
- 75.Iyer SP, Sica RA, Ho pj, et al. EHA Library; 2022. The COBALT-LYM Study of CTX130: a phase 1 dose escalation study of CD70-targeted allogeneic CRISPR-CAS9–engineered CAR T cells in patients with relapsed/refractory (R/R) T-cell malignancies; p. S262. 357126. [Google Scholar]
- 76.Png YT, Vinanica N, Kamiya T, Shimasaki N, Coustan-Smith E, Campana D. Blockade of CD7 expression in T cells for effective chimeric antigen receptor targeting of T-cell malignancies. Blood Adv. 2017;1(25):2348–2360. doi: 10.1182/bloodadvances.2017009928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gomes-Silva D, Srinivasan M, Sharma S, et al. CD7-edited T cells expressing a CD7-specific CAR for the therapy of T-cell malignancies. Blood. 2017;130(3):285–296. doi: 10.1182/blood-2017-01-761320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rasaiyaah J, Georgiadis C, Preece R, Mock U, Qasim W. TCRalphabeta/CD3 disruption enables CD3-specific antileukemic T cell immunotherapy. JCI Insight. 2018;3(13):e99442. doi: 10.1172/jci.insight.99442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cooper ML, Choi J, Staser K, et al. An “off-the-shelf” fratricide-resistant CAR-T for the treatment of T cell hematologic malignancies. Leukemia. 2018;32(9):1970–1983. doi: 10.1038/s41375-018-0065-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Georgiadis C, Rasaiyaah J, Gkazi SA, et al. Base-edited CAR T cells for combinational therapy against T cell malignancies. Leukemia. 2021;35(12):3466–3481. doi: 10.1038/s41375-021-01282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Diorio C, Murray R, Naniong M, et al. Cytosine base editing enables quadruple-edited allogeneic CART cells for T-ALL. Blood. 2022;140(6):619–629. doi: 10.1182/blood.2022015825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chan JD, Lai J, Slaney CY, Kallies A, Beavis PA, Darcy PK. Cellular networks controlling T cell persistence in adoptive cell therapy. Nat Rev Immunol. 2021;21(12):769–784. doi: 10.1038/s41577-021-00539-6. [DOI] [PubMed] [Google Scholar]
- 83.Pan J, Tan Y, Wang G, et al. Donor-derived CD7 chimeric antigen receptor T cells for T-cell acute lymphoblastic leukemia: first-in-human, phase I trial. J Clin Oncol. 2021;39(30):3340–3351. doi: 10.1200/JCO.21.00389. [DOI] [PubMed] [Google Scholar]
- 84.Wang X, Li S, Gao L, et al. Safety and efficacy results of GC027: The first-in-human, universal CAR-T cell therapy for adult relapsed/refractory T-cell acute lymphoblastic leukemia (r/r T-ALL) J Clin Oncol. 2020;38(suppl 15) 3013-3013. [Google Scholar]
- 85.Chiesa R, Georgiadis C, Ottaviano G, Syed F, et al. Tvt CAR7: Phase 1 clinical trial of base-edited “universal” CAR7 T cells for paediatric relapsed/refractory T-ALL [abstract] Blood. 2022;140(suppl 1) Abstract 2001. [Google Scholar]
- 86.Mock U, Nickolay L, Philip B, et al. Automated manufacturing of chimeric antigen receptor T cells for adoptive immunotherapy using CliniMACS prodigy. Cytotherapy. 2016;18(8):1002–1011. doi: 10.1016/j.jcyt.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 87.Turtle CJ, Hanafi L.-A., Berger C, et al. CD19 CAR–T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J Clin Investig. 2016;126(6):2123–2138. doi: 10.1172/JCI85309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hiwarkar P, Hubank M, Qasim W, et al. Cord blood transplantation recapitulates fetal ontogeny with a distinct molecular signature that supports CD4(+) T-cell reconstitution. Blood Adv. 2017;1(24):2206–2216. doi: 10.1182/bloodadvances.2017010827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu DD, Hong WC, Qiu KY, et al. Umbilical cord blood: a promising source for allogeneic CAR-T cells. Front Oncol. 2022;12:944248. doi: 10.3389/fonc.2022.944248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cael B, Galaine J, Bardey I, et al. Umbilical cord blood as a source of less differentiated T cells to produce CD123 CAR-T cells. Cancers. 2022;14(13):3168. doi: 10.3390/cancers14133168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Themeli M, Kloss CC, Ciriello G, et al. Generation of tumor-targeted human T lymphocytes from induced pluripotent stem cells for cancer therapy. Nat Biotechnol. 2013;31(10):928–933. doi: 10.1038/nbt.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang B, Iriguchi S, Waseda M, et al. Generation of hypoimmunogenic T cells from genetically engineered allogeneic human induced pluripotent stem cells. Nat Biomed Eng. 2021;5(5):429–440. doi: 10.1038/s41551-021-00730-z. [DOI] [PubMed] [Google Scholar]
- 93.van der Stegen SJC, Lindenbergh PL, Petrovic RM, et al. Generation of T-cell-receptor-negative CD8αβ-positive CAR T cells from T-cell-derived induced pluripotent stem cells. Nat Biomed Eng. 2022;6:1284–1297. doi: 10.1038/s41551-022-00915-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mandal M, Clarke R, van der Stegen S, et al. Abstract 3245: FT819 path to IND: first-of-kind off-the-shelf CAR19 T-cell for B cell malignancies. Cancer Res. 2020;80(suppl 16) 3245-3245. [Google Scholar]
- 95.Scholler J, Brady TL, Binder-Scholl G, et al. Decade-long safety and function of retroviral-modified chimeric antigen receptor T cells. Sci Transl Med. 2012;4(132):132ra153. doi: 10.1126/scitranslmed.3003761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Steffin DHM, Muhsen IN, Hill LC, et al. Long-term follow-up for the development of subsequent malignancies in patients treated with genetically modified IECs. Blood. 2022;140(1):16–24. doi: 10.1182/blood.2022015728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302(5644):415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 98.Ott MG, Schmidt M, Schwarzwaelder K, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12(4):401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 99.Braun CJ, Boztug K, Paruzynski A, et al. Gene therapy for Wiskott-Aldrich syndrome--long-term efficacy and genotoxicity. Sci Transl Med. 2014;6(227):227ra233. doi: 10.1126/scitranslmed.3007280. [DOI] [PubMed] [Google Scholar]
- 100.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2003;348(3):255–256. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- 101.Hacein-Bey-Abina S, Garrigue A, Wang GP, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118(9):3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Montini E, Cesana D, Schmidt M, et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat Biotechnol. 2006;24(6):687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- 103.Montini E, Cesana D, Schmidt M, et al. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection. Hum Gene Ther. 2008;19(10) 1074-1074. [Google Scholar]
- 104.Goyal S, Tisdale J, Schmidt M, et al. Acute myeloid leukemia case after gene therapy for sickle cell disease. N Engl J Med. 2022;386(2):138–147. doi: 10.1056/NEJMoa2109167. [DOI] [PubMed] [Google Scholar]
- 105.Fraietta JA, Nobles CL, Sammons MA, et al. Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells. Nature. 2018;558(7709):307–312. doi: 10.1038/s41586-018-0178-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shah NN, Qin H, Yates B, et al. Clonal expansion of CAR T cells harboring lentivector integration in the CBL gene following anti-CD22 CAR T-cell therapy. Blood Adv. 2019;3(15):2317–2322. doi: 10.1182/bloodadvances.2019000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Melenhorst JJ, Chen GM, Wang M, et al. Decade-long leukaemia remissions with persistence of CD4(+) CAR T cells. Nature. 2022;602(7897):503–509. doi: 10.1038/s41586-021-04390-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sheridan C. Off-the-shelf, gene-edited CAR-T cells forge ahead, despite safety scare. Nat Biotechnol. 2021;40(1):5–8. doi: 10.1038/d41587-021-00027-1. [DOI] [PubMed] [Google Scholar]
- 109.Stadtmauer EA, Fraietta JA, Davis MM, et al. CRISPR-engineered T cells in patients with refractory cancer. Science. 2020;367(6481):eaba7365. doi: 10.1126/science.aba7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nahmad AD, Reuveni E, Goldschmidt E, et al. Frequent aneuploidy in primary human T cells after CRISPR-Cas9 cleavage. Nat Biotechnol. 2022;40(12):1807–1813. doi: 10.1038/s41587-022-01377-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wartewig T, Kurgyis Z, Keppler S, et al. PD-1 is a haploinsufficient suppressor of T cell lymphomagenesis. Nature. 2017;552(7683):121–125. doi: 10.1038/nature24649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Webber BR, Lonetree CL, Kluesner MG, et al. Highly efficient multiplex human T cell engineering without double-strand breaks using Cas9 base editors. Nat Commun. 2019;10(1):5222. doi: 10.1038/s41467-019-13007-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Neelapu SS, Nath R, Munoz J, et al. ALPHA study: ALLO-501 produced deep and durable responses in patients with relapsed/refractory non-Hodgkin’s lymphoma comparable to autologous CAR T [abstract] Blood. 2021;138(suppl 1) Abstract 3878. [Google Scholar]
- 114.Mailankody S, Liedtke M, Sidana S, et al. Universal updated phase 1 data validates the feasibility of allogeneic anti-BCMA ALLO-715 therapy for relapsed/refractory multiple myeloma [abstract] Blood. 2021;138(suppl 1) Abstract 651. [Google Scholar]
- 115.Jain N, Roboz GJ, Konopleva M, et al. Preliminary results from the flu/cy/alemtuzumab arm of the phase I BALLI-01 trial of UCART22, an anti-CD22 allogeneic CAR-T cell product, in adult patients with relapsed or refractory (R/R) CD22+ B-cell acute lymphoblastic leukemia (B-ALL) [abstract] Blood. 2021;138(suppl 1) Abstract 1746. [Google Scholar]
- 116.Hu Y, Zhou Y, Zhang M, et al. CRISPR/Cas9-engineered universal CD19/CD22 dual-targeted CAR-T cell therapy for relapsed/refractory B-cell acute lymphoblastic leukemia. Clin Cancer Res. 2021;27(10):2764–2772. doi: 10.1158/1078-0432.CCR-20-3863. [DOI] [PubMed] [Google Scholar]