ABSTRACT
Actin-related proteins (Arps) are classified according to their similarity to actin and are involved in diverse cellular processes. ACTL7B is a testis-specific Arp, and is highly conserved in rodents and primates. ACTL7B is specifically expressed in round and elongating spermatids during spermiogenesis. Here, we have generated an Actl7b-null allele in mice to unravel the role of ACTL7B in sperm formation. Male mice homozygous for the Actl7b-null allele (Actl7b−/−) were infertile, whereas heterozygous males (Actl7b+/−) were fertile. Severe spermatid defects, such as detached acrosomes, disrupted membranes and flagella malformations start to appear after spermiogenesis step 9 in Actl7b−/− mice, finally resulting in spermatogenic arrest. Abnormal spermatids were degraded and levels of autophagy markers were increased. Co-immunoprecipitation with mass spectrometry experiments identified an interaction between ACTL7B and the LC8 dynein light chains DYNLL1 and DYNLL2, which are first detected in step 9 spermatids and mislocalized when ACTL7B is absent. Our data unequivocally establish that mutations in ACTL7B are directly related to male infertility, pressing for additional research in humans.
Keywords: Actl7b, Infertility, Spermatogenesis, Actin-related proteins, LC8 light chains, Mouse
Summary: Actl7b-deficient mice display spermatogenic arrest, which is likely due to loss of the interaction of ACTL7B with DYNLL1/DYNLL2, a spermatid cytoskeleton protein.
INTRODUCTION
Proteins belonging to the superfamily of actin-like/actin-related proteins share up to 60% amino acid identity with conventional actins and act in various cellular processes, including vesicle trafficking, chromatin modulation, microtubule motility and actin filament dynamics (Schafer and Schroer, 1999).
ACTL7B was first described by Chadwick et al. (1999), who identified and characterized two previously unreported actin-like genes, ACTL7A and ACTL7B (previously known as T-ACTIN 2 and T-ACTIN 1), from the familial dysautonomia candidate region on chromosome 9q31 in human. However, neither gene was found to be mutated in individuals with dysautonomia, suggesting that they are not involved in its pathogenesis. Nucleotide alignment showed high level identity between these two genes and a greater than 40% predicted amino acid identity to a variety of actin proteins. In mice, Actl7a and Actl7b were mapped to chromosome 4 (Chadwick et al., 1999; Hisano et al., 2003). It has been proposed these genes arose before the divergence of rodents and primates by retropositioning of a spliced mRNA transcribed from an actin progenitor gene (Hisano et al., 2003). In mouse and human, ACTL7B is expressed exclusively in the testis (Tanaka et al., 2003; Hisano et al., 2003), suggesting a role in spermatogenesis.
In mouse and human, ACTL7B has been found to be expressed post-meiotically in round and elongating spermatids (Hisano et al., 2003; Guo et al., 2018). ACTL7B is detected in the cytoplasm and at lesser amounts in the nucleus of round and elongating spermatids; it seems to be, in contrast to ACTL7A, evicted with excess cytoplasm at the end of spermiogenesis (Tanaka et al., 2003).
Interestingly, five polymorphisms in ACTL7B (and six in ACTL7A) were detected in a cohort of Japanese infertile male individuals, suggesting that ACTL7B plays a role in fertility (Tanaka et al., 2019). Noteworthy, a study comparing two groups of Luzhong mutton sheep with different fecundity identified nine genes, among these ACTL7B (and ACTL7A), that were associated with reduced litter size (Tao et al., 2021). Furthermore, proteomic and phosphoproteomic analysis of prepubertal and pubertal testis of swamp buffalo identified ACTL7B to be more abundant and phosphorylated in the pubertal testis, again suggesting a role in spermatogenesis (Huang et al., 2020). Single nucleotide polymorphisms in the coding sequence of ACTL7B in infertile men have been reported; however, they have not been directly correlated to male infertility (Tanaka et al., 2007, 2019). Furthermore, a study comparing the expression of testis-enriched genes in fertile and teratozoospermic men identified ACTL7B to be among those genes expressed at significantly lower levels in teratozoospermic men (Ahn et al., 2017). Additionally, a recent study, using comparative proteomics on human testicular tissue, identified ACTL7B, both at the protein and mRNA level, among the six proteins/transcripts with the highest discriminating power of obstructive and non-obstructive azoospermia subtypes (Davalieva et al., 2022).
The molecular function of ACTL7B is not fully understood, although its immunolocalization suggests a role in cytoskeletal organization and/or protein trafficking during spermatogenesis. A recent study by Clement et al. described the generation and analysis of Actl7b-null mice, (Clement et al., 2023). Male Actl7b-null mice are infertile and show severe oligoteratozoospermia with malformations of the sperm tails and heads.
Here, we have generated an Actl7b-null allele using CRISPR/Cas9-mediated gene editing to investigate the role of ACTL7B in more detail. While Aclt7b+/− males were unaffected, Actl7b−/− males were infertile, showing morphological sperm abnormalities as described previously (Clement et al., 2023). Spermatogenic abnormalities arose after step 9 of spermiogenesis, leading to disruption of sperm differentiation as well as spermatid phagocytosis and degradation. Co-immunoprecipitation and mass spectrometry analyses revealed interaction of ACTL7B with dynein light chains DYNLL1 and DYNLL2. Loss of ACTL7B leads to mislocalization of DYNLL1 and DYNLL2 in spermatids of Actl7b−/− males highlighting its role in cytoskeletal re-organization.
RESULTS
Generation of Actl7b-deficient mice
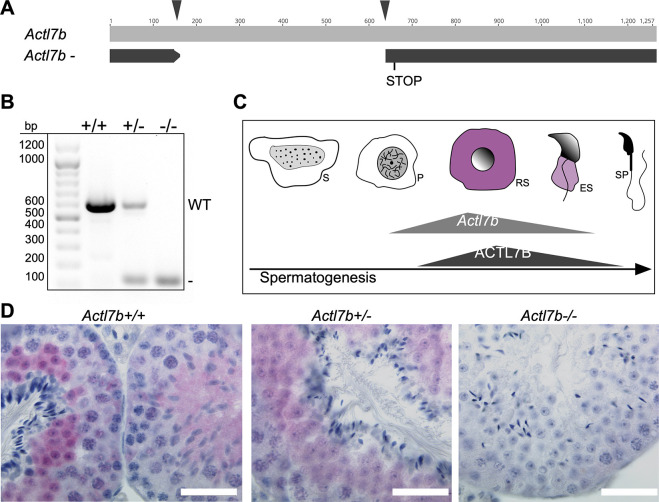
We applied CRSIPR/Cas9-mediated gene editing in zygotes to generate Actl7b-deficient mice. Two guides were used targeting the intron-less coding sequence of Actl7b (Fig. 1A, arrowheads). Founders were backcrossed to C57BL/6J mice and the Actl7b locus was sequenced in the F1 generation. A mouse carrying a 473 bp deletion causing a frameshift was selected to establish an Actl7b-deficient line and animals were analyzed starting from generation N2. A genotyping PCR was established to discriminate the Actl7bΔ from the wild-type allele (Fig. 1B). Actl7b was described to be expressed in round and elongating spermatids in mouse and human (Hisano et al., 2003; Guo et al., 2018; Lukassen et al., 2018) (Fig. 1C, Fig. S1).
Fig. 1.
Establishment of Actl7b-deficient mice. (A) Graphical representation of CRISPR-Cas9-mediated gene editing of the Actl7b locus using two guide RNAs (black arrowheads) targeting the intron-less Actl7b-coding sequence. 473 bp were deleted, causing a frameshift leading to a premature stop. (B) Agarose gel of genotyping polymerase chain reaction of Actl7b+/+, Actl7b+/− and Actl7b−/− mice (wild-type band, 607 bp; KO band, 134 bp). (C) Actl7b expression and ACTL7B immunolocalization during spermiogenesis based on literature (Hisano et al., 2003; Tanaka et al., 2003; Guo et al., 2018). ES, elongating spermatids; P, pachytene spermatocytes; RS, round spermatids; S, spermoatogonia; SP, spermatozoa. (D) Immunohistochemical staining against ACTL7B on Bouin-fixed, paraffin wax-embedded Actl7b+/+, Actl7b+/− and Actl7b−/− testis sections counterstained with Hematoxylin. Scale bars: 20 μm.
We used testis sections from heterozygous (Actl7b+/−), homozygous (Actl7b−/−) and wild-type mice (Actl7b+/+) for immunohistochemical staining against ACTL7B. We demonstrate that ACTL7B localizes to the cytoplasm of round and elongating spermatids in wild-type and heterozygous animals, confirming published data (Tanaka et al., 2003) (Fig. 1D). Of note, the staining is intense in round spermatids and weakens as spermatids elongate. In testis sections of Actl7b+/− mice, ACTL7B signal appeared weaker. No staining was detected in Actl7b−/− testis sections, validating the null allele.
Actl7b deficiency leads to a disruption of spermatogenesis and infertility in male mice
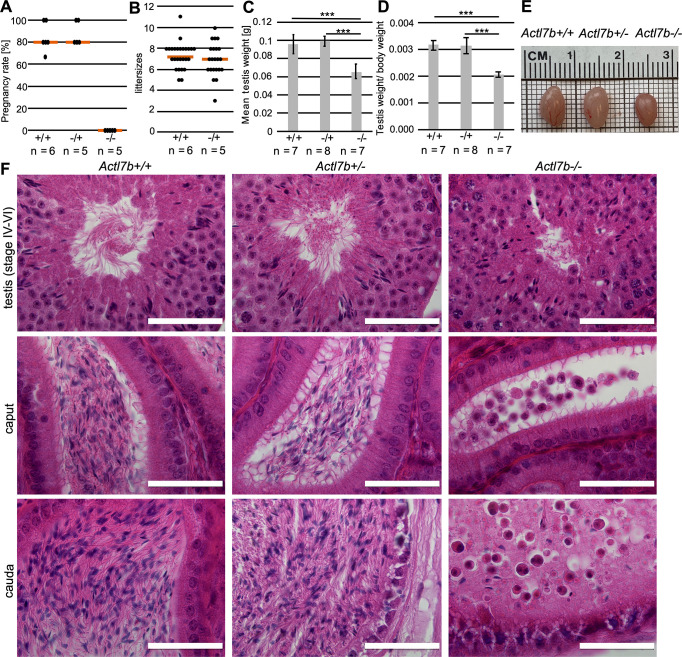
Fertility analysis revealed that Actl7b+/− males produce similar litter sizes and pregnancy frequencies to Actl7b+/+ males (Fig. 2A,B), whereas Actl7b−/− males are infertile. Macroscopic analysis of the reproductive organs showed that testis weight, testis to body weight ratio and testis size of Actl7b−/− males is significantly reduced compared with Actl7b+/− and Actl7b+/+ males (Fig. 2C-E), indicating defects in spermatogenesis. Similar results described by Clement et al. were that homozygous male mice were infertile and showed an approximately 20% reduction in testis weight compared with wild-type controls (Clement et al., 2023).
Fig. 2.
Fertility analysis and reproductive organ morphology. (A) Pregnancy rate of Actl7b+/+, Actl7b+/− and Actl7b−/− males mated with female wild-type C57BL/6J mice (n=number of males). (B) Average litter sizes monitored after mating of Actl7b+/+ and Actl7b+/− males with female wild-type C57BL/6J mice (n=number of males). Five plugs per male were recorded. (C) Mean testis weight of Actl7b+/+, Actl7b+/− and Actl7b−/− males (n=number of males). (D) Testis to body weight ratio of Actl7b+/+, Actl7b+/− and Actl7b−/− males (n=number of males). (E) Photographs of representative testes dissected from Actl7b+/+, Actl7b+/− and Actl7b−/− littermates with similar body weight. (F) Hematoxylin and Eosin staining of testis (stages IV-VI of the epithelial cycle), caput epididymis and cauda epididymis of Actl7b+/+, Actl7b+/− and Actl7b−/− males. Scale bars: 50 μm. Statistical analyses were carried out using a two-tailed, unpaired Student's t-test (***P<0.001). Error bars represent s.d.
Next, histological sections of testis, caput and cauda epididymis were prepared. Sperm production appears normal in Actl7b+/− males (Fig. 2F). Actl7b+/− testis sections containing step 13-15 spermatids do not significantly differ from wild-type testis sections, and epididymides contain mature, morphologically normal sperm. In contrast, spermatogenesis in Actl7b−/− males appears to be disrupted. Caput and cauda epididymides are filled with cell debris, morphologically abnormal spermatids and roundish cells most likely representing immature germ cells (Fig. 2F). Similar observations have been made by Clement et al., who describe the presence of cell debris and many roundish cells with the appearance of round spermatids in cauda epididymides of Actl7b-KO males (Clement et al., 2023). These cells were described to be TUNEL positive, i.e. apoptotic.
Differences between cauda epididymides of wild-type and Actl7b−/− males were already apparent upon inspecting the dissected organs (Fig. S2A). Cauda from Actl7b−/− males appeared smoother and less filled. We isolated sperm from the cauda epididymis from Actl7b+/+, Actl7b+/− and Actl7b−/− males via swim-out. Sperm count was severely reduced in Actl7b−/− with an average of 32,250 sperm from both cauda epididymides (Fig. S2B,C). This corresponds to a 1000-fold reduction in sperm number and differs from the results of Clement et al., who showed an ∼10-fold reduction (Clement et al., 2023). Sperm count was not significantly different between Actl7b+/− and wild-type males. Actl7b+/− sperm appear morphologically normal (Fig. S2D,E), viable (Fig. S2F) and motile (not shown). Although Actl7b−/− males show a pathomorphological phenotype, loss of one allele of Actl7b seems to be phenotypically inconspicuous. In Actl7b−/− mice, daily sperm production (Fig. S2F) and the number of elongating spermatids per seminiferous tubule cross-section (Fig. S2G) were significantly reduced, indicating that the reduction in spermatids originates at least partially from defective spermiogenesis in the testis.
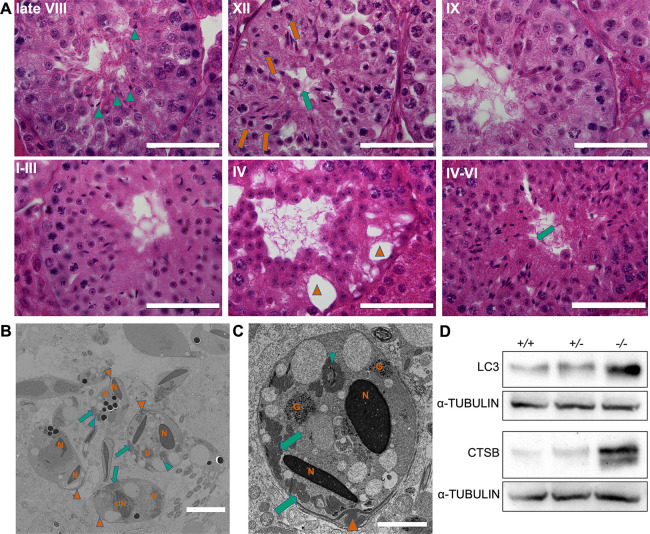
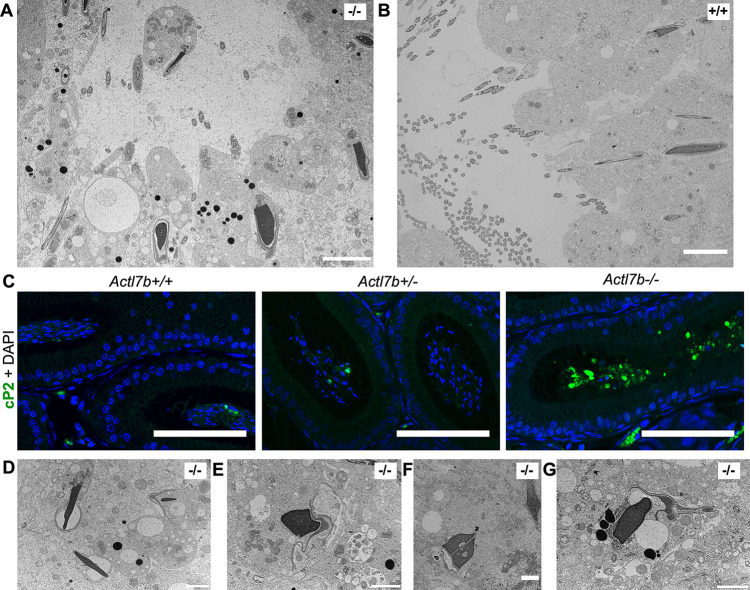
Actl7b−/− seminiferous tubules appear disorganized and germ cell development abnormal (Fig. 3A). Vacuolations in the seminiferous tubules are detected mostly in the basal region of the seminiferous tubules, indicating recent loss of germ cells. During stage VIII, retained abnormally formed elongated spermatids originating from the previous epithelial cycle can be seen. Importantly, we detected round spermatids that seem to be blocked in development and present with dark cytoplasm, indicative of apoptosis/degradation. Immature germ cells seem to be released into the lumen of seminiferous tubules. Clement et al. described the roundish cells being shed into the lumen of seminiferous tubules in their Actl7b-KO mouse model to be TUNEL positive (Clement et al., 2023). Here, we detected vesicles filled with degrading spermatids in Actl7b−/− seminiferous tubules (Fig. 3B,C). These contained condensed nuclei, acrosomal and tail structures, and mitochondria and granular material. This suggests that Sertoli cells have become engulfed and are degrading spermatids. However, TUNEL staining did not reveal significant differences (Fig. S2H). Of note, we found that levels of a mammalian autophagy marker, microtubule-associated protein 1A/1B-light chain 3 (LC3), and of cathepsin B (CTSB) significantly increased in Act7b−/− testis (Fig. 3D). Interestingly, deletion of CTSB in mice leads to inhibition of autophagy and promotion of apoptosis in murine testis, suggesting that CTSB is a regulator between autophagy and apoptosis during spermatid development (Wen et al., 2022). Hence, based on our data we favor autophagy as being the main cause of spermatid degradation.
Fig. 3.
Morphology of Actl7b-deficient seminiferous tubules. (A) Hematoxylin and Eosin staining of Bouin-fixed paraffin wax-embedded testis sections of Actl7b−/− mice. Immature apoptotic germ cells can be seen to be released into the lumen (green arrows). At late stage VIII, elongated spermatids with an abnormal morphology, which were not spermiated, were seen (green arrowheads) and round spermatids blocked in development with dark cytoplasm were found (orange arrows). Vacuolation of seminiferous tubules was detected (orange arrowheads). Scale bars: 50 µm. (B,C) Transmission electron micrographs of vesicles filled with degrading spermatids detected in Actl7b−/− seminiferous tubules. N, condensed nuclei; dN, degraded nucleus; G, granular material. Orange arrowheads indicate acrosomal structures; green arrowheads indicate flagellar cross-sections; green arrows indicate mitochondria. Scale bars: 5 µm in B; 2 µm in C. (D) Western blots on protein extractions from whole Actl7b+/+, Actl7b+/− and Actl7b−/− testis. Anti-LC3 and anti-CSTB were used. α-Tubulin was used as a loading control.
Closer inspection of testicular sections revealed defects in synchronization of the epithelial cycle in Actl7b−/− mice. Cohorts of spermatids in stage IX start elongation but it appears they do not elongate in a synchronous manner (Fig. S3A). Spermatids do not condense properly and seem to localize too close to the basal lamina. Furthermore, cohorts of round spermatids appear abnormal and seem disorganized at stage IX (Fig. S3B). In comparison, wild-type stage IX tubules appear more synchronized. Next, at stage X, morphologically abnormal elongating spermatids are detected, whereas in wild-type stage X tubules, all elongating spermatids appear to be at the same stage and look normal (Fig. S3C). Finally, in stage VII seminiferous tubules of Actl7b−/− males, all elongating spermatids were morphologically abnormal (Fig. S3D). Compared with wild type, Actl7b−/− elongating spermatids do not align properly at the lumen and are very few in number. Substantial numbers of round spermatids are formed in Actl7b−/− testis and spermatogenesis appears normal until this stage. As expected, meiosis was inconspicuous in Actl7b−/− testis and no increased numbers of apoptotic divisions were detected (Fig. S3E).
Spermiogenesis is disrupted in Actl7b−/− males, and Actl7b−/− spermatids show various structural defects
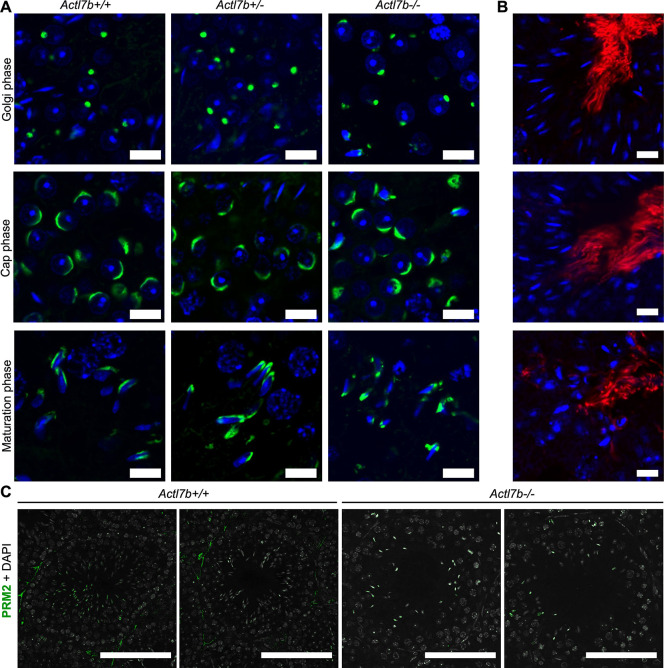
To examine spermiogenesis in Actl7b-deficient males in more detail, we looked at basic parameters, including acrosome biogenesis, DNA condensation, manchette formation and sperm tail formation. Because ACTL7B is first present in round spermatids, effects were expected to manifest from round spermatid stage onwards. In comparison with Actl7b+/− and Actl7b+/+, in Actl7b−/− testis, acrosomal structures are less frequent and disorganized after spermatids start to elongate (Fig. 4A, Fig. S4A,B). In Golgi and Cap phase Actl7b−/− acrosomal structures appear normal, abnormalities become apparent in Maturation phase (Fig. 4A). A signal for ODF2 in the lumen of seminiferous tubules of Actl7b−/−, Actl7b+/− and Actl7b+/+ testis sections indicated that sperm flagellar structures are formed (Fig. 4B, Fig. S4C). These are, however, less frequent and more often found in clusters close to the basal membrane, again suggesting engulfment and degradation of spermatids (Fig. S4C). Staining against PRM2 in the nuclei of elongating spermatids in Actl7b−/−, Actl7b+/− and Actl7b+/+ testis sections indicates that nuclear remodeling and chromatin condensation is initiated (Fig. 4C). Transition proteins are loaded onto the DNA and, later, protamines are detected in the nuclei of spermatids (Fig. S5A,B). Staining against the cleaved domain of PRM2 (cP2) showed that PRM2 localizes to the nucleus of developing spermatids. Finally, in wild-type mice, the remaining full-length unprocessed PRM2 is evicted to the residual bodies (Fig. S5C), as described previously (Arévalo et al., 2022). In contrast, in Actl7b−/− testis sections, residual bodies seem enlarged and less clearly separated from spermatid nuclei. Seemingly, the eviction of cytoplasm fails in large parts of Actl7b−/− spermatids and cP2 filled cytoplasm is retained.
Fig. 4.
Acrosome formation, flagella formation and chromatin condensation in Actl7b-deficient mice. (A) PNA staining of testis of Actl7b+/+, Actl7b+/− and Actl7b−/− males. Acrosomal structures in Golgi, cap and maturation phases are shown. Scale bars: 10 μm. (B) Immunohistochemistry staining against ODF2 on Actl7b+/+ (top) Actl7b+/− (middle) and Actl7b−/− (bottom) testis tissue sections. DAPI was used as the counterstain. Scale bars: 10 μm. (C) Immunohistochemistry staining against PRM2 on Actl7b+/+, Actl7b+/− and Actl7b−/− testis tissue sections. DAPI (in gray) was used as the counterstain. Scale bars: 50 μm.
Transmission electron micrographs reveal that spermatids in Actl7b−/− testes show abnormal morphologies and excess cytoplasm, which is indicative of defective eviction of cytoplasm (Fig. 5A). In comparison, in the lumen of Actl7b+/+ tubules morphologically normal sperm line up to be spermiated (Fig. 5B). Staining against cP2 (to visualize residual cytoplasm) on caput epididymal sections showed that large amounts of cP2 are retained in the cytoplasm of immature germ cells in Actl7b−/− mice (Fig. 5C). This, again, suggests defects in the eviction of cytoplasm. In later developmental stages of Actl7b−/− spermatids in the testis, sperm membranes and acrosomal structures become detached (Fig. 5D,E). Part of the condensed nuclei show inclusions (Fig. 5F) and the overall organization of elongating spermatids appears disorganized (Fig. 5G). Sperm-specific structures fail to assemble correctly. Staining of epididymal sperm with PNA and Mitotracker showed no significant differences between Actl7b+/− and wild-type sperm (Fig. S6A). In contrast, Actl7b−/− epididymal sperm show mislocalization of acrosomal structures and mitochondria. In most Actl7b−/− sperm, PNA and Mitotracker signals were found along the whole tail and in the head region. Of note, Clement et al. reported mislocalization of mitochondria in their Actl7b-KO model (Clement et al., 2023). Clement et al. further describe mislocalization of flagellar proteins in KO sperm and multiple morphological malformations of flagellae. Therefore, we used IC staining against α-tubulin to analyze manchette formation in Actl7b−/− spermatids. Manchettes appear irregular compared with wild type, and Actl7b−/− spermatid head shapes are abnormal (Fig. S6B).
Fig. 5.
Ultrastructural analysis of Actl7b−/− testis. (A) Transmission electron micrograph of a lumen of an Actl7b−/− seminiferous tubule. Scale bar: 5 µm. (B) Transmission electron micrograph of a lumen of an Actl7b+/+ seminiferous tubule. Scale bar: 5 µm. (C) Immunohistochemistry against cP2 on caput sections from Actl7b+/+, Actl7b+/− and Actl7b−/− mice. DAPI was used as a counterstain. Scale bars: 100 µm. (D-G) Images of representative Actl7b−/− spermatids with condensed nuclei. Scale bars: 2 µm.
When analyzing the single steps of spermiogenesis, the first defects become apparent after step 9 (Figs S7 and S8). In part of the developing Actl7b−/− spermatids, chromatin condensation seems to be initiated earlier than in wild-type spermatids. Darker stained chromatin could, however, also be a sign of DNA degradation. Irregular sperm head shaping becomes apparent in step 10 spermatids.
These results lead us to conclude that spermiogenesis is disrupted in Actl7b−/− males. However, the block in development seems to be heterogeneous. Although some germ cells arrest in development at round spermatid stage and appear to be degraded or are released into the lumen, others develop further, form more advanced acrosomal structures, and show hypercondensed chromatin and flagella formation. Apparently, key processes of sperm development are initiated, but seemingly run in an uncoordinated fashion. Finally, abnormally formed spermatids become engulfed and degraded.
ACTL7B interacts with dynein light chains DYNLL1 and DYNLL2
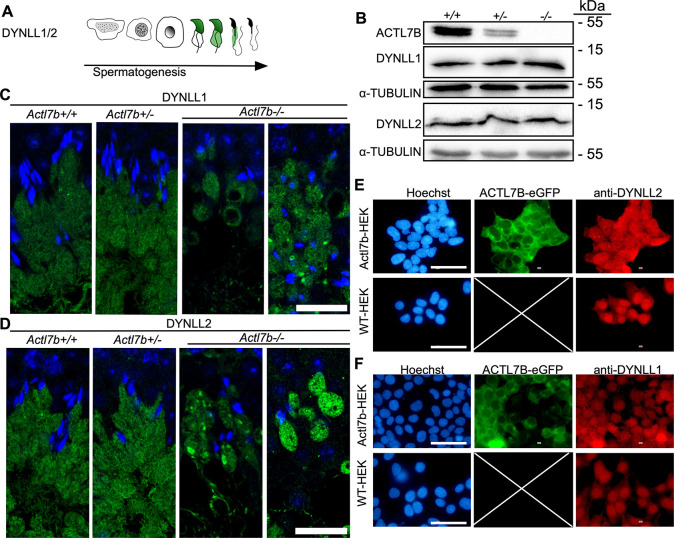
To identify ACTL7B-protein interactions, anti-ACTL7B antibody was coupled to Dynabeads and used for co-immunoprecipitation on protein extracts from whole wild-type testes. Uncoupled beads were used as control. Eluted proteins were identified by mass spectrometry (Table S1). After excluding contaminating proteins such as keratins, proteins identified in the ‘beads only’ control were subtracted from the dataset. Furthermore, a published bead proteome from HeLa cells was used to filter out proteins that nonspecifically bind Dynabeads (Trinkle-Mulcahy et al., 2008). Those included various H2B histone variants. In the co-immunoprecipitation using the anti-ACTL7B-coupled beads, we identified LC8 light chains, dynein light chain 1 (DYNLL1) and its paralog dynein light chain 2 (DYNLL2). Other proteins identified were ribonucleoproteins and ribosomal proteins, which were excluded from further analysis.
We next performed co-immunoprecipitation using anti DYNLL1 and anti-DYNLL2 coupled beads (Fig. 6, Fig. S9). The anti-DYNLL1 antibody appeared not to be suitable for coupling and/or co-immunoprecipitation, as no DYNLL1 was detected in the eluate (not shown). In the eluate of the co-immunoprecipitation using anti-DYNLL2-coupled beads, DYNLL2 and ACTL7B were detected, further supporting the ACTL7B-DYNLL2 interaction.
Fig. 6.
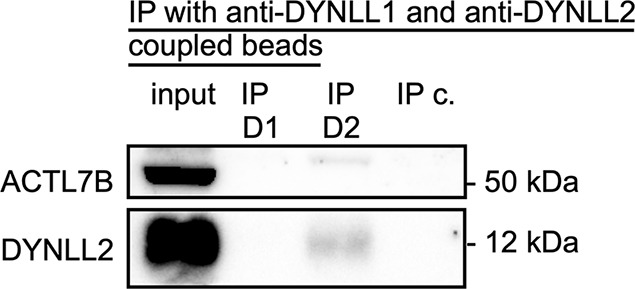
Interaction of ACTL7B with DYNLL1 and DYNLL2. Western blots of the protein input (whole wild-type testis), the IP eluate of the anti-DYNLL1-coupled beads (IP D1), the IP eluate of the anti-DYNLL2-coupled beads (IP D2) and the IP eluate of the beads-only control (IP c.). Anti-ACTL7B and anti-DYNLL2 antibodies were used for western blots.
Next, we generated HEK cells stably expressing Actl7b fused to eGFP (HEKActl7b-eGFP) and performed a pull-down with GFP nanobody-coupled beads followed by mass spectrometry analysis on the eluate. Here, we identified ACTL7B and DYNLL1 enriched in the Actl7b sample using a targeted analysis (Table S2). Of note, DYNLL2 peptides were detected but they were either identical to DYNLL1 or the detection level was low.
DYNLL1 is described to be localized first to the nucleus of elongating spermatids then later in the cytoplasm and residual bodies (Fig. 7A) (Wang et al., 2005). Western blots on protein extractions from Actl7b−/−, Actl7b+/− and Actl7b+/+ testis showed that DYNLL1 and DYNLL2 protein amounts are not reduced in Actl7b−/− testis compared with Actl7b+/− and Actl7b+/+ testes (Fig. 7B, Fig. S10). Of note, ACTL7B levels were reduced in Actl7b+/− testes. Immunofluorescent staining against DYNLL1 and DYNLL2 revealed that both proteins show the identical localization. As described, they localize to the head of early elongating spermatids and were present in the cytoplasm at later developmental stages (Figs S11 and S12). Interestingly, DYNLL1 and DYNLL2 expression correlates with the onset of defects observed in spermatids of Actl7b−/− mice. In Actl7b−/− testis, the first clear differences in DYNLL1 and DYNLL2 staining compared with wild type were seen at around stage I-III of the seminiferous cycle. Both light chains should be homogeneously present throughout the whole cytoplasm, which should be orientated towards the lumen in a stream-like fashion. In Actl7b−/− testes, however, the DYNLL1/2-positive cytoplasm is arranged in almost round sacs (Fig. 7C,D, Figs S11 and S12). Vacuolation of the staining and foci of concentrated DYNLL1/2 were detected. Taken together, these results suggest that the localization of DYNLL1 and DYNLL2 is altered in the absence of ACTL7B, whereas the amount of protein is unchanged. In HEKActl7b-eGFPcells, ACTL7B localized mainly to the cytoplasm, and DYNLL1 and DYNLL2 were localized throughout the whole cytoplasm and the staining appeared fibrous (Fig. 7E,F, Fig. S13). In comparison, DYNLL1 and DYNLL2 localized mainly in the nucleus in wild-type HEK cells and only weakly in the cytoplasm. Furthermore, the staining appeared much more even in wild-type HEK cells. Next, we transfected HEK cells with an expression plasmid containing the murine DYNLL1 or DYNLL2 fused to mCherry. Here, a similar staining pattern can be seen (Fig. S14). This supports the notion that ACTL7B interacts with and controls DYNLL1 and DYNLL2 localization in the cell.
Fig. 7.
Localization of DYNLL1 and DYNLL2 in Actl7b-deficient testis. (A) Graphical depiction of DYNLL1 and DYNLL2 immunolocalization during spermiogenesis based on literature (Wang et al., 2005). (B) Western blots on protein extracts from whole Actl7b+/+, Actl7b+/− and Actl7b−/− testis. Anti-ACTL7B, anti-DYNLL2 and anti-DYNLL2 were used. α-Tubulin was used as loading control. (C) DYNLL1 staining in Actl7b+/+, Actl7b+/− and Actl7b−/− elongating spermatids. DAPI was used as a counterstain. Scale bar: 20 µm. (D) DYNLL2 staining in Actl7b+/+, Actl7b+/− and Actl7b−/− elongating spermatids. DAPI was used as a counterstain. Scale bar: 20 µm. (E) Immunocytochemical staining against DYNLL2 in wild-type and ACTL7B-eGFP-expressing HEK cells. Scale bars: 50 µm. (F) Immunocytochemical staining against DYNLL1 in wild-type and ACTL7B-eGFP-expressing HEK cells. Scale bars: 50 µm. Images shown in E and F are taken from the overviews displayed in Figs S11 and S12.
Of note, F-/G-actin ratios are not significantly different in Actl7b−/− compared with Actl7b+/+ or Actl7b+/− testis, suggesting that actin filament turnover is unlikely to be affected in the absence of ACTL7B (Fig. S15A). Even though the co-immunoprecipitation did not reveal ACTL7B-actin interaction, actin filament arrangement is disturbed in some areas of Actl7b−/− seminiferous tubules (Fig. S15B).
Loss of ACTL7B leads to proteomic changes in Actl7b−/− testis
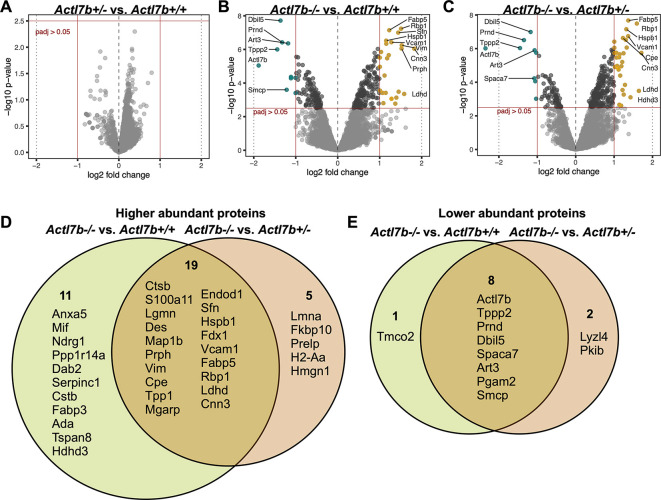
In order to analyze alterations in the testicular proteome in Actl7b-deficient mice, protein samples isolated from whole testes of five Actl7b−/−, five Actl7b+/− and five Aclt7b+/+ mice were used for mass spectrometric analysis (Table S3). Principal component analysis (PCA) showed that the Actl7b−/− samples cluster is separated from the Actl7b+/− and Actl7b+/+ samples clusters (Fig. S16A). Differential abundance (DA) analysis revealed no significant difference in protein abundance in Actl7b+/− compared with Actl7b+/+ samples (Fig. 8A, Fig. S16B). Differentially abundant proteins were detected in Actl7b−/− samples compared with Actl7b+/− and Actl7b+/+ samples, respectively (Fig. 8B,C). Thirty proteins were more abundant and nine proteins were less abundant in Actl7b−/− compared with Actl7b+/+ samples; 24 proteins were more abundant and 10 proteins were less abundant in Actl7b−/− compared with Actl7b+/−, with a stringent log fold change (LFC) of ≥1 (Fig. 8D,E, Fig. S16B). Nineteen of the more abundant proteins were detected in both comparisons (Fig. 8D).
Fig. 8.
Proteomic analysis of Actl7b-deficient testis. (A-C) Volcano plots showing differential abundance (DA) of proteins in Actl7b+/− compared with Actl7b+/+ (A), in Actl7b−/− compared with Actl7b+/− (B) and in Actl7b−/− compared with Actl7b+/+ (C) testis. Proteins showing a significant DA are indicated in teal (less abundant) and yellow (more abundant) (adjusted P-value>0.05). Top DA proteins are labeled with their corresponding gene symbol. (D,E) Venn diagrams showing the overlap of significantly higher (D) and lower (E) abundance proteins in the comparisons of Actl7b−/− with Actl7b+/+, and Actl7b−/− with Actl7b+/+ testis (adjusted P-value≤0.05, LFC≤1).
Several Sertoli cell-expressed proteins, such as the type III intermediate filament proteins vimentin, desmin and peripherin (Mruk and Cheng, 2004; Vogl et al., 2008), as well as fatty acid-binding protein 5 (FABP5) and intracellular retinol-binding protein 1 (RBP1) (Oresti et al., 2013; Griswold, 2022), were found to be more abundant in Actl7b−/− testicular samples. Differential abundance of these mostly Sertoli cell-enriched proteins might indicate a secondary effect caused by arrested and degrading/apoptotic germ cells. Moreover, proteins associated with protein or nucleic acid degradation, as well as early apoptosis, were more abundant in Actl7b−/− testis (ANXA5, CTSB, LGMN, TPP1 and ENDOD1). Of note, CTSB was also shown to be more abundant via western blot (Figs 8D and 3D). Furthermore, VCAM1, which is proposed to function as an adhesion protein in immunoregulation of the testis, was significantly more abundant in Actl7b−/− testis (Sainio-Pöllänen et al., 1997). As expected, ACTL7B was detected among the significantly less abundant proteins in Actl7b−/− compared with Actl7b+/− and Actl7b+/+ samples. Additional spermatocyte- and spermatid-related proteins were detected to be less abundant in Actl7b−/− samples (ART3, SPACA7, SMCP, TPPP2, TMCO2, PGAM2, LYZL4, PRND and PKIB). One example is the prion-like protein doppel (PRND), which is expressed both in Sertoli cells and in spermatids during the final stages of spermiogenesis (Allais-Bonnet and Pailhoux, 2014). Knockout of Prnd leads to male mouse infertility (Behrens et al., 2002; Paisley et al., 2004). Here, immunohistochemical staining against PRND on testis sections revealed that PRND is absent in some areas of Actl7b−/− seminiferous tubules (Fig. S17).
When applying a less stringent LFC of ≥0.5, 193 proteins were more abundant and 59 proteins less abundant in Actl7b−/− compared with Actl7b+/+ samples (Fig. S16B, Table S1). These proteins were used to analyze the enrichment in biological processes (Fig. S18A,B). Proteins that were more abundant in Actl7b−/− compared with Actl7b+/+ showed an enrichment in protein degradation processes, apoptosis, oxidative stress response, cell motility and localization (Fig. S18A). Levels of the autophagy-related protein LC3B, which were shown to be increased in Actl7b−/− testis compared with Actl7b+/+ testis via western blot (Fig. 3D), were also identified as more abundant by mass spectrometry (Map1lc3b; Table S1). Interestingly, N-cadherin (encoded by Cdh2) was more abundant in Actl7b−/− samples. In Sertoli cells, N-cadherin is required for blood-testis-barrier integrity (Jiang et al., 2015). Sertoli cell N-cadherin interacts with actin and vimentin, and is found at the ectoplasmic specializations between Sertoli cells and germ cells, and in the basal compartment of the seminiferous tubules (Mruk and Cheng, 2004). Ezrin, which was also more abundant in Actl7b−/− testis, regulates Sertoli cell-spermatid-adhesion, influences spermatid polarity and is involved in residual body/phagosome transport (Gungor-Ordueri et al., 2014). It has been shown that in cells transfected with a mutated ezrin, the structures of tubulin, actin and vimentin are altered (Zhang et al., 2020). Furthermore, it has been shown that ezrin interacts with VCAM1 in vitro (Barreiro et al., 2002). Immunohistochemical staining against ezrin showed staining surrounding the seminiferous tubules and an intense accumulation of ezrin around germ cells and vacuolations in Actl7b−/− seminiferous tubules, indicating increased germ cell transport and clearance (Fig. S18C). In comparison, ezrin was below the detection threshold for immunohistochemistry in the epithelial tissue in Actl7b+/− and Actl7b+/+ samples, where ezrin was detected only surrounding the seminiferous tubules. On the other hand, less abundant proteins showed an enrichment in male gamete formation, fertilization and reproduction, as well as in microtubule-based movement (Fig. S18B).
ACTL7A and ACTL7B are highly conserved across primates and rodents
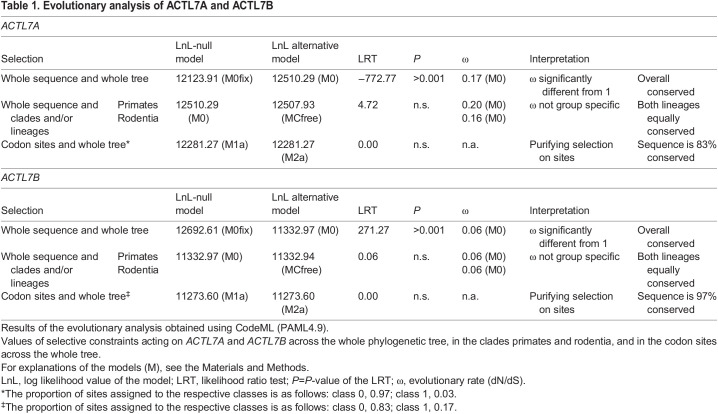
As ACTL7A and ACTL7B are both testis specific and show sequence similarity, we performed evolutionary analysis of both genes to compare their levels of sequence conservation and predict their essentiality. ACTL7A and ACTL7B show 57% amino acid and 52% coding sequence identity in Mus musculus. Of note, human and mouse ACTL7B show 85.9% pairwise amino acid identity. Selective pressures on the codon level were assessed via the nonsynonymous/synonymous substitution rate ratio (ω=dN/dS). This ratio distinguishes between purifying selection (codon sequence conservation) (ω<1), neutral evolution (ω=1) and positive selection (ω>1). Analysis of the selective constrains on ACTL7A and ACTL7B revealed that both are under strong purifying selection across primates and rodents (Table 1, Figs S19 and S20). The evolutionary rates (ω) of the whole sequences across all included species trees were significantly lower than 1 (ACTL7A: ω=0.17, P>0.001; ACTL7B: ω=0.06, P>0.001), with ACTL7B showing an even lower evolutionary rate compared with ACTL7A. When comparing selective pressures between primates and rodents, we found no significant difference in evolutionary rate between the clades. Hence, ACTL7A and ACTL7B are equally highly conserved in both clades. Finally, the selective constraints acting on each codon site were calculated across the whole tree. Ninety-seven percent of the ACTL7B codon sites were conserved, further confirming that ACTL7B is under strong purifying selection. Similarly, 83% of the codon sites were conserved for ACTL7A. These results indicate that changes in the coding sequences, such as mutations, are highly detrimental and are strongly selected against. This usually indicates that the gene and its protein product are highly essential. ACTL7B seems to be more strongly conserved than ACTL7A. Additionally, as both rodents and primate clades, including the murine and human sequences, are equally conserved, we can surmise that ACTL7B is highly essential for both human and mouse spermatogenesis.
Table 1.
Evolutionary analysis of ACTL7A and ACTL7B
DISCUSSION
Here, we have generated an Actl7b-deficient mouse model to analyze the role of ACTL7B in spermatogenesis. We show that, in line with published data (Clement et al., 2023), loss of ACTL7B led to male infertility in mice due to the absence of functional mature sperm. Actl7b−/− spermatids showed an arrest in development, resulting in a wide variety of abnormalities starting after step 9, such as detached acrosomes and membranes, malformed flagella, and impaired eviction of excess cytoplasm. The majority of spermatids were subsequently degraded. A subset of degrading and immature spermatids was released into the lumen of the seminiferous tubules. Vesicles containing degrading spermatids seem to be eliminated by Sertoli cells and levels of autophagy marker proteins are increased in Actl7b−/− testes. Actl7b+/− males showed reduced ACTL7B levels but remained fertile, producing motile viable sperm at concentrations similar to wild type. As the genes of human and mouse ACTL7B are highly similar, we can surmise that ACTL7B variants can lead to failed spermatogenesis and infertility in humans.
Interestingly, the actin-related protein ACTL7B does not seem to interact with the actin cytoskeleton, as we could not find a change in steady-state actin filaments. Rather, ACTL7B seems to link to the microtubule network related functions.
The phenotype of Actl7b deficiency described here is highly similar to the one described by Clement et al., who used homologous recombination in embryonic stem cells to generate and analyze an Actlt7b-KO mouse (Clement et al., 2023). Both models demonstrate severe reductions in epididymal sperm number, germ cell loss in testis, immature germ cell release from testis and multifaceted morphological sperm abnormalities. Of note, although we observed a 1000-fold reduction in sperm count (approximately 32,000,000 in wild type versus 32,000 in KO), Clement et al. reported this reduction to be only 10-fold. Both models were generated by deleting the entire Actl7b-coding sequence (CDS), but the lines were established on C57Bl/6J by us and on C57BL/6N by Clement et al. (2023). This might explain the phenotypic differences (Simon et al., 2013).
By mass spectrometry analyses, we demonstrated that ACTL7B interacts with dynein light chains DYNLL1 and DYNLL2. Of note, ACTL7B is upregulated in round spermatids whereas dynein light chains DYNLL1 and DYNLL2 are detected later, in step 9 spermatids (Wang et al., 2005). This correlates with the onset of the defects in spermatogenesis of Actl7b−/− males, suggesting that the ACTL7B-DYNLL1/2 interaction is crucial and leads to the defects observed. The LC8 family light chains DYNLL1 and its paralog DYNLL2 are highly conserved orthologs among mammals (Rapali et al., 2011). LC8 light chains are presumably involved in dynein complex assembly, thereby indirectly affecting cargo binding. The LC8 light chains are hub proteins that form homodimers with high conformational dynamics of binding grooves, interacting with a wide variety of proteins and functioning in multiple cellular processes such as mitosis, intracellular transport, the stabilization of microtubules, nuclear transport, apoptosis, postsynaptic density and regulation of transcription (Rapali et al., 2011; Nyarko et al., 2011; Jespersen and Barbar, 2020; Hall et al., 2008). They bind intrinsically disordered proteins as dimers, thereby linking subunits in multiprotein complexes (Reardon et al., 2020; Asthana et al., 2012). DYNLL1 was further shown to facilitate dissociation of dynactin from dynein, regulating cargo release (Jin et al., 2014). We hypothesize that DYNLL1 and DYNLL2 promote ACTL7B dimerization and stabilization in multiprotein complexes. Alternatively, ACTL7B might be involved in LC8 transport or activation, as absence of ACTL7B caused mislocalization of the LC8 light chains. Furthermore, we show that expression of Actl7b in HEK cells alters DYNLL1 and DYNLL2 localization in the cell. Effects of the LC8 light chains on the microtubule structure or the dynein 1 motor complex might explain the phenotype seen in Actl7b-deficient mice.
It has been shown that knockdown of the cytoplasmic dynein 1 heavy chain DYNC1H1, causes a disruption of the microtubule structure and polymerization in Sertoli cells in rat (Wen et al., 2018). Furthermore, F-actin organization was perturbed, spermatid polarity was affected, and spermatid transport and release were defective. Finally, phagosome transport was affected. These effects are similar to those described for Actl7b-deficient mice here. Another study identified SPEF2 as interaction partner of DYNC1H1 (Lehti et al., 2017). Germ cell-specific knockout of Spef2 let to multiple spermatid differentiation defects, severely reduced sperm numbers and male mice infertility. It has been proposed that SPEF2 functions as a linker protein, interacting with dynein 1 to facilitate cargo transport along microtubules during spermatid differentiation. ACTL7B might have similar functions. Other dynein light chains have been described in spermatogenesis. For the dynein light chain Tctex-type 4 (DNLT4), 40 different interactors have been identified in human testis, showing the functional variety of dynein light chain interacting proteins (Freitas et al., 2014). In human, absence or lower levels of dynein light chain Tctex-type 1 (DNLT1) have been associated with male infertility (Indu et al., 2015). A detailed analysis of LC8 function in murine spermatogenesis is mandatory in order to reveal the role and consequence of ACTL7B-LC8 interactions.
Mass spectrometric analysis of testicular proteins revealed that Sertoli cell-associated proteins are more abundant in Actl7b−/− testis compared with Actl7b+/− and Actl7b+/+ testis, indicating a reaction to defective spermatids. We show that key autophagy marker proteins show higher levels in Actl7b−/− testes. As TUNEL staining was negative, we favor autophagy as the main cause of spermatid degradation. Even so, apoptosis-related proteins were also found to be more abundant in Actl7b−/− testis. Intermediate filaments (vimentin, desmin and peripherin), ezrin, N-cadherin and VCAM-1 were found to be more abundant. Intermediate filaments usually surround the Sertoli cell nucleus and from there extend to desmosome junctions, which are localized between adjacent Sertoli cells, and between Sertoli cells and germ cells (Johnson, 2014). Ezrin, which accumulated around germ cells and vacuolations, indicating recent germ cell loss in Actl7b−/− testis tissue. Ezrin regulates Sertoli cell-spermatid adhesion as well as phagosome transport (Gungor-Ordueri et al., 2014). N-cadherin localizes to ectoplasmic specializations between Sertoli cells and germ cells (Jiang et al., 2015). It interacts with intermediate filaments and actin. The vascular adhesion molecule VCAM1 interacts with ezrin in vitro (Barreiro et al., 2002). Under inflammation or chronic conditions, VCAM1 expression can be activated by multiple stimuli including pro-inflammatory cytokines and ROS (Kong et al., 2018). VCAM1 expression has been shown to be increased in Sertoli cells that have been exposed to inflammatory mediators in vitro (Riccioli et al., 1995). Taken together, these secondary effects of Actl7b deficiency suggest rapid degradation of abnormal, developmentally blocked spermatids by Sertoli cells. Spermatid degradation correlated with a lower abundance of proteins related to spermatids, male gamete formation and fertility.
ACTL7A and ACTL7B show a high level of sequence identity, and are expressed specifically in the testes of mouse and human, which might suggest functional redundancy (Chadwick et al., 1999; Hisano et al., 2003; Tanaka et al., 2003). However, we have shown that both genes are evolutionarily conserved and under purifying selection in both rodents and primates, suggesting that both genes are required for proper sperm development and function. Furthermore, in murine sperm, ACTL7A is localized to the acrosome and tail, whereas ACTL7B is present in the cytoplasm of round and elongating spermatids (Fu et al., 2012; Tanaka et al., 2003). Although ACTL7B is evicted and detected in residual bodies, ACTL7A is present in the acrosome of mature sperm. Together, the data strongly suggest that ACTL7A and ACTL7B have adapted to different functions. Consequently, the phenotype described for Actl7a mutant mice and humans carrying ACTL7A variants differ from those described here. Homozygous ACTL7A missense mutation causes sperm acrosomal defects and infertility in human and mouse (Xin et al., 2020). ACTL7A-deficient sperm showed reduced levels of PLCζ, a sperm-borne oocyte-activation factor, and artificial oocyte activation overcame the infertility caused by ACTL7A deficiency. Similar phenotypes have recently been described for human and murine sperm carrying homozygous pathogenic variants in ACTL9 (Dai et al., 2021). Indeed, ACTL9 seems to interact with ACTL7A, and both proteins are mislocalized when ACTL9 is mutated. Another recent study identified a homozygous missense variant of ACTL7A in a teratozoospermic patient (Dai et al., 2022). Analysis of a mouse model carrying an equivalent mutation showed that the acrosome and acroplaxome become detached during spermiogenesis. The acroplaxome, ACTL7A and PLCζ are shed and evicted in cytoplasmic droplets. Supporting the results of previous studies, Actl7a-mutated sperm failed to activate the oocyte, leading to infertility. All these studies showed that spermatogenesis is not disrupted when ACTL7A is missing, but acrosome formation is impaired. Hence, the phenotypes of Actl7b and Actl7a deficiency differ greatly. Here, we additionally showed that ACTL7A levels are not elevated in Actl7b−/− mice, further arguing against a compensatory role for ACTL7A in Actl7b−/− mice.
In human, one study identified ACTL7B levels to be significantly lower in teratozoospermic individuals compared with unaffected controls (Ahn et al., 2017). Furthermore, single nucleotide polymorphisms in ACTL7B have been identified in cohorts of infertile individuals (Tanaka et al., 2007, 2019). However, these have not been directly correlated to the infertility. Finally, ACTL7B has been identified to convey a high discriminating power between obstructive and non-obstructive azoospermia subtypes, both at protein and transcript levels, suggesting ACTL7B as a screening marker (Davalieva et al., 2022). Our study clearly shows that mutations in ACTL7B might be directly connected to male infertility, calling for further investigations in human.
MATERIALS AND METHODS
Ethics statement
Animal experiments were performed according to the German law of animal protection and in agreement with the approval of the local institutional animal care committees Landesamt für Natur, Umwelt und Verbraucherschutz, North Rhine-Westphalia (AZ81-0204.2018.A369).
Generation of Actl7b-deficient mice (Mus musculus)
Single guide RNAs (sg1_ts, 5′-CACCCGGACACGGCGTGTCGCAT; sg1_bs, 5′-AAACCATGCGACACGCCGTGTCC; sg2_ts, 5′-CACCAATACGGAAGATCAAGGCG; sg2_bs, 5′-AAACGCGCCTTGATCTTCCGTAT) were designed using the Benchling CRISPR Guide RNA design tool (https://www.benchling.com/crispr/; ENSMUSG00000070980) and tested in embryonic stem cells as described previously (Schneider et al., 2016). The selected guides were ordered as crRNA sequences (Integrated DNA Technologies) and prepared for electroporation as described previously (Arévalo et al., 2022). Briefly, crRNAs were annealed to tracRNA (IDT) (50 nM) and mixed with Cas9 (IDT) in OPTI-MEM (Thermo Fisher Scientific). Potential off-targets of the single guide RNAs were analyzed using the ‘CRISPR-Cas9 guide RNA design checker’ from IDT (https://eu.idtdna.com/site/order/designtool/index/CRISPR_SEQUENCE). The guides had off-target scores of 91/100 and 86/100, respectively. Only two potential off-targets on chromosome 4 were detected (for guide 2), both of which are localized in non-coding areas.
CRISPR-Cas9-mediated gene editing of oocytes was performed as described previously (Arévalo et al., 2022). Six- to eight-week-old B6D2F1 females were superovulated by intraperitoneal injections of 5 i.u. pregnant mare serum (PMS) and 5 i.u. human chorionic gonadotropin (hCG) and mated with B6D2F1 males. Oocytes were isolated at 0.5 dpc and electroporated in OPTI-MEM containing the guide RNA mix using a BioRad Gene Pulser . After recovery and washing, the oocytes were incubated in KSOM (Merck) overlaid with mineral oil at 37°C overnight.
Developing two-cell stage embryos were transferred into the fallopian tube of pseudo-pregnant CD1 foster mice. Offspring was genotyped by PCR and sequenced to identify founder animals. Selected founders were backcrossed to C57BL/6J mice and the F1 generation was sequenced. The Actl7bΔ allele (NM_025271.2:c.159_631del) was further back-crossed to C57BL/6J mice. Starting from the N2 generation, analyses were performed. The allele was registered with Mouse Genome Informatics (Actl7bem1Hsc; 671828).
No significant differences in body weight were monitored for genetically altered mice compared with wild-type mice. Mice were assessed as newborn litter, as litter at weaning and as individual animals every 3 months starting at 8 weeks of age. Nutritional status, posture, coat and orifices, as well as behavior and reaction to handling, were normal in Actl7b-deficient mice.
Genotyping and sequencing of mice
Primers flanking the gene edited region (Actl7b_fwd, 5′-GGGACACAGGTTCCACTCAAC; Actl7b_rev, 5′-AGGTAGTTGGTGAGGTCGCA) were used to amplify both the wild-type and edited allele (cycling conditions: 5 min at 95°C; 35 cycles of 30 s at 95°C, 30 s at 60°C and 45 s at 72°C; 5 min at 72°C). PCR products (wild-type allele, 607 bp; Actl7b−/−, 134 bp) were separated on agarose gels. Samples for sequencing were prepared as described previously (Merges et al., 2022) and sent to GATC/Eurofins for sequencing.
Fertility assessment
Male mice, aged between 8 and 12 weeks, were mated 1:1/1:2 with C57BL/6J females, and females were examined for the presence of a vaginal plug daily. Plug-positive females were separated and monitored for pregnancies and litter sizes. A minimum of five plugs per male were monitored. Pregnancy rate was determined by calculating the percentage of plugs resulting in live-born litter.
Immunohistochemistry and immunofluorescence
Tissues were fixed in Bouin's solution (4°C, overnight), embedded in paraffin wax and 3 µm sections were generated using a microtome (Microm CP60). Heat-mediated antigen retrieval was performed at pH 6 or pH 9 [pH 6: rabbit monoclonal (clone SD08-04) anti-DYNLL1 (Invitrogen, SD08-04, 1:1500), rabbit polyclonal anti-DYNLL2 (Proteintech, 16811-1-AP, 1:1500), rabbit polyclonal anti-ACTL7B (Proteintech, 13537-1-AP, 1:750), mouse anti-PRM2 (Briar Patch Biosciences, Hup2B, 1:200), rabbit polyclonal anti-ODF2 (Proteintech, 12058-1-AP, 1:500), monoclonal (clone 3C12) mouse anti-Erzin (Santa Cruz, sc-58758, 1:100), rabbit polyclonal anti-TNP1 (Abcam, ab73135, 1:1000) and rabbit anti-cP2 (custom antibody, Davids Biotechnologie, 1:500 (Arévalo et al., 2022); pH 9: rabbit anti-actin monoclonal (clone EPR16769) (Abcam, ab179467, 1:500) and rabbit polyclonal anti-PRND (Proteintech, 26947-1-AP, 1:500)]. For slides stained using anti-ACTIN and anti-PRND antibodies, an additional peroxidase blocking step was performed. Slides stained against PRM2, cP2 and TNP1 were additionally treated with decondensation buffer, as described previously (Schneider et al., 2020).
Sections stained using anti-DYNLL1, anti-DYNLL2, anti-TNP1, anti-cP2, anti-PRND, anti-ODF2 and anti-actin were processed using the VectaFluor Horse Anti-Rabbit IgG, DyLight 488 Antibody Kit (Vector Laboratories; DI-1788). Sections stained for ezrin were processed with the VectaFluor Anti-Mouse IgG, DyLight 594 Kit (Vector Laboratories; DI-2794). sections stained for PRM2 were processed with the VectaFluor Duet Immunofluorescence Double Labeling Kit, DyLight 594 Anti-Rabbit, DyLight 488 Anti-Mouse (Vector Laboratories; DK-8828). Sections stained for anti-ACTL7B were processed using the Vectastain ABC-AP Kit (Vector Laboratories; AK-5001) and ImmPACT Vector Red alkaline phosphatase substrate (Vector Laboratories; SK-5105). For all stainings, an extra 30 min blocking step with 5% BSA in PBS was performed. Fluorescent stainings were DAPI counterstained with ProLong Gold antifade reagent (Thermo Fisher Scientific) or ROTI Mount FluorCare DAPI (Carl Roth).
Peanut agglutinin (PNA)-fluorescein isothiocyanite (FITC) Alexa 615 Fluor 488 conjugate (Invitrogen Molecular Probes) was used on deparaffinized sections that were fixed using Bouin's solution. After permeabilization with 0.1% Triton-X 100 for 5 min at room temperature, slides were blocked for 1 h with 1% BSA and incubated for 1 h with PNA (1:200). Slides were mounted with or ROTI Mount FluorCare DAPI (Carl Roth).
TUNEL assay was performed using the TUNEL Assay Kit- HRP-DAB (Abcam, ab206386) according to manufacturer's instructions. Positive control slides were generated by treatment with 1 µg/µl DNase I in TBS/1 mM MgSO4 for 20 min at room temperature, as recommended by the manufacturer.
Testicular sperm were stained using mouse monoclonal (clone DM1A) anti-α-tubulin (Abcam, ab7291, 1:1000) antibodies. After permeabilization with 0.1% Triton-X 100 for 15 min at room temperature, slides were blocked for 1 h with 5% BSA, incubated for 1 h with anti-α-tubulin (1:1000) and mounted with or ROTI Mount FluorCare DAPI (Carl Roth).
PNA-Mito red staining was performed on PFA-fixed (4%, 20 min at room temperature) epididymal sperm. Sperm were incubated with 5 µg/ml Peanut agglutinin (PNA)-fluorescein isothiocyanite (FITC) Alexa 615 Fluor 488 conjugate (Molecular Probes) and 20 nm MitoTracker Red CMXRos (Cell Signaling, 9082) for 45 min at room temperature, washed in PBS and smeared on slides. Slides were mounted with or ROTI Mount FluorCare DAPI (Carl Roth).
Imaging was performed using a confocal Visitron VisiScope and the VisiView Software. Anti-ACTL7B stained sections were counterstained with Haemalum acidic Mayer (Waldeck) and imaged using a Leica DM5500 B microscope. Imaging of testicular and epididymal sperm was performed using a Leica DM5500 B microscope. Staining against DYNLL1 and DYNLL2 was imaged using a LSM 710 (Zeiss).
Protein extraction for immunoblotting and mass spectrometry analysis
Whole testis tissue was homogenized in 1 ml per 100 mg tissue 1:10 RIPA buffer (Cell Signaling Technology) supplemented with Protease Inhibitor (cOmplete ULTRA Tablets, Mini, EASYpack; Roche, Mannheim, Germany). After incubation on ice for 15 min, the samples were sonicated for 5 min using the Bioruptor UCD-200TM-EX (Tosho Denki). Next, the samples were centrifuged for 30 min at 14,000 rpm (20.817 g) at 4°C and the supernatant was used for downstream applications.
Immunoblotting
Protein extracts were separated on a 12% SDS gel with a 5% stacking gel and transferred to PVDF membranes using the Trans-Blot Turbo System (BioRad). Membranes were blocked using 5% milk for 1 h at room temperature. Primary antibodies (rabbit polyclonal anti-ACTL7B; Proteintech, 13537-1-AP), mouse monoclonal (clone DM1A) anti-α-tubulin (Santa Cruz Biotechnology, sc-8035), rabbit monoclonal (clone SD08-04) anti-DYNLL1 (Invitrogen, SD08-04), rabbit polyclonal anti-DYNLL2 (Proteintech, 16811-1-AP), rabbit monoclonal (clone EPR21033) anti-CSTB (Abcam, ab214428) and rabbit polyclonal anti-LC3A/B (ab128025) were diluted 1:1000 in 5% milk and membranes were incubated at 4°C overnight. After washing in TBST, the membranes were incubated with secondary antibodies [polyclonal goat anti-rabbit IgG/HRP (P044801-2; 1:2000), polyclonal rabbit anti-mouse IgG/HRP (P026002-2; 1:1000), Agilent Technologies/Dako] for 1 h at room temperature. After washing in TBST, the signals were detected using WESTARNOVA2.0 chemiluminescent substrate (Cyanagen) and the ChemiDoc MP Imaging system (Bio-Rad). For western blots after Co-IP SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific) was used.
Macroscopic analysis of testis
Sections of Bouin-fixed testis were processed and stained with Hemalum solution acid (Henricks and Mayer, 1965) and Eosin Y solution (Carl Roth) as described previously (Merges et al., 2022).
Periodic acid-Schiff staining
PAS staining was performed as described previously (Schneider et al., 2020). Deparaffinized, re-hydrated slides were incubated for 10 min in periodic acid (0.5%), washed, incubated for 20 min with Schiff's reagent and counterstained. Elongating spermatids per tubule cross-section were counted.
Isolation of epididymal and testicular sperm
Sperm from wild-type and Actl7b+/− males were isolated from the cauda epididymis by swim-out as described previously (Schneider et al., 2016). Epididymal tissue was incised multiple times and incubated in PBS at 37°C for 20-30 min. Sperm count was performed using a Neubauer counting chamber.
Testicular sperm were isolated as described by Kotaja et al. (2004), pipetted onto slides, coverslipped and snap frozen in liquid nitrogen. The cover slip was flipped off and slides were fixed for 5 min in 90% ethanol.
Testicular daily sperm production
Daily sperm production was determined as described by Juma et al. (2017), with modifications. In brief, after removal of the tunica albuginea, testes were homogenized in 400 µl DSP buffer (0.15 M NaCl, 0.1 M NaN3 and 0.05% Triton-X 100 in water). Wild-type and heterozygous samples were adjusted to 4 ml, and KO samples were adjusted to 2 ml final volume using DSP buffer. Elongating spermatids were counted using a Neubauer counting chamber; the result was divided by 4.84.
Transmission electron microscopy
Testis tissue was prepared as described previously (Merges et al., 2022). In brief, the tissue was fixed in 3% glutaraldehyde at 4°C overnight, washed, post-fixed with 2% osmium tetroxide at 4°C for 2 h and washed again. After dehydration and contrasting in 70% (v/v) ethanol 0.5% (m/v) uranyl acetate (1–1.5 h, 4°C), samples were washed with propylenoxide (three times for 10 min at room temperature) and stored in propylenoxide:Epon C (1:1 v/v) at 4°C overnight. Next, the pellets were embedded in Epon C (70°C, 48 h) and ultra-thin sections were prepared. Ultrathin sections were contrasted with UranyLess (Electron Microscopy Sciences) and lead citrate. Images were taken using the Philips CM10 transmission electron microscope equipped with analySiS imaging software and a Zeiss Crossbeam 550 FIB scanning electron microscope equipped with a retractable STEM detector.
Eosin-Nigrosin staining
Staining was performed as described previously (Merges et al., 2022) using 50 μl of sperm swim-out and 50 μl Eosin-Nigrosin stain [0.67 g Eosin Y (color index 45380), 0.9 g sodium chloride, 10 g Nigrosin (color index 50420) and 100 ml deionized H2O]. Two-hundred sperm per animal were analyzed.
Generation of HEKActl7b-eGFPcells
Actl7b CDS was amplified using the Q5 high-fidelity thermostable DNA polymerase (NEB) from mouse testis cDNA using the following primers (GFP-Actl7b_fw, ctcgagctcaagcttcgatggcgacaaagaacag; GFP-Actl7b_rv, ggtaccgtcgactgcagttagcacttgctgtagatgg) and inserted into the pEGFP-C1 vector (Clontech) digested with EcoRI (NEB) using the NEBuilder HiFi DNA Assembly kit (NEB) according to the manufacturer's instructions. The pEGFP-C1-Actl7b plasmid was then linearized with ApaLI (NEB) and transfected into HEK293 cells using Lipofectamine 2000 (ThermoFisher Scientific). Next, cells were selected for 3 weeks with 1 mg/ml geneticin (G418 sulfate, Gibco), trypsinized, diluted to 0.5 cells/100 µl and plated 100 µl/well on a 96-well plate. After 6 weeks, five clones with different degrees of fluorescence were selected, expanded in 0.8 mg/ml geneticin and cryopreserved. Protein lysates of the five clones were tested by western blotting using polyclonal anti-ACTL7B (Invitrogen, PA5-113560, 1:1000). All clones showed different expression levels of the 76 kDa fusion protein in agreement with the fluorescence intensity levels. We chose the clone with the highest fluorescence intensity for further experiments.
Generation of DYNLL1-mCherry and DYNLL2-mCherry expression plasmids
Dynll1 and Dynll2 CDC was amplified from mouse testis cDNA using the following primers: Dynll1_fw, AAAAGAATTCATGTGCGACCGGAAGGC; Dynll1_rv, AAAAGAATTCATGTGCGACCGGAAGGC, TTTTGGATCCTTACCAGATTTGAACAGAAGAATG; Dynll2_fw, AAAAGAATTCATGTCTGACCGGAAGGCAG and Dynll2_rv, TTTTGGATCCTTGCCCGACTTGAGAGGAG). The amplified cDNA was cloned into the p-mCherry-N1 plasmid (Clontech, PT3974-5). Plasmids were sent to GATC/Eurofins (Cologne, Germany) for sequencing.
Transfection of cells
HEK cells were cultured in DMEM (Gibco) with 10% FBS and transfected at 80% confluency. Transfection was performed using 3 μg of expression plasmid with FuGENE HD Transfection Reagent (Promega) according to the manufacturer's instructions. Cells were imaged 12 h later using a Leica DM5500 B microscope.
Co-immunoprecipitation
Proteins from WT C57BL/6J testis were isolated utilizing the T-PER Tissue Protein Extraction Reagent (Thermo Fisher Scientific) supplemented with Halt Protease Inhibitor Single-Use Cocktail EDTA-Free (Thermo Fisher Scientific) according to the manufacturers instructions. Proteins from HEK cells were isolated using the M-PER Mammalian Protein Extraction Reagent (Thermo Fisher Scientific) supplemented with Halt Protease Inhibitor Single-Use Cocktail EDTA-Free (Thermo Fisher Scientific) according to the manufacturers' instructions.
The ACTL7B antibody (Proteintech; 13537-1-AP) was purified using the Amicon Ultra 30K – 0.5 Centrifugal Filter Device (Merck Millipore, UFC503008) according to the manufacturer's instructions. The purified antibody was coupled to beads using the Dynabeads Antibody Coupling Kit (Thermo Fisher Scientific; 14311D), using 7 µg antibody per mg beads. Next, 7.5 mg of antibody-coupled beads and 5 mg empty beads were used with the Dynabeads Co-Immunoprecipitation Kit (Thermo Fisher Scientific; 14321D) according to the manufacturer's instructions. Proteins eluted from the beads in 1 ml HPH EB (0.5 M NH4OH and 0.5 mM EDTA) buffer. 700 µl were lyophilized and sent for mass spectrometry analysis. 300 µl were lyophilized and solubilized in SDS-PAGE sample loading buffer.
DYNLL1 and DYNLL2 antibodies were also purified using Amicon Ultra 30K – 0.5 Centrifugal Filter Device (Merck Millipore). For antibody coupling using the Dynabeads Antibody Coupling Kit (Thermo Fisher Scientific), 10 µg antibody per mg beads were used. 1.5 mg beads were used with the Dynabeads Co-Immunoprecipitation Kit (Thermo Fisher Scientific) for Western Blot applications according to the manufacturer's instructions.
GFP pull-down assay was performed using GFP nanobody-coupled magnetic beads (GFP-Trap Magnetic Particles Kit, Chromotek) according to the manufacturer's instructions. After IP, the bound proteins were eluted in Laemmli's buffer and used for mass spectrometry analysis.
Mass spectrometry
Proteins from whole testis from five wild-type, Actl7b+/− and Actl7b−/− mice were isolated as described and used for mass spectrometric analysis. Peptide preparation and liquid chromatography (LC)-mass spectrometry (MS) were performed at the University of Bonn Core Facility Mass Spectrometry.
Preparation of co-immunoprecipitation samples for LC/MS
50 µg of protein per sample were subjected to in solution preparation of peptides with the iST 96x sample preparation kit (Preomics) according to manufacturer's recommendations.
Whole-cell proteomic analysis: precipitation, proteolysis, peptide labeling and fractionation
Protein lysates with 200 µg protein were mixed with a fourfold volume of chilled acetone (−20°C). After 1 h at −20°C, proteins were sedimented by centrifugation for 15 min at 14,000× g. The supernatant was discarded und pellets air-dried. Pellets were dissolved in 20 µl Lyse buffer (Preomics iST-NHS kit) and protein content was determined by BCA assay. Solutions with 40 µg protein were mixed with 50 µl of the kit's DIGEST solution (3 h, 37°C). 0.25 mg of TMTpro isobaric Mass Tag Labeling reagent (15plex) were added to each sample and incubated at room temperature for 1 h. 10 µl 5% hydroxylamine were used to quench the reaction. The preparation procedure was continued according to the iST-NHS kit instructions. Pooled peptides were dried in a vacuum concentrator, dissolved in 20 mM ammonium formate (pH 10) and fractionated by reverse-phase chromatography at elevated pH with a Reprosil 100 C18 column (3 µm 125×4 mm, Dr Maisch). Sixty fractions were combined into six pools and dried in a vacuum concentrator. Peptides were purified by solid phase extraction (Oasis HLB cartridges, Waters).
LC/MS measurements
Before measurement peptides were re-dissolved in 0.1% formic acid (FA) to yield a 1 g/l solution and separated on a Dionex Ultimate 3000 RSLC nano HPLC system (Dionex). 1 µl was injected onto a C18 analytical column (self-packed 400 mm length and 75 µm inner diameter, ReproSil-Pur 120 C18-AQ, 1.9 µm, Dr Maisch). Peptides were separated during a linear gradient from 5% to 35% solvent B (90% acetonitrile and 0.1% FA) at 300 nl/min. The nanoHPLC was coupled online to an Orbitrap Fusion Lumos mass spectrometer (ThermoFisher Scientific).
Measurement of peptides from co-immunoprecipitated proteins
Gradient length was 90 min. Peptide ions between 300 and 1600 m/z were scanned in the Orbitrap detector every 3 s with a resolution of 120,000 (maximum fill time 50 ms, AGC target 100%). In a top-speed method, peptides were subjected to higher energy collision induced dissociation (1.0 Da isolation, normalized energy 27%) and fragments analyzed in the Orbitrap (resolution 15,000, AGC target 100% and maximum fill time 22 ms). Fragmented peptide ions were excluded from repeat analysis for 20 s.
Targeted measurements
ACTL7B, DYNLL1 and DYNLL2 peptides were analyzed in a targeted mode with 120 min gradient length. MS1 scans were performed every 3 s, peptide precursors were isolated with 1 Da isolation width, fragmented with HCD (28%, maximum injection time 30 ms) and analyzed in Orbitrap.
Measurement of TMT-labelled fractions
Gradient length was 150 min. Peptide ions between 330 and 1600 m/z were scanned in the Orbitrap detector with settings as above. In a top-speed method, peptides were subjected to collision-induced dissociation for identification (CID: 0.7 Da isolation and normalized energy 30%) and fragments analyzed in the linear ion trap with AGC target 50% and maximum fill time 35 ms, rapid mode. Fragmented peptide ions were excluded from repeat analysis for 30 s. Top 10 fragment ions were chosen for synchronous precursor selection and fragmented with higher energy CID (HCD: 3 Da MS2 isolation and 65% collision energy) for detection of reporter ions in the Orbitrap analyzer (range 100-180 m/z, resolution 50,000, maximum fill time 86 ms and AGC target 200%).
Data analysis
Raw data processing and database search were performed with Proteome Discoverer software 2.5.0.400 (Thermo Fisher Scientific). Peptide identification was carried out with an in-house Mascot server version 2.8.1 (Matrix Science). MS data were searched against Mus musculus sequences from the SwissProt database, including isoforms (2022/03, 17132 murine sequences) and contaminants database (cRAP; Mellacheruvu et al., 2013). Precursor ion m/z tolerance was 10 ppm; fragment ion tolerance was 0.5 Da (CID). Tryptic peptides with up to two missed cleavages were searched. C6H11NO-modification of cysteines (delta mass of 113.08406) and TMTpro on N-termini and lysines were set as static modifications. Oxidation was allowed as a dynamic modification of methionine. Mascot results were evaluated by the Percolator algorithm version 3.02.1 (The et al., 2016) as implemented in Proteome Discoverer. Spectra with identifications above 1% q-value were sent to a second round of database search with semi-tryptic enzyme specificity (one missed cleavage allowed). Protein N-terminal acetylation, methionine oxidation, TMTpro and cysteine alkylation were then set as dynamic modifications. Actual FDR values were 0.7% (peptide spectrum matches) and 1.0% (peptides and proteins). Reporter ion intensities (most confident centroid) were extracted from the MS3 level, with SPS mass match>65%.
Data from targeted measurements were analyzed in Skyline (Pino et al., 2020). Validation of MS2 spectra was aided by a spectral library created on the PROSIT server (Gessulat et al., 2019). Protein quantification was carried out at the MS2 level.
Differential abundance analysis
Data for proteins detected in all genotypes and replicates were with more than two peptides were log2 transformed and median normalized. Abundances were analyzed as described previously (Merges et al., 2022). The Bioconductor package proDA (Ahlmann-Eltze and Anders, 2021 preprint) was used [peptide spectrum match (PSM)-level data extracted from Protein Discoverer]. Proteins with LFC>0.5 and LCF>1 (FDR adjusted P<0.05) compared with wild type were analyzed. The R-package ggplot2 (Wickham, 2011) was used to plot the data.
F-actin/G-actin ratio
The F-/G-Actin ratio was analyzed as described previously (Pilo Boyl et al., 2007). Whole-testis tissue was homogenized in 500 µm 1.1×PHEM buffer (600 mM PIPES-Na, 250 mM HEPES-Na, 100 mM EGTA and 20 mM MgCl2; pH 6.9) with 1.2% TritonX-100 and incubated on ice for 15 min. Next, the samples were centrifuged in a swing-out rotor for 10 min at 10,000 g at 4°C. 400 µl of the G-actin congaing supernatant were boiled with 100 µl of 5×SDS loading buffer [110 mM Tris/HCl (pH 6.8), 20% glycerol, 3.8% SDS, 8% β-mercaptoethanol and 0.05% Bromophenol Blue) for 10 min and then cooled on ice. After discarding the rest of supernatant, the pellet was dried and resuspended in 650 µl 1×SDS loading buffer, boiled for 10 min and cooled on ice. Equal volumes of supernatant and pellet fraction were loaded on a 10% acrylamide gel, the gel was blotted on methanol-activated PVDF membrane and anti-β-actin (1:5000; 08-691002; MP Biomedicals) was used to detect the G- and the F-actin fractions. Band quantification was performed using the GE FujiFilm ImageQuant LAS-4000 CH mini Imager (GE Healthcare).
Evolutionary analysis
Evolutionary rates of ACTL7B among rodents and primates were analyzed according to Lüke et al. (2016). ACTL7B-coding sequences were obtained from NCBI GenBank and Ensembl genome browser. Phylogenetic trees of considered species were built according to the ‘Tree of Life web project’ (Letunic and Bork, 2021). The webPRANK software was applied for codon-based phylogeny-aware alignment of orthologous gene sequences (Löytynoja and Goldman, 2010). The tree and alignment were visualized using the ETE toolkit (Huerta-Cepas et al., 2016).
Evolutionary rates and selective pressures were determined using codeML implemented in PAML4.9 (Yang, 1997, 2007). The evolutionary rate is based on the calculation of the nonsynonymous/synonymous substitution rate ratio (ω=dN/dS). It distinguishes between purifying selection (ω<1), neutral evolution (ω=1) and positive selection (ω>1).
Different null and alternative models (M) were applied. The M0 model served as basic model for all performed analyses. Different codon frequency settings were tested for the M0 model of each gene and the setting with the highest likelihood was chosen. To test whether alternative models describe the selective constraints within a dataset better than the null models, likelihood ratio tests (LRT) were performed.
In order to determine the overall evolutionary rate and selective pressure on the coding sequence among all included species, we employed two models: M0 ‘one ratio’, in which all branches were constrained to evolve at the same freely estimated evolutionary rate; and M0fix (fixed ratio) in which the evolutionary rate for all branches was constrained to 1. The M0 model calculates the overall evolutionary rate. A LRT between M0 and M0fix was performed to determine whether the calculated evolutionary rate significantly differs from 1 (neutral).
In order to determine whether the selective pressures differ between rodents and primates, we employed two models: M0 ‘one ratio’; MCfree ‘two-ratio’, which allows the estimation of a free and independent ω for the two marked clades. To test whether the alternative MCfree model presents a better fit for the data, we compared the models log likelihood values by LRT.
To test evolution along coding sequences and infer codon sites under positive or purifying selection, we applied LRT comparing the null model M1a ‘nearly neutral’, which does not allow sites with ω>1, with the alternative model M2a ‘selection’, which does. The models assign the codon sites into different classes: class 0, sites under purifying selection (0>ω>1); class 1, sites evolving neutrally (ω=1); class 2a (only M2a), sites subject to positive selection (ω<1). Bayesian statistics were used to identify those codons that have been subject to either positive selection (if the alternative model is the better fit) or purifying selection (if the null model is the better fit). Only sites with posterior probabilities (Bayes Empirical Bayes) higher than 0.95 to be assigned to class 0 or class 2a were determined to be under purifying or positive selection, respectively.
Statistics
Values are, if not indicated otherwise, given as mean values with standard deviation. Statistical significance was calculated using a two-tailed, unpaired Student's t-test. P<0.05 was considered significant (*P<0.05, **P<0.005, ***P<0.001).
Supplementary Material
Acknowledgements
We thank Gaby Beine, Greta Zech, Andrea Jäger, Angela Egert and Irina Kosterin for excellent technical assistance. Protein identification was performed at the University of Bonn Core Facility Analytical Proteomics, Institute of Biochemistry and Molecular Biology, Medical Faculty, University of Bonn, funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation; Projektnummer 386936527). We thank the Microscopy Core Facility of the Medical Faculty at the University of Bonn for providing support and instrumentation funded by the DFG (388171357). We thank Pietro Pilo Boyl, PhD, for critical discussion and support for construct design to generate the stable cell lines.
Footnotes
Author contributions
Conceptualization: H.S.; Methodology: G.E.M., L.A.; Formal analysis: G.E.M., L.A., D.G.d.R., M.J., W.W.; Investigation: G.E.M., L.A., A.K., K.L.; Resources: C.S.; Data curation: L.A.; Writing - original draft: G.E.M.; Writing - review & editing: G.E.M., L.A., H.S.; Visualization: G.E.M., L.A.; Supervision: H.S.; Project administration: H.S.; Funding acquisition: H.S.
Funding
This study was supported by grants from the Deutsche Forschungsgemeinschaft (SCHO 503/27-1 to H.S.). Open Access funding provided by the University of Bonn. Deposited in PMC for immediate release.
Data availability
Mass spectrometry data have been deposited in PRIDE under accession number PXD038920.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/lookup/doi/10.1242/dev.201593.reviewer-comments.pdf
References
- Ahlmann-Eltze, C. and Anders, S. (2021). proDA: probabilistic dropout analysis for identifying differentially abundant proteins in label-free mass spectrometry. bioRxiv. [Google Scholar]
- Ahn, J., Park, Y.-J., Chen, P., Lee, T. J., Jeon, Y.-J., Croce, C. M., Suh, Y., Hwang, S., Kwon, W.-S., Pang, M.-G.et al. (2017). Comparative expression profiling of testis-enriched genes regulated during the development of spermatogonial cells. PLoS ONE 12, e0175787. 10.1371/journal.pone.0175787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allais-Bonnet, A. and Pailhoux, E. (2014). Role of the prion protein family in the gonads. Front. Cell Dev. Biol. 2, 56. 10.3389/fcell.2014.00056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arévalo, L., Merges, G. E., Schneider, S., Oben, F. E., Neumann, I. S. and Schorle, H. (2022). Loss of the cleaved-protamine 2 domain leads to incomplete histone-to-protamine exchange and infertility in mice. PLoS Genet. 18, e1010272. 10.1371/journal.pgen.1010272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asthana, J., Kuchibhatla, A., Jana, S. C., Ray, K. and Panda, D. (2012). Dynein light chain 1 (LC8) association enhances microtubule stability and promotes microtubule bundling. J. Biol. Chem. 287, 40793-40805. 10.1074/jbc.M112.394353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro, O., Yáñez-Mó, M., Serrador, J. M., Montoya, M. C., Vicente-Manzanares, M., Tejedor, R., Furthmayr, H. and Sánchez-Madrid, F. (2002). Dynamic interaction of VCAM-1 and ICAM-1 with moesin and ezrin in a novel endothelial docking structure for adherent leukocytes. J. Cell Biol. 157, 1233-1245. 10.1083/jcb.200112126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens, A., Genoud, N., Naumann, H., Rulicke, T., Janett, F., Heppner, F. L., Ledermann, B. and Aguzzi, A. (2002). Absence of the prion protein homologue Doppel causes male sterility. EMBO J. 21, 3652-3658. 10.1093/emboj/cdf386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick, B. P., Mull, J., Helbling, L. A., Gill, S., Leyne, M., Robbins, C. M., Pinkett, H. W., Makalowska, I., Maayan, C., Blumenfeld, A.et al. (1999). Cloning, mapping, and expression of two novel actin genes, actin-like-7A (ACTL7A) and actin-like-7B (ACTL7B), from the familial dysautonomia candidate region on 9q31. Genomics 58, 302-309. 10.1006/geno.1999.5848 [DOI] [PubMed] [Google Scholar]
- Clement, T. M., Geyer, C. B., Willis, W. D., Goulding, E. H., Upadhyay, S. and Eddy, E. M. (2023). Actin-related protein ACTL7B ablation leads to OAT with multiple morphological abnormalities of the flagellum and male infertility in mice. Biol. Reprod. 108, 447-464. 10.1093/biolre/ioad001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, J., Zhang, T., Guo, J., Zhou, Q., Gu, Y., Zhang, J., Hu, L., Zong, Y., Song, J., Zhang, S.et al. (2021). Homozygous pathogenic variants in ACTL9 cause fertilization failure and male infertility in humans and mice. Am. J. Hum. Genet. 108, 469-481. 10.1016/j.ajhg.2021.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, J., Chen, Y., Li, Q., Zhang, T., Zhou, Q., Gong, F., Lu, G., Zheng, W. and Lin, G. (2022). Pathogenic variant in ACTL7A causes severe teratozoospermia characterized by bubble-shaped acrosomes and male infertility. Mol. Hum. Reprod. 28, gaac028. 10.1093/molehr/gaac028 [DOI] [PubMed] [Google Scholar]
- Davalieva, K., Rusevski, A., Velkov, M., Noveski, P., Kubelka-Sabit, K., Filipovski, V., Plaseski, T., Dimovski, A. and Plaseska-Karanfilska, D. (2022). Comparative proteomics analysis of human FFPE testicular tissues reveals new candidate biomarkers for distinction among azoospermia types and subtypes. J. Proteomics 267, 104686. 10.1016/j.jprot.2022.104686 [DOI] [PubMed] [Google Scholar]
- Freitas, M. J., Korrodi-Gregório, L., Morais-Santos, F., da Cruz e Silva, E. and Fardilha, M. (2014). TCTEX1D4 interactome in human testis: unraveling the function of dynein light chain in spermatozoa. OMICS 18, 242-253. 10.1089/omi.2013.0133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, J., Wang, Y., Fok, K. L., Yang, D., Qiu, Y., Chan, H. C., Koide, S. S., Miao, S. and Wang, L. (2012). Anti-ACTL7a antibodies: a cause of infertility. Fertil. Steril. 97, 1226-33.e1-8. 10.1016/j.fertnstert.2012.02.023 [DOI] [PubMed] [Google Scholar]
- Gessulat, S., Schmidt, T., Zolg, D. P., Samaras, P., Schnatbaum, K., Zerweck, J., Knaute, T., Rechenberger, J., Delanghe, B., Huhmer, A.et al. (2019). Prosit: proteome-wide prediction of peptide tandem mass spectra by deep learning. Nat. Methods 16, 509-518. 10.1038/s41592-019-0426-7 [DOI] [PubMed] [Google Scholar]
- Griswold, M. D. (2022). Cellular and molecular basis for the action of retinoic acid in spermatogenesis. J. Mol. Endocrinol. 69, T51-T57. 10.1530/JME-22-0067 [DOI] [PubMed] [Google Scholar]
- Gungor-Ordueri, N. E., Tang, E. I., Celik-Ozenci, C. and Cheng, C. Y. (2014). Ezrin is an actin binding protein that regulates sertoli cell and spermatid adhesion during spermatogenesis. Endocrinology 155, 3981-3995. 10.1210/en.2014-1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, J., Grow, E. J., Mlcochova, H., Maher, G. J., Lindskog, C., Nie, X., Guo, Y., Takei, Y., Yun, J., Cai, L.et al. (2018). The adult human testis transcriptional cell atlas. Cell Res. 28, 1141-1157. 10.1038/s41422-018-0099-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, J., Hall, A., Pursifull, N. and Barbar, E. (2008). Differences in dynamic structure of LC8 monomer, dimer, and dimer-peptide complexes. Biochemistry 47, 11940-11952. 10.1021/bi801093k [DOI] [PubMed] [Google Scholar]
- Henricks, D. M. and Mayer, D. T. (1965). Isolation and characterization of a basic keratin-like protein from mammalian spermatozoa. Exp. Cell Res. 40, 402-412. 10.1016/0014-4827(65)90273-9 [DOI] [PubMed] [Google Scholar]
- Hisano, M., Yamada, S., Tanaka, H., Nishimune, Y. and Nozaki, M. (2003). Genomic structure and promoter activity of the testis haploid germ cell-specific intronless genes, Tact1 and Tact2. Mol. Reprod. Dev. 65, 148-156. 10.1002/mrd.10276 [DOI] [PubMed] [Google Scholar]
- Huang, Y.-L., Zhang, P.-F., Fu, Q., He, W.-T., Xiao, K. and Zhang, M. (2020). Novel targets identified by integrated proteomic and phosphoproteomic analysis in spermatogenesis of swamp buffalo (Bubalus bubalis). Sci. Rep. 10, 15659. 10.1038/s41598-020-72353-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas, J., Serra, F. and Bork, P. (2016). ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 33, 1635-1638. 10.1093/molbev/msw046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indu, S., Sekhar, S. C., Sengottaiyan, J., Kumar, A., Pillai, S. M., Laloraya, M. and Kumar, P. G. (2015). Aberrant expression of Dynein light chain 1 (DYNLT1) is associated with human male factor infertility. Mol. Cell. Proteomics 14, 3185-3195. 10.1074/mcp.M115.050005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersen, N. and Barbar, E. (2020). Emerging features of linear motif-binding hub proteins. Trends Biochem. Sci. 45, 375-384. 10.1016/j.tibs.2020.01.004 [DOI] [PubMed] [Google Scholar]
- Jiang, X., Ma, T., Zhang, Y., Zhang, H., Yin, S., Zheng, W., Wang, L., Wang, Z., Khan, M., Sheikh, S. W.et al. (2015). Specific deletion of Cdh2 in Sertoli cells leads to altered meiotic progression and subfertility of mice. Biol. Reprod. 92, 79. 10.1095/biolreprod.114.126334 [DOI] [PubMed] [Google Scholar]
- Jin, M., Yamada, M., Arai, Y., Nagai, T. and Hirotsune, S. (2014). Arl3 and LC8 regulate dissociation of dynactin from dynein. Nat. Commun. 5, 5295. 10.1038/ncomms6295 [DOI] [PubMed] [Google Scholar]
- Johnson, K. J. (2014). Testicular histopathology associated with disruption of the Sertoli cell cytoskeleton. Spermatogenesis 4, e979106. 10.4161/21565562.2014.979106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juma, A. R., Grommen, S. V. H., O'Bryan, M. K., O'Connor, A. E., Merriner, D. J., Hall, N. E., Doyle, S. R., Damdimopoulou, P. E., Barriga, D., Hart, A. H.et al. (2017). PLAG1 deficiency impairs spermatogenesis and sperm motility in mice. Sci. Rep. 7, 5317. 10.1038/s41598-017-05676-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, D.-H., Kim, Y. K., Kim, M. R., Jang, J. H. and Lee, S. (2018). Emerging roles of vascular cell adhesion molecule-1 (VCAM-1) in immunological disorders and cancer. Int. J. Mol. Sci. 19, 1057. 10.3390/ijms19041057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaja, N., Kimmins, S., Brancorsini, S., Hentsch, D., Vonesch, J.-L., Davidson, I., Parvinen, M. and Sassone-Corsi, P. (2004). Preparation, isolation and characterization of stage-specific spermatogenic cells for cellular and molecular analysis. Nat. Methods 1, 249-254. 10.1038/nmeth1204-249 [DOI] [PubMed] [Google Scholar]
- Lehti, M. S., Zhang, F.-P., Kotaja, N. and Sironen, A. (2017). SPEF2 functions in microtubule-mediated transport in elongating spermatids to ensure proper male germ cell differentiation. Development 144, 2683-2693. 10.1242/dev.152108 [DOI] [PubMed] [Google Scholar]
- Letunic, I. and Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293-W296. 10.1093/nar/gkab301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löytynoja, A. and Goldman, N. (2010). webPRANK: a phylogeny-aware multiple sequence aligner with interactive alignment browser. BMC Bioinformatics 11, 579. 10.1186/1471-2105-11-579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukassen, S., Bosch, E., Ekici, A. B. and Winterpacht, A. (2018). Single-cell RNA sequencing of adult mouse testes. Sci. Data 5, 180192. 10.1038/sdata.2018.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüke, L., Tourmente, M., Dopazo, H., Serra, F. and Roldan, E. R. S. (2016). Selective constraints on protamine 2 in primates and rodents. BMC Evol. Biol. 16, 21. 10.1186/s12862-016-0588-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellacheruvu, D., Wright, Z., Couzens, A. L., Lambert, J.-P., St-Denis, N. A., Li, T., Miteva, Y. V., Hauri, S., Sardiu, M. E., Low, T. Y.et al. (2013). The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 10, 730-736. 10.1038/nmeth.2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merges, G. E., Meier, J., Schneider, S., Kruse, A., Fröbius, A. C., Kirfel, G., Steger, K., Arévalo, L. and Schorle, H. (2022). Loss of Prm1 leads to defective chromatin protamination, impaired PRM2 processing, reduced sperm motility and subfertility in male mice. Development 149, dev200330. 10.1242/dev.200330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mruk, D. D. and Cheng, C. Y. (2004). Sertoli-Sertoli and Sertoli-germ cell interactions and their significance in germ cell movement in the seminiferous epithelium during spermatogenesis. Endocr. Rev. 25, 747-806. 10.1210/er.2003-0022 [DOI] [PubMed] [Google Scholar]
- Nyarko, A., Hall, J., Hall, A., Hare, M., Kremerskothen, J. and Barbar, E. (2011). Conformational dynamics promote binding diversity of dynein light chain LC8. Biophys. Chem. 159, 41-47. 10.1016/j.bpc.2011.05.001 [DOI] [PubMed] [Google Scholar]
- Oresti, G. M., García-López, J., Aveldaño, M. I. and Del Mazo, J. (2013). Cell-type-specific regulation of genes involved in testicular lipid metabolism: fatty acid-binding proteins, diacylglycerol acyltransferases, and perilipin 2. Reproduction 146, 471-480. 10.1530/REP-13-0199 [DOI] [PubMed] [Google Scholar]
- Paisley, D., Banks, S., Selfridge, J., Mclennan, N. F., Ritchie, A.-M., Mcewan, C., Irvine, D. S., Saunders, P. T. K., Manson, J. C. and Melton, D. W. (2004). Male infertility and DNA damage in Doppel knockout and prion protein/Doppel double-knockout mice. Am. J. Pathol. 164, 2279-2288. 10.1016/S0002-9440(10)63784-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilo Boyl, P., Di Nardo, A., Mulle, C., Sassoè-Pognetto, M., Panzanelli, P., Mele, A., Kneussel, M., Costantini, V., Perlas, E., Massimi, M.et al. (2007). Profilin2 contributes to synaptic vesicle exocytosis, neuronal excitability, and novelty-seeking behavior. EMBO J. 26, 2991-3002. 10.1038/sj.emboj.7601737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pino, L. K., Searle, B. C., Bollinger, J. G., Nunn, B., Maclean, B. and Maccoss, M. J. (2020). The Skyline ecosystem: Informatics for quantitative mass spectrometry proteomics. Mass Spectrom. Rev. 39, 229-244. 10.1002/mas.21540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapali, P., Szenes, A., Radnai, L., Bakos, A., Pál, G. and Nyitray, L. (2011). DYNLL/LC8: a light chain subunit of the dynein motor complex and beyond. FEBS J. 278, 2980-2996. 10.1111/j.1742-4658.2011.08254.x [DOI] [PubMed] [Google Scholar]
- Reardon, P. N., Jara, K. A., Rolland, A. D., Smith, D. A., Hoang, H. T. M., Prell, J. S. and Barbar, E. J. (2020). The dynein light chain 8 (LC8) binds predominantly “in-register” to a multivalent intrinsically disordered partner. J. Biol. Chem. 295, 4912-4922. 10.1074/jbc.RA119.011653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccioli, A., Filippini, A., De Cesaris, P., Barbacci, E., Stefanini, M., Starace, G. and Ziparo, E. (1995). Inflammatory mediators increase surface expression of integrin ligands, adhesion to lymphocytes, and secretion of interleukin 6 in mouse Sertoli cells. Proc. Natl. Acad. Sci. USA 92, 5808-5812. 10.1073/pnas.92.13.5808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainio-Pöllänen, S., Sundström, J., Erkkilä, S., Hänninen, A., Vainiopää, M., Martikainen, M., Salminen, E., Veräjänkorva, E., Antola, H., Nikula, H.et al. (1997). CD106 (VCAM-1) in testicular immunoregulation. J. Reprod. Immunol. 33, 221-238. 10.1016/S0165-0378(97)00024-7 [DOI] [PubMed] [Google Scholar]
- Schafer, D. A. and Schroer, T. A. (1999). Actin-related proteins. Annu. Rev. Cell Dev. Biol. 15, 341-363. 10.1146/annurev.cellbio.15.1.341 [DOI] [PubMed] [Google Scholar]
- Schneider, S., Balbach, M., Jikeli, J. F., Fietz, D., Nettersheim, D., Jostes, S., Schmidt, R., Kressin, M., Bergmann, M., Wachten, D.et al. (2016). Re-visiting the Protamine-2 locus: deletion, but not haploinsufficiency, renders male mice infertile. Sci. Rep. 6, 36764. 10.1038/srep36764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, S., Shakeri, F., Trötschel, C., Arévalo, L., Kruse, A., Buness, A., Poetsch, A., Steger, K. and Schorle, H. (2020). Protamine-2 deficiency initiates a Reactive Oxygen Species (ROS)-mediated destruction cascade during epididymal sperm maturation in mice. Cells 9, 1789. 10.1101/2020.05.05.075929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, M. M., Greenaway, S., White, J. K., Fuchs, H., Gailus-Durner, V., Wells, S., Sorg, T., Wong, K., Bedu, E., Cartwright, E. J.et al. (2013). A comparative phenotypic and genomic analysis of C57BL/6J and C57BL/6N mouse strains. Genome Biol. 14, R82. 10.1186/gb-2013-14-7-r82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, H., Iguchi, N., Egydio de Carvalho, C., Tadokoro, Y., Yomogida, K. and Nishimune, Y. (2003). Novel actin-like proteins T-ACTIN 1 and T-ACTIN 2 are differentially expressed in the cytoplasm and nucleus of mouse haploid germ cells. Biol. Reprod. 69, 475-482. 10.1095/biolreprod.103.015867 [DOI] [PubMed] [Google Scholar]
- Tanaka, H., Hirose, M., Tokuhiro, K., Matsuoka, Y., Miyagawa, Y., Tsujimura, A., Okuyama, A. and Nishimune, Y. (2007). Single nucleotide polymorphisms: discovery of the genetic causes of male infertility. Soc. Reprod. Fertil. Suppl. 65, 531-534. [PubMed] [Google Scholar]
- Tanaka, H., Miyagawa, Y., Tsujimura, A. and Wada, M. (2019). Genetic polymorphisms within the intronless ACTL7A and ACTL7B genes encoding spermatogenesis-specific actin-like proteins in Japanese males. Int. J. Fertil. Steril. 13, 245-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, L., He, X. Y., Wang, F. Y., Pan, L. X., Wang, X. Y., Gan, S. Q., Di, R. and Chu, M. X. (2021). Identification of genes associated with litter size combining genomic approaches in Luzhong mutton sheep. Anim. Genet. 52, 545-549. 10.1111/age.13078 [DOI] [PubMed] [Google Scholar]
- The, M., Maccoss, M. J., Noble, W. S. and Käll, L. (2016). Fast and accurate protein false discovery rates on large-scale proteomics data sets with percolator 3.0. J. Am. Soc. Mass Spectrom. 27, 1719-1727. 10.1007/s13361-016-1460-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy, L., Boulon, S., Lam, Y. W., Urcia, R., Boisvert, F.-M., Vandermoere, F., Morrice, N. A., Swift, S., Rothbauer, U., Leonhardt, H.et al. (2008). Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol. 183, 223-239. 10.1083/jcb.200805092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogl, A. W., Vaid, K. S. and Guttman, J. A. (2008). The Sertoli cell cytoskeleton. Adv. Exp. Med. Biol. 636, 186-211. 10.1007/978-0-387-09597-4_11 [DOI] [PubMed] [Google Scholar]
- Wang, R.-A., Zhao, M., Meistrich, M. L. and Kumar, R. (2005). Stage-specific expression of dynein light chain-1 and its interacting kinase, p21-activated kinase-1, in rodent testes: implications in spermiogenesis. J. Histochem. Cytochem. 53, 1235-1243. 10.1369/jhc.5A6688.2005 [DOI] [PubMed] [Google Scholar]
- Wen, Q., Tang, E. I., Lui, W.-Y., Lee, W. M., Wong, C. K. C., Silvestrini, B. and Cheng, C. Y. (2018). Dynein 1 supports spermatid transport and spermiation during spermatogenesis in the rat testis. Am. J. Physiol. Endocrinol. Metab. 315, E924-E948. 10.1152/ajpendo.00114.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, Z., Zhu, H., Wu, B., Zhang, A., Wang, H., Cheng, Y., Zhao, H., Li, J., Liu, M. and Gao, J. (2022). Cathepsin B plays a role in spermatogenesis and sperm maturation through regulating autophagy and apoptosis in mice. PeerJ 10, e14472. 10.7717/peerj.14472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, H. (2011). ggplot2. Wiley Interdiscipl. Rev. Comput. Stat. 3, 180-185. 10.1002/wics.147 [DOI] [Google Scholar]
- Xin, A., Qu, R., Chen, G., Zhang, L., Chen, J., Tao, C., Fu, J., Tang, J., Ru, Y., Chen, Y.et al. (2020). Disruption in ACTL7A causes acrosomal ultrastructural defects in human and mouse sperm as a novel male factor inducing early embryonic arrest. Sci. Adv. 6, eaaz4796. 10.1126/sciadv.aaz4796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. (1997). PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13, 555-556. 10.1093/bioinformatics/13.5.555 [DOI] [PubMed] [Google Scholar]
- Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586-1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Zhang, X., Flores, L. R., Keeling, M. C., Sliogeryte, K. and Gavara, N. (2020). Ezrin phosphorylation at T567 modulates cell migration, mechanical properties, and cytoskeletal organization. Int. J. Mol. Sci. 21, 435. 10.3390/ijms21020435 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.