Abstract
Background
SARS-CoV-2 remains a world-wide health issue. SARS-CoV-2-specific immunity is induced upon both infection and vaccination. However, defining the long-term immune trajectory, especially after infection, is limited. In this study, we aimed to further the understanding of long-term SARS-CoV-2-specific immune response after infection.
Results
We conducted a longitudinal cohort study among 93 SARS-CoV-2 recovered individuals. Immune responses were continuously monitored for up to 20 months after infection. The humoral responses were quantified by Spike- and Nucleocapsid-specific IgG levels. T cell responses to Spike- and non-Spike epitopes were examined using both intercellular cytokine staining (ICS) assay and Activation-Induced marker (AIM) assay with quantification of antigen-specific IFNγ production. During the 20 months follow-up period, Nucleocapsid-specific antibody levels and non-Spike-specific CD4 + and CD8 + T cell frequencies decreased in the blood. However, a majority of participants maintained a durable immune responses 20 months after infection: 59% of the participants were seropositive for Nucleocapsid-specific IgG, and more than 70% had persisting non-Spike-specific T cells. The Spike-specific response initially decreased but as participants were vaccinated against COVID-19, Spike-specific IgG levels and T cell frequencies were boosted reaching similar or higher levels compared to 1 month post-infection. The trajectory of infection-induced SARS-CoV-2-specific immunity decreases, but for the majority of participants it persists beyond 20 months. The T cell response displays a greater durability. Vaccination boosts Spike-specific immune responses to similar or higher levels as seen after primary infection.
Conclusions
For most participants, the response persists 20 months after infection, and the cellular response appears to be more long-lived compared to the circulating antibody levels. Vaccination boosts the S-specific response but does not affect the non-S-specific response. Together, these findings support the understanding of immune contraction, and with studies showing the immune levels required for protection, adds to the knowledge of durability of protection against future SARS-CoV-2.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12865-023-00583-y.
Keywords: SARS-CoV-2, Infection, Antigen-specific T cells, Antibodies, Vaccine, Immune durability
Background
In late 2019 Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) emerged, causing the Coronavirus Disease 2019 (COVID-19) pandemic. Now, three years later, the World Health Organisation (WHO) has declared that SARS-CoV-2 no longer constitutes a public health emergency of international concern, but infection remains an ongoing health issue and SARS-CoV-2 variant waves still have a great impact on public health [1]. The humoral and cellular immune responses elicited by SARS-CoV-2 infection can lead to viral clearance, protect against severe disease, and can generally limit viral spread [2–4]. High antibody titers have been correlated with protection against different variants of concern but cannot provide definitive immunity [5–7]. The T cell response is also essential for both achieving viral clearance and limiting disease severity, but the role of the T cell responses is less understood [3, 8].
Early in the pandemic, and following the approval of Spike-based COVID-19 vaccines, great efforts have been put into investigating immunodominant peptide domains within SARS-CoV-2, and a great portion has been identified in the Spike (S) protein [9–12]. Yet, other epitopes outside the S-protein have also showed great potential [9, 10, 12–14], but as vaccines were rolled out the focus on non-S-specific immune characterization declined. The non-S-specific humoral and cellular responses grant an understanding of the adaptive immune contraction over time regardless of vaccination, while the S-specific response enables tracking of the temporal booster effect of vaccination. A decrease in Nucleocapsid (N)-specific antibody levels over time has previously been shown independent of vaccination [10, 15]. The levels of S-specific T cells are also well described, with an upregulation upon infection and vaccination [16, 17]. Some studies have also characterized non-S-specific T cells following infection [15, 17], but there is a lack of longitudinal studies investigating the long-term dynamics of the natural immune response after primary infection, which will help us understand the expected longevity of the T cell response, and thus the necessity of a continued vaccine effort.
Therefore, we selected 93 SARS-CoV-2 recovered participants based on having completed all longitudinal visits, from the previously described CoroNAT cohort [18, 19]. We characterized their adaptive immune responses 1 month after initial SARS-CoV-2 infection and at 3 consecutive time points during a 20-month follow-up. We performed in-depth analyses of the adaptive immune responses involving longitudinal T cell responses towards both S and non-S peptides as well as S-specific and N-specific antibody levels.
Results
The 93 participants from the CoroNAT cohort had completed 4 study visits at approximately 1, 10, 13 and 20 months post primary SARS-CoV-2 infection, respectively (Fig. 1A). The demographics of the participants are shown in Table 1. Briefly, 54% of the cohort were males, and the median age was 48 years (range 20–68 years). The majority of participants (84%) experienced mild SARS-CoV-2 infection (disease severity group 1 + 2), while 16% were hospitalized (disease severity group 3 + 4). COVID-19 vaccines became available during the follow-up period, and the participants received vaccinations according to national guidelines. The vaccine status of the participants can be seen in Supp. Table 1. At 10 months post-infection, 9.7% had received their first vaccine dose, at month 13, 33% had received 1 or 2 vaccine doses and at month 20, 99% had received one, two or three vaccine doses [18, 19].
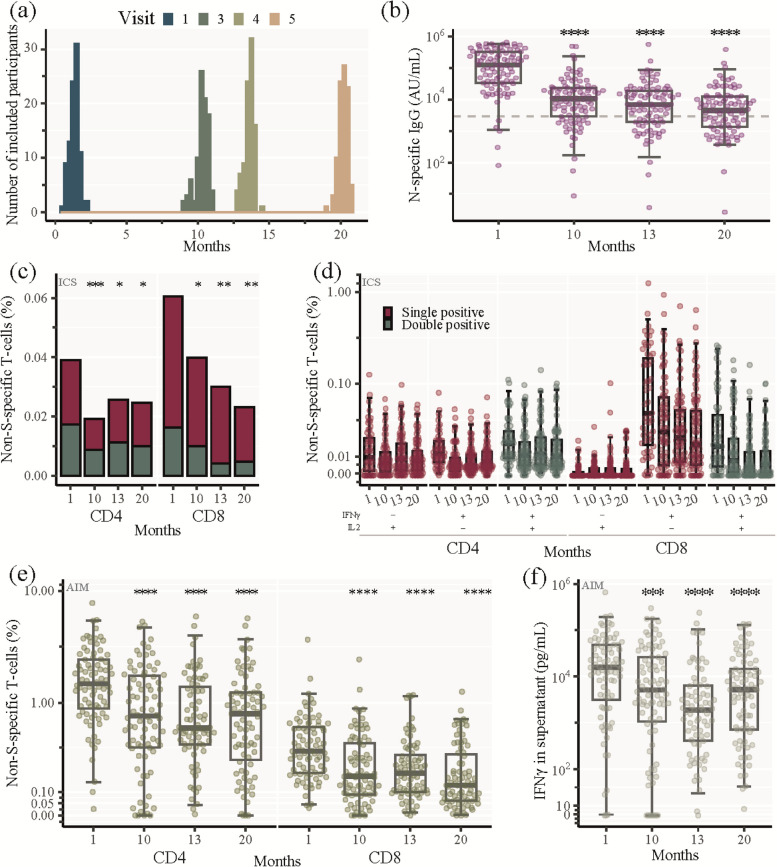
Fig. 1.
SARS-CoV-2 non-Spike-specific longitudinal immune response. a Sampling time of the 93 participants for each visit. Zero months represents the time of infection defined as positive PCR test. b SARS-CoV-2 nucleocapsid (N)-specific IgG analysed by Mesoscale. Being seropositive was defined as IgG levels above 3000 AU/mL (dashed line). c, d Percentage of SARS-CoV-2 non-spike (S)-specific CD4 + and CD8 + memory T cells analysed by ICS. c Total SARS-CoV-2 non-S-specific CD4 + and CD8 + memory T cells at indicated time points. IFNγ only and IL2 only producing cells: red bar, IFNγ and IL2 double producing cells: green bar. Stacked bars represent median values. d Percentage of SARS-CoV-2 non-S-specific CD4 + and CD8 + memory T cells producing either IL2 only (red, left panel), IFNγ only (red, middle panel), or co-producing IFNγ and IL2 (green, right panel) at each visit. e, f Percentage of SARS-CoV-2 non-S-specific CD4 + and CD8 + T cells analysed by AIM. e Total SARS-CoV-2 non-S-specific CD4 + and CD8 + T cells at indicated time points, i.e., cells expressing 2 or 3 activation induced markers (CD69, OX40 or 41BB). f IFNγ production by SARS-CoV-2 non-S-specific cells measured in the supernatant harvested from the cell stimulations in the AIM assay (Analysed by mesoscale). b, d, e, f Box and whisker plots show median values ± IQR and error bars indicate 95% CI. Statistical comparisons were performed using Friedman test and Wilcoxon unpaired signed-ranks test adjusted using Bonferroni with visit 1 as a reference. *P ≤ 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, no asterisk indicates non-significance
Table 1.
Demographics at time of inclusion
| N = 93 | |
|---|---|
| Sex, n (%) | |
| Female | 43 (46%) |
| Male | 50 (54%) |
| Age (years), Median [IQR] | 48 [42, 55] |
| Comorbidities (No. of comorbidities), n (%) | |
| > 1 Comorbidities | 40 (43%) |
| No Comorbidities | 53 (57%) |
| BMI index, n (%) | |
| Normal | 45 (48%) |
| Overweight | 28 (30%) |
| Obese | 20 (22%) |
| Disease severity group, n (%) | |
| 1 | 9 (9.7%) |
| 2 | 69 (74%) |
| 3 + 4 | 15 (16%) |
Table showing demographics at time of inclusion. Disease severity group is divided as follows: 1) Home/outpatient not experiencing any limitations in daily life; 2) Home/outpatient, certain limitations in daily activity level (e.g., fever, bedridden during illness); 3+4) All hospitalized patients regardless of need for supplemental oxygen treatment, and/or ICU admission
SARS-CoV-2 non-spike-specific longitudinal immune response
To investigate the non-vaccine related immune response towards primary SARS-CoV-2 infection we measured SARS-CoV-2 nucleocapsid (N)-specific IgG as well as the frequency of antigen-specific T cells targeting nucleocapsid and other non-S elements. At 1 month post-infection, the median level of N-specific IgG was 127,556 AU/mL which then declined gradually over the course of the study to 4,542 AU/mL after 20 months (Fig. 1B).
We characterized the non-vaccine related cellular response using two separate polychromatic flow cytometry assays, ICS and AIM, after ex vivo stimulation of PBMCs with a non-S peptide pool (34 immunodominant peptides outside the S protein). We first characterized CD4 + and CD8 + memory differentiation in naïve, central memory, effector memory and terminal differentiated subsets, using the ICS assay, and found only minor changes over the 4 visits (Supp. Figure 1). We then measured IFN-γ and IL2 expression in unfractionated memory CD4 + and CD8 + T cells, i.e., central memory, effector memory and terminally differentiated subsets combined (Supp. Figure 2). At 1 month post-infection there were high levels of both CD4 + and CD8 + non-S-specific memory T cells (Median: 0.039% and 0.061%, respectively) (Fig. 1C). After 10, 13 and 20 months post-infection the frequency of non-S-specific cells significantly decreased to 0.019% (p = 0.0007); 0.026% (p = 0.02) and 0.025% (p = 0.01), respectively for CD4 + memory T cells, and 0.040% (p = 0.03); 0.030% (p = 0.004) and 0.023% (p = 0.001), respectively, for CD8 + memory T cells (Fig. 1C).
The cytokine profile of the memory CD4 + T cells consisted of IL2 single (IL2 + /IFNγ-), IFNγ single (IL2-/IFNγ +) and IL2/IFNγ double (IL2 + /IFNγ +) producing cells. The highest frequency of each population was observed at 1 month post-infection, (Median: 0.0093%; 0.012%; and 0.018% for IL2 + / IFNγ-; IFNγ + /IL2-; and IL2 + /IFNγ + CD4 + T cells, respectively) (Fig. 1D). In contrast, the SARS-CoV-2 memory CD8 + T cells consisted primarily of IFNγ single producing cells and to a lesser degree IL2/IFNγ double producing cells. The highest frequency of each population was observed at visit 1 (Median: 0.045% and 0.016% for IFNγ + /IL2- and IL2 + /IFNγ + CD8 + T cells, respectively) (Fig. 1D).
Evaluation of non-S-specific CD4 + and CD8 + T cells using AIM revealed higher levels of non-S-specific T cells than measured by ICS. One month after infection the median frequencies of non-S-specific T cells using AIM were 1.48% for CD4 + T cells and 0.35% for CD8 + T cells. We observed a contraction at 10 months post-infection (median: 0.76% (p < 0.0001) for CD4 + T cells and 0.18% (p < 0.0001) and CD8 + T cells) after which the response remained stable throughout the 20-month follow-up period (Fig. 1E). From the PBMCs stimulated with the non-S peptide pool for the AIM assay, we collected the supernatant and measured the IFNγ-secretion. We found the highest median IFNγ-production 1 month after infection (15,806 pg/mL), which significantly decreased at the subsequent study visits (5,144 pg/mL (p = 0.0004); 1,885 pg/mL (p < 0.0001) and 5,214 pg/mL (p < 0.0001) at month 10, 13 and 20, respectively).
Collectively, we found high levels of both N-specific antibodies and non-S-specific CD4 + and CD8 + T cells in response to primary SARS-CoV-2 infection. Ten months after infection, both the circulating antibody levels and the T cell responses had contracted significantly. However, in the following period up to 20 months, both the humoral and cellular immune responses decreased at a slower rate.
Durability of SARS CoV-2 infection-induced immunity
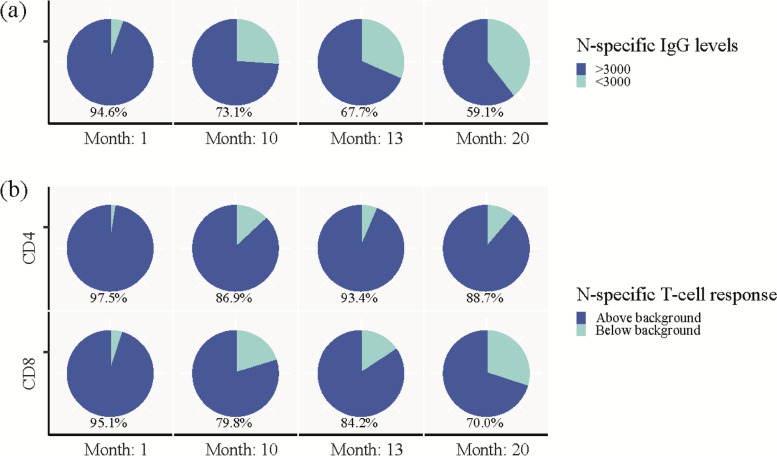
To evaluate the durability of the SARS-CoV-2 immune response, we defined seropositivity as N-specific IgG levels above 3000 AU/mL. We defined a positive AIM response as non-S-specific T cells as levels above 0.107% for CD4 + T cells and 0.078% for CD8 + T cells, in accordance with Dietz el al. [16]. One month post-infection, 94.6% of the participants were seropositive for N-specific IgG. During the study, the number of N-specific seropositive participants decreased to 73.1%, 67.7% and 59.1% at the 10-, 13-, and 20-month study visits, respectively (Fig. 2A). At 1 month post-infection, 97.5% of participants had a positive response for non-S-specific CD4 + T cells, and 88.7% of participants maintained a positive response after 20 months. For non-S-specific CD8 + T cells, 95% of participants had a positive response at the 1-month visit and 70% of participants sustained a positive response at 20 months post-infection (Fig. 2B).
Fig. 2.
Durability of SARS-CoV-2 infection-induced immunity. a Pie charts showing percentage of participants with SARS-CoV-2 nucleocapsid-specific IgG above (dark blue) and below (light blue) 3000 AU/mL at each visit. b Pie charts showing percentage of participants with SARS-CoV-2 non-spike-specific CD4 + (top panel) and CD8 + (lower panel) T cells above (dark blue) or below (light blue) background response analysed by AIM
SARS-CoV-2 S-specific longitudinal immune response
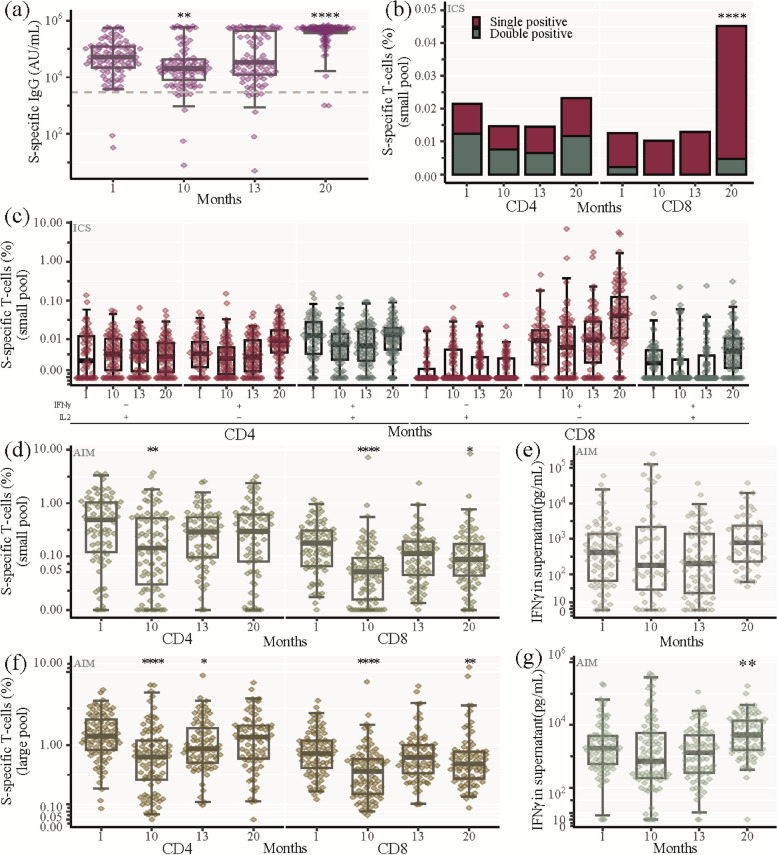
Next, we assessed the strength of the humoral immunological memory response elicited by vaccination. At 1 month post-infection, the median level of S-specific IgG was 51,016 AU/mL and had declined to 20,551 AU/mL at 10 months post-infection (Fig. 3A). As the majority of participants received COVID-19 vaccination median S-specific IgG levels increased to 493,232 AU/mL at month 20 post-infection demonstrating a strong memory response to vaccination with a tenfold increase in IgG levels compared to the levels immediately after infection (Fig. 3A).
Fig. 3.
SARS-CoV-2 Spike-specific longitudinal immune response. a SARS-CoV-2 spike (S)-specific IgG analysed by Mesoscale. Being seropositive was defined as IgG above 3000 AU/mL (dashed line). b, c Percentage of SARS-CoV-2 S-specific CD4 + and CD8 + memory T cells analysed by ICS after stimulation with the S-small peptide pool. b Total SARS-CoV-2 S-specific CD4 + and CD8 + T memory cells at indicated time points. IFNγ only and IL2 only producing cells: red bar, IFNγ and IL2 double producing cells: green bar. Stacked bars represent median values. c Percentage of SARS-CoV-2 S-specific CD4 + and CD8 + memory T cells producing either IL2 only (red, left panel), IFNγ only (red, middle panel), or co-producing IFNγ and IL2 (green, right panel) at each visit. d-g Percentage of SARS-CoV-2 S-specific CD4 + and CD8 + T cells analysed by AIM after stimulation with the S-small (d, e) or the large (f, g) peptide pool, i.e., cells expressing 2 or 3 activation induced markers (CD69, OX40 or 41BB). d SARS-CoV-2 S-specific CD4 + and CD8 + T cells at indicated time points (S-small pool). e IFNγ production by SARS-CoV-2 S-specific cells (S-small pool) (Analysed by mesoscale). f SARS-CoV-2 S-specific CD4 + and CD8 + T cells at indicated time points (S-large pool). g IFNγ production by SARS-CoV-2 S-specific cells (S-large pool) (Analysed by mesoscale). a, c, d-g Box and whisker plots show median values ± IQR and error bars indicate 95% CI. Statistical comparisons were performed using Friedman test and Wilcoxon unpaired signed-ranks test adjusted using Bonferroni with visit 1 as a reference. *P ≤ 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, no asterisk indicates non-significance
To characterize the cellular immunological memory response to vaccination, we first used ICS with ex vivo stimulation of PBMCs with a S-small peptide pool (11 immunodominant S peptides). One month post-infection, both CD4 + and CD8 + S-specific memory T cells (Median: 0.021% and 0.012%, respectively) were detected. These levels declined slightly by 10 and 13 months post-infection (Fig. 3B). At 20 months post-infection, where 99% of the participants had received at least one vaccine dose, the percentage of CD4 + S-specific memory T cells had only increased slightly compared to 1 month post-infection. On the other hand, there was a fourfold increase of CD8 + S-specific memory T cells (Median: 0.045%, p < 0.0001) at the 20-month visit (Fig. 3B). The cytokine profile of the S-specific memory T cells consisted, for CD4 + memory T cells of IL2 + /IFNγ-, IL2-/IFNγ + and IL2 + /IFNγ + producing cells, while the CD8 + memory response consisted primarily of IL2-/IFNγ + producing cells (Fig. 3C).
Additionally, we evaluated the S-specific T cell immunological memory response using the AIM assay after stimulation with either the S-small peptide pool (11 selected peptides) (Fig. 3D, E) or an S-large peptide pool (315 overlapping peptides spanning the entire S protein (Fig. 3F, G). At 1-month post-infection, we observed a median of 0.49% and 0.18% for CD4 + and CD8 + T cells, respectively, using the S-small pool. Using the S-large pool the levels were greater with a median of 1.34% and 0.77% for CD4 + and CD8 + T cells, respectively (Fig. 3D + F). At 10 months post-infection, there was a significant decrease of both S-specific CD4 + and CD8 + T cells (Median: S-small pool: 0.11% and 0.05% for CD4 + and CD8 + T cells, respectively; S-large pool: 0.69% and 0.42% for CD4 + and CD8 + T cells, respectively). At the 20-month follow-up, most participants (92 of 93) had received COVID-19 vaccinations and the frequency of S-specific CD4 + T cells had increased to levels comparable to 1 month after infection (Fig. 3D + F). The frequency of S-specific CD8 + T cells had also increased at the 20-month follow-up relative to the month 10 time-point, but the response did not reach the same level as 1 month post-infection (Fig. 3D + F). We observed a 2.5-fold increase in the S-specific IFNγ-production at 20 months post-infection relative to 1 month post-infection (Fig. 2G), consistent with the increase of IFN producing CD8 + T cells measured by ICS. Lastly, at 10 months post-infection, there was a significant decrease of S-specific IL4 production, which was followed by an increase at 20 months post-infection, to levels comparable to 1 month after infection (Supp. Figure 3).
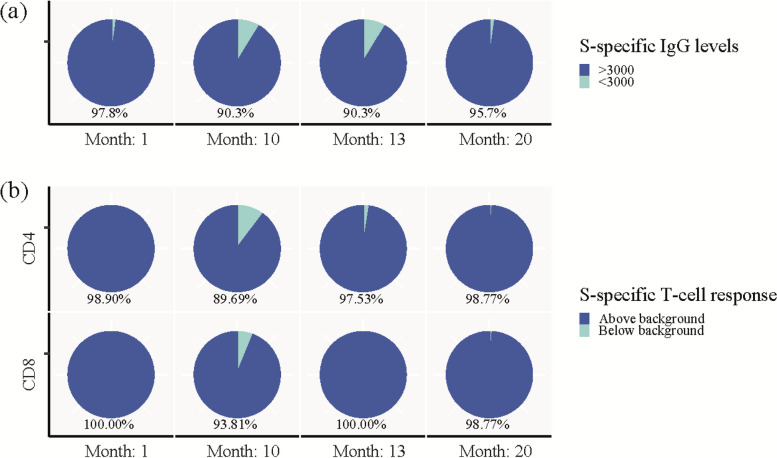
As previously described, we defined being seropositive as S-specific IgG levels above 3000 AU/mL [20, 21] and a positive AIM response as S-specific CD4 + and CD8 + T cells as levels above 0.107% and 0.078%, respectively [16]. We used this to further characterise the impact of vaccination on the S-specific immune response. One month after infection 97.8% were seropositive for S-specific IgG, which decreased to 90.3% at 10 months. Twenty months post-infection, 99% of participants had received vaccination and 95.7% were seropositive (Fig. 4A). With regards to T cell responses, we observed that at 1 month post-infection 98.9% and 100.0% had a S-specific CD4 + and CD8 + T cell response, respectively, while at ten months post-infection, 89.7% and 93.8% had maintained a S-specific CD4 + and CD8 + T cell response, respectively. At 20 months post-infection the frequency of participants with a S-specific T cell response had increased to 98.8% for both T cell subsets (Fig. 4B).
Fig. 4.
Population frequency of Spike-specific immune responses. a Pie charts showing percentage of participants with SARS-CoV-2 spike (S)-specific IgG above (dark blue) and below (light blue) 3000 AU/mL at each visit. b Pie charts showing percentage of participants with SARS-CoV-2 S-specific CD4 + (top panel) and CD8 + (lower panel) T cells above (dark blue) or below (light blue) background response analysed by AIM after stimulation with the S-large pool
Evaluation of SARS-CoV-2 vaccination on humoral and cellular immunity
To perform a direct evaluation of the immune-boosting effect of 2 vaccinations after primary infection, we selected a subset of patients, who had received their first 2 vaccinations between two subsequent visits (n = 65, Supp. Table 2), and compared the humoral and cellular immunological memory responses.
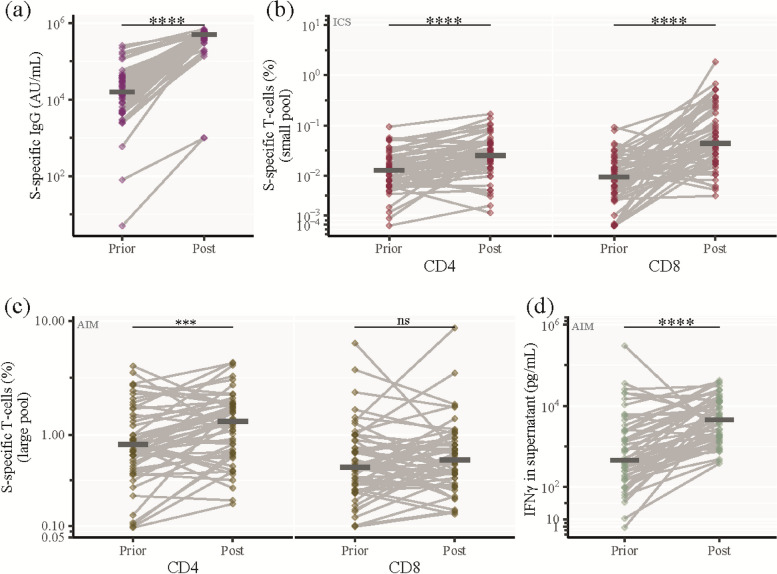
Prior to vaccination, the median level of S-specific IgG was 16,003 AU/mL which increased to 501,794 AU/mL after two vaccinations (p < 0.0001) (Fig. 5A). Using the ICS assay, we also detected a significant increase in both S-specific CD4 + and CD8 + memory T cells from 0.013% to 0.025% (p < 0.0001) and 0.009% to 0.044% (p < 0.0001), respectively (Fig. 5B). When measuring S-specific T cells using the AIM assay, there was no increase in CD8 + T cells, but we found a significant increase in both CD4 + T cells (0.82% to 1.32%, p = 0.0007) and IFNγ production (454 pg/mL to 4,558 pg/mL, p < 0.0001) (Fig. 5C, D, Supp. Figure 4A, B). Additionally, we investigated the non-S-specific response before and after vaccination, but we did not identify any significant increases in N-specific IgG levels or non-S-specific T cells (Supp. Figure 4B-F).
Fig. 5.
Evaluation of SARS-CoV-2 vaccination on humoral and cellular immunity. Analysis of a subset of 65 patients who received their first 2 vaccinations between two subsequent visits (Prior: A visit where the participant had not been vaccinated, Post: The subsequent visit, where the participant had received 2 vaccinations). a SARS-CoV-2 S-specific IgG levels (b) Percentage of SARS-CoV-2 S-specific CD4 + and CD8 + memory T cells analysed by ICS after stimulation with the S-small peptide pool. c Percentage of SARS-CoV-2 S-specific CD4 + and CD8 + T cells analysed by AIM after stimulation with the S-large pool. d IFNγ production by SARS-CoV-2 S-specific cells (S-large pool). Horizontal line shows median. Statistical comparisons were performed using Wilcoxon unpaired signed-ranks test adjusted using Bonferroni. *P ≤ 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
Discussion
In this study, we investigated the longitudinal humoral and cellular response trajectories following primary SARS-CoV-2 infection up to 20 months post-infection. One month after primary infection, more than 94% of all participants seroconverted for both N- and S-specific IgG. Both non-S- and S-specific T cells were also detected in more than 95% of participants, which demonstrated the adaptive immune response to infection. Over the following 20 months after infection, both the levels of non-S-specific humoral and cellular immunity decreased but remained detectable in the majority of participants implying efficient long-term immune memory formation. Greater proportions of participants had a quantifiable persisting T cell response compared to persisting circulating antibody levels when evaluating the responses to non-S antigens. This suggests very long-lasting cellular immune memory similar to what has been observed for the first SARS-CoV-1 outbreak, where antigen specific T cells could be detected in infected individuals for more than a decade post-infection[22, 23]. The S-specific humoral and cellular response also decreased in the first 10 months ensuing the initial infection, but a robust S-specific memory response was then elicited upon vaccination.
These findings confirm results from multiple other studies investigating the longitudinal SARS-CoV-2 immune response, showing a general decline of both the humoral and cellular SARS-CoV-2-specific immune response after 12 months [10, 17, 24] and a strong vaccine-induced S-specific memory response [25]. However, this study includes data on both humoral and cellular immunity 20 months after primary SARS-CoV-2 infection, thus providing new key insights into the longevity of the adaptive immune memory. Additionally, we have investigated both S- and non-S-specific T cells, allowing us to differentiate between the naturally contracting immunity after primary infection and the following vaccine-induced memory response. Gittelman et al. has used T cell receptor (TCR) sequencing to evaluate the breadth and depth of SARS-CoV-2-specific TCR towards both S and other viral proteins. They found a high frequency of SARS-CoV-2 associated TCRs 15 months after infection with increased clonal breadth and depth for the S protein in participants who received vaccination, while they did not see such an increase for the non-S proteins [26]. Our data substantiate the findings by Gittelman et al. both in terms of detecting long-term cellular responses as well as clearly being able to distinguish between the course of immunity induced by primary infection (non-S immunity) versus S-specific immunity boosted by vaccination. Further, we complement this data by providing a more detailed description of the functionality of the T cell response, by measuring the frequency of SARS-CoV-2-specific T cells using two different but complementary flow cytometric T cell assays (ICS and AIM), which includes the antigen-specific production of IFNγ and IL2 in response to SARS-CoV-2 peptide stimulation. These findings are in alignment, showing a natural durability of a non-S-specific response even after 15 and 20 months in the majority of participants.
Even though a reduction of the SARS-CoV-2-specific immune response was observed, the non-S-specific response persisted even after 20 months. During this follow-up period (April 2020 – November 2021) the level of community acquired infections remained relatively low in Denmark with population cumulative infection estimates at 5% [5, 27]. None of the participants reported re-infection of SARS-CoV-2 during the follow-up period, but undiagnosed asymptomatic infections cannot be ruled out. However, given the low level of population infections, the relatively few undiagnosed cases would not interfere with our conclusions. Other studies have shown an increase of antigen-specific T cells without known infection or seroconversion [17, 28–32]. Cross-reactivity to other human coronaviruses does not seem prominent, but some specificity may be preserved in epitopes in the Nucleocapsid region [13, 33]. Further investigations regarding non-S T cell responses may prove helpful in terms of expanding the protection against future SARS-CoV-2 variant waves.
We performed two flow cytometry assays, ICS and AIM, to characterize SARS-CoV-2-specific T cells induced by infection. Due to the nature of the different primary analytes of two assays, the ICS provides an approximate equal detection of the antigen-specific CD4 + and CD8 + T cell response because of the easily detectable intracellular IFNγ and IL2 in each subset, while some of the activation-induced markers used in the AIM assay are more prevalent on CD4 + T cells favouring detection of antigen-specific CD4 + T cells.
This phenomenon explains, why we detected higher levels of CD8 + versus CD4 + non-S-specific T cells at the 1-month time-point using ICS, while detecting higher levels of CD4 + versus CD8 + non-S-specific T cells using AIM. However, since the AIM assay is relying on more broadly expressed surface markers and is not limited to a few selected cytokines, we were still able to detect significantly higher levels of both antigen-specific CD4 + and CD8 + T cells using the AIM assay versus, verifying that AIM is a very useful addition to ICS when estimating the total levels of antigen-specific T cells. Further, when measuring IFNγ from the AIM supernatants, we observed a response pattern similar to that of ICS IFNγ expressing CD8 + T cells, demonstrating the response of antigen-specific CD8 + T cells. After vaccination we observed an induction of S-specific T cells to similar or higher levels as compared to 1 month after infection. Previously, Grifoni et al. compiled various epitope data and showed that CD8 + respond to a high number of immunogenic epitopes spanning the total S protein [34]. Looking at the ICS, the vaccine induced response is especially prominent for CD8 + T cells as well as for the IFNγ production from the AIM supernatants, where levels were increased more than twofold compared to 1 month post-infection, supporting a great vaccine-induced S-specific CD8 + T cell response.
Conclusions
This study shows long-term trajectories of both humoral and cellular immunity after SARS-CoV-2 infection. For most participants, the response persists 20 months after infection, and the cellular response appears to be more long-lived compared to the circulating antibody levels. Vaccination clearly boosts the S-specific response but does not affect the non-S-specific response. Together, this supports the understanding of immune contraction, and with studies showing the immune levels required for protection, adds to the knowledge of durability of protection against future SARS-CoV-2.
Methods
Study design and sample collection
Participants were enrolled at the Department of Infectious Diseases at Aarhus University Hospital, Denmark from April 3rd to July 9th 2020. Participants who had completed a study visit at 1, 10, 13, and 20 months after infection were included (n = 93). All participants had recovered from a PCR-verified SARS-CoV-2 infection. The study was approved by The National Health Ethics Committee (#1–10-72–76-20) and the Danish Data Protection Agency (case number not applicable). All participants provided informed written consent prior to any study activities. A more detailed description of the cohort has been performed by Nielsen et al. and Vibholm et al. [18, 19].
Quantification of SARS-CoV-2 IgG
Serum levels of SARS-CoV-2 Spike- and Nucleocapsid-specific IgG levels were measured using Meso Scale Discovery (MSD) platform (Meso Scale Diagnostics LLC, Maryland, USA) panel 2 IgG kit (K15383U-2). The assays were performed according to the manufacturer’s protocol with a serum dilution of 1:5,000 for IgG measurements. Plates were read on a MESO QuickPlex SQ 120 reader. Raw data were processed by Discovery Workbench 4.0 Software. Quantifications were reported in arbitrary units per mL (AU/mL).
We have previously utilized this analysis to evaluate the variation of Nucleocapsid antibody levels in a large Danish nation-wide prospective vaccination cohort and defined a cut-off for seroconversion to be a value above 3000 AND a > twofold increase above baseline [20, 21]. Using these parameters, we observed positive seroconversion in 96% of participants with a verified PCR-diagnosis [35]. In the present evaluation all individuals had been infected and therefore, seroconversion could not be defined as a twofold increase like in our previous publication. Instead, we have used only the 3000 AU/mL cut-off for the definition of seroconversion to evaluate the level of persistence of nucleocapsid antibodies.
Evaluation of antigen-specific T cells using flow cytometry
Antigen-specific T cells were measured by the two methods i.e., Activation Induced Marker assay (AIM) and Intracellular Cytokine Stain assay (ICS). For both AIM and ICS cryopreserved PBMCs were thawed, washed, and rested at 37˚C/5% CO2 for 3–4 h. Cells were then plated into wells of a 96-well plate, at a total of 1 × 106 cells per well and stimulated with SARS-CoV-2 peptide pools, containing a mix of peptides specific for both CD4 + and CD8 T + cells. The following stimulations were used for the AIM/ICS assays: No exogenous stimulation with Dimethyl sulfoxide (DMSO) as negative control (AIM/ICS), a S peptide large pool (S-large) consisting of 315 overlapping peptides (JPT peptides, #PM-WCPV-S-2, Swiss-Prot ID: P0DTC2, REF PMID: 32,015,508)(AIM); an S peptide small pool (S-small) consisting of 11 immunodominant S peptides [8, 9, 12, 13, 36, 37] (Supp. Table 3) (JPT peptides, custom order)(AIM/ICS); and a non-S peptide pool (non-S) consisting of 34 immunodominant peptides outside the S protein [8, 9, 12, 13, 36, 37] (Supp. Table 3) (JPT peptides, custom order) (AIM/ICS) used at a final concentration of 2 μg/mL of each peptide. Upon antigen stimulation, antigen-specific T cells were defined as cells expressing 2 or 3 activation markers for the AIM assay and cells expressing IFNγ and/or IL2 for the ICS assay (see further AIM/ICS methods below). All samples with viability below 70% measured by flow cytometry or with a CD4 + or CD8 + T cell count below 10,000 were excluded from analyses.
Intracellular cytokine stain assay
The ICS assay was performed essentially as previously published [38, 39]. For each sample, 4 conditions were used: DMSO, S peptide small pool, non-S peptide pool and staphylococcal enterotoxin B as a positive control. PBMCs were stimulated for 16–18 h at 37 °C, 5%CO2 in the presence of secretion inhibitors (Golgistop a final dilution of 1:2,000, and Golgiplug at a final dilution of 1:3,000, BD). After the stimulation, cells were stained with fixable Aqua dead cell stain (Invitrogen# L34966) as well as surface antibodies including CD14-PE-Cy5, CD19-PE-Cy5, CD56-PE-Cy5, CD3-BUV395, CD4-APC-fire750, CD8-BV786, CCR7-PE-dazzle594, CD45RA-Alexa700 and intracellular cytokine staining IFNγ and IL-2 using BD Cytofix/ Cytoperm protocol. Samples were acquired on a BD Fortessa X20 cytometer and data was analysed using FlowJo Version 10.8 using the following gating strategy: Live cells ➔ Single cells (FSC-A/FSC-H) ➔ Single cells (SSC-A/SSC-H) ➔ Lymphocytes (FSC-H/SSC-A) ➔ CD3 + T cells (CD14-CD19-CD56-/CD3 +) ➔ CD4 + or CD8 + T cells and then CCR7 versus CD45RA to define memory subsets within both the CD8 + and CD4 + T cell populations; naïve (CD45RA + CCR7 +), central memory (CM) (CD45RA-CCR7 +), effector memory (EM) (CD45RA-CCR7-) and terminally differentiated (TD) (CD45RA + CCR7-). For cytokine analyses, we defined a gate including all CD4 + /CD8 + memory and effector subsets (CM, EM and TD). Finally, within the CD4 + and CD8 + memory/effector subset, we gated IFNγ and IL2 and the frequency of cells expressing either IFNγ + IL2 + , IFNγ + IL2- or IFNγ-IL2 + was calculated by the “Boolean gating” function in FlowJo (Supp. Figure 2). The frequency of cytokine-producing antigen-specific T cells was determined by subtraction of the background cytokine response in unstimulated control samples from the positive response in the samples stimulated with SARS-CoV-2.
Activation-induced marker assay (AIM assay)
The AIM assay was performed essentially as previously published [16, 40]. For each sample, 4 conditions were used: DMSO, S peptide large pool, S peptide small pool and non-S peptide pool. A subset of samples on each plate were stimulated with staphylococcal enterotoxin B (SEB, 1 μg/mL, #S4881) as a positive control. Following 20 h incubation at 37℃, supernatants were harvested for cytokine quantification and cells were stained with the following dye/antibodies: Fixable Near-IR dead cell stain (Invitrogen, #L34976), CD3-PerCP-Cy5.5 (Biolegend #344808), CD4-BV650 (Biolegend #300536), CD8-BV605 (Biolegend #301040), CD69-APC (Biolegend #310910), OX40-BV421 (Biolegend #350014), and 41BB-PE (Biolegend #309804). Samples were acquired on a MACSQuant16 and data was analysed using FlowJo 10.8 using the following gating strategy: Live cells ➔ Single cells (FSC-A/FSC-H) ➔ Lymphocytes (FSC-H/SSC-A) ➔ CD3 + T cells ➔ CD4 + or CD8 + T cells and the expression of CD69, OX40 and 41BB was determined within CD4 + /CD8 + T cells. The frequency of cells expressing either CD69 + OX40 + 4-1BB + , CD69 + OX40 + , CD69 + 4-1BB + or OX40 + 4-1BB + was calculated by the “Boolean gating” function in FlowJo (Supp. Figure 5). Lastly, the frequency of antigen-specific cells was determined by subtracting the frequency of the non-stimulation condition from each antigen-stimulated condition. Total SARS-CoV-2-specific cells were calculated as a summation of each of the 4 populations (CD69 + OX40 + 4-1BB + , CD69 + OX40 + 4-1BB-, CD69 + OX40-4-1BB + or CD69-OX40 + 4-1BB + T cells). For Figs. 2 and 4 we have defined a positive AIM response in an individual if the response was above 0.107% and 0.078% for SARS-CoV-2 specific CD4 + T cells and CD8 + T cells, respectively, as previously published. The threshold values of 0.107% and 0.078% (CD4 + T cells and CD8 + T cells, respectively) were calculated from the SARS-CoV-2 specific AIM response in SARS-CoV-2 negative individuals, that had not been vaccinated (n = 238) as follows: Threshold for a positive response = the median value plus one standard deviation (SD) [16].
Cytokine detection in AIM assay
Interferon-γ (IFNγ) was measured in supernatants from the AIM assay using Meso Scale Discovery (MSD) platform (Meso Scale Diagnostics LLC, Maryland, USA) V-PLEX Plus Proinflammatory Panel 1 (K15049G) according to the manufacturer’s instructions at a dilution of 1:200. Plates were read on a MESO QuickPlex SQ 120 reader. Raw data were processed by Discovery Workbench 4.0 Software. Quantifications were reported in pg/mL. To investigate the antigen-specific IFNγ response, the measurement from the non-stimulated condition was subtracted from the antigen-stimulated condition.
Data and statistical analysis
Data were shown as individual points with a boxplot indicating median and IQR, and error bars showing 95% CI for the longitudinal study and connected individual points for the vaccine-induced immunity. To assess the statistical change over time both Kruskal–Wallis and Friedman’s test was used to check for differences between visits. Kruskal–Wallis was used on the whole dataset and to account for measurements being repeated, Friedman test was used excluding participant data where the analysis was not quantifiable for one or more visits. Post hoc analysis was done using Wilcoxon rank-sum adjusted using Bonferroni with visit 1 as a reference. Wilcoxon signed-rank test was used to compare the vaccine-induced immune response. P values ≤ 0.05 were considered statistically significant. P values were denoted as follows: * = p ≤ 0.05, ** = p < 0.01, *** = p < 0.001, and **** = p < 0.0001.
Data analysis and visualization was conducted in R (version 4.2.2), and RStudio Desktop (version 2022.12.0 + 353).
Supplementary Information
Additional file 1: Supplementary Figure 1. T cell memory subsets within overall CD4+ and CD8+ T cells. (A) Composition of memory subsets within the overall CD4+ and CD8+ T cells during visits 1,3,4 and 5. Memory subsets were defined as naïve (CD45RA+CCR7+), central memory (CM) (CD45RA-CCR7+), effector memory (EM) (CD45RA-CCR7-) and terminally differentiated (TD) (CD45RA+CCR7-). Individual values and box and whisker plots with median values shown (Box shows IQR, error bars indicate 95% CI). (B) Stacked bar graphs illustrate the movement of memory subsets within the CD4+ and CD8+ T cells during visits 1,3,4 and 5. (A-B).
Additional file 2: Supplementary Figure 2. Gating strategy for flow cytometric intracellular cytokine staining(ICS). (A-C) Shown are dot plots for patient 9 at visit 1. (B) Response to stimulation with negative control (DMSO) and (C) SARS-CoV-2 -peptide pools for effector/memory CD4+ T cells and effector/memory CD8+ T cells. Numbers represent percentage of the shown population that’s within the shown gate. (D) Frequency of cells expressing only one or two cytokines (IFNγ+IL2+, IFNγ+IL2- or IFNγ-IL2+) calculated by the Boolean gating.
Additional file 3: Supplementary Figure 3. IL4 production by SARS-CoV-2 specific cells. IL4 production by SARS-CoV-2 specific cells measured in the supernatant harvested from the cell stimulations in the AIM assay (Analysed by mesoscale) after stimulation with (A) Non-spike peptide pool, (B), Spike-small pool or (C) Spike-large pool. Horizontal line shows median Statistical comparisons were performed using Wilcoxon unpaired signed-ranks test adjusted using Bonferroni. *P ≤ 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, no asterisk indicates non-significance.
Additional file 4: Supplementary Figure 4. Impact of SARS-CoV-2 vaccination on humoral and cellular immunity for S-small and non-S immunity. Analysis of a subset of 65 patients who received their first 2 vaccinations between two subsequent visits (Prior: A visit where the participant had not been vaccinated, Post: The subsequent visit, where the participant had received 2 vaccinations). (A) Percentage of SARS-CoV-2 spike (S)-specific CD4+ and CD8+ T cells analysed by AIM after stimulation with the S-small pool. (B) IFNγ production by SARS-CoV-2 S-specific cells (S-small pool). (C) SARS-CoV-2 nucleocapsid-specific IgG levels (D) Percentage of SARS-CoV-2 S-specific CD4+ and CD8+ memory T cells analysed by ICS after stimulation with the non-S peptide pool. (E) Percentage of SARS-CoV-2 non-S-specific CD4+ and CD8+ T cells analysed by AIM. (F) IFNγ production by SARS-CoV-2 non-S-specific cells. Horizontal line shows median Statistical comparisons were performed using Wilcoxon unpaired signed-ranks test adjusted using Bonferroni. *P ≤ 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Additional file 5: Supplementary Figure 5. Gating strategy for flow activation induced marker assay (AIM). (A-C) Shown are dot plots for patient 9 at visit 1. (B) Response to stimulation with negative control (DMSO) and (C) SARS-CoV-2 -peptide pools for CD4+ T cells and CD8+ T cells. Numbers represent percentage of the shown population that's within the shown gate. (D) Frequency of cells expressing two or three activation markers (CD69+OX40+4-1BB+, CD69+OX40+, CD69+4-1BB+ or OX40+4-1BB+) calculated by the Boolean gating.
Additional file 6: Supplementary Table 1. Vaccine information and time from PCR to visit.
Additional file 7: Supplementary Table 2. Vaccine response.
Additional file 8: Supplementary Table 3. Peptides in the Spike peptide small pool and the Non-spike peptide pool.
Acknowledgements
We would like to thank all the participants who donated samples.
Data sharing statement
Individual participant data cannot be made available due to EU Data Protection Regulations (GDPR). A limited and completely anonymized version of the dataset can be obtained upon request. Study protocols, including laboratory protocols will be available upon request.
Authors’ contributions
AC, EP and MT conceptualized the work. MT, SN, LV, MT and MS contributed to establishing the CoroNAT cohort. LV and RA did clinical visits and collected samples. RA, AG, RO and AH performed laboratory analysis. AH and RO performed the data analysis and visualization. RO and MT supervised and led the study. AH, RO and MT drafted the manuscript and have accessed and verified the underlying data. All authors read and approved the final draft. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Funding
This study was funded by The Novo Nordisk Foundation (grant # NNF21SA0069154).
Declarations
Ethics approval and consent to participate
The study was approved by The National Health Ethics Committee (#1-10-72-76-20) and the Danish Data Protection Agency (case number not applicable). All participants provided informed written consent. All methods were carried out in accordance with relevant guidelines and regulations.
Consent for application
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Statement on the fifteenth meeting of the IHR. Emergency Committee on the COVID-19 pandemic; 2005. https://www.who.int/news/item/05-05-2023-statement-on-the-fifteenth-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-coronavirus-disease-(covid-19)-pandemic. Accessed 15 June 2023.
- 2.Karlsson AC, Humbert M, Buggert M. The known unknowns of T cell immunity to COVID-19. Sci Immunol. 2020;5(53):eabe8063. doi: 10.1126/sciimmunol.abe8063. [DOI] [PubMed] [Google Scholar]
- 3.Moss P. The T cell immune response against SARS-CoV-2. Nat Immunol. 2022;23(2):186–193. doi: 10.1038/s41590-021-01122-w. [DOI] [PubMed] [Google Scholar]
- 4.Qi H. T follicular helper cells in space-time. Nat Rev Immunol. 2016;16(10):612–625. doi: 10.1038/nri.2016.94. [DOI] [PubMed] [Google Scholar]
- 5.Staerke NB, Reekie J, Nielsen H, Benfield T, Wiese L, Knudsen LS, Iversen MB, Iversen K, Fogh K, Bodilsen J, et al. Levels of SARS-CoV-2 antibodies among fully vaccinated individuals with Delta or Omicron variant breakthrough infections. Nat Commun. 2022;13(1):4466. doi: 10.1038/s41467-022-32254-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei J, Pouwels KB, Stoesser N, Matthews PC, Diamond I, Studley R, Rourke E, Cook D, Bell JI, Newton JN, et al. Antibody responses and correlates of protection in the general population after two doses of the ChAdOx1 or BNT162b2 vaccines. Nat Med. 2022;28(5):1072–1082. doi: 10.1038/s41591-022-01721-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aldridge RW, Yavlinsky A, Nguyen V, Eyre MT, Shrotri M, Navaratnam AMD, Beale S, Braithwaite I, Byrne T, Kovar J, et al. SARS-CoV-2 antibodies and breakthrough infections in the Virus Watch cohort. Nat Commun. 2022;13(1):4869. doi: 10.1038/s41467-022-32265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sekine T, Perez-Potti A, Rivera-Ballesteros O, Stralin K, Gorin JB, Olsson A, Llewellyn-Lacey S, Kamal H, Bogdanovic G, Muschiol S, et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183(1):158–168 e114. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng Y, Mentzer AJ, Liu G, Yao X, Yin Z, Dong D, Dejnirattisai W, Rostron T, Supasa P, Liu C, et al. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Immunol. 2020;21(11):1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garanina E, Hamza S, Stott-Marshall RJ, Martynova E, Markelova M, Davidyuk Y, Shakirova V, Kaushal N, Baranwal M, Khaertynova IM, et al. Antibody and T cell immune responses to SARS-CoV-2 peptides in COVID-19 convalescent patients. Front Microbiol. 2022;13:842232. doi: 10.3389/fmicb.2022.842232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao J, Wang L, Schank M, Dang X, Lu Z, Cao D, Khanal S, Nguyen LN, Nguyen LNT, Zhang J, et al. SARS-CoV-2 specific memory T cell epitopes identified in COVID-19-recovered subjects. Virus Res. 2021;304:198508. doi: 10.1016/j.virusres.2021.198508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelde A, Bilich T, Heitmann JS, Maringer Y, Salih HR, Roerden M, Lubke M, Bauer J, Rieth J, Wacker M, et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat Immunol. 2021;22(1):74–85. doi: 10.1038/s41590-020-00808-x. [DOI] [PubMed] [Google Scholar]
- 13.Ferretti AP, Kula T, Wang Y, Nguyen DMV, Weinheimer A, Dunlap GS, Xu Q, Nabilsi N, Perullo CR, Cristofaro AW, et al. Unbiased screens show CD8(+) T cells of COVID-19 patients recognize shared epitopes in SARS-CoV-2 that largely reside outside the spike protein. Immunity. 2020;53(5):1095–1107 e1093. doi: 10.1016/j.immuni.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weingarten-Gabbay S, Klaeger S, Sarkizova S, Pearlman LR, Chen DY, Gallagher KME, Bauer MR, Taylor HB, Dunn WA, Tarr C, et al. Profiling SARS-CoV-2 HLA-I peptidome reveals T cell epitopes from out-of-frame ORFs. Cell. 2021;184(15):3962–3980 e3917. doi: 10.1016/j.cell.2021.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peluso MJ, Deitchman AN, Torres L, Iyer NS, Munter SE, Nixon CC, Donatelli J, Thanh C, Takahashi S, Hakim J, et al. Long-term SARS-CoV-2-specific immune and inflammatory responses in individuals recovering from COVID-19 with and without post-acute symptoms. Cell Rep. 2021;36(6):109518. doi: 10.1016/j.celrep.2021.109518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dietz LL, Juhl AK, Søgaard OS, Reekie J, Nielsen H, Johansen IS, Benfield T, Wiese L, Stærke NB, Jensen T, et al. Impact of age and comorbidities on SARS-CoV-2 vaccine-induced T cell immunity. Commun Med (Lond) 2023;3(1):58. doi: 10.1038/s43856-023-00277-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo L, Wang G, Wang Y, Zhang Q, Ren L, Gu X, Huang T, Zhong J, Wang Y, Wang X, et al. SARS-CoV-2-specific antibody and T-cell responses 1 year after infection in people recovered from COVID-19: a longitudinal cohort study. Lancet Microbe. 2022;3(5):e348–e356. doi: 10.1016/S2666-5247(22)00036-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nielsen SS, Vibholm LK, Monrad I, Olesen R, Frattari GS, Pahus MH, Hojen JF, Gunst JD, Erikstrup C, Holleufer A, et al. SARS-CoV-2 elicits robust adaptive immune responses regardless of disease severity. EBioMedicine. 2021;68:103410. doi: 10.1016/j.ebiom.2021.103410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vibholm LK, Nielsen SSF, Pahus MH, Frattari GS, Olesen R, Andersen R, Monrad I, Andersen AHF, Thomsen MM, Konrad CV, et al. SARS-CoV-2 persistence is associated with antigen-specific CD8 T-cell responses. EBioMedicine. 2021;64:103230. doi: 10.1016/j.ebiom.2021.103230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Staerke NB, Reekie J, Johansen IS, Nielsen H, Benfield T, Wiese L, Sogaard OS, Tolstrup M, Iversen KK, Tarp B, et al. Cohort profile: the Danish National Cohort Study of Effectiveness and Safety of SARS-CoV-2 vaccines (ENFORCE) BMJ Open. 2022;12(12):e069065. doi: 10.1136/bmjopen-2022-069065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sogaard OS, Reekie J, Johansen IS, Nielsen H, Benfield T, Wiese L, Staerke NB, Iversen K, Fogh K, Bodilsen J, et al. Characteristics associated with serological COVID-19 vaccine response and durability in an older population with significant comorbidity: the Danish Nationwide ENFORCE study. Clin Microbiol Infect. 2022;28(8):1126–1133. doi: 10.1016/j.cmi.2022.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng OW, Chia A, Tan AT, Jadi RS, Leong HN, Bertoletti A, Tan YJ. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine. 2016;34(17):2008–2014. doi: 10.1016/j.vaccine.2016.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang F, Quan Y, Xin ZT, Wrammert J, Ma MJ, Lv H, Wang TB, Yang H, Richardus JH, Liu W, Cao WC. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J Immunol. 2011;186(12):7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- 24.Canto ECL, Gomes A, Serrano M, Pereira AHG, Ribeiro R, Napoleao P, Domingues I, Silva C, Fanczal J, Afonso A, et al. Longitudinal SARS-CoV-2 seroprevalence in Portugal and antibody maintenance 12 months after infection. Eur J Immunol. 2022;52(1):149–160. doi: 10.1002/eji.202149619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodda LB, Morawski PA, Pruner KB, Fahning ML, Howard CA, Franko N, Logue J, Eggenberger J, Stokes C, Golez I, et al. Imprinted SARS-CoV-2-specific memory lymphocytes define hybrid immunity. Cell. 2022;185(9):1588–1601 e1514. doi: 10.1016/j.cell.2022.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gittelman RM, Lavezzo E, Snyder TM, Zahid HJ, Carty CL, Elyanow R, Dalai S, Kirsch I, Baldo L, Manuto L, et al. Longitudinal analysis of T cell receptor repertoires reveals shared patterns of antigen-specific response to SARS-CoV-2 infection. JCI Insight. 2022;7(10):e151849. doi: 10.1172/jci.insight.151849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fogh K, Eriksen ARR, Larsen TG, Hasselbalch RB, Bundgaard H, Scharff B, Nielsen SD, Jørgensen CS, Erikstrup C, Østergaard L, et al. A cross-sectional study of SARS-CoV-2 antibodies and risk factors for seropositivity in staff in day care facilities and preschools in Denmark. Microbiol Spectr. 2023;11(1):e0417422. doi: 10.1128/spectrum.04174-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallais F, Velay A, Nazon C, Wendling MJ, Partisani M, Sibilia J, Candon S, Fafi-Kremer S. Intrafamilial exposure to SARS-CoV-2 associated with cellular immune response without seroconversion, France. Emerg Infect Dis. 2021;27(1):113–121. doi: 10.3201/eid2701.203611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z, Yang X, Zhong J, Zhou Y, Tang Z, Zhou H, He J, Mei X, Tang Y, Lin B, et al. Exposure to SARS-CoV-2 generates T-cell memory in the absence of a detectable viral infection. Nat Commun. 2021;12(1):1724. doi: 10.1038/s41467-021-22036-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mok CKP, Zhu A, Zhao J, Lau EHY, Wang J, Chen Z, Zhuang Z, Wang Y, Alshukairi AN, Baharoon SA, et al. T-cell responses to MERS coronavirus infection in people with occupational exposure to dromedary camels in Nigeria: an observational cohort study. Lancet Infect Dis. 2021;21(3):385–395. doi: 10.1016/S1473-3099(20)30599-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jay C, Ratcliff J, Turtle L, Goulder P, Klenerman P. Exposed seronegative: cellular immune responses to SARS-CoV-2 in the absence of seroconversion. Front Immunol. 2023;14:1092910. doi: 10.3389/fimmu.2023.1092910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao Y, Cai C, Grifoni A, Muller TR, Niessl J, Olofsson A, Humbert M, Hansson L, Osterborg A, Bergman P, et al. Ancestral SARS-CoV-2-specific T cells cross-recognize the Omicron variant. Nat Med. 2022;28(3):472–476. doi: 10.1038/s41591-022-01700-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, Rawlings SA, Sutherland A, Premkumar L, Jadi RS, et al. Targets of T Cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501 e1415. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grifoni A, Sidney J, Vita R, Peters B, Crotty S, Weiskopf D, Sette A. SARS-CoV-2 human T cell epitopes: adaptive immune response against COVID-19. Cell Host Microbe. 2021;29(7):1076–1092. doi: 10.1016/j.chom.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baerends EAM, Hvidt AK, Reekie J, Sogaard OS, Staerke NB, Raben D, Nielsen H, Petersen KT, Juhl MR, Johansen IS, et al. SARS-CoV-2 vaccine-induced antibodies protect against Omicron breakthrough infection. iScience. 2023;26(9):107621. doi: 10.1016/j.isci.2023.107621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schulien I, Kemming J, Oberhardt V, Wild K, Seidel LM, Killmer S, Sagar, Daul F, Salvat Lago M, Decker A, et al. Characterization of pre-existing and induced SARS-CoV-2-specific CD8(+) T cells. Nat Med. 2021;27(1):78–85. doi: 10.1038/s41591-020-01143-2. [DOI] [PubMed] [Google Scholar]
- 37.Tarke A, Sidney J, Kidd CK, Dan JM, Ramirez SI, Yu ED, Mateus J, da Silva Antunes R, Moore E, Rubiro P et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. bioRxiv. 2020. Cell Rep Med . 2021;2(2):100204. [DOI] [PMC free article] [PubMed]
- 38.Pillet S, Aubin E, Trepanier S, Bussiere D, Dargis M, Poulin JF, Yassine-Diab B, Ward BJ, Landry N. A plant-derived quadrivalent virus like particle influenza vaccine induces cross-reactive antibody and T cell response in healthy adults. Clin Immunol. 2016;168:72–87. doi: 10.1016/j.clim.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 39.Pillet S, Aubin E, Trepanier S, Poulin JF, Yassine-Diab B, Ter Meulen J, Ward BJ, Landry N. Humoral and cell-mediated immune responses to H5N1 plant-made virus-like particle vaccine are differentially impacted by alum and GLA-SE adjuvants in a Phase 2 clinical trial. NPJ Vaccines. 2018;3:3. doi: 10.1038/s41541-017-0043-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosas-Umbert M, Gunst JD, Pahus MH, Olesen R, Schleimann M, Denton PW, Ramos V, Ward A, Kinloch NN, Copertino DC, et al. Administration of broadly neutralizing anti-HIV-1 antibodies at ART initiation maintains long-term CD8(+) T cell immunity. Nat Commun. 2022;13(1):6473. doi: 10.1038/s41467-022-34171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Figure 1. T cell memory subsets within overall CD4+ and CD8+ T cells. (A) Composition of memory subsets within the overall CD4+ and CD8+ T cells during visits 1,3,4 and 5. Memory subsets were defined as naïve (CD45RA+CCR7+), central memory (CM) (CD45RA-CCR7+), effector memory (EM) (CD45RA-CCR7-) and terminally differentiated (TD) (CD45RA+CCR7-). Individual values and box and whisker plots with median values shown (Box shows IQR, error bars indicate 95% CI). (B) Stacked bar graphs illustrate the movement of memory subsets within the CD4+ and CD8+ T cells during visits 1,3,4 and 5. (A-B).
Additional file 2: Supplementary Figure 2. Gating strategy for flow cytometric intracellular cytokine staining(ICS). (A-C) Shown are dot plots for patient 9 at visit 1. (B) Response to stimulation with negative control (DMSO) and (C) SARS-CoV-2 -peptide pools for effector/memory CD4+ T cells and effector/memory CD8+ T cells. Numbers represent percentage of the shown population that’s within the shown gate. (D) Frequency of cells expressing only one or two cytokines (IFNγ+IL2+, IFNγ+IL2- or IFNγ-IL2+) calculated by the Boolean gating.
Additional file 3: Supplementary Figure 3. IL4 production by SARS-CoV-2 specific cells. IL4 production by SARS-CoV-2 specific cells measured in the supernatant harvested from the cell stimulations in the AIM assay (Analysed by mesoscale) after stimulation with (A) Non-spike peptide pool, (B), Spike-small pool or (C) Spike-large pool. Horizontal line shows median Statistical comparisons were performed using Wilcoxon unpaired signed-ranks test adjusted using Bonferroni. *P ≤ 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, no asterisk indicates non-significance.
Additional file 4: Supplementary Figure 4. Impact of SARS-CoV-2 vaccination on humoral and cellular immunity for S-small and non-S immunity. Analysis of a subset of 65 patients who received their first 2 vaccinations between two subsequent visits (Prior: A visit where the participant had not been vaccinated, Post: The subsequent visit, where the participant had received 2 vaccinations). (A) Percentage of SARS-CoV-2 spike (S)-specific CD4+ and CD8+ T cells analysed by AIM after stimulation with the S-small pool. (B) IFNγ production by SARS-CoV-2 S-specific cells (S-small pool). (C) SARS-CoV-2 nucleocapsid-specific IgG levels (D) Percentage of SARS-CoV-2 S-specific CD4+ and CD8+ memory T cells analysed by ICS after stimulation with the non-S peptide pool. (E) Percentage of SARS-CoV-2 non-S-specific CD4+ and CD8+ T cells analysed by AIM. (F) IFNγ production by SARS-CoV-2 non-S-specific cells. Horizontal line shows median Statistical comparisons were performed using Wilcoxon unpaired signed-ranks test adjusted using Bonferroni. *P ≤ 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Additional file 5: Supplementary Figure 5. Gating strategy for flow activation induced marker assay (AIM). (A-C) Shown are dot plots for patient 9 at visit 1. (B) Response to stimulation with negative control (DMSO) and (C) SARS-CoV-2 -peptide pools for CD4+ T cells and CD8+ T cells. Numbers represent percentage of the shown population that's within the shown gate. (D) Frequency of cells expressing two or three activation markers (CD69+OX40+4-1BB+, CD69+OX40+, CD69+4-1BB+ or OX40+4-1BB+) calculated by the Boolean gating.
Additional file 6: Supplementary Table 1. Vaccine information and time from PCR to visit.
Additional file 7: Supplementary Table 2. Vaccine response.
Additional file 8: Supplementary Table 3. Peptides in the Spike peptide small pool and the Non-spike peptide pool.