Abstract
From our compound library of vitamin K derivatives, we found that some compounds exhibited anti-SARS-CoV-2 activity in VeroE6/TMPRSS2 cells. The common structure of these compounds was menaquinone-2 (MK-2) with either the m-methylphenyl or the 1-naphthyl group introduced at the end of the side chain. Therefore, new vitamin K derivatives having more potent anti-SARS-CoV-2 activity were explored by introducing various functional groups at the ω-position of the side chain. MK-2 derivatives with a purine moiety showed the most potent antiviral activity among the derivatives. We also found that their mechanism of action was the inhibition of RNA-dependent RNA polymerase (RdRp) of SARS-CoV-2. The chemical structures of our compounds were completely different from those of nucleic acid derivatives such as remdesivir and molnupiravir, clinically approved RdRp inhibitors for COVID-19 treatment, suggesting that our compounds may be effective against viruses resistant to these nucleic acid derivatives.
Introduction
More than 2 years have passed since COVID-19 (coronavirus disease 2019) began to spread around the world, but the number of infected people has continued to increase, reaching a cumulative total of approximately 20 million in Japan and more than 600 million worldwide (on October 20, 2022).1 Antiviral drugs for COVID-19 treatment have been developed in Japan, yet their efficacy has not been confirmed in clinical trials. Even outside of Japan, highly effective oral drugs have not been completely developed yet. In addition, the sequelae of COVID-19, which persist for several months or even longer after COVID-19 itself has been cured, are a serious problem.2 Although oral drugs with anti-SARS-CoV-2 activity have been developed, there is no drug that improves the aftereffects of COVID-19.
Drug repositioning, in which existing drugs developed for other purposes and already proven safe are diverted to treat COVID-19, has been examined. For example, remdesivir was developed as an anti-Ebola drug;3 the anthelmintic ivermectin was approved for the treatment of intestinal fecal nematodes and scabies;4 and nelfinavir was used for the treatment of HIV-1 infection.5 Remdesivir was approved for emergency use in the United States in May 2021. It was granted a special exception by the Japanese Ministry of Health, Labor and Welfare in Japan after its efficacy was confirmed in phase III clinical trials in serious patients.6 Dexamethasone, a steroid drug approved for the treatment of interstitial pneumonia and other diseases, is used as a drug to improve excessive immune response called “cytokine storm” or acute respiratory distress syndrome, which causes severe COVID-19.7 However, these drugs are not “optimized” for SARS-CoV-2 and have many shortcomings as agents for treatment of COVID-19, such as insufficient efficacy at ordinary doses, the use of contraindicated drugs, and a variety of side effects.
Although various anti-SARS-CoV-2 drugs have been developed to date, highly effective drugs with confirmed safety do not exist. Patients infected with SARS-CoV-2 must stop taking antihypertensive drugs, antiarrhythmic drugs, cerebral embolization agents, and antiepileptics, when they are treated with nirmatrelvir.8 In addition, it is possible that a variant of SARS-CoV-2 resistant to current drugs may emerge in the future. Thus, the development of novel drugs with chemical structures different from those of existing anti-SARS-CoV-2 drugs is a desirable goal.
We have been investigating compounds with anti-SARS-CoV-2 activity based on liposoluble vitamins. Bioactive lipids, including fat-soluble vitamins, have a variety of biological effects and are expected to be applied to the treatment of many diseases due to their unique mechanisms of action. Vitamin K, a fat-soluble vitamin, has various physiological activities, such as blood coagulation and bone formation. It is also involved in the pathogenesis of various diseases, including atherosclerosis and cancer.9 It is clinically applied as a blood coagulant and osteoporosis treatment.10 Vitamin K is the only fat-soluble vitamin for which no excess has been reported. There are two main types of vitamin K in nature. Vitamin K1 (also called phylloquinone) is mainly synthesized by plants. Vitamin K2 (also called menaquinone-n: MK-n) is produced by microorganisms, such as intestinal bacteria, and is found in animals and fermented foods (Figure 1).11 Another form is menadione (vitamin K3), which is a synthetic product and has no side chain. Vitamin K3 is converted to menaquinone-4 in the tissues of birds and mammals and widely used as a vitamin K additive in animal feed (Figure 1).12
Figure 1.
Chemical structures of vitamin K homologues.
Recently, vitamin K3 (menadione: MD) was found to inhibit the 3-chymotrypsin-like (3CL) protease of SARS-CoV-2, thereby inhibiting viral replication.13 We predicted that vitamin K derivatives, such as vitamins K1 and K2, might also exhibit anti-SARS-CoV-2 activity. We explored compounds with anti-SARS-CoV-2 activity from our compound’s library consisting of approximately 60 vitamin K derivatives synthesized in a program to obtain agonists to nuclear receptor SXR (steroid and xenobiotic receptor)14,15 or LXR (liver X receptor)16 and inducers of neural stem cell differentiation into neurons.17−21 They were mainly three kinds of analogues, which are MK-2 and MK-3 analogues with aromatic ring substituents introduced at the end of their side chains (A), MK-1-MK-4 analogues having two of the same side chains (B), and vitamin K analogues bonded to benzene derivatives via a methylene group (C) (Figure 2). The anti-SARS-CoV-2 activity of the test compounds was evaluated by infecting VeroE6/TEMPRESS2 cells with SARS-CoV-2, adding the above vitamin K derivatives, and then quantifying the number of viable cells by the tetrazolium dye method.22 As a result, anti-SARS-CoV-2 activity was observed in derivatives 1 and 2 in which liposoluble substituents such as m-methylphenyl and 1-naphthyl groups were introduced at the end of the side chain of MK-2 (Figure 2).
Figure 2.
Extraction of vitamin K derivatives exhibited anti-SARS-CoV-2 activity from compound’s library and their common structure.
In particular, compound 1 containing m-methylphenyl groups was reported to induce higher neuronal differentiation activity than natural vitamin K homologues,17 although the anti-SARS-CoV-2 activity of 1 was not sufficient compared to the existing drug remdesivir. Therefore, we explored novel vitamin K analogues prepared by chemical synthesis to improve the activity. We synthesized several analogues in which the properties of the substituents at the ends of the side chains were systematically changed. To increase the interaction with the working protein, the bulkiness or hydrophobicity of the substituent and the introduced heteroatoms was changed.
Results and Discussion
Based on the findings of the active compounds 1 and 2 obtained in our study, we synthesized new compounds by introducing further substituents at the end of the side chain of MK-2. We synthesized 1–13, in which an aromatic ring was introduced at the ω-position of the side chain of MK-2 by a C–C bond to enhance the interaction with the target protein. Compound 3 is an isomer of compound 2, and compounds 4–9 are derivatives of compound 1 prepared by substituting the methyl group of the m-methylphenyl group and modifying the bulkiness and hydrophobicity of the adducts. Among 1–13, compounds 1–3 and 5 were previously synthesized and reported by us with neuronal differentiation activity. Furthermore, compounds 14–19 were synthesized by introducing an aromatic ring containing a nitrogen atom, such as a purine skeleton, via a C–N bond. We adopted Mitsunobu reaction conditions for coupling of the ω-position of the side chain of MK-2 to obtain compounds 14–17, while reductive amination gave compounds 18 and 19. They were expected to enhance interactions including hydrogen bond formation between the compounds and target proteins.
Method for the Synthesis of Compounds 1–13
The synthesis scheme of derivatives 1–13, compounds with substituents and side chains of MK-2 linked by the C–C bond, is shown in Scheme 1. The intermediate 22 was obtained by our reported method using geranyl acetate as the starting material. Using 22 as a common intermediate, various substituents were introduced by the Grignard reaction to give 23–35 in 60–90% yield. Coupling of 23–35 with menadione by the Friedel–Crafts reaction gave desired vitamin K derivatives 1–13 in 15–26% yield.
Scheme 1. Synthesis of Compounds 1–13.
Reagents and conditions: (a) I2, Mg, R1-Br, THF, 70 °C, and 1 h; (b) CuI, THF, 40 °C, 3 h, and 60–90%; (c) Na2S2O4, Et2O, r.t., and 1 h; and (d) BF3·OEt2, AcOEt/1,4-dioxane = 1:2, 70 °C, 3 h, and 15–26%.
Method for the Synthesis of Compounds 14–17
The synthetic scheme for derivatives 14–17 is shown in Scheme 2. MK-2 (36) was first synthesized by coupling of geraniol and menadione by Friedel–Crafts reaction in 39% yield by our reported method.17 Quinone 36 was then converted to a hydroquinone by sodium hydrosulfite and successively protected with methyl groups to afford 37 in 74% yield in two steps. The selenium oxidation of 37 led to alcohol 38, used as an intermediate, in 36% yield, and various nucleobases at the ω-position of the side chain of 38 were introduced by the Mitsunobu reaction to afford 39–42 in 33–42% yield. Compounds 39–42 were finally converted to desired vitamin K derivatives 14–17 with deprotection of methyl groups by ceric ammonium nitrate (CAN) in 48–84% yield (Scheme 2).
Scheme 2. Synthesis of Compounds 14–17.
Reagents and conditions: (a) Na2S2O4, Et2O, r.t., and 1 h; (b) BF3·OEt2, AcOEt/1,4-dioxane = 1:2, 70 °C, 3 h, and 39%; (c) Bu4NI, Na2S2O4, 2 M KOH aq, and THF; (d) (CH3)2SO4 THF/H2O, r.t., 7 h, and 74%; (e) SeO2, salicylic acid, 70% TBHP, CH2Cl2, r.t., 20 h, and 36%; (f) Ph3P, secondary cyclic amine, DIAD, DMF, r.t., 1 h, and 33–42%; and (g) CAN, CH3CN/water, r.t., 0.5 h, and 48–84%.
Method for the Synthesis of Compounds 18 and 19
The synthesis of derivatives 18 and 19 is shown in Scheme 3. The quinone 36 was converted to alcohol 43 with selenium dioxide in 40% yield and then 43 was oxidized to aldehyde 44 with manganese dioxide. Then, primary amines such as aminothiazole and adamantylamine were coupled with primary amines as amino thiazole or adamantylamine by reductive amination to obtain the desired vitamin K derivatives 18 and 19.
Scheme 3. Synthesis of Compounds 18 and 19.
Reagents and conditions: (a) SeO2, salicylic acid, 70% TBHP, CH2Cl2, r.t., 20 h, and 40%; (b) MnO2, CH2Cl2, r.t., 3 h, and 94%; and (c) CH3CO2H, primary amine, NaBH(OAc)3, CH2Cl2, r.t., 3 h, and 4–5%.
Evaluation of Vitamin K Derivatives for Anti-SARS-CoV-2 Activity
The commercially available vitamin K homologues PK, MK-2, and MD, as well as the synthesized vitamin K derivatives 1–19, were evaluated for their anti-SARS-CoV-2 activity in cell cultures. SARS-CoV-2 uses TMPRSS2 to cleave S proteins at the S1/S2 and S′2 sites. Therefore, VeroE6 cells expressing TMPRSS2 (VeroE6/TMPRSS2) are highly susceptible to SARS-CoV-2 infection.23 The cells infected with SARS-CoV-2 were mostly all killed after 3 days. Thus, the inhibitory effect of the compounds on SARS-CoV-2 replication can be determined by cell viability. VeroE6/TMPRSS2 cells (2 × 104 cells/well) were cultured in a microplate at 37 °C for 24 h. Then, the cells were infected or mock-infected with SARS-CoV-2 at a multiplicity of infection (MOI) of 0.01 in the presence of various concentrations of vitamin K derivatives and further incubated. After 3 days, the number of viable cells was determined by the tetrazolium dye (MTT) method. The 50% effective concentration (EC50) and 50% cytotoxic concentration (CC50) of each compound were determined. All experiments with compounds 1–19 were conducted in triplicate.
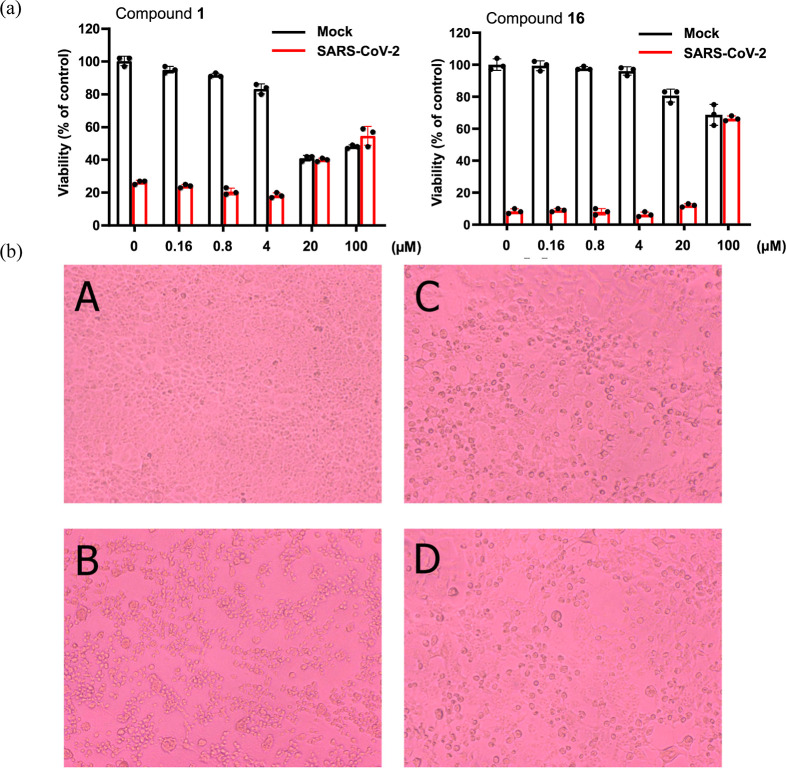
The results for compounds 1 and 16, which showed the highest activity among the compounds, are shown in Figure 3a. This graph shows the cell viability of SARS-CoV-2-infected and uninfected cells when the compounds were dose dependently added (Figure 3a). Regarding compound 1, microscopic observation for the anti-SARS-CoV-2 activity in VeroE6/TMPRSS2 cells is evaluated (Figure 3b). A–D represent the following cell condition: A, SARS-CoV-2-uninfected cells with no addition of compound; B, SARS-CoV-2-infected cells with no addition of compound; C, SARS-CoV-2-uninfected cells with 20 μM compound 1; and D: SARS-CoV-2-infected cells with 20 μM compound 1. These observations confirm that the cells in C and D survived as well as control A. The evaluation results for all compounds are listed in Table 1. Detailed evaluation data are provided in the Experimental Section. From these experimental results, the EC50 and CC50 values for each compound were determined, as well as the theoretical clogP value for liposolubility and the tPSA value for membrane permeability from the chemical structure (Table 1). The highlighted compounds represent active compounds. The EC50 value for remdesivir was 4.2 μM, whereas the most active of our compound was 16. This compound 16 showed no toxicity even at a concentration of 100 μM.
Figure 3.
(a) Anti-SARS-CoV-2 activity of compounds 1 and 16 in VeroE6/TMPRSS2 cells. The cells were cultured in a microplate at 37 °C for 24 h. Then, the cells were infected (orange column) or mock-infected (blue column) with SARS-CoV-2 at a MOI of 0.01 in the presence of various concentrations of the compound and further incubated. After 3 days, the number of viable cells was determined by the MTT method. Data represent mean ± SD in a triplicate experiment. (b) Microscopic observation for the anti-SARS-CoV-2 activity of compound 1 in VeroE6/TMPRSS2 cells. The cells were cultured in a microplate at 37 °C for 24 h. Then, the cells were mock-infected (A and C) or infected with SARS-CoV-2 at a MOI of 0.01 (B and D) in the absence (A and B) or presence of compound 1 at a concentration of 20 μM (C and D) and further incubated. After 3 days, the cells were observed microscopically (magnification: ×100).
Table 1. Anti-SARS-CoV-2 Activity of Vitamin K Derivatives in VeroE6/TMPRSS2 Cellsa.

RDV: remdesivir; EC50: 50% effective concentration based on the inhibition of virus-induced cell destruction; CC50: 50% cytotoxic concentration based on the reduction of mock-infected cell viability; ND: not determined.
Investigation of the Action Mechanism of the Active Compounds
Currently, marketed anti-SARS-CoV-2 drugs mainly target two enzymes essential for viral replication. ensitrelvir and nirmatrelvir target the 3CL protease from SARS-CoV-2,24 while remdesivir and molnupiravir target the RNA-dependent RNA polymerase (RdRp),25 which is cleaved by 3CL protease.26 To investigate the mechanism of action of the active compounds, we examined their inhibitory effect on SARS-CoV-2 3CL protease and RdRp.
1. Inhibitory Effect of Compounds 1, 2, and 16 on RdRp Activity
To investigate the mechanism of action of active compounds 1, 2, and 16, their inhibitory effects on RdRp were examined using a commercially available assay kit. RdRp is an enzyme that replicates viral RNA and plays an important role in the growth of SARS-CoV-2. In the assay kit used in this study, RdRp activity can be evaluated by staining the RNA replicated by RdRp with a fluorescent dye and quantifying the RNA. Thus, the decrease in fluorescence intensity upon drug treatment suggests RdRp inhibition; apart from the remdesivir analogue, EDTA was selected as a positive control to evaluate RdRp inhibition. The results showed that 1, 2, and 16 inhibited RdRp in a concentration-dependent manner like EDTA (Figure 4). On the other hand, remdesivir did not show activity in this assay since it is a prodrug and needs triphosphorylation through hydrolysis of the ester moiety in the body to exert its activity.
Figure 4.

Inhibitory effect of compounds 1, 2, and 16 on SARS-CoV-2 RdRp activity. Values not sharing a common letter in each group are significant differences: p < 0.05 (Tukey’s–Kramer HSD test).
2. Inhibitory Effect of Compounds 1 and 2 on 3CL Protease Activity
The inhibitory effect of the active compounds on 3CL protease was performed using a commercially available kit. The mechanism is based on FRET (fluorescence resonance energy transfer). When a peptide chain with a fluorescent substance attached to both ends is cleaved by 3CL-protease, excitation energy is transferred, and fluorescence is observed. By measuring the fluorescence intensity, we can determine whether the test compound inhibits the activity of 3CL-protease. The results showed that these compounds had no inhibitory effect on 3CL protease activity (Figure 5).
Figure 5.

Inhibitory effect of compounds 1 and 2 at 10 nM on SARS-CoV-2 3CL protease activity.
We examined how the compounds bind to RdRp from these assay results using computational chemistry methods. The crystal structure of RdRp was obtained from the Protein Data Bank (PDB) (PDB code: 7BV2). The crystal structure was optimized for the binding region using the integrated computational chemistry system Molecular Operating Environment (MOE; 2022.02 Chemical Computing Group), and the binding energies were calculated by fitting each ligand. The results showed that compounds 1, 16, and remdesivir triphosphate (RTP) bind as shown in Figure 6, with compound 1 interacting with U10 and compound 16 with Mg1004 (Table 2). Furthermore, the docking score with RdRp was −18.1724 kcal/mol for RTP, whereas it was −8.4788 and −9.0060 kcal/mol for compound 1 and compound 16, respectively. These findings suggest that further structural modifications are needed to develop derivatives with higher RdRp inhibitory activity based on vitamin K.
Figure 6.
Scheme for energetically lowest calculated conformation of each compound and RdRp. Two-dimensional (left) and three-dimensional (right) schematic binding interactions of (a) 1, (b) 16, and (c) triphosphate form of remdesivir (RTP) with RdRp (PDB code: 7BV2) using the MOE software.
Table 2. Predicted Binding-Free Energies of Compounds 1, 16, and RTP with RdRp (PDB: 7BV2) by Using MOE.
| compound | receptor | type of interaction | distance (A) | docking score (kcal/mol) |
|---|---|---|---|---|
| 1 | U10 | H-acceptor | 3.26 | –8.4788 |
| 16 | Mg1004 | metal | 2.23 | –9.0060 |
| RTP | U20 | H-donor | 3.24 | –18.1724 |
| Mg1004 | metal | 1.81 | ||
| Mg1004 | metal | 1.63 | ||
| Mg1005 | metal | 2.26 | ||
| Mg1004 | metal | 2.26 | ||
| Mg1005 | metal | 2.30 | ||
| Mg1004 | ionic | 1.81 | ||
| Mg1004 | ionic | 1.63 | ||
| Mg1005 | ionic | 2.26 | ||
| Mg1004 | ionic | 2.26 | ||
| MG1005 | ionic | 2.30 | ||
| U20 | H-pi | 3.20 |
These results suggest that the vitamin K compounds exhibit anti-SARS-CoV-2 activity through the inhibition of SARS-CoV-2 RdRp activity. The RdRp inhibitors remdesivir and molnupiravir and the 3CL-protease inhibitors nirmatrelvir and ensitrelvir are on the market for the treatment of COVID-19. However, the development of additional new therapeutic agents has the following significance. First, current therapies have limited efficacy. In addition, the therapeutic efficacy of nirmatrelvir and ensitrelvir is reported to be reduced if dosing is started too late. In addition, current therapeutic agents present problems in terms of use restrictions and administration methods. For example, remdesivir must be administered intravenously, and molnupiravir has the potential to cause liver dysfunction. Emergence of drug resistance caused by the mutations of viral genomes is also a problem with current antiviral agents. Thus, the development of new therapeutic agents is expected to increase the number of treatment options and improve the effectiveness of antiviral chemotherapy. Furthermore, the development of drugs effective in preventing sequelae is extremely important since the sequelae have also been a serious problem during the worldwide pandemic of COVID-19.
In this study, we have succeeded in finding a series of compounds that has a completely different chemical structure from existing drugs, and some of the compounds inhibit SARS-CoV-2 replication through the inhibition of viral RdRp. In our previous study, 1 and 2 have been shown to induce neural stem cells to differentiate into neurons.17 COVID-19 causes various complications due to thrombosis in addition to fever and respiratory disorders. Among these side effects, cerebral infarction occurs in about 1% of the infected patients and is one of the causes of COVID-19 death, which is a major problem of serious sequelae. Vitamin K derivatives have been shown to cross the blood–brain barrier and may act on the brain and other central organs; the mechanism of cerebral infarction caused by COVID-19 has not been clarified, but it may be due to the transfer of SARS-CoV-2 to the brain. Therefore, a compound having antiviral activity in the brain while also differentiating neural stem cells into neurons may be able to regenerate the neurons that have been lost due to cerebral infarction.
Conclusions
In conclusion, we successfully obtained novel vitamin K derivatives by introducing various substituents into the side chain. We observed that some of them had anti-SARS-CoV-2 activity. We revealed that significant differences in the antiviral activity occurred by altering the functional group and heteroatom of the side chain. Studies are now in progress to obtain vitamin K analogues with much higher anti-SARS-CoV-2 activity. Further optimization of substituents should make it possible to design and synthesize new compounds with more potent anti-SARS-CoV-2 activity and efficacy against COVID-19-induced cerebral sequelae through the activation of neural stem cells.
Experimental Section
General Material and Methods
All reagents, materials, and solvents were of reagent grade. For all reactions that require heating, an oil bath was used. 1H NMR and 13C NMR spectra were recorded at 400 and 100 MHz, respectively, using CDCl3 as a solvent unless otherwise specified. Chemical shifts are given in parts per million (δ) using tetramethylsilane as an internal standard. High-resolution ESI-MS (ESI-HRMS) was performed with an Exactive mass spectrometer. Column chromatography was carried out with silica gel 60 (70–230 mesh), and preparative thin-layer chromatography was run with silica gel 60F254. All reagents were purchased from commercial suppliers unless otherwise noted. We confirmed by HPLC analysis that the purity of all compounds used in the biological assays satisfied 95% and above.
Synthetic methods of compounds 1, 2, and 5 in Scheme 1 were previously reported by us.17
(2E,6E)-8-(3-Isopropylphenyl)-3,7-dimethylocta-2,6-dien-1-ol (26)
To 1 mL of dehydrated THF were added magnesium (turnings) (272 mg, 11.18 mmol) and iodine (75 mg, 0.59 mmol), and the mixture was heated to 84 °C for 10 min under an Ar atmosphere. A solution of 1-bromo-3-isopropylbenzene (2.23 g, 11.18 mmol) in dehydrated THF (2 mL) was added to the mixture, and the solution was heat-refluxed at 70 °C for 30 min under Ar. After dissolution of the magnesium turnings was confirmed, the mixture was cooled to 50 °C. Then, the solution of compound 22 (300 mg, 1.18 mmol) and copper(I) iodide (67 mg, 0.35 mmol) in dehydrated THF (2 mL) was added to the mixture and refluxed at 50 °C for 3 h under an Ar atmosphere. After completion of the reaction, the reaction mixture was cooled to room temperature, then quenched with saturated NH4Cl aqueous solution, and filtered through Celite. The resulting solution was extracted with ethyl acetate three times. The organic layer was combined, then washed with brine, dried with MgSO4, and concentrated in vacuo. The resulting residue was purified by silica gel chromatography (n-hexane/ethyl acetate = 6:1) to afford compound 26 (283 mg, 1.04 mmol), as a colorless liquid, in 88% yield. 1H NMR (CDCl3, 400 MHz); δ 1.22 (s, 3H, PhCH(CH3)CH3), 1.23 (s, 3H, PhCH(CH3)CH3), 1.53 (s, 3H, CH=CCH3), 1.65 (s, 3H, CH=CCH3), 2.04–2.08 (m, 2H, CH2CH2), 2.12–2.17 (m, 2H, CH2CH2), 2.80–2.91 (m, 1H, PhCH(CH3)CH3), 3.24 (s, 2H, CH=C(CH3)CH2), 4.10 (d, 2H, J = 6.4 Hz, CH2CH=CH2(CH3)), 5.22 (t, 1H, J = 6.4 Hz, CH=CCH3), 5.39 (t, 1H, J = 6.8 Hz, CH=CCH3), 6.95–7.04 (m, 3H, Ph), 7.18 (t, J = 7.6 Hz, 1H, Ph); 13C NMR (CDCl3, 100 MHz): δ 16.4, 24.2, 24.2, 26.5, 34.2, 39.6, 46.4, 59.4, 139.5, 135.0, 128.4, 128.3, 127.2, 126.4, 125.9, 124.0, 123.7, 148.9. HRMS ([M + Na]+) m/z: calcd for C19H28ONa, 295.2032; found, 295.2032.
2-((2E,6E)-8-(3-Isopropylphenyl)-3,7-dimethylocta-2,6-dien-1-yl)-3-methylnaphthalene-1,4-dione (4)
To a solution of menadione (250 mg, 1.45 mmol) in 30 mL of Et2O was added 30 mL of 10% Na2S2O4 (aqueous solution) and stirred at room temperature for 1.5 h under an Ar atmosphere. The reaction mixture turned from yellow to colorless. The reaction mixture was then diluted with Et2O and washed with brine. The organic layer was separated, dried with MgSO4, and concentrated in vacuo. The residue was dissolved in 3 mL of dehydrated ethyl acetate and 6 mL of dehydrated 1,4-dioxane. After compound 26 (100 mg, 0.37 mmol) was added to the solution, a catalytic amount of BF3·EtO2 was added and refluxed at 70 °C for 3 h under an Ar atmosphere. The reaction mixture was cooled to room temperature, water was added, and the reaction mixture was extracted with ethyl acetate. The mixture was washed with brine, and the organic layer was dried with MgSO4 and concentrated in vacuo. After concentration, the residue was purified by silica gel chromatography (n-hexane/ethyl acetate = 20:1) to afford compound 4 (33 mg, 0.08 mmol) as a yellow oil in 21% yield. 1H NMR (CDCl3, 400 MHz): δ 1.21 (s, 3H, PhCH(CH3)CH3), 1.23 (s, 3H, PhCH(CH3)CH3), 1.51 (s, 3H, CH=CCH3), 1.81 (s, 3H, CH=CCH3), 2.03 (q, 2H, J = 7.4 Hz, CH2CH2), 2.11 (q, 2H, J = 7.3 Hz, CH2CH2), 2.19 (s, 3H, 2-CH3 of naphthoquinone), 2.83–2.89 (m, 1H, PhCH(CH3)CH3), 3.20 (s, 2H, CH=C(CH3)CH2), 3.38 (d, 2H, J = 6.8 Hz, CH2CH=CH2(CH3)), 5.04 (t, 1H, J = 7.2 Hz, CH=CCH3), 5.18 (t, 1H, J = 7.0 Hz, CH=CCH3), 6.95–7.08 (m, 3H), 7.17 (t, 1H, J = 7.6 Hz, Ph), 7.68–7.70 (m, 2H, Ph of naphthoquinone), 8.08–8.10 (m, 2H, Ph of naphthoquinone); 13C NMR (CDCl3, 100 MHz): δ 12.8, 15.9, 16.5, 24.1, 24.1, 26.7, 26.7, 34.1, 39.8, 46.3, 119.3, 123.9, 126.0, 126.3, 126.3, 126.4, 127.2, 128.2, 132.2, 132.2, 133.4, 133.5, 134.7, 137.6, 140.3, 143.5, 146.2, 148.8, 184.7, 185.6. HRMS ([M + Na]+) m/z: calcd for C30H34O2Na, 449.2245; found, 449.2447.
(2E,6E)-3,7-Dimethyl-8-(3-(trimethylsilyl)phenyl)octa-2,6-dien-1-ol (28)
Compound 28 was prepared by a similar procedure to that in the case of 26 but with 1-bromo-3-(trimethylsilyl) benzene instead of 1-bromo-3-isopropylbenzene as a reactant. Yield: 64%; colorless oil; 1H NMR (CDCl3, 400 MHz): δ 0.26 (s, 9H, Si(CH3)3), 1.55 (s, 3H, CH=CCH3), 1.67 (s, 3H, CH=CCH3), 2.05–2.09 (m, 2H, CH2CH2), 2.16 (q, 2H, J = 7.6 Hz, CH2CH2), 3.28 (s, 2H, CH=C(CH3)CH2), 4.30 (d, 2H, J = 7.2 Hz, CH2CH=CH2(CH3)), 5.24 (t, 1H, J = 6.9 Hz, CH=CCH3), 5.41 (t, 1H, J = 6.9 Hz, CH=CCH3), 7.14–7.16 (m, 1H, Ph), 7.23–7.36 (m, 3H, Ph); 13C NMR (CDCl3, 100 MHz): δ −0.2, 16.0, 16.4, 26.5, 39.6, 46.4, 59.4, 123.7, 126.0, 127.7, 129.4, 131.0, 134.0, 139.5, 139.5, 139.5, 140.3. HRMS ([M + H]+) m/z: calcd for C19H31OSi, 303.2144; found, 303.2143.
2-((2E,6E)-3,7-Dimethyl-8-(3-(trimethylsilyl)phenyl)octa-2,6-dien-1-yl)-3-methylnaphthalene-1,4-dione (6)
Compound 6 was prepared by a similar procedure to that in the case of 4 but with compound 28 instead of compound 26 as a reactant. Yield: 26%; yellow oil; 1H NMR (CDCl3, 400 MHz): δ 0.24 (s, 9H, Si(CH3)3), 1.51 (s, 3H, CH=CCH3), 1.80 (s, 3H, CH=CCH3), 2.01–2.04 (m, 2H, CH2CH2), 2.11 (q, 2H, J = 7.6 Hz, CH2CH2), 2.19 (s, 3H, 2-CH3 of naphthoquinone), 3.20 (s, 2H, CH=C(CH3)CH2), 3.38 (d, 2H, J = 6.8 Hz, CH2CH=CH2(CH3)), 5.04 (t, 1H, J = 6.8 Hz, CH=CCH3), 5.19 (t, 1H, J = 6.8 Hz, CH=CCH3), 7.10–7.12 (m, 1H, Ph), 7.21–7.33 (m, 3H, Ph), 7.66–7.70 (m, 2H, Ph of naphthoquinone), 8.05–8.10 (m, 2H, Ph of naphthoquinone); 13C NMR (CDCl3, 100 MHz): δ 0, 12.8, 15.9, 16.5, 26.1, 26.7, 39.7, 46.3, 119.3, 126.1, 126.3, 126.4, 127.7, 129.7, 131.0, 131.0, 133.4, 133.4, 133.9, 136.5, 137.8, 139.8, 140.2, 143.5, 146.0, 184.6, 185.6. HRMS ([M + Na]+) m/z: calcd for C30H36O2NaSi, 479.2377; found, 479.2373.
(2E,6E)-8-(3-Ethylphenyl)-3,7-dimethylocta-2,6-dien-1-ol (29)
Compound 29 was prepared by a similar procedure to that for 26 but with 1-bromo-3-ethylbenzene instead of 1-bromo-3-isopropylbenzene as a reactant. Yield: 77%; colorless oil; 1H NMR (CDCl3, 400 MHz): δ 1.22 (t, 3H, J = 5.0 Hz, PhCH2CH3), 1.53 (s, 3H, CH=CCH3), 1.67 (s, 3H, CH=CCH3), 2.03–2.09 (m, 2H, CH2CH2), 2.15 (q, 2H, J = 7.2 Hz, CH2CH2), 2.62 (q, 2H, J = 7.5 Hz, PhCH2CH3), 3.24 (s, 2H, CH2CH=CH2(CH3)), 4.12 (d, 2H, J = 7.6 Hz, CH2C=CH(CH3)), 5.23 (t, 1H, J = 7.2 Hz, CH=CCH3), 5.40 (t, 1H, J = 7.2 Hz, CH=CCH3), 6.97–7.03 (m, 3H, Ph), 7.19 (t, 1H, J = 7.4 Hz, Ph); 13C NMR (CDCl3, 100 MHz): δ 16.8, 17.4, 27.0, 27.4, 30.0, 40.6, 47.3, 60.4, 124.7, 126.6, 126.9, 127.2, 128.5, 129.3, 129.6, 136.0, 140.5, 145.2. HRMS ([M + H]+) m/z: calcd for C18H27O, 259.2062; found, 259.2060.
2-((2E,6E)-8-(3-Ethylphenyl)-3,7-dimethylocta-2,6-dien-1-yl)-3-methylnaphthalene-1,4-dione (7)
Compound 7 was prepared by a similar procedure to that for 4 but with compound 29 instead of compound 26 as a reactant. Yield: 15%; yellow oil; 1H NMR (CDCl3, 400 MHz): δ 1.21 (t, 3H, J = 7.6 Hz, PhCH2CH3), 1.50 (s, 3H, CH=CCH3), 1.80 (s, 3H, CH=CCH3), 2.01–2.05 (m, 2H, CH2CH2), 2.09–2.15 (m, 2H, CH2CH2), 2.19 (s, 3H, 2-CH3 of naphthoquinone), 2.60 (q, 2H, J = 7.6 Hz, PhCH2CH3), 3.20 (s, 2H, CH=C(CH3)CH2), 3.39 (d, 2H, J = 7.6 Hz, CH2CH=CH2(CH3)), 5.04 (t, 1H, J = 6.8 Hz, CH=CCH3), 5.18 (t, 1H, J = 6.8 Hz, CH=CCH3), 6.93–7.04 (m, 3H, Ph), 7.15 (t, 1H, J = 3.6 Hz, Ph), 7.67–7.74 (m, 2H, Ph of naphthoquinone), 8.06–8.11 (m, 2H, Ph of naphthoquinone); 13C NMR (CDCl3, 100 MHz): δ 12.9, 14.4, 15.9, 16.6, 26.2, 26.7, 29.0, 39.8, 46.3, 120.0, 122.4, 126.2, 126.4, 126.5, 128.3, 128.6, 132.3, 132.3, 133.5, 133.6, 134.8, 137.6, 138.0, 140.5, 143.6, 144.2, 146.3, 184.7, 185.7. HRMS ([M + Na]+) m/z: calcd for C29H32O2Na, 435.22945; found, 435.2292.
(2E,6E)-8-(3-Butylphenyl)-3,7-dimethylocta-2,6-dien-1-ol (30)
Compound 30 was prepared by a similar procedure to that for 26 but with compound 36 instead of 1-bromo-3-isopropylbenzene as a reactant. Yield: 72%; colorless oil; 1H NMR (CDCl3, 400 MHz): δ 0.92 (t, 3H, J = 7.8 Hz, PhCH2CH2CH2CH3), 1.34–1.39 (m, 2H, PhCH2CH2CH2CH3), 1.53 (s, 3H, CH=CCH3), 1.57–1.62 (m, 2H, PhCH2CH2CH2CH3), 1.67 (s, 3H, CH=CCH3), 2.07–2.09 (m, 2H, CH2CH2), 2.13–2.19 (m, 2H, CH2CH2), 2.58(t, 2H, J = 7.6 Hz, PhCH2CH2CH2CH3), 3.24 (s, 2H, CH=C(CH3)CH2), 4.13 (d, 2H, J = 7.2 Hz, CH2CH=CH2(CH3)), 5.22 (t, 1H, J = 7.0 Hz, CH=CCH3), 5.41(t, 1H, J = 7.0 Hz, CH=CCH3), 6.97–7.01 (m, 3H, Ph), 7.16–7.19 (m, 1H, Ph); 13C NMR (CDCl3, 100 MHz): δ 14.0, 15.8, 16.2, 22.4, 26.3, 33.7, 35.6, 39.5, 46.2, 59.4, 123.5, 125.7, 125.8, 126.0, 128.0, 129.0, 134.9, 139.6, 140.2, 142.8. HRMS ([M + Na]+) m/z: calcd for C20H30ONa, 309.2189; found, 309.2189.
2-((2E,6E)-8-(3-Butylphenyl)-3,7-dimethylocta-2,6-dien-1-yl)-3-methylnaphthalene-1,4-dione (8)
Compound 8 was prepared by a procedure similar to that in the case of 4 but with compound 30 instead of compound 26 as a reactant. Yield: 19%; yellow oil; 1H NMR (CDCl3, 400 MHz): δ 0.91 (t, 3H, J = 7.4 Hz, Ph(CH2)3CH3), 1.30–1.38 (m, 2H, Ph(CH2)2CH2CH3), 1.50 (s, 3H, CH=CCH3), 1.53–1.60 (m, 2H, PhCH2CH2CH2CH3), 1.80 (s, 3H, CH=CCH3), 2.01–2.05 (m, 2H, CH2CH2), 2.08–2.14 (m, 2H, CH2CH2), 2.19 (s, 3H, 2-CH3 of naphthoquinone), 2.55 (t, 2H, J = 7.8 Hz, PhCH2CH2CH2CH3), 3.19 (s, 2H, CH=C(CH3)CH2), 3.38 (d, 2H, J = 7.2 Hz, CH2CH=CH2(CH3)), 5.03 (t, 1H, J = 6.4 Hz, CH=CCH3), 5.17 (t, 1H, J = 6.8 Hz, CH=CCH3), 6.91–7.01 (m, 3H, Ph), 7.14 (t, 1H, J = 7.8 Hz, Ph), 7.66–7.72 (m, 2H, Ph of naphthoquinone), 8.06–8.10 (m, 2H, Ph of naphthoquinone); 13C NMR (CDCl3, 100 MHz): δ 12.8, 14.1, 15.9, 16.5, 22.5, 26.1, 26.7, 33.8, 35.7, 39.7, 46.2, 119.3, 125.9, 126.0, 126.1, 126.3, 126.4, 128.1, 129.1, 132.2, 132.3, 133.4, 133.4, 134.8, 137.5, 137.5, 140.3, 142.8, 143.4, 184.6, 185.6. HRMS ([M + Na]+) m/z: calcd for C31H36O2Na, 463.2608; found, 463.2605.
(2E,6E)-8-(3-Hexylphenyl)-3,7-dimethylocta-2,6-dien-1-ol (31)
Compound 31 was prepared by a similar procedure to that used for 26 but with 1-bromo-3-hexylbenzene instead of 1-bromo-3-isopropylbenzene as a reactant. Yield: 88%; clear colorless oil; 1H NMR (CDCl3, 400 MHz): δ 0.88(t, 3H, J = 6.2 Hz, Ph(CH2)5CH3), 1.24–1.34 (m, 6H, PhCH2CH2(CH2)3CH3), 1.53 (s, 3H, CH=CCH3), 1.55–1.62 (m, 2H, PhCH2CH2(CH2)3CH3), 1.66 (s, 3H, CH=CCH3), 2.06 (q, 2H, J = 7.6 Hz, CH2CH2), 2.15 (q, 2H, J = 7.6 Hz, CH2CH2), 2.56 (t, 2H, J = 7.8 Hz, PhCH2(CH2)4CH3), 3.24 (s, 2H, CH=C(CH3)CH2), 4.11 (d, 2H, J = 7.6 Hz, CH2CH=CH2(CH3)), 5.22(t, 1H, J = 7.2 Hz, CH=CCH3), 5.40(t, 1H, J = 7.2 Hz, CH=CCH3), 6.95–6.99 (m, 3H, Ph), 7.15–7.19 (m, 1H, Ph); 13C NMR (CDCl3, 100 MHz): δ 14.3, 15.9, 16.4, 22.8, 26.5, 29.2, 31.7, 31.9, 36.1, 39.6, 46.3, 59.3, 123.6, 125.8, 126.1, 126.2, 128.2, 129.1, 130.0, 135.0, 140.3, 142.9. HRMS ([M + Na]+) m/z: calcd for C22H34ONa, 337.2502; found, 337.2501.
2-((2E,6E)-8-(3-Hexylphenyl)-3,7-dimethylocta-2,6-dien-1-yl)-3-methylnaphthalene-1,4-dione (9)
Compound 9 was prepared by a similar procedure to that for 4 but with compound 31 instead of compound 26 as a reactant. Yield: 26%; yellow oil; 1H NMR (CDCl3, 400 MHz): δ 0.88 (t, 3H, J = 6.4 Hz, Ph(CH2)5CH3), 1.29 (m, 6H, PhCH2CH2(CH2)3CH3), 1.50 (s, 3H, CH=CCH3), 1.55–1.62 (m, 2H, PhCH2CH2(CH2)3CH3), 1.80 (s, 3H, CH=CCH3), 2.05 (q, 2H, J = 7.4 Hz, CH2CH2), 2.11 (q, 2H, J = 7.4 Hz, CH2CH2), 2.19 (s, 3H, 2-CH3 of naphthoquinone), 2.54 (t, 2H, J = 7.6 Hz, PhCH2(CH2)4CH3), 3.19 (s, 2H, CH=C(CH3)CH2), 3.37 (d, 2H, J = 8.0 Hz, CH2CH=CH2(CH3)), 5.03 (t, 1H, J = 7.2 Hz, CH=CCH3), 5.17 (t, 1H, J = 7.6 Hz, CH=CCH3), 6.93–7.01 (m, 3H, Ph), 7.13–7.19 (m, 1H, Ph), 7.67–7.70 (m, 2H, Ph of naphthoquinone), 8.05–8.09 (m, 2H, Ph of naphthoquinone); 13C NMR (CDCl3, 100 MHz): δ 12.3, 14.2, 15.9, 16.5, 22.8, 26.0, 26.4, 29.0, 31.7, 31.9, 36.0, 39.8, 46.3, 119.3, 125.9, 126.0, 126.1, 126.3, 126.4, 128.1, 129.1, 131.9, 133.4, 133.5, 134.8, 136.5, 137.6, 140.2, 143.0, 143.5, 146.5, 184.6, 185.6. HRMS ([M + Na]+) m/z: calcd for C33H40O2Na, 491.2921; found, 491.2918.
(2E,6E)-3,7-Dimethyl-8-(3-(trifluoromethyl)phenyl)octa-2,6-dien-1-ol (32)
Compound 32 was prepared by a similar procedure to that for 26 but with 3-bromobenzotrifluoride instead of 1-bromo-3-isopropylbenzene as a reactant. Yield: 82%; clear colorless oil; 1H NMR (CDCl3, 400 MHz): δ 1.44 (s, 3H, CH=CCH3), 1.59 (s, 3H, CH=CCH3), 1.98–2.02 (m, 2H, CH2CH2), 2.07 (q, 2H, J = 7.1 Hz, CH2CH2), 3.23 (s, 2H, CH=C(CH3)CH2), 4.06 (d, 2H, J = 6.8 Hz, CH2CH=CH2(CH3)), 5.16(t, 1H, J = 7.0 Hz, CH=CCH3), 5.34 (t, 1H, J = 6.8 Hz, CH=CCH3), 7.25–7.37 (m, 4H, Ph); 13C NMR (CDCl3, 100 MHz): δ 14.7, 15.2, 25.2, 38.3, 44.9, 52.3, 121.8, 122.7, 124.4, 124.5, 125.8, 127.6, 131.2, 131.2, 132.9, 138.2, 140.3. HRMS ([M + Na]+) m/z: calcd for C17H21F3ONa, 321.1437; found, 321.1436.
2-((2E,6E)-3,7-Dimethyl-8-(3-(trifluoromethyl)phenyl)octa-2,6-dien-1-yl)-3-methylnaphthalene-1,4-dione (10)
Compound 10 was prepared by a procedure similar to that in the case of 4 but with compound 32 instead of compound 26 as a reactant. Yield: 15%; yellow oil; 1H NMR (CDCl3, 400 MHz): δ 1.49 (s, 3H, CH=CCH3), 1.80 (s, 3H, CH=CCH3), 2.04 (q, 2H, J = 7.4, CH2CH2), 2.12 (q, 2H, J = 7.2 Hz, CH2CH2), 2.19 (s, 3H, 2-CH3 of naphthoquinone), 3.26 (s, 2H, CH=C(CH3)CH2), 3.38 (d, 2H, J = 6.8 Hz, CH2CH=CH2(CH3)), 5.04 (t, 1H, J = 6.4 Hz, CH=CCH3), 5.19 (t, 1H, J = 6.8 Hz, CH=CCH3), 7.31 (s, 1H, Ph), 7.29–7.43 (m, 3H, Ph), 7.67–7.72 (m, 2H, Ph of naphthoquinone), 8.05–8.09 (m, 2H, Ph of naphthoquinone); 13C NMR (CDCl3, 100 MHz): δ 12.7, 15.8, 16.4, 26.1, 26.4, 39.6, 45.9, 119.4, 122.9, 125.5, 126.3, 126.3, 127.0, 128.6, 132.0, 132.2, 133.4, 133.4, 133.7, 133.7, 137.3, 141.3, 143.4, 146.1, 184.6, 185.5. HRMS ([M + Na]+) m/z: calcd for C28H27F3O2Na, 475.1855; found, 475.1852.
(2E,6E)-8-(3-Fluorophenyl)-3,7-dimethylocta-2,6-dien-1-ol (33)
Compound 33 was prepared by a similar procedure to that for 26 but with 1-bromo-3-fluorobenzene instead of 1-bromo-3-isopropylbenzene as a reactant. Yield: 68%; clear colorless oil; 1H NMR (CDCl3, 400 MHz): δ 1.53(s, 3H, CH=CCH3), 1.68(s, 3H, CH=CCH3), 2.08(q, 2H, J = 7.0 Hz, CH2CH2), 2.16(q, 2H, J = 7.0 Hz, CH2CH2), 3.26(s, 2H, CH=C(CH3)CH2), 4.15(d, 2H, J = 6.8 Hz, CH2CH=CH2(CH3)), 5.23(t, 1H, J = 7.2 Hz, CH=CCH3), 5.42(t, 1H, J = 7.6 Hz, CH=CCH3), 6.85–6.94 (m, 3H, Ph), 7.18–7.26 (m, 1H, Ph); 13C NMR (CDCl3, 100 MHz): δ 15.8, 16.3, 26.3, 39.5, 46.0, 59.4, 112.8, 115.7, 123.8, 124.6, 126.6, 130.0, 134.2, 139.4, 143.1, 161.8. HRMS ([M + Na]+) m/z: calcd for C16H21FONa, 271.1469; found, 271.1468.
2-((2E,6E)-8-(3-Fluorophenyl)-3,7-dimethylocta-2,6-dien-1-yl)-3-methylnaphthalene-1,4-dione (11)
Compound 11 was prepared by a procedure similar to that in the case of 4 but with compound 33 instead of compound 26 as a reactant. Yield: 18%; yellow oil; 1H NMR (CDCl3, 400 MHz): δ 1.49(s, 3H, CH=CCH3), 1.80 (s, 3H, CH=CCH3), 2.04(q, 2H, J = 7.2 Hz, CH2CH2), 2.12(q, 2H, J = 7.4 Hz, CH2CH2), 2.19(s, 3H, 2-CH3 of naphthoquinone), 3.20(s, 2H, CH=C(CH3)CH2), 3.38(d, 2H, J = 6.8 Hz, CH2CH=CH2(CH3)), 5.04 (t, 1H, J = 6.8 Hz, CH=CCH3), 5.18(t, 1H, J = 7.2 Hz, CH=CCH3), 7.26–7.43 (m, 4H, Ph), 7.67–7.71 (m,, 2H, Ph of naphthoquinone), 8.05–8.09 (m, 2H, Ph of naphthoquinone); 13C NMR (CDCl3, 100 MHz): δ 12.7, 15.7, 16.4, 26.0, 26.4, 39.6, 45.9, 112.7, 115.6, 119.4, 124.4, 126.2, 126.3, 126.7, 129.4, 129.5, 133.3, 134.1, 133.4, 133.9, 133.9, 137.3, 143.4, 146.1, 164.1, 184.5, 185.5. HRMS ([M + Na]+) m/z: calcd for C27H27FO2Na, 425.1887; found, 425.1884.
(2E,6E)-8-(4-Fluorophenyl)-3,7-dimethylocta-2,6-dien-1-ol (34)
Compound 34 was prepared by a procedure similar to that for 26 but with 4-fluorophenyl magnesium bromide solution (1.0 M in THF) (0.94 g, 4.72 mmol) as a reactant. Yield: 60%; clear colorless oil; 1H NMR (CDCl3, 400 MHz): δ 1.52 (s, 3H, CH=CCH3), 1.68 (s, 3H, CH=CCH3), 2.04–2.09 (m, 2H, CH2CH2), 2.15(q, J = 7.5 Hz, 2H, CH2CH2), 3.23(s, 2H, CH=C(CH3)CH2), 4.15(d, 2H, J = 6.8 Hz, CH2CH=CH2(CH3)), 5.21(t, 1H, J = 6.6 Hz, CH=CCH3), 5.42(t, 1H, J = 6.8 Hz, CH=CCH3), 6.93–6.98 (m, 2H, Ph), 7.09–7.12 (m, 2H, Ph); 13C NMR (CDCl3, 100 MHz): δ 15.7, 16.2, 26.2, 39.4, 45.3, 59.4, 114.8, 115.0, 123.6, 126.0, 130.1, 130.2, 134.7, 139.4, 160.2. HRMS ([M + Na]+) m/z: calcd for C16H21FONa, 273.0522; found, 273.0521.
2-((2E,6E)-8-(4-Fluorophenyl)-3,7-dimethylocta-2,6-dien-1-yl)-3-methylnaphthalene-1,4-dione (12)
Compound 12 was prepared by a similar procedure to that in the case of 4 but with compound 34 instead of compound 26 as a reactant. Yield: 20%; yellow oil; 1H NMR (CDCl3, 400 MHz): δ 1.48 (s, 3H, CH=CCH3), 1.57 (s, 3H, CH=CCH3), 2.03 (q, 2H, J = 6.8 Hz, CH2CH2), 2.10 (q, 2H, J = 7.2 Hz, CH2CH2), 2.19 (s. 3H, 2-CH3 of naphthoquinone), 3.18 (s, 2H, CH=C(CH3)CH2), 3.38 (d, 2H, J = 6.8 Hz, CH2CH=CH2(CH3)), 5.03 (t, 1H, J = 6.8 Hz, CH=CCH3), 5.15 (t, 1H, J = 7.2 Hz, CH=CCH3), 6.90–6.94 (m, 2H, Ph), 7.04–7.08 (m, 2H, Ph), 7.67–7.72 (m, 2H, Ph of naphthoquinone), 8.06–8.10 (m, 2H, Ph of naphthoquinone); 13C NMR (CDCl3, 100 MHz): δ 12.7, 15.7, 16.4, 26.0, 26.4, 39.6, 45.3, 114.8, 115.0, 119.3, 126.2, 126.2, 126.3, 130.0, 130.1, 130.6, 132.1, 133.3, 133.4, 134.4, 135.9, 137.3, 143.4, 146.1, 184.5, 185.5. HRMS ([M + Na]+) m/z: calcd for C27H27FO2Na, 425.1887; found, 425.1887.
(2E,6E)-8-(4-Fluoronaphthalen-1-yl)-3,7-dimethylocta-2,6-dien-1-ol (35)
Compound 35 was prepared by a similar procedure to that in the case of 26 but with 1-bromo-4-fluoronaphthalene instead of 1-bromo-3-isopropylbenzene as a reactant. Yield: 60%; clear colorless oil; 1H NMR (CDCl3, 400 MHz): δ 1.61 (s, 6H, 2xCH=CCH3), 2.00 (q, 2H, J = 7.6 Hz, CH2CH2), 2.13 (q, 2H, J = 7.3 Hz, CH2CH2), 3.68 (s, 2H, CH=C(CH3)CH2), 4.08 (d, 2H, J = 7.2 Hz, CH2CH=CH2(CH3)), 5.12 (t, 1H, J = 7.2 Hz, CH=CCH3), 5.32 (t, 1H, J = 7.0 Hz, CH=CCH3), 7.06 (t, 1H, J = 9.2 Hz, naphthalene), 7.19 (t, 1H, J = 5.2 Hz, naphthalene), 7.50 (d, 1H, J = 7.6 Hz, naphthalene), 7.52 (d, 1H, J = 7.6 Hz, naphthalene), 8.00 (m, 1H, naphthalene), 8.12 (m, 1H, naphthalene); 13C NMR (CDCl3, 100 MHz): δ 16.3, 16.5, 26.3, 39.4, 42.5, 59.4, 109.0, 121.1, 121.3, 123.7, 124.5, 124.6, 125.8, 126.3, 126.6, 131.9, 132.0, 133.6, 139.3, 156.6. HRMS ([M + Na]+) m/z: calcd for C20H23FONa, 321.1631; found, 321.1630.
2-((2E,6E)-8-(4-Fluoronaphthalen-1-yl)-3,7-dimethylocta-2,6-dien-1-yl)-3-methylnaphthalene-1,4-dione (13)
Compound 13 was prepared by a similar procedure to that in the case of 4 but with compound 35 instead of compound 26 as a reactant. Yield: 15%; yellow oil; 1H NMR (CDCl3, 400 MHz): δ 1.61 (s, 6H, 2xCH=CCH3), 2.01 (q, 2H, J = 7.2 Hz, CH2CH2), 2.12 (q, J = 7.2 Hz, 2H, CH2CH2), 2.20 (s, 3H, 2-CH3 of naphthoquinone), 3.38 (d, 2H, J = 7.2 Hz, CH2CH=CH2(CH3)), 3.70 (s, 2H, CH=C(CH3)CH2), 5.00 (t, 1H, J = 7.6 Hz, CH=CCH3), 5.12 (t, 1H, J = 7.6 Hz, CH=CCH3), 7.30 (t, 3H, J = 15.2 Hz, naphthalene), 7.52–7.54 (m, 3H, naphthalene), 7.70–7.74 (m, 2H, naphthalene), 8.09–8.14 (m, 2H, naphthalene); 13C NMR (CDCl3, 100 MHz): δ 12.7, 16.38, 16.40, 26.0, 26.6, 39.5, 42.8, 119.2, 125.4, 125.5, 126.3, 126.3, 126.9, 128.5, 132.1, 132.5, 133.3, 133.3, 133.8, 134.0, 136.1, 137.4, 146.1, 184.5. HRMS ([M + Na]+) m/z: calcd for C31H29FO2Na, 475.2049; found, 475.2047.
(E)-2-(3,7-Dimethylocta-2,6-dien-1-yl)-3-methylnaphthalene-1,4-dione (MK-2) (36)
Compound 36 was prepared by a procedure similar to that in the case of 4 but with geraniol instead of compound 26 as a reactant. Yield: 39%; yellow oil; 1H NMR (CDCl3, 400 MHz): δ 1.56 (s, 3H, CH=CCH3), 1.62 (s, 3H, CH=CCH3), 1,79 (s, 3H, CH=CCH3), 1.99 (t, 2H, J = 7.2 Hz, CH2CH2), 2.05 (t, 2H, J = 7.2 Hz, CH2CH2), 2.18 (s, 3H, 2-CH3 of naphthoquinone), 3.35 (d, 2H, J = 6.8 Hz, CH2CH=CH2(CH3)), 5.02 (t, 2H, J = 0.8 Hz, CH=CCH3), 7.66 (m, 2H, naphthoquinone), 8.05 (m, 2H, naphthoquione). 13C NMR (CDCl3, 100 MHz): δ 12.6, 16.4, 17.7, 25.7, 26.0, 26.6, 39.7, 119.2, 124.1, 126.0, 126.3, 132.2, 132.2, 133.3, 133.3, 137.5, 143.4, 146.1, 184.4, 185.4. HRMS ([M + H]+) m/z: calcd for C21H25O2, 309.1849; found, 309.1847.
(E)-2-(3,7-Dimethylocta-2,6-dien-1-yl)-1,4-dimethoxy-3-methylnaphthalene (37)
A solution of compound 36 (4.19 g, 13.57 mmol) and tetra-n-butylammonium iodide (0.59 g, 1.60 mmol) in 30 mL of dehydrated THF and 10 mL of water was stirred at room temperature for 10 min. Sodium hydrosulfate (14.2 g, 81.6 mmol) was dissolved in 40 mL of water, and the solution was stirred at room temperature for 20 min. Dimethyl sulfate (15.5 mL, 0.16 mol) was added dropwise to the solution, and the mixture was stirred at room temperature for 22 h. After the reaction was completed, the reaction mixture was diluted with ethyl acetate and washed with brine. The organic layer was separated, dried with MgSO4, and filtered. The filtrate was then concentrated in vacuo. The residue was purified by silica gel chromatography (n-hexane/ethyl acetate = 30:1) to afford compound 37 (3.40 g, 10.05 mmol) in 74% yield as a yellow oil. 1H NMR (CDCl3, 400 MHz): δ 1.56 (s, 3H, CH=CCH3), 1.63 (s, 3H, CH=CCH3), 1.82 (s, 3H, CH=CCH3), 2.02 (q, 2H, J = 7.4 Hz, CH2CH2), 2.07 (q, 2H, J = 7.6 Hz, CH2CH2), 2.37 (s, 3H, 2-CH3 of naphthoquinone), 3.57 (d, 2H, J = 6.4 Hz, CH2CH=CH2(CH3)), 3.85 (s, 3H, OCH3), 3.87 (s, 3H, OCH3), 5.05 (t, 1H, J = 7.0 Hz, CH=CCH3), 5.11 (t, 1H, J = 6.4 Hz, CH=CCH3), 7.44 (m, 2H, Ph of naphthoquinone), 8.05 (m, 2H, Ph of naphthoquinone); 13C NMR (CDCl3, 100 MHz): δ 12.5, 16.4, 17.8, 25.8, 26.4, 26.5, 39.8, 61.4, 62.3, 121.2, 122.2, 122.4, 123.0, 124.2, 124.3, 125.4, 125.5, 131.0, 131.5, 131.7, 135.7, 149.8, 150.2. HRMS ([M + H]+) m/z: calcd for C23H31O2, 339.2319; found, 339.2316.
(2E,6E)-8-(1,4-Dimethoxy-3-methylnaphthalen-2-yl)-2,6-dimethylocta-2,6-dien-1-ol (38)
To a solution of SeO2 (42.61 mg, 0.38 mmol) and salicylic acid (53.04 mg, 0.38 mmol) in 50 mL of CH2Cl2 was added 70%-tert-butyl hydroperoxide (1.79 g, 13.83 mmol), and the mixture was stirred for 10 h. A solution of compound 37 (1.30 g, 3.84 mmol) in 1 mL of CH2Cl2 was added to the mixture and stirred at 0 °C for 1 h. The mixture was then stirred at room temperature for 23 h. The mixture was diluted with CH2Cl2 and then washed with sat. NaHCO3 aq., water, sat. CuSO4 aq., water, sat. Na2S2O4 aq., and brine. The organic layer was separated, dried over MgSO4, and filtered. The filtrate was then concentrated in vacuo. The residue was purified by silica gel chromatography (n-hexane/ethyl acetate = 4:1) to afford compound 38, a clear colorless liquid, in 490 mg (1.38 mmol) yield, 36% yield. 1H NMR (CDCl3, 400 MHz): δ 1.62 (s, 3H, CH=CCH3), 1.83 (s, 3H, CH=CCH3), 2.04 (q, 2H, J = 7.4 Hz, CH2CH2), 2.13 (q, 2H, J = 7.3 Hz, CH2CH2), 2.37 (s, 3H, 2-CH3 of naphthoquinone), 3.56 (d, 2H, J = 6.4 Hz, CH2CH=CH2(CH3)), 3.87 (s, 3H, OCH3), 3.88 (s, 3H, OCH3), 3.93 (s, 2H, CH2OH), 5.10 (t, 1H, J = 5.2 Hz, CH=CCH3), 5.33 (t, 1H, J = 5.2 Hz, CH=CCH3), 7.68–7.71 (m, 2H, Ph of naphthoquinone and purine), 8.07–8.10 (m, 2H, Ph of naphthoquinone and purine); 13C NMR (CDCl3, 100 MHz): δ 12.5, 14.3, 16.3, 25.8, 26.7, 39.8, 61.4, 62.3, 122.2, 122.4, 123.1, 124.4, 125.4, 125.5, 127.1, 127.4, 127.6, 131.0, 131.3, 135.8, 149.9, 150.2. HRMS ([M + Na]+) m/z: calcd for C23H30O3Na, 377.2087; found, 377.2084.
9-((2E,6E)-8-(1,4-Dimethoxy-3-methylnaphthalen-2-yl)-2,6-dimethylocta-2,6-dien-1-yl)-9H-purine (39)
To 8 mL of dehydrated DMF were added PPh3 (437 mg, 1.67 mmol), purine (200 mg, 1.67 mmol), and compound 38 (197 mg, 0.56 mmol) and stirred at 0 °C for 30 min. Dehydrated DMF (1.0 mL) was dissolved with DIAD (337 mg, 1.67 mmol), and the mixture was added dropwise using a syringe. The reaction was then stirred for 1 h at room temperature. After completion of the reaction, the mixture was extracted with ethyl acetate and then washed with brine. The organic layer was dried with MgSO4 and filtered. After the filtrate was concentrated in vacuo, the residue was purified by silica gel chromatography (n-hexane/ethyl acetate = 1:2) to afford a brown liquid, compound 39 in 83 mg (0.18 mmol) yield, 33% yield. 1H NMR (CDCl3, 400 MHz): δ 1.55 (s, 3H, CH=CCH3), 1.83 (s, 3H, CH=CCH3), 2.05 (q, 2H, J = 7.4 Hz, CH2CH2), 2.16 (q, 2H, J = 7.3 Hz, CH2CH2), 2.38 (s, 3H, 2-CH3 of naphthoquinone), 3.57 (d, 2H, J = 6.4 Hz, CH2CH=CH2(CH3)), 3.87 (s, 3H, OCH3), 3.90 (s, 3H, OCH3), 4.70 (s, 2H, CH=C(CH3)CH2), 5.12 (t, 1H, J = 6.8 Hz, CH=CCH3), 5.38 (t, 1H, J = 7.0 Hz, CH=CCH3), 7.45–7.47 (m, 2H, Ph of naphthoquinone), 7.97 (s, 1H, purine), 8.05 (m, 2H, Ph of naphthoquinone), 8.98 (s, 1H, purine), 9.14 (s, 1H, purine); 13C NMR (CDCl3, 100 MHz): δ 12.5 14.4, 16.4, 26.2, 26.4, 39.0, 50.8, 61.4, 62.2, 116.5, 122.2, 122.3, 123.6, 125.4, 125.6, 126.8, 127.3, 127.6, 129.5, 134.0, 134.9, 140.0, 144.9, 145.4, 148.6, 149.8, 150.2, 152.7. HRMS ([M + Na]+) m/z: calcd for C28H32N4O2Na, 272.0682; found, 272.0681.
2-((2E,6E)-3,7-Dimethyl-8-(9H-purin-9-yl)octa-2,6-dien-1-yl)-3-methylnaphthalene-1,4-dione (14)
Compound 39 (167 mg, 0.37 mmol) was added to 1 mL of CH3CN and cooled to 0 °C. CAN (401 mg, 0.73 mmol) dissolved in 0.5 mL of water was added, and the reaction was stirred for 1 h. After 1 h, the reaction was terminated by addition of brine. After completion of the reaction, the mixture was extracted with ethyl acetate three times and then the combined organic layer was washed with brine The organic layer was dried with MgSO4 and filtered. After the filtrate was concentrated in vacuo, the residue was purified by silica gel chromatography (n-hexane/ethyl acetate = 1:10) to afford compound 14 (76 mg, 0.18 mmol) in 48% yield as a brown oil. 1H NMR (CDCl3, 400 MHz): δ 1.58 (s, 3H, CH=CCH3), 1.79 (s, 3H, CH=CCH3), 2.04 (q, 2H, J = 7.2 Hz, CH2CH2), 2.16 (q, 2H, J = 7.2 Hz, CH2CH2), 2.19 (s, 3H, 2-CH3 of naphthoquinone), 3.37 (d, 2H, J = 6.8 Hz, CH2CH=CH2(CH3)), 4.75 (s, 2H, CH=C(CH3)CH2), 5.03 (t, 1H, J = 6.2 Hz, CH=CCH3), 5.39 (t, 1H, J = 6.0 Hz, CH=CCH3), 7.66–7.71 (m, 2H, Ph of naphthoquinone), 8.06–8.08 (m, 2H, Ph of naphthoquinone), 8.08 (s. 1H, purine), 8.98 (s, 1H, purine), 9.13 (s, 1H, purine); 13C NMR (CDCl3, 100 MHz): δ 12.8, 14.4, 16.4, 26.2, 26.3, 39.0, 50.9, 120.0, 126.3, 126.3, 129.6, 130.1, 132.2, 133.4, 133.5, 134.0, 134.0, 136.5, 143.5, 145.5, 145.9, 148.6, 151.6, 152.7, 184.6, 185.5. HRMS ([M + H]+) m/z: calcd for C26H27N4O2, 427.2129; found, 427.2127.
9-((2E,6E)-8-(1,4-Dimethoxy-3-methylnaphthalen-2-yl)-2,6-dimethylocta-2,6-dien-1-yl)-9H-purin-6-amine (40)
Compound 40 was prepared by a procedure similar to that for 39 but with adenine instead of purine as a reactant. Yield: 42%; brown oil; 1H NMR (CDCl3, 400 MHz): δ 1.55 (s, 3H, CH=CCH3), 1.82 (s, 3H, CH=CCH3), 2.04 (q, 2H, J = 7.6 Hz, CH2CH2), 2.15 (q, 2H, J = 7.3 Hz, CH2CH2), 2.37 (s, 3H, 2-CH3 of naphthoquinone), 3.56 (d, 2H, J = 6.4 Hz, CH2CH=CH2(CH3)), 3.87 (s, 3H, OCH3), 3.89 (s, 3H, OCH3), 4.61 (s, 2H, CH=C(CH3)CH2), 5.12 (t, 1H, J = 6.0 Hz, CH=CCH3), 5.32 (t, 1H, J = 6.4 Hz, CH=CCH3), 7.44–7.48 (m, 2H, Ph of naphthoquinone), 7.67 (s, 1H, purine), 8.02–8.05 (m, 2H, Ph of naphthoquinone), 8.35 (s, 1H, purine); 13C NMR (CDCl3, 100 MHz): δ 12.4, 14.3, 16.4, 26.32, 26.35, 39.0, 50.7, 61.4, 62.2, 119.3, 122.1, 122.2, 123.4, 125.4, 125.5, 126.7, 127.2, 127.5, 129.4, 129.8, 130.6, 134.9, 140.4, 149.7, 150.1, 153.0, 155.6. HRMS ([M + H]+) m/z: calcd for C28H34N5O2, 472.2707; found, 472.2707.
2-((2E,6E)-8-(6-Amino-9H-purin-9-yl)-3,7-dimethylocta-2,6-dien-1-yl)-3-methylnaphthalene-1,4-dione (15)
Compound 15 was prepared by a procedure similar to that for 14 but with 40 instead of 39 as a reactant. Yield: 50%; red brown oil; 1H NMR (CDCl3, 400 MHz): δ 1.58 (s, 3H, CH=CCH3), 1.79 (s, 3H, CH=CCH3), 2.06 (q, 2H, J = 6.4 Hz, CH2CH2), 2.17 (s, 3H, 2-CH3 of naphthoquinone), 2.17–2.20 (q, 4H, J = 6.4 Hz, CH2CH2), 3.36 (d, 2H, J = 7.2 Hz, CH2CH=CH2(CH3)), 4.69 (s, 2H, CH=C(CH3)CH2), 5.02 (t, 1H, J = 6.4 Hz, CH=CCH3), 5.38 (t, 1H, J = 6.0 Hz, CH=CCH3), 7.70 (m, 4H), 8.09 (m, 4H); 13C NMR (CDCl3, 100 MHz): δ 12.8, 14.2, 14.3, 16.3, 21.1, 26.0, 26.2, 38.8, 120.1, 126.3, 126.5, 128.9, 131.0, 132.0, 132.1, 133.5, 133.6, 133.7, 126.4, 143.5, 145.8, 184.6, 185.5. HRMS ([M + H]+) m/z: calcd for C26H27N5O2, 442.2238; found, 442.2234.
6-Chloro-9-((2E,6E)-8-(1,4-dimethoxy-3-methylnaphthalen-2-yl)-2,6-dimethylocta-2,6-dien-1-yl)-9H-purine (41)
Compound 41 was prepared by a procedure similar to that for 39 but with 6-chloro-7H-purine instead of purine as a reactant. Yield: 40%; yellow oil; 1H NMR (CDCl3, 400 MHz): δ 1.57 (s, 3H, CH=CCH3), 1.79 (s, 3H, CH=CCH3), 1.99 (q, 2H, J = 7.6 Hz, CH2CH2), 2.13 (q, 2H, J = 7.3 Hz, CH2CH2), 2.36 (s, 3H, 2-CH3 of naphthoquinone), 3.55 (d, 2H, J = 6.4 Hz, CH2CH=CH2(CH3)), 3.86 (s, 3H, OCH3), 3.88 (s, 3H, OCH3), 4.83 (s, 2H, CH=C(CH3)CH2), 5.08–5.18 (m, 2H, 2xCH=CCH3), 7.43–7.46 (m, 2H, Ph of naphthoquinone), 8.03–8.06 (m, 3H, Ph of naphthoquinone and purine), 8.85 (s, 1H, purine); 13C NMR (CDCl3, 100 MHz): δ 12.5, 14.4, 16.4, 26.3, 26.4, 38.8, 54.0, 61.5, 62.2, 122.2, 122.3, 122.7, 123.7, 125.5, 125.6, 126.8, 127.3, 127.6, 129.5, 129.8, 130.7, 134.6, 143.2, 149.2, 149.8, 150.2, 152.5, 162.0. HRMS ([M + Na]+) m/z: calcd for C28H31ClN4O2Na, 513.2028; found, 513.2026.
2-((2E,6E)-8-(6-Chloro-9H-purin-9-yl)-3,7-dimethylocta-2,6-dien-1-yl)-3-methylnaphthalene-1,4-dione (16)
Compound 16 was prepared by a similar procedure to that for 14 but with 41 instead of 39 as a reactant. Yield: 84%; yellow oil; 1H NMR (CDCl3, 400 MHz): δ 1.57 (s, 3H, CH=CCH3), 1.79 (s, 3H, CH=CCH3), 2.04 (q, J = 7.2 Hz, CH2CH2), 2.16 (q, 2H, J = 7.4 Hz, CH2CH2), 2.20 (s, 3H, 2-CH3 of naphthoquinone), 3.37 (d, 2H, J = 6.8 Hz, CH2CH=CH2(CH3)), 4.74 (s, 2H, CH=C(CH3)CH2), 5.03 (t, 1H, J = 6.2 Hz, CH=CCH3), 5.41 (t, 1H, J = 6.4 Hz, CH=CCH3), 7.68 (m, 2H, Ph of naphthoquinone), 8.08(m, 3H, Ph of naphthoquinone and purine), 8.74(s, 1H, purine); 13C NMR (CDCl3, 100 MHz): δ 12.9, 14.4, 16.4, 26.2, 26.3, 39.0, 51.6, 120.1, 126.3, 129.3, 130.6, 130.9, 131.6, 132.2, 133.5, 133.5, 133.5, 136.6, 143.5, 145.2, 145.3, 146.0, 151.1, 152.1, 184.6, 185.5. HRMS ([M + Na]+) m/z: calcd for C26H25ClN4O2Na, 483.1558; found, 483.1554.
2,6-Dichloro-9-((2E,6E)-8-(1,4-dimethoxy-3-methylnaphthalen-2-yl)-2,6-dimethylocta-2,6-dien-1-yl)-9H-purine (42)
Compound 42 was prepared by a similar procedure to that for 39 but with 2,6-dichloro purine instead of purine as a reactant. Yield: 30%; yellow oil; 1H NMR (CDCl3, 400 MHz): δ 1.57 (s, 3H, CH=CCH3), 1.81 (s, 3H, CH=CCH3), 2.00 (q, 2H, J = 6.8 Hz, CH2CH2), 2.14 (q, 2H, J = 7.3 Hz, CH2CH2), 2.37 (s, 3H, 2-CH3 of naphthoquinone), 3.54 (d, 2H, J = 6.4 Hz, CH2CH=CH2(CH3)), 3.87 (s, 3H, OCH3), 3.89 (s, 3H, OCH3), 4.79 (s, 2H, CH=C(CH3)CH2), 4.92–4.98 (m, 1H, CH=CCH3), 5.08–5.16 (m, 1H, CH=CCH3), 7.41–7.48 (m, 2H, Ph of naphthoquinone), 8.02–8.06 (m, 2H, Ph of naphthoquinone), 8.09 (s, 1H, purine); 13C NMR (CDCl3, 100 MHz): δ 12.5, 14.3, 16.4, 22.0, 26.2, 38.8, 54.1, 61.4, 62.2, 122.2, 122.3, 122.3, 123.6, 125.5, 125.6, 126.8, 127.2, 127.5, 129.2, 129.9, 130.7, 134.6, 143.9, 149.8, 150.1, 150.7, 153.0, 163.5. HRMS ([M + H]+) m/z: calcd for C28H31Cl2N4O2, 525.1824; found, 525.1826.
2-((2E,6E)-8-(2,6-Dichloro-9H-purin-9-yl)-3,7-dimethylocta-2,6-dien-1-yl)-3-methylnaphthalene-1,4-dione (17)
Compound 17 was prepared by a procedure similar to that for 14 but with 42 instead of 39 as a reactant. Yield: 38%; yellow oil; 1H NMR (CDCl3, 400 MHz): δ 1.63 (s, 3H, CH=CCH3), 1.78 (s, 3H, CH=CCH3), 2.00 (q, 2H, J = 7.8 Hz, CH2CH2), 2.15 (q, 2H, J = 7.4 Hz, CH2CH2), 2.19 (s, 3H, 2-CH3 of naphthoquinone), 3.35 (d, 2H, J = 7.2 Hz, CH2CH=CH2(CH3)), 4.91 (s, 2H, CH=C(CH3)CH2), 4.99 (t, 1H, J = 6.8 Hz, CH=CCH3), 5.17 (t, J = 7.2 Hz, CH=CCH3), 7.67–7.69 (m, 2H, Ph of naphthoquinone), 8.05–8.08 (m, 2H, Ph of naphthoquinone), 8.22 (s, 1H, purine); 13C NMR (CDCl3, 100 MHz): δ 12.8, 14.4, 16.4, 26.1, 26.2, 38.8, 54.2, 120.1, 126.2, 126.3, 129.2, 130.1, 130.4, 132.1, 132.2, 133.5, 133.5, 136.5, 137.0, 143.5, 144.0, 145.9, 150.5, 153.2, 163.6, 184.6, 185.5. HRMS ([M + H]+) m/z: calcd for C26H25Cl2N4O2, 495.1349; found, 495.1350.
2-((2E,6E)-8-Hydroxy-3,7-dimethylocta-2,6-dien-1-yl)-3-methylnaphthalene-1,4-dione (43)
Compound 43 was prepared by a procedure similar to that for 38 but with 36 instead of 37 as a reactant. Yield: 40%; yellow oil; 1H NMR (CDCl3, 400 MHz): δ 1.63 (s, 3H, CH=CCH3), 1.79 (s, 3H, CH=CCH3), 2.03 (q, 2H, J = 7.0 Hz, CH2CH2), 2.12 (q, 2H, J = 7.0 Hz, CH2CH2), 2.19 (s, 3H, 2-CH3 of naphthoquinone), 3.36 (d, 2H, J = 6.8 Hz, CH2CH=CH2(CH3)), 3.94 (s, 2H, CH=C(CH3)CH2), 5.02 (t, 1H, J = 7.2 Hz, CH=CCH3), 5.32 (t, 1H, J = 6.8 Hz, CH=CCH3), 7.67–7.72 (m, 2H, Ph of naphthoquinone), 8.06–8.10 (m, 2H, Ph of naphthoquinone); 13C NMR (CDCl3, 100 MHz): δ 12.7, 13.7, 16.4, 26.0, 26.1, 39.3, 68.9, 119.6, 125.5, 126.3, 126.3, 132.2, 132.2, 133.4, 133.5, 135.1, 137.1, 143.5, 146.1, 184.7, 185.5. HRMS ([M + H]+) m/z: calcd for C21H25O3, 325.1804; found, 325.1802.
(2E,6E)-2,6-Dimethyl-8-(3-methyl-1,4-dioxo-1,4-dihydronaphthalen-2-yl)octa-2,6-dienal (44)
To a solution of compound 43 (385 mg, 1.19 mmol) in 40 mL of CH2Cl2 was added MnO2 (3.10 g, 35.60 mmol), and the reaction mixture was stirred at room temperature for 3 h under an Ar atmosphere. After the reaction was completed, the reaction solution was filtered through Celite and extracted with ethyl acetate. The organic layer was dried with MgSO4 and filtered. The filtrate was evaporated in vacuo, and the residue was purified by silica gel chromatography (n-hexane/ethyl acetate = 4:1) to afford compound 44 (358 mg, 1.11 mmol) as a clear colorless liquid in 94% yield. 1H NMR (CDCl3, 400 MHz): δ 1.71 (s, 3H, CH=CCH3), 1.84 (s, 3H, CH=CCH3), 2.17–2.21 (m, 2H, CH2CH2), 2.19 (s, 3H, 2-CH3 of naphthoquinone), 2.45–2.48 (m, 2H, CH2CH2), 3.38 (d, 2H, J = 6.4 Hz, CH2CH=CH2(CH3)), 5.09 (t, 1H, J = 7.0 Hz, CH=CCH3), 6.42 (t, 1H, J = 7.4 Hz, CH=CCH3), 7.69–7.71 (m, 2H, Ph of naphthoquinone), 8.06–8.07 (m, 2H, Ph of naphthoquinone), 9.33 (s, 1H, CHO); 13C NMR (CDCl3, 100 MHz): δ 9.3, 12.8, 16.4, 26.1, 27.5, 38.1, 120.6, 126.3, 126.3, 132.1, 133.5, 133.5, 136.1, 139.5, 139.7, 143.5, 145.8, 153.9, 184.5, 185.3, 195.2. HRMS ([M + H]+) m/z: calcd for C21H23O3, 323.1642; found, 323.1642.
2-((2E,6E)-3,7-Dimethyl-8-(thiazol-2-ylamino)octa-2,6-dien-1-yl)-3-methylnaphthalene-1,4-dione (18)
A mixture of compound 44 (100 mg, 0.31 mmol), 2-aminothiazole (34 mg, 0.34 mmol), acetic acid (22 mg, 0.37 mmol), and MS4 Å in CH2Cl2 (3 mL) was stirred at room temperature for 0.5 h under an Ar atmosphere. The reaction was cooled to 0 °C, sodium triacetoxyborohydride (132 mg, 0.62 mmol) was added, and the reaction was stirred at room temperature for 6 h under an Ar atmosphere. After completion of the reaction, the mixture was concentrated in vacuo, and the residue was dissolved in ethyl acetate and washed with brine. After concentrated in vacuo, the residue was purified by silica gel chromatography (n-hexane/ethyl acetate = 10:1) to afford compound 18, a yellow liquid, in 5.6 mg (0.01 mmol) yield (4%). 1H NMR (CDCl3, 400 MHz): δ 1.64 (s, 3H, CH=CCH3), 1.79 (s, 3H, CH=CCH3), 2.02 (q, 2H, J = 7.6 Hz, CH2CH2), 2.12 (q, 2H, J = 7.4 Hz, CH2CH2), 2.19 (s, 3H, 2-CH3 of naphthoquinone), 3.36 (d, 2H, J = 6.8 Hz, CH2CH=CH2(CH3)), 3.74 (d, 2H, J = 3.6 Hz, CH=C(CH3)CH2), 5.01 (t, 1H, J = 6.4 Hz, CH=CCH3), 5.37 (t, 1H, J = 6.4 Hz, CH=CCH3), 6.46 (d, 1H, J = 3.6 Hz, S–CH=CH–N), 7.09 (d, 1H, J = 3.6 Hz, S–CH=CH–N), 7.68–7.70 (m, 2H, Ph of naphthoquinone), 8.08–8.09 (m, 2H, Ph of naphthoquinone); 13C NMR (CDCl3, 100 MHz): δ 12.7, 14.5, 16.4, 26.1, 26.2, 39.1, 53.8, 119.6, 126.2, 126.3, 127.4, 130.8, 132.1, 133.3, 133.4, 137.0, 143.4, 146.1, 165.9, 184.6. HRMS ([M + H]+) m/z: calcd for C24H27N2O2S, 407.1788; found, 407.1788.
2-((2E,6E)-8-(((3s,5s,7s)-Adamantan-1-yl)amino)-3,7-dimethylocta-2,6-dien-1-yl)-3-methylnaphthalene-1,4-dione (19)
Compound 19 was prepared by a similar procedure to that for 18 but with 1-adamantanamine instead of 2-aminothiazole as a reactant. Yield: 4%; yellow oil; 1H NMR (CDCl3, 400 MHz): δ 1.65–1.70 (m, 7H, adamatan), 1.98–2.18 (m, 5H, adamatane), 2.00–2.04 (m, 2H, CH2CH2), 1.98 (s, 3H, CH=CCH3), 2.07 (q, 2H, J = 6.8 Hz, CH2CH2), 2.12 (s, 3H, CH=CCH3), 2.18 (s, 3H, 2-CH3 of naphthoquinone), 3.35 (d, 2H, J = 6.8 Hz, CH2CH=CH2(CH3)), 3.38 (s, 2H, CH=C(CH3)CH2), 4.99 (t, 1H, J = 7.2 Hz, CH=CCH3), 5.55 (t, 1H, J = 7.4 Hz, CH=CCH3), 7.69–7.70 (m, 2H, Ph of naphthoquinone), 8.06–8.08 (m, 2H, Ph of naphthoquinone); 13C NMR (CDCl3, 100 MHz): δ 12.7, 15.1, 16.4, 26.1, 26.5, 29.1, 35.5, 38.4, 38.5, 48.2, 119.6, 126.2, 126.3, 132.1, 133.4, 136.7, 143.4, 146.0, 184.6, 185.4. HRMS ([M + H]+) m/z: calcd for C31H40NO2, 458.3054; found, 458.3056.
RdRp Inhibition Assay
SARS-CoV-2 RNA polymerase (RdRp) assay kit plus-20: Catalog no. S2RPA020 KE [ProFoldin Co., Ltd.] was purchased and used for the experiment. In accordance with an experiment described by ProFoldin, we evaluated RdRp inhibitory activity by choosing EDTA as a positive control, which is different from the remdesivir analogue. Fluorescence intensity was measured by excitation at a wavelength of 485 nm and detection of emission at a wavelength of 535 nm using a fluorescence plate reader, INFINITE 200 PRO [TECAN, Inc.]. For specific experimental methods, see the Supporting Information.
3CL Protease Inhibition Assay
3CL Protease, Untagged (SARS-CoV-2) Assay Kit: catalogue number 78042-1 [BPS Bioscience] was purchased and used in the experiments; the 3CL protease substrate was an internally quenched 14-mer fluorescent (FRET) peptide (DABCYL-KTSAVLQSGFRKME-EDANS). When the donor (EDANS) and acceptor (DABCYL) fluorophores are in close proximity, the energy released from EDANS is quenched by DABCYL (intact substrate); proteolysis by 3CL results in the peptide substrate being cleaved to produce highly fluorescent peptide fragments (SGFRKME-EDANS). The fluorescence intensity increases in proportion to the activity of 3CL. We evaluated the inhibitory activity of 3CL-protease by choosing GC376, a known inhibitor. Fluorescence intensity was measured by excitation at a wavelength of 360 nm and detection of emission at a wavelength of 460 nm using a fluorescence plate reader, INFINITE 200 PRO [TECAN, Inc.]. For specific experimental methods, see the Supporting Information.
Docking Simulation
The structures of RdRp (PDB code: 7BV2): the nsp12 nsp7 nsp8 complex bound to the template–primer RNA and triphosphate form of RTP were obtained from the Protein Data Bank (PDB). The cocrystallized structure of RdRp was prepared using Molecular Operating Environment (MOE; 2022.02 Chemical Computing Group) to correct structural problems (such as broken bonds or missing loops), add hydrogens, and calculate partial charges. First, alpha PMI, which is fast and best suited to a tight pocket placement method, was employed to place different conformations of the ligand or test reagents into the binding pocket of RdRp, setting the maximum number of placement poses to 1000. The ligand and test reagents were docked in the regions around the most likely ligand binding sites.27 The best 100 conformations were then selected using the London ΔG scoring. The ligand and test reagent structures were then further refined using the induced fit option. This refinement step is an energy minimization using the conventional Amber10 with extended Hückel theory (EHT) molecular mechanics force field applied to account for electronic effects into account. The generalized Born solvation model was used to choose 20 poses at the end of the refinement step to estimate the final energy.
Acknowledgments
We thank the National Institutes of Biomedical Innovation, Health and Nutrition and National Institute of Infectious Diseases for kindly providing VeroE6/TMPRSS2 cells and SARS-CoV-2 (WK-521 strain), respectively.
Glossary
Abbreviations
- COVID-19
coronavirus disease 2019
- MK-2
menaquinone-2
- MD
menadione
- PK
phylloquinone
- RdRp
RNA-dependent RNA polymerase
- IFN
interferon
- EC50
50% effective concentration
- CC50
50% cytotoxic concentration
- 3CL
3-chymotrypsin like
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c04175.
Author Contributions
T.H. and M.O. equally contributed to this work. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Barber R. M.; Sorensen R. J. D.; Pigott D. M.; Bisignano C.; Carter A.; Amlag J. O.; Collins J. K.; Abbafati C.; Adolph C.; Allorant A.; et al. Estimating global, regional, and national daily and cumulative infections with SARS-CoV-2 through Nov 14, 2021: a statistical analysis. Lancet 2022, 399, 2351–2380. 10.1016/s0140-6736(22)00484-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A. D.; Lavelle M.; Boursiquot B. C.; Wan E. Y. Long-term complications of COVID-19. Am. J. Physiol.: Cell Physiol. 2022, 322, C1–C11. 10.1152/ajpcell.00375.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferner R. E.; Aronson J. K. Remdesivir in covid-19. BMJ 2020, 369, m1610. 10.1136/bmj.m1610. [DOI] [PubMed] [Google Scholar]
- López-Medina E.; López P.; Hurtado I. C.; Dávalos D. M.; Ramirez O.; Martínez E.; Díazgranados J. A.; Oñate J. M.; Chavarriaga H.; Herrera S.; Parra B.; Libreros G.; Jaramillo R.; Avendaño A. C.; Toro D. F.; Torres M.; Lesmes M. C.; Rios C. A.; Caicedo I. Effect of Ivermectin on Time to Resolution of Symptoms Among Adults with Mild COVID-19: A Randomized Clinical Trial. JAMA, J. Am. Med. Assoc. 2021, 325, 1426–1435. 10.1001/jama.2021.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musarrat F.; Chouljenko V.; Dahal A.; Nabi R.; Chouljenko T.; Jois S. D.; Kousoulas K. G. The anti-HIV drug nelfinavir mesylate (Viracept) is a potent inhibitor of cell fusion caused by the SARSCoV-2 spike (S) glycoprotein warranting further evaluation as an antiviral against COVID-19 infections. J. Med. Virol. 2020, 92, 2087–2095. 10.1002/jmv.25985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb Y. N. Remdesivir: First Approval. Drugs 2020, 80, 1355–1363. 10.1007/s40265-020-01378-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdin S. M.; Elgendy S. M.; Alyammahi S. K.; Alhamad D. W.; Omar H. A. Tackling the cytokine storm in COVID-19, challenges and hopes. Life Sci. 2020, 257, 118054. 10.1016/j.lfs.2020.118054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond J.; Leister-Tebbe H.; Gardner A.; Abreu P.; Bao W.; Wisemandle W.; Baniecki M.; Hendrick V. M.; Damle B.; Simón-Campos A.; Pypstra R.; Rusnak J. M. Oral Nirmatrelvir for High-Risk, Nonhospitalized Adults with Covid-19. Engl. J. Med. 2022, 386, 1397–1408. 10.1056/NEJMoa2118542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellinge J. W.; Dalgaard F.; Murray K.; Connolly E.; Blekkenhorst L. C.; Bondonno C. P.; Lewis J. R.; Sim M.; Croft K. D.; Gislason G.; Torp-Pedersen C.; Tjønneland A.; Overvad K.; Hodgson J. M.; Schultz C.; Bondonno N. P. Vitamin K Intake and Atherosclerotic Cardiovascular Disease in the Danish Diet Cancer and Health Study. J. Am. Heart Assoc. 2021, 10, e020551 10.1161/jaha.120.020551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder M.; Petsophonsakul P.; Akbulut A. C.; Pavlic A.; Bohan F.; Anderson E.; Maresz K.; Kramann R.; Schurgers L. Vitamin K: Double Bonds beyond Coagulation Insights into Differences between Vitamin K1 and K2 in Health and Disease. Int. J. Mol. Sci. 2019, 20, 896. 10.3390/ijms20040896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther B.; Karl J. P.; Booth S. L.; Boyaval P. Menaquinones, bacteria, and the food supply: the relevance of dairy and fermented food products to vitamin K requirements. Adv. Nutr. 2013, 4, 463–473. 10.3945/an.113.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan S. M.; E Ball M. E.; McDonald E.; Hull G. L. J.; Danaher M.; Cashman K. D. Biofortification of Chicken Eggs with Vitamin K-Nutritional and Quality Improvements. Foods 2020, 9, 1619. 10.3390/foods9111619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R.; Hu Q.; Wang H.; Zhu G.; Wang M.; Zhang Q.; Zhao Y.; Li C.; Zhang Y.; Ge G.; Chen H.; Chen L. Identification of Vitamin K3 and its analogues as covalent inhibitors of SARS-CoV-2 3CLpro. Int. J. Biol. Macromol. 2021, 183, 182–192. 10.1016/j.ijbiomac.2021.04.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhara Y.; Watanabe M.; Nakagawa K.; Wada A.; Ito Y.; Takeda K.; Takahashi K.; Okano T. Synthesis of novel vitamin K2 analogues with modification at the ω-terminal position and their biological evaluation as potent steroid and xenobiotic receptor (SXR) agonists. J. Med. Chem. 2011, 54, 4269–4273. 10.1021/jm200025f. [DOI] [PubMed] [Google Scholar]
- Suhara Y.; Watanabe M.; Motoyoshi S.; Nakagawa K.; Wada A.; Takeda K.; Takahashi K.; Tokiwa H.; Okano T. Synthesis of new vitamin K analogues as steroid and xenobiotic receptor (SXR) agonists: insights into the biological role of the side chain part of vitamin K. J. Med. Chem. 2011, 54, 4918–4922. 10.1021/jm200201k. [DOI] [PubMed] [Google Scholar]
- Nishioka T.; Endo-Umeda K.; Ito Y.; Shimoda A.; Takeuchi A.; Tode C.; Hirota Y.; Osakabe N.; Makishima M.; Suhara Y. Synthesis and In Vitro Evaluation of Novel Liver X Receptor Agonists Based on Naphthoquinone Derivatives. Molecules 2019, 24, 4316. 10.3390/molecules24234316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhara Y.; Hirota Y.; Hanada N.; Nishina S.; Eguchi S.; Sakane R.; Nakagawa K.; Wada A.; Takahashi K.; Tokiwa H.; Okano T. Synthetic Small Molecules Derived from Natural Vitamin K Homologues that Induce Selective Neuronal Differentiation of Neuronal Progenitor Cells. J. Med. Chem. 2015, 58, 7088–7092. 10.1021/acs.jmedchem.5b00999. [DOI] [PubMed] [Google Scholar]
- Kimura K.; Hirota Y.; Kuwahara S.; Takeuchi A.; Tode C.; Wada A.; Osakabe N.; Suhara Y. Synthesis of Novel Synthetic Vitamin K Analogues Prepared by Introduction of a Heteroatom and a Phenyl Group That Induce Highly Selective Neuronal Differentiation of Neuronal Progenitor Cells. J. Med. Chem. 2017, 60, 2591–2596. 10.1021/acs.jmedchem.6b01717. [DOI] [PubMed] [Google Scholar]
- Sakane R.; Kimura K.; Hirota Y.; Ishizawa M.; Takagi Y.; Wada A.; Kuwahara S.; Makishima M.; Suhara Y. Synthesis of novel vitamin K derivatives with alkylated phenyl groups introduced at the ω-terminal side chain and evaluation of their neural differentiation activities. Bioorg. Med. Chem. Lett. 2017, 27, 4881–4884. 10.1016/j.bmcl.2017.09.038. [DOI] [PubMed] [Google Scholar]
- Yoshimura H.; Hirota Y.; Soda S.; Okazeri M.; Takagi Y.; Takeuchi A.; Tode C.; Kamao M.; Osakabe N.; Suhara Y. Study on structure-activity relationship of vitamin K derivatives: Conversion of the naphthoquinone part into another aromatic ring and evaluation of their neuronal differentiation-inducing activity. Bioorg. Med. Chem. Lett. 2020, 30, 127059. 10.1016/j.bmcl.2020.127059. [DOI] [PubMed] [Google Scholar]
- Chen X.; Liu Y.; Furukawa N.; Jin D. Y.; Paul Savage G.; Stafford D. W.; Suhara Y.; Williams C. M.; Tie J. K. A novel vitamin K derived anticoagulant tolerant to genetic variations of vitamin K epoxide reductase. J. Thromb. Haemostasis 2021, 19, 689–700. 10.1111/jth.15209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M.; Toyama M.; Baba M. The chemokine receptor antagonist cenicriviroc inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020, 182, 104902. 10.1016/j.antiviral.2020.104902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S.; Nao N.; Shirato K.; Kawase M.; Saito S.; Takayama I.; Nagata N.; Sekizuka T.; Katoh H.; Kato F.; Sakata M.; Tahara M.; Kutsuna S.; Ohmagari N.; Kuroda M.; Suzuki T.; Kageyama T.; Takeda M. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. U.S.A. 2020, 117, 7001–7003. 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda T.; Nobori H.; Fukao K.; Baba K.; Matsumoto K.; Yoshida S.; Tanaka Y.; Watari R.; Oka R.; Kasai Y.; Inoue K.; Kawashima S.; Shimba A.; Hayasaki-Kajiwara Y.; Tanimura M.; Zhang Q.; Tachibana Y.; Kato T.; Shishido T. Efficacy comparison of 3CL protease inhibitors ensitrelvir and nirmatrelvir against SARS-CoV-2 in vitro and in vivo. J. Antimicrob. Chemother. 2023, 78, 946–952. 10.1093/jac/dkad027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J.; Guo S.; Yi D.; Li Q.; Ma L.; Zhang Y.; Wang J.; Li X.; Guo F.; Lin R.; Liang C.; Liu Z.; Cen S. A cell-based assay to discover inhibitors of SARS-CoV-2 RNA dependent RNA polymerase. Antiviral Res. 2021, 190, 105078. 10.1016/j.antiviral.2021.105078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q.; Xiong Y.; Zhu G. H.; Zhang Y. N.; Zhang Y. W.; Huang P.; Ge G. B. The SARS-CoV-2 main protease (Mpro): Structure, function, and emerging therapies for COVID-19. MedComm. 2022, 3, e151 10.1002/mco2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W.; Mao C.; Luan X.; Shen D. D.; Shen Q.; Su H.; Wang X.; Zhou F.; Zhao W.; Gao M.; Chang S.; Xie Y. C.; Tian G.; Jiang H. W.; Tao S. C.; Shen J.; Jiang Y.; Jiang H.; Xu Y.; Zhang S.; Zhang Y.; et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 2020, 368, 1499–1504. 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.