As in all organisms, metal cations are crucial for nutrition in plants. Several metals, such as copper, iron, zinc, and manganese, act as important cofactors for many enzymes and are essential for both mitochondrial and chloroplast functions. However, when supplied in excess, these essential cations can become toxic, like heavy metals with no generally established function, such as cadmium, lead, or mercury. To maintain micronutrient metal homeostasis and to cope with the deleterious effects of nonessential heavy metals, plants have developed a complex network of metal uptake, chelation, trafficking, and storage processes. Metal transporters are required to maintain metal homeostasis and thus constitute important components of this network (Clemens, 2001; Hall and Williams, 2003).
In recent years, a number of membrane transport protein families have been implicated in metal homeostasis in plants. These include the cation diffusion facilitators (CDF), the Zrt-, Irt-like proteins (ZIP), the cation exchangers (CAX), the copper transporters (COPT), the heavy-metal P-type ATPases (HMA), the natural resistance-associated macrophage proteins (NRAMP), and the ATP-binding cassette (ABC) transporters (Williams et al., 2000; Maser et al., 2001; Cobbett et al., 2003; Hall and Williams, 2003). These transporters are encoded by multigene families. For example, 15 ZIP genes, 12 metal tolerance protein (MTP) genes, and 8 HMA genes are present in the Arabidopsis (Arabidopsis thaliana) genome (Cobbett et al., 2003; Delhaize et al., 2003). However, the transport specificities, patterns of expression, or subcellular localizations of metal transport proteins are still largely unknown. To further understand plant metal homeostasis, it will be necessary to elucidate the contribution of each of these transporters to the uptake, trafficking, and storage of essential metals, as well as to the detoxification of toxic heavy metals.
In this article, we present an overview of our current knowledge of the metal transport function in metal homeostasis and tolerance in eukaryotes, with a special emphasis on plants. We also provide a timely inventory of putative metal transporters in two unicellular algal models, the green alga Chlamydomonas reinhardtii and the red alga Cyanidioschizon merolae. These new data should facilitate functional genomics and molecular analysis of metal homeostasis and tolerance in photosynthetic organisms. Moreover, the comparison of metal transporters from species belonging to the red and green algae with those of the land plant Arabidopsis, as well as with their human and yeast homologs, allows some light to be shed on the molecular evolution of metal homeostasis and tolerance systems.
TWO UNICELLULAR ALGAL MODEL SYSTEMS
The use of a simple model organism is often helpful to dissect the functions and the interactions of various transport systems. The yeasts Saccharomyces cerevisiae and, to a lesser extent, Schizosaccharomyces pombe have been successfully used to elucidate the molecular basis of cellular metal homeostasis (Clemens and Simm, 2003; De Freitas et al., 2003; Eide, 2003). However, investigation of the essential chloroplast metal metabolism is not accessible in yeast models.
The green alga Chlamydomonas reinhardtii Dangeard is a well-known model of a photosynthetic cell. This unicellular eukaryote is widely used for studies of a number of physiological processes, such as photosynthesis, respiration, nitrogen assimilation, flagella motility, and basal body function (Rochaix et al., 1998; Harris, 2001; Silflow and Lefebvre, 2001). Its short life cycle is easily controlled, making this organism a powerful tool for genetic analysis. The past few years have seen a dramatic development of the molecular technologies applicable to C. reinhardtii (Lefebvre and Silflow, 1999; Grossman, 2000; Fuhrmann, 2002). In the near future, the completion of the Chlamydomonas genome project, which includes molecular mapping, whole-genome sequencing, and extended expressed sequence tag (EST) generation programs (Grossman et al., 2003; Kathir et al., 2003; Shrager et al., 2003), will make C. reinhardtii an even more accessible model.
C. reinhardtii has also been used in the past to study metal tolerance (for review, see Hanikenne, 2003) and homeostasis, and especially iron and copper metabolism in the chloroplast (e.g. Merchant, 1998; Moseley et al., 2002a, 2002b). In a recent review, we recommended the use of C. reinhardtii as a photosynthetic eukaryotic model to study metal homeostasis and tolerance (Hanikenne, 2003).
The red alga Cyanidioschyzon merolae De Luca, Taddei, and Varano is a blue-green-colored unicellular alga that is found in sulfur-rich acidic hot springs. Its ultrastructure and other cytological and biochemical features suggest that it is one of the most primitive algae. C. merolae has no rigid cell wall and contains one nucleus, one mitochondrion, one chloroplast with a centrally located nucleoid, one Golgi body, and one microbody (Seckbach, 1994; Seckbach and Ott, 1994; Matsuzaki et al., 2004). Belonging to the extremophilic Cyanidiales, it shares the same photosynthetic pigments and ecological niche as Cyanidium caldarium and Galdieria sulphuraria, but differs by being smaller, structurally simpler, and dividing only by binary fission (no formation of endospores; Ott and Seckbach, 1994; Gross, 2000; Ciniglia et al., 2004). C. merolae was extensively used in studies of mitochondrial and chloroplast divisions, which can be highly synchronized by light/dark cycles (Kuroiwa, 1998). Its nuclear genome is one of the smallest known in photosynthetic eukaryotes. It has just been completely characterized (Matsuzaki et al., 2004), with the mitochondrial (Ohta et al., 1998) and plastid genomes (Ohta et al., 2003) also being available.
According to many phylogenetic studies, red algae and green plants (including green algae) are considered to be sister groups (Cavalier-Smith, 1982; Bhattacharya and Medlin, 1995; Moreira et al., 2000). In particular, red and green plastids are thought to be derived from the same primary endosymbiotic event, the former retaining more cyanobacterial features than the latter (Nozaki et al., 2003; Ohta et al., 2003). Along with fossil evidence (Butterfield, 2000), this supports the idea that red algae are among the most primitive eukaryotes still living today. Red algae are also worth being studied because their plastid seems to be at the origin of the plastids of various algal groups of fundamental importance in marine primary production (diatoms, dinophytes, and brown algae) through a process of secondary endosymbiosis (Moreira and Philippe, 2001; Yoon et al., 2002).
A MINING STRATEGY SUITED FOR MULTIGENE FAMILIES
We used a high-throughput semiautomated approach to identify metal transporters in the recently released genomic sequences of C. reinhardtii (U.S. Department of Energy Joint Genome Institute [JGI; http://www.jgi.doe.gov]) and C. merolae (Matsuzaki et al., 2004). Briefly, numerous BLAST searches were conducted on both algal genomes, using all members of the human, yeast, and Arabidopsis CDF, ZIP, CAX, COPT, HMA, yellow-stripe 1-like (YSL), Fe transporter (FTR), NRAMP, and iron-regulated 1 (IREG1)-like (sub)families, as well as multidrug resistance-associated protein (MRP) and ABC transporter of the mitochondria (ATM)/heavy-metal tolerance (HMT) subfamilies of ABC transporters (see Supplemental Table I). This custom strategy allowed us to identify, respectively, 41 and 25 putative metal transporters in C. reinhardtii and C. merolae. Their distribution among the different transporter families, compared to the three reference organisms, is given in Table I. The identified proteins were first subjected to topology and targeting predictions, then used to mine available EST databases (Tables II and III; Supplemental Tables II–VII). Finally, evolutionary relationships within each transporter family were examined using robust phylogenetic trees (Figs. 1–7). A detailed description of the whole procedure is given in Supplemental Figure 1.
Table I.
Summary of metal transporter family/subfamily sizes in humans, S. cerevisiae, Arabidopsis, C. reinhardtii, and C. merolae
–, Not found.
| Organisms
|
Protein Families/Subfamilies
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CDF | ZIP | CAX | COPT | P-Type ATPases
|
ABC Transporters
|
YSL | FTR | NRAMP | IREG1 | ||
| HMA | MRP | ATM/HMT | |||||||||
| H. sapiens | 9 | 14 | – | 2 | 2 | 12 | 2 | – | – | 2 | 1 |
| S. cerevisiae | 5 | 5 | 1 | 3 | 2 | 6 | 1 | 1 | 1 | 3 | – |
| Arabidopsis | 12 | 17 | 12 | 5 | 8 | 15 | 3 | 9 | – | 6 | 3 |
| C. reinhardtii | 5 | 14 | 4 | 1 | 3 | 7 | 3 | – | 1 | 3 | – |
| C. merolae | 3 | 4 | 2 | 1 | 2 | 2 | 3 | – | 4 | 3 | 1 |
Table II.
Overview of the identified metal transporters in C. reinhardtii
Columns 1 to 6 contain protein name, JGI identification number (transcript), scaffold number (version 2 of the C. reinhardtii genome), protein model length, predicted subcellular localization, and number of EST clones found. Subcellular localization is indicated when both prediction programs (iPSORT and TargetP) agree. Organelle localization corresponds to either chloroplast or mitochondria, and secretory pathway corresponds to ER, Golgi, or plasma membrane localization. For more details, see Supplemental Tables II, IV, and VI. CrMRP1 (AAL35383), CrCds1 (AAQ19847), and CrFTR1 (AAM45938) have been named previously (Im and Grossman, 2002; La Fontaine et al., 2002; Hanikenne et al., 2005). –, Not found.
| Protein Name | JGI ID | Scaffold | Length (Amino Acids) | Predicted Subcellular Localization | No. of EST Clones |
|---|---|---|---|---|---|
| CDF Family | |||||
| CrMTP1 | 168519 | 67 | 640 | Vacuole | 3 |
| CrMTP2 | 153887 | 11 | 568 | Vacuole | 4 |
| CrMTP3 | 153892 | 11 | 318 | Organelle | – |
| CrMTP4 | 154037 | 11 | 295 | Vacuole | – |
| CrMTP5 | 162326 | 29 | 523 | – | 1 |
| ZIP Family | |||||
| CrZIP1 | 153077 | 10 | 300 | Organelle | – |
| CrZIP2 | 157106 | 167 | 745 | Vacuole | 1 |
| CrZIP3 | 160009 | 21 | 310 | Secretory pathway | – |
| CrZIP4 | 171817 | 98 | 334 | – | – |
| CrZIP5 | 171818 | 98 | 428 | Vacuole | 4 |
| CrZIP6 | 158835 | 19 | 413 | Vacuole | 3 |
| CrZIP7 | 167842 | 60 | 715 | – | 1 |
| CrZIP8 | 164608 | 3 | 294 | – | 2 |
| CrZIP9 | 153825 | 119 | 385 | Secretory pathway | 5 |
| CrZIP10 | 164248 | 38 | 289 | Vacuole | 1 |
| CrZIP11 | 164249 | 38 | 388 | Secretory pathway | 1 |
| CrZIP12 | 171130 | 8 | 480 | Secretory pathway | – |
| CrZIP13 putative | 159981 | 21 | 857 | Organelle | 2 |
| CrZIP14 putative | 164406 | 39 | 508 | Organelle | – |
| CAX Family | |||||
| CrCAX1 | 157233 | 16 | 447 | Vacuole | 11 |
| CrCAX2 | 166617 | 52 | 262 | Organelle | 3 |
| CrCAX3 | 158919 | 1 | 478 | Vacuole | – |
| CrCAX4 | 163085 | 31 | 709 | – | – |
| COPT Family | |||||
| CrCOPT1 | 163944 | 36 | 155 | – | – |
| P-Type ATPases | |||||
| HMA subfamily | |||||
| CrHMA1 | 159065 | 1 | 1,189 | Organelle | 1 |
| CrHMA2 | 168288 | 65 | 1,137 | – | 1 |
| CrHMA3 | 161566 | 26 | 937 | Vacuole | – |
| ABC Transporter Family | |||||
| MRP subfamily | |||||
| CrMRP1 | – | 61 | 1,082 | Secretory pathway | 21 |
| CrMRP2 | 153344 | 111 | 1,441 | Vacuole | 16 |
| CrMRP3 | 166481 | 51 | 1,519 | – | 2 |
| CrMRP4 | 165660 | 46 | 1,480 | Organelle | – |
| CrMRP5 | 162457 | 2 | 1,614 | – | – |
| CrMRP6 | – | 142 | 1,524 | – | 2 |
| CrMRP7 | 160938 | 24 | 875 | Vacuole | – |
| ATM/HMT subfamily | |||||
| CrCds1 | – | 122 | 1,062 | – | 7 |
| CrATM/HMT-2 | 158975 | 1 | 666 | Organelle | – |
| CrATM/HMT-3 | 156620 | 15 | 1,170 | – | – |
| FTR Family | |||||
| CrFTR1 | 164771 | 3 | 541 | Secretory pathway | 9 |
| NRAMP Family | |||||
| CrNRAMP1 | 157858 | 17 | 513 | Vacuole | 13 |
| CrNRAMP2 | 167153 | 57 | 287 | – | – |
| CrNRAMP3 | 165437 | 44 | 820 | Secretory pathway | – |
Table III.
Overview of the identified metal transporters in C. merolae
Columns 1 to 6 contain protein name, protein identification number, chromosome number, protein model length, predicted subcellular localization, and number of EST clones found. Subcellular localization is indicated when both prediction programs (iPSORT and TargetP) agree. Organelle localization corresponds to either chloroplast or mitochondria, and secretory pathway corresponds to ER, Golgi, or plasma membrane localization. For more details, see Supplemental Tables III, V, and VII. –, Not found.
| Protein Name | Protein ID | Chromosome | Length (Amino Acids) | Predicted Subcellular Localization | No. of EST Clones |
|---|---|---|---|---|---|
| CDF Family | |||||
| CmMTP1 | CMF058C | VI | 434 | Vacuole | 1 |
| CmMTP2 | CMC075C | III | 420 | – | 2 |
| CmMTP3 | CMT536C | XX | 827 | Secretory pathway | 1 |
| ZIP Family | |||||
| CmZIP1 | CMS155C | XIX | 395 | – | 7 |
| CmZIP2 | CMG102C | VII | 429 | Vacuole | – |
| CmZIP3/GUFA | CMQ444C | XVII | 305 | Secretory pathway | 5 |
| CmZIP4/LIV1 | CMP282C | XVI | 325 | Secretory pathway | 8 |
| CAX Family | |||||
| CmCAX1 | CMQ284C | XVII | 558 | Vacuole | 3 |
| CmCAX2 | CMR378C | XVIII | 335 | Organelle | 2 |
| COPT Family | |||||
| CmCOPT1 | CMS307C | XIX | 430 | Vacuole | 7 |
| P-Type ATPases | |||||
| HMA subfamily | |||||
| CmHMA1 | CMS330C | XIX | 896 | Organelle | – |
| CmHMA2 | CMP215C | XVI | 1,425 | Organelle | 9 |
| ABC Transporter Family | |||||
| MRP subfamily | |||||
| CmMRP1 | CMN251C | XIV | 1,796 | Vacuole | 2 |
| CmMRP2 | CMD133C | IV | 2,055 | Vacuole | 2 |
| ATM/HMT subfamily | |||||
| CmATM/HMT-1 | CMN105C | XIV | 749 | Organelle | 1 |
| CmATM/HMT-2 | CMQ176C | XVII | 787 | Organelle | 3 |
| CmATM/HMT-3 | CMT066C | XX | 710 | – | 7 |
| FTR Family | |||||
| CmFTR1 | CMJ315C | X | 526 | Secretory pathway | 40 |
| CmFTR2 | CML004C | XII | 526 | Secretory pathway | 67 |
| CmFTR3 | CMQ004C | XVII | 431 | Organelle | 14 |
| CmFTR4 | CMN003C | XIV | 388 | Organelle | 1 |
| NRAMP Family | |||||
| CmNRAMP1 | CMJ138C | X | 718 | Vacuole | 11 |
| CmNRAMP2 | CML262C | XII | 594 | Vacuole | 4 |
| CmNRAMP3 | CMT180C | XX | 586 | Vacuole | 6 |
| IREG1 Family | |||||
| CmIREG1 | CMG212C | VII | 616 | Vacuole | 1 |
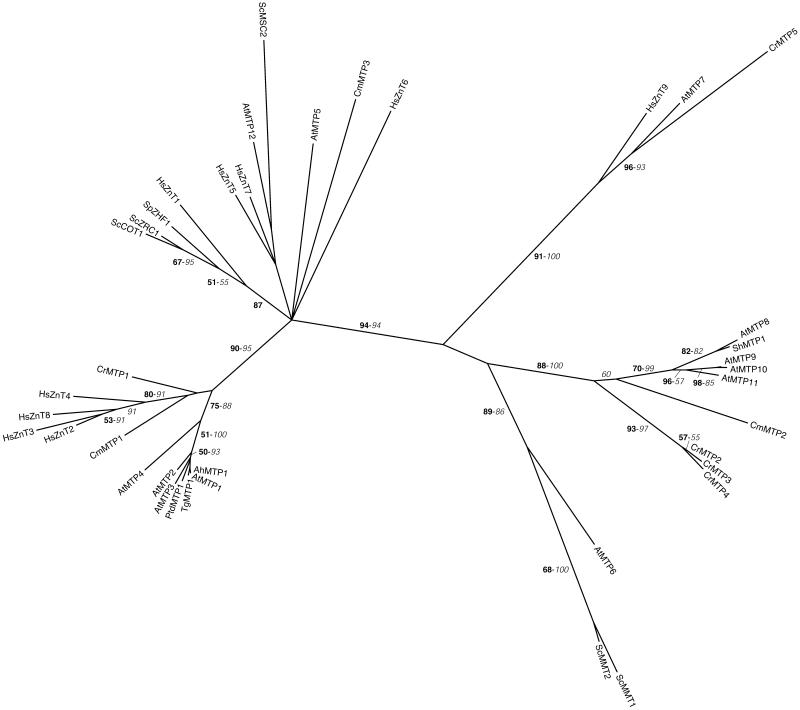
Figure 1.
Phylogenetic tree of the CDF family. The tree is the strict consensus of the five most parsimonious trees (8,358 steps) for which branch lengths were estimated by maximum likelihood (ML) using the VT model of amino acid substitution (log L = −37,883.42). Quartet puzzling support values above 50% from an independent ML reconstruction (10,000 puzzling steps) are printed in bold type and bootstrap percentages above 50% from an independent distance reconstruction (1,000 replicates) are printed in italic type (see Supplemental Fig. 1 for methodological details). Protein accession numbers from organisms other than C. reinhardtii and C. merolae are given in Supplemental Table I, except for AhMTP1 (CAD89013), TgMTP1 (AAS67026), PtdMTP1 (AAR23528), ShMTP1 (AAO38707), and SpZHF1 (NP_593645). Ah, Arabidopsis halleri; Ptd, Populus balsamifera subsp. trichocarpa × Populus deltoids; Sh, Stylosanthes hamata; Sp, Schizosaccharomyces pombe; Tg, Thlaspi goesingense.
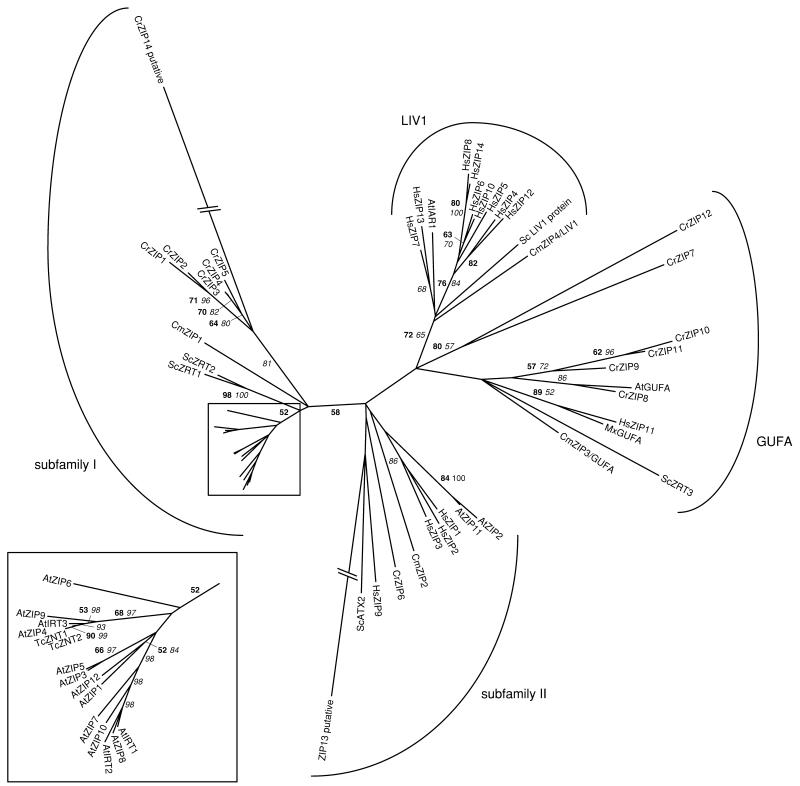
Figure 2.
Phylogenetic tree of the ZIP family. The tree is the most parsimonious tree (10,168 steps) for which branch lengths were estimated by maximum likelihood (ML) using the WAG model of amino acid substitution (log L = −43,192.29). Quartet puzzling support values above 50% from an independent ML reconstruction (25,000 puzzling steps) are printed in bold type and bootstrap percentages above 50% from an independent distance reconstruction (1,000 replicates) are printed in italic type (see Supplemental Fig. 1 for methodological details). CrZIP13 and CrZIP14 positions were ignored in previous steps because they were subject to long-branch attraction artifacts in both the distance and ML reconstructions. The inset is enlarged twice. Protein accession numbers from organisms other than C. reinhardtii and C. merolae are given in Supplemental Table I, except for TcZNT1 (AAF61374), TcZNT2 (AAK69429), and MxGUFA (CAA50380). Mx, Myxococcus xanthus; Tc, Thlaspi caerulescens.
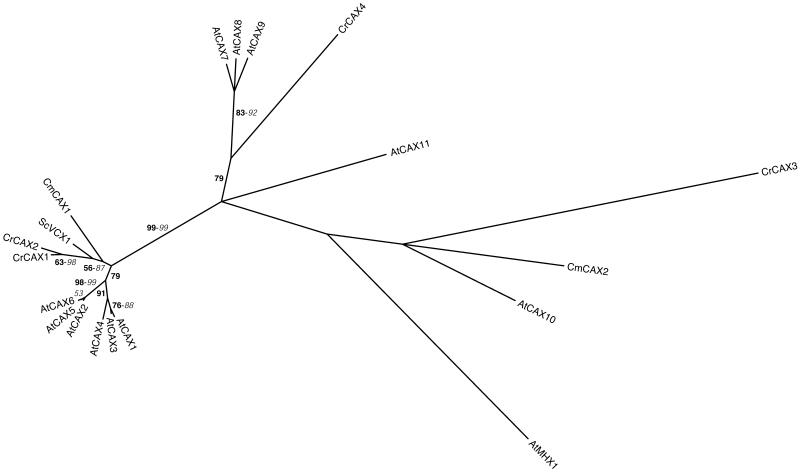
Figure 3.
Phylogenetic tree of the CAX family. The tree is the most parsimonious tree (4,207 steps) for which branch lengths were estimated by maximum likelihood (ML) using the VT model of amino acid substitution (log L = −19,012.92). Quartet puzzling support values above 50% from an independent ML reconstruction (1,000 puzzling steps) are printed in bold type and bootstrap percentages above 50% from an independent distance reconstruction (1,000 replicates) are printed in italic type (see Supplemental Fig. 1 for methodological details). Protein accession numbers from organisms other than C. reinhardtii and C. merolae are given in Supplemental Table I.
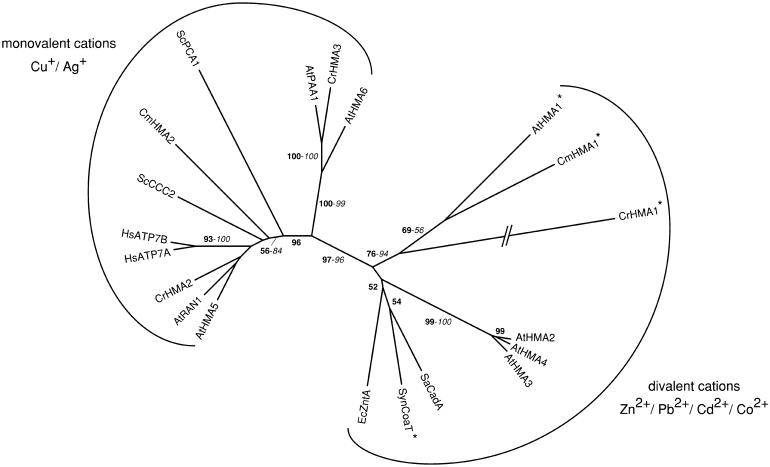
Figure 4.
Phylogenetic tree of the HMA subfamily of P-type ATPases. The tree is the most parsimonious tree (9,500 steps) for which branch lengths were estimated by maximum likelihood (ML) using the WAG model of amino acid substitution (log L = −44,546.19). Quartet puzzling support values above 50% from an independent ML reconstruction (1,000 puzzling steps) are printed in bold type and bootstrap percentages above 50% from an independent distance reconstruction (1,000 replicates) are printed in italic type (see Supplemental Fig. 1 for methodological details). Proteins having the uncharacteristic Ser/Pro/Cys motif in the sixth transmembrane domain are marked by an asterisk. Protein accession numbers from organisms other than C. reinhardtii and C. merolae are given in Supplemental Table I, except for SaCadA (BAC57487), EcZntA (NP_417926), and SynCoaT (S77012). Ec, Escherichia coli; Sa, Staphylococcus aureus; Syn, Synechocystis sp.
Figure 5.
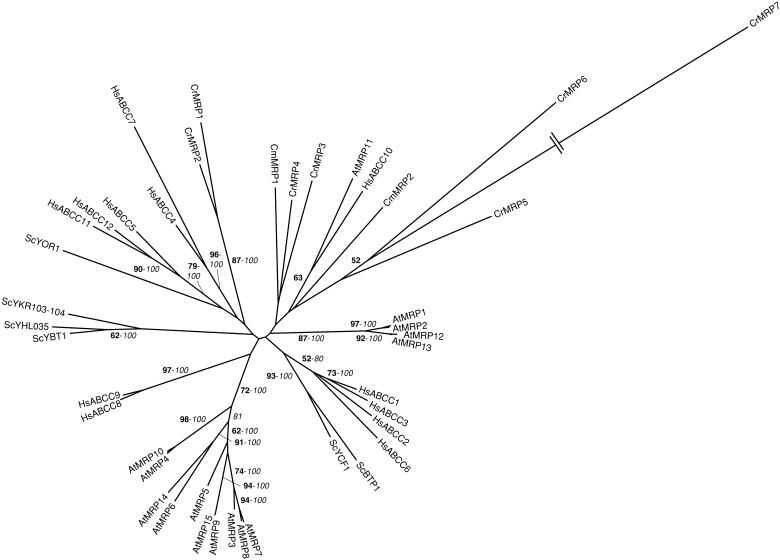
Phylogenetic tree of the MRP subfamily of ABC transporters. The tree is the most parsimonious tree (25,015 steps) for which branch lengths were estimated by maximum likelihood (ML) using the VT model of amino acid substitution (log L = −113,214.98). Quartet puzzling support values above 50% from an independent ML reconstruction (10,000 puzzling steps) are printed in bold type and bootstrap percentages above 50% from an independent distance reconstruction (1,000 replicates) are printed in italic type (see Supplemental Fig. 1 for methodological details). Protein accession numbers from organisms other than C. reinhardtii and C. merolae are given in Supplemental Table I. CrMRP1 (AAL35383) has been named previously by Im and Grossman (2002).
Figure 6.
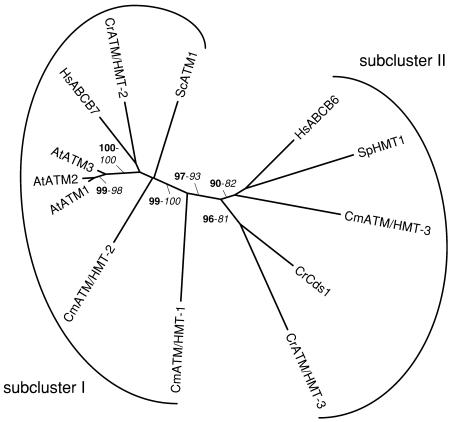
Phylogenetic tree of the ATM/HMT subfamily of ABC transporters. The tree is the most parsimonious tree (3,950 steps) for which branch lengths were estimated by maximum likelihood (ML) using the VT model of amino acid substitution (log L = −20,342.67). Quartet puzzling support values above 50% from an independent ML reconstruction (1,000 puzzling steps) are printed in bold type and bootstrap percentages above 50% from an independent distance reconstruction (1,000 replicates) are printed in italic type (see Supplemental Fig. 1 for methodological details). Protein accession numbers from organisms other than C. reinhardtii and C. merolae are given in Supplemental Table I. CrCds1 (AAQ19847) has been named previously by Hanikenne et al. (2005).
Figure 7.
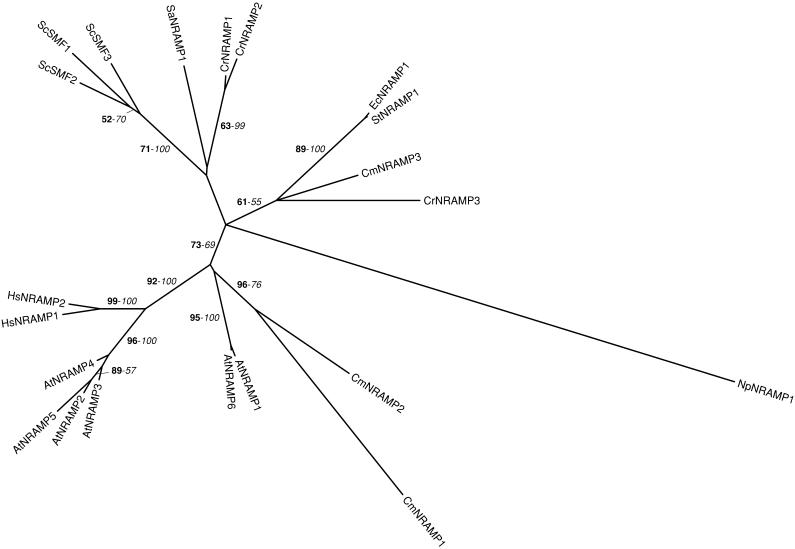
Phylogenetic tree of the NRAMP family. The tree is the strict consensus of the two most parsimonious trees (4,055 steps) for which branch lengths were estimated by maximum likelihood (ML) using the VT model of amino acid substitution (log L = −20,415.07). Quartet puzzling support values above 50% from an independent ML reconstruction (1,000 puzzling steps) are printed in bold type and bootstrap percentages above 50% from an independent distance reconstruction (1,000 replicates) are printed in italic type (see Supplemental Fig. 1 for methodological details). Protein accession numbers from organisms other than C. reinhardtii and C. merolae are given in Supplemental Table I, except for NpNRAMP1 (ZP_00111957), EcNRAMP1 (NP_416893), StNRAMP1 (NP_461349), and SaNRAMP1 (YP_040492). Ec, Escherichia coli; Np, Nostoc punctiforme; Sa, Staphylococcus aureus; St, Salmonella typhymurium.
THE CDF FAMILY
The CDFs form a family of ubiquitous transporters involved in metal homeostasis and tolerance. These proteins catalyze the efflux of transition metal cations, like Zn2+, Cd2+, Co2+, Ni2+, or Mn2+, from the cytoplasm to the outside of the cell or into subcellular compartments. Most CDF proteins possess six putative transmembrane domains, with the N and C termini predicted to be cytoplasmic, and exhibit a His-rich loop region between transmembrane domains IV and V, a signature sequence between transmembrane domains I and II, and a cation efflux domain comprising transmembrane domains I to VI (Paulsen and Saier, 1997; Gaither and Eide, 2001). The plant members of the CDF family are named MTPs.
We have identified, respectively, five and three MTPs in the genome sequences of C. reinhardtii and C. merolae (Tables II and III). CrMTP1 and CmMTP1 are related to zinc transporters of higher plants, humans, and, more distantly, yeasts (Fig. 1). AtMTP1 (formerly ZAT) confers zinc resistance to Arabidopsis when overexpressed (van der Zaal et al., 1999). Homologs of AtMTP1 have been identified in several species, including poplar and the metal hyperaccumulators Arabidopsis halleri and Thlaspi goesingense, and were shown to be involved in zinc homeostasis (Persans et al., 2001; Blaudez et al., 2003; Becher et al., 2004; Drager et al., 2004; Kim et al., 2004). In A. halleri, two AhMTP1 genes, which are particularly highly expressed compared to Arabidopsis, cosegregate with zinc tolerance. The AhMTP1 protein is localized in the vacuolar membrane and likely to mediate the accumulation of zinc in the vacuole (Becher et al., 2004; Drager et al., 2004). In S. cerevisiae, the ScZRC1 and ScCOT1 proteins are located in the vacuolar membrane and export zinc, cobalt, and possibly cadmium from the cytoplasm into the vacuole, playing an essential role in the tolerance to these metals (Gaither and Eide, 2001). In S. pombe, SpZHF1 is an endoplasmic reticulum (ER)-localized protein and is responsible for zinc storage in this compartment. The disruption of the gene results in zinc and cobalt sensitivity but tolerance to cadmium and nickel. SpZHF1 is also required for growth under zinc deficiency (Borrelly et al., 2002; Clemens et al., 2002). Nine MTPs (HsZnT1–9) are found in humans, with HsZnT1 to 7 being responsible for zinc transport in different cell types and organs (for review, see Palmiter and Huang, 2004). Both CrMTP1 and CmMTP1 are predicted to localize to the vacuolar membrane (Tables II and III), similar to their plant and yeast homologs. Interestingly, the three ESTs corresponding to CrMTP1 were only identified in the stress II cDNA library (Supplemental Table IV) prepared from cells exposed to cadmium (Shrager et al., 2003), therefore suggesting that CrMTP1 might be involved cadmium detoxification.
CrMTP2 to 4 and CmMTP2 cluster with AtMTP8 to 11 and with ShMTP1, a subgroup of putative manganese transporters. The ShMTP1 protein of the manganese-tolerant legume Stylosanthes hamata confers manganese resistance to yeast and Arabidopsis when overexpressed. It is localized in the plant vacuolar membrane and was proposed to be involved in manganese sequestration in this organelle (Delhaize et al., 2003). By analogy, AtMTP8 to 11 may have similar substrate specificity, although no experimental data are available. Our phylogenetic analysis suggests that the diversification of this subfamily in C. reinhardtii and Arabidopsis evolved independently through gene duplications. In that respect, it is interesting to note that CrMTP2 to 4 are found on the same genome scaffold very close to each other (Supplemental Table II). Given the important need of manganese to maintain photosynthesis, it is possible that at least one of these proteins is located in the chloroplast, although our prediction analysis only localized these proteins in the vacuole or the mitochondria (Supplemental Tables VI and VII).
CrMTP5 groups with AtMTP7 and HsZnT9, two proteins of unknown function, whereas the remaining CmMTP3 is distantly related to a few other CDF proteins. It is, however, worth mentioning that, similar to ScMSC2, AtMTP12, and HsZnT5 (Fig. 1), CmMTP3 possesses a long N-terminal extension with 6 predicted additional transmembrane domains that share no homology with other CDFs. HsZnT5 is believed to act in the sequestration of zinc in the Golgi complex of different tissues (Palmiter and Huang, 2004), and ScMSC2 is localized in the ER and is essential to maintain proper zinc homeostasis in this compartment (Ellis et al., 2004). Our analysis suggests that CmMTP3 is targeted to the secretory pathway as well (Table III).
Finally, neither C. reinhardtii nor C. merolae seem to possess a homolog of the yeast ScMMT1 and ScMMT2 (Fig. 1), two mitochondrial proteins believed to play a role in iron homeostasis (Li and Kaplan, 1997).
THE ZIP FAMILY
The ZIP protein family forms another ubiquitous transporter family involved in metal homeostasis, generally mediating the influx of metal cations, like zinc, iron, cadmium, or manganese, from outside the cell or from a subcellular compartment into the cytoplasm. ZIPs are predicted to have eight transmembrane domains, with the N and C termini being extracytoplasmic. As a common feature, ZIPs possess a long cytoplasmic loop (the so-called variable region) of variable length and sequence between transmembrane domains III and IV. The variable region very often contains a probable metal-binding His-rich domain. Transmembrane domains IV and V are amphipathic and believed to form a polar cavity required for the cation metal transport, while the loop between transmembrane domains II and III could be the site of initial binding of the substrate (Guerinot and Eide, 1999; Guerinot, 2000; Gaither and Eide, 2001).
Based on sequence conservation, Gaither and Eide (2001) have classified the ZIP proteins into four subfamilies. Subfamily I includes ZIPs from plants and fungi, while subfamily II contains nematode and mammalian proteins. The GUFA subfamily contains proteins of mainly unknown function present in both prokaryotes and eukaryotes, and the LIV1 group is only found in eukaryotes.
We have identified, respectively, 14 and 4 ZIPs in the genomes of C. reinhardtii and C. merolae and, altogether, our findings suggest that these algal ZIPs are mainly localized in the vacuolar or the plasma membrane (Tables II and III).
CrZIP1 to 5 and CmZIP1 cluster with subfamily I, which contains most of the Arabidopsis and S. cerevisae ZIPs (Fig. 2). Interestingly, the proteins of each of the four species form distinct groups within the subfamily. The protein diversification probably occurred independently in the different groups through duplications of an ancestral gene. In that respect, it is noteworthy that CrZIP4 and 5 are found in tandem on the same genome scaffold (Supplemental Table II). The algal proteins seem to branch shortly before the ScZRT1 and ScZRT2 proteins, the yeast high- and low-affinity zinc uptake systems, respectively, located in the plasma membrane (Zhao and Eide, 1996a, 1996b).
A few Arabidopsis subfamily I ZIPs have been characterized through yeast complementation and expression analyses. AtZIP1 to 4 play a role in cellular zinc uptake, AtZIP1, 3, and 4 being induced under zinc-limiting conditions at the transcriptional level (Guerinot, 2000; Gaither and Eide, 2001; Hall and Williams, 2003). In the zinc/cadmium hyperaccumulator Thlaspi caerulescens, TcZNT1 and TcZNT2, two ZIP transporters, are probably involved in zinc hyperaccumulation (Lasat et al., 2000; Pence et al., 2000; Assuncao et al., 2001), while in the zinc hyperaccumulator A. halleri, AhZIP6 and AhZIP9 are constitutively highly expressed compared to Arabidopsis, respectively, in shoots and in roots (Becher et al., 2004; Weber et al., 2004). In Arabidopsis, the AtIRT1 and AtIRT2 proteins are expressed in roots under iron-limiting conditions. In addition to iron, AtIRT1 also transports zinc, manganese, and cadmium, whereas AtIRT2 is only responsible for zinc uptake (Guerinot, 2000; Gaither and Eide, 2001; Hall and Williams, 2003). In Arabidopsis, AtIRT1 represents the major iron uptake system in roots. None of the CrZIPs identified in this article appears to be clearly related to this plant iron uptake system. Our findings are in agreement with the results of Herbik et al. (2002) and La Fontaine et al. (2002), revealing the occurrence in C. reinhardtii of an iron assimilation pathway related to the high-affinity iron uptake pathway of S. cerevisiae (see also below).
CrZIP6 belongs to ZIP subfamily II, together with CmZIP2 and HsZIP1 to 3 (Fig. 2). While HsZIP3 has not been characterized, HsZIP1 and 2 are involved in zinc uptake across the plasma membrane (Eide, 2004). Surprisingly, C. reinhardtii possesses six GUFA-related proteins (CrZIP7–12), whereas only one is found in C. merolae (CmZIP3/GUFA), yeast (ScZRT3), Arabidopsis (AtGUFA), and humans (HsZIP11; Fig. 2). The distant relationship of CrZIP7 and 12 with the GUFA subfamily is only supported by neighbor-joining analysis (data not shown). The GUFA subfamily has been named after the GUFA protein of the bacteria Myxococcus xanthus, a protein of unknown function identified by genome sequencing (McGowan et al., 1993). The yeast protein ScZRT3 is the only GUFA protein that has been functionally characterized. It is localized in the vacuolar membrane and involved in the remobilization of stored zinc under zinc-deficiency conditions (MacDiarmid et al., 2000).
Finally, a LIV1-like protein is found in C. merolae but not in C. reinhardtii. CmZIP4/LIV1 contains a highly conserved motif [(H,E) E(L,F) P(H,Q,A) E(L,I,V,M)(G,S) D(F,L,V)(M,A,V,G)XL(L,I,V), defined by Taylor and Nicholson (2003) as a signature for human LIV1 proteins] in the transmembrane domain V, which is specific to this subfamily of ZIPs. Initially implicated in human metastatic breast cancer, the LIV1 proteins have recently been shown to be capable of zinc transport (Taylor et al., 2003, 2004).
It should be mentioned that the CrZIP13 and 14 proteins are only distantly related to the ZIP family. Due to difficulties in aligning them properly with other ZIPs, their positions in the tree vary according to the phylogenetic method used (data not shown). These proteins may be based on inaccurate gene models and should be considered as possibly distant, putative ZIPs.
Nonetheless, our findings surprisingly reveal the presence of a very high number of putative zinc transporters of the ZIP family in the C. reinhardtii genome compared to yeast and C. merolae. The unicellular C. reinhardtii possesses as many ZIPs as the complex multicellular eukaryotes Arabidopsis and humans, although the ZIP diversification occurred in different subfamilies in each of these organisms (within subfamily I for Arabidopsis, LIV1 for humans, and GUFA and subfamily I for C. reinhardtii; Fig. 2) from possibly four ancestral genes still present in C. merolae.
THE CAX FAMILY
The CAX proteins are divalent cation/H+ antiporters generally containing 10 to 14 transmembrane domains. The AtCAX1 and AtCAX2 proteins have been identified by functional complementation of a S. cerevisiae mutant defective in vacuolar calcium accumulation. Both having 11 putative transmembrane domains, AtCAX1 is a vacuolar high-affinity Ca2+/H+ antiporter, while AtCAX2 displays a lower affinity for Ca2+ (Hirschi et al., 1996) and has been proposed to transport Mn2+ and Cd2+ across the tonoplast of plant cells (Hirschi et al., 2000). Nine other CAX proteins have been identified in the Arabidopsis genome, but have not been functionally characterized yet (Maser et al., 2001). An additional member of this family is the AtMHX1 protein, which is an H+-coupled antiporter that can transport Mg2+ and Zn2+ across the vacuole membrane (Shaul et al., 1999).
We have found, respectively, four and two CAXs in the genomes of C. reinhardtii and C. merolae (Tables II and III). CrCAX1 to 2 and CmCAX1 are related to the yeast ScVCX1 and AtCAX1 to 6 (Fig. 3). CrCAX3 to 4 and CmCAX2 are spread among the other CAXs. Maser et al. (2001) highlighted the difficulty of determining transport properties from sequence information within this family. Experimental data will be necessary to establish whether any of the newly identified CAXs plays a role in transition metal homeostasis in algae. To support this idea, it is worth mentioning that CrCAX1 is expressed in the stress II and stress III EST libraries (Supplemental Table IV), both of which were prepared from metal-stressed cells (Shrager et al., 2003).
Finally, it is interesting to note that neither C. reinhardtii nor C. merolae seem to possess a close AtMHX1 homolog, suggesting that this function is lacking in algae. In Arabidopsis, AtMHX1 is mainly expressed in vascular tissues and believed to participate in Mg2+ and Zn2+ partitioning between plant organs (Shaul et al., 1999). This function would have evolved associated with the emergence of land plants. Alternatively, it might be performed by a more distant member of the CAX family.
THE COPT FAMILY
The COPT proteins form a eukaryotic family of copper transporters (Eide, 1998). COPTs share common topology features, including (1) the presence of three transmembrane domains with an extracellular N terminus and a cytoplasmic C terminus; (2) the presence of two Met-rich regions in the N terminus that may act as a copper scavenger in the extracellular milieu; and (3) an additional Met-rich region in transmembrane domain II, probably involved in copper coordination during transmembrane transport (Petris, 2004). In S. cerevisiae, high-affinity copper uptake, like iron uptake (see below), requires plasma membrane reductases to reduce Cu(II) to Cu(I), the reduction step being mediated by the same reductases that are involved in iron uptake. Cu(I) is then transported by the redundant high-affinity copper transporters ScCTR1 and ScCTR3, via a Cu(I)/2 K(I) antiport mechanism that is highly specific for Cu(I) over other metal ions (Eide, 1998). S. cerevisiae possesses an additional CTR-related protein (ScCTR2). Located in the vacuole, the function of ScCTR2 remains unknown (Eide, 1998; Petris, 2004). Two CTR homologs are found in humans, but only HsCTR1 has been functionally characterized. It is a plasma membrane protein involved in copper uptake in various cell types (Zhou and Gitschier, 1997; Petris, 2004). Sancenon et al. (2003) have recently identified 5 COPT proteins (AtCOPT1 to 5) in Arabidopsis and showed that AtCOPT1, 2, 3, and 5 restore copper uptake in a ctr1 ctr3 yeast mutant. Antisense AtCOPT1 transgenic lines display reduced copper uptake together with increased root length and defects in pollen development (Sancenon et al., 2004). AtCOPT1 thus plays an important role in copper acquisition in Arabidopsis.
We have identified a single COPT protein in the genomes of both C. reinhardtii and C. merolae (Tables II and III). CrCOPT1 is more related to the Arabidopsis COPTs than to the human and yeast transporters, while CmCOPT1 is only distantly related to the other COPTs (data not shown). It should also be mentioned that CmCOPT1 is substantially larger than the plant and human COPTs. CrCOPT1 might be located in the plasma membrane (Supplemental Table VI), while CmCOPT1 is predicted to be localized in the vacuolar membrane (Supplemental Table VII).
THE HMA SUBFAMILY OF P-TYPE ATPASES
P-type ATPases are transporters characterized by the formation of a phosphorylated intermediate in the reaction cycle. These proteins typically contain 8 to 12 transmembrane domains and a large cytoplasmic loop, including ATP-binding and phosphorylation sites. P-type ATPases transport a broad range of small cations, and possibly phospholipids, and have been classified into five subfamilies according to their predicted substrate specificities and phylogenies. The type 1B subfamily proteins (HMAs or CPx ATPases) are involved in heavy-metal transport. HMAs possess eight transmembrane domains, the sixth of which contains a conserved Cys-Pro-Cys/His/Ser motif (CPx motif) believed to be involved in metal cation translocation across the membrane. Identified in a wide range of organisms, HMAs can be divided into two main groups with different substrate specificities (monovalent Cu+/Ag+ cations or divalent Zn2+/Co2+/Cd2+/Pb2+ cations; Axelsen and Palmgren, 2001; Cobbett et al., 2003).
We have identified three and two HMAs in the genomes of C. reinhardtii and C. merolae, respectively (Tables II and III). CrHMA1 and CmHMA1 are related to the divalent cation transporters of bacteria (EcZntA, SaCadA) and Arabidopsis (AtHMA1 to 4), especially to AtHMA1 (Fig. 4). EcZntA and SaCadA are involved in zinc or cadmium excretion and tolerance in Escherichia coli and Staphylococcus aureus, respectively (Nies, 2003). AtHMA2 and AtHMA4 probably play a role in zinc translocation in the plant (Mills et al., 2003; Hussain et al., 2004), while functional studies in yeast suggest that AtHMA3 is a cadmium/lead transporter (Gravot et al., 2004). In addition, AhHMA3 and TcHMA4 are constitutively highly expressed in the zinc hyperaccumulator A. halleri and the zinc/cadmium hyperaccumulator T. caerulescens, respectively, when compared to Arabidopsis. These proteins could be at least partly responsible for the metal hyperaccumulation and hypertolerance in these two species (Becher et al., 2004; Bernard et al., 2004). So far, nothing is known about the function of AtHMA1. According to phylogenetic analysis, it belongs to the divalent cation transporter subfamily of P1B-ATPases but is only distantly related to AtHMA2 to 4 (Cobbett et al., 2003). Interestingly, like AtHMA1, CrHMA1 and CmHMA1 have an uncharacteristic Ser/Pro/Cys motif in the sixth transmembrane domain instead of the common Cys-Pro-Cys/His/Ser motif. This property is shared with the cobalt transporter CoaT of the cyanobacterium Synechocystis and might influence the substrate specificity of the transporter. On that basis, Cobbett et al. (2003) suggested that AtHMA1 might be a cobalt transporter.
Although widespread in prokaryotes, the occurrence of these divalent cation-transporting HMAs in eukaryotes is apparently limited to photosynthetic organisms (Fig. 4). Cobbett et al. (2003) suggested that these proteins might have evolved from one or more, probably two, horizontal gene transfers from the prokaryotic endosymbiont believed to be at the origin of the plastid. The AtHMA2 to 4 genes apparently result from the recent repeated duplication of an ancestral gene (Cobbett et al., 2003). The AtHMA2 to 4 subgroup is absent in C. merolae and C. reinhardtii, indicating that this function might have been lost in both algae.
CrHMA2 and 3 and CmHMA2 cluster with the monovalent cation transporters of the P1B-ATPases and are most probably copper transporters (Fig. 4). In particular, CrHMA2 and CmHMA2 are closely related to yeast (ScCCC2), plant (AtRAN1), and human (HsATP7A and B) transporters known to deliver copper to proteins in the trans-Golgi network (Cobbett et al., 2003; Hall and Williams, 2003). The CrHMA2 protein corresponds to the EST 1021007C06, which has been previously identified, and is believed to be involved, together with the chaperone ATX1, in copper delivery to the secretory pathway (La Fontaine et al., 2002).
CrHMA3 clusters with AtPAA1 and AtHMA6 (Fig. 4). AtPAA1 is a chloroplastic protein mediating the transport of copper into the chloroplast. Arabidopsis paa mutants have a lower chloroplast copper content, accumulate reduced levels of holoplastocyanin, and display reduced chloroplastic copper/zinc superoxide dismutase activity (Shikanai et al., 2003). By analogy, CrHMA3 could play a role in copper delivery to the chloroplast. However, this protein is predicted to localize to the vacuolar membrane (Table II). In some cyanobacteria and green algae, plastocyanin (involved in the transfer of photosynthetic electrons from cytochrome b6/f to PSI) may be functionally replaced by a soluble c-type (c-552 or c-553) cytochrome. Plastocyanin is expressed under copper-replete conditions and replaced by cytochrome c6 when copper availability is limited (Kerfeld and Krogmann, 1998; Merchant, 1998). Whereas higher plants only express plastocyanin, all examined red algae (along with Chrysophyceae and Xanthophyceae) seem to use the c-type cytochrome alternative (Sandmann et al., 1983; Price et al., 1991). Accordingly, C. merolae also lacks plastocyanin (data not shown) and its plastid genome contains a petJ gene encoding a c-553 cytochrome (Ohta et al., 2003). The fact that photosynthesis is not based on plastocyanin in C. merolae may explain the lack of an AtPAA1-like function in this organism (Fig. 4).
THE ABC TRANSPORTER FAMILY
ABC transporters are ubiquitous transporters involved in a large number of physiological processes. This family is one of the largest protein families with 29, 128, and 48 members in S. cerevisiae, Arabidopsis, and humans, respectively. Typical ABC transporters (the so-called full-size transporters) possess two conserved nucleotide-binding folds responsible for ATP hydrolysis alternating with two highly hydrophobic domains (containing 4–6 transmembrane spans) that specify the substrates to be transported. The half-size ABC transporters possess a single copy of each domain and are assumed to function as homo- or heterodimers (Holland et al., 2003).
Based on structural similarities, ABC transporters can be classified in several subfamilies (Decottignies and Goffeau, 1997; Sanchez-Fernandez et al., 2001; Dean et al., 2003). Among them, members of only two subfamilies (MRP and ATM/HMT) have been implicated in metal transport (Table I).
MRPs are full-size ABC transporters mainly acting as glutathione S-conjugate pumps (Rea et al., 1998). In S. cerevisiae, the ScYCF1 protein ensures the transport of bis(glutathionato) cadmium complexes from the cytoplasm into the vacuole. The lack of the transporter determines hypersensitivity to cadmium, arsenate, and mercury (Szczypka et al., 1994; Li et al., 1997; Ghosh et al., 1999; Gueldry et al., 2003). A second vacuolar yeast MRP, named ScBTP1, is also involved in cadmium tolerance, but its role is only marginal compared to ScYCF1 (Klein et al., 2002; Sharma et al., 2002). The overexpression of ScYCF1 in Arabidopsis enhances the tolerance of the plant to lead and cadmium (Song et al., 2003). Among 14 MRP proteins (AtMRP1–14) identified in Arabidopsis (Sanchez-Fernandez et al., 2001; Kolukisaoglu et al., 2002; Martinoia et al., 2002), only AtMRP3 has been implicated in cadmium detoxification and transport. The AtMRP3 gene expression is up-regulated in response to cadmium and this induction is apparently correlated with the accumulation of cadmium in the plant organs (Bovet et al., 2003). Moreover, AtMRP3 complements the cadmium sensitivity of a yeast ycf1 mutant (Tommasini et al., 1998). None of the 4 other cloned and characterized AtMRPs (AtMRP1, 2, 4, and 5) or the 12 HsMRPs (or HsABCC) are involved in metal detoxification, but in the transport of a wide range of substrates (for review, see Dean et al., 2003; Rea et al., 2003).
We have found seven and two MRPs in the genome sequences of C. reinhardtii and C. merolae, respectively (Tables II and III). Although it is obviously a MRP transporter, protein model 155613 is too small (428 amino acid residues) compared to the other members of the family. With a TBLASTN search, we have identified additional ABC transporter domains in close proximity to the gene model on scaffold 142 (Table II; Supplemental Table II). Using three gene prediction software programs (GreenGenie, GeneMark, and GENSCAN), we determined an alternative gene model (encoding CrMRP6, 1,524 amino acid residues), which, albeit imperfect, probably much better reflects the real structure of the gene (data not shown; available upon request). The cloning of the corresponding cDNA will nevertheless be necessary to fully elucidate the gene structure.
CrMRP1 and 2 group with human (HsABCC4, 5, 11, and 12), plant (AtMRP11 and 15), and yeast (ScYOR1) proteins (Fig. 5), all of which lack the N-terminal domain characteristic of the members of the family (Dean et al., 2003; Rea et al., 2003). CrMRP1 is a chloroplastic protein that might be involved in bicarbonate uptake and is regulated by light intensity and CO2 level (Im and Grossman, 2002). The other members of the MRP subfamily identified here, CrMRP3 to 7 and CmMRP1 and 2, are related to AtMRP11 and HsABCC10 (Fig. 5). Based on sequence homology, these algal transporters are probably glutathione S-conjugate pumps. However, to understand the function of these proteins in drug and/or metal tolerance, a detailed analysis (including the determination of the membrane localization and substrate specificity) will be required.
ATM/HMTs are half-size transporters located either in the mitochondrial or vacuolar membranes. Mitochondrial transporters (HsABCB6 and 7, AtATM1–3, and ScATM1) are involved in the export of iron/sulfur clusters from the mitochondrial matrix to the cytoplasm (Kispal et al., 1997; Csere et al., 1998; Allikmets et al., 1999; Kispal et al., 1999; Mitsuhashi et al., 2000; Kushnir et al., 2001). We have recently shown that a C. reinhardtii homolog of these mitochondrial transporters, named CrCds1, plays an essential role in cadmium tolerance, possibly by exporting cadmium out of the mitochondria, thereby protecting the mitochondrial function from cadmium toxicity (Hanikenne et al., 2001, 2005). The ability of mitochondrial ABC transporters to act in cadmium detoxification could be a new property among green organisms (Hanikenne et al., 2005). Out of 7 ESTs corresponding to the Cds1 gene (Supplemental Table IV), 3 were identified in the stress II cDNA library prepared from cells incubated in the presence of cadmium (Shrager et al., 2003), suggesting that the gene might be regulated by the metal. This was indeed demonstrated by northern-blot analysis, showing that Cds1 is strongly induced in the presence of cadmium (Hanikenne et al., 2005). This highlights that the careful analysis of EST data might be used to generate testable hypotheses concerning gene regulation.
The vacuolar SpHMT1 of the fission yeast S. pombe is involved in the transport of cadmium-phytochelatin complexes from the cytoplasm into the vacuole. A mutant strain lacking this transporter is unable to accumulate high-Mr cadmium-phytochelatin complexes in the vacuoles and displays hypersensitivity to cadmium (Ortiz et al., 1992, 1995). Although the existence of such a transport function has been biochemically confirmed in plant vacuolar membranes (Vögeli-Lange and Wagner, 1990; Salt and Rauser, 1995), no functionally related SpHMT1 analog has been identified yet in the Arabidopsis genome. The Arabidopsis proteins most highly similar to SpHMT1 are the ATM transporters (Sanchez-Fernandez et al., 2001; Martinoia et al., 2002).
We have identified two and three ATM/HMTs in the genomes of C. reinhardtii and C. merolae, respectively (Tables II and III). Intriguingly, we could not find any protein model corresponding to CrCds1 in the JGI sequence data but have found by TBLASTN that it falls within scaffold 122 (Table II; Supplemental Table II).
Two subclusters can be distinguished within the ATM/HMT subfamily (Fig. 6). On the one hand, subcluster I, which contains CrATM/HMT-2, CmATM/HMT-1 and -2, only includes mitochondrial transporters (AtATM1–3, ScATM1, HsABCB7) possessing 5 or 6 conserved transmembrane-spanning regions. Since they share structural and targeting predictions with ScATM1 (Supplemental Tables VI and VII), the 3 algal proteins are likely functional homologs of the yeast transporter and might have a role in iron homeostasis of mitochondria. On the other hand, subcluster II, which contains CrCds1, CrATM/HMT-3, and CmATM/HMT-3, includes both mitochondrial (HsABCB6) and vacuolar (SpHMT1) transporters possessing 5 additional transmembrane-spanning segments at the N-terminal end of the protein. Phytochelatins are the main intracellular chelators for cadmium and accumulate in the vacuole in C. reinhardtii (Howe and Merchant, 1992; Hu et al., 2001). Therefore, a functional analog of SpHMT1 should be present in the alga and might possibly be fulfilled by CrATM/HMT-3. We will have to wait for experimental evidence supporting these hypotheses. Although a putative phytochelatin synthase gene (protein model CMI111C) is present in the genome of C. merolae, the occurrence of phytochelatins in this organism is not known. It is thus difficult to speculate on the function of CmATM/HMT-3.
IRON TRANSPORTERS
Two strategies are known for iron uptake in higher plants. Strategy I (or reduction strategy) occurs in all plants, except graminaceous monocots, and involves 3 steps: (1) soil acidification by H+ ATPases to solubilize iron; (2) reduction of ferric iron [Fe(III)] by plasma membrane ferric chelate reductases; and (3) uptake of ferrous iron [Fe(II)] by AtIRT1, a member of the ZIP family (Guerinot and Yi, 1994; Connolly and Guerinot, 2002). We have already mentioned above that neither C. reinhardtii nor C. merolae seem to possess any homologs of IRT1 (Fig. 2). Strategy II (or chelation strategy) is found in graminaceous monocots and involves the release of Fe(III) chelators called phytosiderophores, coupled to the induction of a specific Fe(III) chelate transporter (Guerinot and Yi, 1994; Connolly and Guerinot, 2002). In maize, this transporter, ZmYS1, has recently been cloned (Curie et al., 2001). ZmYS1 belongs to the oligopeptide transporter family and is induced under iron deficiency both at transcriptional and translational levels. It has been shown to cotransport protons and phytosiderophore- or nicotianamine-chelated iron. It is interesting to mention that, although ZmYS1 displays a broad metal specificity (iron, zinc, nickel, copper, and cobalt) in Xenopus oocytes and yeast, it seems to be regulated only by iron availability in planta (Roberts et al., 2004; Schaaf et al., 2004). Moreover, the rice OsYSL2 has been shown to transport iron and manganese chelated by nicotianamine (Koike et al., 2004). Phytosiderophores are not synthesized in Arabidopsis, a strategy I species, but eight YS1 homologs (AtYSL1–8) are found in the plant genome. The AtYSL2 protein has been shown to transport iron and copper chelated by nicotianamine, a precursor of phytosiderophores (DiDonato et al., 2004).
To our knowledge, phytosiderophores or nicotianamine are not found in algae. Nevertheless, we searched for YS1-like proteins in the C. reinhardtii and C. merolae genome sequence but could not identify any homolog. This function may thus have evolved after the emergence of land plants. As soon as genome sequence data become available, it would be interesting to analyze green algae that belong to the evolutionary lineage of higher plants (Charophyta, sensu lato; van den Hoek et al., 1995) in this respect.
In S. cerevisiae, the first step of iron uptake involves, as for the plant strategy I, the reduction of Fe(III) to Fe(II) by ferric reductases. Then occurs high-affinity iron uptake mediated by (1) a multicopper oxidase (ScFET3) that reoxidizes Fe(II) to Fe(III) and (2) an iron permease (ScFTR1) transporting Fe(III) into the cell. ScFET3 and ScFTR1 are induced under iron deficiency and the corresponding proteins form a complex at the plasma membrane. In iron-sufficient conditions, iron uptake is driven by the low-affinity transporter ScFET4 (Radisky and Kaplan, 1999).
Recently, La Fontaine et al. (2002) identified genes encoding a multicopper oxidase (CrFOX1) and an iron permease (CrFTR1) in the C. reinhardtii EST databases and showing, in a photosynthetic organism, the occurrence of an iron assimilation pathway analogous to the high-affinity iron uptake pathway of S. cerevisiae (for review, see Hanikenne, 2003). Although no additional FTR1 homolog was found in the C. reinhardtii genome, we have identified four FTR proteins (CmFTR1–4) in C. merolae (Tables II and III). CmFTR1 and 2 are almost identical in sequence and are highly represented in the EST databases (Table III; Supplemental Table V). Both are predicted to localize in the plasma membrane like ScFTR1 and CrFTR1 (Tables II and III). Interestingly, we could not find any multicopper oxidase homolog in C. merolae (data not shown). Living in an iron- and sulfur-rich milieu with low copper availability (see below), C. merolae might have an alternative Fe(II) oxidase associated with the FTRs and its iron uptake might be independent on copper, in contrast to S. cerevisiae. CmFTR3 and 4 are predicted to reside in the mitochondria (Supplemental Table VII). This putative function is not found in yeast and C. reinhardtii and might represent an independent evolution in C. merolae. Alternatively, such a function might have been lost in other studied unicellular eukaryotes.
Members of the NRAMP family are also known to transport Fe(II) ions. These transporters use the transmembrane proton gradient to facilitate transport of divalent cations and iron in particular. NRAMPs are ubiquitous proteins that possess common structural features, including the presence of 12 transmembrane domains, 2 conserved His residues in transmembrane domain 6, and a transport motif in the intracellular loop between transmembrane domains 8 and 9 (Forbes and Gros, 2001; Mackenzie and Hediger, 2004). In humans, HsNRAMP1 is required for natural resistance to intracellular pathogenic bacteria by transporting Mn(II) and Fe(II) across the phagosomal membranes in the macrophages, thus restricting metal availability for pathogens, whereas HsNRAMP2 (or HsDMT1) plays a role in the uptake of dietary Fe(II) at the apical surface of enterocytes (Forbes and Gros, 2001; Mackenzie and Hediger, 2004). When expressed in Xenopus oocytes, HsNRAMP2 is able to drive the transport of a wide range of divalent metal cations (including cadmium, cobalt, copper, zinc, manganese, and lead) in addition to iron (Mackenzie and Hediger, 2004). It has been suggested that HsNRAMP2 might contribute to cadmium and lead poisoning in iron-deficient human subjects (Bressler et al., 2004).
Six NRAMP proteins are encoded in the genome of Arabidopsis (Maser et al., 2001). Expression of AtNRAMP1, 3, and 4 complements iron or manganese uptake deficiency and increases cadmium sensitivity and accumulation in yeast (Curie et al., 2000; Thomine et al., 2000). In planta, the 3 genes are induced under iron deficiency and the overexpression of AtNRAMP1 leads to an increased resistance to toxic iron concentrations (Curie et al., 2000; Thomine et al., 2000). Furthermore, inactivation of the AtNRAMP3 gene enhances cadmium resistance, while its overexpression leads to cadmium sensitivity (Thomine et al., 2000). Recently, Thomine et al. (2003) showed that AtNRAMP3 is localized in the vacuolar membrane and proposed that the protein functions as a metal (iron, cadmium, manganese, and zinc) exporter from the vacuole into the cytoplasm.
Three NRAMP homologs, named ScSMF1 to 3, are found in S. cerevisiae. These proteins transport a broad range of divalent cations, but have been more specifically implicated in manganese, copper, and, more marginally, iron homeostasis (Radisky and Kaplan, 1999; Cohen et al., 2000). ScSMF1 is localized in the plasma membrane, whereas ScSMF2 resides in the membrane of intracellular vesicles and ScSMF3 in the vacuolar membrane (Supek et al., 1996; Cohen et al., 2000; Luk and Culotta, 2001).
We have identified three NRAMPs in the genomes of both C. reinhardtii and C. merolae (Tables II and III). In a previous report, Rosakis and Koster (2004) have found five NRAMPs in C. reinhardtii. However, due to gaps in the sequence of version 1 of the genome, it is likely that at least some partial NRAMP sequences identified belong to a common open reading frame, as hypothesized by the authors (Rosakis and Koster, 2004). CmNRAMP1 and 2 distantly group with the AtNRAMP1 and 6 proteins, while the other algal NRAMPs are more related to bacterial, cyanobacterial, and yeast proteins than to Arabidopsis and human NRAMP proteins (Fig. 7). As in other organisms, these proteins most probably play a role in the transport of divalent metals across various membrane systems.
Finally, in humans, the HsIREG1 protein (or ferroportin 1) mediates the transport of iron at the basolateral surface of the enterocytes into the blood for delivery to other organs (McKie et al., 2000). A search in the The Arabidopsis Information Resource (TAIR) database (http://www.arabidopsis.org) allows the identification of 3 IREG1-like proteins in Arabidopsis. Although these proteins have not been characterized, they might play a role in iron homeostasis in plants. We could not find any IREG1 homologs in C. reinhardtii or S. cerevisiae (data not shown), whereas a related protein, named CmIREG1, was identified in C. merolae (Table III). An IREG1-like protein then seems to have been present in the common ancestor of photosynthetic and nonphotosynthetic organisms. It might have been subsequently lost in yeast and C. reinhardtii, while an iron efflux function might be important for C. merolae to cope with an environment characterized by high iron availability.
METHODOLOGICAL INSIGHTS
Using a custom analysis pipeline (Supplemental Fig. 1), we have identified members of 10 families or subfamilies of metal transporters in the unicellular algae C. reinhardtii and C. merolae (Table I). Most of the identified proteins have never been described in these organisms. The identification of all members in a protein family may become crucial when characterizing single members of this family. This indeed allows speculation as to possible functional redundancies within multigene families. For example, the AtHMA2 and 4 proteins are closely related to each other in Arabidopsis (Cobbett et al., 2003). Individual mutants exhibit no apparent phenotype and athma2 athma4 double mutants have been more useful to determine the function of these transporters in metal homeostasis (Hussain et al., 2004).
Rosakis and Koster (2004) have recently reported a search for some metal transporters in C. reinhardtii, initiated when version 1 of the genome became available (February 2003 at the JGI Web site). Within each transporter family except the NRAMPs, we have identified a large number of additional members. This validates our high-throughput approach using entire protein families from reference organisms to search genomes instead of using a few already characterized proteins. This is especially relevant for families formed of a constellation of subgroups with considerable sequence divergence, which is often the case among metal transporter families. For example, a search of the C. reinhardtii genome with AtMTP1 (a zinc transporter) does not identify manganese-transporting MTPs.
Nevertheless, our work partly suffers from the limitations of all in silico analyses. Although state-of-the-art modeling methods have been used by JGI to predict C. reinhardtii gene structures, these models are of overall poorer quality and reliability than those obtained for C. merolae. Most probably, the presence of many short introns and exons in C. reinhardtii nuclear genes (Silflow, 1998) is responsible for such mispredictions, whereas the C. merolae genome contains only a very small number of introns (only present in 26 out of 5,331 genes (Matsuzaki et al., 2004). As a consequence, some C. reinhardtii proteins might be inaccurately assigned to subgroups within their families, or may seem artificially distant from their yeast, plant, or human homologs (e.g. see CrHMA1, Fig. 4, or CrMRP7, Fig. 5). The quality of the protein models also has an impact on the results of topology or localization predictions. Notably, models shorter than expected based on their homologs in reference organisms might correspond to truncated proteins lacking targeting peptides (see CrHMA3, for example; Table II). After the completion of the genome project, a major challenge for the Chlamydomonas community will be the cloning of full-length cDNAs to accurately determine gene structures and protein sequences.
Another limitation concerns the reliability of the topology and subcellular localization prediction software itself. First of all, although the best program available was used to predict transmembrane domains (TMHMM, according to Krogh et al., 2001), it has been observed that these algorithms are not able to distinguish transmembrane helices from N-terminal targeting signals (Chen et al., 2002). Moreover, all localization prediction algorithms have been optimized for higher plants or nonphotosynthetic eukaryotes such as yeast, and it is hardly known whether targeting signals are conserved in C. reinhardtii and C. merolae. Franzen et al. (1990) have nevertheless shown that the N-terminal chloroplast-targeting peptides of C. reinhardtii are more related to the yeast and plant mitochondria-targeting signals than those of higher plant chloroplast proteins. The CrCds1 ABC transporter of C. reinhardtii was experimentally localized in the mitochondria (Hanikenne et al., 2005), whereas iPSORT and TargetP predicted a localization in the chloroplast or in the vacuole, respectively (Supplemental Table VI). For all these reasons, in silico predictions of localization should be taken with caution. In some cases, however, they are meaningful when compared to information available for the reference species (e.g. CrMTP1 and CmMTP1 in Tables II and III).
For most of the genes described here, we have identified corresponding ESTs (Supplemental Tables IV and V), indicating that these genes are expressed. As already mentioned, the analysis of these EST data is useful to generate working hypotheses concerning potential gene regulation. On the other hand, genes lacking ESTs could be weakly transcribed in the conditions used for all cDNA library constructions. Moreover, some rare transcripts might have been lost during the normalization procedure. Alternatively, these genes might correspond to pseudogenes where the promoter is no longer functional, as no stop codon interrupting the coding sequence was found in these genes.
PHYLOGENETIC INSIGHTS
Our phylogenetic analyses clearly show that an ancestral gene for almost all families and subfamilies was already present early in eukaryote evolution, with the notable exception of YSL proteins. Therefore, the transition to multicellularity was generally associated with the diversification of existing functions rather than with the appearance of novel gene families. In most cases, it is thus legitimate to use unicellular models to gain a fundamental understanding of cellular metal tolerance and homeostasis.
Two intriguing topics are the considerable diversification among the ZIP protein family in C. reinhardtii and the FTRs in C. merolae (see above), but, as a rule, C. reinhardtii generally appears to be more complex than C. merolae with respect to the number of different metal homeostasis-related transporters (Table I). As a flagellate organism found in water and soils, C. reinhardtii lives under fluctuating environmental conditions and therefore needs the flexibility to adapt. Moreover, C. reinhardtii has a relatively sophisticated life cycle with complicated vegetative and sexual stages, for which specialized functions may have evolved (Harris, 1989). In that respect, it is interesting to note that several metal transporters are represented in the gamete/zygote EST library (Supplemental Table IV). On the contrary, the Cyanidiales are a group of asexual algae (Ciniglia et al., 2004). Although it is apparently a highly primitive organism, the possibility that C. merolae represents a simplification of a once complex ancestral organism cannot be ruled out. C. merolae lives in acidic sulfur- and metal-rich (iron, aluminum, nickel, and various heavy metals) hot springs (Gross, 2000) and is able to face these stable, though extreme, conditions with a relatively limited number of metal transporters when compared to C. reinhardtii. The related unicellular red alga C. caldarium has been shown to accumulate high levels of iron and phosphate in intracellular electron-dense structures (Nagasaka et al., 2003), as well as high concentrations of zinc, manganese, nickel, and copper (Nagasaka et al., 2004).
CONCLUSION
In this update on metal homeostasis and tolerance systems encountered in eukaryotes, we present an inventory of metal transporters found in two unicellular algae, along with topology and targeting predictions, as well as searches of the available EST collections. These data were produced through a carefully designed semiautomated in silico mining strategy that might be useful for the research community, while uncovering the pitfalls of such an approach. Although we mostly speculate on the functional and evolutionary implications unveiled by our findings, this work should provide a basis for further experimental molecular and genomic studies of heavy-metal homeostasis and tolerance in photosynthetic organisms.
Supplementary Material
Acknowledgments
The C. reinhardtii sequence data were produced by the U.S. Department of Energy Joint Genome Institute (http://www.jgi.doe.gov) and are provided for use in this publication/correspondence only.
This work was supported by the European Union Research Training Network Metalhome (contract no. HPRN–CT–2002–00243 to M.H., U.K.) and by the German Federal Ministry of Education and Research (grant no. 0311877 to U.K.). D.B. is a postdoctoral researcher of the Fonds National de la Recherche Scientifique (Belgium).
The online version of this article contains Web-only data.
References
- Allikmets R, Raskind WH, Hutchinson A, Schueck ND, Dean M, Koeller DM (1999) Mutation of a putative mitochondrial iron transporter gene (ABC7) in X-linked sideroblastic anemia and ataxia (XLSA/A). Hum Mol Genet 8: 743–749 [DOI] [PubMed] [Google Scholar]
- Assuncao AGL, Da Costa Martins P, De Folter S, Vooijs R, Schat H, Aarts MGM (2001) Elevated expression of metal transporter genes in three accessions of the metal hyperaccumulator Thlaspi caerulescens. Plant Cell Environ 24: 217–226 [Google Scholar]
- Axelsen KB, Palmgren MG (2001) Inventory of the superfamily of P-type ion pumps in Arabidopsis. Plant Physiol 126: 696–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher M, Talke IN, Krall L, Krämer U (2004) Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator Arabidopsis halleri. Plant J 37: 251–268 [DOI] [PubMed] [Google Scholar]
- Bernard C, Roosens N, Czernic P, Lebrun M, Verbruggen N (2004) A novel CPx-ATPase from the cadmium hyperaccumulator Thlaspi caerulescens. FEBS Lett 569: 140–148 [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Medlin L (1995) The phylogeny of plastids: a review based on comparisons of small-subunit ribosomal RNA coding regions. J Phycol 31: 489–498 [Google Scholar]
- Blaudez D, Kohler A, Martin F, Sanders D, Chalot M (2003) Poplar metal tolerance protein 1 (MTP1) confers zinc tolerance and is an oligomeric vacuolar zinc transporter with an essential leucine zipper motif. Plant Cell 15: 2911–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrelly GP, Harrison MD, Robinson AK, Cox SG, Robinson NJ, Whitehall SK (2002) Surplus zinc is handled by Zym1 metallothionein and Zhf endoplasmic reticulum transporter in Schizosaccharomyces pombe. J Biol Chem 277: 30394–30400 [DOI] [PubMed] [Google Scholar]
- Bovet L, Eggmann T, Meylan-Bettex M, Polier JE, Kammer P, Marin E, Feller U, Martinoia E (2003) Transcript level of AtMRPs after cadmium treatment: induction of AtMRP3. Plant Cell Environ 26: 371–381 [Google Scholar]
- Bressler JP, Olivi L, Cheong JH, Kim Y, Bannona D (2004) Divalent metal transporter 1 in lead and cadmium transport. Ann N Y Acad Sci 1012: 142–152 [DOI] [PubMed] [Google Scholar]
- Butterfield NJ (2000) Bangiomorpha pubescens n. gen., n. sp.: implications for the evolution of sex, multicellularity, and the Mesoproterozoic/Neoproterozoic radiation of eukaryotes. Paleobiology 26: 386–404 [Google Scholar]
- Cavalier-Smith T (1982) The origins of plastids. Biol J Linn Soc 17: 289–306 [Google Scholar]
- Chen CP, Kernytsky A, Rost B (2002) Transmembrane helix predictions revisited. Protein Sci 11: 2774–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciniglia C, Yoon HS, Pollio A, Pinto G, Bhattacharya D (2004) Hidden biodiversity of the extremophilic Cyanidiales red algae. Mol Ecol 13: 1827–1838 [DOI] [PubMed] [Google Scholar]
- Clemens S (2001) Molecular mechanisms of plant metal tolerance and homeostasis. Planta 212: 475–486 [DOI] [PubMed] [Google Scholar]
- Clemens S, Bloss T, Vess C, Neumann D, Nies DH, Zur Nieden U (2002) A transporter in the endoplasmic reticulum of Schizosaccharomyces pombe cells mediates zinc storage and differentially affects transition metal tolerance. J Biol Chem 277: 18215–18221 [DOI] [PubMed] [Google Scholar]
- Clemens S, Simm C (2003) Schizosaccharomyces pombe as a model for metal homeostasis in plant cells: the phytochelatin-dependent pathway is the main cadmium detoxification mechanism. New Phytol 159: 323–330 [DOI] [PubMed] [Google Scholar]
- Cobbett CS, Hussain D, Haydon MJ (2003) Structural and functional relationships between type 1B heavy metal-transporting P-type ATPases in Arabidopsis. New Phytol 159: 315–321 [DOI] [PubMed] [Google Scholar]
- Cohen A, Nelson H, Nelson N (2000) The family of SMF metal ion transporters in yeast cells. J Biol Chem 275: 33388–33394 [DOI] [PubMed] [Google Scholar]
- Connolly EL, Guerinot M (2002) Iron stress in plants. Genome Biol 3: 1024.1–1024.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csere P, Lill R, Kispal G (1998) Identification of a human mitochondrial ABC transporter, the functional orthologue of yeast Atm1p. FEBS Lett 441: 266–270 [DOI] [PubMed] [Google Scholar]
- Curie C, Alonso JM, Le Jean M, Ecker JR, Briat JF (2000) Involvement of NRAMP1 from Arabidopsis thaliana in iron transport. Biochem J 347 (Pt 3): 749–755 [PMC free article] [PubMed] [Google Scholar]
- Curie C, Panaviene Z, Loulergue C, Dellaporta SL, Briat JF, Walker EL (2001) Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409: 346–349 [DOI] [PubMed] [Google Scholar]
- De Freitas J, Wintz H, Kim JH, Poynton H, Fox T, Vulpe C (2003) Yeast, a model organism for iron and copper metabolism studies. Biometals 16: 185–197 [DOI] [PubMed] [Google Scholar]
- Dean M, Rzhetsky A, Allikmets R (2003) Human and Drosophila ABC proteins. In IB Holland, SPC Cole, K Kuchler, CF Higgins, eds, ABC Proteins—From Bacteria to Man. Academic Press, London, pp 47–61
- Decottignies A, Goffeau A (1997) Complete inventory of the yeast ABC proteins. Nat Genet 15: 137–145 [DOI] [PubMed] [Google Scholar]
- Delhaize E, Kataoka T, Hebb DM, White RG, Ryan PR (2003) Genes encoding proteins of the cation diffusion facilitator family that confer manganese tolerance. Plant Cell 15: 1131–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiDonato RJ Jr, Roberts LA, Sanderson T, Eisley RB, Walker EL (2004) Arabidopsis Yellow Stripe-Like2 (YSL2): a metal-regulated gene encoding a plasma membrane transporter of nicotianamine-metal complexes. Plant J 39: 403–414 [DOI] [PubMed] [Google Scholar]
- Drager DB, Desbrosses-Fonrouge AG, Krach C, Chardonnens AN, Meyer RC, Saumitou-Laprade P, Kramer U (2004) Two genes encoding Arabidopsis halleri MTP1 metal transport proteins co-segregate with zinc tolerance and account for high MTP1 transcript levels. Plant J 39: 425–439 [DOI] [PubMed] [Google Scholar]
- Eide DJ (1998) The molecular biology of metal ion transport in Saccharomyces cerevisiae. Annu Rev Nutr 18: 441–469 [DOI] [PubMed] [Google Scholar]
- Eide DJ (2003) Multiple regulatory mechanisms maintain zinc homeostasis in Saccharomyces cerevisiae. J Nutr 133: 1532S–1535S [DOI] [PubMed] [Google Scholar]
- Eide DJ (2004) The SLC39 family of metal ion transporters. Pflugers Arch 447: 796–800 [DOI] [PubMed] [Google Scholar]
- Ellis CD, Wang F, MacDiarmid CW, Clark S, Lyons T, Eide DJ (2004) Zinc and the Msc2 zinc transporter protein are required for endoplasmic reticulum function. J Cell Biol 166: 325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes JR, Gros P (2001) Divalent-metal transport by NRAMP proteins at the interface of host-pathogen interactions. Trends Microbiol 9: 397–403 [DOI] [PubMed] [Google Scholar]
- Franzen LG, Rochaix JD, von Heijne G (1990) Chloroplast transit peptides from the green alga Chlamydomonas reinhardtii share features with both mitochondrial and higher plant chloroplast presequences. FEBS Lett 260: 165–168 [DOI] [PubMed] [Google Scholar]
- Fuhrmann M (2002) Expanding the molecular toolkit for Chlamydomonas reinhardtii—from history to new frontiers. Protist 153: 357–364 [DOI] [PubMed] [Google Scholar]
- Gaither LA, Eide DJ (2001) Eukaryotic zinc transporters and their regulation. Biometals 14: 251–270 [DOI] [PubMed] [Google Scholar]
- Ghosh M, Shen J, Rosen BP (1999) Pathways of As(III) detoxification in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 96: 5001–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravot A, Lieutaud A, Verret F, Auroy P, Vavasseur A, Richaud P (2004) AtHMA3, a plant P(1B)-ATPase, functions as a Cd/Pb transporter in yeast. FEBS Lett 561: 22–28 [DOI] [PubMed] [Google Scholar]
- Gross W (2000) Ecophysiology of algae living in highly acidic environments. Hydrobiologia 1–3: 31–37 [Google Scholar]
- Grossman AR (2000) Chlamydomonas reinhardtii and photosynthesis: genetics to genomics. Curr Opin Plant Biol 3: 132–137 [DOI] [PubMed] [Google Scholar]
- Grossman AR, Harris EE, Hauser C, Lefebvre PA, Martinez D, Rokhsar D, Shrager J, Silflow CD, Stern D, Vallon O, et al (2003) Chlamydomonas reinhardtii at the crossroads of genomics. Eukaryot Cell 2: 1137–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueldry O, Lazard M, Delort F, Dauplais M, Grigoras I, Blanquet S, Plateau P (2003) Ycf1p-dependent Hg(II) detoxification in Saccharomyces cerevisiae. Eur J Biochem 270: 2486–2496 [DOI] [PubMed] [Google Scholar]
- Guerinot ML (2000) The ZIP family of metal transporters. Biochim Biophys Acta 1465: 190–198 [DOI] [PubMed] [Google Scholar]
- Guerinot ML, Eide D (1999) Zeroing in on zinc uptake in yeast and plants. Curr Opin Plant Biol 2: 244–249 [DOI] [PubMed] [Google Scholar]
- Guerinot ML, Yi Y (1994) Iron:nutritious, noxious, and not readily available. Plant Physiol 104: 815–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JL, Williams LE (2003) Transition metal transporters in plants. J Exp Bot 54: 2601–2613 [DOI] [PubMed] [Google Scholar]
- Hanikenne M (2003) Chlamydomonas reinhardtii as a eukaryotic photosynthetic model for studies of heavy metal homeostasis and tolerance. New Phytol 159: 331–340 [DOI] [PubMed] [Google Scholar]
- Hanikenne M, Matagne RF, Loppes R (2001) Pleiotropic mutants hypersensitive to heavy metals and to oxidative stress in Chlamydomonas reinhardtii. FEMS Microbiol Lett 196: 107–111 [DOI] [PubMed] [Google Scholar]
- Hanikenne M, Motte P, Wu MCS, Wang T, Loppes R, Matagne RF (2005) A mitochondrial half-size ABC transporter is involved in cadmium tolerance in Chlamydomonas reinhardtii. Plant Cell Environ (in press)
- Harris EH (1989) The Chlamydomonas Sourcebook. A Comprehensive Guide to Biology and Laboratory Use. Academic Press, New York [DOI] [PubMed]
- Harris EH (2001) Chlamydomonas as a model organism. Annu Rev Plant Physiol Plant Mol Biol 52: 363–406 [DOI] [PubMed] [Google Scholar]
- Herbik A, Bolling C, Buckhout TJ (2002) The involvement of a multicopper oxidase in iron uptake by the green algae Chlamydomonas reinhardtii. Plant Physiol 130: 2039–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschi KD, Korenkov VD, Wilganowski NL, Wagner GJ (2000) Expression of Arabidopsis CAX2 in tobacco. Altered metal accumulation and increased manganese tolerance. Plant Physiol 124: 125–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschi KD, Zhen RG, Cunningham KW, Rea PA, Fink GR (1996) CAX1, an H+/Ca2+ antiporter from Arabidopsis. Proc Natl Acad Sci USA 93: 8782–8786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland IB, Cole SPC, Kuchler K, Higgins CF (2003) ABC Proteins: From Bacteria to Man. Academic Press, London
- Howe G, Merchant S (1992) Heavy metal-activated synthesis of peptides in Chlamydomonas reinhardtii. Plant Physiol 98: 127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Lau KWK, Wu M (2001) Cadmium sequestration in Chlamydomonas reinhardtii. Plant Sci 161: 987–996 [Google Scholar]
- Hussain D, Haydon MJ, Wang Y, Wong E, Sherson SM, Young J, Camakaris J, Harper JF, Cobbett CS (2004) P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell 16: 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im CS, Grossman AR (2002) Identification and regulation of high light-induced genes in Chlamydomonas reinhardtii. Plant J 30: 301–313 [DOI] [PubMed] [Google Scholar]
- Kathir P, LaVoie M, Brazelton WJ, Haas NA, Lefebvre PA, Silflow CD (2003) Molecular map of the Chlamydomonas reinhardtii nuclear genome. Eukaryot Cell 2: 362–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerfeld CA, Krogmann DW (1998) Photosynthetic cytochrome c in cyanobacteria, algae, and plants. Annu Rev Plant Physiol Plant Mol Biol 49: 397–425 [DOI] [PubMed] [Google Scholar]
- Kim D, Gustin JL, Lahner B, Persans MW, Baek D, Yun DJ, Salt DE (2004) The plant CDF family member TgMTP1 from the Ni/Zn hyperaccumulator Thlaspi goesingense acts to enhance efflux of Zn at the plasma membrane when expressed in Saccharomyces cerevisiae. Plant J 39: 237–251 [DOI] [PubMed] [Google Scholar]
- Kispal G, Csere P, Guiard B, Lill R (1997) The ABC transporter Atm1p is required for mitochondrial iron homeostasis. FEBS Lett 418: 346–350 [DOI] [PubMed] [Google Scholar]
- Kispal G, Csere P, Prohl C, Lill R (1999) The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J 18: 3981–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein M, Mamnun YM, Eggmann T, Schuller C, Wolfger H, Martinoia E, Kuchler K (2002) The ATP-binding cassette (ABC) transporter Bpt1p mediates vacuolar sequestration of glutathione conjugates in yeast. FEBS Lett 520: 63–67 [DOI] [PubMed] [Google Scholar]
- Koike S, Inoue H, Mizuno D, Takahashi M, Nakanishi H, Mori S, Nishizawa NK (2004) OsYSL2 is a rice metal-nicotianamine transporter that is regulated by iron and expressed in the phloem. Plant J 39: 415–424 [DOI] [PubMed] [Google Scholar]
- Kolukisaoglu HU, Bovet L, Klein M, Eggmann T, Geisler M, Wanke D, Martinoia E, Schulz B (2002) Family business: the multidrug-resistance related protein (MRP) ABC transporter genes in Arabidopsis thaliana. Planta 216: 107–119 [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Kuroiwa T (1998) The primitive red algae Cyanidium caldarium and Cyanidioschyzon merolae as model system for investigating the dividing apparatus of mitochondria and plastids. Bioessays 20: 344–354 [Google Scholar]
- Kushnir S, Babiychuk E, Storozhenko S, Davey M, Papenbrock J, De Rycke RR, Engler G, Stephan U, Lange H, Kispal G, et al (2001) A mutation of the mitochondrial ABC transporter Sta1 leads to dwarfism and chlorosis in the Arabidopsis mutant starik. Plant Cell 13: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Fontaine S, Quinn JM, Nakamoto SS, Page MD, Gohre V, Moseley JL, Kropat J, Merchant S (2002) Copper-dependent iron assimilation pathway in the model photosynthetic eukaryote Chlamydomonas reinhardtii. Eukaryot Cell 1: 736–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasat MM, Pence NS, Garvin DF, Ebbs SD, Kochian LV (2000) Molecular physiology of zinc transport in the Zn hyperaccumulator Thlaspi caerulescens. J Exp Bot 51: 71–79 [PubMed] [Google Scholar]
- Lefebvre PA, Silflow CD (1999) Chlamydomonas: the cell and its genomes. Genetics 151: 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Kaplan J (1997) Characterization of two homologous yeast genes that encode mitochondrial iron transporters. J Biol Chem 272: 28485–28493 [DOI] [PubMed] [Google Scholar]
- Li ZS, Lu YP, Zhen RG, Szczypka M, Thiele DJ, Rea PA (1997) A new pathway for vacuolar cadmium sequestration in Saccharomyces cerevisiae: YCF1-catalyzed transport of bis(glutathionato) cadmium. Proc Natl Acad Sci USA 94: 42–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk EE, Culotta VC (2001) Manganese superoxide dismutase in Saccharomyces cerevisiae acquires its metal co-factor through a pathway involving the Nramp metal transporter, Smf2p. J Biol Chem 276: 47556–47562 [DOI] [PubMed] [Google Scholar]
- MacDiarmid CW, Gaither LA, Eide D (2000) Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J 19: 2845–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie B, Hediger MA (2004) SLC11 family of H+-coupled metal-ion transporters NRAMP1 and DMT1. Pflugers Arch 447: 571–579 [DOI] [PubMed] [Google Scholar]
- Martinoia E, Klein M, Geisler M, Bovet L, Forestier C, Kolukisaoglu U, Muller-Rober B, Schulz B (2002) Multifunctionality of plant ABC transporters—more than just detoxifiers. Planta 214: 345–355 [DOI] [PubMed] [Google Scholar]
- Maser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, Talke IN, Amtmann A, Maathuis FJ, Sanders D, et al (2001) Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol 126: 1646–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M, Misumi O, Shin IT, Maruyama S, Takahara M, Miyagishima SY, Mori T, Nishida K, Yagisawa F, Yoshida Y, et al (2004) Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428: 653–657 [DOI] [PubMed] [Google Scholar]
- McGowan SJ, Gorham HC, Hodgson DA (1993) Light-induced carotenogenesis in Myxococcus xanthus: DNA sequence analysis of the carR region. Mol Microbiol 10: 713–735 [DOI] [PubMed] [Google Scholar]
- McKie AT, Marciani P, Rolfs A, Brennan K, Wehr K, Barrow D, Miret S, Bomford A, Peters TJ, Farzaneh F, et al (2000) A novel duodenal iron-regulated transporter, IREG1, implicated in the basolateral transfer of iron to the circulation. Mol Cell 5: 299–309 [DOI] [PubMed] [Google Scholar]
- Merchant S (1998) Synthesis of metalloproteins involved in photosynthesis: plastocyanin and cytochromes. In JD Rochaix, M Goldschmidt-Clermont, S Merchant, eds, The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 597–611
- Mills RF, Krijger GC, Baccarini PJ, Hall JL, Williams LE (2003) Functional expression of AtHMA4, a P1B-type ATPase of the Zn/Co/Cd/Pb subclass. Plant J 35: 164–176 [DOI] [PubMed] [Google Scholar]
- Mitsuhashi N, Miki T, Senbongi H, Yokoi N, Yano H, Miyazaki M, Nakajima N, Iwanaga T, Yokoyama Y, Shibata T, et al (2000) MTABC3, a novel mitochondrial ATP-binding cassette protein involved in iron homeostasis. J Biol Chem 275: 17536–17540 [DOI] [PubMed] [Google Scholar]
- Moreira D, Le Guyader H, Philippe H (2000) The origin of red algae and the evolution of chloroplasts. Nature 405: 69–72 [DOI] [PubMed] [Google Scholar]
- Moreira D, Philippe H (2001) Sure facts and open questions about the origin and evolution of photosynthetic plastids. Res Microbiol 152: 771–780 [DOI] [PubMed] [Google Scholar]
- Moseley JL, Allinger T, Herzog S, Hoerth P, Wehinger E, Merchant S, Hippler M (2002. a) Adaptation to Fe-deficiency requires remodeling of the photosynthetic apparatus. EMBO J 21: 6709–6720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley JL, Page MD, Alder NP, Eriksson M, Quinn J, Soto F, Theg SM, Hippler M, Merchant S (2002. b) Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell 14: 673–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaka S, Nishizawa NK, Mori S, Yoshimura EY (2004) Metal metabolism in the red alga Cyanidium caldarium and its relationship to metal tolerance. Biometals 17: 177–181 [DOI] [PubMed] [Google Scholar]
- Nagasaka S, Nishizawa NK, Watanabe T, Mori S, Yoshimura E (2003) Evidence that electron-dense bodies in Cyanidium caldarium have an iron-storage role. Biometals 16: 465–470 [DOI] [PubMed] [Google Scholar]
- Nies DH (2003) Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol Rev 27: 313–339 [DOI] [PubMed] [Google Scholar]
- Nozaki H, Matsuzaki M, Takahara M, Misumi O, Kuroiwa H, Hasegawa M, Shin-i T, Kohara Y, Ogasawara N, Kuroiwa T (2003) The phylogenetic position of red algae revealed by multiple nuclear genes from mitochondria-containing eukaryotes and an alternative hypothesis on the origin of plastids. J Mol Evol 56: 485–497 [DOI] [PubMed] [Google Scholar]
- Ohta N, Matsuzaki M, Misumi O, Miyagishima SY, Nozaki H, Tanaka K, Shin IT, Kohara Y, Kuroiwa T (2003) Complete sequence and analysis of the plastid genome of the unicellular red alga Cyanidioschyzon merolae. DNA Res 10: 67–77 [DOI] [PubMed] [Google Scholar]
- Ohta N, Sato N, Kuroiwa T (1998) Structure and organization of the mitochondrial genome of the unicellular red alga Cyanidioschyzon merolae deduced from the complete nucleotide sequence. Nucleic Acids Res 26: 5190–5198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz DF, Kreppel L, Speiser DM, Scheel G, McDonald G, Ow DW (1992) Heavy metal tolerance in the fission yeast requires an ATP-binding cassette-type vacuolar membrane transporter. EMBO J 11: 3491–3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz DF, Ruscitti T, McCue KF, Ow DW (1995) Transport of metal-binding peptides by HMT1, a fission yeast ABC-type vacuolar membrane protein. J Biol Chem 270: 4721–4728 [DOI] [PubMed] [Google Scholar]
- Ott FD, Seckbach J (1994) A review on the taxonomic position of the algal genus Cyanidium Geitler 1933 and its ecological cohorts Galdieria Merola in Merola et al. 1981 and Cyanidioschyzon De Luca, Taddei and Varano 1978. In J Seckbach, ed, Evolutionary Pathways and Enigmatic Algae: Cyanidium caldarium (Rhodophyta) and Related Cells. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 113–132
- Palmiter RD, Huang L (2004) Efflux and compartmentalization of zinc by members of the SLC30 family of solute carriers. Pflugers Arch 447: 744–751 [DOI] [PubMed] [Google Scholar]
- Paulsen IT, Saier MH Jr (1997) A novel family of ubiquitous heavy metal ion transport proteins. J Membr Biol 156: 99–103 [DOI] [PubMed] [Google Scholar]
- Pence NS, Larsen PB, Ebbs SD, Letham DL, Lasat MM, Garvin DF, Eide D, Kochian LV (2000) The molecular physiology of heavy metal transport in the Zn/Cd hyperaccumulator Thlaspi caerulescens. Proc Natl Acad Sci USA 97: 4956–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persans MW, Nieman K, Salt DE (2001) Functional activity and role of cation-efflux family members in Ni hyperaccumulation in Thlaspi goesingense. Proc Natl Acad Sci USA 98: 9995–10000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petris MJ (2004) The SLC31 (Ctr) copper transporter family. Pflugers Arch 447: 752–755 [DOI] [PubMed] [Google Scholar]
- Price NT, Smith AJ, Sykes AG, Rogers LJ (1991) Cytochrome c-553 from two species of macroalgae. Phytochemistry 30: 2845–2848 [Google Scholar]
- Radisky D, Kaplan J (1999) Regulation of transition metal transport across the yeast plasma membrane. J Biol Chem 274: 4481–4484 [DOI] [PubMed] [Google Scholar]
- Rea PA, Li ZS, Lu YP, Drozdowicz YM, Martinoia E (1998) From vacuolar GS-X pumps to multispecific ABC transporters. Annu Rev Plant Physiol Plant Mol Biol 49: 727–760 [DOI] [PubMed] [Google Scholar]
- Rea PA, Sanchez-Fernandez R, Chen S, Peng M, Klein M, Geisler M, Martinoia E (2003) The plant ABC transporter superfamily: the functions of a few and identities of many. In IB Holland, SPC Cole, K Kuchler, CF Higgins, eds, ABC Proteins—From Bacteria to Man. Academic Press, London, pp 335–355
- Roberts LA, Pierson AJ, Panaviene Z, Walker EL (2004) Yellow stripe1. Expanded roles for the maize iron-phytosiderophore transporter. Plant Physiol 135: 112–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix JD, Goldschmidt-Clermont M, Merchant S (1998) The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas, Vol 7. Kluwer Academic Publishers, Dordrecht, The Netherlands
- Rosakis A, Koster W (2004) Transition metal transport in the green microalga Chlamydomonas reinhardtii—genomic sequence analysis. Res Microbiol 155: 201–210 [DOI] [PubMed] [Google Scholar]
- Salt DE, Rauser WE (1995) MgATP-dependent transport of phytochelatins across the tonoplast of oat roots. Plant Physiol 107: 1293–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancenon V, Puig S, Mateu-Andres I, Dorcey E, Thiele DJ, Penarrubia L (2004) The Arabidopsis copper transporter COPT1 functions in root elongation and pollen development. J Biol Chem 279: 15348–15355 [DOI] [PubMed] [Google Scholar]
- Sancenon V, Puig S, Mira H, Thiele DJ, Penarrubia L (2003) Identification of a copper transporter family in Arabidopsis thaliana. Plant Mol Biol 51: 577–587 [DOI] [PubMed] [Google Scholar]
- Sanchez-Fernandez R, Davies TG, Coleman JO, Rea PA (2001) The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J Biol Chem 276: 30231–30244 [DOI] [PubMed] [Google Scholar]
- Sandmann G, Reck H, Kessler E, Boger P (1983) Distribution of plastocyanin and soluble plastidic cytochrome c in various classes of algae. Arch Microbiol 134: 23–27 [Google Scholar]
- Schaaf G, Ludewig U, Erenoglu BE, Mori S, Kitahara T, von Wiren N (2004) ZmYS1 functions as a proton-coupled symporter for phytosiderophore- and nicotianamine-chelated metals. J Biol Chem 279: 9091–9096 [DOI] [PubMed] [Google Scholar]
- Seckbach J (1994) The natural history of Cyanidium (Geitler 1933): past and present perspectives. In J Seckbach, ed, Evolutionary Pathways and Enigmatic Algae: Cyanidium caldarium (Rhodophyta) and Related Cells. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 99–112
- Seckbach J, Ott FD (1994) Systematic position and phylogenetic status of Cyanidium Geitler 1933. In J Seckbach, ed, Evolutionary Pathways and Enigmatic Algae: Cyanidium caldarium (Rhodophyta) and Related Cells. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 133–143
- Sharma KG, Mason DL, Liu G, Rea PA, Bachhawat AK, Michaelis S (2002) Localization, regulation, and substrate transport properties of Bpt1p, a Saccharomyces cerevisiae MRP-type ABC transporter. Eukaryot Cell 1: 391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul O, Hilgemann DW, de-Almeida-Engler J, Van Montagu M, Inz D, Galili G (1999) Cloning and characterization of a novel Mg2+/H+ exchanger. EMBO J 18: 3973–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T, Muller-Moule P, Munekage Y, Niyogi KK, Pilon M (2003) PAA1, a P-type ATPase of Arabidopsis, functions in copper transport in chloroplasts. Plant Cell 15: 1333–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrager J, Hauser CR, Chang C-W, Harris EH, Davies JP, McDermott J, Tamse R, Zhang Z, Grossman A (2003) Chlamydomonas reinhardtii genome project. A guide to the generation and use of the cDNA information. Plant Physiol 131: 401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silflow CD (1998) Organization of the nuclear genome. In JD Rochaix, M Goldschmidt-Clermont, S Merchant, eds, The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 25–40
- Silflow CD, Lefebvre PA (2001) Assembly and motility of eukaryotic cilia and flagella. Lessons from Chlamydomonas reinhardtii. Plant Physiol 127: 1500–1507 [PMC free article] [PubMed] [Google Scholar]
- Song WY, Ju Sohn E, Martinoia E, Jik Lee Y, Yang YY, Jasinski M, Forestier C, Hwang I, Lee Y (2003) Engineering tolerance and accumulation of lead and cadmium in transgenic plants. Nat Biotechnol 21: 914–919 [DOI] [PubMed] [Google Scholar]
- Supek F, Supekova L, Nelson H, Nelson N (1996) A yeast manganese transporter related to the macrophage protein involved in conferring resistance to mycobacteria. Proc Natl Acad Sci USA 93: 5105–5110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczypka MS, Wemmie JA, Moye-Rowley WS, Thiele DJ (1994) A yeast metal resistance protein similar to human cystic fibrosis transmembrane conductance regulator (CFTR) and multidrug resistance- associated protein. J Biol Chem 269: 22853–22857 [PubMed] [Google Scholar]
- Taylor KM, Morgan HE, Johnson A, Hadley LJ, Nicholson RI (2003) Structure-function analysis of LIV-1, the breast cancer-associated protein that belongs to a new subfamily of zinc transporters. Biochem J 375: 51–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KM, Morgan HE, Johnson A, Nicholson RI (2004) Structure-function analysis of HKE4, a member of the new LIV-1 subfamily of zinc transporters. Biochem J 377: 131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KM, Nicholson RI (2003) The LZT proteins; the LIV-1 subfamily of zinc transporters. Biochim Biophys Acta 1611: 16–30 [DOI] [PubMed] [Google Scholar]
- Thomine S, Lelievre F, Debarbieux E, Schroeder JI, Barbier-Brygoo H (2003) AtNRAMP3, a multispecific vacuolar metal transporter involved in plant responses to iron deficiency. Plant J 34: 685–695 [DOI] [PubMed] [Google Scholar]
- Thomine S, Wang R, Ward JM, Crawford NM, Schroeder JI (2000) Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc Natl Acad Sci USA 97: 4991–4996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommasini R, Vogt E, Fromenteau M, Hortensteiner S, Matile P, Amrhein N, Martinoia E (1998) An ABC-transporter of Arabidopsis thaliana has both glutathione-S conjugate and chlorophyll catabolite transport activity. Plant J 13: 773–780 [DOI] [PubMed] [Google Scholar]
- van den Hoek C, Mann DG, Jahns HM (1995) Algae. An Introduction to Phycology. Cambridge University Press, Cambridge, UK
- van der Zaal BJ, Neuteboom LW, Pinas JE, Chardonnens AN, Schat H, Verkleij JA, Hooykaas PJ (1999) Overexpression of a novel Arabidopsis gene related to putative zinc-transporter genes from animals can lead to enhanced zinc resistance and accumulation. Plant Physiol 119: 1047–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vögeli-Lange R, Wagner GJ (1990) Subcellular localization of cadmium and cadmium-binding peptides in tobacco leaves. Implication of a transport function for cadmium-binding peptides. Plant Physiol 82: 1086–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Harada E, Vess C, Roepenack-Lahaye EV, Clemens S (2004) Comparative microarray analysis of Arabidopsis thaliana and Arabidopsis halleri roots identifies nicotianamine synthase, a ZIP transporter and other genes as potential metal hyperaccumulation factors. Plant J 37: 251–268 [DOI] [PubMed] [Google Scholar]
- Williams LE, Pittman JK, Hall JL (2000) Emerging mechanisms for heavy metal transport in plants. Biochim Biophys Acta 1465: 104–126 [DOI] [PubMed] [Google Scholar]
- Yoon HS, Hackett JD, Pinto G, Bhattacharya D (2002) The single, ancient origin of chromist plastids. Proc Natl Acad Sci USA 99: 15507–15512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Eide D (1996. a) The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc Natl Acad Sci USA 93: 2454–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Eide D (1996. b) The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J Biol Chem 271: 23203–23210 [DOI] [PubMed] [Google Scholar]
- Zhou B, Gitschier J (1997) hCTR1: a human gene for copper uptake identified by complementation in yeast. Proc Natl Acad Sci USA 94: 7481–7486 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.