ABSTRACT
Pathogens must adapt to disparate environments in permissive host species, a feat that is especially pronounced for vector-borne microbes, which transition between vertebrate hosts and arthropod vectors to complete their lifecycles. Most knowledge about arthropod-vectored bacterial pathogens centers on their life in the mammalian host, where disease occurs. However, disease outbreaks are driven by the arthropod vectors. Adapting to the arthropod is critical for obligate intracellular rickettsial pathogens, as they depend on eukaryotic cells for survival. To manipulate the intracellular environment, these bacteria use type IV secretion systems (T4SS) to deliver effectors into the host cell. To date, few rickettsial T4SS translocated effectors have been identified and have only been examined in the context of mammalian infection. We identified an effector from the tick-borne rickettsial pathogen Anaplasma phagocytophilum, HGE1_02492, as critical for survival in tick cells and acquisition by ticks in vivo. Conversely, HGE1_02492 was dispensable during mammalian cell culture and murine infection. We show that HGE1_02492 is translocatable in a T4SS-dependent manner to the host cell cytosol. In eukaryotic cells, the HGE1_02492 localized with cortical actin filaments, which is dependent on multiple sub-domains of the protein. HGE1_02492 is the first arthropod-vector specific T4SS translocated effector identified from a rickettsial pathogen. Moreover, the subcellular target of HGE1_02492 suggests that A. phagocytophilum is manipulating actin to enable arthropod colonization. Based on these findings, we propose the name AteA for Anaplasma (phagocytophilum) tick effector A. Altogether, we show that A. phagocytophilum uses distinct strategies to cycle between mammals and arthropods.
IMPORTANCE
Ticks are the number one vector of pathogens for livestock worldwide and for humans in the United States. The biology of tick transmission is an understudied area. Understanding this critical interaction could provide opportunities to affect the course of disease spread. In this study, we examined the zoonotic tick-borne agent Anaplasma phagocytophilum and identified a secreted protein, AteA, which is expressed in a tick-specific manner. These secreted proteins, termed effectors, are the first proteins to interact with the host environment. AteA is essential for survival in ticks and appears to interact with cortical actin. Most effector proteins are studied in the context of the mammalian host; however, understanding how this unique set of proteins affects tick transmission is critical to developing interventions.
KEYWORDS: rickettsia, Anaplasma, obligate intracellular, effector functions, vector-borne diseases, actin, host-pathogen interactions
INTRODUCTION
Multi-host pathogens often have specific adaptation mechanisms to survive in disparate environments (1). For example, vector-borne microbes must adapt and survive in both vertebrate hosts and arthropod vectors (2). These two environments differ significantly with discrepancies in body temperature, nutrient availability, cell types infected, physiological architecture, and immunological pressures (2). Most of our understanding of tick-borne pathogens focuses on interactions with mammalian hosts, as this is where disease occurs. However, the mammal represents only half of the lifecycle for tick borne pathogens. In contrast, little is known about how tick-vectored pathogens mediate interactions with the arthropod. With over 680 million years of evolution separating ticks from mammals (3), our understanding of mammal-pathogen interactions cannot simply be transposed onto ticks (2).
Adapting to different environments is especially critical for obligate intracellular rickettsial pathogens, which are intimately dependent on both arthropod and vertebrate host cells. One of the most common tick-borne rickettsial human pathogens in the United States is Anaplasma phagocytophilum, which causes human granulocytic anaplasmosis (4). To complete its lifecycle, A. phagocytophilum must transit between Ixodes scapularis ticks and mammalian hosts. Interestingly, A. phagocytophilum host cell tropisms are not equivalent between mammals and ticks. In mammals, the bacterium preferentially infects neutrophils. In contrast, A. phagocytophilum must infect and traverse the tick midgut, travel through the hemocoel, and infect the salivary glands of the arthropod (5, 6). Given the disparities in the host environment and cell tropisms, it is expected that A. phagocytophilum would have unique expression profiles adapted for each situation. Transcriptional studies demonstrated that A. phagocytophilum differentially transcribes 41% of its genes when growth in tick cells was compared to mammalian cells (7, 8). Transposon mutagenesis found that many A. phagocytophilum genes are dispensable for growth in the human monocyte cell line HL60 (9), and several of these same genes are upregulated during tick cell infection. Altogether, this suggests that the tick-specific genes may be involved in arthropod-pathogen interactions (7, 8).
Rickettsial pathogens primarily manipulate host cell biology through effector proteins that are delivered to the host cytosol with a type 4 secretion system (T4SS) (10, 11). T4SS effector molecules subvert host cell defenses and modulate a wide variety of host cell processes (12 – 19). A common target of such effectors can be the actin cytoskeleton, which forms the structural scaffolding of the host cell (20). When compared to the model intracellular bacterium Legionella pneumophila (1), relatively little is known about the effector repertoire from A. phagocytophilum and other rickettsial pathogens. Only five A. phagocytophilum T4SS translocated proteins have been identified to date, and none in the context of tick colonization (16, 17, 21 – 23). The machine learning algorithm OPT4e predicts that A. phagocytophilum encodes 48 candidate effectors (24). Fifteen of these candidate genes show unique expression patterns during mammalian and tick cell infection (7, 8). Putative effector HGE1_02492 (APH_0546 in A. phagocytophilum strain HZ) demonstrated the highest transcriptional increase when grown in tick cell culture, relative to mammalian cells. Herein, we show that HGE1_02492 is critical for growth in tick cells and colonization of ticks in vivo. Further, HGE1_02492 is deliverable by the T4SS into host cell cytosol, where it associates with cortical actin filaments, through multiple sub-domains of the protein. Based on our findings, we propose the name AteA, for Anaplasma (phagocytophilum) tick effector A, and will henceforth refer to HGE1_02492 as AteA. Altogether, we have identified and characterized the first arthropod-vector specific effector from A. phagocytophilum, which targets the eukaryotic cytoskeleton.
RESULTS
A protein of 1,103 amino acids (~120 kDa) is encoded by ateA (HGE1_02492) and appears to be unique to A. phagocytophilum. It has 100% sequence identity with APH_0546 from a different strain of A. phagocytophilum, strain HZ. In silico analysis of AteA predicts that most of the protein is highly disordered, with an N-terminal globular domain (25) and there are two regions containing tandem repeats starting at amino acids 431 and 702 (26).
Expression of ateA is specific to growth in tick cells
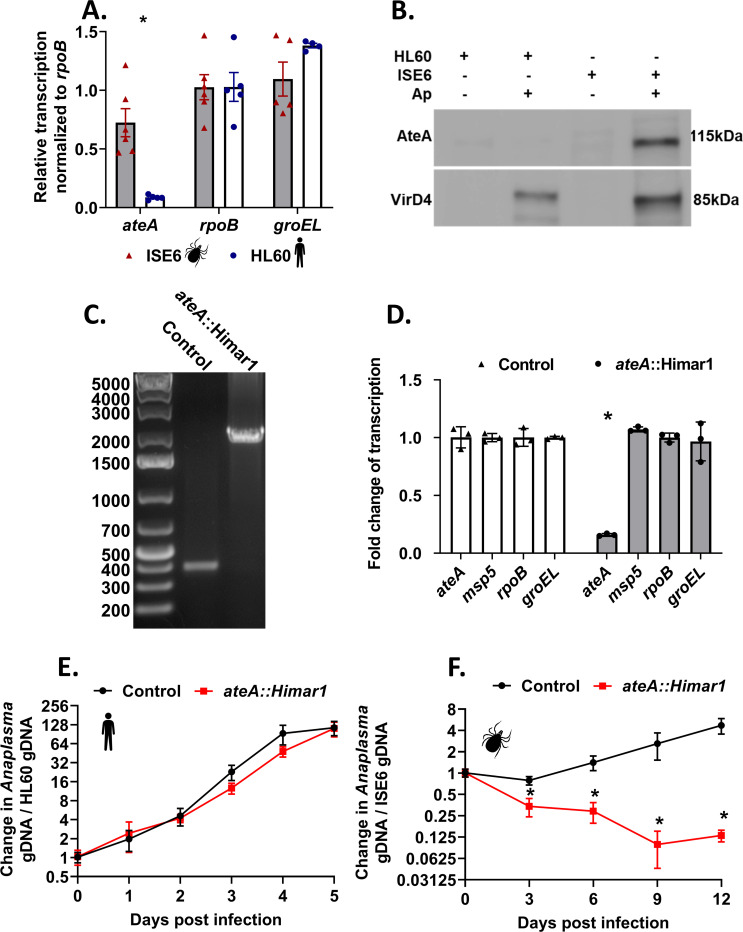
Previous transcriptomics studies using tiling arrays demonstrated that ateA was upregulated during A. phagocytophilum culture in ISE6 tick cells and was minimally expressed during growth in the human monocyte-like cell line HL60 (7, 8). Repetitive sequences, like those found in ateA, can artifactually inflate transcriptional signals in tiling arrays. We, therefore, used qRT-PCR with primers targeting non-repetitive sequences to quantify ateA transcription patterns from A. phagocytophilum grown in HL60 and ISE6 cells. Housekeeping A. phagocytophilum genes, rpoB and groEL, are equally expressed when grown in either mammalian or tick cell lines (7, 27), and were, therefore, used as baseline controls for expression. Expression of ateA was >8-fold higher during growth in tick cells than in mammalian cells as measured by qPCR (Fig. 1A). To examine AteA protein production, mock-infected and peak wild-type A. phagocytophilum infected human HL60 and tick ISE6 cells were examined by western blot for AteA and the T4SS protein VirD4. While VirD4 was detectable during both mammalian and tick cell infection, the AteA protein was only detected from the infected ISE6 lysates (Fig. 1B). Our findings demonstrate that AteA is specifically produced during tick cell infection.
Fig 1.
ateA is essential for A. phagocytophilum survival in tick cells, but dispensable within human cells. (A) A. phagocytophilum gene expression during growth within tick ISE6 and human HL60 cells. Transcription of ateA and housekeeping genes rpoB and groEL. (B) α-AteA (top) and α-VirD4 (bottom) western blot analysis of mock-infected and peak wild-type A. phagocytophilum infected human HL60 and tick ISE6 cells. Lanes were equally loaded with 1 × 105 host cells. (C) Agarose gel electrophoresis of PCR amplicons flanking the transposon insertion site in the ateA gene. (D) Transcriptional analysis from A. phagocytophilum transposon mutants during culture with tick ISE6 cells. Mutants contain the transposon inserted in a neutral intergenic location (control), or within the ateA gene. Transcription measured by qRT-PCR of ateA, msp5, rpoB, and groEL. Transcription normalized to rpoB. Results shown are the mean of three biological replicates with two technical replicates each ±SD. *P < 0.05 (Student’s t-test). (E and F) Growth of A. phagocytophilum ateA or control strain in panel E human HL60 cells and (F) tick ISE6 cells. Bacterial burden was measured as Anaplasma gDNA vs host cell gDNA via qPCR. Data shown are the mean of three biological replicates with two technical replicates each ±SD and is representative of three experimental replicates. *P < 0.05 (Mann-Whitney t-test).
A. phagocytophilum survival in tick cells is dependent on ateA expression
The tick-specific expression pattern of ateA led us to ask if it impacts growth in tick cells. For this experiment, we used several tools available to us, including an ateA insertion mutant that we previously isolated from a A. phagocytophilum Himar1 transposon mutant library (9). As a control strain, we used another mutant, which contains the Himar1 transposon in an intergenic location. This strain has been shown to be phenotypically equivalent to wild-type A. phagocytophilum (27, 28). The purity of the ateA::Himar1 mutant was examined by PCR bridging the insertion site. The product amplified from ateA::Himar1 DNA lacked amplicons of the expected size for the wild type (~400 bp) and contained only the larger (>2,000 bp) product reflecting the Himar1 transposon insertion in ateA (Fig. 1C). Our analysis confirmed that transcription of ateA was significantly decreased in the ateA::Himar1 mutant, but transcription of housekeeping genes, rpoB and groEL, and the conserved Anaplasma gene encoding major surface protein 5 (msp5) were all unaffected (Fig. 1B). To test whether the ateA::Himar1 mutation impacted growth in the ISE6 cell line, both HL60 and ISE6 tick cells were infected with the ateA::Himar1 mutant and growth rates were compared to the intergenic transposon control strain. We found that ateA::Himar1 growth in HL60 cells was comparable to the control strain (Fig. 1E), but ateA::Himar1 growth in ISE6 cells was significantly attenuated, indicating ateA is necessary for survival in tick cells (Fig. 1F).
Expression of ateA is necessary for in vivo tick colonization
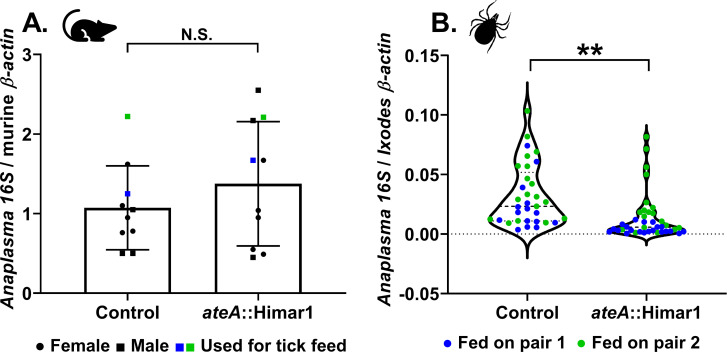
To examine the importance of ateA in vivo, we infected mice with either the control (intergenic transposon strain) or the ateA::Himar1 mutant strain. No colonization defect was observed in mice, indicating that ateA is dispensable during mammalian infection (Fig. 2A). In contrast, larval I. scapularis ticks that fed to repletion on A. phagocytophilum burden-matched mice acquired significantly less of the ateA::Himar1 mutant when compared to the control. This indicates that ateA is critical for A. phagocytophilum colonization of the tick (Fig. 2B).
Fig 2.
ateA is dispensable for murine infection, but mutation attenuates tick acquisition. (A) Anaplasma burden in mouse blood 7 days post infection by intraperitoneal inoculation with 1 × 108 host cell free A. phagocytophilum ateA::Himar1 or control. Blood samples were processed for DNA isolation and bacterial burden was measured by qPCR of A. phagocytophilum 16S rDNA relative to mouse actin by ΔΔCt. Each strain was tested in five male (squares) and five female (circles) mice and samples were tested in duplicate. Mice used for tick feeding are indicated by blue and green symbols. Blue symbols indicate experimental replicate 1. Green symbols indicate experimental replicate 2. (B) Ixodes scapularis larvae were infected by feeding to repletion on A. phagocytophilum burden-matched infected mice. Whole replete I. scapularis larvae were processed for RNA. Bacterial loads were measured by A. phagocytophilum 16S rRNA levels relative to mouse actin via qRT-PCR. Data represent ticks from two burden matched mouse pairs indicated in blue and green for two experimental replicates. Blue symbols indicate experimental replicate 1. Green symbols indicate experimental replicate 2. From each mouse, 17–20 individual ticks were collected as biological replicates, and each qRT-PCR was performed in duplicate. **P < 0.005 (Welch’s t-test).
AteA is a T4SS substrate
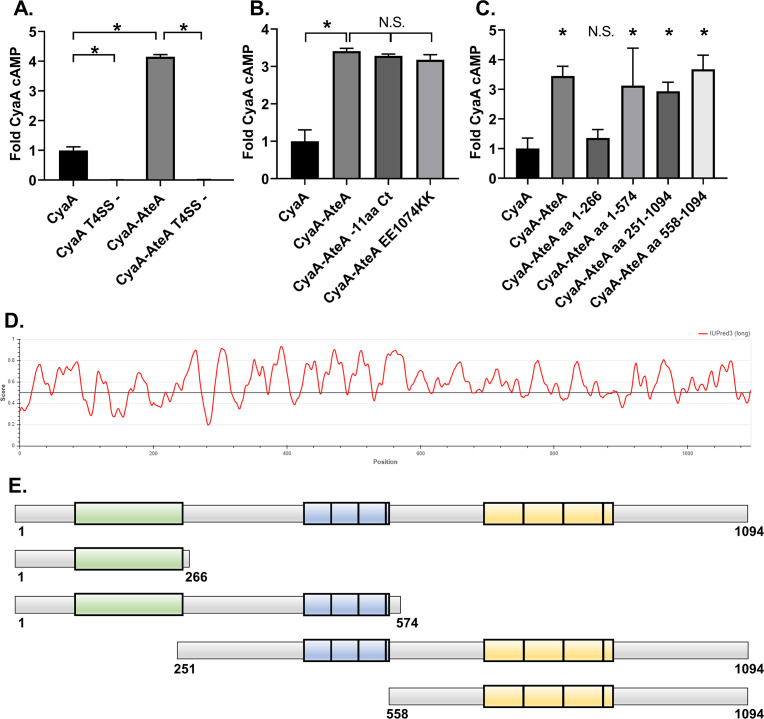
Several translocated effector prediction algorithms (OPT4e [24], S4TE [29], and T4EffPred [30]) predict that AteA is a T4SS substrate. To empirically test if AteA is translocatable by a T4SS, we used a well-established (31) surrogate assay in Legionella pneumophila (32). In this system, the candidate T4SS substrate is fused to adenylate cyclase (CyaA) and expressed in L. pneumophila. Candidate effector translocation is detected by the accumulation of cAMP in host cells during L. pneumophila infection. CyaA-AteA led to significantly greater cAMP than the control (CyaA alone) (Fig. 3A). Secretion was not detected from the T4SS deficient L. pneumophila strain (dotA−), indicating translocation of AteA is T4SS dependent.
Fig 3.
AteA is recognized and secreted by a T4SS. (A–C) THP-1 cells were infected with a L. pneumophila strain expressing the indicated Cya-fusion proteins for 1 h. cAMP concentrations were quantified from infected cell lysates by ELISA and compared as fold change over CyaA alone. (A) CyaA and CyaA-AteA expressed in both wild-type L. pneumophila Lp02 or T4SS-deficient Lp03 strain (T4SS−). (B) C-terminal mutants of CyaA-AteA were expressed in Lp02 and compared to CyaA alone and CyaA-AteA. (C) Truncation constructs of CyaA-AteA diagrammed in panel E were expressed in Lp02 and compared to CyaA alone and CyaA-AteA. (D) IUPred3 order/disorder plot of AteA protein. (E) Diagram of AteA truncation mutants. Potential globular region highlighted in green, two tandem repeat regions highlighted in blue and yellow. (A–C) Error bars represent ±SD of the mean of three biological replicates with two technical replicates each, and graph is representative of two repeated experiments. *P < 0.05 (one-way ANOVA).
A secretion signal common to many T4SS translocated proteins is charged residue at the C-terminus (32, 33). We, therefore, removed 11 C-terminal amino acids from AteA or mutagenized two acidic residues to basic residues in the C-terminus and tested secretion. Neither manipulation strategy affected secretion (Fig. 3B), indicating that a different feature of AteA is being recognized by the T4SS. Intrinsically unstructured regions are another feature common among translocated proteins (34). In silico analysis of AteA predicts that most of the protein is highly disordered, with only the N-terminal portion scoring for a globular structure (25) (Fig. 3D). The disordered region has two prominent tandem repeat segments containing either 40 or 59 amino acid repeat units (26) (Fig. 3E). We tested four large truncation fragments of CyaA-AteA for translocation (Fig. 3E). Truncations retaining a large relative amount of the disordered region were secreted in our assay. The truncation that removed the disordered region, leaving only the N-terminal globular region, was not translocated. Altogether, this suggests that AteA contains multiple internal secretion signals, or that the unstructured nature of the protein itself is being recognized by the T4SS for translocation (Fig. 3C and E).
AteA localizes to the cortical actin cytoskeleton, dependent on multiple domains
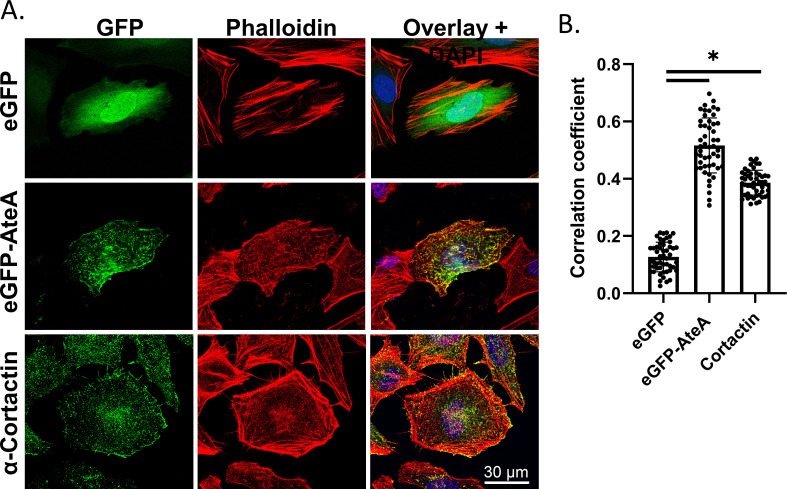
Since AteA is translocated to the host cell, we examined the eukaryotic host cell structures that AteA may be targeting by ectopically expressing a GFP fusion protein (eGFP-AteA) in HeLa cells. Laser-scanning confocal microscopy revealed that eGFP-AteA appeared as branched filamentous structures, resembling the actin cytoskeleton (Fig. 4). Filamentous actin (F-actin) was visualized using fluorescently labeled phalloidin, which revealed that AteA co-localized with actin filaments (Fig. 4A and B). Many pathogens are known to target actin, which alters host cell processes with the goal of promoting replication and survival. Two prominent F-actin morphologies in cells are cortical actin and stress fibers. Cortical actin resembles a branched web of fibers just under the cell surface. Stress fibers appear as linear actin bundles connecting two anchor points across the cell (35). The highly branched appearance of AteA localization resembles the appearance of cortical actin. For comparison, we stained HeLa cell for the cortical-actin binding protein, cortactin, and phalloidin (Fig. 4A and B). The lack of AteA localization with longer linear actin fibers and the similar appearance to cortactin led us to conclude that AteA preferentially associates with the cortical actin network.
Fig 4.
AteA localization resembling cortical actin. (A) Confocal images of HeLa cells transiently transfected to express eGFP or eGFP-AteA (rows 1 and 2). Cortical actin binding protein visualized using α-cortactin mouse antibody and Alexa Fluor 488 anti-mouse. Actin was stained with Alexa Fluor 564 phalloidin 36 h post transfection. (B) Relative co-localization with phalloidin as measured by Pearson’s correlation (n = 20 cells/group). Compared by one-way ANOVA (*P < 0.05 and ns; not significant).
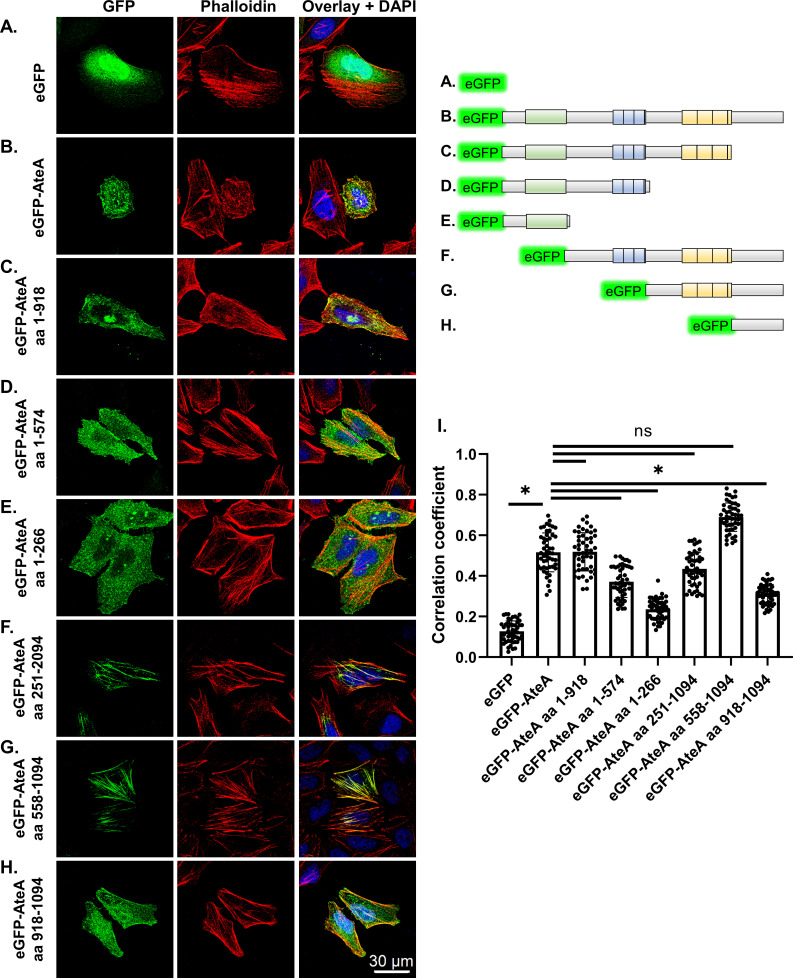
To identify the regions of AteA responsible for its actin co-localization pattern, truncations of the protein were constructed and ectopically expressed in HeLa cells (Fig. 5). Removal of the C-terminal region following the repeat segments (eGFP-AteA aa1-918) did not change the localization pattern relative to the full-length protein (Fig. 5B and C). Truncations that removed the second tandem repeat region (eGFP-AteA aa 1–574) reduced localization with phalloidin (Fig. 5I) and caused dispersed distribution following the topology of the cell surface (Fig. 5D). The N-terminal globular domain alone (eGFP-AteA aa 1–266) showed a similar localization pattern to eGFP-AteA aa 1–574 (Fig. 5E). These results suggested that the second tandem repeat region (residues 703–918) is necessary for localization with actin. When this region is removed, the protein appears to associate with the cell’s cortex. To test how the other regions of AteA impact the localization, we performed the converse experiment, expressing truncations beginning at the N-terminus. AteA lacking the globular N-terminal region (eGFP-AteA aa 251–1094) lost the appearance of highly branched cortical actin but retained association with actin fibers. This indicates that the N-terminal region of AteA is necessary for the cortical localization (Fig. 5F). Interestingly, the actin fibers associated with AteA lacking the N-terminal globular region did not resemble a cortical actin morphology but appeared more branch-like or distorted than typical actin stress fibers (Fig. 5F). Truncations that removed the central region of AteA containing the first set of tandem repeats (eGFP-AteA aa 558–1094) resulted in the loss of the branched pattern altogether. Instead, the protein localized with long linear actin fibers characteristic of stress fibers (Fig. 5G). The C-terminal portion of AteA alone (eGFP-AteA aa 918–1094) did not localize specifically with phalloidin (Fig. 5H1). To examine the AteA specificity to cortical actin, HeLa cells expressing eGFP-AteA and AteA truncations were stained for the cortical actin-binding-protein cortactin (Fig. S1). Only full length AteA (eGFP-AteA) displayed increased co-localization with cortactin relative to eGFP alone (Fig. S1). However, the incomplete correlation between AteA and cortactin localization (Fig. S1G) suggests that AteA localization to cortical actin may not be specific to the protein cortactin. Taken together, our findings indicate that the second tandem repeat region of AteA is responsible for localization to actin fibers, the central region of the protein alters this actin localization pattern, and the N-terminal domain provides specificity to the cortex. Altogether, these regions function in combination to associate AteA with cortical actin.
Fig 5.
Localization to actin is dependent on the second repeat region, but other regions of AteA influence the pattern. (A–H) Upper right: schematic of eGFP-AteA fusion constructs and truncations used in transfections. (A–H) Left: confocal images of HeLa cells transiently transfected to express eGFP, eGFP-AteA, or an eGFP-AteA truncation construct as diagramed. Cells were stained to visualize actin using Alexa Fluor 564 phalloidin at 36–48 h post transfection. (I) Relative co-localization with phalloidin as measured by Pearson’s correlation (n = 50 cells/group from three independent experiments). Compared by one-way ANOVA (*P < 0.05 and ns; not significant).
DISCUSSION
Here, we demonstrate that the A. phagocytophilum gene encoding ateA (i) is highly expressed in the tick environment, (ii) is essential for growth and survival within tick cells, and (iii) is a T4SS translocated substrate that targets the eukaryotic cytoskeleton. Further, ateA is necessary for A. phagocytophilum acquisition by I. scapularis larvae when feeding on an infected host. However, ateA was dispensable for growth in mammalian cell culture, and mutation did not affect bacterial burden in mice. To our knowledge, this is the first description of an arthropod specific rickettsial T4SS translocated effector.
AteA joins the few T4SS effectors identified from A. phagocytophilum (16, 17, 21 – 23). However, machine learning algorithms predict that A. phagocytophilum encodes many more that remain to be tested (24). Among the 48 putative effectors predicted by OPT4e, 15 are differentially transcribed between mammalian and tick cells (7, 8). The three best characterized A. phagocytophilum T4SS translocated effectors, Ats-1 (17), AnkA (15, 16), and HGE14 (23), are all downregulated during growth in tick cells (7, 8), suggesting their contributions may be more important during mammalian infection. Our understanding of how A. phagocytophilum mediates interactions within mammalian cells is limited, but even less is understood about how the bacteria navigate tick cell biology. A full mechanistic understanding of how rickettsial pathogens facilitate their vector-borne life cycle will require effector identification and characterization in the context of both mammalian hosts and arthropod vectors.
While genetic tools among rickettsial organisms remain limited (36), the A. phagocytophilum transposon mutant library (9) allowed us to isolate and test a mutation disrupting ateA. Although maintenance of the library in HL60 cell culture precludes mutation of genes essential for mammalian infection, it has equipped us to test the contributions of genes that are important for growth in the tick. This is the second mutant from these libraries shown to have a tick cell-specific phenotype. Mutation of an outer-membrane O-methyltransferase similarly led to a tick cell-specific infection defect (28). Additionally, transposon mutation of a paralogous T4SS component, virB6-4, partially attenuated growth in both tick and mammalian cells demonstrating this mutant collection also retains some utility for investigating incomplete phenotypes in mammalian cell models (27). We took the ateA::Himar1 mutant beyond cell culture experiments and demonstrated an in vivo phenotype through both murine and tick infections that ateA is important for bacterial acquisition by ticks from a blood meal. Our work with ateA::Himar1 represents the first A. phagocytophilum mutant examined in live ticks.
Due to the difficulties in generating recombinant expression systems in an obligate intracellular bacterium (36), efficient T4SS translocation assays using rickettsial organisms have not yet been developed. However, surrogate systems in Legionella (31), Coxiella (23), and Escherichia (17, 37) have been used to identify rickettsial T4SS substrates. We demonstrated that L. pneumophila recognizes and translocates AteA into the host cell cytosol in a T4SS dependent manner. Motifs at the C-terminus often serve as translocation signals for both the Legionella and rickettsial T4SS, but they are not universally required and alternative signals can be used (32, 33). AteA contains multiple charged residues in the C-terminus that we determined are dispensable for translocation. Instead, the large, disordered region of AteA was sufficient for translocation. This suggests that the T4SS is recognizing the unstructured nature of the protein or unidentified internal secretion signals. Indeed, disordered regions are a common characteristic among bacterial effectors (34). While the specificity of the Legionella T4SS translocation assay cannot be directly projected onto the A. phagocytophilum T4SS, A. phagocytophilum effectors Ats1, AnkA, and HGE14 also contain intrinsically disordered regions suggesting that this may be a common feature recognized for T4SS translocation (25).
Intracellular bacteria exist among the scaffold of the host cell’s cytoskeleton composed of actin, tubulin, and various intermediate filaments. Pathogens manipulate this cytoskeleton network to promote internalization, evade destruction, alter intracellular trafficking, and disseminate within and between cells (20). Our understanding of how A. phagocytophilum interfaces with this cellular scaffold is limited, but differences exist between mammalian and tick cell infection (38, 39). During mammalian infection, the A. phagocytophilum vacuole protein AptA recruits intermediate filaments to the Anaplasma vacuole. However, transcription of aptA is not detectable during A. phagocytophilum infection in ticks (39). Actin polymerization is necessary for A. phagocytophilum entry into mammalian cells, but during tick cell infection actin is phosphorylated and depolymerized, which is not seen during mammalian infection (38). Although ectopic expression of AteA did not appear to lead to actin depolymerization, this possibility will require further study. We found that AteA localized with branched actin at the cell cortex and was dependent on the predicted tandem repeats and the N-terminal globular domain (Fig. 5). Many other intracellular pathogens are known to manipulate cortical actin to attach to the host cell, induce internalization of the bacteria (20, 40), alter endosome maturation (41, 42), block degradation by the lysosome (43), support the pathogen containing vacuole (44), induce extrusion from the host cell (45), and mediate cell-to-cell bacterial transfer (20, 46). Understanding how AteA manipulates the cytoskeleton to further the A. phagocytophilum infection cycle will require further mechanistic dissection.
We demonstrated that A. phagocytophilum ateA is specifically important in the tick environment. While examination of AteA localization in tick cells is desired, ectopic expression in ISE6 cells is difficult as transfection efficiency is extremely low, and we were unable to visualize AteA in tick cells. Further, we were unable to visualize AteA during tick infection by immunofluorescence due to high background binding of the α-AteA antibody to the tick cells (not shown). However, we were able to visualize AteA localization with cortical actin in mammalian cells, which may have been possible because actin is one of the most conserved proteins across all eukaryotes (47). Further, mammalian systems have previously been used to investigate multiple actin-targeting T4SS effectors, despite mammals not being the evolutionarily relevant environment (1, 41, 42, 48, 49). The tick-specific expression of ateA is highlighted by its dispensability during mammalian infection and severe attenuation of the knockout during tick infection. Why A. phagocytophilum requires ateA during tick infection remains unclear, but it may stem from the different cell types infected. In mammals, the bacteria preferentially infect phagocytic neutrophils, while in ticks it infects multiple non-phagocytic cell types. AteA may be required to induce internalization or trafficking within tick cells, which neutrophils perform without manipulation.
In summary, we have identified the first tick-specific translocated effector from A. phagocytophilum and have shown that it targets cortical actin. While most research on A. phagocytophilum focuses on mammalian infection, it is increasingly clear that the mammalian and tick environments are not equivalent. Given that the arthropod vector is the driver of A. phagocytophilum transmission, it is critical to understand how these bacteria survive in the tick. We expect that A. phagocytophilum deploys a unique repertoire of effectors to navigate the tick environment, with ateA being only the first of many to identify.
MATERIALS AND METHODS
Bacterial and eukaryotic cell culture
Escherichia coli was grown using solid and liquid lysogeny broth (LB) medium with the addition of kanamycin or zeocin (25 µg mL−1) antibiotics for selection as needed. L. pneumophila Lp02 and Lp03 (dotA−) strains were cultured using N-(2-acetamido)-2-aminoethanesulfonic acid buffered yeast extract medium and solid charcoal buffered yeast extract agar medium. L. pneumophila cultures were supplemented with 0.4 mg mL−1 iron(III) nitrate, 0.135 mg mL−1 cysteine (50), 0.1 mg mL−1 thymidine, and when appropriate 50 µg mL−1 kanamycin.
HeLa human cervical epithelial cells (American Type Culture Collection [ATCC]; CCL-2) were maintained in Eagle’s Minimum Essential Medium (Corning; 10-010-CV) with 10% fetal bovine serum (FBS: Atlanta biologicals: S11550) and 1× Glutamax (Gibco; 35050061). HL60 human promyelocytic cells (ATCC; CCL-240) and the THP1 human monocyte cell line (ATCC TIB-202) were maintained in Roswell Park Memorial Institute (RPMI) 1640 medium with 10% FBS and 1× Glutamax. Mammalian cell cultures were maintained in a humidified chamber at 37°C with 5% CO2. HL60 density was kept between 5 × 104 and 1 × 106 and limited to less than 20 passages to prevent differentiation or phenotypic drift.
A. phagocytophilum strain HGE1 and mutant lines were cultured in HL60 cells (9). Insertion mutant ateA::Himar1 and control strain were isolated from a previously reported A. phagocytophilum Himar1 transposon mutant library (9). The control strain contains the Himar1 transposon in an intergenic location and has been shown to be phenotypically equivalent to wild type (27, 28). Infection status of HL60 cells was assessed by Diff-Quick Romanowsky–Giemsa staining. A. phagocytophilum were liberated from HL60 cells by 27-gauge needle syringe lysis to generate host-cell-free organisms. Bacterial numbers were estimated as previously described (51, 52).
Tick cells derived from embryonated eggs of the blacklegged tick, I. scapularis (Say), ISE6, were grown in L15C-300 medium with 10% FBS (Sigma; F0926), 10% tryptone phosphate broth (TPB; BD; B260300), and 0.1% lipoprotein cholesterol concentrate (MP Biomedicals; 219147680) (53). Infected ISE6 cell cultures were additionally supplemented with 0.25% NaHCO3 and 25 mM HEPES buffer (Sigma). Tick cell cultures were incubated at 34°C and 1% CO2 (6).
Anaplasma phagocytophilum growth curves
Growth of A. phagocytophilum strains in HL60 and ISE6 cells was evaluated as described previously (27, 28). Briefly, HL60 cells were seeded at 5 × 104 cells per well in 24-well plates. The plate was then infected with 5 × 104 host cell-free A. phagocytophilum per well for an MOI of 1. Triplicate wells were harvested at the time of inoculation and 1-, 2-, 3-, 4-, and 5-days post inoculation. ISE6 cells were seeded at 3 × 105 cells per well in 24-well plates and allowed to adhere to the plate overnight. Host cell-free preparations of A. phagocytophilum strains were done immediately before inoculation and bacteria were suspended in L15C300 supplemented with 0.25% NaHCO3 and 25 mM HEPES. ISE6 plates were inoculated at 3 × 106 A. phagocytophilum per well for an MOI of 10. Twenty-four hours post infection, the tick cell media was exchanged for fresh L15C300 + NaHCO3 +HEPES to remove remaining extracellular bacteria, and three wells were collected for initial timepoint. Triplicate samples were collected at subsequent time points and frozen to be processed for gDNA using a QIAGEN DNeasy blood and tissue kit. Change in bacteria and host cell gDNA copies was assessed by qPCR using iTaq universal SYBR green Supermix (Bio-Rad; 1725125) in duplicate reactions. Bacterial gDNA was measured targeting the single copy A. phagocytophilum gene msp5. HL60 and ISE6 host cell gDNA was measured targeting genes tlr9 (toll-like receptor 9) and crt (calreticulin), respectively (27) (Table S1).
Transcriptional analysis of ateA
Triplicate samples were collected during the HL60 and ISE6 A. phagocytophilum growth curve experiments. Samples were processed to purify RNA using Direct-zol RNA Microprep Kit (ZymoResearch) according to the product protocols for tissue culture samples. The Verso cDNA Synthesis Kit (ThermoFisher) was used to generate cDNA. Transcripts of ateA, rpoB, groEL, and msp5 genes were measured by qPCR using iTaq universal SYBR green Supermix (Bio-Rad; 1725125) according to Bio-Rad specified cycle conditions. Transcription of ateA was compared between experimental groups by ΔΔCt using rpoB as the housekeeping control gene.
Antibody generation and western analysis
Rabbit antibodies against AteA(aa1056-1075): Cys-ERQMPHKTKSVHELAKQLEE and VirD4(aa720-740): Cys-EDEFGEDRPTDDNDSSNGRLK were generated and purified using synthetic peptides from Pacific Immunology, Ramona, CA, USA (National Institutes of Health animal welfare assurance No. A41820-01).
Proteins from mock-infected and peak wild-type A. phagocytophilum infected human HL60 and tick ISE6 cells were separated using 4–15% MP TGX precast gels (Bio-Rad; 4561083). Lanes were equally loaded with 1 × 105 host cells. Proteins were transferred to nitrocellulose membranes and blocked in 5% bovine serum albumin (BSA) in TBS-T (1× Tris-buffered saline + 0.1% Tween 20). Blocked membranes were incubated overnight at 4°C with either primary α-AteA (1:1,000) or α-VirD4 (1:4,000) diluted in TBS-T with 5% BSA. HRP-linked goat α-rabbit IgG secondary antibody (Cell Signaling; 7074) was applied for 2 h at 4°C. Blots were visualized using Pierce ECL western blotting substrate (ThermoFisher; 32209).
Animal infection
Two gender matched groups of 10, 6-week-old C57BL/6 mice (The Jackson Laboratory) were intraperitoneally infected with either the ateA::Himar1 mutant or the control strain at 1 × 107 host cell-free A. phagocytophilum bacteria per mouse. The control strain contains the Himar1 transposon in an intergenic location and has been shown to be phenotypically equivalent to wild type (27, 28). Seven days post infection, 25–50 µL of blood was collected from the lateral saphenous vein. Levels of A. phagocytophilum in the blood was measured by qPCR (16S rRNA relative to mouse β-actin [51, 54]) (Table S1). Uninfected I. scapularis larval ticks were purchased from Oklahoma State University (Stillwater, OK, USA). Ticks were housed at 23°C with 16/8 h light/dark photoperiods in 95–100% humidity. As sources of tick acquisition for A. phagocytophilum, two burden-matched-pairs of mice were selected from the ateA::Himar1 and control strain infected mice. Each mouse was individually housed and infested with 200 naive unfed I. scapularis larvae. Three to seven days post infestation, replete larvae were collected, individually flash frozen with liquid nitrogen, ground with a pestle, dissolved in TRIzol, and processed to purify total RNA according to Direct-zol RNA Microprep Kit protocol. The Verso cDNA Synthesis Kit (ThermoFisher) was used to generate cDNA. Levels of viable A. phagocytophilum in the ticks were measured by quantifying A. phagocytophilum 16S rRNA relative to I. scapularis β-actin transcripts by qRT-PCR (Table S1) by absolute quantification (51, 54).
Plasmid construction
Full length and truncations of the ateA open reading frame were amplified from A. phagocytophilum genomic DNA with Gateway compatible primers (Table S1). Amplicons were introduced into pDONR/Zeo by BP Clonase (Invitrogen). Sequence confirmed inserts were then transferred to destination expression vectors with LR Clonase (Invitrogen). For ectopic expression in mammalian cells, we used a Gateway compatible version of pEGFP-C1 (Clontech) and pEZYegfp (Addgene). To create a Gateway destination vector for use in translocation assays (pJC125DEST), the Gateway attR cassette was inserted into the CyaA translational fusion construct pJC125 (55) at a SmaI restriction site. CyaA fusion constructs were introduced to L. pneumophila by electroporation.
Translocation assay
THP-1 cells were seeded at 5 × 105/mL in 24-well plate and differentiated to macrophage-like cells by treatment with 200 nM phorbol 12-myristate 13-acetate (Sigma) for 18 h. Transformed L. pneumophila cells were grown overnight to an OD600 of 2.0, at which point the bacteria were in post-exponential phase, highly motile, and infectious. Expression of the CyaA fusion proteins was induced by adding 1 mM IPTG for 1 h, and motility was verified by microscopy of wet mounted samples. Cell culture medium was used to dilute L. pneumophila, which was then used to infect THP-1 cells at an MOI of 1. One hour post infection, cAMP was extracted and quantified as previously described (56) using the cAMP Parameter Assay Kit (R&D Systems).
Immunofluorescence
HeLa cells were transfected using FuGENE 6 Transfection Reagent at a 3:1 FuGENE to DNA ratio. Proteins were expressed for 36–48 h, then fixed in 4% paraformaldehyde. Fixed cells were permeabilized with 0.1% Triton X-100 for 15 min and washed three times in phosphate buffered saline (PBS). To visualize actin, cells were incubated with Alexa Fluor 568 phalloidin (ThermoFisher Scientific) in PBS containing 1% BSA for 30 min. To visualize cortical actin, we stained for the cortical actin-binding protein, cortactin, α-cortactin (p80/85) mouse monoclonal antibody 4F11 (MilliporeSigma) followed by Alexa Fluor 488 or 594 anti-mouse (Cell Signalling Technology). After each treatment, cells were washed three times for 5 min with PBS and coverslips were mounted on slides using Vectashield mounting medium with DAPI (4′,6-diamidino-2-phenylindole). Slides were imaged using a Leica SP8 confocal microscope. Co-localization was analyzed per cell selected in the green channel using Fiji ImageJ with Coloc_2 co-localization plug-in. A total of twenty cells were analyzed per condition.
Statistics
All in vitro experiments were performed with three biological replicates, measured in technical duplicate assays, and experiments were repeated three times to ensure reproducibility of findings. In vivo experiments used 10 independently inoculated mice per group. From each mouse, 17–20 ticks were collected. Two burden matched mice pairs were used for experimental replicates of the tick feeding. Data were expressed as means and graphed with standard deviation. Data points were analyzed with a Student’s t-test (Mann-Whitney) or ANOVA for in vitro experiments and in vivo experiments were analyzed with an unpaired Welch’s t-test. Statistical analysis was performed and graphed with GraphPad Prism version 9.0. A P-value of <0.05 was considered statistically significant.
ACKNOWLEDGMENTS
We are grateful to Daniel Voth at the University of Arkansas College of Medicine (Little Rock, AR) and Jean Celli at Paul G. Allen School for Global Health, Washington State University (Pullman, WA), for sharing protocols and plasmid constructs for the CyaA secretion assay. Tamara O’Connor at Johns Hopkins University (Baltimore, MD) generously provided L. pneumophila Lp02 and Lp03 (dotA −) strains. We thank Lisa Price and Nicole Burkhardt at the University of Minnesota (Saint Paul, MN) for their assistance in isolating and verifying strains from the Himar1 transposon collection.
This work was funded through generous support from the National Institutes of Health (NIAID), grant numbers R21AI154023, R21AI151412 to K.A.B., and R01AI042792. B.M.G. was supported by NIH training grant T32-GM008336.
J.M.P. was responsible for conceptualization, methodology, investigation, writing original draft and editing, visualization, and funding acquisition. B.M.G. was involved in methodology, investigation, visualization, reviewing, and editing. D.F. and D.K.S was responsible for methodology, investigation, review, and editing. K.T.S. performed the investigation. C.M.N. and J.D.O. helped with investigation and resources. U.G.M. helped with resources and funding acquisition. K.A.B. contributed for conceptualization, supervision, funding acquisition, review and editing.
Contributor Information
Jason M. Park, Email: Jason.Park@wsu.edu.
Scot P. Ouellette, University of Nebraska Medical Center, Omaha, Nebraska, USA
ETHICS APPROVAL
All animal use protocols were approved by the Washington State University Institutional Animal Care and Use Committee (ASAF #6630). The animals were housed and maintained in an AAALAC-accredited facility at Washington State University in Pullman, WA.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mbio.01711-23.
Localization to cortical actin depends on the N-terminal region of AteA.
Oligonucleotides used in this study.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Park Jason M, Ghosh S, O’Connor TJ. 2020. Combinatorial selection in amoebal hosts drives the evolution of the human pathogen Legionella pneumophila. 4. Nat Microbiol 5:599–609. doi: 10.1038/s41564-019-0663-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Park J.M, Oliva Chávez AS, Shaw DK. 2021. Ticks: more than just a pathogen delivery service. Front Cell Infect Microbiol 11:739419. doi: 10.3389/fcimb.2021.739419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kumar S, Stecher G, Suleski M, Hedges SB. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol 34:1812–1819. doi: 10.1093/molbev/msx116 [DOI] [PubMed] [Google Scholar]
- 4. Eisen RJ, Kugeler KJ, Eisen L, Beard CB, Paddock CD. 2017. Tick-borne zoonoses in the United States: persistent and emerging threats to human health. ILAR J 58:319–335. doi: 10.1093/ilar/ilx005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ueti MW, Reagan JO, Knowles DP, Scoles GA, Shkap V, Palmer GH. 2007. Identification of midgut and salivary glands as specific and distinct barriers to efficient tick-borne transmission of Anaplasma marginale. Infect Immun 75:2959–2964. doi: 10.1128/IAI.00284-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Munderloh UG, Jauron SD, Fingerle V, Leitritz L, Hayes SF, Hautman JM, Nelson CM, Huberty BW, Kurtti TJ, Ahlstrand GG, Greig B, Mellencamp MA, Goodman JL. 1999. Invasion and intracellular development of the human granulocytic ehrlichiosis agent in tick cell culture. J Clin Microbiol 37:2518–2524. doi: 10.1128/JCM.37.8.2518-2524.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nelson CM, Herron MJ, Felsheim RF, Schloeder BR, Grindle SM, Chavez AO, Kurtti TJ, Munderloh UG. 2008. Whole genome transcription profiling of Anaplasma phagocytophilum in human and tick host cells by tiling array analysis. BMC Genomics 9:1–16. doi: 10.1186/1471-2164-9-364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nelson CM, Herron MJ, Wang X-R, Baldridge GD, Oliver JD, Munderloh UG. 2020. Global transcription profiles of Anaplasma phagocytophilum at key stages of infection in tick and human cell lines and granulocytes. Front Vet Sci 7:111. doi: 10.3389/fvets.2020.00111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. O’Conor MC, Herron MJ, Nelson CM, Barbet AF, Crosby FL, Burkhardt NY, Price LD, Brayton KA, Kurtti TJ, Munderloh UG. 2021. Biostatistical prediction of genes essential for growth of Anaplasma phagocytophilum in a human promyelocytic cell line using a random transposon mutant library. Pathog Dis 79:ftab029. doi: 10.1093/femspd/ftab029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gillespie JJ, Phan IQH, Driscoll TP, Guillotte ML, Lehman SS, Rennoll-Bankert KE, Subramanian S, Beier-Sexton M, Myler PJ, Rahman MS, Azad AF, Mobley H. 2016. The Rickettsia type IV secretion system: unrealized complexity mired by gene family expansion. Pathog Dis 74:ftw058. doi: 10.1093/femspd/ftw058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gillespie JJ, Brayton KA, Williams KP, Diaz MAQ, Brown WC, Azad AF, Sobral BW. 2010. Phylogenomics reveals a diverse rickettsiales type IV secretion system. Infect Immun 78:1809–1823. doi: 10.1128/IAI.01384-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beyer AR, Truchan HK, May LJ, Walker NJ, Borjesson DL, Carlyon JA. 2015. The Anaplasma phagocytophilum effector AmpA hijacks host cell SUMOylation. Cell Microbiol 17:504–519. doi: 10.1111/cmi.12380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lehman SS, Noriea NF, Aistleitner K, Clark TR, Dooley CA, Nair V, Kaur SJ, Rahman MS, Gillespie JJ, Azad AF, Hackstadt T. 2018. The rickettsial ankyrin repeat protein 2 is a type IV secreted effector that associates with the endoplasmic reticulum. mBio 9:419–422. doi: 10.1128/mBio.00975-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Voss OH, Gillespie JJ, Lehman SS, Rennoll SA, Beier-Sexton M, Rahman MS, Azad AF. 2020. Risk1, a phosphatidylinositol 3-kinase effector, promotes Rickettsia typhi intracellular survival. mBio 11:e00820-20. doi: 10.1128/mBio.00820-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garcia-Garcia JC, Rennoll-Bankert KE, Pelly S, Milstone AM, Dumler JS. 2009. Silencing of host cell CYBB gene expression by the nuclear effector AnkA of the intracellular pathogen Anaplasma phagocytophilum. Infect Immun 77:2385–2391. doi: 10.1128/IAI.00023-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lin Mingqun, den Dulk-Ras A, Hooykaas PJJ, Rikihisa Y. 2007. Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell Microbiol 9:2644–2657. doi: 10.1111/j.1462-5822.2007.00985.x [DOI] [PubMed] [Google Scholar]
- 17. Niu H, Kozjak-Pavlovic V, Rudel T, Rikihisa Y, Valdivia RH. 2010. Anaplasma phagocytophilum Ats-1 is imported into host cell mitochondria and interferes with apoptosis induction. PLoS Pathog 6:e1000774. doi: 10.1371/journal.ppat.1000774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yan Q, Zhang W, Lin M, Teymournejad O, Budachetri K, Lakritz J, Rikihisa Y. 2021. Iron robbery by intracellular pathogen via bacterial effector–induced ferritinophagy. Proc Natl Acad Sci U S A 118:e2026598118. doi: 10.1073/pnas.2026598118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin M, Liu H, Xiong Q, Niu H, Cheng Z, Yamamoto A, Rikihisa Y. 2016. Ehrlichia secretes Etf-1 to induce autophagy and capture nutrients for its growth through RAB5 and class III phosphatidylinositol 3-kinase. Autophagy 12:2145–2166. doi: 10.1080/15548627.2016.1217369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Colonne PM, Winchell CG, Voth DE. 2016. Hijacking host cell highways: manipulation of the host actin cytoskeleton by obligate intracellular bacterial pathogens. Front Cell Infect Microbiol 6:107. doi: 10.3389/fcimb.2016.00107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tang H, Zhu J, Wu S, Niu H. 2020. Identification and characterization of an actin filament-associated Anaplasma phagocytophilum protein. Microb Pathog 147:104439. doi: 10.1016/j.micpath.2020.104439 [DOI] [PubMed] [Google Scholar]
- 22. Zhu J, He M, Xu W, Li Y, Huang R, Wu S, Niu H. 2019. Development of TEM-1 β-lactamase based protein translocation assay for identification of Anaplasma phagocytophilum type IV secretion system effector proteins. 1. Sci Rep 9:4235. doi: 10.1038/s41598-019-40682-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sinclair SHG, Garcia-Garcia JC, Dumler JS. 2015. Bioinformatic and mass spectrometry identification of Anaplasma phagocytophilum proteins translocated into host cell nuclei. Front Microbiol 6:55. doi: 10.3389/fmicb.2015.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Esna Ashari Z, Brayton KA, Broschat SL. 2019. Prediction of T4SS effector proteins for Anaplasma phagocytophilum using OPT4e, A new software tool. Front Microbiol 10:1391. doi: 10.3389/fmicb.2019.01391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Erdős G, Pajkos M, Dosztányi Z. 2021. IUPred3: prediction of protein disorder enhanced with unambiguous experimental annotation and visualization of evolutionary conservation. Nucleic Acids Res. 49:W297–W303. doi: 10.1093/nar/gkab408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Newman AM, Cooper JB. 2007. XSTREAM: a practical algorithm for identification and architecture modeling of tandem repeats in protein sequences. BMC Bioinformatics 8:382. doi: 10.1186/1471-2105-8-382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Crosby FL, Munderloh UG, Nelson CM, Herron MJ, Lundgren AM, Xiao Y-P, Allred DR, Barbet AF. 2020. Disruption of VirB6 paralogs in Anaplasma phagocytophilum attenuates its growth. J Bacteriol 202:e00301-20. doi: 10.1128/JB.00301-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oliva Chávez AS, Fairman JW, Felsheim RF, Nelson CM, Herron MJ, Higgins L, Burkhardt NY, Oliver JD, Markowski TW, Kurtti TJ, Edwards TE, Munderloh UG. 2015. An O-methyltransferase is required for infection of tick cells by Anaplasma phagocytophilum . PLoS Pathog 11:e1005248. doi: 10.1371/journal.ppat.1005248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Noroy C, Lefrançois T, Meyer DF. 2019. Searching algorithm for type IV Effector proteins (S4Te) 2.0: improved tools for type IV effector prediction, analysis and comparison in proteobacteria. PLoS Comput Biol 15:e1006847. doi: 10.1371/journal.pcbi.1006847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zou L, Nan C, Hu F. 2013. Accurate prediction of bacterial type IV secreted effectors using amino acid composition and PSSM profiles. Bioinformatics 29:3135–3142. doi: 10.1093/bioinformatics/btt554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lockwood S, Voth DE, Brayton KA, Beare PA, Brown WC, Heinzen RA, Broschat SL. 2011. Identification of Anaplasma marginale type IV secretion system effector proteins. PLoS One 6:e27724. doi: 10.1371/journal.pone.0027724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang L, Boyd D, Amyot WM, Hempstead AD, Luo Z-Q, O’Connor TJ, Chen C, Machner M, Montminy T, Isberg RR. 2011. The E block motif is associated with Legionella pneumophila translocated substrates. Cell Microbiol 13:227–245. doi: 10.1111/j.1462-5822.2010.01531.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Christie PJ, Whitaker N, González-Rivera C. 2014. Mechanism and structure of the bacterial type IV secretion systems. Biochim Biophys Acta 1843:1578–1591. doi: 10.1016/j.bbamcr.2013.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marín M, Uversky VN, Ott T. 2013. Intrinsic disorder in pathogen effectors: protein flexibility as an evolutionary hallmark in a molecular arms race. Plant Cell 25:3153–3157. doi: 10.1105/tpc.113.116319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chalut KJ, Paluch EK. 2016. The actin cortex: a bridge between cell shape and function. Dev Cell 38:571–573. doi: 10.1016/j.devcel.2016.09.011 [DOI] [PubMed] [Google Scholar]
- 36. McClure EE, Chávez ASO, Shaw DK, Carlyon JA, Ganta RR, Noh SM, Wood DO, Bavoil PM, Brayton KA, Martinez JJ, McBride JW, Valdivia RH, Munderloh UG, Pedra JHF. 2017. Engineering of obligate intracellular bacteria: progress, challenges and paradigms. Nat Rev Microbiol 15:544–558. doi: 10.1038/nrmicro.2017.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Whitaker N, Berry TM, Rosenthal N, Gordon JE, Gonzalez-Rivera C, Sheehan KB, Truchan HK, VieBrock L, Newton ILG, Carlyon JA, Christie PJ. 2016. Chimeric coupling proteins mediate transfer of heterologous type IV effectors through the Escherichia coli pkm101-encoded conjugation machine. J Bacteriol 198:2701–2718. doi: 10.1128/JB.00378-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sultana H, Neelakanta G, Kantor FS, Malawista SE, Fish D, Montgomery RR, Fikrig E. 2010. Anaplasma phagocytophilum induces actin phosphorylation to selectively regulate gene transcription in Ixodes scapularis ticks. J Exp Med 207:1727–1743. doi: 10.1084/jem.20100276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sukumaran B, Mastronunzio JE, Narasimhan S, Fankhauser S, Uchil PD, Levy R, Graham M, Colpitts TM, Lesser CF, Fikrig E. 2011. Anaplasma phagocytophilum AptA modulates Erk1/2 signalling. Cell Microbiol 13:47–61. doi: 10.1111/j.1462-5822.2010.01516.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hardt W-D, Chen L-M, Schuebel KE, Bustelo XR, Galán JE. 1998. S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell 93:815–826. doi: 10.1016/s0092-8674(00)81442-7 [DOI] [PubMed] [Google Scholar]
- 41. Michard C, Sperandio D, Baïlo N, Pizarro-Cerdá J, LeClaire L, Chadeau-Argaud E, Pombo-Grégoire I, Hervet E, Vianney A, Gilbert C, Faure M, Cossart P, Doublet P. 2015. The Legionella kinase LegK2 targets the ARP2/3 complex to inhibit actin nucleation on phagosomes and allow bacterial evasion of the late endocytic pathway. mBio 6:e00354-15. doi: 10.1128/mBio.00354-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hervet E, Charpentier X, Vianney A, Lazzaroni J-C, Gilbert C, Atlan D, Doublet P. 2011. Protein kinase LegK2 is a type IV secretion system effector involved in endoplasmic reticulum recruitment and intracellular replication of Legionella pneumophila. Infect Immun 79:1936–1950. doi: 10.1128/IAI.00805-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Walpole GFW, Plumb JD, Chung D, Tang B, Boulay B, Osborne DG, Piotrowski JT, Catz SD, Billadeau DD, Grinstein S, Jaumouillé V. 2020. Inactivation of Rho GTPases by Burkholderia cenocepacia induces a WASH-mediated actin polymerization that delays phagosome maturation. Cell Rep 31:107721. doi: 10.1016/j.celrep.2020.107721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kumar Y, Valdivia RH. 2008. Actin and intermediate filaments stabilize the Chlamydia trachomatis vacuole by forming dynamic structural scaffolds. Cell Host Microbe 4:159–169. doi: 10.1016/j.chom.2008.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thomas S, Popov VL, Walker DH. 2010. Exit mechanisms of the intracellular bacterium ehrlichia. PLoS One 5:e15775. doi: 10.1371/journal.pone.0015775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lamason RL, Welch MD. 2017. Actin-based motility and cell-to-cell spread of bacterial pathogens. Curr Opin Microbiol 35:48–57. doi: 10.1016/j.mib.2016.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wickstead B, Gull K. 2011. The evolution of the cytoskeleton. J Cell Biol 194:513–525. doi: 10.1083/jcb.201102065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. He L, Lin Y, Ge Z-H, He S-Y, Zhao B-B, Shen D, He J-G, Lu Y-J. 2019. The Legionella pneumophila effector WipA disrupts host F-actin polymerisation by hijacking phosphotyrosine signalling. Cell Microbiol 21:e13014. doi: 10.1111/cmi.13014 [DOI] [PubMed] [Google Scholar]
- 49. Liu Y, Zhu W, Tan Y, Nakayasu ES, Staiger CJ, Luo Z-Q. 2017. A Legionella effector disrupts host cytoskeletal structure by cleaving actin. PLoS Pathog. 13:e1006186. doi: 10.1371/journal.ppat.1006186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Feeley JC, Gibson RJ, Gorman GW, Langford NC, Rasheed JK, Mackel DC, Baine WB. 1979. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J Clin Microbiol 10:437–441. doi: 10.1128/jcm.10.4.437-441.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shaw DK, Wang X, Brown LJ, Chávez ASO, Reif KE, Smith AA, Scott AJ, McClure EE, Boradia VM, Hammond HL, Sundberg EJ, Snyder GA, Liu L, DePonte K, Villar M, Ueti MW, de la Fuente J, Ernst RK, Pal U, Fikrig E, Pedra JHF. 2017. Infection-derived lipids elicit an immune deficiency circuit in arthropods. Nat Commun 8:14401. doi: 10.1038/ncomms14401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chen G, Wang X, Severo MS, Sakhon OS, Sohail M, Brown LJ, Sircar M, Snyder GA, Sundberg EJ, Ulland TK, Olivier AK, Andersen JF, Zhou Y, Shi G-P, Sutterwala FS, Kotsyfakis M, Pedra JHF. 2014. The tick salivary protein sialostatin L2 inhibits caspase-1-mediated inflammation during Anaplasma phagocytophilum infection. Infect Immun 82:2553–2564. doi: 10.1128/IAI.01679-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Oliver JD, Chávez ASO, Felsheim RF, Kurtti TJ, Munderloh UG. 2015. An Ixodes scapularis cell line with a predominantly neuron-like phenotype. Exp Appl Acarol 66:427–442. doi: 10.1007/s10493-015-9908-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sidak-Loftis LC, Rosche KL, Pence N, Ujczo JK, Hurtado J, Fisk EA, Goodman AG, Noh SM, Peters JW, Shaw DK. 2022. The unfolded-protein response triggers the arthropod immune deficiency pathway mBio 13:e0292222. doi: 10.1128/mbio.02922-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Myeni S, Child R, Ng TW, Kupko JJ, Wehrly TD, Porcella SF, Knodler LA, Celli J. 2013. Brucella modulates secretory trafficking via multiple type IV secretion effector proteins. PLoS Pathog. 9:e1003556. doi: 10.1371/journal.ppat.1003556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Voth DE, Howe D, Beare PA, Vogel JP, Unsworth N, Samuel JE, Heinzen RA. 2009. The Coxiella burnetii ankyrin repeat domain-containing protein family is heterogeneous, with C-terminal truncations that influence DOT/Icm-mediated secretion. J Bacteriol 191:4232–4242. doi: 10.1128/JB.01656-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Localization to cortical actin depends on the N-terminal region of AteA.
Oligonucleotides used in this study.