Abstract
(−)-Menthone is the predominant monoterpene produced in the essential oil of maturing peppermint (Mentha x piperita) leaves during the filling of epidermal oil glands. This early biosynthetic process is followed by a second, later oil maturation program (approximately coincident with flower initiation) in which the C3-carbonyl of menthone is reduced to yield (−)-(3R)-menthol and (+)-(3S)-neomenthol by two distinct NADPH-dependent ketoreductases. An activity-based in situ screen, by expression in Escherichia coli of 23 putative redox enzymes from an immature peppermint oil gland expressed sequence tag library, was used to isolate a cDNA encoding the latter menthone:(+)-(3S)-neomenthol reductase. Reverse transcription-PCR amplification and RACE were used to acquire the former menthone:(−)-(3R)-menthol reductase directly from mRNA isolated from the oil gland secretory cells of mature leaves. The deduced amino acid sequences of these two reductases share 73% identity, provide no apparent subcellular targeting information, and predict inclusion in the short-chain dehydrogenase/reductase family of enzymes. The menthone:(+)-(3S)-neomenthol reductase cDNA encodes a 35,722-D protein, and the recombinant enzyme yields 94% (+)-(3S)-neomenthol and 6% (−)-(3R)-menthol from (−)-menthone as substrate, and 86% (+)-(3S)-isomenthol and 14% (+)-(3R)-neoisomenthol from (+)-isomenthone as substrate, has a pH optimum of 9.3, and Km values of 674 μm, > 1 mm, and 10 μm for menthone, isomenthone, and NADPH, respectively, with a kcat of 0.06 s−1. The recombinant menthone:(−)-(3R)-menthol reductase has a deduced size of 34,070 D and converts (−)-menthone to 95% (−)-(3R)-menthol and 5% (+)-(3S)-neomenthol, and (+)-isomenthone to 87% (+)-(3R)-neoisomenthol and 13% (+)-(3S)-isomenthol, displays optimum activity at neutral pH, and has Km values of 3.0 μm, 41 μm, and 0.12 μm for menthone, isomenthone, and NADPH, respectively, with a kcat of 0.6 s−1. The respective activities of these menthone reductases account for all of the menthol isomers found in the essential oil of peppermint. Biotechnological exploitation of these genes could lead to improved production yields of (−)-menthol, the principal and characteristic flavor component of peppermint.
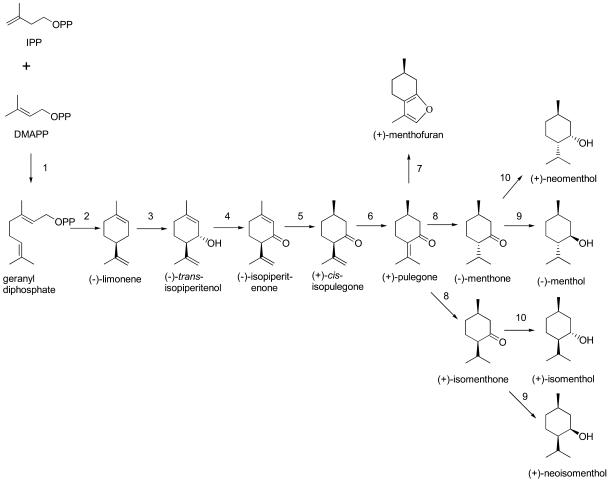
The essential oil of peppermint (Mentha x piperita) is composed principally of C3-oxygenated p-menthane monoterpenes, and the characteristic organoleptic properties of this oil are derived predominantly from (−)-menthol. The more commonly used p-menthane monoterpene numbering system is utilized here in which the exocyclic methyl (C7) is appended to C1, the oxygen is at C3, and the isopropyl group is attached to C4; thus, (−)-menthol is 1R:3R:4S configured. Menthol biosynthesis (Fig. 1) begins with the diversion of the primary isoprenoid intermediates, isopentenyl diphosphate and its allylic isomer, dimethylallyl diphosphate, to geranyl diphosphate (the precursor of all monoterpenes) by the prenyltransferase geranyl diphosphate synthase (GPPS; Burke et al., 1999), and is followed by ionization and cyclization of this C10 intermediate to limonene by the terpene cyclase (−)-(4S)-limonene synthase (Kjonaas and Croteau, 1983). Regiospecific and stereospecific hydroxylation of this monoterpene olefin intermediate is catalyzed by cytochrome P450 limonene-3-hydroxylase to yield (−)-trans-isopiperitenol and is followed by dehydrogenation of this allylic alcohol (by isopiperitenol dehydrogenase) to afford the α,β-unsaturated ketone (-)-isopiperitenone. The endocyclic double bond of this intermediate is reduced by isopiperitenone reductase to yield (+)-cis-isopulegone, which undergoes enzymatic isomerization of the isopropenyl double bond (Δ8,9) to yield the conjugated ketone (+)-pulegone (Δ4,8). The newly formed isopropylidene double bond is reduced by pulegone reductase to yield (−)-menthone and lesser amounts of (+)-isomenthone. Alternatively, in a cytochrome P450-mediated side-reaction, pulegone undergoes C9 hydroxylation and intramolecular cyclization and dehydration to yield (+)-menthofuran. In the final reductive step of the pathway, (−)-menthone and (+)-isomenthone are reduced to (−)-menthol and (+)-neoisomenthol, respectively, or, by a separate reductase, to (+)-neomenthol and (+)-isomenthol, respectively; the latter three monoterpenol isomers are minor constituents of peppermint oil.
Figure 1.
The primary pathway for monoterpene biosynthesis in peppermint. The enzymes responsible for the transformations shown are GPPS (1), (−)-limonene synthase (2), (−)-limonene-3-hydroxylase (3), (−)-trans-isopiperitenol dehydrogenase (4), (−)-isopiperitenone reductase (5), (+)-cis-isopulegone isomerase (6), (+)-menthofuran synthase (7), (+)-pulegone reductase (8), (−)-menthone:(−)-(3R)-menthol reductase (9), and (−)-menthone:(+)-(3S)-neomenthol reductase (10).
Monoterpene biosynthesis in peppermint occurs in the highly specialized secretory cells of epidermal oil glands (McCaskill et al., 1992), and recent evidence suggests this metabolic process is divided into two temporally distinct periods of transcriptional and translational activity. The initial process is characterized by the de novo biosynthesis of the p-menthane monoterpenes (as determined by 14CO2 incorporation) that results in the accumulation of mostly (−)-menthone; once synthesized, only trace levels of monoterpenes are lost to evaporation or catabolism (Gershenzon et al., 2000). In vitro assay of the relevant enzymes of menthone biosynthesis (McConkey et al., 2000) demonstrated that these activities appear coincidentally during leaf expansion and endure for a brief time period (12–20 d post leaf initiation) with peak activity levels correlating with the essential oil secretion (gland filling) phase of gland development (Turner et al., 2000). RNA-blot analysis further showed that maximum transcript accumulation of limonene synthase occurs immediately prior to maximal enzyme activity, suggesting that at least the first committed enzyme of monoterpene biosynthesis is transcriptionally regulated (McConkey et al., 2000). However, the production of (−)-menthol from menthone is not significant until late in leaf development, after de novo monoterpene biosynthesis is essentially complete (Gershenzon et al., 2000); this process occurs in mature oil gland cells during the postsecretory phase (Turner et al., 2000). This second developmental process, termed oil maturation, is characterized by the depletion of the dominant intermediate menthone and is concomitant with increased activity of the menthone reductases and the accumulation of menthol (and lesser amounts of the epimer neomenthol; Croteau and Martinkus, 1979); this process may be accompanied by conjugation of the resulting monoterpenols to acetate esters and glucosides (Martinkus and Croteau, 1981; Croteau and Winters, 1982).
All of the enzymes of menthol biosynthesis have been characterized in cell-free systems, and cDNAs encoding GPPS, limonene synthase, and limonene-3-hydroxylase were isolated using reverse genetic approaches (Colby et al., 1993; Burke et al., 1999; Lupien et al., 1999). The development of a method for isolating intact oil gland secretory cells from peppermint leaf surfaces (Gershenzon et al., 1992) has provided a highly enriched source of terpene biosynthetic enzymes and their corresponding transcripts. These isolated secretory cells were used to prepare a cDNA pool of which 1,250 random clones were partially sequenced to generate an expressed sequence tag (EST) library in which at least 18% of the acquisitions were cDNAs involved in terpene metabolism (Lange et al., 2000). Exploitation of this highly enriched source of cDNAs has led to the cloning and functional expression of isopiperitenone reductase and pulegone reductase (Ringer et al., 2003), menthofuran synthase (Bertea et al., 2001), and, in a companion publication, isopiperitenol dehydrogenase (Ringer et al., 2005).
Because menthol content is paramount to peppermint essential oil quality, considerable interest has focused on the biosynthesis of this compound, the developmental timing of menthone reduction (and its impact on oil yield and composition at harvest), the stereoselectivity of the reduction, and the specificity of the monoterpenol conjugation reactions (Croteau and Hooper, 1979; Croteau and Martinkus, 1979). Clearly, of the remaining pathway genes, the menthone reductases are of principal interest in the context of the oil maturation process, and molecular genetic information about these ketoreductases could provide the tools for understanding, and the means of controlling, menthol production in peppermint. This paper reports the cloning, functional expression, and characterization of the two target menthone reductases, (−)-menthone:(−)-(3R)-menthol reductase (MMR) and (−)-menthone: (+)-(3S)-neomenthol reductase (MNR), and, taken together with previous work (Ringer et al., 2003) and the accompanying publication (Ringer et al., 2005), provides the entire complement of cDNAs encoding the redox enzymes of (−)-menthol biosynthesis in peppermint.
RESULTS
Cloning of Menthone Reductases
Several lines of evidence, including 14CO2 incorporation studies, developmental time courses of relevant enzyme activities, and RNA-blot analyses (Gershenzon et al., 2000; McConkey et al., 2000), indicated that peppermint undergoes two temporally distinct developmental programs during essential oil metabolism. The first is marked by de novo monoterpene biosynthesis and the accumulation of primarily (−)-menthone that occurs during the secretory phase of oil gland development and filling (12–20 d post leaf initiation; Turner et al., 2000). The second program, called the oil maturation process, follows approximately one week later when the oil gland cells are largely in the postsecretory phase of development (i.e. net accumulation of essential oil has ceased) and is characterized by an increase in the enzymatic activity responsible for the ketoreduction of (−)-menthone and the concomitant increase in the production of menthol and its stereoisomers in the essential oil (McConkey et al., 2000). An EST library was available that was prepared from mRNA isolated from oil glands of immature leaves undergoing the first biosynthetic program in which the oil content of menthol and its isomers was low and the menthone content was high. This library yielded a number of cDNAs encoding enzymes of isoprenoid metabolism (Burke et al., 1999; Lupien et al., 1999; Lange et al., 2000; Bertea et al., 2001), including isopiperitenone reductase and pulegone reductase (Ringer et al., 2003), and isopiperitenol dehydrogenase (Ringer et al., 2005), that were acquired by using a novel screen in which the recombinant proteins were expressed and functionally assayed for these reductase activities in situ in Escherichia coli. Similar in situ functional screens were also conducted to search for the menthone reductases. Out of 23 unique, putative full-length reductases tested, the screen revealed but a single cDNA, designated ml472, for which the corresponding expressed protein was capable of reducing menthone and isomenthone to neomenthol and isomenthol, respectively. The acquisition of a lone menthone reductase from this immature oil gland library was entirely consistent with the developmental studies that suggested that the abundance of menthone reductase cDNAs in this library would be low (McConkey et al., 2000). Clearly, the cloning of the menthone reductase cDNAs would be facilitated by using a message pool derived from tissue undergoing the oil maturation program, at which time significant amounts of menthol are produced.
Oil analyses were conducted with developing leaves of peppermint plants during a 40-d period following cutting and regrowth to identify the time during which the rate of menthol production was highest (i.e. indicative of increased transcription of the target reductase). The rate of menthol accumulation was maximum from 25 to 33 d post leaf initiation, with increases of about 2% menthol (relative to the total oil) per day; maximum menthol levels (approximately 25% of total oil) were reached by 40 d (approximately 50% menthol is achieved in commercial production). Therefore, secretory cell clusters were isolated from fully expanded leaves 25 d post emergence, and RNA was extracted, purified, and used for cDNA synthesis.
Since the two NADPH-specific reductases utilize the same substrates (menthone or isomenthone) and differ only in stereochemistry of the reduction to the respective menthol isomers, and because the native reductases have quite similar physical properties (Kjonaas et al., 1982), it was reasoned that the MMR might share significant primary structure with the MNR. Sequence analysis of the cDNA encoding the MNR (see below) indicated it to be a member of the short-chain dehydrogenase/reductase (SDR) superfamily (Kallberg et al., 2002), like the previously acquired isopiperitenone reductase (Ringer et al., 2003), thereby suggesting that a homology-based PCR cloning approach, using primers designed to conserved sequences shared by these two redox enzymes and other SDR family members, could be productive in the search for the MMR (see Fig. 2). Thus, forward primers were designed to the NADPH-binding site (residues 21–30 in MNR) and to a downstream highly conserved, structural domain (residues 97–104 in MNR) found in other SDR family members. Reverse primers of minimum degeneracy were designed to four conserved regions of MNR and isopiperitenone reductase (corresponding to residues 171–176, 266–272, 305–310, and 313–320 of MNR). PCR amplification of the cDNA (synthesized from mRNA isolated from mature oil glands) with the eight primer pairs generated amplicons of the predicted size, and sequence analysis of six independent clones of each showed all but two to be 90% to 98% identical to isopiperitenone reductase. One clone, an approximately 500-bp gene fragment, was 100% identical to the MNR (amino acids 21–176), and the other, an approximately 525-bp gene fragment (corresponding to amino acids 97–272 of MNR) was only 72% identical to isopiperitenone reductase and 82% identical to MNR. The latter sequence was used to design specific primers for the 5′-RACE and 3′-RACE protocols that would exclude amplification of the isopiperitenone reductase and the MNR. Following amplification and cloning of the derived RACE amplicons, sequence from the 5′- and 3′ termini was obtained that included 59 and 104 bp of the respective untranslated regions and provided the information necessary to design new primers that would amplify the complete open reading frame of the cDNA and provide restriction sites for transfer into the expression vectors, pSBET and pET28a. PCR amplification from the 5′-RACE cDNA pool yielded two clone types, one of which was missing 75 internal nucleotides from a region corresponding to amino acids 180 to 204 of the MNR clone. Both the long clone (designated 6-2; two independent clones were sequenced and showed 100% nucleotide identity within the coding region) and short clone (designated 6-3) were expressed and evaluated for reductase function.
Figure 2.
Alignment of the deduced amino acid sequences of the (−)-menthone:(−)-(3R)-menthol reductase (MMR) and (−)-menthone:(+)-(3S)-neomenthol reductase (MNR), isopiperitenone reductase (ISPR) and (−)-trans-isopiperitenol dehydrogenase (ISPD) of peppermint. Highly conserved sequences (between MNR and ISPR) used to synthesize primers for the RT-PCR-RACE-based cloning of MMR are overscored by bars and the conserved motifs discussed in the accompanying paper (Ringer et al., 2005) are underlined by bars. Identical residues and similar residues are shaded black and gray, respectively.
In Vitro Demonstration of Menthone Reductase Activity
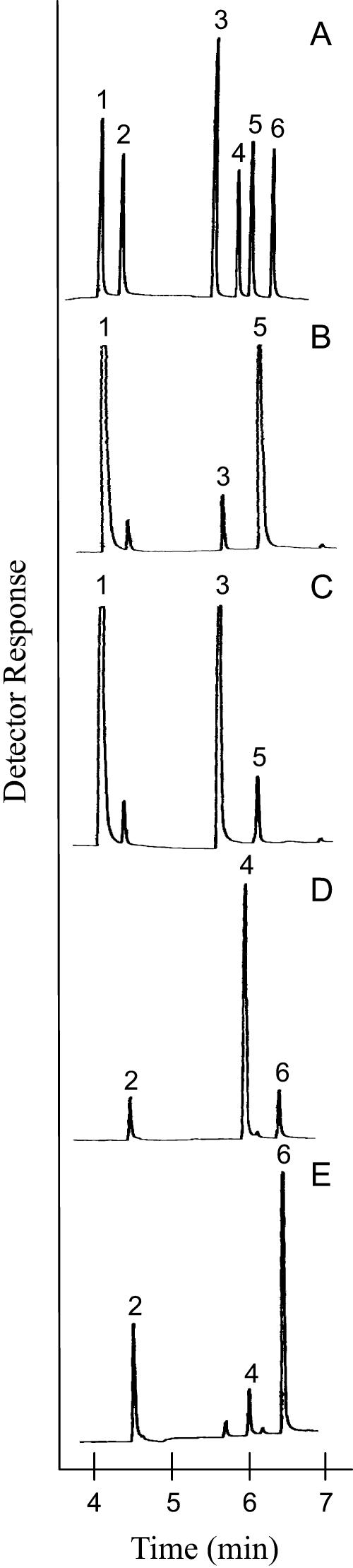
Cell-free assay of the partially purified recombinant protein derived from clone ml472 confirmed that an authentic MNR had been isolated, as this clone yielded a reductase capable of converting (−)-menthone to 94% (+)-(3S)-neomenthol and 6% (−)-(3R)-menthol, and (+)-isomenthone to 86% (+)-(3S)-isomenthol and 14% (+)-(3R)-neoisomenthol in the presence of NADPH (Fig. 3). The long (6-2) and the short (6-3) putative reductase clones obtained from the reverse transcription (RT)-PCR and RACE protocols described above were also expressed in E. coli, and the resulting partially purified recombinant proteins were assayed for menthone reductase activity. The expressed protein obtained from clone 6-3 (the shorter sequence) displayed no detectable reductase activity and was not characterized further. The protein product derived from clone 6-2 (longer version) was functional and, in the presence of NADPH, produced 95% (−)-(3R)-menthol and 5% (+)-(3S)-neomenthol from (−)-menthone, and 87% (+)-(3R)-neoisomenthol and 13% (+)-(3S)-isomenthol from (+)-isomenthone (Fig. 3), thus confirming the identity of this clone as MMR; the identities of the enzyme products were verified by gas chromatography-mass spectrometry (GC-MS) analysis. These two (−)-menthone/(+)-isomenthone reductases account for the production of all of the menthol diastereoisomers found in peppermint oil.
Figure 3.
Gas chromatographic analyses of monoterpenol formation by recombinant MMR with menthone (B) and isomenthone (D) as substrates, and by recombinant MNR with menthone (C) and isomenthone (E) as substrates. The monoterpene standards (A) are menthone (1), isomenthone (2), neomenthol (3), neoisomenthol (4), menthol (5), and isomenthol (6). Assays for the conversion of isomenthone to monoterpenols (D and E) were scaled up 12-fold in time and 16-fold in protein concentration to increase product yield to that comparable for the conversions of menthone to monoterpenols (B and C).
Sequence Analysis and Enzyme Characterization
The MNR cDNA (clone ml472) encodes a protein of 324 amino acids (1,131 nucleotides) with a calculated molecular mass of 35,722, and the MMR cDNA (clone 6-2) encodes a 311 residue protein (1,096 nucleotides) with a deduced molecular mass of 34,070. The molecular mass of each reductase is consistent with the 35-kD size estimate previously obtained by gel permeation chromatography of both native enzymes (Kjonaas et al., 1982). The apparent absence of N-terminal organellar targeting information is consistent with a cytoplasmic localization for both enzymes.
Both reductases exhibited broad pH response curves, with the pH optimum for the MMR at 7.0 with one-half maximal activities at pH 4.5 and 8.5, while the MNR exhibited a pH optimum at 9.3 with one-half maximal activities at pH 6.5 and 10.5. Of the alternate substrates tested [(−)-isopiperitenone, (+)-cis-isopulegone, (+)-pulegone, (+)-menthone, and (−)-carvone], the (3R)- and (3S)-ketoreductases were only able to detectably reduce (+)-menthone to the corresponding monoterpenols, each maintaining the indicated stereoselectivity. NADH served as a much less efficient cofactor than NADPH (less than 20% turnover at saturation under linear reaction conditions), but the stereochemical fidelity of (−)-menthone and (+)-isomenthone reduction was maintained. Oxidation of each of the menthol diastereoisomers in the presence of NADP+ was evaluated, and the only significant conversion was of neomenthol to menthone by the MNR (at the same rate as the forward reaction but with very high Km for the alcohol substrate).
The facile purification and high yield of the His-tagged MMR prompted initial kinetic studies aimed at determining what, if any, effect the N-terminal His tag (consisting of 20 extra amino acids including six His) would have on kinetic parameters. Thus, nonlinear regression analysis of Michaelis-Menten plots was used to obtain Km values and turnover numbers for the His-tagged MMR and its untagged recombinant counterpart for menthone and NADPH as cosubstrates. Although similar Km values were observed for the ketone substrate, an increase by an order of magnitude in the Km value for NADPH with the His-tagged reductase relative to the untagged form (data not shown) was noted. The kinetically compromised His-tagged enzyme was not examined further, and kinetic constants are reported for the untagged recombinant enzymes expressed from pSBET (Table I). The kcat determined for both reductases with isomenthone as substrate are approximately an order of magnitude lower than the turnover numbers reported using menthone (data not shown). The Km values for menthone reported here for the MMR and the MNR (3 μm and 650 μm, respectively) are at odds with those values reported previously for the native peppermint reductases (250 μm and 22 μm, respectively; Kjonaas et al., 1982); this suggests that the kinetic data were transposed in the figures presented in the earlier account.
Table I.
Kinetic parameters for MNR and MMR
Assays to determine Km values utilized are listed as footnotes.
|
Km ± se
|
kcatd
|
|||||
|---|---|---|---|---|---|---|
| Menthonea | Isomenthonea | NADPHb | Monoterpenolc | NADP+ | ||
| μm | s−1 | |||||
| MNR | 674 ± 78 | >1,000 | 10 ± 1 | >1,000e | 5.4 ± 0.5e | 0.06 |
| MMR | 3.0 ± 0.6 | 41 ± 5 | 0.12 ± 0.04 | N.D.f | N.D.f | 0.6 |
NADPH as cofactor.
Menthone as substrate.
Menthol, neomenthol, isomenthol or neoisomenthol as monoterpenol substrate and NADP+ as cofactor.
Turnover numbers were determined using menthone and NADPH.
Neomenthol was the only monoterpenol that afforded sufficient turnover to determine kinetic values.
N.D. indicates the values were not determinable due to insufficient turnover of any of the monoterpenols.
DISCUSSION
The molecular genetic basis of two NADPH-dependent reductases involved in the developmentally late-onset reduction of menthone to menthol and neomenthol is reported. A single acquisition of the MNR was obtained from a novel in situ screen of an immature oil gland cDNA library that is highly enriched in early isoprenoid pathway biosynthetic genes (Lange et al., 2000). A cDNA for the MMR was obtained using an RT-PCR-RACE based method from mRNA prepared from mature oil gland secretory cells isolated from leaves undergoing the late onset oil maturation program.
Not surprisingly, the menthone reductases share 73% identity at the amino acid level, and these carbonyl reductases (neither enzyme possessed double-bond reductase activity; see below) appear to share a common evolutionary ancestry (i.e. 64%–66% amino acid identity) with the mechanistically distinct isopiperitenone reductase (Fig. 2), a Δ1,2-double-bond reductase that does not possess carbonyl reductase activity (Ringer et al., 2003), and both are much more distant from the NAD+-dependent isopiperitenol dehydrogenase (12%–13% amino acid identity; Ringer et al., 2005). All four of these peppermint redox enzymes belong to the SDR based on similar length (300 ± 50 amino acids) and the common presence of several highly conserved sequence motifs, including the N-terminal cofactor-binding domain (GxxxGxG), a downstream structural domain [(N/C)NAG] of undefined function, and a catalytic domain (YxxxK; Kallberg et al., 2002; Ringer et al., 2003). Notably, pulegone reductase, an NADPH-dependent, Δ4,8-double-bond reductase very similar in mechanism to isopiperitenone reductase (Fig. 1), is a member of the medium-chain dehydrogenase/reductase superfamily (MDR; Ringer et al., 2003), and it shares little sequence identity with the peppermint SDRs and contains none of the characteristic SDR motifs noted above. This observation suggests that the origins of isopiperitenone and menthone reductases, and the more distant isopiperitenol dehydrogenase (SDR) and pulegone reductase (MDR), are not related by similar reaction type (i.e. strict carbonyl reduction versus strict double-bond reduction versus alcohol dehydrogenation) but likely have evolved from distinct ancestral redox genes that encoded enzymes with shared binding determinants for these structurally similar monoterpenoid substrates.
The product profiles generated by the recombinant reductases with (−)-menthone (1R, 4S) or (+)-isomenthone (1R, 4R) as substrate are consistent with previous work with the native enzymes that demonstrated stereoselectivity of product formation by MMR for (3R)-configured monoterpenols and by MNR for (3S)-configured monoterpenols (Kjonaas et al., 1982). Furthermore, stereoselectivity was maintained by both recombinant ketoreductases when each was presented with (+)-menthone (1S, 4R) as an alternate substrate, suggesting that hydride transfer from the nicotinamide ring occurs to the si face or to the re face of the prochiral carbonyl group yielding (3R)-monoterpenols (MMR) or (3S)-monoterpenols (MNR), respectively, without consequence of the spatial orientation of the methyl or isopropyl substituents. That catalysis with each of these menthone isomers also generates minor amounts of the oppositely configured alcohol [i.e. (3S)- and (3R)-configured monoterpenols for MMR and MNR, respectively] suggests an active site permissive enough to allow sufficient movement to change orientation of the substrate (or cofactor). Nevertheless, the lack of detectable activity with (+)-cis-isopulegone (1R, 4R), (−)-isopiperitenone (planar at C1, 4R), and (+)-pulegone (1R, planar at C4) as substrate for either enzyme suggests reasonably strict substrate selectivity; further studies are needed to decipher the subtle features of these reductases that underlie substrate recognition and reaction stereochemistry.
The efficient utilization of (−)-menthone and (+)-isomenthone by both menthone reductases is of note because, by contrast, previous studies with the terpenone reductases of menthol biosynthesis, pulegone reductase and isopiperitenone reductase, showed these early pathway double bond reductases to exhibit very strict specificity (Ringer et al., 2003). The broader substrate specificity of the menthone reductases is, nevertheless, of little significance in vivo because principally (−)-menthone accumulates during the secretory phase of peppermint oil gland development [i.e. the (−)-menthone to (+)-isomenthone ratio is approximately 10:1, and only trace levels of other monoterpene ketones occur in the essential oil]. As a consequence, the major monoterpenol produced is (−)-menthol, which often comprises over 50% of the mature oil (with a ratio of menthol to the total of other diastereoisomers of about 20:1).
The menthone reductases were tested for the ability to oxidize monoterpenols in the presence of NADP+, and the only significant reverse reaction was by MNR in the oxidation of neomenthol at rates comparable to the reduction of menthone. Although the Km value for (+)-neomenthol in this reverse reaction to (−)-menthone exceeded 1 mm, the Km for the cofactor was low (5 μm). Thus, over the time frame for oil maturation in planta (several weeks), the reverse oxidation of neomenthol to menthone may be of significance and, coupled with the approximately 2,000-fold lower catalytic efficiency (kcat/Km) for menthone reduction by MNR compared with MMR (Table I), is consistent with the very low neomenthol levels observed in peppermint oil (<3%;Lawrence, 1978).
The availability of DNA probes for, and antibodies directed against, several enzymes of monoterpene biosynthesis in peppermint has permitted evaluation of the regulation (Gershenzon et al., 2000; McConkey et al., 2000) and organization (Turner et al., 1999) of early steps of essential oil formation in developing glands. The cloning of the menthone reductases now provides a means of defining the organization and transcriptional and translational control of these critical enzymes of the essential oil maturation process.
Transgenic manipulation of enzymes involved in de novo monoterpene biosynthesis in peppermint has led to increased essential oil yield and improved oil composition (Mahmoud and Croteau, 2001). In this context, constitutive overexpression of MMR in secretory cells, independent of the normal maturation process, has the potential to increase the conversion of menthone to menthol from the outset of oil gland development. As noted above, the specificity of these ketoreductases for (−)-menthone and (+)-isomenthone as substrates would appear to preclude the possibility of adventitious reduction of the early pathway ketones and suggests the possibility of producing an oil with high menthol content in younger plants, thereby permitting earlier harvest and offering the potential for two harvests of good quality oil per season from this perennial crop.
MATERIALS AND METHODS
Plants, Enzymes, Substrates, and Reagents
Peppermint (Mentha x piperita) L. cv Black Mitcham plants were propagated from stem cuttings and were grown during the summer in a greenhouse with supplemental lighting (14-/10-h photoperiod at approximately 70 μmol photons m−2 s−1) and maintained at 30°C/16°C day/night temperature. Plants were grown in Sunshine Mix LC1 (SunGro Horticulture, Bellevue, WA) with daily watering and fertilized with 200 μg mL−1 nitrogen concentrate 5 times per week. For the purpose of oil gland secretory cell isolation and oil analysis, leaves were harvested at periodic intervals for 40 d following leaf initiation; sampling procedures have been previously described (Gershenzon et al., 2000). Enzymes and reagents were obtained from New England Biolabs (Beverly, MA), Promega (Madison, WI), Stratagene (La Jolla, CA), Sigma Chemical (St. Louis), Research Products International (Mt. Prospect, IL), Fisher Scientific (Fairlawn, NJ), and EM Science (Gibbstown, NJ), and were used according to the manufacturers' instructions. Substrates and standards were obtained from Haarmann and Reimer (Holzminden, Germany), Fluka Chemical (Buchs, Switzerland), and Aldrich Chemical (Milwaukee, WI), or were from our own collection and were purified as necessary (to >99%) by silica gel chromatography using step gradients of diethyl ether in pentane.
Oil Analysis and Product Identification
Simultaneous steam distillation and solvent extraction of 3 to 8 g samples of peppermint leaves were performed using 10 mL of pentane in a condenser-cooled Likens-Nickerson apparatus (J and W Scientific, Folsom, CA) as described (Ringer et al., 2003). Microbial cultures (that expressed the target genes; see below) used for in situ feeding experiments (100 mL) were similarly distilled and extracted. Capillary GC (flame ionization detection) was used for the preliminary identification and quantification of monoterpenes, and employed a Hewlett-Packard model 5890 Series II gas chromatograph (Palo Alto, CA.) using a 30-m × 0.25-mm id fused-silica capillary column with a 0.25-μm film of polyethylene glycol ester coating (AT-1000, Alltech, Deerfield, IL). Cool on-column injection was used with programming from 40°C to 50°C at 40°C/min, 50°C to 120°C at 10°C/min, and 40°C/min to 220°C. A Hewlett-Packard 6890 Series GC-Mass Spectrometer (similar separation conditions, spectra collected at 70 eV and analyzed using Hewlett-Packard Chemstation software) was employed for product identification by comparison of retention times and mass spectra to those of authentic standards.
Oil Gland Secretory Cell Isolation
Oil gland secretory-cell isolation (as a prelude to RNA isolation) was carried out as previously described utilizing a leaf surface abrasion technique (Gershenzon et al., 1992) and a pH 6.6 buffer containing 0.5 m NaH2PO4, 0.2 m sorbitol, 10 mm Suc, 2 mm dithiothreitol, 1% (w/v) polyvinylpyrrolidone-40, 0.6% (w/v) methyl cellulose, and aurintricarboxylic acid (1 mm) and thiourea (5 mm) to inhibit RNase and phenol oxidase activities, respectively (McConkey et al., 2000). Secretory cell clusters, largely free from contaminating mesophyll cells and nonglandular trichomes, were frozen and stored in liquid N2 until use.
RNA Isolation and cDNA Preparation
Sonication of the thawed, isolated secretory cell clusters was performed in a 1:5 (v/v) mixture of the above described isolation buffer and a guanidinium-isothiocyanate-based buffer (Buffer RLT, Qiagen, Valencia, CA) at full power for 30 s using the Virtis model CL4 sonicator (Virtis, Gardiner, NY). Total RNA was purified using the RNeasy kit following the manufacturer's protocol (Qiagen). Approximately 1 to 5 μg of total RNA and an oligo(dT)16-20 were utilized as template and primer, respectively, for RT of mRNA using MMLV-reverse transcriptase (RNaseH−) following the indicated protocol (Promega). After 1 h at 42°C, the RT reaction was heated to 68°C for 3 min to denature RNA secondary structure, and an additional unit of the reverse transcriptase was added to the reaction mixture, which was incubated for 30 min at 42°C. Single-stranded cDNA was stored at −20°C until use.
Cloning of the Menthone Reductases
Sticky-end PCR (Zeng, 1998) was utilized for subcloning the MNR cDNA (designated ml472) from the EST library in pBluescript SK− into pSBET, a T7-based protein expression vector (Schenk et al., 1995). Primers for sticky-end PCR were the gene-specific forward primers 5′-TATGAAACACAACAACACAACACACAC-3′ and 5′-TGAAACACAACAACACAACACACAC-3′, and the vector-specific reverse primers 5′-CGTACCGGGCCCCCCCCTCGAG-3′ and 5′-GATCCGTACCGGGCCCCCCCCTCGAG-3′. All PCR amplifications were performed with 1 unit of Pfu DNA polymerase (Life Technologies, Gaithersburg, MD) with approximately 50 ng template, 0.2 mm dNTPs, 1.25 μm of each primer, and 1× Pfu reaction buffer in 25-μL reactions, using an MJ thermocycler model PTC-100 (MJ Research, Waltham, MA) with an initial 1-min denaturation step at 94°C, followed by 35 to 40 cycles of 94°C for 30 s, 53°C for 1 min, 72°C for 2 min, and final elongation at 72°C for 15 min, unless otherwise indicated. The gel-purified amplicons were ligated into pSBET as described (Zeng, 1998) and transformed into Escherichia coli XL2Blue cells (Stratagene) for sequence analysis or into E. coli BL21 (DE3) cells (Novagen, Madison, WI) for protein expression using standard protocols.
Cloning of the MMR by PCR amplification of the cDNA (see above) involved designing primers based on conserved regions (amino acid sequence) shared by isopiperitenone reductase and MNR. PCR reactions combined one of the following forward primers: 5′-GCRAACARAGGAATCGGG-3′; 5′-AGGAATCGGGTTCGAAATCTGC-3′; and 5′-GATATTCTGGTGAATAATGCAGGA-3′ with one of the following reverse primers: 5′-GGCCCYCCATCAGGCAGCA-3′; 5′-GGAATGAGGGCTTGTGTTA-3′; 5′-GCTTYGTCTCGAGKGAAGAAGCA-3′; and 5′-ATTTATGCRGAAACTCGGGTA-3′ to amplify internal fragments of the MMR. The resulting purified amplicons were cloned using topoisomerase T/A-based cloning methods (Invitrogen), and sequenced.
5′-RACE was accomplished by terminal transferase-mediated tailing of the single-stranded cDNA (obtained above) with dCTP (Frohman et al., 1988), and PCR amplification of the dC-tailed product using forward primer 5′-GGAAACAGCTATGACCATGACGGGIIGGGIIGGGIIGG-3′ and one of the following gene-specific reverse primers: 5′-ATCCGAGTATACGCATTAAC-3′; 5′-TTGTTGCAATTTACCATCAA-3′; 5′-TTCCTCCACCTTCCCTTCAT-3′; 5′-AGGCTGGAGCAGTAAGGTCGA-3′; and 5′-ATTGTTGGTGAATCAGATTT-3′. 3′-RACE utilized a 5′-modified oligo(dT) primer, 5′-CATTATGCTGAGTGATATCCCG(T)18-3′, for RT, followed by PCR amplification using a reverse primer that annealed to the 5′-end of the newly synthesized cDNA, 5′-CATTATGCTGAGTGATATCCCG-3′, and one of the following gene-specific, internal forward primers: 5′-TGAATAATGCAGGATTTACT-3′; 5′-CTTGAGGCAAACATTATTGC-3′; 5′-AACATTATTGCAGCTCAGGGTGG-3′; or 5′-CCTCTCCTGCAAAAATCTGA-3′. Amplification of the RT-PCR-RACE products utilized Taq DNA polymerase with the conditions described above, except that the Mg2+ concentration was 1.5 mm and an additional 1.5 mm dATP was added to the reaction mixture during the final incubation at 72°C. The resulting amplicons were gel purified and cloned into TOPO2.1 vector according to the manufacturer's instructions (Invitrogen).
5′- and 3′-RACE clones were sequenced, and this information was utilized to design a forward primer containing a 5′-NdeI site at the starting Met (5′-GGAATTCCATATGGCAGATACGTTTACCCAA-3′) and a reverse primer containing a 5′-BamHI site downstream of the stop codon (5′-CGCGGATCCTTACTAGATTTAGTACAAGGACAAGGC-3′) for PCR amplification of full-length reductases from the original cDNA. Following restriction digestion, the DNA was directionally ligated into pSBET for protein expression. N-terminal His6-tagged constructs were prepared by BamHI and NdeI restriction digestion of the pSBET constructs followed by ligation into similarly digested pET-28a vector (Invitrogen). Standard procedures were followed for ligations and alkaline lysis-based plasmid preparations.
Sequencing and Bioinformatics
All clones were fully sequenced using Amplitaq DNA polymerase and fluorescence cycle sequencing using an ABI Prism 373 DNA sequencer at the Washington State University Laboratory for Biotechnology and Bioanalysis. Sequences were analyzed and aligned using the GCG 10.0 sequence analysis package (GCG, Madison, WI) and the ClustalX v.1.83 multiple sequence alignment program (Thompson et al., 1997), and sequence comparisons were made using the BLAST algorithm (Altschul et al., 1990) at the National Center for Biotechnology Information (NCBI) Web-site (http://www.ncbi.nlm.nih.gov/BLAST). cDNAs encoding putative redox enzymes were identified for in situ functional screening in E. coli as described (Ringer et al., 2003).
Expression and Assay of Recombinant Menthone Reductases
Expression of recombinant terpene biosynthetic enzymes in E. coli has been described (Williams et al., 1998) and was carried out with the following minor changes. Bacteria harboring the pSBET-reductase plasmid or the pET-reductase plasmid were grown in 5-mL cultures of Luria-Bertani media to an OD600 of 0.8 to 1.0, at which time the cultures were cooled to 10°C, isopropyl-β-d-thiogalactopyranoside was added to 0.25 mm, and growth was continued for 12 to 18 h at 10°C with shaking at 220 rpm. Cells were pelleted by centrifugation for 25 min at 2,500g and 4°C, and then resuspended in 1 mL resuspension buffer (50 mm MOPSO, pH 7, with 10% (v/v) glycerol, 10% (w/v) sorbitol, and 10 mm β-mercaptoethanol) containing 0.4 mg/mL lysozyme. Following incubation on ice for 30 min, the extract was sonicated for 10 s with a Virtis sonicator using a microtip probe at medium power. Cell debris was pelleted by centrifugation at 27,000g at 4°C for 30 min, and the supernatant, containing the operationally soluble reductase, was used for initial characterization.
Preparative scale cultures (1.0 L) were grown and prepared essentially as described above, except that the pelleted E. coli cells were resuspended in 10 mL resuspension buffer and sonicated using the macrotip probe. Purification of the recombinant reductases generated from the pSBET vector was achieved by anion-exchange chromatography using Macro-Prep High Q media (Bio-Rad, Hercules, CA); although neither reductase bound to the matrix, significant purification was achieved and the resulting flow through and/or wash contained the reductase at approximately 90% purity. Purification of the N-terminal His6 tagged constructs utilized Ni-agarose chromatography (Qiagen) in which the recombinant reductase bound to 3 mL of the matrix, and, following a 30-mL wash (40 mm KH2PO4, pH 7, with 0.5 m NaCl, 10 mm β-mercaptoethanol, and 5 mm His), was eluted with a similar buffer containing 155 mm His to yield the reductase at >95% purity.
Preliminary assays contained 50 to 100 μL of the above enzyme preparation (15–50 μg protein) in 2 mL assay buffer (40 mm KH2PO4, pH 7, with 10 mm β-mercaptoethanol) containing 100 μm menthone and 500 μm NADPH. Following incubation with gentle shaking at 31°C for 12 h, 0.5 mL of pentane was added and was vigorously mixed to extract the monoterpene products that were analyzed by GC and GC-MS as described above. In situ functional assays utilized 100-mL E. coli cultures that were grown and induced under the conditions described above, except that 100 μm menthone was added directly to the culture at the time of induction. Following 15 h of shaking (250 rpm) at 15 to 20°C, these cultures were transferred to 0.5-L round-bottom flasks, to which 6.6 μmol of (+)-camphor was added as an internal standard, and the mixture was steam distilled and the distillate analyzed as described above.
The optimum pH was determined in assay buffer (pH 4–12 at 0.5 pH unit increments) containing 25 μm menthone and 500 μm NADPH to which 0.5 μg protein was added prior to initiation of the reaction (6 min at 31°C), with GC-based product analysis as described above. Substrate specificity was evaluated in assay buffer containing 0.5 μg reductase at the pH optimum and 100 μm monoterpenone substrate [(−)-isopiperitenone, (+)-pulegone, (+)-cis-isopulegone, (−)-carvone, or (+)-menthone] and 500 μm NADPH, or 100 μm monoterpenol substrate [(−)-menthol, (+)-isomenthol, (+)-neoisomenthol, or (+)-neomenthol] and 500 μm NADP+, by overnight incubation as described above.
Kinetic Analysis of Recombinant Menthone Reductases
Typical kinetic assays were performed essentially as described (Ringer et al., 2003) under linear reaction conditions (with respect to protein and time) with the following changes. Assay mixtures containing 2 mL of 40 mm KH2PO4 at pH 7 (MMR) or at pH 9.3 (MNR), with 10 mm β-mercaptoethanol, were combined with 500 μm cofactor and concentrations of monoterpene substrate ranging from 2.5 μm to 1 mm, or with 100 μm monoterpene substrate, and concentrations of cofactor ranging from 0.1 μm to 1 mm, and preheated to 31°C before initiation of the reaction by enzyme addition (0.008–1.3 μg protein). The reactions were quenched after 6 to 9 min by addition of 0.5 mL pentane containing camphor as internal standard (10 μm) followed by vigorous mixing and cooling on ice; quantitation of products in the pentane extract was achieved by capillary-GC using the conditions described above with peak area quantification using the internal standard for normalization. The relatively high Km value for neomenthol in the reverse (oxidation) reaction by MNR (>1 mm) precluded accurate quantification of the product by capillary-GC; thus, kinetic constants were determined spectrophotometrically (ΔA340) by observing the appearance of NADPH. Protein concentration of the purified N-terminally His6 tagged MMR was determined spectrophotometrically (A280) using an extinction coefficient of 23,260 m−1 cm−1. Recombinant menthone reductases that lacked the His-tag were quantified by densitometry following separation by SDS-PAGE and Coomassie Blue (R250) staining, using the His6 tagged form as standard. Kinetic constants, representing the average of at least two independent experiments, were determined by nonlinear regression analysis of Michaelis-Menten plots using Enzyme Kinetics, version 1.11 (Trinity Software, Plymouth, NH). The values reported are the means ± se.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY288137 and AY288138.
Acknowledgments
We thank Rachael Parkin, Amanda Grimm, and Derek Pouchnik for DNA sequencing and Julianna Gothard for raising the plants.
This work was supported by the U.S. Department of Energy, by the Mint Industry Research Council, and by the Washington State University Agricultural Research Center (project 0268).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.053306.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Bertea CM, Schalk M, Karp F, Maffei M, Croteau R (2001) Demonstration that menthofuran synthase of mint (Mentha) is a cytochrome P450 monooxygenase: cloning, functional expression, and characterization of the responsible gene. Arch Biochem Biophys 390: 279–286 [DOI] [PubMed] [Google Scholar]
- Burke CB, Wildung M, Croteau R (1999) Geranyl diphosphate synthase: cloning, expression, and characterization of this prenyltransferase as a heterodimer. Proc Natl Acad Sci USA 96: 13062–13067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau R, Hooper CL (1979) Metabolism of monoterpenes: acetylation of (-)-menthol by a soluble enzyme preparation from peppermint (Mentha piperita) leaves. Plant Physiol 61: 737–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau R, Martinkus C (1979) Metabolism of monoterpenes: demonstration of (+)-neomenthyl-β-D-glucoside as a major metabolite of (-)-menthone in peppermint (Mentha piperita). Plant Physiol 64: 169–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau R, Winters JN (1982) Demonstration of the intercellular compartmentation of l-menthone metabolism in peppermint (Mentha piperita) leaves. Plant Physiol 69: 975–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby SM, Alonso WR, Katahira EJ, McGarvey DJ, Croteau R (1993) 4S-Limonene synthase from the oil glands of spearmint (Mentha spicata): cDNA isolation, characterization and bacterial expression of the catalytically active monoterpene cyclase. J Biol Chem 268: 23016–23024 [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin GR (1988) Rapid production of full-length cDNA from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA 85: 8998–9002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenzon JG, McCaskill D, Rajaonarivony JIM, Mihaliak C, Karp F, Croteau R (1992) Isolation of secretory cells from plant glandular trichomes and their use in biosynthetic studies of monoterpenes and other gland products. Anal Biochem 200: 130–138 [DOI] [PubMed] [Google Scholar]
- Gershenzon J, McConkey M, Croteau R (2000) Regulation of monoterpene accumulation in leaves of peppermint. Plant Physiol 122: 205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallberg Y, Oppermann U, Jörnvall H, Persson B (2002) Short-chain dehydrogenases/reductases (SDR): coenzyme-based functional assignments in completed genomes. Eur J Biochem 269: 4409–441712230552 [Google Scholar]
- Kjonaas R, Croteau R (1983) Demonstration that limonene is the first cyclic intermediate in the biosynthesis of oxygenated p-menthane monoterpenes in Mentha piperita and other Mentha species. Arch Biochem Biophys 220: 79–89 [DOI] [PubMed] [Google Scholar]
- Kjonaas R, Martinkus-Taylor C, Croteau R (1982) Metabolism of monoterpenes: conversion of l-menthone to l-menthol and d-neomenthol by stereospecific dehydrogenases from peppermint (Mentha piperita) leaves. Plant Physiol 69: 1013–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BM, Wildung MR, Stauber EJ, Sanchez C, Pouchnik D, Croteau R (2000) Probing essential oil biosynthesis and secretion by functional evaluation of expressed sequence tags from mint glandular trichomes. Proc Natl Acad Sci USA 97: 2934–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence BM (1978) A study of the monoterpene interrelationships in the genus Mentha with special reference to the origin of pulegone and menthofuran. PhD thesis. University of Groningen, Groningen, The Netherlands
- Lupien S, Karp F, Wildung M, Croteau R (1999) Regiospecific cytochrome P450 limonene hydroxylases from mint (Mentha) species: cDNA isolation, characterization, and functional expression of (-)-4S-limonene-3-hydroxylase and (-)-4S-limonene-6-hydroxylase. Arch Biochem Biophys 368: 181–192 [DOI] [PubMed] [Google Scholar]
- Mahmoud SS, Croteau R (2001) Metabolic engineering of essential oil yield and composition in mint by altering expression of deoxyxylulose phosphate reductoisomerase and menthofuran synthase. Proc Natl Acad Sci USA 98: 8915–8920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinkus C, Croteau R (1981) Metabolism of monoterpenes: evidence for compartmentation of l-menthone metabolism in peppermint (Mentha piperita) leaves. Plant Physiol 68: 99–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaskill D, Gershenzon J, Croteau R (1992) Morphology and monterpene biosynthetic capabilities of secretory cell clusters isolated from glandular trichomes of peppermint (Mentha piperita L.). Planta 187: 445–454 [DOI] [PubMed] [Google Scholar]
- McConkey M, Gershenzon J, Croteau R (2000) Developmental regulation of monoterpene biosynthesis in the glandular trichomes of peppermint. Plant Physiol 122: 215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringer KL, Davis EM, Croteau R (2005) Monoterpene metabolism. Cloning, expression and characterization of (−)-isopiperitenol/(−)-carveol dehydrogenase of peppermint and spearmint. Plant Physiol 137: 863–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringer KL, McConkey ME, Davis EM, Rushing GW, Croteau R (2003) Monoterpene double-bond reductases of the (-)-menthol biosynthetic pathway: isolation and characterization of cDNAs encoding (-)-isopiperitenone reductase and (+)-pulegone reductase of peppermint. Arch Biochem Biophys 418: 80–92 [DOI] [PubMed] [Google Scholar]
- Schenk PM, Baumann S, Mattes R, Steinbiss H-H (1995) Improved high-level expression system for eukaryotic genes in Escherichia coli using T7 RNA polymerase and rare ArgtRNAs. Biotechniques 19: 196–200 [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner G, Gershenzon J, Croteau R (2000) Distribution of peltate glandular trichomes on developing leaves of peppermint. Plant Physiol 124: 655–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner G, Gershenzon J, Nielson EE, Froehlich JE, Croteau R (1999) Limonene synthase, the enzyme responsible for monoterpene biosynthesis in peppermint, is localized to leucoplasts of oil gland secretory cells. Plant Physiol 120: 879–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DC, McGarvey DJ, Katahira EJ, Croteau R (1998) Truncation of limonene synthase preprotein provides a fully active ‘pseudomature’ form of this monoterpene cyclase and reveals the function of the amino-terminal arginine pair. Biochemistry 37: 12213–12220 [DOI] [PubMed] [Google Scholar]
- Zeng G (1998) Sticky-end PCR: new method for subcloning. Biotechniques 25: 206–208 [DOI] [PubMed] [Google Scholar]