ABSTRACT
Borrelia burgdorferi, the pathogen of Lyme disease, encodes many conserved proteins of unknown structure or function, including ones that serve essential roles in microbial infectivity. One such protein is BB0238, which folds into a two-domain protein, as we have determined by X-ray crystallography and AlphaFold analysis. The N-terminal domain begins with a helix-turn-helix motif (HTH), previously referred to as a tetratricopeptide repeat (TPR) motif, known to mediate protein-protein interactions. The fold of the C-terminal domain has been seen in proteins with a range of unrelated activities and thus does not infer function. In addition to its previously known binding partner BB0323, another essential borrelial virulence determinant, we show that BB0238 also binds a second protein, BB0108, a borrelial ortholog of the chaperone protein SurA and the peptidyl-prolyl cis/trans isomerase protein PrsA. An in vitro enzymatic assay confirmed the catalytic activity. We also determined the crystal structure of the catalytic domain of BB0108, which revealed the parvulin-type organization of the key catalytic residues. We show that BB0238 influences the proteolytic processing of BB0323, although the TPR/HTH motif is not involved in the process. Instead, we show that the motif stabilizes BB0238 in the host environment and facilitates tick-to-mouse pathogen transmission by aiding spirochete evasion of early host cellular immunity. Taken together, these studies highlight the biological significance of BB0238 and its interactions with multiple B. burgdorferi proteins essential for microbial infection.
IMPORTANCE
Lyme disease is a major tick-borne infection caused by a bacterial pathogen called Borrelia burgdorferi, which is transmitted by ticks and affects hundreds of thousands of people every year. These bacterial pathogens are distinct from other genera of microbes because of their distinct features and ability to transmit a multi-system infection to a range of vertebrates, including humans. Progress in understanding the infection biology of Lyme disease, and thus advancements towards its prevention, are hindered by an incomplete understanding of the microbiology of B. burgdorferi, partly due to the occurrence of many unique borrelial proteins that are structurally unrelated to proteins of known functions yet are indispensable for pathogen survival. We herein report the use of diverse technologies to examine the structure and function of a unique B. burgdorferi protein, annotated as BB0238—an essential virulence determinant. We show that the protein is structurally organized into two distinct domains, is involved in multiplex protein-protein interactions, and facilitates tick-to-mouse pathogen transmission by aiding microbial evasion of early host cellular immunity. We believe that our findings will further enrich our understanding of the microbiology of B. burgdorferi, potentially impacting the future development of novel prevention strategies against a widespread tick-transmitted infection.
KEYWORDS: Borrelia burgdorferi, tick-borne pathogens, immune evasion
INTRODUCTION
Borrelia burgdorferi sensu lato, the agent of Lyme borreliosis, displays striking evolutionary divergence from other trees of life (1) and features a plethora of unique proteins that are relevant to the atypical biology and infectivity of these spirochetes (2). B. burgdorferi (Bb) and related spirochetes, which can evade host immunity and establish long-term infections in a variety of vertebrate hosts, including humans (3), are responsible for the most prevalent vector-borne illnesses in the United States and many other parts of the globe (4, 5). Bb survives in nature through an enzootic infection cycle that incorporates a disparate set of hosts, usually wild mammals, such as mice and deer, and tick vectors belonging to the Ixodes species complex (3). Ticks can transmit B. burgdorferi into the skin of their mammalian hosts, including humans or domesticated animals, during the engorgement process. Bb remains locally at the tick bite site on the dermis for several days to weeks, and then migrates to many internal organs, including the joints, heart, and central nervous system, where a cascade of inflammatory responses can be triggered; this ultimately results in clinical complications, most frequently manifesting as Lyme arthritis, Lyme carditis, and a variety of neurological disorders termed as neuroborreliosis (6). No vaccine exists against Bb infection in humans (7, 8); hence, the disease is treated with a regimen of antibiotics. However, several months to years after antibiotic treatment, a substantial fraction of patients experiences a range of neurological or other clinical symptoms collectively known as post-treatment Lyme disease syndrome, with unknown etiology and pathogenesis and no further treatment options (9, 10). Therefore, a more thorough understanding of the intriguing biology of Bb, including the molecular bases of immune evasion, virulence, and pathogenesis, may contribute to the development of new vaccines or antimicrobials against Lyme disease.
B. burgdorferi features distinctive outer surface and cellular organization dictated by an unusually segmented genome encoding numerous proteins that lack orthologs in other bacterial species, even in related spirochetes (11 – 13). Many of these unique gene-products have been shown to serve essential roles in microbial survival in the tick-rodent infection cycle (3, 14). While a number of these proteins support borrelial infection, either in mammals or in ticks, a conserved gene-product annotated as BB0323 is essential for B. burgdorferi survival throughout its entire infection cycle (15, 16). BB0323 is a multifunctional virulence determinant that undergoes a complex post-translational maturation process via proteolytic events (17), yielding two mature polypeptides with distinct functional roles in spirochete biology and infection (18). The maturation, stability, and functions of BB0323 require multifaceted protein-protein interaction (PPI) events, which support spirochete infectivity (19). These BB0323 PPI events involve multiple Bb proteins, including a protease-chaperone called B. burgdorferi high temperature requirement protein A (BbHtrA) (18, 20) and a membrane-associated protein of unknown structure or function, annotated as BB0238 (21, 22). The deletion of each of these genes renders Bb noninfectious in mice, underscoring their biological significance (15, 21 – 23). Although Bb sensu lato strains feature variable components of extra-chromosomal elements that can be unstable and polymorphic, these PPI-associated genes are encoded by the chromosome and are highly conserved (13). All three proteins also localize to the periplasm (18, 21). However, it remains obscure precisely how these proteins, either independently or as a complex, support B. burgdorferi biology and infection. Structurally, BbHtrA bears homology to the HtrA family of chaperone-proteases with a serine protease domain and two copies of a common structural domain of 80–90 amino acids called the PDZ domain (24), although the amino acid sequence is markedly different in Bb compared to other organisms (23). In contrast, the BB0323 and BB0238 proteins, whose post-translational stabilities are mutually interdependent, bear no significant homology to proteins of known function, except for a C-terminal peptidoglycan-binding motif called LysM in BB0323 (15) and a putative N-terminal PPI motif referred to previously as tetratricopeptide repeat (TPR) in BB0238 (22). The only available 3D protein crystal structure is that of the N-terminal region of BB0323 (25), which exhibits a rod-like structure with an overall fold belonging to the spectrin superfamily, generally absent in bacterial proteins. Spectrins function as linkers between actin and other cytoskeletal proteins in eukaryotes (26, 27). To gain further insight into how these unique borrelial proteins support multiple aspects of their poorly defined yet essential biological functions, we undertook a detailed structure-function analysis of BB0238. We provide evidence that BB0238 function is relevant to immune evasion in mammals, and identify an additional PPI involving a borrelial ortholog of a chaperone/peptidyl-prolyl cis/trans isomerase, BB0108. BB0238 and its partner proteins could represent potential targets for novel antimicrobial strategies against Lyme disease.
RESULTS
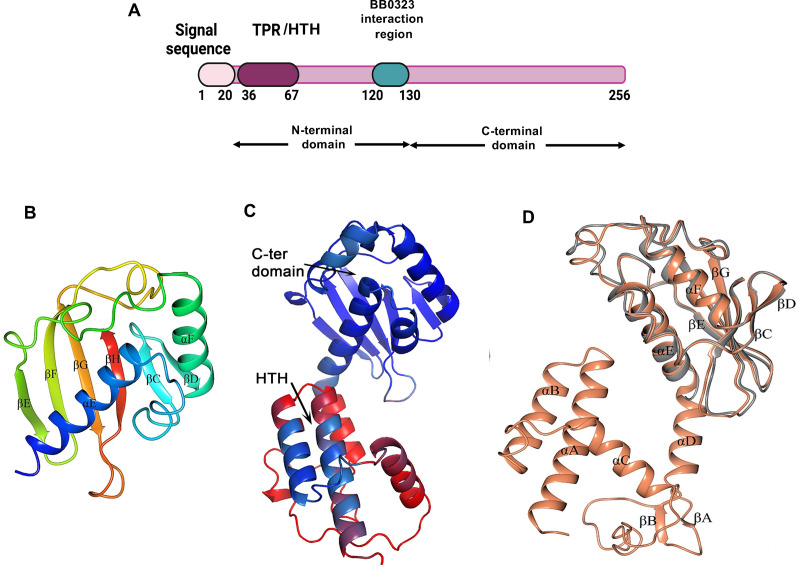
BB0238 features a two-domain structure with a globular carboxyl-terminal domain determined by X-ray crystallography
The B. burgdorferi (Bb) protein BB0238 (Fig. 1A) is essential for spirochete infection of mammalian hosts (21, 22). BB0238 also plays a critical role in maintaining the protein stability of another virulence determinant, BB0323 (21). However, the structure or molecular functions of the protein in spirochete biology and infectivity, including that of its N-terminal region harboring the putative TPR-like motif (22), remain enigmatic. As the primary sequence of BB0238 displays no homology to proteins of known function or structure, we sought to define its crystal structure. Multiple crystallization trials with the full-length protein were unsuccessful; however, one of the truncated versions, ranging amino acids 118–256 (designated as BB0238118-256) and its Se-Met version, formed crystals that diffracted X-ray to resolutions of 2.1 Å and 2.15 Å, respectively (Table 1), which enabled structure determination using the anomalous diffraction of the Se-Met containing protein. However, residues 118–131 were associated with uninterpretable electron density and could not be modeled, including residues 120–130, whose deletion prevents the interaction with BB0323 (19). Henceforth, the crystal structure will be termed as BB0238132-256 (Fig. 1B). The structure reveals a globular domain that adopts an α + β fold comprising two α-helices and six β-strands associated with the nuclear transport factor 2 family, which is present in proteins that have unrelated activities. Thus, the structure cannot inform function assignment in this case.
Fig 1.
Structural organization of BB0238. (A) Schematic representation of full-length BB0238 depicting a signal sequence, with N-terminal and C-terminal domains, and the known interaction site with BB0323. The region encompassing the TPR-like motif, herein denoted as helix-turn-helix region (TPR/HTH), is also indicated. (B) Crystal structure of B. burgdorferi BB0238132-256 (PDB ID 8P33). The structure is spectrum-colored from blue at the N-terminus to red at the C-terminus. (C) A model of full-length BB0238 as predicted by AlphaFold2. The cartoon is shown with coloring according to pLDDT values, with red corresponding to the lowest confidence and blue to the highest confidence. (D) Crystal structure of B. burgdorferi BB0238132-256 (gray) superimposed with the AlphaFold model of BB023824-256 (orange). All α-helices (αA-αF) and β-sheets (βA-βH) are labeled.
TABLE 1.
Data collection and structure refinement statistics a
| Data set | BB0238 native | BB0238 Se-Met | BB0108 PPIase |
|---|---|---|---|
| X-ray diffraction data | |||
| PDB entry | 8P33 | 8P32 | 8SOT |
| Beamline | 14.1 BESSY II, Helmholtz-Zentrum, Berlin | 14.1 BESSY II, Helmholtz-Zentrum, Berlin | 23-ID-B, APS |
| Space group | P 41212 | P 41212 | C2 |
| No. of molecules in asymmetric unit | 1 | 1 | 2 |
| Solvent content (%) | 45 | 45 | 42 |
| a, b, c (Å) | 55.5, 55.5, 95.1 | 55.2, 55.2, 94.5 | 101.9, 38.8, 53.6 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 106.0, 90.0 |
| Wavelength (Å) | 0.91840 | 0.97880 | 1.0332 |
| Resolution (Å) | 47.97–2.10 | 94.53–2.15 | 31.47–1.99 |
| Highest resolution bin (Å) | 2.16–2.10 | 2.21–2.15 | 2.06–1.99 |
| No. of reflections | 64,483 | 205,383 | 42,478 |
| No. of unique reflections | 9,268 | 8,514 | 13,591 |
| Completeness (%) | 100.0 (100.0) | 100.0 (100.0) | 96.47 (92.17) |
| R merge b | 0.02 (0.32) | 0.08 (0.33) | 0.06 (0.27) |
| CC1/2 | 1.00 (0.96) | 0.99 (0.98) | 1.00 (0.92) |
| I/σ (I) | 28.8 (3.8) | 24.1 (9.8) | 13.5 (3.7) |
| Multiplicity | 7.0 (7.3) | 13.1 (11.7) | 3.1 (3.2) |
| Refinement | |||
| R work b | 0.208 (0.250) | 0.201 (0.183) | 0.201 (0.1944) |
| R free b | 0.297 (0.298) | 0.257 (0.266) | 0.247 (0.261) |
| Average B-factor (Å2) | |||
| Overall | 57.0 | 40.0 | 33.0 |
| From Wilson plot | 51.3 | 36.6 | 29.7 |
| No. of atoms | |||
| Protein | 1,015 | 1,015 | 1,761 c |
| Water | 30 | 46 | 88 |
| RMS deviations from ideal | |||
| Bond lengths (Å) | 0.007 | 0.009 | 0.006 |
| Bond angles (o) | 1.473 | 1.610 | 0.81 |
| Ramachandran outliers (%) | |||
| Residues in most favored regions (%) | 93.50 | 95.93 | 99.1 |
| Residues in allowed regions (%) | 6.50 | 4.07 | 0.9 |
| Outliers (%) | 0.00 | 0.00 | 0.00 |
Values in parentheses are for the highest resolution bin.
R merge = Σ hkl [(Σ j | I j – < I > | ) / Σ j | I j | ]; R work = Σ hkl | |F o | – |F c | | / Σ hkl |F o |, where F o and F c are the observed and calculated structure factors of the refined reflections, respectively; R free is computed from randomly selected 5% of reflections omitted from the refinement.
The number of protein atoms include those from a cloning artifact at the C-terminus resulting in additional six residues and one residue at the N-terminus. Electron density enabled modeling of the N-terminus residue in both molecules of the asymmetric unit, six C-terminal residues in the molecule A and four C-terminal residues in the molecule B, as labeled in the coordinates deposited in the PDB.
Lacking an experimental structure of the N-terminal domain, the AlphaFold neural network was employed to predict the structure of the intact protein (28). The model confidence factor, expressed as the predicted local-distance difference test, pLDDT (28), was ~70 and exhibited notable variations between different regions of the molecule (Fig. 1C). The C-terminal domain structure was predicted with the highest confidence (pLDDT values spanning ~83–96), except for residues 188–196 (pLDDT values ranging ~70–80). The BB0238 AlphaFold model aligns very well with the crystal structure of BB0238132-256, demonstrating the ability of an artificial intelligence program, such as AlphaFold, to confidently predict protein structures (Fig. 1D). In contrast, the AlphaFold model of the N-terminal domain features a single helix-turn-helix motif (HTH) predicted with high confidence (pLDDT values of ~67–85), whereas the ensuing linker comprising residues 70–130 displayed an extremely low confidence level (pLDDT values of ~30–60). The site-directed mutations in the current study are located within the high confidence region of the AlphaFold model, and therefore their impact on function can be rationalized based on the structure. Note that the TPR prediction computer program TPRpred (29) and the protein family databases SMART (30) and Pfam (31) do not consider the single HTH motif of BB0238 as a TPR protein. However, a protein containing only a single TPR motif involved in PPIs, TOM20, has been described (32, 33). Therefore, in this article, the segment corresponding to BB023826-64 will be designated as a TPR-like/HTH motif.
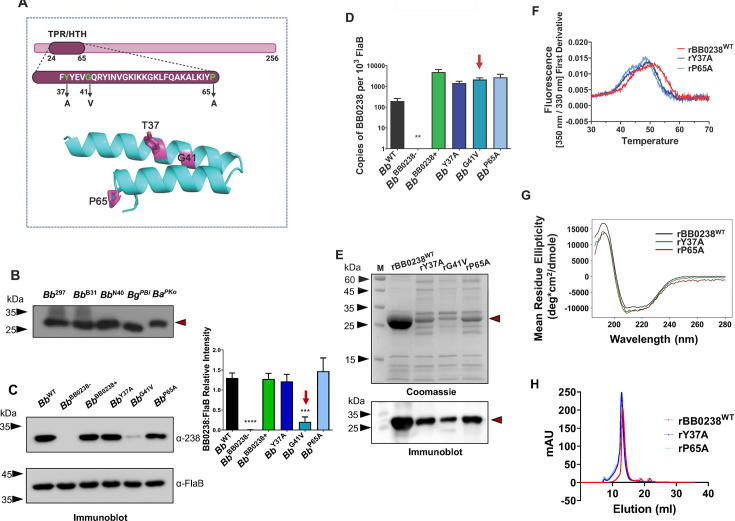
The TPR-like motif in BB0238 contributes to protein stability and structural integrity
To gain further insights into BB0238 structure and function, specifically its TPR-like/HTH motif (Fig. 2A), we used the previously reported point mutants of B. burgdorferi BB0238 (22). Immunoblot analysis confirmed the overall conservation of BB0238 orthologs in representative strains and isolates of Bb sensu lato, at least the sharing of common epitopes (Fig. 2B). Sequence analysis showed that the protein, including the TPR-like motif, is generally conserved within a wide range of spirochete species (Fig. S1). A previous report suggested that the TPR-like motif is important for the stability of the BB0238 protein (22); indeed, the current study substantiates these initial observations, at least for one selected mutation (G41V). Immunoblotting and densitometric analyses of the previously generated TPR-like motif point mutants (22) showed that BB0238Y37A (Bb Y37A) and BB0238P65A were maintained at levels similar to that of the wild-type protein, and each complemented the knockout Bb BB0238− isolate (annotated Bb BB0238+ in Fig. 2C). However, the production of the point mutant BB0238G41V was very low, nearly to the extent of the knockout mutant (Bb BB0238−) (Fig. 2C). The bb0238 transcript levels in all three BB0238 mutants, including the G41V mutant, are comparable to those of the parental or complemented isolates (Fig. 2D), confirming post-transcriptional protein loss. Notably, the recombinant wild-type BB0238 (rBB0238WT) was copiously produced in Escherichia coli, whereas the TPR-like point mutant proteins were produced at much lower levels, with rG41V production being the lowest (Fig. 2E). Note that both the wild-type and mutated proteins were partitioned with the soluble fraction. The reductions in mutant protein production might be attributed to their impaired stability. The extremely low protein abundance precluded further biochemical analysis of the rG41V protein, whereas rY37A and rP65A yielded enough protein to be investigated using nano differential scanning fluorimetry (nanoDSF). These experiments showed that both the rY37A and rP65A point mutants had significantly lower melting temperatures (Tm) compared to the wild-type protein, with ∆Tm of −1.75°C and −2.60°C, respectively (Fig. 2F). Nevertheless, the wild-type and mutant proteins had similar room temperature tryptophan fluorescence ratios measured at 350 nm and 330 nm, respectively, suggesting that by this criterion they were all similarly folded (data not shown). Circular dichroism (CD) studies suggested that the secondary structure contents of the rY37A and rP65A proteins were the same as rBB0238WT (Fig. 2G). Furthermore, the CD analysis indicated that all three BB0238 variants contained more α-helices (~32%) than β-strands (~18%) (Fig. S2), which agrees with the AlphaFold model (Fig. 1). As BB0238 is known to form a homomeric complex (19), we assessed the oligomerization property of the TPR-like motif point mutants. Size exclusion chromatography (SEC) showed that rY37A and rP65A eluted in earlier fractions than the wild type, consistent with the less compact dimeric forms of the mutants, compared with that of rBB0238WT (Fig. 2H). The predicted BB0238 structure fully explains the extreme instability of the G41V mutant (Fig. 2A). Gly41 is located on the αA helix at the closest distance to the opposing αB helix, where the inter-helix space precludes the accommodation of any larger side chain. In contrast, the P65 and Y37 side chains are located on the protein surface. Replacements of Y37 and P65 by smaller residues might reduce protein stability, but not necessarily fold integrity. Overall, these results indicate that the specific composition of these TPR-like amino acids is crucial for the stability of BB0238. Based on the AlphaFold model, the remaining N-terminal domain polypeptide lacks a well-defined structure. Hence, the entire N-terminal domain might be fully (G41V) or partially (Y37A and P65A) unfolded, and vulnerable to protease degradation, as suggested by the SEC profile and the yields of the proteins produced in E. coli (Fig. 2). Together, these experiments reveal that the presence of the TPR-like motif could be important for maintaining structure integrity, which ultimately should impact the biological functions of BB0238.
Fig 2.
Characterization of TPR-like/HTH motif in BB0238. (A) Location of the TPR-like/HTH motif and amino acid sequence. Residues relevant to the point mutants are shown in green within the context of the HTH motif (upper panel). Replaced residues are highlighted in magenta in the lower panel; Tyr37, Gly41, and Pro65 correspond to residues Tyr43, Gly47, and Pro71 in B. burgdorferi 297. (B) Conservation of BB0238. Equal amounts of lysates from B. burgdorferi isolates 297 (Bb 297), B31 (Bb B31), and N40 (Bb N40), as well as B. garinii isolate PBi (Bg PBi) and B. afzelii isolate PKo (Ba PKo), were immunoblotted against BB0238 antibody raised against the recombinant protein from B. burgdorferi B31. Position of BB0238 is shown by red arrow. The molecular weight markers (kDa) are indicated by black arrowheads. (C) Mutations in the TPR-like/HTH motif destabilize BB0238. Lysates of B. burgdorferi 297 (Bb WT), bb0238 mutant (Bb BB0238−), bb0238 complement (Bb BB0238+), and bb0238 point mutants Y37A (Bb Y37A), G41V (Bb G41V), and P65A (Bb P65A) were detected using antiserum against BB0238 (upper left panel). Antiserum against FlaB was used as a loading control (lower left panel). The right panel denotes the densiometric analysis of BB0238 protein shown in the left panel. The red arrow highlights significantly low Bb G41V transcript levels. ***P < 0.01. (D) bb0238 transcript levels. The mRNA levels for isolates, as in panel C, were measured using RT-qPCR. The bb0238 transcript levels do not significantly change between isolates, even in Bb G41V (P > 0.05), except for the null mutant Bb BB0238- where it is undetectable (**). (E) Recombinant BB0238 proteins in E. coli. Isolated proteins in gels were either stained with Coomassie brilliant blue or immunoblotted with anti-BB0238 antibody. (F) NanoDSF thermal profiles for rBB0238WT, as well as rY37A and rP65A point mutants, were determined via the first derivative of intrinsic fluorescence emission ration (350 nm/330 nm). Thermal transition (Tm) decreased significantly (P < 0.0001) with rY37A and rP65A. (G) Mutations in TPR-like/HTH motif do not alter the secondary structure of BB0238. Far-ultraviolet (UV) circular dichroism (CD) spectra of rBB0238WT (black), rY37A (green), and rP65A (red) from 200 to 260 nm are similar in all cases. (H) Quaternary structures of rBB0238WT, rY37A, and rP65A were examined via size exclusion chromatography. The elution profile for rBB0238WT depicted the dimerization of wild-type protein (red). Slightly earlier elution profiles of rY37A and rP65A (dark blue and light blue, respectively) reflected a size change in the dimeric state of the point mutants.
BB0238 modulates the proteolytic processing of BB0323 by the periplasmic chaperone-protease BbHtrA
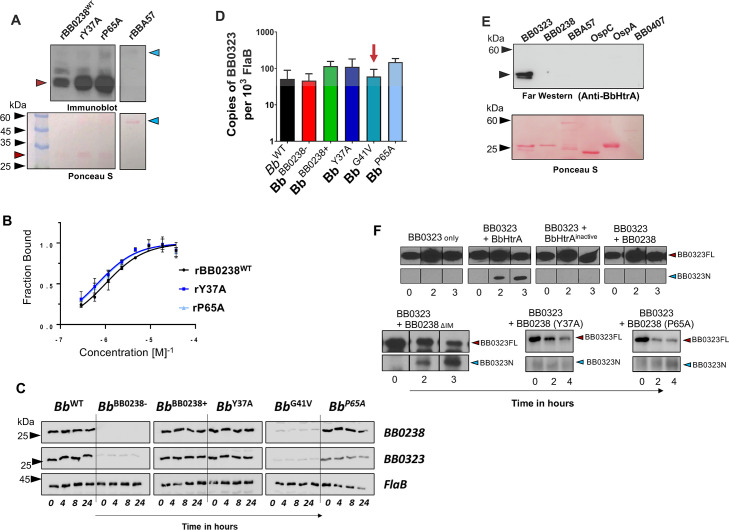
BB0238 interacts with another important borrelial virulence determinant, BB0323, in a manner that is essential for their mutual stability and ultimately microbial infectivity (21). As each of the above TPR-like motif point mutants of BB0238 from Bb isolates lost the ability to infect mice (22), we examined the interaction of these mutants with BB0323. Three independent assays confirmed that, as with the wild-type protein, both mutant proteins bind BB0323 similarly, as shown by far-western blotting (Fig. 3A). Moreover, binding affinity did not change significantly, as evidenced by the comparable binding kinetic constants (K D ) between rBB0323 and wild-type rBB0238, rY37A, or rP65A (1,001 ± 307, 658 ± 246, and 702 ± 326 nM, respectively) (Fig. 3B). Therefore, the only biophysical impact reflected thus far by the TPR-like mutants is on the thermal stability of BB0238 (Fig. 2F). We next examined the effects of the mutations on BB0323 turnover in Bb cells, as the instability of one interacting partner has been shown to impact the turnover of the other (21). A protein turnover assay was performed following erythromycin treatment to inhibit protein synthesis at both 33°C and 37°C, in order to measure protein levels of native BB0238, BB0323, and FlaB (as a loading control) over various time points. The results showed that, except for the BB0238 deletion mutant (Bb BB0238−) and the G41V mutant, all other isolates produced both BB0238 and BB0323 at comparable levels, either at 37°C (Fig. 3C) or 33°C (Fig. S3). The RT-qPCR analysis of transcript levels showed no change in bb0323 mRNA levels in borrelial cells harboring either the mutant or wild-type genes (Fig. 3D). Thus, although the cellular level of BB0323 depends critically on the presence of BB0238, a reduced thermal stability of the BB0238 TPR-like motif does not impair this PPI. Taken together, these results indicate that the selected residues within the BB0238 TPR-like motif, despite impacting thermal stability (Fig. 2F) or overall conformation (Fig. 2H), are unlikely to serve as factors in the interaction between BB0238 and BB0323.
Fig 3.
BB0238 influences proteolytic processing of BB0323, and their interaction does not involve the TPR-like/HTH motif. (A) BB0238-BB0323 interaction assessed by a far-western assay. Recombinant proteins rBB0238WT, rY37A, and rP65A (red arrow) and control protein BBA57 (blue arrows) were separated via SDS-PAGE, transferred to nitrocellulose membrane and probed with 1 µg of BB0323, and interactions were detected via BB0323 antibody. Comparable loading of proteins was evidenced by Ponceau S staining (lower panels). The image is representative of three separate experiments. (B) Kinetics of BB0238-BB0323 interaction assessed by microscale thermophoresis (MST). The MST analysis depicts a titration of fluorescently labeled rBB0238WT, rY37A, and rP65A with bound BB0323. K D values were indicated as 1,001.3 ± 306.8 nM, 658.4 ± 245.7 nM, and 702.1 ± 326.1 nM, respectively, displaying no difference between the groups. (C) BB0238 stabilization of BB0323 was depicted via a protein turnover assay at 37°C. Protein synthesis was inhibited via the addition of erythromycin to B. burgdorferi culture, and levels were detected via immunoblotting using specific antibodies. (D) bb0323 transcript levels in wild-type or mutant B. burgdorferi were compared by RT-qPCR. The red arrow highlights no significant change in bb0323 levels (P > 0.05), even though protein levels were diminished in protein turnover analysis (panel C). (E) Far-western analysis (upper panel). Indicated proteins were probed with BbHtrA and bound protein detected using anti-BbHtrA antibody. Comparable loading of proteins was evidenced by Ponceau S staining (lower panel). (F) BB0238 influences BB0323 proteolytic processing via BbHtrA. The upper panel shows an in vitro digestion assay using BB0323 and catalytically active or inactive BbHtrA and/or BB0238, revealing that the latter protects BB0323 from proteolytic cleavage by BbHtrA (upper panels). BB0323 full-length (FL) protein was incubated either alone, with BbHtrA or with inactive BbHtrA, or in the presence of both BbHtrA and BB0238. Reactions were stopped at the indicated time intervals, and proteolytic processing of BB0323 was detected via immunoblot analysis using anti-BB0323 antibody showing full-length (BB0323FL, 42 kDa, red arrow) and processed protein (BB0323N, 27 kDa, blue arrow). The lower panels denote a repetition of the upper panel experiment, where wild-type BB0238 is replaced by either interaction-deficient BB0238 (BB0238∆IM) or TPR-like/HTH motif mutants.
The precise proteolytic cleavage of BB0323 by a periplasmic serine protease with chaperone activities, BbHtrA, is essential for the infectivity of Lyme disease pathogens (17). We next used the TPR-like mutants to examine whether BB0238 influences the processing of full-length BB0323 (BB0323FL) by BbHtrA. Far-western blotting demonstrated that BbHtrA fails to interact with BB0238 (Fig. 3E). A 3-hour in vitro digestion assay (Fig. 3F) using recombinant proteins was performed to analyze the proteolytic processing of BB0323 by BbHtrA, in the presence or absence of BB0238. The experiments showed that the addition of active BbHtrA, but not of a catalytically inactive enzyme (with the catalytic serine replaced by alanine residue) (18), cleaved BB0323, as seen by the appearance of the smaller N-terminal BB0323 protein (Fig. 3F, upper panels). However, the addition of BB0238 protected BB0323FL from proteolytic cleavage by BbHtrA, as evidenced by the absence of detectable N-terminal BB0323 protein. In contrast, a BB0238 mutant protein lacking the BB0323 interaction motif (BB0238∆IM) (19) or either of the TPR-like mutant proteins (BB0238Y37A or BB0238P65A) failed to protect such proteolytic cleavage (Fig. 3F, lower panels). This suggests that either the BB0323 interaction motif or TPR-like/HTH motifs are not involved in the BB0238-mediated protection of BB0323 proteolysis, although the precise biological significance of this event remains unknown.
BB0238 interacts with borrelial protein BB0108
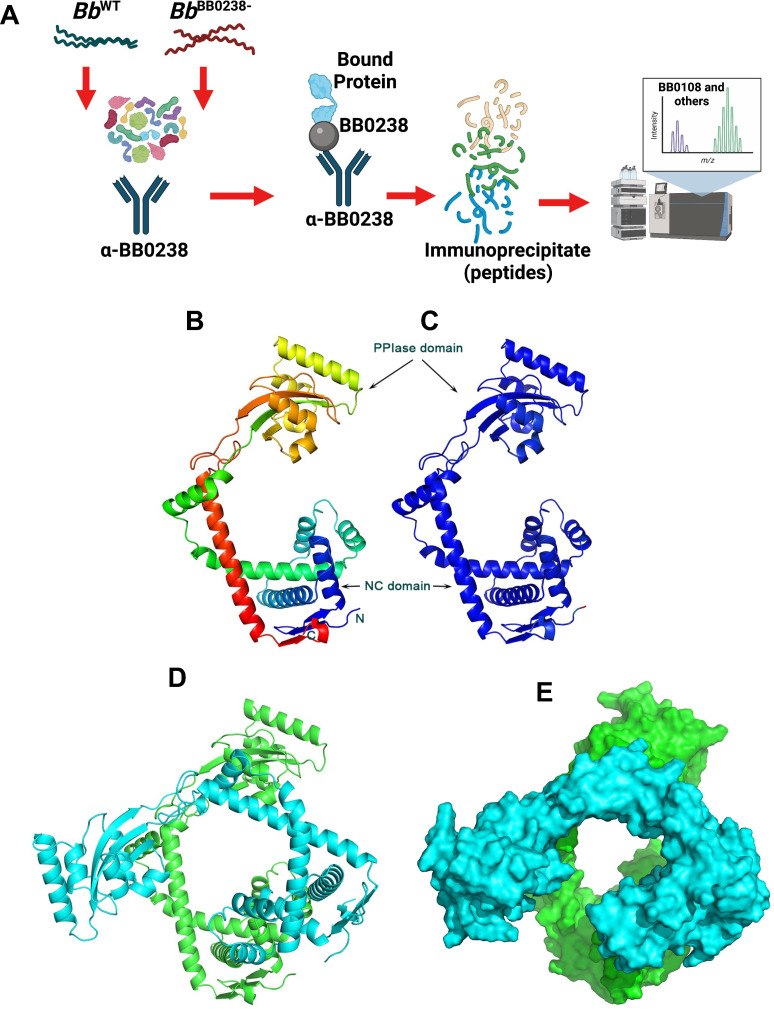
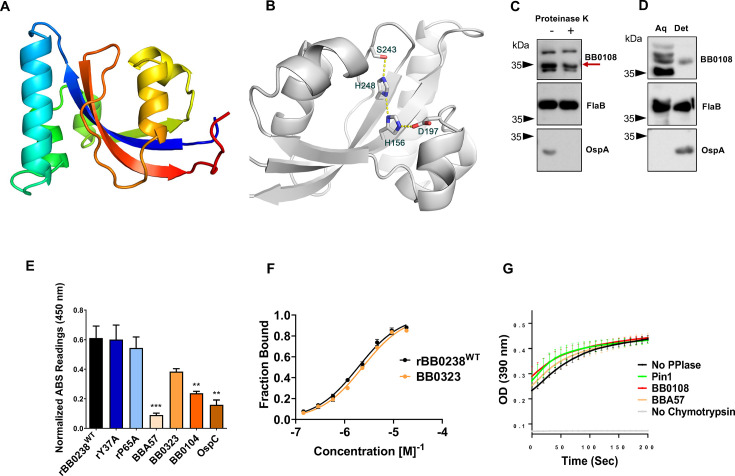
Antiparallel compact HTH structural units, as found in the TPR-like motif of BB0238, are involved in a myriad of PPIs across a wide range of organisms and cellular processes (34). Therefore, we explored whether BB0238 binds any additional spirochete proteins using an immunoprecipitation (IP) assay (Fig. 4A). The IP assays used lysates from the wild-type or bb0238 deletion and point mutants and BB0238 antibodies, followed by liquid chromatography and mass spectrometry (LC-MS/MS) analyses. Several uncharacterized proteins interacted with BB0238 at lower abundance in the bb0238 mutants compared with the wild-type counterpart (Table S1). One such protein, the 317 amino-acid periplasmic BB0108 (excluding a signal sequence), is annotated in sequence databases as a basic membrane protein. The GO molecular function in the UniProt database further annotates the protein as a peptidyl-prolyl cis/trans isomerase. A BLAST search identified sequence homology between BB0108, the bacterial Survival Protein A (SurA), and the foldase protein PrsA. Both SurA and PrsA contain parvulin-type peptidyl-propyl cis/trans isomerase (PPIase) domains. SurA is a PPIase chaperone found within the periplasmic space of many gram-negative bacteria, including Leptospira interrogans (35, 36). The crystal structure of SurA from E. coli (PDB ID: 1M5Y) revealed two parvulin PPIase domains, although only the second domain is enzymatically active. A third domain comprises the N- and C-terminal segments of the polypeptide chain (thus denoted NC domain) (36). The Bacillus subtilis PrsA crystal structure revealed a single enzymatically active parvulin domain and an NC domain (37). BB0108 contains a single parvulin domain and therefore resembles a PrsA. Indeed, a BLAST search of the PDB revealed that the closest sequence homolog with a known structure is PrsA from Bacillus anthracis (PDB entry code 6VJ4, Center for Structural Genomics of Infectious Diseases, unpublished). Approximately 80% of the BB0108 residues were aligned with those of the PrsA sequence with 27% sequence identity.
Fig 4.
BB0238 interacts with a periplasmic protein, BB0108. (A) The schema represents the experimental design of the BB0238 immunoprecipitation assay (IP). IP assays were performed on five separate groups of B. burgdorferi wild-type or bb0238 mutants (null or point mutants), as follows: Bb WT (with no antibody, control), Bb WT (with anti-BB0238), Bb BB0238− (with anti-BB0238), Bb Y37A (with anti-BB0238), and Bb P65A (with anti-BB0238). Eluted BB0238 interacting proteins were identified by liquid chromatography-MS/MS analysis. (B) Prediction of a model of BB0108 monomer by AlphaFold. The rainbow color scheme from blue to red corresponds to the tracing of the polypeptide chain from the N- to the C-terminus. Thus, the NC domain contains both red and blue colors, and the central PPIase domain is depicted in yellow and orange colors. (C) BB0108 monomer with colors corresponding to the confidence of the AlphaFold model from blue (highest confidence) to red (lowest confidence). The high confidence of the model is evidenced by having only blue colors. (D) Cartoon depiction of the AlphaFold model of the BB0108 dimer. Each monomer is shaded in a different color. The green molecule is shown in the same orientation as that of the monomer. (E) Surface representation of the dimer depicts the channel between the two protomers.
The AlphaFold neural network was employed to predict the structure of BB0108 (Fig. 4B). The structure was generated with exceptionally high confidence, as evidenced by the high overall pLDDT and pTMscore values of 95.1 and 0.81, respectively (Fig. 4C). The AlphaFold2 multimer version (trained to recognize oligomeric assemblies) was employed to generate a BB0108 dimer model, because both PrsA and SurA function as dimers. The dimer model was also predicted with high confidence (pLDDT = 92.6 and dimer interface confidence ipTMscore = 0.73) (Fig. 4D). The modes of dimerization of BB0108, PrsA, and SurA are different; they all engage their respective NC domains (data not shown), whereas only the BB0108 AlphaFold model also engages extensively the two parvulin domains in the interface. Consequently, a large channel is formed in the center of the BB0108 dimer that may serve as a chaperone to assist with periplasmic protein folding (Fig. 4E).
We next attempted to determine the structure of BB0108 using X-ray crystallography. Crystals of BB0108 grew readily in multiple forms, yet all diffracted X-ray to low resolution limits of ~7 Å. However, a truncated BB0108 encompassing only the PPIase domain, which was also produced in E. coli, formed crystals that diffracted X-ray to a resolution limit of 2.0 Å. The crystal structure was determined by the Molecular Replacement method using the AlphaFold structure as a search model (Table 1). The refined 2.0 Å resolution structure confirmed the parvulin fold and revealed the active site architecture (Fig. 5A and B). Superposition of the crystal structure and the AlphaFold model resulted in an rmsd value of 0.63 Å, including 100 aligned pairs of Cα atoms. Superposition with the PPIase domains of PrsA and SurA yielded higher rmsd values than the AlphaFold model, as determined by the DALI program (38). PrsA from Bacillus anthracis (PDB entry 6XD8) and the BB0108 PPIase domain yielded an rmsd value of 1.4 Å over 94 aligned Cα atoms, and SurA from E. coli (PDB entry 1M5Y) yielded an rmsd value of 1.9 Å between 103 aligned Cα atoms.
Fig 5.
Crystal structure and characterization of BB0108. (A) The crystal structure of the PPIase domain shown with the rainbow color, as in Fig. 4B. (B) The active site of the BB0108 PPIase domain showing the hydrogen bond network within the catalytic machinery. The side chains are shown in stick models with the atomic colors (carbon—gray, oxygen—red, nitrogen—blue). Electrostatic interactions are shown in yellow dotted lines. (C) BB0108 reflects limited cell surface exposure. Proteinase K assays were performed on wild-type spirochete cultures, which were either treated with (+) or without enzyme (−). Protein levels for BB0108, FlaB, and OspA were determined via the respective serums. (D) BB0108 primarily exists as a soluble protein. Lysates from wild-type spirochetes were partitioned into aqueous (aq) and detergent (det) phases. Protein levels for BB0108, FlaB, and OspA were determined via respective antibodies. (E) BB0108-BB0238 interactions examined by microtiter assay. Wells were coated with 1 μg of recombinant rBB0238WT, rY37A, rP65A, BBA57, BB0323, BB0104 (BbHtrA), and OspC (control). Plates were incubated with 1 μg of BB0108 and detected via BB0108 antibody. The data are representative of three separate experiments, each with three technical replicates, and presented after background signal was subtracted. ***P < 0.001; **P < 0.01. (F) BB0108-BB0238 interactions examined by microscale thermophoresis assay. MST analysis depicting the titration of BB0108 binding to fluorescently labeled rBB0238WT or BB0323 is presented; KD values are 1.8930 ± 0.2368 µM and 2.3293 ± 0.3962 µM, respectively. (G) BB0108 reflects PPIase activity. Enzymatic coupled assay demonstrates PPIase cis/trans isomerization through the chymotrypsin-dependent cleavage of succinyl-Ala-Phe-Pro-Phe-4-nitroanilide, resulting in a color signal read at 390 nm. Readings were taken every 10 seconds over 15 minutes. The background reaction without PPIase enzyme (black), positive control using a known PPIase, Pin1 (green), BB0108 (red), the integral membrane protein BBA57 (orange), and a negative control without chymotrypsin (gray) are shown. Each dot represents the average of four individual assays, with each group performed in duplicate wells; errors bars represent the standard error of mean (SEM).
By analogy to other parvulin PPIases, key active site residues that form a hydrogen bond network and are likely to form the catalytic machinery are His156, Asp197, His248, and Ser243. The coplanar histidine pair is invariant in all parvulin PPIases, whereas Asp197 can only be replaced by a cysteine residue, and the Ser243 position is sometimes occupied by a hydrophobic residue. Mutational analysis of Pin1, which belongs to the PPIase family that contains PrsA and SurA, has shown that the histidine pair is not essential for function, whereas the active site Cys residue, equivalent to Asp197 in BB0108, is essential, and its substitution by an Asp residue maintains enzymatic activity (39, 40).
BB0108 contains a signal sequence; therefore, it is transported, at the least, out of the inner membrane. Limited proteolysis (Fig. 5C) and phase partitioning (Fig. 5D) assays suggested that BB0108 lacks extracellular exposure and is predominantly present as a soluble protein; therefore, it likely resides in the periplasm. Notably, the high PI of the protein (9.3) and the positively charged patches on the surface of the 3D model indicate potential interactions with the membrane. The low level of proteolysis by proteinase K (Fig. 5C), together with a minor partitioning into the detergent phase (Fig. 5D), also suggest that a fraction of the protein could be membrane-associated. Taken together, these data show that BB0108 is predominantly a periplasmic protein, along with BB0238 and the N-terminal of BB0323 (21).
By analogy to SurA and PsrA, BB0108 may function as a chaperone and interact with multiple proteins. Two independent assays confirmed and quantified the BB0108 and BB0238 interaction in vitro. A microtiter plate-based assay tested this hypothesis using several Bb proteins. Based on absorption readings at 450 nm, BB0108 displayed the most abundant interactions with wild-type recombinant BB0238 and its TPR-like motif point mutants, Y37A and P65A (Fig. 5E). BB0108 also interacted with the periplasmic proteins BB0323 and BbHtrA (BB0104), and to a lesser extent, with the integral membrane proteins OspC and BBA57. MST analysis confirmed the interaction of BB0108 with BB0238 and BB0323, and defined K D values of 1.893 ± 0.2368 µM and 2.329 ± 0.3962 µM, respectively (Fig. 5F). An enzymatic assay that couples the cis/trans isomerization of succinyl-Ala-Phe-Pro-Phe-4-nitroanilide substrate to peptide cleavage by chymotrypsin (41) revealed a BB0108 PPIase activity comparable with that of Pin1 (Fig. 5G).
BB0238 TPR-like motif is crucial for tick-to-mouse transmission of spirochetes and establishment of early mammalian infection
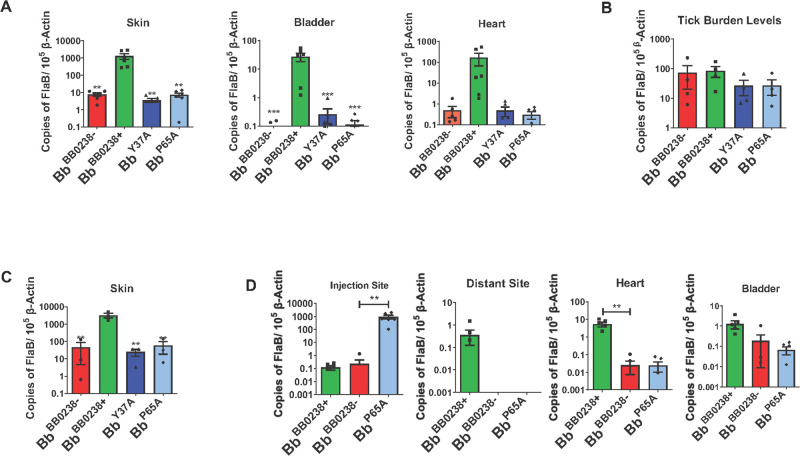
Although the TPR-like motif within BB0238 has been identified as critical for Bb infection of murine hosts (22), it remains unknown whether this region is also important for spirochete survival in ticks or transmission from ticks to a mammalian host. As the bb0238 deletion mutants or the TPR-like point mutants are unable to establish infection in mice and thus cannot be acquired naturally by ticks, the direct introduction of spirochetes into ticks using an established microinjection method was necessary (42). Groups of unfed nymphal Ixodes scapularis ticks were injected with either the null mutant (Bb BB0238−), the complemented isolate (Bb BB0238+), or the TPR-like motif point mutants (Bb Y37A and Bb P65A), and then allowed to engorge on separate groups of naïve C3H mice. Bb infection levels and spirochete viability 14 days following tick feeding were determined via RT-qPCR and culture analysis. The results revealed that the bacterial isolate complemented with BB0238 was present at high levels in all tested tissues, whereas the null mutant and the TPR-like motif point mutants were either undetectable or persisted at almost 100-fold lower levels (Fig. 6A). The culture data also displayed the loss of viability of the TPR-like motif point mutants, as live spirochetes were only reisolated from the biopsies of mice infected with the complemented isolate (Fig. S4). However, none of the bb0238 mutants displayed impairment of survival in ticks (Fig. 6B), suggesting that either BB0238 or its TPR-like motif does not play a role in supporting spirochete persistence in the vector prior to transmission.
Fig 6.
BB0238 TPR-like/HTH motif supports initial infection in the mammalian host, contributing to evasion of the host cellular immune responses. (A) Disruption of the TPR-like/HTH motif attenuates B. burgdorferi transmission. Ticks (5 ticks/mouse) were infected via microinjection with Bb BB0238−, Bb BB0238+, Bb Y37A, or Bb P65A, and then allowed to feed on C3H/HeN mice (3 mice/group). Infection was assessed via RT-qPCR analysis of B. burgdorferi flaB transcripts normalized against mouse Actb levels. (B) Survival of spirochetes within the infected ticks. Microinjected ticks, as indicated in panel A, were examined for spirochete burden using RT-qPCR, by measuring copies of B. burgdorferi flaB transcripts normalized against tick Actb levels. (C) The TPR-like/HTH motif is important for establishing infection within the first 7 days of infection. C3H/HeN mice were subcutaneously injected with spirochetes and infection was assessed by RT-qPCR analysis at the injection dermal site 7 days post-infection (dpi). (D) The TPR-like/HTH motif point mutant, Bb P65A, can survive locally after a hyper-infection inoculum. C3H/HeN mice (3 mice per group) were subcutaneously injected with Bb BB0238+ (105 spirochetes per mouse) or with Bb BB0238− and the Bb P65A point mutant (107 spirochetes per mouse). Infection was assessed by RT-qPCR analysis of pathogen burden at two weeks post-infection. Bars and error bars represent means and standard error of means, respectively. ***P < 0.001; **P < 0.01.
We next investigated when the TPR-like motif is essential in the mammalian phase of spirochete infection, and whether the bactericidal host immune response is responsible for the clearance of the TPR-like motif point mutants. To address this, we subcutaneously injected mice with equal levels of Bb isolates [105 cells/animal of either the null mutant (Bb BB0238−), complemented isolate (Bb BB0238+), or TPR-like motif point mutants (Bb Y37A or Bb P65A)] and examined pathogen levels around the dermal inoculation sites via RT-qPCR at an early timepoint (7 days) post-infection, before the major dissemination of spirochetes to distant organs. Bb levels were significantly lower in all bb0238 mutants (Bb BB0238−, Bb Y37A, and Bb P65A) compared to the complemented isolate (Fig. 6C), raising the possibility that the function of BB0238, including its TPR-like motif, could be relevant to spirochete defenses against the host’s early bactericidal innate immune responses. We have previously demonstrated that a larger inoculum of mutant spirochetes can overcome the phenotypic defects of survival in the host, in cases where the mutated gene confers microbial resistance against innate immunity (43). To examine this, groups of mice were subcutaneously injected with a 100-fold higher inoculum (107 spirochetes/mouse) of either the null mutant (Bb BB0238−) or one of the TPR-like motif point mutants (Bb P65A), whereas a 100-fold lesser inoculum (105 cells/mouse) of the complemented isolate (Bb BB0238+) was injected into a separate group of mice. Two weeks following the inoculation, mice were euthanized, and their spirochete levels were analyzed by collecting tissue samples at the injection site, as well as distant skin sites and organs. The results showed that, by comparison with the Bb BB0238− or Bb BB0238+ isolates, the Bb P65A mutants survived at the injection site at a significantly higher (~1000×) level (Fig. 6D). However, despite their high survivability at the dermal injection site, the TPR-like mutants behaved similarly to the null mutant when analyzed in other collected tissues, as they were unable to disseminate to distant skin sites or persisted poorly in distant organs (Fig. 6D). Overall, these results suggest that BB0238, through its TPR-like motif, supports spirochete survival at the dermal entry site, potentially facilitating the resistance of Bb against the host’s early innate immune responses.
BB0238 supports spirochete defense against host immunity
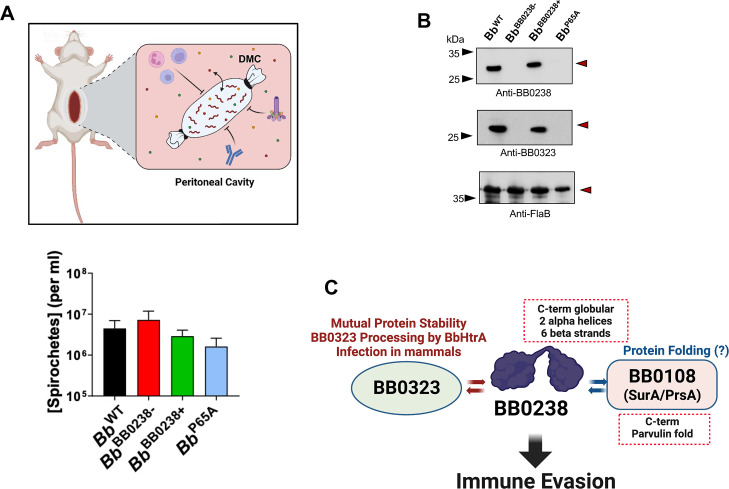
The dialysis membrane chamber (DMC) model of spirochete infection (44, 45) was used to examine directly whether BB0238 supports Bb resistance against immunity, particularly against host cellular microbicidal responses. To accomplish this, either wild-type (Bb WT), null mutant (Bb BB0238−), complemented isolate (Bb BB0238+), or one of the TPR-like mutants (Bb P65A) were introduced in the DMCs, which were then implanted in the peritoneal cavity of separate groups of rats (Fig. 7A). The 8 kDa cutoff of dialysis membranes allows the free passage of host nutrients or small molecules, including bactericidal compounds within the DMC, but prevents the passage of larger (>8 kDa) molecules, such as antibodies or immune cells (Fig. 7A, upper panel). The enumeration of spirochetes after two weeks of incubation in the DMC (Fig. 7A, lower panel) showed that none of the isolates displayed any survival defects, as comparable or significantly higher levels of bb0238 mutants were detectable. Because bb0238 mutants were found to display survival defects when directly inoculated in the skin and at the early stage of infection, before the development of host antibodies, but persist in a protected (DMC) environment at the same level as wild-type isolates, it is possible that BB0238 confers spirochete resistance against host cellular immunity. Notably, the examination of the protein profile of DMC-derived spirochetes via western blotting demonstrated comparable levels of BB0238 and its interaction partner BB0323 in the wild-type or complemented Bb BB0238+ isolates, yet both proteins were absent in the TPR-like Bb P65A mutant or Bb BB0238− (Fig. 7B). These results confirm that neither BB0238 nor BB0323 is essential for normal spirochete growth and survival, while both are indispensable for Bb resistance against host cellular immunity.
Fig 7.
BB0238 contributes to evasion of host cellular immunity. (A) Spirochete survival within dialysis membrane chambers (DMCs). Schematic diagram (upper panel) shows the DMC experimental design, which allows spirochetes to access host nutrients (small circles) while maintaining separation from the cellular host innate immune cells, complement, and antibodies. Lower panel denotes comparable bacterial concentrations within the DMC, as determined after two weeks within the rat peritoneal cavity (two rats/group). No statistical difference was recorded (P > 0.05). (B) Immunoblot analysis of DMC-cultivated spirochetes. Lysates from each of the DMCs were collected and analyzed via western analysis with antisera for BB0238 (upper panel), BB0323 (center panel), and FlaB (lower panel). Arrows indicate migration of proteins. Images represent similar western analysis from two rats of three independent DMC experiments, totaling six animals per group. (C) A diagram summarizing BB0238 structure and functions. BB0238 features a two-domain organization and is involved in multifaceted interactions with BB0323 and BB0108 that ultimately support immune evasion in mammals, promoting spirochete infectivity. Crystallization results are indicated by dotted callout boxes.
Overall, these data highlight the two-domain organization of BB0238, which interacts with BB0323 and BB0108, ultimately assisting in spirochete evasion of host immunity (Fig. 7C).
DISCUSSION
Many B. burgdorferi genes evolved to encode proteins of yet unknown functions that support the atypical biology and infectivity of spirochetes. In the current study, we focused on the structure-function analysis of a protein of unknown functions, BB0238. The X-ray crystallographic structure of the C-terminal domain of BB0238, along with the predicted structure of the full-length protein using AI methods, revealed a TPR-like/HTH motif at the N-terminus and a well-ordered C-terminal domain, connected by a poorly defined long polypeptide segment. Despite the low confidence of the AlphaFold structure prediction within this region, no intrinsic disorder due to amino acid composition in this range is detected (46). It is tempting to speculate that the long linker region between the HTH motif and the C-terminal domain is flexible in its conformation, which would facilitate changes in spatial orientation between the TPR-like/HTH motif and the C-terminal domain depending on its PPI partners, as BB0238 binds multiple spirochete proteins.
The current discovery of the functions of BB0238 in influencing the processing and stability of the BB0323 protein overlaps with the functions of other known transport proteins with a single TPR-like motif. For example, TOM20 is a general import receptor on the outer membrane of mitochondria, and CPR6, a cyclophilin PPIase, is involved in the stabilization of Hsp90 and interaction with the ribosome. Moreover, the BB0238 TPR-like/HTH motif (BB023826-64) superposes well on analogous motifs of other proteins, such as the vacuolar protein sorting-associated protein, VTA1 (data not shown), which is also involved in protein binding and transport (47, 48). We have also discovered that BB0238 preferentially binds the BB0108 protein, a homolog of chaperones with PPIase functions. Binding was confirmed in vitro, although the role of BB0108 in assisting the folding of BB0238 needs further investigation.
In addition to its role in BB0323 stability and spirochete infectivity (21), our findings that BB0238 influences the processing of BB0323 by BbHtrA suggests a regulatory role in the controlled proteolysis of full-length BB0323 to produce mature N- and C-terminal BB0323, each of which possess distinct roles in borrelial fission (18), outer membrane stability (15, 16), and infectivity (15). Recent studies have highlighted that the alteration of the optimal production of mature N-terminal BB0323 in spirochete cells leads to the impaired abundance or stoichiometry of its complex with BB0238, ultimately affecting the infectivity of Bb in mammals (17). The observation that bb0238 mutants, including the Bb P65A mutant, are unable to establish early infection in mice and disseminate to distant skin or organs, despite surviving within the protected environment of host-implanted DMCs, suggests a critical role of BB0238 in spirochete evasion of host innate immunity during the early establishment of infection, involving direct interaction with host cells or large host extracellular elements. However, it is difficult to ascribe spirochete survival to the lack of immune cell contact, as low molecular weight antibacterial peptides can diffuse through the membranes, and these evidently had no effect. Since introduced Bb are sequestered in a membrane in the DMC model, it is entirely possible that neither the inflammatory response nor the immune system was able to detect the presence of antibacterial peptides and mount an effective microbicidal response. Notably, we also observed that the stability of BB0238 and BB0323 differed dramatically in cultured spirochetes versus spirochetes that persist in a DMC model. This could be due to the well-characterized differences in spirochete gene expression in vitro compared to various host environments (such as DMC), suggesting that the protein turnover events of BB0238 vary based on the environment. Bb may use several pathways to evade the innate immune responses of a host and establish early infection; our results underscore the importance of BB0238 in mediating one or more of these responses. As the interaction of BB0238 with BB0323 (or potentially with additional proteins) is essential for mammalian infection, these proteins comprise candidate targets for the development of novel therapeutics against Lyme disease.
In summary, herein we present new insights into the structural and functional aspects of a unique protein of B. burgdorferi with multi-domain organization, BB0238, that is involved in multiplex PPIs involving spirochete proteins, such as BB0323 and BB0108, which are indispensable for the evasion of early host cellular immune responses. Although we present structural evidence that BB0238 domains represent unorthodox similarities to transport and/or chaperone proteins, precisely how the protein supports borrelial biology and virulence remains puzzling. Further exploration would not only enrich our knowledge of unusual spirochete biology, but may also present unique opportunities for the development of new antimicrobials against B. burgdorferi.
MATERIALS AND METHODS
Bacteria, mice, and ticks
Borrelia burgdorferi 297 (Bb WT), bb0238 mutant (Bb BB0238−), complement (Bb BB0238+), and three TPR-like motif point mutants (Bb Y37A, Bb G41V, and Bb P65A) used within this study were kindly provided by John Blevins (22). Four- to six-week-old C3H/HeN mice were purchased from Charles River Laboratories or bred in the laboratory. Ixodes scapularis ticks have been reared and maintained in our laboratory. All studies using animals and pathogens were performed under protocols approved by the Institutional Animal Care and Use Committee (IACUC) and the Institutional Biosafety Committee of the respective institutions.
Production of recombinant proteins and generation of antibodies
For BB0238 protein crystallization studies, four different gene fragments were amplified by polymerase chain reaction (PCR) from the genomic DNA of B. burgdorferi B31 (primer sequences are given in Table S2) to cover the coding region for the full-length protein but without the signal sequence (BB023824-256); a slightly truncated protein from the N-terminus (BB023853-256); the C-terminal domain (BB0238118-256); and the N-terminal domain (BB023824-114). The amplified gene fragments were ligated into the pETm-11 expression vector coding for an N-terminal 6xHis tag separated from the inserted gene by a tobacco etch virus (TEV) protease cleavage site. All four BB0238 variants were expressed in Escherichia coli, as described previously for another B. burgdorferi protein, BB0365 (49).
For SeMet-labeled BB0238118-256 protein expression, a Leu240Met mutant was produced to obtain sufficient anomalous signal during diffraction data collection. SeMet-labeled BB0238118-256 Leu240Met was produced as described previously for BB0689 (50). All BB0238 variants were purified by affinity chromatography using Ni-NTA agarose (Macherey-Nagel, Düren, Germany) gravity-flow column. The elution buffer (300 mM NaCl, 25 mM NaH2PO4, and 300 mM imidazole, pH 7.5) was exchanged into 20 mM Tris (pH 8), 50 mM NaCl, and 10 mM NaH2PO4 using a centrifugal filter unit (Millipore, Burlington, MA, USA), prior to digestion with TEV to remove the 6xHis tag. After 16–20 h of incubation at room temperature, the digested 6xHis tag and the His-tagged TEV protease were removed by using Ni-NTA agarose (Macherey-Nagel, Düren, Germany) gravity-flow column. The eluted BB023824-256, BB023853-256, and BB0238118-256 were buffer exchanged into 10 mM Tris (pH 8) and concentrated to 8.0 mg/mL. To avoid protein precipitation, BB023824-114 was buffer exchanged into 20 mM Tris (pH 8), 50 mM NaCl, and 10 mM NaH2PO4, and concentrated to 8.0 mg/mL.
For structure determination of the BB0108 PPIase domain (amino acid residues 168–276), the DNA encoding this region was amplified by PCR using as a template the full-length BB0108 gene sequence optimized for production in E. coli. The PCR product was digested with BamHI and XhoI, cloned into pGEX-6P-1, and transformed into E. coli BL21 (DE3). The positive clone-containing colony was inoculated into Luria–Bertani (LB) medium supplemented with 100 µg mL−1 ampicillin at 37°C and induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) on reaching an OD600 nm of 0.8. The culture was grown for 16 h at 25°C. The cells were harvested by centrifugation and the pellet was resuspended in lysis buffer (50 mM Tris–HCl pH 7.5, 300 mM NaCl, and 1 mM EDTA) in the presence of 1 mM protease inhibitor cocktail. The cells were lysed by sonication for 10 minutes and the cell debris was removed by high-speed centrifugation for 40 minutes. The lysate was passed through a GST-IGA column pre-equilibrated with equilibration buffer (50 mM Tris–HCl buffer pH 7.5, 300 mM NaCl). Precision Protease amounting to 15 U per milliliter was added to the cleavage buffer (50 mM Tris–HCl buffer pH 7.5, 150 mM NaCl, 1 mM EDTA, and 1 mM DTT) and incubated for 16 h at 277 K. The GST-free PPIase domain was eluted in buffer containing 50 mM Tris–HCl pH 8, 100 mM NaCl. Elution fractions were pooled and purified by size-exclusion chromatography on a Superdex S-200 column (GE Healthcare, USA) pre-equilibrated with 50 mM Tris–HCl buffer pH 8, 100 mM NaCl. The purity of the sample was assessed at each step on 16% SDS–PAGE.
A list of oligonucleotide primers used for the cloning of additional recombinant proteins included in this study is listed in Table S2. Recombinant proteins BB0238 (21), BB0238∆IM (19), BB0323 N2 (18), BB0104 (BbHtrA) and BbHtrAS226A (18), and BBA57 (51) were obtained from previous studies. Recombinant proteins for BB0108 and OspC were cloned using the listed primers (Table S2). All proteins were expressed in E. coli with either pET28a, pGEX-6P-1, or pET303 expression vectors. Standard expressions were performed with 1 mM IPTG at 25°C for 4 h. Point mutations of BB0238 were created using the specified primers (Table S2) and the Phusion Site-Directed Mutagenesis Kit (ThermoScientific). Isolation of pET28a His-Tag proteins was done using Nickel Beads from ProBond (ThermoFisher Scientific). Isolation of pGEX-6P-1 GST-fused proteins was performed following the manufacturer’s instructions (GE Healthcare), resulting in eluted product with GST-tag, and proteins were cleaved using overnight cleavage with Precision Protease. Generation of polyclonal antisera was done as previously described (21), using GST-cleaved recombinant protein.
Site-directed mutagenesis
BB0238118-256 Leu240Met mutant was produced by PCR amplification using pETm-11-BB0238118-256 plasmid as a template and two complementary primers containing the relevant mutation (Table S2). Methylated parental DNA template was degraded by adding endonuclease DpnI to the PCR reaction and incubating for 1 h at 37°C, followed by transformation in E. coli XL1-blue cells and growth overnight at 37°C on LB agar plates containing kanamycin. Separate colonies were picked and transferred into LB medium supplemented with kanamycin. After 24 h, the plasmid DNA was isolated and the presence of Leu240Met mutation was confirmed by DNA sequencing (Table S2).
Crystallization
Crystallization experiments of BB023824-256, BB023853-256, BB0238118-256, BB0238118-256 Leu240Met, and BB023824-114 were set using a Tecan Freedom EVO100 workstation (Tecan Group, Männedorf, Switzerland) in 96-well sitting drop plates by mixing 0.4 µL of protein with 0.4 µL of precipitant. For each BB0238 variant, approximately 400 conditions were screened by using 96-reagent sparse-matrix screens JCSG+, Structure Screen 1&2 (Molecular Dimensions, Newmarket, UK), JBScreen Classic HTS I, and JBScreen Classic HTS II (Jena Bioscience, Jena, Germany). Irregularly shaped crystals for BB023824-256 were obtained in 0.1 M Tris (pH 8.5), 0.2 M magnesium chloride, and 3.4 M 1,6-hexanediol, while hexagonal crystals for BB0238118-256 were obtained in 1.8 M D,L-Malic acid (pH 7), and needle-shaped crystals for BB023824-114 in 0.2 M calcium chloride, 0.1 M sodium acetate (pH 4.6), and 36% MPD. The SeMet-labeled BB0238118-256 Leu240Met protein did not form crystals under the same conditions as the native protein, but after screening and optimizing the crystallization conditions, hexagonal crystals were obtained from a solution containing 0.1 M MES (pH 6.5) and 2.6 M ammonium sulfate. Prior to data collection, the crystals were frozen in liquid nitrogen without additional cryoprotection.
BB0108 PPIase crystal screening was performed at 295K with the nanoliter microdrop robotic system (mosquito) using 96-well sitting drop trays. Following optimization, crystals were obtained in 10% (vol/vol) 2-propanol, 0.1 M Bicine pH 8.5, and 30% (wt/vol) polyethylene glycol 1500 at 15°C using the vapor diffusion method with 25 mg/mL protein. For X-ray data collection, single crystals were mounted on cryo-loops using 25% (vol/vol) glycerol as a cryoprotectant and flash-cooled in liquid nitrogen.
Diffraction data and structure determination
Diffraction data for native BB0238118-256 were collected at the MX beamline instrument BL 14.1 at Helmholtz-Zentrum, Berlin, Germany, and SeMet data for BB0238118-256 were collected under proposal 20210447 at MAX IV Laboratory at the BioMAX beamline, Lund, Sweden. The structure was determined using the SAD method. Reflections were indexed by XDS and scaled by AIMLESS from the CCP4 suite (52 – 54). The initial phases were determined by SHELX C/D/E (55). The crystal unit cell solvent content was determined by MATTHEWS (56). The protein model was built in BUCCANEER (57). The crystal structures were improved by manual rebuilding in COOT (58). Crystallographic refinement was performed using REFMAC5 (59). A summary of the data collection, refinement, and validation statistics for BB0238 is given in Table 1.
Diffraction data of a BB0108 PPIase crystal were acquired at the General Medicine and Cancer Institute Collaborative Access Team (GM/CA-CAT) beamline 23ID-B at the Advance Photon Source (Argonne National Laboratory, Argonne, IL, USA). The data were processed using the program HKL2000 (60). The structure was determined by the molecular replacement method with the program PHASER as implemented in CCP4, using the corresponding coordinates generated by AlphaFold2 as the search model (61). The structure was refined using the program PHENIX (62). Model building and modification were performed using the interactive computer graphics program COOT (63). Table 1 provides a summary of data collection, refinement, and validation statistics.
Protein 3D structure prediction using AlphaFold
AlphaFold v2.1 (28) was used to predict the 3D structure of BB0238 (GenBank: 1195075). Structure prediction with AlphaFold v2.1 was performed according to the default parameters, as indicated at the website <https://github.com/deepmind/alphafold/>, running on AMD Ryzen Threadripper 2990W × 32 Core; 128 GB RAM; 4× NVIDIA GeForce RTX 2080, and using the full databases downloaded on 2021-09-25. For further structural analysis, only the predicted structure with the highest confidence was used [as ranked by using LDDT (pLDDT) scores].
The BB0108 monomer structure was modeled using the AlphaFold2 (28) with the Advanced ColabFold licensed under MIT (64), using the fast mmseq2 multi-sequence alignment method (65). The dimer structure was modeled with the Colab multimer version. The monomer predictions were ranked based on the pLDDT score, and the dimer predictions were ranked based on the pTMscore. The top rank structures are described here; however, consistent with the high confidence of both monomer and dimer BB0108 structures, all the models generated by AlphaFold are essentially the same.
Infection studies with mice and ticks
For studies involving the transmission of B. burgdorferi, nymphal ticks were artificially infected through microinjection with 109 bacteria/mL of either bb0238-complemented (Bb BB0238+), bb0238-mutant (Bb BB0238−), or bb0238 TPR-like motif point mutants (Bb Y37A, Bb G41V, and Bb P65A), as detailed (22). The artificially infected ticks were allowed to infest naïve C3H/HeN mice (5 ticks per mouse, 3 mice per group) until the conclusion of feeding. Fully engorged ticks were collected to confirm infection via reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Burden levels of B. burgdorferi in the skin, heart, and bladder were analyzed by RT-qPCR 12 days after the completion of tick feeding. Further infection studies were utilized to assess the spirochete levels of Bb BB0238+, Bb BB0238−, Bb Y37A, Bb G41V, or Bb P65A isolates within C3H/HeN mice (3 mice per group). Early or hyper-infection experiments were initiated via subcutaneous injection with 105 spirochetes per mouse or 107 spirochetes per mouse, respectively. Skin, heart, joint, and bladder were harvested at either 7 days post-infection (dpi) for early infection or 14 dpi for all other infections, and B. burgdorferi burdens were assessed via RT-qPCR, as detailed (21).
PCR analysis
Primers used for PCR or RT-qPCR are shown in Table S2. For RT-qPCR, RNA was isolated from either B. burgdorferi lysates, mouse tissues, or fed ticks using TRIzol reagent (Invitrogen) and further treated with DNase I (NEB). RNA was used for cDNA synthesis (ThermoScientific), and then analyzed by RT-qPCR using iQ SYBR Green Supermix (Bio-Rad) as previously described (21). The relative expression of target genes was normalized by flaB, mouse actin, or tick actin from Borrelia lysates, mouse tissues, and fed ticks, respectively.
Protein alignment and analysis
Protein sequence alignments and phylogenetic trees for BB0238 were created using Clustal Omega (66) with basic parameters and viewed using Jalview (2.11.2.0). Sequences used for BB0238 alignment were B. burgdorferi B31 (WP_002556837.1), B. burgdorferi Bol26 (WP_002657674.1), B. mayonii (WP_075551947.1), B. japonica (WP_091972713.1), B. bavariensis (WP_215540274.1), B. garinii Far04 (EED29651.1), B. afzelii PKo (ABH01505.1), B. duttonii Ly (ACH93191.1), B. recurrentis (WP_041178099.1), B. parkeri (WP_025407471.1), B. turicatae (WP_041178431.1), B. hermsii (WP_043924407.1), B. miyamotoi (WP_117374734.1), and Spirochaeta cellobiosiphila (WP_037572047.1).
Western blotting and far-western blotting
Immunoblotting was performed via standard procedures, as described in our previous papers (21). Briefly, B. burgdorferi lysate or recombinant protein was resolved using SDS-PAGE and protein was transferred via semi-dry method to nitrocellulose membrane. Blocking was done in phosphate-buffered saline (PBS) with 0.05% Tween-20 (PBSt) and 2% skim milk. One microgram of interacting ligand for far-western was incubated with the membrane in PBSt. Appropriate primary antisera were used and either horseradish peroxidase (HRP)-labeled anti-mouse or anti-rabbit (for anti-FlaB) secondary antibody was used for detection at 1:3,000 and 1:10,000 dilution, respectively, unless otherwise stated.
Enzyme-linked immunosorbent assay
The base protocol was performed as previously described (67). Briefly, for protein interaction, 1 μg of interacting protein, ligand, or control protein was incubated overnight in PBS to coat the wells. After blocking with BSA, 1 μg of the target protein was added to the wells for 1 h and then detected via the corresponding primary antisera (1:3,000) and secondary anti-mouse HRP-labeled antibody (1:10,000). Plates were read at 450 nm wavelength and the average OD reading for three experiments performed in duplicates was presented after removing background signal.
Microscale thermophoresis
Microscale thermophoresis (MST) analysis was performed using a Monolith NT instrument (NanoTemper Technologies), as detailed (68). A 12-point 20 µM 1:1 serial dilution of each target ligand was made in assay buffer (PBS + 0.05% Tween-20, pH 7.4) and mixed 1:1 with 10 nM of target protein previously labeled with NanoTemper Red-tris-NTA dye per the manufacturer’s instructions. Samples were loaded into standard capillaries and run on a Nanotemper NT.Automated at 40% MST power and 10% LED power for 10 seconds. Data from three biological replicates were analyzed using MO.Affinity analysis software using TJump analysis and evaluated in a KD fit model that describes a molecular interaction with 1:1 stoichiometry according to the law of mass action.
Nano-differential scanning fluorescence (nanoDSF)
Thermal unfolding and aggregation data were measured using the Nanotemper Prometheus NT.48 with PR.ThermControl software. Briefly, 0.2 mg/mL of each target protein was resuspended in assay buffer and incubated for 10 minutes at room temperature before loading into Nanotemper standard capillaries. Excitation energy was set so that each target protein gave at least 2,500 counts in the 330 and 350 nm channels, and thermal ramp settings were set to run from 25°C to 90°C at a ∆4°C/minute ramp speed. The Tm, or midpoint of the thermal unfolding sigmoid, was determined by taking the global minimum or maximum of the first derivative of each protein’s thermal unfolding curve, while initial folding values were derived from the ratio of 350:330 nm before the thermal ramp was applied.
Circular dichroism
Briefly, rBB0238WT, rY37A, and rP65A were dialyzed into 20 mM sodium phosphate, 20 mM KCl. Concentrations were determined by A280, and extinction coefficients for rBB0238WT, rY37A, and rP65A were determined. Circular dichroism (CD) spectra were collected using Chirascan CD spectrometer (Applied Photophysics LTD) using a 0.1-mm demountable quartz cell over a wavelength range of 188–280 nm. Representative data shown after repeated scans result in similar data. Data were converted to mean residue molar ellipticity (deg cm2/dmol) after baseline subtraction of the buffer. Secondary structure deconvolution of the CD data was estimated using CDNN deconvolution program (CDNN courtesy of Gerald Bohn, 1997, at the Institut Für Biotechnologie, Martin-Luther Univserität Halle-Wittenberg, through distribution with Applied Photophysics software).
Analytical size exclusion chromatography
Analytical size exclusion chromatography was used to analyze the quaternary structures of rBB0238WT, BB0238 TPR-like motif point mutants (rY37A and rP65A), and rBB0108. A Superose 12 column was calibrated and standard curves were established using known proteins and plotting their apparent molecular masses as a function of their corresponding elution coefficients. The standard curve was then used to approximate the apparent molecular masses of BB0238, BB0238 TPR-like motif point mutants, and BB0108.
Protein turnover assay
The stability of BB0323 was analyzed through a protein turnover assay, as previously described (21); however, it was optimized with erythromycin instead of streptomycin, the complementation antibiotic selective marker. Briefly, Bb WT, Bb BB0238−, Bb BB0238+, and three TPR-like motif point mutants (Bb Y37A, Bb G41V, and Bb P65A) were incubated in BSK medium containing erythromycin (0.4 µg/mL) at 33°C and 37°C for 24 hours. Spirochetes were removed at 0, 4, 8, and 24 hours time points. Lysates were prepped and processed for immunoblotting using anti-BB0323, anti-BB0238, and anti-FlaB antibody.
BB0323 proteolysis assay
Proteolysis of full-length BB0323 (BB0323 FL) by BbHtrA (BB0104) was performed in a 20-µL reaction at 37°C in 50 mM Tris-HCl pH 7, 150 mM NaCl, 1 mM EDTA, and 1 mM DTT buffer, as detailed (18). Five micrograms of BB0323 FL and various versions of BB0238 (rBB0238WT, rBB0238∆IM, rY37A, or rP65A) were incubated in the presence of 900 ng of BbHtrA, BbHtrAS226A, or no protease. The reaction was terminated by SDS-PAGE loading buffer at various time points over the course of 4 hours. Samples were analyzed by SDS-PAGE gel separation and western blot analysis for BB0323 FL protein levels using antisera against BB0323. All experiments were performed in three separate replicates.
Immunoprecipitation of BB0238 and mass spectrometry analysis
Immunoprecipitation (IP) was performed as detailed by the Pierce Co-Immunoprecipitation Kit (Thermo Scientific). In sum, roughly 109 bacteria from two Bb WT cultures, and individual Bb BB0238−, Bb Y37A, and Bb P65A cultures were centrifuged, washed, and lysed according to protocol, with the addition of protease inhibitor cocktail. The lysate supernatant was pre-cleared with resin to reduce background, and then incubated at 4°C for 2 hours with an agarose resin that was previously coupled with purified BB0238 antisera (Melon Gel IgG Purification Kit; ThermoFisher Scientific). One agarose resin prep for a Bb WT culture omitted BB0238 antisera to serve as a negative IP control. After incubation, the resin was washed until the flow-through could not detect any protein via A280 nanodrop measurement. Protein from IP was eluted in two fractions and stored at −80°C until processed for mass spectrometry (MS). MS processing was performed as previously described.
Proteinase K accessibility assay and Triton X-114 phase partitioning assay
Proteinase K digestion assay was performed as previously established (21). In brief, 108 spirochetes were incubated in PBS in the presence or absence of proteinase K (200 µg/mL) for 20 min. Phenylmethylsulfonyl fluoride was added to each sample at a final concentration of 1 mM, then centrifuged, and lysate was immunoblotted using antibodies against BB0108, FlaB, and OspA. Triton X-114 phase partitioning was performed as described elsewhere (21). Briefly, spirochete cultures were resuspended in 800 µL of PBS and sonicated. Using a final concentration of 2% Triton X-114, the detergent phase was separated from the aqueous phase. Protein was precipitated via acetone, centrifuged at 13,000 × g for 15 min, and immunoblotted using antibodies against BB0108, FlaB, and OspA.
Cultivation in dialysis membrane chambers within rat peritoneal cavity
Intraperitoneal cultivation of B. burgdorferi cultures contained in DMCs was performed as previously described (45). In brief, ~9–10 mL of BSKII media containing 103 spirochetes mL−1 was used to fill each DMC. Spirochete-filled chambers were implanted into the peritoneal cavity of 4- to 6-week-old Sprague-Dawley rats. The dialysis chambers were harvested two-weeks post-implantation, and spirochetes counted by darkfield microscopy using a Petroff Hauser counting chamber. The experiment was performed twice, two rats per group per experiment. Each animal is represented by a single point.
Statistics
All numerical results are expressed as the mean ± standard error of the mean (SEM). The significance of differing mean values between groups was evaluated using the Student’s t-test or ANOVA using GraphPad Prism software (GraphPad Software, CA, USA).
ACKNOWLEDGMENTS
We are grateful to John Blevins for providing the B. burgdorferi mutants with point mutations at the key TPR-like motif in BB0238. The authors thank Kathryn Nassar for the assistance with the preparation of this manuscript, in addition to Kazi Antara, Meghna Thakur, and Kavita Sharma for their assistance with some of the experiments. We thank Manfred S. Weiss and Jan Wollenhaupt at the BL14.1 beamline at BESSY II electron storage ring operated by the Helmholtz-Zentrum, Germany, and we are grateful to Ana Gonzalez and Tobias Krojer at the BioMAX beamline at MAX IV Laboratory, Sweden, for their help and support during data collection. We thank the staff at the General Medicine and Cancer Institute Collaborative Access Team (GM/CA-CAT) at the Advanced Photon Source (Argonne National Laboratory, Argonne, IL, USA) for their help during BB0108 data collection.
This study was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH) (Award Numbers R01AI080615, R01AI116620, and P01AI138949 to U.P. and R01AI029735-30 to M.J.C.) and the Intramural Research Program of the National Center for Advancing Translational Sciences, NIH (ZIA TR000355-04 to A.S.).
The authors declare that they have no potential conflicts of interest.
Footnotes
This article is a direct contribution from Utpal Pal, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Jorge L. Benach, Stony Brook University, and Richard T. Marconi, Virginia Commonwealth University Medical Center.
Contributor Information
Utpal Pal, Email: upal@umd.edu.
Victor J. Torres, St. Jude Children's Research Hospital, Memphis, Tennessee, USA
DATA AVAILABILITY
The data that support the findings of this study are available from the corresponding author upon reasonable request. The coordinates and the structure factors for native and SeMet-labeled BB0238 have been deposited in the Protein Data Bank with accession codes 8P33 and 8P32, respectively. The coordinates and the structure factors for BB0108 PPIase domain have been deposited in the Protein Data Bank with accession code 8SOT.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mbio.02135-23.
Fig. S1 to S4; Tables S1 and S2.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hernsdorf AW, Amano Y, Ise K, Suzuki Y, Dudek N, Relman DA, Finstad KM, Amundson R, Thomas BC, Banfield JF. 2016. A new view of the tree of life. Nat Microbiol 1:16048. doi: 10.1038/nmicrobiol.2016.48 [DOI] [PubMed] [Google Scholar]
- 2. Oppler ZJ, O’Keeffe KR, McCoy KD, Brisson D. 2021. Evolutionary genetics of Borrelia. Curr Issues Mol Biol 42:97–112. doi: 10.21775/cimb.042.097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Radolf JD, Caimano MJ, Stevenson B, Hu LT. 2012. Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat Rev Microbiol 10:87–99. doi: 10.1038/nrmicro2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mead PS. 2015. Epidemiology of Lyme disease. Infect Dis Clin North Am 29:187–210. doi: 10.1016/j.idc.2015.02.010 [DOI] [PubMed] [Google Scholar]
- 5. Marques AR, Strle F, Wormser GP. 2021. Comparison of Lyme disease in the United States and Europe. Emerg Infect Dis 27:2017–2024. doi: 10.3201/eid2708.204763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Steere AC, Strle F, Wormser GP, Hu LT, Branda JA, Hovius JW, Li X, Mead PS. 2016. Lyme borreliosis. Nat Rev Dis Primers 2:16090. doi: 10.1038/nrdp.2016.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marconi RT, Earnhart CG. 2010. Lyme disease vaccines, p 467–486. In Samuels DS, Radolf JD (ed), Borrelia, molecular biology, host interaction and pathogenesis. Caister Academic Press, Norfolk, UK. [Google Scholar]
- 8. Gomes-Solecki M, Arnaboldi PM, Backenson PB, Benach JL, Cooper CL, Dattwyler RJ, Diuk-Wasser M, Fikrig E, Hovius JW, Laegreid W, Lundberg U, Marconi RT, Marques AR, Molloy P, Narasimhan S, Pal U, Pedra JHF, Plotkin S, Rock DL, Rosa P, Telford SR, Tsao J, Yang XF, Schutzer SE. 2020. Protective immunity and new vaccines for Lyme disease. Clin Infect Dis 70:1768–1773. doi: 10.1093/cid/ciz872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aucott JN, Crowder LA, Kortte KB. 2013. Development of a foundation for a case definition of post-treatment Lyme disease syndrome. Int J Infect Dis 17:e443–e449. doi: 10.1016/j.ijid.2013.01.008 [DOI] [PubMed] [Google Scholar]
- 10. Koedel U, Fingerle V, Pfister HW. 2015. Lyme neuroborreliosis-epidemiology, diagnosis and management. Nat Rev Neurol 11:446–456. doi: 10.1038/nrneurol.2015.121 [DOI] [PubMed] [Google Scholar]
- 11. Casjens SR, Mongodin EF, Qiu WG, Luft BJ, Schutzer SE, Gilcrease EB, Huang WM, Vujadinovic M, Aron JK, Vargas LC, Freeman S, Radune D, Weidman JF, Dimitrov GI, Khouri HM, Sosa JE, Halpin RA, Dunn JJ, Fraser CM, Stevenson B. 2012. Genome stability of Lyme disease spirochetes: comparative genomics of Borrelia burgdorferi plasmids. PLoS ONE 7:e33280. doi: 10.1371/journal.pone.0033280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schutzer SE, Fraser-Liggett CM, Casjens SR, Qiu W-G, Dunn JJ, Mongodin EF, Luft BJ. 2011. Whole-genome sequences of thirteen isolates of Borrelia burgdorferi. J Bacteriol 193:1018–1020. doi: 10.1128/JB.01158-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb JF, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Weidman J, Venter JC. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nat 390:580–586. doi: 10.1038/37551 [DOI] [PubMed] [Google Scholar]
- 14. Stevenson B, Krusenstjerna AC, Castro-Padovani TN, Savage CR, Jutras BL, Saylor TC. 2022. The consistent tick-vertebrate infectious cycle of the Lyme disease spirochete enables Borrelia burgdorferi to control protein expression by monitoring its physiological status. J Bacteriol 204:e0060621. doi: 10.1128/jb.00606-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang X, Yang X, Kumar M, Pal U. 2009. BB0323 function is essential for Borrelia burgdorferi virulence and persistence through tick-rodent transmission cycle. J Infect Dis 200:1318–1330. doi: 10.1086/605846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stewart PE, Hoff J, Fischer E, Krum JG, Rosa PA. 2004. Genome-wide transposon mutagenesis of Borrelia burgdorferi for identification of phenotypic mutants. Appl Environ Microbiol 70:5973–5979. doi: 10.1128/AEM.70.10.5973-5979.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thakur M, Bista S, Foor SD, Dutta S, Yang X, Ronzetti M, Rana VS, Kitsou C, Linden SB, Altieri AS, Baljinnyam B, Nelson DC, Simeonov A, Pal U. 2022. Controlled proteolysis of an essential virulence determinant dictates infectivity of Lyme disease pathogens. Infect Immun 90:e0005922. doi: 10.1128/iai.00059-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kariu T, Yang X, Marks CB, Zhang X, Pal U. 2013. Proteolysis of BB0323 results in two polypeptides that impact physiologic and infectious phenotypes in Borrelia burgdorferi. Mol Microbiol 88:510–522. doi: 10.1111/mmi.12202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thakur M, Sharma K, Chao K, Smith AA, Herzberg O, Pal U. 2017. A protein-protein interaction dictates borrelial infectivity. Sci Rep 7:2932. doi: 10.1038/s41598-017-03279-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Coleman JL, Crowley JT, Toledo AM, Benach JL. 2013. The HtrA protease of Borrelia burgdorferi degrades outer membrane protein BmpD and chemotaxis phosphatase CheX. Mol Microbiol 88:619–633. doi: 10.1111/mmi.12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kariu T, Sharma K, Singh P, Smith AA, Backstedt B, Buyuktanir O, Pal U. 2015. BB0323 and novel virulence determinant BB0238: Borrelia burgdorferi proteins that interact with and stabilize each other and are critical for infectivity. J Infect Dis 211:462–471. doi: 10.1093/infdis/jiu460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Groshong AM, Fortune DE, Moore BP, Spencer HJ, Skinner RA, Bellamy WT, Blevins JS. 2014. BB0238, a presumed tetratricopeptide repeat-containing protein, is required during Borrelia burgdorferi mammalian infection. Infect Immun 82:4292–4306. doi: 10.1128/IAI.01977-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ye M, Sharma K, Thakur M, Smith AA, Buyuktanir O, Xiang X, Yang X, Promnares K, Lou Y, Yang XF, Pal U. 2016. HtrA, a temperature- and stationary phase-activated protease involved in maturation of a key microbial virulence determinant, facilitates Borrelia burgdorferi infection in mammalian hosts. Infect Immun 84:2372–2381. doi: 10.1128/IAI.00360-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Clausen T, Kaiser M, Huber R, Ehrmann M. 2011. HTRA proteases: regulated proteolysis in protein quality control. Nat Rev Mol Cell Biol 12:152–162. doi: 10.1038/nrm3065 [DOI] [PubMed] [Google Scholar]
- 25. Brangulis K, Akopjana I, Kazaks A, Tars K. 2019. Crystal structure of the N-terminal domain of the major virulence factor BB0323 from the Lyme disease agent Borrelia burgdorferi. Acta Crystallogr D Struct Biol 75:825–830. doi: 10.1107/S2059798319010751 [DOI] [PubMed] [Google Scholar]
- 26. Nicolas A, Delalande O, Hubert JF, Le Rumeur E. 2014. The spectrin family of proteins: a unique coiled-coil fold for various molecular surface properties. J Struct Biol 186:392–401. doi: 10.1016/j.jsb.2014.03.011 [DOI] [PubMed] [Google Scholar]
- 27. Liem RK. 2016. Cytoskeletal integrators: the spectrin superfamily. Cold Spring Harb Perspect Biol 8. doi: 10.1101/cshperspect.a018259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D. 2021. Highly accurate protein structure prediction with AlphaFold. Nat 596:583–589. doi: 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Karpenahalli MR, Lupas AN, Söding J. 2007. TPRpred: a tool for prediction of TPR-, PPR- and SEL1-like repeats from protein sequences. BMC Bioinf 8:2. doi: 10.1186/1471-2105-8-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Letunic I, Khedkar S, Bork P. 2021. SMART: recent updates, new developments and status in 2020. Nucleic Acids Res 49:D458–D460. doi: 10.1093/nar/gkaa937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, Finn RD, Bateman A. 2021. Pfam: the protein families database in 2021. Nucleic Acids Res 49:D412–D419. doi: 10.1093/nar/gkaa913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Abe Y, Shodai T, Muto T, Mihara K, Torii H, Nishikawa S, Endo T, Kohda D. 2000. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell 100:551–560. doi: 10.1016/s0092-8674(00)80691-1 [DOI] [PubMed] [Google Scholar]
- 33. D’Andrea LD, Regan L. 2003. TPR proteins: the versatile helix. Trends Biochem Sci 28:655–662. doi: 10.1016/j.tibs.2003.10.007 [DOI] [PubMed] [Google Scholar]
- 34. Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. 2005. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev 29:231–262. doi: 10.1016/j.femsre.2004.12.008 [DOI] [PubMed] [Google Scholar]
- 35. Giuseppe PO, Von Atzingen M, Nascimento A, Zanchin NI, Guimarães BG. 2011. The crystal structure of the leptospiral hypothetical protein LIC12922 reveals homology with the periplasmic chaperone SurA. J Struct Biol 173:312–322. doi: 10.1016/j.jsb.2010.10.009 [DOI] [PubMed] [Google Scholar]
- 36. Bitto E, McKay DB. 2002. Crystallographic structure of SurA, a molecular chaperone that facilitates folding of outer membrane porins. Struct 10:1489–1498. doi: 10.1016/s0969-2126(02)00877-8 [DOI] [PubMed] [Google Scholar]
- 37. Jakob RP, Koch JR, Burmann BM, Schmidpeter PA, Hunkeler M, Hiller S, Schmid FX, Maier T. 2015. Dimeric structure of the bacterial extracellular foldase PrsA. J Biol Chem 290:3278–3292. doi: 10.1074/jbc.M114.622910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Holm L, Sander C. 1995. Dali: a network tool for protein structure comparison. Trends Biochem Sci 20:478–480. doi: 10.1016/s0968-0004(00)89105-7 [DOI] [PubMed] [Google Scholar]
- 39. Behrsin CD, Bailey ML, Bateman KS, Hamilton KS, Wahl LM, Brandl CJ, Shilton BH, Litchfield DW. 2007. Functionally important residues in the peptidyl-prolyl isomerase Pin1 revealed by unigenic evolution. J Mol Biol 365:1143–1162. doi: 10.1016/j.jmb.2006.10.078 [DOI] [PubMed] [Google Scholar]
- 40. Bailey ML, Shilton BH, Brandl CJ, Litchfield DW. 2008. The dual histidine motif in the active site of Pin1 has a structural rather than catalytic role. Biochem 47:11481–11489. doi: 10.1021/bi800964q [DOI] [PubMed] [Google Scholar]
- 41. Fischer G, Bang H, Mech C. 1984. Determination of enzymatic catalysis for the cis-trans-isomerization of peptide binding in proline-containing peptides. Biomed Biochim Acta 43:1101–1111. [PubMed] [Google Scholar]
- 42. Kariu T, Coleman AS, Anderson JF, Pal U. 2011. Methods for rapid transfer and localization of Lyme disease pathogens within the tick gut. J Vis Exp:2544. doi: 10.3791/2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang X, Qin J, Promnares K, Kariu T, Anderson JF, Pal U. 2013. Novel microbial virulence factor triggers murine Lyme arthritis. J Infect Dis 207:907–918. doi: 10.1093/infdis/jis930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Akins DR, Bourell KW, Caimano MJ, Norgard MV, Radolf JD. 1998. A new animal model for studying Lyme disease spirochetes in a mammalian host-adapted state. J Clin Invest 101:2240–2250. doi: 10.1172/JCI2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Caimano MJ. 2005. Cultivation of Borrelia burgdorferi in dialysis membrane chambers in rat peritonea. Curr Protoc Microbiol Chapter 12:Unit 12C3. doi: 10.1002/9780471729259.mc12c03s00 [DOI] [PubMed] [Google Scholar]
- 46. McGuffin LJ. 2008. Intrinsic disorder prediction from the analysis of multiple protein fold recognition models. Bioinf 24:1798–1804. doi: 10.1093/bioinformatics/btn326 [DOI] [PubMed] [Google Scholar]
- 47. Azmi I, Davies B, Dimaano C, Payne J, Eckert D, Babst M, Katzmann DJ. 2006. Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1. J Cell Biol 172:705–717. doi: 10.1083/jcb.200508166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Han H, Monroe N, Sundquist WI, Shen PS, Hill CP. 2017. The AAA ATPase Vps4 binds ESCRT-III substrates through a repeating array of dipeptide-binding pockets. Elife 6. doi: 10.7554/eLife.31324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Brangulis K, Akopjana I, Petrovskis I, Kazaks A, Jekabsons A, Jaudzems K, Viksna A, Bertins M, Tars K. 2020. Structural analysis of Borrelia burgdorferi priplasmic lipoprotein BB0365 involved in Lyme disease infection. FEBS Lett 594:317–326. doi: 10.1002/1873-3468.13594 [DOI] [PubMed] [Google Scholar]
- 50. Brangulis K, Jaudzems K, Petrovskis I, Akopjana I, Kazaks A, Tars K. 2015. Structural and functional analysis of BB0689 from Borrelia burgdorferi, a member of the bacterial CAP superfamily. J Struct Biol 192:320–330. doi: 10.1016/j.jsb.2015.09.007 [DOI] [PubMed] [Google Scholar]
- 51. Bernard Q, Smith AA, Yang X, Koci J, Foor SD, Cramer SD, Zhuang X, Dwyer JE, Lin Y-P, Mongodin EF, Marques A, Leong JM, Anguita J, Pal U. 2018. Plasticity in early immune evasion strategies of a bacterial pathogen. Proc Natl Acad Sci USA 115:E3788–E3797. doi: 10.1073/pnas.1718595115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Evans PR. 2011. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr D Biol Crystallogr 67:282–292. doi: 10.1107/S090744491003982X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lima FA, Saleta ME, Pagliuca RJ, Eleotério MA, Reis RD, Fonseca Júnior J, Meyer B, Bittar EM, Souza-Neto NM, Granado E. 2016. XDS: a flexible beamline for X-ray diffraction and spectroscopy at the Brazilian synchrotron. J Synchrotron Radiat 23:1538–1549. doi: 10.1107/S160057751601403X [DOI] [PubMed] [Google Scholar]
- 54. Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi: 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sheldrick GM. 2008. A short history of SHELX. Acta Crystallogr A 64:112–122. doi: 10.1107/S0108767307043930 [DOI] [PubMed] [Google Scholar]
- 56. Kantardjieff KA, Rupp B. 2003. Matthews coefficient probabilities: improved estimates for unit cell contents of proteins, DNA, and protein-nucleic acid complex crystals. Protein Sci 12:1865–1871. doi: 10.1110/ps.0350503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cowtan K. 2006. The buccaneer software for automated model building. 1. tracing protein chains. Acta Crystallogr D Biol Crystallogr 62:1002–1011. doi: 10.1107/S0907444906022116 [DOI] [PubMed] [Google Scholar]
- 58. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 59. Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53:240–255. doi: 10.1107/S0907444996012255 [DOI] [PubMed] [Google Scholar]
- 60. Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. doi: 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 61. Storoni LC, McCoy AJ, Read RJ. 2004. Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr 60:432–438. doi: 10.1107/S0907444903028956 [DOI] [PubMed] [Google Scholar]
- 62. Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. 2012. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68:352–367. doi: 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. 2022. ColabFold: making protein folding accessible to all. Nat Methods 19:679–682. doi: 10.1038/s41592-022-01488-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mirdita M, Steinegger M, Söding J. 2019. MMseqs2 desktop and local web server app for fast, interactive sequence searches. Bioinf 35:2856–2858. doi: 10.1093/bioinformatics/bty1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sievers F, Higgins DG. 2014. Clustal omega. Curr Protoc Bioinf 48:3. doi: 10.1002/0471250953.bi0313s48 [DOI] [PubMed] [Google Scholar]
- 67. Promnares K, Kumar M, Shroder DY, Zhang X, Anderson JF, Pal U. 2009. Borrelia burgdorferi small lipoprotein Lp6.6 is a member of multiple protein complexes in the outer membrane and facilitates pathogen transmission from ticks to mice. Mol Microbiol 74:112–125. doi: 10.1111/j.1365-2958.2009.06853.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rana VS, Kitsou C, Dutta S, Ronzetti MH, Zhang M, Bernard Q, Smith AA, Tomás-Cortázar J, Yang X, Wu M-J, Kepple O, Li W, Dwyer JE, Matias J, Baljinnyam B, Oliver JD, Rajeevan N, Pedra JHF, Narasimhan S, Wang Y, Munderloh U, Fikrig E, Simeonov A, Anguita J, Pal U. 2023. Dome1-JAK-STAT signaling between parasite and host integrates vector immunity and development. Sci 379:eabl3837. doi: 10.1126/science.abl3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4; Tables S1 and S2.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. The coordinates and the structure factors for native and SeMet-labeled BB0238 have been deposited in the Protein Data Bank with accession codes 8P33 and 8P32, respectively. The coordinates and the structure factors for BB0108 PPIase domain have been deposited in the Protein Data Bank with accession code 8SOT.