Abstract
Genes encoding histone proteins are recurrently mutated in tumor samples, and these mutations may impact nucleosome stability, histone post-translational modification, or chromatin dynamics. The prevalence of histone mutations across diverse cancer types suggest that normal chromatin structure is a barrier to tumorigenesis. Oncohistone mutations disrupt chromatin structure and gene regulatory mechanisms, resulting in aberrant gene expression and the development of cancer phenotypes. Examples of oncohistones include the histone H3 K27M mutation found in pediatric brain cancers that blocks post-translational modification of the H3 N-terminal tail and the histone H2B E76K mutation found in some solid tumors that disrupts nucleosome stability. Oncohistones may comprise a limited fraction of the total histone pool yet cause global effects on chromatin structure and drive cancer phenotypes. Here, we survey histone mutations in cancer and review their function and role in tumorigenesis.
Keywords: cancer, chromatin, histones, point mutations
Introduction
Histone proteins play a fundamental role in protecting and regulating access to DNA during replication, repair, and transcription. The four core histones (H2A, H2B, H3, and H4) are well-conserved basic proteins ranging in size from 102 to 135 amino acids. H2A forms a dimer with H2B, while an H3 dimer interacts with H4 to form the H3/H4 tetramer [1]. Two H2A/H2B dimers bind with one H3/H4 tetramer to form the histone octamer that interacts with ∼147 bp of negatively charged DNA to form a nucleosome [1,2]. Core histone proteins have a positively charged N- or C-terminal tail that protrudes from the nucleosome and histone-fold domains that associate with 27–28 bp of DNA [2]. In addition, the linker histone H1 dynamically interacts with DNA and nucleosomes to condense and stabilize DNA into higher-order structures [3]. Nucleosome formation, histone eviction from DNA and chromatin remodeling, which shifts the distance between nucleosomes, allow for dynamic regulation of chromatin accessibility to gene expression mechanisms.
There are 118 histone genes: 36 H2A genes, 31 H2B genes, 23 H3 genes, 16 H4 genes, and 12 H1 genes, though some are believed to be pseudogenes not transcribed or translated [4,5]. Histone genes are primarily clustered at the HIST1 locus of chromosome 6 with the second-largest cluster on chromosome 1 (Figure 1A) [4,6]. Many histone genes encode proteins that diverge by a few amino acids, which may subtly adjust nucleosome stability or mobility and provide additional regulation for chromatin accessibility and gene expression [4,7].
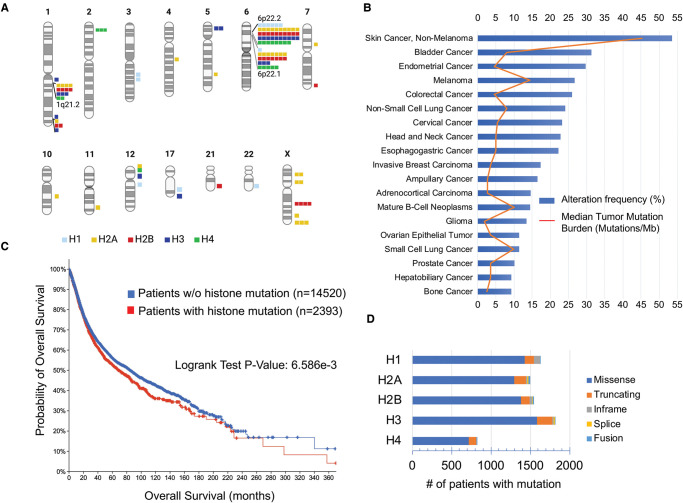
Figure 1. Histones are frequently mutated in cancer and coincide with worse overall survival.
(A) Location of histone genes on each chromosome. (B) Frequency of histone mutations by cancer type in studies with a minimum of 100 cases. (C) Comparison of overall survival for cancer patients with a histone mutation versus cancer patients without a histone mutation in 186 cancer studies of all cancer types. (D) Distribution of mutations among histones.
There are two classes of histones: canonical and variant [8]. Expression of canonical histones is orchestrated by promoter sequences which guide S-phase specific transcription to increase the histone supply necessary to complex with newly synthesized DNA [8,9]. Rather than being polyadenylated, canonical histone genes have a stem loop sequence and a histone downstream element (HDE) in the 3′UTR [10]. The stem loop and HDE are required for efficient translation and catabolism of the canonical histone RNA, restricting its expression to S phase [10]. Notably, canonical histones do not contain introns. Variant histones are expressed in a replication-independent manner throughout the cell cycle [11]. Variant histones may replace canonical histones in the nucleosome to confer distinct functional or structural features, impacting mechanisms such as gene expression and DNA repair [12]. Histones are subject to post-translation modifications that may affect histone stability, histone–DNA interaction, chromatin accessibility or binding of nucleosome interacting proteins to regulate replication, transcription, or repair. Consequently, patterns of histone post-translational modifications create chromatin states associated with either gene expression or gene repression [13–19]. Within the past decade, canonical and variant histone mutations have been revealed as a new class of epigenetic mutations in cancer. [20–26]. Here we catalog mutations of histone H2A, H2B, H3, H4, and H1 in cancer and summarize recent findings for how these mutations may contribute to tumorigenesis.
Histone mutation frequency in cancer
To determine the landscape of recurrent histone mutations in cancer, we surveyed 205 non-redundant cancer studies (65 489 patient samples) using the cBioPortal mirror hosted by the University of Florida Health Cancer Center, which allowed simultaneous search of all histone genes. After excluding 32 622 patient samples not profiled for every histone gene, we found genomic alterations (mutations, structural variants, copy number alterations) of H1 genes in 7% of patients, H2A genes in 11% of patients, H2B genes in 9% of patients, H3 genes in 10% of patients, and H4 genes in 6% of patients. Genomic alterations in any histone gene occurred with a high frequency in a wide range of cancers and were found in more than 50% of non-melanoma skin cancer patients, and up to 30% of patients with tobacco-related cancers, such as bladder urothelial carcinoma, head and neck cancer, esophagogastric adenocarcinoma, or non-small cell lung cancer (Figure 1B). Importantly, cancer patients carrying any histone genomic alteration had a significantly lower overall survival compared with cancer patients without a histone mutation (Figure 1C). In addition, patients carrying genomic alterations of either H2A, H2B, H3, or H4 had a significantly decreased overall survival across all cancers.
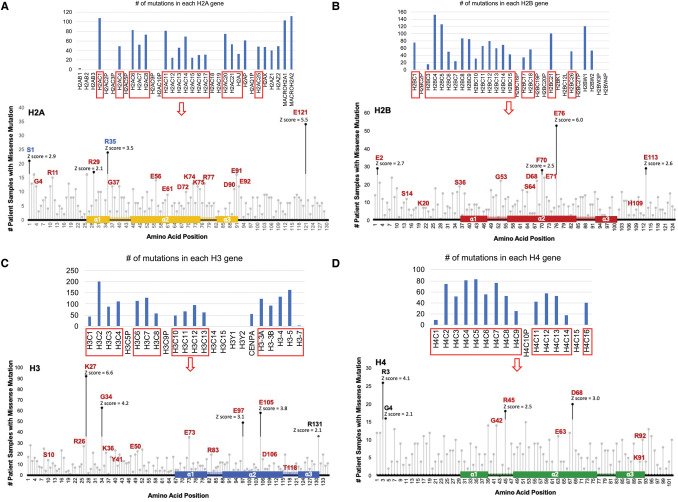
About 85% of recorded mutations in each histone were missense mutations (Figure 1D). To identify which histone amino acids were most often found mutated in cancer patients, we combined cBioportal data from multiple gene paralogs whose amino acid sequences aligned well. We found that most histone consensus amino acid residues had a missense mutation in at least one patient, and on average each residue was mutated in 4–12 patient samples. A Z-score was calculated at each amino acid residue of each histone by subtracting the number of mutations at a particular amino acid from the mean number of mutations across all amino acids and dividing the result by the standard deviation across all amino acids. Any amino acid with a Z-score >2 was considered a significant hotspot of recurrent mutation (Figures 2 and 4). Hotspot mutations were found in both histone tails and the histone-fold domains (Figures 2 and 4). Many histone missense mutations caused a charge swap (E to K mutations) or removed a positive charge (R to C mutations). Mutational hotspots in tumors suggest gain or change of function that promotes tumorigenesis. Indeed, some of these recurrent histone mutations have been previously studied in cancer such as histone H3.3 K27 and H3.3 G34 mutations [20,21,25–27]. Our prior analysis of histone mutations found only two histone mutant hotspots, H2A S1 and R35, had single nucleotide polymorphisms (SNP) present with similar frequency in the SNP database (dbSNP) and cBioPortal, indicating that most of the hotspot mutations are not simply passenger SNPs [25]. Also, some histone residues are rarely found mutated, which may be due to carcinogen or sequence specificity of DNA mutations or reflect that these histone mutations have deleterious effects on cell growth.
Figure 2. Survey of the most frequent core histone mutations in cancer patient samples.
A cross cancer mutation summary was performed using the cBioPortal to search a total of 29 836 non-redundant patient samples across all cancer types, excluding sample not profiled for all histone genes. The number of patients reported to have a missense mutation in the nucleosome core histones H2A, H2B, H3 and H4 at each amino acid residue position across well conserved histone paralogs was graphed by lollipop plot to depict the location of the cumulative missense mutations found in: H2A (A), H2B (B), H3 (C), and H4 (D). Alignment of histone proteins was performed in SnapGene v7.0.1. A Z-score was calculated by subtracting the number of mutations at a particular amino acid from the mean number of mutations across all amino acids and dividing the result by the standard deviation across all amino acids to identify the amino acids most frequently found mutated in each histone. Amino acid positions indicated by black dots are at least two standard deviations above the average number of mutations across all residues. Amino acids displayed in red were observed to have functional defects when mutated by Bagert et al. [27]. Amino Acids in blue were reported to have a similar polymorphism frequency between the general population (dbSNP) and cancer patient (cBioPortal) databases [25].
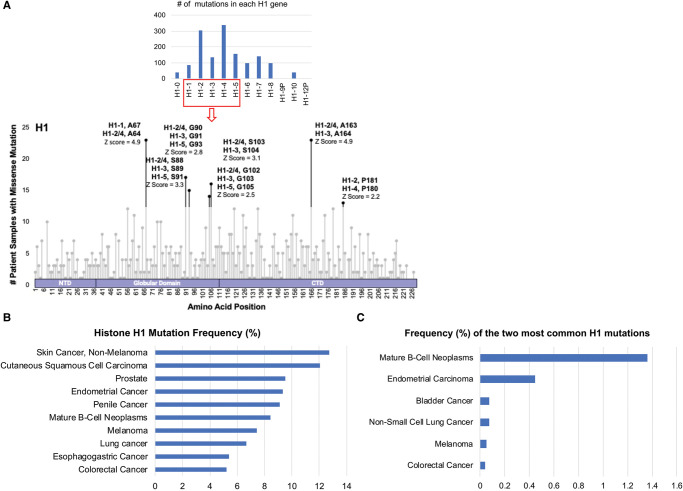
Figure 4. Survey of histone H1 mutations in cancer.
A cross cancer mutation summary was performed using cBioPortal to search 29 836 non-redundant patient samples across all cancer types for mutations in histone H1, excluding patient samples not profiled for all H1 genes. Alignment of H1 proteins was performed in Snapgene v7.0.1. (A) The number of patients reported to have a missense mutation at each amino acid residue position across well conserved histone H1 paralogs was graphed by lollipop plot. Amino acid positions depicted by black dots are at least two standard deviations above the average number of mutations observed across all residues. A Z-score was calculated to classify the most common mutations. (B) Histone H1 alteration frequency by cancer type considering only studies with >100 cases and a frequency of H1 mutation over 5%. (C) Alteration frequency of the two most common H1 mutations at A64/67and A163/164 in cancer reveals that they are predominately found mutated in B-cell neoplasms.
H2A histone mutations in cancer
Histone H2A missense mutations constituted ∼20% of histone missense mutations in cancer (Figure 1D). The most common mutated H2A residue, E121, demonstrated decreased nucleosome sliding via ATP-utilizing chromatin remodeling and assembly factor (ACF) when mutated in vitro and impaired yeast growth (Table 1) [27]. The second most common mutated H2A residue, R29, is a site of post-translational modification by methylation and a point of contact with nucleosomal DNA [2,28]. Mutations at this residue demonstrated increased nucleosome sliding via ACF in vitro, decreased nucleosome thermal stability, increased histone H2A/H2B dimer exchange via histone chaperone nuclear assembly protein-1 (Nap1), and impaired yeast growth (Table 1) [27]. Interestingly, many H2A missense mutations occur in the histone-fold domain or the acidic patch on the histone (Figure 2A). The acidic patch consists of six H2A (E56, E61, E64, D90, E91, E92) and two H2B (E105, E113) amino acids on the nucleosome surface that provide a charged surface for protein binding [29]. Mutations occurring on acidic patch residues increased Nap1-mediated histone H2A/H2B dimer exchange (H2A E56), decreased thermal stability (H2A E56), increased (H2A E56) or decreased (H2A D90, E92) nucleosome sliding via ACF or ISWI, and impaired yeast growth (H2A E56, D90, E91, E92) [27,30,31] (Table 1). Recently, H2A E92K, which reverses the amino acid charge, was found to decrease binding of DNMT3A and the deacetylase SIRT6 to the nucleosome while increasing binding of bromodomain proteins BRD2/3/4 [32]. Mutations in the histone-fold domain of H2A were found to increase nucleosome sliding (H2A K74, K75) and impair yeast growth (H2A D72, K75) [27] (Table 1).
Table 1. Summary of known histone mutations.
| Histone amino acid | Function/phenotype of mutant | References |
|---|---|---|
| H2A | ||
| G4 | 1 | [27] |
| R11 | Unknown | |
| R29 | 1,2,3,4 | [27] |
| G37 | Unknown | |
| E56 | 1,2,3,4 | [27] |
| D72 | 1 | [27] |
| K74 | 3 | [27] |
| K75 | 1,3 | [27] |
| D90 | 1,3 and inhibited chromatin remodeling by ISWI | [27,30,31] |
| E91 | 1 | [27] |
| E92 | 1,3 and inhibited chromatin remodeling by ISWI | [27,30,31] |
| E121 | 1,3 | [27] |
| H2B | ||
| E2 | 1 | [27] |
| S36 | 1 | [27] |
| G53 | 1,3 and decreased DNA interaction, increased migration, increased RNA Pol II passage, altered gene expression. | [27,34,35,37] |
| D68 | 1,2,4, unstable histone octamer, and impaired H2A.Z-H2B incorporation into chromatin | [27,78,89] |
| F70 | 2 | [27] |
| E71 | 1,2,4,5 | [27] |
| E76 | 1,2,4,5 and nucleosome instability, increased H2A–H2B dimer mobility, altered gene expression, increased proliferation and colony formation, increased chromatin accessibility, increased interaction with NAP1L1 and NAP1L2 chaperones. | [24–27,33] |
| E113 | 1,5 and increased chromatin remodeling by ISWI | [27,31] |
| H3 | ||
| R26 | Unknown | |
| K27 | 5 and global loss & focal gain of H3K27me3 modification, suppression of PRC2, altered proliferation, increased H3K27ac histone modification, increased DNA hypomethylation | [20,21,39–45,90] |
| G34 | 5 and increased colony formation, increased infiltration, increased proliferation, DNA hypomethylation, decreased H3K36 methylation in cis, increased H3K27 methylation. | [20,39,59,60,61,62,66,91] |
| K36 | 1,5 and increased colony formation, suppression of SETD2, global decrease in H3K36 di- and trimethylation, tumor formation in vivo, decreased sensitivity to apoptotic stimuli, impaired homologous recombination | [27,62,66–68] |
| E50 | 1,2 | [27] |
| E73 | 1 | [27] |
| E97 | 1,2,4 and enhanced colony formation, nucleosome instability | [24,27,70] |
| E105 | 1 and yeast sin mutant | [27,71] |
| H4 | ||
| G42 | 1,2,3 | [27] |
| R45 | 1,3 and Increased resistance to UV damage, increased nucleotide excision repair, yeast sin mutant, prevents Mg2+ dependent chromatin condensation, loss of interaction with nucleosome DNA, decreased RNA Pol II pausing | [7,27,74–76,77,92] |
| E63 | 1 | [27] |
| D68 | 1,2,4 | [27] |
| R92 | 1,6 | [27,73] |
Phenotype key: .
Impaired yeast growth;
Increased Nap1-mediated histone dimer exchange;
Altered nucleosome sliding;
Decreased thermal stability;
Impaired differentiation of cells;
Hypersensitivity to DNA damaging agents.
H2B histone mutations in cancer
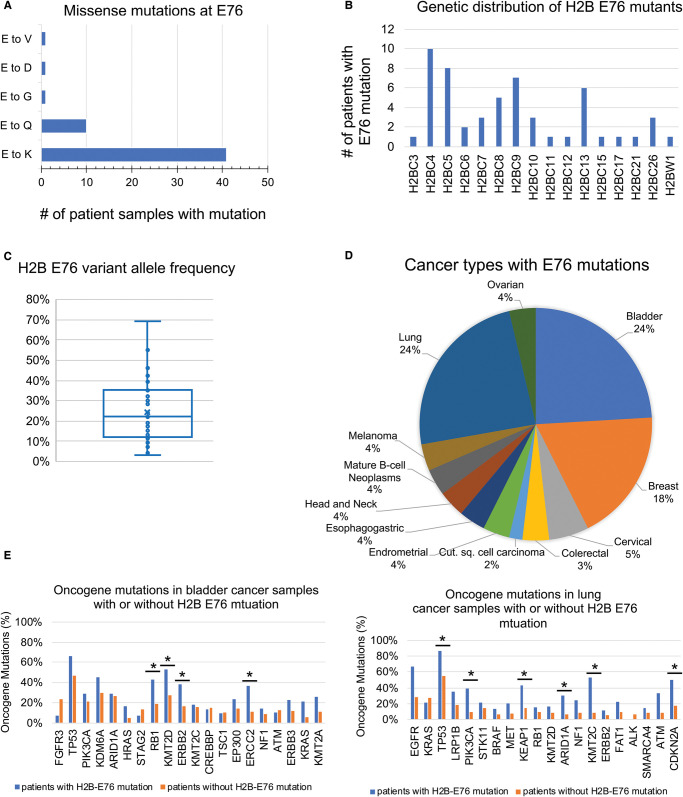
Histone H2B mutations account for ∼21% of all histone missense mutants in cancer samples (Figure 1D). Several H2B mutations have been identified for their roles in destabilizing the nucleosome or weakening histone–DNA interactions [24,25,27,33–35]. The most frequent H2B mutation occurs at residue 76, in which glutamate is often mutated to a lysine or, less commonly, to a glutamine (Figure 3A). In a pan-cancer analysis using data from cBioPortal, we found 41/54 patient samples with a glutamate to lysine mutation (H2B E76K), and 10/54 patient samples with a glutamate to glutamine mutation (H2B E76Q) (Figure 3A). While it is unknown why these missense mutations are most common, only a single nucleotide substitution would change glutamate to lysine (G/C > A/T) or glutamine (G/C > C/G). In our cBioPortal analysis, the most common single base substitutions for the H2B-E76K mutation were C > T or G > A transition mutations with about half carrying a C > T mutation, a signature cancer-related substitution [36]. H2B E76 mutations were distributed across 16 of 22 canonical H2B genes surveyed (Figure 3B). The variant allele frequency (VAF) of H2B E76 mutations in cancer was ∼20%, suggesting these are subclonal mutations contributing to tumor progression rather than initiating mutations (Figure 3C). H2B E76 mutations occur in numerous cancer types such as bladder (24%) and lung (24%) cancers (Figure 3D). We examined whether the top 20 genomic alterations found in bladder or lung cancers coincide with the H2B E76 mutation in patient samples with these cancers. In bladder cancer patient samples, H2B E76 mutations significantly co-occur with genomic alterations in RB1, KMT2D, ERBB2, and ERCC2, while in lung cancer patient samples the E76 mutation coincides with TP53, PIK3CA, KEAP1, ARID1A, KMT2C, and CDKN2A genomic alterations and these mutations may synergize to propel tumor progression (Figure 3E).
Figure 3. Analysis of H2B E76 mutations in cancer.
(A) Distribution of missense mutations at H2B amino acid E76. (B) Frequency of mutation at E76 across H2B genes. (C) H2B E76 variant allele frequency in all cancer types. (D) Distribution of cancer types with H2B E76 mutations. (E) The frequency of genomic alterations in the indicated oncogene was compared in profiled patient samples with (blue bar) or without (orange bar) H2B missense mutations at E76. Oncogene alterations that significantly co-occur with H2B E76 mutations (P < 0.5 and Q < 0.5) are indicated by a *.
X-ray crystallography demonstrated that the H2B E76K mutation repels the H4 R92 residue, disrupting an important H2B/H4 interaction point and destabilizing the histone octamer [24–26]. Thus, H2B E76K is not readily able to form a histone octamer without DNA present [24,25,27]. H2A/H2B dimer exchange is increased in cells expressing H2B E76K and is dependent on the histone chaperone Nap1 (nuclear assembly protein-1) [24,25,27] (Table 1). Others have reported increased interaction between H2B E76K histones and chaperone proteins Nap1l1 and Nap1l2 that may contribute to increased H2A/H2B dimer exchange in H2B E76K expressing cells [27,33]. Unlike other histone mutants, H2B E76K does not affect global histone post-translational modification levels [33].
The H2B E76K oncohistone has been reported to promote chromatin accessibility at promoters and enhancers [25,33]. Multiple studies report that H2B E76K alters gene expression [25,27,33]. These changes in gene expression translate to phenotypic changes such as increased proliferation, increased colony formation and inhibition of differentiation that is observed in cells expressing H2B E76K [24,25,27,33]. Why only certain sets of genes are activated is not certain, but it has been reported that H2B E76K leads to enhanced transcriptional elongation rates, which might favor expression of subsets of genes.
The second most mutated histone H2B residue, H2B E113, is part of the nucleosome acidic patch. Previous studies have demonstrated that H2B E113 is most often mutated to lysine (H2B E113K), which causes increased nucleosome remodeling via ACF or ISWI [27,31]. Like other H2B mutants, H2B E113K has been reported to inhibit differentiation of murine mesenchymal cells and impair yeast growth [27] (Table 1).
Another well-characterized H2B oncohistone is H2B G53D originally found in pancreatic ductal adenocarcinoma (PDAC), glioblastoma multiforme, and lung cancer [35,37]. This residue lies close to the DNA entry/exit point on the nucleosome. Nucleosome pulling and holding assays suggest that the H2B G53D acquired negative charge may interfere with histone–DNA interactions [35]. Though DNA replication is not affected, H2B G53D expressing cells have increased RNA polymerase II passage through mutant nucleosomes [35]. Consistent with a weakened histone–DNA interaction, H2B G53D nucleosomes display increased nucleosome sliding via ACF in vitro [27] (Table 1). Expression of H2B G53D also causes gene expression changes, affecting pathways such as growth factor signaling, cell adhesion and cancer development [34] (Table 1). Based on analysis of CUT&RUN, gene expression, and PRO-seq data from H2B G53D expressing cells, H2B G53D increases active RNA-polymerase II and increased gene expression at a subset of genes [34]. One oncogene found in this subset, ANXA3, was demonstrated to have increased transcription due to H2B G53D, and ANXA3 expression was greater in cancer patients carrying the H2B G53D mutation, supporting the notion that expression of H2B G53D drives expression of the ANXA3 oncogene [34]. In addition to chromatin and gene expression changes, the H2B G53D mutation also promotes gap closure and transwell migration in a pancreatic cell line [35], but impairs yeast growth [27] (Table 1).
A common phenotype for multiple histone H2B mutations such as amino acid E2, S36, G53, D68, E71, E76, or E113, is impaired yeast growth [27] (Table 1). How this translates to human oncogenic phenotypes is not yet certain. In addition, mutations at H2B D68 destabilize the histone octamer, like the H2B E76K nucleosome [25,27]. Furthermore, histone H2A/H2B dimer exchange via Nap1 histone chaperone was also found to be increased when residues H2B D68, F70, or E71 were mutated in vitro [27]. Expression of H2B E71K inhibits differentiation of murine mesenchymal cells, as observed with many other histone mutants [27].
Histone H3 mutations in cancer
Almost 25% of histone missense mutations in the cBioPortal are in Histone H3 (Figure 1D). The first oncohistones identified were mutations in genes H3F3A and H3F3B that encode variant histone H3.3. Recurrent H3.3 mutations affecting two critical amino acids (K27 and G34) have been identified in one-third of pediatric glioblastoma (GBM) and nearly 80% of diffuse intrinsic pontine gliomas (DIPG). Our cBioPortal analysis indicates that the most frequently mutated histone H3 residue is K27, usually in H3.3 (Figure 2C). This amino acid on the N-terminal tail of H3 can be post-translationally modified by acetylation, methylation, ubiquitination, and ADP-ribosylation [38]. H3.3 K27 is commonly mutated to a methionine (H3 K27M), which can no longer be methylated or acetylated, leading to decreased global H3 K27me2/me3 and increased H3 K27 acetylation levels [39–44]. Polycomb repressive complex 2 (PRC2) methylates H3 K27 via its enhancer of zeste homolog 2 (EZH2) catalytic subunit. Initial reports suggested PRC2 became sequestered on the K27M mutant histone, leading to a global decrease in H3K27me3 [39,41,45]. Such experiments showed enhanced co-precipitation of H3 K27M with PRC2 and increased affinity of H3 K27M for PRC2 compared with WT H3 [39,41]. Additionally, H3 K27M was found to have decreased association with corepressor protein CYL, which normally connects PRC2 with neighboring nucleosomes to promote H3K27me3 spreading [46,47]. However, recent reports offer differing results, highlighting that the mechanism of action of H3 K27M may not fully be elucidated. In contrast with the sequestration model: (1) H3K27me3 marks are focally retained in chromatin, primarily on CpG islands and a subset of genes such as the tumor suppressor p16/INK4A [41,42,48–51]; (2) PRC2 has not consistently been shown to have greater affinity for H3 K27M over H3 WT [50,52,53]; (3) H3 K27M does not always co-localize with PRC2 on chromatin, in some cases PRC2 has been excluded [54–56]; (4) H3K27ac marks are enriched on H3 K27M nucleosomes, which further exclude PRC2 [55]; and (5) there is a persistent decrease in PRC2 activity following its transient interaction with H3 K27M [41,50]. Collectively these data indicate that H3K27M disrupts normal patterns of histone methylation, with large regions of H3K27me3 loss but other areas of focal gain which together lead to disruption of normal gene expression contributing to oncogenesis. Accordingly, H3 K27M increases proliferation of cells and the combination of H3 K27M, p53 knockdown, and oncogenic PDGFRA inhibited differentiation of astrocytes [38]. Deposition of K27M at active enhancers of neurodevelopment genes leads to H3K27ac loss, decreased chromatin accessibility and reduced expression of neurodevelopmental genes in neural stem cells [57]. In addition, because DNMT3A/B DNA methyltransferases associate with EZH2, K27M mutant tumors have a substantially different DNA methylation profile from H3.3 WT pediatric high-grade gliomas, with an overall reduction in global DNA methylation and enrichment of hypomethylated regions [41,58].
Another H3 oncohistone is at amino acid G34 on the N-terminal tail. In pediatric glioblastoma, H3.3 G34 is frequently mutated to arginine or valine (H3.3 G34R/V) [20]. In giant cell tumors of bone (GCTB) this residue is most often mutated to tryptophan (H3.3 G34W) [59]. Primary cells derived from GCTB biopsies displayed increased proliferation, invasion, and colony formation [60]. In addition, H3.1 G34 mutations are found in carcinomas [26]. Although not a target of post-translational modification itself, mutations at G34 disrupt histone H3 K36 and H3 K27 methylation in cis (on the same histone tail) [39,59,61,62]. Mutations changing glycine 34 to a bulkier residue have been reported to block histone tail binding by SETD2 and H3K36 methylation [63]. Despite the inhibitory effect of H3.3 G34W on SETD2, genome-wide studies indicate more areas of H3K36me3 gain than loss [64]. Furthermore, H3 G34W has been associated with global DNA hypomethylation as well as specific focal DNA hypermethylation that impairs differentiation in vitro [59]. Mass spectrometry has identified that H3.3 G34W binding partners include SWI/SNF components ARID2, SMARCA1, and HDAC2 [60]. Thus, G34W may recruit proteins from other sites in the genome to alter local chromatin architecture as well as alter the accumulation of H3.3 through aberrant interaction with histone chaperones. Furthermore, in GCTB cell lines H3.3 G34W made up only 3% of histone H3, yet H3K27me3 levels on non-mutant histones H3.3 and H3.1 decreased by 30% to 40%, suggesting an important trans effect of the mutant histone [64]. In the presence of G34W, H3K27me3 and PRC2 were significantly depleted from intergenic regions, and this may play a role in the observed chromatin and gene expression changes observed in H3.3 G34W mutant tumors [64]. In addition, a recent study revealed that H3.3 G34R/V/W germline mutations have distinct and substantial impacts on genome-wide distribution of epigenetic marks and interacting proteins that may account for the phenotypic diversity associated with these mutants [65]. Specifically, G34R greatly depleted H3K36me2/3 in cis but had no obvious effect on H3K27 methylation [65]. In contrast, G34V/W significantly increased H3K27me3 but had little effect on H3K36me2/3 [65]. Furthermore, the G34R-mediated redistribution of H3K36me2 altered the recruitment and redistribution of DNMT3A, which in turn changed the landscape of DNA methylation, leading to deregulated expression of complement genes and suppression of neuronal developmental genes [65].
About one year after the discoveries of the H3.3 K27 and H3.3 G34 oncohistones, the H3.3 K36M N-terminal tail mutation was discovered in chondroblastoma patient samples [66]. H3 K36 is a site of post-translational modification by acetylation, methylation or ubiquitylation [38]. H3 K36M causes a global decrease in H3 K36 di- and trimethylation levels [62,67,68] at least in part by trapping NSD enzymes and suppressing SETD2, the histone methyltransferases responsible for H3 K36 methylation [62,69]. Cells expressing H3.3 K36M displayed increased colony formation and resistance to apoptosis [68]. The H3 K36M mutation impaired adipocyte and osteocyte differentiation of murine mesenchymal cells, and mesenchymal progenitor cells expressing H3 K36M formed an undifferentiated sarcoma after injection into mice [27,67,68].
Though not as thoroughly studied as H3.3 K27, G34, and K36 mutations, H3.1 mutations at amino acids E97 and E105 were found mutated at a high frequency in our pan-cancer analysis (Figure 2C). Both H3 E97 and E105 residues are in the histone-fold domain, as opposed to the N-terminal tail, and are often found mutated to lysine from our cBioPortal results (Figure 2C). H3 E97 mutations increased Nap1-mediated histone H2A/H2B dimer exchange, destabilized the nucleosome, and decreased thermal stability [24,27]. Mutations at both H3 E97 and E105 impair yeast growth, and H3 E97 mutants also increase colony formation [24,27,70]. In yeast, the H3 E105K causes very little change in nucleosome structure and stability yet causes SWI/SNF independence (SIN) i.e. this mutation alleviates the transcriptional defects caused by inactivation of the yeast SWI/SNF complex [71].
Histone H4 mutations in cancer
The only histone to have a single variant form [72], histone H4 binds with histone H3 to form the crucial H3/H4 tetramer and histone H4 interacts with histone H2B to form the histone octamer [1]. The most frequent histone H4 missense mutations in cancer patient samples were located at amino acids R45 and D68. Other, less frequent histone H4 mutations at G42, E63, and R92 were found to impair yeast growth while mutation of H4 at R92 has been reported to increase sensitivity to DNA damaging agents [27,73] (Table 1).
H4 R45 is inserted into the minor groove of the nucleosomal DNA, interacting with DNA via van der Waals forces rather than site specific contacts [1,74]. Based on our cBioPortal search, histone H4 R45 is most often mutated to a glutamine or cysteine. Mutations to this residue has been shown to increase nucleosome sliding, decrease RNA polymerase II pausing, and alter interactions with nucleosomal DNA [27,74,75] (Table 1). In yeast, the H4 R45C mutation functions as a SIN mutant yet still impaired yeast growth [76]. In addition, H4 R45C increased nucleotide excision repair and resistance to UV damage in yeast [27,76]. The R45C mutation does not disrupt nucleosome positioning or the accessibility of nucleosomal DNA but rather has been reported to eliminate Mg2+-dependent, intramolecular folding of nucleosomal arrays, suggesting that SIN histone mutants may alleviate the need for SWI/SNF in vivo by disrupting higher-order chromatin folding [77].
The most frequently mutated amino acid of histone H4 is located at D68 (Figure 2D). This residue forms a stable electrostatic interaction between histones H2B E76 and H4 [24–26]. Histone H4 D68 is commonly mutated to an asparagine and these mutations have been reported to increase Nap1-mediated histone H2A/H2B dimer exchange in vitro [27]. In addition, H4 mutations at D68 impair binding of histone H2A.Z-H2B dimer to chromatin [78].
Histone H1 mutations in cancer
The linker histone H1 binds to DNA exiting the nucleosome core to catalyze condensation of nucleosomal arrays [79]. H1 is a larger, highly mobile histone that interacts with DNA in a more rapid fashion than core histones comprising the nucleosome [79]. Similar to other histones, histone H1 is expressed from multiple genes, though the sequences are less conserved [79]. In our cBioPortal pan-cancer analysis, we found missense mutations in 10 of 12 histone H1 genes (Figure 4A). Histone H1 missense mutations occurred in a broad range of cancers, including skin, prostrate and endometrial cancers as well as mature B-cell neoplasms (Figure 4B). H1 missense mutations were detected in cancer patient samples at nearly every residue, and the two most common ‘hotspot’ mutations were observed at A64/67 and A163/164 (Figure 4A). The two most common histone H1 mutations were overwhelmingly observed in mature B-cell neoplasms (Figure 4C), a cancer type previously reported to carry histone H1 mutations and deletions [22,80–85].
Histone H1 may have a tumor suppressor function. Loss of histone H1 was associated with chromatin decompaction as well as increased H3 K36 methylation and decreased H3 K27 methylation, causing B to A compartment shifts and transcriptional activation [81,86,87]. For instance, genes related to T-cell activation were expressed with H1 loss [81]. Also, H1 depletion by deletion of three H1 genes in pluripotent embryonic stem cells blocked differentiation due to unchecked expression of pluripotency genes [88]. Germinal center B cells may be especially sensitive to H1 dosage or mutation due to their rapid proliferation rate [81]. In Diffuse Large B-cell Lymphoma (DLBCL), samples carrying H1 mutations have been reported to display an increased mutational burden [84]. In an analysis of mutations driving follicular lymphoma initiation and progression, mutations affecting at least one histone H1 gene were observed in 28% of samples with most being missense mutations in the globular domains of H1, H1–2, and H1–4 genes [80]. When either WT or a S101F mutation of histone H1–2 was expressed in H1 knockout murine embryonic stem cells, the S101F mutant was not associated with chromatin to the same degree as wild-type histone, suggesting this is a loss of function mutation that reduces the affinity of H1 with chromatin [80]. In our pan-cancer analysis, histone H1–2 residues G102 and S103 on H1–2 are more often found mutated than S101 (Figure 4A), and these mutations may disrupt a similar region of H1 as S101 necessary for DNA interaction. When GFP-tagged S101F, G102A, and S103F were expressed in 3T3 cells more rapid fluorescence recovery after photobleaching was observed than in cells expressing GFP-tagged wild-type H1, and the dissociation constant of H1 S101F was much higher than wild-type H1 in a mononucleosome binding assay [81]. Thus, H1 globular domain mutations may impair the association of H1 with chromatin, while C-terminal mutants remain able to bind chromatin normally but disrupt compaction [81].
Summary
Histone mutations are pervasive in all cancer types. They may disrupt histone post-translational modifications, protein interactions with the nucleosome acid patch, nucleosome stability, remodeling, or chromatin compaction (Figure 5). Oncohistone mutations disrupt chromatin function to cause a state of gene expression favoring tumorigenesis. Many facets of histone mutations in cancer remain to be elucidated. For instance, histones are encoded by multiple genes with small sequence variations of unknown significance that may provide functional diversity. Various tissues have distinct histone gene expression patterns, but the importance of these patterns is unknown. Explicit histone variants may be required by specific cell types during development or other cellular processes. Histone mutations are rarely found on their own in cancer patient samples but instead often co-occur with well-known oncogenes. It isn't yet clear how histone mutations and destabilization of chromatin structure or function may impact or enable oncogene activity.
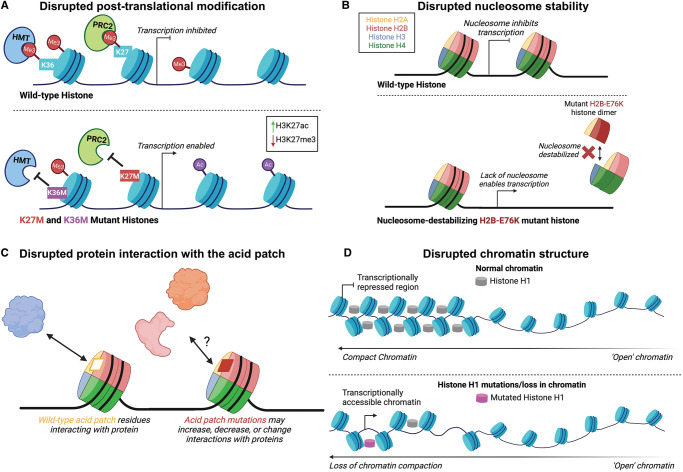
Figure 5. Histone mutations may alter chromatin structure and function by diverse mechanisms.
(A) Histone mutations that affect sites of post-translational modification can block epigenetic writers such as PRC2, resulting in a redistribution of epigenetic marks that regulate gene expression. HMT, Histone methyltransferase; PRC2, Polycomb repressive complex 2. (B) Histone mutations that disrupt intra-nucleosomal histone interactions may destabilize the nucleosome, alleviating nucleosome mediated gene repression. (C) Histone mutations that affect the acid patch may disrupt other proteins from binding to the nucleosome. (D) Mutation of histone H1 can block both association of H1 with chromatin and chromatin compaction, changing chromatin accessibility.
Perspectives
Histone mutations are frequently found in patient samples of all cancer types and associate with worse survival. Yet how these mutations may affect chromatin structure and regulation of gene expression is unknown.
Oncohistone mutations may disrupt nucleosome stability, histone post-translational modifications, or chromatin dynamics to undermine gene regulatory mechanisms, resulting in gene expression programs that favor oncogenesis.
As the functions of oncohistone mutations become better understood, we will gain further insights into how loss of nucleosomal mediated gene repression contributes to tumorigenesis and whether this may be countered therapeutically.
Abbreviations
- ACF
chromatin remodeling and assembly factor
- EZH2
zeste homolog 2
- GCTB
giant cell tumors of bone
- HDE
histone downstream element
- PRC2
Polycomb repressive complex 2
- SIN
SWI/SNF independence
- SNP
single nucleotide polymorphisms
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
R01CA266078 and the UF Health Cancer Center, supported by state appropriations provided in Florida Statute § 381.915 and P30CA247796.
Open Access
Open access for this article was enabled by the participation of University of Florida in an all-inclusive Read & Publish agreement with Portland Press and the Biochemical Society under a transformative agreement with Individual.
Author Contributions
K.N.E.P., J.S. and R.L.B. conceived and wrote the manuscript as well as created the figures, J.D.L. edited the manuscript and contributed to the design and direction of the work.
References
- 1.Cutter, A.R. and Hayes, J.J. (2015) A brief review of nucleosome structure. FEBS Lett. 589, 2914–2922 10.1016/j.febslet.2015.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luger, K., Mader, A.W., Richmond, R.K., Sargent, D.F. and Richmond, T.J. (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- 3.Bednar, J., Garcia-Saez, I., Boopathi, R., Cutter, A.R., Papai, G., Reymer, A.et al. (2017) Structure and dynamics of a 197 bp nucleosome in complex with linker histone H1. Mol. Cell 66, 384–97.e8 10.1016/j.molcel.2017.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh, R., Bassett, E., Chakravarti, A. and Parthun, M.R. (2018) Replication-dependent histone isoforms: a new source of complexity in chromatin structure and function. Nucleic Acids Res. 46, 8665–8678 10.1093/nar/gky768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Susano Pinto, D.M. and Flaus, A. (2019) The human canonical core histone catalogue. bioRxiv 10.1101/720235 [DOI] [Google Scholar]
- 6.Albig, W. and Doenecke, D. (1997) The human histone gene cluster at the D6S105 locus. Hum. Genet. 101, 284–294 10.1007/s004390050630 [DOI] [PubMed] [Google Scholar]
- 7.Flaus, A., Rencurel, C., Ferreira, H., Wiechens, N. and Owen-Hughes, T. (2004) Sin mutations alter inherent nucleosome mobility. EMBO J. 23, 343–353 10.1038/sj.emboj.7600047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rattray, A.M. and Muller, B. (2012) The control of histone gene expression. Biochem. Soc. Trans. 40, 880–885 10.1042/BST20120065 [DOI] [PubMed] [Google Scholar]
- 9.Zheng, L., Roeder, R.G. and Luo, Y. (2003) S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell 114, 255–266 10.1016/s0092-8674(03)00552-x [DOI] [PubMed] [Google Scholar]
- 10.Marzluff, W.F. and Koreski, K.P. (2017) Birth and death of histone mRNAs. Trends Genet. 33, 745–759 10.1016/j.tig.2017.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martire, S. and Banaszynski, L.A. (2020) The roles of histone variants in fine-tuning chromatin organization and function. Nat. Rev. Mol. Cell Biol. 21, 522–541 10.1038/s41580-020-0262-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phillips, E.O.N. and Gunjan, A. (2022) Histone variants: the unsung guardians of the genome. DNA Repair (Amst) 112, 103301 10.1016/j.dnarep.2022.103301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strahl, B.D. and Allis, C.D. (2000) The language of covalent histone modifications. Nature 403, 41–45 10.1038/47412 [DOI] [PubMed] [Google Scholar]
- 14.Akhtar, A. and Becker, P.B. (2000) Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol. Cell 5, 367–375 10.1016/s1097-2765(00)80431-1 [DOI] [PubMed] [Google Scholar]
- 15.Lu, X., Simon, M.D., Chodaparambil, J.V., Hansen, J.C., Shokat, K.M. and Luger, K. (2008) The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure. Nat. Struct. Mol. Biol. 15, 1122–1124 10.1038/nsmb.1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wysocka, J., Swigut, T., Xiao, H., Milne, T.A., Kwon, S.Y., Landry, J.et al. (2006) A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442, 86–90 10.1038/nature04815 [DOI] [PubMed] [Google Scholar]
- 17.Margueron, R., Trojer, P. and Reinberg, D. (2005) The key to development: interpreting the histone code? Curr. Opin. Genet. Dev. 15, 163–176 10.1016/j.gde.2005.01.005 [DOI] [PubMed] [Google Scholar]
- 18.Vettese-Dadey, M., Grant, P.A., Hebbes, T.R., Crane- Robinson, C., Allis, C.D. and Workman, J.L. (1996) Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 15, 2508–2518 10.1002/j.1460-2075.1996.tb00608.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clements, A., Poux, A.N., Lo, W.-S., Pillus, L., Berger, S.L. and Marmorstein, R. (2003) Structural basis for histone and phosphohistone binding by the GCN5 histone acetyltransferase. Mol. Cell 12, 461–473 10.1016/s1097-2765(03)00288-0 [DOI] [PubMed] [Google Scholar]
- 20.Schwartzentruber, J., Korshunov, A., Liu, X.Y., Jones, D.T., Pfaff, E., Jacob, K.et al. (2012) Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231 10.1038/nature10833 [DOI] [PubMed] [Google Scholar]
- 21.Wu, G., Broniscer, A., McEachron, T.A., Lu, C., Paugh, B.S., Becksfort, J.et al. (2012) Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 44, 251–253 10.1038/ng.1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lohr, J.G., Stojanov, P., Lawrence, M.S., Auclair, D., Chapuy, B., Sougnez, C.et al. (2012) Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl Acad. Sci. U.S.A. 109, 3879–3884 10.1073/pnas.1121343109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao, S., Bellone, S., Lopez, S., Thakral, D., Schwab, C., English, D.P.et al. (2016) Mutational landscape of uterine and ovarian carcinosarcomas implicates histone genes in epithelial-mesenchymal transition. Proc. Natl Acad. Sci. U.S.A. 113, 12238–12243 10.1073/pnas.1614120113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arimura, Y., Ikura, M., Fujita, R., Noda, M., Kobayashi, W., Horikoshi, N.et al. (2018) Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome. Nucleic Acids Res. 46, 10007–10018 10.1093/nar/gky661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennett, R.L., Bele, A., Small, E.C., Will, C.M., Nabet, B., Oyer, J.A.et al. (2019) A mutation in histone H2B represents a new class of oncogenic driver. Cancer Discov. 9, 1438–1451 10.1158/2159-8290.CD-19-0393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nacev, B.A., Feng, L., Bagert, J.D., Lemiesz, A.E., Gao, J., Soshnev, A.A.et al. (2019) The expanding landscape of ‘oncohistone’ mutations in human cancers. Nature 567, 473–478 10.1038/s41586-019-1038-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bagert, J.D., Mitchener, M.M., Patriotis, A.L., Dul, B.E., Wojcik, F., Nacev, B.A.et al. (2021) Oncohistone mutations enhance chromatin remodeling and alter cell fates. Nat. Chem. Biol. 17, 403–411 10.1038/s41589-021-00738-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waldmann, T., Izzo, A., Kamieniarz, K., Richter, F., Vogler, C., Sarg, B.et al. (2011) Methylation of H2AR29 is a novel repressive PRMT6 target. Epigenetics Chromatin 4, 11 10.1186/1756-8935-4-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGinty, R.K. and Tan, S. (2021) Principles of nucleosome recognition by chromatin factors and enzymes. Curr. Opin. Struct. Biol. 71, 16–26 10.1016/j.sbi.2021.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dann, G.P., Liszczak, G.P., Bagert, J.D., Muller, M.M., Nguyen, U.T.T., Wojcik, F.et al. (2017) ISWI chromatin remodellers sense nucleosome modifications to determine substrate preference. Nature 548, 607–611 10.1038/nature23671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dao, H.T., Dul, B.E., Dann, G.P., Liszczak, G.P. and Muir, T.W. (2020) A basic motif anchoring ISWI to nucleosome acidic patch regulates nucleosome spacing. Nat. Chem. Biol. 16, 134–142 10.1038/s41589-019-0413-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seath, C.P., Burton, A.J., Sun, X., Lee, G., Kleiner, R.E., MacMillan, D.W.C.et al. (2023) Tracking chromatin state changes using nanoscale photo-proximity labelling. Nature 616, 574–580 10.1038/s41586-023-05914-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang, T.Z.E., Zhu, L., Yang, D., Ding, D., Zhu, X., Wan, Y.C.E.et al. (2021) The elevated transcription of ADAM19 by the oncohistone H2BE76K contributes to oncogenic properties in breast cancer. J. Biol. Chem. 296, 100374 10.1016/j.jbc.2021.100374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wan, Y.C.E., Liu, J., Zhu, L., Kang, T.Z.E., Zhu, X., Lis, J.et al. (2020) The H2BG53D oncohistone directly upregulates ANXA3 transcription and enhances cell migration in pancreatic ductal adenocarcinoma. Signal. Transduct. Target. Ther. 5, 106 10.1038/s41392-020-00219-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wan, Y.C.E., Leung, T.C.S., Ding, D., Sun, X., Liu, J., Zhu, L.et al. (2020) Cancer-associated histone mutation H2BG53D disrupts DNA-histone octamer interaction and promotes oncogenic phenotypes. Signal. Transduct. Target. Ther. 5, 27 10.1038/s41392-020-0131-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bacolla, A., Cooper, D.N. and Vasquez, K.M. (2014) Mechanisms of base substitution mutagenesis in cancer genomes. Genes (Basel) 5, 108–146 10.3390/genes5010108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shah, M.A., Denton, E.L., Arrowsmith, C.H., Lupien, M. and Schapira, M. (2014) A global assessment of cancer genomic alterations in epigenetic mechanisms. Epigenetics Chromatin 7, 29 10.1186/1756-8935-7-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Millan-Zambrano, G., Burton, A., Bannister, A.J. and Schneider, R. (2022) Histone post-translational modifications - cause and consequence of genome function. Nat. Rev. Genet. 23, 563–580 10.1038/s41576-022-00468-7 [DOI] [PubMed] [Google Scholar]
- 39.Lewis, P.W., Muller, M.M., Koletsky, M.S., Cordero, F., Lin, S., Banaszynski, L.A.et al. (2013) Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340, 857–861 10.1126/science.1232245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Funato, K., Major, T., Lewis, P.W., Allis, C.D. and Tabar, V. (2014) Use of human embryonic stem cells to model pediatric gliomas with H3.3K27M histone mutation. Science 346, 1529–1533 10.1126/science.1253799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bender, S., Tang, Y., Lindroth, A.M., Hovestadt, V., Jones, D.T., Kool, M.et al. (2013) Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell 24, 660–672 10.1016/j.ccr.2013.10.006 [DOI] [PubMed] [Google Scholar]
- 42.Chan, K.-M., Fang, D., Gan, H., Hashizume, R., Yu, C., Schroeder, M.et al. (2013) The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 27, 985–990 10.1101/gad.217778.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Venneti, S., Garimella, M.T., Sullivan, L.M., Martinez, D., Huse, J.T., Heguy, A.et al. (2013) Evaluation of histone 3 lysine 27 trimethylation (H3K27me3) and enhancer of Zest 2 (EZH2) in pediatric glial and glioneuronal tumors shows decreased H3K27me3 in H3F3A K27M mutant glioblastomas. Brain Pathol. 23, 558–564 10.1111/bpa.12042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silveira, A.B., Kasper, L.H., Fan, Y., Jin, H., Wu, G., Shaw, T.I.et al. (2019) H3.3 K27M depletion increases differentiation and extends latency of diffuse intrinsic pontine glioma growth in vivo. Acta Neuropathol. 137, 637–655 10.1007/s00401-019-01975-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Justin, N., Zhang, Y., Tarricone, C., Martin, S.R., Chen, S., Underwood, E.et al. (2016) Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat. Commun. 7, 11316 10.1038/ncomms11316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siddaway, R., Canty, L., Pajovic, S., Milos, S., Coyaud, E., Sbergio, S.G.et al. (2022) Oncohistone interactome profiling uncovers contrasting oncogenic mechanisms and identifies potential therapeutic targets in high grade glioma. Acta Neuropathol. 144, 1027–1048 10.1007/s00401-022-02489-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, Y., Yang, X., Gui, B., Xie, G., Zhang, D., Shang, Y.et al. (2011) Corepressor protein CDYL functions as a molecular bridge between polycomb repressor complex 2 and repressive chromatin mark trimethylated histone lysine 27. J. Biol. Chem. 286, 42414–42425 10.1074/jbc.M111.271064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harutyunyan, A.S., Krug, B., Chen, H., Papillon-Cavanagh, S., Zeinieh, M., De Jay, N.et al. (2019) H3k27m induces defective chromatin spread of PRC2-mediated repressive H3K27me2/me3 and is essential for glioma tumorigenesis. Nat. Commun. 10, 1262 10.1038/s41467-019-09140-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jain, S.U., Rashoff, A.Q., Krabbenhoft, S.D., Hoelper, D., Do, T.J., Gibson, T.J.et al. (2020) H3 K27M and EZHIP impede H3K27-methylation spreading by inhibiting allosterically stimulated PRC2. Mol. Cell 80, 726–35.e7 10.1016/j.molcel.2020.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stafford, J.M., Lee, C.H., Voigt, P., Descostes, N., Saldana-Meyer, R., Yu, J.R.et al. (2018) Multiple modes of PRC2 inhibition elicit global chromatin alterations in H3K27M pediatric glioma. Sci. Adv. 4, eaau5935 10.1126/sciadv.aau5935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mohammad, F., Weissmann, S., Leblanc, B., Pandey, D.P., Hojfeldt, J.W., Comet, I.et al. (2017) EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat. Med. 23, 483–492 10.1038/nm.4293 [DOI] [PubMed] [Google Scholar]
- 52.Herz, H.M., Morgan, M., Gao, X., Jackson, J., Rickels, R., Swanson, S.K.et al. (2014) Histone H3 lysine-to-methionine mutants as a paradigm to study chromatin signaling. Science 345, 1065–1070 10.1126/science.1255104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, X., Paucek, R.D., Gooding, A.R., Brown, Z.Z., Ge, E.J., Muir, T.W.et al. (2017) Molecular analysis of PRC2 recruitment to DNA in chromatin and its inhibition by RNA. Nat. Struct. Mol. Biol. 24, 1028–1038 10.1038/nsmb.3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fang, D., Gan, H., Cheng, L., Lee, J.H., Zhou, H., Sarkaria, J.N.et al. (2018) H3.3K27M mutant proteins reprogram epigenome by sequestering the PRC2 complex to poised enhancers. eLife 7, e36696 10.7554/eLife.36696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Piunti, A., Hashizume, R., Morgan, M.A., Bartom, E.T., Horbinski, C.M., Marshall, S.A.et al. (2017) Therapeutic targeting of polycomb and BET bromodomain proteins in diffuse intrinsic pontine gliomas. Nat. Med. 23, 493–500 10.1038/nm.4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sarthy, J.F., Meers, M.P., Janssens, D.H., Henikoff, J.G., Feldman, H., Paddison, P.J.et al. (2020) Histone deposition pathways determine the chromatin landscapes of H3.1 and H3.3 K27M oncohistones. eLife 9, e61090 10.7554/eLife.61090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brien, G.L., Bressan, R.B., Monger, C., Gannon, D., Lagan, E., Doherty, A.M.et al. (2021) Simultaneous disruption of PRC2 and enhancer function underlies histone H3.3-K27M oncogenic activity in human hindbrain neural stem cells. Nat. Genet. 53, 1221–1232 10.1038/s41588-021-00897-w [DOI] [PubMed] [Google Scholar]
- 58.Vire, E., Brenner, C., Deplus, R., Blanchon, L., Fraga, M., Didelot, C.et al. (2006) The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439, 871–874 10.1038/nature04431 [DOI] [PubMed] [Google Scholar]
- 59.Lutsik, P., Baude, A., Mancarella, D., Oz, S., Kuhn, A., Toth, R.et al. (2020) Globally altered epigenetic landscape and delayed osteogenic differentiation in H3.3-G34W-mutant giant cell tumor of bone. Nat. Commun. 11, 5414 10.1038/s41467-020-18955-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lim, J., Park, J.H., Baude, A., Yoo, Y., Lee, Y.K., Schmidt, C.R.et al. (2017) The histone variant H3.3 G34W substitution in giant cell tumor of the bone link chromatin and RNA processing. Sci. Rep. 7, 13459 10.1038/s41598-017-13887-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi, L., Shi, J., Shi, X., Li, W. and Wen, H. (2018) Histone H3.3 G34 mutations alter histone H3K36 and H3K27 methylation in cis. J. Mol. Biol. 430, 1562–1565 10.1016/j.jmb.2018.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, Y., Shan, C.M., Wang, J., Bao, K., Tong, L. and Jia, S. (2017) Molecular basis for the role of oncogenic histone mutations in modulating H3K36 methylation. Sci. Rep. 7, 43906 10.1038/srep43906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang, S., Zheng, X., Lu, C., Li, G.M., Allis, C.D. and Li, H. (2016) Molecular basis for oncohistone H3 recognition by SETD2 methyltransferase. Genes Dev. 30, 1611–1616 10.1101/gad.284323.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Khazaei, S., De Jay, N., Deshmukh, S., Hendrikse, L.D., Jawhar, W., Chen, C.C.L.et al. (2020) H3.3 G34W promotes growth and impedes differentiation of osteoblast-like mesenchymal progenitors in giant cell tumor of bone. Cancer Discov. 10, 1968–1987 10.1158/2159-8290.CD-20-0461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khazaei, S., Chen, C.C.L., Andrade, A.F., Kabir, N., Azarafshar, P., Morcos, S.M.et al. (2023) Single substitution in H3.3G34 alters DNMT3A recruitment to cause progressive neurodegeneration. Cell 186, 1162–78.e20 10.1016/j.cell.2023.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Behjati, S., Tarpey, P.S., Presneau, N., Scheipl, S., Pillay, N., Van Loo, P.et al. (2013) Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat. Genet. 45, 1479–1482 10.1038/ng.2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lu, C., Jain, S.U., Hoelper, D., Bechet, D., Molden, R.C., Ran, L.et al. (2016) Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science 352, 844–849 10.1126/science.aac7272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fang, D., Gan, H., Lee, J.H., Han, J., Wang, Z., Riester, S.M.et al. (2016) The histone H3.3K36M mutation reprograms the epigenome of chondroblastomas. Science 352, 1344–1348 10.1126/science.aae0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abe, S., Nagatomo, H., Sasaki, H. and Ishiuchi, T. (2021) A histone H3.3K36M mutation in mice causes an imbalance of histone modifications and defects in chondrocyte differentiation. Epigenetics 16, 1123–1134 10.1080/15592294.2020.1841873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sakamoto, M., Noguchi, S., Kawashima, S., Okada, Y., Enomoto, T., Seki, M.et al. (2009) Global analysis of mutual interaction surfaces of nucleosomes with comprehensive point mutants. Genes Cells 14, 1271–1330 10.1111/j.1365-2443.2009.01350.x [DOI] [PubMed] [Google Scholar]
- 71.Kurumizaka, H. and Wolffe, A.P. (1997) Sin mutations of histone H3: influence on nucleosome core structure and function. Mol. Cell. Biol. 17, 6953–6969 10.1128/MCB.17.12.6953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Long, M., Sun, X., Shi, W., Yanru, A., Leung, S.T.C., Ding, D.et al. (2019) A novel histone H4 variant H4G regulates rDNA transcription in breast cancer. Nucleic Acids Res. 47, 8399–8409 10.1093/nar/gkz547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hyland, E.M., Cosgrove, M.S., Molina, H., Wang, D., Pandey, A., Cottee, R.J.et al. (2005) Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol. Cell. Biol. 25, 10060–10070 10.1128/MCB.25.22.10060-10070.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Muthurajan, U.M., Bao, Y., Forsberg, L.J., Edayathumangalam, R.S., Dyer, P.N., White, C.L.et al. (2004) Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions. EMBO J. 23, 260–271 10.1038/sj.emboj.7600046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hsieh, F.K., Fisher, M., Ujvari, A., Studitsky, V.M. and Luse, D.S. (2010) Histone Sin mutations promote nucleosome traversal and histone displacement by RNA polymerase II. EMBO Rep. 11, 705–710 10.1038/embor.2010.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nag, R., Gong, F., Fahy, D. and Smerdon, M.J. (2008) A single amino acid change in histone H4 enhances UV survival and DNA repair in yeast. Nucleic Acids Res. 36, 3857–3866 10.1093/nar/gkn311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Horn, P.J., Crowley, K.A., Carruthers, L.M., Hansen, J.C. and Peterson, C.L. (2002) The SIN domain of the histone octamer is essential for intramolecular folding of nucleosomal arrays. Nat. Struct. Biol. 9, 167–171 10.1038/nsb762 [DOI] [PubMed] [Google Scholar]
- 78.Nakabayashi, Y., Harata, M. and Seki, M. (2020) An improved functional analysis of linker-mediated complex (iFALC) strategy. Biochem. Biophys. Res. Commun. 526, 1164–1169 10.1016/j.bbrc.2020.04.039 [DOI] [PubMed] [Google Scholar]
- 79.Soshnev, A.A., Allis, C.D., Cesarman, E. and Melnick A, M. (2021) Histone H1 mutations in lymphoma: a link(er) between chromatin organization, developmental reprogramming, and cancer. Cancer Res. 81, 6061–6070 10.1158/0008-5472.CAN-21-2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Okosun, J., Bodor, C., Wang, J., Araf, S., Yang, C.Y., Pan, C.et al. (2014) Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat. Genet. 46, 176–181 10.1038/ng.2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yusufova, N., Kloetgen, A., Teater, M., Osunsade, A., Camarillo, J.M., Chin, C.R.et al. (2021) Histone H1 loss drives lymphoma by disrupting 3D chromatin architecture. Nature 589, 299–305 10.1038/s41586-020-3017-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Morin, R.D., Mendez-Lago, M., Mungall, A.J., Goya, R., Mungall, K.L., Corbett, R.D.et al. (2011) Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476, 298–303 10.1038/nature10351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reddy, A., Zhang, J., Davis, N.S., Moffitt, A.B., Love, C.L., Waldrop, A.et al. (2017) Genetic and functional drivers of diffuse large B cell lymphoma. Cell 171, 481–94.e15 10.1016/j.cell.2017.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chapuy, B., Stewart, C., Dunford, A.J., Kim, J., Kamburov, A., Redd, R.A.et al. (2018) Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat. Med. 24, 679–690 10.1038/s41591-018-0016-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Krysiak, K., Gomez, F., White, B.S., Matlock, M., Miller, C.A., Trani, L.et al. (2017) Recurrent somatic mutations affecting B-cell receptor signaling pathway genes in follicular lymphoma. Blood 129, 473–483 10.1182/blood-2016-07-729954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Willcockson, M.A., Healton, S.E., Weiss, C.N., Bartholdy, B.A., Botbol, Y., Mishra, L.N.et al. (2021) H1 histones control the epigenetic landscape by local chromatin compaction. Nature 589, 293–298 10.1038/s41586-020-3032-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Torres, C.M., Biran, A., Burney, M.J., Patel, H., Henser-Brownhill, T., Cohen, A.S.et al. (2016) The linker histone H1.0 generates epigenetic and functional intratumor heterogeneity. Science 353, aaf1644 10.1126/science.aaf1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang, Y., Cooke, M., Panjwani, S., Cao, K., Krauth, B., Ho, P.Y.et al. (2012) Histone h1 depletion impairs embryonic stem cell differentiation. PLoS Genet. 8, e1002691 10.1371/journal.pgen.1002691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kawashima, S., Nakabayashi, Y., Matsubara, K., Sano, N., Enomoto, T., Tanaka, K.et al. (2011) Global analysis of core histones reveals nucleosomal surfaces required for chromosome bi-orientation. EMBO J. 30, 3353–3367 10.1038/emboj.2011.241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Castel, D., Philippe, C., Kergrohen, T., Sill, M., Merlevede, J., Barret, E.et al. (2018) Transcriptomic and epigenetic profiling of ‘diffuse midline gliomas, H3 K27M-mutant’ discriminate two subgroups based on the type of histone H3 mutated and not supratentorial or infratentorial location. Acta Neuropathol. Commun. 6, 117 10.1186/s40478-018-0614-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sturm, D., Witt, H., Hovestadt, V., Khuong-Quang, D.A., Jones, D.T., Konermann, C.et al. (2012) Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22, 425–437 10.1016/j.ccr.2012.08.024 [DOI] [PubMed] [Google Scholar]
- 92.Kruger, W., Peterson, C.L., Sil, A., Coburn, C., Arents, G., Moudrianakis, E.N., et al. (1995) Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 9, 2770–2779 10.1101/gad.9.22.2770 [DOI] [PubMed] [Google Scholar]