Abstract
In the current study, we have shown that USP51 promotes colorectal cancer stemness and chemoresistance, and high expression of USP51 predicts survival disadvantage in colorectal cancer patients. Mechanically, USP51 directly binds to Elongin C (ELOC) and forms a larger functional complex with VHL E3 ligase (USP51/VHL/CUL2/ELOB/ELOC/RBX1) to regulate the ubiquitin-dependent proteasomal degradation of HIF1A. USP51 efficiently deubiquitinates HIF1A and activates hypoxia-induced gene transcription. Conversely, the activation of HIF1A under hypoxia transcriptionally upregulates the expression of USP51. Thus, USP51 and HIF1A form a positive feedback loop. Further, we found that the SUMOylation of ELOC at K32 inhibits its binding to USP51. SUMO-specific protease 1 (SENP1) mediates the deSUMOylation of ELOC, promoting the binding of USP51 to ELOC and facilitating the deubiquitination and stabilization of HIF1A by USP51. Importantly, USP51 plays a crucial role in promoting the HIF1A and SENP1-dependent proliferation, migration, stemness, and chemoresistance under hypoxia in colorectal cancer. Together, our data revealed that USP51 is an oncogene stabilizing the pro-survival protein HIF1A, offering a potential therapeutic target for colorectal cancer.
Subject terms: Sumoylation, Oncogenes, Deubiquitylating enzymes
Introduction
Despite recent advances and new treatments, colorectal cancer (CRC) remains a major cause of morbidity and mortality in humans [1]. A common cause of treatment failure is drug resistance. Therefore, there is still an urgent need to study the mechanisms of drug resistance and identify new therapeutic targets in CRC.
Ubiquitin-specific protease 51 (USP51) is known to bind to H2A-H2B to deubiquitinate H2AK13,15ub and regulate DNA damage response [2]. Another recognized substrate of USP51 is ZEB1 [3], a transcription factor that induces epithelial-mesenchymal transition, tumor metastasis, DNA damage response, and therapy resistance [4–6]. Upregulation of USP51 correlates with poor survival in breast cancer patients [3]. However, the role USP51 played in CRC needs to be better characterized.
In this study, we have presented evidence indicating that USP51 binds to Elongin C (ELOC), a subunit of the VHL E3 ubiquitin ligase complex. This binding facilitates the deubiquitination and stabilization of HIF1A by USP51. And interestingly, USP51 was identified as a direct target gene of HIF1A, establishing a positive feedback loop between HIF1A and USP51.
SUMOylation is a reversible post-translational modification and has a crucial role in regulating essential cellular processes, including protein stability, subcellular localization, and protein-protein interactions [7, 8]. In mammalian cells, SUMOylation is mediated by only one E2 enzyme, UBC9, which can conjugate SUMO to the target protein. DeSUMOylation can be mediated by a family of SUMO-specific proteases (SENPs). SENP1 is a deSUMOylating protease that deconjugates a large number of SUMOylated proteins [9] and is deregulated in many types of cancers [10]. In addition to identifying the HIF1A-USP51 positive feedback loop, we also demonstrated that ELOC was post-translational modified by SUMO1. The deSUMOylation of ELOC mediated by SENP1 facilitated the interaction between ELOC and USP51, consequently enhancing the process of USP51-mediated deubiquitination and stabilization of HIF1A.
Results
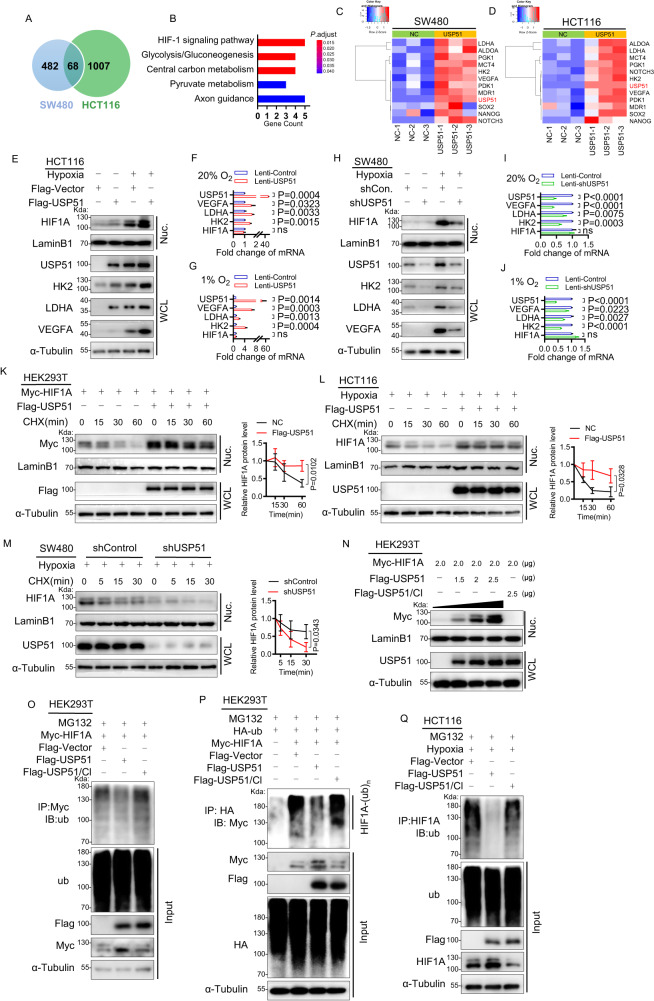
USP51 can effectively deubiquitinate and stabilize HIF1A
To investigate the role USP51 played in colorectal cancer, we first tested the protein level of USP51 in CRC cell lines and found that USP51 was detectable in SW480, DLD-1, and LoVo but not in HCT116 (Supplementary Fig. 1A). We then established SW480 and HCT116 cell lines with overexpression of Flag-USP51. RNA-sequencing showed that, with the overexpression USP51, 68 genes were significantly upregulated in both SW480 and HCT116 (Fig. 1A, Supplementary data 1). KEGG analysis revealed that the HIF-1 signaling pathway was the critical pathway activated by the overexpression of USP51 (Fig. 1B). Downstream target genes of HIF1 such as HK2, LDHA, PDK1, PGK1, MCT4, ALDOA, NOTCH3, MDR1, SOX2, NANOG, and VEGFA were significantly upregulated (Fig. 1C, D). Under normoxia (20% O2), we confirmed the upregulation of HIF1A and two of its downstream genes (HK2 and LDHA) at the protein level in HCT116 cells forced expression of USP51 (Fig. 1E). Forced expression of USP51 in HCT116 cells increased the mRNA of HK2, LDHA, and VEGFA but did not influence the mRNA level of HIF1A (Fig. 1F). Similar results were observed in SW480 cells with USP51 overexpression (Supplementary Fig. 1B, C). Conversely, the knockdown of USP51 in SW480 cells resulted in a notable reduction in the protein level of HK2 and LDHA (Fig. 1H), as well as the mRNA levels of HK2, LDHA and VEGFA (Fig. 1I), while the mRNA level of HIF1A remained unaffected. Because hypoxia is commonly found in malignant tumors, thus, we conducted cell experiments under hypoxic conditions to simulate the in vivo environment. Under hypoxia, overexpressing USP51 in HCT116 cells significantly increased the protein levels of HIF1A, HK2, LDHA, and VEGFA (Fig. 1E), as well as the mRNA levels of HK2, LDHA, and VEGFA (Fig. 1G), while the mRNA level of HIF1A remained unaffected. Similar results were observed in SW480 cells with USP51 overexpression under hypoxia (Supplementary Fig. 1B, D). The knockdown of USP51 in SW480 cells decreased the protein levels of HIF1A, HK2, LDHA, and VEGFA (Fig. 1H), as well as the mRNA levels of HK2, LDHA, and VEGFA (Fig. 1J), while the mRNA level of HIF1A remained unaffected.
Fig. 1. USP51 can effectively prevent HIF1A protein degradation by deubiquitination.
A USP51 was overexpressed in SW480 and HCT116 cells, and 68 genes were upregulated in both SW480-USP51 and HCT116-USP51 cells. B KEGG analysis of the 68 genes upregulated. The heatmaps of USP51 and downstream target genes of HIF1 upregulated by USP51 overexpression in SW480 (C) and HCT116 (D). E Immunoblotting assay analyzed the protein level of USP51, HIF1A, HK2, LDHA, and VEGFA upon USP51 overexpression in HCT116 cells under normoxia or hypoxia. The relative mRNA level of USP51, HIF1A, HK2, LDHA, and VEGFA upon USP51 overexpression in HCT116 cells under normoxia (F) or hypoxia (G). H The protein level of USP51, HIF1A, HK2, LDHA, and VEGFA upon USP51 knockdown in SW480 cells under normoxia or hypoxia. The relative mRNA level of USP51, HIF1A, HK2, LDHA, and VEGFA upon USP51 knockdown in SW480 cells under normoxia (I) or hypoxia (J). K Exogenous USP51 efficiently stables exogenous HIF1A protein in HEK293T cells. L Exogenous USP51 efficiently stables endogenous HIF1A protein in HCT116 cells. M USP51 knockdown accelerated endogenous HIF1A protein degradation in SW480 cells. N Plasmids encoding Flag-USP51 or Flag-USP51/CI were transiently transfected into HEK293T cells stably expressing Myc-HIF1A for 48 h. USP51, but not USP51/CI, prevents the degradation of the HIF1A protein. O Co-expression of HIF1A and USP51, but not USP51/CI, led to a significant reduction in HIF1A ubiquitination. P HEK293T cells co-transfected HA-ub, Myc-HIF1A with Flag-USP51 or Flag-USP51/CI, followed by anti-HA immunoprecipitates and immunoblot analysis with anti-Myc antibody. Q Exogenous USP51, not USP51/CI, reduced endogenous HIF1A ubiquitination. WCL whole cell lysate, Nuc nuclear fraction. Data are expressed as mean ± SD, n = 3 biological replicates. ns not significant.
VEGFA has been demonstrated to stimulate angiogenesis in CRC [11–13], we then investigated whether USP51 played a role in regulating tumor angiogenesis in vivo. Tumor xenografts were obtained upon the subcutaneous injection of SW480 cells, the assessment of angiogenesis was conducted through CD31 staining. As expected, the introduction of shUSP51 led to a notable reduction in microvessel density within the tumors (Supplementary Fig. 1E), paralleling the effect observed with shHIF1A. This result aligns with the in vitro findings, where shUSP51 was observed to cause a reduction in hypoxia-induced VEGFA.
We then investigate the effects of USP51 on preventing HIF1A degradation by cycloheximide (CHX) chase experiment. Myc-HIF1A was stably expressed in HEK293T cells (HEK293T-HIF1A). HEK293T-HIF1A cells were transfected Flag-USP51 or its control vector and then treated with CHX for the indicated times to inhibit novel protein synthesis. Immunoblot analysis revealed that overexpression of USP51 rescued the HIF1A protein level and resulted in a steady level of HIF1A protein (Fig. 1K). We further confirmed the protective effect of USP51 on endogenous HIF1A in HCT116 cells (Fig. 1L). Conversely, the knockdown of USP51 in SW480 cells accelerated HIF1A degradation (Fig. 1M).
To investigate whether the catalytic activity of USP51 is necessary to stabilize HIF1A, HEK293T cells were co-transfected Myc-HIF1A with either Flag-USP51 or catalytically inactive Flag-USP51/CI (C372S, H665R) [2]. We found that overexpression of USP51, but not USP51/CI, prevents the degradation of the HIF1A protein (Fig. 1N). The failure of USP51/CI in preventing HIF1A was further confirmed in HCT116 cells (Supplementary Fig. 1F).
Next, we confirmed that USP51 deubiquitinated HIF1A. HEK293T cells were co-transfected Myc-HIF1A with either Flag-USP51 or Flag-USP51/CI, and anti-Myc immunoprecipitates were probed for the level of ubiquitination using the antibody specifically against ubiquitin. Co-expression of HIF1A and USP51, but not USP51/CI, led to a significant reduction in HIF1A ubiquitination (Fig. 1O). We further confirm the observations in HEK293T cells co-transfected HA-ub, Myc-HIF1A with either Flag-USP51 or Flag-USP51/CI, followed by anti-HA immunoprecipitates and immunoblot analysis with anti-Myc antibody (Fig. 1P). To examine the effect of USP51 on the ubiquitination level of endogenous HIF1A under hypoxia, we transfected either Flag-USP51 or Flag-USP51/CI in HCT116 cells and anti-HIF1A immunoprecipitates were prepared and subjected to immunoblot analysis using a specific antibody against ubiquitin. Overexpression of USP51, not USP51/CI, reduced the HIF1A ubiquitination markedly (Fig. 1Q). And the effect of USP51 on the endogenous HIF1A ubiquitination level was further confirmed in SW480 cells (Supplementary Fig. 1G). Conversely, the knockdown of USP51 in DLD-1 cells led to a marked increase in endogenous HIF1A ubiquitination (Supplementary Fig. 1H).
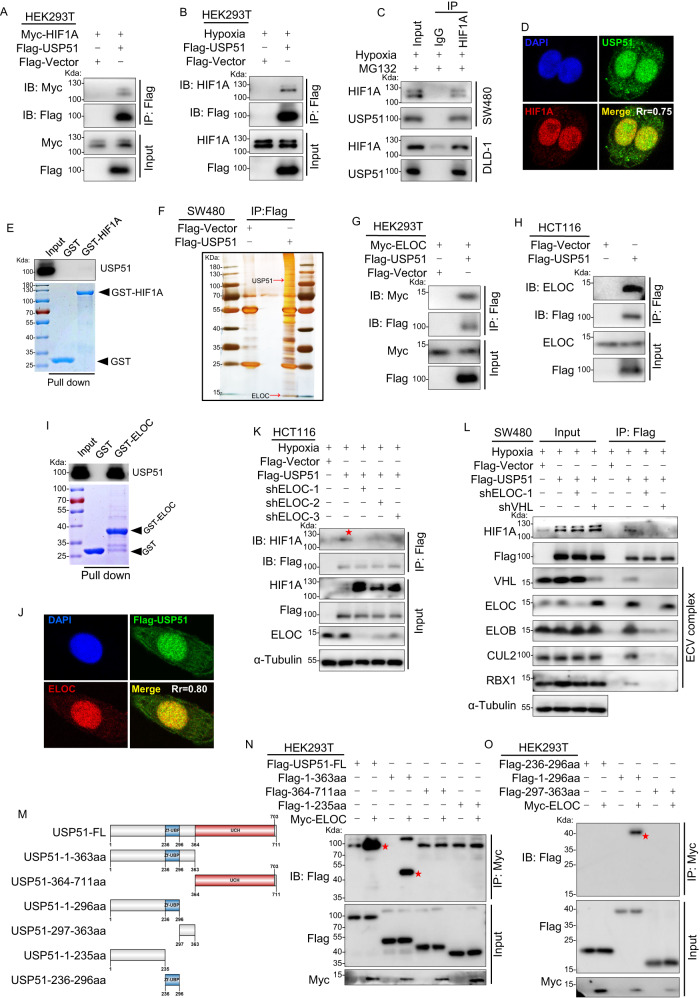
USP51 interacts with HIF1A via directly binding to ELOC
To investigate the relationship between USP51 and HIF1A, we first examined the interaction between USP51 and HIF1A by co-immunoprecipitation. HEK293T cells were co-transfected with Myc-HIF1A and Flag-USP51, and anti-Flag immunoprecipitates were probed using the anti-Myc antibody. Exogenous Myc-HIF1A was detected in the anti-Flag immunoprecipitates (Fig. 2A). Exogenous USP51 could also co-immunoprecipitate endogenous HIF1A in HEK293T cells under hypoxia (Fig. 2B). Next, we confirmed in DLD-1 and SW480 cells that endogenous USP51 could be co-immunoprecipitated with endogenous HIF1A under hypoxia (Fig. 2C). Immunofluorescence analysis also showed the colocalization of endogenous HIF1A and endogenous USP51 in SW480 cells under hypoxia (Fig. 2D). However, a glutathione S-transferase (GST) pull-down assay revealed that USP51 could not bind to HIF1A directly (Fig. 2E).
Fig. 2. USP51 interacts with HIF1A via direct binding to ELOC.
A Exogenous Myc-HIF1A was co-immunoprecipitated with exogenous Flag-USP51 in HEK293T cells. B Exogenous USP51 co-immunoprecipitated endogenous HIF1A in HEK293T cells. C Endogenous USP51 could be co-immunoprecipitated with endogenous HIF1A in SW480 and DLD-1 cells. D Immunofluorescence analysis of the colocalization of endogenous HIF1A and endogenous USP51 in SW480 cells under hypoxia. E USP51 did not directly bind to HIF1A in vitro. GST and GST-HIF1A protein were analyzed by Coomassie Brilliant Blue staining. GST or GST-HIF1A was incubated with purified His-USP51. The bound proteins were pulled down by glutathione-agarose and analyzed by immunoblotting with anti-USP51 antibodies. F Flag-USP51 or Flag-Vector were stably expressed in SW480 cells. Anti-Flag immunoprecipitants were analyzed by silver staining. G Exogenous Myc-ELOC was co-immunoprecipitated with exogenous Flag-USP51 in HEK293T cells. H Exogenous USP51 co-immunoprecipitated endogenous ELOC in HCT116 cells. I USP51 directly binds to ELOC in vitro. GST and GST-ELOC protein were analyzed by Coomassie Brilliant Blue staining. GST or GST-ELOC was incubated with purified His-USP51. The bound proteins were pulled down by glutathione-agarose and analyzed by immunoblotting with anti-USP51 antibodies. J Immunofluorescence showed the colocalization of endogenous ELOC and exogenous Flag-USP51 in SW480 cells. K Knockdown of ELOC abolished co-immunoprecipitation between endogenous HIF1A and exogenous USP51. L Exogenous USP51 co-immunoprecipitated the ECV complex. ELOC knockdown prevented co-immunoprecipitation of HIF1A and ECV subunits. VHL knockdown maintained ELOC co-immunoprecipitation with Flag-USP51. M The schematic of the six Flag-tagged truncated forms of USP51. N, O USP51 full-length and truncations plasmids were co-transfected with Myc-ELOC plasmid into HEK293T cells and Co-IP assays were performed using anti-Myc antibody.
To clarify how USP51 interacts with HIF1A, we performed affinity enrichment mass spectrometry in SW480 cells stably overexpressing Flag-USP51. We found that Flag-USP51 effectively co-immunoprecipitated ELOC (Fig. 2F, Supplementary Fig. 2A, B), a member of the ECV complex, an E3 ligase responsible for ubiquitinating HIF1A protein. To confirm the interaction between USP51 and ELOC, we co-transfected Myc-ELOC and Flag-USP51 in HEK293T cells, and immunoprecipitation analysis showed exogenous ELOC could be co-immunoprecipitated by exogenous USP51 (Fig. 2G). Endogenous ELOC could also be co-immunoprecipitated by exogenous USP51 in HCT116 cells (Fig. 2H). Importantly, a GST pull-down assay revealed that ELOC could directly interact with USP51 (Fig. 2I). Immunofluorescence analysis also showed the colocalization of endogenous ELOC and exogenous USP51 in SW480 cells (Fig. 2J).
Therefore, we hypothesized that USP51 interacts with HIF1A through its direct interaction with ELOC, and USP51 thus binds to the E3 ligase complex (VHL/CUL2/ELOB/ELOC/RBX1, ECV complex). This E3 ligase complex is responsible for tagging HIF1A. To verify this hypothesis, we knocked down ELOC in HCT116 cells transfected with Flag-USP51. Knockdown of ELOC abolished co-immunoprecipitation of HIF1A with Flag-USP51(Fig. 2K). In SW480 cells, the ECV complex could efficiently co-immunoprecipitated by exogenous USP51 (Fig. 2L, lane 6). However, the knockdown of ELOC led to neither HIF1A nor anyone of the subunits of the ECV complex being co-immunoprecipitated by exogenous USP51 efficiently (Fig. 2L, lane 7). Knockdown of VHL, the HIF1A recognition component, also abolished the co-immunoprecipitation of HIF1A with Flag-USP51 but maintained the co-immunoprecipitation of ELOC with Flag-USP51 (Fig. 2L, lane 8).
To determine the specific domains of USP51 that mediate its interaction with ELOC, we generated six Flag-tagged truncated forms of USP51 (Fig. 2M). HEK293T cells were co-transfected with Myc-ELOC and one of the six Flag-tagged truncations of USP51. Anti-Myc immunoprecipitates were probed using the anti-Flag antibody. The results revealed that the N-terminal region(1-296aa) including the zinc finger domain(234-296aa) mediated the interaction between USP51 and ELOC (Fig. 2N, O).
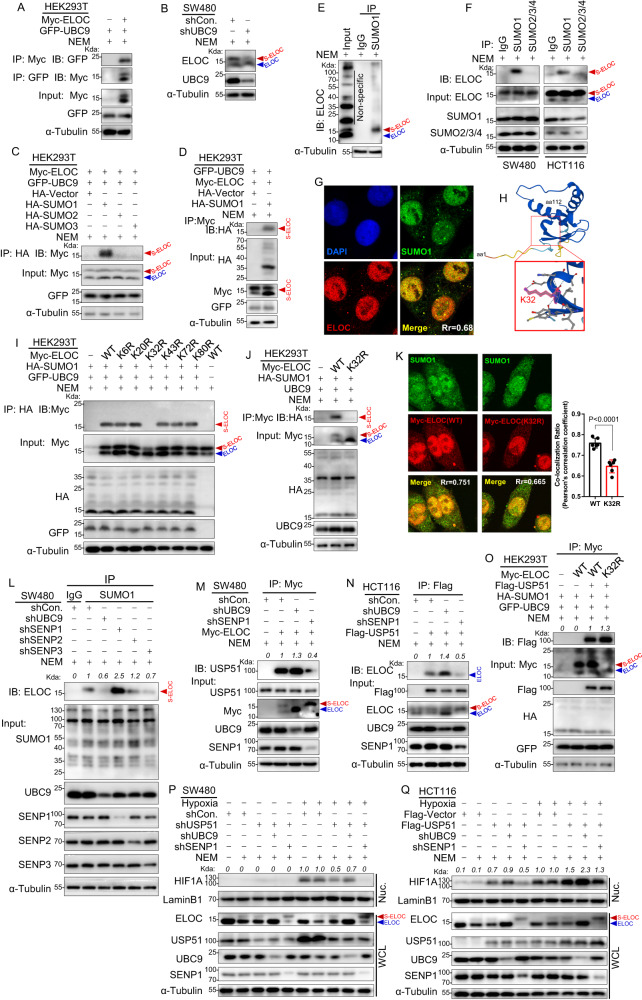
SENP1-mediated ELOC deSUMOylation promotes USP51 binding to ELOC
To explore the potential SUMO modification of ELOC, we generated whole cell lysates from SW480, DLD-1, and HCT116 cells under two conditions: with or without the inclusion of 20 mM N-ethylmaleimide (NEM), an inhibitor of SUMO-specific proteases (SENPs) [14]. Notably, upon adding NEM, immunoblot analysis revealed the emergence of a distinct, persistent band ranging from 15kD to 25kD (Supplementary Fig. 3A), suggestive of a SUMO-ELOC conjugate. This intriguing finding prompted us to investigate whether ELOC indeed serves as a target for protein SUMOylation. We first investigate whether ELOC interacts with UBC9, the sole E2 SUMO-conjugating enzyme in mammalian cells. HEK293T cells were co-transfected with GFP-UBC9 and Myc-ELOC, and the immunoprecipitation and immunoblot analysis confirmed the interaction between ELOC and UBC9 (Fig. 3A). Knockdown of UBC9 in SW480 cells weakened the SUMO-ELOC band (Fig. 3B). Next, we investigated which SUMO protein could be covalently conjugated to ELOC. We individually co-expressed either HA-SUMO1, HA-SUMO2, or HA-SUMO3 with Myc-ELOC in HEK293T cells. The Myc-ELOC band was detected in the immunoprecipitates of anti-HA-SUMO1 (Fig. 3C), not in the immunoprecipitates of anti-HA-SUMO2 or SUMO3, showing that ELOC is conjugated by SUMO1. This was confirmed in HEK293T cells transfected with Myc-ELOC and HA-SUMO1 by probing the anti-Myc immunoprecipitates using the anti-HA antibody (Fig. 3D). Additionally, the endogenous SUMO-ELOC protein could be immunoprecipitated by an anti-SUMO1 antibody, not anti-SUMO2/3/4 (Fig. 3E, F). Colocalization of endogenous SUMO1 and ELOC in DLD-1 cells also supported that ELOC was SUMOylated by SUMO1 (Fig. 3G).
Fig. 3. SENP1-mediated ELOC deSUMOylation promotes USP51 binding to ELOC.
A HEK293T cells were co-transfected with GFP-UBC9 and Myc-ELOC and Co-IP assays were performed using anti-Myc or anti-GFP antibodies. B Knockdown of UBC9 in SW480 cells resulted in the weakening of the suspected SUMO-ELOC band. C Myc-ELOC and GFP-UBC9 were co-transfected in HEK293T cells with HA-SUMO1, HA-SUMO2, or HA-SUMO3. SUMO-ELOC band was detected in the immunoprecipitates of anti-SUMO1. D HEK293T cells were co-transfected with Myc-ELOC, GFP-UBC9 and HA-SUMO1. The Anti-Myc immunoprecipitates were probed using the anti-HA antibody. E The endogenous SUMO-ELOC protein could be immunoprecipitated by an anti-SUMO1 antibody in SW480 cells. F The endogenous SUMO-ELOC protein could be immunoprecipitated by an anti-SUMO1 antibody, not anti-SUMO2/3/4 antibodies, in SW480 and HCT116 cells. G Immunofluorescence analysis of the colocalization of endogenous SUMO1 and endogenous ELOC in DLD-1 cells. H The schematic of K32 lysine residue in ELOC protein. I HEK293T cells were co-transfected with HA-SUMO1 and one of the six mutant forms of Myc-ELOC (K6R, K20R, K32R, K43R, K72R, and K80R). Cell lysates were precipitated with an anti-HA antibody and blotted by an anti-Myc antibody. J HEK293T cells were co-transfected with HA-SUMO1, GFP-UBC9 and Myc-ELOC (WT or K32R). Cell lysates were precipitated with anti-Myc antibody and blotted by an anti-HA antibody. K SW480 cells were co-transfected with Myc-ELOC WT or Myc-ELOC K32R. Immunofluorescence analysis of the colocalization of endogenous SUMO1 and exogenous ELOC. L Either UBC9, SENP1, SENP2, or SENP3 were stably knockdown by shRNA in SW480 cells. Cell lysates were prepared for precipitation with an anti-SUMO1 antibody, and SUMO-ELOC was detected by an anti-ELOC antibody. M Either UBC9 or SENP1 was stably knocked down by shRNA in SW480 cells stably expressed Myc-ELOC. Cell lysates were prepared for precipitation with anti-Myc antibody, and blotted by an anti-USP51 antibody. N Either UBC9 or SENP1 was stably knocked down by shRNA in HCT116 cells stably expressed Flag-USP51. Cell lysates were prepared for precipitation with an anti-Flag antibody, and blotted by an anti-ELOC antibody. O HEK293T cells were co-transfected with GFP-UBC9, HA-SUMO1, Flag-USP51, and Myc-ELOC (WT or K32R). Cell lysates were prepared for precipitation with anti-Myc antibody, and blotted by an anti-Flag antibody. P USP51, UBC9, and SENP1 were stably knockdown by shRNA in SW480 cells under normoxia or hypoxia. Cell lysates were blotted by the indicated antibodies. Q UBC9 and SENP1 were stably knockdown by shRNA in HCT116 cells that stably expressed Flag-USP51 under normoxia or hypoxia. Cell lysates were blotted by the indicated antibodies. Data are expressed as mean ± SD, n = 5 biological replicates. WCL whole cell lysate, Nuc nuclear fraction.
To further identify the SUMO modification sites of ELOC, we used online SUMOylation prediction tools JASSA [15] and found only one lysine residue, K32, with a high predicted score (Fig. 3H, Supplementary Fig. 3B). To confirm the prediction, HEK293T cells were co-transfected with HA-SUMO1 and either of the six mutant forms of Myc-ELOC (K6R, K20R, K32R, K43R, K72R, and K80R). Immunoblot analysis of anti-HA immunoprecipitates using anti-Myc antibody revealed that K32R mutation abolished the SUMO1 modification of ELOC (Fig. 3I). In HEK293T cells, we co-transfected HA-SUMO1 with either Myc-ELOC WT or Myc-ELOC K32R. We probed anti-Myc immunoprecipitates using an anti-HA antibody and found the SUMO-ELOC band in Myc-ELOC WT, but not in Myc-ELOC K32R (Fig. 3J). Immunofluorescence analysis also showed that the colocalization of endogenous SUMO1 and exogenous Myc-ELOC in SW480 cells was disturbed by K32R mutation (Fig. 3K).
Next, we investigated which SENPs mediated the deSUMOlytion of ELOC. In SW480 cells, we observed that the SUMO-ELOC band decreased following the knockdown of UBC9 but increased following the knockdown of SENP1, not SENP2 or SENP3 (Fig. 3L). This observation was further confirmed in HCT116 cells (Supplementary Fig. 3C). In HEK293T cells transfected with HA-SUMO1, GFP-UBC9, and Myc-ELOC, overexpression of SENP1, not SENP2 or SENP3, decreased the SUMO-ELOC band (Supplementary Fig. 3D–F). These results indicate that SENP1 mediated the deSUMOlytion of ELOC.
Next, we investigated the functional regulation linked to the SUMOylation status of ELOC. We initially examined whether the SUMOylation status of ELOC influences its capacity to form a VHL complex alongside other ECV complex subunits. Stable expression of Myc-ELOC WT or Myc-ELOC K32R, a mutant that prevents SUMO1 modification of ELOC, was accomplished in SW480 cells. Notably, both the WT and mutant forms of exogenous ELOC effectively co-immunoprecipitated with the ECV complex (Supplementary Fig. 3G). The outcome suggests that the SUMOylation state of ELOC does not play a role in determining its ability to form a VHL complex. Following that, we hypothesized that the SUMOylation state of ELOC disrupts its interaction with USP51. In SW480 cells, when UBC9 was knocked down, there was an augmentation in the interaction between USP51 and ELOC (Fig. 3M). Conversely, the interaction diminished upon the knockdown of SENP1 (Fig. 3M). We further confirmed the result in HCT116 cells transfected with Flag-USP51 (Fig. 3N). In HEK293T cells transfected with GFP-UBC9 and HA-SUMO1, we expressed Myc-ELOC WT or Myc-ELOC K32R. The K32R mutation in ELOC led to an elevation in the co-immunoprecipitation of Flag-USP51 protein with Myc-ELOC (Fig. 3O). In SW480 cells, UBC9 knockdown attenuated the shUSP51-induced degradation of HIF1A, while SENP1 knockdown enhanced the shUSP51-induced degradation of HIF1A under hypoxia (Fig. 3P), all the while leaving the mRNA level of HIF1A unaffected (Supplementary Fig. 3H). In HCT116 cells, UBC9 knockdown intensified the USP51-induced HIF1A protein accumulation, while SENP1 knockdown mitigated the USP51-induced HIF1A protein accumulation (Fig. 3Q). This effect remained consistent under both normoxia and hypoxia, with the mRNA level of HIF1A unaffected (Supplementary Fig. 3I).
These findings imply that the SUMOylation status of ELOC modulates its interaction with USP51, consequently regulating the deubiquitylation and stabilization of HIF1A. SENP1-mediated ELOC deSUMOylation promotes the binding of USP51 to ELOC and the stabilization of HIF1A. DeSUMOylation of ELOC through SENP1 appears to enhance the affinity between ELOC and USP51, potentially leading to the stabilization of HIF1A.
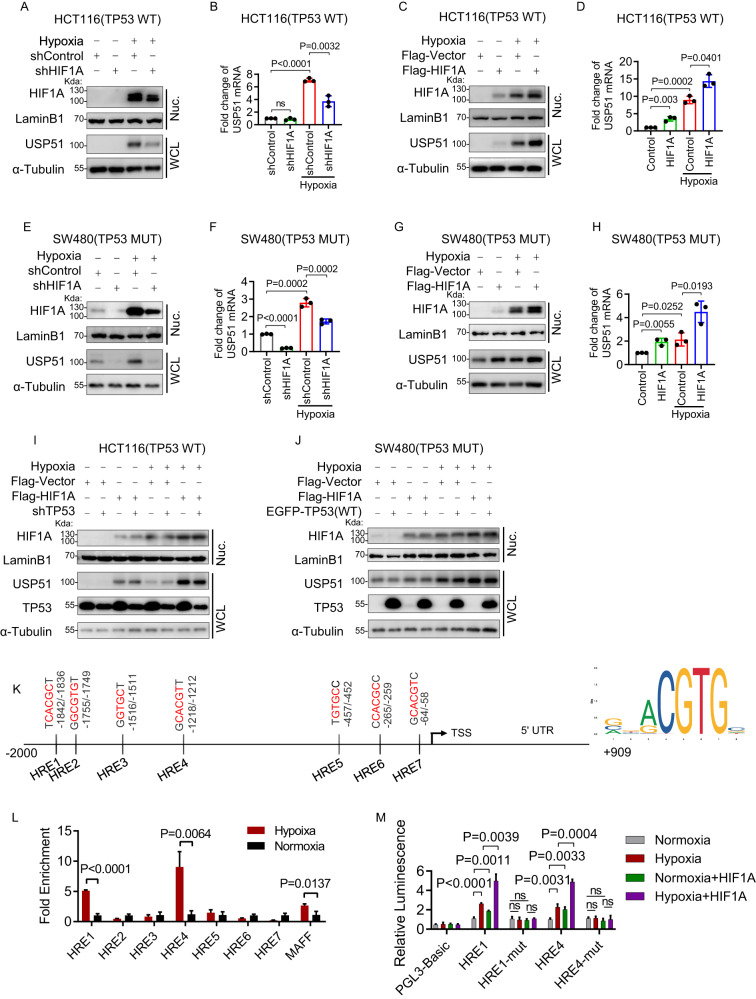
USP51 is a direct target gene of both HIF1A and HIF2A
To investigate whether USP51 is a HIF1A target gene, we exposed SW480(TP53 mutant) and HCT116(TP53 WT) cells to hypoxia or normoxia. We observed a marked increase in USP51 expression under hypoxia in HCT116 cells, both in protein and mRNA level (Fig. 4A–D). Hypoxia-induced USP51 upregulation in HCT116 cells was attenuated upon HIF1A knockdown (Fig. 4A, B) while enhanced upon HIF1A overexpression (Fig. 4C, D). In SW480 cells, USP51 was significantly upregulated under hypoxia both in protein and mRNA levels (Fig. 4E–H). Hypoxia-induced USP51 upregulation in SW480 cells was also attenuated upon HIF1A knockdown (Fig. 4E, F) while enhanced upon HIF1A overexpression (Fig. 4G, H). Under normoxia, HIF1A overexpression also could induced an increase in both protein and mRNA of USP51 (Fig. 4C, D, G, H).
Fig. 4. USP51 is a direct target gene of HIF1A.
A, B HIF1A was stably knocked down by shRNA in HCT116 cells under normoxia or hypoxia. The protein level (A) and mRNA level (B) of USP51 were examined. C, D HIF1A was stably overexpressed in HCT116 cells under normoxia or hypoxia. The protein level (C) and mRNA level (D) of USP51 were examined. E, F HIF1A was stably knocked down by shRNA in SW480 cells under normoxia or hypoxia. The protein level (E) and mRNA level (F) of USP51 were examined. G, H HIF1A was stably overexpressed in SW480 cells under normoxia or hypoxia. The protein level (G) and mRNA level (H) of USP51 were examined. I TP53 was stably knocked down by shRNA in HCT116 (TP53 WT) cells stably expressed Myc-HIF1A or Myc-vector under normoxia or hypoxia. Cell lysates were blotted by the anti-USP51, HIF1A, and TP53 antibodies. J TP53 WT was stably expressed in SW480 (TP53 MUT) cells stably expressed Myc-HIF1A or Myc-vector under normoxia or hypoxia. Cell lysates were blotted by the anti-USP51, HIF1A, and TP53 antibodies. K The schematic of the hypoxia response elements in the USP51 promoter (Left) and the sequence motif (Right) of the HIF binding sites. TSS: transcription start site. L ChIP assay was performed to identify two HIF1A binding sites on the USP51 promoter (HRE1 and HRE4). MAFF promoter was used as a positive control. M Luciferase reporter assays showed the transcription activity of luciferase constructs under different condition. Data are expressed as mean ± SD, n = 3 biological replicates. ns not significant, WCL whole cell lysate, Nuc nuclear fraction.
TP53 knockdown in TP53 WT HCT116 cells did not affect the hypoxia-induced USP51 upregulation (Fig. 4I), and forced expression of WT TP53 in TP53 mutant SW480 cells did not prevent the hypoxia-induced USP51 upregulation (Fig. 4J). These results indicate that USP51 is regulated under hypoxia in a HIF1A-dependent and TP53-independent manner.
Next, we investigated whether USP51 is a direct target of HIF1A, chromatin immunoprecipitation assay, and luciferase reporter assay was used. We searched promoter region (−2000 bp to +909 bp) of human USP51 and found seven hypoxia response element (HRE) sites, 5′-(A/G) CGTG-3′ [16] (Fig. 4K). We performed real-time PCR to analyze the DNA fragments immunoprecipitated by anti-HIF1A antibody. A primer flanking an HRE site in the MAFF promoter was used as a positive control [17]. Under hypoxia, strong enrichments were observed in HRE1 and HRE4 sites (Fig. 4L). Next, we generated luciferase reporter plasmids by introducing these sequences into the pGL3-luciferase reporter plasmid. Mutations in the core HRE sequences were used as a negative control. Under hypoxia, significant luciferase activity increases were observed in reporter plasmids constructed with HRE1 or HRE4, not their mutation sequences (Fig. 4M). Introducing an HIF1A plasmid considerably amplified luciferase activity even under normoxia. This enhancement was notably intensified when the cells were exposed to hypoxic conditions. These results indicate both HRE1 and HRE4 are required for transcriptional activation of the USP51 gene under hypoxia, and HIF1A played an important role in regulating USP51 expression.
SENP1 was reported as a direct target gene of HIF1A in hepatocellular carcinoma cancer [18]. But hypoxia-induced SENP1 upregulation was not observed in DLD-1, SW480, HCT116, and LoVo, four CRC cell lines (Supplementary Fig. 4A).
Next, we investigated whether USP51 was also the target gene of HIF2A. We used shRNA to knockdown HIF2A and noted that the hypoxia-induced increase in both protein and mRNA levels of USP51 was diminished in both HCT116 and SW480 cells (Supplementary Fig. 4B–E). Subsequently, we employed chromatin immunoprecipitation assay and luciferase reporter assay to validate whether USP51 is a direct target of HIF2A. We performed real-time PCR to analyze the DNA fragments immunoprecipitated by anti-HIF2A antibody. A primer flanking an HRE site in DMT1A promoter was used as a positive control [19]. Under hypoxia, strong enrichments were observed in HRE3 and HRE4 sites (Supplementary Fig. 4F). Next, we generated luciferase reporter plasmids by introducing these sequences into the pGL3-luciferase reporter plasmid. Mutations in the core HRE sequences were used as a negative control. Under hypoxia, significant luciferase activity increases were observed in reporter plasmids constructed with HRE3 or HRE4, not their mutation sequences (Supplementary Fig. 4G). Co-transfection with an HIF2A plasmid led to a strong enhancement in luciferase activity under normoxia. This enhancement of luciferase activity by HIF2A was notably intensified when the cells were subjected to hypoxic conditions. These results indicate that USP51 is also a direct target gene of HIF2A. Taken together, these results strongly underscore the significant regulatory role of not only HIF1A but also HIF2A in regulating USP51 expression.
We subsequently questioned whether the regulation of the HIF2A protein involves USP51. Nonetheless, the overexpression of USP51 did not result in a substantial alteration of the HIF2A protein levels, observed both during normoxia and hypoxia in HCT116 cells (Supplementary Fig. 4H). Conversely, the knockdown of USP51 yielded no significant change in the HIF2A protein levels, under both normoxic and hypoxic conditions in SW480 cells (Supplementary Fig. 4I). Additionally, the overexpression of USP51 did not result in a reduction of endogenous HIF1A ubiquitination in either HCT116 or SW480 cells (Supplementary Fig. 4J, K). The findings suggest that HIF2A is not targeted for deubiquitination by USP51.
Both HIF1A and SENP1 are required for USP51 to promote CRC proliferation, migration, and stemness under hypoxia
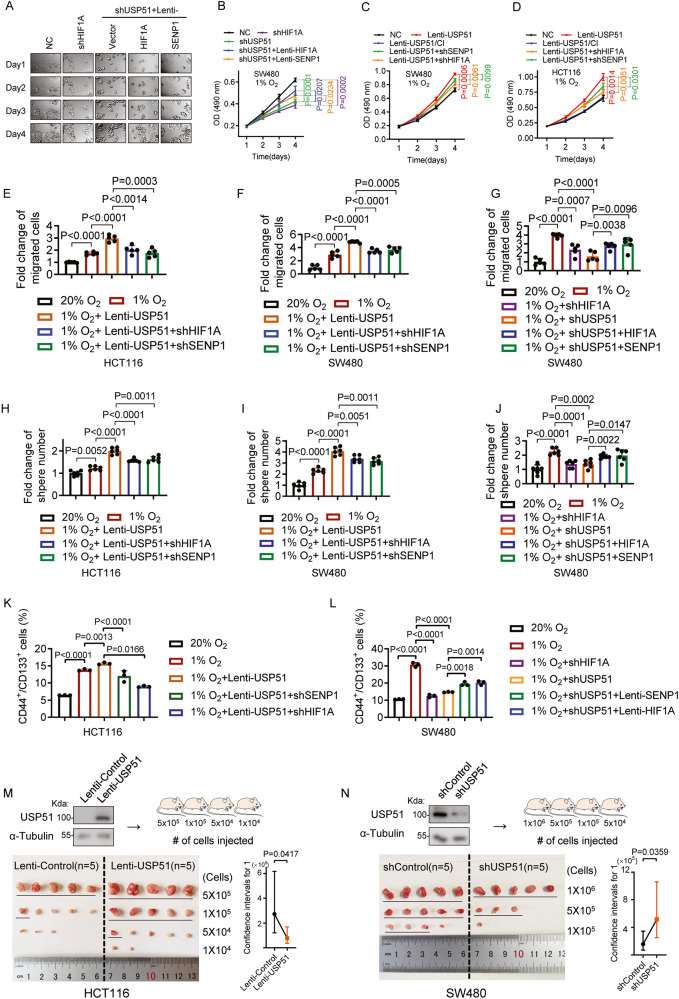
To investigate the role USP51 played in the progression of CRC in hypoxia, we initially established cell lines with stable knockdown of either HIF1A or USP51 in SW480 cells, along with stable overexpression of USP51 in both SW480 and HCT116 cells. In hypoxia, knockdown of USP51 led to a notable reduction in the proliferation of SW480 cells, mirroring the effects observed with HIF1A knockdown (Fig. 5A, B). Notably, when either HIF1A or SENP1 was simultaneously overexpressed, the anti-proliferative effect of USP51 knockdown weakened. The overexpression of USP51, not USP51/CI, considerably promoted the proliferation of SW480 cells under hypoxia (Fig. 5C). However, this pro-proliferative effect was mitigated by the knockdown of either HIF1A or SENP1. The HIF1A/SENP1-dependent pro-proliferation effect of USP51 overexpression was also observed in HCT116 cells (Fig. 5D). Additionally, the overexpression of USP51 enhanced the hypoxia-induced HCT116 and SW480 cell migration, and this enhancement was reduced by the knockdown of either HIF1A or SENP1 (Fig. 5E, F, Supplementary Fig. 5A). The knockdown of USP51 attenuated the hypoxia-induced SW480 cells migration, similar to the effects observed with HIF1A knockdown (Fig. 5G, Supplementary Fig. 5B). However, this weakening was counteracted by the overexpression of either HIF1A or SENP1. In the sphere formation assay, we observed that hypoxia induced more and larger spheres in both SW480 and HCT116 cells (Fig. 5H, I, Supplementary Fig. 5C). The overexpression of USP51 further augmented this effect induced by hypoxia. However, this enhancement was mitigated by the knockdown of either HIF1A or SENP1. Conversely, the knockdown of USP51 markedly reduced the ability of SW480 cells to form spheres under hypoxic conditions, resembling the outcomes seen with HIF1A knockdown (Fig. 5J, Supplementary Fig. 5D). Furthermore, this reduction in sphere formation was mitigated by the overexpression of either HIF1A or SENP1. We further evaluated the impact of USP51 on cell stemness by performing staining for CD44 and CD133, reported markers of CRC stem cells [20]. The results demonstrated that hypoxia led to a significant increase in the proportion of CD44+/CD133+ cells in both SW480 and HCT116 cells populations (Fig. 5K, L, Supplementary Fig. 5E, F). The overexpression of USP51 in HCT116 cells further amplified this effect (Fig. 5K, Supplementary Fig. 5E). However, this enhancement was mitigated by the knockdown of either HIF1A or SENP1. Conversely, the knockdown of USP51 in SW480 cells considerably diminished the ratio of CD44+/CD133+ cells under hypoxic conditions, akin to the effects of HIF1A knockdown (Fig. 5L, Supplementary Fig. 5F). This decline was counteracted by either HIF1A or SENP1 overexpression.
Fig. 5. Both HIF1A and SENP1 are required for USP51 to promote CRC proliferation, migration, and stemness under hypoxia.
A, B Stable knockdown of HIF1A or USP51 was done in SW480 cells. SW480 cells with stable USP51 knockdown were further subjected to overexpression of HIF1A or SENP1. The cells were monitored in Cell Observer under hypoxia (A) and the cell viability was determined by MTT (B). C Stable overexpression of USP51 or USP51/CI was achieved in SW480 cells. In cells with stable USP51 overexpression, knockdown of either SENP1 or HIF1A was performed. The cell viability was determined by MTT. D USP51 or USP51/CI was stably overexpressed in HCT116 cells. SENP1 or HIF1A was stably knocked down in HCT116 cells with stable USP51 overexpression. The cell viability was determined by MTT. USP51 was stably overexpressed in HCT116 (E) and SW480 (F) cells. In cells with stable USP51 overexpression, knockdown of either SENP1 or HIF1A was performed. The cells were used to perform transwell assay. G Stable knockdown of HIF1A or USP51 was done in SW480 cells. SW480 cells with stable USP51 knockdown were further subjected to overexpression of HIF1A or SENP1. The cells were used to perform transwell assay. The HCT116 (H) and SW480 (I) cells described in E and F were used to perform sphere formation assay. J The cells described in G were used to perform sphere formation assay. K The HCT116 cells described in E were used to analyze the expression of CD44 and CD133. L The SW480 cells described in G used to analyze the expression of CD44 and CD133. M USP51 was ectopically expressed in HCT116 cells. A limiting dilution assay was performed. The positive responses were underlined. N USP51 was stably knocked down in SW480 cells. A limiting dilution assay was performed. The positive responses were under lined. Data are expressed as mean ± SD, n = 3 biological replicates in B–D, K, L, n = 5 biological replicates in E–G, n = 6 biological replicates in H–J.
Given the in vitro findings, we next validated these findings in vivo. We performed an in vivo limiting dilution assay to determine how USP51 regulates the stemness capacity of cancer cells. NOD/SCID mice were subcutaneously injected with different dilutions of monolayer cultured indicated cells. The calculation of stem cell frequency showed that the overexpression of USP51 in HCT116 cells led to a 3.5-fold decrease in the number of cells per positive response (Fig. 5M). Conversely, the knockdown of USP51 resulted in a 3.3-fold increase in the number of cells per positive response compared to its control (Fig. 5N), akin to the comparative effect observed with the knockdown of HIF1A, which led to a 5.8-fold increase in the number of cells per positive response (Supplementary Fig. 5G). These results indicate that USP51 promotes the stemness of CRC cells, both in vitro and in vivo.
Both HIF1A and SENP1 are required for USP51 to promote CRC chemoresistance under hypoxia
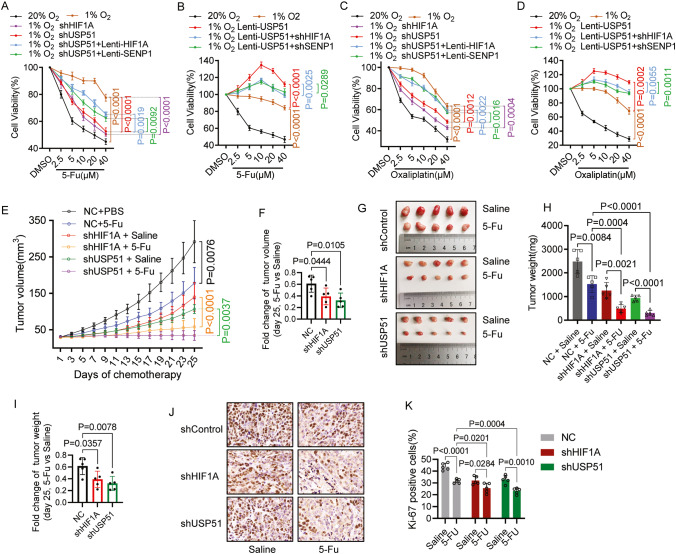
We next investigated whether USP51 promote CRC chemoresistance under hypoxia condition. We observed that hypoxia caused SW480 cells to exhibit increased resistance to fluorouracil (5-FU) treatment (Fig. 6A, B) as well as to Oxaliplatin treatment (Fig. 6C, D). Knockdown of USP51 partially mitigated the hypoxia-induced drug resistance of SW480 cells to 5-FU (Fig. 6A) and Oxaliplatin (Fig. 6C), resembling the impact of HIF1A knockdown. Furthermore, this mitigation by USP51 knockdown was counteracted by the overexpression of either HIF1A or SENP1. Conversely, the overexpression of USP51 amplified the hypoxia-induced drug resistance of SW480 cells to 5-FU (Fig. 6B) and Oxaliplatin (Fig. 6D). However, this reinforcement was partly diminished by the knockdown of either HIF1A or SENP1. These in vitro results indicate that USP51 promote SW480 resistance to 5-FU and Oxaliplatin under hypoxia in HIF1A and SENP1-dependent manner.
Fig. 6. Both HIF1A and SENP1 are required for USP51 to promote CRC chemoresistance under hypoxia.
A, C USP51 or HIF1A was stably knocked down in SW480 cells. Stably USP51 knockdown SW480 cells were overexpressed either HIF1A or SENP1. The cells were exposed to the indicated concentration of 5-FU (A) or Oxaliplatin (C) for 72 h. The cell viability was determined by MTT. B, D USP51 was stably overexpressed in SW480 cells. Stably USP51 overexpressed SW480 cells were subjected to knockdown of either HIF1A or SENP1. The cells were exposed to the indicated concentration of 5-FU (B) or Oxaliplatin (D) for 72 h. The cell viability was determined by MTT. E–K Mice with SW480-Vector, SW480-shHIF1A, and SW480-shUSP51 tumors were administered either saline or 5-FU for 25 days, starting when the tumor volume reached 30mm3. The tumor volumes (E), the fold change of tumor volumes at day 25 (F), images of xenograft models (G), tumor weight (H), fold change of tumor weight at day 25 (I), representative images of Ki-67 immunohistochemical staining (J), and analysis of Ki-67 levels (K) are presented. Data are expressed as mean ± SD, n = 3 biological replicates in A–D and n = 5 biological replicates in E–K.
We next validated these findings in vivo using an SW480 cells xenograft model. USP51 knockdown significantly increased tumor sensitivity to 5-FU treatment with an approximately a 3-fold reduction in tumor volume after 25 days of administration of 5-FU (Fig. 6E, F), compared to a 1.6-fold decrease observed in the control group. This effect of USP51 knockdown mirrored the outcome of HIF1A knockdown, which also resulted in a 2.6-fold reduction in tumor volume. The tumor weight showed a similar change as the volume (Fig. 6G–I). Histological analyses of the harvested tumors revealed that the knockdown of USP51 enhanced the anti-proliferative effect of 5-FU treatment (Fig. 6J, K). This effect paralleled the impact of HIF1A knockdown, as both conditions exhibited notably reduced levels of Ki67 expression in comparison to the control group.
High expression of USP51 predicts survival disadvantage in colorectal cancer
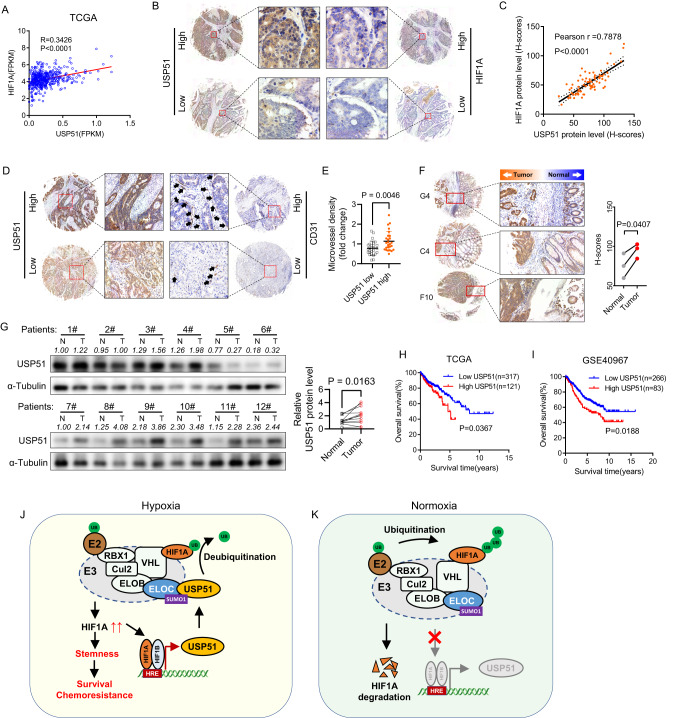
The correlation analysis in human CRC specimens from the TCGA dataset revealed a positive correlation between the mRNA expression of HIF1A and USP51 (Fig. 7A). Immunohistochemical analysis of a tissue microarray containing 76 tumor samples of CRC patients revealed a significant positive correlation between the protein levels of HIF1A and USP51 (Fig. 7B, C). Furthermore, an analysis of a tissue microarray containing 52 tumor samples demonstrated that heightened levels of USP51 protein corresponded with an elevated microvessel density (Fig. 7D, E). Analysis of tissue containing both tumor and normal adjacent tissue revealed that USP51 protein was upregulated in the tumors compared to their normal adjacent tissue (Fig. 7F). Immunoblot analysis of 12 patients’ samples further confirmed that USP51 protein was upregulated in tumors (Fig. 7G). Based on data from both TCGA and GEO (GSE40967), a high mRNA expression of USP51 was indicative of unfavorable overall survival among colorectal cancer patients (Fig. 7H, I).
Fig. 7. High expression of USP51 predicts survival disadvantage in colorectal cancer.
A The correlation between the mRNA expression of HIF1A and USP51 in the TCGA CRC dataset. B Represent images of immunohistochemistry staining of HIF1A and USP51 in CRC tissue microarray. C The correlation between the protein levels of HIF1A and USP51 in CRC tissue microarray. D Represent images of immunohistochemistry staining of USP51 and CD31 in CRC tissue microarray. E The microvessel density differed between samples exhibiting low and high USP51 expression levels. F Immunohistochemical analysis of tissue containing both tumor and normal adjacent tissue. G Immunoblot analysis of patients’ samples. USP51 predicted poor overall survival in colorectal cancer patients according to the TCGA (H) and GEO data (GSE40967) (I). Schematic diagrams illustrating the role USP51 played in regulating the HIF1A pathway under normoxia (J) and hypoxia (K). Data are expressed as mean ± SD, n = 26 biological replicates in E, n = 3 biological replicates in F, n = 12 biological replicates in G.
Discussion
In this study, we demonstrated that USP51 could directly interact with ELOC, a subunit of the VHL E3 ligase complex (VHL/CUL2/ELOC/ELOB/RBX1), facilitating an indirect binding of USP51 to HIF1A. This interaction leads to the stabilization of HIF1A through deubiquitination by USP51. Interestingly, USP51 turned out to be a direct target gene of HIF1A. Thus, USP51 and HIF1A formed a positive feedback loop. Under hypoxia, HIF1A activates the transcription of USP51, speeds up the HIF1A-USP51 loop, contributing to the accumulation of the USP51 and HIF1A protein (Fig. 7J). Under normoxia, HIF1A degrades and slows down the HIF1A-USP51 loop, contributing to the depletion of the USP51 and HIF1A protein (Fig. 7K).
With USP51 recruited to the VHL E3 ligase complex (also called ECV complex), the VHL E3 ligase complex together with USP51 forms a more complicated complex (USP51-ECV). USP51-ECV is a bidirectional function complex, which can ubiquitinate and yet deubiquitinate HIF1A. Under different oxygen concentrations, USP51-ECV may have a different capacity for deubiquitinating HIF1A. Under hypoxia (not absolute absence of oxygen), the HIF1A protein binding USP51-ECV is more likely to be deubiquitinated by USP51 than ubiquitinated by E2 because of the significant upregulation of USP51 under hypoxia. Under normoxia, USP51-ECV exhibits a low deubiquitinating capacity because of the markedly low expression of USP51. In the present study, the knockdown of USP51 under hypoxia inhibits the deubiquitinating ability of USP51-ECV, resulting in the downregulation of the HIF1A protein. While overexpression of USP51 under normoxia promotes the deubiquitinating capacity of USP51-ECV, resulting in the upregulation of the HIF1A protein.
We also demonstrated that ELOC is modified by SUMO1. SUMOylation of ELOC inhibits the interaction between ELOC and USP51. SENP1 mediates the deSUMOylation of ELOC, promoting ELOC-USP51 interaction, enhancing the USP51-mediated deubiquitination and stabilization of HIF1A. SENP1 was reported to promote stemness or chemoresistance in several types of cancer [18, 21–23]. SENP1 could activate the transcriptional activity of HIF1A, and HIF1A reversely activated the transcription of SENP1. Thus, SENP1 and HIF1A formed a positive feedback loop [18]. However, we observed that the SENP1 protein level was not significantly upregulated under hypoxia in CRC cells. But, we did find another association between SENP1 and HIF1A: SENP1 can enhance the USP51-mediated deubiquitination and stabilization of HIF1A, upregulating the HIF1A protein level. This is new evidence that hypoxia collaborates with post-transcriptional modifications, including ubiquitination and SUMOylation, to promote CRC progression.
The TP53 gene is mutated in approximately 60% of all CRC cases [24] and is usually associated with poor outcomes [25]. Previous studies reported that an accumulation of TP53 inhibited HIF activity [26], WT TP53 could block HIF1A-induced USP22 upregulation, inhibiting cancer stemness [27]. But other researchers showed that both WT and mutant TP53 proteins increases HIF transcriptional ability [28]. In fact, the relationship between HIF1A and TP53 is not well-defined. Thus, taking into account the status of TP53 is advisable when investigating HIF1A’s transcription of specific genes. In the present study, we showed that HIF1A-induced USP51 expression is independent of the status of TP53. This result indicates that HIF1A-USP51 interaction may widely present in CRC and deserves attention.
USP51 is not well recognized in cancer previously except for deubiquitinating H2AK13,15ub and ZEB1. Our data provide evidence that USP51 plays a critical role in regulating the HIF1A pathway, thereby acting as a mediator of cellular response to hypoxic conditions. Given the prevalence of hypoxia in solid tumors and its association with hypoxia-induced chemoresistance [29], our study holds meaningful significance as it suggests novel strategies for addressing drug resistance in patients with colorectal cancer.
Materials and methods
Cell cultures and lentiviral vectors transfection
Colorectal cancer cell lines HCT116, DLD-1, SW480, and LoVo were purchased from the National Collection of Authenticated Cell Cultures (Shanghai, China). All the cell lines have been authenticated by STR profiling. All the cell lines were tested monthly for mycoplasma contamination using a Mycoplasma Stain Assay Kit (Beyotime, Shanghai, China). The cells were routinely cultured in Dulbecco’s modified Eagle’s medium (DMEM) (GIBCO, NY, USA) supplemented with 10% fetal bovine serum (GIBCO, NY, USA) at 5% CO2 at 37 °C. The lentiviral vectors pUBi-EGFP-tag were used to overexpress protein. The lentiviral vectors phU6-EGFP-shRNA were used to knockdown the protein. All the lentiviral vectors and plasmids were prepared by GeneChem Co., Ltd (Shanghai, China). All transfections were conducted as per the manufacturer’s protocol. Target sequences in shRNA are shown in Supplementary Data 2.
Cell viability assay
The cell viability was determined by MTT (3-(4,5-Dimethylthial-2-yl)-2,5-Diphenyltetrazalium Bromide) (Sigma-Aldrich, MA, USA). Cells were seeded in a 96-well plate. At the indicated time, 10 μL of 5 mg/L MTT solution was added to each well containing 100 μL medium. After incubation for 4 h, the medium was removed, and each well was added 150 μL of dimethyl sulfoxide. The absorbance was measured at 490 with background subtraction at 630 nm.
Western blot
Whole cells lysis was prepared using RIPA buffer (Beyotime, Shanghai, China) containing protease inhibitors. Nuclear fractions were separated using a Nuclear Extraction Kit (Abcam, ab113474, Cambridge, UK) according to the manufacturer’s instructions. The samples were heat denatured, separated by SDS-PAGE, transferred to PVDF membranes (Invitrogen, MA, USA), and blocked with 5% bovine serum albumin. Then membranes were incubated with primary antibodies at 4 °C overnight and secondary antibodies for 1 h at room temperature. Immunoblots were visualized using an ECL detection reagent (Millipore, MA, USA). The primary and secondary antibody information was present in Supplementary Data 3. The uncropped original western blots were present in Supplementary Data 4.
Real-time reverse transcription PCR (qRT-PCR)
Total RNA was extracted from cells using an RNA Easy Fast Tissue/Cell Kit (TIANGEN, Beijing, China). Reverse transcription reactions were performed using RT Kit (Takara, Tokyo, Japan). PCR was performed using a PCR premix Kit (Takara, Tokyo, Japan). All procedures were conducted as per the manufacturer’s protocol. Each experiment was performed in triplicate. The sequences of primers were summarized in Supplementary Data 5.
RNA-sequencing
SW480 and HCT116 cells were overexpressed Flag-USP51 or Flag-Vector. The RNA-seq was performed on BGISEQ-500 by BGI company (Shenzhen, China). The data were analyzed on Dr. Tom Multi-Omics Data Mining System (https://biosys.bgi.com/). The result is summarized in Supplementary Data 1.
Transwell assays
The assay was performed using SW480 and HCT116 cells and the details have been previously described [30]. In brief, cells were seeded in upper chamber and incubated at 37 °C, 5% CO2 for 24–36 h. The number of the migrated cells was counted in five randomly selected fields. Each experiment was performed in triplicate.
Sphere formation assay
SW480 and HCT116 cells were grown as spheres in DMEM/F-12 (GIBCO, NY, USA) supplemented with B27 (Thermo Scientific, MA, USA), 20 ng/ml epidermal growth factor (Sigma-Aldrich, MA, USA), and 20 ng/ml basic fibroblast growth factor (GIBCO, NY, USA). Viable cells were seeded in 96-wells ultra-low adhesion plates (Corning, NY, USA) at 1000 cells/well density and cultured for ten days. Spheres larger than 50 μm (HCT116) or 40 μm (SW480) in diameter were counted.
Co-Immunoprecipitation (Co-IP) assay and Mass spectrometry
Cells were lysed with IP lysis buffer (Beyotime, Shanghai, China). The lysate was incubated with 5 ug primary antibody at 4 °C overnight and then the magnetic beads (Thermo Scientific, MA, USA) for 15 min at room temperature. The beads were collected with a magnetic stand and the supernatant was discarded. The beads were washed three times. Add low-pH elution buffer and incubate for 5 min at room temperature. The elution was neutralized by adding a Neutralization Buffer. For Co-IP assay, the samples were used for western blot analysis. The primary and secondary antibody information was present in Supplementary Data 3. For mass spectrometry, the protein solution underwent a process of analysis using a Thermo Scientific Q Exactive mass spectrometer by Fitgene Biotechnology Co., Ltd (Guangzhou, China).
Silver staining and Coomassie staining
Silver staining was performed using a Fast Silver Stain Kit (Beyotime, Shanghai, China). Coomassie staining was performed using Coomassie brilliant blue R250 (Beyotime, Shanghai, China).
GST pull-down assay
His-USP51 was expressed in E. coli BL21 (DE3) cells and purified using His-tag Protein Purification Kit (Beyotime, Shanghai, China) following the manufacturer’s protocol. The purified proteins of GST Tag (Proteintech, NJ, USA), GST-HIF1A (Novus Biologicals, CO, USA) or GST-ELOC (Proteintech, NJ, USA) were mixed with glutathione sepharose 4B resin (Cytiva, MA, USA) and purified His-USP51 protein in vitro binding buffer [31]. The sample mixtures were incubated for 2 h at 4 °C. Then the resin was washed three times before being boiled in SDS sample buffer. Bound and input proteins were analyzed by SDS-PAGE and Western blot.
Immunofluorescence and confocal microscopy
The cells were fixed with 4% paraformaldehyde for 10 min and permeabilized with 0.2% Triton X-100 for 10 min. After blocking in 1% BSA for 30 min at room temperature, the cells were incubated at 4 °C overnight with primary antibodies (present in Supplementary Data 3). The dishes were washed three times with phosphate-buffered saline (PBS) for 5 min each and then incubated with secondary antibodies (present in Supplementary Data 3) for 1 h at room temperature. The nuclei were stained by DAPI (Solarbio, Beijing, China) for 5 min. The samples were examined via microscopy (Leica Microsystems, Heidelberg, Germany). All confocal images were analyzed and quantified using Image J v. 1.45 (http://rsb.info.nih.gov/ij/).
Flow cytometry analysis
Cells were incubated with primary antibodies at 4 °C for 1 h. Then cells were washed three times with cold cell staining buffer (abs9475, Absin, Shanghai, China) and incubated with secondary antibody at 4 °C for 1 h. Samples were washed three times and analyzed on a NovoCyte Flow Cytometer (Santa Clara, CA, USA). The primary and secondary antibody information was present in Supplementary Data 3.
Chromatin immunoprecipitation (ChIP) assays
The CHIP assay was performed in SW480 cells exposed to normoxia (20% O2) or hypoxia (1% O2) for 24 h using an anti-HIF1A or anti-HIF2A antibody (present in Supplementary Data 3). 400-800 bp DNA fragments were prepared by 130-Watt Ultrasonic Processor (VCX-130PB, 25%, 10 s on, 10 s off, 10 cycles). The DNA immunoprecipitated was purified using a Purification Kit (TIANGEN, Beijing, China). Quantification of all ChIP samples was performed by real-time PCR using PCR premix Kit (Takara, Tokyo, Japan). The data are presented as the fold enrichment of the antibody signal compared to the negative control IgG. Primers for ChIP were present in Supplementary Data 6.
Luciferase reporter assay
The pGL3-Basic (Promega, Madison, WI, USA), pGL3-HRE1, pGL3-HRE1(mut), pGL3-HRE3, pGL3-HRE3(mut), pGL3-HRE4, pGL3-HRE4(mut) vectors and pRL-TK vector were prepared by GeneChem Co., Ltd (Shanghai, China). 1 ×104 cells were seeded on a 96-well plate. Then 50 ng of pGL3 firefly luciferase vector were transfected with 5 ng of pRLK-TK vector using Lipo3000 (Invitrogen, MA, USA). 24 h after transfection, cells were exposed to normoxia or hypoxia for additional 24 h. The luminescence was detected using the Dual-Luciferase Reporter Gene Assay Kit II (Beyotime, Shanghai, China) and Varioskan Flash (Thermo Scientific, MA, USA). The promoter sequences for luciferase reporter assays were present in Supplementary Data 7.
Human tissue samples and human tissue microarray
The Ethics Committee of the First Affiliated Hospital of Xi’an Jiaotong University approved the experiment. Twelve pairs of human tissue samples for immunoblot analysis were obtained from patients who had undergone surgery for cancer resection at the First Affiliated Hospital of Xi’an Jiaotong University (Xi’an, China) between Dec. 2021 and Jan. 2022. Tissue microarrays consisting of human CRC specimens were purchased from Xi’an Bioaitech, Co., Ltd (D081Co01). The sections were incubated with primary antibodies overnight at 4 °C, followed by secondary antibody. The H-score was calculated using the ∑Pi(i) formula, where i = intensity of staining with a value of 1, 2, or 3 (weak, moderate, or strong, respectively) and Pi is the percentage of stained cells for each intensity. The primary and secondary antibody information was present in Supplementary data 3.
In vivo limiting dilution assay and xenograft tumor models
The Ethics Committee of the First Affiliated Hospital of Xi’an Jiaotong University approved the experiment. All mice were purchased from Shanghai SLAC Laboratory Animal Co., Ltd. (Shanghai, China) and maintained in the Xi’an Jiao Tong University animal facility under specific-pathogen-free (SPF) conditions. Mice were randomly divided into groups. For limiting dilution assay, 8-week-old female NOD/SCID mice were subcutaneously injected with different dilutions of monolayer cultured cells. A tumor >20 mm3 at 21 days post-injection was defined as a positive response. Stem cell frequencies were determined using ELDA [32] and the output are provided in Supplementary Data 8. For investigate angiogenesis, 1 ×106 SW480-shControl cells, SW480-shHIF1A cells or SW480-shUSP51 cells with 0.1 ml PBS were subcutaneously transplanted into the right dorsal flanks of 8-week-old female nude mice. The tumors were obtained 28 days after inoculation. For investigate chemoresistance, 1 ×106 SW480 cells or SW480-shUSP51 cells with 0.1 ml PBS were subcutaneously transplanted into the right dorsal flanks of 8-week-old female nude mice. The treatment was initiated when the tumor volume reached approximately 30 mm3. 5-FU was intraperitoneally injected 2- times/5-day at 25 mg/kg. The control group received the vehicle treatment (saline, 0.2 ml). Tumors were measured every two days and volume was calculated as ½(L×W2). For immunohistochemistry, the tumor tissues were subjected to formalin fixation, followed by embedding in paraffin wax and subsequent sectioning into sections of 4 μm thickness. These sections underwent an overnight incubation at 4 °C with the primary antibodies, followed by HRP-conjugated secondary goat antibody. Sample size for each group: n = 5 animals. The investigators were blinded to group allocation during data collection and analysis.
Statistics analysis
Data are presented as the mean ± SD. The differences among the groups were compared by the two-sided Student’s t test or one-way ANOVA. Correlations were analyzed by using Pearson linear regression analysis. The Kaplan−Meier survival method was used to evaluate overall survival. All statistics were calculated by GraphPad Prism 7. P value < 0.05 indicated statistical significance.
Supplementary information
Supplementary Data 4-uncropped western blot
Author contributions
MM and QZ: Conceptualization, methodology, validation, original draft writing, review & editing. JL: Conducted animal experiments. YZ, XL, and ZC: Analyzed the data. XS: Conceptualization, supervision, project administration. JY: Supervision, project administration, investigation, methodology, resourcing, review & editing of the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (No. 82002547).
Data availability
All data needed to evaluate the conclusions in the paper are present in the paper. Additional data related to this paper may be requested from the corresponding author.
Competing interests
The authors declare no competing interests.
Ethics approval
All related experiments in our study were carried out in accordance with the Helsinki Declaration, and approved by the Ethics Committee of the First Affiliated Hospital of Xi’an Jiaotong University.
Informed consent
Informed consent was acquired from all participants involving human tissues.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xuejun Sun, Email: sunxy@mail.xjtu.edu.cn.
Junhui Yu, Email: yujunhui@mail.xjtu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-023-01228-8.
References
- 1.Lin JS, Perdue LA, Henrikson NB, Bean SI, Blasi PR. Screening for colorectal cancer: updated evidence report and systematic review for the US preventive services task force. JAMA. 2021;325:1978–98. doi: 10.1001/jama.2021.4417. [DOI] [PubMed] [Google Scholar]
- 2.Wang Z, Zhang H, Liu J, Cheruiyot A, Lee JH, Ordog T, et al. USP51 deubiquitylates H2AK13,15ub and regulates DNA damage response. Genes Dev. 2016;30:946–59. doi: 10.1101/gad.271841.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou Z, Zhang P, Hu X, Kim J, Yao F, Xiao Z, et al. USP51 promotes deubiquitination and stabilization of ZEB1. Am J Cancer Res. 2017;7:2020–31. [PMC free article] [PubMed] [Google Scholar]
- 4.Spaderna S, Schmalhofer O, Wahlbuhl M, Dimmler A, Bauer K, Sultan A, et al. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res. 2008;68:537–44. doi: 10.1158/0008-5472.CAN-07-5682. [DOI] [PubMed] [Google Scholar]
- 5.Zhang P, Wei Y, Wang L, Debeb BG, Yuan Y, Zhang J, et al. ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1. Nat Cell Biol. 2014;16:864–75. doi: 10.1038/ncb3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmalhofer O, Brabletz S, Brabletz T. E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev. 2009;28:151–66. doi: 10.1007/s10555-008-9179-y. [DOI] [PubMed] [Google Scholar]
- 7.Seeler JS, Dejean A. SUMO and the robustness of cancer. Nat Rev Cancer. 2017;17:184–97. doi: 10.1038/nrc.2016.143. [DOI] [PubMed] [Google Scholar]
- 8.El-Asmi F, McManus FP, Thibault P, Chelbi-Alix MK. Interferon, restriction factors and SUMO pathways. Cytokine Growth Factor Rev. 2020;55:37–47. doi: 10.1016/j.cytogfr.2020.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Yeh ET. SUMOylation and De-SUMOylation: wrestling with life’s processes. J Biol Chem. 2009;284:8223–7. doi: 10.1074/jbc.R800050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun XX, Chen Y, Su Y, Wang X, Chauhan KM, Liang J, et al. SUMO protease SENP1 deSUMOylates and stabilizes c-Myc. Proc Natl Acad Sci USA. 2018;115:10983–8. doi: 10.1073/pnas.1802932115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hicklin DJ, Ellis LM. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J Clin Oncol. 2005;23:1011–27. doi: 10.1200/JCO.2005.06.081. [DOI] [PubMed] [Google Scholar]
- 12.Xu Z, Guo C, Ye Q, Shi Y, Sun Y, Zhang J, et al. Endothelial deletion of SHP2 suppresses tumor angiogenesis and promotes vascular normalization. Nat Commun. 2021;12:6310. doi: 10.1038/s41467-021-26697-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao D, Hou M, Guan YS, Jiang M, Yang Y, Gou HF. Expression of HIF-1alpha and VEGF in colorectal cancer: association with clinical outcomes and prognostic implications. BMC Cancer. 2009;9:432. doi: 10.1186/1471-2407-9-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang M, Chiu SY, Hsu W. SUMO-specific protease 2 in Mdm2-mediated regulation of p53. Cell Death Differ. 2011;18:1005–15. doi: 10.1038/cdd.2010.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beauclair G, Bridier-Nahmias A, Zagury JF, Saïb A, Zamborlini A. JASSA: a comprehensive tool for prediction of SUMOylation sites and SIMs. Bioinformatics. 2015;31:3483–91. doi: 10.1093/bioinformatics/btv403. [DOI] [PubMed] [Google Scholar]
- 16.Castro-Mondragon JA, Riudavets-Puig R, Rauluseviciute I, Lemma RB, Turchi L, Blanc-Mathieu R, et al. JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022;50:D165–d73. doi: 10.1093/nar/gkab1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moon EJ, Mello SS, Li CG, Chi JT, Thakkar K, Kirkland JG, et al. The HIF target MAFF promotes tumor invasion and metastasis through IL11 and STAT3 signaling. Nat Commun. 2021;12:4308. doi: 10.1038/s41467-021-24631-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui CP, Wong CC, Kai AK, Ho DW, Lau EY, Tsui YM, et al. SENP1 promotes hypoxia-induced cancer stemness by HIF-1α deSUMOylation and SENP1/HIF-1α positive feedback loop. Gut. 2017;66:2149–59. doi: 10.1136/gutjnl-2016-313264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shah YM, Matsubara T, Ito S, Yim SH, Gonzalez FJ. Intestinal hypoxia-inducible transcription factors are essential for iron absorption following iron deficiency. Cell Metab. 2009;9:152–64. doi: 10.1016/j.cmet.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang D, Fu L, Sun H, Guo L, DuBois RN. Prostaglandin E2 promotes colorectal cancer stem cell expansion and metastasis in mice. Gastroenterology. 2015;149:1884–95.e4. doi: 10.1053/j.gastro.2015.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen MC, Nhan DC, Hsu CH, Wang TF, Li CC, Ho TJ, et al. SENP1 participates in Irinotecan resistance in human colon cancer cells. J Cell Biochem. 2021;122:1277–94. doi: 10.1002/jcb.29946. [DOI] [PubMed] [Google Scholar]
- 22.Shangguan X, He J, Ma Z, Zhang W, Ji Y, Shen K, et al. SUMOylation controls the binding of hexokinase 2 to mitochondria and protects against prostate cancer tumorigenesis. Nat Commun. 2021;12:1812. doi: 10.1038/s41467-021-22163-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu YC, Ling TY, Lu SH, Kuo HC, Ho HN, Yeh SD, et al. Chemotherapeutic sensitivity of testicular germ cell tumors under hypoxic conditions is negatively regulated by SENP1-controlled sumoylation of OCT4. Cancer Res. 2012;72:4963–73. doi: 10.1158/0008-5472.CAN-12-0673. [DOI] [PubMed] [Google Scholar]
- 24.Hassin O, Nataraj NB, Shreberk-Shaked M, Aylon Y, Yaeger R, Fontemaggi G, et al. Different hotspot p53 mutants exert distinct phenotypes and predict outcome of colorectal cancer patients. Nat Commun. 2022;13:2800. doi: 10.1038/s41467-022-30481-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–70. doi: 10.1016/S0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 26.Sano M, Minamino T, Toko H, Miyauchi H, Orimo M, Qin Y, et al. p53-induced inhibition of Hif-1 causes cardiac dysfunction during pressure overload. Nature. 2007;446:444–8. doi: 10.1038/nature05602. [DOI] [PubMed] [Google Scholar]
- 27.Ling S, Shan Q, Zhan Q, Ye Q, Liu P, Xu S, et al. USP22 promotes hypoxia-induced hepatocellular carcinoma stemness by a HIF1α/USP22 positive feedback loop upon TP53 inactivation. Gut. 2020;69:1322–34. doi: 10.1136/gutjnl-2019-319616. [DOI] [PubMed] [Google Scholar]
- 28.Madan E, Parker TM, Pelham CJ, Palma AM, Peixoto ML, Nagane M, et al. HIF-transcribed p53 chaperones HIF-1α. Nucleic Acids Res. 2019;47:10212–34. doi: 10.1093/nar/gkz766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang YA, Chen YF, Bao Y, Mahara S, Yatim S, Oguz G, et al. Hypoxic tumor microenvironment activates GLI2 via HIF-1α and TGF-β2 to promote chemoresistance in colorectal cancer. Proc Natl Acad Sci USA. 2018;115:E5990–E5999. doi: 10.1073/pnas.1801348115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mu M, Yu Q, Zhang Q, Guo J, Wang X, Sun X, et al. A pan-cancer analysis of molecular characteristics and oncogenic role of gasdermins. Cancer Cell Int. 2022;22:80. doi: 10.1186/s12935-022-02483-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyamoto T, Hosoba K, Itabashi T, Iwane AH, Akutsu SN, Ochiai H, et al. Insufficiency of ciliary cholesterol in hereditary Zellweger syndrome. Embo j. 2020;39:e103499. doi: 10.15252/embj.2019103499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–8. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data 4-uncropped western blot
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper. Additional data related to this paper may be requested from the corresponding author.