Abstract
A hallmark of Idiopathic Pulmonary Fibrosis is the TGF-β-dependent activation of lung fibroblasts, leading to excessive deposition of collagen proteins and progressive scarring. We have previously shown that synthesis of collagen by lung fibroblasts requires de novo synthesis of glycine, the most abundant amino acid in collagen protein. TGF-β upregulates the expression of the enzymes of the de novo serine/glycine synthesis pathway in lung fibroblasts through mTORC1 and ATF4-dependent transcriptional programs. SHMT2, the final enzyme of the de novo serine/glycine synthesis pathway, transfers a one-carbon unit from serine to tetrahydrofolate (THF), producing glycine and 5,10-methylene-THF (meTHF). meTHF is converted back to THF in the mitochondrial one-carbon (1C) pathway through the sequential actions of MTHFD2 (which converts meTHF to 10-formyl-THF), and either MTHFD1L, which produces formate, or ALDH1L2, which produces CO2. It is unknown how the mitochondrial 1C pathway contributes to glycine biosynthesis or collagen protein production in fibroblasts, or fibrosis in vivo. Here, we demonstrate that TGF-β induces the expression of MTHFD2, MTHFD1L, and ALDH1L2 in human lung fibroblasts. MTHFD2 expression was required for TGF-β-induced cellular glycine accumulation and collagen protein production. Combined knockdown of both MTHFD1L and ALDH1L2 also inhibited glycine accumulation and collagen protein production downstream of TGF-β; however knockdown of either protein alone had no inhibitory effect, suggesting that lung fibroblasts can utilize either enzyme to regenerate THF. Pharmacologic inhibition of MTHFD2 recapitulated the effects of MTHFD2 knockdown in lung fibroblasts and ameliorated fibrotic responses after intratracheal bleomycin instillation in vivo. Our results provide insight into the metabolic requirements of lung fibroblasts and provide support for continued development of MTHFD2 inhibitors for the treatment of IPF and other fibrotic diseases.
INTRODUCTION
Idiopathic Pulmonary Fibrosis (IPF) is a progressive, fatal disease, which has a median survival of 3.5 years and affects approximately 150,000 people in the United States (1, 2). IPF is characterized by the Transforming Growth Factor-β (TGF-β)-dependent activation of lung fibroblasts, leading to the excessive secretion of extracellular matrix proteins, including collagens (3–6). Replacement of healthy lung tissue with matrix proteins leads to progressive loss of lung function and thus, the production of collagens by lung fibroblasts represents an important therapeutic target for the treatment of IPF (6–8).
Metabolic reprogramming during the process of fibroblast activation is an emerging hallmark of IPF and an increasingly-studied target for therapeutic intervention (9, 10). We and others have demonstrated that de novo synthesis of glycine, the most abundant amino acid in collagen protein is required for production of collagen downstream of TGF-β (11–14). This requires the mTOR and ATF4-dependent induction of the enzymes of the de novo serine/glycine synthesis pathway, which converts the glycolytic intermediate 3-phosphoglycerate into the amino acids serine and glycine (13, 15). Inhibition of this pathway reduces TGF-β-induced collagen protein production in vitro, and ameliorates bleomycin-induced lung fibrosis in vivo (11, 12, 16).
The last enzyme of the de novo serine/glycine synthesis pathway, serine hydroxymethyltransferase 2 (SHMT2), is an entry point for carbon into the mitochondrial one-carbon (1C) pathway (17–22). SHMT2 catalyzes the transfer of a 1C group from serine to tetrahydrofolate (THF), producing glycine and 5,10-methylene-THF (meTHF). In order for the SHMT2 reaction to proceed, mitochondrial meTHF must be recycled back to THF by the enzymes of the mitochondrial 1C pathway. First, the bifunctional dehydrogenase/cyclohydrolase, MTHFD2 (meTHF Dehydrogenase 2) catalyzes the conversion of meTHF to 10-formyl-THF. 10-formyl-THF is then converted back to THF by either MTHFD1L (meTHF Dehydrogenase 1 Like), producing formate, or by ALDH1L2 (Aldehyde Dehydrogenase Family 1 Member L2), producing CO2 and reducing NADP+ to NADPH. It is unknown how 1C metabolism contributes to glycine synthesis and collagen protein production downstream of TGF-β signaling.
Here, we demonstrate that TGF-β induces the expression of MTHFD2, MTHFD1L, and ALDH1L2 in an mTORC1 and ATF4-dependent manner. MTHFD2 is required for intracellular glycine accumulation downstream of TGF-β and for collagen protein production in lung fibroblasts. The combined knockdown of MTHFD1L and ALDH1L2 recapitulated the effects of MTHFD2 knockdown; however, the individual knockdown of either enzyme did not affect glycine or collagen accumulation, suggesting that lung fibroblasts have flexibility to utilize either enzyme to regenerate THF. We further show that the inhibition of MTHFD2 using DS18561882 recapitulates the effects of MTHFD2 knockdown on glycine and collagen production downstream of TGF-β. Furthermore, treatment of mice with DS18561882 ameliorates fibrotic responses after intratracheal instillation of bleomycin. Our results demonstrate that MTHFD2 may be a viable therapeutic target for IPF and other fibrotic diseases.
RESULTS
TGF-β induces the expression of mitochondrial 1C pathway enzymes through mTORC1 and ATF4.
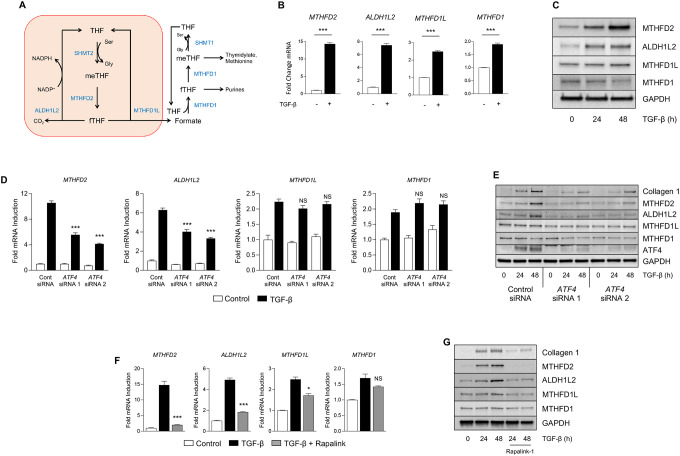
We have previously demonstrated that lung fibroblasts depend on de novo synthesis of glycine to support production of collagen protein downstream of TGF-β (11, 12). Synthesis of glycine from serine produces 1C units which are transferred to THF, producing 5,10-methylene-THF (meTHF), which must be recycled back to THF to support glycine production by SHMT2 (Fig. 1A) (22). To determine how the TGF-β regulates the expression of 1C enzymes, we treated normal human lung fibroblasts (NHLFs) with TGF-β (1ng/mL) and measured mRNA and protein expression of the mitochondrial 1C enzymes MTHFD2, ALDH1L2, and MTHFD1L (Fig. 1B). TGF-β significantly increased the mRNA and protein expression of MTHFD2 and ALDH1L2, with a smaller increase in MTHFD1L expression (Fig. 1B, 1C, S1A–C).
Figure 1. Mitochondrial one-carbon enzymes are induced by TGF-β.
(A) Schematic representation of metabolite flux through the mitochondrial and cytosolic one-carbon pathways. (B) qRT-PCR analysis of 1C enzyme mRNA expression in normal human lung fibroblasts (NHLFs) cultured in the presence or absence of TGF-β for 24 hours. (C) Western blot analysis of 1C enzyme protein expression in NHLFs treated with TGF-β for the indicated intervals. (D) qRT-PCR analysis of 1C enzyme mRNA expression in NHLFs transfected with siRNA targeting ATF4 or nontargeting siRNA. Cells were treated with TGF-β or left untreated for 24 hours. (E) Western blot analysis of 1C enzyme protein expression in NHLFs transfected with siRNA targeting ATF4 or nontargeting siRNA. Cells were treated with TGF-β for the indicated intervals. (F) qRT-PCR analysis of 1C enzyme mRNA expression in NHLFs either left untreated or treated with TGF-β in the presence or absence of the mTORC1 inhibitor Rapalink-1. (G) Western blot analysis of 1C enzyme protein expression in NHLFs treated with TGF-β for the indicated intervals in the presence or absence of Rapalink-1. Bar graphs represent mean ± SEM, n=3. *P<0.05, **P<0.01, ***P<0.001.
Flux through the mitochondrial 1C pathway has been shown to be required to supply 1C units to the cytosolic 1C pathway through formate production and mitochondrial export. Formate overflow from the mitochondria supplies 1C units for generation of purines, thymidylate, and methionine (Fig. 1A) (22). In contrast to the mitochondrial pathway, which uses multiple enzymes, the cytoplasmic 1C pathway is catalyzed only by MTHFD1. We found that while MTHFD1 transcript levels were increased by TGF-β, its protein expression was reduced compared with untreated cells (Fig. 1B, 1C, S1D).
We have previously demonstrated that induction of the serine/glycine synthesis pathway downstream of TGF-β is dependent on mTORC1 and ATF4 (15). Thus, we sought to determine the requirement of these factors for the regulation of 1C enzyme expression downstream of TGF-β. We found that TGF-β-mediated induction of MTHFD2 and ALDH1L2 was inhibited by ATF4 knockdown, while MTHFD1L and MTHFD1 were unaffected (Figure 1D, 1E, S1E–H). Consistent with a role of mTORC1 promoting ATF4 activation, inhibition of mTORC1 with the selective inhibitor Rapalink-1 resulted in similar reduced expression of MTHFD2 and ALDH1L2, with a partial inhibition of MTHFD1L induction downstream of TGF-β (Fig. 1F, 1G, S1I–L).
Mitochondrial 1C metabolism is required for TGF-β-induced collagen protein production in NHLFs.
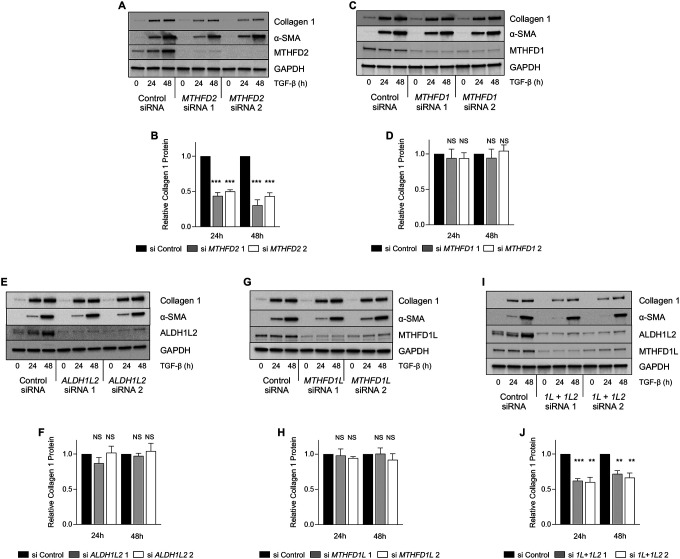
We have previously shown that SHMT2-mediated production of glycine is required to support collagen protein production in lung fibroblasts (11). Because the SHMT reaction requires mitochondrial THF regeneration (Fig. 1A), we sought to determine whether mitochondrial 1C enzymes are required for collagen protein production downstream of TGF-β. Consistent with the specific importance of mitochondrial 1C metabolism for collagen production, knockdown of MTHFD2 was sufficient to inhibit TGF-β-induced collagen protein production (Fig 2A, 2B), while knockdown of the cytosolic enzyme, MTHFD1 did not affect collagen induction downstream of TGF-β (Fig. 2C, 2D). To determine whether the enzymes downstream of MTHFD2 are required TGF-β-induced collagen production, we knocked down either ALDH1L2 (Fig. 2E, 2F), MTHFD1L (Fig. 2G, 2H), or both ALDH1L2 and MTHFD1L (Fig. 2I, 2J). Surprisingly, knockdown of either ALDH1L2 or MTHFD1L did not affect collagen induction in NHLFs treated with TGF-β; however, the combined knockdown of both enzymes did reduce collagen induction, suggesting that while MTHFD2 is required for collagen production downstream of TGF-β, NHLFs have flexibility to use either ALDH1L2 or MTHFD1L in order to regenerate mitochondrial THF.
Figure 2. Mitochondrial one-carbon metabolism is required for TGF-β-induced collagen protein production.
(A) Western blot analysis of collagen and α-smooth muscle actin (α-SMA) protein expression in normal human lung fibroblasts (NHLFs) transfected with siRNA targeting MTHFD2 or nontargeting siRNA. Cells were treated with TGF-β for the indicated intervals. (B) Quantification of relative Collagen 1 levels in MTHFD2 knockdown NHLFs at the indicated intervals. (C) Western blot analysis of collagen and α-SMA protein expression in NHLFs transfected with siRNA targeting MTHFD1 or nontargeting siRNA. Cells were treated with TGF-β for the indicated intervals. (D) Quantification of relative Collagen 1 levels in MTHFD1 knockdown NHLFs at the indicated intervals. (E) Western blot analysis of collagen and α-SMA protein expression in NHLFs transfected with siRNA targeting ALDH1L2 or nontargeting siRNA. Cells were treated with TGF-β for the indicated intervals. (F) Quantification of relative Collagen 1 levels in ALDH1L2 knockdown NHLFs at the indicated intervals. (G) Western blot analysis of collagen and α-SMA protein expression in NHLFs transfected with siRNA targeting MTHFD1L or nontargeting siRNA. Cells were treated with TGF-β for the indicated intervals. (H) Quantification of relative Collagen 1 levels in MTHFD1L knockdown NHLFs at the indicated intervals. (I) Western blot analysis of collagen and α-SMA protein expression in NHLFs transfected with siRNA targeting both ALDH1L2 and MTHFD1L or nontargeting siRNA. Cells were treated with TGF-β for the indicated intervals. (J) Quantification of relative Collagen 1 levels in ALDH1L2/MTHFD1L knockdown NHLFs at the indicated intervals. Bar graphs represent mean ± SEM, n=3. *P<0.05, **P<0.01, ***P<0.001.
Our results suggest that mitochondrial formate production and cytosolic 1C metabolism are dispensable for collagen protein production downstream of TGF-β. Consistent with this, formate supplementation was insufficient to rescue the effects of MTHFD2 knockdown on collagen production by NHLFs (Fig. S2A, S2B). Furthermore, treatment of NHLFs with methotrexate, which reduces cytosolic THF by inhibiting dihydrofolate reductase, had no effect on collagen protein production downstream of TGF-β (Fig. S2C). Finally, to determine whether folate supplementation into the media could rescue MTHFD2 knockdown, we added increasing doses of 5-methyl-THF to the media of MTHFD2 knockdown NHLFs. Consistent with an absolute requirement of mitochondrial THF regeneration for collagen protein production by lung fibroblasts, 5-methyl-THF was unable to rescue the effects of MTHFD2 knockdown (Fig. S2D–S2E). This finding is consistent with previous reports demonstrating high levels of compartmentalization of cellular folate pools with limited transport (23, 24).
Mitochondrial 1C metabolism is required for TGF-β-induced cellular glycine accumulation.
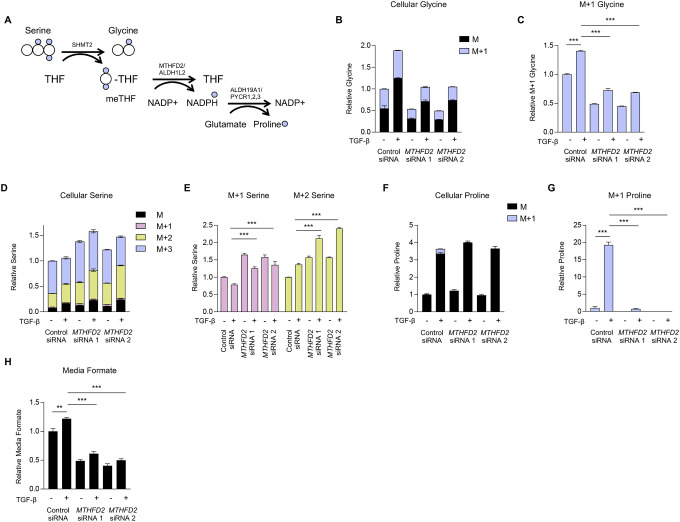
To determine how mitochondrial 1C metabolism contributes to de novo glycine synthesis in NHLFs, we measured cellular serine and glycine levels in control and MTHFD2 knockdown cells using gas chromatography mass spectrometry (GC-MS). Cells were cultured in media containing 13C2-glycine (glycine labeled with 13C on both carbon atoms) to allow us to determine the relative contribution of extracellular (M+2) glycine as well as glycine synthesized de novo from either extracellular glucose or serine (M+0) to total cellular glycine pools (Fig. S3A). As shown in Figure S3B, unlabeled glycine, derived from both extracellular glucose and extracellular serine constituted over half of intracellular glycine in untreated cells. TGF-β increased both total cellular glycine and levels of de novo-synthesized (M+0) glycine in NHLFs (Fig. S3C, S3D). Both total glycine and de novo-synthesized glycine were reduced in MTHFD2 knockdown cells (Fig. S3C, S3D). MTHFD2 knockdown was also associated with a significant increase in cellular serine levels as well as increased levels of cellular serine labeled from extracellular glycine (Fig. S3E–G). This suggests that in the absence of MTHFD2, catabolism of serine is inhibited and reverse flux through SHMT2 is increased, leading to increased serine synthesis from glycine. This was confirmed in SHMT2 knockdown cells which also displayed reduced total glycine and de novo synthesized glycine levels (Fig. S4A–C). While SHMT2 knockdown cells had elevated total serine levels compared with knockdown cells, labeling of serine from extracellular glycine was inhibited in SHMT2 knockdown cells (Fig. S4D–F).
To better parse the relative contributions of the individual mitochondrial 1C enzymes to cellular glycine production, we labeled control and MTHFD2 knockdown cells with 2,3,3-D3-Serine (serine labeled with one deuterium atom on the 2-carbon, and 2 deuterium atoms on the 3-carbon), which has been used to demonstrate the contribution of 1C metabolism to cellular NADPH homeostasis (25, 26) (Fig. 3A). Consistent with our findings in Figure S3, cellular glycine levels, and glycine synthesized from serine were increased in TGF-β-treated cells (Fig. 3B, 3C, S5A). Total cellular serine levels were elevated in MTHFD2 knockdown cells, as were M+1 and M+2 serine resulting from reverse flux through SHMT (Fig. 3D, 3E, S5B). The activity of the pathways downstream of MTHFD2 can be measured by tracing the presence of deuterium atoms into proline. As has been previously demonstrated, the reduction of NADP+ by MTHFD2 and ALDH1L2 leads to labeling of mitochondrial NADPH with deuterium, which can then be transferred to proline during its de novo synthesis (25, 26). Indeed, we found that TGF-β increased cellular proline levels and labeling of cellular proline (M+1) with deuterium from serine (Fig. 3F, 3G, S5C). While labeling of proline was abolished in MTHFD2 knockdowns, TGF-β-induced proline accumulation was not inhibited, demonstrating that while mitochondrial 1C metabolism contributes to mitochondrial NADPH pools, it is not required to support cellular proline production. Formate, which is exported from mitochondria and can be released from the cell, was elevated in the media of control cells after TGF-β treatment, consistent with increased MTHFD1L activity (Fig. 3H). MTHFD2 knockdown reduced formate accumulation in the media both in control and TGF-β-treated NHLFs.
Figure 3. MTHFD2 is required for increased cellular glycine levels downstream of TGF-β.
(A) Schematic representation of metabolite labeling downstream of 2,3,3-D3-Serine. Normal human lung fibroblasts (NHLFs) were transfected with siRNA targeting MTHFD2 or nontargeting siRNA. Cells were labeled with 2,3,3-D3-Serine and treated with TGF-β for 48 hours or left untreated. (B) Analysis of cellular glycine after labeling with 2,3,3-D3-Serine in NHLFs treated with TGF-β or left untreated. (C) Relative levels of M+1 glycine from (B). (D) Analysis of cellular serine after labeling with 2,3,3-D3-Serine in NHLFs treated with TGF-β or left untreated. (E) Relative levels of M+1 and M+2 serine from (D). (F) Analysis of cellular proline after labeling with 2,3,3-D3-Serine in NHLFs treated with TGF-β or left untreated. (G) Relative levels of M+1 and proline from (F). (H) Relative levels of media formate content in NHLFs treated with TGF-β or left untreated. Bar graphs represent mean ± SEM, n=3. *P<0.05, **P<0.01, ***P<0.001.
Lung fibroblasts can use either ALDH1L2 or MTHFD1L to support glycine production.
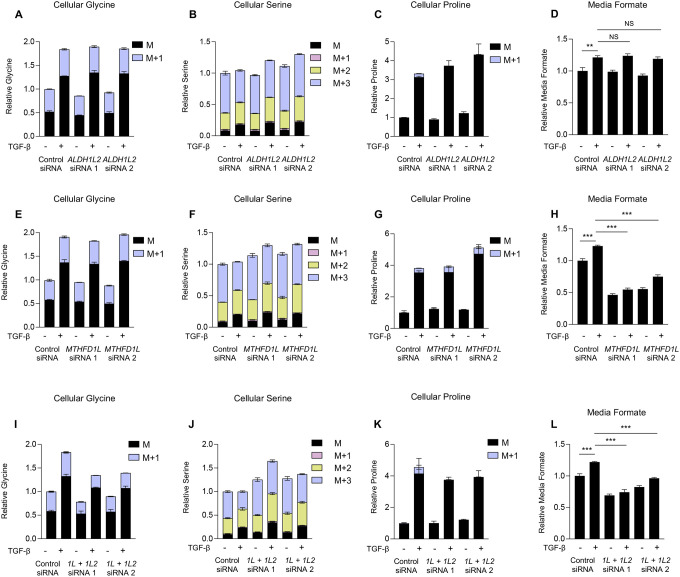
Consistent with a lack of effect of ALDH1L2 or MTHFD1L knockdown on TGF-β-induced collagen production, knockdown of either protein did not prevent TGF-β-induced increases in cellular glycine levels (Fig. 4A, 4E, S6A, S7A) or lead to a significant decrease in glycine derived from 2,3,3-D3-Serine (M+1) (Fig. S6B, S7B). Interestingly, an increase in total serine levels (Fig 4B, 4F, S6C, S7C) and in M+2 serine (Fig. S6D, S7D) was detected after either ALDH1L2 or MTHFD1L knockdown, suggesting a level of inhibition in the mitochondrial one carbon pathway; however, this was insufficient to inhibit TGF-β-induced glycine accumulation. Consistent with the reduction of NADP+ by ALDH1L2, TGF-β-induced proline labeling was abolished in ALDH1L2 knockdowns (Fig. 4C, S6F) while MTHFD1L knockdown had no effect (Fig 4G, S7F). Total proline accumulation was unaffected by either knockdown, consistent with our findings from MTHFD2 knockdowns (Fig. S6E, S7E). Consistent with a role for MTHFD1L in producing formate downstream of MTHFD2, MTHFD1L knockdown inhibited formate accumulation in the media after TGF-β treatment (Fig. 4H), while ALDH1L2 knockdown had no effect (Fig 4D). Our results suggest that while MTHFD2 is essential for elevation of cellular glycine levels downstream of TGF-β, NHLFs have the flexibility to utilize either ALDH1L2 or MTHFD1L to regenerate mitochondrial THF. Consistent with this, combined knockdown of both MTHFD1L and ALDH1L2, inhibited TGF-β-induced production of glycine from serine (Fig. 4I, S8B) and mimicked the effect of MTHFD2 knockdown on serine and proline labeling after TGF-β treatment as well as on media formate accumulation (Fig. 4J–4L, S8). Consistent with a lack of effect of MTHFD1 knockdown on collagen protein production, MTHFD1 knockdown did not affect accumulation or labeling of glycine, serine, or proline downstream of TGF-β (Fig. S9).
Figure 4. Human lung fibroblasts can utilize either ALDH1L2 or MTHFD1L to support glycine synthesis.
Normal human lung fibroblasts (NHLFs) were transfected with the indicated siRNA. Cells were labeled with 2,3,3-D3-Serine and treated with TGF-β for 48 hours or left untreated. (A) Analysis of cellular glycine in ALDH1L2 knockdown NHLFs after labeling with 2,3,3-D3-Serine in the presence or absence of TGF-β. (B) Analysis of cellular serine in ALDH1L2 knockdown NHLFs after labeling with 2,3,3-D3-Serine in the presence or absence of TGF-β. (C) Analysis of cellular proline in ALDH1L2 knockdown NHLFs after labeling with 2,3,3-D3-Serine in the presence or absence of TGF-β. (D) Relative levels of media formate content in ALDH1L2 knockdown NHLFs treated with TGF-β or left untreated. (E) Analysis of cellular glycine in MTHFD1L knockdown NHLFs after labeling with 2,3,3-D3-Serine in the presence or absence of TGF-β. (F) Analysis of cellular serine in MTHFD1L knockdown NHLFs after labeling with 2,3,3-D3-Serine in the presence or absence of TGF-β. (G) Analysis of cellular proline in MTHFD1L knockdown NHLFs after labeling with 2,3,3-D3-Serine in the presence or absence of TGF-β. (H) Relative levels of media formate content in MTHFD1L knockdown NHLFs treated with TGF-β or left untreated. (I) Analysis of cellular glycine in ALDH1L2/MTHFD1L knockdown NHLFs after labeling with 2,3,3-D3-Serine in the presence or absence of TGF-β. (J) Analysis of cellular serine in ALDH1L2/MTHFD1L knockdown NHLFs after labeling with 2,3,3-D3-Serine in the presence or absence of TGF-β. (K) Analysis of cellular proline in ALDH1L2/MTHFD1L knockdown NHLFs after labeling with 2,3,3-D3-Serine in the presence or absence of TGF-β. (L) Relative levels of media formate content in ALDH1L2/MTHFD1L knockdown NHLFs treated with TGF-β or left untreated. Bar graphs represent mean ± SEM, n=3. *P<0.05, **P<0.01, ***P<0.001.
MTHFD2 regulates fibrotic responses in vivo.
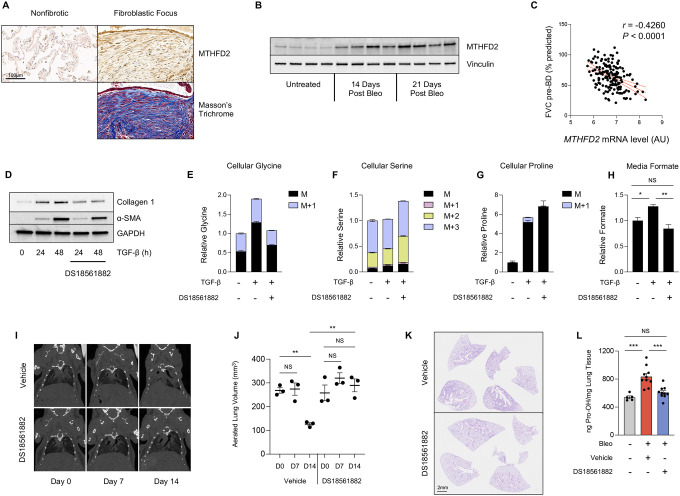
To determine whether mitochondrial one carbon metabolism might play a role in fibrotic processes in vivo, we examined human IPF lung tissue as well as lung tissue from mouse lung after induction of fibrosis with bleomycin. Immunohistochemical analysis demonstrated that MTHFD2 is highly expressed in fibrotic foci of IPF patients when compared with non-fibrotic areas of the same lung (Fig. 5A). MTHFD2 was also highly induced in mouse lung tissue after bleomycin instillation (Fig 5B). Using publicly available data sets of gene expression from IPF patients (27), we found that MTHFD2 expression was negatively correlated with forced vital capacity (FVC), suggesting an involvement of MTHFD2 in disease progression (Fig 5C).
Figure 5. Pharmacologic inhibition of MTHFD2 inhibits fibrotic responses in vivo.
(A) Histological analysis of MTHFD2 protein expression in fibrotic and nonfibrotic areas of paraffin-embedded lung sections from a patient with idiopathic pulmonary fibrosis. (B) Western blot analysis of MTHFD2 protein expression in mouse lung tissue either prior to, or 14 or 21 days after bleomycin instillation. (C) Pearson’s correlation of MTHFD2 mRNA level and forced vital capacity before bronchodilator (FVC-pre-BD) as percentage of what was predicted for each patient. Data is from GSE32537. (D) Western blot analysis of collagen and α-smooth muscle actin protein expression in normal human lung fibroblasts (NHLFs) treated with TGF-β for the indicated intervals in the presence or absence of DS18561882 (5μM). (E-G) Analysis of cellular (E) glycine, (F) serine, and (G) proline in NHLFs after labeling with 2,3,3-D3-Serine in the presence or absence of TGF-β and DS18561882. (H) Relative levels of media formate content in NHLFs treated with TGF-β or left untreated in the presence or absence of DS18561882. (I) Representative coronal microCT images of mice prior to bleomycin instillation (Day 0), 7 days after bleomycin instillation (Day 7), and 14 days after bleomycin instillation (Day 14). Mice began receiving DS18561882 (125 mg/kg) or vehicle 8 days after bleomycin instillation. (J) Quantification of aerated lung volumes from microCT-scanned mice treated as in (I) (mean ± SEM). (K) Histological analysis of mouse lung tissue on day 21 after bleomycin instillation. Beginning on day 8, mice received daily IP injections of either DS18561882 or vehicle. (L) Hydroxyproline content of mouse lungs on day 21 after bleomycin or vehicle (saline) instillation. Beginning on day 8, mice received either DS18561882 or vehicle IP, daily (mean ± SEM). *P<0.05, **P<0.01, ***P<0.001.
DS18561882 is a small molecule inhibitor of MTHFD2 that has been shown to inhibit cancer cell growth and T-cell-mediated inflammation in vivo (28–30). We found that treatment of NHLFs with DS18561882 inhibited TGF-β-induced collagen protein production (Fig. 5D). Furthermore, DS18561882 treatment mimicked the effects of MTHFD2 knockdown on cellular glycine accumulation after TGF-β and labeling from 2,3,3-D3-Serine (Fig. 5E, S10A, S10B). Elevated cellular levels of serine, loss of labeling on proline from 2,3,3-D3-Serine, and inhibition of media formate accumulation were also observed in the presence of DS18561882, suggesting that the effects of DS18561882 are specific to MTHFD2 inhibition (Fig. 5F–5H, S10).
We thus treated mice with DS18561882 to determine if MTHFD2 inhibition can therapeutically inhibit progression of lung fibrosis after bleomycin instillation. Mice were intratracheally instilled with bleomycin, which induces an acute lung injury/inflammatory state lasting approximately one week, followed by a fibrotic phase which lasts 2–3 weeks. To determine the therapeutic efficacy of MTHFD2 inhibition on active fibrotic processes, mice were treated with DS18561882 or vehicle beginning on day 8 after bleomycin instillation. Using micro computed tomography (microCT), we measured lung aeration at day 7 and day 14 after bleomycin instillation. All mice had similar levels of lung aeration prior to and at day 7 after bleomycin (Fig 5I, 5J, S10G). Mice receiving vehicle displayed decreased aeration on day 14 concomitant with the induction of fibrosis. Mice receiving DS18561882 beginning on day 8 did not exhibit a significant drop in lung aeration on day 14. Endpoint analysis at day 21 post bleomycin revealed greatly reduced degree of fibrosis in DS18561882-treated mice (Fig 5K, S10H). Lung hydroxyproline levels, an indicator of collagen content, were also significantly reduced in mice treated with DS18561882 (Fig 5L). Together, our results suggest that pharmacologic targeting of MTHFD2 may be a viable therapeutic avenue for IPF.
DISCUSSION
Metabolic reprogramming in lung fibroblasts is an emerging mechanism required to support matrix production and the development of fibrotic disease. We have previously demonstrated that TGF-β induces metabolic reprogramming in lung fibroblasts characterized by increased levels of de novo serine and glycine synthesis (11, 12, 15). Glycine constitutes one third of the primary structure of collagens, and we and others have found that de novo glycine synthesis supports collagen synthesis downstream of TGF-β (11–16). Because glycine synthesis by SHMT2 is linked with the mitochondrial one-carbon pathway, we sought to determine the roles and requirements of 1C metabolism in glycine synthesis and collagen protein production downstream of TGF-β.
We found that TGF-β significantly increases the expression of mitochondrial 1C enzymes in lung fibroblasts. Consistent with our previous findings that the enzymes of the serine/glycine synthesis pathway are regulated by mTORC1 and ATF4, we found that MTHFD2 and ALDH1L2 are induced by TGF-β through these same mechanisms. This is consistent with previous reports of regulation of these enzymes by mTORC1 and ATF4 downstream of insulin signaling (31–33). MTHFD1L has been shown to be regulated by transcription factors including NRF2 and c-myc (34–36). While the mechanism of MTHFD1L regulation by TGF-β remains to be determined, it is clear that metabolic pathways that support amino acid biosynthesis are among the primary targets of TGF-β-mediated transcriptional responses.
Our results demonstrate that MTHFD2 is a critical regulator of glycine synthesis and collagen protein production in lung fibroblasts. While these cells appear to have the flexibility to use either MTHFD1L or ALDH1L2 to regenerate mitochondrial THF for the SHMT2 reaction, MTHFD2 is essential for mitochondrial THF regeneration, and inhibition of MTHFD2 results in reduced cellular glycine levels and collagen protein production. Our results suggest that loss of MTHFD2 function not only inhibits de novo glycine synthesis, but promotes the reversal of the SHMT2 reaction, potentially making MTHFD2 a more effective therapeutic target for fibrotic disease. While we cannot rule out potential additional roles for formate production by MTHFD1L and activity of the cytoplasmic 1C pathway in lung fibroblasts, our findings demonstrate that these are dispensable for collagen protein production. The cytoplasmic 1C pathway has been shown to reverse flux after inhibition of the mitochondrial one-carbon pathway (25), and may contribute to some of the de novo glycine synthesis that we observed after MTHFD2 or SHMT2 knockdown; however, this is not sufficient to maintain elevated cellular glycine levels after TGF-β treatment. Our current and previous findings show that SHMT1 and MTHFD1 expression are decreased by TGF-β treatment (11), which may limit the ability of lung fibroblasts to utilize the reverse cytoplasmic 1C pathway in the absence of MTHFD2.
Our findings demonstrate that while the mitochondrial 1C pathway contributes to proline biosynthesis via NADP+ reduction, loss of MTHFD2 or ALDH1L2 is insufficient to decrease cellular proline levels downstream of TGF-β. While it would have been extremely elegant for the two most abundant amino acids in collagen to be linked in their mitochondrial production via an NADPH/NADP+ redox cycle, other sources of mitochondrial NADPH are sufficient to support proline production in the absence of one-carbon flux. NAD kinase 2 (NADK2) has recently been shown be required for proline synthesis (37, 38), and thus, phosphorylation of mitochondrial NAD(H) may be the primary mechanism by which NADPH is produced to support proline synthesis.
MTHFD2 is highly expressed during development, but exhibits low or absent expression in most adult tissues (39). MTHFD2 expression is highly upregulated in a variety of cancers and is an independent prognostic indicator in breast and pancreatic cancer (39–41). Our findings demonstrate that MTHFD2 is highly expressed in fibrotic lungs and that increased MTHFD2 expression correlates with loss of lung function in IPF patients. Due to the role of one-carbon metabolism in supporting cancer cell growth, inhibitors of this pathway are in development (42, 43). DS18561882 has been shown to inhibit tumor cell growth and T-cell mediated inflammation in mice (28–30). Our findings demonstrate that DS18561882 mimics the effect of MTHFD2 knockdown on glycine accumulation and collagen production after TGF-β. Furthermore, DS18561882 was effective at inhibiting fibrotic responses after bleomycin instillation. While effects of DS18561882 on targets other than lung fibroblasts cannot be ruled out, MTHFD2 inhibition did not appear to have toxic effects on mice during the course of the experiment. Together, our findings suggest that MTHFD2 may be a viable target for fibrosis of the lung and other tissues.
MATERIALS AND METHODS
Fibroblast Culture.
Normal human lung fibroblasts (NHLFs) (Lonza, CC-2512)) were maintained in Fibroblast Growth Medium 2 (PromoCell, C023020). Cells were plated at 1×105 on 12 well plates for experiments. For experiments not involving metabolic labeling, 24 hours after plating, cells were serum starved in DMEM (Gibco, 11054020) supplemented with 0.1% bovine serum albumin and 2mM glutamine for 24 hours prior to treatment with 1ng/mL TGF-β (Peprotech, 100–21C). DS18561882 (MedChemExpress, HY-130251) was added at the time of TGF-β treatment. For serine and glycine labeling experiments, MEM (Gibco, 11095080) was supplemented with MEM vitamin solution (Gibco, 1120052). For glycine labeling, 400μM 13C2 glycine (Cambridge Isotope Laboratories, CLM-1017) and 400μM unlabeled L-Serine (Sigma, 84959) were added. For serine labeling, 400μM 2,3,3-D3-L-Serine (Cambridge Isotope Laboratories, DLM-582) and 400μM unlabeled (Glycine Sigma, 50046).
siRNA Knockdowns.
For siRNA knockdowns, 1×106 NHLFs were transfected with 250 pmol ON-TARGETplus siRNA (Dharmacon) using an Amaxa Nucleofector 2b set to program A-024. Cells were plated on 10cm dishes for 24 hours and then replated for experiments as above. Dharmacon product numbers: ATF4 (si1: J-005125–10, si2: J-005125–11), MTHFD2 (si1: J-009340–09, si2: J009349–11), MTHFD1L (si1: J-009949–09, si2: J-009949–12), ALDH1L2 (si1: J-026918–09, si2: J-026918–12), MTHFD1 (si1: J-009577–05, si2: J-009577–06), SHMT2 (si1: J-004906–05, si2: J004906–06).
Western Blotting.
Cells were lysed, and electrophoresis was performed as we previously described (44). Wells were lysed in 100μL Urea Sample Buffer (8M deionized urea, 1% SDS, 10% Glycerol, 60mM Tris pH 6.8, 0.1% pyronin-Y, 5% β-mercaptoethanol). Lysates were run through a 28 gauge needle and were electrophoresed on Criterion gels (Bio-Rad) and transferred to nitrocellulose using a Trans-Blot Turbo (Bio-Rad) set to the Mixed MW program. Primary antibodies used were: α-SMA (Sigma, A2547), ALDH1L2 (Proteintech, 21391–1-AP), ATF4 (Proteintech, 10835–1-AP), Collagen 1 (Abcam, ab138492), GAPDH (Cell Signaling, 2118), MTHFD1 (Proteintech, 10794–1-AP), MTHFD1L (Proteintech, 18112–1-AP), MTHFD2 (Proteintech, 12270–1-AP), Vinculin (Cell Signaling, 13901). Blots were imaged using a ChemiDoc Touch (Bio-Rad). Densitometry was performed using ImageJ.
Quantitative PCR.
RNA as isolated using GenElute Mammalian Total RNA Miniprep Kit (Sigma RTN350) and reverse transcribed using iScript Reverse Transcription Supermix (Bio-Rad, 1708841). Quantitative mRNA expression was determined by real-time RT-PCR using iTaq Universal SYBR Green Supermix (Bio-Rad, 172–5124). Primers used were: MTHFD2 (F: 5’-AAACACATCTGTCTGGTATGGT-3’, R: 5’-TGGTTAGGTCACAACTAGGAGTC-3’), ALDH1L2 (F: 5’-GCACTAATTGGCCAGAGCCT-3’, R: 5’-AGCCAGAGGGTCAGCTTTTC-3’), MTHFD1L (F: 5’-TTTGGTCGGAACGATGAGCA-3’, R: 5’-GTCCTGTGAGAGCCTTGTCC-3’), MTHFD1 (F: 5’-ACCCGGCCCTGTTTTTATGA-3’, R: 5’-TCCCAGTGGGCCTGAATAGA-3’), RPL13 (F: 5’-GTCGTACGCTGTGAAGGCAT-3’, R: 5’-GGAAAGCCAGGTACTTCAACTT-3’).
Gas Chromatography-Mass Spectrometry.
NHLFs grown for 48 hours in the presence or absence of TGF-β were washed in blood bank saline (Thermo, 23–293-184) and metabolites were extracted in 600μL ice cold 80% Methanol (Fisher, A456 (MeOH), W6 (H2O)). The solution was vortexed and centrifuged at 21,000 × g for 20 minutes. 400 μLof each extract was transferred to a new tube and dried under nitrogen. Dried metabolites were derivatized in 16μL Methoxamine reagent (Thermo, TS-45950) for 1hr at 37°C. 20 μL of 1% N-tert-butyldimethylsilyl-N-methyltrifluoroacetamide (Sigma, 394882) for 1hr at 60°C. Derivatized samples were analyzed with an 8890 gas chromatograph with an HP-5MS column (Agilent) coupled with a 5977B Mass Selective Detector mass spectrometer (Agilent). Helium was used as the carrier gas at a flow rate of 1.2ml/min. One microliter of each sample was injected in split mode (1:8) at 280°C. After injection, the GC oven was held at 100°C for 1 min and increased to 300°C at 3.5°C/min. The oven was then ramped to 320°C at 20°C/min and held for 5 minutes. The MS system was operated under electron impact ionization at 70eV and the MS source was operated at 230°C and quadrupole at 150°C. The detector was used in scanning mode, and the scanned ion range was 100–650 m/z. Peak ion chromatograms for metabolites of interest were extracted at their specific m/z with Mass Hunter Quantitative Analysis software (Agilent Technologies). Ions used for quantification of metabolite levels were as follows: Glycine m/z 246, Serine m/z 390, Proline m/z 258. Mass isotopomer distributions were determined by integrating appropriate ion fragments for each metabolite using IsoCor (45) to correct for natural abundance.
Formate and Hydroxyproline Measurement.
Formate content of media was measured using a Formate Assay Kit (Abcam, ab111748) on 50 μL of culture media following manufacturer’s instructions. For hydroxyproline measurements, 50 mg of cryogenically pulverized lung tissue was homogenized in 500 μL ultrapure H2O and hydroxy proline content was measured using a Hydroxyproline Assay Kit (Sigma-Aldrich, MAK357) following manufacturer’s instructions.
Bleomycin-Induced Pulmonary Fibrosis.
The protocol for the use of animals was approved by the University of Chicago Institutional Animal Care and Use Committee. Male C57Bl/6J mice (8–12 weeks old; Jackson Laboratory) were intubated and bleomycin (0.7U/kg, TEVA Pharmaceuticals) was instilled intratracheally. Mice were sacrificed on the indicated days and perfused lung tissue was either snap frozen for hydroxyproline measurement or processed for histology. Mice receiving MTHFD2 inhibitor were IP injected with DS18561882 (MedChemExpress, HY-130251) beginning on day 8 after bleomycin instillation. Drug was administered at 100mg/kg in 150μL of 10% DMSO, 40% PEG300, 5% Tween-80, 45% saline.
Micro Computed Tomography.
MicroCT was performed on an X-Cube CT imager (Molecubes, Ghent, Belgium). Animals were anesthetized via inhalation of 2% isoflurane. MicroCT scan was performed using a respiratory-gated protocol to reduce motion artifacts. A window delay of 30% and window width of 50% was used to capture the end-expiration phase of the breathing cycle. Voltage was set to 50 kVp and current at 350 mAS. For quantitative 3D assessment of lung area, HU ranges [−900, −500] were considered as normally-aerated lung tissue, and HU ranges [−500, −100] were defined as poorly-aerated. Volumetric microCT images were reconstructed in an 800 × 800 × 749 format with isotropic voxel dimensions of 50 × 50 × 50 μm3. Images were processed using VivoQuant 4 patch 1 (InviCRO).
Immunohistochemistry.
The collection and use of human lung specimens were approved by the University of Chicago Institutional Review Board. After deparaffinization and rehydration, tissue sections were treated with antigen retrieval buffer (DAKO, S1699) in a steamer for 20 min. Rabbit polyclonal anti-MTHFD2 antibody (Proteintech, 12270–1-AP, 1:400) was applied on tissue sections for a 1-h incubation at room temperature in a humidity chamber. Following a TBS wash, tissue sections were incubated with biotinylated anti-rabbit IgG (1:200, Vector Laboratories, BA-1000) for 30 min at room temperature. The antigen-antibody binding was detected by Elite kit (PK-6100, Vector Laboratories) and DAB (DAKO, K3468) system. Tissue sections were briefly immersed in hematoxylin for counterstaining and were covered with cover glasses. Whole slide images were analyzed using ImageJ.
Analysis of gene expression and patient data.
Processed gene expression datasets from GSE32537 were downloaded from Gene Expression Omnibus. Available clinical data was correlated to individual gene expression profiles using Pearson’s correlation analysis using GraphPad Prism v10.0.2.
Statistical analysis.
Data were analyzed in Prism 10 (GraphPad Software, Inc). All data are shown as mean ± standard error of the mean (SEM). Significance was determined by unpaired two-tailed Student’s t test (for comparisons between two samples), or by one or two-way ANOVA using Tukey’s multiple comparison test. * P < 0.05, ** P < 0.01, *** P < 0.001.
Supplementary Material
Funding
R01ES010524 and W81XWH-22-1-0387 (GMM), and R01HL151680 (RBH).
Funding Statement
R01ES010524 and W81XWH-22-1-0387 (GMM), and R01HL151680 (RBH).
Footnotes
Author contributions
Conception and design: RBH, AM, GMM
Acquisition of data: AYM, YT, JCHS, KDS, TC, KAS, PSW, ORS, BC, RBH
Analysis and interpretation of data: AYM, RCA, JCHS, AM, GMM, RBH
Manuscript writing: RBH, AM, RCA, GMM
Final approval of manuscript: AYM, RCA, YT, JCHS, KDS, TC, KAS, PSW, ORS, BC, AM, GMM, RBH
REFERENCES
- 1.Glassberg MK. Overview of idiopathic pulmonary fibrosis, evidence-based guidelines, and recent developments in the treatment landscape. Am J Manag Care 2019; 25: S195–S203. [PubMed] [Google Scholar]
- 2.Martinez FJ, Collard HR, Pardo A, Raghu G, Richeldi L, Selman M, Swigris JJ, Taniguchi H, Wells AU. Idiopathic pulmonary fibrosis. Nat Rev Dis Primers 2017; 3: 17074. [DOI] [PubMed] [Google Scholar]
- 3.Wynn TA, Ramalingam TR. Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat Med 2012; 18: 1028–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolters PJ, Collard HR, Jones KD. Pathogenesis of idiopathic pulmonary fibrosis. Annu Rev Pathol 2014; 9: 157–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheppard D. Transforming growth factor beta: a central modulator of pulmonary and airway inflammation and fibrosis. Proc Am Thorac Soc 2006; 3: 413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scotton CJ, Chambers RC. Molecular targets in pulmonary fibrosis: the myofibroblast in focus. Chest 2007; 132: 1311–1321. [DOI] [PubMed] [Google Scholar]
- 7.Staab-Weijnitz CA. Fighting the Fiber: Targeting Collagen in Lung Fibrosis. Am J Respir Cell Mol Biol 2021. [DOI] [PubMed] [Google Scholar]
- 8.Sivakumar P, Ntolios P, Jenkins G, Laurent G. Into the matrix: targeting fibroblasts in pulmonary fibrosis. Curr Opin Pulm Med 2012; 18: 462–469. [DOI] [PubMed] [Google Scholar]
- 9.Hamanaka RB, Mutlu GM. Metabolic requirements of pulmonary fibrosis: role of fibroblast metabolism. FEBS J 2021; 288: 6331–6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selvarajah B, Azuelos I, Anastasiou D, Chambers RC. Fibrometabolism-An emerging therapeutic frontier in pulmonary fibrosis. Sci Signal 2021; 14. [DOI] [PubMed] [Google Scholar]
- 11.Nigdelioglu R, Hamanaka RB, Meliton AY, O’Leary E, Witt LJ, Cho T, Sun K, Bonham C, Wu D, Woods PS, Husain AN, Wolfgeher D, Dulin NO, Chandel NS, Mutlu GM. Transforming Growth Factor (TGF)-beta Promotes de Novo Serine Synthesis for Collagen Production. J Biol Chem 2016; 291: 27239–27251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamanaka RB, Nigdelioglu R, Meliton AY, Tian Y, Witt LJ, O’Leary E, Sun KA, Woods PS, Wu D, Ansbro B, Ard S, Rohde JM, Dulin NO, Guzy RD, Mutlu GM. Inhibition of Phosphoglycerate Dehydrogenase Attenuates Bleomycin-induced Pulmonary Fibrosis. Am J Respir Cell Mol Biol 2018; 58: 585–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Selvarajah B, Azuelos I, Plate M, Guillotin D, Forty EJ, Contento G, Woodcock HV, Redding M, Taylor A, Brunori G, Durrenberger PF, Ronzoni R, Blanchard AD, Mercer PF, Anastasiou D, Chambers RC. mTORC1 amplifies the ATF4-dependent de novo serineglycine pathway to supply glycine during TGF-beta1-induced collagen biosynthesis. Sci Signal 2019; 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schworer S, Berisa M, Violante S, Qin W, Zhu J, Hendrickson RC, Cross JR, Thompson CB. Proline biosynthesis is a vent for TGFbeta-induced mitochondrial redox stress. EMBO J 2020; 39: e103334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Leary EM, Tian Y, Nigdelioglu R, Witt LJ, Cetin-Atalay R, Meliton AY, Woods PS, Kimmig LM, Sun KA, Gokalp GA, Mutlu GM, Hamanaka RB. TGF-beta Promotes Metabolic Reprogramming in Lung Fibroblasts via mTORC1-dependent ATF4 Activation. Am J Respir Cell Mol Biol 2020; 63: 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamanaka RB, O’Leary EM, Witt LJ, Tian Y, Gokalp GA, Meliton AY, Dulin NO, Mutlu GM. Glutamine Metabolism Is Required for Collagen Protein Synthesis in Lung Fibroblasts. Am J Respir Cell Mol Biol 2019; 61: 597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ducker GS, Rabinowitz JD. One-Carbon Metabolism in Health and Disease. Cell Metab 2017; 25: 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer 2013; 13: 572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brosnan ME, Brosnan JT. Formate: The Neglected Member of One-Carbon Metabolism. Annu Rev Nutr 2016; 36: 369–388. [DOI] [PubMed] [Google Scholar]
- 20.Yang M, Vousden KH. Serine and one-carbon metabolism in cancer. Nat Rev Cancer 2016; 16: 650–662. [DOI] [PubMed] [Google Scholar]
- 21.Newman AC, Maddocks ODK. One-carbon metabolism in cancer. Br J Cancer 2017; 116: 1499–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tibbetts AS, Appling DR. Compartmentalization of Mammalian folate-mediated one-carbon metabolism. Annu Rev Nutr 2010; 30: 57–81. [DOI] [PubMed] [Google Scholar]
- 23.Trent DF, Seither RL, Goldman ID. Compartmentation of intracellular folates. Failure to interconvert tetrahydrofolate cofactors to dihydrofolate in mitochondria of L1210 leukemia cells treated with trimetrexate. Biochem Pharmacol 1991; 42: 1015–1019. [DOI] [PubMed] [Google Scholar]
- 24.Horne DW, Patterson D, Cook RJ. Effect of nitrous oxide inactivation of vitamin B12-dependent methionine synthetase on the subcellular distribution of folate coenzymes in rat liver. Arch Biochem Biophys 1989; 270: 729–733. [DOI] [PubMed] [Google Scholar]
- 25.Ducker GS, Chen L, Morscher RJ, Ghergurovich JM, Esposito M, Teng X, Kang Y, Rabinowitz JD. Reversal of Cytosolic One-Carbon Flux Compensates for Loss of the Mitochondrial Folate Pathway. Cell Metab 2016; 23: 1140–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao X, Lee K, Reid MA, Sanderson SM, Qiu C, Li S, Liu J, Locasale JW. Serine Availability Influences Mitochondrial Dynamics and Function through Lipid Metabolism. Cell Rep 2018; 22: 3507–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang IV, Coldren CD, Leach SM, Seibold MA, Murphy E, Lin J, Rosen R, Neidermyer AJ, McKean DF, Groshong SD, Cool C, Cosgrove GP, Lynch DA, Brown KK, Schwarz MI, Fingerlin TE, Schwartz DA. Expression of cilium-associated genes defines novel molecular subtypes of idiopathic pulmonary fibrosis. Thorax 2013; 68: 1114–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawai J, Toki T, Ota M, Inoue H, Takata Y, Asahi T, Suzuki M, Shimada T, Ono K, Suzuki K, Takaishi S, Ohki H, Matsui S, Tsutsumi S, Hirota Y, Nakayama K. Discovery of a Potent, Selective, and Orally Available MTHFD2 Inhibitor (DS18561882) with in Vivo Antitumor Activity. J Med Chem 2019; 62: 10204–10220. [DOI] [PubMed] [Google Scholar]
- 29.Sugiura A, Andrejeva G, Voss K, Heintzman DR, Xu X, Madden MZ, Ye X, Beier KL, Chowdhury NU, Wolf MM, Young AC, Greenwood DL, Sewell AE, Shahi SK, Freedman SN, Cameron AM, Foerch P, Bourne T, Garcia-Canaveras JC, Karijolich J, Newcomb DC, Mangalam AK, Rabinowitz JD, Rathmell JC. MTHFD2 is a metabolic checkpoint controlling effector and regulatory T cell fate and function. Immunity 2022; 55: 65–81 e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu Z, Kiang KM, Li N, Liu J, Zhang P, Jin L, He X, Zhang S, Leung GK. Folate enzyme MTHFD2 links one-carbon metabolism to unfolded protein response in glioblastoma. Cancer Lett 2022; 549: 215903. [DOI] [PubMed] [Google Scholar]
- 31.Ben-Sahra I, Hoxhaj G, Ricoult SJH, Asara JM, Manning BD. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science 2016; 351: 728–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torrence ME, MacArthur MR, Hosios AM, Valvezan AJ, Asara JM, Mitchell JR, Manning BD. The mTORC1-mediated activation of ATF4 promotes protein and glutathione synthesis downstream of growth signals. Elife 2021; 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park Y, Reyna-Neyra A, Philippe L, Thoreen CC. mTORC1 Balances Cellular Amino Acid Supply with Demand for Protein Synthesis through Post-transcriptional Control of ATF4. Cell Rep 2017; 19: 1083–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee D, Xu IM, Chiu DK, Lai RK, Tse AP, Lan Li L, Law CT, Tsang FH, Wei LL, Chan CY, Wong CM, Ng IO, Wong CC. Folate cycle enzyme MTHFD1L confers metabolic advantages in hepatocellular carcinoma. J Clin Invest 2017; 127: 1856–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pikman Y, Puissant A, Alexe G, Furman A, Chen LM, Frumm SM, Ross L, Fenouille N, Bassil CF, Lewis CA, Ramos A, Gould J, Stone RM, DeAngelo DJ, Galinsky I, Clish CB, Kung AL, Hemann MT, Vander Heiden MG, Banerji V, Stegmaier K. Targeting MTHFD2 in acute myeloid leukemia. J Exp Med 2016; 213: 1285–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Satoh K, Yachida S, Sugimoto M, Oshima M, Nakagawa T, Akamoto S, Tabata S, Saitoh K, Kato K, Sato S, Igarashi K, Aizawa Y, Kajino-Sakamoto R, Kojima Y, Fujishita T, Enomoto A, Hirayama A, Ishikawa T, Taketo MM, Kushida Y, Haba R, Okano K, Tomita M, Suzuki Y, Fukuda S, Aoki M, Soga T. Global metabolic reprogramming of colorectal cancer occurs at adenoma stage and is induced by MYC. Proc Natl Acad Sci U S A 2017; 114: E7697–E7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu J, Schworer S, Berisa M, Kyung YJ, Ryu KW, Yi J, Jiang X, Cross JR, Thompson CB. Mitochondrial NADP(H) generation is essential for proline biosynthesis. Science 2021; 372: 968–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tran DH, Kesavan R, Rion H, Soflaee MH, Solmonson A, Bezwada D, Vu HS, Cai F, Phillips JA 3rd, DeBerardinis RJ, Hoxhaj G. Mitochondrial NADP(+) is essential for proline biosynthesis during cell growth. Nat Metab 2021; 3: 571–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nilsson R, Jain M, Madhusudhan N, Sheppard NG, Strittmatter L, Kampf C, Huang J, Asplund A, Mootha VK. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat Commun 2014; 5: 3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noguchi K, Konno M, Koseki J, Nishida N, Kawamoto K, Yamada D, Asaoka T, Noda T, Wada H, Gotoh K, Sakai D, Kudo T, Satoh T, Eguchi H, Doki Y, Mori M, Ishii H. The mitochondrial one-carbon metabolic pathway is associated with patient survival in pancreatic cancer. Oncol Lett 2018; 16: 1827–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu F, Liu Y, He C, Tao L, He X, Song H, Zhang G. Increased MTHFD2 expression is associated with poor prognosis in breast cancer. Tumour Biol 2014; 35: 8685–8690. [DOI] [PubMed] [Google Scholar]
- 42.Dekhne AS, Hou Z, Gangjee A, Matherly LH. Therapeutic Targeting of Mitochondrial One-Carbon Metabolism in Cancer. Mol Cancer Ther 2020; 19: 2245–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cuthbertson CR, Arabzada Z, Bankhead A 3rd, Kyani A, Neamati N. A Review of Small-Molecule Inhibitors of One-Carbon Enzymes: SHMT2 and MTHFD2 in the Spotlight. ACS Pharmacol Transl Sci 2021; 4: 624–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamanaka RB, Mutlu GM. PFKFB3, a Direct Target of p63, Is Required for Proliferation and Inhibits Differentiation in Epidermal Keratinocytes. J Invest Dermatol 2017; 137: 1267–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Millard P, Delepine B, Guionnet M, Heuillet M, Bellvert F, Letisse F. IsoCor: isotope correction for high-resolution MS labeling experiments. Bioinformatics 2019; 35: 4484–4487. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.