Abstract
Originally extracted from Momordica charantia seeds, the antiviral and anti-tumor activities of Momordica anti-HIV protein MAP30 have become well known. Although MAP30 has been reported to possess antiviral activity against several human viruses, the current understanding of the MAP30-mediated antiviral response is mainly derived from the previous research work on anti-HIV herbal medicines; the mechanistic insight of its effects on other viruses remains largely unknown. In this study, we showed that both ectopically expressed and purified recombinant MAP30 (rMAP30) impeded Epstein-Barr virus Nuclear Antigen 1 (EBNA1)-mediated transcription from the viral latent replication origin. Mechanistically, in vivo and in vitro studies revealed that MAP30 caused EBNA1 to dissociate from the cognate binding sites, which disrupted downstream EBNA1-dependent viral epigenome accumulation and cell maintenance of Epstein-Barr virus (EBV)-associated neoplastic cells. Finally, mutational analysis indicated that the N-terminal ricin A homologous domain shared by ricin-like proteins was implicated in the anti-EBV response. Our study provides evidence to support that MAP30 has a unique property to combat EBV latent infection, suggesting a potential to develop this herbal protein to be an alternative medicine for EBV associated diseases.
1. Introduction
Medicinal plants have been used as alternative or complementary therapies since ancient times, several of which have been routinely used as both medicines and as food sources, such as Momordica charantia (the bitter gourd). A member of the Cucurbitaceae family, Momordica charantia is a vegetable whose multifunctional health benefits are well known, whether eaten or used to treat various diseases [1]. Momordica anti-HIV protein MAP30 is one of the most bioactive components found in medicinal plants and its immunomodulatory, anti-tumor, and antiviral activities have been widely reported [2,3]. Cumulative studies have shown that MAP30 can be utilized for both anti-cancer and anti-virus treatments, despite the limited understanding of its underlying mechanisms [[4], [5], [6], [7]]. The understanding of MAP30-mediated antiviral response has largely relied on major research studies of its effect on HIV, although its antiviral activities for hepatitis B and herpes complex viruses have also been documented [8]. A comprehensive understanding of the natural antiviral activity produced by MAP30 will require more mechanistic studies on its antiviral strategies against other viruses.
The Epstein-Barr virus was first detected in a case of Burkitt's lymphoma in 1958 and was subsequently identified as the first known human oncogenic virus [9]. Unlike other tumor viruses that can be prevented by effective vaccines or treated with a specific antiviral drug, no current antiviral strategies have been shown to be effective against EBV [10]. Like all herpesviruses, EBV can establish a biphasic latent and lytic infection cycle in host cells [11]. It can also cause disease in both lymphocytic and epithelial cells.
B cells are considered to be the primary host cells for EBV to establish a lifelong infection because of their prominent capability to drive B-cell transformation in vitro [12,13]. Apart from vaccine biotechnology, the development of anti-EBV drugs mainly focuses on those key viral factors which are actively involved in either lytic or latent infection [10,14,15]. Nevertheless, ablating EBV latency is key to eradicating viral entities from the host.
Although EBV infection of B cells in vivo results in four different types of latent infection (I, IIa and IIb, and III), the EBV-mediated transformation of B cells into indefinitely proliferating lymphoblastoid cell lines (LCL) in vitro only produce a latency III program [16,17]. Among viral latency-associated genes, EBNA1 is uniquely detected in most EBV-positive neoplastic cells [18]. The specific involvement of EBNA1 in episome formation and maintenance means that this viral protein is a potential drug target for anti-EBV compound discoveries [15,19,20]. Binding to the latent origin of replication (oriP) has been shown to be a prerequisite for EBNA1 to perform its episome dependent functions [21]. Several small compounds exhibit specific anti-EBV activities by effectively blocking EBNA1 functioning, thereby eradicating EBV from the host cells [[22], [23], [24]].
MAP30 has been shown to target HIV integrase, reverse transcriptase, and the hepatitis B virus antigens, HBsAg and HBeAg, suggesting that it could drive distinct mechanisms to terminate the viral infectious cycle [5,25,26]. In this study, we explored the unknown antiviral activity of MAP30 for EBV using a well-established EBNA1/oriP-Luc reporter platform, and the mechanistic insight of the newly identified anti-EBV response was investigated and presented.
2. Materials and methods
2.1. The DNA recombination procedure and recombinant protein production
The synthetic cDNAs for MAP30 and its mutant derivatives were purchased from Yao Hong biotechnology Inc, Taiwan. The MAP30 DNA fragments were released by digesting XhoI and BamHI and subcloned into the expression vectors, pSG5-Flag [27] and pTrcHis A (Invitrogen™ V36020), respectively. PSG5-Flag-derived expression vectors were used to perform transfection-mediated luciferase and/or β-gal reporter assays [28], while pTri-HisA-derived vectors were used to produce and purify the recombinant proteins. The produced Flag-tagged (DYKDDDDK) expression plasmids of EBNA1(FEBNA1), MAP30(FMAP30), and GAP31(FGAP31) were used in the transfection mediated luciferase reporter assay whenever necessary. For the recombinant protein purification, bacterial cells of E. coli BL21 (DE3) transformed with separate His-tagged MAP30 plasmids were cultured at 37 °C. Isopropyl β-D-thiogalactoside (IPTG: Sigma 16758) was added to the medium to make a final concentration of 0.50 mM soon as optical density at 600 nm (OD600) reached approximately 0.5.
2.2. Cell transfection and luciferase activity reporter assays
Both BJAB and AKATA(−) are negative Burkitt's lymphoma (BL) cell lines, whereas AKATA and CCL87 are EBV-positive BL cell lines, and LCL1 and LCL2 are EBV-transformed lymphoblastoid cell lines (LCL) [[29], [30], [31]]. Cells were cultured in RPMI1640 media supplemented with 10 % fetal bovine serum (Gibco™ 26140079), 100 units of penicillin, 100 μg of streptomycin, and 292 μg/ml of l-glutamine (Gibco™ 10378016). BJAB cells stably expressed the FEBNA1/oriP-Luc reporter system, while the platform for transfection-mediated EBNA1/oriP-luc dependent transcription assay platform has been described previously [32,33].
In total, 1x107 BJAB cells were co-transfected with 10 μg of the FEBNA1 expression vector, 5 μg of oriP-Luc reporter plasmid, and either 1 μg of CMV-β Gal internal control plasmid or the indicated amount of FMAP30/or mutant expression vectors using Gene Pulser Xcell Electroporation Systems (BioRad) at setting of 220 V and 960 μF. Alternatively, the transfected FMAP30 plasmids were replaced by the indicated amount of MAP30 recombinant proteins (rMAP30) or mutants. The above experimental procedure was also conducted for the control group using 10 μg of the EBNA2 expression vector, 5 μg of LMP1-Luc reporter plasmid, and 1 μg of internal control CMV-β Gal plasmid. The fold activation produced by EBNA1/oriP-Luc dependent transcription activity was determined as the observed luciferase activity normalized by the β-gal activity which was elicited by the internal CMV-βgal control plasmid.
2.3. Chromatin immunoprecipitation (ChIP), in vitro DNA/protein binding, and the measurements of EBV genome accumulation
A total of 5x106 LCL or AKATA cells treated with 0–100 ng/mL rMAP30, or its mutants, were subjected to a ChIP assay using the antibodies, EBNA1 (Nova biologicals 0211), EBNA2 (PE2: Abcam 90543), and control IgG (R&D 1-001-A) following the manufacturer's protocol (Pierce 26157). The amount of ChIPed DNA was quantified by real-time quantitative PCR (qPCR) using a pair of primers for oriP and a glyceraldehyde 3-phosphate dehydrogenase (GAPDH) control, respectively. The primers for oriP-DNA were 5′-GCTAAACGAAGGAG AATGAAGAAG-3’ (Forward) and 5′-CAGTGGCT GAAGATCAAGGAG-3’ (Reverse), while the primers used to quantify the genomic DNA of GAPDH were 5′-AAGGTGAAGGTCGGA GTCAA-3’ (Forward) and 5′-AATGAAGGGGTCATTGATGG-3’ (Reverse).
For the in vitro oriP-DNA and EBNA1 binding assay, the oriP-DNA fragments were amplified by PCR using a pair of biotin-labeled or unlabeled primers. The primer sequences were the same as the ChIP primers. The control LMP1-promoter DNA fragments were amplified by PCR using a set of biotin-labeled primers, 5′CTGCCGCCAACGACCTC3 and 5′-GTACGGGTACAGATTT-3’. The lysates from 1x107 cells of an LCL and 50 μg of biotin-conjugated DNA were prepared for each binding assay. The cell lysates mixed with the indicated biotin-labeled or non-biotin-labeled DNA were used to perform a streptavidin agarose pulldown assay [34]. A total of 5x106 LCL or AKATA cells treated with the indicated amount of rMAP30, or its mutants, were harvested, and the total genomic DNA was isolated using a DNA Extraction/Genomic DNA Purification kit in accordance with the manufacturer's protocols (Geneaid GT300).
2.4. Protein affinity binding, immunoblot analysis, and image quantitation
Equal volume (50 μl) of Ni-NTA resin bound rMAP30 or its mutants was used to pull down 500 μl of lymphoblastoid cell lysate which was prepared by adding 5x106 of an LCL into lysis buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1%NP-40 and 5 mM EDTA). A 4 °C binding procedure was set in a rotator for 30 min, the pulldown matrices were analyzed by performing an immune blot analysis. The proteins of interest were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and identified by immunoblotting analysis using the following antibodies: DYKDDDDK tag (Abnova MAB7373; 1:1000), EBNA1 (Santa Cruz sc-81581; 1:200), Actin (Abnova MAB12828; 1:2000), EBNA2 (Millipore MABE8; 1:500), and His-tag (Merck 05–531). The proteins were detected and visualized using an ECL detection kit (PerkinElmer PK-NEL105). When necessary, the protein expression levels on the immunoblotting image were taken and quantified with the UVP GelSolo and ImageJ.
2.5. Real-time quantitative PCR (qPCR)
The qPCR was performed on the QuantStudio 3 Real-Time PCR System using the SYBR Green master mix (Thermo Scientific™ K0171). The enrichment of EBNA1 at the oriP-DNA was determined as a percentage of input DNA. The relative quantity of the EBV epigenome was determined by the following formula: 2–[ΔCt (oriP) -ΔCt (GAPDH)]
The EBV genome accumulation in each sample was calculated by expressing the oriP-DNA amplified signal to that of the GAPDH DNA. The above ratio in the control group was set as 1 and the detected values of genome accumulation in other samples were calculated as the relative level to control.
2.6. Cell growth and inhibition analyses
A total of 5x103 LCL or 2x103 BL cells per 200 μL were seeded in triplicate in a 96-well plate and treated with either the indicated concentration of rMAP30 or its mutants for nine days. The medium was refreshed and replenished every three days and cell numbers were counted by performing a trypan blue exclusion assay using Cellometer Vision CBA (Nexcelom). The cells were counted 0, 1, 3, 5, 7, and 9 days after the treatments.
2.7. Statistical analysis
The data were represented as the mean ± the standard error of the mean (SEM) from three independent experiments. The student's t-test was used to perform the statistical comparisons, with a p-value of <0.05 denoting statistical significance.
3. Results
3.1. MAP30 vigorously impairs EBNA1-mediated transcription from a mini-episome reporter system
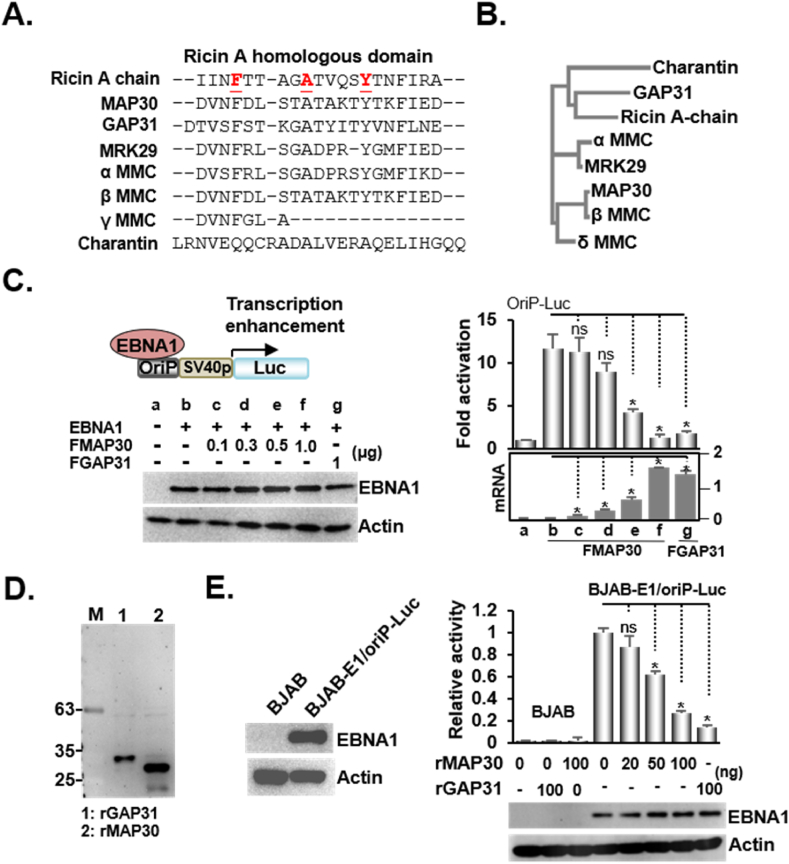
The discovery of ribosome-inactivating proteins (RIPs) was mainly attributed to earlier research work on ricin and abrin [35]. We used the Clustal Omega multiple sequence alignment program to generate alignments between the N-terminal ricin homolog derived from several RIPs (Fig. 1A). Three amino residues—phenylalanine, alanine, and tyrosine—were identified as the most conserved residues residing within the N-termini of the RIPs. Phylogenetic analysis revealed that MAP30 and GAP31, two well-known antiviral RIPs, had evolved distant from the ricin origin (Fig. 1B). According to a new anti-EBV activity of GAP31 has been recently identified, MAP30 was first assessed for its impact on an EBNA1 driven transcription enhancement of an oriP-Luc reporter plasmid [32,36]. As expected, 1 μg of transfected FGAP31 blocked approximately 85 % of EBNA1/oriP-Luc-induced luciferase activity. The dose-dependent inhibiting effects produced by 0.1–1 μg of transfected FMAP30 resulted in a 10–90 % reduction in the transcription of EBNA1/oriP-Luc driven luciferase (Fig. 1C and sFig1A). The increasing levels of plasmid-expressed FMAP30 mRNA were correlated with the increasing amounts of transfected plasmids, suggesting ectopic FMAP30 expression elicited an anti-EBV effect.
Fig. 1.
MAP30 inhibits EBNA1/oriP-Luc mediated transcription enhancement A). This figure shows the N-terminal alignment of the ricin-like proteins and the ricin N-terminal peptide encompassing amino residues from 8 to 22 was used as the template. The conserved residues of phenylalanine, alanine, and tyrosine are underlined and shown in boldface marked with red color. B). The phylogenetic tree depicts the lines of evolutionary descent of N-termini from different ricin-like proteins. C). A transfection-mediated EBNA1/oriP-Luc induced luciferase reporter assay was performed using the Flag-tagged DYKDDDDK expression plasmids of EBNA1(FEBNA1), MAP30(FMAP30), and GAP31(FGAP31), and the reporter plasmids, oriP-Luc, and β-Gal internal control. The resulting fold activation in each sample was determined by expressing the luciferase activity to β-Gal activity. The relative mRNA levels produced by 0.1–1 μg FMAP30 or 1 μg of GAP31 to GAPDH internal control and the immune blot images for FEBNA1 and actin control are also shown. D). The recombinant proteins rGAP31 and rMAP30 were purified and resolved by Western blot. A student t-test was performed to compare the resulting fold activation in each FMAP30 co-transfected sample to the EBNA1/oriP-Luc transfected sample. A p-value of <0.05 is considered statistically significant and an asterisk (*) is marked, whereas non-significance (ns) is noted when p > 0.05. The same approach was employed for all the necessary comparisons in all the following statistical analyses described in the study. E). Cells derived from an EBNA1/oriP-Luc reporter cell line (BJAB-E1/oriP-Luc) were treated with the indicated amount of rGAP31 or rMAP30 and subjected to a luciferase activity assay.
Next, rMAP30 was used to show that the protein levels from 0 to 100 ng/mL caused a 0–80 % reduction in endogenous luciferase activity produced by a BJAB-FEBNA1/oriP-Luc reporter cell line, whereas 100 ng/mL of the rGAP31 positive control shut down approximately 85 % of the same reporter system (Fig. 1D and E and sFig. 1B-D). Here, the study suggested that MAP30 could induce an antiviral action against EBV by blocking EBNA1/oriP-mediated transcription.
3.2. The conserved residues within the N-terminal ricin A homologous domain is linked to MAP30 mediated anti-EBV response
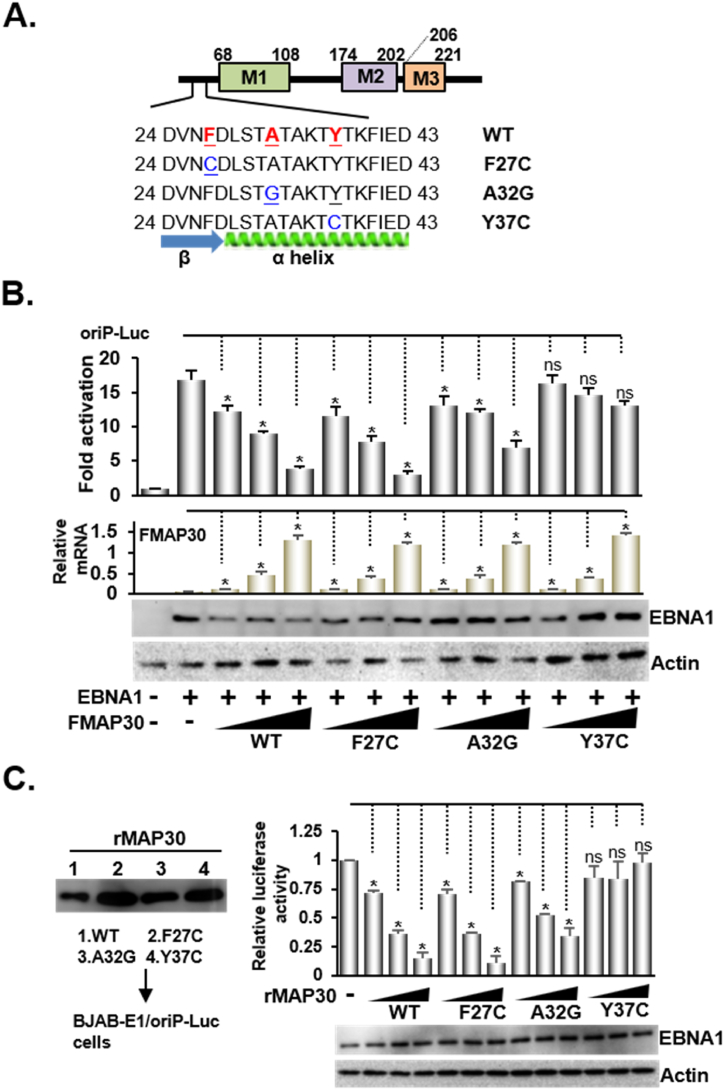
As there was a significant matching with conserved hydrophobic amino acid, plasmids of FMAP30 mutants, F27C, A32G, and Y37C were used to test whether the homologous N-terminus contributed to the anti-EBV response (Fig. 2A and sFig.2A). An increasing amount of FMAP30 or F27C mutant from 0.2 to 1 μg produced a dose-dependent mRNA expression and caused a 20–80 % reduction in EBNA1/oriP-Luc-induced luciferase activity. A32G caused a 25–50 % reduction in EBNA1/oriP-Luc-dependent reporter activity compared with FMAP30, while Y37C had a null effect (Fig. 2B and sFig2B). The rMAP30 or the mutants were further assessed for their effects on endogenous EBNA1/oriP-Luc induced transcription using a BJAB-FEBNA1-oriP-Luc reporter cell line. Similar to the transfection-mediated reporter assays, 20–100 ng/mL of rMAP30 or rF27C led to a 30–85 % reduction endogenous EBNA1/oriP-Luc-induced luciferase activity, and rA32G caused a 20–50 % reduction, while rY37C had no effect (Fig. 2C and sFig.2C-D). The results suggested that the N-terminal ricin homologous domain could has a role in MAP30 mediated anti-EBV response.
Fig. 2.
The N-terminal ricin A homologous domain of MAP30 is implicated in anti-EBV response A). This figure shows the schematic diagram of N-terminal peptides and mutants of MAP30. B). The increasing amount of the indicated expression vectors from 0.1 to 1 μg was co-transfected with EBNA1, oriP-Luc, and β-Gal control into BJAB cells. The mRNAs produced by each FMAP30 or mutant were determined by qPCR and the protein expression from the transfected EBNA1 plasmid was identified by Western blot analysis. C). A BJAB-EBNA1/oriP-Luc reporter cell line was applied to perform a luciferase activity assay after the cells were treated with rMAP30 or a mutant for 16 h. The purified recombinant proteins for MAP30 or mutants were resolved by immunoblotting analysis. The expression protein levels of EBNA1 or actin control in protein drug treated cells was determined by western blots.
3.3. rMAP30 caused EBNA1 dissociation from the cognate binding sites
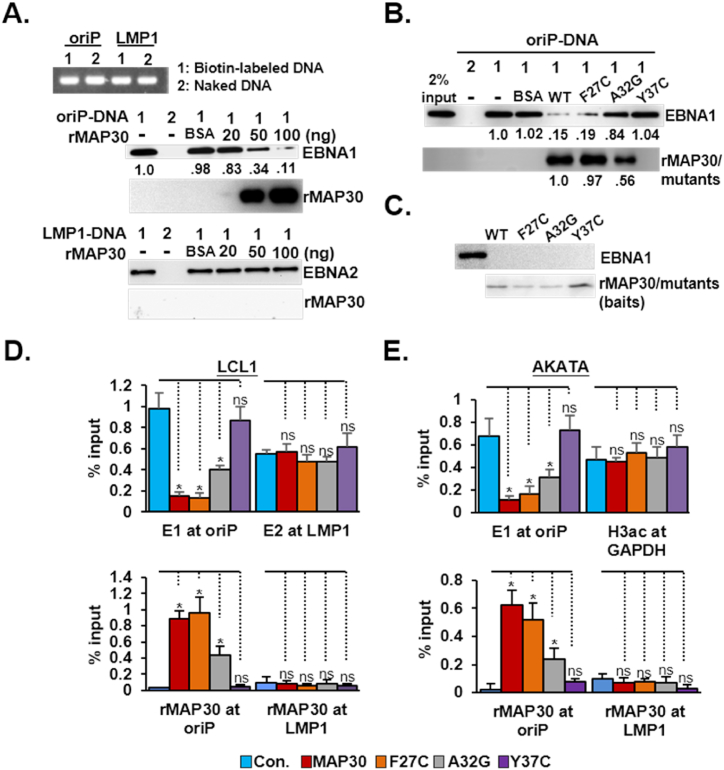
The binding of EBNA1 to the cognate sites residing within oriP is known to be a prerequisite for EBNA1-driven episome-dependent events. Consequently, we evaluated whether MAP30 affected this DNA-protein interaction. Cell lysates from lymphoblastoid cells were used to perform a previously established in vitro streptavidin agarose-mediated DNA pulldown assay [34]. The results showed that 20–100 ng/mL of rMAP30 progressively produced a 17–89 % blocking effect on EBNA1/oriP-DNA binding, whereas rMAP30 had the same treatments did not affect EBNA2/LMP-1 DNA binding (Fig. 3A and sFig. 3A-B). When 100 ng/mL of rMAP30 and its mutant derivatives were applied, both rMAP30 and rF27C effectively inhibited 85 % and 81 % of EBNA1/oriP-DNA binding, respectively, compared with the BSA control, rA32G inhibited 16 % of oriP-DNA-bound EBNA1, and rY37C had no effect (Fig. 3B and sFig. 3C). In addition to blocking EBNA1/oriP-DNA binding, this study noted that 20–100 ng/mL of rMAP30 treatments resulted in the increasing enrichment of rMAP30 at the oriP-DNA. Again, LMP-1 DNA was not detected in LMP-1DNA bound matrices (Fig. 3A and sFig3A-B). The amount of oriP-DNA-bound rF27C was similar to that of rMAP30; the rA32G only retained 50 % binding potency compared with rMAP30, while Y37C did not associate with oriP-DNA (Fig. 3A and B and sFig. 3A-C). The His-tagged affinity pulldown assay showed that neither rMAP30 nor its’ mutants could pulldown endogenously expressed EBNA1 from the lymphoblastoid cell lysates (Fig. 3C and sFig. 3D), suggesting rMAP30 and its mutants did not bind to EBNA1.
Fig. 3.
MAP30 effectively impairs EBNA1/oriP-DNA binding A). PCR-amplified biotin-labeled or naked oriP-DNA and LMP1-DNA were used to perform a streptavidin agarose-mediated DNA pulldown assay. Each DNA sample was mixed with LCL-derived cell lysates in the presence of 20, 50, 100 ng/mL rMAP30, or 100 ng BSA/mL controls. Samples were subjected to a streptavidin agarose pulldown procedure, and the amount of oriP-DNA-bound EBNA1 or LMP-1 DNA-bound EBNA2 was determined by immunoblotting analysis and quantified by ImageJ whenever necessary. B). The same experimental procedure described above was also performed with three MAP30 mutants: rF27C, rA32G, and rY37C. C) Similar amount of Ni-NTA resin bound rMAP30 or its mutants were used as bait proteins to perform an in vitro protein affinity binding assay in the context of lymphoblastoid cell lysates. The bound matrices were used to perform immune blot analysis for EBNA1, rMAP30, and its mutants, respectively. D). LCL1 cells or E) AKATA cells treated with rMAP30 or mutants were used to perform a ChIP assay using antibodies for EBNA1, EBNA2, or His-tag. Cells treated with BSA were used as the control. The amount of ChIPed DNA was quantified by qPCR and results were presented as a percentage of the input DNA. E1:EBNA1; E2:BNA2.
Next, the ChIP assay was used to validate whether the rMAP30 treatments led to the dissociation of EBNA1 from the oriP of an endogenous EBV genome in the context of lymphosblastoid cells, and AKATA cells, respectively. qPCR identified that rMAP30 or rF27C treatments caused an 80–85 % reduction of EBNA1-bound oriP-DNA, rA32G impaired approximately 45–55 % of EBNA1 abundance, and rF37C and the BSA control had no effect (Fig. 3D and E). The rMAP30 or rF27C bound oriP-DNA ranged from 0.5 to 0.9 % of input DNA, and rA32G bound oriP-DNA ranged from 0.3 to 0.4 % was identified by qPCR, whereas no specific enrichment of rY37C was observed. The enrichment of EBNA2 at the LMP1 promoter in LCL1, and of H3ac at the GAPDH promoter in the AKATA(−) cells were used as controls in ChIP assays, respectively. Neither the abundance of EBNA2 nor the H3ac at the cognate sites was affected by treatments of rMAP30 or its mutants. Overall, our experiments suggested that rMAP30 effectively blocked EBNA1/oriP-DNA binding both in vitro and in vivo could be resulted from its binding competition with EBNA1.
3.4. MAP30 declines EBV genome accumulation and EBV-dependent cell proliferation
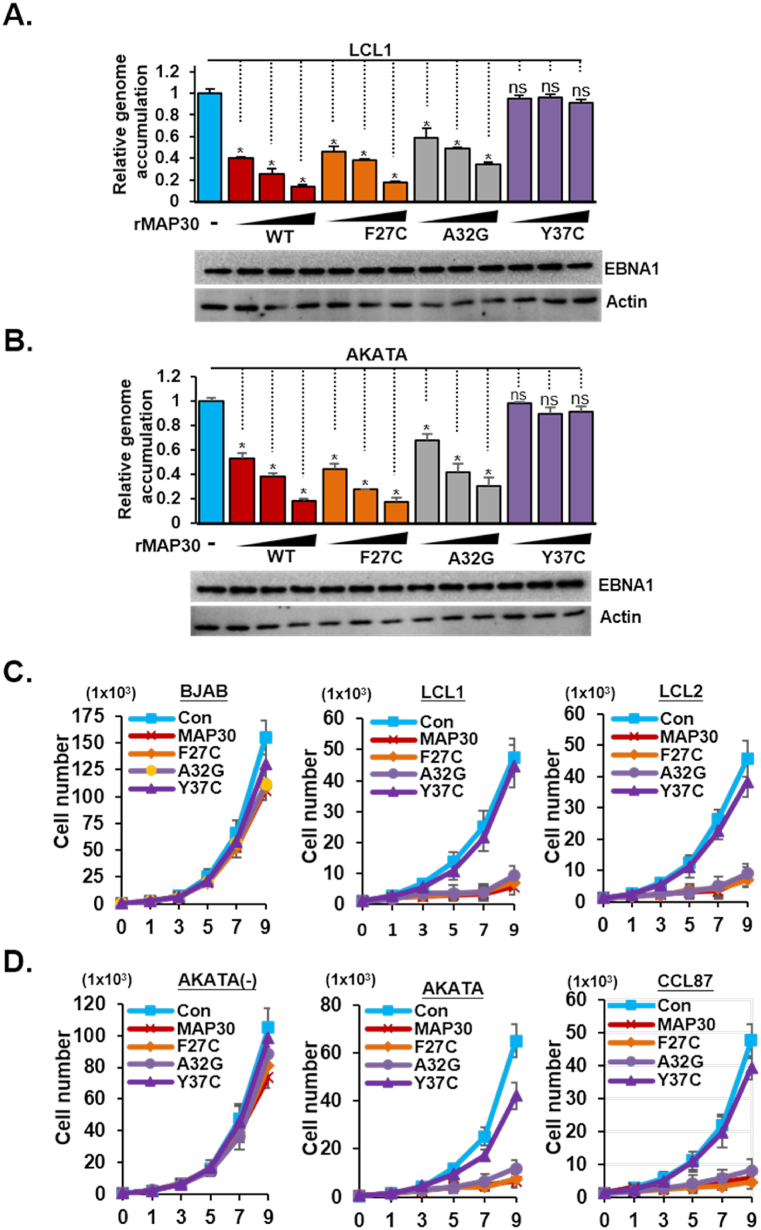
The fundamental role of EBNA1 in the EBV latency program is known to ultimately contribute to viral genome maintenance and the EBV-dependent cell outgrowth of EBV-positive neoplastic cells [28,37]. First, 20–100 ng/mL of rMAP30 or rF27C caused an EBV genome accumulation of ∼50–80 %, while the same treatments using rA32G caused a ∼30–65 % reduction of EBV genome accumulation in the context of LCL1, and AKATA cells, respectively. Once again, Y37C had no effect (Fig. 4 A-B and sFig. 4A-B). Next, a cell proliferation assay was performed to show that rMAP30, rY27C, and A32G impaired approximately 80–90 % of cell growth in the EBV positive neoplastic cells compared with the BSA control after nine days of protein drug treatments. The rY37C consistently behaved as a null mutant (Fig. 4C and D). The rMAP30 and its mutants exhibited extremely minor effects on cell proliferation of EBV-negative BLs, BJAB, and AKATA(−). These results reflected that the specific EBNA1 targeting effect produced by MAP30 could causally lead to the abrogation of EBV persistency in host cells.
Fig. 4.
MAP30 disrupts EBV genome maintenance and blocks cell proliferation of EBV-associated neoplastic cells A-B) Cells derived from LCL1 or AKATA were treated with 10, 50, or 100 ng/mL of rMAP30 or a mutant, while cells treated with 100 ng/mL of BSA were used as the control group. EBV genome accumulation in each sample was determined by qPCR and the amount of detected EBV DNA in the control group was set to 1. The EBV genome accumulation in each drug treated sample was calculated as the relative detected signal to the control group. The immune blots for EBNA1 and actin control are also shown. C-D) Two LCLs (LCL1 and LCL2) and two EBV(+) BL cell lines (AKATA and CCL87) were used to perform a cell proliferation assay. Two EBV-negative BL cell lines, BJAB and AKATA(−), were used as controls. Cells treated with 100 ng/mL of rMAP30, mutants, or control BSA were counted 0,1, 3, 5, 7, and 9 days after treatments.
4. Discussion
Despite having its origins in ancient times, herbal medicine is becoming increasingly popular, with the number of published research papers on the subject steadily increasing each year. Momordica charantia is a common vegetable that has been proven to possess beneficial pharmacological functions, which are attributable to its various bioactive components [38]. Proteins and peptides are the main functional components in the flesh and seeds of Momordica charantia and MAP30 is the main ingredient showing antiviral activity in in vitro studies [25]. It is important to note that the antiviral activity of MAP30 was observed in its ability to selectively destroy lymphocytes and macrophages infected by HIV while exerting minimal cytotoxicity on uninfected cells [26,39]. GAP31, a ricin-like protein distantly related to MAP30 produced anti-HIV effects by driving similar mechanisms, which suggests that herbal antiviral proteins have been evolutionally preserved in a particular group of herbal medicine plants [26].
In this study, we identified a new type of antiviral activity against EBV exhibited by MAP30. MAP30 is a single-chain, type I RIP that possesses an enzymatic peptide of glycosidase and an N-terminal anti-HIV peptide [40]. Our data showed that MAP30 had a specific inhibiting effect on EBNA1-mediated transcription from a mini-EBV episome reporter using either transfected plasmids or purified rMAP30. Significantly, MAP30 could bind to the oriP-DNA in both an in vitro streptavidin agarose-mediated DNA pulldown analysis and in a ChIP assay. This finding is reminiscent of its well-known unique potency in binding to HIV-1 long-terminal repeat DNA [26]. The enrichment of rMAP30 at oriP-DNA may lead to alterations of DNA topology after rMAP30 binding, thereby ultimately promoting the dissociation of EBNA1 from its cognate sites. In addition, rMAP30 may compete with EBNA1 to directly bind with oriP-DNA.
According to its prevalent expression in most EBV-associated neoplasms, EBNA1 is one of the most promising drug targets for anti-EBV compound discovery [41]. EBNA1 targeting methods are expected to neutralize EBV latency from host cells by ablating epigenome maintenance. We showed that 100ng/per ml of rMAP30 can cause an approximate 85 % reduction of EBV genome accumulation and cell proliferation in the context of lymphoblastoid or BL cells. Moreover, this study demonstrated that the N-terminal ricin homolog conserve residue, tyrosine 37, was critical to the MAP30-mediated anti-EBV response. MAP30 with a Y37C mutation had a null DNA binding activity for oriP-DNA, which nullified its EBNA1 blocking activity. The previously reported GAP31 derived anti-HIV peptide also contains the ricin like domain [42]. Our study implies that the N-terminal peptide of ricin-like proteins is likely to have evolved specific strategies to trigger an antiviral response against different virus types. The future work underling how ricin like domain peptide could exhibit anti-viral activities against distinct types of virus will be a key to understand the mechanistic insight of RIP mediated anti-viral response. It should be noted that although our study suggested rMAP30 could impair EBNA1 dependent function to abrogate EBV dependent cell proliferation of AKATA BL cells, this remains a controversial interpretation due to an AKATA derived EBV(−) BL cells AKATA(−) exhibited an EBV independent cell proliferating phenotypes [43]. Further study toward deciphering EBNA1 dependent cell outgrowth of EBV positive lymphoma cells would be a key to answer the above concern.
Among the seven known cancer viruses, EBV is the only one with no specific antiviral strategy [44]. We have presented a complete set of studies to address how MAP30 triggers an anti-EBV response by targeting EBNA1. These data further reflect that the EBNA1 targeting effect produced by MAP30 could significantly abolish the cell maintenance of EBV-associated neoplastic cells. Advances in the screening of MAP30-derived antiviral peptides could assist in discovering the most potent anti-EBV peptide.
Author contribution statement
Wei-Hang Huang: Performed the experiments; Analyzed and interpreted the data.
Wen-Min Su: Performed the experiments; Contributed reagents, materials, analysis tools or data.
Chung-Wei Wang: Yue-Hao Fang: Yuan-Wei Jian: Performed the experiments.
Hao-Jen Hsu: Contributed reagents, materials, analysis tools or data.
Chih-Wen Peng: Conceived and designed the experiments; Wrote the paper.
Data availability statement
Data will be made available on request.
Declaration of competing interest
The authors declare that they do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
Acknowledgements
The research work is supported by MOST109-2320-B-259 -002 -MY3 and MOST110-2320-B-259 -002 -MY3 from the National Science and Technology Council (Taiwan), 111T2561-08, 110P1136–1.6, and 111C1101–6.9 from National Dong Hwa University to C.W.P., TCMMP110-01-02 from Buddhist Tzu Chi Medical Foundation, MOST 111-2314-B-303 -005 and 110-2314-B-303 -009 from the Minister of Science and Technology (Taiwan) to W.H.H. The founder has no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e21486.
Appendix A. Supplementary data
The following is/are the supplementary data to this article.
References
- 1.Lucas E.A., et al. In: Bioactive Foods in Promoting Health. Watson R.R., Preedy V.R., editors. Academic Press; San Diego: 2010. Chapter 35 - health benefits of bitter melon (Momordica charantia) pp. 525–549. [Google Scholar]
- 2.Bourinbaiar A.S., Lee-Huang S. Potentiation of anti-HIV activity of anti-inflammatory drugs, dexamethasone and indomethacin, by MAP30, the antiviral agent from bitter melon. Biochem. Biophys. Res. Commun. 1995;208(2):779–785. doi: 10.1006/bbrc.1995.1405. [DOI] [PubMed] [Google Scholar]
- 3.Lee-Huang S., et al. Anti-HIV and anti-tumor activities of recombinant MAP30 from bitter melon. Gene. 1995;161(2):151–156. doi: 10.1016/0378-1119(95)00186-a. [DOI] [PubMed] [Google Scholar]
- 4.Huang P.L., et al. Proteolytic fragments of anti-HIV and anti-tumor proteins MAP30 and GAP31 are biologically active. Biochem. Biophys. Res. Commun. 1999;262(3):615–623. doi: 10.1006/bbrc.1999.1213. [DOI] [PubMed] [Google Scholar]
- 5.Fan J.M., et al. Inhibition on Hepatitis B virus in vitro of recombinant MAP30 from bitter melon. Mol. Biol. Rep. 2009;36(2):381–388. doi: 10.1007/s11033-007-9191-2. [DOI] [PubMed] [Google Scholar]
- 6.Bourinbaiar A.S., Lee-Huang S. The activity of plant-derived antiretroviral proteins MAP30 and GAP31 against herpes simplex virus in vitro. Biochem. Biophys. Res. Commun. 1996;219(3):923–929. doi: 10.1006/bbrc.1996.0334. [DOI] [PubMed] [Google Scholar]
- 7.Chan D.W., et al. MAP30 protein from Momordica charantia is therapeutic and has synergic activity with cisplatin against ovarian cancer in vivo by altering metabolism and inducing ferroptosis. Pharmacol. Res. 2020;161 doi: 10.1016/j.phrs.2020.105157. [DOI] [PubMed] [Google Scholar]
- 8.Puri M., et al. Ribosome inactivating proteins (RIPs) from Momordica charantia for anti viral therapy. Curr. Mol. Med. 2009;9(9):1080–1094. doi: 10.2174/156652409789839071. [DOI] [PubMed] [Google Scholar]
- 9.Burkitt D. A sarcoma involving the jaws in African children. Br. J. Surg. 1958;46(197):218–223. doi: 10.1002/bjs.18004619704. [DOI] [PubMed] [Google Scholar]
- 10.Cui X., Snapper C.M. Epstein barr virus: development of vaccines and immune cell therapy for EBV-associated diseases. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.734471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen J.I. Epstein-Barr virus infection. N. Engl. J. Med. 2000;343(7):481–492. doi: 10.1056/NEJM200008173430707. [DOI] [PubMed] [Google Scholar]
- 12.Henderson E., et al. Efficiency of transformation of lymphocytes by Epstein-Barr virus. Virology. 1977;76(1):152–163. doi: 10.1016/0042-6822(77)90292-6. [DOI] [PubMed] [Google Scholar]
- 13.Tangye S.G. Genetic susceptibility to EBV infection: insights from inborn errors of immunity. Hum. Genet. 2020;139(6–7):885–901. doi: 10.1007/s00439-020-02145-3. [DOI] [PubMed] [Google Scholar]
- 14.Kerr J.R. Epstein-Barr virus (EBV) reactivation and therapeutic inhibitors. J. Clin. Pathol. 2019;72(10):651–658. doi: 10.1136/jclinpath-2019-205822. [DOI] [PubMed] [Google Scholar]
- 15.Soldan S.S., et al. EBNA1 inhibitors have potent and selective antitumor activity in xenograft models of Epstein-Barr virus-associated gastric cancer. Gastric Cancer. 2021;24(5):1076–1088. doi: 10.1007/s10120-021-01193-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatton O.L., et al. The interplay between Epstein-Barr virus and B lymphocytes: implications for infection, immunity, and disease. Immunol. Res. 2014;58(2–3):268–276. doi: 10.1007/s12026-014-8496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price A.M., Luftig M.A. To be or not IIb: a multi-step process for Epstein-Barr virus latency establishment and consequences for B cell tumorigenesis. PLoS Pathog. 2015;11(3) doi: 10.1371/journal.ppat.1004656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frappier L. Ebna1. Curr Top Microbiol Immunol. 2015;391:3–34. doi: 10.1007/978-3-319-22834-1_1. [DOI] [PubMed] [Google Scholar]
- 19.Deakyne J.S., et al. Structural and functional basis for an EBNA1 hexameric ring in epstein-barr virus episome maintenance. J. Virol. 2017;91(19) doi: 10.1128/JVI.01046-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jakhmola S., et al. Identification of potential inhibitors against epstein-barr virus nuclear antigen 1 (EBNA1): an insight from docking and molecular dynamic simulations. ACS Chem. Neurosci. 2021;12(16):3060–3072. doi: 10.1021/acschemneuro.1c00350. [DOI] [PubMed] [Google Scholar]
- 21.Bochkarev A., et al. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein EBNA 1. Cell. 1995;83(1):39–46. doi: 10.1016/0092-8674(95)90232-5. [DOI] [PubMed] [Google Scholar]
- 22.Messick T.E., et al. Structure-based design of small-molecule inhibitors of EBNA1 DNA binding blocks Epstein-Barr virus latent infection and tumor growth. Sci. Transl. Med. 2019;11(482) doi: 10.1126/scitranslmed.aau5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang M.S., et al. Roscovitine inhibits EBNA1 serine 393 phosphorylation, nuclear localization, transcription, and episome maintenance. J. Virol. 2011;85(6):2859–2868. doi: 10.1128/JVI.01628-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee E.K., et al. Small molecule inhibition of Epstein-Barr virus nuclear antigen-1 DNA binding activity interferes with replication and persistence of the viral genome. Antivir. Res. 2014;104:73–83. doi: 10.1016/j.antiviral.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee-Huang S., et al. Map 30: a new inhibitor of HIV-1 infection and replication. FEBS Lett. 1990;272(1–2):12–18. doi: 10.1016/0014-5793(90)80438-o. [DOI] [PubMed] [Google Scholar]
- 26.Lee-Huang S., et al. Inhibition of the integrase of human immunodeficiency virus (HIV) type 1 by anti-HIV plant proteins MAP30 and GAP31. Proc Natl Acad Sci U S A. 1995;92(19):8818–8822. doi: 10.1073/pnas.92.19.8818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harada S., Kieff E. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J. Virol. 1997;71(9):6611–6618. doi: 10.1128/jvi.71.9.6611-6618.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y.L., et al. Nucleolin is important for Epstein-Barr virus nuclear antigen 1-mediated episome binding, maintenance, and transcription. Proc Natl Acad Sci U S A. 2014;111(1):243–248. doi: 10.1073/pnas.1321800111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clements G.B., Klein G., Povey S. Production by EBV infection of an EBNA-positive subline from an EBNA-negative human lymphoma cell line without detectable EBV DNA. Int. J. Cancer. 1975;16(1):125–133. doi: 10.1002/ijc.2910160114. [DOI] [PubMed] [Google Scholar]
- 30.Takada K., et al. An Epstein-Barr virus-producer line Akata: establishment of the cell line and analysis of viral DNA. Virus Gene. 1991;5(2):147–156. doi: 10.1007/BF00571929. [DOI] [PubMed] [Google Scholar]
- 31.Shen C.L., et al. Ribosome protein L4 is essential for epstein-barr virus nuclear antigen 1 function. Proc Natl Acad Sci U S A. 2016;113(8):2229–2234. doi: 10.1073/pnas.1525444113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y.L., Tsai H.L., Peng C.W. EGCG debilitates the persistence of EBV latency by reducing the DNA binding potency of nuclear antigen 1. Biochem. Biophys. Res. Commun. 2012;417(3):1093–1099. doi: 10.1016/j.bbrc.2011.12.104. [DOI] [PubMed] [Google Scholar]
- 33.Kang M.S., Hung S.C., Kieff E. Epstein-Barr virus nuclear antigen 1 activates transcription from episomal but not integrated DNA and does not alter lymphocyte growth. Proc Natl Acad Sci U S A. 2001;98(26):15233–15238. doi: 10.1073/pnas.211556598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu C.D., et al. The nuclear chaperone nucleophosmin escorts an Epstein-Barr Virus nuclear antigen to establish transcriptional cascades for latent infection in human B cells. PLoS Pathog. 2012;8(12) doi: 10.1371/journal.ppat.1003084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olsnes S. The history of ricin, abrin and related toxins. Toxicon. 2004;44(4):361–370. doi: 10.1016/j.toxicon.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 36.Shen C.L., et al. GAP31 from an ancient medicinal plant exhibits anti-viral activity through targeting to Epstein-Barr virus nuclear antigen 1. Antivir. Res. 2019;164:123–130. doi: 10.1016/j.antiviral.2019.02.015. [DOI] [PubMed] [Google Scholar]
- 37.Wang L., et al. Epstein-Barr virus nuclear antigen 1 (EBNA1) protein induction of epithelial-mesenchymal transition in nasopharyngeal carcinoma cells. Cancer. 2014;120(3):363–372. doi: 10.1002/cncr.28418. [DOI] [PubMed] [Google Scholar]
- 38.Wang S., et al. Momordica charantia: a popular health-promoting vegetable with multifunctionality. Food Funct. 2017;8(5):1749–1762. doi: 10.1039/c6fo01812b. [DOI] [PubMed] [Google Scholar]
- 39.Schreiber C.A., et al. The antiviral agents, MAP30 and GAP31, are not toxic to human spermatozoa and may be useful in preventing the sexual transmission of human immunodeficiency virus type 1. Fertil. Steril. 1999;72(4):686–690. doi: 10.1016/s0015-0282(99)00302-7. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y.X., et al. Anti-HIV and anti-tumor protein MAP30, a 30 kDa single-strand type-I RIP, shares similar secondary structure and beta-sheet topology with the A chain of ricin, a type-II RIP. Protein Sci. 2000;9(1):138–144. doi: 10.1110/ps.9.1.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li N., et al. Development of drugs for Epstein-Barr virus using high-throughput in silico virtual screening. Expert Opin Drug Discov. 2010;5(12):1189–1203. doi: 10.1517/17460441.2010.524640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee-Huang S., et al. Human immunodeficiency virus type 1 (HIV-1) inhibition, DNA-binding, RNA-binding, and ribosome inactivation activities in the N-terminal segments of the plant anti-HIV protein GAP31. Proc Natl Acad Sci U S A. 1994;91(25):12208–12212. doi: 10.1073/pnas.91.25.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimizu N., et al. Isolation of Epstein-Barr virus (EBV)-negative cell clones from the EBV- positive Burkitt's lymphoma (BL) line Akata: malignant phenotypes of BL cells are dependent on EBV. J. Virol. 1994;68(9):6069–6073. doi: 10.1128/jvi.68.9.6069-6073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chakravorty S., Afzali B., Kazemian M. EBV-associated diseases: current therapeutics and emerging technologies. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1059133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.